Abstract
The chronicity of Brucella abortus infection in humans and animals depends on the organism's ability to escape host defenses by gaining entry and surviving inside the macrophage. Although no human vaccine exists for Brucella, vaccine development in other bacteria has been based on deletions of selective nutritional as well as regulatory systems. Our goal is to develop a vaccine for Brucella. To further this aim, we have used a green fluorescent protein (GFP) reporter system to identify constitutively and intracellularly induced B. abortus genes. Constitutively producing gfp clones exhibited sequence homology with genes associated with protein synthesis and metabolism (initiation factor-1 and tRNA ribotransferase) and detoxification (organic hydroperoxidase resistance). Of greater interest, clones negative for constitutively produced gfp in agar were examined by fluorescence microscopy to detect promoter activity induced within macrophages 4 and 24 h following infection. Bacterial genes activated in macrophages 4 h postinfection appear to be involved in adapting to intracellular environmental conditions. Included in this group were genes for detoxification (lactoglyglutathione lyase gene), repair (formamidopyrimidine-DNA glycosylase gene), osmotic protection (K+ transport gene), and site-specific recombination (xerD gene). A gene involved in metabolism and biosynthesis (deoxyxylulose 5′ phosphate synthase gene) was also identified. Genes activated 24 h following infection were biosynthesis- and metabolism-associated genes (iron binding protein and rhizopine catabolism). Identification of B. abortus genes that are activated following macrophage invasion provides insight into Brucella pathogenesis and thus is valuable in vaccine design utilizing selective targeted deletions of newly identified Brucella genes.
Brucella spp. are gram-negative facultative intracellular bacteria and the causative agent of human and animal brucellosis, a persistent disease that is difficult to diagnose and treat. The bacteria penetrate the mucosa of the nasal, oral, or pharyngeal cavities and are phagocytized by host macrophages, where survival and replication occurs. Following a variable bacteremic stage, the organisms localize in the reticuloendothelial system. This intracellular localization likely accounts for the persistence of Brucella infections. Thus, the identification of bacterial proteins that contribute to the replication and survival of the bacteria is critical in understanding the protective mechanisms and pathogenesis of the disease.
Although several virulence factors are known, the basis for virulence in Brucella infections remains poorly understood. Early studies on virulence factors were directed at the structural features of the outer membrane (37). The outer membrane contains two recognized virulence factors: lipopolysaccharide (LPS) and outer membrane proteins. LPS was recognized for its virulence role when naturally occurring isolates lacking LPS showed reduced survival. However, the role of outer membrane proteins in survival and virulence remains unknown.
Recent attention has focused on the interaction between the bacterium and macrophages. The use of the gfp reporter system to identify genes of other bacteria that are differentially expressed in host cells has been documented (4, 42). Using murine macrophages and the gfp expression system, several Brucella suis genes have been identified that are induced 24 to 48 h after intracellular infection (20). The majority of the genes identified were involved in metabolic pathways. Alternatively, using signature-tagged mutagenesis (STM), others have identified a number of genes involved in virulence 48 h postinfection (13). Here, using the gfp expression system, we have identified 33 Brucella genes induced 4 h following intracellular infection of macrophages and 13 genes induced by 24 h of infection. Twenty-two Brucella genes were constitutively induced in agar. This report identifies Brucella genes that encode proteins associated with stress, as well as global bacterial regulatory mechanisms, induced following early intracellular infection. These findings provide an approach to vaccine design through selective targeted deletions of newly identified Brucella genes.
MATERIALS AND METHODS
Bacteria and cell lines.
Brucella abortus strain S2308 (National Animal Disease Center, Ames, Iowa) was grown in 12-by-75-mm tubes on a shaker platform in 4 ml of brucella broth (Difco) or on plates of brucella broth containing 1.5% agar. Cultures were grown at 37°C for 3 to 4 days.
The mouse macrophage cell line RAW 264.7 (ATCC TIB71), which has been used for the study of Brucella (5), was maintained at 37°C with 5% CO2 in supplemented RPMI 1640 (including 10% fetal bovine serum, 0.2 mM l-glutamine, penicillin [100 U/ml], and streptomycin [100 μg/ml]) (Sigma, St. Louis, Mo.). Cells were plated in eight-well chamber slides (Nalge Nunc International, Naperville, Ill.) in supplemented RPMI 1640 without antibiotics 1 day prior to infection.
Plasmid construction.
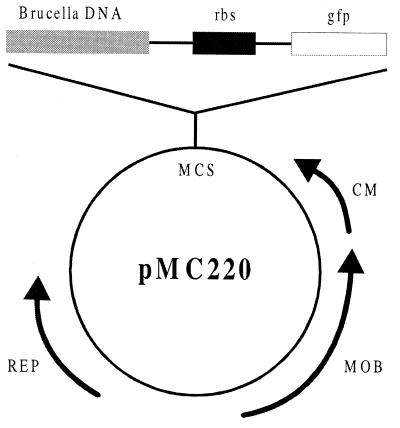
Plasmid KEN containing the T7 (gene 10) ribosomal binding site (RBS) and wild-type gfp gene was obtained from Stanley Falkow (Stanford University). The RBS was inserted into the pGVFPuv (Clontech Laboratories, Palo Alto, Calif.) by ligating the KpnI/NcoI fragment of pKEN into the KpnI/NcoI site of pGVFPuv. The resulting plasmid, termed pMR219-1, contained the RBS and gfpuv. The SmaI/StuI fragment was removed from pMR219-1 and inserted into the EcoRV site of pSL118. The KpnI/SmaI fragment, which contained the RBS, gfpuv, and a BglII site, was removed and ligated into the SmaI/KpnI site of the low-copy-number plasmid, BBR1MCS, and the resulting plasmid was termed pMC220 (Fig. 1). Plasmid MC220Tac was created by inserting the Tac promoter into the BglII site for use as a positive control.
