ABSTRACT
By chance, we discovered a window of extracellular magnesium (Mg2+) availability that modulates the division frequency of Bacillus subtilis without affecting its growth rate. In this window, cells grown with excess Mg2+ produce shorter cells than do those grown in unsupplemented medium. The Mg2+-responsive adjustment in cell length occurs in both rich and minimal media as well as in domesticated and undomesticated strains. Of other divalent cations tested, manganese (Mn2+) and zinc (Zn2+) also resulted in cell shortening, but this occurred only at concentrations that affected growth. Cell length decreased proportionally with increasing Mg2+ from 0.2 mM to 4.0 mM, with little or no detectable change being observed in labile, intracellular Mg2+, based on a riboswitch reporter. Cells grown in excess Mg2+ had fewer nucleoids and possessed more FtsZ-rings per unit cell length, consistent with the increased division frequency. Remarkably, when shifting cells from unsupplemented to supplemented medium, more than half of the cell length decrease occurred in the first 10 min, consistent with rapid division onset. Relative to unsupplemented cells, cells growing at steady-state with excess Mg2+ showed an enhanced expression of a large number of SigB-regulated genes and the activation of the Fur, MntR, and Zur regulons. Thus, by manipulating the availability of one nutrient, we were able to uncouple the growth rate from the division frequency and identify transcriptional changes that suggest that cell division is accompanied by the general stress response and an enhanced demand to sequester and/or increase the uptake of iron, Mn2+, and Zn2+.
IMPORTANCE The signals that cells use to trigger cell division are unknown. Although division is often considered intrinsic to the cell cycle, microorganisms can continue to grow and repeat rounds of DNA replication without dividing, indicating that cycles of division can be skipped. Here, we show that by manipulating a single nutrient, namely, Mg2+, cell division can be uncoupled from the growth rate. This finding can be applied to investigate the nature of the cell division signal(s).
KEYWORDS: Bacillus, LB, magnesium, manganese, SigB, UppS, zinc, cell division, morphology, undecaprenyl
INTRODUCTION
The abundant cation Mg2+ is perhaps most appreciated for its role as an enzymatic cofactor, as it supports catalysis in hundreds of biochemical reactions (1–3). However, Mg2+ has diverse biological functions and is also critical for the assembly of ribosomes (4, 5), chelation, the stabilization of ATP and other polyphosphates (6), the regulation of phosphate uptake (7, 8), osmotic adaptation (9), the setting of the circadian period in plants (10), and the support of envelope integrity in bacteria (11–13). Due to the central place of Mg2+ in physiology, cells must be able to respond rapidly when availability fluctuates, as these fluctuations sometimes occur over several orders of magnitude. In human serum, where Mg2+ concentration is tightly controlled, 0.7 to 1.0 mM is considered homeostatic (14). In contrast, Mg2+ in the digestive tract of animals or in soil and aquatic environments is much more variable, ranging from micromolar to millimolar levels.
Whereas free-living bacteria can adapt to large fluctuations in extracellular Mg2+, they keep intracellular levels relatively constant. Under replete conditions, cell-associated Mg2+ in E. coli is estimated to be 20 to 100 mM (15, 16), of which only 1 to 10 mM is considered labile (16–19). Of the remaining pool, approximately half is associated with nucleic acid, proteins, and ribosomes. The other half is found complexed with the enzymatically relevant form of ATP, the relatively stable chelate Mg2+-ATP (20). Not surprisingly, ATP synthesis is tightly coordinated with Mg2+ availability (6). In fact, cells will scavenge Mg2+ from ribosomes at the expense of protein synthesis before allowing intracellular Mg2+ to fall to levels that are insufficient to support ATP chelation (6, 9, 21).
Intracellular Mg2+ is acquired using importer proteins that, in Gram-negatives, are often regulated by PhoPQ two-component systems (22, 23). These systems sense and respond to changes in both external Mg2+ availability and cellular demand primarily by regulating the transcription of Mg2+ transporters. B. subtilis lacks a homologous two-component system and controls the expression of its major Mg2+ importer (MgtE) via a riboswitch. The M-box riboswitch attenuates the transcription of mgtE when intracellular Mg2+ is sufficient by forming a terminator (4). Two other importers, YfjQ and CitM, also contribute to Mg2+ uptake (24). YfjQ is a minor importer, whereas CitM is a symporter that allows for the cotransport of Mg2+ and citrate (25, 26). Under hyperosmotic conditions and concomitant with potassium influx, B. subtilis can also efflux Mg2+ through an exporter called MpfA (27).
Aside from its role in supporting basic physiological functions, Mg2+ is known to suppress phenotypes associated with the inactivation of cell envelope-related genes. The provision of 10 to 25 mM Mg2+ (more than an order of magnitude higher than the concentrations found in typical media) restores both viability and rod shape to strains with deletions in the morphogenes mreB, mbl, mreBH, mreC, and mreD (28–30). Mg2+ restores a regular rod shape to strains with deletions in ponA (encoding PBP1A) and lytE (a major d, l-endopeptidase) (11, 31) as well as to strains with mutations in teichoic acid synthesis genes (12, 32, 33). A ΔglmR mutant, which is unable to upregulate gluconeogenesis, is inviable when grown on a gluconeogenic carbon source (34). Remarkably, millimolar Mg2+ can suppress this lethality (34). Mg2+ even increases resistance to the cell wall-targeting antibiotic methicillin (35). Thus, Mg2+ elicits cellular changes that allow it to function as a general suppressor of a wide variety of envelope-related defects.
The mechanism by which Mg2+ promotes envelope integrity is unclear. B. subtilis grown with higher levels of Mg2+ display lower levels of amidated meso-diaminopimelic acid in their peptidoglycan (PG) (36); however, reduced amidation itself is unlikely to account for Mg2+ rescue, as a mutant lacking the modification (ΔasnB) also has shape defects that are suppressed by Mg2+ (36). Mg2+ also reduces the dysregulated d, l-endopeptidase activity that is associated with deletion of mreB (37); it is unknown whether the impact of Mg2+ on endopeptidase activity is direct or is a consequence of other effects that Mg2+ has on the cell.
Here, we investigate a phenotype not previously associated with Mg2+. We identify a window in which increasing Mg2+ availability increases the frequency of cell division without affecting the growth rate. Our results suggest that as Mg2+ availability decreases, cells prioritize the maintenance of cell elongation and growth over cell division. This prioritization of cell resources, which transcriptional profiling suggests is accompanied by changes in metal homeostasis and in the general stress response, results in longer cells with more nucleoids and fewer Z-rings.
RESULTS
Mg2+ supplementation leads to cell shortening in rich media in both domestic and undomesticated B. subtilis strains.
In 2017, we obtained a new bottle of premade LB-Lennox powder from Sigma. While examining micrographs of membrane-stained B. subtilis 168 cells that were grown in liquid LB made from the new powder (LB*), we noted that the cells appeared qualitatively longer, compared to cells grown in previous lots of LB. LB is a rich medium consisting of only NaCl, tryptone, and yeast extract. We reasoned that the longer cells most likely resulted from a difference in trace element content. Mg2+ was a priority candidate both because tryptone-based media, such as LB, tend to be low in Mg2+ (37, 38) and because prior studies had shown that low Mg2+ media results in B. subtilis filamentation (39–41). We could not directly compare our results to those of published studies, as their microscopy methods did not include the visualization of septa present along filaments. To test whether the longer cells could be the result of low Mg2+ content in the new LB, we supplemented the medium with 10.0 mM MgCl2 and imaged cells following membrane staining. As shown in Fig. 1A and B, cells grown with supplemental Mg2+ were 36% shorter on average, consistent with the possibility that the cells were longer in the new medium due to the reduced Mg2+ content.
FIG 1.
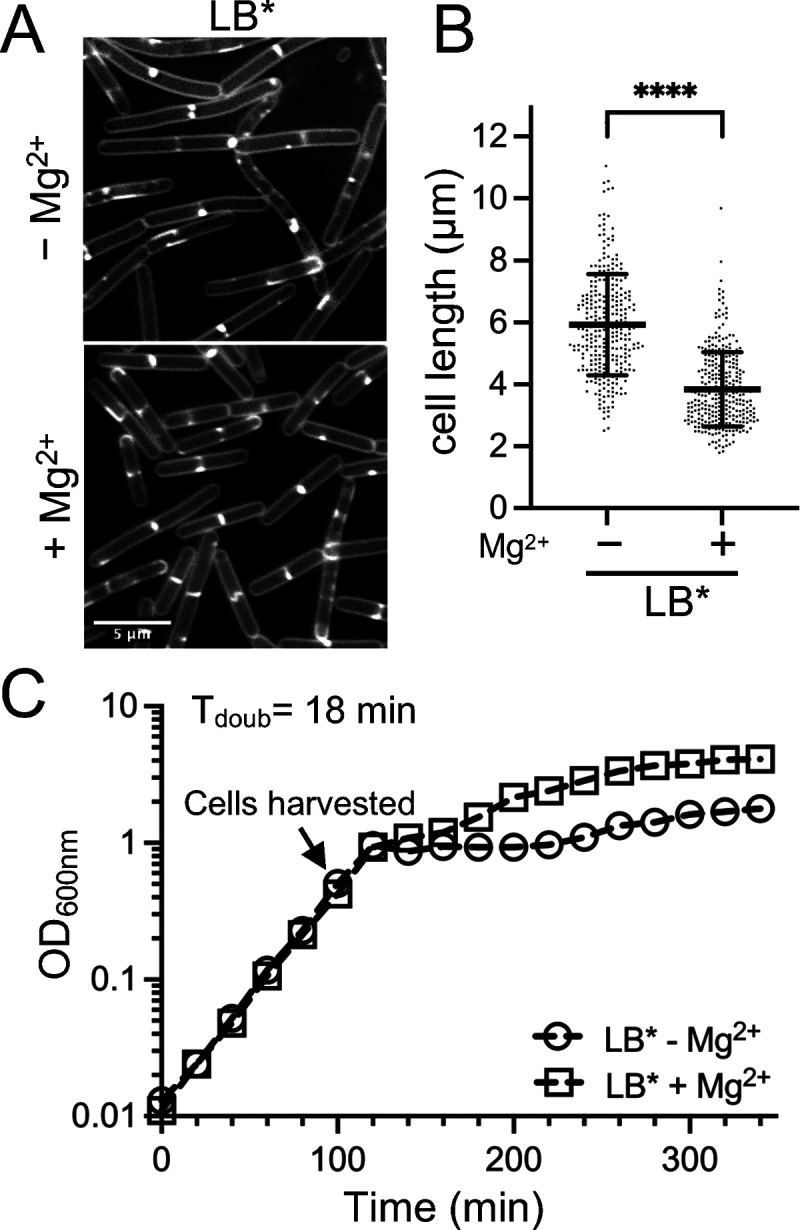
Cell length following growth in LB. WT B. subtilis 168 (BJH004) was grown at 37°C to the mid-exponential phase in LB* medium without or with 10.0 mM MgCl2 supplementation. LB* indicates that the phenotype was lot-specific and is not generalizable to all LB. (A) Micrographs following the membrane staining and epifluorescence microscopy (scaled identically). (B) Scatterplots showing the distribution of cell lengths quantitated for 300 cells from each condition. The bars represent the means of 300 cells ± the SDs. (C) Representative growth curves. ****, P ≤ 0.0001.
Since the growth rate can affect the cell size (42–48), we next assessed whether the Mg2+ addition affected the doubling time of the cells in the new LB. We found that the growth rates of cells cultured without and with 10.0 mM MgCl2 were identical during the exponential stage (Fig. 1C), the same phase of growth used for cell imaging. From these results, we concluded that there is a growth rate-independent effect of Mg2+ availability on cell length, at least in the window tested.
