Abstract
Direct cellular reprogramming has recently gained attention of cancer researchers for the possibility to convert undifferentiated cancer cells into more differentiated, postmitotic cell types. While a few studies have attempted reprogramming of glioblastoma (GBM) cells toward a neuronal fate, this approach has not yet been used to induce differentiation into other lineages and in vivo data on reduction in tumorigenicity are limited. Here, we employ cellular reprogramming to induce astrocytic differentiation as a therapeutic approach in GBM. To this end, we overexpressed key transcriptional regulators of astroglial development in human GBM and GBM stem cell lines. Treated cells undergo a remarkable shift in structure, acquiring an astrocyte-like morphology with star-shaped bodies and radial branched processes. Differentiated cells express typical glial markers and show a marked decrease in their proliferative state. In addition, forced differentiation induces astrocytic functions such as induced calcium transients and ability to respond to inflammatory stimuli. Most importantly, forced differentiation substantially reduces tumorigenicity of GBM cells in an in vivo xenotransplantation model.
The current study capitalizes on cellular plasticity with a novel application in cancer. We take advantage of the similarity between neural developmental processes and cancer hierarchy to mitigate, if not completely abolish, the malignant nature of tumor cells and pave the way for new intervention strategies.
Introduction
Glioblastoma (GBM) is the most aggressive and common form of malignant brain tumor, representing 56% of all gliomas (1, 2). The prognosis is unfavorable, with less than 5% of patients surviving 5 years after diagnosis and most of them dying from relapse, rather than from primary treatment failure (3). The high intra- and inter-tumor heterogeneity as well as complex and dynamic tumor microenvironment results in combinations of undifferentiated and differentiated cellular populations. Undifferentiated tumor stem cell resilience to therapy is one of the major causes of GBM relapse (4).
Inducing cellular differentiation represents a potential strategy to reduce tumor heterogeneity and decrease the presence of undifferentiated, stem cell–like tumor clones, with the aim of rendering a tumor less aggressive. Direct cell reprogramming represents a fast and efficient way of influencing cell fate decisions. This strategy may be employed for therapeutic purposes, primarily by decreasing tumorigenic potential and secondarily by creating a homogenous cell population easier to target with available therapeutic approaches. Direct reprogramming has been used to convert human GBM cells into neuronal-like cells, leading to decreased tumorigenesis (5–7). However, reprogramming to other neural lineages has not been extensively explored.
The aim of the current study is to convert GBM cells/stem cells into astrocyte-like cells, by induced overexpression of key transcriptional regulators of glial development: Nfia, Nfib, and Sox9. These factors are essential for glial differentiation, and their expression persists in mature astrocytes (8). Mouse fibroblasts can be reprogrammed into astrocyte-like cells by overexpression of Nfia, Nfib, and Sox9 (9) and we recently described efficient induction of astrocytes from human embryonic stem cells by ectopic expression of Nfib alone or together with Sox9 (10).
These findings highlight essential roles of these factors as key regulators of astrocytic differentiation. In glioma, higher expression levels of these factors correlate with lower grade tumors and more differentiated cells (11, 12). Hence, downregulation of these regulators may play a role in evading differentiation, thereby sustaining high proliferative activity.
We employ combinations of Sox9/Nfia/Nfib for one-step conversion of GBM cells and GBM stem cells (GSC) into astrocyte-like cells. By mimicking glial development, we interfere with cell fate, subsequently impairing the malignant phenotype.
Our results show that Nfia, Nfib, and Sox9 exert a pro-differentiation role in GBM, similar to the function they have in inducing astrocyte differentiation, ultimately interfering with tumor evolution in vivo.
Materials and Methods
Cell culture
U87 GBM (male patient; Age at sampling, 44Y; RRID:CVCL_0022) cell line was a kind gift of Dr. Mattias Belting, Lund University, Sweden. The cells were maintained in DMEM supplemented with 1 mmol/L sodium pyruvate, 10 mmol/L HEPES, 1% nonessential aminoacids (NEAA; Thermo Fisher Scientific Inc., Waltham, MA), 10% FBS (Biochrom AB, Berlin, Germany) and 1% antibiotic–antimycotic solution (AAS; Sigma-Aldrich, Stockholm, Sweden). U3054 (female patient; Age at sampling, 60Y; RRID:CVCL_IR83) Patient-derived GBM cells were acquired from the Human Glioblastoma Cell Culture resource (www.hgcc.se) at the Department of Immunology, Genetics and Pathology, Uppsala University, Uppsala, Sweden and cultured under stem cell conditions, in Neurobasal/DMEM F:12 Glutamax medium (1:1 v/v) supplemented with B27 (without vitamin A), N2 (Thermo Fisher Scientific Inc., Waltham, MA), 10 ng/mL EGF, 10 ng/mL FGF (PeproTech Nordic, Stockholm, Sweden), and 1% AAS (Sigma-Aldrich, Stockholm, Sweden) on laminin-coated Corning Primaria Tissue Culture Dishes (Thermo Fisher Scientific Inc., Waltham, MA).
To generate primary GBM cell lines, fresh surgically resected tissue from GBM tumor front of 2 patients were placed in artificial CSF, immediately transferred to the lab and dissociated into a cell suspension. Dissociated cells were grown in serum-free condition, i.e., Neurobasal medium (Gibco) added with 1 mmol/L sodium pyruvate (Gibco), 1× B-27 supplement (Gibco), 1× N-2 supplement (Gibco), 1× NEEA (Gibco). Cells were supplemented with EGF (Thermo Fisher Scientific) and FGF (Thermo Fisher Scientific) 20 ng/mL and grown on Poly-l-ornithine (Merck) and Laminin (Gibco) coated dishes.
Cell lines were kept at 37°C, 5% CO2 for no longer than 6 weeks and all cells were used before passage 30.
The isolated and expanded cell line LU-GBM1 was obtained from primary surgery of a male patient, aged 62 years, and molecular analysis revealed a IDHwt, 1p19q non-deleted, p53wt tumor with preserved ATRX expression and non-methylated MGMT promotor. The LU-GBM2 cell line was collected from surgery of a recurring tumor in a female patient, 57 years of age. This GBM had a IDHwt, 1p19q non-deleted, KRASwt, CTNNB1wt, BRAFwt profile and a non-methylated MGMT promotor.
Written informed consent was obtained from each patient with GBM to generate cell lines according to the ethical permit 2018/37 issued by the Regional Ethics Committee in Lund, Sweden. Studies were conducted in accordance with the Declaration of Helsinki and the ethical permit to perform these studies were reviewed and approved by the head of the Department of Neurosurgery, Skåne University Hospital, Lund, Sweden.
All cell lines were tested at the end of each experimental procedure (35 days upon thawing) and prior to freezing using the MycoAlert PLUS Mycoplasma Detection Kit (Lonza). Primary lines LU-GBM1 and LU-GBM2 were tested at culturing Day 7 and Day 4 respectively, U87 and U3054 were tested at Day 4.
Lentiviral particle production
Lentiviral particles were produced in HEK 293T cells by CaCl2 mediated transfection using 3rd generation LV packaging plasmids, as previously described (13). Briefly, pMD2.G, pRSV-Rev and pMDLg/pRRE helper vectors (Addgene, #12251 #12253 #12259) were co-transfected using calcium chloride with vectors containing DNAs for reverse tetracycline-controlled transactivator (rtTA; Addgene, #20342), Nfia-Blast (Addgene, #117270), Nfib-Hygro (Addgene, #117271), Sox9-Puro (Addgene, #117269), after 16 hours the medium was changed, and viruses harvested 48 hours later. All lentiviral vectors used murine variants of the transcription factors.
