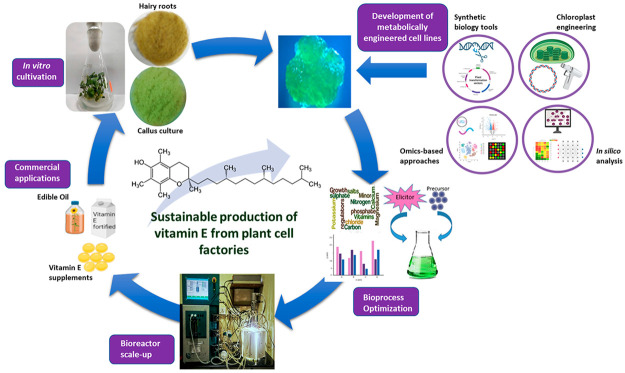
Abstract
Vitamin E is a dietary supplement synthesized only by photosynthetic organisms and, hence, is an essential vitamin for human well-being. Because of the ever-increasing demand for natural vitamin E and limitations in existing synthesis modes, attempts to improve its yield using plant in vitro cultures have gained traction in recent years. With inflating industrial production costs, integrative approaches to conventional bioprocess optimization is the need of the hour for multifold vitamin E productivity enhancement. In this review, we briefly discuss the structure, isomers, and important metabolic routes of biosynthesis for vitamin E in plants. We then emphasize its vital role in human health and its industrial applications and highlight the market demand and supply. We illustrate the advantages of in vitro plant cell/tissue culture cultivation as an alternative to current commercial production platforms for natural vitamin E. We touch upon the conventional vitamin E metabolic pathway engineering strategies, such as single/multigene overexpression and chloroplast engineering. We highlight the recent progress in plant systems biology to rationally identify metabolic bottlenecks and knockout targets in the vitamin E biosynthetic pathway. We then discuss bioprocess optimization strategies for sustainable vitamin E production, including media/process optimization, precursor/elicitor addition, and scale-up to bioreactors. We culminate the review with a short discussion on kinetic modeling to predict vitamin E production in plant cell cultures and suggestions on sustainable green extraction methods of vitamin E for reduced environmental impact. This review will be of interest to a wider research fraternity, including those from industry and academia working in the field of plant cell biology, plant biotechnology, and bioprocess engineering for phytochemical enhancement.
1. Introduction
The year 2022 marks the centennial year of research in vitamin E. In 1922, Evans and Bishop first identified an unknown molecule as an essential factor for rat reproduction.1 It was later named vitamin E because its discovery succeeded that of vitamin D. In nature, vitamin E was found to occur as a group of eight isomeric molecules, tocopherols, and tocotrienols, collectively called tocochromanols, and exclusively synthesized by photosynthetic organisms, such as plants and algae. Following the report of vitamin E as a crucial element in rat reproduction, it was established that the molecule has antioxidant activity in vitro, plays critical physiological functions in humans, and is a vital element for survival.2 It is not synthesized in the human body and, hence, is an essential supplement satisfied by plant sources only through diet. Deficiency in vitamin E, though rare, results in ataxia, a loss in motor control, and a case of nonalcoholic fatty liver disease.3 Though vitamin E occurs as a mixture of tocochromanols in plants, α-tocopherol is reported to exhibit the highest biological activity because of preferential retention and distribution in the human body.4 Thus, it can be stated that the term “vitamin E” corresponds to only α-tocopherol and not all the tocochromanols,2 in contrast to most existing literature. Any reference to vitamin E corresponds to the naturally occurring RRR - α-tocopherol and its synthetic forms,5 as addressed henceforth in this review.
Apart from being an essential dietary supplement, there are several physiological studies on the role of vitamin E in treating cancer, cardiovascular, and ocular diseases.6−8 However, these have not been strongly supported by clinical trials.2 Vitamin E has been reported to have a positive effect when taken as a supplement by specific populations, such as patients on hemodialysis or diabetic individuals (along with haptoglobin genotype 2-2) with cardiovascular diseases9 and aged people with respiratory diseases.3 With the growing demand for natural cosmetic formulations, it is also one of the key ingredients in skin creams and hair formulations.10 Furthermore, it is used as an additive in animal feeds because of its nutritive value.11 With the growing need to combat lifestyle diseases and the inclination toward natural products, the market size of vitamin E is now on the rise.
It is currently extracted from whole plants or chemically synthesized to meet this rising demand. Extraction from whole plants may be disadvantageous because of its occurrence as different isomers in different parts of the plant, which vary with seasonal changes at different geographical locations. Since the primary sources are food crops, repeated extraction may lead to competition for food. Furthermore, chemical synthesis of the molecule yields a racemic mixture instead of the bioactive RRR - α-tocopherol.12 Other strategies, including production from plant cell cultures, algae, and genetically modified bacterial cultures, have been reviewed previously.13−15
This review aims to highlight the advantages of vitamin E production using plant cell and tissue cultures, with emphasis on rational strategies that can be employed to enhance productivity and overcome the limitations in current commercial production. This approach can pave the way for its sustainable commercial production using plant cultures that cater to the booming market.
2. Tocochromanols
2.1. Structure, Classification and Properties
Tocochromanols collectively refer to a group of amphipathic organic compounds characterized by a chromanol ring (polar headgroup) and isoprenyl (C16) side chain (lipophilic tail). Depending on the saturation in their C16 side chain, they are further classified as tocopherols (saturated) and tocotrienols (unsaturated). Within these two classes, different structural isomers exist, namely the α-, β-, γ- and δ-tocopherols and tocotrienols, which differ in the methylation pattern of their aromatic head (Figure 1).
Figure 1.
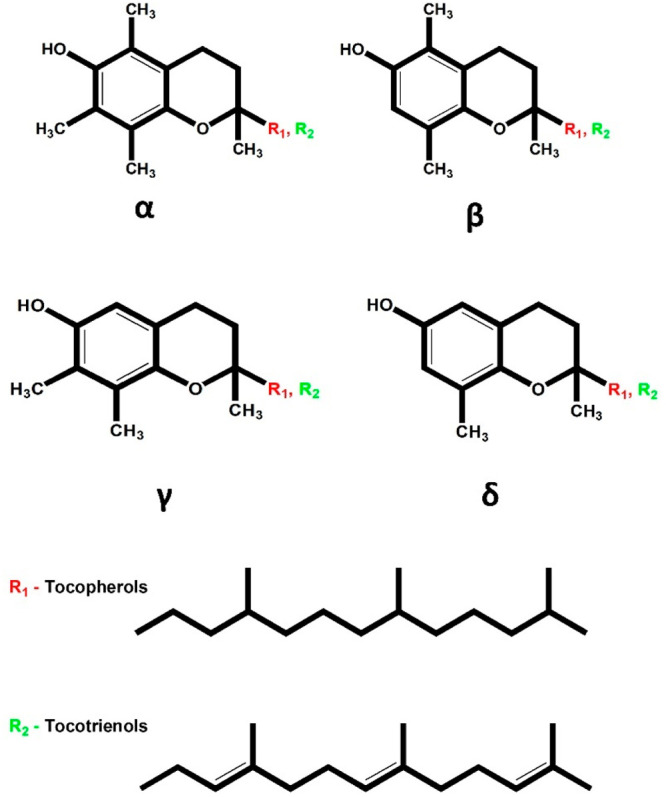
Structure of tocopherols and tocotrienols. The tocopherols and tocotrienols vary in the degree of unsaturation in their isoprenyl side chain.
Among the eight isomeric forms, the human hepatic α-tocopherol transfer protein (α-TTP) selectively binds to α-tocopherol.16 The unbounded isomeric forms are catabolized by cytochrome P450 and oxidized by omega-hydroxylase.4 Thus, because of the preferential retention of α-tocopherol in the human body, it is the most bioactive molecule among its isomeric counterparts. Also, the naturally occurring RRR form of α-tocopherol is the most bioactive compared with the synthetic derivative, which is a racemic mixture.17 Vitamin E molecules are soluble in alcohol and sensitive to heat and light. α-Tocopherol has an absorption maximum of 292 nm when soluble in ethanol.12
2.2. Biosynthesis in Plants
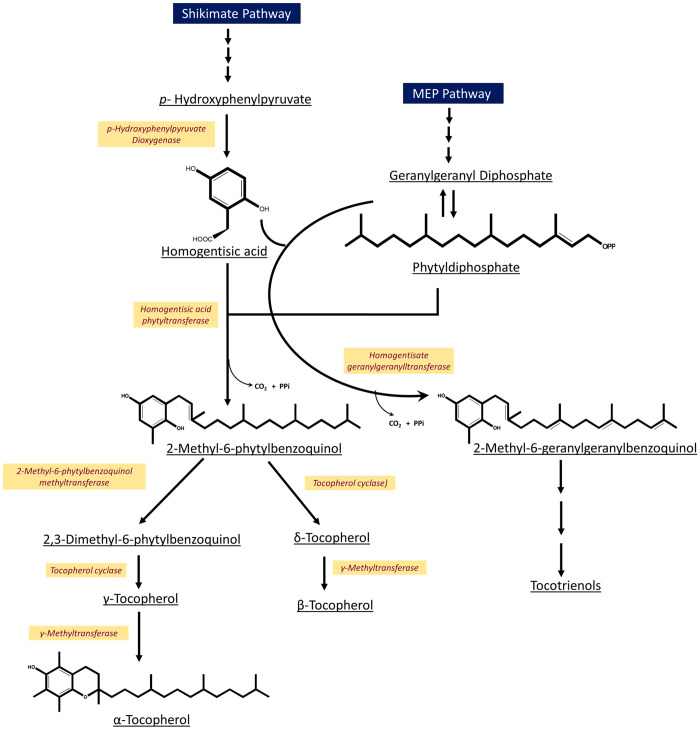
Photosynthetic organisms ubiquitously synthesize vitamin E. In plants, the biosynthesis of tocochromanols takes place in the plastids. The polar headgroup is formed via the aromatic amino acid metabolism (cytosolic shikimate pathway), and the hydrophobic tail is synthesized via the plastidic methylerythritol phosphate (MEP) pathway (Figure 2). Since the MEP pathway contributes to the plastidic isoprenoid precursor pool, vitamin E biosynthesis is also interconnected with other metabolic pathways such as the biosynthesis of chlorophylls, phylloquinones, carotenoids, gibberellins, and plastoquinones. The first committed step in the biosynthesis is the formation of homogentisic acid (HGA) thereby contributing to the headgroup, from p-hydroxyphenylpyruvate (HPP), which is catalyzed by the enzyme p-hydroxyphenylpyruvate dioxygenase (HPPD). The subcellular localization of this step has been much debated since some plant HPPDs, such as in carrot, Arabidopsis, and soybean, lack a chloroplast transit peptide, and Western blot shows cytosolic localization.18 Subsequent steps take place in the plastid. Prenylation of HGA with phytyl diphosphate (PDP) by homogentisate phytyl transferase (HPT/VTE2) or with geranylgeranyl diphosphate (GGDP) by homogentisate geranylgeranyl transferase (HGGT) results in the formation of 2-methyl-6-phytylbenzoquinol (MPBQ) and 2-methyl-6-geranylgeranylbenzoquinol (MGGBQ), which are the precursors of tocopherols and tocotrienols, respectively. A second methyl group is added to MPBQ by a methyl transferase enzyme VTE3 to form 2,3-dimethyl-5-phytyl-1,4-benzoquinone (DMPBQ). Tocopherol cyclase (TC/VTE1) then converts MPBQ and DMPBQ to form δ- and γ-tocopherols. In the final conversion step, γ-tocopherol methyltransferase (γ-TMT/VTE4) methylates the sixth carbon in the chromanol ring of δ- and γ-tocopherols to form β- and α-tocopherols, respectively.
