ABSTRACT
The anaerobic bioremediation of polychlorinated biphenyls (PCBs) is largely impeded by difficulties in massively enriching PCB dechlorinators in short periods of time. Tetrachloroethene (PCE) is often utilized as an alternative electron acceptor to preenrich PCB-dechlorinating bacteria. In this study, resuscitation promoting factor (Rpf) was used as an additive to enhance the enrichment of the microbial communities involved in PCE/PCBs dechlorination. The results indicated that Rpf accelerates PCE dechlorination 3.8 to 5.4 times faster than control cultures. In Aroclor 1260-fed cultures, the amendment of Rpf enables significantly more rapid and extensive dechlorination of PCBs. The residual high-chlorinated PCB congeners (≥5 Cl atoms) accounted for 36.7% and 59.8% in the Rpf-amended cultures and in the corresponding controls, respectively. This improvement was mainly attributed to the enhanced activity of the removal of meta-chlorines (47.7 mol % versus 14.7 mol %), which did not appear to affect dechlorination pathways. The dechlorinators, including Dehalococcoides in Chloroflexi and Desulfitobacterium in Firmicutes, were greatly enriched via Rpf amendment. The abundance of nondechlorinating populations, including Methanosarcina, Desulfovibrio, and Bacteroides, was also greatly enhanced via Rpf amendment. These results suggest that Rpf serves as an effective additive for the rapid enrichment of active dechlorinating cultures so as to provide a new approach by which to massively cultivate bioinoculants for accelerated in situ anaerobic bioremediation.
IMPORTANCE The resuscitation promoting factor (Rpf) of Micrococcus luteus has been reported to resuscitate and stimulate the growth of functional microorganisms that are involved in the aerobic degradation of polychlorinated biphenyls (PCBs). However, few studies have been conducted to investigate the role of Rpf on anaerobic microbial populations. In this study, the enhancement of Rpf on the anaerobic microbial dechlorination of PCE/PCBs was discovered. Additionally, the Rpf-responsive populations underlying the enhanced dechlorination were uncovered. This report reveals the rapid enrichment of active dechlorinating cultures via Rpf amendment, and this sheds light on massively enriching PCB dechlorinators in short periods of time. The enhanced in situ anaerobic bioremediation of PCBs could be expected by supplementing Rpf.
KEYWORDS: resuscitation promoting factor, Aroclor 1260, rapid enrichment, reductive dechlorination, Rpf-responsive populations
INTRODUCTION
Polychlorinated biphenyls (PCBs), the most widely-known, persistent organic pollutants, are a global problem due to their highly toxicity, stability and bioaccumulation potential (1, 2). Various types of PCBs (209 congeners) have frequently been detected in soils, sediments, and aquatic environments, and they pose a significant health risk to humans as well as a global threat to ecosystems (3, 4). There is growing evidence that microbial degradation, as a cost-effective and environmentally sustainable means, is crucial for reducing the risks associated with PCBs. Microorganisms can degrade PCBs through anaerobic reductive dechlorination and oxidative degradation processes. A large number of studies have reported the natural transformation of PCBs by indigenous anaerobic and aerobic microorganisms (2, 5, 6). Most highly chlorinated PCBs, such as Aroclor mixtures, are resistant to aerobic degradation unless chlorine removal occurs via anaerobic microbial dechlorination. However, anaerobic microbial dechlorination proceeds in the natural environment at an extremely low rate due to the low growth rates and low environmental adaptability of dechlorinators (7).
Considerable attempts have been conducted to accelerate the microbial dechlorination of PCBs via the addition of cosubstrates and/or bacteria with efficient dechlorinating abilities, which are known as biostimulation and bioaugmentation (4, 8, 9). Krumins et al. (8) demonstrated that supplementing pentachloronitrobenzene and a culture containing Dehalococcoides ethenogenes strain 195 could enhance the dechlorination of PCB116. Chen and He (10) showed the accelerated onset of Aroclor 1260 dechlorination via preculturing dechlorinators with tetrachloroethene (PCE) or trichloroethene (TCE) as alternative electron acceptors. Previous studies confirmed that PCE, as a priming organohalide, could enhance the enrichment of PCB-dechlorinators in microcosms (5, 11, 12). Thus, PCE-fed cultures could be used as seeding inoculants for the rapid enrichment of Aroclor 1260-dechlorinating cultures. Xu et al. (13) found that the addition of 20% fresh waste-activated sludge could achieve the fastest dechlorination of PCBs in a soil microcosm. It should be noted that the isolation of highly efficient PCB-dechlorinating bacteria is critical to massively cultivating bioinoculants for bioaugmentation. Many investigators have devoted effort toward the cultivation of anaerobic PCB-dehalorespiring bacteria, but the progress has been slow. Since the identification of the PCB-dechlorinating strain o-17 by Cutter et al. (14), only 7 strains (DF-1, 195, CBDB1, JNA, CG1, CG4, and CG5), belonging to Dehalobium chlorocoercia and Dehalococcoides mccartyi, have been isolated and characterized (15–19).
Although the anaerobic microbial dechlorination of halogenated organic contaminants has been extensively documented, there is little research focused on the viable but nonculturable (VBNC) or dormant states of the functional populations that are involved in dechlorination. Kim et al. (20) suggested that the VBNC state and persister state describe the same dormant phenotype. The VBNC/dormant state is regarded as a long-term survival strategy under unfavorable conditions, and it has been identified in more than 100 species of microorganisms and with a wide spectrum of environmental stresses (21, 22). Tripathi et al. (11) indicated that identifying the VBNC/dormant state of pollutant-degraders is necessary to evaluate their bioremediation potential. Fida et al. (23) found that the culturability and phenanthrene-degrading activity of Novosphingobium sp. strain LH128 decreased rapidly due to entering a VBNC-like state after its inoculation into soil. Xie et al. (24) reported that the phenol-degrading strain Candida sp. LN1 entered into a VBNC state that was induced by high phenol concentrations. Importantly, the VBNC state of PCB/biphenyl-degrading Rhodococcus biphenylivorans strain TG9T was verified under low temperature and oligotrophic conditions (25). Thus, the huge potential and important roles of VBNC/dormant microorganisms in pollutant removal is worthy of further investigations.
Quorum sensing (QS) signals control the behaviors of microorganisms, especially those in the VBNC/dormant state, in order to promote their adaptation and survival in adverse environments, which is a promising method by which to activate functional microbial populations for better bioremediation performance (22, 26). Notably, the resuscitation promoting factor (Rpf) from Micrococcus luteus, known as the QS signaling compound, has been reported to resuscitate and stimulate the growth of the majority of the members in the phyla Actinobacteria, Proteobacteria, and Firmicutes (27, 28). The Rpf, working at picomolar concentrations, possesses muralytic activity that is thought to remodel the cell walls of VBNC cells in order to facilitate cell division and regrowth (21, 26). Following the report about the enhanced biphenyl/PCBs degradation by Rpf-containing extracellular organic matter (EOM) from M. luteus (29), the Rpf/EOM from M. luteus was used to enhance the biodegradation of many refractory organic pollutants, such as phenol (30), anthraquinone dye reactive blue 19 (31), lubricant oils (32), and Aroclor 1242 (27). When Rpf/EOM was applied to enhance the bioremediation of PCB-contaminated soils and sediments, 28 species of PCB-degrading bacteria, belonging to 16 genera, were resuscitated (27, 29). Moreover, the resuscitated strains obtained via the addition of Rpf/EOM exhibited excellent performance in the degradation of pollutants, which provides important microbial resources for bioaugmentation (33–35). Although Rpf has been extensively used to promote bacterial growth and culturability under aerobic conditions, few studies have looked at the stimulation and resuscitation functions of Rpf on anaerobic microbial populations.
Therefore, in the current study, the effect of Rpf on the enrichment of anaerobic PCE/PCB-dechlorinating cultures was assessed by comparing the dechlorination performances in Rpf-amended cultures and in the corresponding controls. To elucidate the Rpf-responsive populations underlying the enhanced dechlorination, the changes in the microbial communities and in the microbial gene copy numbers of cultures in the presence and absence of Rpf were analyzed from PCE-fed stages to the Aroclor 1260-fed stage. This report expands the knowledge on the potential of anaerobic microbial dechlorination.
RESULTS
Effect of Rpf on the enrichment of anaerobic PCE/PCB-dechlorinating cultures.
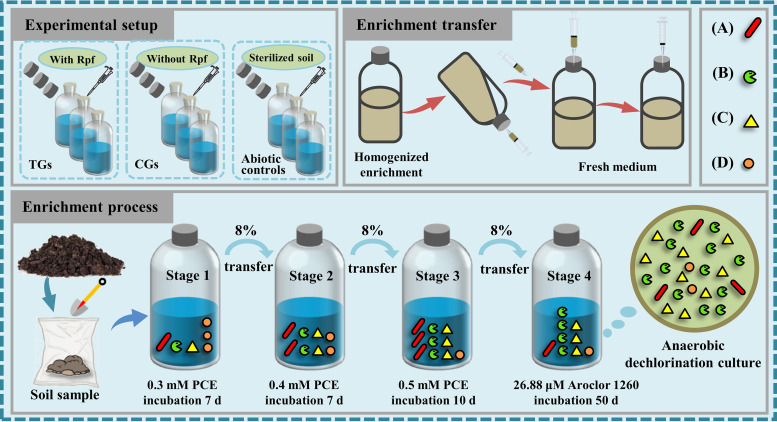
(i) PCE dechlorination. The enrichment process of anaerobic PCE/PCB-dechlorinating cultures is depicted in Fig. 1. From stage 1 to stage 3 (S1 to S3) of the enrichment process, the effect of Rpf on the removal of PCE was investigated. As shown in Fig. 2, PCE dechlorination started quickly in cultures with Rpf amendment. More than 73% (73.4% to 79.1%) of the PCE was removed in the treatment groups (TGs) with Rpf amendment after 3 days of incubation, whereas less than 57% (28.1% to 56.5%) of the PCE was removed in the control groups (CGs) without Rpf amendment. Similarly, a shorter lag phase was observed in the TGs, compared with the CGs; the cell growth in the TGs reached the stationary phase within the first 3 days of each enrichment stage (Fig. S1). At the end of each stage, the residual PCE amounts in the TGs (<0.03 mM) were all significantly (P < 0.05) lower than those in the CGs (>0.12 mM). Notably, the PCE dechlorination of the enrichment cultures was fitted well by a pseudo first-order model (Fig. S2; Table S1). From S1 to S3, the PCE-dechlorination kinetic constants increased from 0.428 day−1 to 0.729 day−1 in the TGs and from 0.111 day−1 to 0.135 day−1 in the CGs. The results clearly show that Rpf amendment significantly enhanced the PCE-dechlorinating capabilities of the enrichment cultures.
