Abstract
Despite being frequently observed in cancer cells, chromosomal instability (CIN) and its immediate consequence, aneuploidy, trigger adverse effects on cellular homeostasis that need to be overcome by anti-stress mechanisms. As such, these safeguard responses represent a tumor-specific Achilles heel, since CIN and aneuploidy are rarely observed in normal cells. Recent data have revealed that epitranscriptomic marks catalyzed by RNA-modifying enzymes change under various stress insults. However, whether aneuploidy is associated with such RNA modifying pathways remains to be determined. Through an in silico search for aneuploidy biomarkers in cancer cells, we found TRMT61B, a mitochondrial RNA methyltransferase enzyme, to be associated with high levels of aneuploidy. Accordingly, TRMT61B protein levels are increased in tumor cell lines with an imbalanced karyotype as well as in different tumor types when compared to control tissues. Interestingly, while TRMT61B depletion induces senescence in melanoma cell lines with low levels of aneuploidy, it leads to apoptosis in cells with high levels. The therapeutic potential of these results was further validated by targeting TRMT61B in transwell and xenografts assays. We show that TRM61B depletion reduces the expression of several mitochondrial encoded proteins and limits mitochondrial function. Taken together, these results identify a new biomarker of aneuploidy in cancer cells that could potentially be used to selectively target highly aneuploid tumors.
Subject terms: Cancer, Biomarkers
Introduction
Aneuploidy, the immediate consequence of chromosomal instability (CIN) that describes an imbalanced karyotype, is frequently detected in solid and hematopoietic tumors [1, 2]. Accumulated evidence indicates that particular aneuploidy conditions drive carcinogenesis [3]. Significantly, the genetic diversity associated with CIN and aneuploidy might lie behind the development of drug resistance, high metastatic incidence and low survival rates in many cancer patients [4–8]. Paradoxically, aneuploidy is not only associated with cancer promotion but also with the suppression of malignant growth [9]. In the short term, karyotypic changes normally results in a poor cellular fitness due to different stress factors, such as proteoxic and genotoxic burden, overloaded endoplasmic reticulum, augmented glycolytic flux and mitochondrial activity, and an increase in reactive oxygen species (ROS) [10–14]. However, cancer cells are generally characterized by an aneuploid and proliferative phenotype, which suggests that specific timely gains or losses of chromosomes should ultimately promote adaptation once the initial challenge of gene imbalance has been overcome. In fact, different studies have shown that aneuploidy could facilitate the selection of cells with specific karyotypic compositions that allow them to tolerate high stress conditions and adapt to an ever-changing environment [15–19]. These aneuploidy-induced responses include upregulation of autophagy, DNA repair mechanisms, and antioxidant levels as well as more efficient bioenergetic and biosynthetic processes [13, 20–23]. Importantly, aneuploidy might make cancer cells more sensitive to certain antitumoral therapies that exploit some of these aneuploidy coping mechanisms [24, 25]. Despite this knowledge, the mechanism whereby aneuploidy overcomes deleterious effects and promotes cell selection is not fully understood [9].
Over 160 RNA modifications involving post-transcriptional changes in the chemical composition of different classes of nuclear and mitochondrial RNAs have been described to date, with tRNAs and rRNAs containing the most numerous and chemically diverse modified ribonucleosides [26, 27]. These modifications range from the addition of a single methyl group in a nitrogenous base or sugar residue to more complex molecular transformations. In this way, RNA function can be fine-tuned at different levels depending on the type, location and target of the modification (reviewed in [28, 29]). Thus, certain RNA modifications are required for proper processing, folding, and stability during RNA biogenesis, while others play a crucial role in RNA-driven protein synthesis by modulating translation efficiency and fidelity [30–33]. Many of these RNA composition changes are neither static nor stable [34, 35]; rather, they are dynamic marks introduced and removed in a reversible process catalyzed by both epitranscriptomic “writer” and “eraser” enzymes, respectively, which can be potentially modulated in response to different cellular stress cues [34, 35]. However, this stress-induced reprogramming of RNA modifications and, specifically, its likely association with aneuploidy and therapeutic utility have yet to be explored.
In an in silico bioinformatics study, we identified the RNA methyltransferase enzyme TRNA Methyltransferase 61B (TRMT61B) as a potential aneuploidy biomarker. In this study, we examine in detail the connection between this molecule and highly aneuploid cancers and explore its potential therapeutic value. Although it is encoded by the nuclear genome, TRMT61B is predominantly localized in the mitochondrial compartment, where it catalyzes methylation at the N1 position of specific adenosine residues (referred to as m1A) present in the 3 major types of mitochondrial RNA (mt-RNA) [36–38]. Although the molecular consequences of TRMT61B-mediated methylation on RNA biology have received some attention, its biological function is still poorly understood [36–38]. Very recent studies suggest that this enzyme is involved in protein synthesis regulation, which is consistent with the role of TRMT61B mtRNA targets as critical players in the mitochondrial translation mechanism [36–38]. However, a large body of research is required to address this issue in depth.
Our in silico and in vitro assays reveal a strong direct correlation between the degree of aneuploidy and TRMT61B protein levels in both NCI-60 cancer cell line collection and a more specific panel of human melanoma cell lines. Furthermore, TRMT61B is overexpressed in different tumor types compared to normal tissues, and is positively associated with aneuploidy in certain human cancers. We also demonstrate that cancer cells with high levels of aneuploidy are addicted to TRMT61B, since its elimination, using shRNA or CRISPR technology, has an antiproliferative effect and is detrimental to mitochondrial function. Finally, xenotransplant experiments performed in both immunodeficient mice and zebrafish embryos show that TRMT61B deficient cells with high aneuploidy levels are less effective in tumor formation. These findings heighten interest in this enzyme as a potential cancer target.
Results
TRMT61B as a novel cancer and aneuploidy biomarker
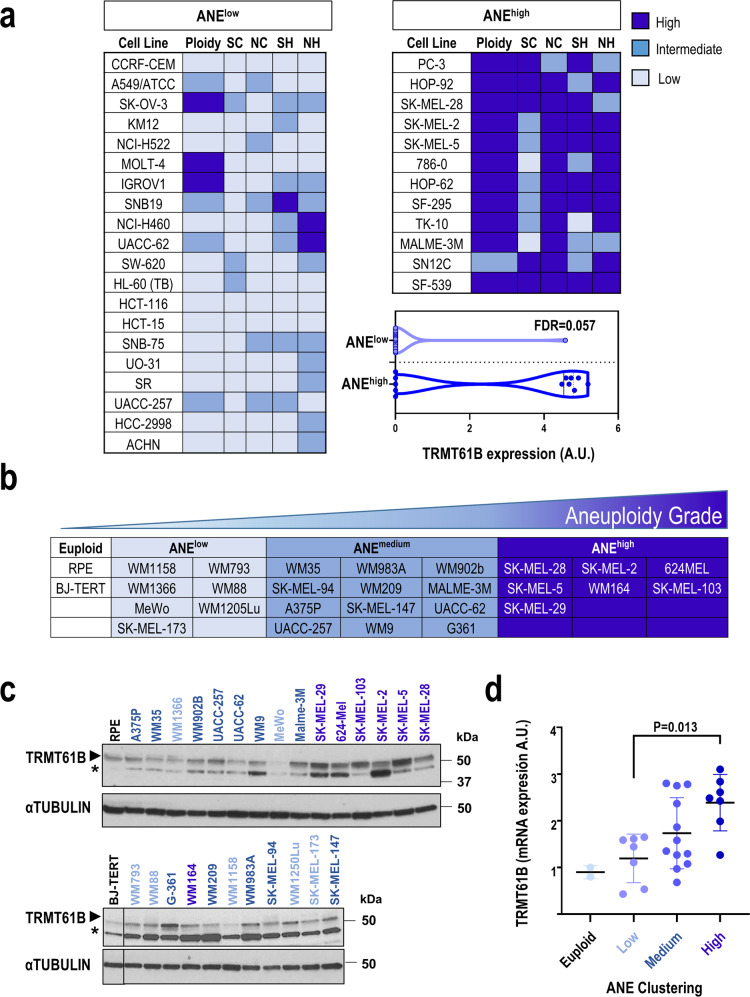
To discover novel molecular traits related to high aneuploidy, we used publicly available data on the NCI-60 human cancer cell line panel [39–46]. Using DNA ploidy information and 4 different structural and numerical karyotypic features (Supplementary Table 1) [44], we performed unsupervised hierarchical analysis to stratify the NCI-60 cell lines included in the panel into 3 groups: low, intermediate, and high aneuploidy levels (Sup. Figure 1a). We compared the proteomic signatures [43] of the group with the highest (ANEhigh) vs. the lowest (ANElow) aneuploidy levels (Fig. 1a) to identify proteins strongly associated with aneuploidy. Taking FDR < 0.15 as the threshold of significance, the mitochondrial RNA methyltransferase TRMT61B was the only protein found to be differentially altered, being overexpressed in the ANEhigh group (Fig. 1a). Interestingly, mRNA levels of TRMT61B were also significantly upregulated (FDR = 0.011) in ANEhigh NCI-60 cancer cell lines vs. the ANElow cell lines.
Fig. 1. TRMT61B protein expression positively correlates with aneuploidy levels in human cell lines.
a Following bioinformatics analysis, tumor cell lines included in the NCI-60 panel were grouped into 3 clusters according to different karyotypic features (SC, SH, NC, NH, and ploidy) ranging from low (ANElow) to high (ANEhigh) aneuploidy levels. Cell lines belonging to the most extreme groups in terms of aneuploidy levels are listed. Differential protein expression analysis in cell lines included in these 2 clusters allowed us to identify the TRMT61B gene, a mitochondrial RNA methyltransferase, whose protein expression levels were upregulated in ANEhigh vs. ANElow (FDR = 0.057). SC: Structural Complexity; SH: Structural Heterogeneity; NC: Numerical Complexity; NH: Numerical Heterogeneity. b A collection of 26 human melanoma cell lines and 2 euploid controls were karyotypically characterized for some of the foregoing karyotypic parameters (SC, NC, and ploidy), divided in 4 clusters with null (euploid group) or increased rate of aneuploidy (ANEhigh, ANEmedium, and ANElow). c TRMT61B expression was analyzed by western blot for each of the cell lines in three independent experiments. αTUBULIN was used as loading control for the normalization of TRMT61B protein expression. One representative western blot is shown. d As shown in the dotplot, ANEhigh shows statistically significant upregulation of TRMT61B protein expression compared to ANElow group. Triplicates were considered in the quantification analysis. Error bars represent standard error. *P < 0.05; **0.001 < P < 0.01; ***P < 0.001 (Student’s t-test; unpaired, 2-tailed). One-way ANOVA for multiple comparisons was used to determine statistically significant differences. Asterisks indicate non-specific bands. Lighter to darker blue color code indicates lower to higher aneuploidy grades.
To validate these in silico findings, a large collection of 26 primary and metastatic melanoma cell lines was karyotypically characterized and subjected to immunoblotting to determine TRMT61B expression (Fig. 1b, c). Cell lines were clustered in 4 groups in accordance with their structural and numerical chromosome alterations and their ploidy (Supplementary Table 2, Sup. Figure 1b) (euploid, ANElow, ANEmedium and ANEhigh). The euploid and ANEhigh groups had the lowest and highest aneuploidy levels, respectively (Supplementary Table 2, Fig. 1b). Quantification of TRMT61B expression showed a positive correlation between aneuploidy grade and TRMT61B protein levels (Fig. 1d).
