Abstract
Gut-microbial butyrate is a short-chain fatty acid (SCFA) of significant physiological importance than the other major SCFAs (acetate and propionate). Most butyrate producers belong to the Clostridium cluster of the phylum Firmicutes, such as Faecalibacterium, Roseburia, Eubacterium, Anaerostipes, Coprococcus, Subdoligranulum, and Anaerobutyricum. They metabolize carbohydrates via the butyryl-CoA: acetate CoA-transferase pathway and butyrate kinase terminal enzymes to produce most of butyrate. Although, in minor fractions, amino acids can also be utilized to generate butyrate via glutamate and lysine pathways. Butyrogenic microbes play a vital role in various gut-associated metabolisms. Butyrate is used by colonocytes to generate energy, stabilizes hypoxia-inducible factor to maintain the anaerobic environment in the gut, maintains gut barrier integrity by regulating Claudin-1 and synaptopodin expression, limits pro-inflammatory cytokines (IL-6, IL-12), and inhibits oncogenic pathways (Akt/ERK, Wnt, and TGF-β signaling). Colonic butyrate producers shape the gut microbial community by secreting various anti-microbial substances, such as cathelicidins, reuterin, and β-defensin-1, and maintain gut homeostasis by releasing anti-inflammatory molecules, such as IgA, vitamin B, and microbial anti-inflammatory molecules. Additionally, butyrate producers, such as Roseburia, produce anti-carcinogenic metabolites, such as shikimic acid and a precursor of conjugated linoleic acid. In this review, we summarized the significance of butyrate, critically examined the role and relevance of butyrate producers, and contextualized their importance as microbial therapeutics.
Keywords: butyrate producers, microbial homeostasis, gut epithelial barrier, immunomodulation, gut inflammation, colorectal cancer, gut-organ axis
Role of butyrate-producing gut-commensals
The human gut harbors an enormous number of microbes, approximately 38 × 1012 in total (Sender et al., 2016), comprising genetic material that is comparable to the human genome itself (Manson et al., 2008). This complex gut microbiome contains both aerobic and anaerobic commensal microbes, but anaerobic microbes constitute 99% of the gut microbiota (Nagpal et al., 2017). The gut environment is predominantly anaerobic, providing a suitable ecological niche for anaerobic commensals. The gut microbiome is host-specific, and even among healthy individuals, it varies with geographical location, race, ethnicity, and diet (Gupta et al., 2017). These host-specific gut communities interact with each other through a number of metabolites, which in turn promote gut health (Lin and Zhang, 2017; Krautkramer et al., 2021). Gut microbes also affect the overall health of the host by participating in various metabolic pathways, regulating gene expression, and synthesizing beneficial bioactive compounds, such as short-chain fatty acids (SCFAs), amines, secondary bile acids, and vitamins. In the gut, SCFAs are the major beneficial metabolites produced by gut microbes through metabolizing indigestible dietary fibers. SCFAs are fatty acids with fewer than six carbon atoms and comprise three major forms, i.e., acetate (60%), propionate (20%), and butyrate (20%) (Chambers et al., 2018). Among them, butyrate has been considered of significant importance, as it is involved in several functions of physiological importance, such as trans-epithelial transport, amelioration of mucosal inflammation, alleviation of oxidative stress, enforcement of the epithelial barrier, and protection against colorectal cancer (CRC) (Hamer et al., 2008). The microbial origin butyrate is mainly synthesized by certain anaerobic commensal microbes belonging to the Clostridium cluster (Clostridium_IV and Clostridium_XIVa) of the phylum Firmicutes (Manson et al., 2008). In addition, it is also known that certain commensals convert bacterial metabolites such as lactate and acetate into butyrate via the acetyl-CoA pathway (Bui et al., 2015; Belzer et al., 2017).
In the gut, colon is the primary site of fermentation of indigestible fibers by fibrolytic, butyrate-producing microbes, such as Roseburia intestinalis, Faecalibacterium prasunitzi, and Eubacterium, which are sensitive to the presence of oxygen (Manson et al., 2008). Colonic butyrate is actively transported to colonocytes by monocarboxylate transporters, where the majority (~70%) of transported butyrate is used to generate energy via the citric acid cycle. Non-metabolized butyrate, on the other hand, is transported to the hepatic portal system (Zheng et al., 2017) where butyrate acts as an energy source for hepatocytes, and from there, it is transported to peripheral tissue and systemic circulation. The concentration of butyrate in portal circulation is around 30 μM, and falls near 0.2–15 μM in the systemic circulation, which is almost 2% of the colonic butyrate concentration (Dalile et al., 2019).
The lower level of butyrate producers is continuously found to be associated with various ailments, such as Roseburia in colorectal cancer and inflammatory bowel disease (Sun et al., 2020; Wu et al., 2022), butyrate-producing Coprococcus in pregnant preeclampsia patients (Altemani et al., 2021), and Faecalibacterium in gut inflammation (Fujimoto et al., 2013). Therefore, the level of butyrate producers should be considered to be of therapeutic importance, which has even promoted its oral administration in various studies (Vieira et al., 2012; Chen et al., 2018; Liu et al., 2019). Additionally, butyrate producers are present in the human gut, and their proportion can be enhanced by selecting a suitable diet and healthy lifestyle, thus facilitating the maintenance of overall gut health.
Microbial butyrate and its fate in the gut
Studies suggest that initial butyrate-producing communities, i.e., initial butyrate producers in infant gut, such as Clostridiaceae, Lachnospiraceae, and Ruminococcaceae spp., might be introduced into the human gastrointestinal tract via resistant microbial endospores (Appert et al., 2020). A recent study on a Swiss-cohort confirmed that Eubacterium hallii, a member of the family Lachnospiraceae, is one of the earliest butyrate producers in the gut of infants (Schwab et al., 2017). This is also supported by a study on Swiss, Venezuela, Malawi, and USA populations, which confirmed the human milk oligosaccharide metabolizing ability of Eubacterium Hallii (Schwab et al., 2017). The majority of butyrate producers are gram-positive and come under Clostridium clusters IV and XIVa of the phylum Firmicutes (Manson et al., 2008; Table 1). These microbial communities comprise a significant population of butyrate-producers, including various butyrogenic species of Eubacterium, Faecalibacterium, and Roseburia (Manson et al., 2008; Louis and Flint, 2009). Among all butyrate producers, Faecalibacterium prausnitzii is most abundant in fecal samples (~ 5%) (Miquel et al., 2013), and its proportion can increase up to 13–17.6% (Manson et al., 2008). Other major butyrate producers in fecal gut microbiota are Eubacterium rectale, Eubacterium Hallii, and Roseburia intestinalis, which can constitute up to ~13% (Rivière et al., 2016), 2.4% (mean, 0.6%), and 0.9–5% (mean, 2.3%), respectively (Hold et al., 2003). In smaller fractions, various other butyrate producers are also present in the gut, which produce butyrate by utilizing different dietary oligosaccharides, polysaccharides, and metabolic intermediates (Table 1). Although the majority of butyrate-producing microbes belong to the phylum Firmicutes, studies have suggested that certain members of the phyla Actinobacteria, Bacteroidetes, Fusobacteria, and Proteobacteria can also produce butyrate (Vital et al., 2014). During fermentation, butyrate producers cause substrate-level phosphorylation of the dietary substrate to generate energy in the form of ATP, which results in the formation of multiple end-products, including butyrate (Louis and Flint, 2009). In the human gut, the majority of microbial butyrate is synthesized from carbohydrate metabolism via butyryl-CoA: acetate CoA-transferase pathway (but) and butyrate kinase (buk) pathway, of which the but-pathway is predominant (Vital et al., 2013); (but) and (buk) are derived from the genes encoding enzymes involved in the terminal steps of microbial butyrate synthesis (Altemani et al., 2021). Radioisotope analysis of human fecal microbiota has shown that the majority of butyrate in the gut is produced from carbohydrates through the Embden-Meyerhof-Parnas pathway (glycolysis) via acetyl-CoA (Miller and Wolin, 1996; Louis and Flint, 2009; Figure 1). During this process, two molecules of acetyl-CoA combine to form a butyrate molecule (Miller and Wolin, 1996), and the transformation of crotonyl-CoA to butyryl-CoA is the main energy generation step (Tsukuda et al., 2021; Figure 1). In addition to carbohydrates, in minor fraction, butyrate can also be synthesized from proteins via glutamate, lysine, glutarate, and 4-aminobutyrate pathways (Louis and Flint, 2017; Vital et al., 2017; Mallott and Amato, 2022). Furthermore, butyrate is transported into colonocytes in the gut epithelium via monocarboxylate transporter 1 (MCT1) (Cuff et al., 2002), where it participates in various activities, including stabilization of hypoxia-inducible factor (HIF), inhibition of histone deacetylase (HDAC), and regulation of specific G-protein coupled receptors, which will be discussed later.
Table 1.
Major butyrate producers in the human gut and their relevance.
| Butyrate producer | |||||
|---|---|---|---|---|---|
| Phylum | Sub-cluster | Genus | Species | Relevance | Reference |
| Firmicutes | Clostridium IV Or Clostridium leptum group | Faecalibacterium | F. prasunitzi | Most abundant butyrate producer | Louis and Flint (2009) |
| Subdoligranulum | S. variabile | Metabolizes calprotectin | Kamp et al. (2022) | ||
| Anaerotruncus | A. colihominis | Degrade mucin | Raimondi et al. (2021) | ||
| Ruminococcus | R. bromii | Key fermenter of resistant starch | Ze et al. (2012) | ||
| R. callidus | Degrades complex polysaccharides such as starch or xylan | Chassard et al. (2012) | |||
| R. champanellensis | Most efficient cellulolytic bacterium in human colon | Chassard et al. (2012) | |||
| Clostridium XIVa or Clostridium coccoides group | Roseburia | R. intestinalis | Major Xylan degrader in human gut | Leth et al. (2018), Mirande et al. (2010) | |
| R. faecis | Utilizes fructose, glucose, maltose, cellobiose, raffinose, xylose, sorbitol, melibiose and amylopectin starch; but not Arabinose, and sucrose | Duncan et al. (2006) | |||
| R. hominis | Utilizes arabinose, fructose, glucose, maltose, cellobiose, xylose and glycerol; but not Sucrose, sorbitol, oat spelt xylan, amylopectin starch and inulin (dahlia) | Duncan et al. (2006) | |||
| R. inulinivorans | Utilizes inulin (dahlia), fructose, glucose, and maltose cellobiose, and amylopectin; but not rabinose, raffinose, xylose, glycerol, sorbitol and oat spelt xylan | Duncan et al. (2006) | |||
| Anaerostipes | A. caccae | Utilizes Lactate to produce butyrate | Duncan et al. (2004) | ||
| A. hadrus | Utilizes D-Lactate (not L-Lactose) and acetate to produce butyrate | Allen-Vercoe et al. (2012) | |||
| A. butyraticus | Utilizes fructooligosaccharide (FOS) to produce butyrate | Endo et al. (2022) | |||
| A. rhamnosivorans | Utilizes lactate and acetate for butyrate generation | Bui et al. (2019) | |||
| Butyrivibrio | B. fibrisolvens | Utilizes cellulose | Rodríguez Hernáez et al. (2018), Paillard et al. (2007) | ||
| Eubacterium | E. rectale | Metabolizes sulfonated monosaccharide (sulfoquinovose) present in green vegetables; Dahlia inulin is specifically catabolized | Hanson et al. (2021) | ||
| E. ramulus | Metabolizes variety of flavonoids | Schneider and Blaut (2000), Braune et al. (2001) | |||
| E. hallii | Utilizes glucose and the intermediates acetate and lactate, for butyrate generation | Engels et al. (2016a) | |||
| E. limosum | Transformation of 8-prenylanringenin (phyto-estrogen) from iso-xanthohumol | Possemiers et al. (2008) | |||
| Coprococcus | C. cactus | Metabolizes fructose; cross-feed on fermentation products (acetate, lactate) to produce butyrate | Reichardt et al. (2014), Alessi et al. (2020) | ||
| C. eutactus | Metabolizes β-glucan, cellobiose and lichenan | Alessi et al. (2020) | |||
| C. comes | Metabolizes glucose | Alessi et al. (2020) | |||
| Anaerobutyricum | A. soehngenii | Utilizes D-and L-lactate and acetate to produce butyrate | Gilijamse et al. (2020) | ||
Figure 1.