Figure 2.
Vitamin E biosynthetic pathway in plants.
2.3. Role of Vitamin E in Plants
Vitamin E occur in the chloroplast envelope of plants and are stored in the stroma. Their primary role is to quench reactive oxygen species in response to stress (induced by salinity, drought, temperature, light, infection by pathogens, and presence of heavy metals). They help prevent lipid peroxidation by scavenging the lipid peroxy radicals in the thylakoid membranes of the chloroplast by donating a proton from its polar head.19 This protection from lipid peroxidation is crucial for maintaining photosynthetic membrane integrity.20 They also aid in stress tolerance, along with other antioxidants, such as ascorbate and glutathione. Tocopherols are lipid-soluble, whereas ascorbate and glutathione are water-soluble antioxidants, and this antioxidant triad scavenges the oxygen radicals in a coordinated manner.21−23 Ascorbate and glutathione regenerate tocopherols from their α-cromanoxy radical form, in addition to glutathione regenerating ascorbate by acting as the electron donor to reduce dehydroascorbate, the oxidized form of ascorbate. This regeneration of tocopherols allows a single molecule to participate in multiple scavenging reactions.
Further, tocopherols modulate reactive oxygen species accumulation by scavenging singlet oxygen (photoprotective function) produced by excess energy captured by photosynthesis during stress conditions, which could cause oxidation of carbohydrates, proteins, and lipids in the cells.24 Consequently, they get converted to tocopherol quinone, a potential antioxidant in vitro,17 and activate the cyclic electron flow around photosystem II. In addition, tocopherols prevent oxidation of polyunsaturated fatty acids in nonphotosynthetic sites, such as seeds. Tocopherol also prevents acidification of the thylakoid lumen by inhibiting the unrestricted accumulation of protons in the thylakoid membrane and regulates phyto-hormonal levels in plants, thereby acting as a cell signaling molecule.20
2.4. Role of Vitamin E in Human Health
Humans do not synthesize vitamin E, an essential supplement in the diet. According to the National Institutes of Health, Office of Dietary Supplements, United States, the recommended dietary allowance is 15 mg of natural RRR- α-tocopherol for all adults and 19 mg of natural RRR - α-tocopherol for lactating mothers. Deficiency in vitamin E is very rare but can occur in premature babies with less birth weight.25 It can also occur in adults who have fat malabsorption disorder and can occur because of genetic inheritance.26 This deficiency leads to ataxia with vitamin E deficiency (AVED), a neurodegenerative disease that impairs movement coordination. Secondary deficiency complications include liver failure and digestive tract diseases. Vitamin E is therapeutically effective in patients with nondiabetic nonalcoholic steatohepatitis (NASH) and nonalcoholic fatty liver disease (NAFLD).27 α-Tocopherol is also found to accumulate in the nonhepatic tissues in the body, such as the mitochondrial and endoplasmic reticulum membranes in the heart and lungs, which are more prone to free radical production.28
In a regular diet, vitamin E is an antioxidant in the body in some instances, but it also has independent cell signaling and gene regulatory functions. It has been subjected to further research regarding whether its beneficial health-promoting activities in the body are due to antioxidant activity or other physiological molecular mechanisms.27 It is known to act as an anti-inflammatory agent,29 and enhanced immune responses were reported after a supplemental daily intake of 200 mg in the elderly.28 It is also reported that vitamin E regulates gene expression in mast cells and monocytes, thereby positively impacting the immune system.2
Furthermore, vitamin E plays a key role in gene regulation in brain cells, and its deficiency can lead to a dysfunctional central nervous system. Studies have proven a positive impact on cognitive impairment diseases, such as Alzheimer’s disease, Parkinson’s disease,30 and epilepsy.31 Other dysfunctions, such as male32 and female infertility;33 osteoporosis;34 and lifestyle diseases, such as diabetes mellitus,35 have had a promising positive impact from vitamin E supplementation in some clinical trials. There has also been evidence from physiological studies that it plays a beneficial role in preventing cancer.6,36 In fact, studies have showcased that a Mediterranean diet has high proportions of vitamin E and C, which leads to lower incidences of cancer.37 Nevertheless, there are also reports of contrasting results showing no significant effect on the treatment of cancer, thereby implying the necessity of further research.9,38
2.5. Diverse Applications of Vitamin E
The global annual production of vitamin E for numerous industrial applications is around 70 kilotonnes.39 In the biomedical industry, it is used as an antioxidant stabilizer and antimicrobial agent in prosthetic materials and employed for accelerated wound healing and tissue regeneration because of its cell proliferative potential.39 Furthermore, it has been used widely as a drug delivery vector for bioavailability drugs, such as paclitaxel,40 and is also being used as a solubilizing agent for excipients in solid dosages of pharmaceuticals.41
In the food industry, vitamin E is an antioxidant and antimicrobial agent in food preservation and packaging for the prolonged shelf life of canned foods.42 Because of its capability to prevent lipid oxidation, which may lead to a reduction in organoleptic properties and food quality, it is included in food as an additive. It is widely used in skin and hair care formulations because of its photoprotective functions.43 Besides human welfare applications, vitamin E is provided as nutritional feed additives for cattle and poultry because it has been shown to enhance the visual appeal and flavor of meat.44 The role of vitamin E in plants, human health, and other diverse applications has been summarized in Table 1.
Table 1. Role of Vitamin E in Plants, Human Health, and Its Applications.
| role in plants | role in human health | applications |
|---|---|---|
| • quenches reactive oxygen species in response to stress | • protects against AVEDa,27 | • antioxidant stabilizer and antimicrobial agent in prosthetic materials40 |
| • prevents lipid peroxidation in thylakoid membrane19 | • effective in nondiabetic patients for NASH and NAFLDa,27 | • drug delivery vector for bioavailability drugs40 |
| • photoprotective function by scavenging singlet oxygen24 | • cell signaling27 | • solubilizing agent for excipients in solid dosages of pharmaceuticals41 |
| • prevents oxidation of polyunsaturated fatty acids in nonphotosynthetic sites20 | • gene regulation27 | • antioxidant and antimicrobial agent in food preservation and packaging42 |
| • acts as a signaling molecule20 | • regulates platelet aggregation3 | • used in clinical and cosmetic dermatology for its photoprotective function43 |
| • activation of protein kinase C3 | • nutritional feed additives for cattle and poultry44 | |
| • anti-inflammatory (low-degree inflammation)29 | ||
| • breakdown of lipids and, hence, effective in reducing obesity2 | ||
| • cytoprotective function—degradation of harmful xenobiotics27 | ||
| • beneficial role in preventing cardiovascular diseases,7,9 ocular pathologies,8 and cancer6,36 |
AVED, ataxia with vitamin E deficiency; NASH, nonalcoholic steatohepatitis; NAFLD, nonalcoholic fatty liver disease.
3. Sources, Market Demand, and Limitations in Current Methods of Production
Though vitamin E deficiency is rare, consumption of the recommended amount is crucial for general well-being. With changes in the modern diet, there has been a decline in the consumption of healthy foods.45,46 It is observed that the levels of vitamin E in healthy human blood have reduced drastically because of lifestyle changes. Hence, there has been a rise in market demand for vitamin E as a supplement. Steps have also been taken to fortify food crops to meet the daily demands of humans.17 Furthermore, there has been an increase in the need for vitamin E production to cater to the demands of the cosmetic, biomedical, food, and animal feed industry. There has also been a rise in animal meat consumption, which has expanded its requirement as a food additive.
Currently, vitamin E supplements in the markets fall into three categories and suffer from the following limitations.
