Abstract
Dysregulation of gut homeostasis is associated with irritable bowel syndrome (IBS), a chronic functional gastrointestinal disorder affecting approximately 11.2% of the global population. The poorly understood pathogenesis of IBS has impeded its treatment. Here, we report that the E3 ubiquitin ligase tripartite motif-containing 27 (TRIM27) is weakly expressed in IBS but highly expressed in inflammatory bowel disease (IBD), a frequent chronic organic gastrointestinal disorder. Accordingly, knockout of Trim27 in mice causes spontaneously occurring IBS-like symptoms, including increased visceral hyperalgesia and abnormal stool features, as observed in IBS patients. Mechanistically, TRIM27 stabilizes β-catenin and thus activates Wnt/β-catenin signaling to promote intestinal stem cell (ISC) self-renewal. Consistent with these findings, Trim27 deficiency disrupts organoid formation, which is rescued by reintroducing TRIM27 or β-catenin. Furthermore, Wnt/β-catenin signaling activator treatment ameliorates IBS symptoms by promoting ISC self-renewal. Taken together, these data indicate that TRIM27 is critical for maintaining gut homeostasis, suggesting that targeting the TRIM27/Wnt/β-catenin axis could be a potential treatment strategy for IBS. Our study also indicates that TRIM27 might serve as a potential biomarker for differentiating IBS from IBD.
Keywords: TRIM27, Wnt/β-catenin signaling, ISC self-renewal, IBS
Subject terms: Mechanisms of disease, Target identification
Introduction
Dysregulation of gut homeostasis has been associated with two of the most common gastrointestinal disorders, namely, irritable bowel syndrome (IBS) and inflammatory bowel disease (IBD), which affect 11.2% and 0.3% of the worldwide population, respectively [1, 2]. IBD, which includes Crohn’s disease (CD) and ulcerative colitis (UC), is usually characterized by organic intestinal lesions, and an increasing number of studies have suggested that IBD results from a complex interplay among genetic variability, the host immune system and environmental factors [3]. On the other hand, IBS has been considered to be a functional intestinal disorder and is divided into four different subtypes: constipation-predominant IBS (IBS-C), diarrhea-predominant IBS (IBS-D), IBS with mixed bowel habits (IBS-M) and unclassified IBS (IBS-U). Among these subtypes, IBS-D appears to be the most common, accounting for 37–62% of IBS cases [4]. Compared with IBD, IBS is a largely neglected but much more common gastrointestinal disorder, and its symptoms, such as increased gut permeability and dysregulation of the gut microbiota, overlap with those of IBD [5]. However, to date, the pathophysiology and underlying molecular mechanisms, especially genetic susceptibility to IBS, remain poorly understood, which impedes the precise diagnosis and rational treatment of this disease.
Gut homeostasis depends mainly on the constant regeneration of the intestinal epithelium, which is achieved via a tightly regulated balance between intestinal stem cell (ISC) self-renewal and differentiation [6]. The intestinal immune system and gut microbiota also contribute to the maintenance of gut homeostasis [7]. Thus, dysregulation of the intestinal epithelial barrier, intestinal immune system or gut microbiota can cause a variety of intestinal disorders, such as IBS. To date, the majority of previous studies have focused mainly on the regulatory roles of the gut microbiota in IBS. A recent genome-wide association study suggested that gut microbiota has genetic associations with but not mechanistic implications in the development of IBS [8]. However, to date, the regulatory functions and molecular mechanisms of specific genetic factors involved in the occurrence and development of IBS remain poorly understood. Ubiquitination is a critical posttranslational modification related to various intestinal inflammatory diseases [9, 10]. Tripartite motif (TRIM) proteins constitute one of the largest subfamilies of Really Interesting New Gene (RING) E3 ubiquitin ligases that mediate the ubiquitination of numerous protein substrates to regulate diverse biological processes, including cell development and differentiation. Notably, TRIM proteins also participate in multiple biological processes independent of their E3 ubiquitin ligase activity. Dysfunction of both their E3 ubiquitin ligase activity-dependent and E3 ubiquitin ligase activity-independent roles is involved in a variety of diseases, such as inflammatory and infectious diseases [11, 12]. However, their physiological functions and regulatory mechanisms in the maintenance of gut homeostasis remain largely undefined.
In this study, by analyzing the expression levels of TRIM family members in intestinal tissues of IBS and IBD patients, we identified the E3 ubiquitin ligase TRIM27 to be highly expressed in IBD patients but weakly expressed in IBS patients compared with healthy controls. By establishing Trim27-knockout mouse models, we found that Trim27 deficiency causes impaired self-renewal of Lgr5-positive (Lgr5+) ISCs due to the inactivation of Wnt/β-catenin signaling, followed by disruption of the tridirectional relationship among intestinal epithelial integrity, the intestinal immune system and the gut microbiota, eventually leading to spontaneously occurring IBS-D-like symptoms in mice. Furthermore, the Wnt/β-catenin signaling activator SKL2001 restores the self-renewal ability of ISCs and ameliorates IBS-D-like symptoms in Trim27-deficient mice. Together, our data reveal a key genetic factor, TRIM27, in ISC self-renewal and gut homeostasis maintenance and indicate a potential treatment strategy for IBS, especially IBS-D, by targeting the TRIM27/Wnt/β-catenin axis.
Results
TRIM27 deficiency induces spontaneous IBS-like symptoms
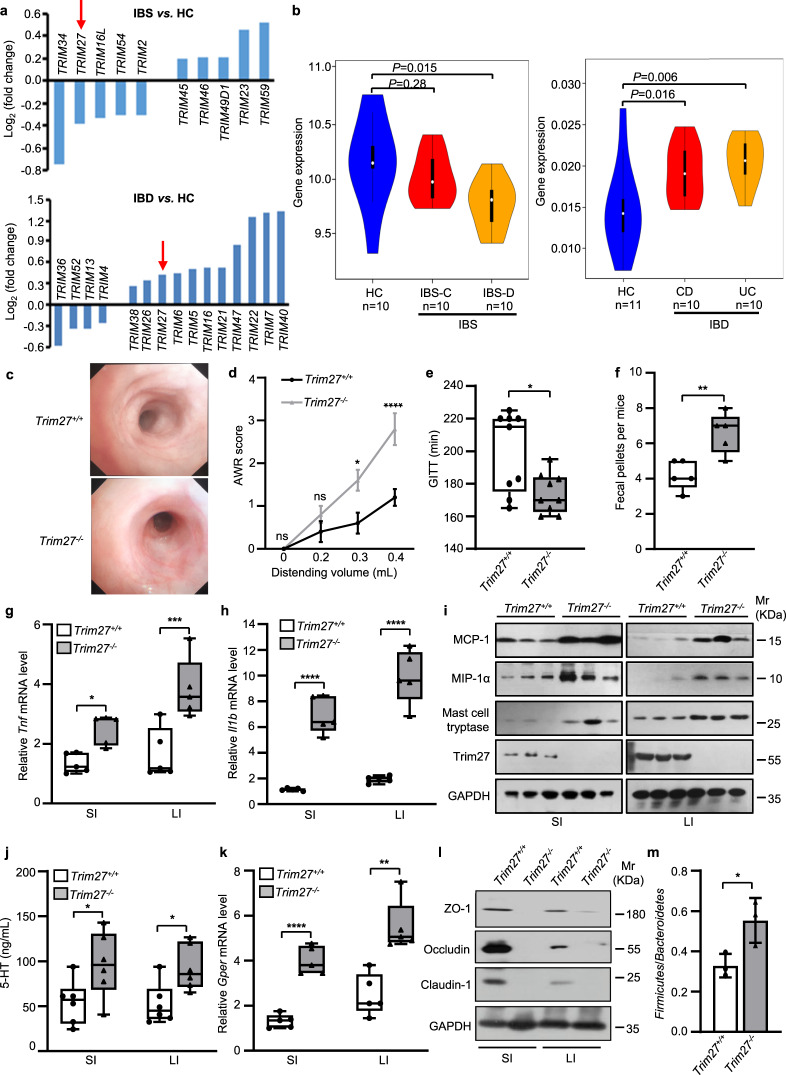
To investigate the regulatory roles of TRIM family proteins in gut homeostasis, we collected publicly available intestinal tissue microarray data of IBS (including IBS-C and IBS-D) and IBD (including CD and UC) patients for gene expression analysis. We screened for TRIM proteins with altered gene expression levels to identify potential genetic factors linked to IBS and IBD. Among the identified proteins, TRIM34, TRIM40, TRIM7 and TRIM27 exhibited significant changes in gene expression. However, TRIM34, TRIM40 and TRIM7 expression changed only in patients with IBS or only in patients with IBD, while TRIM27 was the only protein whose gene expression level changed in both patients with IBS and patients with IBD. More interestingly, TRIM27 showed opposite gene expression patterns in those two diseases with overlapping symptoms (Fig. 1a). Specifically, TRIM27 showed a high expression level in the intestinal tissues of IBD patients, consistent with a previous study [13], but it exhibited low expression in the intestinal tissues of patients with IBS, especially IBS-D (Fig. 1b). These results suggest that TRIM27 may play a precise regulatory role in maintaining gut homeostasis. Previous studies have shown that TRIM27 is involved in various biological processes, such as the antiviral immune response [14], cancer development [15] and liver homeostasis [11]. However, the physiological function of TRIM27 in the regulation of gut homeostasis remains largely unknown.
Fig. 1.
TRIM27 deficiency induces spontaneous IBS-like symptoms. a Changes in the expression levels of TRIM family genes in patients with IBS and IBD compared with healthy controls (HCs), as determined by microarray analysis. b Comparison of TRIM27 gene expression in patients with IBS-C and IBS-D with that in HCs (left) and in patients with CD and UC with that in HCs (right). c Endoscopic images of colons from 16-week-old Trim27+/+ and Trim27-/- mice (n = 5). d The AWR scores of 16-week-old Trim27+/+ and Trim27-/- mice in response to graded colorectal distention (0, 0.2, 0.3 and 0.4 mL). e GITT of 16-week-old Trim27+/+ and Trim27-/- mice. f The number of fecal pellets excreted from Trim27+/+ and Trim27-/- mice every hour. g, h Quantitative PCR (qPCR) analysis of Tnf (g) and Il1b (h) mRNA levels in the small intestine (SI) and large intestine (LI) from 16-week-old Trim27+/+ and Trim27-/- mice. i Immunoblot analysis of MCP-1, MIP-1α, mast cell tryptase, Trim27 and GAPDH protein levels in the SI and LI from 16-week-old Trim27+/+ and Trim27-/- mice. j Enzyme-linked immunosorbent assay (ELISA) of 5-HT in the SI and LI from 16-week-old Trim27+/+ and Trim27-/- mice. k qPCR analysis of the G protein-coupled estrogen receptor (Gper) mRNA level in the SI and LI from 16-week-old Trim27+/+ and Trim27-/- mice. l Immunoblot analysis of ZO-1, Occludin, Claudin-1 and GAPDH protein levels in the SI and LI from 16-week-old Trim27+/+ and Trim27-/- mice. m The ratio of the Firmicutes to Bacteroidetes in the colonic contents from 16-week-old Trim27+/+ and Trim27-/- mice. In b, d, e, f, g, h, j, k and m, statistical analyses were performed using the Wilcoxon rank-sum test with the Benjamini–Hochberg method (b), two-way ANOVA with Sidak’s multiple comparisons test (d) and unpaired two-tailed Student’s t test (e–h, j, k, m). The data are shown as the means ± SEMs (d, n = 5; m, n = 3) or in box-and-whisker plots displaying the medians, interquartile ranges (boxes) and minima and maxima (whiskers) (e, n = 9; f, g, h, k, n = 5; j, n = 6). P > 0.05, not significant (ns); *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001
Given that a decreased TRIM27 gene expression level is associated with IBS, we generated mice with global knockout of Trim27 (Trim27-/- mice) to investigate the physiological function of TRIM27 in maintaining gut homeostasis (Supplementary Fig. 1a–c). We found that Trim27-/- mice did not exhibit symptoms that are common in IBD patients, such as weight loss, growth retardation and bloody stool (Supplementary Fig. 1d, e). Furthermore, endoscopic examination showed no intestinal mucosal changes, such as mucosal thickening, ulceration or ischemic lesions, in Trim27-/- mice (Fig. 1c), while those symptoms are commonly observed in the IBD mouse model [16]. Moreover, the brains, hearts, lungs, livers, spleens and kidneys showed no morphological or histopathological abnormalities (Supplementary Fig. 1e, f) in 16-week-old Trim27+/+, Trim27 heterozygous-knockout (Trim27+/-) and Trim27-/- mice. However, compared with Trim27+/+ mice, Trim27-/- mice showed decreases in the crypt numbers, crypt depth and villus height in the small and large intestines (Supplementary Fig. 2a–h), suggesting functional dysregulation of gut homeostasis. Furthermore, Trim27-/- mice exhibited a marked increase in the abdominal withdrawal reflex (AWR) score (Fig. 1d), which indicates the result of an involuntary motor reflex assay to assess visceral hypersensitivity [17]. In addition, the gastrointestinal transit time (GITT), a biomarker for evaluating gastrointestinal motility [18], was significantly decreased in Trim27-/- mice (Fig. 1e). Consistent with this finding, an increased number of fecal pellets was observed in Trim27-/- mice compared with Trim27+/+ mice (Fig. 1f). Together, these results indicate that Trim27-/- mice exhibit IBS-like symptoms, including increased visceral hyperalgesia and abnormal stool features, as observed in IBS patients [19, 20], suggesting TRIM27 as a potential regulator of gut homeostasis maintenance.
