Abstract
Phospholipid extrusion by ABC subfamily A (ABCA) exporters is central to cellular physiology, although the specifics of the underlying substrate interactions and transport mechanisms remain poorly resolved at the molecular level. Here we report cryo‐EM structures of lipid‐embedded human ABCA7 in an open state and in a nucleotide‐bound, closed state at resolutions between 3.6 and 4.0 Å. The former reveals an ordered patch of bilayer lipids traversing the transmembrane domain (TMD), while the latter reveals a lipid‐free, closed TMD with a small extracellular opening. These structures offer a structural framework for both substrate entry and exit from the ABCA7 TMD and highlight conserved rigid‐body motions that underlie the associated conformational transitions. Combined with functional analysis and molecular dynamics (MD) simulations, our data also shed light on lipid partitioning into the ABCA7 TMD and localized membrane perturbations that underlie ABCA7 function and have broader implications for other ABCA family transporters.
Keywords: ABCA7, Alzheimer's disease, cryo‐EM, exporter, flippase
Subject Categories: Structural Biology
Snapshots of open, closed and intermediate states show that human ABCA7 expands and contracts like a pair of bellows to extrude phospholipids from the membrane.

Introduction
ABCA family exporters mediate efflux of phospholipids and sterols from cells, contributing to membrane homeostasis, bilayer structure and asymmetry, and the formation of serum lipoproteins, among other key physiological processes (Albrecht & Viturro, 2007). Their dysfunction therefore underlies several human diseases (Allikmets et al, 1997; Brooks‐Wilson et al, 1999; Akiyama et al, 2005). The molecular details governing the ABCA exporter substrate transport cycle are not fully resolved. To fill this knowledge gap, here we present the structural and functional analysis of human ABCA7, whose dysfunction has been strongly linked to Alzheimer's disease (AD) (Hollingworth et al, 2011; Lambert et al, 2013; Le Guennec et al, 2016; Swerdlow, 2016; Bellenguez et al, 2017; Almeida et al, 2018; Ma et al, 2018; De Roeck et al, 2019), in a lipid environment. Deficient ABCA7 activity leads to alterations in both brain lipid profiles and fatty acid and phospholipid biosynthetic pathways (Aikawa et al, 2021), impaired memory, and reduced immune responses (Serhan, 2014; Astudillo et al, 2019). Both in vitro lipid flipping (Quazi & Molday, 2013) and lipid extrusion to apolipoproteins by cells over‐expressing ABCA7 (Tomioka et al, 2017) have been demonstrated, although the correlation between the two processes, if any, remains unclear. To date, no direct structural information exists for ABCA7, and molecular insights into how its dysfunction alters inflammatory and immune responses, lipid homeostasis, and phagocytosis, which all contribute to AD progression (Popp et al, 2012; Testa et al, 2016; Tonnies & Trushina, 2017; Newcombe et al, 2018; Czubowicz et al, 2019) are missing.
ABCA7 encodes a 2,146 amino acid membrane transporter found in many tissues and blood, hippocampal neurons, macrophages, and microglia (Iwamoto et al, 2006; Sleegers, 2020). Like the phospholipid and sterol exporter ABCA1 and retinal importer ABCA4, with which it shares 54 and 59% sequence similarity, respectively, ABCA7 comprises two halves assembled as a full transporter. Each half consists of a TMD, with the first two transmembrane helices (TMs) of each separated by a large extracellular domain (ECD), and a nucleotide binding domain (NBD) attached to a cytoplasmic regulatory domain (RD). To visualize its ATP‐dependent conformational cycle in a lipid environment, we resolved the structures of human ABCA7 in multiple conformations in lipid and detergent environments using cryo‐EM and probed its lipid interactions using ATPase assays and MD simulations. Our data allow us to directly visualize lipid partitioning into the TMDs and the associated conformational changes in ABCA7 that provide insights into its mechanisms of substrate entry and export that likely hold true for other members of the ABCA family and formulate a model for its potential role in inter‐bilayer lipid translocation as well as lipid extrusion from the extracellular leaflet.
Results
Dependence of ABCA7 ATPase activity on its lipid environment
Human ABCA7 expressed in a tetracycline inducible stable HEK293 cell line was reconstituted in liposomes and nanodiscs comprising 80% brain polar lipids (BPL) and 20% cholesterol (Chol) and its ATPase activity was measured (Fig 1A, Appendix Fig S1A). Although ATP hydrolysis was slowest in nanodiscs, it followed Michaelis–Menten kinetics similar to ABCA7 in detergent or liposomes comprising the same lipid/cholesterol composition and Michaelis constant (KM) values for all three were in the 0.5–0.8 mM range (Appendix Table S1). ATPase rates for a hydrolysis‐deficient mutant carrying E965Q and E1951Q substitutions (ABCA7EQ) were drastically reduced compared to wildtype in both nanodiscs and detergents, demonstrating that the observed activity was specific (Appendix Fig S1A).
Figure 1. Functional characterization and structure of human ABCA7 incorporated in nanodiscs.
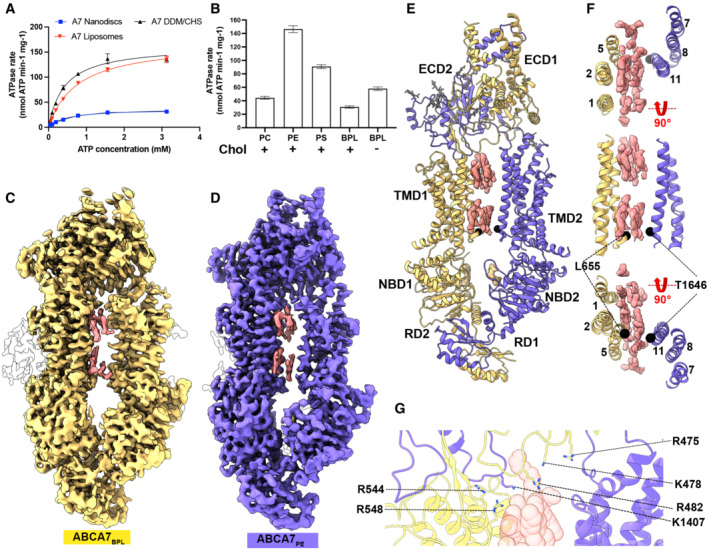
-
A, B(A) ATPase activity of ABCA7 at different ATP concentrations and (B) different nanodisc phospholipid/cholesterol compositions. Cholesterol (20%) presence is indicated by + or −. Experimental replicates (n) = 3 and error bars represent standard deviation (s.d.).
-
C, D(C) Cryo‐EM map of ABCA7BPL (yellow) at 3.6 Å resolution and (D) ABCA7PE at 4.0 Å resolution (purple). Density for protein is shown in yellow (0.025 contour) and that for modeled lipid acyl chains (0.035 contour) shown in pink (TMD luminal lipids) and white (peripherally associated lipids).
-
EABCA7PE shown in ribbon format with half 1 colored yellow and half 2 purple, along with density for TMD lipids (pink 0.025 contour) and bound nucleotide (yellow 0.035 contour). Acyl chains and glycans are shown as pink and gray sticks, respectively.
-
FTMD lumen with density for TMD lipids (pink 0.025 contour) viewed from the extracellular side (top), membrane plane (middle) and cytoplasmic side (bottom). Cα atoms for the cytoplasmic gate are shown as black spheres.
-
GView of the TMD‐ECD interface with select residues oriented toward lipids (transparent red spheres) shown.
To analyze the effect of different lipid headgroups on ATPase activity, we compared ATPase rates of ABCA7 in BPL/Chol nanodiscs to those in which BPL was replaced by either brain polar phosphatidylethanolamine (PE), phosphatidylserine (PS), or phosphatidylcholine (PC), as well a 100% BPL nanodiscs. As seen in Fig 1B, ATPase rates were highest in PE and PS nanodiscs followed by 100% BPL nanodiscs lacking cholesterol and, lastly, PC nanodiscs that showed a marginal increase. These results largely match previous findings (Quazi & Molday, 2013), although differences exist in extent of ATPase rate stimulation, specifically the high ATPase stimulation with PE that are likely due to the divergent lipid formulations used for reconstitution. Overall, they demonstrate that lipids modulate ABCA7 activity in a species dependent manner and that cholesterol has an inhibitory effect on ATPase activity of ABCA7 at least in nanodisc reconstituted samples.
Cryo‐EM structures of ABCA7 in nanodiscs reveal an asymmetric, open TMD
To visualize ABCA7 in a lipid environment, we determined its cryo‐EM structures in BPL/Chol nanodiscs (ABCA7BPL) at 3.6 Å resolution (Fig 1C, Appendix Fig S1B–E). Despite the addition of the non‐hydrolysable ATP analog adenosine‐5′‐o‐(3‐thio‐triphosphate) (ATP𝛾S), the transporter adopted an open conformation with separated NBDs and a wide open TMD pathway. We observed density features consistent with a patch of ordered lipids from both bilayer leaflets traversing the width of TMD as expanded upon below. A second, higher‐resolution 3D class displayed more complete density for the ECD tunnel region but comparatively lower quality of EM density for TMD2‐NBD2 and the entire RD, indicating greater conformational disorder, which was therefore not analyzed further (Appendix Fig S1D and E). Analysis of TMD‐ECD interfaces in the ABCA7BPL structures revealed more extensive contacts between the ECD and TMD1 (buried surface area (BSA) of 790 Å2) compared to TMD2 (330 Å2 BSA). Both TMD1 and TMD2 made contacts with the opposite ECD subunits, leading to a domain swapped arrangement. The TMD1‐ECD2 interface (BSA 565 Å2) was significantly larger than that of TMD1‐ECD1 (BSA 223 Å2) and both interfaces comprised an extensive network of polar and electrostatic interactions. The RD adopted a domain‐swapped arrangement similar to that in ABCA4 structures (Liu et al, 2021), with RD1 and RD2 associated with the opposite NBDs. To see whether structural changes could rationalize the enhanced ATPase activity observed in ABCA7PE compared to ABCA7BPL, we also determined the cryo‐EM structure of the former to 4.0 Å resolution (Fig 1D, Appendix Fig S2). Interestingly, both structures were identical to each other (r.m.s.d 0.31 Å). However, the ABCA7PE sample appears more conformational homogeneous, with only a single high‐resolution 3D class resolved (Appendix Fig S2) and more homogeneous density for TMD lipids compared to ABCA7BPL despite its comparatively lower resolution, as described below. For both ABCA7BPL and ABCA7PE, greater positional disorder was observed for TMD2‐NBD2 compared to TMD1 and NBD1 as indicated by relatively weaker EM density quality (Fig EV1A).
Figure EV1. Variation in local resolution and TMD electrostatics.
