Abstract
Early embryonic development depends on proper utilization and clearance of maternal transcriptomes. How these processes are spatiotemporally regulated remains unclear. Here we show that nuclear RNA‐binding protein Rbm14 and maternal mRNAs co‐phase separate into cytoplasmic condensates to facilitate vertebrate blastula‐to‐gastrula development. In zebrafish, Rbm14 condensates were highly abundant in blastomeres and markedly reduced after prominent activation of zygotic transcription. They concentrated at spindle poles by associating with centrosomal γ‐tubulin puncta and displayed mainly asymmetric divisions with a global symmetry across embryonic midline in 8‐ and 16‐cell embryos. Their formation was dose‐dependently stimulated by m6A, but repressed by m5C modification of the maternal mRNA. Furthermore, deadenylase Parn co‐phase separated with these condensates, and this was required for deadenylation of the mRNAs in early blastomeres. Depletion of Rbm14 impaired embryonic cell differentiations and full activations of the zygotic genome in both zebrafish and mouse and resulted in developmental arrest at the blastula stage. Our results suggest that cytoplasmic Rbm14 condensate formation regulates early embryogenesis by facilitating deadenylation, protection, and mitotic allocation of m6A‐modified maternal mRNAs, and by releasing the poly(A)‐less transcripts upon regulated disassembly to allow their re‐polyadenylation and translation or clearance.
Keywords: deadenylation, embryogenesis, maternal mRNA, maternal‐to‐zygotic transition, phase separation
Subject Categories: Development, RNA Biology
The RNA binding protein Rbm14 regulates maternal‐to‐zygotic transition in zebrafish and mouse via formation of cytoplasmic condensates with m6A‐modified maternal mRNAs and the RNA deadenylase Parn.
Introduction
All metazoan embryogenesis depends on maternal mRNAs, transcriptomes preserved in the cytoplasm of oocytes, for protein synthesis before full zygotic genome activation (ZGA). Such a process, termly maternal‐to‐zygotic transition (MZT), occurs gradually during early embryonic development but differs dramatically across species in kinetics, even among vertebrates (Jukam et al, 2017; Eckersley‐Maslin et al, 2018; Vastenhouw et al, 2019). In zebrafish (Danio rerio), for instance, the 10 rapid, synchronous rounds of embryonic cleavages during the initial 3 h post‐fertilization (hpf) depend on proteins translated from maternal mRNAs. Following a gradual decrease in maternal mRNA levels during this period of time, transcriptional activities of the zygotic genome emerge weakly from 2 hpf (64‐cell stage; minor ZGA) and strongly from 3 hpf (1k‐cell stage; the major ZGA). The embryo then develops into the blastula period marked by elongated, asynchronous cell cycles. The epiboly, that is, inward and vegetal pole‐ward migrations of the single‐layer cells in the blastula, generates the gastrula, accompanied by the completion of ZGA and full clearance of maternal mRNAs. In mammals such as house mouse (Mus musculus), three rounds of embryonic cleavages generate the 8‐cell embryo containing eight totipotent blastomeres, followed by cell differentiations (Takaoka & Hamada, 2012; Rivera‐Perez & Hadjantonakis, 2014). The majority of maternal mRNAs, however, are degraded by the middle 2‐cell stage, whereas the minor ZGA occurs in 1‐cell embryos quickly after fertilization, followed by the major ZGA in the 2‐cell stage (Jukam et al, 2017; Eckersley‐Maslin et al, 2018; Vastenhouw et al, 2019). Whether maternal mRNAs are indeed required differently across species or have common functions in embryonic cell differentiations despite the seemingly irregularity remains unclear.
In sharp contrast to zygotic mRNAs transcribed on demands, maternal transcripts are pre‐loaded in full into oocytes. Therefore, early embryos must possess unique, exquisite mechanisms to spatiotemporally control the translational activity and stability of their maternal mRNAs. For instance, maternal mRNAs tend to be initially converted into translationally incompetent forms through deadenylation, followed by re‐activations through cytoplasmic polyadenylation (Salles et al, 1994; Kim & Richter, 2006; Aanes et al, 2014; Despic & Neugebauer, 2018; Winata et al, 2018). Furthermore, although deadenylation is usually the major rate‐limiting step of mRNA decay, deadenylated maternal mRNAs can be kept stable for hours prior to degradation (Salles et al, 1994; Kim & Richter, 2006; Weill et al, 2012; Winata et al, 2018; Vastenhouw et al, 2019). How the deadenylated maternal mRNAs are protected from immediate degradation but allowed for timely re‐activation and clearance, however, is poorly understood.
Maternal transcripts are degraded through both ZGA‐dependent and ZGA‐independent pathways during MZT (Giraldez et al, 2006; Walser & Lipshitz, 2011; Despic & Neugebauer, 2018). Different transcripts can display distinct degradation kinetics (Mishima & Tomari, 2016; Vejnar et al, 2019). N 6‐methyladenosine (m6A)‐ and 5‐methylcytosine (m5C)‐RNA modifications, ploy‐A/U, and codon usages are cis‐elements that impact maternal mRNA metabolism, whereas RNA‐binding proteins (RBPs) and miRNAs can act as trans‐factors (Bazzini et al, 2016; Mishima & Tomari, 2016; Morgan et al, 2017; Zhao et al, 2017; Despic & Neugebauer, 2018; Vastenhouw et al, 2019; Yang et al, 2019). For instance, in zebrafish zYthdf2 binds to N6‐methyladenosine (m6A)‐modified maternal mRNAs to facilitate their clearance, whereas the zygotic gene‐transcribed microRNA miR‐430 binds to 3′ untranslated regions of its target maternal mRNAs and recruits the CCR4/NOT deadenylase to chop off the poly(A) tails for exonuclease‐mediated RNA degradation (Giraldez et al, 2006; Zhao et al, 2017). On the other hand, zYbx1 recognizes 5‐methylcytosine (m5C)‐modified maternal mRNAs and recruits the poly(A)‐binding protein zPabpc1a to protect their poly(A) tails from deadenylation (Yang et al, 2019). Interestingly, both m6A and m5C modifications increase after fertilization and peak at around 2 or 3 hpf (Zhao et al, 2017; Yang et al, 2019; Liu et al, 2022). Whether these two types of epigenetic modifications act independently or could work jointly in some way is unknown.
Emerging evidence indicates that liquid–liquid phase separation (LLPS), a process in which biological macromolecules demix into liquid‐like condensates through multivalent interactions, underlies the formation of various membrane‐less organelles or intracellular compartments (Banani et al, 2017; Zhang et al, 2020; Alberti & Hyman, 2021). A large number of RNA‐binding proteins containing intrinsically disordered regions (IDRs) are able to undergo LLPS with RNA to form biological condensates important for various aspects of mRNA metabolisms, cell cycle regulation, and differentiation (Hentze et al, 2018; Rhine et al, 2020; Roden & Gladfelter, 2021). Rbm14, also called CoAA, is a multifunctional nuclear RNA‐binding protein abundant in paraspeckles and also involved in transcriptional regulation and pre‐mRNA splicing (Auboeuf et al, 2004; Hennig et al, 2015; Firmin et al, 2017; Xiao et al, 2019). We have previously shown that zebrafish contains two functionally‐redundant Rbm14 paralogues (zRbm14a and zRbm14a‐b) capable of both LLPS through the C‐terminal IDR and cell fate regulation (Xiao et al, 2019). Interestingly, zRbm14 is located in the cytoplasm in early embryos but the function is unknown (Xiao et al, 2019). In this study, we demonstrated that zRbm14 co‐phase separates with m6A‐modified maternal mRNAs and zParn deadenylase into cytoplasmic condensates in blastomeres to deadenylate maternal mRNAs, protect them from premature degradations, and segregate them in mitosis. Furthermore, our studies suggest that Rbm14 plays conserved roles in regulating vertebrate maternal mRNA metabolisms for proper cell fate specifications and blastula development.
Results
zRbm14 decorates robust cytoplasmic condensates in zebrafish cleavage‐stage embryos
We have previously shown that zRbm14a is expressed in embryos from the one‐cell stage (Xiao et al, 2019). Detailed examinations indicated that zRbm14a levels per embryo were upregulated slightly from 1 hpf and further from 2 hpf (Fig 1A–C). In comparison, α‐tubulin levels per embryo increased mainly at 6 hpf (Fig 1B and C), correlated with massive transcriptions of zygotic genes (Fig 1A; Mathavan et al, 2005; Vastenhouw et al, 2019).
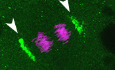
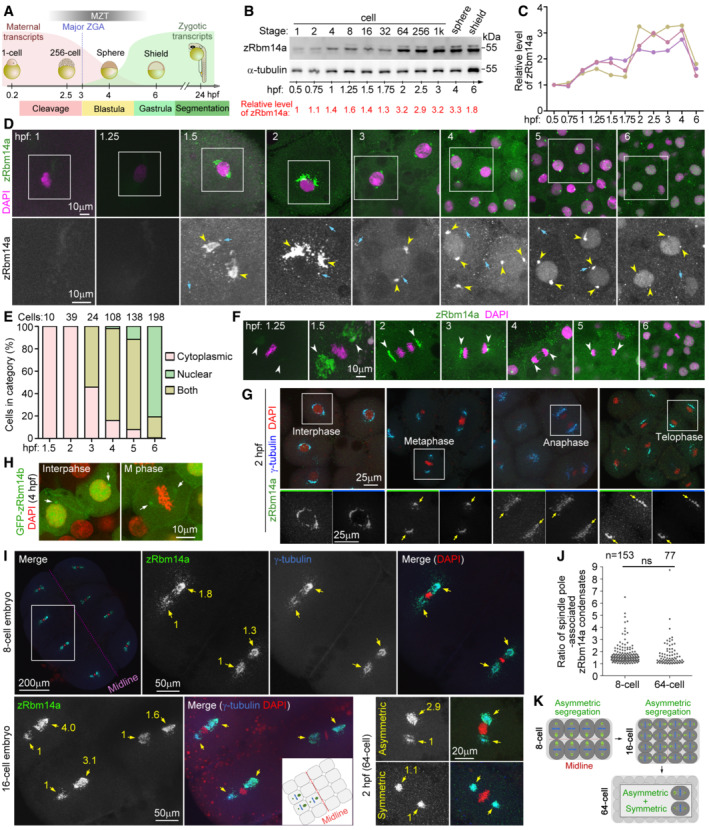
Figure 1. zRbm14a decorates cytoplasmic condensates implicated in cell fate regulations in early embryos.
-
AA schematic diagram of zebrafish early embryonic development (Aanes et al, 2014; Winata et al, 2018; Vastenhouw et al, 2019). The red and green curves illustrate levels of maternal and zygotic RNA transcripts, respectively. Hpf, hours post‐fertilization; MZT, maternal‐to‐zygotic transition; ZGA, zygotic gene activation. The diagrams are not drawn to scale.
-
B, CExpression profile of zRbm14a during early embryonic development. Lysates from 5 embryos were loaded in each lane and subjected to immunoblotting (B). α‐tubulin served as an internal control. The relative levels of zRbm14a (B, C) are shown as band intensities normalized to those of α‐tubulin. Quantification results from three independent experiments are shown in (C).
-
D, ESubcellular localizations of zRbm14a. The representative confocal micrographs (D) were acquired from the animal pole of zebrafish embryos. Exposures were kept the same during the imaging for comparison. DAPI labels nuclear DNA. Framed regions were magnified to show details. Arrows denote representative cytoplasmic puncta, whereas arrowheads point to perinuclear condensates. Subcellular localizations (E) were scored from the indicated number of cells from 4 to 6 embryos.
-
FSpindle‐pole enrichment of zRbm14a condensates (arrowheads) in mitotic cells. Exposures were kept the same during the imaging for comparison.
-
GzRbm14a condensates colocalized with γ‐tubulin puncta. The framed blastomeres at the indicated cell cycle stages were magnified to show details. Arrows point to spindle poles.
-
HSubcellular localizations of zRbm14b resembled those of zRbm14a. Zebrafish embryos were microinjected with 400 pg of GFP‐Rbm14b mRNA per egg at the 1‐cell stage and fixed at 4 hpf.
-
IAsymmetric segregations of zRbm14a condensates in mitotic blastomeres by associating with centrosomal γ‐tubulin puncta (arrows). A typical 8‐cell mitotic embryo was entirely represented, with the two blastomeres in the framed region magnified to show details. The cartoon inset illustrates relative positions of the three blastomeres in the 16‐cell embryo. Numbers in yellow are relative fluorescent intensities of spindle pole‐associated zRbm14a condensates, with the smaller ones set to 1. Refer to Movies EV1 and EV2 for 3D‐reconstructed images of the two representative cells in 2‐hpf embryos.
-
JQuantification results on the ratio of zRbm14a condensates associated with spindle pole pairs in mitotic blastomere.
-
KSummarizing illustrations showing segregation patterns of zRbm14 condensates (green) in mitotic blastomeres. Condensates in 8‐ and 16‐cell embryos are segregated mainly in an asymmetric pattern identical to that of centrosomes (Rathbun et al, 2020), that is, larger centrosome contains more zRbm14 condensates than the smaller centrosome does in a cell and is located closer to the embryonic midline. In 64‐cell embryos, blastomeres display both asymmetric and symmetric segregations of the condensates, though the distribution pattern of these cells is currently unclear. The diagrams are not drawn to scale.
Data information: Micrographs were maximum intensity‐projected images. Quantification results in (J), from 20 mitotic 8‐cell embryos and 29 2‐hpf embryos, are presented as mean ± SD plus sample dots. Unpaired two‐sided Student's t‐test: ns, no significance.
Source data are available online for this figure.
Intriguingly, immunofluorescent signals of zRbm14a emerged as punctate cytoplasmic condensates. The condensates tended to gather into two prominent perinuclear clusters that peaked at approximately 2 hpf in cleavage‐stage embryos (Fig 1D). After the onset of the major ZGA from 3 hpf, the perinuclear clusters shrank markedly into spots, accompanied by increased nuclear localization of zRbm14a (Fig 1D and E). zRbm14a became mainly located in the nucleus in gastrulas at 6 hpf (Fig 1D and E), correlated with the completion of MZT (Aanes et al, 2014; Despic & Neugebauer, 2018). Notably, the condensates were not disassembled in mitosis. Rather, they clustered at spindle pole regions and were segregated into daughter cells (Fig 1F). Interestingly, when γ‐tubulin, a centrosomal protein critical for microtubule nucleation (Moritz et al, 1995), was co‐immunostained, we observed that the spindle pole‐enriched Rbm14a condensates co‐localized nicely with γ‐tubulin‐positive puncta in 2‐hpf blastomeres (Fig 1G). The perinuclear clusters of zRbm14 condensates in interphase blastomeres also co‐localized with γ‐tubulin‐positive puncta (Fig 1G). These results suggest that zRbm14 condensates are enriched to the centrosomal/spindle pole regions through associations with γ‐tubulin‐containing granules.
Due to the lack of an antibody, we examined subcellular localization of GFP‐Rbm14b, expressed through microinjection of its in vitro transcribed mRNA (Xiao et al, 2019). Similar to endogenous zRbm14a, GFP‐zRbm14b was distributed in the cytoplasm distributions with one or two perinuclear puncta in interphase cells at 4 hpf, in addition to nuclear localization (Fig 1H). It was also concentrated at the spindle pole in mitotic cells (Fig 1H). As Rbm14 is usually a typical nuclear protein (Auboeuf et al, 2004; Hennig et al, 2015; Xiao et al, 2019), the peculiar cytoplasmic localization and behavior of zRbm14 suggest a unique role in early embryos.
zRbm14 condensates display asymmetric and symmetric mitotic segregations
Symmetric and asymmetric cell divisions are known to govern the self‐renewal and differentiation of pluripotent stem cells by equally or differentially allocating cell fate determinants such as signaling molecules through proper spindle orientation and positioning (Knoblich, 2001; Venkei & Yamashita, 2018; Lechler & Mapelli, 2021). The centrosome can also function as cell fate determinants: mouse neural stem cells inherited the mother or the daughter centrosome assume different fates (Wang et al, 2009; Venkei & Yamashita, 2018). Interestingly, centrosome pairs in mitotic blastomeres of 8‐ and 16‐cell zebrafish embryos have recently been shown to be asymmetric in size when measured as areas containing γ‐tubulin puncta (Rathbun et al, 2020). Furthermore, the larger centrosome tends to be positioned towards the embryonic midline, a process affected by protein kinases PLK1 and PLK4 (Rathbun et al, 2020). As zRbm14a condensates in some mitotic cells appeared to be more abundant at one spindle pole (Fig 1F and G), we investigated whether the asymmetric mitotic centrosomes could also serve as carriers to partition zRbm14a condensates unequally (asymmetrically) for the regulation of cell fates.
We imaged 8‐ and 16‐cell mitotic embryos and observed that zRbm14a condensates still tightly colocalized with γ‐tubulin puncta at spindle poles (Fig 1I). Quantifications on 8‐cell embryos indicated that, in 90.8% of blastomeres (n = 153), immunofluorescent intensity of zRbm14a condensates at the spindle pole close to the embryonic midline exceeded that at the opposite pole (Fig 1J). Furthermore, the difference was over 1.5‐ and 2‐fold in 50 and 20.3% of the blastomeres, respectively (Fig 1J). Interestingly, even in mitotic 2‐hpf (64‐cell) embryos, asymmetric distributions of the condensates still existed: 42.9 and 23.4% of mitotic blastomeres contained spindle‐pole condensates that differed by ≥ 1.5‐ and 2‐fold in intensities, respectively (Fig 1J). 3D reconstructions revealed that the condensates tended to be preferentially concentrated at the spindle pole oriented towards the animal‐pole surface (Movies EV1 and EV2). Therefore, zRbm14 condensates are allocated asymmetrically or symmetrically into daughter blastomeres along with centrosomes in cleavage‐stage embryos (Fig 1K), suggesting a role of them in cell fate regulation.
zRbm14 condensates enrich and sequester maternal mRNAs for their allocations into daughter blastomeres
zRbm14 is a potential cell fate regulator with two RNA‐recognition motifs (RRMs; Loerch & Kielkopf, 2015) at the N‐terminus (Fig 2A; Xiao et al, 2019). Its mammalian orthologues are proved to be RNA‐binding proteins (Auboeuf et al, 2004; Hennig et al, 2015; Firmin et al, 2017). We thus suspected that its cytoplasmic condensates might concentrate certain maternal mRNAs and deliver them equally or differentially into daughter blastomeres for cell fate regulations. We initially stained embryos with the RNA‐specific dye Pyronin Y (Darzynkiewicz et al, 2004; Eddaoudi et al, 2018) and indeed observed enhanced Pyronin Y staining in the condensates (Fig 2B). RNase treatment abolished the Pyronin Y staining while the zRbm14a condensates remained (Fig 2B), further indicating that the Pyronin Y signals come from RNA.
