Abstract
As the world continues to experience the effects of SARS‐CoV‐2, there is evidence to suggest that the sequelae of viral infection (the post‐COVID‐19 condition; PCC) at both an individual and population level will be significant and long‐lasting. The history of pandemics or epidemics in the last 100 years caused by members of the RNA virus family, of which coronaviruses are a member, provides ample evidence of the acute neurological effects. However, except for the H1N1 influenza pandemic of 1918/1919 (the Spanish flu) with its associated encephalitis lethargica, there is little information on long‐term neurological sequelae. COVID‐19 is the first pandemic that has occurred in a setting of an aging population, especially in several high‐income countries. Its survivors are at the greatest risk for developing neurodegenerative conditions as they age, rendering the current pandemic a unique paradigm not previously witnessed. The SARS‐CoV‐2 virus, among the largest of the RNA viruses, is a single‐stranded RNA that encodes for 29 proteins that include the spike protein that contains the key domains required for ACE2 binding, and a complex array of nonstructural proteins (NSPs) and accessory proteins that ensure the escape of the virus from the innate immune response, allowing for its efficient replication, translation, and exocytosis as a fully functional virion. Increasingly, these proteins are also recognized as potentially contributing to biochemical and molecular processes underlying neurodegeneration. In addition to directly being taken up by brain endothelium, the virus or key protein constituents can be transported to neurons, astrocytes, and microglia by extracellular vesicles and can accelerate pathological fibril formation. The SARS‐CoV‐2 nucleocapsid protein is intrinsically disordered and can participate in liquid condensate formation, including as pathological heteropolymers with neurodegenerative disease‐associated RNA‐binding proteins such as TDP‐43, FUS, and hnRNP1A. As the SARS‐CoV‐2 virus continues to mutate under the immune pressure exerted by highly efficacious vaccines, it is evolving into a virus with greater transmissibility but less severity compared with the original strain. The potential of its lingering impact on the nervous system thus has the potential to represent an ongoing legacy of an even greater global health challenge than acute infection.
Keywords: coronavirus, influenza virus, liquid condensates, RNA metabolism, RNA virus
The consequences of nervous system exposure by the SARS‐CoV‐2 virus in an aging population are discussed, including the potential acceleration or advancement of neurodegenerative processes. Pathophysiological mechanisms include the ongoing manifestations of acute immune‐mediated inflammation but also chronic immune dysregulation. While there is little evidence to suggest viral replication in the central nervous system, the presence of the ACE2 receptor and coreceptors such as Neuropilin‐1 suggest the ability for uptake. The evidence that the exosomal transport of components of the viral protein, and in particular, of the intrinsically disordered nucleocapsid protein, can lead to neuronal protein expression is discussed. Created with BioRender.com
Abbreviations
- 3CLpro
chymotrypsin‐like protease
- ACE‐2
angiotensin‐converting enzyme 2
- ALS
amyotrophic lateral sclerosis
- ARDS
acute respiratory distress syndrome
- BCECs
brain capillary endothelia‐like cells
- CD4 cells
cluster of differentiation 4 cells
- CD8 cells
cluster of differentiation 8 cells
- CERAD
Consortium to Establish a Registry for Alzheimer's Disease
- DaT‐SPECT
dopamine transporter single photon emission computerized tomography
- E protein
envelop glycoprotein
- EIF
eukaryotic translation initiation factor
- FUS
RNA‐binding protein fused in sarcoma
- hnRNP1A
heterogeneous nuclear riboprotein A1
- IFITM1 & M2
interferon‐induced protein with tetratricopeptide repeats 1 & 2
- IFN
interferon
- IFNγ
interferon gamma
- IL‐4/IL‐6
interleukin 4/interleukin 6
- LIN28B
protein lin‐28 homolog B
- M protein
membrane glycoprotein
- MAVS
mitochondrial antiviral signaling protein
- MCI
minimal cognitive impairment
- MERS
Middle East respiratory syndrome
- MERS‐CoV
Middle East respiratory syndrome‐coronavirus
- MOV10
Moloney leukemia virus 10 like
- MRI
magnetic resonance imaging
- mRNA
messenger RNA
- N protein
nucleocapsid protein
- NFL
low molecular weight neurofilament protein
- NLRP3
NOD‐, LRR‐, and pyrin domain‐containing protein 3
- NMD
nonsense‐mediated decay
- NPC
nuclear pore complex
- NRP‐1
neuropilin‐1
- NSPs
nonstructural proteins
- ORFs
open reading frames
- PABPC1
poly(A)‐binding protein cytoplasmic 1
- PAMPs
pathogen‐associated molecular patterns
- PASC
postacute sequelae of COVID
- PCC
post‐COVID‐19 condition
- PD
Parkinson's disease
- PET
positron emission tomography
- PLpro
papain‐like protease
- pp1a
polyprotein 1a (containing NSPs 1–11)
- pp1b
polyprotein 1b (containing NSPs 1–16)
- PRR
pattern recognition receptors
- qRT‐PCR
quantitative reverse transcription‐polymerase chain reaction
- RBD
receptor binding domain
- RBPs
RNA‐binding proteins
- RNA
ribonucleic acid
- RTC
replication/transcription complex
- S protein
spike protein
- SARS‐CoV
Severe Acute Respiratory Syndrome (SARS)‐associated coronavirus
- SARS‐CoV‐2
Severe Acute Respiratory Syndrome (SARS)‐associated coronavirus‐2
- SG
stress granule
- STAT
signal transducer and activator of transcription
- TDP‐43
Transactive response DNA‐binding protein 43 kDa
- Th cells
T helper cells
- TMPRSS2
transmembrane protein serine 2
- TNFα
Tumor necrosis factor‐alpha
- UPR
unfolded protein response
- WHO
World Health Organization
As we enter a phase of the severe acute respiratory syndrome‐coronavirus‐2 (SARS‐CoV‐2) pandemic in which the virus becomes endemic, there is increasing evidence that its health and socioeconomic consequences will be significant and long‐lasting. Among the former, there are early warnings that the function of the nervous system, and in particular the central nervous system, will be significantly impaired in a subset of those infected by the virus. This includes residual cognitive impairment consistent with a dysexecutive syndrome and the possibility of either the acceleration of preexisting neurodegenerative disorders or an increase in the incidence rates of those which disproportionately affect aging populations. To understand the basis of such consequences, it is helpful to examine the lessons learned from previous pandemics, the interrelationship between COVID‐19 (the acute clinical manifestations of SARS‐CoV‐2 infection) and population aging, as well as the evolving evidence of the virus's impact on the central nervous system, both directly and indirectly. In doing so, it becomes evident that the consequences of the pandemic will be long‐lasting and will herald an acceleration of both the prevalence and processes of adult‐onset neurodegeneration.
1. PREVIOUS RNA VIRUS PANDEMICS AND LESSONS LEARNED FROM HISTORY
RNA viruses known to infect vertebrates arose from a common RNA virus ancestor (Riboviria) which has two major phylogenetic orders (Figure 1). The first order, nidovirales, is characterized by positive sense, nonsegmented, single‐stranded RNA and contains three families (among a broad number of families) that give rise to vertebrate infections. These include Arteriviridae, Coronaviridae, and Tobaniviridae virus families. Within the Coronaviridae family, the Betacoronavirus virus genus has caused several human epidemics or pandemics over the last century, including those due to Middle East respiratory syndrome‐coronavirus (MERS‐CoV), SARS‐CoV, and SARS‐CoV‐2 viruses. The second order, Articulavirales, is characterized by negative sense, segmented, single‐stranded RNA and contains two families that give rise to vertebrate infections. This includes Amnoonviridae and Orthomyxoviridae families, with the latter giving rise to the Alphainfluenzavirus species that include H1N1, H2N2, and H3N2 viruses, each of which has been associated with pandemics. The difference between a positive sense (also known as a sense strand) and a negative sense (also known as an antisense strand) RNA virus is that a positive sense RNA virus comprises viral mRNA that can be directly translated into proteins. In contrast, a negative sense RNA virus is comprised of RNA complementary to the viral mRNA and thus cannot be directly translated into proteins but requires the presence of an RNA‐dependent RNA polymerase to yield a translationally competent mRNA.
FIGURE 1.
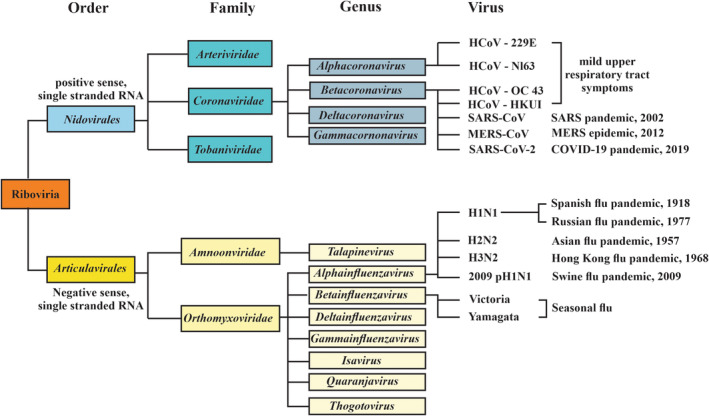
Phylogenetic tree of selected Riboviria associated with human pandemics or epidemics (Agrawal et al., 2021; Fung & Liu, 2021; Gulyaeva & Gorbalenya, 2021; Socan et al., 2014). Only viral families of the Nidovirales order that are associated with vertebrate infections have been shown. Of these three (Arteriviridae, Coronaviridae, and Tobaniviridae), only viruses derived from the Betacoronavirus group have been associated with significant human infections, including the SARS epidemic of 2002, the MER epidemic of 2012, and the COVID‐19 pandemic of 2019. Alphainfluenzavirus (influenza A) and Betainfluenzavirus (influenza B) species members of the Orthomyxoviridae family have given rise to the Spanish flu pandemic of 1918, the Asian flu pandemic of 1957, the Hong Kong flu pandemic of 1968, the Russian flu of 1977, and the swine flu of 2009. Less pathogenic viruses of the Coronaviridae family give rise to approximately 15–30% of cases of the common cold (mild upper respiratory tract symptoms) annually and include human coronaviruses (HCoV) HCoV‐229 E, HCoV‐N163, HCoV‐OC43, and HCoV‐HKUI.
Although pandemics have been documented for several centuries, in the last 100 years RNA viruses have given rise to either pandemics or epidemics, including the Spanish flu (the 1918–1919 H1N1 pandemic) considered to be the ‘mother of all pandemics’ which affected roughly one‐third of the world's population with estimates of 50–100 million human deaths (approximately 2.7–4.5% of the world's population). This 100‐year timeline also includes the Asian flu pandemic of 1957 (H2N2 virus; approximately 1 million deaths), the Hong Kong flu pandemic of 1968 (H3N2 virus; approximately 1 million deaths), the Russian flu pandemic of 1977 (H1N1 virus; an estimated 700 000 deaths), the SARS epidemic of 2002 (SARS‐CoV virus; an estimated 774 deaths), the swine flu pandemic of 2009 (2009 pH1N1 virus; an estimated 284 000 deaths), the MERS epidemic of 2012 (MERS‐CoV virus; an estimated 858 deaths), and the current COVID‐19 pandemic (SARS‐CoV‐2 viral origin) (Agrawal et al., 2021; da Costa et al., 2020; Fung & Liu, 2021; Rozo & Gronvall, 2015). As of the time of writing, there have been an estimated >159 million COVID‐19 cases reported worldwide with more than 6.3 million deaths—placing it among the more lethal of the RNA virus‐mediated pandemics, second only to the 1918 H1N1 pandemic. Thus, since 1918, the world has experienced at least eight outbreaks of pandemics or epidemics related to RNA viruses with varying degrees of lethality. The appearance of pandemics related to Coronaviridae seems, however, to be more recent (post 2000 AD), raising important considerations regarding how significant climate changes may have altered the intricate relationship and balance between animal, environmental, and human health (the One Health paradigm) to the extent that zoonotic transmissions of this group of viruses to humans have become increasingly frequent (Petrovan et al., 2021; Weaver et al., 2022).
The neurological consequences of an RNA virus pandemic have been well delineated for the Alphainfluenzaviruses, but less so for those illnesses arising from Betacoronavirus. Although direct causality has been challenging to prove, for the former the acute stage of the viral infection has been associated with a prominent impact on the nervous system (Cardenas et al., 2014; Henry et al., 2010; Robinson & Busl, 2020; Tsai & Baker, 2013). Most commonly, this has been in the form of either an encephalopathy or encephalitis, the latter of which can vary from mild to a fulminant, fatal acute necrotizing encephalopathy. Acute inflammatory neuropathies such as the Guillain‐Barre syndrome (distinct from that associated with Influenza vaccination), polyradiculopathies, as well as transverse myelitis, amyotrophy, bulbar and muscle paralysis have also been observed, as have flairs of multiple sclerosis following influenza A (Oikonen et al., 2011). There is, however, no clear evidence that any of these acute syndromes were the harbinger of a more progressive neurodegenerative syndrome with the sole exception of that suggested to occur following encephalitis lethargica.
