Abstract
Rhinovirus causes many types of respiratory illnesses, ranging from minor colds to exacerbations of asthma. Moraxella catarrhalis is an opportunistic pathogen that is increased in abundance during rhinovirus illnesses and asthma exacerbations and is associated with increased severity of illness through mechanisms that are ill-defined. We used a co-infection model of human airway epithelium differentiated at the air-liquid interface to test the hypothesis that rhinovirus infection promotes M. catarrhalis adhesion and survival on the respiratory epithelium. Initial experiments showed that infection with M. catarrhalis alone did not damage the epithelium or induce cytokine production, but increased trans-epithelial electrical resistance, indicative of increased barrier function. In a co-infection model, infection with the more virulent rhinovirus-A and rhinovirus-C, but not the less virulent rhinovirus-B types, increased cell-associated M. catarrhalis. Immunofluorescent staining demonstrated that M. catarrhalis adhered to rhinovirus-infected ciliated epithelial cells and infected cells being extruded from the epithelium. Rhinovirus induced pronounced changes in gene expression and secretion of inflammatory cytokines. In contrast, M. catarrhalis caused minimal effects and did not enhance RV-induced responses. Our results indicate that rhinovirus-A or C infection increases M. catarrhalis survival and cell association while M. catarrhalis infection alone does not cause cytopathology or epithelial inflammation. Our findings suggest that rhinovirus and M. catarrhalis co-infection could promote epithelial damage and more severe illness by amplifying leukocyte inflammatory responses at the epithelial surface.
Keywords: Moraxella catarrhalis, rhinovirus, airway epithelium, co-infection, asthma
Introduction
Rhinovirus (RV) infections can cause respiratory illnesses ranging from mild colds to lower respiratory tract infections with wheezing. The RV-A and RV-C types are particularly associated with wheezing illnesses and hospitalizations due to asthma exacerbations (Bizzintino et al., 2011; Lee et al., 2012). Moraxella catarrhalis is an opportunistic pathogen that can cause otitis media, sinusitis, and exacerbations of chronic obstructive pulmonary disease. Moraxella catarrhalis abundance is also increased in nasal secretions of children during RV upper respiratory and wheezing illnesses (Kloepfer et al., 2014; Bashir et al., 2018). RV infections precede increased bacterial detection during these illnesses, and their concurrent or sequential detection is associated with an increased incidence of wheezing illnesses and asthma exacerbations (Kloepfer et al., 2014).
Respiratory viruses can promote secondary bacterial infections by inducing receptors that increase bacterial binding, which is the first step in bacterial invasiveness. In studies using several respiratory viruses, virus infections increased the expression of respiratory epithelial receptors that can be used by Haemophilus influenzae ( Sajjan et al., 2006; Avadhanula et al., 2006) and Streptococcus pneumoniae ( Avadhanula et al., 2006). Moraxella catarrhalis has several outer membrane proteins (OMPs) (e.g., UspA1, UspA2, and Hemagglutinin [Hag/MID]) that can bind to multiple cellular proteins, including carcinoembryonic antigen-related cell adhesion molecule (CEACAM) and matrix proteins, including fibronectin and laminin (Balder et al., 2009). CEACAM1, CEACAM5 (carcinoembryonic antigen or CEA), and 6 are expressed in the respiratory epithelium in varying co-expression patterns (Klaile et al., 2013). Secreted CEACAM5 and 6 are detected in normal bronchial mucus (Matsuoka et al., 1990; Klaile et al., 2013). CEACAM1 can serve as a receptor for the respiratory pathogens M. catarrhalis, non-typeable Haemophilus influenzae, and Neisseria meningitidis and the urogenital pathogen N. gonorrhoeae ( Virji et al., 2000; Hill et al., 2005). In addition to bacterial binding, CEACAM engagement also triggers endocytosis and transcytosis of N. gonorrhoeae allowing epithelial barrier function to remain intact (Wang et al., 1998). Virus-induced interferon responses may promote respiratory tract colonization by pathobionts, including M. catarrhalis, by dampening innate immune responses necessary for bacterial clearance (Sun and Metzger, 2008; de Steenhuijsen Piters et al., 2022). Finally, viral infections can disturb epithelial cell barrier function, and this activity can promote invasiveness. However, the specific mechanisms that cause RV-M. catarrhalis co-infections to increase the abundance of bacteria in the airways and the severity of illness are currently unknown.
In this study, we tested the hypothesis that RV infection promotes M. catarrhalis adhesion and survival on the respiratory epithelium by increasing the production of M. catarrhalis OMPs and upregulating CEACAM production on airway epithelial cells. To test this hypothesis, we used an in vitro model of differentiated bronchial epithelial cells and identified effects of RV co-infection on M. catarrhalis cell association, abundance, and cellular responses.
Results
Moraxella catarrhalis adherence and survival on differentiated airway epithelial cells
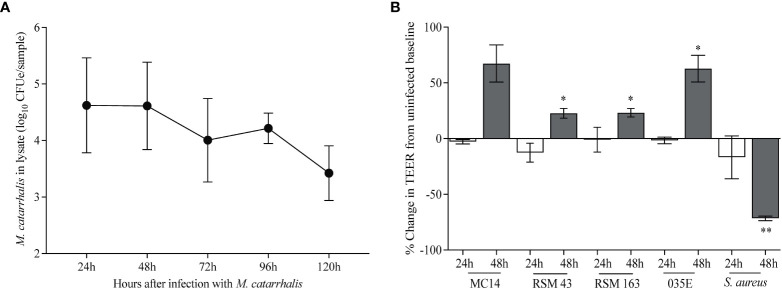
To determine the effects of M. catarrhalis on airway epithelial cells, we infected cultures of airway epithelial cells differentiated at air-liquid interface (ALI) with M. catarrhalis strain MC14 and monitored bacterial abundance and cellular responses. Over a 5-day incubation period, the number of cell-associated bacteria decreased by ~3 log units ( Figure 1A ). We did not observe any adverse effects of M. catarrhalis on bronchial epithelial cell morphology or ciliary motion throughout infection. Trans-epithelial resistance (TEER), a measure of epithelial barrier function, did not change at 24 h and tended to increase by 48 h post-infection ( Figure 1B ). This is in contrast to the effect of another common respiratory tract pathogen, S. aureus, which caused a decrease in TEER indicating damage to the epithelial barrier.
Figure 1.
M. catarrhalis survival on airway epithelium. Fully differentiated epithelial cell cultures from a single donor were infected with (A) M. catarrhalis strain MC14, and the infection was allowed to persist over five days. Cell-associated M. catarrhalis CFUe was quantified at each timepoint using quantitative PCR. The result is a combination of four independent experiments. Error bars represent geometric mean ± SD. (B) Percentage change in TEER at 24 and 48 h following infection with either M. catarrhalis MC14, RSM43, RSM163, 035E or a clinical isolate of S. aureus. Bars represent the mean ± SEM from 3 independent experiments (* P < 0.05, ** P < 0.01 vs. uninfected control; paired t-test).
Rhinovirus effects on M. catarrhalis association with epithelial cells
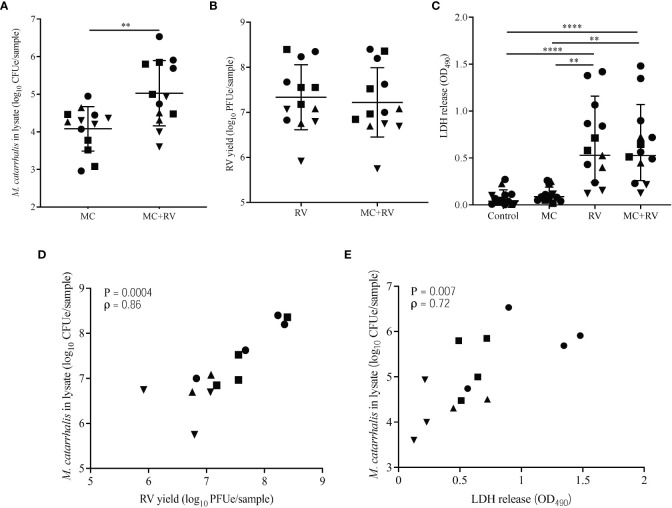
To determine the effects of RV infection on M. catarrhalis adherence and survival, we inoculated the apical surface of differentiated bronchial epithelium with RV-A16, followed 2 hours later by infection with a clinical isolate of M. catarrhalis (strain MC14). Repeat experiments in cells from four epithelial cell donors demonstrated that RV-A16 infection significantly increased cell-associated M. catarrhalis (geometric means for MC = 2.2×104, MC+RV = 4.9×105; P = 0.003) ( Figure 2A ). Notably, M. catarrhalis did not influence RV replication ( Figure 2B ). RV infection caused significant cytotoxicity, as measured by LDH release (mean for control = 0.08, MC = 0.11, RV = 0.67, MC+RV = 0.64; control vs RV or MC+RV, P < 0.0001) ( Figure 2C ) that was not altered by M. catarrhalis. Both RV replication (ρ = 0.86, P = 0.0004) ( Figure 2D ) and cytotoxicity (ρ = 0.72, P = 0.007) ( Figure 2E ) were positively correlated with cell-associated M. catarrhalis. These findings suggest that RV replication promotes M. catarrhalis attachment or persistence. It is of note that of the four BEC donors that we analyzed, two of them (denoted by circle and squares), had greater bacterial burden with RV co-infection than the other donors. These donors also had more RV replication and RV-induced cytotoxicity. These observations suggest that some individuals may be more susceptible to M. catarrhalis colonization during RV infection.
