Abstract
Objective:
Transcriptomic changes in joint tissues during the development of osteoarthritis (OA) are of interest for the discovery of biomarkers and mechanisms of disease. The objective of this study was to use the rat medial meniscus transection (MMT) model to discover stage and tissue-specific transcriptomic changes.
Design:
Sham or MMT surgeries were performed in mature rats. Cartilage, menisci and synovium were scored for histopathological changes at 2, 4 and 6 weeks post-surgery and processed for RNA-sequencing. Differentially expressed genes (DEG) were used to identify pathways and mechanisms. Published transcriptomic datasets from animal models and human OA were used to confirm and extend present findings.
Results:
The total number of DEGs was already high at 2 weeks (723 in meniscus), followed by cartilage (259) and synovium (42) and declined to varying degrees in meniscus and synovium but increased in cartilage at 6 weeks. The most upregulated genes included tenascins. The ‘response to mechanical stimulus’ and extracellular matrix-related pathways were enriched in both cartilage and meniscus. Pathways that were enriched in synovium at 4 weeks indicate processes related to synovial hyperplasia and fibrosis. Synovium also showed upregulation of IL-11 and several MMPs. The mechanical stimulus pathway included upregulation of the mechanoreceptors PIEZO1, PIEZO2 and TRPV4 and nerve growth factor. Analysis of data from prior RNA-sequencing studies of animal models and human OA support these findings.
Conclusion:
These results indicate several shared pathways that are affected during OA in cartilage and meniscus and support the role of mechanotransduction in OA pathogenesis.
Keywords: osteoarthritis, RNA sequencing, mechanotransduction
Introduction
Osteoarthritis (OA) can be initiated by acute joint trauma, leading to posttraumatic OA (PTOA)1 or by a series of risk factors, including excessive mechanical load2, 3, which during aging manifest their effects through decompensation of cellular homeostasis mechanisms4 and subsequent joint inflammation, pain and structural damage to the joint tissues. Based on pathological, histopathological, joint imaging, cellular and molecular studies, it is apparent that the disease process ultimately affects all joint tissues5. The temporal relationship and the types of changes in the different joint tissues vary among patient subpopulations and among the different animal models. Despite their limitations, animal models allow controlled approaches to systematically examine tissue level changes and with the availability of Omics technologies to obtain global insight into changes in cellular activities during OA initiation and progression.
The rat medial meniscus transection (MMT) model is a frequently used OA model6. A full thickness cut in the medial meniscus leads to joint instability and progressive OA development characterized by proteoglycan loss, cartilage fibrillation, chondrocyte death, eventual damage to the subchondral bone and formation of osteophytes7. The initial damage to the cartilage, as defined by fibrillation and loss of proteoglycans, is localized to the medial side of knee joint after MMT. In the later stages, there is significant cartilage damage and, in some cases, complete loss of the cartilage layer with exposure of the subchondral bone. Large medial tibial plateau osteophytes are also present by 3 weeks8, 9. MMT also results in a time-dependent increase in the hind paw weight distribution between the arthritic knee and the contralateral control knee throughout the duration of the 3-week protocol which, coupled with histologic changes, is indicative of a chronic OA-like pain8.
The goals of the present study were to profile transcriptomic changes by using RNA-sequencing (RNA-seq) in cartilage, menisci and synovium during the development of experimental OA in the rat MMT model. The data presented here identify genes altered during OA progression that are shared between all three tissues as well as identify those unique to each. Specifically, these data identify Tnn as a potential candidate for further studies, as this gene was differentially expressed across all stages of OA in all three tissues. Moreover, these data identify mechanotransduction and extracellular matrix pathways as critically dysregulated during OA progression, thus supporting a role for these in pathogenesis.
Methods
Rat MMT model
All animal procedures were approved by the Scripps Research Institutional Animal Care and Use Committee. Male Lewis rats (n=42 total) were assigned to the following groups: 24 rats with MMT surgery on the right and sham surgery (SS) on the left knee for knee tissue collection and RNA sequencing at 2,4 and 6 weeks (n= 8 animals per time point); 18 rats with MMT surgery on the right and sham surgery on the left knee for knee histology at 2, 4 and 6 weeks (n= 6 animals per time point). Animals were anesthetized with a ketamine/xylazine cocktail (50mg/kg Ketaved - Henry Schein, and 5mg/kg Xyalzine - AniSed respectively) by intraperitoneal injection, administered appropriate analgesics (Flunixin, 5mg/kg subcutaneous injection), and the surgical site shaved and disinfected. A small incision was made on the medial right hind knee to expose the collateral ligament. The collateral ligament was transected, and the joint capsule opened sufficiently to expose the medial meniscus. The meniscus was transected completely, then the skin incision was closed, and pressure applied to the wound to prevent hematoma development. Animals were allowed to recover on a heated blanket and were returned to their home cage. Sham procedures were carried out in a similar fashion except the medial meniscus was not cut. Due to serious joint infection, one sample at 2 weeks post-surgery was excluded from our study.
Histopathology
Rats were euthanized 2 (n=6), 4 (n=6) and 6 (n=6) weeks after MMT. Because of joint infection, one sample at 2 weeks post-surgery was excluded. Formalin-fixed rat knees were decalcified, bisected in the frontal plane, and embedded in paraffin (both halves in a single block). All blocks were sectioned for 3 levels (spaced at approx. 160 μm) with one slide per level stained with hematoxylin and eosin (H&E) and one stained with toluidine blue. The slides were scored by a veterinary pathologist (Inotiv, Bolder, CO) for cartilage degeneration, osteophytes, medial tibial bone damage and sclerosis, synovitis and medial tibial collagen degeneration as described in detail10. The total cartilage degeneration width was measured on tibial plateau affected by any type of degeneration (matrix fibrillation/loss, proteoglycan loss with or without chondrocyte death). The largest osteophyte was measured from base to edge at the thickest point and then given a score based on that measurement. The calcified cartilage and subchondral bone were scored to capture changes in these areas which are closely associated with changes in the non-calcified cartilage. The most severe lesion was scored in each section was scored. The synovial histopathology scoring considers numbers of synovial lining cell layers, proliferation of subsynovial tissue, and infiltration of inflammatory cells and the measurements of these parameters were then expressed as a score10.
One-way ANOVA with the post-hoc test was used to compare cartilage degeneration width among 3 groups. For nonparametric data, analyses were performed with Kruskal-Wallis test with multiple comparisons using Dunn’s corrections. GraphPad PRISM 9 was used for above-mentioned statistical analyses.
Tissue collection and RNA isolation
Rats were euthanized 2 (n=8), 4 (n=8) and 6 (n=8) weeks after MMT. Cartilage from both the medial and lateral sides of tibial plateaus and femoral condyles, medial and lateral menisci, and synovium of both knees were collected. Upon opening the joint capsule, synovial membranes were carefully resected to minimize the amount of sub-synovial adipose tissue. All arthroscopically visible synovium tissue was collected from the medial and lateral sides of the joint capsule and femoral condyles, distal of the meniscus and directly adjacent to the tibial plateau at all time points. Due to the very thin layer of the synovium, it was not possible to collect exclusively the lining layer without some subsynovial adipose tissue. Tissues were homogenized with 350 μl TRIZOL using a homogenizer, for three 30 s intervals interrupted by 3 min breaks. The homogenized tissue was added to 70 μl 100% ethanol. This mixture was vortexed, and the 100 μl supernatant was collected, centrifuged, and added to the mRNeasy Mini kit column (Qiagen) following manufacturer’s instructions. Total RNA was eluted in 12 μl of RNase-free water. The RNA integrity was determined using an Agilent Bioanalyzer and only samples with values >5 were used for RNA-seq.
High-throughput RNA-seq library construction and sequencing
The high throughput RNA-seq assay used was part of an early access kit from iGenomX Inc. Briefly, total RNA (50 ng) was placed in a solution containing 1.4X First Strand Buffer and barcoded oligo dT primer and incubated at 80°C for 10 minutes in 7 μL total volume, subsequently added was 4.5 mM DTT, barcoded template-switching oligonucleotide, 1.8 U/μL RNaseOUT (ThermoFisher), dNTP mix and Reverse Transcriptase in 11 μL final volume. Reactions were incubated 50 minutes at 42°C, followed by 5 minutes at 85°C. This was followed by addition of 2 μL reaction clean-up mix and incubated at 37°C for 30 minutes and 80°C for 20 minutes. Samples were pooled together, and cDNA products were purified by 1.2X SPRI bead clean-up procedure with SPRI bead mixture. cDNA products were taken directly into a PCR reaction containing 1X KAPA HiFi reaction mixture and 0.5 μM barcoded PCR primers of Illumina p5 and p7 sequences) in 100 μL final volume. PCR products were purified using 1X SPRI bead clean-up mix, PCR products were purified on 2% agarose gel, products approximately 350 bp - 800 bp were excised and isolated using agarose gel dissolving buffer and clean-up and concentrator column (Zymo Research). Purified PCR products were quantified using Qubit dsDNA HS assay kit, analyzed on Agilent Tapestation, and sequenced on 100 cycle P2 flow cells with an Illumina NextSeq2000 sequencing instrument using paired-end sequencing (read1: 26 bases, read2: 94 bases, i5 and i8 indexes used 8 base reads).
RNA-seq data analysis
The high throughput RNA-seq data was initially processed with in-house custom scripts using BBTools (https://sourceforge.net/projects/bbmap/) to obtain demultiplexed reads for each sample based on the inline barcodes. The 5’-end 10-base barcodes and 3’-end 12-base UMI information from read1 fastq were first incorporated into read2 fastq as part of sequence ID using UMI-tools11. The read2 sequences (from the pre-deduplictated dataset) were analyzed using nf-core/RNA-seq pipeline v1.4.2 (implemented on Nextflow v20.07.1), which is an open-source and available at https://github.com/nf-core/rnaseq as part of the nf-core project12. Read2s were trimmed for adapters using trimGalore! v0.6.4 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/) and aligned to the rat genome (Rattus norvegicus) ENSEMBL build Rnor 6.0 using STAR v2.6.1d13. Gene-level assignment was then performed using featureCounts v1.6.414. The gene expression matrix with raw gene counts was then used for differential gene expression analysis using the Bioconductor DESeq2 R package v1.20.015. The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value (padj) <0.05 found by DESeq2 were assigned as significantly differentially expressed genes (DEGs). The uniquely mapped alignments resulting from alignments to the reference genome were deduplicated removing the PCR duplicates. The transcript UMI for each gene was counted for each sample using UMI-tools. Further, statistical analysis of pathways and gene ontology terms were carried out for gene lists showing significant differences between the compared groups using iPathwayGuide (Advaita Bioinformatics, Plymoth, MI, USA; https://advaitabio.com/ipathwayguide/).
The pathway enrichment (KEGG) and GO analyses, including biological process, from different gene lists was carried out by using DAVID (Database for Annotation, Visualization, and Integrated Discovery) (http://david.abcc.ncifcrf.gov/). For GO classification and KEGG, the cutoff conditions were set as FDR = 0.05. The GO graphs of all enriched GO terms and volcano plots were drawn by ggplot2 package in R environment. Heatmaps were generated in R package ‘pheatmap’. The DEGs were mapped to the STRING database (https://string-db.org/) to establish protein-protein interaction (PPI) network. The network visualization of PPI was carried out by Cytoscape.
Utilization of RNA-seq datasets from normal and OA human knee cartilage tissues
Our previous RNA-seq dataset from normal and OA human knee cartilage tissues was utilized (GEO accession number: GSE114007)16. Normal human knee cartilage tissues were from 13 males and 5 females (age 18 to 61, mean 37 ± 3) without history of joint disease or trauma. OA-affected cartilage was harvested from tissues removed during knee replacement surgery from 8 males and 12 females (age 51 to 82, mean 66 ± 2). Genes with log2 CPM > 3 in one or more samples were included in the analysis16. A FDR was calculated based on the moderated t-statistic using the Benjamini-Hochberg method17.