1. Synthetic vitamin E or dl-α-tocopherol or all-rac-α-tocopherol is sold chiefly in its acetate ester (α-tocopherol acetate) form to increase its stability. It is produced by the condensation of isophytol, a C20 alcohol, and trimethylhydroquinone and is a racemic mixture of eight stereoisomers. Though the cost of the product is less, as it is produced from the raw materials of fossil fuel origin, raw material costs are approximately 70% of the production costs, and hence, manufacturers are on the lookout for cheaper sources of raw materials. Also, the natural d-α-tocopherol is 1.36 times more active than this all-rac-α-tocopherol form because of the stereoselectivity and specific chiral recognition of the enzymes and receptors in the body only with the RRR -form, and the isoforms cannot interconvert within the human body.28,47
2. Esters of natural vitamin E (in the form of α-tocopherol) -these are also termed semisynthetic because they are synthesized by methylation of plant oil derived from highly fractionated β- and γ-tocopherols. The vegetable oil deodorized distillate (VODD) left out of the vegetable oil refining process, such as from soybean and sunflower, is used to produce these semisynthetic forms. Limited vegetable oil availability and variation of tocopherol concentration with geographical location are leading to a significant supply and demand imbalance for natural vitamin E produced from VODD.48
3. Mixed tocopherols used in supplements are less fractionated forms derived from plant oils. They suffer from the same limitations discussed above, including less activity of the mixed tocopherols than the RRR-α-form and limited raw material availability.
Direct extraction from whole plants/plant oils can be a solution for the production of vitamin E (natural α-tocopherol). According to the United States Department of Agriculture Food Composition databases, oilseeds such as wheat germ, hazelnut, sunflower, safflower, almonds, and peanuts are rich in α-tocopherol. However, extraction from such plant sources yields a mixture of tocopherols in varying concentrations, and there is a demand for pure α-tocopherol sources. Tocopherols also vary in their isoforms and levels between and within plant species. For example, green leafy regions of plants are rich in α-tocopherol, but total tocopherol content is low. Conversely, seeds have a high abundance of tocopherols but are rich in γ-tocopherol, though α-tocopherol is high in sunflower seeds.17 Thus, the occurrence varies with plant species and also tends to be in low amounts. The concentration of each isoform may vary with geographical location, climatic conditions, quality of soil, environment, susceptibility to pathogen attack, crop year, and genetic factors like genotype.
A research report by Valuates Reports,49 published in January 2022, estimates the global vitamin E market size is close to USD 670 million in 2022. The report predicted that the market will witness over 4.7% compound annual growth rate and a market size of USD 882.2 million by 2028. The Asia Pacific region is expected to witness a steady rise in the market. The top companies holding major shares in the global vitamin E market include DSM (Cargill), BASF, NHU, Nenter and Co., Inc and Zhejiang Medicine Company. Some key vitamin E commercial products and their production methods are summarized in Table 2. The major challenge in this growing industry is the demand for consistent natural vitamin E supply.
Table 2. Examples of Commercially Produced Vitamin E.
| market name | method of production | application | company/country |
|---|---|---|---|
| novotol | vegetable-oil-sourced natural vitamin E | food supplements; snack bars and beverages; and cosmetics, such as sunscreens and lip balms | ADM, USA |
| vitamin E | synthetic production of dl-α-tocopherol | dietary supplement | BASF, Germany |
| dl-α-tocopherol | synthetic production | dietary supplements, infant nutrition, and cosmetics | Zhejiang NHU, China |
| vitapherole E org | mixed tocopherols rich in RRR α-tocopherol extracted from sunflower oil | fortification of food supplements and soft drinks | Vitae Naturals, Spain |
| d-α-tocopherol powder 50 | mixed tocopherols obtained from natural plant extraction | food fortification and nutrient supplement tablets | Mitsubishi Chemical Corporation, Japan |
| Naturall-e | natural extraction from sunflower and soy plants | antioxidant and dietary supplement | Matrix Life Science, India |
4. Strategies for Multifold Enhancement of Vitamin E Content in Plant Cultures
With the growing market demand, sustainable production methods of vitamin E are the need of the hour. Challenges in chemical synthesis have been overcome by successfully producing precursor molecules, such as isophytol, through fermentation of genetically engineered Saccharomyces cerevisiae and using them to synthesize vitamin E and reach an annual output of 30 000 tonnes.50 However, because of the nature of synthesis, racemic mixture of α-tocopherol is produced. Biotechnological methods of vitamin E production have been reviewed earlier,14 where α-tocopherol production from plant and algal cell cultures has been compared. Though the existing lab-scale studies showcase lower α-tocopherol yield in plant cultures than algal cultures, there is immense potential to improve and overcome current challenges faced by the “natural” vitamin E market using in vitro plant cell cultivation via rational metabolic engineering and process optimization.
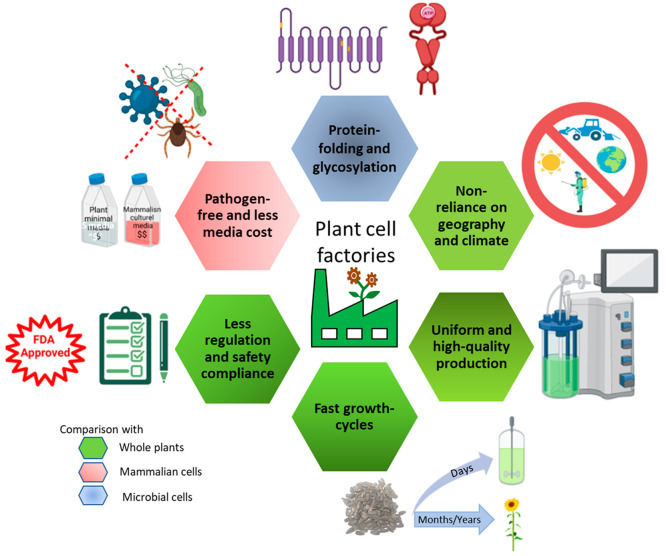
Plant cell cultures are increasingly used for continuous, uniform, and large-scale production of commercial biologics, such as therapeutic enzymes; nontherapeutic proteins, including growth factors; industrial enzymes; biopolymers; and secondary metabolites with many industrial applications.51−55 Plant in vitro cultures retain genetic information from their parent source and exhibit similar product profiles in culture. Bioreactor cultivation of plant cell/tissue cultures can be adopted to produce phytochemicals because they can be grown at a large scale throughout the year, independent of geography and climate, and under controlled conditions in less space and time. Unlike natural plants, controlled conditions in bioreactors enable consistency and stability in the quality and quantity of product profiles from plant cells. In vitro cultures also ensure pathogen-free cultivation and shorter production cycles than whole plants. Plant cells can, furthermore, be subjected to genetic engineering to synthesize complex bioactive proteins, such as antibodies and enzymes, with post-translational modifications, which is not possible with prokaryotic production platforms (Figure 3).
Figure 3.
Advantages of plant cell cultures over whole-plant, mammalian, and microbial-based production platforms (figure icons created using BioRender.com).
However, plant cell cultivations can suffer from limitations, like low product yield, under in vitro conditions; lower growth rates; and scale-up limitations compared with microbial processes.56 Therefore, bioprocess optimization and metabolic engineering strategies (with control over the biosynthetic route) are generally applied in plant cell cultures for improved productivity of biomass and the desired metabolite. An integrative approach could be beneficial to achieve multifold enhancement. The strategies discussed in Section 4.1 can be used for the development of α-tocopherol-enhanced metabolically engineered/chloroplast-transformed plants/cell lines. These engineered whole plants/cell lines can be used as an explant for generating in vitro cultures or parent culture for cell suspension initiation, respectively. This ensures elevated concentrations of α-tocopherol in the starting material before the application of bioprocess strategies, as discussed in Section 4.2, for multifold productivity enhancement.
4.1. Stable and High-Yielding Cell Line Development by Metabolic Pathway Engineering
4.1.1. Overexpression and Deletion of Single/Multiple Genes
The ever-increasing demand for natural vitamin E has led to increased efforts in developing whole plants (major crops) and plant cells (in vitro) with higher vitamin E content via crop breeding and genetic engineering.15,57,58 In the past decade, plant metabolic engineering has witnessed a paradigm shift in the context of enhancing important plant-derived metabolite yields, including vitamin E.59−71 Metabolic engineering is “the direct improvement of production, formation, or cellular properties through the modification of specific biochemical reactions or the introduction of new ones using recombinant DNA technology”.72 It requires a systems-wide understanding of plant metabolism. This has been aided by the advances in low-cost, high-throughput genome sequencing technologies that have enabled the rapid sequencing of plant genomes in recent years73 and the development of integrated -omics approaches to comparatively annotate, analyze, and study the metabolic phenotype in acute detail.
The successful introduction of genes in plants for conventional metabolic engineering requires target recombinant transgenes under the control of suitable constitutive/tissue-specific/inducible promoters, such as CaMV 35S/2A11/APase, along with any required organelle targeting (plastid, endoplasmic reticulum) and polyadenylation signals. Appropriate vectors, such as pCAMBIA/pRI with multiple cloning sites, are required for transgene integration, in addition to removable/integrated selectable markers and reporters, including antibiotics hygromycin and kanamycin and fluorescent protein tags, such as green and red fluorescent protein, for transformant selection. Furthermore, suitable transformation systems for gene delivery and expression in plant cells, such as Agrobacterium-mediated, particle bombardment, and biolistic methods, are necessary. Agrobacterium-mediated transformation is one of the most commonly used methods for plant transformation74−76 because it is highly effective for the stable integration of the transgene into the plant genome with low transgene copy number.74
In the context of vitamin E metabolic engineering to elevate α-tocopherol content, earlier attempts were directed toward increasing the total tocochromanols content by increasing the flux upstream of the tocopherol biosynthetic pathway (from HGA formation until they branch into various tocopherols), which overexpressed the biosynthetic enzymes in the pathway to convert other forms of tocopherols into α-tocopherol and downregulated competing pathways that redirect flux away from tocopherol biosynthesis, such as deletion of homogentisate dioxygenase, that metabolizes homgentisic acid to central metabolic intermediates.77
As can be seen from Table 3, the most commonly used strategy is the overexpression of the enzymes involved in the first and final conversion steps, HPPD (PDS1) and γ-tocopherol methyltransferase (VTE4), respectively, which results in an increased content of α-tocopherol in the majority of the target species. Also, multiple genes (in the vitamin E biosynthetic pathway) overexpression strategy led to relatively enhanced α-tocopherol content compared with single gene overexpression studies that suggested existing bottlenecks in multiple steps of the pathway.