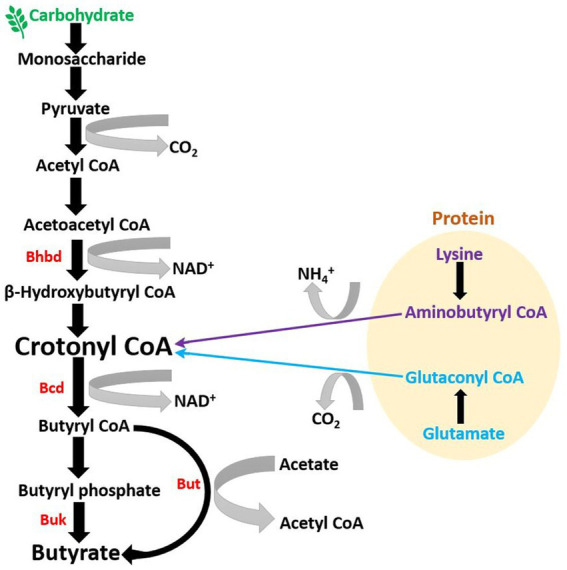
Microbial pathway to generate butyrate in gut: Majority of butyrate in the colon is generated by the metabolization of dietary fibers, primarily of carbohydrate origin (BHBD, β-hydroxybutyryl-CoA dehydrogenase; Bcd, butyryl-CoA dehydrogenase; But, butyryl-CoA: acetate CoA-transferase; Buk, butyrate kinase).
Impact of butyrate producers on neighboring gut microbial communities
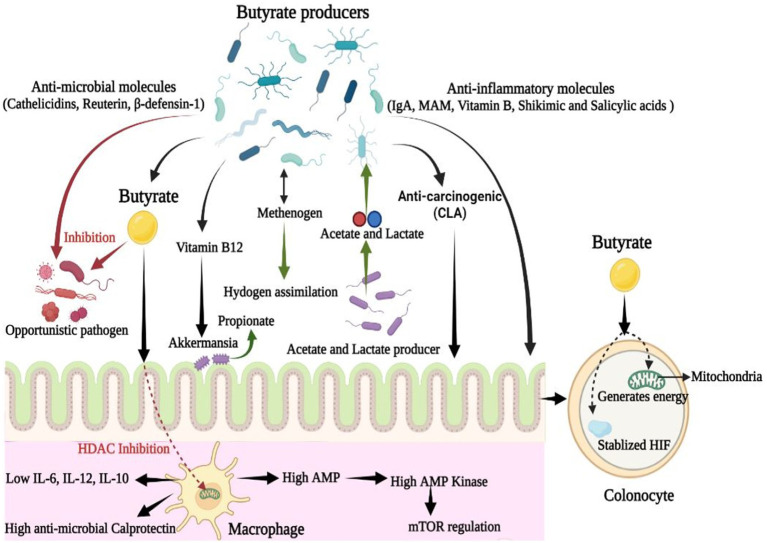
In the gut, butyrate-producing microbial communities play a crucial role in maintaining a healthy gut environment as they restrict the entry and establishment of other microbes, especially pathogenic microbes. Butyrate is used by colonocytes to generate energy which increases epithelial oxygen consumption (Litvak et al., 2018). As a result, the presence of butyrate producing bacteria helps maintain an anaerobic environment in the gut, which further prevents the colonization of opportunistic aerobic pathogens, such as Salmonella and E. coli (Manson et al., 2008; Parada Venegas et al., 2019). Butyrate also regulates the production of cathelicidins, a polycationic peptide that participates in mammalian innate immunity and exhibits broad-spectrum antimicrobial activity against potential gut pathogens (van Vliet et al., 2010; Kościuczuk et al., 2012; van Harten et al., 2018). Moreover, butyrate-producing bacteria such as E. hallii produces reuterin, a broad-spectrum antimicrobial agent with yeast inhibition activity (Engels et al., 2016b) while metabolizing glycerol to 3-hydroxypropionaldehyde (Figure 2). These anti-microbial agents limit the incursion or abundance of potential pathogens and thus, help maintain a healthy gut microbiome.
Figure 2.
Dynamic role of butyrate producing microbial communities in gut: Along with butyrate, butyrate-producing communities also produce various bioactive molecules that are anti-microbial, anti-inflammatory, and anti-carcinogenic in nature. These molecules are of therapeutic importance in alleviating gut-associated disorders and maintaining gut-homeostasis (CLA, Conjugated Linoleic Acid; IL, Interleukin; MAM, Microbial Anti-inflammatory Molecule).
Butyrate produced in the gut shapes the gut microbial community via regulating IgA secretion and by limiting the hyperresponsiveness of macrophages toward colonic commensals to maintain their abundance (Chang et al., 2014; Isobe et al., 2020). Butyrate regulates colonic macrophages present in the lamina propria by inhibiting HDAC, and limits the generation of proinflammatory IL-12 and IL-6, as well as antimicrobial nitric oxide from lipopolysaccharide-stimulated macrophages (Chang et al., 2014; Kibbie et al., 2021). Butyrate enhances the GPCR-independent antimicrobial activity of macrophages via metabolites, as evidenced by a study that showed that macrophages grown in the presence of microbial butyrate upregulated the expression of antimicrobial protein calprotectin but showed lowered expression of anti-inflammatory IL-10 (Schulthess et al., 2019; Jukic et al., 2021; Figure 2). Additionally, microbial butyrate significantly enhances the ability of macrophages to eliminate possible pathogens, such as Salmonella enterica and Citrobacter rodentium (Flemming, 2019). Thus, butyrate bolsters gut defense against invasive pathogens without causing tissue-damaging inflammation or hyper-responsiveness. Butyrate-induced macrophages also exhibit higher levels of AMP, an inducer of AMP-kinase (AMPK), which inhibits mammalian target of rapamycin (mTOR), the master regulator protein kinase of autophagy, which is associated with cancer, insulin resistance, and other diseases (Schulthess et al., 2019; Figure 2).
In vitro and in vivo studies have also shown that butyrate producers participate in vitamin biosynthesis, especially vitamin B complex biosynthesis. For example, Eubacterium hallii produces vitamin B12, which is symbiotically utilized by Akkermansia to produce propionate (Belzer et al., 2017; Pham et al., 2021; Figure 2). The vitamin B complex acts as an essential cofactor in various metabolic activities and is also associated with the regulation of immunological homeostasis in the host (Yoshii et al., 2019). A cross-feeding relationship is also reported between butyrogenic genera, such as Faecalibacterium, Roseburia, Anaerostipes, Eubacterium, and probiotic Bifidobacterium (Rivière et al., 2016). For example, Bifidobacterium produces lactate and acetate, which are further utilized by butyrogenic microbes, such as E. Hallii, to generate butyrate; this in turn supports the abundance of Bifidobacterium (Louis and Flint, 2009; Schwab et al., 2017). Similarly, Anaerostipes hadrus and Anaerobutyricum hallii, members of the family Lachnospiraceae, utilize lactate and acetate to produce butyrate in the gut (Duncan et al., 2004).
Importance of butyrate producers in maintaining the gut epithelial barrier
The intestinal epithelium is a single-layer structure covered by a mucous layer and functions as the first line of defense against gut pathogens. The cells of intestinal epithelium are interconnected with tight junctions. The intestinal epithelium contains mucous-secreting goblet cells that provide barrier protection by secreting mucus, which also functions as a reservoir of immunoglobulin IgA and antimicrobial peptides (Martens et al., 2018). The mucous layer is composed of mucin, and in colon MUC2 is the primary mucin-producing gene (Martens et al., 2018). The mucous layer adhering to the gut epithelium is thick and limits the microbial growth near the epithelial layer, whereas the outer mucous layer is less dense and suitable for the growth of different commensals, such as Akkermansia muciniphila, Faecalibacterium, and Eubacterium rectale (Maier et al., 2015; Martens et al., 2018). Some harmful microbes can decrease mucus thickness by degrading it, thereby allowing pathogens to enter the gut; for example, Vibrio cholerae secretes hemagglutinin protease that possesses mucolytic activity. Cholera-causing bacteria also secrete zonula occludens toxin, which further hampers epithelial integrity by acting on tight junctions (Martens et al., 2018). Another microbe, Clostridium perfringens, disrupts tight junctions by secreting endotoxins (Saitoh et al., 2015). Additionally, decreased abundance of butyrate producers leads to compromised defense and dysfunctional gut epithelium as observed in the case of Clostridium difficile infection (Antharam et al., 2013).
Faecalibacterium, a major butyrate producer in the human gut, enhances mucus formation by increasing goblet cell differentiation and expression of genes related to mucin glycosylation (Wrzosek et al., 2013). Furthermore, clinical studies have demonstrated rapid recovery in patients with cholera after oral administration of resistant starch, a butyrate precursor (Canani et al., 2011). In addition, butyrate produced by bacteria in the gut accelerates mitochondria-dependent oxygen consumption in gut epithelial cells, which stabilizes HIF. Butyrate itself also inhibits HIF-prolyl hydroxylase that degrades HIF (Wang et al., 2021). Stabilized HIF regulates the tight junction protein claudin-1, MUC2 expression, and generation of antimicrobial peptide beta defensin-1 (DEFB1) (Zheng et al., 2017; Wang et al., 2021). Butyrate also regulates the immunological aspect of barrier function as it tightens the intestinal epithelial cell barrier via inducing anti-inflammatory cytokine IL-10RA-dependent suppression of claudin-2 protein, which forms paracellular channels in tight junctions and increases gut permeability (Zheng et al., 2017; Zhu et al., 2019). A recent study also demonstrated the role of butyrate in the regulation of actin-binding protein synaptopodin (SYNPO), which is expressed in gut epithelial tight junctions and is crucial for gut-barrier integrity (Wang et al., 2020).
Protective role of butyrate producers against bowel inflammation
Based on their severity, inflammatory diseases of the gut can be categorized into irritable bowel syndrome (IBS) and inflammatory bowel disease (IBD). IBS is characterized by cramps, bloating, diarrhea, and/or constipation (Camilleri et al., 2016). There are no biological markers to confirm it; moreover, this condition does not pose major discomfort to the patients. Normally, IBS patients are identified using a questionnaire prepared by medical staff (Werlang et al., 2019). In contrast, IBD is a generic term for more severe conditions, such as Crohn’s disease and ulcerative colitis (Franzosa et al., 2019), which cause inflammation and ulcers in the intestine, rectal bleeding, anemia, and diarrhea. Incidentally, decreased butyrate levels have often been reported in both IBS and IBD. In the case of IBD, butyrate producers play important roles as they increase mucus production from goblet cells to strengthen the intestinal mucous barrier and regulate the expression of tight junction proteins via butyrate to restrict the harmful penetration through the gut (Pozuelo et al., 2015; Pascal et al., 2017; Dalile et al., 2019; Schirmer et al., 2019). Similarly, in the case of IBS lower number of butyrate producers result in a reduced availability of butyrate and thus decrease the gut permeability (Camilleri et al., 2016).