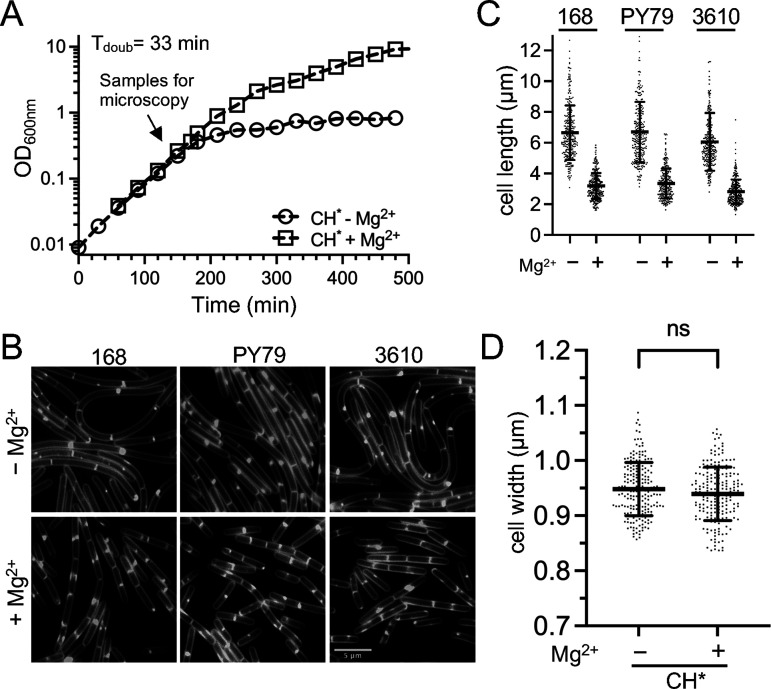
The phenotype that we initially observed was specific to only one batch of LB. So, we next investigated whether the Mg2+ responsive phenotype could be observed in another rich medium. CH (casein hydrolysate) is an amino acid-based medium (49) that is commonly used in B. subtilis studies. We found that B. subtilis grew robustly in CH without the standard addition of Mg2+ and Mn2+ salts (Fig. 2A) and designated this medium CH*. Similar to the results with LB*, the cells grown in CH* exhibited a Mg2+-responsive phenotype. B. subtilis 168 cells cultured in CH* with 10.0 mM Mg2+ were approximately 2-fold reduced in average cell length, compared to those cultured in CH* only (Fig. 2B and C). No difference in doubling time (Fig. 2A) or cell width (Fig. 2D) was detected between CH* and CH* supplemented with Mg2+.
FIG 2.
Cell length of three B. subtilis strains following growth in CH* medium. WT B. subtilis 168 (BJH004), PY79 (BJH001), and 3610 (BJH403) were grown at 37°C to the mid-exponential phase in CH* supplemented with 10.0 mM MgCl2 as indicated. (A) Representative growth curves for the WT. (B) Representative micrographs following the membrane staining and epifluorescence microscopy (scaled identically). (C) Scatterplots showing the distribution of cell with quantitated for 300 cells from each condition. The bars represent the means of 300 cells ± the SDs. (D) Scatterplots showing the distribution of cell widths for 200 cells grown without or with 10.0 mM MgCl2. The bars represent the means of 200 cells ± the SDs. ns, P > 0.05.
Next, we investigated whether the Mg2+-responsive phenotype observed in the B. subtilis 168 cells was present in two other commonly utilized B. subtilis strains: the SPβ-cured laboratory strain PY79 (50) and the undomesticated strain NCIB 3610 (50). Independent of strain, the addition of 10.0 mM MgCl2 to CH* consistently resulted in cells that were approximately 2-fold reduced in length, compared to CH* alone (Fig. 2B and C). These results suggest that the effect of Mg2+ is likely to be a phenomenon that is generalizable to the species.
Mn2+ and Zn2+ supplementation also elicit cell shortening.
We wondered whether the growth rate-independent cell shortening was specific to Mg2+ or could also be induced by providing other metals in excess. For these experiments, we again utilized CH*. Cells were grown as before, but instead of supplementing cultures with MgCl2, salts of Ca2+, Cu2+, Fe2+, Fe3+, Mn2+, and Zn2+ were added. Of the metals tested, only Mn2+ and Zn2+ elicited cell shortening similar to that observed for Mg2+ (Fig. 3; Fig. S1); however, Zn2+ strongly reduced the growth rate at the concentrations required to observe the cell shortening effect (0.2 mM) (Fig. S1). While Mn2+ did not reduce the doubling time of cells once the culture reached the exponential phase, the precultures containing Mn2+ remained in lag for several hours longer than did the unsupplemented cultures. Due to the pleiotropic effects of Zn2+ and Mn2+ on the growth, we chose to focus on Mg2+ for the remainder of the study.
FIG 3.
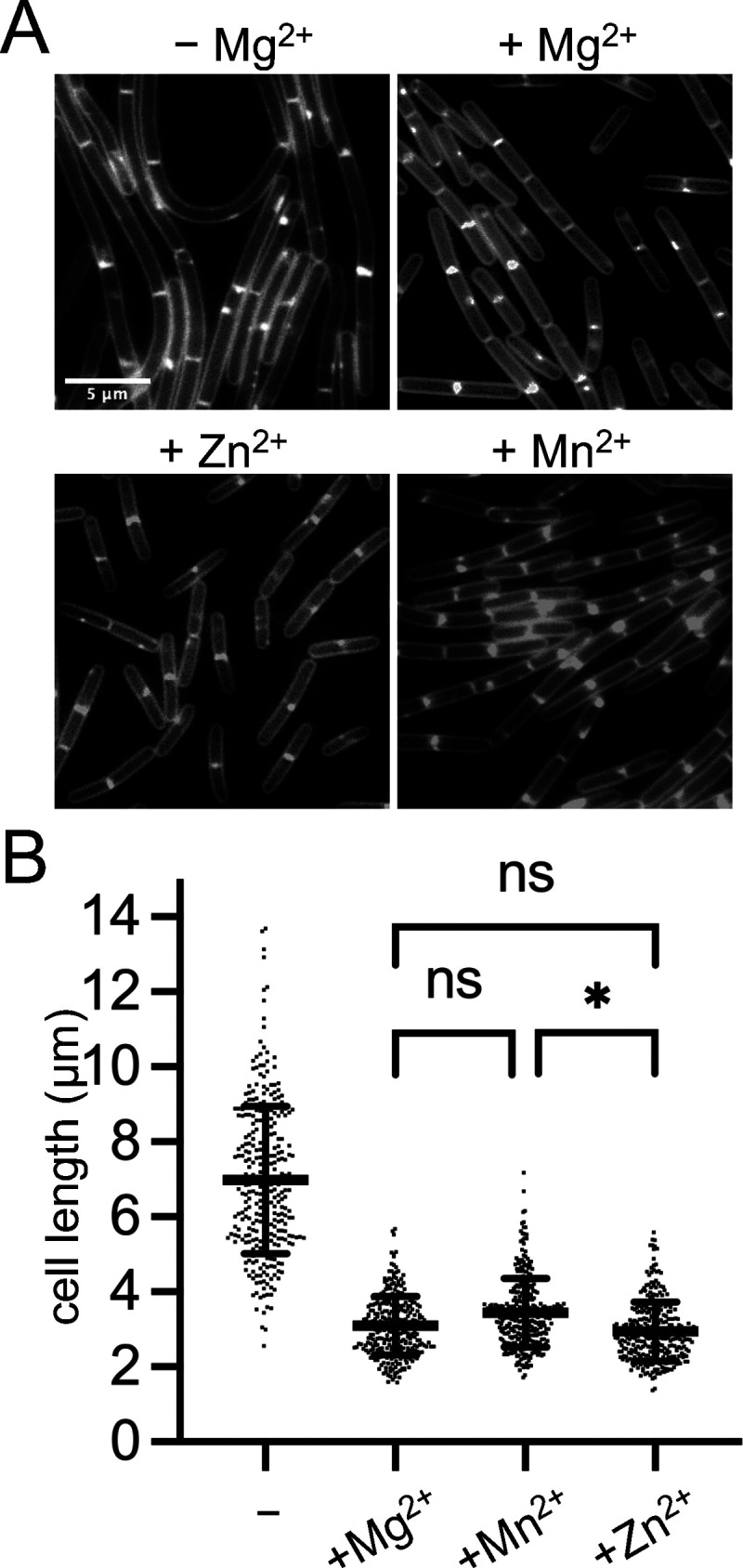
Effect of divalent metals on cell length. WT 168 (BJH004) cells were cultured in CH* without or with the following concentrations of divalent cation salts: 0.1 mM MnSO4, 0.2 mM ZnCl2, 10.0 mM MgCl2. (A) Micrographs following the membrane staining and epifluorescence microscopy (scaled identically). (B) Scatterplots showing the distribution of cell lengths quantitated for 300 cells from each condition. The bars represent the means of 300 cells ± the SDs. *, 0.01 < P ≤ 0.05; ns, P ≥ 0.05. For all pairwise comparisons to the control without additional metal (−), a P value of <0.0001 was calculated.
The Mg2+-responsive phenotype occurs in minimal medium and does not require the addition of exogenous amino acids.
Minimal media (MM) allows for the manipulation of individual medium components but generally results in a slower doubling time because it requires that cells undertake extensive de novo synthesis. To test whether Mg2+ could modulate cell length in a defined medium, we utilized a phosphate-buffered glucose MM. We began with a base medium that contained 13 amino acids and 50.0 μM Mg2+ (MM-13aa). In MM-13aa, the doubling time for the B. subtilis 168 prototroph (functional trpC+) was 53 min, both with and without the addition of 10.0 mM MgCl2 (Fig. 4A). The cell length in MM-13aa was assessed using microscopy and quantitation. As shown in Fig. 4B and C, the cells grown with supplemental Mg2+ were 45% shorter than those grown in MM-13aa only.
FIG 4.
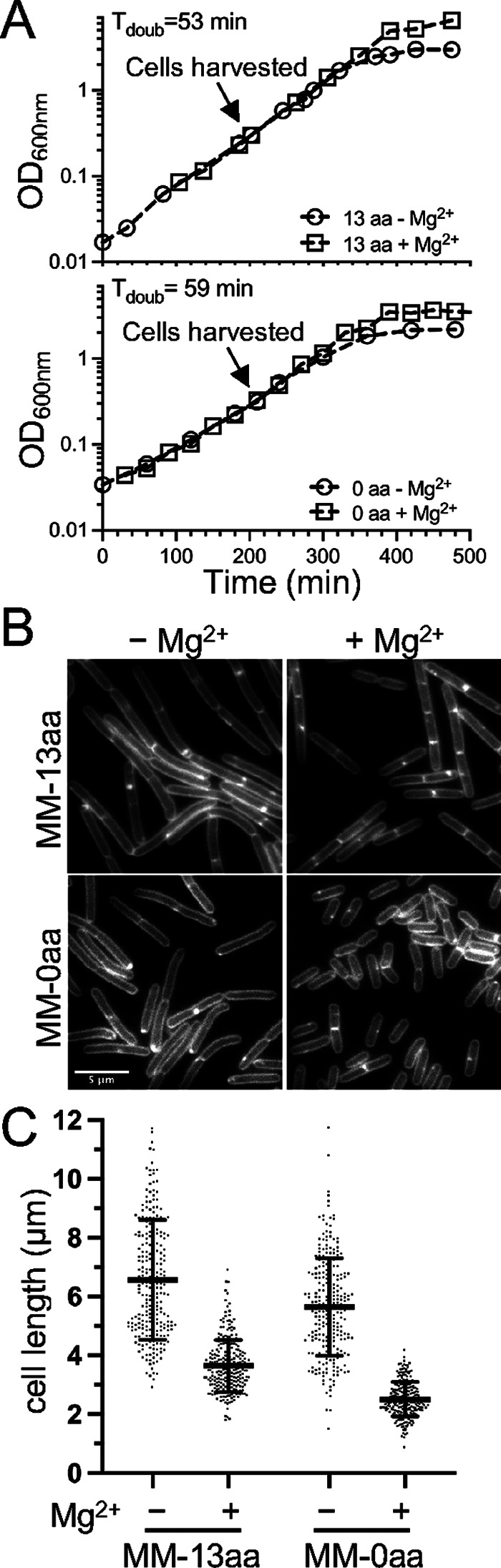
Cell length of B. subtilis following growth in minimal medium (MM). WT B. subtilis 168 (trpC prototroph) (BTG169) was cultured in a base MM containing 50.0 μM Mg2+ supplemented with 10.0 mM MgCl2 and amino acids as indicated. (A) Representative growth curves. (B) Micrographs following the membrane staining and epifluorescence microscopy (scaled identically). (C) Scatterplots showing the distribution of cell lengths quantitated for 250 cells from each condition. The bars represent the means of 250 cells ± the SDs.
