Abstract
Background
Inflammatory bowel diseases (IBDs) involve an aberrant host response to intestinal microbiota causing mucosal inflammation and gastrointestinal symptoms. Patient-reported outcomes (PROs) are increasingly important in clinical care and research. Our aim was to examine associations between PROs and fecal microbiota in patients 0 to 22 years of age with IBD.
Methods
A longitudinal, prospective, single-center study tested for associations between microbial community composition via shotgun metagenomics and PROs including stool frequency and rectal bleeding in ulcerative colitis (UC) and abdominal pain and stool frequency in Crohn’s disease (CD). Mucosal inflammation was assessed with fecal calprotectin. A negative binomial mixed-effects model including clinical characteristics and fecal calprotectin tested for differentially abundant species and metabolic pathways by PROs.
Results
In 70 CD patients with 244 stool samples, abdominal pain correlated with increased relative abundance of Haemophilus and reduced Clostridium spp. There were no differences relative to calprotectin level. In 23 UC patients with 76 samples, both rectal bleeding and increased stool frequency correlated with increased Klebsiella and reduced Bacteroides spp. Conversely, UC patients with lower calprotectin had reduced Klebsiella. Both UC and CD patients with active symptoms exhibited less longitudinal microbial community stability. No differences in metabolic pathways were observed in CD. Increased sulfoglycolysis and ornithine biosynthesis correlated with symptomatic UC.
Conclusions
Microbial community composition correlated with PROs in both CD and UC. Metabolic pathways differed relative to PROs in UC, but not CD. Data suggest that microbiota may contribute to patient symptoms in IBD, in addition to effects of mucosal inflammation.
Keywords: patient-reported outcomes, pediatrics, microbiome
What is already known?
Enteric microbiota shifts have previously been identified in patients with inflammatory bowel disease (IBD) relative to healthy control subjects, but the influence of these shifts on disease activity and IBD symptomatology is not well understood.
What is new here?
Microbial shifts correlated with patient-reported outcomes while controlling for fecal calprotectin as a surrogate marker of mucosal inflammation in both ulcerative colitis and Crohn’s disease, suggesting that microbial community composition may influence patient symptoms.
How can this study help patient care?
Microbial shifts, regardless of inflammatory status, may directly contribute to the patient experience of IBD and may be a future diagnostic and therapeutic target.
Introduction
Inflammatory bowel diseases (IBDs) including Crohn’s disease (CD) and ulcerative colitis (UC), are relapsing, remitting, and often progressive inflammatory diseases affecting the gastrointestinal tract with increasing incidence in pediatric patients over the past decades.1 Symptoms are heterogeneous and commonly include diarrhea, rectal bleeding, abdominal pain, and poor growth. Studies of gut microbiome composition in IBD have revealed intestinal and fecal microbial shifts associated with both CD and UC including reduced overall microbial community diversity, depletion of taxa including Bacteroides and Ruminococcus, and strain-level depletion in butyrate-producing organisms such as Faecalbacterium prausnitzii and Roseburia hominis.2-4 Increased relative abundance of Escherichia and Klebsiella have been associated with active IBD, and Collinsella and Veillonella have been specifically associated with increased risk of surgery in CD in children.4-6 Despite these findings, the exact role of microbial community composition on IBD pathogenesis and symptomatology remains unclear.
Optimal medical therapy for IBD targets gastrointestinal inflammation to alleviate symptoms and reduce the risk of complications such as fistulae, stricturing, and intra-abdominal abscesses. Despite evidence of mucosal healing by laboratory and fecal testing, some patients continue to experience debilitating gastrointestinal symptoms. Food and Drug Administration guidelines recommend combined biomarker and endoscopic endpoints in therapeutic clinical trials for IBD as well as patient-reported outcome (PROs).7,8 PROs, typically obtained via standardized, validated, PRO measures, are exclusively PR factors and do not include physician interpretation of patient status. In UC, a concise 2-item PRO score (UC PRO2) incorporating patient report of rectal bleeding and stool frequency has been validated against the Ulcerative Colitis Clinical Disease Activity Index and Mayo Clinic Score as well as combined with endoscopic endpoints to assess clinical disease activity with excellent performance.9 The UC PRO2 metric is already in use in clinical trials.10,11 Similarly, in CD, the use of PR stool frequency or liquid stool and abdominal pain (CD PRO2), measures derived from the Crohn’s Disease Activity Index, have been measured against the Simple Endoscopic Score with good performance and are also in use in clinical trials.11-14
Microbial therapies such as fecal microbial transplantation and antibiotic therapy have been used to date with some success in treating IBD.15-17 Targeted, personalized microbial therapies are not yet available. Novel treatment modalities to target microbial shifts may both improve disease remission as well as improve symptomatology and patient quality of life. This study aimed to evaluate the relationship between PROs, mucosal inflammation as measured by fecal calprotectin, and microbial and metabolic shifts in the fecal microbiome of pediatric IBD patients using shotgun metagenomic testing.
Methods
Study Population and Design
We conducted a single site prospective study at a tertiary care center. Patients with a diagnosis of IBD were enrolled from September 2016 to March 2018. Inclusion criteria included a diagnosis of CD or UC. Patients were enrolled regardless of time since diagnosis. UC patients with a history of colectomy were excluded. Crohn’s patients with a history of surgery were not excluded. No other exclusion criteria were implemented. Patients were asked to submit a total of 6 stool samples at minimum 4 weeks apart and return questionnaires about symptoms with each sample returned. Previous analyses of this cohort were undertaken regarding C. difficile history and progression to surgery and reported in 2020.18
Patients were instructed to return each fecal sample divided between 2 containers: one containing a proprietary preservative by OMNIgene (DNA Genotek, Ottawa, ON, Canada), which was used for metagenomic sequencing, and one sterile tube, which was used for fecal calprotectin measurement. Samples were stored at -80°C upon receipt and thawed at the time of processing. Stool samples were returned in person or via express mail depending on the family’s preference and remained at room temperature until delivery as per OMNIgene GUT instructions. If a mail service was used, stools were shipped same day or next day if collected Monday through Thursday morning and shipped on Monday if collected Thursday afternoon through Sunday.
