Sun et al. found that STAT3, activated by IL-10 and IL-21, regulates the terminal differentiation of effector CD8+ T cells in cancer and also plays an important role in effector CD8+ T cells in acute infection.
Abstract
In cancer, persistent antigens drive CD8+ T cell differentiation into exhausted progenitor (Texprog) and terminally exhausted (Texterm) cells. However, how the extrinsic and intrinsic regulatory mechanisms cooperate during this process still remains not well understood. Here, we found that STAT3 signaling plays essential roles in promoting intratumor Texterm cell development by enhancing their effector functions and survival, which results in better tumor control. In tumor microenvironments, STAT3 is predominantly activated by IL-10 and IL-21, but not IL-6. Besides, STAT3 also plays critical roles in the development and function of terminally differentiated effector CD8+ T cells in acute infection. Mechanistically, STAT3 transcriptionally promotes the expression of effector function-related genes, while it suppresses those expressed by the progenitor Tex subset. Moreover, STAT3 functions in collaboration with BATF and IRF4 to mediate chromatin activation at the effector gene loci. Thus, we have elucidated the roles of STAT3 signaling in terminally differentiated CD8+ T cell development, especially in cancer, which benefits the development of more effective immunotherapies against tumors.
Introduction
In acute infection and vaccination, effector CD8+ T cells develop into two subpopulations: one subpopulation is KLRG1+IL-7Rlow short-lived effector CD8+ T cells, also termed terminally differentiated effector CD8+ T cells, and another, KLRG1−IL-7Rhi memory precursors, which can subsequently develop into memory CD8+ T cells after the clearance of antigens (Joshi et al., 2007; Rutishauser et al., 2009). However, in cancer and chronic infection, persistent antigenic stimulation drives CD8+ T cells into an exhaustion state (McLane et al., 2019; Pauken and Wherry, 2015). Exhausted CD8+ T cells (Tex) are functionally distinct from effector and memory CD8+ T cells observed in acute infections and are characterized by elevated expression of inhibitory receptors (PD-1, LAG3, TIGIT, etc.), poor effector functions, altered epigenetic and transcriptional landscapes, and skewed metabolic status (McLane et al., 2019). Similar to the classification of effector CD8+ T cells, in cancer, tumor-specific Tex cells are mainly classified into two subpopulations, named progenitors (Texprog) and terminally differentiated CD8+ T (Texterm) cells (Miller et al., 2019; Siddiqui et al., 2019). Tumor-specific Texprog cells are stem-like and highly express TCF1 transcription factor, which persist long term and are capable of differentiating into Texterm cells for long-term tumor control (Miller et al., 2019; Siddiqui et al., 2019). A distinguishing feature of memory CD8+ T cells is their ability to rapidly expand and efficiently generate effector functions in response to antigen re-stimulation. Although Texprog cells are stem-like, their memory potentials are progressively lost and not at the levels of memory CD8+ T cells generated in acute infection (Angelosanto et al., 2012; Huang et al., 2022; Kaech and Cui, 2012; McLane et al., 2019). In contrast, tumor-specific TIM-3+ Texterm cells have characteristics of potent cytotoxicity, active proliferative capacity, and high-level apoptosis (Miller et al., 2019; Siddiqui et al., 2019). However, the transcriptional and epigenetic mechanisms underlying CD8+ T cell differentiation and maintenance, especially Texterm cells in cancer, remain elusive, and the cell-intrinsic and -extrinsic factors controlling Tex cell differentiation also need to be further identified.
T cell differentiation is generally regulated by three signals: signal 1 from antigen receptors, signal 2 from costimulatory or inhibitory molecules, and signal 3 from inflammatory cytokines (McLane et al., 2019). During chronic infection and cancer, persistent T cell receptor stimulation sustains the activation of NFAT and induces high-level expression of TOX and Nr4a1, which limit T cell function and promote T cell exhaustion (Chen et al., 2019; Khan et al., 2019; Liu et al., 2019; Martinez et al., 2015; Scott et al., 2019). In addition, elevated expression of coinhibitory receptors (e.g., PD-1 and CTLA-4) in Tex cells also enforces exhaustion, and the blockade of their signaling has shown clinical benefits in various human cancer types (Callahan et al., 2016; Kamphorst et al., 2017). Moreover, it has been reported that diminished TCR and costimulatory signaling in established chronic infection preferentially programs Tex cells into TCF1+ memory-like subset (Snell et al., 2018). Importantly, in addition to TCR and costimulatory/inhibitory molecules, Tex cell generation is also shaped by a broad range of inflammatory cytokines (McLane et al., 2019). Among them, JAK-STAT cytokines are critical in regulating T cell development in immune responses (Dong, 2021). For example, STAT1 is critical in promoting T cell expansion and antitumor response in cancer (Ryan et al., 2020). Consistently, STAT1 upstream cytokines, including type I IFN and IL-27, function to drive TIM-3+ Tex cell differentiation in both cancer and chronic infection (Chihara et al., 2018; Wu et al., 2016; Yang et al., 2012). In addition, IL-2–STAT5 axis functions as an inducer of exhaustion and promotes the generation of TIM-3+ Tex cells within tumors (Liu et al., 2021).
STAT3 is a cytoplasmic transcription factor belonging to the signal transducer and activators of transcription (STAT) family (Darnell, 1997). It can be phosphorylated and activated by multiple cytokines (e.g., IL-6, IL-10, IL-21, and IL-23), and it transcriptionally regulates the fate decisions of various CD4+ T lymphocyte lineages (Dong, 2021). The IL-6/IL-23–STAT3 axis induces RORγt expression during Th17 cell development (Chang et al., 2020; Yang et al., 2007), and the IL-6/IL-21–STAT3 pathway also promotes Bcl6 expression during Tfh cell differentiation (Nurieva et al., 2009; Xu et al., 2019). However, the role of STAT3 in regulating CD8+ T cell differentiation and function remains controversial and lacks mechanistic understanding. More importantly, its role in tumor-specific Tex cell development still remains unclear. STAT3 was long considered to promote memory CD8+ T cell development (Cui et al., 2011; Hanna et al., 2021; Olson and Jameson, 2011; Siegel et al., 2011). A previous study showed that Stat3 deficiency impaired the maturation and self-renewal of IL-7RhiKLRG1low memory CD8+ T cells in acute LCMV virus–infected mice (Cui et al., 2011). The development and maintenance of human central memory CD8+ T cells were also diminished in autosomal-dominant hyper-IgE syndrome patients with STAT3 mutations (Siegel et al., 2011). Moreover, in the chronic lymphocytic leukemia model, Stat3 deletion lowered the frequencies of memory-like PD-1intTCF-1+ CD8+ T cells (Hanna et al., 2021), and the leptin-induced STAT3 activation in obesity-driven breast cancer inhibited IFN-γ production and CD8+ T cell effector function (Zhang et al., 2020). However, contrary to these findings, another study showed that Stat3 deficiency decreased KLRG-1+ effector CD8+ T cells in HSV-1 infection (Yu et al., 2013) and reduced the expression of Blimp1, BATF, and Granzyme B in IL-21–stimulated CD8+ T cells in vitro (Xin et al., 2015). Thus, the role of STAT3 signaling in CD8+ T cell differentiation and function, especially in the context of tumor antigen–driven exhaustion, urgently needs to be clarified via a detailed mechanistic approach, and its upstream inducers within the tumors also need to be systematically identified.
In this study, we illustrated the function of STAT3 signaling in terminally differentiated CD8+ T cells, especially in cancer. Activated by IL-10 and IL-21, STAT3 played critical roles in promoting tumor-specific Texterm cell development and their effector functions in cancer. It was also intrinsically critical in promoting the development and function of terminally differentiated effector CD8+ T cells in acute infection. Mechanistically, STAT3 transcriptionally promoted the expression of Texterm-associated genes and inhibited that of Texprog genes, thus enforcing the Texterm transcriptional program. STAT3 functioned by epigenetically activating multiple Texterm-related gene loci. In addition, STAT3 cooperated with BATF and IRF4 to mediate Texterm cell differentiation. Altogether, this study has mechanistically clarified the role of STAT3 signaling in terminally differentiated CD8+ T cells, especially in cancer, which may benefit the development of novel cancer immunotherapies.
Results
STAT3 promotes the effector function of CD8+ T cells
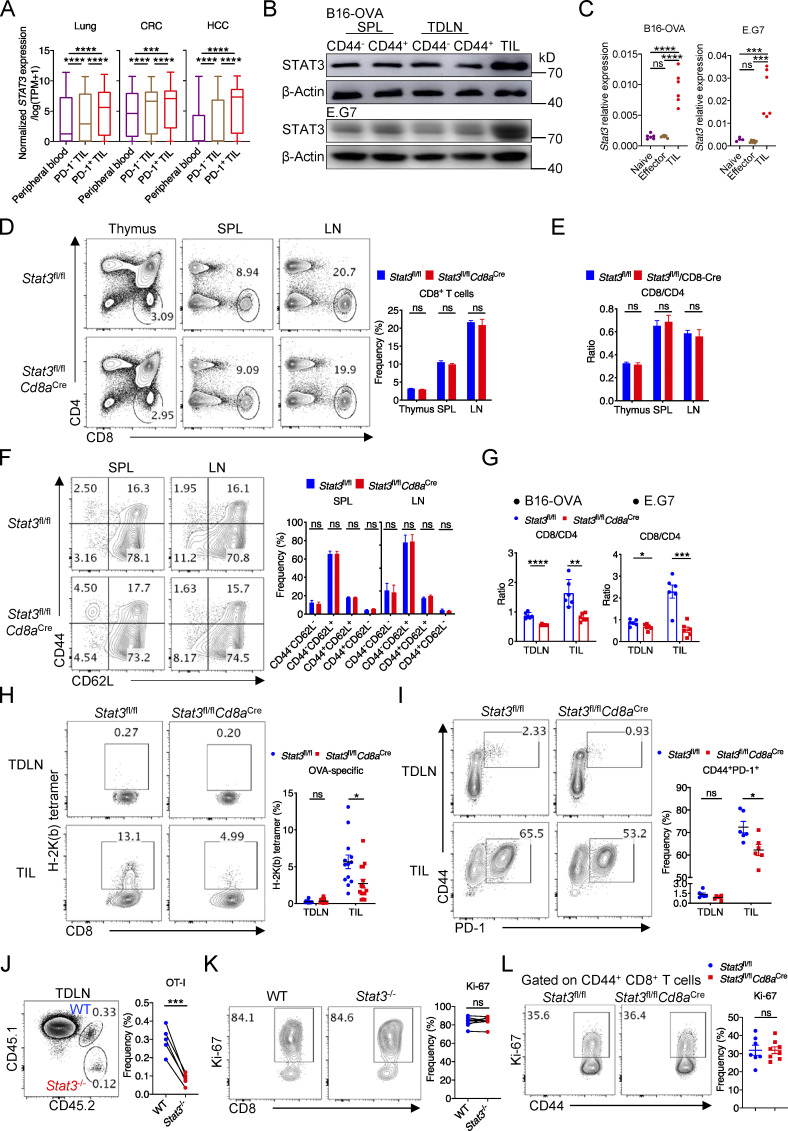
To first understand the function of STAT3 in the regulation of anti-cancer CD8+ T cells, we analyzed public single-cell RNA sequencing (scRNA-seq) data of human patients with lung cancer, colorectal cancer (CRC), and hepatocellular carcinoma (HCC; Guo et al., 2018; Zhang et al., 2018; Zheng et al., 2017). We found that STAT3 expression was elevated in tumor-infiltrating CD8+ T cells, especially in PD-1+ Tex cells, as compared with peripheral CD8+ T cells in the blood (Fig. S1 A). High levels of Stat3 expression were also found in tumor-infiltrating CD8+ T cells in murine B16-OVA and E.G7 tumor models at both transcriptional and protein levels (Fig. S1, B and C).
Figure S1.
STAT3 is intrinsically critical for the effector function of CD8+ T cells. (A) mRNA level of STAT3 in CD8+ T cells from peripheral blood or tumors in human cancer patients (GEO accession nos. GSE99254, GSE108989, and GSE98638). scRNA-seq data of CD8+ T cells in tumors and peripheral blood from lung, CRC, and HCC patients were previously processed and uploaded by others (http://crc.cancer-pku.cn/, http://hcc.cancer-pku.cn/, http://lung.cancer-pku.cn/; Guo et al., 2018; Zhang et al., 2018; Zheng et al., 2017), and we obtained the STAT3 expression values from the above websites. The expression unit was normalized expression/log2(TPM+1). We compared the STAT3 expression values in CD8+ T cells from peripheral blood, and in PD-1–negative (PDCD1 expression value = 0) and –positive (PDCD1 expression value >0) CD8+ T cells from tumors. (B) STAT3 protein expression in CD8+ T cells from spleens, TDLNs, and tumors in B16-OVA (upper plot) and E.G7 (bottom plot) models. (C) mRNA levels of Stat3 in CD8+ T cells from TDLNs and tumors in B16-OVA (left panel) and E.G7 (right panel) models. (D) Frequencies of CD8+ T cells in thymuses, spleens, and lymph nodes in 6- to 8-wk-old Stat3fl/fl (n = 5) and Stat3fl/flCd8aCre (n = 5) mice. (E) Ratios of CD8+ T cells to CD4+ T cells in the thymuses, spleens, and lymph nodes from 6- to 8-wk-old Stat3fl/fl (n = 5) and Stat3fl/flCd8aCre (n = 5) mice. (F) Representative plots (left panel) and quantifications (right panel) of CD44 and CD62L expression on CD8+ T cells in the spleens and lymph nodes from 6- to 8-wk-old Stat3fl/fl (n = 5) and Stat3fl/flCd8aCre (n = 5) mice. (G) Ratios of CD8+ T cells to CD4+ T cells in TDLNs and tumors. (H) Representative plots (left panel) and quantifications (right panel) of H-2K(b) tetramer+ CD8+ T cells among total CD8+ T cells from TDLNs and E.G7 tumors. (I) Representative plots (left panel) and quantifications (right panel) of CD44+ PD-1+ CD8+ T cells among total CD8+ T cells from TDLNs and E.G7 tumors. (J) OT-I TIL frequencies among total CD8+T cells from TDLNs in B16-OVA model on day 20 after tumor inoculation. (K) Representative plots (left panel) and quantifications (right panel) of Ki-67 expression in OT-I TILs from B16-OVA tumors on day 20 after tumor inoculation. (L) Representative plots (left panel) and quantifications (right panel) of Ki-67 expression in splenic CD44+ CD8+ T cells on day 8 after LM-OVA infection. Data are pooled from three independent experiments (H), or are representative of two (B–F, G [left], J, and L) or three (G [right], I, and K) independent experiments. Data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by one-way ANOVA (A and C), unpaired two-tailed Student’s t test (D–I and L) or paired two-tailed Student’s t test (J and K). Source data are available for this figure: SourceData FS1.
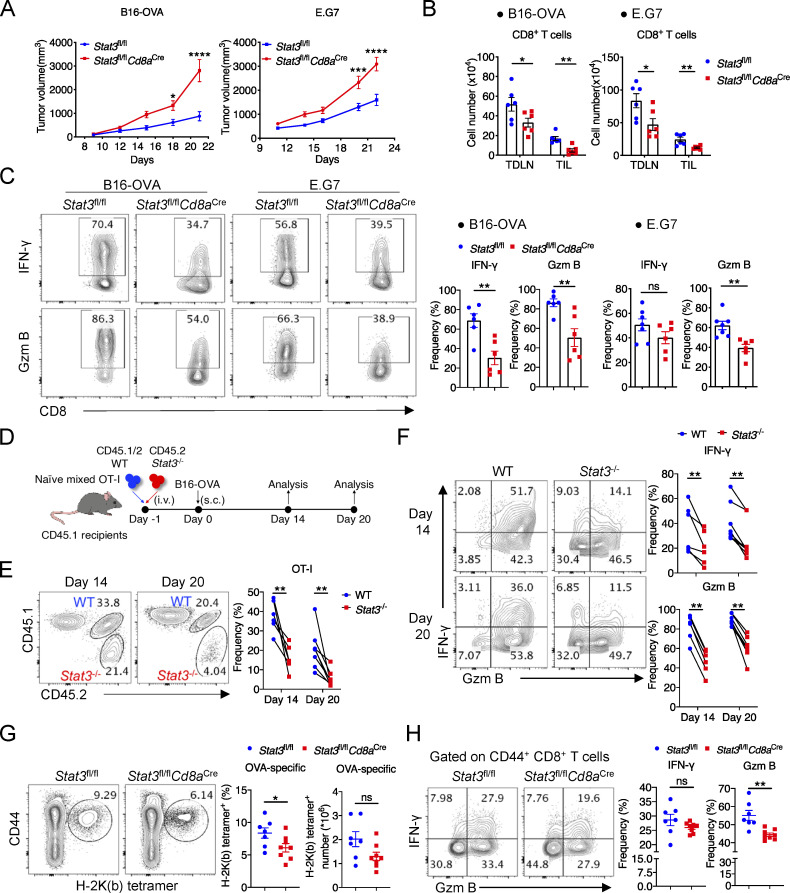
To investigate the function of STAT3 in anti-tumor CD8+ T cells, we generated Stat3fl/flCd8aCre mice, in which Stat3 was specifically deleted in mature CD8+ T cells (Maekawa et al., 2008; Takeda et al., 1998). CD8+ T cell development was intact in the thymus and peripheral lymphoid organs in 6–8-wk-old Stat3fl/flCd8aCre mice as compared with Stat3fl/fl mice (Fig. S1, D and E), and their homeostasis in peripheral spleens and lymph nodes was not significantly affected by the deficiency of Stat3 (Fig. S1 F). To further explore the roles of STAT3 in CD8+ T cell function in tumor models, we inoculated Stat3fl/flCd8aCre mice, as well as Stat3fl/fl control mice, with B16-OVA or E.G7 tumor cells. We found that Stat3 deficiency in CD8+ T cells resulted in enhanced growth of B16-OVA and E.G7 tumors (Fig. 1 A). As a result of Stat3 deficiency, the numbers of CD8+ T cells decreased substantially in both tumor-draining lymph nodes (TDLNs) and tumors (Fig. 1 B), and so did the ratio of CD8+ T cells to CD4+ T cells (Fig. S1 G). More importantly, the frequencies of OVA-specific tumor-infiltrating CD8+ T cells (TILs) and CD44+PD-1+ antigen–experienced CD8+ TILs were significantly reduced in E.G7-bearing Stat3fl/flCd8aCre mice (Fig. S1, H and I). In addition, the cytotoxic function of PD-1+–exhausted Stat3−/− CD8+ T cells was dramatically reduced as indicated by decreased production of IFN-γ and Granzyme B in both B16-OVA and E.G7 models (Fig. 1 C). These data indicated a critical role of STAT3 in antitumor response of CD8+ T cells.