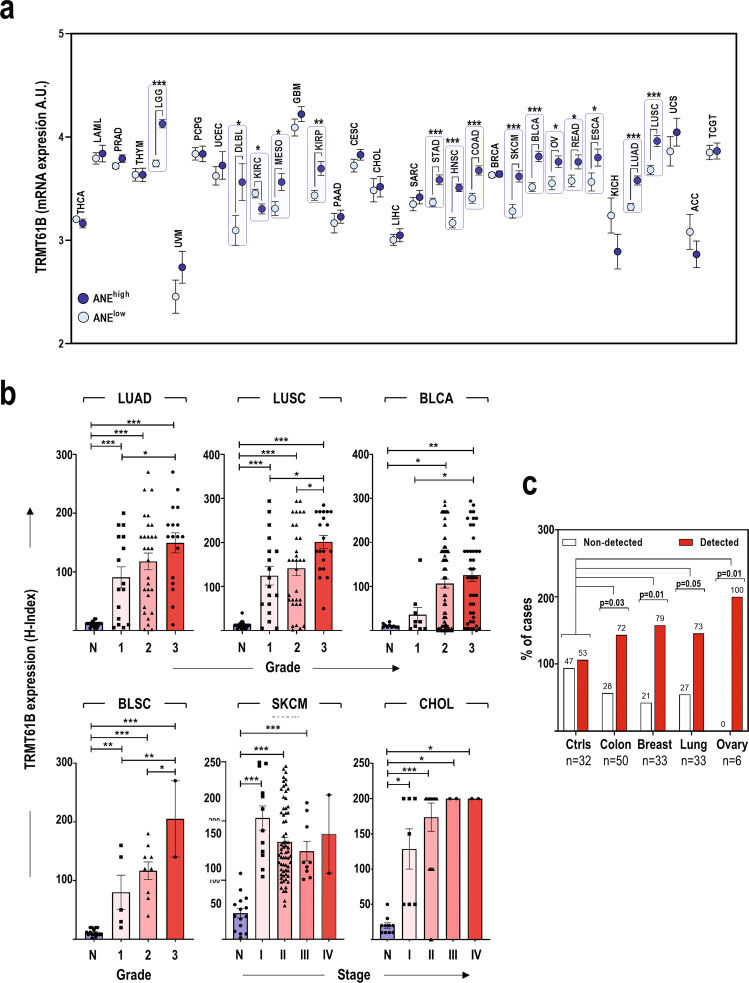
To explore the clinical relevance of these findings, we analyzed TRMT61B expression in different cancer types. First, we investigated TRMT61B mRNA expression in silico using publicly available information. TRMT61B mRNA is frequently over-expressed in a wide range of cancer types from the TCGA tumor collection (Sup. Figure 2a). Next, we studied the potential connection between TRMT61B mRNA expression and aneuploidy scores of the same TCGA tumor collection previously characterized in terms of aneuploidy levels (Supplementary Table 3) [15]. A significant positive association between both variables was detected in 14 out of the 33 tumor types analyzed (Fig. 2a). Although no significant differences were found among the rest of tumor types, a positive and significant correlation between TRMT61B mRNA expression and aneuploidy grade was observed when all the TCGA tumors were individually considered in the analysis (Sup. Figure 2b).
Fig. 2. TRMT61B expression is positively associated with cancer and aneuploidy levels in human cancers.
a Examination of TRMT61B expression in the TCGA collection reveals significantly increased mRNA levels of this RNA methyltransferase in samples with higher aneuploidy scores in 14 out of the 33 tumor types. b TRMT61B protein levels are elevated in lung (LUAD and LUSC) and bladder (BLCA, BLSC) cancer as well as in melanoma (SKCM) and cholangiocarcinomas (CHOL) when compared with the corresponding healthy tissues (N). A significant direct relationship with histopathological grade was also detected in some tumor types (BLCA, BLSC, LUAD, and LUSC), especially in the case of the highest tumor grade. c TRMT61B protein is detected in plasmas from cancer patients in a higher percentage of cases than in control plasmas. Error bars represent standard error in all the cases. *P < 0.05; **0.001 < P < 0.01; ***P < 0.001 (Student’s t-test; unpaired, 2-tailed). One-way ANOVA for multiple comparisons was used to determine statistically significant differences in (b). Chi-square test was used in (c) to compare control vs cancer groups and calculate P values. Tumor abbreviations: THCA Thyroid carcinoma, LAML Acute Myeloid Leukemia, PRAD Prostate adenocarcinoma, THYM Thymoma, LGG Brain Lower Grade Glioma, UVM Uveal Melanoma, PCPG Pheochromocytoma and Paraganglioma, UCEC Uterine Corpus Endometrial Carcinoma, DLBC Lymphoid Neoplasm Diffuse Large B-cell Lymphoma, KIRC Kidney renal clear cell carcinoma, MESO Mesothelioma, GBM Glioblastoma multiforme, KIRP Kidney renal papillary cell carcinoma, PAAD Pancreatic adenocarcinoma, CESC Cervical squamous cell carcinoma and endocervical adenocarcinoma, CHOL Cholangiocarcinoma, LIHC Liver hepatocellular carcinoma, SARC Sarcoma, STAD Stomach adenocarcinoma, HNSC Head and Neck squamous cell carcinoma, COAD Colon adenocarcinoma, BRCA Breast invasive carcinoma, SKCM Skin Cutaneous Melanoma, BLCA Bladder Urothelial Carcinoma, BLSC Bladder Squamous Cell Carcinoma, OV Ovarian serous cystadenocarcinoma, READ Rectum adenocarcinoma, ESCA Esophageal carcinoma, KICH Kidney Chromophobe, LUAD Lung adenocarcinoma, LUSC Lung squamous cell carcinoma, UCS Uterine Carcinosarcoma, ACC Adrenocortical carcinoma, TGCT Testicular Germ Cell Tumors.
These bioinformatics results prompted us to evaluate TRMT61B protein levels in published datasets and tissue microarrays (TMAs) that represent cancers with different aneuploidy grades (Fig. 2b, Sup. Figure 2c). Using proteomic information deposited in the UALCAN platform [47, 48], we analyzed TRMT6B protein expression in 10 different types of human cancer (Sup. Figure 2c). A significant upregulation of TRMT61B levels was detected in the neoplastic lesions compared with healthy tissues in the majority (6 out of 10) of the cancer types (Sup. Figure 2c). TMA analysis validated and extended this in silico observation. TRMT61B protein upregulation was detected in lung squamous cell carcinomas and adenocarcinomas, bladder cell carcinomas and squamous cell carcinomas, skin cutaneous melanoma, and cholangiocarcinomas (Fig. 2b; Sup. Figure 3). Remarkably, a significant positive correlation was observed between TRM61B expression and histopathological tumor grade in most TMAs (Fig. 2b). Finally, we explored the potential of TRMT61B as a cancer biomarker in liquid biopsies from plasma of both healthy and cancer patients. Although TRMT61B was found in 53% of controls, it was also detected in more than 70% of plasmas from colon, ovary, lung, and breast cancer patients (Fig. 2c).
In summary, these results indicate that there is a positive association tendency between TRMT61B expression, higher aneuploidy and cancer development in a wide range of cells and tumors.
TRMT61B overexpression does not enhance tumor growth and does not alter the chromosomal landscape
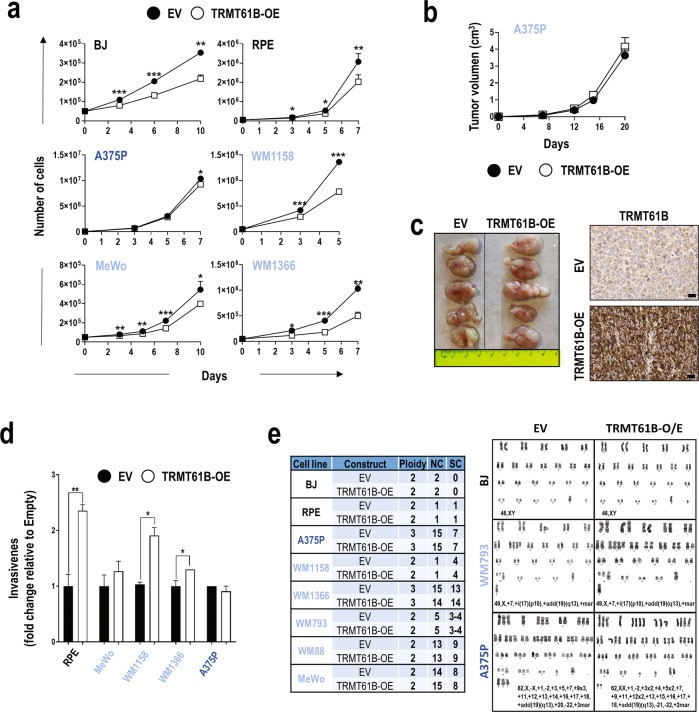
The fact that tumor cells with increased aneuploidy tend to express larger amounts of TRMT61B suggests that an initial abnormal up-regulation of TRMT61B might induce aneuploidy and, subsequently, tumor growth. To test this hypothesis, euploid and ANElow/medium melanoma cell lines expressing reduced protein amounts of TRMT61B were infected with a lentiviral construct encoding a FLAG-tagged, wild-type TRMT61B (Sup. Figure 4a–d). Next, we analyzed the 2 main features associated with malignant transformation, cell proliferation, and invasiveness. Elevated TRMT61B levels were associated with a slight, though significant, decrease in cell proliferation rates (Fig. 3a). However, xenografting showed no significant change in the tumor growth capacity of cells overexpressing TRMT61B when compared with their respective controls (Fig. 3b, c). Invasiveness, however, showed the opposite effect in 3 out of the 5 cell lines analyzed (Fig. 3d). Importantly, an examination of the karyotype of all these cell lines overexpressing TRMT61B showed no gross chromosomal rearrangements or changes in aneuploidy levels (Fig. 3e). These findings show that TRMT61B over-expression per se is not sufficient to induce either aneuploidy or consistent tumoral features.
Fig. 3. TRMT61B overexpression fails to produce karyotype abnormalities, causes milder effects on cell biology, and does not promote tumor growth in vivo.
a Proliferation of the indicated cell lines is slightly reduced after ectopic expression of TRMT61B (white squares) compared to controls (black circles). b In vivo tumor formation of injected A375P cells overexpressing (TRMT61B-OE) or not overexpressing (EV) TRMT61B does not result in significant differences in tumor growth. Twelve NSG immunodeficient mice were engrafted with each of the two groups of A375P cells. c Image showing the tumor size of both TRMT61B genetic conditions at the end of the experiment (left). Micrographs show representative immunostaining for TRMT61B detection in TRMT61B-OE and control xenografted cells (right). Scale bars, 100 μm. d In vitro evaluation by transwell assays of the invasive capacity of MeWo, RPE, A375P, WM1158, WM1366, and A375P cells showing base line (EV) or increased (TRMT61B-OE) levels of TRMT61B. e Table shows ploidy, numerical (NC), and structural (SC) changes observed in BJ, RPE, MeWo, A375P, WM1158, WM1366, and A375P cells 30 to 40 days after TRMT61B OE was initiated. No changes were detected in the karyotype status of all the cell lines regardless of TRMT61B expression levels. Representative karyotypes are shown for BJ, WM793, and A375P cells overexpressing (TRMT61B-OE) or not (EV) TRMT61B. Lighter to darker blue color code indicates lower to higher aneuploidy grades in all the panels. Error bars represent standard deviation in (a, d) and standard error in (b). *P < 0.05; **0.001 < P < 0.01; *** P < 0.001 (Student’s t-test; unpaired, 2-tailed).
Since TRMT61B overexpression could also be a consequence of the aneuploidy phenotype, we examined TRMT61B protein expression in trisomic, tetrasomic and tetraploid cell lines derived from 2 different human chromosomally stable and diploid cell lines (HCT116 and RPE). As shown in Supplementary Figure 4e, TRMT61B induction was detected in 4 out of the 10 aneuploidy scenarios analyzed. Therefore, although overexpression of TRMT61B was associated with some cell lines with supernumerary chromosomes, we cannot conclude that this is a constant in all aneuploidy contexts.
