Abstract
Activating mutations in mitogen-activated protein kinase kinase 1 (MAP2K1) are involved in a variety of cancers and may be classified according to their RAF dependence. Sensitivity to combined BRAF and MEK treatments is associated with co-mutations of MAP2K1 and BRAF; however, the significance of less frequent MAP2K1 mutations is largely unknown. The transforming potential and drug sensitivity of 100 MAP2K1 variants were evaluated using individual assays and the mixed-all-nominated-in-one method. In addition, A375, a melanoma cell line harboring the BRAF V600E mutation, was used to evaluate the function of the MAP2K1 variants in combination with active RAF signaling. Among a total of 67 variants of unknown significance, 16 were evaluated as oncogenic or likely oncogenic. The drug sensitivity of the individual variants did not vary with respect to BRAF inhibitors, MEK inhibitors (MEKi), or their combination. Sensitivity to BRAF inhibitors was associated with the RAF dependency of the MAP2K1 variants, whereas resistance was higher in RAF-regulated or independent variants compared with RAF-dependent variants. Thus, the synergistic effect of BRAF and MEKis may be observed in RAF-regulated and RAF-dependent variants. MAP2K1 variants exhibit differential sensitivity to BRAF and MEKis, suggesting the importance of individual functional analysis for the selection of optimal treatments for each patient. This comprehensive evaluation reveals precise functional information and provides optimal combination treatment for individual MAP2K1 variants.
Introduction
The mitogen-activated protein kinase (MAPK) signaling pathway (RAS/RAF/MEK/ERK pathway) is one of the most widely studied signal transduction cascades. It regulates various cellular processes including proliferation, differentiation, motility, transcriptional regulation, survival, and apoptosis (1). It is activated by upstream receptor tyrosine kinases, and deregulation of the MAPK pathway contributes to malignant transformation and tumor growth. Further, strength of the downstream ERK signal defines the negative feedback (2, 3). Recurrent mutations within the KRAS, NRAS, BRAF, and MAP2K1 genes are commonly found in human malignancies (4) including melanoma (5, 6), lung cancer (7, 8), gastric cancer (9), colon cancer (10, 11), and ovarian cancer (12, 13).
MAP2K1, also known as MAPK/ERK kinase 1 (MEK1), is a serine/threonine and tyrosine kinase that is activated by RAF kinases. The structure of human MAP2K1 protein consists of a central protein kinase domain (68–381 amino acids) containing an activation loop and a proline-rich domain (PRD; ref. 14). It is activated through phosphorylation of S218 and S222; thus, mutations affecting these sites abolish MEK and ERK activation (15). MEK1 is also deregulated by feedback phosphorylation on the T292 site of the PRD by ERK1 and ERK2 activation (16).
MEK inhibitors (MEKi) inhibit cell proliferation and the growth of BRAF V600E-mutated cancer cells (17). Trametinib as a monotherapy results in increased progression-free survival compared with dacarbazine in metastatic melanoma harboring BRAF mutations (18). This was followed by the development of combination therapy with BRAFi and MEKi in melanoma (19–22), colorectal cancer (23) and non–small cell lung cancer (24). Currently available MEKis exhibit high selectivity because they are not ATP-competitive, rather they bind to an allosteric site adjacent to the ATP pocket (25). Combination treatments with MEKis and ATP-competitive ERK inhibitors are currently undergoing clinical trial (Phase I to III; ref. 26).
Recent studies have revealed various mechanisms of innate, adaptive, and acquired resistance to MEKi and BRAFi. First, the dysregulation of cyclin D1 (CCND1) and cyclin-dependent kinases 4 and 6 (CDK4 and CDK6) results in resistance to MAPK pathway inhibition (27). Second, MEKi induces negative feedback of the EGFR-HER3 and PI3K-dependent signaling pathways and reactivates the MAPK pathway (28, 29). Third, acquired resistance mutations occur in MAP2K1 during exposure to MEKi. For instance, MAP2K1 P124 L was identified in a metastatic melanoma with a BRAF V600E mutation after treatment with selumetinib (2). MAP2K1 L115P, F129L, and V211D decrease the affinity of MEKi binding (10, 25, 30–32). Amplification of BRAF V600E is also one mechanism of acquired resistance to MEKi (32).
Several MAP2K1 mutations have been identified as oncogenic by an evaluation of the transforming activity or the phosphorylation of MEK and ERK (9, 13, 33–40). A previous study established three different classes within MAP2K1 mutations according to RAF-dependency (41). RAF-dependent class I mutants (D67N, P124L/S, L177V) are phosphorylated and activated by RAF, RAF-regulated class II mutants (F53_Q58del, F53L, Q56P, K57E/N, C121S, L177M, E203K) are partially phosphorylated by RAF but retain its kinase activity independent of RAF, and RAF-independent class III mutants (I98_I103del, I99_K104del, E102_I103del, I103_K104del) auto-phosphorylate and activate downstream signals. Other uncommon mutations represent variants of unknown significance (42–48).
In the current study, we evaluated non-synonymous MAP2K1 mutations, which are recurrent in the COSMIC database (v84), and determined their pathogenicity as well as their RAF-dependency, which correlated with sensitivity to BRAF and MEKis. We used the mixed-all-nominated-in-one (MANO) method, which is a high-throughput functional assay developed in our laboratory (49–51).
Materials and Methods
Cell lines and culture conditions
Human embryonic kidney (HEK) 293T cells, 3T3 mouse fibroblasts, A375 human skin malignant melanoma cells, and A2058 human skin malignant melanoma cells were purchased from the ATCC (Manassas, VA) and cultured in DMEM-F12 supplemented with 10% FBS, 2 mmol/L glutamine, and 1% each of penicillin and streptomycin (all from Thermo Fisher Scientific, Waltham, MA). Ba/F3 cells were maintained in RPMI1640 medium (Thermo Fisher Scientific) supplemented with 10% FBS, 2 mmol/L glutamine, and 1% each of penicillin and streptomycin, and mouse IL3 (20 U/mL; Merck, Darmstadt, Germany). HT-29 human colon colorectal adenocarcinoma cells were purchased from the ATCC (Manassas, VA) and cultured in McCoy's 5A (modified) medium supplemented with 10% FBS, 2 mmol/L glutamine, and 1% each of penicillin and streptomycin. All cell lines were used for the experiments within ten passages from thawing. Cell line authentication and Mycoplasma testing were not carried out within 6 months.
Plasmid construction
The pCX6 vector was produced by inserting random 10-bp DNA barcode sequences upstream of the start codon of the genes of interest into the pCX4 vector (52). The full-length wild-type (WT) cDNA for human MAP2K1 (NM_002755), GFP, EGFR E746_A750del, KRAS G12V were cloned into the pCX6 vector. Recurrent 99 variants of MAP2K1 reported in the COSMIC database (v84) were selected for this study. MAP2K1 variant cDNAs were constructed using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA) with corresponding mutation-specific primers. cDNA fragments of the MAP2K1 fusion partners were constructed (Integrated DNA Technologies, Coralville, IA) and joined with fragments of the MAP2K1s using the NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs, Ipswich, MA). All plasmids were verified by Sanger sequencing. Three clones with specific barcodes were constructed for each variant to obtain triplicate data for each individual assay.
Transfection and transduction
The recombinant plasmids were transduced with the packaging plasmids (Takara Bio, Shiga, Japan) into HEK293T cells to obtain recombinant retroviral particles. The 3T3 cells were infected in 96-well plates with ecotropic recombinant retroviruses using 4 μg⁄mL Polybrene (Merck) for 24 hours. A375 cells were infected in 96-well plates with amphotropic retroviruses using 4 μg/mL Polybrene for 24 hours. Ba/F3 cells were plated in retronectin-coated (Takara Bio) 12-well plates and infected with the retroviruses in RPMI1640 medium containing 20 U/mL IL3. Two days after transfection, cells were cultured with 100 μg/mL of zeocin (Thermo Fisher Scientific) for 7 days.
Focus formation assay
The 3T3 cells expressing various MAP2K1 variants were cultured for 2 weeks by changing the medium to DMEM/F12 supplemented with 5% bovine serum (Thermo Fisher Scientific) for 2 weeks after the cells were nearly confluent. The cells were stained with Giemsa solutions. Focus formation assay was scored visually as follows: 1, no focus was observed; 2, focus of the transformed cells was partially observed; 3, diffuse transformed foci were observed; and 4, round-shaped, spheroid-formed and anchorage-independent foci were observed. Focus formation score (FFS) was defined by integrating the results of three technical triplicate experimental batches of the assays.