Figure 1.
STAT3 promotes the effector function of CD8+ T cells. (A) Mean volumes of B16-OVA tumors (left panel) in Stat3fl/fl (n = 6) and Stat3fl/flCd8aCre (n = 6) mice, or E.G7 tumors (right panel) in Stat3fl/fl (n = 7) and Stat3fl/flCd8aCre (n = 6) mice. (B) Numbers of CD8+ T cells in TDLNs and tumors from Stat3fl/fl and Stat3fl/flCd8aCre mice. (C) Representative plots (left panel) and quantifications (right panel) of IFN-γ and Granzyme B production in CD44+PD-1+ CD8+ TILs from B16-OVA (left panel) or E.G7 (right panel) tumors. (D) Experimental design for adoptive cotransfer of naive WT and Stat3−/− OT-I cells and B16-OVA tumor–challenge assay. (E) OT-I TIL frequencies among total CD8+TILs from B16-OVA tumors on day 14 and day 20 after tumor inoculation. (F) Representative plots (left panel) and quantifications (right panel) of IFN-γ and Granzyme B production in OT-I TILs from B16-OVA tumors. (G) Frequencies of OVA-specific CD8+ T cells and their numbers (P = 0.0590) in LM-OVA–infected Stat3fl/fl and Stat3fl/flCd8aCre mice on day 8. (H) Representative plots (left panel) and quantifications (right panel) of IFN-γ and Granzyme B production in splenic CD44+ CD8+ T cells on day 8 after LM-OVA infection. Data are representative of two (A [left], B [left], C [left], G, and H) or three (A [right], B [right], C [right], and D–F) independent experiments. Data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by two-way ANOVA (A), unpaired two-tailed Student’s t test (B, C, G, and H), or paired two-tailed Student’s t test (E and F).
To further understand the function of STAT3 in tumor-specific CD8+ T cells and to rule out the impact of tumor volumes, we crossed Stat3fl/flCd8aCre mice into CD45.2 OT-I strain and cotransferred equal numbers of their naive OT-I cells and CD45.1/2 WT OT-I cells into CD45.1 recipients with B16-OVA tumors (Fig. 1 D). Consistent with the above results, the frequencies of Stat3−/− OT-I cells among total CD8+ TILs were dramatically reduced as compared with WT OT-I cells on both day 14 and day 20 after inoculation (Fig. 1 E). The percentages of Stat3−/− OT-I cells were also substantially decreased in TDLNs on day 20 (Fig. S1 J). Importantly, IFN-γ and Granzyme B production was significantly defective in Stat3−/− OT-I TILs (Fig. 1 F). However, Ki-67 expression in OT-I TILs was not affected by Stat3 deficiency (Fig. S1 K). These results demonstrated that STAT3 was intrinsically required for the homeostasis and cytotoxic function of CD8+ T cells in cancer.
We next examined STAT3 functions in effector CD8+ T cells using an acute infection model. We infected Stat3fl/flCd8aCre mice and their littermate Stat3fl/fl mice with Listeria-OVA (LM-OVA) bacteria and analyzed them on day 8. Consistent with the data in tumor models, reduced percentages and numbers of OVA-specific CD8+ T cells were observed in spleens from infected Stat3fl/flCd8aCre mice (Fig. 1 G), and the percentages of Granzyme B–producing cells were significantly decreased by Stat3 deletion in splenic CD8+ T cells (Fig. 1 H). However, the frequency of Ki-67–expressing cells was comparable in Stat3fl/flCd8aCre and Stat3fl/fl mice (Fig. S1 L). These results altogether indicated a critical role of STAT3 in the homeostasis and function of terminally differentiated CD8+ T cells in both tumor and acute infection models.
STAT3 promotes the development of terminally differentiated CD8+ T cells
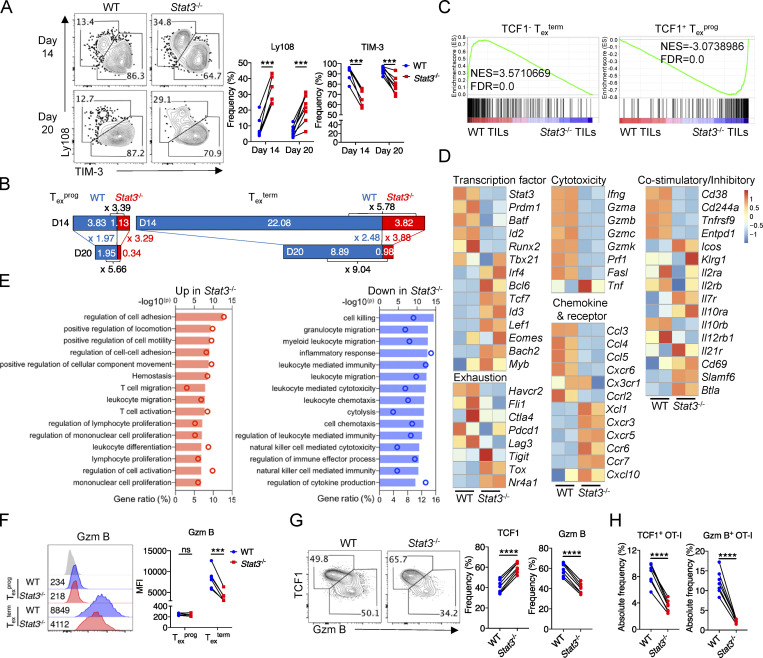
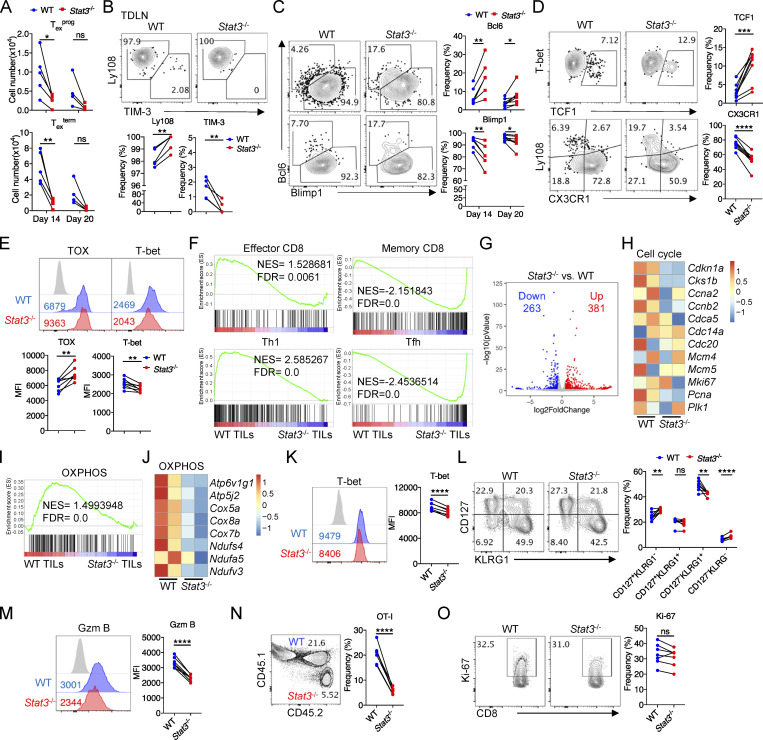
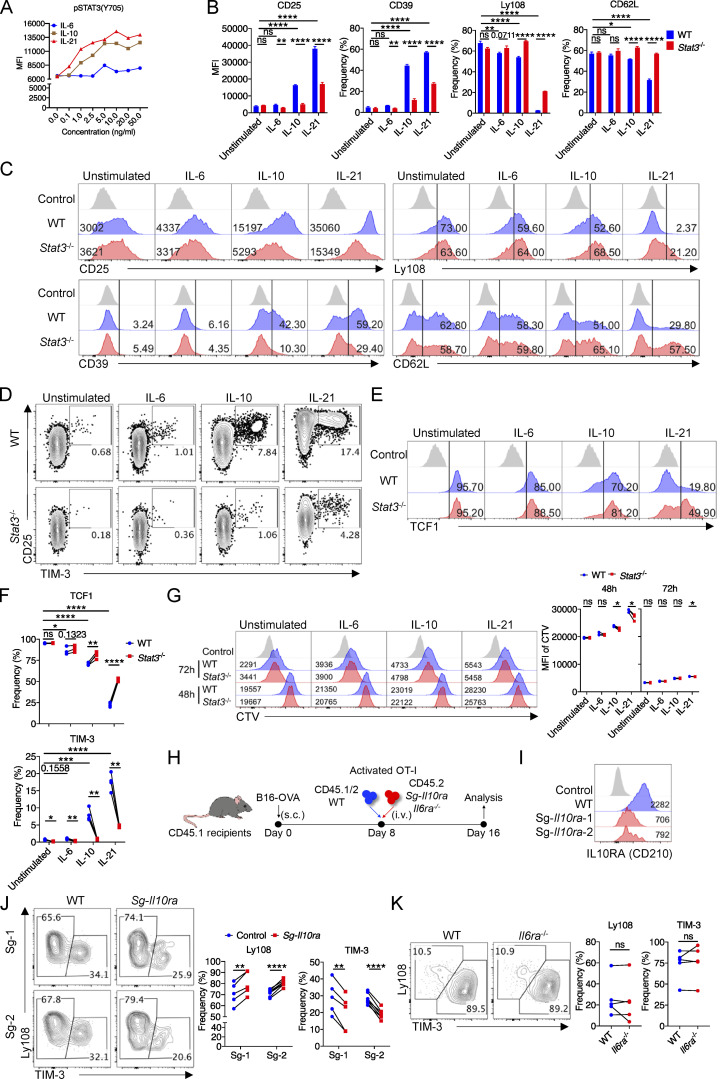
Since Texterm vs. Texprog cells mediate the cytotoxic function and long-term maintenance of Tex cells, respectively (Miller et al., 2019), we next investigated whether STAT3 plays any role in the regulation of tumor-specific Tex cell fates. By analyzing cotransferred WT and Stat3−/− OT-I cells from B16-OVA tumors, we found that the percentages of Ly108−TIM-3+ Texterm cells were dramatically reduced, while those of Ly108+TIM-3− Texprog cells were significantly increased by the deficiency of Stat3 on both day 14 and day 20 (Fig. 2 A). Importantly, both the Texterm and Texprog TIL numbers significantly decreased as a result of the deficiency in Stat3 on day 14 and day 20, and Texterm cells were reduced to a greater extent than Texprog cells (Fig. 2, B and A). Moreover, by comparing the numbers of Texterm and Texprog TILs between day 14 and day 20, we found that the maintenance of Texterm and Texprog cells was both decreased by the deletion of Stat3, especially the Texterm cells (Fig. 2 B). Consistently, decreased frequencies of TIM-3+ cells in Stat3−/− OT-I cells were also observed in TDLNs, while almost all OT-I cells were maintained as Ly108+ progenitor cells and a lesser number was TIM-3+ Texterm cells in TDLNs, as previously reported (Schenkel et al., 2021; Fig. S2 B). In addition, the frequencies of TCF1+ and Bcl6+Blimp1− cells in Stat3−/− OT-I TILs became higher than those in WT OT-I TILs, while the percentages of CX3CR1+Ly108− and Bcl6−Blimp1+ cells in Stat3−/− OT-I TILs significantly decreased (Fig. S2, C and D). Moreover, the mean fluorescence intensities of TOX staining in Stat3−/− OT-I TILs increased, while those of T-bet decreased as compared with WT OT-I TILs (Fig. S2 E). These results indicated that STAT3 might function to promote Texterm vs. Texprog cell differentiation, and it was also critical for the maintenance of Tex cells, especially the Texterm cells.
Figure 2.
STAT3 promotes the development of terminally differentiated CD8+ T cells. (A) Representative plots (left panel) and quantifications (right panel) of Ly108 and TIM-3 expression on OT-I cells from B16-OVA tumors on day 14 and day 20 after tumor inoculation. (B) Stacked bar graphs showing the numbers (× 104) of Ly108+TIM-3− Texprog and Ly108−TIM-3+ Texterm cells based on experimental results in Fig. S2 A. The width of the stacked bar represents cell numbers (× 104). (C) GSEA results for identifying gene signatures of WT and Stat3−/− OT-I TILs compared with other T cell subsets (GEO accession no. GSE114631). NES, normalized enrichment score; FDR, false discovery rate q-value. (D) Heatmaps illustrating the relative expression of signature genes in WT and Stat3−/− OT-I TILs. (E) Lists of top 15 Gene Ontology (GO) biological pathways for DEGs of WT and Stat3−/− OT-I TILs. P value presented as −log10(P value). Columns designate −log10(P value) and circles designate gene ratio (%). (F) Representative plots (left panel) and quantification (right panel) of Granzyme B production level in TIM-3− Texprog and TIM-3+ Texterm OT-I cells from B16-OVA tumors. (G) Representative plots (left panel) and quantifications (right panel) of TCF1 and Granzyme B expression in splenic OT-I cells on day 8 after LM-OVA infection. (H) Absolute frequencies of TCF1+ and Granzyme B+ OT-I cells in the spleen, which were the product of OT-I cell frequencies and TCF1+ or Granzyme B+ cell ratios. Data are representative of two (G and H) or three (A–C and F) independent experiments. Data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by paired two-tailed Student’s t test (A and F–H).
Figure S2.
STAT3 promotes the development of terminally differentiated CD8+ T cells. (A) Numbers of Texprog (upper panel) and Texterm (bottom panel) OT-I cells from B16-OVA tumors on day 14 and day 20 after tumor inoculation. (B) Representative plots (upper panel) and quantifications (bottom panel) of Ly108 and TIM-3 expression on OT-I cells from TDLNs in B16-OVA tumor model on day 20 after tumor inoculation. (C) Representative plots (left panel) and quantifications (right panel) of Bcl6 and Blimp1 expression in OT-I cells from B16-OVA tumors on day 14 and day 20 after tumor inoculation. (D) Representative plots (left panel) and quantifications (right panel) of TCF1 and CX3CR1 expression in/on OT-I cells from B16-OVA tumors on day 20 after tumor inoculation. (E) Representative plots (upper panel) and quantifications (bottom panel) of TOX and T-bet expression in OT-I cells from B16-OVA tumors on day 20 after tumor inoculation. (F) GSEA results for identifying gene signatures of WT and Stat3−/− OT-I TILs compared with other T cell subsets (GEO accession nos. GSE65660 and GSE8678). NES, normalized enrichment score; FDR, false discovery rate q-value. (G) Volcanic plot showing DEGs in WT and Stat3−/− OT-I TILs. (H) Heatmap illustrating the relative expression of signature genes related to cell cycle pathway in WT and Stat3−/− OT-I TILs. (I) GSEA result for identifying gene signatures of WT and Stat3−/− OT-I TILs compared to OXPHOS feature genes (GSEA: MM3893). NES, normalized enrichment score; FDR, false discovery rate q-value. (J) Heatmap illustrating the relative expression of signature genes related to OXPHOS pathway in WT and Stat3−/− OT-I TILs. (K) Representative plots (left panel) and quantification (right panel) of T-bet expression in splenic OT-I cells on day 8 after LM-OVA infection. (L) Representative plots (left panel) and quantifications (right panel) of CD127 and KLRG1 expression on splenic OT-I cells on day 8 after LM-OVA infection. (M) Representative plots (left panel) and quantification (right panel) of Granzyme B expression in TCF1−Granzyme B+ splenic OT-I cells on day 8 after LM-OVA infection. (N) OT-I cell frequencies among total splenic CD8+T cells on day 8 after LM-OVA infection. (O) Representative plots (left panel) and quantification (right panel) of Ki-67 expression in splenic OT-I cells on day 8 after LM-OVA infection. Data are representative of two (K–O) or three (A–E) independent experiments. Data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by paired two-tailed Student’s t test (A–E and K–O).
To further analyze tumor-specific Stat3−/− CD8+ TILs at the transcriptional level, we performed RNA-seq for cotransferred WT and Stat3−/− OT-I TILs from B16-OVA tumors on about day 16. Consistent with the above flow cytometry results, GSEA (Gene Set Enrichment Analysis) results revealed that the transcriptome of WT OT-I TILs was relatively more similar with TCF1− Texterm subset, while Stat3−/− OT-I TILs were more transcriptionally related to TCF1+ Texprog cells (Fig. 2 C). In addition, signature genes of effector CD8+ T cells and Th1 cells were more enriched in WT OT-I TILs, while those of memory CD8+ T cells and Tfh cells were relatively concentrated in Stat3−/− OT-I TILs (Fig. S2 F). The expression of 263 genes was significantly downregulated in Stat3−/− OT-I TILs, while that of 381 genes was elevated by the deficiency of Stat3 (Fig. S2 G and Table S1). Specifically, the mRNA expression of Texterm-related surface molecules (Havcr2, Entpd1, Cx3cr1, etc.) and transcriptional factors (Prdm1, Id2, Batf, etc.) was significantly downregulated, while the expression of numerous Texprog-associated surface molecules (Cxcr5, Ccr7, Slamf6, Btla, etc.) and transcriptional factors (Tcf7, Id3, Bcl6, Bach2, etc.) was upregulated in Stat3−/− OT-I TILs (Fig. 2 D), which were consistent with their protein expression. In addition, pathway analyses revealed that several cell killing–related pathways were downregulated in Stat3−/− OT-I TILs (Fig. 2 E), consistent with the decreased expression of cytotoxic molecules (Gzmb and Ifng) at both transcriptional and protein levels. Moreover, multiple pathways involved in lymphocyte differentiation, activation, adhesion, and migration were also significantly affected by the deficiency of Stat3 (Fig. 2 E). However, the expression of many cell cycle–related genes was comparable in WT and Stat3−/− OT-I TILs at the transcriptional level (Fig. S2 H), which was consistent with the above Ki-67 staining results (Fig. S1 K), indicating that STAT3 might have limited effect on CD8+ T cell proliferation in tumors. In addition, GSEA analysis revealed that genes involved in the OXPHOS pathway were relatively enriched in WT OT-I cells as compared with Stat3−/− OT-I TILs, and mRNA levels for some of them decreased dramatically in Stat3−/− OT-I TILs (Fig. S2, I and J). These results suggested that STAT3 might be critical in promoting OXPHOS and maintaining the mitochondrial fitness of Tex cells within tumors, which was consistent with its functions previously found in regulating cellular respiration and homeostasis (Huynh et al., 2019; Wegrzyn et al., 2009).