Emerging evidence indicates that IBS, especially the IBS-D subtype, is characterized by low-grade intestinal mucosal inflammation, intestinal barrier impairment, visceral hypersensitivity, altered gastrointestinal motility and gut dysbiosis [4, 21–24]. The low-grade intestinal inflammation in IBS-D manifests as increased levels of proinflammatory cytokines (including TNF-α and IL-1β) and chemokines (including monocyte chemoattractant protein-1 (MCP-1) and macrophage inflammatory protein-1α (MIP-1α)) in the intestines [25]. Consistent with this finding, the levels of Tnf, Il1b, MCP-1 and MIP-1α were found to be significantly higher in both the small and large intestines in Trim27-/- mice than in Trim27+/+ mice (Fig. 1g–i). Tryptase from mast cells and 5-HT (5-hydroxytryptamine) from enterochromaffin cells are key components leading to visceral hypersensitivity in IBS pathophysiology [26]. As expected, the levels of both tryptase and 5-HT were increased in the intestine of Trim27-/- mice compared with Trim27+/+ mice (Fig. 1i, j). A recent study implied that the G protein-coupled estrogen receptor (GPER), a crucial factor involved in the regulation of gastrointestinal motility and visceral pain, is usually upregulated in IBS-D patients [21]. Consistent with this finding, Trim27-/- mice showed higher expression of GPER in intestinal tissues than did Trim27+/+ mice (Fig. 1k). Furthermore, Trim27-/- mice showed decreased levels of zonula occludens-1 (ZO-1), occludin and claudins (Fig. 1l), which are the three most important tight junction proteins that play vital roles in maintaining cell polarity and intestinal epithelial barrier integrity [27], and the compromised intestinal barrier has been reported to be a contributor to the development of IBS-D symptoms [28]. In addition, the composition of the normal microbiota was altered in Trim27-/- mice, with an increase in the ratio of Firmicutes to Bacteroidetes, consistent with results from clinically reported IBS patients (Fig. 1m) [29, 30]. Collectively, these findings indicate that Trim27-/- mice exhibit IBS-D-like symptoms. Thus, TRIM27 might be a critical genetic factor for IBS.
TRIM27 promotes the development and regeneration of intestinal tissues
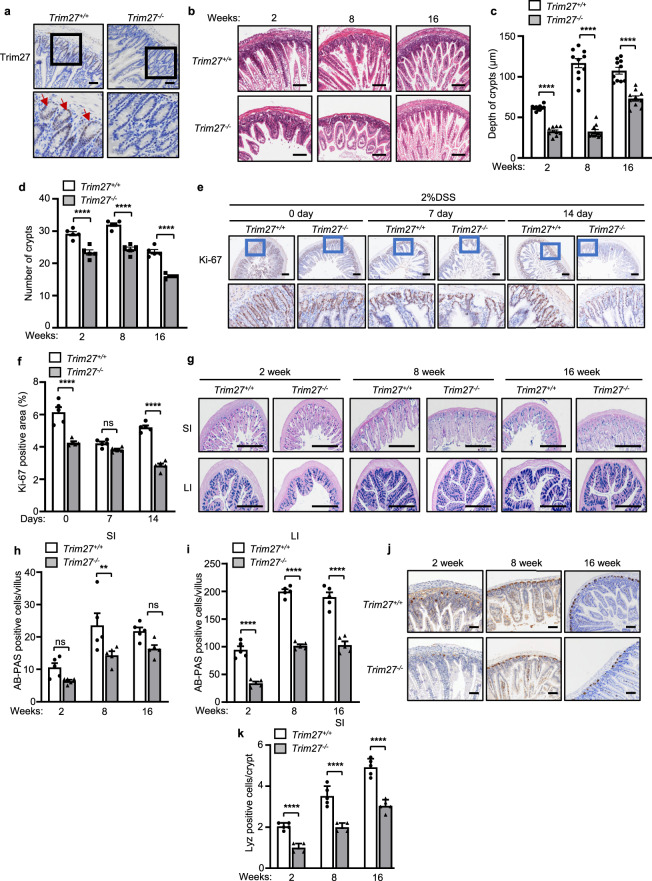
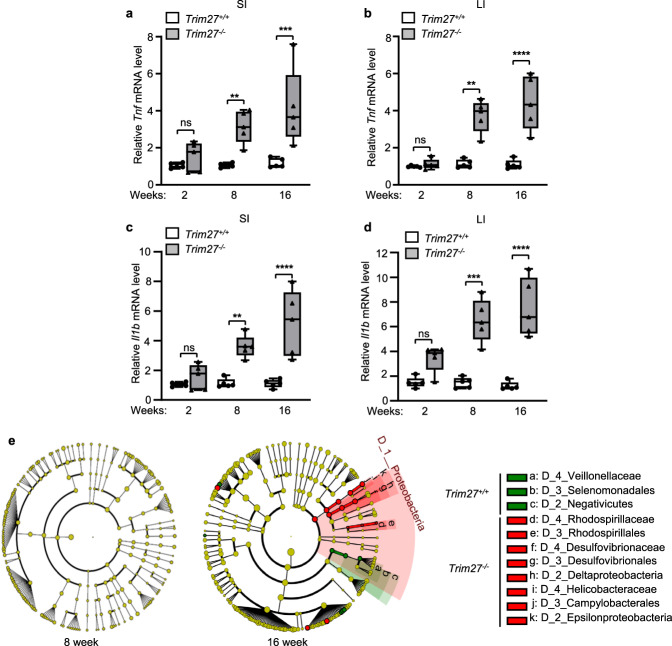
To further investigate the underlying mechanism by which TRIM27 maintains gut homeostasis, we conducted immunohistochemical analysis of Trim27 expression in mouse intestinal tissues. We found that TRIM27 was highly expressed in intestinal crypts (Fig. 2a), which are responsible for the development and regeneration of the intestinal epithelium following injury or inflammation. We then carried out hematoxylin and eosin (H&E) staining to assess pathologic changes in tissue. Interestingly, histopathological analysis demonstrated a significant decrease in crypt numbers and the crypt depth in small intestines from 2–16-week-old Trim27-/- mice compared with small intestines from Trim27+/+ mice (Fig. 2b–d). Crypts are critical to the repair and renewal of the intestinal barrier. We thus hypothesized that the decreases in the number and depth of crypts may lead to defects in intestinal injury repair and to dysplasia of intestinal epithelial tissues in Trim27-/- mice. As expected, our Ki-67 staining data showed that Trim27 deficiency reduced cell proliferation in mice stimulated with 2% dextran sulfate sodium (DSS) (Fig. 2e, f). Furthermore, using Alcian blue-periodic acid–Schiff (AB-PAS) and lysozyme staining to analyze the main cell types, including goblet cells and Paneth cells, in the intestinal epithelial lining, we found that the proportions of both goblet and Paneth cells were significantly decreased in the intestines of 2–16-week-old Trim27-/- mice (Fig. 2g–k). It has been reported that the intestinal epithelium is the interface between the microbiota and host and that a decrease in the number of intestinal epithelial cell subtypes is frequently related to the occurrence of intestinal inflammation and dysbiosis of the gut microbiota [31]. As expected, we found that the levels of proinflammatory cytokines, including Tnf and Il1b, were significantly increased in the intestines of 8- and 16-week-old Trim27-/- mice (Fig. 3a–d). Moreover, at 16 weeks, Trim27-/- mice exhibited an increase in the relative abundance of Proteobacteria at the phylum level, which is an important characteristic of gut microbiota dysbiosis [29]. In parallel, the amounts of bacteria reported to be abundant in IBS-D patients, such as Desulfovibrionaceae at the family level [32], were also significantly increased in Trim27-/- mice (Fig. 3e). Taken together, these findings indicate that deficiency of TRIM27 causes defects in the development and regeneration of intestinal tissues, followed by enhanced intestinal inflammation and gut dysbiosis over time.
Fig. 2.
TRIM27 promotes the development and regeneration of intestinal tissues. a Immunohistochemical analysis of Trim27 expression in the SI from Trim27+/+ and Trim27-/- mice. Bottom, enlarged images of the regions outlined in the upper panels. The red arrows indicate Trim27-positive cells. Scale bars, 150 μm. b H&E staining of the SI from 2–16-week-old Trim27+/+ and Trim27-/- mice. Scale bars, 150 μm. c, d Crypt depth (c) and number of crypts per mm (d). At least 20 randomly selected crypts (c) or randomly selected fields (d) from the SI of each mouse were analyzed, and the average was calculated. Ten mice per group were analyzed. e Ki-67 staining of the SI from 16-week-old Trim27+/+ and Trim27-/- mice after treatment with 2% DSS (top). Bottom, enlarged images of the regions outlined in the upper panels. Scale bars, 150 μm. f Ki-67-positive area in the intestinal tissues described in e. Five mice per group were analyzed. g Alcian blue-periodic acid–Schiff (AB-PAS) staining of the SI and LI from 2–16-week-old Trim27+/+ and Trim27-/- mice. Scale bars, 500 μm. h, i The number of AB-PAS-positive goblet cells per villus in the SI (h) and LI (i) as described in g. At least 20 randomly selected villi from the small intestine of each mouse were analyzed, and the average was calculated. Five mice per group were analyzed. j Immunohistochemical analysis of lysozyme expression in small intestinal tissue sections from 2–16-week-old Trim27+/+ and Trim27-/- mice. Scale bars, 100 μm. k The number of lysozyme-positive Paneth cells per crypt in the SI as described in j. At least 20 randomly selected crypts from the SI of each mouse were analyzed, and the average was calculated. Five mice per group were analyzed. In c, d, f, h, i and k, statistical analyses were performed using two-way ANOVA with Sidak’s multiple comparisons test. The data are presented as the means ± SEMs (c, n = 10; d, f, h, i, k, n = 5). P > 0.05, not significant (ns); **P < 0.01; ****P < 0.0001
Fig. 3.