In further understanding this and its relevancy to the current SARS‐CoV‐2 pandemic and the potential for long‐term neurological sequelae, the 1918–1919 H1N1 pandemic (also known as the Spanish flu) is highly informative. This pandemic differed in many ways from the current pandemic but also bears striking similarities. It occurred in the setting of World War I, and indeed, the 1918 H1N1 influenza strain was thought to attack the Axis troops in advance of attacking the Alliance troops, contributing to the decision of the Kaiser to opt for earlier peace negotiations. While global travel was more limited than in current times, the 1918 influenza pandemic was driven by the return of troops from the war theater with the infection already at play. It was named the Spanish flu because Spain first recognized and reported the epidemic nature of the infection and began public health measures rapidly (Martini et al., 2019).
At the time, the Spanish flu was considered to be avian‐like influenza derived from an entirely unknown source (Taubenberger, 2006). The pandemic lasted approximately 2 years, occurred in multiple waves, affecting a third of the world's population (disproportionately amongst the poorest), and most profoundly affected those between the ages of 15 and 40 years with men outnumbering women. Approximately 50% of the deaths occurred in this age range in contrast to the current pandemic in which this age group has been largely spared death despite being epidemiologically the most heavily affected (Liang et al., 2021; Simonsen et al., 1998; Taubenberger, 2006). This is an important observation in the context of global aging in that it is this specific age group that can be argued to be at risk for either the acceleration of latent or the acquisition of new neurodegenerative disease as a consequence of SARS‐CoV‐2 infection.
Encephalitis lethargica was an acute neurological syndrome, rarely observed prior to the Spanish flu, with a mean age of onset of 31 years which spared no age group and which was characterized by the sudden onset of high fever, ophthalmoplegia, mental confusion, and lethargy which in some cases was severe (Dourmashkin, 1997; Hoffman & Vilensky, 2017; Stallybass, 1923). von Economo recognized three distinct variants of this acute syndrome: a somnolent‐ophthalmoplegic variant, a hyperkinetic variant, and a variant that more closely resembled parkinsonism which he termed the amyostatic‐akinetic variant (Dickman, 2001). Of those who survived this acute event, approximately 80% developed an atypical Parkinson's disease (Dourmashkin, 1997). Both an early onset variant (within 6 months and predominantly affecting young individuals) and a late onset variant (onset latency post encephalitis lethargica ranging from 1 to 20 years and predominantly affecting older individuals (> age 40 years)) were observed (Hudson & Rice, 1990). In those individuals with late‐onset parkinsonism, neuropathological features were largely restricted to the substantia nigra and basal ganglia, with calcification and hyaline degeneration of blood vessels, severe neuronal cell loss with gliosis, and a lack of inflammatory features (Geddes et al., 1993; McAlpine, 1923, 1926). Involvement of the upper and lower motor neurons, consistent with a diagnosis of amyotrophic lateral sclerosis (ALS) was described, although rarely (Hudson & Rice, 1990).
However, the notion that the H1N1 influenza A virus caused encephalitis lethargic and the subsequent neurodegenerative states remain controversial (Davis et al., 2014; Jang et al., 2009; McCall et al., 2008). Causation has been considered based on the closely linked time course. It is a valuable lesson that documentation of causation should not be ignored as we progress through the current pandemic in an era with advanced technologies and methods that can be applied to prove causation.
2. SARS‐COV‐2 AND THE POST‐COVD‐19 SYNDROME
Much has been written about the genomic structure of SARS‐CoV‐2, its mutations, the critical interaction between the S1 component of the spike protein with its primary receptor angiotensin‐converting enzyme 2 (ACE‐2) and the subsequent impact of viral infection/replication on the immune system (Li, Jia, et al., 2022; Majumdar & Niyogi, 2021). However, in understanding the potential long‐term effects of the virus on neuronal function, and in particular its nonstructural proteins (NSPs), an overview is helpful.
SARS‐CoV‐2 is amongst the largest of the RNA viruses with a molecular weight of 30 kb and a genome size of 29 903 bp that encodes for 14 open reading frames (ORFs), of which those at the 5′‐end encode for NSPs while those at the 3′‐end encode structural and accessory proteins (Figure 2). The largest ORFs (ORF1a and ORF1b, respectively) account for approximately two‐thirds of the viral genome and are the first to be translated into two large overlapping polyproteins (pp1a which yields NSPs 1–11 and pp1ab which yields NSPs 1–16) upon cellular entry. A feature of coronaviruses is that a ribosomal frameshift is required for the translation of ORF1ab into a polyprotein that encompasses both ORF1a and 1b (pp1ab) whereas typically the stop codon at the termination of ORF1a would lead to the cessation of translation (yielding pp1a in the absence of a ribosomal frameshift) (Bhatt et al., 2021; Brierley et al., 1989; Krichel et al., 2020; Plant & Dinman, 2008; Sun, Tang, et al., 2021). While there is an initial use of the host ribosomes to synthesize SARS‐CoV‐2 proteins, the failure of a programmed −1 ribosomal frameshift in which a subset of ribosomes backtracks by one nucleoside followed by the continuation of translational elongation, leads to the failure of synthesis of pp1ab and a failure thus to synthesize the RNA‐dependent RNA polymerase required for the coronavirus mRNA replication. While much of the focus has been on the spike (S1 & S2) protein given its crucial role as the ligand for ACE2, the NSPs encoded by ORF1a and 1b play a crucial role in not only the initiation of viral RNA translation and in evading the host immune response but are increasingly recognized as critical players in driving many of the neurological consequences of SARS‐CoV‐2 infection (Gusev et al., 2022). The remaining SARS‐CoV‐2 genome encodes structural (spike (S) glycoprotein, small envelop glycoprotein (E), membrane glycoprotein (M), nucleocapsid protein (N)) and auxiliary proteins (ORFs 3a, 3b, 6, 7a, 7b, 8, 9b, 10 encoding for proteins orf3a, orf3b, orf6, orf7a, orf8, orf9b, and orf10) (Figure 2, Table 1).
FIGURE 2.
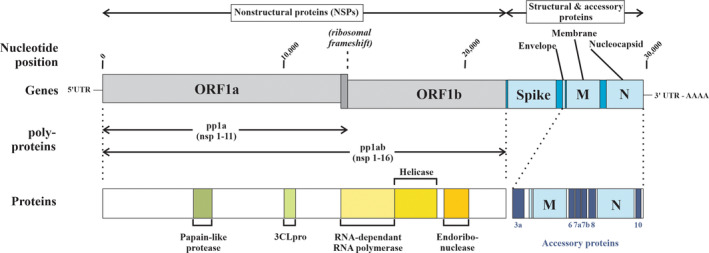
Schematic representation of the genomic organization of SARS‐CoV‐2 and major proteins encoded for (Chilamakuri & Agarwal, 2021; Majumdar & Niyogi, 2021; Rastogi et al., 2020). The nonstructural protein‐coding segments (NSPs) ORF1a and ORF1b occupy approximately two‐thirds of the genomic sequence of the SARS‐CoV‐2 gene and encode for two polyproteins, pp1a and pp1b. Whereas ORF1a translation terminates due to a stop codon and yields pp1a containing NSPs 1‐11, the translation of pp1ab is critically dependent on a ribosomal frameshift. The absence of this leads to a failure of pp1ab generation which includes the RNA‐dependent RNA polymerase, helicase, and endoribonuclease necessary for viral replication. The structural protein segment of the SARS‐CoV‐2 gene encodes for the spike protein (including both the S1 and S2 subunits), the envelope, membrane, and nucleocapsid proteins, and accessory proteins involved in virus maintenance. In addition, both the NSP and structural protein segments encode for numerous additional proteins, with 16 NSPs in total encoded by ORF1a and ORF1b 3CLpro, Chymotrypsin‐like protease; NSPs, Nonstructural proteins; ORF, Open reading frame; pp1a, Polyprotein 1a; pp1ab, Polyprotein 1ab.
TABLE 1.
SARS‐CoV‐2 nonstructural (NSPs), structural, and accessory proteins functions and postulated linkage to the process of neurodegeneration
| Protein | Function in SARS‐CoV‐2 replication | Postulated mechanism contributing to neurodegeneration | References |
|---|---|---|---|
| Nsp1 | Suppresses apoptosis; promotes host mRNA degradation; suppresses host gene expression; binds to the ribosome small subunit and blocks host translation; and suppresses the innate immune antiviral response | Unknown | (Gusev et al., 2022; Rohaim et al., 2021) |
| Nsp2 | Disrupts intracellular host response by mechanism not yet defined | Unknown | (Gusev et al., 2022; Rohaim et al., 2021) |
| Nsp3 | Component of RTC to facilitate mRNA transcript translation while suppressing host protein synthesis; interacts with N protein; promotes cytokine expression; PLPro/deubiqutinase domain cleaves polyproteins pp1a and pp1ab | Unknown | (Gusev et al., 2022; Rohaim et al., 2021) |
| Nsp4 | Transmembrane scaffold protein with a role in vesicle formation; assembly of replicative structures | Mitochondrial dysfunction | (Gusev et al., 2022; Rohaim et al., 2021) |
| Nsp5 | 3CLpro cleaves polyproteins pp1a and pp1ab; role in RNA replication and double‐membrane formation | Unknown | (Gusev et al., 2022; Rohaim et al., 2021) |
| Nsp6 | Autophagosome formation; potential transmembrane scaffold protein | Vesicle trafficking | (Gusev et al., 2022; Rohaim et al., 2021) |
| Nsp7 | Primer‐independent RNA polymerase activity; forms a complex with Nsp8 | Vesicle trafficking | (Gusev et al., 2022; Rohaim et al., 2021) |
| Nsp8 | Primase activity; forms complex with Nsp7 | RNA processing; enhanced liquid phase transition; Mitochondrial dysfunction | (Gusev et al., 2022; Rohaim et al., 2021) |
| Nsp9 | Single‐stranded RNA‐binding protein phosphatase | Unknown | (Gusev et al., 2022; Rohaim et al., 2021) |
| Nsp10 | Co‐factor for Nsp16 and Nsp14 in the RTC | Vesicle trafficking | (Gusev et al., 2022; Rohaim et al., 2021) |
| Nsp11 | Necessary for replication; possible role in a ribosomal frameshift | Unknown | (Gusev et al., 2022) |
| Nsp12 | RNA‐dependant RNA polymerase (a key component of RTC); coding sequence contains the ribosomal frameshift site | Unknown | (Gusev et al., 2022; Rohaim et al., 2021) |
| Nsp13 | RNA helicase (a key component of RTC); 5′ triphosphatase | Vesicle trafficking | (Gusev et al., 2022; Rohaim et al., 2021) |
| Nsp14 | Exoribonuclease activity and N7 methyl transferase | Unknown | (Gusev et al., 2022; Rohaim et al., 2021; Tahir, 2021) |
| Nsp15 | Endoribonuclease; role in evading host dsRNA sensors | Vesicle trafficking | (Gusev et al., 2022; Rohaim et al., 2021) |
| Nsp16 | 2’‐O‐methyltransferase (shields viral RNA from MDA5 recognition) | Unknown | (Gusev et al., 2022; Rohaim et al., 2021) |
| Spike | Binding to ACE2 and coreceptors, virus–cell membrane fusion protein | Lipid modifications; UPR | (Bosch et al., 2003; Gusev et al., 2022) |
| Nucleocapsid | Viral genome packaging, RBP, cell surface expressed N protein binding to chemokines with the ability to block chemotaxis of immune effector cells | RNA processing through RBP interactions; enhanced liquid phase transition | (Cubuk et al., 2021; Lopez‐Munoz et al., 2022) |
| orf3a | Viroporin involved in virus replication and release; stimulation of pro‐IL‐1B gene transcription, activation of NLRP3 inflammasome (Bianchi et al., 2021; Zhang et al., 2022) | Vesicle trafficking; programmed cell death and necrosis | (Gusev et al., 2022; Redondo et al., 2021) |
| orf3b | IFN antagonist | Unknown | (Gusev et al., 2022; Konno et al., 2020; Redondo et al., 2021) |
| orf3c | Unknown | Unknown | (Redondo et al., 2021) |
| orf3d | Unknown | Unknown | (Redondo et al., 2021) |
| orf6 | Binding to NPC to inhibit STAT1 cytosolic‐nuclear translocation with blockage of IFN signaling | Unknown | (Gusev et al., 2022; Miorin et al., 2020; Redondo et al., 2021) |
| orf7a | Transmembrane protein with IFN‐1 response antagonism function | Unknown | (Gusev et al., 2022; Redondo et al., 2021) |
| orf7b | Transmembrane protein, function unknown | Unknown | (Gusev et al., 2022; Redondo et al., 2021) |
| orf8 | Apoptosis, antagonism of IFN‐1 response, evasion of host innate immune response | Unknown | (Gusev et al., 2022; Redondo et al., 2021) |
| orf9b | Suppression of innate immunity by targeting MAVS signalosome | Mitochondrial dysfunction | (Gusev et al., 2022; Redondo et al., 2021) |
| orf9c | Membrane‐associated protein that suppresses antiviral response by interference with IFN signaling and antigen presentation | Unknown | (Dominguez Andres et al., 2020; Redondo et al., 2021) |
| orf10 | Suppression of innate immune response with impairment of IFN signaling, antigen processing and presentation, complement signaling, IL‐6 signaling; degradation of MAVS | Unknown | (Li, Hou, et al., 2022; Redondo et al., 2021) |
Abbreviations: 3CLpro, chymotrypsin‐like protease; IFN, interferon; MAVS, mitochondrial antiviral signaling protein; NLRP3, NOD‐, LRR‐ and pyrin domain‐containing protein 3; NPC, nuclear pore complex; NSP, nonstructural protein; ORF, open reading frame; PLpro, papain‐like protease; pp1a, polyprotein 1a; pp1ab, polyprotein 1ab; RBP, RNA‐binding protein; RTC, replication/transcription complex; STAT, signal transducer and activator of transcription 1; UPR, unfolded protein response.