To further analyze the specific roles of STAT3 in regulating the cytotoxic function of Texprog and Texterm cells, we assessed the Granzyme B expression in Stat3−/− Texprog and Texterm cells. Granzyme B expression levels were significantly reduced in Stat3-deficient Texterm cells, while those in Texprog cells were not affected, as compared with the corresponding WT cells (Fig. 2 F), indicating that STAT3 might play more important roles in promoting the cytolytic function of Texterm cells.
To further analyze the role of STAT3 in antigen-specific CD8+ T cells in acute infection, we next adoptively cotransferred naive CD45.1/2 WT and CD45.2 Stat3−/− OT-I cells at 1:1 ratio into CD45.1 recipients 1 d prior to LM-OVA infection. Consistent with the results in tumors, TCF1−Granzyme B+ cell frequencies were significantly reduced and TCF1+Granzyme B− cell percentages were increased in the absence of Stat3 (Fig. 2 G). T-bet expression levels were also downregulated in splenic Stat3−/− OT-I cells (Fig. S2 K). Consistently, the percentages of CD127−KLRG1+ short-lived effector CD8+ T cells were reduced and those of CD127+KLRG1− memory precursors were increased in the deficiency of Stat3 (Fig. S2 L). Moreover, the expression levels of Granzyme B in TCF1−Granzyme B+ Stat3−/− OT-I cells were also significantly reduced (Fig. S2 M). These results support the essential role of Stat3 in the development of cytolytic effector CD8+ T cells. Consistently, Stat3 deficiency resulted in lower percentages of OT-I cells in the spleen, and both TCF1+Granzyme B− and TCF1−Granzyme B+ OT-I cell numbers were decreased by Stat3 deletion (Fig. 2 H and Fig. S2 N). However, the proliferation of OT-I cells was unaffected in the absence of Stat3 as indicated by the similar percentages of Ki-67–expressing cells (Fig. S2 O).
These results altogether indicated that STAT3 played critical roles in regulating the differentiation, survival, and function of CD8+ T cells in cancer and acute infection, especially in favoring terminally differentiated CD8+ T cell fitness and effector functions.
IL-10 and IL-21 promote STAT3 activation in Texterm cells in cancer
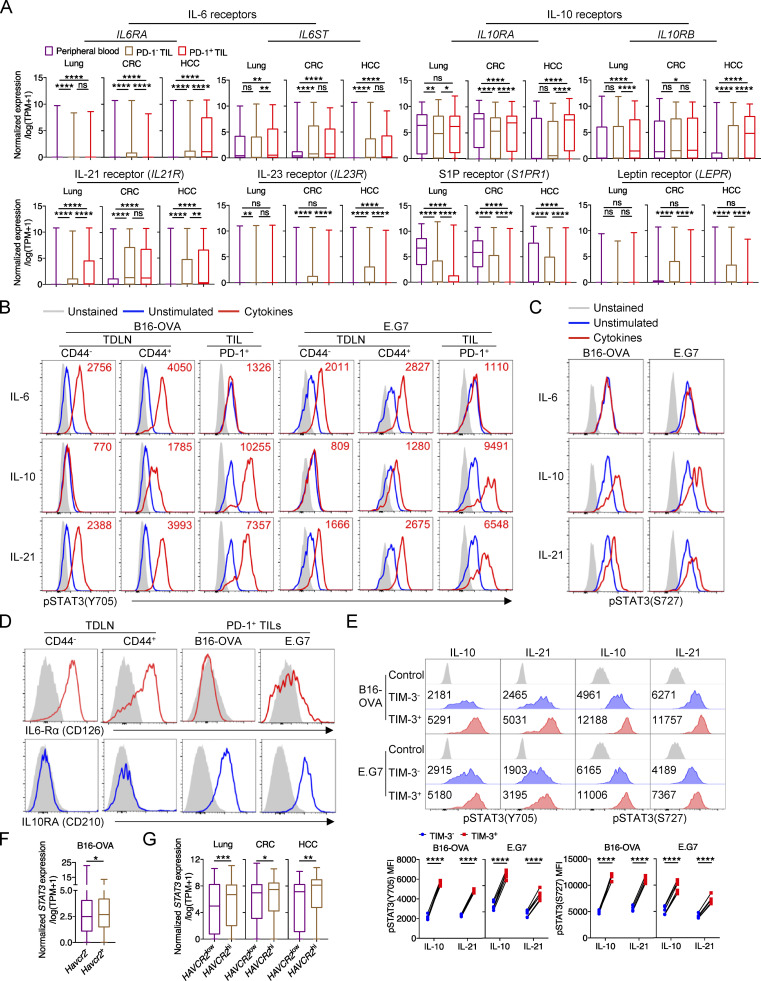
Multiple STAT3-activating cytokines have been implicated in tumor immunity (Cui et al., 2021; Guo et al., 2021; Hanna et al., 2021; Yu et al., 2014; Yu et al., 2009). Thus, to identify the upstream extracellular inducers for STAT3 activation within tumor-infiltrating CD8+ T cells, we first analyzed the expression of receptors for its canonical inducing cytokines in CD8+ TILs from public single-cell datasets of human cancer patients (Guo et al., 2018; Zhang et al., 2018; Zheng et al., 2017). We found that IL-10 receptors (IL10RA and IL10RB) were highly expressed in tumor-infiltrating CD8+ T cells in lung cancer, CRC, and HCC patients (Fig. 3 A). IL21R expression was also elevated in PD-1+ CD8+ TILs as compared with peripheral CD8+ T cells (Fig. 3 A). However, IL6RA was barely expressed in CD8+ TILs in lung cancer and CRC patients although high mRNA levels of IL6ST (encoding GP130) were found in CD8+ TILs (Fig. 3 A). In addition, IL23R and S1PR1 were also almost not expressed in tumor-infiltrating CD8+ T cells (Fig. 3 A). Moreover, although leptin was reported to regulate CD8+ T cells via STAT3 signaling in murine breast cancer (Zhang et al., 2020), its receptor was hardly expressed in CD8+ TILs in human lung cancer, CRC, and HCC patients at the mRNA level (Fig. 3 A). In addition, growth factor receptors, GPCRs, TLRs, and CSF receptors also had limited mRNA expression in tumor-infiltrating CD8+ T cells (data not shown). Thus, IL-10 and IL-21 might be two candidate upstream inducers for STAT3 activation in CD8+ TILs within human tumors.
Figure 3.
IL-10 and IL-21 promote STAT3 activation in Texterm cells in cancer. (A) mRNA levels of cytokine receptors in CD8+ T cells from peripheral blood and tumors in human cancer patients (GEO accession nos. GSE99254, GSE108989, and GSE98638). (B) Representative plots of phosphorylated STAT3 at the Y705 site in CD8+ T cells from TDLNs and B16-OVA tumors (left panel) or E.G7 tumors (right panel). Cells were stimulated by IL-6, IL-10, or IL-21 for 30 min in vitro. (C) Representative plots of phosphorylated STAT3 at the S727 site in CD8+ T cells from B16-OVA (left panel) and E.G7 (right panel) tumors. Cells were stimulated by IL-6, IL-10, or IL-21 for 30 min in vitro. (D) IL6-Rα (CD126) and IL10RA (CD210) expression on CD8+ T cells from TDLNs and tumors. (E) Representative plots (upper panel) and quantifications (bottom panel) of pSTAT3(Y705) and pSTAT3(S727) in PD-1+TIM-3− and PD-1+TIM-3+ CD8+ TILs from B16-OVA (left panel) or E.G7 (right panel) tumors. Cells were stimulated by IL-10 or IL-21 for 30 min in vitro. (F) mRNA level of Stat3 in Pdcd1hiHavcr2− and Pdcd1hiHavcr2+ CD8+ T cells from B16-OVA tumors (GEO accession no. GSE122675). Pdcd1hi, Pdcd1 expression value >1; Havcr2+, Havcr2 expression value >0; Havcr2−, Havcr2 expression value = 0. (G) mRNA level of STAT3 in PDCD1hiHAVCR2low and PDCD1hiHAVCR2hi CD8+ T cells from tumors in human cancer patients (GEO accesssion nos. GSE99254, GSE108989, and GSE98638). PDCD1hi, PDCD1 expression value >5; PDCD1hi cells were ranked by HAVCR2 expression value, top 50% are HAVCR2hi, and the other 50% are HAVCR2low. Data are representative of three (B–D) or two (E) independent experiments. Data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by one-way ANOVA (A), paired two-tailed Student’s t test (E) and unpaired two-tailed Student’s t test (F and G).
Next, we further examine the ability of IL-10 and IL-21 in inducing STAT3 phosphorylation and activation in murine tumor-infiltrating CD8+ T cells. Consistent with the above human T cell data, IL-10 and IL-21, but not IL-6, induced STAT3 phosphorylation at both Tyr705 (pY705) and Ser727 (pS727) sites in CD8+ TILs from both B16-OVA and E.G7 tumors (Fig. 3, B and C). Importantly, pY705 levels in CD8+ TILs were much higher than those in CD44− and CD44+ CD8+ T cells from TDLNs (Fig. 3 B), consistent with elevated STAT3 expression in CD8+ TILs mentioned above (Fig. S1, B and C). In addition, though IL-6 failed to induce pSTAT3 in tumor-infiltrating Tex cells, it potently induced STAT3 phosphorylation in CD44− and CD44+ CD8+ T cells from TDLNs, while IL-10 was not able to induce pSTAT3 in naive CD8+ T cells (Fig. 3 B). The differential abilities of IL-6 and IL-10 to induce pSTAT3 were consistent with the expression of their receptors. IL-6Rα expression was highly enriched in naive CD8+ T cells but was downregulated during activation and was diminished in Tex cells within tumors (Fig. 3 D; Yang et al., 2016). Conversely, IL10RA expression was gradually upregulated in CD8+ T cells during activation and differentiation and highly expressed in tumor-infiltrating CD8+ T cells (Fig. 3 D; Emmerich et al., 2012).
We further compared the phosphorylated levels of STAT3 in Texprog and Texterm subsets and found that IL-10 and IL-21 induced relatively higher levels of pSTAT3 at both Y705 and S727 sites in Texterm cells as compared with those in Texprog cells from B16-OVA and E.G7 tumors (Fig. 3 E), which was consistent with higher Stat3 expression in Pdcd1hiHavcr2+ CD8+ TILs than in Pdcd1hiHavcr2− CD8+ TILs observed from single-cell data of B16-OVA–infiltrated CD8+ T cells (Fig. 3 F). Consistently, the mRNA level of STAT3 was also increased in PDCD1hiHAVCR2hi CD8+ TILs as compared with that in PDCD1hiHAVCR2low CD8+ TILs in human lung cancer, CRC, and HCC patients (Fig. 3 G). These results suggest that STAT3 signaling might more preferentially regulate Texterm TILs, consistent with the above results on the potent regulation of Texterm cell maintenance and effector function by STAT3 (Fig. 2, A and F).
IL-10 and IL-21 signaling intrinsically promotes the development of Texterm cells
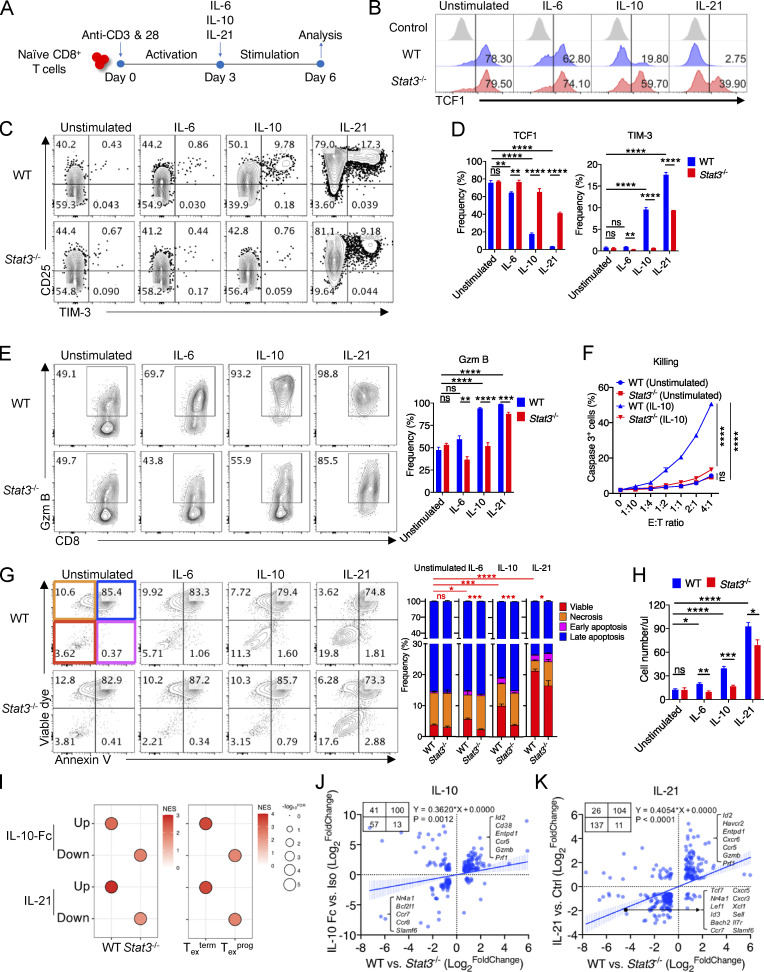
Next, to understand the activation of STAT3 signaling in CD8+ TILs, we functionally analyzed and compared the roles of IL-10 and IL-21 signaling. We stimulated in vitro–activated WT and Stat3-deficient CD8+ T cells with IL-10 or IL-21 for another 3 d, with IL-6 as the control (Fig. 4 A). Results showed that IL-10 and IL-21 significantly induced the generation of CD25hiTIM-3+ subsets in vitro as compared with unstimulated control, while TCF1 expression was dramatically decreased by the stimulation of both IL-10 and IL-21 (Fig. 4, B–D). Importantly, IL-10 failed in inducing the generation of CD25hiTIM-3+ population in Stat3−/− CD8+ T cells, indicating that the induction of CD25hiTIM-3+ cells by IL-10 was STAT3-dependent (Fig. 4, C and D). IL-21 induced increased frequencies of CD25hiTIM-3+ cells in WT CD8+ T cells than in Stat3−/− CD8+ T cells, with a great portion of CD25hiTIM-3+ cells found in Stat3-deficient CD8+ T cells (Fig. 4, C and D), indicating that IL-21 inducing TIM-3+ cell development partially, not totally, relied on STAT3 signaling, which might be due to IL-21 activation of STAT1 and STAT5 in addition to STAT3 signaling in CD8+ T cells (Delgoffe and Vignali, 2013; Mujib et al., 2012). It should also be noticed that IL-21 induced a much higher percentage of CD25hiTIM-3+ cells than IL-10 or IL-6, and TCF1+ cell percentages were also reduced the most in IL-21–stimulated CD8+ T cells (Fig. 4, B–D), associated with higher pSTAT3 levels following IL-21 treatment of in vitro–activated CD8+ T cells as compared with IL-6 and IL-10 treatment (Fig. S3 A). IL-6 had limited effects in promoting CD25hiTIM-3+ cell development or inhibiting TCF1 expression in activated CD8+ T cells (Fig. 4, B–D), perhaps due to dramatically downregulated IL6-Rα expression in the process of priming, with little or no expression in CD8+ T cells activated in vitro (Yang et al., 2016). Moreover, the expression of CD25 and CD39 was significantly upregulated, while that of Ly108 and CD62L was downregulated in WT CD8+ T cells exposed to IL-10 and IL-21 as compared with unstimulated control or Stat3−/− CD8+ T cells (Fig. S3, B and C). These results suggest that the IL-10/21–STAT3 axis plays critical roles in promoting Texterm cell generation after activation.
Figure 4.
IL-10 and IL-21 signaling intrinsically promotes the development of Texterm cells. (A) Experimental design for in vitro culture of WT and Stat3−/− CD8+ T cells with the stimulation of IL-6, IL-10, or IL-21. (B–D) Representative plots (B and C) and quantifications (D) of TCF1 and TIM-3 expression in/on in vitro–stimulated WT and Stat3−/− CD8+ T cells (n = 4 in each group). (E) Representative plots (left panel) and quantification (right panel) of Granzyme B production in in vitro–stimulated WT and Stat3−/− CD8+ T cells (n = 4 in each group). (F) Killing assay of WT and Stat3−/− OT-I cells to B16-OVA tumor cells indicated by intracellular cleaved caspase 3 (n = 3 in each group). Activated CD8+ T cells were stimulated with IL-10 for 3 d or not. (G) Representative plots (left panel) and quantifications (right panel) of viable dye and Annexin V staining on in vitro–stimulated WT and Stat3−/− CD8+ T cells (n = 4 in each group). (H) Numbers of in vitro–stimulated WT and Stat3−/− CD8+ T cells (n = 4 in each group). (I) GSEA results for comparing the enrichment of IL-10-Fc– or IL-21–regulated feature genes in the transcriptomes of WT and Stat3−/− OT-I TILs (left panel), or Texterm and Texprog cells (right panel) (GEO accession nos. GSE168990, GSE143903, and GSE114631). NES, normalized enrichment score; FDR, false discovery rate q-value. FDR presented as −log10(FDR). (J) Correlation analysis for DEGs identified in IL-10-Fc– and STAT3-relevant RNA-seq (GEO accession no. GSE168990). DEGs were filtered by fold change >1.5 and P value <0.05. (K) Correlation analysis for DEGs identified in IL-21– and STAT3-relevant RNA-seq (GEO accession no. GSE143903). DEGs were filtered by fold change >1.5 and P value <0.05. Data are representative of three (B–E and H) or two (F and G) independent experiments. Data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired two-tailed Student’s t test (D, E, G, and H) and two-way ANOVA (F).
Figure S3.