FIG 1.
The enrichment process of anaerobic PCE/PCB-dechlorinating cultures. (A) PCE dechlorinators without PCB-dechlorinating ability. (B) PCE/PCBs dechlorinators. (C) Nondechlorinating populations which work synergistically with dechlorinators. (D) Other microorganisms not involved in PCE/PCBs dechlorination.
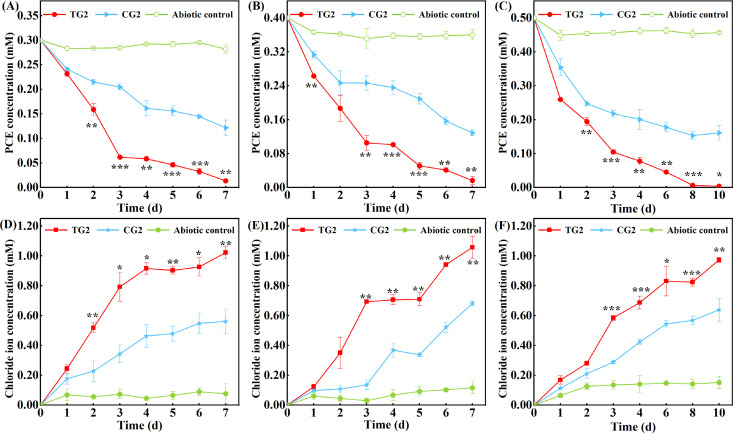
FIG 2.
Comparison of the PCE-dechlorinating capability of PCE-fed cultures with and without Rpf amendment (TGs and CGs) in stage 1 (panels A and D, 0.3 mM PCE), stage 2 (panels B and E, 0.4 mM PCE) and stage 3 (panels C and F, 0.5 mM PCE). Abiotic controls indicating PCE dechlorination in the sterilized soil incubated in parallel. (A–C) PCE concentration changes during incubation. (D and E) Chloride ion accumulation during PCE dechlorination. The PCE and chloride ion in cultures were analyzed by gas chromatography and ion chromatography, respectively. Statistical significance is indicated by asterisks. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Meanwhile, the difference in chloride ion release between the TGs and CGs further demonstrated that Rpf could accelerate PCE dechlorination (Fig. 2). A rapid release of chloride ions in TG1 to TG3 occurred during days 1 to 3 of incubation, and this increased from 0.12 to 0.24 mM to 0.58 to 0.79 mM. The chloride ion releases at the ends of the enrichment stages were 1.5 to 2.0 times higher in the TGs (0.97 to 1.06 mM) than in the CGs (0.56 to 0.68 mM). The measured chloride ion concentrations in both the TGs and CGs were lower than the theoretical values that were calculated from the PCE reduction, assuming a complete conversion of PCE to ethene (Fig. S3). The results suggest the generation of partially dechlorinated intermediates during PCE dechlorination. It should be noted that the releases of chloride ions were greater than the theoretically expected values during the later periods of S2 and S3. For example, releases of chloride ions occurred during days 5 to 7 in TG2 and days 8 to 10 in TG3 were 0.35 mM and 0.15 mM, respectively, whereas the calculated values were 0.14 mM and 0.01 mM, respectively. These findings suggest that the further dechlorination of partially dechlorinated intermediates occurred during the later period of enrichment stages.
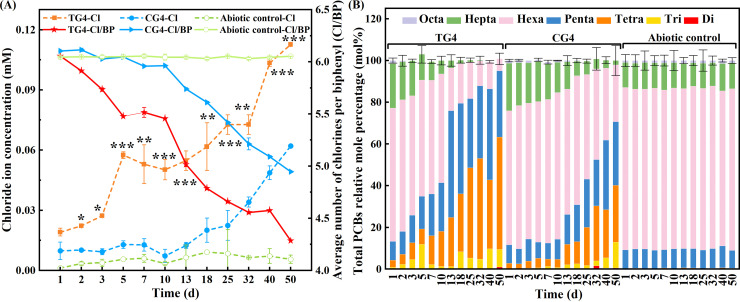
(ii) Aroclor 1260 dechlorination. The effect of Rpf on the dechlorination performance in Aroclor 1260-fed cultures was investigated at stage 4 (S4) of the enrichment process. Compared to CG4, the dechlorination of Aroclor 1260 was significantly enhanced via Rpf amendment (Fig. 3; Fig. S4). After 50 days of incubation, 64.2% and 51.2% of the total chlorine of Aroclor 1260 was removed in TG4 and CG4, respectively. The average number of chlorines per biphenyl (Cl/BP) was reduced from 6.38 to 4.24 in TG4 and 4.96 in CG4 (Fig. 3A). The average chlorine removal rate in TG4 could reach 1.15 μM d−1, which was 1.51 times that of CG4 (0.76 μM·d−1). Meanwhile, the changes in the mole percentages of di-, tri-, tetra-, penta-, hexa-, hepta-, and octa-chlorinated biphenyls (CBs) were compared in TG4 and CG4 (Fig. 3B). The reduction in the octa- and hepta-CBs reached 100% and 21.1% in TG4 after 3 days, whereas 20.7% and 3.5% reductions were observed in CG4. A 60.8% reduction of hexa-CBs was achieved in TG4 between days 10 and 13, which was 4.2 times higher than that observed in CG4 (14.6%). After the incubation of TG4 for 50 days, 90.9% and 98.7% of the hexa- and hepta-CBs were mainly dechlorinated to tetra- and penta-CBs, which were increased from 3.4 mol% to 53.8 mol% and from 9.0 mol% to 31.8 mol% of total PCBs, respectively. Correspondingly, 57.5% and 91.4% of the hexa- and hepta-CBs were mainly converted to tetra-CBs (from 2.4 mol% to 27.3 mol%) and penta-CBs (from 8.8 mol% to 30.5 mol%) in CG4. Some tetra- and penta-CB congeners, including PCBs 70, 87, 92, 105, 114, 123, and 126, were reduced by 100% after 50 days in both the TGs and CGs. The mole percentages of the high-chlorinated PCB congeners (≥5 Cl atoms) accounted for 36.7% in TG4 after 50 days of incubation, whereas 59.8% still existed in CG4. The abiotic control exhibited no obvious difference from the original Aroclor 1260 after 50 days (Fig. 3; Fig. S4). These results further confirm that the amendment of Rpf could achieve a more rapid and extensive dechlorination of Aroclor 1260.
FIG 3.
Comparison of the Aroclor 1260 dechlorinating capability of cultures with and without Rpf amendment (TG4 and CG4) in stage 4. Abiotic controls indicating Aroclor 1260 dechlorination in the sterilized soil incubated in parallel. (A) Chloride ion accumulation and the average number of chlorines per biphenyl (Cl/BP) during Aroclor 1260 dechlorination. (B) Changes in the proportion (mol%) of PCBs with different chlorination degrees. The chloride ion and PCBs in cultures were analyzed by ion chromatography and gas chromatography, respectively. Statistical significance is indicated by asterisks. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Effect of Rpf on PCB dechlorination pathways.
