Abstract
Surface topography modification with nano- or micro- textured structures has been an efficient approach to inhibit microbial adhesion and biofilm formation and thereby to prevent biomaterial associated infection without modification of surface chemistry/bulk properties of materials and without causing antibiotic resistance. This manuscript focuses on submicron textured patterns with ordered arrays of pillars on polyurethane (PU) biomaterial surfaces in an effort to understand the effects of surface pillar features and surface properties on adhesion and colonization responses of two Staphylococcal strains. Five submicron patterns with a variety of pillar dimensions were designed and fabricated on PU film surfaces and bacterial adhesion and biofilm formation of Staphylococcal strains (S. epidermidis RP62A and S. aureus Newman D2C) were characterized. Results show that all submicron textured surface significantly reduced bacterial adhesion and inhibited biofilm formation, and bacterial adhesion linearly decreased with the reduction in top surface area fraction. Surface wettability did not show a linear correlation with bacterial adhesion, suggesting that surface contact area dominates bacterial adhesion. From this, it appears that the design of textured patterns should minimize surface area fraction to reduce the bacterial interaction with surfaces but in a way that ensures the mechanical strength of pillars in order to avoid collapse. These findings may provide a rationale for design of polymer surfaces for antifouling medical devices.
Keywords: microbial infection, bacterial adhesion, topography modification, textured surface, Staphylococcus, wettability
Graphical Abstract
1. INTRODUCTION
Infections associated with implantable medical devices are the most common cause of healthcare-associated infections, with 50–70% of all healthcare-associated infections attributed to medical device induced infections1,2. Bacterial colonization and subsequent biofilm formation on biomaterial surfaces is central to the processes of device associated infections. Biofilm formation confers broad protection to its bacterial inhabitants from the deleterious effects of antimicrobials and the immune response, so that it is extremely difficult to treat biofilms by use of antibiotics alone. In addition, antibiotic resistance continues to be a problem3,4. For example, methicillin resistant Staphylococcus aureus (MRSA) now accounts for about 60% of clinical S. aureus isolates from intensive care units in the United States5 and in other countries6,7, and up to 93% of MRSA isolates were found to be multidrug-resistant8. For these reasons, interdisciplinary efforts in recent years have been focused on developing new generation biomaterials and technologies for combatting microbial infections through either reducing biofilm formation without antibiotics9 or increasing bacterial susceptibility to antibiotics10.
Bacterial adhesion to surfaces is a crucial step in the pathogenesis of biomaterial associated infection. Controlling the interfacial chemical and physical properties, and thus modulating the interactions of bacteria and material surfaces, has been an important approach for the development of high-performance biomaterials for medical devices. In recent years the importance of surface topographic modification utilizing micro- or nano- scale topographies has come to the fore in the area of antimicrobial and anti-biofouling materials11–14, and some of the designs of these topographies were inspired by the naturally existing antifouling surfaces such as plant leaves15, shark skin16, and insect wings17. By mimicking these natural surfaces, many topographic patterns have been developed and created, and have been shown to be a promising stratgegy for designing next generation antifouling materials for implantable medical devices without causing antibiotic resistence18–22. These topographic features, ranging from the micro- to the nano- scale, influence bacterial sensing and attachment to material surfaces, thereby hindering biofilm formation13,23. For example, the Sharklet™ micropatterned surface, a microtopography feature inspired by shark skin, exhibited significantly less bacterial colonization and lower rate of bacterial migration over the device surface than the standard of care smooth surface, and was proposed to be applied to Foley catheters for reducing catheter-associated urinary tract infection24. The behavior of this material was attributed to physical impediments that limit bacterial contact thus reducing biofilm formation25. Whereas micro-scale topographies mainly affect bacterial interactions with surfaces, some nano-scale topographies were created to inhibit biofilms by killing bacteria through mechanical forces26. These bactericidal nanofeatures were inspired from insect wings, e.g. cicada wings, where bacterial cells were not repelled but were penetrated by the nanopillar arrays present on the wing surface, resulting in bacterial death27,28. Inspired by these nanofeatures, strong bactericidal effects were found on vertically aligned high aspect ratio carbon nanotube29, and black silicon30 against both Gram-negative and Gram-positive bacteria. Wu et al. reported that gold nanostructured surfaces fabricated by templated electrodeposition and forming a variety of geometries including nanopillars, nanorings or nanonuggets all exhibited strong bactericidal effects (>99%) on MRSA31. In addition to these effects of surface topography on bacterial attachment, micro- and nano-scale topographies may also influence physicochemical forces, hydrodynamics, wettability, surface condition, or chemical gradients, leading to cell ordering, segregation, and removal from surfaces32.
Although micro- and nano-scale topographic features have shown effective inhibition of bacterial adhesion, other topographic features such as shape, size, and distribution of topography influence bacterial attachment and colonization. Bacterial cells may sense and respond to 3D surface topography, leading to different levels of adhesion on surfaces23. It is a common observation that bacterial adhesion decreases as the size of topographic pattern gets smaller by reducing contact area33–35, while the other properties, e.g., wettability, may become important in some cases. We have previously developed textured surfaces with ordered submicron pillars and created patterns on poly(urethane urea) (PUU) biomaterial films with pillar geometries of 400/400/600 and 500/500/600 nm (diameter of pillars/space distance between pillars/height of pillars) for reducing bacterial adhesion and biofilm formation. These pillar patterns feature small pillar diameter and spaces between pillars which are smaller than the dimension of cells, thereby reducing the accessible surface contact area for bacterial interaction. Results showed that these submicron textured patterns inhibit S. epidermidis and S. aureus adhesion by up to 90% under shear in physiological related solutions36. We further developed micro-scale pillars with dimensions of 2.5/1.5/1.2 μm and 4.0/1.5/1.2 μm and found that the micro-scale textured patterns influence on bacterial adhesion is dependent on surface wettability, where hydrophobic micro-scale textured patterns inhibit bacterial adhesion while hydrophilic textured surfaces increased bacterial adhesion37, demonstrating that surface wettability is important for bacterial attachment on micro-scale topographic surfaces. However, results also showed that both hydrophobic and hydrophilic submicron textured surfaces significantly reduced bacterial adhesion, strongly suggesting that submicron scale patterns minimize bacterial attachment by reducing the contact area. However, the above findings are still not sufficient for rational design of antifouling materials, especially for polymeric materials for medical devices. For example, decreasing the diameter of pillars reduces surface contact area, but it may also decrease the mechanical strength of pillars, resulting in pillar collapse and defects in patterns. Therefore, further studies on the effect of pattern geometry on bacterial adhesion are necessary to find the relationship between pattern geometry and bacterial attachment and colonization, and better provide the rationale for the design of medical device surfaces.
In this study we focus on submicron pillar patterns and adhesion/colonization of Staphylococcal strains as Staphylococci are the most commonly diagnosed microorganisms in health care-associated infections38. We designed a series of pillar patterns with different pillar size, spacing, height, and shape and fabricated these patterns on polyurethane biomaterial surfaces. Through analysis of surface topographic features and the bacterial adhesion/biofilms on these surfaces, a relation of topography properties and bacterial responses is revealed, and the findings may provide the rationale for design of polymer surfaces for antifouling medical devices.
2. MATERIALS AND METHODS
2.1. Materials.
Polyurethane urea (PUU) (BioSpan® MS/0.4) was purchased from Polymer Technology Group (Berkeley, CA), and was supplied at a concentration of 22 wt% in dimethylacetamide (DMAC). The PUU contains 4,4’-methylene di(p-phenyl isocyanate) hard segment, poly(tetramethylene oxide) soft segment, ethylene diamine chain extender and the chain is end-capped with 2000 molecular weight poly(dimethlysiloxane) at 0.4% by weight. Phosphate buffered saline (PBS, 0.01M, pH 7.4, Sigma) was prepared using purified water (18 MΩ) from a Millipore Simplicity 185 system.