Fecal calprotectin was performed via enzyme-linked immunosorbent assay (Bühlmann Laboratories AG, Schönenbuch, Switzerland). A value of ≤100 µg/g for fecal calprotectin was considered a marker of mucosal healing in UC, and a level of ≤250 µg/g was considered a marker of mucosal healing for CD.19 Endoscopic assessment of mucosal healing was not performed to validate fecal calprotectin levels as part of this study. Data were analyzed with regard to mucosal healing as a binary measure as well as with regard to fecal calprotectin as a continuous value. Demographic data including patient sex, age, race, ethnicity, IBD diagnosis and phenotype, duration of IBD, surgical history, and medication history were collected via the electronic medical record (Epic Systems, Verona, WI, USA).
Patient-Reported Outcomes
With each stool sample, patients, regardless of diagnosis, were provided with both a Pediatric Ulcerative Colitis Activity Index (PUCAI) assessing symptoms over the past 48 hours and the Short Pediatric Clinical Disease Activity Index questionnaire assessing symptoms over the past 7 days.20 For patients with UC, in alignment with UC PRO2, stool frequency and rectal bleeding were assessed via the PUCAI questionnaire to determine clinical disease activity.11 A score of 0, 1, 2, or 3 was assigned depending on degree of rectal bleeding (Supplemental Figure 1). Any patient with a score ≥1 (any report of rectal bleeding) was deemed to have clinically active disease. For stool frequency, a score of 0, 1, 2, or 3 was assigned dependent on the reported stool frequency. Any patient with a score ≥1 (3-5 stools) was deemed to have clinically active disease. Patients with either or both rectal bleeding and increased stool frequency were determined to have overall PR active disease.
For those with CD, in alignment with CD PRO2, a similar process was used. A score of 0, 1, or 2 was assigned based on abdominal pain severity (Supplemental Figure 1). Those with a score of 2 (abdominal pain that was “moderate/severe: daily, affects activity, nocturnal”) were deemed to have clinically active disease. For stool frequency, a score of 0, 1, or 2 was assigned dependent on reported stool frequency. Any patient with a score ≥1 (up to 2 semiformed with small blood, or 2-5 liquid) was determined to have clinically active disease. Patient with either or both moderate to severe abdominal pain and increased stool frequency were determined to have overall PR active disease. CD patients with isolated abdominal pain marked as “mild, brief, not interfering with activities” (score = 1) were not considered to have clinically active disease or overall PR active symptoms.
Shotgun Metagenome Sequencing
DNA was extracted from approximately 0.1 g of stool aliquot using the PowerFecal DNA isolation kit by MO BIO (MO BIO Laboratories, Carlsbad, CA, USA) per manufacturer recommendations. After DNA samples were diluted to approximately 200 ng/mL, sequencing libraries were created using the Nextera XT protocol (Illumina, San Diego, CA, USA). Sequencing was performed on an Illumina NextSeq500 machine using 150-bp DNA paired end reads to a depth of approximately 4 G base pairs per sample. Raw sequence data were de-multiplexed and converted to fastq format for downstream processing.
Taxonomic and Functional Profiling
Raw sequence reads were extracted and de-multiplexed using the Illumina program bcl2fastq. Raw reads were then filtered and trimmed for quality control using the program Sickle.21 Paired-end sequencing reads from each sample were assigned to the lowest common ancestor using Kraken2.22 Sequence alignment with Kraken2 was performed against a custom genome database consisting of the human genome and approximately 40 054 bacterial, fungal, viral, and parasitic genomes as described in detail previously.18 In brief, the database was derived initially from all bacteria, fungi, and viruses in the RefSeq genome database (accessed November 27, 2017) as well as in the human genome database (GR38Ch). Manual curation was used to add additional Bacteroides, Parabacteroides, and Clostridia genomes including draft genomes from the National Center for Biotechnology Information Assemblies and PATRIC, and additional fungal and viral genome sequences were recovered from the previous 2 resources, as well as from dedicated viral and fungal databases. Species or genera that contributed <0.01% of overall mapped reads or were present in <10% of the samples were removed.
Functional profiling was performed by first removing human reads by aligning against the human reference genome. Filtered reads were then aligned sequentially to annotated nucleotide and peptide databases within the HUMAnN2 pipeline and aggregated to MetaCyc pathways.23 The number of reads aligning to gene families and metabolic pathways were normalized to counts per million reads for analysis.
Statistical Analysis
Patient demographic and clinical characteristics were described using median (interquartile range [IQR]) for continuous variables and frequency and percentage for categorical variables according to PROs. Odds ratios (ORs) and 95% confidence intervals for PROs according to fecal calprotectin levels were obtained using generalized linear mixed-effects models with a logit link function to model the binary response. A random subject-specific intercept was used to account for the repeated measures within patients. Models were fit using the glmmTMB package (version 1.0.2)24 in R version 4.0.2 (R Foundation for Statistical Computing, Vienna, Austria) and included disease duration in years, disease location (colonic only, ileal or ileocolonic for CD, rectal only, left-sided disease or pancolitis for UC), IBD medications (no biologic vs. biologic therapy), antibiotics taken within 60 days of sample collection (yes/no), fecal calprotectin, and history of surgery (yes/no) in CD patients as model covariates. ORs were reported for a 1-SD increase in fecal calprotectin levels to facilitate interpretation.
Shannon diversity was calculated for each sample using phyloseq (version 1.32.0)25 after subsampling to the lowest observed read depth (1 316 647 reads UC; 201 626 reads CD). Differences in Shannon diversity according to PROs were estimated using linear mixed-effects regression incorporating a random subject-specific intercept via the lme4 package (version 1.1.23)26 with formal testing performed via the Satterthwaite approximation to the model degrees of freedom as implemented in the lmerTest package (version 3.1.2).27 A random subject-specific intercept was used to account for the repeated measures within patients. Model covariates included disease duration, disease location, IBD medications, antibiotics taken within 60 days, fecal calprotectin, and history of surgery (CD only). Principal component analysis (PCA) was conducted on the species count matrix after variance stabilizing transformation as implemented in the DESeq2 package (version 1.24.0)28 and visualized using the plot ordination function in phyloseq to assess the unsupervised clustering of samples based on species community composition.