TRIM27 deficiency causes enhanced intestinal inflammation and gut dysbiosis over time. a, b qPCR analysis of Tnf mRNA expression in the SI (a) and LI (b) from 2–16-week-old Trim27+/+ and Trim27-/- mice. c, d qPCR analysis of Il1b mRNA expression in the SI (c) and LI (d) from 2–16-week-old Trim27+/+ and Trim27-/- mice. e Taxonomic cladogram generated via linear discriminant analysis effect size (LEfSe) analysis showing the bacterial taxa (phylum, class, order and family) in 8-week-old (left) and 16-week-old (right) Trim27+/+ and Trim27-/- mice. Green and red indicate an increased abundance in Trim27+/+ and Trim27-/- mice, respectively. In a–d, statistical analyses were performed using two-way ANOVA with Sidak’s multiple comparisons test. The data are shown as box-and-whisker plots displaying the medians, interquartile ranges (boxes) and minima and maxima (whiskers) (a–d, n = 5). P > 0.05, not significant (ns); **P < 0.01; ***P < 0.001; ****P < 0.0001
TRIM27 promotes the activation of Wnt/β-catenin signaling in intestinal crypts
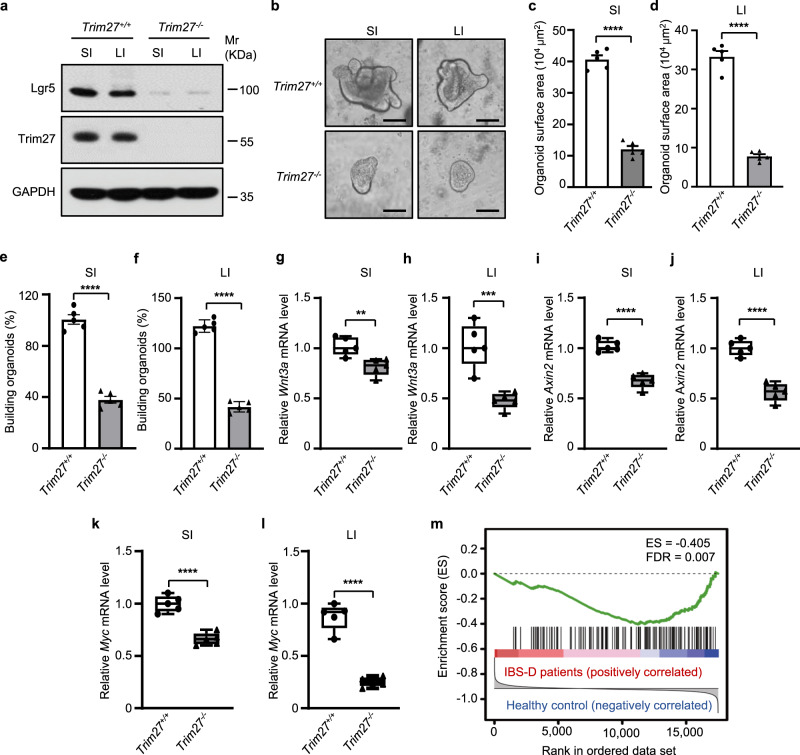
ISCs are the main cell type in crypts and are required for the development and regeneration of the intestinal epithelium [33]. We thus examined the expression of Lgr5, which is the main marker of ISCs in crypts, and found that the expression of Lgr5 was significantly decreased in Trim27-/- mice compared with Trim27+/+ mice (Fig. 4a). To further confirm that TRIM27 promotes ISC proliferation, we additionally conducted a pulse-chase assay with EdU labeling in Trim27+/+ and Trim27-/- mice to examine the proliferation of ISCs in vivo, and we observed decreased proliferation of Lgr5+ ISCs in Trim27-/- mice compared with Trim27+/+ mice (Supplementary Fig. 3a, b). Consistent with this finding, the organoid-forming efficiency of crypts harvested from Trim27-/- mice was impaired (Fig. 4b–f). Since the Wnt and Notch signaling pathways are critical in promoting ISC proliferation [34, 35], we sought to determine whether the Wnt and Notch pathways are suppressed in ISCs of Trim27-/- mice. Our data showed that the expression of Wnt target genes (such as Wnt3a, Axin2 and Myc) was significantly downregulated in crypts from Trim27-/- mice (Fig. 4g–l). Furthermore, gene set enrichment analysis (GSEA) revealed a negative correlation between Wnt signaling pathway activity and IBS-D (Fig. 4m). However, the expression of Notch target genes was not influenced by TRIM27 (Supplementary Fig. 3c–h), and no correlation between Notch signaling pathway activity and IBS-D was revealed by GSEA (Supplementary Fig. 3i). Collectively, these results indicate that low expression of TRIM27 might result in inhibition of Wnt/β-catenin signaling in Lgr5+ ISCs in IBS-D patients.
Fig. 4.
TRIM27 promotes the activation of Wnt/β-catenin signaling in intestinal crypts. a Immunoblot analysis of the Lgr5 protein level in intestinal tissues from 16-week-old Trim27+/+ and Trim27-/- mice. b Representative images of small intestinal and large intestinal organoids formed by crypts of 16-week-old Trim27+/+ and Trim2-/- mice. Scale bars, 200 μm. c, d Quantification of the surface area of small intestinal (c) and large intestinal (d) organoids formed by crypts as described in b. Five randomly selected organoids were analyzed per group. e, f The small intestinal (e) and large intestinal (f) organoid-forming efficiency of crypts as described in b. Five randomly selected microscopic fields were analyzed per group. g, h qPCR analysis of Wnt3a mRNA expression in intestinal crypts isolated from the SI (g) and LI (h) of 16-week-old Trim27+/+ and Trim27-/- mice. i, j qPCR analysis of Axin2 mRNA expression in intestinal crypts isolated from the SI (i) and LI (j) of mice as described in g. k, l qPCR analysis of Myc mRNA expression in intestinal crypts isolated from the SI (k) and LI (l) of mice as described in g. m Gene set enrichment analysis (GSEA) results for the Wnt signaling pathway in IBS-D. FDR, false discovery rate. In c–l, statistical analyses were performed using unpaired two-tailed Student’s t test. The data are presented as the means ± SEMs (c–f, n = 5) or in box-and-whisker plots displaying the medians, interquartile ranges (boxes) and minima and maxima (whiskers) (g–l, n = 5). **P < 0.01; ***P < 0.001; ****P < 0.0001
TRIM27 stabilizes β-catenin to activate Wnt/β-catenin signaling in ISCs
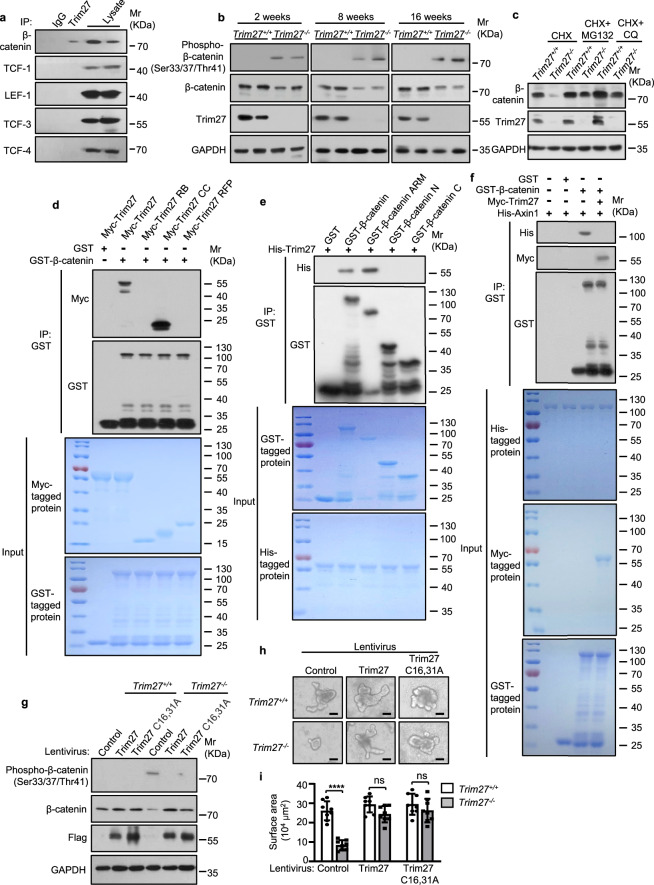
As reported, Wnt/β-catenin signaling is activated by the transport of β-catenin from the cytoplasm to the nucleus followed by its interaction with nuclear lymphoid enhancer factor (LEF)/T-cell factor (TCF) transcription factors [36]. Thus, we speculated that TRIM27 regulates the Wnt/β-catenin signaling pathway by interacting with β-catenin or LEF/TCF transcription factors. Through a coimmunoprecipitation assay, we found that TRIM27 interacted with β-catenin but not with LEF/TCF transcription factors in the cytoplasm (Fig. 5a and Supplementary Fig. 4a). Moreover, an increased protein level of phospho-β-catenin (Ser33/37/Thr41), which can trigger ubiquitination-mediated proteasomal degradation of β-catenin, was detected in intestinal crypts from 2–16-week-old Trim27-/- mice. Consistent with this finding, the total β-catenin protein level was decreased in Trim27-/- mouse crypts (Fig. 5b). Moreover, both the proteasomal inhibitor MG132 and the autophagy inhibitor chloroquine significantly rescued the decrease in β-catenin expression in Trim27 knockout crypts, indicating that TRIM27 inhibits both the proteasomal and autophagic degradation of β-catenin (Fig. 5c). Furthermore, TRIM27 mainly suppressed K48-linked polyubiquitination and only slightly suppressed K63-linked polyubiquitination of β-catenin (Supplementary Fig. 4b). In addition, TRIM27 did not affect the mRNA level of β-catenin (Supplementary Fig. 4c). Taken together, these results indicate that TRIM27 stabilizes β-catenin by inhibiting its proteasomal and autophagic degradation.
Fig. 5.
TRIM27 stabilizes β-catenin to activate Wnt/β-catenin signaling in ISCs. a Immunoprecipitation (IP) of β-catenin, TCF-1, LEF-1, TCF-3 and TCF-4 by Trim27 in crypts from 16-week-old Trim27+/+ mice. Crypts were lysed and immunoprecipitated with an anti-Trim27 antibody. b Immunoblot analysis of phospho-β-catenin (Ser33/37/Thr41), β-catenin, Trim27 and GAPDH protein levels in crypts from 2–16-week-old Trim27+/+ and Trim27-/- mice. c Immunoblot analysis of β-catenin, Trim27 and GAPDH protein levels in crypts from 16-week-old Trim27+/+ and Trim27-/- mice after treatment with cycloheximide (CHX, 10 μM), CHX in combination with MG132 (5 μM) or CHX in combination with chloroquine (CQ, 20 μM) for 8 h. d Pulldown of Myc-tagged Trim27 (5 μg) or its truncations (3 μg each) by GST (3 μg) or GST-β-catenin (10 μg each). e Pulldown of His-tagged Trim27 (5 μg each) by GST (3 μg), GST-β-catenin (10 μg), GST-β-catenin ARM (8 μg), GST-β-catenin N (4 μg) or GST-β-catenin C (3 μg). f Pulldown of His-tagged Axin1 (10 μg each) by GST-β-catenin (10 μg each) in the presence or absence of Myc-tagged Trim27 (5 μg each). g Immunoblot analysis of phospho-β-catenin (Ser33/37/Thr41), β-catenin, Trim27 and GAPDH protein levels in crypts infected with control, Trim27 or Trim27 C16,31A lentivirus. h Representative images of organoids formed by crypts from 16-week-old Trim27+/+ and Trim27-/- mice in the presence of control, Trim27 or Trim27 C16,31A lentivirus. Scale bars, 200 μm. i Quantification of the surface area of organoids formed by crypts as described in h. Eight randomly selected organoids were analyzed per group. In i, statistical analyses were performed using two-way ANOVA with Sidak’s multiple comparisons test. The data are presented as the means ± SEMs (i, n = 8). P > 0.05, not significant (ns); ****P < 0.0001
We then explored the molecular mechanism by which TRIM27 suppresses the phosphorylation of β-catenin to inhibit its degradation. We found that TRIM27 directly bound to the conserved Armadillo (ARM) repeat in β-catenin through its coiled-coil (CC) domain (Fig. 5d, e and Supplementary Fig. 4d). Axin, which negatively regulates Wnt/β-catenin signaling by forming a complex with glycogen synthase kinase-3β (GSK-3β) and casein kinase 1 (CK1) to promote the phosphorylation and degradation of β-catenin, directly interacts with the ARM repeat of β-catenin [37]. We thus speculated that TRIM27 might inhibit the phosphorylation and degradation of β-catenin by competitively binding to the ARM repeat of β-catenin to block its interaction with Axin. Indeed, Axin interacted with β-catenin directly, and this interaction was abolished by ectopic expression of TRIM27 (Fig. 5f). We further confirmed that TRIM27 interacted with β-catenin to increase its stability in an E3 ubiquitin ligase activity-independent manner (Fig. 5g). TRIM27 in intestinal crypts from Trim27-/- mice was then reconstituted with Trim27 or its E3 ubiquitin ligase activity-dead mutant (Trim27 C16,31A) [13, 38] by treatment with recombinant lentivirus expressing Trim27 or Trim27 C16,31A, respectively. As expected, reconstitution with either Trim27 or Trim27 C16,31A restored the organoid-forming capacity of crypts (Fig. 5h, i). Together, these results suggest that TRIM27 activates Wnt/β-catenin signaling by stabilizing the β-catenin protein to promote ISC self-renewal.