2.2. Design and fabrication of submicron textured surfaces.
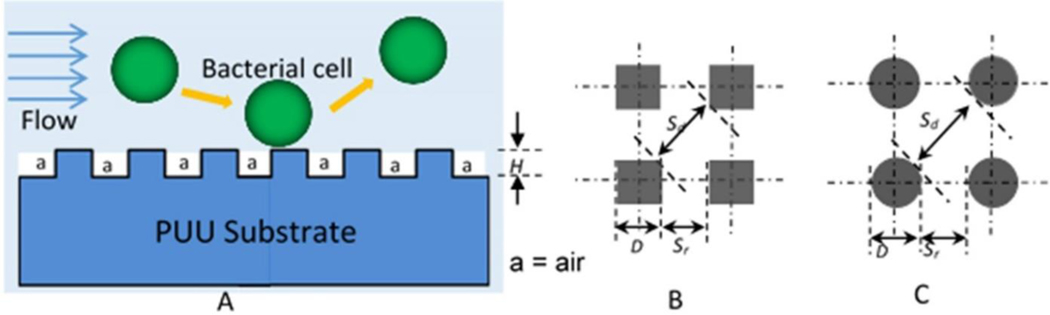
Textured surfaces consist of ordered arrays of pillars with different dimensions, but all smaller than the size of bacterial cells (Figure 1). These textured patterns reduce surface contact area for the interaction of cells and surfaces, resulting in lower adhesion of cells which are then easily removed by flow (Figure 1A). Two shapes of pillars (square and round) are uniformly distributed on surfaces. The square shape pillars include: Pattern 1= 500/300/500 (S), Pattern 2 = 700/700/300 (S), and Pattern 3 = 500/500/600 (S) nm, and round shape pillars are: Pattern 4 = 400/400/600 (R), Pattern 5 = 500/500/600 (R) nm (D/Sr/H) (Table 1). The pillar distributions are illustrated in Figures 1B and 1C. Fabrication of textured patterns on PUU surfaces was performed by a two-stage soft lithography replication process39. Briefly, a master pattern was first fabricated on a silicon wafer using electron beam lithography (Penn State Naofabrication Lab) and a silicone negative mold was then cast against master pattern. Poly(dimethylsiloxane) (PDMS, Sylgard 184 Dow Corning) was prepared with the mixture of base and curing agent at a ratio of 10:1. PDMS was degassed in a vacuum desiccator and was poured over the Si master pattern. The polymer was further degassed to ensure conformation of the silicone to the features and then cured overnight at 65°C. PDMS was then gently peeled from the master pattern. PUU replicas were prepared by spin casting MS/0.4 onto the PDMS mold in one thin layer first, and then adding additional layers until the desired film thickness was reached. Each layer was degassed and cured at room temperature under vacuum. Finally, the PUU was cured for an additional 24 h at 65°C. The PUU replica was then gently separated from the PDMS mold and stored in a desiccator before use. Similarly, smooth control molds were prepared by casting PDMS over a smooth Si wafer and a smooth PUU replica was created from this smooth silicone mold using the same procedures as for fabrication of textured PUU replica.
Figure 1.
Scheme of textured patterns, (A) textured patterns with ordered of arrays of pillars, (B) square shaped pillar distribution, (C) round shaped pillar distribution. D=size of pillar, Sr= space distance between two pillar in row, Sd =space distance between pillars in diagonal, and H= height of pillars.
Table 1.
Design parameters of textured pillars on wafer
| Pattern # | Pattern (D/Sr/H) (nm) | Pillar Shape | Pillar diameter (D) (nm) | Pillar Spacing (Sr) (nm ) | Pillar height (H) (nm) | Aspect ratio (H/D) |
|---|---|---|---|---|---|---|
|
| ||||||
| 1 | 500/300/500 | Square | 500 | 300 | 500 | 1.00 |
| 2 | 700/700/300 | Square | 700 | 700 | 300 | 0.43 |
| 3 | 500/500/600 | Square | 500 | 500 | 600 | 1.20 |
| 4 | 400/400/600 | Round | 400 | 400 | 600 | 1.50 |
| 5 | 500/500/600 | Round | 500 | 500 | 600 | 1.20 |
2.3. Textured surface topography characterization.
Atomic force microscopy (AFM) was used to characterize surface textures of the PUU replicas. A Multimode AFM with a Nanoscope IIIa control system (software version 5.12r3, Veeco, Santa Barbra, CA) was operated in tapping mode in air using high aspect ratio (5:1) Si probes (AR5T-NCHR-10, Nanoworld®, Switzerland ). AFM images were treated and analyzed by off-line AFM software (version 5.12r3, Veeco, Santa Barbra, CA) to measure the size and height of pillars. The images were further analyzed by Image J software to measure the pillar top surface area and total surface area fractions. The pillar top surface area is defined as the area of the uppermost pillar surface, while the surface fraction is the ratio of pillar top surface area to the total projected 2-D surface area of the region. Pillar sidewall areas are not considered in any of these surface area calculations as these regions are deemed to be inaccessible to bacteria due to the dimensions.
2.4. Surface wettability.
The water wettability of each PUU sample was determined as the water contact angle and measured by sessile drop measurements using a Krüss contact angle goniometer. All measurements were made using water as a probe liquid. Contact angles were measured by a minimum of eight independent measurements and are presented as mean ± standard deviation.
2.5. Pillar collapse assay.
The ability of pillars to resist collapse was assessed by soaking films in mixed solvent of methyl ethyl ketone (MEK) and methanol (7:3 in v/v) at room temperature for 2 h. After evaporation of solvent, the textured film surfaces were examined by AFM to determine if pillars had collapsed.
2.6. Bacterial adhesion and biofilm formation assay.
Bacterial strains S. epidermidis RP62A (ATCC 35984) and S. aureus Newman D2C (ATCC 25904) were selected to study bacterial adhesion and biofilm formation on textured and smooth PUU surfaces. Bacteria were cultured in tryptic soy broth (TSB) for S. epidermidis or brain heart infusion (BHI) for S. aureus in 250 mL flasks containing ~110 mL media on an orbital shaker at 250 rpm at 37 °C for 24 h. Bacterial cells were collected by centrifuging at 1500 g for 10 min. The pellet was washed with PBS and then resuspended in PBS, and the concentration of bacteria was measured at wavelength of 600 nm and adjusted to a final concentration of 1×108 cfu/mL for bacterial adhesion experiments.
Bacterial adhesion was performed in petri dishes (100 × 25 mm). Polymer samples were punched from each polymer film with diameter of 10 mm and attached to the bottom of dishes. Three replicates of each type of material surface (n=3) were tested. Each dish contains 40 mL bacterial solution and is placed in an incubator at 37°C for 1 h with shaking at 90 rpm. After adhesion, bacterial solutions were replaced with 40 mL PBS for 5× to remove non-adherent bacterial cells and samples were then fixed in 2.5 % glutaraldehyde for 2 h. After rinsing with PBS, samples were stained with Hoechst 33258 (Invitrogen) and analyzed under a fluorescence optical microscope with 100× objective lens (area 87×66 μm2) (Nikon, Eclipse 80i). Bacterial adhesion was imaged from 15 randomly selected locations of each sample and bacterial cells were manually counted. Bacterial adhesion was quantified by the number of cells per 2-D surface area and presented as bacteria/104 μm2. To further observe the individual bacterial adhesion behavior, samples were rinsed with DI water after fixation, dried in air, and then scanned by AFM operating in tapping mode.
A CDC biofilm reactor (Biosurface Technologies, Bozeman, MT) was used for evaluation of resistance to biofilm formation based on standard ASTM E2562 – 17. All culturing medium, the CDC bioreactor and coupon holders were autoclaved before use. Polymer samples were cut into round pieces with diameter of 12 mm and attached to coupon holders. The reactor containing 600 mg/L TSB medium (total ~ 400 mL volume) was inoculated with ~1×107 CFU/mL bacteria at 37°C with stirring at 150 rpm. Every 24 h, 100 mL of the culturing medium was replaced with PBS and 4 mL of sterilized TSB medium (30 g/mL) added for nutrients supplement. All procedures were performed carefully to ensure the system remained sterile while replacing medium. Bacterial growth in the medium was examined by agar plating method every day. Additionally, we observed no contamination in these colonies from the agar plating. After either 10 or 14 days, samples were lightly rinsed in sterile PBS buffer to remove non-adherent material, fixed in 2.5 % glutaraldehyde for 2 h and then stained with 100 μg/mL wheat germ agglutinin-FITC (Sigma) for 30 min. The samples were then examined by fluorescence optical microscopy using a 20× objective lens resulting in an observation area of 434×330 μm2. Each sample (n=3) was also scanned by a 10× objective lens to find the biofilms locations, and 9 locations having large or significant biofilms were selected for assessing biofilm formation. The images were analyzed using ImageJ program to characterize the areas of individual biofilms and total biofilm coverage on surfaces using our own definition of biofilms being > 10 μm2.
2.7. Data Analysis.
Statistical analysis was performed using OriginLab Origin 2020b. All data is reported as the mean ± standard deviation. Means of experimental data were compared by a 2-sample t-test or by ANOVA as appropriate, and differences were considered statistically significant for p< 0.05. Significance is denoted with one symbol denoting p< 0.05, two symbols denoting p < 0.01, and three symbols denoting p< 0.001. Bacterial adhesion data was fit to surface area fraction and water contact angle using a general linear model.
