Abstract
The locations of the catalytic and receptor-binding domains of the Pasteurella multocida toxin (PMT) were investigated. N- and C-terminal fragments of PMT were cloned and expressed as fusion proteins with affinity tags. Purified fusion proteins were assessed in suitable assays for catalytic activity and cell-binding ability. A C-terminal fragment (amino acids 681 to 1285) was catalytically active. When microinjected into quiescent Swiss 3T3 cells, it induced changes in cell morphology typical of toxin-treated cells and stimulated DNA synthesis. An N-terminal fragment with a His tag at the C terminus (amino acids 1 to 506) competed with full-length toxin for binding to surface receptors and therefore contains the cell-binding domain. The inactive mutant containing a mutation near the C terminus (C1165S) also bound to cells in this assay. Polyclonal antibodies raised to the N-terminal PMT region bound efficiently to full-length native toxin, suggesting that the N terminus is surface located. Antibodies to the C terminus of PMT were microinjected into cells and inhibited the activity of toxin added subsequently to the medium, confirming that the C terminus contains the active site. Analysis of the PMT sequence predicted a putative transmembrane domain with predicted hydrophobic and amphipathic helices near the N terminus over the region of homology to the cytotoxic necrotizing factors. The C-terminal end of PMT was predicted to be a mixed α/β domain, a structure commonly found in catalytic domains. Homology to proteins of known structure and threading calculations supported these assignments.
The Pasteurella multocida toxin (PMT) is an extremely potent mitogen for Swiss 3T3 cells, other fibroblast cell lines, and early-passage cultures (15, 39). The toxin is produced by some strains of P. multocida and is responsible for the loss of nasal turbinate bone associated with porcine atrophic rhinitis (33). Furthermore, experimental nasal infection with toxigenic P. multocida leads to proliferation of bladder epithelium (17).
PMT interacts with host cell signaling pathways and results in production of inositol triphosphates and diacyl glycerol, with mobilization of Ca2+ from intracellular stores and subsequent activation of protein kinase C (48, 49). PMT activates PLCβ via a Gq-mediated pathway (31, 56, 57), and this heterotrimeric G protein may be the direct target of PMT. The toxin stimulates Ras-dependent ERK activation via transactivation of the epidermal growth factor receptor (44). PMT also induces cytoskeletal rearrangments, with the formation of actin stress fibers and focal adhesions, and causes tyrosine phosphorylation of paxillin and focal adhesion kinase (24). This occurs via activation of the Rho protein and its effector p160/ROCK (51).
There is considerable evidence that PMT is an intracellularly acting toxin. There is a pronounced lag between the addition of toxin to cells and any cellular effects (39). Its action is also inhibited by neutralizing antibody or methylamine added early but not late after toxin. PMT undergoes a conformational change at low pH, which affects its protease sensitivity and circular dichroism spectra (46, 47). This suggests that PMT may be trafficked and perhaps processed through a low-pH compartment. By analogy with other large intracellularly acting toxins, it is predicted to comprise domains for receptor binding, membrane translocation, and catalytic activity.
PMT is a monomeric 146-kDa protein. It has been purified, cloned, and sequenced (3, 26, 27, 34). PMT shares significant homology with the cytotoxic necrotizing factors (CNFs) of Escherichia coli (9, 32). The homology is highest toward the N termini of both toxins. In CNF, the N terminus is known to contain the domains for binding and internalization of the toxin (28). The C terminus of CNF is homologous to the C terminus of the Bordetella dermonecrotic toxin (DNT) (36, 52), and in both toxins this region possesses catalytic activity (22, 28). CNF and DNT have similar enzymatic activities: each modifies small GTP binding proteins of the Rho family by deamidation or transglutamination, respectively, of a specific glutamine residue (11, 16, 29, 30, 42, 43), whereas PMT has a different mode of action. The sequence homologies strongly suggest that PMT has a molecular organization similar to those of CNF and DNT.
In support of this hypothesis, our group previously reported the construction of a mutant near the C terminus of PMT (C1165S) that was completely inactive in cell assays and that had lost all toxicity in vivo (53). This mutation did not grossly affect the structure of the molecule since it had circular dichroism spectra and protease sensitivity patterns similar to those of the wild-type toxin and therefore is probably near the active site. In contrast, it has been reported that the N terminus of PMT possesses catalytic activity, since microinjection of an N-terminal peptide led to a response in voltage-clamped Xenopus oocytes (55). The reasons for this discrepancy remain unclear.
The aim of the present study was to clarify the location of the functional domains of PMT. We demonstrate definitively that PMT has a molecular organization similar to that of CNF and DNT, with the cell-binding and/or internalization domain at the N terminus and the domain for mitogenic activity at the C terminus.
MATERIALS AND METHODS
Materials.
All chemical reagents were from Sigma, Ltd., unless otherwise stated. Restriction enzymes and DNA-modifying enzymes were purchased from Promega Corporation or New England Biolabs and used according to the manufacturer's instructions. Oligonucleotides were synthesized by Sigma Genosys. Chromatography reagents and [3H]thymidine (code no. TRK296) were purchased from Amersham Pharmacia Biotech.
Bacterial strains, plasmid vectors, and growth conditions.
E. coli JM109 was used as a general purpose host for cloning and expression of toxin fragments cloned as glutathione S-transferase (GST) fusions. E. coli XL1-Blue was the host for pTox2 (which expresses full-length recombinant PMT from its own promoter [53]) and for the corresponding mutant toxin plasmid, pC1165S. Novablue (DE3) was the host for expression of the His-tagged PMT fragment. Plasmid vectors pGEX-4T-1 and pGEX-4T-3 were from Amersham Pharmacia Biotech, pET30a was from Novagen, and pGEM-T was from Promega Corporation.
E. coli strains were grown routinely in Luria-Bertani (LB) broth or on LB agar aerobically at 37°C unless otherwise stated. Antibiotics were added as appropriate at the following concentrations: tetracycline, 20 μg/ml; ampicillin, 50 μg/ml; and kanamycin, 25 μg/ml.
DNA isolation and sequence analysis.
Plasmid DNA for use in subcloning was isolated by using Wizard kits (Promega). For sequence analysis, DNA was further purified by using Qiaquick columns (Qiagen). DNA sequencing was performed with a Beckman CEQ2000 automated DNA sequencer.
Site-directed mutagenesis.
Mutagenesis was performed by using the Quikchange mutagenesis kit (Stratagene), according to the manufacturer's instructions. For reversal of the C1165S mutation, the following oligonucleotides were used: 5′-GGAAGCTGGCTCTTGTGATTCAGTAAG and 5′-CTTACTGAATCACAAGAGCCAGCTTCC.
Construction of expression plasmids.
Plasmids for expression of GST- and His-tagged fusions of PMT and its fragments were constructed as follows.
(i) pGST/681-C.
A fragment encoding PMT residues 681 to 1285 was amplified by PCR from pTox2 by using oligonucleotides 8012 (5′-TACATAACAGTAATATC) and PMTSacAS (5′-AGCGAGCTCGCGCTAATAAGATCGATTGC), gel purified by using Geneclean (Anachem), and A-tailed by using Taq polymerase. This fragment was ligated into pGEM-T (Promega) to produce pGEM/681-C. The insert from this was excised by digestion with BstZI, followed by gel purification, and ligated to pGEX-4T-3 digested with NotI.
(ii) pGST/849-C.
A fragment encoding residues 849 to 1285 was PCR amplified from pTox2 by using primers 8013 (5′-TTGCCACACGTAACTTT) and PMTSacAS, gel purified, and A-tailed, before ligation into pGEM-T. The 1.6-kb insert was excised from this plasmid by digestion with BstZI and then ligated to pGEX-4T-3 digested with NotI.