Following interaction of the S protein receptor binding domain (RBD) with ACE2, the S protein undergoes proteolytic cleavage at the S1/S2 boundary by transmembrane protein serine 2 (TMPRSS2) which is activated by the host protein convertase furin, furin‐like proteases, or by cathepsins after endocytosis (for a proportion of SARS‐CoV‐2 virus that is directly internalized by endosomes). Typically, the fusion of the viral envelope to the host cell membrane is followed by endocytosis of the viral genome, the release of the viral genome into the cytoplasm, and the synthesis of a replication/transcription complex which generates a negative‐strand viral RNA template (Figure 3). The process of internalization can be enhanced by coreceptors or alternative receptors, including neuronally‐expressed Neuropilin‐1 (NRP‐1) which has been shown to enhance SARS‐CoV‐2 uptake in the central nervous system, heparin sulfate proteoglycans, and C‐type lectins (Daly et al., 2020; Gusev et al., 2022). The importance of such coreceptors or alternate receptors is to enhance SARS‐CoV‐2 infection of cells with low ACE2 expression. NSPs generated by translation of the genomic RNA utilize membranes from the endoplasmic reticulum and Golgi to synthesize new double‐membrane vesicles in which viral replication and transcription are compartmentalized (thus evading detection), and then packaging of new virions for exocytosis and release of the newly synthesized virus into the extracellular space.
FIGURE 3.
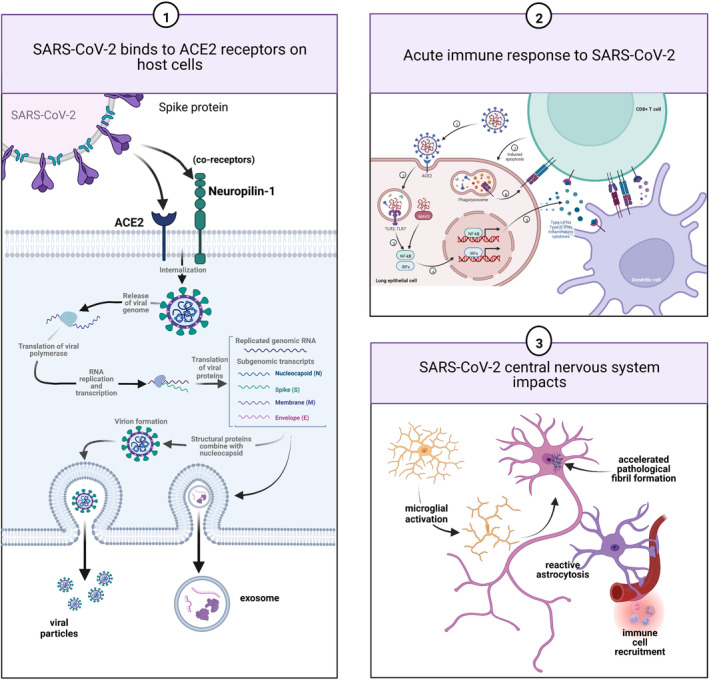
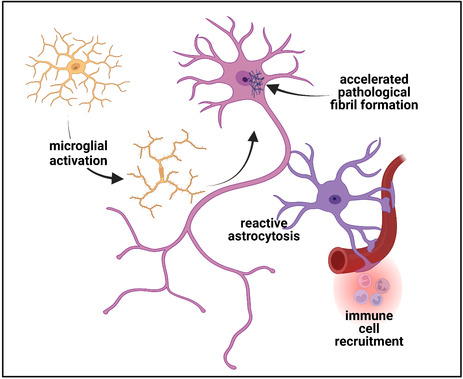
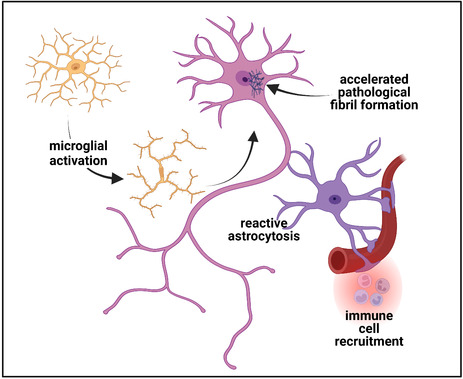
Schematic representation of the pathobiology of long‐term neuronal injury following COVID‐19. Following endocytosis of the SARS‐CoV‐2 virus which can be facilitated by coreceptors such as Neuropilin‐1, replication of the virus proceeds to yield intact virions which are then exocytosed. Components of the virus can, however, also be packaged into exosomes that are then exocytosed and can then act to induce the synthesis of SARS‐CoV‐2 proteins at a distance or disrupt cellular homeostasis (panel 1). Failure of the acute immune response can lead to viral persistence while in a subgroup of patients, an IFN‐mediated cytokine storm with consequent persistent tissue damage in addition to long‐lasting immune upregulation can occur (panel 2). Neuronal injury can be the manifestation of multiple concurrent processes including the residual in microvascular disease with microthrombi and infarction and persistent perivascular inflammation and CNS immune dysregulation (panel 3). Microglial and astrocytic activation can be long‐lasting and contribute directly to neuronal injury and dysfunction. In vitro evidence is supportive of a role for SARS‐CoV‐2 proteins, and in particular, the nucleocapsid protein in which there are three intrinsically disordered domains, in accelerating the formation of neuronal cytoplasmic inclusions (NCIs). Such NCI's can be composed of cytoskeletal proteins or through enhanced liquid–liquid phase shifting with condensate formation, of an array of RNA‐binding proteins. Created with BioRender.com
This carefully choreographed internalization of SARS‐CoV‐2, the release of its RNA into the cytosol, the process of new virion synthesis and its release, is subject to intense scrutiny by the host immune system, the purpose of which is to recognize its presence and then limit its entry, translation, replication and assembly (Diamond & Kanneganti, 2022; Gu et al., 2022; Primorac et al., 2022; Sette & Crotty, 2021). Like most viruses, SARS‐CoV‐2 possesses an array of mechanisms to evade both the innate and adaptive host immune response; the latter of which is required for viral clearance (Merad et al., 2022). The initial innate immune response functions to restrict viral replication, generate an inflammatory response at the site of local tissue invasion, and prime the adaptive response. Immune cells sense the viral presence through pattern recognition receptors (PRR) that recognize pathogen‐associated molecular patterns (PAMPs) and trigger the synthesis of pro‐inflammatory cytokines and chemokines in addition to activation of the direct antiviral mechanisms (Diamond & Kanneganti, 2022). Central to the innate immunity is a primary antiviral interferon beta response and activation of dendritic cells which prime CD4+ T cells to differentiate into T helper (Th) cells (key to the production of IFN‐gamma and generation of antibody‐mediated immunity). This pro‐inflammatory state is contributed to by an array of cells including dendritic cells, neutrophils, monocytes, and tissue‐resident cells. CD8+ T cells are recruited to kill infected cells. The adaptive immune response, which includes T cell and antibody‐mediated immunity, is reduced with aging, potentially contributing to the increased severity of COVID‐19 amongst the aged.
The SARS‐CoV‐2 virus mounts a massive blockade of the IFN response through a broad array of NSP functions with a high degree of redundancy (Table 1) (Gusev et al., 2022; Setaro & Gaglia, 2021). This includes NSP14 (N‐7‐methyltransferase) and NSP16 (2‐O′‐methyltransferase) modification of the viral RNA 5′ end to mimic host mRNA and thus preventing it from being recognized by PRR and IFIT1 (interferon‐induced protein with tetratricopeptide repeats 1), the inhibition of PRR and IFN synthesis (NSP1) and the inhibition of IFN transmembrane transport (NSP8, NSP9) (Gu et al., 2022; Gusev et al., 2022; Tahir, 2021).
Viral invasion is further driven by a significant blunting or delay of the adaptive immune response, the extent of which correlates with increased disease severity (Moss, 2022). With increasing disease severity, lymphopenia can become prominent and can include reductions in the numbers of dendritic cells necessary for antigen presentation, in addition to the reduction in a broad range of T cells due to sequestration in injured tissues or T cell apoptosis (including both CD8+ cells and CD4+ cells) which can be long‐lasting (Cox & Brokstad, 2020). In more severe cases, a cytokine storm with marked increases in IFNγ and TNFα expression leads to uncontrolled immune cell activation which in turn leads to immune cell exhaustion, dysfunction, and apoptosis. An autoimmune response to the SARS‐CoV‐2 virus has been well recognized although its mechanism(s) of induction is less certain. Possibilities include molecular mimicry (e.g., both S and E proteins possess regions analogous to human proteins), epitope spreading, or presentation of cryptic antigens leading to autoimmune antibody generation (Yazdanpanah & Rezaei, 2022). Such autoantibodies contribute directly to several acute neurological syndromes observed in COVID‐19, including Guillain‐Barre syndrome, Miller‐Fisher syndrome, and acute thrombotic thrombocytopenic purpura (with attendant brain hemorrhaging). They can also contribute to the induction of IFN autoantibodies, further blunting the immune response.
Among those who recover from COVID‐19, a significant proportion exhibit a persistent altered immune response marked by ongoing chronic inflammation, in addition to significant tissue damage (Khazaal et al., 2022). Viral persistence has been observed in the gastrointestinal tract, central nervous system, and other ACE2‐expressing organ systems, potentially contributing to a more prolonged disease course.
These broad‐based immunological defects are believed to underlie a new clinical entity termed the post‐COVID‐19 condition (PCC, also known as Long COVID, postacute sequelae of COVID (PASC), or chronic COVID). Common features amongst the different definitions of PCC include the persistence or development of signs and symptoms after a defined period of time (ranging from 4 weeks after the “initial phase” of the infection (Department of Health and Human Service guidelines (Department of Health and Human Services, Office of the Assistant Secretary for Health., 2022 . National Research Action Plan on Long COVID, 200 Independence Ave SW, Washington, DC 20201.)); 12 weeks duration with onset either during or after an infection consistent with COVID‐19 (United Kingdom (“National Institute for Health and Care Excellence. COVID‐19 rapid guideline: managing the long‐term effects of COVID‐19,” 2022)); or 3 months following the onset of an infection consistent with probable or documented SARS‐CoV‐2 (WHO (WHO, 2021)), that the symptoms are of a certain duration (2 months in the WHO criteria), and that they occur in the absence of an alternative diagnosis. Symptoms and signs may improve over time, or they may worsen. Reflective of the multisystem involvement is the recognition that over 200 different symptoms have been noted, with the most common systems clustering being constitutional (fatigue, limb weakness, generalized pain), nervous system/mental health (short‐term memory loss, slowing down in thinking, difficulty concentrating, ‘brain fog’, cognitive dysfunction, anxiety/depression, insomnia, sleep disorder, disorders of taste and smell, headache), and cardiorespiratory impairment (breathlessness, chest pain, palpitations, persistent cough) (Anaya et al., 2021; Davis et al., 2021; Graham et al., 2021; Groff et al., 2021; Group, 2022; Nalbandian et al., 2021). While the risk factors for developing PCC in the general population remain to be fully understood, in those individuals who were hospitalized, risk factors associated with failure to recover at 1 year include female sex, obesity (BMI ≥30 kg/m2) and invasive mechanical ventilation (Group, 2022).