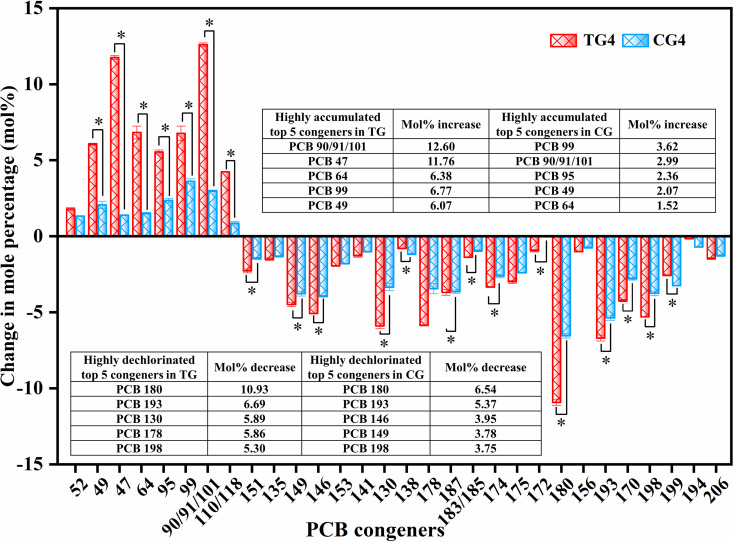
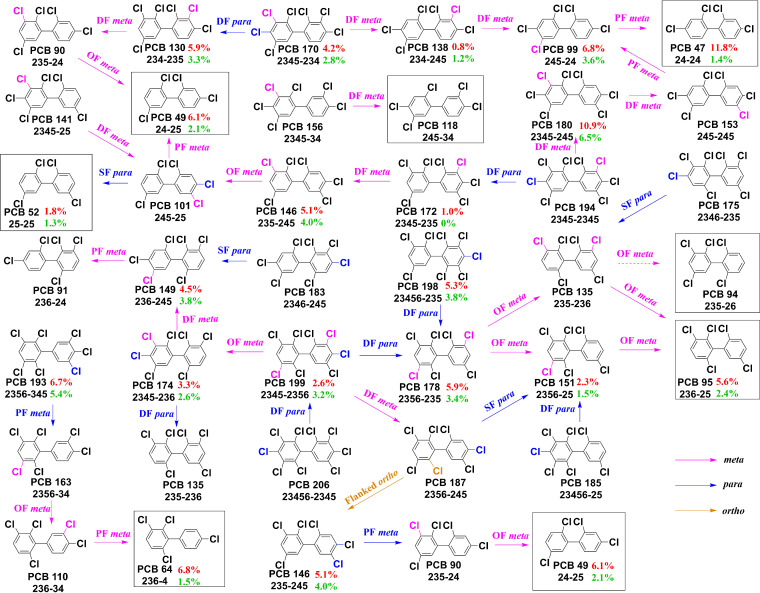
The main dechlorination metabolites of Aroclor 1260 in TG4 and CG4 were compared to reveal the effect of Rpf on the PCB dechlorination pathways (Table S2; Table 1; Fig. 4). After 50 days of incubation, the predominant dechlorination products both in TG4 and CG4 were PCBs 47, 49, 52, 64, 95, 99, 90/91/101, and 110/118 (Fig. S4 and S5), which may have been produced from PCBs 90, 99, 101, 110, 130, 135, 138, 141, 146, 149, 151, 153, 156, and 163 by attacking the ortho-flanked (OF), para-flanked (PF) or double-flanked (DF) meta-chlorines (i.e., 34-, 234-, 235-, 236-, 245-, 2345- and 2356-chlorophenyl rings), and single-flanked (SF) para-chlorines (245- and 2346-chlorophenyl rings). There were no apparent changes in the major metabolites by amended Rpf, except for changes in the mole percentages of dechlorination products and in PCB 172 (2345-235-CB) production. The dominant dechlorination pathways were proposed based on the profile changes in individual PCB congeners (Fig. 5). The dechlorination patterns in TG4 and CG4 include PCB dechlorination Process H, which attacks flanked para- and DF meta-chlorines (i.e., 34-, 234-, 245- and 2345-chlorophenyl rings), Process T, which attacks the flanked meta-chlorines of the 2345-group chlorines (i.e., 2345-chlorophenyl ring), Process H′ which attacks the flanked para- and meta-chlorines of the 23- and 234-group chlorines (i.e., 34-, 234, 245- and 2345-chlorophenyl ring), and Process N, which attacks flanked meta-chlorines (i.e., 234-, 236-, 245-, and 2345-chlorophenyl rings). Compared with CG4, a pathway by which PCB 194 (2345-2345-CB) was dechlorinated to PCB 172 by attacking DF para chlorines was only found in TG4. Meanwhile, PCB 180 (2345-345-CB), comprising 11.0 mol % of the total PCB congeners in Aroclor 1260, decreased significantly to 0.1 mol% in TG4 and 3.5 mol % in CG4. Notably, the most predominant final products, PCBs 47, 49, 64, 95, and 99 were increased by 11.76, 6.07, 6.83, 5.55, and 6.77 mol %, respectively, after 50 days in the TG4, and these were significantly higher than the levels observed in the CG4 (1.39, 2.07, 1.52, 2.36, and 3.62 mol%) (Table 1; Fig. 5). These PCB congeners (PCBs 47, 49, 64, 95, and 99) accounting for 47.7 mol % and 14.7 mol % of the total products in TG4 and CG4, respectively, were meta-dechlorination products. There was no significant difference in the mol % of the para-dechlorination product PCB 52 between TG4 (1.80 mol %) and CG4 (1.34 mol %). In addition, PCB dechlorination via the removal of ortho-chlorines may have occurred in the enrichment cultures (Fig. 5) because PCB 49 may have been produced from PCB 187 through flanked ortho-chlorine (2356-chlorophenyl rings), PF meta-chlorine (245-chlorophenyl rings), and OF meta-chlorine (235-chlorophenyl rings) removal. These results suggest that the amendment of Rpf enhances the activity of meta-chlorine removal without affecting the dechlorination pathways.
TABLE 1.
Final dechlorination products of Aroclor 1260 in TG4 and CG4 after 50 days of incubation
| Parent | Final product(s)a | Change in mol% after 50 db |
|
|---|---|---|---|
| TG4 | CG4 | ||
| PCB 101 | PCB 52 | 1.80 ± 0.09 | 1.34 ± 0.02 |
| PCB 90/101 | PCB 49 | 6.07 ± 0.07* | 2.07 ± 0.24* |
| PCB 99 | PCB 47 | 11.76 ± 0.13* | 1.39 ± 0.03* |
| PCB 110 | PCB 64 | 6.83 ± 0.42* | 1.52 ± 0.06* |
| PCB 135/ 151 | PCB 95 | 5.55 ± 0.13* | 2.36 ± 0.14* |
| PCB 138/153 | PCB 99 | 6.77 ± 0.47* | 3.62 ± 0.17* |
| PCB146/130/149/141 | PCB 90/91/101 | 12.60 ± 0.12* | 2.99 ± 0.06* |
| PCB 156/163 | PCB 110/118 | 4.24 ± 0.03* | 0.86 ± 0.11* |
| PCB 178/185/187 | PCB 151 | –2.27 ± 0.13* | −1.45 ± 0.07* |
| PCB 174/175/178 | PCB 135 | −1.51 ± 0.10 | –1.32 ± 0.05 |
| PCB 183/174 | PCB 149 | –4.49 ± 0.13* | –3.78 ± 0.15* |
| PCB 187/172 | PCB 146 | –5.07 ± 0.00* | –3.95 ± 0.05* |
| PCB 180 | PCB 153 | –1.94 ± 0.01 | –1.80 ± 0.02 |
| PCB 141 | PCB 141 | –1.29 ± 0.08 | –1.01 ± 0.01 |
| PCB 170 | PCB 130 | –5.89 ± 0.18* | –3.34 ± 0.23* |
| PCB 170 | PCB 138 | –0.80 ± 0.03* | –1.17 ± 0.05* |
| PCB 198/199 | PCB 178 | –5.86 ± 0.04* | –3.44 ± 0.33* |
| PCB 199 | PCB 187 | –3.69 ± 0.19 | –3.63 ± 0.11 |
| PCB 183/185 | PCB 183/185 | –1.38 ± 0.03* | –0.95 ± 0.06* |
| PCB 199 | PCB 174 | –3.34 ± 0.02* | −2.61 ± 0.11* |
| PCB 175 | PCB 175 | −2.99 ± 0.10 | –2.39 ± 0.01 |
| PCB 194 | PCB 172 | –0.95 ± 0.08* | 0.00 ± 0.00* |
| PCB 194 | PCB 180 | –10.93 ± 0.19* | –6.54 ± 0.15* |
| PCB 156 | PCB 156 | –1.00 ± 0.03 | –0.74 ± 0.05 |
| PCB 193 | PCB 193 | –6.69 ± 0.19* | –5.37 ± 0.15* |
| PCB 170 | PCB 170 | –4.22 ± 0.07* | –2.79 ± 0.08* |
| PCB 198 | PCB 198 | –5.30 ± 0.02* | −3.75 ± 0.15* |
| PCB 206 | PCB 199 | –2.56 ± 0.04* | –3.23 ± 0.03* |
| PCB 194 | PCB 194 | –0.16 ± 0.02 | –0.70 ± 0.01 |
| PCB 206 | PCB 206 | −1.46 ± 0.06 | −1.26 ± 0.06 |
The products were analyzed by gas chromatography-mass spectrometry (GC-MS) based on the standards of Aroclor 1260 and Aroclor 1242.
An asterisk indicates a statistically significant difference between TG4 and CG4 (P < 0.05).
FIG 4.
Changes in the mol % of PCB congeners after 50 days of incubation in TG4 and CG4. Congeners less than 0.5 mol % at any time point were excluded. Statistical significance is indicated by an asterisk. *, P < 0.05.
FIG 5.
Dechlorination pathways of Aroclor 1260. The dechlorination metabolites were detected based on gas chromatography-mass spectrometry analysis. Solid arrows indicate identified metabolites, and dashed arrows show proposed metabolites. The changes in the mol % of each PCB congener in TG4 and CG4 are shown in red and green, respectively. The dominant end products are boxed.
Microbial community shifts in response to Rpf amendment.
To uncover the changes in the microbial community in response to the amendment of Rpf, high-throughput sequencing was performed for CG1–CG4 and TG1–TG4. The clean Q20/Q30 reads (Table S3) and rarefaction curves (Fig. S6) demonstrated the high quality of the sequencing data. The microbial alpha diversity in the Rpf-amended cultures was relatively higher than those observed in corresponding CGs, except for the cultures at S1 (Table S4). As indicated by a principal coordinates analysis (PCoA), the Aroclor 1260-fed cultures and PCE-fed cultures were well-separated, and the two coordinate axes explained 29.2% and 17.8% of the variation, respectively (Fig. S7). It also demonstrated that differentiation in the taxonomic composition and the functional profile of the microbial community occurred with the amendment of Rpf.