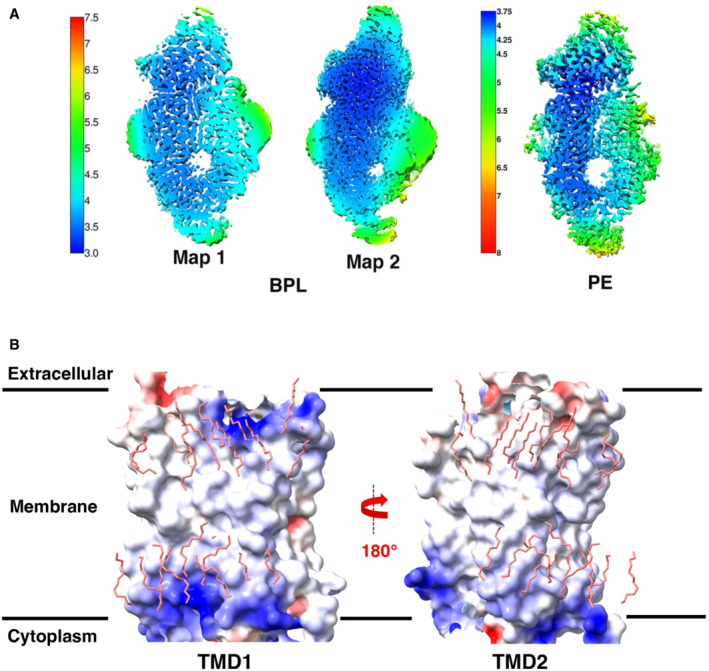
- Local resolution maps of ABCA7BPL and ABCA7PE structures. Color keys are indicated on the left of each set of maps with numbers representing resolution (Å).
- Analysis of TMD electrostatics of ABCA7PE with electrostatic potential maps for TMD1 (left) and TMD2 (right) shown along with modeled lipids (pink sticks).
The ABCA7 TMD lumen is accessible to an ordered file of bilayer lipids
Our ABCA7BPL/PE structures are distinguished from available structures of ABCA1, ABCA3, and ABCA4 (all resolved in a detergent environment) by the wider open TMD lumen that is almost completely occupied by lipids. The resolution and quality of the EM density for these features precluded a determination of the exact identity of the lipids, which were therefore only modeled as individual acyl chains. The observed lipids are continuous with the surrounding membrane except at the cytoplasmic/lower leaflet near residues L655 and T1646 from TM5 and TM11, respectively (Fig 1E and F). EM density for the modeled lipids is recognizable by gaps between the extracellular/upper and cytoplasmic leaflets and the two acyl chains of a single file of phospholipids in the two lipid leaflets. Toward the extracellular end, ECD residues R475, K478, R482, K1407, and TMD1 residues R544 and R548 form a cluster of positively charged side chains oriented towards the luminal lipids (Fig 1G). Overall, the luminal lipids are more closely associated with TMD1, with residues from TM2, TM5, and TM11 within 5 Å of the modeled acyl chains compared to only residues from TM11 from TMD2. TMD interfaces with the observed lipids display differences in electrostatics (Fig EV1B) that may contribute to orientation of luminal lipids toward TMD1. To gain insight into the role the identified positively charged ECD residues may play in functional lipid interactions of ABCA7, we generated an ABCA7 mutant harboring R475A, K478A, and R482A substitutions (ABCA7AAA). While this mutant displayed similar expression profiles and biochemical behavior as the wildtype protein (Fig EV2A), it displayed significantly lower ATPase activity in liposomes (Fig EV2B). A qualitative analysis of annular lipids co‐purifying with ABCA7 and ABCA7AAA by thin layer chromatography (TLC) revealed no discernable differences between the two, with PE, PS, and PC phospholipids detected in detergent purified samples (Fig EV2C).
Figure EV2. Purification and characterization of ABCA7 and ABCA7AAA .
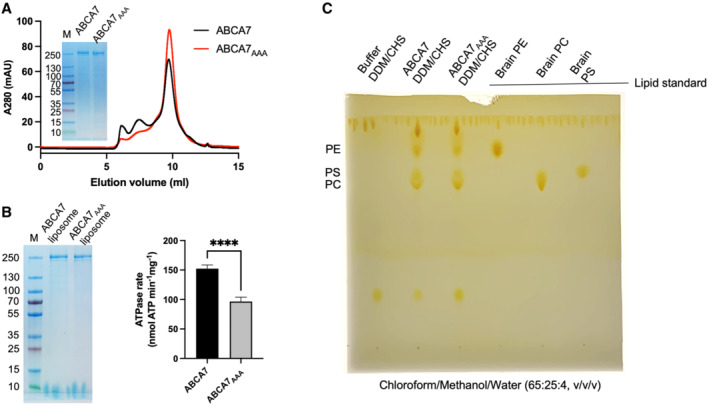
- SEC chromatograms and SDS‐PAGE analysis of ABCA7 and ABCA7AAA in DDM/CHS.
- SDS‐PAGE gel and ATPase activity comparison of ABCA7 and ABCA7AAA reconstituted in liposomes. Experimental replicates (n) = 6 and error bars represent standard deviation (s.d.). Statistical significance by unpaired, two‐tailed t‐test P‐value of < 0.0001 is indicated by ****.
- Detection of endogenous lipids co‐purified with ABCA7 and ABCA7AAA by thin‐layer chromatography (TLC).
Source data are available online for this figure.
To compare the architecture of ABCA7 in the presence or absence of lipids, we also determined its structure in the detergent digitonin (Fig 2A and B, Appendix Fig S3A–E). This ABCA7DIGITONIN structure revealed a similar overall conformation to that seen in structures of ABCA1 (Qian et al, 2017) and ABCA4 (Liu et al, 2021; Scortecci et al, 2021; Xie et al, 2021) with a more complete map for the ECD compared to ABCA7BPL/PE. No EM density for TMD lipids was visible, and only isolated density features attributable to detergent molecules associated with TMD1 were observed, although only at a contour level below where much of the surrounding detergent micelle was visible (Fig 2A and C). This is in contrast to ABCA7BPL/PE structures where TMD lipid density was much stronger than that of the bulk nanodisc (Fig 2D and E), further validating our assignment of bilayer lipids. Compared to structures in nanodiscs, the TMDs in this ABCA7DIGITONIN structure were arranged more symmetrically with respect to each other with a narrower lumen. Interestingly, EM density for TMD2 was still weaker than that of TMD1, in contrast to structures of ABCA4 (Liu et al, 2021; Scortecci et al, 2021; Xie et al, 2021) and ABCA1 (Qian et al, 2017; Appendix Fig S3E) indicating that a more mobile TMD2‐NBD2 pair may be a unique feature of ABCA7.
Figure 2. Cryo‐EM structure of ABCA7DIGITONIN .
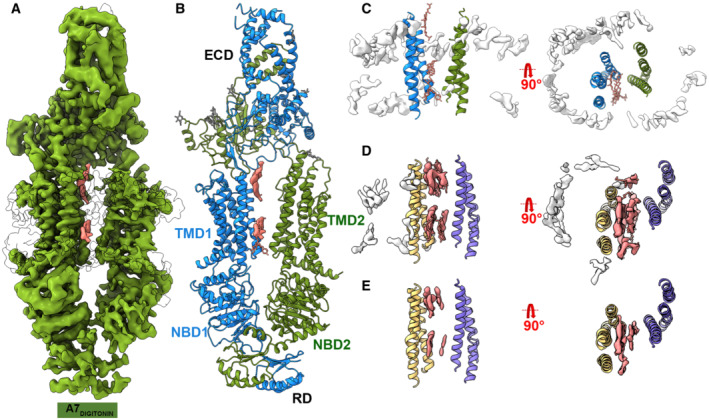
- Cryo‐EM map of human ABCA7DIGITONIN at 3.9 Å with density for protein shown in green (0.013 contour) and extraneous density likely belonging to detergent shown in pink (0.013 contour).
- Cryo‐EM structure of human ABCA7DIGITONIN shown in ribbon format with each half colored differently (blue and green).
- TMD lumen of ABCA7DIGITONIN with density for bulk micelle shown in white at 0.015 contour where detergent density inside TMDs disappears. Sticks for unmodeled detergent molecules are shown for which density is visible at the lower contour of 0.013.
- TMD lumen of ABCA7PE with density (0.035 contour) for TMD lipids (pink) and peripherally associated ordered lipids (white) shown.
- Same as (D) with higher density contour of 0.046 where density of peripherally associated lipids is absent but that of luminal lipids remains.
Closed TMD lumen and exit pocket for lipid extrusion in ATP bound ABCA7
To gain insight into possible mechanisms of lipid extrusion from the ABCA7 TMD, we used the hydrolysis‐deficient ABCA7EQ mutant reconstituted in BPL/Chol nanodiscs and determined its structure in a closed, ATP‐bound state at 3.7 Å resolution (Fig 3A, Appendix Fig S4). This ABCA7EQ‐ATP structure revealed closely interacting TMD‐NBD pairs and a largely occluded TMD lumen (Fig 3B). At the extracellular end of the TMD lumen, we observed a small central opening to the extracellular space/ECD that may represent an “exit pocket” (Fig 3C and D) akin to that seen in the yeast pleiotropic drug resistance transporter Pdr5 (Harris et al, 2021) that could likely accommodate two acyl chains (Fig 3D). TMD closure involves formation of a 4‐TM bundle comprising TMs 2, 5, 8, and 11 that occludes the cytoplasmic bilayer leaflet. The exit pocket is comprised largely of hydrophobic residues, except for R548, which is part of a cluster of positively charged residues including R475, K478, R482, and K1407 identified in our ABCA7BPL/PE structures that may aid in directing a negatively charged phospholipid headgroup toward the ECD. R678, previously oriented away from the TMD lipids in the open conformation is now seen to swing toward the exit pocket. In general, side‐chain density for this group of basic residues was poor compared to that in ABCA7BPL/PE structures, indicating greater disorder. TMD closure is accompanied by a rearrangement of both TMD‐ECD interfaces compared to the open state structures (Appendix Fig S5). Compared to the ABCA7BPL structure, both TMD1 and TMD2 interfaces with ECD1 increased from BSAs of 190 to 331 Å2 and from 76 to 285 Å2, respectively. Conversely, the BSAs of TMD1 and TMD2 with ECD2 decreased from 543 to 275 Å2 and from 317 to 192 Å2, respectively. As expected, the ABCA7EQ‐ATP structure shows a canonical NBD sandwich dimer with bound ATP (Fig 3E). In contrast to the ATP bound ABCA4 structure, RD density was too weak for accurate placement and was removed in the final model. However, contoured at lower thresholds, the available density features were more compatible with a rigid body shift rather than a conformational rearrangement of RD, with RD2 appearing to maintain contact with NBD1, but RD1 disengaged from NBD2.