MANO method
A retroviral vector is used to enable stable integration of individual variants into the genome of the assay cells (such as 3T3 cells or A375 human melanoma cells) along with the 10 bp barcode sequences. Individually transduced assay cells are mixed and pooled in a competitive manner and cultured to assess transformation potential, cell proliferation, or drug sensitivity in vitro. At the end of the cell culture assay, genomic DNA was extracted from the cell lysates using the QIAmp DNA mini kit (Qiagen, Hilden, Germany) and PCR amplification was performed using the primers, 5′-TGGAAAGGACCTTACACAGTCCTG-3′ and 5′-GACTCGTTGAAGGGTAGACTAGTC-3′ (primer sequences are shown in Supplementary Table S1). The amplicons were purified on AMPure beads (Beckman Coulter, Bera, CA), and sequencing libraries were prepared using the NEBNext Q5 Hor Start HiFi PCR Master Mix (NEB) according to the manufacturer's instructions. Quality check of the libraries was assessed using a Qubit 2.0 fluorometer (Thermo Fisher Scientific) and the Agilent 2200 TapeStation System (Agilent). The libraries were then sequenced using an Illumina MiSeq system and the Reagent Kit V2 (300 cycles), and 150 bp paired-end reads were created (the sequencing primer loaded into the MiSeq cartridge is described in Supplementary Table S2). The barcode sequences 5′-CTAGACTGCCXXXXXXXXXXGGATCACTCT-3′ (where X denotes any nucleotide) were included in the sequencing results and the number of each barcode in each mutant was quantified.
Cell proliferation assay of MAP2K1 using the MANO method
The 3T3 cells expressing each MAP2K1 mutant were combined together at 2 days after infection, and the mixed cell population was maintained in 1.5% or 10% FBS for 14 to 17 days. Cell pellets were stored every 3 days and the experiment was executed in triplicate for both cell lines. We denote the timing at 3 days after cell mixing as “day 0″, and the cell mixture acquired on day 0 was used as a reference control to normalize the signal of each cell clone. The signal from each cell pellet collected every 3 days was designated as 100 × (average read number across replicates)/(average read number of the mixed cell population on day 0). The fold change in the ratio of MAP2K1-mutant cell number on day 14 or 17 relative to day 0 was compared with that of the WT MAP2K1 cell number to perform a paired t test. MAP2K1 mutants whose fold change increased significantly (P < 0.05) were regarded as activating mutant candidates.
Evaluation of sensitivity of MAP2K1 variants to BRAF or MEK inhibitors using the MANO method
The 3T3 cells expressing each MAP2K1 mutant was cultured in DMEM/F12 medium with 1.5% FBS. The transformed 3T3 cells were then incubated for 5 days with the indicated concentrations of the MEKi, trametinib (0.1 nmol/L to 10 nmol/L). A375 cells expressing each MAP2K1 mutant were cultured in DMEM/F12 medium with 10% FBS. Transduced A375 cells were mixed in equal amounts and incubated for 5 days with the indicated concentrations of each BRAF inhibitor, which included vemurafenib, dabrafenib, or encorafenib (0.1 nmol/L to 10 nmol/L); and each MEKi: cobimetinib, trametinib, or binimetinib (0.1 nmol/L to 10 nmol/L). The experiments were performed in triplicate. We determined the number of each barcode sequences using the MANO method. Considering the different doubling times of the integrated cells, DMSO-treated cell mixtures were used as the reference control for normalizing each cell clone signal. The relative growth inhibition of each cell clone was calculated as 100 × (average read number across triplicates)/(average read number of the DMSO control). Three BRAF inhibitors and three MEKis used in the drug sensitivity assay were purchased commercially: vemurafenib (LC Laboratories), dabrafenib (Selleckchem), encorafenib (Selleckchem), cobimetinib (Selleckchem), trametinib (LC Laboratories), binimetinib (Selleckchem), ulixertinib (ref. 53; Selleckchem), and GDC-0994 (ref. 54; Selleckchem).
Cell viability assay
A375 cells and HT-29 cells expressing MAP2K1 variants were seeded in 96-well plates at a density of 3.0 × 103 cells/well in DMEM/F12 medium or McCoy's 5A medium with 10% FBS, MEKi at 10 concentrations and BRAF inhibitor at 6 concentrations ranging from 0.1 nmol/L to 10 μmol/L for 5 days. The 3T3 cells expressing the MAP2K1 variants were cultured in 96-well plates in DMEM/F12 medium with 10% FBS (e.g., GFP and WT) or 1.5% FBS (e.g., transformed variants), and MEKi at 10 concentrations ranging from 0.1 nmol/L to 10 μmol/L for 5 days. Subsequently, after adding 10 μL of PrestoBlue (Thermo Fisher Scientific) to the plates and further incubation for 3 hours, fluorescence was measured at 0.1 seconds (excitation 530 nm, emission 590 nm). The fluorescence intensity of the wells without cells was used as negative controls, and dose–response curves were fit to the observed cell viabilities using the drc package in R language. The two-parameter sigmoidal function LL.2 was used with the following settings: y0 (response without drug) = 0, robust = “mean,” method = “Nelder-Mead.” The IC50 was defined as the inflection point in the dose–response curve.
Combinatorial synergy analysis
Drug synergy assays were performed on the basis of the zero interaction potency (ZIP) model (55). ZIP δ scores (the excessed inhibition rate over the estimation) were calculated by the R package Synergyfinder. The average ZIP scores across all of the combination doses were calculated.
Western blot analysis
Cells were treated with the indicated concentrations of MEKis in DMEM/F12 with 10% FBS for 4 hours. The cells were then lysed in 1% NP-40 lysis buffer containing protease and phosphatase inhibitors for 15 minutes on ice. The lysates were subjected to 7.5% SDS-PAGE, and to immunoblotting using primary antibodies against MEK1/2 (1:1,000, #4694), p44/42 ERK1/2 (1:1,000, #4695), phospho-p44/42 ERK1/2 (1:2,000, #4370), and GAPDH (#2118) from Cell Signaling Technology (Danvers, MA). The secondary antibody was horseradish peroxidase–linked anti-rabbit IgG (1:10,000, NA934V; GE Healthcare, Chicago, IL).
Statistical analysis
Statistical significance was evaluated using a Student t test for comparisons between two mutants in vitro (each MAP2K1 mutant versus WT MAP2K1 or GFP). For all comparisons, P < 0.05 was considered statistically significant.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper.
Code availability
The references of all source codes are included within Materials and Methods.
Results
Selection of MAP2K1 mutations in the COSMIC database
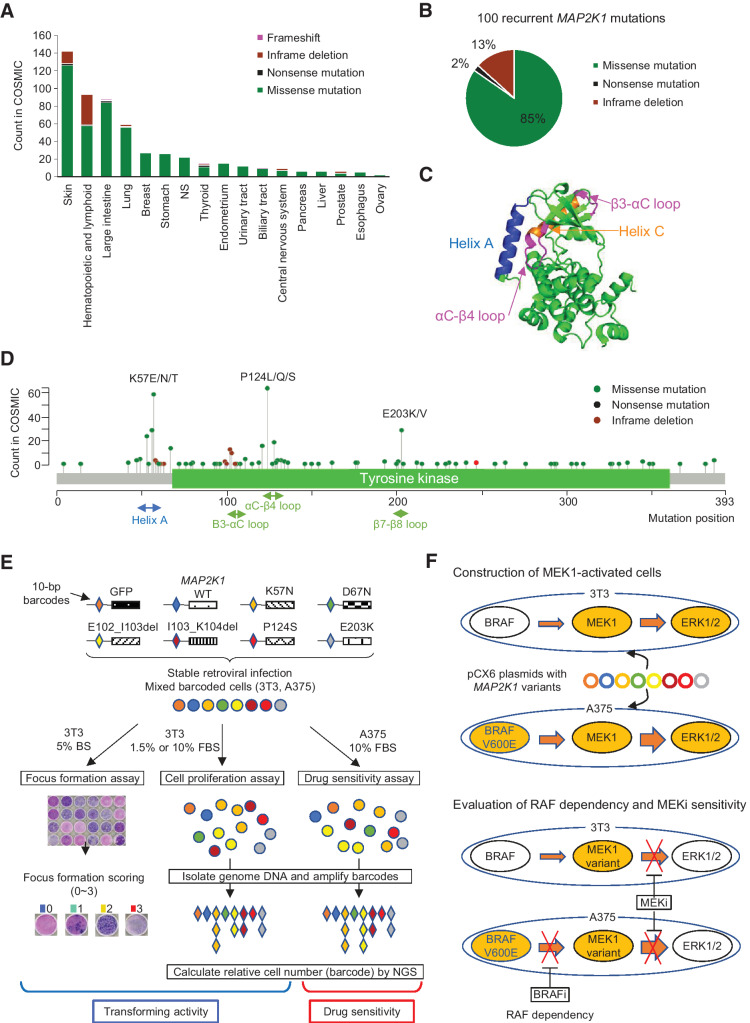
First, we selected 99 non-synonymous mutations in MAP2K1 recurrently identified in the COSMIC data base v84 (Supplementary Table S3). COSMIC currently contains the largest number of MAP2K1 variants compared with the AACR Project GENIE cBioPortal (https://genie.cbioportal.org/) or MSK-IMPCT data set. MAP2K1 mutations are found in a variety of cancer types with the highest frequency in malignant melanoma, followed by hematologic and lymphoid tumors, and colorectal cancer (Fig. 1A). The 99 recurrent mutations consist of missense mutations (85%), in-frame deletions (13%), and nonsense mutations (2%) (Fig. 1B).