Forced differentiation of GBM cells
On Day 3, 4×105 cells were plated on laminin-coated primaria 6-well dishes (Corning Inc., New York) and 24 hours later transduced with 0.5 μL/vector/mL of culture medium, carrying doxycycline-inducible vectors combined with rtTA-expressing lentivirus in the presence of polybrene (8 μg/mL; Merck KGaA, Darmstadt, Germany). Cells infected with rtTA vector only served as control. Transgenes were induced at Day 0 by doxycycline (2.5 μg/mL; Sigma-Aldrich, Stockholm, Sweden) and medium switched to astrocyte differentiation medium (DMEM F:12 Glutamax medium, N2, AAS, NEAA, and 10% FBS). On Day 2, cells were subjected to 5 days (puromycin) or 7 days (blasticidin/hygromycin) of antibiotic selection (Thermo Fisher Scientific Inc., Waltham, MA). After selection, culture medium was gradually switched to serum-free medium. On Day 9, cells were incubated for 2 days with basic fibroblast growth factor (10 ng/mL), and subsequently cultured in astrocyte maturation medium, enriched with hCNTF (5 ng/mL), hBMP4 (10 ng/mL) (PeproTech Nordic, Stockholm, Sweden), and forskolin (10 μmol/L; STEMCELL Technologies, Cambridge, UK). Experiments were conducted at 31 dpi. The U3054 MG cell line was cultured as described above except that amount of virus used for transduction was 0.15 μL/vector/ml and differentiation medium consisted of Neurobasal/DMEM F:12 Glutamax medium (1:1 v/v) supplemented with B27 (without vitamin A), N2, and 1% AAS.
iA induction and culturing
Human iPSC-derived astrocytes (iPSC-iAs) were generated as previously described (10). Briefly, iPSCs at ∼80% confluency were dissociated with Accutase on Day 2, and 3.5×105 cells were plated on Matrigel-coated 6-well plates with mTeSR-1 supplemented with 10 μmol/L ROCK inhibitor (STEMCELL Technologies, Y-27632). The following day (Day 1), medium was replaced with fresh mTeSR-1 medium, and 1 μL of rtTA, Sox9, and Nfib lentivirus was added to each well.
On Day 0, medium was replaced with fresh mTeSR-1 medium containing 2.5 μg/mL doxycycline, which was kept in the medium throughout experiments. On Days 1 and 2 iAs were cultured in expansion medium (DMEM/F12, 10% FBS, 1% N2). Between Day 3 and 5, expansion medium was gradually changed to FGF medium (Neurobasal, 2% B27, 1% NEEA, 1% GlutaMax, 1% FBS, 8 ng/mL FGF, 5 ng/mL CNTF, 10 ng/ML BMP4). 48 hours of puromycin (1.25 μg/mL) selection and 5 days of Hygromycin (200 μg/mL) selection was performed.
On Day 7, iAs were dissociated using Accutase for 10 minutes. 4×105 iAs were plated on Matrigel-coated 6-well plates in FGF medium. On Day 8, a full medium change was performed to remove dead cells and debris. From Day 9 FGF medium was changed to Maturation medium (1:1 DMEM/F12 and Neurobasal, 1% N2, 1% Na Pyruvate, 10 μg/μL NAC, 10 μg/μL hbEGF, 10 ng/mL CNTF, 10 ng/mL BMP4, 500 ug/mL cAMP) and half of the medium was changed every 2 to 3 days.
Immunocytochemistry and quantification
4×104 cells were plated onto poly-L-ornithine/laminin-coated coverslips (Sigma-Aldrich, Stockholm, Sweden) in 24-well plates or 5×105 cells in 6-well culture plates (Thermo Fisher Scientific Inc., Waltham, MA), respectively.
Cells were fixed for 30 minutes with 4% paraformaldehyde, permeabilized for 5 minutes with PBS-0.25% Triton X-100 (referred to as T-PBS; Sigma-Aldrich, Stockholm, Sweden) and blocked for 20 minutes with 5% goat and/or donkey serum (Jackson ImmunoResearch Inc., West Grove, PA). Samples were incubated for 2.5 hours with primary antibodies against glial fibrillary acidic protein (GFAP; 1:500; Synaptic Systems, 173 004) and Ki67 (1:500; Abcam, ab16667), followed by 30 minutes incubation with suitable secondary antibodies. Slides were mounted wet and cover-slipped with PVA-DABCO solution (Sigma-Aldrich, Stockholm, Sweden). Staining procedures were performed at room temperature and cell nuclei were counterstained with Hoechst 33342 (Thermo Fisher Scientific Inc., Waltham, MA). Nonspecific antibody binding was assessed by incubating cells with secondary antibodies only. Images used for quantification were taken at 10x or 20x magnification on an epifluorescence Olympus IX73SC Microscope equipped with pE-300white LED Microscope Illuminator (CoolLED Limited), Olympus UC90 Color Camera - 9MP and CellSens acquisition software (Olympus Sverige AB, Solna, Sweden). Cells from 5 random fields per well were counted using ImageJ software (NIH, Bethesda, Maryland). Illumination and camera exposure settings were maintained constant along different experimental groups throughout experiments.
Single-cell gene expression analysis
Cells were sorted on a BD FACSAria III (BD Biosciences, San Jose, CA) depositing single cells in each well of 96-well plates containing lysis buffer, centrifuged at 3,000 rpm for 1 minute and stored at −80°C. Sorting strategy included FSC/SCC, singlets and live/dead cells discrimination using the DRAQ7 fluorescent probe (Abcam plc, Cambridge, UK). Single-cell gene expression analysis was performed on a Fluidigm Biomark 48.48 Dynamic Array as previously reported (14) with minimal modifications. Namely, reverse transcription and specific target amplification was carried out on a Bio-Rad iQTM5 thermocycler (Bio-Rad Laboratories Inc., Solna, Sweden) with the following parameters: 50°C for 60 minutes, 95°C for 2 minutes, 25 cycles of 95°C for 15 seconds, and 60°C for 4 minutes. TaqMan Gene Expression Assays (Thermo Fisher Scientific Inc., Waltham, MA) used are listed in Supplementary Table S1.
Single-cell data were visualized with Fluidigm Real-Time PCR Analysis software and plots generated using Fluidigm R package SINGuLAR Analysis Toolset. Only cells showing amplification of at least 2 housekeeping genes were included in the analysis. Probes for detection of Nfia, NfiB, and Sox9 were human-specific designed to avoid the introduction of the exogenous variants in the transcriptomic analysis.
Reazurin proliferation assay
In vitro cell proliferation was assessed by plating 1×104 cells in 96-well plates and 24 hours later analyzed by Presto Blue fluorimetric assay (Thermo Fisher Scientific Inc., Waltham, MA) according to the manufacturer's instructions.
Cytokine stimulation assay
2×105 cells were plated onto poly-L-ornithine/laminin-coated 6-well plates and the day after treated for 8 hours with 10 ng/mL of IL1β (PeproTech Nordic, Stockholm, Sweden) or medium alone as control. Total RNA was extracted using RNeasy Mini Kit and traces of DNA were removed using RNase-Free DNase Set (QIAGEN AB, Sollentuna, Sweden) according to the manufacturer's instructions. Single stranded cDNA was obtained with the qScript cDNA Synthesis Kit (Quanta BioSciences Inc., Gaithersburg, MD) and a total of 1 ng/μL was used to perform qPCR using TaqMan Gene Expression Assays (Thermo Fisher Scientific Inc., Waltham, MA) for the following genes: GAPDH (Hs02786624_g1), CXCL8 (Hs00174103_m1), CXCL10 (Hs00171042_m1), CCL5 (Hs00982282_m1), and IL6 (Hs00174131_m1). qPCR was carried out on a StepOnePlus thermocycler (Thermo Fisher Scientific Inc., Waltham, MA) according to the manufacturer's instructions.
Calcium imaging
Cells grown in poly-L-ornithine/laminin-coated 6-well plates were bulk-loaded for 20 minutes at 37°C in DMEM/F-12 without phenol red containing Fluo-4/AM (12.5 mg/mL), pluronic acid (0.05%), DMSO (0.1%), and HEPES (1%).
Live calcium imaging was performed with 10x magnification on an epifluorescence Olympus IX73SC Microscope equipped with pE-300white LED Microscope Illuminator (CoolLED Limited), Olympus UC90 Color Camera - 9MP and CellSens acquisition software (Olympus Sverige AB, Solna, Sweden).