(iii) pGST/PMT.
The complete PMT gene was PCR amplified from pTox2 by using primers PMTGST (5′-CGCGGATCCATGAAAACAAAACATTTTTTTAACTC) and PMTSacAS, gel purified, A-tailed, and ligated to pGEM-T. The insert was excised from this plasmid by digestion with BamHI and SalI and ligated to pGEX-4T-1 digested with the same enzymes.
(iv) pGST/1-506.
This was prepared from pGST/PMT by deletion of residues 507 to 1285 by digestion with XhoI and self-ligation.
(v) p1-506/His.
This contains the PMT N terminus with a hexahistidine (His) tag at its C terminus. The N terminus of PMT, including ca. 180 nucleotides upstream, was excised from pTox2 by digestion with XhoI and BamHI and gel purified. This was ligated to pET30a which had been digested with the same enzymes and transformed into E. coli JM109. Plasmid DNA containing the insert was digested with XhoI and treated with mung bean nuclease before self-ligation. This placed the His tag in the same reading frame as the PMT fragment.
Expression and purification of fusion proteins.
GST fusion expression plasmids were transformed into E. coli JM109. The bacteria were inoculated into 15 ml of L broth containing ampicillin and cultured overnight at 30°C. They were subcultured into 300 ml of L broth containing ampicillin and grown for 90 to 120 min at 30°C, before the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to 0.2 mM, and culture continued for 5 to 6 h. Cells were harvested by centrifugation at 4,000 × g in a Heraeus Biofuge Primo centrifuge, pellets were resuspended in 15 ml of phosphate-buffered saline (PBS), and the cells were lysed by sonication for three rounds of 20-s pulses at a duty cycle of 50% on a Vibra-Cell sonicator (Sonics and Materials, Inc.). Cell debris was removed by centrifugation at 7,000 × g for 10 min. GST fusion proteins were purified from the supernatants by using glutathione-Sepharose beads (Amersham Pharmacia Biotech) according to the manufacturer's instructions. For some applications, the GST tag was cleaved by using a thrombin cleavage capture kit (Novagen) according to the manufacturer's instructions.
E. coli Novablue (DE3/p1-506/His) was inoculated into 15 ml of L broth with kanamycin and tetracycline and grown overnight at 30°C. This was subcultured into 300 ml of L broth containing kanamycin and cultured for 90 to 120 min. Expression was induced and cells were harvested as for the GST fusion method and sonicated in 10 mM Tris-HCl (pH 7.4)–300 mM NaCl. Cleared supernatants were purified on 1 ml of HiTrap columns (Amersham Pharmacia Biotech) loaded with 0.1 M NiSO4 according to the manufacturer's instructions. Fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The fusion protein generally eluted with 100 mM imidazole.
Both GST- and His-tagged fusion proteins were dialyzed overnight at 4°C against PBS or other buffers as appropriate and then concentrated by using Microcon 10 columns (Millipore) before storage at 4°C for up to 1 week. Protein content was quantitated by use of the BCA protein assay (Pierce).
Deletion mutagenesis.
Random deletion mutants of GST/681-C were prepared by using the Erase-a-Base kit (Promega Corporation) according to the manufacturer's instructions. pGST/681-C was first digested with SalI, protected with phosphorothioate, and then digested with XhoI before deletions were generated with ExoIII. Deletants were cultured to logarithmic phase in 10-ml volumes of L broth, and expression was induced with 0.2 mM IPTG for 2 h, followed by preparation of crude lysates. Expression of polypeptides of the required molecular weight was detected by SDS-PAGE, followed by immunoblotting using PMT polyclonal antisera.
Expression and purification of rPMT and C1165S.
The untagged recombinant PMT (rPMT) and its mutant were purified as previously described (53). Glycerol was added to 50% (vol/vol), and preparations were stored at −20°C for up to 1 year.
PAGE.
Proteins were separated in denatured form on 4% stacking and 8% resolving gels (25). Proteins were visualized by a silver-staining technique as described previously (14).
Cell culture.
Swiss 3T3 cells were cultured in Dulbecco modified Eagle medium (DMEM; Sigma) supplemented with 10% fetal calf serum and containing penicillin G (100 U/ml) and streptomycin (100 μg/ml) at 37°C in 10% CO2. Cells were passaged before reaching confluency.
DNA synthesis assay.
DNA synthesis was measured by determining the incorporation of [3H]thymidine into quiescent Swiss 3T3 cells as described previously (6). Briefly, cells were plated at 2.5 × 104/ml in 24-well plates and incubated for 7 to 8 days until quiescent. Cells were then washed twice in PBS and incubated at 37°C in a 10% CO2 atmosphere in 1 ml of DMEM-Waymouth medium, 1:1 (vol/vol), containing 0.037 MBq of [3H]thymidine per ml and various concentrations of PMT in triplicate. After 40 h, DNA synthesis was assessed by measuring the level of [3H]thymidine incorporated into the acid-precipitable material.
Competition assay for cell binding.
The cell binding of PMT fragments was assessed by determining their ability to compete with PMT in DNA synthesis assays. Swiss 3T3 cells plated at 2.5 × 104 per well in 24-well plates were incubated for 7 days. Cells were washed twice with serum-free DMEM, and then 1 ml of serum-free DMEM was added to each well. Blocking peptides were added at a range of concentrations, and cells were incubated at 37°C for 30 min. PMT was added to 5 ng/ml, and cells incubated for a further 3 h. Wells were washed twice with DMEM, and 1 ml of serum-free 1:1 (vol/vol) DMEM-Waymouth medium containing [3H]thymidine was added. The DNA synthesis assay was continued as described above.
Schild plot analysis: estimation of toxin binding affinity.
The ability of the inactive mutant, C1165S, to compete with PMT was used to estimate its apparent binding affinity for cell surface receptors with the Schild plot method (41). The assay was performed essentially as for the competition assay described above, except that a range of concentrations of wild-type toxin and C1165S mutant were added together to washed, quiescent cells in the presence of [3H]thymidine, and incubation was continued for 40 h.
Microinjection.
The cytoplasm of quiescent Swiss 3T3 cells plated on coverslips was injected by using an Olympus IMT-2 inverted microscope fitted with Narishige MO-202 micromanipulators. Proteins for injection were prepared in KCl microinjection buffer (10 mM Tris-HCl [pH 7.4], 140 mM KCl, 8 mM NaCl, 1 mM MgCl2) at 1 to 2 mg/ml. Nonspecific rabbit immunoglobulin G (IgG; 1 mg/ml) was coinjected to allow detection of injected cells. Each dish of cells was subjected to microinjection for a maximum of 15 min, and then the medium was changed to serum-free 1:1 (vol/vol) DMEM-Waymouth medium containing 1:100 bromodeoxyuridine (BrdU) labeling reagent (5-bromo-2′-deoxyuridine labeling and detection kit 1 [Boehringer]). Injected cells were incubated at 37°C for 40 h. Cells were then washed twice in PBS, fixed for 7 min in 3.7% (wt/vol) paraformaldehyde in PBS, and then placed in cold 1:1 (vol/vol) acetone-methanol for 4 min at 4°C. The detection of BrdU was achieved by incubation in PBS containing 1:10 anti-BrdU monoclonal antibody (Boehringer) according to the manufacturer's instructions for 1 h at 37°C, followed by 1:10 anti-mouse-FITC. Injected cells were detected by using anti-rabbit-IgG-Cy3 (or anti-rabbit-IgG-FITC for morphology experiments) (Sigma), added with the anti-mouse antibody at 1:100. After being washed with PBS, coverslips were mounted in Vectashield anti-fading reagent (Vector Laboratories) and then examined with an Olympus BH2 fluorescence microscope. For antibody injection experiments, injected cells were treated in the same way except that they were subsequently placed in medium containing 1 or 5 ng of PMT/ml.