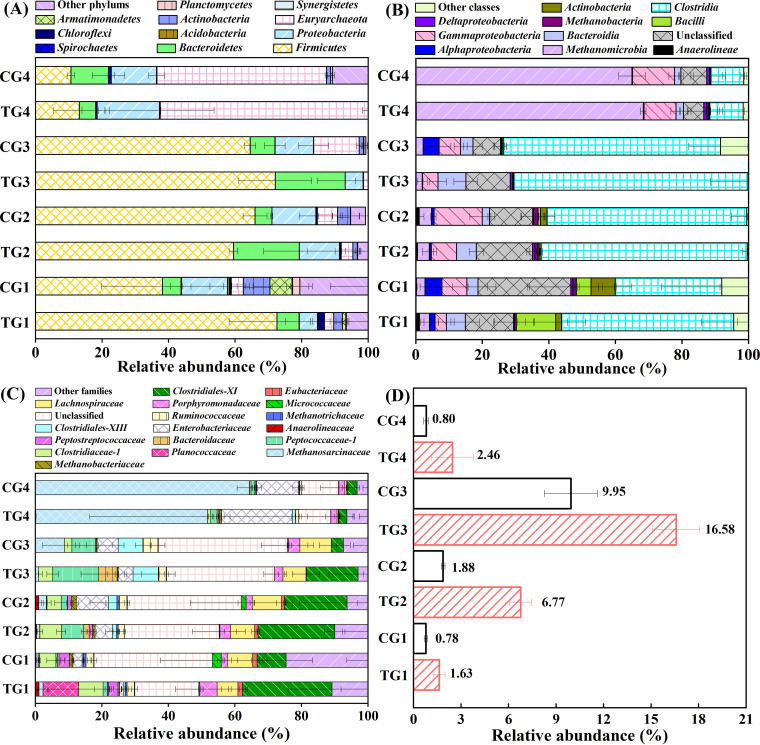
The analysis of the microbial community at the phylum, class, and family levels in the TGs and CGs are illustrated in Fig. 6. At the phylum level (Fig. 6A), Firmicutes was the most dominant phylum in TG1–TG3 and CG1–CG3, which accounted for 59.5 to 72.6% and 38.1 to 66.0%, respectively. Proteobacteria and Bacteroidetes, as the second largest phyla, jointly accounted for 12.2%, 31.8%, and 26.4% in TG1, TG2, and TG3 as well as for 19.4%, 18.2%, and 19.0% in CG1, CG2, and CG3, respectively. At S4, the most predominated phylum was Euryarchaeota, followed by Proteobacteria, which accounted for 62.5% and 18.8% in TG4 as well as for 51.0% and 13.6% in CG4, respectively. Firmicutes, as the third largest phylum, was still higher in TG4 (15.7%) than in CG4 (10.6%). Additionally, a higher relative abundance of Chloroflexi was observed in the TGs (0.02% to 1.90%) compared to the CGs (0.03% to 0.49%). These results reveal an increasing abundance in the phyla Firmicutes, Chloroflexi, Proteobacteria, Euryarchaeota, and Bacteroidetes with the amendment of Rpf.
FIG 6.
Taxonomic distribution of microbial phyla, classes, and families in cultures with and without Rpf amendment (TGs and CGs). The abundances of phyla (A) and classes (B) are greater than 0.1%. The abundance of families (C) is greater than 0.05%. (D) Comparison of the relative abundance of the family Peptococcaceae in TGs and CGs. Each bar represents the average value of three biological replicates in each group.
At the class level (Fig. 6B), Clostridia in phylum Firmicutes, as the most dominant class in the PCE-fed cultures, increased from 51.8% to 70.4% in the TGs from S1 to S3, whereas an increase from 32.0% to 65.0% was observed in the corresponding CGs. Meanwhile, compared with the CGs, the TGs demonstrated a higher relative abundance of Bacteroidia (5.7 to 8.4% versus 2.2 to 3.7%) and a lower relative abundance of Gammaproteobacteria (3.4 to 7.5% versus 6.5 to 14.5%). At S4, Methanomicrobia was predominant, and it was followed by Clostridia and Gammaproteobacteria, and these accounted for 68.3%, 10.0%, and 9.6% in TG4 as well as for 64.9%, 9.9%, and 12.6% in CG4, respectively. In particular, a higher relative abundance of Deltaproteobacteria was observed in the TGs (0.7 to 0.9%) from S2 to S4, compared with the CGs (0.1 to 0.8%). The taxonomic composition at the family level further verified the Rpf-responsive populations (Fig. 6C). Notably, family Peptococcaceae within Clostridia (Fig. 6D) had significantly higher abundance in the TGs, in comparison with the CGs (1.63 to 16.53% versus 0.78 to 9.95%).
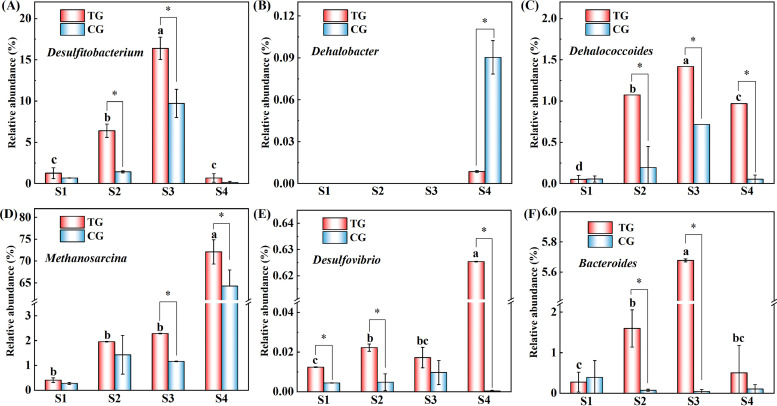
The results of a taxonomic composition at genus level further confirmed that Desulfitobacterium was greatly enriched with the amendment of Rpf (Fig. 7). The relative abundance amounted to 0.7 to 16.4% in TG1–TG4 and 0.2 to 9.7% in CG1–CG4. Especially, Desulfitobacterium dehalogenans, accounting for 0.7 to 30.0% in the TGs, was not detected in the CGs (Fig. S8). Meanwhile, Dehalococcoides had a higher relative abundance in TG2–TG4, compared with the corresponding cultures without Rpf amendment, which were 1.07% versus 0.19%, 1.42% versus 0.71%, and 0.97% versus 0.14%, respectively. In addition, as the major nondechlorinating populations, the genera Methanosarcina, Desulfovibrio, and Bacteroides were also greatly abundant in the Rpf-amended cultures. The abundance of Methanosarcina increased from S1 to S4, and it reached the highest value in TG4 (72.1%) and CG4 (64.3%). Similarly, Desulfovibrio achieved the highest abundance in TG4 (0.6%), which was significantly higher than the level observed in CG4 (<0.1%). However, the highest abundances of Bacteroides was obtained in S3, which were 5.5% and 0.1% in TG3 and CG3, respectively. These results were consistent with the microbial population that was involved in the dechlorination of PCE/PCBs at the phylum level.
FIG 7.
Comparison of the relative abundances of the major genera involved in PCE/PCBs dechlorination in cultures with and without Rpf amendment (TGs and CGs). (A–C) PCE/PCB-dechlorinating bacteria. (D–F) The synergistic genera for enhancing dechlorination performance. Statistical significance is indicated by an asterisk. *, P < 0.05. Different letters indicate statistically significant differences (P < 0.05) among the four enrichment stages in the TGs.
Quantification of microbial genes.
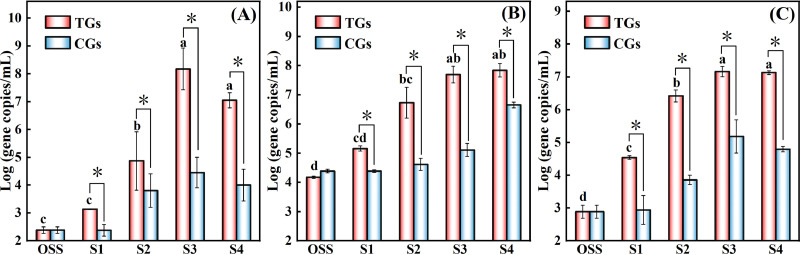
To further verify the effect of Rpf on the enrichment of the PCE/PCB-dechlorinating cultures. The 16S rRNA gene copy numbers of the bacteria, archaea and Dehalococcoides in the enrichment cultures were quantified via quantitative polymerase chain reaction (qPCR). As shown in Fig. 8, there were statistically significant differences in the copy numbers between the TGs and the CGs for all of the genes that were tested. The bacteria in the TGs had a shorter lag phase in their growth, and they rapidly increased from 1.37 × 103 to 1.52 × 108 copies/mL during S1–S3, but they decreased to 1.12 × 107 copies/mL at S4 (Fig. 8A). The copy numbers of the bacterial 16S rRNA gene was significantly lower in the CGs, and these were 2.84 × 104 and 9.98 × 103 copies/mL at S3 and S4, respectively. The total archaeal 16S rRNA gene copy numbers in both the TGs and the CGs showed a continuous increase from S1 to S4 (Fig. 8B). There were also more archaeal genes in the TGs (1.45 × 105 to 6.98 × 107 copies/mL), compared to the CGs (2.44 × 104 to 4.51 × 106 copies/mL). More importantly, the 16S rRNA gene copy numbers of Dehalococcoides were revealed to be significantly higher in the TGs, and these were approximately 40 to 365 times than those observed in the CGs (Fig. 8C). Meanwhile, Dehalococcoides were significantly enriched from S1 to S4 in the TGs, and the gene numbers achieved 1.45 × 107 copies/mL and 1.34 × 107 copies/mL in TG3 and TG4, respectively. The efficient enrichment of Dehalococcoides is the main factor contributing to the improvement of the PCE/PCB-dechlorinating capability of the Rpf-amended cultures. These findings are in line with the results of a microbial community analysis, which uncovered the Rpf-responsive microbial populations that were involved in the enhanced dechlorination of PCE/PCBs.
FIG 8.
Comparison of the 16S rRNA gene copy numbers of bacteria (A), archaea (B), and Dehalococcoides (C) in original soil samples (OSS) and in PCE/PCB-fed cultures with/without Rpf amendment (TGs/CGs) in stage 1 (S1, 0.3 mM PCE), stage 2 (S2, 0.4 mM PCE), stage 3 (S3, 0.5 mM PCE) and stage 4 (S4, 26.88 μM Aroclor 1260). An asterisk indicates the statistical significance of the differences between the TGs and CGs. *, P < 0.05. Different lowercase letters above the error bars indicate statistically significant differences (P < 0.05) among the four enrichment stages in the TGs.
