Abstract
In contrast to conventional vaccines, DNA and other subunit vaccines exclusively utilize host cell molecules for transcription and translation of proteins. The adenine plus thymine content of Plasmodium falciparum gene sequences (∼80%) is much greater than that of Homo sapiens (∼59%); consequently, codon usage is markedly different. We hypothesized that modifying codon usage of P. falciparum genes encoded by DNA vaccines from that used by the parasite to those resembling mammalian codon usage would lead to increased P. falciparum protein expression in vitro in mouse cells and increased antibody responses in DNA-vaccinated mice. We synthesized gene fragments encoding the receptor-binding domain of the 175-kDa P. falciparum erythrocyte-binding protein (EBA-175 region II) and the 42-kDa C-terminal processed fragment of the P. falciparum merozoite surface protein 1 (MSP-142) using the most frequently occurring codon in mammals to code for each amino acid, and inserted the synthetic genes in DNA vaccine plasmids. In in vitro transient-expression assays, plasmids containing codon-optimized synthetic gene fragments (pS plasmids) showed greater than fourfold increased protein expression in mouse cells compared to those containing native gene fragments (pN plasmids). In mice immunized with 0.5, 5.0, or 50 μg of the DNA plasmids, the dose of DNA required to induce equivalent antibody titers was 10- to 100-fold lower for pS than for pN plasmids. These data demonstrate that optimizing codon usage in DNA vaccines can improve protein expression and consequently the immunogenicity of gene fragments in DNA vaccines for organisms whose codon usage differs substantially from that of mammals.
Malaria is a major cause of illness and death throughout the world, accounting for 150 to 270 million cases and 1.5 to 2.7 million deaths annually. DNA vaccination has recently emerged as a promising approach to development of vaccines for a wide range of pathogens, including malaria (7). In murine models, vaccination with DNA encoding antigens expressed in either the preerythrocytic or erythrocytic stages of the parasite has protected mice from challenge with infective sporozoites (2, 5, 18). Immunization of human volunteers with a DNA plasmid encoding the major coat protein of the sporozoite, the circumsporozoite protein of Plasmodium falciparum, induced antigen-specific cytotoxic T-lymphocyte (CTL) responses (25). However, the first generation of DNA vaccines did not induce optimal protective responses. In the murine model, protection is incomplete, and in humans, although a CTL response was induced, no antibody response was made (25). One approach to improving the response to DNA vaccines is to maximize the expression of malaria proteins from the vaccine plasmids.
A major obstacle to optimal expression of P. falciparum genes in transfected cells in the mammalian host may be the dramatic differences in codon usage between P. falciparum and mammals. The A+T content in the genome of P. falciparum is 80%, compared to 59% in humans. Each amino acid, with the exception of methionine and tryptophan, can be encoded by two to six different synonymous codons. The frequencies at which these synonymous codons are used depend on the level of protein expression and also differ among organisms. In general, highly expressed genes are biased towards codons that are recognized by the most abundant tRNA species in the organism (10). One measure of this bias is the codon adaptation index (CAI) (19), which measures the extent to which the codons used to encode each amino acid in a particular gene are those which occur most frequently in a reference set of highly expressed genes from an organism. A number of studies have found that there is a good correlation between the codon bias of a gene and its level of expression (1, 3, 6, 20, 26). Furthermore, a recent study showed a correlation between the CAI (based on mammalian codon usage) of a series of synthetic gene segments encoding the same T-cell epitope from Plasmodium yoelii and the level of expression in in vitro transfection assays and of T-cell responses in mice (15). Because the native sequences of Plasmodium genes have very low CAIs in mammalian cells, it is to be expected that expression of these native sequences will be suboptimal. We therefore synthesized gene segments encoding two P. falciparum vaccine candidate antigens using a set of codons designed to maximize the mammalian CAI and tested their in vitro expression and murine immunogenicity.
We chose two leading malaria vaccine candidate antigens. The first molecule is the 175-kDa P. falciparum erythrocyte-binding protein EBA-175, which is a parasite ligand that binds to its erythrocyte receptor sialic acids on glycophorin A for invasion of erythrocytes (22). A domain within EBA-175, identified as region II (RII), has been identified as the receptor-binding domain (24). Antibodies directed against RII block invasion of P. falciparum strains which have the ability to invade erythrocytes by distinct pathways in vitro (17). Immunization of mice, rabbits, and Aotus monkeys with an RII DNA vaccine plasmid encoded by the native gene (pNRII) induces RII-specific antibodies that block EBA-175 binding to erythrocytes and inhibit parasite growth in vitro (23). Aotus monkeys immunized against RII by a DNA prime/protein boost approach control blood-stage challenge infections (11).