Butyrate maintains the anaerobic environment in the colon by enhancing colonocyte oxygen consumption and stabilizing HIF, while its absence facilitates the buildup of potentially harmful bacteria and molecules, such as Salmonella, E. coli, and nitric oxide (NO), respectively (Parada Venegas et al., 2019). The reduced proportion of butyrate producers is also associated with a decreased count of methanogens, which disposes of the excess hydrogen (H2) produced in the form of CH4 during dietary fermentation, one of the possible reasons for the bloating experienced by IBS and IBD patients (Pozuelo et al., 2015; Chong et al., 2019). Studies have reported that among SCFAs, butyrate alone is responsible for gut motility, possibly via regulating serotonin, and can be used to increase propulsive gut movement, making it a suitable microbial therapeutic for patients with IBS (Vincent et al., 2018). An induced-colitis study in a murine model confirmed the decrease in butyrate-producing Clostridium clusters and reduced butyrate levels in the gut, which facilitated gut epithelial oxygenation and growth of Salmonella enterica serovar Typhimurium (S. Typhimurium), a known cause of foodborne gut inflammation and diarrhea (Rivera-Chávez et al., 2016; Anderson and Kendall, 2017; Litvak et al., 2019). Similarly, a reduced proportion of butyrate producers in the gut increases the expansion of aerobic Enterobacteriaceae, which is a common marker of gut dysbiosis (Matamouros et al., 2018; Parada Venegas et al., 2019). Studies have demonstrated a decreased count of butyrate-producing Faecalibacterium and Roseburia in the gut of ulcerative colitis patients (Sartor, 2011; Franzosa et al., 2019). On the other hand, the culture supernatant of Faecalibacterium was reportedly effective against IBD (Crohn’s disease) and colitis in murine models, and Faecalibacterium was found to secrete an anti-inflammatory peptide (MAM, m.wt. 15 KDa), which inhibits pro-inflammatory NF-κB signaling to arrest colitis (Quévrain et al., 2016). Additionally, Faecalibacterium inhibits colitis by producing anti-inflammatory shikimic and salicylic acids (Miquel et al., 2015). In another study, a combination of six different butyrate producers (B. pullicaecorum 25–3 T, F. prausnitzii, Roseburia hominis, Roseburia inulinivorans, Anaerostipes caccae, and E. hallii) reportedly enhanced butyrate production in IBD fecal microbiota by 5–10% and enhanced higher gut-barrier integrity, as examined in the Caco-2 cell line (Geirnaert et al., 2017). Similarly, patients with Clostridium difficile infection, which has a high mortality rate and increases the chances of acquiring hospital-acquired diarrhea, also exhibited a significant depletion in butyrate producers such as Roseburia, Anaerostipes, Blautia, and Faecalibacterium, along with lowered butyrate levels (Antharam et al., 2013). By contrast, in the case of mucositis, microbial butyrate enhances mucosal healing to accelerate the recovery of inflamed gut epithelium by stimulating the migration of gut epithelial cells (van Vliet et al., 2010).
By acting as a ligand, microbial butyrate participates in anti-inflammatory reactions to cease the inflammation and maintain gut homeostasis through the aryl hydrocarbon receptor (AhR) and various G-protein coupled receptors (GPCRs) such as GPR109a, GPR43, and GPR41 (Marinelli et al., 2019; Yip et al., 2021). AhR and GPCRs are transcription factors that control the transcriptional machinery of various immunoregulators following their activation. AhR exhibits the anti-inflammatory effect by enhancing anti-inflammatory IL-10 secreting B and Th2 cells, with a decline in pro-inflammatory Th1 and Th17 cells (Dong and Perdew, 2020; Abdulla et al., 2021). Among GPCRs, butyrate-activated GPR109a promotes differentiation of Treg cells and enhances anti-inflammatory IL-10 producing Th2 cells and plasma levels of IL-10, which in turn inhibits pro-inflammatory IL-17 (Akitsu and Iwakura, 2018; Martens et al., 2018). Upon butyrate activation, GPR43 reduces CD4 T-cell proliferation and limits the secretion of pro-inflammatory cytokines such as IL-17 and IL-22 (Kibbie et al., 2021). In addition, butyrogenic clostridia such as Clostridium butyricum limit IBD-associated inflammation by increasing Treg cell differentiation through microbial butyrate, which exerts its effects via transforming growth factor-β (TGF-β) (Ihara et al., 2017).
Relevance of butyrate producers in CRC and tumorigenesis
Colorectal cancer (CRC) begins with a growth of the inner lining of the colon and rectum, which can later transform into cancerous polyps (Das et al., 2017; Salmerón et al., 2022). Evidence has shown that alterations in the gut microbiota are closely associated with CRC progression (Xie et al., 2020). Microbiome profiles of CRC patients exhibit a decrease in major butyrate-producing genera, including Roseburia, Clostridiales, Faecalibacterium, and members of the Lachnospiraceae family, and administration of butyrate-producing Clostridium butyricum was effective in decreasing the proliferation of cancerous cells and enhancing cancer cell apoptosis (Zou et al., 2018; Stoeva et al., 2021). Similarly, a lower abundance of Eubacterium ventriosum is a potential biomarker for CRC patients (Mukherjee et al., 2020), and its administration in CRC patients has been patented,1 indicating its significant therapeutic importance. Additionally, gut commensals such as Butyricicoccus pullicaecorum, Butyrivibrio fibrisolvens, Ruminococcus bromii, and members of the family Lachnospiraceae also produce sodium butyrate upon fermenting dietary fibers, which inhibits CRC cell proliferation by regulating immune cells such as natural killer cells and macrophages, and causes apoptosis (Xi et al., 2021).
Luminal butyrate inhibits CRC mainly through HDAC inhibition by inactivating oncogenic pathways, such as mitogen-activated protein kinase (MAPK), Akt/ERK signaling, Wnt signaling pathway, and TGF-β signaling (Li et al., 2017; Geng et al., 2021). Butyrate-mediated inhibition of HDAC3 blocks the activation of Akt and ERK1/2, which are required for CRC cell migration and invasion (Li et al., 2017). Similarly, Wnt is a hydrophobic glycoprotein ligand that participates in various cellular processes, and aberration in Wnt signaling can cause CRC (Patel et al., 2019). An aberrant Wnt pathway can be suppressed by the butyrate-dependent activation of GPR109, as exhibited by Clostridium butyricum, but further investigation is required to confirm its direct or indirect role (Chen D. et al., 2020). Similarly, TGF-β is an immunosuppressive cytokine that regulates cell proliferation, differentiation, growth, and apoptosis, and any decrease in the inhibitory activity of TGF-β can lead to cancer, including CRC (Ku et al., 2007). Recent in vivo findings have reported significant expression of TGF-β after ingestion of dietary sodium butyrate, which can help combat CRC (Liu et al., 2014). Usually, cancer cells have a higher glucose demand and metabolic rate to support accelerated cell growth, which makes glycolysis inhibitors a promising anticancer drug candidate (Figure 3). Besides being an HDAC inhibitor, microbial butyrate differentially inhibits glucose transport, glycolysis, and DNA synthesis in cancerous colonocytes via inhibiting GLUT1 and glucose-6-phosphate dehydrogenase (G6PD) through the GPR109a-AKT pathway (Geng et al., 2021). GLUT1 is a glucose transporter, while G6PD is a key enzyme that produces ribose-5-phosphate for nucleotide synthesis (Geng et al., 2021). Microbial butyrate also inhibits CRC by increasing the 2-oxoglutarate level, which in turn downregulates proinflammatory cytokines such as IL-6, IL-22, IL1-β, and TNF-α (Wang et al., 2021). Furthermore, colonic butyrogenic microbes such as Roseburia and Butyrivibrio metabolize linoleic acid to produce the precursor of conjugated linoleic acid (CLA) (Devillard et al., 2007; Louis and Flint, 2009), which induces apoptosis and has been reported as an effective anti-carcinogenic molecule in various studies, including CRC (den Hartigh, 2019). Roseburia species, which are among the most active linoleic acid metabolizers, also produce vaccenic acid, which is known to be beneficial for the host (Devillard et al., 2007).
Figure 3.
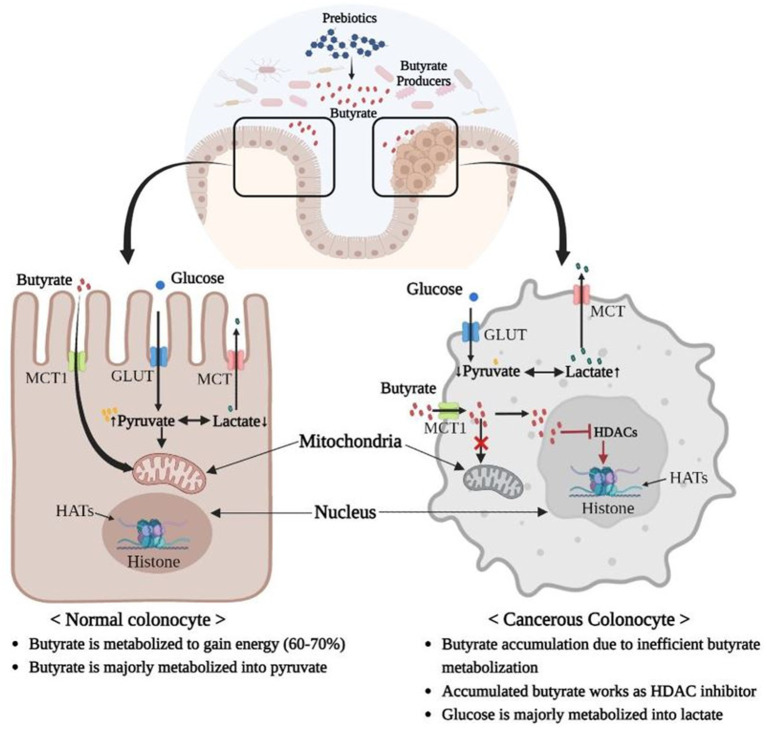
Warburg Effect: Inefficient butyrate metabolization by mitochondria of cancerous colonocytes leads to accumulation of butyrate, which in turn acts as an HDAC inhibitor and induces cancer. Additionally, majority of glucose is converted into lactate in cancerous colonocytes owing to their higher glycolysis rates, which is less energy efficient compared to phosphorylation of pyruvate in mitochondria via the TCA cycle. Therefore, cancerous colonocytes need higher glucose inflow and a higher rate of glycolysis to survive (MCT, Monocarboxylate Transporter; GLUT, Glucose Transporter; HDAC, Histone deacetylase, HAT; Histone acetyltransferase).
In contrast, some studies have reported an association between microbiota-derived butyrate and CRC upregulation (Okumura et al., 2021). This is a butyrate-paradox, wherein butyrate can act differently in normal and cancerous colonocytes. This is due to a metabolic shift of cancerous cells toward glycolysis, also called Warburg effect. In colonocyte mitochondria, butyrate is not metabolized to the same extent as in normal cells, and therefore, accumulates in the nucleus where it inhibits HDAC (Bultman and Jobin, 2014; Bultman, 2016; Hajjar et al., 2021; Figure 3). A similar paradox was observed in the microbial regulation of the PI3/Akt pathway, which is a major signaling cascade involved in the regulation of normal cellular activities, such as cell proliferation, growth, motility, and survival; however, its aberrant activation is associated with cancer (Luo et al., 2003; Prossomariti et al., 2020). Studies have reported that the PI3-Akt pathway is activated in 60–70% of CRC patients, and inhibitors of this pathway are considered therapeutic (Malinowsky et al., 2014). In the dysbiotic gut of CRC patients, the abundance of rare Porphyromonas species, such as P. gingivalis and P. asaccharolytica, may promote CRC via butyrate-mediated activation of the PI3/Akt pathway (Okumura et al., 2021).
Relevance in gut-organ axis
Butyrate producers are associated with various gut-organ axes, such as the gut-brain, gut-lung, gut-liver, gut, kidney, and gut-heart axes (Ahlawat and Asha, 2021). In such complex relationships, butyrate producers act as microbial regulators and exert their effects through their metabolites. As in the gut-brain axis, microbiota-induced expression of AhR in gut neurons allows them to respond to the environment of the gut lumen while simultaneously connecting their functional output to the gut (Obata et al., 2020). As stated earlier, butyrate acts as a ligand for AhR, making butyrate producers a relevant community in the gut-brain axis. Studies have identified the antidepressant effects of the butyrate-producing genera Butyricimonas and Coprococcus and their depletion in depressed individuals (Yang et al., 2017; Valles-Colomer et al., 2019). Similarly, Faecalibacterium and Coprococcus are robustly associated with better mental health (Valles-Colomer et al., 2019). Metagenomic analysis of fecal samples from a Belgian cohort identified butyrate-producing Alistipes and Roseburia as potential producers of serotonin (Valles-Colomer et al., 2019), which is a neurotransmitter expressed abundantly in the gut where it regulates bowel movement, secretion (McLean et al., 2007), and glucose homeostasis (Singh et al., 2022). Studies also confirmed the gut-lung axis, as it’s been found that gut dysbiosis is closely related to the occurrence of asthma and pulmonary diseases. In infants reduced gut microbial diversity is reported to increases the risk of asthma and infectious respiratory diseases (Bisgaard et al., 2011; Abrahamsson et al., 2014). Specially, reduced abundance of butyrogenic Faecalibacterium in the gut is closely related with the increased risk of atopy and asthma (Dang and Marsland, 2019). In addition, during a viral infection such as influenza, through GPCR41 receptors, microbial butyrate enhances the Ly6C-monocytes in the lungs, which differentiate into alternatively activated macrophages (AAMs) that alleviate the immunopathological response in the lungs by limiting the neutrophil influx into the airways (Dang and Marsland, 2019).