FIG. 1.
The engineered pMC220 vector with the promoterless gfpuv gene (gfp), the RBS, and an inserted fragment of B. abortus DNA. The plasmid, known to transform B. abortus, has been modified to contain a random library of 0.3 to 3 kb Sau3AI DNA fragments of B. abortus upstream of the inserted T7 (gene 10) RBS optimally placed for transcriptional fusion of the promoterless gfpuv gene. Abbreviations: MCS, multiple cloning site; CM, chloramphenicol acetyltransferase gene conferring chloramphenicol resistance; MOB, gene required for plasmid mobilization; REP, gene required for plasmid replication.
Library construction.
To construct the B. abortus library, genomic DNA of B. abortus was partially digested with Sau3AI and size fractionated on a 1% agarose gel. Fragments ranging from 0.3 to 3 kb were excised and isolated using a gel extraction kit (Qiagen Inc., Valencia, Calif.). Plasmid MC220 was digested with BglII, treated with calf intestine phosphatase for 30 min at 37°C, and then incubated at 75°C for 10 min to stop the reaction. B. abortus fragments were ligated into pMC220, and the resulting library was expanded in Escherichia coli UltraMAX DH5α competent cells (Life Technologies, Rockville, Md.) prior to electroporation into Brucella competent cells. To prepare B. abortus competent cells, bacteria were grown for 42 to 45 h, chilled on ice for 10 min, and then centrifuged. The pellet was washed three times with double-distilled water, resuspended in 10% glycerol, and diluted 1:10,000 in brucella broth. After 48 h of culture, the competent cells were resuspended at 5 × 108 cells/ml and frozen in 50-μl aliquots. The library construct (0.25 μg/2 μl) was mixed with 50 μl of competent cells and incubated on ice for 30 min. Electroporation was performed using a Gene Pulser II (Bio-Rad, Hercules, Calif.) at 1.5 kV, 400 Ω, and 25 μF. Brucella broth (1 ml) was added, and cultures were incubated on a shaker platform for 4 h at 37°C. Colonies were plated on agar plates containing chloramphenicol (20 μg/ml) and incubated at 37°C for 3 days. Master plates were created by subcloning individual colonies in six-by-six grid plates.
Screening.
Colonies that were positive on agar were identified using a handheld UV light (wavelength, 350 nm). To identify colonies possessing promoter activity inside the macrophage, RAW cells were plated in eight-well chamber slides. Cells were cultured overnight in 1 ml of supplemented RPMI 1640 without antibiotics. For rapid screening, three to six clones from the master plates were combined in each chamber, but colonies exhibiting green fluorescence on agar were excluded. Cultures were incubated at 37°C with 5% CO2. After 4 or 24 h, medium was removed, and cells were washed twice with phosphate-buffered saline (PBS) containing gentamicin (Sigma) and then fixed in 4% paraformaldehyde for 30 min at room temperature. The fixative was removed, and the slide was detached from the chamber. For cultures infected for 24 h, the bacteria were removed by washing after 1.5 h and medium containing gentamicin was added. After an additional 22.5 h, cultures were fixed as described above. Slides were washed with PBS and examined by microscopy (Carl Zeiss, Halbergmoos, Germany) for fluorescence. Macrophages containing fluorescent bacteria were digitally recorded using a charge-coupled device camera and Axiovert version 2.0 software. Colonies identified as positive were reexamined individually using isolates from the master plates to identify the positive colony. Brucella containing pMC220Tac electroporated into macrophages was used as a positive control for screening Brucella colonies at 4 and 24 h.
Sequencing of selected clones.
DNA was isolated from selected colonies and transformed into E. coli DH5α. A single clone was selected, grown overnight, and digested with SacI to determine insert size. DNA was sequenced by the UW Biotechnology Center as previously described (21). Data were analyzed using PE-Biosystems Sequencing Analysis version 3.1.
RT-PCR.
Bacteria from 10 ml of exponentially growing cultures of B. abortus strain S2308 containing pMC220Tac were pelleted; resuspended in a solution containing 900 μl of Tris-EDTA, 100 μl of 10% sodium dodecyl sulfate, and 5 μl of proteinase K (20 μg/ml; Promega, Madison, Wis.); and incubated at 37°C for 1 h. Total RNA was isolated using the RNeasy Mini Kit according to the manufacturer's protocol (Qiagen). RNA was treated with 1 μl of RQ1 RNase-free DNase (Promega) in a 1× reaction mixture of RQ1 DNase buffer (Promega) plus 1 μl of rRNasin (Promega) for 20 min at 37°C. After digestion, RNA was reisolated using an RNeasy Mini Kit according to the manufacturer's protocol for RNA cleanup. The DNase-treated RNA was heat denatured and reverse transcribed as previously described (20). Sense and antisense primers for the tested genes are shown in Table 1. The reverse transcription (RT) mix was diluted 1:10 in DNase-, RNase-free water (Life Technologies), and 2 μl was used as the cDNA template. The PCR master mix for each reaction was mixed according to the FailSafe PCR PreMix selection kit (Epicentre, Madison, Wis.), which contained 1 μl of 50 μM sense primer, 1 μl of 50 μM antisense primer, 2 μl of cDNA template, 0.5 μl of FailSafe PCR Enzyme Mix (Epicentre), and DNase-, RNase-free water in a total volume of 25 μl. Twenty-five microliters of FailSafe PCR 2× PreMix was added to the PCR master mix solution. PCR was performed for 30 cycles at 94°C for 60 s, 60°C for 60 s, and 72°C for 60 s followed by 10 min at 72°C. Products were analyzed on 1.5% agarose gel, with molecular mass standards (Gene Ruler 50bp DNA Ladder; MBI Fermentas, Amherst, N.Y.). PCR amplification products were purified using a QIAquick PCR Purification Kit (Qiagen) and confirmed by sequencing.