IL-10 and IL-21 signaling intrinsically promotes the development of Texterm cells. (A) Phosphorylation levels of STAT3 at the Y705 site in in vitro activated CD8+ T cells with the stimulation of IL-6, IL-10, or IL-21 for 30 min. (B and C) Quantifications (B) and representative plots (C) and of CD25, CD39, Ly108, and CD62L expression on in vitro–stimulated WT and Stat3−/− CD8+ T cells. (D–F) Representative plots (D and E) and quantifications (F) of TIM-3 and TCF1 expression on/in in vitro cocultured WT and Stat3−/− CD8+ T cells with the stimulation of IL-6, IL-10, or IL-21 for 3 d. (G) Proliferation of in vitro cocultured WT and Stat3−/− CD8+ T cells marked by CTV dilution. (H) Experimental design for adoptive cotransfer of WT and Sg-Il10ra or Il6ra−/− OT-I cells and B16-OVA tumor–challenge assay. (I) Representative plots of IL10RA (CD210) expression on WT and Sg-Il10ra OT-I TILs from B16-OVA tumors. (J) Representative plots (left panel) and quantifications (right panel) of Ly108 and TIM-3 expression on WT and Sg-Il10ra OT-I TILs from B16-OVA tumors. (K) Representative plots (left panel) and quantifications (right panel) of Ly108 and TIM-3 expression on WT and Il6ra−/− OT-I TILs from B16-OVA tumors. Data are representative of three (B–F, I, and K) or two (A, G, and J) independent experiments. Data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired two-tailed Student’s t test (B) and paired two-tailed Student’s t test (F, G, J, and K).
Next, to further explore whether the IL-10/21–STAT3 axis regulated CD8+ T cell differentiation via an intrinsic and direct way, we cocultured naive WT and Stat3−/− CD8+ T cells in vitro and stimulated them with cytokines for another 3 d after activation. We found in the coculture system that IL-10 and IL-21 significantly induced CD25hiTIM-3+ cell generation and inhibited TCF1 expression in WT, but less in cocultured Stat3−/− CD8+ T cells and unstimulated CD8+ T cells (Fig. S3, D–F). These data indicated that the function of IL-10 and IL-21 in Stat3-deficient CD8+ T cells could not be rescued by the factors in the supernatant of IL-10/21–treated WT CD8+ T cells. Hence, the IL-10/21–STAT3 axis intrinsically regulates CD8+ T cell fate decision after activation.
Moreover, IL-10/21 signaling significantly promoted Granzyme B expression in activated WT CD8+ T cells as compared with Stat3−/− CD8+ T cells and unstimulated control (Fig. 4 E). To further assess the function of STAT3 in the regulation of CD8+ T cell killing on targeted tumor cells, we used IL-10 to activate STAT3 in OT-I cells primed in vitro for 3 d, then performed a killing assay with B16-OVA tumor cells as targets. IL-10–stimulated WT OT-I cells had a greater cytolytic capacity for tumor cells than IL-10–treated Stat3−/− OT-I cells and unstimulated control (Fig. 4 F), indicating that active STAT3 signaling could efficiently enhance CD8+ T cell–mediated tumor killing. In addition, we found IL-10/21 signaling dramatically enhanced the proportion of viable cells in WT CD8+ T cells in vitro, resulting in greater numbers of live CD8+ T cells as compared with Stat3−/− CD8+ T cells and unstimulated control (Fig. 4, G and H), indicating a critical role of STAT3 in promoting CD8+ T cell survival. However, IL-10/21–STAT3 axis did not enhance the proliferative activity of activated CD8+ T cells, illustrated by CellTrace Violet (CTV) dilution, in IL-10– and IL-21–stimulated WT and Stat3−/− CD8+ T cells (Fig. S3 G).
In vivo, by analyzing cotransferred WT and Il10ra−/− OT-I cells within B16-OVA tumors, we found the frequencies of Ly108−TIM-3+ Texterm cells were decreased by the deficiency of Il10r as previously reported, while the percentages of Ly108+TIM-3− Texprog cells were increased (Fig. S3, H–J). IL-10-Fc was shown to further expand tumor-specific TIM-3+ cells in B16F10 tumors (Guo et al., 2021), which was consistent with the results obtained in vivo by the deletion of Stat3. Moreover, Il21r deficiency impaired the differentiation and function of cytolytic Tex cells in B16F10 and KP-HELLO solid tumor models (Cui et al., 2021; Zander et al., 2019), which was also consistent with the results of Stat3-deficiency. In contrast, Il6ra deficiency in activated OT-I cells did not affect the development of Texterm vs. Texprog cells within tumors as expected (Fig. S3, H and K).
To further comprehensively analyze the functional relationships between IL-10- and IL-21–STAT3 signaling, we compared the RNA-seq data of WT and Stat3−/− OT-I cells from B16-OVA tumors (Fig. S2 G) with previously reported RNA-seq results of tumor-specific PD-1+TIM-3+ CD8+ TILs from IL-10-Fc–treated B16F10-bearing mice (Guo et al., 2021) and in vitro IL-21–stimulated activated CD8+ T cells (Hermans et al., 2020). GSEA results revealed that both IL-10-Fc– and IL-21–induced signature genes were relatively enriched in tumor-infiltrating WT Tex cells, while their suppressed feature genes were significantly concentrated in Stat3−/− CD8+ TILs (Fig. 4 I). In addition, both IL-10-Fc– and IL-21–stimulated CD8+ T cells were more similar to Texterm cells in their gene expression, while their inhibited genes were relatively more enriched in Texprog cells (Fig. 4 I; Siddiqui et al., 2019). Moreover, by overlapping IL-10-Fc– and STAT3-regulated genes, we found their regulated genes were significantly correlated (Fig. 4 J). Specifically, IL-10 upregulated the expression of numerous Texterm-related genes (Id2, Entpd1, Gzmb, etc.) as STAT3 did, while it inhibited the expression of multiple Texprog-related genes (Ccr7, Slamf6, etc.), which were simultaneously repressed by STAT3 (Fig. 4 J). IL-21 and STAT3 also had identical functions in promoting the expression of Texterm-related genes (Id2, Havcr2, Gzmb, etc.) and in suppressing those of Texprog-related genes (Tcf7, Id3, Cxcr5, etc.; Fig. 4 K).
These results, therefore, indicate that IL-10 and IL-21 are two inducers of STAT3 activation, and thereby regulate Tex cells within tumors. STAT3 signaling not only plays critical roles in promoting Texterm cell maintenance but is also essential for Texterm cell development.
STAT3 transcriptionally regulates Tex cell differentiation
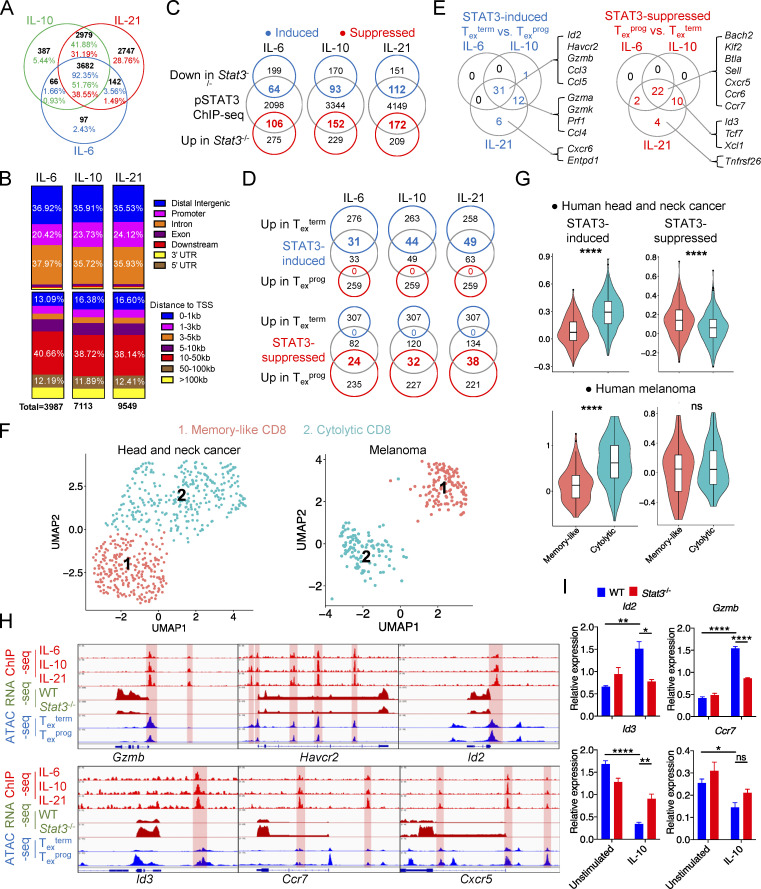
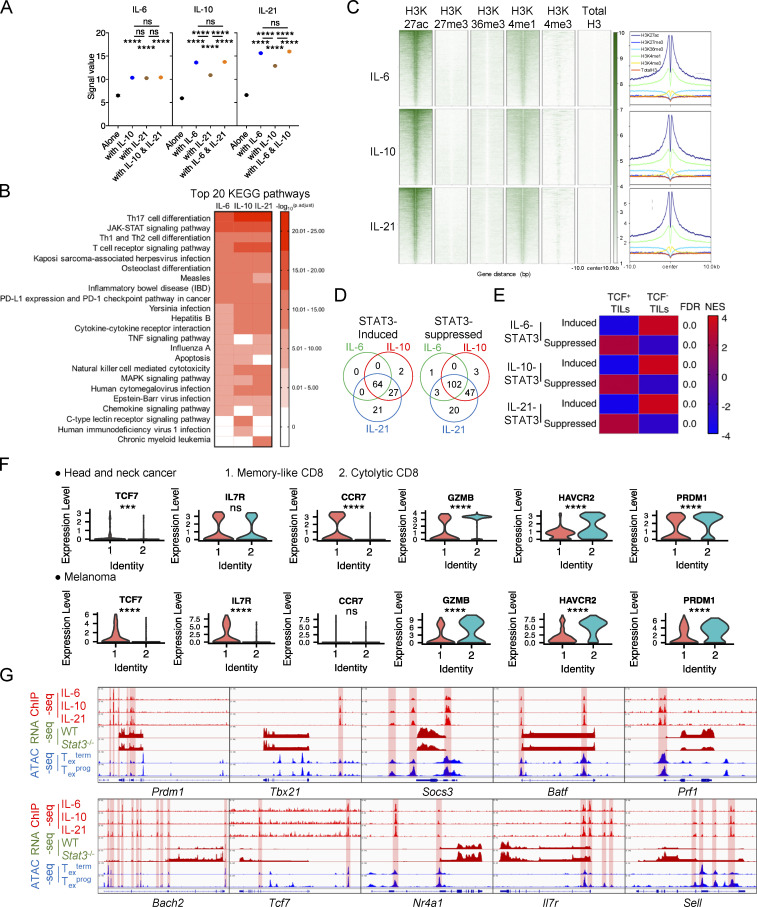
To systematically understand the transcriptional mechanisms whereby STAT3 regulates CD8+ T cell differentiation, we further analyzed the genome-wide occupancy of phosphorylated STAT3 in activated CD8+ T cells. Since pY705 was predominantly responsible for the nuclear translocation of STAT3 dimer and the binding to DNA consensus sequence (Huynh et al., 2019), we performed chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq) analysis using in vitro activated CD8+ T cells stimulated by IL-6, IL-10, or IL-21 to induce phosphorylation at Y705. Results showed that IL-6–, IL-10–, and IL-21–induced pSTAT3 significantly bound to 3,987, 7,113, and 9,549 genomic sites, respectively (Fig. 5, A and B; and Table S2). Almost all sites occupied by pSTAT3 induced by IL-6 and IL-10 were also bound by pSTAT3 induced by IL-21 (Fig. 5 A). Stronger binding signals could be detected in IL-6–, IL-10–, and IL-21–induced pSTAT3 cobinding sites as compared with their own unshared binding sites (Fig. S4 A). Thus, IL-6/10/21–induced pSTAT3 bound to a shared genomic profile, and the binding affinities in activated CD8+ T cells correlated with the pSTAT3 levels induced by these three cytokines (Fig. S3 A and Fig. S4 A). Consistently, genes bound by IL-6/10/21–induced pSTAT3 were significantly enriched in biological pathways related to T cell differentiation, T cell receptor signaling, cytotoxicity, and other immunological pathways (Fig. S4 B).
Figure 5.
STAT3 transcriptionally regulates Tex cell differentiation. (A) Venn diagram of pSTAT3-binding peaks in IL-6–, IL-10–, and IL-21–stimulated CD8+ T cells across the whole mouse genome (mm10). (B) Genetic feature distribution (upper panel) or distribution relative to TSS (bottom panel) of pSTAT3-binding peaks in CD8+ T cells. (C) Venn diagrams of pSTAT3-binding genes and DEGs in WT or Stat3−/− CD8+ OT-I TILs. (D) Venn diagrams of STAT3-regulated genes and DEGs in Texterm or Texprog TILs (GEO accession no. GSE114631). (E) Representative STAT3-regulated feature genes of Texterm (left panel) or Texprog (right panel) cells. (F) UMAP plots identifying tumor-infiltrating memory-like and cytolytic CD8+ T cell clusters from human head and neck cancer (left panel) or melanoma (right panel) patient (GEO accession nos. GSE103322 and GSE120575). (G) Average expression of STAT3-induced (left panel) or -suppressed (right panel) signature gene clusters in memory-like or cytolytic tumor-infiltrating CD8+ T cells. STAT3-induced or -suppressed signature genes are listed in Table S3. (H) pSTAT3 ChIP-seq tracks aligned with RNA-seq tracks of WT and Stat3−/− CD8+ TILs, and ATAC-seq tracks of Texprog or Texterm cells at the specific gene loci (GEO accession no. GSE123236). (I) mRNA levels of specific genes in in vitro activated WT and Stat3−/− CD8+ T cells with the stimulation of IL-10 for 2 d or not. Data are representative of three (I) independent experiments. Data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired two-tailed Student’s t test (G and I).
Figure S4.
STAT3 transcriptionally regulates Tex cell differentiation. (A) Signal values of pSTAT3-binding peaks in IL-6–, IL-10–, and IL-21–stimulated CD8+ T cells. (B) Top 20 KEGG pathways for pSTAT3-binding genes in in vitro activated CD8+ T cells with the stimulation of IL-6, IL-10, or IL-21. (C) Heatmaps and peak plots for read density profiles of H3K27ac, H3K27me3, H3K36me3, H3K4me1, and H3K4me3 occupations centered on pSTAT3-binding peaks (GEO accession no. GSE54191). Values were normalized to the total number of reads. (D) Venn diagrams of STAT3-regulated genes in in vitro activated CD8+ T cells with the stimulation of IL-6, IL-10, or IL-21. (E) GSEA results for comparing the enrichment of IL-6/10/21-STAT3 regulated genes in the transcriptomes of TCF1− Texterm and TCF1+ Texprog cells (GEO accession no. GSE114631). NES, normalized enrichment score; FDR, false discovery rate q-value. FDR presented as −log10(FDR). (F) Violin plots illustrating the mRNA amounts of signature genes between tumor-infiltrating memory-like and cytolytic CD8+ T cells. (accession nos. GSE103322 and GSE120575). (G) pSTAT3 ChIP-seq tracks aligned with RNA-seq tracks of WT and Stat3−/− CD8+ TILs, and ATAC-seq tracks of Texprog or Texterm cells at the specific gene loci (GEO accession no. GSE123236). Data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired two-tailed Student’s t test (A and F).
Further analyses revealed that pSTAT3-binding sites were mainly distributed in the distal intergenic, intron, and promoter regions (Fig. 5 B), and above 38% of binding sites fell into 10–50 kb upstream or downstream of the transcription start sites (TSS), with another 13–17% and 11–13% binding sites located at 0–1 and 50–100 kb away from TSS, respectively (Fig. 5 B). Consistently, density maps and peak plots showed that extensive H3K27ac and H3K4me1 modifications, markers for actively transcribed regions and enhancers, respectively (Kurachi et al., 2014), were localized within ±10 kb regions centered around pSTAT3-binding sites (Fig. S4 C). These results indicated that pSTAT3 mostly bound to enhancers or other distal regulatory elements to regulate gene transcription, consistent with its role in shaping active enhancer landscape found in CD4+ T cells (Vahedi et al., 2012), while another major portion of STAT3 binding was also in promoter and intron regions.
To identify pSTAT3-regulated genes in tumor-specific Tex cells, we integrated pSTAT3 ChIP-seq data with RNA-seq data of WT and Stat3−/− OT-I TILs and got STAT3-induced and STAT3-suppressed genes (Fig. 5 C, Fig. S2 G, Fig. S4 D, and Table S2). To pinpoint STAT3-regulated genes that were directly involved in tumor-infiltrating Tex cell fate decision, we overlapped STAT3-regulated genes with the signature genes of tumor-specific TCF1+ Texprog or TCF1− Texterm cells from B16-OVA tumors, respectively (Siddiqui et al., 2019). Interestingly, numerous STAT3-induced genes were identified as Texterm signature genes, including Id2, Havcr2, Entpd1, Gzmb, and Cxcr6, whereas Texprog feature genes were not found (Fig. 5, D and E). Moreover, a broad range of Texprog signature genes (Tcf7, Id3, Sell, Btla, Cxcr5, Ccr7, etc.) were directly inhibited by STAT3 while none of the Texterm feature genes were suppressively bound by STAT3 (Fig. 5, D and E). Consistently, GSEA results revealed that IL-6–, IL-10–, and IL-21-STAT3–induced genes were all relatively enriched in TCF1− Texterm subset, while their inhibited genes were significantly concentrated in the TCF1+ Texprog subset (Fig. S4 E). To investigate the relevance of STAT3-regulated genes in tumor-infiltrating CD8+ T cells from human cancers, we utilized published scRNA-seq data from human head and neck cancer and melanoma patients (Puram et al., 2017; Sade-Feldman et al., 2018). A memory-like CD8+ T cell subset was identified with higher expression of memory hallmark genes such as TCF7, IL7R, and CCR7 (Fig. 5 F and Fig. S4 F). Another subset showed high-level transcripts of effector and cytotoxicity signature genes, including GZMB, HAVCR2, and PRDM1, and thus was identified as a cytolytic subset (Fig. 5 F and Fig. S4 F). In line with the above murine data, STAT3-induced genes showed higher transcript expression in the cytolytic subset in both human head and neck cancer and melanoma patients (Fig. 5 G and Table S3). Altogether, these results indicated the potent transcriptional functions of STAT3 signaling in regulating Tex cell differentiation, especially in promoting effective Texterm cell development.