TRIM27 maintains Lgr5+ ISC homeostasis
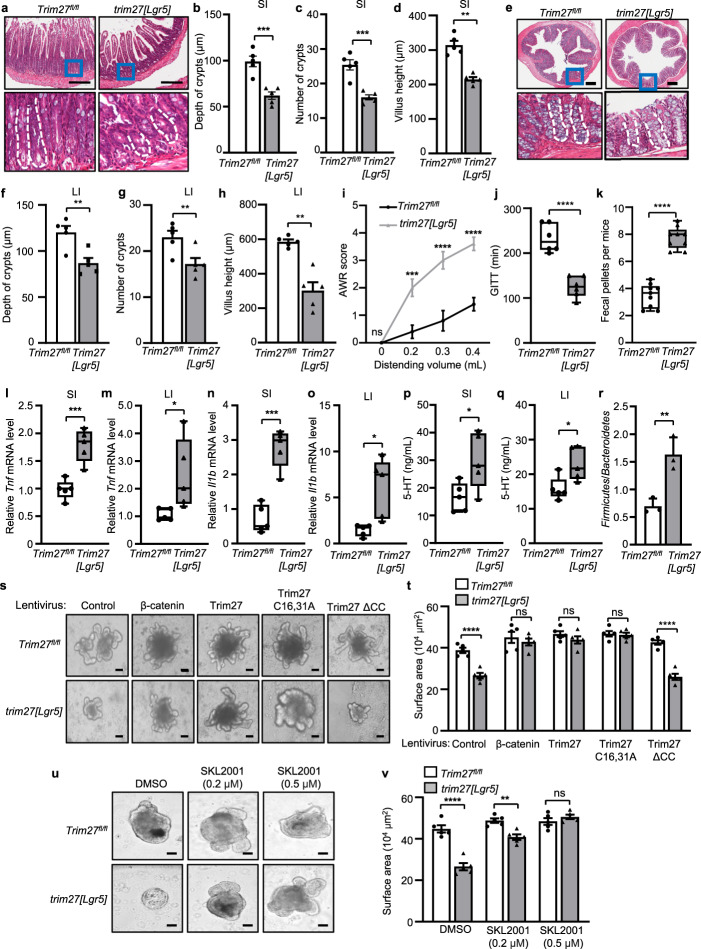
To confirm the regulatory role of TRIM27 in ISCs, we generated Trim27flox/flox (Trim27fl/fl) mice (Supplementary Fig. 5a) and then generated mice with ISC-specific Trim27 knockout by introducing Lgr5-creERT2 into Trim27fl/fl mice (designated trim27[Lgr5] mice) (Supplementary Fig. 5b). Histological analysis showed that the Trim27 protein level was significantly decreased in the ISCs of trim27[Lgr5] mice (Supplementary Fig. 5c). Interestingly, trim27[Lgr5] mice exhibited phenotypes similar to those of Trim27-/- mice, including the decreased crypt number, crypt depth and villus height in the small and large intestines (Fig. 6a–h); increased AWR scores (Fig. 6i); a decreased GITT (Fig. 6j); and an increased number of fecal pellets (Fig. 6k). In addition, trim27[Lgr5] mice exhibited decreased numbers of goblet and Paneth cells (Supplementary Fig. 5d–g); reduced ISC proliferation (Supplementary Fig. 5h, i); elevated Tnf, Il1b, 5-HT and tryptase levels (Fig. 6l–q and Supplementary Fig. 5j); and dysbiosis of the gut microbiota (Fig. 6r). These results further confirmed that TRIM27 plays a key role in maintaining Lgr5+ ISC homeostasis. Furthermore, crypts were isolated from trim27[Lgr5] mice and treated with recombinant lentiviruses expressing β-catenin, Trim27, Trim27 C16,31A or Trim27 ΔCC. As expected, expression of β-catenin, Trim27 or Trim27 C16,31A but not Trim27 ΔCC rescued the disruption of organoid formation from intestinal crypts isolated from trim27[Lgr5] mice (Fig. 6s, t). In addition, intestinal crypts were treated with SKL2001, a Wnt/β-catenin signaling activator that can disrupt the interaction of β-catenin with Axin [39] without affecting the interaction between β-catenin and TRIM27 (Supplementary Fig. 5k), and we found that SKL2001 promoted organoid formation from intestinal crypts isolated from trim27[Lgr5] mice (Fig. 6u, v). Thus, TRIM27 promotes Lgr5+ ISC self-renewal in a manner dependent on β-catenin but independent of its own E3 ubiquitin ligase activity.
Fig. 6.
TRIM27 maintains Lgr5+ ISC homeostasis. a H&E staining of the SI from 16-week-old Trim27fl/fl and trim27[Lgr5] mice. Bottom, enlarged images of the regions outlined in the upper panels. Scale bars, 300 μm. The white dashed lines mark crypt borders. b–d Crypt depth (b), crypt number per mm (c) and villus height (d) in the SI from mice as described in a. At least 20 randomly selected crypts (b, d) or randomly selected fields (c) from the SI of each mouse were analyzed, and the average was calculated. Five mice per group were analyzed. e H&E staining of the LI from 16-week-old Trim27fl/fl and trim27[Lgr5] mice. Bottom, enlarged images of the regions outlined in the upper panels. Scale bars, 300 μm. The white dashed lines mark crypt borders. f–h Crypt depth (f), crypt number per mm (g) and villus height (h) in the LI from mice as described in e. At least 20 randomly selected crypts (f, h) or randomly selected fields (g) from the LI of each mouse were analyzed, and the average was calculated. Five mice per group were analyzed. i The AWR scores of 16-week-old Trim27fl/fl and trim27[Lgr5] mice in response to graded colorectal distention (0, 0.2, 0.3 and 0.4 mL). j GITT of 16-week-old Trim27fl/fl and trim27[Lgr5] mice. k The number of fecal pellets excreted from Trim27fl/fl and trim27[Lgr5] mice every hour. l, m qPCR analysis of Tnf mRNA expression in the SI (l) and LI (m) from 16-week-old Trim27fl/fl and trim27[Lgr5] mice. n, o qPCR analysis of Il1b mRNA expression in the SI (n) and LI (o) from mice as described in l. p, q ELISA of 5-HT in the SI (p) and LI (q) from 16-week-old Trim27fl/fl and trim27[Lgr5] mice. r The ratio of Firmicutes to Bacteroidetes in the colonic contents from 16-week-old Trim27fl/fl and trim27[Lgr5] mice. s Representative images of organoids formed by crypts of 16-week-old Trim27fl/fl and trim27[Lgr5] mice in the presence of control-, β-catenin-, Trim27-, Trim27 C16,31A or Trim27 ΔCC lentivirus. Scale bars, 200 μm. t Quantification of the surface area of organoids formed by crypts from mice as described in s. Five randomly selected organoids were analyzed per group. u Representative images of organoids formed by crypts of 16-week-old Trim27fl/fl and trim27[Lgr5] mice treated with DMSO or various concentrations of SKL2001. Scale bars, 200 μm. v Quantification of the surface area of organoids formed by crypts from mice as described in u. Five randomly selected organoids were analyzed per group. In b–d, f–r, t and v, statistical analyses were performed using unpaired two-tailed Student’s t test (b–d, f–h and j–r) or two-way ANOVA with Sidak’s multiple comparisons test (i, t and v). The data are presented as the means ± SEMs (b–d, f–i, t and v, n = 5; r, n = 3) or in box-and-whisker plots displaying the medians, interquartile ranges (boxes) and minima and maxima (whiskers) (j, n = 6; k, n = 9; l–q, n = 5). P > 0.05, not significant (ns); *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001
Wnt/β-catenin signaling activation or probiotic supplementation alleviates TRIM27 deficiency-induced IBS symptoms
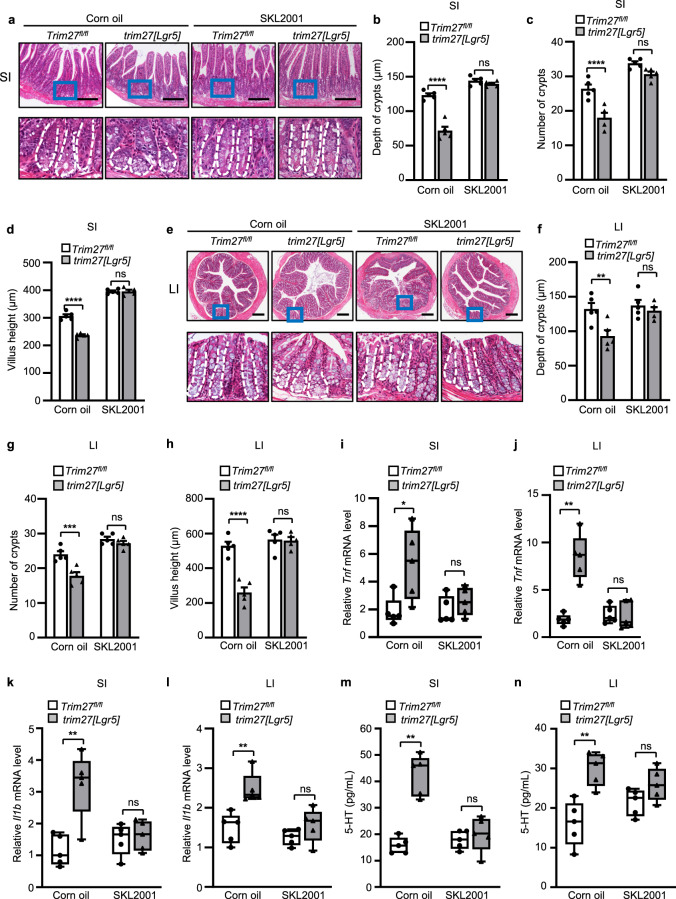
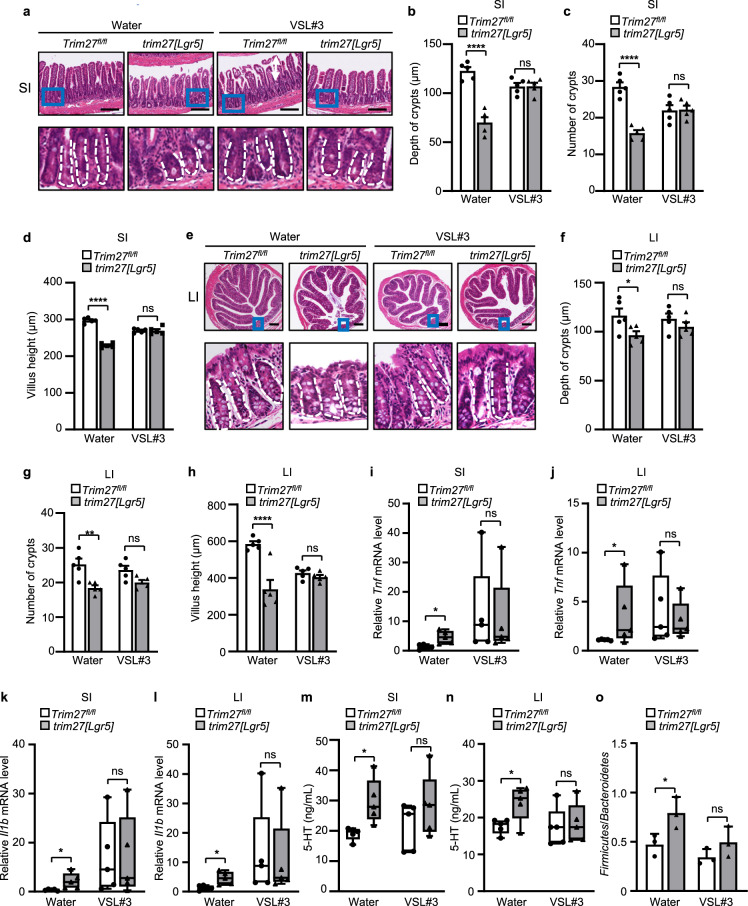
Based on the rationale that TRIM27 maintains Lgr5+ ISC self-renewal and gut homeostasis by activating Wnt/β-catenin signaling, we then treated Trim27fl/fl and trim27[Lgr5] mice with the Wnt/β-catenin signaling activator SKL2001. As expected, SKL2001 alleviated the IBS phenotype in trim27[Lgr5] mice, including the decreased crypt number, crypt depth and villus height in the small and large intestines (Fig. 7a–h) as well as the reduced proliferation of ISCs (Supplementary Fig. 6a, b). Consistent with this finding, SKL2001 attenuated the Trim27 deficiency-mediated upregulation of proinflammatory cytokines (including Tnf and Il1b) (Fig. 7i–l) and increase in the level of 5-HT in intestinal tissues (Fig. 7m, n). Thus, these results indicate that SKL2001 is a potentially attractive drug for alleviating IBS-D symptoms. Furthermore, since knockout of TRIM27 induces gut microbiota dysbiosis, we further speculated that restoring gut eubiosis should also be an effective strategy for relieving IBS-D-like symptoms in trim27[Lgr5] mice. Administration of probiotics can restore the disrupted gut microbiota to restore intestinal homeostasis [40]. We then found that treatment with VSL#3, a probiotic preparation containing a combination of eight different probiotic bacteria [41], alleviated the IBS-D-like symptoms in trim27[Lgr5] mice, including the decreased crypt number, crypt depth and villus height in the small and large intestines (Fig. 8a–h); elevated Tnf, Il1b and 5-HT levels (Fig. 8i–n); and dysbiosis of the gut microbiota (Fig. 8o). Taken together, these findings indicate that both targeting the TRIM27/Wnt/β-catenin axis and maintaining gut eubiosis can ameliorate Trim27 deficiency-induced IBS-D symptoms (Fig. 9).
Fig. 7.
