Abstract
Dosage compensation for X-linked genes in mammals is accomplished by inactivating one of the two X chromosomes in females. X-chromosome inactivation (XCI) occurs during development, coupled with cell differentiation. In somatic cells, XCI is random, whereas in extraembryonic tissues, XCI is imprinted in that the paternally inherited X chromosome is preferentially inactivated. Inactivation is initiated from an X-linked locus, the X-inactivation center (Xic), and inactivity spreads along the chromosome toward both ends. XCI is established by complex mechanisms, including DNA methylation, heterochromatinization, and late replication. Once established, inactivity is stably maintained in subsequent cell generations. The function of an X-linked regulatory gene, Xist, is critically involved in XCI. The Xist gene maps to the Xic, it is transcribed only from the inactive X chromosome, and the Xist RNA associates with the inactive X chromosome in the nucleus. Investigations with Xist-containing transgenes and with deletions of the Xist gene have shown that the Xist gene is required in cis for XCI. Regulation of XCI is therefore accomplished through regulation of Xist. Transcription of the Xist gene is itself regulated by DNA methylation. Hence, the differential methylation of the Xist gene observed in sperm and eggs and its recognition by protein binding constitute the most likely mechanism regulating imprinted preferential expression of the paternal allele in preimplantation embryos and imprinted paternal XCI in extraembryonic tissues. This article reviews the mechanisms underlying XCI and recent advances elucidating the functions of the Xist gene in mice and humans.
X-CHROMOSOME INACTIVATION
General Features
In female mammals, dosage compensation of X-linked genes between males and females occurs by genetic inactivation of one of the two X chromosomes (96). The choice of X chromosome to be inactivated is random in somatic cells, i.e., either the paternally or maternally inherited X chromosome is inactivated in a given cell. Once established, the inactivity is clonally maintained, and spontaneous, unprogrammed reactivation is extremely rare. The inactive X chromosome is distinguished from the active X chromosome by the following characteristics: (i) overall transcriptional inactivation (53) (apart from certain X-linked genes which escape inactivation and the Xist gene); (ii) heterochromatic condensation at interphase of the cell cycle, sometimes visible as the Barr body (6); (iii) late replication during S phase (155); (iv) DNA methylation of cytosine residues at CpG dinucleotides in the 5′ region of X-linked genes (reviewed in reference 116); (v) hypoacetylation of histone H4 (65); and (vi) expression of the Xist (X-inactive specific transcript) gene located at the X-inactivation center (15, 17, 19, 20).
These characteristics are associated, causally and/or consequentially, with the process of X-chromosome inactivation (XCI), which occurs early in development. Both the paternally and maternally inherited X chromosomes are active in preimplantation embryos after fertilization, and XCI occurs sequentially, coupled with cell differentiation, first in the extraembryonic trophectoderm and primitive endoderm of the blastocyst and then in the fetal precursor cells arising from the inner cell mass (ICM) around the time of implantation (113, 158). XCI is a multistep process, comprising (i) counting of the number of X chromosomes with respect to the number of autosome complements, (ii) choice of a single X chromosome to remain active per autosome complement, (iii) initiation of inactivation of an additional X chromosome(s), (iv) spreading of inactivity along the length of the inactive X chromosome(s), and (v) stabilization and maintenance of inactivity throughout future cell divisions. These steps in the XCI process are discussed further below.
Theoretically, the choice involved in XCI could be either the choice of X chromosome to be active or the choice of X chromosome to be inactive. The observation of a single active X chromosome in cases of X-chromosome aneuploidy, e.g., X+X−X−AA and X+X−X−X−AA (X+, active X chromosome; X−, inactive X chromosome; A, haploid set of autosomes), suggests that one X chromosome is always chosen to be active rather than different numbers of X chromosomes being chosen to be inactive. Cells count the number of autosome complements for determining the number of X chromosomes to be active, since tetraploid cells have two active X chromosomes, i.e., X+X+X−X−AAAA.
XCI is initiated from the X-inactivation center (Xic), which is required in cis for the X chromosome to be inactivated (145). The evidence for the existence of the Xic is derived from observations on X-autosome translocations; the X chromosome devoid of a particular segment (putative Xic) does not undergo inactivation (135, 145). Once XCI is initiated, inactivity spreads from the Xic in both directions along the chromosome.
The spreading process has been probed by using transgenes. Some X-linked transgenes (transgene integrated on the X chromosome) are inactivated on the inactive X chromosome (37, 39, 159, 160), whereas others escape inactivation (49, 164). In addition, the strength of inactivation of the transgene may depend on tissue-specific transgene expression; e.g., a transgene containing the α-fetoprotein gene was shown to be inactivated on the inactive X chromosome in somatic tissues but to remain active in the yolk sac (where α-fetoprotein is normally expressed) of the developing mouse conceptuses (82). Whether a transgene is inactivated on the inactive X chromosome may depend on its size, site of insertion, mode of insertion, and tissue specificity of expression, as well as on the structure of the transgene itself. On the other hand, it has been shown that X-linked phosphoglycerate kinase-1 (Pgk-1) gene-containing transgenes on autosomes are ubiquitously expressed (100, 132), which suggests that inactivation is not intrinsic to the X-linked genes themselves. This is a striking contrast to the dosage compensation mechanism in Drosophila, where X-linked genes translocated to autosomes show inactivation (reviewed in reference 95).
Several X-linked genes have been shown to escape XCI in both humans and mice (reviewed in reference 42). Although the mechanism of escape from inactivation is unknown, the phenomenon indicates that spreading of inactivation can be interrupted and then resume and that local control is also involved. More X-linked genes escape XCI in humans than in mice, and this difference is considered to be responsible for the more severe phenotype and reproductive failure in human XO females, known as Turner’s syndrome or haploinsufficiency (reviewed in reference 167). In contrast, XO female mice are fertile, although they produce a smaller number of eggs with poorer developmental potential compared to those from XX mice (24, 25).
Maintenance (or stabilization) of inactivity may involve multiple mechanisms, such as changes in Xist gene structure or Xist transcription, some function of Xist RNA, X-chromosome DNA methylation, histone H4 hypoacetylation, heterochromatinization, late replication during S phase, and nuclear compartmentalization (reviewed in references 116, 120, and 143). Among these, late replication and heterochromatinization seem to be more universal mechanisms since they are also observed for non-X-linked genes and are found in other species, such as yeast and Drosophila (reviewed in references 59, 62, and 111).
Another locus affecting the process of XCI is the X-chromosome-controlling element (Xce) (30). The Xce is mapped within the Xic (31), but is separable from Xist (151), and affects the likelihood of an X chromosome being inactivated. To date, three different alleles of the Xce locus, Xcea, Xceb, and Xcec, have been characterized in Mus species (32, 67). In Xcea/Xceb heterozygotes, the X chromosome carrying Xcea is more likely to be inactivated than the X chromosome carrying Xceb. In Xceb/Xcec heterozygotes, the X chromosome carrying Xceb is more likely to be inactivated than the X chromosome carrying Xcec. In Xcea/Xcec heterozygotes, nonrandom (skewed) XCI is most prominent. Therefore, the relative strengths of the three Xce alleles, with respect to likelihood of remaining active, are Xcea < Xceb < Xcec. Recently, a fourth allele, Xced, carried in Mus spretus, has been identified and seems to be stronger than Xcec (34). The strength of the Xce has been inversely correlated with the degree of Xist expression from the inactive X chromosome in that the level of Xist RNA is markedly lower in Mus spretus mice which carry the Xced allele than in C57BL/10 mice which carry the Xceb allele (17).
Activity of the X Chromosomes in Female Development
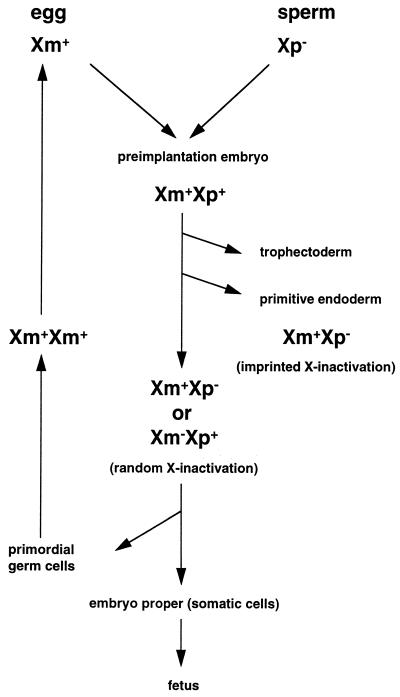
The activity of the X chromosomes throughout female mouse development is illustrated in Fig. 1.
FIG. 1.
Activity of X chromosomes in female mouse development. The maternally inherited X chromosome (Xm) in the egg is active, and the paternally inherited X chromosome (Xp) in the sperm is inactive. Soon after fertilization, Xp is reactivated and both Xm and Xp are active (Xm+Xp+) in preimplantation embryos. In the first delineating cell lineages, the trophectoderm and the primitive endoderm, which differentiate at 3.5 and 4.5 dpc, respectively, Xp is preferentially inactivated (Xm+Xp−; imprinted X inactivation). In the epiblast cells, which will give rise to the germ cells and somatic cells of the embryo proper, either Xm or Xp is inactivated (Xm+Xp− or Xm−Xp+; random X inactivation). In the primordial germ cells, the inactive X chromosome is reactivated at around 12.5 dpc, when the female germ cells enter meiosis; hence, both X chromosomes are active (Xm+Xm+) throughout oogenesis. + denotes the active X chromosome, and − denotes the inactive X chromosome.
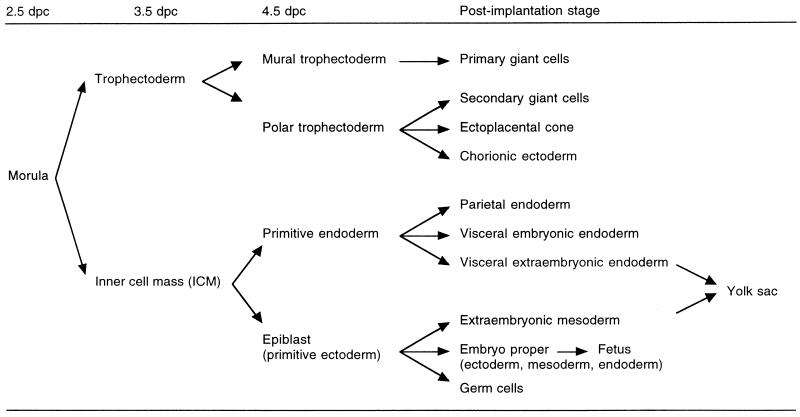
It has been shown cytologically (157) and enzymatically (44, 80, 112) that both the maternally and paternally inherited X chromosomes are active in female mouse preimplantation embryos. Then XCI occurs sequentially in development, coupled with cell differentiation (113, 158). XCI first occurs in the trophectoderm and the primitive endoderm which are the first two cell lineages that differentiate, at 3.5 and 4.5 days post- coitum (dpc), respectively (Fig. 2). In these two extraembryonic lineages and their derivative tissues, XCI is nonrandom in that the paternally inherited X chromosome is preferentially inactivated (46, 47, 55, 126, 156, 163) (summarized in Table 1). In the primitive ectoderm (epiblast), which will give rise to the embryo proper (somatic cells and germ cells) and yolk sac mesoderm, XCI is initiated at around the time of implantation and is random in that either the paternal or the maternal X chromosome is inactivated in a given cell.
FIG. 2.
Diagrammatic representation of cell differentiation during mouse development. The trophectoderm is the first cell lineage, which differentiates at 3.5 dpc. Then the primitive endoderm differentiates, at 4.5 dpc. In the derivatives of these two extraembryonic cell lineages, XCI is nonrandom, in that the paternally inherited X chromosome is preferentially inactivated (see Table 1 for summary). In contrast, XCI is random in the epiblast, which gives rise to the yolk sac mesoderm, the embryo proper, and the primordial germ cells; i.e., either the paternal or the maternal X chromosome is inactivated in a given cell.
TABLE 1.
Summary of studies on paternal XCI in mouse extraembryonic tissues
| Study | Method | Embryonic stage (dpc) | Tissue |
|---|---|---|---|
| Takagi and Sasaki (156) | Allocyclic X | 8.5 | Chorion |
| West et al. (163) | PGK-1 isozyme | 13.5 | Yolk sac (endoderm) |
| Frels et al. (46) | PGK-1 isozyme | 8.5 | Chorionic ectoderm |
| Frels and Chapman (47) | PGK-1 isozyme | 9.5 | Mural trophoblast |
| Papaioannou and West (126) | PGK-1 isozyme | 12.5 and 17.5 | Parietal endoderm |
| Harper et al. (55) | PGK-1 isozyme | 6.5 | Extraembryonic ectoderm |
| Harper et al. (55) | PGK-1 isozyme | 3.5 + in vitro culture | Outgrowth of blastocyst |
In female germ cells, XCI is also random, and it has been shown, by a study of X-inactivation cell mosaicism, that the somatic tissues and the germ cells are derived from a common pool of cells after XCI has occurred and that the germ cells are derived from a sizable pool of precursor cells (104, 105). This original work (115) challenged the popular view that the continuity of the totipotency of the germ line necessarily required that the germ cells be set aside very early in development and from very few cells. It also means that the “germ line” itself includes the early stages of preimplantation development through to the epiblast of the implanted embryo when the primordial germ cells are delineated. A few years later, Monk et al. (117) provided further evidence in support of this view and showed that the ground state, with respect to erasure of epigenetic methylation programs, was not a property of the gametes at the time of fertilization but, rather, a property of the totipotent ICM, epiblast cells and germ cells at the time of implantation.