Figure 1.
Spectrum of MAP2K1 variants, the structure of the MEK1 protein, and a schematic overview of the MANO method. A, Patterns of MAP2K1 mutations in various cancers. The bar charts illustrate the number of tumor samples with MAP2K1 mutations in the COSMIC database for each cancer type. Mutation types are coded in green (missense mutation), black (nonsense mutation), light green (inframe deletion), and pink (frameshift). B, Circle chart showing the percentage of MAP2K1 mutation types selected in this study. C, Categorization of MEK1 mutations in the crystal structure location. Helix A (blue), Helix C (orange) as well as β3-αC loop and αC-β4 loop (pink) are highlighted. D, Distribution of MAP2K1 mutations detected in the COSMIC database. E, Schematic representation of the MANO method. 3T3 cells and A375 cells were infected with recombination retrovirus-expressing MAP2K1 variants with individual 10 bp barcodes. The transforming activity of MAP2K1 variants in 3T3 cells was evaluated using a focus formation assay. Equal numbers of stabilized, transduced cells were mixed and cultured with two types of medium in 3T3 cells or treated with drugs in A375 cells. Genomic DNA was extracted from the cells at the end of each experiment. The barcode sequences were PCR-amplified and subjected to deep sequencing to quantitate the relative abundance. F, Construction of MEK-dependent cells and evaluation of MEKi sensitivity against MEK1 variants. Both cell lines were transfected with MAP2K1 variants by retrovirus using pCX6 plasmid to generate MEK-dependent cell lines. MEK1-dependent 3T3 cells were assessed for drug sensitivity and RAF dependency, and sensitivity to MEKis was evaluated in A375 cells transfected with MAP2K1 variants, with or without RAF inhibitors.
The location of mutations in the protein structure of MAP2K1 is depicted as a lolliplot (Fig. 1C and D). Mutations in the negative regulatory helix A, αC-β4 loop and β7-β8 loop, lead to the RAF-dependent or RAF-regulated MEK1 functions frequently observed in hematologic, skin, and colorectal cancers (56). In contrast, in-frame deletions in the β3-αC loop, which are mainly observed in Langerhans cell histiocytosis and malignant melanoma, belong to the previously reported RAF-independent class (57).
Establishment of MANO method for A375
Previous studies with the MANO method used Ba/F3 and 3T3 cell lines for the functional assay (49, 50). Whereas highly oncogenic variants of MAP2K1 can transform Ba/F3 cells, not all MAP2K1 oncogenic variants abrogate IL3 dependency in Ba/F3 cells. Therefore, 3T3 cells were used to evaluate the oncogenicity of MAP2K1 variants in the presence of physiologic RAF signaling. The loss of contact inhibition, one aspect of transforming activity, promoted by MAP2K1 variants was assessed using a focus formation assay (Fig. 1E).
In addition, we used A375, a melanoma cell line harboring a BRAF V600E mutation, to measure the function of MAP2K1 variants in combination with active RAF signaling (Fig. 1E). In the A375 cell model, treatment with BRAF inhibitors determines RAF-dependency (Supplementary Fig. S1), while treatment with MEKis assesses the sensitivities of MAP2K1 variants to MEKis (Fig. 1F).
Focus formation assay of MAP2K1 variants
The focus formation assay revealed strong transforming activities in the variants of the helix A domain (Q56_G61delinsR, Q56P, K57N/T, K57_G61del, V60E) and β3-αC loop deletion (H100_I103delinsPL, E102_K103del, I103_K104del; Supplementary Fig. S2). Moderate transforming activities were observed in some helix A (F53C/I/L/V, K57E), helix-C (I99_K104del, I103N, P105_106del, I111S, L115P, H119Y, C121S) and β7-β8 loop (E203K) mutations.
P124L/S, the most recurrent MAP2K1 mutations, exhibited only a partial focus formation, while the WT MAP2K1 did not form any foci. We then evaluated the transforming potential of each variant with the FFS scoring system, which integrates the results of three technical triplicate and two biological duplicate experimental batches of the focus formation assays. These results suggest that the MAP2K1 mutations conferred different transforming activities (Supplementary Fig. S2).
Cell proliferation assay of MAP2K1 variants
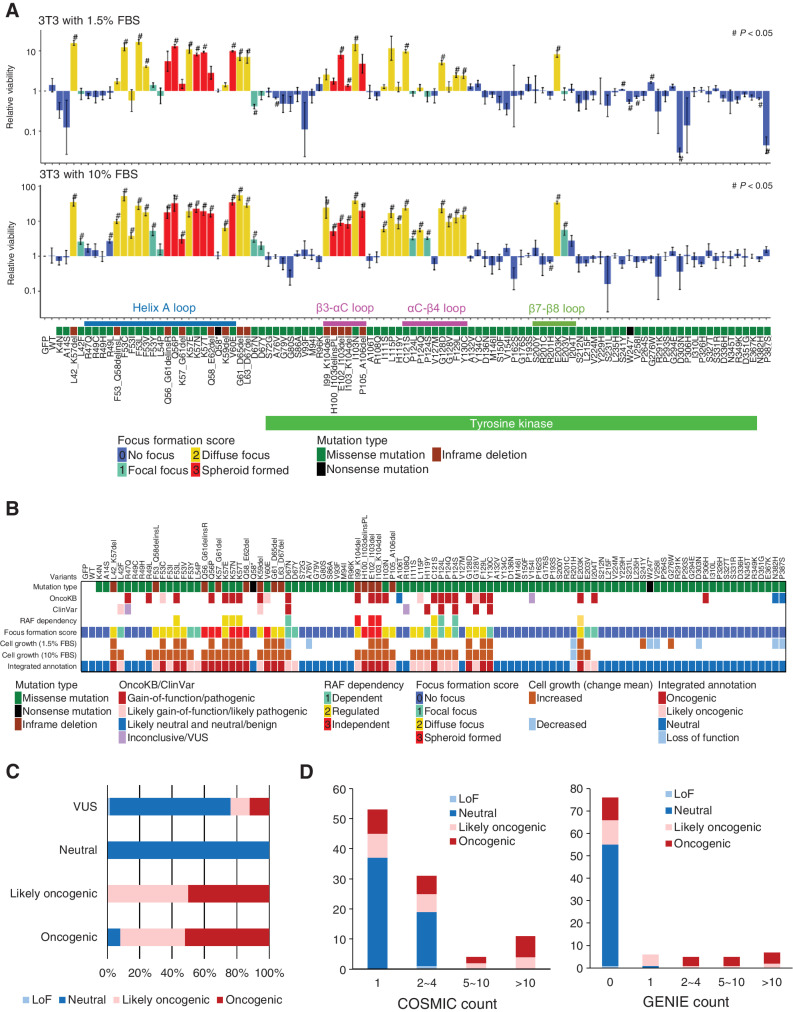
In addition to the loss of contact inhibition, cell proliferative capacity was evaluated using the MANO method. The MANO method compared the number of 3T3 cells expressing each MAP2K1 variant at day 0 and day 14 (10% FBS) or day 17 (1.5% FBS) to the rate of growth of the cells expressing MAP2K1 WT (Fig. 2A; Supplementary Fig. S3A and S3B; Supplementary Table S4). Protein expression of several MAP2K1 variants were evaluated by western blotting. Similar level (2.5–3.0 fold change) of protein expressions in MAP2K1 overexpressed cells compared with GFP introduced cells was observed (Supplementary Fig. S3C). The same variants were introduced into 293T cells (HEK cell line) to evaluate oncogenicity in normal human cells. The oncogenic variants annotated by the focus formation assay of 3T3 cells indicated significant increase in 293T cell growth compared with WT (Supplementary Fig. S3D).