Figure 2.
Rhinovirus increases M. catarrhalis survival and cell adhesion. ALI cultures from one donor were infected apically with RV-A16 for 2 hours followed by M. catarrhalis strain MC14. Cell-associated M. catarrhalis CFU counts from infected ALI cultures were determined every 24 h for 5 days. Data representative of 4 independent experiments. Symbols represent the geometric mean ± SD (panels A-D). Cultures from four different donors were infected with RV-A16 (2 hours), and then with M. catarrhalis for 48 hours. Cell lysates were analyzed for (A) M. catarrhalis genomic DNA abundance (CFUe), (B) RV RNA abundance (PFUe), and (C) cytotoxicity of epithelial cells (LDH release). Each symbol represents a separate cell line. Bars represent geometric mean ± SD (for CFUe and PFUe) or mean ± SEM (for LDH release). Results are from at least two independent experiments per donor (unpaired t-test or Mann-Whitney test; ** P<0.01, **** P<0.0001). Correlations were tested between cell-associated M. catarrhalis and (D) RV-A16 replication or (E) LDH release (Spearman’s correlation).
Cellular response to M. catarrhalis
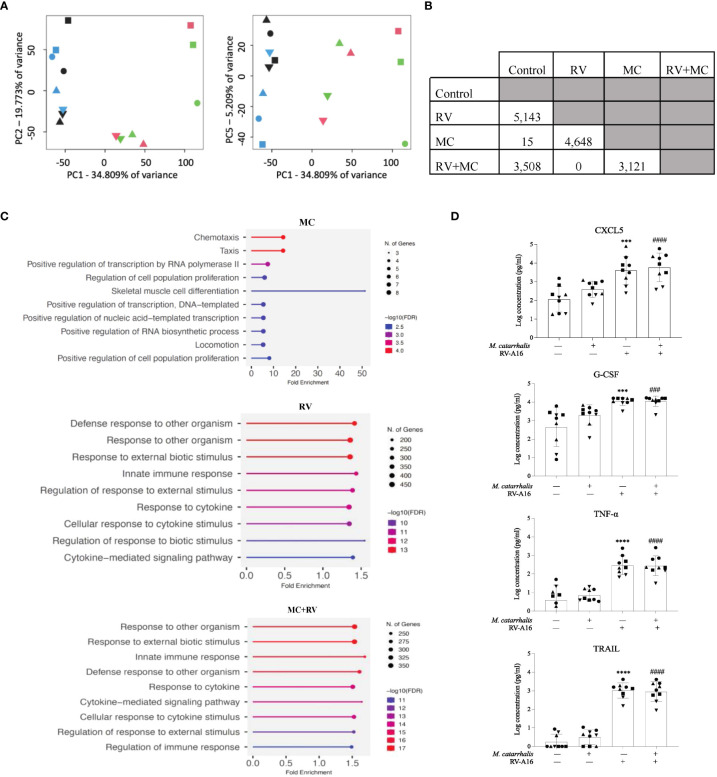
We next tested whether RV and/or M. catarrhalis infection influenced epithelial gene expression. Using principal component analysis (PCA), PC1 was related to RV infection and PC5 was related to M. catarrhalis infection ( Figure 3A ). RV induced a robust transcriptional response (5143 mRNAs), while M. catarrhalis alone upregulated only 15 mRNAs. There were no significant differences in gene expression between cells infected with RV vs. RV+MC ( Figure 3B ). Gene Ontology (GO) enrichment analysis of significantly expressed genes revealed that RV induced a number of innate immune and antiviral pathways. Figure 3C shows the top 10 enriched pathways for each treatment. M. catarrhalis downregulated the expression of 15 genes that are associated with chemotaxis and cell proliferation. Among them were the transcription factors early growth response (EGR) 1, 2, and 3, which are involved in mitogenesis and fibrinogenesis suggesting that M. catarrhalis could inhibit epithelial regeneration.
Figure 3.
Gene expression and cytokine analyses of ALI cultures infected with RV and M. catarrhalis. ALI cultures from epithelial cell were infected with RV-A16, M. catarrhalis or both as in previous experiments. Principal component analysis of RNA-seq data indicates clustering of samples by treatment. (A) PC1 separates RV treated samples from non-RV treated samples while PC5 separates M. catarrhalis treated from M. catarrhalis untreated samples. The treatment applied to ALI cultures is represented by symbol color: control (blue), RV (green), MC (black), and MC+RV (red). The donors are coded by symbol shape. (B) Differentially expressed genes in ALI cultures from a single epithelial cell donor infected with RV, M. catarrhalis (MC) or both (RV+MC). RNA-seq analysis, conducted using linear regression modeling, indicated differentially expressed genes between each group except between the RV treated and the RV+M. catarrhalis treated samples. For each comparison, the number indicates the number of differentially expressed genes after multiple testing correction per analysis (FDR < 0.05). (C) Gene enrichment analysis of differentially expressed genes following infection with RV, M. catarrhalis (MC) or both (RV+MC) using ShinyGO 0.76 (Ge et al., 2020). The pathways were sorted by -log10 (FDR) values with aunpn FDR cutoff of 0.05. The dotplots show the top 10 enriched pathways for each treatment. (D) Cytokine levels were measured in the medium of ALI cultures infected with RV-A16 and M. catarrhalis. Bars represent geometric mean + SD. #Control vs M. catarrhalis + RV; *Control vs RV; (n = minimum of two per donor; ordinary one-way ANOVA; three symbols P < 0.001, four symbols P < 0.0001).
We next tested whether M. catarrhalis altered the epithelial immune responses to RV. RV-A16 infection alone significantly increased secretion of chemokines (CXCL5, CXCL10, eotaxin-3, and G-CSF) and proinflammatory cytokines (IL-1β, IL-6, and TNF-α). RV-A16 infection also led to the secretion of TNF-related apoptosis-inducing ligand (TRAIL) which activates cell death by binding to the TRAIL receptor (Girkin et al., 2017). Figure 3D is a representation of the cytokines that were measured. As expected, RV-A16 infection also led to the secretion of the anti-viral cytokine IL-28A (IFN-λ2). Moraxella catarrhalis alone did not significantly induce cytokine secretion from the epithelium and did not alter RV-induced cytokine secretion.
Effects of RV species and bacterial isolate on cell-associated M. catarrhalis
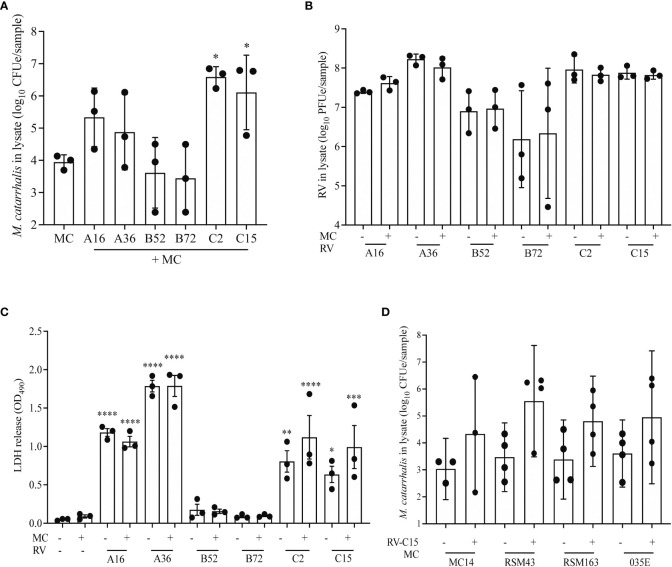
RV-A and RV-C cause more severe respiratory illness and are more often associated with increased detection of M. catarrhalis in vivo compared to RV-B (Bashir et al., 2018). To evaluate the differential effect of RV species on replication and M. catarrhalis cell adhesion and cell association, we infected fully differentiated ALI cultures from a single bronchial epithelial cell donor with two types of each RV species (A16, A35, B52, B72, C2, and C15). Compared to RV uninfected cells, both RV-C types significantly increased cell-associated M. catarrhalis while RV-B types did not (P = 0.0003 vs RV-C2; P = 0.03 vs RV-C15) ( Figure 4A ). M. catarrhalis did not affect RV replication ( Figure 4B ). Compared to RV-B types, RV-A and C types also caused more cytotoxicity as indicated by the increase in LDH levels ( Figure 4C ; one-way ANOVA; A16 and A36 P < 0.0001, C2 P = 0.002, C15 P = 0.02 vs B72). To examine strain-specific effects on cell association, we next tested four different strains of M. catarrhalis (three clinical isolates and laboratory strain 035E). RV-C15 infection induced increased adhesion of all of the bacterial strains to AECs. None of the strains caused significant epithelial cell death or augmented RV-driven cell death ( Figure 4D ).
Figure 4.