To test whether the Mg2+-responsive phenotype was dependent on the presence of amino acids in the medium, the experiments were repeated in the base MM without amino acid supplementation. This modification increased the doubling time to 59 min, but the growth rate again remained unchanged, both with and without Mg2+ supplementation (Fig. 4A). Similar to the pattern observed in the media that contained amino acids (LB, CH*, and MM-13aa), the cells were shorter in the Mg2+ supplemented medium, compared to those in the unsupplemented control (Fig. 4B and C). These results suggest that the Mg2+-responsive phenotype is generalizable to both complex and defined media and does not depend on the addition of amino acids.
Mg2+-responsive cell length changes follow a dose-response curve.
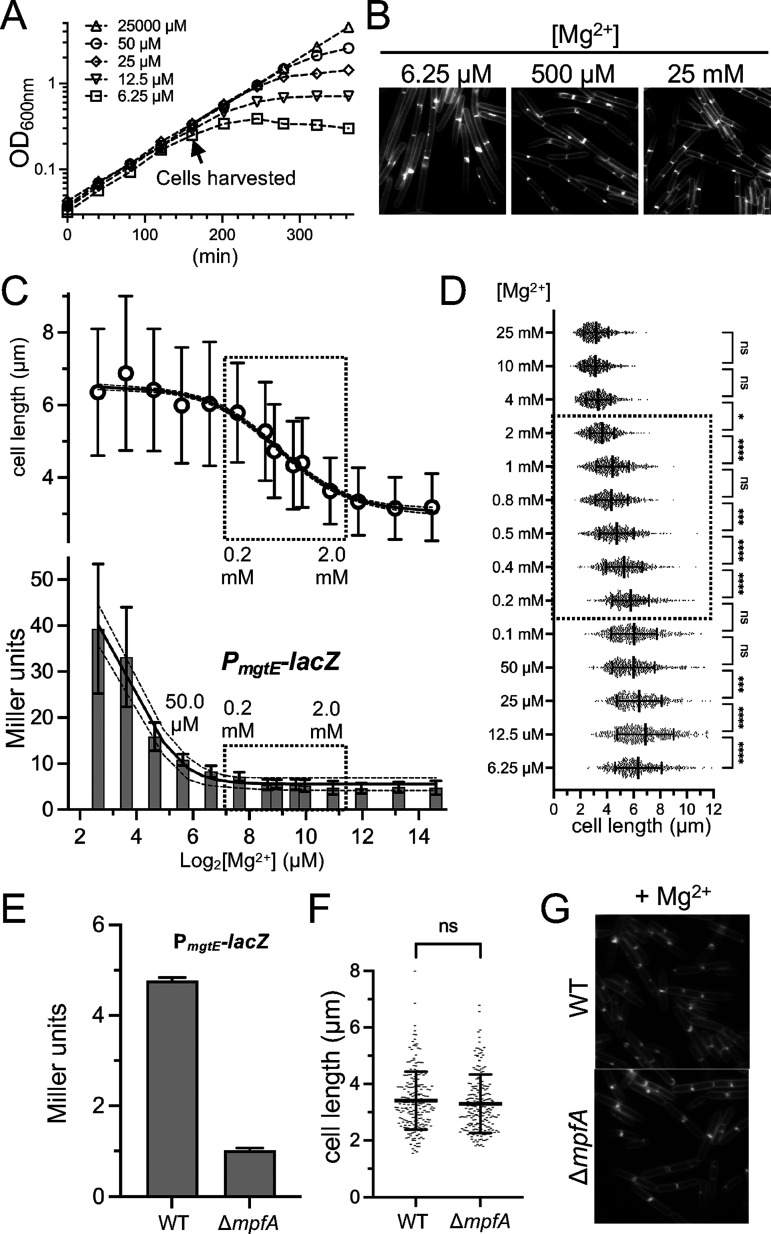
Next, we wanted to know whether cell length decreased proportionally with increasing extracellular Mg2+ or, alternatively, there was a threshold at which cells underwent a switch in cell length. To investigate this, we cultured cells in media across a range of Mg2+ concentrations, from 6.25 μM MgCl2 (at which differences in the growth rate begin to emerge at the densities at which we collected the cells) (Fig. 5A) up to 25.0 mM. Cells from each condition were imaged, and the cell lengths were quantified (Fig. 5). We found that cell shortening was neither directly proportional to Mg2+ availability nor a switch. Instead, cells became progressively shorter from 12.5 μM to 4.0 mM, with the largest decrease occurring between 0.2 μM and 2.0 mM (Fig. 5C and D).
FIG 5.
Correlation analysis of extracellular Mg2+ availability with cell length and with a reporter of intracellular Mg2+. WT cells harboring a reporter for PmgtE-lacZ (BTG182) were grown in MM-13aa with the indicated concentrations of supplemental MgCl2. (A) Representative growth curves and (B) representative micrographs following the membrane staining (scaled identically) from across the range of Mg2+ concentrations examined. (C) Mean cell lengths for 500 cells ± the SD for each condition (top) and the β-galactosidase activity assay for the mgtE riboswitch transcriptional reporter (bottom). (D) Scatterplots showing the distribution of cell lengths at each Mg2+ concentration. The bars represent the mean cell lengths for 500 cells ± the SDs. (E–G) WT (BTG182) or the ΔmpfA mutant (BTG333) harboring a reporter for PmgtE-lacZ was grown in MM-13aa supplemented with 10.0 mM MgCl2. Samples were taken during the exponential growth phase. (E) β-galactosidase activity assay. The bars represent the means from 2 biological replicates ± the SDs. (F) Scatterplots showing the distribution of cell lengths. The bars represent the mean cell lengths for 200 cells ± the SDs. (G) Representative micrographs of cells stained with TMA and scaled identically. *, 0.01 < P ≤ 0.05; **, 0.001 < P ≤ 0.01; ***, 0.0001 < P ≤ 0.001; ****, P < 0.0001; (ns) P ≥ 0.05.
Cells in the 0.2 μM to 2.0 mM window grew at equivalent doubling times (Fig. 5A), suggesting that the cells were not yet experiencing significant intracellular Mg2+ limitation. To independently assess this, we introduced a promoter fusion (PmgtE-lacZ) that reports on changes in internal Mg2+ availability. The promoter region includes a riboswitch that terminates transcription when the intracellular Mg2+ levels are sufficient (51, 52). Using this reporter, Dann et al. observed an approximately 10-fold induction of LacZ activity following the extended growth of cells with 5.0 μM Mg2+ (growth rate-limiting), compared to a Mg2+ excess (2.5 mM in their study) (Fig. 1B in reference [51]). Consistent with these findings, we observed that LacZ activity was also induced in medium containing 6.25 μM Mg2+ (Fig. 5B). Notably, this induction coincided with the time when the growth rate effects began to be observable (Fig. 5A).
The steepest decline in cell-shortening occurred between 0.2 and 2.0 mM MgCl2. To our surprise, no substantial differences in PmgtE-lacZ activity were detectable in this window, suggesting that the cell shortening is unlikely to be attributable to increased levels of labile intracellular Mg2+. We considered the possibility that the riboswitch of the PmgtE-lacZ reporter may already be saturated (terminator form) in this range and thus may be insensitive to further increases. To test this, we deleted the gene for the Mg2+ exporter protein MpfA, which was previously shown to increase intracellular Mg2+ (53). Even when cells were grown in 10.0 mM MgCl2, the ΔmpfA mutant displayed an additional 4-fold reduction in LacZ activity, compared to the wild-type (Fig. 5E), indicating that the reporter retained sensitivity. This result suggests that if there are changes in intracellular Mg2+ in the range in which we observe the most dramatic decreases in cell length, then the changes are not detectable with our reporter. Notably, even though intracellular Mg2+ can be inferred to have increased in the ΔmpfA mutant, compared to the wild-type, no differences in average cell length were observed (Fig. 5F and G). Collectively, these results suggest that under our growth conditions, changes in intracellular labile Mg2+ are neither necessary nor sufficient to elicit cell shortening. At the same time, we do not exclude the possibility that the cells may be responding to changes in intracellular Mg2+ that are not detectable by PmgtE-lacZ, such as an increase in the nonlabile pool. An alternative possibility is that the response is driven by differences in extracellular, or at least extracytoplasmic, Mg2+.
Cell shortening following a shift from lower to higher Mg2+ is rapid.
In the experiments above, the cell lengths were determined during steady-state growth, and care was taken to collect the cells at equivalent densities, as Mg2+ continues to be depleted from the medium with time. To assess the transition from longer to shorter, we grew cells in CH* and in MM-13aa, and we monitored the lengths of the cells at 10 min intervals after the addition of 10.0 mM MgCl2 (Fig. 6). For both media, the mean cell length continued to decrease for 40 to 50 min, at which point the cells achieved a mean length similar to those of cells always grown with excess Mg2+ (Fig. 6). Remarkably, 54% and 40% of the total decrease in the mean cell length occurred within the first 10 min of adding Mg2+ in the CH* and MM-13aa media, respectively (Fig. 6). Thus, the initial response to Mg2+ is relatively rapid and is well below the doubling time of the cells. After the initial rapid decrease, a more gradual decline in average cell length was observed. These results suggest that the adjustment to higher Mg2+ may occur through two distinct mechanisms: a dramatic initial decrease in cell length resulting from rapid division onset and a second, more protracted period in which the average cell length decreases more incrementally. These observations suggest that the initial response may have a more biophysical or enzymatic basis, whereas the second requires biosynthesis and outgrowth to achieve a new steady-state length.
FIG 6.
