Abstract
Aims
Lung tissue from COVID‐19 patients shares similar histomorphological features with chronic lung allograft disease, also suggesting activation of autoimmune‐related pathways in COVID‐19. To more clearly understand the underlying spectrum of pathophysiology in COVID‐19 pneumonia, we analysed mRNA expression of autoimmune‐related genes in post‐mortem lung tissue from COVID‐19 patients.
Methods and results
Formalin‐fixed, paraffin‐embedded lung tissue samples of 18 COVID‐19 patients and eight influenza patients were used for targeted gene expression profiling using NanoString technology. Multiplex immunofluorescence for tryptase and chymase was applied for validation. Genes related to mast cells were significantly increased in COVID‐19. This finding was strengthened by multiplex immunofluorescence also showing a significant increase of tryptase‐ and chymase‐positive cells in COVID‐19. Furthermore, receptors for advanced glycation end‐products (RAGE) and pro‐platelet basic protein (PPBP) were up‐regulated in COVID‐19 compared to influenza. Genes associated with Type I interferon signalling showed a significant correlation to detected SARS‐CoV2 pathway‐related genes. The comparison of lung tissue samples from both groups based on the presence of histomorphological features indicative of acute respiratory distress syndrome did not result in finding any specific gene or pathways.
Conclusion
Two separate means of measuring show a significant increase of mast cells in SARS‐CoV‐2‐infected lung tissue compared to influenza. Additionally, several genes involved in fibrosis and thrombosis, among which are RAGE and PPBP, are up‐regulated in COVID‐19. As mast cells are able to induce thrombosis and fibrosis, they may play an important role in the pathogenesis of COVID‐19.
Keywords: autoimmunity, COVID‐19, fibrosis, gene expression profiling, genomics, mast cells, thrombosis
This study depicts the role of mast cells in COVID‐19 pneumonia.
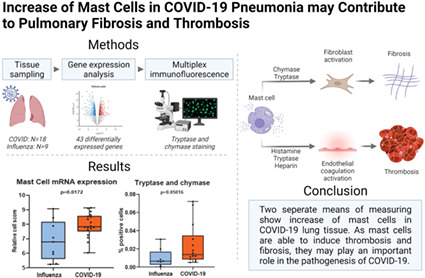
Introduction
The ongoing pandemic of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) has led to a massive number of infections resulting in coronavirus disease‐2019 (COVID‐19). As of May 2022, the World Health Organisation reported approximately 520 million confirmed cases worldwide, with more than 6.2 million deaths. COVID‐19 has a vastly higher mortality compared to other viral lung diseases, such as influenza, which has an in‐hospital mortality of only 5.8% compared to 16.9% for COVID‐19. 1 Symptoms of COVID‐19 vary from mild respiratory symptoms to progressive respiratory deterioration, resulting in acute respiratory distress syndrome (ARDS). 2 , 3 Histology of lung tissue obtained from autopsy on COVID‐19 patients shows features indicative of ARDS, including diffuse alveolar damage (DAD), acute fibrinous and organising pneumonia (AFOP), organising pneumonia (OP) and microvascular thrombotic occlusions. 4 , 5 , 6 , 7 These histomorphological features have also been observed to the same extent in lung tissue obtained from patients with severe influenza, but are not limited to viral lung diseases only. 8 , 9 Chronic rejection after lung transplantation, especially chronic lung allograft dysfunction (CLAD) with a restrictive allograft syndrome phenotype, also shows AFOP, OP and DAD. 10 This similarity poses the question of whether autoimmune‐related inflammatory reaction plays an important role in COVID‐19.
To more clearly understand the pathophysiology behind COVID‐19, measuring differential protein and gene expression in lung tissue of COVID‐19 pneumonia can be an important tool. Recent ‘omics’ studies, including proteomics, transcriptomics and genomics, found up‐regulation of various markers in serum involved in complement system pathways, metabolic suppression, coagulation and inflammation processes. 11 , 12 , 13 , 14 Some studies have identified specific markers in serum to be up‐regulated in COVID‐19 compared to influenza; namely, cytokines and interferons such as CCL2, CXCL10 and IL10. 15 , 16 Interestingly, CCL2 is also elevated in bronchoalveolar lavage (BAL) fluid within the local allograft milieu during primary graft dysfunction. 17
Mudd and colleagues, however, found no significant difference in expression of proinflammatory genes predicting death in serum of COVID‐19 patients compared to influenza. 18 Interestingly, when comparing serum RNA‐seq analyses with lung tissue, minimal concordance in detected biomarkers was found. 19
In light of the above‐mentioned similarities with CLAD, we chose to employ an autoimmune‐specific expression panel in lung tissue to delineate which autoimmune‐related pathways were activated in COVID‐19. To our knowledge, this is the first study in which specifically autoimmune‐related genes were measured in post‐mortem lung tissue. Comparison to influenza as a control group was chosen, as both diseases show similar histomorphological features in autopsy findings but have different mortality rates.
Methods
Tissue procurement
Tissue samples were obtained from patients who died due to COVID‐19 during the first months of the COVID‐19 pandemic, March 2020 to June 2020. Table 1 shows an overview of patient characteristics, medical history and clinical presentation before death or lung transplantation (details are shown in Supporting information, Table S1). Post‐mortem lung tissue from COVID‐19 patients was derived from eight full autopsies performed in the Erasmus Medical Center in Rotterdam, the Netherlands. Additionally, nine surgical biopsies of post‐mortem lung tissue were included, which were performed at the St Antonius Hospital in Nieuwegein, the Netherlands. Additionally, one patient was included who underwent lung transplantation in the Erasmus Medical Center due to progressive respiratory deterioration after COVID‐19 infection. Lung tissue was fixated in formalin for 24 h. In the Erasmus Medical Center, post‐mortem lung tissue was fixated in formalin for 2 weeks, due to risk of infection. Formalin‐fixed paraffin‐embedded (FFPE) post‐mortem lung tissue from influenza patients served as a non‐matched control group. Autopsy reports between 1990 and 2020 were automatically selected if the conclusion stated that the cause of death was (partially) due to influenza. Lung tissue samples from nine consecutive influenza full autopsies were ultimately selected. Consent was given from the next of kin to utilise post‐mortem organ tissue from autopsies for research purposes.
Table 1.