In keeping with a progressive dedifferentiation in early development approaching a ground state in the epiblast cells and the primordial germ cells arising from them, the inactive X chromosome is reactivated in the primordial germ cells at around the time of entry into meiosis, i.e., 12.5 to 13.5 dpc. Following reactivation, both X chromosomes continue to be active throughout oogenesis (66, 81, 104, 114).
Activity of the X Chromosome in Male Development
In males, the single X chromosome is active in all somatic cells. However, it is transcriptionally inactivated during spermatogenesis. At meiosis, the X chromosome pairs with the Y chromosome in the XY body, or the sex vesicle, where the X and Y chromosomes are sequestered away from the autosomes. In some instances, transcriptional silencing of the X-linked genes during spermatogenesis is compensated by expression of autosomal genes, e.g., the X-linked phosphoglycerate kinase-1 (Pgk-1) gene by the autosomal Pgk-2 gene (102) and the X-linked pyruvate dehydrogenase E1α subunit (Pdha-1) gene by the autosomal Pdha-2 gene (38).
The inactive X chromosome carried in sperm is reactivated soon after fertilization in female preimplantation embryos (44, 80, 112).
X-CHROMOSOME INACTIVATION AND GENOMIC IMPRINTING
Genomic imprinting is a phenomenon in which expression of a gene locus is determined by its parental origin: either the paternal or the maternal allele is expressed and the other is repressed. Genomic imprinting demonstrates that the parental chromosomal complements are not equivalent with respect to their potential for expression and implies some form of marking (imprint or blueprint) distinguishing certain genes in the egg and in the sperm. These marks must be imposed as modifications to the DNA of the imprinted genes during gametogenesis (reviewed in reference 121). The imprinting mark distinguishing the imprinted gene in either eggs or sperm is perpetuated into early development after fertilization, so that parental alleles are differentially expressed in the progeny embryos and offspring.
Imprinting is a form of transgenerational change in gene expression. A gene may be switched on or off in successive generations as it passes through the male or female parent. Therefore, in the germ line of the progeny, the imprint must be erased to be reestablished once again, depending on the sex of progeny, as the male or female parental imprint in sperm and eggs, respectively. For example, a maternally transmitted allelic imprint in male progeny undergoes erasure and new modification during spermatogenesis to become the paternal allelic imprint.
Genomic imprinting may explain the requirement for both the paternal and maternal sets of chromosomes for normal development. Pronuclear transplantation experiments (97, 103, 154) have shown that neither androgenetic (paternal chromosomes only) nor gynogenetic (maternal chromosomes only) embryos develop to term. Androgenetic embryos show proliferative growth of the extraembryonic tissues and poor growth of the fetus, whereas gynogenetic embryos show poor growth of the extraembryonic tissues and comparatively normal growth of the fetus. In the human, an aberration in embryonic development during pregnancy, known as the hydatidiform mole, is the hyperplastic growth of the extraembryonic trophoblast with reduced or absent fetal growth, which resembles the growth pattern of mouse androgenetic embryos. In fact, the complete form of the hydatidiform mole (no fetal components) is androgenetic in origin, with karyotype 46, XpXp in 96% of cases and karyotype 46, XpY in the remaining 4% of cases (71, 124). The incomplete hydatidiform mole (with fetal components) is, in most cases, due to triploidy with one set of maternal and two sets of paternal genomic complements and karyotype 69, XmXpXp or XmXpY.
The chromosomal regions containing putative imprinted genes have been identified by investigations of abnormal phenotype and developmental lethality associated with regions of uniparental disomy in progeny derived from breeding mice carrying translocations (33). Once such an imprinted chromosomal region is identified, specific genes can be cloned from the chromosomal region. These specific genes can be tested for imprinted expression by using a sequence polymorphism (often found between species in an interspecific cross) or by observing a parent-of-origin-specific effect of a deletion or mutation created in a candidate imprinted gene. Cattanach and Jones (35) have reported that imprinted chromosomal regions in mice may be limited to only six autosomes (chromosomes 2, 6, 7, 11, 12, and 17). Recently, however, a newly identified chromosomal region, Irlgs3, which contains at least one imprinted gene, Grf1, has been mapped to chromosome 9 by the technique of restriction landmark genomic scanning with methylation-sensitive enzymes (RLGS-M) (130). Thus, an increasing number of regions containing imprinted genes have been identified and, to date, around 20 imprinted genes have been identified in humans and mice.
Preferential paternal XCI in mice (46, 47, 55, 126, 156, 163) was one of the first examples of genomic imprinting in mammals and was well characterized before specific imprinted genes were identified. Imprinted XCI depends on the timing of XCI and/or the tissue involved; preferential paternal XCI is observed only in the extraembryonic trophectoderm and the primitive endoderm, which are the first differentiated tissues, delineating at 3.5 and 4.5 dpc, respectively (Fig. 2). This suggests that the imprint for nonrandom XCI is present at these stages of development and is recognized in these tissues and that the imprint is erased by the time XCI occurs in the fetal cells (reviewed in reference 118). Alternatively, the gametic imprint may not be recognized by fetal cells (irrespective of timing).
The imprint for nonrandom XCI should be present on either the paternal or the maternal X chromosome. Shao and Takagi (149) reported that a supernumerary maternally derived X chromosome (Xm) in XmXmXpAmAp (where A refers to a haploid set of autosomes) causes early embryonic death and hypothesized that this is due to two active X chromosomes (Xm+Xm+Xp−AmAp) in the extraembryonic tissues. Their results suggest that the maternally derived X chromosome is resistant to inactivation in those lineages. Recently, Marahrens et al. (98) have shown that embryos inheriting an inactivation-resistant (owing to a deleted Xist locus) X chromosome from the father also die, presumably due to their inability to inactivate the normal maternal X chromosome (hence both Xm and Xp are active) in the extraembryonic tissues. However, Rastan et al. (134) showed that XCI does occur in the extraembryonic tissues of diploid XmXm parthenogenones, which can develop up to the somite stage (72). In addition, XpO embryos are viable (26, 27, 45, 126), suggesting that Xp is not necessarily inactivated in the extraembryonic tissues. Therefore, these relatively imprecise studies do not help at this stage to elucidate the possible mechanisms of the differential marking of parental X chromosomes in gametes and extraembryonic tissues.
In contrast to mice, it is less clear whether the paternally derived X chromosome is preferentially inactivated in human extraembryonic tissues. Ropers et al. (144) and Harrison (56) claimed that preferential paternal XCI does occur in the human extraembryonic tissues, but Migeon and Do (108) and Migeon et al. (109) reported that there is no evidence for paternal XCI in these tissues. All these studies are based on the glucose-6-phosphate dehydrogenase (G6PD) isozyme assay. Harrison (56) used term placentas heterozygous for G6PD isozymes and showed that the maternal G6PD allele is preferentially expressed in isolated trophoblasts. Recently, we have used a DNA methylation assay to show that preferential paternal XCI also occurs in human trophoblastic cells obtained at 10 to 12 weeks of gestation (50). Our results support the conclusion of Harrison (56) and, in addition, strongly suggest that preferential inactivation of the paternally inherited allele seen in term placentas is not due to cell selection during development against cells with the paternal X chromosome active but is due to primary nonrandom XCI occurring early in development (50).
Many studies have sought the underlying parental allelic differences of imprinted genes. DNA modification by CpG methylation has been most extensively studied, since highly sensitive procedures are available for the detection of this modification throughout development. It is also clear that DNA methylation plays an important role in the regulation of gene expression and, moreover, that it is subject to significant changes during development (117; reviewed in references 118, 121, 139, 141).
X-CHROMOSOME INACTIVATION AND DNA METHYLATION
DNA Methylation of the Mammalian Genome
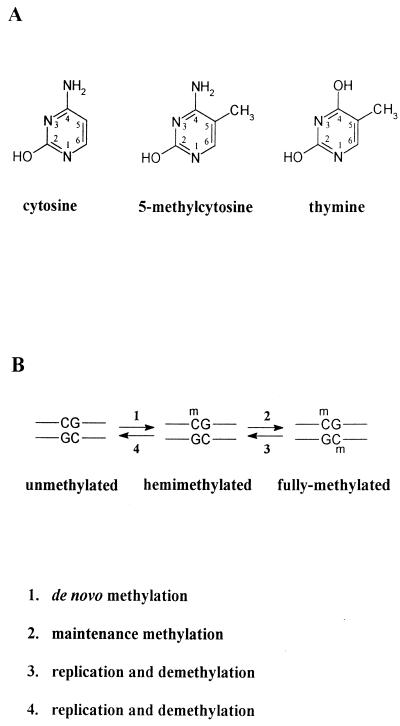
DNA methylation is an epigenetic modification in that it is superimposed on the DNA base sequence and affects the genetic potential of that sequence. In mammals, DNA methylation is found as 5-methylcytosine (5mC) occurring at CpG dinucleotides (Fig. 3A). Theoretically, CpG and GpC dinucleotides should be found at equal frequencies in bulk genomic DNA; i.e., the CpG/GpC ratio should be 1. However, overall, CpG dinucleotides (unmethylated and methylated CpG combined) are found at only 20 to 25% of the expected frequency (43, 48). This lower than expected frequency of CpG dinucleotides is considered to be due to a tendency to mutation transition of the 5mC residue to thymine by deamination at position 4 of the carbon ring (i.e., from CpG to TpG) throughout evolution (Fig. 3A).
FIG. 3.
Methylation of cytosine residues in the mammalian genome. (A) Structure of the cytosine residue and its methylated derivative, 5-methylcytosine, which is methylated at the 5 position of the carbon ring of the cytosine residue. The structure of the thymine residue is also shown, to illustrate that mutational deamination at the 4 position of 5-methylcytosine results in thymine. (B) After replication, daughter strands of fully methylated DNA are hemimethylated (reaction 3) and the original pattern of DNA methylation is maintained by the DNA methyltransferase (reaction 2), which preferentially methylates the cytosine residues at hemimethylated CpG sites. Further replication without methylation of the hemimethylated DNA results in fully unmethylated DNA (reaction 4). De novo methylation (reaction 1) is also considered to be mediated by the DNA methyltransferase, although the efficiency of de novo methylation is low.
Approximately 70% of all CpGs are methylated (reviewed in references 12, 89, and 138). The significance of DNA methylation as a mechanism of gene regulation has been controversial since well-researched systems, such as Drosophila and Caenorhabditis elegans, are known to regulate gene transcription without detectable DNA methylation being present (152, 162). Nevertheless, recent experiments have shown that DNA methylation is essential for normal mouse development (91, 93), and it is well documented that DNA methylation plays an important role in the regulation of gene expression, XCI, tumor growth, and genomic imprinting in mammals (reviewed in references 12, 83, 92, 139, and 140).
Conversion of cytosine to 5mC is catalyzed by the enzyme DNA (cytosine-5)-methyltransferase (DNA MTase; EC 2.1.1.37) (9, 54; reviewed in reference 2), which transfers a methyl moiety from S-adenosylmethionine to the 5 position of cytosine. Mammalian DNA MTase is composed of 1,573 amino acid residues and has a relative mass (Mr) of 190,000 (10). Amino acids 72 to 92 contain the signal for entry into the nucleus, and the presence of amino acids 207 to 455 in the N-terminal domain is essential for the DNA MTase to be associated with replication foci (88).
Specific patterns of DNA methylation are faithfully maintained during DNA replication by the DNA MTase, and therefore the enzyme is also called a maintenance methylase (reviewed in reference 138). Usually, cytosine residues at CpG dinucleotides are symmetrically methylated on both strands. After replication, cytosine residues in CpG doublets are methylated on one strand only (hemimethylated; Fig. 3B, reaction 3). Hemimethylated CpG sites are the preferred substrate of the DNA MTase, which methylates the cytosine on the daughter strand (9, 54) to create fully methylated sites (reaction 2). Specific methylation patterns may also be lost, presumably when the DNA MTase is scarce or the methylation site is inaccessible (reaction 4). A marked loss of methylation occurs early in development (69, 117, 150; see below). This loss in methylation is associated with a marked decrease in the DNA methyltransferase enzyme activity in preimplantation embryos from the eight-cell stage to the blastocyst stage (119).
The DNA MTase also catalyzes de novo methylation but with lower efficiency (1, 11) (Fig. 3B, reaction 1). However, it is not clear whether the DNA MTase recognizes a specific sequence motif for its de novo methylation activity or whether it methylates any CpG dinucleotide accessible due to an open chromatin structure, e.g., during DNA replication or displacement of nucleosomes.
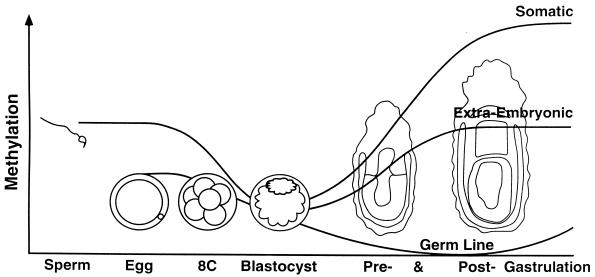
During development, dynamic demethylation and de novo methylation occur (69, 117, 150). Figure 4 illustrates these changes in DNA methylation early in development, as shown by Monk et al. (117). Gametic DNA is globally demethylated, descending toward the ground state of absence of methylation and erasure of the bulk of the original gametic methylation patterns of paternal and maternal genetic complements by the blastocyst stage. Stage- and tissue-specific methylation patterns are created by de novo methylation during the process of implantation and gastrulation, whereas demethylation in the germ line continues at this stage.