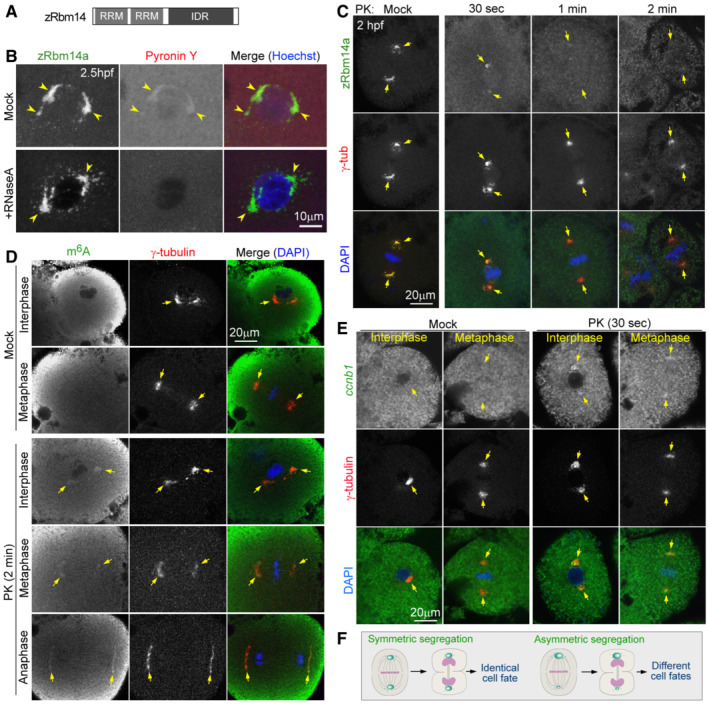
Figure 2. zRbm14 condensates enrich maternal mRNAs, including cyclin B1 mRNA.
-
AA diagram of zRbm14. RRM, RNA‐recognition motif; IDR, intrinsically‐disordered region.
-
BzRbm14a condensates contained RNA. Zebrafish embryos fixed at 2.5 hpf were treated with RNase A or mock‐treated, followed by immunofluorescent staining to label endogenous Rbm14a and fluorescent staining with Pyronin Y and Hoechst 33342 to respectively label RNA and nuclear DNA. Maximum intensity‐projected confocal images are presented. Arrowheads point to representative condensates.
-
CzRbm14a in condensates was sensitive to proteinase K (PK) digestion, whereas γ‐tubulin was resistant to the digestion. Maximum intensity‐projected confocal images are presented. Arrows point to spindle poles.
-
D, EPK digestion exposed m6A‐modified maternal RNAs (D) and maternal ccnb1 mRNA (E) in zRbm14 condensates of both interphase and mitotic blastomeres in 2‐hpf embryos. As the high background fluorescent signals, probably due to high levels of dispersed RNAs in the cytoplasm, tended to obscure the spindle‐pole RNA signals in maximum intensity‐projected images, confocal images of a representative single optical section are shown.
-
FSpeculative illustrations for possible cell fate‐regulatory functions of zRbm14 condensates (green) by asymmetrically or symmetrically allocating maternal mRNAs in mitosis. The diagrams are not drawn to scale.
We next investigated whether the zRbm14a condensates could enrich m6A‐mRNAs because zebrafish maternal mRNAs are abundant in m6A modification (Zhao et al, 2017). Our initial immunostaining of 2‐hpf embryos with the anti‐m6A antibody did not generate obvious immunofluorescent signals over the background (see the mock‐treated samples in Fig 2D as examples). We reasoned that this might be due to inaccessibility of the antibody to m6A‐mRNAs sequestered inside the condensates. We thus sought to degrade zRbm14 with proteinase K (PK) to expose the mRNAs and found that PK digestion for 2 min eliminated the immunofluorescent signals of zRbm14a condensates but only mildly affected those of γ‐tubulin (Fig 2C). We then used γ‐tubulin as a marker for positions of zRbm14 condensates. To our excitement, we observed m6A immunofluorescent signals in centrosomal/spindle pole regions specifically in PK‐treated interphase and mitotic embryos (Fig 2D).
The cell cycle progression of cleavage‐stage oviparous embryos requires cell‐cycle regulators translated from maternal mRNAs. Among them, cyclins (such as cyclin B) need to be synthesized at defined cell cycle stages, followed by proteolytic degradation (Murray & Kirschner, 1989; Murray, 2004; Petrachkova et al, 2019). As maternal ccnb1 mRNA is also a typical m6A‐RNA in zebrafish (Zhao et al, 2017) and enriched at spindle poles in Xenopus early embryos (Groisman et al, 2000), we performed fluorescent in situ hybridization (FISH) and indeed observed zebrafish ccnb1 mRNA signals in γ‐tubulin‐indicated zRbm14 condensates in PK‐treated but not mock‐treated embryos (Fig 2E). These results further strengthen our speculation that cytoplasmic zRbm14 condensates probably recruit, sequester, and allocate maternal mRNAs for cell fate specifications (Fig 2F).
m6A‐modified RNA markedly promotes LLPS of zRbm14 through co‐phase separation
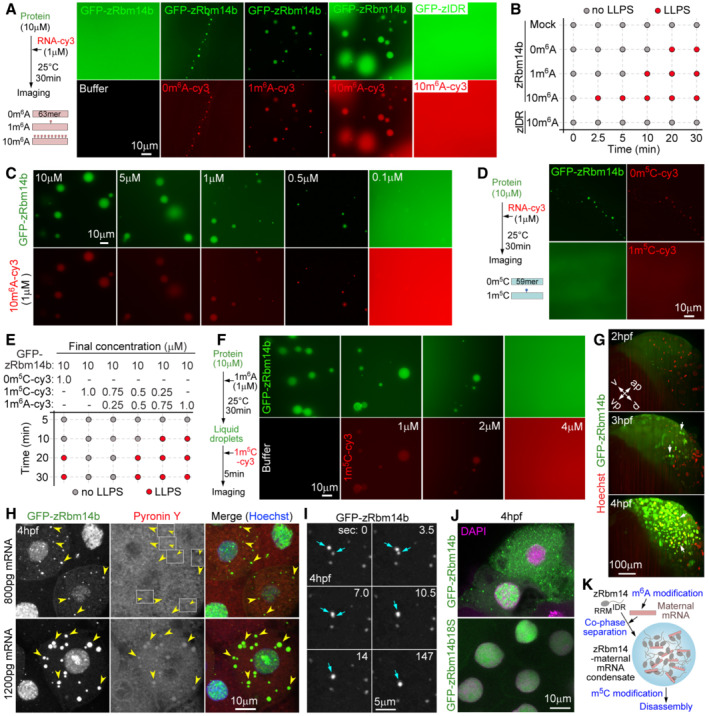
Next, we performed in vitro assays to clarify how zRbm14 condensates recruited maternal mRNAs. zRbm14a and zRbm14b are functionally redundant (Xiao et al, 2019). As we have previously expressed and purified polyhistidine‐green fluorescent protein (His‐GFP)‐tagged zRbm14b and its phase‐separation domain (zIDR) from Escherichia coli and characterized their phase separation properties (Xiao et al, 2019), we chose to use zRbm14b for these assays. We prepared three cy3‐labeled RNA fragments containing 0, 1, or 10 m6A residues (namely 0m6A‐cy3, 1m6A‐cy3, and 10m6A‐cy3; Fig EV1B and C) following a previous literature (Ries et al, 2019) to mimic maternal RNAs of different extent of m6A modification. Interestingly, although His‐GFP‐zRbm14b was unable to phase separate at a 10‐μM concentration (Figs 3A and EV1A; Xiao et al, 2019), 1 μM of each RNA fragment triggered its LLPS through co‐phase separation (Fig 3A). Among the three RNA fragments, 10m6A‐cy3 was the most potent one, judged by the number and size of liquid droplets (Fig 3A). Even 10m6A‐cy3, however, was unable to induce LLPS of zIDR (Fig 3A), indicating that these RNAs exert the phase separation‐promoting effect by binding to the RRM region of zRbm14b.
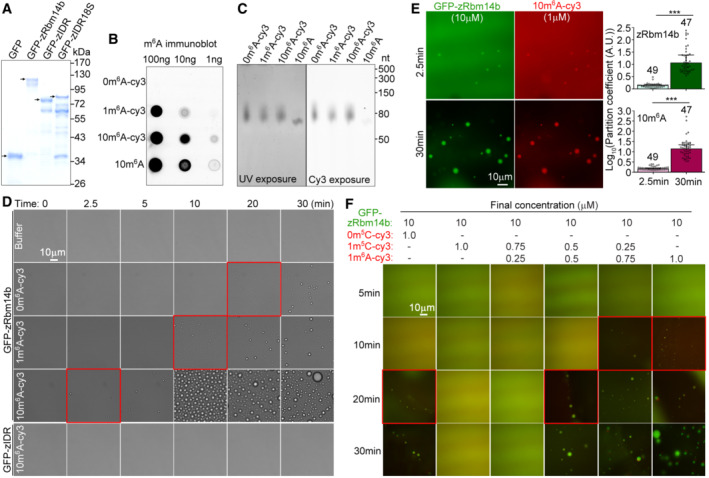
Figure EV1. Proteins and 63‐mer RNA fragments used for in vitro phase separation assays and time‐dependent effect of 10m6A‐cy3 on LLPS of zRbm14b (related to Fig 3).
-
APurified proteins from Escherichia coli. The proteins were subjected to SDS–PAGE and stained with Coomassie Blue. Arrowheads indicate the full‐length band of each protein.
-
B, CValidation of the quality of RNA fragments. In vitro transcribed 63‐mer RNA fragments were subjected to immuno‐northern dot blotting using an anti‐m6A antibody (B) or resolved in 15% urea‐denaturing PAGE followed by fluorescent imaging to visualize RNA (UV exposure) and the cy3 label (cy3 exposure) (C).
-
DRepresentative differential interference contrast images at the indicated time points after the addition of the indicated RNA fragments or buffer into GFP‐zRbm14b or GFP‐zIDR. The final concentrations of RNAs and proteins were 1 μM and 10 μM, respectively. Imaging was performed at 25°C. Red squares highlight the time of initial LLPS in each group. The results are summarized in Fig 3B.
-
EKinetics of 10m6A‐induced LLPS of zRbm14b. 10m6A‐cy3 was mixed with His‐GFP‐zRbm14b at the indicated final concentrations and imaged at 25°C. Partition coefficients were respectively calculated for 49 or 47 droplets from three independent experiments and pooled together in the histograms.
-
FRepresentative fluorescent images showing effects of m6A‐ and m5C‐RNA modifications on the LLPS of zRbm14b. Imaging was performed at 25°C. Red squares highlight the time of initial LLPS in each group. The results are summarized in Fig 3E.
Data information: Fluorescent micrographs in (E) and (F) were taken manually under a wide‐field microscope. Sometimes some condensates moved during imaging, resulting in a positional shift between their GFP and cy3 images. Quantification results are presented as mean ± SD. Unpaired two‐sided Student's t‐test: ***P < 0.001.
Source data are available online for this figure.
Figure 3. Maternal mRNAs co‐phase separate with zRbm14 into cytoplasmic condensates with efficacies inversely regulated by m6A and m5C modifications.
-
ARNA fragments co‐phase separated with GFP‐zRbm14b by binding to the RRM region. Note that the co‐phase separation is correlated with markedly reduced background fluorescence.
-
BLLPS‐promoting effect of the RNA was positively correlated with the number of m6A residues. Refer to Fig EV1D for representative images.
-
C10m6A markedly reduced the critical concentration of GFP‐Rbm14b for LLPS.
-
Dm5C modification repressed the LLPS‐promoting effect of RNA.
-
EAntagonizing effect of m6A‐ and m5C‐RNA modifications on the LLPS of zRbm14b. Refer to Fig EV1F for representative images.
-
F1m5C disassembled pre‐existing 1m6A‐zRbm14b condensates in a dose‐dependent manner. Note that 1m5C‐cy3 also entered the liquid droplets.
-
GExpression and dynamics of GFP‐zRbm14b in a representative zebrafish embryo. Nuclei were labeled with Hoechst 33342. Arrows point to typical nuclei strongly positive for GFP‐zRbm14b. Ap, animal pole; vp, vegetal pole; d, dorsal region; v, ventral region. Please refer to Movie EV3.
-
HPyronin Y staining indicated that cytoplasmic GFP‐zRbm14b condensates enriched RNA. 1‐cell zebrafish embryos were microinjected with the indicated amount of GFP‐zRbm14b mRNA per embryo to induce ectopic GFP‐zRbm14b condensates and imaged at 4 hpf. Arrowheads point to representative ectopic condensates. Insets were magnified by twofold to show Pyronin Y‐positive signals of representative condensates. Note that the different nuclear distributions of GFP‐Rbm14b and Pyronin Y fluorescence indicate no leaky signals between the two channels.
-
IFusion of GFP‐zRbm14b condensates (arrows). 4‐hpf embryos were live imaged with a spinning disk microscope at 3.5‐s intervals.
-
JPhase separation‐defective mutant zRbm14b18S displayed only nuclear localization.
-
KA summarizing illustration. zRbm14 and maternal mRNAs co‐phase separate into cytoplasmic condensates. The diagrams are not drawn to scale.
Data information: Micrographs for in vitro assays (A, C, D, and F) were taken manually under a wide‐field microscope. Sometimes some condensates moved during imaging, resulting in a positional shift between their GFP and cy3 images. Maximum intensity‐projected confocal images are presented in (G–J).
Time‐lapse imaging further indicated that, while 10‐μM His‐GFP‐zRbm14b did not undergo LLPS in 30 min, liquid droplets emerged in 20 min in the presence of 1‐μM 0m6A‐cy3 (Figs 3B and EV1D). In total, 1 μM of 1m6A‐cy3 and 10m6A‐cy3 further shortened the time of LLPS to 10 and 2.5 min, respectively (Figs 3B and EV1D). In comparison, 10m6A still failed to induce the LLPS of His‐GFP‐zIDR in 30 min (Figs 3B and EV1D). Furthermore, at 30 min, partition coefficients of both His‐GFP‐Rbm14b and 10m6A increased by more than 10‐fold (Fig EV1E).
When 1 μM of 10m6A‐cy3 was mixed with different concentrations of His‐GFP‐zRbm14b for 30 min, the size of liquid droplets reduced following the reduction of the protein concentration (Fig 3C). Despite this, LLPS of Rbm14b was clearly observed at 0.5 μM (Fig 3C). Even at 0.1 μM of His‐GFP‐zRbm14b, some liquid droplets were still observed over the strong fluorescent background (Fig 3C).
These results suggest that RNA facilitates the LLPS through multivalent interactions with the RRM regions of zRbm14 molecules, whereas the m6A modification further potentiates the efficacy of RNA in a dose‐dependent manner.
m5C modification represses the LLPS‐promoting effects of RNA and m6A‐modified RNA
We next examined whether the m5C modification, which is known to protect maternal mRNAs from MZT‐dependent decay (Yang et al, 2019), could also impact the LLPS of zRbm14. We synthesized two cy3‐labeled RNA fragments of cap1 respectively containing zero and one m5C modification (namely 0m5C‐cy3 and 1m5C‐cy3; Yang et al, 2019). Interestingly, while 0m5C‐cy3 (1 μM) induced the LLPS of 10‐μM GFP‐zRbm14b and co‐phase separated into the condensates, 1‐μM 1m5C‐cy3 had no stimulatory effect (Fig 3D and E), indicating that the m5C modification represses the LLPS‐promoting effect of the RNA. Furthermore, similar to 0m6A (Figs 3B and EV1D), 1‐μM 0m5C induced the LLPS of 10‐μM zRbm14b in 20 min (Figs 3E and EV1F), suggesting that the LLPS‐promoting effect of RNA is largely sequence‐independent.
To assess whether m5C modification could antagonize m6A modification, we examined how mixtures of 1m6A and 1m5C impacted the LLPS of zRbm14b. When the total concentration of RNA was kept constant (1 μM), 1m6A‐cy3 and 1m5C‐cy3 mixed at a 1:3 stoichiometry still failed to induce the LLPS in 30 min, whereas the mixture of a 1:1 stoichiometry induced the LLPS in 20 min (Figs 3E and EV1F). Moreover, the mixture of a 3:1 stoichiometry induced the LLPS in 10 min, similar to 1m6A‐cy3 alone (Figs 3E and EV1F). When increasing amounts of 1m5C‐cy3 were added into pre‐formed liquid droplets of zRbm14b and 1m6A, 1m5C‐cy3 entered the droplets and rapidly induced their disassembly: in 5 min 2‐μM 1m5C‐cy3 markedly reduced the number of the droplets, while 4‐μM 1m5C‐cy3 completely disrupted the droplets (Fig 3F). Therefore, the m5C modification antagonizes the LLPS‐promoting effect of the m6A modification in a dose‐dependent manner, possibly by impairing multivalent interactions among RNA and zRbm14b molecules.
zRbm14b undergoes LLPS in vivo with RNA
To verify the phase separation of zRbm14 in vivo, we micro‐injected in vitro‐transcribed mRNA into 1‐cell embryos to express GFP‐zRbm14b (Xiao et al, 2019). Time‐lapse microscopy indicated that GFP‐zRbm14b was visible in the cytoplasm at 2 hpf and underwent cytoplasmic‐to‐nuclear translocation from 3 hpf (Fig 3G and Movie EV3), similar to endogenous zRbm14a (Fig 1D and E). We examined embryos at 4 hpf due to their widespread high‐level expressions of GFP‐zRbm14b (Fig 3G). Furthermore, endogenous cytoplasmic zRbm14 condensates were no longer abundant at 4 hpf and mainly appeared as one or two juxtanuclear puncta in interphase cells (Fig 1D), allowing attribution of ectopic condensates to the overexpression of GFP‐zRbm14b. We found that ectopic GFP‐zRbm14b condensates formed at 800‐pg mRNA and were generally much larger in size at 1,200‐pg mRNA (Fig 3H), suggesting a dose‐dependent effect. The condensates were positively stained for RNA‐specific dye Pyronin Y (Fig 3H), suggesting their enrichment of RNA. Large condensates were more prominent for the Pyronin signals (Fig 3H), probably due to their large volumes: for spherical condensates, volumes are a cubic function of the radius. Clear fusion events among the puncta were observed in live imaging (Fig 3I), confirming their liquid property. Furthermore, in contrast to the prominent cytoplasmic distribution of GFP‐zRbm14b, GFP‐zRbm14b18S, a previously constructed LLPS‐defective mutant due to Y‐to‐S mutations in its IDR (Xiao et al, 2019), only displayed nuclear localization (n > 100 cells; Fig 3J), indicating that LLPS retains zRbm14 in the cytoplasm.
