ABSTRACT
Recent advances in next-generation sequencing technologies (NGS) coupled with machine learning have demonstrated the potential of microbiome-based analyses in applied areas such as clinical diagnostics and forensic sciences. Particularly in forensics, microbial markers in biological stains left at a crime scene can provide valuable information for the reconstruction of crime scene cases, as they contain information on bodily origin, the time since deposition, and donor(s) of the stain. Importantly, microbiome-based analyses provide a complementary or an alternative approach to current methods when these are limited or not feasible. Despite the promising results from recent research, microbiome-based stain analyses are not yet employed in routine casework. In this review, we highlight the two main gaps that need to be addressed before we can successfully integrate microbiome-based analyses in applied areas with a special focus on forensic casework: one is a comprehensive assessment of the method’s strengths and limitations, and the other is the establishment of a standard operating procedure. For the latter, we provide a roadmap highlighting key decision steps and offering laboratory and bioinformatic workflow recommendations, while also delineating those aspects that require further testing. Our goal is to ultimately facilitate the streamlining of microbiome-based analyses within the existing forensic framework to provide alternate lines of evidence, thereby improving the quality of investigations.
KEYWORDS: bioinformatics, biological stains, forensic science, human microbiome, machine learning, next-generation sequencing
INTRODUCTION
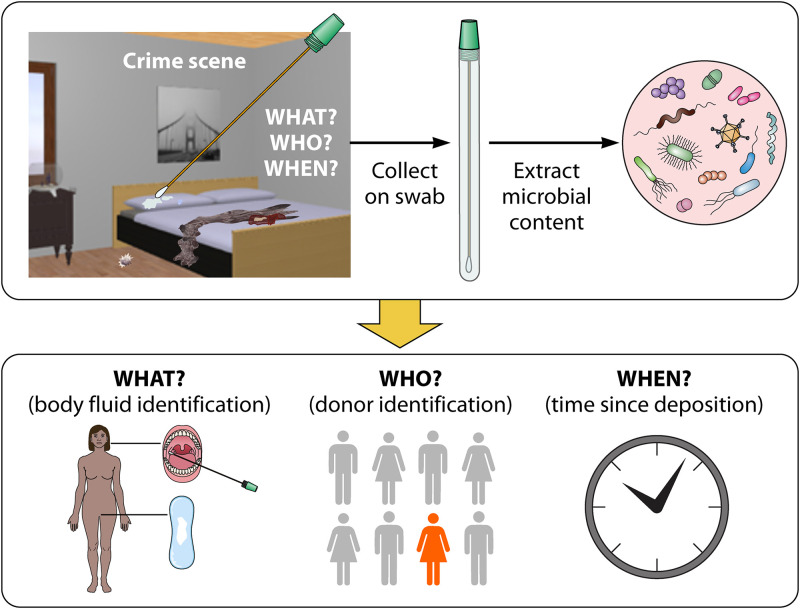
The wealth of microbiome sequencing studies from the last decade has opened new avenues in forensic investigations, most notably by demonstrating that microbial communities are rich sources of information for criminal investigations (1–4). For instance, changes in the microbial composition in different parts of a cadaver are indicative of the time since death, known as the postmortem interval (PMI) (1, 3, 5). Environmental samples collected from an object, such as soil from socks and shoes, can be linked to a geographic location (1, 6). Particularly, one application of microbiome forensics, the analysis of biological stains, has witnessed much progress in recent years and may soon be ready for integration in forensic laboratories. The microbes in such samples, typically traces, may reveal information on the persons involved (who), through the identification of the donor(s); the nature of the activities that took place (what), through the examination of the type of cellular material (e.g. vaginal, oral, skin, saliva); and the time of occurrence (when), through estimation of the time of stain deposition (Fig. 1) (7). Overall, such information can be critical for reconstruction of crime scene events and for corroborating testimonies (8). Some examples of case investigations using microbiome-based analyses as evidence are summarized in Table 1.
FIG 1.
Microbiome-based analyses of human biological stains in crime scenes. Here, we illustrate a hypothetical crime scene involving biological evidence found on bedsheets: questions on who was involved, what, and when the crime occurred are being investigated. The stains were collected on swabs, and the microbial content could be analyzed to determine the bodily origin of the stain (BFI), identify the individual (ID), and estimate time since deposition (TsD) (The figure was created using 3D Home Design software [Planner 5D] and BioRender).
TABLE 1.
Summary of cases where microbiome-based analyses were conducted as part of the forensic investigation
| Nature of case | Microbiome analysed | Case report | Year | Country | References |
|---|---|---|---|---|---|
| Postmortem interval (PMI) | Swabs from external auditory canal, eyes, nares, mouth, umbilicus, and rectum | No | 2015 | The United States of America | (62) |
| Sexual assault | Vaginal microbiome from object used for vaginal penetration | Yes | 2017 | France | (61) |
| Sexual assault | Vaginal microbiome from digits of suspect used for digital penetration | Yes | 2017 | France | (61) |
| Sexual assault | Fecal microbiome from tissue | Yes | 2018 | The Netherlands | (21) |
| House robbery | Fecal, skin, and vaginal microbiome from swab | Yes | 2018 | The Netherlands | (21) |
| Linking suspect to a location | Soil microbiome from suspect’s sock | No | 2020 | The United States of America | (6) |
The utility of microbes for two applications, body fluid and tissue identification (BFI) and individual identification (ID), became apparent as key characteristics of the human microbiota were revealed in sequencing studies. These microbes, and particularly bacteria, have been shown to form diverse and intricate communities that vary in composition across habitats, as well as across individuals (9, 10). These notable characteristics have been uncovered through studies harnessing the power of next-generation sequencing (NGS) technologies, which have facilitated the examination of microbial diversity in healthy tissues by bypassing the need for laboratory cultivation. The two main methods used are marker gene amplicon sequencing, whereby one or more markers are targeted for PCR amplification and NGS, and whole-metagenome shotgun sequencing, whereby sequences are recovered from across the genomes present, including those of the host, through NGS (11). Most human microbiome studies to date have focused on bacteria, typically targeting relatively short stretches of the prokaryotic 16S rRNA gene through amplicon sequencing, with an increase in the use of shotgun sequencing over time and recent attention to other microbes such as fungi and archaea (12–14). What these studies highlight, in addition to tissue and individual specificity, is that the temporal stability of bacterial communities within the human body is dependent on the body site itself and on variables including lifestyle. Outside the human body, community composition also changes over time, and preliminary work has indicated patterns that can be exploited for the inference of time since deposition (TsD) (15).