Patient characteristics
| COVID‐19 (n = 18) | Influenza (n = 9) | |
|---|---|---|
| Age, years (SD) | 67.5 (21) | 39 (48) |
| Sex | ||
| Male (%) | 11 (61) | 2 (22) |
| Female (%) | 7 (39) | 7 (78) |
| Comorbidities | ||
| Obesity (%) | 6 (33) | 0 (0) |
| Cardiovascular disease (%) | 7 (39) | 2 (22) |
| Malignancy (%) | 2 (11) | 3 (33) |
| Complications during admittance | ||
| Acute renal insufficiency (%) | 8 (44) | 0 (0) |
| Thrombotic complication (%) | 10 (56) | 4 (44) |
| Co‐infection (%) | 4 (22) | 4 (44) |
FFPE blocks were sectioned by microtome (4 μm) and stained with haematoxylin and eosin. Lung tissue slides per autopsy varied between two and 20 slides, of which one slide representative of the overall histology was selected by an experienced pulmonary pathologist for analysis of pattern‐based histomorphological diagnosis. Slides were classified by DAD phase as either ‘acute’ or ‘organising’ based on histomorphological features in the majority of affected alveoli (Figure 1). Hyaline membranes, oedema, intra‐alveolar erythrocytes and AFOP were features indicative for acute DAD; proliferations of fibroblasts, intra‐alveolar organising fibrosis and squamous metaplasia were indicative for organising DAD. Presence of (micro)thrombi was determined if present in at least one of the lung tissue slides per autopsy. Then, one paraffin block of which the corresponding slide had the highest presence of these histomorphological features was selected for gene expression profiling.
Figure 1.
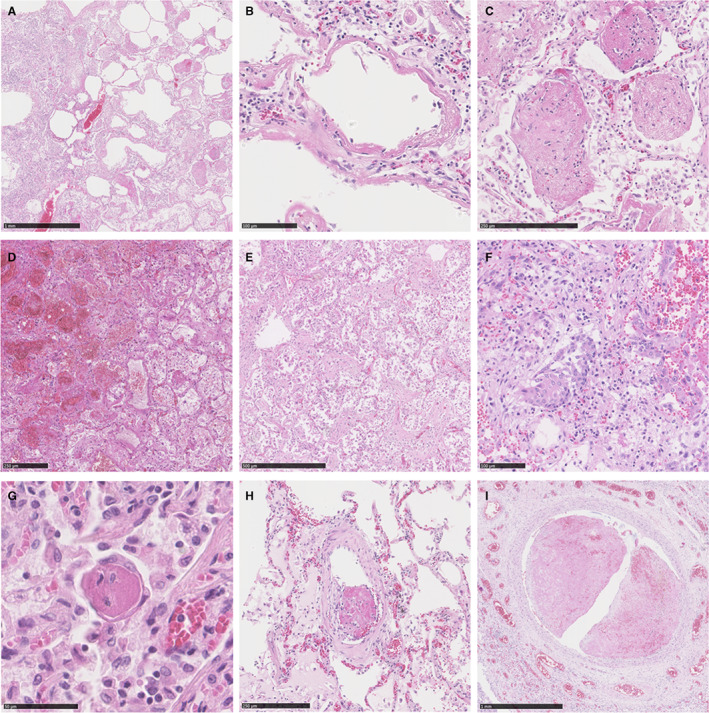
Histology of lung tissue of severe COVID‐19 and influenza. An overview of COVID‐19 lung tissue (A) displays features of acute DAD consisting of intra‐alveolar dispositions of hyaline membranes (B) and scattered ‘balls’ of fibrin indicative for AFOP (C). Lung tissue of influenza samples show features of acute and organising DAD. Extensive intra‐alveolar haemorrhage and macrophages (D) are indicative for acute DAD, while patchy distribution of fibroblastic proliferations (E) and squamous metaplasia (F) are features fitting organising DAD. Displayed features indicative for acute DAD, organising DAD and AFOP were interchangeable between COVID‐19 and influenza. G–I, Thrombi of various sizes from COVID‐19 samples. Microthrombi (G) as well as thrombi in small or large arteries (H, I) were more often seen in COVID‐19 than influenza. AFOP, acute fibrinous and organising pneumonia; DAD, diffuse alveolar damage.
RNA isolation and quality control
Ten sections of 10 μm thickness were mounted on glass slides for RNA isolation. One slide per case was used for RNA isolation, which was a consecutive slide from the same FFPE block used for histomorphological examination. The FFPE samples were deparaffinised in xylene for 2 min followed by a sequential of 90–70% ethanol, then washed in distilled water for 4 min. Tissue samples were scraped from glass slides and transferred to Eppendorf tubes with 240 μl digestion buffer. RNA isolation was performed with the RNeasy FFPE kit (Qiagen, Valencia, CA, USA), following the manufacturer's instructions. RNA was eluted in 14 μl RNase‐free water and stored at −80°C until profiled. The quality and quantity of RNA samples were measured by the Agilent 2100 Bioanalyser (Agilent Technologies, Santa Clara, CA, USA). In case of insufficient quantity or quality, RNA was re‐isolated from additional sections of the sample. A minimum corrected concentration of 60 ng/μl was required for inclusion in autoimmune‐related gene expression profiling. Corrected concentration was calculated using the percentage of fragments between 300 and 4000 nucleotides (RNA) multiplied by the concentration of all fragments.
NanoString gene expression profiling
The autoimmune profiling panel (XT_Hs_AIProfiling_CSO) and coronavirus panel plus, provided by NanoString Technologies (Seattle, WA, USA), were used for gene expression profiling. The autoimmune panel consists of 770 genes, including internal reference genes. The coronavirus panel consists of 10 genes and enables measurement of the SARS‐CoV‐2 virus. For each sample, 300 ng of RNA was hybridised with probes of the panels for 17 h at 65°C using the SimpliAmp Thermal Cycler provided by Applied Biosciences (Waltham, MA, USA). The excess‐unhybridised probes were washed in the PrepStation of the nCounter® FLEX System (GLMX_SCT0002). Gene counts were performed by scanning 490 field‐of‐view using the nCounter digital scanner.
Data analysis
Data were analysed using the nSolver™ software version 4.0 and the advanced analysis module version 2.0 (NanoString). A quality control step was performed to check the technical performance of gene expression profiling. Raw data were normalised using the most stable housekeeping genes (Supporting information, Table S2) using the geNorm algorithm in the advanced analysis module. The differentially expressed genes were identified using the advanced analysis module version 2.0. Log2 normalised counts were used to calculate relative change throughout the samples.
Cell types and pathway scores
The SARS‐CoV‐2 pathway score was defined using the average count of SARS‐CoV‐2‐related genes included in the coronavirus panel plus. The SARS‐CoV‐2 pathway included SARS‐CoV‐2 genes expressed above the detection limit in ≥ 50% of the samples. Cell scores were calculated based on expression of predefined genes. Genes that did not correlate (P > 0.05) with other genes within the same cell type were discarded. Cell type abundances were defined as the average log2 expression of their characteristic genes. Genes used for characterisation of the pathway score and cell types are shown in Supporting information, Table S3.