RV-A and RV-C, but not RV-B, cause increased adherence of M. catarrhalis. Differentiated cell cultures from two epithelial cell donors were infected with two representative types from each RV species (A16, A35, B52, B72, C2 and C15) (A–C) followed by the addition of M. catarrhalis strain MC14. After co-infection the following parameters were measured: (A) Adherent M. catarrhalis in cell lysate (P vs. uninfected control [“MC”]), (B) RV replication (P value vs B72), and (C) cytotoxicity (P vs. uninfected control [“MC-/RV-”]). Bars represent geometric mean ± SD. Results are from 3 independent experiments (ordinary one-way ANOVA; * P < 0.05, ** P<0.01, ***P<0.001, **** P<0.0001). (D) Following infection with RV-C15, four strains of M. catarrhalis (MC14, RSM43, RSM163, and 035E) were added to the culture. After co-infection, quantitative PCR was used to quantify the cell-associated M. catarrhalis. Bars represent geometric means ± SD (n = minimum of 3 per strain; unpaired t-test).
To identify the factors that contribute most to M. catarrhalis cell association, we used a mixed-effects model that included RV species, RV replication, epithelial cell donor, and cytotoxicity. In this model M. catarrhalis cell association was significantly related to RV species (C > A > B, P = 0.003) and RV replication (P = 0.05).
Moraxella catarrhalis adheres to epithelial cells infected with RV
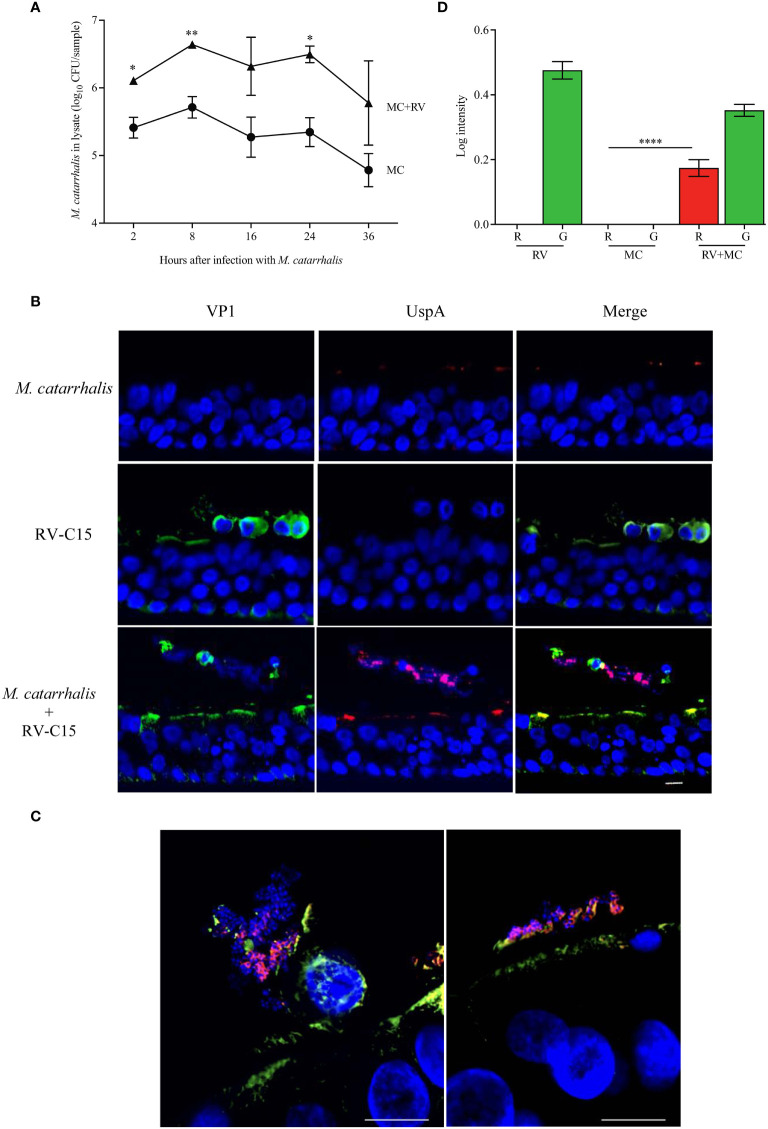
As we were able to show that RV-C species had the greatest effect on M. catarrhalis cell association and epithelial outcomes such as cytotoxicity, we used RV-C15 as a representative to look at the timeline of infection. To determine how soon M. catarrhalis binds to RV-infected cells, we infected ALI cultures with RV-C15 for 16 h (approximately one replication cycle) and then added M. catarrhalis for 2-36 h. Moraxella catarrhalis had increased cell adhesion starting 2 h post-infection and persisting for at least 36 h post-infection ( Figure 5A ). As M. catarrhalis showed maximal adhesion at 8 h after infection, we used this time point to evaluate conditions affecting bacterial adhesion in subsequent experiments.
Figure 5.
M. catarrhalis adheres to epithelial cells undergoing cell death due to RV-C15 infection. (A) ALI cultures were infected with RV-C15 (16 hours) and then with M. catarrhalis MC14. Live CFU counts were obtained by plating dilutions of apical washes taken after incubation with M. catarrhalis for 2, 8, 16, 24, and 36 hours. Symbols represent geometric mean ± SD (n = 4; multiple unpaired t-tests. For (B, C), differentiated epithelial cell cultures were infected with RV-C15 (16 hours), M. catarrhalis MC14 (8 hours), or sequentially with RV-C15 (16 hours) followed by M. catarrhalis (8 hours). Representative histological sections were imaged (M. catarrhalis, red; RV-C15, green; DNA, blue) using an (B) 20✕ and a (C) 100✕ silicone immersion objective (n = 4; scale bar = 10 μm). (D) Intensity analysis of red (R = UspA) and green (G = VP1) signals from histological sections obtained in (B). Bars represent geometric mean ± SD (n = 4, ****P < 0.0001).
We next used immunohistochemistry to visualize M. catarrhalis-cellular interactions. Cells were infected with RV-C15 for 16 h and then incubated with M. catarrhalis for 8 h. Without RV infection, few bacteria adhered to the epithelium. In contrast, M. catarrhalis attached to the apical surface of RV-infected cells, particularly cells that stained positive for RV and that were extruded from the infected epithelium and are dying ( Figures 5B, C ). While M. catarrhalis has been reported to internalize into lung epithelial cell lines and primary cells by macropinocytosis (Slevogt et al., 2007), we did not observe internalized bacteria in RV-infected or uninfected cells. Analysis of the immunofluorescent signal intensities showed a significantly higher signal for UspA in the presence of RV-C15 co-infection (P < 0.0001) ( Figure 5D ).
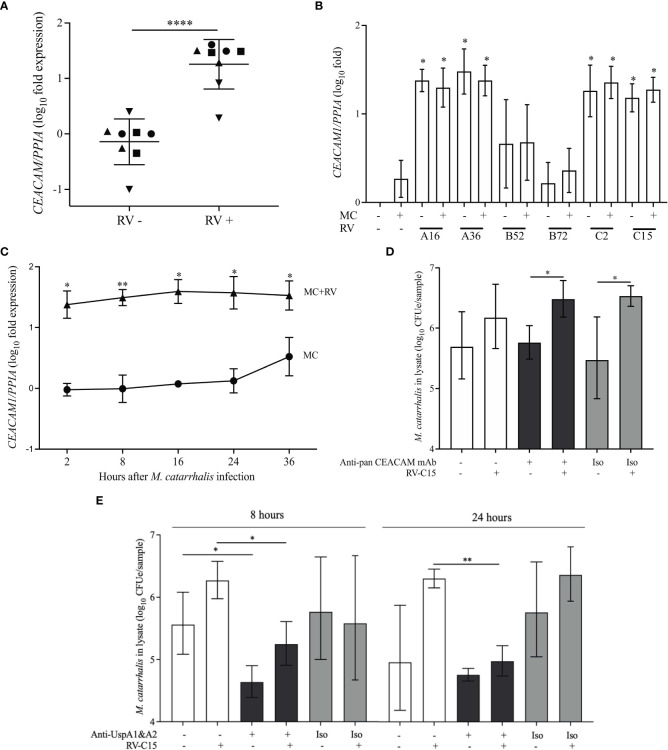
UspA is involved in the adhesion of M. catarrhalis to the respiratory epithelium
M. catarrhalis contains several OMPs that are important for adhesion. These include lipooligosaccharide (Spaniol et al., 2008), UspA1 (Lafontaine et al., 2000; Spaniol et al., 2008), UspA2 (Lafontaine et al., 2000), and Hag/MID (Bullard et al., 2005). Human airway epithelial cells express CEACAM1, CEACAM5, and CEACAM6 (Klaile et al., 2013) and UspA1 has been shown to bind to the N-terminus of CEACAM1 (Hill and Virji, 2003). M. catarrhalis adhesion can be prevented by using monoclonal antibodies that bind to the N-terminus of CEACAM1 (Hill and Virji, 2003; Green et al., 2011). RV-A16 infection significantly increased the expression of CEACAM in the epithelial cell donors with increased M. catarrhalis cell association ( Figure 5A ). Infection with RV-A and RV-C induced the expression of CEACAM1 by ~30-fold, while RV-B and M. catarrhalis did not have an effect on CEACAM1 expression ( Figure 5B ). Blocking CEACAM with mouse monoclonal antibodies D14HD11 that bind to the N-terminus of all CEACAMs, that have previously been used to inhibit Neisseria meningitidis adhesion (Green et al., 2011), did not inhibit M. catarrhalis cell adhesion ( Figure 6D ) despite being significantly upregulated beginning at 18 hours post-infection with RV-C15 ( Figure 5C ). We also used rat monoclonal antibodies YTH71.3 to CEACAM that have previously been used to inhibit Moraxella catarrhalis (Hill and Virji, 2003), but did not see any effect (data not shown). Moraxella catarrhalis did not influence CEACAM1 expression ( Figures 5B, C ). This is in contrast to a previous study that showed induction of CEACAM1 by M. catarrhalis (Klaile et al., 2013). In addition, fluorescent microscopy demonstrated that bacterial adhesion did not colocalize with CEACAM staining of RV-infected epithelial cells (Pearson’s correlation coefficient range = -0.09 to 0.14; P value range = 0.1 to 0.8) ( Figures S1A, B ). In contrast, blocking UspA1 and A2 with mouse monoclonal antibodies significantly reduced RV-induced M. catarrhalis adherence to airway epithelium at 24 h ( Figure 6E ). Interestingly, UspA1 and UspA2 showed minimal induction even at 36 h post-infection with RV-C15 infection ( Figures S2A, B ), showing that the increased adhesion of M. catarrhalis was not due to the increased expression of the outer membrane adhesion protein UspA.