DISCUSSION
Enhanced dechlorination performance by Rpf amendment.
The excellent performance of the Rpf-amended cultures in dechlorination revealed that Rpf could efficiently accelerate the enrichment of highly active PCE/PCB-dechlorinating cultures. The enrichment of highly active dechlorinating cultures is critical for the enhancement of the bioremediation of halogenated organic contaminants (9, 36). Chen et al. (36) reported that the PCE-dechlorination kinetic constants of an enriched PCE-dechlorinating culture increased from 0.057 day−1 in M-1 (generation 1) to 0.322 day−1 in M-11 (generation 11) at an initial PCE concentration of 0.7 mM, which was significantly lower than that of the Rpf-amended culture in this study (0.729 day−1 at 0.5 mM PCE). Tang et al. (37) found that the PCE-dechlorination kinetic constants of enriched consortium amended with rhamnolipids and Tween 80 as surfactants were 0.660 day−1 and 0.780 day−1, respectively. Although the maximum kinetic constant (0.780 day−1) is slightly higher than that in the Rpf-amended cultures (0.729 day−1), the initial PCE concentration was significantly lower than that used in this study (0.03 mM versus 0.5 mM). The further dechlorination of the chlorinated intermediates in TG1–TG3, based on an analysis of chloride ion release, was consistent with the results of previous studies that found that the PCE dechlorination products (DCE and TCE) could be further dechlorinated to vinyl chloride and/or nontoxic ethene (5, 9).
Although no study has yet reported the effect of Rpf on the anaerobic microbial echlorination of PCBs, the enhanced aerobic biodegradation of penta-, tetra-, and tri-CBs in Aroclor 1242 was found via Rpf addition (27). Ye et al. (28) also demonstrated that supplementation with Rpf enhanced Aroclor 1242 degradation by R. biphenylivorans strain TG9T. Moreover, the isolates obtained via Rpf addition exhibited excellent performances in the degradation of PCBs (33, 34). Thus, Rpf could be used as a bioremediation-enhancing technology for PCB contaminants. The Aroclor 1260-dechlorinating capability of TG4 was compared with reported cultures in literature (Table S5), and this revealed the high performance of Rpf-amended cultures in chlorine removal. For example, at the same initial concentration (26.88 μM) of Aroclor 1260, Xu et al. (13) found that the average Cl/BP dropped from 6.38 to 6.06 in waste-activated sludge after 180 days of incubation. There was no obvious dechlorination of Aroclor 1260 (53.80 μM) that occurred in sediment microcosms after 120 days of incubation (5).
The higher meta-chlorine removal activity that was observed in the Rpf-amended cultures played a key role in the enhancement of the dechlorination of PCB congeners. The dechlorination of Aroclor 1260 in the Rpf-amended cultures was mainly in Process N, and this was followed by Process H and Process H′, and these results were was consistent with the results of the culturing systems reported by Li et al. (38). Furthermore, the dechlorination pathways of Aroclor 1260 that were obtained in this study were similar to those of reported PCB-dechlorinating cultures or microcosms (13, 19, 38, 39). The extensive dechlorination of highly chlorinated PCBs was primarily attributed to the ability to remove chlorines from meta- and para-positions (9). Wang et al. (19) reported that three unique D. mccartyi strains predominantly dechlorinated PCB congeners via the removal of meta- and para-chlorines, and the most extensive dechlorination was found by removing chlorines predominantly from meta-positions. Xu et al. (13) also demonstrated that the preferential removal of chlorines at meta-positions achieved the most extensive dechlorination of Aroclor 1260 in digestion sludge. In addition, the observed rare ortho dechlorination indicated the potential of enrichment cultures for the full dechlorination of some PCBs (40).
Rpf-responsive populations underlying the enhanced dechlorination.
The changes in microbial populations that occurred in the Rpf-amended cultures were consistent with the results of previous studies that demonstrated microbial community shifts with the addition of Rpf (21, 27, 30). Similarly, Wan et al. (41) found changes in community composition in cultures amended with extracellular secretions. The Rpf-responsive populations agree well with the results of previous studies at the phylum level, demonstrating these phyla to be important populations that are involved in the dechlorination of PCE/PCBs (7, 12, 42). It has been well-established that members of the phyla Firmicutes and Chloroflexi, as the main PCB-dechlorinating bacteria, could utilize chlorinated congeners as electron acceptors (43). Wang and He (44) found that Dehalobacter in the phylum Firmicutes and Dehalococcoides in Chloroflexi were responsible for PCB dechlorination in culture AD14. Zhang et al. (9) demonstrated that Desulfitobacterium within the phylum Firmicutes possessed the capability to dechlorinate chlorinated compounds. Krzmarzick et al. (42) reported that Chloroflexi populations were capable of respiring a diverse array of halogenated chemicals. To date, several isolates belonging to D. mccartyi and D. chlorocoercia in the phylum Chloroflexi have been identified as dechlorinators of PCE/PCBs (16, 17, 19, 40). Notably, nondechlorinating populations in the phyla Euryarchaeota, Proteobacteria, and Bacteroidetes that coexisted with PCE/PCB-dechlorinating bacteria were reported to play roles in the provision of H2 and acetate or in the elimination of toxic substances (39, 45, 46). In addition, the resuscitation and stimulation functions of Rpf on several members of the phyla Proteobacteria, Bacteroidetes, and Firmicutes have been established in previous studies (21, 27, 30). It can be concluded that the amendment of Rpf can promote the enrichment of dechlorinators of PCE/PCBs and their synergistic populations.
These results were further confirmed by an analysis of the taxonomic compositions at the class, family, and genus levels. Several halorespiring bacteria in Clostridia and Deltaproteobacteria have been found in dechlorinating cultures (43, 45). Notably, the family Peptococcaceae in the TGs was mainly composed (>99%) of the genera Desulfitobacterium and Desulfonispora (Fig. S9). These results are in accordance with those of previous studies that reported members of the two genera that had the ability to dechlorinate PCE/PCBs (9, 12, 43). Additionally, Peptococcaceae showed a significant increase in the PCE-fed cultures from S1 to S3, but they decreased in the PCB-fed cultures. This phenomenon may be attributed to the fact that some species of Desulfitobacterium and Desulfonispora only possessed the ability of PCE-dechlorination and did not possess the ability of PCB dechlorination. In concordance with these findings, Qiu et al. (12) found that several populations within Desulfitobacterium that were involved in PCE dechlorination did not possess PCB dechlorination activity. The significant enrichment of Desulfitobacterium was observed in S2 and S3 (Fig. S10), and this result was in agreement with the hypothesis in Peptococcaceae. It has been reported that D. dehalogenans is able to completely dehalogenate all of the flanking chlorines from hydroxylated PCBs (17). The enrichment of D. dehalogenans in the TGs indicated the enhanced dechlorinating capability of the cultures with the amendment of Rpf. The ability of Dehalococcoides to dechlorinate PCBs mainly through meta- and para-chlorine removal has been well-recognized (9, 19). These results further explained that the chlorine removal in the TGs was predominantly from the meta- and para-positions.
In addition, the significantly higher copy numbers of the Dehalococcoides gene that were achieved in the TGs further verify the results of the Rpf-responsive population analysis. Previous study demonstrated that the 16S rRNA gene copies of Dehalococcoides in culture AD14 increased from 1.14 × 105 copies/mL to 7.04 × 106 copies/mL, with the average number of Cl/BP reducing from 6.38 to 5.87 (44). Compared with AD14, TG4 showed a higher abundance of Dehalococcoides, which achieved 1.34 × 107 copies/mL, with Cl/BP reducing from 6.38 to 4.24. Interestingly, the genus Dehalobacter was only found with lower abundance in the PCB-fed cultures. Wang and He (44) found that the abundance of Dehalobacter greatly increased after the removal of 41.13 μM chlorine from Aroclor 1260. The Dehalococcoides and Dehalobacter, as obligate organohalides-respiring bacteria (OHRB), have been identified in a majority of the PCB-dechlorinating enrichment cultures (12, 13, 44). Meanwhile, the synergistic interactions of Dehalococcoide, Methanosarcina, and Desulfovibrio for the effective dechlorination of PCB have been confirmed by Wang et al. (39), who found that Methanosarcina in Euryarchaeota and Desulfovibrio in Proteobacteria help D. mccartyi by mediating acetate sources and H2 sources. Qiu et al. (12) indicated that Methanosarcina were the predominant methanogens in the PCE/PCB-dechlorinating cultures. Bacteroides species that commonly exist in Dehalococcoides-dominated cultures were also found to enhance the dechlorination performance of Dehalococcoides (45, 46). Therefore, the amendment of Rpf could shape the microbial community toward beneficial populations for the enhancement of the dechlorination of PCE/PCBs.
Conclusions and future perspectives.
In summary, this study demonstrated the rapid enrichment of active dechlorinating cultures via Rpf amendment. Significantly enhanced performances of Rpf-amended cultures in the dechlorination of PCE/PCBs were confirmed, compared with the corresponding cultures without Rpf. The increase of PCB dechlorination was mainly attributable to the enhanced meta-chlorine removal activity. The predominant Rpf-responsive populations involved in the dechlorination of PCE/PCBs were Dehalococcoides in Chloroflexi and Desulfitobacterium in Firmicutes. Rpf also enhanced the abundance of the genera Methanosarcina, Desulfovibrio, and Bacteroides, which synergistically interact with dechlorinating bacteria to improve anaerobic dechlorination.