The second vaccine target is the 42-kDa carboxy-terminal fragment of merozoite surface protein 1 (MSP-1) of P. falciparum. MSP-1 is synthesized as an approximately 200-kDa precursor protein and processed into several smaller fragments which reside on the merozoite surface. The 42-kDa MSP-1 C-terminal fragment (MSP-142) is further processed to a 33-kDa and a 19-kDa fragment at the time of merozoite invasion (4). Immunization of mice or monkeys with either the native full-length MSP-1 or the 19-kDa or 42-kDa carboxy-terminal fragment has induced protective immunity (12).
Here we report on the construction, in vitro expression, and immunogenicity of DNA vaccines using codon-optimized synthetic gene fragments encoding EBA-175 RII and MSP-142.
MATERIALS AND METHODS
DNA vaccine construction.
The DNA vaccine containing the native gene fragment for EBA-175 RII has been described previously (23). Synthetic DNA sequences encoding the 616 amino acids of EBA-175 RII (3D7 strain) and the 376 amino acids of MSP-142 (FVO strain) were designed by reverse translation of the amino acid sequence using DNAStar Lasergene99 (DNAStar, Inc. Madison, Wis.). The reverse translation used the most abundant codon for each amino acid found in a compendium of highly expressed human genes (8) (Table 1) except for S, which was reverse translated as UCC rather than AGC (the two codons are used at approximately equal frequency in highly expressed human genes). Within the back-translations encoding RII and MSP-142, BamHI or BamHI and Bg1II restriction enzyme sites, respectively, were removed by changing individual nucleotides without altering the amino acid sequence. The RII and MSP-142 genes were synthesized by Operon Technologies, Inc. (Alameda, Calif.), and by Retrogen (San Diego, Calif.), respectively.
TABLE 1.
Comparison of most abundant codon for each amino acid used by P. falciparum and highly expressed H. sapiens genes
| Amino acid | Most abundant codon
|
|
|---|---|---|
| P. falciparum | H. sapiens | |
| A | GCA | GCC |
| R | AGA | CGC |
| N | AAU | AAC |
| D | GAU | GAC |
| C | UGU | UGC |
| Q | CAA | CAG |
| E | GAA | GAG |
| G | GGU | GGC |
| H | CAU | CAC |
| I | AUU | AUC |
| L | UUA | CUG |
| K | AAA | AAG |
| Ma | AUG | AUG |
| F | UUU | UUC |
| P | CCA | CCC |
| S | AGU | AGC |
| T | ACA | ACC |
| Wa | UGG | UGG |
| Y | UAU | UAC |
| V | GUA | GUG |
Encoded by single codons.
The synthetic RII and synthetic and native MSP-142 genes were amplified by PCR and cloned into the DNA vaccine plasmid VR1020 (14). The synthetic gene fragment encoding RII was PCR amplified using the following primers: sense, 5′-ATCGGGATCCGGCCGCAACACCTCCTCC-3′, and antisense, 5′-ATCGGGATCCTCAGGAGGTCTGCTCGTTGTT-3′, and directly cloned into a Bg1II restriction site in plasmid VR1020. The native and synthetic gene fragments encoding the 42-kDa fragment of MSP-1 were PCR amplified using the following primers: sense, 5′-ATGGATCCGGAGAAGCAGTAACTCCTTCCGTAATT-3′, and antisense, 5′-GATGGATCCTTAAATGAAACTGTATAATATTAACAT-3′, and sense, 5′-GGTACCGGATCCGCCGTGACCCCCTCCGTGATCGAC-3′, and antisense, 5′-GATCTGGATCCTTAGATGAAGGAGTACAGGATCAG-3′, respectively. The PCR-amplified gene fragments of MSP-142 were cloned into a BamHI restriction site in plasmid VR1020. The DNA sequence of the junction site of the inserted gene or the complete insert was determined to ensure that an open reading frame was maintained. EBA-175 RII (pSRII) and MSP-142 (Synthetic, pSMSP-142, and native, pNMSP-142) DNA vaccines were purified by methods described previously (23). Endotoxin levels detected using either the Kinetic-QCL endotoxin detection assay (BioWhittaker, Walkersville, Md.) or the Limulus amebocyte assay (Associates of Cape Cod, Cape Cod, Mass.) were less than 10 endotoxin units/mg of plasmid DNA.