Figure 3. Cryo‐EM structure of ABCA7EQ‐ATP .
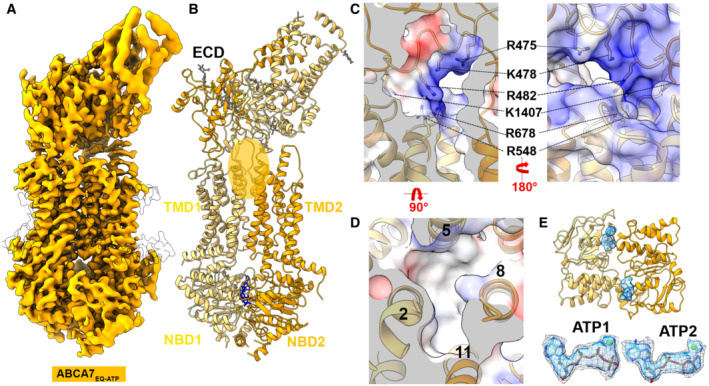
- Cryo‐EM map of human ABCA7EQ‐ATP in BPL/Chol nanodiscs at 3.7 Å resolution (orange density, 0.02 contour).
- Cryo‐EM structure of ABCA7EQ‐ATP shown in ribbon format with half 1 and half 2 colored yellow and orange, respectively. The transparent orange oval demarcates the observed exit pocket.
- Central slice of electrostatic surface map showing surface details of exit pocket in two different orientations.
- 4‐TM bundle forming the exit pocket.
- NBDs viewed from the extracellular side with density for bound nucleotide (sticks colored by element) shown in blue (0.02 contour). Two ATP molecules are shown with the density at two different contours 0.02 (mesh) and 0.035 (blue) at the bottom.
Rigid‐body motions of the TMDs define ABCA7 conformational transitions
Despite significant overall conformational differences, the individual TMDs in ABCA7BPL/PE, ABCA7DIGITONIN, and ABCA7EQ‐ATP remain largely unaltered (Fig 4A and D). Moreover, the TMD‐NBD pairs move as rigid body units from the fully open ABCA7BPL structure to that of ABCA7DIGITONIN, which we assert is akin to an intermediate open state between the fully open ABCA7BPL/PE state and the closed ABCA7EQ‐ATP states. The transition from open to intermediate open involves a rigid‐body rotation of 9° and translation of 2 Å for TMD2‐NBD2. The transition to the closed state involves a further 6° rotation and 15 Å translation of TMD2‐NBD2 (Fig 4B and C), albeit with a greater alteration in the NBDs owing to the expected rearrangement of the rec‐A like domain and helical domains associated with NBD dimerization. Rigid‐body movements were also observed for the entire RD. The RD of ABCA7DIGITONIN maintained molecular interactions with both NBD1 (RD2) and NBD2 (RD1), whereas the RD of ABCA7BPL/PE separated from NBD2 while maintaining contact with NBD1. Finally, the ECD base region transitioned through a rigid body rotation of 34° and translation of 6.3 Å going from the open to closed states, whereas the tunnel and lid regions displayed greater heterogeneity.
Figure 4. Comparison of open and closed conformations of ABCA7.
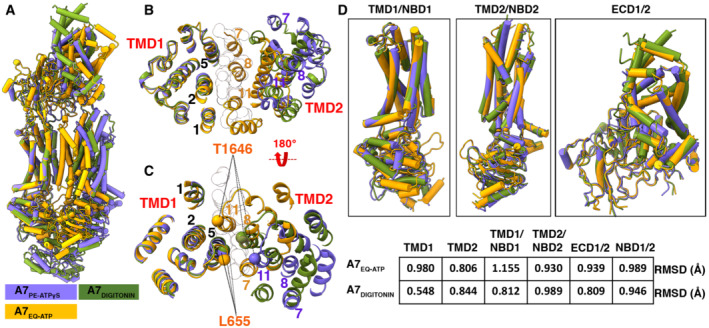
- Overall structural alignment of the three ABCA7 conformations.
- Overall alignment of the three ABCA7 conformations showing only TMD1 and TMD2 viewed from the extracellular side using the TMD1‐NBD1 pair as an alignment reference. TMs lining the TMD pathway are numbered, and modeled lipid acyl chains are shown as outlined white spheres.
- Same as panel (B), viewed from the cytoplasmic side with Cα atoms of gate forming residues shown as spheres.
- Individual alignments of rigid body pairs TMD1‐NBD1, TMD2‐NBD2, and ECD along with the Root mean square deviations (RMSD) of aligned atoms of ABCA7PE versus ABCA7EQ‐ATP and ABCA7DIGITONIN.
To establish whether the concerted TMD‐NBD rigid body motions outlined above may extend to other ABC transporters of the Type‐V ABC transporter/Type II ABC exporter fold (Thomas et al, 2020), we extended our analysis of individual TMD‐NBD pairs to members of the G family, using ABCG2 and ABCG5/G8 as examples (Fig EV3A; Manolaridis et al, 2018; Orlando & Liao, 2020). As shown in Fig EV3B, the TMD‐NBD pairs from both the apo and ATP‐bound closed states of ABCG2, as well as from both halves of ABCG5/G8, all shared a similar overall architecture. Despite the divergent topologies of ABCG and ABCA transporters, with the former are arranged in an NBD‐TMD configuration compared to TMD‐NBD for the latter, the individual TMD‐NBD pairs from ABCA7 shared very strong structural similarities with those of ABCG2, further establishing the role of rigid body movements of the TMDs to affect large‐scale conformational changes in type V ABC transporters.
Figure EV3. Conservation of ABCA and ABCG family structural elements.
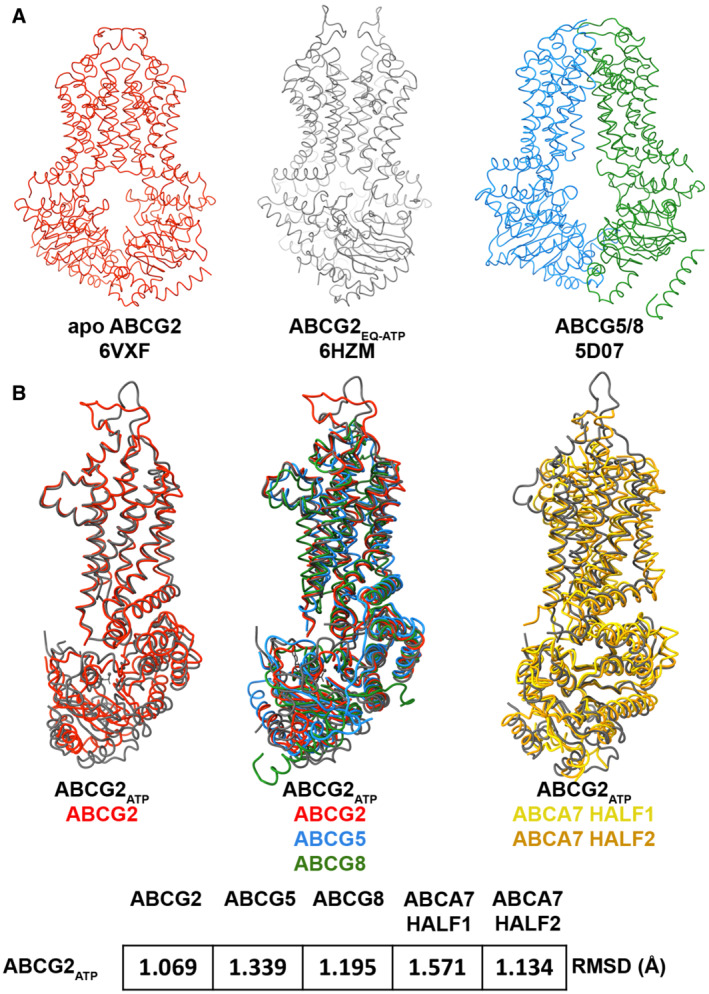
- Ribbon representation of select ABCG family transporter structures and their respective PDB IDs including apo open ABCG2 (red), closed ATP‐bound structure of ABCG2EQ (black), and ABCG5/G8 (blue and green, respectively).
- Alignment of NBD‐TMD pairs from open and closed conformations of ABCG2 (left), ABCG2 and ABCG5/G8 (center), and closed ABCG2 with TMD‐NBD pairs from ABCA7 half 1 (gold) and half 2 (orange) along with the root mean square deviations (RMSD) of aligned atoms shown at the bottom.
Lipid partitioning in the ABCA7 TMD captured with MD simulations
To gain additional molecular insight into the TMD‐mediated lipid partitioning, membrane perturbation, and lipid extrusion from ABCA7, we performed multi‐𝜇s MD simulations of the open conformation of ABCA7PE, with positional restraints on protein Cα atoms to maintain the conformation captured in our cryo‐EM structure. The transporter was embedded into two distinct lipid bilayers, one containing PE/Chol (4:1) and the other PC/Chol (4:1) (Figs 5 and EV4), chosen to mimic experimental conditions in our ATPase assays of samples with highest and lowest ATPase rate modulations. Each simulation was performed for 2 μs using a system including four copies of the protein (Fig 5A), resulting in an aggregate 8 μs sampling of lipid–protein interactions for each lipid composition. Interestingly, during the simulations, we observed PE and PC phospholipids penetrating the TMD cavity in both the cytoplasmic and extracellular leaflets in all four protein replicas (Fig 5A, insets). We quantified the number of phospholipids in this bilayer patch within the TMD cavity by counting the lipid headgroups (Fig EV4C, Appendix Fig S6A and B). We observed a distinct elevation of lipid headgroups in both bilayer leaflets within the TMD, causing the thickening and thinning of the extracellular and cytoplasmic leaflets, respectively. To further quantify ABCA7‐mediated deformation of the lipid bilayer bridging the TMDs, the average heights of phospholipid headgroups were calculated with respect to the midplane of the bilayer. We captured a tendency for lipids to move towards the cytoplasmic gate resulting in the formation of an elevated region of phospholipids within the TMD lumen (Figs 5B–D and EV4A and B). Interestingly, our MD simulations showed an accumulation of phospholipid headgroups in the vicinity of residues R475, K478, R482, R544, and R548, with R482 and R548 displaying the most frequent lipid contacts (Fig EV4D and E), corroborating our structural observations, and providing a rationale for the lipid‐elevation effect in the extracellular leaflet. Analysis of the cytoplasmic leaflet highlighted a continuous distribution of phospholipids with the surrounding membrane, except near TM5 and TM11 around residues L655 and T1646 (Fig 5C and D), which correlated well with the bilayer‐like density in the ABCA7BPL and ABCA7PE cryo‐EM maps (Fig 1C and D) that we modeled with acyl chains. Steric hindrance from this cytoplasmic gate may contribute to the observed lipid elevation.