Table 3. Overexpression Strategies Reported for Successful Enhancement of α-Tocopherol Content in Plants.
| overexpressed enzyme(s) | species (source of transgene)a | fold increase in α-tocopherol compared with wild type [final content (units) or activity in α-tocopherol equivalents (α-TE)]b |
|---|---|---|
| HPPD (PDS1) | Arabidopsis leaves (At)81 | 1.44 [18 pmol mg–1 (FW)] |
| Arabidopsis leaves (Mi)82 | 2.58 [31 μg g–1 (FW)] | |
| tomato leaves (Mi)82 | 2.67 [11.5 μg g–1 (FW)] | |
| tomato fruits (Mi)82 | 5.0 [10.3 μg g–1 (FW)] | |
| potato leaves (At)83 | 2.12 [15.68 μg g–1 (FW)] | |
| potato tubers (At)83 | 3.66 [1030.7 ng g–1 (FW)] | |
| lettuce leaves (Ls)84 | 4.0 [25.6 μg g–1 (FW)] | |
| Brassica napus (Sy)85 | 1.5 [252 mg kg–1 oil) | |
| HPT (VTE2) | tomato leaves (Md)86 | 3.6 (36.83 μg/g FW) |
| tomato fruits (Md)86 | 1.7 (4.57 μg/g FW) | |
| potato tubers (At)83 | 2.0 [578.6 ng g–1 (FW)] | |
| tobacco leaves (At)87 | 5.4 [12.09 μg g–1 (FW)] | |
| Arabidopsis seeds (At)88 | 1.33 (5.2 mg α-TE 100 g–1 tissue) | |
| tomato mature leaves (Sy)89 | 2.5 [500 ng mg–1 (DW)] | |
| MPBQ (VTE3) | soybean (At)90 | 2.5 [75 ng mg–1 (seed)] |
| TC (VTE1) | lettuce leaves (At)91 | 2.24 [4.7 μg g–1 (FW)] |
| tobacco leaves (At)87 | 4.0 [8.96 μg g–1 (FW)] | |
| γ-TMT (VTE4) | soybean (At)90 | 8.0 [240 ng mg–1 (seed)] |
| Arabidopsis seed (At)92 | 93.4 [342 ng mg–1 (seed)] | |
| Arabidopsis seeds (At)88 | 9.1 (35.6 mg α-TE 100 g–1 tissue) | |
| soybean seed (Pf)93 | 10.4 (450.1 pmol mg–1 tissue) | |
| soybean seed (Bn)94 | 11.1 [340 μg g–1 (DW)] | |
| Brassica juncea seeds (At)95 | 6.6 [367 ng mg–1 (seed)] | |
| lettuce leaves (At)96 | 3.1 [0.37 mg g–1 (DW)] | |
| Perilla frutescens leaves (At)97 | 1.8 (87.8 μg g–1 tissue) | |
| Perilla frutescens seeds (Pf)98 | 22.5 [1.31 mg g–1 (seed)] | |
| tobacco seeds (At)23 | 9.6 [120 μg g–1 (FW)] | |
| VTE2+VTE1 | tobacco leaves (At+At)87 | 7.1 [15.90 μg g–1 (FW)] |
| VTE3+VTE4 | soybean (At+At)90 | 9.1 [273 ng mg–1 (seed)] |
| VTE2+VTE4 | Arabidopsis seeds (At+At)88 | 12.0 (47.4 mg α-TE 100 g–1 tissue) |
| VTE1+VTE4 | tobacco leaves (At+At)79 | 1.2 [14.8 μg g–1 (FW)] |
| VTE2+VTE1+VTE4 | tomato mature leaves (Sy+Sy+At)89 | 4.0 [900 ng mg–1 (DW)] |
At, Arabidopsis thaliana; Mi, Mangifera indica; Ls, Lactus sativa; Sy, Synechocystis; Pf, Perilla frutescens; Bn, Brassica napus.
FW, fresh weight; DW, dry weight.
However, the traditional metabolic engineering approaches do not always yield straightforward results across all plant species. For instance, co-overexpression of At-HPPD and MPBQ MT resulted in a 3-fold enhancement in γ-tocopherol with other tocochromanols in undetectable levels in maize kernels, which suggests that γ-TMT step is rate-controlling78 in that species. Thus, the traditional approach to the engineering of plant metabolism is more time-consuming and restricted to intuitive candidates in the core pathways related to the product.
In addition to nuclear transformation, because of its advantages of biosafety, the potential to express high levels of protein, a lower scope for epigenetic variation, and instability in the transgene, chloroplast DNA transformation is on the rise. Several studies have exploited this technology for vitamin E enhancement. Tocopheryl methyltransferase was overexpressed in tobacco and lettuce to increase vitamin E content by plastid transformation.79 This caused an increase in the total tocopherol with a major percentage elevation of α-tocopherol. In another similar study, in high saline conditions and exposure to metal stress, the overexpression of tocopheryl methyltransferase resulted in a 2.4 -fold, and 2-fold higher α-tocopherol content compared with wild-type respectively.23 In addition to single gene overexpression, the multigene engineering of Homogentisate phytyltransferase, tocopheryl cyclase, and tocopheryl methyltransferase in tobacco and tomato plants was successfully performed by synthetic operon development.80 This multigene overexpression resulted in a 10-fold increase in the total tocochromanol content.
4.1.2. Computational-Model-Guided Engineering of Vitamin E Metabolism
Conventional metabolic engineering requires knowledge of the basic biochemical pathways of interest, with most engineering attempts focusing only on overexpression and/or downregulation of closely related reactions to the target metabolic pathway. However, systems metabolic engineering for strain improvement enables the prediction of known and novel perturbation targets that are sometimes not directly connected to the pathway of interest.
Reconstruction of genome-scale metabolic models (GSMMs) that can mimic different metabolic states of an organism facilitate exploration of metabolic capabilities in silico, thereby aiding in the selection of suitable experimental strategies, thus saving time and cost over the traditional hit and trial approach in experimentation. GSMMs comprise a comprehensive list of all possible reactions within a cell identified using the genome annotation of the organism aided by prior knowledge on biochemical pathways and comparative analysis with closely related organisms across different databases.99 The commonly used databases for data collation for genome-scale metabolic network reconstruction include genome databases CMR,100 PlantGDB,101 Gramene,102 SEED,103 GenBank,104 and BioCyc;105 biochemical databases KEGG,106 BRENDA,107 UniParc,108 TransportDB,109 and MetaCyc;105 organism-specific databases like AraCyc,110 HumanCyc,111 EcoCyc;112 etc. The collated reaction information is represented with details on stoichiometry, directionality (reversibility), catalyzing enzymes with corresponding genes coding for them (collectively abbreviated as GPRs for genes–protein–reaction information), and regulatory information, if available. Several tools and software packages, such as Model SEED,113 Pathway Tools,114 and Path2Models,115 among others, facilitate this iterative and otherwise labor and time-intensive reconstruction process in an automated/semiautomated manner. Most draft reconstructions require extensive manual curation before further simulation to check for accuracy of the annotation, compartmentation, mass/charge balances, missing links, dead-end metabolites (that are only produced or consumed), and blocked reactions.116 Several software packages, such as the COBRA toolbox,117 CellNetAnalyzer,118 OptFlux open-source software platform,119 and RAVEN toolbox,120 to name only a few, are available for the analysis of the reconstructed metabolic networks. The GSMMs are usually exchanged in the SBML121 and packages extending SBML-FBC,122 MFAML,123 and BioPAX124 formats. In particular, the SBML format is widely used with the COBRA toolbox. Manually curated and automated draft genome-scale models of various species can be found in BioModels,125 Model SEED113 (with further analysis in the associated KBase126), and BiGG Models.127 In addition, the integration of data from transcriptomics, proteomics, metabolomics, and fluxomics studies using advanced computational and statistical methods in recent times has improved the predictive power of the metabolic model reconstructions.128
For systems analysis of large metabolic networks, such as in plants, constraint-based approaches are powerful techniques. The approach exploits the fact that biological systems survive and grow under certain restrictions or constraints, such as diffusion rates of molecules inside a cell, that affect nutrient uptake and enzyme concentrations, packing of DNA, and other macromolecules inside the cell and environmental conditions, such as pH, temperature, osmolarity, and self-imposed regulation of biological processes, including transcription, translation, and enzyme activity. One of the most popular constraint-based techniques is flux balance analysis (FBA), which is widely used in predicting an organism’s growth or production of a target metabolite by calculating the allowable flux distribution or rate of metabolite flow through a metabolic network129,130 under given constraints.
In plant metabolic model reconstruction, the power of prediction is further complicated by tissue-specific reactions, diurnal cycles, temporal storage of metabolites, lack of experimental data for compartmentation, exchange or transport of metabolites between compartments and tissues using intracellular transporters, presence of parallel metabolic pathways, and the highly interconnected and complex secondary metabolism.131 Recent advances in plant systems biology have helped understand the metabolic capabilities at multiple levels of organization, such as single cells,132−134 tissues,135−137 and whole plants.138 Genome-scale metabolic reconstructions are now available for Arabidopsis;132,133,135,139 industrially relevant crops, such as rice,140−142 tomato,143 maize;144 a C4GEM model applicable for C4 plants maize, sorghum, and sugar cane;145 and the C4 plant Setaria italica,138 with multiomics data integrated into the reconstructed model. The PlantSEED interface in the ModelSEED database also enables automated annotation and reconstruction of plant primary metabolism with the draft reconstructions of many plant species publicly available.103 A list of some of the plant metabolic reconstructions and their applications is summarized in the Supporting Information Table 1.