Within-subject Bray-Curtis dissimilarly from the time of first sample collection was calculated using the first distances function in QIIME2 (version 2020.8.0) to examine the stability of the community composition over time. Differences in within-subject Bray-Curtis dissimilarly between those ever vs never experiencing a PR outcome meeting the criteria for clinically active disease were tested using linear mixed-effects regression as described previously for Shannon diversity.
Negative binomial mixed models incorporating random subject-specific intercepts were fit using the glmmTMB package to identify differentially abundant species according to PROs. A random subject-specific intercept was used to account for the repeated measures within patients. Models included, sex, disease duration, disease location, biologic therapy, antibiotics taken within 60 days, and history of surgery as model covariates. Fecal calprotectin as a continuous covariate was included if it was not the variable of interest. A scaling factor as estimated by the geometric mean of pairwise ratios methods as implemented in the GMPR package (version 0.1.3) was included as a model offset to account for differences in sequencing depth.29 Winsorization of outlying counts was performed by truncating values at the 97% percentile, as it has been shown to improve the performance of count-based models for microbiome data.30 Species seen in fewer than 20% of samples were filtered prior to testing, and the Benjamini-Hochberg false discovery rate (FDR) correction applied to control the proportion of false positive results. Log2 fold changes were estimated from the model predicted values. The negative binomial mixed models framework was also used to identify differentially abundant MetaCyc pathways31 according to PR symptoms, with total counts per million used as the model offset to directly model the relative abundance given the normalized counts.
Ethical Considerations
This study was approved by the Cincinnati Children’s Hospital Medical Center Institutional Review Board. Informed consent was obtained by the patient (18 years of age and above) or by the legal guardian. Patients 11 to 17 years of age provided assent.
Results
Longitudinal PROs in UC
There were 23 UC patients who submitted 76 total stool samples (Supplemental Figure 2). At first sample collection, 9 (39%) patients reported active disease and 14 (61%) did not (Table 1). Analyses of the PROs at first sample collection showed 6 (67%) had increased stool frequency, 7 (78%) had rectal bleeding, and 4 (44%) had both. Although abdominal pain was not used as a PRO in those with UC, these data were collected. Only 1 patient with UC had moderate-to-severe pain at first sample collection and ever, and this patient also had increased stool frequency and rectal bleeding. There were no material differences in demographics, disease location, or medication use between those with or without active symptoms. Patients were subsequently classified as ever active if at any sample collection they reported active symptoms or never active if they never reported active symptoms throughout the study. A total of 12 (52%) UC patients reported symptoms at any time. With the first reported active sample, 9 (75%) had increased stool frequency, 10 (83%) had rectal bleeding, and 7 (58%) had both (Supplemental Table 1).
Table 1.
Clinical and Demographic Characteristics of Ulcerative Colitis and Crohn’s Disease Patients Grouped by Patient-Reported Disease Activity at Study Enrollment
| Ulcerative Colitis (n = 23) | P Value | Crohn’s Disease (n = 70) | P Value | |||
|---|---|---|---|---|---|---|
| Patient-Reported Active (n = 9 [39%]) | Patient-Reported Nonactive (n = 14 [61%]) | Patient-Reported Active (n = 21 [30%]) | Patient-Reported Nonactive (n = 49 [70%]) | |||
| Increased stool frequency | 6 (67) | 0 (0%) | 18 (86) | 0(0) | ||
| Abdominal pain | 1 (11) | 0 (0) | 8 (38) | 0(0) | ||
| Rectal bleeding | 7 (78) | 0 (0) | 11 (52) | 3 (6) | ||
| Mucosal healing (fecal calprotectin ≤100 µg/g) | 1 (11) | 5 (36) | .34 | |||
| Mucosal healing (fecal calprotectin ≤250 µg/g | 12 (57) | 29 (59) | 1.0 | |||
| Fecal calprotectin, µg/g | 364 (104-825) | 141 (92-629) | .4 | 204 (90-484) | 128 (83-393) | .62 |
| Disease location | ||||||
| Left side | 2 (22) | 3 (21) | 1.0 | |||
| Pancolitis | 7 (78) | 11 (79) | ||||
| Ileal | 0 (0) | 8 (16) | .38 | |||
| Ileocolonic | 20 (95) | 34 (69) | ||||
| Colon only | 1 (5) | 7 (14) | ||||
| Age at sample collection, y | 16.05 (15.24-16.56) | 15.05 (12.84-17.14) | .82 | 15.7 (13.2-17.3) | 13.9 (12.4-15.8) | .10 |
| Male | 5 (56) | 10 (71) | .66 | 10 (48) | 32 (65) | .19 |
| Caucasian | 8 (89) | 14 (100) | .39 | 19 (90) | 44 (90) | 1.0 |
| Disease duration, y | 2.5 (0.5-3.7) | 1.8 (0.8-4.2) | .73 | 2.1 (0.5-4.7) | 3.1 (1.4-6.3) | .08 |
| IBD medication | ||||||
| On biologic | 3 (33) | 7 (50) | .67 | 13 (62) | 35 (71) | .58 |
| Antibiotic within 60 d | 2 (22) | 2 (14) | 1.0 | 3 (14) | 5 (10) | .69 |
| History of surgery | 2 (10) | 11 (22) | .32 | |||
Values are n (%) or median (interquartile range).
Abbreviation: IBD, inflammatory bowel disease.
Longitudinal PROs in CD
A total of 70 CD patients provided 244 stool samples within the study period (Supplemental Figure 2). Of the 70 patients, there were 21 (30%) with active symptoms at first sample collection, which included 8 (38%) with moderate-to-severe abdominal pain, 18 (86%) with increased stool frequency, and 12 (57%) with both (Table 1). No material difference in demographics, time since IBD diagnosis, or clinical characteristics including use of a biologic medication were found between patients with and without active symptoms at first sample collection. When CD patients were classified as ever vs never having active symptom throughout the study, there were 31 (44%) patients who ever reported active symptoms. Of those, 26 (84%) had increased stool frequency, 10 (32%) had moderate-to-severe abdominal pain, and 17 (55%) had both (Supplemental Table 1). Thirteen (19%) CD patients reported rectal bleeding with the first stool sample collection, though this was not used as a PRO for CD patients. Of those, 11 (85%) of 13 patients had either increased stools or abdominal pain indicating overall PR active symptoms while 2 (15%) of 13 did not.