Of those individuals who report symptoms following either confirmed or probable COVID‐19, recovery can be protracted with self‐reporting studies suggesting that a majority of patients will have residual symptoms, including fatigue in 80% or more, post‐exertional malaise in over 70% and cognitive dysfunction in over 50% (Davis et al., 2021). A relapsing–remitting course has been reported (Davis et al., 2021; Tran, Porcher, et al., 2022). Beyond the immediate physical and psychosocial impact, the socioeconomic impact is and will be significant due to the percentage of individuals either not able to return to work, or if able to do so, at a reduced level largely due to memory and cognitive impairment (Cutler, 2022; Davis et al., 2021).
3. THE IMPACT OF AN AGING POPULATION
This issue of the prevalence of the post‐COVID‐19 syndrome and its associated clinical sequelae is rapidly becoming an impending crisis in the context of population aging. Epidemiological data consistently demonstrates that the peak age of onset for COVID‐19 remains between the age of 20 and 50. However, the mortality rates are highest amongst those who contracted the infection in the age group of 70–90. This is in striking contrast to the “W‐shaped” mortality curves of the Spanish flu with peaks in the very young and very old, and a third peak in the 15 through 40 years of age. In blunt terms, unlike the Spanish flu, the proportion of the population with the highest SARS‐COV‐2 virus infection rates survived and are thus at risk for its long‐term consequences. At a population level, this cohort is most likely to be affected by the post‐COVID‐19 syndrome and have a significant impact on the health system and socioeconomic consequences (Groff et al., 2021).
This is highlighted by a further epidemiological difference between the Spanish flu and COVID‐19 in that the latter is occurring in the context of population aging in which the number of centenarians (>100 years of age) is rapidly increasing, particularly in high‐income countries with low mortality rates (Robine & Cubaynes, 2017; Statistics Canada, 2022). In Canada, the population of older adults (aged 65+ years) surpassed the number of children (age 0–14) in 2016 as the number of older adults continues to increase from 17.2% of the population in 2018 to projections of 29.5% in 2068 (Statistics Canada, 2019). Between 2016 and 2021, the centenarian population grew at the fastest rate, increasing by 16% with projections to rise from approximately 9500 individuals in 2021 to slightly less than 80 000 individuals by 2061, of which the majority will be women.
An understanding of the natural history of cognitive function amongst this cohort of aging individuals is thus core to understanding the potential long‐term impact of COVID‐19. It has been estimated that 40% of centenarians will have dementia (Corrada et al., 2010; Lobo et al., 2011). But what of the remaining 60%? In a prospective 4‐year clinicopathological study of 340 centenarians (median age 100.5 years; range 100.2–101.7) between 2013 and 2019 (and thus preceding the current pandemic), only a slight decline in cognition driven by a minor decline in memory function was observed (Beker et al., 2021). For those patients undergoing a neuropathological study (n = 44), although varying stages of β‐amyloid deposition (Thal phases), neurofibrillary tangles (Braak Stage), and neuritic plaques (CERAD score) were observed, there was no clear association between the decline and the extent of underlying neuropathology although lower levels of tau pathology were associated with higher cognitive performance.
In short, the world's population has been dramatically aging in a manner that was not present during the Spanish flu, or indeed for many of the subsequent RNA virus‐associated pandemics for which neurological sequelae were observed. The prevalence of dementia (and neurodegenerative syndromes writ large) increases with aging, often with a prolonged subclinical latency as the neuropathological load gradually accumulates. The premise of the concern is that those individuals who have been infected by SARS‐CoV‐2 and have survived are largely amongst the population who are at the greatest risk of either an accelerated or a de novo neurodegenerative state.
4. THE NEUROLOGICAL AND NEUROPATHOLOGICAL IMPACTS OF SARS‐COV‐2 VIRUS INFECTION
The acute neurological consequences of SARS‐CoV‐2 infection can vary widely, but in those who manifest with an encephalopathy, associated features can include agitation, confusion, signs of diffuse corticospinal tract dysfunction in the presence of minimal magnetic resonance imaging findings (MRI) such as leptomeningeal enhancement, bilateral frontotemporal hypoperfusion, and rarely evidence of microvascular disease (Helms et al., 2020). Less common features can include seizures, cerebrovascular events, myoclonus, tremor, ataxia, chorea, an acute necrotizing hemorrhagic encephalopathy, and Guillain‐Barré syndrome (Lingor et al., 2022; Salari et al., 2021).
At the time of hospital discharge, a significant proportion of COVID‐19 patients with acute respiratory distress syndrome (ARDS) will have a persistent dysexecutive function (Lingor et al., 2022). These features (including deficits in attention, executive functioning, phonemic fluency, memory encoding, memory recall, and poorly organized movements suggestive of impaired praxis) were all significantly more likely to be evident on average 7.6 months postdiagnosis in those patients hospitalized for COVID‐19 than those treated as outpatients (Becker et al., 2021). When the cross‐sectional cognitive performance was assessed using an online tool (The Great British Intelligence Test) amongst predominantly United Kingdom COVID‐19 patients who had recovered (both hospitalized and nonhospitalized) between January and December 2020, significant cognitive deficits were observed, again consistent with a dysexecutive syndrome (Hampshire et al., 2021). In a 2‐year retrospective cohort study using an international data registry with neurological conditions ascertained through ICD10 codes, the risk of cognitive disorder, dementia (including vascular dementia, dementia in other diseases, unspecified dementia, Alzheimer's disease, frontotemporal dementia, and dementia with Lewy bodies), psychotic disorder and epilepsy or seizures was still increased at 2 years whereas other neurological symptoms/diagnosis associated with PCC had resolved (Taquet et al., 2022). No difference in 2‐year risk was observed between the patients infected by delta or omicron variants, whereas both were greater than the risk associated with the alpha variant.
While in general features of cognitive dysfunction are more severe in hospitalized patients, and in particular, those requiring mechanical ventilation, there is consistent evidence that COVID‐19 patients who were treated as outpatients, and are thus considered to have had a mild course, are not spared these neurological deficits which are often present to the same degree as that observed in the acute or subacute phase (Alemanno et al., 2021; Ali et al., 2022; Del Brutto et al., 2021; Graham et al., 2021; Hampshire et al., 2021; Jaywant et al., 2021; Pistarini et al., 2021; Woo et al., 2020). Neuropsychological, clinical, functional neuroimaging and electroencephalographic evidence consistently support either frontal or frontal network predominance of the cognitive deficits (Toniolo et al., 2021). Equally disconcerting, in a recent United Kingdom Biobank study of participants undergoing regular MRI studies and cognitive testing, those cases who tested positive for SARS‐CoV‐2 between two successive images (n = 401; age 58.9 ± 7.0 yrs at first scan with the second scan on average 141 days following diagnosis) demonstrated significant reductions in global brain size and cortical thinning in the orbitofrontal and parahippocampal gyrus, with evidence of greater cognitive decline (both Trails A and Trails B testing) than those cases in which no infection had occurred (n = 384; age 60.2 ± 7.4 yrs at first scan) (Douaud et al., 2022). No differences were observed between those patients hospitalized (n = 15) and nonhospitalized (n = 386) patients.
Noteworthy in the context of the H1N1 influenza A‐associated encephalitis lethargica, cases of an acute asymmetric hypokinetic‐rigid syndrome in the setting of an acute encephalopathy have been reported, as have cases arising posthospitalization and as the initial manifestation of SARS‐CoV‐2 infection (Boura & Chaudhuri, 2022; Cohen et al., 2020; Maramattom & Kishore, 2022; Mendez‐Guerrero et al., 2020; Morassi et al., 2021; Schneider et al., 2022). Asymmetric putamen dopamine uptake using either [123I]‐ioflupane dopamine transporter single photon emission computerized tomography (DaT‐SPECT) scanning or 18F‐fluordopa positron emission tomography (PET) imaging has been observed in such cases. A case of asymmetric akinetic‐rigid Parkinsonism onsetting 3 weeks after COVID‐19 in the absence of encephalopathy has also been observed (Schneider et al., 2022).
With respect to amyotrophic lateral sclerosis (ALS), the most common adult‐onset degenerative disorder of the motor system, the observations of increased rates of deterioration following COVID‐19 fall into two categories: those related to a failure of the patients to be able to access the full extent of multidisciplinary care, treatments, and interventions (De Marchi et al., 2021; Esselin et al., 2021), and those in which an acceleration of disease progression can be more directly linked to SARS‐CoV‐2 infection (Li & Bedlack, 2021).
In the acute stages of the illness, the most common neuropathological feature is that of a hypoxic–ischemic injury ranging from mild to severe in virtually all patients accompanied in the majority by vascular pathology including infarcts of various ages across multiple brain regions, but in particular the isocortex, striatum, hippocampus, corpus callosum, and brainstem (Mukerji & Solomon, 2021; Thakur et al., 2021). Immune‐complex‐mediated microvascular pathology with complement activation underlies breakdown in the blood–brain barrier, microthomboses, and perivascular injury (Lee et al., 2022). Most brains also demonstrate diffuse microglia activation, including microglial nodules in association with CD3+ and CD8+ T cells, the latter most commonly observed in the brainstem. Perivascular lymphocytic infiltration, generally minimal, has been observed in the majority of brains. In those with ongoing cognitive difficulties, increased plasma IL‐4, IL‐6, and CD70 levels have been observed, suggesting an ongoing role for neuroinflammation (“National Institute for Health and Care Excellence. COVID‐19 rapid guideline: managing the long‐term effects of COVID‐19,” 2022; Sun, Abriola, et al., 2021).
With respect to the presence of SARS‐CoV‐2 expression in the brain, there is a general consensus that if present at all, it is at exceptionally low levels as observed by qRT‐PCR paired with the failure to observe viral RNA in affected tissue using in situ hybridization against an array of SAR‐CoV‐2 sequences (Thakur et al., 2021).
5. PUTATIVE MECHANISMS OF THE INDUCTION OF NEURODEGENERATION
Although the exact mechanism(s) by which SARS‐CoV‐2 attacks the nervous system, both acutely and chronically, are still being investigated, there are a number of pathways that have been postulated, none of which are mutually exclusive (Crook et al., 2021; Jha et al., 2021; Spudich & Nath, 2022). Such effects may be due to direct nervous system infection or a remote effect of primary infection elsewhere. The concept that the brain is protected from direct SARS‐CoV‐2 infection due to the absence of ACE2 protein expression has been refuted by a number of observations that suggest low‐level ACE2 expression and the presence of coreceptors such as NRP‐1 (Daly et al., 2020; Jha et al., 2021; Li et al., 2020). The ability for SARS‐CoV‐2 to cross the blood–brain barrier has been confirmed in both human postmortem tissue and in vitro using human‐induced pluripotent stem cells differentiated into brain capillary endothelial‐like cells (BCECs) (Krasemann et al., 2022; Paniz‐Mondolfi et al., 2020). In the latter, the process of active uptake, replication, and transcellular transport can be inhibited by antibodies to the spike protein, ACE2 or NRP‐1, as well as the inhibition of TMPRSS2 (Krasemann et al., 2022). Upon SARS‐CoV‐2 infection, BCECs significantly upregulated genes associated with the innate immune type I IFN responses, including upregulation of IFN‐induced transmembrane proteins 1 and 2 (IFITM1 and IFITM2, respectively), mirroring the transcriptional profile of the neurovascular unit of COVID‐19 brains.
Exosomes, membrane‐bound vesicles ranging in size from 40 to 100 nm in diameter that are capable of shuttling cellular constituents including proteins, microRNA, and RNA between cells are emerging as having a potentially critical role as a vehicle by which SARS‐CoV‐2 or key components of its proteome are transported from the primary site of viral infection to the central nervous system. Lung‐derived exosomes from SARS‐CoV‐2‐affected patients have been observed to contain transcription factors that could be constructed into an exosomal network linked to neuronal gene dysregulation in Alzheimer's disease and Parkinson's disease (PD) (Ahmed et al., 2021). Having arrived at their recipient cell, the efficiency of fusion of the exosomal membrane to the receptor membrane is significantly enhanced by the presence of the spike glycoprotein, as is the induction of pathological aggregate formation in the recipient cell (Liu et al., 2021).
Both the S1 and nucleocapsid protein have been suggested to increase the propensity of pathological fibril formation, a cornerstone of many neurodegenerative processes. Importantly, the S1 protein RBD contains several heparin‐binding sites that are predicted to seed the aggregation of misfolded proteins (Tavassoly et al., 2020). In silico modeling of the interaction of this domain with several proteins associated with pathological fibril formation (amyloid beta, α‐synuclein, tau, prion protein, and Tar DNA‐binding protein of 43 kDa [TDP‐43]) has supported this by demonstrating significant binding affinities between the S1 RBD and each protein (Idrees & Kumar, 2021). In vitro, the nucleocapsid protein has been shown to significantly increase the propensity of α‐synuclein to aggregate and form amyloid fibrils, suggesting a mechanism by which the pathology of PD may be accelerated (Semerdzhiev et al., 2022). Preliminary reports of persistent nucleocapsid protein immunoreactivity in brain tissue are thus of some concern (Chertow et al., under review, DOI: https://doi.org/10.21203/rs.3.rs‐1139035/v1). In individuals with PCC who report ongoing neurological impairment, neuronal‐enriched extracellular vesicles (including exosomes) from plasma are enriched in markers of neurodegeneration, including amyloid, low molecular weight neurofilament subunit protein (NFL), total tau, pathologically phosphorylated tau (pThr181tau) and neurogranin, suggesting ongoing neural injury and neuroinflammation (Sun, Abriola, et al., 2021). The composition of such vesicles, thought to be shed from degenerating neurons, mirrors that observed in individuals with mild cognitive impairment (MCI) who progress to Alzheimer's disease (Winston et al., 2016).