Results
Histopathological changes in the rat MMT model
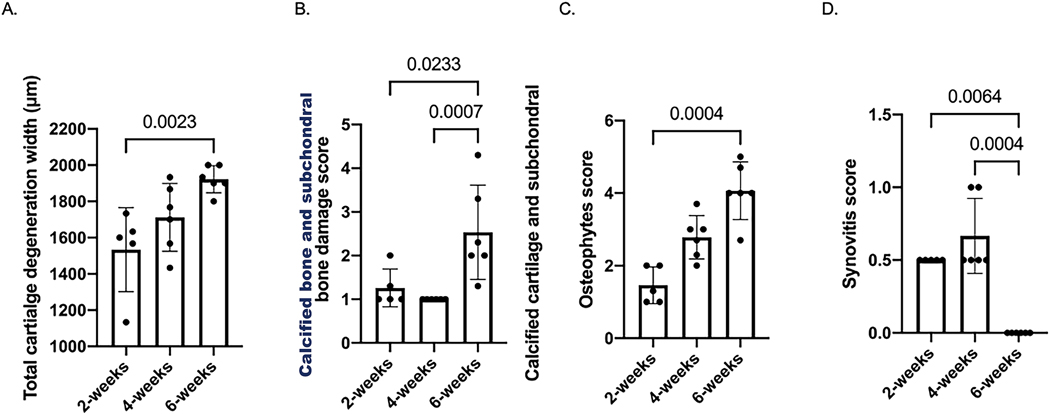
Cartilage degeneration lesions were more severe in knee joints at 6 weeks than those at 2 weeks (Fig. 1A). Knee joints at 6 weeks post-surgery displayed a significantly higher calcified cartilage and subchondral bone damage score than those at 2 and 4 weeks (Fig. 1B). Osteophytes scores were significantly increased from 2 to 6 weeks post-surgery (Fig. 1C). Synovitis scores were only modestly elevated and knee joints at 6 weeks post-surgery showed a significant reduction in synovitis score compared to both 2 and 4 weeks (Fig. 1D). Representative histology images are included in Suppl. Figure 1.
Fig. 1. OARSI scoring of histological sections of knees with MMT.
A: Total cartilage degeneration width. B: Calcified cartilage and subchondral bone damage score. C: Osteophyte score. D: Synovitis score.
Differentially expressed genes in cartilage
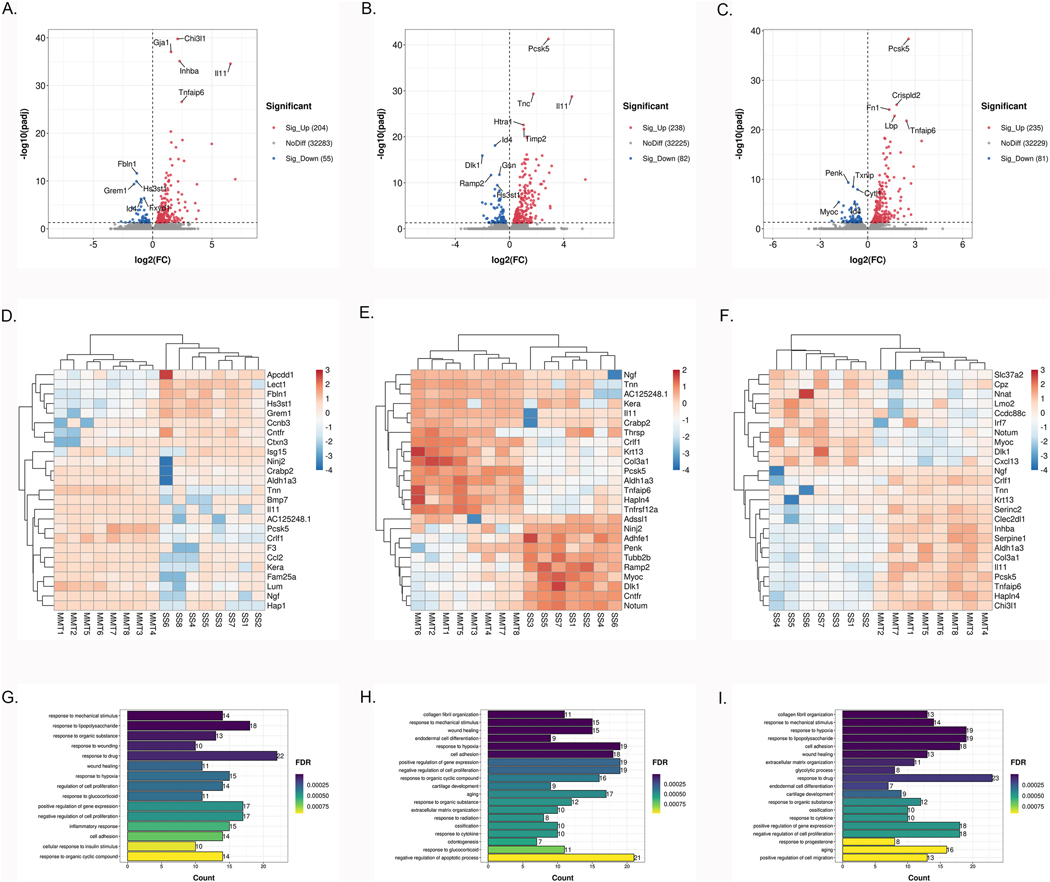
We first determined the genes that were differentially expressed (DE) in the knee cartilage between SS and MMT surgery at 2, 4 and 6 weeks post-surgery. Principal component analysis (PCA) showed good separation between sham and MMT at all 3 time points (Supplementary Fig. 2A–C). As presented in the volcano plot (Fig. 2A), 259 genes were significantly DE between SS and MMT cartilage with 204 being upregulated and 55 downregulated at 2 weeks (Full results in Supplementary Table 1). Among the 259 DE genes, 31 had a log2FC of ⩾ 2 (29 upregulated and 2 downregulated). The most significantly upregulated gene was Chi3l1 (log2FC 2.10, padj 1.76*10−40), encoding human glycoprotein of cartilage (YKL-40), which is a major secretory product of chondrocytes and synovial cells and a biomarker of cartilage turnover and synovitis18. As presented in the volcano plot (Fig. 2B), 238 genes were upregulated and 82 downregulated at 4 weeks (Full results in Supplementary Table 1). Ordered by adjusted p value, the top five upregulated or downregulated DEGs were Pcsk5, Tnc, Il11, Htra1, Timp2, and Id4, Dlk1, Gsn, Ramp2, Hs3st1, respectively. At 6 weeks, there were 316 DEGs between SS and MMT cartilage. As presented in the volcano plot (Fig. 2C), 235 genes were upregulated in the MMT cartilage, while 81 were downregulated (Full results in Supplementary Table 1). The most significant upregulated genes were Pcsk5, Crispld2, Fn2, Lbp and Tnfaip6, while the most downregulated genes were Penk, Txnip and Cytl1. The heatmap of 15 upregulated and 10 downregulated genes with the highest fold change in cartilage at all 3 time points are shown in Fig. 2D–F. These individual gene levels analyses show that the largest upregulation is for ECM-related genes but also for Ngf and Il11 at 2, 4 and 6 weeks post-surgery.
Fig. 2. DEGs and biological processes in articular cartilage between sham-operated and MMT-operated knees.
A–C: Volcano plot of genome-wide mRNA expression between two groups at 2 (A), 4 (B), 6 (C) weeks post-surgery (padj<0.05). D–F: Heatmap of 25 DEGs with the highest fold change in articular cartilage at 2 (D), 4 (E), 6 (F) weeks post-surgery. G–I: The top biological processes for up-regulated DEGs between two groups at 2 (G), 4 (H), 6 (I) weeks post-surgery.
Pathways and biological processes in cartilage
We explored whether the DEGs were enriched for particular pathways or processes, using DAVID. The results at 2 weeks demonstrated significantly enriched Gene Ontology (GO) terms regarding processes involved in ‘response to mechanical stimulus’ (G0:0009612; 14 genes) (FDR 1.50*10−7), characterized by upregulation of Ngf (log2FC 4.98, padj 1.70*10−18) and Ccl2 (log2FC 3.87, padj 1.60*10−4). GO analysis revealed the biological processes upregulated at 4 weeks were related to ‘collagen fibril organization’ (G0:0030199; 11 genes) (FDR 4.94*10−8), characterized by enhancement of Col3a1 (log2FC 2.28, padj 1.95*10−10) and Lum (log2FC 1.88, padj 8.64*10−6) and ‘response to mechanical stimulus’ (G0:0009612; 15 genes) (FDR 4.94*10−8), characterized by upregulation of Ngf (log2FC 2.92, padj 6.68*10–13), Chi3l 1 (log2FC 2.12, padj 2.89*10−7) and Tnc - (log2FC 1.75, padj 4.29*10−30). In contrast, the biological processes downregulated at 4 weeks post-surgery were related to ‘osteoblast differentiation’ (G0:0001649; 9 genes) (FDR 1.31*10−5), characterized by depletion of Myoc (log2FC −1.77, padj 3.80*10−6) and Penk (log2FC −1.70, padj 6.62*10−3). Finally, GO analysis revealed the biological processes upregulated at 6 weeks post-surgery were ‘collagen fibril organization’ (13 genes) (FDR 7.03*10−11), characterized by enhancement Col3a1 (log2FC 2.03, padj 9.37*10−9) and Tnxb (log2FC 1.66, padj 3.89*10−11), ‘response to mechanical stimulus’ (14 genes) (FDR 4.54*10−7), characterized by upregulation of Ngf (log2FC 2.73, padj 6.96*10−10), Chi3l1 (log2FC 2.30, padj 5.90*10−11), Col3a1 (log2FC 2.03, padj 9.37*10–9) and Fos (log2FC 1.41, padj 9.77*10–3) and ‘response to hypoxia’ (G0:0001666; 19 genes) (FDR 4.55*10−6), characterized by expression of Mmp3 (log2FC 1.42, padj 2.68*10−18), Nr4a2 (log2FC 1.32, padj 1.18*10−3) and Serpina1 (log2FC 1.20, padj 3.68*10−15). The only downregulated biological process was ‘osteoblast differentiation’ (8 genes) (FDR 4.52*10−4), characterized by depletion of Myoc (log2FC −1.83, padj 4.09*10−6) and Penk (log2FC −1.23, padj 4.69*10−10). The top upregulated biological processes (FDR<0.001) are illustrated in Fig. 2G–I. The top biological processes (FDR<0.001) and KEGG pathways (FDR<0.05) are listed in Supplementary Table 2.