FIG. 4.
Changes in DNA methylation in early development. Sperm DNA is overall more methylated than egg DNA but less methylated than somatic DNA. During preimplantation development, gamete-specific patterns of methylation are erased by the genome-wide demethylation, tending toward a ground state of absence of methylation in the ICM of the blastocyst and in the primordial germ cells. Around the time of implantation and gastrulation, stage- and tissue-specific patterns of DNA methylation are created de novo. In the extraembryonic tissues, the methylation level is globally lower than that in somatic tissues. It is hypothesized that demethylation continues in the germ line until the onset of establishment of new egg- or sperm-specific patterns of methylation, including the differences in methylation between the gametes that will survive as imprints in the next generation. Imprinting can be viewed as differences in gametic DNA modification (differences between sperm and egg) that survive erasure during preimplantation development and thus are perpetuated into the soma. Reprinted from reference 121 with permission of the publisher.
The genome-wide demethylation occurring in preimplantation embryos is thought to be a process of erasure of the methylation patterns associated with differential programming of oogenesis and spermatogenesis (117; reviewed in reference 121) and is correlated with the decrease in activity of the DNA MTase (119). However, the process may be more complicated in that demethylation at preimplantation stages seems to be site specific; certain CpG sites within imprinted genes are more resistant to demethylation so that they persist until the blastocyst stage (16, 51, 70, 150). We could view a genetic imprint as differential parental gametic modification which survives erasure during preimplantation development and persists through implantation and gastrulation to be perpetuated into the soma (121).
Recently, Yoder et al. (165) presented a unique view on the role of DNA methylation in development. Based on the observation that the majority of methylated CpG dinucleotides lie in transposable elements (transposons), such as L1 and Alu repeats, which are abundantly present in and around genes, they hypothesize that a primary role of CpG methylation is suppression of these transposons, through inactivation of their potential promoter activity by methylation, to maintain the integrity and function of the genes. They consider that transposons are detected, by some cellular mechanism scanning the genome at the time of or soon after implantation, due to their characteristic repetitive DNA sequences, and therefore CpGs in transposon sequences are specifically methylated. In their hypothesis, nontransposon CpGs, which often appear as CpG clusters at the 5′ end of genes (known as CpG islands [see below]), escape de novo methylation and remain unmethylated. Their view may well explain why CpG islands of genes are methylation free (except for imprinted genes and genes on the inactive X chromosome), regardless of their transcriptional status. However, as the authors admit, there must be other underlying mechanisms for de novo methylation in postimplantation embryos, since this hypothesis alone cannot explain differential allelic methylation observed for imprinted genes and genes on the active and inactive X chromosomes (see below). Neither does this hypothesis explain the observed differences in methylation pattern between sperm and eggs (117; see above).
Three related forms of DNA MTase (Mr, 190,000, 175,000, and 150,000) have been identified to date, and a detailed characterization has suggested that they are proteins encoded by a single gene with different extents of deletion of the carboxyl terminus (10). Li et al. (91) introduced targeted mutations into the DNA MTase gene to investigate the role of the enzyme and DNA methylation during development. Mice carrying the mutated allele(s) of the DNA MTase gene die at around midgestation, depending on the severity of deficiency of the enzyme activity (91). Investigations of expression patterns of imprinted genes, insulin-like growth factor 2 (Igf2), Igf2 receptor (Igf2r), and H19, in these DNA MTase-deficient mice have shown that all three genes are aberrantly expressed and that the extents of demethylation of normally methylated CpG sites are different among these genes (93). H19, which is normally expressed only from the maternal allele (7), shows biallelic expression, and the differentially methylated region on the paternal allele is demethylated. Igf2, which is normally expressed only from the paternal allele (41), is not expressed from either allele. Igf2r, which is normally expressed only from the maternal allele (5), shows an unchanged level of expression and methylation (at the CpG island of the region 2 on the expressed maternal allele) in embryos with a less severe type of mutation of the DNA MTase but shows no detectable expression and substantial demethylation in a more severe type of mutation. These results clearly show the biological significance of DNA methylation in development and in the regulation of gene expression and imprinting. They also indicate that the sensitivity to demethylation is site and/or gene specific.
DNA Methylation, CpG Islands, and Gene Expression
Approximately 30% of all CpG dinucleotides of the mammalian genome are unmethylated, and these unmethylated CpG dinucleotides make up only 1 to 2% of the whole genome. Unmethylated CpG dinucleotides are often found as a cluster (12, 48); if the cluster is 0.5 to 2 kb long, GC-rich (65% or more), and of the expected frequency of CpGs (CpG/GpC ratio of ca. 1; no CpG suppression), it is called a CpG island (reviewed in reference 12). The CpG island is often associated with the 5′ region and/or the promoter region of a gene, and its methylation pattern is correlated with gene activity for X-linked genes and imprinted genes (reviewed in references 12, 13, and 139).
In principle, CpG islands are not methylated (they are therefore also called methylation-free islands). For example, CpG islands of ubiquitously expressed housekeeping genes are unmethylated. Some tissue-specific genes also have CpG islands, and these are normally unmethylated in both expressing and nonexpressing tissues (reviewed in reference 12). Exceptions are observed for X-linked genes on the inactive X chromosome (reviewed in references 12 and 116) and certain tissue-specific genes, such as the α-globin gene and the Thy-1 gene, in in vitro-cultured cells (3). Since CpG islands are usually associated with the promoter region and the 5′ end of a gene, binding of transcription factors and the chromatin structure around the transcription start site may inhibit the access of the DNA MTase and protect the CpG islands from de novo methylation. This could be the case for housekeeping genes and tissue-specific genes in the expressing tissue. However, for tissue-specific genes in nonexpressing tissues, other mechanisms, such as binding of a tissue-specific repressor protein or packaging into a closed chromatin structure, must be operative to maintain the methylation-free status.
The repressive effect of DNA methylation on the regulation of gene expression involves the formation of an inactive chromatin structure (28, 75, 76), which is thought to be mediated by proteins which specifically bind to methylated DNA. To date, in addition to the DNA MTase, several such proteins have been identified (Table 2); methylated DNA-binding protein (MDBP) (63), methyl-CpG binding proteins, MeCP1 (106) and MeCP2 (90, 107), MDBP-2-H1 (a member of the histone H1 family) (68, 127), the protein which binds to the mouse sex-limited protein (Slp) promoter (166), and the methylation-dependent protein which binds to an Xist promoter sequence in mice (64). MDBP requires high sequence specificity (77) and MeCP1 binds only to long DNA fragments with more than 15 methylated CpG dinucleotides (106). By contrast, MeCP2 is able to bind at a single methyl-CpG site (90) and is essential for mouse embryonic development (161). However, the level of expression of MeCP2 is very low in undifferentiated embryonic stem (ES) cells (107), and these cells can survive with no MeCP2 proteins (161), indicating that MeCP2 is required during or after differentiation. The methylation-dependent protein binding to the Xist promoter, identified in our laboratory, is sequence specific and preferentially binds to the methylated promoter sequence (64; see below).
TABLE 2.
Methylated DNA-binding proteins
| Proteina | Molecular mass (kDa) | Binding sequencea | Reference(s) |
|---|---|---|---|
| DNA MTase | 190, 175, 150 | NDb | 10 |
| MeCP1 | 120 | >15 mCpGs | 106 |
| MeCP2 | 84 | A single mCpG site | 90, 107 |
| MDBP | 250 | 5′-ATmCGTCAmCGGmCGAT-3′ | 63, 77 |
| MDBP-2-H1 | 21 (monomer), 42 (dimer) | 5′-TTCACCTTmCGCTATGAGGGGGATCATACTGG-3′ | 68, 127 |
| Slp promoter binding protein | ND | (5′-TTCmCGGGC-3′)2 | 166 |
| Xist promoter binding protein | 100 | 5′-GmCGCmCGmCGG-3′c | 64 |
mC, 5-methylcytosine residue; MeCP, methyl-CpG binding protein; MDBP, methylated DNA-binding protein; H1, histone H1.
ND, not determined.
Xist promoter-binding protein preferentially binds to the DNA sequence when at least one of the three CpG sites is methylated.
DNA Methylation and X-Chromosome Inactivation
The X chromosome contains a large number of housekeeping genes. As described above, CpG islands of housekeeping genes are, in general, unmethylated. However, CpG islands of most X-linked genes, including housekeeping genes, are methylated on the inactive X chromosome, and this DNA methylation is involved in the maintenance of inactivity of genes on the inactive X chromosome (reference 116 and references therein). It has been well established that genes on the inactive X chromosome are at least partially reactivated in cultured cells by treatment with an inhibitor of the DNA MTase, 5-azacytidine (reviewed in reference 116 and references therein).
Although DNA methylation is considered to play a role in the maintenance of inactivity, it is controversial whether it is causally involved in initiation and spreading of inactivity along the X chromosome in development. It has been reported that the timing of DNA methylation for the Pgk-1 gene and the G6pd gene is coincident with XCI (52, 153). An earlier report suggested that methylation was a later consequence of genetic inactivation, since methylation of the Hprt gene occurs several days after XCI in development (94). The discrepancy of these results may be explained by (i) the difference in the nature and sensitivity of assays used (PCR-based or Southern blotting)—the Southern blot procedure used for the detection of methylation of the Hprt gene would have missed a minority of unmethylated alleles whereas the PCR procedure used for the Pgk-1 gene and the G6pd gene detected a minority of methylated alleles; (ii) the purity of the tissues examined—the Southern blot approach used whole conceptuses with hypomethylated extraembryonic tissues present; and/or (iii) the fact that methylation may occur at different times for specific genes located at different loci on the X chromosome; interestingly, methylation of the G6pd gene, which is located between Pgk-1 and Hprt, occurs slightly after methylation of the Pgk-1 gene but much earlier than that of the Hprt gene (although these data were determined by different procedures), suggesting that methylation spreads from the Xic in linear fashion throughout the X chromosome (52).
THE XIST GENE
Characterization
An X-linked regulatory gene, Xist (X-inactive specific transcript; Xist in the mouse, XIST in the human), was identified by the unique characteristic that it is expressed exclusively from the inactive X chromosome (15, 17, 19) and is thus implicated in the XCI process itself. Further evidence for a regulatory role is that the Xist gene maps to the Xic (15, 17, 20). In this review, unless specifically stated as the mouse Xist gene or the human XIST gene, the term “Xist” is used to indicate both the mouse and the human genes.
The transcript is a large, polyadenylated RNA molecule, 17 kb for the human XIST transcript and 15 kb for the mouse Xist transcript, but there is no extended open reading frame for protein translation, suggesting that Xist functions as an RNA molecule (18, 21). In keeping with this idea, Xist RNA is localized in the nucleus (18, 21) and RNA FISH (fluorescent in situ hybridization) studies have shown that Xist RNA is associated with the inactive X chromosome at interphase (21, 36, 86, 125).
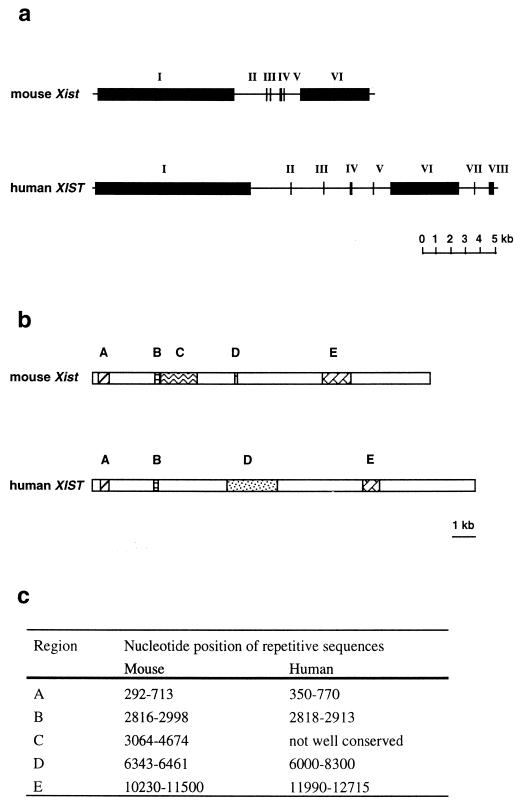
The human and mouse Xist genes consist of eight and six exons, respectively (18, 21) (Fig. 5a), with conserved repetitive sequences in exons 1 and 6 (18, 21) (Fig. 5b). The most conserved repetitive sequence resides at the 5′ end of the gene in exon 1 (nucleotides [nt] 350 to 770 for XIST and 292 to 713 for Xist). This region (Fig. 5b, region A) consists of nine and eight repeats of consensus sequence, 5′-GCCCATCGGGGCCTCGGATACCTGC-3′, in the human and mouse Xist genes, respectively (18, 21). This repeat shows homology to a sequence of Xlsirts, which is essential for the transfer of Xlsirts to the cytoplasm (78), and to a sequence of the latency-associated transcript of herpes simplex virus, which is required for efficient reactivation of the latent herpes simplex virus (14). It has also been reported that a protein binds to this repetitive sequence in human XIST RNA (23). Hendrich et al. (58) determined the DNA sequence of this repetitive region in mice carrying different Xce alleles, since the level of Xist expression correlates with strength of Xce (17). The major difference in DNA sequence identified is the number of adenine (A) residues between the fourth and fifth repeat sequence; the Xcea Xist allele (BALB/c and C3H mice) has 24 uninterrupted A residues, the Xceb Xist allele (C57BL/6) has 23 A residues with a single cytosine insertion, and the Xcec Xist allele (M. m. musculus and At10) has 40 to 44 uninterrupted A residues (58). However, it remains to be clarified whether the difference in the repetitive DNA sequence of Xist is the basis for the relative strength of Xce, since Simmler et al. (151), analyzing segregation patterns between polymorphic microsatellite markers surrounding Xist and the Xce allele, have shown that Xce and Xist are separable gene loci.