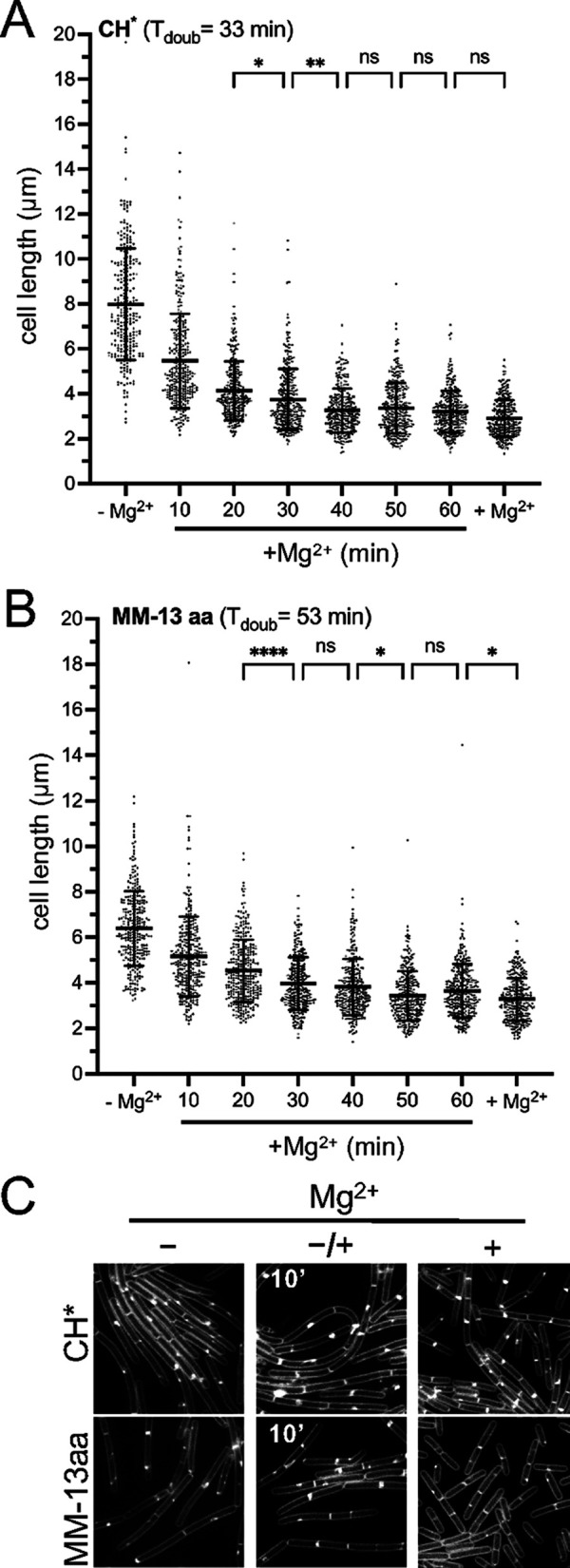
The B. subtilis cell length response to a change in extracellular Mg2+ is fast and is independent of the growth rate. WT 168 (BJH004) was cultured in the indicated media without Mg2+ supplementation (–Mg2+). Following addition of 10.0 mM MgCl2, the cells were imaged at 10 min intervals. (A and B) The bars represent the mean cell lengths for 300 cells ± the SDs. (C) Representative micrographs. The membranes are stained with TMA, and the images are scaled identically. *, 0.01 < P ≤ 0.05; **, 0.001 < P ≤ 0.01; ****, P < 0.0001; (ns) P ≥ 0.05.
Mg2+ modulates the frequency of Z-ring assembly.
A rod-shaped bacterium can increase cell length without altering other dimensions by elongating faster, dividing less frequently, or both. Based on the optical density, the growth rate of the cells in our experiments was equivalent, both with and without Mg2+ supplementation. Although absorbance readings are the most widely accepted method for monitoring cell growth, we considered the possibility that the populations that we were comparing might absorb light differently enough to mask differences in mass accumulation. To independently test this, we examined the abundance of the constitutively expressed protein SigA via a western blot analysis. In both CH* and MM-13aa, the SigA levels were equivalent, with and without Mg2+ supplementation, when cells were normalized to each other using OD600 values (Fig. S2A). As an additional control, we compared the dry weights of samples grown in MM-13aa with and without excess Mg2+ (Fig. S2B). The dry weights were indistinguishable in the two conditions, further supporting the conclusion that the optical density provides an accurate approximation of the cell mass. These results strongly support the conclusion that Mg2+ supplemented cells become shorter as a result of more frequent cell divisions, and, conversely, that cells divide less often under lower Mg2+ conditions.
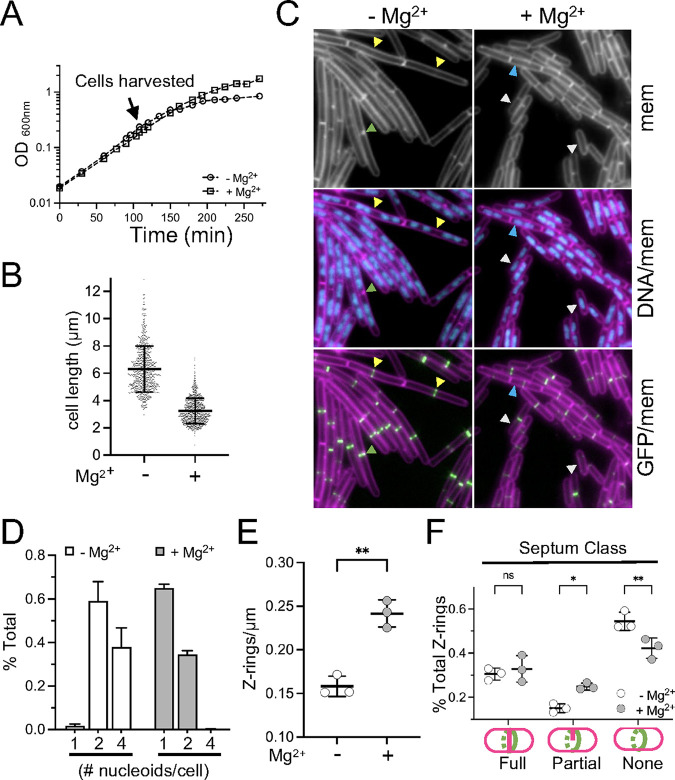
A delay in septation could occur before or after the assembly of the divisome. To assess this, we grew wild-type harboring a GFP fusion to ZapA, an early-arriving cell division protein that colocalizes with FtsZ as part of the “Z-ring” (54, 55). Cells expressing PxylA-GFP-zapA grew equivalently, with and without 10.0 mM MgCl2 (Fig. 7A), and retained the Mg2+-responsive reduction in cell length (Fig. 7B and C). Epifluorescence microscopy was performed, and images were captured of membranes, DNA (nucleoids), and Z-rings from both conditions (Fig. 7B). Overlays of the micrographs were used to quantitate both the number of distinct nucleoids per cell (Fig. 7D) and the number of Z-rings per unit cell length (Fig. 7E).
FIG 7.
Mg2+ modulates the frequency of Z-ring assembly. WT cells harboring Pxyl-GFP-zapA (BTG186) were cultured in CH* with 2.0 mM xylose. 10.0 mM MgCl2 was added as indicated. (A) Representative growth curves. (B) Scatterplots showing the distribution of cell lengths. The bars represent the mean cell lengths of 800 cells ± the SDs. (C) Representative micrographs following the staining of membranes and nucleoids with FM4-64 and DAPI, respectively. The images are scaled identically. The arrowheads indicate examples of cells with four nucleoid masses and Z-rings that lack septa (yellow), 1 nucleoid mass (white), 2 nucleoid masses (blue), or partial septa (green). (D) Fraction of cells with the indicated number of nucleoids from three independent biological replicates. (E) Average number of Z-rings per unit cell length ± SD. Each circle represents the mean of 300 cells from three independent biological replicates per condition. (F) Average fraction of cells with coalesced ZapA-GFP that presented the indicated septum type ± the SD. Each circle represents the mean of 300 cells from three independent biological replicates per condition. *, 0.01 < P ≤ 0.05, **, 0.001 < P ≤ 0.01; (ns) P ≥ 0.05.
Regardless of the condition, both populations possessed a large proportion of cells with two nucleoids (35% and 59% for the samples with and without supplemental Mg2+, respectively). The most striking differences became apparent when comparing the proportions of cells with one or four nucleoids. While only 2% of the cells grown in CH* had one distinct nucleoid mass, this proportion increased 30-fold (to 65%) in the Mg2+-supplemented medium. Conversely, cells with four nucleoids were relatively frequent (38%) in the base medium but were rare (<1%) in the Mg2+-supplemented cultures (Fig. 7D). These results indicate that division is more frequent when Mg2+ is in excess. Consistent with this idea, Z-rings were more frequently observed along the lengths of Mg2+-supplemented cells, and fewer Z-rings were observed along the lengths of cells grown in unsupplemented CH* (Fig. 7E). Moreover, the Z-rings that did form were less likely to be associated with a partial (incomplete) septum (Fig. 7F). Similar results were observed when FtsZ was tracked directly with an FtsZ-GFP fusion (Fig. S3). We conclude that there is a window of Mg2+ availability in which the cell division frequency can be modulated before the growth rate is impacted.
Overexpressing undecaprenyl pyrophosphate synthetase (UppS) results in the loss of the Mg2+-responsive phenotype and in constitutively short cells.
In the CH* shifting experiment, more than half of the decrease in the average cell length occurred within 10 min of adding the Mg2+ (Tdoub = 33 min) (Fig. 6), suggesting that a majority of the cells were generally poised to divide but were somehow inhibited. In a 1969 study, Garrett showed that Mg2+, in the same ranges of interest as were examined in our study see Table 2 in reference [56]), caused B. subtilis (W23) to accumulate the cytoplasmic PG precursor UDP-MurNAc-pentapeptide (56). This result suggests that the next step in the PG synthesis pathway, namely, the generation of Lipid I via the conjugation of the MurNAc-pentapeptide to Und-P (Fig. S4A), is sensitive to Mg2+ availability.
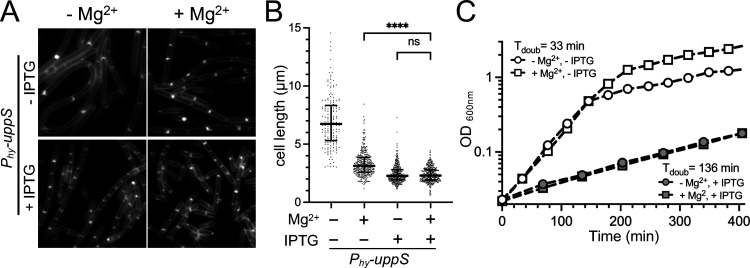
Und-P is generated through the dephosphorylation of Und-PP, the latter of which is either synthesized de novo by UppS or is regenerated following the transfer of Und-P-linked precursors to acceptor molecules (Fig. S4A). To test whether increasing the Und-PP pools from the de novo synthesis pathway would promote more frequent division, we overexpressed uppS from an IPTG-inducible promoter. uppS overexpression resulted in shorter cells, compared to the uninduced control in CH*, and the overexpressing cells were equivalently short, irrespective of Mg2+ supplementation (Fig. 8A and B). In contrast, overexpressing mraY, which encodes the enzyme that reversibly transfers phospho-MurNAc-pentapeptide to Und-P (Fig. S4A) (57), did not shorten the cells, and the cells remained responsive to Mg2+ (Fig. S4B and 4C). In contrast, cells overexpressing bcrC, which encodes the major Und-PP phosphatase (58–60) (Fig. S4A), were 37% shorter in CH*, compared to the uninduced control (Fig. S4C and D), consistent with enhanced division. However, unlike uppS, bcrC-overexpressing cells were still able to decrease in length following the addition of Mg2+ (Fig. S4D). The growth rate of the cells overexpressing uppS was slower, but it was equivalent with and without Mg2+ supplementation (Fig. 8C), suggesting that the cell length decrease is not simply attributable to the lower growth rate. At the same time, we cannot exclude the possibility that slow growth reduces the cells to a length that makes differences too subtle to be measured with our assay. Together, these results suggest that the availability of Und-P is rate-limiting for cell division in the unsupplemented medium.
FIG 8.
UppS overexpression results in constitutively short cells. WT cells harboring Phy-uppS (BTG708) were cultured in CH* with MgCl2 (10.0 mM) and IPTG (0.5 mM) added as indicated. (A) Representative micrographs of cells stained with TMA and scaled identically. (B) Scatterplots showing the distribution of cell lengths quantitated for 300 cells from each condition. The bars represent the mean cell length ± the SD. (C) Representative growth curves. ****, P < 0.0001; (ns) P > 0.05.