To further analyze the specific occupancy of pSTAT3 on open chromatin regions (OCRs) of its target genes in tumor-infiltrating CD8+ T cells, we aligned pSTAT3 ChIP-seq tracks with transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) tracks of OVA-specific Texterm and Texprog cells from B16-OVA tumors (Miller et al., 2019), as well as RNA-seq tracks of the above mentioned WT and Stat3−/− OT-I cells from B16-OVA tumors. We found that pSTAT3 occupied the promoter regions of multiple Texterm-related genes, including Id2, Batf, Gzmb, and Prf1, which might lead to their enhanced transcriptional activities (Fig. 5 H and Fig. S4 G). Also pSTAT3 occupied the promoters of Texprog-related genes, including Cxcr5 and Il7r, accompanied by their reduced transcripts (Fig. 5 H and Fig. S4 G). Moreover, pSTAT3 was found to directly bind to multiple distal intergenic regions of Texprog-associated genes, including Id3, Tcf7, Ccr7, Cxcr5, and Il7r along with decreased transcriptional activities (Fig. 5 H and Fig. S4 G). In addition, pSTAT3 also inductively bound to the introns of Havcr2 and Prdm1 genes, and suppressively bound to those of Ccr7, Bach2, and Sell genes, and these loci were all accessible in tumor-specific Tex cells (Fig. 5 H and Fig. S4 G).
To further confirm the transcriptional regulation of STAT3 on the identified target genes, we used IL-10 to activate STAT3 signaling in CD8+ T cells primed in vitro and then analyzed their mRNA levels. We found that Id2 and Gzmb expressions were dramatically upregulated in IL-10–stimulated WT CD8+ T cells as compared with IL-10–stimulated Stat3−/− CD8+ T cells and unstimulated control (Fig. 5 I). In the meantime, Id3 and Ccr7 expressions were significantly downregulated by IL-10 in WT CD8+ T cells, as expected (Fig. 5 I).
Thus, these results demonstrate that STAT3 signaling promotes Texterm cell differentiation via transcriptionally upregulating the expression of their signature genes, including Id2, Havcr2, Gzmb, etc. In the meantime, STAT3 also inhibits Texprog cell fate commitment by directly suppressing the expression of their fate-associated genes, including Id3, Ccr7, Tcf7, etc. It should be noticed that although IL-6 transiently induces STAT3 activation in CD8+ T cells in vitro, it fails in constantly regulating CD8+ T cell differentiation via STAT3 signaling after activation due to the diminishing IL6-Rα expression (Fig. 4, B–D; Yang et al., 2016).
STAT3 regulates the epigenetic landscape of Tex cells in cancer
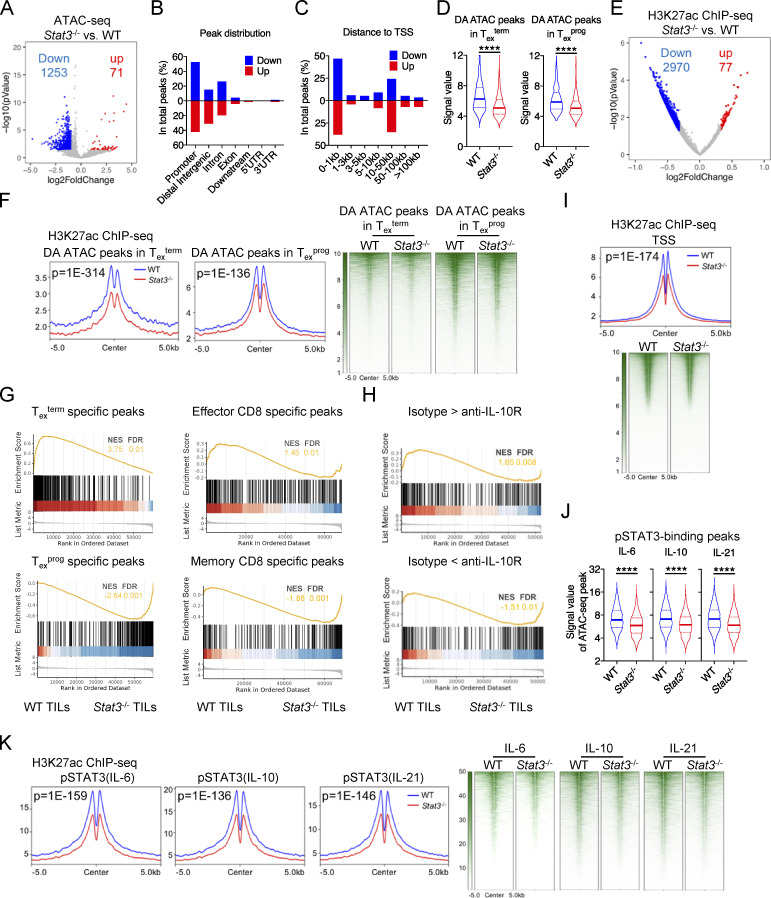
Since Tex cells exhibit specific epigenetic landscapes (Miller et al., 2019), we further explored whether STAT3 has a function in regulating the chromatin accessibilities of Tex cells using ATAC-seq on cotransferred WT and Stat3−/− OT-I cells from B16-OVA tumors (Fig. 1 D). Results showed that the chromatin accessibilities of Tex cells were globally reduced by the deficiency of Stat3 (Fig. 6 A). 1,253 OCRs had reduced accessibilities across the whole genome in Stat3−/− OT-I TILs, with only 71 OCRs showing increased accessibilities in Stat3−/− OT-I TILs (Fig. 6 A and Table S4), suggesting a critical role of STAT3 mainly in activating the chromatin accessibility of CD8+ T cells during T cell exhaustion in cancer. The altered OCRs were mainly distributed in the promoter, distal intergenic, and intron regions (Fig. 6 B), and mainly located at 0–1 kb and 10–50 kb distance from TSS (Fig. 6 C).
Figure 6.
STAT3 regulates the epigenetic landscape of Tex cells in cancer. (A) Volcanic plot showing OCR peak changes between WT and Stat3−/− OT-I TILs. (B) Genetic feature distribution of changed OCR peaks between WT and Stat3−/− OT-I TILs. (C) Distribution relative to TSS of changed OCR peaks between WT and Stat3−/− OT-I TILs. (D) Violin plots showing the signal values of OCR peaks in WT and Stat3−/− OT-I TILs at Texterm or Texprog DA peaks (GEO accession no. GSE123236). (E) Volcanic plot showing differentially H3K27ac modified regions (DMRs) between IL-10–stimulated WT and Stat3−/− CD8+ T cells. DMRs were filtered by fold change >1.25 and P value <0.05. (F) Peak plots (left panel) and heatmaps (right panel) showing the deposition of H3K27ac modification centered on Texterm or Texprog DA peaks in WT and Stat3−/− OT-I TILs (GEO accession no. GSE123236). P value by Fisher test was added in the plot. (G and H) PSEA results for comparing the enrichment of signature OCR peaks in other CD8+ T cell subsets in WT and Stat3−/− OT-I TILs (GEO accession nos. GSE123236, GSE86797, and GSE116389). NES, normalized enrichment score; FDR, false discovery rate q-value. (I) Peak plot (upper panel) and heatmap (bottom panel) showing the deposition of H3K27ac modification centered on TSS in WT and Stat3−/− OT-I TILs. P value by Fisher test was added in the plot. (J) Violin plots showing the signal values of OCR peaks in WT and Stat3−/− OT-I TILs at pSTAT3-binging sites. (K) Peak plots (left panel) and heatmaps (right panel) showing the deposition of H3K27ac modification centered on pSTAT3-binding peaks in WT and Stat3−/− OT-I TILs. P value by Fisher test was added in the plot. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired two-tailed Student’s t test (D and J).
To further investigate the regulation of STAT3 on the chromatin structures of Texterm or Texprog signature genes, we compared the accessibilities of the differentially accessible peaks (DA peaks) in WT and Stat3−/− T cells (Miller et al., 2019). We found that the overall accessibilities of both Texterm and Texprog DA peaks were reduced in Stat3−/− Tex cells as compared with WT Tex cells (Fig. 6 D). To further illustrate the epigenetic regulation by STAT3, we also analyzed the genome-wide profiling of H3K27ac modification in in vitro IL-10–stimulated WT and Stat3−/− CD8+ T cells. Consistent with ATAC-seq results, the H3K27ac modification was globally reduced in IL-10–stimulated Stat3−/− CD8+ T cells as compared with IL-10–stimulated WT CD8+ T cells (Fig. 6 E and Table S5). Moreover, Stat3 deficiency decreased H3K27ac modification profiling around both the Texterm and Texprog DA Peaks (Fig. 6 F). Importantly, peak set enrichment analysis (PSEA) results revealed that the epigenetic signatures of Texterm and effector CD8+ T cells were strongly enriched in WT OT-I TILs, while signature OCRs in Texprog and memory CD8+ T cells were significantly concentrated in Stat3−/− OT-I TILs (Fig. 6 G). Thus, these results indicated that STAT3 exerted a critical function in the epigenetic regulation of Tex cell chromatin profiles, especially in favoring Texterm epigenetic program. Moreover, the feature OCRs with reduced accessibilities in anti-IL-10R–treated PD-1hi CD8+ T cells in the leukemia model (Hanna et al., 2021) were relatively enriched in WT CD8+ T cells, while those with increased accessibilities were significantly associated with Stat3−/− CD8+ T cells (Fig. 6 H), revealing the identical role of IL-10 and STAT3.
Moreover, we found that H3K27ac modification profiling around TSS in Stat3−/− CD8+ T cells declined as compared with that in WT CD8+ T cells (Fig. 6 I), and the chromatin accessibilities and H3K27ac modifications around pSTAT3-binding peaks were also significantly reduced as a result of Stat3 deficiency (Fig. 6, J and K), indicating that the STAT3 binding to target gene loci might be critical for the openness and transcriptional activation in these sites.
STAT3 cooperates with BATF and IRF4 to mediate Texterm cell development
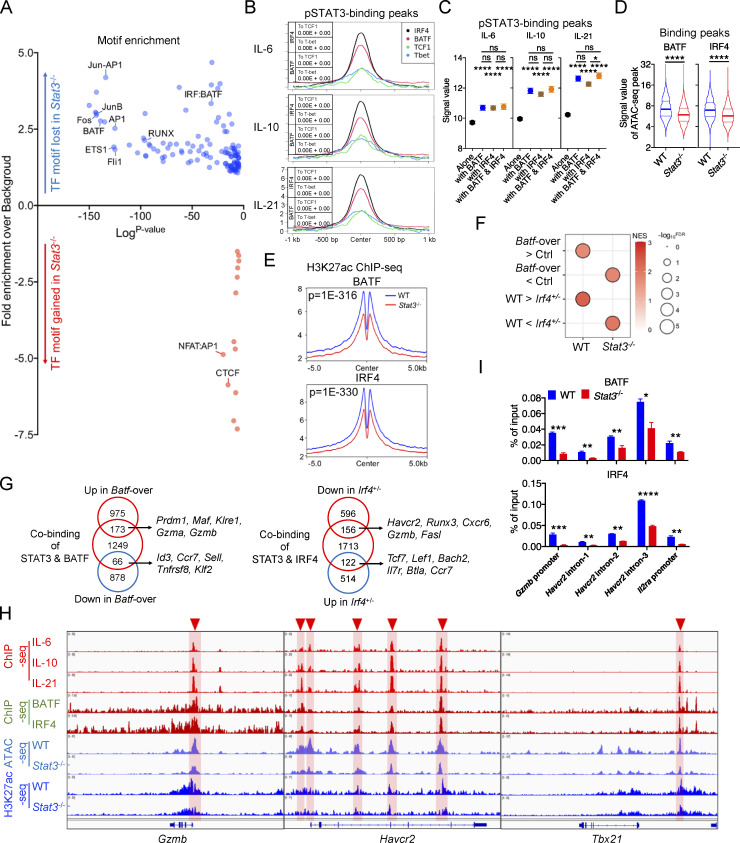
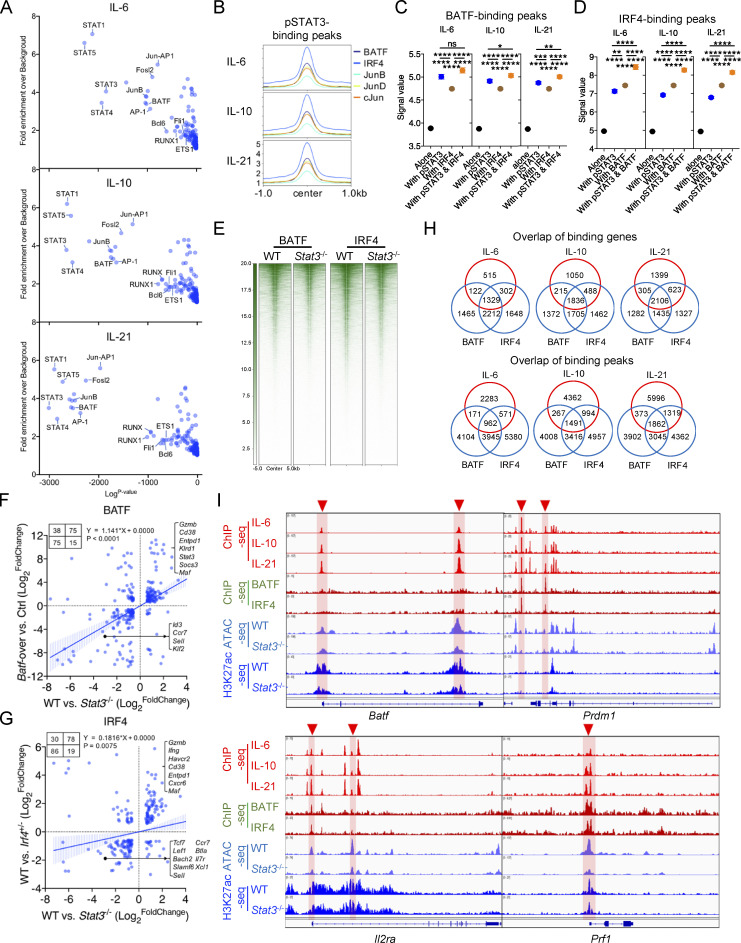
In addition to the above results, known motif enrichment analyses for STAT3-regulated OCRs revealed that Stat3 deficiency resulted in decreased enrichment for binding motifs of numerous AP-1, ETS, and RUNX family members (Fig. 7 A). Moreover, plenty of STAT, AP-1, ETS, and RUNX family members were also significantly enriched in pSTAT3-occupied peaks (Fig. S5 A). These indicated that there might exist significant crosstalk between STAT3 and other transcription factors. AP-1 transcription factors play critical roles in T cell activation, differentiation, and exhaustion (Philip and Schietinger, 2021). To further explore the relationships between STAT3 and AP-1 family members, we compared their binding profiles in detail using previously published datasets (Kurachi et al., 2014). Density maps and peak plots revealed that a great deal of AP-1–occupied peaks was shared with pSTAT3-binding sites around the genome, especially BATF and IRF4 (Fig. S5 B), and compared with TCF1 and T-bet binding, that of BATF and IRF4 was significantly enriched at pSTAT3-occupied sites (Fig. 7 B). Moreover, increased pSTAT3 occupancy was found at genomic sites cobound by BATF or IRF4 or both as compared with pSTAT3 unshared binding sites (Fig. 7 C). Similarly, greater binding signals of BATF or IRF4 were also detected in their cobinding sites with pSTAT3 than in their own unshared occupied peaks (Fig. S5, C and D). Importantly, the overall chromatin accessibilities and H3K27ac modifications located at BATF- or IRF4-occupied sites were significantly reduced in Stat3−/− Tex cells (Fig. 7, D and E; and Fig. S5 E).
Figure 7.
STAT3 cooperates with BATF and IRF4 to mediate Texterm cell development. (A) Enrichment of known transcription factor–binding motifs in DA OCR peaks between WT and Stat3−/− OT-I TILs. The x and y axes represent the logP value and fold change of motif enrichment separately. Targeted motifs were compared to the whole genome background to calculate the P value and fold change. (B) Peak plots for read density profiles of BATF, IRF4, TCF1, and T-bet occupations centered on pSTAT3-binding peaks in CD8+ T cells (GEO accession nos. GSE54191, GSM5016615, and GSE96724). Values were normalized to the total number of reads. P value by Chi-squared test was added in the plot. (C) Signal values of pSTAT3-binding peaks in IL-6–, IL-10–, and IL-21–stimulated CD8+ T cells at BATF- or IRF4-binding sites or not. (D) Violin plots showing the signal values of OCR peaks in WT and Stat3−/− OT-I TILs at BATF- or IRF4-binging sites. (E) Peak plots showing the deposition of H3K27ac modification centered on BATF- or IRF4-binging sites in WT and Stat3−/− OT-I TILs. P value by Fisher test was added in the plot. (F) GSEA results for comparing the enrichment of BATF- and IRF4-regulated feature genes in the transcriptomes of WT and Stat3−/− OT-I TILs (GEO accession nos. GSE154745 and GSE84820). NES, normalized enrichment score; FDR, false discovery rate q-value. FDR presented as −log10(FDR). (G) Venn diagrams of pSTAT3 and BATF-cobinding genes and BATF-regulated genes (left panel), or of pSTAT3 and IRF4-cobinding genes and IRF4-regulated genes (right panel) in CD8+ T cells. (H) Aligned ChIP-seq tracks of pSTAT3, BATF, and IRF4 in CD8+ T cells, ATAC-seq tracks of WT and Stat3−/− OT-I TILs, and H3K27ac ChIP-seq tracks of WT and Stat3−/− CD8+ T cells at the specific gene loci. (I) ChIP assays were performed with in vitro IL-10–stimulated WT and Stat3−/− CD8+ T cells with anti-BATF or anti-IRF4. The relative amount of immune-precipitated DNA was detected by real-time PCR and normalized relative to the input control. Data are representative of two (I) independent experiments. Data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired two-tailed Student’s t test (C, D, and I).
Figure S5.