Treatment with a Wnt/β-catenin signaling activator alleviates TRIM27 deficiency-induced IBS symptoms. a H&E staining of the SI from 16-week-old Trim27fl/fl and trim27[Lgr5] mice fed corn oil or SKL2001. Bottom, enlarged images of the regions outlined in the upper panels. Scale bars, 300 μm. The white dashed lines mark crypt borders. b–d Crypt depth (b), crypt number per mm (c) and villus height (d) in the SI from mice as described in a. At least 20 randomly selected crypts (b, d) or randomly selected fields (c) from the SI of each mouse were analyzed, and the average was calculated. Five mice per group were analyzed. e H&E staining of the LI from 16-week-old Trim27fl/fl and trim27[Lgr5] mice fed corn oil or SKL2001. Bottom, enlarged images of the regions outlined in the upper panels. Scale bars, 300 μm. The white dashed lines mark crypt borders. f–h Crypt depth (f), crypt number per mm (g) and villus height (h) of the LI from mice as described in e. At least 20 randomly selected crypts (f, h) or randomly selected fields (g) from the LI of each mouse were analyzed, and the average was calculated. Five mice per group were analyzed. i, j qPCR analysis of Tnf mRNA expression in the SI (i) and LI (j) from mice as described in a. k, l qPCR analysis of Il1b mRNA expression in the SI (k) and LI (l) from mice as described in a. m, n ELISA of 5-HT in the SI (m) and LI (n) from mice as described in a. In b–d and f–n, statistical analyses were performed using two-way ANOVA with Sidak’s multiple comparisons test. The data are presented as the means ± SEMs (b–d and f–h, n = 5) or in box-and-whisker plots displaying the medians, interquartile ranges (boxes) and minima and maxima (whiskers) (i–n, n = 5). P > 0.05, not significant (ns); *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001
Fig. 8.
Probiotic supplementation alleviates TRIM27 deficiency-induced IBS symptoms. a H&E staining of the SI from 16-week-old Trim27fl/fl and trim27[Lgr5] mice fed VSL#3 daily. Bottom, enlarged images of the regions outlined in the upper panels. Scale bars, 300 μm. The white dashed lines mark crypt borders. b–d Crypt depth (b), crypt number per mm (c) and villus height (d) in the SI from mice as described in a. At least 20 randomly selected crypts (b, d) or randomly selected fields (c) from the SI of each mouse were analyzed, and the average was calculated. Five mice per group were analyzed. e H&E staining of the LI from 16-week-old Trim27fl/fl and trim27[Lgr5] mice fed VSL#3 daily. Bottom, enlarged images of the regions outlined in the upper panels. Scale bars, 300 μm. The white dashed lines mark crypt borders. f–h Crypt depth (f), crypt number per mm (g) and villus height (h) in the LI from mice as described in e. At least 20 randomly selected crypts (f, h) or randomly selected fields (g) from the LI of each mouse were analyzed, and the average was calculated. Five mice per group were analyzed. i, j qPCR analysis of Tnf mRNA expression in the SI (i) and LI (j) from mice as described in a. k, l qPCR analysis of Il1b mRNA expression in the SI (k) and LI (l) from mice as described in a. m, n ELISA of 5-HT in the SI (m) and LI (n) from mice as described in a. o The ratio of Firmicutes to Bacteroidetes in the colonic contents from mice as described in a. In b–d and f–o, statistical analyses were performed using two-way ANOVA with Sidak’s multiple comparisons test. The data are presented as the means ± SEMs (b–d and f–h, n = 5; o, n = 3) or in box-and-whisker plots displaying the medians, interquartile ranges (boxes) and minima and maxima (whiskers) (i–n, n = 5). P > 0.05, not significant (ns); *P < 0.05; **P < 0.01; ****P < 0.0001
Fig. 9.
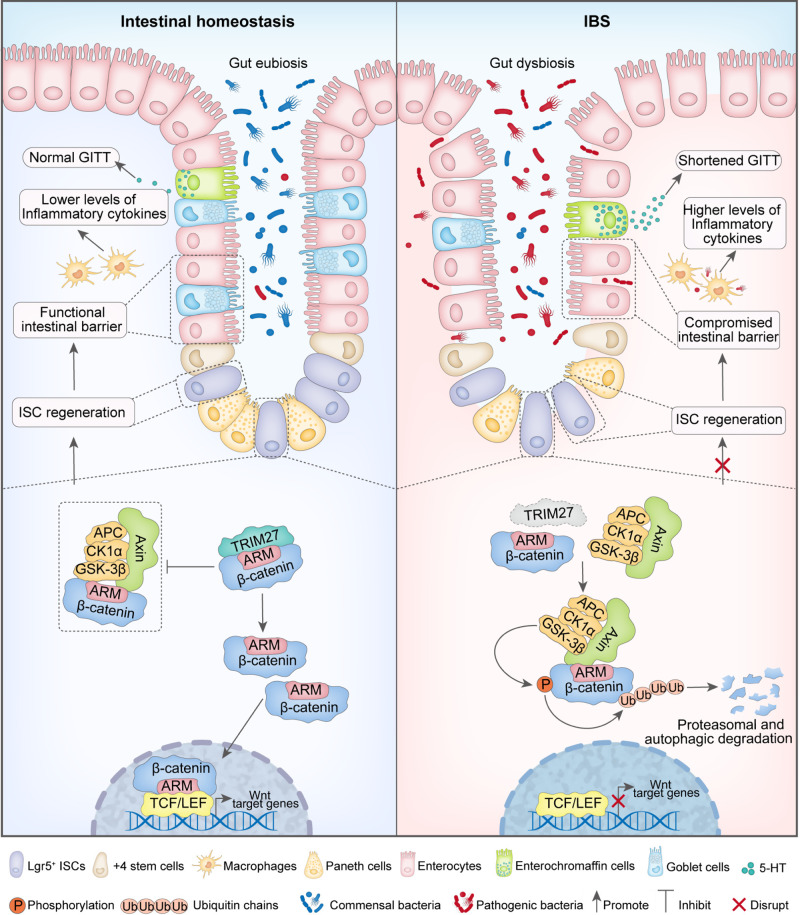
Proposed model showing the mechanism underlying TRIM27-mediated IBS symptoms. In the setting of intestinal homeostasis, TRIM27 competitively binds to the conserved Armadillo (ARM) repeat domain of β-catenin to disrupt the interaction between Axin and β-catenin, thus promoting the stabilization of β-catenin, followed by its transport from the cytoplasm to the nucleus to activate Wnt/β-catenin signaling pathway-mediated self-renewal of Lgr5+ ISC and the ensuing development and regeneration of the intestinal epithelium to maintain intestinal epithelial barrier integrity and gut homeostasis. In the absence of TRIM27, Axin directly interacts with the ARM domain of β-catenin to promote its phosphorylation and ubiquitination-mediated proteasomal and autophagic degradation, leading to suppression of the Wnt/β-catenin signaling pathway and defects in Lgr5+ ISC self-renewal. In turn, the defective self-renewal of Lgr5+ ISCs causes the disruption of intestinal epithelial barrier integrity and gut homeostasis, leading to the occurrence of IBS symptoms, including reduced goblet and Paneth cell numbers, elevated Tnf, Il1b and 5-hydroxytryptamine (5-HT) levels, a decreased gastrointestinal transit time (GITT) and dysregulation of the gut microbiota
Discussion
IBS is an extremely common chronic functional gastrointestinal disorder that prevents patients from participating in routine activities; however, our understanding of its etiology remains limited, which seriously limits its precise diagnosis and rational treatment. A variety of factors, such as genetic predisposition, stress, anxiety, and changes in the gut-brain axis and gut microbiota, have been connected with the pathogenesis of IBS [8]. However, the detailed molecular mechanisms, especially the genetic factors, and their regulatory mechanisms in IBS remain poorly understood. Here, we reveal that TRIM27 is a critical genetic protective factor for IBS, especially IBS-D (the major subtype of IBS). Specifically, we show that TRIM27 is expressed at low levels in IBS-D, and by establishing Trim27-knockout mouse models, we further demonstrate that Trim27 deficiency causes spontaneous IBS-D-like symptoms in mice. Although the symptoms of IBS-D overlap with those of IBD, there are still some differences between these two types of disease. For example, IBD patients exhibit weight loss, growth retardation, bloody stool and other extracolonic symptoms, all of which are absent in IBS patients [42]. Furthermore, the composition of the gut microbiota in IBS and IBD patients is different. For example, the ratio of Firmicutes to Bacteroidetes is usually increased in patients with IBS but decreased in patients with IBD [29]. Data from our group and others show that TRIM27 is highly expressed in the intestinal tissues of IBD patients [13], and our data further show that TRIM27 is expressed at low levels in IBS patients, especially IBS-D patients. Collectively, these data indicate that TRIM27 might function as a bidirectional regulator of gut homeostasis and might serve as a potential biomarker for differentiating IBS (a functional gastrointestinal disorder) from IBD (an organic gastrointestinal disorder). Because of the poorly understood etiology and pathogenesis of IBS, there has historically been a lack of suitable animal models for basic and clinical studies of IBS. The currently available IBS animal models are established by exposing mice to variable stressors (such as trinitrobenzenesulfonic acid treatment, cold and water avoidance) [43]. However, to date, a genetically based model in which animals spontaneously develop IBS symptoms that mimic the clinical manifestations in IBS patients is still lacking. Mice in our Trim27-knockout mouse model spontaneously develop a series of symptoms similar to those of IBS-D patients; thus, our model provides an important tool for further mechanistic and drug development studies for IBS.
Gut homeostasis is maintained mainly by intestinal epithelial barrier integrity, which requires coordination of ISC self-renewal and differentiation, and dysregulation of ISC self-renewal and differentiation is associated with a variety of intestinal disorders [44, 45]. However, to date, the mechanism by which ISCs are regulated in IBS patients and the physiological relevance of ISCs remain unexplored. Here, we reveal that TRIM27 maintains gut homeostasis by promoting ISC self-renewal. Mechanistically, TRIM27 activates Wnt/β-catenin signaling by stabilizing β-catenin through both the proteasomal and autophagic pathways in Lgr5+ ISCs to promote ISC self-renewal. It should be pointed out that although TRIM27 can function as an E3 ubiquitin ligase, TRIM27 exerts this effect independent of its E3 ubiquitin ligase activity. In this context, TRIM27 stabilizes β-catenin by competitively binding to the ARM repeat of β-catenin to block its interaction with Axin. Our finding is consistent with that of a recent study showing that β-catenin is downregulated in IBS patients but highly expressed in IBD and colorectal cancer patients [46], and our study further provides the molecular mechanism underlying the previous observation. It is worth mentioning that data from these two studies help explain why IBD but not IBS is usually a driver of the carcinogenic process [47, 48]. It is also important to note that Wnt/β-catenin signaling, which plays a key role in promoting cell proliferation, could perform a bidirectional regulatory function in the maintenance of gut homeostasis. On the one hand, as revealed in our study, Wnt/β-catenin signaling is required for gut homeostasis by promoting Lgr5+ ISC self-renewal; on the other hand, as shown by others, overactivated Wnt/β-catenin signaling leads to colorectal cancer under certain conditions [49, 50]. Thus, appropriate activation of Wnt/β-catenin signaling is crucial to maintaining gut homeostasis. It should also be noted that compared with mice with global Trim27 knockout, mice with stem cell-specific conditional Trim27 knockout exhibited more obvious IBS symptoms, suggesting that in addition to regulating ISC functions, TRIM27 may also play certain roles in other intestinal cell types, such as immune cells, to affect intestinal homeostasis. As reported, immune cells such as macrophages and dendritic cells (DCs) located in the gut are crucial for the maintenance of intestinal homeostasis by shaping a healthy microbial community and reinforcing epithelial barrier function [51], and uncontrolled activation of intestinal immune cells such as macrophages, DCs and T cells leads to the occurrence of IBD [52]. It has also been reported that TRIM27 is involved in antiviral defense processes in immune cells such as macrophages [14], but its regulatory role in intestinal immune cells remains unclear. Based on these previous observations, together with our finding that trim27[Lgr5] mice showed more obvious IBS symptoms than Trim27-/- mice, we speculate that TRIM27 may also function in immune cells to regulate intestinal homeostasis, a possibility that warrants further investigation in the future.
Gut homeostasis is maintained by a tridirectional relationship among intestinal epithelial integrity, the intestinal immune system and the gut microbiota. Previous studies have focused mainly on the regulatory roles of the gut microbiota in intestinal homeostasis, but the genetic factors and their regulatory mechanisms involved in this process are poorly understood. Our data reveal that TRIM27 deficiency-mediated dysfunction of Lgr5+ ISCs leads to the disruption of intestinal epithelial barrier integrity, followed by enhanced intestinal inflammation and gut dysbiosis. Thus, contrary to the previous idea that IBS is caused by gut dysbiosis [53, 54], our data indicate that, instead of being an initiating factor of IBS, gut dysbiosis may instead be an outcome of defective ISC homeostasis under certain conditions, for example, aberrant expression of genetic factors such as TRIM27. Clinically, IBS patients are usually treated with symptom-relieving drugs, such as antibiotics, probiotics and serotonin receptor ligands [42]. Targeting pro- and anti-inflammatory cytokines (such as TNF-α and IL-10, respectively) is another strategy adopted for IBS treatment [55]. However, these currently available treatment strategies for IBS only ameliorate certain symptoms without addressing the pathology of the disease from the root, and these kinds of symptom-based therapy tend to cause disease recurrence [56]. Here, we reveal that TRIM27 is an upstream regulator that maintains gut homeostasis by promoting Wnt/β-catenin signaling-dependent ISC renewal to prevent IBS. Together with the finding that activating Wnt/β-catenin signaling with SKL2001 ameliorates Trim27 deficiency-induced IBS symptoms, our data indicate that TRIM27 is a critical genetic protective factor for IBS. Together, our findings provide new insights into the pathogenesis of IBS by revealing a crucial role of TRIM27 in maintaining gut homeostasis. Our study identifies a potential treatment for IBS by targeting the TRIM27/Wnt/β-catenin axis and suggests that TRIM27 might serve as a biomarker for differentiating IBS from IBD.