Figure 6.
UspA1 and UspA2 contribute to the adhesion of M. catarrhalis to BECs To evaluate RV-induced CEACAM1 expression in AECs, differentiated ALI cultures (A) from four epithelial cell donors were infected with RV-A16 for 2 hours and then with M. catarrhalis MC14 for 48 hours. Cell donors are represented by different symbols, and expression is relative to donor 1 (circles) (n = 2 per donor; unpaired t-test test). (B) ALI cultures from donor 1 were infected with RV species (A16, A35, B52, B72, C2 and C15) for 2 hours and then with M. catarrhalis MC14 for 48 hours. CEACAM1 expression was evaluated by qPCR. For (C) ALI cultures were infected with RV-C15 for 16 hours and then with M. catarrhalis. CEACAM1 expression was evaluated at 2, 8, 16, 24, and 36 hours following M. catarrhalis infection (n = 4, multiple unpaired t-tests). (D) Following RV-C15 infection for 16 hours, ALI cultures were incubated with mouse anti-pan CEACAM antibodies for 1 hour before adding M. catarrhalis MC14 for 8 hours. Cell-associated M. catarrhalis were quantified by qPCR. Bars represent geometric mean ± SD (n = 4, unpaired t-test). (E) M. catarrhalis strain MC14 was incubated with mouse anti-UspA antibodies for 1 hour and the mixture was added onto the apical surface of the epithelium for 8 or 24 hours following RV-C15 infection. Cell-associated M. catarrhalis was quantified by qPCR. White bars = no antibodies, black bars = anti-UspA antibodies, grey bars = isotype control (Iso). Bars represent geometric mean ± geometric SD (n = 4; unpaired t-test). *P < 0.05, **P<0.01, ****P<0.0001.
Discussion
Using a fully differentiated in vitro model of the human airway epithelium, we demonstrated that M. catarrhalis, an opportunistic pathogen of the human respiratory tract, was well tolerated and did not cause epithelial damage ( Figure 1 ). During co-infection with RV-A or RV-C, there was a significant increase in cell-associated M. catarrhalis ( Figures 2 , 4 ). This suggests that the increased replication rates for RV-A and RV-C compared to RV-B species leads to more epithelial damage which supports the cell association of M. catarrhalis ( Nakagome et al., 2014). These findings support clinical observations in which infections with RV-A or RV-C, which cause more severe illness than RV-B, were associated with increased detection in nasal secretions of respiratory pathogens like M. catarrhalis, H. influenzae, and S. pneumoniae ( Bashir et al., 2018). The epithelial cytokine and gene expression responses were driven by RV with little contribution from M. catarrhalis ( Figure 3 ). We also found that M. catarrhalis preferentially associates with RV-infected cells that may be undergoing cell death ( Figure 5B, C ) and that the outer membrane adhesion protein UspA is involved in the attachment of M. catarrhalis to respiratory epithelial cells ( Figure 5 ). Analysis of the immunofluorescent signal intensities showed a significantly higher signal for UspA in the presence of RV-C15 co-infection (P < 0.0001) ( Figure 5D ).
Our observations are consistent with other studies showing that viral infections can induce the adhesion of bacterial pathogens. Respiratory syncytial virus infection increases the virulence of S. pneumoniae by directly binding to bacteria and upregulating genes required for virus binding and bacterial invasiveness (Smith et al., 2014). Influenza virus also directly interacts with H. influenzae, M. catarrhalis, S. aureus, and S. pneumoniae, increases adhesion to respiratory epithelial cells, and induces middle ear translocation of bacteria to promote otitis media (Rowe et al., 2019). Moraxella catarrhalis produces several adhesins, including UspA1, UspA2, UspA2H, and Hag, and these molecules mediate binding to airway tissues. UspA1 and UspA2 are relatively conserved and can be detected in most nasopharyngeal isolates of M. catarrhalis ( Meier et al., 2002). UspA1 binds to extracellular matrix proteins laminin (Tan et al., 2006) and fibronectin (Tan et al., 2005) as well as to CEACAM1, which is a transmembrane protein expressed by airway epithelial cells (Hill and Virji, 2003; Conners et al., 2008). UspA2, also binds to extracellular matrix proteins such as collagen (Singh et al., 2016), laminin (Tan et al., 2006), fibronectin (Tan et al., 2005), and vitronectin (Singh et al., 2010), while the hybrid UspA2H has mixed binding properties. Hag/MID can bind to collagen (Balder et al., 2009). In our studies, RV-A and RV-C infection induced CEACAM1 but blocking CEACAM did not inhibit M. catarrhalis binding to airway epithelial cells ( Figures 6B, D ). The respiratory epithelium expresses CEACAM1, 5, and, 6 (Klaile et al., 2013). Soluble CEACAM5 and CEACAM6 can be detected in normal bronchial mucus (Matsuoka et al., 1990; Klaile et al., 2013), and secreted isoforms could inhibit adhesion to CEACAM expressed on the surface of the respiratory epithelium. As RV can increase the synthesis of fibronectin (Wang et al., 2009) and damage the epithelial barrier (Looi et al., 2018) allowing access to fibronectin and other matrix proteins (Singh et al., 2016), it is also possible that M. catarrhalis binds to these proteins which become more abundant following epithelial damage.
We observed that M. catarrhalis formed clusters around dying cells on the apical surface of the epithelium and in cell debris in the mucin layer ( Figure 6 ). Therefore, it is possible that cellular damage also leads to the release of nutrients providing an environment conducive for M. catarrhalis growth as seen in the GI tract, where colonocytes undergoing apoptosis release small molecules that are a source of nutrition for bacterial growth, a process known as death-induced nutrient release (Anderson et al., 2021).
Upper airway colonization with M. catarrhalis is widespread in healthy preschool children (Verhaegh et al., 2011), and M. catarrhalis increases in abundance during RV infections and asthma exacerbations (Kloepfer et al., 2014; Bashir et al., 2018). Bacterial invasion of host epithelia is a strategy used by bacteria for persistence and dissemination leading to more severe and recurrent disease (Alexander and Hudson, 2001; Asmat et al., 2014; Murphy et al., 2004). UspA1 and lipooligosaccharide (LOS) also contribute to cellular invasion (Spaniol et al., 2008). Our observations indicate that M. catarrhalis, by itself, does not invade or damage the respiratory epithelium or induce epithelial cytokine responses. In contrast, RV infection caused the secretion of cytokines, including chemoattractants for neutrophils (CXCL5 and CXCL10) and eosinophils (CCL26 [eotaxin-3]) and the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL), which has been linked in mouse models to airway hyperactivity, cellular infiltration and anti-viral IFN responses (Girkin et al., 2017). Co-infection with RV and M. catarrhalis did not cause a synergistic cytokine response, suggesting that M. catarrhalis induces minimal responses from airway epithelial cells, with or without RV infection. Nevertheless, RV infection induces neutrophil chemotactic factors and the influx and activation of neutrophils. Strong inflammatory and neutrophilic responses have been observed in a mouse model of airway M. catarrhalis infection with more intense responses when infection occurred in house dust mite sensitized animals (Alnahas et al., 2017). In a cohort of asymptomatic neonates, whose upper airways were colonized with M. catarrhalis or other pathogens, cytokine analysis of the nasal mucosal lining fluid revealed a mixed Th1/Th2/Th17-type response with increased levels of IL-8 and IL-17, which cause neutrophil recruitment and activation (Følsgaard et al., 2013). This suggests that neutrophilic inflammation, induced by RV and amplified by M. catarrhalis, could worsen the RV-induced epithelial damage and increase illness severity. In our experiments, RV-C caused significantly more cell association than other RV species, which corresponds with clinical observations (Bashir et al., 2018). This raises the possibility that RV-C induces a unique environment that is preferred by M. catarrhalis.
