Abstract
The maintenance of a healthy mitochondrial network and the ability to adjust organelle population in response to internal or external stimuli are essential for the function and the survival of eukaryotic cells. Over the last two decades several studies have demonstrated the paramount importance of mitophagy, a selective form of autophagy that removes damaged and/or superfluous organelles, in organismal physiology. Post-mitotic neuronal cells are particularly vulnerable to mitochondrial damage, and mitophagy impairment has emerged as a causative factor in multiple neurodegenerative pathologies, including Alzheimer's disease and Parkinson's disease among others. Although mitochondrial turnover is a multifaceted process, neurons have to tackle additional complications, arising from their pronounced bioenergetic demands and their unique architecture and cellular polarisation that render the degradation of distal organelles challenging. Mounting evidence indicates that despite the functional conservation of mitophagy pathways, the unique features of neuronal physiology have led to the adaptation of compartmentalised solutions, which serve to ensure seamless mitochondrial removal in every part of the cell. In this review, we summarise the current knowledge concerning the molecular mechanisms that mediate mitophagy compartmentalisation and discuss their implications in various human pathologies.
Key words: Cell death, energy metabolism, mitochondria, mitophagy, neurodegeneration, neuron
Introduction
Mitochondria are highly dynamic eukaryotic organelles, known as the major source of cellular ATP, generated through the tricarboxylic acid cycle and oxidative phosphorylation (OXPHOS). Nonetheless, they are also involved in other essential cellular processes, including calcium (Ca2+) homoeostasis, iron metabolism and the maintenance of redox balance among others (Refs 1–3). Therefore, the maintenance of proper mitochondrial function is pivotal for cellular homoeostasis and viability, and cells have evolved a wide arsenal of quality control mechanisms to sense and respond to aberrations in their activity. Such mechanisms include the control of mitochondrial biogenesis, the mitochondrial unfolded protein response, the integrated stress response, fission and fusion events (collectively termed mitochondrial dynamics) and the selective autophagic removal of damaged or superfluous organelles via mitophagy. The latter has gained a lot of focus, as it has been shown to be an important contributor to key life processes, such as cellular homoeostasis, stress response and differentiation, as well as organismal development, survival, healthspan and longevity. Especially in neurons, mitophagy defects have been associated with a variety of pathological conditions, including Alzheimer's Disease (AD), Parkinson's Disease (PD), Huntington's Disease (HD) and Amyotrophic Lateral Sclerosis (ALS) among others (Table 1) (Refs 4, 5).
Table 1.
Mitophagy pathways or molecules affected in major neurodegenerative diseases
| Disease | Mitophagy pathway/molecule | Reference |
|---|---|---|
| Parkinson's Disease | ↓PINK1 activity | (Ref. 197) |
| ↓Parkin activity | (Ref. 198) | |
| ↑LRKK2 activity | (Ref. 110) | |
| ↓Miro translocation to Mito-ER contact sites | (Ref. 199) | |
| Alzheimer's Disease | ↓PINK1/Parkin proteins in mitochondria | (Refs 200, 201) |
| ↓NIX, BNIP3, FUNDC1, OPTN mRNA levels | (Ref. 202) | |
| ↓DISC1, BCL2L13, FUNDC1, PINK1 protein levels | (Refs 176, 187) | |
| Huntington's Disease | ↓OPTN, SQSTM1, NBR1 interaction with LC3 | (Ref. 42) |
| ↑DRP1 activity | (Ref. 203) | |
| Amyotrophic Lateral Sclerosis | ↓TBK1 activity | (Ref. 43) |
| ↓OPTN and SQSTM1 interaction with Ub or LC3 | (Refs 44–46) | |
| ↓SQSTM1 promoter activity | (Ref. 47) | |
| ↑DRP1 protein levels | (Ref. 204) | |
| ↓Miro protein levels | (Ref. 205) |
↑ – increased compared to control, ↓ – decreased compared to control.
Neurons are terminally differentiated, highly polarised cells with distinct subcellular compartments; small cell body, long axonal processes, multiple dendritic branches and numerous synapses. While they are overall energy-demanding cells, synapses are considered to be the major sites of energy consumption, and the positioning of mitochondria near them is required for the maintenance of neurotransmission (Refs 6, 7). At the same time, tight regulation of local Ca2+ fluxes is essential for neuronal function and the maintenance of axon potential. To deal with different mitochondrial demands between compartments, neurons rely heavily on the binding of mitochondria to cytoskeletal components that can mediate both local movements and bidirectional transport to or from distal processes. Long-distance transport is mainly performed along precisely oriented microtubule tracks, and mediated by motor proteins that bind mitochondria either directly or indirectly via the action of adaptor molecules (Ref. 8). In axonal processes, microtubules are strictly organised with their plus-ends oriented away and their minus-ends towards the soma (Ref. 9). Therefore, binding of mitochondria to kinesin motor proteins drive anterograde transport, while binding to dynein motor proteins promote retrograde movement. Conversely, movement along microtubules is inhibited by the action of protein-tethers that anchor mitochondria on cytoskeletal components and immobilise them (Ref. 8). Among those, syntaphilin (SNPH) mediates the immobilisation of mitochondria to axonal processes, and its release has been suggested to remobilise stressed mitochondria and retrieve them back to the soma (Refs 10, 11). Although in dendrites microtubule polarity is mixed to a degree that depends on neuronal type, species and the position within the dendrite, mitochondrial transport can be performed using the same molecular mechanisms (Ref. 9). On the other hand, short range distribution and docking of mitochondria seems to be primarily mediated by myosin/actin-based mechanisms (Refs 12–14).
The ability to transport healthy and damaged mitochondria among their compartments liberates neuronal cells from the necessity to locally deploy all available quality control mechanisms in order to sustain mitochondrial homoeostasis. Instead, the bulk of mitochondria quality control can be performed in the soma and proximal regions, where transcripts of relevant nuclear genes are readily available, while organelle transport can mediate their gathering and redistribution according to subcellular demands. However, accumulating evidence suggests that similar events can also occur distally. Mitochondrial biogenesis for instance has been assumed to be restricted in the soma, but recent data argue in favour of local mitochondrial replication, away from the cell body (Ref. 15). Similar implications have been stipulated in the case of mitophagy, but so far data have been contradictory. This review aims to summarise the current knowledge concerning the compartmentalisation of mitophagy in neurons and discuss the associated mechanisms, along with their possible implications in human pathologies.
Mitophagy mechanisms
The termed mitophagy was first introduced in 2005 to describe the selective removal of mitochondria by the autophagic machinery, which includes three basic steps: the recognition of damaged organelles, the generation of a nascent double-layered autophagic membrane around them and the delivery of the resulting autophagosomes to lysosomes for degradation. (Ref. 16). Since then, a lot of effort has been put in the investigation of the molecular pathways that mediate mitochondrial elimination and govern its regulation, with special interest in the signals that classify an organelle as damaged. Accumulating evidence underlines the existence of multiple mitophagy regulators that respond to various intracellular and/or environmental stimuli and often communicate with each other to exert a fine-tuned mitochondrial quality control. Depending on the involvement of ubiquitin molecules, these pathways can be classified in two large categories: ubiquitin-dependent and ubiquitin-independent mechanisms.
Ubiquitin-dependent mechanisms
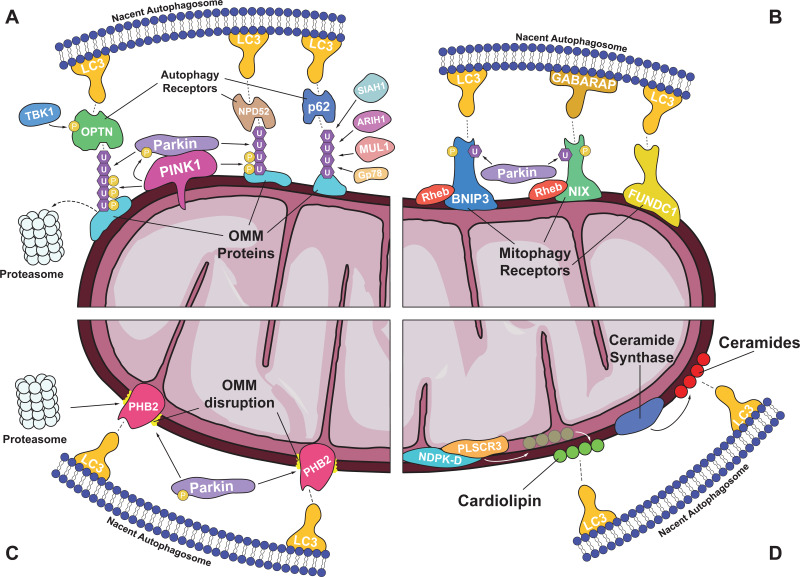
The pathway that involves the action of the phosphatase and tensin homologue (PTEN)-induced putative kinase protein 1 (PINK1) and the E3 ubiquitin ligase Parkin, constitutes the most extensively studied ubiquitin-dependent mitophagy signalling cascade (Fig. 1a). Both proteins were originally identified as mitochondrial quality control regulators by early studies in Drosophila melanogaster, which reported their function in a common pathway, and their mutated forms have been associated with familial cases of PD and other neurodegenerative diseases (Refs 17–19). Under non-stressed conditions, the serine/threonine kinase PINK1 is imported into the inner mitochondrial membrane (IMM) of healthy organelles, where it is proteolytically cleaved, predominantly by PARL protease (Refs 20, 21). Its truncated form is then released to the cytoplasm, where it is rapidly ubiquitinated and degraded by the proteasome, ultimately resulting in low basal protein levels (Refs 20, 21). However, the dissipation of membrane potential in damaged organelles impairs mitochondrial import, leading to the accumulation of PINK1 on the outer mitochondrial membrane (OMM) (Refs 20, 21). In turn, PINK1 phosphorylates cytoplasmic Parkin, stimulating its translocation to the mitochondrial surface and activating its E3 ubiquitin ligase activity (Refs 21–23). At the same time, PINK1 phosphorylates pre-existing or Parkin-attached ubiquitin molecules on various OMM proteins. Binding of Parkin to such phospho-ubiquitin moieties impairs its autoinhibition and further facilitates its direct activation by PINK1 phosphorylation, establishing a feed-forward mechanism for the generation of ubiquitin chains that amplifies the initial signal and ensures mitophagy execution (Refs 21, 24–27). Once activated, Parkin mediates the poly-ubiquitination of various OMM proteins, including the fusion-promoting mitofusins (MFNs) and the kinesin adaptor protein Miro, triggering their proteasomal degradation (Refs 28, 29). The removal of such proteins from the OMM is believed to reduce mitochondrial motility, release mitochondria from contact sites with the endoplasmic reticulum (ER) and promote organelle fission, thereby creating an environment that favours mitophagy (Refs 28, 30, 31). Moreover, PINK1 stimulates the activity of the fission-promoting dynein related protein 1 (DRP1) to induce mitochondrial network fragmentation and the subsequent isolation of defective organelles leading eventually to their engulfment by autophagosomes (Ref. 32). However, the diversity of Parkin substrates suggests that the identity of ubiquitinated OMM proteins is of secondary importance compared to the density of ubiquitin moieties for the induction of mitophagy (Ref. 33).
Fig. 1.