Figure 2.
Transforming activity and functional annotation of MAP2K1 variants. A, Fold change from day 0 to day 14 (10% FBS; bottom) or day 17 (1.5% FBS; top) of 3T3 cells with respective MAP2K1 variants in the mixed cell population was computed using the MANO method and shown on a logarithmic scale as relative proliferation. Mutations with a relative proliferation significantly different from GFP (#) are shown (paired t test, P < 0.05). The color of the bars indicates the FFS based on the focus formation assay and sorted according to the amino acid position. The error bars quantify the mean fold change across the three replicates and indicate the standard error. B, For each of 100 MAP2K1 variants (at the top of this figure), the mutation types and clinical significance annotated in OncoKB and ClinVar databases are presented in the top three rows. Below that, RAF dependency is shown according to Y. Gao and colleagues (41). Cell growth (1.5% or 10% FBS) is shown by increased (brown) and decreased (light blue) mutations (#) with significant differences in the cell growth assay (shown in Fig. 3A), respectively. C, The oncogenicity evaluated by the method is compared with that of OncoKB. OncoKB annotations are shown below the stacked bar chart. D, The results of the focus formation assay and growth competition assay by the MANO method were summarized to annotate the oncogenicity of the variants according to the classification described in the method. The oncogenicity evaluated by this method is compared with the variant count number of COSMIC or AACR Project GENIE. Error bars indicate SD.
The results of the cell proliferation assay using the MANO method and the focus formation assay were correlated (Supplementary Fig. S3E). The variants with strong transforming activity (FFS = 2, 3) exhibited a significant growth advantage in both 1.5% and 10% FBS, whereas there were no significant differences in cell proliferation between FFS 2 and 3. In contrast, the FFS 1 mutation exhibited a significant growth advantage under 10% FBS conditions, although growth was significantly slower compared with that of FFS 2 (Supplementary Fig. S3E). Several FFS 0 mutations (A76V, W247*, V258I, D303N, N382H, P387S) revealed slower growth compared with the WT in 1.5% FBS.
Integrated annotation of oncogenicity of MAP2K1 variants
The results of focus formation assay and growth competition assay by the MANO method were summarized to annotate the oncogenicity of variants according to the following classification (Supplementary Table S3): oncogenic as FFS 1 or higher and significantly faster growth compared with WT; likely oncogenic as FFS 1 or higher, but not significantly faster growth compared with WT; loss-of-function (LoF) as FFS 0 and significantly slower growth compared with WT; and neutral as none of above.
The concordance between the annotation of the current study and that of OncoKB and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) was evaluated (Fig. 2B and C). The results of gain-of-function (GoF)/pathogenic and likely gain-of-function/likely pathogenic in OncoKB and ClinVar, were mostly consistent with that of our annotation. A discrepancy was observed in the evaluation of R47Q, R49L, and P306H, which are considered GoF in OncoKB, whereas they did not show any activity in our assay. Our integrated annotation was compared with variant counts in COSMIC and GENIE. The recurrent variants in COSMIC and GENIE tended to be annotated as oncogenic or likely oncogenic in our annotation, thus validating our assessment (Fig. 2D).
By combining the results of the proliferative capacity and susceptibility tests, we created a four-level classification. The agreement rate between the new classification and the existing classification was generally consistent. From this experiment, 67 VUSs were functionally analyzed. We annotated 16 new variants (e.g., helix-A variants; F53I/V/Y etc.) as oncogenic and 51 variants as neutral or LoF.
Sensitivity of MAP2K1 variants to MEK1 and BRAF inhibitors
The drug sensitivity of the transformed MAP2K1 variants in 3T3 cells was evaluated by the MANO method. First, 3T3 cells with 29 mutants that survived 1.5% FBS culture were treated with various concentrations of trametinib, a MEKi. As shown in Supplementary Fig. S4 and Supplementary Table S5, trametinib was not effective to the β3-αC loop deletion (I99_K104del, E102_I03del, and P105_A106del) which are RAF-independent variants resistant to allosteric inhibition, and to L115P substitution, a known resistant mutation that abrogates the MEKi binding pocket. The other variants were sensitive to trametinib (IC50 < 0.5 nmol/L). EGFR E746_A750del and KRAS G12V were used as control variants. There were no remarkable differences in the sensitivity among helix A variants.
Next, A375 and HT-29, BRAF V600E–positive melanoma, and colon adenocarcinoma cell lines were used to evaluated the function of the MAP2K1 variants in combination with active RAF signaling. The most frequent MAP2K1 variants concurrent with BRAF V600 mutations is MAP2K1 P124L/Q/S in the COSMIC database, followed by P387S and G128D (Supplementary Fig. S5; Supplementary Table S6).
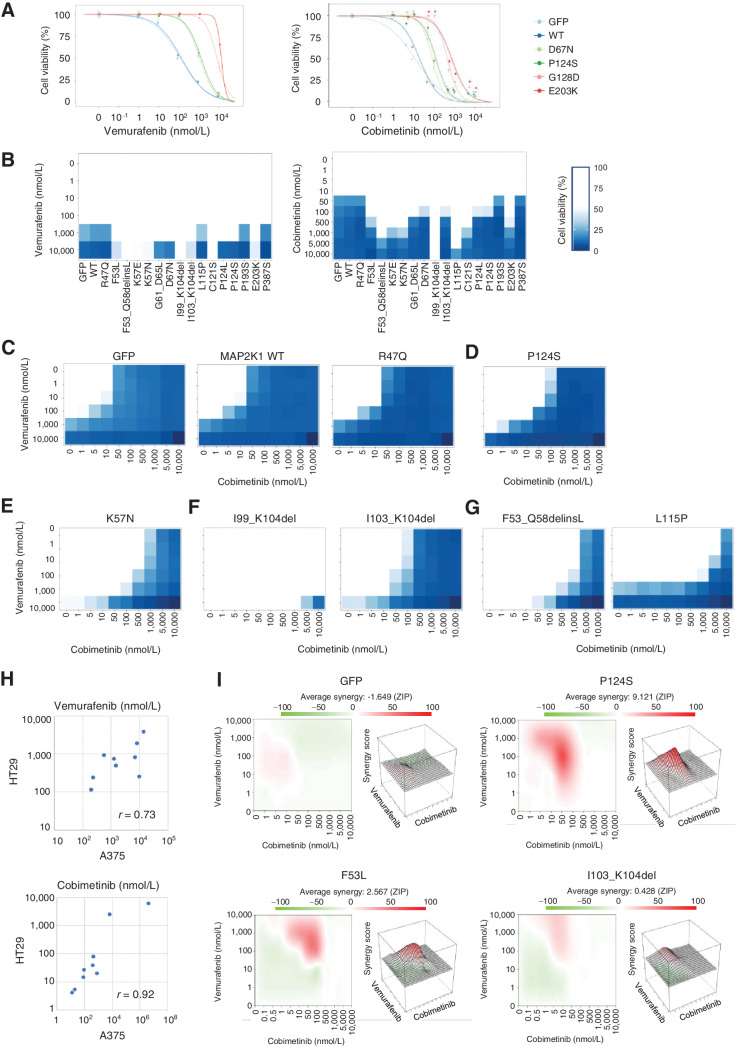
For a pilot study, five MAP2K1 variants including P124S and G128D were introduced into A375 and HT-29 to assess the sensitivities to a BRAF inhibitor (vemurafenib) and a MEKi (cobimetinib; Fig. 3A; Supplementary Fig. S6). The vemurafenib sensitivity of A375 changed to moderately resistant by expressing two MAP2K1 variants (D67N and P124S) and to highly resistant by G128D and E203K, whereas it was not changed by MAP2K1 WT or GFP. Considering that the BRAF-dependency of the MAP2K1 variants was evaluated by examining the effect of BRAF inhibition, D67N and P124S are supposed to be BRAF-regulated variants and G128D and E203K are BRAF-independent variants.
Figure 3.