Materials and methods
Bacterial strains, mammalian cell lines and plasmids
Escherichia coli DH5α and BL21 (DE3) were used for genetic manipulation and protein overexpression. For protein expression in mammalian HEK293T cells (ATCC CRL-3216, RRID: CVCL_0063), Trim27 or β-catenin was cloned into pcDNA6A (with a Myc tag). Bacterial expression plasmids were constructed by cloning cDNA into pET30a (with a His6 tag) or pGEX-6P-1 (with a GST tag). All the plasmids were sequenced at the Beijing Genomics Institute (BGI) for verification. The plasmids used in this study are listed in Supplementary Table 1.
Antibodies
All of the antibodies were used according to the manufacturer’s instructions. The following commercial antibodies were used: anti-Myc (Santa Cruz Biotechnology, sc-40; RRID: AB_2857941), 1:2000 for immunoblotting; anti-GST (ZSGB-BIO, TA-03; RRID: AB_2756893), 1:5000 for immunoblotting; anti-His (ZSGB-BIO, TA-02; RRID: AB_2801388), 1:2000 for immunoblotting; anti-HA (Abcam, ab236632; RRID:AB_2864361), 1:2000 for immunoblotting; anti-MCP-1 (Abcam, ab25124; RRID: AB_448636), 1:2000 for immunoblotting; anti-MIP-1α (Abcam, ab179638), 1:2000 for immunoblotting; anti-Mast cell tryptase (Abcam, ab151757), 1:2000 for immunoblotting; anti-Trim27 (Abcam, ab78393; RRID: AB_2042796), 1:2000 for immunoblotting and 1:200 for immunocytochemistry; anti-ZO-1 (Invitrogen, 402200; RRID: AB_2533456), 1:2000 for immunoblotting; anti-Occludin (Invitrogen, 710192; RRID: AB_2532617), 1:1000 for immunoblotting; anti-Claudin-1 (Invitrogen, 374900; RRID: AB_2533323), 1:1000 for immunoblotting; anti-Ki-67 (Novus, NB500-170; RRID: AB_10001977), 1:2000 for immunoblotting and 1:200 for immunocytochemistry; anti-β-catenin (Abcam, ab32572; RRID: AB_725966), 1:5000 for immunoblotting; anti-phospho-β-catenin (Ser33/37/Thr41), (Cell signaling technology, 9561T), 1:1000 for immunoblotting; anti-TCF-1 (Proteintech, 14464-1; RRID: AB_2878061), 1:5000 for immunoblotting; anti-LEF-1 (Proteintech, 14972-1; RRID: AB_2265677), 1:5000 for immunoblotting; anti-TCF-3 (Proteintech, 14519-1; RRID: AB_2287033), 1:5000 for immunoblotting; anti-TCF-4 (Proteintech, 13838-1; RRID: AB_2199812), 1:5000 for immunoblotting; Anti-Axin1 (Cell signaling technology, 2087; RRID:AB_2274550), 1:1000 for immunoblotting; anti-GAPDH (Santa Cruz Biotechnology, sc-25778; RRID: AB_10167668), 1:5000 for immunoblotting; anti-Lgr5 (Origene, TA503316; RRID: AB_2622395) 1:1000 for immunoblotting and 1:200 for immunocytochemistry; anti-rabbit IgG H&L (HRP) (ZSGB-BIO, ZB-2301; RRID: AB_2747412), 1:10,000 for immunoblotting; and anti-mouse IgG H&L (HRP) (ZSGB-BIO, ZB-2305; RRID: AB_2747415), 1:10,000 for immunoblotting.
Mice
Lgr5-creERT2 mice were a kind gift from Shu Zhu (University of Science and Technology of China) [57]. Trim27-/- and Trim27fl/fl mice were generated by Biocytogen (Beijing, China) using the CRISPR‒Cas9 system. Mice on the C57BL/6 genetic background were used in all experiments. Trim27fl/fl mice were first crossed with Lgr5-creERT2 mice to generate F1 trim27fl/+-Lgr5-cre mice. Trim27fl/+-Lgr5-cre mice were then crossed with Trim27fl/fl mice to generate trim27fl/fl-Lgr5-cre (trim27[Lgr5]) mice as intestinal stem cell-specific trim27-knockout mice and Trim27fl/fl mice as controls. Cre was activated by 1 intraperitoneal injection of 0.1 mg/g body weight tamoxifen (Sigma, T5648). Tamoxifen powder was dissolved in 900 μL of corn oil (Sigma, C8267) and 100 μL of ethanol by sonication. The trim27[Lgr5] mice received tamoxifen injections. All mice were bred and maintained in specific pathogen-free-grade cages using standard humane animal husbandry protocols. Female mice that were 2–16 weeks old during the course of the experiments were used. The sample size was based on empirical data from pilot experiments. All experimental protocols were performed in accordance with the instructional guidelines of the China Council on Animal Care and were approved by the Biomedical Research Ethics Committee of the Institute of Microbiology, Chinese Academy of Sciences (SQIMCAS2020039).
Endoscopy
Mice were anesthetized using an intraperitoneal injection of Nembutal, and a high-resolution microendoscope imaging system (R. Wolf) was used to determine the endoscopic condition of the mice.
AWR measurement
A distension balloon catheter (6-Fr, 2 mm external diameter) was placed in the descending colons of the mice. Each 10 s distention period was followed by a 20 s resting period. Mice were subjected to each level of distention (0, 0.2, 0.3 and 0.4 mL) two times, and the balloon was deflated and withdrawn after measuring the AWR. The visceral sensory responses to colorectal distention (CRD) were quantified by scoring the AWR. The AWR score was assigned as follows: 0, no behavioral response to CRD; 1, brief head movement followed by immobility; 2, contraction of abdominal muscles; 3, lifting of the abdomen; 4, body arching and lifting of the pelvic structures [58].
Expression and purification of recombinant proteins
GST- and His-tagged fusion proteins were purified as described previously [59]. For the expression and purification of Myc-tagged proteins, including β-catenin, TRIM27 and TRIM27 truncations, HEK293T cells were transfected with the indicated plasmids prior to immunoprecipitation using Anti-Myc Affinity Gel (Yeasen, 20587ES03).
Histological evaluation
The intestinal tissues of each mouse, including the small intestine (10–15 cm from the cecum) and the large intestine (5–7 cm from the cecum), were dissected, washed in PBS to remove intestinal contents, and fixed with 10% neutral-buffered formalin. The fixed intestinal tissues were then embedded in paraffin, cut perpendicularly to the surface at a thickness of 4 µm and stained with hematoxylin and eosin. The depth and number of crypts were evaluated for all of the slides from the intestines of each mouse, and the values were averaged.
Quantitative PCR (qPCR), immunoblot analysis and enzyme-linked immunosorbent assay (ELISA)
The intestinal tissues of each mouse, including the small intestine (10–15 cm from the cecum) and the large intestine (5–7 cm from the cecum), were dissected and washed in PBS to remove intestinal contents and were then sonicated in PBS and centrifuged at 900 × g for 5 min. Then, the supernatants were collected to analyze the production of 5-HT with a mouse 5-HT ELISA Kit (SAB, EK12351) according to the manufacturer’s instructions. The cell pellets were washed three times with PBS and harvested to analyze gene expression by qPCR or immunoblotting. Specifically, total RNA was extracted from isolated intestinal cells and was reverse transcribed into cDNA using Hieff First Strand cDNA Synthesis Super Mix (YEASEN, 11103ES70). The cDNA was then analyzed by qPCR using Hieff qPCR SYBR Green Master Mix (YEASEN, 11202ES08) on an ABI 7500 system (Applied Biosystems). Each experiment was performed with triplicate samples and repeated at least three times. The data were analyzed by the 2−ΔΔCT method and were normalized to the expression of the control gene GAPDH. For immunoblot analysis, cells were lysed in Cell Lysis Buffer (Beyotime Biotechnology, P0013) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF; Beyotime Biotechnology, ST505). Proteins were separated by SDS‒PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, PIVH00010). The membranes were blocked with 5% nonfat dry milk in Tris-buffered saline (20 mM Tris-HCl, pH 7.4; 150 Mm NaCl) with 0.1% Tween 20 (Sigma‒Aldrich, P1379) (TBST) for 1 h at room temperature (RT) and subsequently incubated overnight at 4 °C with primary antibodies. Following three washes with TBST for 10 min each, the membranes were incubated with goat anti-mouse IgG/HRP (ZSGB-BIO, ZB-2305) or goat anti-rabbit IgG/HRP (ZSGB-BIO, ZB-2301) for 1 h at RT. After three washes with TBST, the membranes were treated with Immobilon Western Chemiluminescent HRP Substrate (Millipore, WBKLS0500) and exposed to X-ray film.
GITT measurement
Mice were transferred to individual cages and orally administered 0.3 mL of 0.5% methylcellulose solution containing 6% carmine red (Mreda, M042735). The cage floor was covered with white paper for easy detection of carmine in feces. The mice were then allowed access to food and water ad libitum until the first red fecal pellet appeared. The GITT was calculated as the time period between oral gavage and the appearance of the first red fecal pellet.
Microarray and gene set enrichment analysis
For microarray analysis of TRIM family gene expression in the intestinal tissues of IBD and IBS patients, publicly available microarray data (E-MTAB-5811 for IBS analysis, GSE4183 for IBD analysis) were obtained from the EMBL-EBI ArrayExpress database and NCBI Gene Expression Omnibus, respectively. Specifically, the publicly available microarray data for IBS included data for 20 IBS patients (10 IBS-C and 10 IBS-D, 5 men and 5 women each, aged 18–55 years old) and 10 age/sex-matched healthy controls (HCs). The HCs had no personal or family history of IBS or other chronic pain conditions. Additional exclusion criteria for all subjects included infectious or inflammatory disorders; active psychiatric illness over the past 6 months; use of corticosteroids in the past 6 months; use of narcotics, antidepressants or other medications that could affect neuroendocrine function in the past 2 months; and current tobacco or alcohol abuse. The publicly available microarray data for IBD included data for 20 IBD patients (10 CD, 4 women and 6 men, aged 19–66 years old; 10 UC, 2 women and 8 men, aged 27–51 years old) and 11 HCs (five women and six men, aged 25–84 years old). The HCs had no significant pathological findings on endoscopic or histological examination. All endoscopic biopsies from which the publicly available IBS and IBD microarray data were derived were taken from a defined area of the sigmoid colon (at 20–30 cm measured during withdrawal). For analysis of the publicly available IBS and IBD microarray data, data from the microarrays were processed and normalized to gene counts using the rma function in the affy (v 1.68.0) package [60], and the values for the probes associated with the same gene symbol were then summed. Differentially expressed genes were identified using the Linear Models for Microarray Analysis (limma; v3.46.0) package in R (v4.0) after log2 transformation [61]. An adjusted P value threshold (Benjamini–Hochberg) of 0.05 was used to determine statistical significance. For gene set enrichment analysis (GSEA) of E-MTAB-5811 microarray data, Wnt and Notch pathways were retrieved from the Molecular Signatures Database (MSigDB; http://www.gsea-msigdb.org/gsea/msigdb/index.jsp, v6.1) [62, 63], and GSEA software (v.4.0.3) was then used for further analysis [63, 64]. An adjusted P value threshold (false discovery rate) of 0.05 was used to determine statistical significance.
Bacterial 16S rRNA sequencing and data analysis
Bacterial DNA was extracted using PowerMax Stool/Soil) DNA Isolation Kits (MoBio Laboratories, Carlsbad). Then, the V4 region of the bacterial 16S rRNA gene was amplified by PCR using the designated primers and Phusion High-Fidelity PCR Master Mix. After the individual quantification step, amplicons in the samples were sequenced on the Illumina HiSeq 4000 platform (Guheinfo, China). Quantitative Insights Into Microbial Ecology (QIIME; v 1.9.0, http://qiime.org/) was used to process the sequencing data [65]. Sequences with ≥97% similarity were assigned to the same operational taxonomic units (OTUs). Quality control and taxonomic classification of the OTUs were conducted with Vsearch (v2.4.4, https://github.com/torognes/vsearch) to search the representative sequences according to the Greengenes database [66]. An OTU table was further generated to record the abundance of each OTU in each sample and the taxonomic classification of these OTUs. Differences in bacterial abundances were calculated using linear discriminant analysis (LDA) effect size (LEfSe) software (v 1.1.1) [67]. LDA values of greater than two were considered to be significant. The 16S rRNA gene sequences are available in the NCBI Sequence Read Archive (SRA) database under accession number SRP355336.