Together with the previous in vivo and in vitro results (Figs 2 and 3A–F), we propose that m6A modifications in zebrafish early embryos (Zhao et al, 2017) promote the formation of cytoplasmic zRbm14‐maternal mRNA condensates, whereas m5C modifications (Yang et al, 2019) facilitate their disassembly (Fig 3K). As m6A and m5C modifications are antagonizing ways impacting the stability of maternal mRNAs (Zhao et al, 2017; Yang et al, 2019), our results thus far point to a role of zRbm14 in proper maternal mRNA metabolism.
Maternally expressed zRbm14 facilitates the blastula‐to‐gastrula development by facilitating cell cycle and differentiation
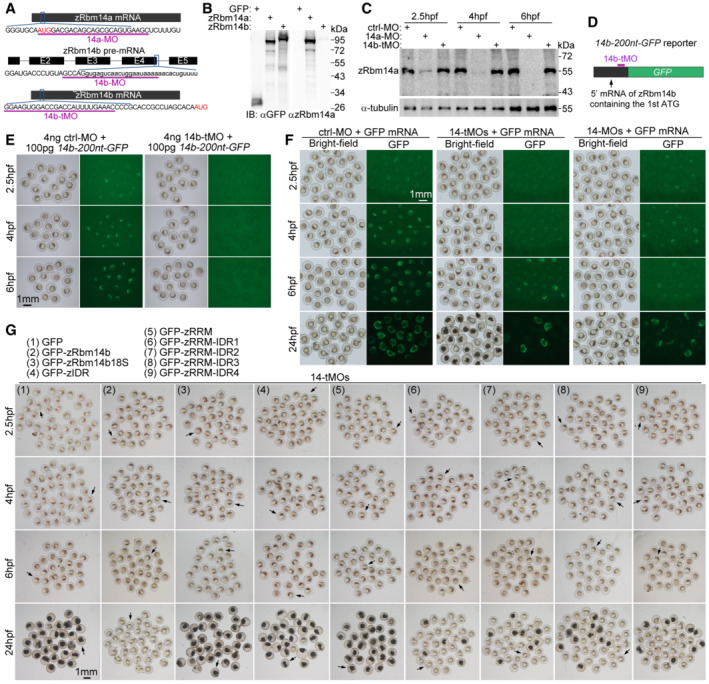
To understand physiological roles of the zRbm14‐maternal mRNA condensate, we investigated phenotypes of early zebrafish embryos lacking zRbm14. We have previously shown that zRbm14a and zRbm14b are functionally redundant and need to be concomitantly depleted to induce severe dorsalized phenotypes in the segmentation stage (Xiao et al, 2019). Nevertheless, as the previously‐used morpholino oligonucleotides 14a‐MO and 14b‐MO were designed to block translation initiation of zRbm14a mRNA and pre‐mRNA splicing of zRbm14b, respectively (Fig EV2A; Xiao et al, 2019), 14b‐MO is only able to repress zRbm14b expression from its zygotic mRNA but not maternal mRNA. We reasoned that this is why zebrafish embryos co‐injected with 14a‐MO and 14b‐MO (14‐MOs), termed herein as zygotic zRbm14 morphants, did not display prominent earlier defects. We thus designed a new MO (14b‐tMO) to block translation initiation so that the expression from maternal zRbm14b mRNA could also be repressed (Fig EV2A). Immunoblotting with our antibody specific to zRbm14a (Fig EV2B) confirmed that zRbm14a was efficiently depleted in embryos from 2.5 to 6 hpf by micro‐injected 14a‐MO, but not 14b‐tMO or ctrl‐MO (Fig EV2C). Due to the lack of an antibody to zRbm14b, we confirmed the efficiency of 14b‐tMO by using a reporter mRNA containing the translation initiation region of zRbm14b mRNA (Fig EV2D and E).
Figure EV2. zRbm14 functions in the blastula‐to‐gastrula development through both RNA‐binding and phase separation (related to Figs 4 and 5A–C).
-
ADiagrams showing target sequences of the indicated MOs on zRbm14 mRNAs. Translation initiation codons (AUG) are shown in red. Lowercase letters represent intron sequences. 14a‐MO and 14b‐tMO were designed to respectively repress the translation of both maternal and zygotic mRNAs of zRbm14a and zRbm14b. 14b‐MO was designed to interfere with the splicing of zRbm14b pre‐mRNA and thus only interfered with the zygotic transcript. The efficacies of both 14a‐MO and 14b‐MO were confirmed previously using 24‐hpf morphants (Xiao et al, 2019).
-
BOur anti‐zRbm14a antibody only weakly cross‐reacted with zRbm14b. HEK293T cell lysates expressing GFP, GFP‐zRbm14a, or GFP‐zRbm14b were immunoblotted with antibodies against GFP and zRbm14a, respectively.
-
C14a‐MO efficiently repressed the translation of zRbm14a mRNA in early embryos. Zebrafish 1‐cell embryos were each microinjected with 4 ng of MO and collected at the indicated time points. Lysates from 5 embryos were loaded in each lane. α‐tubulin served as an internal control.
-
D, E14b‐tMO efficiently repressed the translation of zRbm14b mRNA in early embryos. 14b‐200 nt‐GFP, a reporter mRNA in vitro transcribed from a construct containing a 5′ 200‐nucleotide zRbm14b cDNA fragment followed by an in‐frame GFP cDNA (D), was used to assess the efficacy of 14b‐tMO. In total, 100 pg of the reporter mRNA were co‐injected with 4 ng of MO into each 1‐cell embryo. The embryos were imaged at the indicated time points (E). Note that the embryos co‐injected with 14b‐tMO failed to express GFP.
-
FA typical set of zebrafish morphants. In total, 8 ng of ctrl‐MO, 14‐tMOs (4 ng 14a‐MO + 4 ng 14b‐tMO), or 14‐MOs (4 ng 14a‐MO + 4 ng 14b‐MO) were co‐injected with 100 pg of in vitro transcribed GFP mRNA (as a tracer) into each 1‐cell embryo to generate control, maternal zRbm14, and zygotic zRbm14 morphants, respectively. GFP‐positive embryos were collected at 2.5 hpf and imaged sequentially at the indicated time points. Representative embryos and quantification results are presented in Fig 4A and B.
-
GA typical set of zebrafish embryos in rescue experiments. 800 pg of in vitro transcribed mRNA for one of the indicated proteins were co‐injected with 14‐tMOs (4 ng 14a‐MO + 4 ng 14b‐tMO) into 1‐cell embryos. GFP‐positive morphants were imaged at the indicated time points. Representative embryos (arrows) and quantification results are presented in Fig 5B and C.
Source data are available online for this figure.
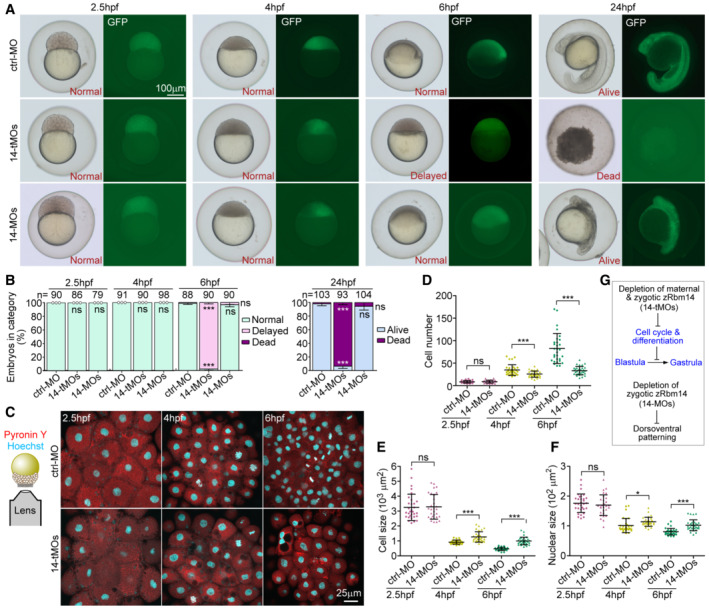
While zygotic zRbm14 morphants are viable to at least 72 hpf (Xiao et al, 2019), embryos co‐injected with 14a‐MO and 14b‐tMO (14‐tMOs), termed herein as maternal zRbm14 morphants, displayed severe embryonic lethality at 24 hpf (Figs 4A and B, and EV2F). At 6 hpf, while 98.9% of ctrl‐MO‐injected embryos (control morphants) and 96.6% of zygotic zRbm14 morphants completed 50% epiboly (Kimmel et al, 1995), 97.8% of maternal zRbm14 morphants displayed little epiboly movement and morphologically resembled 4‐hpf sphere embryos (Figs 4A and B, and EV2F). All three groups of embryos, however, were morphologically similar at 2.5 and 4 hpf (Figs 4A and B, and EV2F).
Figure 4. Maternal zRbm14 morphants display impaired cell cycle, blastula arrest, and embryonic death.
-
A, BPhenotypes of zRbm14 morphants. Totally 8 ng of ctrl‐MO, 14‐tMOs, or 14‐MOs were co‐injected with 100 pg of in vitro transcribed GFP mRNA (as a tracer) into 1‐cell embryos to generate control, maternal zRbm14, and zygotic zRbm14 morphants, respectively. Representative embryos, imaged under a dissecting microscope, are shown in (A) and phenotyping results in (B). Total numbers of embryos analyzed are listed over histograms. Sample dots are included for quantification results with no error bars. Please also refer to Fig EV2.
-
C–FMaternal zRbm14 morphants displayed reduced cell proliferation in MZT. Embryos stained with Hoechst 33342 and Pyronin Y to mark the nucleus and the cytoplasm, respectively, were imaged from the animal pole to cover a 10‐μm z‐depth with a confocal microscope (C). Maximum intensity‐projected images (C) from 30 embryos in each condition were used to quantify cell number (D), cell size (E), and nuclear size (F). Each sample dot represents the average value from each embryo.
-
GSummary of the results.
Data information: Quantification results are presented as mean ± SD from three independent experiments. Student's t‐test against ctrl‐MO‐injected populations: ns, no significance (P > 0.05); *P < 0.05; ***P < 0.001.
Source data are available online for this figure.
As rapid cell divisions of early zebrafish embryos require translations from maternal mRNAs (Winata et al, 2018), we examined cell size and number to assess whether the cell cycles and differentiations were affected in maternal zRbm14 morphants. We labeled the nucleus and the cytoplasm respectively with Hoechst 33342 and Pyronin Y and quantified cell images acquired from the animal pole to cover a 10‐μm‐deep region (Fig 4C). Comparing to ctrl‐MO embryos, maternal zRbm14 morphants displayed significantly reduced cell number, accompanied by increased cell size and nuclear size, at both 4 and 6 hpf, but not 2.5 hpf (Fig 4C–F). At 6 hpf, for instance, the average cell number in the imaged region of maternal zRbm14 morphants decreased by 60% and the average cell size increased by 2.2‐fold as compared to the control embryos (Fig 4D and E). Furthermore, the average cell number, size, and nuclear size of the 6‐hpf maternal zRbm14 morphants were similar to those of control 4‐hpf embryos (Fig 4C–F), further confirming that maternal zRbm14 morphants are arrested in the blastula stage. Therefore, zRbm14 (i.e., both zRbm14a and zRbm14b) expressed from maternal mRNAs is critical for proper cell cycle and differentiation to enable embryonic development into gastrula, whereas zRbm14 expressed from zygotic genes mainly contributes to proper dorsoventral patterning (Fig 4G; Xiao et al, 2019).
zRbm14 functions by forming maternal mRNAs‐containing condensates
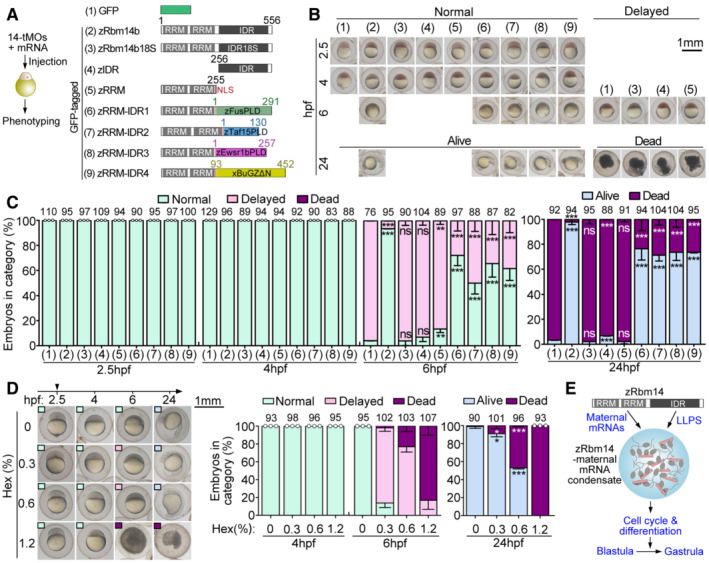
To rule out possible off‐target effect of 14t‐MOs and understand molecular mechanisms underlying the functions of maternal zRbm14, we co‐injected in vitro transcribed mRNAs with 14‐tMOs to express GFP‐tagged zRbm14b and various constructs (Fig 5A) as done previously (Xiao et al, 2019). Comparing to the blastula arrest at 6 hpf (96% on average) and lethality at 24 hpf (97%) of maternal zRbm14 morphants expressing GFP, 94% of the morphants expressing GFP‐zRbm14b developed normally into shield embryos at 6 hpf and 98% reached the segmentation stage at 24 hpf (Figs 5A–C and EV2G), confirming the specificity of 14t‐MOs. In contrast, neither the 18S mutant nor constructs lacking either the IDR or the RRM region displayed prominent rescue effects (Figs 5A–C and EV2G), suggesting the essentialness of both the phase‐separation and RNA‐binding properties of zRbm14.
Figure 5. zRbm14 functions by enriching maternal RNA into its phase‐separated condensates.
-
AExperimental scheme and diagrams of constructs used in the rescue experiments. A nuclear localization signal (NLS) was added to the zRRM and zRRM‐IDR1‐4 constructs to maintain their nuclear localization capacity. Prion‐like domains (PLD) of zFus, zTaf15, and zEwsr1b (Wang et al, 2018) and the low complexity region of Xenopus BuGZ (xBuGZΔN; Jiang et al, 2015) were used to replace the IDR of zRbm14b (zIDR; Xiao et al, 2019). RRM, RNA‐recognition motif.
-
B, CPhenotypes of embryos. Typical embryos, cropped from representative experimental groups in Fig EV2G, are shown in (B). Their phenotypes served as criteria for quantifications in (C). Only GFP‐positive embryos viable at 2.5 hpf were collected and imaged for analyses. Total embryo numbers analyzed are listed over histograms.
-
DHexanediol (Hex) treatment from 2.5 hpf blocked the blastula development. Representative images are presented for each group of embryos, with phenotypes indicated by insets of colored boxes. Total embryo numbers analyzed are listed over histograms.
-
EA summarizing model. zRbm14 binds to maternal mRNAs through the N‐terminal RRM‐containing region and phase separates into cytoplasmic condensates through its IDR. Such maternal mRNA‐zRbm14 condensates facilitate proper cell cycle and differentiation for embryonic development into the gastrula stage.
Data information: Quantification results (mean ± SD) were from three independent experiments. Sample dots are included for results with no error bars and thus excluding t‐test. Unpaired two‐sided Student's t‐test against control group (group 1): ns, no significance; **P < 0.01; ***P < 0.001.
Source data are available online for this figure.
If the IDR of zRbm14 indeed mainly functions as a phase separation domain in cleavage‐stage embryos, it would be functionally interchangeable with a phase‐separation IDR from other proteins. To clarify this, we performed rescue experiments with hybrid constructs (zRRM‐IDR1 to zRRM‐IDR4; Fig 5A; Xiao et al, 2019) consisting of the RRM region of zRbm14b and a phase‐separation IDR from zebrafish Fus, Taf15, Ewsr1b, or Xenopus BuGZ (Jiang et al, 2015; Wang et al, 2018). We observed that at least 50% of the morphants developed to shield embryos at 6 hpf regardless of the hybrid expressed. Furthermore, at least 71% of the 24‐hpf morphants became viable and developed mostly to the segmentation stage (Figs 5A–C and EV2G). Such an IDR interchangeability not only clarifies the molecular role of zRbm14 IDR but further strengthens the importance of LLPS for the functions of maternal zRbm14 as well.
To further assess the importance of LLPS in the blastula‐to‐gastrula development, we used 1,6‐hexanediol (Hex), a chemical specifically dissolving liquid‐featured assemblies (Kroschwald et al, 2017), to induce acute disassembly of intracellular liquid condensates, including those of zRbm14, from 2.5 hpf. Embryos treated with different concentrations of Hex (up to 1.2% w/v) developed normally to 4 hpf but displayed a dose‐dependent abnormality at 6 hpf, including approximately 1‐h epiboly delay (84% at 0.3% Hex), severe epiboly delay or arrest (77% at 0.6% Hex and 17% at 1.2% Hex), and embryonic death (23% at 0.6% Hex and 83% at 1.2% Hex; Fig 5D). At 24 hpf, the embryos also displayed dose‐dependent developmental retardation and lethality (Fig 5D). The high sensitivity of the embryos to Hex strengthens the importance of liquid condensates in the blastula‐to‐gastrula development.