Polyclonal antisera.
Antisera against PMT fragments were raised in sheep. Purified proteins (150 to 200 μg) mixed with Freund incomplete adjuvant in 1-ml volumes were administered at 28-day intervals by subcutaneous injection. The proteins used were GST fusion proteins from which the GST tag had been removed by thrombin cleavage and 1-506/His. Test bleeds of ca. 100 ml were removed after the second, third, and fourth injections and stored at −70°C. A rabbit polyclonal antibody against the formalinized whole toxin was also prepared. The antisera were purified on 1-ml protein G columns equilibrated with 20 mM sodium phosphate (pH 7.0; Amersham Pharmacia Biotech) according to the manufacturer's instructions. Antibody was eluted in 0.1 M glycine (pH 2.7) and neutralized with a few drops of 1 M Tris (pH 9.0). IgG-containing fractions were pooled and dialyzed against PBS at 4°C overnight. The protein concentration was adjusted to 2 mg/ml, and aliquots were stored at −20°C.
Immunoblot analysis.
Proteins separated by SDS-PAGE were transferred to Hybond ECL membranes for 1 h at 100 V. Membranes were blocked in TBS–0.5% milk powder for 30 min and then placed in Tween (0.2% [vol/vol])–Tris-buffered saline (20 mM Tris, 500 mM NaCl [pH 7.5]) (TTBS) containing primary antibody at the required dilution. After three washes in TTBS, membranes were incubated in alkaline phosphatase-conjugated secondary antibody in TTBS, followed by three more washes. Blots were developed by using an AP conjugate substrate kit (Bio-Rad).
Immunoprecipitation of PMT.
Purified PMT (1 μg) was incubated with 20 μl of a 2-mg/ml concentration of polyclonal antisera for 20 min on ice. Immune complexes were precipitated by the addition of 15 μl of protein G-Sepharose beads (30 min at 4°C). The supernatant was retained for analysis, and the beads were eluted with Laemmli buffer containing 8 M urea. Samples were analyzed for PMT by immunoblot with anti-PMT polyclonal antibody.
Neutralization of PMT by antisera.
Swiss 3T3 cells were plated at 2.5 × 104 cells per well in 24-well plates and incubated for 7 days until confluent and quiescent. PMT (1 ng/ml) was preincubated with a range of concentrations of antisera in 1 ml of serum-free 1:1 (vol/vol) DMEM-Waymouth medium containing [3H]thymidine as described above (1 h at room temperature), and this was placed onto the cells after they were washed twice in PBS. After 40 h of incubation, DNA synthesis was assayed as described above.
Proteolysis of PMT.
Purified PMT (ca. 300 μg) was incubated with 0.5 μg of endoproteinase Asp-N (Sigma) for 2 h at 37°C. Fragments were separated by SDS-PAGE on a 6% gel and transferred to a Sequi-Blot polyvinylidene difluoride membrane (Bio-Rad). The membrane was stained in Coomassie blue for 1 min and then destained in several changes of 50% (vol/vol) methanol for a few minutes, rinsed in water, and air dried. Protein bands of interest were excised and subjected to N-terminal amino acid sequencing by using an Applied Biosystems sequencer (model 473A).
Sequence analysis.
The amino acid sequence of PMT (27) corresponds to that of the sequence database entry with accession number S12998 (TOXA_PASMU). This sequence was analyzed, along with that of a related sequence, CNF1 (9). MALIGN (20) was used to align the two sequences. The secondary structure prediction method of profile-fed neural network systems (PHD; Heidelberg) (37, 38) was used. The presence of domains was predicted by using both the alignment with CNF and the PHD results. A search for the presence of local similarity between regions of PMT and a database of protein sequences available in the Protein Data Bank was performed by using the sequence search tool at the GenQuest(Q) server at the John Hopkins University Bioinformatics Web Server. Sequences that aligned were analyzed by the Smith-Waterman method. Further predictions of structure were made by using the programs THREADER and THREADER 2 (21; http://globin.bio.warwick.ac.uk/∼threader).
RESULTS
Expression of PMT fragments and production of polyclonal antisera.
To identify the regions of PMT responsible for different biological functions, several PMT fragments were cloned with affinity tags to aid their subsequent purification (Fig. 1). The C-terminal fusion proteins, GST/681-C and GST/849-C were found in the insoluble fraction when induction was carried out at 37°C (data not shown), but growth at 30°C led to expression of soluble proteins, so this temperature was routinely used for expression of all fusion proteins. The four proteins were expressed well and were readily purified by standard methods. Figure 1B shows a typical gel of purified fusion proteins. The four fusion proteins were inactive in standard assays for PMT activity, that is, the ability to induce actin stress fibers or induce thymidine incorporation when placed on quiescent Swiss 3T3 cells (not shown).
FIG. 1.
(A) Composition of the recombinant polypeptides used in this study relative to full-length PMT (residues 1 to 1285). Open bar, regions derived from the PMT gene; solid bar, location of the GST tag; gray bar, location of the His tag. (B) Purification of recombinant proteins. Purified fusion proteins were separated by SDS–8% PAGE and detected by silver staining. Lanes: 1, GST/1-506; 2, GST/849-C; 3, GST/681-C; 4, 1-506/His. (C) Immunoblot showing the binding of anti-PMT and anti-peptide polyclonal antibodies to recombinant peptides. The antibody is indicated below each blot. For each blot, the lanes contained the following proteins: lane 1, PMT; lane 2, GST/1-506; lane 3, GST/681-C.
Polyclonal antisera were raised against formalinized PMT and against fragments 1-506, 681-C, and 849-C (cleaved from the GST fusion proteins with thrombin; these antisera are subsequently referred to as anti-1-506, anti-681-C, and anti-849-C) and 1-506/His as described in Materials and Methods. The antibodies recognized the appropriate PMT fragments by Western blot analysis (Fig. 1C).
Localization of the catalytic domain.
To locate the catalytic domain of PMT, purified GST fusion proteins were microinjected into the cytoplasm of quiescent Swiss 3T3 cells. Biological activity was assessed in two ways: the ability to induce cell morphology typical of PMT treated cells and the ability to induce DNA synthesis, which was detected by BrdU incorporation.
Microinjection was carried out on quiescent Swiss 3T3 cells that were subsequently placed into serum-free medium. Under these conditions uninjected cells become contracted as shown in Fig. 2A. In contrast, when PMT was added to the medium, the cells were stimulated and showed the normal, elongated appearance of growing fibroblasts (Fig. 2B). Microinjected cells, after transfer to serum-free medium, were therefore examined by staining for the coinjected rabbit IgG to determine whether any of the injected PMT fragments led to a toxin-treated morphology. Microinjection of buffer or the N-terminal fusion protein GST/1-506 led to untreated, contracted cell morphology in most cells (Fig. 2C and D), whereas injection with GST/681-C led to the normal, elongated morphology of toxin-treated cells (Fig. 2E). The smaller C-terminal polypeptide GST/849-C was inactive and led to a contracted morphology (not shown). These effects were reproducibly observed.
FIG. 2.
Effects of PMT and of microinjected PMT fragments on the morphology of quiescent Swiss 3T3 cells. (A and B) Representative phase contrast micrographs of Swiss 3T3 cells incubated in serum-free DMEM-Waymouth (1:1 [vol/vol]) for 24 h without (A) or with (B) 5 ng of PMT/ml. (C to E) Representative fluorescence micrographs of cells microinjected with buffer (C), GST/1-506 (D), and GST/681-C (E) as described in Materials and Methods and stained for rabbit IgG to detect injected cells.