Our study’s main strength was the co-infection model using fully differentiated respiratory epithelium, providing a multicellular model of the cellular responses to bacterial and viral infection. We also tested multiple types of RV and different strains of M. catarrhalis to understand their similarities and differences. The RV were all cloned from clinical isolates, and three of the four M. catarrhalis strains used in the study were isolated from clinical samples to mimic conditions in vivo. Our study also has limitations that should be considered in interpreting these results. The small number of cell donors in our study limited the power to explore individual variability. Second, multicellular models containing epithelial and immune cells are needed to test the effects of M. catarrhalis adhesion on leukocyte-mediated inflammation. Although we did not observe an epithelial cytokine response to M. catarrhalis infection, RV infection recruits neutrophils and other leukocytes into the airway. It is possible that interactions between epithelial cells, adherent bacteria and neutrophils could amplify inflammatory responses at the epithelial surface to increase the severity of illness. In this study we only looked at a possible interaction between CEACAM and UspA proteins. It is possible that use of polyclonal blocking antibodies instead of monoclonal antibodies as used in this study, may give better blockage of the bacterial binding epitopes of CEACAM. We have not looked at possible interactions between other OMPs and extracellular matrix proteins and therefore cannot exclude the possibility that these proteins may play a role in RV-induced cell association of M. catarrhalis.
In conclusion, our study revealed that M. catarrhalis increases association with respiratory epithelial cells during RV infection by adhering to infected cells and those extruded from the epithelial surface. These findings suggest that virus-induced cell death promotes adherence and survival of this common bacterial pathogen. The lack of M. catarrhalis-induced inflammation or cytotoxicity further suggests that this bacterium could intensify the severity of viral illness by focusing leukocyte-mediated inflammation at the epithelial surface. This suggests a possible mechanism by which RV and M. catarrhalis co-infection increases the severity of respiratory illness in children.
Methods
Bacteria and viruses
Moraxella catarrhalis strain MC14 was isolated from a clinical specimen by the University of Wisconsin Clinical Pathology Laboratory. In addition, M. catarrhalis strains RSM43 and RSM163 were isolated from the nasal secretions of children with acute respiratory illnesses (Kloepfer et al., 2014), and strain 035E was originally isolated from the middle ear fluid of a patient with otitis media (Unhanand et al., 1992). For epithelial inoculation, we cultured M. catarrhalis from a frozen glycerol stock onto chocolate agar plates that were incubated at 37 °C in a 5% CO2 environment for 48 h. Bacteria were re-streaked onto fresh chocolate agar plates and cultured for a further 24 h.
RV strains were cloned from clinical isolates and grown as described previously (Nakagome et al., 2014). The types used in the study were RV-A16, RV-A36, RV-B52, RV-B72, RV-C2, and RV-C15 (Bochkov et al., 2011; Lee et al., 2012; Nakagome et al., 2014).
Isolation of genomic DNA from M. catarrhalis isolates
Moraxella catarrhalis was cultured overnight in 3 ml brain-heart-infusion (BHI) broth at 37 °C. The cultures were centrifuged at 21,130 × g for 5 min at ambient temperature and washed in 1 ml of Buffer 1 (150 mM NaCl, 10 mM EDTA, 20 mM Tris-HCl, pH 8). The cells were resuspended in 1 mL of fresh Buffer 1 with 10 µL of 100 mg/mL RNase A and 163 µl of 10% [wt/vol] SDS. The samples were incubated at 37 °C for 90 min. Subsequently, 20 µl of 20 mg/ml proteinase K were added and the samples were incubated at 65 °C for 20 min. After an additional incubation at 37 °C for 16 h to ensure complete lysis, genomic DNA was purified using standard phenol-chloroform extraction and precipitated with isopropanol. Genomic libraries for Illumina MiSeq 2×150-bp paired-end sequencing were prepared and sequenced by the University of Wisconsin-Madison Biotechnology Center. The raw reads were corrected using fastp 0.20.0 (Chen et al., 2018) and draft genomes were generated using SPAdes v3.11.0 with default parameters (Prjibelski et al., 2020).
Culture of airway epithelium at air-liquid interface
Human bronchial and tracheal epithelial cells were obtained from residual tissue from lungs destined for transplantation in collaboration with the University of Wisconsin - Health Lung Transplant Program. The protocol was reviewed by the University of Wisconsin Institutional Review Board and was deemed “not human subjects research.” Cryopreserved aliquots of cells were thawed and expanded as monolayers in PneumaCult-Ex Plus Medium (StemCell Technologies) supplemented with 0.1% [vol/vol] gentamicin (Sigma) and 0.1% [vol/vol] fluconazole (Novaplus). Once the cells reached 80% confluence, they were transferred to 12-well plates with Transwell semi-permeable inserts (Corning 3460) and were allowed to differentiate in PneumaCult-ALI medium (StemCell Technologies) supplemented with 0.1% [vol/vol] gentamicin (Sigma) and 0.1% [vol/vol] fluconazole (Novaplus) at the air-liquid interface for at least 21 days when ciliary motion was observed. The culture medium was changed to an antibiotic-free medium containing 0.05% [vol/vol] hydrocortisone 48 hours prior to RV infection. All ALI cultures were used between 28 – 35 days of air lifting.
RV and M. catarrhalis infection of epithelial cultures
The apical surface of the epithelium was washed with pre-warmed phosphate-buffered saline (PBS) supplemented with 100 mg/l of Ca2+ and Mg2+and then infected with 107 PFU of RV (in 50 μl of antibiotic-free culture medium) at a multiplicity of infection (MOI) of 10. The cells were incubated at 34 °C for 2 h, after which the apical surface was washed (3×) with pre-warmed PBS (containing Ca2+ and Mg2+). For infection of differentiated cultures, 3-4 colonies of M. catarrhalis were picked from the growth plates and used to make a bacterial suspension in pre-warmed PBS at a concentration of 2 McFarland units (~6×108 CFU/ml). The apical surface was then infected with 50 μl of the M. catarrhalis suspension (~3×107 CFU) and incubated at 37 °C for 48 h.
For time-course experiments in Figure 1A , differentiated respiratory epithelium was infected with 107 PFU of RV-A16 at 34 °C for 2 h and then with 3×107 CFU of M. catarrhalis. At specified timepoints after M. catarrhalis infection, the apical surface was washed with 0.5 ml of pre-warmed PBS (with Ca2+ and Mg2+) and cell lysates were collected in 350 µl RLT Plus Buffer from AllPrep DNA/RNA Mini Kit (Qiagen) containing 0.5% [vol/vol] Reagent DX (Qiagen) for quantitative PCR. To quantify cell associated live bacteria in Figure 5A , ALI cultures were infected with 107 PFU of RV-C15 at 34 °C for 2 h and then for at 37 °C for a further 16 h before infecting with 3×107 CFU of M. catarrhalis for specified durations. The apical surface washed with 0.5 ml of pre-warmed PBS (with Ca2+ and Mg2+). Cell associated bacteria were quantified by adding 100 μl of 1% [vol/vol] saponin on to the apical epithelial surface at RT for 15 min, washing with 0.5 ml of warmed PBS (with Ca2+ and Mg2+) and plating the serially diluted washes on chocolate agar. The colony counts were done 48 h after plating.
For trans-epithelial electrical resistance (TEER) measurements, the apical surface of ALI cultures were infected with 50 μl of the M. catarrhalis suspension (~3×107 CFU) that was prepared as described earlier and incubated at 37 °C for 24 or 48 h. As a control, a clinical isolate of Staphylococcus aureus was cultured from a frozen glycerol stock onto blood agar plates that were incubated at 37 °C in a 5% CO2 environment for 48 h. Bacteria were re-streaked onto fresh blood agar plates and cultured for a further 24 h. For infection of ALI cultures, a colony of S. aureus was picked from the growth plates and used to make a bacterial suspension in pre-warmed PBS at a concentration of 2 McFarland units (~6×108 CFU/ml). The apical surface of the ALI cultures were then infected with 50 μl of the suspension (~3×107 CFU) and incubated at 37 °C for 24 or 48 h.
Gentamycin protection assay for quantification of intracellular M. catarrhalis
The apical surface of the epithelium was washed with pre-warmed phosphate-buffered saline (PBS) supplemented with 100 mg/l of Ca2+ and Mg2+and then infected with 107 PFU of RV (in 50 μl of antibiotic-free culture medium) at an MOI of 10. The cells were incubated at 34 °C for 2 h, after which the apical surface was washed (3×) with pre-warmed PBS (containing Ca2+ and Mg2+). For infection of differentiated cultures, 3-4 colonies of M. catarrhalis were picked from the growth plates and used to make a bacterial suspension in pre-warmed PBS at a concentration of 2 McFarland units (~6×108 CFU/ml). The suspension was diluted 100-fold to obtain a suspension with a concentration of 6×106 CFU/ml. The apical surface was then infected with 50 μl of the 6×106 CFU/ml M. catarrhalis suspension (~3×105 CFU) and incubated at 37 °C for 48 h. Following the incubation period, the apical surface was washed with 0.5 ml of pre-warmed PBS and 100 µl of 100 µg/ml of gentamicin was added to the apical surface and incubated at 37 °C for 1 hour. The apical surface was washed with 0.5 ml of pre-warmed PBS and 100 µl of 1% saponin was added to the apical surface for 15 minutes at room temperature. The apical surface was washed again with 0.5 ml of pre-warmed PBS and serial dilutions of the washes were plated to determine live intracellular CFU counts.
TEER measurement
Culture medium in outer wells of the Transwell plate were removed and replaced with 1 ml of warmed DMEM/F12 medium (Gibco) and 0.5 ml of DMEM/F12 in the insert (on apical surface of cells). The cells and medium were allowed to incubate at RT for 15 min. The TEER was measured using an epithelial Voltohmmeter (EVOM2; World Precision Instruments, USA).