Overview of mitophagy mechanisms. (A) Ubiquitin-mediated mitophagy. Dissipation of mitochondrial membrane potential leads to the accumulation of PINK1 kinase on the surface of damaged organelles, where it phosphorylates and activates the E3 ubiquitin ligase Parkin. Ubiquitin moieties attached to OMM proteins, either by Parkin or by other ubiquitin ligases (e.g., MUL1, SIAH1, ARIH1 and Gp78), are stabilised by PINK1-mediated phosphorylation, and are ultimately recognised by autophagy receptors (e.g., OPTN, NPD52 and p62) that drive autophagosome formation through their interaction with LC3 protein. The action of such receptors is enhanced by their TBK1-mediated phosphorylation. Alternatively, ubiquitinated OMM proteins can be degraded by the proteasome, altering mitochondrial dynamics and motility to favor mitophagy. (B) OMM receptor-mediated mitophagy. OMM proteins can act as mitophagy receptors (e.g., BNIP3, NIX and FUNDC1) and induce autophagosome formation through their direct interaction with LC3/GABARAP. The activation of most mitophagy receptors involves their phosphorylation and is facilitated by the translocation of Rheb small GTPase to mitochondria. Further induction of their activity can be achieved by their Parkin-mediated ubiquitination, establishing a cross-talk mechanism between different pathways. (C) IMM receptor-mediated mitophagy. IMM proteins (e.g., PHB2) can act as mitophagy receptors by interacting with LC3 upon the disruption of OMM, due to Parkin-mediated ubiquitination and proteasomal degradation of OMM proteins. (D) Lipid-mediated mitophagy. The presence of certain lipids (e.g., cardiolipin and ceramides) on the OMM can induce mitophagy by driving autophagosome formation through their interaction with LC3. Cardiolipin is externalised from the IMM by the action of PLSCR3 and NDPK-D, while ceramides and their analogues are freshly generated on the OMM by ceramide synthases.
Beyond the action of Parkin, additional E3 ubiquitin ligases have also been reported to mediate ubiquitination of OMM proteins under mitophagy-inducing conditions, such as Gp78, SIAH1, MUL1 and ARIH1 (Refs 34–37). Regardless of the ubiquitination mechanism, polyubiquitinylated proteins on the OMM have been shown to act as an ‘eat me’ signal recognised by several autophagy adaptors, including optineurin (OPTN), nuclear dot protein 52 (NDP52), sequestosome 1 (SQSTM1/p62), TAX1 binding protein 1 (TAX1BP1) and neighbour of BRCA1 gene 1 (NBR1) (Ref. 38) (Fig. 1a). These adaptor proteins interact directly with ubiquitin chains, through their ubiquitin-binding domain, and with microtubule-associated protein 1A/1B-light chain 3 (LC3), through their LC3-interacting region (LIR), driving autophagosome formation around ubiquitin-tagged mitochondria (Refs 38, 39). Phosphorylation by TANK binding kinase 1 (TBK1) can further modulate the activity of such autophagy receptors, enhancing their affinity for both ubiquitin chains and LC3 (Refs 38, 40, 41). Interestingly, the binding of OPTN to ubiquitin stimulates its phosphorylation by TBK1, suggesting the existence of additional feed-forward mechanisms that enhance the recruitment of the autophagosomal machinery to defective organelles (Ref. 41). Such adaptor proteins seem to have a prominent role in HD, since the polyQ tract in mutant huntingtin (HTT) has been reported to impair their interaction with LC3. As a result, cargo recognition downstream of PINK1/Parkin is impaired, leading to the accumulation of damaged organelles and increased oxidative stress, which contribute to disease pathogenesis (Ref. 42) (Table 1). Likewise, mutations that reduce the function or the expression of TBK1, OPTN or SQSTM1 have been reported to play a causative role in mitophagy disfunctions that contribute to the pathogenesis of ALS (Refs 43–47) (Table 1).
Furthermore, both PINK1 and Parkin have been implicated in some manifestations of an alternative mitochondrial quality control mechanism that involves the generation of Mitochondria Derived Vesicles (MDVs), small vesicular carriers that bud from mildly damaged mitochondria. MDVs are considered to participate in the detoxification system, as they have been shown to transport large assemblies of aggregated or oxidised mitochondrial proteins and lipids to peroxisomes, lysosomes or multivesicular bodies for degradation or export (Refs 48, 49). This mechanism for removing damaged mitochondrial content functions independently of autophagy, does not always necessitate mitochondrial fission and results in the targeted removal of damaged mitochondrial parts, instead of entire organelles. Although the exact mechanism of MDV generation is not clear, targeted recruitment of PINK1/Parkin to specific parts of the organelle is thought to represent the first response, aiming to mitigate mitochondrial dysfunction by ejecting damaged molecules, while sustained insults and depolarisation lead to the outspread of PINK1 and Parkin localisation on the entire mitochondrial surface, activating eventually the process of mitophagy (Refs 48, 49). Despite the undisputable ability of the PINK/Parkin pathway to induce mitochondrial clearance, it is important to highlight that most studies concerning the role of both proteins in mitophagy have been performed in experimental systems that overexpress the corresponding genes. Such artificial conditions could create an unrealistic environment that facilitates mitophagy, resulting in findings that do not relate to physiological conditions. In line with this scepticism, PINK1 or Parkin deficient mice do not exhibit substantial phenotypes related to PD, while both proteins appear dispensable for basal mitophagy in vivo (Refs 50–53). Nonetheless, their role seems to be important in stress- and age-induced mitophagy (Refs 54–56). Additional work is necessary to delineate the contribution of the this pathway and of other ubiquitin mediated mechanisms in the regulation of mitophagy under various physiological and pathological conditions.
Ubiquitin independent mechanisms
Besides ubiquitin-dependent mitophagy, recent studies have identified several regulatory mechanisms that drive autophagosome formation around mitochondria without involving ubiquitination. These mechanisms depend on the presence of mitochondrial receptors that are constitutively localised on the OMM and interact directly with LC3 or GABA receptor associated protein (GABARAP) through their LIR motif, to initiate autophagosomal formation and subsequent mitochondrial elimination (Ref. 57) (Fig. 1b). In contrast to the PINK1/Parkin pathway that appears to orchestrate mitophagy after acute organelle damage, mitophagy receptors seem to be mainly involved in the reorganisation of the mitochondrial network in response to metabolic rewiring, due to environmental stress or during cellular reprograming and differentiation. BCL2 interacting protein 3 (BNIP3) and NIP3-like protein X (NIX/BNIP3L) are two mitophagy receptors that were initially identified as cell death-promoting proteins, but later studies revealed their central role in the selective degradation of mitochondria (Ref. 58). The expression levels of both proteins are induced by hypoxia-inducible factor 1α (HIF1α) and have been found to mediate hypoxia-induced mitophagy (Ref. 59). Additionally, NIX plays a critical role in the autophagic clearance of mitochondria during erythrocyte maturation or somatic cell reprogramming, in the remodelling of the mitochondrial network during differentiation in cardiac progenitor cells or oligodendrocytes of the optic nerve, and in the cytoprotective induction of mitophagy in neurons during recovery from cerebral ischaemia (Refs 60–64). Despite being mainly associated with basal or programmed mitophagic events, NIX has also been implicated in the clearance of depolarised mitochondria, by promoting GABARAP recruitment and binding on its LIR motif (Ref. 65). The mechanism of NIX activation is not well understood, but it has been proposed to involve phosphorylation of serine residues near its LIR motif, which enhances affinity to LC3, as well as the translocation of Rheb small GTPase to mitochondria that promotes the interaction of NIX with LC3, to induce autophagosomal formation (Refs 66–68). On the other hand, C-terminus phosphorylation inhibits NIX dimerisation and severely reduces its ability to drive mitophagy (Ref. 69). Similar to NIX, BNIP3 interacts with Rheb GTPase, which, in turn, phosphorylates its LIR motif and promotes its homodimerisation and the induction of mitophagy (Refs 70–72). Nonetheless, the function of these mitophagy receptors is not redundant, since NIX overexpression is not sufficient to compensate for BNIP3 depletion and activate excessive mitophagy during ischaemic stroke (Ref. 73).
Other examples of mitophagy receptors include FUN14 domain-containing 1 (FUNDC1), BCL2 like 13 (BCL2L13) and FKBP propyl isomerase 8 (FKBP8) (Ref. 74). Similar to NIX and BNIP3, FUNDC1 is also involved in hypoxia-induced mitophagy, although its activation is not mediated by HIF1-dependent transcriptional induction, but rather on post-transcriptional modifications (Refs 75, 76). Beyond hypoxia, additional physiological or stress induced responses have been reported to involve FUNDC1-mediated mitophagy, such as the remodelling of the mitochondrial network during cardiac progenitor cell differentiation, the clearance of paternal mitochondria in the nematode Caenorhabditis elegans and the removal of depolarised organelles following exposure to uncoupling agents (Refs 62, 77, 78). BCL2L13 is the functional homologue of yeast Atg32, an OMM protein that is essential for mitochondrial turnover during respiratory growth. Atg32 interacts with the LC3 homologue Atg8 to initiate autophagosome formation, and its activity is stimulated by phosphorylation (Ref. 79). Likewise, FKBP8 was identified as a mitophagy receptor due to its affinity for Atg8 in yeast two hybrid assays, while later work revealed its implication in hypoxia- or iron depletion-induced mitophagy in mammalian cells (Refs 80, 81). Interestingly, NIX, BNIP3 and FUNDC1 facilitate the translocation of Parkin to the mitochondrial surface, while NIX ubiquitination by Parkin promotes the generation of autophagosomes around damaged organelles, suggesting a crosstalk between different mitophagy pathways (Refs 82–85). Moreover, BNIP3 and FUNDC1 can also interfere with mitochondrial dynamics to favour fragmentation of mitochondria and, thereby, promote mitophagy, by recruiting the fission-promoting DRP1 and triggering the dissociation of the optic atrophy 1 (OPA1) complex that mediates fusion of the IMM (Refs 84, 86, 87). Similarly, BCL2L13 and FKBP8 are able to promote mitochondrial fragmentation even in the absence of DRP1, although the function of the latter depends on OPA1 (Refs 79, 81).
Although mitophagy receptors are located on the OMM, similar functions can be performed by molecules that normally reside on the IMM, upon their exposure to the mitochondrial surface. Prohibitin 2 (PHB2) is an IMM protein that induces mitophagy in a Parkin-dependent manner, as its interaction with LC3 is possible only after OMM disruption, which is achieved by Parkin-mediated ubiquitination and subsequent proteasomal degradation of OMM proteins (Refs 88, 89) (Fig. 1c). PHB2 is essential for the clearance of paternal mitochondria in C. elegans, while its interaction with p62 and LC3 is required for bile-acid induced mitophagy in cholestatic liver cells (Refs 88, 90). Moreover, PHB2 has been shown to stabilise PINK1 and promote the recruitment of Parkin and OPTN to mitochondria, by negatively regulating the stability and the activity of PARL protease upon membrane potential dissipation or misfolded protein aggregation (Ref. 91). Likewise, Cardiolipin (CL), a mitochondrion-specific phospholipid, is mainly distributed along the IMM, but is externalised to the OMM of damaged organelles and serves as a mitophagy receptor (Refs 92, 93). In contrast to PHB2 though, externalisation of CL is not the result of OMM disruption, but depends on the action of the mitochondrial phospholipid scramblase-3 (PLSCR3) and the hexameric intermembrane space protein complex of mitochondrial nucleoside diphosphate kinase D (NDPK-D) (Ref. 93) (Fig. 1d). Once on the mitochondrial surface, CL induces autophagosome formation by directly interacting with LC3 (Refs 93, 94). Moreover, NDPK-D activity is inhibited by its association with OPA1, while DRP1 directly binds to CL to promote organelle fission, establishing a direct link between CL translocation and mitochondrial dynamics (Refs 94, 95). Accumulating evidence suggests that additional lipids, such as ceramides and their analogues, can function as mitophagy receptors and promote mitochondrial clearance (Refs 96, 97). Indeed, DRP1 activation results in the p17/PERMIT-mediated translocation of newly translated ceramide synthases from the ER to the OMM, through mitochondrial associated membranes, and the subsequent generation of ceramide that induces LC3 recruitment and autophagosome formation (Ref. 98) (Fig. 1d). While ceramides have been mainly associated with cell death in cancer cells, they have also been implicated in the induction of mitophagy in cardiomyocytes during lipotoxicity (Refs 96, 97). Taken together these observations have sketched a complex network of factors and pathways that regulate and mediate the clearance of mitochondria. Significant effort is still necessary in order to unravel the interplay between the numerous receptors and their interconnections with different mitophagy pathways that coordinate organelle degradation under different physiological or pathological conditions.