Together, we conclude that maternal zRbm14 functions in the blastula‐to‐gastrula development by forming maternal mRNAs‐containing cytoplasmic condensates (Fig 5E).
zRbm14 is required for timely clearance of maternal mRNAs
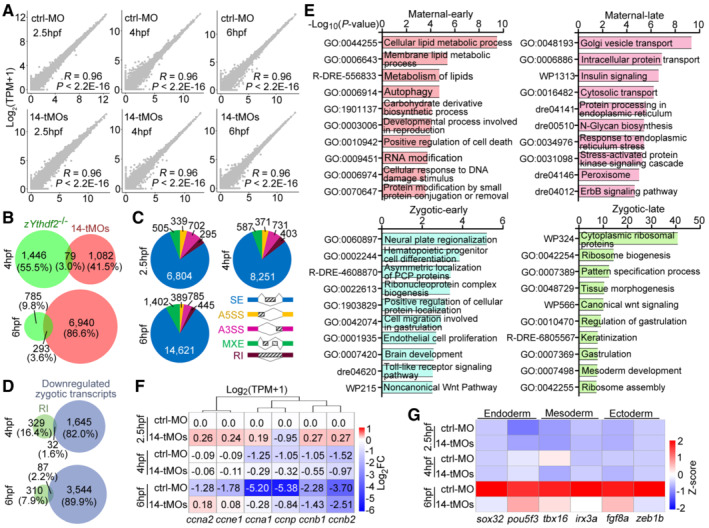
For insights on how zRbm14 impacted maternal mRNAs, we performed RNA‐seq on control and maternal zRbm14 morphants at 2.5, 4, and 6 hpf, respectively, and profiled the transcriptomes using RNA‐seq results from two independent replicates of samples (Fig EV3A). Principal component analysis (PCA) revealed that transcriptomic differences between the morphants and control (ctrl‐MO) embryos became increasingly prominent from 4 hpf (Fig 6A). Furthermore, transcriptomes of the 6‐hpf morphants were more closely related to those of the 4‐hpf control embryos (Fig 6A), confirming from the transcriptome level their arrest at the mid‐blastula stage. The altered transcripts, which were those with an absolute fold change by > 2 fold between ctrl‐MO and 14‐tMOs samples, were involved in RNA metabolic processes, RNP and ribosome biogenesis, and RNA processing at 2.5 hpf, morphogenesis and pattern formation at 4 hpf, and signaling and cell fate commitment at 6 hpf (Fig 6B). Cumulative curves indicated that massive mRNA clearance occurred between 4 and 6 hpf in the control embryos (Fig 6C), consistent with previous reports (Aanes et al, 2014; Zhao et al, 2017; Yang et al, 2019). In contrast, the 6‐hpf zRbm14 morphants displayed a prominent stabilization of transcripts (Fig 6C).
Figure EV3. Transcriptome analysis of zebrafish embryos (related to Fig 6).
-
AScatter plots showing correlations of independent replicates of RNA‐seq results. The Pearson correlation coefficient (R) and P values are shown.
-
BzRbm14 and zYthdf2 were largely responsible for the decay of different mRNAs. Only a small portion of transcripts accumulated in maternal zRbm14 morphants by more than 2‐fold overlapped with those accumulated in maternal zYthdf2−/− embryos (Zhao et al, 2017). Note that the zRbm14 depletion interfered with the levels of much more transcripts than the zYthdf2 deficiency at 6 hpf.
-
CDifferential alternative splicing events in the zRbm14 morphants. SE, skipped exon; A3SS, alternative 3′ splicing site; A5SS, alternative 5′ splicing site; MXE, mutually exclusive exon; RI, retained intron.
- D
-
ETop 10 enriched terms of differentially expressed genes (DEGs, fold change > 2) between control and maternal zRbm14 morphants at 6 hpf.
-
FRelative transcript levels of cyclin genes. FCs were calculated from RNA‐seq results, relative to the levels in 2.5‐hpf ctrl‐MO embryos, of two independent sets of samples. Only transcripts with TPM > 500 and classified as “maternal transcripts” (Fig 6D) were analyzed. Note the increasingly reduced clearance of the transcripts following time in maternal zRbm14 morphants. ccna1 and ccna2, zebrafish cyclin A genes; ccnb1 and ccnb2, cyclin B genes; ccne1, cyclin E gene; and ccnp, cyclin P gene.
-
GExpression profile of zygotic transcripts critical for the differentiation of three germ layers. Sox32 (also named casanova), pou5f3 (homolog of mammalian Oct4), tbx16 (spadetail), irx3a (encoding Iro3), fgf8a (encoding Fgf8a), and zeb1b (encoding Kheper) were selected from our pools of zygotic transcripts (Fig 6D) according to literature (Kudoh et al, 2004; Schier & Talbot, 2005; Perez‐Camps et al, 2016).
Data information: P values of the sequencing data in (A) were calculated by using two‐sided Wilcoxon and Mann–Whitney tests. P values in (E) were calculated by using Metascape. A P value of < 0.05 is considered statistically significant.
Source data are available online for this figure.
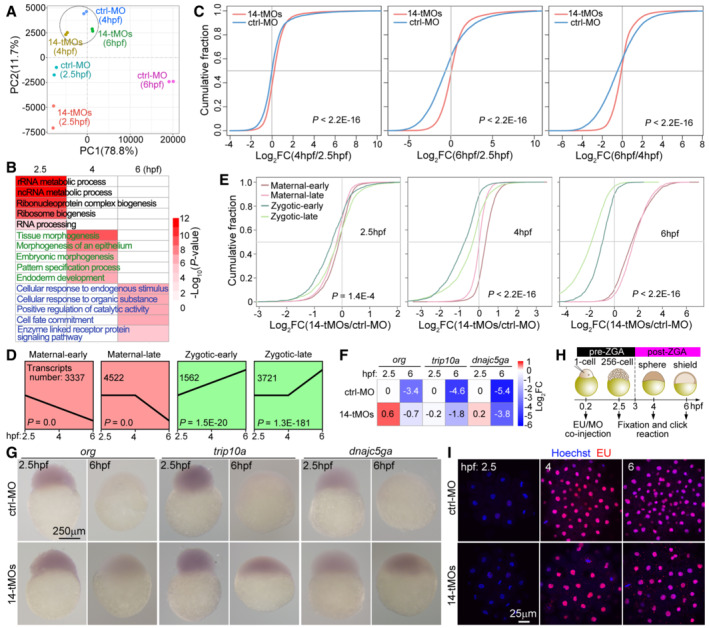
Figure 6. Depletion of maternal zRbm14 hinders maternal RNA clearance and MZT.
-
AVariance in gene expression profiles of control (ctrl‐MO) and maternal zRbm14 (14‐tMOs) morphants. Principal component analysis (PCA) was performed using two biological replicates.
-
BRepresentative gene ontology (GO) biological processes enriched for differential transcripts between control and maternal zRbm14 morphants.
-
CCumulative distribution showing fold changes (FCs) in transcript abundance in control morphants (blue curves) and maternal zRbm14 morphants (red curves) between the indicated stages. Transcript abundances were averaged from two independent experiments. P values were calculated using two‐sided Wilcoxon and Mann–Whitney tests.
-
DExpression patterns of four clusters of representative transcripts, categorized from RNA‐seq results of ctrl‐MO embryos. The transcripts in the maternal‐early and the zygotic‐early clusters respectively displayed continual degradation or accumulation from 2.5 hpf, whereas those in the maternal‐late and zygotic‐late clusters respectively started degradation or accumulation from 4 hpf.
-
ECumulative distribution showing FCs of transcript abundance of the indicated clusters between control and maternal zRbm14 morphants.
-
FRelative transcript levels of three typical maternal genes. FCs were calculated relatively to the levels in 2.5‐hpf ctrl‐MO embryos. org, oogenesis‐related; trip10a, thyroid hormone receptor interactor 10a; dnajc5ga, DnaJ (Hsp40) homolog, subfamily C, member 5 gamma a.
-
GWhole‐mount in situ RNA hybridization of org, trip10a, and dnajc5ga. A representative embryo is shown for each group of at least 26 embryos.
-
H, IMaternal zRbm14 morphants displayed normal ZGA onset. MO(s) and 5‐ethynyl uridine (EU) were co‐injected and treated as illustrated (H). The diagrams are not drawn to scale. EU incorporated into nascent transcripts were visualized through a click reaction. Hoechst 33342 was used to counterstain the nucleus. A representative maximum intensity‐projected confocal micrograph, taken from the animal pole, is shown for each group of 10 embryos (I).
Data information: P values of the sequencing data in (C) and (E) were calculated by using two‐sided Wilcoxon and Mann–Whitney tests. P values in (D) were calculated by using Short Time‐series Expression Miner (STEM) software. A P value of < 0.05 is considered statistically significant.
Source data are available online for this figure.
When four classes of transcripts in the control embryos were selected through statistical analysis to represent “maternal early,” “maternal late,” “zygotic early,” and “zygotic late” genes, respectively, based on different expression profiles (Fig 6D; Aanes et al, 2014; Zhao et al, 2017), we observed increasing accumulations of maternal transcripts in maternal zRbm14 morphants from 2.5 to 6 hpf (Fig 6E), indicating a failure in maternal RNA decay. For instance, 66.0% or more of the transcripts in the maternal‐early or maternal‐late class exhibited at least 2‐fold accumulations in the 6‐hpf morphants. By contrast, only 1.3% or less of the transcripts in the zygotic‐early or zygotic‐late class increased by at least two folds (Fig 6E). Interestingly, 33.7% of transcripts (1,876 out of 5,566) accumulated in 4‐hpf and/or 6‐hpf maternal zRbm14 morphants (≥ 2‐fold) overlapped with m6A‐modified transcripts reported previously (Zhao et al, 2017). These transcripts, however, differed largely from those accumulated in maternal zYthdf2 −/− embryos (Fig EV3B; Zhao et al, 2017). Therefore, zRbm14 facilitates the timely decay of a maternal mRNA population that differs largely from the population targeted by zYthdf2.
Maternal RNA transcripts are degraded through both ZGA‐dependent and ZGA‐independent pathways during MZT (Bashirullah et al, 1999; Giraldez et al, 2006; Walser & Lipshitz, 2011; Despic & Neugebauer, 2018), with different transcripts displaying distinct degradation kinetics (Mishima & Tomari, 2016; Vejnar et al, 2019). We thus examined abundance changes of three well‐characterized maternal transcripts, each degraded through a distinct pathway: org transcripts are degraded through the maternal‐dependent pathway, dnajc5ga through the zygotic miR‐430‐dependent pathway, and trip10a through the zygotic‐dependent but miR‐430‐independent pathway (Vejnar et al, 2019). All of them displayed ≥ 2‐fold accumulations in 6‐hpf maternal zRbm14 morphants in our RNA‐seq results (Fig 6F). Whole‐mount in situ hybridization further confirmed their striking accumulations in 6‐hpf maternal zRbm14 morphants (Fig 6G). zRbm14 thus appears to function in a pathway commonly required for timely degradation of maternal transcripts.
Depletion of zRbm14 impairs MZT without influencing ZGA onset
In contrast to the global accumulation of maternal mRNAs, zygotic transcripts displayed progressive downregulations from 2.5 to 6 hpf (Fig 6E). As zRbm14 gradually entered the nucleus during this period of time (Figs 1D and E, and 3G; Movie EV3; Xiao et al, 2019), we evaluated the contribution of zRbm14‐mediated pre‐mRNA splicing (Auboeuf et al, 2004; Xiao et al, 2019) to the phenotype. Compared to 2.5 hpf, differential alternative splicing events between the zRbm14‐depleted and control samples increased by 1.2‐fold at 4 hpf and 2.0‐fold at 6 hpf (Fig EV3C). Despite this, the number of retained intron(s) (RI), which would potentiate mRNA instability (Ge & Porse, 2014), was low comparing to the total events (Fig EV3C). Accordingly, the number of genes showing RI (e.g., 397 at 6 hpf) was much smaller than that of zygotic genes downregulated in maternal zRbm14 morphants (e.g., 3,631 at 6 hpf; Fig EV3D). Furthermore, both populations only slightly overlapped (Fig EV3D). Therefore, we attribute the downregulation of zygotic transcripts mainly to MZT, but not to RI‐induced mRNA degradation.
To gain further insights into the contribution of differential maternal and zygotic transcripts to developmental defects of maternal zRbm14 morphants (Fig 4), we performed gene ontology (GO) enrichment analysis. The altered maternal transcripts mainly influenced biological processes such as lipid metabolism, autophagy, intracellular transport, and signaling, whereas the altered zygotic transcripts mainly influenced ribosome biogenesis, cell proliferation, differentiation, and gastrulation (Fig EV3E). Consistently, the timely clearance of multiple maternal cyclin transcripts was impaired in maternal zRbm14 morphants, especially at 6 hpf (Fig EV3F), which would contribute to their cell proliferation defects (Fig 4C–F). Furthermore, while zygotic transcripts of multiple germ‐layer regulators (Kudoh et al, 2004; Schier & Talbot, 2005; Perez‐Camps et al, 2016) were highly upregulated in control embryos at 6 hpf, their levels were low in maternal zRbm14 morphants (Fig EV3G), indicating defective endoderm, mesoderm, and ectoderm differentiations.
To clarify whether the downregulation of zygotic transcripts was due to a failure in ZGA onset, we co‐injected 5‐ethynyl uridine (EU) into 1‐cell embryos to label nascent zygotic transcripts (Fig 6H; Chan et al, 2019). We observed similar timing of the major ZGA (between 2.5 and 4 hpf) in both control and maternal zRbm14 morphants (Fig 6I). Therefore, the depletion of zRbm14 impairs MZT by hindering the extent, but not the timing, of ZGA due to defects in maternal mRNA decay.
zRbm14 facilitates deadenylation of its target maternal mRNAs
Why did maternal mRNAs fail to decay in the zRbm14‐depleted embryos? As poly(A) tail shorting by deadenylase is the major rate‐limiting step in mRNA degradation (Garneau et al, 2007; Mishima & Tomari, 2016; Despic & Neugebauer, 2018), we examined the poly(A) status of representative maternal org, trip10a, and dnajc5ga transcripts that were confirmed to be accumulated in maternal zRbm14 morphants (Fig 6G). PCR analysis revealed that the total transcript levels of the three genes in zRbm14‐depleted embryos generally exceeded those in control embryos at 4 and 6 hpf (Figs 7B and C, and EV4A and B), consistent with the deep sequencing and in‐situ hybridization results (Fig 6F and G). Furthermore, poly(A)‐length assays (Fig 7A; Kusov et al, 2001) revealed that the levels of poly(A)‐containing transcripts also significantly increased (Figs 7B and C, and EV4A and B). For instance, the poly(A)‐transcripts of org at 4 hpf exceeded those in control embryos by 3.5‐fold (Fig 7B and C); the poly(A)‐transcripts of trip10a increased at all three time points (3.1‐fold at 2.4 hpf, 1.6‐fold at 4 hpf, and 2.9‐fold at 6 hpf), whereas those of dnajc5ga increased at 2.5‐hpf (4.6‐fold) and 4‐hpf (2.6‐fold; Figs 7B and C, and EV4A and B). In comparison, neither total transcripts nor poly(A)‐transcripts of actb1 (β‐actin gene), which was not a target gene of zRbm14, were significantly altered between the two groups of embryos (Fig EV4A and B). Therefore, the depletion of zRbm14 impairs the deadenylation of its target maternal mRNAs, suggesting a role of its condensates in the deadenylation of their associated mRNAs.
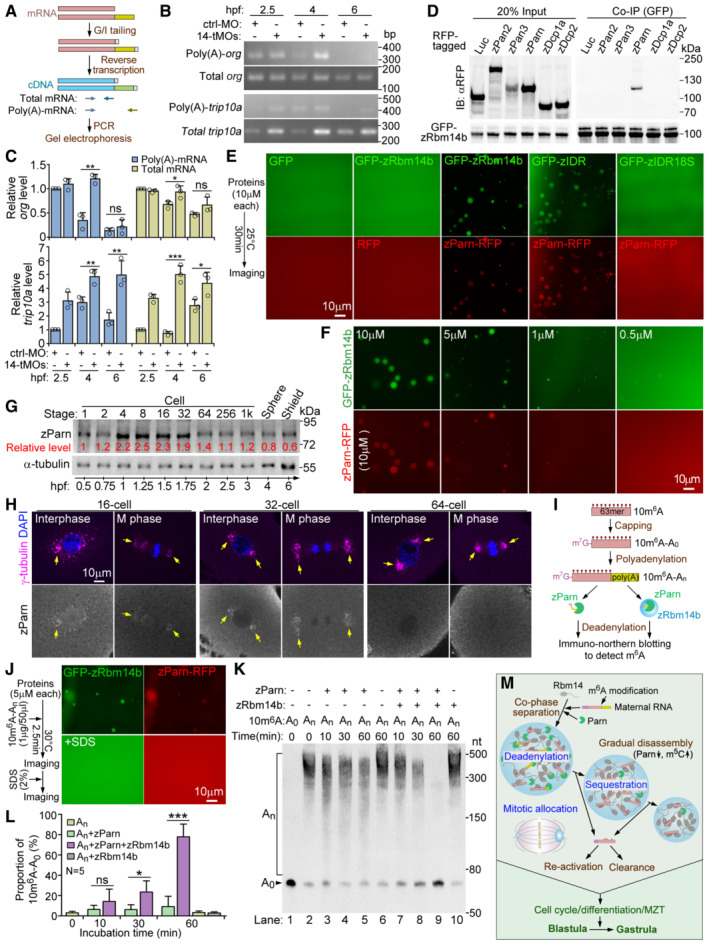
Figure 7. zParn co‐phase separates into zRbm14 condensates to efficiently deadenylate their content maternal mRNAs.
-
AExperimental scheme for poly(A) length assays. Two sets of PCR primers (colored arrows) were used to amplify total transcripts and poly(A)‐transcripts, respectively, for a certain gene as illustrated.
-
B, CDepletion of zRbm14 resulted in accumulation of poly(A)‐containing maternal mRNAs. Total RNAs purified from the indicated zebrafish embryos were subjected to poly(A) length assays. One set of representative PCR results for org and trip10a (B) and quantification results from three independent experiments (C) are presented. As the PCR products of the poly(A)‐transcripts mainly emerged as a single band, only intensities of this major band were measured. Refer to Fig EV4A and B for additional results.
-
DzParn associated with zRbm14b. GFP‐zRbm14b was co‐expressed respectively with the indicated RFP fusion proteins in HEK293T cells. Co‐immunoprecipitations (co‐IP) were performed using anti‐GFP beads. Luciferase (Luc)‐RFP served as a negative control.
-
E, FzParn markedly promoted LLPS of zRbm14b by interplaying with zIDR. Purified His‐tagged RFP or zParn‐RFP was mixed with His‐tagged GFP or GFP‐zRbm14b or mutants (E). Parn‐RFP was mixed with decreasing concentrations of GFP‐zRbm14b (F).