RNA-seq analysis of cells grown with and without excess Mg2+.
To further explore the Mg2+-responsive phenotype, we used an RNA-seq analysis to determine whether there were relative changes in transcription between the short and long cells that could provide insight into the mechanism. RNA-seq was performed using samples collected from exponential-phase cells that were growing in CH* or in CH* supplemented with 10.0 mM MgCl2. Genes expressed either more or less in the Mg2+-supplemented CH*, relative to CH* alone, were categorized based on known regulatory information that was retrieved from SubtiWiki (Tables S1 and S2) (61, 62).
As expected, based on the riboswitch data (Fig. 5), the transcriptional changes related to Mg2+ homeostasis were not significant. To our surprise, a number of other regulons that are related to metal homeostasis showed significant and internally consistent relative shifts in transcription. In particular, the profiles indicated the upregulation of the genes repressed by Fur (Fe2+ acquisition, sequestration, and sparing), MntR (Mn2+ uptake), and Zur (Zn2+ acquisition). In addition, there was enhanced expression of a large number of genes, consistent with a response to enhanced general stress (SigB). A subset of genes from two different prophages (PBSX and SPβ) were also expressed at higher levels but not in a manner consistent with prophage induction (Table S1). Conversely, and consistent with the observed upregulated genes, we observed the downregulation of the CzrA repressed genes cadA, czcD, and trkA (Zn2+ efflux/resistance to toxic metals), mneP (MntR-activated Mn2+ efflux), a large number of genes under the control of various cell envelope stress sigma factors (SigM/X/W/V), and those activated by the two-component response regulator YvrHb (63). Notably, several regulatory genes showed decreased transcription, including rsiX (encoding a SigX anti-sigma factor), sigX, and abh (a regulator of the transition state). Although we screened strains deleted for genes that showed differential expression via RNA-seq, those that were part of an annotated two-component system, and a strain that was cured of all prophages, each of the mutants retained the Mg2+-responsive phenotype (Tables S1–S3). Thus, although metal homeostasis and general stress response genes are clearly impacted by the changes in Mg2+ availability, we currently lack data that link any of these changes to enhanced division.
DISCUSSION
The findings in this study surprised us in several ways. First, the discovery of the cell division phenotype was itself unanticipated. Although LB is known to be low in Mg2+ and is generally discouraged by cell physiologists for various reasons (nicely outlined in a Small Things Considered blogpost by Hiroshi Nikaido [64]), LB has also been the default medium for the routine propagation of B. subtilis and E. coli for decades - not necessarily a context in which one expects to observe a novel phenotype. We cannot definitively say whether or not the phenotype observed in LB (Fig. 1) was due to the unusually low level of Mg2+ in the batch, as the powder was exhausted, and its bottle and lot number were discarded long before we appreciated the significance of the phenotype. Nonetheless, the data strongly implicated Mg2+ and motivated further experiments, which ultimately demonstrated that reduced Mg2+ availability impacts division before the growth rate and cell elongation.
The finding itself makes sense intuitively, as cell division is not a prerequisite for the replication of genetic material, growth, or survival. In fact, many bacteria are known to subsist as filamentous forms that divide infrequently. An extreme version of the filamentous lifestyle was recently discovered in the marine organism Thiomargarita magnifica, which displays average lengths of nearly a centimeter (65). This remarkable bacterium was found to harbor approximately 37,000 copies of its genome per millimeter of cell length - evidence of a dramatic uncoupling between growth and cell division. It is notable that many of the longest bacteria ever recorded, including the giant bacterium Beggiatoa that was discovered by Winogradsky, are sulfur-oxidizers (66). The evidence for an intimate relationship between sulfur metabolism and cell division is not restricted to giant bacteria. E. coli mutants with a reduced ability to convert ATP and the sulfur-containing amino acid methionine into S-adenosyl methionine (SAM) are inhibited for cell division but continue to grow and replicate DNA (67). The capacity of cells to uncouple division from other biosynthetic processes makes sense from an evolutionary perspective; it could provide cells with a mechanism by which to continue generating new “units” as resources deplete, while at the same time reserving the option to rapidly separate into individual cells when conditions are favorable.
Additional surprising results were the degree and type of the transcriptional changes that occurred between the supplemented and unsupplemented growth conditions. We expected that if changes occurred at all, then they would be associated with envelope synthesis or Mg2+ uptake or efflux. Instead, we observed changes in genes related to the general stress response and to iron, Mn2+, and Zn2+ homeostasis. More specifically, we found that when Mg2+ is more available, the expression of the SigB regulon is enhanced, and the Fur, MntR, and Zur regulons undergo shifts consistent with acquisition, sequestration, and/or sparing responses.
At first, the metal responses were perplexing, as we are accustomed to thinking about most metal regulons in terms of responses to starvation or toxicity, conditions that were not present in our experiments. However, the responses are relative, not absolute, and we can think of several other ways to interpret the data. The first is to think of the changes as homeostatic adjustments. For example, the data indicate that in the higher Mg2+ condition, the cells receive signals consistent with the need to acquire, sequester, and spare iron, as well as to increase internal Mn2+ and Zn2+. The expression of genes for surfactin biosynthesis (srfABCD) and the SigB general stress response are also increased. Iron sequestration and iron-sparing responses would reduce the possibility of Fenton reactions (68). Mn2+ and Zn2+ both reduce internal reactive oxygen species (69–75), and surfactin and SigB both combat oxidative/energy stress, the former by reducing proton motive force (76, 77). The overall profile suggests that, compared to the unsupplemented cells, the Mg2+-supplemented cells exhibit changes consistent with the prevention of oxidative stress.
The second possibility, which is not mutually exclusive, is that at least some of the transcriptional changes are associated with events before, after, and/or during cell division itself. Cell cycle variations are typically obscured when samples are collected from pooled populations of asynchronously growing cells. The samples collected for RNA-seq were also growing asynchronously; however, because the Mg2+-supplemented populations were essentially “enriched” for dividing cells (Fig. 7; Fig. S3), the transcriptional changes associated with division should also be enriched. The idea that cells may adjust metal pools not only to respond to different environmental stressors but also as a regulated part of the life cycle or development is intriguing, and we think it merits further inquiry.
We noted one anomaly in the RNA-seq data that we do not understand. In the Zur regulon, we see more RNA corresponding to yczL but relatively little or no increase in other genes found in the predicted (78) folEB-yciB-yczL-zagA transcript (Table S1). The unexpected differential expression of the yczL region could be attributable to differences in RNA stability, rather than transcription itself. The gene upstream of yczL (yciB) is proceeded by four instances of putative cotranslational coupling, the last of which could lead to the translation of yczL. It may be worth exploring whether there is a functional consequence to the predicted coupling, such as the alteration of transcriptional readthrough, transcript stability, or the effects on the expression of the downstream gene zagA.
We do not know whether any of the transcriptional changes relate directly to the Mg2+-responsive phenotype or are incidental. We are uncertain whether differential expression would still occur if cells were grown at maximal “shortness” but different Mg2+ concentrations, such as the 4.0 mM to 25.0 mM range shown in Fig. 5. The cell shortening phenotype was still detected in a prophage-cured strain (Table S3). So, at the least, the prophage-related changes appear to be incidental. Consistent with this conclusion, others have documented differential expression from various B. subtilis prophage loci as having resulted from variation the in growth regime (79).
Visually screening a number of deletion strains also failed to implicate any single gene knockouts in the Mg2+-responsive phenotype (Tables S1–S3). The only condition identified that showed an apparent loss of Mg2+-responsiveness in CH* was the overexpression of uppS (Fig. 8). However, as these cells grew slowly, we could not confidently exclude the possibilities that (i) cell shortening occurred but fell outside the detection limit of our assay and/or (ii) that the low growth rate imposes an upper limit on the division frequency that is dominant over the effects of Mg2+.
Still, it is plausible that exogenous Mg2+ somehow enhances the availability of Und-P to the divisome. UppS utilizes Mg2+ as a cofactor (80–82), so we considered the possibility that UppS activity could be affected by altering the availability of Mg2+. Although we do not exclude this possibility, several observations argue against it. First, E. coli UppS achieves maximal turnover (kcat) around 0.5 mM MgCl2 and half-maximal turnover at 49.0 μM MgCl2 (80). The levels of labile, intracellular Mg2+ in B. subtilis (bulk measurements) are estimated to be between 0.8 and 3.7 mM in media containing 1.6 mM MgCl2 (83). Notably, we still observe cell shortening at this concentration (Fig. 5). Thus, assuming that the B. subtilis and E. coli enzymes possess similar properties, there is likely sufficient Mg2+ to support full UppS activity when Mg2+ is not limiting the growth rate. Second, based on in vitro transcription termination measurements, the PmgtE riboswitch transitions from an anti-terminator conformation to a terminator conformation between 2.0 and 3.0 mM Mg2+ (51). We did not see significant induction of the PmgtE-lacZ reporter at the concentrations of Mg2+ at which the most dramatic changes in cell length were observed (Fig. 5C). This suggests that the pool of intracellular Mg2+ does not vary dramatically at the exogenous ranges that we focused on. Moreover, this is consistent with the observations made by others that intracellular Mg2+ does not vary significantly during exponential growth (83). Third, cell elongation also requires Und-P, and elongation was not affected in our conditions. Of course, it is possible that division is more sensitive to Und-P availability. At least in E. coli, there is some evidence that cells have a propensity to filament before bulging when Und-P is limiting (84). Finally, the rapid onset of division that we observed in the shifting experiment suggests that the cells have the capacity to divide without significant biosynthesis. Capturing these initial divisions in real time with a flow cell would provide more control and resolution regarding their timing.
One idea that is purely speculative is that the rapid division onset is driven by the more frequent flipping of Und-P to the cytoplasmic face of the membrane and/or the enhanced conversion of Und-PP to Und-P. There is also a pool of undecaprenyl-OH in some Gram-positives (85–88), so increasing Und-P availability through undecaprenyl-OH phosphorylation may be another route to enhancing pools. If the mechanism is not dependent on changes in cytoplasmic Mg2+ or on the activity of cytoplasmic enzymes such as UppS, there are also extracytoplasmic enzymes to consider. For example, the enzymes for wall teichoic acid and PG synthesis compete for Und-P, and there is at least one report that wall teichoic acid synthesis is favored over PG synthesis when Mg2+ is limited (89).
In summary, we show that the frequency of cell division can be uncoupled from the growth rate by manipulating a single nutrient, namely, Mg2+. Though we did not arrive at a mechanism, we hope that the observations documented here will inspire further studies in additional bacteria as well as more consideration of medium selection during experimentation. Rich media are convenient but have the considerable weakness of variable formulation with regard to cofactor, trace metal, and amino acid abundance. Not only can this lead to confusing and irreproducible results but also (as we unintentionally discovered), rich media may be masking some important biology.
MATERIALS AND METHODS
General methods.
The strains and the details of the strain construction can be found in the supplemental material (Table S4; Text S1). Cells were stored at −80°C in 15% glycerol (vol/vol). Strains were streaked for isolation on lysogeny broth, Lennox (LB) containing 1.5% (wt/vol) bactoagar (Sigma) and were incubated overnight (approximately 16 h) at 37°C. Cultures for experiments were begun with colonies from same-day plates by inoculating single colonies into a 20 mm glass tube that contained 5 mL of the indicated medium. The tube was incubated at 37°C in a roller drum until the exponential stage (OD600 < 0.6). For the microscopy and RNA-seq experiments, the exponential-stage cultures (see above) were back-diluted in 25 mL of fresh medium in a 250 mL baffled flask to an optical density that would allow the cells to reach the desired growth stage (exponential) after no fewer than four doublings. The flasks were placed in a shaking water bath that was set to 280 rpm and 37°C.