As a result of these attributes, microbial markers may complement routine forensic tests or provide a replacement when the routine tests do not yield reliable information. Especially in cases where samples are degraded and human cellular components are limited, the abundance of microbes and their general resilience may be an advantage (16, 17). For example, at some body sites, bacterial cells outnumber human cells (18). In the case of human DNA profiling, the current gold standard approach involves fragment length analyses of 17 to 25 highly variable human short tandem repeat (STR) loci and searches for matches among potential donors or in extensive databases comprising human profiles (19). Nonetheless, there are cases where stains contain low human DNA quantity and quality that yield incomplete profiles or increase the probability for spurious matches. Some rarer cases involve monozygotic twins who cannot be distinguished through DNA profiling alone. In such situations, microbial markers could yield further clues. Moreover, microbiome-based analyses could also provide new avenues to access information from a biological stain with potentially higher performance than the traditional body fluid and tissue test. This option is of particular interest because current tests often destroy valuable biological evidence, typically test for only one body fluid, and have some limitations in terms of specificity and/or sensitivity (20).
To date, much progress has been made showcasing the utility of microbes for stain analyses, particularly in proof-of-concept studies. Nonetheless, there are critical gaps that need to be addressed before this novel method can be integrated in routine casework. Some of these gaps mirror those in other applied settings, such as clinical diagnostics. In this review, we examine the current state of forensic microbiome analyses of stains, with a special focus on BFI, which we deem to be close to maturity. We draw attention to two crucial issues that need to be thoroughly addressed: on the one hand, delineating the strengths and limitations of a novel method in the field, and on the other hand, establishing a standardized protocol that can be employed in forensic laboratories. For the latter, we provide general recommendations that can serve as a roadmap in applied fields, while also highlighting specific points which merit further consideration and forensic benchmarking.
ADVANCES IN MICROBIOME-BASED STAIN ANALYSES
What: body fluid and tissue identification.
The association between microbial composition and body site has prompted forensic scientists to develop new tools for BFI. Initial endeavors focused on limited sets of bacterial markers, such as the microarray with 389 bacterial DNA probes developed by Benschop et al. (8). This array was successfully applied in two crime cases in the Netherlands to determine the bodily origin of samples obtained from tissue paper and from penile skin. The microbial composition of these samples was deemed most similar to that of fecal samples (21), and the evidence was presented in court.
More recent forensic investigations, however, have shifted to 16S rRNA gene amplicon data, as the sequencing costs have continued to decrease and more complex statistical analyses have become feasible in forensic laboratories. A number of these studies have explored the reliability of microbiome signatures in settings that are common in crime scenes. In the first forensic study using 16S rRNA amplicon data, Hanssen et al. (22) explored the detection of saliva and skin including “mixture” cases in which saliva was deposited on skin. Across 144 samples from 6 different donors, they were able to correctly classify 94% of these. Dobay et al. (23) focused on the effects of aging, exposing samples from skin, vaginal fluid, menstrual blood, semen, and saliva to indoor conditions for 30 days. Their results showed that most test and exposed samples could be grouped according to bodily origin, thus indicating a preservation of tissue-specific microbial signatures over time. In further explorations of the robustness of microbial signatures, test samples and mock samples comprising skin, saliva, and vaginal fluid (n = 110 [test] and n = 41 [mock]), as well as test and mock blood samples originating from four different sources (menstrual, nasal, venous, and finger prick; n = 180 and n = 45) were evaluated (24, 25). The mock samples were deposited on various substrates such as nylon, fabric, glass, plastic, chewing gum, and paper and exposed at different temperatures and for different periods of time. Interestingly, higher misclassifications (37.8%) were found for aged mock samples and for venous blood samples, which generally carry low bacterial loads (22–25). Hanssen et al. (22) and Diez-Lopez et al. (24, 25) employed machine learning (ML) methods trained on Human Microbiome Project (HMP) samples, paving the way for further leveraging of such algorithms. Overall, despite the limited number of mock samples per condition, these studies highlighted the utility of 16S rRNA amplicon data for BFI.
Who: individual identification.
Although human microbiome composition varies primarily across body sites, which represent distinct ecological niches, at a given body site individuals display variation shaped by host genetics and lifestyle (26). Such interindividual variations can potentially be used to assist the identification of donors, for example, by matching the bacterial composition found on an object to that of an individual. Although microbial markers are not expected to match forensic human STRs in their statistical power, 16S rRNA amplicon data have been used to link bacterial community composition patterns from keyboards, air filters, and dormitory surfaces to individuals (27, 28). Shotgun data have also been used to obtain higher individualizing power in a data set comprising a larger number of individuals (29). However, shotgun sequence data pose challenges to forensic samples, as adequate coverage of the microbial markers of interest is necessary but not economical or practical when the proportion of background human DNA is large. To address this challenge, Schmedes et al. (30) developed an alternative approach that is specific to a given body site: focusing on skin samples, they identified the most variable markers from common taxa, which could then be sequenced through a targeted approach. By mining publicly available metagenomic data from 12 donors sampled at three different time points, the authors were able to select a set of 286 clade-specific markers from 3 bacterial families as well as several phages that could accurately distinguish among 8 donors. Most of the 286 markers of this panel (hidSkinPlex) were specific to Propionibacterium, which was present in all skin samples examined. These are promising insights and open the way for the development of multiplexes for other body sites, or alternatively, a combined multiplex for multiple body sites.
When: time since deposition.
Another potential application of microbiome analyses of human stains is the estimation of TsD. It is analogous to the determination of the PMI of an individual, as they both require the investigation of time-dependent changes. While various methods for PMI estimation may be employed (including rigor mortis, state of decomposition, or entomology analyses), there are no forensically validated methods for stain TsD (15, 31). However, a recent RNA sequencing study by Salzmann et al. (15) of mock samples (saliva, blood, semen, menstrual blood, and vaginal fluid) deposited and aged for up to 1.5 years, both indoors and outdoors, found around 30 specific bacterial orders that were highly informative for TsD estimation. For saliva samples alone, the proof-of-principle study of Diez-Lopez et al. (2021) identified four abundant bacterial species that can be targeted through quantitative PCR to determine the ages of stains. This targeted approach requires a priori knowledge of the body site but involves a simpler workflow than amplicon or metagenomic next-generation sequencing. Further work investigating forensically relevant settings, such as the effect of different substrates and mixed stains with one or more body fluids, is needed to demonstrate the most suitable approaches for estimation of TsD.