Quantitative PCR
Nucleic acids from the epithelium (± bacteria) were harvested from the inserts using 350 µl RLT Plus Buffer from AllPrep DNA/RNA Mini Kit (Qiagen) containing 0.5% [vol/vol] Reagent DX (Qiagen). The lysate was transferred to PowerBead tubes (Qiagen) for bead beating (5 min, 50 oscillations/s, Qiagen TissueLyser LT). RNA and DNA were extracted as separate fractions (AllPrep® DNA/RNA Mini Kit). Moraxella catarrhalis copB and RV RNA levels were quantified by real-time PCR using specific primers ( Table 1 ) as previously described (Greiner et al., 2003; Bochkov et al., 2011). The CFUe measurements were calibrated by using a standard curve. To generate the standard curve, M. catarrhalis strain MC14 was cultured overnight in tryptic soy broth. The suspension was serially diluted and plated to obtain CFU counts. Two milliliters of the original suspension were centrifuged at 13,000 rpm for 30 minutes. The resulting bacterial pellet was lysed by adding warmed (55 °C) Solution PM1 followed by DNA extraction (AllPrep® Power Viral Kit, Qiagen). The extracted DNA was serially diluted to make a standard curve corresponding to 106 to 102 CFU.
Table 1.
Primers and probes used in the study.
| Name | Sequence 5’ ➔ 3’ | Description |
|---|---|---|
| RV1A16 | CCTCCGGCCCCTGAAT | RV |
| R848 | AAACACGGACACCCAAAGTAGT | RV |
| UspA1-F | AGGGATCCAACGACGGTCCAAGA TGG | UspA1 |
| UspA1-R | AGGGATCCCCTGCCACCTAAAGCCTTG | UspA1 |
| UspA2-F | CGGGATCCCTTCTCCCC CTAAAAATCGCTG | UspA2 |
| UspA2-R | AGGGATCCCGCTGTATGCCGCTAC TCGCAGCT | UspA2 |
| copB-F | GTGAGTGCCGCTTTACAACC | copB |
| copB-R | TGTATCGCCTGCCAAGACAA | copB |
| copB probe | NED-TGCTTTTGCAGCTGTTAGCCAGCCTAA-MGB-NFQ | copB |
| CEACAM1-F | TGCTCTGATAGCAGTAGCCCT | CEACAM1 |
| CEACAM1-R | TGCCGGTCTTCCCGAAATG | CEACAM1 |
| PPIA-F | CCCACCGTGTTCTTCGACATT | PPIA |
| PPIA-R | GGACCCGTATGCTTTAGGATGA | PPIA |
For the quantification of mRNA expression, cDNA was prepared from total RNA extracted from ALI culture lysates using TaqMan™ Reverse Transcription Reagents (Applied Biosystems). CEACAM1 mRNA expression was measured using specific primers (PrimerBank ID 329112546c1). UspA1 and UspA2 expression were quantified using specific primers as described previously (Lafontaine et al., 2001) (Wang and Seed, 2003). Gene expression was quantified using Power SYBR Green PCR Master Mix (Applied Biosystems) on an Applied Biosystems™ 7500 Real-time PCR system and normalized against the expression of the stable housekeeping gene PPIA (PrimerBank ID 114520617c1). The expression was normalized against copB. All primers used in the study are given in Table 1 .
Gene expression analysis
RNA was converted to cDNA using the SMART-Seq v4 Ultra Low Input RNA kit for sequencing (Takara Bio cat. 634898). Standard library preparation was completed using the Illumina Nextera XT DNA Library Preparation Kit and library quality and concentration were assessed using an Agilent 2100 bioanalyzer. Indexed samples were pooled and sequenced on the Illumina Hi-Seq 4000 with 100 base-pair, paired-end sequencing to a minimum of 10 million mapped reads per sample. RNA-seq reads were mapped to genome assembly GRCh37(hg19) reference sequence using STAR (version 2.6.1) (Dobin et al., 2013). Read counts were adjusted to counts per million and normalized using TMM normalization (Robinson and Oshlack, 2010). PCA was conducted in R (version 3.6.2) using prcomp. voom was used to adjust for significant technical covariates (Law et al., 2014). Genes significantly associated with RV or M. catarrhalis treatment were identified using Limma linear mixed modeling in R (Limma R package version 3.5 https://bioconductor.org/packages/release/bioc/html/limma.html). An FDR-adjusted p value of 0.05 was used as a significance threshold, using the method of Benjamini and Hochberg (Hochberg and Benjamini, 1990).
Cytotoxicity measurement
Cellular cytotoxicity was estimated by measuring extracellular lactate dehydrogenase using a CytoTox 96® Non-Radioactive Cytotoxicity Assay, (Promega) following manufacturer’s instructions.
Cellular responses to infection
The cytokine levels in the culture medium in the basal compartment of the ALI culture system were measured by multiplex ELISA using a MILLIPLEX MAP 10-plex Human Cytokine/TH17 Mag Kit (Millipore Sigma). Epithelial cell gene expression was assessed by using 2×150 bp paired-end NovaSeq platform at the University of Wisconsin – Madison Biotechnology Center’s Gene Expression Center Core Facility (Madison, WI) (Research Resource Identifier – RRID : SCR_017757) for RNA library preparation and the DNA Sequencing Facility (RRID : SCR_017759) for sequencing.
Gene enrichment analysis of differentially expressed genes was done using the ShinyGO 0.76 online software (Ge et al., 2020). The pathways were sorted by -log10 (FDR) values with an FDR cutoff of 0.05.
Blocking cellular or bacterial surface proteins
To block UspA1 and A2, we incubated 50 μl of a 2 McFarland unit M. catarrhalis suspension (~3×107 CFU) with an equal volume of undiluted culture supernatant from the mouse hybridoma 17C7 (ATCC HB-11093) (Helminen et al., 1994; Aebi et al., 1997), (37 °C, 1 h). The mixture was then added to the apical surface of differentiated airway epithelial cells, with or without RV infection. Moraxella catarrhalis was quantified by qPCR at 8 and 24 h post-infection.
To block CEACAM, airway epithelial cells were infected with RV-C15 for 16 h. After washing with 0.5 ml of pre-warmed PBS, mouse anti-pan CEACAM antibody D14HD11 (ab4567, Abcam; 100 μl of 100 ng/ml) or of rat monoclonal anti-CEACAM antibody YTH71.3 (Santa Cruz Biotechnology; 100ul of 100 ng/ml) was added to the apical surface and incubated at 37 °C for 2 h. We washed the cells three times and then incubated them with 50 μl of a 2 McFarland unit M. catarrhalis (~3×107 CFU, 37 °C, 8 h or 24 h).
Immunohistochemistry
Mature ALI cultures were fixed in 10% [vol/vol] neutral-buffered formalin, embedded in paraffin wax and sectioned. The sections were deparaffinized by heating (60 °C, 20 min) and then transferring to 3 changes of xylenes. The sections were rehydrated using graded ethanol solutions (100%, 95%, 80%, 70% and 50% [vol/vol]) and deionized water. Antigen retrieval was performed by heating at 80 °C in a water bath for 2.5 h. Sections were permeabilized (1% normal goat serum with 0.4% [vol/vol] Triton X-100, 5 min), blocked (PBS with 0.1%[vol/vol] Tween 20 and 5% [vol/vol] goat serum), and then incubated (overnight, 4 °C) with antibodies to RV-C15 VP1 (1:200) or mouse anti-pan CEACAM antibody D14HD11 (ab4567, Abcam) (1:2000) diluted in 1% [vol/vol] normal goat serum containing 0.1% [vol/vol] Tween 20. After washing (PBS with 0.1% [vol/vol] Tween 20, 5 min), sections were incubated with the secondary antibody (Alexa Fluor® 488 goat anti-mouse), blocked for 1 h with 5% mouse serum in PBS, and incubated with the primary antibody to M. catarrhalis UspA (Helminen et al., 1994; Aebi et al., 1997)(undiluted culture supernatant from the mouse hybridoma 17C7 - ATCC HB-11093, overnight, 4 °C). After washing three times, the second secondary antibody (Alexa Fluor® 555 goat anti-mouse) was added (1 h, ambient temperature). The sections were counterstained with 4’,diamidine-2’-phenylindole dihydrochloride at 1:1000 for 15 min before mounting with FluorSave Reagent (EMD Millipore). Images were acquired on a Nikon A1 R confocal microscope (20× objective) and on a Nikon AX R point scanning confocal system equipped with 4 high-sensitivity GaAsP detectors using a Plan Apo lambda S 100× silicone immersion objective (NA 1.35) at 2048 x 2048 pixel density and 4x scan zoom. The 100× images were processed with Denoise.ai after acquisition. Image analysis was done using RGB Profile Plot ( Figure 5D ) and Colocalization Finder (Figure S1B) plugins on ImageJ v1.53t.
Statistical analysis
Statistical analyses were performed using GraphPad Prism v.9.3.1 (GraphPad Software, Inc). Student’s ordinary t-test or the Mann-Whitney test were used to compare two groups. When more than two groups were compared, ordinary one-way ANOVA was used for the analysis. Correlation coefficients were calculated using Spearman’s rho statistic. To identify factors that significantly predict M. catarrhalis cell association, linear regression modeling with random slope and random intercept using SAS procedure Mixed was conducted. The likelihood ratio test (LRT) was used to for model selection. A p-value < 0.05 was considered statistically significant. SAS software (v.9.4, SAS Institute, Cary, NC) was used to develop the mixed-effects model.
Data availability statement
The datasets presented in this study can be found in online repositories. The name of the repository/repositories and accession number(s) can be found below: PRJNA875903.
Author contributions
JG, RB-S and ED conceived the study. ED and RB-S planned and conducted experiments. RS provided clinical bacterial isolates and performed bacterial whole genome sequencing. YB provided virus preparations. BH and CO planned and performed epithelial gene expression studies and analysis. CK, TM, and CC provided guidance on study design, experimental methods and data interpretation. ED, RB-S, and ZZ performed statistical analysis. All authors contributed to the final draft of the manuscript and approved the final version. ED and RB-S contributed equally to this study.
Acknowledgments
The authors thank the University of Wisconsin-Madison, Department of Surgery, Histology Core Lab, Dr. Susan Thibeault Ph.D., CCC-SLP, PI of the DOS Histology Core, along with certified Histotechnician, Sierra Raglin HTL (ASCP), and Lab Supervisor, Sara Dutton Sackett, Ph.D. for their assistance with immunohistochemistry, Lance Rodenkirch of the University of Wisconsin-Madison Imaging Core and Evdokia Menelaou, Ph.D. of Nikon Instruments Inc. for assistance with confocal imaging.
Funding Statement
This work was funded by the National Institutes of Health Centers of Excellence for Translational Research (U19 AI142720). This work was also partially funded by NIH grants R01 AI19641 (TM) and UL1 TR001412 to the University at Buffalo.
Abbreviations
AEC, Airway epithelial cells; ALI, Air-liquid interface; CCL26, Chemokine ligand 26; CEACAM, Carcinoembryonic antigen-related cell adhesion molecule; CEA, Carcinoembryonic antigen; CFU, Colony-forming unit; CFUe, Colony-forming unit equivalent; CXCL, C-X-C motif chemokine ligand; EGR, Early growth response; G-CSF, Granulocyte colony-stimulating factor; IFN, Interferon; IL, Interleukin; LOS, Lipooligosaccharide; OMP, Outer membrane protein; PFU, Plaque forming unit; PFUe, Plaque forming unit equivalent; RV, Rhinovirus; TEER, Trans-epithelial electrical resistance; TNF-α, Tumor necrosis factor-alpha; TRAIL, TNF-related apoptosis-inducing ligand; Usp, Ubiquitous surface protein.
Conflict of interest
JG has received fees from AstraZeneca and Meissa Vaccines Inc. and has stock options in Meissa Vaccines Inc. outside of the current work.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2022.1060748/full#supplementary-material
References
- Aebi C., Maciver I., Latimer J. L., Cope L. D., Stevens M. K., Thomas S. E., et al. (1997). A protective epitope of moraxella catarrhalis is encoded by two different genes. Infect. Immun. 65, 4367–4377. doi: 10.1128/iai.65.11.4367-4377.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander E. H., Hudson M. C. (2001). Factors influencing the internalization of staphylococcus aureus and impacts on the course of infections in humans. Appl. Microbiol. Biotechnol. 56, 361–366. doi: 10.1007/s002530100703 [DOI] [PubMed] [Google Scholar]
- Alnahas S., Hagner S., Raifer H., Kilic A., Gasteiger G., Mutters R., et al. (2017). IL-17 and TNF-α are key mediators of. Front. Immunol. 8, 1562. doi: 10.3389/fimmu.2017.01562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson C. J., Medina C. B., Barron B. J., Karvelyte L., Aaes T. L., Lambertz I., et al. (2021). Microbes exploit death-induced nutrient release by gut epithelial cells. Nature 596, 262–267. doi: 10.1038/s41586-021-03785-9 [DOI] [PubMed] [Google Scholar]
- Asmat T. M., Agarwal V., Saleh M., Hammerschmidt S. (2014). Endocytosis of streptococcus pneumoniae via the polymeric immunoglobulin receptor of epithelial cells relies on clathrin and caveolin dependent mechanisms. Int. J. Med. Microbiol. 304, 1233–1246. doi: 10.1016/j.ijmm.2014.10.001 [DOI] [PubMed] [Google Scholar]
- Avadhanula V., Rodriguez C. A., Devincenzo J. P., Wang Y., Webby R. J., Ulett G. C., et al. (2006). Respiratory viruses augment the adhesion of bacterial pathogens to respiratory epithelium in a viral species- and cell type-dependent manner. J. Virol. 80, 1629–1636. doi: 10.1128/JVI.80.4.1629-1636.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balder R., Krunkosky T. M., Nguyen C. Q., Feezel L., Lafontaine E. R. (2009). Hag mediates adherence of moraxella catarrhalis to ciliated human airway cells. Infect. Immun. 77, 4597–4608. doi: 10.1128/IAI.00212-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir H., Grindle K., Vrtis R., Vang F., Kang T., Salazar L., et al. (2018). Association of rhinovirus species with common cold and asthma symptoms and bacterial pathogens. J. Allergy Clin. Immunol. 141, 822–824.e829. doi: 10.1016/j.jaci.2017.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizzintino J., Lee W. M., Laing I. A., Vang F., Pappas T., Zhang G., et al. (2011). Association between human rhinovirus c and severity of acute asthma in children. Eur. Respir. J. 37, 1037–1042. doi: 10.1183/09031936.00092410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkov Y. A., Palmenberg A. C., Lee W. M., Rathe J. A., Amineva S. P., Sun X., et al. (2011). Molecular modeling, organ culture and reverse genetics for a newly identified human rhinovirus c. Nat. Med. 17, 627–632. doi: 10.1038/nm.2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard B., Lipski S. L., Lafontaine E. R. (2005). Hag directly mediates the adherence of moraxella catarrhalis to human middle ear cells. Infect. Immun. 73, 5127–5136. doi: 10.1128/IAI.73.8.5127-5136.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y., Gu J. (2018). Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conners R., Hill D. J., Borodina E., Agnew C., Daniell S. J., Burton N. M., et al. (2008). The moraxella adhesin UspA1 binds to its human CEACAM1 receptor by a deformable trimeric coiled-coil. EMBO J. 27, 1779–1789. doi: 10.1038/emboj.2008.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Steenhuijsen Piters W. A. A., Watson R. L., de Koff E. M., Hasrat R., Arp K., Chu M. L. J. N., et al. (2022). Early-life viral infections are associated with disadvantageous immune and microbiota profiles and recurrent respiratory infections. Nat. Microbiol. 7, 224–237. doi: 10.1038/s41564-021-01043-2 [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. (2013). STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Følsgaard N. V., Schjørring S., Chawes B. L., Rasmussen M. A., Krogfelt K. A., Brix S., et al. (2013). Pathogenic bacteria colonizing the airways in asymptomatic neonates stimulates topical inflammatory mediator release. Am. J. Respir. Crit. Care Med. 187, 589–595. doi: 10.1164/rccm.201207-1297OC [DOI] [PubMed] [Google Scholar]
- Ge S. X., Jung D., Yao R. (2020). ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 36, 2628–2629. doi: 10.1093/bioinformatics/btz931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girkin J. L., Hatchwell L. M., Collison A. M., Starkey M. R., Hansbro P. M., Yagita H., et al. (2017). TRAIL signaling is proinflammatory and proviral in a murine model of rhinovirus 1B infection. Am. J. Physiol. Lung Cell Mol. Physiol. 312, L89–L99. doi: 10.1152/ajplung.00200.2016 [DOI] [PubMed] [Google Scholar]
- Green L. R., Monk P. N., Partridge L. J., Morris P., Gorringe A. R., Read R. C. (2011). Cooperative role for tetraspanins in adhesin-mediated attachment of bacterial species to human epithelial cells. Infect. Immun. 79, 2241–2249. doi: 10.1128/IAI.01354-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner O., Day P. J., Altwegg M., Nadal D. (2003). Quantitative detection of moraxella catarrhalis in nasopharyngeal secretions by real-time PCR. J. Clin. Microbiol. 41, 1386–1390. doi: 10.1128/JCM.41.4.1386-1390.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helminen M. E., Maciver I., Latimer J. L., Klesney-Tait J., Cope L. D., Paris M., et al. (1994). A large, antigenically conserved protein on the surface of moraxella catarrhalis is a target for protective antibodies. J. Infect. Dis. 170, 867–872. doi: 10.1093/infdis/170.4.867 [DOI] [PubMed] [Google Scholar]
- Hill D. J., Edwards A. M., Rowe H. A., Virji M. (2005). Carcinoembryonic antigen-related cell adhesion molecule (CEACAM)-binding recombinant polypeptide confers protection against infection by respiratory and urogenital pathogens. Mol. Microbiol. 55, 1515–1527. doi: 10.1111/j.1365-2958.2005.04487.x [DOI] [PubMed] [Google Scholar]
- Hill D. J., Virji M. (2003). A novel cell-binding mechanism of moraxella catarrhalis ubiquitous surface protein UspA: Specific targeting of the n-domain of carcinoembryonic antigen-related cell adhesion molecules by UspA1. Mol. Microbiol. 48, 117–129. doi: 10.1046/j.1365-2958.2003.03433.x [DOI] [PubMed] [Google Scholar]