Genome-scale model reconstructions have also been used for predicting targets for vitamin E enhancement. A subcellular compartmentalized tissue-specific model of Arabidopsis thaliana was developed135 through the integration of omics data, which can be utilized for predicting the feasibility of metabolic engineering strategies. The authors applied computational methods to predict the increase of vitamin E levels up to 366% with knockout studies of 71 enzymes. The same model was adapted to simulate the metabolism of sunflower in another study.146 A constraint-based approach called flux-scanning-based enforced objective flux (FSEOF)147 was used to identify and rank the enzyme targets, which can be overexpressed to enhance vitamin E. The best strategy to overexpress p-hydroxyphenylpyruvate dioxygenase was experimentally validated by initiating sunflower suspension cultures with the metabolically engineered cell line. Bioprocess optimization strategies were applied to the culture leading to a 10-fold increase in the α-tocopherol productivity compared with the untransformed cell lines. Thus, the application of rational computational strategies gives the leverage to choose optimum targets for overexpression, and further application of bioprocess strategies, as discussed in Section 4.2, can result in multifold enhancement of vitamin E.
4.2. Bioprocess Optimization for Yield and Productivity Enhancement
4.2.1. Explant Type, Media, And Culture Condition Optimization
Initially, the choice of the plant and the type of explant for callus induction is vital in order to optimize in vitro production of vitamin E. The importance of suitability of the plant tissue for manipulation in vitro has been reviewed earlier.15 Sunflower cell cultures have been majorly used for in vitro α-tocopherol production since it comprises almost 91% of total tocochromanols in the plant. Callus/suspension cultures induced from safflower are also commonly studied for the same application. The potential of using in vitro callus generated from soybeans has also recently been showcased.148 Though authors of the study obtained a higher percentage of tocopherols in the callus than the seed, the callus line was predominated by γ-tocopherol. An analysis of callus generated from tocopherol-rich leaf regions showed a better percentage concentration of α-tocopherol. This highlights the importance of the choice of explant type for better tocopherol yields in vitro.
Plant growth basal medium and growth regulators, such as various auxins and cytokinins, including naphthalene acetic acid, 6-benzylaminopurine, indole acetic acid, kinetin, 2,4-dichlorophenoxy acetic acid, and thidiazuron, affect growth and secondary metabolite production in plant cells.149 Their different combinations and concentrations have varying effects on the secondary metabolite production in plant cell suspension cultures.149,150 Hence, the right medium conditions are needed for optimum α-tocopherol productivity. Besides nutrients and growth regulators, parameters, such as illumination, pH, agitation, and temperature, influence growth and product titers.
Media and culture conditions were optimized for improved α-tocopherol in safflower (Carthamus tinctorius) callus cultures.151 The authors employed a revised tobacco medium supplemented with indole-3-butyric acid, kinetin, and casamino acid (RT-B2K medium), which enhanced the tocopherol content. Furthermore, a combination of inositol with casamino acid was investigated to be the most effective for the growth of safflower callus and α-tocopherol compared with inositol added alone. This shows that a systematic study of the interaction of medium components is crucial for obtaining optimum levels of vitamin E in plant cell cultures. In the same RT-B2K medium, the rate-limiting factors influencing α-tocopherol productivity in safflower cell cultures were studied with a cultivation period of 35 days.47 It was observed that specific productivity of α-tocopherol increased linearly with dissolved oxygen concentration up to 9 ppm, but a minimum threshold is required for the production of α-tocopherol. The study also demonstrated that the release of conditioning factors (molecules that are released from the cells alongside its growth) from the safflower cell during the growth phase enhances α-tocopherol productivity. This was further confirmed by the maintenance of cell suspension in a specific conditioned media along with the spent media in which cells were cultured. The growth and α-tocopherol productivity increased linearly with an increase in concentration of the conditioned media, and a lack of these factors resulted in slow growth and decreased productivity.
As an extension of the previous study,47 factors influencing α-tocopherol productivity in safflower cell suspension cultures were studied with an engineering outlook.152 A controlled dissolved oxygen concentration at 8 mg of O2 L–1 was maintained in combination with the addition of a conditioning factor (supernatant of culture withdrawn at an interval of 3 or 5 days that is readded into the reactor). The controlled maintenance of the dissolved oxygen concentration elevated oxidative stress in the cells and was shown to be a stimulating factor for α-tocopherol productivity and a specific production rate. Repeated batch operation was another strategy used in the same study where 10% v/v inoculum was transferred to a fresh medium, which ensured high dissolved oxygen concentration and sufficient conditioning factor for enhanced α-tocopherol. It was reported that a large dilution rate and small inoculum size hinders tocopherol productivity. This mode of operation (with the addition of conditioning factors) and the optimum dissolved oxygen resulted in 9 mg per 100 g of dry cell α-tocopherol yield, which was significantly higher than in safflower seeds.
Another interesting observation was that the α-tocopherol titer was high in green callus.153 A fresh green callus line developed in the RT-B2K medium under light conditions of 7000 lx and 16/8 light–dark cycle produced 3.3 times more α-tocopherol than in dark conditions. Similarly, α-tocopherol was hypothesized to be more abundant in photosynthetic tissues and lower in nongreen callus.154 This hypothesis was investigated by cultivating sunflower cell suspension cultures in photomixotrophic conditions. It was observed that there is a direct relation between tocopherol synthesis and photosynthesis, and hence, there was an elevation in tocopherol levels (up to 4-fold) when the culture was grown in continuous white fluorescent light with reduced sucrose (3 g L–1) concentration. Sucrose, a primary carbon source in plant cell culture media, depresses photosynthetic rate.155 Hence, light exposure in reduced sucrose was found to positively influence tocopherol productivity by altering the cell’s photosynthetic capability. Though culturing cells in a photoautotrophic condition by supplying carbon dioxide as the sole carbon source can be a strategy, the chloroplasts are more localized when the culture is grown in photomixotrophic conditions. It was also observed that there were elevated levels of geranylgeranyl pyrophosphate synthase, a crucial enzyme for tocopherol synthesis. The tocopherol biosynthesis machinery is localized in the chloroplasts of photosynthetic tissues, and hence, there could be a direct relevance to light and tocopherol levels.
In another study, where a cell suspension of sunflower was grown in photomixotrophic conditions (light + sucrose, along with growth hormones, naphthalene, acetic acid, and 6-benzylaminopurine), there was a significant rise (230%) in the levels of α-tocopherol production in comparison with heterotrophic conditions (without light).156 This strengthens the evidence that α-tocopherol production relies on light conditions in addition to the medium nutrients supplied. Photomixotrophic cultures grew slowly but synthesized more chlorophyll and a higher concentration of α-tocopherol than heterotrophic cultures. This positive correlation between chlorophyll content and α-tocopherol was also observed in other published reports.19,21,157
Furthermore, it was demonstrated that the α-tocopherol production in hairy root cultures of sunflower increased up to 2-fold in the presence of 16/8 light/dark conditions compared with completely dark conditions.158 This result points out that the effect of light on tocopherol is also relevant in hairy root cultures where greening due to the presence of chlorophyll is not observed.
In contrast, α-tocopherol concentration was found to be higher in Rosa damascene callus cultures in dark conditions compared with cultures exposed to light when supplemented with methyl jasmonate as elicitor and phenyl alanine as precursor in independent experiments.159
It was also observed that culture age plays a major role in tocopherol synthesis. Prolonged subculture intervals were shown to affect the α-tocopherol production positively.156,157 The authors point out that this could be due to stress-induced α-tocopherol enhancement. Older cultures were found to produce more α-tocopherol with prolonged exposure to light than their younger counterparts.
4.2.2. Addition of Precursors
Precursor feeding involves the supplementation of the biosynthetic pathway intermediates, which can relieve the bottlenecks restricting the desired metabolite production.160,161
Studying the effect of precursors on α-tocopherol production in safflower, Furuya et al. demonstrated that the addition of phytol enhanced α-tocopherol production. However, when phytol was added in combination with homogentisic acid, there was no significant effect on α-tocopherol.151 The potential of l-phenylalanine, d- and l-tyrosine, p-coumaric acid, p-hydroxyphenylpyruvic acid, and homogentisic acid as precursors of the aromatic ring and geranylgeraniol, phytol, and isophytol as precursors of the side chain was evaluated in safflower.153 The precursors of the aromatic chain were not effective in achieving an increase in the tocopherol levels, but phytol as a precursor of the side chain resulted in an effective increase up to 5 times that of the control, which is similar to previous observation.151 In sunflower cell cultures, the addition of homogentisic acid enhanced α-tocopherol production by 30%, whereas there was no enhancement in α-tocopherol with phytol as the sole precursor.157 This was similar to the results of another precursor study of sunflower cell cultures162 but contrasting to the previously discussed precursor addition in safflower.151,153 Thus, the precursor addition is species specific despite the common vitamin E biosynthesis pathway.
4.2.3. Addition of Elicitors
Elicitors are compounds that induce secondary metabolite production in plant cells. They can be classified as abiotic (temperature, ultraviolet radiation, osmotic pressure, heavy metals, antibiotics, endogenous plant signaling molecules such as jasmonic acid) or biotic (autoclaved fungal extracts and fungal-derived polysaccharides, such as chitins or glucans) depending on their origin. Several defense-related gene expressions are regulated directly or indirectly by elicitors, thereby enhancing the production and accumulation of secondary metabolites upon exogenous addition to culture medium.165,166
The addition of the elicitor, methyl jasmonate, to the callus cultures of Amaranthus caudatus increased the α-tocopherol levels by 5-fold with respect to the control cultures.163 Similarly, the effect of addition of jasmonic acid in varying concentrations during the stationary phase of sunflower and Arabidopsis cultures was observed on α-tocopherol production.164 Using 5 μM of jasmonic acid with a treatment time of 72 h was found to have the best effect on sunflower and Arabidopsis cell cultures. It has been reported that methyl jasmonate could promote tyrosine aminotransferase activity in A. caudatus, a key enzyme in α-tocopherol biosynthesis, and jasmonic acid can upregulate α-tocopherol biosynthesis by enhancing gene expression of p-hydroxyphenylpyruvate dioxygenase and homogentisate phytyltransferase in sunflower and Arabidopsis cell cultures.