Negative regulators of mitophagy
Although recent years have seen the emergence of impaired mitophagy as a common denominator of various human pathologies, it is becoming increasingly evident that excessive organelle clearance can also have detrimental effects on cellular and organismal homoeostasis (Refs 74, 99, 100). This realisation has spurred novel interest for the discovery of proteins that negatively regulate the process. Targeted inhibition of these proteins can serve as a therapeutic approach that enhances mitophagy, while their induction has therapeutic potential in pathological conditions related to unrestrained organelle turnover. Nevertheless, manipulation of such regulators should be approached with great caution, taking into account possible effects stemming from their involvement in other forms of selective or general autophagy, as well as in additional cellular processes.
Concerning ubiquitin-mediated mechanisms, the action of Parkin and other ubiquitin ligases in promoting mitochondrial clearance is antagonised by deubiquitinating enzymes (DUBs), such as USP30, USP35 and USP15, that mediate ubiquitin chain disassembly and reduce the overall ubiquitin content on the mitochondrial surface (Refs 101–103). Interestingly, depletion of UPS30 has been reported to enhance basal mitophagy in a PINK1-dependent but Parkin-independent manner, an observation that implicates its action in the regulation of basal ubiquitin levels, upstream of any mitophagy-inducing phospho-ubiquitination events (Ref. 104). Therefore, UPS30 (and possibly other DUBs) can determine the threshold for mitophagy initiation by reducing the amount of PINK1 substrates on the OMM. Conversely, UPS30 has been identified as a target of Parkin-mediated ubiquitination that leads to its proteasomal degradation (Refs 101, 103), suggesting that the activation of the PINK1/Parkin pathway suppresses counteracting de-ubiquitination mechanisms, to ensure mitophagy robustness and prevent energy waste by repetitive rounds of ubiquitination and de-ubiquitination. It is therefore possible that the local balance between the activity of ubiquitination and de-ubiquitination machineries ultimately determines which organelles will be deemed damaged and proceed to degradation. Not all deubiquitinating enzymes though have a negative impact on mitophagy. USP8 for instance, has been reported to have an opposing function and induce mitophagy by directly de-ubiquitinating Parkin and, thus, promoting its translocation to impaired mitochondria (Ref. 105).
A similar mitophagy-inhibiting role has been described for PTEN-L, the long isoform of phosphatase and tensin homologue (PTEN), which has been shown to counteract PINK1 and dephosphorylate ubiquitin molecules on the OMM, thus inhibiting the activation of Parkin and its translocation to mitochondria (Ref. 106). Since phosphorylated ubiquitin molecules and polyubiquitin chains on the OMM are resistant to hydrolytic removal (Ref. 107), their dephosphorylation by PTEN-L is also expected to promote the action of DUBs and favour the reduction of the overall ubiquitin content on the mitochondrial surface, ultimately serving as a general blockade of the feedforward phospho-ubiquitination loop that activates mitophagy. Moreover, there are several studies that frame the short isoform of PTEN (or canonical PTEN) as a negative regulator of mitophagy, albeit in a more indirect manner, though the modulation of PI3K-AKT or TLR4-JNK signalling, RAB7A activity and AMPK-CREB transcriptional control (Refs 108, 109). Nonetheless, a lot of work is still necessary in order to fully elucidate the role of PTEN in the regulation mitophagy.
Beyond the regulation of mitochondrial ubiquitin content, other proteins have been found to exert a negative effect on mitophagy through different mechanisms. Among them, Leucine-Rich Repeat Kinase 2 (LRRK2) is of particular interest, since LRRK2 mutations are the most common monogenic cause of PD (Ref. 110). Data from FRET imaging have suggested that LRKK2 kinase activity inhibits mitophagy by interfering with protein-protein interactions that involve PINK1 and DRP1 on mitochondria (Ref. 111). Moreover, LRKK2 has been reported to phosphorylate RAB10, a member of the Rab small GTPase family, which contains key regulators of vesicular trafficking (Ref. 112). In turn, unphosphorylated RAB10 was found to bind OPTN and induce its translocation to depolarised mitochondria, facilitating their clearance, a response that was not recapitulated by a RAB10 phosphomimetic variant (Ref. 113). Interestingly, the two most common LRKK2 mutations in PD patients (G2019S and R1441C) enhance RAB10 phosphorylation and exhibit impaired mitophagy, while genetic knockout or pharmacological inhibition of LRKK2 appears sufficient to rescue mitophagy defects in both primary skin fibroblasts derived from PD patients with relevant mutations, and tissues with clinical relevance of a mouse GS2019S model, without affecting general autophagy (Refs 113–115). Additionally, the same mutation has been reported to impair the removal of Miro1 from damaged mitochondria and delay organelle clearance, indicating a more complex role for LRKK2 in the regulation of mitophagy (Ref. 116).
A more indirect role has been described for poly-ADP-ribose polymerase 1 (PARP1), a DNA repair enzyme that consumes NAD+ as a substrate for ADP-ribosylation. PARP1 activation depletes cellular NAD+ and attenuates the activity of the NAD+-dependent deacetylase SIRT1. This impairment of SIRT1 has been shown to reduce the activity of peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α/PPARGC1A), and thus decrease the expression of the uncoupling protein UPC2. As a result, mitochondrial membrane potential is increased, leading to PINK1 cleavage and the inhibition of mitophagy (Refs 117, 118). An additional negative regulatory mechanism has been described in yeast, where the PP2A-like protein phosphatase Pgp1 counteracts the phosphorylation of Atg32 mitophagy receptor by casein kinase 2 (CK2) to inhibit mitophagy. Accordingly, deletion of Pgp1 results in the constitutive interaction of Atg32 with Atg11, a cytosolic adaptor protein for selective autophagy, and accelerates mitophagy (Ref. 119). Given the functional and structural conservation of the PP2A family from yeast to mammals, it is possible that a similar mechanism restrains the phosphorylation of BCL2L13 or other mitophagy receptors, to negatively regulate mitophagy in mammalian cells. Supporting this notion, mammalian PP2A has been shown to regulate autophagy by dephosphorylating the autophagy activating kinase ULK1 upon amino acid starvation (Ref. 120). However, evidence for its involvement in selective mitophagy is still lacking.
Mitophagy compartmentalisation in neuronal cells
Although mitochondrial turnover is taking place in nearly all eukaryotic cells, underscoring its pivotal role for cellular and tissue homoeostasis, the post-mitotic nature of mature neurons provides an extra challenge. Dividing cells can reshuffle their mitochondrial content by inducing organelle biogenesis, coupled with cell division to ‘dilute’ dysfunctional organelles with healthy ones in daughter cells. Besides, most tissues have the ability to face a decline in mitochondrial quality surveillance system by inducing the death of problematic cells, and replenish their cell content by stimulating mitosis of healthy ones. These options however do not exist in neurons, which have to maintain the functionality of their mitochondrial network throughout the lifespan of the organism, which can reach several decades. The aforementioned mitophagy pathways have been mostly studied in non-neuronal cells under stressful conditions, leaving many answered questions concerning their implication in basal mitochondrial turnover of neurons, the answers to which are only just starting to emerge. Moreover, the complex architecture of neuronal cells poses additional challenges in mitochondrial clearance that small and simply organised cells do not have to face, since a number of mitochondria is located at distal neuronal compartments, away from the main cell body. Can such distal organelles be locally degraded or do they have travel back to the soma where mature lysosomes are relatively more abundant? Are all neuronal mitochondria subjected to the same quality control mechanisms regardless of their topology, or do different mitophagy pathways prevail in each compartment? The next part of this review summarises our current knowledge concerning these issues.
Mitophagy in somatodendritic regions
The cell body, along with proximal dendritic regions, are considered to be the main sites of mitophagy in neuronal cells, as there resides the vast majority of mature lysosomes (Refs 121, 122). Indeed, the inhibition of lysosomal activity has been reported to result in the accumulation of organelles in neuronal somata (Ref. 123). Still, it is not clear which is the prevalent pathway that mediates this process, as relevant results have been contradictive. In vitro studies in neuronal cell lines and primary neurons have reported that Parkin selectively translocates to depolarised mitochondria in the soma and proximal dendritic regions to induce autophagosomal organelle clearance after exposure to various damaging agents, suggesting that the function of the pathway is conserved across cell types (Refs 123–126). However, in vivo data do not support a major role for PINK1 and Parkin in mitochondrial turnover in neuronal cell bodies under physiological conditions. Studies in both mice and Drosophila model systems have shown that although basal mitophagy appears to be a widespread phenomenon in the nervous system, it is minimally affected by PINK1 deletion (Refs 52, 53, 127–129). Deficiency of PINK1 or Parkin though has been shown to impair the age-dependent rise in mitophagy events that is observed in D. melanogaster dopaminergic neurons, and lead to the accumulation of abnormal mitochondria (Refs 56, 130). This defect could account for the occurrence of PD-like phenotypes in PINK1 and Parkin Drosophila mutants, that are not observed in the corresponding murine models, as it is possible that mice deploy additional quality control mechanisms to sustain mitochondrial quality during ageing, which are not activated or even present in flies (Refs 50, 51, 131–133). The apparent discrepancy between in vivo and in vitro data has been at least partially attributed to the physiologically irrelevant conditions of in vitro experimental systems, which include the overexpression of Parkin and the administration of depolarising agents, often coupled with additional insults, to induce mitophagy (Refs 123–126). Interestingly, even under extreme in vitro conditions, artificial overexpression of Parkin seems to be a prerequisite for its recruitment to neuronal mitochondria, while the percentage of treated neurons that necessitates Parkin for the removal or the repair of depolarised organelles is rather dispensable (Refs 123, 126, 134). Moreover, in the apparently rare cases of its induction, the kinetics of Parkin-induced mitophagy in neurons are much slower than what has been reported for non-neuronal cells (Refs 123, 126, 134).
A molecular mechanism that could account for this delayed response has recently been described in mouse cortical neurons treated with low dosage of antimycin A, to induce mild depolarisation. Under these conditions, the mitochondrial E3 ubiquitin ligase 1 (MUL1) was reported to ubiquitinate and destabilise MFN2. In turn, MFN2 destabilisation promotes ER-mitochondrial (ER-Mito) tethering, which impairs the translocation of Parkin to slightly damaged organelles and protects them from degradation (Ref. 135). Upregulation of MFN2, due to MUL1 depletion, was shown to result in the release of mitochondria from ER-Mito contact sites and trigger an upsurge in cytoplasmic Ca2+ levels that activates calcineurin and stimulates DRP1-mediated mitochondrial fragmentation and clearance (Ref. 135). MUL1 is therefore proposed to act as an early checkpoint that prevents the degradation of neuronal mitochondria under mild stress. On the other hand, results from D. melanogaster dopaminergic neurons, mouse cortical neurons and human induced pluripotent stem cells (iPSCs)-derived dopaminergic neurons suggest that under acute exposure to challenging conditions, ubiquitination of MNF2 by either MUL1 or Parkin, and its subsequent degradation, releases mitochondria from the ER and induces mitophagy (Refs 31, 136). These observations are in line with the dual role of MFN2 in the tethering of mitochondria to ER, as it has been reported to both promote and impair their juxtaposition (Refs 137–140). It is possible that a mild decrease of MFN2 levels promotes ER-Mito interactions and inhibits mitophagy, but its depletion below a critical threshold has the opposite result. Although additional interactions could influence the outcome in a cell-type specific manner, evidence for their existence remains obscure. Interestingly, the ubiquitination levels of the kinesin adaptor protein Miro1 were recently correlated with the rate of Parkin translocation to damaged mitochondria in cultured mouse hippocampal neurons, while conditional Miro1 knockout was shown to impair MFN2 ubiquitination and its subsequent degradation in the same cells in vivo (Ref. 141). Whether differences in the ubiquitination status of Miro1 between neuronal and non-neuronal cells are involved in the delayed response of neurons to mitochondrial damage remains to be further investigated.