The LB was made by dissolving 20 g of Difco LB Broth, Lennox (product number 240230) in 1 L ddH2O, and this was followed by sterilization in an autoclave. The CH* utilized is a modified form of CH (49). The CH* (1L) contained 10.0 g casein acid hydrolysate (Acumedia, Lot No. 104,442B), 3.7 g l-glutamic acid (25.0 mM), 1.6 g l-asparagine monohydrate (10.0 mM), 1.25 g l-alanine (14.0 mM), 1.36 g (10.0 mM) anhydrous KH2PO4, 1.34 g (25.0 mM) NH4Cl, 0.11 g Na2SO4 (0.77 mM), 0.1 g NH4NO3 (1.25 mM), 0.001 g FeCl3•6H2O (3.7 μM), and ddH2O. The pH was adjusted to 7.0 with 10.0 N NaOH before the medium was sterilized in an autoclave. After autoclaving, sterile CaCl2 was added to 0.2 mM, and l-sterile tryptophan was added to 0.1 mM. The base minimal medium (MM) contained 15.0 mM (NH4)2SO4, 5.0 mM KH2PO4, 50.0 mM Tris-HCl [pH 7.5], 27.0 mM KCl, 0.05 mM FeCl3, 0.01 mM MnSO2, 0.01 mM ZnSO4, 0.2% glucose (wt/vol), 27.0 mM sodium citrate, and 10.2 mM CaCl2. 50.0 μM MgCl2 was added to create the base medium used in the MM experiments, except for those in Fig. 5, which required final concentrations below 50.0 μM. In these experiments, MgCl2 was added to the final concentrations indicated in the figure. For the MM-13aa, 10× filter-sterilized amino acids [pH 7.5] were added to a 1× final concentration. The 10× mixture consisted of the following concentrations of l-amino acids: 250.0 mM glutamate; 100.0 mM (each) alanine, asparagine, and proline; 50.0 mM (each) aspartic acid, phenylalanine, glycine, isoleucine, leucine, threonine, and valine; 10.0 mM (each) histidine and tryptophan. The amino acids were added to increase the growth rate but were selected based on their availability in our laboratory, not on logic.
β-galactosidase assays.
The B. subtilis strains were grown in 250 mL baffled flasks with 25 mL of the indicated medium at 37°C and 280 rpm to the target OD600. The optical density was recorded, and 1 mL of culture was harvested via centrifugation at 21,130 × g for 1 min at room temperature, removing the supernatant via aspiration. The pellets were immediately stored at −80°C. To perform the assay, the pellets were thawed, resuspended in 0.5 mL Z-buffer (40.0 mM NaH2PO4, 60.0 mM Na2HPO4, 1.0 mM MgSO4, 10.0 mM KCl, 40.0 mM β-mercaptoethanol, and 0.2 mg/mL lysozyme), and incubated at 30°C for 15 min. 100 μL of 4.0 mg/mL 2-nitrophenyl-β-d-galactopyranoside (ONPG) in Z-buffer was added, and the samples were incubated at 30°C until pale yellow. The reaction was stopped with 250 μL 1.0 M Na2CO3, and the reaction time was recorded. The sample was vortexed for 5 sec and centrifuged at 21,130 × g for 3 min at room temperature in a tabletop centrifuge. The supernatant (minimum volume of 0.8 mL) was transferred to a 1 mL cuvette, and the OD420 and OD550 absorbance readings were recorded. The β-galactosidase specific activity in Miller Units was calculated using the following formula: (OD420 − [1.75× OD550]) / (time [min] × volume × OD600) × 1,000.
Western blot analysis.
1 mL of culture was collected and spun at 21,130 × g for 1 min at room temperature at the exponential stage, and the OD600 value at the time of sampling was recorded. The pellet was resuspended in the lysis buffer (20.0 mM Tris [pH 7.5], 10.0 mM EDTA, 1 mg/mL lysozyme, 10 μg/mL DNase I, 100 μg/mL RNase A, 1.0 mM PMSF, 1 μL protease inhibitor cocktail [Sigma P8465-5ML] resuspended in 1 mL lysis buffer) to give a final OD600 equivalent of 15. The samples were incubated at 37°C for 10 min, and this was followed by the addition of an equal volume of sodium dodecyl sulfate (SDS) sample buffer (0.25 M Tris [pH 6.8], 4% [wt/vol] SDS, 20% [wt/vol], 20% glycerol [vol/vol], 10.0 mM EDTA, and 10% β-mercaptoethanol [vol/vol]). The samples were heated for 5 min at 100°C prior to loading. The proteins were separated on 12% SDS-PAGE polyacrylamide gels, transferred to a nitrocellulose membrane at 100 V for 60 min, and then blocked in 1× PBS containing 0.05% (vol/vol) Tween 20 and 5% (wt/vol) dry milk powder. The blocked membranes were probed overnight at 4°C with anti-SigA (1:20,000, rabbit, gift from Fujita Masaya, University of Houston, Houston, TX) diluted in 1× PBS with 0.05% (vol/vol) Tween 20 and 5% (wt/vol) milk powder. The membranes were washed three times with 1× PBS containing 0.05% (vol/vol) Tween 20, transferred to 1× PBS with 0.05% (vol/vol) Tween 20 and 5% (wt/vol) milk powder containing 1:5,000 horseradish peroxidase-conjugated goat anti-mouse IgG secondary antibodies (AbCam, ab205719), and incubated on a shaking platform for 1 h at room temperature. The membranes were washed 3× with 1× PBS containing 0.05% (vol/vol) Tween 20, and signals were detected using the SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher) and a Bio-Rad Gel Doc Imaging System.
Fluorescence microscopy.
1 mL of cultured cells at the exponential stage were harvested, concentrated via centrifugation at 6,010 × g for 1 min, and resuspended in 5 μL 1× PBS with either 1-(4-(trimethylamino)phenyl)-6-phenylhexa-1,3,5-triene (TMA-DPH) (50.0 μM) or N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide (FM4-64) (6 μg/mL) and 4′,6-diamidino-2-phenylindole (DAPI) (2 μg/mL). All dyes were purchased from Thermo Fisher. For the Zap-GFP and FtsZ-GFP experiments, the cells were mounted on 1% (wt/vol) agarose pads made with PBS [pH 7.4] and overlaid with an untreated glass coverslip. Otherwise, the cells were mounted on glass sides with poly-l-lysine-treated coverslips prior to imaging.
The cells were imaged on a Nikon Ti-E microscope using a CFI Plan Apo Lambda DM 100× objective and an X-Cite XYLIS 365 nm illumination system (Excelitas Technologies). The filter cubes utilized were C-FL UV-2E/C (DAPI), C-FL Texas Red HC HISN Zero Shift, and GFP HC HISN Zero Shift. Micrographs were acquired using a CoolSNAP HQ2 monochrome camera with NIS Elements Advanced Research, and they were analyzed in ImageJ (90).
Cell length measurements.
The TMA-DPH micrographs were analyzed in ImageJ (90). For each cell, a line that extended from pole to pole was placed. One compartment between two bright, solid septa with homogenous signals were counted as one intact cell. Cells with bright partial septa or septa interrupted with discrete dark areas were counted as cells with partial septa.
Quantitation of Z-rings per unit cell length.
The cell length lines (see above) were overlapped with the GFP micrographs of the Z-rings, and the number of Z-rings along the cell length line was counted. When two cells shared a pole and a Z-ring, each Z-ring was counted as 0.5/cell. The Z-ring number per μm was calculated by dividing the Z-ring total number by the total cell length.
Nucleoid number per cell.
The cell length line was overlapped with the DAPI micrograph, and the cells were classified based on the number of nucleoids overlapping the length label. The number of nucleoids per cell was calculated by dividing the number of cells containing each number of nucleoids (1, 2, or 4) by the total number of cells counted.
Classification of septa in cells containing coalesced Z-rings.
Micrographs of the GFP channel (ZapA-GFP or FtsZ-GFP) were overlaid with the corresponding TMA-DPH micrograph. The cells were classified as having no, partial, or full septa. The fraction of each septum type was determined by dividing the number of cells in each class by the total number of cells counted.
Statistical analysis and data plotting.
The graphs were generated and the statistical analysis was performed using GraphPad Prism version 9.4.0 for Mac (GraphPad Software, San Diego, California, USA, www.graphpad.com). The statistical analyses in the following figures were performed via one-way analyses of variance (ANOVA) followed by Tukey’s multiple-comparison tests, assuming Gaussian distributions with equal standard deviations (SDs): Fig. 3B, 5E, 6A, 6B, and 8B as well as Fig. S2B and S3C. The statistical analyses in the following figures were performed via two-way ANOVA followed by Tukey’s multiple-comparison tests, fitting a full model: Fig. 7F and Fig. S3D. The statistical analyses in the following figures were performed via two-tailed unpaired t tests, assuming a Gaussian distribution and that both populations have the same SD: Fig. 1B, 2D, 6B, and 7E.
RNA-seq.
For each condition, samples were collected from three independent biological replicates grown in either CH* medium or CH* medium with 10.0 mM MgCl2. The cells were precultured in the same medium used for the experiment. 0.5 mL cells were collected at the exponential stage (OD600 ~of approximately 0.2) via centrifugation at 21,130 × g for 2 min at room temperature. RNA was collected using the RNAprotect Bacteria Reagent and an RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. RNA sequencing was performed by the Texas A&M AgriLife Research Genomics and Bioinformatics Service (College Station). The total RNA were prepared for sequencing using TruSeq Stranded Total RNA and Ribo-Zero Gold (Illumina). The RNA-seq was performed using a HISeq 4000 platform (Illumina). The RNA-seq data were processed through HISAT2 (91) and StringTie (92). A differential expression analysis was done using the Deseq2 R package (93). Regulons were assigned using the GinTool (62) starter package obtained from L.W. Hamoen (University of Amsterdam, Swammerdam Institute for Life Sciences). The cutoff for inclusion in the final analysis was set as genes with an absolute log2-fold change (log2FC) value of ≥0.585 and an adjusted P-value of ≤0.05. The adjusted P-values that are reported as 1.0 in the raw data indicate that the gene could not be included or excluded as statistically significant (generally due to insufficient reads in either the control or experimental samples).
Data availability.
The raw RNA-seq data may be accessed through the NCBI Gene Expression Omnibus at the following link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE219221.
ACKNOWLEDGMENTS
We thank Veronica Chemelewski for the critical reading of the manuscript, Morgan Chapman for the generation of the Phy-uppS strain, Tatiana Castro Padovani for the assistance with visually screening deletion mutants for the loss of Mg2+-responsiveness, and Leendert Hamoen for the sharing of the Gintool starter package. This work was supported by T. Guo teaching every semester and by scrap funds from the startup account of J.K. Herman. We dedicate this work to Ry Young in honor of his retirement.
Footnotes
Supplemental material is available online only.
Contributor Information
Jennifer K. Herman, Email: jkherman@tamu.edu.