Differentially expressed genes in meniscus
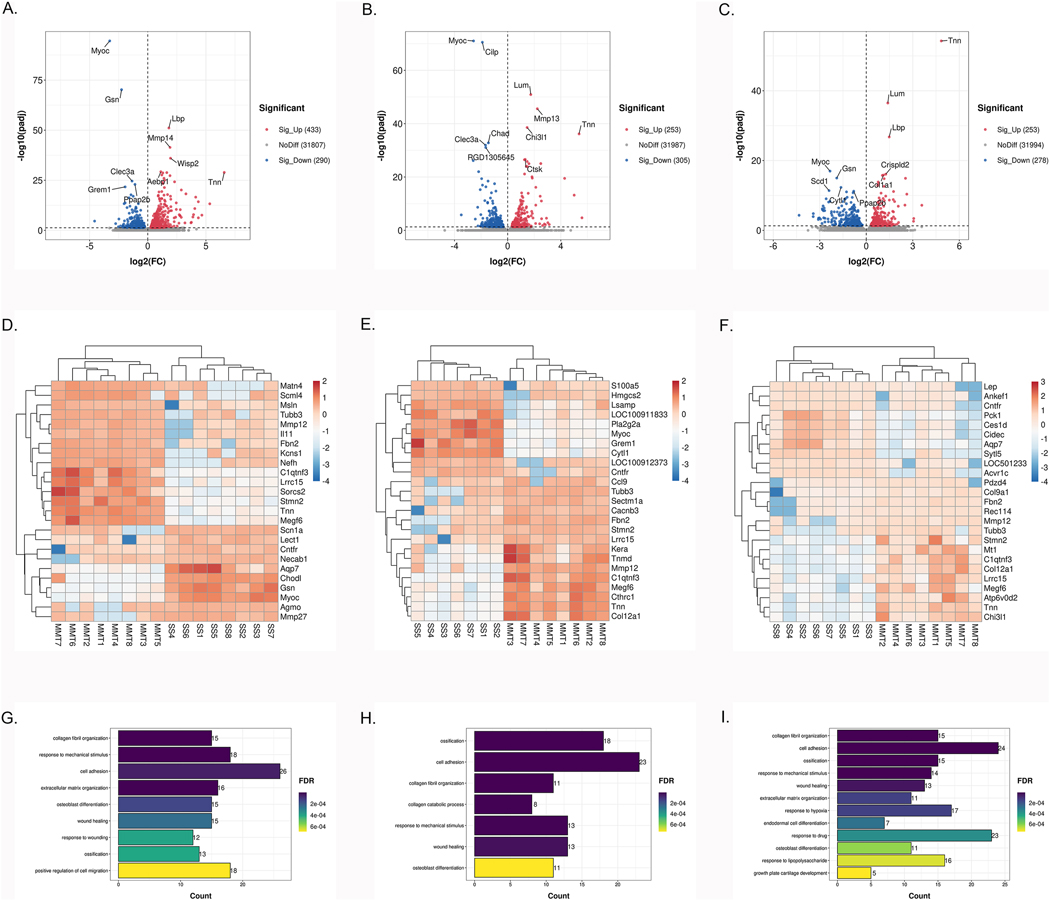
We next determined the genes that were DE in the knee meniscus between SS and MMT at 2, 4 and 6 weeks post-surgery. PCA showed good separation between SS and MMT at all 3 time points (Supplementary Fig. 2D–F). A total of 723 DEGs were observed in the SS versus MMT menisci at 2 weeks with 433 genes upregulated, and 290 genes downregulated in MMT menisci (Fig. 3A, full results in Supplementary Table 3). The most significantly upregulated genes were Lbp (log2FC 1.82, padj 7.97*10−52), Mmp14 (log2FC 1.92, padj 3.92*10−42) and Wisp2 (log2FC 1.96, padj 9.94*10−37), while the most significantly downregulated genes were Myoc (log2FC - 3.27, padj 2.82*10−95), Gsn (log2FC −2.25, padj 6.40*10−71) and Clec3a (log2FC −1.35, padj 2.41*10−25). At 4 weeks there were 558 DEGs (253 upregulated and 305 downregulated) (Fig. 3B, full results in Supplementary Table 3). The most significantly upregulated genes were Lum (log2FC 1.74, padj 1.23*10−51), Mmp13 (log2FC 2.23, padj 2.61*10−46) and Chi3l1 (log2FC 1.46, padj 2.81*10−39). The most significantly downregulated genes were Myoc (log2FC −2.57, padj 9.62*10−72), Cilp (log2FC −1.90, padj 3.02*10−71) and Chad (log2FC −1.45, padj 1.59*10−33). At 6 weeks, we found 531 significant DEGs between the SS and MMT meniscus (Fig. 3C, full results in Supplementary Table 3). The top significantly upregulated genes were Tnn (log2FC 4.84, padj 5.26*10−55), Lum (log2FC 1.39, padj 3.17*10−37) and Lbp (log2FC 1.48, padj 1.72*10−27), while the downregulated genes were Myoc (log2FC −2.34, padj 1.06*10−17), Gsn (log2FC −1.90, padj 1.03*10−15) and Cytl1 (log2FC −1.63, padj 4.88*10−13). The heatmap of 15 upregulated and 10 downregulated genes with the highest fold change in meniscus at the 3 time points are shown in Fig. 3D–F.
Fig. 3. DEGs and biological processes in meniscus between sham-operated and MMT-operated knees.
A-C: Volcano plot of genome-wide mRNA expression between two groups at 2 (A), 4 (B), 6 (C) weeks post-surgery (padj<0.05). D–F: Heatmap of 25 DEGs with the highest fold change in menisci at 2 (D), 4 (E), 6 (F) weeks post-surgery. G–I: The top biological processes for up-regulated DEGs between two groups at 2 (G), 4 (H), 6 (I) weeks post-surgery.
Pathways and biological processes in meniscus
GO analysis revealed that the biological processes upregulated in MMT menisci were ‘collagen fibril organization’ (15 genes) (FDR 1.38*10−10), ‘ossification’ (G0:0001503 18 genes) (FDR 1.26*10−11) and ‘collagen fibril organization’ (15 genes) (FDR 8.63*10−14) at 2 (Fig. 3G), 4 (Fig. 3H) and 6 (Fig. 3I) weeks, respectively (Full results in Supplementary Table 4). In contrast, the biological processes downregulated in MMT menisci were ‘brown fat cell differentiation’ (GO:0050873 8 genes) (FDR 8.21*10−4), ‘angiogenesis’ (G0:0001525 16 genes) (FDR 2.84*10−4) and ‘response to drug’ (G0:0042493 26 genes) (FDR 1.12*10−4) at 2, 4 and 6 weeks, respectively (Supplementary Table 4). KEGG further confirmed the enrichment of genes for ECM-receptor interaction, focal adhesion, regulation of lipolysis in adipocytes, and vascular smooth muscle contraction at each of the respective time points (Supplementary Table 4). The top biological processes (FDR<0.001) and KEGG pathways (FDR<0.05) are listed in Supplementary Table 4.
Differentially expressed genes in synovium
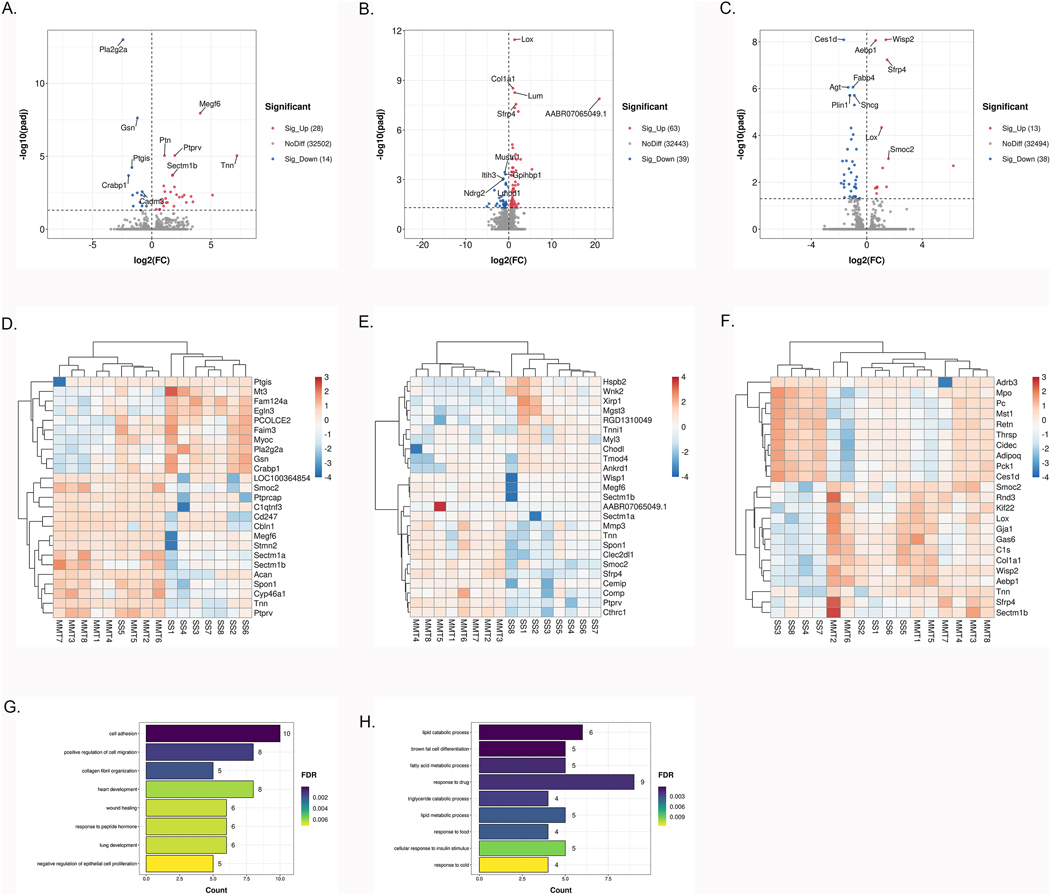
Finally, we determined the genes that were DE in the knee synovium between SS and MMT at 2, 4 and 6 weeks post-surgery. PCA showed good separation between SS and MMT at all 3 time points (Supplementary Fig. 2G–I). At 2 weeks, we identified 42 DEGs in synovium, with 28 genes upregulated and 14 genes downregulated (Fig. 4A, full results in Supplementary Table 5). At 4 weeks, among the 102 DEGs, 63 genes were upregulated and 39 were downregulated in MMT synovium compared with SS synovium (Fig. 4B, full results in Supplementary Table 5). At 6 weeks, we identified 13 upregulated DEGs and 38 downregulated DEGs (Fig. 4C, Supplementary Table 5). Tnn was the most upregulated gene at 2 and 6 weeks and the second most upregulated gene at 4 weeks. Notably, besides Tnn only few known inflammation genes were upregulated (C1s at 4 weeks). The heatmap of 15 upregulated and 10 downregulated genes with the highest fold change in synovium at all 3 time points are shown in Fig. 4D–F.
Fig. 4. DEGs and biological processes in synovium between sham-operated and MMT-operated knees.
A-C: Volcano plot of genome-wide mRNA expression between two groups at (A), 4 (B), 6 (C) weeks post-surgery (padj<0.05). D–F: Heatmap of 25 DEGs with the highest fold change in synovium at 2 (D), 4 (E), 6 (F) weeks post-surgery. G–I: The top biological processes for up-regulated DEGs between two groups at 2 (G), 4 (H), 6 (I) weeks post-surgery.
Pathways and biological processes in synovium
KEGG and GO functional enrichment analyses revealed that genes upregulated in MMT synovium at 4 weeks were enriched in ‘ECM-receptor interaction’ (rno:04512 6 genes) (FDR 1.34*10−3) and ‘cell adhesion’ (GO:0007155; 10 genes) (FDR 8.67*10−5) (Fig. 4G, full results in Supplementary Table 6). GO biological process revealed that downregulated genes in MMT synovium at 6 weeks were associated with ‘lipid catabolic process’ (Fig. 4H, full results in Supplementary Table 6). Furthermore, the KEGG pathway analysis of downregulated genes at 6 weeks were significantly enriched in the ‘PPAR signaling pathway’ (rno:03320; 7 genes) (FDR 1.73*10−5) and ‘AMPK signaling pathway’ (rno:04152; 7 genes) (FDR 1.74*10−4). The top biological processes (FDR<0.01) and KEGG pathways (FDR<0.05) are listed in Supplementary Table 6.