Multiplex immunofluorescence for mast cells
For multiplex immunofluorescence analysis, one slide was cut from the same FFPE blocks used for gene expression profiling. Due to lack of material, four COVID‐19 FFPE blocks were excluded. Multiplex immunofluorescence was performed with an automated, validated and accredited staining system (Ventana Ultra Discovery; Ventana Medical Systems, Tucson, AZ, USA) using the universal procedure protocol (Supporting information, Table S4). In brief, following deparaffinisation and heat‐induced antigen retrieval with CC1 (#950‐224; Ventana) for 32 min at 100°C, slides were incubated with CMA1 for 32 min at 37°C. Detection was carried out using Ultramap anti‐rabbit horseradish peroxidase (HRP) (#760‐4315; Ventana) followed by 8 min cyanin 5 (Cy5) (#760‐238; Ventana) for visualisation. Antibody heat deactivation was performed using cell conditioning 2 buffer (CC2) (#950‐123; Ventana) for 20 min at 100°C. Secondly, tryptase was incubated for 32 min at 37°C followed by Ultramap anti‐mouse HRP (#760‐4313; Ventana), followed by 4 min FAM (#760‐243; Ventana) for visualisation. Slides were washed in phosphate‐buffered saline and coverslipped with 4′,6‐diamidino‐2‐phenylindole (DAPI) in vectashield. Full scans of the slides were made with a Zeiss Axxio Imager M2 (Zeiss, Oberkochen, Germany). As distribution and quantity of positive‐stained cells by immunofluorescence varied between cases, areas of 1 mm2 with the highest density of stained cells by immunofluorescence were selected to determine the distribution of tryptase‐, chymase‐ or double‐positive cells. Selected hot‐spots corresponded with areas of high inflammation on the haematoxylin and eosin (H&E) slide. Automated cell counting in the said area was then performed by QuPath (version 0.3.1, developed at the University of Edinburgh). Lastly, cells were considered tryptase‐, chymase‐ or double‐positive based on the intensity of the corresponding immunofluorescence staining.
Statistical analysis
P‐values < 0.05 were considered statistically significant. Correction for multiple testing was performed using Benjamin–Hochberg procedures. Pearson's correlation was used to test correlation between the SARS‐CoV‐2 pathway score and differentially expressed genes. Wilcoxon's rank‐sum exact test was used to test for difference in stained cells by multiplex immunofluorescence, as well as differences in continuous data between COVID‐19 and influenza stated in the patient characteristics. Statistical analyses were performed in R software (version 4.1.2). Figures were created in GraphPad Prism (version 8.0.2).
Results
Differences between COVID‐19 and severe influenza
No significant differences in time to death from symptom onset (P = 0.824), time of hospitalisation (P = 0.861) and intubation time (P = 0.791) were found between COVID‐19 and severe influenza patients (Table 1). Gene expression analysis was first performed by comparing samples of COVID‐19 to severe influenza (Figure 2). A total of 43 genes were significantly differentially expressed between COVID‐19 and influenza (P = 0.05), including RAGE (receptor for advanced glycation end‐products), PPBP (pro‐platelet basic protein) and MDM2 (proto‐oncogene). Although none of the individual differentially expressed genes were significantly different after correction for multiple testing, the data trended towards a higher expression of specific genes in COVID‐19, as 16 of the 43 differentially expressed genes correlated with the SARS‐CoV‐2 pathway score (P < 0.05) (Table 2). Expression of AGER was borderline significantly correlated with the SARS‐CoV‐2 pathway score (0.20, P = 0.058).
Figure 2.
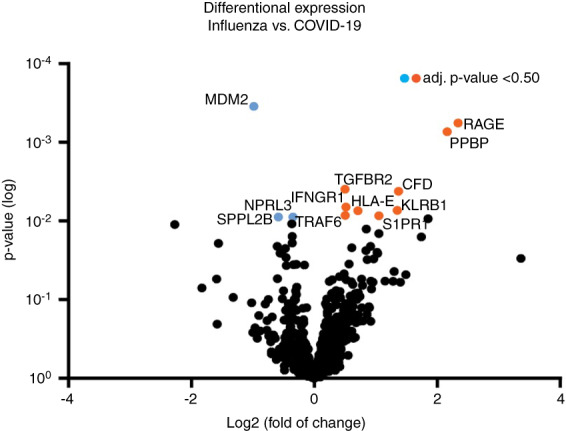
Volcano plot displaying each gene's –log10 (P‐value) and log2 fold change. Twelve of the 43 differentially expressed genes had an adjusted P‐value of <0.50. Genes with a negative fold change are up‐regulated in influenza (blue), whereas genes with positive fold change are up‐regulated in COVID‐19 (orange). [Color figure can be viewed at wileyonlinelibrary.com]
Table 2.
Correlation scores of differentially expressed genes to the SARS‐CoV‐2 pathway score with corresponding P‐values
| Gene | Correlation | P‐value |
|---|---|---|
| IFIH1 | 0.812 | <0.001 |
| RSAD2 | 0.784 | <0.001 |
| IFI44L | 0.766 | <0.001 |
| S1PR1 | 0.756 | <0.001 |
| ISG20 | 0.73 | <0.001 |
| IL18R1 | 0.728 | <0.001 |
| HLA‐E | 0.728 | <0.001 |
| IFIT1 | 0.728 | <0.001 |
| CXCL13 | 0.668 | 0.002 |
| BST2 | 0.665 | 0.002 |
| TNFRSF10B | 0.624 | 0.005 |
| GBP3 | 0.547 | 0.018 |
| TGFBR2 | 0.539 | 0.02 |
| TEK | 0.533 | 0.022 |
| CFLAR | 0.514 | 0.028 |
| MYC | 0.486 | 0.039 |
Genes related to mast cells were significantly up‐regulated in COVID‐19 compared to influenza (P = 0.0172), which were TPSAB1/B2, CPA3 and HDC (Figure 3). Sets of genes related to other cell types or pathways of the immune system showed no significant difference. COVID‐19 samples were analysed for presence of SARS‐CoV‐2‐related genes. Seven of 11 genes were expressed in > 50% of the COVID‐19 cases and were therefore included for pathway analysis with individually expressed genes (SARS‐CoV‐2‐E/G/S, SARS‐COV‐2‐ORF1a/3a/7a/8).
Figure 3.