The individual sensitivity of MAP2K1 variants to combination therapy of BRAF and MEK inhibitors in A375 cells. A, A375 cells transduced with four MAP2K1 variants, WT, and GFP were treated with DMSO, a BRAF inhibitor (vemurafenib, right), or a MEKi (cobimetinib, left) at the concentrations indicated for 5 days. Cell viability was measured using the PrestoBlue cell viability assay. The relative viability of the treated cells was measured in comparison with drug-free treatment. Data were plotted as the mean ± SD (n = 3). B, A375 cells transduced with 16 variants, WT, and GFP were treated with DMSO, a BRAF inhibitor (left), or MEKis (right) for 5 days. The relative viability of the results is illustrated using color shading heatmap compared with drug-free treatment. Data were plotted as the mean ± SD (n = 3). C–G, Results of cell viability assay using combination treatment with two drugs at different concentrations for each individual variant color shading heatmap. C, Parental (GFP) and no function. D, RAF dependent. E, RAF regulated. F, RAF independent. G, Others. H, The variant sensitivities evaluated in A375 and HT-29 were well correlated (r = 0.73 for vemurafenib and r = 0.92 for cobimetinib). I, The synergistic effect of the combination therapy of vemurafenib and cobimetinib, and the average synergy of the four variants of GFP, P124S, F53L, and I103_K104del were calculated (Materials and Methods). Those with high synergy are highlighted in red in the 2D figure on the left and are highly represented in the 3D landscape on the right.
In contrast, cobimetinib was used directly to evaluate the sensitivities of MAP2K1 variants. D67N and P124S were moderately resistant, and G128D and E203K were highly resistant to cobimetinib. The variant sensitivities against vemurafenib and cobimetinib assessed in HT-29 were changed to a lesser extent compared with that of A375.
Next, we expanded the cell viability assays to 18 MAP2K1 mutants using inhibitors consisting of three-drug combinations that are currently clinically available for the treatment of melanoma: vemurafenib + cobimetinib (VC; Fig. 3B–G; Supplementary Fig. S7), dabrafenib + trametinib (DT; Supplementary Fig. S8), and encorafenib + binimetinib (EB; Supplementary Fig. S9). The relative sensitivities of the MAP2K1 variants to BRAF or MAP2K1 inhibitors were well correlated among the inhibitors. The Fleiss’ kappa coefficient was used to evaluate the consistency of the classifications between the experiments with different drugs. The classification of drug sensitivity determined for individual variants among the three combinational therapies were completely consistent (Supplementary Table S7).
The representative sensitivities of nonfunctional (R47Q), RAF-dependent (P124S), RAF-regulated (K57N), RAF-independent (I99_K104del and I103_K104del), and others are shown in Fig. 3B–G. Notably, both being located in the β3-αC loop, the sensitivities of I99_K104del and I103_K104del were very different (Fig. 3F). L115P was sensitive to BRAF inhibitors, but highly resistant to MEKis (Fig. 3G).
Additional number of variants were also introduced into HT-29 cells to confirm the variant drug sensitivities in a different cell context. MEKi resistant variants observed in A375 such as L115P and I99_K104del were also evaluated as resistant in HT-29. The variant sensitivities evaluated in A375 and HT-29 were well correlated (r = 0.73 for vemurafenib and r = 0.92 for cobimetinib; Fig. 3H; Supplementary Fig. S10).
We determined the synergistic effects of the two drugs using an interaction potency model (Fig. 3I; ref. 55). Minimal synergy was observed in A375 with nonfunctional variants (e.g., GFP) for the combination of vemurafenib and cobimetinib, whereas strong synergy was observed in P124S and F53 L at a concentration of vemurafenib 100 nmol/L + cobimetinib 50 nmol/L and vemurafenib 100 nmol/L + cobimetinib 100 nmol/L, respectively. I103_K104del, a RAF-independent β3-αC loop deletion, exhibited only a small synergistic effect.
Inhibition of the downstream ERK signaling pathway by MEKis was determined by immunoblot analysis (Supplementary Fig. S11). ERK phosphorylation of A375 with D67N, P124S, F53L, E203K, and I103_K104 del were inhibited by 1 μmol/L trametinib for 4 hours, whereas that with I99_K104del was unchanged.
We further tested the vemurafenib and cobimetinib treatment to A2058 cell line harboring BRAF V600E and MAP2K1 P124S. Combination therapy has additive effect but synergistic effect. It was noted that dose–response curve of vemurafenib and cobimetinib monotherapy did not reveal sigmoid curves but linear effect, suggesting the cells is not completely dependent on the BRAF/MAPK pathway (Supplementary Fig. S12).
Sensitivity of MAP2K1 variants to MEK1 and BRAF inhibitors using the MANO method
A mixture of A375 cells expressing 99 MAP2K1 variants was treated with a combination therapy of three different BRAF and MEKis at various concentrations to assess the drug sensitivity using the MANO method. Expression of GFP or MAP2K1 K99_I104del conferred the cells sensitive or resistant to the treatments, respectively, confirming the validity of the assay (Fig. 4; Supplementary Figs. S13–S15). The Fleiss’ kappa coefficient for classification with the three-drug sets was 0.812, which was highly consistent (P < 0.001; Supplementary Table S8). The consistency of the classification between the MANO method and the individual experiments was evaluated using Cohen's kappa coefficient. The Cohen's kappa coefficients for VC, DT, and EB were as high as 0.882, 0.882, and 0.771, respectively (Supplementary Tables S7 and S8). This suggests that the drug sensitivities of the individual variants are similar among the three combination therapies. Fleiss's and Cohen's kappa coefficients were calculated with the irr package (v0.84.1).
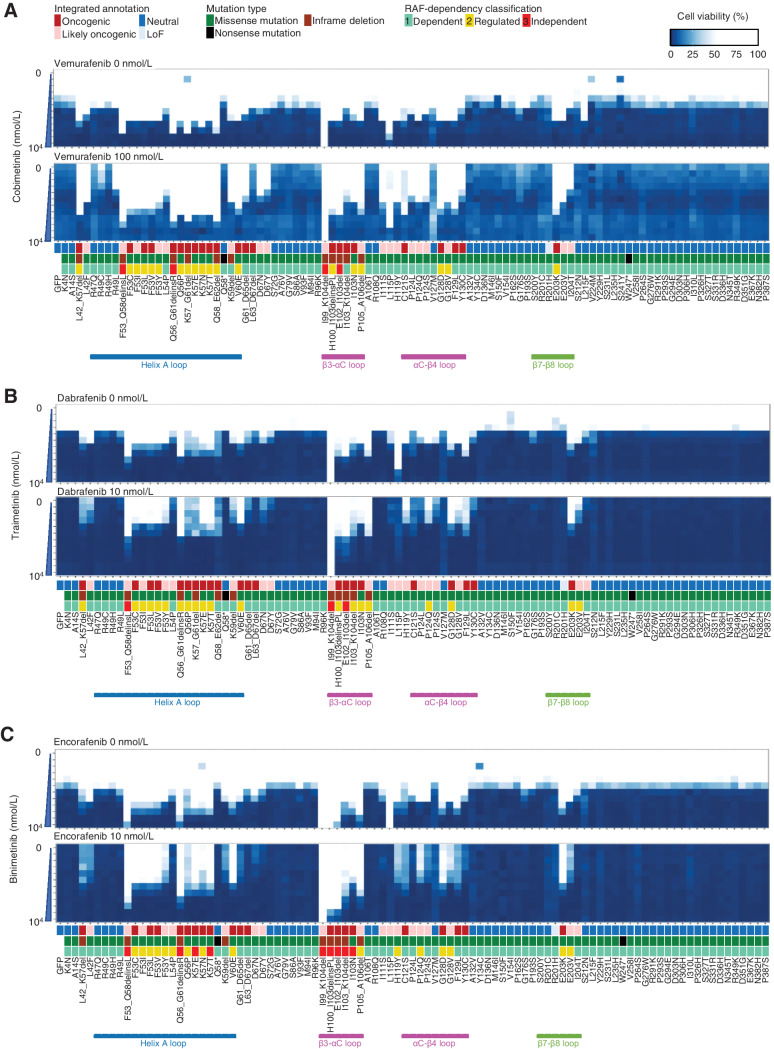
Figure 4.