-
G, HzParn was highly expressed from 4‐ to 32‐cell stages and enriched in zRbm14 condensates. Immunoblotting (G) and immunostaining (H) were performed using a rabbit anti‐zebrafish Parn antibody. Lysates from 5 embryos were loaded in each lane in (G). α‐tubulin served as internal controls. Representative single optical sections are presented in (H). Chromatin DNA was stained with DAPI. Arrows point to typical regions abundant in condensates.
-
IExperimental scheme for deadenylation assays. The 5′ and 3′ ends of 10m6A were capped and polyadenylated in vitro to generate a test mRNA, Cap‐10m6A‐An, for the assays.
-
JzRbm14b and zParn efficiently co‐phase separated in deadenylation experiments. The addition of SDS disrupted the condensates to terminate the deadenylation reaction.
-
K, LCo‐phase separation with zRbm14b markedly enhanced the deadenylation activity of zParn. RNAs extracted from the indicated samples were subjected to immuno‐northern blotting using an antibody against m6A (K). Relatively proportions of Cap‐10m6A‐A0 in total RNAs (L) were quantified from five sets of independent results.
-
MA summarizing model. Refer to main text for details.
Data information: Micrographs for in vitro assays (E, F, and J) were taken manually under a wide‐field microscope. Sometimes some condensates moved during imaging, resulting in a positional shift between their GFP and RFP images. Quantification results are presented as mean ± SD. Unpaired two‐sided Student's t‐test against control group: ns, no significance; *P < 0.05; **P < 0.01; ***P < 0.001. Note that the t‐test cannot be performed for the 2.5‐hpf samples in (C) because the relative mRNA levels of control samples were set as 1 and thus have no error bars.
Source data are available online for this figure.
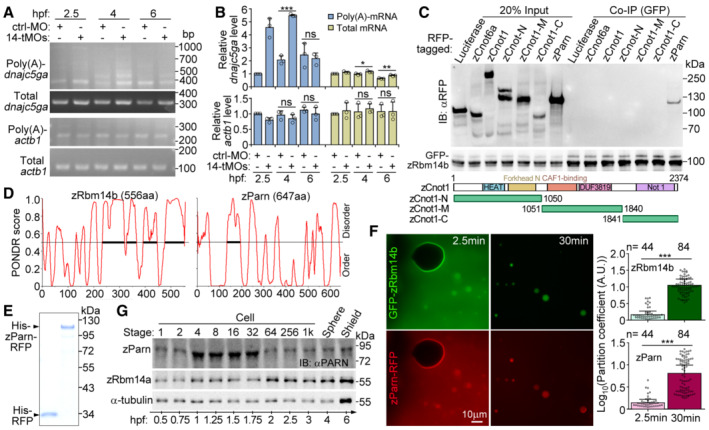
Figure EV4. Relationships between zRbm14 and zParn (related to Fig 7).
-
A, BDepletion of zRbm14 resulted in an accumulation of poly(A)‐containing maternal transcripts of dnajc5ga. Total RNAs purified from the indicated zebrafish embryos were subjected to poly(A) length assays as illustrated in Fig 7A. Transcripts of actb1 (β‐actin mRNA), which was not a target of zRbm14 based on Fig 6D and E, served as a negative control. One set of representative PCR results (A) and quantification results from three independent experiments (B) are presented. The major band of poly(A)‐actb1 was quantified, as in Fig 7B. As poly(A)‐dnajc5ga emerged as multiple bands, their total intensity was measured.
-
CzParn, but not components of the CCR4‐NOT complex, associated with zRbm14b. GFP‐zRbm14b was co‐expressed with the indicated RFP‐fusion proteins in HEK293T cells. Co‐immunoprecipitations (co‐IP) were performed with anti‐GFP beads. Luciferase served as a negative control. zCnot6a, zebrafish CCR4a orthologue; zCnot1, zebrafish Not1 orthologue. Diagrams of zCnot1 mutants are provided.
-
DSecondary structure prediction for zRbm14b and zParn using Predictor of Natural Disordered Regions (PONDR). Bold line indicates predicted IDR.
-
EPurified RFP and zParn‐RFP from E. coli. The proteins were subjected to SDS–PAGE, followed by Coomassie blue staining.
-
FEfficient co‐phase separation of 10‐μM zParn and 10‐μM zRbm14b. Purified zParn‐RFP was mixed with GFP‐zRbm14b and imaged under a wide‐field microscope at 2.5 and 30 min, respectively. Partition coefficients were respectively calculated for 44 or 84 droplets from three independent experiments and pooled together in the histograms.
-
GExpression profile of zParn during early embryogenesis. Lysates from 5 embryos were loaded in each lane. Immunoblotting was performed with an anti‐human PARN antibody. α‐tubulin and zRbm14a served as internal controls.
Data information: Quantification results are presented as mean ± SD, with sample dots. Unpaired two‐sided Student's t‐test: ns, no significance; *P < 0.05; **P < 0.01; ***P < 0.001. Note that the t‐test cannot be performed for the 2.5‐hpf samples in (B) because the relative mRNA levels of the control samples were set as 1 and thus have no error bars.
Source data are available online for this figure.
We noticed that, in control embryos, the levels of poly(A)‐transcripts and total transcripts varied differently for each gene following the embryonic development (Figs 7B and C, and EV4A and B). From 2.5 to 4 hpf, for instance, the levels of poly(A)‐transcripts of org dropped by 65% on average. Those of its total transcripts, that is, both poly(A)‐containing and poly(A)‐less transcripts, however, dropped by only 32% (Fig 7B and C), suggesting a selective stabilization of deadenylated org transcripts. In addition, poly(A)‐transcripts of trip10a and dnajc5ga increased by 3.0‐ and 2.1‐fold, respectively, from 2.5 to 4 hpf, whereas their total transcripts were moderately reduced (trip10a) or unchanged (dnajc5ga; Figs 7B and C, and EV4A and B), suggesting that a large fraction of transcripts that were already deadenylated at 2.5 hpf becomes polyadenylated at 4 hpf. These results further suggest that zRbm14 condensates can protect deadenylated mRNAs from premature degradation for re‐activation through cytoplasmic polyadenylation.
Deadenylase zParn co‐phase separates into zRbm14 condensates in early embryos
As deadenylation and stabilization followed by cytoplasmic polyadenylation or clearance are unique mechanisms controlling the translational competence and decay of maternal mRNAs in developing zebrafish embryos (Aanes et al, 2014; Despic & Neugebauer, 2018; Winata et al, 2018), our results to date implied that zRbm14 condensates might recruit certain deadenylase to deadenylate their target mRNAs and then protect them from degradation before the mRNAs were released for re‐activation or clearance. Co‐immunoprecipitation (co‐IP) indicated that, among the three major deadenylases (Parn, Pan2‐Pan3, and the CCR4‐NOT complex; Garneau et al, 2007; Despic & Neugebauer, 2018), only zParn associated with zRbm14b (Figs 7D and EV4C). In contrast, the Dcp1‐Dcp2 decapping enzyme, which removes the m7G cap of deadenylated mRNAs to trigger their degradation by 5′–3′ exonuclease (Garneau et al, 2007; Despic & Neugebauer, 2018), did not co‐immunoprecipitate with zRbm14b (Fig 7D).
We then investigated whether zRbm14b condensates could recruit zParn. Compared to zRbm14b, zParn had only a short IDR (Fig EV4D). In total, 10 μM of purified zParn‐RFP was incapable of LLPS alone but facilitated the LLPS of 10‐μM GFP‐Rbm14b through co‐phase separation (Figs 7E, and EV4E and F). zParn‐RFP also similarly stimulated the LLPS of 10‐μM GFP‐zIDR but not GFP‐zIDR18S (Fig 7E), an LLPS‐defective mutant (Xiao et al, 2019). When 10 μM of zParn‐RFP was mixed with different concentrations of GFP‐Rbm14b, co‐phase separation was clearly observed in as low as 1‐μM GFP‐Rbm14b (Fig 7F). Therefore, zParn promotes the LLPS of zRbm14 through an interaction with zIDR and is accordingly partitioned into zRbm14 condensates.
To understand when zParn would impact zRbm14 condensates in zebrafish embryos, we examined its expression profile. Immunoblotting using two independent rabbit antibodies consistently revealed that zParn levels increased by approximately 2‐fold between the 4‐ and 32‐cell stages (Figs 7G and EV4G). When 16‐ and 32‐cell embryos were co‐immunostained for γ‐tubulin, which served as a marker for zRbm14 condensates because the anti‐zRbm14a antibody was also from rabbit, a fraction of zParn was observed to co‐localize nicely with centrosomal γ‐tubulin in both interphase and mitotic blastomeres (Fig 7H). In 64‐cell embryos, however, zParn no longer displayed such a localization (Fig 7H), consistent with its downregulation at this stage (Figs 7G and EV4G). Therefore, zParn is enriched in cytoplasmic zRbm14‐maternal mRNA condensates between 4‐ and 32‐cell stages, indeed suggesting a role in their formation and function.
Co‐phase separated zParn displays potent deadenylation activities
To assess the deadenylation activity of zParn in zRbm14‐maternal mRNA condensates, we synthesized an mRNA substrate, Cap‐10m6A‐An, composed of 5′ capped 10m6A with a 3′ poly(A) tail of varying length (Fig 7I). As 10 μM of zParn‐RFP deadenylated Cap‐10m6A‐An too rapidly in pilot experiments, we used 5‐μM zParn‐RFP and confirmed its rapid co‐phase separation with 5‐μM GFP‐Rbm14b in the deadenylation buffer containing Cap‐10m6A‐An (Fig 7J). We used SDS to disrupt the condensates to terminate the deadenylation reaction (Fig 7J).
Immuno‐northern blotting revealed that the mRNA sample contained only a residual portion (3.1%) of the poly(A)‐less form, Cap‐10m6A‐A0 (Fig 7K, lanes 1–2, and L). The presence of zParn‐RFP gradually increased the proportion of Cap‐10m6A‐A0, reaching 9.3% on average in 60 min (Fig 7K, lanes 3–5, and L), whereas the mock treatment or GFP‐zRbm14b had no effect (Fig 7K, lanes 6, 10, and L). In sharp contrast, the proportion of Cap‐10m6A‐A0 increased to 77.9% in reactions containing both zParn‐RFP and GFP‐zRbm14b (Fig 7K, lanes 7–9, and L), indicating that co‐phase separated zParn displays more than 10‐fold deadenylation activity over free zParn.
Taken together, we propose that maternal m6A‐mRNAs and zParn co‐phase separate with zRbm14 by interacting with its RRM and IDR regions. The resultant cytoplasmic condensates efficiently deadenylate the maternal mRNAs by the 32‐cell stage, sequester the poly(A)‐less mRNAs, and allocate them into daughter cells during mitosis (Fig 7M). The condensates are gradually disassembled, due to downregulation of zParn and upregulation of the m5C modification, resulting in a stepwise release of their deadenylated mRNAs for either re‐activation by cytoplasmic polyadenylation or clearance by degradation enzymes to eventually facilitate the blastula‐to‐gastrula transition (Fig 7M; Weill et al, 2012; Winata et al, 2018; Vastenhouw et al, 2019).
Rbm14 forms spindle pole‐enriched cytoplasmic condensates in mouse cleavage‐stage embryos
Next, we explored whether the roles of Rbm14 are conserved in mammals. Distinct from zebrafish, maternal mRNAs in mouse are mostly degraded by the middle 2‐cell stage, whereas ZGA initiates soon after fertilization (Fig 8A; Eckersley‐Maslin et al, 2018; Vastenhouw et al, 2019). Mouse Rbm14 was rapidly synthesized after fertilization and displayed both cytoplasmic and nuclear distributions in the 1‐cell stage (Fig 8B). Punctate cytoplasmic condensates peaked in blastomeres from the 2‐ to 8‐cell stages (Fig 8B). Furthermore, in all four mitotic blastomeres identified out of 350 embryos at the 2‐cell stage, Rbm14 displayed striking accumulations at spindle poles (Fig 8B, arrows, and Movie EV4). Cytoplasmic Rbm14 reduced obviously in morulae and became undetectable in blastocysts (Fig 8B). These results suggest the conservation of Rbm14‐maternal mRNA condensates in vertebrate.
Figure 8. Functions of Rbm14 in MZT and the blastula‐to‐gastrula development are conserved in vertebrate.
-
AIllustration of MZT in mouse embryos. Most maternal mRNAs are degraded during 1‐ and 2‐cell stages, during which period zygotic gene transcriptions are also gradually activated through two waves of ZGA (minor and major; Eckersley‐Maslin et al, 2018).
-
BMouse Rbm14 formed cytoplasmic puncta in blastomeres (1‐ to 8‐cell). Maximum intensity‐projected confocal immunofluorescent micrographs are presented. Framed regions are magnified to show details of cytoplasmic Rbm14 puncta. Arrows point to Rbm14 condensates accumulated at spindle poles. Refer to Movie EV4 for corresponding 3D‐reconstructed images.
-
CSummary of mouse progeny obtained by inbreeding Rbm14+/− mice.
-
DRelative transcript levels of Rbm14 in the indicated E4.5 blastocysts (S2–S10; refer to Fig EV5C) based on RNA deep Sequencing results, normalized to those of Gapdh. Six blastocysts were considered as Rbm14−/− embryos because they contained only trace amount of Rbm14 transcript reminiscent of maternal mRNA.
-
EPCA of transcriptomes of the indicated embryos. Blastocysts similar in transcriptomes are encircled.
-
FHeat map of differentially expressed genes.
-
GHeat map of representative lineage‐specific transcripts. TE, trophectoderm; EPI, epiblast; PrE, primitive endoderm.
-
H, ISevere down‐regulation of Gata6 and Nanog in Rbm14‐depleted mouse blastocysts. Representative immunofluorescent images (H) were from single‐option sections. DAPI stained nuclear DNA. As Gata6, Nanog, and Rbm14 localize to the nucleus in blastocysts, their relative intensity in a cell (I) was quantified as their nuclear immunofluorescent intensity normalized with fluorescent intensity of the nuclear DNA. Five brightest cells were quantified for each embryo.
Data information: Quantification results in (D) and (I) are presented as mean ± SD with sample dots. Unpaired two‐sided Student's t‐test: ns, no significance; **P < 0.01; ***P < 0.001.
Source data are available online for this figure.
Rbm14 deficiency results in blastocyst arrest and early embryonic lethality due to defective MZT
We generated Rbm14‐deficient mice through a Cre/loxP gene knockout strategy (Fig EV5A). When Rbm14 +/− mice were mated, no Rbm14 −/− neonatal mice were identified from over 400 offspring, despite normal Mendelian ratios of Rbm14 +/+ and Rbm14 +/− littermates (Fig 8C). We further examined embryos of different stages and found that genotyping by PCR (Fig EV5A) failed to identify any Rbm14 −/− embryos at E8.5 or later (Fig 8C). Approximately one‐third of yolk sacs at E8.5 contained only empty deciduae, deciduae without embryos or with only remnants of resorbing embryos (Fig EV5B). Nevertheless, immunostaining identified 20 Rbm14‐negative embryos out of 65 E4.5 blastocysts (Fig 8C). Therefore, Rbm14‐deficient mice also fail to develop into the gastrula stage, even though their maternal transcripts are intact.
Figure EV5. Generation of Rbm14‐deficient mice (related to Fig 8).
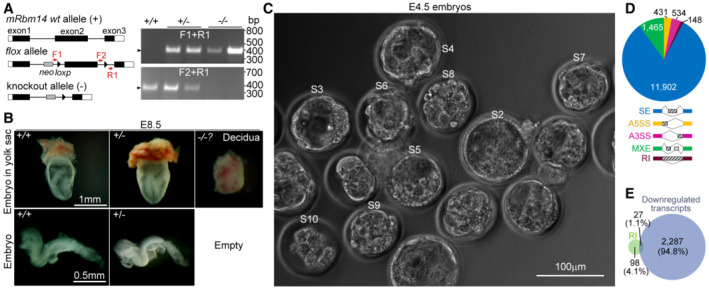
-
AGene knock‐out strategy and representative genotyping PCR results. The exon 2 of Rbm14 was knocked out by crossing Rbm14flox/flox mice with EIIa‐Cre mice. Positions of two forward primers (F1 and F2) and the reverse primer (R1) for genotyping are indicated. Neo, neomycin resistance gene. The PCRs were performed using genomic DNAs from E8.5 embryos or remnants of resorbing Rbm14−/− embryos.
-
BMorphologies of mouse E8.5 embryos. No Rbm14−/− embryos were identified except for empty deciduae.
-
CE4.5 embryos used for RNA deep sequencing. Zygotic embryos were collected from the oviduct of superovulated Rbm14+/− female mice after mating with Rbm14+/− male mice and cultured to E4.5. Nine viable embryos (designated S2–S10) were picked as specimens for RNA extraction and deep sequencing.
-
DDifferential alternative splicing events in Rbm14−/− embryos. SE, skipped exon; A3SS, alternative 3′ splicing site; A5SS, alternative 5′ splicing site; MXE, mutually exclusive exon; RI, retained intron.
-
EVenn diagrams between the genes showing differential RI and genes downregulated in the Rbm14−/− embryos (Fig 8F).
Source data are available online for this figure.
We cultured E1.5 embryos into E4.5 blastocysts in vitro (Wang et al, 2017) and picked nine (Fig EV5C) for RNA deep‐sequencing. Five of them displaying low levels of Rbm14 transcripts and clustered closely in 3D‐PCA graphs were considered as Rbm14 −/− embryos, whereas the remaining three with at least 8‐fold higher levels of the transcripts as Rbm14 +/+ or Rbm14 +/− embryos (Fig 8D and E). The heatmap of 2,315 differentially expressed genes indicated that, comparing to the wild‐type or heterozygous embryos, most genes were strikingly downregulated in the Rbm14 −/− embryos (Fig 8F). To assess the contribution of splicing defects‐induced mRNA degradation (Ge & Porse, 2014) to the global downregulation of zygotic transcripts, we analyzed differential alternative splicing events between the two groups of embryos and identified only 148 differential RI events in the Rbm14 −/− embryos (Fig EV5D). Moreover, the 125 genes showing RI only slightly overlapped with the 2,314 genes down‐regulated in the Rbm14 −/− embryos (Fig EV5E). We thus conclude that the depletion of Rbm14 in mouse embryos also impairs MZT.