TABLE 1.
Sense and antisense primers for RT-PCR-amplified genes
| Clone | Target | Primers | PCR product (bp) |
|---|---|---|---|
| None | gfp | 5′-AAGGAGAAGAACTTTTCACT-3′, 5′-TAATGGTCTGCTAGTTGAAC-3′ | 522 |
| 17B2 | Hypothetical protein gene | 5′-GCTATACCAATTCCGGCCCC-3′, 5′-AACGGGCTTCGACCATTCGG-3′ | 98 |
| 19E4 | IF-1 | 5′-ATGGCGAAAGAAGAAGTCCT-3′, 5′-ACTAGAACCTTGTCACCGGC-3′ | 164 |
| 30C4 | ohr | 5′-CTTTACACGACCCAGTCCAC-3′, 5′-AAAGCTGTTCGGGATTGGTG-3′ | 138 |
Nucleotide sequence accession numbers.
Accession numbers for sequences submitted to GenBank (http://www.ncbi.nlm.nih.gov) are indicated in Tables 2 to 4.
TABLE 2.
Genes associated with 4-h Brucella infection of macrophagesa
| Clone | Gene | Product description | % Similarity | Species | Accession no. |
|---|---|---|---|---|---|
| 15D5 | xerD | Integrase recombinase | 62 | E. coli | AF279676 |
| 25C5 | glxI | Lactoylglutathione lyase | 77 | N. meningitidis | AF291852 |
| cspE | Negative regulator of cold shock protein | 67 | E. coli | —b | |
| 32C5 | fpg | Formamidopyrimidine DNA glycosylase | 82 | R. meliloti | AF279677 |
| fadB1 | Enoyl-CoA hydratase | 84 | R. meliloti | —b | |
| 36D5 | trkH | K+ transport system | 61 | Vibrio alginolyticus | AF288526 |
| 24E3 | mtgA | Monofunctional biosynthesis peptidoglycan transglycosylase | 57 | H. influenzae | AF287157 |
| 13C3/10B2 | dxs/tkt | Deoxyxylulose 5′ phosphate synthase/transketolase | 67 | Arabidopsis thaliana | AF288528 |
| 8B4 | p39 | Cytoplasmic protein p39 | 96 | B. abortus | AF360361c |
An additional 25 clones were identified, but no sequence similarity was determined using GenBank.
—, two gene fragments were present within the same clone.
Previously identified by other workers (10).
TABLE 4.
Genes constitutively produced in agara
| Clone | Gene | Product description | % Similarity | Species | Accession no. |
|---|---|---|---|---|---|
| 30C4 | ohr | Organic hydroperoxide resistance | 67 | Xanthomonas campestris | AF288739 |
| 19E4 | IF-1 | Translation initiation factor | 87 | Rickettsia prowazekii | AF288531 |
| 17B2 | NIc | Hypothetical protein | 65 | C. beijerinckii | AF293418 |
| 36C2 | putA | Ribotransferase protein dehydrogenase | 86 | Agrobacter tumefaciens | AF291853 |
| tgt | Queuine tRNA | 66 | R. prowazeki | —b | |
| 33F5 | NI | ATP binding transporter | 66 | A. tumefaciens | AF291423 |
An additional 17 clones were identified, but no sequence similarity was determined using GenBank.
—, two gene fragments were present within the same clone.
NI, not identified.
RESULTS
Identification of genes associated with 4-h Brucella infection of macrophages.
To determine whether Brucella could distinguish the macrophage environment as distinct from agar, nonfluorescent bacterial colonies, containing pMC220 with a Brucella gene fragment, were added to macrophages. After 4 h, the cells were washed with PBS containing gentamicin and then fixed with paraformaldehyde. Using fluorescence microscopy (Fig. 2), 34 clones were identified that were activated within the macrophage environment but not activated in agar. Of these clones, 9 possessed similarity with other bacterial sequences in GenBank ranging from 57 to 96% (Table 2) and were in the correct orientation. Although all inserts were unique, the dxs/tkt gene was identified twice, supporting the importance of this bacterial gene to intracellular survival. Twenty-five clones contained sequences that could not be identified. Bacteria outside of the macrophages failed to fluoresce, excluding the possibility of promoter activation by the medium.
FIG. 2.
The macrophage cell line RAW 267.4 infected with B. abortus s2308 that has been transformed with the plasmid pMC220 expressing B. abortus DNA fragment and the GFP, clone 39B6. Note individual GFP-positive bacteria are contained within the cytoplasm of the macrophages.
Identification of genes associated with 24-h Brucella infection of macrophages.
Clones, negative on agar and after 4 h infection, were added to macrophages for 24 h. After fixation with paraformaldehyde, slides were examined by fluorescent microscopy for gfp induction. Thirteen clones were identified. Three clones exhibited sequence similarity to sequences in GenBank (Table 3).
TABLE 3.