BRINGING MICROBIOME ANALYSES TO FORENSIC CASEWORK: WHAT IS MISSING?
In order to make the leap from research to application, two critical issues need to be addressed before microbiome-based analyses can be integrated in forensic laboratories. First, it is imperative that we determine the strengths and weaknesses of this novel method, in other words, in what contexts we may expect it to work well and with what degree of accuracy. The assessment of this reliability and accuracy requires larger sample sizes and more conditions representing forensic settings. For BFI, considerable progress has been made in the testing of conditions such as substrates, temperatures, and ageing. Nonetheless, systematic, and larger-scale analyses across conditions will facilitate the establishment of a solid reference data basis for each body site. In the case of ID and TsD, we are close to the stage where research efforts can start moving from proof-of-concept to testing forensic conditions. For this move to happen, we first need to establish whether shotgun data, 16S rRNA amplicon data, or the targeted amplification of genetic markers from specific bacterial taxa is most appropriate, and whether the approach should be body site specific or generalized across body sites. For ID, it will be particularly useful to test data sets that include larger numbers of subjects from more diverse geographical locations and across more time intervals. This latter aspect will enable us to determine the extent to which microbial signatures change over time and whether there is a time limit after which attempting to match the microbial composition in a stain and on an individual is no longer useful. Overall, comprehensive sample designs will benefit from a collaborative research network involving multiple laboratories as well as the sharing of resources, such as a microbiome database.
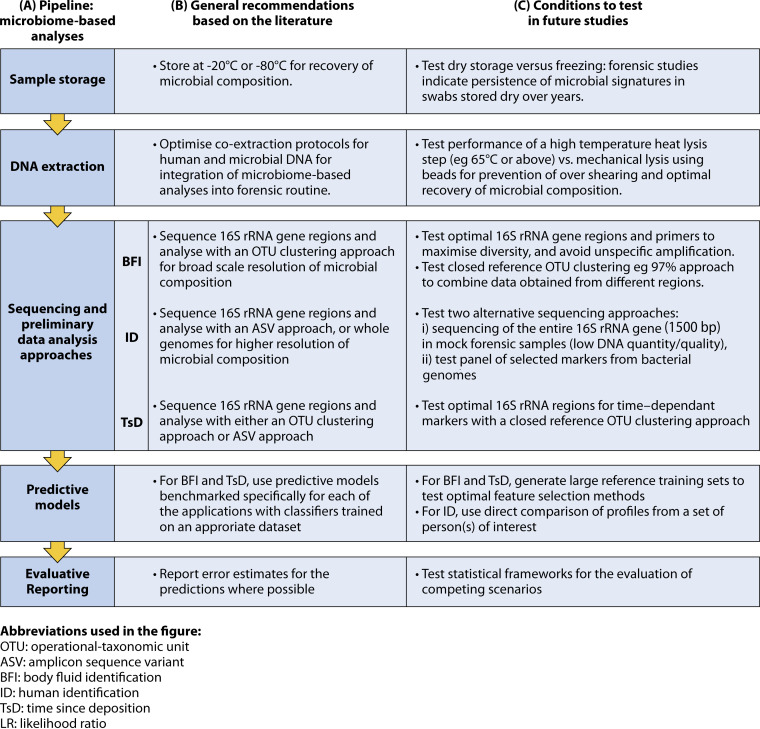
Second, as with any novel method in forensics or the clinic, the workflow needs to be established as a standardized operating procedure (SOP) that is validated and its quality assessed through accreditation. Each of the individual steps from sample collection to the choice of sequencing approach, and to the generation of prediction probabilities must be carefully selected. As seen in the plethora of human microbiome sequencing studies, the approaches utilized have varied considerably and are also evolving as recommendations are updated. As a result, selection of an optimal pipeline is challenging but necessary for consistency and reproducibility. We propose that the forensic SOP be guided by both benchmarking studies and hands-on tests with mock forensic samples and that it be updated when necessary. Some of the laboratory and bioinformatic choices have been highlighted in previous human microbiome publications, including those focusing on clinical applications. Here, we provide a short summary of some of the most important aspects that need to be established, highlighting the specific forensic challenges. A summary of the workflow, including recommendations for individual steps, as well as aspects that require further investigation, is provided in Fig. 2.
FIG 2.
Roadmap highlighting recommendations to test in order to bring microbiome-based analyses to forensics. (A) Main steps of the microbiome-based analyses of forensic stains for BFI, ID, and TsD. (B) General recommendations for each step, based on microbiome literature. (C) Some of the conditions to be tested at each step, for optimal integration of microbiome analyses in forensics.
LABORATORY WORKFLOW
Sample collection, storage, and DNA extraction.
Current forensic protocols for sample collection, storage, and DNA extraction are generally adapted to human DNA profiling, which is common practice in forensic laboratories. Here, it will be useful to evaluate what modifications or new protocols are required to also include the possibility to conduct microbial DNA sequencing. For this evaluation, forensic scientists may draw on the extensive benchmarking studies conducted to optimize the study of human microbiota at body sites such as the gut, vagina, urine, and skin (32–37). A limitation here is that recommendations often vary according to body site, evolving as further tests are carried out. Therefore, specific studies with forensic mock samples will be necessary to establish general protocols that are applicable to samples from different body sites.
For collection and storage, forensic laboratories may not have much choice in practice, as this step is often conducted by police investigators with their own labor, time, and cost constraints. The investigators generally retrieve objects or parts thereof, e.g., undergarments, or use forensically validated swabs to collect evidence. The handing over of biological evidence to the forensic laboratory may take place immediately or sometime after, when the analyses are deemed necessary. Until then, samples are often stored in dry and dark conditions. Nonetheless, if microbiome-based stain analyses prove their utility, there may be opportunities to make recommendations with a view to improving the recovery of microbial community composition, particularly for trace samples containing low DNA quantity and quality. For example, investigation of storage practices for microbiome analyses indicate that freezing at −20°C or −80°C is preferable over room temperature to preserve microbiome community composition (33, 34, 38, 39). However, recent forensic studies indicated that samples stored in dry and dark conditions for years still had microbial signatures indicative of body site (24).