In vitro transfection studies.
Mouse melanoma cells (VM92) were transiently transfected with plasmids separately with Lipofectamine following the manufacturer's suggestions (Life Technologies, Gaithersburg, Md.). Recombinant RII that was secreted into culture supernatant or MSP-142 localized within the cell cytosol was detected and quantitated as a chemiluminescent signal by using specific RII monoclonal antibody (MAb) R217 (D. L. Narum, unpublished data) or MSP-142-specific conformation-dependent MAb 5.2 (21) and a commercially obtained chemiluminescence-linked Western blot kit (Western-Light, Tropix, Bedford, Mass.) according to the manufacturer's protocol. Chemiluminescent signals were detected by exposure of the processed membrane to autoradiographic film (Hyperfilm-ECL; Amersham Life Sciences Inc., Cleveland, Ohio). Quantitation of enhanced chemiluminescence Western blots was performed on a Molecular Imager FX (Bio-Rad, Hercules, Calif.), and units of intensity are reported as counts per square millimeter.
Animals, immunizations, and antibody assays.
Groups of outbred ICR (Harlan Sprague Dawley, Inc., Indianapolis, Ind.) or CDI (Charles River, Raleigh, N.C.) mice were inoculated intradermally in the tail with a total of 100 μg of plasmid DNA thrice at 3-week intervals and bled 2 weeks after the final boost essentially as previously described (23). Sera were assessed for RII- or MSP142-specific antibodies by enzyme-linked immunosorbent assay (ELISA) and indirect immunofluorescent antibody test (IFAT) as previously described (13, 23). The mouse experiments reported herein were conducted according to the principles set forth in the Guide for the Care and Use of Laboratory Animals (Institute of Laboratory Animal Resources, National Research Council, National Academy Press, 1996).
Analysis of codon usage.
The most commonly used codons for H. sapiens and P. falciparum were determined from the Codon Usage Tabulated from Genbank website (www.kazusa.or.jp/codon/) (16). CAIs (19) were calculated using the Codon W software (J. Peden; available as freeware at www.molbiol.ox.ac.uk/cu/codonW.html) with a user-defined set of codon relative adaptiveness values (wcodon) based on codon usage in a compendium of highly expressed human genes (8).
Statistical analysis.
Comparisons between antibody responses were performed by two-tailed t test on log-transformed titers except when all members of a group had no detectable response, in which case the Mann-Whitney test was used.
RESULTS AND DISCUSSION
Codon optimization.
In order to test the hypothesis that codon optimization would improve the expression and immunogenicity of malaria DNA vaccine plasmids, we designed codon-optimized gene fragments encoding EBA-175 RII and MSP-142. There are large differences between the most frequently used codons in P. falciparum genes and in highly expressed human genes (Table 1). For every amino acid which can be encoded by more than one codon, the most frequently used codon in P. falciparum is different from that used in humans. Approximately one in three nucleotides was changed to optimize the sequences, and the G + C content was raised from approximately 26% to approximately 56% in the optimized sequences (Table 2). The CAI of the codon-optimized pSRII and pSMSP142 genes was increased from <0.28 to ≥0.98 (maximum possible value, 1.0).
TABLE 2.
Comparison of native and optimized vaccine candidate sequences
| Gene | No. of native nucleotides altered/ total (% altered) | G+C content (%), native/optimized | CAI, native/ optimized |
|---|---|---|---|
| EBA-175 RII | 591/1,848 (32.0) | 27.3/56.7 | 0.276/0.988 |
| MSP-142 | 370/1,128 (32.8) | 25.7/56.8 | 0.245/0.980 |
In vitro protein expression.