3. RESULTS
3.1. Fabrication and characterization of textured surfaces.
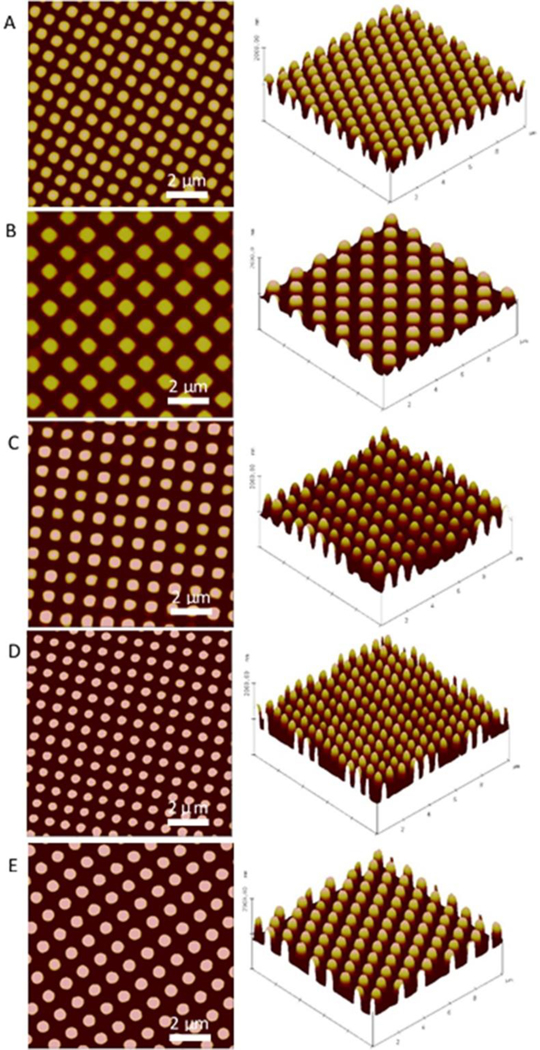
Submicron textured PUU film surfaces were fabricated by a two-stage soft lithography replication process. Each pattern was first fabricated on a silicon wafer using electron beam lithography and a negative PDMS mold was then produced by casting against master pattern. The primary advantage to the two-stage technique is that we can prepare as many as 30 replicates from each silicone negative, while utilizing the wafer directly for fabrication often leads to damage when the first replicate is made making it prohibitively expensive. Our prior study shows the pillar yield can reach ~99.8% on PUU film surfaces36, indicative of high efficiency in reproducing textured surfaces. Further, previous surface chemistry analysis of PUU replicates by XPS showed that this fabrication process through the PDMS negative mold only affected material surface topography and not PUU surface chemistry40. In this study, we used AFM to characterize the surface topography of textured films, and observed no defects of pillars on each patterned surface, as shown in Figure 2.
Figure 2.
2D and 3D AFM topography images of (A) Pattern 1 500/300/500 (S), (B) Pattern 2 700/700/300 (S), and (C) Pattern 3 500/500/600 (S) with square pillars, (D) Pattern 4 400/400/600 (R) and (E) Pattern 5 500/500/600 (R) with round pillars. All samples use polyurethane urea MS/0.4 as substrate. (S=square-shaped, R=Round-shaped)
The geometry of ordered arrays of pillars on textured PUU film surfaces was analyzed by AFM and ImageJ software, and results are tabulated in Table 2. Pillar heights for the variety of patterns ranged from 300–600 nm, consistent with the desired height of pillars as designed. The distances of adjacent pillars within rows and in diagonal of all patterns are smaller than 1000 nm, except for the pattern 700/700/300 nm for which pillar diagonal distance is 1012.7±25.9 nm. As the dimension of staphylococcus bacterial cells is normally larger than 1 μm, such design of separation distances between pillars ensures a reduction in available contact area when bacterial cells contact the surface with minimal opportunities for bacteria to become lodged in the spaces between pillars. The total pillar top surface areas varied with pillar dimensions and separation distances between pillars. Pattern 4 (400/400/600 nm) yields the smallest effective surface area fraction (24.8%) while Pattern 1 (500/300/500 nm) generates the largest surface area fraction (36.6%) among these patterns.
Table 2.
Characterization of PUU submicron textured pillar patterns
| Pattern # | Pattern (D/Sr/H) (nm) Pillar shape | Pillar height (nm) | Distance in row (Sr) (nm ) | Distance in diagonal (Sd,) (nm) | Pillar top surface area (μm2) | Surface area fraction (%) |
|---|---|---|---|---|---|---|
|
| ||||||
| 1 | 500/300/500 Square | 467.7 ±10.2 | 319.5 ±21.7 | 576.5 ±17.2 | 0.243 ±0.018 | 36.6 |
| 2 | 700/700/300 Square | 307.6 ±13.2 | 622.5 ±21.7 | 1012.7 ±25.9 | 0.478 ±0.045 | 29.1 |
| 3 | 500/500/600 Square | 565.8 ±21.1 | 481.6 ±23.5 | 799.6 ±32.5 | 0.291 ±0.029 | 28.9 |
| 4 | 400/400/600 Round | 585.6 ±18.5 | 374.7 ±20.5 | 677.2 ±20.4 | 0.155 ±0.010 | 24.8 |
| 5 | 500/500/600 Round | 601.8 ±9.8 | 453.8 ±22.2 | 850.6 ±37.2 | 0.279 ±0.030 | 26.7 |
| Smooth | -- | -- | -- | -- | 100.0 | |
Note: Surface area fraction is the ratio of total pillar top surface area and total 2-D projected surface area including both the pillar top surface and bottom surface between pillars, but not the pillar sidewalls.
3.2. Surface wettability of textured PUU surfaces.
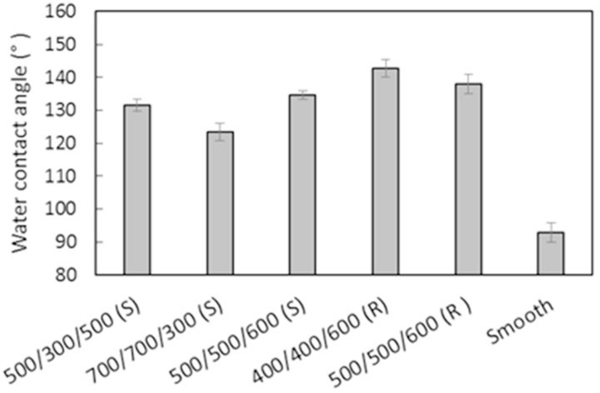
The smooth Biospan® MS/0.4 PUU surface is hydrophobic with water contact angle of 92.8±3°. The surface texturing modification dramatically increases water contact angles due to air captured in the spaces between pillars (Figure 3). The increase in water contact angles varied with surface area fractions and pillar sizes based on Cassie-Baxter model41. Pattern 4 (400/400/600 nm) produced the most hydrophobic surface with contact angle of 142.7±2.6°, about 50° higher than the smooth surfaces, while Pattern 2 (700/700/300 nm) increases the water contact angle only ~30.6° more than the smooth surface, suggesting that reducing the top surface fraction and the sizes of pillars yields the most hydrophobic surfaces.
Figure 3.
Surface wettability of submicron textured surfaces (S=square, R=round). Data is mean ± standard deviation.
3.3. Mechanical integrity of pillars.
Figure 4 shows the AFM topography images of textured PUU films with different patterns after soaking in MEK/MeOH solvent for 2 h. No pillar collapse was observed on the 700/700/300 (S) pattern which has the smalled aspect ratio of pillars (H/D = 0.43) (Table 1). A small number of radomly collapsed pillars were detected on patterns of 500/300/500 (S) and 500/500/600 (S), however, all pillars were found collapsed on 400/400/600 (R) pattern having the largest aspect ratio of 1.40, while most of the pillars were collapsed on 500/500/600 (R) pattern with only a few pillars left standing. It is interesting to see the collapsed pillars are likely to touch each other over every 2–4 adjacent pillars and form a new pattern with only a few of random pillars still standing on pattern 5, 500/500/600 (R). Results show that the mechanical integrity of pillars depends on the size and aspect ratio of pillars, and square shaped pillars are stronger than round shaped pillars. For example, pattern 500/500/600 (S) and pattern 500/500/600 (R) have the similar size and height of pillars, but fewer pillars were collapsed on square shaped pillar pattern.
Figure 4.
AFM topography images of textured films after soaking in MEK/MeOH for 2 h. Patterns of (A) 500/300/500 (S), (B) 700/700/300 (S), (C) 500/500/600 (S), (D) 400/400/600 (R) and (E) 500/500/600 (R).