Continuous DE gene expression changes during OA development in each tissue
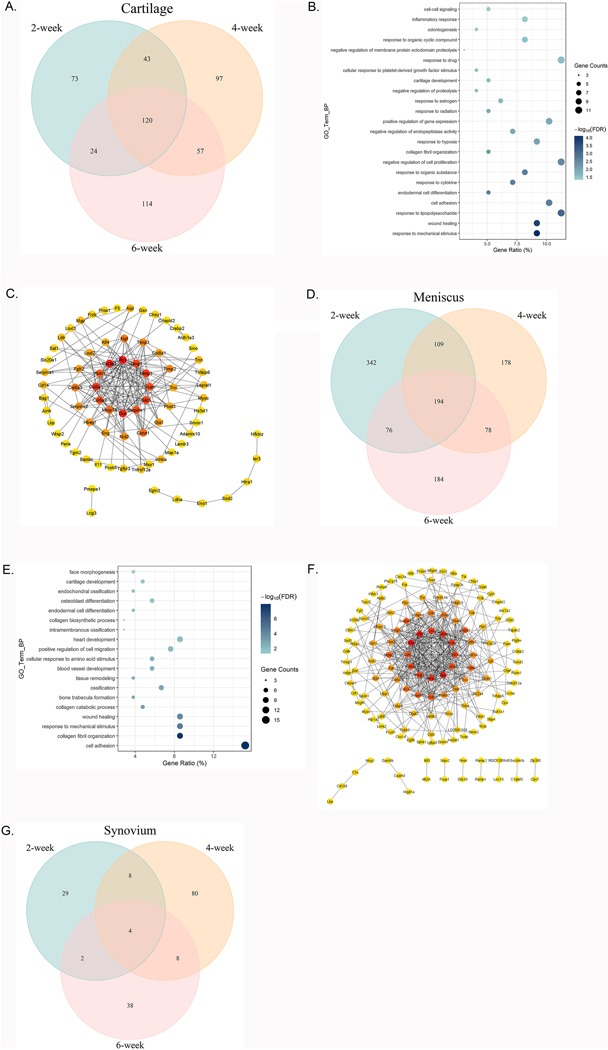
To investigate the genes that were differentially expressed at all stages of OA progression, we intersected the DEG lists from 2, 4 and 6 weeks post-surgery for each tissue type (Supplementary Tables 1, 3 and 5). In cartilage from knees with MMT, intersection of these three lists revealed 120 genes were differentially expressed at all three time points (Fig. 5A). Of these, 99 genes were upregulated and 21 were downregulated. GO pathway analysis revealed 23 biological processes, including those commonly implicated in OA development (Fig. 5B). The most abundant changes were in upregulated pathways related to the ‘response to mechanical stimulus’ (9 genes, FDR 7.84*10−5), ‘wound healing’ (9 genes, FDR 1.74*10−4) and ‘response to lipopolysaccharide’ (11 genes, FDR 7.82*10−4). Additionally, the downregulated genes at all points were only significantly enriched for the terms ‘osteoblast differentiation’ (5 genes, FDR 0.003) and ‘positive regulation of gene expression’ (6 genes, FDR 0.008). KEGG pathway analysis indicated that genes upregulated in MMT knees at the three time points were significantly enriched in ‘ECM-receptor interaction’ (9 genes, FDR 5.77*10−6), ‘focal adhesion’ (10 genes, FDR 2.25*10−4), ‘PI3K-Akt signaling pathway’ (10 genes, FDR 0.007), ‘amoebiasis’ (6 genes, FDR 0.015) and ‘complement and coagulation cascades’ (5 genes, FDR 0.020). Network analysis was performed using STRING and Cytoscape. Each node represents a protein and the edges their regulatory relationships. Multi-edged nodes are described as hubs. We observed 201 edges between 120 nodes (Fig. 5C). The interactions revealed hubs that are placed at the center of the network. Fn1 was the node with the highest number of edges, followed by Col3a1, Dcn, Col5a1, Mmp3, Sdc1 and Timp1.
Fig. 5. DEG, biological processes and protein–protein interactions in cartilage, meniscus, and synovium.
A: Comparison of DEGs in articular cartilage of MMT-operated knees versus sham-operated knees among three time points post-surgery. B: Gene oncology functional clustering of DEGs commonly changed at each time point for biological processes. Node color represents FDR as indicated by the color bar. Node size represents gene counts. C: Network representation of the protein-protein interactions between genes that were significantly regulated at each time point. Only genes with at least one interaction partner are represented. Nodes are colored according to the degree. Genes with no identified interactions are excluded from the graph. D: Comparison of DEGs in meniscus of MMT-operated knees versus sham- operated knees among three time points post-surgery. E: Gene oncology functional clustering of DEGs commonly changed at each time point for biological processes. Node color represents FDR as indicated by the color bar. Node size represents gene counts. F: Network representation of the protein-protein interactions between genes that were significantly regulated at each time point. Only genes with at least one interaction partner are represented. Nodes are colored according to the degree. Genes with no identified interactions are excluded from the graph. G: Comparison of DEGs in the synovium of MMT-operated knees versus sham-operated knees at 62, 4, and 6 weeks post-surgery.
We then performed the same intersection analysis in meniscus, which revealed 194 genes that were significantly changed across all 3 time points (Fig. 5D). Of these, 105 genes were upregulated and 89 were downregulated. As in cartilage, functional analysis was carried out for these DEGs. GO analysis revealed the biological processes that were upregulated at all stages in OA progression included ‘cell adhesion’, ‘collagen fibril organization’, ‘response to mechanical stimulus’, ‘wound healing’ and ‘collagen catabolic process’ (Fig. 5E). However, there was only one significantly downregulated biological process – ‘regulation of blood pressure’. KEGG also showed enrichment related to ‘ECM-receptor interaction’ (12 genes, FDR 1.13*10−10), ‘focal adhesion’ (11 genes, FDR 7.57*10−6), ‘protein digestion and absorption’ (8 genes, FDR 1.39*10−5) and ‘PI3K-Akt signaling pathway’ (11 genes, 2.87*10−4). We then performed network analysis as described above. This analysis showed 451 edges between 193 nodes (Fig. 5F). We found Col1a1, Col3a1, Bgn, Col5a1, Mmp2, Lox and Acan occupy central positions in the interaction network.
Finally, we performed the intersection analysis in synovium from MMT knees, which revealed only 4 genes were significantly changed at all time points, including Tnn, Smoc2, Sectm1b and Lox (Fig. 5G).
Time-specific gene expression changes during OA development in each tissue and in all joint tissues
Tnn was the only gene that was DE in all 3 tissues at all 3 time points (Table 1).
Table 1.
Summary of continuously overlapping genes among three tissues and overlapping genes among three tissues at each time point.
| Time points | Tissue | Overlapping genes |
|---|---|---|
| 2, 4 and 6 weeks | Cartilage, meniscus and synovium | Tnn |
| Cartilage and meniscus | Tnn, Crabp2, Crlf1, Tnfaip6, Serpine1, Chi3l 1, Lbp, Tnc, Col3a1, Timp1, Gja1, Mt2A, Sdc1, Crispld2, Tnfrsf12a, Chsy1, Wisp2, Clec2dl1, Col5a1, Sod2, Htra1, C1qtnf5, Tspan2, Rab15, Col6a3, Col8a1, Mmp3, Col5a2, Pmepa1, Mmp14, Vcan, Aebp1, Txnip, Fxyd1, Gsn, Frzb, Id4, Cytl1, Myoc, Hs3st1 | |
| Meniscus and synovium | Tnn, Lox, Sectm1b | |
| Cartilage and synovium | Tnn | |
| 2 weeks | Cartilage, meniscus and synovium | Tnn, Ptn, Crlf1, Mmp14, Gsn, Myoc |
| 4 weeks | Cartilage, meniscus and synovium | Tnn, Mmp3, Sfrp4, Clec2dl1, Wisp2, Lum, Lox, Tnfaip6, Col12a1, Gja1, C1s, Mmp2, Mmp14, Bgn, Vcan, Hif1a, Timp1, Aebp1, Col5a1, Nbl1, Col6a3 |
| 6 weeks | Cartilage, meniscus and synovium | Tnn, Sectm1b, Wisp2, Lox, Gja1, Aebp1, Pla2g16, Myoc, Nnat |
At 2 weeks, there were 6 genes (Tnn, Ptn, Crlf1, Mmp14, Gsn and Myoc) that were DE in all three tissues, 21 at 4 weeks and 9 at 6 weeks (Tnn, Sectm1b, Wisp2, Lox, Gja1, Aebp1, Pla2g16, Myoc and Nnat). By comparing the DEGs in cartilage and meniscus, we found 40 genes shared by these two tissues at all time points (Table 1). At 2 weeks, there were 108 overlapping genes between cartilage and meniscus, 32 genes between meniscus and synovium, and 7 genes between cartilage and synovium.
At 4 weeks, there were 103 overlapping genes between cartilage and meniscus, 56 genes between meniscus and synovium, and 23 genes between cartilage and synovium.
At 6 weeks, 110 overlapping genes were DE between cartilage and meniscus, which were enriched in ‘collagen fibril organization’ (12 genes, FDR 2.65*10−13), ‘extracellular matrix organization’ (10 genes, FDR 2.30*10−6), ‘response to mechanical stimulus’ (10 genes, FDR 2.85*10−6), ‘cell adhesion’ (13 genes, FDR 2.25*10−6) and ‘wound healing’ (10 genes, FDR 8.25*10−6), 45 genes between meniscus and synovium, which were enriched in ‘response to drug’ (11 genes, FDR 3.93*10−4), ‘fatty acid metabolic process’ (5 genes, FDR 0.005), ‘lipid catabolic process’ (5 genes, FDR 0.006) and ‘triglyceride catabolic process’ (4 genes, FDR 0.006), and 10 genes between cartilage and synovium (Tnn, Sectm1b, Wisp2, Lox, Gja1, Aebp1, Pla2g16, Myoc, Enpp2 and Nnat).
Supplementary Table7 shows biological processes that are not only shared by multiple time points and tissues, but also those that are tissue- and time-specific.
Shared DE genes in the rat MMT model with a prior DNA array studies of rat MMT
To compare and validate the present findings, we used data from a prior study by Mitchell et al. that was methodologically most similar to the present study, which identified 475 DEGs in the cartilage of rat MMT at one or more time points when compared with the time-matched sham control group19. We compared these DEGs with the 120 genes differentially expressed in cartilage at all 3 time points in our study. There were 34 genes commonly changed between two studies (Table 2). When we compared this to the DEGs in cartilage only at week 6 in our study, there were 57 genes involving ‘response to lipopolysaccharide’, ‘response to cytokine’ and ‘response to mechanical stimulus’ (Table 2).
Table 2.