- Hochberg Y., Benjamini Y. (1990). More powerful procedures for multiple significance testing. Stat. Med. 9, 811–818. doi: 10.1002/sim.4780090710 [DOI] [PubMed] [Google Scholar]
- Klaile E., Klassert T. E., Scheffrahn I., Müller M. M., Heinrich A., Heyl K. A., et al. (2013). Carcinoembryonic antigen (CEA)-related cell adhesion molecules are co-expressed in the human lung and their expression can be modulated in bronchial epithelial cells by non-typable haemophilus influenzae, moraxella catarrhalis, TLR3, and type I and II interferons. Respir. Res. 14, 85. doi: 10.1186/1465-9921-14-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloepfer K. M., Lee W. M., Pappas T. E., Kang T. J., Vrtis R. F., Evans M. D., et al. (2014). Detection of pathogenic bacteria during rhinovirus infection is associated with increased respiratory symptoms and asthma exacerbations. J. Allergy Clin. Immunol. 133, 1301–1307, 1307.e1301-1303. doi: 10.1016/j.jaci.2014.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine E. R., Cope L. D., Aebi C., Latimer J. L., McCracken G. H., Jr., Hansen E. J. (2000). The UspA1 protein and a second type of UspA2 protein mediate adherence of moraxella catarrhalis to human epithelial cells in vitro . J. Bacteriol 182, 1364–1373. doi: 10.1128/JB.182.5.1364-1373.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine E. R., Wagner N. J., Hansen E. J. (2001). Expression of the moraxella catarrhalis UspA1 protein undergoes phase variation and is regulated at the transcriptional level. J. Bacteriol 183, 1540–1551. doi: 10.1128/JB.183.5.1540-1551.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law C. W., Chen Y., Shi W., Smyth G. K. (2014). Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29. doi: 10.1186/gb-2014-15-2-r29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. M., Lemanske R. F., Evans M. D., Vang F., Pappas T., Gangnon R., et al. (2012). Human rhinovirus species and season of infection determine illness severity. Am. J. Respir. Crit. Care Med. 186, 886–891. doi: 10.1164/rccm.201202-0330OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looi K., Buckley A. G., Rigby P. J., Garratt L. W., Iosifidis T., Zosky G. R., et al. (2018). Effects of human rhinovirus on epithelial barrier integrity and function in children with asthma. Clin. Exp. Allergy 48, 513–524. doi: 10.1111/cea.13097 [DOI] [PubMed] [Google Scholar]
- Matsuoka Y., Endo K., Kawamura Y., Yoshida T., Saga T., Watanabe Y., et al. (1990). Normal bronchial mucus contains high levels of cancer-associated antigens, CA125, CA19-9, and carcinoembryonic antigen. Cancer 65, 506–510. doi: [DOI] [PubMed] [Google Scholar]
- Meier P. S., Troller R., Grivea I. N., Syrogiannopoulos G. A., Aebi C. (2002). The outer membrane proteins UspA1 and UspA2 of moraxella catarrhalis are highly conserved in nasopharyngeal isolates from young children. Vaccine 20, 1754–1760. doi: 10.1016/S0264-410X(02)00030-0 [DOI] [PubMed] [Google Scholar]
- Murphy T. F., Brauer A. L., Schiffmacher A. T., Sethi S. (2004). Persistent colonization by haemophilus influenzae in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 170, 266–272. doi: 10.1164/rccm.200403-354OC [DOI] [PubMed] [Google Scholar]
- Nakagome K., Bochkov Y. A., Ashraf S., Brockman-Schneider R. A., Evans M. D., Pasic T. R., et al. (2014). Effects of rhinovirus species on viral replication and cytokine production. J. Allergy Clin. Immunol. 134, 332–341. doi: 10.1016/j.jaci.2014.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prjibelski A., Antipov D., Meleshko D., Lapidus A., Korobeynikov A.. (2020). Using SPAdes de novo assembler. Current Protocols in Bioinformatics 70, e102. doi: 10.1002/cpbi.102 [DOI] [PubMed] [Google Scholar]
- Robinson M. D., Oshlack A. (2010). A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25. doi: 10.1186/gb-2010-11-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe H. M., Meliopoulos V. A., Iverson A., Bomme P., Schultz-Cherry S., Rosch J. W. (2019). Direct interactions with influenza promote bacterial adherence during respiratory infections. Nat. Microbiol. 4, 1328–1336. doi: 10.1038/s41564-019-0447-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajjan U. S., Jia Y., Newcomb D. C., Bentley J. K., Lukacs N. W., LiPuma J. J., et al. (2006). H. influenzae potentiates airway epithelial cell responses to rhinovirus by increasing ICAM-1 and TLR3 expression. FASEB J. 20, 2121–2123. doi: 10.1096/fj.06-5806fje [DOI] [PubMed] [Google Scholar]
- Singh B., Alvarado-Kristensson M., Johansson M., Hallgren O., Westergren-Thorsson G., Mörgelin M., et al. (2016). The respiratory pathogen moraxella catarrhalis targets collagen for maximal adherence to host tissues. mBio 7, e00066. doi: 10.1128/mBio.00066-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B., Blom A. M., Unal C., Nilson B., Mörgelin M., Riesbeck K. (2010). Vitronectin binds to the head region of moraxella catarrhalis ubiquitous surface protein A2 and confers complement-inhibitory activity. Mol. Microbiol. 75, 1426–1444. doi: 10.1111/j.1365-2958.2010.07066.x [DOI] [PubMed] [Google Scholar]
- Slevogt H., Seybold J., Tiwari K. N., Hocke A. C., Jonatat C., Dietel S., et al. (2007). Moraxella catarrhalis is internalized in respiratory epithelial cells by a trigger-like mechanism and initiates a TLR2- and partly NOD1-dependent inflammatory immune response. Cell Microbiol. 9, 694–707. doi: 10.1111/j.1462-5822.2006.00821.x [DOI] [PubMed] [Google Scholar]
- Smith C. M., Sandrini S., Datta S., Freestone P., Shafeeq S., Radhakrishnan P., et al. (2014). Respiratory syncytial virus increases the virulence of streptococcus pneumoniae by binding to penicillin binding protein 1a. a new paradigm in respiratory infection. Am. J. Respir. Crit. Care Med. 190, 196–207. doi: 10.1164/rccm.201311-2110OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spaniol V., Heiniger N., Troller R., Aebi C. (2008). Outer membrane protein UspA1 and lipooligosaccharide are involved in invasion of human epithelial cells by moraxella catarrhalis. Microbes Infect. 10, 3–11. doi: 10.1016/j.micinf.2007.09.014 [DOI] [PubMed] [Google Scholar]
- Sun K., Metzger D. W. (2008). Inhibition of pulmonary antibacterial defense by interferon-gamma during recovery from influenza infection. Nat. Med. 14, 558–564. doi: 10.1038/nm1765 [DOI] [PubMed] [Google Scholar]
- Tan T. T., Forsgren A., Riesbeck K. (2006). The respiratory pathogen moraxella catarrhalis binds to laminin via ubiquitous surface proteins A1 and A2. J. Infect. Dis. 194, 493–497. doi: 10.1086/505581 [DOI] [PubMed] [Google Scholar]
- Tan T. T., Nordström T., Forsgren A., Riesbeck K. (2005). The respiratory pathogen moraxella catarrhalis adheres to epithelial cells by interacting with fibronectin through ubiquitous surface proteins A1 and A2. J. Infect. Dis. 192, 1029–1038. doi: 10.1086/432759 [DOI] [PubMed] [Google Scholar]
- Unhanand M., Maciver I., Ramilo O., Arencibia-Mireles O., Argyle J. C., McCracken G. H., et al. (1992). Pulmonary clearance of moraxella catarrhalis in an animal model. J. Infect. Dis. 165, 644–650. doi: 10.1093/infdis/165.4.644 [DOI] [PubMed] [Google Scholar]
- Verhaegh S. J. C., Snippe M. L., Levy F., Verbrugh H. A., Jaddoe V. W. V., Hofman A., et al. (2011). Colonization of healthy children by moraxella catarrhalis is characterized by genotype heterogeneity, virulence gene diversity and co-colonization with haemophilus influenzae. Microbiol. (Reading) 157, 169–178. doi: 10.1099/mic.0.042929-0 [DOI] [PubMed] [Google Scholar]
- Virji M., Evans D., Griffith J., Hill D., Serino L., Hadfield A., et al. (2000). Carcinoembryonic antigens are targeted by diverse strains of typable and non-typable haemophilus influenzae. Mol. Microbiol. 36, 784–795. doi: 10.1046/j.1365-2958.2000.01885.x [DOI] [PubMed] [Google Scholar]
- Wang J., Gray-Owen S. D., Knorre A., Meyer T. F., Dehio C. (1998). Opa binding to cellular CD66 receptors mediates the transcellular traversal of neisseria gonorrhoeae across polarized T84 epithelial cell monolayers. Mol. Microbiol. 30, 657–671. doi: 10.1046/j.1365-2958.1998.01102.x [DOI] [PubMed] [Google Scholar]
- Wang J. H., Kwon H. J., Jang Y. J. (2009). Rhinovirus enhances various bacterial adhesions to nasal epithelial cells simultaneously. Laryngoscope 119, 1406–1411. doi: 10.1002/lary.20498 [DOI] [PubMed] [Google Scholar]
- Wang X., Seed B. (2003). A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 31, e154. doi: 10.1093/nar/gng154 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The name of the repository/repositories and accession number(s) can be found below: PRJNA875903.