SARS‐CoV‐2 can also disrupt neuronal RNA metabolism at multiple levels, including by the recruitment of host RNA‐binding proteins (RBPs) into its interactome. By examining the network of RBPs that interact directly with the SARS‐CoV‐2 transcript and comparing these to published gene databases for neurological disorders, a common pool of 9 RBPs that interact with the SARS‐CoV‐2 transcript and are associated with cancer and neurological disorders has been characterized (Prasad et al., 2022; Schmidt et al., 2021). Among the most highly linked neurological dysfunctions were dementia, depressive disorder, and anxiety. This pool of RBPs is associated with RNA degradation, stabilization, and translational enhancement and includes EIFA1, EIF4B, and EIF 5A (eukaryotic translation initiative factors A1, 4B & 5A, respectively), the suppressor of miRNA biogenesis LIN28B (protein lin‐28 homolog B), the RNA helicase MOV10 (Moloney leukemia virus 10 like), the poly(A) binding protein PABPC1 (poly(A) binding protein cytoplasmic 1) involved in promoting ribosome recruitment and translational initiation as well as poly(A) shortening in nonsense‐mediated decay (NMD), ribosomal protein RPL18A, and RPS3, RPS10 (ribosomal proteins S3 and S10, respectively). hnRNPA1 (heterogeneous nuclear ribonucleoprotein A1) was highly linked to the brain disease pool. Of these, several are involved in stress granules or processing bodies (EIF4B, MOV10, RPS3, PABPC1).
The importance of alterations in RNA metabolism in neurodegeneration, and in particular ALS, has been highlighted in recent years (Aulas & Vande Velde, 2015; Strong, 2010). Key to this has been an alteration in the formation of liquid condensates driven by the liquid–liquid phase separation of intrinsically disordered proteins, yielding specific subcellular compartments not delineated by membranes. It is of note therefore that the SARS‐CoV‐2 N protein possesses three intrinsically disordered domains suggesting that it can participate in liquid condensates (Cubuk et al., 2021). In a computational analysis, 77 potential interactors were found that included 33 associated with stress granules amongst which was hnRNPA1 (Moosa & Banerjee, 2020). In vitro, the N protein can form liquid condensates on its own, but it co‐phase separates with RBPs whose metabolism is disrupted in ALS, including TDP‐43, fused in sarcoma (FUS), hnRNPA2 and hnRNPA1 (Li, Lu, et al., 2022; Perdikari et al., 2020). No evidence of co‐phase separation was found for S, NSP1, Nsp8, or orf7α proteins (Li, Jia, et al., 2022). The kinetics of phase separation was altered to not only enhance the rate of stress granule (SG) assembly following an in vitro stress but also to delay SG disassembly. Further, the presence of an ALS‐associated FUS mutation (FUSP525L) accelerated liquid condensate formation.
6. THERAPEUTIC INTERVENTIONS
At this point in time, there are no specific therapies that can arrest or prevent the neurological features of PCC. There is conflicting evidence with respect to the role of vaccination postinfection in reducing both symptom number and severity. In a preliminary, nonpeer‐reviewed analysis of 910 adult long COVID patients in the French ComPaRe long COVID cohort, Tran and colleagues have reported a doubling of those in remission by 120 days (16.6% of the vaccinated group had become asymptomatic compared with 7.5% of the unvaccinated group) (Tran, Perrodeau, Saldanha, Pane, & Ravaud, 2022). The median time between symptom onset and baseline study was 10.7 months (IQR 6.4–12.4) and 60.1% of the patients had a confirmed SARS‐CoV‐2 infection. When scored for a possible 53 symptoms at 120 days following baseline, the vaccinated group reported a score of 13.0 (+/− 9.4) symptoms while the unvaccinated group scored 14.8 (+/− 9.8) symptoms. Using an aggregate score of 6 measures of disease impact (long COVID IT score), vaccinated patients reported less disease impact. For all measures, there was no difference between those with a confirmed diagnosis and those without. A similar strong effect of a single vaccination (either of the two available mRNA or the single inactivated viral vaccine) either before developing COVID‐19 (a 7–10‐fold reduction in the likelihood of infection) or within 4–12 weeks after (a three3 fold reduction) was observed in a retrospective analysis of over 240 000 American patients (analysis prior to the emergence of the delta variant) (Simon et al., 2021). More recently, the potential role of a second vaccine dose postinfection to promote a more sustained improvement has been suggested in a large community‐based population survey undertaken in the United Kingdom (Ayoubkhani et al., 2022).
However, the role of vaccination in reducing the probability of developing the post‐COVID‐19 condition, or in reducing its severity or the number of symptoms, remains controversial with several studies have shown no impact on these outcomes (Wisnivesky et al., 2022; Wynberg et al., 2022).
A recent US Department of Veterans Affairs study using ICD10 coding of healthcare databases found that the use of oral nirmatrelvir within 5 days of a SARS‐CoV‐2 positive test result was associated with an absolute risk reduction for at least one PCC symptom, including neurocognitive changes (not further defined), of 2.32 (95% confidence interval of 1.73, 2.91) in treated patients (n = 9217) vs untreated (n = 47 213), irrespective of vaccine or booster status (Xie et al., 2022). In practical terms, this translates into the need to treat 100 patients to see an added benefit of approximately 2 patients not developing PACS symptoms. Of note was the sex bias of males over females (87.6% male, 12.4% female) raising questions as to the generalizability of these modest results.
7. SUMMARY AND CONCLUSIONS
It is early days in our understanding of the neurodegenerative consequences of the SARS‐CoV‐2 infection and too soon to ascertain whether a “wave” of neurodegenerative diseases will be its long‐term legacy. What is clear however is that while SARS‐CoV‐2 can acutely affect the central nervous system, no single mechanism can account for the broad range of persistent manifestations observed in PCC (Figure 3). Similarly, the nature of the insult to the brain is also linked to the severity of the inflammation and/or immunopathology in each individual, and of the microvascular status. With chronicity, however, there are likely different mechanisms at play. The epidemiological data would suggest that the group at risk for this are those who are most likely to survive as older adults and those who will become centenarians. It thus behooves us to direct the same degree of energy and intent that was directed toward dealing with the acute infection to understanding and treating its long‐term sequelae.
AUTHOR CONTRIBUTIONS
Michael J. Strong is the sole author of the paper and is responsible for all aspects of its conceptualization and writing.
CONFLICT OF INTEREST
The author reports no conflicts of interest.
ACKNOWLEDGMENTS
The author is grateful for the careful review and critique of this article by Professors Eric Arts (Western University), Christian Baron (Univeristé de Montréal), Charu Kaushic (McMaster University), and Jane Rylett (Western University). The author is also grateful to the Honorable Kirsty Dunan PC MP for providing insights into the post‐Spanish Flu complications. Crystal Mclellan provided assistance with BioRender.
Strong, M. J. (2022). SARS‐CoV‐2, aging, and Post‐COVID‐19 neurodegeneration. Journal of Neurochemistry, 00, 1–16. 10.1111/jnc.15736
DATA AVAILABILITY STATEMENT
Data sharing is not applicable as this article is a literature review. There is no accompanying data.
REFERENCES
- Agrawal, A. , Gindodiya, A. , Deo, K. , Kashikar, S. , Fulzele, P. , & Khatib, N. (2021). A comparative analysis of the Spanish flu 1918 and COVID‐19 pandemics. The Open Public Health Journal, 14(14), 128–134. [Google Scholar]
- Ahmed, S. , Paramasivam, P. , Kamath, M. , Sharma, A. , Rome, S. , & Murugesan, R. (2021). Genetic exchange of lung‐derived exosome to brain causing neuronal changes on COVID‐19 infection. Molecular Neurobiology, 58(10), 5356–5368. 10.1007/s12035-021-02485-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alemanno, F. , Houdayer, E. , Parma, A. , Spina, A. , Del Forno, A. , Scatolini, A. , Angelone, S. , Brugliera, L. , Tettamanti, A. , Beretta, L. , & Iannaccone, S. (2021). COVID‐19 cognitive deficits after respiratory assistance in the subacute phase: A COVID‐rehabilitation unit experience. PLoS One, 16(2), e0246590. 10.1371/journal.pone.0246590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali, S. T. , Kang, A. K. , Patel, T. R. , Clark, J. R. , Perez‐Giraldo, G. S. , Orban, Z. S. , Lim, P. H. , Jimenez, M. , Graham, E. L. , Batra, A. , Liotta, E. M. , & Koralnik, I. J. (2022). Evolution of neurologic symptoms in non‐hospitalized COVID‐19 "long haulers". Annals of Clinical Translational Neurology, 9(7), 950–961. 10.1002/acn3.51570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anaya, J. M. , Rojas, M. , Salinas, M. L. , Rodriguez, Y. , Roa, G. , Lozano, M. , Rodriguez‐Jimenez, M. , Montoya, N. , Zapata, E. , Post, C. , Monsalve, D. M. , Acosta‐Ampudia, Y. , & Ramirez‐Santana, C. (2021). Post‐COVID syndrome. A case series and comprehensive review. Autoimmunity Reviews, 20(11), 102947. 10.1016/j.autrev.2021.102947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulas, A. , & Vande Velde, C. (2015). Alterations in stress granule dynamics driven by TDP‐43 and FUS: A link to pathological inclusions in ALS? Frontiers in Cellular Neuroscience, 9, 423. 10.3389/fncel.2015.00423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayoubkhani, D. , Bermingham, C. , Pouwels, K. B. , Glickman, M. , Nafilyan, V. , Zaccardi, F. , Khunti, K. , Alwan, N. A. , & Walker, A. S. (2022). Trajectory of long covid symptoms after covid‐19 vaccination: Community based cohort study. BMJ, 377, e069676. 10.1136/bmj-2021-069676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker, J. H. , Lin, J. J. , Doernberg, M. , Stone, K. , Navis, A. , Festa, J. R. , & Wisnivesky, J. P. (2021). Assessment of cognitive function in patients after COVID‐19 infection. JAMA Network Open, 4(10), e2130645. 10.1001/jamanetworkopen.2021.30645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beker, N. , Ganz, A. , Hulsman, M. , Klausch, T. , Schmand, B. A. , Scheltens, P. , Sikkes, S. A. M. , & Holstege, H. (2021). Association of Cognitive Function Trajectories in centenarians with postmortem neuropathology, physical health, and other risk factors for cognitive decline. JAMA Network Open, 4(1), e2031654. 10.1001/jamanetworkopen.2020.31654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, P. R. , Scaiola, A. , Loughran, G. , Leibundgut, M. , Kratzel, A. , Meurs, R. , Dreos, R. , O'Connor, K. M. , McMillan, A. , Bode, J. W. , Thiel, V. , Gatfield, D. , Atkins, J. F. , & Ban, N. (2021). Structural basis of ribosomal frameshifting during translation of the SARS‐CoV‐2 RNA genome. Science, 372(6548), 1306–1313. 10.1126/science.abf3546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi, M. , Borsetti, A. , Ciccozzi, M. , & Pascarella, S. (2021). SARS‐Cov‐2 ORF3a: Mutability and function. International Journal of Biological Macromolecules, 170, 820–826. 10.1016/j.ijbiomac.2020.12.142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch, B. J. , van der Zee, R. , de Haan, C. A. , & Rottier, P. J. (2003). The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. Journal of Virology, 77(16), 8801–8811. 10.1128/jvi.77.16.8801-8811.