Images were taken with a 1,688×1,352 pixel size, 16-bit depth at a frequency of 1 frame per second. Illumination and camera exposure settings were maintained constant throughout experimental sets.
Ca2+ imaging data was analyzed using Fiji-ImageJ. Metadata and the image data of the raw images were read with GraphPad Prism9. Ca2+ signals were presented as relative fluorescence changes (dF/F0) from specified regions of interest (ROI). Single ROIs were manually selected on the basis of fluorescence maximum intensity projection and bright-field images overlay.
Calcium elevation events were detected with thresholds of 3 times of standard deviation of the baseline. The baseline was set as the mean value of the time interval preceding the stimulus.
In experiments that presented spontaneous Ca2+ oscillations in the baseline time window, the F0 was calculated in the time interval between spontaneous transients. Image series and single cells with obvious motion were excluded.
Transplantation
Treated and control cells were prepared into single-cell suspension in culture medium. Six- to 8-week-old male NSG mice were anesthetized with isofluorane inhalation and stereotaxically injected with 1 μL single-cell suspension (50,000 U87; 200,000 U3054 cells) in the right hemisphere using a pulled glass pipette. A small burr hole was made at following coordinates: 1.0 mm anterior and 2.0 mm lateral to bregma and the glass pipette was inserted through the burr hole 2.0 mm ventral to dura for injection.
NSG mice were given water mixed with 1 g/L doxycycline and 10% sucrose beginning 2 days before transplantation and continued until sacrifice.
IHC and quantification
Animals were perfused with PBS followed by 4% paraformaldehyde, brains dissected and post-fixed in 25mL paraformaldehyde overnight followed by cryoprotection in 30% sucrose in PBS overnight at +4°C. Microtome sections (50 μm) were prepared and stored in cryopreservative (glycerol:ethylene glycol:PBS 1:1:2) at −20°C. For staining, free-floating sections were permeabilized overnight (1%triton-X-100, 10% serum in PBS) at +4°C, incubated in primary antibody overnight (1% tritonX-100, 10% serum in PBS) at +4°C and for 2 hours in secondary antibody (0.5% triton-X-100, 10% serum in PBS) at room temperature. Sections were counterstained with DAPI for 10 minutes at room temperature and mounted with PVA:DABCO, polyvinyl alcohol (Sigma-Aldrich)-based mounting media containing DABCO (Sigma-Aldrich) anti-fading reagent.
The following antibodies were used: rabbit anti-Ki67 (1:500; Abcam, ab16667), guinea pig anti-GFAP (1:500; Synaptic Systems, 173 004), mouse anti-HuNu (1:250; Millipore, mab1281), rat anti-CD68 (1:300; Bio-Rad, MCA1957), rabbit anti-IBA1 (1:400; Wako Chemicals, 019–19741).
For tumor volume measurements, images were collected at 4× or 10× magnification on an epifluorescence Olympus BX61 Microscope equipped with Olympus DP80 Color Camera - 9MP and CellSens acquisition software (Olympus Sverige AB, Solna, Sweden). For single-cell quantifications images were taken using a Zeiss LSM 780 confocal microscope at 20× or 40× magnification (Carl Zeiss Microscopy GmbH, Jena, Germany).
Tumor volume quantification
Assessment of tumor volumes was based on serial brain sectioning. The entire perfused brain was dissected and cut in sections to include the entire tumor mass in subsequent analyses. The tumor mass was then identified by staining single sections with the anti-HuNu antibody. For each slice containing the tumor mass, the corresponding volume was calculated by measuring the surface covered by the HuNu stained area and multiplying it by the slice thickness. The overall volume was finally determined by summing all the single-sections values per each tumor.
Data analysis
Data are presented as the mean ± SEM. Statistical analyses were performed using GraphPad Prism. Parametric or nonparametric tests were performed as appropriate using Student t test or Mann–Whitney U test upon normality test assessment with a Shapiro–Wilk test. For cytokine stimulation, statistical analysis was carried out by one-tailed ratio-paired t test comparing reprogrammed and control groups. Significance was set at P < 0.05.
Data availability
The data generated in this study are available upon request from the corresponding author.
Results
Gliogenic transcription factors induce astrocytic morphology in GBM cells
We reasoned that gliogenic transcription factors could be exploited to direct GBM cells towards an astrocytic fate, with the intent to reduce their tumorigenicity.
To this end, we used lentiviral vectors for inducible and selectable expression of Nfia, Nfib, and Sox9 to force differentiation of GBM cells (Fig. 1A). We screened efficacy of these factors, alone or cooperatively, in inducing astrocytic-like phenotypes in the human GBM cell line U87 and the human GSC line U3054 (15). At 2 days post-infection (dpi), cells were selected for their specific antibiotic resistance combinations and, starting from 14 dpi, cells were cultured in differentiation medium (Fig. 1A).
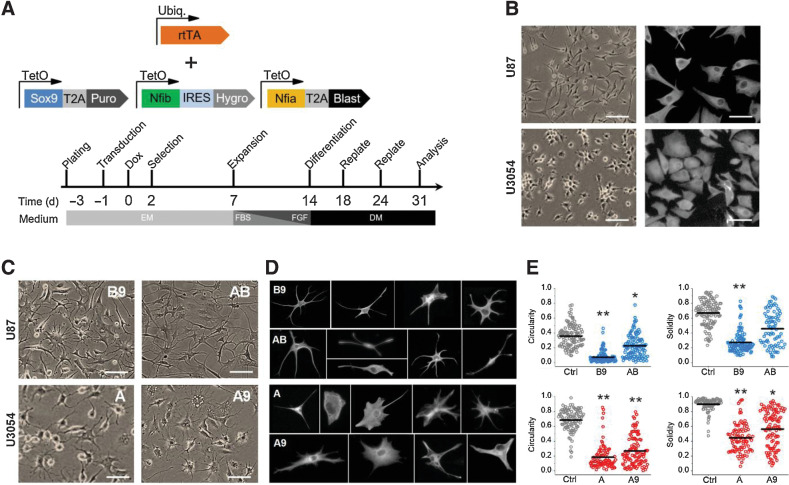
Figure 1.
Gliogenic transcription factors induce morphologic changes in U87 and U3054 cells. A, Schematics of lentiviral vectors used and timeline of experiments. B, Representative bright-field (left, scale bar, 50 μm) and vimentin staining (right, scale bar, 20 μm) images of untreated U87 and U3045 cell lines. C, Representative bright-field images of cancer cell lines upon conversion with different transcription factors combinations. A, Nfia; B, Nfib; 9, Sox9. Scale bar, 50 μm. D, Representative images of individual reprogrammed cells cultured at low density and stained for vimentin, showing their morphologic heterogeneity. Each cell was cropped and placed in the image. E, Morphometric analysis of Solidity, calculated as (Area)/(Convex area) and Circularity, calculated 4π×(Area)/(Perimeter)2 (black bars indicate the population mean). n = 4 independent experiments for each experimental condition (U87, top; U3054, bottom). *, P < 0.05; **, P < 0.01. DM, differentiation medium; EM, expansion medium; Ubiq, ubiquitin.
Strikingly, in all examined cases, forced expression of gliogenic factors induced strong morphologic remodeling based on bright-field microscopy and vimentin immunocytochemical staining. Morphologic analysis revealed that U87 cells had the best conversion performance when infected with either Nfib/Sox9 (B9) or Nfia/Nfib (AB) transcription factors combinations, whereas conversion of the U3054 GSC line was more effective upon overexpression of Nfia (A) or Nfia/Sox9 (A9).
Starting by 14 dpi, both cell types showed a progressive loss of the pleomorphic/epithelial morphology that characterizes rtTA-only transduced controls (Fig. 1B), and by 30 dpi, most cells assumed a highly heterogeneous morphology, resembling that of astrocytes in vitro and in vivo (Fig. 1C and D). We quantified the overall morphologic complexity using two shape descriptors, circularity and solidity, as metrics for roundness and concavity/convexity, respectively. Following conversion, U87 showed morphologic reshaping indicating a substantial increase in morphologic complexity. While this effect was milder and partial for AB treated cells, the B9 group showed a markedly more robust effect. We obtained similar results for U3054 cells, with both A and A9 conditions showing significant effect on cellular morphology (Fig. 1E).