The sensitivity of MAP2K1 variants to BRAF and MEK inhibitors using the MANO method. A375 cells with MAP2K1 variants, GFP were treated with DMSO or BRAF inhibitors (vemurafenib, dabrafenib, and encorafenib) and MEKis (cobimetinib, trametinib, and binimetinib) at the indicated concentrations. Concentrations of the BRAF and MEK inhibitors were varied by 6 (0 to 10,000 nmol/L) and 12 steps (0 to 10,000 nmol/L), respectively. The relative viability of the treated cells with each drug versus DMSO-treated cells was measured, and the results are presented using a color-coded scale. Data are presented as the mean ± SD (n = 3). A, Combination therapy with cobimetinib alone (top) and vemurafenib 100 nmol/L in addition to cobimetinib (bottom). B, Combination therapy with trametinib alone (top) and dabrafenib 10 nmol/L in addition to trametinib (bottom). C, Combination therapy with binimetinib alone (top) and encorafenib 10 nmol/L in addition to trametinib (bottom).
Cobimetinib monotherapy was effective at 5 nmol/L for the non-oncogenic variants, at 50 nmol/L for the RAF-dependent D67N and P124S variants, and 100 to 500 nmol/L for the RAF-regulated (F53 L and K57) and independent (E102_I103del, I103_K104del) variants. F53_Q58delinsL, a helix A deletion, I99_K104del, and L115P were highly resistant to cobimetinib (IC50 > 1,000 nmol/L; Fig. 4A; Supplementary Fig. S13; Supplementary Table S9).
The RAF-dependency of the MAP2K1 variants was investigated by determining the sensitivity to BRAF inhibitors. Because RAF signaling of A375 is abolished at 100 nmol/L of vemurafenib, vemurafenib treatment changed the RAF-dependent mutants into highly sensitive to cobimetinib (IC50 became ∼10 nmol/L). In contrast, vemurafenib treatment slightly changed the IC50 of RAF-regulated mutants to cobimetinib (IC50 became 100–500 nmol/L), whereas it did not change that of the RAF-independent mutants (Fig. 4A). Similar results were obtained when trametinib/dabrafenib or binimetinib/encorafenib was treated (Fig. 4B and C; Supplementary Figs. S14 and S15).
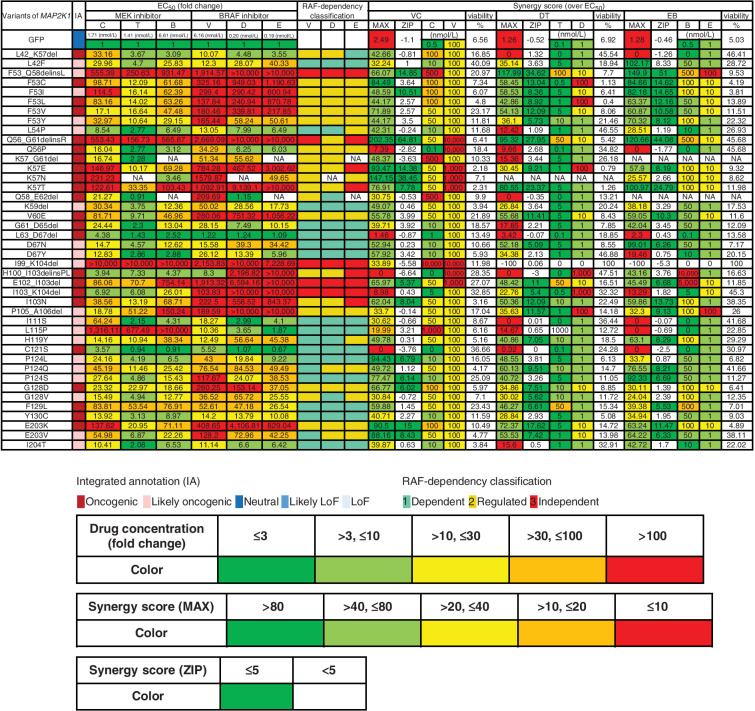
Summary data for the estimated drug sensitivities determined by the MANO method is shown in Fig. 5 and Supplementary Fig. S16. In general, the RAF-dependent group revealed weak oncogenicity, moderate drug sensitivity, and moderate synergistic effects in combination, whereas RAF-regulated mutants (such as in helix A, αC-β4 loop, and β7-β8) tended to be moderately resistant to the drugs, but revealed high synergy upon combination treatments. RAF-independent mutants were highly resistant and did not exhibit synergy with the inhibitors.
Figure 5.
Assessment of drug sensitivity and synergy for oncogenic MAP2K1 variants. The drug sensitivities of the indicated oncogenic MAP2K1 variant mutants were categorized into five levels; ≤ 3 (green); > 3, ≤ 10 (yellow-green); > 10, ≤ 30 (yellow); > 30, ≤ 100 (orange); and > 100 (red) based on the fold change (to GFP) of the IC50 value for each drug against each mutant. The maximum synergistic effect among the combinations of concentrations above the IC50 was calculated and concentrations of BRAF and MEK inhibitors in the case shown on the right side were categorized into five levels; > 80 (green); > 40, ≤ 80 (yellow-green); > 20, ≤ 40 (yellow); > 10, ≤ 20 (orange); and ≤ 10 (red). The concentration of the drug in each case is displayed (nmol/L) and colored on the basis of the fold change of IC50 for GFP. The mean of the synergy effect is shown in the ZIP, where ≥ 5 (green) is defined as having a synergy effect. B, binimetinib; C, cobimetinib; D, dabrafenib; DT, dabrafenib and trametinib; E, encorafenib; EB, encorafenib and binimetinib; EC50, half maximal effective concentration; NA, not applicable; T, trametinib; V, vemurafenib, VC, vemurafenib and cobimetinib.
The 51 variants evaluated as non-oncogenic using 3T3 out of 67 VUSs were further confirmed in A375 cells and exhibited no effect on drug sensitivity (Supplementary Fig. S16). Among the 16 mutations considered to be oncogenic, three mutations (L54P, D67Y, E203V) were RAF-dependent or -regulated variants considering the data with the focal focus formation assay and the sensitivity to BRAF inhibitors. Although another three mutations (K59del, G61_D65del, L63_D67del) exhibited high transforming activity, they may also be classified as RAF-dependent, because their activities are inhibited by RAF inhibitors. The other mutations are classified as RAF-independent (Fig. 5).
To further explore effective treatments to the mutations resistant to MEKis, two ERK inhibitors (ulixertinib and GDC-0994) were administered to A375 with GFP and five MAP2K1 variants. Both ulixertinib and GDC-0994 were less effective in MAPK1 mutants compared with the WT (Supplementary Fig. S17).
Discussion
Three types of active MAP2K1 mutations (RAF-independent, RAF-regulated, and RAF-dependent; ref. 41) respond differently to BRAF and MEKis, depending on the degree of dependence on BRAF. Therefore, we hypothesized that it was possible to classify them by measuring their response to the inhibitors.
There are two advantages of using the A375 cell line. First, because it is active in RAF signaling, the BRAF dependencies of the MAP2K1 variants can be properly evaluated in A375. Second, because it is a cancer cell line, it may be used to evaluate the drug sensitivity of non-oncogenic variants, which is difficult to assess in normal cells such as 3T3 or Ba/F3 cells. This modified MANO method is applicable to the evaluation of other oncogenes in combination such as BRAF variant-assessments with RAS mutations. BRAF is known to be classified by RAS-dependency (58).
The sensitivity of MAP2K1 mutants to MEKis was associated with RAF-dependency; however, certain MAP2K1 variants exhibited unexpected drug sensitivity. Interestingly, the IC50 of I103_K104del, which is considered RAF-independent, was lower compared with the other RAF-independent variants and similar to that of the RAF-regulated variants.
Although no significant differences among the three combination therapies were observed, certain variants are preferable for specific inhibitors. For instance, encorafenib appears to be effective against L115P, whereas cobimetinib and trametinib may be good candidates for I103_K104del. To our knowledge, this is the first study to comprehensively evaluate the drug sensitivities of MAP2K1 mutations, although kinase activity and RAF-dependency of 17 mutations have been described previously (41, 59).
BRAF V600E and RAF-dependent MAP2K1 mutants coexist in primary tumors, but are unlikely to cause resistance (35). In contrast, the acquired RAF-dependent and RAF-regulated mutations, together with ERK activation, result in resistance to RAF and MEKis. Moreover, trametinib is known to be ineffective against E102_I103 deletion in colorectal cancer (57), whereas RAF-regulated and RAF-independent mutations in hematologic tumors, such as Langerhans cell histiocytosis and hairy cell leukemia, respond to MEKis (43, 60, 61).
Currently available MEKis, all of which are non–ATP-competitive allosteric inhibitors, are generally effective against RAF-regulated and -dependent mutations; however, we identified several highly resistant variants to MEKis. The use of ATP-competitive MEKis (41) and inhibitors of other pathways may be useful to overcome such resistance.