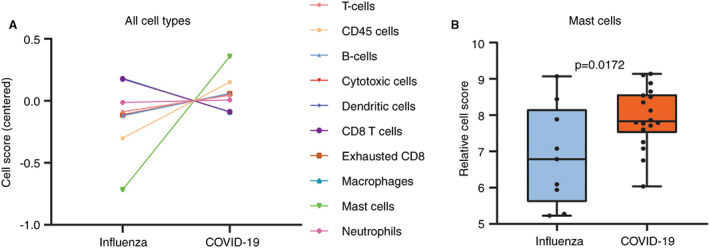
Cell type scores and mRNA expression of mast cell related genes. A, Plots of cell scores in influenza and COVID‐19. Mast cells were the only types of cells which were found to be differentially expressed between influenza and COVID‐19 with statistical significance (P = 0.0172). B, Distribution of normalised data of genes related to mast cells in influenza (blue) and COVID‐19 (orange). Included genes for this comparison were TPSAB1/B2, CPA3 and HDC. [Color figure can be viewed at wileyonlinelibrary.com]
Differences based on histomorphology
To determine whether there is a correlation between gene expression and phase of DAD (acute or organising) independent of the type of pulmonary disease, lung tissue slides from both disease groups were divided into two (Supporting information, Table S5). The acute phase of DAD was seen in 11 of 18 COVID‐19 cases compared to seven of nine influenza cases. AFOP was present in seven of 18 COVID‐19 cases and seven of nine influenza cases. Microthrombi were present in 10 of 18 COVID‐19 cases compared to four of nine influenza cases. Gene expression was performed between groups based on the DAD phase, regardless of disease (18 acute DAD and nine organising DAD cases); 36 differentially expressed genes were found. Similarly, comparison of gene expression between 14 cases with AFOP and 13 cases without AFOP showed 42 differentially expressed genes (Figure 4). The differentially expressed genes in both analyses did not differ significantly after corrective statistical analysis. No significant differences in pathway expression or cell types were found.
Figure 4.
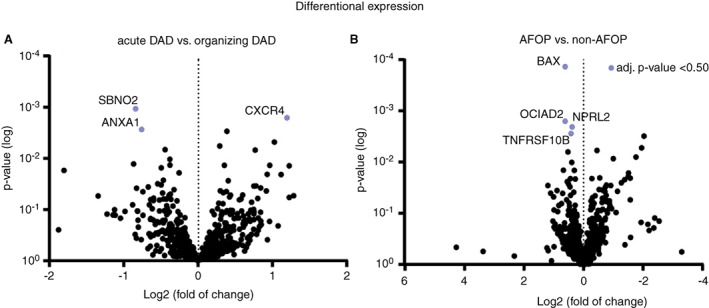
Volcano plots displaying each gene's –log10 (P‐value) and log2 fold change based upon predominant DAD phase (A) and presence of AFOP (B). Genes with a negative fold change (left side) are up‐regulated in groups with either acute DAD or AFOP. Highlighted genes have an adjusted P‐value >0.50. AFOP, acute fibrinous and organising pneumonia; DAD, diffuse alveolar damage. [Color figure can be viewed at wileyonlinelibrary.com]
Multiplex immunofluorescence
Multiplex immunofluorescence staining for tryptase and chymase was performed on both groups of lung tissue to analyse distribution of different mast cell types between COVID‐19 and influenza (Figures 5 and 6). Percentage of either tryptase‐, chymase‐ or double‐positive‐stained cells of the total detected cells in these hot‐spots were compared between both groups. Hot‐spots of COVID‐19 lung tissue showed a significantly higher percentages of double‐positive stained cells compared to influenza lung tissue (P = 0.050). Percentages of either tryptase‐ or chymase‐positive cells were also elevated in COVID‐19 compared to influenza, although without statistical significance (respective P‐values were 0.082 and 0.148).
Figure 5.
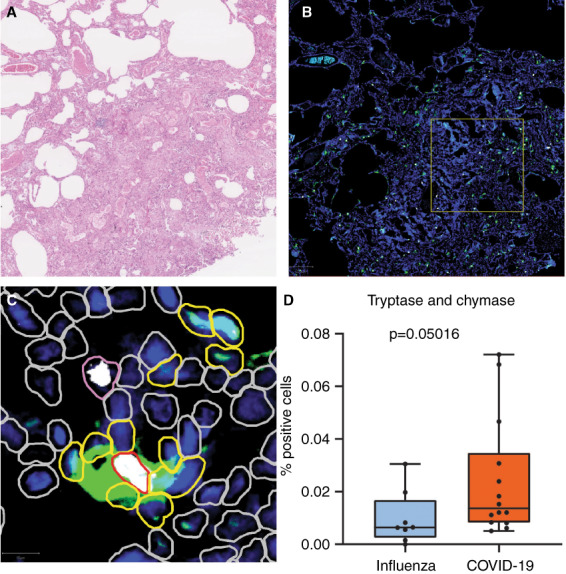
Multiplex immunofluorescence of tryptase and chymase. A, Haematoxylin and eosin staining of lung tissue. This case depicts lung tissue affected by COVID‐19. B, Multiplex immunofluorescence of the same area. Tryptase is stained green and chymase is stained white. Cell nuclei are stained blue by 4′,6‐diamidino‐2‐phenylindole (DAPI). The yellow square depicts the hot‐spot selected for automated cell count. C, Enhanced view of hot‐spot. Cells detected by QuPath are encircled. Yellow border: tryptase‐positive cells. Pink border: chymase‐positive cells. Red border: tryptase‐ and chymase‐positive cells. Grey border: tryptase‐ and chymase‐negative cells. D, Box‐plot of tryptase‐ and chymase‐positive cells; the y‐axis depicts the percentage of double‐positive cells from the total cells.
Figure 6.