Based on the results of this study, Rpf amendment provides a promising approach by which to enhance anaerobic microbial dechlorination, and it thereby offers a potential strategy by which to rapidly dechlorinate halogenated organic contaminants. In future work, environmental factors should be considered when evaluating the feasibility of Rpf on implementing engineered bioremediation. In addition, despite the enhancement of Rpf on anaerobic microbial dechlorination being revealed in this work, not much is known about the effect of Rpf on the culturability of anaerobic dechlorinators. Further research regarding the effects of Rpf during the isolation and cultivation of pure PCE/PCB-dechlorinating strains should urgently be carried out for a better understanding of the fates of the microorganisms that are involved in anaerobic dechlorination. Importantly, to gain more insight into the enhancement of Rpf on the anaerobes involved in dichlorination, further specific studies aimed at elucidating the VBNC/dormant states of PCE/PCBs dechlorinators and their synergistic populations are needed. Such studies should also include the cultivation of various pure PCB-dechlorinating strains under different VBNC-inducing conditions, although it is notoriously difficult to grow them as pure cultures. There remains much work to reveal the mechanism of Rpf that is involved in enhanced anaerobic dichlorination.
MATERIALS AND METHODS
Sample collection and Rpf preparation.
PCB-contaminated soils (80 to 100 cm depth) were collected from an e-waste-dismantling region (121.38° E, 28.56° N) in Taizhou (Zhejiang Province, China). The homogenized soil sample was stored anaerobically in presterilized serum bottles and was transported immediately to a laboratory. The basic chemical composition of the soil sample, as well as the concentrations of pollutants (heavy metals and PCBs), were measured as described (4, 27, 47). The properties of the soil are detailed in Table S6.
The Rpf protein was prepared according to the methods described in previous studies (30, 48). Briefly, the rpf gene was amplified from the DNA of M. luteus NCIMB 13267, using the primers rpf-F (5′-GCGCGGATCCATGGACACCATGACTCTCTTCACC-3′) and rpf-R (5′-GATCAAGCTTTCAGGCCTGCGGCAGGACGAGCTCC-3′) (restriction sites are presented in bold). The purified PCR products were cloned into NedI/BamHI-treated vector pET-28a and were expressed in Escherichia coli BL21(DE3). The recombinant cells were inoculated in super optimal broth medium supplemented with kanamycin and induced by isopropyl-β-d-thiogalactopyranoside. Cell pellets were resuspended in Tris-HCl (25 mM) for sonication. The supernatant obtained via crude extract centrifugation was applied to a Ni-NTA-agarose column, and then the eluate was dialyzed overnight at 4°C. The obtained Rpf protein (0.348 mg/mL) was stored at −20°C until use.
Enrichment of anaerobic PCE/PCB-dechlorinating cultures.
The anaerobic mineral salts medium (AMSM) shown in Table S7 was prepared as described by Sohn and Häggblom (49), and it was supplemented with acetate (10 mM) as the sole carbon source, l-cysteine (0.2 mM) and Na2S (0.2 mM) as the reducing agents, and 5 × 104 Pa H2 as the electron donor. The prepared AMSM (150 mL) was added to each 250 mL serum bottle and was then sealed with butyl rubber stoppers. Chlorinated compounds (PCE or Aroclor 1260, as indicated) were spiked as electron acceptors (10). PCE was added neatly to a final concentration of 0.3 to 0.5 mM. Aroclor 1260 stock solution (50 mg of total PCBs per mL) in acetone was spiked into the medium to obtain a final concentration of 26.88 μM (or 10 mg/L) in the cultures. The solvent acetone that was introduced to the cultures (0.02%, vol/vol) can be neglected.
The enrichment process of anaerobic PCE/PCB-dechlorinating cultures was divided into 4 stages (S1 to S4). At S1, the soil was inoculated (6%, wt/vol) in AMSM with 0.3 mM PCE. Afterward, the cultures from the end of previous stage were transferred (8%, vol/vol) to fresh AMSM that contained 0.4 to 0.5 mM PCE (S2 and S3) or 26.88 μM Aroclor 1260 (S4). From S1 to S3, the PCE concentration was increased from 0.3 mM to 0.5 mM in increments of 0.1 mM. The cultures in each stage were statically incubated in the dark at 30°C until either the PCE/Aroclor 1260 was completely depleted or the cell growth reached the stationary phase. During the enrichment process, two different groups were set up for the checking the effects of Rpf. The TG in each stage was conducted with a final concentration of 0.696 mg/L purified, amended Rpf protein. A CG without the amendment of Rpf was performed in parallel. The enrichment cultures of the TG in S1 to S4 were named TG1, TG2, TG3, and TG4, and the corresponding enrichment cultures in the CG were named CG1, CG2, CG3, and CG4. Sterilized soil, which was autoclaved three times at 121°C for 4 h, was incubated in parallel as an abiotic control.
Analytical techniques.
The residual amount of PCE, dechlorination of Aroclor 1260, cell growth, and release of chloride ions were monitored in the enrichment cultures. PCE and PCBs were analyzed as described (9, 12, 27). In brief, headspace samples of chloroethenes were injected manually, using a gastight Luer Lock syringe (Hamilton, Reno, NV, USA), into a gas chromatography system (GC, Agilent 8890N) that was equipped with a flame ionization detector (FID) and a GS-GasPro column (30 m × 0.32 mm × 0.25 μm). The oven temperature was initially held at 80°C for 1 min, increased at 35°C/min to 220°C, and held for 2 min. The injector and detector temperatures were 250°C and 315°C, respectively. The residual Aroclor 1260 in each culture was extracted using n-hexane, and it was quantified using an Agilent 8890N GC with an electron capture detector (ECD) and a column capillary HP-5 (30 m × 0.32 mm × 0.25 μm) (1). The injector and detector temperatures were set at 250°C and 300°C, respectively. The temperature program was as follows: 80°C for 2 min, 80°C to 196°C at a rate of 8°C/min, 196°C to 228°C at a rate of 2°C/min, 228°C to 250°C at a rate of 8°C/min, and maintenance at 250°C for 12 min. Finally, the temperature was increased to 290°C at a rate of 10°C/min, and it was held for 3 min. The removal efficiency (Et, %) and the dechlorination kinetic constant (k, day−1) of PCE were calculated using the equations Et = (Ca − Ct)/C0 × 100% and k = (lnC0 − lnCt)/t, respectively, in which C0 was the initial PCE concentration, Ca and Ct were the PCE concentration in the abiotic control and in the inoculated culture at time t, t represented the reaction time, and k represented the rate constant (50). The PCB-dechlorinating activity of the enrichment cultures in S4 was evaluated by the changes of the mol % of the PCB congeners. The Cl/BP was calculated as the product of the average number of chlorines and the concentration of each congener, divided by the sum of the total concentrations of all of the congeners (10, 13, 44, 51). The average chlorine removal rate was calculated using the formula of (N0 − Nt) × m/t, in which N0 is the initial Cl/BP, Nt is the final Cl/BP, m is the initial total PCBs moles, and t is the incubation time (9, 10, 38). The cell growth was assessed by measuring the optical density at 600 nm (OD600) on a spectrophotometer (TU-1810 UV/VIS, Purkinje, China). The measurement was completed within 30 min to minimize air exposure. In addition, the chloride ion concentration of the cell free culture supernatant was quantified using a Dionex ICS-900 ion chromatography system that was equipped with an IonPac AS11-HC (4 mm × 250 mm) column. To subtract the background chloride concentrations, the abiotic and growth medium controls were established simultaneously (6, 34).
The Aroclor 1260 dechlorination pathways of the enrichment cultures in S4 were investigated. The culture samples were collected after 50 days of incubation. The dechlorination metabolites in each culture were analyzed using a GC-mass spectrometry system (GC-MS, Agilent 7890/5975). The GC column and temperature program were the same as described above. The injector, ion source, and quadrupole temperatures were 270, 230, and 150°C, respectively. The identification of the Aroclor 1260 and its dechlorination products were conducted by comparing the retention times to standards (Tables S8 and S9; Fig. S11).
Microbial community analyses of enrichment cultures.
The enrichment cultures at the end of S1, S2, S3, and S4 were sampled for microbial community analyses. Total genomic DNA was extracted using a DNeasy PowerSoil Pro Kit (Qiagen, Germany). The concentration of purified DNA was quantified by a Qubit 4.0 Fluorometer (Thermo Fisher Scientific, USA). The V3-V4 region of the 16S rRNA gene was amplified using the primer sets 341 F (5′-CCTAYGGGRBGCASCAG-3′) and 806 R (5′-GGACTACNNGGGTATCTAAT-3′) and was then sequenced on an Illumina NovaSeq platform (27).
The raw reads were checked and filtered to ensure their quality (12). The high-quality reads were subsequently analyzed using USEARCH (version 11.2.64). Operational taxonomic units (OTUs) were defined at 97% sequence similarity, using unoise3 in USEARCH. The taxonomic assignment of the representative sequences for each OTU was performed using a Ribosomal Database Project (RDP) classifier with a confidence cutoff of 80%. The alpha diversity indices, which included Chao1, Dominance, Equitability, Richness, Simpson, and Shannon_2, were calculated using USEARCH alpha_div. A PCoA based on Bray-Curtis was performed to examine the dissimilarities in the community composition (27).
Real-time quantitative PCR analyses.
The occurrence and abundance of bacteria, archaea, and Dehalococcoides were measured in the original soil sample (OSS) and in enrichment cultures in each stage using a LightCycler 96 real-time PCR system (Dice TP 600, TaKaRa, Japan). The primer sets Eub338/Eub518, F515/R806, and DhcF/DhcR that were used in this study are shown in Table S10. All three reactions were run as follows: denaturation at 95°C for 10 min, 45 cycles of 95°C for 10 s, annealing at 57 to 58°C for 30 s (Table S10), and extension at 72°C for 30 s. Each qPCR contained 10.4 μL SYBR Green PCR Master Mix, 2.0 μL template DNA, 0.5 μL each primer, and 6.6 μL nuclease-free water. The qPCR standard curves were created as described by Chow et al. (52). In brief, the PCR products of individual genes (bacterial 16S rRNA gene, archaeal 16S rRNA gene, and Dehalococcoides 16S rRNA gene) that were amplified with the primers that are specified in Table S10 were ligated into a pEASY-T1 vector and transformed into E. coli Trans1-T1 competent cells. A calibration curve was obtained via the serial dilutions of known plasmid DNA concentrations. The standard curves (Fig. S12) spanned a range of 102 to 109 gene copies per μL of template DNA. The experiments were performed in triplicate, along with nuclease-free water as a negative control.