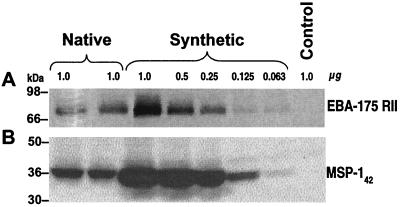
We next asked whether the greatly increased CAI of the Plasmodium gene fragments in the optimized codon plasmids would be associated with increased protein expression. Comparison of protein levels expressed by DNA vaccine plasmids pNRII and pSRII or pNMSP-142 and pSMSP-142 revealed that levels from optimized-codon gene fragments were greater than levels from native genes (Fig. 1). A comparison of the in vitro expression levels comparing secreted RII or cell-based MSP-142 for pNRII and pNMSP-142 to the expression levels of pSRII and pSMSP-142 is shown in Fig. 1A and B. The results indicated that 0.25 μg of pSRII or pSMSP-142 expressed amounts of protein comparable to 1 μg of pNRII or pNMSP-142, respectively. For example, quantitating RII expression by correlating the levels of RII protein with band intensities demonstrated that 0.125 and 0.25 μg of pSRII expressed 1,919 and 4,361 counts/mm2, respectively, compared to 1 μg of pNRII, which expressed an average of 3,298 counts/mm2 for the duplicate samples (Fig. 1A). This represents a greater than fourfold increase in protein expression. The proteins encoded by the vaccine plasmids retained conformational epitopes, because RII and MSP-142 proteins produced in in vitro transfection experiments were recognized by RII-specific and MSP-1-specific growth-inhibitory MAbs R217 (Narum, unpublished) and 12.10 (4), respectively, which recognize reduction-sensitive epitopes (data not shown).
FIG. 1.
Protein expression in mouse melanoma cells transfected with various quantities of plasmids expressing native or synthetic RII of EBA-175 or MSP-142. pSRII and pSMSP-142 expressed greater quantities of recombinant protein than pNRII and pNMSP-142 (A and B, respectively).
In vivo immunogenicity.
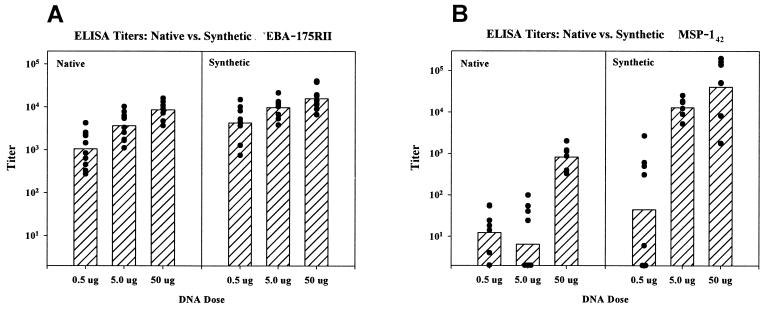
Outbred ICR mice were immunized with 0.5, 5.0, and 50 μg of EBA-175 RII plasmids. At each dose of DNA tested, mice immunized with pSRII had statistically significantly higher ELISA titers against recombinant RII (Fig. 2A) than mice immunized with pNRII (P = 0.024, 0.003, and 0.003 at 50, 5.0, and 0.5 μg, respectively, by two-tailed t-test on log-transformed titers). Similar results were obtained against whole parasites in IFAT tests against parasite-infected erythrocytes (data not shown).
FIG. 2.
Antibody titers in outbred mice immunized with DNA vaccines expressing native or synthetic EBA-175 RII or MSP142. ELISA results are reported as the interpolated reciprocal dilution estimated to give an optical density of 0.5. Group sizes were as follows: for pSRII and pNRII, 10 ICR mice/group; for pSMSP-142 and pNMSP-142, 8 CD-1 mice/group, except for 50-μg pNMSP-142, which had 6 mice. Bars represent geometric mean titers, and solid circles represent individual responses.
In outbred CD-1 mice, pSMSP-142 was dramatically more immunogenic than the corresponding pNMSP-142 (Fig. 2B). At each DNA dose, the ELISA titers induced by pSMSP-142 were higher than those induced by pNMSP-142. Essentially no antibodies were detected with the native construct at the two lower DNA doses, while the synthetic construct induced geometric mean ELISA titers of 44 (0.5 μg), 12,883 (5 μg), and 40,310 (50 μg). At the 50-μg dose, the geometric mean ELISA titer induced by pSMSP-142 was approximately 50-fold higher than that induced by pNMSP-142 (P < 0.001, by two-tailed t test on log-transformed titers; 95% confidence interval for fold increase, 9 to 268). Similar results were obtained in IFAT assays (data not shown). The geometric mean IFAT titer in sera from mice immunized with 0.5 μg of pSRII was similar to that induced by 50.0 μg of pNRII. Thus, the same antibody response was induced by 100-fold less of the plasmid expressing the codon-optimized synthetic gene.