PROs and Fecal Calprotectin
The median fecal calprotectin at first sample collection was equal to 204 (IQR, 90-484) µg/g in CD patients with active symptoms, compared with 128 (IQR, 83-393) µg/g in those without active symptoms (P = .62). There was no association between fecal calprotectin and overall PR disease activity or individual symptoms of frequent stools and abdominal pain in those with CD. The median fecal calprotectin at first sample collection was equal to 364 (IQR, 104-825) µg/g in UC patients with active symptoms, compared with 141 (IQR, 92-629) µg/g in those without active symptoms (P = .4). In UC, higher fecal calprotectin was associated with rectal bleeding (OR, 4.93; 95% confidence interval, 1.18-20.64; P = .03), but no association was seen with increased stool frequency or overall PR active symptoms (Table 2).
Table 2.
Odds Ratio and 95% Confidence Intervals for Patient-Reported Outcomes According to Fecal Calprotectin Level in Crohn’s Disease and Ulcerative Colitis
| Odds Ratio | 95% Confidence Interval | P Value | |
|---|---|---|---|
| Crohn’s disease | |||
| Overall patient-reported active disease | 1.20 | 0.77-1.87 | .416 |
| Abdominal pain | 1.09 | 0.21-5.76 | .918 |
| Increased stool frequency | 1.30 | 0.83-2.03 | .256 |
| Ulcerative colitis | |||
| Overall patient-reported active disease | 2.85 | 0.87-9.35 | .085 |
| Rectal bleeding | 4.93 | 1.18-20.64 | .029 |
| Increased stool frequency | 1.88 | 0.62-5.64 | .263 |
Microbial Community Composition in UC
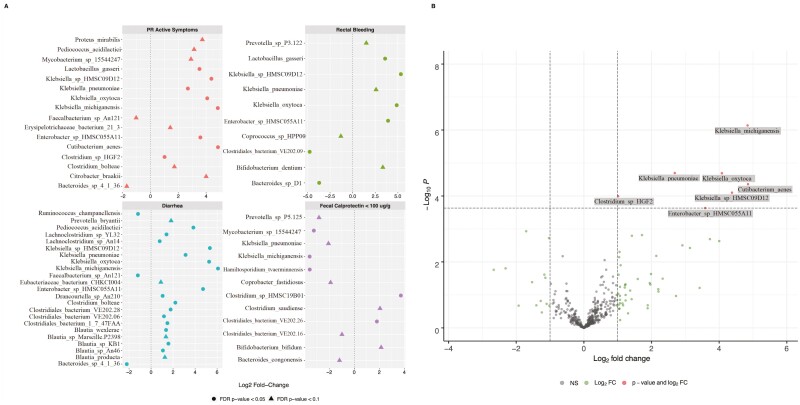
Analysis of the microbiome in those with UC showed no material differences in overall Shannon diversity (Supplemental Figure 3) or beta diversity as assessed by ordinations of the first 2 PCA axes (Supplemental Figure 4) when examining PROs, mucosal healing, or fecal calprotectin level as a continuous variable. Examination for differentially abundant species in UC with an FDR P value of .05 found that those with overall PR active symptoms had increases in multiple Klebsiella species and Cutibacterium acnes and reduction in Bacteroides species (Figure 1). When analyzed by specific PROs, those with UC and rectal bleeding had increased Klebsiella and Enterobacter species and reduced Bacteroides and Clostridiales species. Those with UC and increased stool frequency had increased Klebsiella, Lachnoclostridium, Clostridiales, Blautia, and Enterobacter and reduced Ruminococcus, Faecalbacterium, and Bacteroides. Conversely, UC patients with mucosal healing (fecal calprotectin <100 µg/g) had reduced Klebsiella and increased Bifidobacteria and Bacteroides. When fecal microbial shifts were examined relative to fecal calprotectin as a continuous variable, reduction in Staphylococcus aureus, Streptococcus lutetiensis, and Turicibater were observed (Supplemental Figure 5).
Figure 1.
Microbial shifts in ulcerative colitis (UC) patients. A, Log2 fold changes (FCs) for shifts in relative abundance of microbiota in UC patients based on overall patient-reported active symptoms, individual patient-reported outcomes of diarrhea and rectal bleeding, and fecal calprotectin <100 µg/g, as a surrogate marker of mucosal healing. B, The Log2 FCs and P values obtained from negative binomial mixed-effects regression fit to 76 samples from 23 patients for active symptoms. FDR, false discovery rate; NS, not significant.
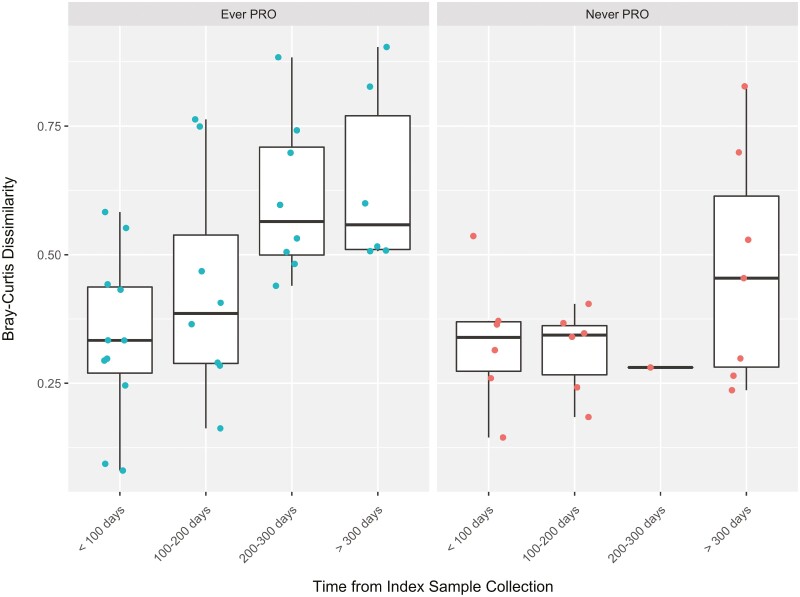
Examination of the microbial community stability over time relative to the index (first) sample collection revealed that within 53 paired dissimilarities in 19 UC patients, those who were ever active had increased dissimilarity (ie, less stable) in the microbial community composition over time (within-subject Bray-Curtis dissimilarity = 0.16 ± 0.06; P = .03). A patient had to submit at least 2 samples in the study to be included in this analysis (Figure 2).