Figure 5. MD Simulations of ABCA7 in Lipid Bilayers.
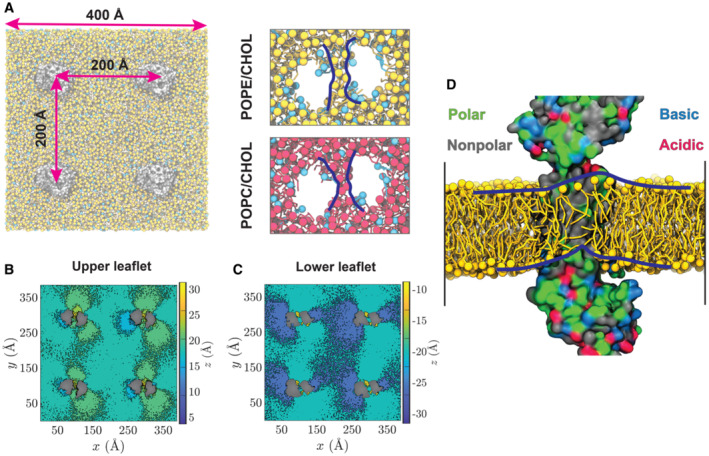
-
ALeft: A representative simulation system with four copies of ABCA7 (silver), taken from the POPE/cholesterol (yellow/cyan) lipid patch. Right: the phospholipid belt (blue lines) formed in (top) the POPE/cholesterol (yellow/cyan) and (bottom) POPC/cholesterol (red/cyan) membranes (t = 2 μs).
-
B, CHeatmaps representing the average height (z values) of POPC headgroups with respect to the membrane midplane in the extracellular/upper (B) and cytoplasmic/lower (C) leaflets. Phospholipids are observed to climb the protein and form an elevated configuration within the TMD lumen.
-
DSnapshot of lipids partitioned in the TMD lumen at t = 2 μs. Polar, nonpolar, basic, and acidic residues are colored green, gray, blue, and red, respectively. TMD2 is hidden for a clearer view of the luminal dome‐like lipid configuration (outlined by blue lines).
Figure EV4. Lipid configuration induced by protein copies in a bilayer and criteria used for identifying luminal lipids in MD simulations.
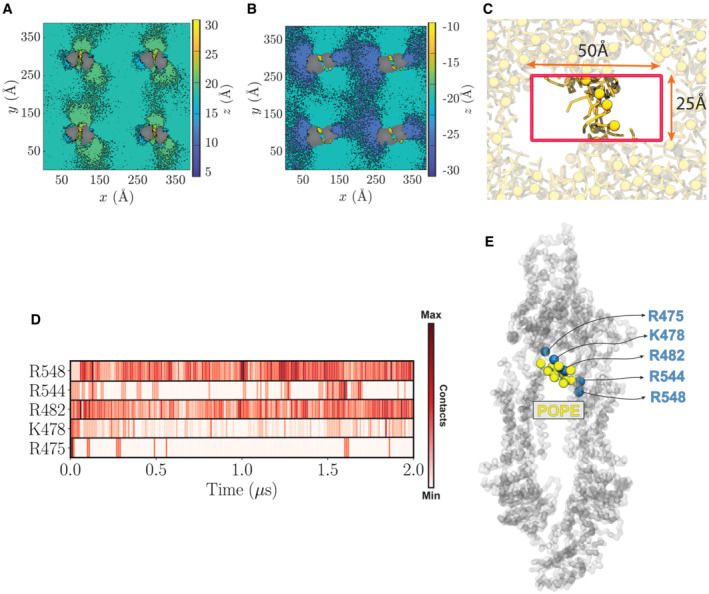
-
A, BPOPE headgroup height calculated for extracellular (A) and cytoplasmic (B) leaflets.
-
CDimension and position of the box used to calculate the number of phospholipids (yellow) partitioned in the TMD leaflets. The box is centered at the protein center and has dimensions of 50 and 25 Å in x and y directions, respectively. The ABCA7 transporter is hidden for clarity.
-
DContact map, based on the number of POPE headgroups in proximity of each residue throughout the simulation.
-
EAccumulation of POPE headgroups (yellow spheres) in close proximity of most frequently contacted residues (blue spheres) throughout the simulation. All protein residues are represented as spheres (Cα atoms).
Discussion
A mechanistic model derived from our results is shown in Fig 6, akin to a bellows like mechanism where TMD expansion/opening (ABCA7BPL/PE) draws in bilayer lipids and its contraction/closure (ABCA7EQ‐ATP) expels them back out into the bilayer or up into the ECD. Our ABCA7AAA data indicate the mutated ECD residues may play a role in extrusion of extracellular leaflet lipids. As this process is linked to NBD dimerization‐induced TMD closure, its disruption in ABCA7AAA may account for its lower ATPase activity. The identified residues are partially conserved in ABCA1, ABCA4, and ABCA7, with R678 and K1407 being the notable exceptions (Appendix Fig S7) and could be similarly important for substrate interactions. In ABCA7, R475 mutations have been identified in AD (Cuyvers et al, 2015) patients, and mutation of R638 in ABCA1 (equivalent to R548 in ABCA7) is associated with reduced serum HDL (Cohen et al, 2004; Appendix Fig S8). Our data suggest these pathological effects could be at least partially due to disrupted lipid interactions. The upward protrusion of both bilayer leaflet lipids within the TMD lumen observed in MD simulations is in line with this model of lipid extrusion from the extracellular leaflet. Favorable interaction of these positively charged side chains with the phospholipid headgroups may, upon TMD closure, aid in isolating phospholipids in the identified exit pocket, primed for extrusion toward the ECD. Similarly, the upward protrusion of extracellular leaflet lipids seen in our MD simulations may prime lipids for translocation from the cytoplasmic to extracellular leaflet.
Figure 6. Proposed model of ABCA7 conformational transitions and lipid interactions.
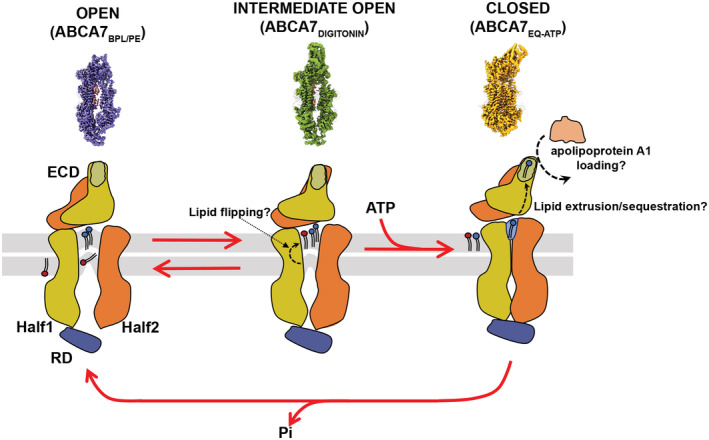
The proposed model displays the fully open conformation (left) with ordered lipids (red and blue) in the TMD, the intermediate open (middle) with the upward protrusion of the upper and lower leaflets with black dash arrows indicating movement of lipids under the influence of TMD conformation, and the closed conformation (right) exhibiting the lipid exit pocket for extrusion to the ECD where they may be sequestered and reside until an acceptor molecule is available. Question marks denote speculated mechanisms. The membrane bilayer leaflets are shown in gray. The surface maps on the upper panel represent three different states of ABCA7 the mechanistic model is derived from. Differences in ECD appearance are meant to show different orientations and not large‐scale structural transitions.
Membrane deformations similar to those highlighted here have also been observed in the signal peptidase complex (Liaci et al, 2021), and further evidence the influence membrane proteins have on local membrane bilayer structure. ABCA7 could thus play a role in changing its local membrane lipid environment by flipping cytoplasmic leaflet lipids to the extracellular leaflet and also extruding lipids back out into the bulk bilayer environment and toward the ECD. Whether it does so with explicit specificity is yet to be fully understood. Our observation of enhanced ATPase activity of the ABCA7PE sample may be, in part, due to the smaller PE headgroup, which has been shown to aid in folding and stabilization of membrane proteins (Raja, 2011) and easier passage into and out of the TMD upon the transporter's conformational cycling. Lipid translocation, whether from the cytoplasmic to extracellular leaflet, or extrusion from the extracellular leaflet may take advantage of the existing asymmetric bilayer lipid distribution. Our proposed model is in line with this and rationalizes the reported higher levels of PC extrusion (Tomioka et al, 2017) and higher levels of PS translocation from the cytoplasmic to extracellular leaflet (Quazi & Molday, 2013), considering the former headgroup is predominantly found in the extracellular leaflet while the latter is enriched in the cytoplasmic leaflet. In line with this, the reported defects in the phagocytotic activity of cells that express dysfunctional ABCA7 (Tanaka et al, 2010; Abe‐Dohmae, 2012) could be related to ABCA7's influence on the asymmetric distribution of bilayer lipids (Zachowski, 1993), as enrichment of PS at the extracellular surface is linked to phagocytosis and phagocytosis‐associated proteins within the membrane (Fadok et al, 2001; Hirt & Leist, 2003; Gabande‐Rodriguez et al, 2020).
While it is still unclear whether TMD–lipid interactions in other ABC transporters, e.g., ABCA1, are similar to those reported here, or the reported patterns are unique to ABCA7, our data and the proposed mechanistic model are in line with the purported function of ABCA1 both as an extracellular leaflet lipid extruder (Segrest et al, 2022) and as a cytoplasmic leaflet lipid flippase. Additional insights from structure–function studies of ABCA transporters are required to address such open questions as how the opposite directions of substrate transport in exporters like ABCA1 and ABCA7 and the importer ABCA4 is achieved considering similarities in the ATP‐bound closed and open state structures of ABCA family transporter structures (Fig EV5), and what the exact mechanisms of ABCA transporter interactions with apolipoproteins are.
Figure EV5. Structure alignment of ABCA7, ABCA1, ABCA3 and ABCA4.