Statistical analyses.
All of the experiments were performed in triplicate, and the data were expressed as means and standard deviations (SD) that were calculated from three independent experiments. A one-way analysis of variance (ANOVA) followed by a least significant difference (LSD) test was used to determine the statistical significance of the differences between the TGs and the CGs. The statistical significance (P < 0.05) of the differences between the four enrichment stages in the TGs was also determined via a one-way ANOVA, using both Tukey’s and the Waller-Duncan post hoc tests. All of the statistical calculations were conducted using SPSS 25.0 software (SPSS Inc., Chicago, IL, USA).
Data availability.
The raw sequences were deposited in the Sequence Read Archive of The National Center for Biotechnology Information (NCBI), under the project accession number PRJNA842152.
ACKNOWLEDGMENTS
We gratefully acknowledge the financial support provided by the Natural Science Foundation of Zhejiang Province, China (grant no. LY21D010006), the National Natural Science Foundation of China (grant no. 42277025), and the Jinhua Science and Technology Bureau, China (grant no. 2022-4-033).
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Faqian Sun, Email: faqian@zju.edu.cn.
Isaac Cann, University of Illinois Urbana-Champaign.
REFERENCES
- 1.Huang CC, Zeng YH, Luo X, Ren Z, Lu QH, Tian YK, Gao ST, Wang SQ, Harrad S, Mai BX. 2020. Tracing the sources and microbial degradation of PCBs in field sediments by a multiple-line-of-evidence approach including compound-specific stable isotope analysis. Water Res 182:115977. 10.1016/j.watres.2020.115977. [DOI] [PubMed] [Google Scholar]
- 2.Needham TP, Payne RB, Sowers KR, Ghosh U. 2019. Kinetics of PCB microbial dechlorination explained by freely dissolved concentration in sediment microcosms. Environ Sci Technol 53:7432–7441. 10.1021/acs.est.9b01088. [DOI] [PubMed] [Google Scholar]
- 3.Huang CC, Zeng YH, Luo XJ, Ren ZH, Tian YK, Mai BX. 2021. Comprehensive exploration of the ultraviolet degradation of polychlorinated biphenyls in different media. Sci Total Environ 755:142590. 10.1016/j.scitotenv.2020.142590. [DOI] [PubMed] [Google Scholar]
- 4.Xu Y, Gregory KB, VanBriesen JM. 2016. Microbial-catalyzed reductive dechlorination of polychlorinated biphenyls in Hudson and Grasse River sediment microcosms: determination of dechlorination preferences and identification of rare ortho removal pathways. Environ Sci Technol 50:12767–12778. 10.1021/acs.est.6b03892. [DOI] [PubMed] [Google Scholar]
- 5.Xu GF, Lu QH, Yu L, Wang SQ. 2019. Tetrachloroethene primes reductive dechlorination of polychlorinated biphenyls in a river sediment microcosm. Water Res 152:87–95. 10.1016/j.watres.2018.12.061. [DOI] [PubMed] [Google Scholar]
- 6.Pathiraja G, Egodawatta P, Goonetilleke A, Te'o VSJ. 2019. Effective degradation of polychlorinated biphenyls by a facultative anaerobic bacterial consortium using alternating anaerobic aerobic treatments. Sci Total Environ 659:507–514. 10.1016/j.scitotenv.2018.12.385. [DOI] [PubMed] [Google Scholar]
- 7.Yu H, Feng CH, Liu XP, Yi XY, Ren Y, Wei CH. 2016. Enhanced anaerobic dechlorination of polychlorinated biphenyl in sediments by bioanode stimulation. Environ Pollut 211:81–89. 10.1016/j.envpol.2015.12.039. [DOI] [PubMed] [Google Scholar]
- 8.Krumins V, Park J-W, Son E-K, Rodenburg LA, Kerkhof LJ, Häggblom MM, Fennell DE. 2009. PCB dechlorination enhancement in Anacostia River sediment microcosms. Water Res 43:4549–4558. 10.1016/j.watres.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Zhang DD, Li XK, Zhang CF, Xiao ZX, Li YH, Liang YP, Dang HY. 2021. Electrostimulated bio-dechlorination of a PCB mixture (Aroclor 1260) in a marine-originated dechlorinating culture. Environ Pollut 291:118157. 10.1016/j.envpol.2021.118157. [DOI] [PubMed] [Google Scholar]
- 10.Chen C, He JZ. 2018. Strategy for the rapid dechlorination of polychlorinated biphenyls (PCBs) by Dehalococcoides mccartyi strains. Environ Sci Technol 52:13854–13862. 10.1021/acs.est.8b03198. [DOI] [PubMed] [Google Scholar]
- 11.Tripathi V, Edrisi SA, Chen B, Gupta VK, Vilu R, Gathergood N, Abhilash P. 2017. Biotechnological advances for restoring degraded land for sustainable development. Trends Biotechnol 35:847–859. 10.1016/j.tibtech.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Qiu L, Fang W, He H, Liang Z, Zhan Y, Lu Q, Liang D, He Z, Mai B, Wang S. 2020. Organohalide-respiring bacteria in polluted urban rivers employ novel bifunctional reductive dehalogenases to dechlorinate polychlorinated biphenyls and tetrachloroethene. Environ Sci Technol 54:8791–8800. 10.1021/acs.est.0c01569. [DOI] [PubMed] [Google Scholar]
- 13.Xu GF, Zhao XJ, Zhao SY, He JZ. 2021. Acceleration of polychlorinated biphenyls remediation in soil via sewage sludge amendment. J Hazard Mater 420:126630. 10.1016/j.jhazmat.2021.126630. [DOI] [PubMed] [Google Scholar]
- 14.Cutter LA, Watts JE, Sowers KR, May HD. 2001. Identification of a microorganism that links its growth to the reductive dechlorination of 2, 3, 5, 6-chlorobiphenyl. Environ Microbiol 3:699–709. 10.1046/j.1462-2920.2001.00246.x. [DOI] [PubMed] [Google Scholar]
- 15.Wu QZ, Watts JE, Sowers KR, May HD. 2002. Identification of a bacterium that specifically catalyzes the reductive dechlorination of polychlorinated biphenyls with doubly flanked chlorines. Appl Environ Microbiol 68:807–812. 10.1128/AEM.68.2.807-812.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fennell DE, Nijenhuis I, Wilson SF, Zinder SH, Häggblom MM. 2004. Dehalococcoides ethenogenes strain 195 reductively dechlorinates diverse chlorinated aromatic pollutants. Environ Sci Technol 38:2075–2081. 10.1021/es034989b. [DOI] [PubMed] [Google Scholar]
- 17.Adrian L, Dudková V, Demnerová K, Bedard DL. 2009. “Dehalococcoides” sp. strain CBDB1 extensively dechlorinates the commercial polychlorinated biphenyl mixture Aroclor 1260. Appl Environ Microbiol 75:4516–4524. 10.1128/AEM.00102-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.LaRoe SL, Fricker AD, Bedard DL. 2014. Dehalococcoides mccartyi strain JNA in pure culture extensively dechlorinates Aroclor 1260 according to polychlorinated biphenyl (PCB) dechlorination process N. Environ Sci Technol 48:9187–9196. 10.1021/es500872t. [DOI] [PubMed] [Google Scholar]
- 19.Wang SQ, Chng KR, Wilm A, Zhao S, Yang KL, Nagarajan N, He JZ. 2014. Genomic characterization of three unique Dehalococcoides that respire on persistent polychlorinated biphenyls. Proc Natl Acad Sci USA 111:12103–12108. 10.1073/pnas.1404845111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JS, Chowdhury N, Yamasaki R, Wood TK. 2018. Viable but non-culturable and persistence describe the same bacterial stress state. Environ Microbiol 20:2038–2048. 10.1111/1462-2920.14075. [DOI] [PubMed] [Google Scholar]
- 21.Cai YW, Liu JY, Li GY, Wong PK, An TC. 2022. Formation mechanisms of viable but nonculturable bacteria through induction by light-based disinfection and their antibiotic resistance gene transfer risk: a review. Crit Rev Environ Sci Technol 52:3651–3688. 10.1080/10643389.2021.1932397. [DOI] [Google Scholar]
- 22.Tripathi S, Chandra R, Purchase D, Bilal M, Mythili R, Yadav S. 2022. Quorum sensing-a promising tool for degradation of industrial waste containing persistent organic pollutants. Environ Pollut 292:118342. 10.1016/j.envpol.2021.118342. [DOI] [PubMed] [Google Scholar]
- 23.Fida TT, Moreno-Forero SK, Breugelmans P, Heipieper HJ, Röling WFM, Springael D. 2017. Physiological and transcriptome response of the polycyclic aromatic hydrocarbon degrading Novosphingobium sp. LH128 after inoculation in soil. Environ Sci Technol 51:1570–1579. 10.1021/acs.est.6b03822. [DOI] [PubMed] [Google Scholar]
- 24.Xie MQ, Xu LN, Zhang R, Zhou Y, Xiao YY, Su XM, Shen CF, Sun FQ, Hashmi MZ, Lin HJ, Chen JR. 2021. Viable but nonculturable state of yeast Candida sp. strain LN1 induced by high phenol concentrations. Appl Environ Microbiol 87:e01110-21. 10.1128/AEM.01110-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Su XM, Sun FQ, Wang YL, Hashmi MZ, Guo L, Ding LX, Shen CF. 2015. Identification, characterization and molecular analysis of the viable but nonculturable Rhodococcus biphenylivorans. Sci Rep 5:18590. 10.1038/srep18590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lennon JT, Jones SE. 2011. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol 9:119–130. 10.1038/nrmicro2504. [DOI] [PubMed] [Google Scholar]