Blastocysts contain the first three cell lineages generated from totipotent blastomeres: trophectoderm (TE), epiblast (EPI), and primitive endoderm (PrE; Takaoka & Hamada, 2012; Rivera‐Perez & Hadjantonakis, 2014). Representative markers of the three lineages were mostly downregulated as well (Fig 8G). We examined Gata6 and Nanog, two typical markers respectively for PrE and EPI cells in the inner cell mass (ICM), through immunofluorescent staining and confirmed their low expression levels in E4.5 blastocysts negative for Rbm14 (Fig 8H and I). We further examined Oct4, the ICM marker, and found its existence in 100% of Rbm14‐negative blastocysts (Fig 8H and I), indicating its normal expression in morula embryos. Thus, Rbm14 deficiency impairs the differentiation of TE, EPI, and PrE, explaining the blastocyst arrest of Rbm14‐deficient embryos.
Discussion
We demonstrate that Rbm14 is critical for vertebrate blastula‐to‐gastrula development by regulating mRNA metabolism (Figs 4, 6 and 8). Our results suggest that, in zebrafish early embryos, zRbm14 phase separates into cytoplasmic condensates to actively deadenylate maternal mRNAs and protect them from premature enzymatic decay (Fig 7M). Maternal mRNAs (Fig 1A; Aanes et al, 2014; Winata et al, 2018; Vastenhouw et al, 2019), especially transcripts with m6A modifications, which maximize at around 2 hpf (Zhao et al, 2017), interact with the RNA‐binding region of zRbm14 to promote the condensate formation through co‐phase separation, whereas m5C modifications, which peak at around 3 hpf (Yang et al, 2019; Liu et al, 2022), negatively regulate the condensate formation (Figs 2 and 3). Deadenylase zParn promotes the condensate formation through co‐phase separation as well by interacting with zIDR (Fig 7D–F). Furthermore, zParn massively deadenylates maternal mRNAs in the condensates from the 4‐ to 32‐cell stages (Figs 7 and EV4) to probably avoid deregulated protein synthesis. Deadenylated maternal mRNAs are sequestered in the condensates from premature degradation, probably by excluding decapping and degradation enzymes (Garneau et al, 2007; Despic & Neugebauer, 2018) in a way analogous to the sequestration against the anti‐m6A antibody (Fig 2). The gradual disassembly of the condensates following the embryonic development (Fig 1D and F) releases poly(A)‐less maternal mRNAs stepwise for cytoplasmic polyadenylation enzymes‐induced re‐activation or degradation enzymes‐induced clearance (Weill et al, 2012; Despic & Neugebauer, 2018; Winata et al, 2018; Vastenhouw et al, 2019), depending on genes and developmental stages (Figs 6A–G, 7A–C, and EV4A and B). For instance, among multiple cyclin mRNAs whose timely clearances required zRbm14 (Fig EV3F), we confirmed that ccnb1 mRNA is sequestered in zRbm14 condensates (Fig 2E), and ccnb1 mRNA has been shown to depend on cytoplasmic polyadenylation for re‐activation in Xenopus early embryos (Groisman et al, 2000; Kim & Richter, 2006). Interestingly, zebrafish embryos lacking cytoplasmic polyadenylation are also arrested in blastula (Winata et al, 2018), similar to maternal zRbm14 morphants (Fig 4), indicating the requirement of both the zRbm14 condensate‐mediated deadenylation pathway and the cytoplasmic polyadenylation pathway for the blastula‐to‐gastrula development.
It should be noted that maternal mRNAs accumulated in maternal zRbm14 morphants (Fig 6E) may not all be the direct targets of zRbm14. Some of them may only be due to the developmental delay of the morphants (Fig 4). Furthermore, as thousands of alternative splicing events occurred in the morphants (Fig EV3C), splicing defects of zygotic transcripts encoding proteins involved in the zygotic program of maternal mRNA clearance (Lee et al, 2013; Despic & Neugebauer, 2018) might also contribute to the accumulation of some maternal mRNAs. Future studies will thus be required to further discriminate bona fide target maternal mRNAs of zRbm14 and understand how the regulations on them impact early embryogenesis.
Rbm14 appears to be a unique m6A reader that uses m6A modifications to strengthen its LLPS. Unlike YTHDF proteins, whose LLPS abilities are only enhanced by multiple m6A sites (Gao et al, 2019; Ries et al, 2019), LLPS of zRbm14 was promoted by a single m6A modification and further by multiple m6A modifications (Figs 3A–C and EV1D). zRbm14 also mainly regulated different sets of m6A‐modified maternal mRNAs as zYthdf2 does (Fig EV3B; Zhao et al, 2017). Furthermore, unlike zRbm14, Ythdf2 appears to use the CCR4‐NOT complex to deadenylate its target mRNAs (Du et al, 2016). As maternal zYthdf2 −/− zebrafish embryos display only a mild gastrulation delay and can develop into the segmentation period (Zhao et al, 2017), zRbm14 appears to be more important in early embryogenesis. Interestingly, RNA‐recognition motif (RRM)‐containing proteins hnRNPA2B1, hnRNPC, and hnRNPG also preferentially bind to m6A‐modified RNAs (Liu et al, 2015, 2017; Wu et al, 2018; Zaccara et al, 2019). Among them, only hnRNPA2B1 is documented to undergo LLPS (Wang et al, 2018). Furthermore, m6A modifications are shown to enhance the accessibility of RNA to hnRNPC and hnRNPG by inducing RNA conformational changes, or structural switches (Liu et al, 2015, 2017). A similar mechanism might also underly the effect of m6A modifications on the LLPS of zRbm14. In this context, the m6C modification might reverse the process to make RNAs less accessible to zRbm14, therefore impairing multivalent interactions among zRbm14b and RNA molecules to repress their co‐phase separation (Fig 3D–F). Future studies, however, are required to clarify these issues.
zRbm14 condensates also serve as carriers to distribute their content maternal mRNAs in mitosis by associating with centrosomal γ‐tubulin puncta (Figs 1, 2, and 7M). In transcription‐silenced blastomeres, maternal transcriptomes are the major cell fate determinants. As maternal transcriptomes are hierarchically aliquoted into blastomeres by serial embryonic cleavages, progeny blastomeres inherited with equal or different aliquots of the transcriptome would theoretically assume distinct developmental potentials. Indeed, different maternal mRNAs have been documented to associate with centrosomes in different cells of early mollusk embryos and divide asymmetrically into daughter cells (Lambert & Nagy, 2002). In addition to zebrafish (Fig 2), spindle pole‐associated maternal transcripts are also reported in early embryos of Drosophila and Xenopus (Groisman et al, 2000; Lecuyer et al, 2007). We thus speculate that the polarized asymmetric segregation of zRbm14 condensates in 8‐ and 16‐cell embryos (Fig 1I–K; Rathbun et al, 2020) might accordingly enable daughter blastomeres close to the midline to acquire more copies of the condensates‐associated maternal transcripts, forming a bilateral mRNA gradient. Subsequent asymmetric and symmetric segregations of zRbm14 condensates, as exampled in 64‐cell embryos (Fig 1I–K), might further increase the difference in maternal mRNA contents among blastomeres to assume different fates. Therefore, we propose that the deadenylation, sequestration, mitotic allocation, and stepwise release of maternal mRNAs by zRbm14 condensates collectively facilitate cell cycle, differentiation, and MZT of zebrafish embryos (Fig 7M). Details, however, still await future investigations. How zRbm14 condensates achieve their tight associations with γ‐tubulin puncta (Fig 1) also remains to be clarified.
Our investigations in mouse suggest that some maternal m6A‐mRNAs have evolutionarily‐conserved essential functions in the blastula‐to‐gastrula development. As zRbm14 requires both the RNA‐binding region and the IDR for its embryonic functions (Fig 5), it would be reasonable to assume that the mouse condensates also enrich, deadenylate, sequester, and allocate maternal mRNAs. Consistently, although the mouse condensates are dispersed in interphase cells due to the lack of the centrosome in early mouse embryos till gastrula (Howe & FitzHarris, 2013), they are highly concentrated at spindle poles (Fig 8B and Movie EV4). Therefore, the persistence of some Rbm14 condensates to the blastula (zebrafish) or morula (mouse) stage (Figs 1D and 8B) suggests their persistent protection of a small fraction of deadenylated mRNAs. Furthermore, the common blastula arrest and early differentiation defects of mouse Rbm14‐deficient embryos and maternal zRbm14 morphants (Figs 4, 8, and EV3G) can be attributed to the loss of functions of these long‐life maternal mRNAs. Firstly, as most murine maternal mRNAs are eliminated by the 2‐cell stage (Fig 8A; Jukam et al, 2017; Eckersley‐Maslin et al, 2018; Vastenhouw et al, 2019) and mouse Rbm14‐deficient oocytes contain intact maternal Rbm14 transcripts, Rbm14 condensates in 1‐ and 2‐cell Rbm14‐deficient embryos (Fig 8B) should be largely normal and capable of deadenylate their content maternal mRNAs. Accordingly, Rbm14 deficiency is expected to cause premature elimination, rather than stabilization, of the long‐life maternal mRNAs. Secondly, although target maternal mRNAs of zRbm14 condensates are stabilized in zebrafish maternal zRbm14 morphants (Fig 6), the zRbm14 condensates‐mediated mitotic segregation mechanism (Figs 1 and 2) is completely lost. Therefore, the blastula arrest and differentiation defects in zebrafish (Figs 4 and EV3G) can still be attributed to the lack of functions of the long‐life maternal mRNAs. It will thus be interesting to identify them and understand how they regulate full ZGA and the blastula development (Figs 4, 6, and 8) in future researches.
Our findings help to understand how phase separation‐induced condensates orchestrate spatiotemporally with machineries of deadenylation, m6A and m5C epigenetic modifications, and cytoplasmic polyadenylation to elegantly regulate maternal mRNA metabolisms and functions in early embryogenesis. It will be important to identify additional components of the Rbm14‐maternal mRNA condensates and elaborate their functions and interplays with other pathways. Notably, in Xenopus oocytes, Parn‐mediated maternal mRNA deadenylation is coupled to cytoplasmic polyadenylation machinery (Korner et al, 1998; Kim & Richter, 2006; Moraes et al, 2006; Weill et al, 2012). Multiple components of the machinery are concentrated on spindle poles as well in Xenopus early embryos (Groisman et al, 2000). In addition, Rbm14 is identified as a candidate spindle matrix protein from Xenopus egg extracts (Ma et al, 2009; Zheng, 2010) and can also co‐phase separate with the essential spindle matrix protein BuGZ (Jiang et al, 2015; Xiao et al, 2019). It will thus also be interesting to clarify whether the mitotic allocation of Rbm14 condensates (Figs 1, 2 and 8B) is related to the spindle matrix.
Materials and Methods
Plasmids
For antibody production, the plasmid encoding GST‐tagged full‐length zebrafish rbm14a (GenBank accession NM_001115144) was from our previous work (Xiao et al, 2019).
For rescue experiments in zebrafish, the plasmids encoding EGFP cDNA, GFP‐tagged full‐length zRbm14b (NM_212808), zRbm14b18S (18 Y‐to‐S mutations including Y260S, Y269S, Y290S, Y298S, Y316S, Y321S, Y350S, Y357S, Y363S, Y382S, Y425S, Y454S, Y455S, Y458S, Y459S, Y462S, Y468S, and Y471S), zIDR (nucleotides 766–1668), zIDR18S, zRRM (nucleotides 1–765), xBuGZΔN (Jiang et al, 2015), and the IDRs of zFus (NM_201083; nucleotides 1–873 starting from the initiation codon), zEwsr1b (NM_212630; nucleotides 1–771), and zTaf15 (NM_001079973; nucleotides 1–390) were described previously (Xiao et al, 2019).
For co‐IP, the GFP‐tagged full‐length zRbm14 plasmid was from our published work (Xiao et al, 2019). The RFP cDNA was amplified from pmRFP‐C1 by PCR and cloned into pCS2 between BamHI and XhoI sites to form pCS2‐RFP. The luciferase cDNA was amplified from pcDNA3‐FLAG‐Luciferase; the full‐length zCnot1 (NM_001079951), zCnot1‐N (nucleotides 1–3150), zCont1‐M (nucleotides 3151–5520), zCnot1‐C (nucleotides 5521–7122), full‐length zCnot6a (NM_212660), zPan2 (XM_001919965), zPan3 (NM_001105692), zParn (NM_201088), zDcp1a (NM_182893), and zDcp2 (NM_200152) were amplified by PCR from zebrafish embryo cDNA, then inserted into pCS2‐RFP at the BamHI site.
To express His‐tagged proteins in E. coli, plasmids encoding His‐GFP, His‐GFP‐zRbm14b, His‐GFP‐zIDR, His‐GFP‐zIDR18S were from our published work (Xiao et al, 2019). To generate His‐RFP, the RFP cDNA was amplified from pmRFP‐C1 by PCR and cloned into pET28a between the BamHI and NotI sites to form pET28a‐RFP. The cDNAs coding for full‐length zParn were amplified by PCR and inserted at the BamHI site of pET28a‐RFP. Purified His‐zParn‐RFP was used as antigen to generate rabbit anti‐zParn antibody.
For in situ hybridization, full‐length zebrafish org (NM_001100070), trip10a (NM_001007403), and dnajc5ga (NM_199623) were amplified from zebrafish embryo cDNA and inserted into pcDNA3 between BamHI and EcoRI sites. Full‐length zebrafish ccnb1 (NM_131513) was amplified from zebrafish embryo cDNA and inserted into pcDNA3 between BamHI and EcoRI sites.
To verify 14b‐tMO depletion efficiency, zebrafish rbm14b‐200 nt (NM_212808, nucleotides 1–200 of mRNA N‐terminus) was amplified from zebrafish embryo cDNA, GFP was amplified from pEGFP‐C1, then inserted into pCS2 between EcoRI and XhoI sites in‐frame.
All the plasmids were subjected to sequencing confirmation. Detailed information, including the sequences of PCR primers, is listed in Appendix Table S1.
Cell culture and transfection
The human HEK293T cell line was maintained in DMEM (Life Technologies) with 10% fetal bovine serum (FBS), 2 mM L‐glutamine at 37°C in a water‐saturated atmosphere of 5% CO2. Cultured cells were routinely tested for mycoplasma contamination. To express exogenous proteins, HEK293T cells were transfected using polyethylenimine (PEI, Polysciences 23966‐2) as described (Duan et al, 2021).
Animal use and care
All animal procedures were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China).
Mouse husbandry, pre‐implantation embryos culture, and genotyping
For wild‐type mouse embryo collection during MZT, C57BL/6 female mice were superovulated by injecting 5–7 IU of pregnant mare serum gonadotropin (PMSG), followed by 5–7 IU of human chorionic gonadotropin (hCG) 48 h later. Superovulated female mice were mated to male mice. Zygotes were collected from the oviduct through a brief treatment with hyaluronidase to wash clean of cumulus cells and then were cultured in KSOM medium at 37°C under 5% CO2 in the air to E4.5 as described (Wang et al, 2017).
Rbm14 flox/lox mice were generated by the West China Center of Medical Sciences, Sichuan University (Chengdu, China). Rbm14 −/− mice were generated by crossing Rbm14 flox/lox mice with EIIa‐Cre mice (Lakso et al, 1996), followed by inbreeding. For genotyping, genomic DNA was extracted from embryos, embryo remnants, or postnatal mouse tails at indicated stages. In brief, the DNA was amplified by denaturation at 95°C for 5 min, followed by 30 s at 95°C, 30 s at 55°C, and 30 s at 72°C for 30 cycles and a final 10‐min incubation at 72°C, using the primers for genotyping listed in Appendix Table S1.
Zebrafish embryo maintenance, microinjection, and drug treatment
Zebrafish embryos in the AB background were maintained at 28.5°C and staged as described (Kimmel et al, 1995). MOs (Gene Tools; sequences are listed in Appendix Table S1) were dissolved in nuclease‐free water and injected at 2 nl (containing 8 ng MO) per embryo at the one‐cell stage using a Narishige IM300 micro‐injector. One‐hundred picograms of GFP mRNA, in vitro‐transcribed from pCS2‐GFP, were co‐injected as an injection marker. In rescue experiments, 2 nl of MOs solution containing 800 pg of in vitro‐transcribed mRNA were injected into 1‐cell stage embryos. For dose expression of zRbm14b, 400–1,200 pg of in vitro‐transcribed GFP‐zRbm14b mRNA were injected at the 1‐cell stage.
GFP‐positive embryos were collected at 2.5 hpf for further investigation. Embryos were photographed under an Olympus SZX16 stereo microscope or ZEISS Axio Zoom.V16 Microscope with a SPOT Insight digital camera.
Zebrafish embryos were treated with several drugs (details are listed in Appendix Table S1), basically followed as described (Chan et al, 2019). For nascent RNA labeling, 5‐Ethynyl Uridine (EU, Click chemistry tools 1261) was diluted in the nuclease‐free water and injected into 1‐cell stage embryos. Embryos were collected and fixed at 2.5, 4, and 6 hpf. EU signals were revealed with a click reaction kit (Beyotime, C0081S) to be labeled with Alexa Fluor 647. To disrupt LLPS during MZT, 1,6‐Hexanediol (Sangon Biotech, A601513; Kroschwald et al, 2017) was added in zebrafish culturing medium from 2.5 hpf with a working concentration range from 0.3 to 1.2%.
In vitro transcription for zebrafish microinjection
In vitro transcription was performed using linearized plasmids and mMESSAGE mMACHINE Kit (Ambion, AM1340). Transcribed mRNAs were purified using MEGAclear™ Purification Kit (Ambion, AM1908) and dissolved in RNase‐free water. mRNA concentrations were quantified by using a NanoDrop 2000 spectrophotometer (Thermo Fisher). Restriction enzymes used to linearize the plasmids and RNA polymerases used for mRNA syntheses are listed in Appendix Table S1.
Co‐immunoprecipitation
For co‐IP, HEK293T cells were harvested 48 h after transfection with ice‐cold lysis buffer [20 mM Tris–HCl, 100 mM KCl, 0.5% NP‐40, 1 mM EDTA, 10% glycerol, 50 mM NaF, 10 mM sodium pyrophosphate, 1 mM Na3VO4, 1 mM PMSF, and Phosphatase Inhibitor Cocktail (Sigma, p0044), pH 7.4], followed by sonication on ice (10–15 cycles with 3‐s pulse‐on and 7‐s pulse‐off, 30% amplitude). After centrifugation at 16,363 g for 10 min, the supernatant was incubated with anti‐GFP beads (ChromoTek, gta‐20) for 2 h at 4°C under rotary agitation. The mixture was washed three times with lysis buffer, three times with wash buffer (20 mM Tris–HCl, 150 mM KCl, 0.5% NP‐40, 1 mM EDTA, 10% glycerol, 50 mM NaF, 10 mM sodium pyrophosphate, pH 7.4), and then boiled in 2 × SDS loading buffer (100 mM Tris–HCl, 4% SDS, 2% glycerol, bromophenol blue, pH 6.8).