Suppression of TRMT61B expression induces several antiproliferative effects that are highly dependent on the aneuploidy grade
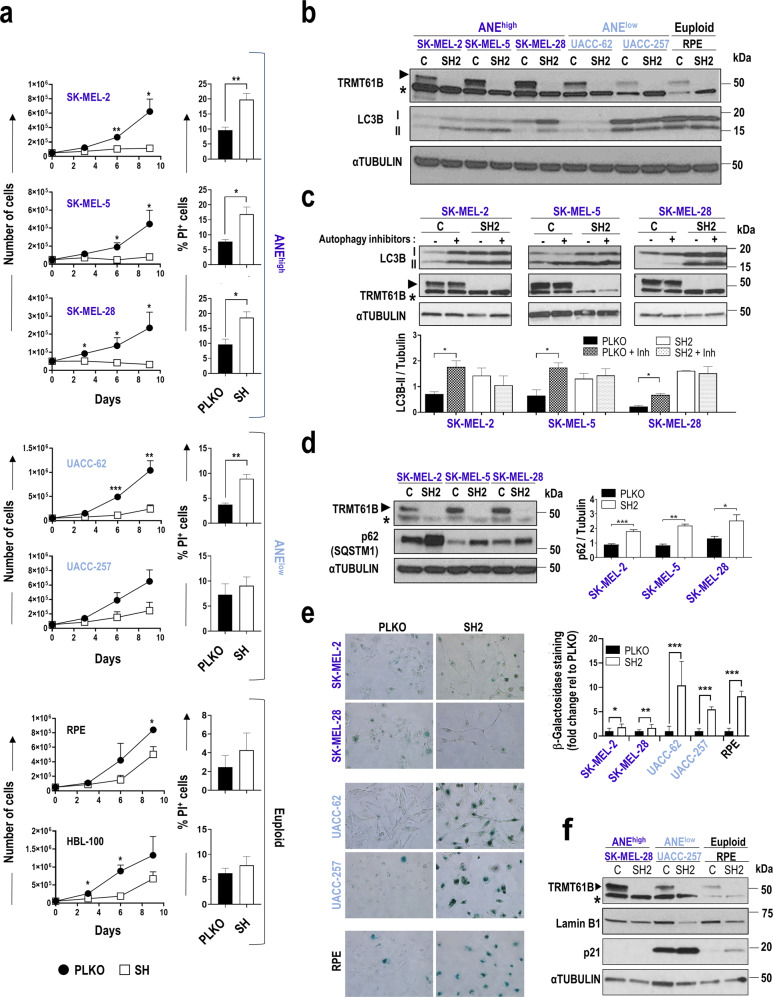
To determine the impact of TRMT61B removal in a highly aneuploid context, we performed loss of function assays, targeting TRMT61B expression via lentiviral shRNA interference (Sup. Figure 5). We decided to focus on a single cancer type – melanoma - which was well represented in the ANElow and ANEhigh groups of the NCI-60 collection and melanoma cell line panel previously analyzed (Fig. 1a, b). Four different TRMT61B-specific shRNAs and the corresponding scramble construction were analyzed in several melanoma cell lines. Since all of them effectively reduced TRMT61B expression (Sup. Figure 6), they were used indistinctly in the different procedures. In a first set of experiments, 3 ANEhigh and 2 ANElow melanoma cell lines together with 2 non-tumoral euploid cell lines, were silenced for TRMT61B expression and subsequently subjected to cell viability assays (Fig. 4 and Sup. Figure 6a). Our results showed that TRMT61B depletion reduces cell proliferation in all cases. Notably, ANEhigh cells were more prominently and consistently affected since proliferation stopped completely 16 days after lentiviral infection (Fig. 4a).
Fig. 4. Antiproliferative effects induced by TRMT61B knock-down in cell lines with different aneuploid levels.
a Proliferation capacity (left graphs) of cell lines exhibiting different aneuploidy levels (euploid, low and high) according to the in silico analysis, is greatly compromised after TRMT61B interference with a concomitant apoptotic response (right columns), especially in ANEhigh cells. Results are the mean of 4 independent experiments performed with 3 different shRNAs (SH1, SH2 and SH4). Apoptosis is analyzed using PI staining and is the average of 4 measurements taken at different time points (0, 3, 6, and 9 days after cell plating) during each of the experiments. b Western blot analysis of the autophagy marker LC3BII in cell lines of the 3 ANE groups with or without TRMT61B expression. A representative blot showing accumulation of LC3BII in ANEhigh cells in response to TRMT61B knockdown is shown. c The biological significance of LC3BII induction detected in ANEhigh cells after TRMT61B abrogation was determined by exposing both control and TRMT61B depleted cells to the autophagy inhibitory cocktail PepstatinA and E64d (5 μg/ml, 7 h). Measurement of the autophagy flux indicates blockade or delay of the autophagy cascade in the absence of TRMT61B. The histogram shows the quantification of three independent experiments. d Increased expression of p62 protein levels after TRMT61B silencing in ANEhigh cells confirms the impairment of the autophagy process. Quantification of 3 different experiments is represented in the histogram adjacent to the blot. e β-galactosidase staining (left panels) and quantification (right histogram) corresponding to control and melanoma cell lines with baseline or downregulated TRMT61B levels show a prominent senescence response in the ANElow and euploid clusters. Two independent experiments were carried out in the case of SK-MEL-28, UACC-257, and RPE cell lines using two different shRNAs (SH1 and SH4). A representative experiment is shown. f p21 up-regulation and LMNA B1 down-regulation, 2 well-known senescence features, are also associated with TRMT61B knockdown in ANElow and euploid cell lines. Lighter to darker blue color code indicates lower to higher aneuploidy grades in all the panels. Error bars represent standard error in (a, c, d) and standard deviation in (e). *P < 0.05; **0.001 < P < 0.01; ***P < 0.001 (Student’s t-test; unpaired, 2-tailed).
We then analyzed whether the absence of TRMT61B prevents cell growth by triggering apoptosis. Whereas euploid and ANElow cells were barely affected, ANEhigh cells were characterized by a significant and consistent increase in the percentage of propidium iodide (PI) positive cells and the amounts of cleaved caspase-3 (Fig. 4a, Sup. Figure 6a, Sup. Fig. 7a). TRM61B-deficient cells treated with the pan-caspase inhibitor Z-VAD-FMK strongly reduced PI staining, particularly 72 hours after drug administration (Sup. Fig. 7b). Importantly, most of these and previous results were also replicated in an independent collection of melanoma cell lines (Sup. Figure 6b, Sup. Fig. 7a, Sup. Fig. 8a). Taking all the analyzed cell lines together, a more pronounced and extended apoptotic induction was observed in the high aneuploidy cluster (6/6 cell lines) compared with that of the low-medium aneuploidy (2/7 cell lines) and euploid groups (0/2 cell lines) (Fig. 4a, Sup. Figure 6, Sup. Fig. 8a).
Since autophagy has recently been proposed as an important cell survival mechanism for cells with chromosomal aberrations [22, 25, 49–52], we investigated this process in response to TRMT61B depletion. Interestingly, the absence of TRMT61B led to an accumulation of the autophagy marker LC3B-II in ANEhigh cell lines (Fig. 4b, Sup. Figure 6a, Sup. Fig. 8b bottom graph). LC3B-II levels, however, remained unchanged in the ANElow and euploid cell lines despite TRMT61B elimination (Fig. 4b, Sup. Figure 6a, Sup. Fig. 8b). To properly understand the biological significance of this finding, we treated both TRMT61B depleted and non-depleted ANEhigh cells with the autophagy inhibitor cocktail E64d and pepstatin A, a known disruptor of autophagy flux [53]. As expected, blockage of the autophagy cascade clearly induces LC3B-II in a TRMT61B-proficient setting (Fig. 4c). However, the treatment fails to produce a similar effect in a TRMT61B-deficient context (Fig. 4c), suggesting that TRMT61B depletion inhibits or slows down the autophagy process in an ANEhigh context. These results were confirmed by the significant increase in p62, a selective substrate of autophagy [53, 54], detected in response to TRMT61B elimination (Fig. 4d).
Finally, considering that euploid and some ANElow-medium cell lines show partially compromised cell proliferation (without exhibiting a significantly higher PI staining) after TRMT61B knockdown, we looked for additional antiproliferative mechanisms that could inhibit cell proliferation in these cells (Fig. 4a, Sup. Figure 6a, Sup. Fig. 8a). We evaluated senescence response as a potential candidate by measuring β-galactosidase activity as well as p21 and LAMIN B1 protein expression. In line with a senescence scenario, while β-galactosidase activity and p21 levels were upregulated, LAMIN B1 was downregulated once TRMT61B had been silenced (Fig. 4e, f; Sup. Fig. 8c, d). These results suggest that senescence activation is the main anti-proliferative mechanism in euploid and ANElow-medium cells, and play a secondary role in cancer cell lines with high aneuploidy levels.
Structural and functional mitochondrial defects due to TRMT61B deficiency are enhanced in ANEhigh melanoma contexts
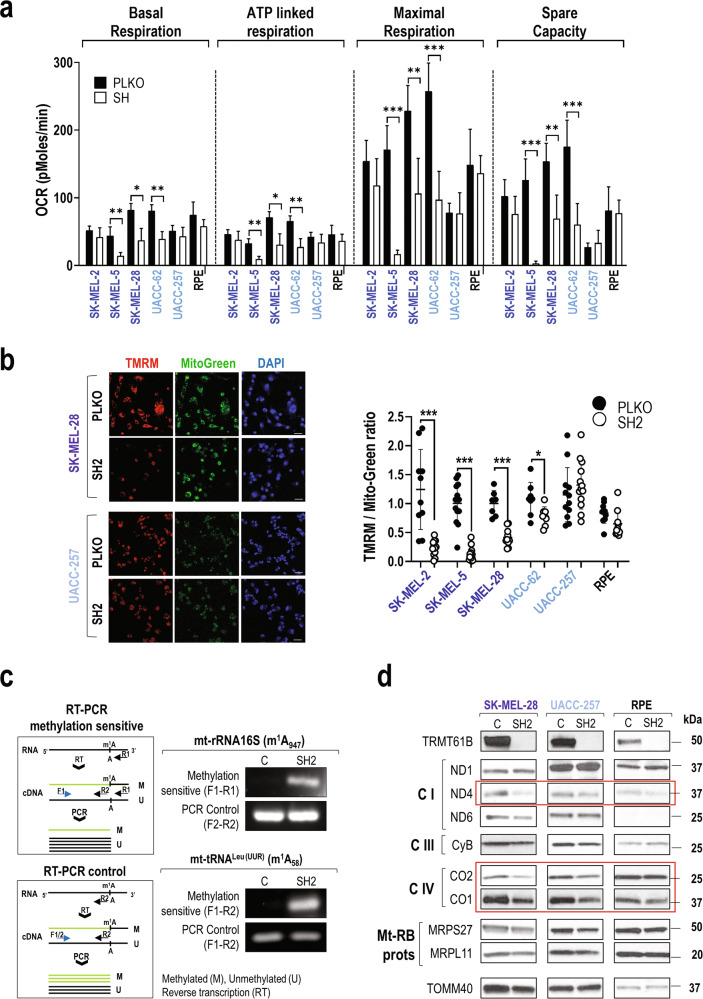
As TRMT61B exerts its function in the mitochondrial compartment [36], we studied the impact of TRMT61B downregulation at different levels in mitochondrial structure and physiology. As shown in Supplementary Figure 9a, mitochondria appear morphologically altered and partially devoid of crests in cells infected with shRNAs specific for TRMT61B. Since ATP generation through oxidative phosphorylation (OXPHOS) driven by the electron transport chain complex (ETC) is one of the most prominent mitochondrial functions, we analyzed the consequences of TRMT61B loss in the respiration metabolism by seahorse analysis [55]. As shown in Fig. 5a, mitochondrial respiration in 2 out of 3 ANEhigh and 1 out of 2 ANElow cell lines was greatly compromised after TRMT61B depletion. No differences were detected in UAC-257 (ANElow) and RPE (euploid) cells. Importantly, no glycolytic compensation was detected (Sup. Fig. 9b). These results were validated in a second set of melanoma cell lines that were subjected to seahorse analysis (Sup. Fig. 9c). In summary, when TRMT61B is silenced, the majority (5 out of 6) of the ANEhigh cell lines and the minority (2 out of 6) of the ANElow cell lines displayed a statistically significant reduction in the major mitochondrial respiration parameters. We next measured the mitochondrial membrane potential (ΔΨm), a good indicator of mitochondrial health. In line with previous bioenergetic results, we found that TRMT61B deficient ANEhigh cells showed a dramatic decrease in ΔΨm compared to their respective controls, whereas almost no signs of mitochondrial depolarization were detected in ANElow cell lines devoid of TRMT61B (Fig. 5b, Sup. Fig. 9d).
Fig. 5. TRMT61B is required for mitochondrial functionality more prominently in ANEhigh cells.
a Seahorse analysis of OXPHOS (oxidative phosphorylation) reveals a statistically significant reduction in ATP- linked, baseline, and maximal respiration as well as spare capacity (difference between maximal and baseline respiration) in 2 out of 3 ANEhigh (SK-MEL-28 and SK-MEL-5) and 1 out of 2 ANElow(UACC-62) cell lines, after TRMT61B elimination, suggesting that this RNA methyltransferase is required for proper stress response. Continuous OCR values (pmoles/min) are shown. Mitochondrial functions were analyzed as explained in Materials and Methods. Five independent experiments were carried out between 5 and 7 days after TRMT61B silencing using two different shRNAs (SH1 and SH4). b In the same group of cells also including SK-MEL-2 cell line, TMRM staining, a fluorescent probe accumulated in active mitochondria, reveals a clear loss of mitochondrial membrane potential after TRMT61B downregulation, as shown by a diminished signal detected after quantification (right dotplot). Conversely, RPE and UACC-257 cells do not display significant differences in TMRM staining irrespective of TRMT61B status. Representative fluorescence images showing TMRM (red), Mitotracker (green), and DAPI (blue) (left). One representative experiment is shown. Scale bars, 50 μm. c PCR strategy to determine methylation levels (top left) of the indicated mt-RNA types based on the ability of N1-methyladenosine (m1A) to block reverse transcription. Amplification levels of the resultant cDNA by semi-quantitative PCR indirectly estimate the presence of the desired methylation mark. As expected, TRMT61B silencing reduced specific m1A formation (residues in brackets) in 2 known mt-RNA targets according to the PCR product detected. A second PCR test (bottom left) insensitive to m1A methylation is also described and used as a loading control. d Protein expression levels of several mitochondrial encoded genes that are part of the electron transport chain (Complexes I, III, and IV) as well as nuclear-encoded mitoribosomal proteins (Mt-Rb) were analyzed by western blot in mitochondrial protein extracts derived from the indicated cell lines expressing baseline or reduced levels of TRMT61B. The mitochondrial marker TOMM40 was used as a loading control, since its expression is not expected to be altered. Genes highlighted in red are the most affected when TRMT61B is removed. Lighter to darker blue color code indicates lower to higher aneuploidy grades in all the panels. Error bars represent standard error in a and standard deviation in b. *P < 0.05; ***P < 0.001 (Student’s t-test; unpaired, 2-tailed).