However, the slow kinetics of mitochondrial elimination in neuronal cells extend beyond the rate of Parkin translocation. Recent work in primary hippocampal neurons during mild stress revealed that Parkin-dependent recruitment of OPTN and autophagophore formation around damaged mitochondria occurs at a rate comparable to non-neuronal cells. However, the acidification of the resulting mitophagosomes and the turnover of engulfed organelles is a considerably slow process compared to other cell types (Ref. 142). While the reason behind this delay in acidification remains elusive, the sequestration of damaged organelles is expected to protect neurons by preventing the dissemination of damaged components and the generation of harmful excessive reactive oxygen species (ROS). Further supporting this notion, defective engulfment of damaged mitochondria, due to an ALS-linked dominant OPTN mutation, results in an overall decrease of mitochondrial homoeostasis (Ref. 142). Conversely, the delayed induction of mitophagy could be of vital importance to the survival of neurons, due to their high dependence on OXPHOS, compared to the more glycolytic metabolism of other cell types. Thus, delaying mitophagy would prevent ATP depletion and allow alternative quality control mechanisms to act and repair damaged organelles. Moreover, low ATP levels caused by mitochondrial depolarisation have been proposed to directly impede the energy consuming process of PINK1/Parkin-mediated mitophagy in neurons, by limiting both de novo synthesis of PINK1 and the phosphorylation of its targets (Ref. 134).
Collectively these observations indicate that neurons reserve PINK1/Parkin-mediated mitophagy as the last resort for the clearance of terminally damaged organelles, while the maintenance of the mitochondrial network under basal conditions or mild stress is mediated by alternative quality surveillance pathways (Fig. 2A, B). These might include the action of the parallel MUL1-MFN2 pathway, as MUL1 overexpression suppresses PINK1 and Parkin mutant phenotypes in Drosophila dopaminergic neurons, while its depletion aggravates these phenotypes and leads to synthetic lethality in flies, or to the degeneration of mouse cortical neurons (Ref. 136). Moreover, accumulating evidence also suggests key roles for receptor-mediated mitophagy. CL-mediated removal of damaged organelles has been reported in rat primary cortical neurons after exposure to damaging agents, while CL metabolism in the brain is emerging as a critical factor in the pathogenesis of many neurodegenerative conditions (Refs 93, 143). The function of both NIX and FUNDC1 appears to be involved in mitophagy induction that protects neurons during recovery from cerebral ischaemia (Refs 64, 144), while the upregulation of BNIP3 leads to the toxic overactivation of mitophagy in response to ischaemic stroke, further supporting that proper regulation of different mitophagy mechanisms is vital for neuronal cells viability (Ref. 73). Additionally, NIX has been proposed to mediate the compensatory mechanism that preserves mitochondrial function and clearance in cells of asymptomatic Parkin mutation carriers, while genetic or pharmacological induction of its expression has been shown to restore mitophagy in cells derived from PINK1- or Parkin-related PD patients (Ref. 145). Recently, BNIP3 overexpression was shown to induce mitophagy and prevent the accumulation of dysfunctional organelles in the brain of aged flies (Ref. 146). Intriguingly, neuron-specific upregulation of BNIP3 was able to delay the onset of age-dependent phenotypes in muscle and intestinal cells, suggesting that mitochondrial function in the brain can regulate the rate of organismal ageing through a cell non-autonomous mechanism (Ref. 146). However, the mechanistic insights that mediate such systemic responses are still elusive.
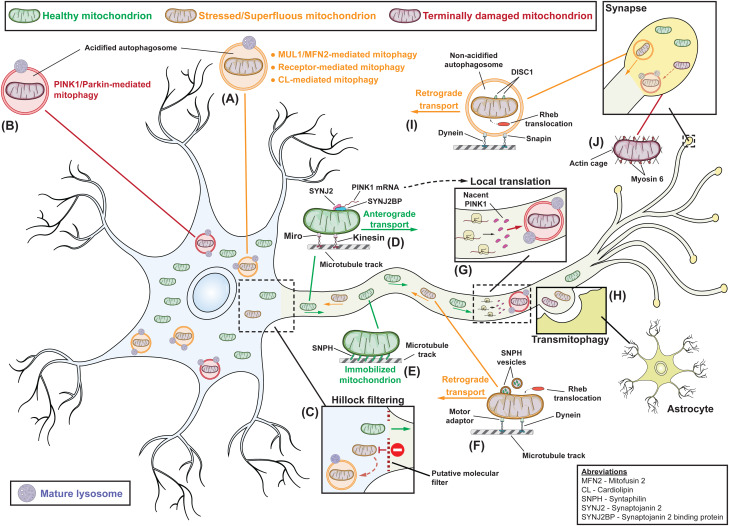
Fig. 2.
Putative integrated model for the compartmentalisation of mitophagy in neurons. The turnover of stressed or superfluous mitochondria in the main cell body of neurons is predominantly mediated by PINK1-independent mechanisms, which involve the action of mitophagy receptors, cardiolipin and the ubiquitination of MFN2 by MUL1 (A), while terminally damaged organelles are cleared through the PINK1/Parkin pathway (B). Quality control mechanisms at the axonal hillock (C) filter organelles and only allow healthy mitochondria to travel anterograde along microtubule tracks (D), whereas damaged ones are targeted for degradation. Along the axon, the anchoring protein syntaphilin (SNPH) antagonises motor proteins to promote the immobilisation of functional organelles at areas of high mitochondrial demand (E). Rheb small GTPase translocates on stressed axonal mitochondria, which release the anchoring protein SNPH and are remobilised in order to travel retrograde and be degraded in the soma (F). Terminally damaged axonal organelles are degraded locally through the PINK1/Parkin pathway, which is supported by local PINK1 mRNA translation (G). Damaged mitochondria that fail to travel retrograde or be locally degraded can be externalised and cleared by neighbouring astrocytes though the process of transmitophagy (H). At synapses, Rheb translocation induces DISC1-mediated formation of autophagosomes around stressed mitochondria, that remain non-acidified and interact with the dynein adaptor protein Snapin to travel back to the soma (I). Terminally damaged synaptic mitochondria acquire myosin 6, which promotes the assembly of stable actin cages and immobilises organelles to enable their local Parkin-mediated turnover (J). Somatodendritic regions are marked with light blue, axonal regions with light green and synapses with yellow background.
Mitophagy in axonal processes
The complex architecture and the polarised nature of neurons create an extra challenge in the autophagic turnover of mitochondria; while most enzymatically active and degradative lysosomes are located in their soma, a large portion of their mitochondrial content is located at distal compartments (Refs 121, 122). This problem becomes even more pronounced for motoneurons that can project very long axons, often extending beyond one metre. How do distal organelles meet the lysosomal machinery? Analysis in Purkinje cells of mice expressing the mito-QC mitophagy reporter suggests that mitophagic events in axonal projections are extremely scarce, and the wide majority of basal mitochondrial turnover occurs in somatodendritic regions (Ref. 147). Likewise, mild treatment of mouse cortical neurons with depolarising agents has been reported to induce the recruitment of Parkin to mitochondria specifically in cell bodies and proximal regions (Ref. 123), while OPTN translocation to depolarised mitochondria has been shown to transpire mainly in the soma of rat primary hippocampal neurons under mild oxidative stress, with very limited occurrences in distal compartments (Ref. 142). Moreover, in vivo studies in mutant PINK1 or Parkin flies did not detect any structural defects that would indicate the accumulation of dysfunctional organelles in the axons of motoneurons when the pathway is impaired, but on the contrary, they reported a reduction in axonal mitochondrial content (Refs 128, 129). At the same time, loss of either protein was shown to disturb the morphology of somatic mitochondria and induce the accumulation of hyperfused organelles in cell bodies of the same neurons (Refs 128, 129). These results support the notion that Parkin-mediated quality control in the soma is essential for the entry of mitochondria in the axons and underline the existence of a ‘gatekeeper’ filtering mechanism at the hillock that only allows organelles with specific qualitative criteria to enter (Fig. 2c). Evidence for the molecular identity of this filter though is yet to be presented.
As far as organelle turnover is concerned, the aforementioned data have shaped a model where damaged mitochondria are transferred back to the soma, where mature lysosomes are predominately located, in order to be degraded. Early evidence in favour of this model comes from a study using the voltage-sensitive dye JC-1 in chicken dorsal root ganglion (DRG) neurons, where axonal mitochondria with low membrane potential were reported to travel towards the cell body, while the inhibition of the electron transport chain (ETC) with antimycin A was shown to increase retrograde mitochondrial transport (Ref. 148). More recent work uncovered a crucial role for the static anchor protein SNPH, which counteracts the motility-promoting function of motor proteins and immobilises mitochondria on specific positions along microtubule tracks (Ref. 149) (Fig. 2d, e). Low-dosage treatment with antimycin A, was shown to induce the release of SNPH from mildly stressed axonal mitochondria of mouse cortical neurons, in the form of cargo vesicles that are generated independently of both Parkin and DRP1 (Ref. 11). In turn, the release of SNPH was shown to remobilise damaged organelles and facilitate their retrograde transport, ultimately mediating the recovery of mitochondrial membrane in axonal organelles (Ref. 11) (Fig. 2f). Moreover, the small GTPase Rheb has been proposed to sense membrane potential dissipation and induce clearance of axonal mitochondria under both physiological and stressed conditions (Ref. 150). Recent studies in mouse cortical neurons indicate that treatment with antimycin A or the depolarising agent CCCP, triggers Rheb translocation to depolarised axonal mitochondria initiating the formation of mitophagosomes and their dynein-mediated transport to the soma, where they are acidified by fusion with degradative lysosomes (Ref. 150). Notably, Parkin is dispensable for Rheb-associated mitophagy initiation within axons, whereas the function of NIX is required, highlighting its pivotal role of in the turnover of distal mitochondria (Ref. 150). In addition to Rheb, the small GTPase Rhes was shown to promote mitochondrial removal in the striatum of mice in response to 3-nitropropionic acid (3-NP), an inhibitor of the respiratory complex II, suggesting that additional small GTPases might perform similar functions in a cell type-specific manner (Ref. 151). A recent study utilised ischaemic mouse primary cortical neurons combined with the fluorescence-shifting mitochondrial protein mitoDentra2 to investigate the fate of axonal mitochondria. Interestingly, retrograde moving axonal organelles were shown to ultimately fuse with lysosomes in the neuronal cell body in response to oxygen and glucose deprivation underscoring that those motile mitochondria are targeted for autophagic clearance upon stress conditions (Ref. 152).