These findings show that overexpression of gliogenic transcription factors induce astrocytic morphology in human GBM cells.
Converted cells increase GFAP expression and decrease their proliferative potential
Next, we assessed whether the morphologic changes observed were accompanied by other phenotypic changes indicative of an astrocytic fate and reduced tumor aggressiveness. First, we analyzed expression of the common astrocyte marker GFAP by immunocytochemistry. Importantly, GFAP expression has been shown to inversely correlate with tumor grade, with significantly lower levels in grade IV, compared with grade II or III astrocytoma (16–18). In this respect, GFAP levels and intracellular localization serves as a molecular marker for cellular differentiation and reduced tumorigenicity.
To delineate the overall cytoarchitecture with higher sensitivity, GFAP stainings were co-labeled with the actin marker Phalloidin. Non-treated U87 and U3054 cell lines presented negligible expression levels of GFAP, with no sign of intracellular structural organization of GFAP intermediate filaments.
U87 cells infected with either AB or B9 transcription factor combinations showed marked upregulation of GFAP. Similarly, A and A9 treated U3054 cultures showed significant increases in the number of GFAP positive cells, although the effect appeared less pronounced as compared with U87 cells (Fig. 2A and B). Importantly, in all examined conditions, cells showed a subcellular localization of GFAP consistent with the structural role in organization of intermediate filaments. Expression levels were particularly high and the intracellular distribution of GFAP was most distinct in the B9 and A groups for U87 and U3054 cell types respectively. The spatial organization of GFAP filaments in these cells followed the cytoskeletal organization, reflecting the overall morphologic reshaping. Consistently, combined Phalloidin staining indicated the contribution of GFAP positive intermediate filaments in the cytoskeletal structuration (Supplementary Fig. S1)
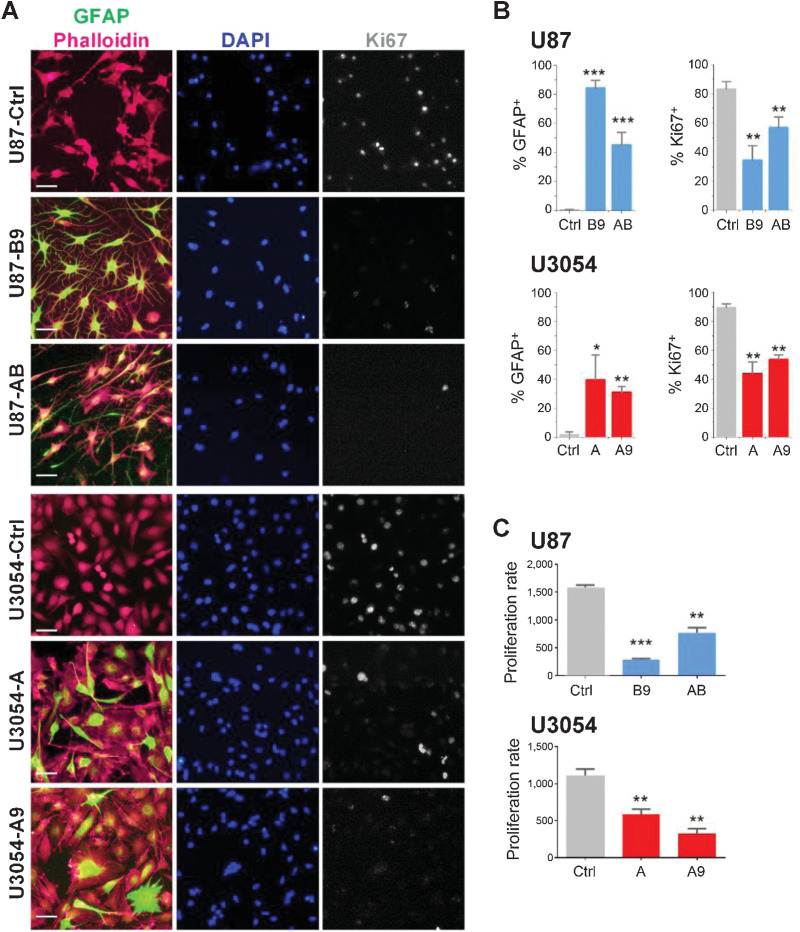
Figure 2.
Differentiated cells gain GFAP expression and decrease proliferative potential. A, Representative fields of U87 and U3054 cells immunocytochemistry stainings for GFAP and Ki67 cells; nuclei and actin filament are counterstained with DAPI and Phalloidin, respectively. Scale bars, 50 μm. B, Quantification of GFAP+ and KI67+ out of DAPI+ cells for treated and untreated conditions. n = 4 independent experiments for each experimental condition. C, Proliferation assay of U87 (top) and U3054 (bottom). Cells (x-axis) are plotted versus relative fluorescence units (RFU; y-axis). *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data are shown as mean ± SEM. n = 3 independent experiments for each experimental condition.
To analyze if changes in morphology and GFAP expression were accompanied by reduction in proliferation rate, we assessed Ki67 expression under control and reprogramming conditions. Control U87 and U3054 cell exhibited high proliferation rate, consistently accompanied by robust Ki67 expression, with over 80% of the cells positive in both lines. Upon differentiation with all protocols tested, both cell lines exhibited marked and significant reduction in Ki67 labeling (Fig. 2B).
In support of downregulated proliferative capacity, in vitro resazurin-based proliferation assays confirmed that both cell types reduced their proliferation in all experimental conditions (Fig. 2C).
Different combinations of transcription factors give rise to discrete populations of astrocyte-like cells
To further confirm the validity of our protocols in inducing an astrocyte phenotype and to assess potential heterogeneity in converted cell populations, we analyzed gene expression changes at the single cell level. To this end, we took advantage of the FluidigmC1 platform to profile expression of several genes involved in glial specification and maturation as well as cell cycle and proliferation.
We found that several astrocytic, oligodendrocytic, stem- and progenitor cell, as well as proliferation marker genes, were differentially expressed in converted U87 and U3054 cells compared with controls (Fig. 3A and B). Consistent with a shift in fate, we found that both reprogrammed U87 and U3054 cells increased expression of genes typical of mature astrocytes, such as GFAP, ALDOC, and FABP7. In addition, the U3054 cancer stem cell line displayed a reduction in oligodendrocyte progenitor (SOX10) and stem cell (PAX6) gene expression, and in OLIG2, a transcription factor involved in cancer stem cell maintenance (19) and oligodendrocyte differentiation, as well as negatively contributing to astrocyte development (20). Consistent with entry into a postmitotic phase (21), expression of proliferative genes KI67 and TOP2A decreased regardless of cell type and reprogramming cocktail.
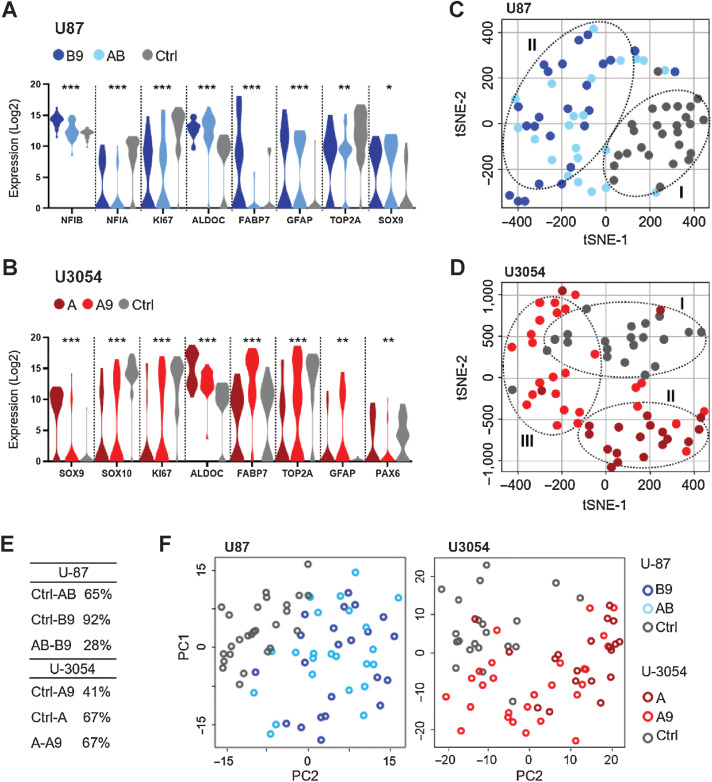
Figure 3.