3.4. Bacterial adhesion on textured PUU surfaces.
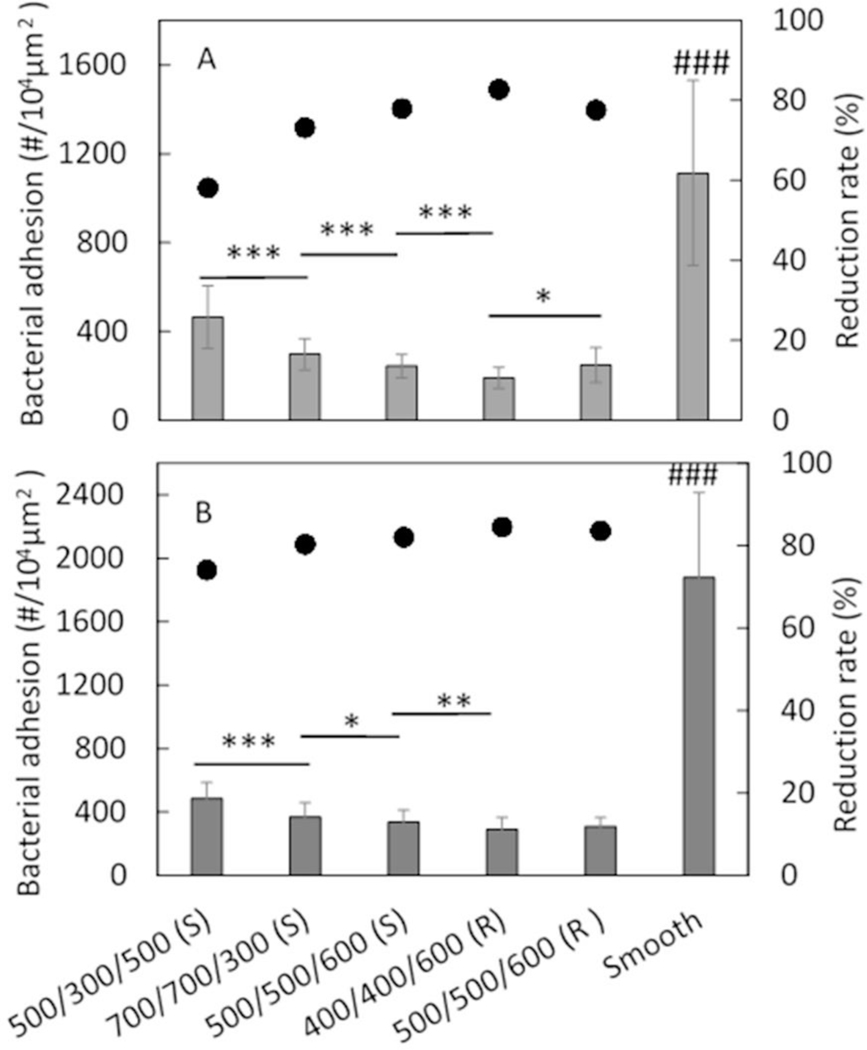
Bacterial adhesion on textured surfaces was assessed in PBS under near static conditions with slow shaking, and examined by optical fluorescence microscopy. The adhesion of bacterial strains S. epidermidis RP62A and S. aureus Newman D2C on smooth control and a variety of textured PUU surfaces is illustrated in Figure 5. A significant reduction in bacterial adhesion was observed on all textured surfaces compared to smooth surfaces (p<0.001), demonstrating that submicron textured surfaces inhibit bacterial adhesion. The adhesion reduction rates of textured surfaces varied over the ranges of 58.2–82.7% for S. epidermidis (Figure 5A) and 74.1–84.5% for S. aureus (Figure 5B), depending on textured patterns. Generally, the reduction rates of bacterial adhesion were in the order of Pattern 1 <Pattern 2 < Pattern 3 and Pattern 5 < Pattern 4. No significance was observed between Patterns 3 and 5 which have same size of pillars but with different shape of pillars, suggesting shape of pillar is not important for bacterial adhesion rates in this study.
Figure 5.
Bacterial adhesion on textured surfaces in PBS for 1 h with (A) S. epidermidis RP62A, and (B) S. aureus Newman D2C (S=square, R=round). (*: p<0.05; **: p<0.01; ***: p<0.001, ###: p<0.001 comparing to all textured surfaces.) Data is shown as mean ± standard deviation for n=3 samples. Bar and left axis show the bacterial adhesion, and dots and the right axis show the reduction rate compared to smooth PUU control.
The statistical analysis of adhesion data was further performed among the textured surfaces and results show that texture pattern significantly influences bacterial adhesion. Among textured patterns, Pattern 4 (400/400/600 (R)) exhibits the lowest bacterial adhesion and Pattern 1 (500/300/500 (S)) shows higher bacterial adhesion than all of other patterns. This trend is generally consistent with surface area fraction and surface wettability of textured patterns except for the wettability of Pattern 2 (Table 2 and Figure 3), suggesting that surface area fraction and surface wettability may be two main factors influencing bacterial adhesion for the topography modified surfaces.
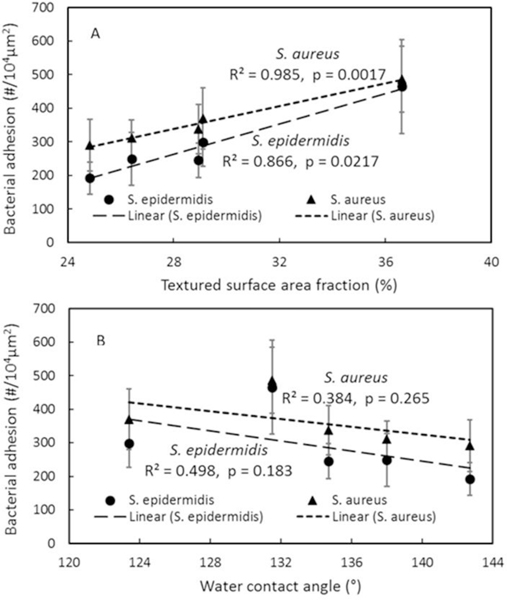
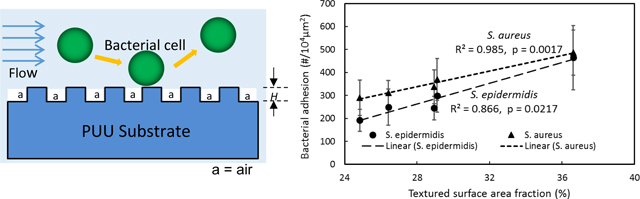
To further understand the correlation of surface area fraction and hydrophobicity with bacterial adhesion, adhesion data were linearly fit against these two factors as shown in Figure 6. It is interesting to see the bacterial adhesion linearly increased with surface area fraction with R2 value of 0.866 and p value of 0.0217 for S. epidermidis, however, the linear correlation between adhesion and water contact angle is not significant (R2 = 0.498, p =0.183). Similar results were also observed from S. aureus adhesion on textured surfaces (Figure 6).
Figure 6.
Correlation of bacterial adhesion with (A) textured surface fraction, (B) textured surface water contact angle.
3.5. Bacterial adhesion behaviors on textured surfaces.
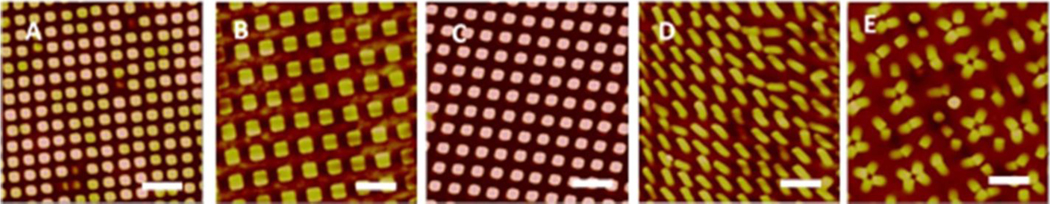
Staphylococcus bacterial adhesion on textured surfaces was also characterized by AFM in air. The AFM images show that individual bacteria or small clusters of 2–4 cells often adhered on the top surface areas of adjacent pillars to reach the maximum contact area on all textured surfaces (Figure 7). Only a few cells were observed to be captured in the gap spaces between pillars such as on Pattern 2 surface with larger pillar diagonal distance, although some cells appear to be deformed (indicated by blue arrow in Figure 7B). We also observed some cells sitting on the pillar but the cell height is less than the pillar height, and it seems possible that these bacteria are experiencing pillar penetration through the cell, e.g., cell on Pattern 4 (Figure 7D), indicating that the sharp polymer pillars may also have some bactericidal effects and inhibit biofilm formation.
Figure 7.