STAT3 cooperates with BATF and IRF4 to mediate Texterm cell development. (A) Enrichment of known transcription factor–binding motifs in pSTAT3-binding peaks. The x and y axes represent the logP value and fold change of the motif enrichment separately. Targeted motifs were compared to the whole genome background to calculate P value and fold change. (B) Peak plots showing the read density profiles of BATF, IRF4, JunB, JunD, and c-Jun occupations centered on pSTAT3-binding peaks in CD8+ T cells (GEO accession no. GSE54191). Values were normalized to the total number of reads. (C) Signal values of BATF-binding peaks in CD8+ T cells at pSTAT3- or IRF4-binding sites or not (GEO accession no. GSE54191). (D) Signal values of IRF4-binding peaks in CD8+ T cells at pSTAT3- or BATF-binding sites or not (GEO accession no. GSE54191). (E) Heatmaps showing the deposition of H3K27ac modification centered on BATF- or IRF4-binding peaks in WT and Stat3−/− OT-I TILs (accession GEO: GSE54191). (F) Correlation analysis for DEGs identified in BATF- and STAT3-relevant RNA-seq (GEO accession no. GSE154745). DEGs were filtered by fold change >1.5 and P value <0.05. (G) Correlation analysis for DEGs identified in IRF4- and STAT3-relevant RNA-seq (GEO accession no. GSE84820). DEGs were filtered by fold change >1.5 and P value <0.05. (H) Venn diagram of pSTAT3-, BATF-, and IRF4-binding peaks and genes. (I) Aligned ChIP-seq tracks of pSTAT3, BATF, and IRF4 in CD8+ T cells, ATAC-seq tracks of WT and Stat3−/− OT-I TILs, and H3K27ac ChIP-seq tracks of WT and Stat3−/− CD8+ T cells at the specific gene loci. Data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired two-tailed Student’s t test (C and D).
BATF and IRF4 have been demonstrated to play vital roles in promoting the expansion and cytolytic function of exhausted effector CD8+ T cells in chronic infection and cancer (Chen et al., 2021; Man et al., 2017; Seo et al., 2021; Xin et al., 2015), and STAT3 could directly promote BATF expression in CD8+ T cells (Fig. 2 D and Fig. S4 G; Xin et al., 2015). To further explore the functional relationships between STAT3 and BATF or IRF4 in CD8+ T cells during their exhaustion, we next compared their regulated transcriptomes. GSEA results revealed that both BATF- and IRF4-induced signature genes were significantly enriched in WT TILs, while their inhibited genes were relatively concentrated in Stat3−/− TILs (Fig. 7 F; Man et al., 2017; Seo et al., 2021). By overlapping their regulated genes identified from their RNA-seq data, we found that a set of Texterm feature genes (Entpd1, Gzmb, Havcr2, Cxcr6, etc.) were coupregulated by STAT3 and BATF, or by STAT3 and IRF4 (Fig. S5, F and G), and multiple Texprog signature genes (Id3, Ccr7, Tcf7, Slamf6, etc.) were downregulated jointly by STAT3 and BATF or IRF4 (Fig. S5, F and G). This implied that the function of STAT3 in regulating Tex cell differentiation might be in collaboration with BATF or IRF4. To further explore the transcriptional program coordinated by these three factors, we overlapped their binding genes identified from their ChIP-seq data and found they co-occupied to numerous gene loci (Fig. S5 H; Kurachi et al., 2014). We further overlapped their cobinding genes with BATF- or IRF4-regulated genes identified from their RNA-seq data respectively, and found that STAT3 occupied numerous genes directly regulated by BATF or IRF4 (Fig. 7 G and Table S6). For example, STAT3, BATF, and IRF4 all occupied the promoter of Gzmb gene, and both the chromatin accessibility and H3K27ac modification were reduced at this locus in Stat3−/− CD8+ T cells (Fig. 7 H). In addition, these three transcription factors were shown to cobind to the intron and promoter regions of Havcr2 gene and the distal enhancer of Tbx21 gene, and Stat3 deficiency declined the accessibilities and H3K27ac modifications at these gene loci (Fig. 7 H). Similar occupancies and epigenetic changes were also found in Prdm1, Il2ra, and Prf1 gene loci as shown (Fig. S5 I).
It was previously reported that STAT3-binding efficiency was greatly diminished in Irf4-deficient CD4+ T cells (Kwon et al., 2009). To further investigate whether STAT3 had any effect on the occupation of BATF and IRF4 around the genome, we determined the binding of BATF and IRF4 to their target genes in Stat3−/− CD8+ T cells. ChIP-qPCR results showed that the binding of BATF and IRF4 to the loci of Gzmb, Havcr2, and Il2ra genes was significantly reduced in IL-10–stimulated Stat3−/− as compared with IL-10–stimulated WT CD8+ T cells (Fig. 7 I). Thus, these results altogether indicate that STAT3 might function cooperatively with BATF and IRF4 in regulating Texterm cell development and function.
Discussion
The molecular mechanisms whereby the extrinsic factors regulate CD8+ T cell differentiation and function, especially in cancer, are still not well understood. In this study, we found that Stat3 deficiency in CD8+ T cells impaired their effector functions and worsened tumor controls. IL-10 and IL-21 activate STAT3 signaling to promote Texterm cell development and survival and enhance their effector functions in cancer. Moreover, STAT3 also plays a similar role in promoting terminally differentiated effector CD8+ T cell development and function in acute infection. Mechanistically, STAT3 directly promotes the expression of Texterm-related genes, while it suppressed those of the Texprog cells. Moreover, STAT3 also regulates the chromatin accessibility for Texterm fate commitment, in collaboration with BATF and IRF4.
STAT3 is a polyfunctional transcription factor that can be activated by various extrinsic and intracellular molecules (Kane et al., 2014). During CD8+ T cell priming and exhaustion, multiple cytokines are involved in STAT3 activation (Guo et al., 2021; Xin et al., 2015). IL-6 specifically induces STAT3 activation in naive CD8+ T cells and has a role in CD8+ T cell activation (Bottcher et al., 2014), but with limited functions in Tex cells within tumors. In contrast, IL-10 specifically functions in the process of T cell exhaustion but not early activation in tumor models, and the IL-21–STAT3 axis acts on CD8+ T cells during both activation and exhaustion. Moreover, both IL-10 and IL-21 are previously shown to be enriched in the tumor microenvironment, mainly derived from CD4+ T cells (Cui et al., 2021; Sawant et al., 2019; Snell et al., 2018; Zander et al., 2019). Besides IL-10 and IL-21, there may exist other mediators inducing STAT3 phosphorylation in Tex cells within tumors, which needs to be further investigated (Yu et al., 2014). For example, IL-27, a member of the IL-6/IL-12 family regulating CD8+ T cell exhaustion in chronic infection and cancer, is shown to induce STAT3, besides STAT1, activation in T cells (Chihara et al., 2018; Hirahara et al., 2015; Huang et al., 2019; Owaki et al., 2008). In addition, some microRNAs and even hypoxia may also induce STAT3 activation in immune cells within tumors (Gao et al., 2017; Yu et al., 2014). Moreover, besides canonical homodimer, STAT3 can also interact with STAT1, STAT4, or STAT5 and form heterodimers in response to cytokines and CSF signaling (Delgoffe and Vignali, 2013), which implies the other potential mechanisms of STAT3 activation and function.
IL-10 and IL-21 functions, though have been reported in previous studies (Cui et al., 2011; Elsaesser et al., 2009; Fröhlich et al., 2009; Guo et al., 2021; Hanna et al., 2021; Ren et al., 2020; Xin et al., 2015; Yi et al., 2009; Zander et al., 2022), still remain controversial and lack mechanistic analysis. Briefly, IL-10 is shown to promote memory CD8+ T cell development in acute LCMV infection and the leukemia model (Cui et al., 2011; Hanna et al., 2021), but is also reported to enhance TIM-3+ CD8+ T cell development and expansion in B16 melanoma models (Guo et al., 2021; Sawant et al., 2019). IL-21 is shown not only to be critical in memory CD8+ T cell maturation in acute LCMV infection (Cui et al., 2011) but also functions to enhance CX3CR1+ or Granzyme B+ effector CD8+ T cell development in chronic LCMV infection and tumor models (Cui et al., 2021; Snell et al., 2018; Zander et al., 2022; Zander et al., 2019). Here, we systematically determined the functions of IL-10 and IL-21 in STAT3 signaling by phenotypical and transcriptional analyses. Both IL-10 and IL-21 had critical roles in promoting Texterm cell development, at least partially, in a STAT3-dependent manner. In addition, IL-10 and IL-21 also exert potent functions in promoting cytotoxicity, protecting against apoptosis, and enhancing the fitness of CD8+ T cells via STAT3 signaling. In addition to Texterm cells, STAT3 signaling also plays an important role in Texprog cells in cancer, although it is not absolutely required for their development. Considering the importance of Texprog cells in the long-term persistence of antitumor immunity and their ability to develop into Texterm cells (Miller et al., 2019), it is possible that in the absence of STAT3, Texterm cells were defective, at least in part, secondary to impaired Texprog cells, especially at a later stage.
In acute infection, CD8+ T cells differentiate into TCF1+ memory precursors and Granzyme B–producing terminally differentiated CD8+ T cells, similar to tumor-specific Texprog and Texterm cells, respectively, in their transcriptomes and functions to some extent. However, there are differences in the microenvironment between cancer and acute infection, resulting in many differences between tumor-specific Tex cells and effector or memory CD8+ T cells (McLane et al., 2019). One major difference is the increased and sustained expression of inhibitory receptors, including PD-1, TIM-3, TIGIT, 2B4, and LAG3, which are induced by chronic TCR stimulation (McLane et al., 2019). Terminally differentiated CD8+ T cells arise from TCF-1+ stem-like T cells in tumor microenvironments, in which STAT3 plays critical roles. Its role in the differentiation of effector T cells in acute infection requires further investigation.
There are two functional phosphorylated sites in the STAT3 protein located at Tyr705 and Ser727 residues (You et al., 2015). Phosphorylation of Tyr705 residue dominantly leads to the dimerization and nuclear translocation of STAT3 protein for the binding and transcriptional regulation of target genes (You et al., 2015). Phosphorylation at Ser727 residue mainly mediates mitochondrial localization and favors mitochondrial functions and OXPHOS (Huynh et al., 2019; Wegrzyn et al., 2009; You et al., 2015). pS727 can also facilitate transcriptional regulation by STAT3 (Wen et al., 1995). In this study, both Tyr705 and Ser727 residues are deleted in mature CD8+ T cells in Stat3fl/flCd8aCre mice (Takeda et al., 1998). By performing ChIP-seq for STAT3(pY705)-binding sites in the whole genome, we demonstrated that pSTAT3 directly bound to the loci of numerous Texterm-related genes (e.g., Id2, Prdm1, and Havcr2) and transcriptionally promoted their expression for Texterm cell development in cancer. Moreover, pSTAT3 also directly regulated the expression of Gzmb and Prf1 via binding to their promoters, thereby enhancing cytotoxic responses. In addition, STAT3 was also shown to be critical in promoting OXPHOS of Tex cells within tumors in this study. It is previously demonstrated that promoting OXPHOS in Texterm cells dramatically enhances their expansion and cytotoxicity (Guo et al., 2021). Thus, it is possible that besides transcriptional regulation, STAT3 may also function via S727 phosphorylation to further promote Texterm cell fitness and function, which needs to be further investigated.
The cooperation of STAT3 with AP-1 transcription factors (e.g., BATF and IRF4) has been reported in B cells and CD4+ T cells (Ciofani et al., 2012; Kwon et al., 2009; Xu et al., 2019). It is demonstrated that IRF4 cooperates with STAT3 to regulate IL-21–dependent gene expression in B cells (Kwon et al., 2009; Li and Leonard, 2018) and the IL-21–STAT3–BATF axis is required for sustaining the effector function of CD8+ T cells in chronic infections (Xin et al., 2015). Here, we analyzed the functional relationships between STAT3 and BATF/IRF4 in Tex cell differentiation by multiomics approaches. STAT3 had extensive co-occupancy in Tex fate-related gene loci with BATF and IRF4 across the whole genome in CD8+ T cells. Moreover, it epigenetically promoted chromatin accessibility for the efficient binding of BATF and IRF4 to target loci in activated CD8+ T cells. Hence, these data revealed a complex regulatory network involving STAT3 and AP-1 factors (e.g., BATF and IRF4) in the fate determination of CD8+ T cells during exhaustion.
Harnessing STAT3 signaling in improving cancer immunotherapies has been attempted a few times. For example, it is shown that fusing a STAT3-binding motif from IL-21 receptor into CD19 CAR against acute B lymphoblastic leukemia, with the goal of activating STAT3 signaling, results in superior antitumor effects (Kagoya et al., 2018). Recombinant human IL-6/IL-6Rα fusion protein and constitutively active GP130 or p40 subunit from IL-23/IL-12 both improve CAR-T cells in antitumor efficacy against B cell leukemia and solid tumors (Jiang et al., 2020; Ma et al., 2020). Moreover, it is reported that IL-21–primed polyclonal antigen–specific CD8+ T cells combined with CTLA-4 blockade induces long-term remission in a melanoma patient (Chapuis et al., 2016), and human CAR-modified CD8+ T cells treated with affinity-enhanced IL-10 variants, with greater STAT3 activation, display robust antitumor activities in vitro against leukemic cells (Gorby et al., 2020). Therefore, our research has provided a mechanistic basis for a STAT3-enhancing strategy in cancer immunotherapies and may benefit the further development of novel T cell– and cytokine-based therapies in the future.
Materials and methods
Mice
C57BL/6, CD45.1, Tcrbd−/−, and OT-I (TCR specific for E.G7 and B16-OVA KbOVA257-264) congenic mice were maintained in-house. Stat3fl/fl (Takeda et al., 1998) and Il6rafl/fl mice (McFarland-Mancini et al., 2010) were crossed with Cd8aCre mice (Maekawa et al., 2008) and then were bred with OT-I mice. Rosa26-Cas9 mice were bred with OT-I mice. All animal experiments conducted at Tsinghua University were approved by the Institutional Animal Care and Use Committee. 6–8-wk-old, male and female, age and sex-matched mice were used for all experiments.
Listeria monocytogenes (LM) infection
Erythromycin-resistant LM expressing OVA257-264 (LM-OVA) were grown in brain heart infusion media supplemented with 5 μg/ml erythromycin. 1 × 105 CFU of LM-OVA were resuspended in 100 μl PBS and injected intravenously (i.v.) into 6–8-wk-old mice. The infected mice were analyzed on day 8 after injection.
Tumor inoculation
B16-OVA cells were cultured in a DMEM medium with 10% FBS, penicillin, and streptomycin. E.G7 (EL4-OVA) cells were cultured in RPMI 1640 medium with 10% FBS, penicillin, and streptomycin. 1 × 106 E.G7 or 2 × 105 B16-OVA cells were resuspended in 100 μl PBS and injected subcutaneously (s.c.) into 6- to 8-wk-old mice. Tumor growth was monitored every 2 or 3 d. Tumor volume was calculated by the following formula: tumor volume = π × length × width2/6.
Isolation of TILs
E.G7 and B16-OVA tumors were digested with 1 mg/ml collagenase D supplemented with 10 U/ml DNase I for 40 min at 37°C prior to centrifugation on a discontinuous Percoll gradient (GE Healthcare).
Flow cytometry
Single cells were suspended in PBS and incubated with antibodies against surface molecules for 30 min at 4°C. Dead cells were excluded based on viability dye staining (Fixable viability dye eF506; eBioscience). For intracellular cytokine detection, cells were stimulated with PMA (50 ng/ml; Sigma-Aldrich) and ionomycin (500 ng/ml; Sigma-Aldrich) in the presence of Brefeldin A (Golgiplug, BD Bioscience) for 5 h prior to staining. For intracellular staining, surface-stained cells were fixed and permeabilized with the Foxp3/Transcription Factor Staining Buffer Set (eBiosciences), followed by incubation with fluorochrome-conjugated antibodies.
For the detection of phosphorylated STAT3, cells were cultured in a fresh medium supplied with cytokines for 30 min, resuspended in cold PBS, and stained with antibodies against surface molecules. Then the cells were fixed with Phosflow Lyse/Fix buffer (BD Bioscience), followed by 90% methanol permeabilization and staining with antibodies against phosphorylated STAT3 in PBS. For analysis, the cells were acquired on an LSRFortessa (BD) flow cytometer, and data were analyzed using FlowJo 10.4.
Naive CD8+ T cell adoptive transfer
CD8+ T cells were purified from spleens and lymph nodes using Dynabeads FlowComp Mouse CD8 Kit (Invitrogen). Naive CD8+CD25−CD44lowCD62Lhi T cells were FACS-sorted from purified CD8+ T cells on a FACS Aria (BD) flow cytometer. The purity of the sorted cells was >99%. Then 0.3 million FACS-sorted naive CD45.1/2 WT and CD45.2 Stat3−/− OT-I cells were mixed at a 1:1 ratio and transferred (i.v.) into CD45.1 recipients 1 d prior to B16-OVA tumor inoculation.
In vitro activation and stimulation
FACS-sorted naive CD8+ T cells were activated with the plate-bound anti-CD3 (clone 145-2C11, 5 μg/ml; BioXCell) and anti-CD28 (clone 37.51, 1 μg/ml; BioXCell) for 3 d. Then activated CD8+ T cells were cultured at 37°C for another 3 d in fresh medium with 10 ng/ml IL-6, 10 ng/ml IL-10, or 10 ng/ml IL-21.
Il10ra knockout
Single-guide RNA targeting Il10ra were designed on the CHOPCHOP website as sg-Il10ra-1 (5′- TGATCTCACAACGTTCACCC-3′) and sg-Il10ra-2 (5′-GATCTGTATCACCGAAGCTA-3′). Single-guide RNA was cloned into pWKO retroviral expression vectors which also encoded IRES-BFP cassettes. Empty vectors were used as controls. Retrovirus plasmids and pCL-ECO (packaging plasmid) were transfected into the Phoenix cell line using calcium phosphate. Retrovirus supernatant was harvested 48 h after transfection. FACS-sorted naive CD8+ T cells were activated with the plate-bound anti-CD3 (5 μg/ml) and anti-CD28 (5 μg/ml) for 48 h. Activated CD8+ T cells were infected with retrovirus in the presence of polybrene (1 μg/ml) with spin (1,800 rpm, 32°C) for 90 min, and medium was replaced 4 h after infection.