We next wondered whether lack of TRMT61B affects mitochondrial gene expression since the 13 protein-coding genes encoded by the mitochondrial genome are involved in the formation of the ETC complex together with several TRMT61B mt-RNA targets [36–38, 56]. On this basis, mitochondrial fractionation was performed on 1 representative cell line of each of the ANE groups 7–10 days after TRMT61B silencing, and mitochondrial RNA and proteins were studied. As expected, m1A marks in 2 of the best characterized TRMT61B RNA substrates, mt-RNA 16S and mt-tRNAleu(UUR) [36, 37], were greatly diminished when TRMT61B was reduced (Fig. 5c). Western blot analysis revealed significant reduction in the expression levels of some mitochondrial encoded genes, such as CO2, CO1 and ND4 (Fig. 5d, Sup. Fig. 9e). Taken together, these data show that TRMT61B is needed, especially in ANEhigh cells, for efficient mitochondrial genome expression linking TRMT61B downregulation with mitochondrial membrane potential and OXPHOS deficiencies.
Strong anticancer effects induced by in vivo TRMT61B suppression in melanoma cell lines with high aneuploidy levels
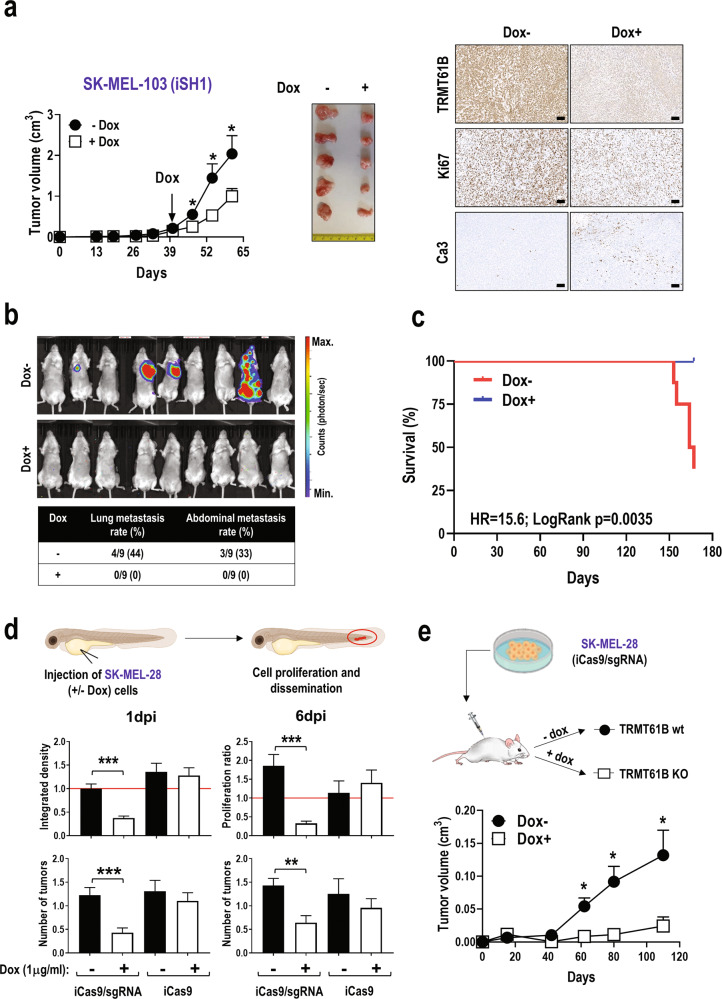
Our results suggest that TRMT61B abrogation could be a valuable antitumoral target in a high aneuploidy context by negatively affecting mitochondrial function and cell viability. To explore this hypothesis, first, we analyzed the invasive capacity of TRMT61B deficient cells from the 3 aneuploidy groups (euploid, ANElow, and ANEhigh). Regardless of the degree of aneuploidy, downregulation of TRMT61B significantly diminishes cell mobility through a matrigel basement membrane matrix suggesting a potential negative impact on metastatic capability (Sup. Fig. 10a). Next, we examined the in vivo effect of TRMT61B depletion on tumor growth by subcutaneous injection into immunocompromised mice of ANEhigh (SK-MEL-103) and ANEmedium (A375P) cell lines genetically modified to knockdown TRMT61B expression in response to doxycycline (Sup. Fig. 10b, c). In the case of SK-MEL-103 cells, in vivo administration of doxycycline, after tumor onset, significantly attenuates tumor growth (Fig. 6a). Furthermore, tumors in doxycycline treated animals showed decreased Ki67 and increased cleaved caspase-3 staining, supporting the antitumoral effect of TRMT61B silencing (Fig. 6a). To promote metastasis, primary tumors were surgically resected and animals were followed up using in vivo bioluminescence imaging (Fig. 6b). Lung and abdominal metastatic lesions only started to appear 110 days after tumor removal in the untreated group, with no sign of disease in the doxycycline-treated, TRMT61B-silenced mice (Fig. 6b). Control animals showed significant lifespan shortening in contrast to the prolonged survival observed in the TRMT61B silenced cohort (Fig. 6c). Lastly, inducible elimination of TRMT61B in the ANEmedium cell line A375P slightly reduced tumor growth (Sup. Fig. 10d), which highlights the specific antineoplastic utility of TRMT61B downregulation in an ANEhigh tumor context.
Fig. 6. TRMT61B abrogation diminishes the tumorigenic potential of ANEhigh melanoma cell lines.
a Tumor growth in NSG mice injected with SK-MEL-103 cells consecutively transduced with lentiviral vectors harboring a luciferase reporter gene and a doxycycline inducible shRNA (iSH1) targeting TRMT61B expression. Monitoring tumor volume by external caliper indicates a statistically significant reduction in tumor growth after doxycycline administration (left). Two different experiments were carried out with 10 mice per group (doxycycline-treated and untreated). Representative macroscopic images and immunohistochemical staining of TRMT61B, Ki67, and cleaved caspase-3 in tumor sections from each mouse cohort are shown (right). Scale bars, 100 μm. b Representative bioluminescence images of doxycycline-treated and untreated NSG immunodeficient mice obtained 150 days after SK-MEL-103 tumor removal by surgical excision in order to facilitate metastatic spread. The color scale represents the bioluminescence levels (photons per second) emitted from the thoracic and abdominal region of xenografted mice (ventral view) ranging from low (blue), to medium (green) and high (red). The table below summarizes the incidence of metastatic lesions detected in both group of animals. Metastatic rates are calculated as the proportion of affected vs. total mice. c The Kaplan-Meier survival curve shows a reduced lifespan of Dox- mice in comparison with the Dox+ group. The log rank test was used to evaluate survival differences between both groups, and the HR indicates an increased risk of death in the untreated group. d Integrated density (at 1 day post injection, 1dpi) and proliferation ratio (at 6 days post-injection, 6dpi) of SK-MEL-28 cells (both Cas9/sgRNA and Cas9 alone, treated or not with doxycycline) injected into the circulation of the zebrafish embryos (top histograms). The red line at 6dpi represents a threshold below which cell proliferation stops. Integrated density at 1dpi is a total measure of the fluorescence of the fish tails. Average tumor number present in the tail of each condition tested at 1 and 6dpi is also shown (bottom histograms). nreplica = 20 embryos/condition (3 replica), ntotal = 60 embryos/condition; Error bars represent standard error. e SK-MEL-28 cells containing CRISPR/Cas9 elements were subcutaneously injected in nude mice being treated or not with doxycycline from the very beginning to schedule a preventive strategy. As is shown in the graph, doxycycline administration causes a statistically significant tumor growth retardation. One experiment was performed with 12 animals per group. Error bars represent standard error in all the panels. *P < 0.05; **0.001 < P < 0.01; ***P < 0.001 (Student’s t-test; unpaired, 2-tailed).
As a complementary strategy, we used a doxycycline inducible CRISPR/Cas9-mediated gene editing approach to abolish TRMT61B expression in SK-MEL-28 cells (Sup. Fig. 11). Remarkably, iCas9/sgRNA Dox+ cells recapitulate all the features found in TRMT61B knockdown cells analyzed in previous experiments (Sup. Fig. 11, Sup. Fig. 12a, b). Both iCas9/sgRNA and (control) iCas9 cells were injected in zebrafish embryos to monitor their tumorigenic behavior (Fig. 6d). Zebrafish embryos harboring pretreated iCas9/sgRNA Dox+ cells, showed reduced tumor burden and fluorescence signal at the end of the experiment (6dpi) in the continuous presence of doxycycline, while embryos bearing control cells (iCas9/sgRNA Dox-, iCas9 Dox- and iCas9 Dox+) maintained or increased the initial values of both parameters (Fig. 6d). We also carried out similar in vivo experiments by subcutaneously injecting iCas9/sgRNA SK-MEL-28 cells in immunodeficient mice (Fig. 6e). Similar to the zebrafish experiments, doxycycline administration causes conspicuous tumor growth retardation (Fig. 6e). In sharp contrast, xenograft experiments performed with the ANElow cell line UACC-257, genetically modified with the same CRISPR/Cas9 inducible system to abrogate TRMT61B expression, resulted in the absence of significant tumor growth differences between doxycycline treated and non-treated animals (Sup. Fig. 13a, b).
These results indicate that TRMT61B function is critical for in vivo tumor formation and the metastatic capacity of ANEhigh melanoma cell lines and highlights its potential effectiveness as a druggable target for future anticancer interventions.
Discussion
Aneuploidy is a characteristic feature of most cancer cells and distinguishes them from their normal counterparts [1, 2, 9]. It is widely accepted that genetic alterations associated with aneuploidy promotion deregulate a different variety of cell functions and pathways [57–60]. As a consequence, aneuploid cells need to overcome different stresses imposed by gene dosage imbalance and protein stoichiometry changes [19]. The discovery and functional characterization of new molecular players associated with aneuploidy development and aneuploidy-tolerating mechanisms will deepen our understanding on the underlying mechanisms involved in the cancer-aneuploidy connection and also help to define new anti-cancer treatments. Our research into novel aneuploidy-associated genes allowed us to identify TRMT61B. The protein expression of this RNA modifier correlates with the extent of chromosomal alterations in a large collection of human cancer cell lines included in the NCI-60 panel. This interesting finding was experimentally and computationally validated in several human melanoma cell lines and TCGA tumor types, showing the potential of TRMT61B as an aneuploidy biomarker in a wide spectrum of tumor types. We have also detected a strong and frequent positive association between TRMT61B protein expression and cancer. Moreover, in some tumor types (lung and bladder cancers; Fig. 2b), we observed a positive correlation between TRMT61B protein expression and tumor grade. TRMT61B had previously been associated with estrogen receptor-negative breast cancer as a novel susceptibility locus [61] and as an overexpressed transcript that might indicate poor clinical outcome in gastric cancer [62]. Ours are the first results to correlate increased TRMT61B protein expression with cancer pathogenesis and malignancy in a wide range of cancer types. Further studies are needed to accurately determine the tumors and histopathological features in which TRMT61B expression can be used as a cancer grade and/or aneuploidy marker.