Since the morphologic effects were difficult to quantitate, the ability of the fragments to stimulate DNA synthesis when introduced into the cytoplasm was assessed. A double immunofluorescence method was set up to allow the simultaneous detection of injected cells (by using anti-rabbit IgG antibody) and BrdU-positive nuclei. This involved fixing the rabbit IgG in the cytoplasm by using limited formaldehyde exposure, followed by permeabilization with cold acetone-methanol (1:1) to allow the anti-BrdU antibody to enter the nuclei. The results showed that injection of GST/681-C led to induction of DNA synthesis in some cells (Fig. 3E to H), whereas most cells injected with buffer or GST/1-506 were not stimulated. GST/849-C was also inactive in this assay (data not shown). The graph in Fig. 4A shows the percentage of injected cells that were stimulated to undergo DNA synthesis. These values represent the totals of three separate experiments. Overall, 44% of cells injected with GST/681-C were BrdU positive. This percentage varied from ca. 30 to 50% from experiment to experiment, which probably reflected the activity and concentration of the fragments and the condition of the cells. The percentage of BrdU-positive cells with GST/681-C was significantly higher (P < 0.001) than that obtained when cells were microinjected with the N-terminal protein GST/1-506, with GST/849-C or with buffer alone.
FIG. 3.
Effects of PMT fragments microinjected into quiescent Swiss 3T3 cells on induction of DNA synthesis. Representative fluorescence micrographs are shown. (A, C, E, and G) Injected cells, detected by use of anti-rabbit IgG-Cy3. (B, D, F, and H) The same fields of cells stained for BrdU. The cells were injected with KCl microinjection buffer (A and B), GST/1-506 (C and D), or GST/681-C (E to H). Panels G and H are at a twofold-higher magnification than panels A to F. The arrows indicate microinjected cells that were BrdU positive.
FIG. 4.
(A) Graph showing the percentage of cells microinjected with GST fusion proteins that were BrdU positive. The results show the totals from three separate experiments. At least 40 cells were injected with each protein. Results for the fragments were compared to results for buffer alone by using the chi-square test (✽✽, P < 0.001). (B) Inhibition of DNA synthesis induced by PMT in Swiss 3T3 cells microinjected with different antisera. Cells were incubated in the presence of 5 or 1 ng of PMT/ml as shown. The x axis shows the four antisera used. Results for the PMT antibodies were compared to those with preimmune serum by using the chi-square test (✽, P < 0.01; ✽✽, P < 0.001).
To locate the upstream limit of the catalytic domain progressive deletions of GST/681-C were made from residue 681, and four mutants with small deletions were identified in which the PMT fragment was in frame with the GST tag. Sequence analysis showed that these consisted of residues 716, 720, 782, and 797 to the C terminus. These fusion proteins were purified, but only 716-C and 720-C were highly expressed. These were injected into quiesced Swiss 3T3 cells, and both led to normal, toxin-affected elongated morphology (not shown). Thus, the catalytic domain is located within the 720-C-terminal region.
Antibodies to the PMT fragments were microinjected into the cytoplasm of quiescent Swiss 3T3 cells to determine whether they could inhibit the activity of toxin subsequently added to the media. The antibodies to the two C-terminal fragments (anti-681-C and anti-849-C) inhibited PMT induction of BrdU incorporation significantly more than preimmune sera or the N-terminal antibody (anti-1-506) (Fig. 4B). This inhibition was most marked in the presence of the lower concentration of PMT (1 ng of toxin/ml compared to 5 ng/ml). These results confirmed that the C terminus of PMT is trafficked to the cytoplasm and is essential for mitogenic activity.
Mutant C1165S binds specifically to Swiss 3T3 cells.
Previous work from this laboratory showed that mutation of cysteine 1165 to serine led to a complete loss of activity without grossly affecting the structure of the toxin molecule (53). The essential role of this residue was verified by reversal of the mutation to cysteine by site-directed mutagenesis. This led to restoration of activity to wild-type levels in the thymidine incorporation assay (data not shown).
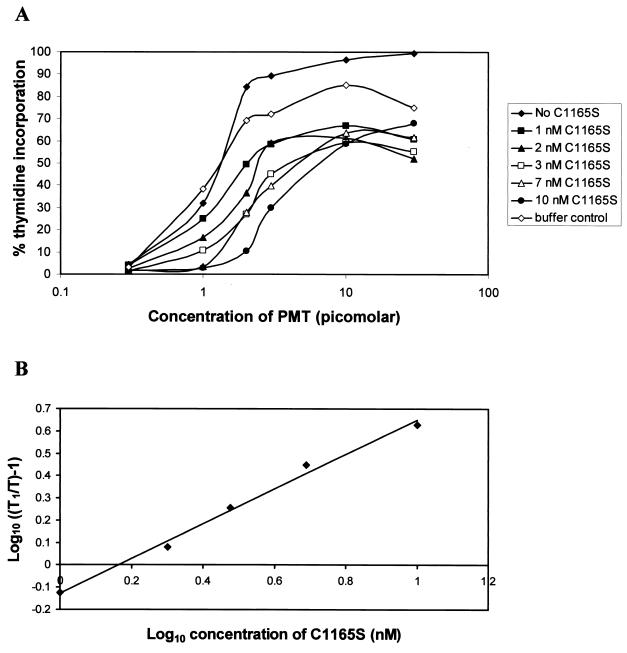
Competition experiments between the inactive C1165S mutant toxin and wild-type toxin showed that C1165S competes with native PMT for interaction with the target cell, since it inhibited PMT-induced DNA synthesis (see Fig. 5A). The addition of increasing concentrations of mutant toxin to cells decreased the induction of DNA synthesis by PMT in a dose-dependent manner, leading to a progressive shift of the dose-response curves to the right (Fig. 5A). Interestingly, the curves in the presence of C1165S did not reach the maximal stimulation level achieved with PMT alone, which suggests that the mutant has an irreversible effect, possibly blocking toxin at a later stage of the intoxication process as well as inhibiting cell binding. The curve for the buffer control shows the effect of adding 1 μl of toxin storage buffer (0.2 M NaCl; 25 mM Tris, pH 6.5; 50% [vol/vol] glycerol) instead of mutant toxin. This volume is equivalent to that added with the highest concentration (10 nM) of C1165S. The buffer had only a slight effect on the activity of PMT. These results show that this mutation does not prevent cell binding and is therefore likely to affect catalytic activity.
FIG. 5.
Schild plot analysis for the determination of the affinity of PMT for its cell surface receptor. (A) Competition curves showing the inhibition of PMT activity by the addition of several fixed concentrations of C1165S. The results are expressed as the percentage of tritiated thymidine incorporation induced by 10% fetal calf serum. The buffer control curve was obtained by using the same volume of toxin storage buffer (0.2 M NaCl; 25 mM Tris, pH 6.5; 50% [vol/vol] glycerol) as was present for the highest concentration of C1165S (10 nM). (B) Data were plotted according to the method of Schild (41). T, concentration of PMT to obtain 45% of the thymidine incorporation induced by 10% fetal calf serum; T1, concentration of PMT after addition of a given amount of C1165S which gave the same activity. The dissociation constant, Kd, for the toxin was calculated from the point at which the line intersects the x axis to be ca. 1.5 nM.
Analysis of the competition results by using the Schild plot method (41) (Fig. 5B), as described for other toxins such as diphtheria toxin (DT) and CNF1 (5, 19), showed that the apparent affinity between PMT and its cell receptor (Kd) is ca. 1.5 nM.
Localization of the receptor-binding domain.