A study investigated the effect of stress due to oxygen levels on α-tocopherol in cultures of Arabidopsis thaliana.167 When cultures were grown in a shake flask in static condition for 24 h, elevation of α-tocopherol of up to 62% was observed due to the anoxic stress experienced by the cells. To have control over the oxygen supply, the cell suspension of A. thaliana was cultivated in a stirred tank bioreactor. The cells were subjected to an anoxic shock by sparging the vessel with nitrogen. There was a 14% increase in α-tocopherol, which shows that the reactive oxygen species producing pathways are activated in conditions of anoxia, and hence to combat the stress, the cells produce α-tocopherol in higher amounts without having any significant effect on the cell viability. Addition of methyl jasmonate as an elicitor also found to have an effect in the improvement in α-tocopherol levels. In the callus cultures of Rosa damascene, methyl jasmonate at a concentration of 5 μM was able to enhance α-tocopherol up to 4-fold.159
Cyclodextrins, a common excipient in medicines, was used as an elicitor in suspension cultures of carrots,168 safflower, and mung bean169 for enhanced α-tocopherol. Specifically, in carrot cell suspension cultures, it was reported that cyclodextrin was also effective in promoting the extracellular accumulation of tocopherol, which could help reduce the downstream processing cost and effort significantly.
It is well known that nanoparticles are effective agents in elicitation of in vitro cultures for improved production of secondary metabolites. A study of the effect of nanoparticles in the enhancement of α-tocopherol in Argania spinosa demonstrated that titanium dioxide and silicon dioxide had up to 4.5-fold and 4.7-fold increase in production, respectively.170 Safflower cultures have showed an increase in α-tocopherol as a response to salinity stress by the addition of abiotic elicitor sodium chloride.171 The addition of this cheaper elicitor (sodium chloride) was found to be effective in sunflower162 and cucumber172 suspension cultures for α-tocopherol enhancement, as well.
When sunflower cell suspension cultures were exposed to hypoxia, there was an increase in the α-tocopherol levels compared to untreated cultures.15 In contrast, it was found that increased dissolved oxygen concentration has a positive influence on the α-tocopherol production up to 9 ppm safflower cell suspension culture cultivated in a bubble column reactor.47 This could mean that both hypoxia and hyperoxia cause a surge in reactive oxygen species levels, causing an increase in α-tocopherol production.
Cadmium chloride, generally toxic to plants, was added in grape cell suspension cultures to study its effect on α-tocopherol.173 In contrast to the use of oil seed crops as a starting material, this study focused on α-tocopherol production from grape cultures. It was observed that α-tocopherol concentration enhanced by 1.7-fold (up to 1.45 μg g–1 DW) at 1.0 mM concentration of cadmium chloride. However, when the concentration of the elicitor was increased to 1.5 mM, a dip in the α-tocopherol yield was observed, which could be due to the negative effect of the elicitor on biomass growth. Also, cadmium effectively increased the α-tocopherol levels in safflower callus cultures acting as an elicitor.174 Exposure to ultraviolet radiation has also shown an effective increase in α-tocopherol concentrations in in vitro cultures. High tocopherol concentrations were observed when grape callus cultures were subjected to ultraviolet-C radiation for 10 min.175
In another study, sunflower hairy root cultures were treated with an increased concentration of reactive red dye, which is one of the significant components in textile wastewater.158 It was reported that the α-tocopherol levels improved to a maximum of 49.3 μg g–1 DW. The dye from the wastewater stream triggered the release of reactive oxygen species, and the antioxidant α-tocopherol levels elevated to combat the stress.
Thus, it is essential that the starting material and type of explant used, the growth hormone and its concentration, the sucrose content and light intensity, and the addition of a suitable precursor and elicitor play a crucial role in enhancing the α-tocopherol productivity in cell cultures. The various bioprocess strategies employed and their effect on the in vitro cultures are highlighted in Table 4.
Table 4. Summary of the Bioprocess Optimization Strategies Employed for Enhancement of α-Tocopherol in In Vitro Cultures.
| species | culture type | strategy employed | enhancement in tocopherol levels (yield/titer/productivity of α-tocopherol)a | control with which yield enhancement was compared | |
|---|---|---|---|---|---|
| Helianthus annuus | callus157 | media optimization | naphthaleneacetic acid (0.5 mg L–1) and 6-benzylaminopurine (0.5 mg L–1) | 2.2-fold (15 μg g–1 FW) | callus grown in Murashige and Skoog basal medium |
| callus157 | media optimization | synergy of naphthaleneacetic acid (0.5 mg L–1), 6-benzylaminopurine (0.5 mg L–1), casamino acids (0.1% w/v), and myo-inositol (0.1% w/v) | 3.6-fold (19 μg g–1 FW) | callus grown in Murashige and Skoog basal medium + naphthaleneacetic acid (0.5 mg L–1) and 6-benzylaminopurine (0.5 mg L–1) | |
| cell suspension157 | media optimization | casamino acids (0.1% w/v) and myo-inositol (0.1% w/v) | 28% (13 μg g–1 FW) | suspension culture grown in absence of casamino acids and myo-inositol | |
| cell suspension154 | photomixotrophic conditions | white fluorescent light (125 μmol photons m–2 s–1) with reduced sucrose (3 g L–1) concentration | 4-fold (18 μg g–1 FW) | culture grown in white light with 30 g L–1 sucrose concentration | |
| cell suspension156 | photomixotrophic conditions | light (125 μmol m–2 s–1) + sucrose (15 g L–1) | 230% (77 μg g–1 DW) | culture grown in heterotrophic conditions (without light) | |
| cell suspension164 | elicitor addition | 72 h treatment of jasmonic acid (5 μM) | 49% (11 μg g–1 FW) | culture grown in absence of elicitor | |
| cell suspension157 | precursor addition | 72 h treatment of homogentisic acid (100 mg L–1) | 30% (16 μg g–1 FW) | culture grown in absence of precursor | |
| cell suspension15 | elicitor exposure | hypoxic conditions | 1.4-fold (140% concentration of control) | culture grown in normal conditions | |
| cell suspension162 | elicitor addition | 48 h treatment sodium chloride (50 mg L–1) | 2.3-fold (46 μg L–1 d–1) | culture grown in absence of elicitor | |
| hairy roots158 | elicitor exposure | 32 h treatment reactive red dye (110 mg L–1) | up to a maximum of 49 μg g–1 DW | ||
| hairy roots158 | light treatment | 16/8 light/dark conditions | 2-fold | culture grown in dark conditions | |
| safflower | callus151 | media optimization | revised tobacco medium with indole-3-butyric acid (2 ppm), kinetin (0.1 ppm), and casamino acid (0.1% w/v) | 5-fold (38 μg g–1 DW) | callus grown in Murashige and Skoog medium with 2,4-D and kinetin |
| suspension culture153 | light treatment | 7000 lx and 16/8 light/dark conditions | 3.3 times (340 μg g–1 DW) | culture grown in dark with optimal media conditions151 | |
| callus176 | media optimization | casein (0.1% w/v) with 2,4-dichlorophenoxy acetic acid (0.3 mg L–1) and 6-benzylaminopurine (1.8 mg L–1) | up to 4.17 μg g–1 FW | culture without addition of casein | |
| cell suspension152 | synergy of media and aeration rate | 8 mg O2 L–1 + addition of conditioning factor | up to 0.09 mg g–1 DW | higher than parent safflower seeds | |
| cell suspension153 | precursor addition | phytol (100 mg L–1) | 5 times (1.4 mg g–1 DW) | culture without addition of precursor | |
| cell suspension171 | elicitor addition | 4 week treatment of dried fungal mats of Trametes versicolor (50 mg L–1) | 12.7-fold (2 mg g–1 FW) | culture without addition of elicitor | |
| cell suspension171 | elicitor addition | 4 week treatment of sodium chloride (50–70 mg L–1) | 1.24-fold (200 μg g–1 FW) | culture without addition of elicitor | |
| callus175 | elicitor exposure | 10 min exposure to UV -C radiation | positive effect | culture without exposure to elicitor | |
| Vitis vinifera | cell suspension173 | elicitor addition | 6 day treatment of cadmium chloride (1 mM) | 1.7-fold (1.45 μg g–1 DW) | culture without addition of elicitor |
| Rosa damascene | callus159 | precursor addition | 21 day treatment of phenyl alanine (100 μM) in dark light | 2-fold (11 ppm) | culture without addition of precursor |
| callus159 | elicitor addition | 21 day treatment of methyl jasmonate (5 μM) in dark light | 4-fold (20 ppm) | culture without addition of elicitor | |
| Argania spinosa | cell suspension170 | precursor addition | 10 day treatment of tyrosine (276 μM) | 2-fold (0.6 mg g–1 DW) | culture without addition of precursor |
| cell suspension170 | elicitor addition | 10 day treatment of titanium dioxide (5 ppm) and tyrosine (276 μM) | 4.5 times (2.7 mg g–1 DW) | culture without addition of elicitor | |
| cell suspension170 | elicitor addition | 10 day treatment of silicon dioxide (5 ppm) and tyrosine (276 μM) | 4.7 times (2.8 mg g–1 DW) | culture without addition of elicitor | |
| Amaranthus caudatus | callus163 | elicitor addition | 72 h treatment of methyl jasmonate (100 μM) | 5-fold (11 μg g–1 DW) | culture without addition of elicitor |
| Arabidopsis | cell suspension164 | elicitor addition | 72 h treatment of jasmonic acid (5 μM) | 66% (9 μg g–1 FW) | culture without addition of elicitor |
| cell suspension167 | exposure to elicitation | anoxic stress in shake flask without mixing for 24 h | 62% (12 μg g–1 FW) | culture without exposure to elicitation | |
| cell suspension167 | exposure to elicitation | 4 h anoxic stress in stirred tank reactor by nitrogen sparging | 14% (8 μg g–1 FW) | culture without exposure to elicitation | |
| Dacus carota | cell suspension168 | elicitor addition | 21 day treatment of cyclodextrin (10–70 mM) | up to a maximum of 172 μg g–1 DW | |
| Cucumis sativus | cell suspension172 | elicitor addition | 72 h treatment of sodium chloride (150 mM) | 50% (4 μg g–1 protein) | culture without exposure to elicitation |
FW, fresh weight; DW, dry weight.