The choice of the next step, DNA extraction, is more critical, as it has a greater effect on DNA yield and microbial diversity than collection and storage methods (40–42). Here forensic geneticists have greater leverage in decision-making and should select protocols that are suitable for both human and microbial DNA. These need to incorporate cell lysis steps adapted to the more rigid cell walls of some microorganisms, such as Gram-positive bacteria, to obtain a representative picture of microbial composition. Numerous human and environmental microbiome studies use commercial kits, such as the PowerFecal and PowerSoil kits (Qiagen), which include a bead beating step originally designed to homogenize and mechanically lyse the microbes in stool and soil. As this step may be too aggressive for the low-biomass samples typically found in crime scenes, another possibility is to make use of a heat lysis step as an alternative (43, 44). A list of the most popular kits and recommended modifications is provided in Table S1 in the supplemental material.
Amplicon sequence data or shotgun sequence data?
Another key issue in the microbiome analyses of stains is determining the most appropriate sequencing strategy, which will depend on the specific application, and more specifically, on the level of genetic variability required to resolve the forensic question. Here, we focus on two main types of data generated through next-generation sequencing techniques: amplicon sequencing and shotgun sequencing.
For BFI and TsD, we recommend focusing on the 16S rRNA gene, as it is so far the most cost-effective. Shotgun sequencing has the drawback of a larger proportion of background human DNA at some body sites and is still more expensive from both laboratory and computational perspectives. One question to be resolved, however, is which 16S rRNA gene variable region(s) to target for amplification, as these regions display biases in the recovery of specific bacterial taxa. Even for the same 16S rRNA region, the sequences of the primers employed have an effect on the taxa recovered, e.g., depending on the number and positions of degenerate bases (45, 46). For BFI, this issue may not be so critical, as the microbiome differences across body sites are robust, as shown by studies targeting different 16S rRNA gene regions. Some technical aspects to consider when choosing the target region are amplicon lengths and unspecific amplification of the eukaryotic 18S rRNA gene regions. Smaller amplicon lengths are preferred, due to faster data acquisition and the challenges faced when sequencing longer fragments from degraded samples. However, the trade-off is a smaller number of polymorphic sites and thus lower resolution to distinguish taxa. While the V1-V3 region enables comparison with the most comprehensive catalogue of global body sites generated so far, the HMP data, the V4 is much shorter, which is an advantage for degraded samples. A comprehensive analysis of the different 16S rRNA regions and their polymorphisms is summarized in Johnson et al. (47). A promising option is to leverage bioinformatic methods that allow combining data from different 16S rRNA gene regions, thus alleviating some of the issues with 16S rRNA gene region(s) selection.
In the future, however, if the hurdles associated with shotgun sequencing are overcome, this method may be of great utility. Shotgun sequencing opens opportunities to exploit a wide range of metagenomic markers beyond bacteria, like fungi, viruses, and the human host. Additionally, shotgun sequence data can provide not only taxonomic profiles but also microbial functional profiles that are inferred directly through gene annotation (48). Together, community composition and direct functional profiling could yield stronger evidence for the identification of a body site. Another advantage of shotgun sequence data is that human DNA would also be available for questions related to the donor. With the impending changes in legislation across many countries, it will be possible to utilize human coding single-nucleotide polymorphisms to make inferences on phenotypic traits, such as biogeographical ancestry, hair, eye, and skin color, all of which can then be used as investigative leads by the police (49, 50).
For ID, the literature is equivocal: matching a microbial community from a sample to a donor is a more challenging task, and to evaluate the individualizing power of sequence data, larger data sets than those generally tested so far are required. Nonetheless, we recommend forensic scientists to start with 16S rRNA gene regions, despite the reduced diversity and uniqueness of the sequences. Although the short V4 region of the 16S rRNA gene has been shown to be suboptimal compared to metagenomic shotgun when distinguishing among hundreds of individuals (29), the 16S rRNA gene may still offer untapped potential in matching a sample to a person of interest. Sequencing longer amplicons such as the V1-V3 region, or even the entire 16S rRNA gene, using third-generation sequencing techniques like the MinION, can provide finer resolution (47). Higher distinction is also afforded by the analyses of reads inferred to be biological variants, or amplicon sequence variants (ASVs), instead of the representatives of clustered reads, that is, operational taxonomic units (OTUs). An alternate possibility is to follow the approach of Schmedes et al. (30), by developing a panel comprising highly variable and common microbial markers for each type of forensic tissue or fluid of interest. Further analyses are necessary, but if amplicon sequence data turn out to be sufficiently individualizing, it will be the most cost-effective approach facilitating the parallel analysis of BFI and ID.
BIOINFORMATICS WORKFLOW
Read processing and preliminary data exploration.
In this section, we provide a brief overview of the most commonly used approaches for the initial raw read processing and preliminary exploration of microbiome data, with a focus on bacterial sequences. We also underscore some of the questions that need to be resolved for the establishment of a SOP. Among these, a general one is the selection of software from among the large number of open source tools currently available, in the context of frequent updates and improvements to available programs as the field of microbiome research evolves. While an SOP is necessary in the applied fields of forensics and the clinic, we currently recommend workflows to be updated regularly as new best practices are benchmarked, and at the same time, to evaluate whether previous results still hold.
For amplicon sequence data from the 16S rRNA gene, the bioinformatics workflow typically includes quality control and merging of reads when these are paired-end sequences. Next, the reads are processed further, with steps including denoising or clustering of reads and removal of chimeras, in order to generate bacterial abundance tables for downstream statistical analyses. It is possible to choose and combine different programs specifically designed for each step, which in some cases results in more control of the various parameters. However, for both forensic and clinical applications, ease of use is preferred, and thus comprehensive software that includes as many steps as possible is an advantage. In addition, a certain degree of automation is preferred, but it will be necessary for scientists to carry out visual inspection of the data and quality checks in order to choose appropriate parameters for the filtering and trimming of reads. It is also important to include both positive and negative controls in the study design, as these are fundamental to quality control (40).
Notably, one of the most critical bioinformatic decisions relates to a key step: read processing. Here, it is possible to cluster the reads into OTUs, the classical approach, or to adopt an error model-based approach to retain the biological variants, or ASVs. When clustering OTUs, erroneous reads are minimized by collapsing reads into representative sequences using a threshold of similarity, e.g., 97%, 98%, or 99%. This procedure can be done using a curated reference database comprising microbial sequences (closed-reference clustering), using only the sequences of the data set (de novo clustering), or by performing closed-reference clustering followed by de novo clustering for unclustered sequences (open-reference clustering) (51). Closed-reference clustering with the same reference database and version allows the comparison of different data sets as well as different 16S rRNA regions, but it results in the loss of biological variants not present in the reference database. By contrast, ASVs can easily be compared for data sets sequencing the same region and provide finer-scale resolution down to single nucleotides (52).