Regarding the potential causal relationship between TRMT61B and aneuploidy, we have shown that supraphysiological expression of TRMT61B failed to induce detectable karyotype changes (Fig. 3e). These results suggest that TRMT61B is not an aneuploidy inducer. However, we detected increased TRMT61B in some aneuploid scenarios derived from control, diploid cell lines (Sup. Figure 4e). Therefore, it is tempting to hypothesize that TRMT61B might be an adaptive or tolerance-inducing molecule in an early aneuploidy, tumorigenesis stage. This is consistent with studies showing that RNA modifications are not cancer drivers, but instead play a role in different cancer hallmarks, such as survival, proliferation, self-renewal, differentiation, stress adaptation, and invasion [63, 64]. Taken together, our data suggest that TRMT61B upregulation reflects a gene addiction scenario that can be therapeutically exploited, as has been shown in several studies in which RNA-modifying enzymes help to maintain cell proliferation and tumor progression [64].
In line with this hypothesis, our TRMT61B loss-of-function experiments induced an antiproliferative phenotype due to several cytotoxic and cytostatic responses that depend on aneuploidy levels. But, how is this limited proliferation connected to the biological function of TRMT61B? We are afraid this is still an open question with no clear answer. Taking into account its capacity to regulate tRNAs, mRNAs, and rRNAs [36–38], TRMT61B up-regulation might help cancer cells to cope with aneuploidy by fine-tuning the expression of mitochondrial genes. In fact, TRMT61B depleted cells exhibit reduced expression levels of several mitochondrial encoded genes associated with a severe reduction of m1A epitranscriptomic marks in, at least, 2 of the best-known TRMT61B RNA targets. Surprisingly, although the magnitude of the expression changes detected in the affected mitochondrial genes of ANEhigh and ANElow cells is quite similar, the latter together with non-tumoral cells show much less respiratory and mitochondrial membrane potential alterations following TRMT61B abrogation. This strong and significant effect in high aneuploid cancer cells could be a consequence of a higher dependency on the generation of energy in an oxygen-dependent manner. In contrast to past assumptions that aerobic glycolysis is the main energy source in cancer, many tumors reprogram their metabolism by increasing mitochondrial activity to respond to the new energetic and anabolic needs of several stressors, such as aneuploidy [23, 65, 66]. In the particular case of melanoma, despite the general consensus that glycolysis plays a predominant metabolic role, emerging data have shown the coexistence of mitochondrial energy metabolism, which could be a promising therapeutic target [67–69]. Interestingly, like respiratory ATP depletion mediated by TRMT61B abrogation, some studies have shown that chemical inhibition of OXPHOS reduces tumor growth and metastasis [70, 71].
Besides ATP production, other key cell functions, such as redox balance maintenance, programmed cell death or autophagy require the critical participation of mitochondria [72–74] and, therefore, could be mediating the TRMT61B role in aneuploidy-cancer phenotypes. In this regard, recent evidence has shown that aneuploid cells show robust activation of the autophagy machinery to maintain metabolic and oxidative stresses within tolerable levels, even though lysosomal function can be partially compromised [22, 49–51]. In line with this, aneuploidy cells are more sensitive to autophagy inhibitors than euploid counterparts [22, 25, 52]. Interestingly, our experiments show that TRMT61B depletion in ANEhigh cell lines hampers or delays autophagy flux and causes apoptosis. However, it is unclear whether TRMT61B is directly or indirectly involved in autophagy regulation, which is also linked to mitochondrial respiration. Ideally, all our findings need to be explored further in animal models for TRMT61B.
In this study, we have demonstrated that TRMT61B in highly aneuploidy cancer cells critically regulates mitochondrial function and that TRMT61B depletion leads to autophagy impairment, reduced levels of key ETC members, and apoptosis. Therefore, targeting this epitranscriptomic modulator could effectively disrupt the cellular stress responses critically required for aneuploid cell homeostasis and paves the way to the development of antitumoral treatments that specifically target more aggressive tumors.
Methods
The materials and reagents used in this work are summarized in Supplementary Table 4. Full length original western blots for the results showed in this work are provided in Supplementary Fig. 14.
Cell lines and culture conditions
MeWo (A), A375P (A), 624-MEL (A) and WM35 (A) were a gift from Amparo Cano’s Laboratory. SK-MEL-2 (A), SK-MEL-5 (A), SK-MEL-28 (A), SK-MEL-173, SK-MEL-147, SK-MEL-103, SK-MEL-29, UACC-62(A), UACC-257 (A), SK-MEL-94, WM1158, WM1205Lu, WM793, WM1366 (A), WM88, WM902B (A), WM209, G-361, Malme-3M(A), WM983A, WM9 (A) and WM164 (A) were kindly provided by Marisol Soengas’ group. BJ-TERT and RPE-TERT (A) were obtained from Marcos Malumbres’ laboratory. Finally, tetraploid, trisomic, and tetrasomic RPE and HCT116 previously reported [75, 76] cell lines (RPE-1, RPT1, RPT3, RPT4, RPE 21/3, RPE 5/3;12/3, HCT116, HPT1, HPT2, HPT4, HCT116 5/4 and HCT116 3/3) were supplied by Zuzana Storchová’s group. Since most of them were already authenticated by previous users, we confirmed the identity of those cell lines (with an “A” in brackets) more relevant for the study by STR DNA profiling. All cell lines were routinely tested for mycoplasma contamination, grown in DMEM medium supplemented with 10% FBS (Fetal Bovine Serum) and 1% penicillin-streptomycin, and cultured at 37 °C under a 5% CO2 atmosphere. Cells subjected to doxycycline treatment were exposed to a final concentration of 1–2 μg/ml during a minimum period of 4 days.
Karyotype analysis
Exponentially growing cells were treated with 0.5 mg/ml of the microtubule-depolymerizing drug colchicine for 4–6 h, then centrifuged with a table-top centrifuge, swollen in 40% medium/60% tap water for 15 min at 37 °C, fixed with Carnoy solution (75% methanol and 25% acetic acid) and spread on a glass slide. Metaphase chromosomes were GTG-Banded by a conventional trypsin-Giemsa technique and karyotypes (20–40 metaphases per cell line) were analyzed using a capture and analysis Cytogenetic software (Citovysion Version 7.7, Leica Biosystems Richmont, inc.).
Aneuploidy-based clustering of NCI-60 collection and melanoma cell lines
For the NCI-60 collection, we used the karyotype information previously reported by Roschke et al. [44]. In that work, ten metaphases were analyzed for each cell line using SKY technology. Five different parameters were characterized: ploidy, structural complexity (SC), numerical complexity (NC), structural heterogeneity (SH), and numerical heterogeneity (NH). In all the cases but ploidy, the values were used to differentiate three categories (low, intermediate, and high levels). In our work, we transformed this qualitative information into a numerical value category: low to 1, intermediate to 2, and high to 3 (Supplementary Table 1). All those values were added to ploidy (ranging from 2 to 5) to generate a numerical matrix that allowed us to cluster the cell lines using unsupervised hierarchical UPGMA clustering. Consensus clustering was performed using ConsensusClusterPlus R package in order to determine cluster count and membership by stability evidence. The optimal number of clusters was determined using Delta Gini coefficient.
For the melanoma cell lines we have grown and kariotypically characterized in our work, on the basis of all of the clonal (at least in 20% of the metaphases) aberrations found in all of the analyzed metaphases, we created a composite karyotype for each cell line. A total of three parameters were considered: poidy, numerical (NC) and structural chromosome complexity (SC). Information obtained for the cell lines divided them in three groups (high, intermediate, and low NC and SC levels) that were transformed in 1, 2, and 3 values, respectively. These processed values together with the ploidy content (ranging from 2 to 5) were aggregated and aneuploidy scores were assigned to each cell line (Supplementary Table 2). According to their aneuploidy score, cell lines were clustered with an unsupervised clustering (UPGMA) approach using city-block distance. For the analysis of the aforementioned karyotypic features, we follow the criteria described by Roschke and coworkers based on the ISCN convention [44].
Bioinformatic and pan-cancer computational analyses
For the identification of differentially expressed proteins in the NCI-60 cell lines, raw protein expression [43] was obtained from the CellMiner™ data set (https://discover.nci.nih.gov/cellminer/loadDownload.do). Differential protein expression analysis was performed using limma R package. The significance threshold for differentially expressed proteins were established at FDR < 0.15.
In the TCGA database, TRMT61B expression levels were calculated as log2(TPM+1) transformed counts for 9131 tumor samples (belonging to 33 different cancer types) profiled by RNAseq. For these same tumor samples, aneuploidy scores were called from Taylor AM et al., 2018 [15]. Tumors were subdivided into top and bottom quartiles based on their aneuploidy scores and compared for TRMT61B expression. Processed TCGA expression data were kindly provided by CNIO Bioinformatics Unit (Supplementary Table 3). UALCAN platform [47] was consulted to perform in-depth analysis of cancer proteomic expression data (http://ualcan.path.uab.edu). The UALCAN web resource facilitates the P values for the differential expression between comparison groups assessed using t-tests on log-transformed values [48].
Seahorse extracellular flux analysis
Mitochondrial respiration in SK-MEL-28, SK-MEL-2, SK-MEL-5, UACC-257, UACC-62, and RPE cells was characterized by extracellular flux analysis using Agilent Seahorse XF96 Analyzer (Agilent Seahorse Bioscience, Santa Clara, CA, USA), as an indicator of the functional bioenergetic capacity of the mitochondria and overall cellular health in response to TRMT61B downregulation.
To this end, the indicated cell lines were subjected to Agilent Seahorse XF Cell Mito Stress Test, and oxygen consumption rate (OCR) was measured in function of time and added ETC disruptors according to the manufacturer´s indications (Ref: https://www.agilent.com/cs/library/usermanuals/public/XF_Cell_Mito_Stress_Test_Kit_User_Guide.pdf). In short, 5–7 days after the last lentiviral infection, control, and TRMT61B interfered cells were harvested and seeded in hexaplicates at the optimized cell density (3 × 105 for SK-MEL-2, SK-MEL-28, and UACC-62; 2.5 × 105 for SK-MEL-5; 1.5 × 105 for UACC-257 and RPE) in a Seahorse XF96 Cell Culture Microplate (Agilent Seahorse XFe96 FluxPak: 102416-100) and were allowed to adhere for 24 h in a 37 °C humidified incubator with 5% CO2. The four corners were left only with medium for background correction. On the day of the analysis, the culture medium was replaced with 175 μl assay medium (bicarbonate-free DMEM supplemented with 2 mM Gln, 1 mM Pyruvate, and 10 mM Glucose), and cells were incubated at 37 °C for 60 min before measurement in a non-CO2 incubator. OCR was determined at basal conditions and after addition of different ETC disruptors: oligomycin (1 µM) was used to block ATP synthase, carbonyl-cyanide-4-(trifluoromethoxy) phenyhydrazone (FCCP, 0.25 µM) was used to make the inner mitochondrial membrane permeable for protons and allow maximum electron flux through the electron transport chain, and a mix of rotenone (0.5 µM) and antimycin A (0.5 µM) were used together to inhibit complexes I and III, respectively. Basal respiration is determined as the oxygen consumption rate used at basal conditions in the presence of glucose and pyruvate. ATP-linked respiration is the difference between the basal respiration and the resulting OCR after inhibition of the ATP synthase by oligomycin. Maximal respiration is the maximal OCR attained by addition of the uncoupler FCCP and shows the maximum rate of respiration that the cell can achieve. Spare respiratory capacity indicates the capability of the cell to respond to an energetic demand and is the difference between the maximal and the basal respiration. The results shown in Fig. 5 are derived from 5 independent experiments and those represented in Supplementary Fig. 9 are representative of the assays performed.
For the analysis of the glycolytic capacity of the cells XF Glycolysis stress kit was used (Ref: https://www.agilent.com/cs/library/usermanuals/public/XF_Glycolysis_Stress_Test_Kit_User_Guide.pdf). Briefly, cells were seeded and handled as previously described. For the assay, growth media was replaced by minimal media (bicarbonate-free DMEM supplemented with 2 mM Gln) 1 h before the analysis. Glucose (10 mM) was injected, followed by oligomycin (1 μM), which as an ATP inhibitor permits the readout of the maximal ECAR (extracellular acidification rate). Finally, 2-DG (50 mM) was added to completely block glycolysis. The level of glycolysis is calculated as the ECAR rate reached after injection of a saturating amount of glucose. The glycolytic capacity is the maximum ECAR rate reached after shutting down oxidative phosphorylation by oligomycin. The glycolytic reserve is calculated as the difference between the glycolytic capacity and the glycolysis. Data represented in Supplementary Fig. 9 come from 5 independent experiments. Reagent details are included in Supplementary Table 4.