However, there is a series of evidence that puts this model into question. Autophagosomes have been shown to be generated distally, with their maturation taking place within the confinements of axonal processes in mouse DRG neurons (Ref. 153). At same time, delivery of enzymatically active degradative lysosomes from the soma to distal axons has been observed in mouse cortical neurons, rendering the local degradation of axonal organelles a plausible scenario (Ref. 154). Moreover, the assessment of mitochondrial membrane potential in chicken DRG neurons did not uncover any differences between anterograde and retrograde moving axonal organelles (Ref. 155), while treatment with ETC inhibitors and depolarising agents has often been reported to prevent mitochondrial movement, by inducing the degradation of Miro1 (Refs 29, 152, 156). This immobilisation has been proposed to isolate defective mitochondria and promote their in-situ clearance, thus avoiding the dissemination of their damaged components to healthy mitochondrial population via fusion events, or the spreading of excessive ROS to other cellular compartments. Indeed, selective impairment of axonal mitochondria in rat hippocampal neurons either by local administration of antimycin A or by expressing the light-inducible mitochondria-targeted photosensitizer KillerRed, has been shown to trigger local autophagosome formation, engulfing defective axonal mitochondria in a PINK1- and Parkin-dependent manner (Ref. 157). Interestingly, the same study reported that Parkin is not required for the initial remodelling of the mitochondrial network after depolarisation, and its translocation is restricted to a limited subset of damaged organelles. These observations indicate the existence of additional mechanisms that restore mitochondrial health in axons highlighting Parkin availability as a limiting factor for local mitophagy initiation. The latter is in line with the particular needs of the PINK1/Parkin pathway that is expected to necessitate a continuous supply of newly synthesised PINK1 protein from the cell body. However, the rapid cleavage of the protein in healthy mitochondria would make its transport to distal parts of the axon problematic. A mechanism to overcome this challenge was described recently in human iPSCs-derived neurons, where Pink1 mRNA was found to be tethered on mitochondria via the action of synaptojanin 2 (SYNJ2) and synaptojanin 2 binding protein (SYNJ2BP) (Fig. 2d). Through this tethering, Pink1 transcripts were shown to hitchhike their way to distal processes and mediate the local translation of nascent PINK1, thereby, enabling the PINK1/Parkin-mediated distal mitophagy (Ref. 158) (Fig. 2g). Interestingly, this mechanism for Pink1 mRNA distribution appears to be neuron-specific, due to low levels of SYNJ2 expression in non-neuronal cells.
The integration of these data has shaped a proposed model for the degradation of distal mitochondria, according to which basal turnover occurs via their transport to the soma, where they meet with mature lysosomes (Fig. 2f), but severely damaged organelles are immobilised in axons and eventually trigger the PINK1/Parkin machinery that mediates their local engulfment by autophagosomal membranes and their subsequent degradation (Fig. 2g). Further evidence for this model stems from the observations that overexpression of SNPH results in the enrichment of Parkin-positive immobilised mitochondria in distal axonal processes of mouse cortical neurons and inhibits mitochondrial retrograde transport and degradation in the soma after oxygen deprivation (Refs 123, 152). Therefore, it seems that distal mitochondria that carry damage but fail to travel retrograde to the soma will eventually recruit Parkin. However, the scarcity of PINK1/Parkin events predicted by this model fails to explain the absence of organelle accumulation in distal parts of the axon, despite the anterograde bias in the ratio of anterograde to retrograde transport that has been reported in various systems (Refs 29, 159, 160). Moreover, the use of the time-sensitive MitoTimer fluorescent probe in primary mouse hippocampal neurons has revealed the accrual of old proteins in stationary distal mitochondria (Ref. 161). If aged mitochondria do not travel back to the soma, how do they get replenished by new ones? A possible solution could be provided by the cell non-autonomous mechanism of transmitophagy that has been described in mouse retinal ganglion cells (RGCs) and nigrostriatal dopaminergic neurons (DAn) of chemically induced PD rodent models (Refs 162, 163). According to these studies, pools of damaged mitochondria can be transported to adjacent astrocytes, where they get degraded (Fig. 2h). Coupled with the observation that the mitophagy promoting small GTPase Rhes can travel from cell to cell and interact with NIX in the murine striatum, these data suggest the existence of an intercellular mitochondrial quality control mechanism in the nervous system (Ref. 151). Similar conclusions have been recently drawn from co-cultures of murine primary cortical neurons with astrocytes or human iPSCs-derived neurons with astrocyte-like cells, indicating that transmitophagy is a common strategy adopted by various types of neurons (Ref. 164). This alternative way to remove damaged mitochondria from the axons could be essential for neuronal survival under pathological conditions that involve defects in mitophagy. In agreement with this scenario, astrocyte clearance of neuronal mitochondria is induced in co-cultured cells from AD mouse models, or iPSCs from symptomatic AD patients (Ref. 164), probably acting as compensatory mechanism, while the reduced capacity of astrocytes to execute transmitophagy has been suggested to participate in the pathogenesis of PD (Ref. 163). Further investigation on the field of axonal mitochondria clearance is expected to provide valuable information on the pathogenesis and the treatment of neurodegenerative diseases.
Mitophagy of synaptic organelles
Synaptic dysfunction is a common denominator of many late-onset neurodegenerative disorders and it is believed to have a significant impact on disease progression (Ref. 165). There is a plethora of studies that highlights the contribution of synaptic failure in the manifestations of PD (Ref. 166). At the same time, the toxic effect of Αβ oligomers on synaptic activity is considered as one of the main events that underlie cognitive deficits related to AD (Ref. 167). The high metabolic status of synapses and their role in neuronal physiology renders the presence of healthy mitochondria essential for their function, due to their role in ATP generation and Ca2+ buffering. It is therefore not surprising that dysfunctional mitophagy is starting to emerge as a causative factor of synaptic failure and neuronal cell death (Refs 4, 168). However, the molecular mechanisms that sustain mitochondrial homoeostasis at synapses remain elusive. Evidence from multiple experimental systems suggest that the majority of synaptic mitochondria in mature neurons are stationary and their immobilisation is further enhanced upon synaptic activity (Refs 169–172). This reduced mobility might contribute to the increased wear of synaptic mitochondria, which have been reported to display decreased capacity for energy production and to become more susceptible to Ca2+-induced depolarisation with age, compared to axonal organelles (Ref. 173). Such observations suggest that clearance of synaptic organelles requires their remobilisation in order to travel back to the soma, without however excluding the contribution of local degradation mechanisms.
The importance of mitochondria remobilisation for the maintenance of synaptic activity was recently reported in a mouse AD model that exhibits synapse loss at early disease stages due to the expression of mutant human amyloid precursor protein (APP). Observations from synaptic regions in the brain of these animals revealed a marked increase in the number of Rheb-associated mitochondria that display features of early mitophagic events. However, despite the induction of mitophagy, retrograde movement of organelles in the corresponding axons was impaired leading to the accumulation of non-acidified mitophagosomes at synapses (Ref. 150). Interestingly, deficiency of the dynein adaptor protein Snapin was sufficient to mimic this phenotype, while its overexpression was able to alleviate the accumulation of mitophagosomes and mitigate synaptic loss in the brain of AD mice, highlighting the importance of retrograde transport in both physiological and pathological conditions (Ref. 150). A putative role in the clearance of dysfunctional synaptic organelles has also been suggested for disrupted in schizophrenia 1 (DISC1), a protein that is present in multiple subcellular compartments, but localises predominantly at mitochondria, and is enriched in synaptic terminals (Refs 174, 175). DISC1 has been associated with psychiatric disorders and AD pathology, and was recently identified as a potent inducer of mitophagy in response to treatment with Αβ oligomers or CCCP (Ref. 176). Mechanistically, DISC1 serves as a mitophagy receptor and interacts directly with LC3 through its LIR motif to promote autophagosomal formation (Ref. 176). Moreover, DISC1 overexpression was reported to induce mitophagy and, thereby, prevent both mitochondrial dysfunction and decreased density of dendritic spines upon Αβ oligomers supplementation in mouse primary cortical neurons (Ref. 176). Notably, DISC1-mediated mitophagy acts as a major contributor of energy homoeostasis at synapses, since its activation alleviates Αβ-induced impairment of synaptic plasticity and improves cognitive function in a mouse AD model (Ref. 176). Furthermore, DISC1 has been also reported to interact directly with SNPH, inhibit its anchoring ability and, thereby, promote the mobility of axonal organelles (Ref. 177). Therefore, it is tempting to speculate that both Rheb and DISC1 act synergistically to enhance the remobilisation of aged and/or damaged mitochondria that are anchored in synaptic regions, subsequently leading to their degradation through receptor-mediated mitophagy (Fig. 2i). Despite all indications though, direct evidence for the regulation of synaptic mitophagy by DISC1 is still lacking and additional work is required to support such a model.
An opposing role that could promote the local degradation of mitochondria at synaptic regions might be performed by myosin 6, which has been identified as a mitophagy modulator in non-neuronal cells. Particularly, myosin 6 interacts with Parkin-generated poly-ubiquitin chains and promotes the assembly of stable actin cages around damaged mitochondria mediating their isolation from the healthy mitochondrial network and their subsequent removal (Ref. 178). The implication of myosin 6 in mitophagy is further supported by its interaction with established mitophagy receptors that act downstream of Parkin, such as OPTN, NDP52 and TAX1BP1 (Refs 179, 180). Given the well-established role of the protein in synapse formation and function, it is plausible that its recruitment to dysfunctional organelles orchestrates their local Parkin-mediated turnover to sustain synaptic homoeostasis (Ref. 181) (Fig. 2j). Supporting this hypothesis, myosin 6 knockout mice exhibit alterations in synaptic structure and gliosis in the brain, phenotypes related to neurodegenerative disorders (Ref. 182). Although such local turnover events are yet to be confirmed, these observations suggest that despite the contribution of additional regulators, mitophagy at synapses shares the same mechanistic principles with proximal axonal regions, and while retrograde transport is the predominant pathway for the clearance of organelles, severe damage can induce Parkin-mediated local degradation. However, further investigation is deemed necessary to unravel the mechanistic insights of mitophagy at synapses.
Concluding remarks
A plethora of studies in the past few years has highlighted the essential contribution of mitophagy in the maintenance of a healthy mitochondrial network, a feature of paramount importance for the function and the survival of neuronal cells. Although the basic principles and molecular constituents of mitophagy appear to be mechanistically conserved across cell types, accumulative data has shown that neurons differentiate on the kinetics of the process and have adopted compartment-specific variations, in order to mitigate challenges that arise from their unique structural and functional characteristics. Despite the recent advances in our knowledge though, there are gaps in the implicated molecular interactions and several questions that still remain to be addressed. Is mitophagy organised in the same way in all neurons? Do different subtypes exhibit unique features? Are perturbations in the compartmentalisation of mitophagy related to the pathogenesis of neurodegenerative disorders? Future studies focusing on these subjects will greatly deepen our understanding on neuronal mitophagy and help design targeted therapeutic interventions.
Failure to achieve proper turnover of mitochondria leads to a progressive accumulation of dysfunctional organelles that is considered a hallmark of ageing, as well as a causative factor in many age-related neurodegenerative disorders (Refs 100, 183, 184). Such observations have spurred great interest in the discovery of natural or synthetic compounds that stimulate mitophagy, and indeed pharmacological upregulation of the process has been shown to protect against various neurodegenerative pathologies (Refs 4, 117, 185, 186). Induction of mitophagy by treatment with the microflora-derived metabolite urolithin A (UA) has been reported to protect against Αβ-induced neurotoxicity and extend the lifespan of the nematode C. elegans (Refs 187, 188). Likewise, UA supplementation has been found to inhibit Αβ and tau pathologies in transgenic AD model mice (APP/PS1), whilst reversing their cognitive defects (Ref. 187). At the same time, the increase of nicotinamide adenine dinucleotide (NAD+) levels by direct supplementation or by administration of precursor molecules, such as nicotinamide mononucleotide (NMN) or nicotinamide riboside (NR), has been shown to alleviate pathologies of multiple neurodegenerative disorders, including AD, PD and HD in various model systems (Refs 187, 189, 190). Although data from clinical trials are still insufficient to provide safe conclusions, initial observations suggest that UA, NMN and NR can be safely administered to humans, without the occurrence of adverse side effects, further highlighting their therapeutical potential (Refs 190, 191). Nevertheless, the pathways that are affected downstream of such mitophagy inducing compounds are still largely unexplored. Deciphering their mechanism of action should constitute a major goal in the field, that will maximise their therapeutic potential and improve our perception of mitophagy regulatory mechanisms under both normal and challenged conditions. In this effort, animal models of neurodegenerative diseases and in vivo tools for mitophagy assessment, such as mitoRosella, mitoKeima and mito-QC, will be valuable tools.