Summary of overlapping DEGs in cartilage of present study and a prior study by Mitchell et al.
| Gene list | Biological process (FDR<0.01) | Hits | FDR | |
|---|---|---|---|---|
| Continuous DEGs in cartilage at all time points and study by Mitchell et al. | Tnn, Il11, Crabp2, Pcsk5, Crlfl, Aldh1a3, Tnfaip6, Inhba, Serpinel, Chi3l1, Lbp, Serpinal, Tnc, Col3a1, Cdh13, Cd14, Tnfrsf12a, Tgm2, Ldlr, Wisp2, Tektl, Col5a1, Sod2, Timp3, Tspan2, Junb, Tesc, Klf4, Mmp3, Mmp14, Loxl2, Ccndl, Tgfbr3, Myoc | response to mechanical stimulus | 6 | 6.67*10−4 |
| response to hypoxia | 7 | 0.002 | ||
| response to lipopolysaccharide | 7 | 0.002 | ||
| response to cytokine | 5 | 0.004 | ||
| response to estrogen | 5 | 0.005 | ||
|
| ||||
| DEGs in cartilage at 6 weeks and study by Mitchell et al. | Il11, Tnn, Pcsk5, Tnfaip6, Chi3l1, Crlf1, Inhba, Aldh1a3, Col3a1, Serpine1, Tekt1, Lbp, Tnfrsf12a, Crabp2, Mmp3, Tesc, Fos, Wisp2, Timp3, Serpina1, Loxl2, Pdgfc, Cd14, Slc6a12, Cdh13, Ldlr, Sod2, Tspan2, Ccnd1, Col5a1, Junb, Sqle, Tgm2, Trpv4, Col6a2, Egfl6, Cyr61, Tnc, Ptn, Slc2a1, Tnfrsf21, P4ha2, Mmp14, Klf4, Blvrb, RT1, Adhfel, Ramp2, Myoc | response to lipopolysaccharide | 10 | 1.30*10−4 |
| response to cytokine | 7 | 1.99*10−4 | ||
| response to mechanical stimulus | 7 | 1.99*10−4 | ||
| response to organic substance | 7 | 0.001 | ||
| positive regulation of gene expression | 9 | 0.002 | ||
| response to hypoxia | 8 | 0.002 | ||
| response to estradiol | 7 | 0.004 | ||
| positive regulation of cell migration | 7 | 0.004 | ||
Shared DE genes in the rat MMT model and in human OA
We previously performed RNA-seq analysis on human knee articular cartilage isolated from normal (n=18) and OA (n=20) donors, in which we identified 1332 DEGs16. To determine similarities with the rat MMT, we compared the 120 genes that were differentially expressed at all 3 timepoints in rat cartilage to the 1332 DEGs identified in the OA patients. This comparison resulted in 46 genes commonly altered in rat MMT model and OA patients. 36 of these 46 genes shared the same expression pattern (up vs downregulated) in the MMT model and in human OA (Table 3). Pathway analysis of these 36 genes revealed 4 enriched biological processes, including ‘response to mechanical stimulus’ (Col3a1, Cited2, Tnc, Txnip, Timp3 and Ngf; FDR 8.65*10−4), ‘wound healing’ (Col3a1, Serpinel, Tnc, Timpl and Eng; FDR 0.026), ‘negative regulation of endopeptidase activity’ (Serpine2, Serpine1, Timp3, Timp1 and Ngf; FDR 0.032) and ‘negative regulation of catalytic activity’ (Txnip, Timp3, Timp1 and Cd55; FDR 0.038). Furthermore, we identified 16 DEGs commonly changed in the cartilage and meniscus in MMT model and human OA cartilage (Crlf1, Tnfaip6, Serpine1, Tnc, Col3a1, Timp1, Col5a1, Htra1 and Tspan2).
Table 3.
DEGs shared by the articular cartilage of the rat MMT models at 2, 4 and 6 weeks and human OA cartilage.
| Rat cartilage | Human cartilage | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 2 weeks | 4 weeks | 6 weeks | |||||||
| SYMBOL | log2(FC) | padj | log2(FC) | padj | log2(FC) | padj | SYMBOL | log2(FC) | adj.P.Val |
| Il1 1 | 6.560 | 2.67E-35 | 4.595 | 1.65E-29 | 3.404 | 1.81E-18 | IL11 | 2.990 | 1.50E-02 |
| Ngf | 4.981 | 1.70E-18 | 2.924 | 6.68E-13 | 2.734 | 6.96E-10 | NGF | 2.781 | 1.09E-02 |
| Crlfl | 2.510 | 2.70E-19 | 2.405 | 1.20E-16 | 2.291 | 2.88E-03 | CRLF1 | 2.139 | 5.12E-05 |
| Tnfaip6 | 2.452 | 2.60E-27 | 2.142 | 1.54E-14 | 2.447 | 1.49E-22 | TNFAIP6 | 4.384 | 9.77E-07 |
| Serpinel | 2.181 | 2.55E-15 | 1.917 | 2.50E-13 | 1.947 | 2.63E-08 | SERPINE1 | 1.424 | 3.73E-02 |
| Lamb3 | 2.003 | 3.84E-03 | 1.534 | 3.68E-03 | 1.301 | 3.98E-02 | LAMB3 | 1.240 | 7.25E-03 |
| Tnc | 1.894 | 3.01E-04 | 1.754 | 4.29E-30 | 0.757 | 1.47E-02 | TNC | 3.646 | 4.07E-05 |
| Col3a1 | 1.647 | 5.23E-14 | 2.280 | 1.95E-10 | 2.032 | 9.37E-09 | COL3A1 | 2.731 | 1.75E-04 |
| Timpl | 1.577 | 8.32E-19 | 1.268 | 7.40E-17 | 0.905 | 5.27E-06 | TIMP1 | 1.575 | 2.69E-02 |
| Tgm2 | 1.349 | 1.11E-10 | 1.384 | 5.21E-09 | 0.866 | 2.01E-03 | TGM2 | 1.452 | 2.86E-03 |
| Tppp3 | 1.304 | 6.79E-05 | 1.154 | 1.46E-07 | 1.183 | 3.89E-05 | TPPP3 | 3.061 | 5.43E-05 |
| Nid2 | 1.245 | 4.90E-08 | 0.934 | 1.91E-04 | 1.069 | 8.92E-04 | NID2 | 1.174 | 1.69E-02 |
| Vwa1 | 1.224 | 8.99E-07 | 1.480 | 2.29E-12 | 1.305 | 6.12E-13 | VWA1 | 1.028 | 6.89E-03 |
| Serpine2 | 1.172 | 2.07E-12 | 0.929 | 8.39E-15 | 0.988 | 6.06E-16 | SERPINE2 | 2.132 | 3.71E-05 |
| Col5a1 | 1.130 | 1.09E-08 | 1.221 | 8.26E-16 | 0.919 | 5.76E-07 | COL5A1 | 1.680 | 2.71E-05 |
| Timp3 | 1.113 | 5.24E-04 | 1.071 | 8.08E-10 | 1.212 | 2.50E-11 | TIMP3 | 1.140 | 8.94E-05 |
| Htra1 | 1.099 | 5.29E-08 | 1.028 | 2.66E-23 | 1.163 | 1.59E-13 | HTRA1 | 2.712 | 1.43E-07 |
| Eng | 1.059 | 4.31E-07 | 0.912 | 3.80E-06 | 0.787 | 9.00E-03 | ENG | 1.066 | 1.71E-02 |
| Tspan2 | 1.030 | 4.92E-10 | 1.157 | 1.37E-12 | 0.988 | 6.86E-06 | TSPAN2 | 1.894 | 1.51E-04 |
| Col6a3 | 0.955 | 3.90E-07 | 0.971 | 5.73E-12 | 0.936 | 4.70E-10 | COL6A3 | 1.280 | 1.96E-02 |
| F5 | 0.936 | 4.28E-03 | 0.815 | 2.36E-02 | 0.755 | 1.24E-02 | F5 | 1.334 | 3.40E-03 |
| Col8a1 | 0.876 | 9.03E-03 | 1.324 | 4.48E-10 | 1.421 | 7.77E-07 | COL8A1 | 2.591 | 8.76E-03 |
| Cd55 | 0.776 | 1.01E-02 | 0.871 | 2.61E-03 | 0.806 | 2.38E-02 | CD55 | 1.828 | 2.58E-06 |
| Col5a2 | 0.766 | 1.62E-04 | 0.967 | 3.15E-13 | 0.752 | 7.54E-05 | COL5A2 | 1.947 | 2.03E-04 |
| Mapla | 0.765 | 4.39E-02 | 0.555 | 7.49E-03 | 0.540 | 2.61E-03 | MAP1A | 1.200 | 3.98E-04 |
| Cxcl14 | 0.706 | 1.54E-07 | 0.773 | 5.73E-12 | 1.095 | 6.49E-19 | CXCL14 | 2.368 | 3.07E-02 |
| Ccndl | 0.673 | 8.27E-04 | 0.864 | 3.17E-07 | 0.927 | 2.32E-09 | CCND1 | 2.102 | 2.29E-06 |
| Prosi | 0.633 | 3.28E-02 | 0.693 | 2.26E-03 | 0.733 | 2.32E-02 | PROS1 | 1.087 | 5.46E-04 |
| Aebpi | 0.601 | 2.37E-02 | 0.903 | 4.20E-07 | 0.865 | 6.08E-08 | AEBP1 | 1.403 | 3.37E-05 |
| Smoci | 0.558 | 2.24E-02 | 0.671 | 4.73E-03 | 0.795 | 1.72E-03 | SMOC1 | 1.812 | 1.14E-04 |
| Txnip | −0.552 | 1.96E-02 | −0.637 | 2.54E-03 | −0.926 | 3.22E-09 | TXNIP | −2.311 | 3.79E-05 |
| Klf15 | −0.691 | 2.93E-02 | −0.938 | 6.82E-03 | −0.930 | 4.90E-03 | KLF15 | −2.241 | 1.15E-04 |
| Cited2 | −0.711 | 1.24E-04 | −0.633 | 2.25E-04 | −0.566 | 5.88E-04 | CITED2 | −1.655 | 3.59E-05 |
| Cytll | −1.072 | 4.44E-02 | −0.783 | 1.09E-08 | −0.651 | 1.14E-08 | CYTL1 | −1.886 | 1.06E-02 |
| Myoc | −1.087 | 1.10E-04 | −1.766 | 3.80E-06 | −1.834 | 4.09E-06 | MYOC | −3.446 | 4.07E-05 |
| Fblnl | −1.329 | 2.52E-12 | −1.116 | 7.18E-08 | −0.729 | 1.01E-02 | FBLN1 | −1.743 | 2.79E-03 |
| Il11 | 6.560 | 2.67E-35 | 4.595 | 1.65E-29 | 3.404 | 1.81E-18 | IL11 | 2.990 | 1.50E-02 |
| Ngf | 4.981 | 1.70E-18 | 2.924 | 6.68E-13 | 2.734 | 6.96E-10 | NGF | 2.781 | 1.09E-02 |
| Crlfl | 2.510 | 2.70E-19 | 2.405 | 1.20E-16 | 2.291 | 2.88E-03 | CRLF1 | 2.139 | 5.12E-05 |
| Tnfaip6 | 2.452 | 2.60E-27 | 2.142 | 1.54E-14 | 2.447 | 1.49E-22 | TNFAIP6 | 4.384 | 9.77E-07 |
| Serpine1 | 2.181 | 2.55E-15 | 1.917 | 2.50E-13 | 1.947 | 2.63E-08 | SERPINE1 | 1.424 | 3.73E-02 |
| Lamb3 | 2.003 | 3.84E-03 | 1.534 | 3.68E-03 | 1.301 | 3.98E-02 | LAMB3 | 1.240 | 7.25E-03 |
| Tnc | 1.894 | 3.01E-04 | 1.754 | 4.29E-30 | 0.757 | 1.47E-02 | TNC | 3.646 | 4.07E-05 |
| Col3a1 | 1.647 | 5.23E-14 | 2.280 | 1.95E-10 | 2.032 | 9.37E-09 | COL3A1 | 2.731 | 1.75E-04 |
| Timp1 | 1.577 | 8.32E-19 | 1.268 | 7.40E-17 | 0.905 | 5.27E-06 | TIMP1 | 1.575 | 2.69E-02 |
| Tgm2 | 1.349 | 1.11E-10 | 1.384 | 5.21E-09 | 0.866 | 2.01E-03 | TGM2 | 1.452 | 2.86E-03 |
| Tppp3 | 1.304 | 6.79E-05 | 1.154 | 1.46E-07 | 1.183 | 3.89E-05 | TPPP3 | 3.061 | 5.43E-05 |
| Nid2 | 1.245 | 4.90E-08 | 0.934 | 1.91E-04 | 1.069 | 8.92E-04 | NID2 | 1.174 | 1.69E-02 |
| Vwa1 | 1.224 | 8.99E-07 | 1.480 | 2.29E-12 | 1.305 | 6.12E-13 | VWA1 | 1.028 | 6.89E-03 |
| Serpine2 | 1.172 | 2.07E-12 | 0.929 | 8.39E-15 | 0.988 | 6.06E-16 | SERPINE2 | 2.132 | 3.71E-05 |
| Col5a1 | 1.130 | 1.09E-08 | 1.221 | 8.26E-16 | 0.919 | 5.76E-07 | COL5A1 | 1.680 | 2.71E-05 |
| Timp3 | 1.113 | 5.24E-04 | 1.071 | 8.08E-10 | 1.212 | 2.50E-11 | TIMP3 | 1.140 | 8.94E-05 |
| Htra1 | 1.099 | 5.29E-08 | 1.028 | 2.66E-23 | 1.163 | 1.59E-13 | HTRA1 | 2.712 | 1.43E-07 |
| Eng | 1.059 | 4.31E-07 | 0.912 | 3.80E-06 | 0.787 | 9.00E-03 | ENG | 1.066 | 1.71E-02 |
| Tspan2 | 1.030 | 4.92E-10 | 1.157 | 1.37E-12 | 0.988 | 6.86E-06 | TSPAN2 | 1.894 | 1.51E-04 |
| Col6a3 | 0.955 | 3.90E-07 | 0.971 | 5.73E-12 | 0.936 | 4.70E-10 | COL6A3 | 1.280 | 1.96E-02 |
| F5 | 0.936 | 4.28E-03 | 0.815 | 2.36E-02 | 0.755 | 1.24E-02 | F5 | 1.334 | 3.40E-03 |
| Col8a1 | 0.876 | 9.03E-03 | 1.324 | 4.48E-10 | 1.421 | 7.77E-07 | COL8A1 | 2.591 | 8.76E-03 |
| Cd55 | 0.776 | 1.01E-02 | 0.871 | 2.61E-03 | 0.806 | 2.38E-02 | CD55 | 1.828 | 2.58E-06 |
| Col5a2 | 0.766 | 1.62E-04 | 0.967 | 3.15E-13 | 0.752 | 7.54E-05 | COL5A2 | 1.947 | 2.03E-04 |
| Map1a | 0.765 | 4.39E-02 | 0.555 | 7.49E-03 | 0.540 | 2.61E-03 | MAP1A | 1.200 | 3.98E-04 |
| Cxcl14 | 0.706 | 1.54E-07 | 0.773 | 5.73E-12 | 1.095 | 6.49E-19 | CXCL14 | 2.368 | 3.07E-02 |
| Ccnd1 | 0.673 | 8.27E-04 | 0.864 | 3.17E-07 | 0.927 | 2.32E-09 | CCND1 | 2.102 | 2.29E-06 |
| Pros1 | 0.633 | 3.28E-02 | 0.693 | 2.26E-03 | 0.733 | 2.32E-02 | PROS1 | 1.087 | 5.46E-04 |
| Aebp1 | 0.601 | 2.37E-02 | 0.903 | 4.20E-07 | 0.865 | 6.08E-08 | AEBP1 | 1.403 | 3.37E-05 |
| Smoc1 | 0.558 | 2.24E-02 | 0.671 | 4.73E-03 | 0.795 | 1.72E-03 | SMOC1 | 1.812 | 1.14E-04 |
| Txnip | −0.552 | 1.96E-02 | −0.637 | 2.54E-03 | −0.926 | 3.22E-09 | TXNIP | −2.311 | 3.79E-05 |
| Klf15 | −0.691 | 2.93E-02 | −0.938 | 6.82E-03 | −0.930 | 4.90E-03 | KLF15 | −2.241 | 1.15E-04 |
| Cited2 | −0.711 | 1.24E-04 | −0.633 | 2.25E-04 | −0.566 | 5.88E-04 | CITED2 | −1.655 | 3.59E-05 |
| Cytl1 | −1.072 | 4.44E-02 | −0.783 | 1.09E-08 | −0.651 | 1.14E-08 | CYTL1 | −1.886 | 1.06E-02 |
| Myoc | −1.087 | 1.10E-04 | −1.766 | 3.80E-06 | −1.834 | 4.09E-06 | MYOC | −3.446 | 4.07E-05 |