Analysis of mitochondrial membrane potential
4000–5000 cells lacking or not TRMT61B were seeded in triplicates in 96 well plates. 24 hours later, cells were incubated at 37 °C for 30’ with 50 nM teramethylrhodamine methyl ester (TMRM), a cationic red-orange fluorescence dye that accumulates in active mitochondria with intact membrane potential, in combination with Hoechst (10 μg/ml) and Mitotracker Green (50 nM) for nuclear staining and mitochondrial mass evaluation, respectively. At least 8 different images per cell line and condition were acquired using a SP5 confocal microscope. The 560 nm laser line was used to excite the TMRM and the 488 nm to excite Mitrotracker Green. Emitted fluorescence was measured above 580 nm for TMRM and between 500 and 540 nm for Mitotracker Green. TMRM and Mitotracker Green quantification (calculated as mean intensity) of 2–3 different fields per well was carried out by Image J software and TMRM/Mitotracker Green ratio was determined. Reagents are listed in Supplementary Table 4.
Detection of TRMT61B mediated m1A in mitochondrial tRNAleu(UUR) and 16S rRNA
For semi-quantitative RT-PCR based detection of m1A58 and m1A947 in human mitochondrial tRNAleu(UUR) and 16S rRNA, respectively, 100 ng of small RNA containing samples were annealed with specific reverse primers (tRNAleu(UUR) –R1 and 16S rRNA-R1 for the methylated sensitive PCR reaction or tRNAleu(UUR) –R2 and 16S rRNA-R2 for the control PCR strategy) and retrotranscribed. The resultant cDNA was used as template in semi-quantitative PCR using the primer combinations tRNAleu(UUR) –F1 + tRNAleu(UUR) –R2 or 16S rRNA-F1 + 16S rRNA-R1. Similar to other RNA modifications, the presence of m1A58 and m1A947 can partially or totally block reverse transcription. Therefore, considering that R1 primers target the downstream region of the methylated adenosine, cDNAs generated by R1 dependent reverse transcription are less efficiently synthesized and amplified by subsequent PCR reactions using the above primer combinations. In contrast, PCR amplification using tRNAleu(UUR) –F1 + tRNAleu(UUR) –R2 or 16S rRNA-F2 + 16S rRNA-R2 primer pairs is not altered by the presence or absence of m1A in cDNAs derived from R2 mediated retrotranscription, since R2 annealing occurs upstream of the methylated adenosine. This last condition was used as a reference. Primer sequences are provided in Supplementary Table 4.
RNA extraction, reverse transcription and semi-quantitative PCR
For total RNA extraction, isolated mitochondrial fractions were lysed in Trizol followed by standard phenol-chloroform extraction protocol. Small and long RNAs were purified using miR Neasy kit (see Supplementary Table 4) following the manufacturer’s instructions. For reverse transcription, total amount of RNA isolated from cells was adjusted to 100 ng/μl in RNase-free water. Then, we mixed 1 μl of total RNA with 1 μl dNTPs 10 mM and 2 μl 20 μM reverse primer R1 or reverse primer R2, heat-denatured the mixture at 65 °C for 5 min, and cooled it rapidly on ice for at least 5 min. With the mixture still on ice, reverse transcriptase was added. Reverse transcription was performed in a total reaction volume of 20 μl at 55 °C for 45 min and then heat-inactivated at 70 °C for 15 min. We subjected 1 μl synthesized cDNA to semiquantitative PCR with the 2X Green Master Mix supplemented with DMSO (10%) and 0.5 μl of each of the corresponding forward and reverse primers (20 μM) to a final volume of 20 μl. PCR program includes the following time and temperature parameters: denaturation (Temp: 95 °C. Time: 5 min on the initial stage; 30 s on rest), annealing (Temp: 50 °C for tRNAleu(UUR) –F1 + tRNAleu(UUR) –R2 and 58 °C for 16S rRNA-F1/F2 + 16S rRNA-R1/R2. Time: 30 s) and extension (Temp: 72 °C. Time: 30 s; 2 min on the last cycle). 26 cycles were performed in all the cases.
Mitochondrial isolation
Around 20–25 × 106 cells growing in four 15 cm plates were collected in 5.2 ml of fractionation buffer (20 mM HEPES (pH 7.4), 10 mM KCl, 2 mM MgCl2, 1 mM EDTA, 1 mM EGTA, and protease inhibitor cocktail). Then, cell suspension was passed through a 27-gauge needle 10 times, left on ice for 20 min, and centrifuged at 720 g during 5 min to pellet nuclei and cell debris. 90% of the supernatant containing cytoplasm, membranes and mitochondria was transferred (the remaining 10% was discarded to minimize contamination) into a fresh tube and centrifuged at 3000 g during 15 min to separate mitochondria (pellet) from cytoplasm fraction (supernatant). Nuclear and mitochondrial fractions were subjected to an additional wash involving syringe-based homogenization and centrifugation at the previously recommended speeds. Cytoplasm was also centrifuged once again (3000 g during 15 min) and pellet was discarded. Mitochondrial fractions were equally divided for ulterior RNA and protein analysis and preserved at −80 °C. WB analysis of different nuclear (LaminA/C), mitochondrial (TOMM20), and cytosolic (a-tubulin) markers confirmed an efficient subcellular fractionation with reduced contamination levels in each of the fractions.
Cell viability assays
Cells from the different cell lines reported along the present study were seeded at 500–1000 cells per well in 96-well (always performed in tetraplicates), black-walled, clear-bottomed tissue culture plates (see Supplementary Table 4), incubated at 37 °C and monitored every 2–3 days for cell death and proliferation during a minimum period of 7 days. For total and dead nuclei counting assays, Hoechst 33342 and Propidium Iodide were added at 10 μg/ml and 1 μg/ml final concentration, respectively, and incubated for 15–30 min in the dark at 37 °C. High-throughput quantification was performed by automated imaging system (Cytell, GE Healthcare, Buckinghamshire, UK) equipped with built-in enumeration software and suitable fluorescence filters. For evaluation of the antiapoptotic effect of Z-Vad-FM capase-3 inhibitor, 2000–3000 SK-MEL-28 cells lacking or not TRMT61B were seeded in tetraplicates and cell viability was analyzed 48 or 72 h after drug addiction. Reagents are detailed in Supplementary Table 4.
Invasion assay
Invasion assays of the indicated melanoma and control cell lines were performed using transwell matrigel coated inserts (see Supplementary Table 4) as previously described in Salvador et al., 2017 with minimal changes 77. In brief, 0.7 ml DMEM supplemented with 10% FBS was added to the well of the plate as attractant. Then, proper cell numbers per cell line were seeded by triplicates on the upper part of each chamber in low serum conditions (0.5% FBS). After 48 h, non-invading cells on the upper surface of the membrane were wiped with a cotton swab, and invading cells on the lower surface of the nucleopore membrane were fixed (1% glutaraldehyde), stained with crystal violet (0.5% in PBS) and counted by examination of five microscopic fields per insert. For each cell line, numbers were normalized to the corresponding PLKO-scramble or empty vector (EV) values.
Senescence analysis
β-Galactosidase activity was measured according to the manufacturer’s instructions (see Supplementary Table 4). β-Galactosidase+ cells were determined using a LEICA DM IL LED Inverted Phase Contrast Microscope and the ImageJ 2.0.0 software (NIH, USA) was used for further analysis. 10 different fields were examined per cell line and condition. Percentages of senescence cells were calculated per each cell line and normalized to the corresponding control values. Reagents are listed in Supplementary Table 4.
Measurement of autophagic flux
Control and TRMT61B deficient SKMEL28, SKMEL2 and SKMEL5 cell lines were treated during 7 hours with the lysosomal protease inhibitor cocktail E64d (5 μg/ml) and pepstatin A (5 μg/ml) or DMSO vehicle alone. Overall LC3B-II levels were determined by western blot analysis and measured from the scanned film by densitometric analysis using ImageJ software (NIH, USA). The ratio of the LC3B-II protein level to the αTUBULIN was reported as the result of LC3B-II quantification for each experimental condition.
Expression constructs, shRNAs and Lentiviral transduction
TRMT61B depletion by shRNA was achieved by using the PLKO.1 system. All lentiviral plasmids were commercially obtained from Sigma Aldrich. Lentiviruses were obtained as previously shown in Salvador et al., 2017 with minor changes [77]. Very briefly, 5 μg PLKO.1 shRNA plasmids or scramble versions were co-transfected with 5 μg secondary packaging plasmids into HEK293T cells. A similar strategy was followed for the generation of luciferase reporter lentivirus. Media was harvested at 48 and 72 h post-transfection and stored at −80 °C. Target cells were seeded at 8 × 105 cells per 10 cm plate and 5 ml of virus with 8 μg/ml polybrene was added. Cells were subjected to 2–3 consecutive infections to enhance TRMT61B downregulation. Puromycin was used for selection (2–5 μg/ml). In general, 5–6 days after the last infection round, cells were plated for the different experiments. For generation of the doxycycline-inducible lentiviral construction, modified SH1 containing a 7 nucleotide loop (SH1-7NT) was cloned in EZ-Tet-pLKO-puro vector following the protocol described in Frank et al., 2017 [78]. TRMT61B overexpression construct was obtained from genecopoeia plasmid collection and virus generation was conducted with the third-generation packaging system. Reagents and shRNA sequences are shown in Supplementary Table 4.
TRMT61B knock-out by CRISPR/Cas9 gene edition
For CRISPR/Cas9 knockout of TRMT61B, SKMEL28 and UACC-257 cells were transfected with inducible endonuclease Cas9 construction (gently provided by Dr. Iain Cheeseman) and selected in the presence of G418 (1 mg/ml) for 21 days. Then, lentiviral plasmids containing sgRNAs targeting exon 1 of TRMT61B locus were transduced into target cells. 3–4 days after puromycin selection, individual clones were isolated by single-cell cloning, expanded, and tested for TRMT61B deletion by WB analysis in response to doxycycline treatment. sgRNA sequences and plasmids are listed in Supplementary Table 4.
Transmission Electron Microscopy
For electron microscopy ultrastructural analysis, cells were fixed with a mixture of 2.5% glutaraldehyde and 2% PFA phosphate buffer (Na2HPO4/NaHPO4, 0.1 M, pH 7.4) for 1 h at 4 °C and then washed three times in phosphate buffer. Postfixation was performed with a 1% osmium tetroxide and 1% potassium ferricyanide and 0.15% tannic acid. Samples were treated with 2% uranyl acetate and dehydrated in increasing concentrations of ethanol (50, 75, 90, 95, and 100%). Finally, infiltration in epoxy-resin was done using increasing concentrations of resin (25, 50, 75, and 100%) and polymerization was performed at 60 °C. Ultrathin sections of the samples were stained with saturated uranyl acetate and 2% lead citrate following standard procedures and sections were imaged with a Tecnai 12 FEI microscope (FEI, Hillsboro, Oregon, USA) operated at 120 kV. Quantification of the indicated mitochondrial parameters was performed by using Image J software.
Immunoblots
Western blotting was performed using standard methods. In brief, cells were lysed in RIPA buffer (50 mM Tris, pH7.4, 150 mM NaCl, 1% NP40, 0.1% SDS, 0.5% DOC, protease inhibitors, 30 mM NaF, 1 mM Na3VO4 and 0.1 mM PMSF). Protein concentration was determined by the BCA assay. Equivalent amounts of protein were separated on precast polyacrylamide midi gels and electroblotted onto nitrocellulose membranes. Dry transfer apparatus was run according to manufacturer’s recommendations (Transfer-Blot Turbo System, BioRad). After blocked with 5% milk, blots were then probed with primary (overnight incubation at 4 °C) and the appropriate secondary antibodies (1 h at room temperature) diluted in 5% milk. Reactions for protein visualization were detected by enhanced chemiluminescence assay. Where appropriate, densitometric analysis of the immunostaining results was performed by using Image J software. Antibodies and reagents are listed in Supplementary Table 4.
Immunofluorescence
Cells were fixed with 4% PFA at RT during 20 min, washed three times with PBS, permeabilized for 10 min with 0.3% TritonX-100, blocked 1 h at 37 °C, and immunostained during 1 h with primary and secondary antibodies diluted in blocking solution (3% BSA). Finally, coverslips were mounted on slides and imaged using a Sp5 confocal microscope. Reagents are indicated in Supplementary Table 4.
Histology and immunohistochemical staining
For histopathological analysis, formalin-fixed paraffin embedded (FFPE) blocks were serially sectioned (3 μm thick) and stained with hematoxylin and eosin (H&E). Additional serial sections were used for immunohistochemical (IHC) studies. Immunohistochemistry was performed on tumor xenografts as well as on commercial (SKCM, LUAD, LUSC, BLCA, and BLSC; see Supplementary Table 4) and homemade (CHOL) tissue microarrays (TMAs). Tissue sections were processed in an Autostainer Link (Dako), with antigen retrieval at pH 9.0 and incubation with TRMT61B, ki67, and cleaved capase-3 primary antibodies (Supplementary Table 4). Antigens were visualized using 3,3-diaminobenzidine tetrahydrochloride plus (DAB + ) in all cases except for SKCM immunohistochemistry in which magenta chromogen was used. A semi-quantitative histology score (H-score) was calculated considering the intensity of the staining [negative (0), mild (+1), moderate (+2) and strong (+3)] and the area containing a positive signal. All the quantification analysis and hitological examination were performed blinded under the supervision of an expert pathologic team.