Despite promising results though, there is significant evidence that a disproportionate reduction of organelle numbers can have detrimental effects in the physiology of neurons and ultimately trigger cell death (Refs 100, 192, 193). Congruently, BNIP3-mediated mitophagy has been reported to promote neuronal cell death both in vivo and in vitro after ischaemic stroke, while excessive Rhes- and NIX-mediated mitophagy has been shown to cause degeneration of striatum neurons in the brain of 3-NP-treated mice, emulating lesions related to HD (Refs 73, 151, 194). Additionally, exaggerated mitophagy was recently found to mediate mitochondrial depletion from the axons of retinal ganglion cells (RGCs) that express the mutated forms of the IMM fusion promoting protein OPA1 (Ref. 195). In turn, decreased mitochondrial number in the axonal region was identified as the main cause of neurodegeneration that leads to the development of autosomal dominant optical atrophy (ADOA), a genetic disorder characterised by bilateral visual loss in early childhood, and secondary multi-systemic pathologies, which include deafness, ataxia and myopathies (Refs 195, 196). Interestingly, mitochondria were found to co-accumulate with autophagic vesicles close to the axonal hillock of ADOA RGCs. Moreover, inhibition of autophagy or mitophagy was able to restore axonal mitochondrial content in both primary RGCs and C. elegans GABAergic neurons modelling ADOA, and protect RGC-specific Opa1 knockout mice from exhibiting visual loss (Ref. 195). These observations indicate that local mitophagy close to the hillock could be part of the putative filtering machinery that ensures the health of axonal organelles, since the degradation of those that fail quality control seems necessary in order to prevent their anterograde travel. Furthermore, the prevention of visual loss by curtailing autophagy suggests that, despite their defects, Opa1 depleted mitochondria are competent to support neuronal function and survival if they are spared from degradation and allowed to travel, rendering the function of a quality-reassuring mechanism detrimental. In this context, a better understanding of the mechanisms that allow or inhibit anterograde mitochondrial transfer along the axon will provide valuable information with therapeutical potential in neurodegenerative conditions. Moreover, such observations make evident that both the dysfunction and the overstimulation of mitophagy can lead to neuronal pathologies and any intervention that aims to interfere with organelle turnover should take into account the importance of maintaining the proper equilibrium between mitochondrial biogenesis and degradation.
Acknowledgements
We apologise to those colleagues whose work has not been cited in this review due to space limitations.
Financial support
This work is supported by the Fondation Santé (Grant number, 18376).
Conflict of interest
The authors declare no conflicts of interest.
References
- 1.Pathak T and Trebak M (2018) Mitochondrial Ca(2+) signaling. Pharmacology & Therapeutics 192, 112–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paul BT et al. (2017) Mitochondria and iron: current questions. Expert Review of Hematology 10, 65–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shadel GS and Horvath TL (2015) Mitochondrial ROS signaling in organismal homeostasis. Cell 163, 560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lou G et al. (2020) Mitophagy and neuroprotection. Trends in Molecular Medicine 26, 8–20. [DOI] [PubMed] [Google Scholar]
- 5.Yan X et al. (2020) Abnormal mitochondrial quality control in neurodegenerative diseases. Frontiers in Cellular Neuroscience 14, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun T et al. (2013) Motile axonal mitochondria contribute to the variability of presynaptic strength. Cell Reports 4, 413–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris JJ, Jolivet R and Attwell D (2012) Synaptic energy use and supply. Neuron 75, 762–777. [DOI] [PubMed] [Google Scholar]
- 8.Barnhart EL (2016) Mechanics of mitochondrial motility in neurons. Current Opinion in Cell Biology 38, 90–99. [DOI] [PubMed] [Google Scholar]
- 9.Kelliher MT, Saunders HA and Wildonger J (2019) Microtubule control of functional architecture in neurons. Current Opinion in Neurobiology 57, 39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang JS et al. (2008) Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell 132, 137–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin MY et al. (2017) Releasing syntaphilin removes stressed mitochondria from axons independent of mitophagy under pathophysiological conditions. Neuron 94, 595–610 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quintero OA et al. (2009) Human Myo19 is a novel myosin that associates with mitochondria. Current Biology 19, 2008–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pathak D, Sepp KJ and Hollenbeck PJ (2010) Evidence that myosin activity opposes microtubule-based axonal transport of mitochondria. Journal of Neuroscience 30, 8984–8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sung JY et al. (2008) WAVE1 Controls neuronal activity-induced mitochondrial distribution in dendritic spines. Proceedings of the National Academy of Sciences of the United States of America 105, 3112–3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cardanho-Ramos C and Morais VA (2021) Mitochondrial biogenesis in neurons: how and where. International journal of molecular sciences 22, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemasters JJ (2005) Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation research 8, 3–5. [DOI] [PubMed] [Google Scholar]
- 17.Clark IE et al. (2006) Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 441, 1162–1166. [DOI] [PubMed] [Google Scholar]
- 18.Park J et al. (2006) Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 441, 1157–1161. [DOI] [PubMed] [Google Scholar]
- 19.Quinn PMJ et al. (2020) PINK1/PARKIN Signalling in neurodegeneration and neuroinflammation. Acta Neuropathologica Communications 8, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sekine S and Youle RJ (2018) PINK1 Import regulation; a fine system to convey mitochondrial stress to the cytosol. BMC Biology 16, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harper JW, Ordureau A and Heo JM (2018) Building and decoding ubiquitin chains for mitophagy. Nature Reviews Molecular Cell Biology 19, 93–108. [DOI] [PubMed] [Google Scholar]
- 22.Kondapalli C et al. (2012) PINK1 Is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating serine 65. Open Biology 2, 120080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shiba-Fukushima K et al. (2012) PINK1-mediated Phosphorylation of the parkin ubiquitin-like domain primes mitochondrial translocation of Parkin and regulates mitophagy. Scientific Reports 2, 1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koyano F et al. (2014) Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166. [DOI] [PubMed] [Google Scholar]
- 25.Kane LA et al. (2014) PINK1 Phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. Journal of Cell Biology 205, 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aguirre JD et al. (2017) Structure of phosphorylated UBL domain and insights into PINK1-orchestrated parkin activation. Proceedings of the National Academy of Sciences of the United States of America 114, 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ordureau A et al. (2014) Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Molecular Cell 56, 360–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka A et al. (2010) Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. Journal of Cell Biology 191, 1367–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X et al. (2011) PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147, 893–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shlevkov E et al. (2016) Miro phosphorylation sites regulate Parkin recruitment and mitochondrial motility. Proceedings of the National Academy of Sciences of the United States of America 113, E6097–E6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLelland GL et al. (2018) Mfn2 ubiquitination by PINK1/parkin gates the p97-dependent release of ER from mitochondria to drive mitophagy. Elife 7, e32866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pryde KR et al. (2016) PINK1 Disables the anti-fission machinery to segregate damaged mitochondria for mitophagy. Journal of Cell Biology 213, 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarraf SA et al. (2013) Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496, 372–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu M et al. (2013) Regulation of mitophagy by the Gp78 E3 ubiquitin ligase. Molecular Biology of the Cell 24, 1153–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szargel R et al. (2016) The PINK1, synphilin-1 and SIAH-1 complex constitutes a novel mitophagy pathway. Human Molecular Genetics 25, 3476–3490. [DOI] [PubMed] [Google Scholar]
- 36.Ambivero CT et al. (2014) Mulan E3 ubiquitin ligase interacts with multiple E2 conjugating enzymes and participates in mitophagy by recruiting GABARAP. Cellular Signalling 26, 2921–2929. [DOI] [PubMed] [Google Scholar]
- 37.Villa E et al. (2017) Parkin-independent mitophagy controls chemotherapeutic response in cancer cells. Cell Reports 20, 2846–2859. [DOI] [PubMed] [Google Scholar]
- 38.Lazarou M et al. (2015) The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gatica D, Lahiri V and Klionsky DJ (2018) Cargo recognition and degradation by selective autophagy. Nature Cell Biology 20, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heo JM et al. (2015) The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Molecular Cell 60, 7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richter B et al. (2016) Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proceedings of the National Academy of Sciences of the USA 113, 4039–4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Franco-Iborra S et al. (2021) Mutant HTT (huntingtin) impairs mitophagy in a cellular model of Huntington disease. Autophagy 17, 672–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oakes JA, Davies MC and Collins MO (2017) TBK1: a new player in ALS linking autophagy and neuroinflammation. Molecular Brain 10, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maruyama H et al. (2010) Mutations of optineurin in amyotrophic lateral sclerosis. Nature 465, 223–226. [DOI] [PubMed] [Google Scholar]
- 45.Wong YC and Holzbaur EL (2014) Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proceedings of the National Academy of Sciences of the USA 111, E4439–E4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Teyssou E et al. (2013) Mutations in SQSTM1 encoding p62 in amyotrophic lateral sclerosis: genetics and neuropathology. Acta Neuropathologica 125, 511–522. [DOI] [PubMed] [Google Scholar]
- 47.Du Y, Wooten MC and Wooten MW (2009) Oxidative damage to the promoter region of SQSTM1/p62 is common to neurodegenerative disease. Neurobiology of Disease 35, 302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugiura A et al. (2014) A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO Journal 33, 2142–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Picca A et al. (2020) Generation and release of mitochondrial-derived vesicles in health, aging and disease. Journal of Clinical Medicine 9, 1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Goldberg MS et al. (2003) Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. Journal of Biological Chemistry 278, 43628–43635. [DOI] [PubMed] [Google Scholar]
- 51.Perez FA and Palmiter RD (2005) Parkin-deficient mice are not a robust model of parkinsonism. Proceedings of the National Academy of Sciences of the United States of America 102, 2174–2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee JJ et al. (2018) Basal mitophagy is widespread in Drosophila but minimally affected by loss of Pink1 or parkin. Journal of Cell Biology 217, 1613–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McWilliams TG et al. (2018) Basal mitophagy occurs independently of PINK1 in mouse tissues of high metabolic demand. Cell Metabolism 27, 439–449 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim YY et al. (2019) Assessment of mitophagy in mt-Keima Drosophila revealed an essential role of the PINK1-Parkin pathway in mitophagy induction in vivo. FASEB Journal 33, 9742–9751. [DOI] [PubMed] [Google Scholar]
- 55.Sliter DA et al. (2018) Parkin and PINK1 mitigate STING-induced inflammation. Nature 561, 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cornelissen T et al. (2018) Deficiency of parkin and PINK1 impairs age-dependent mitophagy in Drosophila. Elife 7, e35878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choubey V, Zeb A and Kaasik A (2021) Molecular mechanisms and regulation of mammalian mitophagy. Cells 11, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J and Ney PA (2009) Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death & Differentiation 16, 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bellot G et al. (2009) Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Molecular and Cellular Biology 29, 2570–2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schweers RL et al. (2007) NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proceedings of the National Academy of Sciences of the United States of America 104, 19500–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiang G et al. (2017) BNIP3L-dependent Mitophagy accounts for mitochondrial clearance during 3 factors-induced somatic cell reprogramming. Autophagy 13, 1543–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lampert MA et al. (2019) BNIP3L/NIX and FUNDC1-mediated mitophagy is required for mitochondrial network remodeling during cardiac progenitor cell differentiation. Autophagy 15, 1182–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yazdankhah M et al. (2021) BNIP3L-mediated Mitophagy is required for mitochondrial remodeling during the differentiation of optic nerve oligodendrocytes. Autophagy 17, 3140–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan Y et al. (2017) BNIP3L/NIX-mediated Mitophagy protects against ischemic brain injury independent of PARK2. Autophagy 13, 1754–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Novak I et al. (2010) Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Reports 11, 45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rogov VV et al. (2017) Phosphorylation of the mitochondrial autophagy receptor Nix enhances its interaction with LC3 proteins. Scientific Reports 7, 1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poole LP et al. (2021) ULK1 Promotes mitophagy via phosphorylation and stabilization of BNIP3. Scientific Reports 11, 20526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Melser S et al. (2013) Rheb regulates mitophagy induced by mitochondrial energetic status. Cell Metabolism 17, 719–730. [DOI] [PubMed] [Google Scholar]