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boura, I. , & Chaudhuri, K. R. (2022). Coronavirus disease 2019 and related parkinsonism: The clinical evidence thus far. Movement Disorders Clinical Practice, 9, 584–593. 10.1002/mdc3.13461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley, I. , Digard, P. , & Inglis, S. C. (1989). Characterization of an efficient coronavirus ribosomal frameshifting signal: Requirement for an RNA pseudoknot. Cell, 57(4), 537–547. 10.1016/0092-8674(89)90124-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas, G. , Soto‐Hernandez, J. L. , Diaz‐Alba, A. , Ugalde, Y. , Merida‐Puga, J. , Rosetti, M. , & Sciutto, E. (2014). Neurological events related to influenza A (H1N1) pdm09. Influenza and Other Respiratory Viruses, 8(3), 339–346. 10.1111/irv.12241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilamakuri, R. , & Agarwal, S. (2021). COVID‐19: Characteristics and therapeutics. Cell, 10(2), 206. 10.3390/cells10020206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, M. E. , Eichel, R. , Steiner‐Birmanns, B. , Janah, A. , Ioshpa, M. , Bar‐Shalom, R. , Paul, J. J. , Gaber, H. , Skrahina, V. , Bornstein, N. M. , & Yahalom, G. (2020). A case of probable Parkinson's disease after SARS‐CoV‐2 infection. Lancet Neurology, 19(10), 804–805. 10.1016/S1474-4422(20)30305-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrada, M. M. , Brookmeyer, R. , Paganini‐Hill, A. , Berlau, D. , & Kawas, C. H. (2010). Dementia incidence continues to increase with age in the oldest old: The 90+ study. Annals of Neurology, 67(1), 114–121. 10.1002/ana.21915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, R. J. , & Brokstad, K. A. (2020). Not just antibodies: B cells and T cells mediate immunity to COVID‐19. Nature Reviews. Immunology, 20(10), 581–582. 10.1038/s41577-020-00436-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crook, H. , Raza, S. , Nowell, J. , Young, M. , & Edison, P. (2021). Long covid‐mechanisms, risk factors, and management. BMJ, 374, n1648. 10.1136/bmj.n1648 [DOI] [PubMed] [Google Scholar]
- Cubuk, J. , Alston, J. J. , Incicco, J. J. , Singh, S. , Stuchell‐Brereton, M. D. , Ward, M. D. , Zimmerman, M. I. , Vithani, N. , Griffith, D. , Wagoner, J. A. , Bowman, G. R. , Hall, K. B. , Soranno, A. , & Holehouse, A. S. (2021). The SARS‐CoV‐2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nature Communications, 12(1), 1936. 10.1038/s41467-021-21953-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler, D. M. (2022). The costs of long COVID. JAMA Forum, 3(5), e221809. [DOI] [PubMed] [Google Scholar]
- da Costa, V. G. , Saivish, M. V. , Santos, D. E. R. , de Lima Silva, R. F. , & Moreli, M. L. (2020). Comparative epidemiology between the 2009 H1N1 influenza and COVID‐19 pandemics. Journal of Infection and Public Health, 13(12), 1797–1804. 10.1016/j.jiph.2020.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly, J. L. , Simonetti, B. , Klein, K. , Chen, K. E. , Williamson, M. K. , Anton‐Plagaro, C. , Shoemark, D. K. , Simon‐Gracia, L. , Bauer, M. , Hollandi, R. , Greber, U. F. , Horvath, P. , Sessions, R. B. , Helenius, A. , Hiscox, J. A. , Teesalu, T. , Matthews, D. A. , Davidson, A. D. , Collins, B. M. , … Yamauchi, Y. (2020). Neuropilin‐1 is a host factor for SARS‐CoV‐2 infection. Science, 370(6518), 861–865. 10.1126/science.abd3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, H. E. , Assaf, G. S. , McCorkell, L. , Wei, H. , Low, R. J. , Re'em, Y. , Redfield, S. , Austin, J. P. , & Akrami, A. (2021). Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. EClinicalMedicine, 38, 101019. 10.1016/j.eclinm.2021.101019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, L. E. , Koster, F. , & Cawthon, A. (2014). Neurologic aspects of influenza viruses. Handbook of Clinical Neurology, 123, 619–645. 10.1016/B978-0-444-53488-0.00030-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marchi, F. , Gallo, C. , Sarnelli, M. F. , De Marchi, I. , Saraceno, M. , Cantello, R. , & Mazzini, L. (2021). Accelerated early progression of amyotrophic lateral sclerosis over the COVID‐19 pandemic. Brain Sciences, 11(10), 1291. 10.3390/brainsci11101291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Brutto, O. H. , Wu, S. , Mera, R. M. , Costa, A. F. , Recalde, B. Y. , & Issa, N. P. (2021). Cognitive decline among individuals with history of mild symptomatic SARS‐CoV‐2 infection: A longitudinal prospective study nested to a population cohort. European Journal of Neurology, 28(10), 3245–3253. 10.1111/ene.14775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Department of Health and Human Services, Office of the Assistant Secretary for Health . (2022). National Research Action Plan on Long COVID, 200 Independance Ave SW, Washington, DC 20201.
- Diamond, M. S. , & Kanneganti, T. D. (2022). Innate immunity: The first line of defense against SARS‐CoV‐2. Nature Immunology, 23(2), 165–176. 10.1038/s41590-021-01091-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickman, M. S. (2001). von Economo encephalitis. Archives of Neurology, 58(10), 1696–1698. 10.1001/archneur.58.10.1696 [DOI] [PubMed] [Google Scholar]
- Dominguez Andres, A. , Feng, Y. , Campos, A. R. , Yin, J. , Yang, C. C. , James, B. , Murad, R. , Kim, H. , Deshpande, A. J. , Gordon, D. E. , Krogan, N. , Pippa, R. , & Ronai, Z. A. (2020). SARS‐CoV‐2 ORF9c is a membrane‐associated protein that suppresses antiviral responses in cells. bioRxiv. 10.1101/2020.08.18.256776 [DOI] [Google Scholar]
- Douaud, G. , Lee, S. , Alfaro‐Almagro, F. , Arthofer, C. , Wang, C. , McCarthy, P. , Lange, F. , Andersson, J. L. R. , Griffanti, L. , Duff, E. , Jbabdi, S. , Taschler, B. , Keating, P. , Winkler, A. M. , Collins, R. , Matthews, P. M. , Allen, N. , Miller, K. L. , Nichols, T. E. , & Smith, S. M. (2022). SARS‐CoV‐2 is associated with changes in brain structure in UK biobank. Nature, 604(7907), 697–707. 10.1038/s41586-022-04569-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dourmashkin, R. R. (1997). What caused the 1918‐30 epidemic of encephalitis lethargica? Journal of the Royal Society of Medicine, 90(9), 515–520. 10.1177/014107689709000916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esselin, F. , De La Cruz, E. , Pageot, N. , Juntas‐Morales, R. , Alphandery, S. , & Camu, W. (2021). Increased worsening of amyotrophic lateral sclerosis patients during Covid‐19‐related lockdown in France. Amyotroph Lateral Scler Frontotemporal Degener, 22(7–8), 505–507. 10.1080/21678421.2021.1883669 [DOI] [PubMed] [Google Scholar]
- Fung, T. S. , & Liu, D. X. (2021). Similarities and dissimilarities of COVID‐19 and other coronavirus diseases. Annual Review of Microbiology, 75, 19–47. 10.1146/annurev-micro-110520-023212 [DOI] [PubMed] [Google Scholar]
- Geddes, J. F. , Hughes, A. J. , Lees, A. J. , & Daniel, S. E. (1993). Pathological overlap in cases of parkinsonism associated with neurofibrillary tangles: A study of recent cases of postencephalitic parkinsonism and comparison with progressive supranuclear palsy and Guamanian parkinsonism‐dementia complex. Brain, 116(1), 281–302. [DOI] [PubMed] [Google Scholar]
- Graham, E. L. , Clark, J. R. , Orban, Z. S. , Lim, P. H. , Szymanski, A. L. , Taylor, C. , DiBiase, R. M. , Jia, D. T. , Balabanov, R. , Ho, S. U. , Batra, A. , Liotta, E. M. , & Koralnik, I. J. (2021). Persistent neurologic symptoms and cognitive dysfunction in non‐hospitalized Covid‐19 "long haulers". Annals of Clinical Translational Neurology, 8(5), 1073–1085. 10.1002/acn3.51350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groff, D. , Sun, A. , Ssentongo, A. E. , Ba, D. M. , Parsons, N. , Poudel, G. R. , Lekoubou, A. , Oh, J. S. , Ericson, J. E. , Ssentongo, P. , & Chinchilli, V. M. (2021). Short‐term and long‐term rates of Postacute sequelae of SARS‐CoV‐2 infection: A systematic review. JAMA Network Open, 4(10), e2128568. 10.1001/jamanetworkopen.2021.28568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Group, P.‐C. C . (2022). Clinical characteristics with inflammation profiling of long COVID and association with 1‐year recovery following hospitalisation in the UK: A prospective observational study. The Lancet Respiratory Medicine, 10(8), 761–775. 10.1016/S2213-2600(22)00127-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, W. , Gan, H. , Ma, Y. , Xu, L. , Cheng, Z. J. , Li, B. , Zhang, X. , Jiang, W. , Sun, J. , Sun, B. , & Hao, C. (2022). The molecular mechanism of SARS‐CoV‐2 evading host antiviral innate immunity. Virology Journal, 19(1), 49. 10.1186/s12985-022-01783-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulyaeva, A. A. , & Gorbalenya, A. E. (2021). A nidovirus perspective on SARS‐CoV‐2. Biochemical and Biophysical Research Communications, 538, 24–34. 10.1016/j.bbrc.2020.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev, E. , Sarapultsev, A. , Solomatina, L. , & Chereshnev, V. (2022). SARS‐CoV‐2‐specific immune response and the pathogenesis of COVID‐19. International Journal of Molecular Sciences, 23(3), 1716. 10.3390/ijms23031716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampshire, A. , Trender, W. , Chamberlain, S. R. , Jolly, A. E. , Grant, J. E. , Patrick, F. , Mazibuko, N. , Williams, S. C. , Barnby, J. M. , Hellyer, P. , & Mehta, M. A. (2021). Cognitive deficits in people who have recovered from COVID‐19. EClinicalMedicine, 39, 101044. 10.1016/j.eclinm.2021.101044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms, J. , Kremer, S. , Merdji, H. , Clere‐Jehl, R. , Schenck, M. , Kummerlen, C. , Collange, O. , Boulay, C. , Fafi‐Kremer, S. , Ohana, M. , Anheim, M. , & Meziani, F. (2020). Neurologic features in severe SARS‐CoV‐2 infection. The New England Journal of Medicine, 382(23), 2268–2270. 10.1056/NEJMc2008597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry, J. , Smeyne, R. J. , Jang, H. , Miller, B. , & Okun, M. S. (2010). Parkinsonism and neurological manifestations of influenza throughout the 20th and 21st centuries. Parkinsonism & Related Disorders, 16(9), 566–571. 10.1016/j.parkreldis.2010.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman, L. A. , & Vilensky, J. A. (2017). Encephalitis lethargica: 100 years after the epidemic. Brain, 140(8), 2246–2251. 10.1093/brain/awx177 [DOI] [PubMed] [Google Scholar]
- Hudson, A. J. , & Rice, G. P. (1990). Similarities of guamanian ALS/PD to post‐encephalitic parkinsonism/ALS: Possible viral cause. The Canadian Journal of Neurological Sciences, 17(4), 427–433. 10.1017/s0317167100031024 [DOI] [PubMed] [Google Scholar]