Tina M. Henkin, Ohio State University
REFERENCES
- 1.Jahnen-Dechent W, Ketteler M. 2012. Magnesium basics. Clin Kidney J 5:i3–i14. 10.1093/ndtplus/sfr163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sissi C, Palumbo M. 2009. Effects of magnesium and related divalent metal ions in topoisomerase structure and function. Nucleic Acids Res 37:702–711. 10.1093/nar/gkp024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meneely KM, Sundlov JA, Gulick AM, Moran GR, Lamb AL. 2016. An open and shut case: the interaction of magnesium with MST enzymes. J Am Chem Soc 138:9277–9293. 10.1021/jacs.6b05134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein DJ, Moore PB, Steitz TA. 2004. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA 10:1366–1379. 10.1261/rna.7390804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akanuma G, Kobayashi A, Suzuki S, Kawamura F, Shiwa Y, Watanabe S, Yoshikawa H, Hanai R, Ishizuka M. 2014. Defect in the formation of 70S ribosomes caused by lack of ribosomal protein L34 can be suppressed by magnesium. J Bacteriol 196:3820–3830. 10.1128/JB.01896-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pontes MH, Sevostyanova A, Groisman EA. 2015. When too much ATP is bad for protein synthesis. J Mol Biol 427:2586–2594. 10.1016/j.jmb.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruna RE, Kendra CG, Groisman EA, Pontes MH. 2021. Limitation of phosphate assimilation maintains cytoplasmic magnesium homeostasis. Proc Natl Acad Sci USA 118. 10.1073/pnas.2021370118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin X, Wu Orr M, Wang H, Hobbs EC, Shabalina SA, Storz G. 2019. The small protein MgtS and small RNA MgrR modulate the PitA phosphate symporter to boost intracellular magnesium levels. Mol Microbiol 111:131–144. 10.1111/mmi.14143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wendel BM, Pi H, Kruger L, Herzberg C, Stulke J, Helmann JD. 2022. A central role for magnesium homeostasis during adaptation to osmotic stress. mBio 13:e0009222. 10.1128/mbio.00092-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Melo JRF, Gutsch A, Caluwe T, Leloup JC, Gonze D, Hermans C, Webb AAR, Verbruggen N. 2021. Magnesium maintains the length of the circadian period in Arabidopsis. Plant Physiol 185:519–532. 10.1093/plphys/kiaa042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray T, Popham DL, Setlow P. 1998. Bacillus subtilis cells lacking penicillin-binding protein 1 require increased levels of divalent cations for growth. J Bacteriol 180:4555–4563. 10.1128/JB.180.17.4555-4563.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuoka S, Chiba M, Tanimura Y, Hashimoto M, Hara H, Matsumoto K. 2011. Abnormal morphology of Bacillus subtilis ugtP mutant cells lacking glucolipids. Genes Genet Syst 86:295–304. 10.1266/ggs.86.295. [DOI] [PubMed] [Google Scholar]
- 13.Schirner K, Errington J. 2009. The cell wall regulator σI specifically suppresses the lethal phenotype of mbl mutants in Bacillus subtilis. J Bacteriol 191:1404–1413. 10.1128/JB.01497-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blaine J, Chonchol M, Levi M. 2015. Renal control of calcium, phosphate, and magnesium homeostasis. Clin J Am Soc Nephrol 10:1257–1272. 10.2215/CJN.09750913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moncany ML, Kellenberger E. 1981. High magnesium content of Escherichia coli B. Experientia 37:846–847. 10.1007/BF01985672. [DOI] [PubMed] [Google Scholar]
- 16.Theillet FX, Binolfi A, Frembgen-Kesner T, Hingorani K, Sarkar M, Kyne C, Li C, Crowley PB, Gierasch L, Pielak GJ, Elcock AH, Gershenson A, Selenko P. 2014. Physicochemical properties of cells and their effects on intrinsically disordered proteins (IDPs). Chem Rev 114:6661–6714. 10.1021/cr400695p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alatossava T, Jutte H, Kuhn A, Kellenberger E. 1985. Manipulation of intracellular magnesium content in polymyxin B nonapeptide-sensitized Escherichia coli by ionophore A23187. J Bacteriol 162:413–419. 10.1128/jb.162.1.413-419.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyrrell J, McGinnis JL, Weeks KM, Pielak GJ. 2013. The cellular environment stabilizes adenine riboswitch RNA structure. Biochemistry 52:8777–8785. 10.1021/bi401207q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Outten CE, O'Halloran TV. 2001. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488–2492. 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 20.Maguire ME, Cowan JA. 2002. Magnesium chemistry and biochemistry. Biometals 15:203–210. 10.1023/a:1016058229972. [DOI] [PubMed] [Google Scholar]
- 21.Pontes MH, Yeom J, Groisman EA. 2016. Reducing ribosome biosynthesis promotes translation during low Mg(2+) stress. Mol Cell 64:480–492. 10.1016/j.molcel.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groisman EA. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol 183:1835–1842. 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groisman EA, Duprey A, Choi J. 2021. How the PhoP/PhoQ system controls virulence and Mg(2+) homeostasis: lessons in signal transduction, pathogenesis, physiology, and evolution. Microbiol Mol Biol Rev 85:e0017620. 10.1128/MMBR.00176-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wakeman CA, Goodson JR, Zacharia VM, Winkler WC. 2014. Assessment of the requirements for magnesium transporters in Bacillus subtilis. J Bacteriol 196:1206–1214. 10.1128/JB.01238-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krom BP, Warner JB, Konings WN, Lolkema JS. 2000. Complementary metal ion specificity of the metal-citrate transporters CitM and CitH of Bacillus subtilis. J Bacteriol 182:6374–6381. 10.1128/JB.182.22.6374-6381.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krom BP, Huttinga H, Warner JB, Lolkema JS. 2002. Impact of the Mg(2+)-citrate transporter CitM on heavy metal toxicity in Bacillus subtilis. Arch Microbiol 178:370–375. 10.1007/s00203-002-0465-8. [DOI] [PubMed] [Google Scholar]
- 27.Smith RL, Maguire ME. 1998. Microbial magnesium transport: unusual transporters searching for identity. Mol Microbiol 28:217–226. 10.1046/j.1365-2958.1998.00810.x. [DOI] [PubMed] [Google Scholar]
- 28.Rogers HJ, Thurman PF, Buxton RS. 1976. Magnesium and anion requirements of rodB mutants of Bacillus subtilis. J Bacteriol 125:556–564. 10.1128/jb.125.2.556-564.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawai Y, Asai K, Errington J. 2009. Partial functional redundancy of MreB isoforms, MreB, Mbl and MreBH, in cell morphogenesis of Bacillus subtilis. Mol Microbiol 73:719–731. 10.1111/j.1365-2958.2009.06805.x. [DOI] [PubMed] [Google Scholar]
- 30.Leaver M, Errington J. 2005. Roles for MreC and MreD proteins in helical growth of the cylindrical cell wall in Bacillus subtilis. Mol Microbiol 57:1196–1209. 10.1111/j.1365-2958.2005.04736.x. [DOI] [PubMed] [Google Scholar]
- 31.Carballido-Lopez R, Formstone A, Li Y, Ehrlich SD, Noirot P, Errington J. 2006. Actin homolog MreBH governs cell morphogenesis by localization of the cell wall hydrolase LytE. Dev Cell 11:399–409. 10.1016/j.devcel.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 32.Wagner PM, Stewart GC. 1991. Role and expression of the Bacillus subtilis rodC operon. J Bacteriol 173:4341–4346. 10.1128/jb.173.14.4341-4346.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lazarevic V, Soldo B, Medico N, Pooley H, Bron S, Karamata D. 2005. Bacillus subtilis alpha-phosphoglucomutase is required for normal cell morphology and biofilm formation. Appl Environ Microbiol 71:39–45. 10.1128/AEM.71.1.39-45.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorke B, Foulquier E, Galinier A. 2005. YvcK of Bacillus subtilis is required for a normal cell shape and for growth on Krebs cycle intermediates and substrates of the pentose phosphate pathway. Microbiology (Reading) 151:3777–3791. 10.1099/mic.0.28172-0. [DOI] [PubMed] [Google Scholar]
- 35.Wecke J, Madela K, Fischer W. 1997. The absence of D-alanine from lipoteichoic acid and wall teichoic acid alters surface charge, enhances autolysis and increases susceptibility to methicillin in Bacillus subtilis. Microbiology (Reading) 143:2953–2960. 10.1099/00221287-143-9-2953. [DOI] [PubMed] [Google Scholar]
- 36.Dajkovic A, Tesson B, Chauhan S, Courtin P, Keary R, Flores P, Marliere C, Filipe SR, Chapot-Chartier MP, Carballido-Lopez R. 2017. Hydrolysis of peptidoglycan is modulated by amidation of meso-diaminopimelic acid and Mg(2+) in Bacillus subtilis. Mol Microbiol 104:972–988. 10.1111/mmi.13673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tesson B, Dajkovic A, Keary R, Marliere C, Dupont-Gillain CC, Carballido-Lopez R. 2022. Magnesium rescues the morphology of Bacillus subtilis mreB mutants through its inhibitory effect on peptidoglycan hydrolases. Sci Rep 12:1137. 10.1038/s41598-021-04294-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Christensen DG, Orr JS, Rao CV, Wolfe AJ. 2017. Increasing growth yield and decreasing acetylation in Escherichia coli by optimizing the carbon-to-magnesium ratio in peptide-based media. Appl Environ Microbiol 83. 10.1128/AEM.03034-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Webb M. 1951. The influence of magnesium on cell division. IV. The specificity of magnesium. J Gen Microbiol 5:480–484. 10.1099/00221287-5-3-480. [DOI] [PubMed] [Google Scholar]
- 40.Webb M. 1951. The influence of magnesium on cell division. V. The effect of magnesium on the growth of bacteria in chemically-defined media of varying complexity. J Gen Microbiol 5:485–495. 10.1099/00221287-5-3-485. [DOI] [PubMed] [Google Scholar]
- 41.Webb M. 1951. The influence of magnesium on cell division. VI. The action of certain hydrolytic enzymes on the filamentous and chain forms of gram-positive rod-shaped organisms. J Gen Microbiol 5:496–501. 10.1099/00221287-5-3-496. [DOI] [PubMed] [Google Scholar]
- 42.Yaginuma H, Kawai S, Tabata KV, Tomiyama K, Kakizuka A, Komatsuzaki T, Noji H, Imamura H. 2014. Diversity in ATP concentrations in a single bacterial cell population revealed by quantitative single-cell imaging. Sci Rep 4:6522. 10.1038/srep06522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Westfall CS, Levin PA. 2018. Comprehensive analysis of central carbon metabolism illuminates connections between nutrient availability, growth rate, and cell morphology in Escherichia coli. PLoS Genet 14:e1007205. 10.1371/journal.pgen.1007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vadia S, Tse JL, Lucena R, Yang Z, Kellogg DR, Wang JD, Levin PA. 2017. Fatty acid availability sets cell envelope capacity and dictates microbial cell size. Curr Biol 27:1757–1767. 10.1016/j.cub.2017.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taheri-Araghi S, Bradde S, Sauls JT, Hill NS, Levin PA, Paulsson J, Vergassola M, Jun S. 2015. Cell-size control and homeostasis in bacteria. Curr Biol 25:385–391. 10.1016/j.cub.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sargent MG. 1975. Control of cell length in Bacillus subtilis. J Bacteriol 123:7–19. 10.1128/jb.123.1.7-19.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vadia S, Levin PA. 2015. Growth rate and cell size: a re-examination of the growth law. Curr Opin Microbiol 24:96–103. 10.1016/j.mib.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaechter M, Maaloe O, Kjeldgaard NO. 1958. Dependency on medium and temperature of cell size and chemical composition during balanced grown of Salmonella typhimurium. J Gen Microbiol 19:592–606. 10.1099/00221287-19-3-592. [DOI] [PubMed] [Google Scholar]
- 49.Sterlini JM, Mandelstam J. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J 113:29–37. 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeigler DR, Pragai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, Bai R, Wyss M, Perkins JB. 2008. The origins of 168, W23, and other Bacillus subtilis legacy strains. J Bacteriol 190:6983–6995. 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dann CE, 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. 2007. Structure and mechanism of a metal-sensing regulatory RNA. Cell 130:878–892. 10.1016/j.cell.2007.06.051. [DOI] [PubMed] [Google Scholar]
- 52.Ramesh A, Winkler WC. 2010. Magnesium-sensing riboswitches in bacteria. RNA Biol 7:77–83. 10.4161/rna.7.1.10490. [DOI] [PubMed] [Google Scholar]
- 53.Pi H, Wendel BM, Helmann JD. 2020. Dysregulation of magnesium transport protects Bacillus subtilis against manganese and cobalt intoxication. J Bacteriol 202. 10.1128/JB.00711-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gamba P, Veening JW, Saunders NJ, Hamoen LW, Daniel RA. 2009. Two-step assembly dynamics of the Bacillus subtilis divisome. J Bacteriol 191:4186–4194. 10.1128/JB.01758-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gueiros-Filho FJ, Losick R. 2002. A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes Dev 16:2544–2556. 10.1101/gad.1014102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Garrett AJ. 1969. The effect of magnesium ion deprivation on the synthesis of mucopeptide and its precursors in Bacillus subtilis. Biochem J 115:419–430. 10.1042/bj1150419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bouhss A, Crouvoisier M, Blanot D, Mengin-Lecreulx D. 2004. Purification and characterization of the bacterial MraY translocase catalyzing the first membrane step of peptidoglycan biosynthesis. J Biol Chem 279:29974–29980. 10.1074/jbc.M314165200. [DOI] [PubMed] [Google Scholar]
- 58.Radeck J, Lautenschlager N, Mascher T. 2017. The Essential UPP phosphatase pair BcrC and UppP connects cell wall homeostasis during growth and sporulation with cell envelope stress response in Bacillus subtilis. Front Microbiol 8:2403. 10.3389/fmicb.2017.02403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bernard R, El Ghachi M, Mengin-Lecreulx D, Chippaux M, Denizot F. 2005. BcrC from Bacillus subtilis acts as an undecaprenyl pyrophosphate phosphatase in bacitracin resistance. J Biol Chem 280:28852–28857. 10.1074/jbc.M413750200. [DOI] [PubMed] [Google Scholar]
- 60.Inaoka T, Ochi K. 2012. Undecaprenyl pyrophosphate involvement in susceptibility of Bacillus subtilis to rare earth elements. J Bacteriol 194:5632–5637. 10.1128/JB.01147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pedreira T, Elfmann C, Stulke J. 2022. The current state of SubtiWiki, the database for the model organism Bacillus subtilis. Nucleic Acids Res 50:D875–D882. 10.1093/nar/gkab943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.FvdK BW, Kes MB, Luirink J, Hamoen LW. 2022. Transcriptome analyses using regulon, functional category, and operon information with GINtool. Biorxiv. 10.1101/2022.10.24.513545. [DOI]
- 63.Serizawa M, Kodama K, Yamamoto H, Kobayashi K, Ogasawara N, Sekiguchi J. 2005. Functional analysis of the YvrGHb two-component system of Bacillus subtilis: identification of the regulated genes by DNA microarray and northern blot analyses. Biosci Biotechnol Biochem 69:2155–2169. 10.1271/bbb.69.2155. [DOI] [PubMed] [Google Scholar]
- 64.Nikaido H. 2009. Small things considered: the limitations of LB medium. https://schaechter.asmblog.org/schaechter/2009/11/the-limitations-of-lb-medium.html.