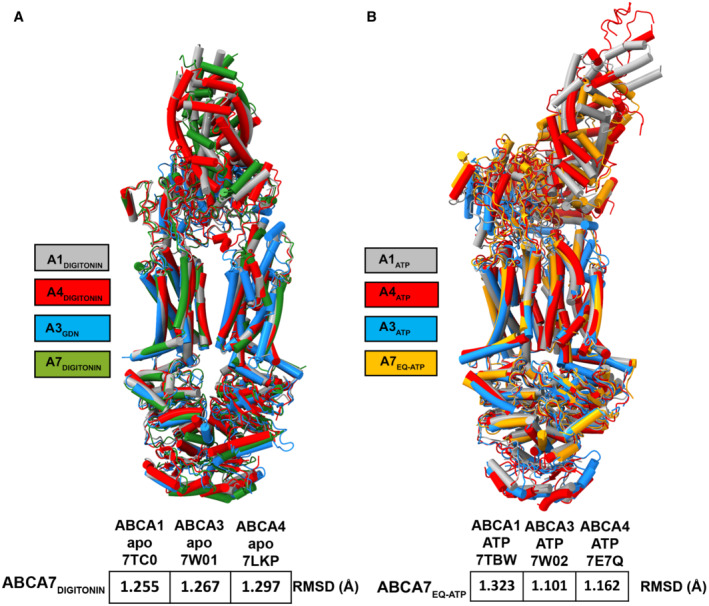
-
A, BSuperposition of (A) ABCA7 (green, this manuscript), ABCA1 (gray, PDB: 7TC0), ABCA3 (blue, PDB: 7W01), and ABCA4 (red, PDB: 7LKP) structures in detergent and (B) ABCA7 (orange, this manuscript), ABCA1 (gray, PDB: 7TBW), ABCA3 (blue, PDB: 7W02), and ABCA4 (red, PDB: 7E7Q) structures in ATP‐bound states. Color keys are indicated on the left and the RMSD of aligned atoms of ABCA7 versus ABCA1, ABCA3, and ABCA4 are displayed at the bottom of each set of structures.
Materials and Methods
Protein purification
We utilized the Flp‐In TREX system (Thermo Fisher Scientific) for tetracycline inducible expression of human ABCA7. In short, a codon optimized synthetic gene construct (GeneArt/Thermo Fisher Scientific) of isoform 1 of ABCA7 (Uniprot ID Q8IZY2‐1), harboring a C‐terminal eYFP‐Rho1D4 tag (Molday & Molday, 2014) with a 3C/precision protease site between the protein and purification tags, was cloned into a PCDNA5.1 FRT/TO vector between BamHI and NotI restriction sites and a stable cell line was generated as per manufacturer's protocol (Flp‐In™ T‐Rex™ Core Kit, Thermo Fisher Scientific). The resulting HEK293 based stable cells were grown and maintained in adherent cell culture in Dulbecco's Modified Eagle Medium (DMEM, Thermo Fisher Scientific) supplemented with 9% fetal bovine serum (FBS, Gibco) and a penicillin/streptomycin mixture (Thermo Fisher Scientific) at 37°C with 5% carbon dioxide (CO2) under humidified conditions. For protein production, cells were induced with 0.6 μg ml−1 tetracycline at a confluency of 80% in fresh DMEM supplemented with 2% FBS under otherwise identical conditions for an additional 72 h before being washed with phosphate‐buffered saline (PBS), harvested, and flash frozen in liquid nitrogen.
For purification, thawed cells were resuspended in a lysis buffer (Buffer L) comprising 25 mM Hepes pH 7.5, 150 mM sodium chloride (NaCl), 20% glycerol, 1 complete EDTA‐free protease inhibitor tablet (Roche) per 50 ml Buffer L, 800 μM phenylmethylsulfonyl fluoride (PMSF), 20 μg ml−1 soybean trypsin inhibitor (both Sigma), and mechanically cracked using a dounce homogenizer before addition of a 0.5%/0.1% w:v mixture of dodecyl maltoside (DDM) and cholesteryl hemisuccinate (CHS) (both Anatrace). Protein extraction was allowed to proceed for 90 min at 4°C with gentle agitation, after which, the suspension was centrifuged at 48,000 r.c.f for 30 min and the supernatant applied to rho‐1D4 antibody (University of British Columbia) coupled Sepharose resin (Cytiva). Binding was allowed to proceed for 3 h before the unbound fraction was discarded and beads rinsed with 4 × 10 bed volumes (BVs) of wash buffer (25 mM Hepes pH 7.5, 150 mM NaCl, 20% glycerol, 0.02%/0.004% w:v DDM/CHS). Protein was eluted by incubation with 3 BVs elution buffer (wash buffer supplemented with either 3C protease (1:10 w:w 3C:ABCA7) or 0.25 mg ml−1 1D4 peptide (GenScript)) for 2–18 h.
The EQ variant of ABCA7 contained two site mutations, E965Q and E1951Q. The E965Q site was created within the ABCA7 construct using site‐directed mutagenesis by PCR with the primers: A7eq1for 5′‐GGTCATCCTGGATCAACCTACAGCAGGCGTGG‐3′ and A7eq1rev 5′‐GCCTGCTGTAGGTTGATCCAGGATGACCACC‐3′. E1951Q was generated using a synthesized dsDNA block and the enzymes NheI and BsiWI. ABCA7EQ with C‐terminal eYFP‐Rho1D4 tag was then transferred to a pCAG vector (addgene) using KpnI and NotI restriction sites. The HEK293T cells were grown in Dulbecco's Modified Eagle Medium (DMEM, Scientific) supplemented with 9% FBS (Gibco), penicillin/streptomycin mixture (Scientific), and antimycotic (Gibco) at 37°C and 5% CO2 under humidified conditions. A mixture of 37.5 μg of ABCA7EQ plasmid and 75 μg of polyethyleneimine (PEI, Sigma) was incubated for 15 min at room temperature before being applied to 15 ml plates of HEK293T cells at a confluency of 60–80% to initiate transfection and expression. The cells were further cultured for 72 h before being washed with PBS, harvested, and flash frozen in liquid nitrogen. ABCA7EQ was purified using the same approach of ABCA7 and eluted by incubation with 3 BVs elution buffer containing either 3C protease for ATPase assay or with 0.25 mg ml−1 of 1D4 peptide and an additional 2 mM ATP and 10 mM MgCl2 for nanodisc reconstitution.
The ABCA7AAA variant harboring 3 ECD residue mutations (R475A, K478A, and R482A) was generated by swapping in gene fragments (IDT gBlocks) into the synthetic ABCA7 construct using XbaI and XmaI‐restriction sites. A stable cell line of ABCA7AAA was generated as described for the original construct. Expression, purification, and lipid reconstitution of the mutant followed the same approach as that for the original ABCA7 construct.
Membrane scaffold protein D1 (MSP1D1, addgene) and apoA1 were purified using established protocols for MSP (Ritchie et al, 2009) with the following modifications: A synthetic construct of apoA1 bearing a 3C protease cleavable N‐terminal deca‐histidine tag (GeneArt/Thermo Fisher Scientific) was cloned into a pET28a vector (Addgene) and transformed in Escherichia coli BL21 DE3 cells (New England Biolabs). One‐liter cultures of Terrific Broth (TB) supplemented with 50 μg ml−1 kanamycin were grown from 10 ml overnight cultures from single colonies grown in LB. Cells were grown to an OD600 of 0.8 in a shaking incubator at 37°C and induced with 1 mM isopropyl β‐d‐1‐thiogalactopyranoside (IPTG). Protein expression was allowed to proceed at 20°C for 12 h. Cells were centrifuged at 12,000 r.c.f, and pellets were flash frozen in liquid nitrogen and stored at −80°C until required. Frozen pellets were resuspended in 8 ml/gram cell pellet resuspension buffer comprising 25 mM Hepes pH 7.5, 150 mM NaCl, and 1 mM phenylmethylsulfonyl fluoride (PMSF) and sonicated. The suspension was spun down at 16,000 r.c.f at 4°C for 30 min, and the supernatant was applied 5 ml Ni‐NTA resin (Qiagen)/L culture medium. After discarding the flowthrough, the resin was washed with 25 mM Hepes pH 7.5, 150 mM NaCl, 1 mM PMSF, and 20 mM imidazole until a pre‐established baseline A280 reading was achieved. ApoA1 was eluted in 4 BVs of 25 mM Hepes pH 7.5, 150 mM NaCl, 1 mM PMSF, and 200 mM imidazole, concentrated using a 10 kDa molecular weight cutoff (MWCO) Amicon filter (Millipore‐Sigma) and desalted using a PD10 column (Cytiva) into 25 mM Hepes pH 7.5, 150 mM NaCl. The concentration of apoA1 was adjusted to 1 mg ml−1 for flash freezing in liquid nitrogen and storage at −80°C.
Nanodisc and proteoliposome preparation
For nanodisc reconstitution, peptide eluted or 3C cleaved ABCA7 was mixed with MSP1D1 and a mixture of BPL (brain polar lipid extract from Avanti) and cholesterol (80:20 w:w) with 0.5%/0.1% DDM:CHS using a 1:10:350 (ABCA7:MSPD1:lipid mix) molar ratio in nanodisc buffer (25 mM Hepes pH 7.5, 150 mM NaCl) that contained up to 4% glycerol for 30 min at room temperature (RT). Nanodisc reconstitution was induced by removing detergent with 0.8 mg ml−1 pre‐washed Biobeads SM‐2 (Bio‐Rad) for 2 h with gentle agitation at RT. For different phospholipid compositions, BPL was replaced with brain PE, PS, or PC (all from Avanti Polar Lipids). For structural studies, nanodisc‐reconstituted ABCA7 bearing the eYFP‐Rho1D4 tag was bound to rho‐1D4 resin for an additional 2 h, washed with 4 × 10 BVs of nanodisc buffer, and eluted with 3C protease for 2 h at 4°C. The eluted ABCA7 nanodiscs were concentrated using a 100,000 MWCO kDa Amicon filter and further purified by size exclusion chromatography using a G4000swxl column (TOSOH biosciences) equilibrated with nanodisc buffer at 4°C. Appendix Fig S1B shows a SEC chromatogram for pure ABCA7 placed into 80% BPL/20% Chol nanodiscs, while a SEC chromatogram for ABCA7 in 80% PE/20% Chol nanodiscs is in Appendix Fig S2A. Generally, three fractions were pooled from the main resultant peak.
ABCA7EQ was reconstituted in nanodiscs for cryo‐EM preparation using the same approach of ABCA7, except that an additional 2 mM ATP (Sigma) and 10 mM MgCl2 were present until the end of the purification procedure prior to grid preparation. A SEC chromatogram for ABCA7EQ‐ATP in BPL/Chol nanodiscs is shown in Appendix Fig S4A, where the trace is affected by the additional ATP added during the run.
ABCA7 proteoliposomes were generated by mixing detergent purified ABCA7 or ABCA7AAA with preformed liposomes at a protein: lipid ratio of 1:10 w:w. Liposomes were prepared by extruding a 20 mg ml−1, 80:20 w:w BPL/Chol lipid mixture 11 times using a previously described protocol (Geertsma et al, 2008). Briefly, detergent purified ABCA7 and liposomes were supplemented with a final concentration of 0.14 and 0.3% Triton X100 (Sigma), respectively, then incubated for 30 min at RT before being mixed and incubated for 60 min. Detergent was removed by adding 40 mg fresh Biobeads SM2 (Bio‐Rad) per ml reaction mixture in five successive incubation steps of 30 min at RT, 60 min at 4°C, overnight at 4°C, and two 60 min incubations at 4°C with gentle agitation. The suspension was centrifuged at 278,088 rcf for 20 min in an ultracentrifuge. The supernatant was removed, and the liposomal pellet was washed once with reconstitution buffer containing 150 mM NaCl, 25 mM Hepes pH 7.5. The ABCA7‐liposome suspension was then centrifuged to remove the supernatant, and the proteoliposomes were resuspended in reconstitution buffer at a final concentration of 0.5–1 mg ml−1 for ATPase assays.