Here, we propose using closed-reference OTUs for BFI and testing the individualizing power of ASVs for ID. For the first application, OTUs provide sufficient resolution, at least for most of the body sites examined so far. Additionally, using closed-reference clustering when generating OTUs will enable forensic scientists to incorporate publicly available data from different studies, irrespective of the variable region of the 16S rRNA amplicon sequenced, which is an advantage when generating a reference database to train predictive models. For ID, ASVs are expected to be more useful than OTUs (clustered with the widely used 97% identity), as indicated in several recent studies matching skin samples to donors (28, 53). Richardson et al. (28) tested two different methods to produce ASVs: denoising and error learning with DADA2 versus minimal entropy decomposition (MED), which was based on the oligotyping method of Eren et al. (54). Their results showed higher performance of MED compared to denoised data. Further studies addressing the individualizing power of ASVs, and also OTUs clustered at higher thresholds such as 99%, are sorely needed.
For shotgun data, the analyses are more complex and computationally more intensive than for amplicon sequence data, and they are not commonly used in forensic microbiome-based analyses. Therefore, we provide here only a very brief overview of the key steps and decisions (for a detailed review see reference 14). In shotgun data processing, the trimmed and quality filtered reads are used for taxonomic classification either by assigning the reads directly (read based) or by assembling reads into contigs and then assigning to genomes in a reference database. For forensic purposes, given the limitations of DNA quantity and quality, assemblies are expected to be fragmented, and thus a read-based approach is preferred. To increase computational speed, reads can be matched to specific marker genes instead of genomes, as done for the HMP shotgun data from human body sites using MetaPhlAn’s custom database of clade-specific markers. A range of different tools is available for functional profiling, including HUMANn2, which was part of the HMP pipeline. An overview of the bioinformatic steps for amplicon sequencing and shotgun sequencing are provided in the supplemental material (Fig. S2).
Predictive modeling.
The most critical analysis after raw read data processing is the application of a predictive model to infer the bodily origin, donor, and/or time since deposition of the stain. The selection of a suitable model is based on two key criteria: its (high) prediction accuracy and its forensic interpretability. Research so far indicates that numerous ML algorithms satisfy the first criterion, while the second criterion needs further investigation.
The most popular ML methods include supervised learning approaches, which require data sets for which the variable of interest is known (labeled data), e.g., healthy versus diseased state or the bodily origin. These data sets are used to generate a predictive model that is then applied to unlabeled data (55, 56). For BFI, a wide range of benchmarked supervised methods have yielded high prediction accuracies using 16S rRNA gene data. For example, Diez-Lopez et al. (24) used deep neural networks to classify their test and mock skin, oral, and vaginal samples, as well as blood samples originating from different body sites (25). Hanssen et al. (22) evaluated linear discriminant analyses to detect saliva deposited on skin. Using shotgun data, Torres et al. (57) compared linear support vector machines, radial support vector machines, decision trees such as random forests, and gradient boosting, all of which performed similarly well. For ID and TsD, various regression models (15, 30, 31) have been tested on small subsets of individuals, with promising results, although investigation of larger data sets is sorely needed.
Arguably, more important than the ML algorithm itself is the nature of the training data set that is used to generate the predictive model. A study by Tackmann et al. (58) on body site classification underscored the role of the training data set. For their study, the authors generated a large heterogeneous data set comprising 15,082 samples from 5 body sites, from 57 published amplicon and shotgun sequencing studies. To be able to compare the reads, they were mapped to a reference 16S rRNA gene database and clustered into OTUs. Two random forest classifiers (RFCs) were tested: one was trained on the entire database (global RFC) and one on data from a single study (single RFC). The global RFC resulted in markedly higher prediction accuracies (58).
Putting together a training data set from publicly available sources is, however, not a trivial task. For example, for BFI, it is imperative that body site labels for samples obtained from portals such as NCBI and SRA be accurate and unambiguous in order to prevent erroneous predictions. In the case of BFI and TsD, scientists investigating forensic settings will need to collaborate and share data to achieve sample sizes that are adequate for ML training, with inclusion of relevant samples like mixtures and diluted samples. In recent years, several new online resources targeting specific types of applications have emerged, providing access to curated data as well as microbial read abundance tables. A few examples include the Forensic Microbiome Database (http://fmd.jcvi.org/index.php) for geolocation, Microbiome Db (https://microbiomedb.org/mbio/app) for the interrogation of data from specific experiments, and Qiita (https://qiita.ucsd.edu/) for general web-based bioinformatic analyses of multiomics data. To date, however, a customized database for BFI or TsD is not available. For ID, generation of reference databases may not be feasible, particularly given the temporal variability of an individual’s microbiome. Rather, the comparison of a microbial profile obtained from a sample with that from a person of interest is the more likely approach.
The suitability of a predictive model in forensics also depends on whether it is biologically interpretable and whether scientists can explain what the prediction was based on. In the case of many of the supervised learning approaches detailed earlier, such as RFCs, the features are biological sequences associated with a given taxon, which is easily interpretable as evidence. A different aspect of interpretability to consider is that clear guidelines will be necessary when reading the results of the predictive model, for instance, through the establishment of thresholds for the acceptance of a probability value, e.g., above 75% (59).
Reporting evidence for evaluation in court and in diagnostic settings.
A crucial challenge with the introduction of microbiome-based analyses in forensic casework is the reporting of outcomes in an interpretable manner, particularly results obtained from an unconventional analytical method, such as machine learning model predictions. In this section, we provide an overview of existing avenues for a reporting framework, while raising issues that require further investigation.
For DNA evidence from STR profiles, the DNA Commission of the International Society for Forensic Genetics and the European Network of Forensic Science Institutes recommend reporting to be in the form of likelihood ratios (LRs), in order to assess how much more likely the evidence is for one scenario compared to another (60), based on the following equation: LR = Pr(E|Hp, I)/Pr(E|Hd, I), where Pr denotes the probability of E, the evidence. Hp and Hd denote the prosecutor’s and the defense’s propositions, respectively. As an example, let us assume that DNA profiles have been obtained from a stain and from a suspect, and that E refers to the probability of a match between these two. The numerator refers to the probability of the DNA profile match given the prosecutor’s proposition (that the suspect is the perpetrator) and the denominator is the probability of the DNA profile match given the defense’s proposition (that another person is the perpetrator).