Figure 2.
Microbial temporal variation in ulcerative colitis patients. Within-subject Bray-Curtis dissimilarity was calculated from the time of first sample collection in ulcerative colitis patients. A total of 53 paired dissimilarities were calculated on 19 patients. The Bray-Curtis dissimilarity was increased in samples from patients ever reporting a patient-reported outcome (PRO) (0.16 ± 0.06; P = .03).
Microbial Community Composition in CD
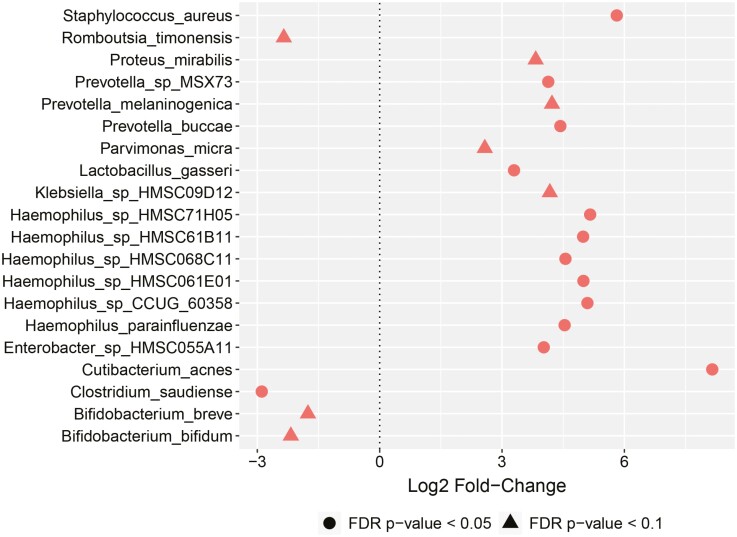
Similarly, analysis of the microbiome composition in patients with CD showed no differences in Shannon diversity (Supplemental Figure 6) or beta diversity as measured by PCA (Supplemental Figure 7). Examination of differentially abundant species in those with CD showed no microbial shifts associated with overall PR disease activity, increased stool frequency, or mucosal healing (fecal calprotectin ≤250 µg/g). In those with CD and abdominal pain, increased Haemophilus, Staphylococcus, Enterobacter, Prevotella, and Cutibacterium and reduced Bifidobacteria and Clostridium were observed (Figure 3). Reduction in Veillonella strains was found in association with increased in fecal calprotectin level as a continuous variable (Supplemental Figure 5). Although rectal bleeding was not used as a PRO in CD, given that 19% of patients reported this symptom with first stool collection, examination of microbial shifts associated with rectal bleeding was performed and no differences were seen.
Figure 3.
Microbial shifts in Crohn’s disease (CD) patients. Depiction of log2 fold changes (FCs) for shifts in relative abundance of microbiota in CD patients based on abdominal pain. No microbial shifts were observed for CD patients with overall patient-reported active disease, increased stool frequency, or fecal calprotectin ≤250 µg/g; thus, no additional plots are provided. Log2 FCs and P values were obtained from negative binomial mixed-effects regression fit to 244 samples from 70 patients. FDR, false discovery rate;
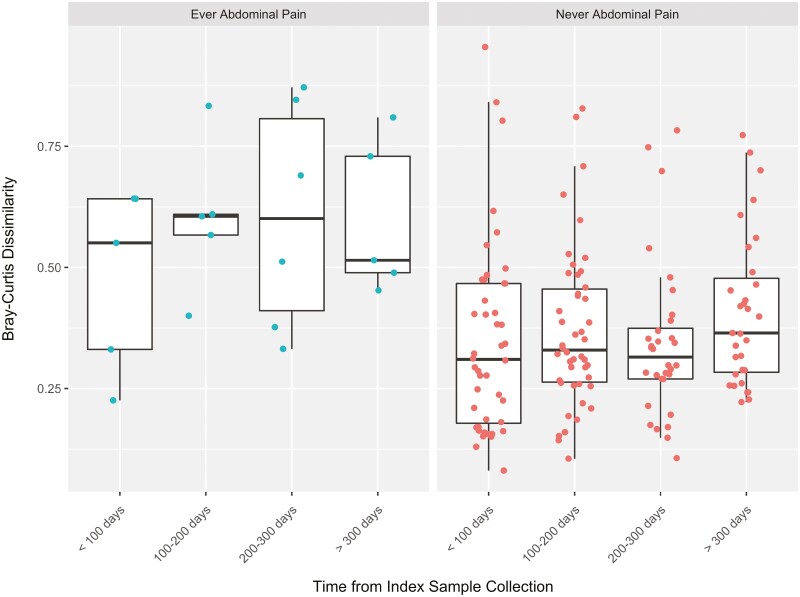
Examination of the microbial community composition over time relative to the index (first) sample collection revealed that within 147 paired dissimilarities in 50 CD patients, those who ever had abdominal pain had less stability or increased dissimilarity in the microbial community composition over time (within-subject Bray-Curtis dissimilarity = 0.20 ± 0.06; P = .002). A patient had to submit at least 2 samples in the study to be included in this analysis (Figure 4).
Figure 4.
Microbial temporal variation in Crohn’s disease patients. Within-subject Bray-Curtis dissimilarity was calculated from the time of first sample collection in Crohn’s disease patients. A total of 147 paired dissimilarities were calculated on 50 patients. The Bray-Curtis dissimilarity was increased in samples from patients ever reporting abdominal pain (0.20 ± 0.06; P = .002).