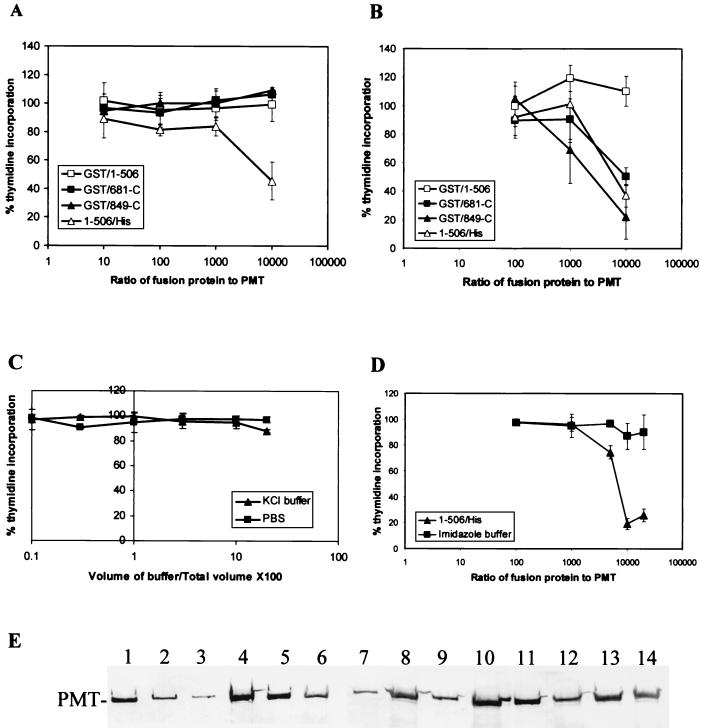
To localize the receptor-binding domain of PMT to the N or C terminus of the molecule, the ability of the recombinant polypeptides to bind to cells was determined by using the competition assay (Fig. 6A to D). The C-terminal fragments GST/681-C and GST/849-C showed variable results in this assay. This was found to be dependent on the storage buffer. When dialyzed into KCl microinjection buffer (see Materials and Methods), the C-terminal fragments were unable to block PMT activity even at concentrations 10,000 times that of PMT (Fig. 6A). However, in PBS, both proteins inhibited toxin activity (Fig. 6B). This inhibition is probably artifactual since we have observed that these C-terminal fragments form insoluble aggregates under certain conditions, for example, during expression at 37°C. Furthermore, large aggregates of GST/681-C have been observed bound to cells by indirect immunofluoresence by using anti-GST antibody for detection (not shown). Buffer controls showed that neither KCl nor PBS had a significant inhibitory effect on PMT activity in this assay, even when added to 20% of the total volume of medium in the well (the volume of peptides never exceeded 10% of the total volume) (Fig. 6C).
FIG. 6.
(A to D) Inhibition of the mitogenic effect of PMT by recombinant proteins. The results are expressed as the percentage of tritiated thymidine incorporation induced by 10% fetal calf serum. Error bars represent the standard deviations obtained from at least three determinations at each point. The x axis values (A, B, and D) represent the ratio of protein concentration in the blocking peptide sample relative to the toxin concentration (i.e., 5 ng/ml). (A) Competition results for PMT fragments prepared in KCl microinjection buffer. (B) Competition results for blocking peptides prepared in PBS. (C) Effect of KCl buffer and PBS on the activity of 5 ng of PMT/ml. The x axis represents the volume of buffer added relative to the total volume of medium expressed as a percentage. (D) Competition results for 1-506/His in 100 mM imidazole buffer. The buffer controls were obtained by using equivalent volumes of imidazole buffer at each point instead of peptide. (E) Immunoprecipitation of PMT by different polyclonal antibodies. The antibodies used for immunoprecipitation were as follows: lanes 1 to 2, PBS buffer control; lanes 3 to 4, rabbit anti-PMT; lanes 5 to 6, preimmune sheep sera; lanes 7 to 8, anti-1-506; lanes 9 to 10, anti-1-506/His; lanes 11 to 12, anti-681-C; lanes 13 to 14, anti-849-C. Lanes 1, 3, 5, 7, 9, 11, and 13 contain supernatant; lanes 2, 4, 6, 8, 10, 12, and 14 contain protein eluted from beads.
The N-terminal fragment GST/1-506 did not bind to cells and was unable to compete with toxin. Similar results were obtained with protein stored in PBS or in KCl microinjection buffer (Fig. 6A and B). However, the same PMT fragment with a fusion tag on the C terminus (1-506/His) instead of the N terminus blocked PMT in a dose-dependent manner in both buffers (Fig. 6A and B). A significant competition was observed with an excess of 10,000-fold compared with active PMT. To confirm that this effect was not due to residual imidazole remaining from the purification procedure, the ability of 1-506/His to block PMT was assayed by using the polypeptide in 100 mM imidazole buffer. It effectively competed with whole toxin, whereas equivalent volumes of the buffer alone had no effect on DNA synthesis (Fig. 6D). These results localized the receptor-binding domain to residues 1-506 at the N terminus of the toxin. The GST tag at the N terminus of GST/1-506 presumably either prevented folding of the receptor-binding site or masked the binding site so that it was not surface located.
The relative abilities of the anti-PMT polyclonal antibodies to bind to and immunoprecipitate whole PMT were compared (Fig. 6E). PMT polyclonal antibody immunoprecipitated toxin most efficiently, followed by anti-1-506 and anti-1-506/His antisera. Antibodies to the C-terminal fragments bound poorly to whole toxin. These results suggest a surface location for the N terminus of PMT.
The ability of these antibodies to neutralize PMT in the thymidine incorporation assay was tested by coincubation for 1 h before placement on the cells (not shown). PMT polyclonal antibody neutralized PMT activity, at an antibody concentration of ≥0.3 μg of antibody/ng of PMT, but none of the four anti-fragment antibodies were neutralizing at concentrations up to 10 μg of antibody/ng of PMT (results not shown), suggesting that they do not contain antibodies that react with the binding site.
PMT has previously been shown to be highly resistant to most proteases at neutral pH (46). One protease, Asp-N, was found to cleave PMT at pH 7, producing three fragments of ca. 90, 87, and 55 kDa (46). Amino-terminal sequencing of these fragments showed that the toxin was cleaved between residues 473 and 474, and between 509 and 510. Thus, the region toward the end of the identified receptor-binding domain (which may also contain the translocation domain [see below]) is likely to be in an exposed loop typical of interdomain regions.
Secondary structure analysis based on the primary amino acid sequence.
The sequence identity between PMT and CNF1 was nearly 22% after sequence alignment with MALIGN (20). The presence of conserved and similar residues suggest a higher conservation in a region corresponding to residues 250 to 530 of the PMT sequence and is predicted to be significant by using the algorithm of Doolittle et al. (7). The alignment with CNF1 and the PHD results are consolidated in Fig. 7 to show the prominent features. These include four helix-rich regions at positions 110 to 220, 379 to 498, 590 to 720, and 820 to 875. Regions 225 to 285 and 515 to 590 are potentially beta rich, while regions 50 to 100, 285 to 350, and 889 to 1280 are alternating α/β folds. The assignment of these regions is supported by weak homology to proteins of known structure (Fig. 7). For example, the predicted helix-rich region between residues 593 and 720 is supported by marginal homology to the highly helical protein tropinin C (40).
FIG. 7.
Structural predictions of PMT. (A) The location of predicted α, β, and mixed α/β regions is shown based on the PHD analysis of the PMT amino acid sequence aligned with CNF. The predicted hydrophobic region was described by Lax et al. (27). The location of the eight cysteine (C) residues is shown. The narrow solid bars indicate the location and extent of regions of weak homology to proteins of known structure (see the text): the broader bar indicates homology to CNF. The arrow indicates the start (amino acid 917) of the analysis by the THREADER 2 program.
The predicted helix at residues 379 to 498 is of particular interest since this region corresponds closely to a predicted hydrophobic domain from residues 370 to 470 (27), is significantly similar to CNF, and is predicted to contain a buried helix between residues 400 to 420. Weak homology to predominantly alpha regions of the protein C inhibitor (23) and asparaginase (50) supports the helical assignment.