Adapting this format for microbiome-based analyses of stains will depend on the application, that is, whether it is BFI, ID, or TsD. For BFI predictions, LRs can be calculated for body site categories according to the two competing hypotheses being tested, i.e., the probability of a stain originating from one or more sites against the others. Importantly, the denominator of the LR is limited by the body site categories that are included in the classifier training. Alternatively, the prediction probabilities for each of the body sites can be ranked in the order of highest prediction probability. For TsD, where the prediction probabilities may correspond to a time point or a time interval estimate, it is unclear whether LRs will be suitable. In any case, the prediction probabilities for BFI and TsD should be provided together with some form of error rate estimate, as well as caveats pertinent to the analysis. For ID, the similarities in the microbial composition of a sample and potential donors can be reported in the form of a LR when there are several persons of interest. However, it will be more challenging to assess matches to random individuals in a population, as done in STR profiling, since databases for microbial markers are impractical. Overall, current reporting guidelines provide a structure and starting point, but the establishment of a suitable framework for microbiome-based prediction results requires modifications specific to the novel method itself and will have to be developed with forensic expertise.
Conclusion.
Microbiome-based analyses offer enormous potential in applied areas such as forensic investigation, especially for BFI, individual ID, and TsD. However, successful integration of microbiome analyses in forensic casework requires comprehensive investigations of reliability and accuracy in typical forensic settings including, for instance, finding stains on substrates, stains originating from multiple donors, and stains comprising multiple tissues and/or fluids. Importantly, a consensus for the standardization and validation of laboratory and bioinformatic pipelines is needed, with particular focus on steps that are seen to impact overall results. In the case of laboratory work, these steps are DNA extraction protocols and choice of the sequencing approach (amplicon versus shotgun). In the case of the bioinformatic pipeline, the choice and training of the predictive model plays a major role and has to be validated. Here, the shift to collaborative studies across laboratories with a systematic approach will enable the generation of a comprehensive interlaboratory training data set. Such studies will enable the inclusion of diverse individuals with different backgrounds and lifestyles. Such a pipeline is not only crucial for forensic applications but is also necessitated in other applied areas, such as clinical diagnostics and therapeutics. The approaches and gaps highlighted in this review together with our recommendations can therefore be used for other applied areas beyond forensics.
ACKNOWLEDGMENTS
We thank Mario Gysi, Venus Kallupurackal, Pamela Voegeli, and Adelgunde Kratzer for their assistance with practical and technical forensic questions related to casework. We are thankful for the funding received from the Swiss Government Excellence Scholarship, Emma Luis Kessler funds, and SNF grant 31003A_182499.
Footnotes
Supplemental material is available online only.
Contributor Information
Meghna Swayambhu, Email: meghna.swayambhu@irm.uzh.ch.
Natasha Arora, Email: natasha.arora@irm.uzh.ch.
Knut Rudi, Norwegian University of Life Sciences.
REFERENCES
- 1.Metcalf JL, Xu ZZ, Bouslimani A, Dorrestein P, Carter DO, Knight R. 2017. Microbiome tools for forensic science. Trends Biotechnol 35:814–823. 10.1016/j.tibtech.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Robinson JM, Pasternak Z, Mason CE, Elhaik E. 2020. Forensic applications of microbiomics: a review. Front Microbiol 11:608101. 10.3389/fmicb.2020.608101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clarke TH, Gomez A, Singh H, Nelson KE, Brinkac LM. 2017. Integrating the microbiome as a resource in the forensics toolkit. Forensic Sci Int Genet 30:141–147. 10.1016/j.fsigen.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Hampton-Marcell JT, Lopez JV, Gilbert JA. 2017. The human microbiome: an emerging tool in forensics. Microb Biotechnol 10:228–230. 10.1111/1751-7915.12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belk A, Xu ZZ, Carter DO, Lynne A, Bucheli S, Knight R, Metcalf JL. 2018. Microbiome data accurately predicts the postmortem interval using random forest regression models. Genes 9:104. 10.3390/genes9020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cocking JH, Turley SR, Fofanov VY, Samuels-Crow K, Hungate B, Mau RL, Keim PS, Gregory Caporaso J, Hepp C. 2020. Forensic analysis of soil microbiomes: linking evidence to a geographic location. bioRxiv. 10.1101/2020.07.10.198044. [DOI]
- 7.Fernández-Rodríguez A, González-Candelas F, Arora N, Moran-Gilad J, Yagel Y. 2021. Omics for forensic and post-mortem microbiology, p 219–240. In Moran-Gilad J, Yagel Y (ed), Application and integration of omics-powered diagnostics in clinical and public health microbiology. Springer, Cham, Switzerland. [Google Scholar]
- 8.Benschop CCG, Quaak FCA, Boon ME, Sijen T, Kuiper I. 2012. Vaginal microbial flora analysis by next generation sequencing and microarrays; can microbes indicate vaginal origin in a forensic context? Int J Legal Med 126:303–310. 10.1007/s00414-011-0660-8. [DOI] [PubMed] [Google Scholar]
- 9.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science 326:1694–1697. 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, Fierer N, Knight R. 2011. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 108(Suppl 1):4516–4522. 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. 2007. The human microbiome project. Nature 449:804–810. 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bradford LL, Ravel J. 2017. The vaginal mycobiome: a contemporary perspective on fungi in women’s health and diseases. Virulence 8:342–351. 10.1080/21505594.2016.1237332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, Kong HH, Segre JA, NIH Intramural Sequencing Center Comparative Sequencing Program . 2013. Topographic diversity of fungal and bacterial communities in human skin. Nature 498:367–370. 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quince C, Walker AW, Simpson JT, Loman NJ, Segata N. 2017. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol 35:833–844. 10.1038/nbt.3935. [DOI] [PubMed] [Google Scholar]