The gut microbiome is also involved in the gut-liver axis because the liver receives approximately 70% blood supply from the gut, and even shows the presence of higher microbial liposaccharide (LPS) levels in the portal and hepatic circulation during chronic liver ailments (Compare et al., 2012). Microbial butyrate maintains the integrity of the gut barrier and inhibits the inflow of antigens (LPS). In murine studies, butyrate supplementation in the form of tributyrin was found to be effective in alleviating alcohol-induced liver injury (Cresci et al., 2017; Singhal et al., 2021). Alcohol-induced dysbiosis significantly reduces the members of Firmicutes and Lachnospiraceae with a lower abundance of butyrate-producing genera such as Anaerostipes, Coprococcus, and Roseburia (Singhal et al., 2021). A study based on a large human population (n = 1,148) also identified a significantly lower abundance of the genus Faecalibacterium in patients with non-alcoholic fatty liver disease (NAFLD) (Iino et al., 2019). Additionally, the butyrate-producing strain (MIYAIRI 588) of Clostridium butyricum reportedly suppresses oxidative stress and hepatic inflammatory indices in NAFLD (Endo et al., 2013).
Metabolites of protein fermentation, such as choline, phosphatidylcholine, and carnitine, are metabolized by the gut microbiota into trimethylamine, which is further converted into trimethylamine-N-oxide (TMAO) in the liver by hepatic flavin-containing monooxygenase (FMO) (Tong et al., 2022). TMAO is known to cause chronic kidney disease (CKD) and induces cardiovascular diseases such as atherosclerosis and coronary heart disease (Evenepoel et al., 2017). Although, a study also suggested that a low dose of TMAO might reduce cardiac dysfunction (Huc et al., 2018). Other than that, butyrate can lower the circulating cholesterol through reverse cholesterol transport by stimulating secretion of apoA-IV-containing lipoprotein (Chen W. et al., 2020). In addition, butyrate also enhances the secretion of glucagon-like peptide-1 (GLP-1) from the gut, which decreases blood pressure (Yadav et al., 2013). While, in CKD, the levels of uremic toxins such as indoxyl sulfate and p-cresyl sulfate are abnormally high, which can also lead to hypertension (Chen et al., 2019). Studies have reported decreased abundance of major butyrate producers such as Roseburia, Faecalibacterium, and Coprococcus in CKD patients (Jiang et al., 2017; Yang et al., 2018). In a murine study, CKD treatment with traditional medicine was found to be mediated by the butyrate-producing microbe Lachnospiraceae-NK4A136 via the gut-kidney axis (Tong et al., 2022). In addition to maintaining gut integrity to limit the level of uremic toxins, butyrate improves renal inflammation and dysfunction in patients with CKD.
Impact of selective dietary interventions to enhance butyrate producers
Prebiotic administration positively affects butyrate producers, as they metabolize prebiotics into butyrate. Prebiotics are also beneficial in treating diarrhea and cholera, as prebiotic (e.g., resistant starch) administration accelerates recovery via microbial butyrate (Canani et al., 2011). Indigestible dietary fibers are commonly used as prebiotics, but other bioactive molecules, such as polyphenols, can also function as prebiotics to generate butyrate. Polyphenol intervention significantly increases the abundance of butyrate producers such as Faecalibacterium and members of the Ruminococcaceae family (Del Bo et al., 2021). Among other polyphenols, the impact of catechins, anthocyanins, and proanthocyanidins as prebiotics is more evident because they increase the abundance of Roseburia and Faecalibacterium spp. (Alves-Santos et al., 2020). Other phenolic compounds such as caffeic acid, chlorogenic acid, and rutin are also reported to increase microbial butyrate (Catalkaya et al., 2020). Additionally, the microbial accessibility of different prebiotics also varies among butyrate producers; therefore, the administration of different prebiotics can selectively enrich specific butyrate producers (Table 2). Other than prebiotics, synbiotic treatments can also be administered to promote butyrate production in the gut (Gurry, 2017). Synbiotics contain a combination of prebiotics and probiotics, and their synergistic effects are more prominent than those of prebiotics and probiotics used individually (Singh et al., 2021). Synbiotic treatment with Bacillus subtilis DSM 32315 and L-Alanyl-L-glutamine improved butyrate levels and enhanced the major butyrate producers such as Faecalibacterium prausnitzii, both in vitro and in humans (tom Dieck et al., 2022). Similarly, another study reported the prevalence of butyrate-producing Eubacterium and Pseudobutyrivibrio upon synbiotic administration of fiber-enriched yogurt (Jaagura et al., 2022).
Table 2.
Impact of different fiber and bioactive metabolites on various gut butyrate producers.
| Dietary substance | Monomer unit | Affected microbe | Model | Reference |
|---|---|---|---|---|
| Human milk oligosaccharides (HMOs) | β-d-galactose (Gal), β-d-glucose (Glc), β-d-N-acetyglucosamine (GlcNAc), α-l-fucose (Fuc), and the sialic acid α-d-N-acetylneuraminic acid (Sia) | Roseburia↑ Eubacterium↑ | Human | Pichler et al. (2020) |
| Inulin | D-Fructose | Faecalibacterium ↑; | Human; Humanized mice | Healey et al. (2018), Van den Abbeele et al. (2011) |
| Roseburia intestinalis ↑ | ||||
| Eubacterium rectale ↑ | ||||
| Anaerostipes caccae ↑ | ||||
| Xylan | D-xylose | Roseburia intestinalis ↑ | In vitro | Leth et al. (2018) |
| Fructooligosaccharide | D-fructose | Faecalibacterium ↑ | Human | Tandon et al. (2019) |
| Ruminococcus ↑ | ||||
| Oscillospira ↑ | ||||
| Galacto-oligosaccharides | Galactose | Anaerostipes caccae ↑ | Murine | Sato et al. (2008) |
| Polyphenols | Phenol | Anaerobutyricum hallii↑ | Human | Del Bo et al. (2021) |
| Butyricicoccus spp.↑ | ||||
| Faecalibacterium prausnitzii↑ | ||||
| Pectin | Galacturonic acid | Faecalibacterium↑ | In vitro | Bang et al. (2018), Chung et al. (2016) |
| Eubacterium eligens | ||||
| Guar gum (Galactomannan polysaccharide) | Galactose and Mannose | Clostridium coccoides group↑ | Human | Ohashi et al. (2015) |
| Roseburia/Eubacterium rectale group↑ | ||||
| Anaerobutyricum halli↑ | ||||
| Butyrate-producing bacterium strain SS2/1↑ | ||||
| Alginate | D-mannuronic acid and L-guluronic acid | Bacteroides ovatus ↑ | In vitro | Li et al. (2016) |
| Bacteroides xylanisolvens ↑ | ||||
| Arabinoxylan | D-xylosyl | Roseburia/Eubacterium rectale group↑ | Murine | Damen et al. (2011) |
| Stachyose | Galactose, Glucose, and Fructose | Faecalibacterium | In vitro | Zhao et al. (2021) |
| Lactulose | Galactose and Fructose | Anaerostipes | In vitro | Bothe et al. (2017) |
Strain and strategies for tomorrow
Butyrate-producing gut microbes are of significant therapeutic importance and are believed to be niche-specific next-generation probiotics. Multiple butyrate-producing probiotic strains of Clostridium butyricum (Stoeva et al., 2021) and Butyricicoccus pullicaecorum (Geirnaert et al., 2014; Boesmans et al., 2018) have been used as they exhibit good bile tolerance, viability, and metabolic activity (Table 3). Microbes of interest or butyrate producers can also be genetically manipulated to increase their butyrate-producing capacity. For example, heterologous genes required for butyrate production from acetyl-CoA can be introduced by inactivating the gene encoding the conversion of acetyl-CoA to acetate and the gene encoding the aldehyde/alcohol dehydrogenase for ethanol production or simply disrupting a CoA transferase gene, which may be an alternative route for acetate production (Ueki et al., 2014; Suo et al., 2018). Additionally, a co-culture strategy, that is an interactive microbial population of more than two microbes, can also be implemented to achieve higher levels of butyrate and increased abundance of butyrate producers in the gut. Co-culture of F. prausnitzii and Bifidobacterium catenulatum with fructooligosaccharides as an energy source resulted in a higher viable cell count and butyrate production (Kim et al., 2020). Moreover, butyrate producers of animal origin (ruminants), such as cellulose-degrading Ruminococcus albus and R. flavefaciens (Flint et al., 2008; Chassard et al., 2012), can also be considered to study their impact on human hosts.
Table 3.
Butyrate producers that can be used as microbial therapeutic to maintain microbial homeostasis and gut health.
| Microbes | Model | Reference |
|---|---|---|
| Butyricicoccus pullicaecorum 25-3T | Human | Boesmans et al. (2018) |
| Faecalibacterium prausnitzii A2-165 | Murine | Martín et al. (2015) |
| Eubacterium Hallii DSM 3353 | Human | Engels et al. (2016a) |
| Eubacterium Hallii DSM 17630 | Human | Engels et al. (2016a) |
| Eubacterium limosum KIST612 | Bio-fermenter | Litty and Müller (2021) |
| Co-culture of Clostridium hylemonae DSM 15053; or Coprococcus comes ATCC 27758; or Roseburia hominis A2-183; or Eubacterium rectale ATCC 33656; or Eubacterium biforme DSM 3989 and Clostridium ljungdahlii | Dynamic metabolic modelling | Li and Henson (2021) |
| Butyricicoccus pullicaecorum 1.20; Roseburia hominis DSM 16839; Roseburia inulinivorans DSM 16841; Anaerostipes caccae DSM 14662; Eubacterium hallii DSM 3353 | Fed batch fermenter and Caco-2 cell line | Geirnaert et al. (2017) |
| Clostridium butyricum (CGMCC0313.1) | Murine | Pan et al. (2019) |
| Clostridium butyricum (MIYAIRI 588) | Murine | Endo et al. (2013), Pan et al. (2019) |
| Clostridium butyricum Prazmowski | Murine | Wu et al. (2022) |
| Ruminococcus albus | Caco-2 cell line | Park et al. (2017) |
Conclusion
The present review critically examined all aspects of butyrate-producing gut microbial communities and their possible impact on host health to better understand their therapeutic significance. We considered the significance of butyrate producers and butyrate in the gut to understand their importance as microbial therapeutics. Although butyrate is an important metabolite, butyrate producers are much more important as they actively control the gut microbiome via various anti-microbial and anti-inflammatory molecules, and by synthesizing vitamin B. Butyrate-producing microbial communities inhibit cancer growth by secreting anti-carcinogenic substances and regulate tumorigenesis via butyrate. Butyrate producers are promising next-generation probiotics, and their counts in the gut can be regulated by dietary interventions to benefit the host. Moreover, butyrate producers can also be genetically manipulated to enhance butyrate synthesis, making them suitable microbial therapeutic agents. We also see the possibility of introducing new butyrate communities to the gut, which are alien to the human gut, to study their impact and to analyze any possible health effects. However, detailed studies are required to cease all safety concerns regarding the introduction of animal or soil origin butyrate producers in the human gut.
Author contributions
VS conceptualized, analyzed, and wrote the draft. GL and HS participated in writing and project management. HK and EK supervised the manuscript. TU and J-HS supervised, reviewed, and approved the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by Korea Basic Science Institute (National research Facilities and Equipment center) grant funded by the Ministry of Education (2021R1A6C101A416), and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2016R1A6A1A03012862). This research was also supported by the project to train professional personnel in biological materials by the Ministry of Environment.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank the KNU NGS Core Facility (Kyungpook National University, Daegu, South Korea) for providing the facilities.
Footnotes
References
- Abdulla O. A., Neamah W., Sultan M., Alghetaa H. K., Singh N., Busbee P. B., et al. (2021). The ability of AhR ligands to attenuate delayed type hypersensitivity reaction is associated with alterations in the gut microbiota. Front. Immunol. 12:684727. doi: 10.3389/fimmu.2021.684727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamsson T., Jakobsson H., Andersson A. F., Björkstén B., Engstrand L., Jenmalm M. (2014). Low gut microbiota diversity in early infancy precedes asthma at school age. Clin. Exp. Allergy 44, 842–850. doi: 10.1111/cea.12253, PMID: [DOI] [PubMed] [Google Scholar]
- Ahlawat S., Asha S. K. K. (2021). Gut–organ axis: a microbial outreach and networking. Lett. Appl. Microbiol. 72, 636–668. doi: 10.1111/lam.13333, PMID: [DOI] [PubMed] [Google Scholar]
- Akitsu A., Iwakura Y. (2018). Interleukin-17-producing γδ T (γδ17) cells in inflammatory diseases. Immunology 155, 418–426. doi: 10.1111/imm.12993, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessi A. M., Gray V., Farquharson F. M., Flores-López A., Shaw S., Stead D., et al. (2020). β-Glucan is a major growth substrate for human gut bacteria related to Coprococcus eutactus. Environ. Microbiol. 22, 2150–2164. doi: 10.1111/1462-2920.14977, PMID: [DOI] [PubMed] [Google Scholar]
- Allen-Vercoe E., Daigneault M., White A., Panaccione R., Duncan S. H., Flint H. J., et al. (2012). Anaerostipes hadrus comb. nov., a dominant species within the human colonic microbiota; reclassification of Eubacterium hadrum Moore et al. Anaerobe 18, 523–529. doi: 10.1016/j.anaerobe.2012.09.002, PMID: [DOI] [PubMed] [Google Scholar]
- Altemani F., Barrett H. L., Gomez-Arango L., Josh P., David McIntyre H., Callaway L. K., et al. (2021). Pregnant women who develop preeclampsia have lower abundance of the butyrate-producer Coprococcus in their gut microbiota. Pregnancy Hypertens. 23, 211–219. doi: 10.1016/j.preghy.2021.01.002, PMID: [DOI] [PubMed] [Google Scholar]
- Alves-Santos A. M., Sugizaki C. S. A., Lima G. C., Naves M. M. V. (2020). Prebiotic effect of dietary polyphenols: a systematic review. J. Funct. Foods 74:104169. doi: 10.1016/j.jff.2020.104169 [DOI] [Google Scholar]
- Anderson C. J., Kendall M. M. (2017). Salmonella enterica serovar Typhimurium strategies for host adaptation. Front. Microbiol. 8:1983. doi: 10.3389/fmicb.2017.01983, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antharam V. C., Li E. C., Ishmael A., Sharma A., Mai V., Rand K. H., et al. (2013). Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol. 51, 2884–2892. doi: 10.1128/JCM.00845-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appert O., Garcia A. R., Frei R., Roduit C., Constancias F., Neuzil-Bunesova V., et al. (2020). Initial butyrate producers during infant gut microbiota development are endospore formers. Environ. Microbiol. 22, 3909–3921. doi: 10.1111/1462-2920.15167, PMID: [DOI] [PubMed] [Google Scholar]
- Bang S.-J., Kim G., Lim M. Y., Song E.-J., Jung D.-H., Kum J.-S., et al. (2018). The influence of in vitro pectin fermentation on the human fecal microbiome. AMB Express 8:98. doi: 10.1186/s13568-018-0629-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belzer C., Chia L. W., Aalvink S., Chamlagain B., Piironen V., Knol J., et al. (2017). Microbial metabolic networks at the mucus layer lead to diet-independent butyrate and vitamin B12 production by intestinal symbionts. MBio 8, e00770–e00717. doi: 10.1128/mBio.00770-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgaard H., Li N., Bonnelykke K., Chawes B. L. K., Skov T., Paludan-Müller G., et al. (2011). Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J. Allergy Clin. Immunol. 128:e5, 646–652.e5. doi: 10.1016/j.jaci.2011.04.060 [DOI] [PubMed] [Google Scholar]
- Boesmans L., Valles-Colomer M., Wang J., Eeckhaut V., Falony G., Ducatelle R., et al. (2018). Butyrate producers as potential next-generation probiotics: safety assessment of the administration of Butyricicoccus pullicaecorum to healthy volunteers. mSystems 3, e00094–e00018. doi: 10.1128/mSystems.00094-18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothe M. K., Maathuis A. J. H., Bellmann S., Van der Vossen J. M. B. M., Berressem D., Koehler A., et al. (2017). Dose-dependent prebiotic effect of lactulose in a computer-controlled in vitro model of the human large intestine. Nutrients 9:767. doi: 10.3390/nu9070767, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braune A., Gütschow M., Engst W., Blaut M. (2001). Degradation of quercetin and luteolin by Eubacterium ramulus. Appl. Environ. Microbiol. 67, 5558–5567. doi: 10.1128/AEM.67.12.5558-5567.2001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui T. P. N., Ritari J., Boeren S., De Waard P., Plugge C. M., De Vos W. M. (2015). Production of butyrate from lysine and the Amadori product fructoselysine by a human gut commensal. Nat. Commun. 6, 1–10. doi: 10.1038/ncomms10062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui T. P. N., Schols H. A., Jonathan M., Stams A. J., De Vos W. M., Plugge C. M. (2019). Mutual metabolic interactions in co-cultures of the intestinal Anaerostipes rhamnosivorans with an acetogen, methanogen, or pectin-degrader affecting butyrate production. Front. Microbiol. 10:2449. doi: 10.3389/fmicb.2019.02449, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman S. J. (2016). The microbiome and its potential as a cancer preventive intervention. Seminars Oncol. 43, 97–106. doi: 10.1053/j.seminoncol.2015.09.001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bultman S. J., Jobin C. (2014). Microbial-derived butyrate: an oncometabolite or tumor-suppressive metabolite? Cell Host Microbe 16, 143–145. doi: 10.1016/j.chom.2014.07.011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilleri M., Oduyebo I., Halawi H. (2016). Chemical and molecular factors in irritable bowel syndrome: current knowledge, challenges, and unanswered questions. American journal of physiology-gastrointestinal and liver. Physiology 311, G777–G784. doi: 10.1152/ajpgi.00242.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canani R. B., Di Costanzo M., Leone L., Pedata M., Meli R., Calignano A. (2011). Potential beneficial effects of butyrate in intestinal and extraintestinal diseases. World J. Gastroenterol. 17, 1519–1528. doi: 10.3748/wjg.v17.i12.1519, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalkaya G., Venema K., Lucini L., Rocchetti G., Delmas D., Daglia M., et al. (2020). Interaction of dietary polyphenols and gut microbiota: microbial metabolism of polyphenols, influence on the gut microbiota, and implications on host health. Food Front. 1, 109–133. doi: 10.1002/fft2.25 [DOI] [Google Scholar]
- Chambers E. S., Preston T., Frost G., Morrison D. J. (2018). Role of gut microbiota-generated short-chain fatty acids in metabolic and cardiovascular health. Curr. Nutr. Rep. 7, 198–206. doi: 10.1007/s13668-018-0248-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P. V., Hao L., Offermanns S., Medzhitov R. (2014). The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. 111, 2247–2252. doi: 10.1073/pnas.1322269111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassard C., Delmas E., Robert C., Lawson P. A., Bernalier-Donadille A. (2012). Ruminococcus champanellensis sp. nov., a cellulose-degrading bacterium from human gut microbiota. Int. J. Syst. Evol. Microbiol. 62, 138–143. doi: 10.1099/ijs.0.027375-0 [DOI] [PubMed] [Google Scholar]
- Chen Y.-Y., Chen D.-Q., Chen L., Liu J.-R., Vaziri N. D., Guo Y., et al. (2019). Microbiome–metabolome reveals the contribution of gut–kidney axis on kidney disease. J. Transl. Med. 17, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Jin D., Huang S., Wu J., Xu M., Liu T., et al. (2020). Clostridium butyricum, a butyrate-producing probiotic, inhibits intestinal tumor development through modulating Wnt signaling and gut microbiota. Cancer Lett. 469, 456–467. doi: 10.1016/j.canlet.2019.11.019, PMID: [DOI] [PubMed] [Google Scholar]
- Chen G., Ran X., Li B., Li Y., He D., Huang B., et al. (2018). Sodium butyrate inhibits inflammation and maintains epithelium barrier integrity in a TNBS-induced inflammatory bowel disease mice model. EBioMedicine 30, 317–325. doi: 10.1016/j.ebiom.2018.03.030, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Zhang S., Wu J., Ye T., Wang S., Wang P., et al. (2020). Butyrate-producing bacteria and the gut-heart axis in atherosclerosis. Clin. Chim. Acta 507, 236–241. doi: 10.1016/j.cca.2020.04.037, PMID: [DOI] [PubMed] [Google Scholar]
- Chong P. P., Chin V. K., Looi C. Y., Wong W. F., Madhavan P., Yong V. C. (2019). The microbiome and irritable bowel syndrome–a review on the pathophysiology, current research and future therapy. Front. Microbiol. 10:1136. doi: 10.3389/fmicb.2019.01136, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W. S. F., Walker A. W., Louis P., Parkhill J., Vermeiren J., Bosscher D., et al. (2016). Modulation of the human gut microbiota by dietary fibres occurs at the species level. BMC Biol. 14:3. doi: 10.1186/s12915-015-0224-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compare D., Coccoli P., Rocco A., Nardone O. M., De Maria S., Cartenì M., et al. (2012). Gut–liver axis: the impact of gut microbiota on non alcoholic fatty liver disease. Nutr. Metab. Cardiovasc. Dis. 22, 471–476. doi: 10.1016/j.numecd.2012.02.007 [DOI] [PubMed] [Google Scholar]
- Cresci G. A., Glueck B., McMullen M. R., Xin W., Allende D., Nagy L. E. (2017). Prophylactic tributyrin treatment mitigates chronic-binge ethanol-induced intestinal barrier and liver injury. J. Gastroenterol. Hepatol. 32, 1587–1597. doi: 10.1111/jgh.13731, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuff M. A., Lambert D. W., Shirazi-Beechey S. P. (2002). Substrate-induced regulation of the human colonic monocarboxylate transporter, MCT1. J. Physiol. 539, 361–371. doi: 10.1113/jphysiol.2001.014241, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalile B., Van Oudenhove L., Vervliet B., Verbeke K. (2019). The role of short-chain fatty acids in microbiota–gut–brain communication. Nat. Rev. Gastroenterol. Hepatol. 16, 461–478. doi: 10.1038/s41575-019-0157-3, PMID: [DOI] [PubMed] [Google Scholar]
- Damen B., Verspreet J., Pollet A., Broekaert W. F., Delcour J. A., CMJMN C., et al. (2011). Prebiotic effects and intestinal fermentation of cereal arabinoxylans and arabinoxylan oligosaccharides in rats depend strongly on their structural properties and joint presence. Mol. Nutr. Food Res. 55, 1862–1874. doi: 10.1002/mnfr.201100377 [DOI] [PubMed] [Google Scholar]
- Dang A. T., Marsland B. J. (2019). Microbes, metabolites, and the gut–lung axis. Mucosal Immunol. 12, 843–850. doi: 10.1038/s41385-019-0160-6, PMID: [DOI] [PubMed] [Google Scholar]
- Das V., Kalita J., Pal M. (2017). Predictive and prognostic biomarkers in colorectal cancer: a systematic review of recent advances and challenges. Biomed. Pharmacother. 87, 8–19. doi: 10.1016/j.biopha.2016.12.064, PMID: [DOI] [PubMed] [Google Scholar]
- Del Bo C., Bernardi S., Cherubini A., Porrini M., Gargari G., Hidalgo-Liberona N., et al. (2021). A polyphenol-rich dietary pattern improves intestinal permeability, evaluated as serum zonulin levels, in older subjects: the MaPLE randomised controlled trial. Clin. Nutr. 40, 3006–3018. doi: 10.1016/j.clnu.2020.12.014, PMID: [DOI] [PubMed] [Google Scholar]