Genes associated with 24-h Brucella infection of macrophagesa
| Clone | Gene(s) | Product description | % Similarity | Species | Accession no. |
|---|---|---|---|---|---|
| 35F6 | ssuBAC | Aliphatic sulfonate transport permease protein | 51 | B. subtilis | AF288529 |
| 39B6 | mocC | Rhizopine catabolism | 67 | R. leguminosarum | AF288530 |
| 41B1 | fbpA | Iron binding protein precursor | 74 | Mannheimia haemolytica | AF290976 |
An additional 11 clones were identified, but no sequence similarity was determined using GenBank.
Identification of constitutively produced Brucella genes.
DNA from 22 of the 32 clones that constitutively produced gfp were isolated and sequenced. Five of the identified clones demonstrated significant similarity to sequences within the GenBank database (Table 4). Similarities ranged from 66 to 87%. The remaining 17 clones possessed no significant similarity with known sequences.
Verification of gene transcription.
To verify the active transcription of genes identified by GFP expression, RT-PCR was performed for selected genes. Three genes that were induced on agar were chosen: hypothetical protein, IR-1, and ohr. Reactions were performed with and without reverse transcriptase, and gfp expression was used as a positive control. Bands of appropriate sizes were observed as shown in Fig. 3. The bands were isolated and sequenced. Sequence analysis confirmed that all three genes initially identified by gfp expression were actively transcribed. Attempts to verify the active transcription of genes expressed within the macrophage were unsuccessful. Insufficient amounts of bacterial RNA were obtained following 4-h infection of macrophages.
FIG. 3.
RT-PCR analysis of constitutively produced genes. RT-PCR was performed using DNase-treated total RNA from B. abortus transformed with pMC22Tac. Transcription was done in the presence (lanes 2, 4, 6, and 8) or absence (lanes 3, 5, 7, and 9) of reverse transcriptase and IF-1 primers (lanes 4 and 5), hypothetical protein primers (lane 6 and 7), or ohr primers (lanes 8, 9). GFP primers (lanes 2 and 3) were included as a positive control. Gene Ruler 50bp DNA Ladder was used as the molecular mass standard (lane 1).
DISCUSSION
Intracellular bacterial infection requires significant interaction between host and pathogen and comprises four steps: adherence, invasion, establishment, and dissemination. To survive and multiply in host cells, bacteria sense the environment, activate various genes, and secrete proteins that facilitate bacterial adaptation. However, the mechanisms used by Brucella to sense and respond to changes within the intracellular environment are not understood. To identify genes activated during infection, Brucella gene fragments (0.3 to 3 kb) were inserted upstream of the promoterless gfp gene. The resulting plasmids were electroporated into B. abortus. Using this promoter expression strategy, bacterial genes activated under different environmental conditions could be identified.
To survive, intracellular bacteria must respond to a variety of environmental conditions. When bacteria enter a different environment, virulence genes may be expressed that enable bacteria to survive. Environmental cues that cause expression of virulence genes are often physical and chemical factors such as temperature, pH, and reactive oxygen molecules (15). We have identified two clones, 25C5 and 32C5, whose genes are induced within 4 h of macrophage infection, and these genes appear to play a role in response to such factors.
Lactoglyglutathione lyase (GlxI) is part of the glyoxalase pathway. This metabolic pathway functions in detoxification by converting methylglyoxal, a product of glycolysis, into d-lactic acid (40). S-Lactoglyglutathione, the product of glyoxalase activity, activates KefB and KefC. These components protect against the toxic effect of electrophiles by decreasing intracellular pH (24). Methylglyoxal is required for growth in some environmental situations; however, overproduction of methylglyoxal can lead to cell death (12). Cells that overproduce glyoxalase I have an increased tolerance for methylglyoxal and increased rates of detoxification. Null mutant strains exhibit low rates of detoxification and die rapidly (24). Thus, the induction of glxI may result in enhanced proliferation and survival of intracellular Brucella. An additional gene, cspE, was identified from the sequence of clone 25C5. cspE encodes the cold shock protein E. Bacteria respond to an abrupt decrease in temperature by producing cold shock proteins, synthesizing proteins involved in transcription and translation, and repressing heat shock proteins. CspE is produced at 37°C and functions as a negative regulator to prevent the expression of cold shock gene cspA (3). Expression of csp genes in Myxococcus xanthus is not regulated by temperature, but these genes are considered important for this soil dwelling bacterium to survive under conditions of exposure to environmental changes. Although the role of cspE in Brucella virulence is unknown, the protein encoded by the cspE gene may serve to regulate other bacterial genes during stress of intracellular infection.
Formamidopyrimidine-DNA glycosylase (the fpg gene encodes formamidopyrimidine-DNA glycosylase) is a pyrimidine base excision repair protein that functions to protect cells from mutagenic effects. Fpg prepares the damaged DNA for excision by exonuclease III and IV (2). Interestingly, this repair mechanism is important for E. coli damaged by H2O2 under low-iron conditions (14). Thus, Brucella exposed to H2O2 under low-iron conditions as present in the macrophage phagosome may activate this DNA repair mechanism. Also, downstream of fpg was an additional gene, fadB1, encoding enoyl coenzyme A (enoyl-CoA) hydratase. Enoyl-CoA is the product of a metabolism-associated gene that is utilized in the enzymatic catabolic pathway to oxidize fatty acids to produce acetyl-CoA, which enters the trichloroacetic acid cycle. This same fpg and fadB1 gene organization can be found in Rhizobium meliloti, a closely related bacterium of plants (25).
In addition to adapting to a changing environment, bacteria must be able to survive and replicate. Needed genes are expressed while unnecessary genes are downregulated. We have identified four replication or metabolism associated genes.
Deoxyxylulose 5′ phosphate synthase is a transketolase-like enzyme within the recently identified mevalonate-independent pathway (39) that catalyzes the synthesis of d-1-deoxyxylulose 5′ phosphate and 2-C-methyl-d-erythritol 4-phosphate leading to the formation of isopentenyl diphosphate, the precursor of isoprenoids, thiamine, and pyridoxal biosynthesis (23). Synthesis of d-1-deoxyxylulose 5′ phosphate is a first step in isoprenoid biosynthesis of antioxidants. Interestingly, one of these antioxidants, 2-C-methyl-d-erythritol-2,4-cyclopyrophosphate, accumulates in some bacteria during heat shock and in the presence of O2−-generating compounds from macrophages (30). Thus, Brucella may utilize deoxyxylulose 5′ phosphate synthase to catabolize the accumulation of antioxidant compounds that accumulate during initial shock and in the presence of O2− -generating compounds within the phagosome of the macrophage.
Site-specific recombinase (XerD) is a site-specific recombinase that functions with a second recombinase, XerC, to catalyze DNA strand cleavage to ensure normal chromosome or plasmid DNA segregation during cell division. These recombinases can remove the potential damage of homologous recombination (6). In E. coli, a two-component regulatory system is essential for Xer site-specific recombination in plasmids under anaerobic conditions. Such anaerobic or stress-like conditions might exist within the macrophage. Upregulation of XerD may be an early step in the process of replication.
Monofunctional biosynthetic peptidoglycan transglycosylase (MtgA) synthesizes linear glycan chains from lipid-linked precursors and functions in peptidoglycan biosynthesis. MtgA has the amino-terminal domain of the class A high-molecular-weight penicillin binding protein but not the carboxy transpeptidase domain and, thus, is not a penicillin binding protein (B. G. Spratt, J. Zhou, M. Taylor, and M. J. Merrick, Letter, Mol. Microbiol. 19:639–640, 1996). The activation of the mtgA gene to a detectable level in macrophages and not agar is uncertain but may be in response to damage to the peptidoglycan layer of the bacterium occurring in the phagosome.
The mocC (mannityl opine catabolism) gene encodes an oxidoreductase that is utilized in the catabolism of mannopine (18). At least four genes, mocABC and mocR, appear to play a role in the catabolism of this compound. This gene is homologous to the mocC gene of Rhizobium leguminosarum. Other genes common to Rhizobium and Brucella that are involved in pathogenesis have been identified (16, 22). In R. leguminosarum, MocC catabolizes rhizopine, an opine-like compound utilized by bacteria to enhance survival. Thus, it is possible that MocC enhances the intracellular survival of Brucella.
In order to survive in a changing environment, bacteria must adapt to variations in nutrient availability. An array of transport systems are used to scavenge for needed substrates. In response to the intracellular macrophage environment, four transport systems were induced in Brucella.
Bacteria that encounter an increase in osmolarity respond initially by taking up large amounts of K+. This response to osmotic stress quickly leads to an increase in the synthesis of glutamate, which is necessary for maintenance of the steady-state K+ pool (17). Three different K+ uptake mechanisms function in E. coli: Trk, Kup, and Kdp. The Trk system is composed of four proteins: TrkA, TrkE, TrkG, and TrkH. Although only TrkA is essential for K+ uptake, mutations in TrkH reduce the binding of TrkA. TrkG and TrkH appear to play similar roles since either, but not both, is necessary in Trk activity (35).
Iron is present in limited quantities in the human host since most of the iron is complexed with various proteins. Additionally, during an infection, the intracellular and extracellular iron is reduced (43). Bacteria need iron for growth and thus have developed elaborate mechanisms to obtain iron. The periplasmic protein-dependent transporter, fbpA, carries iron through the periplasm to the cytoplasmic membrane. Transcriptional regulation of fbpA in Neisseria gonorrhoeae and E. coli is related to the level of available iron (36); thus, fbpA in Brucella may serve to scavenge intracellular iron.
The Brucella p39 gene encodes a putative cytoplasmic protein (10). Based on comparative modeling, the predicted role of p39 is as a periplasmic substrate-binding component of a binding-protein-dependent transport system (9). Recent studies suggest that bacterial periplasmic substrate-binding proteins, in addition to their function in transport and chemotaxis, are implicated in protein folding and protection from stress in the periplasm (34). Although p39 is an immunodominant protein, deletion mutants did not alter virulence in mice (41). However, p39, in conjunction with adjuvant CpG, was capable of inducing a T helper 1 (Th1) response in mice (1). The ability of p39 to induce a Th1 immune response makes this protein an appealing candidate for a vaccine against brucellosis.
The 35F6 sequence demonstrated similarity to the aliphatic sulfonate transport permease protein (ssuBAC). The ssuBAC genes are required for the utilization of sulfur from aliphate sulfonates. The B, A, and C forms appear to comprise an ABC-type transport system. The ssuBAC genes are only expressed during sulfate or cysteine starvation (11).
The five clones sequenced from constitutively produced gfp function in detoxification, electron transfer, and protein synthesis. The organic hydroperoxide resistance (ohr) gene (28) functions in the breakdown of toxic organic peroxides resulting from respiration. A second gene, IF-1, is essential for bacterial survival and likely functions in the initiation phase of protein synthesis (8). Clone 17B2 exhibited sequence similarity with a hypothetical protein identified in Clostridium beijerinckii. This open reading frame (open reading frame 1) is just upstream of the peptide deformylase (fms) and formyltransferase (fmt) operon (26). The function of open reading frame 1 has not been identified. PutA is a membrane protein which catalyzes the conversion of proline to glutamate (38). Although some bacteria utilize an osmoregulated proline transport system, proline is not an osmoprotectant in gram-negative bacteria (17). PutA is required for growth on proline as a carbon source. A second gene identified from the sequence of 36C2 was queuine tRNA ribotransferase. This enzyme is involved in the biosynthesis of the hypermodified tRNA nucleoside queuosine (Q) (33). Lastly, clone 33F5 exhibited homology with a gene encoding an ABC associated with mannopine transport (29).
Of the 1,500 clones examined for gfp expression, 2.1% of the clones exhibited constitutive fluorescence in agar, while gfp was induced 4 h following infection of macrophages in 2.2% of the clones. Of the 288 clones examined 24 h after infection of macrophages, 4.5% of the clones induced gfp. Thus, our system provides the opportunity to examine the bacterial genes expressed under different environmental conditions. Visually, the fluorescence intensity varied among clones grown on agar as well as inside macrophages. However, clones reexamined on different occasions possessed a similar fluorescence intensity, suggesting stability of promoter activity.
The gfp reporter system has been useful in identifying genes activated in intracellular bacteria. Using this system, others have identified several B. suis genes activated 24 to 48 h following macrophage infection (20). The majority of the identified genes encoded nucleotide-binding proteins and transport system proteins. Additionally, a glutaredoxin-like gene was identified (20). This gene may play a role in maintaining a reducing environment inside the macrophage. The role of these genes in virulence has not been confirmed. No constitutively expressed genes were identified. An alternative and complementary approach to identifying virulence genes is STM. STM identifies mutant strains defective for growth within a host cell. Using this method, a type IV secretion system and two LysR transcriptional regulator systems were identified along with several genes involved in metabolism and biosynthesis (13).
For intracellular bacteria, survival and replication within phagocytic cells are the key to pathogenesis. Successful strategies for intracellular survival include the ability to survive in acidified membrane-bound vesicles (32), inhibition of macrophage apoptosis (27), prevention of phagosome-lysosome fusion (7) and detoxification and repair mechanisms. Although the ability of B. abortus to prevent phagosome-lysosome fusion (31) has been documented, the mechanisms for Brucella intracellular survival remain poorly understood. B. abortus likely utilizes a number of strategies to counter the hostile macrophage environment. Within the context of the macrophage, bacterial genes are likely activated in an orchestrated fashion as the macrophage displays an array of potentially bactericidal pathways.
Our GFP reporter system allows the simultaneous identification of constitutively and intracellularly induced genes. We have identified several potential antioxidant defense mechanisms that appear to function in early intracellular survival of B. abortus. Genes identified later in the infectious process were metabolism associated, while constitutively produced genes functioned in detoxification, electron transfer, and protein synthesis. Experiments are under way to verify the role of these genes in virulence. Responses to the environment, nutritional requirements, and metabolic and regulatory systems are important aspects of bacterial pathogenesis. An understanding of the expression of bacterial genes, both in vivo and in vitro, will provide valuable information for vaccine development.
ACKNOWLEDGMENTS
This work was supported by USDA grant 98-35204-6760, CAPES-Brazil fellowship BEX1239/98-8, NIH grant R01-AI48490, and BARD grants US-2781-96 and US-2968-98C.
Appreciation is expressed to Ivin Chen and Darby Brown for colony screening.
Linda Eskra and Aurea Canavessi contributed equally to this work.
REFERENCES
- 1.Al-Mariri A, Tibor A, Mertens P, De Bolle X, Michel P, Godefroid J, Walravens K, Letesson J J. Protection of BALB/c mice against Brucella abortus 544 challenge by vaccination with bacterioferritin or P39 recombinant proteins with CpG oligodeoxynucleotides as adjuvant. Infect Immun. 2001;69:4816–4822. doi: 10.1128/IAI.69.8.4816-4822.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asagoshi K, Yamada T, Terato H, Ohyama Y, Monden Y, Arai T, Nishimura S, Aburatani H, Lindahl T, Ide H. Distinct repair activities of human 7,8-dihydro-8-oxoguanine DNA glycosylase and formamidopyrimidine DNA glycosylase for formamidopyrimidine and 7,8-dihydro-8-oxoguanine. J Biol Chem. 2000;275:4956–4964. doi: 10.1074/jbc.275.7.4956. [DOI] [PubMed] [Google Scholar]
- 3.Bae W, Phadtare S, Severinov K, Inouye M. Characterization of Escherichia coli cspE, whose product negatively regulates transcription of cspA, the gene for the major cold shock protein. Mol Microbiol. 1999;31:1429–1441. doi: 10.1046/j.1365-2958.1999.01284.x. [DOI] [PubMed] [Google Scholar]
- 4.Barker L P, Brooks D M, Small P L. The identification of Mycobacterium marinum genes differentially expressed in macrophage phagosomes using promoter fusions to green fluorescent protein. Mol Microbiol. 1998;29:1167–1177. doi: 10.1046/j.1365-2958.1998.00996.x. [DOI] [PubMed] [Google Scholar]
- 5.Barthel R, Feng J, Piedrahita J A, McMurray D N, Templeton J W, Adams L G. Stable transfection of the bovine NRAMP1 gene into murine RAW264.7 cells: effect on Brucella abortus survival. Infect Immun. 2001;69:3110–3119. doi: 10.1128/IAI.69.5.3110-3119.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blake J A, Ganguly N, Sherratt D J. DNA sequence of recombinase-binding sites can determine Xer site-specific recombination outcome. Mol Microbiol. 1997;23:387–398. doi: 10.1046/j.1365-2958.1997.2261600.x. [DOI] [PubMed] [Google Scholar]
- 7.Buchmeier N A, Heffron F. Inhibition of macrophage phagosome-lysosome fusion by Salmonella typhimurium. Infect Immun. 1991;59:2232–2238. doi: 10.1128/iai.59.7.2232-2238.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cummings H S, Hershey J W. Translation initiation factor IF1 is essential for cell viability in Escherichia coli. J Bacteriol. 1994;176:198–205. doi: 10.1128/jb.176.1.198-205.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Fay K, Tibor A, Lambert C, Vinals C, Denoel P, De B X, Wouters J, Letesson J J, Depiereux E. Structure and function prediction of the Brucella abortus P39 protein by comparative modeling with marginal sequence similarities. Protein Eng. 1999;12:217–223. doi: 10.1093/protein/12.3.217. [DOI] [PubMed] [Google Scholar]
- 10.Denoel P A, Vo T K, Tibor A, Weynants V E, Trunde J M, Dubray G, Limet J N, Letesson J J. Characterization, occurrence, and molecular cloning of a 39-kilodalton Brucella abortus cytoplasmic protein immunodominant in cattle. Infect Immun. 1997;65:495–502. doi: 10.1128/iai.65.2.495-502.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eichhorn E, van der Ploeg J R, Leisinger T. Deletion analysis of the Escherichia coli taurine and alkanesulfonate transport systems. J Bacteriol. 2000;182:2687–2695. doi: 10.1128/jb.182.10.2687-2695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferguson G P, Totemeyer S, MacLean M J, Booth I R. Methylglyoxal production in bacteria: suicide or survival? Arch Microbiol. 1998;170:209–218. doi: 10.1007/s002030050635. [DOI] [PubMed] [Google Scholar]
- 13.Foulongne V, Bourg G, Cazevieille C, Michaux-Charachon S, O'Callaghan D. Identification of Brucella suis genes affecting intracellular survival in an in vitro human macrophage infection model by signature-tagged transposon mutagenesis. Infect Immun. 2000;68:1297–1303. doi: 10.1128/iai.68.3.1297-1303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galhardo R S, Almeida C E, Leitao A C, Cabral-Neto J B. Repair of DNA lesions induced by hydrogen peroxide in the presence of iron chelators in Escherichia coli: participation of endonuclease IV and Fpg. J Bacteriol. 2000;182:1964–1968. doi: 10.1128/jb.182.7.1964-1968.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guiney D G. Regulation of bacterial virulence gene expression by the host environment. J Clin Investig. 1997;99:565–569. doi: 10.1172/JCI119196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iñón de Iannino N, Briones G, Tolmasky M, Ugalde R A. Molecular cloning and characterization of cgs, the Brucella abortus cyclic β(1-2) glucan synthetase gene: genetic complementation of Rhizobium meliloti ndvB and Agrobacterium tumefaciens chvB mutants. J Bacteriol. 1998;180:4392–4400. doi: 10.1128/jb.180.17.4392-4400.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kempf B, Bremer E. Uptake and synthesis of compatible solutes as microbial stress responses to high-osmolality environments. Arch Microbiol. 1998;170:319–330. doi: 10.1007/s002030050649. [DOI] [PubMed] [Google Scholar]
- 18.Kim K S, Chilton W S, Farrand S K. A Ti plasmid-encoded enzyme required for degradation of mannopine is functionally homologous to the T-region-encoded enzyme required for synthesis of this opine in crown gall tumors. J Bacteriol. 1996;178:3285–3292. doi: 10.1128/jb.178.11.3285-3292.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ko J, Splitter G A. Brucella abortus tandem repeated ATP-binding proteins, BapA and BapB, homologs of Haemophilus influenzae LktB, are not necessary for intracellular survival. Microb Pathog. 2000;29:245–253. doi: 10.1006/mpat.2000.0389. [DOI] [PubMed] [Google Scholar]
- 20.Kohler S, Ouahrani-Bettache S, Layssac M, Teyssier J, Liautard J P. Constitutive and inducible expression of green fluorescent protein in Brucella suis. Infect Immun. 1999;67:6695–6697. doi: 10.1128/iai.67.12.6695-6697.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee L G, Spurgeon S L, Heiner C R, Benson S C, Rosenblum B B, Menchen S M, Graham R J, Constantinescu A, Upadhya K G, Cassel J M. New energy transfer dyes for DNA sequencing. Nucleic Acids Res. 1997;25:2816–2822. doi: 10.1093/nar/25.14.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.LeVier K, Phillips R W, Grippe V K, Roop II R M, Walker G C. Similar requirements of a plant symbiont and a mammalian pathogen for prolonged intracellular survival. Science. 2000;287:2492–2493. doi: 10.1126/science.287.5462.2492. [DOI] [PubMed] [Google Scholar]
- 23.Lois L M, Campos N, Putra S R, Danielsen K, Rohmer M, Boronat A. Cloning and characterization of a gene from Escherichia coli encoding a transketolase-like enzyme that catalyzes the synthesis of D-1-deoxyxylulose 5-phosphate, a common precursor for isoprenoid, thiamin, and pyridoxol biosynthesis. Proc Natl Acad Sci USA. 1998;95:2105–2110. doi: 10.1073/pnas.95.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacLean M J, Ness L S, Ferguson G P, Booth I R. The role of glyoxalase I in the detoxification of methylglyoxal and in the activation of the KefB K+ efflux system in Escherichia coli. Mol Microbiol. 1998;27:563–571. doi: 10.1046/j.1365-2958.1998.00701.x. [DOI] [PubMed] [Google Scholar]