Metabolic Pathway Analysis
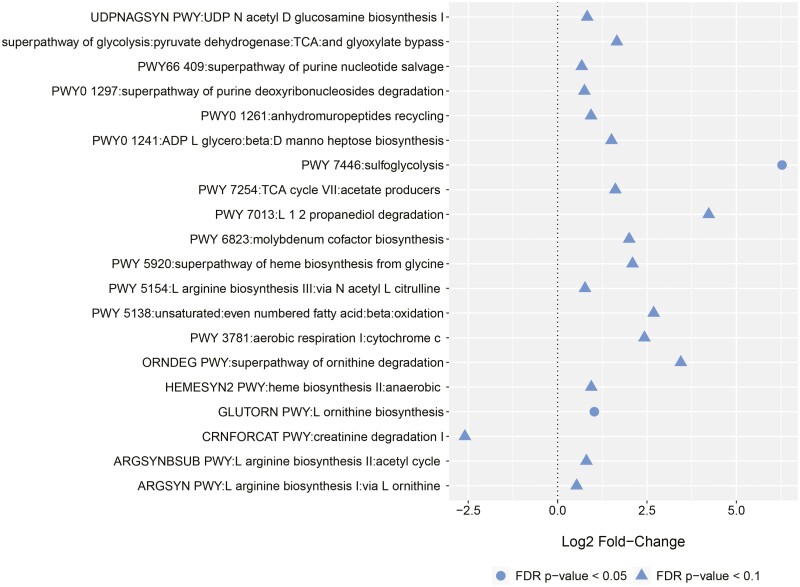
Metabolic pathway analysis found no differences in those with CD when stratified by overall PR active symptoms, increased stool frequency, abdominal pain, or fecal calprotectin level. In UC patients, increases in sulfoglycolysis and ornithine biosynthesis were associated with overall PR active symptoms (Figure 5). No differences were found based on individual PROs.
Figure 5.
Shifts in metabolic pathways in ulcerative colitis patients based on overall patient-reported active disease. Log2 fold changes and false discovery rate (FDR)–corrected P values were obtained from negative binomial mixed-effects regression fit to 76 samples from 23 patients.
Discussion
Despite advancing knowledge about the human microbiome and its role in disease pathogenesis, the association between fecal microbial shifts and the development, progression, and experience of IBD is not well understood. Via shotgun metagenomic sequencing of stool samples, our study demonstrated that microbial shifts correlate with PROs in children and young adults with IBD, while controlling for fecal calprotectin level as a surrogate marker of mucosal inflammation. In UC, microbial shifts were observed with overall disease activity, increased stool frequency, and mucosal healing indirectly measured by fecal calprotectin. In CD patients, microbial shifts were associated with abdominal pain but not with increased stool frequency and not with mucosal healing. In both UC and CD, those with active symptoms at any time point in the study had decreased microbial community stability over time compared with those who never reported symptoms during the study period. Metabolic pathways also differed relative to PROs in UC but not in CD. Together these findings suggest that microbial shifts and microbial community volatility may be associated with patient symptoms independent of mucosal inflammation in pediatric IBD.
This study replicated microbial shifts previously identified in association with IBD and also identified novel patterns. Controlling for fecal calprotectin allowed us to identify microbial shifts not directly due to mucosal inflammation. With this methodology, we interestingly also replicated microbial community composition findings previously reported in patients with irritable bowel syndrome (IBS), who by definition lack gut mucosal inflammation. For example, Klebsiella, which was increased in our UC patients with overall PR active disease, increased stool frequency, and rectal bleeding, and which was reduced in those with fecal calprotectin <100 µg/g, has been previously associated with more a more severe disease course in children with UC.32Lactobacillus gasseri was elevated in CD patients with abdominal pain and UC patients with both overall PR active disease and rectal bleeding. L. gasseri is typically found in intestinal or vaginal flora33 and is contained in many over-the-counter probiotics. It is possible that those with increased symptoms were more likely to be prescribed or electively take probiotics, thus skewing the results. Our finding of microbial community instability in both UC and CD patients with PR active disease is also in line with prior studies showing increased temporal microbial variation in those with active IBD relative to control groups.3,34 In our Crohn’s analyses, we found elevation in multiple Haemophilus strains. A small study of 22 pediatric patients with IBS similarly found increases in H. parainfluenzae compared with healthy control subjects.35 Similarly, reduction in Bacteroides, which we found in UC patients with diarrhea, has been observed in IBS cohorts.36 Our replication of microbial shifts previously reported in IBS patients lends support to our conclusion that microbial shifts may indeed contribute to patient symptoms even in the absence of inflammation.
In contrast to existing literature, our study found an inverse association of Veillonella and elevated fecal calprotectin in CD (Supplemental Figure 5). Increases in Veillonella species have been previously associated with active, refractory, and complicated IBD.5 Additional analyses represented via scatter plots showed that this association was driven by a few outliers with markedly elevated fecal calprotectin levels (Supplemental Figure 8).
Our study utilized shotgun metagenomic sequencing, which enables precise analysis of the human microbiome and metabolic pathways, which in turn may have an amplified impact on the clinical course of IBD.37,38 Our analyses identified increased ornithine biosynthesis in UC patients with overall PR active disease. Prior studies have identified increased tissue metabolism of L-arginine as part of the arginine-ornithine pathway in murine colitis models and humans with active UC.39 In a microbial-induced colitis model, this was modeled by altered enzymatic gene expression in the arginine-ornithine pathway, leading to increased nitric oxide production and altered mucosal healing, which was attenuated with L-arginine supplementation.40 Interestingly, mouse models have recently demonstrated that upregulation of ornithine metabolism by Clostridiodes difficile may confer a competitive advantage allowing asymptomatic C. difficile colonization.41 Enriched sulfoglycolysis pathways were also observed in in our UC cohort of patients with PR active disease. Little is known about the role of sulfoglycolysis in IBD, but bacteria such as Escherichia coli possess this pathway, which breaks down sulfoquinovose from plant material to provide energy.31 Notably, Klebsiella, which we found to be increased in UC patients with PR active disease, has also been found to utilize sulfoquinovose as a sole carbon source.42 Thus, microbial advantages in utilizing certain amino acids may influence overall community microbial composition, which in turn can have amplified effects on host health. Direct measurement of microbial functional contributions via RNA metatranscriptomics and direct measurement of metabolites may provide even further insight into the impact of microbial shifts on disease symptomatology and progression.