5. Bioreactor Cultivation of Plant Cell Cultures for Large-Scale Production of Vitamin E
For the large-scale production of vitamin E from plant cultures, it is important to gauge productivity in bioreactor cultivations. The cultivation of plant cells in bioreactors has always not been straightforward, and there are limited success stories of bioreactor cultivation for plant cell cultures compared with microbial and mammalian cell cultivations. The challenges in the scale-up of plant cell cultures in bioreactors are due to their specific morphology, which results in aggregation, an increase in viscosity, non-Newtonian behavior, mixing, and oxygen transfer issues, all of which may affect culture growth and productivity.177 The choice of a suitable bioreactor for a particular plant species is critical for the successful scale-up of plant cell cultures from shake flask to industrial bioreactors.178 A systematic approach to understanding the nature of the plant cell cultures and consequent bioprocess optimization and scale-up would be a need of the hour.
Sunflower cultures grown in a shake flask were observed to form cell aggregates with increasing cultivation time, thereby leading to the heterogeneity of the culture.179 Another study reported a significant reduction in mean growth rates of highly aggregated sunflower cell suspension cultures ranging from 0.42 d–1 in a shake flask to 0.16 d–1 in a 3 L stirred tank bioreactor.180 It is key, here, to note that although a large aggregate size of plant cells is undesirable for bioprocessing, it may correspond to increased secondary metabolite levels.181 Thus, the design of bioreactors that consider the heterogeneity and inherent nature of cell aggregation is essential.
Plant cells are generally sensitive to shear. Some cultures can grow in an air-lift reactor with a reduced shear environment, whereas some are successfully scaled up in a conventional stirred tank bioreactor. The design of a bioreactor and optimization of operating parameters with knowledge of the shear threshold of the plant is necessary. The shear sensitivity of safflower cell cultures was studied using computational fluid dynamic approaches to quantify the shear threshold of the culture in both the shake flask and stirred tank bioreactor.182,183 This approach proved to be less time-consuming, economical, and very useful in optimizing the bioreactor design because shear rates are challenging to measure in experiments.
The mixing time for the cultivation of plant cells is the time taken to achieve a significant homogeneity in nutrient and oxygen mass transfer to all the aggregated cells in the suspension. Some reports claim that wave bioreactors and orbital shaking bioreactors provide better mixing at low shear rates and are more suitable for plant cell cultures.184,185 With the advent of bag bioreactors, the use of disposable reactors is on the rise, and they are known for their ease in scale-up for plant cell suspension cultures. Bag bioreactors are known to have less flotation, reduced foam formation, and low power input.177 Though there have been studies of mixing time with water in different culture vessels, it is crucial to account for the rheology of plant cell cultures. Sodium carboxymethyl cellulose solution was used to mimic sunflower cell cultures (up to shear rates of 10 s–1), and the mixing times in a shake flask, wave-mixed bag bioreactor, and orbitally mixed bag bioreactor were studied.177 It was observed that the wave bag bioreactors showed low diagonal mixing and high mixing times, even with increasing agitation rates. However, mixing was not limited in shake flasks and orbital shaking bioreactors for increasing plant cell densities up to 8 g L–1.
In the context of the mass transfer requirement of plant cell cultures, the maximum specific oxygen demand of sunflower cell cultures was determined to be 2 mmol L–1 h–1 by noninvasive real-time monitoring of oxygen concentrations in the shake flask.179 This corresponds to a minimum volumetric mass transfer coefficient requirement of 8 h–1. This was well met for plant cell densities up to 8 g L–1 cultivated in shake flasks, orbital shaking, and wave-mixed reactors by controlling the agitation rate, as studied in the previously discussed work.177 However, both the wave-mixed and orbital shaking reactors showed oxygen limitations at very high viscosities of plant cell cultures. Upon analysis of the kinetics of the plant cells, it was revealed that the cells have low oxygen demands upon reaching the stationary phase. This shows that the prevailing oxygen limitation is insignificant for the plant cell growth. Hence, an integrative study of the culture kinetics with rheology, mass transfer, and mixing time is, thus, essential.
In another study, a sunflower cell suspension culture was grown in a 5 L stirred tank bioreactor with a silicon tube with fine holes to improve aeration and two Rushton impellers.186 This modification enhanced growth and α-tocopherol productivity by reducing the flotation of cells and increasing oxygen stress due to improved aeration, respectively.
Thus, it is important to develop an optimum bioreactor design that is species specific for commercial scale α-tocopherol production.
6. Kinetic Modeling for Prediction of Vitamin E Production
Understanding the complex behavior of plant metabolism can open new avenues for optimization and enhancement strategies. Generally, a model is developed with inputs from experimental data from a real system with sufficient simplifications to establish theoretical forecasts. Further, a comparison is made with experiments and appropriate revisions of the model or the theory as applicable.
In bioprocess, models can be structured or unstructured. Structured models describe each cell as distinguished compartments, whereas unstructured models consider the cell as a black box. Similarly, models can be segregated where every cell is a unique entity in a population, unlike unsegregated models, which do not differentiate single cells. Mathematical modeling of plant in vitro cultures has been extensively reviewed.187 However, modeling for vitamin E production, in particular, has not been well studied.
In a two-decade-old study,188 a structured mathematical model was developed to predict the growth of safflower cell suspension and the production kinetics of α-tocopherol in the cultures. The model focused on the compartmentalization and dependence of α-tocopherol formation on respiratory intermediates. This model was able to show that inoculum size was directly proportional to α-tocopherol levels in the culture. The authors also determined that casamino acids played a key role in α-tocopherol productivity, which is in line with experimental observations.151,153 Thus, there is a promising scope for developing mathematical models that can help understand and predict the kinetic behavior of the cell culture and the production of vitamin E under different conditions in silico.
7. Downstream Processing of Vitamin E from Plant Cultures
Followed by large-scale production of plant cell cultures, it is important to develop sustainable extraction methods for vitamin E from plant cultures. Because of the controlled growth of in vitro cultures, downstream processing is easier than with whole plant extraction because the product is pathogen- and adulterant-free. Popular methods, including solvent extraction by saponification with stabilization by pyrogallol, have been used in multiple studies.146,157,164,167,171 For sustainable and efficient extraction of heat-labile α-tocopherol, supercritical fluid extraction (SFE) is a strategy with bright prospects.43 SFE optimization has been done previously for β-carotene recovery from broccoli byproducts.189 A significant amount of α-tocopherol was recovered from the byproduct, though the concentration was less, which revealed the importance of optimization of SFE for α-tocopherol in plant in vitro cultures. There have also been studies where SFE using carbon dioxide was used to extract sunflower oil from seeds with a high percentage of tocopherols. A recent publication has reviewed the advantages of SFE of tocopherols compared with traditional methods, which is a promising read.190 The prospects of using the SFE-based extraction of metabolites from plant cultures are yet to be explored but could prove to be an efficient method.
8. Conclusions
Among the many prospective avenues available to meet the demands of the natural vitamin E market, in vitro production using plant cell cultures is an attractive option. Strategies to divert fluxes toward maximum α-tocopherol productivity can be identified using omics-based systems modeling workflows and applied through the metabolic engineering of nuclear and chloroplast genome in vitro plant cultures. The rationally engineered cell lines can be integrated with bioprocess optimization strategies to enable multifold enhancement of the widely popular nutraceutical vitamin E. As the next step, sustainable production of the high-yielding transgenic plant cell/hairy root strains can be scaled up to the industrial level with the knowledge of the engineering characteristics of the cultures. Furthermore, the green technologies for efficient solvent extraction can bring down the environmental impact on the overall production process of vitamin E, thereby making it sustainable. Moreover, this systematic framework can also be applied to enhance the production of other diverse high-value, low-volume phytochemicals in plant cell cultures to facilitate the transition from bench to industry.
Acknowledgments
Financial assistance from the Department of Biotechnology (Project no. BT/PR5835/PID/6/688/2012) and Department of Science and Technology (DST) (Project no. SR/FT/LS-52/2012), Government of India, is gratefully acknowledged. M.V.M. would like to acknowledge the Ministry of Education, Government of India, for the Prime Minister’s Research Fellowship (PMRF). A.S. thanks DST for the INSPIRE fellowship for the included PhD thesis work.
Glossary
Abbreviations
- MEP
methylerythritol phosphate
- HGA
homogentisic acid
- HPP
p-hydroxyphenylpyruvate
- HPPD
p-hydroxyphenylpyruvate dioxygenase
- PDP
phytyl diphosphate
- HPT/VTE2
homogentisate phytyl transferase
- GGDP
geranylgeranyl diphosphate
- HGGT
homogentisate geranylgeranyl transferase
- MPBQ
2-methyl-6-phytylbenzoquinol
- MGGBQ
2-methyl-6-geranylgeranylbenzoquinol
- DMPBQ
2,3-dimethyl-5-phytyl-1,4-benzoquinone
- DMGGBQ
2,3-dimethyl-5-geranylgeranyl-1,4-benzoquinone
- TC/VTE1
tocopherol cyclase
- γ-TMT/VTE4
γ-tocopherol methyltransferase
- AVED
ataxia with vitamin E deficiency
- NASH
nonalcoholic steatohepatitis
- NAFLD
nonalcoholic fatty liver disease
- VODD
vegetable oil deodorized distillate
- GSMM
genome-scale metabolic models
- FBA
flux balance analysis
- FVA
flux variability analysis
- FW
fresh weight
- DW
dry weight
- SFE
supercritical fluid extraction
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c05819.