- den Hartigh L. J. (2019). Conjugated linoleic acid effects on cancer, obesity, and atherosclerosis: a review of pre-clinical and human trials with current perspectives. Nutrients 11:370. doi: 10.3390/nu11020370, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devillard E., McIntosh F. M., Duncan S. H., Wallace R. J. (2007). Metabolism of linoleic acid by human gut bacteria: different routes for biosynthesis of conjugated linoleic acid. J. Bacteriol. 189, 2566–2570. doi: 10.1128/JB.01359-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong F., Perdew G. H. (2020). The aryl hydrocarbon receptor as a mediator of host-microbiota interplay. Gut Microbes 12:1859812. doi: 10.1080/19490976.2020.1859812, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S. H., Aminov R. I., Scott K. P., Louis P., Stanton T. B., Flint H. J. (2006). Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces. Int. J. Syst. Evol. Microbiol. 56, 2437–2441. doi: 10.1099/ijs.0.64098-0, PMID: [DOI] [PubMed] [Google Scholar]
- Duncan S. H., Louis P., Flint H. J. (2004). Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 70, 5810–5817. doi: 10.1128/AEM.70.10.5810-5817.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo H., Niioka M., Kobayashi N., Tanaka M., Watanabe T. (2013). Butyrate-producing probiotics reduce nonalcoholic fatty liver disease progression in rats: new insight into the probiotics for the gut-liver axis. PLoS One 8:e63388. doi: 10.1371/journal.pone.0063388, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo A., Tanno H., Kadowaki R., Fujii T., Tochio T. (2022). Extracellular fructooligosaccharide degradation in Anaerostipes hadrus for co-metabolism with non-fructooligosaccharide utilizers. Biochem. Biophys. Res. Commun. 613, 81–86. doi: 10.1016/j.bbrc.2022.04.134, PMID: [DOI] [PubMed] [Google Scholar]
- Engels C., Ruscheweyh H.-J., Beerenwinkel N., Lacroix C., Schwab C. (2016a). The common gut microbe Eubacterium hallii also contributes to intestinal propionate formation. Front. Microbiol. 7:713. doi: 10.3389/fmicb.2016.00713, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels C., Schwab C., Zhang J., Stevens M. J. A., Bieri C., Ebert M.-O., et al. (2016b). Acrolein contributes strongly to antimicrobial and heterocyclic amine transformation activities of reuterin. Sci. Rep. 6:36246. doi: 10.1038/srep36246, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evenepoel P., Poesen R., Meijers B. (2017). The gut–kidney axis. Pediatr. Nephrol. 32, 2005–2014. doi: 10.1007/s00467-016-3527-x [DOI] [PubMed] [Google Scholar]
- Flemming A. (2019). Butyrate boosts microbicidal macrophages. Nat. Rev. Immunol. 19:135. doi: 10.1038/s41577-019-0132-9, PMID: [DOI] [PubMed] [Google Scholar]
- Flint H. J., Bayer E. A., Rincon M. T., Lamed R., White B. A. (2008). Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol. 6, 121–131. doi: 10.1038/nrmicro1817, PMID: [DOI] [PubMed] [Google Scholar]
- Franzosa E. A., Sirota-Madi A., Avila-Pacheco J., Fornelos N., Haiser H. J., Reinker S., et al. (2019). Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 4, 293–305. doi: 10.1038/s41564-018-0306-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto T., Imaeda H., Takahashi K., Kasumi E., Bamba S., Fujiyama Y., et al. (2013). Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn’s disease. J. Gastroenterol. Hepatol. 28, 613–619. doi: 10.1111/jgh.12073 [DOI] [PubMed] [Google Scholar]
- Geirnaert A., Calatayud M., Grootaert C., Laukens D., Devriese S., Smagghe G., et al. (2017). Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci. Rep. 7:11450. doi: 10.1038/s41598-017-11734-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geirnaert A., Steyaert A., Eeckhaut V., Debruyne B., Arends J. B. A., Van Immerseel F., et al. (2014). Butyricicoccus pullicaecorum, a butyrate producer with probiotic potential, is intrinsically tolerant to stomach and small intestine conditions. Anaerobe 30, 70–74. doi: 10.1016/j.anaerobe.2014.08.010, PMID: [DOI] [PubMed] [Google Scholar]
- Geng H.-W., Yin F.-Y., Zhang Z.-F., Gong X., Yang Y. (2021). Butyrate suppresses glucose metabolism of colorectal cancer cells via GPR109a-AKT signaling pathway and enhances chemotherapy. Front. Mol. Biosci. 8:634874. doi: 10.3389/fmolb.2021.634874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilijamse P. W., Hartstra A. V., Levin E., Wortelboer K., Serlie M. J., Ackermans M. T., et al. (2020). Treatment with Anaerobutyricum soehngenii: a pilot study of safety and dose–response effects on glucose metabolism in human subjects with metabolic syndrome. npj Biofilms Microbiomes 6:16. doi: 10.1038/s41522-020-0127-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V. K., Paul S., Dutta C. (2017). Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front. Microbiol. 8:1162. doi: 10.3389/fmicb.2017.01162, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurry T. (2017). Synbiotic approaches to human health and well-being. Microb. Biotechnol. 10, 1070–1073. doi: 10.1111/1751-7915.12789, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar R., Richard C. S., Santos M. M. (2021). The role of butyrate in surgical and oncological outcomes in colorectal cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 320, G601–G608. doi: 10.1152/ajpgi.00316.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer H. M., Jonkers D., Venema K., Vanhoutvin S., Troost F. J., Brummer R. J. (2008). Review article: the role of butyrate on colonic function. Aliment. Pharmacol. Ther. 27, 104–119. doi: 10.1111/j.1365-2036.2007.03562.x, PMID: [DOI] [PubMed] [Google Scholar]
- Hanson B. T., Dimitri Kits K., Löffler J., Burrichter A. G., Fiedler A., Denger K., et al. (2021). Sulfoquinovose is a select nutrient of prominent bacteria and a source of hydrogen sulfide in the human gut. ISME J. 15, 2779–2791. doi: 10.1038/s41396-021-00968-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Healey G., Murphy R., Butts C., Brough L., Whelan K., Coad J. (2018). Habitual dietary fibre intake influences gut microbiota response to an inulin-type fructan prebiotic: a randomised, double-blind, placebo-controlled, cross-over, human intervention study. Br. J. Nutr. 119, 176–189. doi: 10.1017/S0007114517003440, PMID: [DOI] [PubMed] [Google Scholar]
- Hold G. L., Schwiertz A., Aminov R. I., Blaut M., Flint H. J. (2003). Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl. Environ. Microbiol. 69, 4320–4324. doi: 10.1128/AEM.69.7.4320-4324.2003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huc T., Drapala A., Gawrys M., Konop M., Bielinska K., Zaorska E., et al. (2018). Chronic, low-dose TMAO treatment reduces diastolic dysfunction and heart fibrosis in hypertensive rats. Am. J. Phys. Heart Circ. Phys. 315, H1805–H1820. doi: 10.1152/ajpheart.00536.2018 [DOI] [PubMed] [Google Scholar]
- Ihara S., Hirata Y., Koike K. (2017). TGF-β in inflammatory bowel disease: a key regulator of immune cells, epithelium, and the intestinal microbiota. J. Gastroenterol. 52, 777–787. doi: 10.1007/s00535-017-1350-1, PMID: [DOI] [PubMed] [Google Scholar]
- Iino C., Endo T., Mikami K., Hasegawa T., Kimura M., Sawada N., et al. (2019). Significant decrease in Faecalibacterium among gut microbiota in nonalcoholic fatty liver disease: a large BMI-and sex-matched population study. Hepatol. Int. 13, 748–756. doi: 10.1007/s12072-019-09987-8, PMID: [DOI] [PubMed] [Google Scholar]
- Isobe J., Maeda S., Obata Y., Iizuka K., Nakamura Y., Fujimura Y., et al. (2020). Commensal-bacteria-derived butyrate promotes the T-cell-independent IgA response in the colon. Int. Immunol. 32, 243–258. doi: 10.1093/intimm/dxz078, PMID: [DOI] [PubMed] [Google Scholar]
- Jaagura M., Part N., Adamberg K., Kazantseva J., Viiard E. (2022). Consumption of multi-fiber enriched yogurt is associated with increase of Bifidobacterium animalis and butyrate producing bacteria in human fecal microbiota. J. Funct. Foods 88:104899. doi: 10.1016/j.jff.2021.104899 [DOI] [Google Scholar]
- Jiang S., Xie S., Lv D., Wang P., He H., Zhang T., et al. (2017). Alteration of the gut microbiota in Chinese population with chronic kidney disease. Sci. Rep. 7:2870. doi: 10.1038/s41598-017-02989-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukic A., Bakiri L., Wagner E. F., Tilg H., Adolph T. E. (2021). Calprotectin: from biomarker to biological function. Gut 70, 1978–1988. doi: 10.1136/gutjnl-2021-324855, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamp K., Li N., Lachance D. M., Saad K., Tolentino E., Yoo L., et al. (2022). Interpersonal variability in gut microbial calprotectin metabolism. Gastro Hep Adv. 1, 853–856. doi: 10.1016/j.gastha.2022.05.007, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibbie J. J., Dillon S. M., Thompson T. A., Purba C. M., McCarter M. D., Wilson C. C. (2021). Butyrate directly decreases human gut lamina propria CD4 T cell function through histone deacetylase (HDAC) inhibition and GPR43 signaling. Immunobiology 226:152126. doi: 10.1016/j.imbio.2021.152126, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Jeong Y., Kang S., You H. J., Ji G. E. (2020). Co-culture with Bifidobacterium catenulatum improves the growth, gut colonization, and butyrate production of Faecalibacterium prausnitzii: in vitro and in vivo studies. Microorganisms 8:788. doi: 10.3390/microorganisms8050788, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kościuczuk E. M., Lisowski P., Jarczak J., Strzałkowska N., Jóźwik A., Horbańczuk J., et al. (2012). Cathelicidins: family of antimicrobial peptides. A review. Mol. Biol. Rep. 39, 10957–10970. doi: 10.1007/s11033-012-1997-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautkramer K. A., Fan J., Bäckhed F. (2021). Gut microbial metabolites as multi-kingdom intermediates. Nat. Rev. Microbiol. 19, 77–94. doi: 10.1038/s41579-020-0438-4, PMID: [DOI] [PubMed] [Google Scholar]
- Ku J.-L., Park S.-H., Yoon K.-A., Shin Y.-K., Kim K.-H., Choi J.-S., et al. (2007). Genetic alterations of the TGF-β signaling pathway in colorectal cancer cell lines: a novel mutation in Smad3 associated with the inactivation of TGF-β-induced transcriptional activation. Cancer Lett. 247, 283–292. doi: 10.1016/j.canlet.2006.05.008, PMID: [DOI] [PubMed] [Google Scholar]
- Leth M. L., Ejby M., Workman C., Ewald D. A., Pedersen S. S., Sternberg C., et al. (2018). Differential bacterial capture and transport preferences facilitate co-growth on dietary xylan in the human gut. Nat. Microbiol. 3, 570–580. doi: 10.1038/s41564-018-0132-8, PMID: [DOI] [PubMed] [Google Scholar]
- Li Q., Ding C., Meng T., Lu W., Liu W., Hao H., et al. (2017). Butyrate suppresses motility of colorectal cancer cells via deactivating Akt/ERK signaling in histone deacetylase dependent manner. J. Pharmacol. Sci. 135, 148–155. doi: 10.1016/j.jphs.2017.11.004, PMID: [DOI] [PubMed] [Google Scholar]
- Li X., Henson M. A. (2021). Dynamic metabolic modelling predicts efficient acetogen–gut bacterium cocultures for CO-to-butyrate conversion. J. Appl. Microbiol. 131, 2899–2917. doi: 10.1111/jam.15155, PMID: [DOI] [PubMed] [Google Scholar]
- Li M., Li G., Shang Q., Chen X., Liu W., Xe P., et al. (2016). In vitro fermentation of alginate and its derivatives by human gut microbiota. Anaerobe 39, 19–25. doi: 10.1016/j.anaerobe.2016.02.003, PMID: [DOI] [PubMed] [Google Scholar]
- Lin L., Zhang J. (2017). Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 18:2. doi: 10.1186/s12865-016-0187-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litty D., Müller V. (2021). Butyrate production in the acetogen Eubacterium limosum is dependent on the carbon and energy source. Microb. Biotechnol. 14, 2686–2692. doi: 10.1111/1751-7915.13779, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak Y., Byndloss M. X., Baumler A. J. (2018). Colonocyte metabolism shapes the gut microbiota. Science 362:eaat9076. doi: 10.1126/science.aat9076, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litvak Y., Mon K. K., Nguyen H., Chanthavixay G., Liou M., Velazquez E. M., et al. (2019). Commensal Enterobacteriaceae protect against Salmonella colonization through oxygen competition. Cell Host Microbe 25:e5, 128–139.e5. doi: 10.1016/j.chom.2018.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Chang G., Huang J., Wang Y., Ma N., Roy A. C., et al. (2019). Sodium butyrate inhibits the inflammation of lipopolysaccharide-induced acute lung injury in mice by regulating the toll-like receptor 4/nuclear factor kappaB signaling pathway. J. Agric. Food Chem. 67, 1674–1682. doi: 10.1021/acs.jafc.8b06359, PMID: [DOI] [PubMed] [Google Scholar]