A recent study identified MAP2K1 mutations involved in acquired resistance to covalent KRAS G12C inhibitors (62). Therefore, MAP2K1 variants combined with KRAS G12C is another topic drawing attention and the system developed in this study is applicable to evaluate their function in combination.
The potential limitations of this study include the following. First, retroviral transduction of cell lines with MAP2K1 mutations is predicted to result in elevated MEK1 protein overexpression compared with that of endogenous MAP2K1 expression. However, MAP2K1 overexpression itself did not reveal any transforming activity in 3T3 cells, thus variant oncogenicity may be evaluated in comparison with the overexpressed WT. Second, we primarily used A375 for functional analysis and there remains a possibility of cell context-dependent or cell line–specific drug sensitivity. Although the variant sensitivities evaluated in A375 and HT-29 were well correlated, the combination therapy against BRAF V600E and MAP2K1 P124S was not completely concordant between A375 and A2058. Third, the functional annotation was based on preclinical data and is not fully supported by clinical data. As there is little recurrence of mutations for MAP2K1 mutations, further clinical data should be thoroughly aggregated to construct optimal and personalized treatment for tumors harboring MAP2K1 mutations in the clinic.
In conclusion, a comprehensive evaluation of MAP2K1 was successfully performed using the MANO method, and the drug sensitivities of individual variants to combination treatment with MEK and BRAF inhibitors were provided. We also identified several resistant variants for which novel MEKis are needed. The approach used in this study has revealed different sensitivity to each inhibitor for different mutants both single or in combination, thus it will drive new drug development as well as personalized medicine for individual patients.
Supplementary Material
Figure S1 to S17
Table S1 to S9
Acknowledgments
We thank A. Maruyama and M. Kitami for technical assistance. This study was financially supported in part through grants from the Practical Research for Innovative Cancer Control under grant number JP20ck0106536 from the Japan Agency for Medical Research and Development (AMED).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC Section 1734.
This article is featured in Highlights of This Issue, p. 153
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Disclosures
T. Koyama reports other support from Sysmex; grants from Chugai, Daiichi Sankyo, Eli Lilly, Takeda, and Novartis; and grants from Noile-Immune outside the submitted work. K. Sunami reports personal fees from Chugai Pharma; and grants from Sysmex outside the submitted work. H. Kage reports grants from Konica Minolta, Inc. outside the submitted work. H. Mano reports a patent for a method for evaluating multiple different genes of interest pending. S. Kohsaka reports grants from Konica Minolta, Boehringer Ingelheim, AstraZeneca, Eisai, Chordia Therapeutics, and TransThera Sciences; and grants from CIMIC outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
S. Mizuno: Conceptualization, data curation, formal analysis, validation, investigation, methodology, writing–original draft. M. Ikegami: Software, formal analysis, visualization, methodology. T. Koyama: Resources, writing–review and editing. K. Sunami: Resources, writing–review and editing. D. Ogata: Resources, writing–review and editing. H. Kage: Resources, writing–review and editing. M. Yanagaki: Investigation. H. Ikeuchi: Investigation. T. Ueno: Data curation, software, formal analysis, methodology. M. Tanikawa: Writing–review and editing. K. Oda: Writing–review and editing. Y. Osuga: Writing–review and editing. H. Mano: Conceptualization, supervision, funding acquisition, methodology, writing–review and editing. S. Kohsaka: Conceptualization, supervision, funding acquisition, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Balmanno K, Cook SJ. Tumor cell survival signaling by the ERK1/2 pathway. Cell Death Differ 2009;16:368–77. [DOI] [PubMed] [Google Scholar]
- 2. Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci USA 2009;106:20411–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dougherty MK, Muller J, Ritt DA, Zhou M, Zhou XZ, Copeland TD, et al. Regulation of Raf-1 by direct feedback phosphorylation. Mol Cell 2005;17:215–24. [DOI] [PubMed] [Google Scholar]
- 4. Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949–54. [DOI] [PubMed] [Google Scholar]
- 5. Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell 2012;150:251–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet 2011;43:1219–23. [DOI] [PubMed] [Google Scholar]
- 7. Marks JL, Gong Y, Chitale D, Golas B, McLellan MD, Kasai Y, et al. Novel MEK1 mutation identified by mutational analysis of epidermal growth factor receptor signaling pathway genes in lung adenocarcinoma. Cancer Res 2008;68:5524–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arcila ME, Drilon A, Sylvester BE, Lovly CM, Borsu L, Reva B, et al. MAP2K1 (MEK1) mutations define a distinct subset of lung adenocarcinoma associated with smoking. Clin Cancer Res 2015;21:1935–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C, et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet 2011;44:133–9. [DOI] [PubMed] [Google Scholar]
- 10. Wang H, Daouti S, Li WH, Wen Y, Rizzo C, Higgins B, et al. Identification of the MEK1(F129L) activating mutation as a potential mechanism of acquired resistance to MEK inhibition in human cancers carrying the B-RafV600E mutation. Cancer Res 2011;71:5535–45. [DOI] [PubMed] [Google Scholar]
- 11. Giannakis M, Hodis E, Jasmine Mu X, Yamauchi M, Rosenbluh J, Cibulskis K, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet 2014;46:1264–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Estep AL, Palmer C, McCormick F, Rauen KA. Mutation analysis of BRAF, MEK1, and MEK2 in 15 ovarian cancer cell lines: implications for therapy. PLoS One 2007;2:e1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Grisham RN, Sylvester BE, Won H, McDermott G, DeLair D, Ramirez R, et al. Extreme outlier analysis identifies occult mitogen-activated protein kinase pathway mutations in patients with low-grade serous ovarian cancer. J Clin Oncol 2015;33:4099–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bromberg-White JL, Andersen NJ, Duesbery NS. MEK genomics in development and disease. Brief Funct Genomics 2012;11:300–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zheng CF, Guan KL. Activation of MEK family kinases requires phosphorylation of two conserved Ser/Thr residues. EMBO J 1994;13:1123–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eblen ST, Slack-Davis JK, Tarcsafalvi A, Parsons JT, Weber MJ, Catling AD. Mitogen-activated protein kinase feedback phosphorylation regulates MEK1 complex formation and activation during cellular adhesion. Mol Cell Biol 2004;24:2308–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature 2006;439:358–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med 2012;367:107–14. [DOI] [PubMed] [Google Scholar]
- 19. Ascierto PA, McArthur GA, Dréno B, Atkinson V, Liszkay G, Di Giacomo AM, et al. Cobimetinib combined with vemurafenib in advanced BRAFV600-mutant melanoma (coBRIM): updated efficacy results from a randomized, double-blind, phase III trial. Lancet Oncol 2016;17:1248–60. [DOI] [PubMed] [Google Scholar]
- 20. Robert C, Karaszewska B, Schachter J, Rutkowski P, Mackiewicz A, Stroiakovski D, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med 2015;372:30–9. [DOI] [PubMed] [Google Scholar]
- 21. Dummer R, Ascierto PA, Gogas HJ, Arance A, Mandala M, Liszkay G, et al. Overall survival in patients with BRAF-mutant melanoma receiving encorafenib plus binimetinib versus vemurafenib or encorafenib (COLUMBUS): a multicenter, open-label, randomized, phase III trial. Lancet Oncol 2018;19:1315–27. [DOI] [PubMed] [Google Scholar]
- 22. Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 2012;367:1694–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N Engl J Med 2019;381:1632–43. [DOI] [PubMed] [Google Scholar]
- 24. Planchard D, Smit EF, Groen HJM, Mazieres J, Besse B, Helland Å, et al. Dabrafenib plus trametinib in patients with previously untreated BRAFV600E-mutant metastatic non–small cell lung cancer: an open-label, phase II trial. Lancet Oncol 2017;18:1307–16. [DOI] [PubMed] [Google Scholar]
- 25. Caunt CJ, Sale MJ, Smith PD, Cook SJ. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat Rev Cancer 2015;15:577–92. [DOI] [PubMed] [Google Scholar]