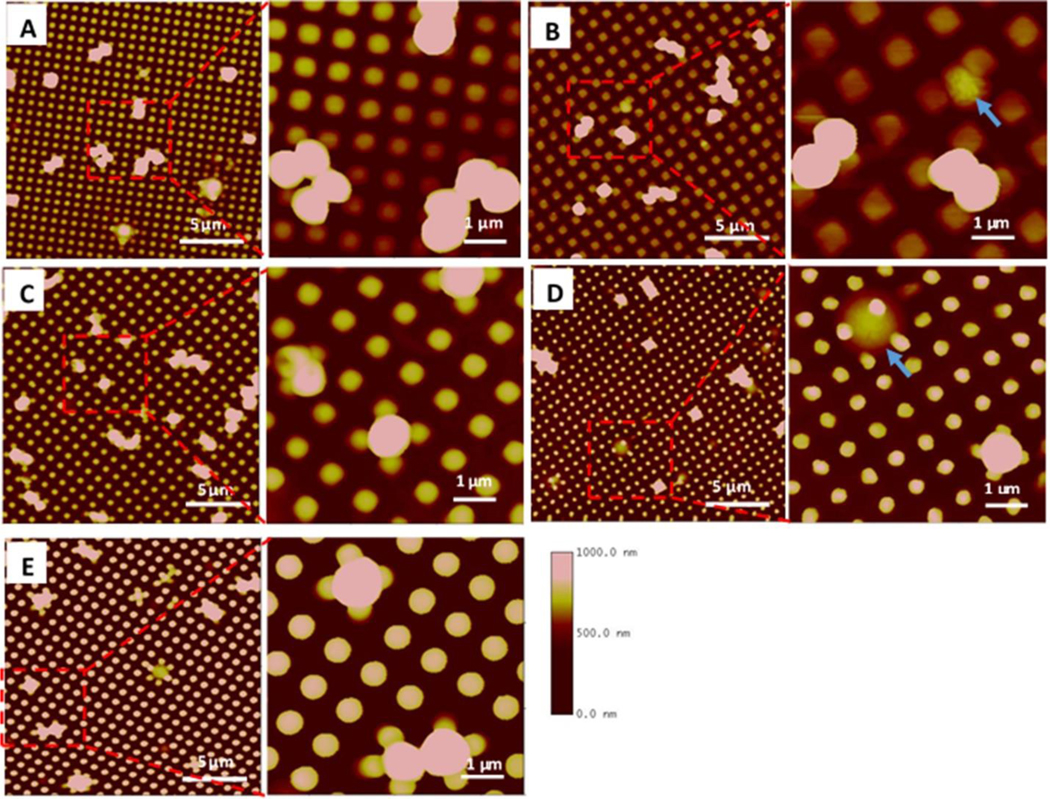
AFM images of S. aureus adhesion on textured surfaces. Square shaped pillars: (A) Pattern 1, 500/300/500 nm, (B) Pattern 2, 700/700/300 nm, (C) Pattern 3, 500/500/600 nm. Round shaped pillars: (D) Pattern 4, 400/400/600 nm, (E) Pattern 5, 500/500/600 nm. Blue arrows show bacterial cells captured in pattern 700/700/300 nm or penetrated by the pillar in 400/400/600 nm textures.
3.6. Biofilm formation inhibition by textured surfaces.
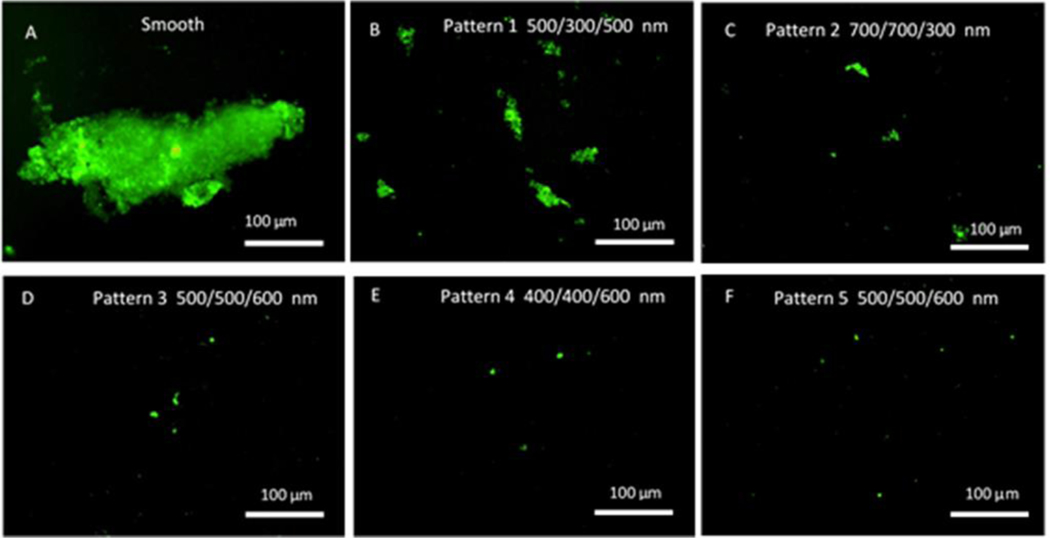
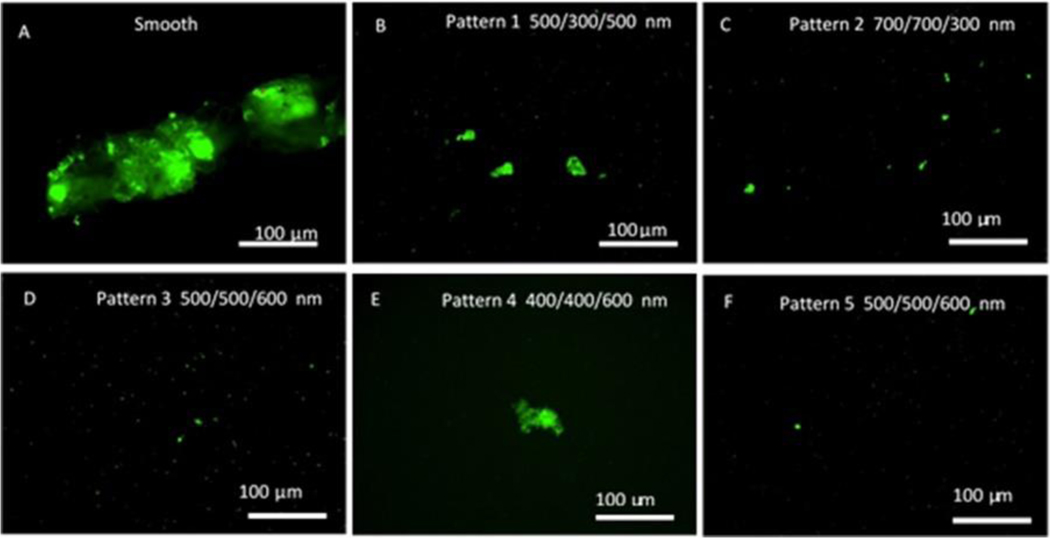
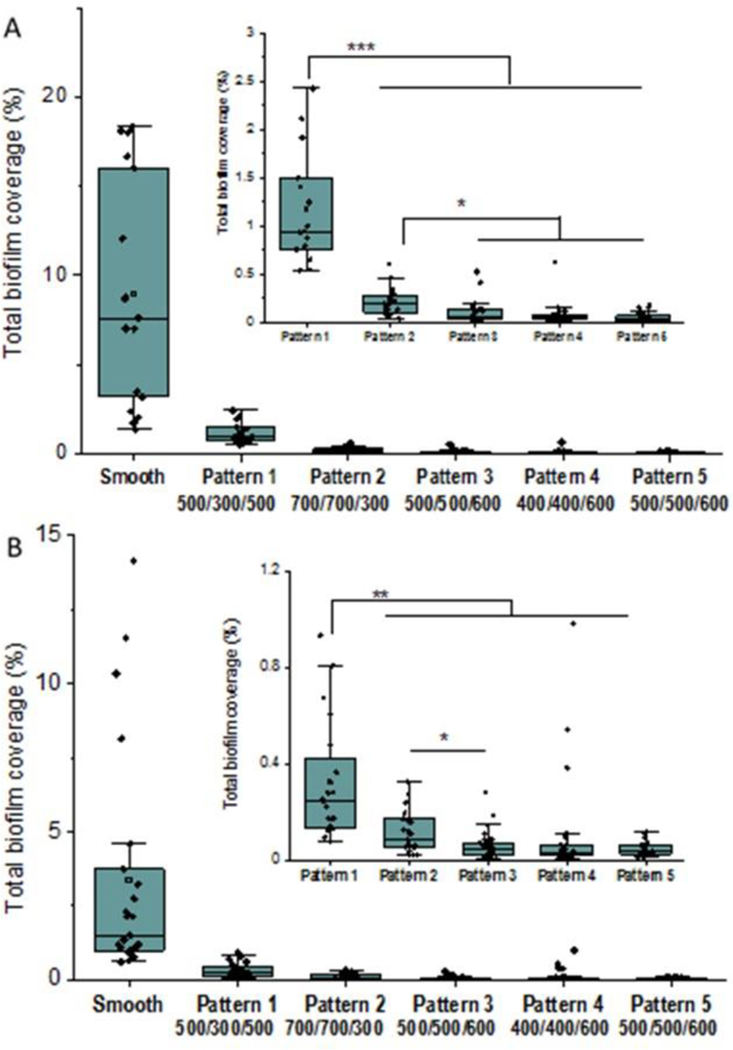
Biofilm formation assays were carried out in a CDC biofilm reactor. The CDC biofilm reactor has been utilized in many studies to mimic the environment for bacterial growth and biofilm formation on the material surfaces under moderate fluid sheer stress42. Bacterial adhesion and biofilms can be assessed by enumeration of cells43 or through imaging by microscopy techniques44,45. In this study all textured and smooth films were included in the same reactor to ensure all samples were exposed to the same environment and conditions and sample surfaces were imaged by fluorescent optical microscopy after labeling. Figures 8 and 9 illustrate representative fluorescence optical microscopy images of material surfaces after exposure to S. epidermidis culture for 10 days and S. aureus culture for 14 days, respectively. Large areas of biofilm were observed on PUU MS/0.4 smooth surfaces for both strains while only small patches or bacterial clusters were occasionally observed on textured surfaces. For example, a large biofilm of S. epidermidis RP62A with area of ~350 μm×100 μm was observed on the smooth surface (Fig. 8A) and the largest biofilm patch observed on Pattern 1 surface was only about 56 μm×13 μm (Fig. 8B). Results suggest the textured surfaces significantly inhibited biofilm formation compared to the smooth surfaces. To compare the biofilm patches formed on the textured surfaces, we defined 10 μm2 as biofilms in this study. This was based on our observations of bacterial aggregates/clusters vs larger structures that appear to take on more characteristics of a biofilm. Total biofilm coverages on material surfaces were analyzed by ImageJ program and results are illustrated in Figure 10. Results show that smooth PUU surfaces were covered with biofilms at 9.0±6.4% after exposure to S. epidermidis RP62A for 10 days while the biofilm coverages on textured surfaces were in the range of 0.05 – 1.2%. Specifically, the biofilm coverages on Patterns 3, 4, and 5 surfaces varied in the range of 0.05–0.1%, an approximately 2 log reduction in coverage compared to smooth surfaces. Comparing the biofilm coverages on textured surfaces, the average biofilm coverages are in the order of Pattern 1 > Pattern 2 > Pattern 3, 4 and 5 (Figure 10A). Similar results were obtained on material surfaces exposed to S. aureus Newman D2C culture for 14 days although the biofilm coverages were generally smaller than for S. epidermidis. The average biofilm coverage of S. aureus on smooth surfaces was about 3.4±3.9% while the average coverages of biofilms on textured surfaces varied in the range of 0.05–0.3%. The textured surfaces exhibited similar trends in biofilm coverages as observed for the textured materials with S. epidermidis (Figure 10B). However, it should be noted that some large biofilms were observed on one sample of Pattern 4. This may be due to defects in pillars on this material after long exposure to culture medium.
Figure 8.
Representative fluorescence optical microscopy images of S. epidermidis RP62A biofilms on smooth and textured surfaces in CDC biofilm reactor for 10 days.
Figure 9.
Representative fluorescence optical microscopy images of S. aureus Newman biofilms on smooth and textured surfaces in CDC biofilm reactor for 14 days.
Figure 10.