The C-terminal region from positions 889 to 1220 was predicted to be an alternating α/β fold (Fig. 7). This designation was supported by marginal homology, in particular to an alternating α/β region of flavodoxin (45). There was also marginal homology with part of isocitrate dehydrogenase (18), which has an alternating α/β structure, and with short regions of sheet structure in FKBP12 protein (1) and in plastocyanin (4). The program THREADER II was run from residue 917 to the C terminus (residue 1285) to see whether the sequence would fit a known protein structure. The top 10 scores are shown in Table 1. The best score is within the highest category of significance (>3.50) and corresponds to a d-alanine–d-alanine ligase (10), which has essentially an alternating α/β structure. These findings suggest that the C terminus of PMT folds into a structurally discrete domain with a mixed α/β structure.
TABLE 1.
Analysis of the C terminus of PMT by using THREADER 2a
| Score | Protein identifier | % Alignment
|
Protein | |
|---|---|---|---|---|
| Structural | Sequence | |||
| 3.51 | 2dln | 88.9 | 73.7 | d-Alanine–d-alanine ligase |
| 3.36 | 2ran | 95.9 | 82.1 | Annexin V |
| 3.32 | 1ann | 90.8 | 77.5 | Annexin IV |
| 3.31 | 1ala | 90.2 | 77.2 | Annexin V |
| 3.29 | 1ypi | 96.8 | 64.8 | Triosephosphate isomerase |
| 3.29 | 1agx | 93.7 | 84.0 | Glutaminase-asparaginase |
| 3.29 | 1ald | 84.8 | 83.5 | Aldolase |
| 3.15 | 1ral | 91.9 | 76.7 | 3α-hydroxysteroid hydrogenase |
| 3.13 | 1add | 86.8 | 82.1 | Adenosine deaminase |
| 3.11 | 1fba | 85.6 | 83.5 | Flavodoxin |
The PMT sequence from amino acid 917 to the C terminus was analyzed by using the THREADER 2 program. The top 10 scores are displayed.
DISCUSSION
The work presented here demonstrates that the C terminus of PMT (residues 681 to 1285) possesses catalytic activity. Microinjection of this fragment led to the typical cell morphology associated with toxin treatment. More importantly, this peptide stimulated quiescent cells to undergo DNA synthesis, showing that it is mitogenic, the most striking and distinctive activity of this toxin.
The identification of the catalytic domain was supported by the inhibitory effect of antibodies to the C terminus after microinjection into the cytoplasm of Swiss 3T3 cells. We also showed that the inactive mutant C1165S (53) competes with native PMT for interaction with cells, and this provided further indirect evidence that this C-terminal point mutation affected enzymatic activity. The C terminus was predicted to be folded into an alternating α/β structure that is commonly found in the catalytic domain of proteins. Threading calculations to fit this region to known structures lend further support to this idea, since the highest scoring protein has an α/β structure. Interestingly, the PMT fragment GST/849-C was not catalytically active, although it contained the predicted α/β domain.
Our assignment of catalytic activity to the C terminus of PMT is supported by a very recent publication (2) that showed that a slightly larger C-terminal fragment (residues 581 to 1285) could induce cytoskeletal changes typical of PMT and induce inositol phosphate production after electroporation into embryonic bovine lung cells. In contrast, Wilson et al. (56) showed that microinjection of PMT into voltage clamped Xenopus oocytes activated the inositol triphosphate signaling pathway, leading to Ca2+-induced Cl− currents, and that this response was also observed after microinjection of an N-terminal fragment (residues 1 to 568 [55]). However, the two-peak response obtained with whole toxin was not clearly observed with this fragment, and the response may have been an artifact due to the hydrophobic domain present in this fragment inserting into the membrane. Indeed, when deletions were made from residue 568 into the predicted hydrophobic helical region, the response was gradually lost, with residues 1 to 400 being completely inactive.
The C-terminal peptides are predicted to contain a hydrophobic region (27) and were found in the insoluble fraction when their expression was induced at 37°C but were soluble when expressed at 30°C. Storage in PBS led to these peptides apparently competing with toxin in binding competition assays, suggesting that they either bound to cells or aggregated with toxin molecules. This did not occur when the peptides were prepared in potassium chloride microinjection buffer. Interestingly, this buffer resembles the physiological environment of the cytoplasm, where the catalytic domain would function.
This is the first report showing that the N terminus of PMT (residues 1 to 506) binds to cells, effectively competing with full-length toxin. The inhibitory effect of placing a fusion tag on the N terminus of this fragment suggests that the binding site did not fold properly in this construct and may therefore be very close to the N terminus. This would be analogous to CNF, in which the N-terminal 190 amino acids are sufficient for binding (8). Antibodies to the N terminus of PMT bound strongly to full-length toxin, showing that this domain is surface located, as would be expected for a receptor-binding domain. The receptor for PMT has not yet been identified, although there is some evidence that it binds to a ganglioside-type receptor (35). We showed, by Schild plot analysis, that PMT has an apparent binding affinity for surface receptors on Swiss 3T3 cells of ca. 1.5 × 10−9 M. This is comparable to the binding affinity of other toxins such as diphtheria toxin (DT) (19). Interestingly, the apparent binding affinity of CNF to Hep2 cells was higher by ca. 2 orders of magnitude (5) in spite of the homology between PMT and CNF, indicating diverse toxin-receptor interactions. The localization of the binding domain of PMT will facilitate the identification of the receptor and allow different types of cells to be analyzed for their ability to bind toxin.
The region of highest homology between PMT and the related toxin CNF corresponds to PMT residues 250 to 530 (which corresponds to CNF residues 200 to 465). This region in CNF is not essential for cell binding or catalytic activity and is assumed to contain the membrane translocation domain (28). This region of PMT, which was predicted to be hydrophobic and helical, is likely to have a similar role.
Limited digestion of toxins with proteases has frequently been used to separate functional domains; for example, DT is cleaved into its constituent A and B fragments by trypsin (12). PMT is highly resistant to proteolysis at neutral pH (46), but Asp-N did cleave the toxin in two positions (after residues 473 and 509) just downstream of the proposed translocation domain. Similarly, the Bordetella DNT, which shares homology with CNF, as well as a similar domain organization, was reported to be cleaved into two fragments by trypsin, an N-terminal fragment of 60 kDa and a C-terminal fragment of 90 kDa (22). It remains unknown whether PMT, DNT, or CNF is cleaved during the intoxication process. However, it is notable that PMT has a motif for cleavage by the mammalian protease furin (54) at residues 515 to 520 (KIRLVR), and this is a possible site for in vivo cleavage of PMT. Furin is involved in the cleavage and activation of several bacterial toxins, including protective antigen from Bacillus anthracis, Pseudomonas exotoxin A, and DT (13).
The development of assays for intracellular activity and binding and the localization of the functional domains of PMT will allow us to investigate its molecular mode of action in more detail.
ACKNOWLEDGMENTS
We thank Mark Munson (King's College London) for DNA sequencing and Arthur Moir (University of Sheffield) for amino-terminal sequencing. We also thank Richard Pickersgill (Queen Mary and Westfield College, London) for performing the THREADER analysis.
This work was supported by Wellcome Trust project grant no. 049649.