FIG. 5.
Structure of the Xist gene. (a) The mouse Xist gene and the human XIST gene comprise six and eight exons, respectively. (b) Schematic representation of the Xist RNA. The mouse Xist RNA is 15 kb long, and the human XIST RNA is 17 kb long. Corresponding conserved repetitive sequences in exons 1 (regions A, B, C, and D) and 6 (region E) are indicated by similarly hatched regions. The numbers of repeats are different between the mouse and human. (c) Nucleotide positions of the repetitive sequences are shown. Data are from references 18 and 21.
Developmental Patterns of Xist Expression and Regulation by DNA Methylation
Xist expression in somatic cells.
Xist is expressed only from the inactive X chromosome in somatic cells (15, 17, 19). The amount of Xist RNA produced from an established inactive X chromosome is calculated as 1 × 103 to 2 × 103 molecules/cell in adult female kidney cells (29). Given that the Xist RNA remains associated with the inactive X chromosome, it may be noted that 103 molecules of a 15-kb Xist RNA are not sufficient to cover the entire length of the X chromosome (1.5 × 108 bp).
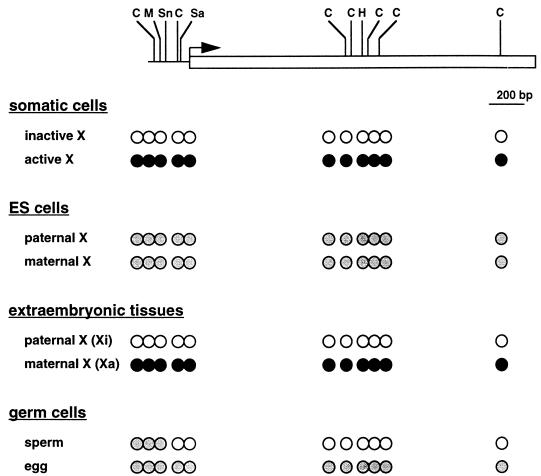
DNA methylation of the Xist gene in somatic cells correlates with its activity; the active Xist allele on the inactive X chromosome is unmethylated, whereas the inactive Xist allele on the active X chromosome is methylated (58, 123) (see also Fig. 6). Further evidence for the role of DNA methylation in the control of Xist expression in somatic cells is shown by studies with DNA MTase-deficient mice; the normally silent, and methylated, Xist allele on the single, active X chromosome in the male is induced to be expressed in the DNA MTase-deficient mice (8, 125).
FIG. 6.
DNA methylation of the Xist gene. The horizontal line indicates the Xist promoter region. The open rectangle indicates the 5′ portion of the exon 1. The hooked arrow denotes the transcription start site. The methylation-sensitive restriction enzyme sites so far tested are shown: C, HhaI (CfoI); M, MluI; Sn, SnaBI, Sa, SacII; H, HpaII. The methylation status of the methylation-sensitive restriction sites is shown. Open circles indicate lack of methylation; solid circles indicate complete methylation; shaded circles indicate mosaic methylation in that a particular site is methylated with a certain probability on each allele. It is hypothesized that the promoter and the 5′ end of exon 1 are mosaically methylated in eggs. Xi, inactive X chromosome; Xa, active X chromosome. Data for somatic cells and extraembryonic tissues are from reference 123; data for ES cells are from references 123 and 146; and data for germ cells are from references 4, 123, 124a, 168, and 169.
Xist expression in embryonic stem cells.
Since Xist is involved in early steps of random and imprinted XCI (98, 128) (see below), the patterns of Xist expression, with regard to the initiation of random XCI, have been studied in differentiating ES cells. Both X chromosomes are active in undifferentiated ES cells, and random XCI occurs when they are induced to differentiate (136). Even though both X chromosomes are active, reverse transcriptase PCR studies showed that Xist is expressed in undifferentiated ES cells. However, the level of expression is much lower than in somatic cells (8, 29, 73). RNA FISH analyses confirmed these findings and also demonstrated low-level Xist expression in XY ES cells, as in XX ES cells (125). In undifferentiated female (XX) ES cells, where Xist expression is very low, CpG sites in the promoter region and the 5′ end of the body of the Xist gene are mosaically methylated (146); i.e., any particular CpG site on each allele is methylated with a certain probability in a population of molecules of the Xist gene (Fig. 6).
When ES cells are induced to differentiate, Xist expression on the X chromosome chosen to remain active is silenced (down-regulated) whereas Xist expression on the X chromosome chosen to be inactive is enhanced (up-regulated) (8, 29, 73, 146). As differentiation proceeds, the mosaically methylated Xist alleles become methylated on the active X chromosome and unmethylated on the inactive X chromosome (8, 146). In male DNA MTase-deficient ES cells, the level of Xist expression is significantly increased during differentiation and is inversely correlated with the degree of methylation (8).
Xist expression in extraembryonic tissues.
In the extraembryonic tissues, Xist expression occurs only from the paternal allele (imprinted), and this correlates with preferential inactivation of the paternal X chromosome in these tissues (73) (see below). The active Xist allele on the inactive paternal X chromosome is unmethylated, whereas the inactive allele on the active maternal X chromosome is methylated (123) (Fig. 6).
Xist expression in germ cells.
Xist expression in germ cells is also correlated with the presence of the inactive X chromosome. In female primordial germ cells, Xist expression ceases at around 12.5 to 13.5 dpc (101), when the oocyte enters meiosis and the inactive X chromosome is reactivated (114). It has been hypothesized that the promoter region and the 5′ end of exon 1 of the Xist gene are methylated during oogenesis (4, 168; see below).
In male germ cells, Xist is expressed during spermatogenesis (101, 142, 147), when the single X chromosome is inactive, although the presence of the Xist gene and hence the expression of Xist do not appear to be required for silencing of the X chromosome in spermatogenesis (98) (see below).
Since the Xist gene itself is not required for production of viable, fertile sperm, the role of DNA methylation for the regulation of Xist expression in spermatogenesis may not be significant (although the methylation pattern created in spermatogenesis is, in fact, important for imprinted Xist expression in preimplantation embryos [see below]). It has been shown that Xist expression during spermatogenesis can be detected, by reverse transcriptase PCR, at as early as 3 days postpartum (dpp) (73) whereas demethylation of the promoter and the 5′ end of exon 1 is only detected between 9 and 21 dpp by Southern blot analysis (123). However, a later study involving a PCR-based methylation assay has shown that demethylation at the 5′ end of the first exon occurs even before birth, at between 18.5 and 21.5 dpc (4). The discrepancy with respect to the timing of demethylation detected by the two different methods (Southern blot and PCR-based methods) is, as described above, probably due to the difference in the purity of the samples and the sensitivity of the methods, and demethylation in spermatogenesis may be occurring throughout the perinatal period.
Xist expression in preimplantation embryos.
In mouse preimplantation embryos, Xist expression is imprinted in that the paternal allele is expressed whereas the maternal allele is repressed (73). This preferential paternal allele expression precedes preferential (imprinted) paternal X-inactivation in the extraembryonic tissues, suggesting a causal role for the early Xist expression in the imprinting of XCI. The mark for imprinted Xist expression is either erased or not acted upon when both the maternal and paternal alleles are expressed around the time of implantation, correlating with the timing of when random XCI occurs in the fetal precursor cells.
DNA methylation is considered to play a crucial role in the regulation of imprinted Xist expression in mouse preimplantation embryos. This is discussed below.
Mechanism of Imprinting of Mouse Xist
DNA methylation.
The imprinted Xist expression in preimplantation embryos must be controlled by a gametic imprint (differential marking in sperm and eggs), since analyses of androgenetic and gynogenetic embryos show Xist expression only in the androgenetic embryos (74, 84, 85). It has been proposed that DNA methylation is involved in the regulation of imprinted expression in preimplantation embryos (4, 168). The promoter region (168) and the 5′ end of the first exon (4) are differentially methylated at certain CpG sites, which are hypomethylated in sperm and methylated in oocytes (Fig. 6). More importantly, the methylation of the maternal Xist allele survives the genome-wide demethylation occurring in preimplantation development (4, 117, 168). This strongly suggests that the differential methylation in sperm and oocytes is a candidate gametic imprint for the repression of the maternal allele of Xist, leading to preferential paternal allele expression in preimplantation embryos. Later experiments have suggested that methylation of the promoter region in oocytes is mosaic, as seen for undifferentiated ES cells (169), and recent investigations involving bisulfite genomic sequencing supported these findings (124a) (Fig. 6).
Protein binding.
In keeping with the mosaic methylation of specific CpG sites in the Xist gene promoter in oocytes, Huntriss et al. (64) have recently identified a 100-kDa protein which binds to the mouse Xist promoter in a methylation-dependent and sequence-specific manner. This protein, identified in ES cell nuclear extracts, recognizes the GC-rich sequence 5′-GCGCCGCGG-3′ (nt −44 to −36 from the transcription start site) encompassing three CpG sites differentially methylated in sperm and oocytes [the HhaI (5′-GCGC-3′) and SacII (5′-CCGCGG-3′) sites in Fig. 6] and binds to this sequence only when it is methylated at at least one of the three CpG sites it contains (64). Moreover, this sequence is important for transcription in its unmethylated form (64). Therefore, it is hypothesized that this repressor protein is also present in preimplantation embryos and is responsible for imprinted Xist expression, i.e., repression of the maternal allele and expression of the paternal allele, by binding to the mosaically methylated promoter sequence of the maternal Xist allele. This hypothesis is further supported by comparison of the mouse and human Xist promoter sequences and of their expression patterns in preimplantation embryos, but this is discussed below.
Functional Studies of Xist and X-Chromosome Inactivation Using Xist Gene Deletions and Transgenesis
The role played by Xist in XCI has been extensively investigated in the past few years, although it is still unknown exactly how this gene functions.
Most reports on the role of XIST in humans are descriptive rather than the results of experimental manipulation. Migeon et al. (110) have shown that an X chromosome lacking the XIST locus due to a spontaneously occurring deletion is not inactivated, suggesting that XIST is required in cis for the X chromosome to be inactivated. However, it has also been reported that the continued presence of XIST is not required to maintain the inactivity of the X chromosome in leukemic cells of a female patient (133) and in cultured mouse/human somatic cell hybrids (22); both cases involve human X chromosomes which have lost chromosomal regions containing the XIST gene. Muscatelli et al. (122) have reported on a male patient whose single X chromosome has a duplication of the chromosomal region containing the Xic (where XIST is mapped). This X chromosome remains active, indicating that the duplicated XIST-containing chromosomal regions on the same X chromosome are not counted separately, since otherwise we would expect XCI to occur. This contrasts with the findings with Xist-containing yeast artificial chromosome (YAC) transgenes in mice, where multiple copies of transgene Xics seem to be counted as a separate Xic (61, 86, 87) (see below). It is possible that the duplicated region spanning the human XIST gene lacks an essential sequence required for XCI and therefore is not counted as two intact Xics.
In mice, more direct experimental strategies have been used to investigate the functional role of Xist; these include (i) experiments involving knockout (deletion) of the Xist gene and (ii) transgenic experiments with the Xist gene. The findings reported in published work are summarized in Tables 3 and 4, respectively.
TABLE 3.
Summary of Xist knockout experiments
| Study | Deletiona | Studied in: | Findings |
|---|---|---|---|
| Penny et al. (128) | 7 kb of exon 1 and 36-bp promoterb | ES cells and chimeric embryos | Differentiated ES cells: X bearing the mutant Xist remains active and X bearing the normal Xist is inactivated in 60% of cells; both Xs are active in the remaining cells |
| Chimeric embryos: X bearing the mutant Xist remains active, and X bearing the normal Xist is inactivated in the ES cell-derived cells | |||
| Marahrens et al. (98) | Exons 1–5c | Mice | Germ cells: male with the mutant Xist is able to produce viable, fertile sperm |
| Somatic tissues: X bearing the mutant Xist is not inactivated, but dosage compensated by inactivation of X bearing the normal Xist; cells with both Xs active may be eliminated during development. | |||
| Extraembryonic tissues: suggested that lethal when Xp bears the mutant Xist, possibly due to inability to inactivate Xp; viable when Xm bears the mutant Xist |
In both studies, the Xist gene is replaced with the neomycin resistance gene by homologous recombination.
An Xist deletion was created on 129-derived X chromosome in a female XX ES cell line (PGK12.1).
The deletion was created on 129-derived X chromosome in a male XY ES cell line (J1).
TABLE 4.