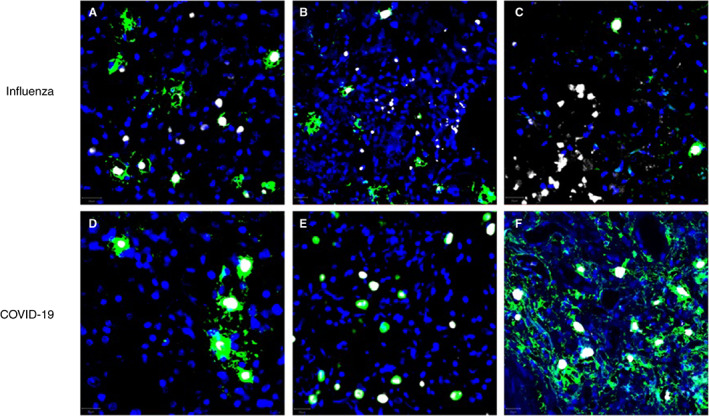
Examples of multiplex immunofluorescence from different influenza (A–C) and COVID‐19 (D–F) tissue samples. Tryptase and chymase are stained green and white, respectively; nuclei are stained blue by 4′,6‐diamidino‐2‐phenylindole (DAPI). Note that the vast majority of tryptase‐positive cells in COVID‐19 samples are also positive for chymase, while a substantial amount of tryptase‐positive cells are negative for tryptase in influenza samples. [Color figure can be viewed at wileyonlinelibrary.com]
Discussion
Using two separate means of measuring (gene expression analysis and multiplex immunofluorescence), we show that mast cells are significantly increased in COVID‐19 lung tissue. Several studies have shown mast cell activity in both viral lung diseases, although without direct comparison. 20 , 21 , 22 To our knowledge, only one study directly compared mast cell infiltration in post‐mortem lung biopsies from influenza and COVID‐19 patients with the use of immunohistochemistry and found an increase of mast cells in COVID‐19, similar to our results. 23 Furthermore, it has been described that COVID‐19 hyper‐inflammation is consistent with mast‐cell‐driven inflammation, and may have therapeutic implications. 21 , 22 Our study focused upon autoimmune‐related genes, as autoimmune‐related hyper‐inflammation and the concurrent cytokine storm are described as important drivers for disease progression. 24 The outcome of increased mast cells activity in COVID‐19 is strengthened by our multiplex immunofluorescence data, as COVID‐19 lung tissue has a higher density of tryptase‐ and chymase‐positive cells compared to influenza. Both tryptase and chymase are secreted by mast cells, and are known to be fibrogenic factors. 25 , 26 Interestingly, tryptase‐ and chymase‐positive mast cells are also increased in CLAD and other diseases with involvement of pulmonary fibrosis, 27 , 28 , 29 suggesting that both diseases have similar underlying biological processes. The increase of mast cells may also affect the relatively high occurrence of pulmonary embolisms in COVID‐19, as mast cells were found to induce thrombosis through activation of clotting factors and platelets. 30 , 31 Taken together, these results suggest that tryptase‐ and chymase‐positive mast cells may play a role in pulmonary fibrosis and embolisms in COVID‐19.
Up‐regulated genes in COVID‐19
Among the individual genes up‐regulated in COVID‐19, the gene with the highest fold change of up‐regulated in COVID‐19 lung tissue was RAGE, also known as AGER. RAGE is a proinflammatory receptor and has been implicated in the pathogenesis of various lung diseases, including chronic obstructive pulmonary disease (COPD), asthma and pulmonary fibrosis. 32 , 33 RAGE may not be highly specific for identifying COVID‐19, as advanced glycation end‐products (AGE) are also associated with diabetes, atherosclerosis, inflammation and obesity. 34 , 35 , 36 Also, the aforementioned comorbidities are risk factors for developing severe COVID‐19. Nonetheless, there is considerable evidence that RAGE is related to pathogenic processes in COVID‐19. First, the soluble form of the receptor (sRAGE) and damage‐associated molecular patterns (DAMP) ligands of RAGE such as HMGB1 and S100A derivatives were elevated in serum of COVID‐19 patients, with higher concentrations corresponding with increased severity. 37 , 38 Secondly, HMGB1 was found to be associated with the formation of NETs (neutrophil extracellular traps), which has been demonstrated in COVID‐19 patients. 39 , 40 , 41 , 42 Lastly, AGE–RAGE interaction induces histamine production in mast cells, which further stimulates fibroblast migration in lungs. 43 , 44
Another interesting up‐regulated gene in COVID‐19 lung tissue is PPBP, also known as pro‐platelet basic protein or chemokine C‐X‐C motif ligand 7 (CXCL7). PPBP is present in platelets after thrombus formation followed by neutrophil attraction, and is up‐regulated in the serum of COVID‐19 patients in a grade‐dependent manner for predicting intubation. 45 , 46 , 47 Several studies suggest that the incidence of thrombotic complications in hospitalised patients with COVID‐19 patients is higher than in patients hospitalised with influenza. Therefore, it is not surprising that a platelet‐derived protein would be up‐regulated in COVID‐19, as opposed to influenza. 48 , 49 These findings, combined with our data, suggest that both RAGE and PPBP are potential biomarkers to predict severity of COVID‐19. As RAGE and PPBP levels have been measured before in bronchoalveolar fluid in previous studies, future research should focus upon measuring these markers in bronchoalveolar fluid from patients in the early stage of COVID‐19 to predict disease severity. 50 , 51 Prognostic biomarkers may eventually influence the treatment modality of choice of treatment before severe disease progression.
SARS‐CoV‐2 pathway correlated genes
None of the predetermined pathway genes showed a significant difference in expression between COVID‐19 and influenza lungs. Pathway expression of Type I interferon signalling genes showed no significant difference in expression (P = 0.327). However, a variety of Type I interferons and corresponding antiviral markers were correlated with the SARS‐CoV‐2‐pathway score. This suggests that the expression of Type I interferons is correlated with viraemia in the lung tissue. IFIH1, an intracellular sensor of viral RNA that triggers an innate immune response, had the highest correlation with the SARS‐CoV‐2 pathway. Additional genes correlated with the SARS‐CoV‐2 pathway were other Type I interferons IFI44L and IFIT1, which appeared in bronchoalveolar fluid from patients with severe COVID‐19. 52 Biomarkers involved in Type I interferon immunoregulation, such as RSAD2, ISG20, IL18R1 and BST2, also showed a significant correlation with the SARS‐CoV‐2 pathway. Multiple studies have demonstrated Type I interferons to be up‐regulated in the serum of severe COVID‐19 patients. 53 , 54 Moreover, mast cells secrete Type I interferons in response to a variety of viral infections, including both influenza and COVID‐19. 55 , 56 , 57
Up‐regulated genes in influenza
MDM2 stands out as being up‐regulated in lung tissue of influenza patients. MDM2 is considered to be a main negative regulator of p53 and is involved in various intracellular signalling pathways, including tumour suppression, cell nucleus stress response and interferon Type I regulation. 58 , 59 Studies have found that influenza‐infected cells are associated with accumulation of MDM2, as influenza causes cells to produce viral non‐structural protein 1, linked to nucleolar stress. 60 , 61 Regarding COVID‐19, it was hypothesised that SARS‐CoV‐2 would also interfere with MDM2/p53 regulation, as other coronaviruses, SARS‐CoV and MERS‐CoV, induce low Type I interferon levels. 62 , 63 , 64 , 65 Despite down‐regulation of MDM2 in COVID‐19 lung tissue, which implies dysregulation of MDM2/p53 balance, our data oppose these hypotheses, as various Type I interferons were up‐regulated in COVID‐19.