Immunohistochemistry
Immunohistochemical staining for Trim27, Ki-67 (a cell proliferation marker), Lgr5 (an intestinal stem cell marker) and lysozyme (a Paneth cell marker) was performed in this study. In brief, formalin-fixed tissue sections were embedded in paraffin by standard methods. Sections (5 mm) were deparaffinized in xylene and rehydrated with graded ethanols, and antigen retrieval was performed by boiling in citrate buffer (pH 6) for 20 min. The slides were then treated with 3% hydrogen peroxide for 5 min and washed with PBS prior to the addition of primary antibodies. Afterward, the slides were washed with PBS, incubated with horseradish peroxidase-conjugated secondary antibodies at RT for 30 min, visualized using 3,3′-diaminobenzidine tetrahydrochloride with 0.05% H2O2 for 3 min and counterstained with Mayer’s hematoxylin. The percentage of Ki-67-positive area was analyzed with QuPath software [68]. At least five randomly selected fields from the small intestine of each mouse were analyzed, and the average was calculated.
AB-PAS staining
Formalin-fixed tissue sections were embedded in paraffin by standard methods, cut perpendicularly to the surface at a thickness of 4 µm and stained with Alcian blue and periodic acid–Schiff reagent using an AB-PAS staining kit (Solarbio, G1285) according to the manufacturer’s instructions.
Determination of crypt number, crypt depth and villus height
The crypt number, crypt depth and villus height were independently and objectively analyzed and quantitated with ImageJ (v 1.37) software in a blinded manner from coded digital photographs of crypts. The height in pixels from the bottom to the top of a crypt or villus in the small and large intestines, respectively, was measured. These measurements in pixels were converted to length measurements (in μm).
Pulse-chase assay with EdU labeling
Mice were repeatedly treated with EdU (10 mg/kg; Yeasen, 40284ES50) intraperitoneally every 8 h for 4 days, and the small intestines were harvested and fixed with 10% neutral-buffered formalin 0.5 days after the last EdU treatment. Then, the formalin-fixed tissue sections were embedded in paraffin and sectioned. Sections on glass slides were stained with Lgr5 for 10–30 min. DAPI Staining Solution (Beyotime, C1002) was used to visualize nuclei and to mount the slides. The slides were then scanned using an Aperio Versa 200 slide scanner (Leica Biosystems). The EdU+Lgr5+ cells in each intestinal section were counted using ImageJ (v 1.37) software.
Intestinal crypt culture (organoid formation)
The intestinal tissues of each mouse, including the small intestine (10–15 cm from the cecum) and the large intestine (5–7 cm from the cecum), were harvested and transferred into Petri dishes with ice-cold PBS. The intestinal contents were removed by gentle swirling, and the tissues were cut into 2 mm strips. The tissue pieces were then cleaned by gentle pipetting in ice-cold PBS. This washing step was repeated several times until a clear supernatant was visible. The tissue pieces were incubated in Gentle Cell Dissociation Reagent (Stemcell, 07174) for 30 min at RT to loosen crypts from the surrounding tissues. The tissue pieces were then washed in 5 mL of DMEM/F12 (Stemcell, 36254) and gently sheared using a 10 mL pipette. This step was repeated three times, and the loosened crypts were collected at each step. The resulting suspension was filtered through a 70 μm filter (Corning, 352350) and centrifuged at 300 × g for 5 min. The pellet was resuspended in 10 mL of DMEM/F12 and centrifuged at 150 × g for 2 min. Then, the pellet was resuspended in 200 µL of DMEM/F12 and a corresponding volume of Matrigel Matrix (Corning, 356231). Twelve-well plates were heated at 37 °C for 30 min before use. Fifty microliters of Matrigel containing intestinal crypts was placed at the center of each well and allowed to solidify for 15 min at 37 °C in 5% CO2. IntestiCult™ Organoid Growth Medium (Stemcell, 06005) heated to 37 °C in a water bath was added to each well (700 µL/well). The plates were incubated at 37 °C in 5% CO2 with a medium change every 3 days. After 14 days, organoids were counted under a light microscope. The size of the organoids was evaluated by quantifying the surface area of a horizontal cross-section of organoids acquired from multiple random nonoverlapping images using ImageJ software.
Pulldown assay
Recombinant proteins were added to 20 μL of glutathione resin (for GST-tagged proteins) in 500 μL of binding buffer (50 mM Tris, pH 7.5; 150 mM NaCl; 5 mM DTT; and 0.1% NP-40) supplemented with 1% protease inhibitors for 1 h at 4 °C. The beads were washed five times with binding buffer and further incubated with the prey protein supplemented with 0.1 mg/mL BSA (1:1 molar ratio of GST-tagged protein to prey protein). After 2 h of incubation at 4 °C, the beads were washed and subjected to immunoblot analysis.
In vivo ubiquitination assays
In vivo ubiquitination assays were performed using TRIM27+/+ and TRIM27-/- HEK293T cells. For knockdown of the TRIM27 gene in HEK293T cells, 50 nM of small interfering RNA was transfected into cells in the presence of Lipofectamine RNAiMAX (Invitrogen, 13778) according to the manufacturer’s instructions. Twelve hours after the first transfection, the cells were retransfected as described above and were then incubated for an additional 24 h. The small interfering RNA sequences were as follows: TRIM27 siRNA, forward-ggagaaaatccaagaattauu and reverse- aauucuuggauuuucuccuu; Luciferase siRNA (NC siRNA), forward-uucuccgaacgugucacguuu and reverse-acgugacacguucggagaauu. For ubiquitination assays, cells were transfected with Myc-β-catenin, HA, HA-Ub, HA-Ub (K48 only) or HA-Ub (K63 only) for 24 h. Then, the cells were lysed in Cell Lysis buffer and centrifuged at 12,000 × g. Prewashed anti-Myc beads were incubated with the supernatant for 4 h at 4 °C to pull down Myc-tagged β-catenin and its bound proteins. The beads were then washed with Cell Lysis buffer and analyzed by immunoblotting with anti-HA or anti-Myc antibodies.
Lentivirus production and the indicated lentivirus-reconstituted organoid formation
Control, β-catenin, Trim27 and Trim27 mutant recombinant lentiviruses were produced by Genechem (Beijing, China). Purified crypts were infected with the indicated recombinant lentiviruses at a multiplicity of infection of 15, and the complexes were then spinoculated at 400 × g and 20 °C for 1 h, plated in IntestiCult Organoid Growth Medium (Stemcell, 06005) supplemented with the same volume of Matrigel Matrix (Corning, 356231) and 8 μg/mL polybrene (Santa Cruz Biotechnology, sc-134220), and incubated in a culture incubator. The plates were maintained at 37 °C in 5% CO2 with a medium change every 3 days. After 14 days, organoids were counted under a light microscope.
Mouse model
For dextran sulfate sodium (DSS) treatment, mice aged 12–16 weeks were given 2% DSS (30–50 kDa) (YEASEN, 60316ES60) in sterile water for 5 days, and the DSS-containing water was then changed to sterile water for the duration of the experiment. Then, the mice were euthanized, and the intestinal tissues were harvested from each mouse on days 0, 7 and 14 after treatment with 2% DSS. For SKL2001 treatment, mice aged 12–16 weeks were orally fed SKL2001 (200 µg in 100 µL of corn oil) (Selleck, S88219) or corn oil (100 µL) daily for 14 days. Then, the mice were euthanized, and the intestinal tissues, including the small intestine (10–15 cm from the cecum) and the large intestine (5–7 cm from the cecum), were harvested from each mouse for further analysis. For probiotic treatment, mice aged 16 weeks were divided into two groups (n = 5). One group of mice was orally fed VSL#3 daily for 30 days. The daily dose of VSL#3 was 1 × 108 colony-forming units (CFUs) per mouse in 200 μL of PBS. The other group of mice (control group) was orally fed sterile water. Then, the mice were euthanized, and the small intestine (10–15 cm from the cecum), the large intestine (5–7 cm from the cecum) and the colon contents were collected from each mouse for further analysis.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8.0 software. Data were analyzed by unpaired two-tailed Student’s t test or two-way ANOVA with Sidak’s test to correct for multiple comparisons, as indicated in the corresponding figure legends. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001 were considered to be statistically significant; P > 0.05 (not significant (ns)) denoted a nonsignificant difference between groups. All experiments were performed with at least two biological replicates with similar results. Representative micrographs are shown.
Supplementary information
Acknowledgements
We thank Junfeng Hao (Core Facility for Protein Research, Institute of Biophysics, Chinese Academy of Sciences, Beijing) for helping with the histological analysis and Zhihua Liu and Hongying Wang (National Cancer Center/National Clinical Research Center for Cancer/Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College) for helping with the endoscopy experiment. This work was supported by the National Key Research and Development Project of China (2021YFA1300200 to CHL and LZ, 2022YFC2302900 to CHL and JW), the National Natural Science Foundation of China (81825014 to CHL, 31830003 to CHL, 82022041 to JW and 81871616 to JW), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB29020000 to CHL), Youth Innovation Promotion Association CAS (2018118 to JW) and the State Key Laboratory of Proteomics (SKLP-K202001 to LZ and SKLPO202003 to JW).
Author contributions
CHL and LZ conceived and supervised the study. CHL and JW designed the experiments. JW, DZ and PG performed the majority of the experiments. ZLei, ZLu, QC, YZ, LQ, YY, XZ, BL and SZ helped with some of the experiments and data analysis. CHL, JW and ZLei analyzed the data and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Data availability
The 16S rRNA gene sequences are available in the NCBI Sequence Read Archive (SRA) database under accession number SRP355336. Publicly available microarray data (E-MTAB-5811, GSE1710) were downloaded from the EMBL-EBI ArrayExpress database and NCBI Gene Expression Omnibus. Any additional information required to reanalyze the data reported in this paper is available from the corresponding author upon request.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Jing Wang, Dongdong Zhao, Zehui Lei, Pupu Ge.
Contributor Information
Lingqiang Zhang, Email: zhanglq@nic.bmi.ac.cn.
Cui Hua Liu, Email: liucuihua@im.ac.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-022-00963-1.