| Fbln1 | −1.329 | 2.52E-12 | −1.116 | 7.18E-08 | −0.729 | 1.01E-02 | FBLN1 | −1.743 | 2.79E-03 |
Discussion
Overview of DEGs
The present study is the first to analyze global transcriptomic profiles in three joint tissues, including cartilage, meniscus and synovium at multiple timepoints following either sham or MMT surgery. The goals of the study were two-fold: 1) to determine temporal patterns of gene expression during OA progression and 2) to reveal both shared and tissue-specific changes in gene expression. Compared to cartilage and synovium, meniscus showed the largest number of DEGs at all time points (2, 4 and 6 weeks post-surgery). Additionally, of the three time points, the DEG count was highest at the 2-week time point. Both observations appear to reflect, at least in part, a response to the surgical meniscus injury. The number of DEGs in cartilage increased from 2 to 4 weeks and then remained largely stable, which is in line with the significantly increasing cartilage degeneration width from 2 to 6 weeks in our study. This is also consistent with histological changes in cartilage becoming detectable by 3–4 weeks9. Synovium showed a much lower number of DEGs which increased from week 2 to week 4 and decreased by week 6. This is highly consistent with the low level of histological changes in this model in our study and prior publications20, 21. It is also notable that there was no inflammation signature in the DEGs. Synovium also showed several enriched and downregulated pathways related to adipose tissues. This is likely due to inclusion of sub-synovial fat tissue during collection of the synovial lining.
Individual DEGs
Largely due to the limited number of DEGs in synovium, the only gene that changed in all tissues at all time points was Tnn (TN-N, also known as Tenascin-W), an extracellular matrix glycoprotein. The expression of Tnn was low in sham control knee joints (Supplementary Tables 1,3,5); however, it showed the largest increase of all genes at 2 weeks post-MMT surgery to levels, similar to those of other tenascins that have previously been reported to be upregulated in OA22, 23. We also found enhancement of Tnc, but depletion of Tnxb in cartilage and meniscus at all time points (except for Tnxb in cartilage at 2 weeks). Our findings are similar to Tnn dysregulation throughout the time course of DMM in mice as reported earlier24. TN-C is also reported to be significantly enhanced in the knee cartilage and synovial fluid in OA patients22, 23. TN-C and TN-N are largely undetectable in adult tissue, but they are often upregulated under pathological stresses, including angiogenesis and wound healing25. The functions of TN-N in OA knee joints are still unclear, but it potentially induces inflammatory mediators and promotes degradation of matrix in OA cartilage22, 23, possibly by activating both caspase-1 and TLR4/NF-kB/NLRP3 pathways26.
Focusing on cartilage and meniscus – which each had a substantial number of DEGs at all timepoints - several other genes of interest were identified in this study including Tnfaip6, Htra1, Aebp1 and Cytl1. Tnfaip6, also known as Tsg-6, encodes a secretory protein that is produced in response to inflammatory mediators, which is thought to play an anti-inflammatory role in arthritis and protect against destruction of joint cartilage27. Htra1 is involved in the degradation of ECM through inducing expression of MMPs and aggrecanases via MEK signaling pathway28. Adipocyte enhancer binding protein 1 (AEBP1) was originally discovered as a transcriptional repressor that binds to the AE-1 element of the ap-2 gene, coding for the fatty acid binding protein (FABP4)29. Through upregulation of sonic Hedgehog and NF-kappa B (NF-kB) pathway, AEBP1 functions as a pro-inflammatory mediator and promoted mammary cell hyperplasia30. Cytl1 is involved mainly in the maintenance of cartilage homeostasis, and loss of Cytl1 function is associated with OA progression31.
We next looked for dysregulation of genes known to be altered in multiple rat OA models. At all time-points, cartilage and meniscus showed increased expression of ECM components (Col3a1, Col5a1, Col5a2, Col6a3, Col8a1), which play important roles in OA pathology32, 33. We also found that the expression of the matrix-degrading enzymes including Mmps (Mmp3, Mmp14), Ctsb and Ctsk were all increased in MMT cartilage and meniscus at two or more time points, which have long been associated with the onset and progression of OA34–36. Further, we identified dysregulation of lipopolysaccharide and cytokine responsive genes (Serpina1, Serpine1, Lbp, Timp1, Timp2), which are commonly changed throughout cartilage degradation induction37–41 and an early microarray data of cartilage in rat OA model42. Several mediators of inflammation were increased (IL-11 in synovium, CCL2 and CCL7 in meniscus, MIF in cartilage) and members of the TNF receptor superfamily (TNFRSF9, TNFRSF21 and TNFRSF26 in cartilage). These results suggest OA-related changes occurred at the transcriptomic level in our model.
Pathways
The present data show downregulation of protective pathways and upregulation of pathogenic pathways during OA progression in all tissue types. We observed the pathways involved in cartilage ECM synthesis and metabolism, including ‘collagen fibril organization’ and ‘extracellular matrix organization’, were significantly enriched in cartilage and meniscus at all time points. The downregulated KEGG pathways in meniscus at 6 weeks include AMPK and PPAR signaling pathway, which have been suggested to play a protective role in OA pathogenesis43, 44. The upregulated KEGG pathways include ECM-receptor interaction, focal adhesion, PI3K-Akt and HIF-1 signaling in cartilage at each time point and meniscus at 6 weeks, which are classical pathways for maintaining the normal cartilage metabolism and are involved in the OA development45, 46. Inflammation is an established pathogenic pathway in OA47. In the present GO and KEGG pathway analyses, the inflammation-related pathways that were upregulated, included ‘TNF signaling pathway’ (cartilage at 2 weeks), ‘response to lipopolysaccharide’ (cartilage at 2, 4, 6 weeks, meniscus at 6 weeks), ‘inflammatory response’ (cartilage at 2 weeks) and ‘complement and coagulation cascade’ (cartilage at 2 and 4 weeks). ynovium did not show upregulation of inflammation-related pathways, in part due to the small number of DEGs. This is consistent with a prior DNA array study of synovium in the rat MMT model where ECM, but not inflammation-related genes were the most differentially expressed48. These results support a role of inflammation, mainly through innate immune mechanisms mainly in cartilage and meniscus and to a lesser extent in synovium.
Mechanical stimulation
‘Response to mechanical stimulus’ was identified as an actively regulated biological process in cartilage and meniscus at all time points in rat MMT model and in human OA cartilage. Interestingly, complete joint immobilization has been shown to protect mice from surgically-induced OA and prevents expression of OA-associated genes. Partial joint immobilization reveals that different types of mechanical load induce different sets of genes41. The key function of cartilage and meniscus is to respond to mechanical loads with changes in metabolic activity by physiological ion homeostasis, especially the mechanically induced Ca2+ influx and activation of the Ca2+-related signaling pathways49. While several channels are expressed in chondrocytes that could affect calcium signaling, two types of mechanosensitive channels, TRPV4, Piezo1 and Piezo2, have been identified in chondrocytes50. We found persistently increased expression of Piezo1 in cartilage at all time points and Trpv4 at 4 and 6 weeks post- MMT. In addition, Piezo2 was upregulated in the MMT meniscus at 2 and 6 weeks. Piezo1 expression was significantly increased in IL-1 stimulated porcine chondrocytes and human osteoarthritic cartilage, which amplified mechanically-induced deformation microtrauma51. Inhibiting Piezos with GsMTx4, a PIEZO-blocking peptide, protected chondrocytes from mechanically induced cell death52. TRPV4 expression and activation by pharmacological agonist increased SOX9 reporter activity in the murine chondrogenic ATDC5 cells53. The loss of TRPV4 resulted in the development of obesity-induced, age- and sex-dependent OA54, 55. Furthermore, TRPV4 may synergize with PIEZOs to regulate chondrocyte response to different magnitudes of strain52. Additionally, we found continuously decreasing expression of stimuli-responsive transcriptional regulator, Cited2, in PTOA cartilage. Cited2 is chondroprotective by suppressing MMP13 in human chondrocytes56, 57. Interestingly, we observed consistent upregulation of Ngf in MMT and human OA cartilage, which has also been documented in cartilage from mice with the meniscus destabilization model of OA58. TRPV4 contributes to the sensation of pain, where TRPV4 sensitization by intracellular signaling leads to pain behaviors in mice59. TRPV1 is involved in the development of NGF-induced persistent mechanical hypersensitivity in rats60. Furthermore, Piezo2 also promotes NGF induced sensitization of afferent neurons in bone61. These findings indicate that the increased expression of mechanoreceptors coupled with the increased NGF expression in the MMT model contributes to the sensitization to mechanical stimulation and related pain behaviors. Collectively, these findings support the hypothesis that early upregulation of PIEZO already 2 weeks after MMT sensitizes to mechano-stimulus plus abnormal loading due to MMT leads to abnormal mechanosignaling and consequent production of OA mediators, including ECM-related factors which would also suggest a potential link between the mechanosignaling and ECM pathways.