Histopathological similarities
To determine whether the similar histomorphological features between COVID‐19 and influenza were caused by interchangeable biological processes, gene expression in both diseases was compared according to the presence of AFOP or DAD phases. However, differentially expressed genes were either non‐specific chemokines or involved in rudimentary metabolic processes. Groups based on the presence of these features show no specific markers to be up‐regulated, suggesting that COVID‐19 and severe influenza have no similar biological processes behind characteristic histomorphological features for ARDS.
Similarities between COVID‐19 and CLAD
As an interesting side note, our data showed that TNFRSF10B was also found to be correlated with the COVID‐19 pathway. TNFRSF10B, as well as TNFRSF10A, transduces an apoptosis signal when activated by TNFRSF10. 66 , 67 Interestingly, TNFRSF10A and TNFRSF10 were also up‐regulated in bronchoalveolar lavage fluid from patients with CLAD. CLAD is an autoimmune inflammatory lung disease with similar histomorphological features to COVID‐19, including AFOP. 68 , 69 In addition, TNFRSF10B was up‐regulated in lung tissue with AFOP when compared to lung tissue without this particular feature. Up‐regulation of TNFRSF10B was also found in serum of patients receiving renal transplantation. 70 This further implies that the histopathological similarities of CLAD and COVID‐19 are caused by a similar biological process.
Limitations
This study was subject to a few limitations. First, data were derived from a relatively small sample cohort due to the limited number of available autopsies on influenza and COVID‐19 patients from the first wave of COVID‐19. This may have also affected the differences in age and gender between both groups. However, the cohort of COVID‐19 lung tissue selected for this study is unique, as it is derived from unvaccinated patients. In most cases, prior vaccination will influence the immune response due to the presence of neutralising antibodies. As the worldwide COVID‐19 vaccination rate increases, the availability of lung tissue from non‐vaccinated patients will be scarce, be it from autopsies, biopsies or resections. Secondly, none of the genes showed a statistical difference between both groups after corrective statistics, although this result was expected, because both groups have relatively similar pathogenesis, histomorphological lung tissue and clinical outcome. Nevertheless, if substantiated it would be considered highly relevant for disease‐specific processes. Thirdly, treatment details were not available for all influenza patients. Disease‐specific differences in gene expression may be influenced by medication such as immunosuppressant or anticoagulation. Although we do not expect a substantial difference in treatment type between cohorts, we could not rule out its effect in our reported results with certainty. Nonetheless, such limitations are inherent to the retrospective study design. Lastly, lung tissue from COVID‐19 patients were collected in two different medical centres with different protocols for fixation. The fixation process may affect the quality of retrieved RNA. However, RNA quantity and quality control and comparison of gene expression between centres verified that the fixation process had no influence on outcomes.
Conclusions
Using two separate means of measurement, we found mast cells to be increased in lung tissue of patients with severe COVID‐19 compared to influenza. As mast cells are able to induce both thrombosis and pulmonary fibrosis, the increase of mast cells may play an important role in the pathogenesis of COVID‐19. Furthermore, we identified several genes linked to fibrosis and thrombosis that are up‐regulated in severe COVID‐19, among which are RAGE and PPBP. We demonstrated that influenza and COVID‐19 show similar histomorphological patterns, although no up‐regulation of specific autoimmune‐related pathways was found when combining patients with these patterns in COVID‐19 and influenza. Our finding may have a clinical implication, as the understanding of underlying pathophysiology is essential for new targeted immunosuppressive therapy development. We suggest that future studies should focus upon whether these biomarkers could be detected in bronchoalveolar lavage fluid of COVID‐19 to predict the severity of COVID‐19 and potentially adjust treatment decisions.
Conflicts of interest
The authors declare that they have no conflicts of interest and no source of support to declare.
Supporting information
Table S1. Patient characteristics. 18 COVID‐19 patients and 9 influenza patients were included. Age is displayed in years. M = male, F = Female, ICU = intensive care unit. *Not applicable. ***Medical reports were unavailable.
Table S2. SARS‐CoV‐2 pathway characterization.
Table S3. Marker genes used for cell type characterization.
Table S4. Multiplex immunofluorescence for mast cells antibody information.
Table S5. Phase of diffuse alveolar damage (DAD) and presence of acute fibrinous and organizing pneumonia (AFOP).
Data availability statement
The data sets used and/or analysed during the current study are available from the corresponding author upon reasonable request.
References
- 1. Piroth L, Cottenet J, Mariet AS et al. Comparison of the characteristics, morbidity, and mortality of COVID‐19 and seasonal influenza: a nationwide, population‐based retrospective cohort study. Lancet Respir. Med. 2021; 9; 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dhama K, Khan S, Tiwari R et al. Coronavirus disease 2019‐COVID‐19. Clin. Microbiol. Rev. 2020; 33; e00028‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mohanty SK, Satapathy A, Naidu MM et al. Severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) and coronavirus disease 19 (COVID‐19) ‐ anatomic pathology perspective on current knowledge. Diagn. Pathol. 2020; 15; 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Xu Z, Shi L, Wang Y et al. Pathological findings of COVID‐19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020; 8; 420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chong WH, Saha BK, Chopra A. Does COVID‐19 pneumonia signify secondary organizing pneumonia? A narrative review comparing the similarities between these two distinct entities. Heart Lung 2021; 50; 667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Menter T, Haslbauer JD, Nienhold R et al. Post‐mortem examination of COVID‐19 patients reveals diffuse alveolar damage with severe capillary congestion and variegated findings in lungs and other organs suggesting vascular dysfunction. Histopathology 2020; 77; 198–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lax SF, Skok K, Zechner P et al. Pulmonary arterial thrombosis in COVID‐19 with fatal outcome: results from a prospective, single‐center, clinicopathologic case series. Ann. Intern. Med. 2020; 173; 350–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McMullen P, Pytel P, Snyder A et al. A series of COVID‐19 autopsies with clinical and pathologic comparisons to both seasonal and pandemic influenza. J. Pathol. Clin. Res. 2021; 7; 459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hariri LP, North CM, Shih AR et al. Lung histopathology in coronavirus disease 2019 as compared with severe acute respiratory sydrome and H1N1 influenza: a systematic review. Chest 2021; 159; 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vanstapel A, Verleden SE, Weynand B et al. Late‐onset ‘acute fibrinous and organising pneumonia’ impairs long‐term lung allograft function and survival. Eur. Respir. J. 2020; 56; 1902292. [DOI] [PubMed] [Google Scholar]