Conversion induces gene expression changes resulting in distinct populations. A and B, Violin plot of gene expression for selected differentially expressed genes presented in the order of P values for U87 and U3054. Corrected ANOVA *, P < 0.05; **, P < 0.01; ***, P < 0.001. C and D, Integrated tSNE analyses of Fluidigm gene expression data from single cells. Note two distinct cell populations for U87 and three for U3054. E, ANOVA pairwise summary of gene expression: percentage of differentially expressed genes (genes with P < 0.05) between all experimental groups taken in pairs. F, PCA of single-cell qPCR data from U87 and U3054 confirms cluster segregation of treated and untreated cell groups for both U87 and U3054. For each experimental group 20 < n < 25 cells were analyzed.
To visualize the relationship between different cell populations, we used t-distributed stochastic neighbor embedding (tSNE) analysis. Interestingly, this analysis revealed distinct distribution of differentiated cells. Irrespective of cell of origin and induction protocol, converted cells clustered together and segregated from controls (Fig. 3C–E), with the exception of the A9 group, which did not exhibit any distinctive pattern. Furthermore, principal component analysis (PCA) loading plots confirmed the clustered separation showed by the tSNE analysis (Fig. 3F).
In summary, we demonstrate that diverse combinations of transcription factors are required to induce an astroglial transcriptomic signature in two different GBM lines, resulting in the generation of two distinct clustered populations.
GBM-derived astrocyte-like cells respond to cytokine stimulation
Astrocytes have been shown to take part in receiving and conveying inflammatory signals, and to respond by undergoing changes in their immune profile (22). In contrast, GBM cells can alter their immune response by downregulating co-stimulatory molecule expression in the surrounding environment, actively contributing to immune suppression and overall GBM progression, resulting in treatment resistance (23).
Therefore, we investigated whether reprogrammed GBM cells display immunoresponsive behavior by stimulating cells with IL1β and analyzing cytokine expression. Strikingly, treatments triggered significantly increased expression, as compared with controls, of CCL5, CXCL8, CXCL10, and IL6, typical pro-inflammatory cytokines, in reprogrammed U87 and U3054 cell lines (Fig. 4).
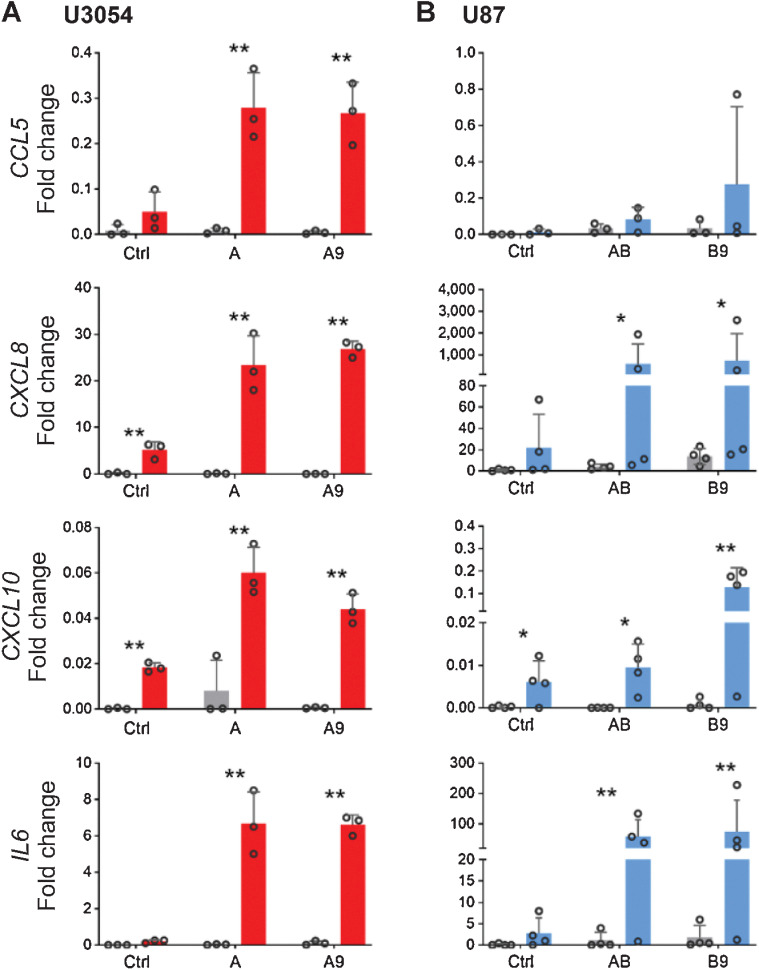
Figure 4.
Induced reactive response in converted cells. Cytokine expression in U3054 (A) and U87 (B), upon stimulation. Shown is relative magnitude (fold increase) in CCL5, CXCL8, CXCL10, and IL6 expression after 8-hour stimulation with 10 ng/mL IL1β. The relative magnitude in increase is normalized to GAPDH expression and relative to control. One-tailed ratio paired t test comparing cells treated or untreated with respective transcription factor protocols. *, P < 0.05; **, P < 0.01. n ≥ 3 independent experiments for each experimental condition.
These data show that converted cells respond to inflammatory stimuli and indicate that forced differentiation can induce astrocyte functionality in GBM cells.
Taken together, our results highlight a particular efficacy for B9 and A protocols for U87 and U3054 cell lines respectively, in triggering cellular conversion. Considering this, we decided to focus subsequent experiments on these two protocols.
GBM-derived astrocyte-like cells exhibit ATP-evoked oscillatory calcium transients
To further explore functionality of reprogrammed GBM cells, we examined intracellular calcium dynamics. Astrocytes are known to dynamically regulate intracellular calcium levels. Stimulated intracellular calcium activity follows specific spatiotemporal dynamics, resulting in typical responses at the single cell level (24). Calcium oscillation is a known plastic signaling system through which astrocyte networks functionally interact with neurons and respond to local stimuli (25, 26).
We tested this functional activity using calcium imaging and analyzing capability of single cells to respond to ATP, before and after conversion. Cells were loaded with Fluo4 at dpi31 and stimulation was conducted by administering single puffs of ATP to the cell culture during imaging (Fig. 5A and B).
Figure 5.
Converted cells exhibit an increase in the ATP-induced response. A, Pseudocolored Fluo4 fluorescence images from U87-B9 and U3054-A cells. Scale bar, 50 μm. B, Representative ΔF/F0 traces showing the intracellular calcium activity upon ATP stimulation in responsive cells. C, Traces of Fluo4 fluorescence changes in U87 and U3054 cells stimulated by 100 μmol/L ATP administration. Average traces are represented as solid lines. Shaded areas indicate the standard deviation of the mean. D, Quantification of the percentage of cells showing ATP response signals, percentage of cells with oscillatory behavior of such responsive population, and quantification of the peak amplitude of the ATP-evoked calcium elevations. n = 5 independent experiments for each experimental condition. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Bar graphs show mean frequency (± SEM).
Upon stimulation, we found significant differences between converted and non-converted cells, in both U87 and U3054 cell lines. Only a limited fraction of rtTA infected U87 cells responded to ATP stimuli, showing a stereotyped profile characterized by single intracellular transients with overall low amplitude to the peak (Fig. 5C and D). In stark contrast, the vast majority of B9 reprogrammed U87 cells responded to the same stimulus with a dramatically different profile, characterized by higher amplitudes, and a significant fraction of the responsive cells showing oscillatory calcium dynamics, to a degree comparable to both adult human (27) and stem cell derived astrocytes (Supplementary Fig. S2).