ABCA7DIGITONIN preparation
For ABCA7DIGITONIN purification, ABCA7 was extracted from the cell in a lysis buffer (Buffer L) using the same approach as above, and the supernatant was applied to rho‐1D4 resin for a 3‐h binding period. The resin was then rinsed with the 4 × 10 BVs of wash buffer containing 25 mM Hepes pH 7.5, 150 mM NaCl, 20% glycerol (v/v), and 0.06% digitonin (w/v). Protein was eluted by incubation with 3 BVs of elution buffer composed of wash buffer supplemented with 3C protease (1:10 w:w 3C:ABCA7). Interestingly, we obtained better particle distribution and ice quality with addition of a 1:2.5 molar excess of apoA1, prepared in house, to 3C cleaved ABCA7 prior to grid preparation. The mixture was concentrated using a 100,000 kDa MWCO Amicon filter (Millipore) and further purified by size exclusion chromatography using a G4000swxl column (TOSHOH biosciences) equilibrated with a buffer containing 25 mM Hepes pH 7.5, 150 mM NaCl, 0.035% digitonin (w/v), as shown in Appendix Fig S3A. Peak fractions were pooled and concentrated for cryo‐EM grid preparation.
ATPase assays
ATPase assays were based on a molybdate‐based colorimetric assay (Chifflet et al, 1988). Protein concentrations used were in the range of 0.05–0.1 mg ml−1. Assays were started by the addition of either 2 mM ATP, except for experiments in Fig 1B where 6.25 mM ATP was used, in the presence of 10 mM magnesium chloride (MgCl2), incubated for 30 min at 37°C, then stopped by addition of 6% SDS. The assay was also performed in the presence of ABCA7 inhibitors as additives, such as ATPγS or sodium orthovanadate. For ATP KM measurements, a range of ATP concentrations was used. Statistical analysis was done using GraphPad Prism 9. ATPase rates were measured using simple linear regression, and the KM of detergent, liposome, and nanodisc reconstituted ABCA7 were determined from the fit to the Michaelis–Menten equation of the corresponding rates. All assays were performed in replicates (n) of three independent experimental setups except for the ABCA7 and ABCA7AAA comparison (n = 6). Statistical significance was calculated by unpaired, two‐tailed t‐test, with P‐values of < 0.05, 0.01, 0.001, 0.0001 indicated by *, **, ***, ****, respectively. ABCA7 concentrations were measured using gel densitometry analyzed in ImageStudio Lite (LI‐COR Biosciences) based on detergent purified ABCA7 standards with known concentrations determined by A280 measurements. All reaction components were mixed with ABCA7 in detergent or reconstituted in nanodiscs and liposome in the absence of ATP, incubated for 10 min at 37°C prior to addition of ATP to start the reaction.
Thin‐layer chromatography (TLC)
For TLC analysis of ABCA7, lipid extraction was carried out on detergent purified protein by mixing 0.15 mg of protein with chloroform and methanol at a 1:2:2 chloroform:methanol:protein volumetric ratio. The lipids in the lower chloroform phase were collected and dried under argon gas. The lipids were further dissolved in 20 μl of chloroform and 10 μl sample volume was applied to a silica TLC plate (Sigma) together with lipid standards including L‐α‐phosphatidylethanolamine (Brain PE), L‐α‐phosphatidylcholine (Brain PC), L‐α‐phosphatidylserine (Brain PS) (Avanti) that were previously used for nanodisc preparation. The different lipids were separated under a mobile phase (chloroform:methanol:water at 65:25:4 volumetric ratio) and stained with iodine vapor (Sigma) for visualization.
Cryo‐electron microscopy grid preparation
For ABCA7 reconstituted in nanodiscs comprising 80:20 w:w BPL/cholesterol and PE/cholesterol, SEC purified samples were mixed with 5 mM ATPγS (TOCRIS) and 5 mM MgCl2 for 20 min at room temperature and concentrated to 0.5–1.0 mg ml−1. ABCA7EQ‐ATP nanodisc samples were prepared similarly except that the SEC buffer also contained 2 mM ATP and 10 mM MgCl2. For ABCA7DIGITONIN, SEC purified samples were concentrated to between 2 and 5 mg ml−1. 4 μl samples were applied to glow discharged Quantifoil R1.2/1.3 grids (Electron Microscopy Sciences, Hatfield, PA, USA) using a Vitrobot Mark IV (Thermo Fisher Scientific) with a 4 s blotting time and 0 blotting force under > 90% humidity at 4°C, then plunge frozen into liquid ethane. For nanodisc reconstituted ABCA7EQ‐ATP and ABCA7DIGITONIN, two sample droplets were applied to glow discharged grids to obtain more particles per hole.
Cryo‐electron microscopy data collection and processing
Grids were clipped as per manufacturer guidelines, and cryo‐EM data was collected using a Titan Krios electron microscope operating at 300 kV and equipped with a Falcon 3EC direct electron detector (Thermo Fisher Scientific.). Automated data collection was carried out using EPU 2.8.0.1256REL software package (Thermo Fisher Scientific) over multiple sessions in counting mode at a nominal magnification of 96,000×, corresponding to a calibrated pixel size of 0.895 Å for nanodisc reconstituted ABCA7BPL. Image stacks comprising 60 frames were collected at a defocus range of −0.6 to −2.6 μm and estimated dose rate of 1 electron/Å2/frame and further processed in Relion‐3.1 (beta). Motion correction was done using Motioncor2 (Relion implementation) (Zheng et al, 2017), and contrast transfer function (CTF) correction was performed using Gctf 1.06 (Zhang, 2016). A summary of the overall data processing scheme and the quality was presented in Appendix Fig S1C–E. In brief, 11,802 micrographs were used for template free picking of 6,725,108 particles, followed by particle extraction at a 3× binned pixel size of 2.685 Å/pix. The data set was processed in two batches. After 2–3 rounds of 2D classification 1,259,324 particles from Set 1 and 1,088,487 particles from Set 2 were selected for independent 3D classification steps (number of classes (K) = 8 for both). The structure of human ABCA1 (EMDB6724) was used as a 3D reference for an initial 3D classification of a subset of the total data to yield an initial sub‐nanometer resolution map of ABCA7 that was used as a 3D reference for the full data sets. After 1 round of 3D classification, both sets of data yielded a similar ensemble of classes. A total of 113,291 particles from similar looking classes (black boxes) were subjected to an additional round of classification (K = 3), ~80% of which fell into a high‐resolution class that yielded a 3.6 Å map after refinement and particle polishing steps. Similarly, 124,114 particles from a second set of two similar classes (red boxes in Appendix Fig S1D) were selected for subsequent refinement, particle polishing, and post processing to yield a 3.1 Å map. All resolution estimates were based on the gold standard 0.143 cutoff criterion (Scheres & Chen, 2012). Data sets of ABCA7PE, ABCA7EQ‐ATP, and ABCA7DIGITONIN were collected at a nominal magnification of 96,000× corresponding to a calibrated pixel size 0.889 Å, and image stacks containing 40 frames were collected a defocus range of −0.8 to −2.6 μm with an estimated dose rate of 1 electron/Å2/frame and further processed using the same software versions as for the ABCA7BPL data set unless otherwise indicated.
For ABCA7PE, a total of 2,849,251 particles were picked from 9,218 ctf corrected (Gctf) micrographs in Relion ver. 3.1 in two batches at a threefold binned pixel size of 2.667 Å/pixel (Appendix Fig S2B–D). Of these, 783,655 particles and 413,330 particles were selected from Batch 1 and Batch 2, respectively, and independently subjected to 3D classification (K = 8) using a low pass filtered (60 Å) map of our ABCA7BPL structure as a reference. A single highest resolution class from each was selected, and their particles combined and subjected to an additional round of 2D classification. 2,024,649 particles were subjected to another round of 3D classification (K = 8). A single, highest resolution class containing 50,704 particles was refined to 5.4 Å. Particles were re‐extracted using refined coordinates and unbinned (pixel size 0.889 Å/pixel) and subjected to a round of 3D refinement, Bayesian Polishing, and postprocessing/B‐factor sharpening to yield a final map 4.0 Å resolution and its local resolution filtered variant calculated using Relion's own algorithm. Local resolution maps are shown for ABCA7BPL (Map1 & Map2) and ABCA7PE in Fig EV1.
For the ABCA7DIGITONIN data set (Appendix Fig S3B–E), a total of 7,437,149 particles were picked from 16,213 ctf corrected and motion corrected micrographs and extracted at a 3‐fold binned pixel size of 2.667 Å/pixel. After 2D classification, 1,220,497 particles were subjected to 3D classification (K = 8) using a low pass filtered (60 Å) map of our ABCA7BPL structure as a reference. A single class comprising 324,727 particles was refined to 5.4 Å. The refined coordinates were then used to re‐extract unbinned particles (0.889 Å/pixel), subjected to 3D refinement, and Bayesian polishing. Further 3D classification (K = 5) was performed and two similar classes containing 149,590 particles were picked for 3D refinement and postprocessing/B‐factor Sharpening to yield a 3.9 Å map.
For the ABCA7EQ‐ATP data set, an initial set of 2,660,267 particles was picked from 4,914 ctf corrected micrographs (Appendix Fig S4B–E). After 2D classification, 469,397 particles were subjected to initial model building. This model was used as a 3D reference to perform 3D classification (k = 5). A single class comprising 174,415 particles was refined to 5.4 Å, the corresponding particles unbinned and re‐refined. The nanodisc density was subtracted within Relion, followed by 3D classification (k = 3). The highest resolution class comprising 51,780 particles was refined to 4.3 Angstroms and used for 3D reference‐based particle picking for a larger data set comprising the initial 4,914 movies and a new set of 4,474 ctf corrected micrographs. A total of 3,773,280 particles were picked and subjected to multiple rounds of 2D classification in Relion 4.0‐beta. In total, 898,916 particles were subjected to 3D classification (K = 3) and a single highest resolution class consisting of 407,424 particles was refined to 5.4 Å. The refined coordinates were then used to re‐extract the respective particles without binning (0.889 Å/pixel) and refined again before 3D classification (K = 5). A single class comprising 177,230 particles was selected and subjected to 3D refinement, Bayesian polishing, and postprocessing/B‐factor sharpening to yield a final map at 3.7 Å.