Immunoblotting
To prepare zebrafish embryo lysates, embryos were collected at indicated stages, deyolked as described (Link et al, 2006), then washed in phosphate‐buffered saline (PBS) once, lysed with 2 × SDS loading buffer. Zebrafish embryo lysates were resolved on 10% SDS–PAGE gels. The immunoprecipitates were resolved on 3–12% SDS–PAGE gels.
Proteins separated by SDS–PAGE were transferred to nitrocellulose membranes (General Electric Company). Blots were pre‐blocked with 5% non‐fat milk diluted in TBST (50 mM Tris–HCl, 150 mM NaCl, 0.05% Tween‐20, pH 7.5) for 1 h and then incubated with primary antibodies. After thorough washing with TBST, membranes were incubated with secondary IgG‐HRP antibodies. After extensive washes in TBST, protein bands were visualized with enhanced chemiluminescent reagent (PerkinElmer) and exposed to Mini Chemiluminescent Imager (MiniChemi 610 Plus, Beijing Sage Creation Science Co).
The primary and secondary antibodies are listed in Appendix Table S1.
Immunofluorescence of embryos
For whole embryo immuno‐staining, zebrafish embryos were fixed at indicated stages with 4% fresh PFA in PBS at 4°C overnight. After fixation, embryos were dehydrated in MeOH/PBS graded series (from 25, 50, 75 to 100%), then were stored in 100% MeOH at −20°C overnight. On the next day, embryos were rehydrated in MeOH/PBS graded series (from 75, 50, 25 to 0%) and were blocked with blocking buffer (5% goat serum, 1% BSA, 1% DMSO, 0.5% Triton X‐100 in PBS) for 1 h at R.T. Embryos were incubated with primary antibodies in blocking buffer overnight at 4°C. After three times of wash (PBS with 0.5% Triton X‐100), the incubation with secondary antibodies was carried out in blocking buffer overnight at 4°C. DNA was stained with DAPI (1 μg/ml) with the secondary antibody incubation. Followed by three times of wash (PBS with 0.5% Triton X‐100), embryos were placed on 29 mm glass‐bottomed dishes (Cellvis 191207) and embedded in 1% agarose, low gelling temperature (Sigma, A9414) with the animal pole facing the glass bottom.
To stain RNA in zebrafish, all buffers were RNase‐free. Embryos were co‐stained with Pyronin‐Y (MedChem Express, HY‐D0971) and Hoechst 33324 (Life Technologies, H3570) in PBS with 0.5% Triton X‐100 for 1 h at 4°C, basically as described (Darzynkiewicz et al, 2004). To diminish the RNA signal, RNase A (Life Technologies 12091021) was added to the mixture for 2 h at 37°C after fixation and permeabilizing. The RNase A‐digested embryos and mock‐treated embryos were washed three times by PBS, then followed with the same procedure of Pyronin‐Y and Hoechst 33342 staining.
For proteinase K treatment, rehydrated embryos were incubated with proteinase K (Sigma, P6556) in PBS at 37°C for indicated times. After 3‐time washes with PBS, embryos were re‐fixed with 4% PFA at R.T. for 20 min to eliminate the proteinase activity.
Mouse embryos were fixed and permeabilized synchronously with 1% PFA and 0.2% Triton X‐100 in PBS at R.T. for 1 h, then were blocked with 1% BSA in PBS for 1 h. The primary antibody was incubated (in 1% BSA in PBS) overnight at 4°C. After three times of wash (PBS with 0.5% Triton X‐100), the secondary antibody was carried out (in 1% BSA in PBS) at R.T. for 1 h. DNA was stained with DAPI (1 μg/ml) at R.T. for 1 h. Followed by three times of wash (PBS with 0.5% Triton X‐100), mouse embryos were mounted with ProLong Diamond anti‐fade mounting medium.
Confocal microscopy images were captured on a Leica TCS SP8 WLL system with a 63×/1.40 or 40×/1.30 Oil objective and processed with maximum intensity projections. 3D images were re‐constructed by Imaris (version 7.6.5).
The details of dyes, drugs, primary and secondary antibodies are listed in Appendix Table S1.
Whole‐mount in situ hybridization
Plasmids harboring the cDNAs for zebrafish org, trip10a, dnaj5ga, and ccnb1 were linearized as indicated in Appendix Table S1. Digoxigenin‐UTP‐labeled antisense RNA probes were generated by in vitro transcription using DIG RNA Labeling Kit (Roche 11175025910). Whole‐mount in situ hybridization was carried out as previously described (Cao et al, 2012; Xiao et al, 2019). Zebrafish embryos were then immersed in glycerol and photographed under an Olympus SZX16 stereo microscope.
For co‐staining of ccnb1 mRNA with γ‐tubulin, the in situ hybridization was performed firstly, basically as described (Welten et al, 2006). The primary antibody anti‐DIG‐POD was co‐incubated with the anti‐γ‐tubulin antibody. The fluorescence signals of ccnb1 mRNA were visualized with a TSA Signal Amplification kit (PerkinElmer, NEL744) with 1:100 dilution at R.T. for 1 h. After three times of wash, the samples were incubated with the secondary antibody and DAPI (to stain nuclear DNA or chromosomes) following the immunostaining procedure.
Fluorescent live imaging
Zebrafish embryos were injected at 1‐cell stage with 800 pg in vitro‐transcribed mRNA of GFP‐zRbm14b, then were cultured to indicated stages. Before imaging, the DNA of the live embryo was stained by NucBlue™ Live ReadyProbes™ Reagent Kit (Life Technologies, R37605) according to the instruction for 30 min under culturing conditions.
For time‐lapse studies of GFP‐zRbm14b localization during MZT, live embryos at 2 hpf were embedded in 2% agarose, low gelling temperature, then sucked into a capillary glass tube (Brand 701904), surrounded with culturing medium. Images were acquired by a light‐sheet microscope (LiTone LBS, Light Innovation Technology Ltd.) with a 25×/1.1 Water objective (Nikon, CFI75) at 28.5°C for 2 h. With a 121 s‐interval and a 1 μm z‐step, images of 500 μm depth were deconvoluted and z‐projected with maximum intensity by the microscope software (LitScan 2.0).
To capture droplet fusion events, decorticated live zebrafish embryos at 4 hpf were placed on glass‐bottomed dishes, embedded in 1% agarose, low gelling temperature, then covered with culture medium. Live images were taken with a spinning disk confocal microscope (Nikon CSU W1 Sora on Ti2E) with a 60×/1.40 Oil objective at 28.5°C. Images were captured every 3.5 s with a 1 μm Z‐step. Images of 7 μm depth were deconvoluted and z‐projected with maximum intensity with Nikon software (NIS‐Elements 5.21).
Protein purification
For protein purification, proteins of His‐tagged GFP, GFP‐zRbm14b, GFP‐zIDR, GFP‐zIDR18S; His‐tagged RFP, zParn‐RFP, zPabpc1a‐RFP were expressed in the E. coli BL21‐CondonPlus (DE3) strain after 1 mM IPTG induction for 12–16 h at 16°C. Proteins were purified using Ni‐NTA resin (Qiagen) as we described previously (Xiao et al, 2019). The bound proteins were eluted using 500 mM imidazole in NTA buffer and concentrated to 1–20 mg/ml in DEPC‐treated PBS. Purified proteins were aliquoted, snap‐frozen in liquid nitrogen, and stored at −80°C.
Methylated RNAs preparation and fluorescence labeling
To test the role of m5C methylation‐modified RNAs on Rbm14 LLPS, the 59‐nt RNA from cap1 mRNA either containing one m5C modification (cap1‐m 5 C) or no m5C methylation (cap1‐C) were kind gifts from Dr. Yungui Yang (Yang et al, 2019). Detailed sequences were listed in Appendix Table S1. The Alexa Fluro 546 dyes were in vitro‐labeled to these oligos using the ULYSIS Nucleic Acid Labeling kit (Life Technologies, U21652).
To test the role of m6A methylated RNAs on Rbm14 LLPS, three 63 nt RNA oligos of identical sequence from Dr. Samie R. Jaffrey's published work were used with a similar labeling method (Ries et al, 2019). RNA sequences were shown in Appendix Table S1. In brief, three pairs of DNA oligos containing the T7 promoter sequence and either 1 A or 10 A nucleotides were synthesized at TSINGKE Biological Technology (Nanjing, China) and formed double‐stranded DNA templates through denatured at 95°C for 5 min then annealed at R.T. for 1 h. RNAs were in vitro‐transcribed and fluorescence‐labeled following the manufacturer's instruction of the HighYield T7 Cy3 RNA Labeling Kit (Jena Bioscience, RNT‐101‐CY3). To generate 1 or 10 m6A‐modified versions of RNAs, the ATP was replaced by m6‐ATP (Jena Bioscience, NU‐1101 S) by using the in vitro transcription kit. After transcription, DNA templates were digested using 1 μl Turbo™ DNase (Life Technologies) for 15 min at 37°C.
All the labeled RNAs were purified with the NucAway Spin Columns (Life Technologies, AM10070) from the labeling reagent into the Nuclease‐free Water and quantified by a Nanodrop2000 spectrophotometer.
The integrity and fluorescence labeling efficiency of transcribed RNAs were confirmed with 15% TBE‐urea denaturing gel electrophoresis and fluorophore excitation by a cy3 laser (Fujifilm, FLA‐9000 scanner) and a UV light after Ethidium Bromide (EB) stained. An RNA ladder (NEB, N0364S) was loaded as a reference.
The m6A modification level was confirmed by dot blotting assay with an m6A antibody (Appendix Table S1).
Phase separation assay
Purified proteins and RNAs (with fluorescence or not) were thawed on ice and diluted into ice‐cold PBS buffer. In total, 5 μl of the protein–protein mixtures (10 + 10 μM) or protein–RNA mixtures (10 + 1 μM) were loaded into a flow chamber, consisting of a coverslip on top of a glass slide, separated slightly with two pieces of double‐sided adhesive tape as described (Jiang et al, 2015; Xiao et al, 2019). Followed by incubation at 25°C for the indicated time, samples were imaged immediately. Fluorescence images were obtained by an Olympus BX51 fluorescence microscope with a 60×/1.25 Oil objective. Differential interference contrast (DIC) images were captured by an Olympus BX71 fluorescence microscope with a 60×/1.42 Oil objective.
Partition coefficient (PC) measurement was performed as described (Ries et al, 2019). For partition coefficient experiments of protein‐RNA mixtures, cy3‐labeled 10 m6A‐RNA (1 μM) were mixed with His‐GFP‐zRbm14b proteins (10 μM) in DEPC‐treated PBS buffer at 25°C. For partition coefficients of fluorescent protein–protein droplets, His‐GFP‐zRbm14b (10 μM) were mixed with His‐zParn‐RFP (10 μM) in PBS buffer at 25°C. Fluorescence images were captured at 2.5 or 30 min, respectively. Partition coefficients of His‐GFP‐zRbm14b were calculated by measuring the ratio of zRbm14b fluorescence intensity in droplet over its intensity located in the adjacent solution. Partition coefficients of His‐zParn‐RFP or 10 m6A‐RNA‐cy3 were calculated according to their fluorescence intensity in droplets over the adjacent solution, respectively.
In vitro deadenylation assay
For the generation of 10 m6A‐modified mRNA substrates (An), the capped 63 nt 10 m6A‐RNAs were in vitro transcribed by the m6A RNA in vitro transcription kit (Jena Bioscience, NU‐1101S) with the m7G(5′)ppp(5′)G Cap Analog (Ambion, AM8048) addition. The capped 10 m6A‐RNA was purified from free m6‐ATP into Nuclease‐free water by sodium acetate (pH 5.2) and ethanol precipitation. The poly(A) tails were added by Yeast Poly(A) Polymerase Tailing Kit (Jena Bioscience, RNT‐004). The m6A‐modified mRNA substrates were purified by sodium acetate (pH 5.2) and ethanol precipitation then were dissolved in Nuclease‐free water.
For in vitro deadenylation assay, 5 μM of purified His‐zParn‐RFP protein, 5 μM His‐GFP‐zRbm14b protein, and 1 μg 10 m6A‐modified mRNA substrates were incubated at 30°C in a 50 μl volume reaction containing 20 mM HEPES (pH 6.8), 1 mM magnesium acetate, 2 mM spermidine (MCE, HY‐B1776), 1 mM DTT, 1 U/μl RNase inhibitor (Life Technologies 10777‐019), basically followed as described (Korner & Wahle, 1997). At indicated time points, 2% SDS was added into the aliquot to disrupt LLPS. RNA was extracted from the reaction mixtures using TRIzol Reagent (Life Technologies), followed by immuno‐northern blotting with an m6A antibody (Appendix Table S1) basically as described (Mishima & Abe, 2019). The deadenylation level was quantified by measuring the signal intensity ratio of A0/(A0 + An) from five independent experiments.
Zebrafish mRNA deep sequencing and data analysis
Total RNAs of pooled 30 embryos were extracted using TRIzol Reagent (Life Technologies). The quality of RNAs, library construction, and sequencing were all performed by GENEWIZ (Suzhou, China). Briefly, RNAs were quantified and qualified by Agilent 2100 Bioanalyzer (Agilent Technologies, USA), NanoDrop, and 1% agarose gel. In total, 1 μg total RNA with a RIN value above 6.5 was used for the following library preparation. Next‐generation sequencing library preparations were constructed according to the manufacturer's protocol (NEBNext® UltraTM RNA Library Prep Kit for Illumina®). Then libraries with different indices were multiplexed and loaded on an Illumina HiSeq instrument according to the manufacturer's instructions (Illumina, USA). Sequencing was carried out using a 2 × 150 bp paired‐end (PE) configuration; image analysis and base calling were conducted by the HiSeq Control Software (HCS) + OLB + GAPipeline‐1.6 (Illumina) on the HiSeq instrument.
For data analysis, reference genome sequences and gene annotation files were downloaded from ENSEMBL (Danio_rerio.GRCz10.86). Hisat2 (v2.1.0; Kim et al, 2015) was used to index reference genome sequence. Raw data were pre‐processed by Fastp (v0.12.1; Chen et al, 2018) with default parameters. Cleaned data were then aligned to the reference genome via software Hisat2 (v2.1.0). FeatureCounts (v1.5.3; Liao et al, 2014) was used to count the reads numbers mapped to each gene.
We performed the cumulative distribution to display the mRNA changes of control and maternal zRbm14 morphants over time as described (Yang et al, 2019).The Wilcoxon and Mann–Whitney tests were used to calculate P values for the fold changes (log2) at different time points. The fold changes (log2) were calculated by edgeR (3.28.0; Robinson et al, 2010). To classify the gene expression patterns during MZT, we used Short Time‐series Expression Miner software (STEM, v1.3.13; Ernst & Bar‐Joseph, 2006) with parameters as follows: maximum unit change in model profiles between time points is 1; profiles number < 20; fold change of DEGs > 2. Eight profiles were categorized, only four of them were significant. According to their patterns during MZT, the four profiles were respectively named maternal early (degradation from 2.5 hpf), maternal late (degradation after 4 hpf), zygotic early (onset at 2.5 hpf), and zygotic late (onset at 4 hpf). The cumulative distribution and Wilcoxon and Mann–Whitney test were applied to display the difference between maternal and zygotic clustered transcripts. The PCA and Spearman's correlation coefficient of biological replicates were performed with R packages. Transcripts with absolute fold change > 2 between samples at each stage were considered as altered transcripts. Gene ontology (GO) enrichment analysis of genes was carried out by using Metascape (https://metascape.org/) with default parameters (Zhou et al, 2019). GO terms with P value < 0.05 were considered significantly enriched. To compare the upregulated mRNA transcripts in zYthdf2 −/− embryos and maternal zRbm14 morphants, transcripts changed above 2‐fold were selected by comparing zebrafish zYthdf2 −/− embryos with WT embryos from published data (Zhao et al, 2017). For zRbm14, a threshold was set with maxTPM > 1 among three time points (2.5, 4, and 6 hpf) to exclude low‐expressed genes. Then, genes above 2‐fold change in maternal zRbm14 morphants comparing with control morphants were selected as zRbm14 targets. The Venn diagram was generated with the BioVenn web tool.
Poly(a) tail‐length assay
Total RNAs were extracted from equal numbers of 2.5‐, 4‐, and 6‐hpf embryos using TRIzol Reagent (Life Technologies). G/I tailing, reverse transcription, and PCR amplifications were performed by using USB Poly(A) Tail‐Length Assay Kit (USB, 76455), following manufacturer's instructions. The PCR products were resolved in 2.5% agarose gels. Primer sequences are listed in Appendix Table S1.
Mouse mRNA deep sequencing and data analysis
Total RNAs of each E4.5 mouse embryo were extracted following the Geo‐seq methods (Chen et al, 2017). cDNAs were quantified and qualified by Agilent 2100 Bioanalyzer (Agilent Technologies, USA). In total, 5 ng PCR products were used for the following library preparation. Next‐generation sequencing library preparations were constructed according to the manufacturer's protocol (Nextera XT DNA Sample Preparation Kit, Illumina, FC‐131‐1096). Then, libraries with different indices were multiplexed and loaded on an HiSeq instrument according to the manufacturer's instructions (Illumina, USA). Sequencing was carried out using a 2 × 150 bp paired‐end (PE) reads set.
For data analysis, we pre‐processed the raw data by Fastp (v0.12.1; v0.12.1; Chen et al, 2018) with default parameters. Then cleaned data were then aligned to the reference genome (Mus_musculus. GRCm38) via Hisat2 (v2.1.0; Kim et al, 2015). FeatureCounts (v1.5.3; Liao et al, 2014) was used to count the reads numbers mapped to each gene. The expression levels were determined based on TPM (Transcripts Per Kilobase per Million mapped reads).
To analyze differentially expressed genes (DEG), 2,315 genes were selected with maxTPM > 1 and minTPM > 0 among the nine samples to exclude low‐expressed genes. TPM of each gene was normalized to Gapdh, then DEG was called using edgeR (fold change ≥ 2 and FDR ≤ 0.001; Robinson et al, 2010).