A similar trend was observed in parallel experiments conducted on U3054 cells. Indeed, the overall number of responsive cells significantly increased following forced differentiation, and responsive cells also exhibited higher ΔF/F0 amplitude to the peak in response transients.
Collectively, these results provide further evidence that differentiated cells do not only adopt morphologic and phenotypic, but also functional properties of astrocytes.
Primary GBM cells are responsive to forced astrocytic differentiation
Tumor cell lines are fundamental models to investigate cancer biology, but their clinical relevance is debated. Although driver mutations are generally retained, prolonged cell culturing can introduce secondary genetic and epigenetic changes that could alter responsiveness to reprogramming. Therefore, we tested efficiency of A and B9 combinations on two primary GBM cell lines recently derived in our laboratories, LU-GBMB1 and LU-GBMB2. Both lines were cultured under standard stem cell conditions and positive for Sox2 and Nestin (28). As for U87 and U3054, we evaluated conversion by GFAP and Ki67 immunocytochemistry to measure astrocytic differentiation and proliferation. In contrast to U87 and U3054, LU-GBMB1 and LU-GBMB2 contained a fraction of cells positive for GFAP. However, upon differentiation both lines showed significant increase in the fraction of GFAP expressing cells using both transcription factor combinations. Consistently, both lines also showed a significant reduction in number of Ki67 positive cells for both conditions, with the exception for the NfiB-Sox9 combination in LU-GBMB2, which did not reach significance (Supplementary Fig. S3). These experiments support our previous findings and suggest the applicability of forced astrocytic differentiation on different GBM cell lines, including primary cultures.
Converted GBM cells retain their astrocyte-like signature and reduce their tumorigenic properties in vivo
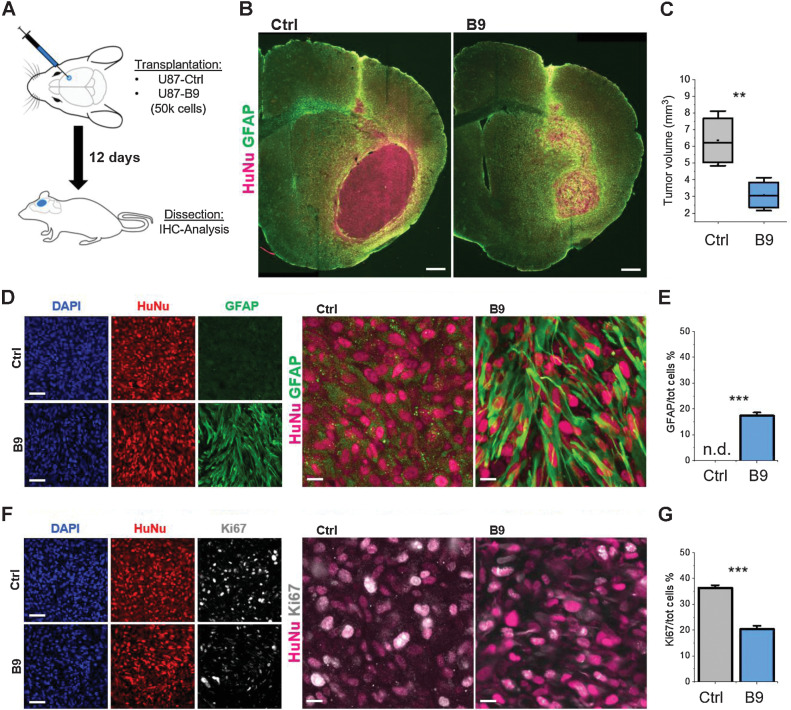
To provide proof of principle of a therapeutic potential, we stereotactically injected control or dpi7 B9 reprogrammed U87 cells intracerebrally into immunocompromised mice and evaluated tumor volume, phenotype, and proliferation at Day 12 after injection (Fig. 6A). We chose this endpoint because mice injected with control cells started exhibiting abnormal behavior and begun to die shortly after.
Figure 6.
Forced differentiation reduces in vivo tumorigenesis. A, Schematic representation of the transplantation procedure. B, Brain slices from animals transplanted with either non-reprogrammed or reprogrammed U87 cells stained for the human nuclei marker HuNu (magenta) and GFAP (green). Scale bar, 200 μm. C, Volumetric quantification of the tumor mass based on HuNu IHC staining. D, High magnification images of GFAP-HuNu staining from sections shown in B. Scale bars, 50 μm (left) and 10 μm (right). E, Relative population of GFAP-expressing cells in the U87-B9 and control groups. F, High magnification images of Ki67-HuNu staining. Scale bars, 50 μm (left) and 10 μm (right). G, Quantification of the relative number of Ki67-positive cells over the total HuNu-positive cells in the U87-B9 and control groups. n = 5 mice for each experimental condition. **, P < 0.01; ***, P < 0.001. Bar graphs show mean frequency (± SEM). n.d., not detected.
Brain sections were immunostained for GFAP, Ki67 and the human-specific nuclear marker HuNu. We measured the tumor volumes in B9 and control transplanted mice, by using HuNu immunostaining to delineate the tumor (Fig. 6B). This volumetric measurement showed a striking effect of B9 treatment, with a very marked and significant reduction in tumor volume (Fig. 6C). Decreased tumor volume was accompanied by a significant fraction of HuNu-GFAP–positive cells (Fig. 6D and E), suggesting a close relationship between astrocytic differentiation and reduction of tumorigenicity. As further proof of the low-proliferative state of converted U87-B9 cells, we observed a dramatic reduction in Ki67-positive cells (Fig. 6F and G). Similar results were also obtained with A treated U3054 cells. We analyzed these at Day 30 after injection, due to their slower in vivo proliferation rate, and observed a significant increase in GFAP expressing cells, together with a substantial reduction in Ki67 positive cells (Supplementary Fig. S4).
GBMs are known to be highly immunosuppressive (29) while astrocytes have important immunologic function (30). Therefore, we evaluated the host immune response induced by control and reprogrammed cells in vivo by assessing tumor-induced astro- and microglial responses. Interestingly, converted cells increased the recruitment of both astrocytes and activated microglia at the tumor-host tissue border, as compared with control cells (Supplementary Fig. S5).
These results represent an in vivo confirmation of the effectiveness of forced gliogenic differentiation in directing tumor cells towards the astrocytic fate, and a proof of its potential in dampening GBM tumorigenesis.
Discussion
In this study, we demonstrate for the first time that overexpression of key gliogenic transcription factors can convert GBM cells into astrocyte-like cells reducing their malignant properties both in vitro and in vivo. It is important to notice that the approach taken here is based on direct reprogramming, but whether the mechanism could be considered as reprogramming per se or rather forced differentiation is, due to the limited but existing potential of some GBM cells to spontaneously differentiate to astrocytes, debatable and a matter of definitions.
Nonetheless, converted cells display typical astrocyte morphology, increased expression of astrocytic markers, acquired astrocytic functions, and decrease their proliferative potential. Most importantly, forced differentiation reduces tumorigenicity of GBM cells in a xenotransplantation setting.
The role of Sox9, Nfia and Nfib, in astrocyte specification and differentiation is well established (8). Specifically, Sox9 governs both initial stages of neurogenesis and the subsequent gliogenic switch (31, 32), while Nfia and Nfib are involved at the onset of gliogenesis, differentially regulating astrocyte development (33).