- 11. Shen B, Yi X, Sun Y et al. Proteomic and metabolomic characterization of COVID‐19 patient sera. Cell 2020; 182; 59–72.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. D'Alessandro A, Thomas T, Dzieciatkowska M et al. Serum proteomics in COVID‐19 patients: altered coagulation and complement status as a function of IL‐6 level. J. Proteome Res. 2020; 19; 4417–4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Messner CB, Demichev V, Wendisch D et al. Ultra‐high‐throughput clinical proteomics reveals classifiers of COVID‐19 infection. Cell Syst. 2020; 11; 11–24.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zeng HL, Chen D, Yan J et al. Proteomic characteristics of bronchoalveolar lavage fluid in critical COVID‐19 patients. FEBS J. 2021; 288; 5190–5200. [DOI] [PubMed] [Google Scholar]
- 15. Hou X, Zhang X, Wu X et al. Serum protein profiling reveals a landscape of inflammation and immune signaling in early‐stage COVID‐19 infection. Mol. Cell. Proteomics 2020; 19; 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McClain MT, Constantine FJ, Henao R et al. Dysregulated transcriptional responses to SARS‐CoV‐2 in the periphery support novel diagnostic approaches. Nat. Commun. 2021; 12; 1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Der Hovanessian V, Palchevskiy V, Weigt SS et al. The CCL2/CCR2 Axis in primary graft dysfunction and bronchiolitis obliterans syndrome following lung transplantation. J. Heart Lung Transplant. 2013; 32; S28–S29. [Google Scholar]
- 18. Mudd PA, Crawford JC, Turner JS et al. Distinct inflammatory profiles distinguish COVID‐19 from influenza with limited contributions from cytokine storm. Sci. Adv. 2020; 6; eabe3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moni MA, Quinn JMW, Sinmaz N, Summers MA. Gene expression profiling of SARS‐CoV‐2 infections reveal distinct primary lung cell and systemic immune infection responses that identify pathways relevant in COVID‐19 disease. Brief. Bioinform. 2021; 22; 1324–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Graham AC, Temple RM, Obar JJ. Mast cells and influenza A virus: association with allergic responses and beyond. Front. Immunol. 2015; 6; 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Afrin LB, Weinstock LB, Molderings GJ. Covid‐19 hyperinflammation and post‐Covid‐19 illness may be rooted in mast cell activation syndrome. Int. J. Infect. Dis. 2020; 100; 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gebremeskel S, Schanin J, Coyle KM et al. Mast cell and eosinophil activation are associated with COVID‐19 and TLR‐mediated viral inflammation: implications for an anti‐Siglec‐8 antibody. Front. Immunol. 2021; 12; 650331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Motta Junior JDS, Miggiolaro A, Nagashima S et al. Mast cells in alveolar septa of COVID‐19 patients: a pathogenic pathway that may link interstitial edema to Immunothrombosis. Front. Immunol. 2020; 11; 574862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang C, Wu Z, Li JW, Zhao H, Wang GQ. Cytokine release syndrome in severe COVID‐19: interleukin‐6 receptor antagonist tocilizumab may be the key to reduce mortality. Int. J. Antimicrob. Agents 2020; 55; 105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bagher M, Larsson‐Callerfelt AK, Rosmark O, Hallgren O, Bjermer L, Westergren‐Thorsson G. Mast cells and mast cell tryptase enhance migration of human lung fibroblasts through protease‐activated receptor 2. Cell Commun. Signal 2018; 16; 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pejler G. The emerging role of mast cell proteases in asthma. Eur. Respir. J. 2019; 54; 1900685. [DOI] [PubMed] [Google Scholar]
- 27. Banga A, Han Y, Wang X, Hsieh FH. Mast cell phenotypes in the allograft after lung transplantation. Clin. Transplant. 2016; 30; 845–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pesci A, Bertorelli G, Gabrielli M, Olivieri D. Mast cells in fibrotic lung disorders. Chest 1993; 103; 989–996. [DOI] [PubMed] [Google Scholar]
- 29. Miranda E, Dunmore R, Rassl D et al. Tryptase+ mast cells associate with fibrotic regions in the lungs of idiopathic pulmonary fibrosis patients; a multiplex staining approach. Eur. Respir. J. 2016; 48(Suppl 60); PA3412. [Google Scholar]
- 30. Karhausen J, Choi HW, Maddipati KR et al. Platelets trigger perivascular mast cell degranulation to cause inflammatory responses and tissue injury. Sci. Adv. 2020; 6; eaay6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seidel H, Hertfelder HJ, Oldenburg J, Kruppenbacher JP, Afrin LB, Molderings GJ. Effects of primary mast cell disease on hemostasis and erythropoiesis. Int. J. Mol. Sci. 2021; 22; 8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sanders KA, Delker DA, Huecksteadt T et al. RAGE is a critical mediator of pulmonary oxidative stress, alveolar macrophage activation and emphysema in response to cigarette smoke. Sci. Rep. 2019; 9; 231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Oczypok EA, Perkins TN, Oury TD. All the ‘RAGE’ in lung disease: the receptor for advanced glycation endproducts (RAGE) is a major mediator of pulmonary inflammatory responses. Paediatr. Respir. Rev. 2017; 23; 40–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Henning C, Glomb MA. Pathways of the Maillard reaction under physiological conditions. Glycoconj. J. 2016; 33; 499–512. [DOI] [PubMed] [Google Scholar]
- 35. Gaens KH, Goossens GH, Niessen PM et al. Nε‐(carboxymethyl)lysine‐receptor for advanced glycation end product axis is a key modulator of obesity‐induced dysregulation of adipokine expression and insulin resistance. Arterioscler. Thromb. Vasc. Biol. 2014; 34; 1199–1208. [DOI] [PubMed] [Google Scholar]