REFERENCES
- 1.Becker J W, Rotonda J, McKeever B M, Chan H K, Marcy A I, Wiederrecht G, Hermes J D, Springer J P. FK-506-binding protein: three-dimensional structure of the complex with the antagonist L-685,818. J Biol Chem. 1993;268:11335–11339. [PubMed] [Google Scholar]
- 2.Busch C, Orth J, Djouder N, Aktories K. Biological activity of a C-terminal fragment of Pasteurella multocida toxin. Infect Immun. 2001;69:3628–3634. doi: 10.1128/IAI.69.6.3628-3634.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buys W E C M, Smith H E, Kamps A M I E, Kamp E M, Smits M A. Sequence of the dermonecrotic toxin of Pasteurella multocida ssp. multocida. Nucleic Acids Res. 1990;18:2815–2816. doi: 10.1093/nar/18.9.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collyer C A, Guss J M, Sugimura Y, Yoshizaki F, Freeman H C. Crystal structure of plastocyanin from a green alga, Enteromorpha prolifer. J Mol Biol. 1990;211:617–632. doi: 10.1016/0022-2836(90)90269-R. [DOI] [PubMed] [Google Scholar]
- 5.Contamin S, Galmiche A, Doye A, Flatau G, Benmerah A, Boquet P. The p21 Rho-activating toxin cytotoxic necrotizing factor 1 is endocytosed by a clathrin-independent mechanism and enters the cytosol by an acidic-dependent membrane translocation step. Mol Biol Cell. 2000;11:1775–1787. doi: 10.1091/mbc.11.5.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dicker P, Rozengurt E. Phorbol esters and vasopressin stimulate DNA synthesis by a common mechanism. Nature. 1980;287:607–612. doi: 10.1038/287607a0. [DOI] [PubMed] [Google Scholar]
- 7.Doolittle R F, Feng D F, Johnson M S, McClure M A. Relationships of human protein sequences to those of other organisms. Cold Spring Harbor Symp Quant Biol. 1986;51:447–455. doi: 10.1101/sqb.1986.051.01.054. [DOI] [PubMed] [Google Scholar]
- 8.Fabbri A, Gauthier M, Boquet P. The 5′ region of cnf1 harbours a translational regulatory mechanism for CNF1 synthesis and encodes the cell-binding domain of the toxin. Mol Microbiol. 1999;33:108–118. doi: 10.1046/j.1365-2958.1999.01453.x. [DOI] [PubMed] [Google Scholar]
- 9.Falbo V, Pace T, Picci L, Pizzi E, Caprioli A. Isolation and nucleotide sequence of the gene encoding cytotoxic necrotizing factor 1 of Escherichia coli. Infect Immun. 1993;61:4909–4914. doi: 10.1128/iai.61.11.4909-4914.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan C, Moews P C, Walsh C T, Knox J R. Vancomycin resistance: structure of d-alanine:d-alanine ligase at 2.3 Å resolution. Science. 1994;266:439–443. doi: 10.1126/science.7939684. [DOI] [PubMed] [Google Scholar]
- 11.Flatau G, Lemichez E, Gauthier M, Chardin P, Paris S, Fiorentini C, Boquet P. Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature. 1997;387:729–733. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- 12.Gill D M, Donnius L L. Observation on the structure of diphtheria toxin. J Biol Chem. 1971;246:1485–1491. [PubMed] [Google Scholar]
- 13.Gordon V M, Leppla S H. Proteolytic activation of bacterial toxins: role of bacterial and host cell proteases. Infect Immun. 1994;62:333–340. doi: 10.1128/iai.62.2.333-340.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heukeshoven J, Dernick R. Simplified method for silver staining of proteins in polyacrylamide gels and the mechanism of silver staining. Electrophoresis. 1985;6:103–112. [Google Scholar]
- 15.Higgins T E, Murphy A C, Staddon J M, Lax A J, Rozengurt E. Pasteurella multocida toxin is a potent inducer of anchorage-independent cell growth. Proc Natl Acad Sci USA. 1992;89:4240–4244. doi: 10.1073/pnas.89.10.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horiguchi Y, Inoue N, Masuda M, Kashimoto T, Katahira J, Sugimoto N, Matsuda M. Bordetella bronchiseptica dermonecrotizing toxin induces reorganization of actin stress fibers through deamidation of Gln-63 of the GTP-binding protein Rho. Proc Natl Acad Sci USA. 1997;94:11623–11626. doi: 10.1073/pnas.94.21.11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoskins I C, Thomas L H, Lax A J. Nasal infection with Pasteurella multocida causes proliferation of bladder epithelium in gnotobiotic pigs. Vet Rec. 1997;140:22. doi: 10.1136/vr.140.1.22. [DOI] [PubMed] [Google Scholar]
- 18.Hurley J H, Thorsness P E, Ramalingham V, Helmers N H, Koshland D E, Stroud R M. Structure of a bacterial enzyme regulated by phosphorylation, isocitrate dehydrogenase. Proc Natl Acd Sci USA. 1989;86:8635–8639. doi: 10.1073/pnas.86.22.8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ittelson T R, Gill D M. Diphtheria toxin: specific competition for cell receptors. Nature. 1973;242:330–332. doi: 10.1038/242330b0. [DOI] [PubMed] [Google Scholar]
- 20.Johnson M S, Overington J P, Blundell T L. Alignment and searching for common protein folds using a databank of structural templates. J Mol Biol. 1993;231:735–752. doi: 10.1006/jmbi.1993.1323. [DOI] [PubMed] [Google Scholar]
- 21.Jones D T, Taylor W R, Thornton J M. A new approach to protein fold recognition. Nature. 1992;358:86–89. doi: 10.1038/358086a0. [DOI] [PubMed] [Google Scholar]
- 22.Kashimoto T, Katahira J, Cornejo W R, Masuda M, Fukuoh A, Matsuzawa T, Ohnishi T, Horiguchi Y. Identification of functional domains of Bordetella dermonecrotizing toxin. Infect Immun. 1999;67:3727–3732. doi: 10.1128/iai.67.8.3727-3732.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhn L A, Griffin J H, Fisher C L, Greengard J S, Bouma B N, Espana F, Tainer J A. Elucidating the structural chemistry of glycosaminoglycan recognition by protein C inhibitor. Proc Natl Acad Sci USA. 1990;87:8506–8510. doi: 10.1073/pnas.87.21.8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lacerda H M, Lax A J, Rozengurt E. Pasteurella multocida toxin, a potent intracellularly acting mitogen, induces p125FAK and paxillin tyrosine phosphorylation, actin stress fiber formation and focal contact assembly in Swiss 3T3 cells. J Biol Chem. 1996;271:439–445. doi: 10.1074/jbc.271.1.439. [DOI] [PubMed] [Google Scholar]
- 25.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;277:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Lax A J, Chanter N. Cloning of the toxin gene from Pasteurella multocida and its role in atrophic rhinitis. J Gen Microbiol. 1990;136:81–87. doi: 10.1099/00221287-136-1-81. [DOI] [PubMed] [Google Scholar]
- 27.Lax A J, Chanter N, Pullinger G D, Higgins T H, Staddon J M, Rozengurt E. Sequence analysis of the potent mitogenic toxin of Pasteurella multocida. FEBS Lett. 1990;277:59–64. doi: 10.1016/0014-5793(90)80809-w. [DOI] [PubMed] [Google Scholar]
- 28.Lemichez E, Flatau G, Bruzzone M, Boquet P, Gauthier M. Molecular localization of the Escherichia coli cytotoxic necrotizing factor CNF1 cell-binding and catalytic domains. Mol Microbiol. 1997;24:1061–1070. doi: 10.1046/j.1365-2958.1997.4151781.x. [DOI] [PubMed] [Google Scholar]