These three transcription factors have also been shown to play important roles as fate regulators in the developmental evolution of gliomas. In astrocytomas, higher expression of Nfia and Nfib correlate with higher differentiation status and lower grade tumors (11, 12). On the other hand, Sox9 expression has been shown to correlate with glioma severity, and Sox9 has been proposed as a regulator of gliomagenesis (34). However, although Sox9 is suggested to promote rather than suppress cancer cell proliferation (35), our findings demonstrate that Sox9 overexpression, in combination with either Nfia or Nfib, can reduce tumorigenicity by driving GBM cells to differentiate towards an astrocytic fate both in vitro and in vivo. Indeed, we found that two combinations of these factors (Nfia and Sox9-Nfib) efficiently convert GBM cells to astrocyte-like cells. These differential requirements are in line with previous findings that Sox9 and Nfib are among the original factors identified to reprogram mouse (9) and human (36) fibroblasts to astrocytes and sufficient to efficiently induce astrocyte differentiation in human pluripotent cells (10). Nfia on the other hand, have been shown to be crucial in the gliogenic switch and in driving differentiation of human neural stem cells to astrocytes (37). This is intriguing considering the high reprogramming efficiency shown by Nfia alone on GSC lines. The different requirements on reprogramming factors of these two cell lines might thus reflect differences in GBM subtypes, cell of origin, driving mutations or developmental stages.
Taking the substantial differences between the four cell lines in consideration, it is striking how reprogramming converges to a similar outcome, with cells bearing morphologic and transcriptional features of normal astrocytes and displaying similar functional properties.
Several lines of evidence support our conclusion that conversion induces an astrocyte fate in GBM cells. First, cells undergo substantial morphologic change, mimicking human adult astrocyte in vitro and in vivo morphology (38). Second, we demonstrated a marked upregulation of canonical astrocyte markers such as GFAP at transcriptional and protein level, and a consistent reduction in proliferation rate.
It is noteworthy that GFAP transcriptional activity and protein levels undergo substantial changes in human astrocytoma (17). Indeed, loss of expression and decrease in amount of GFAP filaments might indicate a general dedifferentiation in brain tumors (16). Third, cells display typical astrocyte functions such as regulated calcium signaling and response to inflammatory stimuli similar to normal and stem cell derived astrocytes (22, 39, 40). Moreover, our transcriptomic analysis did not detect expression of pluripotency, neural stem cell, neuronal or oligodendrocytic genes in treated cells. This is consistent with our previous studies showing close to complete astrocytic induction from human pluripotent stem cells (10) and direct conversion of human fibroblasts to astrocytes using the same transcription factors (36). Taken together, it is unlikely that forced differentiation induces alternative fates other than astroglial.
Conversion markedly reduced proliferation in vitro and in vivo, and tumor growth in vivo. It is worth noting that doxycycline, used to induce transgene expression in this study, have been shown to affect tumor growth (41) and could potentially also selectively affect astrocytes. However, we controlled for a potential effect by equally treating all samples, including controls, with doxycycline.
Further work is needed to address the stability of converted GBM cells. Although we did not observe any alternative cell-fate in vitro, the possible effect of hypoxia/necrosis in cell cultures remains to be investigated. In addition, interactions between cancer and immune cells could potentially alter the state of GBM cells in vivo, driving transitions to different cell-fates in the long-term. In this regard, decreased immune cell and macrophage populations in NSG mice, absence of hypoxia and relative short survival in in vivo experiments could mask or prevent tumor evolution such as the transition to a mesenchymal subtype of GBM (42).
The perspective to develop a new therapy for GBMs based on reprogramming builds on early studies of forced differentiation (43, 44). However, these were mainly dependent on growth factors and small molecules, which would be difficult if not impossible to translate into effective therapies due to a lack of specific effects and difficulties in localized delivery. A transcription factor-based reprogramming approach, as described here, has a clearer path towards the clinic as a concept of viral vector-mediated gene therapy, which is already in GBM clinical trials (45). The potential of lentiviral-based gene therapy approaches has been widely explored, and already led to successful clinical applications (46). However, further work is needed to identify which viral-mediated delivery system would be optimal for in vivo differentiation of preestablished tumors.
Other issues needing to be addressed include the high inter- and intratumoral cell type heterogeneity in GBM. The cell lines used here reflect heterogeneity in terms of growth rate and invasiveness in vivo. Successful reprogramming of these different lines, indicate that a reprogramming strategy might indeed have potential to convert heterogeneous cell populations towards similar phenotypic fate.
Further supporting our approach as a potential therapeutic option, previous studies have described reprogramming of GBM cells to neuron-like cells (5–7). However, addition of new neurons to the brain poses a risk of altering excitability and inducing seizures. In addition, so far, no neuronal reprogramming study has convincingly shown reduced tumorigenicity in vivo. In contrast, our results show strikingly reduced tumor volume and proliferation as well as upregulation of GFAP in vivo. Oligodendrocytic reprogramming may represent an alternative option, but so far most protocols rely on a transient progenitor stage, which could mimic tumor dedifferentiation, and risks related to excessive myelin supply (47) raise concerns about the suitability of this strategy.
Overall, the potential of astrocytes as antigen-presenting cells and their active role in the immunologic function represents a strategic added value for further exploration of this strategy.
Additional work is needed to increase confidence in the robustness of astrocytic differentiation as a therapeutic option for GBM and rule out potential negative effects such as excessive gliosis and alteration of neuronal activity. Nonetheless, we believe that our results represent a promising step in the preclinical development of a potential gene-therapy approach, based on reprogramming towards astrocytic fate, as a new therapeutic approach for GBM.
Our findings raise the question whether targeted therapies can be applicable to a heterogeneous type of tumor such as GBM and if priority should be given on decreasing intra and inter-tumor heterogeneity as such. A clinical scenario in which viral-mediated transcription factor-induced differentiation can be offered to patients with non-resectable or partially resected GBM can be envisaged. Future exciting prospects include the possibility to convert undifferentiated GSCs, resistant to conventional treatments, into more well-differentiated cells, unable to cause tumor recurrence.
Supplementary Material
Supplementary figures and table
Acknowledgments
We are grateful to Dr. Zaal Kokaia for sharing laboratory equipment. The authors also thank the StemTherapy Imaging Core Facility and FACS Core Facility at the Lund Stem Cell Center for excellent technical assistance.
F. Trovato and J. Li were supported by Elsa Schmitz´ foundation and J. Li by the Segerfalk foundation (501100006686); I. Canals by a grant from the Swedish Society of Medical Research (S20–0003); H. Ahlenius by grants from the Swedish Cancer Society (20 0957 PjF); and J. Bengzon by grants from the Swedish state under the agreement between the Swedish government and the county councils, the ALF-agreement, Region Skåne, and donations from Viveca Jeppsson and Maj-Britt and Allan Johansson.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
F. Trovato: Data curation, formal analysis, funding acquisition, investigation, visualization, writing-original draft. F.R. Stefani: Conceptualization, data curation, formal analysis, investigation. J. Li: Investigation. O.G. Zetterdahl: Investigation. I. Canals: Conceptualization, supervision. H. Ahlenius: Conceptualization, resources, supervision, funding acquisition, validation, investigation, methodology, writing-original draft, project administration. J. Bengzon: Conceptualization, resources, supervision, funding acquisition, validation, writing-original draft, project administration.