Total biofilm coverage on material surfaces (n=3 samples) exposed to (A) S. epidermidis for 10 days (B) S. aureus for 14 days. (*: p<0.05; **: p<0.01; ***: p<0.001). Inset graphs show only the textured surfaces for easier comparison.
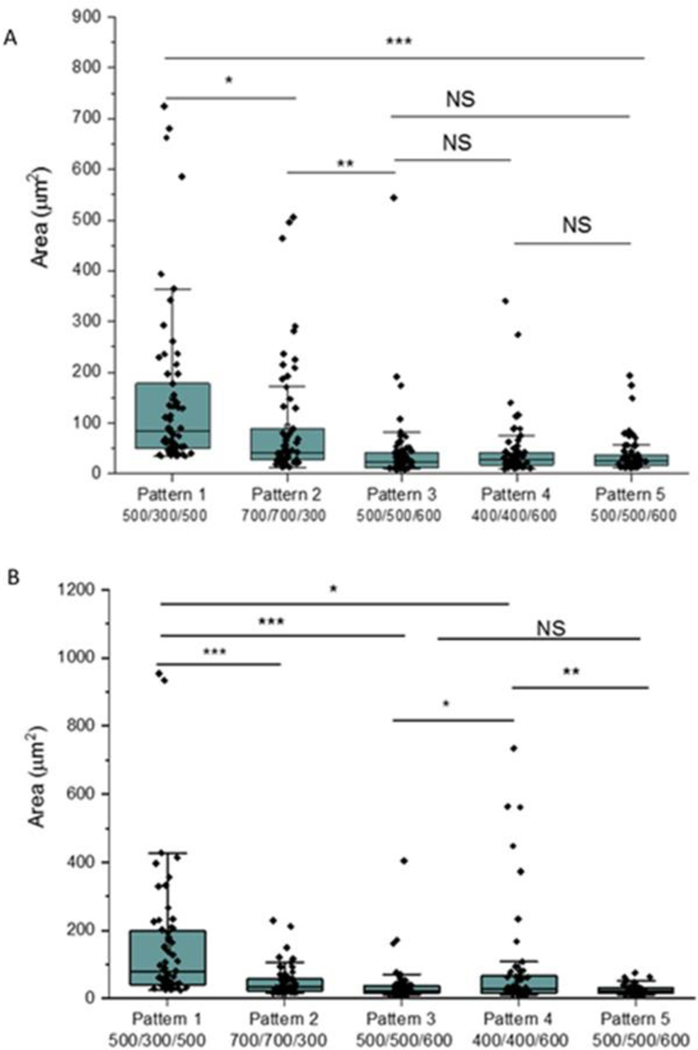
To further understand the biofilm inhibition on textured surfaces, a total of 60 individual biofilm patches with the largest areas were selected from the images of each pattern that were analyzed for total biofilm coverage. The distributions of the individual biofilm areas are shown in Figure 11. For S. epidermidis, the largest area for biofilm patches on Pattern 1 was ~725 μm2, and the mean area was 149±163 μm2, significantly higher than the biofilms observed on other textured surfaces. The largest areas of biofilm patches on Patterns 2 and 3 were found to be 506 μm2 and 544 μm2, and mean areas were 93±115 μm2 and 42±74 μm2, respectively. Pattern 4 and Pattern 5 exhibited less biofilm formation, and the biofilm patches were 44 ± 57 μm2 and 36 ±37 μm2 with largest areas of 341 and 194 μm2, respectively (Figure 10A). Similar results were found for S. aureus biofilms on textured surfaces. Pattern 1 shows larger biofilm formation than other patterns. Pattern 5 exhibited the smallest biofilm formation, with the average area of 25±14 μm2, but no significant difference was observed comparing to Pattern 3, suggesting the shape of pillar is not important in inhibition of biofilm formation. As previously mentioned, some larger biofilms were observed on Pattern 4 surfaces, possibly due to the defects of pillars with long time exposure (Figure 11B). Taken together, the data show that Pattern 3 and Pattern 5 appear the best textures for long term inhibition of Staphylococcus bacterial biofilm formation.
Figure 11.
Image analysis of area sizes for individual biofilm patches or bacterial clusters on textured surfaces (n=3 samples). (A) S. epidermidis for 10 days and (B) S. aureus for 14 days. (*: p<0.05; **: p<0.01; ***: p<0.001; NS=not significant)
4. DISCUSSION
Bacterial associated microbial infection due to the pathogen bacterial adhesion and biofilm formation is surface dependent. Surface topography modification with micro- or nano-sized structures has been a promising approach to mitigate biofilm development and control microbial infection11,46–50. Our previous studies36,37 have shown that the pillar textured surfaces, either submicron or micron size, can significantly reduce bacterial adhesion, but the micron textured surfaces are largely dependent on surface wettability. Also, we found that submicron textured surfaces have higher efficiency than micron size patterns in controlling bacterial adhesion, especially under low shear stress37. Thus, in this study, we focus on bacterial adhesion on submicron textured surface modification with ordered pillars and explore the structure of pillar patterns to provide the rationale for the design of such biomaterial surfaces.
Pillar geometry including shape, size, height, and spacing (Sr and Sd in Fig. 1) is important in design of textured surface modification. These parameters determine the surface accessible contact area and wettability that influence bacterial interactions at interfaces. Submicron pillar textured surfaces reduce accessible surface contact area for staphylococcal bacterial interactions since the space between pillars are smaller than the dimension of bacterial cells. In this study, Pattern 4 with smallest top surface contact area shows the highest reduction of bacterial adhesion while Pattern 1 having highest surface contact area exhibits the highest bacterial adhesion among a variety of patterns (Fig. 5). The trend of bacterial adhesion is consistent with the order of surface contact area of patterns and verified with linear correlation between surface contact area and adhesion (p<0.05, Fig. 6A), suggesting that surface contact area correlates with bacterial adhesion, i.e., the smaller the surface contact area, the lower the bacterial adhesion.