Summary of Xist transgenic experiments
| Study | Transgene | Integrationa | Copy no. | Studied in: | Findingsb |
|---|---|---|---|---|---|
| Lee et al. (86) | 450-kb YAC | Autosome in male ES cells | 3–24 | ES cells and fibroblasts | Tg Xist is expressed exclusively (53–76% of cells) or mutually (24–44% of cells) with endogenous Xist upon differentiation. Expression of Tg Xist represses the downstream reporter gene. Tg Xist RNA coats an autosomal region flanking the Tg Xist in cis. |
| Lee and Jaenisch (87) | 450-kb YAC | Chromosome 12 in male ES cells | 24 | ES cells and fibroblasts | Tg Xist RNA coats the autosomal region in cis. Tg Xist inactivates a region of 50 cM around the integration site and causes the Ch.12 Tg to be late replicated and hypoacetylated. |
| Herzing et al. (61) | 35-kb YAC | Autosome in male ES cells | 2–8 | ES cells | Endogenous Xist expression is induced in 30% of male differentiating ES cells. Expression of Tg Xist represses the downstream reporter gene. |
| Heard et al. (57) | 460-kb YAC | Autosome by pronuclear injection | 1–2 | Transgenic mice | Tg Xist is not expressed in males and females. Tg Xist is methylated. Reporter gene downstream of Tg Xist is expressed. |
| Matsuura et al. (99) | 350-kb YAC | Autosome or Y by pronuclear injection | 3–4 | Transgenic mice | Autosomal: Tg Xist is not expressed in males (female not tested) and is methylated. |
| Y-linked: Tg Xist is expressed, and some copies are hypomethylated. Mice are phenotypically normal. | |||||
| Hendrich et al. (60) (human XIST) | 0.2–1.2-kb promoter | Autosome by pronuclear injection | NRc | Transgenic mice | Tg Xist promoter is active in 9.5–13.5-dpc embryos in 9 of 11 lines to variable extents. DNA methylation was not investigated. |
| Goto et al. (51) | 233-bp promoter | Autosome by pronuclear injection | 1–71 | Transgenic mice | Tg Xist promoter is inactive and methylated in somatic tissues but active and hypomethylated in the testis. Activity of Tg Xist promoter in preimplantation embryos is correlated with the extent of methylation in sperm. |
In all studies, transgenes were integrated at a single site.
cM, centimorgan; Tg Xist, transgene Xist; Ch.12 Tg, chromosome 12 with the Xist transgene integrated.
NR, not reported.
Xist knockout mutations.
Two knockout experiments have been reported (98, 128) (summarized in Table 3). Penny et al. (128) deleted the minimal promoter region and the first exon of the Xist gene to create a null mutation in XX ES cells which are heterozygous for the Xce allele. They showed that the X chromosome bearing the null mutation does not undergo inactivation in the differentiating XX ES cells. Their results show that the intact Xist gene and/or Xist RNA is required in cis for the X chromosome to be inactivated. In addition, it would seem that the deletion (the promoter region and 7 kb of exon 1 of the Xist gene) does not contain the sequence used for counting, i.e., selection of one active X chromosome, since the counting function, if it were present in the deleted region, might be expected to select the one intact Xist locus on the normal X chromosome and choose this X chromosome to be active.
Marahrens et al. (98) replaced exons 1 to 5 of the Xist gene with the neomycin resistance gene, creating a null mutation but leaving the promoter region and the 5′ end of the first exon intact in male XY ES cells. In these cells, Xist RNA is not produced. However, it was confirmed, by RNA FISH with a probe hybridizing with the intact region of the first exon, that the remaining promoter was functional in the transcription of truncated Xist-neomycin resistance gene hybrid RNA and to a similar extent to that in wild-type XY ES cells. These authors have shown that (i) the Xist-deficient X chromosome is not inactivated; (ii) the Xist structural gene is not required to produce viable, fertile sperm, and (iii) paternal transmission of the mutated Xist is lethal, suggesting that Xist is necessary in cis for the paternally inherited X chromosome to be inactivated in the extraembryonic tissues.
The results of Marahrens et al. (98) also suggest that the action of Xist is, in fact, mediated in cis by the transcribed Xist RNA and that the mere act of transcription at the Xist locus is not sufficient. This contrasts with a report of deletion of the H19 gene, in which transcription of either the endogenous H19 structural gene, or the replaced luciferase reporter gene, driven by the intact H19 promoter results in the normal pattern of imprinting of the neighboring Igf2 gene (148).
Xist-containing transgenes.
Transgene experiments (summarized in Table 4) have also supported a significant role for Xist in XCI. In male (XY karyotype) ES cells, an autosomally integrated 450-kb Xist-containing YAC shows some properties of the Xic in that the YAC transgene can inactivate limited chromosomal regions surrounding its integration site (86, 87). However, the inactivation spreads only up to 50 centimorgans from the integration site (87). Again with male ES cells, Herzing et al. (61) have shown that an autosomally integrated YAC, which contains only the Xist gene and 9 kb of 5′- and 6 kb of 3′-flanking sequences, is sufficient to show the properties of the Xic in that the transgene can inactivate a downstream reporter gene in the YAC construct. In both studies, multiple copies of the YAC transgenes are integrated in an autosome in male XY ES cells (Table 4). The level of Xist expression correlates with the copy number in the study of Lee et al. (86) but not in that of Herzing et al. (61). The difference may be explained by the actual copy numbers (3 to 24 versus 2 to 8 copies) and the sizes and structures of the YAC transgenes (450 versus 35 kb) (Table 4). It is possible that a larger transgene unit will be better insulated from the regulation on transcription imposed by the surrounding sequences.
Lee et al. (86) and Herzing et al. (61) have both reported that the Xist-containing YAC transgenes used in their studies also contain the information required for counting because the endogenous Xist allele on the single, active X chromosome in the XY ES cells is chosen to be up-regulated in a proportion of the cells following induction of differentiation. Lee et al. (86) have reported that the YAC transgene induces cell death in the male ES cells, probably due to activation of the endogenous Xist gene and inactivation of the single X chromosome in these cells.
Transgenic mice harboring Xist-containing YAC transgenes have been created by Heard et al. (57) and Matsuura et al. (99) (Table 4). Heard et al. (57) reported that the Xist gene in an autosomally integrated 460-kb YAC transgene construct is not expressed but that a lacZ reporter gene, which is ligated downstream of the Xist in the YAC, is expressed. They concluded that essential sequences for Xist expression might be absent in their construct and therefore that the construct could not cause inactivation of the region surrounding the integration site. Similarly, Matsuura et al. (99) reported that the Xist gene in an autosomally integrated 350-kb YAC transgene construct is not expressed. However, in one transgenic line, in which a 240-kb YAC transgene (due to spontaneous deletion of the 110-kb 3′-region of the 350-kb YAC) is integrated in a heterochromatic region of the long arm of the Y chromosome, the Xist gene is expressed. This indicates that the 240-kb Xist-containing YAC transgene has the information required for expression of the Xist gene in vivo in certain circumstances. This contradicts the conclusion by Heard et al. (57). These large Xist-containing YAC transgenes include the 9-kb 5′-flanking and 6-kb 3′-flanking sequences shown to be sufficient for initiating inactivation of the region surrounding the autosomal integration site in differentiating ES cells (61), but neither expression of the transgene Xist nor inactivation of the surrounding autosomal region was observed with the autosomally integrated transgenes in adult mice.
The differences between the results obtained with ES cells by Lee et al. (86) and Herzing et al. (61) and those obtained with adult transgenic mice by Heard et al. (57) and Matsuura et al. (99) may be explained by the differences in the copy number and/or by the differences in the methods of introduction of the YACs into ES cells and oocytes. For example, pronuclear microinjection of a large YAC transgene may alter the nature of the Xist gene and its flanking sequences, resulting in permanent silencing of Xist. It is also possible that in the transgenic mice, the cells in which the transgene expresses Xist and initiates inactivation of the autosomal genes around the integration site were eliminated or that fetuses where this occurred died in utero. In the case of the Y-linked transgene (99), which is the only YAC transgene expressed in transgenic mice, the relatively smaller size (240 kb) and/or the structure of the integration site may have allowed expression. Alternatively, the region around the integration site may not contain essential genes.
Although these knockout and YAC transgene studies indicate that the Xist gene and the 15 kb of surrounding sequences have some properties of the Xic, i.e., counting, spreading, and maintenance, it remains to be investigated how Xist expression itself is regulated. To this end, further transfection and transgene studies have been carried out with Xist promoter-reporter gene constructs (see below).
Xist promoter methylation and function.
Deletion and mutation analyses of Xist promoter-reporter gene constructs in transfection assays have defined the Xist minimal promoter sequence: nucleotide (nt) sequences −81 to +1 for the mouse Xist (129) and −93 to +31 for the human XIST (60). Both the mouse Xist and the human XIST minimal promoters contain the binding sequences for ubiquitous transcription factors, TATA box-binding protein, and Sp1 (60, 79, 129). An additional binding site for the initiation protein YY1 has also been identified within the human XIST minimal promoter (60). Transient transfection assays have shown that the Xist minimal promoter is active to a similar extent in male and female cultured somatic cells (60, 129). These results indicate that repression of Xist (on the single active X chromosome) in male somatic cells is not due to the absence of transcription factors but is due to the presence of some mechanism to silence the Xist gene.
We have investigated a 233-bp fragment of the mouse Xist promoter (nt position −220 to +13) by using a reporter construct, incorporating the minimal promoter sequence linked to the firefly luciferase gene in both microinjected mouse embryos and transgenic mice (51). To investigate the role of DNA methylation in Xist expression, we have created six lines of transgenic mice (Table 4). In all six lines, the 233-bp promoter fragment is inactive and methylated in somatic tissues in both males and females. However, in the male germ cells in all six lines, the promoter transgene is active and hypomethylated, although the degree of hypomethylation is variable among the lines.
Hendrich et al. (60) have recently reported a transgenic-mouse study of three different human XIST promoter fragments (nt −1187 to +31, −221 to +31, and −211 to +767) linked to the luciferase reporter gene. They found that the promoter fragments are active to variable extents in both male and female 9.5- to 13.5-dpc embryos in 9 of 11 transgenic lines. Their results contrast with those obtained with the mouse Xist transgenes by Goto et al. (51) and others (57, 99) in that the human XIST promoter transgenes are active in most transgenic mouse lines whereas the mouse Xist transgenes are inactive in all cases of autosomally integrated transgenes (51, 57, 99). It is possible that the presence of the exogenous (transgene) mouse Xist promoter interferes with some function of the endogenous mouse Xist promoter in development, and therefore that the transgene Xist promoter is preferentially inactivated and methylated (51, 57, 99) or cells with an active transgene Xist promoter are eliminated during development. In the case of the human XIST promoter transgene in transgenic mice, such an interference may not occur between the human XIST promoter and the mouse Xist promoter.
In our study, we specifically addressed whether imprinted expression of the endogenous Xist gene in preimplantation embryos is controlled by DNA methylation (51). In microinjection experiments, we have shown that the 233-bp Xist promoter fragment (−220 to +13) is sufficient to drive transcription of the luciferase reporter gene at the two-cell stage when microinjected into one-cell embryos. This promoter activity is repressed by in vitro methylation of the construct with site-specific DNA methylases prior to microinjection, consistent with the hypothesis that DNA methylation regulates the expression of the endogenous Xist gene in preimplantation embryos. To further investigate the role of DNA methylation as a gametic imprint for imprinted Xist expression, the activity and degree of methylation of the transgene were studied in the male germ cells of the testis and in morula-stage embryos in the established transgenic mouse lines. We have shown that the degree of methylation of the transgene Xist promoter in the germ cells (and sperm) does not correlate with the promoter activity in these cells in the testis (the promoter is active even when it is methylated) but does correlate with the activity in morula-stage embryos following transmission of the transgene from the father. Only when the transgene is extensively hypomethylated in sperm is it active, after fertilization, in transgenic morulae (51). In addition, we have found that certain CpG sites within the transgene Xist promoter are more resistant to the genome-wide demethylation occurring between the eight-cell and blastocyst stages than are neighboring CpG sites, suggesting that methylation of CpG sites within the transgene Xist promoter can be locally and independently regulated. Significantly, one of the transgene promoter CpG sites surviving the genome-wide demethylation is present in the sequence 5′-GCGCCGCGG-3′ (−44 to −36), which is differentially methylated in sperm and eggs (168) and whose methylation is recognized by the methylation-dependent repressor protein identified by Huntriss et al. (64). These results strongly support our hypothesis that the gametic methylation pattern governs the imprinted expression of the endogenous Xist gene in preimplantation embryos.
Human XIST Regulation in Comparison with Mouse Xist Regulation
Although the mouse and human Xist genes are positionally and structurally conserved, it is not yet known whether the two genes share all aspects of their functions in XCI. For example, the continued presence of the human XIST gene is not required for maintenance of the inactivity of the established X chromosome (22, 133), but it is not known whether this is also the case in mice. A transgene containing the mouse Xist gene and a 15-kb flanking sequence, integrated on an autosome, is counted as a separate Xic in differentiating ES cells (61), but the duplicated XIST-containing chromosomal regions, on the same X chromosome, are not counted separately in humans (122).
A most striking difference in the characteristics of the mouse and human Xist genes is their expression patterns in preimplantation embryos. In mouse preimplantation embryos, Xist expression is imprinted, with the paternal allele being preferentially expressed (73). By contrast, in human preimplantation embryos, XIST expression is not demonstrably imprinted, in that both the paternally and maternally inherited alleles are expressed (40, 137). As to the molecular basis underlying imprinted mouse Xist expression in preimplantation embryos, we have strong evidence that the promoter sequence 5′-GCGCCGCGG-3′ (−44 to −36) is one of the key CpG sites, whose methylation in gametes controls the promoter activity in preimplantation embryos (51, 64, 168) (see above). In humans, where XIST expression in preimplantation embryos is not imprinted (40, 137), this mouse promoter sequence is not conserved but is replaced by 5′-GCCCCCCCT-3′ (−45 to −37), thus containing no methylatable CpG sites (60) (for detailed sequence comparison, refer to reference 60).
Plenge et al. (131) have recently reported that a single-base-pair mutation within this sequence, from 5′-GCCCCCCCT-3′ to 5′-GCGCCCCCT-3′, in a short XIST promoter-reporter construct causes a two- to fivefold decrease in promoter activity in a transfection assay and that the X chromosome carrying the mutation is preferentially inactivated (skewed XCI) in somatic cells of females from two unrelated families. Their results indicate that this promoter sequence in humans not only is required for normal transcription but also affects some step(s) (choice, initiation, spreading or maintenance) of XCI in development.