Some limitations apply to our study. We employed MMT as only one model of post-traumatic OA in this study rather than primary age-related OA. For economic reasons, we included a sham surgery control group but not a group of animals with normal knees that were not subjected to any surgery. While it has been demonstrated that sham surgery does induce gene expression changes42, 62, the sham surgery is a suitable control as it eliminates genes that are differentially expressed due to the surgical manipulation or altered mechanical loading and not due to the OA-like changes that are induced by MMT. Future studies using a nonsurgical PTOA model, or an aging model would better resolve gene expression differences and eliminate DEGs that are related to the surgical trauma. The bulk RNA sequencing performed here was most likely only able to detect genes with large expression differences. Single cell RNA-sequencing analyses have potential to detect genes whose expression differences are restricted to specific cell subpopulations63. While mechanotransduction is a notable pathway, other pathways, such as extracellular matrix-related pathways or innate immune response are involved independently or by interaction with mechanotransduction.
In conclusion, this study identified overlapping genes that are differentially expressed across three tissues in knee joints and at all stages during OA development, and ‘response to mechanical stimulus’ and ECM pathways are enriched in cartilage and meniscus in the rat MMT model and in human OA cartilage, underscoring their role in OA pathogenesis.
Supplementary Material
Supplementary Figure 1. Histology of rat knees. Representative images are shown for the medial compartment of the knee joint following sham surgery (4 weeks) or MMT (2, 4, 6 weeks post-surgery). No pathological changes are evident on the knee with sham surgery. At 2 weeks post MMT, there is reduced cellularity on the tibial cartilage, and a structural defect in the superficial zone. At 4 weeks post MMT, there is a larger structural defect on the medial tibia, affecting the mid zone and full thickness cartilage clefts. A marginal osteophyte is also apparent. At 6 weeks post MMT, the structural defect on the tibia is wider, the marginal osteophyte is larger, and the medial femur also shows a structural cartilage defect reaching into the mid-zone and complete cell loss in that area. High resolution versions of these images are available as eSlides.
Supplementary Figure 2. Principal component analysis of the data was performed and the first and second (PC1 and PC2) principal components are shown. There are comparisons between MMT and sham controls in cartilage at 2 (A), 4 (B) and 6 (C) weeks, in meniscus at 2 (D), 4 (E), and 6 (F) weeks, and in synovium at 2 (G), 4 (H) and 6 (I) weeks. Red: Sham surgery (SM); Blue: MMT surgery (MT).
Supplementary Table 1. All DEGs in articular cartilage in MMT joints at 2, 4, and 6 weeks. Fold change (log2 scale) values and adjusted P values (padj) are shown in the table relative to sham controls.
Supplementary Table 2. Summary of actively regulated Gene Ontology terms (FDR<0.001) and KEGG pathways (FDR<0.05) of genes differentially expressed in cartilage between MMT and sham control models for each time point.
Supplementary Table 3. All DEGs in meniscus in MMT joints at 2, 4, and 6 weeks. Fold change (log2 scale) values and adjusted P values (padj) are shown in the table relative to sham controls.
Supplementary Table 4. Summary of actively regulated Gene Ontology terms (FDR<0.001) and KEGG pathways (FDR<0.05) of genes differentially expressed in meniscus between MMT and sham control models for each time point.
Supplementary Table 5. All DEGs in synovium in MMT joints at 2, 4, and 6 weeks. Fold change (log2 scale) values and adjusted P values (padj) are shown in the table relative to sham controls.
Supplementary Table 6. Summary of actively regulated Gene Ontology terms (FDR<0.001) and KEGG pathways (FDR<0.05) of genes differentially expressed in synovium between MMT and sham control models for each time point.
Supplementary Table 7. Summary of Gene Ontology terms (FDR<0.001) that are shared by multiple time points and tissues or are tissue-/time-specific.
Acknowledgements:
We thank Dr. Gogce Crynen (Scripps Genomics Core) for her support with data analysis.
Funding:
This study was supported by NIH grants AG049617 and AG059418. Y.H. was supported by a grant from the China Scholarship Council.
Footnotes
Data and materials availability: All data are included in the manuscript and supplemental material. The RNA-sequencing data will be deposited into Gene Expression Omnibus (GEO) upon acceptance of the manuscript.
Competing interests: No financial support or other benefits have been obtained from any commercial sources for this study and the authors declare that they have no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Watt FE. Posttraumatic osteoarthritis: what have we learned to advance osteoarthritis? Curr Opin Rheumatol 2021; 33: 74–83. [DOI] [PubMed] [Google Scholar]
- 2.Loughlin J. Genetics of osteoarthritis. Current Opinion in Rheumatology 2011; 23: 479–483. [DOI] [PubMed] [Google Scholar]
- 3.Driban JB, Harkey MS, Barbe MF, Ward RJ, MacKay JW, Davis JE, et al. Risk factors and the natural history of accelerated knee osteoarthritis: a narrative review. Bmc Musculoskeletal Disorders 2020; 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lotz MK, Carames B. Autophagy and cartilage homeostasis mechanisms in joint health, aging and OA. Nature Reviews Rheumatology 2011; 7: 579–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: A disease of the joint as an organ. Arthritis and Rheumatism 2012; 64: 1697–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashraf S, Mapp PI, Walsh DA. Contributions of Angiogenesis to Inflammation, Joint Damage, and Pain in a Rat Model of Osteoarthritis. Arthritis and Rheumatism 2011; 63: 2700–2710. [DOI] [PubMed] [Google Scholar]
- 7.Bendele AM. Animal models of osteoarthritis. J Musculoskelet Neuronal Interact 2001; 1: 363–376. [PubMed] [Google Scholar]
- 8.Bove SE, Laemont KD, Brooker RM, Osborn MN, Sanchez BM, Guzman RE, et al. Surgically induced osteoarthritis in the rat results in the development of both osteoarthritis-like joint pain and secondary hyperalgesia. Osteoarthritis and Cartilage 2006; 14: 1041–1048. [DOI] [PubMed] [Google Scholar]
- 9.Janusz MJ. Induction of osteoarthritis in the rat by surgical tear of the meniscus: Inhibition of joint damage by a matrix metalloproteinase inhibitor (vol 10, pg 785, 2002) (vol 10, pg 905, 2002). Osteoarthritis and Cartilage 2003; 11: 299–299. [DOI] [PubMed] [Google Scholar]
- 10.Gerwin N, Bendele AM, Glasson S, Carlson CS. The OARSI histopathology initiative - recommendations for histological assessments of osteoarthritis in the rat. Osteoarthritis Cartilage 2010; 18 Suppl 3: S24–34. [DOI] [PubMed] [Google Scholar]
- 11.Smith T, Heger A, Sudbery I. UMI-tools: modeling sequencing errors in Unique Molecular Identifiers to improve quantification accuracy. Genome Research 2017; 27: 491–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, et al. The nf-core framework for community-curated bioinformatics pipelines. Nature Biotechnology 2020; 38: 276–278. [DOI] [PubMed] [Google Scholar]
- 13.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014; 30: 923–930. [DOI] [PubMed] [Google Scholar]
- 15.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 2014; 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisch KM, Gamini R, Alvarez-Garcia O, Akagi R, Saito M, Muramatsu Y, et al. Identification of transcription factors responsible for dysregulated networks in human osteoarthritis cartilage by global gene expression analysis. Osteoarthritis and Cartilage 2018; 26: 1531–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behavioural Brain Research 2001; 125: 279–284. [DOI] [PubMed] [Google Scholar]
- 18.Zivanovic S, Rackov LP, Vojvodic D, Vucetic D. Human cartilage glycoprotein 39- biomarker of joint damage in knee osteoarthritis. International Orthopaedics 2009; 33: 1165–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei T, Kulkarni NH, Zeng QQ, Helvering LM, Lin X, Lawrence F, et al. Analysis of early changes in the articular cartilage transcriptisome in the rat meniscal tear model of osteoarthritis: pathway comparisons with the rat anterior cruciate transection model and with human osteoarthritic cartilage. Osteoarthritis and Cartilage 2010; 18: 992–1000. [DOI] [PubMed] [Google Scholar]
- 20.Kloefkorn HE, Allen KD. Quantitative histological grading methods to assess subchondral bone and synovium changes subsequent to medial meniscus transection in the rat. Connective Tissue Research 2017; 58: 373–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mapp PI, Avery PS, McWilliams DF, Bowyer J, Day C, Moores S, et al. Angiogenesis in two animal models of osteoarthritis. Osteoarthritis and Cartilage 2008; 16: 61–69. [DOI] [PubMed] [Google Scholar]
- 22.Hasegawa M, Yoshida T, Sudo A. Role of tenascin-C in articular cartilage. Modern Rheumatology 2018; 28: 215–220. [DOI] [PubMed] [Google Scholar]
- 23.Hasegawa M, Yoshida T, Sudo A. Tenascin-C in Osteoarthritis and Rheumatoid Arthritis. Frontiers in Immunology 2020; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardiner MD, Vincent TL, Driscoll C, Burleigh A, Bou-Gharios G, Saklatvala J, et al. Transcriptional analysis of micro-dissected articular cartilage in post-traumatic murine osteoarthritis. Osteoarthritis and Cartilage 2015; 23: 616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aubert A, Mercier-Gouy P, Aguero S, Berthier L, Liot S, Prigent L, et al. Latent TGF-beta Activation Is a Hallmark of the Tenascin Family. Frontiers in Immunology 2021; 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu MY, Ye ZS, Zhao X, Guo HZ, Gong XH, Huang RC. Deficiency of tenascin-C attenuated cardiac injury by inactivating TLR4/ NLRP3/caspase-1 pathway after myocardial infarction. Cellular Signalling 2021; 86. [DOI] [PubMed] [Google Scholar]
- 27.Milner CM, Day AJ. TSG-6: a multifunctional protein associated with inflammation. Journal of Cell Science 2003; 116: 1863–1873. [DOI] [PubMed] [Google Scholar]