We have demonstrated that codon optimization of two different P. falciparum antigen genes leads to substantial improvement in in vitro expression by mammalian cells and comparable increases in antibody responses in outbred mice, which is consistent with similar codon optimization studies reported for genes encoded by the human immunodeficiency virus (1, 26). There were two characteristics to the improved immunogenicity. At any single dose, the codon-optimized constructs induced higher antibody titers than the native constructs. In addition, 10- to 50-fold less of the synthetic construct was required to induce antibody titers comparable to those induced by the native construct at the highest DNA dose tested. By enabling the use of reduced DNA doses for individual antigen genes, codon optimization may allow the combination of large numbers of plasmids encoding different vaccine targets into a single vaccine cocktail. This will be essential for any vaccine strategy based on taking advantages of the hundreds of potential new vaccine targets emerging from the Malaria Genome Project (9). Furthermore, to date, while DNA vaccines have been shown to induce T-cell responses in humans (25), they have not been effective in eliciting antibody responses. This may be a function of quantity of protein, and improving protein expression may lead to improved immune responses even at the highest doses of DNA.
Codon optimization may improve expression of P. falciparum genes in mammalian cells by a number of possible mechanisms. For example, the use of common mammalian codons may prevent the depletion of rare tRNAs or may kinetically improve the rate of translation by avoiding the requirement for tRNAs present at stable but low concentrations, or the global reduction in A + T content may affect mRNA stability in mammalian cells or the rate at which mRNA is exported to the cytoplasm. A clearer understanding of the mechanism of improved expression would allow more rational design of further optimized synthetic genes in the future.
In summary, we report that plasmids carrying P. falciparum synthetic gene fragments encoding EBA-175 RII and MSP-142 with mammalian codon usage gave increased levels of protein expression and enhanced immunogenicity in outbred mice compared to plasmids with native gene fragments. Preclinical studies in Aotus monkeys comparing these synthetic and native DNA vaccine plasmids are under way, and plasmids suitable for human use are being manufactured.
ACKNOWLEDGMENTS
D.L.N. and S.K. contributed equally to this work.
We gratefully acknowledge Peter Hobart (Vical, Inc., San Diego, Calif.) for providing the DNA vaccine plasmid, Trevor Jones (Malaria Program, NMRC) for assistance with the statistical analysis, and Tony Holder (NIMR, Mill Hill, London) for supplying anti-MSP-1 MAb 12.10.
This work was supported by Phase II Small Business Innovative Research grant AI36758-02 awarded to B.K.L.S. and by funds from the Naval Medical Research Center Work Unit (STOF 6.2.622787A.0101.870.EFX).
REFERENCES
- 1.Andre S, Seed B, Eberle J, Schraut W, Bultmann A, Haas J. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J Virol. 1998;72:1497–1503. doi: 10.1128/jvi.72.2.1497-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker S I, Wang R, Hedstrom R C, Aguiar J C, Jones T R, Hoffman S L, Gardner M J. Protection of mice against Plasmodium yoelii sporozoite challenge with P. yoelii merozoite surface protein 1 DNA vaccines. Infect Immun. 1998;66:3457–3461. doi: 10.1128/iai.66.7.3457-3461.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bennetzen J L, Hall B D. Codon selection in yeast. J Biol Chem. 1982;257:3026–3031. [PubMed] [Google Scholar]
- 4.Blackman M J, Heidrich H G, Donachie S, McBride J S, Holder A A. A single fragment of a malaria merozoite surface protein remains on the parasite during red cell invasion and is the target of invasion- inhibiting antibodies. J Exp Med. 1990;172:379–382. doi: 10.1084/jem.172.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doolan D L, Sedegah M, Hedstrom R C, Hobart P, Charoenvit Y, Hoffman S L. Circumventing genetic restriction of protection against malaria with multigene DNA immunization: CD8+ cell-, interferon gamma-, and nitric oxide-dependent immunity. J Exp Med. 1996;183:1739–1746. doi: 10.1084/jem.183.4.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gouy M, Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982;10:7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gurunathan S, Klinman D M, Seder R A. DNA vaccines: immunology, application, and optimization. Annu Rev Immunol. 2000;18:927–974. doi: 10.1146/annurev.immunol.18.1.927. [DOI] [PubMed] [Google Scholar]