References
- 1. Louis DN, Perry A, Wesseling P, Brat DJ, Cree IA, Figarella-Branger D, et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol 2021;23:1231–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ostrom QT, Patil N, Cioffi G, Waite K, Kruchko C, Barnholtz-Sloan JS. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2013–2017. Neuro Oncol 2020;22:iv1–iv96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Koshy M, Villano JL, Dolecek TA, Howard A, Mahmood U, Chmura SJ, et al. Improved survival time trends for glioblastoma using the SEER 17 population-based registries. J Neurooncol 2012;107:207–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer 2013;108:479–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao J, He H, Zhou K, Ren Y, Shi Z, Wu Z, et al. Neuronal transcription factors induce conversion of human glioma cells to neurons and inhibit tumorigenesis. PLoS One 2012;7:e41506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oh J, Kim Y, Che L, Kim JB, Chang GE, Cheong E, et al. Regulation of cAMP and GSK3 signaling pathways contributes to the neuronal conversion of glioma. PLoS One 2017;12:e0178881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Su Z, Zang T, Liu ML, Wang LL, Niu W, Zhang CL. Reprogramming the fate of human glioma cells to impede brain tumor development. Cell Death Dis 2014;5:e1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature 2010;468:214–22. [DOI] [PubMed] [Google Scholar]
- 9. Caiazzo M, Giannelli S, Valente P, Lignani G, Carissimo A, Sessa A, et al. Direct conversion of fibroblasts into functional astrocytes by defined transcription factors. Stem Cell Reports 2015;4:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Canals I, Ginisty A, Quist E, Timmerman R, Fritze J, Miskinyte G, et al. Rapid and efficient induction of functional astrocytes from human pluripotent stem cells. Nat Methods 2018;15:693–6. [DOI] [PubMed] [Google Scholar]
- 11. Song HR, Gonzalez-Gomez I, Suh GS, Commins DL, Sposto R, Gilles FH, et al. Nuclear factor IA is expressed in astrocytomas and is associated with improved survival. Neuro Oncol 2010;12:122–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stringer BW, Bunt J, Day BW, Barry G, Jamieson PR, Ensbey KS, et al. Nuclear factor one B (NFIB) encodes a subtype-specific tumor suppressor in glioblastoma. Oncotarget 2016;7:29306–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Quist E, Ahlenius H, Canals I. Transcription factor programming of human pluripotent stem cells to functionally mature astrocytes for monocultures and cocultures with neurons. Methods Mol Biol 2021;2352:133–48. [DOI] [PubMed] [Google Scholar]
- 14. Sanchez-Freire V, Ebert AD, Kalisky T, Quake SR, Wu JC. Microfluidic single-cell real-time PCR for comparative analysis of gene expression patterns. Nat Protoc 2012;7:829–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xie Y, Bergström T, Jiang Y, Johansson P, Marinescu VD, Lindberg N, et al. The human glioblastoma cell culture resource: validated cell models representing all molecular subtypes. EBioMedicine 2015;2:1351–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang S, Chen G, Dang Y, Chen LH. Overexpression of DcR3 and its significance on tumor cell differentiation and proliferation in glioma. The Scientific World Journal 2014;2014:605236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Qin JB, Liu Z, Zhang H, Shen C, Wang XC, Tan Y, et al. Grading of gliomas by using radiomic features on multiple magnetic resonance imaging (MRI) sequences. Med Sci Monit 2017;23:2168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sereika M, Urbanaviciute R, Tamasauskas A, Skiriute D, Vaitkiene P. GFAP expression is influenced by astrocytoma grade and rs2070935 polymorphism. J Cancer 2018;9:4496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ligon KL, Huillard E, Mehta S, Kesari S, Liu H, Alberta JA, et al. Olig2-regulated lineage-restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron 2007;53:503–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rowitch DH. Glial specification in the vertebrate neural tube. Nat Rev Neurosci 2004;5:409–19. [DOI] [PubMed] [Google Scholar]
- 21. Sofroniew MV, Vinters HV. Astrocytes: biology and pathology. Acta Neuropathol 2010;119:7–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liddelow SA, Barres BA. Reactive astrocytes: production, function, and therapeutic potential. Immunity 2017;46:957–67. [DOI] [PubMed] [Google Scholar]
- 23. Nduom EK, Weller M, Heimberger AB. Immunosuppressive mechanisms in glioblastoma. Neuro Oncol 2015;17:vii9–vii14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Semyanov A, Henneberger C, Agarwal A. Making sense of astrocytic calcium signals — from acquisition to interpretation. Nat Rev Neurosci 2020;21:551–64. [DOI] [PubMed] [Google Scholar]
- 25. MacDonald CL. Calcium dynamics in astrocytes: from oscillations to Alzheimer's. Diss Abstr Int 2012.
- 26. Pasti L, Volterra A, Pozzan T, Carmignoto G. Intracellular calcium oscillations in astrocytes: a highly plastic, bidirectional form of communication between neurons and astrocytes in situ. J Neurosci 1997;17:7817–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dong Q, Liu Q, Li R, Wang A, Bu Q, Wang KH, et al. Mechanism and consequence of abnormal calcium homeostasis in Rett syndrome astrocytes. Elife 2018;7:e33417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li J, Ek F, Olsson R, Belting M, Bengzon J. Glioblastoma CD105+ cells define a SOX2− cancer stem cell–like subpopulation in the preinvasive niche. Acta Neuropathol Commun 2022;10:126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Himes BT, Geiger PA, Ayasoufi K, Bhargav AG, Brown DA, Parney IF. Immunosuppression in glioblastoma: current understanding and therapeutic implications. Front Oncol 2021;11:770561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Colombo E, Farina C. Astrocytes: key regulators of neuroinflammation. Trends Immunol 2016;37:608–20. [DOI] [PubMed] [Google Scholar]
- 31. Esain V, Postlethwait JH, Charnay P, Ghislain J. FGF-receptor signaling controls neural cell diversity in the zebrafish hindbrain by regulating olig2 and sox9. Development 2010;137:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev 2003;17:1677–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Deneen B, Ho R, Lukaszewicz A, Hochstim CJ, Gronostajski RM, Anderson DJ. The transcription factor NFIA controls the onset of gliogenesis in the developing spinal cord. Neuron 2006;52:953–68. [DOI] [PubMed] [Google Scholar]
- 34. Swartling FJ, Ferletta M, Kastemar M, Weiss WA, Westermark B. Cyclic GMP-dependent protein kinase II inhibits cell proliferation, Sox9 expression, and Akt phosphorylation in human glioma cell lines. Oncogene 2009;28:3121–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang L, He S, Yuan J, Mao X, Cao Y, Zong J, et al. Oncogenic role of SOX9 expression in human malignant glioma. Med Oncol 2012;29:3484–90. [DOI] [PubMed] [Google Scholar]
- 36. Quist E, Trovato F, Avaliani N, Zetterdahl OG, Gonzalez-Ramos A, Hansen MG, et al. Transcription factor-based direct conversion of human fibroblasts to functional astrocytes. Stem Cell Reports 2022;17:1620–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tchieu J, Calder EL, Guttikonda SR, Gutzwiller EM, Aromolaran KA, Steinbeck JA, et al. NFIA is a gliogenic switch enabling rapid derivation of functional human astrocytes from pluripotent stem cells. Nat Biotechnol 2019;37:267–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Oberheim NA, Takano T, Han X, He W, Lin JHC, Wang F, et al. Uniquely hominid features of adult human astrocytes. J Neurosci 2009;29:3276–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature 2017;541:481–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Araque A, Carmignoto G, Haydon PG, Oliet SHR, Robitaille R, Volterra A. Gliotransmitters travel in time and space. Neuron 2014;81:728–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhong W, Chen S, Zhang Q, Xiao T, Qin Y, Gu J, et al. Doxycycline directly targets PAR1 to suppress tumor progression. Oncotarget 2017;8:16829–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hara T, Chanoch-Myers R, Mathewson ND, Myskiw C, Atta L, Bussema L, et al. Interactions between cancer cells and immune cells drive transitions to mesenchymal-like states in glioblastoma. Cancer Cell 2021;39:779–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kang TW, Choi SW, Yang SR, Shin TH, Kim HS, Yu KR, et al. Growth arrest and forced differentiation of human primary glioblastoma multiforme by a novel small molecule. Sci Rep 2014;4:5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee C, Robinson M, Willerth SM. Direct reprogramming of glioblastoma cells into neurons using small molecules. ACS Chem Neurosci 2018;9:3175–85. [DOI] [PubMed] [Google Scholar]
- 45. Banerjee K, Núñez FJ, Haase S, McClellan BL, Faisal SM, Carney SV, et al. Current approaches for glioma gene therapy and virotherapy. Front Mol Neurosci 2021;14:621831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Naldini L, Trono D, Verma IM. Lentiviral vectors, two decades later. Science 2016;353:1101–2. [DOI] [PubMed] [Google Scholar]
- 47. Almeida RG, Pan S, Cole KLH, Williamson JM, Early JJ, Czopka T, et al. Myelination of neuronal cell bodies when myelin supply exceeds axonal demand. Curr Biol 2018;28:1296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and table
Data Availability Statement
The data generated in this study are available upon request from the corresponding author.