Model building and refinement
Model building was done in coot 0.9.5 (Emsley et al, 2010) using a combination of Map 1 and its local resolution filtered variant and Map 2. Both Map 1 and Map 2 displayed significant conformational heterogeneity in the second half of ABCA7, with the quality of density in Map 1 allowing placement of a TMD2 model guided in part by the homologous ABCA1 structure. Density attributed to inter‐TMD phospholipids was clearest in Map 1. Map 2 revealed very poor and discontinuous density for TMD2‐NBD2 but significantly better density for TMD1‐NBD1 and the majority of ECD1 and ECD2, allowing for de novo model building. The model for ECD was also guided by the presence of nine glycosylation sites (N78, N98, N312, N340, N1335, N1381, N1386, N1457, and N1518) as well as 4 disulfide bond pairs. Density for the lid region of the ECD was missing in both maps. Initial model building for both NBDs was guided by structures of the homologous transporters TM287 (Hohl et al, 2012), ABCG2, and ABCA1, as well as our ABCA7EQ‐ATP structure, which showed better resolution for the NBDs and the alphafold (Jumper et al, 2021) predicted structure of ABCA7. We observed extra density at the nucleotide binding sites for both NBDs despite their open conformation that was modeled as ATPγS. The structure of the RD was based on a homology model of the predicted RD structure in ABCA4 (Liu et al, 2021). The quality of density for the ABCA7 RD allowed rigid body placement for the entire domain. Restrained real space refinement of the model was carried out in Phenix 1.19.1 (Adams et al, 2010) using automatically generated secondary structure restraints. Structural figures were prepared in UCSF Chimera v. 1.13.1 (Pettersen et al, 2004), ChimeraX v. 1.2.5 (Pettersen et al, 2021), and PyMOL 2.4.1 (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC).
The model for ABCA7PE was generated by rigid body placement of the ABCA7BPL model followed by real space refinement in phenix as described above. The model for ABCA7DIGITONIN was generated by rigid body fitting each TMD, NBD, ECD, and RD into its postprocessed map followed my manual adjustment of sidechains as allowed for the map. The corrected model was real space refined against the postprocessed map. The model for ABCA7EQ‐ATP was built starting with a homology model based on the ATP‐bound structure of ABCA4 (PDB 7LKZ), followed by manual adjustment of the structure as required and permitted by the map. The ECD was replaced by a rigid body fitted model of the ECD from the ABAC7Digitonin structure. This model was then refined against both the postprocessed map, and its local resolution filtered counterpart. Model building and refinement statistics for all data sets are provided in Appendix Table S2.
MD simulations
We employed MD simulations to capture the arrangement and dynamics of the lipid bilayer induced by the experimentally derived open conformation of ABCA7PE, which was used as the starting model in all the simulations. For system setup, a C‐terminal carboxylate capping group, an N‐terminal ammonium capping group, and all the hydrogen atoms were added using the psfgen plugin of vmd (Visual Molecular Dynamics) (Humphrey et al, 1996). The resulting all‐atom (AA) model was then converted to a coarse‐grained (CG) Martini 2.2 model using the martinize protocol (http://www.cgmartini.nl/), using an elastic network on atom pairs within a 10‐Å cutoff. The Orientations of Proteins in Membranes (opm) database (Lomize et al, 2006) was used to identify and align the transmembrane region of the protein with the membrane normal. The protein was embedded in two distinct lipid bilayers (palmitoyl‐oleoyl‐phosphatidyl‐ethanolamine (POPE) and cholesterol with a molar ratio of 4:1 (POPE/Chol), and palmitoyl‐oleoyl‐phosphatidyl‐choline (POPC) and cholesterol with a molar ratio of 4:1 (POPC/Chol), respectively). The protein secondary structure was defined from the AA model and was maintained throughout the CG simulations by the applied elastic network. To increase the sampling of lipid–protein interactions and improve statistics, four independent copies of the CG protein were placed at a distance of 200 Å in a large lipid bilayer (400 × 400 Å2). The system was then solvated and ionized with 150 mM salt using insane (Wassenaar et al, 2015).
The systems were simulated employing GROMACS 2021.3 (Hess et al, 2008; de Jong et al, 2013). A 20‐fs timestep was employed in all the simulations. The temperature was maintained at 310 K with a velocity‐rescaling thermostat (Bussi et al, 2007) employing a coupling time constant of 1 ps. A semi‐isotropic 1 bar pressure was maintained using the Berendsen barostat (Berendsen et al, 1984) with a compressibility and relaxation time constant of 3 × 10−4 bar and 5 ps, respectively. The systems were energy minimized for 1,000 steps, followed by short equilibration runs of 18 ns, while restraints were applied to lipid bilayer headgroups and protein backbones. During this time, the restraints on bilayer headgroups were reduced gradually from k = 200 kJ mol−1 nm−2 to zero, whereas the protein backbones' restraints (k = 1,000 kJ mol−1 nm−2) were kept constant. Each system was then simulated for 2 μs, with restraints only applied to the protein backbones (to maintain the cryo‐EM captured conformation), resulting in an aggregate sampling of 8 μs (4 copies × 2 μs). All the systems were simulated following the same MD protocol.
All the molecular images were generated using VMD (Humphrey et al, 1996). The membrane deformation induced by ABCA7 was quantified by calculating the z distance of the lipid phosphate moieties (PO4 bead type in martini) with respect to the bilayer midplane, over the last 1 μs of each trajectory. The generated histogram (binned in 2 × 2 Å2 bins) in each leaflet illustrates the spatial distribution of the height of the lipid head groups within each leaflet. We quantified the differential movement of POPE and POPC within the protein lumen by calculating the number of phospholipids located within the TMDs. If the PO4 bead of a phospholipid was within 22.5 and 12.5 Å in x and y, respectively, with respect to a protein's center in the x‐y plane (membrane plane), then the phospholipid was considered to be within the TMD lumen (Fig EV4C).
Author contributions
Le Thi My Le: Formal analysis; validation; investigation; visualization; methodology; writing – original draft; writing – review and editing. James Robert Thompson: Formal analysis; validation; investigation; visualization; methodology; writing – original draft; writing – review and editing. Sepehr Dehghani‐Ghahnaviyeh: Formal analysis; methodology; writing – review and editing. Shashank Pant: Formal analysis; validation; methodology; writing – review and editing. Phuoc Xuan Dang: Methodology. Takahisa Kanikeyo: Investigation; writing – review and editing. Emad Tajkhorshid: Resources; software; formal analysis; validation; writing – review and editing. Jarrod Bradley French: Resources; validation; methodology; writing – review and editing. Amer Alam: Conceptualization; resources; software; formal analysis; supervision; funding acquisition; validation; investigation; visualization; methodology; writing – original draft; project administration; writing – review and editing.
Disclosure and competing interests statement
SP is currently an employee of Loxo Oncology @ Lilly and is a shareholder of stock in Eli Lilly and Co. The rest of the authors declare no competing interests.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View and Appendix
PDF+
Acknowledgements
We would like to thank Dr. Kaspar Locher at ETH, Zurich, Switzerland, for providing the synthetic gene construct of ABCA7. We would also like to thank the cryo‐EM and shared instruments core facilities at the Hormel Institute for help with experimental setup, and Dr. Rhoderick Brown, Dr. Jeppe Olsen, and Dr. Devanshu Kurre for critical reading and discussion during manuscript preparation. This work was supported in part by the Hormel Foundation (Institutional research funds to AA), the National Institutes of Health (NIH) award 1R21‐AG069180‐01A1 (to AA), NIH award R35GM124898 (to JBF), and the Cure Alzheimer's fund (to TK). The computational component of the project was supported by the NIH awards P41‐GM104601 (to ET) and R01‐GM123455 (to ET). We also acknowledge computing resources provided by Blue Waters at National Center for Supercomputing Applications (NCSA), by eXtreme Science and Engineering Discovery Environment (XSEDE) (grant MCA06N060 to ET), and by Microsoft Azure.
The EMBO Journal (2023) 42: e111065
Data availability
The cryo‐EM Maps have been deposited at the Electron Microscopy Databank (EMDB) under accession codes EMD‐28041 (ABCA7BPL Map 1), EMD‐28044 (ABCA7BPL Map 2), EMD‐28047 (ABCA7PE), EMD‐28451 (ABCA7EQ‐ATP), and EMD‐28050 (ABCA7DIGITONIN). The associated atomic coordinates have been deposited at the Protein Data bank (PDB) under accession codes 8EDW (ABCA7BPL), 8EE6 (ABCA7PE), 8EOP (ABCA7EQ‐ATP), and 8EEB (ABCA7DIGITONIN).