- Idrees, D. , & Kumar, V. (2021). SARS‐CoV‐2 spike protein interactions with amyloidogenic proteins: Potential clues to neurodegeneration. Biochemical and Biophysical Research Communications, 554, 94–98. 10.1016/j.bbrc.2021.03.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang, H. , Boltz, D. A. , Webster, R. G. , & Smeyne, R. J. (2009). Viral parkinsonism. Biochimica et Biophysica Acta, 1792(7), 714–721. 10.1016/j.bbadis.2008.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaywant, A. , Vanderlind, W. M. , Alexopoulos, G. S. , Fridman, C. B. , Perlis, R. H. , & Gunning, F. M. (2021). Frequency and profile of objective cognitive deficits in hospitalized patients recovering from COVID‐19. Neuropsychopharmacology, 46(13), 2235–2240. 10.1038/s41386-021-00978-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha, N. K. , Ojha, S. , Jha, S. K. , Dureja, H. , Singh, S. K. , Shukla, S. D. , Chellappan, D. K. , Gupta, G. , Bhardwaj, S. , Kumar, N. , Jeyaraman, M. , Jain, R. , Muthu, S. , Kar, R. , Kumar, D. , Goswami, V. K. , Ruokolainen, J. , Kesari, K. K. , Singh, S. K. , & Dua, K. (2021). Evidence of coronavirus (CoV) pathogenesis and emerging pathogen SARS‐CoV‐2 in the nervous system: A review on neurological impairments and manifestations. Journal of Molecular Neuroscience, 71(11), 2192–2209. 10.1007/s12031-020-01767-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazaal, S. , Harb, J. , Rima, M. , Annweiler, C. , Wu, Y. , Cao, Z. , Abi Khattar, Z. , Legros, C. , Kovacic, H. , Fajloun, Z. , & Sabatier, J. M. (2022). The pathophysiology of long COVID throughout the renin‐angiotensin system. Molecules, 27(9), 2903. 10.3390/molecules27092903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno, Y. , Kimura, I. , Uriu, K. , Fukushi, M. , Irie, T. , Koyanagi, Y. , Sauter, D. , Gifford, R. J. , Consortium, U.‐C. , Nakagawa, S. , & Sato, K. (2020). SARS‐CoV‐2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Reports, 32(12), 108185. 10.1016/j.celrep.2020.108185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann, S. , Haferkamp, U. , Pfefferle, S. , Woo, M. S. , Heinrich, F. , Schweizer, M. , Appelt‐Menzel, A. , Cubukova, A. , Barenberg, J. , Leu, J. , Hartmann, K. , Thies, E. , Littau, J. L. , Sepulveda‐Falla, D. , Zhang, L. , Ton, K. , Liang, Y. , Matschke, J. , Ricklefs, F. , … Pless, O. (2022). The blood‐brain barrier is dysregulated in COVID‐19 and serves as a CNS entry route for SARS‐CoV‐2. Stem Cell Reports, 17(2), 307–320. 10.1016/j.stemcr.2021.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichel, B. , Falke, S. , Hilgenfeld, R. , Redecke, L. , & Uetrecht, C. (2020). Processing of the SARS‐CoV pp1a/ab nsp7‐10 region. The Biochemical Journal, 477(5), 1009–1019. 10.1042/BCJ20200029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M. H. , Perl, D. P. , Steiner, J. , Pasternack, N. , Li, W. , Maric, D. , Safavi, F. , Horkayne‐Szakaly, I. , Jones, R. , Stram, M. N. , Moncur, J. T. , Hefti, M. , Folkerth, R. D. , & Nath, A. (2022). Neurovascular injury with complement activation and inflammation in COVID‐19. Brain, 145(7), 2555–2568. 10.1093/brain/awac151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J. , Jia, H. , Tian, M. , Wu, N. , Yang, X. , Qi, J. , Ren, W. , Li, F. , & Bian, H. (2022). SARS‐CoV‐2 and emerging variants: Unmasking structure, function, infection, and immune escape mechanisms. Frontiers in Cellular and Infection Microbiology, 12, 869832. 10.3389/fcimb.2022.869832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, M. Y. , Li, L. , Zhang, Y. , & Wang, X. S. (2020). Expression of the SARS‐CoV‐2 cell receptor gene ACE2 in a wide variety of human tissues. Infectious Diseases of Poverty, 9(1), 45. 10.1186/s40249-020-00662-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , & Bedlack, R. (2021). COVID‐19‐accelerated disease progression in two patients with amyotrophic lateral sclerosis. Muscle & Nerve, 64(3), E13–E15. 10.1002/mus.27351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X. , Hou, P. , Ma, W. , Wang, X. , Wang, H. , Yu, Z. , Chang, H. , Wang, T. , Jin, S. , Wang, X. , Wang, W. , Zhao, Y. , Zhao, Y. , Xu, C. , Ma, X. , Gao, Y. , & He, H. (2022). SARS‐CoV‐2 ORF10 suppresses the antiviral innate immune response by degrading MAVS through mitophagy. Cellular & Molecular Immunology, 19(1), 67–78. 10.1038/s41423-021-00807-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , Lu, S. , Gu, J. , Xia, W. , Zhang, S. , Zhang, S. , Wang, Y. , Zhang, C. , Sun, Y. , Lei, J. , Liu, C. , Su, Z. , Yang, J. , Peng, X. , & Li, D. (2022). SARS‐CoV‐2 impairs the disassembly of stress granules and promotes ALS‐associated amyloid aggregation. Protein & Cell, 13(8), 602–614. 10.1007/s13238-022-00905-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, S. T. , Liang, L. T. , & Rosen, J. M. (2021). COVID‐19: A comparison to the 1918 influenza and how we can defeat it. Postgraduate Medical Journal, 97(1147), 273–274. 10.1136/postgradmedj-2020-139070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingor, P. , Demleitner, A. F. , Wolff, A. W. , & Feneberg, E. (2022). SARS‐CoV‐2 and neurodegenerative diseases: What we know and what we don't. Journal of Neural Transmission (Vienna), 129, 1155–1167. 10.1007/s00702-022-02500-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Hossinger, A. , Heumuller, S. E. , Hornberger, A. , Buravlova, O. , Konstantoulea, K. , Muller, S. A. , Paulsen, L. , Rousseau, F. , Schymkowitz, J. , Lichtenthaler, S. F. , Neumann, M. , Denner, P. , & Vorberg, I. M. (2021). Highly efficient intercellular spreading of protein misfolding mediated by viral ligand‐receptor interactions. Nature Communications, 12(1), 5739. 10.1038/s41467-021-25855-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo, A. , Lopez‐Anton, R. , Santabarbara, J. , de‐la‐ Camara, C. , Ventura, T. , Quintanilla, M. A. , Roy, J. F. , Campayo, A. J. , Lobo, E. , Palomo, T. , Rodriguez‐Jimenez, R. , Saz, P. , & Marcos, G. (2011). Incidence and lifetime risk of dementia and Alzheimer's disease in a southern European population. Acta Psychiatrica Scandinavica, 124(5), 372–383. 10.1111/j.1600-0447.2011.01754.x [DOI] [PubMed] [Google Scholar]
- Lopez‐Munoz, A. D. , Kosik, I. , Holly, J. , & Yewdell, J. W. (2022). Cell surface SARS‐CoV‐2 nucleocapsid protein modulates innate and adaptive immunity. Science Advances, 8(31), eabp9770. 10.1126/sciadv.abp9770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar, P. , & Niyogi, S. (2021). SARS‐CoV‐2 mutations: The biological trackway towards viral fitness. Epidemiology and Infection, 149, e110. 10.1017/S0950268821001060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maramattom, B. V. , & Kishore, A. (2022). Acute akinetic‐rigid syndrome in COVID‐19 encephalitis. Acta Neurologica Belgica, 122(3), 847–849. 10.1007/s13760-022-01892-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martini, M. , Gazzaniga, V. , Bragazzi, N. L. , & Barberis, I. (2019). The Spanish influenza pandemic: A lesson from history 100 years after 1918. Journal of Preventive Medicine and Hygiene, 60(1), E64–E67. 10.15167/2421-4248/jpmh2019.60.1.1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAlpine, D. (1923). The pathology of the parkinsoniam syndrome following encephalitis lethargica, with a note on the occurrence of calcification in this disease. Brain, 46(3), 255–280. [Google Scholar]
- McAlpine, D. (1926). The anatomo‐pathological basis of the parkinsonian syndrome following epidemic encephalitis. Brain, 49(4), 525–556. [Google Scholar]
- McCall, S. , Vilensky, J. A. , Gilman, S. , & Taubenberger, J. K. (2008). The relationship between encephalitis lethargica and influenza: A critical analysis. Journal of Neurovirology, 14(3), 177–185. 10.1080/13550280801995445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez‐Guerrero, A. , Laespada‐Garcia, M. I. , Gomez‐Grande, A. , Ruiz‐Ortiz, M. , Blanco‐Palmero, V. A. , Azcarate‐Diaz, F. J. , Rabano‐Suarez, P. , Alvarez‐Torres, E. , de Fuenmayor‐Fernandez de la Hoz, C. P. , Vega Perez, D. , Rodriguez‐Montalban, R. , Perez‐Rivilla, A. , Sayas Catalan, J. , Ramos‐Gonzalez, A. , & Gonzalez de la Aleja, J. (2020). Acute hypokinetic‐rigid syndrome following SARS‐CoV‐2 infection. Neurology, 95(15), e2109–e2118. 10.1212/WNL.0000000000010282 [DOI] [PubMed] [Google Scholar]
- Merad, M. , Blish, C. A. , Sallusto, F. , & Iwasaki, A. (2022). The immunology and immunopathology of COVID‐19. Science, 375(6585), 1122–1127. 10.1126/science.abm8108 [DOI] [PubMed] [Google Scholar]
- Miorin, L. , Kehrer, T. , Sanchez‐Aparicio, M. T. , Zhang, K. , Cohen, P. , Patel, R. S. , Cupic, A. , Makio, T. , Mei, M. , Moreno, E. , Danziger, O. , White, K. M. , Rathnasinghe, R. , Uccellini, M. , Gao, S. , Aydillo, T. , Mena, I. , Yin, X. , Martin‐Sancho, L. , … Garcia‐Sastre, A. (2020). SARS‐CoV‐2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proceedings of the National Academy of Sciences of the United States of America, 117(45), 28344–28354. 10.1073/pnas.2016650117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosa, M. M. , & Banerjee, P. R. (2020). Subversion of host stress granules by coronaviruses: Potential roles of pi‐rich disordered domains of viral nucleocapsids. Journal of Medical Virology, 92, 2891–2893. 10.1002/jmv.26195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morassi, M. , Palmerini, F. , Nici, S. , Magni, E. , Savelli, G. , Guerra, U. P. , Chieregato, M. , Morbelli, S. , & Vogrig, A. (2021). SARS‐CoV‐2‐related encephalitis with prominent parkinsonism: Clinical and FDG‐PET correlates in two patients. Journal of Neurology, 268(11), 3980–3987. 10.1007/s00415-021-10560-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss, P. (2022). The T cell immune response against SARS‐CoV‐2. Nature Immunology, 23(2), 186–193. 10.1038/s41590-021-01122-w [DOI] [PubMed] [Google Scholar]
- Mukerji, S. S. , & Solomon, I. H. (2021). What can we learn from brain autopsies in COVID‐19? Neuroscience Letters, 742, 135528. 10.1016/j.neulet.2020.135528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbandian, A. , Sehgal, K. , Gupta, A. , Madhavan, M. V. , McGroder, C. , Stevens, J. S. , Cook, J. R. , Nordvig, A. S. , Shalev, D. , Sehrawat, T. S. , Ahluwalia, N. , Bikdeli, B. , Dietz, D. , Der‐Nigoghossian, C. , Liyanage‐Don, N. , Rosner, G. F. , Bernstein, E. J. , Mohan, S. , Beckley, A. A. , … Wan, E. Y. (2021). Post‐acute COVID‐19 syndrome. Nature Medicine, 27(4), 601–615. 10.1038/s41591-021-01283-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Institute for Health and Care Excellence . (2022). COVID‐19 rapid guideline: managing the long‐term effects of COVID‐19. version 1.14, published on 01.03.2022. [PubMed]
- Oikonen, M. , Laaksonen, M. , Aalto, V. , Ilonen, J. , Salonen, R. , Eralinna, J. P. , Panelius, M. , & Salmi, A. (2011). Temporal relationship between environmental influenza A and Epstein‐Barr viral infections and high multiple sclerosis relapse occurrence. Multiple Sclerosis, 17(6), 672–680. 10.1177/1352458510394397 [DOI] [PubMed] [Google Scholar]
- Paniz‐Mondolfi, A. , Bryce, C. , Grimes, Z. , Gordon, R. E. , Reidy, J. , Lednicky, J. , Sordillo, E. M. , & Fowkes, M. (2020). Central nervous system involvement by severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2). Journal of Medical Virology, 92(7), 699–702. 10.1002/jmv.25915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdikari, T. M. , Murthy, A. C. , Ryan, V. H. , Watters, S. , Naik, M. T. , & Fawzi, N. L. (2020). SARS‐CoV‐2 nucleocapsid protein phase‐separates with RNA and with human hnRNPs. The EMBO Journal, 39(24), e106478. 10.15252/embj.2020106478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovan, S. O. , Aldridge, D. C. , Bartlett, H. , Bladon, A. J. , Booth, H. , Broad, S. , Broom, D. M. , Burgess, N. D. , Cleaveland, S. , Cunningham, A. A. , Ferri, M. , Hinsley, A. , Hua, F. , Hughes, A. C. , Jones, K. , Kelly, M. , Mayes, G. , Radakovic, M. , Ugwu, C. A. , … Sutherland, W. J. (2021). Post COVID‐19: A solution scan of options for preventing future zoonotic epidemics. Biological Reviews of the Cambridge Philosophical Society, 96(6), 2694–2715. 10.1111/brv.12774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistarini, C. , Fiabane, E. , Houdayer, E. , Vassallo, C. , Manera, M. R. , & Alemanno, F. (2021). Cognitive and emotional disturbances due to COVID‐19: An exploratory study in the rehabilitation setting. Frontiers in Neurology, 12, 643646. 10.3389/fneur.2021.643646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plant, E. P. , & Dinman, J. D. (2008). The role of programmed‐1 ribosomal frameshifting in coronavirus propagation. Frontiers in Bioscience, 13, 4873–4881. 10.2741/3046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad, K. , Gour, P. , Raghuvanshi, S. , & Kumar, V. (2022). The SARS‐CoV‐2 targeted human RNA binding proteins network biology to investigate COVID‐19 associated manifestations. International Journal of Biological Macromolecules, 217, 853–863. 10.1016/j.ijbiomac.2022.07.200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primorac, D. , Vrdoljak, K. , Brlek, P. , Pavelic, E. , Molnar, V. , Matisic, V. , Erceg Ivkosic, I. , & Parcina, M. (2022). Adaptive immune responses and immunity to SARS‐CoV‐2. Frontiers in Immunology, 13, 848582. 10.3389/fimmu.2022.848582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi, M. , Pandey, N. , Shukla, A. , & Singh, S. K. (2020). SARS coronavirus 2: From genome to infectome. Respiratory Research, 21(1), 318. 10.1186/s12931-020-01581-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo, N. , Zaldivar‐Lopez, S. , Garrido, J. J. , & Montoya, M. (2021). SARS‐CoV‐2 accessory proteins in viral pathogenesis: Knowns and unknowns. Frontiers in Immunology, 12, 708264. 10.3389/fimmu.2021.708264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robine, J. M. , & Cubaynes, S. (2017). Worldwide demography of centenarians. Mechanisms of Ageing and Development, 165(Pt B), 59–67. 10.1016/j.mad.2017.03.004 [DOI] [PubMed] [Google Scholar]