TRMT61B detection in plasma samples by ELISA
Determination of TRMT61B levels in 1:10 diluted plasma samples from patients with colorectal, breast, lung, or ovarian cancer was assessed by ELISA (Supplementary Table 4) following manufacturers’ instructions. Briefly, reagents were brought to room temperature and 100 µl of the standard, control, and plasma samples were added. The plate was then carefully sealed and incubated for 2 h at 37 °C. After washing three times with 350 µl of the wash buffer solution, 100 µl of the detection reagent A was added to each well and incubated for 1 h at 37 °C. After another three washing steps, 100 µl of the detection reagent B were added to each well and incubated for 1 h at 37 °C. Then, wells were washed five and signal developed at 37 °C with 90 µl of TMB substrate per well. Reaction was stopped with 50 µl of stop solution per well and measured at 450 nm on The Spark multimode microplate reader (Tecan Trading AG, Switzerland).
Bioluminescence imaging (BLI)
For BLI, mice were injected intraperitoneally with 100 μl of D-Luciferin (Caliper Life Sciences). Then, mice were anesthetized with isoflurane, placed supine, with their abdomen toward the camera, and imaged, following the manufacturer’s recommendations, for 30 seconds using the Xenogen IVIS 200 BLI system. Animals were evaluated every 15 days for metastasis appearance during 5 months.
Xenograft experiments
SK-MEL-103 (2.5–3 × 106), A375P (5 × 105), UACC-257 (5 × 105) and SK-MEL-28 cells (2.5–3 × 106) in 100 μL of PBS were subcutaneously injected in one of the flanks of immunocompromised female NSG (SK-MEL-103, UACC-257, and A375P) or nude mice in the case of SK-MEL-28 cell line. Tumor dimensions were measured using a caliper and tumor volume was calculated according to the formula V = 4/3πR2r. Doxycycline was administrated every 2 days in the drinking water (1 mg/ml) with 3% of sucrose from the very beginning (SK-MEL-28 injected cells) or when tumor size reaches 0.15–0.2 cm3 (SK-MEL-103, UACC-257 and A375P injected cells).
Zebrafish embryos were obtained by mating adult zebrafish (Danio rerio, wild type), maintained in aquaria with a ratio of 1 fish per liter of water, with 14 h/10 h light/dark cycle and a temperature of 28.5 °C according to published procedures [79]. 106 10-day doxycycline pretreated or untreated SK-MEL-28 cells harboring iCas9 alone or in combination with sgRNA were resuspended in 10 μl of PBS with 2% of polyvinylpyrrolidone (to avoid aggregation) and injected into the circulation of 48 hpf embryos (100–200 cells per embryo) using a microinjector (IM-31 Electric Microinjector, Narishige) with an output pressure of 34 kPA and 30 ms of injection time per injection. Inoculated embryos were incubated at a temperature of 34 °C in the presence or absence of doxycycline (1 μg/ml) and images were captured using a fluorescence stereomicroscope (AZ-100, Nikon) at 1, 4, and 6 dpi (only 1 and 6 days are shown) to measure dissemination and proliferation of the injected cells in the caudal hematopoietic tissue (CHT) of the zebrafish embryos in each of the conditions assayed. The image analysis of the injected embryos was carried out using the Quantifish software v2.1 (University College London, London, UK) to obtain the proliferation ratio of the cells in the region of the caudal hematopoietic tissue of the embryos, where the cells proliferate and metastasize. This program measures in each of the images provided the intensity of the fluorescence and the area of the positive pixel above a certain threshold of the cells. With these parameters, an integrated density value is obtained allowing the researcher to compare different times between images to reach a proliferation ratio. A ratio above and below 1 means cell proliferation and cell death, respectively. Apart from that, the number of tumors present in each of the images corresponding to the tails of the embryos was calculated. Tumor counting was performed based on tumor size and separation inside the tail of the fish.
In all the in vivo experiments, immunodeficient mice and zebrafish embryos were randomly and blindly distributed into groups.
Statistics
Statistical analyses p-values were generated using Student’s t-test (unpaired, 2-tailed) to determine differences between the means of two groups. Pair-wise multiple comparisons were performed with one-way ANOVA with Tukey post-tests. In all figures, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001 and ns P > 0.05 p-value. Error bars were calculated as standard error unless otherwise indicated. Sample size and number of experiments are indicated in each of the Figures and/or in the corresponding sections of Material and Methods. Kaplan-Meier survival curve as well as HR and logRank calculations were performed with Prism-GraphPad software.
Supplementary information
Acknowledgements
We are deeply thankful to Oscar Fernández-Capetillo, María Salazar, and David Santamaría for their advice and scientific review of the manuscript. We acknowledge master thesis’ students Martín Salamini and Sarai Araujo for their support in the performance of several in vitro experiments. We thank the research groups led by Zuzana Strochova, Iain Chesseman, Bon-Kyoung Koo, Amparo Cano, Marisol Soengas, and Marcos Malumbres for providing us with cell lines and reagents used in this study. We would also like to thank Daniel Luque and María del Carmen Terrón, members of the Electron Microscopy Unit from ISCIII, for their technical assistance. All the illustrations included in the figures were created with the help of biorender.com.
Author contributions
AM acquired, analyzed, and interpreted most of the data; BV-M, IH, DG-D, and SA assist with several in vitro experiments; BV-M, IH and CE helped with mouse xenograft experiments; BV-M prepared cell cultures for karyotype analyses; CE performed mitochondrial ultrastructural analysis; AC, MGC, ARM, and SV carried out the Seahorse studies; RIRM provided liver samples and performed the subsequent histological analysis; EC is our in-house pathologist and assisted with histological analysis of TMAs and xenograft tumors; RB and MGA performed ELISA assays and collected plasma from normal and colon patients; JC provided additional plasma samples from breast, ovary and lung tumor bearing patients; AM-R performed karyotype analysis; PC-S and LS designed and performed the zebrafish studies. GG-L carried out bioinformatics and clustering analyses. IPC and AM developed the study concept, obtained funding, interpreted the data, and drafted/edited the manuscript; and all the authors edited the manuscript.
Funding
AM was a recipient of a postdoctoral fellowship from the Fundación Española Contra el Cáncer (AECC). This study was supported by grants from the Spanish Ministry of Science and Innovation (to IPC, SAF2016-76929-R) and from the Acción Estratégica de Salud Intramural (to A.M., PI17CIII/00010).
Data availability
The materials described in the manuscript, including all relevant raw data, will be freely available to any researcher wishing to use them for non-commercial purposes, without breaching participant confidentiality.
Competing interests
The authors declare no competing interests.
Ethics statement
All the experiments of the study were carried out in accordance with the Declaration of Helsinki. All mouse studies were performed in compliance with the institutional guidelines for the welfare of experimental animals approved by the Use Committee for Animal Care from Instituto de Salud Carlos III (ISCIII) and La Comunidad de Madrid and in accordance with the guidelines for Ethical Conduct in the Care and Use of Animals as stated in The International Guiding Principles for Biomedical Research involving Animals, developed by the Council for International Organizations of Medical Sciences (CIOMS). All the procedures used in the zebrafish experiments as well as fish care were performed in agreement with the Animal Care and Use Committee of the University of Santiago de Compostela and the standard protocols of Spain (Directive 2012-63-UE). At the final point of the experiments, zebrafish embryos were euthanized by tricaine overdose. Liver samples were obtained from patients having undergone surgical resection at the University Hospital of Salamanca, Spain. The study was approved by the Ethics Committee for Clinical Research of Salamanca (July 18, 2018) and informed written consent for the samples to be used for biomedical research was obtained from each patient. Plasma samples were obtained from the biobank of the Hospital Clínico San Carlos or provided by Hospital Universitari de Sant Joan (Tarragona, Spain) after approval of the Ethical Review Boards of these institutions. All subjects in the study gave their written informed consent to participate and all experiments were performed in accordance with relevant guidelines and regulations.
Footnotes
Edited by JP Medema
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Alberto Martín, Email: martin.alberto77@gmail.com.
Ignacio Pérez de Castro, Email: iperez@isciii.es.
Supplementary information
The online version contains supplementary material available at 10.1038/s41418-022-01044-6.
References
- 1.Weaver BA, Cleveland DW. Does aneuploidy cause cancer? Curr Opin Cell Biol. 2006;18:658–67. doi: 10.1016/j.ceb.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 2.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nat Genet. 2003;34:369–76. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 3.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 4.Swanton C, Nicke B, Schuett M, Eklund AC, Ng C, Li Q, et al. Chromosomal instability determines taxane response. Proc Natl Acad Sci. 2009;106:8671–6. doi: 10.1073/pnas.0811835106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakhoum SF, Ngo B, Laughney AM, Cavallo JA, Murphy CJ, Ly P, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature. 2018;553:467–72. doi: 10.1038/nature25432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ippolito MR, Martis V, Martin S, Tijhuis AE, Hong C, Wardenaar R, et al. Gene copy-number changes and chromosomal instability induced by aneuploidy confer resistance to chemotherapy. Dev Cell. 2021;56:2440–54.e6. doi: 10.1016/j.devcel.2021.07.006. [DOI] [PubMed] [Google Scholar]
- 7.Lukow DA, Sausville EL, Suri P, Chunduri NK, Wieland A, Leu J, et al. Chromosomal instability accelerates the evolution of resistance to anti-cancer therapies. Dev Cell. 2021;56:2427–39.e4. doi: 10.1016/j.devcel.2021.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duesberg P, Stindl R, Hehlmann R. Origin of multidrug resistance in cells with and without multidrug resistance genes: Chromosome reassortments catalyzed by aneuploidy. Proc Natl Acad Sci USA. 2001;98:11283–8. doi: 10.1073/pnas.201398998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vasudevan A, Schukken KM, Sausville EL, Girish V, Adebambo OA, Sheltzer JM. Aneuploidy as a promoter and suppressor of malignant growth. Nat Rev Cancer. 2021;21:89–103. doi: 10.1038/s41568-020-00321-1. [DOI] [PubMed] [Google Scholar]
- 10.Santaguida S, Amon A. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol. 2015;16:473–85. doi: 10.1038/nrm4025. [DOI] [PubMed] [Google Scholar]
- 11.Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, et al. Effects of aneuploidy on cellular physiology and cell division in haploid. Yeast. 2007;317:916–24. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 12.Zhu J, Tsai HJ, Gordon MR, Li R. Cellular stress associated with aneuploidy. Dev Cell. 2018;44:420–31. doi: 10.1016/j.devcel.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaukat Z, Liu D, Choo A, Hussain R, O’Keefe L, Richards R, et al. Chromosomal instability causes sensitivity to metabolic stress. Oncogene. 2015;34:4044–55. doi: 10.1038/onc.2014.344. [DOI] [PubMed] [Google Scholar]