References
- 1.Sperber AD. Epidemiology and burden of irritable bowel syndrome: an international perspective. Gastroenterol Clin North Am. 2021;50:489–503. doi: 10.1016/j.gtc.2021.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2017;390:2769–78. doi: 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 3.Kaplan GG, Windsor JW. The four epidemiological stages in the global evolution of inflammatory bowel disease. Nat Rev Gastroenterol Hepatol. 2021;18:56–66. doi: 10.1038/s41575-020-00360-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanning N, Edwinson AL, Ceuleers H, Peters SA, De Man JG, Hassett LC, et al. Intestinal barrier dysfunction in irritable bowel syndrome: a systematic review. Ther Adv Gastroenterol. 2021;14:1756284821993586. doi: 10.1177/1756284821993586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spiller R, Major G. IBS and IBD – separate entities or on a spectrum? Nat Rev Gastroenterol Hepatol. 2016;13:613–21. doi: 10.1038/nrgastro.2016.141. [DOI] [PubMed] [Google Scholar]
- 6.Santos AJM, Lo YH, Mah AT, Kuo CJ. The intestinal stem cell niche: homeostasis and adaptations. Trends Cell Biol. 2018;28:1062–78. doi: 10.1016/j.tcb.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ahlawat S, Kumar P, Mohan H, Goyal S, Sharma KK. Inflammatory bowel disease: tri-directional relationship between microbiota, immune system and intestinal epithelium. Crit Rev Microbiol. 2021;47:254–73. doi: 10.1080/1040841X.2021.1876631. [DOI] [PubMed] [Google Scholar]
- 8.Eijsbouts C, Zheng T, Kennedy NA, Bonfiglio F, Anderson CA, Moutsianas L, et al. Genome-wide analysis of 53,400 people with irritable bowel syndrome highlights shared genetic pathways with mood and anxiety disorders. Nat Genet. 2021;53:1543–52. doi: 10.1038/s41588-021-00950-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu L, Li Y, Zhou L, Yang G, Wang Y, Han J, et al. Role of RING-Type E3 ubiquitin ligases in inflammatory signalling and inflammatory bowel disease. Mediators Inflamm. 2020;2020:5310180. doi: 10.1155/2020/5310180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai X, Luo Y, Zhang Y, Lin Y, Wu B, Cao Z, et al. The deubiquitinase OTUD1 inhibits colonic inflammation by suppressing RIPK1-mediated NF-kappaB signaling. Cell Mol Immunol. 2022;19:276–89. [DOI] [PMC free article] [PubMed]
- 11.Chen SY, Zhang HP, Li J, Shi JH, Tang HW, Zhang Y, et al. Tripartite motif-containing 27 attenuates liver ischemia/reperfusion injury by suppressing transforming growth factor beta-activated kinase 1 (TAK1) by TAK1 binding protein 2/3 degradation. Hepatology. 2021;73:738–58. doi: 10.1002/hep.31295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gushchina LV, Kwiatkowski TA, Bhattacharya S, Weisleder NL. Conserved structural and functional aspects of the tripartite motif gene family point towards therapeutic applications in multiple diseases. Pharm Ther. 2018;185:12–25. doi: 10.1016/j.pharmthera.2017.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zurek B, Schoultz I, Neerincx A, Napolitano LM, Birkner K, Bennek E, et al. TRIM27 negatively regulates NOD2 by ubiquitination and proteasomal degradation. PLoS One. 2012;7:e41255. doi: 10.1371/journal.pone.0041255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Q, Hou J, Zhou Y, Yang Y, Xie B, Cao X. Siglec1 suppresses antiviral innate immune response by inducing TBK1 degradation via the ubiquitin ligase TRIM27. Cell Res. 2015;25:1121–36. doi: 10.1038/cr.2015.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang HX, Xu ZS, Lin H, Li M, Xia T, Cui K, et al. TRIM27 mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis. Nat Commun. 2018;9:3441. doi: 10.1038/s41467-018-05796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang R, Li H, Wu J, Cai ZY, Li B, Ni H, et al. Gut stem cell necroptosis by genome instability triggers bowel inflammation. Nature. 2020;580:386–90. doi: 10.1038/s41586-020-2127-x. [DOI] [PubMed] [Google Scholar]
- 17.Hong KB, Seo H, Lee JS, Park Y. Effects of probiotic supplementation on post-infectious irritable bowel syndrome in rodent model. BMC Complement Alter Med. 2019;19:195. doi: 10.1186/s12906-019-2610-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kodani M, Fukui H, Tomita T, Oshima T, Watari J, Miwa H. Association between gastrointestinal motility and macrophage/mast cell distribution in mice during the healing stage after DSSinduced colitis. Mol Med Rep. 2018;17:8167–72. doi: 10.3892/mmr.2018.8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilkinson JM, Gill MC. Irritable bowel syndrome: questions and answers for effective care. Am Fam Physician. 2021;103:727–36. [PubMed] [Google Scholar]
- 20.Zhou XY. Visceral hypersensitive rats share common dysbiosis features with irritable bowel syndrome patients. World J Gastroenterol. 2016;22:5211–27. doi: 10.3748/wjg.v22.i22.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zielinska M, Fichna J, Bashashati M, Habibi S, Sibaev A, Timmermans JP, et al. G protein-coupled estrogen receptor and estrogen receptor ligands regulate colonic motility and visceral pain. Neurogastroenterol Motil. 2017;29:e13025. doi: 10.1111/nmo.13025. [DOI] [PubMed] [Google Scholar]
- 22.Grabauskas G, Wu X, Gao J, Li JY, Turgeon DK, Owyang C. Prostaglandin E2, produced by mast cells in colon tissues from patients with irritable bowel syndrome, contributes to visceral hypersensitivity in mice. Gastroenterology. 2020;158:2195–207.e2196. doi: 10.1053/j.gastro.2020.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pimentel M, Lembo A. Microbiome and its role in irritable bowel syndrome. Dig Dis Sci. 2020;65:829–39. doi: 10.1007/s10620-020-06109-5. [DOI] [PubMed] [Google Scholar]
- 24.Blonska A, Konrad P, Chojnacki J, Chojnacki C. [Evaluation of oro-cecal transit time in patients with irritable bowel syndrome with cereal products intolerance] Pol Merkur Lekarski. 2017;42:116–20. [PubMed] [Google Scholar]
- 25.Yoshimoto T, Oshima T, Huang X, Tomita T, Fukui H, Miwa H. Microinflammation in the intestinal mucosa and symptoms of irritable bowel syndrome. J Gastroenterol. 2021;57:62–69. doi: 10.1007/s00535-021-01838-4. [DOI] [PubMed] [Google Scholar]
- 26.Uranga JA, Martinez V, Abalo R. Mast cell regulation and irritable bowel syndrome: effects of food components with potential nutraceutical use. Molecules. 2020;25:4314. doi: 10.3390/molecules25184314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bazzoni G, Martınez-Estrada OM, Orsenigo F, Cordenonsi M, Citi S, Dejana E. Interaction of junctional adhesion molecule with the tight junction components ZO-1, cingulin, and occludin. J Biol Chem. 2000;275:20520–6. doi: 10.1074/jbc.M905251199. [DOI] [PubMed] [Google Scholar]
- 28.Linsalata M, Riezzo G, Clemente C, D’Attoma B, Russo F. Noninvasive biomarkers of gut barrier function in patients suffering from diarrhea predominant-IBS: an update. Dis Markers. 2020;2020:2886268. doi: 10.1155/2020/2886268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vich Vila A, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med. 2018;10:eaap8914. [DOI] [PubMed]
- 30.Vervier K, Moss S, Kumar N, Adoum A, Barne M, Browne H, et al. Two microbiota subtypes identified in irritable bowel syndrome with distinct responses to the low FODMAP diet. Gut. 2021;71:1821–30. doi: 10.1136/gutjnl-2021-325177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fritz T, Niederreiter L, Adolph T, Blumberg RS, Kaser A. Crohn’s disease: NOD2, autophagy and ER stress converge. Gut. 2011;60:1580–8. doi: 10.1136/gut.2009.206466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee SM, Kim N, Yoon H, Kim YS, Choi SI, Park JH, et al. Compositional and functional changes in the gut microbiota in irritable bowel syndrome patients. Gut Liver. 2021;15:253–61. doi: 10.5009/gnl19379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metcalfe C, Kljavin NM, Ybarra R, de Sauvage FJ. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell. 2014;14:149–59. doi: 10.1016/j.stem.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 34.Farin HF, Jordens I, Mosa MH, Basak O, Korving J, Tauriello DVF, et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature. 2016;530:340–3. doi: 10.1038/nature16937. [DOI] [PubMed] [Google Scholar]
- 35.Zha JM, Li HS, Lin Q, Kuo WT, Jiang ZH, Tsai PY, et al. Interleukin 22 expands transit-amplifying cells while depleting Lgr5(+) stem cells via inhibition of Wnt and Notch signaling. Cell Mol Gastroenterol Hepatol. 2019;7:255–74. doi: 10.1016/j.jcmgh.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–99. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 37.Ikeda S. Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3beta and beta-catenin and promotes GSK-3beta-dependent phosphorylation of beta-catenin. EMBO J. 1998;17:1371–84. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasegawa N, Iwashita T, Asai N, Murakami H, Iwata Y, Isomura T, et al. A RING finger motif regulates transforming activity of the rfp/ret fusion gene. Biochem Biophys Res Commun. 1996;225:627–31. doi: 10.1006/bbrc.1996.1221. [DOI] [PubMed] [Google Scholar]
- 39.Gwak J, Hwang SG, Park HS, Choi SR, Park SH, Kim H, et al. Small molecule-based disruption of the Axin/beta-catenin protein complex regulates mesenchymal stem cell differentiation. Cell Res. 2012;22:237–47. doi: 10.1038/cr.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delgado S, Sanchez B, Margolles A, Ruas-Madiedo P, Ruiz L. Molecules produced by probiotics and intestinal microorganisms with immunomodulatory activity. Nutrients. 2020;12:391. doi: 10.3390/nu12020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akutko K, Stawarski A. Probiotics, prebiotics and synbiotics in inflammatory bowel diseases. J Clin Med. 2021;10:2466. doi: 10.3390/jcm10112466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szalwinska P, Wlodarczyk J, Spinelli A, Fichna J, Wlodarczyk M. IBS-symptoms in IBD patients-manifestation of concomitant or different entities. J Clin Med. 2020;10:31. [DOI] [PMC free article] [PubMed]
- 43.Johnson AC, Farmer AD, Ness TJ, Greenwood-Van Meerveld B. Critical evaluation of animal models of visceral pain for therapeutics development: a focus on irritable bowel syndrome. Neurogastroenterol Motil. 2020;32:e13776. doi: 10.1111/nmo.13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dotti I, Mora-Buch R, Ferrer-Picon E, Planell N, Jung P, Masamunt MC, et al. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut. 2017;66:2069–79. doi: 10.1136/gutjnl-2016-312609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du G, Xiong L, Li X, Zhuo Z, Zhuang X, Yu Z, et al. Peroxisome elevation induces stem cell differentiation and intestinal epithelial repair. Dev Cell. 2020;53:169–84. doi: 10.1016/j.devcel.2020.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Fredericks E, Dealtry G, Roux S. beta-Catenin regulation in sporadic colorectal carcinogenesis: not as simple as APC. Can J Gastroenterol Hepatol. 2018;2018:4379673. doi: 10.1155/2018/4379673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keller DS, Windsor A, Cohen R, Chand M. Colorectal cancer in inflammatory bowel disease: review of the evidence. Tech Coloproctol. 2019;23:3–13. doi: 10.1007/s10151-019-1926-2. [DOI] [PubMed] [Google Scholar]
- 48.Wu S, Yuan C, Liu S, Zhang Q, Yang Z, Sun F, et al. Irritable bowel syndrome and long-term risk of cancer: a prospective cohort study among 0.5 million adults in UK Biobank. Am J Gastroenterol. 2022;117:785–93. doi: 10.14309/ajg.0000000000001674. [DOI] [PubMed] [Google Scholar]
- 49.Schmitt M, Schewe M, Sacchetti A, Feijtel D, van de Geer WS, Teeuwssen M, et al. Paneth cells respond to inflammation and contribute to tissue regeneration by acquiring stem-like features through SCF/c-Kit signaling. Cell Rep. 2018;24:2312–28.e2317. doi: 10.1016/j.celrep.2018.07.085. [DOI] [PubMed] [Google Scholar]
- 50.Zhang H, Lin M, Dong C, Tang Y, An L, Ju J, et al. An MST4-pbeta-Catenin(Thr40) signaling axis controls intestinal stem cell and tumorigenesis. Adv Sci (Weinh) 2021;8:e2004850. doi: 10.1002/advs.202004850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kayama H, Okumura R, Takeda K. Interaction between the microbiota, epithelia, and immune cells in the intestine. Annu Rev Immunol. 2020;38:23–48. doi: 10.1146/annurev-immunol-070119-115104. [DOI] [PubMed] [Google Scholar]
- 52.Neurath MF. Targeting immune cell circuits and trafficking in inflammatory bowel disease. Nat Immunol. 2019;20:970–9. doi: 10.1038/s41590-019-0415-0. [DOI] [PubMed] [Google Scholar]
- 53.Mars RAT, Yang Y, Ward T, Houtti M, Priya S, Lekatz HR, et al. Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome. Cell. 2020;182:1460–73.e1417. doi: 10.1016/j.cell.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ford AC, Sperber AD, Corsetti M, Camilleri M. Irritable bowel syndrome. Lancet. 2020;396:1675–88. doi: 10.1016/S0140-6736(20)31548-8. [DOI] [PubMed] [Google Scholar]
- 55.Kumar S, Singh P, Kumar A. Targeted therapy of irritable bowel syndrome with anti-inflammatory cytokines. Clin J Gastroenterol. 2021;15:1–10. doi: 10.1007/s12328-021-01555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Enck P, Aziz Q, Barbara G, Farmer AD, Fukudo S, Mayer EA, et al. Irritable bowel syndrome. Nat Rev Dis Prim. 2016;2:16014. doi: 10.1038/nrdp.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 58.Lu Y, Huang J, Zhang Y, Huang Z, Yan W, Zhou T, et al. Therapeutic effects of berberine hydrochloride on stress-induced diarrhea-predominant irritable bowel syndrome rats by inhibiting neurotransmission in colonic smooth muscle. Front Pharm. 2021;12:596686. doi: 10.3389/fphar.2021.596686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang J, Li BX, Ge PP, Li J, Wang Q, Gao GF, et al. Mycobacterium tuberculosis suppresses innate immunity by coopting the host ubiquitin system. Nat Immunol. 2015;16:237–45. doi: 10.1038/ni.3096. [DOI] [PubMed] [Google Scholar]
- 60.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 61.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–25. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 65.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rognes T, Flouri T, Nichols B, Quince C, Mahe F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bankhead P, Loughrey MB, Fernández JA, Dombrowski Y, McArt DG, Dunne PD, et al. QuPath: Open source software for digital pathology image analysis. Sci Rep. 2017;7:16878. doi: 10.1038/s41598-017-17204-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA gene sequences are available in the NCBI Sequence Read Archive (SRA) database under accession number SRP355336. Publicly available microarray data (E-MTAB-5811, GSE1710) were downloaded from the EMBL-EBI ArrayExpress database and NCBI Gene Expression Omnibus. Any additional information required to reanalyze the data reported in this paper is available from the corresponding author upon request.