Small surface contact area can be reached through reducing pillar diameter and increasing the separation distance between pillars. However, smaller sizes of pillars will weaken the mechanical integrity of pillars, especially for polymeric materials, which can result in pillar collapse and defects on patterns. The pillar strength depends on pillar height aspect ratio (H/D), shape, and material. Pillars with high aspect ratio are easy to collapse. It appears the square-shaped pillars with low aspect ratio (less than 1.20) are strong enough to prevent collapse for textured patterns such as Patterns 1, 2, and 3 (Fig. 4) in the presence of organic solvent. Pillars on Patterns 4 and 5 with round shape and high aspect ratio are weak and collapsed in solvent. This weakness may cause the pillars to collapse during exposure to bacterial culture medium resulting in large area biofilm patches formed in these areas although the initial bacterial adhesion is low, as seen in Pattern 4 (Fig. 11).
Increasing pillar spacing also decreases the surface contact area, however, large pillar spaces greater than bacterium dimension may increase the opportunity for bacterial cells to access and to be trapped in the gaps between pillars. We observed bacterial cells trapped in spaces between pillars on Pattern 2, having a separation distance of 700 nm, while no cells were trapped in other patterns (Fig. 7), suggesting the pillar separation should be at least less than 700 nm for textured patterns. Both pillar collapse and larger pillar spacing increase bacterial colonization and biofilm formation. Therefore, the design of textured patterns needs to consider minimizing the surface contact area but at the same time ensuring the mechanical integrity of pillars, while also avoiding giving opportunity for cells to be trapped in inter-pillar spaces.
Surface wettability is another important factor influencing bacterial adhesion. Surface texturing dramatically increases the surface hydrophobicity due to the air trapped in the spaces between features. The super-hydrophobic property of textured surfaces is often regarded to be a main reason contributing to self-cleaning and low adhesion properties of natural or biomimetic artificial surfaces49,51. In this study, all textured surfaces exhibited lower adhesion, suggesting that while the increase in hydrophobicity of materials may help to prevent the interaction of bacterium and material surface and mitigate the bacterial adhesion, there is no direct correlation between hydrophobicity and adhesion observed among these textured surfaces, suggesting that the small increase in hydrophobicity of textured materials by the changes of pillar size and distribution is less important than the surface contact area (Fig. 6B). This is consistent with observations in our previous study, where we found that submicron textured surfaces were significantly resistant to bacterial adhesion regardless whether the textured surface was hydrophobic or hydrophilic, while the super-hydrophobic property was effective in inhibiting bacterial adhesion on micro-scale textured surfaces37.
5. CONCLUSIONS
Polyurethane biomaterial surfaces were textured with ordered arrays of pillars in a variety of submicron size patterns and pillar shapes. All textured surfaces dramatically increased surface hydrophobicity due to air trapped in spaces between pillars, and present reduced surface contact area for interactions between bacteria and surfaces, thereby significantly reducing the adhesion of S. epidermidis and S. aureus while also inhibiting biofilm formation. Among the textured patterns, Pattern 4 having dimensions of 400/400/600 nm had the lowest fractional surface area coverage and also exhibited the lowest bacterial adhesion, but Patterns 3 and 5 with dimensions of 500/500/600 nm were the most efficient in inhibiting biofilm formation for long term exposure times. Surface contact area appears to be the dominant factor influencing bacterial adhesion. Design of textured surfaces for reducing biomaterial associated microbial infection should consider ways to minimize the surface contact area such as reducing pillar size and increasing the spaces between pillars, but at the same time retaining pillar integrity to avoid defects in patterns resulting from either missing pillars or collapse of pillars from insufficient strength.
ACKNOWLEDGEMENT
The authors acknowledge financial support from NIH R01 HL153231 and R21 AI139706.
Footnotes
CONFLICT OF INTEREST
The authors declare no potential conflict of interest.
REFERENCES
- 1.Darouiche RO. Treatment of Infections Associated with Surgical Implants. New England Journal of Medicine 2004;350(14):1422–1429. [DOI] [PubMed] [Google Scholar]
- 2.VanEpps JS, Younger JG. Implantable Device-Related Infection. Shock 2016;46(6):597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. Pharmacy and Therapeutics 2015;40(4):277. [PMC free article] [PubMed] [Google Scholar]
- 4.Martens E, Demain AL. The antibiotic resistance crisis, with a focus on the United States. J Antibiot 2017;70(5):520–526. [DOI] [PubMed] [Google Scholar]
- 5.Noskin GA, Rubin RJ, Schentag JJ, Kluytmans J, Hedblom EC, Smulders M, Lapetina E, Gemmen E. The burden of Staphylococcus aureus infections on hospitals in the United States: an analysis of the 2000 and 2001 Nationwide Inpatient Sample Database. Arch Intern Med 2005;165(15):1756–61. [DOI] [PubMed] [Google Scholar]
- 6.Cheng H, Yuan W, Zeng F, Hu Q, Shang W, Tang D, Xue W, Fu J, Liu J, Liu N and others. Molecular and phenotypic evidence for the spread of three major methicillin-resistant Staphylococcus aureus clones associated with two characteristic antimicrobial resistance profiles in China. J Antimicrob Chemother 2013;68(11):2453–7. [DOI] [PubMed] [Google Scholar]
- 7.Upreti N, Rayamajhee B, Sherchan SP, Choudhari MK, Banjara MR. Prevalence of methicillin resistant Staphylococcus aureus, multidrug resistant and extended spectrum β-lactamase producing gram negative bacilli causing wound infections at a tertiary care hospital of Nepal. Antimicrob Resist Infect Control 2018;7(121):018–0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kot B, Wierzchowska K, Piechota M, Grużewska A. Antimicrobial Resistance Patterns in Methicillin-Resistant Staphylococcus aureus from Patients Hospitalized during 2015–2017 in Hospitals in Poland. Medical Principles and Practice 2020;29(1):61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li X, Wu B, Chen H, Nan K, Jin Y, Sun L, Wang B. Recent developments in smart antibacterial surfaces to inhibit biofilm formation and bacterial infections. Journal of Materials Chemistry B 2018;6(26):4274–4292. [DOI] [PubMed] [Google Scholar]
- 10.Ipe DS, Kumar PTS, Love RM, Hamlet SM. Silver Nanoparticles at Biocompatible Dosage Synergistically Increases Bacterial Susceptibility to Antibiotics. Frontiers in Microbiology 2020;11(1074). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham M, Cady N. Nano and Microscale Topographies for the Prevention of Bacterial Surface Fouling. Coatings 2014;4(1):37–59. [Google Scholar]
- 12.Rigo S, Cai C, Gunkel-Grabole G, Maurizi L, Zhang X, Xu J, Palivan CG. Nanoscience-Based Strategies to Engineer Antimicrobial Surfaces. Advanced Science 2018;5(5):1700892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Echeverria C, Torres MDT, Fernández-García M, de la Fuente-Nunez C, Muñoz-Bonilla A. Physical methods for controlling bacterial colonization on polymer surfaces. Biotechnology Advances 2020;43:107586. [DOI] [PubMed] [Google Scholar]
- 14.Yang K, Shi J, Wang L, Chen Y, Liang C, Yang L, Wang L-N. Bacterial anti-adhesion surface design: surface patterning, roughness and wettability: A review. Journal of Materials Science & Technology 2021:in press.