- Liu W., Yang Y., Zhang J., Gatlin D. M., Ringø E., Zhou Z. (2014). Effects of dietary microencapsulated sodium butyrate on growth, intestinal mucosal morphology, immune response and adhesive bacteria in juvenile common carp (Cyprinus carpio) pre-fed with or without oxidised oil. Br. J. Nutr. 112, 15–29. doi: 10.1017/S0007114514000610 [DOI] [PubMed] [Google Scholar]
- Louis P., Flint H. J. (2009). Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol. Lett. 294, 1–8. doi: 10.1111/j.1574-6968.2009.01514.x, PMID: [DOI] [PubMed] [Google Scholar]
- Louis P., Flint H. J. (2017). Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 19, 29–41. doi: 10.1111/1462-2920.13589 [DOI] [PubMed] [Google Scholar]
- Luo J., Manning B. D., Cantley L. C. (2003). Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell 4, 257–262. doi: 10.1016/S1535-6108(03)00248-4 [DOI] [PubMed] [Google Scholar]
- Maier E., Anderson R. C., Roy N. C. (2015). Understanding how commensal obligate anaerobic bacteria regulate immune functions in the large intestine. Nutrients 7, 45–73. doi: 10.3390/nu7010045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinowsky K., Nitsche U., Janssen K. P., Bader F. G., Späth C., Drecoll E., et al. (2014). Activation of the PI3K/AKT pathway correlates with prognosis in stage II colon cancer. Br. J. Cancer 110, 2081–2089. doi: 10.1038/bjc.2014.100, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallott E. K., Amato K. R. (2022). Butyrate production pathway abundances are similar in human and nonhuman primate gut microbiomes. Mol. Biol. Evol. 39. doi: 10.1093/molbev/msab279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manson J. M., Rauch M., Gilmore M. S. (2008). “The commensal microbiology of the gastrointestinal tract” in GI Microbiota and Regulation of the Immune System. Advances in Experimental Medicine and Biology. eds. Huffnagle G. B., Noverr M. C., vol. 635 (New York, NY: Springer; ), 15–28. [Google Scholar]
- Marinelli L., Martin-Gallausiaux C., Bourhis J.-M., Beguet-Crespel F., Blottière H. M., Lapaque N. (2019). Identification of the novel role of butyrate as AhR ligand in human intestinal epithelial cells. Sci. Rep. 9, 1–14. doi: 10.1038/s41598-018-37019-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens E. C., Neumann M., Desai M. S. (2018). Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat. Rev. Microbiol. 16, 457–470. doi: 10.1038/s41579-018-0036-x, PMID: [DOI] [PubMed] [Google Scholar]
- Martín R., Miquel S., Chain F., Natividad J. M., Jury J., Lu J., et al. (2015). Faecalibacterium prausnitzii prevents physiological damages in a chronic low-grade inflammation murine model. BMC Microbiol. 15:67. doi: 10.1186/s12866-015-0400-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matamouros S., Hayden H. S., Hager K. R., Brittnacher M. J., Lachance K., Weiss E. J., et al. (2018). Adaptation of commensal proliferating Escherichia coli to the intestinal tract of young children with cystic fibrosis. Proc. Natl. Acad. Sci. 115, 1605–1610. doi: 10.1073/pnas.1714373115, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean P. G., Borman R. A., Lee K. (2007). 5-HT in the enteric nervous system: gut function and neuropharmacology. Trends Neurosci. 30, 9–13. doi: 10.1016/j.tins.2006.11.002, PMID: [DOI] [PubMed] [Google Scholar]
- Miller T. L., Wolin M. J. (1996). Pathways of acetate, propionate, and butyrate formation by the human fecal microbial flora. Appl. Environ. Microbiol. 62, 1589–1592. doi: 10.1128/aem.62.5.1589-1592.1996, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel S., Leclerc M., Martin R., Chain F., Lenoir M., Raguideau S., et al. (2015). Identification of metabolic signatures linked to anti-inflammatory effects of Faecalibacterium prausnitzii. MBio 6, e00300–e00315. doi: 10.1128/mBio.00300-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miquel S., Martín R., Rossi O., Bermúdez-Humarán L. G., Chatel J. M., Sokol H., et al. (2013). Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 16, 255–261. doi: 10.1016/j.mib.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Mirande C., Kadlecikova E., Matulova M., Capek P., Bernalier-Donadille A., Forano E., et al. (2010). Dietary fibre degradation and fermentation by two xylanolytic bacteria Bacteroides xylanisolvens XB1AT and Roseburia intestinalis XB6B4 from the human intestine. J. Appl. Microbiol. 109, 451–460. doi: 10.1111/j.1365-2672.2010.04671.x, PMID: [DOI] [PubMed] [Google Scholar]
- Mukherjee A., Lordan C., Ross R. P., Cotter P. D. (2020). Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health. Gut Microbes 12:1802866. doi: 10.1080/19490976.2020.1802866, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal R., Tsuji H., Takahashi T., Nomoto K., Kawashima K., Nagata S., et al. (2017). Ontogenesis of the gut microbiota composition in healthy, full-term, vaginally born and breast-fed infants over the first 3 years of life: a quantitative bird’s-eye view. Front. Microbiol. 8:1388. doi: 10.3389/fmicb.2017.01388, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata Y., Castaño Á., Boeing S., Bon-Frauches A. C., Fung C., Fallesen T., et al. (2020). Neuronal programming by microbiota regulates intestinal physiology. Nature 578, 284–289. doi: 10.1038/s41586-020-1975-8, PMID: [DOI] [PubMed] [Google Scholar]
- Ohashi Y., Sumitani K., Tokunaga M., Ishihara N., Okubo T., TJBM F. (2015). Consumption of partially hydrolysed guar gum stimulates Bifidobacteria and butyrate-producing bacteria in the human large intestine. Benef Microbes 6, 451–455. doi: 10.3920/BM2014.0118 [DOI] [PubMed] [Google Scholar]
- Okumura S., Konishi Y., Narukawa M., Sugiura Y., Yoshimoto S., Arai Y., et al. (2021). Gut bacteria identified in colorectal cancer patients promote tumourigenesis via butyrate secretion. Nat. Commun. 12:5674. doi: 10.1038/s41467-021-25965-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillard D., McKain N., Chaudhary L. C., Walker N. D., Pizette F., Koppova I., et al. (2007). Relation between phylogenetic position, lipid metabolism and butyrate production by different Butyrivibrio-like bacteria from the rumen. Antonie Van Leeuwenhoek 91, 417–422. doi: 10.1007/s10482-006-9121-7, PMID: [DOI] [PubMed] [Google Scholar]
- Pan L. L., Niu W., Fang X., Liang W., Li H., Chen W., et al. (2019). Clostridium butyricum strains suppress experimental acute pancreatitis by maintaining intestinal homeostasis. Mol. Nutr. Food Res. 63:e1801419. doi: 10.1002/mnfr.201801419, PMID: [DOI] [PubMed] [Google Scholar]
- Parada Venegas D., De la Fuente M. K., Landskron G., González M. J., Quera R., Dijkstra G., et al. (2019). Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10:277. doi: 10.3389/fimmu.2019.00277, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lee J., Yeom Z., Heo D., Lim Y.-H. (2017). Neuroprotective effect of Ruminococcus albus on oxidatively stressed SH-SY5Y cells and animals. Sci. Rep. 7:14520. doi: 10.1038/s41598-017-15163-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal V., Pozuelo M., Borruel N., Casellas F., Campos D., Santiago A., et al. (2017). A microbial signature for Crohn’s disease. Gut 66, 813–822. doi: 10.1136/gutjnl-2016-313235, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S., Alam A., Pant R., Chattopadhyay S. (2019). Wnt signaling and its significance within the tumor microenvironment: novel therapeutic insights. Front. Immunol. 10:2872. doi: 10.3389/fimmu.2019.02872, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham V. T., Dold S., Rehman A., Bird J. K., Steinert R. E. (2021). Vitamins, the gut microbiome and gastrointestinal health in humans. Nutr. Res. 95, 35–53. doi: 10.1016/j.nutres.2021.09.001 [DOI] [PubMed] [Google Scholar]
- Pichler M. J., Yamada C., Shuoker B., Alvarez-Silva C., Gotoh A., Leth M. L., et al. (2020). Butyrate producing colonic Clostridiales metabolise human milk oligosaccharides and cross feed on mucin via conserved pathways. Nat. Commun. 11:3285. doi: 10.1038/s41467-020-17075-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Possemiers S., Rabot S., Espín J. C., Bruneau A., Philippe C., González-Sarrías A., et al. (2008). Eubacterium limosum activates isoxanthohumol from hops (Humulus lupulus L.) into the potent phytoestrogen 8-prenylnaringenin in vitro and in rat intestine. J. Nutr. 138, 1310–1316. doi: 10.1093/jn/138.7.1310, PMID: [DOI] [PubMed] [Google Scholar]
- Pozuelo M., Panda S., Santiago A., Mendez S., Accarino A., Santos J., et al. (2015). Reduction of butyrate-and methane-producing microorganisms in patients with Irritable Bowel Syndrome. Sci. Rep. 5, 1–12. doi: 10.1038/srep12693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prossomariti A., Piazzi G., Alquati C., Ricciardiello L. (2020). Are Wnt/β-catenin and PI3K/AKT/mTORC1 distinct pathways in colorectal cancer? Cell. Mol. Gastroenterol. Hepatol. 10, 491–506. doi: 10.1016/j.jcmgh.2020.04.007, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quévrain E., Maubert M. A., Michon C., Chain F., Marquant R., Tailhades J., et al. (2016). Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut 65, 415–425. doi: 10.1136/gutjnl-2014-307649, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimondi S., Musmeci E., Candeliere F., Amaretti A., Rossi M. (2021). Identification of mucin degraders of the human gut microbiota. Sci. Rep. 11:11094. doi: 10.1038/s41598-021-90553-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt N., Duncan S. H., Young P., Belenguer A., McWilliam Leitch C., Scott K. P., et al. (2014). Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 8, 1323–1335. doi: 10.1038/ismej.2014.14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Chávez F., Zhang L. F., Faber F., Lopez C. A., Byndloss M. X., Olsan E. E., et al. (2016). Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of salmonella. Cell Host Microbe 19, 443–454. doi: 10.1016/j.chom.2016.03.004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivière A., Selak M., Lantin D., Leroy F., De Vuyst L. (2016). Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front. Microbiol. 7:979. doi: 10.3389/fmicb.2016.00979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez Hernáez J., Cerón Cucchi M. E., Cravero S., Martinez M. C., Gonzalez S., Puebla A., et al. (2018). The first complete genomic structure of Butyrivibrio fibrisolvens and its chromid. Microb Genom. 4:e000216. doi: 10.1099/mgen.0.000216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh Y., Suzuki H., Tani K., Nishikawa K., Irie K., Ogura Y., et al. (2015). Structural insight into tight junction disassembly by Clostridium perfringens enterotoxin. Science 347, 775–778. doi: 10.1126/science.1261833, PMID: [DOI] [PubMed] [Google Scholar]
- Salmerón A. M., Tristán A. I., Abreu A. C., Fernández I. (2022). Serum colorectal cancer biomarkers unraveled by NMR metabolomics: past, present, and future. Anal. Chem. 94, 417–430. doi: 10.1021/acs.analchem.1c04360, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor R. (2011). Key questions to guide a better understanding of host–commensal microbiota interactions in intestinal inflammation. Mucosal Immunol. 4, 127–132. doi: 10.1038/mi.2010.87, PMID: [DOI] [PubMed] [Google Scholar]