- 15.Salzmann AP, Arora N, Russo G, Kreutzer S, Snipen L, Haas C. 2021. Assessing time dependent changes in microbial composition of biological crime scene traces using microbial RNA markers. Forensic Sci Int Genet 53:102537. 10.1016/j.fsigen.2021.102537. [DOI] [PubMed] [Google Scholar]
- 16.Claessen D, Errington J. 2019. Cell wall deficiency as a coping strategy for stress. Trends Microbiol 27:1025–1033. 10.1016/j.tim.2019.07.008. [DOI] [PubMed] [Google Scholar]
- 17.Helinski DR, Clewell DB. 1971. Circular DNA. Annu Rev Biochem 40:899–942. 10.1146/annurev.bi.40.070171.004343. [DOI] [PubMed] [Google Scholar]
- 18.Sender R, Fuchs S, Milo R. 2016. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 164:337–340. 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 19.Butler JM, Willis S. 2020. Interpol review of forensic biology and forensic DNA typing 2016–2019. Forensic Sci Int Synergy 2:352–367. 10.1016/j.fsisyn.2019.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Virkler K, Lednev IK. 2009. Analysis of body fluids for forensic purposes: from laboratory testing to non-destructive rapid confirmatory identification at a crime scene. Forensic Sci Int 188:1–17. 10.1016/j.forsciint.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Quaak FCA, van de Wal Y, Maaskant-van Wijk PA, Kuiper I. 2018. Combining human STR and microbial population profiling: two case reports. Forensic Sci Int Genet 37:196–199. 10.1016/j.fsigen.2018.08.018. [DOI] [PubMed] [Google Scholar]
- 22.Hanssen EN, Avershina E, Rudi K, Gill P, Snipen L. 2017. Body fluid prediction from microbial patterns for forensic application. Forensic Sci Int Genet 30:10–17. 10.1016/j.fsigen.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 23.Dobay A, Haas C, Fucile G, Downey N, Morrison HG, Kratzer A, Arora N. 2019. Microbiome-based body fluid identification of samples exposed to indoor conditions. Forensic Sci Int Genet 40:105–113. 10.1016/j.fsigen.2019.02.010. [DOI] [PubMed] [Google Scholar]
- 24.Díez López C, Vidaki A, Ralf A, Montiel González D, Radjabzadeh D, Kraaij R, Uitterlinden AG, Haas C, Lao O, Kayser M. 2019. Novel taxonomy-independent deep learning microbiome approach allows for accurate classification of different forensically relevant human epithelial materials. Forensic Sci Int Genet 41:72–82. 10.1016/j.fsigen.2019.03.015. [DOI] [PubMed] [Google Scholar]
- 25.Díez López C, Montiel González D, Haas C, Vidaki A, Kayser M. 2020. Microbiome-based body site of origin classification of forensically relevant blood traces. Forensic Sci Int Genet 47:102280. 10.1016/j.fsigen.2020.102280. [DOI] [PubMed] [Google Scholar]
- 26.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fierer N, Lauber CL, Zhou N, McDonald D, Costello EK, Knight R. 2010. Forensic identification using skin bacterial communities. Proc Natl Acad Sci USA 107:6477–6481. 10.1073/pnas.1000162107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richardson M, Gottel N, Gilbert JA, Lax S, Bailey MJ. 2019. Microbial similarity between students in a common dormitory environment reveals the forensic potential of individual microbial signatures. mBio 10:e01054-19. 10.1128/mBio.01054-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franzosa EA, Huang K, Meadow JF, Gevers D, Lemon KP, Bohannan BJM, Huttenhower C. 2015. Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci USA 112:E2030–E2938. 10.1073/pnas.1423854112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmedes SE, Woerner AE, Novroski NMM, Wendt FR, King JL, Stephens KM, Budowle B. 2018. Targeted sequencing of clade-specific markers from skin microbiomes for forensic human identification. Forensic Sci Int Genet 32:50–61. 10.1016/j.fsigen.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 31.Díez López C, Kayser M, Vidaki A. 2021. Estimating the time since deposition of saliva stains with a targeted bacterial DNA approach: a proof-of-principle study. Front Microbiol 12:647933. 10.3389/fmicb.2021.647933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollock J, Glendinning L, Wisedchanwet T, Watson M. 2018. The madness of microbiome: attempting to find consensus “best practice” for 16S microbiome studies. Appl Environ Microbiol 84. 10.1128/AEM.02627-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nel Van Zyl K, Whitelaw AC, Newton-Foot M. 2020. The effect of storage conditions on microbial communities in stool. PLoS One 15:e0227486. 10.1371/journal.pone.0227486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bai G, Gajer P, Nandy M, Ma B, Yang H, Sakamoto J, Blanchard MH, Ravel J, Brotman RM. 2012. Comparison of storage conditions for human vaginal microbiome studies. PLoS One 7:e36934. 10.1371/journal.pone.0036934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bundgaard-Nielsen C, Ammitzbøll N, Isse YA, Muqtar A, Jensen A-M, Leutscher PDC, Arenholt LTS, Hagstrøm S, Sørensen S. 2020. Voided urinary microbiota is stable over time but impacted by post void storage. Front Cell Infect Microbiol 10:435. 10.3389/fcimb.2020.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manus MB, Kuthyar S, Perroni-Marañón AG, de la Mora AN, Amato KR. 2021. Comparing different sample collection and storage methods for field-based skin microbiome research. Am J Hum Biol 34:e23584. 10.1002/ajhb.23584. [DOI] [PubMed] [Google Scholar]
- 37.Ezzy AC, Hagstrom AD, George C, Hamlin AS, Pereg L, Murphy AJ, Winter G. 2019. Storage and handling of human faecal samples affect the gut microbiome composition: a feasibility study. J Microbiol Methods 164:105668. 10.1016/j.mimet.2019.105668. [DOI] [PubMed] [Google Scholar]
- 38.Jung CE, Chopyk J, Shin JH, Lukacz ES, Brubaker L, Schwanemann LK, Knight R, Wolfe AJ, Pride DT. 2019. Benchmarking urine storage and collection conditions for evaluating the female urinary microbiome. Sci Rep 9:13409. 10.1038/s41598-019-49823-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu W-K, Chen C-C, Panyod S, Chen R-A, Wu M-S, Sheen L-Y, Chang S-C. 2019. Optimization of fecal sample processing for microbiome study: the journey from bathroom to bench. J Formosan Med Assoc 118:545–555. 10.1016/j.jfma.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 40.Sinha R, Abnet CC, White O, Knight R, Huttenhower C. 2015. The microbiome quality control project: baseline study design and future directions. Genome Biol 16:276. 10.1186/s13059-015-0841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costea PI, Zeller G, Sunagawa S, Pelletier E, Alberti A, Levenez F, Tramontano M, Driessen M, Hercog R, Jung F-E, Kultima JR, Hayward MR, Coelho LP, Allen-Vercoe E, Bertrand L, Blaut M, Brown JRM, Carton T, Cools-Portier S, Daigneault M, Derrien M, Druesne A, de Vos WM, Finlay BB, Flint HJ, Guarner F, Hattori M, Heilig H, Luna RA, van Hylckama Vlieg J, Junick J, Klymiuk I, Langella P, Le Chatelier E, Mai V, Manichanh C, Martin JC, Mery C, Morita H, O'Toole PW, Orvain C, Patil KR, Penders J, Persson S, Pons N, Popova M, Salonen A, Saulnier D, Scott KP, Singh B, et al. . 2017. Towards standards for human fecal sample processing in metagenomic studies. Nat Biotechnol 35:1069–1076. 10.1038/nbt.3960. [DOI] [PubMed] [Google Scholar]