Alternative splicing (AS) events in zebrafish and mouse RNA‐seq data were analyzed as described previously (Xiao et al, 2019). For the identification of differential AS events across samples, rMATS (4.0.2) were used with the parameter ‐c = 0.00001. The differential AS events were calculated at the threshold FDR < 0.05 and ¦Δψ¦ ≥ 5%.
Statistical analysis
The two‐tailed unpaired Student's t‐test was performed to calculate P values using GraphPad Prism version 5.0 (San Diego). Differences were considered significant when P < 0.05. Only results from three independent experiments were subjected to the t‐test. No blinding was used in quantifications.
Author contributions
Yue Xiao: Funding acquisition; investigation; writing – original draft; writing – review and editing. Jiehui Chen: Investigation; writing – original draft; writing – review and editing. Suming Yang: Investigation. Honghua Sun: Investigation. Lele Xie: Investigation. Jinsong Li: Resources; supervision. Naihe Jing: Resources; supervision. Xueliang Zhu: Conceptualization; resources; supervision; funding acquisition; writing – original draft; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Source Data for Expanded View
PDF+
Source Data for Figure 1
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Source Data for Figure 7
Source Data for Figure 8
Acknowledgements
The authors thank Dr. Yungui Yang for kindly providing the m5C modified RNAs, Xiumin Yan for stimulating discussions, and institutional core facilities for zebrafish, cell biology, and molecular biology for instrumental and technical supports. This work was supported by National Key R&D Program of China (2017YFA0503500 to XZ), Strategic Priority Research Program of Chinese Academy of Sciences (XDB19020102 to XZ), National Natural Science Foundation of China (32000556 to YX), and Key Project of Zhejiang Provincial Natural Science Foundation (E1BH3B0401 to YX).
The EMBO Journal (2023) 42: e111364
Contributor Information
Naihe Jing, Email: njing@sibcb.ac.cn.
Xueliang Zhu, Email: xlzhu@sibcb.ac.cn.
Data availability
The datasets produced in this study are available in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo), accessible under GSE179035, or in source data.
References
- Aanes H, Collas P, Alestrom P (2014) Transcriptome dynamics and diversity in the early zebrafish embryo. Brief Funct Genomics 13: 95–105 [DOI] [PubMed] [Google Scholar]
- Alberti S, Hyman AA (2021) Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat Rev Mol Cell Biol 22: 196–213 [DOI] [PubMed] [Google Scholar]
- Auboeuf D, Dowhan DH, Li X, Larkin K, Ko L, Berget SM, O'Malley BW (2004) CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol Cell Biol 24: 442–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK (2017) Biomolecular condensates: organizers of cellular biochemistry. Nat Rev Mol Cell Biol 18: 285–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashirullah A, Halsell SR, Cooperstock RL, Kloc M, Karaiskakis A, Fisher WW, Fu W, Hamilton JK, Etkin LD, Lipshitz HD (1999) Joint action of two RNA degradation pathways controls the timing of maternal transcript elimination at the midblastula transition in Drosophila melanogaster . EMBO J 18: 2610–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzini AA, Del Viso F, Moreno‐Mateos MA, Johnstone TG, Vejnar CE, Qin Y, Yao J, Khokha MK, Giraldez AJ (2016) Codon identity regulates mRNA stability and translation efficiency during the maternal‐to‐zygotic transition. EMBO J 35: 2087–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Shen Y, Zhu L, Xu Y, Zhou Y, Wu Z, Li Y, Yan X, Zhu X (2012) Mir‐129‐3p controls cilia assembly by regulating CP110 and Actin dynamics. Nat Cell Biol 14: 697–706 [DOI] [PubMed] [Google Scholar]
- Chan SH, Tang Y, Miao L, Darwich‐Codore H, Vejnar CE, Beaudoin JD, Musaev D, Fernandez JP, Benitez MDJ, Bazzini AA et al (2019) Brd4 and P300 confer transcriptional competency during zygotic genome activation. Dev Cell 49: 867–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JSS, Tam P, Han J, Peng G, Jing N (2017) Spatial transcriptomic analysis of cryosectioned tissue samples with geo‐seq. Nat Protoc 12: 547–565 [DOI] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J (2018) Fastp: an ultra‐fast all‐in‐one FASTQ preprocessor. Bioinformatics 34: i884–i890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Juan G, Srour EF (2004) Differential staining of DNA and RNA. Curr Protoc Cytom Chapter 7: Unit 7.3 [DOI] [PubMed] [Google Scholar]
- Despic V, Neugebauer KM (2018) RNA tales – how embryos read and discard messages from mom. J Cell Sci 131: jcs201996 [DOI] [PubMed] [Google Scholar]
- Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, Wu L (2016) YTHDF2 destabilizes m(6)A‐containing RNA through direct recruitment of the CCR4‐NOT deadenylase complex. Nat Commun 7: 12626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S, Li H, Zhang Y, Yang S, Chen Y, Qiu B, Huang C, Wang J, Li J, Zhu X et al (2021) Rabl2 GTP hydrolysis licenses BBSome‐mediated export to fine‐tune ciliary signaling. EMBO J 40: e105499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersley‐Maslin MA, Alda‐Catalinas C, Reik W (2018) Dynamics of the epigenetic landscape during the maternal‐to‐zygotic transition. Nat Rev Mol Cell Biol 19: 436–450 [DOI] [PubMed] [Google Scholar]
- Eddaoudi A, Canning SL, Kato I (2018) Flow cytometric detection of G0 in live cells by Hoechst 33342 and Pyronin Y staining. Methods Mol Biol 1686: 49–57 [DOI] [PubMed] [Google Scholar]
- Ernst J, Bar‐Joseph Z (2006) STEM: a tool for the analysis of short time series gene expression data. BMC Bioinformatics 7: 191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firmin FF, Oger F, Gheeraert C, Dubois‐Chevalier J, Vercoutter‐Edouart AS, Alzaid F, Mazuy C, Dehondt H, Alexandre J, Derudas B et al (2017) The RBM14/CoAA‐interacting, long intergenic non‐coding RNA Paral1 regulates adipogenesis and coactivates the nuclear receptor PPARgamma. Sci Rep 7: 14087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Pei G, Li D, Li R, Shao Y, Zhang QC, Li P (2019) Multivalent m(6)a motifs promote phase separation of YTHDF proteins. Cell Res 29: 767–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau NL, Wilusz J, Wilusz CJ (2007) The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 8: 113–126 [DOI] [PubMed] [Google Scholar]
- Ge Y, Porse BT (2014) The functional consequences of intron retention: alternative splicing coupled to NMD as a regulator of gene expression. Bioessays 36: 236–243 [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF (2006) Zebrafish MiR‐430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79 [DOI] [PubMed] [Google Scholar]
- Groisman I, Huang YS, Mendez R, Cao QP, Theurkauf W, Richter JD (2000) CPEB, maskin, and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell 103: 435–447 [DOI] [PubMed] [Google Scholar]
- Hennig S, Kong G, Mannen T, Sadowska A, Kobelke S, Blythe A, Knott GJ, Iyer KS, Ho D, Newcombe EA et al (2015) Prion‐like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J Cell Biol 210: 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentze MW, Castello A, Schwarzl T, Preiss T (2018) A brave new world of RNA‐binding proteins. Nat Rev Mol Cell Biol 19: 327–341 [DOI] [PubMed] [Google Scholar]
- Howe K, FitzHarris G (2013) A non‐canonical mode of microtubule organization operates throughout pre‐implantation development in mouse. Cell Cycle 12: 1616–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wang S, Huang Y, He X, Cui H, Zhu X, Zheng Y (2015) Phase transition of spindle‐associated protein regulate spindle apparatus assembly. Cell 163: 108–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukam D, Shariati SAM, Skotheim JM (2017) Zygotic genome activation in vertebrates. Dev Cell 42: 316–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Richter JD (2006) Opposing polymerase‐deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell 24: 173–183 [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF (1995) Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310 [DOI] [PubMed] [Google Scholar]
- Knoblich JA (2001) Asymmetric cell division during animal development. Nat Rev Mol Cell Biol 2: 11–20 [DOI] [PubMed] [Google Scholar]
- Korner CG, Wahle E (1997) Poly(a) tail shortening by a mammalian poly(a)‐specific 3′‐exoribonuclease. J Biol Chem 272: 10448–10456 [DOI] [PubMed] [Google Scholar]
- Korner CG, Wormington M, Muckenthaler M, Schneider S, Dehlin E, Wahle E (1998) The deadenylating nuclease (DAN) is involved in poly(a) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J 17: 5427–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Simon A (2017) Hexanediol: a chemical probe to investigate the material properties of membrane‐less compartments. Matters 10.19185/matters.201702000010 [DOI] [Google Scholar]
- Kudoh T, Concha ML, Houart C, Dawid IB, Wilson SW (2004) Combinatorial Fgf and bmp signalling patterns the gastrula ectoderm into prospective neural and epidermal domains. Development 131: 3581–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusov YY, Shatirishvili G, Dzagurov G, Gauss‐Muller V (2001) A new G‐tailing method for the determination of the poly(a) tail length applied to hepatitis a virus RNA. Nucleic Acids Res 29: E57‐7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H (1996) Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA 93: 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JD, Nagy LM (2002) Asymmetric inheritance of centrosomally localized mRNAs during embryonic cleavages. Nature 420: 682–686 [DOI] [PubMed] [Google Scholar]
- Lechler T, Mapelli M (2021) Spindle positioning and its impact on vertebrate tissue architecture and cell fate. Nat Rev Mol Cell Biol 22: 691–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM (2007) Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131: 174–187 [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, Giraldez AJ (2013) Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal‐to‐zygotic transition. Nature 503: 360–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30: 923–930 [DOI] [PubMed] [Google Scholar]
- Link V, Shevchenko A, Heisenberg CP (2006) Proteomics of early zebrafish embryos. BMC Dev Biol 6: 1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T (2015) N(6)‐methyladenosine‐dependent RNA structural switches regulate RNA‐protein interactions. Nature 518: 560–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T (2017) N6‐methyladenosine alters RNA structure to regulate binding of a low‐complexity protein. Nucleic Acids Res 45: 6051–6063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Huang T, Chen W, Ding C, Zhao T, Zhao X, Cai B, Zhang Y, Li S, Zhang L et al (2022) Developmental mRNA m(5)C landscape and regulatory innovations of massive m(5)C modification of maternal mRNAs in animals. Nat Commun 13: 2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loerch S, Kielkopf CL (2015) Dividing and conquering the family of RNA recognition motifs: a representative case based on hnRNP L. J Mol Biol 427: 2997–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Tsai MY, Wang SS, Lu BW, Chen R, Yates JR, Zhu XL, Zheng YX (2009) Requirement for Nudel and dynein for assembly of the Lamin B spindle matrix. Nat Cell Biol 11: 247–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathavan S, Lee SG, Mak A, Miller LD, Murthy KR, Govindarajan KR, Tong Y, Wu YL, Lam SH, Yang H et al (2005) Transcriptome analysis of zebrafish embryogenesis using microarrays. PLoS Genet 1: 260–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima E, Abe T (2019) Immuno‐northern blotting: detection of modified RNA using gel separation and antibodies to modified nucleosides. Methods Mol Biol 1870: 179–187 [DOI] [PubMed] [Google Scholar]
- Mishima Y, Tomari Y (2016) Codon usage and 3' UTR length determine maternal mRNA stability in zebrafish. Mol Cell 61: 874–885 [DOI] [PubMed] [Google Scholar]
- Moraes KCM, Wilusz CJ, Wilusz J (2006) CUG‐BP binds to RNA substrates and recruits PARN deadenylase. RNA 12: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan M, Much C, DiGiacomo M, Azzi C, Ivanova I, Vitsios DM, Pistolic J, Collier P, Moreira PN, Benes V et al (2017) mRNA 3′ uridylation and poly(a) tail length sculpt the mammalian maternal transcriptome. Nature 548: 347–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA (1995) Microtubule nucleation by gamma‐tubulin‐containing rings in the centrosome. Nature 378: 638–640 [DOI] [PubMed] [Google Scholar]
- Murray AW (2004) Recycling the cell cycle: cyclins revisited. Cell 116: 221–234 [DOI] [PubMed] [Google Scholar]
- Murray AW, Kirschner MW (1989) Cyclin synthesis drives the early embryonic cell cycle. Nature 339: 275–280 [DOI] [PubMed] [Google Scholar]
- Perez‐Camps M, Tian J, Chng SC, Sem KP, Sudhaharan T, Teh C, Wachsmuth M, Korzh V, Ahmed S, Reversade B (2016) Quantitative imaging reveals real‐time Pou5f3‐Nanog complexes driving dorsoventral mesendoderm patterning in zebrafish. Elife 5: e11475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrachkova T, Wortinger LA, Bard AJ, Singh J, Warga RM, Kane DA (2019) Lack of cyclin B1 in zebrafish causes lengthening of G2 and M phases. Dev Biol 451: 167–179 [DOI] [PubMed] [Google Scholar]
- Rathbun LI, Aljiboury AA, Bai X, Hall NA, Manikas J, Amack JD, Bembenek JN, Hehnly H (2020) PLK1‐ and PLK4‐mediated asymmetric mitotic centrosome size and positioning in the early zebrafish embryo. Curr Biol 30: 4519–4527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhine K, Vidaurre V, Myong S (2020) RNA droplets. Annu Rev Biophys 49: 247–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries RJ, Zaccara S, Klein P, Olarerin‐George A, Namkoong S, Pickering BF, Patil DP, Kwak H, Lee JH, Jaffrey SR (2019) M(6)a enhances the phase separation potential of mRNA. Nature 571: 424–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera‐Perez JA, Hadjantonakis AK (2014) The dynamics of morphogenesis in the early mouse embryo. Cold Spring Harb Perspect Biol 7: a015867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roden C, Gladfelter AS (2021) RNA contributions to the form and function of biomolecular condensates. Nat Rev Mol Cell Biol 22: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salles FJ, Lieberfarb ME, Wreden C, Gergen JP, Strickland S (1994) Coordinate initiation of drosophila development by regulated polyadenylation of maternal messenger RNAs. Science 266: 1996–1999 [DOI] [PubMed] [Google Scholar]
- Schier AF, Talbot WS (2005) Molecular genetics of axis formation in zebrafish. Annu Rev Genet 39: 561–613 [DOI] [PubMed] [Google Scholar]
- Takaoka K, Hamada H (2012) Cell fate decisions and axis determination in the early mouse embryo. Development 139: 3–14 [DOI] [PubMed] [Google Scholar]
- Vastenhouw NL, Cao WX, Lipshitz HD (2019) The maternal‐to‐zygotic transition revisited. Development 146: dev161471 [DOI] [PubMed] [Google Scholar]
- Vejnar CE, Abdel Messih M, Takacs CM, Yartseva V, Oikonomou P, Christiano R, Stoeckius M, Lau S, Lee MT, Beaudoin JD et al (2019) Genome wide analysis of 3' UTR sequence elements and proteins regulating mRNA stability during maternal‐to‐zygotic transition in zebrafish. Genome Res 29: 1100–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkei ZG, Yamashita YM (2018) Emerging mechanisms of asymmetric stem cell division. J Cell Biol 217: 3785–3795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walser CB, Lipshitz HD (2011) Transcript clearance during the maternal‐to‐zygotic transition. Curr Opin Genet Dev 21: 431–443 [DOI] [PubMed] [Google Scholar]
- Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, Shi SH (2009) Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature 461: 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Li MY, Qu C, Miao WY, Yin Q, Liao J, Cao HT, Huang M, Wang K, Zuo E et al (2017) CRISPR‐Cas9‐mediated genome editing in one blastomere of two‐cell embryos reveals a novel Tet3 function in regulating neocortical development. Cell Res 27: 815–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Choi JM, Holehouse AS, Lee HO, Zhang X, Jahnel M, Maharana S, Lemaitre R, Pozniakovsky A, Drechsel D et al (2018) A molecular grammar governing the driving forces for phase separation of prion‐like RNA binding proteins. Cell 174: 688–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill L, Belloc E, Bava FA, Mendez R (2012) Translational control by changes in poly(a) tail length: Recycling mRNAs. Nat Struct Mol Biol 19: 577–585 [DOI] [PubMed] [Google Scholar]
- Welten MC, de Haan SB, van den Boogert N, Noordermeer JN, Lamers GE, Spaink HP, Meijer AH, Verbeek FJ (2006) ZebraFISH: fluorescent in situ hybridization protocol and three‐dimensional imaging of gene expression patterns. Zebrafish 3: 465–476 [DOI] [PubMed] [Google Scholar]
- Winata CL, Lapinski M, Pryszcz L, Vaz C, Bin Ismail MH, Nama S, Hajan HS, Lee SGP, Korzh V, Sampath P et al (2018) Cytoplasmic polyadenylation‐mediated translational control of maternal mRNAs directs maternal‐to‐zygotic transition. Development 145: dev159566 [DOI] [PubMed] [Google Scholar]
- Wu B, Su S, Patil DP, Liu H, Gan J, Jaffrey SR, Ma J (2018) Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat Commun 9: 420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Chen J, Wan Y, Gao Q, Jing N, Zheng Y, Zhu X (2019) Regulation of zebrafish dorsoventral patterning by phase separation of RNA‐binding protein Rbm14. Cell Discov 5: 37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Wang L, Han X, Yang WL, Zhang M, Ma HL, Sun BF, Li A, Xia J, Chen J et al (2019) RNA 5‐Methylcytosine facilitates the maternal‐to‐zygotic transition by preventing maternal mRNA decay. Mol Cell 75: 1188–1202 [DOI] [PubMed] [Google Scholar]
- Zaccara S, Ries RJ, Jaffrey SR (2019) Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol 20: 608–624 [DOI] [PubMed] [Google Scholar]
- Zhang H, Ji X, Li P, Liu C, Lou J, Wang Z, Wen W, Xiao Y, Zhang M, Zhu X (2020) Liquid‐liquid phase separation in biology: mechanisms, physiological functions and human diseases. Sci China Life Sci 63: 953–985 [DOI] [PubMed] [Google Scholar]
- Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, Ho RK, He C (2017) M(6)A‐dependent maternal mRNA clearance facilitates zebrafish maternal‐to‐zygotic transition. Nature 542: 475–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y (2010) A membranous spindle matrix orchestrates cell division. Nat Rev Mol Cell Biol 11: 529–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK (2019) Metascape provides a biologist‐oriented resource for the analysis of systems‐level datasets. Nat Commun 10: 1523 [DOI] [PMC free article] [PubMed] [Google Scholar]