- 65.Volland JM, Gonzalez-Rizzo S, Gros O, Tyml T, Ivanova N, Schulz F, Goudeau D, Elisabeth NH, Nath N, Udwary D, Malmstrom RR, Guidi-Rontani C, Bolte-Kluge S, Davies KM, Jean MR, Mansot JL, Mouncey NJ, Angert ER, Woyke T, Date SV. 2022. A centimeter-long bacterium with DNA contained in metabolically active, membrane-bound organelles. Science 376:1453–1458. 10.1126/science.abb3634. [DOI] [PubMed] [Google Scholar]
- 66.Larkin JM, Strohl WR. 1983. Beggiatoa, Thiothrix, and Thioploca. Annu Rev Microbiol 37:341–367. 10.1146/annurev.mi.37.100183.002013. [DOI] [PubMed] [Google Scholar]
- 67.Newman EB, Budman LI, Chan EC, Greene RC, Lin RT, Woldringh CL, D'Ari R. 1998. Lack of S-adenosylmethionine results in a cell division defect in Escherichia coli. J Bacteriol 180:3614–3619. 10.1128/JB.180.14.3614-3619.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Winterbourn CC. 1995. Toxicity of iron and hydrogen peroxide: the Fenton reaction. Toxicol Lett 82–83:969–974. 10.1016/0378-4274(95)03532-x. [DOI] [PubMed] [Google Scholar]
- 69.Aguirre JD, Culotta VC. 2012. Battles with iron: manganese in oxidative stress protection. J Biol Chem 287:13541–13548. 10.1074/jbc.R111.312181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.MacCain WJ, Kannan S, Jameel DZ, Troutman JM, Young KD. 2018. A defective undecaprenyl pyrophosphate synthase induces growth and morphological defects that are suppressed by mutations in the isoprenoid pathway of Escherichia coli. J Bacteriol 200. 10.1128/JB.00255-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gaballa A, Helmann JD. 2002. A peroxide-induced zinc uptake system plays an important role in protection against oxidative stress in Bacillus subtilis. Mol Microbiol 45:997–1005. 10.1046/j.1365-2958.2002.03068.x. [DOI] [PubMed] [Google Scholar]
- 72.Murdoch CC, Skaar EP. 2022. Nutritional immunity: the battle for nutrient metals at the host-pathogen interface. Nat Rev Microbiol 20:657–670. 10.1038/s41579-022-00745-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Si M, Zhao C, Burkinshaw B, Zhang B, Wei D, Wang Y, Dong TG, Shen X. 2017. Manganese scavenging and oxidative stress response mediated by type VI secretion system in Burkholderia thailandensis. Proc Natl Acad Sci USA 114:E2233–E2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Inaoka T, Matsumura Y, Tsuchido T. 1999. SodA and manganese are essential for resistance to oxidative stress in growing and sporulating cells of Bacillus subtilis. J Bacteriol 181:1939–1943. 10.1128/JB.181.6.1939-1943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Eide DJ. 2011. The oxidative stress of zinc deficiency. Metallomics 3:1124–1129. 10.1039/c1mt00064k. [DOI] [PubMed] [Google Scholar]
- 76.Arjes HA, Vo L, Dunn CM, Willis L, DeRosa CA, Fraser CL, Kearns DB, Huang KC. 2020. Biosurfactant-mediated membrane depolarization maintains viability during oxygen depletion in Bacillus subtilis. Curr Biol 30:1011–1022. 10.1016/j.cub.2020.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rodriguez Ayala F, Bartolini M, Grau R. 2020. The stress-responsive alternative sigma factor SigB of Bacillus subtilis and its relatives: an old friend with new functions. Front Microbiol 11:1761. 10.3389/fmicb.2020.01761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nicolas P, Mader U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, Becher D, Bisicchia P, Botella E, Delumeau O, Doherty G, Denham EL, Fogg MJ, Fromion V, Goelzer A, Hansen A, Hartig E, Harwood CR, Homuth G, Jarmer H, Jules M, Klipp E, Le Chat L, Lecointe F, Lewis P, Liebermeister W, March A, Mars RA, Nannapaneni P, Noone D, Pohl S, Rinn B, Rugheimer F, Sappa PK, Samson F, Schaffer M, Schwikowski B, Steil L, Stulke J, Wiegert T, Devine KM, Wilkinson AJ, van Dijl JM, Hecker M, Volker U, Bessieres P, et al. 2012. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science 335:1103–1106. 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 79.Ng W. 2021. Categorization of prophage genes in Bacillus subtilis 168 and assessing their relative importance through RNA-seq gene expression analysis. Biorxiv. https://www.biorxiv.org/content/10.1101/2021.10.26.466030v1.
- 80.Guo RT, Ko TP, Chen AP, Kuo CJ, Wang AH, Liang PH. 2005. Crystal structures of undecaprenyl pyrophosphate synthase in complex with magnesium, isopentenyl pyrophosphate, and farnesyl thiopyrophosphate: roles of the metal ion and conserved residues in catalysis. J Biol Chem 280:20762–20774. 10.1074/jbc.M502121200. [DOI] [PubMed] [Google Scholar]
- 81.Chang SY, Ko TP, Chen AP, Wang AH, Liang PH. 2004. Substrate binding mode and reaction mechanism of undecaprenyl pyrophosphate synthase deduced from crystallographic studies. Protein Sci 13:971–978. 10.1110/ps.03519904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chang SY, Ko TP, Liang PH, Wang AH. 2003. Catalytic mechanism revealed by the crystal structure of undecaprenyl pyrophosphate synthase in complex with sulfate, magnesium, and triton. J Biol Chem 278:29298–29307. 10.1074/jbc.M302687200. [DOI] [PubMed] [Google Scholar]
- 83.McCluskey K, Boudreault J, St-Pierre P, Perez-Gonzalez C, Chauvier A, Rizzi A, Beauregard PB, Lafontaine DA, Penedo JC. 2019. Unprecedented tunability of riboswitch structure and regulatory function by sub-millimolar variations in physiological Mg2. Nucleic Acids Res 47:6478–6487. 10.1093/nar/gkz316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jorgenson MA, Kannan S, Laubacher ME, Young KD. 2016. Dead-end intermediates in the enterobacterial common antigen pathway induce morphological defects in Escherichia coli by competing for undecaprenyl phosphate. Mol Microbiol 100:1–14. 10.1111/mmi.13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Barreteau H, Magnet S, El Ghachi M, Touze T, Arthur M, Mengin-Lecreulx D, Blanot D. 2009. Quantitative high-performance liquid chromatography analysis of the pool levels of undecaprenyl phosphate and its derivatives in bacterial membranes. J Chromatogr B Analyt Technol Biomed Life Sci 877:213–220. 10.1016/j.jchromb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 86.Higashi Y, Siewert G, Strominger JL. 1970. Biosynthesis of the peptidoglycan of bacterial cell walls. XIX. Isoprenoid alcohol phosphokinase. J Biol Chem 245:3683–3690. 10.1016/S0021-9258(18)62980-1. [DOI] [PubMed] [Google Scholar]
- 87.Gough DP, Kirby AL, Richards JB, Hemming FW. 1970. The characterization of undecaprenol of Lactobacillus plantarum. Biochem J 118:167–170. 10.1042/bj1180167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Umbreit JN, Stone KJ, Strominger JL. 1972. Isolation of polyisoprenyl alcohols from Streptococcus faecalis. J Bacteriol 112:1302–1305. 10.1128/jb.112.3.1302-1305.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Anderson RG, Hussey H, Baddiley J. 1972. The mechanism of wall synthesis in bacteria. The organization of enzymes and isoprenoid phosphates in the membrane. Biochem J 127:11–25. 10.1042/bj1270011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rasband WS. 1997. 2018. ImageJ, U.S. National Institutes of Health, Bethesda, Maryland. USA. https://imagej.nih.gov/ij/. [Google Scholar]
- 91.Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. 2019. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 37:907–915. 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33:290–295. 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4, Table S1 to S3 images, Tables S4 to S6, and Text S1. Download jb.00375-22-s0001.pdf, PDF file, 13.4 MB (13.7MB, pdf)
Tables S1 to S3 and raw data. Download jb.00375-22-s0002.xlsx, XLSX file, 0.8 MB (850.3KB, xlsx)
Data Availability Statement
The raw RNA-seq data may be accessed through the NCBI Gene Expression Omnibus at the following link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE219221.