- 42.Gill C, van de Wijgert JHHM, Blow F, Darby AC. 2016. Evaluation of lysis methods for the extraction of bacterial DNA for analysis of the vaginal microbiota. PLoS One 11:e0163148. 10.1371/journal.pone.0163148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kia E, Wagner Mackenzie B, Middleton D, Lau A, Waite DW, Lewis G, Chan Y-K, Silvestre M, Cooper GJS, Poppitt SD, Taylor MW. 2016. Integrity of the human faecal microbiota following long-term sample storage. PLoS One 11:e0163666. 10.1371/journal.pone.0163666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Videnska P, Smerkova K, Zwinsova B, Popovici V, Micenkova L, Sedlar K, Budinska E. 2019. Stool sampling and DNA isolation kits affect DNA quality and bacterial composition following 16S rRNA gene sequencing using MiSeq Illumina platform. Sci Rep 9:13837. 10.1038/s41598-019-49520-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Darwish N, Shao J, Schreier LL, Proszkowiec-Weglarz M. 2021. Choice of 16S ribosomal RNA primers affects the microbiome analysis in chicken ceca. Sci Rep 11:11848. 10.1038/s41598-021-91387-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fouhy F, Clooney AG, Stanton C, Claesson MJ, Cotter PD. 2016. 16S rRNA gene sequencing of mock microbial populations: impact of DNA extraction method, primer choice and sequencing platform. BMC Microbiol 16:13. 10.1186/s12866-016-0738-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson JS, Spakowicz DJ, Hong B-Y, Petersen LM, Demkowicz P, Chen L, Leopold SR, Hanson BM, Agresta HO, Gerstein M, Sodergren E, Weinstock GM. 2019. Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat Commun 10:5029. 10.1038/s41467-019-13036-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun S, Jones RB, Fodor AA. 2020. Inference-based accuracy of metagenome prediction tools varies across sample types and functional categories. Microbiome 8:46. 10.1186/s40168-020-00815-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xavier C, de la Puente M, Mosquera-Miguel A, Freire-Aradas A, Kalamara V, Vidaki A, E Gross T, Revoir A, Pośpiech E, Kartasińska E, Spólnicka M, Branicki W, E Ames C, M Schneider P, Hohoff C, Kayser M, Phillips C, Parson W, VISAGE Consortium . 2020. Development and validation of the VISAGE AmpliSeq basic tool to predict appearance and ancestry from DNA. Forensic Sci Int Genet 48:102336. 10.1016/j.fsigen.2020.102336. [DOI] [PubMed] [Google Scholar]
- 50.Schneider PM, Prainsack B, Kayser M. 2019. The use of forensic DNA phenotyping in predicting appearance and biogeographic ancestry. Dtsch Arztebl Int 51–52:873–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Navas-Molina JA, Peralta-Sánchez JM, González A, McMurdie PJ, Vázquez-Baeza Y, Xu Z, Ursell LK, Lauber C, Zhou H, Song SJ, Huntley J, Ackermann GL, Berg-Lyons D, Holmes S, Caporaso JG, Knight R. 2013. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol 531:371–444. 10.1016/B798-0-12-407863-5.00019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Callahan BJ, McMurdie PJ, Holmes SP. 2017. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 11:2639–2643. 10.1038/ismej.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hampton-Marcell JT, Larsen P, Anton T, Cralle L, Sangwan N, Lax S, Gottel N, Salas-Garcia M, Young C, Duncan G, Lopez JV, Gilbert JA. 2020. Detecting personal microbiota signatures at artificial crime scenes. Forensic Sci Int 313:110351. 10.1016/j.forsciint.2020.110351. [DOI] [PubMed] [Google Scholar]
- 54.Eren AM, Borisy GG, Huse SM, Mark Welch JL. 2014. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci USA 111:E2875–E2874. 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knights D, Costello EK, Knight R. 2011. Supervised classification of human microbiota. FEMS Microbiol Rev 35:343–359. 10.1111/j.1574-6976.2010.00251.x. [DOI] [PubMed] [Google Scholar]
- 56.Knights D, Parfrey LW, Zaneveld J, Lozupone C, Knight R. 2011. Human-associated microbial signatures: examining their predictive value. Cell Host Microbe 10:292–296. 10.1016/j.chom.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tan-Torres AL, Jr, Brooks JP, Singh B, Seashols-Williams S. 2021. Machine learning clustering and classification of human microbiome source body sites. Forensic Sci Int 328:111008. 10.1016/j.forsciint.2021.111008. [DOI] [PubMed] [Google Scholar]
- 58.Tackmann J, Arora N, Schmidt TSB, Rodrigues JFM, von Mering C. 2018. Ecologically informed microbial biomarkers and accurate classification of mixed and unmixed samples in an extensive cross-study of human body sites. Microbiome 6:192. 10.1186/s40168-018-0565-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gill P, Hicks T, Butler JM, Connolly E, Gusmão L, Kokshoorn B, Morling N, van Oorschot RAH, Parson W, Prinz M, Schneider PM, Sijen T, Taylor D. 2018. DNA commission of the International Society for Forensic Genetics: assessing the value of forensic biological evidence. Guidelines highlighting the importance of propositions. Part I: evaluation of DNA profiling comparisons given (sub-) source propositions. Forensic Sci Int Genet 36:189–202. 10.1016/j.fsigen.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 60.Champod C, Biedermann A, Vuille J, Willis S, De Kinder J. 2016. ENFSI guideline for evaluative reporting in forensic science: a primer for legal practitioners. Crim Law Justice Wkly 180:189–193. [Google Scholar]
- 61.Gouello A, Dunyach-Remy C, Siatka C, Lavigne J-P. 2021. Analysis of microbial communities: an emerging tool in forensic sciences. Diagnostics (Basel) 12:1. 10.3390/diagnostics12010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pechal JL, Schmidt CJ, Jordan HR, Benbow ME. 2017. Frozen: thawing and its effect on the postmortem microbiome in two pediatric cases. J Forensic Sci 62:1399–1405. 10.1111/1556-4029.13419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download aem.01325-22-s0001.pdf, PDF file, 0.3 MB (345.4KB, pdf)