- 69.Marinković M, Šprung M and Novak I (2021) Dimerization of mitophagy receptor BNIP3L/NIX is essential for recruitment of autophagic machinery. Autophagy 17, 1232–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li Y et al. (2007) Bnip3 mediates the hypoxia-induced inhibition on mammalian target of rapamycin by interacting with Rheb. Journal of Biological Chemistry 282, 35803–35813. [DOI] [PubMed] [Google Scholar]
- 71.Zhu Y et al. (2013) Modulation of serines 17 and 24 in the LC3-interacting region of Bnip3 determines pro-survival mitophagy versus apoptosis. Journal of Biological Chemistry 288, 1099–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hanna RA et al. (2012) Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. Journal of Biological Chemistry 287, 19094–19104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi RY et al. (2014) BNIP3 Interacting with LC3 triggers excessive mitophagy in delayed neuronal death in stroke. CNS Neuroscience & Therapeutics 20, 1045–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Palikaras K, Lionaki E and Tavernarakis N (2018) Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nature Cell Biology 20, 1013–1022. [DOI] [PubMed] [Google Scholar]
- 75.Liu L et al. (2012) Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nature Cell Biology 14, 177–185. [DOI] [PubMed] [Google Scholar]
- 76.Chen G et al. (2014) A regulatory signaling loop comprising the PGAM5 phosphatase and CK2 controls receptor-mediated mitophagy. Molecular Cell 54, 362–377. [DOI] [PubMed] [Google Scholar]
- 77.Lim Y et al. (2019) Fndc-1 contributes to paternal mitochondria elimination in C. elegans. Developmental Biology 454, 15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu W et al. (2016) FUNDC1 Regulates mitochondrial dynamics at the ER-mitochondrial contact site under hypoxic conditions. EMBO Journal 35, 1368–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Murakawa T et al. (2015) Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nature Communications 6, 7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bhujabal Z et al. (2017) FKBP8 Recruits LC3A to mediate Parkin-independent mitophagy. EMBO Reports 18, 947–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yoo SM et al. (2020) FKBP8 LIRL-dependent mitochondrial fragmentation facilitates mitophagy under stress conditions. FASEB Journal 34, 2944–2957. [DOI] [PubMed] [Google Scholar]
- 82.Gao F et al. (2015) The mitochondrial protein BNIP3L is the substrate of PARK2 and mediates mitophagy in PINK1/PARK2 pathway. Human Molecular Genetics 24, 2528–2538. [DOI] [PubMed] [Google Scholar]
- 83.Ding WX et al. (2010) Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. Journal of Biological Chemistry 285, 27879–27890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee Y et al. (2011) Mitochondrial autophagy by Bnip3 involves Drp1-mediated mitochondrial fission and recruitment of Parkin in cardiac myocytes. American Journal of Physiology. Heart and Circulatory Physiology 301, H1924–H1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sugo M et al. (2018) Syntaxin 17 regulates the localization and function of PGAM5 in mitochondrial division and mitophagy. EMBO Journal 37, e98899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Landes T et al. (2010) The BH3-only Bnip3 binds to the dynamin Opa1 to promote mitochondrial fragmentation and apoptosis by distinct mechanisms. EMBO Reports 11, 459–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chen M et al. (2016) Mitophagy receptor FUNDC1 regulates mitochondrial dynamics and mitophagy. Autophagy 12, 689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wei Y et al. (2017) Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell 168, 224–238, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yoshii SR et al. (2011) Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. Journal of Biological Chemistry 286, 19630–19640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Xiao Y et al. (2018) PHB2 Interacts with LC3 and SQSTM1 is required for bile acids-induced mitophagy in cholestatic liver. Cell death & disease 9, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yan C et al. (2020) PHB2 (prohibitin 2) promotes PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis. Autophagy 16, 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schlame M (2008) Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes. Journal of Lipid Research 49, 1607–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Chu CT et al. (2013) Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nature Cell Biology 15, 1197–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kagan VE et al. (2016) NDPK-D (NM23-H4)-mediated externalization of cardiolipin enables elimination of depolarized mitochondria by mitophagy. Cell Death & Differentiation 23, 1140–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bustillo-Zabalbeitia I et al. (2014) Specific interaction with cardiolipin triggers functional activation of dynamin-related protein 1. PLoS ONE 9, e102738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sentelle RD et al. (2012) Ceramide targets autophagosomes to mitochondria and induces lethal mitophagy. Nature Chemical Biology 8, 831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Law BA et al. (2018) Lipotoxic very-long-chain ceramides cause mitochondrial dysfunction, oxidative stress, and cell death in cardiomyocytes. FASEB Journal 32, 1403–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Oleinik N et al. (2019) Mitochondrial protein import is regulated by p17/PERMIT to mediate lipid metabolism and cellular stress. Science Advances 5, eaax1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Doblado L et al. (2021) Mitophagy in human diseases. International journal of molecular sciences 22, 3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Doxaki C and Palikaras K (2021) Neuronal mitophagy: friend or Foe? Frontiers in Cell and Developmental Biology 8, 611938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bingol B et al. (2014) The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 510, 370–375. [DOI] [PubMed] [Google Scholar]
- 102.Cornelissen T et al. (2014) The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Human Molecular Genetics 23, 5227–5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang Y et al. (2015) Deubiquitinating enzymes regulate PARK2-mediated mitophagy. Autophagy 11, 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marcassa E et al. (2018) Dual role of USP30 in controlling basal pexophagy and mitophagy. EMBO Reports 19, e45595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Durcan TM et al. (2014) USP8 regulates mitophagy by removing K6-linked ubiquitin conjugates from parkin. EMBO Journal 33, 2473–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang L et al. (2018) PTEN-L is a novel protein phosphatase for ubiquitin dephosphorylation to inhibit PINK1-Parkin-mediated mitophagy. Cell Research 28, 787–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gersch M et al. (2017) Mechanism and regulation of the Lys6-selective deubiquitinase USP30. Nature Structural & Molecular Biology 24, 920–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang L, Lu G and Shen HM (2020) The long and the short of PTEN in the regulation of mitophagy. Frontiers in Cell and Developmental Biology 8, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Eldeeb MA et al. (2022) The role of PTEN-L in modulating PINK1-Parkin-mediated mitophagy. Neurotoxicity Research 40, 1103–1114. [DOI] [PubMed] [Google Scholar]
- 110.Rocha EM et al. (2022) LRRK2 and idiopathic Parkinson's disease. Trends in Neurosciences 45, 224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bonello F et al. (2019) LRRK2 impairs PINK1/Parkin-dependent mitophagy via its kinase activity: pathologic insights into Parkinson's disease. Human Molecular Genetics 28, 1645–1660. [DOI] [PubMed] [Google Scholar]
- 112.Eguchi T et al. (2018) LRRK2 and its substrate Rab GTPases are sequentially targeted onto stressed lysosomes and maintain their homeostasis. Proceedings of the National Academy of Sciences of the USA 115, E9115–E9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wauters F et al. (2020) LRRK2 Mutations impair depolarization-induced mitophagy through inhibition of mitochondrial accumulation of RAB10. Autophagy 16, 203–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Singh F et al. (2021) Pharmacological rescue of impaired mitophagy in Parkinson's disease-related LRRK2 G2019S knock-in mice. Elife, 10, e67604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tasegian A et al. (2021) Impact of type II LRRK2 inhibitors on signaling and mitophagy. Biochemical Journal 478, 3555–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hsieh CH et al. (2016) Functional impairment in miro degradation and mitophagy is a shared feature in familial and sporadic Parkinson's disease. Cell Stem Cell 19, 709–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fang EF et al. (2014) Defective mitophagy in XPA via PARP-1 hyperactivation and NAD(+)/SIRT1 reduction. Cell 157, 882–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wan W et al. (2022) Regulation of mitophagy by sirtuin family proteins: a vital role in aging and age-related diseases. Frontiers in Aging Neuroscience 14, 845330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Furukawa K et al. (2018) The PP2A-like protein phosphatase Ppg1 and the far complex cooperatively counteract CK2-mediated phosphorylation of Atg32 to inhibit mitophagy. Cell Reports 23, 3579–3590. [DOI] [PubMed] [Google Scholar]
- 120.Wong PM et al. (2015) Regulation of autophagy by coordinated action of mTORC1 and protein phosphatase 2A. Nature Communications 6, 8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yap CC et al. (2018) Degradation of dendritic cargos requires Rab7-dependent transport to somatic lysosomes. Journal of Cell Biology 217, 3141–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ferguson SM (2019) Neuronal lysosomes. Neuroscience Letters 697, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cai Q et al. (2012) Spatial parkin translocation and degradation of damaged mitochondria via mitophagy in live cortical neurons. Current Biology 22, 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Vives-Bauza C et al. (2010) PINK1-dependent Recruitment of Parkin to mitochondria in mitophagy. Proceedings of the National Academy of Sciences of the USA 107, 378–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Joselin AP et al. (2012) ROS-dependent regulation of Parkin and DJ-1 localization during oxidative stress in neurons. Human Molecular Genetics 21, 4888–4903. [DOI] [PubMed] [Google Scholar]
- 126.Rakovic A et al. (2013) Phosphatase and tensin homolog (PTEN)-induced putative kinase 1 (PINK1)-dependent ubiquitination of endogenous Parkin attenuates mitophagy: study in human primary fibroblasts and induced pluripotent stem cell-derived neurons. Journal of Biological Chemistry 288, 2223–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sterky FH et al. (2011) Impaired mitochondrial transport and Parkin-independent degeneration of respiratory chain-deficient dopamine neurons in vivo. Proceedings of the National Academy of Sciences of the USA 108, 12937–12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Devireddy S et al. (2015) The organization of mitochondrial quality control and life cycle in the nervous system in vivo in the absence of PINK1. Journal of Neuroscience 35, 9391–9401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sung H et al. (2016) Compartmentalized regulation of Parkin-mediated mitochondrial quality control in the Drosophila nervous system in vivo. Journal of Neuroscience 36, 7375–7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Burman JL et al. (2012) Analysis of neural subtypes reveals selective mitochondrial dysfunction in dopaminergic neurons from parkin mutants. Proceedings of the National Academy of Sciences of the USA 109, 10438–10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Whitworth AJ et al. (2005) Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson's disease. Proceedings of the National Academy of Sciences of the USA 102, 8024–8029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yang Y et al. (2006) Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proceedings of the National Academy of Sciences of the USA 103, 10793–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kitada T et al. (2007) Impaired dopamine release and synaptic plasticity in the striatum of PINK1-deficient mice. Proceedings of the National Academy of Sciences of the USA 104, 11441–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Van Laar VS et al. (2011) Bioenergetics of neurons inhibit the translocation response of Parkin following rapid mitochondrial depolarization. Human Molecular Genetics 20, 927–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Puri R et al. (2019) Mul1 restrains Parkin-mediated mitophagy in mature neurons by maintaining ER-mitochondrial contacts. Nature Communications 10, 3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yun J et al. (2014) MUL1 Acts in parallel to the PINK1/parkin pathway in regulating mitofusin and compensates for loss of PINK1/parkin. Elife 3, e01958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.de Brito OM and Scorrano L (2008) Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610. [DOI] [PubMed] [Google Scholar]