- 25.Margolin W, Bramhill D, Long S R. The dnaA gene of Rhizobium meliloti lies within an unusual gene arrangement. J Bacteriol. 1995;177:2892–2900. doi: 10.1128/jb.177.10.2892-2900.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meinnel T, Blanquet S. Characterization of the Thermus thermophilus locus encoding peptide deformylase and methionyl-tRNAfMet formyltransferase. J Bacteriol. 1994;176:7387–7390. doi: 10.1128/jb.176.23.7387-7390.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monack D M, Raupach B, Hromockyj A E, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mongkolsuk S, Praituan W, Loprasert S, Fuangthong M, Chamnongpol S. Identification and characterization of a new organic hydroperoxide resistance (ohr) gene with a novel pattern of oxidative stress regulation from Xanthomonas campestris pv. phaseoli. J Bacteriol. 1998;180:2636–2643. doi: 10.1128/jb.180.10.2636-2643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oger P, Kim K S, Sackett R L, Piper K R, Farrand S K. Octopine-type Ti plasmids code for a mannopine-inducible dominant-negative allele of traR, the quorum-sensing activator that regulates Ti plasmid conjugal transfer. Mol Microbiol. 1998;27:277–288. doi: 10.1046/j.1365-2958.1998.00671.x. [DOI] [PubMed] [Google Scholar]
- 30.Ostrovsky D, Diomina G, Lysak E, Matveeva E, Ogrel O, Trutko S. Effect of oxidative stress on the biosynthesis of 2-C-methyl-D-erythritol-2,4-cyclopyrophosphate and isoprenoids by several bacterial strains. Arch Microbiol. 1998;171:69–72. doi: 10.1007/s002030050680. [DOI] [PubMed] [Google Scholar]
- 31.Pizarro-Cerda J, Moreno E, Sanguedolce V, Mege J L, Gorvel J P. Virulent Brucella abortus prevents lysosome fusion and is distributed within autophagosome-like compartments. Infect Immun. 1998;66:2387–2392. doi: 10.1128/iai.66.5.2387-2392.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rathman M, Sjaastad M D, Falkow S. Acidification of phagosomes containing Salmonella typhimurium in murine macrophages. Infect Immun. 1996;64:2765–2773. doi: 10.1128/iai.64.7.2765-2773.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reuter K, Ficner R. Sequence analysis and overexpression of the Zymomonas mobilis tgt gene encoding tRNA-guanine transglycosylase: purification and biochemical characterization of the enzyme. J Bacteriol. 1995;177:5284–5288. doi: 10.1128/jb.177.18.5284-5288.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richarme G, Caldas T D. Chaperone properties of the bacterial periplasmic substrate-binding proteins. J Biol Chem. 1997;272:15607–15612. doi: 10.1074/jbc.272.25.15607. [DOI] [PubMed] [Google Scholar]
- 35.Schlosser A, Meldorf M, Stumpe S, Bakker E P, Epstein W. TrkH and its homolog, TrkG, determine the specificity and kinetics of cation transport by the Trk system of Escherichia coli. J Bacteriol. 1995;177:1908–1910. doi: 10.1128/jb.177.7.1908-1910.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sebastian S, Genco C A. FbpC is not essential for iron acquisition in Neisseria gonorrhoeae. Infect Immun. 1999;67:3141–3145. doi: 10.1128/iai.67.6.3141-3145.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith L D, Ficht T A. Pathogenesis of Brucella. Crit Rev Microbiol. 1990;17:209–230. doi: 10.3109/10408419009105726. [DOI] [PubMed] [Google Scholar]
- 38.Surber M W, Maloy S. The PutA protein of Salmonella typhimurium catalyzes the two steps of proline degradation via a leaky channel. Arch Biochem Biophys. 1998;354:281–287. doi: 10.1006/abbi.1998.0697. [DOI] [PubMed] [Google Scholar]
- 39.Takahashi S, Kuzuyama T, Watanabe H, Seto H. A 1-deoxy-D-xylulose 5-phosphate reductoisomerase catalyzing the formation of 2-C-methyl-D-erythritol 4-phosphate in an alternative nonmevalonate pathway for terpenoid biosynthesis. Proc Natl Acad Sci USA. 1998;95:9879–9884. doi: 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thornalley P J. Glutathione-dependent detoxification of alpha-oxoaldehydes by the glyoxalase system: involvement in disease mechanisms and antiproliferative activity of glyoxalase I inhibitors. Chem-Biol Interact. 1998;111-112:137–151. doi: 10.1016/s0009-2797(97)00157-9. [DOI] [PubMed] [Google Scholar]
- 41.Tibor A, Jacques I, Guilloteau L, Verger J M, Grayon M, Wansard V, Letesson J J. Effect of P39 gene deletion in live Brucella vaccine strains on residual virulence and protective activity in mice. Infect Immun. 1998;66:5561–5564. doi: 10.1128/iai.66.11.5561-5564.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valdivia R H, Hromockyj A E, Monack D, Ramakrishnan L, Falkow S. Applications for green fluorescent protein (GFP) in the study of host-pathogen interactions. Gene. 1996;173:47–52. doi: 10.1016/0378-1119(95)00706-7. [DOI] [PubMed] [Google Scholar]
- 43.Wooldridge K G, Williams P H. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol Rev. 1993;12:325–348. doi: 10.1111/j.1574-6976.1993.tb00026.x. [DOI] [PubMed] [Google Scholar]