- 28.Tiaden AN, Klawitter M, Lux V, Mirsaidi A, Bahrenberg G, Glanz S, et al. Detrimental Role for Human High Temperature Requirement Serine Protease A1 (HTRA1) in the Pathogenesis of Intervertebral Disc (IVD) Degeneration. Journal of Biological Chemistry 2012; 287: 21335–21345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majdalawieh AF, Massri M, Ro HS. AEBP1 is a Novel Oncogene: Mechanisms of Action and Signaling Pathways. Journal of Oncology 2020; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holloway RW, Bogachev O, Bharadwaj AG, McCluskey GD, Majdalawieh AF, Zhang L, et al. Stromal Adipocyte Enhancer-binding Protein (AEBP1) Promotes Mammary Epithelial Cell Hyperplasia via Proinflammatory and Hedgehog Signaling. Journal of Biological Chemistry 2012; 287: 39171–39181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu SP, Kuek V, Bennett S, Xu HZ, Rosen V, Xu JK. Protein Cytl1: its role in chondrogenesis, cartilage homeostasis, and disease. Cellular and Molecular Life Sciences 2019; 76: 3515–3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hosseininia S, Weis A, Rai J, Kim L, Funk S, Dahlberg LE, et al. Evidence for enhanced collagen type III deposition focally in the territorial matrix of osteoarthritic hip articular cartilage. Osteoarthritis and Cartilage 2016; 24: 1029–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chou CH, Lee CH, Lu LS, Song IW, Chuang HP, Kuo SY, et al. Direct Assessment of Articular Cartilage and Underlying Subchondral Bone Reveals a Progressive Gene Expression Change in Human Osteoarthritic Knees. Osteoarthritis and Cartilage 2013; 21: S12–S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai R, Wu ZT, Chu HY, Lu J, Lyu AP, Liu J, et al. Cathepsin K: The Action in and Beyond Bone. Frontiers in Cell and Developmental Biology 2020; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baici A, Lang A, Zwicky R, Muntener K. Cathepsin B in osteoarthritis: Uncontrolled proteolysis in the wrong place. Seminars in Arthritis and Rheumatism 2005; 34: 24–28. [DOI] [PubMed] [Google Scholar]
- 36.Oppenheimer H, Gabay O, Meir H, Haze A, Kandel L, Liebergall M, et al. 75-kd sirtuin 1 blocks tumor necrosis factor a-mediated apoptosis in human osteoarthritic chondrocytes. Arthritis and Rheumatism 2012; 64: 718–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plsikova Matejova J, Spakova T, Harvanova D, Lacko M, Filip V, Sepitka R, et al. A .Preliminary Study of Combined Detection of COMP, TIMP-1, and MMP-3 in Synovial Fluid: Potential Indicators of Osteoarthritis Progression. Cartilage 2021; 13: 1421S–1430S [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao ZT, Wang SQ, Wang JQ. Exploring the osteoarthritis-related genes by gene expression analysis. Eur Rev Med Pharmacol Sci 2014; 18: 3056–3062. [PubMed] [Google Scholar]
- 39.Wei T, Kulkarni NH, Zeng QQ, Helvering LM, Lin X, Lawrence F, et al. Analysis of early changes in the articular cartilage transcriptisome in the rat meniscal tear model of osteoarthritis: pathway comparisons with the rat anterior cruciate transection model and with human osteoarthritic cartilage. Osteoarthritis Cartilage 2010; 18: 992–1000. [DOI] [PubMed] [Google Scholar]
- 40.Cai P, Jiang T, Li B, Qin X, Lu Z, Le Y, et al. Comparison of rheumatoid arthritis (RA) and osteoarthritis (OA) based on microarray profiles of human joint fibroblast-like synoviocytes. Cell Biochem Funct 2019; 37: 31–41. [DOI] [PubMed] [Google Scholar]
- 41.Burleigh A, Chanalaris A, Gardiner MD, Driscoll C, Boruc O, Saklatvala J, et al. Joint immobilization prevents murine osteoarthritis and reveals the highly mechanosensitive nature of protease expression in vivo. Arthritis Rheum 2012; 64: 2278–2288. [DOI] [PubMed] [Google Scholar]
- 42.Appleton CT, Pitelka V, Henry J, Beier F. Global analyses of gene expression in early experimental osteoarthritis. Arthritis Rheum 2007; 56: 1854–1868. [DOI] [PubMed] [Google Scholar]
- 43.Zhu XB, Chen F, Lu K, Wei A, Jiang Q, Cao WS. PPAR gamma preservation via promoter demethylation alleviates osteoarthritis in mice. Annals of the Rheumatic Diseases 2019; 78: 1420–1429. [DOI] [PubMed] [Google Scholar]
- 44.Jiang WZ, Liu H, Wan RX, Wu YJ, Shi ZJ, Huang WH. Mechanisms linking mitochondrial mechanotransduction and chondrocyte biology in the pathogenesis of osteoarthritis. Ageing Research Reviews 2021; 67. [DOI] [PubMed] [Google Scholar]
- 45.Sun K, Luo J, Guo J, Yao X, Jing X, Guo F. The PI3K/AKT/mTOR signaling pathway in osteoarthritis: a narrative review. Osteoarthritis and Cartilage 2020; 28: 400–409. [DOI] [PubMed] [Google Scholar]
- 46.Bouaziz W, Sigaux J, Modrowski D, Devignes CS, Funck-Brentano T, Richette P, et al. Interaction of HIF1 alpha and beta-catenin inhibits matrix metalloproteinase 13 expression and prevents cartilage damage in mice. Proceedings of the National Academy of Sciences of the United States of America 2016; 113: 5453–5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chow YY, Chin KY. The Role of Inflammation in the Pathogenesis of Osteoarthritis. Mediators of Inflammation 2020; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salazar-Noratto GE, De Nijs N, Stevens Y, Gibson G, Guldberg RE. Regional gene expression analysis of multiple tissues in an experimental animal model of post-traumatic osteoarthritis. Osteoarthritis and Cartilage 2019; 27: 294–303. [DOI] [PubMed] [Google Scholar]
- 49.Zhang K, Wang LF, Liu ZC, Geng B, Teng YJ, Liu XN, et al. Mechanosensory and mechanotransductive processes mediated by ion channels in articular chondrocytes: Potential therapeutic targets for osteoarthritis. Channels 2021; 15: 339–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu BY, Jin Y, Ma XH, Wang CY, Guo Y, Zhou D. The potential role of mechanically sensitive ion channels in the physiology, injury, and repair of articular cartilage. Journal of Orthopaedic Surgery 2020; 28. [DOI] [PubMed] [Google Scholar]
- 51.Lee W, Nims RJ, Savadipour A, Zhang QJ, Leddy HA, Liu F, et al. Inflammatory signaling sensitizes Piezo1 mechanotransduction in articular chondrocytes as a pathogenic feed-forward mechanism in osteoarthritis. Proceedings of the National Academy of Sciences of the United States of America 2021; 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee W, Leddy HA, Chen Y, Lee SH, Zelenski NA, McNulty AL, et al. Synergy between Piezo1 and Piezo2 channels confers high-strain mechanosensitivity to articular cartilage. Proceedings of the National Academy of Sciences of the United States of America 2014; 111: E5114–E5122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muramatsu S, Wakabayashi M, Ohno T, Amano K, Ooishi R, Sugahara T, et al. Functional gene screening system identified TRPV4 as a regulator of chondrogenic differentiation. Journal of Biological Chemistry 2007; 282: 32158–32167. [DOI] [PubMed] [Google Scholar]
- 54.Clark AL, Votta BJ, Kumar S, Liedtke W, Guilak F. Chondroprotective Role of the Osmotically Sensitive Ion Channel Transient Receptor Potential Vanilloid 4 Age- and Sex-Dependent Progression of Osteoarthritis in Trpv4-Deficient Mice. Arthritis and Rheumatism 2010; 62: 2973–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Conor CJ, Leddy HA, Benefield HC, Liedtke WB, Guilak F. TRPV4-mediated mechanotransduction regulates the metabolic response of chondrocytes to dynamic loading. Proceedings of the National Academy of Sciences of the United States of America 2014; 111: 1316–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He Z, Leong DJ, Xu L, Hardin JA, Majeska RJ, Schaffler MB, et al. CITED2 mediates the cross-talk between mechanical loading and IL-4 to promote chondroprotection. Ann N Y Acad Sci 2019; 1442: 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He Z, Leong DJ, Zhuo Z, Majeska RJ, Cardoso L, Spray DC, et al. Strain-induced mechanotransduction through primary cilia, extracellular ATP, purinergic calcium signaling, and ERK1/2 transactivates CITED2 and downregulates MMP-1 and MMP-13 gene expression in chondrocytes. Osteoarthritis Cartilage 2016; 24: 892–901. [DOI] [PubMed] [Google Scholar]
- 58.Driscoll C, Chanalaris A, Knights C, Ismail H, Sacitharan PK, Gentry C, et al. Nociceptive Sensitizers Are Regulated in Damaged Joint Tissues, Including Articular Cartilage, When Osteoarthritic Mice Display Pain Behavior. Arthritis Rheumatol 2016; 68: 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Darby WG, Grace MS, Baratchi S, McIntyre P. Modulation of TRPV4 by diverse mechanisms. International Journal of Biochemistry & Cell Biology 2016; 78: 217–228. [DOI] [PubMed] [Google Scholar]
- 60.Mills CD, Nguyen T, Tanga FY, Zhong C, Gauvin DM, Mikusa J, et al. Characterization of nerve growth factor-induced mechanical and thermal hypersensitivity in rats. European Journal of Pain 2013; 17: 469–479. [DOI] [PubMed] [Google Scholar]
- 61.Nencini S, Morgan M, Thai J, Jobling AI, Mazzone SB, Ivanusic JJ. Piezo2 Knockdown Inhibits Noxious Mechanical Stimulation and NGF-Induced Sensitization in A-Delta Bone Afferent Neurons. Front Physiol 2021; 12: 644929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loeser RF, Olex AL, McNulty MA, Carlson CS, Callahan M, Ferguson C, et al. Disease progression and phasic changes in gene expression in a mouse model of osteoarthritis. PLoS One 2013; 8: e54633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chou CH, Jain V, Gibson J, Attarian DE, Haraden CA, Yohn CB, et al. Synovial cell cross-talk with cartilage plays a major role in the pathogenesis of osteoarthritis. Sci Rep 2020; 10: 10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Histology of rat knees. Representative images are shown for the medial compartment of the knee joint following sham surgery (4 weeks) or MMT (2, 4, 6 weeks post-surgery). No pathological changes are evident on the knee with sham surgery. At 2 weeks post MMT, there is reduced cellularity on the tibial cartilage, and a structural defect in the superficial zone. At 4 weeks post MMT, there is a larger structural defect on the medial tibia, affecting the mid zone and full thickness cartilage clefts. A marginal osteophyte is also apparent. At 6 weeks post MMT, the structural defect on the tibia is wider, the marginal osteophyte is larger, and the medial femur also shows a structural cartilage defect reaching into the mid-zone and complete cell loss in that area. High resolution versions of these images are available as eSlides.
Supplementary Figure 2. Principal component analysis of the data was performed and the first and second (PC1 and PC2) principal components are shown. There are comparisons between MMT and sham controls in cartilage at 2 (A), 4 (B) and 6 (C) weeks, in meniscus at 2 (D), 4 (E), and 6 (F) weeks, and in synovium at 2 (G), 4 (H) and 6 (I) weeks. Red: Sham surgery (SM); Blue: MMT surgery (MT).
Supplementary Table 1. All DEGs in articular cartilage in MMT joints at 2, 4, and 6 weeks. Fold change (log2 scale) values and adjusted P values (padj) are shown in the table relative to sham controls.
Supplementary Table 2. Summary of actively regulated Gene Ontology terms (FDR<0.001) and KEGG pathways (FDR<0.05) of genes differentially expressed in cartilage between MMT and sham control models for each time point.
Supplementary Table 3. All DEGs in meniscus in MMT joints at 2, 4, and 6 weeks. Fold change (log2 scale) values and adjusted P values (padj) are shown in the table relative to sham controls.
Supplementary Table 4. Summary of actively regulated Gene Ontology terms (FDR<0.001) and KEGG pathways (FDR<0.05) of genes differentially expressed in meniscus between MMT and sham control models for each time point.
Supplementary Table 5. All DEGs in synovium in MMT joints at 2, 4, and 6 weeks. Fold change (log2 scale) values and adjusted P values (padj) are shown in the table relative to sham controls.
Supplementary Table 6. Summary of actively regulated Gene Ontology terms (FDR<0.001) and KEGG pathways (FDR<0.05) of genes differentially expressed in synovium between MMT and sham control models for each time point.
Supplementary Table 7. Summary of Gene Ontology terms (FDR<0.001) that are shared by multiple time points and tissues or are tissue-/time-specific.