In vitro killing assay
Naive WT and Stat3−/− OT-I cells were activated in vitro for 3 d and then stimulated with IL-10 for another 3 d. B16-OVA tumor cells were labeled with CFSE at 37°C for 5 min, mixed with CD8+ T cells at 1:10, 1:4, 1:2, 1:1, and 2:1 ratios, and incubated at 37°C for 5 h. Intracellular cleaved caspase 3-PE in CFSE+ B16-OVA tumor cells were measured by flow cytometry.
In vitro proliferation assay
Naive WT and Stat3−/− CD8+ T cells were mixed and activated in vitro for 3 d and then labeled with CTV at 37°C for 5 min. CTV-labeled CD8+ T cells were stimulated by IL-6, IL-10, or IL-21 in anti-CD3/anti-CD28–bound plate. 2 or 3 d later, CTV in CD8+ T cells was measured by flow cytometry.
RNA processing and real-time quantitative PCR (RT-qPCR) analysis
Total cellular RNA was extracted from CD8+ T cells with Magzol reagent (Magen). M-MLV reverse transcription kit (Invitrogen) was used for generating cDNA. Gene expression at the mRNA level was measured using the SYBR real-time kit (Bio-Rad Laboratories). For RT-qPCR analysis, the data shown were normalized to the expression of the reference gene Actb. The following genes were amplified with primers as described: Actb, 5′-TGGAATCCTGTGGCATCCATGAAAC-3′ and 5′-TAAAACGCAGCTCAGTAACAGTCCG-3′; Id3, 5′- CTGTCGGAACGTAGCCTGG-3′ and 5′-GTGGTTCATGTCGTCCAAGAG-3′; Ccr7, 5′-CTTTCTTGTATGCCTTCATC-3′ and 5′- GGTTAAGCAGTTTCTTAGGT-3′; Id2, 5′-ATGAAAGCCTTCAGTCCGGTG-3′ and 5′-AGCAGACTCATCGGGTCGT-3′; and Gzmb, 5′-CCACTCTCGACCCTACATGG-3′ and 5′-GGCCCCCAAAGTGACATTTATT-3′.
RNA-seq and ATAC-seq
Cotransferred WT and Stat3−/− OT-I cells were FACS-sorted from B16-OVA tumors at about day 16 after inoculation for RNA-seq and ATAC-seq. For RNA-seq, total RNA was extracted and sent to BGI Genomics for library construction. The library products were sequenced via Illumina Hiseq4000 by BGI Genomics. Low-quality reads and adaptor sequences were removed by Trim Galore v0.4.4. The clean reads were aligned to mm10 by bowtie2 with default parameter (Langmead and Salzberg, 2012), and uniquely mapping reads were summarized by featureCounts (from Subread package). Differentially expressed genes (DEGs) were identified by at least twofold change and a false discovery rate (FDR) adjusted P value of 0.05 by using DESeq2. ClusterProfiler (R package) was used for pathway analysis (Yu et al., 2013).
For ATAC-seq, FACS-sorted OT-I TILs were processed and analyzed as indicated (Chang et al., 2020). PSEA was performed as indicated (Khan et al., 2019).
ChIP-seq and ChIP-qPCR
FACS-sorted naive CD8+ T cells were activated with the plate-bound anti-CD3 (5 μg/ml) and anti-CD28 (1 μg/ml) for 3 d. For pSTAT3 ChIP-seq, the activated WT CD8+ T cells were cultured at 37°C for 30 min in fresh medium with 10 ng/ml IL-6, 10 ng/ml IL-10, or 10 ng/ml IL-21. For H3K27ac ChIP-seq, the activated WT and Stat3−/− CD8+ T cells were stimulated by 10 ng/ml IL-10 for another 3 d. For BATF and IRF4 Chip experiment, the activated WT and Stat3−/− CD8+ T cells were stimulated by 10 ng/ml IL-10 for another 3 d. Then CD8+ T cells were fixed by 1% paraformaldehyde, followed by digestion and sonication. Chromatin from 1 × 107 lived cells was used for each ChIP experiment. 7.5 µg anti-pSTAT3(Y705) antibody, 1 µg anti-H3K27ac antibody, 7.5 µg anti-BATF antibody, or 7.5 µg anti-IRF4 antibody was used.
The ChIP experiment was performed as indicated (Xu et al., 2019). Sequencing reads were mapped to the mouse genome mm10 by bowtie2 (Langmead and Salzberg, 2012). PCR duplicates were removed using Picard MarkDuplicates. The uniquely mapped reads were used to call peak with MACS2 using a P value cutoff of 0.01 (Zhang et al., 2008). ChIPseeker was used for peak annotation (Yu et al., 2015). Overlapped peaks were identified by bedtools intersect. Deeptools was used for comparing profiles of different ChIPseq samples (including those from Gene Expression Omnibus [GEO] datasets; Ramirez et al., 2016). HOMER was used for motif discovery.
The enriched heatmaps of pSTAT3 ChIP-seq signal profiles were generated with Deeptools (V3.5.0; Ramirez et al., 2016). The computeMatrix was used to calculate scores for the top 3,000 peak sites of pSTAT3 with the following parameters: reference-point--referencePoint center. The plotHeatmap was used to create the heatmap for scores associated with the genomic regions with the following parameters: --zMin 1 --zMax 10 --kmeans 1.
For ChIP-qPCR analysis, the following DNA fragments were amplified with primers as described: Gzmb promoter, 5′-TACCATAGGCTACAAACCCCACCC-3′ and 5′-CGTCAAGAGTGTTGTCCTTGCTC-3′; Havcr2 intron-1, 5′-TGAGTGCAACCTGGGTTGC-3′ and 5′-TTCACCACATGCAATCTCTAAAGTG-3′; Havcr2 intron-2, 5′-AACTTTTGTGTGTATGGCTGC-3′ and 5′-GTAAGAGGCCAACATGGTCAG-3′; Havcr2 intron-3, 5′-GAGGACAGCCTCTTGTTCTAGTC-3′ and 5′-GTTAGGAGAGCACATTCAGACATG-3′; and Il2ra promoter, 5′-TGACAGACTGAGAGGCCTGA-3′ and 5′-TGGGTCAACCCCTTTACTGC-3′.
scRNA-seq analysis
Gene expression information was downloaded from the website for public human scRNA-seq database (Lung, http://lung.cancer-pku.cn; HCC, http://hcc.cancer-pku.cn/; and CRC, http://crc.cancer-pku.cn/; Guo et al., 2018; Zhang et al., 2018; Zheng et al., 2017). The expression unit was normalized expression/log(TPM+1). PD-1hi cells were filtered using an expression value cutoff of 5.00.
Statistical analysis
Statistical analysis was performed with Graph Prism 8.2.0 (GraphPad software) and presented as mean ± SEM unless specifically indicated. For two group comparisons, statistical analysis was performed with unpaired or paired two-tailed Student’s t tests. For multiple group comparisons, statistical analysis was performed with one-way ANOVA followed by Tukey’s multiple comparisons test. Tumor growth data were compared by two-way ANOVA followed by Tukey’s multiple comparisons test. P values <0.05 were considered statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). Statistical details for each experiment can be found in the figures and the legends.
Online supplemental material
Fig. S1 shows data further supporting the results in Fig. 1. Fig. S2 shows data further supporting the results in Fig. 2. Fig. S3 shows data further supporting the results in Fig. 4. Fig. S4 shows data further supporting the results in Fig. 5. Fig. S5 shows data further supporting the results in Fig. 7. Table S1 lists the DEGs in WT vs. Stat3−/− OT-I cells from B16-OVA tumors. Table S2 lists STAT3-binding and -regulated genes. Table S3 lists STAT3-induced or -suppressed feature genes. Table S4 lists DA peaks in WT vs. Stat3−/− OT-I cells from B16-OVA tumors. Table S5 lists differentially H3K27ac modified regions (DMRs) between IL-10–stimulated WT and Stat3−/− CD8+ T cells. Table S6 lists STAT3- and BATF/IRF4-coregulated genes.
Supplementary Material
lists DEGs in WT vs. Stat3−/− OT-I cells from B16-OVA tumors.
lists STAT3-binding and -regulated genes.
lists STAT3-induced or -suppressed feature genes.
lists DA peaks in WT vs. Stat3−/− OT-I cells from B16-OVA tumors.
lists differentially H3K27ac modified regions (DMRs) between IL-10–stimulated WT and Stat3−/− CD8+ T cells.
lists STAT3 and BATF/IRF4 coregulated genes.
contains original blots for Fig. S1.
Acknowledgments
We thank Dr. Wenwen Zeng (Institute for Immunology, Tsinghua University, Beijing, China) for providing Il6rafl/fl mice.
This work was supported by grants from the Natural Science Foundation of China (31991173, 31821003, and 31991170), the National Key Research and Development Program of China (2021YFC2302403), Tsinghua University Spring Breeze Fund (2020Z99CFG008), Innovative Research Team of High-level Local Universities in Shanghai, Tsinghua University-Xiamen Chang Gung Hospital Joint Research Center for Anaphylactic Disease, and Shanghai Jiao Tong University-Bennu Biotherapeutics Joint Laboratory for Cell Therapy.
Author contributions: Q. Sun and C. Dong designed the project and analyzed the data. Q. Sun conducted the experiments. D. Liu, B. Pan, X. Chi, P. Wei, D. Cai. W. Xu, K. Wei, Z. Zhao, and Y. Fu helped with the experiments. X. Zhao and R. Li analyzed the bioinformatics data. Q. Sun, C. Dong, L. Ni, and B. Xie prepared the manuscript.
Data availability
Data from RNA-seq of WT vs. Stat3−/− OT-I cells transferred into B16-OVA–bearing mice (Fig. 2) are available in the GEO database under accession no. GSE217375. Data from ATAC-seq of WT vs. Stat3−/− OT-I cells transferred into B16-OVA–bearing mice (Fig. 6) are available in the GEO database under accession no. GSE217372. ChIP-seq of pSTAT3 in CD8+ T cells (Fig. 5) and ChIP-seq of H3K27ac in WT vs. Stat3−/− CD8+ T cells (Fig. 6) are available in the GEO database under accession no. GSE217374.
References
- Angelosanto, J.M., Blackburn S.D., Crawford A., and Wherry E.J.. 2012. Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. J. Virol. 86:8161–8170. 10.1128/JVI.00889-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher, J.P., Schanz O., Garbers C., Zaremba A., Hegenbarth S., Kurts C., Beyer M., Schultze J.L., Kastenmüller W., Rose-John S., and Knolle P.A.. 2014. IL-6 trans-signaling-dependent rapid development of cytotoxic CD8+ T cell function. Cell Rep. 8:1318–1327. 10.1016/j.celrep.2014.07.008 [DOI] [PubMed] [Google Scholar]
- Callahan, M.K., Postow M.A., and Wolchok J.D.. 2016. Targeting T cell Co-receptors for cancer Therapy. Immunity. 44:1069–1078. 10.1016/j.immuni.2016.04.023 [DOI] [PubMed] [Google Scholar]
- Chang, D., Xing Q., Su Y., Zhao X., Xu W., Wang X., and Dong C.. 2020. The conserved non-coding sequences CNS6 and CNS9 control cytokine-induced rorc transcription during T helper 17 cell differentiation. Immunity. 53:614–626.e4. 10.1016/j.immuni.2020.07.012 [DOI] [PubMed] [Google Scholar]
- Chapuis, A.G., Lee S.M., Thompson J.A., Roberts I.M., Margolin K.A., Bhatia S., Sloan H.L., Lai I., Wagener F., Shibuya K., et al. 2016. Combined IL-21-primed polyclonal CTL plus CTLA4 blockade controls refractory metastatic melanoma in a patient. J. Exp. Med. 213:1133–1139. 10.1084/jem.20152021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., López-Moyado I.F., Seo H., Lio C.J., Hempleman L.J., Sekiya T., Yoshimura A., Scott-Browne J.P., and Rao A.. 2019. NR4A transcription factors limit CAR T cell function in solid tumours. Nature. 567:530–534. 10.1038/s41586-019-0985-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y., Zander R.A., Wu X., Schauder D.M., Kasmani M.Y., Shen J., Zheng S., Burns R., Taparowsky E.J., and Cui W.. 2021. BATF regulates progenitor to cytolytic effector CD8+ T cell transition during chronic viral infection. Nat. Immunol. 22:996–1007. 10.1038/s41590-021-00965-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihara, N., Madi A., Kondo T., Zhang H., Acharya N., Singer M., Nyman J., Marjanovic N.D., Kowalczyk M.S., Wang C., et al. 2018. Induction and transcriptional regulation of the co-inhibitory gene module in T cells. Nature. 558:454–459. 10.1038/s41586-018-0206-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciofani, M., Madar A., Galan C., Sellars M., Mace K., Pauli F., Agarwal A., Huang W., Parkhurst C.N., Muratet M., et al. 2012. A validated regulatory network for Th17 cell specification. Cell. 151:289–303. 10.1016/j.cell.2012.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, C., Wang J., Fagerberg E., Chen P.M., Connolly K.A., Damo M., Cheung J.F., Mao T., Askari A.S., Chen S., et al. 2021. Neoantigen-driven B cell and CD4 T follicular helper cell collaboration promotes anti-tumor CD8 T cell responses. Cell. 184:6101–6118.e13. 10.1016/j.cell.2021.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, W., Liu Y., Weinstein J.S., Craft J., and Kaech S.M.. 2011. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity. 35:792–805. 10.1016/j.immuni.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgoffe, G.M., and Vignali D.A.. 2013. STAT heterodimers in immunity: A mixed message or a unique signal? JAKSTAT. 2:e23060. 10.4161/jkst.23060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, C. 2021. Cytokine regulation and function in T cells. Annu. Rev. Immunol. 39:51–76. 10.1146/annurev-immunol-061020-053702 [DOI] [PubMed] [Google Scholar]
- Darnell, J.E.Jr. 1997. STATs and gene regulation. Science. 277:1630–1635. 10.1126/science.277.5332.1630 [DOI] [PubMed] [Google Scholar]
- Elsaesser, H., Sauer K., and Brooks D.G.. 2009. IL-21 is required to control chronic viral infection. Science. 324:1569–1572. 10.1126/science.1174182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerich, J., Mumm J.B., Chan I.H., LaFace D., Truong H., McClanahan T., Gorman D.M., and Oft M.. 2012. IL-10 directly activates and expands tumor-resident CD8(+) T cells without de novo infiltration from secondary lymphoid organs. Cancer Res. 72:3570–3581. 10.1158/0008-5472.CAN-12-0721 [DOI] [PubMed] [Google Scholar]
- Fröhlich, A., Kisielow J., Schmitz I., Freigang S., Shamshiev A.T., Weber J., Marsland B.J., Oxenius A., and Kopf M.. 2009. IL-21R on T cells is critical for sustained functionality and control of chronic viral infection. Science. 324:1576–1580. 10.1126/science.1172815 [DOI] [PubMed] [Google Scholar]
- Gao, P., Niu N., Wei T., Tozawa H., Chen X., Zhang C., Zhang J., Wada Y., Kapron C.M., and Liu J.. 2017. The roles of signal transducer and activator of transcription factor 3 in tumor angiogenesis. Oncotarget. 8:69139–69161. 10.18632/oncotarget.19932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorby, C., Sotolongo Bellón J., Wilmes S., Warda W., Pohler E., Fyfe P.K., Cozzani A., Ferrand C., Walter M.R., Mitra S., et al. 2020. Engineered IL-10 variants elicit potent immunomodulatory effects at low ligand doses. Sci. Signal. 13:eabc0653. 10.1126/scisignal.abc0653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X., Zhang Y., Zheng L., Zheng C., Song J., Zhang Q., Kang B., Liu Z., Jin L., Xing R., et al. 2018. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 24:978–985. 10.1038/s41591-018-0045-3 [DOI] [PubMed] [Google Scholar]
- Guo, Y., Xie Y.Q., Gao M., Zhao Y., Franco F., Wenes M., Siddiqui I., Bevilacqua A., Wang H., Yang H., et al. 2021. Metabolic reprogramming of terminally exhausted CD8+ T cells by IL-10 enhances anti-tumor immunity. Nat. Immunol. 22:746–756. 10.1038/s41590-021-00940-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna, B.S., Llaó-Cid L., Iskar M., Roessner P.M., Klett L.C., Wong J.K.L., Paul Y., Ioannou N., Öztürk S., Mack N., et al. 2021. Interleukin-10 receptor signaling promotes the maintenance of a PD-1int TCF-1+ CD8+ T cell population that sustains anti-tumor immunity. Immunity. 54:2825–2841.e10. 10.1016/j.immuni.2021.11.004 [DOI] [PubMed] [Google Scholar]
- Hermans, D., Gautam S., García-Cañaveras J.C., Gromer D., Mitra S., Spolski R., Li P., Christensen S., Nguyen R., Lin J.X., et al. 2020. Lactate dehydrogenase inhibition synergizes with IL-21 to promote CD8+ T cell stemness and antitumor immunity. Proc. Natl. Acad. Sci. USA. 117:6047–6055. 10.1073/pnas.1920413117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirahara, K., Onodera A., Villarino A.V., Bonelli M., Sciumè G., Laurence A., Sun H.W., Brooks S.R., Vahedi G., Shih H.Y., et al. 2015. Asymmetric action of STAT transcription factors drives transcriptional outputs and cytokine specificity. Immunity. 42:877–889. 10.1016/j.immuni.2015.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Q., Wu X., Wang Z., Chen X., Wang L., Lu Y., Xiong D., Liu Q., Tian Y., Lin H., et al. 2022. The primordial differentiation of tumor-specific memory CD8+ T cells as bona fide responders to PD-1/PD-L1 blockade in draining lymph nodes. Cell. 185:4049–4066.e25. 10.1016/j.cell.2022.09.020 [DOI] [PubMed] [Google Scholar]
- Huang, Z., Zak J., Pratumchai I., Shaabani N., Vartabedian V.F., Nguyen N., Wu T., Xiao C., and Teijaro J.R.. 2019. IL-27 promotes the expansion of self-renewing CD8+ T cells in persistent viral infection. J. Exp. Med. 216:1791–1808. 10.1084/jem.20190173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh, J., Chand A., Gough D., and Ernst M.. 2019. Therapeutically exploiting STAT3 activity in cancer - using tissue repair as a road map. Nat. Rev. Cancer. 19:82–96. 10.1038/s41568-018-0090-8 [DOI] [PubMed] [Google Scholar]
- Jiang, Z., Liao R., Lv J., Li S., Zheng D., Qin L., Wu D., Chen S., Long Y., Wu Q., et al. 2020. IL-6 trans-signaling promotes the expansion and anti-tumor activity of CAR T cells. Leukemia. 35:1380–1391. 10.1038/s41375-020-01085-1 [DOI] [PubMed] [Google Scholar]