In mice, Huntriss et al. (64) have shown that mutations of the corresponding mouse sequence, in its unmethylated form, similarly result in a three- to fivefold reduction in promoter activity in cultured cells. Given the result in humans as obtained by Plenge et al. (131), it is not difficult to see that a change in promoter activity, induced by either base pair mutation or DNA methylation, affects the XCI process in mouse development.
The role played by human XIST in preferential paternal XCI in the extraembryonic tissues is not clear. Since XIST expression in human preimplantation embryos is not detectably imprinted, it is not yet known whether preferential paternal XCI in the human extraembryonic tissues is preceded by preferential expression of the paternal XIST allele, as it is in mice (40, 50, 56, 137).
CONCLUSION AND FUTURE DIRECTIONS
We describe fundamental features of XCI and recent advances in molecular analyses and characterization of the Xic, with particular emphasis on the Xist gene. The process of XCI provides an excellent model system for investigation of the regulation of gene activity. XCI shows a variety of features of gene regulation, such as DNA methylation, histone modification, replication timing, formation of inactive chromatin structure, compartmentalization, and genomic imprinting. Studies on the Xist gene will eventually shed light on the mechanisms of XCI. Future studies will involve the identification of functionally important regions of the Xist gene and Xist RNA, dissection of genomic loci surrounding the Xist gene within and around the Xic, characterization of higher-order gene structure around the Xist gene, and elucidation of the subnuclear organization of the inactive X chromosome, in association with Xist RNA, the nuclear membrane, and as yet unidentified proteins, within the nucleus.
ACKNOWLEDGMENTS
We are grateful to present and former members of the Molecular Embryology Unit, Institute of Child Health, particularly Maurizio Zuccotti, Roberto Lorenzi, Alan Thornhill, Elizabeth Wright, Elisabeth Christians, Robert Daniels, John Huntriss, Amarjit Purewal, Enrique Perez, and James Adjaye, for helpful comments and discussions.
REFERENCES
- 1.Adams R L P, Burdon R H, McKinnon K, Rinaldi A. Stimulation of de novo methylation following limited proteolysis of mouse ascites DNA methylase. FEBS Lett. 1983;163:194–198. doi: 10.1016/0014-5793(83)80817-5. [DOI] [PubMed] [Google Scholar]
- 2.Adams R L P. Eukaryotic DNA methyltransferases—structure and function. Bioessays. 1995;17:139–145. doi: 10.1002/bies.950170209. [DOI] [PubMed] [Google Scholar]
- 3.Antequera F, Boyes J, Bird A P. High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell. 1990;62:503–514. doi: 10.1016/0092-8674(90)90015-7. [DOI] [PubMed] [Google Scholar]
- 4.Ariel M, Robinson E, McCarrey J R, Cedar H. Gamete-specific methylation correlates with imprinting of the murine Xist gene. Nat Genet. 1995;9:312–315. doi: 10.1038/ng0395-312. [DOI] [PubMed] [Google Scholar]
- 5.Barlow D P, Stoger R, Herrmann B G, Saito K, Schweiter N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349:84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- 6.Barr M L, Bertram E G. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature. 1949;163:676–677. doi: 10.1038/163676a0. [DOI] [PubMed] [Google Scholar]
- 7.Bartolomei M S, Zemei S, Tilghman S M. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–155. doi: 10.1038/351153a0. [DOI] [PubMed] [Google Scholar]
- 8.Beard C, Li E, Jaenisch R. Loss of methylation activates Xist in somatic but not in embryonic cells. Genes Dev. 1995;9:2325–2334. doi: 10.1101/gad.9.19.2325. [DOI] [PubMed] [Google Scholar]
- 9.Bestor T H, Ingram V M. Two DNA methyltransferase from murine erythroleukemia cells: purification, sequence specificity, and mode of interaction with DNA. Proc Natl Acad Sci USA. 1983;80:5559–5563. doi: 10.1073/pnas.80.18.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bestor T H, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding DNA methyltransferase of mouse cells. J Mol Biol. 1988;203:971–983. doi: 10.1016/0022-2836(88)90122-2. [DOI] [PubMed] [Google Scholar]
- 11.Bestor T H. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 1992;11:2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bird A. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 13.Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 14.Bloom D C, Hill J M, Devi-Rao G, Wagner E K, Feldman L T, Stevens J G. A 348-base-pair region in the latency-associated transcript facilitates herpes simplex virus type 1 reactivation. J Virol. 1996;70:2449–2459. doi: 10.1128/jvi.70.4.2449-2459.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borsani G, Tonlorenzi R, Simmler M C, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, Willard H F, Avner P, Ballabio A. Characterization of a murine gene expressed from the inactive X chromosome. Nature. 1991;351:325–329. doi: 10.1038/351325a0. [DOI] [PubMed] [Google Scholar]
- 16.Brandeis M, Kafri T, Ariel M, Chaillet J R, McCarrey J, Razin A, Cedar H. The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J. 1993;12:3669–3677. doi: 10.1002/j.1460-2075.1993.tb06041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brockdorff N, Ashworth A, Kay G F, Cooper P, Smith S, McCabe V M, Norris D P, Penny G D, Patel D, Rastan S. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature. 1991;351:329–331. doi: 10.1038/351329a0. [DOI] [PubMed] [Google Scholar]
- 18.Brockdorff N, Ashworth A, Kay G F, McCabe V M, Norris D P, Cooper P J, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and localized in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- 19.Brown C J, Ballabio A, Rupert J L, Lafreniere R G, Grompe M, Tonlorenzi R, Willard H F. A gene from the region of the human X inactivation center is expressed exclusively from the inactive X chromosome. Nature. 1991;349:38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 20.Brown C J, Lafreniere R G, Powers V E, Sebastio G, Ballabio A, Pettigrew A L, Ledbetter D H, Levy E, Craig I W, Willard H F. Localization of the X inactivation centre on the human X chromosome in Xq13. Nature. 1991;349:82–84. doi: 10.1038/349082a0. [DOI] [PubMed] [Google Scholar]
- 21.Brown C J, Hendrich B D, Rupert J L, Lafreniere R G, Xing Y, Lawrence J, Willard H F. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 22.Brown C J, Willard H F. The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature. 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- 23.Brown C J, Baldry S E L. Evidence that heteronuclear proteins interact with XIST RNA in vitro. Somatic Cell Mol Genet. 1996;22:403–417. doi: 10.1007/BF02369896. [DOI] [PubMed] [Google Scholar]
- 24.Burgoyne P S, Biggers J D. The consequences of X-dosage deficiency in the germ line: impaired development in vitro of preimplantation embryos from XO mice. Dev Biol. 1976;51:109–117. doi: 10.1016/0012-1606(76)90126-3. [DOI] [PubMed] [Google Scholar]
- 25.Burgoyne P S, Baker T G. Oocyte depletion in XO mice and their XX sibs from 12 to 200 days post coitum. J Reprod Fertil. 1981;61:207–212. doi: 10.1530/jrf.0.0610207. [DOI] [PubMed] [Google Scholar]
- 26.Burgoyne P S, Evans E P, Holland K. XO monosomy is associated with reduced birthweight and lowered weight gain in the mouse. J Reprod Fertil. 1983;68:381–385. doi: 10.1530/jrf.0.0680381. [DOI] [PubMed] [Google Scholar]
- 27.Burgoyne P S, Tam P P L, Evans E P. Retarded development of XO conceptuses during early pregnancy in the mouse. J Reprod Fertil. 1983;68:387–393. doi: 10.1530/jrf.0.0680387. [DOI] [PubMed] [Google Scholar]
- 28.Buschhausen G, Wittig B, Graessmann M, Graessmann A. Chromatin structure is required to block transcription of the methylated herpes simplex thymidine kinase gene. Proc Natl Acad Sci USA. 1987;84:1177–1181. doi: 10.1073/pnas.84.5.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buzin C H, Mann J R, Singer-Sam J. Quantitative RT-PCR assays show Xist RNA levels are low in mouse female adult tissues, embryos and embryoid bodies. Development. 1994;120:3529–3536. doi: 10.1242/dev.120.12.3529. [DOI] [PubMed] [Google Scholar]
- 30.Cattanach B M, Pollard C E, Perez J N. Controlling elements in the mouse X chromosome. I. Interaction with the X-linked genes. Genet Res Camb. 1969;14:223–235. doi: 10.1017/s0016672300002068. [DOI] [PubMed] [Google Scholar]
- 31.Cattanach B M, Perez J N, Pollard C E. Controlling elements in the mouse X chromosome. II. Location in the linkage map. Genet Res Camb. 1970;15:189–195. doi: 10.1017/s0016672300001518. [DOI] [PubMed] [Google Scholar]
- 32.Cattanach B M, Williams C E. Evidence of non-random X chromosome activity in the mouse. Genet Res Camb. 1972;19:220–240. doi: 10.1017/s001667230001449x. [DOI] [PubMed] [Google Scholar]
- 33.Cattanach B M, Kirk M. Differential activity of maternally and paternally derived chromosome regions in mice. Nature. 1985;315:496–498. doi: 10.1038/315496a0. [DOI] [PubMed] [Google Scholar]
- 34.Cattanach B M, Rasberry C. Identification of the Mus spretus Xce allele. Mouse Genome. 1991;89:565–566. [Google Scholar]
- 35.Cattanach B M, Jones J. Genetic imprinting in the mouse: implications for gene regulation. J Inherited Metab Dis. 1994;17:403–420. doi: 10.1007/BF00711356. [DOI] [PubMed] [Google Scholar]
- 36.Clemson C M, McNeil J A, Willard H F, Lawrence J B. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collick A, Reik W, Barton S C, Surani M A H. CpG methylation of an X-linked transgene is determined by somatic events postfertilization and not germline imprinting. Development. 1988;104:235–244. doi: 10.1242/dev.104.2.235. [DOI] [PubMed] [Google Scholar]
- 38.Dahl H-H, Brown R M, Hutchinson W M, Maragos C, Brown G K. A testis-specific form of the human pyruvate dehydrogenase E1α subunit is coded for by an intronless gene on chromosome 4. Genomics. 1990;8:225–232. doi: 10.1016/0888-7543(90)90275-y. [DOI] [PubMed] [Google Scholar]
- 39.Dandolo L, Stewart C L, Mattei M-G, Avner P R. Inactivation of an X-linked transgene in murine extraembryonic and adult tissues. Development. 1993;118:641–649. doi: 10.1242/dev.118.2.641. [DOI] [PubMed] [Google Scholar]
- 40.Daniels R, Zuccotti M, Kinis T, Serhal P, Monk M. XIST expression in human oocytes and preimplantation embryos. Am J Hum Genet. 1997;61:33–39. doi: 10.1086/513892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DeChiara T M, Robertson E J, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- 42.Disteche C M. Escape from X inactivation in human and mouse. Trends Genet. 1995;11:17–22. doi: 10.1016/s0168-9525(00)88981-7. [DOI] [PubMed] [Google Scholar]
- 43.Edwards Y H. CpG islands in genes showing tissue-specific expression. Philos Trans R Soc London Ser B. 1990;326:207–215. doi: 10.1098/rstb.1990.0005. [DOI] [PubMed] [Google Scholar]
- 44.Epstein C J, Smith S, Travis B, Tucker G. Both X chromosomes function before visible X-chromosome inactivation in female mouse embryos. Nature. 1978;274:500–503. doi: 10.1038/274500a0. [DOI] [PubMed] [Google Scholar]
- 45.Frels W I, Chapman V M. Paternal X chromosome expression in extraembryonic membranes of XO mice. J Exp Zool. 1979;210:553–560. doi: 10.1002/jez.1402100319. [DOI] [PubMed] [Google Scholar]
- 46.Frels W I, Rossant J, Chapman V M. Maternal X chromosome expression in mouse chorionic ectoderm. Dev Genet. 1979;1:123–132. [Google Scholar]
- 47.Frels W I, Chapman V M. Expression of the maternally derived X chromosome in the mural trophoblast of the mouse. J Embryol Exp Morphol. 1980;56:179–190. [PubMed] [Google Scholar]
- 48.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 49.Goldman M A, Stokes K R, Idezerda R L, McKnight G S, Hammer R E, Brinster R L, Gartler S M. A chicken transferrin gene in transgenic mice escapes X-chromosome inactivation. Science. 1987;236:593–595. doi: 10.1126/science.2437652. [DOI] [PubMed] [Google Scholar]
- 50.Goto T, Wright E, Monk M. Paternal X-chromosome inactivation in human trophoblastic cells. Mol Hum Reprod. 1997;3:77–80. doi: 10.1093/molehr/3.1.77. [DOI] [PubMed] [Google Scholar]
- 51.Goto T, Christians E, Monk M. Expression of an Xist promoter-luciferase construct during spermatogenesis and in preimplantation embryos—regulation by DNA methylation. Mol Reprod Dev. 1998;49:356–367. doi: 10.1002/(SICI)1098-2795(199804)49:4<356::AID-MRD2>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 52.Grant M, Zuccotti M, Monk M. Methylation of CpG sites of two X-linked genes coincides with X-inactivation in the female mouse embryo but not in the germ line. Nat Genet. 1992;2:161–166. doi: 10.1038/ng1092-161. [DOI] [PubMed] [Google Scholar]
- 53.Graves J A M, Gartler S M. Mammalian X chromosome inactivation: testing the hypothesis of transcriptional control. Somatic Cell Mol Genet. 1986;12:275–280. doi: 10.1007/BF01570786. [DOI] [PubMed] [Google Scholar]
- 54.Gruenbaum Y, Cedar H, Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982;295:620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- 55.Harper M I, Fosten M, Monk M. Preferential paternal X inactivation in extraembryonic tissues of early mouse embryos. J Embryol Exp Morphol. 1982;67:127–135. [PubMed] [Google Scholar]
- 56.Harrison K B. X-chromosome inactivation in the human cytotrophoblast. Cytogenet Cell Genet. 1989;52:37–41. doi: 10.1159/000132835. [DOI] [PubMed] [Google Scholar]
- 57.Heard E, Kress C, Mongelard F, Courtier B, Rougeulle C, Ashworth A, Vourc’h C, Babinet C, Avner P. Transgenic mice carrying an Xist-containing YAC. Hum Mol Genet. 1996;5:441–450. doi: 10.1093/hmg/5.4.441. [DOI] [PubMed] [Google Scholar]
- 58.Hendrich B D, Brown C J, Willard H F. Evolutionary conservation of possible functional domains of the human and murine XIST genes. Hum Mol Genet. 1993;6:663–672. doi: 10.1093/hmg/2.6.663. [DOI] [PubMed] [Google Scholar]
- 59.Hendrich B D, Willard H F. Epigenetic regulation of gene expression: the effect of altered chromatin structure from yeast to mammals. Hum Mol Genet. 1995;4:1765–1777. doi: 10.1093/hmg/4.suppl_1.1765. [DOI] [PubMed] [Google Scholar]
- 60.Hendrich B D, Plenge R M, Willard H F. Identification and characterization of the human XIST gene promoter: implications for models of X chromosome inactivation. Nucleic Acids Res. 1997;25:2661–2671. doi: 10.1093/nar/25.13.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Herzing L B K, Romer J T, Horn J M, Ashworth A. Xist has properties of the X-chromosome inactivation centre. Nature. 1997;386:272–275. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- 62.Holmquist G P. Role of replication time in the control of tissue-specific gene expression. Am J Hum Genet. 1987;40:151–173. [PMC free article] [PubMed] [Google Scholar]
- 63.Huang L-H, Wang R, Gama-Sosa M A, Shenoy S, Ehrlich M. A protein from human placental nuclei binds preferentially to 5-methylcytosine-rich DNA. Nature. 1984;308:293–295. doi: 10.1038/308293a0. [DOI] [PubMed] [Google Scholar]
- 64.Huntriss J, Lorenzi R, Purewal A, Monk M. A methylation-dependent DNA-binding activity recognising the methylated promoter region of the mouse Xist gene. Biochem Biophys Res Commun. 1997;235:730–738. doi: 10.1006/bbrc.1997.6876. [DOI] [PubMed] [Google Scholar]
- 65.Jeppesen P, Turner B M. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- 66.Johnston P G. X chromosome activity in female germ cells of mice heterozygous for Searle’s translocation T(X;16)16H. Genet Res Camb. 1981;37:317–322. doi: 10.1017/s0016672300020322. [DOI] [PubMed] [Google Scholar]
- 67.Johnston P G, Cattanach B M. Controlling elements in the mouse. IV. Evidence of non-random X-inactivation. Genet Res Camb. 1981;37:151–160. doi: 10.1017/s0016672300020127. [DOI] [PubMed] [Google Scholar]
- 68.Jost J-P, Hofsteenge J. The repressor MDBP-2 is a member of the histone H1 family that binds preferentially in vitro and in vivo methylated nonspecific DNA sequences. Proc Natl Acad Sci USA. 1992;89:9499–9503. doi: 10.1073/pnas.89.20.9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kafri T, Ariel M, Brandeis M, Shemer R, Urven L, McCarrey J, Cedar H, Razin A. Developmental pattern of gene-specific DNA methylation in the mouse embryo and germ line. Genes Dev. 1992;6:705–714. doi: 10.1101/gad.6.5.705. [DOI] [PubMed] [Google Scholar]
- 70.Kafri T, Gao X, Razin A. Mechanistic aspects of genome-wide demethylation in the preimplantation mouse embryo. Proc Natl Acad Sci USA. 1993;90:10558–10562. doi: 10.1073/pnas.90.22.10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kajii T, Ohama K. Androgenetic origin of hydatidiform mole. Nature. 1977;268:633–634. doi: 10.1038/268633a0. [DOI] [PubMed] [Google Scholar]
- 72.Kaufman M H, Barton S C, Surani M A H. Normal postimplantation development of mouse parthenogenetic embryos to the forelimb bud stage. Nature. 1977;265:53–55. doi: 10.1038/265053a0. [DOI] [PubMed] [Google Scholar]
- 73.Kay G F, Penny G D, Patel D, Ashworth A, Brockdorff N, Rastan S. Expression of Xist during mouse development suggests a role in the initiation of X chromosome inactivation. Cell. 1993;72:171–182. doi: 10.1016/0092-8674(93)90658-d. [DOI] [PubMed] [Google Scholar]
- 74.Kay G F, Barton S C, Surani M A, Rastan S. Imprinting and X chromosome counting mechanisms determine Xist expression in early mouse development. Cell. 1994;77:639–650. doi: 10.1016/0092-8674(94)90049-3. [DOI] [PubMed] [Google Scholar]
- 75.Keshet I, Yisraeli J, Cedar H. Effect of regional DNA methylation on gene expression. Proc Natl Acad Sci USA. 1985;82:2560–2564. doi: 10.1073/pnas.82.9.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Keshet I, Lieman-Hurwitz J, Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986;44:535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- 77.Khan R, Zhang X-Y, Supakar P C, Ehrlich K C, Ehrlich M. Human methylated DNA-binding protein: determinants of a pBR322 recognition site. J Biol Chem. 1988;263:14374–14383. [PubMed] [Google Scholar]
- 78.Kloc M, Spohr G, Etkin L D. Transcription of repetitive RNA sequences with the germ plasm in Xenopus oocytes. Science. 1993;262:1712–1714. doi: 10.1126/science.7505061. [DOI] [PubMed] [Google Scholar]
- 79.Komura J, Sheardown S A, Brockdorff N, Singer-Sam J, Riggs A D. In vitro ultraviolet and dimethyl sulfate footprinting of the 5′ region of the expressed and silent Xist alleles. J Biol Chem. 1997;272:10975–10980. doi: 10.1074/jbc.272.16.10975. [DOI] [PubMed] [Google Scholar]
- 80.Kratzer P G, Gartler S M. HGPRT activity changes in preimplantation mouse embryos. Nature. 1978;274:503–504. doi: 10.1038/274503a0. [DOI] [PubMed] [Google Scholar]
- 81.Kratzer P G, Chapman V M. X chromosome reactivation in oocytes of Mus caroli. Proc Natl Acad Sci USA. 1981;78:3093–3097. doi: 10.1073/pnas.78.5.3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krumlauf R, Chapman V M, Hammer R E, Brinster R, Tilghman S M. Differential expression of α-fetoprotein genes on the inactive X chromosome in extraembryonic and somatic tissues of a transgenic mouse line. Nature. 1986;319:224–226. doi: 10.1038/319224a0. [DOI] [PubMed] [Google Scholar]
- 83.Laird P W, Jaenisch R. DNA methylation and cancer. Hum Mol Genet. 1994;3:1487–1495. doi: 10.1093/hmg/3.suppl_1.1487. [DOI] [PubMed] [Google Scholar]
- 84.Latham K E, Doherty A S, Scott C D, Schultz R M. Igf2r and Igf2 gene expression in androgenetic, gynogenetic, and parthenogenetic preimplantation mouse embryos: absence of regulation by genomic imprinting. Genes Dev. 1994;8:290–299. doi: 10.1101/gad.8.3.290. [DOI] [PubMed] [Google Scholar]
- 85.Latham K E, Rambhatla L. Expression of X-linked genes in androgenetic, gynogenetic, and normal mouse preimplantation embryos. Dev Genet. 1995;17:212–222. doi: 10.1002/dvg.1020170306. [DOI] [PubMed] [Google Scholar]
- 86.Lee J T, Strauss W M, Dausman J A, Jaenisch R. A 450 kb transgene displays properties of the mammalian X-inactivation center. Cell. 1996;86:83–94. doi: 10.1016/s0092-8674(00)80079-3. [DOI] [PubMed] [Google Scholar]
- 87.Lee J T, Jaenisch R. Long-range cis effects of ectopic X-inactivation centres on a mouse antosome. Nature. 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- 88.Leonhardt H, Page A W, Weier H-U, Bestor T H. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- 89.Lewis J, Bird A. DNA methylation and chromatin structure. FEBS Lett. 1991;285:155–159. doi: 10.1016/0014-5793(91)80795-5. [DOI] [PubMed] [Google Scholar]
- 90.Lewis J D, Meehan R R, Henzel W J, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 91.Li E, Bestor T H, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 92.Li E, Beard C, Forster A C, Bestor T H, Jaenisch R. DNA methylation, genomic imprinting, and mammalian development. Cold Spring Harbor Symp Quant Biol. 1993;58:297–305. doi: 10.1101/sqb.1993.058.01.035. [DOI] [PubMed] [Google Scholar]
- 93.Li E, Beard C, Jaenisch R. Role for DNA methylation in genomic imprinting. Nature. 1993;366:362–365. doi: 10.1038/366362a0. [DOI] [PubMed] [Google Scholar]
- 94.Lock L, Takagi N, Martin G R. Methylation of the Hprt gene on the inactive X occurs after chromosome inactivation. Cell. 1987;48:39–46. doi: 10.1016/0092-8674(87)90353-9. [DOI] [PubMed] [Google Scholar]
- 95.Lucchesi J C, Manning J E. Gene dosage compensation in Drosophila melanogaster. Adv Genet. 1987;24:371–429. doi: 10.1016/s0065-2660(08)60013-9. [DOI] [PubMed] [Google Scholar]
- 96.Lyon M F. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 97.Mann J R, Lovell-Badge R H. Inviability of parthenogenones is determined by pronuclei, not egg cytoplasm. Nature. 1984;310:66–67. doi: 10.1038/310066a0. [DOI] [PubMed] [Google Scholar]
- 98.Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 99.Matsuura S, Episkopou V, Hamvas R, Brown S D M. Xist expression from an Xist YAC transgene carried on the mouse Y chromosome. Hum Mol Genet. 1996;5:451–459. doi: 10.1093/hmg/5.4.451. [DOI] [PubMed] [Google Scholar]
- 100.McBurney M W, Staines W A, Boekeleheide K, Parry D, Jardine K, Pickavance L. Murine Pgk-1 promoter drives widespread but not uniform expression in transgenic mice. Dev Genet. 1994;200:278–293. doi: 10.1002/aja.1002000403. [DOI] [PubMed] [Google Scholar]
- 101.McCarrey J R, Dilworth D D. Expression of Xist in mouse germ cells correlates with X-chromosome inactivation. Nat Genet. 1992;2:200–203. doi: 10.1038/ng1192-200. [DOI] [PubMed] [Google Scholar]
- 102.McCarrey J R, Berg W M, Paragioudakis S J, Zhang P L, Dilworth D D, Arnold B L, Rossi J J. Differential transcription of Pgk genes during spermatogenesis in the mouse. Dev Biol. 1992;154:160–168. doi: 10.1016/0012-1606(92)90056-m. [DOI] [PubMed] [Google Scholar]
- 103.McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- 104.McMahon A, Fosten M, Monk M. Random X-chromosome inactivation in female primordial germ cells in the mouse. J Embryol Exp Morphol. 1981;64:251–258. [PubMed] [Google Scholar]
- 105.McMahon A, Fosten M, Monk M. X-chromosome inactivation mosaicism in the three germ layers and the germ line of the mouse embryo. J Embryol Exp Morphol. 1983;74:207–220. [PubMed] [Google Scholar]
- 106.Meehan R R, Lewis J D, McKay S, Kleiner E L, Bird A P. Identification of a mammalian protein that binds specifically to DNA containing methylated CpGs. Cell. 1989;58:499–507. doi: 10.1016/0092-8674(89)90430-3. [DOI] [PubMed] [Google Scholar]
- 107.Meehan R R, Lewis J D, Bird A. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Migeon B R, Do T T. In search of non-random X inactivation: studies of fetal membranes heterozygous for glucose-6-phosphate dehydrogenase. Am J Hum Genet. 1979;31:581–585. [PMC free article] [PubMed] [Google Scholar]
- 109.Migeon B R, Wolf S F, Axelman J, Kaslow D C, Schmidt M. Incomplete X chromosome dosage compensation in chorionic villi of human placenta. Proc Natl Acad Sci USA. 1985;82:3390–3394. doi: 10.1073/pnas.82.10.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Migeon B R, Luo S, Jani M, Jeppesen P. The severe phenotype of females with ring X chromosomes is associated with inability of these chromosomes to undergo X inactivation. Am J Hum Genet. 1994;55:497–504. [PMC free article] [PubMed] [Google Scholar]
- 111.Moehrle A, Paro R. Spreading the silence: epigenetic transcriptional regulation during Drosophila development. Dev Genet. 1994;15:478–484. doi: 10.1002/dvg.1020150606. [DOI] [PubMed] [Google Scholar]
- 112.Monk M, Harper M. X-chromosome activity in preimplantation mouse embryos from XX and XO mothers. J Embryol Exp Morphol. 1978;46:53–64. [PubMed] [Google Scholar]
- 113.Monk M, Harper M. Sequential X chromosome inactivation coupled with cellular differentiation in early mouse embryos. Nature. 1979;281:311–313. doi: 10.1038/281311a0. [DOI] [PubMed] [Google Scholar]
- 114.Monk M, McLaren A. X-chromosome activity in foetal germ cells of the mouse. J Embryol Exp Morphol. 1981;63:75–84. [PubMed] [Google Scholar]
- 115.Monk M. A stem cell model for cellular and chromosomal differentiation in early mouse development. Differentiation. 1981;19:71–76. doi: 10.1111/j.1432-0436.1981.tb01131.x. [DOI] [PubMed] [Google Scholar]
- 116.Monk M. Methylation and the X chromosome. Bioessays. 1986;4:204–208. doi: 10.1002/bies.950040505. [DOI] [PubMed] [Google Scholar]
- 117.Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–382. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- 118.Monk M. Genomic imprinting. Genes Dev. 1988;2:921–925. doi: 10.1101/gad.2.8.921. [DOI] [PubMed] [Google Scholar]
- 119.Monk M, Adams R L P, Rinaldi A. Decrease in DNA methylation activity during preimplantation development in the mouse. Development. 1991;112:189–192. doi: 10.1242/dev.112.1.189. [DOI] [PubMed] [Google Scholar]
- 120.Monk M. The X chromosome in development in mouse and man. J Inherited Metab Dis. 1992;15:499–513. doi: 10.1007/BF01799608. [DOI] [PubMed] [Google Scholar]
- 121.Monk M. Epigenetic programming of differential gene expression in development and evolution. Dev Genet. 1995;17:188–197. doi: 10.1002/dvg.1020170303. [DOI] [PubMed] [Google Scholar]
- 122.Muscatelli F, Lena D, Mattei M G, Fontes M. A male with two contiguous inactivation centers on a single X chromosome: study of X inactivation and XIST expression. Hum Mol Genet. 1992;1:115–119. doi: 10.1093/hmg/1.2.115. [DOI] [PubMed] [Google Scholar]
- 123.Norris D P, Patel D, Kay G F, Penny G D, Brockdorff N, Sheardown S A, Rastan S. Evidence that random and imprinted Xist expression is controlled by preemptive methylation. Cell. 1994;77:41–51. doi: 10.1016/0092-8674(94)90233-x. [DOI] [PubMed] [Google Scholar]
- 124.Ohama K, Kajii T, Okamoto E, Fukuda Y, Imaizumi K, Tsukahara M, Kobayashi K, Hagiwara K. Dispermic origin of XY hydatidiform moles. Nature. 1981;292:551–552. doi: 10.1038/292551a0. [DOI] [PubMed] [Google Scholar]
- 124a.Oswald, J., M. Monk, and J. Walker. Unpublished data.