- Sato T., Matsumoto K., Okumura T., Yokoi W., Naito E., Yoshida Y., et al. (2008). Isolation of lactate-utilizing butyrate-producing bacteria from human feces and in vivo administration of Anaerostipes caccae strain L2 and galacto-oligosaccharides in a rat model. FEMS Microbiol. Ecol. 66, 528–536. doi: 10.1111/j.1574-6941.2008.00528.x, PMID: [DOI] [PubMed] [Google Scholar]
- Schirmer M., Garner A., Vlamakis H., Xavier R. J. (2019). Microbial genes and pathways in inflammatory bowel disease. Nat. Rev. Microbiol. 17, 497–511. doi: 10.1038/s41579-019-0213-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider H., Blaut M. (2000). Anaerobic degradation of flavonoids by Eubacterium ramulus. Arch. Microbiol. 173, 71–75. doi: 10.1007/s002030050010, PMID: [DOI] [PubMed] [Google Scholar]
- Schulthess J., Pandey S., Capitani M., Rue-Albrecht K. C., Arnold I., Franchini F., et al. (2019). The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 50:e7, 432–445.e7. doi: 10.1016/j.immuni.2018.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab C., Ruscheweyh H.-J., Bunesova V., Pham V. T., Beerenwinkel N., Lacroix C. (2017). Trophic interactions of infant bifidobacteria and Eubacterium hallii during L-fucose and fucosyllactose degradation. Front. Microbiol. 8:95. doi: 10.3389/fmicb.2017.00095, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R., Fuchs S., Milo R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14:e1002533. doi: 10.1371/journal.pbio.1002533, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V., Muthuramalingam K., Kim Y. M., Park S., Kim S. H., Lee J., et al. (2021). Synbiotic supplementation with prebiotic Schizophyllum commune derived β-(1,3/1,6)-glucan and probiotic concoction benefits gut microbiota and its associated metabolic activities. Appl. Biol. Chem. 64:7. doi: 10.1186/s13765-020-00572-4 [DOI] [Google Scholar]
- Singh V., Park Y.-J., Lee G., Unno T., Shin J.-H. (2022). Dietary regulations for microbiota dysbiosis among post-menopausal women 1. Crit. Rev. Food Sci. Nutr., 1–16. doi: 10.1080/10408398.2022.2076651 [DOI] [PubMed] [Google Scholar]
- Singhal R., Donde H., Ghare S., Stocke K., Zhang J., Vadhanam M., et al. (2021). Decrease in acetyl-CoA pathway utilizing butyrate-producing bacteria is a key pathogenic feature of alcohol-induced functional gut microbial dysbiosis and development of liver disease in mice. Gut Microbes 13:1946367. doi: 10.1080/19490976.2021.1946367, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeva M. K., Garcia-So J., Justice N., Myers J., Tyagi S., Nemchek M., et al. (2021). Butyrate-producing human gut symbiont, Clostridium butyricum, and its role in health and disease. Gut Microbes 13, 1–28. doi: 10.1080/19490976.2021.1907272, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Zhao C., Guven E. C., Paoli P., Simal-Gandara J., Ramkumar K. M., et al. (2020). Dietary polyphenols as antidiabetic agents: advances and opportunities. Food Front. 1, 18–44. doi: 10.1002/fft2.15 [DOI] [Google Scholar]
- Suo Y., Ren M., Yang X., Liao Z., Fu H., Wang J. (2018). Metabolic engineering of clostridium tyrobutyricum for enhanced butyric acid production with high butyrate/acetate ratio. Appl. Microbiol. Biotechnol. 102, 4511–4522. doi: 10.1007/s00253-018-8954-0, PMID: [DOI] [PubMed] [Google Scholar]
- Tandon D., Haque M. M., Gote M., Jain M., Bhaduri A., Dubey A. K., et al. (2019). A prospective randomized, double-blind, placebo-controlled, dose-response relationship study to investigate efficacy of fructo-oligosaccharides (FOS) on human gut microflora. Sci. Rep. 9, 1–15. doi: 10.1038/s41598-019-41837-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- tom Dieck H., Schön C., Wagner T., Pankoke H. C., Fluegel M., Speckmann B. (2022). A synbiotic formulation comprising Bacillus subtilis DSM 32315 and L-Alanyl-L-glutamine improves intestinal butyrate levels and lipid metabolism in healthy humans. Nutrients 14:143. doi: 10.3390/nu14010143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong L., Feng Q., Lu Q., Zhang J., Xiong Z. (2022). Combined 1H NMR fecal metabolomics and 16S rRNA gene sequencing to reveal the protective effects of Gushudan on kidney-yang-deficiency-syndrome rats via gut-kidney axis. J. Pharm. Biomed. Anal. 217:114843. doi: 10.1016/j.jpba.2022.114843, PMID: [DOI] [PubMed] [Google Scholar]
- Tsukuda N., Yahagi K., Hara T., Watanabe Y., Matsumoto H., Mori H., et al. (2021). Key bacterial taxa and metabolic pathways affecting gut short-chain fatty acid profiles in early life. ISME J. 15, 2574–2590. doi: 10.1038/s41396-021-00937-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki T., Nevin K. P., Woodard T. L., Lovley D. R., Lee S. Y. (2014). Converting carbon dioxide to butyrate with an engineered strain of Clostridium ljungdahlii. MBio 5, e01636–e01614. doi: 10.1128/mBio.01636-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valles-Colomer M., Falony G., Darzi Y., Tigchelaar E. F., Wang J., Tito R. Y., et al. (2019). The neuroactive potential of the human gut microbiota in quality of life and depression. Nat. Microbiol. 4, 623–632. doi: 10.1038/s41564-018-0337-x, PMID: [DOI] [PubMed] [Google Scholar]
- Van den Abbeele P., Gérard P., Rabot S., Bruneau A., El Aidy S., Derrien M., et al. (2011). Arabinoxylans and inulin differentially modulate the mucosal and luminal gut microbiota and mucin-degradation in humanized rats. Environ. Microbiol. 13, 2667–2680. doi: 10.1111/j.1462-2920.2011.02533.x, PMID: [DOI] [PubMed] [Google Scholar]
- van Harten R., van Woudenbergh E., van Dijk A., Haagsman H. (2018). Cathelicidins: immunomodulatory antimicrobials. Vaccines 6:63. doi: 10.3390/vaccines6030063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet M. J., Harmsen H. J., de Bont E. S., Tissing W. J. (2010). The role of intestinal microbiota in the development and severity of chemotherapy-induced mucositis. PLoS Pathog. 6:e1000879. doi: 10.1371/journal.ppat.1000879, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira E. L., Leonel A. J., Sad A. P., Beltrao N. R., Costa T. F., Ferreira T. M., et al. (2012). Oral administration of sodium butyrate attenuates inflammation and mucosal lesion in experimental acute ulcerative colitis. J. Nutr. Biochem. 23, 430–436. doi: 10.1016/j.jnutbio.2011.01.007, PMID: [DOI] [PubMed] [Google Scholar]
- Vincent A. D., Wang X.-Y., Parsons S. P., Khan W. I., Huizinga J. D. (2018). Abnormal absorptive colonic motor activity in germ-free mice is rectified by butyrate, an effect possibly mediated by mucosal serotonin. Am. J. Physiol. Gastrointest. Liver Physiol. 315, G896–G907. doi: 10.1152/ajpgi.00237.2017 [DOI] [PubMed] [Google Scholar]
- Vital M., Howe A. C., Tiedje J. M. (2014). Revealing the bacterial butyrate synthesis pathways by analyzing (meta) genomic data. MBio 5, e00889–e00814. doi: 10.1128/mBio.00889-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vital M., Karch A., Pieper D. H., Shade A. (2017). Colonic butyrate-producing communities in humans: an overview using omics data. mSystems 2, e00130–e00117. doi: 10.1128/mSystems.00130-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vital M., Penton C. R., Wang Q., Young V. B., Antonopoulos D. A., Sogin M. L., et al. (2013). A gene-targeted approach to investigate the intestinal butyrate-producing bacterialcommunity. Microbiome 1:8. doi: 10.1186/2049-2618-1-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. X., Henen M. A., Lee J. S., Vögeli B., Colgan S. P. (2021). Microbiota-derived butyrate is an endogenous HIF prolyl hydroxylase inhibitor. Gut Microbes 13:1938380. doi: 10.1080/19490976.2021.1938380, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. X., Lee J. S., Campbell E. L., Colgan S. P. (2020). Microbiota-derived butyrate dynamically regulates intestinal homeostasis through regulation of actin-associated protein synaptopodin. Proc. Natl. Acad. Sci. 117, 11648–11657. doi: 10.1073/pnas.1917597117, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werlang M. E., Palmer W. C., Lacy B. E. (2019). Irritable bowel syndrome and dietary interventions. Gastroenterol. Hepatol. 15, 16–26. [PMC free article] [PubMed] [Google Scholar]
- Wrzosek L., Miquel S., Noordine M. L., Bouet S., Joncquel Chevalier-Curt M., Robert V., et al. (2013). Bacteroides thetaiotaomicron and Faecalibacterium prausnitzii influence the production of mucus glycans and the development of goblet cells in the colonic epithelium of a gnotobiotic model rodent. BMC Biol. 11:61. doi: 10.1186/1741-7007-11-61, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Zhou B., Pang X., Song X., Gu Y., Xie R., et al. (2022). Clostridium butyricum, a butyrate-producing potential probiotic, alleviates experimental colitis through epidermal growth factor receptor activation. Food Funct. 13, 7046–7061. doi: 10.1039/D2FO00478J, PMID: [DOI] [PubMed] [Google Scholar]
- Xi Y., Jing Z., Wei W., Chun Z., Quan Q., Qing Z., et al. (2021). Inhibitory effect of sodium butyrate on colorectal cancer cells and construction of the related molecular network. BMC Cancer 21:127. doi: 10.1186/s12885-021-07845-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y.-H., Chen Y.-X., Fang J.-Y. (2020). Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct. Target. Ther. 5:22. doi: 10.1038/s41392-020-0116-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav H., Lee J.-H., Lloyd J., Walter P., Rane S. G. (2013). Beneficial metabolic effects of a probiotic via butyrate-induced GLP-1 hormone secretion*. J. Biol. Chem. 288, 25088–25097. doi: 10.1074/jbc.M113.452516, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Qu Y., Fujita Y., Ren Q., Ma M., Dong C., et al. (2017). Possible role of the gut microbiota–brain axis in the antidepressant effects of (R)-ketamine in a social defeat stress model. Transl. Psychiatry 7, 1–11. doi: 10.1038/s41398-017-0031-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Richards E. M., Pepine C. J., Raizada M. K. (2018). The gut microbiota and the brain–gut–kidney axis in hypertension and chronic kidney disease. Nat. Rev. Nephrol. 14, 442–456. doi: 10.1038/s41581-018-0018-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip W., Hughes M. R., Li Y., Cait A., Hirst M., Mohn W. W., et al. (2021). Butyrate shapes immune cell fate and function in allergic asthma. Front. Immunol. 12:628453. doi: 10.3389/fimmu.2021.628453, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshii K., Hosomi K., Sawane K., Kunisawa J. (2019). Metabolism of dietary and microbial vitamin B family in the regulation of host immunity. Front. Nutr. 6:48. doi: 10.3389/fnut.2019.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ze X., Duncan S. H., Louis P., Flint H. J. (2012). Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J. 6, 1535–1543. doi: 10.1038/ismej.2012.4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Liu W., Pi X. (2021). In vitro effects of stachyose on the human gut microbiota. Starch 73:2100029. doi: 10.1002/star.202100029 [DOI] [Google Scholar]
- Zheng L., Kelly C. J., Battista K. D., Schaefer R., Lanis J. M., Alexeev E. E., et al. (2017). Microbial-derived butyrate promotes epithelial barrier function through IL-10 receptor–dependent repression of claudin-2. J. Immunol. 199, 2976–2984. doi: 10.4049/jimmunol.1700105, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Han J., Li L., Wang Y., Li Y., Zhang S. (2019). Claudin family participates in the pathogenesis of inflammatory bowel diseases and colitis-associated colorectal cancer. Front. Immunol. 10:1441 doi: 10.3389/fimmu.2019.01441, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou S., Fang L., Lee M.-H. (2018). Dysbiosis of gut microbiota in promoting the development of colorectal cancer. Gastroenterol. Rep. 6, 1–12. doi: 10.1093/gastro/gox031 [DOI] [PMC free article] [PubMed] [Google Scholar]