- 138.Naon D et al. (2016) Critical reappraisal confirms that Mitofusin 2 is an endoplasmic reticulum-mitochondria tether. Proceedings of the National Academy of Sciences of the United States of America 113, 11249–11254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Filadi R et al. (2015) Mitofusin 2 ablation increases endoplasmic reticulum-mitochondria coupling. Proceedings of the National Academy of Sciences of the USA 112, E2174–E2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Wang PT et al. (2015) Distinct mechanisms controlling rough and smooth endoplasmic reticulum contacts with mitochondria. Journal of Cell Science 128, 2759–2765. [DOI] [PubMed] [Google Scholar]
- 141.Lopez-Domenech G et al. (2021) Loss of neuronal Miro1 disrupts mitophagy and induces hyperactivation of the integrated stress response. EMBO Journal 40, e100715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Evans CS and Holzbaur EL (2020) Degradation of engulfed mitochondria is rate-limiting in Optineurin-mediated mitophagy in neurons. Elife 9, e50260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Falabella M et al. (2021) Cardiolipin, mitochondria, and neurological disease. Trends in Endocrinology and Metabolism 32, 224–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cai Y et al. (2021) FUNDC1-dependent Mitophagy induced by tPA protects neurons against cerebral ischemia-reperfusion injury. Redox Biology 38, 101792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Koentjoro B, Park JS and Sue CM (2017) Nix restores mitophagy and mitochondrial function to protect against PINK1/Parkin-related Parkinson's disease. Scientific Reports 7, 44373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Schmid ET, Pyo J-H and Walker DW (2022) Neuronal induction of BNIP3-mediated mitophagy slows systemic aging in Drosophila. Nature Aging 2, 494–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.McWilliams TG et al. (2016) mito-QC illuminates mitophagy and mitochondrial architecture in vivo. Journal of Cell Biology 214, 333–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Miller KE and Sheetz MP (2004) Axonal mitochondrial transport and potential are correlated. Journal of Cell Science 117, 2791–2804. [DOI] [PubMed] [Google Scholar]
- 149.van Bergeijk P et al. (2015) Optogenetic control of organelle transport and positioning. Nature 518, 111–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Han S et al. (2020) Mitophagy regulates integrity of mitochondria at synapses and is critical for synaptic maintenance. EMBO Reports 21, e49801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sharma M et al. (2019) Rhes, a striatal-enriched protein, promotes mitophagy via Nix. Proceedings of the National Academy of Sciences of the USA 116, 23760–23771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zheng Y et al. (2019) Somatic autophagy of axonal mitochondria in ischemic neurons. Journal of Cell Biology 218, 1891–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Maday S, Wallace KE and Holzbaur EL (2012) Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. Journal of Cell Biology 196, 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Farfel-Becker T et al. (2019) Neuronal soma-derived degradative lysosomes are continuously delivered to distal axons to maintain local degradation capacity. Cell Reports 28, 51–64, e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Verburg J and Hollenbeck PJ (2008) Mitochondrial membrane potential in axons increases with local nerve growth factor or semaphorin signaling. Journal of Neuroscience 28, 8306–8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liu S et al. (2012) Parkinson's disease-associated kinase PINK1 regulates Miro protein level and axonal transport of mitochondria. PLoS Genetics 8, e1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ashrafi G et al. (2014) Mitophagy of damaged mitochondria occurs locally in distal neuronal axons and requires PINK1 and Parkin. Journal of Cell Biology 206, 655–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Harbauer AB et al. (2022) Neuronal mitochondria transport Pink1 mRNA via synaptojanin 2 to support local mitophagy. Neuron 110, 1516–1531, e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pilling AD et al. (2006) Kinesin-1 and dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Molecular Biology of the Cell 17, 2057–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Plucinska G et al. (2012) In vivo imaging of disease-related mitochondrial dynamics in a vertebrate model system. Journal of Neuroscience 32, 16203–16212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ferree AW et al. (2013) MitoTimer probe reveals the impact of autophagy, fusion, and motility on subcellular distribution of young and old mitochondrial protein and on relative mitochondrial protein age. Autophagy 9, 1887–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Davis CH et al. (2014) Transcellular degradation of axonal mitochondria. Proceedings of the National Academy of Sciences of the USA 111, 9633–9638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Morales I et al. (2020) Neuroglial transmitophagy and Parkinson's disease. Glia 68, 2277–2299. [DOI] [PubMed] [Google Scholar]
- 164.Lampinen R et al. (2022) Neuron-astrocyte transmitophagy is altered in Alzheimer's disease. Neurobiology of Disease 170, 105753. [DOI] [PubMed] [Google Scholar]
- 165.Taoufik E et al. (2018) Synaptic dysfunction in neurodegenerative and neurodevelopmental diseases: an overview of induced pluripotent stem-cell-based disease models. Open Biology 8, 180138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Picconi B, Piccoli G and Calabresi P (2012) Synaptic dysfunction in Parkinson's disease. Advances in Experimental Medicine and Biology 970, 553–572. [DOI] [PubMed] [Google Scholar]
- 167.Karisetty BC et al. (2020) Amyloid-β peptide impact on synaptic function and neuroepigenetic gene control reveal new therapeutic strategies for Alzheimer's disease. Frontiers in Molecular Neuroscience 13, 577622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Palikaras K and Tavernarakis N (2020) Regulation and roles of mitophagy at synapses. Mechanisms of Ageing and Development 187, 111216. [DOI] [PubMed] [Google Scholar]
- 169.Sheng ZH (2014) Mitochondrial trafficking and anchoring in neurons: new insight and implications. Journal of Cell Biology 204, 1087–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Obashi K and Okabe S (2013) Regulation of mitochondrial dynamics and distribution by synapse position and neuronal activity in the axon. European Journal of Neuroscience 38, 2350–2363. [DOI] [PubMed] [Google Scholar]
- 171.Lewis TL Jr et al. (2016) Progressive decrease of mitochondrial motility during maturation of cortical axons in vitro and in vivo. Current Biology 26, 2602–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Silva CA et al. (2021) Activity-dependent regulation of mitochondrial motility in developing cortical dendrites. Elife 10, e62091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lores-Arnaiz S et al. (2016) Brain cortex mitochondrial bioenergetics in synaptosomes and non-synaptic mitochondria during aging. Neurochemical Research 41, 353–363. [DOI] [PubMed] [Google Scholar]
- 174.James R et al. (2004) Disrupted in schizophrenia 1 (DISC1) is a multicompartmentalized protein that predominantly localizes to mitochondria. Molecular and Cellular Neuroscience 26, 112–122. [DOI] [PubMed] [Google Scholar]
- 175.Hayashi-Takagi A et al. (2010) Disrupted-in-Schizophrenia 1 (DISC1) regulates spines of the glutamate synapse via Rac1. Nature Neuroscience 13, 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wang ZT et al. (2019) Disrupted-in-schizophrenia-1 protects synaptic plasticity in a transgenic mouse model of Alzheimer's disease as a mitophagy receptor. Aging Cell 18, e12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Park C et al. (2016) Disrupted-in-schizophrenia 1 (DISC1) and syntaphilin collaborate to modulate axonal mitochondrial anchoring. Molecular Brain 9, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kruppa AJ et al. (2018) Myosin VI-dependent actin cages encapsulate Parkin-positive damaged mitochondria. Developmental Cell 44, 484–499, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Tumbarello DA et al. (2012) Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nature Cell Biology 14, 1024–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hu S et al. (2019) Structure of myosin VI/Tom1 complex reveals a cargo recognition mode of myosin VI for tethering. Nature Communications 10, 3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kneussel M and Wagner W (2013) Myosin motors at neuronal synapses: drivers of membrane transport and actin dynamics. Nature Reviews Neuroscience 14, 233–247. [DOI] [PubMed] [Google Scholar]
- 182.Osterweil E, Wells DG and Mooseker MS (2005) A role for myosin VI in postsynaptic structure and glutamate receptor endocytosis. Journal of Cell Biology 168, 329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Bakula D and Scheibye-Knudsen M (2020) MitophAging: mitophagy in aging and disease. Frontiers in Cell and Developmental Biology 8, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chen G, Kroemer G and Kepp O (2020) Mitophagy: an emerging role in aging and age-associated diseases. Frontiers in Cell and Developmental Biology 8, 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Georgakopoulos ND, Wells G and Campanella M (2017) The pharmacological regulation of cellular mitophagy. Nature Chemical Biology 13, 136–146. [DOI] [PubMed] [Google Scholar]
- 186.Xie C et al. (2022) Amelioration of Alzheimer's disease pathology by mitophagy inducers identified via machine learning and a cross-species workflow. Nature Biomedical Engineering 6, 76–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Fang EF et al. (2019) Mitophagy inhibits amyloid-beta and tau pathology and reverses cognitive deficits in models of Alzheimer's disease. Nature Neuroscience 22, 401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ryu D et al. (2016) Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nature Medicine 22, 879–888. [DOI] [PubMed] [Google Scholar]
- 189.Hou Y et al. (2018) NAD(+) supplementation normalizes key Alzheimer's features and DNA damage responses in a new AD mouse model with introduced DNA repair deficiency. Proceedings of the National Academy of Sciences of the United States of America 115, E1876–E1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lautrup S et al. (2019) NAD + in brain aging and neurodegenerative disorders. Cell Metabolism 30, 630–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Andreux PA et al. (2019) The mitophagy activator urolithin A is safe and induces a molecular signature of improved mitochondrial and cellular health in humans. Nature Metabolism 1, 595–603. [DOI] [PubMed] [Google Scholar]
- 192.Palikaras K and Tavernarakis N (2014) Mitochondrial homeostasis: the interplay between mitophagy and mitochondrial biogenesis. Experimental Gerontology 56, 182–188. [DOI] [PubMed] [Google Scholar]
- 193.Palikaras K, Lionaki E and Tavernarakis N (2015) Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 521, 525–528. [DOI] [PubMed] [Google Scholar]
- 194.Guyot MC et al. (1997) Quantifiable bradykinesia, gait abnormalities and Huntington's disease-like striatal lesions in rats chronically treated with 3-nitropropionic acid. Neuroscience 79, 45–56. [DOI] [PubMed] [Google Scholar]
- 195.Zaninello M et al. (2020) Inhibition of autophagy curtails visual loss in a model of autosomal dominant optic atrophy. Nature Communications 11, 4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Yu-Wai-Man P et al. (2010) Multi-system neurological disease is common in patients with OPA1 mutations. Brain 133, 771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kawajiri S et al. (2011) Genetic mutations and functions of PINK1. Trends in Pharmacological Sciences 32, 573–580. [DOI] [PubMed] [Google Scholar]
- 198.Arkinson C and Walden H (2018) Parkin function in Parkinson's disease. Science 360, 267–268. [DOI] [PubMed] [Google Scholar]
- 199.Berenguer-Escuder C et al. (2019) Variants in Miro1 cause alterations of ER-mitochondria contact sites in fibroblasts from Parkinson's disease patients. Journal of Clinical Medicine 8, 2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Vaillant-Beuchot L et al. (2021) Accumulation of amyloid precursor protein C-terminal fragments triggers mitochondrial structure, function, and mitophagy defects in Alzheimer's disease models and human brains. Acta Neuropathologica 141, 39–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Martín-Maestro P et al. (2016) PARK2 Enhancement is able to compensate mitophagy alterations found in sporadic Alzheimer's disease. Human Molecular Genetics 25, 792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Martín-Maestro P et al. (2017) Slower dynamics and aged mitochondria in sporadic Alzheimer's disease. Oxidative Medicine and Cellular Longevity 2017, 9302761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Song W et al. (2011) Mutant huntingtin binds the mitochondrial fission GTPase dynamin-related protein-1 and increases its enzymatic activity. Nature Medicine 17, 377–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Liu W et al. (2013) Mitochondrial fusion and fission proteins expression dynamically change in a murine model of amyotrophic lateral sclerosis. Current Neurovascular Research 10, 222–230. [DOI] [PubMed] [Google Scholar]
- 205.Zhang F et al. (2015) Miro1 deficiency in amyotrophic lateral sclerosis. Frontiers in Aging Neuroscience 7, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]