- 27.Su XM, Li S, Xie MQ, Tao LQ, Zhou Y, Xiao YY, Lin HJ, Chen JR, Sun FQ. 2021. Enhancement of polychlorinated biphenyl biodegradation by resuscitation promoting factor (Rpf) and Rpf-responsive bacterial community. Chemosphere 263:128283. 10.1016/j.chemosphere.2020.128283. [DOI] [PubMed] [Google Scholar]
- 28.Ye Z, Li HX, Jia YY, Fan JH, Wan JX, Guo L, Su XM, Zhang Y, Wu WM, Shen CF. 2020. Supplementing resuscitation-promoting factor (Rpf) enhanced biodegradation of polychlorinated biphenyls (PCBs) by Rhodococcus biphenylivorans strain TG9T. Environ Pollut 263:114488. 10.1016/j.envpol.2020.114488. [DOI] [PubMed] [Google Scholar]
- 29.Su XM, Shen H, Yao XY, Ding LX, Yu CN, Shen CF. 2013. A novel approach to stimulate the biphenyl-degrading potential of bacterial community from PCBs-contaminated soil of e-waste recycling sites. Bioresour Technol 146:27–34. 10.1016/j.biortech.2013.07.028. [DOI] [PubMed] [Google Scholar]
- 30.Su XM, Wang YY, Xue BB, Zhang YG, Mei RW, Zhang Y, Hashmi MZ, Lin HJ, Chen JR, Sun FQ. 2018. Resuscitation of functional bacterial community for enhancing biodegradation of phenol under high salinity conditions based on Rpf. Bioresour Technol 261:394–402. 10.1016/j.biortech.2018.04.048. [DOI] [PubMed] [Google Scholar]
- 31.Cai JF, Pan AD, Li YL, Xiao YY, Zhou Y, Chen CJ, Sun FQ, Su XM. 2021. A novel strategy for enhancing anaerobic biodegradation of an anthraquinone dye reactive blue 19 with resuscitation-promoting factors. Chemosphere 263:127922. 10.1016/j.chemosphere.2020.127922. [DOI] [PubMed] [Google Scholar]
- 32.Bodor A, Bounedjoum N, Feigl G, Duzs Á, Laczi K, Szilágyi Á, Rákhely G, Perei K. 2021. Exploitation of extracellular organic matter from Micrococcus luteus to enhance ex situ bioremediation of soils polluted with used lubricants. J Hazard Mater 417:125996. 10.1016/j.jhazmat.2021.125996. [DOI] [PubMed] [Google Scholar]
- 33.Su XM, Li S, Cai JF, Xiao YY, Tao LQ, Hashmi MZ, Lin HJ, Chen JR, Mei RW, Sun FQ. 2019. Aerobic degradation of 3,3′,4,4′-tetrachlorobiphenyl by a resuscitated strain Castellaniella sp. SPC4: kinetics model and pathway for biodegradation. Sci Total Environ 688:917–925. 10.1016/j.scitotenv.2019.06.364. [DOI] [PubMed] [Google Scholar]
- 34.Lin QH, Zhou XR, Zhang SS, Gao JL, Xie MQ, Tao LQ, Sun FQ, Shen CF, Hashmi MZ, Su XM. 2022. Oxidative dehalogenation and mineralization of polychlorinated biphenyls by a resuscitated strain Streptococcus sp. SPC0. Environ Res 207:112648. 10.1016/j.envres.2021.112648. [DOI] [PubMed] [Google Scholar]
- 35.Han Z, Lin QH, Zhang SS, Zhou XR, Li S, Sun FQ, Shen CF, Su XM. 2023. High PCBs mineralization capability of a resuscitated strain Bacillus sp. LS1 and its survival in PCB-contaminated soil. Sci Total Environ 856:159224. 10.1016/j.scitotenv.2022.159224. [DOI] [PubMed] [Google Scholar]
- 36.Chen KZ, Liu ZF, Wang XM, Yu CG, Ye JX, Yu CN, Wang FE, Shen CF. 2021. Enhancement of perchloroethene dechlorination by a mixed dechlorinating culture via magnetic nanoparticle-mediated isolation method. Sci Total Environ 786:147421. 10.1016/j.scitotenv.2021.147421. [DOI] [PubMed] [Google Scholar]
- 37.Tang SY, Song X, Wang Q, Wang S. 2020. Effects of two surfactants on microbial diversity of a PCE-degrading microbial consortium. Chemosphere 261:127685. 10.1016/j.chemosphere.2020.127685. [DOI] [PubMed] [Google Scholar]
- 38.Li XK, Xu Q, Cheng YJ, Chen CL, Shen CF, Zhang CF, Zheng DQ, Zhang DD. 2022. Effect of microplastics on microbial dechlorination of a polychlorinated biphenyl mixture (Aroclor 1260). Sci Total Environ 831:154904. 10.1016/j.scitotenv.2022.154904. [DOI] [PubMed] [Google Scholar]
- 39.Wang SQ, Chen C, Zhao SY, He JZ. 2019. Microbial synergistic interactions for reductive dechlorination of polychlorinated biphenyls. Sci Total Environ 666:368–376. 10.1016/j.scitotenv.2019.02.283. [DOI] [PubMed] [Google Scholar]
- 40.Fagervold SK, May HD, Sowers KR. 2007. Microbial reductive dechlorination of aroclor 1260 in Baltimore harbor sediment microcosms is catalyzed by three phylotypes within the phylum Chloroflexi. Appl Environ Microbiol 73:3009–3018. 10.1128/AEM.02958-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wan JX, Chen C, Chen JW, Miao QY, Liu YD, Ye JX, Chen KZ, Jin YY, Tang XJ, Shen CF. 2019. Acceleration of perchloroethylene dechlorination by extracellular secretions from Microbacterium in a mixed culture containing Desulfitobacterium. Environ Pollut 245:651–657. 10.1016/j.envpol.2018.10.008. [DOI] [PubMed] [Google Scholar]
- 42.Krzmarzick MJ, Crary BB, Harding JJ, Oyerinde OO, Leri AC, Myneni SC, Novak PJ. 2012. Natural niche for organohalide-respiring Chloroflexi. Appl Environ Microbiol 78:393–401. 10.1128/AEM.06510-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Praveckova M, Brennerova MV, Holliger C, De Alencastro F, Rossi P. 2016. Indirect evidence link PCB dehalogenation with Geobacteraceae in anaerobic sediment-free microcosms. Front Microbiol 7:933. 10.3389/fmicb.2016.00933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang SQ, He JZ. 2013. Dechlorination of commercial PCBs and other multiple halogenated compounds by a sediment-free culture containing Dehalococcoides and Dehalobacter. Environ Sci Technol 47:10526–10534. [DOI] [PubMed] [Google Scholar]
- 45.Macbeth TW, Cummings DE, Spring S, Petzke LM, Sorenson KS. 2004. Molecular characterization of a dechlorinating community resulting from in situ biostimulation in a trichloroethene-contaminated deep, fractured basalt aquifer and comparison to a derivative laboratory culture. Appl Environ Microbiol 70:7329–7341. 10.1128/AEM.70.12.7329-7341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ismaeil M, Yoshida N, Katayama A. 2018. Bacteroides sedimenti sp. nov., isolated from a chloroethenes-dechlorinating consortium enriched from river sediment. J Microbiol 56:619–627. 10.1007/s12275-018-8187-z. [DOI] [PubMed] [Google Scholar]
- 47.Zhou QX, Gibson C, Foy R. 2000. Long-term changes of nitrogen and phosphorus loadings to a large lake in north-west Ireland. Water Res 34:922–926. 10.1016/S0043-1354(99)00199-2. [DOI] [Google Scholar]
- 48.Xie MQ, Li YL, Xu LN, Zhang SS, Ye HY, Sun FQ, Mei RW, Su XM. 2021. Optimization of bacterial cytokine protein production by response surface methodology for environmental bioremediation. RSC Adv 11:36105–36115. 10.1039/d1ra03565g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sohn SY, Häggblom MM. 2016. Reductive dehalogenation activity of indigenous microorganism in sediments of the Hackensack River, New Jersey. Environ Pollut 214:374–383. 10.1016/j.envpol.2016.04.022. [DOI] [PubMed] [Google Scholar]
- 50.Chen F, Ye Y, Fan BL, Lv M, Liang B, Liu WZ, Cheng HY, Chen YL, Liu Y, Wang YH, Wang AJ, Li ZL. 2022. Simultaneous removal of tetrachloroethylene and nitrate with a novel sulfur-packed biocathode system: the synergy between bioelectrocatalytic dechlorination and sulfur autotrophic denitrification. Chem Eng J 439:135793. 10.1016/j.cej.2022.135793. [DOI] [Google Scholar]
- 51.Kaya D, Imamoglu I, Sanin FD, Payne RB, Sowers KR. 2017. Potential risk reduction of Aroclor 1254 by microbial dechlorination in anaerobic Grasse River sediment microcosms. J Hazard Mater 321:879–887. 10.1016/j.jhazmat.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 52.Chow WL, Cheng D, Wang SQ, He JZ. 2010. Identification and transcriptional analysis of trans-DCE-producing reductive dehalogenases in Dehalococcoides species. ISME J 4:1020–1030. 10.1038/ismej.2010.27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S10 and Fig. S1 to S12. Download aem.01951-22-s0001.pdf, PDF file, 4.3 MB (4.3MB, pdf)
Data Availability Statement
The raw sequences were deposited in the Sequence Read Archive of The National Center for Biotechnology Information (NCBI), under the project accession number PRJNA842152.