One strength of our study was the use of PROs returned with each stool sample collected. The use of mail-in questionnaires fully eliminated provider interpretation of patient symptoms. While the PRO2 for both UC and CD are derived from existing clinical disease activity indices including Mayo Clinical Score or Ulcerative Colitis Clinical Disease Activity Index (UC) and Crohn’s Disease Activity Index (CD), we used the questionnaire for the Short Pediatric Clinical Disease Activity Index, which combines report of liquid stools and blood in stools into one question. Although this language may be confounding, each patient with CD also returned a PUCAI questionnaire, hence report of blood, liquid stools, and stool frequency could be cross-referenced for consistency, and ultimately rectal bleeding was not used as a PRO in CD patients. Concise, simple PROs will increasingly be used as clinical and study endpoints involving IBD, and it is important that pediatric patients are included in this evolution.
Despite several strengths including shotgun metagenomic sequencing and the use of PROs, our study also has several limitations. First, we were not able to assess for mucosal healing via endoscopy. While a fecal calprotectin cutoff of 250 µg/g is an established threshold for determining inflammation in CD, this threshold may misclassify those with isolated small bowel disease.43 Similarly, differences in the fecal microbiome and mucosal microbiome have been observed4,44 and microbial shifts and metabolic pathway variation in tissue could be misrepresented by fecal sample collection alone. In the UC group, there was a relatively low number of patients included with several samples included per patient. We used mixed-effects models to account for the repeated measures design and an FDR-corrected P value of .05 to account for the lack of independence and limit false positive results.
Conclusions
Our knowledge of the pathophysiology of IBD has expanded over recent decades, leading to increasingly effective therapeutic options. Still, many patients continue to experience debilitating gastrointestinal symptoms even when serum and fecal markers have normalized. PROs are increasingly utilized in clinical care and clinical trials and provide direct insight into the patient experience of disease, rather than through a physician’s filter. The association of microbial shifts with specific PROs in IBD, while controlling for fecal calprotectin, suggests that microbial shifts may underlie and in fact drive symptoms regardless of inflammation. Further studies are needed to determine if specific microbial shifts, metabolic pathway variation, and metabolites are reliably associated with PROs and, better yet, if these shifts can be modified with personalized, targeted microbial therapy to improve patient outcomes in IBD.
Supplementary Material
Acknowledgments
We thank Heidi Andersen, MD, for her contribution to data acquisition.
Contributor Information
Jennifer Hellmann, Division of Gastroenterology, Hepatology, and Nutrition, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA; Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, USA.
Allison Ta, Division of Gastroenterology, Hepatology, and Nutrition, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA; Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, USA.
Nicholas J Ollberding, Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, USA; Division of Biostatistics and Epidemiology, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA.
Ramona Bezold, Division of Gastroenterology, Hepatology, and Nutrition, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA.
Kathleen Lake, Division of Gastroenterology, Hepatology, and Nutrition, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA.
Kimberly Jackson, Division of Gastroenterology, Hepatology, and Nutrition, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA.
Kelsie Dirksing, Division of Gastroenterology, Hepatology, and Nutrition, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA.
Erin Bonkowski, Division of Gastroenterology, Hepatology, and Nutrition, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA.
David B Haslam, Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, USA; Division of Infectious Disease, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA.
Lee A Denson, Division of Gastroenterology, Hepatology, and Nutrition, Cincinnati Children’s Hospital Medical Center, Cincinnati, OH, USA; Department of Pediatrics, University of Cincinnati College of Medicine, Cincinnati, OH, USA.
Funding
This work was supported by the National Institutes of Health (Grant Nos. T32 DK007727, P30 DK078392, R01 HD94862 [to L.D.], and T32 ESO10957-14).
Conflicts of Interest
The authors have nothing to disclose.
Data Availability
Metagenomic shotgun data depleted of human reads will be submitted to the Short Read Archive (https://www.ncbi.nlm.nih.gov/sra) and will be made available for public release upon publication of the manuscript.
References
- 1. Benchimol EI, Fortinsky KJ, Gozdyra P, et al. Epidemiology of pediatric inflammatory bowel disease: a systematic review of international trends. Inflamm Bowel Dis. 2011;17(1):423-439. [DOI] [PubMed] [Google Scholar]
- 2. Machiels K, Joossens M, Sabino J, et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 2014;63(8):1275-1283. [DOI] [PubMed] [Google Scholar]
- 3. Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569(7758):655-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15(3):382-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kugathasan S, Denson LA, Walters TD, et al. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet 2017;389(10080):1710-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lewis JD, Chen EZ, Baldassano RN, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe. 2015;18(2):489-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. U.S. Department of Health and Human Services FDA Center for Drug Evaluation and Research; U.S. Department of Health and Human Services FDA Center for Biologics Evaluation and Research; U.S. Department of Health and Human Services FDA Center for Devices and Radiological Health. Guidance for industry: patient-reported outcome measures: use in medical product development to support labeling claims: draft guidance. Health Qual Life Outcomes 2006;4:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. U.S. Food and Drug Administration. Ulcerative Colitis: Clinical Trial Endpoints Guidance for Industry. Accessed. June 1, 2021. https://www.fda.gov/files/drugs/published/Ulcerative-Colitis--Clinical-Trial-Endpoints-Guidance-for-Industry.pdf
- 9. Jairath V, Khanna R, Zou GY, et al. Development of interim patient-reported outcome measures for the assessment of ulcerative colitis disease activity in clinical trials. Aliment Pharmacol Ther. 2015;42(10):1200-1210. [DOI] [PubMed] [Google Scholar]
- 10. Sandborn WJ, Ferrante M, Bhandari BR, et al. Efficacy and safety of mirikizumab in a randomized phase 2 study of patients with ulcerative colitis. Gastroenterology 2020;158(3):537-549.e10. [DOI] [PubMed] [Google Scholar]
- 11. Dragasevic S, Sokic-Milutinovic A, Stojkovic Lalosevic M, et al. Correlation of Patient-Reported Outcome (PRO-2) with endoscopic and histological features in ulcerative colitis and Crohn’s disease patients. Gastroenterol Res Pract 2020;2020:2065383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lewis JD, Rutgeerts P, Feagan BG, et al. Correlation of stool frequency and abdominal pain measures with Simple Endoscopic Score for Crohn’s Disease. Inflamm Bowel Dis. 2020;26(2):304-313. [DOI] [PubMed] [Google Scholar]
- 13. Feagan BG, Sandborn WJ, Danese S, et al. Ozanimod induction therapy for patients with moderate to severe Crohn’s disease: a single-arm, phase 2, prospective observer-blinded endpoint study. Lancet Gastroenterol Hepatol 2020;5(9):819-828. [DOI] [PubMed] [Google Scholar]
- 14. Higgins PDR. Development of the Crohn’s disease, Patient-Reported Outcomes (CD-PRO) Questionnaire in adults. Presented at the University of Michigan Third Gastroenterology Regulatory Endpoints and the Advancement of Therapeutics Workshop; March 31, 2015; Ann Arbor, Michigan.