- 15.Fu J, Zhang H, Guo Z, Feng D-q, Thiyagarajan V, Yao H. Combat biofouling with microscopic ridge-like surface morphology: a bioinspired study. Journal of The Royal Society Interface 2018;15(140):20170823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung KK, Schumacher JF, Sampson EM, Burne RA, Antonelli PJ, Brennan AB. Impact of engineered surface microtopography on biofilm formation of Staphylococcus aureus. Biointerphases 2007;2(2):89–94. [DOI] [PubMed] [Google Scholar]
- 17.Ivanova EP, Nguyen SH, Guo Y, Baulin VA, Webb HK, Truong VK, Wandiyanto JV, Garvey CJ, Mahon PJ, Mainwaring DE and others. Bactericidal activity of self-assembled palmitic and stearic fatty acid crystals on highly ordered pyrolytic graphite. Acta Biomaterialia 2017;59(Supplement C):148–157. [DOI] [PubMed] [Google Scholar]
- 18.Sun T, Qing G, Su B, Jiang L. Functional biointerface materials inspired from nature. Chemical Society Reviews 2011;40(5):2909–2921. [DOI] [PubMed] [Google Scholar]
- 19.Sullivan T, O’Callaghan I. Recent Developments in Biomimetic Antifouling Materials: A Review. Biomimetics 2020;5(4):58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damodaran VB, Murthy SN. Bio-inspired strategies for designing antifouling biomaterials. Biomaterials Research 2016;20(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsiapalis D, De Pieri A, Biggs M, Pandit A, Zeugolis DI. Biomimetic Bioactive Biomaterials: The Next Generation of Implantable Devices. ACS Biomaterials Science & Engineering 2017;3(7):1172–1174. [DOI] [PubMed] [Google Scholar]
- 22.Faustino CMC, Lemos SMC, Monge N, Ribeiro IAC. A scope at antifouling strategies to prevent catheter-associated infections. Advances in Colloid and Interface Science 2020;284:102230. [DOI] [PubMed] [Google Scholar]
- 23.Lee SW, Phillips KS, Gu H, Kazemzadeh-Narbat M, Ren D. How microbes read the map: Effects of implant topography on bacterial adhesion and biofilm formation. Biomaterials 2021;268:120595. [DOI] [PubMed] [Google Scholar]
- 24.Reddy ST, Chung KK, McDaniel CJ, Darouiche RO, Landman J, B BA . Micropatterned Surfaces for Reducing the Risk of Catheter-Associated Urinary Tract Infection: An In Vitro Study on the Effect of Sharklet Micropatterned Surfaces to Inhibit Bacterial Colonization and Migration of Uropathogenic Escherichia coli. Journal of Endourology 2011;25(9):1547–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schumacher JF, Long CJ, Callow ME, Finlay JA, Callow JA, Brennan AB. Engineered nanoforce gradients for inhibition of settlement (attachment) of swimming algal spores. Langmuir 2008;24(9):4931–4937. [DOI] [PubMed] [Google Scholar]
- 26.Linklater DP, Baulin VA, Juodkazis S, Crawford RJ, Stoodley P, Ivanova EP. Mechano-bactericidal actions of nanostructured surfaces. Nature Reviews Microbiology 2021;19(1):8–22. [DOI] [PubMed] [Google Scholar]
- 27.Pogodin S, Hasan J, Baulin Vladimir A, Webb Hayden K, Truong Vi K, Phong Nguyen The H, Boshkovikj V, Fluke Christopher J, Watson Gregory S, Watson Jolanta A and others. Biophysical Model of Bacterial Cell Interactions with Nanopatterned Cicada Wing Surfaces. Biophysical Journal 2013;104(4):835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivanova EP, Hasan J, Webb HK, Truong VK, Watson GS, Watson JA, Baulin VA, Pogodin S, Wang JY, Tobin MJ and others. Natural Bactericidal Surfaces: Mechanical Rupture of Pseudomonas aeruginosa Cells by Cicada Wings. Small 2012;8(16):2489–2494. [DOI] [PubMed] [Google Scholar]
- 29.Linklater DP, De Volder M, Baulin VA, Werner M, Jessl S, Golozar M, Maggini L, Rubanov S, Hanssen E, Juodkazis S and others. High Aspect Ratio Nanostructures Kill Bacteria via Storage and Release of Mechanical Energy. Acs Nano 2018;12(7):6657–6667. [DOI] [PubMed] [Google Scholar]
- 30.Ivanova EP, Hasan J, Webb HK, Gervinskas G, Juodkazis S, Truong VK, Wu AHF, Lamb RN, Baulin VA, Watson GS and others. Bactericidal activity of black silicon. Nature communications 2013;4(1):2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu S, Zuber F, Brugger J, Maniura-Weber K, Ren Q. Antibacterial Au nanostructured surfaces. Nanoscale 2016;8(5):2620–2625. [DOI] [PubMed] [Google Scholar]
- 32.Cheng Y, Feng G, Moraru CI. Micro- and Nanotopography Sensitive Bacterial Attachment Mechanisms: A Review. Frontiers in Microbiology 2019;10(191). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng G, Cheng Y, Wang S-Y, Borca-Tasciuc DA, Worobo RW, Moraru CI. Bacterial attachment and biofilm formation on surfaces are reduced by small-diameter nanoscale pores: how small is small enough? Npj Biofilms And Microbiomes 2015;1:15022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Helbig R, Gunther D, Friedrichs J, Ro, Lasagni A, Werner C. The impact of structure dimensions on initial bacterial adhesion. Biomaterials Science 2016;4(7):1074–1078. [DOI] [PubMed] [Google Scholar]
- 35.Lu N, Zhang W, Weng Y, Chen X, Cheng Y, Zhou P. Fabrication of PDMS surfaces with micro patterns and the effect of pattern sizes on bacteria adhesion. Food Control 2016;68:344–351. [Google Scholar]
- 36.Xu L-C, Siedlecki CA. Submicron-textured biomaterial surface reduces staphylococcal bacterial adhesion and biofilm formation. Acta Biomaterialia 2012;8(1):72–81. [DOI] [PubMed] [Google Scholar]
- 37.Xu L-C, Siedlecki CA. Staphylococcus epidermidis adhesion on hydrophobic and hydrophilic textured biomaterial surfaces. Biomedical Materials 2014;9(3):035003. [DOI] [PubMed] [Google Scholar]
- 38.Wenzel RP. Health care-associated infections: Major issues in the early years of the 21st century. Clinical Infectious Diseases 2007;45:85–88. [DOI] [PubMed] [Google Scholar]
- 39.Xia Y, Whitesides GM. Soft lithography. Annual Review of Materials Science 1998;28(1):153–184. [Google Scholar]
- 40.Milner KR, Snyder AJ, Siedlecki CA. Sub-micron texturing for reducing platelet adhesion to polyurethane biomaterials. Journal of Biomedical Materials Research Part A 2006;76(3):561–570. [DOI] [PubMed] [Google Scholar]
- 41.Lima AC, Mano JF. Micro-/nano-structured superhydrophobic surfaces in the biomedical field: part I: basic concepts and biomimetic approaches. Nanomedicine 2015;10(1):103–119. [DOI] [PubMed] [Google Scholar]
- 42.Goeres DM, Loetterle LR, Hamilton MA, Murga R, Kirby DW, Donlan RM. Statistical assessment of a laboratory method for growing biofilms. Microbiology 2005;151(3):757–762. [DOI] [PubMed] [Google Scholar]
- 43.Hopkins SP, Pant J, Goudie MJ, Schmiedt C, Handa H. Achieving Long-Term Biocompatible Silicone via Covalently Immobilized S-Nitroso-N-acetylpenicillamine (SNAP) That Exhibits 4 Months of Sustained Nitric Oxide Release. ACS Applied Materials & Interfaces 2018;10(32):27316–27325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agostinho A, James G, Wazni O, Citron M, Wilkoff BD. Inhibition of Staphylococcus aureus Biofilms by a Novel Antibacterial Envelope for Use with Implantable Cardiac Devices. Clinical and Translational Science 2009;2(3):193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Williams DL, Woodbury KL, Haymond BS, Parker AE, Bloebaum RD. A modified CDC biofilm reactor to produce mature biofilms on the surface of peek membranes for an in vivo animal model application. Curr Microbiol 2011;62(6):1657–63. [DOI] [PubMed] [Google Scholar]
- 46.Scardino AJ, Hudleston D, Peng Z, Paul NA, de Nys R. Biomimetic characterisation of key surface parameters for the development of fouling resistant materials. Biofouling 2009;25(1):83–93. [DOI] [PubMed] [Google Scholar]
- 47.Scardino AJ, Zhang H, Cookson DJ, Lamb RN, de Nys R. The role of nano-roughness in antifouling. Biofouling 2009;25(8):757–767. [DOI] [PubMed] [Google Scholar]
- 48.Barthlott W, Neinhuis C. Purity of the sacred lotus, or escape from contamination in biological surfaces. Planta 1997;202(1):1–8. [Google Scholar]
- 49.Bhushan B, Jung YC. Natural and biomimetic artificial surfaces for superhydrophobicity, self-cleaning, low adhesion, and drag reduction. Progress in Materials Science 2011;56(1):1–108. [Google Scholar]
- 50.Khalid S, Gao A, Wang G, Chu PK, Wang H. Tuning surface topographies on biomaterials to control bacterial infection. Biomaterials Science 2020;8(24):6840–6857. [DOI] [PubMed] [Google Scholar]
- 51.Kavitha SA, Deeksha P, Deepika G, Nishanthini J, Hikku GS, Antinate Shilpa S, Jeyasubramanian K, Murugesan R. Super-hydrophobicity: Mechanism, fabrication and its application in medical implants to prevent biomaterial associated infections. Journal of Industrial and Engineering Chemistry 2020;92:1–17. [Google Scholar]