- 29.Lerm M, Selzer J, Hoffmeyer A, Rapp U R, Aktories K, Schmidt G. Deamidation of Cdc42 and Rac by Escherichia coli cytotoxic necrotizing factor: activation of c-jun N-terminal kinase in HeLa cells. Infect Immun. 1999;67:496–503. doi: 10.1128/iai.67.2.496-503.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuda M, Betancourt L, Matsuzawa T, Kashimoto T, Takao T, Shimonishi Y, Horiguchi Y. Activation of Rho through a cross-link with polyamines catalyzed by Bordetella dermonecrotizing toxin. EMBO J. 2000;19:521–530. doi: 10.1093/emboj/19.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy A C, Rozengurt E. Pasteurella multocida toxin selectively facilitates phosphatidylinositol 4,5-bisphosphate hydrolysis by bombesin, vasopressin, and endothelin. J Biol Chem. 1992;267:25296–25303. [PubMed] [Google Scholar]
- 32.Oswald E, Sugai M, Labigne A, Wu H C, Fiorentini C, Boquet P, O'Brien A D. Cytotoxic necrotizing factor type 2 produced by virulent Escherichia coli modifies the small GTP-binding proteins Rho involved in assembly of actin stress fibers. Proc Natl Acad Sci USA. 1994;91:3814–3818. doi: 10.1073/pnas.91.9.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pedersen K B, Barfod K. The aetiological significance of Bordetella bronchiseptica and Pasteurella multocida in atrophic rhinitis of swine. Nord Vet-Med. 1981;33:513–522. [PubMed] [Google Scholar]
- 34.Petersen S K. The complete nucleotide sequence of the Pasteurella multocida toxin gene and evidence for a transcriptional repressor, TxaR. Mol Microbiol. 1990;4:821–830. doi: 10.1111/j.1365-2958.1990.tb00652.x. [DOI] [PubMed] [Google Scholar]
- 35.Pettit R K, Ackermann M R, Rimler R B. Receptor-mediated binding of Pasteurella multocida dermonecrotic toxin to canine osteosarcoma and monkey kidney (vero) cells. Lab Investig. 1993;69:94–100. [PubMed] [Google Scholar]
- 36.Pullinger G D, Adams T E, Mullan P B, Garrod T I, Lax A J. Cloning, expression, and molecular characterization of the dermonecrotic toxin gene of Bordetella spp. Infect Immun. 1996;64:4163–4171. doi: 10.1128/iai.64.10.4163-4171.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 38.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 39.Rozengurt E, Higgins T E, Chanter N, Lax A J, Staddon J M. Pasteurella multocida toxin: potent mitogen for cultured fibroblasts. Proc Natl Acad Sci USA. 1990;87:123–127. doi: 10.1073/pnas.87.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Satyshur K A, Rao S T, Pyzalska D, Drendel W, Greaser M, Sundaralingham M. Refined structure of chicken skeletal muscle troponin C in the two-calcium state at 2-Å resolution. J Biol Chem. 1988;263:1628–1647. [PubMed] [Google Scholar]
- 41.Schild H O. Drug antagonism and pAx. Pharmacol Rev. 1957;9:242–246. [PubMed] [Google Scholar]
- 42.Schmidt G, Goehring U-M, Schirmer J, Lerm M, Aktories K. Identification of the C-terminal part of Bordetella dermonecrotic toxin as a transglutaminase for Rho GTPases. J Biol Chem. 1999;274:31875–31881. doi: 10.1074/jbc.274.45.31875. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt G, Selzer J, Lerm M, Aktories K. The Rho-deamidating cytotoxic necrotizing factor 1 from Escherichia coli possesses transglutaminase activity. J Biol Chem. 1998;273:13669–13674. doi: 10.1074/jbc.273.22.13669. [DOI] [PubMed] [Google Scholar]
- 44.Seo B, Choy E W, Maudesley S, Miller W E, Wilson B A, Luttrell L M. Pasteurella multocida toxin stimulates mitogen-activated protein kinase via Gq/11-dependent transactivation of the epidermal growth factor receptor. J Biol Chem. 2000;275:2239–2245. doi: 10.1074/jbc.275.3.2239. [DOI] [PubMed] [Google Scholar]
- 45.Smith W W, Burnett R M, Darling G D, Ludwig M L. Structure of the semiquinone form of flavodoxin from Clostridium MP. J Mol Biol. 1977;117:195–225. doi: 10.1016/0022-2836(77)90031-6. [DOI] [PubMed] [Google Scholar]
- 46.Smyth M G, Pickersgill R W, Lax A J. The potent mitogen Pasteurella multocida toxin is highly resistant to proteolysis but becomes susceptible at lysosomal pH. FEBS Lett. 1995;360:62–66. doi: 10.1016/0014-5793(95)00077-m. [DOI] [PubMed] [Google Scholar]
- 47.Smyth M G, Sumner I G, Lax A J. Reduced pH causes structural changes in the potent mitogenic toxin of Pasteurella multocida. FEMS Microbiol Lett. 1999;180:15–20. doi: 10.1111/j.1574-6968.1999.tb08772.x. [DOI] [PubMed] [Google Scholar]
- 48.Staddon J M, Barker C J, Murphy A C, Chanter N, Lax A J, Michell R H, Rozengurt E. Pasteurella multocida toxin, a potent mitogen, increases inositol 1,4,5-triphosphate and mobilises Ca2+ in Swiss 3T3 cells. J Biol Chem. 1991;266:4840–4847. [PubMed] [Google Scholar]
- 49.Staddon J M, Chanter N, Lax A J, Higgins T E, Rozengurt E. Pasteurella multocida toxin, a potent mitogen, stimulates protein kinase C-dependent and independent protein phosphorylation in Swiss 3T3 cells. J Biol Chem. 1990;265:11841–11848. [PubMed] [Google Scholar]
- 50.Swain A L, Jaskólski M, Housset D, Mohana Rao J K, Wlodawer A. Crystal structure of Escherichia colil-asparaginase, an enzyme used in cancer therapy. Proc Natl Acad Sci USA. 1993;90:1474–1478. doi: 10.1073/pnas.90.4.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas W, Pullinger G D, Lax A J, Rozengurt E. Escherichia coli cytotoxic necrotizing factor and Pasteurella multocida toxin induce focal adhesion kinase autophosphorylation and Src association. Infect Immun. 2001;69:5931–5935. doi: 10.1128/IAI.69.9.5931-5935.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walker K E, Weiss A A. Characterization of the dermonecrotic toxin in members of the genus Bordetella. Infect Immun. 1994;62:3817–3828. doi: 10.1128/iai.62.9.3817-3828.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ward P N, Miles A J, Sumner I G, Thomas L H, Lax A J. Activity of the mitogenic Pasteurella multocida toxin requires an essential C-terminal residue. Infect Immun. 1998;66:5636–5642. doi: 10.1128/iai.66.12.5636-5642.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe T, Murakami K, Nakayama K. Positional and additative effects of basic amino acids on processing of precursor proteins within the constitutive secretory pathway. FEBS Lett. 1993;320:215–218. doi: 10.1016/0014-5793(93)80589-m. [DOI] [PubMed] [Google Scholar]
- 55.Wilson B A, Ponferrada V G, Vallance J E, Ho M. Localization of the intracellular activity domain of Pasteurella multocida toxin to the N terminus. Infect Immun. 1999;67:80–87. doi: 10.1128/iai.67.1.80-87.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson B A, Zhu X, Ho M, Lu L. Pasteurella multocida toxin activates the inositol triphosphate signalling pathway in Xenopus oocytes via Gqα-coupled phospholipase C-β1. J Biol Chem. 1997;272:1268–1275. doi: 10.1074/jbc.272.2.1268. [DOI] [PubMed] [Google Scholar]
- 57.Zywietz A, Gohla A, Schmelz M, Schultz G, Offermans S. Pleiotropic effects of Pasteurella multocida toxin are mediated by Gq-dependent and -independent mechanisms: involvement of Gq but not G11. J Biol Chem. 2001;276:3840–3845. doi: 10.1074/jbc.M007819200. [DOI] [PubMed] [Google Scholar]