- 26. Savoia P, Fava P, Casoni F, Cremona O. Targeting the ERK signaling pathway in melanoma. Int J Mol Sci 2019;20:1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smalley KS, Lioni M, Dalla Palma M, Xiao M, Desai B, Egyhazi S, et al. Increased cyclin D1 expression can mediate BRAF inhibitor resistance in BRAF V600E–mutated melanomas. Mol Cancer Ther 2008;7:2876–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Turke AB, Song Y, Costa C, Cook R, Arteaga CL, Asara JM, et al. MEK inhibition leads to PI3K/AKT activation by relieving a negative feedback on ERBB receptors. Cancer Res 2012;72:3228–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mirzoeva OK, Collisson EA, Schaefer PM, Hann B, Hom YK, Ko AH, et al. Subtype-specific MEK-PI3 kinase feedback as a therapeutic target in pancreatic adenocarcinoma. Mol Cancer Ther 2013;12:2213–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci USA 2009;106:4519–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gao Y, Maria A, Na N, da Cruz Paula A, Gorelick AN, Hechtman JF, et al. V211D mutation in MEK1 causes resistance to MEK inhibitors in colon cancer. Cancer Discov 2019;9:1182–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moriceau G, Hugo W, Hong A, Shi H, Kong X, Yu CC, et al. Tunable-combinatorial mechanisms of acquired resistance limit the efficacy of BRAF/MEK cotargeting but result in melanoma drug addiction. Cancer Cell 2015;27:240–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dudley DT, Pang L, Decker SJ, Bridges AJ, Saltiel AR. A synthetic inhibitor of the mitogen-activated protein kinase cascade. Proc Natl Acad Sci USA 1995;92:7686–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Choi YL, Soda M, Ueno T, Hamada T, Haruta H, Yamato A, et al. Oncogenic MAP2K1 mutations in human epithelial tumors. Carcinogenesis 2012;33:956–61. [DOI] [PubMed] [Google Scholar]
- 35. Carlino MS, Fung C, Shahheydari H, Todd JR, Boyd SC, Irvine M, et al. Preexisting MEK1P124 mutations diminish response to BRAF inhibitors in metastatic melanoma patients. Clin Cancer Res 2015;21:98–105. [DOI] [PubMed] [Google Scholar]
- 36. Brannon AR, Vakiani E, Sylvester BE, Scott SN, McDermott G, Shah RH, et al. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol 2014;15:454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Emery CM, Monaco KA, Wang P, Balak M, Freeman A, Meltzer J, et al. BRAF-inhibitor associated MEK mutations increase RAF-dependent and -independent enzymatic activity. Mol Cancer Res 2017;15:1431–44. [DOI] [PubMed] [Google Scholar]
- 38. Rizos H, Menzies AM, Pupo GM, Carlino MS, Fung C, Hyman J, et al. BRAF inhibitor resistance mechanisms in metastatic melanoma: spectrum and clinical impact. Clin Cancer Res 2014;20:1965–77. [DOI] [PubMed] [Google Scholar]
- 39. Diamond EL, Durham BH, Haroche J, Yao Z, Ma J, Parikh SA, et al. Diverse and targetable kinase alterations drive histiocytic neoplasms. Cancer Discov 2016;6:154–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Greger JG, Eastman SD, Zhang V, Bleam MR, Hughes AM, Smitheman KN, et al. Combinations of BRAF, MEK, and PI3K/mTOR inhibitors overcome acquired resistance to the BRAF inhibitor GSK2118436 dabrafenib, mediated by NRAS or MEK mutations. Mol Cancer Ther 2012;11:909–20. [DOI] [PubMed] [Google Scholar]
- 41. Gao Y, Chang MT, McKay D, Na N, Zhou B, Yaeger R, et al. Allele-specific mechanisms of activation of MEK1 mutants determine their properties. Cancer Discov 2018;8:648–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Izquierdo E, Yuan L, George S, Hubank M, Jones C, Proszek P, et al. Development of a targeted sequencing approach to identify prognostic, predictive and diagnostic markers in pediatric solid tumors. Oncotarget 2017;8:112036–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Diamond EL, Durham BH, Ulaner GA, Drill E, Buthorn J, Ki M, et al. Efficacy of MEK inhibition in patients with histiocytic neoplasms. Nature 2019;567:521–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wagle N, Van Allen EM, Treacy DJ, Frederick DT, Cooper ZA, Taylor-Weiner A, et al. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov 2014;4:61–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhao F, Sucker A, Horn S, Heeke C, Bielefeld N, Schrors B, et al. Melanoma lesions independently acquire T-cell resistance during metastatic latency. Cancer Res 2016;76:4347–58. [DOI] [PubMed] [Google Scholar]
- 46. Gannon HS, Kaplan N, Tsherniak A, Vazquez F, Weir BA, Hahn WC, et al. Identification of an "Exceptional Responder" cell line to MEK1 inhibition: clinical implications for MEK-targeted therapy. Mol Cancer Res 2016;14:207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brown NA, Furtado LV, Betz BL, Kiel MJ, Weigelin HC, Lim MS, et al. High prevalence of somatic MAP2K1 mutations in BRAF V600E–negative Langerhans cell histiocytosis. Blood 2014;124:1655–8. [DOI] [PubMed] [Google Scholar]
- 48. Ng PK, Li J, Jeong KJ, Shao S, Chen H, Tsang YH, et al. Systematic functional annotation of somatic mutations in cancer. Cancer Cell 2018;33:450–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kohsaka S, Nagano M, Ueno T, Suehara Y, Hayashi T, Shimada N, et al. A method of high-throughput functional evaluation of EGFR gene variants of unknown significance in cancer. Sci Transl Med 2017;9:eaan6566. [DOI] [PubMed] [Google Scholar]
- 50. Nagano M, Kohsaka S, Ueno T, Kojima S, Saka K, Iwase H, et al. High-throughput functional evaluation of variants of unknown significance in ERBB2. Clin Cancer Res 2018;24:5112–22. [DOI] [PubMed] [Google Scholar]
- 51. Ikegami M, Kohsaka S, Ueno T, Momozawa Y, Inoue S, Tamura K, et al. High-throughput functional evaluation of BRCA2 variants of unknown significance. Nat Commun 2020;11:2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Akagi T, Sasai K, Hanafusa H. Refractory nature of normal human diploid fibroblasts with respect to oncogene-mediated transformation. Proc Natl Acad Sci USA 2003;100:13567–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kumar R, Suresh PS, Rudresh G, Zainuddin M, Dewang P, Kethiri RR, et al. Determination of ulixertinib in mice plasma by LC-MS/MS and its application to a pharmacokinetic study in mice. J Pharm Biomed Anal 2016;125:140–4. [DOI] [PubMed] [Google Scholar]
- 54. Blake JF, Burkard M, Chan J, Chen H, Chou KJ, Diaz D, et al. Discovery of (S)-1-(1-(4-Chloro-3-fluorophenyl)-2-hydroxyethyl)-4-(2-((1-methyl-1H-pyrazol-5-yl)amino)pyrimidin-4-yl)pyridin-2(1H)-one (GDC-0994), an extracellular signal-regulated kinase 1/2 (ERK1/2) inhibitor in early clinical development. J Med Chem 2016;59:5650–60. [DOI] [PubMed] [Google Scholar]
- 55. Yadav B, Wennerberg K, Aittokallio T, Tang J. Searching for drug synergy in complex dose-- response landscapes using an interaction potency model. Comput Struct Biotechnol J 2015;13:504–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Yuan J, Ng WH, Tian Z, Yap J, Baccarini M, Chen Z, et al. Activating mutations in MEK1 enhance homodimerization and promote tumorigenesis. Sci Signal 2018;11:eaar6795. [DOI] [PubMed] [Google Scholar]
- 57. Wang C, Sandhu J, Fakih M. A case of class 3 MEK1 mutated metastatic colorectal cancer with a non-durable tumor marker response to MEK and ERK inhibitors. J Gastrointest Oncol 2019;10:1140–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yao Z, Yaeger R, Rodrik-Outmezguine VS, Tao A, Torres NM, Chang MT, et al. Tumors with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature 2017;548:234–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hanrahan AJ, Sylvester BE, Chang MT, Elzein A, Gao J, Han W, et al. Leveraging systematic functional analysis to benchmark an in silico framework distinguishes driver from passenger MEK mutants in cancer. Cancer Res 2020;80:4233–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Caeser R, Collord G, Yao WQ, Chen Z, Vassiliou GS, Beer PA, et al. Targeting MEK in vemurafenib-resistant hairy cell leukemia. Leukemia 2019;33:541–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gounder MM, Solit DB, Tap WD. Trametinib in histiocytic sarcoma with an activating MAP2K1 (MEK1) mutation. N Engl J Med 2018;378:1945–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Awad MM, Liu S, Rybkin II, Arbour KC, Dilly J, Zhu VW, et al. Acquired resistance to KRAS(G12C) inhibition in cancer. N Engl J Med 2021;384:2382–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 to S17
Table S1 to S9
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper.