Abstract
Various gram-negative animal and plant pathogens use a novel, sec-independent protein secretion system as a basic virulence mechanism. It is becoming increasingly clear that these so-called type III secretion systems inject (translocate) proteins into the cytosol of eukaryotic cells, where the translocated proteins facilitate bacterial pathogenesis by specifically interfering with host cell signal transduction and other cellular processes. Accordingly, some type III secretion systems are activated by bacterial contact with host cell surfaces. Individual type III secretion systems direct the secretion and translocation of a variety of unrelated proteins, which account for species-specific pathogenesis phenotypes. In contrast to the secreted virulence factors, most of the 15 to 20 membrane-associated proteins which constitute the type III secretion apparatus are conserved among different pathogens. Most of the inner membrane components of the type III secretion apparatus show additional homologies to flagellar biosynthetic proteins, while a conserved outer membrane factor is similar to secretins from type II and other secretion pathways. Structurally conserved chaperones which specifically bind to individual secreted proteins play an important role in type III protein secretion, apparently by preventing premature interactions of the secreted factors with other proteins. The genes encoding type III secretion systems are clustered, and various pieces of evidence suggest that these systems have been acquired by horizontal genetic transfer during evolution. Expression of type III secretion systems is coordinately regulated in response to host environmental stimuli by networks of transcription factors. This review comprises a comparison of the structure, function, regulation, and impact on host cells of the type III secretion systems in the animal pathogens Yersinia spp., Pseudomonas aeruginosa, Shigella flexneri, Salmonella typhimurium, enteropathogenic Escherichia coli, and Chlamydia spp. and the plant pathogens Pseudomonas syringae, Erwinia spp., Ralstonia solanacearum, Xanthomonas campestris, and Rhizobium spp.
“Things derive their being and nature by mutual dependence and are nothing in themselves.”
Nagarjuna, 2nd century Buddhist philosopher
“Such is the power, sometimes called malignant, sometimes benign, that Anastasia, the treacherous city, possesses; if for 8 hours a day you work as a cutter of agate, onyx, chrysoprase, your labour which gives form to desire takes from desire its form, and you believe you are enjoying Anastasia wholly when you are only its slave.”
Italo Calvino, Invisible Cities
The elucidation of the molecules and mechanisms underlying bacterial pathogenesis in humans, animals, and plants is a major focus of microbiological research which yields practical applications ranging from refined diagnostics to new antibiotics and improved vaccines. In addition to the pursuit of these practical purposes, recent research on bacterial pathogenesis has allowed insight into the complex beauty of highly adapted interactions between pathogens and their hosts at the cellular and molecular levels. The relative ease of genetic and biochemical analyses of bacteria and widely used tissue culture models of bacterial infection have dramatically increased our knowledge about the molecular components involved in pathogen-host cell interactions, leading to the emergence of cellular microbiology as a new area of microbiological investigation. Importantly, the availability of suitable animal and plant models has allowed the assessment of the contribution of bacterial pathogenicity factors analyzed in vitro to the ultimate outcome of disease.
Genetic analyses of bacterial virulence factors has shown that pathogens are distinguished from their nonpathogenic relatives by the presence of specific pathogenicity genes, often organized in so-called pathogenicity islands, clusters of genes which apparently have been acquired during evolution via horizontal genetic transfer. Thus, distantly related pathogens have turned out to harbor closely related virulence genes. This point has become particularly apparent for a set of approximately 20 genes which together encode a pathogenicity mechanism termed type III secretion. Type III secretion enables gram-negative bacteria to secrete and inject pathogenicity proteins into the cytosol of eukaryotic host cells. Fascinatingly, while the type III secretion apparatus is conserved in pathogens as distantly related as Yersinia and Erwinia, the secreted proteins differ entirely, illustrating how one bacterial pathogenicity mechanism can give rise to a multitude of diseases that range from bubonic plague in humans to fire blight in fruit trees.
Secretion of bacterial pathogenicity proteins by the type III pathway and their injection into the cytosol of animal or plant cells initiates a sophisticated “biochemical cross-talk” (defined by J. E. Galán) between pathogen and host. The injected proteins often resemble eukaryotic factors with signal transduction functions and are capable of interfering with eukaryotic signalling pathways. Redirection of cellular signal transduction may result in disarmament of host immune responses or in cytoskeletal reorganization, establishing subcellular niches for bacterial colonization and facilitating a highly adapted pathogenic strategy of “stealth and interdiction” (defined by J. B. Bliska) of host defense communication lines.
This review comprehensively describes the type III protein secretion systems known to date, which are present in the animal pathogens Yersinia spp., Shigella flexneri, Salmonella typhimurium, enteropathogenic Escherichia coli (EPEC), Pseudomonas aeruginosa, and Chlamydia spp. and in the plant pathogens Pseudomonas syringae, Erwinia spp., Ralstonia (formerly Pseudomonas) solanacearum, Xanthomonas campestris, and Rhizobium spp. For a shorter and more concise overview of type III secretion, refer to the recent review by C. A. Lee (264).
Pathways of Protein Secretion in Gram-Negative Bacteria
Interaction of bacterial pathogens with host cells is particularly characterized by factors that are located on the bacterial surface or are secreted into the extracellular space. Although the secreted bacterial proteins are numerous and diverse and exhibit a wide variety of functions that include proteolysis, haemolysis, cytotoxicity, and protein phosphorylation and dephosphorylation, only a few pathways exist by which these proteins are transported from the bacterial cytoplasm to the extracellular space. Thus, four pathways of protein secretion (types I to IV) have been described in gram-negative bacteria (114, 122, 391, 447). A fifth system for macromolecular secretion is involved in conjugal transfer of plasmids, T-DNA transfer by Agrobacterim tumefaciens, and secretion of Bordetella pertussis toxin. The last system, which could be named type V secretion, is the least well characterized and comprises only three members so far (481).
In this context, the term “secretion” is used to describe the active transport of proteins from the cytoplasm across the inner and outer membranes into the bacterial supernatant or onto the surface of the bacterial cell. Secretion is distinguished from export, which refers to the transport of proteins from the cytoplasm into the periplasmic space (362, 391).
Type II and type IV sec-dependent secretion pathways.
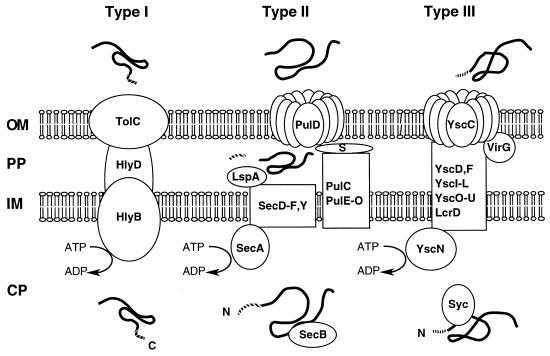
Type II and IV protein secretion pathways involve a separate step of transport across the inner membrane prior to transport across the cell envelope. While these pathways differ in the way in which the proteins are transported across the outer membrane, export to the periplasm occurs via the sec system in both cases. (Secretion of pertussis toxin by B. pertussis, which may be categorized as type V secretion [see above], also involves the sec pathway.) A signature of sec-dependent protein export is the presence of a short (about 30 amino acids [aa]), mainly hydrophobic amino-terminal signal sequence in the exported protein. The signal sequence aids protein export and is cleaved off by a periplasmic signal peptidase when the exported protein reaches the periplasm. In E. coli, the sec pathway comprises a number of inner membrane proteins (SecD to SecF, SecY), a cytoplasmic membrane-associated ATPase (SecA) that provides the energy for export, a chaperone (SecB) that binds to presecretory target proteins, and the periplasmic signal peptidase (see Fig. 1). A number of accessory proteins are also required for normal function (323, 362).
FIG. 1.
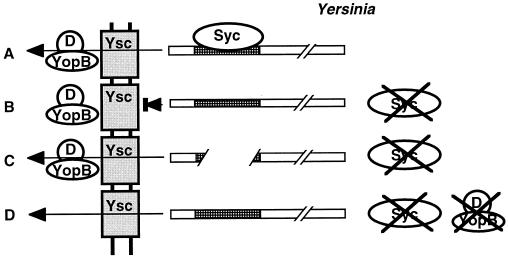
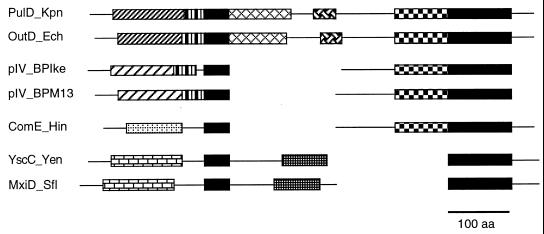
Schematic overview of the type I, II, and III secretion systems as exemplified by alpha-hemolysin secretion by E. coli (type I), pullulanase secretion by Klebsiella oxytoca (type II), and Yop secretion by Yersinia (type III). OM, outer membrane; PP, periplasm; IM, inner membrane; CP, cytoplasm. ATP hydrolysis by HlyB, SecA, and YscN is indicated. The localization of the secretion signals is shown in the secreted proteins (shaded). N, amino terminus; C, carboxy terminus. For type III secretion, the secretion signal may reside in the 5′-region of the mRNA encoding the secreted protein. Type II and type III secretion involve cytoplasmic chaperones (SecB and Syc, respectively) which bind to presecretory proteins. In type II secretion, the amino-terminal signal sequence is cleaved off by a periplasmic peptidase (LspA) after export of the protein via the sec pathway. Type II and type III secretion share a homologous multimeric outer membrane component (PulD, YscC), while the accessory proteins PulS and VirG, which facilitate outer membrane insertion of PulD and YscC, respectively, differ in the two systems. See the text for further details.
In type II secretion, transport across the outer membrane requires an additional set of inner and outer membrane proteins. In the case of pullulanase secretion by Klebsiella oxytoca, the best-studied example of type II secretion, 14 additional secretion factors, which are encoded by a continuous gene cluster, are necessary and sufficient for secretion. At least seven of these proteins are located in the cytoplasmic membrane, while PulS and PulD are outer membrane proteins (362). Other examples of type II secretion (reviewed in reference 203) include the out pathway of Erwinia spp. for the secretion of pectic enzymes and cellulases, the xcp-encoded secretion of elastase, exotoxin A, phospholipase C, and other proteins by Pseudomonas aeruginosa, amylase and protease secretion by Aeromonas hydrophila exe, and secretion of polygalacturonase and other proteins by Xanthomonas campestris xps. Thus, type II secretion is the primary pathway for the secretion of extracellular degradative enzymes by gram-negative bacteria. Furthermore, parts of the type II secretion pathway have homologs in other transport systems, for example in the secretion and assembly of N-methyl-Phe (type 4) pili of P. aeruginosa and other bacteria and in DNA transfer systems of Haemophilus influenzae and Bacillus subtilis (203). Most notably, however, the outer membrane component of the pullulanase secretion system, PulD, is conserved in a variety of gram-negative protein transport systems (see below).
The type IV secretion pathway (reviewed in reference 122) comprises a group of so-called autotransporters, including gonococcal immunoglobulin A and other proteases, the vacuolating cytotoxin of Helicobacter pylori, a family of outer membrane proteins in B. pertussis, and the secreted proteins SepA and EspC from S. flexneri and EPEC, respectively. As in type II secretion, these proteins are exported from the cytoplasm via the sec pathway, involving the cleavage of an amino-terminal signal peptide. However, the information required for transport across the outer membrane resides entirely within the secreted protein. Apparently, these autotransporters form a pore in the outer membrane through which they pass, and autoproteolytic cleavage releases the proteins into the supernatant.
Type I sec-independent pathway.
In contrast to the type II and IV secretion pathways, type I and type III secretion are independent of the sec system and thus do not involve amino-terminal processing of the secreted proteins. Furthermore, protein secretion via the latter pathways occurs in a continuous process without the distinct presence of periplasmic intermediates.
Type I secretion (reviewed in references 114 and 459) is exemplified by the E. coli alpha-hemolysin secretion system. Other members of this group are the adenylate cyclase secretion system of B. pertussis, leukotoxin secretion by Pasteurella haemolytica, and the protease secretion systems from P. aeruginosa and Erwinia chrysanthemi. Type I secretion requires three secretory proteins: an inner membrane transport ATPase (termed ABC protein for ATP-binding cassette), which provides the energy for protein secretion; an outer membrane protein, which is exported via the sec pathway; and a membrane fusion protein, which is anchored in the inner membrane and spans the periplasmic space (see Fig. 1). The genes encoding the secretion apparatus and the secreted protein are usually clustered.
The proteins which are secreted via the type I pathway are not subject to proteolytic cleavage, and the secretion signal is located within the carboxy-terminal 60 aa of the secreted protein. The secretion signal appears to be specific for subfamilies of the secretion system; i.e., the proteases are only poorly secreted via the hemolysin system and vice versa. The nature of the protease family secretion signal may be mainly conformational (459), while for E. coli alpha-hemolysin several dispersed key residues which are essential irrespective of a specific secondary structure and could facilitate recognition by the secretion apparatus have been identified (77).
Type III sec-independent pathway.
Like the type I secretion pathway, type III secretion is independent of the sec system. (Assembly of the type III secretion apparatus, however, probably requires the sec pathway, since several components of the type III secretion apparatus carry sec-characteristic amino-terminal signal sequences.) The type III secretion apparatus is composed of approximately 20 proteins, most of which are located in the inner membrane, and type III secretion requires a cytoplasmic, probably membrane-associated ATPase. Interestingly, most of the inner membrane proteins are homologous to components of the flagellar biosynthesis apparatus of both gram-negative and gram-positive bacteria, while an outer membrane protein of the type III secretion apparatus is homologous to PulD, the outer membrane secretin of the type II secretion pathway. Although type III secretion does not include distinct periplasmic intermediates of the secreted proteins, transport through the inner membrane is genetically separable from secretion through the outer membrane, since a mutant of the outer membrane PulD homolog of P. syringae was shown to accumulate considerable amounts of a secreted protein in the periplasm (72). As in type I and type II secretion, the genes encoding the type III secretion apparatus are clustered.
As in type I secretion, the proteins secreted via the type III pathway are not subjected to amino-terminal processing during secretion. The signal for secretion has long been thought to reside within the amino-terminal 15 to 20 aa of the secreted proteins, since this region is necessary for secretion and suffices to direct the secretion of hybrid fusion proteins. However, the amino-terminal sequences of proteins secreted via the type III pathway do not share any recognizable structural similarities that could function as a common secretion signal, and exhaustive mutational analysis of some secreted proteins has revealed a high degree of tolerance for sequence changes within the amino terminus without loss of secretion. Therefore, it has recently been proposed that the secretion signal resides in the 5′ region of the mRNA which encodes the secreted proteins (19). Interestingly, the secreted proteins require small cytoplasmic proteins with chaperone functions to protect the secreted factors from premature interaction with other components of the secretion system. In contrast to type I secretion, which is a true secretory system in that the secreted enzymes are active in the extracellular space, type III secretion systems appear to be dedicated machineries for the translocation of pathogenicity proteins into the cytosol of eukaryotic cells. Accordingly, protein secretion—at least in some cases—is regulated by contact with the surface of a target cell. In accordance with the homology of the type III secretion apparatus to flagellar biosynthesis factors, some type III secretion systems assemble supermolecular structures on the bacterial surface, which could be involved in protein translocation into eukaryotic cells (158, 374). Figure 1 gives an overview of types I, II, and III secretion.
CELLULAR AND MOLECULAR IMPACT OF TYPE III SECRETION IN BACTERIAL PATHOGENESIS
A variety of diverse gram-negative pathogens use type III secretion as a conserved and at the same time highly adapted virulence mechanism (Fig. 2). Although these pathogens use additional virulence factors (not discussed here), type III secretion is an essential basic virulence determinant. While the mechanism of protein secretion is conserved, the secreted proteins themselves are highly divergent, and the variety of diseases caused by these pathogens in different hosts is reflected by the multitude of type III secreted proteins. Many of the secreted proteins interact directly with host cell components to alter host cell signal transduction, and most of the secreted proteins act inside the eukaryotic cytosol into which they are translocated by the type III secretion mechanism. This section gives a short overview of the pathogenesis of bacteria which require type III secretion systems for virulence. The emphasis is put on the pathogenicity properties which are directly attributable to or correlated with type III secretion at the cellular and molecular levels.
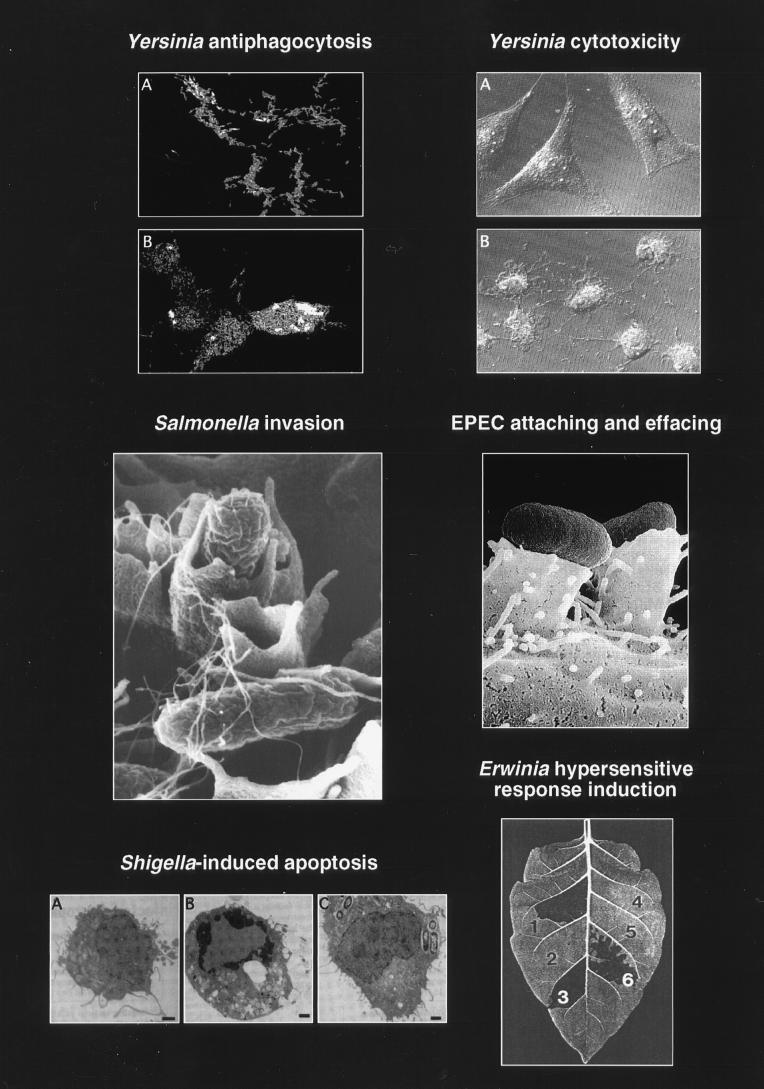
FIG. 2.
Selected phenotypic effects of type III secretion pathogenicity mechanisms on host cells and host tissue. (Top left) Panel B shows how Y. pseudotuberculosis injects (translocates) YopH (immunostained, light) into the cytosol of HeLa cells. Translocation of YopH into macrophages results in inhibition of phagocytosis. Panel A shows a type III secretion mutant: YopH is detected only in association with the bacteria. Reprinted with permission from reference 345. (Top right) Panel B shows the cytotoxic effect of Y. pseudotuberculosis on cultured HeLa cells. Translocation of YopE leads to a collapse of the cytoskeleton. Panel A shows uninfected HeLa cells. Reprinted with permission from reference 383. (Middle left) Invasion of a polarized HEp-2 epithelial cell via the induction of membrane ruffling by S. typhimurium. This figure has previously appeared on the cover of Mol. Microbiol. 1995, vol. 18 no. 3. Reprinted with permission. (Middle right) Pseudopod (pedestal) formation induced by EPEC on HeLa epithelial cells. Reprinted with permission from reference 378. (Bottom left) Induction of apoptosis in macrophages infected with S. flexneri. Panel A shows an uninfected macrophage. Panel B shows an apoptotic macrophage infected with wild-type S. flexneri. Panel C shows a macrophage infected with a S. flexneri type III secretion mutant. Reprinted with permission from reference 509. (Bottom right) Induction of localized tissue necrosis (HR) in a tobacco leaf at sites of infiltration with Erwinia spp. (area 1), buffer alone (area 4), type III secretion mutants (areas 2 and 5), and complemented mutants (areas 3 and 6). Reprinted with permission from reference 34.
Yersinia Species
Three Yersinia species are pathogenic for humans and rodents. Y. pestis, the causative agent of bubonic plague, enters the host through flea bites or by inhalation, and invades and multiplies in regional lymph nodes corresponding to the infection point. Subsequent dissemination via the lymphatic system and bacteremia with necrotic and hemorrhagic lesions in many organs lead to death of the human or rodent host within 2 to 3 days after infection. Similar pathological symptoms are caused in rodents by Y. enterocolitica and Y. pseudotuberculosis. In humans, Y. enterocolitica causes a broad range of gastrointestinal syndromes, while Y. pseudotuberculosis, which is the least pathogenic of the three species for humans, may (rarely) cause a self-limiting gastroenteritis (86). Both Y. enterocolitica and Y. pseudotuberculosis usually enter their hosts via the oral route of infection and cross the intestinal barrier through specialized epithelial cells called M cells (179). M cells are associated with the follicle-associated epithelium overlying lymphoid follicles throughout the intestine and are especially concentrated in aggregates of lymphoid follicles called Peyer’s patches (411). Like Y. pestis, Y. enterocolitica and Y. pseudotuberculosis exhibit a marked tropism for lymphatic tissue and persist and multiply in Peyer’s patches, leading to high bacterial titers in these organs 12 to 24 h after infection. Subsequently, the bacteria colonize liver and spleen, and bacterial multiplication in these organs leads to death of the animal 3 to 4 days after infection (206, 429).
All three pathogenic Yersinia spp. exhibit a characteristic ability to resist the host primary immune defense, most notably by inhibiting their own uptake by professional phagocytes (66). Consequently, these pathogens are found mainly in extracellular locations during infection (179, 180, 412). The antiphagocytic effect is mediated by the Yersinia type III secretion system and specifically requires a protein with tyrosine phosphatase activity (169) called YopH (20, 112, 379). After contact of the bacteria with a macrophage, YopH is injected into the cytosol of the target cell (Fig. 2), where it catalyzes a rapid and specific dephosphorylation of several macrophage proteins, whose transient tyrosine phosphorylation appears to be required for normal phagocytosis (20). While earlier studies had observed YopH-dependent overall dephosphorylation of phosphotyrosine proteins in macrophages (50, 51), a more recent analysis identified the cytoskeleton-associated protein paxillin, which is involved in Fc receptor-mediated phagocytosis (165), as a target for YopH in macrophages. In addition to the antiphagocytic effect, Y. pseudotuberculosis resists killing by macrophages by an efficient inhibition of the Fc receptor-mediated oxidative burst, a phenotype which also requires the phosphatase activity of YopH (49). In HeLa cells, YopH was demonstrated to specifically dephosphorylate focal adhesion kinase and the focal adhesion-associated protein p120cas, and dephosphorylation of these proteins appears to inhibit bacterial uptake by HeLa cells via inhibition of peripheral focal complex formation (48, 344).
In addition to inhibition of phagocytosis, Yersinia spp. are cytotoxic for cultured epithelial cells (Fig. 2) (355), and cytotoxicity also may contribute to the inactivation of macrophages (381). Cytotoxicity is specifically mediated by YopE, another protein secreted by the type III secretion pathway, which, like YopH, is injected into the eukaryotic cytosol (380, 381). YopE does not affect the integrity of the target cell membrane but, rather, causes a collapse of the cytoskeleton via disruption of actin microfilaments (380). The mechanism of actin disruption is probably indirect, since isolated Yops do not affect actin microfilament polymerization or stability in vitro (380).
Mutants with mutations in yopE and yopH of Y. pseudotuberculosis do not reach the same titers in mouse Peyer’s patches as the wild type does, and they are cleared from these organs 4 days after oral infection (206). Therefore, both mutants are avirulent when administered orally (59, 130, 131). However, while a yopH mutant is highly attenuated even after intraperitoneal or intravenous (i.v.) infection (59, 381), a yopE mutant exhibits only a partial virulence defect when injected i.v. (381). Thus, YopE appears to function primarily at an early step in Yersinia pathogenesis whereas YopH is required throughout the infectious process.
In addition to YopE and YopH, Yersinia species secrete and translocate other virulence determinants into the cytosol of target cells by the type III secretion mechanism. A protein with serine/threonine kinase activity (YpkA) is required for later stages of infection (149). A ypkA mutant colonizes Peyer’s patches similarly to the wild-type strain but is unable to colonize the spleen (150). Similarly, YopM, a translocated protein with thrombin binding activity (56, 268, 368), is required for later stages of mouse infection.
Taken together, the type III secretion mechanism enables Yersinia spp. to inject a number of essential virulence determinants into the cytosol of host target cells. The injected proteins appear to interfere with host cell signal transduction pathways and other cellular processes, allowing Yersinia spp. to obstruct the primary immune response and to establish a systemic infection.
Interestingly, suppression of several proinflammatory cytokines in various cell types appears to be another Yersinia virulence mechanism which has been associated with type III secretion and Yop translocation (44, 325, 407). Suppression of cytokines may account for the fact that infections with pathogenic Yersinia spp. proceed without eliciting a strong inflammatory immune response (324, 325, 412). Recently, the inhibition of an inflammatory host response was tentatively attributed to a low and regulated level of translocation of Yop proteins into target cells (205). A mutant with a mutation in a gene encoding the type III secreted protein YopK (206) was found to translocate significantly larger amounts of Yop proteins. Unexpectedly, the mutant was nevertheless impaired in later stages of mouse infection (206). The virulence defect of this mutant was interpreted to mean that this strain may elicit a stronger and eventually assertive host immune response due to an elevated level of Yop translocation (205).
Shigella flexneri
In contrast to pathogenic Yersinia spp., which are found mostly extracellularly during infection of mammalian hosts, Shigella spp. occupy predominantly intracellular locations. Although the pathogenic strategies of yersiniae and shigellae and the diseases caused by these organisms differ entirely, both pathogens use type III secretion systems as a key virulence mechanism.
Shigella spp. cause diarrheal disease with a wide range of clinical symptoms, the most severe form being bacillary dysentery, a bloody diarrhea originating from the colon. Shigellosis is endemic in developing countries, but outbreaks also occur in industrialized nations, especially under conditions of poor hygiene. Children under 5 years of age are the most susceptible victims, with over half a million deaths occurring annually worldwide (275, 292, 508).
Bacterial invasion of the colonic mucosa (258, 435) constitutes an essential step in the pathogenesis of shigellosis. The following model of the cellular basis of shigellosis has been derived from the integration of a variety of in vivo and in vitro observations (147). Similarly to other enteropathogens, shigellae gain initial access to the intestinal epithelium by invading M cells (342, 393, 460). After transcytosis through M cells, the bacteria encounter resident macrophages of the lymphoid tissue underlying the follicle-associated epithelium and resist killing by macrophages by the induction of cell apotosis (Fig. 2) (509, 510). Apoptosis of macrophages in turn leads to the release of significant quantities of interleukin-1 (506), which results in recruitment of polymorphonuclear leukocytes (PMNs) and massive infiltration of the infected tissue with these cells. S. flexneri is unable to invade a polarized epithelial cell monolayer from the apical pole, but transmigration of PMNs across the epithelium destabilizes the integrity of the intestinal barrier and allows S. flexneri to reach the basolateral epithelial cell pole, which is readily invaded by the bacteria (341, 342). Further bacterial invasion and lateral spreading of the bacteria within the epithelium result in the tissue destruction typically associated with Shigella infection (338). The ability of PMNs to kill S. flexneri suggests that these cells may not only contribute initially to the severe tissue damage characteristic of shigellosis but also ultimately participate in clearance and resolution of infection (284).
In in vitro tissue culture infection experiments, S. flexneri provokes its own uptake into nonpolarized epithelial cells by the induction of cytoskeletal rearrangements, resulting in the formation of localized membrane protrusions at the site of bacterial contact with the cell (Fig. 2). These so-called membrane ruffles coalesce around the entering bacterium, leading to its phagocytosis in a membrane-bound vacuole (2). In vivo invasiveness, as well as the induction of membrane ruffles on cultured epithelial cells, depends on the Shigella type III secretion system and has been extensively characterized at the biochemical level.
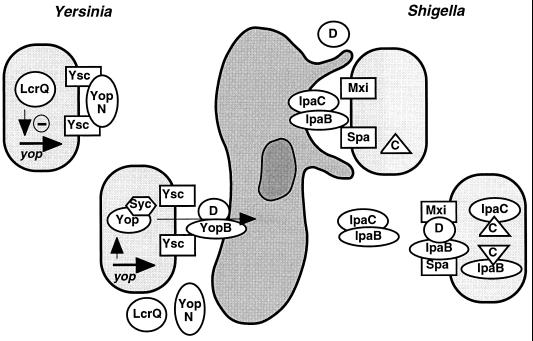
Four proteins which are secreted via the type III pathway, IpaA, IpaB, IpaC, and IpaD, are specifically involved in bacterial invasiveness (200, 304, 442), and two of them, IpaB and IpaC, are sufficient to induce membrane ruffling on epithelial cells when immobilized on the surface of latex beads (301). A potential receptor for IpaB and IpaC on target cells is the cell adhesion molecule α5β1 integrin (433). IpaB, IpaC, and IpaD specifically bind to α5β1 integrin and colocalize with the integrin in Chinese hamster ovary (CHO) cell infection (462). Overexpression of α5β1 integrin in CHO cells led to enhanced S. flexneri invasiveness, and titration of Ipa proteins by extracellular addition of α5β1 integrin decreased invasion in a concentration-dependent manner (462). Ligand-induced stimulation and clustering of integrins is known to cause protein tyrosine phosphorylation of a number of cytoskeletal proteins (78, 220), and protein tyrosine phosphorylation is required for efficient uptake of S. flexneri into CHO cells (462). Therefore, binding of the Ipa proteins to α5β1 integrin may be the primary signal which leads to the induction of membrane ruffles and ultimately results in bacterial internalization.
Induction of membrane ruffling by S. flexneri involves the localized accumulation of a variety of cytoskeletal and signal transduction molecules at the site of bacterial attachment. Shigella attachment induces the accumulation of actin and several actin-binding proteins (myosin, plastin [2], and cortactin [93]). Furthermore, the focal adhesion plaque-associated proteins paxillin, α-actinin (300), vinculin (442), talin, and α5β1 integrin (462) accumulate at the site of bacterial entry. Recently, it was shown that overexpression of vinculin, a protein which is supposed to link the cytoskeleton to the cell membrane, results in increased uptake of S. flexneri by ASML cells and that this effect depends on IpaA (442). Wild-type bacteria were seen to closely associate with a coat of cytoskeletal proteins (F-actin, vinculin, and α-actinin) during the entry process, and although recruitment of these proteins to the entry structure is independent of IpaA, tight association of the proteins with the bacteria was abolished in an ipaA mutant. Accordingly, the ipaA mutant is only partially deficient in invasion, while IpaB to IpaD mutants, which do not induce actin polymerization, are noninvasive. IpaA specifically binds to vinculin and thus appears to modulate bacterial invasion by “optimizing” the impact of invasive S. flexneri on host cell cytoskeletal rearrangements (442).
Entry of S. flexneri also leads to differential recruitment of the protein tyrosine kinase pp60c-src (93) and the GTP-binding protein rho (3) to the entry structure. The focal adhesion kinase pp125FAK, paxillin (462), and cortactin (93) have been identified as targets for protein tyrosine phosphorylation (93, 300). rho is a central signal transducer in Shigella invasiveness, since inhibition of rho activity results in complete loss of Shigella-induced cytoskeletal rearrangements and of Shigella invasiveness (3, 301, 463). Consistent with the involvement of rho in Shigella invasion, rho-dependent activation of protein kinase C was observed upon Shigella invasion, and inhibition of protein kinase C with various inhibitors greatly reduced Shigella invasiveness (463).
Shortly after internalization, the bacteria lyse the phagocytic membrane and gain access to the cytoplasm (398). In macrophages, S. flexneri induces apoptosis after escape from the phagosome (437, 509). Like invasion of epithelial cells, the ability to lyse the endocytic vacuole and to induce apoptosis in macrophages depends on the S. flexneri type III secretion system. While vacuolar lysis appears to require all three type III secreted Ipa proteins (200, 304), induction of apoptosis relies specifically on IpaB (507), which triggers the apoptotic pathway by direct binding to interleukin-1β-converting enzyme (ICE) or a homologous protease (75). The relevance of ICE activation for S. flexneri-induced apoptosis was elegantly shown by reversible inhibition of ICE, which resulted in reversible inhibition of S. flexneri cytotoxicity for macrophages (75). In addition to the induction of apoptosis, binding of IpaB to ICE leads to increased production of mature interleukin-1β by activated ICE (75), which, in vivo, results in attraction of PMN to the site of bacterial infection (see above).
Salmonella typhimurium
Salmonellae are the only species described so far to contain two type III secretion systems, which are encoded by two distinct gene clusters termed SPI-1 and SPI-2 (for Salmonella pathogenicity island). These two type III secretion systems appear to play different roles during pathogenesis, with SPI-1 being required for initial penetration of the intestinal mucosa and SPI-2 necessary for subsequent systemic stages of infection.
Salmonella spp. infect a variety of vertebrate hosts and cause a broad spectrum of diseases (including gastroenteritis, bacteremia, and enteric fever) which originate from enteric infection. S. typhi causes systemic typhoid fever in humans and constitutes a major health problem in underdeveloped regions of the world, with an estimated 16 to 17 million cases and 0.6 million deaths annually (335), while S. typhimurium and S. enteritidis are increasing causes of food poisoning in industrialized countries (309). Several Salmonella spp. are specifically adapted to a particular host (including S. typhi [humans], S. pullorum [poultry], S. dublin [cattle], and S. arizonae [reptiles]), while others (S. typhimurium, S. enteritidis, and S. choleraesius) exhibit a broader host spectrum but may cause different diseases in different hosts. For example, S. typhimurium causes systemic and lethal infection in susceptible mice at very low doses of infection, while in humans even high doses of this organism only cause a self-limiting gastroenteritis (309).
In systemic infections in mice, Salmonella penetrates the mucosa of the small intestine by preferentially adhering to and invading M cells of the Peyer’s patches (79). The bacteria eventually destroy the invaded M cells and the adjacent epithelium (92, 228, 249), thus gaining access to the underlying lymphoid tissue. M-cell invasion is an active event, since the bacteria induce ruffles of the apical cell membrane, which eventually engulf the entering organism (434). Salmonella-induced M-cell membrane ruffles are similar to the localized membrane ruffling observed on cultured epithelial cells after contact with the bacteria (Fig. 2), and, as with epithelial cell invasion by Shigella (see above), the Salmonella-induced ruffles involve rearrangements of the actin cytoskeleton (124, 134). Despite the morphological similarity of membrane ruffles induced by Shigella and Salmonella spp., invasion of epithelial cells by S. typhimurium appears to differ from that by S. flexneri. For example, while Shigella flexneri invasion is inhibited by various protein kinase C inhibitors, S. typhimurium invasion is not (463). Therefore, Salmonella invasion appears to be independent of the rho-controlled signal transduction pathway. In contrast, S. typhimurium requires CDC42 for invasion (73). Furthermore, S. typhimurium does not induce phosphorylation of the focal adhesion kinase p125FAK or of paxillin (463). A further difference, which may be significant to in vivo invasiveness, is the fact that in contrast to Shigella flexneri, S. typhimurium efficiently invades polarized epithelial cells via the apical cell pole (156).
Induction of membrane ruffles and invasion of M cells and cultured epithelial cells require the type III secretion system encoded by SPI-1 (228, 340), and in parallel to the similar invasive capacities of S. typhimurium and Shigella flexneri, the S. typhimurium type III secreted invasion proteins (called Sip or Ssp) are homologous to the Ipa invasins from Shigella flexneri (216, 233, 234). However, S. typhimurium SPI-1 mutants are only slightly attenuated for mouse virulence after oral infection and show no defect in virulence when injected by the intraperitoneal route (143, 227, 340). Thus, it seems possible that invasion-negative mutants are still taken up—at least to some degree—by the naturally phagocytic M cells and that this uptake is sufficient to cause systemic disease. The role of the SPI-1-encoded type III secretion systems in mouse typhoid is therefore restricted to the early stages of infection. Interestingly, tissue tropism of S. typhimurium toward M cells appears to be mediated by species-specific fimbriae (lpf) (36). Like SPI-1 mutants, mutants with mutations in lpf show only a slight virulence defect when administered by the oral infection route, which may be due to SPI-1-mediated invasion of the intestinal epithelium at sites other than M cells. However, combination of lpf and SPI-1 mutations significantly increased the 50% lethal dose for mice (37). Thus, SPI-1 and lpf appear to function synergistically in invasion of the intestinal barrier.
In the lymphoid tissue underneath invaded M cells, salmonellae are phagocytosed by residential macrophages. In contrast to Shigella flexneri, salmonellae do not escape from the phagocytic vacuole. Instead, they survive and replicate inside the vacuole and probably use macrophages as vehicles to disseminate via the host lymphoid system. The bacteria accumulate and massively replicate in the liver, spleen, and bone marrow, organs which are rich in phagocytic cells, leading to organ failure, bacterial sepsis, and death 4 to 6 days after infection. The ability to survive and replicate in professional phagocytes is therefore thought to be an essential virulence determinant of salmonellae that cause systemic infection (16, 119, 273, 311, 331, 406, 453).
Besides its requirement for epithelial cell invasion, the SPI-1-encoded type III secretion system recently has been shown to affect the interaction of Salmonella spp. with cultured murine macrophages (74). However, SPI-1 does not affect the ability to persist and replicate in macrophages but, rather, mediates a cytotoxic effect. Similar to induction of apoptosis by Shigella flexneri, SPI-1 encoded effector proteins induce apoptosis in cultured macrophages (74, 315). However, since SPI-1 mutants are only slightly attenuated for mouse virulence when administered orally (see above), the cytotoxic effect appears to play only a minor role in early stages of infection.
It is interesting that both shigellae and salmonellae use homologous secreted proteins (Ipa and Sip/Ssp, respectively) for similar phenotypes: invasion of epithelial cells and induction of apoptosis in macrophages. The SipB protein from S. typhi, which is highly similar to the respective S. typhimurium protein, even transcomplemented a Shigella flexneri ipaB mutant for invasiveness (197). However, transcomplementation was only partial, indicating that IpaB and SipB are not fully interchangeable. Surprisingly, although Sip/SspB does not allow salmonellae to escape from the phagocytic vacuole, SipB from S. typhi restored the ability of a Shigella flexneri ipaB mutant to lyse the vacuole. Furthermore, Shigella flexneri appears to require vacuolar lysis to be able to induce apoptosis via IpaB, while Salmonella spp., although similarly inducing apoptosis, do not exhibit the same requirement (74). These differences may be due to the fact that Sip/Ssp proteins are translocated across the cell membrane into the eukaryotic cytosol via the type III secretion mechanism (83), while membrane translocation of Ipa proteins has not been reported.
The SPI-1-encoded type III secretion system appears to play a major role in nontyphoidal Salmonella infections of the intestinal epithelium. A characteristic feature of Salmonella-induced enteritis is an intense intestinal secretory and inflammatory response including the induction of PMN transmigration through the intestinal epithelium (295). Transepithelial signalling requires the SPI-1-encoded type III secretion system and has been particularly associated with the type III secreted protein SopB of S. dublin (151).
The second Salmonella type III secretion system, encoded by SPI-2, has been identified by virtue of its large impact on S. typhimurium mouse virulence (195, 410). However, mutants of SPI-2 have no (196) or only a slight (333) effect on bacterial survival and replication in cultured macrophages and are invasive for epithelial cells like the wild-type strain (196). Therefore, although the SPI-2-encoded type III secretion system in Salmonella spp. most significantly affects virulence, its cellular and molecular impact on pathogenesis remains elusive.
Enteropathogenic Escherichia coli
EPEC strains form one of several categories of diarrheagenic E. coli strains, which are distinguished from other E. coli strains by their ability to inflict characteristic lesions in small intestine enterocytes, with gross cytoskeletal damage and loss of brush border microvilli (423). In addition, EPEC strains are characterized by their clustered pattern of adherence to epithelial cells, which results in the formation of microcolonies on the cell surface (403), and by their inability to produce Shiga toxin. EPEC strains are a leading cause of diarrhea in infants in the developing world (101, 270).
After the initial adherence to epithelia, EPEC strains attach intimately to the epithelial cell surface, leading to the effacement of microvilli beneath the bacteria. The resulting characteristic histopathologic finding is known as attaching and effacing (A/E) lesions (316). At the zone of contact between bacteria and the epithelial cell surface, cup-like pseudopod structures appear, which form progressively elongating pedestals carrying individual bacteria on their tops (Fig. 2) (378). The pedestal surface conforms to the curvature of the bacterium and maintains a distance of less than 10 nm across much of the bacterial surface (316). Pedestals consist of a densely packed microfilamentous structure (297) and contain cytoskeletal proteins including actin (248), α-actinin (123), and myosin light chain (285), in addition to talin and ezrin (123), proteins that link actin filaments to the membrane (65, 367).
Intimate attachment, effacing of microvilli, and formation of pedestals require a bacterial adhesin (called intimin) and EPEC type III secretion. Although intimin is not secreted by the type III secretion pathway, the encoding gene (eaeA) is located within the gene cluster that encodes the EPEC type III secretion system (100, 224, 225), and intimin functions synergistically with type III secretion in pedestal and A/E lesion formation. Intimin specifically binds to a 90-kDa protein (originally named Hp90) which is located in the eukaryotic membrane in direct proximity to the attached bacteria. It was recently discovered that this protein is of bacterial origin and is secreted by the type III secretion mechanism (239). The 549-aa protein, which was named Tir (translocated intimin receptor), is inserted into the eukaryotic membrane. Two other type III secreted proteins, EspA and EspB, which also may be translocated into the eukaryotic cytosol (104, 240, 242), are required for membrane insertion of Tir (239). Intimin was shown to directly bind Tir, demonstrating that EPEC strains transfer their own receptor for intimate attachment into eukaryotic cells.
Concomitant with pedestal formation, adherent EPEC strains induce tyrosine phosphorylation of several proteins in the eukaryotic cell, including Hp90/Tir (376, 378) and phospholipase C-γ1 (241). While Tir phosphorylation has been thought to facilitate intimin binding (378), activation of phospholipase C-γ1 induces inositol triphosphate and Ca2+ fluxes (30, 133), which might be involved in the cytoskeletal rearrangements involved in pedestal formation. EPEC-induced tyrosine phosphorylation and host cell signalling also depend on the type III secretion of EspA, EspB, and EspC (132, 223, 240, 242, 259, 376). (It should be noted, however, that the signal transduction events leading to A/E lesion formation may be more complicated than implied by this attractive model, since actin accumulation beneath adherent EPEC cells was observed even in the absence of detectable tyrosine phosphorylation due to inhibition of protein tyrosine kinases with genistein [364]. Furthermore, a mutant impaired in Esp secretion in vitro and in the induction of tyrosine phosphorylation of Hp90 was still capable of causing A/E lesions like the wild type [364].)
Pseudomonas aeruginosa
P. aeruginosa is an opportunistic pathogen capable of infecting immunocompromised individuals, including patients with extensive burns or wounds, cystic fibrosis, and leukemia (53). Recently, secretion of exoenzyme S (ExoS) and the related ExoT (80), two of several virulence determinants of P. aeruginosa (328), has been found to occur via a type III secretion pathway (135, 491). ExoS and ExoT are proteins with ADP-ribosyltransferase activity (221), whose preferred eukaryotic target proteins include members of the H-Ras and K-Ras family of GTP-binding proteins and vimentin, an intermediate filament protein (80). The enzymatic activity of ExoS is absolutely dependent on a eukaryotic cofactor protein, FAS (81), which is a member of a large family of regulatory proteins that play key roles in cell growth and differentiation (142). Although the function of ADP-ribosylation in P. aeruginosa pathogenesis is unclear, the production of ExoS is associated with epithelial cell damage and dissemination of P. aeruginosa within infected hosts (23, 231, 327). It is interesting that ExoS is partially similar to YopE from Yersinia spp. (see “The cytotoxin YopE” below) and that the type III secretion systems of the two pathogens are highly similar (see “Type III secretion system genes in Yersinia species” and “P. aeruginosa type III secretion system genes” below) (135). Indeed, ExoS can be secreted and translocated into mammalian cells via the Yersinia type III secretion pathway and induces host cell cytoskeletal damage similar to that induced by YopE (140). Several other proteins which are secreted by P. aeruginosa and which are highly similar to secreted Yersinia proteins that function in the regulation of secretion and translocation have been identified.
Chlamydia Species
Type III secretion genes have been recently identified in Chlamydia psittaci and other chlamydiae (211). Although these genes have not been shown to be functional, type III secretion could play a major role in host cell interactions of these highly host-adapted bacteria (171). Chlamydiae are a worldwide major cause of sexually transmitted diseases and infectious blindness, and they cause respiratory tract infections and are associated with atherosclerosis; their basic pathogenicity mechanisms remain elusive (171). These obligate intracellular bacteria exhibit a biphasic developmental cycle with a dormant extracellular form and a replicating intracellular form (319). Intracellular bacteria reside within a large vacuole, which, soon after infection, redistributes from the cellular to the nuclear periphery by an F-actin-dependent mechanism (281). The chlamydial inclusion expands by acquiring host membrane components and includes chlamydial proteins. Among the latter, IncA is translocated to the outer surface of the Chlamydia-containing vacuole and may be phosphorylated by a host enzyme (371, 372). IncA does not contain a sec signal sequence, and it has been speculated that this protein may be translocated via the type III secretion pathway (211). Whether type III secretion is involved in tyrosine phosphorylation of host cell proteins observed early after infection (45) remains to be elucidated.
Plant-Pathogenic Bacteria
One of the exciting surprises that came with the discovery of type III secretion systems was the finding that a variety of gram-negative phytopathogenic bacteria use type III secretion for pathogenesis, just like the unrelated animal pathogens do. Type III secretion systems are conserved in the four major genera of plant-pathogenic bacteria, Erwinia, Pseudomonas, Ralstonia, and Xanthomonas (60, 85), and are required for the common ability of these pathogens to cause disease in susceptible host plants. The plant diseases caused by these pathogens range from fire blight of rosaceous plants and soft rot caused by various Erwinia spp., bacterial spot disease of pepper and tomato (Xanthomonas campestris), and bacterial speck (P. syringae), to bacterial wilt of solanaceous plants (Ralstonia solanacearum). Interestingly, genes encoding a putative type III secretion system have also been found in Rhizobium spp. (139, 189), where they are involved in cultivar-specific nodulation of leguminous plants (299).
Type III secretion is also essential for the induction of a defense reaction called the hypersensitive response (HR) in resistant plants that are not normally hosts for the particular pathogen. The HR is characterized by localized tissue necrosis and the production of phenolics and antimicrobial agents at the site of bacterial contact (245, 277), which prevent further spread of the infecting bacteria in the plant. The HR is an active response to bacterial infection and requires plant de novo gene expression and protein synthesis, a calcium flux across membranes, and ATPase activity (188). Although the HR is microscopically small under natural conditions, it is macroscopically visible and readily assessed in the laboratory when plant tissue is infiltrated with large numbers of phytopathogenic bacteria (Fig. 2). The bacterial genes required for pathogenicity in susceptible plants and for the elicitation of an HR in resistant plants have operationally been defined as hrp (for hypersensitive response and pathogenicity) (276), and this name is still maintained as a global label of the plant-pathogenic type III secretion systems. In some plant pathogens, proteinaceous elicitors of plant disease and/or HR have been identified and were shown to be secreted via the type III secretion pathway (26, 34, 188, 472). For example, the structurally similar type III secreted proteins from E. amylovora and P. syringae elicit an HR in nonhost plants when infiltrated into plant tissue in a purified form, and mutants with mutations in the encoding genes are avirulent on host plants (188, 472).
Whether bacterial infection leads to plant disease or to an HR resistance phenotype is determined largely by the presence of a matching pair of a dominant resistance gene (R) in the host and a so-called avirulence gene (avr) in the pathogen (424, 455). Disruption of an avr gene usually allows the pathogen to provoke disease in previously resistant plants, while introduction of an R gene into a susceptible plant confers resistance to the pathogen carrying the matching avr locus. The requirement for a matching R-avr gene pair for HR induction has led to the postulation of the gene-for-gene hypothesis (127). In the gene-for-gene concept, the plant R gene is thought to encode a receptor for the pathogen Avr protein, and interaction between these two proteins may trigger a signalling cascade which ultimately results in elicitation of an HR (29, 260, 424).
Interestingly, like the elicitors of plant disease, the action of several bacterial avirulence proteins is phenotypically dependent on functional type III secretion (162, 247, 348, 363). However, secretion of an avirulence protein has not been directly observed. Nevertheless, several avr-encoded proteins have been shown to elicit an HR in an R-gene-dependent manner when expressed inside host cells (162, 267, 446). It is thus likely that the Avr proteins are injected into the target cell via the type III secretion mechanism (29) like the virulence factors injected by Yersinia spp. and other animal pathogens.
R genes from several plant species and numerous bacterial avirulence genes have been cloned (29, 455). As a common structural motif, most R-gene-encoded proteins carry leucine-rich repeats which could mediate interactions with other proteins and therefore could function in signal recognition and transduction (336). In the case of the tomato R gene Pto, the encoded protein is a serine/threonine kinase. Pto confers resistance to bacterial speck disease caused by P. syringae pv. tomato expressing the corresponding AvrPto protein (287). (On the basis of host range, plant-pathogenic bacterial species are subdivided into pathovars [pv.]. Thus, P. syringae pv. tomato causes disease on susceptible tomato plants, while P. syringae pv. phaseolicola causes disease in beans.) Direct interaction between AvrPto and Pto has recently been demonstrated, providing a physical explanation for the gene-for-gene hypothesis and for the specificity of plant-pathogen interactions (408, 436). Pto interacts with and phosphorylates another serine/threonine kinase (Pti1), which also functions in HR induction (504). Thus, after AvrPto is translocated by the type III secretion mechanism into the plant cell cytosol, binding of AvrPto to Pto might stimulate Pto kinase activity to trigger a phosphorylation cascade involved in activating the HR.
SECRETED PROTEINS
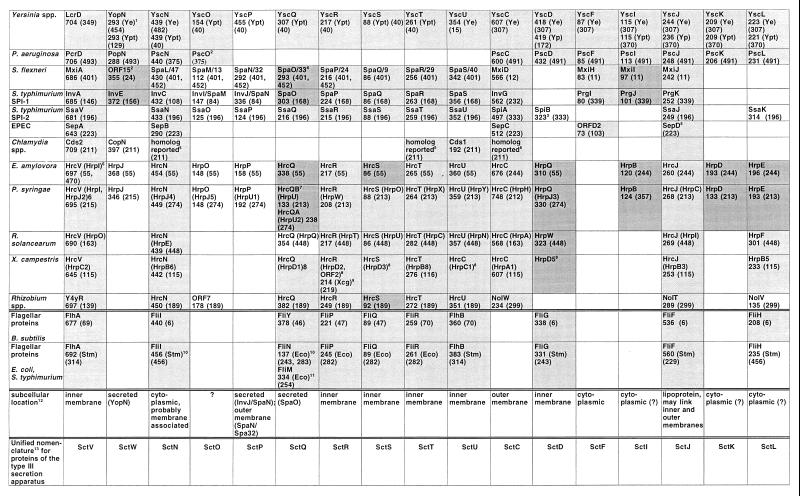
This section describes structural and functional aspects of the virulence proteins which are secreted—and in many cases injected (translocated) into the eukaryotic cytosol—via the type III secretion pathway. These proteins greatly vary in size, structure, and function and account for the species-specific pathogenicity phenotypes associated with type III secretion. Interestingly, several of the secreted proteins are similar to eukaryotic proteins, implying that the pathogens may have acquired the respective genes from their eukaryotic hosts during evolution. A number of secreted proteins do not exhibit direct antihost functions but, rather, are accessory in that they function in secretion and translocation of the actual virulence factors. (These accessory proteins are discussed in detail under “Translocation of proteins into the eukaryotic cytosol” and “Regulation of type III secretion by contact with eukaryotic cells” below). In several cases, the genes encoding secreted proteins are located outside the gene clusters which encode the type III secretion apparatus, but usually the former genes are transcriptionally coregulated with the latter ones. Several secreted proteins have homologs in different type III secretion systems. Thus, while the Salmonella invasion proteins are homologous to S. flexneri invasins, another secreted Salmonella protein contains two domains which each are similar to a different Yersinia antihost factor, respectively. Some secreted factors are even shared by Salmonella, Yersinia, and the plant pathogen X. campestris pv. vesicatoria. The secreted proteins are summarized in Table 1.
TABLE 1.
Proteins which are secreted via the type III secretion pathway
| Organism | Secreted proteina (reference) | Biochemical activity; interaction with host or other proteins | Effect on host cell; function | Remarks |
|---|---|---|---|---|
| Yersinia spp. | YopE, 219 (131) | Cytotoxic, F-actin disruption | Translocated | |
| YopH, 468 (59, 305) | PTPase dephosphorylates paxillin, FAK, p130cas | Inhibition of phagocytosis | Translocated | |
| YpkA, 732 (149) | Protein serine/threonine kinase | Translocated | ||
| YopM, 367 (56, 269) | Binds to thrombin | Translocated | ||
| YopJ/P, 264/288 (150, 313) | Induction of apoptosis in macrophages | Similar to plant pathogen AvrX and AvrA from S. typhimurium | ||
| YopR, 165 (14, 307) | ||||
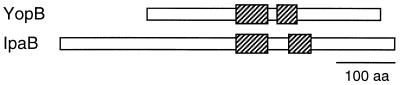
| YopB, 401 (173) | Pore-forming translocase | Translocation | ||
| YopD, 306 (173) | Translocation | |||
| YopK, 182 (206) | Modulation of translocation efficiency | |||
| YopN, 293 (129, 454) | Putative surface sensor, regulation of secretion | |||
| LcrV, 326 (41, 343, 359) | Suppression of immune response | Also regulatory function | ||
| LcrG, 95 (41, 359) | Regulation of secretion | |||
| LcrQ/YscM1/YscM2, 115 (307, 370, 422) | Involved in feedback transcriptional regulation | |||
| P. aeruginosa | ExoS, 453 (253) | ADP-ribosyltransferase | Cytotoxic, F-actin disruption | |
| ExoT, 457 | ADP-ribosyltransferase | |||
| ExoU, 687 (120) | Cytotoxic | |||
| PcrV, 294 (493) | ||||
| PopB, 392 (493) | Translocation (?) | |||
| PopD, 295 (493) | Translocation (?) | |||
| PopN, 288 (493) | Regulation of secretion (?) | |||
| S. flexneri | IpaB, 580 (450) | Binds to α5β1 integrin and to ICE protease | Induction of membrane ruffling, phagosomal lysis, induction of apoptosis | Additional function in regulation of secretion |
| IpaC, 382 (450) | Binds to α5β1 integrin | Induction of membrane ruffling, phagosomal lysis | ||
| IpaD, 332 (450) | Binds to α5β1 integrin | Cell invasion, phagosomal lysis | Regulation of secretion | |
| IpaA, 633 (451) | Binds to vinculin | Modulation of cell invasion | ||
| VirA, 400 (443) | Intercellular spread | |||
| S. typhimurium SPI-1 | AvrA, 302 (183) | |||
| Sip/SspB, 593 (234) | Cell invasion, induction of apoptosis | Translocation | ||
| Sip/SspC, 409 (216, 234) | Cell invasion, induction of apoptosis | Translocation | ||
| Sip/SspD, 338 (216, 233) | Cell invasion, induction of apoptosis | Translocation, regulation of secretion (?) | ||
| Sip/SspA, 684 (233) | ||||
| SopB, 561 (151) | Involved in intestinal transepithelial signalling, translocated | |||
| SopE, 240 (485) | Translocated | |||
| SptP, 544 (235) | PTPase | |||
| EPEC | EspA, 198 (242) | A/E lesion formation | ||
| EspB, 321 (104) | A/E lesion formation | |||
| EspC, 381 (259) | A/E lesion formation | |||
| Tir, 549 (239) | Binds to EPEC intimin | Receptor for EPEC attachment | Inserted in eukaryotic cell membrane, modified by tyrosine phosphorylation | |
| P. syringae | AvrPtob, 164 (390) | Binds to Pto kinase | Elicitation of HR | Host range determination, translocated (?) |
| HrpA, 113 (357) | Structural component of Hrp pilus | |||
| HrpZ, 341 (188) | Elicitation of HR | |||
| E. amylovora | HrpN, 385 (472) | Elicitation of HR | ||
| R. solanacearum | PopA1, 344 (26) | Elicitation of HR |
The size of the protein in amino acids is shown after the protein name. See text for further details and references.
Many more Avr proteins from various plant pathogens (recently summarized in reference 455) have been cloned and might be secreted and translocated via type III secretion pathways.
Virulence Proteins Secreted by Pathogenic Yersinia Species
The designation of the secreted virulence factors of Yersinia spp. as Yersinia outer proteins (Yops) originates from the fact that these proteins were first detected in outer membrane preparations of Y. enterocolitica (355) and Y. pseudotuberculosis (428). However, it was later observed that the Yops are secreted into the supernatant of Y. enterocolitica and Y. pseudotuberculosis (190, 191) and that their appearance in the supernatant does not result from cell lysis or from membrane vesiculation (191). Yops exhibit low solubility due to their generally hydrophobic character and tend to form amazingly abundant macromolecular filamentous aggregates in the supernatant of an induced culture (308), a phenomenon which has also been observed for proteins secreted by S. flexneri (337) and S. typhimurium (216, 485). Therefore, the localization of Yops in the outer membrane is probably an artifact of copurification of insoluble Yop aggregates with outer membrane fractions (308).
Interestingly and in contrast to Y. enterocolitica and Y. pseudotuberculosis, the Yops expressed by Y. pestis (392, 428, 484) are rapidly degraded by the surface protease Pla (392, 417), which is encoded by a Y. pestis-specific 9.5-kb plasmid (416). This plasmid and specifically the Pla protease are required for the pathogenesis of plague (418). However, although most Y. pestis Yops are subjected to degradation by Pla, undegraded YopM and YopN are secreted into the bacterial growth medium (368, 427).
The Yops—like the Yersinia type III secretion system—are encoded on a 70-kb virulence plasmid present in all three pathogenic Yersinia spp. (see “Type III secretion system genes in Yersinia species” below). The yop genes are often organized in monocistronic operons, and their expression is induced after a temperature shift from 25 to 37°C, but yop expression at 37°C remains repressed when the bacterial culture medium contains millimolar concentrations of Ca2+ (355). The Yops encoded by the plasmids from Y. pestis and Y. pseudotuberculosis are almost identical in sizes and isoelectric points, whereas several of the Y. enterocolitica Yops exhibit some size differences (57, 90). Nevertheless, the Yops of all three Yersinia spp. are immunologically related (57, 58, 191, 294, 415).
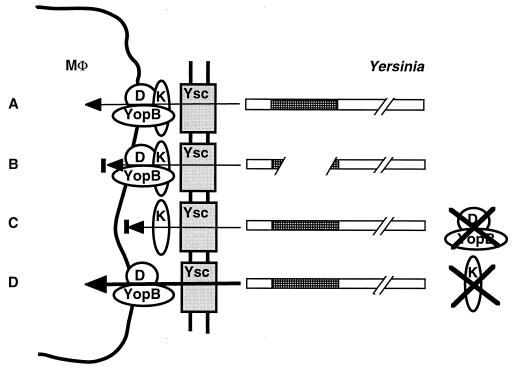
A total of 13 Yops and other proteins secreted by the Yersinia type III secretion system have been characterized to date. They can be roughly grouped into (i) proteins with direct antihost functions, the majority of which are translocated into eukaryotic cells (YopE, YopH, YopM, YpkA); (ii) translocatory proteins involved in the translocation process (YopB, YopD, YopK, and perhaps YopR); and (iii) regulatory proteins, which mediate the cell contact-dependent induction of yop gene expression and Yop secretion (YopN, LcrG, LcrV, and LcrQ).
The cytotoxin YopE.
Pathogenic Yersinia spp. exhibit a strong cytotoxic effect when adhering to the surface of eukaryotic cells, but cytotoxicity is not observed prior to adherence or when bacterial supernatant containing secreted Yops is added to epithelial cells (380, 381). Since cytotoxicity depends on the presence of YopE, Rosqvist et al. analyzed the localization of this protein before and after cell contact. They found that before cell contact, no YopE was secreted into the supernatant (due to a high concentration of Ca2+ in the tissue culture medium, which inhibits Yop expression and secretion in vitro [see “Negative control by Ca2+ via feedback regulation” below]), while after bacterial cell contact, YopE was expressed and all detectable YopE was present in the cytosol of the target cell (383). Similarly, it was demonstrated by the use of a YopE hybrid protein fused to the calmodulin-dependent adenylate cyclase CyaA from B. pertussis that YopE is translocated by adherent Y. enterocolitica into the cytosol of cultured epithelial cells (421) as well as into cultured murine macrophages (420). In this elegant approach, the fusion protein, which contained the amino-terminal 130 aa of YopE fused to the adenylate cyclase domain of CyaA (YopE130-Cya), catalyzes the formation of cyclic AMP from ATP in a calmodulin-dependent manner (420). Since calmodulin is present in the cytosol of eukaryotic cells but absent from bacterial cells and from the tissue culture supernatant, the observed dramatic accumulation of cyclic AMP in this assay system is indicative of transport of YopE-CyaA out of the bacterial cell and translocation of the fusion protein into the eukaryotic cytosol (421). Interestingly, translocalized YopE protein is not evenly distributed inside the target cell but is enriched in the perinuclear region (345, 383).
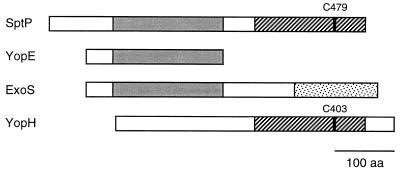
YopE is a 219-aa protein, which is highly similar in all three pathogenic Yersinia spp. (130, 131). As mentioned above, YopE functions early in Yersinia infection. While the secretion and translocation domains of YopE are amino-terminally located (see the sections on the secretion signal and the translocation domains of YopE and YopH, below), the domain responsible for the cytotoxic effect is probably situated in the carboxy-terminal one-third, since a truncated YopE lacking this part did not exhibit cytotoxicity although it was still normally secreted into the supernatant (381). YopE is similar to the amino-terminal half of exotoxin S of Pseudomonas aeruginosa (80) and to the amino-terminal half of SptP from S. typhimurium (235) (Fig. 3).
FIG. 3.
Homologies between the protein tyrosine phosphatase SptP of S. typhimurium, the cytotoxin YopE and the tyrosine phosphatase YopH from Yersinia spp., and the P. aeruginosa ExoS ADP-ribosyltransferase. Identically shaded boxes indicate regions of sequence similarities. The catalytic cysteine residue in the carboxy-terminal parts of SptP and YopH are shown. The predicted catalytic domain of ExoS is located in the carboxy-terminal quarter of the protein (stippled) (253).
The protein tyrosine phosphatase YopH.
A computer similarity search by Guan and Dixon (169) identified the 468-aa YopH (59, 305) to be homologous in its carboxy-terminal half to eukaryotic protein tyrosine phosphatases; accordingly, YopH was shown to be a specific (169) and by far the most active tyrosine phosphatase known (501). The catalytic domain is especially conserved, and exchange of the catalytic residue Cys403 to Ala (C403A) completely abolishes phosphatase activity (169). YopH confers upon Y. pseudotuberculosis the ability to resist phagocytosis by cultured macrophages (20, 112, 379) in vitro, and the antiphagocytic effect is dependent on its protein tyrosine phosphatase (PTPase) activity (20). Furthermore, the YopHC403A mutant is avirulent in mice (51), demonstrating the importance of YopH PTPase activity for Yersinia pathogenesis. Like yopE, yopH is transcribed as a monocistronic operon and is regulated at the transcriptional level by temperature and Ca2+ (59, 305). The yopH genes are highly conserved between Y. enterocolitica and Y. pseudotuberculosis. However, the sequence similarity stops abruptly 240 bp upstream and 175 bp downstream of the gene, showing that the homology between the virulence plasmids of the two species is confined to blocks of conserved DNA sequences (305). In addition to its homology to eukaryotic PTPases, YopH has a bacterial homolog in the carboxy-terminal half of the S. typhimurium type III secreted SptP, which also exhibits strong PTPase activity (235) (see below) (Fig. 3).
Like YopE, YopH is translocated into the eukaryotic cytosol after contact of Y. pseudotuberculosis with cultured cells (Fig. 2) (174, 345, 420). After cell contact, the phosphotyrosine phosphatase activity of YopH is present exclusively in the eukaryotic cytosol (345), where YopH is evenly distributed throughout the target cell (345). YopH translocation by Y. enterocolitica into cultured murine macrophages was also demonstrated by the Yop-Cya fusion approach (420) (see above).
The protein kinase YpkA.
Since Yersinia species secrete a protein phosphatase, Galyov et al. reasoned that there may also be a protein kinase in the supernatant of a Y. pseudotuberculosis culture. Indeed, the authors detected kinase activity and cloned the gene by sequence analysis of a small and (as of then) uncharacterized region of the Y. pseudotuberculosis virulence plasmid (149). YpkA is an autophosphorylating protein kinase with homology to eukaryotic protein kinases (149). The amino-terminal putative catalytic domain of YpkA contains structural motifs, called subdomains I to XI, which are common to all eukaryotic protein kinases, and the consensus sequence Asp-Ile-Lys-Pro-Gly-Asn in subdomain VI indicates that YpkA exhibits specificity for phosphorylation of serine and/or threonine (178). The 732-aa YpkA is transcribed as an operon together with the downstream YopJ protein. The operon is transcriptionally regulated analogously to the other Yops, but the level of expression is lower than that observed for YopH and YopE (150). Because YpkA is only weakly expressed and because the strong cytotoxic effects of YopE and YopH mask the effects of YpkA on HeLa cells, a yopE yopH mutant which overexpressed YpkA was used to demonstrate YpkA translocation (174). Interestingly, translocated YpkA was found to be associated with the inner surface of the plasma membrane of the target cell (174).
YopJ/P is required for apoptosis induction and is homologous to Salmonella and Xanthomonas proteins.
The 264-aa YopJ is transcribed as an operon together with YpkA in Y. pseudotuberculosis (150). (YopJ is called YopP in Y. enterocolitica [313].) Although the protein has characteristics of other Yops (150), YopJ/P is not required for virulence in mice (150, 431). Recently, YopJ/P was shown to be responsible for the induction of apoptosis in cultured murine macrophages (313). Interestingly, YopJ/P shows similarity to the plant avirulence factor AvrRxv from X. campestris (479) and to the avrA-encoded protein from S. typhimurium (183).
The thrombin binding factor YopM.
In addition to YopH and YpkA, YopM is the third secreted Yersinia virulence factor that is similar to eukaryotic proteins. The protein is homologous to the thrombin binding domain of the α chain of human platelet surface glycoprotein Ib (GPIbα) and also to a portion of von Willebrand factor (269). GPIbα is involved in cross-linking platelets by binding thrombin and von Willebrand factor, causing platelets to aggregate and initiate blood clotting at sites of blood vessel injuries. In addition, thrombin activates platelets, causing them to release a variety of inflammatory mediators (474). Purified YopM was shown to bind thrombin and to inhibit platelet aggregation in vitro (368). Although Y. pestis and Y. enterocolitica yopM mutants were strongly attenuated after i.v. infection of mice (269, 321), the significance of thrombin binding for Y. pestis pathogenesis is not clear. It is conceivable that YopM might compete with platelets for thrombin binding in vivo and that the resulting prevention of blood clot formation could contribute to the dissemination of the bacteria throughout the body. In addition, YopM could inhibit platelet activation in vivo and thereby might mute the local inflammatory response to the bacteria (368). More recently, it was demonstrated that YopM is also translocated into the cytosol of macrophages (56). To date, the 367-aa protein has been characterized in Y. pestis (269) and in Y. enterocolitica (56). Interestingly, YopM shares a region of 180 aa with IpaH from S. flexneri (187), a protein with unknown function which is encoded in multiple copies on the Shigella virulence plasmid, and with a protein of S. typhimurium, which is regulated by the global Salmonella virulence gene regulator PhoP (308a). Furthermore, a homolog of YopM was recently identified by sequence analysis of the Rhizobium spp. sym plasmid (139).
The V antigen LcrV.
LcrV, which appears to function in the regulation of secretion (see the section on regulation of type III secretion in Yersinia species, below), nevertheless has a direct antihost function. LcrV (virulence-associated or V antigen) is the oldest known secreted Yersinia virulence protein. Its association with the Y. pestis virulence phenotype was first described by Burrows and Bacon in 1956 (66). Although it has been difficult to assess the direct contribution of LcrV to virulence because of a global regulatory impact of various LcrV mutants (41, 358), a mutant with an internal deletion of LcrV (with residues 108 to 125 deleted), which is impaired in secretion of the protein but otherwise exhibits wild-type regulation, is avirulent in mice (414), suggesting that LcrV exerts an essential direct antihost function after being secreted. Interestingly and in contrast to other Yops, an amino-terminally deleted LcrV is partially secreted, indicating that an LcrV secretion signal is internally located or that LcrV secretion might functionally differ from the secretion of other Yops (414). A purified protein A-LcrV hybrid suppresses the production of inflammatory cytokines by infected cells (324, 325), and LcrV is protective for mice in active and passive immunizations against Y. pestis (262, 263, 317, 318).
Proteins Secreted by the Pseudomonas aeruginosa Type III Pathway
As mentioned above, P. aeruginosa secretes several proteins with high similarity to Yersinia type III secreted factors (135). The two related ADP-ribosyltransferases ExoS (453 aa) and ExoT (457 aa), which have 75% aa sequence identity, both carry an amino-terminal half similar to YopE, while the catalytic domain of the proteins resides in the carboxy terminus (135). Another secreted factor which is associated with epithelial cell damage is the 687-aa ExoU (120). ExoU is coregulated with other type III secreted proteins and is highly similar in its first 6 aa to ExoS and ExoT, suggesting that ExoU might also be secreted via the type III secretion pathway. Furthermore, proteins which show 40 to 60% sequence identity to Yersinia LcrV (PcrV, 294 aa), YopB (PopB, 392 aa), YopD (PopD, 295 aa), and YopN (PopN, 288 aa) are secreted by P. aeruginosa (493) and may be involved in protein translocation by P. aeruginosa as their homologs are in Yersinia spp.
Proteins Secreted by the S. flexneri Type III Pathway
IpaB, IpaC, IpaD, and IpaA.
As in Yersinia spp., the Shigella type III secretion system is encoded on a large virulence plasmid (see the section on the S. flexneri invasion gene cluster, below). The comparison of protein profiles from virulence plasmid-bearing minicells of Shigella spp. and enteroinvasive E. coli (EIEC) strains, which also carry the respective plasmid, led to the identification of a number of plasmid-encoded proteins common to both enteropathogens (176, 177). Expression of these proteins correlated with the ability of Shigella minicells to invade cultured HeLa cells (177), and, like the expression of the invasive phenotype, the synthesis of the plasmid-encoded proteins occurred only at 37°C, not at 30°C (176). Four of these proteins are the predominant Shigella antigens recognized by sera from human shigellosis patients and from monkeys infected with S. flexneri (67, 176, 330). The proteins were named IpaA to IpaD for invasion plasmid antigens (67), and the encoding genes were isolated by direct expression cloning (67, 236, 290) and transposon mutagenesis of the virulence plasmid (33, 402). A 31-kb region comprising the S. flexneri type III secretion system was identified in these analyses.
IpaB (580 aa), IpaC (382 aa), IpaD (332 aa) (399, 450, 494), and IpaA (633 aa) (451) are encoded in the ipgC-ipaBCDA operon located at the left end of the 31-kb invasion gene cluster (see Fig. 9). While mutations in ipaB, ipaC, and ipaD completely abolish the ability of Shigella spp. to enter epithelial cells (200, 304), several investigators have observed no phenotype for ipaA (33, 304, 400). However, an ipaA mutant was recently shown to exhibit a slight (10-fold) invasion defect (442), which appears to result from a reduced reorganization of cytoskeletal proteins by the ipaA mutant (see the section on Shigella flexneri, above). As mentioned above, the role of IpaB to IpaD is not confined to invasion, but these proteins are also required for lysis of the phagocytic membrane (200, 304, 507), and IpaB induces apoptosis in infected macrophages (75, 507).
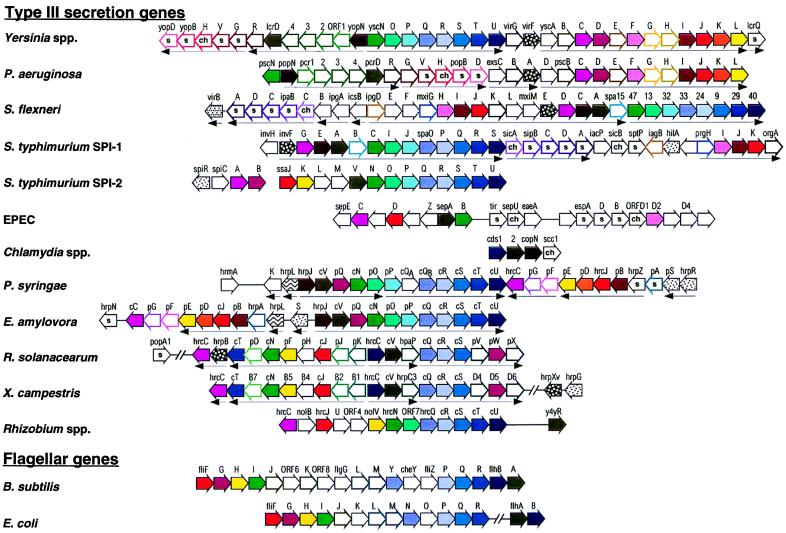
FIG. 9.
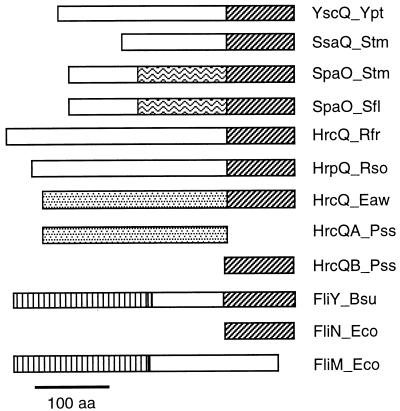
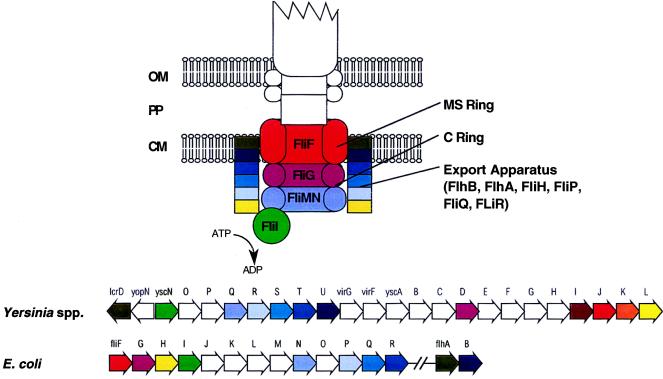
Genetic organization of type III secretion systems and of flagellum biosynthesis genes from B. subtilis and E. coli. Homologies of encoded proteins are indicated by color code (see the text and Fig. 10). The type III secretion systems of animal and plant pathogens are grouped according to genetic similarities. Solid arrows indicate broadly conserved genes, while genes which are conserved only between subgroups are outlined by thicker, colored lines. A thin black outline indicates that no homolog of the respective gene has been identified so far. The filling patterns indicate genes which encode transcription factors (see the text and Table 3). A small s inside a gene symbol indicates secretion of the encoded protein, while the genes which encode chaperonic proteins are labeled ch. Transcriptional units are indicated by arrows underneath the genes where known (see the text for references). For the plant type III secretion systems, the hrc and hrp gene designations are sometimes replaced with c and p, respectively. The genetic maps are drawn according to the references given together with the respective proteins in Fig. 10. See the text for further details.
After secretion into the external medium, IpaB and IpaC form a soluble complex, as shown by coimmunoprecipitation of the two proteins with antibodies against either one of them (303). In addition, this complex contains a protein of 72 kDa which was absent from the IpaB-IpaC complex isolated from the supernatant of an ipaA mutant and therefore probably is IpaA (303). IpaD is not stably associated with the IpaB-IpaC-IpaA complex (301, 302). Ménard et al. demonstrated that latex beads coated with the Ipa complex via antibodies to IpaC induce membrane ruffling and actin rearrangement in HeLa epithelial cells and that the coated beads are phagocytosed by these cells (301). Beads coated with IpaB-IpaC but lacking either IpaA or IpaD had the same effects on HeLa cells (301). It was further shown that affinity-purified IpaC (tagged with six amino-terminal His residues) specifically binds to Henle 407 cells in a concentration-dependent manner and that preincubation of Henle 407 cells with purified IpaC increases the invasion rate of S. flexneri and even promotes the uptake of a noninvasive derivative while purified IpaD does not exhibit either of these effects (286). Taken together, these results indicate that IpaB and IpaC are sufficient to induce the cellular responses leading to the uptake of Shigella into nonphagocytic cells. Together with IpaB, IpaD plays a regulatory role in protein secretion, which is discussed under “Cell contact-induced Ipa secretion: a one-step process modulated by IpaB and IpaD” below.
VirA.
In addition to the four Ipa proteins, at least six other polypeptides have been detected in concentrated supernatants of in vitro-grown S. flexneri (11) and about 15 proteins were detected in the supernatant of a mutant which exhibited enhanced type III secretion (337). None of the characterized secreted proteins carries an amino-terminal signal sequence, and all are secreted without amino-terminal processing. One of the additional secreted proteins, VirA, has been identified and characterized.
The virA locus was identified in a screen for transposon mutants of S. flexneri defective in invasion and intercellular spread (402, 443). The levels of IpaB through IpaD, whether intracellular, present on the bacterial surface, or secreted, were not affected by the virA mutation (443), indicating that the role of VirA in invasion and intercellular spread is independent of the Ipa invasins. The virA gene is located on the Shigella virulence plasmid ca. 24 kb away from the invasion gene cluster and encodes a hydrophilic protein of 400 aa which does not contain an amino-terminal signal sequence. The VirA protein was detected in the soluble cytoplasmic fraction as well as in the supernatant fraction of in vitro-grown S. flexneri, and its secretion is dependent on the type III secretion pathway. Like other genes of the type III secretion system, transcription of virA is under the positive, temperature-regulated control of the VirF-VirB system (see “Temperature regulation mediated by VirF-VirB” below). Interestingly, transcription of virA was reported to be greatly induced after S. flexneri had escaped from the phagocytic vacuole, suggesting that the primary role of VirA may be in intercellular spread rather than in invasion (443).
Proteins Secreted by S. typhimurium
S. typhimurium secretes at least 25 polypeptides larger than 14 kDa into the supernatant of an in vitro-grown culture in Luria-Bertani medium (339), and several of these proteins are absent from the supernatants of type III secretion mutants located in SPI-1 (216). Thirteen of the secreted proteins have been identified so far, and most or all of them require the type III secretion apparatus encoded in SPI-1 for their secretion. As yet, nothing is known about the proteins secreted by the SPI-2-encoded secretion apparatus. Some mutants with mutations in SPI-2, however, show a defect in protein secretion similar to that in SPI-1 mutants (196). These SPI-2 mutants are also partially impaired in epithelial cell invasion and macrophage cytotoxicity, two virulence phenotypes that are associated with proteins secreted by SPI-1. It is unclear how certain mutations in SPI-2 can affect SPI-1-dependent secretion.
Homology between the IpaA to IpaD and Sip/SspA to SspD proteins of S. typhimurium.
The most prominent proteins secreted by S. typhimurium are homologous to Shigella flexneri IpaA through IpaD proteins and accordingly have been named Salmonella invasion proteins SipA through SipD (233, 234) or, alternatively, Salmonella secreted proteins SspA through SspD (216). As in S. flexneri, the sip/ssp genes are clustered (see Fig. 9), and Sip/SspB (593 aa), Sip/SspC (409 aa), and Sip/SspD (338 aa) are required for bacterial invasiveness, while no phenotype has been found for a mutant with a mutation in sip/sspA encoding a protein of 684 aa (216, 233, 234). As in Shigella flexneri, secretion of the Sip/Ssp proteins depends on a functional type III secretion apparatus (encoded by SPI-1), and the proteins are secreted without amino-terminal processing. Also similarly to S. flexneri, it was observed that Sip/SspB is oversecreted in mutants that lack Sip/SspD (216, 233), suggesting that these proteins may have similar secretion-modulatory functions to those in S. flexneri (see “Regulation of type III secretion in S. flexneri” and “Hierarchical type III secretion in S. typhimurium” below).
The sequence similarities between IpaB, IpaC, and IpaD and the respective Sip/Ssp proteins range from 28% (B proteins), 32% (C proteins) to 40% (D proteins). Interestingly, the proteins are dissimilar at their amino-terminal regions of 80 to 130 aa while the similarity between IpaB and Sip/SspB increases to 65% in the central domain of the proteins (aa 300 to 450 of Sip/SspB). This central region of SipB also exhibits 28% identity to the respective region in the Yersinia YopB protein. YopB has a membrane-disrupting activity and is involved in protein translocation by Yersinia spp., and both IpaB and Sip/SspB are implicated in similar phenotypes (see “Translocation of proteins into the eukaryotic cytosol” below). Therefore, the sequence similarities between these proteins may be of functional significance.
After contact of S. typhimurium with epithelial cells, Sip/SspB and Sip/SspC are translocated across the eukaryotic membrane into the cytosol of the target cell via the type III secretion pathway (83). Translocation was also observed in the presence of the invasion inhibitor cytochalasin D, which blocks actin polymerization (126) and phagocytosis of bacteria (121), showing that the Sip/Ssp proteins are internalized independent of bacterial invasion. Interestingly, mutants with individual mutations in the sipB, sipC, and sipD genes prevented translocation of SipB and SipC, suggesting that all three proteins function together as protein translocases (83). In contrast, sipA did not affect translocation, which corresponds to its lack of impact on invasion. Also, the sptP-encoded secreted protein tyrosine phosphatase did not affect translocation or invasion (see below).
In addition to their role in invasion of epithelial cells, secretion of the Sip/SspB to Sip/SspD proteins is required for the induction of apoptosis in cultured macrophages, while Sip/SspA has no influence on host cell killing (74, 315).
SptP, a tyrosine phosphatase with homology to Yersinia YopE and YopH.
One of the structurally most fascinating secreted proteins discovered so far is the Salmonella SptP. The 544-aa protein, encoded downstream of the sip/ssp locus, shows homology in its amino-terminal part to YopE and ExoS from P. aeruginosa, while the carboxy-terminus of SptP corresponds to the carboxy-terminal part of YopH (Fig. 3) (235). Like YopH, SptP is a highly active PTPase, which shares a conserved catalytic domain with eukaryotic PTPases, and replacement of the catalytic residue Cys479 with Ser results in an inactive protein. However, despite the similarities to major virulence determinants of Yersinia and P. aeruginosa, SptP is not required for invasion of epithelial cells, nor does it influence phagocytosis by macrophages, and an sptP mutant is as virulent for mice as is the wild-type strain. Nevertheless, the mutant was somewhat less efficient in colonizing the spleens of mice which were coinfected with the wild-type strain (235).
AvrA is homologous to Yersinia and plant pathogen type III secreted proteins.
The 302-aa AvrA protein is secreted without amino-terminal processing via the SPI-1-encoded type III secretion pathway. The protein is encoded downstream from orgA within SPI-1 (183). Although an avrA mutant did not exhibit any defect in epithelial cell invasion, macrophage cytotoxicity, or mouse virulence, the protein is interesting in that it has significant sequence similarity to an avirulence protein from the plant pathogen X. campestris pv. vesicatoria (479) and to YopJ/P from Yersinia spp. (see above).
SopB and SopE proteins.
In addition to the proteins mentioned above, Salmonella dublin was found to secrete at least five other proteins which were labeled Salmonella outer proteins SopA through SopE (485). The proteins were found in massive amounts as mostly insoluble aggregates in the supernatant of a S. dublin strain which was inactivated for expression of the Sip/Ssp proteins and flagellin (encoded by fliM). Interestingly, the inactivation of Sip/Ssp and flagellin led not only to highly increased secretion of Sop proteins but also to an about fivefold increase in the expression of at least one Sop, SopE (485). The reason for increased Sop expression and secretion in the sip/ssp fliM mutant is unclear but may indicate an interesting feedback regulatory mechanism.
SopB (561 amino acids) is highly similar to an S. flexneri protein, IpgD, of unknown function, which is encoded within the S. flexneri type III secretion gene cluster. SopB is not required for bacterial invasiveness, but the mutant shows a significant defect in induction of an intestinal inflammatory response (151). SopE is a 240-aa protein which also has no or only a minor impact on bacterial invasiveness (485). Both SopB and SopE are secreted without amino-terminal processing, and both proteins appear to be translocated into the cytosol of HeLa cells. In agreement with the results of Collazo et al. described above, translocation of SopB and SopE was found to require the Sip/SspB protein (151, 485).
Secreted Proteins of Enteropathogenic E. coli
A total of 5 (240) to 11 (223) polypeptides, ranging in size from 110 to 19 kDa, are present in the supernatant of EPEC E2348/69 grown in tissue culture medium. At least seven of these proteins were recognized by a rabbit antiserum raised against the bulk of secreted EPEC proteins (223). Furthermore, the EPEC secreted proteins were shown to be highly immunogenic in humans (223). With the exception of the 110-kDa EspC protein, which is a type IV secretion pathway autotransporter (see “Introduction”), none of the antigenic proteins were detected in the supernatant of a type III secretion mutant (223).
Tir, a translocated bacterial receptor for intimin.
A protein which migrates at approximately 72 kDa is secreted under culture conditions which especially induce EPEC type III secretion (238, 239). Tir is encoded by the EPEC type III secretion gene cluster and was shown to be identical to a previously observed protein which is tyrosine phosphorylated upon EPEC attachment (239). Tir contains two potential transmembrane domains and was localized in the eukaryotic cell membrane, where it functions as a receptor for the bacterial attachment factor intimin (see “Enteropathogenic Escherichia coli” above). After transfer into the eukaryotic membrane, Tir is phosphorylated on C-terminally located tyrosine residues, and this phosphorylation may be involved in the second function of Tir, namely, as an actin nucleator and in transmission of signals to the eukaryotic cell (238, 239, 376).
EspA, EspB, and EspD.
Kenny and Finlay determined the amino-terminal sequences of five of the EPEC secreted proteins (240). Of the proteins whose secretion depends on the type III secretion pathway, one was identified as the previously described 37-kDa EspB (99, 104, 240). The encoding gene is located at the “right” end of the EPEC type III secretion gene cluster (see Fig. 9). The protein is required for host cell signalling (132), i.e., tyrosine phosphorylation of Tir and accumulation of tyrosine phosphorylated proteins and actin filaments beneath adherent bacteria (see “Enteropathogenic Escherichia coli” above). The 321-aa EspB shows a 20-aa stretch near its amino terminus which contains 18 serine or threonine residues. In addition, the central part of the protein contains a highly conserved pyridoxal-phosphate binding site which is found in several aminotransferases involved in biosynthetic pathways (104). The significance of these observations, however, remains to be determined. Like all other proteins that exit the bacterial cell via the type III pathway, EspB is secreted without amino-terminal processing.
Like EspB, both EspA (198 aa) (240, 242) and EspD (381 aa) (259) require the type III pathway for their secretion. The genes encoding EspA and EspD are located immediately upstream of espB. Secretion of all three proteins is required for intimate attachment and host cell signalling, and at least EspA and EspB are required to insert Tir in the eukaryotic membrane (239). The EPEC secreted proteins were found to associate tightly with host cells, and EspB became resistant to digestion by proteinase K, suggesting that this protein may be translocated into the host cell cytoplasm (242).
Another protein (39 kDa) whose secretion depends on the type III pathway has a similar amino terminus to the amino terminus of E. coli glyceraldehyde-3-phosphate dehydrogenase (14 of 16 residues identical). The significance of the secretion of this protein is unknown.
Plant-Pathogenic Bacterial Proteins Secreted by the Type III Pathway
Secretion of proteins by the type III secretion pathway is required for the pathogenicity of various gram-negative plant pathogens and for the determination of the bacterial host range (see “Plant-pathogenic bacteria” above). Several secreted proteins which cause disease on susceptible plants and induce an HR defense reaction in resistant plants have been identified. These proteins have been named harpins, a name derived from the hrp gene designation for the plant pathogen type III secretion pathway. Other proteins, termed avirulence proteins (Avr), determine bacterial host range according to the gene-for-gene concept. Avr proteins act inside the host cell and therefore are likely to be translocated into the target cell, but their secretion and translocation has not directly been demonstrated. Furthermore, several proteins have been observed to be secreted by the type III pathway into the supernatant of an in vitro-grown culture of Pseudomonas syringae (499).
Erwinia harpins.
HarpinEa, from Erwinia amylovora, was the first harpin discovered. The small (385-aa), heat-stable, glycine-rich, hydrophilic protein was shown to elicit an HR in tobacco leaves when purified from either E. amylovora or Escherichia coli expressing the hrp gene cluster (472). The protein was found in association with the bacterial membrane fraction and is secreted without amino-terminal processing. The encoding gene, named hrpN, is located at the left side of the hrp cluster (see Fig. 9). Demonstrating the requirement for harpinEa in E. amylovora pathogenicity, an hrpN mutant does not elicit an HR in tobacco and does not cause fire blight in rosaceous plants (472).
In contrast to the E. amylovora hrpN mutant, a mutant with a mutation in the corresponding E. chrysanthemi hrpN gene is not completely impaired in its ability to cause disease but exhibits a reduced frequency of infection in witloof chicory leaves (34). Nevertheless, the mutant is impaired in its ability to elicit an HR in various nonhost plants. Like harpinEa, the 340-aa harpinEch is glycine rich, and the two proteins are highly similar in their carboxy-terminal one-third (34).
Harpins from P. syringae pathovars.
HarpinPss from P. syringae pv. syringae (named HrpZPss) is even less exclusively required for pathogenicity and HR elicitation than is harpinEch in E. chrysanthemi. Thus, although the purified protein elicits an HR in nonhost plants (7, 188), a mutant with a deletion of the encoding gene in P. syringae pv. syringae still induces an HR, albeit at reduced levels compared to the wild type. However, when the same mutation was introduced into the hrp gene cluster expressed in E. coli, the resultant strain no longer induced an HR. Thus, HrpZPss contributes significantly to but is not essential for P. syringae pv. syringae virulence, and P. syringae pv. syringae must contain more HR elicitors (7). As with the respective Erwinia genes, hrpZ is located at one side of the hrp gene cluster in P. syringae pv. syringae (see Fig. 9). Interestingly, a mutant with a mutation in hrmA, a gene located downstream of the other end of the P. syringae pv. syringae hrp gene cluster, does not induce an HR in tobacco leaves, although the mutant secretes normal amounts of HrpZ (7). In the wild-type strain, more than half of the total amount of HrpZPss is found in the supernatant of an overnight culture, and secretion is dependent on the type III pathway (188).
HrpZPss (341 aa) is a glycine-rich (13.5% Gly), mildly thermostable, hydrophilic protein (188) that exhibits interesting structural features. It contains three direct repeats of 7, 5, and 4 identical amino acids and five additional repeats of 24 similar amino acids. Throughout the protein, 14 relatively evenly spaced short hydrophobic regions and 9 probable α-helices are found (7). These repeated structural motifs appear to confer functional redundancy, since various purified amino- and carboxy-terminal parts of the protein are sufficient to elicit an HR (7, 188).
hrpZ genes have also been identified in P. syringae pv. glycinea and tomato (357). The HrpZPsg and HrpZPss proteins have 79% identity, while HrpZPst is 63% identical to HrpZPss. Interestingly, the gene encoding HrpZPst may have undergone horizontal gene transfer and interspecies recombination between P. syringae pv. tomato and Ralstonia solanacearum, since HrpZPst exhibits a 24-aa glycine-rich insertion which is almost identical to an internal part from the otherwise unrelated PopA1, the “harpin” from R. solanacearum (see below) (357). Although P. syringae pv. syringae, glycinea, and tomato have narrow host ranges, purified HrpZ proteins of these pathovars cause an HR in tomato leaves but show no effect when infiltrated in leaves of other host and nonhost plants. Thus, the HrpZ proteins do not directly appear to determine the host range of these P. syringae pathovars (357).
AvrPto from P. syringae pv. tomato binds to tomato Pto kinase.
As described in the section on plant-pathogenic bacteria (above), the avirulence factor AvrPto from P. syringae pv. tomato elicits an HR in tomato plants by direct binding to the cytoplasmic Pto kinase (408, 436). AvrPto is a small (164-aa), mostly hydrophilic protein. Although secretion of the protein has not been demonstrated in vitro, AvrPto depends functionally on an intact type III secretion apparatus (390).
PopA1 from R. solanacearum.
PopA1 (for Pseudomonas out protein), whose secretion by R. solanacearum (formerly Pseudomonas solanacearum) depends on a functional type III pathway (26), exhibits structural features similar to the harpins discussed above: the protein is relatively small (344 aa), glycine rich, and heat stable, and, like HrpZ from P. syringae, it does not contain any tyrosine residues, a feature which was speculated to allow these proteins to escape H2O2-mediated cross-linking of tyrosine residues in plant cell walls during the defense reaction (64, 188). Purified PopA1 exhibits HR-inducing activity on plants (tobacco and some petunia lines) which naturally develop an HR resistance reaction in response to infection with R. solanacearum. However, PopA1 has no activity on tomato, the natural host plant of R. solanacearum (26). Therefore, the protein appears to act like an avirulence protein rather than a direct inducer of plant disease (for a definition of avirulence factors, see “Plant-pathogenic bacteria” above). However, in contrast to classical avirulence genes, popA mutants retain their ability to elicit an HR in tobacco and other nonhost plants and also still cause disease in tomato (26).
After secretion, the PopA1 protein is successively processed at the amino terminus (a deletion of 9 aa results in PopA2; a shortening by 93 aa results in PopA3). The reason for this processing is not apparent, especially since PopA3 exhibits the same biological activity as PopA1. The popA gene is located approximately 3 kb to the left end of the hrp gene cluster in R. solanacearum (see Fig. 9) (26). Expression of the gene is dependent on the transcriptional activator hrpB (26) (see “Transcriptional regulation in R. solanacearum and X. campestris” below), and therefore popA is coregulated with other genes of the type III secretion system and belongs to the hrp regulon (155).
TRANSLOCATION OF PROTEINS INTO THE EUKARYOTIC CYTOSOL
Translocation of bacterial virulence proteins into eukaryotic target cells appears to be a common feature of most if not all type III secretion systems (83, 383, 408, 436, 485). The term “translocation” is used to describe the transport of proteins from the bacterial cell through the eukaryotic plasma membrane into the cytosol of the target cell, while “secretion” refers to the transport of proteins from the bacterial cytoplasm to the extracellular space. Although translocation requires the ability to secrete proteins, secretion and translocation are functionally separate events, and the information required for translocation differs from that required for secretion. Thus, the secreted proteins contain dedicated and distinct secretion and translocation domains, and translocation involves additional accessory proteins, which are themselves secreted via the type III secretion pathway. Translocation occurs only at sites of close contact between the pathogen and the target cell. Protein secretion in vitro may thus represent only an experimental artifact which occurs under laboratory conditions that mimic the regulatory conditions encountered by the bacteria after contact with the surface of a target cell.
Translocation of Yop Proteins by Yersinia Species
The mechanism of Yop translocation by pathogenic Yersinia spp. has been analyzed in some detail and involves (i) the type III secretion apparatus (see the section on the proteins that constitute the type III secretion apparatus, below), (ii) the translocated antihost proteins (see “Virulence proteins secreted by pathogenic Yersinia species” above), (iii) a surface-exposed control protein that functions in concert with a number of secretion regulatory factors (see “Regulation of type III secretion in Yersinia species” below), and (iv) a set of dedicated translocator proteins, which are discussed here.
General characteristics of translocation of Yersinia Yop proteins.
Translocation of Yop proteins is an active process which requires live bacteria (380), protein synthesis (345, 384) and the adherence of bacteria to the target cell (345, 381, 383, 421). In fact, it was already observed by Portnoy et al. in one of the earliest studies on Y. enterocolitica cytotoxicity that close contact of the bacteria with tissue culture cells was required for the bacteria to exert a cytotoxic effect (355). No cytotoxic activity is observed when secreted and concentrated Yop proteins or cell-free lysates of whole cells are added to epithelial cells (355, 380), nor do nonadherent mutants (yadA [192]) exhibit cytotoxicity, i.e., Yop translocation (141, 380, 421). Furthermore, cytochalasin D does not influence Yop translocation (174, 420, 421), demonstrating that neither bacterial internalization nor movement of the cytoskeleton is involved in Yop translocation.
With respect to the kinetics of Yop translocation, several conflicting observations have been made. Translocation of YopH and YpkA into cultured HeLa cells was analyzed by the addition of gentamicin, which effectively inactivates extracellular bacteria (449), at various time points after infection of cells with Y. pseudotuberculosis (174, 345). Translocation of YpkA was blocked only when gentamicin was added prior to 30 min postinfection (174), while addition of the drug 1 h after infection still inhibited YopH translocation (345), suggesting that the translocation process requires 30 min or longer. However, these experiments used immunofluorescence to detect the translocated protein. When analyzing the pattern of protein tyrosine phosphorylation induced in cultured macrophages after contact with Y. pseudotuberculosis, Andersson et al. observed a YopH-dependent dephosphorylation of several proteins within 1 min after bacterial cell contact (20), implying very rapid translocation of enzymatically active amounts of YopH. In this respect, it is of further interest that the proteins secreted by EPEC also exhibit an immediate effect on host cells. Thus, killing of extracellularly adherent EPEC as early as 5 to 10 min after the initial contact with epithelial cells still allowed the bacteria to trigger the full program of A/E lesion formation (see “Enteropathogenic Escherichia coli” and “Secreted proteins of enteropathogenic E. coli” above) (377), again indicating rapid protein translocation.
Translocation of the Yops is polarized; i.e., it occurs only at the zone of contact between a bacterium and the target cell surface, while no Yops are released into the surrounding medium (383). The molecular basis of this phenomenon, which involves the surface control protein YopN, is discussed in the section on regulation of type III secretion in Yersinia species (below).
The protein translocases YopB and YopD.
Translocation of YopE, YopH, YopM, and YpkA across the eukaryotic plasma membrane requires YopB and YopD as protein translocases (56, 174, 175, 345, 383, 420, 421) and involves YopK as a modulatory factor (205). The Yops which enable and modulate translocation are not subject to translocation themselves, but, rather, remain located at the bacterial surface during the translocation step (205). Nevertheless, YopB, YopD, and YopK are transported to the bacterial surface via the type III secretion pathway and thus require the secretion apparatus for functionality.
YopB and/or YopD mutants are avirulent (175, 186) and are specifically affected in the translocation step, while secretion of other Yops is not affected in these mutants (41, 174, 175, 321, 380, 421, 466). Accordingly, after cell contact of a yopBD mutant, Yops accumulate at the zone of contact between the pathogen and the target cell (345, 383). To function in translocation, YopB and YopD have to be supplied by the same bacterial cell which translocalizes a Yop, since a mutant lacking YopB and YopD could not be transcomplemented for translocation by coinfection with a strain providing YopB and YopD but lacking the translocated Yop (420). Taken together, these data suggest that YopBD-mediated translocation immediately follows secretion and occurs at contact points between bacteria and eukaryotic cells (383, 420).
YopB and YopD are encoded in the lcrGVH-yopBD operon, shown at the left side of the type III secretion gene cluster in Fig. 9. Transcription of the operon is induced by temperature and repressed in the presence of Ca2+ (41, 359). Both YopB and YopD are potential transmembrane proteins and thus differ from the predicted globular pattern of other Yops (173). The 401-aa YopB contains two hydrophobic regions of 44 and 35 aa, separated by 15 aa, in the central part of the protein (173), a structural motif reminiscent of pore-forming toxins of the RTX family (475). YopD (306 aa) has one central hydrophobic region of 31 aa and a carboxy-terminal 14-aa amphipathic helix (173). Because of their putative transmembrane structure, YopD and YopB possibly form pores in the eukaryotic cell membrane through which other Yops are translocated into the cytosol of the target cell. Indeed, it was shown that a multiple yop mutant strain of Y. pseudotuberculosis, which expressed YopB, induced a pore with an apparent size of 1.2 to 3.5 nm in sheep erythrocytes after contact of the bacteria with the target cell and that purified YopB disrupted membranes in vitro (175). Whether YopB and YopD directly interact with translocated Yops during the translocation step remains to be determined. However, it is interesting that a strain which expressed YopE in addition to YopB exhibited reduced lytic activity, implying that during translocation the occupancy of the YopB-induced pore by translocated Yops may maintain membrane integrity (175).
YopB is 24% identical (47% similar) (173) to IpaB from S. flexneri and to Sip/SspB from Salmonella spp., which also contain two central hydrophobic regions (Fig. 4). The similarities between YopB, IpaB, and Sip/SspB may be of significant functional importance, since (i) like YopB in Yersinia spp., IpaB is required for the contact-hemolytic activity of S. flexneri (200); (ii) S. typhimurium translocates proteins into eukaryotic cells, a process which—together with Sip/SspC and Sip/SscD—requires Sip/SspB (83, 485); and (iii) Yersinia YopE can be translocated into epithelial cells by S. typhimurium expressing the Yersinia yopE gene (382). Thus, like YopB, the homologous proteins probably act as membrane translocators. Whether P. syringae, which also probably translocates proteins via the type III secretion pathway (408, 436), and other organisms contain similar translocator proteins remains to be determined.
FIG. 4.
Similarity of hydrophobic regions (shaded boxes) in the putative membrane translocator proteins YopB and IpaB (173).
Modulatory role of YopK in translocation.
Like YopB and YopD, YopK (206) is not translocated into eukaryotic cells but, rather, appears to be located in the vicinity of cell-associated bacteria during the infection process (205). An avirulent (206, 321, 429) yopK mutant (see “Yersinia species” above) exhibits several interesting phenotypes which, together, led Holmström et al. to suggest that YopK controls the amount of Yop translocation by negatively influencing the size of the YopB-induced pore in the target cell membrane: a yopK mutant exhibits increased YopB-dependent cell lytic activity, an elevated level of Yop translocation, and, consequently, a stronger cytotoxic effect (205). In contrast, a strain which overexpresses YopK is impaired in Yop translocation and exhibits a much reduced lytic activity, with the apparent size of the YopB-induced pore being smaller than 1.2 nm (205). Like the other Yops, YopK is secreted by the type III secretion pathway and the gene is regulated by temperature and Ca2+ (206). YopK is expressed at levels considerably lower than the other Yops (206), a phenomenon which may be associated with its regulatory role in translocation.
The secretion signal may reside in the mRNA.
In addition to the type III secretion apparatus and the translocator proteins discussed above, information which is located within the individual anti-host Yop factors is required for their secretion and translocation. As in other type III secretion systems, the amino terminus of the secreted Yops is required for secretion (306). For both YopH and YopE, deletion analysis and reporter protein fusions have been used to determine the minimal regions of the genes and proteins required for their secretion and translocation. Two detailed analyses both employed Yopn-Cya fusions with various numbers (n) of amino-terminal amino acids of either YopE (404, 420) or of YopH (420) fused to the catalytic domain of B. pertussis adenylate cyclase (see “Virulence proteins secreted by pathogenic Yersinia species” above). Sory et al. found that a YopE15-Cya fusion and a YopH17-Cya fusion were still effectively secreted by Y. enterocolitica into the supernatant (420), and replacement of the first 6 aa of YopH with the amino-terminal 12 aa of β-galactosidase resulted in a nonsecreted hybrid protein (306). Schesser et al. showed that 11 amino-terminal amino acids of YopE fused to Cya were sufficient to expose the fusion protein on the surface of Y. pseudotuberculosis (404). Site-directed replacement of any of the first 7 aa of YopE11-Cya with glycine led to a significant decrease in the ability to export the fusion protein, while individual replacement of aa 8 to 10 with glycine had no drastic effect (404). In the case of YopN, the first 15 aa were sufficient for secretion (19). Thus, it has long been thought that the secretion signal resides in the very amino terminus of the secreted Yops.
A new perspective on the problem has recently been introduced by Anderson and Schneewind, who suggested that the secretion signal may reside in the mRNA encoding a type III secreted protein (19). The authors introduced nucleotide insertions and deletions in the second codon of both YopE and YopN, respectively, which shifted the original reading frame to the two other frames, and scored the secretion of the respective reporter fusions to neomycin phosphotransferase (Npt). Except for a shift in YopE to the third reading frame, which generated a very hydrophobic amino-terminal peptide, all the mutations allowed secretion of the respective constructs. The authors also individually substituted aa 2 to 15 of both YopE and YopN with alanine and found that exchange of serine 4 to alanine in YopE abolished secretion and translation of the protein. Selection for spontaneous suppressor mutants of YopE4S-A-Npt identified several intragenic suppressors, which all contained a CCC to CCA transversion in codon 12. This suppressor mutation restored the translation and secretion of the fusion protein without altering the amino acid sequence. Since the amino termini of the proteins appear very tolerant to mutational changes without loss of function and since the isolated suppressor at codon 12 in YopE does not affect the peptide sequence, the authors concluded that the signal recognized by the secretion apparatus is mRNA rather than the peptide sequence. Furthermore, since secretion-deficient mutants were also impaired in translation, the authors suggested that secretion and translation may be coupled. Indeed, the 5′ ends of the YopE and YopN mRNAs both are predicted to form stem-loop structures which bury the AUG translational start codon in a base pair duplex while positioning codons 2 and 4 within a loop. Mutations that abolish translation either are located within the predicted loops or affect its adjacent base pairs and could affect recognition of the structure by an RNA-binding protein. Since secretion appears to be cotranslational, the authors further suggested that the mRNA assumes a translatable conformation only after contact with the secretion apparatus.
Translocation domains of YopE and YopH.
In contrast to the short regions required for secretion, at least 50 amino-terminal amino acids of YopE and approximately 70 aa of YopH are required for these proteins to be translocated into eukaryotic cells (404, 420). Thus, a YopH71-Cya fusion is effectively translocated into macrophages while a YopH56-Cya fusion is too short for translocation (420). In agreement with these results, Persson et al. observed that a YopHΔ49–154 deletion, which exhibits wild-type PTPase activity and is secreted like wild-type YopH, was no longer translocated (345). In a similar approach, it was demonstrated that a fusion protein consisting of the amino-terminal 40 aa of YopM fused to CyaA is secreted but not translocated whereas a YopM100-Cya fusion is both secreted and translocated (56). YopE, YopH, and YopM are, thus, modular proteins consisting of a translocation domain and an effector domain (Fig. 5). In this respect, it is interesting that the flagellar hook protein FlgE from Caulobacter crescentus, which is exported without being processed via the type III secretion-related flagellum-specific transport pathway (see “Proteins that constitute the type III secretion apparatus” below), also carries an amino-terminal secretion signal, albeit not located at the very amino terminus but, rather, comprising aa 38 to 58 (250). Figure 6 summarizes the mechanism and factors involved in Yop translocation.
FIG. 5.
Translocation domains (shaded boxes) of YopE and YopH as determined by reporter gene fusions. The approximate boundaries of the domains are shown in amino acids. Secretion requires the 5′ region of yop genes (codons 1 to 15). The secretion signal may reside in the mRNA (see the text for details).
FIG. 6.
(A) Secretion and translocation of a Yop from Yersinia spp. into the cytosol of a macrophage (MΦ). (B) Deletion of the translocation domain (ca. aa 15 to 50) results in loss of translocation. (C) YopB and YopD are required for translocation but not for secretion. (D) A YopK mutant exhibits enhanced translocation. See the text for details.
Protein Translocation by Other Type III Secretion Systems
As mentioned above and in the section on proteins secreted by S. typhimurium (above), translocation has also been demonstrated for proteins secreted by the S. typhimurium SPI-1-encoded type III secretion system (83, 485) and probably occurs in other systems as well (408, 436). No detailed analysis of the minimal secretion and translocation signals has been conducted for other type III secretion systems, so far. However, in the cases analyzed, the secretion signal was localized to the very amino terminus of the secreted proteins (306, 308, 491). The Y. pseudotuberculosis YopE protein can be secreted and translocated by S. typhimurium, while Y. pseudotuberculosis and S. typhi can secrete the Shigella flexneri IpaB protein (197, 382) and ExoS from P. aeruginosa is secreted and translocated by Y. pseudotuberculosis (140), indicating a conserved secretion mechanism. Since these proteins do not exhibit any recognizable structural or physicochemical similarities, the secretion signal may indeed by RNA.
Hrp Pilus and Type III Secretion-Related Surface Structures of S. typhimurium
Does translocation of type III secreted proteins into eukaryotic cells, at least in some systems, involve the assembly of a supermolecular surface structure functioning as a pipeline for protein translocation? At least for P. syringae, the answer to this question is probably yes, since P. syringae pv. tomato produces a pilus-like surface structure which is induced when bacteria are grown on solid, type III secretion gene-inducing medium at a temperature below 25°C (374). The filamentous surface appendages are 6 to 8 nm in diameter and were observed on strains deficient in type IV pilus production but were absent from type III secretion mutant strains (374). Two of the proteins which are found in the supernatant of an in vitro-grown P. syringae culture (499) were shown to be present in a purified pilus fraction; one of these was identified as HrpA, an 11-kDa hydrophilic protein, which is encoded at the right end of the hrp gene cluster in P. syringae (see Fig. 9). HrpA is secreted without amino-terminal processing and is required for bacterial pathogenicity and for two avr genes to trigger genotype-specific HR in resistant plants (374). Thus, it is possible that P. syringae employs this type III secretion-dependent pilus structure to translocate proteins into the target cell. A gene encoding a protein homologous to HrpA is also present in the Erwinia amylovora type III secretion system (244), implying that this pathogen also expresses a pilus structure.
Another novel macromolecular surface structure related to type III secretion was observed on S. typhimurium in epithelial cell infection experiments (158). Shortly after S. typhimurium contacted the surface of polarized cells, the bacteria were seen to express pilus-like surface appendages which covered the entire bacterial cell. These relatively short, fat appendages were approximately 60 nm in diameter and 0.1 to 0.3 mm long and were not seen on in vitro-grown bacteria. Interestingly, the appendages were not seen on bacteria which actually invaded the cell through a membrane ruffle, suggesting that appendages are only transiently expressed. Although the transient expression of surface appendages depends on functional type III secretion (158), further analyses have not been able to demonstrate the direct relation between appendage formation and expression of the SPI-1-encoded type III secretion system (M. A. Jepson, personal communication).
In general, it seems possible that proteins secreted by the type III secretion pathway assemble into specific, pathogenesis-associated macromolecular structures, since macromolecular aggregation has been observed for several of these proteins (216, 308, 337, 485), and many proteins secreted via the type III pathway are predicted to form coiled-coil structures which could be involved in the respective protein-protein interactions (84, 334).
TYPE III SECRETORY CHAPERONES (BODYGUARDS)
Many of the type III secreted proteins in different species require small, usually acidic, cytoplasmic accessory proteins, which bind specifically to individual secreted proteins, for their secretion and presecretory stabilization. Although the type III secretory chaperones do not target the secreted proteins to the secretion apparatus (141, 483), they appear to prevent premature interactions of different secreted proteins with each other and/or with parts of the secretion and translocation machinery (303, 483). According to this preventive and protective function, the type III secretion system chaperones have also been named bodyguards (483). However, a specific secretory role for these chaperones has recently been observed in some mutants (76).
Yop-Specific Chaperones and Their Role in Yop Secretion
SycE/YerA and SycH.
YopE and YopH each require an additional factor to be efficiently secreted. For YopE, this factor is named YerA (Yop regulator A) in Y. pseudotuberculosis (131) and SycE (specific YopE chaperone) in Y. enterocolitica (467). sycE/yerA (131, 467) and sycH mutants (345, 466) exhibit strongly reduced secretion of YopE or YopH, respectively, whereas secretion of other Yops is not affected in these mutants. The defect in YopE secretion is not due to reduced yopE transcription, which is not affected by SycE/YerA (467). The defect in YopE secretion in sycE/yerA mutants may be partially due to reduced YopE translation, since YopE does not accumulate in the cytoplasm (467) and the half-life of YopE is reduced in the sycE/yerA strain (141). However, for YopH, SycH clearly affects secretion, since YopH accumulates in the cytoplasm of the sycH mutant (466). Since radiolabelled SycE or SycH specifically binds to nitrocellulose-blotted YopE or YopH, respectively (467), Wattiau and Cornelis suggested that SycE and SycH might act as Yop-specific chaperones which are involved in posttranscriptional stabilization of the Yop proteins prior to secretion (467). The interaction of YopE with its chaperone was shown to also occur in vivo by copurification of YopE with a biologically active YerA-GST fusion protein (141).
It is important to note that although secretion of YopE is strongly reduced in the sycE/yerA mutant (131, 467), the yerA strain of Y. pseudotuberculosis still translocated 2 to 3% of the wild-type level of YopE. Furthermore, in contrast to type III secretion mutants, the yerA mutant exhibited an albeit delayed cytotoxic effect (141). Therefore, SycE/YerA does not appear to be absolutely necessary for the secretion and translocation process but, rather, seems to function in presecretory stabilization of YopE.
Both SycE and SycH are similar in that they are small (14.7 and 15.7 kDa, respectively), acidic (pI 4.4 and 4.9, respectively) and cytoplasmic (141, 467). In addition, SycE was shown to form homodimers (467), and it is not secreted or translocated into the target cell cytoplasm (141). Despite the lack of overall sequence homology between SycE and SycH, these proteins exhibit similar predicted physicochemical properties and secondary structures. In particular, the hydrophobic moment plots, which are almost identical for these proteins, show two major hydrophobic peaks at the carboxy termini of SycE and SycH (466). The amino acid sequence of the last peak in both proteins contains a conserved motif, Leu-Xaa6-Asn-Xaa6-Leu-Xaa6-Leu-Xaa6-Leu, which resembles a leucine zipper and, according to a secondary-structure prediction, might be arranged as an α-helix (466).
SycE/YerA is encoded immediately upstream of yopE and is transcribed in the opposite orientation to yopE in all three pathogenic Yersinia spp. (131), while sycH is located downstream from yopH in Y. enterocolitica and is transcribed in the opposite direction to yopH (466). (In Y. pseudotuberculosis, however, sycH is not located near yopH [345].) Transcription of the sycE gene is induced by temperature in dependency of VirF (see “Transcriptional regulation of Yop expression” below) and is partially repressed in the presence of Ca2+ in Y. enterocolitica (467), while no Ca2+ repression of YerA expression was observed in Y. pseudotuberculosis (141).
SycE and SycH bind to the translocation domains of their cognate Yop.
Woestyn et al. mapped the domains of YopE and YopH which are required for binding of their respective chaperones (483). By copurification of SycE with various YopEn-Cya fusions (n indicates the number of amino-terminal YopE residues present in the fusion), these authors determined that the first 50 aa of YopE (YopE50-Cya fusion) are indispensable for SycE binding (483). In a similar approach, Schesser et al. found that a YopE49-Cya fusion was already impaired in YerA binding (404). Therefore, the boundary of the SycE/YerA binding domain in YopE is at residue 50. Deletion of aa 2 to 14 from native YopE (yielding YopEΔ2–14), however, had no effect on SycE binding, but deletion of aa 36 to 46 completely abolished binding of the chaperone. For YopH, it was shown that a YopH69-PhoA fusion bound SycH (466), and the SycH-binding domain of YopH was determined to lie within aa 20 to 69, while aa 2 to 19 is not required for SycH binding (483).
Surprisingly, a YopE40-Cya fusion and YopHΔ20–69, both impaired in binding their cognate chaperone, were effectively secreted into the supernatants of sycE and sycH mutants, respectively. In contrast, fusions of YopE (YopE130-Cya) and YopH (YopH YopH69-PhoA) capable of binding their chaperone both required the respective chaperone for effective secretion (483). (Comparable results were obtained for the YopE-YerA interaction in Y. pseudotuberculosis [404] and for secretion of YopE by Y. enterocolitica [76].) These results demonstrate that the chaperone binding domains in YopE and YopH differ from the domains required for secretion of these Yops. In fact, the chaperone binding domains are located immediately C-terminal of the secretion domains and coincide with the domains required for YopE and YopH translocation (Fig. 5 and 7). Since the translocation domain may facilitate interaction of the Yop with other proteins during the secretion and translocation process, the chaperones (or bodyguards) may prevent this interaction prior to secretion and thus appear to function as anti-association factors (483). Deletion of the translocation domain would abolish that interaction and thus abolish the requirement for the chaperone to protect the translocation domain. Since YopB and YopD are required for the translocation process and may interact with the Yops during translocation (see “The protein translocases YopB and YopD” above), Woestyn et al. tested whether a yopBD sycH double mutant behaved differently from a sycH mutant with respect to the secretion of native YopH. In accordance with their assumptions, YopH was secreted in larger amounts by the double mutant than by the sycH mutant but not as efficiently as by a yopBD strain (483). These results indicate that SycH, and probably SycE in the case of YopE, may protect the translocation domain of YopH from premature interaction with YopB and YopD (and/or other proteins) in order to allow functional YopH secretion (Fig. 7).
FIG. 7.
Schematic representation of the function of the Yop-specific chaperones in secretion. (A) The Yop-specific chaperone binds to the translocation domain of its cognate Yop protein. (B) In the absence of the respective chaperone, secretion of the Yop is impaired. (C) Deletion of the translocation domain abolishes the requirement for the chaperone for secretion. (D) In the absence of the chaperone and YopB/D, secretion is (partially) restored, indicating that the chaperone functions to protect premature interaction of the secreted Yop with YopB/D. See the text for further details.
The secretion of YopE and YopH derivatives which are impaired in binding their partner chaperone also demonstrates that the chaperones do not facilitate the recognition of different Yops by the secretion apparatus. Rather, Yop secretion functions independently of the chaperones. This notion is additionally supported by the fact that after contact with a target cell, a yopD yerA double mutant accumulated YopE in discrete spots at the zone of interaction between the bacteria and eukaryotic cell surface in the same way as a yopD mutant did (141, 383). Therefore, targeting of YopE to the putative secretion and translocation loci does not require the chaperone.
LcrH/SycD.
A third putative Yop chaperone was found to be encoded by lcrH (466), which previously had been shown to play an important regulatory role in the low-calcium-response (see “Regulation of type III secretion in Yersinia species” below). Like SycE and SycH, LcrH, which is encoded in proximity to YopBD in the lcrGVH-yopBD operon, is a small (19-kDa) cytoplasmic protein with a predicted pI of 4.5 (359). An lcrH mutant specifically lacks YopB and YopD in the supernatant, and YopD could not be detected intracellularly in the mutant (466). Coimmunoprecipitation of YopD and LcrH demonstrated the capability of the proteins to bind to each other. Therefore, LcrH is considered to be the YopD- and possibly YopB-specific chaperone and was renamed SycD (466). However, in contrast to mutations in either sycE/yerA or sycH, which affect the secretion only of their cognate Yop, an lcrH/sycD mutation has a global regulatory effect on yop expression. Despite its overall similarities to SycE and SycH, SycD does not contain the leucine zipper-similar sequence motif in its carboxy terminus. Nevertheless, the C terminus of SycD displays a high hydrophobic moment (466).
IpgC from S. flexneri Inhibits the Cytoplasmic Interaction of the Secreted Ipa Proteins
In S. flexneri, the invasion plasmid gene ipgC, located upstream of ipaB (see Fig. 9), encodes a small (17-kDa) cytoplasmic protein which is required for S. flexneri invasiveness and functions as a chaperone for the virulence factors IpaB and IpaC (303). Ménard et al. showed that IpgC independently binds to either of the two Ipa proteins in the bacterial cytoplasm (303). In the absence of the chaperone, the two Ipa proteins interact with each other in the cytoplasm as they do after secretion into the supernatant (see “Proteins secreted by the S. flexneri type III pathway” above), and the resulting IpaBC complex is degraded in the cytoplasm. The main target for proteolytic degradation appears to be IpaB, since IpaC is relatively stable in the absence of both IpgC and IpaB, and binding of the chaperone to IpaB is sufficient to prevent proteolysis of IpaB. Interestingly, deletion of the two hydrophobic domains in the carboxy-terminal region of IpaB, which are homologous to similarly located domains in YopB and may be involved in pore formation in eukaryotic cell membranes (Fig. 4), abolished the requirement for IpgC to stabilize IpaB. Taken together, IpgC appears to act as an antiassociation factor which prevents the deleterious formation of the IpaBC complex before secretion (303). Interestingly, IpgC exhibits some similarity (27% identity) to the YopBD chaperone LcrH/SycH.
Homologs of Yop Chaperones in Other Bacteria
Homologs of the Yop chaperones have also been identified in other bacteria (469). SycH is 27% identical to YeaE (225), a putative chaperone encoded immediately upstream of the EPEC eaeA gene, while SycE is 46% identical to the ORF1-encoded protein (492), a putative chaperone for the P. aeruginosa ExoS enzyme. Recently, a homolog of SycE termed Scc1 (146 aa, pI 5.2) has been detected in Chlamydia psittaci (211). A third group of homologous chaperones includes SycD, IpgC from Shigella flexneri (303, 450), SicA from Salmonella spp. (197, 234, 322), and an uncharacterized open reading frame ORFo163 from E. coli (GenBank accession no. 887802). Except for the Yop chaperones and IpgC, the other putative chaperones have not been functionally analyzed so far.
REGULATION OF TYPE III SECRETION BY CONTACT WITH EUKARYOTIC CELLS
According to the role of type III secretion systems in translocation of virulence proteins into eukaryotic cells, several type III secretion systems are activated by contact of the bacteria with the surface of eukaryotic target cells. Interestingly, however, the regulatory proteins required for cell contact-activated protein secretion differ in Yersinia spp. and S. flexneri, the two organisms for which this phenomenon has been unambiguously demonstrated (302, 383). Furthermore, the respective regulatory proteins are either not present or not conserved in other bacterial type III secretion systems. Thus, some type III secretion systems comprise accessory factors which are not directly involved in the core secretion mechanism but function in a sophisticated manner to regulate secretion.
An important difference between protein secretion by Yersinia spp. and S. flexneri is that yersiniae secrete and translocate proteins in a polarized manner (i.e., the secreted factors are translocated into the cytosol of the target cells but are not secreted into the tissue culture medium [345, 383, 421]), whereas, in contrast, S. flexneri secretes Ipa proteins into the surrounding medium after contact with a target cell (302). In addition, Yersinia spp. and S. flexneri exhibit important differences in presecretory conditions. Thus, while S. flexneri stores secreted proteins in the cytoplasm prior to contact with a target cell (302), pathogenic Yersinia spp. even keep expression of the secreted factors repressed until they encounter a target cell (347). This section discusses the regulation of secretion in Yersinia and Shigella spp., while transcriptional regulation of type III secretion systems is discussed in a later section. This section also discusses the few known mechanistic aspects of type III secretion in other pathogens.
Regulation of Type III Secretion in Yersinia Species
As mentioned above, Yersinia spp. coregulate the expression of type III secreted proteins with their secretion; i.e., expression is derepressed only when the secretion channels are opened. (Basically, this regulation is achieved by secretion of a factor which negatively regulates expression [see the section on transcriptional regulation of type III secretion genes, below].) The secretion channels are opened by contact of the bacteria with a target cell. However, efficient secretion can also be artificially induced in vitro by removal of Ca2+ from the growth medium, since Ca2+ appears to be required to keep the secretion channels shut in the absence of target cells. Indeed, regulation of secretion by Ca2+ had been discovered long before the regulation by target cell contact, and artificial induction of secretion in the absence of Ca2+ has been extensively used to analyze the Yersinia type III secretion mechanism and its complex regulation. For reasons of clarity, I discuss almost all factors involved in Ca2+ regulation under “Transcriptional regulation of type III secretion systems” (below), while the present section describes the only factor which has been shown directly to regulate polarized Yop translocation in vivo.
The surface-exposed sensor protein YopN regulates polarized Yop translocation.
Rosqvist et al. used immunoblotting to analyze the location of YopE after contact of Y. pseudotuberculosis with HeLa cells. They found that all detectable YopE was present in the cytosol of the target cells, while no YopE was detected in the tissue culture supernatant. However, when a yopN mutant was used to infect HeLa cells, some YopE was detected in the cell fraction but most of it was released into the supernatant (383). Similar results were obtained for the location of YopH after target cell infection, which was analyzed by measuring the enzymatic PTPase activity of the protein. While wild-type bacteria translocated more than 99% of all detectable YopH into HeLa cells, a yopN mutant released approximately 50% of the protein into the surrounding medium (345). (Note that in contrast to these results, Boland et al. observed a significant release [about 50%] of various Yop protein-reporter fusions into the tissue culture supernatant of PU5-1.8 macrophages [56]. The reason for this discrepancy is not clear. One possible explanation is the use of reporter protein fusions instead of the native Yop proteins.) Thus, the 293-aa YopN, which is called LcrE in Y. pestis (129, 454), appears to control in the polarized secretion and translocation of Yop proteins into eukaryotic cells. Residual protein translocation in the absence of YopN demonstrates that YopN is not directly involved in the translocation step.
YopN is exposed on the bacterial surface of in vitro-grown bacteria (129), and under low-calcium conditions this protein is secreted, together with the other Yop proteins, via the type III secretion pathway (57, 129). In contrast to other Yops, however, YopN is not translocated into eukaryotic cells (56). Since yopN/lcrE mutants secrete large quantities of Yops even under high-Ca2+ conditions (129, 381), YopN is thought to function as a regulatory plug that blocks Yersinia type III secretion channels in the presence of Ca2+ or in the absence of eukaryotic cells. Thus, YopN appears to be a surface-exposed sensor protein which controls type III secretion in response to a signal on eukaryotic cells, facilitating a localized opening of the secretion channels at sites of cell contact, while secretion channels which do not directly contact the cell surface are kept shut. In fact, the physiological signal repressing secretion at noncontact zones may be extracellular Ca2+, which could be required to maintain a certain active conformation of YopN (383). Interaction of YopN with an unknown component on the eukaryotic cell surface could either replace Ca2+ or, more probably, change the conformation of YopN so that secretion is allowed at the points of contact.
As mentioned above, other factors besides YopN are also involved in the regulation of type III secretion in Yersinia spp., and some of these factors may interact with YopN and with the core secretion apparatus to regulate secretion (see “Transcriptional regulation of Yop expression” below).
Regulation of Type III Secretion in S. flexneri
The mechanism by which S. flexneri regulates type III secretion after contact of the bacteria with eukaryotic cells differs entirely from the mechanism functional in Yersinia spp. However, although S. flexneri uses different factors from Yersinia spp. to regulate secretion, the S. flexneri regulatory proteins are also secreted via the type III pathway. While in Yersinia spp. the secreted Yop proteins are expressed only when secretion is activated, S. flexneri accumulates Ipa proteins in the cytoplasm and secretes these presynthesized proteins when the bacterium contacts a eukaryotic cell. Ipa proteins are also partially secreted in the absence of cells, indicating that the type III secretion channels are somewhat leaky or that in vitro culture conditions can mimic conditions that induce Ipa secretion in vivo. It is unclear whether secretion of Ipa proteins, i.e., their transport from the cytoplasm into the external medium, happens in a single step or is a two-step process involving intermediate deposition of the proteins on the bacterial surface before their cell contact-induced release. The following section discusses the data supporting either of these two modes of Ipa secretion.
Cell contact-induced Ipa secretion: a one-step process modulated by IpaB and IpaD.
During in vitro growth of S. flexneri, ∼90% of the IpaB to IpaD proteins accumulate in the bacterial cytoplasm. Upon contact with HeLa epithelial cells, however, almost all of the cytoplasmically stored Ipa proteins are secreted into the surrounding medium within the first 15 min after cell contact (302). Cell contact-induced secretion is regulated by IpaB and IpaD. During in vitro growth, a small portion of the total amount of these two proteins is associated with the bacterial membranes, where they form a complex as a transient intermediate in their own secretion. The localization of the IpaBD complex in the inner or outer membrane was not unambiguously resolved, but cofractionation of IpaB and IpaD with the outer membrane protein OmpC in a sucrose gradient flotation suggested that the IpaBD complex is located in the outer membrane (302). The membrane-associated IpaBD complex appears to constitute a plug of the secretion channels, since ipaD or ipaB mutants constitutively secrete other Ipa proteins and additional polypeptides in large amounts via the type III pathway (302, 304, 337). In the presence of an inducing signal, provided by contact of the Shigella cell with the surface of an epithelial cell, the IpaBD complex presumably dissociates, releasing the block of the secretion channels and allowing rapid secretion of Ipa proteins in high local concentrations into the surrounding medium. The fact that IpaD is indispensable for bacterial invasiveness (304), although it is not part of the IpaBC complex that induces membrane ruffling (see the section on proteins secreted by the S. flexneri type III pathway, above), may be due to a need for high local concentrations of IpaB and IpaC at the site of bacterial cell contact to induce membrane ruffling. Such high concentrations may be achieved only if bacterial cells are preloaded with a larger amount of Ipa proteins at the moment of cell contact.
Cell contact-induced Ipa release: a two-step model for Ipa secretion involving Spa32.
The alternative model for Ipa secretion is based on the observation of localization of IpaB, IpaC, and IpaD on the bacterial surface prior to cell contact-induced release into the medium (464). All three Ipa proteins are detectable by whole-cell enzyme-linked immunosorbent assay and immunofluorescence on the surface of in vitro-grown S. flexneri, and the ratio of surface-exposed to cytoplasmic Ipa was found to be 1:2 for each of these proteins (464). No respective immunological signal can be obtained with type III secretion mutants (401, 452, 464) (with the exception of spa32 [see below]), demonstrating that the Ipa proteins are transported to the bacterial surface via the type III secretion pathway. Upon contact with monkey kidney epithelial cells (MK2 cells), 80 to 90% of the three surface-localized Ipa proteins are released into the medium within 10 min (464, 465). However, in contrast to Ménard et al. (see above), Watarai et al. found that the amount of cytoplasmic Ipa proteins decreases slowly: even after 30 min of cell contact, ∼75% of the starting material was still found in the cytoplasm (464).
The two-step model for Ipa secretion is supported by the phenotype of a noninvasive mutant with a mutation in spa32, which is part of the type III secretion gene cluster and encodes an outer membrane protein. A nonpolar mutant with a mutation in spa32 differs from other noninvasive type III secretion mutants in that it is still able to expose IpaB, IpaC, and IpaD on the bacterial surface but is impaired in the subsequent release of these proteins. The invasiveness of the spa32 mutant was restored by the addition of cell-free, Ipa-containing growth medium in the infection assay, and this effect was blocked by further adding specific IpaBCD antibodies. Addition of the same Ipa-containing medium did not restore the invasiveness of another type III secretion mutant (464). Taken together, these data suggest that the Ipa invasins must be exposed on the bacterial surface but that subsequent cell contact-induced release of the Ipas is also required to promote invasion.
Different secretion signals may account for differences in the two models for Ipa secretion.
The two models for Ipa secretion are not mutually exclusive, although some data supporting one model are hard to reconcile with the other. The most obvious data difference between the two models is the amount and speed of cell contact-induced Ipa secretion. Perhaps, and for lack of a better explanation, the observed differences are due to the different cell lines used in both studies. Ipa secretion is also induced, albeit to different extents, by various other conditions including extracellular matrix proteins such as fibronectin, laminin, and collagen type IV (464), some unknown soluble factor(s) present in fetal bovine serum (302), Congo Red (337), and specific bile salts (352). Thus, secretion of cytoplasmic Ipa proteins may be induced by a signal present in different concentrations under the different conditions tested, or, more likely, by various signals to different extents, and the amount of induced secretion may be modulated by a more or less stringent release of the IpaB-IpaD membrane complex as proposed by Ménard et al. (302). The other contradiction in the data supporting either of the two models is the localization of Ipa proteins, most notably of IpaC, in in vitro-growing bacteria, either solely in the cytoplasm (302) or, at about 50% of their total amount, on the bacterial surface (464). While IpaC can clearly be detected on the surface by immunological techniques (see above), a xylene extraction of whole cells and a flotation sucrose gradient did not allow detection of IpaC in any membrane-associated fraction (302). These differences may be resolved if similar experimental conditions, especially with respect to bacterial growth conditions, and more refined techniques were applied to investigate this point. Nevertheless, the two models agree that the actual release of the Ipa proteins from the bacteria, either from the cytoplasm or from the surface, is induced by contact with eukaryotic cells and is essential for invasion.
It is interesting that Ipa secretion is not induced after contact of S. flexneri with the apical pole of polarized Caco-2 cells, an observation that correlates with the inability of S. flexneri to invade polarized epithelial cells from the apical pole (see “Shigella flexneri” above). However, when the tight junctions between polarized cells were permeabilized, allowing the bacteria to reach the basolateral pole of the cells, Ipa secretion and cell invasion were both strongly enhanced (464). Another point worth mentioning is that cell contact-induced secretion of Ipa proteins takes place preferentially at the site of septum formation of actively dividing bacterial cells (320). Figure 8 gives a schematic overview of the regulation of secretion by contact with a eukaryotic cell in Yersinia and Shigella spp.
FIG. 8.
Schematic representation of regulation of type III secretion by contact with a eukaryotic cell (shown in the middle) in Yersinia and Shigella spp. For Yersinia spp., prior to cell contact (upper left), the type III secretion channels (Ysc) are kept shut by YopN, and cytoplasmic accumulation of the negative regulatory factor LcrQ leads to transcriptional repression of yop genes (see the text). After cell contact, YopN is released, allowing rapid secretion of LcrQ, which in turn relieves the block from yop gene expression. Anti-host Yop proteins, protected by cognate chaperones (Syc), are translocated into the eukaryotic cell via the type III secretion machinery and YopB/D. For Shigella spp. (only the model of regulation via IpaB/D is shown [see the text for further details]), before contact with a eukaryotic cell (lower right), the S. flexneri type III secretion channels (Mxi and Spa) are kept shut by a complex of IpaB and IpaD. Ipa proteins accumulate in the cytoplasm and are protected from interaction and proteolytic degradation by the chaperone IpgC (C in a triangle). IpaB and IpaC, however, are partially secreted and form a protein-protein complex in the bacterial supernatant. After target cell contact, the IpaB/D block is released and high local concentrations of the IpaB/C complex induce membrane ruffling and uptake of Shigella spp.
Hierarchical Type III Secretion in S. typhimurium
Compared to type III secretion by Yersinia spp. and S. flexneri, little is known about the secretion mechanism in S. typhimurium. However, an interesting observation concerns the fact that some S. typhimurium proteins that are secreted via the type III secretion pathway are themselves required for the secretion of others. Thus, InvJ/SpaN (336 aa) (84, 168) and SpaO (303 aa) (168) are both secreted into the supernatant of an in vitro-grown bacterial culture (84, 271) but are also required for secretion of the Sip/Ssp invasion proteins (see “Proteins secreted by S. typhimurium” above) (82). In contrast, a mutation in sipD does not affect the secretion of InvJ/SpaN (82). Therefore, a hierarchy exists in the type III secretion process where some secreted proteins are required for the secretion of others. The genes encoding InvJ/SpaN and SpaO are both located within the type III secretion gene cluster, and both proteins are required for bacterial invasion (82, 84). InvJ/SpaN is slightly similar to Spa32 from Shigella flexneri, while SpaO is homologous to the YscQ family of type III secretion apparatus proteins (see the section on proteins that constitute the type III secretion apparatus, below). In contrast to Spa32, however, InvJ/SpaN is not required for the surface presentation of Sip/SspB (82). Thus, although the type III secretion systems are rather similar between S. typhimurium and Shigella flexneri, the details of the secretion mechanisms appear to differ between these organisms.
GENETIC AND TRANSCRIPTIONAL ORGANIZATION OF TYPE III SECRETION GENES
The genes encoding type III secretion systems—especially those genes which encode the secretion apparatus—are clustered. In some organisms, these gene clusters are located on plasmids which are unique to the pathogen and are not found in nonpathogenic relatives (Yersinia spp., S. flexneri, and R. solanacearum). In other pathogens (S. typhimurium, EPEC, P. aeruginosa, P. syringae, E. amylovora, R. fredii, and X. campestris), the type III secretion gene clusters are located on the chromosomes and often appear to have been acquired during evolution, since related nonpathogenic bacteria lack these pathogenicity islands but share corresponding adjacent sequences.
Figure 9 shows the genetic organization of the 12 known type III secretion systems in 11 different pathogens as well as the organization of genes encoding homologous proteins in the flagellar biosynthesis systems of B. subtilis and S. typhimurium. The color code represents homologies of the encoded proteins (see below). For some of the type III secretion systems, the available sequence information probably represents only parts of the complete set of genes. This is particularly true for the gene clusters from S. typhimurium SPI-2, EPEC, and P. aeruginosa, which are known to extend beyond the region shown in Fig. 9 (102, 193a, 491) but for which comprehensive sequence information has not been published. A complete set of genes required to encode a functional type III secretion system is likely represented by the gene clusters from Yersinia spp. and S. flexneri. The cloned type III secretion gene clusters from EPEC (297) and P. syringae pv. syringae (214, 272) conferred upon nonpathogenic hosts the ability to elicit the respective type III secretion-related virulence phenotype, demonstrating the “completeness” of the set of type III secretion genes within the respective clusters.
Table 2 gives an overview of the genomic location of some type III secretion gene clusters and their relative G+C contents as compared to the G+C content of the rest of the respective genomes.
TABLE 2.
Overview of localization, relative G+C contents, and lengths of pathogenicity islands which encode type III secretion system gene clusters
| Organism | Location | Relative G+C content of type III gene cluster (%) (reference) | Medium relative G+C content of rest of genome (%) (reference) | Length (kb) (reference) |
|---|---|---|---|---|
| Yersinia spp. | 70-kb plasmid | 44 | 46–50 | ca. 20 |
| P. aeruginosa | Chromosomal, 55 min (493) | 67.5 (491) | 69 (491) | ? |
| S. typhimurium SPI-1 | Chromosomal centisome 63 | 42–47 (63) | 52 (332) | ca. 40 (312) |
| S. typhimurium SPI-2 | Chromosomal centisome 30.7 | 42–45 (194, 332) | 52 (332) | ? |
| S. flexneri | 220-kb plasmid | 34 (332) | 51 (332) | ca. 31 |
| EPEC | Chromosomal, centisome 82 | 38.4 (102) | 51 (332) | 35.5 |
| Chlamydia spp. | ? | 40 (211) | 41–44% (211) | ? |
| P. syringae | Chromosomal (365) | ca. 61a | ? | 22 (365) |
| E. amylovora | Chromosomal | ca. 58a | ? | ca. 25 (470) |
| R. solanacearum | Megaplasmid | 69.1a | ? | ca. 27 (448) |
| X. campestris | Chromosomal | 64.6a | ? | ca. 25 (61) |
| Rhizobium spp. | sym plasmid (139) | 56 (139) | 58.5% for the sym plasmid (139) | 17.7 (139) |
Determined from published sequences.
Type III Secretion System Genes in Yersinia Species
The type III secretion system of pathogenic Yersinia spp. is encoded on large virulence plasmids, which are of similar sizes (between 66 and 72 kb) in all three virulent species (39, 117, 153, 154, 160, 193, 356, 505). The virulence plasmids from Y. pestis and Y. pseudotuberculosis are similarly structured (57, 356) and are functionally interchangeable (484), while the Y. enterocolitica plasmid is structurally different (57, 356). However, a block of approximately 20 kb, which contains the genes encoding the core type III secretion apparatus, is remarkably conserved in all three species. Sequence analyses have revealed different and overlapping parts of the DNA sequence of that region in all three organisms. The genes of that region which have been sequenced in more than one species encode proteins with high degrees of similarity (15, 40, 41, 129, 172, 307, 370, 482). (When combined, the known sequences from the three species include the entire 20 kb.) The 31 genes of this region are organized in at least eight transcriptional units, comprising lcrGVH-yopBD (41, 173, 321), yopN (129, 498), lcrDR (31, 349), yscN-U (40), virG (13), virF (89, 130, 497), yscA-L (172, 307), and lcrQ (307, 370). (The gene corresponding to lcrQ in Y. enterocolitica, yscM1, is part of the yscA-LM1 operon [307, 422].) The genetic and operon structure of this region is shown in Fig. 9.
The components of the type III secretion apparatus are encoded by yscA to yscL and yscN to yscU (mnemonic for Yersinia secretion), and lcrD (for low calcium response). In addition, a number of proteins which regulate transcription and secretion (virF, lcrR, lcrGVH, yopN, and lcrQ) and the translocator proteins YopB and YopD are encoded in the 20-kb virulence gene cluster. The direct anti-host Yop proteins are encoded outside this region, and their genes are scattered around the virulence plasmids and distributed differently on the plasmids of the three pathogenic Yersinia species (15, 90, 345).
In Y. pseudotuberculosis, the 12 genes yscA to yscL are organized as an operon (14, 172, 370), while in Y. enterocolitica the downstream gene yscM1 also belongs to this transcription unit (307, 422). A temperature-induced and Ca2+-regulated promoter (see the section on transcriptional regulation of Yop expression, below) is located upstream of yscA (172, 307), while in Y. pestis, an additional internal Ca2+ regulated promoter was detected within yscF (172). Of the 12 genes in the yscA to yscL operon, 8 have unequivocally been shown to be required for Yop secretion: yscD (307), yscE to yscG, and yscI to yscK (14) in Y. enterocolitica and yscC, yscD, and yscG in Y. pestis (351). yscH, surprisingly, turned out to encode a secreted virulence factor, YopR (14). The gene downstream of yscL, which is called lcrQ in Y. pseudotuberculosis and yscM1 in Y. enterocolitica (422), encodes a key negative regulatory factor of the Yersinia type III secretion system (see the section on transcriptional regulation of Yop expression, below). In Y. pseudotuberculosis, the gene is separated from yscL by 120 bp (222 bp in Y. enterocolitica) and is probably transcribed as a monocistronic operon in Y. pseudotuberculosis (370). Clearly, LcrQ/YscM1 is not a constituent of the general type III secretion apparatus, since Yop secretion in the absence of Ca2+ is not affected by lcrQ/yscM1 (14, 370).
A second large operon of type III secretion genes comprises yscN to yscU (40, 118, 454). Two promoters have been identified upstream of yscN, one within the reading frame of the upstream gene lcrE, which is transcribed in the opposite direction of yscN to yscU (454). A potential transcriptional terminator lies 360 bp downstream of yscU (13, 15). Mutations that abolish Yop secretion have been described in yscR, yscS, and yscT in Y. pseudotuberculosis (40), in yscN (482) and yscU (15) in Y. enterocolitica, and in yscQ and yscR in Y. pestis (118).
The yopN, lcrD, and lcrR genes are probably cotranscribed, since their reading frames overlap (31, 349). Further reading frames in Y. enterocolitica are found downstream of yopN. LcrD is a conserved and essential component of the secretion apparatus, while YopN and probably LcrR are secretion regulatory factors (see “Regulation of type III secretion in Yersinia species” above). The five genes at the “left” end of the Yersinia type III secretion gene cluster, lcrGVH-yopBD (Fig. 9) are organized as an operon (41, 359). The proteins encoded by lcrGV function mainly in regulation of secretion in Yersinia spp., while the secreted proteins YopB and YopD and their putative chaperone LcrH play specific roles in protein translocation. The two genes which are located in between the yscN to yscU and yscA yscL operons, virG (13), and virF (87), encode an outer membrane secretion factor and an AraC-homologous transcriptional activator, respectively.
P. aeruginosa Type III Secretion System Genes
The chromosomal type III secretion system in P. aeruginosa has only recently been discovered by characterization of ExoS secretion-deficient mutants (491). The part of the secretory apparatus which has been identified so far is encoded by 12 genes, 11 of which are arranged colinear to the Yersinia yscB to yscL genes (Fig. 9). Except for their homologs in Yersinia spp., the pscB-, pscE-, and pscGH-encoded proteins do not have homologs in other type III secretion systems. In P. aeruginosa, the exsD-pscB to pscL genes are transcribed as a single operon (491). The exsD-encoded protein is unique to P. aeruginosa, but the preceding operon (exsCBA [490]) encodes homologs of Yersinia VirG (ExsB) and VirF (ExsA). Like the Yersinia VirF, the ExsA protein is a positive regulatory factor of the exsD-pscB to pscL operon (490), which belongs to the AraC family of transcriptional regulators (136). Yahr et al. reported that transposon insertions located immediately upstream of the exsCBA operon also led to an ExoS secretion-deficient phenotype, and by homology to Yersinia spp., it is likely that this region contains additional genes of the P. aeruginosa type III secretion system. (A number of P. aeruginosa type III secretion genes homologous to respective Yersinia sp. genes have recently been described [135] and are included in Fig. 9.)
S. flexneri Invasion Gene Cluster
As in Yersinia spp., the Shigella type III secretion system is encoded in one continuous stretch of DNA and is located on a large (220-kb) virulence plasmid (290, 400) which is present in all invasive isolates of Shigella (396, 397) as well as in EIEC (177, 184, 394). Transfer of the virulence plasmid from S. flexneri to noninvasive E. coli K-12 conferred invasiveness upon the latter strain (395), demonstrating that all essential invasion determinants are encoded on the virulence plasmid.
The Shigella invasion gene cluster contains 32 genes comprising a total of 31 kb (338). The ipaBCDA genes (32, 450, 451, 494), located on one end of the invasion gene cluster (shown on the left in Fig. 9), encode the secreted invasins (IpaB, IpaC, and IpaD) (304) and IpaA (304, 399). The type III secretion apparatus is encoded by the 20 mxi-spa genes (mnemonic for membrane expression of Ipa and surface presentation of Ipa antigens) (8–12, 22, 24, 399, 401, 452), 13 of which were shown to be required for Ipa secretion and invasiveness, while no functional data are available for 6 genes, mxiHI and mxiKLME (300). Of the mxi-spa genes analyzed, only spa15 turned out to be dispensable for Ipa secretion and Shigella invasiveness (401).
In addition to the secretion apparatus and the secreted invasins, the invasion gene cluster contains a regulatory gene, virB, located downstream of the ipa operon, which encodes an activator of invasion gene transcription (4, 68, 461, 495). Furthermore, the ipa and mxi-spa genes are linked by a block of seven genes (icsB, ipgA to ipgF), only three of which (ipgABC) are required for invasion (399). ipgC encodes an Ipa-specific chaperone (see “IpgC from S. flexneri inhibits the cytoplasmic interaction of the secreted Ipa proteins” above) (303). icsB does not affect bacterial invasiveness but is required for the spread of intracellular S. flexneri to adjacent cells (9), while the function of ipgD, which encodes a 60-kDa secreted protein, and of ipgE and ipgF is not clear (8).
The invasion genes are organized in two blocks transcribed in divergent directions (Fig. 9). The ipgDEF, mxi, and spa genes are likely to be organized in two large operons comprising (i) ipgDEF and mxiG to mxiA, and (ii) spa47 to spa40 (401, 452). A temperature-regulated promoter was identified 47 bp upstream of spa47 within the spa15 coding sequence (401), and spa9 to spa40 may also be transcribed by an internal promoter (452). The transcriptional organization of the ipgDEF-mxi gene cluster has not been analyzed in detail. However, it is likely that these genes form an operon, since the promoter for a mxiA-lacZ gene fusion lies further than 6 kb upstream of mxiA (21, 22) and this temperature-regulated promoter is close to the start of the ipgDEF-mxi cluster (401, 438).
The nine genes transcribed oppositely from the ipg-mxi-spa cluster, icsB, ipgABC, ipaBCDA, and virB, are organized in at least three transcriptional units comprising (i) icsB (9), (ii) ipgBC and ipaBCDA (399), and (iii) virB (440). ipaD and ipaA also appear to be cotranscribed from a weaker promoter located within the 50-bp intergenic region between ipaB and ipaD (399). Temperature-regulated promoters were identified upstream of icsB (9) and ipgB (399, 438). virB is transcribed from its own, temperature-regulated promoter (440) (see “Transcriptional regulation of S. flexneri invasion genes” below).
Invasion Genes of S. typhimurium Pathogenicity Island 1
Parts of the Salmonella type III secretion system encoded by SPI-1 were independently identified by various investigators via the isolation of noninvasive mutants (43, 125, 197, 426), complementation of a noninvasive S. typhimurium strain (143), isolation of oxygen-regulated invasion genes (227), isolation of oxygen-deregulated hyperinvasive mutants (266), isolation of genes regulated by the global Salmonella virulence regulatory system PhoPQ (38), subtractive isolation of Salmonella-specific sequences (168), identification of secretion- and invasion-deficient mutants (216), and interspecies complementation (110). SPI-1 comprises more than 30 genes clustered in approximately 40 kb located between mutS and fhlA at centisome 63 in the S. typhimurium chromosome (312). SPI-1 is a chromosomal insert in S. typhimurium, since in the respective chromosomal region in E. coli K-12, mutS and fhlA are located immediately adjacent to each other (312). SPI-1 probably originates from a heterologous source, since the G+C content of the gene cluster is 42 to 47% (63), as opposed to an average of 52% for the vast majority of the S. typhimurium chromosome. SPI-1 in S. typhimurium does not carry repeated sequence elements at its boundaries (312), which may explain its relative genetic stability in many Salmonella species (63, 159). However, in S. choleraesuis, a sequence element highly similar to IS3 was identified downstream of the invH gene (17) located at the “left” end of the gene cluster (Fig. 9). The invA gene of S. typhimurium was shown by hybridization analyses to be present and highly conserved in 91 strains of 37 different Salmonella serovars, and invA mutants of S. typhi, S. enteritidis, S. gallinarum, and S. dublin were noninvasive, suggesting that all invasive Salmonella strains probably use the inv secretion system (145).
The type III secretory apparatus is encoded by the inv-spa gene cluster and the prgHIJK-orgA genes, while the sip/sspBCDA genes and sptP encode secreted targets of that apparatus. The sicA gene (called sipE in S. typhi [197]) probably functions as a chaperone for the Sip/SspBCDA-secreted proteins. (It was reported that the sptP gene is preceded in S. typhi by a gene encoding another putative type III secretory chaperone, tentatively labelled sicB in Fig. 9 [352a].) Two regulatory proteins, InvF and HilA, are also encoded by SPI-1. Additional secreted targets (485) as well as additional regulatory factors (38, 226) of the SPI-1-encoded type III secretion system are located in unlinked chromosomal locations.
The transcriptional organization of the genes in SPI-1 has only been partially determined. However, overlapping reading frames, short intergenic sequences, and homologous genetic arrangement of the S. flexneri type III secretion system genes imply the following transcriptional structure. The invFGEABCIJ-spaOPQRS genes may form one large operon (84, 108, 168, 232), while invH is transcribed in the opposite orientation (17). The sicA-sip/sspBCDA genes (and probably the downstream iacP) probably form another operon (197, 216, 233, 234). The transcriptional organization of the genes located between iacP and prgH, namely, sptP (235), iagB, hilA (27), and ORF1 (339), has not been determined. The prgHIJK genes are organized as an operon (339), and the downstream orgA is independently transcribed (339). Several kilobases of uncharacterized DNA are located downstream from orgA. It was recently reported that this region encodes a secreted protein (AvrA) which requires the SPI-1-encoded the type III pathway for secretion. AvrA is 56% identical to YopJ from Y. pseudotuberculosis and also exhibits homology to the avirulence protein AvrRxv from X. campestris pv. vesicatoria (183).
The Second S. typhimurium Type III Secretion System, Encoded by SPI-2
The second Salmonella type III secretion system was identified in S. typhimurium by application of a clever strategy which allows the identification of virulence genes via negative selection of individually tagged transposon mutants (195, 410). The genes encoding this type III secretion system are clustered, and the encoding region has been termed Salmonella pathogenicity island 2 (SPI-2). Part of SPI-2 was independently identified by subtractive isolation of Salmonella-specific DNA sequences (333).
The SPI-2 gene cluster is located at centisome 30.7 in the S. typhimurium chromosome and is inserted adjacent to the tRNAVal gene. The G+C content of the island is about 42% (194), and the region is present in all Salmonella species except for the more distantly related S. bongori. Since S. bongori contains SPI-1, however, it is suggested that SPI-2 was acquired via horizontal gene transfer by a Salmonella strain already harbouring SPI-1 (35).
The genes for which sequence information has been published are shown in Fig. 9 (196, 333). The relative location of the spiAB genes, encoding proteins homologous to YscC and YscD, respectively, and a gene encoding a protein with homology to response regulators (SpiR) (333), is not clear.
Locus of Enterocyte Effacement, Encoding the EPEC Type III Secretion System
The EPEC type III secretion system is encoded by a 35.5-kb continuous stretch of chromosomal DNA (102, 296) called LEE (locus of enterocyte effacement). LEE is also present in other, partially unrelated bacterial pathogens that cause an A/E phenotype (Citrobacter rodentium, Hafnia alvei, enterohemorrhagic E. coli O157:H7, and rabbit EPEC) (296), suggesting that this cluster of virulence genes has spread horizontally during evolution. The notion of a heterologous origin of LEE is supported by its G+C content of 38.4% (102), which is low in comparison to the 50 to 51% of the E. coli chromosome. It is possible that the LEE was acquired by transposition, since one end of the locus (shown at the right side of Fig. 9) comprises ca. 450 bp with up to 89% sequence identity to parts of transposase-encoding insertion elements from Shigella sonnei (103). The respective region in the LEE, however, does not constitute a complete IS element and is interrupted by multiple stop codons in all reading frames. LEE was found to be inserted in the chromosome of EPEC E2348/69 16 bp downstream of the selenocystein tRNA locus (selC) relative to the respective position at min 82 of the E. coli K-12 chromosome (296). The LEE insertion point corresponds exactly to the position where a virulence locus of ca. 70 kb (termed pathogenicity island I [pai I]) is inserted in the chromosome in uropathogenic E. coli (52, 246). Although pai I is structurally and functionally unrelated to LEE (170), both pathogenicity islands carry a similar (71% identity) 93-bp sequence at their right junction to the adjacent DNA (296), which in the LEE is located downstream of the DNA sequence homologous to IS elements. However, in contrast to pai I, which carries a 16-bp direct repeat sequence at both ends, LEE shows no repeat of right-end sequences at its left end. Since additional small deletions relative to the corresponding E. coli K-12 sequence are found adjacent to the left-end junction of LEE (296), it is conceivable that a left-end repeat sequence was also deleted from LEE. Taken together, these data indicate that LEE might have been transposable but that due to the accumulation of mutations, this feature was lost during evolution.
Although the entire LEE was reported to contain at least 41 open reading frames, 12 of which are homologous to components of other type III secretion systems (102), detailed information on the genetic structure and sequence is available only for the right half of the locus (Fig. 9). This region contains five genes, called sepA to sepD (mnemonic for secretion of E. coli proteins) and ORFD2, encoding proteins which are homologous to type III secretion apparatus proteins (103, 223). This part of the LEE also contains the genes encoding intimin (eaeA) (see the section on enteropathogenic Escherichia coli, above) (225, 296), the secreted receptor protein Tir (239), and three secreted proteins (espA [mnemonic for EPEC secreted protein] [242], espB [104], and espD [259]). I also include the relative positions of some other genes reported to encode homologs of type III secretory proteins (102) for which no sequence information is available yet.
Another gene located at the right end of the LEE (ORFD4) encodes a protein (103) which exhibits sequence similarity to eukaryotic proteins including human and rat nuclear pore complex protein nup153 (298, 432) and verprolin of Saccharomyces cerevisiae, which is involved in maintenance of the cytoskeleton (98). These proteins have common proline-rich regions similar to src homology 3 (SH3) protein binding domains (369), and two SH3 domain-homologous regions are present in the protein deduced from the sequence of ORFD4 (103). Whether this gene plays a role in EPEC pathogenesis remains to be determined.
Furthermore, three genes in the LEE were reported to encode proteins with structural similarities to secretory chaperones from other type III secretion systems: sepU (102), located immediately upstream of eaeA; ORFD1, downstream from espB (103); and sepE, located downstream from sepC (102, 364).
Chlamydial Type III Secretion Genes
In Chlamydia psittaci, a few type III secretion genes have been identified (211). A cluster of four genes encodes homologs of YscU (called Cds1, for contact-dependent secretion), LcrD (Cds2), YopN (termed CopN), and the chaperone SycE (termed Scc1) (211) (Fig. 9). The genes located immediately downstream of this gene cluster encode proteins which are most probably unrelated to type III secretion. Further type III secretion genes were found in the chlamydial genome sequencing project: homologs of YscC, YscN, and YscT (211). Interestingly, the G+C content of the chlamydial type III secretion genes is low (40%) and corresponds approximately to the G+C content of the chlamydial genome (211).
hrp Gene Cluster of P. syringae
P. syringae pv. phaseolicola was the first plant pathogen in which a hrp gene cluster was identified by complementation of a nonpathogenic mutant (276). Later, this chromosomally located (365) gene cluster was found to encode a type III secretion system. The system comprises at least 28 genes in a continuous stretch of approximately 24 kb (72). Two secreted factors (HrpA [499] and HrpZ [188]) are encoded at the “right” end (Fig. 9). Regulatory proteins (HrpS and HrpR [166, 487], which both belong to the NtrC family of two-component system response regulators, and an alternative sigma factor [HrpL] [487]) are encoded at the ends of the cluster, respectively. The genes encoding the secretion apparatus are organized in four transcription units (72) comprising hrpJcVpQcNpO (274), hrpPcQabRSTU (274), hrpFGcCpTV (212, 213), and hrpAZBcJpDE (213, 357).
E. amylovora
The Erwinia type III secretion system is encoded in approximately 25 kb of chromosomal DNA which comprise at 22 genes organized in at least six transcriptional units (470) (Fig. 9). The type III secretion apparatus is encoded in three large operons comprising hrpJ to hrcU (55), hrpA to hrpE, and hrpF to hrpV (244). In between the hrpA to hrpE and hrpJ to hrcU operons are several genes, two of which encode transcription factors of the hrp genes: the alternative sigma factor HrpL (471) and the NtrC-like transcriptional regulator HrpS/WtsA (138). A secreted protein, HrpN (harpin) (472), is encoded in a different operon (244) at the left end of the gene cluster, as shown in Fig. 9.
R. solanacearum
In R. solanacearum, the hrp gene cluster comprises approximately 27 kb on a megaplasmid (25, 62). The 18 genes which are required for secretion of the PopA1 protein (26) and to elicit the HR phenotype are organized in four transcription units comprising hrpBA, hrpK to hrpC, hrpNO to hpaP, and hrpQ to hrpX (25) (Fig. 9). The hrpB gene encodes a global transcriptional activator of the hrp genes which belongs to the AraC family (155).
X. campestris
The chromosomally located (61) X. campestris hrp gene cluster contains 21 genes in 25 kb (60). Transcriptional units comprise hrpA1, hrpB1 to hrpB8, hrpC1 to hrpC3, and hrpD1 to hrpD6 (60). (Furthermore, three genes located downstream from hrpD6, called hrpE12 and hrpF, are involved in HR induction but have not been further characterized [61].) As shown in Fig. 9, the majority of the hrpA- to hrpD-encoded proteins are homologous to secretion apparatus components. However, seven genes (hrpB1, hrpB2, hrpB4, hrpB7, hrpC3, hrpD4, and hrpD6) encode proteins which are conserved only in the closely related type III secretion system of R. solanacearum.
Rhizobium Species
Genes with homology to type III secretion systems have been identified in R. fredii (189, 299) and on the large sym plasmid of Rhizobium sp. strain NGR234 (139). Compared to most other type III secretion system genes, the G+C content of the type III system-encoding region is relatively high (56%) and does not differ significantly from the G+C content of the rest of the sym plasmid. Interestingly, proteins with homology to YopM and to avirulence factors from P. syringae and X. campestris, which could represent potential secreted proteins of the Rhizobium type III secretion system, have also been identified on the sym plasmid (139).
Gene and Protein Families
The genes encoding the secretion apparati can be grouped into (i) genes that are found in virtually all type III secretion systems, (ii) genes that are present only in pairs of closely related type III secretion systems, and (iii) genes that are unique to individual systems. The proteins encoded by the first group are listed according to family membership in Fig. 10. Since the Yersinia type III secretion system was the first to be described and remains the best characterized, I name the protein families according to the designation of the Yersinia protein; e.g., the group of type III secretion proteins that are homologous to LcrD will be called the LcrD family. Figure 10 also lists the lengths of the proteins in amino acids and respective references.
FIG. 10.
Broadly conserved proteins of the type III secretion apparatus and of the flagellum biosynthesis systems in B. subtilis and E. coli/S. typhimurium. The lengths of the proteins in amino acids given underneath the protein designation. Numbers in parentheses indicate references. Subgroups of proteins which exhibit significant similarity are indicated by different grades of shading. See the text for further details. 1, Abbreviations in parentheses denote the organism. Ye, Y. enterocolitica; Ypt, Y. pseudotuberculosis; Yp, Y. pestis. 2, Sequence from Shigella sonnei. In S. flexneri, only part of the respective MxiC sequence is available. 3, Sequence incomplete. 4, In SpaO and other members of the YscQ family, only the carboxy-terminal 80 aa is conserved, while SpaO also shares an internal domain (aa 147 to 194) with SpaO from S. typhimurium. 5, Sequence not available. 6, Older designations are given in parentheses. 7, HrcQB corresponds to the conserved carboxy-terminal domain of other YscQ-family members, while the HrcQA corresponds to the amino-terminal domain of the conserved HrcQ from E. amylovora (Fig. 11). 8, U. Bonas, unpublished. 9, Xcg, X. campestris pv. glycinea. 10, Stm, S. typhimurium; Eco, E. coli. 11, FliM of E. coli is homologous to the amino terminus of B. subtilis FliY. See the text and Fig. 11 for further details. 12, In most cases, the subcellular location is predicted according to the physicochemical properties of the protein or inferred from the location of homologous flagellar biosynthesis proteins. 13, Unified nomenclature for conserved proteins of the type III secretion apparatus. Sct stands for secretion and cellular translocation.
The main criterion used to group the proteins into families was the similarity of their sequences. As a working basis, two proteins were considered significantly similar if the probability of nonidentity of their sequences is ≥10−5 (as determined by the BLAST program from the National Center for Biotechnology Information server [18]). For an easy overview, the subgroups of proteins that exhibit significant sequence similarities are highlighted by different shadings in Fig. 10.
The sequence similarities between members of each protein family show a considerable degree of variation, and the degree of similarity variations differ between protein families. In general, the better-conserved protein families (LcrD, YscC, YscF, YscJ, YscN, YscR, YscS, YscT, and YscU) show sequence similarity variations between 30 and 70%, while the less well conserved families (YopN, YscD, YscF, YscI, YscK, YscL, and YscQ) show values of below 20% to about 50%. In some cases, the sequence similarities within one family are so low that they may not be considered significant in a comparison that does not take into account other considerations. Nevertheless, even in the less well conserved protein families, the lengths of the proteins are usually similar and the encoding genes are found in similar genetic neighborhoods. Considering that type III secretion genes most probably have been acquired as intact genetic blocks during evolution, one might speculate that some of the proteins have been conserved while others underwent considerable change, perhaps facilitating the adaptation to different hosts or the secretion of different virulence factors. Thus, the relative location of a gene may have been conserved between different type III secretion systems, while the encoded proteins, although derived from a common ancestor, may have changed to a degree that would not support the notion of homology anymore if solely judged by sequence comparison. This case is demonstrated by the genes and proteins that are grouped together with YscI to YscL in Fig. 9 and 10. While the lengths of the proteins in a family are similar in most cases, significant sequence similarities only exist between subgroups of proteins within these families (Yersinia and P. aeruginosa, S. flexneri and S. typhimurium SPI-1, E. amylovora and P. syringae). However, in contrast to the members of the YscI, YscK, and YscL families, all proteins in the YscJ family are significantly similar to each other (BLAST probabilities for pairwise sequence comparisons are below 10−20). In the YscL family, SsaK from S. typhimurium SPI-2, HrpE from E. amylovora and P. syringae, and NolV from R. fredii are not homologous to the rest of the family members, although the two HrpE proteins are homologous to each other. Furthermore, the flagellar biosynthesis protein FliH from B. subtilis is homologous to YscL, while no significant sequence similarity exists between the FliH proteins from B. subtilis and S. typhimurium. Nevertheless, the genes encoding these proteins are identically or similarly organized in different type III secretion systems and in the flagellar gene clusters, and a common ancestry of the respective proteins is likely.
A further and even more drastic example of sequence dissimilarities but conserved genetic position are the genes and proteins which are listed together with YscO and YscP in Fig. 10. The proteins in these two groups have various lengths and sequences and differ even between closely related type III secretion systems such as those of S. typhimurium SPI-1 and S. flexneri or of E. amylovora and P. syringae. Nevertheless, the respective genes are located within a genetic block (represented by yscN to yscU) that appears to have been largely preserved during evolution in various type III secretion systems (Fig. 9), and the proteins encoded by yscN and yscR to yscU are highly conserved. Since it is rather unlikely that several different type III secretion systems independently have incorporated two genes between the yscN and yscQ homologs, the proteins in the YscO and YscP groups can instead by considered as having evolved from common ancestors. In addition to the YscO and YscP families, the proteins of the YscQ family show a high degree of sequence variation. However, sequence dissimilarities are restricted to their amino-terminal halves of the proteins, while the carboxy-terminal domains are conserved and are also present in the flagellar biosynthesis proteins FliY of B. subtilis and FliN of S. typhimurium (Fig. 11). In Salmonella spp., the proteins that group with YscO (SpaM/InvI), YscP (SpaN/InvJ), and YscQ (SpaO) are highly variable even within the eight subspecies of S. enterica (84, 271). (In the eight subspecies of S. enterica, SpaM/InvI and SpaN/InvJ vary by 18 and 34%, respectively, while other invasion proteins differ by only 2 to 5%, a rate that corresponds to the changes in housekeeping proteins. In addition, SpaN is even variable in length [63]. An elevated level of sequence variation was also observed for SpaO from the eight S. enterica subspecies [271]. This variation is unlikely to be related to adaptations to specific hosts, since the host-adapted Salmonella serovars S. typhi [human], S. choleraesuis [swine], S. dublin [cattle], and S. gallinarium [fowl] showed only very little sequence variation in these proteins [63].) It is interesting that the SpaN/InvJ and SpaO proteins are secreted (84, 271) and that one of the corresponding proteins (SpaN/32) from S. flexneri is exposed on the bacterial surface (464). Therefore, Li et al. have proposed a relationship between the evolutionary rate of change and the subcellular localization of these type III secretion system proteins (271). According to this hypothesis, proteins that are exposed on the bacterial surface or secreted into the supernatant may be subjected to unusually high rates of change due to selective pressures like the need for antigen variation or the adaptation to different hosts. It is an interesting possibility that most or all of the proteins which group with YscO, YscP, and YscQ are secreted or surface exposed in the various pathogens.
FIG. 11.
Schematic representation of homologous domains (shown by corresponding patterns; open areas have no similarity) in YscQ family and flagellar proteins. The N termini of the proteins are at the left side.
A Unified Nomenclature
For historical reasons, the nomenclature of type III secretion genes is not unified. In the prototypic type III secretion system from Yersinia spp., most of the genes encoding the secretion apparatus are named ysc, for Yersinia secretion. Accordingly, the respective genes which have recently been found in P. aeruginosa were named psc (491). In S. flexneri, however, the secretory genes are called mxi and spa (for membrane expression of Ipa and surface presentation of Ipa, respectively). In S. typhimurium, the genes in SPI-1 have been designated inv according to their requirement for epithelial cell invasion (143) and spa due to homology to S. flexneri genes (168). Furthermore, some S. typhimurium SPI-1 genes are called prg, because they were identified by virtue of their transcriptional repression by the global virulence regulator PhoP (38), or org, because they are regulated in response to oxygen tension (227). Secretory genes in S. typhimurium SPI-2 have been termed ssa (for secretion system apparatus) and given suffixes according to homologies to Yersinia genes (410). In EPEC, the secretory genes are named sep for secretion of E. coli proteins (223). A more unified nomenclature has recently been introduced for the type III secretion genes of plant pathogens, which have been renamed according to their homologies to conserved Yersinia spp. genes. Most of the plant pathogen secretory genes had originally been designated hrp according to their requirement to elicit a hypersensitive plant response and pathogenicity (276). However, the broadly conserved hrp genes were renamed hrc (hrp conserved) according to their homologies to respective Yersinia genes (54). (In Fig. 9 the latest nomenclature for the hrc genes is used, while in Fig. 10 the older hrp names are also given.)
It is likely that the many current bacterial sequencing projects and searches for bacterial virulence genes will lead to the identification of several more type III secretion systems. To facilitate future communication, I suggest renaming the known proteins that constitute the type III secretion apparatus and applying a unified nomenclature to all newly identified systems. For reasons mentioned above, it appears reasonable to take the nomenclature basis from the Yersinia system. However, since the name ysc refers specifically to Yersinia, I suggest calling type III secretion apparatus genes sct, for secretion and cellular translocation, and using the suffixes from the Yersinia system as shown in Fig. 10. It appears reasonable—and evolutionarily justified—to label homologous proteins from different species with the same name and add a suffix specifying the species when necessary; e.g., YscN from Y. enterocolitica should be renamed to SctNYen. I further suggest renaming LcrD and its homologs to SctV and renaming YopN and its homologs to SctW. The names of type III secretion system proteins that do not have homologs in other systems, the names of secreted virulence factors, and the names of chaperones and regulators should not be changed. The unified nomenclature is shown in Fig. 10.
(Editor’s Note: The gene names in this section are suggestions and have not been reviewed by the Nomenclature Committee of the ASM Publications Board.)
Comparison of the Genetic Organization of Type III Secretion Genes
Several striking features of the genetic organization of different type III secretion systems are apparent (Fig. 9): (i) the genes encoding these systems are clustered; (ii) the majority of genes in each system encode proteins which have homologs in other type III secretion systems, while each gene cluster also contains genes which are unique to the respective system; (iii) blocks of corresponding genes are conserved in genetic order between different systems; and (iv) the overall genetic organization of type III secretion gene clusters is conserved within various subgroups. In this section, these features and some of their possible evolutionary implications are discussed.
Although the genes encoding type III secretion systems are clustered, the genetic order within these clusters is partially variable between individual systems. Interestingly, two blocks of genes appear to be conserved in gene order in the majority of type III secretion systems. One is exemplified by the Yersinia yscN to yscU genes, with genes corresponding to yscN to yscU present in S. typhimurium SPI-1 and SPI-2, S. flexneri, P. syringae, and E. amylovora. Furthermore, a similar gene cluster is present in R. fredii, which contains only one gene located between those corresponding to yscN and yscQ instead of two. In R. solanacearum and X. campestris, the order of genes corresponding to yscN to yscU is partially conserved (Fig. 9). (Furthermore, and in contrast to other systems, the genes encoding homologs of LcrD [HrpO in R. solanacearum and HrpC2 in X. campestris] are located immediately downstream from the YscU homologs in the respective organisms [Fig. 9].) Part of the yscN to yscU homologous gene block is also present in similar gene order in flagellar biosynthesis systems, as shown for B. subtilis and S. typhimurium in Fig. 9.
A second block of genes which, although somewhat less well conserved in gene order and protein sequences than the yscN to yscU homologous genes, is exemplified by the Yersinia yscI to yscL genes and can be found in the same order in P. aeruginosa (pscI to pscL), P. syringae (hrpB to hrpE), and E. amylovora (hrpB to hrpE). In S. flexneri (mxiIJ), S. typhimurium (prgIJ), R. solanacearum (hrpI and hrpF), and X. campestris (hrpB3 and hrpB5), two of four of these genes are linked. Interestingly, one of the proteins encoded in this genetic block (YscJ and its homologs) is highly conserved in all type III secretion systems and has a homolog (FliF) in flagellar biosynthesis systems as well. (For a detailed comparison and a description of the known properties of these and other proteins, see “Proteins that constitute the type III secretion apparatus” below.) In contrast to the homologs of YscJ, the other genes in the cluster corresponding to yscI to yscL encode proteins which are only partially conserved between species.
A comparison of the overall genetic organization between different type III secretion systems reveals several subgroups which exhibit similarities in the order of larger genetic blocks. These groups comprise (i) S. flexneri and S. typhimurium SPI-1, (ii) Yersinia spp. and P. aeruginosa, (iii) E. amylovora and P. syringae, and (iv) R. solanacearum and X. campestris. The type III secretion systems of S. typhimurium SPI-2, EPEC, Chlamydia spp. and R. fredii cannot be convincingly assigned to one of the four groups, in part because of the lack of complete sequence information. (The proteins encoded by S. typhimurium SPI-2 are generally more closely related to the respective Yersinia proteins than to those of S. typhimurium SPI-1 or S. flexneri). Although blocks of genes are conserved within the four subgroups, genetic rearrangements have occurred within the various systems. This is particularly apparent for the two highly similar type III secretion systems of Yersinia spp. and P. aeruginosa, which appear to differ mostly by an inversion of the genetic block pscN-popD. Also, S. flexneri and S. typhimurium SPI-1 are very similar, with two major conserved genetic blocks (mxiG to mxiK and mxiE to spa40 in S. flexneri and prgH to orgA and invF to spaS in S. typhimurium), which are localized in the opposite orientation relative to each other. The same is true for the hrp genes in P. syringae and E. amylovora. In the type III secretion systems of R. solanacearum and X. campestris, the genetic order is almost completely conserved, implying that one of these pathogens may have acquired the secretion system directly from the other during recent evolution. Supporting the notion of a common ancestry of those type III secretion systems which exhibit conserved gene orders are the relatively high similarities among various proteins of these systems.
The positions of two genes, exemplified by the Yersinia genes lcrD and yscC, do not appear to be particularly highly conserved beyond closely related subgroups, but the genes are nevertheless present in all type III secretion systems. The lcrD homologous genes generally group with the genes which have homologs in flagellar biosynthesis genes, and the members of the LcrD family of proteins have a high degree of sequence similarity in their amino-terminal half with the FlhA flagellar proteins (see “Proteins that constitute the type III secretion apparatus” below). (Interestingly, in the systems where homologs of YopN are present, the encoding genes are always located adjacent to lcrD family genes.) The members of the YscC family have no sequence similarity to any flagellar protein. Rather, these outer membrane proteins are homologous to a component of the type II secretion system and also have homologs in other transport systems (see “Proteins that constitute the type III secretion apparatus” below).
Evolutionary Implications
Type III secretion systems appear to have been acquired as intact genetic blocks by horizontal gene transfer during evolution. However, although the genes encoding the secretion apparatuses are clustered, the genes encoding the secreted proteins and some transcriptional regulators are often located in unlinked positions, indicating that these secreted targets and regulators have evolved independently of the core secretion apparatus and that these different components of type III secretion systems have adapted to form functional units.
According to similarities of genetic organization, the type III secretion systems can be divided into at least four groups (see above). Within these groups, the order of large genetic blocks is highly conserved and the respectively grouped proteins exhibit higher degrees of similarity to each other than to other homologous proteins. Thus, close evolutionary relations of the type III secretion systems within these four groups are implied. In most cases, the G+C contents of type III secretion genes (Table 2) are in the 40 to 45% range and are lower than the G+C contents of the surrounding genomes. Since the low-G+C Chlamydia spp. carry a recently discovered type III secretion system, it appears possible that chlamydiae are the evolutionary ancestors of type III secretion. However, this notion needs further clarification.
The majority of proteins which have been found in all type III secretion systems have homologs in flagellar biosynthesis proteins, specifically in proteins that are involved in the export of flagellar structural components across the cytoplasmic membrane (see “Proteins that constitute the type III secretion apparatus” below). However, the pore-forming factors that are involved in type III secretion through the outer membrane, YscC and its homologs, are homologous to outer membrane components of type II secretion and other transport pathways. Thus, type III secretion systems apparently evolved from the combination of two different transport pathways.
TRANSCRIPTIONAL REGULATION OF TYPE III SECRETION SYSTEMS
Expression of type III secretion systems responds to environmental conditions which usually correspond to the conditions encountered by the bacteria during infection of a host. In many cases, transcription of type III secretion genes is controlled by multicomponent regulatory networks which integrate a diverse set of environmental cues, probably to restrict the energy-consuming expression of 20 or more proteins to the place and time when they are needed. In addition, expression of several type III secretion systems appears to be controlled by unidentified host factors and is activated by contact of the bacteria with eukaryotic tissue. The transcriptional control systems are diverse in various pathogens, although two-component response regulators and AraC-like transcriptional activators are common themes. Furthermore, some systems involve histone-like proteins which regulate gene expression in response to temperature and osmolarity by controlling DNA superhelicity. Other systems require alternative sigma factors. In many cases, the genes encoding the transcription factors are located in unlinked positions, sometimes even on different genetic elements (plasmid versus chromosomal). The known transcription factors involved in expression of type III secretion systems are listed in Table 3.
TABLE 3.
Transcriptional regulators of type III secretion systems
| Organism | AraC-like protein(s) | Two-component response regulators | Alternative sigma factors | Histone-like proteins | Other factors |
|---|---|---|---|---|---|
| Yersinia spp. | VirF/LcrF (87, 89, 130, 497) | YmoA (91) | |||
| P. aeruginosa | ExsA (136) | ||||
| Shigella spp. | MxiE (12), VirF (388) | CpxR (326), OmpR (42) | H-NS (210) | VirB (68, 438, 461, 495) | |
| S. typhimurium SPI-1 | InvF (232) | HilA (27), SirA (226), PhoP (38, 310) | |||
| S. typhimurium SPI-2 | SpiR (333, 410) | ||||
| EPEC | PerA/BfpT (161, 439) | ||||
| E. amylovora | HrpS (138) | HrpL (471) | |||
| P. syringae pv. syringae | HrpR, HrpS (166, 167, 487) | HrpL (487) | |||
| R. solanacearum | HrpB (155) | ||||
| X. campestris | HrpXv (476) | HrpG (478) |
Transcriptional Regulation of Yop Expression
Expression of the Yersinia type III secretion system, especially of the secreted Yop proteins, is transcriptionally activated by contact of the bacteria with the surface of a eukaryotic cell (346) (see “Regulation of type III secretion in Yersinia species” above). The discovery of this important fact was the result of almost 40 years of investigation into a Yersinia-specific in vitro phenomenon called the low-calcium response (LCR) (160, 201, 255). The LCR is characterized by the fact that the Yersinia type III secretion system is expressed only at 37°C in the absence of millimolar concentrations of Ca2+. At 26°C (or at 37°C in the presence of Ca2+), Yop expression remains repressed. Temperature activation of Yop expression and its repression by Ca2+ are two distinct regulatory events. Temperature control directly acts on the transcriptional level of yop expression (90, 261), while high [Ca2+] inhibits the secretion of Yop proteins via the type III secretion pathway (128). Nevertheless, Ca2+ inhibition of Yop secretion leads to transcriptional repression of yop genes (129, 130, 430) by a negative-feedback mechanism.
Positive control by temperature.
Positive control of expression of the Yersinia type III secretion system is regulated by an increase in growth temperature from 26 to 37°C (88, 89, 261). Transcription of yopH is induced 240-fold, while transcription of the type III secretion apparatus genes yscA to yscL, yscN to yscU, and lcrGVH to yopBD is induced 9- to 18-fold (160, 261). The induction of yopH transcription is detectable 30 min after the temperature upshift, reaches a maximum after 2 h, and dramatically decreases after 3 h (308). Similar kinetics of induction were observed for yopE (130) and the yscA to yscL operon (307).
Temperature control of yopH (261), yopE (204), lcrGVH to yopBD (41, 359), and yscA to yscL (261, 307) requires the transacting factor VirF in Y. enterocolitica, which is called LcrF in the other two pathogenic Yersinia spp. (87, 89, 130, 497). Genes which are coordinately regulated by VirF have been grouped as the yop regulon (90). The 271-aa VirF protein belongs to the AraC family of transcriptional regulators, which are involved in the regulation of metabolic pathways but also of virulence gene expression in various gram-negative pathogens (148). The carboxy termini of these proteins comprise a helix-turn-helix motif DNA binding domain. VirF forms dimers in vitro (261) and binds to multiple sites in the promoter regions of yopH (261), yopE, lcrGVH to yopBD, and yscA to yscL (468). The binding sites typically consist of two 13-bp AT-rich direct repeats, but they vary between genes, so that the consensus derived from the comparison of 13 half-sites (TTTTaGYcTtTat; nucleotides with more than 60% conservation are capitalized, Y indicates C or T) is somewhat degenerate (468).
In Y. enterocolitica, transcription of the virF gene itself is thermoregulated, with no mRNA detectable at 26°C (87). (In contrast, the homologous lcrF gene is constitutively expressed in Y. pestis [204].) Thermoregulation of virF in Y. enterocolitica involves a negatively acting chromosomal factor, ymoA (Yersinia modulator A) (91). A ymoA mutation leads to partial derepression of virF transcription and hence of transcription of yopH and yopE at 26°C (91). However, thermoregulation of yop genes is not controlled only by the amount of VirF, since overexpression of VirF was not sufficient to induce yop gene expression at 26°C in Y. enterocolitica (261). Furthermore, although YmoA appears to function via VirF, yop genes are expressed independently of VirF in the ymoA mutant. Thus, VirF is required but not sufficient for thermal induction of Yersinia type III secretion gene expression. YmoA resembles the histone-like proteins HU and integration host factor (IHF) of E. coli in that it is small (8 kDa) and rich in charged amino acids (36%) (91). Because of its histone-like structure, YmoA is likely to influence DNA conformation, and changes in DNA superhelicity have been shown to coincide with Y. enterocolitica virF transcription and Yop expression (373). Thus, thermoregulation of yop genes appears to be controlled independently of VirF/LcrF, possibly by changes in DNA superhelicity in the yop promoters, which are controlled at least in part by YmoA.
Negative control by Ca2+ via feedback regulation.
As mentioned above, Yop expression and secretion remain repressed at 37°C when the growth medium contains millimolar concentrations of Ca2+. Although it was originally assumed that Ca2+ acts as a signal which controls a transcriptional regulator of yop gene expression, it turned out that Ca2+ control is mediated by a negative-feedback control mechanism which allows transcription of yop genes only when type III secretion is functional and induced. Because mutations of the Yop secretion apparatus negatively affect yop transcription (15, 40), the negative-feedback mechanism has long been postulated (89), but a better understanding of the molecular basis of this phenomenon, which in Y. pseudotuberculosis involves the secretion of a negatively acting factor, LcrQ (346), has only recently become possible (Fig. 8). Interestingly, two LcrQ homologous proteins, called YscM1 (307) and YscM2 (422), exist in Y. enterocolitica, and both proteins function together to facilitate negative-feedback regulation (422).
LcrQ of Y. pseudotuberculosis (370) is encoded downstream of the yscA to yscL operon at the “rightmost” end of the type III secretion system region (Fig. 9) and is transcribed as a monocistronic unit (370). LcrQ acts negatively on yop gene expression, since a knockout mutation in Y. pseudotuberculosis leads to constitutive expression of Yops at 37°C (370). In contrast, a mutant with a nonpolar insertion mutation in yscM1 of Y. enterocolitica exhibited Ca2+ dependency like the wild type (14). However, when the gene encoding YscM2, a protein with 57% identity to YscM1 and LcrQ, was mutated in addition to yscM1, the resultant strain exhibited the same deregulated phenotype as the lcrQ mutant of Y. pseudotuberculosis (422). Furthermore, overexpression of LcrQ, YscM1, or YscM2 resulted in repression of yop gene transcription (14, 370, 422). Thus, LcrQ, YscM1, and YscM2 act in a concentration-dependent manner, where higher intracellular concentrations of the proteins lead to repression of yop transcription.
LcrQ, YscM1, and probably YscM2 are secreted via the type III secretion pathway (346, 422). The 115-aa proteins show similarity to the first 128 aa of YopH (370, 422), which is located immediately downstream of lcrQ/yscM1 and has probably emerged from a gene duplication event during evolution. Interestingly, the region of YopH homologous to LcrQ/YscM1 and YscM2 contains both the YopH secretion and translocation domains (see “Translocation of Yop proteins by Yersinia species” above). As for other Yops, secretion of LcrQ is tightly regulated by Ca2+ (346). However, unlike transcriptional regulation of yop genes, transcription of lcrQ is only slightly repressed under high-Ca2+ conditions (346), indicating that a pool of intracellular LcrQ is maintained under conditions which repress secretion. When an lcrQ mutant was combined with a mutant in the type III secretion apparatus, Petterson et al. found that Yop expression was restored to low-calcium levels, demonstrating that LcrQ acts negatively on yop transcription (346). Similar results were obtained for a Y. enterocolitica yscM1 yscM2 double mutant (422). In the wild type, all intracellular LcrQ is secreted rapidly (within 3 min) after removal of Ca2+, and Yop expression increases 5 to 10 min after Ca2+ removal (346). Thus, conditions which inhibit type III secretion (high [Ca2+] or mutations in the secretion apparatus) lead to intracellular accumulation of LcrQ and to transcriptional repression of yop genes, while under secretion-inducing conditions, LcrQ is exported from the cytoplasm, relieving transcriptional repression and allowing yop gene expression. LcrQ does not contain any recognizable DNA binding motif and therefore does not appear to be a transcriptional regulator by itself. Instead, LcrQ may act via regulation of VirF (see above) at a posttranscriptional level (347).
The model for yop gene regulation via secretion of a negatively acting factor shows striking similarities to a regulatory model of flagellum biosynthesis in S. typhimurium (217). In flagellum biosynthesis, the intracellular negative regulator FlgM inhibits the expression of late flagellum biosynthesis genes by blocking the activity of a sigma factor required for transcription of those genes. After the assembly of a functional flagellar export structure is completed, FlgM is secreted through this structure into the supernatant, allowing the expression of late genes. The similarities are even more astounding since the flagellar export system is homologous to the type III secretion system.
In the Y. pseudotuberculosis lcrQ mutant and in the Y. enterocolitica yscM1 yscM2 double mutant, Yop expression is derepressed. However, both mutants also exhibit an unusual Yop secretion phenotype, in that secretion of most of the Yops is repressed when Ca2+ is present whereas LcrV and YopD are released into the supernatant in large amounts under these conditions. (The yscM1 or yscM2 single mutants do not secrete any Yops in the presence of Ca2+ [14, 422].) In the absence of Ca2+, however, the Yop secretion pattern of the lcrQ and the yscM1 yscM2 double mutants is indistinguishable from the secretion pattern of the wild type (370, 422). These data indicate that the role of LcrQ and its homologs in Ca2+ regulation is not only on the level of yop transcription but also of Yop secretion.
Yersinia low-calcium response.
In addition to the in vitro activation of expression and secretion of large quantities of Yops, the absence Ca2+ at 37°C results in cessation of bacterial growth (201, 255). In medium containing millimolar concentrations of Ca2+, however, bacterial replication is not affected and Yop expression and secretion remain repressed. (Yop synthesis and secretion precede growth restriction [430], and growth restriction appears to be a consequence of an ordered shutdown of macromolecular synthesis [500]. It is conceivable that growth restriction is an indirect consequence of the massive production [10 to 20% of total protein] of the Yops [90, 308]. However, specific mutations have been described which do not require Ca2+ for growth at 37°C but still express and secrete most of the Yops normally [see below]. Hence, a specific mechanism may link Yop secretion to growth control in pathogenic Yersinia spp.) The LCR is specific to pathogenic Yersinia spp. and does not occur in plasmid-cured strains (39, 117, 153, 154) or in avirulent strains with mutations in plasmid encoded virulence genes (160, 354). Thus, LCR-coupled growth restriction has allowed the identification of the Yersinia type III secretion system and of regulatory proteins by the isolation of a variety of LCR mutants (88, 89, 160, 354, 497, 498). The epistatic relationships between these mutants can be used to infer functional relations between different secretory and regulatory factors.
Two classes of LCR mutants have been obtained. The first class is LCR−; i.e., the mutants are able to grow at 37°C in the absence of Ca2+. These mutants either are impaired in the transcriptional activation of yop and secretion apparatus genes (90, 261) or constitutively repress yop expression. Due to the negative-feedback mechanism which links yop expression to functional type III secretion, mutations in genes encoding the type III secretion apparatus exhibit a LCR− phenotype (89). The second class of mutations cause a LCR constitutive phenotype. These mutants exhibit growth restriction and express Yops at 37°C, regardless of whether Ca2+ is present. However, the LCR-constitutive mutants do not exhibit the LCR at 26°C. While the wild-type LCR phenotype is referred to as Ca2+ dependency (CD), the LCR− mutants are called Ca2+ independent (CI) and LCR constitutive mutants are called Ca2+ blind (CB) and temperature sensitive for growth (TS). All of these mutations map to the 20-kb region which encodes the Yersinia type III secretion system. While the secretion-regulatory protein YopN, which when mutated leads to a CB phenotype, has already been discussed (see “Regulation of type III secretion in Yersinia species”), other factors which affect Yop secretion in response to Ca2+ are discussed in the following section. Interestingly, proteins which appear to affect the regulation of secretion are encoded by the lcrGVH-yopBD operon, which is located at one end of the Yersinia type III secretion gene cluster and is absent from most other type III secretion systems.
lcrGVH-yopBD operon. (i) LcrG.
Similar to YopN/LcrE, LcrG also regulates Yop secretion; i.e., in a lcrG mutant Yop secretion is derepressed even in the presence of Ca2+ (413). Like YopN, the LcrG protein itself is secreted via the type III pathway and, thus, requires an intact secretion apparatus to exert its regulatory function (351). However, while YopN is localized on the bacterial surface before its release into the supernatant under low-[Ca2+] conditions, nonsecreted LcrG is located in the cytoplasm (329). It is conceivable that under high-[Ca2+] conditions YopN/LcrE and LcrG synergistically act as plugs, with YopN/LcrE blocking the distal part of the secretion channels and LcrG blocking the proximal part in the cytoplasm (329). The YopN/LcrE-LcrG block would be removed in the absence of Ca2+ or in the presence of an appropriate in vivo signal. YopN/LcrE and LcrG differ in their effect on secretion of LcrV, another regulatory factor. Thus, while LcrV is only partially secreted by the wild type and a lcrG mutant, it is released in larger amounts into the supernatant of a yopN/lcrE mutant (414). LcrG was shown by chemical cross-linking and copurification to form a stable heterodimeric complex with LcrV, and this complex could have a regulatory function in Yop secretion (329) (see below).
(ii) LcrV.
The function of LcrV, encoding the Yersinia V antigen, has been difficult to assess and is still not fully understood. Besides its apparent function as a direct antihost factor (414) involved in immunosuppression in Yersinia infection (324, 325) (see “The V antigen LcrV” above), LcrV has a complex regulatory function in Yop secretion. Most puzzlingly, lcrV mutants differ from mutations in other genes in the lcrHGV to yopBD operon in that they are not deregulated for Yop secretion but, rather, constitutively repress yop expression at the transcriptional level (CI phenotype [see above]) (41, 358, 414). Yop secretion, however, is still functional in the mutant and is subject to normal Ca2+ regulation (414). YopN/LcrE, LcrG, and LcrH/SycD (see below) are epistatic to LcrV in regulation of Yop expression and secretion (41, 414). Therefore, since YopN/LcrE, LcrG, and LcrH/SycD negatively regulate Yop expression and secretion in the presence of Ca2+, LcrV might function as an inhibitor of a negative regulatory factor of the LCR (414).
LcrV of Y. pestis was found by chemical cross-linking and copurification to form a cytoplasmic complex with LcrG (329). Nilles et al. derived a model of regulation of Yop secretion in which LcrV, by binding to LcrG, counteracts the negative effect of LcrG on Yop secretion (329). If LcrV inhibits LcrG as a secretory plug, an lcrV mutant would be CI because of constitutive blockage of the secretion channels. On the other hand, LcrV would not be required in an LcrG− strain. Hence, a lcrG mutation is epistatic to lcrV (329). This model is attractive in that it provides a possible explanation for the observed phenotypes of various single and combined lcr mutations.
LcrV is itself secreted by the type III secretion mechanism (350, 351). Surprisingly, a deletion analysis of various regions of the 326-aa protein demonstrated that deletion of an internal fragment (aa 108 to 125) abolished secretion of the protein while deletion of amino-terminal portions had no significant effect on LcrV secretion (414). This is a rather atypical for a protein secreted by the type III secretion pathway, since in all other cases analyzed, the secretion signal lies within the amino-terminal portion of the proteins. While deletion of residues 108 to 125 from LcrV abolished secretion of the protein, Ca2+ regulation of yop expression was still intact in the deletion mutant (414), demonstrating that LcrV does not have to be secreted to exert its regulatory effect. Interestingly, only about 50% of total LcrV was found to be secreted by the wild type under low-Ca2+ conditions, whereas LcrV was almost completely released into the supernatant by a yopN/lcrE mutant, regardless whether Ca2+ was present (414). The Δ108–125 mutant of LcrV, even when overproduced, was not secreted by a yopN/lcrE mutant. According to the model in which YopN/LcrE acts as a plug at the proximal part of a type III secretion channel, LcrV would first have to be transported through the secretion channel before its YopN/LcrE-dependent release into the supernatant can occur. The mutant LcrV Δ108–125 appears to not be transported any more and thus becomes independent of YopN/LcrE.
(iii) LcrH/SycD.
While mutants with mutations in yopN and lcrG are deregulated for both Yop secretion and yop transcription, an lcrH mutant is derepressed for yop transcription while secretion is still repressed by Ca2+ (41, 360, 414). Nevertheless, the lcrH mutant is phenotypically CB, and it has been argued that LcrH could be at the bottom of the Ca2+-dependent regulatory loop, perhaps acting at or close to the transcriptional repressor (41, 360). Indeed, an lcrH mutation is epistatic to all other mutants of Ca2+ regulatory proteins, and overexpression of the negative regulatory factors YopN/LcrE (129), LcrG (413), and LcrQ (370) caused no change of the CB/TS phenotype of lcrH mutants. Although lcrH could encode the yop repressor, the protein does not appear to be a direct DNA binding protein. In contrast to the other proteins encoded in the lcrGVH-yopBD operon, LcrH is not secreted (360).
Another role for LcrH emerged when Wattiau et al. discovered the Yop-specific chaperones (466, 467). LcrH/SycD appears to act as a chaperone specific for YopB and YopD (see “Yop-specific chaperones and their role in Yop secretion” above). It is therefore not surprising that similar phenotypes to that of the lcrH mutant were observed for mutants with mutations in yopB and yopD (41, 173). Thus, yopBD mutants show deregulated yop transcription, but Yop secretion remains tightly controlled by Ca2+ (41). However, the CB/TS phenotype of yopBD mutations is less severe than for the lcrH mutation (for yopBD mutants, a larger fraction of cells grow at 37°C than for lcrH, and the bacteria growing at 37°C are CI) (41). Interestingly, when LcrH was expressed in trans, it shifted yopBD mutants from a phenotypic Ca2+ blindness (CB/TS) to Ca2+ independence (CI) while no phenotypic effect of LcrH overexpression was observed in the wild-type strain (41). These observations were interpreted in favor of a role for LcrH as a factor acting in functional proximity to the transcriptional yop repressor. To explain the fact that overproduction of LcrH shifted various lcrGVH-yopBD mutants from CB/TS to CI but had no phenotypic effect on the wild-type strain, Bergman et al. assumed that an unidentified product of the lcrGVH-yopBD operon, which is present in the wild type but absent from the mutants, acts negatively on LcrH (41). LcrH/SycD also influences the type III secretion mechanism. An lcrH mutant of Y. pestis, which still responded to Ca2+ regulation of Yop secretion, nevertheless showed constitutive secretion of LcrV (414). Since the lcrH mutant analyzed was polar on yopBD, Skrzypek and Straley were not able to determine whether LcrH, YopB, or YopD was responsible for the abnormal pattern of LcrV secretion (414).
Taken together, the proteins encoded by yopN/lcrE and the lcrGVH-yopBD operon regulate type III secretion in a complex way that appears to involve interactions of these proteins with each other and with the type III secretion apparatus, while the effect of these proteins on yop transcription probably is indirect and mediated via type III secretion of LcrQ, a key element in a negative regulatory feedback mechanism which links yop expression to functional type III secretion. It is conceivable that the YopN and the lcrGVH-yopBD-encoded proteins form a complex that somehow may span and plug the secretion channels. This could explain why the function of the secretion regulatory proteins depends on a functional type III secretion apparatus while mutations affecting individual regulatory factors lead to abnormal secretion of other regulatory proteins, indicating disintegration of the regulatory complex.
Other factors affecting Yop secretion.
(i) LcrR.
LcrR (146 aa) is encoded upstream of the lcrGVH-yopBD operon and is probably transcribed together with lcrD (31). An lcrR insertion mutant was found to be phenotypically CB/TS and showed constitutive Yop expression (31). LcrR may be required for posttranscriptional regulation of LcrG expression, and the CB/TS phenotype of lcrR may result from the absence of LcrG (see above). Transcription of lcrR is induced by temperature, but no Ca2+ repression was observed (31).
(ii) VirG and YscF.
Another protein involved in regulation of Yop secretion is VirG, which is encoded immediately upstream of the transcriptional activator VirF in the center of the Yersinia type III secretion gene cluster (13). The 131-aa VirG is a lipoprotein with a typical 15-aa signal sequence and is probably located in the outer membrane (13). Like other mutations affecting the type III secretion apparatus, a virG mutation results in a CI phenotype. However, the mutant still secretes Yops under low-Ca2+ conditions, albeit at a reduced level. Specifically, secretion of YopB, YopD, and LcrV is strongly diminished in the virG mutant. VirG was recently reported to be required for the insertion of the type III secretion apparatus secretin YscC (see “Proteins that constitute the type III secretion apparatus” below) into the outer membrane. From these and the above results, a tentative picture emerges in which YopB, YopD, and LcrV may interact with VirG-YscC. VirG is similar to ExsB from P. aeruginosa (136, 137).
A similar secretion phenotype to that observed for the virG mutant is exhibited by a strain expressing a modified form of the secretion apparatus protein YscF (14). This mutant, called yscFmod, secretes all Yops like the wild-type except for YopB and YopD (the secretion of LcrV was not analyzed in this mutant.) The yscFmod mutant expresses an amino-terminally modified YscF, in which the first 12 aa of the native protein is replaced with 8 aa from a plasmid fusion (14). These results could indicate that YscF, which probably is a cytoplasmic protein (see “Proteins that constitute the type III secretion apparatus” below), may be required for proper VirG function. Like the virG mutant, the yscFmod mutant is phenotypically CI, indicating that growth restriction is not necessarily a consequence of global Yop expression and secretion and that growth restriction in the absence of Ca2+ is coupled to the ability to secrete YopB, YopD, and probably LcrV.
(iii) YopR.
When systematically mutagenizing the Y. enterocolitica yscA to yscM operon which encodes part of the Yersinia type III secretion system, Allaoui et al. found that mutations in yscE to yscG and yscI to yscK led to generally impaired Yop secretion. However, a yscH mutant secreted all Yops except for an 18-kDa protein. This protein was demonstrated to be encoded by yscH and was shown by immunological analysis to be YopR (14). A determination of the 50% lethal dose after intraperitoneal injection in mice showed that the yscH mutant was 10-fold less virulent than the wild type, demonstrating a role for YopR in virulence (14). It is, however, not clear to date whether YopR belongs to the group of translocated Yersinia anti-host virulence factors or whether the protein has accessory functions in regulation of Yop secretion and translocation. However, the location yscH in an operon encoding part of the secretion apparatus rather suggest an accessory function for YopR.
Integrated model of regulation of yop gene transcription.
yop transcription is induced after target cell contact, and this fact correlates with the reported complete secretion of LcrQ after cell contact (346). Considering the role of LcrQ and its two homologs YscM1 and YscM2 as a negative regulatory factor in yop transcription, the following model emerges from the summary of 40 years of investigation into the Yersinia low-calcium response (Fig. 8). Thermal induction at 37°C leads to expression of the type III secretion system in pathogenic Yersinia species at a level allowing the synthesis of a functional secretion apparatus. The host’s body temperature thus provides a global environmental signal priming secretion competence. At this stage, the secretion channels remain shut and expression of Yop proteins remains repressed. Opening of the secretion channels is triggered by contact of the pathogen with the surface of a host cell, removing the repression of yop gene expression by secretion of LcrQ/YscM1. In vitro, the conditions triggering Yop secretion are mimicked by the removal of Ca2+ ions from the growth medium. Ca2+ thus functions as the signal which keeps secretion channels shut and may do so during the infectious process, since the extracellular Ca2+ concentration in body fluids is in the millimolar range.
Transcriptional Regulation in P. aeruginosa
As mentioned above, the P. aeruginosa type III secretion system is highly similar to the Yersinia system. Accordingly, the P. aeruginosa type III gene cluster contains a trans-regulatory locus encoding an AraC-type regulator (ExsA) which exhibits 56% sequence identity to Yersinia VirF. ExsA activates P. aeruginosa type III secretion gene transcription by specifically binding to promoters upstream of pscN, the popN to pcrR operon, the exsD to pscB to pscL operon, and the exsC to exsA operon. In addition, ExsA activates transcription of genes encoding the secreted proteins ExoS, ExoT, and ExoU (135, 208). As in Yersinia spp., expression of P. aeruginosa secreted proteins is activated under low-Ca2+ conditions (135). The ExsA binding site has been determined for several promoters and comprises the core consensus sequence TNAAAANA, located 51 or 52 bp upstream of the transcriptional start site (208).
Transcriptional Regulation of S. flexneri Invasion Genes
Shigella virulence is regulated by a number of environmental conditions, most notably by growth temperature but also by osmolarity and external pH. Bacteria are invasive only when grown at 37°C, and the analysis of temperature-regulated gene fusions has led to the identification of mutations in the S. flexneri type III secretion system (209). Temperature regulation, which leads to 50- to 100-fold changes in gene expression (209), is subject to positive and negative control by a cascade of transcriptional regulators encoded by the virulence plasmid as well as the chromosome (291).
Temperature regulation mediated by VirF-VirB.
Activation of type III secretion gene transcription is dependent on the virF-virB dual transcriptional control system (4). VirB coordinately activates transcription of major promoters in the type III secretion gene cluster in response to changes in growth temperature (401, 438). The 36-kDa VirB of S. flexneri (also called ipaR in S. flexneri [68] and in S. dysenteriae [495] or invE in S. sonnei [461]), encoded downstream from the ipa operon and transcribed from its own promoter (68, 440, 461, 495), is homologous to a family of DNA binding proteins involved in plasmid partitioning such as ParB of phage P1 (1). Transcription of virB in turn is controlled by VirF (388, 389, 402), a member of the AraC family of transcriptional regulators (105, 148), which activates virB expression by specific binding to a DNA region upstream of the virB promoter (440). Interestingly, virF is located approximately 40 kb apart from the type III secretion gene cluster on the virulence plasmid (388). Expression of VirB from an unregulated promoter overcame the effect of a virF mutation and rendered bacteria invasive at 30°C (438). However, VirF function is not sufficient to induce temperature-dependent virB transcription, since overproduction of VirF increased the level of virB transcript only at 37°C but had no strong effect at 30°C (438). In addition, transcription of virF is only slightly affected by growth temperature, indicating that virB transcription is the major target for temperature regulation of type III secretion genes (438).
It should be noted that VirF also activates expression of the icsA gene (4), which is essential for intra- and intercellular spread of S. flexneri after invasion of epithelial cells. However, this activation is not mediated by VirB (4). Thus, VirF is the major virulence gene activator in Shigella spp. identified so far. The downstream regulator VirB activates a subset of these genes required for bacterial internalization in response to changes in growth temperature. Interestingly, the mxiE gene of the S. flexneri type III secretion gene cluster (12) encodes another AraC-like regulator, which is similar to a protein (InvF) encoded in a similar position in S. typhimurium SPI-1 (see below). The function of MxiE in S. flexneri regulation is unknown.
Temperature regulation of VirB expression by H-NS.
The mechanism of temperature regulation of virB transcription involves the chromosomally encoded, histone-like protein H-NS, which represses virB at 30°C (210, 293). The 16-kDa H-NS protein specifically binds to the promoter region of virB immediately downstream of the VirF binding site and is capable of blocking VirF-activated transcription of virB in an in vitro transcription assay (440). H-NS is involved in thermo- and osmoregulation of a large number of genes in gram-negative bacteria, and the mechanism of hns regulation probably involves control of DNA topology (106, 198, 199, 218). Thus, H-NS mediated changes in DNA superhelicity in the virB promoter region may represent the molecular mechanism of temperature regulation of virB. Indeed, virB transcription can be activated by VirF only from negatively supercoiled, not from relaxed, DNA (440), and the requirement of negatively supercoiled DNA for virB transcription was elegantly demonstrated by placing a T7 polymerase-dependent promoter immediately upstream of the virB promoter region. Induction of transcription from the T7 promoter led to the introduction of a local negatively supercoiled domain in the virB promoter region, which allowed virB activation by VirF and expression of bacterial invasiveness at 30°C (441).
Alterations in DNA superhelicity may also account for the regulation of Shigella type III secretion gene transcription by changes in osmolarity of the growth medium. High osmolarity increases the expression of a mxiC-lacZ fusion three- to fourfold (42, 353), while mxiC and icsB are transcriptionally repressed in low-osmolarity medium, even at the inducing temperature of 37°C (353). As with the repression of type III secretion gene expression at low temperature, repression under low-osmolarity conditions was relieved by inactivation of the chromosomal hns gene (353).
Other regulatory factors.
In addition to temperature and osmolarity, Shigella virulence gene transcription and invasiveness are controlled by extracellular pH. A change in the pH of the growth medium from 7.4 to 6.0 decreases invasiveness approximately 10-fold, and this regulation appears to be mediated by a 10-fold decrease in virF transcription at the lower pH (326). A mutant with deregulated virF transcription at low pH affected the two-component regulatory locus cpxRA, and the response regulator CpxR was reported to be essential for virF expression (326). Another factor required for type III secretion gene expression is the two component system OmpR-EnvZ (42). Figure 12 shows an overview of the regulatory factors affecting expression of type III secretion gene transcription in Shigella spp.
FIG. 12.
Schematic overview of selected transcriptional regulatory networks in several type III secretion systems. + and − indicate transcriptional activation and repression, respectively. In some cases, regulatory conditions and signals are indicated in parentheses. 2C RR, the protein is a homolog to two-component system response regulators. The modes of action of LcrQ in Yersinia spp. and of OmpR in S. flexneri are unknown. See the text for details.
Transcriptional Regulation of Type III Secretion Genes in S. typhimurium
Type III secretion genes in the S. typhimurium SPI-1 gene cluster are subject to coordinate regulation by a number of environmental cues. Reporter gene fusions to seven genes in S. typhimurium SPI-1, invF, sspC, sspA, hilA, prgH, prgK, and orgA (Fig. 9), were shown to be regulated by the same environmental conditions (28). These genes are expressed at high levels in vitro only under low-oxygen, high-osmolarity, and slightly alkaline (pH 8) conditions. If any one of these conditions is different, type III secretion gene expression is repressed 10- to 200-fold (28) and bacterial invasiveness in vitro is significantly reduced (111, 144, 265, 405). The listed conditions correspond to the environment in the lumen of the distal ileum, the site where salmonellae initiate infection (71, 97). Therefore, the simultaneous requirement for several environmental cues may allow salmonellae to activate SPI-1-encoded type III secretion genes only when expression of these genes is beneficial in promoting intestinal invasion. In contrast to type III secretion genes in other enteropathogens, transcription of SPI-1 genes in S. typhimurium is not regulated by temperature. However, changes in DNA supercoiling have been shown to affect the expression of type III secretion genes (144), and H-NS globally affects Salmonella mouse virulence (185), although the effect of H-NS on type III secretion gene expression has not been determined.
Several factors that are involved in transcriptional regulation of SPI-1-encoded type secretion genes have been identified. Two of these, HilA (27) and InvF (232), are encoded within the pathogenicity island (Fig. 9), while SirA (226) and PhoP (38, 310) are encoded in different and unlinked chromosomal locations. HilA is required for the expression of all genes in SPI-1 (27, 28). The protein contains a DNA binding and transcription activation domain which is similar to respective domains in the phosphorylated response-regulator family of transcriptional regulators (425) but is otherwise not similar to this family. Thus, HilA does not carry an amino-terminal phosphoryl-acceptor domain, which in other members of the family is phosphorylated by a membrane-bound sensor kinase responding directly to environmental changes (425). Neither does HilA contain a membrane-spanning region, which in other family members (ToxR, CadC) is thought to provide the sensory function and/or interact with other proteins to achieve modulation (94, 96). Therefore, HilA may not be directly involved in sensing environmental conditions that modify type III secretion gene expression. This notion is supported by the fact that expression of hilA itself responds to the same environmental conditions as expression of its target genes. Furthermore, expression of hilA does not appear to be autoregulated (28). It is thus unclear how environmental conditions that affect type III secretion gene expression are sensed and how the signal is transmitted to activate expression of HilA.
HilA may bind directly to the invF and prgH promoters, since the regulatory effect observed in S. typhimurium could be reconstituted in E. coli (28). However, regulation of the genes encoding the secreted invasion proteins sip/sspBCDA by HilA may be mediated through activation of invF expression. InvF, the first gene of the inv-spa gene cluster, encodes a protein with similarity to the AraC family of transcriptional activators (232). The protein is required for activation of sip/sspBCDA but does not affect the expression of other type III secretion genes in SPI-1 (109, 232). As mentioned above, InvF is homologous to S. flexneri MxiE.
SirA is another protein required for transcriptional activation of type III secretion genes and thus for invasion (226). The protein is similar to phosphorylated response regulators, most notably to the pathogenicity factor GacA from Pseudomonas spp. (226). Interestingly, SirA is almost identical to UvrY, an E. coli protein of unknown function encoded immediately downstream from the DNA repair protein UvrC. Unlike in other two-component regulatory systems, no corresponding sensor-kinase is encoded in proximity to sirA. SirA may exert its effect on type III secretion gene expression via regulation of hilA expression.
After passage of salmonellae through the intestinal epithelium, the bacteria are phagocytosed by residential macrophages. In the macrophage phagosome, the global Salmonella virulence response regulator PhoP is activated (310). PhoP appears to act negatively on expression of hilA and shuts off expression of the SPI-1-encoded secretion system (28, 38). A constitutive mutant of PhoP renders SPI-1 type III secretion genes repressed and leads to a protein secretion- and invasion-deficient phenotype (38, 339). PhoP is activated after phosphorylation by the corresponding membrane-bound sensor kinase, PhoQ, which in turn responds to changes in extracellular concentrations of divalent cations (152). Thus, low Ca2+ concentrations in the phagocytic vacuole may be the signal that globally repressed SPI-1 encoded type III secretion. The regulatory network affecting SPI-1 type secretion gene expression is summarized in Fig. 12.
SirA and PhoP both are present in E. coli K-12, while the SPI-1 gene cluster was probably acquired by S. typhimurium after the evolutionary split from E. coli (312). Therefore, transcriptional regulators which preexisted in the common E. coli-Salmonella ancestor appear to have adapted to the regulation of type III secretion genes.
SPI-2, which encodes the second Salmonella type III secretion system, encodes a two-component sensor kinase-response regulator pair (333, 410) (Fig. 9). The respective genes have been reported to be required for SPI-2 gene expression (111a, 193a). (See Addendum in Proof.)
Regulation of A/E Phenotype Expression in EPEC
As in shigellae and yersiniae, expression of the virulence phenotype in EPEC depends on a growth temperature of 37°C, while adherence to epithelial cells, protein secretion (240), and consequently A/E lesion formation are impaired when the bacteria are pregrown at 28°C (377). Induction of A/E lesion formation also depends on the bacterial growth phase with the highest activity occurring during early logarithmic growth (377). Furthermore, maximal secretion requires the presence of sodium bicarbonate and calcium and is stimulated by millimolar concentrations of Fe(NO3)3 (238) and an NH4+ concentration of more than 20 mM (361). Interestingly, protein secretion in vitro was observed only in tissue culture medium but not in Luria-Bertani medium or in M9 minimal medium (240), although growth in these media had no effect on host cell signalling and the induction of A/E lesion formation (377). Thus, tissue culture medium appears to contain a compound capable of activating Esp secretion in vitro, while secretion of Esps in vivo may be activated by contact of the bacteria with epithelial cells.
As in other type III secretion systems, activation of virulence gene expression in EPEC requires an AraC-homologous transcriptional activator protein, PerA/BfpT (161, 439). The protein is encoded in the per locus (161, 439), which, in contrast to the chromosomal type III secretion system, is located on a virulence plasmid, again implying evolutionary adaptation of transcriptional regulation of type III secretion genes (see above). The per locus contains four genes, perA to perD, three of which (perABC, also designated bfpTVW [439]) are presumably organized as an operon. PerA/BfpT is 31% identical to the virulence gene activator VirF from S. flexneri. PerB/BfpV shows weak similarity to some eukaryotic DNA binding proteins (161). PerC/BfpW and PerD have no homologs in the published databases (161, 439).
The genes of the per locus are required for transcriptional activation of type III secretion system genes (eaeA and espB) (161) and protein secretion (240) and also are involved in activation of a plasmid-encoded adhesin (361, 439; see also reference 207). Furthermore, overexpression of the protein(s) encoded in the per locus leads to oversecretion of at least 20 proteins by EPEC. Therefore, the per locus encodes a global regulator of virulence gene expression in EPEC. Purified PerA/BfpT was shown to bind directly to DNA sequences upstream of bfpA (encoding the bundle-forming pilus adhesin) and eaeA (439) (see “Enteropathogenic Escherichia coli” above). Nevertheless, full activation of virulence gene expression requires the presence of all three genes, perABC/bfpTVW (161, 439). Further experiments are needed done to elucidate the mechanism of trans-activation of EPEC virulence genes.
Transcriptional Regulation in P. syringae and E. amylovora Involves an Alternative Sigma Factor
Expression of type III secretion genes is induced early after contact of the bacteria with plant tissue (489). Induction in planta of various genes in the type III secretion gene cluster of P. syringae ranges from 5- to 70-fold (366, 489) and increases from 1 to at least 6 h after infection. Induction, albeit to a lesser extent, is also observed in minimal salts medium, while complex nitrogen sources, high pH, high osmolarity, and some carbon sources have a repressive effect on hrp gene expression (366, 489). Thus, induction of hrp gene expression after contact with plant tissue results from an alteration in nutritional conditions and may also involve in addition a specific plant factor(s).
Transcriptional activation of hrp genes is controlled by a multicomponent regulatory cascade that involves two activators of the response regulator family, HrpR and HrpS, and an alternative sigma factor (166, 167, 487). HrpR and HrpS, encoded at the “right” end of the type III secretion gene cluster in Fig. 9, are both similar to each other and to the NtrC subfamily of response regulators, most of which interact with the ς54 RNA polymerase holoenzyme. Indeed, a sequence motif which may function in interaction with ς54 (RpoN) is conserved in both HrpR and HrpS. Neither of these proteins carries an amino-terminal phosphoreceiver domain that modulates activity in other two-component response regulators, implying that the activity of these proteins may not be controlled by phosphorylation.
HrpR and HrpS activate transcription of the sigma factor gene hrpL, located at the other end of the type III secretion gene cluster. (In accordance with the requirement of RpoN for HrpRS activity, hrpL is expressed from a RpoN promoter sequence [487].) HrpL is homologous to the ECF (extracytoplasmic functions) subfamily of sigma factors (278), which comprises RpoE of E. coli and AlgU of P. aeruginosa. HrpL, in turn, is required for transcription of the type III secretion genes, and a conserved promoter sequence recognized by HrpL (TGGAACCNAN14CCACNNA) has been detected in all hrp promoters as well as in the hrmA and avr promoters of genes encoding potential type III secreted proteins (222, 488). The fact that both HrpR and HrpS are required for HrpL expression probably results from transcriptional activation of hrpS by HrpR, which binds to an activatory site upstream of the hrpS promoter (166). Figure 12 gives an overview over the regulatory network affecting type III secretion gene expression in P. syringae.
As in P. syringae, type III secretion genes in E. amylovora are negatively regulated by environmental conditions, including high concentrations of ammonium sulfate and other nitrogen sources, high pH, some carbon sources, and nicotinic acid, and are activated in planta. Furthermore, hrp genes are transcriptionally activated 2- to 10-fold at lower temperatures (18 versus 30°C). Transcriptional activation in the plant does not exceed in vitro activation, suggesting that no specific plant component is required as a signal (473). Interestingly, in resistant nonhost tobacco, hrp genes are induced earlier after infection and much more strongly than in pear, a host plant. However, whether this observation gives a clue to the mechanism of HR induction requires further analysis.
E. amylovora contains an ECF family alternative sigma factor (HrpL), which is highly similar to HrpL from P. syringae and is required for transcription of type III secretion genes including hrpN, encoding the secreted harpin protein (471). Expression of HrpL is activated by HrpS, a response-regulatory protein highly similar to HrpS from P. syringae (138, 471). However, unlike in P. syringae, no second response regulator which would correspond to HrpR has been found in Erwinia spp. In further contrast to P. syringae, the hrpS and hrpL genes are located near to each other in the center of the type III secretion gene cluster in Erwinia spp. (Fig. 9).
Transcriptional Regulation in R. solanacearum and X. campestris
As in P. syringae and E. amylovora, the type III secretion systems from R. solanacearum and X. campestris are highly similar in genetic organization and in terms of conservation of protein sequences. Also, both systems are regulated in response to environmental stimuli (growth in minimal media) by AraC-like transcriptional activators, which even can partially replace each other (476). In R. solanacearum, the respective gene, hrpB (155), is located between hrcT and hrcC at one end of the type III secretion gene cluster (to the left in Fig. 9), while this gene is absent from the respective position in X. campestris. However, in X. campestris, the AraC-like activator (HrpXv) is encoded in an unlinked position (476).
In both pathogens, the genes encoding the AraC-like regulators are themselves subject to transcriptional activation by growth in minimal medium. However, while partial autoregulation was observed in P. solanacearum (155), transcription of hrpXv in X. campestris does not require its own product (476). The upstream regulator was identified in X. campestris and is a protein (HrpG) encoded next to hrpXv with homology to response regulators of the OmpR subfamily (478) (Fig. 12). Interestingly, activation of expression of the hrcC (hrpA1) gene in X. campestris, which encodes an essential part of the type III secretion apparatus, is independent of HrpXv (477) but is activated by HrpG (478). Accordingly, the corresponding hrpB-hrcC locus (Fig. 9) in R. solanacearum might also be subject to activation by an HrpG homolog.
PROTEINS THAT CONSTITUTE THE TYPE III SECRETION APPARATUS
This section discusses structural and functional features and the subcellular locations of the broadly conserved proteins that constitute the type III secretion apparatus. For the vast majority of these proteins, their requirement for type III secretion or for the respective, secretion-associated phenotypes has been demonstrated. On the other hand, few structural data exist beyond the protein sequences, and the subcellular locations have only been determined in some instances. Therefore, the available data are combined for each protein family. (The criteria used for grouping proteins into families are discussed under “Genetic and transcriptional organization of type III secretion genes” above.)
While the majority of secretion apparatus proteins are localized in the inner membrane, at least one outer membrane protein (YscC and its homologs) is present in all type III secretion systems and at least one other common protein (YscJ family) possibly connects the inner and the outer membrane. It is intriguing that most of the type III secretion proteins which are associated with the inner membrane have homologs in flagellar biosynthesis systems, while YscC is homologous to outer membrane translocator proteins from type II secretion systems and from phage assembly and extrusion pathways. This section therefore also describes some structural and functional aspects of flagellar biosynthesis and phage extrusion and compares the structural data for flagellar proteins and phage exporters with those of the respective homologs in type III secretion systems.
The LcrD Family of Inner Membrane Transport Proteins
The proteins of the LcrD family are located in the inner membrane and could form a central protein-conducting channel. The proteins are characterized by a hydrophobic amino-terminal region predicted to form at least six, more probably eight, membrane-spanning helices and a large hydrophilic carboxy-terminal domain (349).
A number of deletions have been introduced into LcrD, which were all found to result in an inactive protein as measured by analysis of Yop expression and secretion (350). Interestingly, a mutant protein carrying a deletion within the conserved large second cytoplasmic loop (aa 159 to 167) could not be complemented with the wild-type protein and exhibited a dominant negative effect on Yop expression and secretion when expressed in trans. In contrast, deletions of aa 192 to 343 and 618 to 644 abolished Yop expression and secretion but were complementable (350). Plano and Straley further selected point mutations which resulted in varying negative effects on LcrD function, some of which affect residues which are almost completely conserved within the LcrD family (L35P, G191S, D240G, and R261C), suggesting that these residues might also be of functional importance in other members of the family (351).
Sequence similarities between individual members of the LcrD family vary between 36 and 66%. The amino-terminal domain that contains the membrane-spanning segments is highly conserved, while the cytoplasmic carboxy-terminal domain is more variable. Interestingly, some proteins of the LcrD family can functionally replace each other in heterologous complementation. Thus, MxiA from S. flexneri partially complements an S. typhimurium invA strain for invasiveness, while no complementation was obtained with Yersinia LcrD (157). However, the invA mutant was partially complementable with a chimeric protein in which the carboxy-terminal cytoplasmic domain of LcrD was replaced with the respective region from InvA (157). Therefore, the amino-terminal membrane-spanning regions of LcrD and InvA are functionally interchangeable, and the carboxy-terminal cytoplasmic domains of these proteins may specify interactions with other components of the type III secretion system in a species-specific way.
The members of the LcrD family are shown in Fig. 10. All of these proteins are essential for functional type III secretion and/or the related phenotypes in the various type III secretion systems (21, 22, 115, 143, 146, 163, 215, 223, 470). In flagellum biosynthesis systems, the LcrD family is homologous to the inner membrane protein FlhA from S. typhimurium (314), Bacillus subtilis (69), and homologs from several other organisms.
The YscN Family of Cytoplasmic ATPases
Homologs of YscN are present in all type III secretion systems described to date and are among the most highly conserved type III secretory proteins. The proteins show similarity to the α- and β-subunits of the F1 component of the bacterial F0F1 proton-translocating ATPase (457) and to the catalytic subunits of eukaryotic as well as archaebacterial ATPases (482). The water-soluble F1 component of F0F1 ATPase of E. coli couples the synthesis of ATP with a proton flux through the membrane-bound component F0 (409). Three regions around the catalytic domains are especially highly conserved within the members of this protein family: the ATP/GTP-binding domains A and B (Walker boxes A and B [458]), located in the central part of the proteins, and the Mg2+-binding motif (496), located between Walker boxes A and B. Functionality of the respective catalytic domains in YscN of Y. enterocolitica and InvC of S. typhimurium was confirmed by an in-frame deletion of 8 aa acids within Walker box A in YscN (Δ169–177) (482) and replacement of the conserved residue Lys-165 with Glu in InvC (108). Purified InvC was shown to catalyze ATP hydrolysis, while the K165E mutant is enzymatically inactive (108).
Overexpression of InvC K165E in S. typhimurium and of the mutated YscN in Y. enterocolitica did not have a negative transdominant effect on invasion and protein secretion, respectively (108, 482), indicating that, unlike the F1 component of F0F1 ATPase, the ATPases of the type III secretion system do not require dimerization for proper function. The subcellular location has not been determined for any of the members of the YscN family, but by homology to the soluble F1 component of F0F1 ATPase, it is likely that the YscN ATPases are cytoplasmic proteins. YscN proteins may interact with membrane-bound components of the type III secretion apparatus to energize secretion or to provide the energy for the assembly of the secretion apparatus, as has been shown for the flagellum biosynthesis homolog FliI (113).
Flagellar Export Apparatus in Relation to Proteins of Type III Secretion Systems
Like the members of the LcrD and YscN families, those of the YscQ to YscU and YscJ families show significant sequence similarities to proteins of the flagellar biosynthesis systems of both gram-negative and gram-positive organisms. In addition, weak similarities to flagellar proteins are found for YscD and YscL and their homologs. By similarity to the flagellar assembly apparatus, all of the respective proteins from type III secretion systems are likely to be located in or associated with the inner membrane and may form the cytoplasmic gate (and perhaps a periplasmic extension?) of the type III secretion channel. Since the flagellum biosynthesis proteins that are homologous to type III secretion proteins are specifically involved in the flagellum-specific protein export pathway, it is likely that some of the homologous proteins may play similar roles in both systems. For some of the flagellar proteins, their role in assembly of flagellar structural components is known, and several interactions between these proteins have been detected. Therefore, these data may provide hints as to how the respective components of type III secretion systems might interact.
Assembly of the flagellar basal body and protein export apparatus.
Flagella, the supramolecular bacterial surface structures which mediate bacterial motility, are composed of three major structural components, the basal body, the hook, and the filament. The basal body anchors the filamentous flagella to the bacterial cell wall, while the hook links the basal body and the filament. The basal body is composed out of several structural subunits, called the MS ring, which is located in the cytoplasmic membrane; the periplasmically located P ring; and the L ring, which is located in the outer membrane. These three rings are mounted on a central axial structure, the rod (Fig. 13). Once the MS and C rings are formed, the rod, the P and L rings, the hook, and the filament are added on. The flagellar proteins, which are synthesized in the cytoplasm, are exported to the periplasm, the outer membrane, or the extracellular space, where their assembly finally occurs (251). Only the structural components of the P and L rings (FlgI and FlgH, respectively) are synthesized as precursor proteins with cleavable signal sequences and are transported across the cytoplasmic membrane by the sec pathway (229). The other flagellar proteins do not carry amino-terminal leader peptides and are thus exported from the cytoplasm by a sec-independent flagellum-specific export pathway (257, 279). It is within the proteins involved in this transport pathway where the most significant homologies between flagellar biosynthesis factors and type III secretion proteins are found. Unfortunately, the flagellum-specific export pathway is the least well characterized component of flagellar biosynthesis (5). For a recent review of flagellar structure and function, see reference 280.
FIG. 13.
Homologies between type III secretory proteins and factors which form the flagellum-specific export pathway. Part of the Yersinia type III secretion gene cluster is shown in comparison to the flagellar E. coli genes and the location of the respective E. coli proteins in the flagellum basal body. Homologies are indicated by the color code. OM, outer membrane; PP, periplasmic space; CM, cytoplasmic membrane. ATP hydrolysis by the cytoplasmic ATPase FliI is indicated.
Proteins involved in the flagellum-specific export pathway have been identified by various analyses of conditional and knockout mutants. As a general feature, these mutants affect the assembly of flagella beyond MS and C ring formation. The proteins involved comprise all flagellum biosynthetic factors which have homologs in the type III secretion system (Fig. 10 and 13): FlhA (230, 251, 456), FlhB (251, 256), FliG (251), FliI (230, 251, 456), FliM and FliN (230, 251), FliP, FliQ, and FliR (251).
FliF and the YscJ family.
During flagellar morphogenesis, the first structural component which is discernible by electron microscopy is the MS ring (251). It is formed from only one structural protein, FliF (444), which in S. typhimurium is 560 aa and comprises at least three hydrophobic regions that potentially form transmembrane helices (229). FliF shares a domain with members of the YscJ family (aa 134 to 212 in FliF and 108 to 186 in YscJ). (The homologies discussed in this review were determined by BLAST [http://www.ncbi.nlm.nih.gov/cgi-bin/BLAST] [18] and PRODOM [http://protein.toulouse.inra.fr/prodom/prodom.html] [164, 419] computer analysis. Potential transmembrane domains and the subcellular locations of the amino and carboxy termini are predicted according to TOPPRED2 [http://www.biokemi.su.se/~server/toppred2/toppredServer.cgi].) Since the amino- and carboxy-terminal parts of FliF, which do not show homology to YscJ, are thought to form the M-ring structure (445), it is unlikely that YscJ and its homologs form an M-ring-like structure in type III secretion systems. However, FliF interacts with other proteins, some of which are also homologous to type III secretory factors (see below). Therefore, it is conceivable that the FliF-like domain in YscJ proteins specifies an interaction with other components of the type III secretion apparatus.
The members of the YscJ family carry amino-terminal signal sequences and are thus probably exported by the sec pathway. MxiJ from S. flexneri was shown to undergo lipid modification (11), and the presence of a characteristic lipid attachment site motif (Leu-Xaa-Gly-Cys) (486) at the end of the signal sequences of most of the YscJ family members indicates that these proteins are lipoproteins. In addition to the hydrophobic signal sequence, these proteins carry a hydrophobic stretch of 20 to 30 aa at their C-terminal ends, which is followed by basic residues. This domain may function as a stop-transfer signal, which could anchor the proteins to the inner membrane, while the amino-terminus might be linked to the outer membrane by the lipid moiety (11). Therefore, the proteins of the YscJ family could function as a bridge across the periplasmic space that connects the protein secretion channels in the inner and in the outer membrane.
FliM, FliN, FliG, and the YscQ and YscD families.
After MS-ring formation in flagellar biosynthesis, a bell-shaped structure appears at the cytoplasmic rim of the MS ring, which is called the C ring (107). The C ring comprises FliN (502), FliG, and FliM (503). These three proteins associate with the MS ring (FliF) and the bacterial membrane in S. typhimurium, where FliG functions as a linker between FliF and a complex formed by FliM and FliN (252, 289). No apparent homolog of FliM is present in type III secretion systems.
FliN carries a carboxy-terminal domain of 60 aa that is homologous to a respectively located domain in the members of the YscQ family of type III secretion proteins, while FliG has weak similarity to HrpJ3 from P. syringae (274), which belongs to the YscD family. According to the FliF-FliG-NM interaction in the flagellar system and to the similarities between FliF-YscJ, FliG-YscD, and FliN-YscQ, one might speculate that the homologs of YscJ may interact with the proteins of the YscD and YscQ families. It is important to note that sequence similarities between the proteins of the YscQ-FliN group are restricted to their 80 C-terminal amino acids. Furthermore, FliN of gram-negative bacteria is much shorter than the proteins of the YscQ family and corresponds to the region which is conserved in this family (Fig. 11). Interestingly, the homologous flagellar protein from B. subtilis, FliY, corresponds to FliN in its carboxy-terminal one-third, while the amino-terminal part of FliY is homologous to FliM (46). Finally, the variable and the conserved regions in the YscQ family members, which are usually organized as two domains of a single protein, can be split up into two polypeptides (72) (compare HrcQ from E. amylovora and HrcQA and HrcQB from P. syringae in Fig. 11). Taken together, it is conceivable that the role which is occupied by FliM in flagellar protein export may be replaced by the amino-terminal portions of YscQ and its homologs in type III secretion systems.
The high conservation of the C-terminal portion of YscQ family members, together with the variability of their amino termini, suggest that these proteins might constitute a link between the common and the species-specific components of type III secretion systems. The potential membrane topology for members of the YscQ family is not clear. Although the overall nature of the protein is hydrophilic, at least one potential transmembrane domain (aa 127 to 147) resides in the amino-terminal part. A transmembrane domain is also predicted in similar positions in SpaO from S. flexneri and S. typhimurium and in HrcQ from E. amylovora. For proteins of the YscD family, two transmembrane domains can be predicted in the amino-terminal half of the proteins (aa 81 to 101 and 122 to 142 in YscD of Y. enterocolitica) and the amino-terminus could be located in the periplasm. However, this membrane topology is not detected in the flagellar proteins FliG from B. subtilis or S. typhimurium.
FlhB and the YscU family.
FlhB, another component of the flagellum-specific export pathway, is homologous to the YscU family of inner membrane proteins. Not much is known about the function of FlhB. However, some point mutations in flhB act as suppressors of a fliK mutant, a strain unable to control the length of the hook (202, 256, 480). In wild-type bacteria, a completely assembled hook allows the export of the anti-sigma factor FlgM, which in turn allows expression of late flagellar biosynthesis genes (217). Since export of FlgM, which is impaired in fliK, is reconstituted in a fliK strain carrying flhB suppressor mutations, it has been proposed that FlhB may negatively regulate FlgM export prior to completion of the hook (256). Accordingly, the members of the YscU family might function in regulation of secretion.
FlhB is an inner membrane protein and comprises, like the members of the YscU family, several amino-terminal transmembrane domains (314). Four transmembrane domains were detected in YscU, and the amino and carboxy termini were localized to the cytoplasm (15). The cytoplasmically located ends, as well as the cytoplasmic loop between the transmembrane domains II and III, show blocks of highly conserved amino acids in all members of the family. Therefore, it has been speculated that YscU might interact with another conserved component of the type III secretion apparatus (15), potentially with the cytoplasmic ATPases of the YscN family, which, like YscU, are also highly conserved in all type III secretion systems.
FliPQR and the YscRST families.
FliP, FliQ, and FliR are three hydrophobic proteins which are homologous to the members of the YscR, YscS, and YscT families, respectively. FliP (245 aa), is an integral membrane protein which was reported to be associated with the basal body (280). A hydrophilic domain of about 70 aa, which is only weakly conserved between the various YscR homologous proteins, is located in the central part of the proteins. FliP is predicted to form five hydrophobic transmembrane segments and a periplasmically located amino terminus (282), while for the homologous YscR, four transmembrane segments and a periplasmic location of the amino and carboxy termini are predicted. However, in contrast to the proteins of the YscR family, FliP of E. coli is synthesized as a preprotein with a 21-aa signal sequence which is processed during export (282). It is thus likely that the amino terminus of the mature FliP protein is in the periplasm. Therefore, the central hydrophilic and variable region in the proteins of the YscR family are probably located in the periplasm.
The 89-aa protein FliQ and its homologs carry two transmembrane domains. The localization of the amino terminus is not clear. FliR could form at least five, more probably six, membrane-spanning segments (282). Similar predictions are obtained for YscS and its homologs in the type III secretion systems. The location of the N termini of these proteins is probably cytoplasmic.
YscO, YscP, and Similar Proteins
Proteins encoded by genes which are located at positions corresponding to yscO and yscP (Fig. 9) are given in Fig. 10. In the various type III secretion systems, these positions appear to be occupied by genes which encode proteins of various sizes with little or no sequence similarities. Since InvJ/SpaN of S. typhimurium is secreted and the corresponding Spa32 from S. flexneri is exposed on the bacterial surface, it is conceivable that these proteins underwent evolutionary change due to the need for host adaptation or antigen variation (see “Gene and protein families” above). The roles of InvJ/SpaN and Spa32 in protein secretion are discussed under “Regulation of type III secretion by contact with eukaryotic cells” above).
Exposure of the Spa32 protein in the outer membrane of S. flexneri requires an oxidoreductase encoded by dsbA (465). Like a spa32 strain, a dsbA mutant of S. flexneri is impaired in Ipa secretion and epithelial cell invasion (465). Replacement of either of the two cysteine residues Cys-19 and Cys-292 in Spa32 with Ser resulted in the same phenotype as observed for the spa32 and the dsbA mutants (465), indicating that the formation of an intramolecular disulfide bond is required for surface exposure of Spa32. However, these results probably have no significance for other type III secretion systems, since the numbers and positions of cysteine residues in other proteins that group with Spa32 are highly variable (no Cys in SpaN/InvJ, 6 Cys in YscP).
The YscF, YscI, YscK, and YscL Families
YscF and its homologs are short proteins (73 to 87 aa) with an overall hydrophilic character and are therefore probably located in the cytoplasm. The proteins are present only in the mammalian pathogens; they are absent from or have not been detected in plant pathogens. The proteins of the YscL family are mainly hydrophilic. Homologs of YscL are present in almost all type III secretion systems except for those of EPEC (where it may not have been detected yet owing to limited sequence information), S. flexneri and S. typhimurium SPI-1. In contrast to SPI-1, the S. typhimurium SPI-2 systems contains a protein, SsaK, which shows weak similarity to YscL. Furthermore, NolV of R. fredii is somewhat similar to YscL. Proteins of the YscI and YscK families are hydrophilic and may thus be located in the cytoplasm. These proteins are also present only in subgroups of type III secretion systems and are well conserved only within these subgroups (Fig. 10).
YopN and Similar Proteins
YopN is an important player in the regulation of type III secretion by Yersinia spp. (see the section on regulation of type III secretion in Yersinia species, above). The protein is localized on the bacterial surface (129) and keeps type III secretion channels shut in the presence of millimolar concentrations of Ca2+ and in the absence of contact of the bacteria with the surface of a mammalian cell (383). In the presence of an appropriate secretion signal, YopN is secreted into the supernatant by the Yersinia type III secretion system (57, 129). Some proteins from other type III secretion systems (InvE from S. typhimurium, MxiC from S. flexneri, and the HrpJ proteins from E. amylovora and P. syringae) exhibit weak similarity to YopN. As in other groups, the S. flexneri and S. typhimurium proteins and the two HrpJ proteins constitute two subgroups of significantly similar proteins. All five proteins in the YopN group are mainly hydrophilic. Except for YopN, no further structural nor functional data exist for these proteins.
The YscC Family and Its Homologs in Phage Extrusion and Type II Secretion
YscC and its homologs are the only components in type III secretion systems which are clearly located in the outer membrane (351, 477). These proteins are likely to form a channel through which proteins are secreted across the outer membrane. In all cases analyzed, the proteins of the YscC family are required for type III secretion and for type III secretion-dependent virulence phenotypes (12, 158, 172, 232, 333, 410).
The members of the YscC family represent a subfamily of the GspD family of bacterial and phage proteins involved in the transport of large molecules across the outer membrane. The subfamilies of the GspD family are functionally distinct in that they are involved in different transport pathways. (i) the secretion of virulence proteins via the type III secretion pathway (YscC family), (ii) the secretion of extracellular enzymes via the type II secretion pathway (exemplified by PulD of Klebsiella oxytoca [95]), (iii) the extrusion and assembly of filamentous phages (pIV family [386]), and (iv) the export of pilus subunits in the assembly of type IV pili as exemplified by PilQ of Pseudomonas aeruginosa (288).
A common feature of all GspD family members are two conserved domains in the carboxy-terminal half of the proteins, while the amino-termini are only conserved within family subgroups but do not share similarity between different subgroups (Fig. 14). The subgroup-specific amino termini may specify the interaction of the GspD family members with components of the different transport systems (387). Another common feature of the GspD family members is the presence of a signal sequence which allows export of the proteins from the cytoplasm by the sec pathway. In addition to the sec system, PulD of K. oxytoca requires another component of the type II secretion pathway, PulS. PulS is an outer membrane-anchored lipoprotein with chaperone function that protects PulD from degradation and promotes its localization to the outer membrane (181, 182). No PulS-homologous protein exists in type III secretion systems, indicating that the mechanism of targeting YscC family proteins to the outer membrane differs from the respective mechanism in the pullulanase system. Recently, it was reported that the VirG protein of Y. enterocolitica is required for correct localization of YscC in the outer membrane (89a).
FIG. 14.
Conserved domains in selected proteins of the large GspD family. The generally conserved domains are shown in black and are aligned with each other, while domains which are conserved in subfamilies are shown in corresponding shadings. Kpn, Klebsiella pneumoniae; Ech, Erwinia chrysanthemi; BP, bacteriophage; Hin, Haemophilus influenzae; Yen, Yersinia enterocolitica; Sfl, Shigella flexneri.
Considering the diameter of filamentous phages (65 Å [386]), the outer membrane pores formed by pIV and its homologs must be large. The required size of these pores may be achieved by multimerization of GspD family proteins. In the case of pIV of filamentous phages, these multimers are composed of 10 to 12 monomers (237). Furthermore, pIV and OutD of Erwinia chrysanthemi, which belong to two different subfamilies of the PulD family, have been observed to form heteromultimers when coexpressed in E. coli (237), indicating that the conserved carboxy termini may facilitate multimerization. No heteromultimers were detected, however, when HrpA1 (X. campestris pv. vesicatoria) and pIV were coexpressed in E. coli (477). A channel of the size capable of conducting a 65-Å particle must be gated in order to not allow uncontrolled diffusion of macromolecules. With respect to this notion, it is interesting that a point mutation of pIV at Gly-355, a position which is highly conserved throughout the entire GspD family, rendered E. coli sensitive to antibiotics and detergents that are normally excluded by the outer membrane. The mutant pIV still formed multimers but was impaired in promoting phage assembly (385). Thus, Gly-355 could be involved in a gating mechanism.
CONCLUDING REMARKS
Type III secretion systems are dedicated protein translocation machineries that allow bacterial pathogenicity proteins to be delivered directly into the cytosol of eukaryotic host cells. The identification of type III secretion has opened a new and rapidly expanding chapter of research into the molecular factors and mechanisms underlying bacterial pathogenesis. Analysis of the biochemistry of type III secreted pathogenicity factors has yielded fascinating insights into sophisticated and highly adapted bacterium-host interactions which lead to remodeling of the host cell biochemistry and signal transduction pathways to facilitate bacterial infection, colonization, and replication within the host. In contrast, analysis of the mechanism of type III secretion has been dominated so far by sequence analyses of the respective encoding genes, and several important questions about the biochemistry and function of the secretion apparatus remain to be answered. Is the signal for secretion in the many different systems indeed located in the RNA, and if so, which part of the secretion machinery interacts with the RNA? Do the secreted proteins interact with the secretion apparatus? What is the structure of the secretion apparatus; i.e., which components of the secretion apparatus interact with each other? Which minimal set of proteins constitutes a functional secretion apparatus and what is the role of species-specific components? What is the nature of eukaryotic cell surface signals that activate type III secretion systems, and with which components of the secretion system do they interact? Are macromolecular bacterial surface structures involved in protein secretion and protein translocation?
It appears likely that several, if not many more type III secretion systems will be identified in the near future. The resulting increase in sequence information should greatly facilitate the analysis of evolutionary relations of type III secretion systems. More importantly, like the horizontal acquisition of a type III secretion system by a bacterium facilitated an “evolutionary quantum leap” (E. A. Groisman) in bacterial pathogenesis, the identification of a new type III secretion system will lead to tremendous new insights into pathogenicity mechanisms.
Both animal and plant cells are targets for type III-dependent protein translocation, and heterologous proteins are readily translocated via the type III secretion and translocation signals. Consequently, type III secretion could be a useful tool for the targeted delivery of engineered proteins to influence cellular signal transduction and other processes, with a wide range of conceivable applications. Type III secretion may prove especially suited for targeted delivery of highly toxic or labile compounds, since prior to delivery, these compounds would be contained and protected inside the bacterial cell. Even the expression of toxic compounds prior to delivery could be avoided by use of the Yersinia feedback mechanism which links transcription to activated type III secretion. Finally, it should not remain unmentioned that type III secretion systems may provide targets for new drugs which specifically attenuate bacterial pathogens without affecting the commensal flora. In summary, the analysis of type III secretion systems facilitates the understanding of the molecular mechanisms and evolution of bacterial pathogenicity and may yield important practical applications.
ACKNOWLEDGMENTS
I am indebted to Samuel Miller and Werner Goebel, in whose laboratories the manuscript was prepared. I thank Catherine Lee for critical reading of the manuscript. I also thank my colleagues who patiently supported this work and Eva Ng for suggesting the introductory quotes.
Financial support was by a personal grant from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie, Germany.
ADDENDUM IN PROOF
It was recently shown that the S. typhimurium SPI-2-encoded response regulator SpiR/SsrB mediates an approximately 400-fold induction of expression of a SPI-2 gene after the bacteria are phagocytosed by a macrophage (R. H. Valdivia and S. Falkow, Science 277:2007–2011, 1997).
REFERENCES
- 1.Abeles A L, Friedman S A, Austin S J. Partition of unit-copy miniplasmids to daughter cells. III. The DNA sequence and functional organization of the P1 partition region. J Mol Biol. 1985;185:261–272. doi: 10.1016/0022-2836(85)90402-4. [DOI] [PubMed] [Google Scholar]
- 2.Adam T, Arpin M, Prévost M-C, Gounon P, Sansonetti P J. Cytoskeletal rearrangements and the functional role of T-plastin during entry of Shigella flexneri into HeLa cells. J Cell Biol. 1995;129:367–381. doi: 10.1083/jcb.129.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adam T, Giry M, Boquet P, Sansonetti P. Rho-dependent membrane folding causes Shigella entry into epithelial cells. EMBO J. 1996;15:3315–3321. [PMC free article] [PubMed] [Google Scholar]
- 4.Adler B, Sasakawa C, Tobe T, Makino S, Komatsu K, Yoshikawa M. A dual transcriptional activation system for the 230 kb plasmid genes coding for virulence-associated antigens of Shigella flexneri. Mol Microbiol. 1989;3:627–635. doi: 10.1111/j.1365-2958.1989.tb00210.x. [DOI] [PubMed] [Google Scholar]
- 5.Aizawa S-I. Flagellar assembly in Salmonella typhimurium. Mol Microbiol. 1996;19:1–5. doi: 10.1046/j.1365-2958.1996.344874.x. [DOI] [PubMed] [Google Scholar]
- 6.Albertini A M, Caramori T, Crabb W D, Scoffone F, Galizzi A. The flaA locus of Bacillus subtilis is part of a large operon coding for flagellar structures, motility functions, and an ATPase-like polypeptide. J Bacteriol. 1991;173:3573–3579. doi: 10.1128/jb.173.11.3573-3579.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alfano J R, Bauer D W, Milos T M, Collmer A. Analysis of the role of the Pseudomonas syringae pv. syringae HrpZ harpin in elicitation of the hypersensitive response in tobacco using functionally non-polar hrpZ deletion mutations, truncated HrpZ fragments, and hrmA mutations. Mol Microbiol. 1996;19:715–728. doi: 10.1046/j.1365-2958.1996.415946.x. [DOI] [PubMed] [Google Scholar]
- 8.Allaoui A, Ménard R, Sansonetti P J, Parsot C. Characterization of the Shigella flexneri ipgD and ipgF genes, which are located in the proximal part of the mxi locus. Infect Immun. 1993;61:1707–1714. doi: 10.1128/iai.61.5.1707-1714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allaoui A, Mounier J, Prévost M-C, Sansonetti P J, Parsot C. icsB, a Shigella flexneri virulence gene necessary for the lysis of protrusions during intercellular spread. Mol Microbiol. 1992;6:1605–1616. doi: 10.1111/j.1365-2958.1992.tb00885.x. [DOI] [PubMed] [Google Scholar]
- 10.Allaoui A, Sansonetti P J, Ménard R, Barzu S, Mounier J, Phalipon A, Parsot C. MxiG, a membrane protein required for secretion of Shigella spp. Ipa invasins: involvement in entry into epithelial cells and intercellular dissemination. Mol Microbiol. 1995;17:461–470. doi: 10.1111/j.1365-2958.1995.mmi_17030461.x. [DOI] [PubMed] [Google Scholar]
- 11.Allaoui A, Sansonetti P J, Parsot C. MxiJ, a lipoprotein involved in secretion of Shigella Ipa invasins, is homologous to YscJ, a secretion factor of the Yersinia Yop proteins. J Bacteriol. 1992;174:7661–7669. doi: 10.1128/jb.174.23.7661-7669.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allaoui A, Sansonetti P J, Parsot C. MxiD, an outer membrane protein necessary for the secretion of the Shigella flexneri Ipa invasins. Mol Microbiol. 1993;7:59–68. doi: 10.1111/j.1365-2958.1993.tb01097.x. [DOI] [PubMed] [Google Scholar]
- 13.Allaoui A, Scheen R, Lambert de Rouvroit C, Cornelis G R. VirG, a Yersinia enterocolitica lipoprotein involved in Ca2+ dependency, is related to ExsB of Pseudomonas aeruginosa. J Bacteriol. 1995;177:4230–4237. doi: 10.1128/jb.177.15.4230-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allaoui A, Schulte R, Cornelis G R. Mutational analysis of the Yersinia enterocolitica virC operon: characterization of yscE, F, G, I, J, K required for Yop secretion and yscH encoding YopR. Mol Microbiol. 1995;18:343–355. doi: 10.1111/j.1365-2958.1995.mmi_18020343.x. [DOI] [PubMed] [Google Scholar]
- 15.Allaoui A, Woestyn S, Sluiters C, Cornelis G R. YscU, a Yersinia enterocolitica inner membrane protein involved in Yop secretion. J Bacteriol. 1994;176:4534–4542. doi: 10.1128/jb.176.15.4534-4542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alpuche-Aranda C M, Berthiaume E P, Mock B, Swanson J A, Miller S I. Spacious phagosomes formation within mouse macrophages correlates with Salmonella serotype pathogenicity and host susceptibility. Infect Immun. 1995;63:4456–4462. doi: 10.1128/iai.63.11.4456-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altmeyer R M, McNern J K, Bossio J C, Rosenshine I, Finlay B B, Galán J E. Cloning and molecular characterization of a gene involved in Salmonella adherence and invasion of cultured epithelial cells. Mol Microbiol. 1993;7:89–98. doi: 10.1111/j.1365-2958.1993.tb01100.x. [DOI] [PubMed] [Google Scholar]
- 18.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Anderson D M, Schneewind O. A mRNA signal for the type III secretion of Yop proteins by Yersinia enterocolitica. Science. 1997;278:1140–1143. doi: 10.1126/science.278.5340.1140. [DOI] [PubMed] [Google Scholar]
- 20.Andersson K, Carballeira N, Magnusson K-E, Persson C, Stendahl O, Wolf-Watz H, Fällmann M. YopH of Yersinia pseudotuberculosis interrupts early phosphotyrosine signalling associated with phagocytosis. Mol Microbiol. 1996;20:1057–1069. doi: 10.1111/j.1365-2958.1996.tb02546.x. [DOI] [PubMed] [Google Scholar]
- 21.Andrews G P, Hromockyj A E, Coker C, Maurelli A T. Two novel virulence loci, mxiA and mxiB, in Shigella flexneri 2a facilitate excretion of invasion plasmid antigens. Infect Immun. 1991;59:1997–2005. doi: 10.1128/iai.59.6.1997-2005.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrews G P, Maurelli A T. mxiA of Shigella flexneri 2a, which facilitates export of invasion plasmid antigens, encodes a homolog of the low-calcium-response protein, LcrD, of Yersinia pestis. Infect Immun. 1992;60:3287–3295. doi: 10.1128/iai.60.8.3287-3295.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Apodaca G, Bomsel M, Lindstedt R, Engel J, Frank D, Mostov K E, Wiener-Kronish J. Characterization of Pseudomonas aeruginosa-induced MDCK cell injury: glycosylation-defective host cells are resistant to bacterial killing. Infect Immun. 1995;63:1541–1551. doi: 10.1128/iai.63.4.1541-1551.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arakawa E, Kato J-I, Ito K-I, Watanabe H. Comparison and high conservation of nucleotide sequences of spa-mxi regions between S. sonnei and S. flexneri—identification of a new gene coding plausible membrane protein. 1995. GenBank accession no. D50601. [Google Scholar]
- 25.Arlat M, Gough C L, Zischek C, Barberis P A, Trigalet A, Boucher C A. Transcriptional organization and expression of the large hrp gene cluster of Pseudomonas solanacearum. Mol Plant-Microbe Interact. 1992;5:187–193. doi: 10.1094/mpmi-5-187. [DOI] [PubMed] [Google Scholar]
- 26.Arlat M, Van Gijsegem F, Huet J C, Pernollet J C, Boucher C A. PopA1, a protein which induces a hypersensitivity-like response on specific Petunia genotypes, is secreted via the Hrp pathway of Pseudomonas solanacearum. EMBO J. 1994;13:543–553. doi: 10.1002/j.1460-2075.1994.tb06292.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bajaj V, Hwang C, Lee C A. hilA is a novel ompR/toxR family member that activates the expression of Salmonella typhimurium invasion genes. Mol Microbiol. 1995;18:715–727. doi: 10.1111/j.1365-2958.1995.mmi_18040715.x. [DOI] [PubMed] [Google Scholar]
- 28.Bajaj V, Lucas R L, Hwang C, Lee C A. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol. 1996;22:703–714. doi: 10.1046/j.1365-2958.1996.d01-1718.x. [DOI] [PubMed] [Google Scholar]
- 29.Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar S P. Signaling in plant-microbe interactions. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 30.Baldwin T J, Ward W, Aitken A, Knutton S, Williams P H. Elevation of intracellular free calcium levels in HEp-2 cells infected with enteropathogenic Escherichia coli. Infect Immun. 1991;59:1599–1604. doi: 10.1128/iai.59.5.1599-1604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barve S S, Straley S C. lcrR, a low Ca2+-response locus with dual Ca2+-dependent functions in Yersinia pestis. J Bacteriol. 1990;172:4661–4671. doi: 10.1128/jb.172.8.4661-4671.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baudry B, Kaczorek M, Sansonetti P J. Nucleotide sequence of the invasion plasmid antigens B and C genes (ipaB and ipaC) of Shigella flexneri. Microb Pathog. 1988;4:345–357. doi: 10.1016/0882-4010(88)90062-9. [DOI] [PubMed] [Google Scholar]
- 33.Baudry B, Maurelli A T, Clerc P, Sadoff J C, Sansonetti P J. Localization of plasmid loci necessary for the entry of Shigella flexneri into HeLa cells, and characterization of one locus encoding four immunogenic polypeptides. J Gen Microbiol. 1987;133:3403–3413. doi: 10.1099/00221287-133-12-3403. [DOI] [PubMed] [Google Scholar]
- 34.Bauer D W, Wei Z M, Beer S V, Collmer A. Erwinia chrysanthemi harpinEch: an elicitor of the hypersensitive response that contributes to soft-rot pathogenesis. Mol Plant-Microbe Interact. 1995;8:484–491. doi: 10.1094/mpmi-8-0484. [DOI] [PubMed] [Google Scholar]
- 35.Bäumler A. The record of horizontal gene transfer in Salmonella. Trends Microbiol. 1997;5:318–322. doi: 10.1016/S0966-842X(97)01082-2. [DOI] [PubMed] [Google Scholar]
- 36.Bäumler A J, Tsolis R M, Heffron F. The lpf fimbrial operon mediates adhesion of Salmonella typhimurium to murine Peyer’s patches. Proc Natl Acad Sci USA. 1996;93:279–283. doi: 10.1073/pnas.93.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bäumler A J, Tsolis R M, Valentine P J, Ficht T A, Heffron F. Synergistic effect of mutations in invA and lpfC on the ability of Salmonella typhimurium to cause murine typhoid. Infect Immun. 1997;65:2254–2259. doi: 10.1128/iai.65.6.2254-2259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Behlau I, Miller S I. A PhoP-repressed gene promotes Salmonella typhimurium invasion of epithelial cells. J Bacteriol. 1993;175:4475–4484. doi: 10.1128/jb.175.14.4475-4484.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ben-Gurion R, Shafferman A. Essential virulence determinants of different Yersinia species are carried on a common plasmid. Plasmid. 1981;5:183–187. doi: 10.1016/0147-619x(81)90019-6. [DOI] [PubMed] [Google Scholar]
- 40.Bergman T, Erickson K, Galyov E, Persson C, Wolf-Watz H. The lcrB (yscN/U) gene cluster of Yersinia pseudotubercolosis is involved in Yop secretion and shows high homology to the spa gene cluster of Shigella flexneri and Salmonella typhimurium. J Bacteriol. 1994;176:2619–2626. doi: 10.1128/jb.176.9.2619-2626.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergman T, Håkansson S, Forsberg Å, Norlander L, Macellaro A, Bäckman A, Bölin I, Wolf-Watz H. Analysis of the V antigen lcrGVH-yopBD operon of Yersinia pseudotubercolosis: evidence for a regulatory role of LcrH and LcrV. J Bacteriol. 1991;173:1607–1616. doi: 10.1128/jb.173.5.1607-1616.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bernardini M L, Fontaine A, Sansonetti P J. The two-component regulatory system ompR-envZ controls the virulence of Shigella flexneri. J Bacteriol. 1990;172:6274–6281. doi: 10.1128/jb.172.11.6274-6281.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Betts J, Finlay B B. Identification of Salmonella typhimurium invasiveness loci. Can J Microbiol. 1992;38:852–857. doi: 10.1139/m92-138. [DOI] [PubMed] [Google Scholar]
- 44.Beuscher H U, Roedel F, Forsberg Å, Roellinghoff M. Bacterial evasion of host immune defense: Yersinia enterocolitica encodes a suppressor for tumor necrosis factor alpha expression. Infect Immun. 1995;63:1270–1277. doi: 10.1128/iai.63.4.1270-1277.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Birkelund S, Johnson H, Christiansen G. Chlamydia trachomatis serovar L2 induces protein tyrosine phosphorylation during uptake by HeLa cells. Infect Immun. 1994;62:4900–4908. doi: 10.1128/iai.62.11.4900-4908.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bischoff D S, Ordal G W. Identification and characterization of FliY, a novel component of the Bacillus subtilis flagellar switch complex. Mol Microbiol. 1992;6:2715–2723. doi: 10.1111/j.1365-2958.1992.tb01448.x. [DOI] [PubMed] [Google Scholar]
- 47.Bischoff D S, Weinreich M D, Ordal G W. Nucleotide sequences of Bacillus subtilis flagellar biosynthetic genes fliP and fliQ and identification of a novel flagellar gene, fliZ. J Bacteriol. 1992;174:4017–4025. doi: 10.1128/jb.174.12.4017-4025.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Black D S, Bliska J B. Identification of p130Cas as a substrate of Yersinia YopH (Yop51), a bacterial protein tyrosine phosphatase that translocates into mammalian cells and targets focal adhesions. EMBO J. 1997;16:2730–2744. doi: 10.1093/emboj/16.10.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bliska J B, Black D S. Inhibition of the Fc receptor-mediated oxidative burst in macrophages by Yersinia pseudotuberculosis tyrosine phosphatase. Infect Immun. 1995;63:681–685. doi: 10.1128/iai.63.2.681-685.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bliska J B, Clemens J C, Dixon J E, Falkow S. The Yersinia tyrosine phosphatase: specificity of a bacterial virulence determinant for phosphoproteins in the J7774A.1 macrophage. J Exp Med. 1992;176:1625–1630. doi: 10.1084/jem.176.6.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bliska J B, Guan K, Dixon J E, Falkow S. Tyrosine phosphate hydrolysis of host proteins by an essential Yersinia virulence determinant. Proc Natl Acad Sci USA. 1991;88:1187–1191. doi: 10.1073/pnas.88.4.1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Blum G, Ott M, Lischewski A, Ritter A, Imrich H, Tschape H, Hacker J. Excision of large DNA regions termed pathogenicity islands from tRNA-specific loci in the chromosome of an Escherichia coli wild-type pathogen. Infect Immun. 1994;62:606–614. doi: 10.1128/iai.62.2.606-614.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bodey G P, Bolivar R, Fainstein V, Jadeja L. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 1983;5:279–313. doi: 10.1093/clinids/5.2.279. [DOI] [PubMed] [Google Scholar]
- 54.Bogdanove A J, Beer S V, Bonas U, Boucher C A, Collmer A, Coplin D L, Cornelis G R, Huang H C, Hutcheson S W, Panopoulos N J, Van Gijsegem F. Unified nomenclature for broadly conserved hrp genes of phytopathogenic bacteria. Mol Microbiol. 1996;20:681–683. doi: 10.1046/j.1365-2958.1996.5731077.x. [DOI] [PubMed] [Google Scholar]
- 55.Bogdanove A J, Wei Z-M, Zhao L, Beer S V. Erwinia amylovora secretes harpin via a type III pathway and contains a homolog of yopN of Yersinia spp. J Bacteriol. 1996;178:1720–1730. doi: 10.1128/jb.178.6.1720-1730.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boland A, Sory M P, Iriarte M, Kerbourch C, Wattiau P, Cornelis G R. Status of YopM and YopN in the Yersinia Yop virulon: YopM of Y. enterocolitica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB, D, N delivery apparatus. EMBO J. 1996;15:5191–5201. [PMC free article] [PubMed] [Google Scholar]
- 57.Bölin I, Forsberg Å, Norlander L, Skurnik M, Wolf-Watz H. Identification and mapping of temperature-inducible, plasmid-encoded proteins of Yersinia spp. Infect Immun. 1988;56:343–348. doi: 10.1128/iai.56.2.343-348.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bölin I, Portnoy D A, Wolf-Watz H. Expression of the temperature-inducible outer membrane proteins of yersiniae. Infect Immun. 1985;48:234–240. doi: 10.1128/iai.48.1.234-240.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bölin I, Wolf-Watz H. The plasmid-encoded Yop2b protein of Yersinia pseudotuberculosis is a virulence determinant regulated by calcium and temperature at the level of transcription. Mol Microbiol. 1988;2:237–245. doi: 10.1111/j.1365-2958.1988.tb00025.x. [DOI] [PubMed] [Google Scholar]
- 60.Bonas U. hrp genes of phytopathogenic bacteria. Curr Top Microbiol Immunol. 1994;192:79–98. doi: 10.1007/978-3-642-78624-2_4. [DOI] [PubMed] [Google Scholar]
- 61.Bonas U, Schulte R, Fenselau S, Minsavage G V, Staskawicz B J, Stall R E. Isolation of a gene cluster from Xanthomonas campestris pv. vesicatoria that determines pathogenicity and the hypersensitive response on pepper and tomato. Mol Plant-Microbe Interact. 1991;4:81–88. [Google Scholar]
- 62.Boucher C A, Van Gijsegem F, Barberis P A, Arlat M, Zischek C. Pseudomonas solanacearum genes controlling both pathogenicity on tomato and hypersensitivity on tobacco are clustered. J Bacteriol. 1987;169:5626–5632. doi: 10.1128/jb.169.12.5626-5632.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boyd E F, Li J, Ochman H, Selander R K. Comparative genetics of the inv-spa invasion gene complex of Salmonella enterica. J Bacteriol. 1997;179:1985–1991. doi: 10.1128/jb.179.6.1985-1991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bradley D J, Kjelvom P, Lamb C J. Elicitor- and wound-induced oxidative cross-linking of a proline-rich plant cell wall protein: a novel, rapid defense response. Cell. 1992;70:21–30. doi: 10.1016/0092-8674(92)90530-p. [DOI] [PubMed] [Google Scholar]
- 65.Burridge K, Nuckolls G, Otey C, Pavalko F, Simon K, Turner C. Actin-membrane interaction in focal adhesions. Cell Differ Dev. 1990;32:337–342. doi: 10.1016/0922-3371(90)90048-2. [DOI] [PubMed] [Google Scholar]
- 66.Burrows T, Bacon G A. The basis of virulence in Pasteurella pestis: an antigen determinig virulence. Br J Exp Pathol. 1956;37:481–493. [PMC free article] [PubMed] [Google Scholar]
- 67.Buysse J M, Stover C K, Oaks E V, Venkatesan M, Kopecko D J. Molecular cloning of invasion plasmid antigen (ipa) genes from Shigella flexneri: analysis of ipa gene products and genetic mapping. J Bacteriol. 1987;169:2561–2569. doi: 10.1128/jb.169.6.2561-2569.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buysse J M, Venkatesan M, Mills J A, Oaks E V. Molecular characterization of a trans-acting, positive effector (ipaR) of invasion plasmid antigen synthesis in Shigella flexneri serotype 5. Microb Pathog. 1990;8:197–211. doi: 10.1016/0882-4010(90)90047-t. [DOI] [PubMed] [Google Scholar]
- 69.Carpenter P B, Ordal G W. Bacillus subtilis FlhA: a flagellar protein related to a new family of signal-transducing receptors. Mol Microbiol. 1993;7:735–743. doi: 10.1111/j.1365-2958.1993.tb01164.x. [DOI] [PubMed] [Google Scholar]
- 70.Carpenter P B, Zuberi A R, Ordal G W. Bacillus subtilis flagellar proteins FliP, FliQ, FliR and FlhB are related to Shigella flexneri virulence factors. Gene. 1993;137:243–245. doi: 10.1016/0378-1119(93)90014-t. [DOI] [PubMed] [Google Scholar]
- 71.Carter P B, Collins F M. The route of enteric infection in normal mice. J Exp Med. 1974;139:1189–1203. doi: 10.1084/jem.139.5.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Charkowski A O, Huang H C, Collmer A. Altered localization of HrpZ in Pseudomonas syringae pv. syringae hrp mutants suggests that different components of the type III secretion pathway control protein translocation across the inner and outer membranes of gram-negative bacteria. J Bacteriol. 1997;179:3866–3874. doi: 10.1128/jb.179.12.3866-3874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen L M, Hobbie S, Galán J E. Requirement of CDC42 for Salmonella-induced cytoskeletal and nuclear responses. Science. 1996;274:2115–2118. doi: 10.1126/science.274.5295.2115. [DOI] [PubMed] [Google Scholar]
- 74.Chen L M, Kaniga K, Galán J E. Salmonella spp. are cytotoxic for cultured macrophages. Mol Microbiol. 1996;21:1101–1115. doi: 10.1046/j.1365-2958.1996.471410.x. [DOI] [PubMed] [Google Scholar]
- 75.Chen Y, Smith M R, Thirumalai K, Zychlinsky A. A bacterial invasin induces macrophage apoptosis by binding directly to ICE. EMBO J. 1996;15:3853–3860. [PMC free article] [PubMed] [Google Scholar]
- 76.Cheng L W, Anderson D M, Schneewind O. Two independent type III secretion mechanisms for YopE in Yersinia enterocolitica. Mol Microbiol. 1997;24:757–765. doi: 10.1046/j.1365-2958.1997.3831750.x. [DOI] [PubMed] [Google Scholar]
- 77.Chervaux C, Holland I B. Random and directed mutagenesis to elucidate the functional importance of helix II and F-989 in the C-terminal secretion signal of Escherichia coli hemolysin. J Bacteriol. 1996;178:1232–1236. doi: 10.1128/jb.178.4.1232-1236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clark E A, Brugge J S. Integrin and signal transduction pathways: the road taken. Science. 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- 79.Clark M A, Jepson M A, Simmons N L, Hirst B H. Preferential interaction of Salmonella typhimurium with mouse Peyer’s patch M cells. Res Microbiol. 1994;145:543–552. doi: 10.1016/0923-2508(94)90031-0. [DOI] [PubMed] [Google Scholar]
- 80.Coburn J. Pseudomonas aeruginosa exoenzyme S. Curr Top Microbiol Immunol. 1992;175:133–143. doi: 10.1007/978-3-642-76966-5_7. [DOI] [PubMed] [Google Scholar]
- 81.Coburn J, Kane A V, Feig L, Gill D M. Pseudomonas aeruginosa exoenzyme S requires a eukaryotic protein for ADP-ribosyltransferase activity. J Biol Chem. 1991;266:6438–6446. [PubMed] [Google Scholar]
- 82.Collazo C M, Galán J E. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect Immun. 1996;64:3524–3531. doi: 10.1128/iai.64.9.3524-3531.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Collazo C M, Galán J E. The invasion-associated type III secretion system of Salmonella typhimurium directs the translocation of Sip proteins into the host cell. Mol Microbiol. 1997;24:747–756. doi: 10.1046/j.1365-2958.1997.3781740.x. [DOI] [PubMed] [Google Scholar]
- 84.Collazo C M, Zierler M K, Galán J E. Functional analysis of the Salmonella typhimurium invasion genes invI and invJ and identification of a target of the protein secretion apparatus encoded in the inv locus. Mol Microbiol. 1995;15:25–38. doi: 10.1111/j.1365-2958.1995.tb02218.x. [DOI] [PubMed] [Google Scholar]
- 85.Collmer A, Bauer D W. Erwinia chrysanthemi and Pseudomonas syringae: plant pathogens trafficking in extracellular virulence proteins. Curr Top Microbiol Immunol. 1994;192:43–78. doi: 10.1007/978-3-642-78624-2_3. [DOI] [PubMed] [Google Scholar]
- 86.Cornelis G, Laroche Y, Balligand G, Sory M-P, Wauters G. Yersinia enterocolitica, a primary model for bacterial invasivness. Rev Infect Dis. 1987;9:64–87. doi: 10.1093/clinids/9.1.64. [DOI] [PubMed] [Google Scholar]
- 87.Cornelis G, Sluiters C, Lambert de Rouvroit C, Michiels T. Homology between VirF, the transcriptional activator of the Yersinia virulence regulon, and AraC, the Escherichia coli arabinose operon regulator. J Bacteriol. 1989;171:254–262. doi: 10.1128/jb.171.1.254-262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cornelis G, Sory M P, Laroche Y, Derclaye I. Genetic analysis of the plasmid region controlling virulence in Yersinia enterocolitia O:9 by Mini-Mu insertions and lac gene fusions. Microb Pathog. 1986;1:349–359. doi: 10.1016/0882-4010(86)90067-7. [DOI] [PubMed] [Google Scholar]
- 89.Cornelis G, Vanooteghem J-C, Sluiters C. Transcription of the yop regulon from Y. enterocolitica requires trans-acting pYV and chromosomal genes. Microb Pathog. 1987;2:367–379. doi: 10.1016/0882-4010(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 89a.Cornelis, G. R. Personal communication.
- 90.Cornelis G R, Biot T, Lambert de Rouvroit C, Michiels T, Mulder B, Sluiters C, Sory M-P, Van Bouchaute M, Vanooteghem J-C. The Yersinia yop regulon. Mol Microbiol. 1989;3:1455–1459. doi: 10.1111/j.1365-2958.1989.tb00129.x. [DOI] [PubMed] [Google Scholar]
- 91.Cornelis G R, Sluiters C, Delor I, Geib D, Kaniga K, Lambert de Rovroit C, Sory M-P, Vanooteghem J-C, Michiels T. ymoA, a Yersinia enterocolitica chromosomal gene modulating the expression of virulence functions. Mol Microbiol. 1991;5:1023–1034. doi: 10.1111/j.1365-2958.1991.tb01875.x. [DOI] [PubMed] [Google Scholar]
- 92.Daniels J J, Autenrieth I B, Ludwig A, Goebel W. The gene slyA of Salmonella typhimurium is required for destruction of M cells and intracellular survival but not for invasion or colonization of the murine small intestine. Infect Immun. 1996;64:5075–5084. doi: 10.1128/iai.64.12.5075-5084.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dehio C, Prévost M-C, Sansonetti P J. Invasion of epithelial cells by Shigella flexneri induces tyrosine phosphorylation of cortactin by a pp60c-src-mediated signalling pathway. EMBO J. 1995;14:2471–2482. doi: 10.1002/j.1460-2075.1995.tb07244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dell C L, Neely M N, Olson E R. Altered pH and lysine signalling mutants of cadC, a gene encoding a membrane-bound transcriptional activator of the Escherichia coli cadBA operon. Mol Microbiol. 1994;14:7–16. doi: 10.1111/j.1365-2958.1994.tb01262.x. [DOI] [PubMed] [Google Scholar]
- 95.d’Enfert C, Reyss I, Wandersman C, Pugsley A P. Protein secretion by gram-negative bacteria. Characterization of two membrane proteins required for pullulanase secretion by Escherichia coli K-12. J Biol Chem. 1989;264:17462–11748. [PubMed] [Google Scholar]
- 96.DiRita V J. Co-ordinate expression of virulence genes by ToxR in Vibrio cholerae. Mol Microbiol. 1992;6:451–458. doi: 10.1111/j.1365-2958.1992.tb01489.x. [DOI] [PubMed] [Google Scholar]
- 97.Donaldson R M J. The relation of enteric bacterial populations to gastrointestinal function and disease. In: Sleisinger M H, Fordtran J S, editors. Gastrointestinal disease: pathophysiology, diagnosis, and management. Philadelphia, Pa: The W. B. Saunders Co.; 1978. pp. 79–92. [Google Scholar]
- 98.Donnelly S F, Pocklington M J, Pallotta D, Orr E. A proline-rich protein, verprolin, involved in cytoskeletal organization and cellular growth in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1993;10:585–596. doi: 10.1111/j.1365-2958.1993.tb00930.x. [DOI] [PubMed] [Google Scholar]
- 99.Donnenberg M S, Calderwood S B, Donohue-Rolfe A, Keusch G T, Kaper J B. Construction and analysis of TnphoA mutants of enteropathogenic Escherichia coli unable to invade HEp-2 cells. Infect Immun. 1990;58:1565–1571. doi: 10.1128/iai.58.6.1565-1571.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Donnenberg M S, Kaper J B. Enteropathogenic Escherichia coli. Infect Immun. 1992;60:3953–3961. doi: 10.1128/iai.60.10.3953-3961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Donnenberg M S, Kaper J B, Finlay B B. Interactions between enteropathogenic Escherichia coli and host epithelial cells. Trends Microbiol. 1997;5:109–114. doi: 10.1016/S0966-842X(97)01000-7. [DOI] [PubMed] [Google Scholar]
- 103.Donnenberg M S, Lai L-C, Taylor K A. The locus of enterocyte effacement pathogenicity island of enteropathogenic Escherichia coli encodes secretion functions and remnants of transposons at its extreme right end. Gene. 1997;184:107–114. doi: 10.1016/s0378-1119(96)00581-1. [DOI] [PubMed] [Google Scholar]
- 104.Donnenberg M S, Yu J, Kaper J B. A second chromosomal gene necessary for intimate attachment of enteropathogenic Escherichia coli to epithelial cells. J Bacteriol. 1993;175:4670–4680. doi: 10.1128/jb.175.15.4670-4680.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dorman C J. The VirF protein from Shigella flexneri is a member of the AraC transcription factor superfamily and is highly homologous to Rns, a positive regulator of virulence genes in enterotoxigenic Escherichia coli. Mol Microbiol. 1992;6:1575. doi: 10.1111/j.1365-2958.1992.tb00879.x. [DOI] [PubMed] [Google Scholar]
- 106.Dorman C J, Bhriain N N, Higgins C F. DNA supercoiling and environmental regulation of virulence gene expression in Shigella flexneri. Nature. 1990;344:789–792. doi: 10.1038/344789a0. [DOI] [PubMed] [Google Scholar]
- 107.Driks A, DeRosier D J. Additional structures associated with bacterial flagellar basal body. J Mol Biol. 1990;211:669–672. doi: 10.1016/0022-2836(90)90063-R. [DOI] [PubMed] [Google Scholar]
- 108.Eichelberg K, Ginocchio C C, Galán J E. Molecular and functional characterization of the Salmonella typhimurium invasion genes invB and invC: homology of InvC to the F0F1 ATPase family of proteins. J Bacteriol. 1994;176:4501–4510. doi: 10.1128/jb.176.15.4501-4510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Eichelberg K, Kaniga K, Galán J E. Abstracts of the 96th General Meeting of the American Society for Microbiology 1996. Washington, D.C: American Society for Microbiology; 1996. Transcriptional regulation of Salmonella secreted virulence determinants, abstr. B-40. [Google Scholar]
- 110.Elsinghorst E A, Baron L S, Kopecko D J. Penetration of human intestinal epithelial cells by Salmonella: molecular cloning and expression of Salmonella typhi invasion determinants in Escherichia coli. Proc Natl Acad Sci USA. 1989;86:5173–5177. doi: 10.1073/pnas.86.13.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ernst R K, Dombroski D M, Merrik J M. Anaerobiosis, type 1 fimbriae, and growth phase are factors that affect invasion of HEp-2 cells by Salmonella typhimurium. Infect Immun. 1990;58:2014–2016. doi: 10.1128/iai.58.6.2014-2016.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111a.Falkow, S. Personal communication.
- 112.Fällmann M, Andersson K, Håkansson S, Magnusson K-E, Stendahl O, Wolf-Watz H. Yersinia pseudotuberculosis inhibits Fc receptor-mediated phagocytosis in J774 cells. Infect Immun. 1995;63:3117–3124. doi: 10.1128/iai.63.8.3117-3124.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fan F, Macnab R M. Enzymatic characterization of FliI. An ATPase involved in flagellar assembly in Salmonella typhimurium. J Biol Chem. 1996;271:31981–31988. doi: 10.1074/jbc.271.50.31981. [DOI] [PubMed] [Google Scholar]
- 114.Fath M J, Kolter R. ABC transporters: bacterial exporters. Microbiol Rev. 1993;57:995–1017. doi: 10.1128/mr.57.4.995-1017.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fenselau S, Balbo I, Bonas U. Determinants of pathogenicity in Xanthomonas campestris pv. vesicatoria are related to proteins involved in secretion in bacterial pathogens of animals. Mol Plant-Microbe Interact. 1992;5:390–396. doi: 10.1094/mpmi-5-390. [DOI] [PubMed] [Google Scholar]
- 116.Fenselau S, Bonas U. Sequence and expression analysis of the hrpB pathogenicity operon of Xanthomonas campestris pv. vesicatoria which encodes eight proteins with similarity to components of the Hrp, Ysc, Spa, and Fli secretion systems. Mol Plant-Microbe Interact. 1995;8:845–854. doi: 10.1094/mpmi-8-0845. [DOI] [PubMed] [Google Scholar]
- 117.Ferber D M, Brubaker R R. Plasmids in Yersinia pestis. Infect Immun. 1981;31:839–841. doi: 10.1128/iai.31.2.839-841.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Fields K A, Plano G V, Straley S C. A low-Ca2+ response (LCR) secretion (ysc) locus lies within the lcrB region of the LCR plasmid in Yersinia pestis. J Bacteriol. 1994;176:569–579. doi: 10.1128/jb.176.3.569-579.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fields P I, Swanson R V, Haidaris C G, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Finck-Barbançon V, Goranson J, Zhu L, Sawa T, Wiener-Kronish J P, Fleiszig S M, Wu C, Mende-Mueller L, Frank D W. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol Microbiol. 1997;25:547–557. doi: 10.1046/j.1365-2958.1997.4891851.x. [DOI] [PubMed] [Google Scholar]
- 121.Finlay B B, Falkow S. Comparison of the invasion strategies used by Salmonella cholerae-suis, Shigella flexneri, and Yersinia enterocolitica to enter cultured animal cells: endosome acidification is not required for bacterial invasion or intracellular replication. Biochimie. 1988;70:1089–1099. doi: 10.1016/0300-9084(88)90271-4. [DOI] [PubMed] [Google Scholar]
- 122.Finlay B B, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Finlay B B, Rosenshine I, Donnenberg M S, Kaper J B. Cytoskeletal composition of attaching and effacing lesions associated with enteropathogenic Escherichia coli adherence to HeLa cells. Infect Immun. 1992;60:2541–2543. doi: 10.1128/iai.60.6.2541-2543.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Finlay B B, Ruschkowski S. Cytoskeletal rearrangements accompanying Salmonella entry into epithelial cells. J Cell Sci. 1991;99:283–296. doi: 10.1242/jcs.99.2.283. [DOI] [PubMed] [Google Scholar]
- 125.Finlay B B, Starnbach M N, Francis C L, Stocker B A D, Chatfield S, Dougan G, Falkow S. Identification and characterization of TnphoA mutants of Salmonella that are unable to pass through a polarized MDCK epithelial cell monolayer. Mol Microbiol. 1988;2:757–766. doi: 10.1111/j.1365-2958.1988.tb00087.x. [DOI] [PubMed] [Google Scholar]
- 126.Flanagan M D, Lin S. Cytochalasins block actin filament elongation by binding to high affinity sites associated with F-actin. J Biol Chem. 1980;255:835–838. [PubMed] [Google Scholar]
- 127.Flor H H. Current status of the gene-for-gene concept. Annu Rev Phytopathol. 1971;9:275–296. [Google Scholar]
- 128.Forsberg Å, Bölin I, Norlander L, Wolf-Watz H. Molecular cloning and expression of calcium-regulated, plasmid-coded proteins of Y. pseudotuberculosis. Microb Pathog. 1987;2:123–137. doi: 10.1016/0882-4010(87)90104-5. [DOI] [PubMed] [Google Scholar]
- 129.Forsberg Å, Viitanen A-M, Skurnik M, Wolf-Watz H. The surface-lactated YopN protein involved in calcium signal transduction in Yersinia pseudotuberculosis. Mol Microbiol. 1991;5:977–986. doi: 10.1111/j.1365-2958.1991.tb00773.x. [DOI] [PubMed] [Google Scholar]
- 130.Forsberg Å, Wolf-Watz H. The virulence protein Yop5 of Yersinia pseudotuberculosis is regulated at transcriptional level by plasmid-pIB1-encoded trans-acting elements controlled by temperature and calcium. Mol Microbiol. 1988;2:121–133. doi: 10.1111/j.1365-2958.1988.tb00013.x. [DOI] [PubMed] [Google Scholar]
- 131.Forsberg Å, Wolf-Watz H. Genetic analysis of the yopE region of Yersinia spp.: identification of a novel conserved locus, yerA, regulating yopE expression. J Bacteriol. 1990;172:1547–1555. doi: 10.1128/jb.172.3.1547-1555.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Foubister V, Rosenshine I, Donnenberg M S, Finlay B B. The eaeB gene of enteropathogenic Escherichia coli is necessary for signal transduction in epithelial cells. Infect Immun. 1994;62:3038–3040. doi: 10.1128/iai.62.7.3038-3040.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Foubister V, Rosenshine I, Finlay B B. A diarrheal pathogen, enteropathogenic Escherichia coli (EPEC), triggers a flux of inositol phosphates in infected epithelial cells. J Exp Med. 1994;179:993–998. doi: 10.1084/jem.179.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Francis C L, Starnbach M N, Falkow S. Morphological and cytoskeletal changes in epithelial cells occur immediately upon interaction with Salmonella typhimurium grown under low-oxygen conditions. Mol Microbiol. 1992;6:3077–3087. doi: 10.1111/j.1365-2958.1992.tb01765.x. [DOI] [PubMed] [Google Scholar]
- 135.Frank D W. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol Microbiol. 1997;26:621–629. doi: 10.1046/j.1365-2958.1997.6251991.x. [DOI] [PubMed] [Google Scholar]
- 136.Frank D W, Iglewski B H. Cloning and sequence analysis of a trans-regulatory locus required for exoenzyme S synthesis in Pseudomonas aeruginosa. J Bacteriol. 1991;173:6460–6468. doi: 10.1128/jb.173.20.6460-6468.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Frank D W, Nair G, Herbert P S. Construction and characterization of chromosomal insertion mutations of the Pseudomonas aeruginosa exoenzyme S trans-regulatory locus. Infect Immun. 1994;62:554–563. doi: 10.1128/iai.62.2.554-563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Frederick R D, Majerczak D R, Coplin D L. Erwinia stewartii WtsA, a positive regulator of pathogenicity gene expression, is similar to Pseudomonas syringae pv. phaseolicola HrpS. Mol Microbiol. 1993;9:477–485. doi: 10.1111/j.1365-2958.1993.tb01709.x. [DOI] [PubMed] [Google Scholar]
- 139.Freiberg C, Fellay R, Bairoch A, Broughton W J, Rosenthal A, Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 140.Frithz-Lindsten E, Du Y, Rosqvist R, Forsberg A. Intracellular targeting of exoenzyme S of Pseudomonas aeruginosa via type III-dependent translocation induces phagocytosis resistance, cytotoxicity and disruption of actin microfilaments. Mol Microbiol. 1997;25:1125–1139. doi: 10.1046/j.1365-2958.1997.5411905.x. [DOI] [PubMed] [Google Scholar]
- 141.Frithz-Lindsten E, Rosqvist R, Johansson L, Forsberg Å. The chaperone-like protein YerA of Yersinia pseudotuberculosis stabilizes YopE in the cytoplasm but is dispensable for targeting to the secretion loci. Mol Microbiol. 1995;16:635–647. doi: 10.1111/j.1365-2958.1995.tb02426.x. [DOI] [PubMed] [Google Scholar]
- 142.Fu H, Coburn J, Collier R J. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc Natl Acad Sci USA. 1993;90:2320–2324. doi: 10.1073/pnas.90.6.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Galán J E, Curtiss R., III Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Galán J E, Curtiss R., III Expression of Salmonella typhimurium genes required for invasion is regulated by changes in DNA supercoiling. Infect Immun. 1990;58:1879–1885. doi: 10.1128/iai.58.6.1879-1885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Galán J E, Curtiss R., III Distribution of the invA, -B, -C, and -D genes of Salmonella typhimurium among other Salmonella serovars: invA mutants of Salmonella typhi are deficient for entry into mammalian cells. Infect Immun. 1991;59:2901–2908. doi: 10.1128/iai.59.9.2901-2908.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Galán J E, Ginocchio C, Costeas P. Molecular and functional characterization of the Salmonella invasion gene invA: homology of InvA to members of a new protein family. J Bacteriol. 1992;174:4338–4349. doi: 10.1128/jb.174.13.4338-4349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Galán J E, Sansonetti P J. Molecular and cellular bases of Salmonella and Shigella interactions with host cells. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Riley M, Reznikoff W S, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2757–2773. [Google Scholar]
- 148.Gallegos M T, Michan C, Ramos J L. The XylS/AraC family of regulators. Nucleic Acids Res. 1993;21:807–810. doi: 10.1093/nar/21.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Galyov E E, Håkansson S, Forsberg Å, Wolf-Watz H. A secreted protein kinase of Yersinia pseudotuberculosis is an indispensable virulence determinant. Nature. 1993;361:730–732. doi: 10.1038/361730a0. [DOI] [PubMed] [Google Scholar]
- 150.Galyov E E, Håkansson S, Wolf-Watz H. Characterization of the operon encoding the YpkA Ser/Thr protein kinase and the YopJ protein of Yersinia pseudotuberculosis. J Bacteriol. 1994;176:4543–4548. doi: 10.1128/jb.176.15.4543-4548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Galyov E E, Wood M W, Rosqvist R, Mullan P B, Watson P R, Hedges S, Wallis T S. A secreted effector protein of Salmonella dublin is translocated into eukaryotic cells and mediates inflammation and fluid secretion in infected ileal mucosa. Mol Microbiol. 1997;25:903–912. doi: 10.1111/j.1365-2958.1997.mmi525.x. [DOI] [PubMed] [Google Scholar]
- 152.Garcia Vescovi E, Soncini F C, Groisman E A. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 153.Gemski P, Lazere J R, Casey T. Plasmid associated with pathogenicity and calcium dependency of Yersinia enterocolitica. Infect Immun. 1980;27:682–685. doi: 10.1128/iai.27.2.682-685.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Gemski P, Lazere J R, Casey T, Wohlhieter J A. Presence of a virulence-associated plasmid in Yersinia pseudotuberculosis. Infect Immun. 1980;28:1044–1047. doi: 10.1128/iai.28.3.1044-1047.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Genin S, Gough C L, Zischek C, Boucher C A. Evidence that the hrpB gene encodes a positive regulator of pathogenicity genes from Pseudomonas solanacearum. Mol Microbiol. 1992;6:3065–3076. doi: 10.1111/j.1365-2958.1992.tb01764.x. [DOI] [PubMed] [Google Scholar]
- 156.Ginnochio C, Pace J, Galán J E. Identification and molecular characterization of a Salmonella typhimurium gene involved in triggering the internalization of Salmonellae into cultured epithelial cells. Proc Natl Acad Sci USA. 1992;89:5976–5980. doi: 10.1073/pnas.89.13.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ginocchio C C, Galán J E. Functional conservation among members of the Salmonella typhimurium InvA family of proteins. Infect Immun. 1995;63:729–732. doi: 10.1128/iai.63.2.729-732.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ginocchio C C, Olmsted S B, Wells C L, Galán J E. Contact with epithelial cells induces the formation of surface appendages on Salmonella typhimurium. Cell. 1994;76:717–724. doi: 10.1016/0092-8674(94)90510-x. [DOI] [PubMed] [Google Scholar]
- 159.Ginocchio C C, Rahn K, Clarke R C, Galán J E. Naturally occurring deletions in the centisome 63 pathogenicity island of environmental isolates of Salmonella spp. Infect Immun. 1997;65:1267–1272. doi: 10.1128/iai.65.4.1267-1272.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Goguen J D, Yother J, Straley S C. Genetic analysis of the low calcium response in Yersinia pestis Mu d1 (Ap lac) insertion mutants. J Bacteriol. 1984;160:842–848. doi: 10.1128/jb.160.3.842-848.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gómez-Duarte O G, Kaper J B. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect Immun. 1995;63:1767–1776. doi: 10.1128/iai.63.5.1767-1776.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Gopalan S, Bauer D W, Alfano J R, Loniello A O, He S Y, Collmer A. Expression of the Pseudomonas syringae avirulence protein AvrB in plant cells alleviates its dependence on the hypersensitive response and pathogenicity (Hrp) secretion system in eliciting genotype-specific hypersensitive cell death. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Gough C L, Genin S, Zischek C, Boucher C A. hrp genes of Pseudomonas solanacearum are homologous to pathogenicity determinants of animal pathogenic bacteria and are conserved among plant pathogenic bacteria. Mol Plant-Microbe Interact. 1992;5:384–389. doi: 10.1094/mpmi-5-384. [DOI] [PubMed] [Google Scholar]
- 164.Gouzy J, Corpet F, Kahn D. Graphical interface for ProDom domain families. Trends Biochem Sci. 1996;21:493. doi: 10.1016/s0968-0004(96)30040-6. [DOI] [PubMed] [Google Scholar]
- 165.Greenberg S, Chang P, Silverstein S C. Tyrosine phosphorylation of the γ subunit of Fcγ receptors, p72syk, and paxillin during Fc receptor-mediated phagocytosis in macrophages. J Biol Chem. 1994;269:3897–3902. [PubMed] [Google Scholar]
- 166.Grimm C, Aufsatz W, Panopoulos N J. The hrpRS locus of Pseudomonas syringae pv. phaseolicola constitutes a complex regulatory unit. Mol Microbiol. 1995;15:155–165. doi: 10.1111/j.1365-2958.1995.tb02230.x. [DOI] [PubMed] [Google Scholar]
- 167.Grimm C, Panopoulos N J. The predicted protein product of a pathogenicity locus from Pseudomonas syringae pv. phaseolicola is homologous to a highly conserved domain of several procaryotic regulatory proteins. J Bacteriol. 1989;171:5031–5038. doi: 10.1128/jb.171.9.5031-5038.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Groisman E A, Ochman H. Cognate gene clusters govern invasion of host epithelial cells by Salmonella typhimurium and Shigella flexneri. EMBO J. 1993;12:3779–3787. doi: 10.1002/j.1460-2075.1993.tb06056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Guan K, Dixon J E. Protein tyrosine phosphatase activity of an essential virulence determinant in Yersinia. Science. 1990;249:553–556. doi: 10.1126/science.2166336. [DOI] [PubMed] [Google Scholar]
- 170.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbiol evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 171.Hackstadt T, Fischer E R, Scidmore M A, Rockey D D, Heinzen R A. Origins and functions of the chlamydial inclusion. Trends Microbiol. 1997;5:288–293. doi: 10.1016/S0966-842X(97)01061-5. [DOI] [PubMed] [Google Scholar]
- 172.Haddix P L, Straley S C. Structure and regulation of the Yersinia pestis yscBCDEF operon. J Bacteriol. 1992;174:4820–4828. doi: 10.1128/jb.174.14.4820-4828.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Håkansson S, Bergman T, Vanooteghem J-C, Cornelis G, Wolf-Watz H. YopB and YopD constitute a novel class of Yersinia Yop proteins. Infect Immun. 1993;61:71–80. doi: 10.1128/iai.61.1.71-80.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Håkansson S, Galyov E E, Rosqvist R, Wolf-Watz H. The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane. Mol Microbiol. 1996;20:593–603. doi: 10.1046/j.1365-2958.1996.5251051.x. [DOI] [PubMed] [Google Scholar]
- 175.Håkansson S, Schesser K, Persson C, Galyov E E, Rosqvist R, Homble F, Wolf-Watz H. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell plasma membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 1996;15:5812–5823. [PMC free article] [PubMed] [Google Scholar]
- 176.Hale T L, Oaks E V, Formal S B. Identification and antigenic characterization of virulence-associated, plasmid-coded proteins of Shigella spp. and enteroinvasive Escherichia coli. Infect Immun. 1985;50:620–629. doi: 10.1128/iai.50.3.620-629.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Hale T L, Sansonetti P J, Schad P A, Austin S, Formal S B. Characterization of virulence plasmids and plasmid-associated outer membrane proteins in Shigella flexneri, Shigella sonnei, and Escherichia coli. Infect Immun. 1983;40:340–350. doi: 10.1128/iai.40.1.340-350.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hanks S K, Quinn A M, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 179.Hanski C, Kutschka U, Schmoranzer H P, Naumann M, Stallmach A, Hahn H, Menge H, Riecken E O. Immunohistochemical and electron microscopic study of interaction of Yersinia enterocolitica serotype O8 with intestinal mucosa during experimental enteritis. Infect Immun. 1989;57:673–678. doi: 10.1128/iai.57.3.673-678.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Hanski C, Naumann M, Grützkau A, Pluschke G, Friedrich B, Hahn H, Riecken E O. Humoral and cellular defense against intestinal murine infection with Yersinia enterocolitica. Infect Immun. 1991;59:1106–1111. doi: 10.1128/iai.59.3.1106-1111.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hardie K R, Lory S, Pugsley A P. Insertion of an outer membrane protein in Escherichia coli requires a chaperone-like protein. EMBO J. 1996;15:978–988. [PMC free article] [PubMed] [Google Scholar]
- 182.Hardie K R, Seydel A, Guilvout I, Pugsley A P. The secretin-specific, chaperone-like protein of the general secretory pathway: separation of proteolytic protection and piloting functions. Mol Microbiol. 1996;22:967–976. doi: 10.1046/j.1365-2958.1996.01539.x. [DOI] [PubMed] [Google Scholar]
- 183.Hardt W D, Galan J E. A secreted Salmonella protein with homology to an avirulence determinant of plant pathogenic bacteria. Proc Natl Acad Sci USA. 1997;94:9887–9892. doi: 10.1073/pnas.94.18.9887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Harris J R, Wachmuth I K, Davies B R, Cohen M L. High molecular weight plasmid correlates with Escherichia coli invasiveness. Infect Immun. 1982;37:1295–1298. doi: 10.1128/iai.37.3.1295-1298.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Harrison J A, Pickard D, Higgins C F, Khan A, Chatfield S N, Ali T, Dorman C J, Hormaeche C E, Dougan G. Role of hns in the virulence phenotype of pathogenic salmonellae. Mol Microbiol. 1994;13:133–140. doi: 10.1111/j.1365-2958.1994.tb00408.x. [DOI] [PubMed] [Google Scholar]
- 186.Hartland E L, Green S P, Phillips W A, Robins-Browne R M. Essential role of YopD in inhibition of the respiratory burst of macrophages by Yersinia enterocolitica. Infect Immun. 1994;62:4445–4453. doi: 10.1128/iai.62.10.4445-4453.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hartman A B, Venkatesan M, Oaks E V, Buysse J M. Sequence and molecular characterization of a multicopy invasion plasmid antigen gene, ipaH, of Shigella flexneri. J Bacteriol. 1990;172:1905–1915. doi: 10.1128/jb.172.4.1905-1915.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.He S Y, Huang H C, Collmer A. Pseudomonas syringae pv. syringae harpinPss: a protein that is secreted via the hrp pathway and elicits the hypersensitive response in plants. Cell. 1993;73:1255–1266. doi: 10.1016/0092-8674(93)90354-s. [DOI] [PubMed] [Google Scholar]
- 189.He X-T, Krishnan H B, Pueppke S G. Genes controlling cultivar-specific nodulation of soybean by Rhizobium fredii are homologous genes encoding type III secretion systems in other bacteria. 1997. GenBank accession no. L12251. [Google Scholar]
- 190.Heesemann J, Algermissen B, Laufs R. Genetically manipulated virulence of Yersinia enterocolitica. Infect Immun. 1984;46:105–110. doi: 10.1128/iai.46.1.105-110.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Heesemann J, Gross U, Schmidt N, Laufs R. Immunochemical analysis of plasmid-encoded proteins released by enteropathogenic Yersinia sp. grown in calcium-deficient media. Infect Immun. 1986;54:561–567. doi: 10.1128/iai.54.2.561-567.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Heesemann J, Grüter L. Genetic evidence that the outer membrane protein YOP1 of Yersinia enterocolitica mediates adherence and phagocytosis resistance to human epithelial cells. FEMS Microb Lett. 1987;40:37–41. [Google Scholar]
- 193.Heesemann J, Keller C, Morawa R, Schmidt N, Siemens H J, Laufs R. Plasmids of human strains of Yersinia enterocolitica: molecular relatedness and possible importance for pathogenesis. J Infect Dis. 1983;147:107–115. doi: 10.1093/infdis/147.1.107. [DOI] [PubMed] [Google Scholar]
- 193a.Hensel, M. Personal communication.
- 194.Hensel M, Shea J E, Baumler A J, Gleeson C, Blattner F, Holden D W. Analysis of the boundaries of Salmonella pathogenicity island 2 and the corresponding chromosomal region of Escherichia coli K-12. J Bacteriol. 1997;179:1105–1111. doi: 10.1128/jb.179.4.1105-1111.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 196.Hensel M, Shea J E, Raupach B, Monack D, Falkow S, Gleeson C, Kubo T, Holden D W. Functional analysis of ssaJ and ssaK/U operon, 13 genes encoding components of the type III secretion apparatus of Salmonella pathogenicity island 2. Mol Microbiol. 1997;24:155–167. doi: 10.1046/j.1365-2958.1997.3271699.x. [DOI] [PubMed] [Google Scholar]
- 197.Hermant D, Ménard R, Arricau N, Parsot C, Popoff M Y. Functional conservation of the Salmonella and Shigella effectors of entry into epithelial cells. Mol Microbiol. 1995;17:781–789. doi: 10.1111/j.1365-2958.1995.mmi_17040781.x. [DOI] [PubMed] [Google Scholar]
- 198.Higgins C F, Dorman C J, Stirling D A, Waddell L, Booth I R, May G, Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988;52:569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- 199.Higgins C F, Hinton J C, Hulton C S, Owen-Hughes T, Pavitt G D, Seirafi A. Protein H1: a role for chromatin structure in the regulation of bacterial gene expression and virulence? Mol Microbiol. 1990;4:2007–2012. doi: 10.1111/j.1365-2958.1990.tb00559.x. [DOI] [PubMed] [Google Scholar]
- 200.High N, Mounier J, Prévost M-C, Sansonetti P J. IpaB of Shigella flexneri causes entry into epithelial cells and escape from the phagocytic vacuole. EMBO J. 1992;11:1991–1992. doi: 10.1002/j.1460-2075.1992.tb05253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Higushi K, Kupferberg L L, Smith J L. Studies on the nutrition and physiology of Pasteurella pestis. III. Effects of calcium ions on the growth of virulent and avirulent strains of Pasteurella pestis. J Bacteriol. 1959;77:317–321. doi: 10.1128/jb.77.3.317-321.1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hirano T, Yamaguchi S, Oosawa K, Aizawa S. Roles of FliK and FlhB in determination of flagellar hook length in Salmonella typhimurium. J Bacteriol. 1994;176:5439–5449. doi: 10.1128/jb.176.17.5439-5449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hobbs M, Mattick J S. Common components in the assembly of type 4 fimbriae, DNA transfer systems, filamentous phage and protein-secretion apparatus: a general system for the formation of surface-associated protein complexes. Mol Microbiol. 1993;10:233–243. doi: 10.1111/j.1365-2958.1993.tb01949.x. [DOI] [PubMed] [Google Scholar]
- 204.Hoe N P, Minion C, Goguen J D. Temperature sensing in Yersinia pestis: regulation of yopE transcription by lcrF. J Bacteriol. 1992;174:4275–4286. doi: 10.1128/jb.174.13.4275-4286.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Holmström A, Petterson J, Rosqvist R, Håkansson S, Tafazoli F, Fällman M, Magnusson K E, Wolf-Watz H, Forsberg Å. YopK of Yersinia pseudotuberculosis controls translocation of Yop effectors across the eukaryotic cell membrane. Mol Microbiol. 1997;24:73–91. doi: 10.1046/j.1365-2958.1997.3211681.x. [DOI] [PubMed] [Google Scholar]
- 206.Holmström A, Rosqvist R, Wolf-Watz H, Forsberg Å. Virulence plasmid-encoded YopK is essential for Yersinia pseudotuberculosis to cause systemic infection in mice. Infect Immun. 1995;63:2269–2276. doi: 10.1128/iai.63.6.2269-2276.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hook E W. Salmonella species (including typhoid fever) In: Mandell G L, Douglas R G Jr, Bennett J E, editors. Principles and practice and infectious diseases. 3rd ed. New York, N.Y: Churchill Livingstone, Inc.; 1990. pp. 1700–1716. [Google Scholar]
- 208.Hovey A K, Frank D W. Analyses of the DNA-binding and transcriptional activation properties of ExsA, the transcriptional activator of the Pseudomonas aeruginosa exoenzyme S regulon. J Bacteriol. 1995;177:4427–4436. doi: 10.1128/jb.177.15.4427-4436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hromockyj A E, Maurelli A T. Identification of Shigella invasion genes by isolation of temperature-regulated inv::lacZ operon fusions. Infect Immun. 1989;57:2963–2970. doi: 10.1128/iai.57.10.2963-2970.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hromockyj A E, Tucker S C, Maurelli A T. Temperature regulation of Shigella virulence: identification of the repressor gene virR, an analogue of hns, and partial complementation by tyrosyl transfer RNA (tRNA1(Tyr)) Mol Microbiol. 1992;6:2113–2124. doi: 10.1111/j.1365-2958.1992.tb01385.x. [DOI] [PubMed] [Google Scholar]
- 211.Hsia R-C, Pannekeok Y, Ingerowski E, Bavoil P M. Type III secretion genes identify a putative virulence locus of Chlamydia. Mol Microbiol. 1997;25:351–359. doi: 10.1046/j.1365-2958.1997.4701834.x. [DOI] [PubMed] [Google Scholar]
- 212.Huang H-C, He S Y, Bauer D W, Collmer A. The Pseudomonas syringae pv. syringae 61 hrpH product, an envelope protein required for elicitation of the hypersensitive response in plants. J Bacteriol. 1992;174:6878–6885. doi: 10.1128/jb.174.21.6878-6885.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Huang H-C, Lin R-H, Chang C-J, Collmer A, Deng W-L. The complete hrp gene cluster of Pseudomonas syringae pv. syringae 61 includes two blocks of genes required for harpinPss secretion that are arranged colinearly with Yersinia ysc homologs. Mol Plant-Microbe Interact. 1995;8:733–746. doi: 10.1094/mpmi-8-0733. [DOI] [PubMed] [Google Scholar]
- 214.Huang H-C, Schuurink R, Denny T P, Atkinson M M, Baker C J, Yucel I, Hutcheson S W, Collmer A. Molecular cloning of a Pseudomonas syringae pv. syringae gene cluster that enables Pseudomonas fluorescens to elicit the hypersensitive response in tobacco plants. J Bacteriol. 1988;170:4748–4756. doi: 10.1128/jb.170.10.4748-4756.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Huang H-C, Xiao Y, Lin R-H, Lu Y, Hutcheson S W, Collmer A. Characterization of the Pseudomonas syringae pv. syringae 61 hrpJ and hrpI genes: homology of HrpI to a superfamily of proteins associated with protein translocation. Mol Plant-Microbe Interact. 1993;6:515–520. doi: 10.1094/mpmi-6-515. [DOI] [PubMed] [Google Scholar]
- 216.Hueck C J, Hantman M J, Bajaj V, Johnston C, Lee C A, Miller S I. Salmonella typhimurium secreted invasion determinants are homologous to Shigella Ipa proteins. Mol Microbiol. 1995;18:479–490. doi: 10.1111/j.1365-2958.1995.mmi_18030479.x. [DOI] [PubMed] [Google Scholar]
- 217.Hughes K T, Gillen K L, Semon M J, Karlinsey J E. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 218.Hulton C S, Seirafi A, Hinton J C, Sidebotham J M, Waddell L, Pavitt G D, Owen-Hughes T, Spassky A, Buc H, Higgins C F. Histone-like protein H1 (H-NS), DNA supercoiling, and gene expression in bacteria. Cell. 1990;63:631–642. doi: 10.1016/0092-8674(90)90458-q. [DOI] [PubMed] [Google Scholar]
- 219.Hwang I, Lim S M, Shaw P D. Cloning and characterization of pathogenicity genes from Xanthomonas campestris pv. glycines. J Bacteriol. 1992;174:1923–1931. doi: 10.1128/jb.174.6.1923-1931.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Hynes R O. Integrins: versatility, modulation, and signaling in cell adhesion. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 221.Iglewski B H, Sadoff J, Bjorn M J, Maxwell E S. Pseudomonas aeruginosa exoenzyme S: an adenosine diphosphate ribosyltransferase distinct from toxin A. Proc Natl Acad Sci USA. 1978;75:3211–3215. doi: 10.1073/pnas.75.7.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Innes R W, Bent A F, Kunkel B N, Bisgrove S R, Staskawicz B J. Molecular analysis of avirulence gene avrRpt2 and identification of a putative regulatory sequence common to all known Pseudomonas syringae avirulence genes. J Bacteriol. 1993;175:4859–4869. doi: 10.1128/jb.175.15.4859-4869.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Jarvis K G, Girón J A, Jerse A E, McDaniel T K, Donnenberg M S, Kaper J B. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc Natl Acad Sci USA. 1995;92:7996–8000. doi: 10.1073/pnas.92.17.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223a.Jepson, M. A. Personal communication.
- 224.Jerse A E, Gicquelais K G, Kaper J B. Plasmid and chromosomal elements involved in the pathogenesis of attaching and effacing Escherichia coli. Infect Immun. 1991;59:3869–3875. doi: 10.1128/iai.59.11.3869-3875.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Jerse A E, Yu J, Tall B D, Kaper J B. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc Natl Acad Sci USA. 1990;87:7839–7843. doi: 10.1073/pnas.87.20.7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Johnston C, Pegues D A, Hueck C J, Lee C A, Miller S I. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol. 1996;22:715–727. doi: 10.1046/j.1365-2958.1996.d01-1719.x. [DOI] [PubMed] [Google Scholar]
- 227.Jones B D, Falkow S. Identification and characterization of a Salmonella typhimurium oxygen-regulated gene required for bacterial internalization. Infect Immun. 1994;62:3745–3752. doi: 10.1128/iai.62.9.3745-3752.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Jones B D, Ghori N, Falkow S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer’s patches. J Exp Med. 1994;180:15–23. doi: 10.1084/jem.180.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Jones C J, Homma M, Macnab R M. L-, P-, and M-ring proteins of the flagellar basal body of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol. 1989;171:3890–3900. doi: 10.1128/jb.171.7.3890-3900.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Jones C J, Macnab R M. Flagellar assembly in Salmonella typhimurium: analysis with temperature-sensitive mutants. J Bacteriol. 1990;172:1327–1339. doi: 10.1128/jb.172.3.1327-1339.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kang P J, Hauser A R, Apodaca G, Fleiszig S M, Wiener-Kronish J, Mostov K, Engel J N. Identification of Pseudomonas aeruginosa genes required for epithelial cell injury. Mol Microbiol. 1997;24:1249–1262. doi: 10.1046/j.1365-2958.1997.4311793.x. [DOI] [PubMed] [Google Scholar]
- 232.Kaniga K, Bossio J C, Galán J E. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol Microbiol. 1994;13:555–568. doi: 10.1111/j.1365-2958.1994.tb00450.x. [DOI] [PubMed] [Google Scholar]
- 233.Kaniga K, Trollinger D, Galán J E. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J Bacteriol. 1995;177:7078–7085. doi: 10.1128/jb.177.24.7078-7085.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Kaniga K, Tucker S, Trollinger D, Galán J E. Homologs of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J Bacteriol. 1995;177:3965–3971. doi: 10.1128/jb.177.14.3965-3971.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Kaniga K, Uraill J, Bliska J B, Galán J E. A secreted protein tyrosine phosphatase with modular effector domains in the bacterial pathogen Salmonella typhimurium. Mol Microbiol. 1996;21:633–641. doi: 10.1111/j.1365-2958.1996.tb02571.x. [DOI] [PubMed] [Google Scholar]
- 236.Kato J, Ito K, Nakamura A, Watanabe H. Cloning of regions required for contact hemolysis and entry into LLC-MK2 cells from Shigella sonnei form I plasmid: virF is a positive regulator gene for these phenotypes. Infect Immun. 1989;57:1391–1398. doi: 10.1128/iai.57.5.1391-1398.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Kazmierczak B I, Mielke D L, Russel M, Model P. pIV, a filamentous phage protein that mediates phage export across the bacterial cell envelope, forms a multimer. J Mol Biol. 1994;238:187–198. doi: 10.1006/jmbi.1994.1280. [DOI] [PubMed] [Google Scholar]
- 238.Kenny B, Abe A, Stein M, Finlay B B. Enteropathogenic Escherichia coli protein secretion is induced in response to conditions similar to those in the gastrointestinal tract. Infect Immun. 1997;65:2606–2612. doi: 10.1128/iai.65.7.2606-2612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Kenny B, DeVinney R, Stein M, Reinscheid D J, Frey E A, Finlay B B. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 240.Kenny B, Finlay B B. Protein secretion by enteropathogenic Escherichia coli is essential for transducing signals to epithelial cells. Proc Natl Acad Sci USA. 1995;92:7991–7995. doi: 10.1073/pnas.92.17.7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Kenny B, Finlay B B. Intimin-dependent binding of enteropathogenic Escherichia coli to host cells triggers novel signaling events, including tyrosine phosphorylation of phospholipase C-γ1. Infect Immun. 1997;65:2528–2536. doi: 10.1128/iai.65.7.2528-2536.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Kenny B, Lai L-C, Finlay B B, Donnenberg M S. EspA, a protein secreted by enteropathogenic Escherichia coli, is required to induce signals in epithelial cells. Mol Microbiol. 1996;20:313–323. doi: 10.1111/j.1365-2958.1996.tb02619.x. [DOI] [PubMed] [Google Scholar]
- 243.Kihara M, Homma M, Kutsukake K, Macnab R M. Flagellar switch of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol. 1989;171:3247–3257. doi: 10.1128/jb.171.6.3247-3257.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Kim J F, Wei Z-M, Beer S V. The hrpA and hrpC operons of Erwinia amylovora encode components of a type III pathway that secretes harpin. J Bacteriol. 1997;179:1690–1697. doi: 10.1128/jb.179.5.1690-1697.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Klement Z. Hypersensitivity. In: Mount M S, Lacy G H, editors. Phythopathogenic prokaryotes. New York, N.Y: Academic Press, Inc.; 1982. pp. 149–177. [Google Scholar]
- 246.Knapp S, Hacker J, Jarchau T, Goebel W. Large, unstable inserts in the chromosome affect virulence properties of uropathogenic Escherichia coli O6 strain 536. J Bacteriol. 1986;168:22–30. doi: 10.1128/jb.168.1.22-30.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Knoop V, Staskawicz B, Bonas U. Expression of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria is not under the control of hrp genes and is independent of plant factors. J Bacteriol. 1991;173:7142–7150. doi: 10.1128/jb.173.22.7142-7150.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Knutton S, Baldwin T, Williams P H, McNeish A S. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun. 1989;57:1290–1298. doi: 10.1128/iai.57.4.1290-1298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Kohbata S, Yokobata H, Yabuuchi E. Cytopathogenic effect of Salmonella typhi GIFU 10007 on M cells of murine ileal Peyer’s patches in ligated ileal loops: an ultrastructural study. Microbiol Immunol. 1986;30:1225–1237. doi: 10.1111/j.1348-0421.1986.tb03055.x. [DOI] [PubMed] [Google Scholar]
- 250.Kornacker M G, Newton A. Information essential for cell-cycle-dependent secretion of the 591-residue Caulobacter hook protein is confined to a 21-amino-acid sequence near the N-terminus. Mol Microbiol. 1994;14:73–85. doi: 10.1111/j.1365-2958.1994.tb01268.x. [DOI] [PubMed] [Google Scholar]
- 251.Kubori T, Shimamoto N, Yamaguchi S, Namba K, Aizawa S-I. Morphological pathway of flagellar assembly in Salmonella typhimurium. J Mol Biol. 1992;226:433–446. doi: 10.1016/0022-2836(92)90958-m. [DOI] [PubMed] [Google Scholar]
- 252.Kubori T, Yamaguchi S, Aizawa S. Assembly of the switch complex onto the MS ring complex of Salmonella typhimurium does not require any other flagellar proteins. J Bacteriol. 1997;179:813–817. doi: 10.1128/jb.179.3.813-817.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Kulich S M, Yahr T L, Mende-Mueller L M, Barbieri J T, Frank D W. Cloning the structural gene for the 49-kDa form of exoenzyme S (exoS) from Pseudomonas aeruginosa strain 388. J Biol Chem. 1994;269:10431–10437. [PubMed] [Google Scholar]
- 254.Kuo S C, Koshland D E., Jr Sequence of the flaA (cheC) locus of Escherichia coli and discovery of a new gene. J Bacteriol. 1986;166:1007–1012. doi: 10.1128/jb.166.3.1007-1012.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Kupferberg L L, Higuchi K. Role of calcium ions in the stimulation of growth of virulent strains of Pasteurella pestis. J Bacteriol. 1958;76:120–121. doi: 10.1128/jb.76.1.120-121.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Kutusake K, Minamino T, Yokoseki T. Isolation and characterization of FliK-independent flagellation mutants from Salmonella typhimurium. J Bacteriol. 1994;176:7625–7629. doi: 10.1128/jb.176.24.7625-7629.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Kuwajima G, Kawagishi I, Homma M, Asaka J, Kondo E, Macnab R M. Export of N-terminal fragment of Escherichia coli flagellin by a flagellum-specific pathway. Proc Natl Acad Sci USA. 1989;86:4953–4957. doi: 10.1073/pnas.86.13.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.LaBrec E H, Schneider H, Magnani T J, Formal S B. Epithelial cell penetration as an essential step in the pathogenesis of bacillary dysentery. J Bacteriol. 1964;88:1503–1518. doi: 10.1128/jb.88.5.1503-1518.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Lai L C, Wainwright L A, Stone K D, Donnenberg M S. A third secreted protein that is encoded by the enteropathogenic Escherichia coli pathogenicity island is required for transduction of signals and for attaching and effacing activities in host cells. Infect Immun. 1997;65:2211–2217. doi: 10.1128/iai.65.6.2211-2217.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Lamb C J. Plant disease resistance genes in signal perception and transduction. Cell. 1994;76:419–422. doi: 10.1016/0092-8674(94)90106-6. [DOI] [PubMed] [Google Scholar]
- 261.Lambert de Rouvroit C, Sluiters C, Cornelis G R. Role of the transcriptional activator, VirF in the expression of the pVY plasmid genes of Yersinia enterocolitica. Mol Microbiol. 1992;6:395–409. [PubMed] [Google Scholar]
- 262.Lawton W D, Erdman R L, Surgalla M J. Biosynthesis and purification of V and W antigen in Pasteurella pestis. J Immunol. 1963;91:179–184. doi: 10.21236/ad0299868. [DOI] [PubMed] [Google Scholar]
- 263.Leary S E C, Williamson E D, Griffin K F, Russel P, Eley S M, Titball R W. Active immunization with recombinant V antigen from Yersinia pestis protects mice against plague. Infect Immun. 1995;63:2854–2858. doi: 10.1128/iai.63.8.2854-2858.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Lee C A. Type III secretion systems: machines to deliver bacterial proteins into eukaryotic cells? Trends Microbiol. 1997;5:148–156. doi: 10.1016/S0966-842X(97)01029-9. [DOI] [PubMed] [Google Scholar]
- 265.Lee C A, Falkow S. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc Natl Acad Sci USA. 1990;87:4304–4308. doi: 10.1073/pnas.87.11.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Lee C A, Jones B D, Falkow S. Identification of a Salmonella typhimurium invasion locus by selection for hyperinvasive mutants. Proc Natl Acad Sci USA. 1992;89:1847–1851. doi: 10.1073/pnas.89.5.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Leister R T, Ausubel F M, Katagiri F. Molecular recognition of pathogen attack occurs inside of plant cells in plant disease resistance specified by the Arabidopsis genes RPS2 and RPM1. Proc Natl Acad Sci USA. 1996;93:15497–11502. doi: 10.1073/pnas.93.26.15497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Leung K Y, Reisner B S, Straley S C. YopM inhibits platelet aggregation and is necessary for virulence of Yersinia pestis in mice. Infect Immun. 1990;58:3262–3271. doi: 10.1128/iai.58.10.3262-3271.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Leung K Y, Straley S C. The yopM gene of Yersinia pestis encodes a released protein having homology to the human platelet surface protein GPIbα. J Bacteriol. 1989;171:4623–4631. doi: 10.1128/jb.171.9.4623-4632.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Levine M M, Edelman R. Enteropathogenic Escherichia coli of classical serotypes associated with infant diarrhea: epidemiology and pathogenesis. Epidemiol Rev. 1984;6:31–51. doi: 10.1093/oxfordjournals.epirev.a036274. [DOI] [PubMed] [Google Scholar]
- 271.Li J, Ochman H, Groisman E A, Boyd E F, Solomon F, Nelson K, Selander R K. Relationship between evolutionary rate and cellular location among the Inv/Spa invasion proteins of Salmonella enterica. Proc Natl Acad Sci USA. 1995;92:7252–7256. doi: 10.1073/pnas.92.16.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Li T-H, Benson S A, Hutcheson S W. Phenotypic expression of the Pseudomonas syringae pv. syringae 61 hrp/hrm gene cluster in Escherichia coli MC4100 requires a functional porin. J Bacteriol. 1992;174:1742–1749. doi: 10.1128/jb.174.6.1742-1749.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Libby S J, Goebel W, Ludwig A, Buchmeier N, Bowe F, Fang F C, Guiney D G, Songer J G, Heffron F. A cytolysin encoded by Salmonella is required for survival within macrophages. Proc Natl Acad Sci USA. 1994;91:489–493. doi: 10.1073/pnas.91.2.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Lidell M C, Hutcheson S W. Characterization of the hrpJ and hrpU operons of Pseudomonas syringae pv. syringae Pss61: similarity with components of enteric bacteria involved in flagellar biogenesis and demonstration of their role in HairpinPSS secretion. Mol Plant-Microbe Interact. 1994;7:488–497. doi: 10.1094/mpmi-7-0488. [DOI] [PubMed] [Google Scholar]
- 275.Lindberg A A, Pál T. Strategies for development of potential candidate Shigella vaccines. Vaccine. 1993;11:168–179. doi: 10.1016/0264-410x(93)90014-o. [DOI] [PubMed] [Google Scholar]
- 276.Lindgren P B, Peet R C, Panopoulos N J H. Gene cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity on bean plants and hypersensitive response on nonhost plants. J Bacteriol. 1986;168:512–522. doi: 10.1128/jb.168.2.512-522.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Lindsay W P, Lamb C J, Dixon R A. Microbial recognition and activation of plant defense systems. Trends Microbiol. 1993;1:181–187. doi: 10.1016/0966-842x(93)90088-9. [DOI] [PubMed] [Google Scholar]
- 278.Lonetto M A, Brown K L, Rudd K E, Buttner M J. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase sigma factors involved in the regulation of extracytoplasmic functions. Proc Natl Acad Sci USA. 1994;91:7573–7577. doi: 10.1073/pnas.91.16.7573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Macnab R M. Genetics and biogenesis of bacterial flagella. Annu Rev Genet. 1992;26:131–158. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 280.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Riley M, Reznikoff W S, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 281.Majeed M, Kihlström E. Mobilization of F-actin and clathrin during redistribution of Chlamydia trachomatis to an intracellular site in eukaryotic cells. Infect Immun. 1991;59:4465–4472. doi: 10.1128/iai.59.12.4465-4472.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Malakooti J, Ely B, Matsumura P. Molecular characterization, nucleotide sequence, and expression of the fliO, fliP, fliQ, and fliR genes of Escherichia coli. J Bacteriol. 1994;176:189–197. doi: 10.1128/jb.176.1.189-197.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Malakooti J, Komeda Y, Matsumura P. DNA sequence analysis, gene product identification, and localization of flagellar motor components of Escherichia coli. J Bacteriol. 1989;171:2728–2734. doi: 10.1128/jb.171.5.2728-2734.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Mandic-Mulec I, Weiss J, Zychlinsky A. Shigella flexneri is trapped in polymorphonuclear leukocyte vacuoles and efficiently killed. Infect Immun. 1997;65:110–115. doi: 10.1128/iai.65.1.110-115.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Manjarrez-Hernandez H A, Baldwin T J, Aitken A, Knutton S, Williams P H. Intestinal epithelial cell protein phosphorylation in enteropathogenic Escherichia coli diarrhoea. Lancet. 1992;339:521–523. doi: 10.1016/0140-6736(92)90340-9. [DOI] [PubMed] [Google Scholar]
- 286.Marquart M E, Picking W L, Picking W D. Soluble invasion plasmid antigen C (IpaC) from Shigella flexneri elicits epithelial cell responses related to pathogen invasion. Infect Immun. 1996;64:4182–4187. doi: 10.1128/iai.64.10.4182-4187.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Martin G B, Brommonschenkel S H, Chunwongse J, Frary A, Ganal M W, Spivey R, Wu T, Earle E D, Tanksley S D. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 288.Martin P R, Hobbs M, Free P D, Jeske Y, Mattick J S. Characterization of pilQ, a new gene required for the biogenesis of type 4 fimbriae in Pseudomonas aeruginosa. Mol Microbiol. 1993;9:857–868. doi: 10.1111/j.1365-2958.1993.tb01744.x. [DOI] [PubMed] [Google Scholar]
- 289.Marykwas D L, Schmidt S A, Berg H C. Interacting components of the flagellar motor of Escherichia coli revealed by the two-hybrid system in yeast. J Mol Biol. 1996;256:564–576. doi: 10.1006/jmbi.1996.0109. [DOI] [PubMed] [Google Scholar]
- 290.Maurelli A T, Baudry B, d’Hauteville H, Hale T, Sansonetti P J. Cloning of plasmid DNA sequences involved in invasion of HeLa cells by Shigella flexneri. Infect Immun. 1985;49:164–171. doi: 10.1128/iai.49.1.164-171.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Maurelli A T, Hromockyj A E, Bernardini M L. Environmental regulation of Shigella virulence. Curr Top Microbiol Immunol. 1992;180:95–116. doi: 10.1007/978-3-642-77238-2_5. [DOI] [PubMed] [Google Scholar]
- 292.Maurelli A T, Sansonetti P J. Genetic determinants of Shigella pathogenicity. Annu Rev Microbiol. 1988;42:127–150. doi: 10.1146/annurev.mi.42.100188.001015. [DOI] [PubMed] [Google Scholar]
- 293.Maurelli A T, Sansonetti P J. Identification of a chromosomal gene controlling temperature-regulated expression of Shigella virulence. Proc Natl Acad Sci USA. 1988;85:2820–2824. doi: 10.1073/pnas.85.8.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Mazza G, Karu A E, Kingsbury D T. Immune response to plasmid- and chromosome-encoded Yersinia antigens. Infect Immun. 1985;48:676–685. doi: 10.1128/iai.48.3.676-685.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.McCormick B A, Miller S I, Delp-Archer C, Madara J T. Transepithelial signaling to neutrophils by Salmonella: a novel virulence mechanism for gastroenteritis. Infect Immun. 1995;63:2302–2309. doi: 10.1128/iai.63.6.2302-2309.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.McDaniel T K, Jarvis K G, Donnenberg M S, Kaper J B. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc Natl Acad Sci USA. 1995;92:1664–1668. doi: 10.1073/pnas.92.5.1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.McDaniel T K, Kaper J B. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol Microbiol. 1997;23:399–407. doi: 10.1046/j.1365-2958.1997.2311591.x. [DOI] [PubMed] [Google Scholar]
- 298.McMorrow I, Bastos R, Horton H, Burke B. Sequence analysis of a cDNA encoding a human nuclear pore complex protein, hnup153. Biochim Biophys Acta. 1994;1217:219–223. doi: 10.1016/0167-4781(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 299.Meinhardt L W, Krishnan H B, Balatti P A, Pueppke S G. Molecular cloning and characterization of a sym plasmid locus that regulates cultivar-specific nodulation of soybean by Rhizobium fredii USDA257. Mol Microbiol. 1993;9:17–29. doi: 10.1111/j.1365-2958.1993.tb01665.x. [DOI] [PubMed] [Google Scholar]
- 300.Ménard R, Dehio C, Sansonetti P J. Bacterial entry into epithelial cells: the paradigm of Shigella. Trends Microbiol. 1996;4:220–226. doi: 10.1016/0966-842X(96)10039-1. [DOI] [PubMed] [Google Scholar]
- 301.Ménard R, Prévost M-C, Gounon P, Sansonetti P, Dehio C. The secreted Ipa complex of Shigella flexneri promotes entry into mammalian cells. Proc Natl Acad Sci USA. 1996;93:1254–1258. doi: 10.1073/pnas.93.3.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Ménard R, Sansonetti P, Parsot C. The secretion of the Shigella flexneri Ipa invasins is activated by epithelial cells and controlled by IpaB and IpaD. EMBO J. 1994;13:5293–5302. doi: 10.1002/j.1460-2075.1994.tb06863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Ménard R, Sansonetti P, Parsot C, Vasselon T. Extracellular association and cytoplasmic partitioning of the IpaB and IpaC invasins of S. flexneri. Cell. 1994;79:515–525. doi: 10.1016/0092-8674(94)90260-7. [DOI] [PubMed] [Google Scholar]
- 304.Ménard R, Sansonetti P J, Parsot C. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993;175:5899–5906. doi: 10.1128/jb.175.18.5899-5906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Michiels T, Cornelis G. Nucleotide sequence and transcription analysis of yop51 from Yersinia enterocolitica W22703. Microb Pathog. 1988;5:449–459. doi: 10.1016/0882-4010(88)90006-x. [DOI] [PubMed] [Google Scholar]
- 306.Michiels T, Cornelis G R. Secretion of hybrid proteins by the Yersinia Yop export system. J Bacteriol. 1991;173:1677–1685. doi: 10.1128/jb.173.5.1677-1685.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Michiels T, Vanooteghem J-C, Lambert de Rouvroit C, China B, Gustin A, Boudry P, Cornelis G R. Analysis of virC, an operon involved in the secretion of Yop proteins by Yersinia enterocolitica. J Bacteriol. 1991;173:4994–5009. doi: 10.1128/jb.173.16.4994-5009.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Michiels T, Wattiau P, Brasseur R, Ruysschaert J-M, Cornelis G. Secretion of Yop proteins by yersiniae. Infect Immun. 1990;58:2840–2849. doi: 10.1128/iai.58.9.2840-2849.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308a.Miller, S. I. Personal communication.
- 309.Miller S I, Hohmann E L, Pegues D A. Salmonella (including Salmonella typhi) In: Mandell G L, Bennett J E, Raphael D, editors. Principles and practice of infectious diseases. Vol. 2. New York, N.Y: Churchill Livingstone, Inc.; 1995. pp. 2013–2033. [Google Scholar]
- 310.Miller S I, Kukral A M, Mekalanos J J. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci USA. 1989;86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Miller S I, Mekalanos J J. Constitutive expression of the PhoP regulon attenuates Salmonella virulence and survival within macrophages. J Bacteriol. 1990;172:2485–2490. doi: 10.1128/jb.172.5.2485-2490.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Mills D M, Bajaj V, Lee C A. A 40 kilobase chromosomal fragment encoding Salmonella typhimurium invasion genes is absent from the corresponding region of the Escherichia coli K-12 chromosome. Mol Microbiol. 1995;15:749–759. doi: 10.1111/j.1365-2958.1995.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 313.Mills S D, Boland A, Sory M P, van der Smissen P, Kerbourch C, Finlay B B, Cornelis G R. Yersinia enterocolitica induces apoptosis in macrophages by a process requiring functional type III secretion and translocation mechanisms and involving YopP, presumably acting as an effector protein. Proc Natl Acad Sci USA. 1997;94:12638–12643. doi: 10.1073/pnas.94.23.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Minamino T, Iino T, Kutsukake K. Molecular characterization of the Salmonella typhimurium flhB operon and its protein products. J Bacteriol. 1994;176:7630–7637. doi: 10.1128/jb.176.24.7630-7637.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Monack D M, Raupach B, Hromockyj A E, Falkow S. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci USA. 1996;93:9833–9838. doi: 10.1073/pnas.93.18.9833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Moon H W, Whipp S C, Argenzio R A, Levine M M, Giannella R A. Attaching and effacing activities of rabbit and human enteropathogenic Escherichia coli in pig and rabbit intestines. Infect Immun. 1983;41:1340–1351. doi: 10.1128/iai.41.3.1340-1351.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Motin V L, Nakajima R, Smirnov G B, Brubaker R R. Passive immunity to yersinia mediated by anti-recombinant V antigen and protein A-V antigen fusion peptide. Infect Immun. 1994;62:4192–4201. doi: 10.1128/iai.62.10.4192-4201.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Motin V L, Nedialkov Y A, Brubaker R R. V antigen-polyhistidine fusion peptide: binding to LcrH and active immunity against plague. Infect Immun. 1996;64:4313–4318. doi: 10.1128/iai.64.10.4313-4318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Moulder J W. Interaction of chlamydiae and host cells in vitro. Microbiol Rev. 1991;55:143–190. doi: 10.1128/mr.55.1.143-190.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Mounier J, Bahrani F K, Sansonetti P J. Secretion of Shigella flexneri Ipa invasins on contact with epithelial cells and subsequent entry of the bacterium into cells are growth stage dependent. Infect Immun. 1997;65:774–782. doi: 10.1128/iai.65.2.774-782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Mulder B, Michiels T, Simonet M, Sory M-P, Cornelis G. Identification of additional virulence determinants on the pYV plasmid of Yersinia enterocolitica W227. Infect Immun. 1989;57:2534–2541. doi: 10.1128/iai.57.8.2534-2541.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Mullan, P. B., A. V. Gautier, M. W. Wood, M. H. Edwards, B. V. Jones, and E. E. Galyov. Sip invasins of Salmonella dublin are involved in macrophage cytotoxicity and mouse virulence. GenBank accession no. U66877.
- 323.Murphy C K, Beckwith J. Export of proteins to the cell envelope in Escherichia coli. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Riley M, Reznikoff W S, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 967–978. [Google Scholar]
- 324.Nakajima R, Brubaker R R. Association between virulence of Yersinia pestis and suppression of gamma interferon and tumor necrosis factor alpha. Infect Immun. 1993;61:23–31. doi: 10.1128/iai.61.1.23-31.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Nakajima R, Motin V L, Brubaker R R. Suppression of cytokines in mice by protein A-V antigen fusion peptide and restoration of synthesis by active immunization. Infect Immun. 1995;63:3021–3029. doi: 10.1128/iai.63.8.3021-3029.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Nakayama S, Watanabe H. Involvement of cpxA, a sensor of a two-component regulatory system, in the pH-dependent regulation of expression of Shigella sonnei virF gene. J Bacteriol. 1995;177:5062–5069. doi: 10.1128/jb.177.17.5062-5069.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Nicas T I, Bradley J, Lochner J E, Iglewski B H. The role of exoenzyme S in infections with Pseudomonas aeruginosa. J Infect Dis. 1985;152:716–721. doi: 10.1093/infdis/152.4.716. [DOI] [PubMed] [Google Scholar]
- 328.Nicas T I, Iglewski B H. The contribution of exoproducts to virulence of Pseudomonas aeruginosa. Can J Microbiol. 1985;31:387–392. doi: 10.1139/m85-074. [DOI] [PubMed] [Google Scholar]
- 329.Nilles M L, Williams A W, Skrzypek E, Straley S C. Yersinia pestis LcrV forms a stable complex with LcrG and may have a secretion-related regulatory role in the low-Ca2+ response. J Bacteriol. 1997;179:1307–1316. doi: 10.1128/jb.179.4.1307-1316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Oaks E V, Hale T L, Formal S B. Serum immune response to Shigella protein antigens in rhesus monkeys and humans infected with Shigella spp. Infect Immun. 1986;53:57–63. doi: 10.1128/iai.53.1.57-63.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.O’Brien A D, Scher I, Formal S B. Effect of silica on the innate resistance of inbred mice to Salmonella typhimurium infection. Infect Immun. 1979;25:513–520. doi: 10.1128/iai.25.2.513-520.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Ochman H, Lawrence J G. Pholygenetics and the amelioration of bacterial genomes. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Riley M, Reznikoff W S, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 2627–2637. [Google Scholar]
- 333.Ochman H, Soncini F C, Solomon F, Groisman E A. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996;93:7800–7804. doi: 10.1073/pnas.93.15.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Pallen M, Dougan G, Frankel G. Coiled-coil domains in proteins secreted by type III secretion systems. Mol Microbiol. 1997;25:423–425. doi: 10.1046/j.1365-2958.1997.4901850.x. [DOI] [PubMed] [Google Scholar]
- 335.Pang T, Bhutta Z A, Finlay B B, Altwegg M. Typhoid fever and other salmonellosis: a continuing challenge. Trends Microbiol. 1995;3:253–255. doi: 10.1016/s0966-842x(00)88937-4. [DOI] [PubMed] [Google Scholar]
- 336.Parker J E, Coleman M J. Molecular intimacy between proteins specifying plant-pathogen recognition. Trends Biochem Sci. 1997;22:291–296. doi: 10.1016/s0968-0004(97)01089-x. [DOI] [PubMed] [Google Scholar]
- 337.Parsot C, Ménard R, Gounon P, Sansonetti P J. Enhanced secretion through the Shigella flexneri Mxi-Spa translocon leads to assembly of extracellular proteins into macromolecular structures. Mol Microbiol. 1995;16:291–300. doi: 10.1111/j.1365-2958.1995.tb02301.x. [DOI] [PubMed] [Google Scholar]
- 338.Parsot C, Sansonetti P J. Invasion and the pathogenesis of Shigella infections. Curr Top Microbiol Immunol. 1996;209:25–42. doi: 10.1007/978-3-642-85216-9_2. [DOI] [PubMed] [Google Scholar]
- 339.Pegues D A, Hantman M J, Behlau I, Miller S I. PhoP/PhoQ transcriptional repression of Salmonella typhimurium invasion genes: evidence for a role in protein secretion. Mol Microbiol. 1995;17:169–181. doi: 10.1111/j.1365-2958.1995.mmi_17010169.x. [DOI] [PubMed] [Google Scholar]
- 340.Penheiter K L, Mathur N, Giles D, Fahlen T, Jones B D. Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer’s patches. Mol Microbiol. 1997;24:697–709. doi: 10.1046/j.1365-2958.1997.3741745.x. [DOI] [PubMed] [Google Scholar]
- 341.Perdomo J J, Gounon P, Sansonetti P J. Polymorphonuclear leukocyte transmigration promotes invasion of colonic epithelial monolayer by Shigella flexneri. J Clin Invest. 1994;93:633–643. doi: 10.1172/JCI117015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Perdomo O J J, Cavaillon J M, Huerre M, Ohayon H, Gounon P, Sansonetti P J. Acute inflammation causes epithelial invasion and mucosal destruction in experimental shigellosis. J Exp Med. 1994;180:1307–1319. doi: 10.1084/jem.180.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Perry R D, Harmon P A, Bowmer W S, Straley S C. A low-Ca2+ response operon encodes the V antigen of Yersinia pestis. Infect Immun. 1986;54:428–434. doi: 10.1128/iai.54.2.428-434.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Persson C, Carballeira N, Wolf-Watz H, Fällman M. The PTPase YopH inhibits uptake of Yersinia, tyrosine phosphorylation of p130Cas and FAK, and the associated accumulation of these proteins in peripheral focal adhesions. EMBO J. 1997;16:2307–2318. doi: 10.1093/emboj/16.9.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Persson C, Nordfelth R, Holmström A, Håkansson S, Rosqvist R, Wolf-Watz H. Cell-surface-bound Yersinia translocate the protein tyrosine phosphatase YopH by a polarized mechanism into the target cell. Mol Microbiol. 1995;18:135–150. doi: 10.1111/j.1365-2958.1995.mmi_18010135.x. [DOI] [PubMed] [Google Scholar]
- 346.Petterson J, Nordfelth R, Dubinina E, Bergman T, Gustafsson M, Magnusson K E, Wolf-Watz H. Modulation of virulence factor expression by pathogen target cell contact. Science. 1996;273:1231–1233. doi: 10.1126/science.273.5279.1231. [DOI] [PubMed] [Google Scholar]
- 347.Petterson J, Nordfelth R, Wolf-Watz H. Contact with eukaryotic cells: a new signal triggering bacterial gene expression. Response to G. R. Cornelis. 1997. Trends Microbiol. 5:44–45. [DOI] [PubMed] [Google Scholar]
- 348.Pirhonen M U, Lidell M C, Rowley D L, Lee S W, Jin S, Liang Y, Silverstone S, Keen N T, Hutcheson S W. Phenotypic expression of Pseudomonas syringae avr genes in E. coli is linked to the activities of the hrp-encoded secretion system. Mol Plant-Microbe Interact. 1996;9:252–260. doi: 10.1094/mpmi-9-0252. [DOI] [PubMed] [Google Scholar]
- 349.Plano G V, Barve S S, Straley S C. LcrD, a membrane-bound regulator of the Yersinia pestis low-calcium response. J Bacteriol. 1991;173:7293–7303. doi: 10.1128/jb.173.22.7293-7303.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Plano G V, Straley S C. Multiple effects of lcrD mutations in Yersinia pestis. J Bacteriol. 1993;175:3536–3545. doi: 10.1128/jb.175.11.3536-3545.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Plano G V, Straley S C. Mutations in yscC, yscD, and yscG prevent high-level expression and secretion of V antigen and Yops in Yersinia pestis. J Bacteriol. 1995;177:3843–3854. doi: 10.1128/jb.177.13.3843-3854.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Pope L M, Reed K E, Payne S M. Increased protein secretion and adherence to HeLa cells by Shigella spp. following growth in the presence of bile salts. Infect Immun. 1995;63:3642–3648. doi: 10.1128/iai.63.9.3642-3648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352a.Popoff, M. Personal communication.
- 353.Porter M E, Dorman C J. A role for H-NS in the thermo-osmotic regulation of virulence gene expression in Shigella flexneri. J Bacteriol. 1994;176:4187–4191. doi: 10.1128/jb.176.13.4187-4191.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Portnoy D A, Blank H F, Kingsbury D T, Falkow S. Genetic analysis of essential plasmid determinants of pathogenicity in Yersinia pestis. J Infect Dis. 1983;148:297–304. doi: 10.1093/infdis/148.2.297. [DOI] [PubMed] [Google Scholar]
- 355.Portnoy D A, Moseley S L, Falkow S. Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect Immun. 1981;31:775–782. doi: 10.1128/iai.31.2.775-782.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Portnoy D A, Wolf-Watz H, Bölin I, Beeder A B, Falkow S. Characterization of common virulence plasmids in Yersinia species and their role in the expression of outer membrane proteins. Infect Immun. 1984;43:108–114. doi: 10.1128/iai.43.1.108-114.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Preston G, Huang H-C, He S Y, Collmer A. The HrpZ proteins of Pseudomonas syringae pvs. syringae, glycinea, and tomato are encoded by an operon containing Yersinia ysc homologs and elicit the hypersensitive response in tomato but not soybean. Mol Plant-Microbe Interact. 1995;8:717–732. doi: 10.1094/mpmi-8-0717. [DOI] [PubMed] [Google Scholar]
- 358.Price S B, Cowan C, Perry R D, Straley S C. The Yersinia pestis V antigen is a regulatory protein necessary for Ca2+-dependent growth and maximal expression of low-Ca2+ response virulence genes. J Bacteriol. 1991;173:2649–2657. doi: 10.1128/jb.173.8.2649-2657.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Price S B, Leung K Y, Barve S S, Straley S C. Molecular analysis of lcrGVH, the antigen operon of Yersinia pestis. J Bacteriol. 1989;171:5646–5653. doi: 10.1128/jb.171.10.5646-5653.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Price S B, Straley S C. lcrH, a gene necessary for virulence of Yersinia pestis and for the normal response of Y. pestis to ATP and calcium. Infect Immun. 1989;57:1491–1498. doi: 10.1128/iai.57.5.1491-1498.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Puente J L, Bieber D, Ramer S W, Murray W, Schoolnik G K. The bundle-forming pili of enteropathogenic Escherichia coli: transcription regulation by environmental signals. Mol Microbiol. 1996;20:87–100. doi: 10.1111/j.1365-2958.1996.tb02491.x. [DOI] [PubMed] [Google Scholar]
- 362.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Puri N, Jenner C, Bennett M, Stewart R, Mansfield J, Lyons N, Taylor J. Expression of avrPphB, an avirulence gene from Pseudomonas syringae pv. phaseolicola, and the delivery of signals causing the hypersensitive reaction in bean. Mol Plant-Microbe Interact. 1997;10:247–256. doi: 10.1094/MPMI.1997.10.2.247. [DOI] [PubMed] [Google Scholar]
- 364.Rabinowitz R P, Lai L-C, Jarvis K, McDaniel T K, Kaper J B, Stone K D, Donnenberg M S. Attaching and effacing of host cells by enteropathogenic Escherichia coli in the absence of detectable tyrosine kinase mediated signal transduction. Microb Pathog. 1996;21:157–171. doi: 10.1006/mpat.1996.0051. [DOI] [PubMed] [Google Scholar]
- 365.Rahme L G, Mindrinos M N, Panopoulos N J. Genetic and transcriptional organization of the hrp cluster of Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1991;173:575–586. doi: 10.1128/jb.173.2.575-586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Rahme L G, Mindrinos M N, Panopoulos N J. Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1992;174:3499–3507. doi: 10.1128/jb.174.11.3499-3507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Rees J G, Ades S E, Singer S J, Hynes R O. Sequence and domain structure of talin. Nature. 1990;347:685–689. doi: 10.1038/347685a0. [DOI] [PubMed] [Google Scholar]
- 368.Reisner B S, Straley S C. Yersinia pestis YopM: thrombin binding and overexpression. Infect Immun. 1992;60:5242–5252. doi: 10.1128/iai.60.12.5242-5252.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Ren R, Mayer B J, Cichetti P, Baltimore D. Identification of a ten-amino acid prolin-rich SH3 binding site. Science. 1993;259:1157–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 370.Rimpiläinen M, Forsberg Å, Wolf-Watz H. A novel protein, LcrQ, involved in the low-calcium response of Yersinia pseudotubeculosis shows extensive homology to YopH. J Bacteriol. 1992;174:3355–3363. doi: 10.1128/jb.174.10.3355-3363.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Rockey D D, Grosenbach D, Hruby D E, Peacock M G, Heinzen R A, Hackstadt T. Chlamydia psittaci IncA is phosphorylated by the host cell and is exposed on the cytoplasmic face of the developing inclusion. Mol Microbiol. 1997;24:217–228. doi: 10.1046/j.1365-2958.1997.3371700.x. [DOI] [PubMed] [Google Scholar]
- 372.Rockey D D, Heinzen R A, Hackstadt T. Cloning and characterization of a Chlamydia psittaci gene coding for a protein localized in the inclusion membrane of infected cells. Mol Microbiol. 1995;15:617–626. doi: 10.1111/j.1365-2958.1995.tb02371.x. [DOI] [PubMed] [Google Scholar]
- 373.Rohde J R, Fox J M, Minnich S A. Thermoregulation in Yersinia enterocolitica is coincident with changes in DNA supercoiling. Mol Microbiol. 1994;12:187–199. doi: 10.1111/j.1365-2958.1994.tb01008.x. [DOI] [PubMed] [Google Scholar]
- 374.Roine E, Wei W, Yuan J, Nurmiaho-Lassila E L, Kalkkinen N, Romantschuk M, He S Y. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Rooney, P. J., T. L. Yahr, and D. W. Frank. Genbank accession no. AF010151.
- 376.Rosenshine I, Donnenberg M S, Kaper J B, Finlay B B. Signal transduction between enteropathogenic Escherichia coli (EPEC) and epithelial cells: EPEC induces tyrosine phosphorylation of host cell proteins to initiate cytoskeletal rearrangement and bacterial uptake. EMBO J. 1992;11:3551–3560. doi: 10.1002/j.1460-2075.1992.tb05438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Rosenshine I, Ruschkowski S, Finlay B B. Expression of attaching/effacing activity by enteropathogenic Escherichia coli depends on growth phase, temperature, and protein synthesis upon contact with epithelial cells. Infect Immun. 1996;64:966–973. doi: 10.1128/iai.64.3.966-973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Rosenshine I, Ruschkowski S, Stein M, Reinscheid D J, Mills S D, Finlay B B. A pathogenic bacterium triggers epithelial signals to form a functional bacterial receptor that mediates actin pseudopod formation. EMBO J. 1996;15:2613–2624. [PMC free article] [PubMed] [Google Scholar]
- 379.Rosqvist R, Bölin I, Wolf-Watz H. Inhibition of phagocytosis in Yersinia pseudotuberculosis: a virulence plasmid-encoded ability involving the Yop2b protein. Infect Immun. 1988;56:2139–2143. doi: 10.1128/iai.56.8.2139-2143.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Rosqvist R, Forsberg Å, Wolf-Watz H. Intracellular targeting of the Yersinia YopE cytotoxin in mammalian cells induces actin microfilament disruption. Infect Immun. 1991;59:4562–4569. doi: 10.1128/iai.59.12.4562-4569.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Rosqvist R, Forsberg Å, Rimpilainen M, Bergman T, Wolf-Watz H. The cytotoxic protein YopE of Yersinia obstructs the primary host defence. Mol Microbiol. 1990;4:657–667. doi: 10.1111/j.1365-2958.1990.tb00635.x. [DOI] [PubMed] [Google Scholar]
- 382.Rosqvist R, Håkansson S, Forsberg Å, Wolf-Watz H. Functional conservation of the secretion and translocation machinery for virulence proteins of yersiniae, salmonellae and shigellae. EMBO J. 1995;14:4187–4195. doi: 10.1002/j.1460-2075.1995.tb00092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Rosqvist R, Magnusson K-E, Wolf-Watz H. Target cell contact triggers expression and polarized transfer of Yersinia YopE cytotoxin into mammalian cells. EMBO J. 1994;13:964–972. doi: 10.1002/j.1460-2075.1994.tb06341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Rosqvist R, Wolf-Watz H. Virulence-plasmid associated HeLa cell induced cytotoxicity of Yersinia pseudotuberculosis. Microb Pathog. 1986;1:229–240. doi: 10.1016/0882-4010(86)90047-1. [DOI] [PubMed] [Google Scholar]
- 385.Russel M. Mutants at conserved positions in gene IV, a gene required for assembly and secretion of filamentous phages. Mol Microbiol. 1994;14:357–369. doi: 10.1111/j.1365-2958.1994.tb01296.x. [DOI] [PubMed] [Google Scholar]
- 386.Russel M. Moving through the membranes with filamentous phages. Trends Microbiol. 1995;3:223–228. doi: 10.1016/s0966-842x(00)88929-5. [DOI] [PubMed] [Google Scholar]
- 387.Russel M, Kazmierczak B. Analysis of the structure and subcellular location of filamentous phage pIV. J Bacteriol. 1993;175:3998–4007. doi: 10.1128/jb.175.13.3998-4007.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Sakai T, Sasakawa C, Makino S, Yoshikawa M. DNA sequence and product analysis of the virF locus responsible for Congo red binding and cell invasion in Shigella flexneri 2a. Infect Immun. 1986;54:395–402. doi: 10.1128/iai.54.2.395-402.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Sakai T, Sasakawa C, Yoshikawa M. Expression of four virulence antigens of Shigella flexneri is positively regulated at the transcriptional level by the 30 kiloDalton VirF protein. Mol Microbiol. 1988;2:589–597. doi: 10.1111/j.1365-2958.1988.tb00067.x. [DOI] [PubMed] [Google Scholar]
- 390.Salmeron J M, Staskawicz B J. Molecular characterization and hrp dependence of the avirulence gene avrPto from Pseudomonas syringae pv. tomato. Mol Gen Genet. 1993;239:6–16. doi: 10.1007/BF00281595. [DOI] [PubMed] [Google Scholar]
- 391.Salmond G P C, Reeves P J. Membrane traffic wardens and protein secretion in Gram-negative bacteria. Trends Biochem Sci. 1993;18:7–12. doi: 10.1016/0968-0004(93)90080-7. [DOI] [PubMed] [Google Scholar]
- 392.Sample A K, Brubaker R R. Post-translational regulation of Lcr plasmid-mediated peptides in pesticinogenic Yersinia pestis. Microb Pathog. 1987;3:239–248. doi: 10.1016/0882-4010(87)90057-x. [DOI] [PubMed] [Google Scholar]
- 393.Sansonetti P J, Arondel J, Cantey J R, Prevost M C, Huerre M. Infection of rabbit Peyer’s patches by Shigella flexneri: effect of adhesive or invasive bacterial phenotypes on follicle-associated epithelium. Infect Immun. 1996;64:2752–2764. doi: 10.1128/iai.64.7.2752-2764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Sansonetti P J, d’Hauteville H, Ecobichon C, Pourcel C. Molecular comparison of virulence plasmids in Shigella and enteroinvasive Escherichia coli. Ann Inst Pasteur Microbiol. 1983;134A:295–318. [PubMed] [Google Scholar]
- 395.Sansonetti P J, Hale T L, Dammin G J, Kapfer C, Collins H H J, Formal S B. Alterations in the pathogenicity of Escherichia coli K-12 after transfer of plasmid and chromosomal genes from Shigella flexneri. Infect Immun. 1983;39:1392–1402. doi: 10.1128/iai.39.3.1392-1402.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Sansonetti P J, Kopecko D J, Formal S B. Shigella sonnei plasmids: evidence that a large plasmid is necessary for virulence. Infect Immun. 1981;34:75–83. doi: 10.1128/iai.34.1.75-83.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Sansonetti P J, Kopecko D J, Formal S B. Involvement of a plasmid in the invasive ability of Shigella flexneri. Infect Immun. 1982;35:852–860. doi: 10.1128/iai.35.3.852-860.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Sansonetti P J, Ryter A, Clerc P, Maurelli A T, Mounier J. Multiplication of Shigella flexneri within HeLa cells: lysis of the phagocytic vacuole and plasmid-mediated contact hemolysis. Infect Immun. 1986;51:461–469. doi: 10.1128/iai.51.2.461-469.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Sasakawa C, Adler B, Tobe T, Okada N, Nagai S, Komatsu K, Yoshikawa M. Functional organization and nucleotide sequence of virulence region-2 on the large virulence plasmid in Shigella flexneri 2a. Mol Microbiol. 1989;3:1191–1201. doi: 10.1111/j.1365-2958.1989.tb00269.x. [DOI] [PubMed] [Google Scholar]
- 400.Sasakawa C, Kamata K, Sakai T, Makino S, Yamada M, Okada N, Yoshikawa M. Virulence-associated genetic regions comprising 31 kilobases of the 230-kilobase plasmid in Shigella flexneri 2a. J Bacteriol. 1988;170:2480–2484. doi: 10.1128/jb.170.6.2480-2484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Sasakawa C, Komatsu K, Tobe T, Suzuki T, Yoshikawa M. Eight genes in region 5 that form an operon are essential for invasion of epithelial cells by Shigella flexneri 2a. J Bacteriol. 1993;175:2334–2346. doi: 10.1128/jb.175.8.2334-2346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Sasakawa C, Makino S, Kamata K, Yoshikawa M. Isolation, characterization, and mapping of Tn5 insertions into the 140-megadalton invasion plasmid defective in the mouse Sereny test in Shigella flexneri 2a. Infect Immun. 1986;54:32–36. doi: 10.1128/iai.54.1.32-36.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Scaletzky I C A, Silva M L M, Trabulsi L R. Distinctive patterns of adherence of enteropathogenic Escherichia coli to HeLa cells. Infect Immun. 1984;45:534–536. doi: 10.1128/iai.45.2.534-536.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Schesser K, Frihtz-Lindsten E, Wolf-Watz H. Delineation and mutational analysis of the Yersinia pseudotuberculosis YopE domains which mediate translocation across bacterial and eukaryotic membranes. J Bacteriol. 1996;178:7227–7233. doi: 10.1128/jb.178.24.7227-7233.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Schiemann D A, Shope S R. Anaerobic growth of Salmonella typhimurium results in increased uptake by Henle 407 epithelial and mouse peritoneal cells in vitro and repression of a major outer membrane protein. Infect Immun. 1991;59:437–440. doi: 10.1128/iai.59.1.437-440.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Schmitt C K, Darnell S C, Tesh V L, Stocker B A, O’Brien A D. Mutation of flgM attenuated virulence of Salmonella typhimurium, and mutation of fliA represses the attenuated phenotype. J Bacteriol. 1994;176:368–377. doi: 10.1128/jb.176.2.368-377.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Schulte R, Wattiau P, Hartland E L, Robins-Browne R M, Cornelis G R. Differential secretion of interleukin-8 by human epithelial cell lines upon entry of virulent or nonvirulent Yersinia enterocolitica. Infect Immun. 1996;64:2106–2113. doi: 10.1128/iai.64.6.2106-2113.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Scofield S R, Tobias C M, Rathjen J P, Chang J H, Lavelle D T, Michelmore R W, Staskawicz B J. Molecular basis of gene-for-gene specificity in bacterial speck disease of tomato. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 409.Senior A E. ATP synthesis by oxidative phosphorylation. Physiol Rev. 1988;68:177–231. doi: 10.1152/physrev.1988.68.1.177. [DOI] [PubMed] [Google Scholar]
- 410.Shea J E, Hensel M, Gleeson C, Holden D. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Siebers A, Finlay B B. M cells and the pathogenesis of mucosal and systemic infections. Trends Microbiol. 1996;4:22–29. doi: 10.1016/0966-842x(96)81501-0. [DOI] [PubMed] [Google Scholar]
- 412.Simonet M, Richard S, Berche P. Electron microscopic evidence for in vivo extracellular localization of Yersinia pseudotubersulosis harboring the pYV plasmid. Infect Immun. 1990;58:841–845. doi: 10.1128/iai.58.3.841-845.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Skrzypek E, Straley S C. LcrG, a secreted protein involved in negative regulation of the low-calcium response in Yersinia pestis. J Bacteriol. 1993;175:3520–3528. doi: 10.1128/jb.175.11.3520-3528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Skrzypek E, Straley S C. Differential effects of deletions in lcrV on secretion of V antigen, regulation of the low-Ca2+ response, and virulence of Yersinia pestis. J Bacteriol. 1995;177:2530–2542. doi: 10.1128/jb.177.9.2530-2542.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Skurnik M. Expression of antigens encoded by the virulence plasmid of Yersinia enterocolitica under different growth conditions. Infect Immun. 1985;47:183–190. doi: 10.1128/iai.47.1.183-190.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Sodeinde O A, Goguen J D. Genetic analysis of the 9.5-kilobase virulence plasmid of Yersinia pestis. Infect Immun. 1988;56:2743–2748. doi: 10.1128/iai.56.10.2743-2748.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Sodeinde O A, Sample A K, Brubaker R R, Goguen J D. Plasminogen activator/coagulase gene of Yersinia pestis is responsible for degradation of plasmid-encoded outer membrane proteins. Infect Immun. 1988;56:2749–2752. doi: 10.1128/iai.56.10.2749-2752.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Sodeinde O A, Subrahmanyam Y V B K, Stark K, Quan T, Bao Y, Goguen J D. A surface protease and the invasive character of plague. Science. 1992;258:1004–1007. doi: 10.1126/science.1439793. [DOI] [PubMed] [Google Scholar]
- 419.Sonnhammer E L L, Kahn D. Modular arrangement of proteins as inferred from analysis of homology. Protein Sci. 1994;3:482–492. doi: 10.1002/pro.5560030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Sory M-P, Boland A, Lambermont A, Cornelis G R. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc Natl Acad Sci USA. 1995;92:11998–12002. doi: 10.1073/pnas.92.26.11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Sory M-P, Cornelis G R. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol. 1994;14:583–594. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 422.Stainier I, Iriarte M, Cornelis G R. YscM1 and YscM2, two Yersinia enterocolitica proteins causing downregulation of yop transcription. Mol Microbiol. 1997;26:833–843. doi: 10.1046/j.1365-2958.1997.6281995.x. [DOI] [PubMed] [Google Scholar]
- 423.Staley T E, Jones E W, Corley L D. Attachment and penetration of Escherichia coli into intestinal epithelium of the ileum in newborn pigs. Am J Pathol. 1969;56:371–392. [PMC free article] [PubMed] [Google Scholar]
- 424.Staskawicz B J, Ausubel F M, Baker B J, Ellis J G, Jones J D. Molecular genetics of plant disease resistance. Science. 1995;268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- 425.Stock J B, Surette M G, Levit M, Park P. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 25–51. [Google Scholar]
- 426.Stone B J, Garcia C M, Badger J L, Hassett T, Smith R I F, Miller V L. Identification of novel loci affecting entry of Salmonella enteritidis into eukaryotic cells. J Bacteriol. 1992;174:3945–3952. doi: 10.1128/jb.174.12.3945-3952.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Straley S C. The plasmid-encoded outer-membrane proteins of Yersinia pestis. Rev Infect Dis. 1988;10:323–326. doi: 10.1093/cid/10.supplement_2.s323. [DOI] [PubMed] [Google Scholar]
- 428.Straley S C, Brubaker R R. Cytoplasmic and membrane proteins of yersiniae cultivated under conditions simulating mammalian intracellular environment. Proc Natl Acad Sci USA. 1981;78:1224–1228. doi: 10.1073/pnas.78.2.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Straley S C, Cibull M L. Differential clearance of host-pathogen interactions of YopE− and YopK−YopL−Yersinia pestis in BALB/c mice. Infect Immun. 1989;57:1200–1210. doi: 10.1128/iai.57.4.1200-1210.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Straley S C, Plano G V, Skrzypek E, Haddix P L, Fields K A. Regulation by Ca2+ in the Yersinia low-Ca2+ response. Mol Microbiol. 1993;8:1005–1010. doi: 10.1111/j.1365-2958.1993.tb01644.x. [DOI] [PubMed] [Google Scholar]
- 431.Straley S C, Skrzypek E, Plano G V, Bliska J B. Yops of Yersinia spp. pathogenic for humans. Infect Immun. 1993;61:3105–3110. doi: 10.1128/iai.61.8.3105-3110.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Sukegawa J, Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell. 1992;72:29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]
- 433.Takada Y, Strominger J L, Hemler M E. Fibronectin receptor structures in the VLA family of heterodimers. Nature. 1987;326:607–609. doi: 10.1038/326607a0. [DOI] [PubMed] [Google Scholar]
- 434.Takeuchi A. Electron microscopic studies of experimental Salmonella infection I. Penetration into the intestinal epithelium by Salmonella typhimurium. Am J Pathol. 1967;50:109–136. [PMC free article] [PubMed] [Google Scholar]
- 435.Takeuchi A, Formal S B, Sprinz H. Experimental acute colitis in the rhesus monkey following peroral infection with Shigella flexneri. An electron microscope study. Am J Pathol. 1968;52:503–529. [PMC free article] [PubMed] [Google Scholar]
- 436.Tang X, Frederick R D, Zhou J, Halterman D A, Jia Y, Martin G B. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 437.Thirumalai K, Kim K-S, Zychlinsky A. IpaB, a Shigella flexneri invasin, colocalizes with interleukin-1β-converting enzyme in the cytoplasm of macrophages. Infect Immun. 1997;65:787–793. doi: 10.1128/iai.65.2.787-793.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Tobe T, Nagai S, Okada N, Adler B, Yoshikawa M, Sasakawa C. Temperature-regulated expression of invasion genes in Shigella flexneri is controlled through the transcriptional activation of the virB gene on the large plasmid. Mol Microbiol. 1991;5:887–893. doi: 10.1111/j.1365-2958.1991.tb00762.x. [DOI] [PubMed] [Google Scholar]
- 439.Tobe T, Schoolnik G K, Sohel I, Bustamante V H, Puente J L. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol Microbiol. 1996;21:963–975. doi: 10.1046/j.1365-2958.1996.531415.x. [DOI] [PubMed] [Google Scholar]
- 440.Tobe T, Yoshikawa M, Mizuno T, Sasakawa C. Transcriptional control of the invasion regulatory gene virB of Shigella flexneri: activation by VirF and repression by H-NS. J Bacteriol. 1993;175:6142–6149. doi: 10.1128/jb.175.19.6142-6149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 441.Tobe T, Yoshikawa M, Sasakawa C. Thermoregulation of virB transcription in Shigella flexneri by sensing of changes in local DNA superhelicity. J Bacteriol. 1995;177:1094–1097. doi: 10.1128/jb.177.4.1094-1097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Tran Van Nhieu G, Ben-Ze’ev A, Sansonetti P J. Modulation of bacterial entry into epithelial cells by association between vinculin and the Shigella IpaA invasin. EMBO J. 1997;16:2717–2729. doi: 10.1093/emboj/16.10.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Uchiya K, Tobe T, Komatsu K, Suzuki T, Watarai M, Fukuda I, Yoshikawa M, Sasakawa C. Identification of a novel virulence gene, virA, on the large plasmid of Shigella, involved in invasion and intercellular spreading. Mol Microbiol. 1995;17:241–250. doi: 10.1111/j.1365-2958.1995.mmi_17020241.x. [DOI] [PubMed] [Google Scholar]
- 444.Ueno T, Oosawa K, Aizawa S. M ring, S ring and proximal rod of the flagellar basal body of Salmonella typhimurium are composed of subunits of a single protein, FliF. J Mol Biol. 1992;227:672–677. doi: 10.1016/0022-2836(92)90216-7. [DOI] [PubMed] [Google Scholar]
- 445.Ueno T, Oosawa K, Aizawa S. Domain structures of the MS ring component protein (FliF) of the flagellar basal body of Salmonella typhimurium. J Mol Biol. 1994;236:546–555. doi: 10.1006/jmbi.1994.1164. [DOI] [PubMed] [Google Scholar]
- 446.Van den Ackerveken G, Marois E, Bonas U. Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell. 1996;87:1307–1316. doi: 10.1016/s0092-8674(00)81825-5. [DOI] [PubMed] [Google Scholar]
- 447.Van Gijsegem F, Genin S, Boucher C. Conservation of secretion pathways for pathogenicity determinants of plant and animal bacteria. Trends Microbiol. 1993;1:175–180. doi: 10.1016/0966-842x(93)90087-8. [DOI] [PubMed] [Google Scholar]
- 448.Van Gijsegem F, Gough C, Zischek C, Niqueux E, Arlat M, Genin S, Barberis P, German S, Castello P, Boucher C. The hrp gene locus of Pseudomonas solanacearum, which controls the production of a type III secretion system, encodes eight proteins related to components of the bacterial flagellar biogenesis complex. Mol Microbiol. 1995;15:1095–1114. doi: 10.1111/j.1365-2958.1995.tb02284.x. [DOI] [PubMed] [Google Scholar]
- 449.Vaudaux P, Waldvogel F A. Gentamicin antibacterial activity in presence of human polymorphonuclear leukocytes. Antimicrob Agents Chemother. 1979;16:743–749. doi: 10.1128/aac.16.6.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Venkatesan M, Buysse J M, Kopecko D J. Characterization of invasion plasmid antigen (ipaBCD) genes from Shigella flexneri. Proc Natl Acad Sci USA. 1988;85:9317–9321. doi: 10.1073/pnas.85.23.9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Venkatesan M M, Buysse J M. Nucleotide sequence of invasion plasmid antigen gene ipaA from Shigella flexneri 5. Nucleic Acids Res. 1991;18:1648. doi: 10.1093/nar/18.6.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Venkatesan M M, Buysse J M, Oaks E V. Surface presentation of Shigella flexneri invasion plasmid antigens requires the products of the spa locus. J Bacteriol. 1992;174:1990–2001. doi: 10.1128/jb.174.6.1990-2001.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Vidal S M, Malo D, Vogan K. Natural resistance to infection with intracellular parasites: Isolation of a candidate for Bcg. Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 454.Viitanen A-M, Toivanen P, Skurnik M. The lcrE gene is part of an operon in the lcr region of Yersinia enterocolitica O:3. J Bacteriol. 1990;172:3152–3162. doi: 10.1128/jb.172.6.3152-3162.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Vivian A, Gibbon M J. Avirulence genes in plant-pathogenic bacteria: signals or weapons? Microbiology. 1997;143:693–704. doi: 10.1099/00221287-143-3-693. [DOI] [PubMed] [Google Scholar]
- 456.Vogler A P, Homma M, Irikura V M, Macnab R M. Salmonella typhimurium mutants defective in flagellar filament regrowth and sequence similarity of FliI to F0F1, vacuolar, and archaebacterial ATPase subunits. J Bacteriol. 1991;173:3564–3572. doi: 10.1128/jb.173.11.3564-3572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Walker J E, Saraste M, Gay N J. The unc operon nucleotide sequence, regulation and structure of ATP-synthase. Biochim Biophys Acta. 1984;768:164–200. doi: 10.1016/0304-4173(84)90003-x. [DOI] [PubMed] [Google Scholar]
- 458.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Wandersman C. Secretion across the bacterial outer membrane. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Riley M, Reznikoff W S, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 955–966. [Google Scholar]
- 460.Wassef J, Keren D F, Mailloux J L. Role of M cells in initial bacterial uptake and in ulcer formation in the rabbit intestinal loop model in shigellosis. Infect Immun. 1989;57:858–863. doi: 10.1128/iai.57.3.858-863.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Watanabe H, Arakawa E, Ito K, Kato J, Nakamura A. Genetic analysis of an invasion region by use of a Tn3-lac transposon and identification of a second positive regulator gene, invE, for cell invasion of Shigella sonnei: significant homology of invE with ParB of plasmid P1. J Bacteriol. 1990;172:619–629. doi: 10.1128/jb.172.2.619-629.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Watarai M, Funato S, Sasakawa C. Interaction of Ipa proteins of Shigella flexneri with α5β1 integrin promotes entry of the bacteria into mammalian cells. J Exp Med. 1996;183:991–999. doi: 10.1084/jem.183.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Watarai M, Kamata Y, Kozaki S, Sasakawa C. rho, a small GTP-binding protein, is essential for Shigella invasion of epithelial cells. J Exp Med. 1997;185:281–292. doi: 10.1084/jem.185.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Watarai M, Tobe T, Yoshikawa M, Sasakawa C. Contact of Shigella with host cells triggers release of Ipa invasins and is an essential function of invasiveness. EMBO J. 1995;14:2461–2470. doi: 10.1002/j.1460-2075.1995.tb07243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Watarai M, Tobe T, Yoshikawa M, Sasakawa C. Disulfide oxidoreductase activity of Shigella flexneri is required for release of Ipa proteins and invasion of epithelial cells. Proc Natl Acad Sci USA. 1995;92:4927–4931. doi: 10.1073/pnas.92.11.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Wattiau P, Bernier B, Deslee P, Michiels T, Cornelis G R. Individual chaperones required for Yop secretion in Yersinia. Proc Natl Acad Sci USA. 1994;91:10493–10497. doi: 10.1073/pnas.91.22.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Wattiau P, Cornelis G R. SycE, a chaperone-like protein of Yersinia enterocolitica involved in the secretion of YopE. Mol Microbiol. 1993;8:123–131. doi: 10.1111/j.1365-2958.1993.tb01209.x. [DOI] [PubMed] [Google Scholar]
- 468.Wattiau P, Cornelis G R. Identification of DNA sequences recognized by VirF, the transcriptional activator of the Yersinia yop regulon. J Bacteriol. 1994;176:3878–3884. doi: 10.1128/jb.176.13.3878-3884.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Wattiau P, Woestyn S, Cornelis G R. Customized secretion chaperones in pathogenic bacteria. Mol Microbiol. 1996;20:255–262. doi: 10.1111/j.1365-2958.1996.tb02614.x. [DOI] [PubMed] [Google Scholar]
- 470.Wei Z-M, Beer S V. HrpI of Erwinia amylovora functions in secretion of hairpin and is a member of a new protein family. J Bacteriol. 1993;175:7958–7967. doi: 10.1128/jb.175.24.7958-7967.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 471.Wei Z-M, Beer S V. hrpL activates Erwinia amylovora hrp gene transcription and is a member of the ECF subfamily of sigma factors. J Bacteriol. 1995;177:6201–6210. doi: 10.1128/jb.177.21.6201-6210.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Wei Z M, Laby R J, Zumoff C H, Bauer D W, He S Y, Collmer A, Beer S V. Harpin, elicitor of the hypersensitive response produced by the plant pathogen Erwinia amylovora. Science. 1992;257:85–88. doi: 10.1126/science.1621099. [DOI] [PubMed] [Google Scholar]
- 473.Wei Z M, Sneath B J, Beer S V. Expression of Erwinia amylovora hrp genes in response to environmental stimuli. J Bacteriol. 1992;174:1875–1882. doi: 10.1128/jb.174.6.1875-1882.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Weksler B B. Platelets. In: Gallin J I, Goldstein I M, Snyderman R, editors. Inflammation: basic principles and clinical correlates. New York, N.Y: Raven Press; 1988. pp. 543–557. [Google Scholar]
- 475.Welch R A. Pore-forming cytolysins of gram-negative bacteria. Mol Microbiol. 1991;5:521–528. doi: 10.1111/j.1365-2958.1991.tb00723.x. [DOI] [PubMed] [Google Scholar]
- 476.Wengelnik K, Bonas U. HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J Bacteriol. 1996;178:3462–3469. doi: 10.1128/jb.178.12.3462-3469.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Wengelnik K, Marie C, Russel M, Bonas U. Expression and localization of HrpA1, a protein of Xanthomonas campestris pv. vesicatoria essential for pathogenicity and induction of the hypersensitive reaction. J Bacteriol. 1996;178:1061–1069. doi: 10.1128/jb.178.4.1061-1069.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Wengelnik K, Van den Ackerveken G, Bonas U. HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two-component response regulators. Mol Plant-Microbe Interact. 1996;9:704–712. doi: 10.1094/mpmi-9-0704. [DOI] [PubMed] [Google Scholar]
- 479.Whalen M C, Wang J F, Carland F M, Heiskell M E, Dahlbeck D, Minsavage G V, Jones J B, Scott J W, Stall R E, Staskawicz B J. Avirulence gene avrRxv from Xanthomonas campestris pv. vesicatoria species resistance on tomato line Hawaii 7998. Mol Plant-Microbe Interact. 1993;6:616–627. doi: 10.1094/mpmi-6-616. [DOI] [PubMed] [Google Scholar]
- 480.Williams A W, Yamaguchi S, Togashi F, Aizawa S I, Kawagishi I, Macnab R M. Mutations in fliK and flhB affecting flagellar hook and filament assembly in Salmonella typhimurium. J Bacteriol. 1996;178:2960–2970. doi: 10.1128/jb.178.10.2960-2970.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Winans S C, Burns D L, Christie P J. Adaptation of a conjugal transfer system for the export of pathogenic macromolecules. Trends Microbiol. 1996;4:64–68. doi: 10.1016/0966-842X(96)81513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Woestyn S, Allaoui A, Wattiau P, Cornelis G R. YscN, the putative energizer of the Yersinia Yop secretion machinery. J Bacteriol. 1994;176:1561–1569. doi: 10.1128/jb.176.6.1561-1569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Woestyn S, Sory M-P, Boland A, Lequenne O, Cornelis G R. The cytosolic SycE and SycH chaperones of Yersinia protect the region of YopE and YopH involved in translocation across eukaryotic cell membranes. Mol Microbiol. 1996;20:1261–1271. doi: 10.1111/j.1365-2958.1996.tb02645.x. [DOI] [PubMed] [Google Scholar]
- 484.Wolf-Watz H, Portnoy D A, Bölin I, Falkow S. Transfer of the virulence plasmid of Yersinia pestis to Yersinia pseudotuberculosis. Infect Immun. 1985;48:241–243. doi: 10.1128/iai.48.1.241-243.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Wood M W, Rosqvist R, Mullan P B, Edwards M H, Galyov E E. SopE, a secreted protein of Salmonella dublin, is translocated into the target eukaryotic cell via a sip-dependent mechanism and promotes bacterial entry. Mol Microbiol. 1996;22:327–338. doi: 10.1046/j.1365-2958.1996.00116.x. [DOI] [PubMed] [Google Scholar]
- 486.Wu H C, Tokunaga M. Biogenesis of lipoproteins in bacteria. Curr Top Microbiol Immunol. 1986;125:127–157. doi: 10.1007/978-3-642-71251-7_9. [DOI] [PubMed] [Google Scholar]
- 487.Xiao Y, Heu S, Yi J, Lu Y, Hutcheson S W. Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J Bacteriol. 1994;176:1025–1036. doi: 10.1128/jb.176.4.1025-1036.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Xiao Y, Hutcheson S W. A single promoter sequence recognized by a newly identified alternate sigma factor directs expression of pathogenicity and host range determinants in Pseudomonas syringae. J Bacteriol. 1994;176:3089–3091. doi: 10.1128/jb.176.10.3089-3091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Xiao Y, Lu Y, Heu S, Hutcheson S W. Organization and environmental regulation of the Pseudomonas syringae pv. syringae 61 hrp cluster. J Bacteriol. 1992;174:1734–1741. doi: 10.1128/jb.174.6.1734-1741.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Yahr T L, Frank D W. Transcriptional organization of the trans-regulatory locus which controls exoenzyme S synthesis in Pseudomonas aeruginosa. J Bacteriol. 1994;176:3832–3838. doi: 10.1128/jb.176.13.3832-3838.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Yahr T L, Goranson J, Frank D W. Exoenzyme S of Pseudomonas aeruginosa is secreted by a type III pathway. Mol Microbiol. 1996;22:991–103. doi: 10.1046/j.1365-2958.1996.01554.x. [DOI] [PubMed] [Google Scholar]
- 492.Yahr T L, Hovey A K, Kulich S M, Frank D W. Transcriptional analysis of the Pseudomonas aeruginosa exoenzyme S structural gene. J Bacteriol. 1995;177:1169–1178. doi: 10.1128/jb.177.5.1169-1178.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Yahr T L, Mende-Mueller L M, Friese M B, Frank D W. Identification of type III secreted products of the Pseudomonas aeruginosa exoenzyme S regulon. J Bacteriol. 1997;179:7165–7168. doi: 10.1128/jb.179.22.7165-7168.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 494.Yao R, Palchaudhuri S. Nucleotide sequence of the ipaBCD structural genes of Shigella dysenteriae. Mol Microbiol. 1991;5:2217–2221. doi: 10.1111/j.1365-2958.1991.tb02151.x. [DOI] [PubMed] [Google Scholar]
- 495.Yao R, Palchaudhuri S. Nucleotide sequence and transcriptional regulation of a positive regulatory gene of Shigella dysenteriae. Infect Immun. 1992;60:1163–1169. doi: 10.1128/iai.60.3.1163-1169.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 496.Yoshida M, Allison W S, Esch F S, Futai M. The specificity of carboxyl group modification during the inactivation of the Escherichia coli F1-ATPase with dicyclohexyl [14C] carbodiimide. J Biol Chem. 1982;257:10033–10037. [PubMed] [Google Scholar]
- 497.Yother J, Chamess T W, Goguen J D. Temperature-controlled plasmid regulon associated with the low calcium response in Yersinia pestis. J Bacteriol. 1986;165:443–447. doi: 10.1128/jb.165.2.443-447.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Yother J, Goguen J D. Isolation and characterization of Ca2+-blind mutants of Yersinia pestis. J Bacteriol. 1985;164:704–711. doi: 10.1128/jb.164.2.704-711.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Yuan J, He S Y. The Pseudomonas syringae Hrp regulation and secretion system controls the production and secretion of multiple extracellular proteins. J Bacteriol. 1996;178:6399–6402. doi: 10.1128/jb.178.21.6399-6402.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 500.Zahorchak R J, Charnetzky W T, Little R V, Brubaker R R. Consequences of Ca2+ deficiency on macromolecular synthesis and adenylate energy charge in Yersinia pestis. J Bacteriol. 1979;139:792–799. doi: 10.1128/jb.139.3.792-799.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Zhang Z-Y, Clemens J C, Schubert H L, Stuckey J A, Fischer M W F, Hume D M, Saper M A, Dixon J E. Expression, purification, and physicochemical characterization of a recombinant Yersinia protein tyrosine phosphatase. J Biol Chem. 1992;267:23759–23766. [PubMed] [Google Scholar]
- 502.Zhao R, Pathak N, Jaffe H, Reese T S, Khan S. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J Mol Biol. 1996;261:195–208. doi: 10.1006/jmbi.1996.0452. [DOI] [PubMed] [Google Scholar]
- 503.Zhao R, Schuster S C, Khan S. Structural effects of mutations in Salmonella typhimurium flagellar switch complex. J Mol Biol. 1995;251:400–412. doi: 10.1006/jmbi.1995.0443. [DOI] [PubMed] [Google Scholar]
- 504.Zhou J, Loh Y T, Bressan R A, Martin G B. The tomato gene Pti1 encodes a serine/threonine kinase that is phosphorylated by Pto and is involved in the hypersensitive response. Cell. 1995;83:925–935. doi: 10.1016/0092-8674(95)90208-2. [DOI] [PubMed] [Google Scholar]
- 505.Zink D L, Feeley J C, Wells J G, Vanderzant C, Vickery J C, Roofs W D, O’Donovan G A. Plasmid-mediated tissue invasiveness in Yersinia enterocolitica. Nature. 1980;283:224–226. doi: 10.1038/283224a0. [DOI] [PubMed] [Google Scholar]
- 506.Zychlinsky A, Fitting C, Cavaillon J M, Sansonetti P J. Interleukin 1 is released by murine macrophages during apoptosis induced by Shigella flexneri. J Clin Invest. 1994;94:1328–1332. doi: 10.1172/JCI117452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Zychlinsky A, Kenny B, Ménard R, Prévost M-C, Holland I B, Sansonetti P J. IpaB mediates macrophage apoptosis induced by Shigella flexneri. Mol Microbiol. 1994;11:619–627. doi: 10.1111/j.1365-2958.1994.tb00341.x. [DOI] [PubMed] [Google Scholar]
- 508.Zychlinsky A, Perdomo J J, Sansonetti P J. Molecular and cellular mechanisms of tissue invasion by Shigella flexneri. Ann N Y Acad Sci. 1994;730:197–208. doi: 10.1111/j.1749-6632.1994.tb44249.x. [DOI] [PubMed] [Google Scholar]
- 509.Zychlinsky A, Prévost M-C, Sansonetti P J. Shigella flexneri induces apoptosis in infected macrophages. Nature. 1992;358:167–169. doi: 10.1038/358167a0. [DOI] [PubMed] [Google Scholar]
- 510.Zychlinsky A, Thirumalai K, Arondel J, Cantey J R, Aliprantis A O, Sansonetti P J. In vivo apoptosis in Shigella flexneri infections. Infect Immun. 1996;64:5357–5365. doi: 10.1128/iai.64.12.5357-5365.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]