- 36. Anderson MM, Requena JR, Crowley JR, Thorpe SR, Heinecke JW. The myeloperoxidase system of human phagocytes generates Nepsilon‐(carboxymethyl)lysine on proteins: a mechanism for producing advanced glycation end products at sites of inflammation. J. Clin. Invest. 1999; 104; 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lim A, Radujkovic A, Weigand MA, Merle U. Soluble receptor for advanced glycation end products (sRAGE) as a biomarker of COVID‐19 disease severity and indicator of the need for mechanical ventilation, ARDS and mortality. Ann. Intensive Care 2021; 11; 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Leisman DE, Mehta A, Thompson BT et al. Alveolar, endothelial, and organ injury marker dynamics in severe COVID‐19. Am. J. Respir. Crit. Care Med. 2022; 205; 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Thierry AR, Roch B. Neutrophil extracellular traps and by‐products play a key role in COVID‐19: pathogenesis, risk factors, and therapy. J. Clin. Med. 2020; 9; 2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ng H, Havervall S, Rosell A et al. Circulating markers of neutrophil extracellular traps are of prognostic value in patients with COVID‐19. Arterioscler. Thromb. Vasc. Biol. 2021; 41; 988–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zou L, Ruan F, Huang M et al. SARS‐CoV‐2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020; 382; 1177–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roy D, Ramasamy R, Schmidt AM. Journey to a receptor for advanced glycation end products connection in severe acute respiratory syndrome coronavirus 2 infection: with stops along the way in the lung, heart, blood vessels, and adipose tissue. Arterioscler. Thromb. Vasc. Biol. 2021; 41; 614–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kohyama T, Yamauchi Y, Takizawa H, Kamitani S, Kawasaki S, Nagase T. Histamine stimulates human lung fibroblast migration. Mol. Cell. Biochem. 2010; 337; 77–81. [DOI] [PubMed] [Google Scholar]
- 44. Sick E, Brehin S, André P et al. Advanced glycation end products (AGEs) activate mast cells. Br. J. Pharmacol. 2010; 161; 442–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rayes J, Bourne JH, Brill A, Watson SP. The dual role of platelet‐innate immune cell interactions in thrombo‐inflammation. Res. Pract. Thromb. Haemost. 2020; 4; 23–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bdeir K, Gollomp K, Stasiak M et al. Platelet‐specific chemokines contribute to the pathogenesis of acute lung injury. Am. J. Respir. Cell Mol. Biol. 2017; 56; 261–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yatim N, Boussier J, Chocron R et al. Platelet activation in critically ill COVID‐19 patients. Ann. Intensive Care 2021; 11; 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stals M, Grootenboers M, van Guldener C et al. Risk of thrombotic complications in influenza versus COVID‐19 hospitalized patients. Res. Pract. Thromb. Haemost. 2021; 5; 412–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lachant D, Roto D, Rappaport S, Prasad P, Lachant N, White RJ. Venous thromboembolism associates with SARS‐CoV‐2 more than seasonal influenza. Thromb. Res. 2021; 205; 40–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Uchida T, Shirasawa M, Ware LB et al. Receptor for advanced glycation end‐products is a marker of type I cell injury in acute lung injury. Am. J. Respir. Crit. Care Med. 2006; 173; 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hasan MZ, Islam S, Matsumoto K, Kawai T. Meta‐analysis of single‐cell RNA‐seq data reveals phenotypic switching of immune cells in severe COVID‐19 patients. Comput. Biol. Med. 2021; 137; 104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shaath H, Vishnubalaji R, Elkord E, Alajez NM. Single‐cell transcriptome analysis highlights a role for neutrophils and inflammatory macrophages in the pathogenesis of severe COVID‐19. Cell 2020; 9; 2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Reyes L, Morrison T, AJM H, Watts ER, Arienti S et al. A type I IFN, prothrombotic hyperinflammatory neutrophil signature is distinct for COVID‐19 ARDS. Wellcome Open Res. 2021; 6; 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Trouillet‐Assant S, Viel S, Gaymard A et al. Type I IFN immunoprofiling in COVID‐19 patients. J. Allergy Clin. Immunol. 2020; 146; 206–8 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Oldford SA, Salsman SP, Portales‐Cervantes L et al. Interferon α2 and interferon γ induce the degranulation independent production of VEGF‐A and IL‐1 receptor antagonist and other mediators from human mast cells. Immun. Inflamm. Dis. 2018; 6; 176–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kempuraj D, Selvakumar GP, Ahmed ME et al. COVID‐19, mast cells, cytokine storm, psychological stress, and neuroinflammation. Neuroscientist 2020; 26; 402–414. [DOI] [PubMed] [Google Scholar]
- 57. Murdaca G, Di Gioacchino M, Greco M et al. Basophils and mast cells in COVID‐19 pathogenesis. Cell 2021; 10; 2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature 1997; 387; 296–299. [DOI] [PubMed] [Google Scholar]
- 59. Muñoz‐Fontela C, Macip S, Martínez‐Sobrido L et al. Transcriptional role of p53 in interferon‐mediated antiviral immunity. J. Exp. Med. 2008; 205; 1929–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pizzorno A, Dubois J, Machado D et al. Influenza a viruses alter the stability and antiviral contribution of host E3‐ubiquitin ligase Mdm2 during the time‐course of infection. Sci. Rep. 2018; 8; 3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yan Y, Du Y, Wang G, Li K. Non‐structural protein 1 of H3N2 influenza a virus induces nucleolar stress via interaction with nucleolin. Sci. Rep. 2017; 7; 17761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zauli G, Tisato V, Secchiero P. Rationale for considering oral idasanutlin as a therapeutic option for COVID‐19 patients. Front. Pharmacol. 2020; 11; 1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chen J, Subbarao K. The immunobiology of SARS. Annu. Rev. Immunol. 2007; 25; 443–472. [DOI] [PubMed] [Google Scholar]
- 64. Yuan L, Chen Z, Song S et al. p53 degradation by a coronavirus papain‐like protease suppresses type I interferon signaling. J. Biol. Chem. 2015; 290; 3172–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Li G, Fan Y, Lai Y et al. Coronavirus infections and immune responses. J. Med. Virol. 2020; 92; 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. NCBI . TNFRSF10B TNF receptor superfamily member 10b [Homo sapiens (human)] . 2021. Available at: https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=8795.
- 67. NCBI . TNFRSF10A TNF receptor superfamily member 10a [Homo sapiens (human)] . 2021. Available at: https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=8797
- 68. Verleden SE, Von der Thüsen J, Roux A et al. When tissue is the issue: a histological review of chronic lung allograft dysfunction. Am. J. Transplant. 2020; 20; 2644–2651. [DOI] [PubMed] [Google Scholar]
- 69. Iasella CJ, Hoji A, Popescu I et al. Type‐1 immunity and endogenous immune regulators predominate in the airway transcriptome during chronic lung allograft dysfunction. Am. J. Transplant. 2021; 21; 2145–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Song CJ, Liu XS, Zhu Y et al. Expression of TRAIL, DR4, and DR5 in kidney and serum from patients receiving renal transplantation. Transplant. Proc. 2004; 36; 1340–1343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Patient characteristics. 18 COVID‐19 patients and 9 influenza patients were included. Age is displayed in years. M = male, F = Female, ICU = intensive care unit. *Not applicable. ***Medical reports were unavailable.
Table S2. SARS‐CoV‐2 pathway characterization.
Table S3. Marker genes used for cell type characterization.
Table S4. Multiplex immunofluorescence for mast cells antibody information.
Table S5. Phase of diffuse alveolar damage (DAD) and presence of acute fibrinous and organizing pneumonia (AFOP).
Data Availability Statement
The data sets used and/or analysed during the current study are available from the corresponding author upon reasonable request.