References
- Abe‐Dohmae SYS (2012) ABCA7: a potential mediator between cholesterol homeostasis and the host defense system. Clin Lipidol 7: 677–687 [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse‐Kunstleve RW et al (2010) PHENIX: a comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aikawa T, Ren Y, Holm ML, Asmann YW, Alam A, Fitzgerald ML, Bu G, Kanekiyo T (2021) ABCA7 regulates brain fatty acid metabolism during LPS‐induced acute inflammation. Front Neurosci 15: 647974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama M, Sugiyama‐Nakagiri Y, Sakai K, McMillan JR, Goto M, Arita K, Tsuji‐Abe Y, Tabata N, Matsuoka K, Sasaki R et al (2005) Mutations in lipid transporter ABCA12 in harlequin ichthyosis and functional recovery by corrective gene transfer. J Clin Invest 115: 1777–1784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht C, Viturro E (2007) The ABCA subfamily—gene and protein structures, functions and associated hereditary diseases. Pflugers Arch 453: 581–589 [DOI] [PubMed] [Google Scholar]
- Allikmets R, Shroyer NF, Singh N, Seddon JM, Lewis RA, Bernstein PS, Peiffer A, Zabriskie NA, Li Y, Hutchinson A et al (1997) Mutation of the Stargardt disease gene (ABCR) in age‐related macular degeneration. Science 277: 1805–1807 [DOI] [PubMed] [Google Scholar]
- Almeida JFF, Dos Santos LR, Trancozo M, de Paula F (2018) Updated meta‐analysis of BIN1, CR1, MS4A6A, CLU, and ABCA7 variants in Alzheimer's disease. J Mol Neurosci 64: 471–477 [DOI] [PubMed] [Google Scholar]
- Astudillo AM, Balboa MA, Balsinde J (2019) Selectivity of phospholipid hydrolysis by phospholipase A2 enzymes in activated cells leading to polyunsaturated fatty acid mobilization. Biochim Biophys Acta Mol Cell Biol Lipids 1864: 772–783 [DOI] [PubMed] [Google Scholar]
- Bellenguez C, Charbonnier C, Grenier‐Boley B, Quenez O, Le Guennec K, Nicolas G, Chauhan G, Wallon D, Rousseau S, Richard AC et al (2017) Contribution to Alzheimer's disease risk of rare variants in TREM2, SORL1, and ABCA7 in 1779 cases and 1273 controls. Neurobiol Aging 59: 220.e1–220.e9 [DOI] [PubMed] [Google Scholar]
- Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 91: 3684–3690 [Google Scholar]
- Brooks‐Wilson A, Marcil M, Clee SM, Zhang LH, Roomp K, van Dam M, Yu L, Brewer C, Collins JA, Molhuizen HO et al (1999) Mutations in ABC1 in Tangier disease and familial high‐density lipoprotein deficiency. Nat Genet 22: 336–345 [DOI] [PubMed] [Google Scholar]
- Bussi G, Donadio D, Parrinello M (2007) Canonical sampling through velocity rescaling. J Chem Phys 126: 014101 [DOI] [PubMed] [Google Scholar]
- Chifflet S, Torriglia A, Chiesa R, Tolosa S (1988) A method for the determination of inorganic phosphate in the presence of labile organic phosphate and high concentrations of protein: application to lens ATPases. Anal Biochem 168: 1–4 [DOI] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH (2004) Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science 305: 869–872 [DOI] [PubMed] [Google Scholar]
- Cuyvers E, De Roeck A, Van den Bossche T, Van Cauwenberghe C, Bettens K, Vermeulen S, Mattheijssens M, Peeters K, Engelborghs S, Vandenbulcke M et al (2015) Mutations in ABCA7 in a Belgian cohort of Alzheimer's disease patients: a targeted resequencing study. Lancet Neurol 14: 814–822 [DOI] [PubMed] [Google Scholar]
- Czubowicz K, Jesko H, Wencel P, Lukiw WJ, Strosznajder RP (2019) The role of ceramide and Sphingosine‐1‐phosphate in Alzheimer's disease and other neurodegenerative disorders. Mol Neurobiol 56: 5436–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong DH, Singh G, Bennett WF, Arnarez C, Wassenaar TA, Schafer LV, Periole X, Tieleman DP, Marrink SJ (2013) Improved parameters for the martini coarse‐grained protein force field. J Chem Theory Comput 9: 687–697 [DOI] [PubMed] [Google Scholar]
- De Roeck A, Van Broeckhoven C, Sleegers K (2019) The role of ABCA7 in Alzheimer's disease: evidence from genomics, transcriptomics and methylomics. Acta Neuropathol 138: 201–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K (2010) Features and development of coot. Acta Crystallogr D Biol Crystallogr 66: 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadok VA, de Cathelineau A, Daleke DL, Henson PM, Bratton DL (2001) Loss of phospholipid asymmetry and surface exposure of phosphatidylserine is required for phagocytosis of apoptotic cells by macrophages and fibroblasts. J Biol Chem 276: 1071–1077 [DOI] [PubMed] [Google Scholar]
- Gabande‐Rodriguez E, Keane L, Capasso M (2020) Microglial phagocytosis in aging and Alzheimer's disease. J Neurosci Res 98: 284–298 [DOI] [PubMed] [Google Scholar]
- Geertsma ER, Nik Mahmood NA, Schuurman‐Wolters GK, Poolman B (2008) Membrane reconstitution of ABC transporters and assays of translocator function. Nat Protoc 3: 256–266 [DOI] [PubMed] [Google Scholar]
- Harris A, Wagner M, Du D, Raschka S, Nentwig LM, Gohlke H, Smits SHJ, Luisi BF, Schmitt L (2021) Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5. Nat Commun 12: 5254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: algorithms for highly efficient, load‐balanced, and scalable molecular simulation. J Chem Theory Comput 4: 435–447 [DOI] [PubMed] [Google Scholar]
- Hirt UA, Leist M (2003) Rapid, noninflammatory and PS‐dependent phagocytic clearance of necrotic cells. Cell Death Differ 10: 1156–1164 [DOI] [PubMed] [Google Scholar]
- Hohl M, Briand C, Grutter MG, Seeger MA (2012) Crystal structure of a heterodimeric ABC transporter in its inward‐facing conformation. Nat Struct Mol Biol 19: 395–402 [DOI] [PubMed] [Google Scholar]
- Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V et al (2011) Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nat Genet 43: 429–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14: 27–38 [DOI] [PubMed] [Google Scholar]
- Iwamoto N, Abe‐Dohmae S, Sato R, Yokoyama S (2006) ABCA7 expression is regulated by cellular cholesterol through the SREBP2 pathway and associated with phagocytosis. J Lipid Res 47: 1915–1927 [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A et al (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim‐Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier‐Boley B et al (2013) Meta‐analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 45: 1452–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Guennec K, Nicolas G, Quenez O, Charbonnier C, Wallon D, Bellenguez C, Grenier‐Boley B, Rousseau S, Richard AC, Rovelet‐Lecrux A et al (2016) ABCA7 rare variants and Alzheimer disease risk. Neurology 86: 2134–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaci AM, Steigenberger B, Telles de Souza PC, Tamara S, Grollers‐Mulderij M, Ogrissek P, Marrink SJ, Scheltema RA, Forster F (2021) Structure of the human signal peptidase complex reveals the determinants for signal peptide cleavage. Mol Cell 81: e3911 [DOI] [PubMed] [Google Scholar]
- Liu F, Lee J, Chen J (2021) Molecular structures of the eukaryotic retinal importer ABCA4. Elife 10: e63524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI (2006) OPM: orientations of proteins in membranes database. Bioinformatics 22: 623–625 [DOI] [PubMed] [Google Scholar]
- Ma FC, Wang HF, Cao XP, Tan CC, Tan L, Yu JT (2018) Meta‐analysis of the association between variants in ABCA7 and Alzheimer's disease. J Alzheimers Dis 63: 1261–1267 [DOI] [PubMed] [Google Scholar]
- Manolaridis I, Jackson SM, Taylor NMI, Kowal J, Stahlberg H, Locher KP (2018) Cryo‐EM structures of a human ABCG2 mutant trapped in ATP‐bound and substrate‐bound states. Nature 563: 426–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molday LL, Molday RS (2014) 1D4: a versatile epitope tag for the purification and characterization of expressed membrane and soluble proteins. Methods Mol Biol 1177: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcombe EA, Camats‐Perna J, Silva ML, Valmas N, Huat TJ, Medeiros R (2018) Inflammation: the link between comorbidities, genetics, and Alzheimer's disease. J Neuroinflammation 15: 276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando BJ, Liao M (2020) ABCG2 transports anticancer drugs via a closed‐to‐open switch. Nat Commun 11: 2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, Ferrin TE (2021) UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci 30: 70–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp J, Lewczuk P, Kolsch H, Meichsner S, Maier W, Kornhuber J, Jessen F, Lutjohann D (2012) Cholesterol metabolism is associated with soluble amyloid precursor protein production in Alzheimer's disease. J Neurochem 123: 310–316 [DOI] [PubMed] [Google Scholar]
- Qian H, Zhao X, Cao P, Lei J, Yan N, Gong X (2017) Structure of the human lipid exporter ABCA1. Cell 169: 1228–1239 [DOI] [PubMed] [Google Scholar]
- Quazi F, Molday RS (2013) Differential phospholipid substrates and directional transport by ATP‐binding cassette proteins ABCA1, ABCA7, and ABCA4 and disease‐causing mutants. J Biol Chem 288: 34414–34426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja M (2011) Do small headgroups of phosphatidylethanolamine and phosphatidic acid lead to a similar folding pattern of the K(+) channel? J Membr Biol 242: 137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG (2009) Chapter 11 – reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol 464: 211–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres SH, Chen S (2012) Prevention of overfitting in cryo‐EM structure determination. Nat Methods 9: 853–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scortecci JF, Molday LL, Curtis SB, Garces FA, Panwar P, Van Petegem F, Molday RS (2021) Cryo‐EM structures of the ABCA4 importer reveal mechanisms underlying substrate binding and Stargardt disease. Nat Commun 12: 5902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segrest JP, Tang C, Song HD, Jones MK, Davidson WS, Aller SG, Heinecke JW (2022) ABCA1 is an extracellular phospholipid translocase. Nat Commun 13: 4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serhan CN (2014) Pro‐resolving lipid mediators are leads for resolution physiology. Nature 510: 92–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleegers K (2020) Expression of ABCA7 in Alzheimer's disease. Acta Neuropathol 139: 941–942 [DOI] [PubMed] [Google Scholar]
- Swerdlow RH (2016) Rare ABCA7 variants in Alzheimer disease: guilt by association. Neurology 86: 2118–2119 [DOI] [PubMed] [Google Scholar]
- Tanaka N, Abe‐Dohmae S, Iwamoto N, Fitzgerald ML, Yokoyama S (2010) Helical apolipoproteins of high‐density lipoprotein enhance phagocytosis by stabilizing ATP‐binding cassette transporter A7. J Lipid Res 51: 2591–2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa G, Staurenghi E, Zerbinati C, Gargiulo S, Iuliano L, Giaccone G, Fanto F, Poli G, Leonarduzzi G, Gamba P (2016) Changes in brain oxysterols at different stages of Alzheimer's disease: their involvement in neuroinflammation. Redox Biol 10: 24–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Aller SG, Beis K, Carpenter EP, Chang G, Chen L, Dassa E, Dean M, Duong Van Hoa F, Ekiert D et al (2020) Structural and functional diversity calls for a new classification of ABC transporters. FEBS Lett 594: 3767–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka M, Toda Y, Manucat NB, Akatsu H, Fukumoto M, Kono N, Arai H, Kioka N, Ueda K (2017) Lysophosphatidylcholine export by human ABCA7. Biochim Biophys Acta Mol Cell Biol Lipids 1862: 658–665 [DOI] [PubMed] [Google Scholar]
- Tonnies E, Trushina E (2017) Oxidative stress, synaptic dysfunction, and Alzheimer's disease. J Alzheimers Dis 57: 1105–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenaar TA, Ingolfsson HI, Bockmann RA, Tieleman DP, Marrink SJ (2015) Computational lipidomics with insane: a versatile tool for generating custom membranes for molecular simulations. J Chem Theory Comput 11: 2144–2155 [DOI] [PubMed] [Google Scholar]
- Xie T, Zhang Z, Fang Q, Du B, Gong X (2021) Structural basis of substrate recognition and translocation by human ABCA4. Nat Commun 12: 3853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachowski A (1993) Phospholipids in animal eukaryotic membranes: transverse asymmetry and movement. Biochem J 294: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K (2016) Gctf: real‐time CTF determination and correction. J Struct Biol 193: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA (2017) MotionCor2: anisotropic correction of beam‐induced motion for improved cryo‐electron microscopy. Nat Methods 14: 331–332 [DOI] [PMC free article] [PubMed] [Google Scholar]