- 15. Paramsothy S, Paramsothy R, Rubin DT, et al. Faecal microbiota transplantation for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis 2017;11(10):1180-1199. [DOI] [PubMed] [Google Scholar]
- 16. Plichta DR, Graham DB, Subramanian S, et al. Therapeutic opportunities in inflammatory bowel disease: mechanistic dissection of host-microbiome relationships. Cell 2019;178(5):1041-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Breton J, Kastl A, Hoffmann N, et al. Efficacy of combination antibiotic therapy for refractory pediatric inflammatory bowel disease. Inflamm Bowel Dis. 2019;25(9):1586-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hellmann J, Andersen H, Fei L, et al. Microbial shifts and shorter time to bowel resection surgery associated with C. difficile in pediatric Crohn’s disease. Inflamm Bowel Dis. 2020;26(8):1212-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cannatelli R, Bazarova A, Zardo D, et al. Fecal calprotectin thresholds to predict endoscopic remission using advanced optical enhancement techniques and histological remission in IBD patients. Inflamm Bowel Dis. 2021;27(5):647.-. [DOI] [PubMed] [Google Scholar]
- 20. Kappelman MD, Crandall WV, Colletti RB, et al. Short pediatric Crohn’s disease activity index for quality improvement and observational research. Inflamm Bowel Dis. 2011;17(1):112-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Joshi NA, Fass JN.. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files (version 1.33). Accessed August 1, 2020. https://github.com/najoshi/sickle [Google Scholar]
- 22. Lu J, Breitwieser FP, Thielen P, et al. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci. 2017;3(3):e104. [Google Scholar]
- 23. Franzosa EA, McIver LJ, Rahnavard G, et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods. 2018;15(11):962.-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mollie E, Brooks KK, van Benthem KJ, et al. glmmTMB balances speed and flexibility among packages for zero-inflated generalized linear mixed modeling. R Journal 2017;9(2):378-400. [Google Scholar]
- 25. McMurdie PJ, Holmes S.. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bates D, Machler M, Bolker BM, et al. Fitting linear mixed-effects models using lme4. J Stat Soft 2015;67(1):1-48. [Google Scholar]
- 27. Kuznetsova A, Brockhoff PB, Christensen RHB.. lmerTest package: tests in linear mixed effects models. J Stat Soft 2017;82(13):1-26. [Google Scholar]
- 28. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen L, Reeve J, Zhang L, et al. GMPR: a robust normalization method for zero-inflated count data with application to microbiome sequencing data. PeerJ. 2018;6:e4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen J, King E, Deek R, et al. An omnibus test for differential distribution analysis of microbiome sequencing data. Bioinformatics 2018;34(4):643-651. [DOI] [PubMed] [Google Scholar]
- 31. Caspi R, Billington R, Fulcher CA, et al. The MetaCyc database of metabolic pathways and enzymes. Nucleic Acids Res. 2018;46(D1):D633-D639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schirmer M, Denson L, Vlamakis H, et al. Compositional and temporal changes in the gut microbiome of pediatric ulcerative colitis patients are linked to disease course. Cell Host Microbe. 2018;24(4):600-610.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lauer E, Kandler O.. Lactobacillus gasseri sp. nov., a new species of the subgenus Thermobacterium. Zentralbl Bakteriol A. 1980;1(2):75-78. [Google Scholar]
- 34. Clooney AG, Eckenberger J, Laserna-Mendieta E, et al. Ranking microbiome variance in inflammatory bowel disease: a large longitudinal intercontinental study. Gut 2021;70(3):499-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saulnier DM, Riehle K, Mistretta TA, et al. Gastrointestinal microbiome signatures of pediatric patients with irritable bowel syndrome. Gastroenterology 2011;141(5):1782-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rajilić-Stojanović M, Biagi E, Heilig HG, et al. Global and deep molecular analysis of microbiota signatures in fecal samples from patients with irritable bowel syndrome. Gastroenterology 2011;141(5):1792-1801. [DOI] [PubMed] [Google Scholar]
- 37. Durazzi F, Sala C, Castellani G, et al. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci Rep. 2021;11(1):3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Anyansi C, Straub TJ, Manson AL, et al. Computational methods for strain-level microbial detection in colony and metagenome sequencing data. Front Microbiol. 2020;11:1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Coburn LA, Horst SN, Allaman MM, et al. L-arginine availability and metabolism is altered in ulcerative colitis. Inflamm Bowel Dis. 2016;22(8):1847-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gobert AP, Cheng YL, Akhtar M, et al. Protective role of arginase in a mouse model of colitis. J Immunol. 2004;173(3):2109-2117. [DOI] [PubMed] [Google Scholar]
- 41. Pruss KM, Enam F, Battaglioli E, et al. Oxidative ornithine metabolism supports non-inflammatory C. difficile colonization. Nat Metab. 2022;4(1):19-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roy AB, Hewlins MJ, Ellis AJ, et al. Glycolytic breakdown of sulfoquinovose in bacteria: a missing link in the sulfur cycle. Appl Environ Microbiol. 2003;69(11):6434-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gecse KB, Brandse JF, van Wilpe S, et al. Impact of disease location on fecal calprotectin levels in Crohn’s disease. Scand J Gastroenterol. 2015;50(7):841-847. [DOI] [PubMed] [Google Scholar]
- 44. Altomare A, Putignani L, Del Chierico F, et al. Gut mucosal-associated microbiota better discloses inflammatory bowel disease differential patterns than faecal microbiota. Dig Liver Dis. 2019;51(5):648-656. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Metagenomic shotgun data depleted of human reads will be submitted to the Short Read Archive (https://www.ncbi.nlm.nih.gov/sra) and will be made available for public release upon publication of the manuscript.