- 14.Clemente-Ruiz M, Murillo-Maldonado JM, Benhra N, Barrio L, Pérez L, Quiroga G, et al. Gene dosage imbalance contributes to chromosomal instability-induced tumorigenesis. Dev Cell. 2016;36:290–302. doi: 10.1016/j.devcel.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, et al. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell. 2018;33:676–89.e3. doi: 10.1016/j.ccell.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen G, Rubinstein B, Li R. Whole chromosome aneuploidy: Big mutations drive adaptation by phenotypic leap. BioEssays. 2012;34:893–900. doi: 10.1002/bies.201200069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen G, Mulla WA, Kucharavy A, Tsai HJ, Rubinstein B, Conkright J, et al. Targeting the adaptability of heterogeneous aneuploids. Cell. 2015;160:771–84. doi: 10.1016/j.cell.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selmecki AM, Maruvka YE, Richmond PA, Guillet M, Shoresh N, Sorenson AL, et al. Polyploidy can drive rapid adaptation in yeast. Nature. 2015;519:349–51. doi: 10.1038/nature14187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaya A, Mariotti M, Tyshkovskiy A, Zhou X, Hulke ML, Ma S, et al. Molecular signatures of aneuploidy-driven adaptive evolution. Nat Commun. 2020;11:588. doi: 10.1038/s41467-019-13669-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dephoure N, Hwang S, O’Sullivan C, Dodgson SE, Gygi SP, Amon A, et al. Quantitative proteomic analysis reveals posttranslational responses to aneuploidy in yeast. Elife. 2014;3:1–27. doi: 10.7554/eLife.03023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong HWS, Shaukat Z, Wang J, Saint R, Gregory SL. JNK signaling is needed to tolerate chromosomal instability. Cell Cycle. 2014;13:622–31. doi: 10.4161/cc.27484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu D, Shaukat Z, Xu T, Denton D, Saint R, Gregory S. Autophagy regulates the survival of cells with chromosomal instability. Oncotarget. 2016;7:63913–23. doi: 10.18632/oncotarget.11736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newman DL, Gregory SL. Co-operation between aneuploidy and metabolic changes in driving tumorigenesis. Int J Mol Sci. 2019;20:4611. doi: 10.3390/ijms20184611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohen-Sharir Y, McFarland JM, Abdusamad M, Marquis C, Bernhard SV, Kazachkova M, et al. Aneuploidy renders cancer cells vulnerable to mitotic checkpoint inhibition. Nature. 2021;590:486–91. doi: 10.1038/s41586-020-03114-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou L, Jilderda LJ, Foijer F. Exploiting aneuploidy-imposed stresses and coping mechanisms to battle cancer. Open Biol. 2020;10:200148. doi: 10.1098/rsob.200148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boccaletto P, MacHnicka MA, Purta E, Pitkowski P, Baginski B, Wirecki TK, et al. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–7. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCown PJ, Ruszkowska A, Kunkler CN, Breger K, Hulewicz JP, Wang MC, et al. Naturally occurring modified ribonucleosides. Wiley Interdiscip Rev RNA. 2020;11:e1595. doi: 10.1002/wrna.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li S, Mason CE, The pivotal regulatory landscape of RNA modifications. 2014;15:127–50. [DOI] [PubMed]
- 30.Yarian C, Townsend H, Czestkowski W, Sochacka E, Malkiewicz AJ, Guenther R, et al. Accurate translation of the genetic code depends on tRNA modified nucleosides. J Biol Chem. 2002;277:16391–5. doi: 10.1074/jbc.M200253200. [DOI] [PubMed] [Google Scholar]
- 31.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–33. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sloan KE, Warda AS, Sharma S, Entian KD, Lafontaine DLJ, Bohnsack MT. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol. 2017;14:1138–52. doi: 10.1080/15476286.2016.1259781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2016;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huber SM, Leonardi A, Dedon PC, Begley TJ. The versatile roles of the tRNA epitranscriptome during cellular responses to toxic exposures and environmental stress. Toxics. 2019;7:17. doi: 10.3390/toxics7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Endres L, Dedon PC, Begley TJ. Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol. 2015;12:603–14. doi: 10.1080/15476286.2015.1031947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chujo T, Suzuki T. Trmt61B is a methyltransferase responsible for 1-methyladenosine at position 58 of human mitochondrial tRNAs. RNA. 2012;18:2269–76. doi: 10.1261/rna.035600.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bar-Yaacov D, Frumkin I, Yashiro Y, Chujo T, Ishigami Y, Chemla Y, et al. Mitochondrial 16S rRNA is methylated by tRNA methyltransferase TRMT61B in All Vertebrates. PLoS Biol. 2016;14:e1002557. doi: 10.1371/journal.pbio.1002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X, Xiong X, Zhang M, Wang K, Chen Y, Zhou J, et al. Base-resolution mapping reveals distinct m1A Methylome in nuclear- and mitochondrial-encoded transcripts. Mol Cell. 2017;68:993–1005.e9. doi: 10.1016/j.molcel.2017.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monks A, Zhao Y, Hose C, Hamed H, Krushkal J, Fang J, et al. The NCI transcriptional pharmacodynamics workbench: A tool to examine dynamic expression profiling of therapeutic response in the NCI-60 cell line panel. Cancer Res. 2018;78:6807–17. doi: 10.1158/0008-5472.CAN-18-0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reinhold WC, Sunshine M, Liu H, Varma S, Kohn KW, Morris J, et al. CellMiner: a web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the NCI-60 cell line set. Cancer Res. 2012;72:3499–511. doi: 10.1158/0008-5472.CAN-12-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reinhold WC, Varma S, Sunshine M, Elloumi F, Ofori-Atta K, Lee S, et al. RNA sequencing of the NCI-60: Integration into CellMiner and CellMiner CDB. Cancer Res. 2019;79:3514–24. doi: 10.1158/0008-5472.CAN-18-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer. 2006;6:813–23. [DOI] [PubMed]
- 43.Gholami AMM, Hahne H, Wu Z, Auer FJJ, Meng C, Wilhelm M, et al. Global proteome analysis of the NCI-60 cell line panel. Cell Rep. 2013;4:609–20. doi: 10.1016/j.celrep.2013.07.018. [DOI] [PubMed] [Google Scholar]
- 44.Roschke AV, Tonon G, Gehlhaus KS, McTyre N, Bussey KJ, Lababidi S, et al. Karyotypic complex NCI-60 drug-screen panel. 2003;63:8634–47. [PubMed] [Google Scholar]
- 45.Liu H, D’Andrade P, Fulmer-Smentek S, Lorenzi P, Kohn KW, Weinstein JN, et al. mRNA and microRNA expression profiles of the NCI-60 integrated with drug activities. Mol Cancer Ther. 2010;9:1080–91. doi: 10.1158/1535-7163.MCT-09-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reinhold WC, Varma S, Sunshine M, Rajapakse V, Luna A, Kohn KW, et al. The NCI-60 methylome and its integration into CellMiner. Cancer Res. 2017;77:601–12. [DOI] [PMC free article] [PubMed]
- 47.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–58. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen F, Chandrashekar DS, Varambally S, Creighton CJ. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat Commun. 2019;10:5679. doi: 10.1038/s41467-019-13528-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stingele S, Stoehr G, Storchova Z. Activation of autophagy in cells with abnormal karyotype. Autophagy. 2013;9:246–8. doi: 10.4161/auto.22558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol. 2012;8:608. doi: 10.1038/msb.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Santaguida S, Vasile E, White E, Amon A. Aneuploidy-induced cellular stresses limit autophagic degradation. Genes Dev. 2015;29:2010–21. doi: 10.1101/gad.269118.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang Y-CC, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011;144:499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Arozena AA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lippai M, Low P. The role of the selective adaptor p62 and ubiquitin-like proteins in autophagy. Biomed Res Int. 2014;2014:1–11. doi: 10.1155/2014/832704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hüttemann M, Lee I, Samavati L, Yu H, Doan JW. Regulation of mitochondrial oxidative phosphorylation through cell signaling. Biochim Biophys Acta - Mol Cell Res. 2007;1773:1701–20. doi: 10.1016/j.bbamcr.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 56.Taanman JW. The mitochondrial genome: Structure, transcription, translation and replication. Biochim Biophys Acta - Bioenerg. 1999;1410:103–23. doi: 10.1016/S0005-2728(98)00161-3. [DOI] [PubMed] [Google Scholar]
- 57.Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Curr Biol. 2010;20:R285–95. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lepage CC, Morden CR, Palmer MCL, Nachtigal MW, McManus KJ. Detecting chromosome instability in cancer: Approaches to resolve cell-to-cell heterogeneity. Cancers (Basel) 2019;11:226. doi: 10.3390/cancers11020226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carter SL, Eklund AC, Kohane IS, Harris LN, Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–8. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 60.Stirling PC, Bloom MS, Solanki-Patil T, Smith S, Sipahimalani P, Li Z, et al. The complete spectrum of yeast chromosome instability genes identifies candidate cin cancer genes and functional roles for astra complex components. PLoS Genet. 2011;7:e1002057. doi: 10.1371/journal.pgen.1002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Couch FJ, Kuchenbaecker KB, Michailidou K, Mendoza-Fandino GA, Nord S, Lilyquist J, et al. Identification of four novel susceptibility loci for oestrogen receptor negative breast cancer. Nat Commun. 2016;7:11375. doi: 10.1038/ncomms11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li J, Zuo Z, Lai S, Zheng Z, Liu B, Wei Y, et al. Differential analysis of RNA methylation regulators in gastric cancer based on TCGA data set and construction of a prognostic model. J Gastrointest Oncol. 2021;12:1384–97. [DOI] [PMC free article] [PubMed]
- 63.Nombela P, Miguel-López B, Blanco S. The role of m6A, m5C and Ψ RNA modifications in cancer: Novel therapeutic opportunities. Mol Cancer. 2021;20:18. doi: 10.1186/s12943-020-01263-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barbieri I, Kouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020;20:303–22. doi: 10.1038/s41568-020-0253-2. [DOI] [PubMed] [Google Scholar]
- 65.Molina JR, Sun Y, Protopopova M, Gera S, Bandi M, Bristow C, et al. An inhibitor of oxidative phosphorylation exploits cancer vulnerability. Nat Med. 2018;24:1036–46. doi: 10.1038/s41591-018-0052-4. [DOI] [PubMed] [Google Scholar]
- 66.Vasan K, Werner M, Chandel NS. Mitochondrial metabolism as a target for cancer therapy. Cell Metab. 2020;32:341–52. doi: 10.1016/j.cmet.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barbi de Moura M, Vincent G, Fayewicz SL, Bateman NW, Hood BL, Sun M, et al. Mitochondrial respiration–an important therapeutic target in melanoma. PLoS One. 2012;7:e40690. doi: 10.1371/journal.pone.0040690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ho J, de Moura MB, Lin Y, Vincent G, Thorne S, Duncan LM, et al. Importance of glycolysis and oxidative phosphorylation in advanced melanoma. Mol Cancer. 2012;11:76. doi: 10.1186/1476-4598-11-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aminzadeh‐gohari S, Weber DD, Catalano L, Feichtinger RG, Kofler B, Lang R. Targeting mitochondria in melanoma. Biomolecules. 2020;10:1–21. doi: 10.3390/biom10101395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schöckel L, Glasauer A, Basit F, Bitschar K, Truong H, Erdmann G, et al. Targeting mitochondrial complex I using BAY 87-2243 reduces melanoma tumor growth. Cancer Metab. 2015;3:11. doi: 10.1186/s40170-015-0138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sundstrøm T, Prestegarden L, Azuaje F, Aasen SN, Røsland GV, Varughese JK, et al. Inhibition of mitochondrial respiration prevents BRAF-mutant melanoma brain metastasis. Acta Neuropathol Commun. 2019;7:55. doi: 10.1186/s40478-019-0712-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr Biol. 2006;16:R551–60. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 73.Rambold AS, Lippincott-Schwartz J. Mechanisms of mitochondria and autophagy crosstalk. Cell Cycle. 2011;10:4032–8. doi: 10.4161/cc.10.23.18384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Okamoto K, Kondo-Okamoto N. Mitochondria and autophagy: critical interplay between the two homeostats. Biochim Biophys Acta. 2012;1820:595–600. doi: 10.1016/j.bbagen.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 75.Donnelly N, Passerini V, Dürrbaum M, Stingele S, Storchová Z. HSF 1 deficiency and impaired HSP 90‐dependent protein folding are hallmarks of aneuploid human cells. EMBO J. 2014;33:2374–87. doi: 10.15252/embj.201488648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kuznetsova AY, Seget K, Moeller GK, de Pagter MS, de Roos JADM, Dürrbaum M, et al. Chromosomal instability, tolerance of mitotic errors and multidrug resistance are promoted by tetraploidization in human cells. Cell Cycle. 2015;14:2810. doi: 10.1080/15384101.2015.1068482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Salvador F, Martin A, López-Menéndez C, Moreno-Bueno G, Santos V, Vázquez-Naharro A, et al. Lysyl oxidase-like protein LOXL2 promotes lung metastasis of breast cancer. Cancer Res. 2017;77:5846–59. doi: 10.1158/0008-5472.CAN-16-3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frank SB, Schulz VV, Miranti CK. A streamlined method for the design and cloning of shRNAs into an optimized Dox-inducible lentiviral vector. BMC Biotechnol. 2017;17:24. doi: 10.1186/s12896-017-0341-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cabezas-Sainz P, Guerra-Varela J, Carreira MJ, Mariscal J, Roel M, Rubiolo JA, et al. Improving zebrafish embryo xenotransplantation conditions by increasing incubation temperature and establishing a proliferation index with ZFtool. BMC Cancer. 2018;18:3. doi: 10.1186/s12885-017-3919-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The materials described in the manuscript, including all relevant raw data, will be freely available to any researcher wishing to use them for non-commercial purposes, without breaching participant confidentiality.