- 8.Haas J, Park E C, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996;6:315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 9.Hoffman S L, Rogers W O, Carucci D J, Venter J C. From genomics to vaccines: malaria as a model system. Nat Med. 1998;4:1351–1353. doi: 10.1038/3934. [DOI] [PubMed] [Google Scholar]
- 10.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 11.Jones T R, Narum D L, Gozalo A S, Aguiar J, Fuhrmann S R, Liang H, Haynes J D, Moch J K, Lucas C, Luu T, Magill A J, Hoffman S L, Sim B K L. Protection of aotus monkeys by Plasmodium falciparum EBA-175 region II DNA prime-protein boost immunization regimen. J Infect Dis. 2001;183:303–312. doi: 10.1086/317933. [DOI] [PubMed] [Google Scholar]
- 12.Kumar S, Kaslow D, Hoffman S L. Overview of malaria vaccine development efforts. Handb Exp Pharmacol. 1999;133:397–442. [Google Scholar]
- 13.Kumar S, Yadava A, Keister D B, Tian J H, Ohl M, Perdue-Greenfield K A, Miller L H, Kaslow D C. Immunogenicity and in vivo efficacy of recombinant Plasmodium falciparum merozoite surface protein-1 in Aotus monkeys. Mol Med. 1995;1:325–332. [PMC free article] [PubMed] [Google Scholar]
- 14.Luke C J, Carner K, Liang X, Barbour A G. An OspA-based DNA vaccine protects mice against infection with Borrelia burgdorferi. J Infect Dis. 1997;175:91–97. doi: 10.1093/infdis/175.1.91. [DOI] [PubMed] [Google Scholar]
- 15.Nagata T, Uchijima M, Yoshida A, Kawashima M, Koide Y. Codon optimization effect on translational efficiency of DNA vaccine in mammalian cells: analysis of plasmid DNA encoding a CTL epitope derived from microorganisms. Biochem Biophys Res Commun. 1999;261:445–451. doi: 10.1006/bbrc.1999.1050. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from the international DNA sequence databases: its status 1999. Nucleic Acids Res. 1999;27:292. doi: 10.1093/nar/27.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Narum D L, Haynes J D, Fuhrmann S, Moch K, Liang H, Hoffman S L, Sim B K L. Antibodies against the Plasmodium falciparum receptor binding domain of EBA-175 block invasion pathways that do not involve sialic acids. Infect Immun. 2000;68:1964–1966. doi: 10.1128/iai.68.4.1964-1966.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sedegah M, Hedstrom R, Hobart P, Hoffman S L. Protection against malaria by immunization with plasmid DNA encoding circumsporozoite protein. Proc Natl Acad Sci USA. 1994;91:9866–9870. doi: 10.1073/pnas.91.21.9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharp P M, Li W H. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharp P M, Li W H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol. 1986;24:28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- 21.Siddiqui W A, Tam L Q, Kan S C, Kramer K J, Case S E, Palmer K L, Yamaga K M, Hui G S. Induction of protective immunity to monoclonal-antibody-defined Plasmodium falciparum antigens requires strong adjuvant in Aotus monkeys. Infect Immun. 1986;52:314–318. doi: 10.1128/iai.52.1.314-318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sim B K L. EBA-175: an erythrocyte-binding ligand of Plasmodium falciparum. Parasitol Today. 1995;11:213–217. doi: 10.1016/0169-4758(95)80080-8. [DOI] [PubMed] [Google Scholar]
- 23.Sim B K L, Narum D L, Liang H, Fuhrmann S R, Obaldia III N, Gramzinski R, Aguiar J, Haynes J D, Moch J K, Hoffman S L. Induction of biologically active antibodies in mice, rabbits, and monkeys by Plasmodium falciparum EBA-175 region II DNA vaccine. Mol Med, 2001;7:247–254. [PMC free article] [PubMed] [Google Scholar]
- 24.Sim B K L, Chitnis CE, Wasniowska K, Hadley TJ, Miller LH. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science. 1994;264:1941–1944. doi: 10.1126/science.8009226. [DOI] [PubMed] [Google Scholar]
- 25.Wang R, Doolan D L, Le T P, Hedstrom R C, Coonan K M, Charoenvit Y, Jones T R, Hobart P, Margalith M, Ng J, Weiss W R, Sedegah M, de Taisne C, Norman J A, Hoffman S L. Induction of antigen-specific cytotoxic T lymphocytes in humans by a malaria DNA vaccine. Science. 1998;282:476–480. doi: 10.1126/science.282.5388.476. [DOI] [PubMed] [Google Scholar]
- 26.zur Megede J, Chen M C, Doe B, Schaefer M, Greer C E, Selby M, Otten G R, Barnett S W. Increased expression and immunogenicity of sequence-modified human immunodeficiency virus type 1 gag gene. J Virol. 2000;74:2628–2635. doi: 10.1128/jvi.74.6.2628-2635.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]