- 125.Panning B, Jaenisch R. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 1996;10:1991–2002. doi: 10.1101/gad.10.16.1991. [DOI] [PubMed] [Google Scholar]
- 126.Papaioannou V E, West J D. Relationship between the parental origin of the X chromosomes, embryonic cell lineages and X chromosome expression in mice. Genet Res Camb. 1981;37:183–197. doi: 10.1017/s0016672300020152. [DOI] [PubMed] [Google Scholar]
- 127.Pawlak A, Bryans M, Jost J-P. An avian 40 KDa nucleoprotein binds preferentially to a promoter sequence containing one single pair of methylated CpG. Nucleic Acids Res. 1991;19:1029–1034. doi: 10.1093/nar/19.5.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Penny G D, Kay G F, Sheardown S A, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- 129.Pillet N, Bonny C, Schorderet D F. Characterization of the promoter region of the mouse Xist gene. Proc Natl Acad Sci USA. 1995;92:12515–12519. doi: 10.1073/pnas.92.26.12515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Plass C, Shibata H, Kalcheva I, Mullins L, Kotelevsteva N, Mullins J, Kato R, Sasaki H, Hirotsune S, Okazaki Y, Held W A, Hayashizaki Y, Chapman V M. Identification of Grf1 on mouse chromosome 9 as an imprinted gene by RLGS-M. Nat Genet. 1996;14:106–109. doi: 10.1038/ng0996-106. [DOI] [PubMed] [Google Scholar]
- 131.Plenge R M, Hendrich B D, Schwartz C, Arena J F, Naumova A, Sapienza C, Winter R M, Willard H F. A promoter mutation in the XIST gene in two unrelated families with skewed X-chromosome inactivation. Nat Genet. 1997;17:353–356. doi: 10.1038/ng1197-353. [DOI] [PubMed] [Google Scholar]
- 132.Pravtcheva D D, Adra C N, Ruddle F H. Timing of paternal PGK-1 expression in embryos of transgenic mice. Development. 1991;111:1109–1120. doi: 10.1242/dev.111.4.1109. [DOI] [PubMed] [Google Scholar]
- 133.Rack K A, Chelly J, Gibbons R J, Rider S, Benjamin D, Lafreniére R G, Oscier D, Hendriks R W, Craig I W, Willard H F, Monaco A P, Buckle V J. Absence of the XIST gene from late-replicating isodicentric X chromosome in leukaemia. Hum Mol Genet. 1994;7:1053–1059. doi: 10.1093/hmg/3.7.1053. [DOI] [PubMed] [Google Scholar]
- 134.Rastan S, Kaufman M H, Handyside A H, Lyon M F. X-chromosome inactivation in extraembryonic membranes of diploid parthenogenetic mouse embryos demonstrated by differential staining. Nature. 1980;288:172–173. doi: 10.1038/288172a0. [DOI] [PubMed] [Google Scholar]
- 135.Rastan S. Non-random X-chromosome inactivation in mouse X-autosome translocation embryos—location of the inactivation centre. J Embryol Exp Morphol. 1983;78:1–22. [PubMed] [Google Scholar]
- 136.Rastan S, Robertson E J. X-chromosome deletions in embryo-derived (EK) cell lines associated with lack of X-chromosome inactivation. J Embryol Exp Morphol. 1985;90:379–388. [PubMed] [Google Scholar]
- 137.Ray P F, Winston R M L, Handyside A H. XIST expression from the maternal X chromosome in human male preimplantation embryos at the blastocyst stage. Hum Mol Genet. 1997;6:1323–1327. doi: 10.1093/hmg/6.8.1323. [DOI] [PubMed] [Google Scholar]
- 138.Razin A, Riggs A D. DNA methylation and gene function. Science. 1980;210:604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- 139.Razin A, Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991;55:451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Razin A, Cedar H. DNA methylation and genomic imprinting. Cell. 1994;77:473–476. doi: 10.1016/0092-8674(94)90208-9. [DOI] [PubMed] [Google Scholar]
- 141.Razin A, Shemer R. DNA methylation in early development. Hum Mol Genet. 1995;4:1751–1755. doi: 10.1093/hmg/4.suppl_1.1751. [DOI] [PubMed] [Google Scholar]
- 142.Richler C, Soreq H, Wahrman J. X-inactivation in mammalian testis is correlated with inactive X-specific transcription. Nat Genet. 1992;2:192–195. doi: 10.1038/ng1192-192. [DOI] [PubMed] [Google Scholar]
- 143.Riggs A D, Pfeifer G P. X-chromosome inactivation and cell memory. Trends Genet. 1992;8:169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- 144.Ropers H-H, Wolff G, Hitzeroth H W. Preferential X inactivation in human placenta membranes: is the paternal X inactive in early embryonic development of female mammals? Hum Genet. 1978;43:265–273. doi: 10.1007/BF00278833. [DOI] [PubMed] [Google Scholar]
- 145.Russell L B. Mammalian X-chromosome action: inactivation limited in spread and in region of origin. Science. 1963;140:876–978. doi: 10.1126/science.140.3570.976. [DOI] [PubMed] [Google Scholar]
- 146.Sado T, Tada T, Takagi N. Mosaic methylation of Xist gene before chromosome inactivation in undifferentiated female mouse embryonic stem and embryonic germ cells. Dev Dyn. 1996;205:421–434. doi: 10.1002/(SICI)1097-0177(199604)205:4<421::AID-AJA6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 147.Salido E, Yen P H, Mohandas T K, Shapiro L J. Expression of the X-inactivation-associated gene XIST during spermatogenesis. Nat Genet. 1992;2:196–199. doi: 10.1038/ng1192-196. [DOI] [PubMed] [Google Scholar]
- 148.Schmidt J V, Tilghman S M. Abstracts of the Meeting of Genomic Imprinting—Its Role in Development and Disease. Cambridge, United Kingdom: British Society for Developmental Biology; 1997. Analysis of enhancer competition by gene targeting, abstr; p. 81. [Google Scholar]
- 149.Shao C, Takagi N. An extra maternally derived X chromosome is deleterious to early mouse development. Development. 1990;110:969–975. doi: 10.1242/dev.110.3.969. [DOI] [PubMed] [Google Scholar]
- 150.Shemer R, Birger Y, Dean W L, Reik W, Riggs A D, Razin A. Dynamic methylation adjustment and counting as part of imprinting mechanisms. Proc Natl Acad Sci USA. 1996;93:6371–6376. doi: 10.1073/pnas.93.13.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Simmler M-C, Cattanach B M, Rasberry C, Rougeulle C, Avner P. Mapping the murine Xce locus with (CA)n repeats. Mamm Genome. 1993;4:523–530. doi: 10.1007/BF00364788. [DOI] [PubMed] [Google Scholar]
- 152.Simpson V J, Johnson T E, Hammen R F. Caenorhabditis elegans DNA does not contain 5-methylcytosine at any time during development or aging. Nucleic Acids Res. 1986;14:6711–6719. doi: 10.1093/nar/14.16.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Singer-Sam J, Grant M, LeBon J M, Okuyama K, Chapman V, Monk M, Riggs A D. Use of HpaII-polymerase chain reaction assay to study DNA methylation in the Pgk-1 CpG island of mouse embryos at the time of X-chromosome inactivation. Mol Cell Biol. 1990;10:4987–4989. doi: 10.1128/mcb.10.9.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Surani M A H, Barton S C, Norris M L. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Science. 1984;308:548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- 155.Takagi N, Oshimura M. Fluorescence and Giemsa banding studies of the allocyclic X chromosome in embryonic and adult mouse cells. Exp Cell Res. 1973;78:127–135. doi: 10.1016/0014-4827(73)90046-3. [DOI] [PubMed] [Google Scholar]
- 156.Takagi N, Sasaki M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256:640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- 157.Takagi N, Wake N, Sasaki M. Cytogenetic evidence for preferential inactivation of the paternally derived X chromosome in XX mouse blastocysts. Cytogenet Cell Genet. 1978;20:240–248. doi: 10.1159/000130856. [DOI] [PubMed] [Google Scholar]
- 158.Takagi N, Sugawara O, Sasaki M. Regional and temporal changes in the pattern of X-chromosome replication during the early post-implantation development of the female mouse. Chromosoma. 1982;85:275–286. doi: 10.1007/BF00294971. [DOI] [PubMed] [Google Scholar]
- 159.Tam P P L, Williams E A, Tan S-S. Expression of an X-linked HMG-lacZ transgene in mouse embryos: implication of chromosomal imprinting and lineage-specific X-chromosome activity. Dev Genet. 1994;15:491–503. doi: 10.1002/dvg.1020150608. [DOI] [PubMed] [Google Scholar]
- 160.Tam P P L, Zhou S X, Tan S-S. X-chromosome activity of the mouse primordial germ cells revealed by the expression of an X-linked lacZ transgene. Development. 1994;120:2925–2932. doi: 10.1242/dev.120.10.2925. [DOI] [PubMed] [Google Scholar]
- 161.Tate P, Skarnes W, Bird A. The methyl-CpG binding protein MeCP2 is essential for embryonic development in the mouse. Nat Genet. 1996;12:205–208. doi: 10.1038/ng0296-205. [DOI] [PubMed] [Google Scholar]
- 162.Urieli-Shoval S, Gruenbaum Y, Sedat J, Razin A. The absence of detectable methylated bases in Drosophila melanogaster. FEBS Lett. 1982;146:148–152. doi: 10.1016/0014-5793(82)80723-0. [DOI] [PubMed] [Google Scholar]
- 163.West J D, Frels W I, Chapman V M, Papaioannou V E. Preferential expression of the maternally derived X chromosome in the mouse yolk sac. Cell. 1977;12:873–882. doi: 10.1016/0092-8674(77)90151-9. [DOI] [PubMed] [Google Scholar]
- 164.Wu H, Fasser R, Schnieke A, Barker D, Lee K-H, Chapman V, Francke U, Jaenisch R. An X-linked human collagen transgene escapes X inactivation in a subset of cells. Development. 1992;116:687–695. doi: 10.1242/dev.116.3.687. [DOI] [PubMed] [Google Scholar]
- 165.Yoder J A, Walsh C P, Bestor T H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 166.Yokomori N, Moore R, Negishi M. Sexually dimorphic DNA demethylation in the promoter of the Slp (sex-limited protein) gene in mouse liver. Proc Natl Acad Sci USA. 1995;92:1302–1306. doi: 10.1073/pnas.92.5.1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zinn A R, Page D C, Fisher E M C. Turner syndrome: the case of the missing sex chromosome. Trends Genet. 1993;9:90–93. doi: 10.1016/0168-9525(93)90230-f. [DOI] [PubMed] [Google Scholar]
- 168.Zuccotti M, Monk M. Methylation of the mouse Xist gene in sperm and eggs correlates with imprinted Xist expression and paternal X-inactivation. Nat Genet. 1995;9:316–320. doi: 10.1038/ng0395-316. [DOI] [PubMed] [Google Scholar]
- 169.Zuccotti, M., and M. Monk. Unpublished data.