- Joshi, N.S., Cui W., Chandele A., Lee H.K., Urso D.R., Hagman J., Gapin L., and Kaech S.M.. 2007. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 27:281–295. 10.1016/j.immuni.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech, S.M., and Cui W.. 2012. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12:749–761. 10.1038/nri3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagoya, Y., Tanaka S., Guo T., Anczurowski M., Wang C.H., Saso K., Butler M.O., Minden M.D., and Hirano N.. 2018. A novel chimeric antigen receptor containing a JAK-STAT signaling domain mediates superior antitumor effects. Nat. Med. 24:352–359. 10.1038/nm.4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamphorst, A.O., Wieland A., Nasti T., Yang S., Zhang R., Barber D.L., Konieczny B.T., Daugherty C.Z., Koenig L., Yu K., et al. 2017. Rescue of exhausted CD8 T cells by PD-1–targeted therapies is CD28-dependent. Science. 355:1423–1427. 10.1126/science.aaf0683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane, A., Deenick E.K., Ma C.S., Cook M.C., Uzel G., and Tangye S.G.. 2014. STAT3 is a central regulator of lymphocyte differentiation and function. Curr. Opin. Immunol. 28:49–57. 10.1016/j.coi.2014.01.015 [DOI] [PubMed] [Google Scholar]
- Khan, O., Giles J.R., McDonald S., Manne S., Ngiow S.F., Patel K.P., Werner M.T., Huang A.C., Alexander K.A., Wu J.E., et al. 2019. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature. 571:211–218. 10.1038/s41586-019-1325-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurachi, M., Barnitz R.A., Yosef N., Odorizzi P.M., DiIorio M.A., Lemieux M.E., Yates K., Godec J., Klatt M.G., Regev A., et al. 2014. The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat. Immunol. 15:373–383. 10.1038/ni.2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, H., Thierry-Mieg D., Thierry-Mieg J., Kim H.P., Oh J., Tunyaplin C., Carotta S., Donovan C.E., Goldman M.L., Tailor P., et al. 2009. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 31:941–952. 10.1016/j.immuni.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P., and Leonard W.J.. 2018. Chromatin accessibility and interactions in the transcriptional regulation of T cells. Front. Immunol. 9:2738. 10.3389/fimmu.2018.02738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X., Wang Y., Lu H., Li J., Yan X., Xiao M., Hao J., Alekseev A., Khong H., Chen T., et al. 2019. Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature. 567:525–529. 10.1038/s41586-019-0979-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., Zhou N., Zhou L., Wang J., Zhou Y., Zhang T., Fang Y., Deng J., Gao Y., Liang X., et al. 2021. IL-2 regulates tumor-reactive CD8+ T cell exhaustion by activating the aryl hydrocarbon receptor. Nat. Immunol. 22:358–369. 10.1038/s41590-020-00850-9 [DOI] [PubMed] [Google Scholar]
- Ma, X., Shou P., Smith C., Chen Y., Du H., Sun C., Porterfield Kren N., Michaud D., Ahn S., Vincent B., et al. 2020. Interleukin-23 engineering improves CAR T cell function in solid tumors. Nat. Biotechnol. 38:448–459. 10.1038/s41587-019-0398-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa, Y., Minato Y., Ishifune C., Kurihara T., Kitamura A., Kojima H., Yagita H., Sakata-Yanagimoto M., Saito T., Taniuchi I., et al. 2008. Notch2 integrates signaling by the transcription factors RBP-J and CREB1 to promote T cell cytotoxicity. Nat. Immunol. 9:1140–1147. 10.1038/ni.1649 [DOI] [PubMed] [Google Scholar]
- Man, K., Gabriel S.S., Liao Y., Gloury R., Preston S., Henstridge D.C., Pellegrini M., Zehn D., Berberich-Siebelt F., Febbraio M.A., et al. 2017. Transcription factor IRF4 promotes CD8+ T cell exhaustion and limits the development of memory-like T cells during chronic infection. Infect. Immun. 47:1129–1141.e5. 10.1016/j.immuni.2017.11.021 [DOI] [PubMed] [Google Scholar]
- Martinez, G.J., Pereira R.M., Äijö T., Kim E.Y., Marangoni F., Pipkin M.E., Togher S., Heissmeyer V., Zhang Y.C., Crotty S., et al. 2015. The transcription factor NFAT promotes exhaustion of activated CD8⁺ T cells. Immunity. 42:265–278. 10.1016/j.immuni.2015.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarland-Mancini, M.M., Funk H.M., Paluch A.M., Zhou M., Giridhar P.V., Mercer C.A., Kozma S.C., and Drew A.F.. 2010. Differences in wound healing in mice with deficiency of IL-6 versus IL-6 receptor. J. Immunol. 184:7219–7228. 10.4049/jimmunol.0901929 [DOI] [PubMed] [Google Scholar]
- McLane, L.M., Abdel-Hakeem M.S., and Wherry E.J.. 2019. CD8 T cell exhaustion during chronic viral infection and cancer. Annu. Rev. Immunol. 37:457–495. 10.1146/annurev-immunol-041015-055318 [DOI] [PubMed] [Google Scholar]
- Miller, B.C., Sen D.R., Al Abosy R., Bi K., Virkud Y.V., LaFleur M.W., Yates K.B., Lako A., Felt K., Naik G.S., et al. 2019. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20:326–336. 10.1038/s41590-019-0312-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujib, S., Jones R.B., Lo C., Aidarus N., Clayton K., Sakhdari A., Benko E., Kovacs C., and Ostrowski M.A.. 2012. Antigen-independent induction of Tim-3 expression on human T cells by the common γ-chain cytokines IL-2, IL-7, IL-15, and IL-21 is associated with proliferation and is dependent on the phosphoinositide 3-kinase pathway. J. Immunol. 188:3745–3756. 10.4049/jimmunol.1102609 [DOI] [PubMed] [Google Scholar]
- Nurieva, R.I., Chung Y., Martinez G.J., Yang X.O., Tanaka S., Matskevitch T.D., Wang Y.H., and Dong C.. 2009. Bcl6 mediates the development of T follicular helper cells. Science. 325:1001–1005. 10.1126/science.1176676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, J.A., and Jameson S.C.. 2011. Keeping STATs on memory CD8+ T cells. Immunity. 35:663–665. 10.1016/j.immuni.2011.11.006 [DOI] [PubMed] [Google Scholar]
- Owaki, T., Asakawa M., Morishima N., Mizoguchi I., Fukai F., Takeda K., Mizuguchi J., and Yoshimoto T.. 2008. STAT3 is indispensable to IL-27-mediated cell proliferation but not to IL-27-induced Th1 differentiation and suppression of proinflammatory cytokine production. J. Immunol. 180:2903–2911. 10.4049/jimmunol.180.5.2903 [DOI] [PubMed] [Google Scholar]
- Pauken, K.E., and Wherry E.J.. 2015. SnapShot: T cell exhaustion. Cell. 163:1038–1038.e1. 10.1016/j.cell.2015.10.054 [DOI] [PubMed] [Google Scholar]
- Philip, M., and Schietinger A.. 2021. CD8(+) T cell differentiation and dysfunction in cancer. Nat. Rev. Immunol. 22:209–223. 10.1038/s41577-021-00574-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puram, S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S., Rodman C., Luo C.L., Mroz E.A., Emerick K.S., et al. 2017. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 171:1611–1624 e24. 10.1016/j.cell.2017.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez, F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., and Manke T.. 2016. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44:W160–W165. 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, H.M., Kolawole E.M., and Ren M.. 2020. IL-21 from high-affinity CD4 T cells drives differentiation of brain-resident CD8 T cells during persistent viral infection. Sci. Immunol. 5:eabb5590. 10.1126/sciimmunol.abb5590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser, R.L., Martins G.A., Kalachikov S., Chandele A., Parish I.A., Meffre E., Jacob J., Calame K., and Kaech S.M.. 2009. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 31:296–308. 10.1016/j.immuni.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, N., Anderson K., Volpedo G., Hamza O., Varikuti S., Satoskar A.R., and Oghumu S.. 2020. STAT1 inhibits T-cell exhaustion and myeloid derived suppressor cell accumulation to promote antitumor immune responses in head and neck squamous cell carcinoma. Int. J. Cancer. 146:1717–1729. 10.1002/ijc.32781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade-Feldman, M., Yizhak K., Bjorgaard S.L., Ray J.P., de Boer C.G., Jenkins R.W., Lieb D.J., Chen J.H., Frederick D.T., Barzily-Rokni M., et al. 2018. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 175:998–1013.e20. 10.1016/j.cell.2018.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawant, D.V., Yano H., Chikina M., Zhang Q., Liao M., Liu C., Callahan D.J., Sun Z., Sun T., Tabib T., et al. 2019. Adaptive plasticity of IL-10+ and IL-35+ Treg cells cooperatively promotes tumor T cell exhaustion. Nat. Immunol. 20:724–735. 10.1038/s41590-019-0346-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel, J.M., Herbst R.H., Canner D., Li A., Hillman M., Shanahan S.L., Gibbons G., Smith O.C., Kim J.Y., Westcott P., et al. 2021. Conventional type I dendric cells maintain a reservoir of proliferative tumor-antigen specific TCF-1(+) CD8(+) T cells in tumor-draining lymph nodes. Immunity. 54:2338–2353.e6. 10.1016/j.immuni.2021.08.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, A.C., Dündar F., Zumbo P., Chandran S.S., Klebanoff C.A., Shakiba M., Trivedi P., Menocal L., Appleby H., Camara S., et al. 2019. TOX is a critical regulator of tumour-specific T cell differentiation. Nature. 571:270–274. 10.1038/s41586-019-1324-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H., González-Avalos E., Zhang W., Ramchandani P., Yang C., Lio C.J., Rao A., and Hogan P.G.. 2021. BATF and IRF4 cooperate to counter exhaustion in tumor-infiltrating CAR T cells. Nat. Immunol. 22:983–995. 10.1038/s41590-021-00964-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui, I., Schaeuble K., Chennupati V., Fuertes Marraco S.A., Calderon-Copete S., Pais Ferreira D., Carmona S.J., Scarpellino L., Gfeller D., Pradervand S., et al. 2019. Intratumoral Tcf1+PD-1+CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade Immunotherapy. Immunity. 50:195–211.e10. 10.1016/j.immuni.2018.12.021 [DOI] [PubMed] [Google Scholar]
- Siegel, A.M., Heimall J., Freeman A.F., Hsu A.P., Brittain E., Brenchley J.M., Douek D.C., Fahle G.H., Cohen J.I., Holland S.M., and Milner J.D.. 2011. A critical role for STAT3 transcription factor signaling in the development and maintenance of human T cell memory. Immunity. 35:806–818. 10.1016/j.immuni.2011.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell, L.M., MacLeod B.L., Law J.C., Osokine I., Elsaesser H.J., Hezaveh K., Dickson R.J., Gavin M.A., Guidos C.J., McGaha T.L., and Brooks D.G.. 2018. CD8+ T cell priming in established chronic viral infection preferentially directs differentiation of memory-like cells for sustained immunity. Immunity. 49:678–694.e5. 10.1016/j.immuni.2018.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, K., Kaisho T., Yoshida N., Takeda J., Kishimoto T., and Akira S.. 1998. Stat3 activation is responsible for IL-6-dependent T cell proliferation through preventing apoptosis: Generation and characterization of T cell-specific stat3-deficient mice. J. Immunol. 161:4652–4660 [PubMed] [Google Scholar]
- Vahedi, G., Takahashi H., Nakayamada S., Sun H.W., Sartorelli V., Kanno Y., and O’Shea J.J.. 2012. STATs shape the active enhancer landscape of T cell populations. Cell. 151:981–993. 10.1016/j.cell.2012.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn, J., Potla R., Chwae Y.-J., Sepuri N.B., Zhang Q., Koeck T., Derecka M., Szczepanek K., Szelag M., Gornicka A., et al. 2009. Function of mitochondrial Stat3 in cellular respiration. Science. 323:793–797. 10.1126/science.1164551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, Z., Zhong Z., and Darnell J.E. Jr. 1995. Maximal activation of transcription by statl and stat3 requires both tyrosine and serine phosphorylation. Cell. 82:241–250. 10.1016/0092-8674(95)90311-9 [DOI] [PubMed] [Google Scholar]
- Wu, T., Ji Y., Moseman E.A., Xu H.C., Manglani M., Kirby M., Anderson S.M., Handon R., Kenyon E., Elkahloun A., et al. 2016. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci. Immunol. 1:eaai8593. 10.1126/sciimmunol.aai8593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, G., Schauder D.M., Lainez B., Weinstein J.S., Dai Z., Chen Y., Esplugues E., Wen R., Wang D., Parish I.A., et al. 2015. A critical role of IL-21-induced BATF in sustaining CD8-T-cell-mediated chronic viral control. Cell Rep. 13:1118–1124. 10.1016/j.celrep.2015.09.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, W., Zhao X., Wang X., Feng H., Gou M., Jin W., Wang X., Liu X., and Dong C.. 2019. The transcription factor Tox2 drives T follicular helper cell development via regulating chromatin accessibility. Immunity. 51:826–839.e5. 10.1016/j.immuni.2019.10.006 [DOI] [PubMed] [Google Scholar]
- Yang, R., Masters A.R., Fortner K.A., Champagne D.P., Yanguas-Casás N., Silberger D.J., Weaver C.T., Haynes L., and Rincon M.. 2016. IL-6 promotes the differentiation of a subset of naive CD8+ T cells into IL-21-producing B helper CD8+ T cells. J. Exp. Med. 213:2281–2291. 10.1084/jem.20160417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X.O., Panopoulos A.D., Nurieva R., Chang S.H., Wang D., Watowich S.S., and Dong C.. 2007. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 282:9358–9363. 10.1074/jbc.C600321200 [DOI] [PubMed] [Google Scholar]
- Yang, Z.Z., Grote D.M., Ziesmer S.C., Niki T., Hirashima M., Novak A.J., Witzig T.E., and Ansell S.M.. 2012. IL-12 upregulates TIM-3 expression and induces T cell exhaustion in patients with follicular B cell non-Hodgkin lymphoma. J. Clin. Invest. 122:1271–1282. 10.1172/JCI59806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi, J.S., Du M., and Zajac A.J.. 2009. A vital role for interleukin-21 in the control of a chronic viral infection. Science. 324:1572–1576. 10.1126/science.1175194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, L., Wang Z., Li H., Shou J., Jing Z., Xie J., Sui X., Pan H., and Han W.. 2015. The role of STAT3 in autophagy. Autophagy. 11:729–739. 10.1080/15548627.2015.1017192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, C.R., Dambuza I.M., Lee Y.J., Frank G.M., and Egwuagu C.E.. 2013. STAT3 regulates proliferation and survival of CD8+ T cells: Enhances effector responses to HSV-1 infection, and inhibits IL-10+ regulatory CD8+ T cells in autoimmune uveitis. Mediators Inflamm. 2013:359674. 10.1155/2013/359674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G., Wang L.G., and He Q.Y.. 2015. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 31:2382–2383. 10.1093/bioinformatics/btv145 [DOI] [PubMed] [Google Scholar]
- Yu, H., Lee H., Herrmann A., Buettner R., and Jove R.. 2014. Revisiting STAT3 signalling in cancer: New and unexpected biological functions. Nat. Rev. Cancer. 14:736–746. 10.1038/nrc3818 [DOI] [PubMed] [Google Scholar]
- Yu, H., Pardoll D., and Jove R.. 2009. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer. 9:798–809. 10.1038/nrc2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander, R., Kasmani M.Y., Chen Y., Topchyan P., Shen J., Zheng S., Burns R., Ingram J., Cui C., Joshi N., et al. 2022. Tfh-cell-derived interleukin 21 sustains effector CD8+ T cell responses during chronic viral infection. Immunity. 55:475–493.e5. 10.1016/j.immuni.2022.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander, R., Schauder D., Xin G., Nguyen C., Wu X., Zajac A., and Cui W.. 2019. CD4+ T cell Help is required for the formation of a cytolytic CD8+ T cell subset that protects against chronic infection and cancer. Immunity. 51:1028–1042.e4. 10.1016/j.immuni.2019.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., Yue C., Herrmann A., Song J., Egelston C., Wang T., Zhang Z., Li W., Lee H., Aftabizadeh M., et al. 2020. STAT3 activation-induced fatty acid oxidation in CD8+ T effector cells is critical for obesity-promoted breast tumor growth. Cell Metabol. 31:148–161.e5. 10.1016/j.cmet.2019.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Yu X., Zheng L., Zhang Y., Li Y., Fang Q., Gao R., Kang B., Zhang Q., Huang J.Y., et al. 2018. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 564:268–272. 10.1038/s41586-018-0694-x [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, C., Zheng L., Yoo J.K., Guo H., Zhang Y., Guo X., Kang B., Hu R., Huang J.Y., Zhang Q., et al. 2017. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 169:1342–1356.e16. 10.1016/j.cell.2017.05.035 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists DEGs in WT vs. Stat3−/− OT-I cells from B16-OVA tumors.
lists STAT3-binding and -regulated genes.
lists STAT3-induced or -suppressed feature genes.
lists DA peaks in WT vs. Stat3−/− OT-I cells from B16-OVA tumors.
lists differentially H3K27ac modified regions (DMRs) between IL-10–stimulated WT and Stat3−/− CD8+ T cells.
lists STAT3 and BATF/IRF4 coregulated genes.
contains original blots for Fig. S1.
Data Availability Statement
Data from RNA-seq of WT vs. Stat3−/− OT-I cells transferred into B16-OVA–bearing mice (Fig. 2) are available in the GEO database under accession no. GSE217375. Data from ATAC-seq of WT vs. Stat3−/− OT-I cells transferred into B16-OVA–bearing mice (Fig. 6) are available in the GEO database under accession no. GSE217372. ChIP-seq of pSTAT3 in CD8+ T cells (Fig. 5) and ChIP-seq of H3K27ac in WT vs. Stat3−/− CD8+ T cells (Fig. 6) are available in the GEO database under accession no. GSE217374.