- Robinson, C. P. , & Busl, K. M. (2020). Neurologic manifestations of severe respiratory viral contagions. Critical Care Explorations, 2(4), e0107. 10.1097/CCE.0000000000000107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohaim, M. A. , El Naggar, R. F. , Clayton, E. , & Munir, M. (2021). Structural and functional insights into non‐structural proteins of coronaviruses. Microbial Pathogenesis, 150, 104641. 10.1016/j.micpath.2020.104641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozo, M. , & Gronvall, G. K. (2015). The reemergent 1977 H1N1 strain and the gain‐of‐function debate. mBio, 6(4), e01013‐15. 10.1128/mBio.01013-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salari, M. , Zaker Harofteh, B. , Etemadifar, M. , Sedaghat, N. , & Nouri, H. (2021). Movement disorders associated with COVID‐19. Parkinsons Dis, 2021, 3227753–3227711. 10.1155/2021/3227753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, N. , Lareau, C. A. , Keshishian, H. , Ganskih, S. , Schneider, C. , Hennig, T. , Melanson, R. , Werner, S. , Wei, Y. , Zimmer, M. , Ade, J. , Kirschner, L. , Zielinski, S. , Dolken, L. , Lander, E. S. , Caliskan, N. , Fischer, U. , Vogel, J. , Carr, S. A. , … Munschauer, M. (2021). The SARS‐CoV‐2 RNA‐protein interactome in infected human cells. Nature Microbiology, 6(3), 339–353. 10.1038/s41564-020-00846-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, S. A. , Hennig, A. , & Martino, D. (2022). Relationship between COVID‐19 and movement disorders: A narrative review. European Journal of Neurology, 29(4), 1243–1253. 10.1111/ene.15217 [DOI] [PubMed] [Google Scholar]
- Semerdzhiev, S. A. , Fakhree, M. A. A. , Segers‐Nolten, I. , Blum, C. , & Claessens, M. (2022). Interactions between SARS‐CoV‐2 N‐protein and alpha‐synuclein accelerate amyloid formation. ACS Chemical Neuroscience, 13(1), 143–150. 10.1021/acschemneuro.1c00666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setaro, A. C. , & Gaglia, M. M. (2021). All hands on deck: SARS‐CoV‐2 proteins that block early anti‐viral interferon responses. Current Research in Virological Science, 2, 100015. 10.1016/j.crviro.2021.100015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette, A. , & Crotty, S. (2021). Adaptive immunity to SARS‐CoV‐2 and COVID‐19. Cell, 184(4), 861–880. 10.1016/j.cell.2021.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, M. A. , Luginbuhl, R. D. , & Parker, R. (2021). Reduced incidence of long‐COVID symptoms related to administration of COVID‐19 vaccines both before COVID‐19 diagnosis and up to 12 weeks after. medRxiv. 10.1101/2021.11.17.21263608 [DOI] [Google Scholar]
- Simonsen, L. , Clarke, M. J. , Schonberger, L. B. , Arden, N. H. , Cox, N. J. , & Fukuda, K. (1998). Pandemic versus epidemic influenza mortality: A pattern of changing age distribution. The Journal of Infectious Diseases, 178(1), 53–60. 10.1086/515616 [DOI] [PubMed] [Google Scholar]
- Socan, M. , Prosenc, K. , Ucakar, V. , & Berginc, N. (2014). A comparison of the demographic and clinical characteristics of laboratory‐confirmed influenza B Yamagata and Victoria lineage infection. Journal of Clinical Virology, 61(1), 156–160. 10.1016/j.jcv.2014.06.018 [DOI] [PubMed] [Google Scholar]
- Spudich, S. , & Nath, A. (2022). Nervous system consequences of COVID‐19. Science, 375(6578), 267–269. 10.1126/science.abm2052 [DOI] [PubMed] [Google Scholar]
- Stallybass, C. O. (1923). Encephalitis lethargica: Some observations on a recent outbreak. Lancet, 202(5226), 922–925. [Google Scholar]
- Statistics Canada . (2019). Population projections for Canada (2018 to 2068), Provinces and Territories (2018 to 2043). Catalogue no. 91‐520‐X .
- Statistics Canada . (2022). Census in Brief. A portrait of Canada's growing population aged 85 and older from the 2021 census. (978‐0‐660‐43041‐6).
- Strong, M. J. (2010). The evidence for altered RNA metabolism in amyotrophic lateral sclerosis (ALS). Journal of the Neurological Sciences, 288(1–2), 1–12. [DOI] [PubMed] [Google Scholar]
- Sun, B. , Tang, N. , Peluso, M. J. , Iyer, N. S. , Torres, L. , Donatelli, J. L. , Munter, S. E. , Nixon, C. C. , Rutishauser, R. L. , Rodriguez‐Barraquer, I. , Greenhouse, B. , Kelly, J. D. , Martin, J. N. , Deeks, S. G. , Henrich, T. J. , & Pulliam, L. (2021). Characterization and biomarker analyses of Post‐COVID‐19 complications and neurological manifestations. Cell, 10(2), 386. 10.3390/cells10020386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. , Abriola, L. , Niederer, R. O. , Pedersen, S. F. , Alfajaro, M. M. , Silva Monteiro, V. , Wilen, C. B. , Ho, Y. C. , Gilbert, W. V. , Surovtseva, Y. V. , Lindenbach, B. D. , & Guo, J. U. (2021). Restriction of SARS‐CoV‐2 replication by targeting programmed −1 ribosomal frameshifting. Proceedings of the National Academy of Sciences of the United States of America, 118(26), e2023051118. 10.1073/pnas.2023051118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahir, M. (2021). Coronavirus genomic nsp14‐ExoN, structure, role, mechanism, and potential application as a drug target. Journal of Medical Virology, 93(7), 4258–4264. 10.1002/jmv.27009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taquet, M. , Sillett, R. , Zhu, L. , Mendel, J. , Camplisson, I. , Dercon, Q. , & Harrison, P. J. (2022). Neurological and psychiatric risk trajectories after SARS‐CoV‐2 infection: An anaysis of 2‐year retrospective cohort studies including 1 284 437 patients. Lancet Psychiatry, 1‐13, 815–827. 10.1016/S2215-0366(22)00260-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger, J. K. (2006). The origin and virulence of the 1918 "Spanish" influenza virus. Proceedings of the American Philosophical Society, 150(1), 86–112. Retrieved from https://www.ncbi.nlm.nih.gov/pubmed/17526158 [PMC free article] [PubMed] [Google Scholar]
- Tavassoly, O. , Safavi, F. , & Tavassoly, I. (2020). Seeding brain protein aggregation by SARS‐CoV‐2 as a possible long‐term complication of COVID‐19 infection. ACS Chemical Neuroscience, 11(22), 3704–3706. 10.1021/acschemneuro.0c00676 [DOI] [PubMed] [Google Scholar]
- Thakur, K. T. , Miller, E. H. , Glendinning, M. D. , Al‐Dalahmah, O. , Banu, M. A. , Boehme, A. K. , Boubour, A. L. , Bruce, S. S. , Chong, A. M. , Claassen, J. , Faust, P. L. , Hargus, G. , Hickman, R. A. , Jambawalikar, S. , Khandji, A. G. , Kim, C. Y. , Klein, R. S. , Lignelli‐Dipple, A. , Lin, C. C. , … Canoll, P. (2021). COVID‐19 neuropathology at Columbia University Irving medical center/New York Presbyterian hospital. Brain, 144(9), 2696–2708. 10.1093/brain/awab148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toniolo, S. , Di Lorenzo, F. , Scarioni, M. , Frederiksen, K. S. , & Nobili, F. (2021). Is the frontal lobe the primary target of SARS‐CoV‐2? Journal of Alzheimer's Disease, 81(1), 75–81. 10.3233/JAD-210008 [DOI] [PubMed] [Google Scholar]
- Tran, V.‐T. , Perrodeau, E. , Saldanha, J. , Pane, I. , & Ravaud, P. (2022). Efficacy of COVID‐19 vaccination on the symptoms of patients with long COVID: A target trial emulation using data from the ComPaRe e‐cohort in France. Lancet (available at SSRN: https://ssrn.com/abstract=3932953 or 10.2139/ssrn.3932953) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran, V. T. , Porcher, R. , Pane, I. , & Ravaud, P. (2022). Course of post COVID‐19 disease symptoms over time in the ComPaRe long COVID prospective e‐cohort. Nature Communications, 13(1), 1812. 10.1038/s41467-022-29513-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai, J. P. , & Baker, A. J. (2013). Influenza‐associated neurological complications. Neurocritical Care, 18(1), 118–130. 10.1007/s12028-012-9796-8 [DOI] [PubMed] [Google Scholar]
- Weaver, A. K. , Head, J. R. , Gould, C. F. , Carlton, E. J. , & Remais, J. V. (2022). Environmental factors influencing COVID‐19 incidence and severity. Annual Review of Public Health, 43, 271–291. 10.1146/annurev-publhealth-052120-101420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . (2021). A clinical case definition of post COVID‐19 condition by a Delphi consensus. Retrieved from. https://www.who.int/publications/i/item/WHO‐2019‐nCoV‐Post_COVID‐19_condition‐Clinical_case_definition‐2021.1 [DOI] [PMC free article] [PubMed]
- Winston, C. N. , Goetzl, E. J. , Akers, J. C. , Carter, B. S. , Rockenstein, E. M. , Galasko, D. , Masliah, E. , & Rissman, R. A. (2016). Prediction of conversion from mild cognitive impairment to dementia with neuronally derived blood exosome protein profile. Alzheimers Dement (Amst), 3, 63–72. 10.1016/j.dadm.2016.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisnivesky, J. P. , Govindarajulu, U. , Bagiella, E. , Goswami, R. , Kale, M. , Campbell, K. N. , Meliambro, K. , Chen, Z. , Aberg, J. A. , & Lin, J. J. (2022). Association of Vaccination with the persistence of Post‐COVID symptoms. Journal of General Internal Medicine, 37(7), 1748–1753. 10.1007/s11606-022-07465-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, M. S. , Malsy, J. , Pottgen, J. , Seddiq Zai, S. , Ufer, F. , Hadjilaou, A. , Schmiedel, S. , Addo, M. M. , Gerloff, C. , Heesen, C. , Wiesch, S. Z. J. , & Friese, M. A. (2020). Frequent neurocognitive deficits after recovery from mild COVID‐19. Brain Communications, 2(2), fcaa205. 10.1093/braincomms/fcaa205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynberg, E. , Han, A. X. , Boyd, A. , van Willigen, H. D. G. , Verveen, A. , Lebbink, R. , van der Straten, K. , Kootstra, N. , van Gils, M. J. , Russell, C. , Leenstra, T. , de Jong, M. D. , de Bree, G. J. , Prins, M. , & Group, R. E. S . (2022). The effect of SARS‐CoV‐2 vaccination on post‐acute sequelae of COVID‐19 (PASC): A prospective cohort study. Vaccine, 40(32), 4424–4431. 10.1016/j.vaccine.2022.05.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, Y. , Choi, T. , & Al‐Aly, Z. (2022). Nirmatrelvir and the risk of Post‐acute sequelae of COVID‐19. medRxiv. 10.1101/2022.11.03.22281783 [DOI] [Google Scholar]
- Yazdanpanah, N. , & Rezaei, N. (2022). Autoimmune complications of COVID‐19. Journal of Medical Virology, 94(1), 54–62. 10.1002/jmv.27292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. , Ejikemeuwa, A. , Gerzanich, V. , Nasr, M. , Tang, Q. , Simard, J. M. , & Zhao, R. Y. (2022). Understanding the role of SARS‐CoV‐2 ORF3a in viral pathogenesis and COVID‐19. Frontiers in Microbiology, 13, 854567. 10.3389/fmicb.2022.854567 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable as this article is a literature review. There is no accompanying data.