Summary of plant metabolic reconstructions and their applications (PDF)
Author Present Address
‡ Berkeley Lab, Emeryville, California 94608, United States
Author Contributions
§ These authors contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Evans H. M.; Bishop K. S. On the existence of a hitherto unrecognized dietary factor essential for reproduction. Science 1922, 56, 650–651. 10.1126/science.56.1458.650. [DOI] [PubMed] [Google Scholar]
- Azzi A. Many Tocopherols, One Vitamin E. Mol. Aspects Med. 2018, 61, 92–103. 10.1016/j.mam.2017.06.004. [DOI] [PubMed] [Google Scholar]
- Khadangi F.; Azzi A. Vitamin E – The Next 100 Years. Iubmb Life 2018, 71 (4), 411–415. 10.1002/iub.1990. [DOI] [PubMed] [Google Scholar]
- Peh H. Y.; Tan W.S. D.; Liao W.; Wong W.S. F. Vitamin E Therapy beyond Cancer: Tocopherol versus Tocotrienol. Pharmacol. Ther. 2016, 162, 152–169. 10.1016/j.pharmthera.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Frei B.; Traber M.G. The New US Dietary Reference Intakes for Vitamins C and E. Redox Rep. 2001, 6 (1), 5–9. 10.1179/135100001101535978. [DOI] [PubMed] [Google Scholar]
- Jiang Q. Natural Forms of Vitamin E as Effective Agents for Cancer Prevention and Therapy. Adv. Nutr. 2017, 8 (6), 850–867. 10.3945/an.117.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A.; Lee J. C.-Y. Vitamin E: Where Are We Now in Vascular Diseases?. Life 2022, 12 (2), 310. 10.3390/life12020310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Araque A.; Aranda A. G.; Pardo C. L.; Aragues A. R. Los Antioxidantes En El Proceso de Patologías Oculares. Nutr. Hosp. 2017, 34 (2), 469–478. 10.20960/nh.420. [DOI] [PubMed] [Google Scholar]
- Vardi M. Y.; Levy N. S.; Levy A. P. Vitamin E in the Prevention of Cardiovascular Disease: The Importance of Proper Patient Selection. J. Lipid Res. 2013, 54 (9), 2307–2314. 10.1194/jlr.R026641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Yang G.; Luo M.; Lan Q.; Shi X.; Deng H.; Wang N.; Xu X.; Zhang C. Serum Vitamin E Levels and Chronic Inflammatory Skin Diseases: A Systematic Review and Meta-Analysis. PloS 2021, 16 (12), e0261259–e0261259. 10.1371/journal.pone.0261259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idamokoro E. M.; Falowo A. B.; Oyeagu C. E.; Afolayan A. J. Multifunctional Activity of Vitamin E in Animal and Animal Products: A Review. Anim. Sci. J. 2020, 91 (1), e13352. 10.1111/asj.13352. [DOI] [PubMed] [Google Scholar]
- Niki E.; Abe K. Chapter 1 Vitamin E: Structure, Properties and Functions. Vitam. E 2019, 1–11. 10.1039/9781788016216-00001. [DOI] [Google Scholar]
- Valentin H. E.; Qi Q. Biotechnological Production and Application of Vitamin E: Current State and Prospects. Appl. Microbiol. Biotechnol. 2005, 68 (4), 436–444. 10.1007/s00253-005-0017-7. [DOI] [PubMed] [Google Scholar]
- Almagro L.; Sabater-Jara A. B.; Belchí-Navarro S.; Pedreño M. A. Recent Trends in the Biotechnological Production of Tocopherols Using in Vitro Cultures. Phytochem. Rev. 2021, 20, 1193–1207. 10.1007/s11101-021-09742-8. [DOI] [Google Scholar]
- Caretto S.; Nisi R.; Paradiso A.; De Gara L. Tocopherol Production in Plant Cell Cultures. Mol. Nutr. Food Res. 2010, 54 (5), 726–730. 10.1002/mnfr.200900397. [DOI] [PubMed] [Google Scholar]
- DellaPenna D.; Last R. L. Progress in the Dissection and Manipulation of Plant Vitamin E Biosynthesis. Physiol. Plant. 2006, 126 (3), 356–368. 10.1111/j.1399-3054.2006.00611.x. [DOI] [Google Scholar]
- Fritsche S.; Wang X.; Jung C. Recent Advances in Our Understanding of Tocopherol Biosynthesis in Plants: An Overview of Key Genes, Functions, and Breeding of Vitamin E Improved Crops. Antioxidants 2017, 6 (4), 99. 10.3390/antiox6040099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellaud S.; Mène-Saffrané L. Metabolic Origins and Transport of Vitamin E Biosynthetic Precursors. Front. Plant Sci. 2017, 8, 1959–1959. 10.3389/fpls.2017.01959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer M. J. The Antioxidant Effects of Thylakoid Vitamin E (α-Tocopherol). Plant Cell Environ. 1992, 15 (4), 381–392. 10.1111/j.1365-3040.1992.tb00988.x. [DOI] [Google Scholar]
- Munné-Bosch S. A-Tocopherol: A Multifaceted Molecule in Plants. Vitam. Horm. Ser. 2007, 76, 375–392. 10.1016/S0083-6729(07)76014-4. [DOI] [PubMed] [Google Scholar]
- Caretto S.; Paradiso A.; D’Amico L.; De Gara L. Ascorbate and Glutathione Metabolism in Two Sunflower Cell Lines of Differing α-Tocopherol Biosynthetic Capability. Plant Physiol. Biochem. 2002, 40 (6), 509–513. 10.1016/S0981-9428(02)01419-5. [DOI] [Google Scholar]
- Hussain N.; Irshad F.; Jabeen Z.; Shamsi I. H.; Li Z.; Jiang L. Biosynthesis, Structural, and Functional Attributes of Tocopherols in Planta; Past, Present, and Future Perspectives. J. Agric. Food Chem. 2013, 61 (26), 6137–6149. 10.1021/jf4010302. [DOI] [PubMed] [Google Scholar]
- Jin S.; Daniell H. Expression of γ-Tocopherol Methyltransferase in Chloroplasts Results in Massive Proliferation of the Inner Envelope Membrane and Decreases Susceptibility to Salt and Metal-Induced Oxidative Stresses by Reducing Reactive Oxygen Species. Plant Biotechnol. J. 2014, 12 (9), 1274–1285. 10.1111/pbi.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zingg J.-M. Vitamin E: A Role in Signal Transduction. Annu. Rev. Nutr. 2015, 35 (1), 135–173. 10.1146/annurev-nutr-071714-034347. [DOI] [PubMed] [Google Scholar]
- Gomez-Pomar E.; Hatfield E.; Garlitz K.; Westgate P. M.; Bada H. S. Vitamin E in the Preterm Infant: A Forgotten Cause of Hemolytic Anemia. Am. J. Perinatol. 2018, 35 (3), 305–310. 10.1055/s-0037-1607283. [DOI] [PubMed] [Google Scholar]
- Morrissey P. A.; Kiely M. Vitamin E: Physiology and Health Effects. Encyclopedia of Human Nutrition 2013, 390–397. 10.1016/B978-0-12-375083-9.00279-8. [DOI] [Google Scholar]
- Galli F.; Azzi A.; Birringer M.; Cook-Mills J. M.; Eggersdorfer M.; Frank J.; Cruciani G.; Lorkowski S.; Ozer N. K. Vitamin E: Emerging Aspects and New Directions. Free Radic. Biol. Med. 2017, 102, 16–36. 10.1016/j.freeradbiomed.2016.09.017. [DOI] [PubMed] [Google Scholar]
- Rizvi S.; Raza S. T.; Ahmed F.; Ahmad A.; Abbas S.; Mahdi F. The Role of Vitamin E in Human Health and Some Diseases. Sultan Qaboos Univ. Med. J. 2014, 14 (2), 157–165. [PMC free article] [PubMed] [Google Scholar]
- Wallert M.; Börmel L.; Lorkowski S. Inflammatory Diseases and Vitamin E-What Do We Know and Where Do We Go?. Mol. Nutr. Food Res. 2021, 65 (1), 2000097. 10.1002/mnfr.202000097. [DOI] [PubMed] [Google Scholar]
- Regner-Nelke L.; Nelke C.; Schroeter C. B.; Dziewas R.; Warnecke T.; Ruck T.; Meuth S. G. Enjoy Carefully: The Multifaceted Role of Vitamin E in Neuro-Nutrition. Int. J. Mol. Sci. 2021, 22 (18), 10087. 10.3390/ijms221810087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehvari J.; Motlagh F. G.; Najafi M.; Ghazvini M. R. A.; Naeini A. A.; Zare M. Effects of Vitamin E on Seizure Frequency, Electroencephalogram Findings, and Oxidative Stress Status of Refractory Epileptic Patients. Adv. Biomed. Res. 2016, 5 (1), 36–36. 10.4103/2277-9175.178780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabetian S.; Jahromi B. N.; Vakili S.; Forouhari S.; Alipour S. The Effect of Oral Vitamin E on Semen Parameters and IVF Outcome: A Double-Blinded Randomized Placebo-Controlled Clinical Trial. BioMed. Res. Int. 2021, 2021, 5588275. 10.1155/2021/5588275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutalip S. S. M.; Ab-Rahim S.; Rajikin M. H. Vitamin E as an Antioxidant in Female Reproductive Health. Antioxidants 2018, 7 (2), 22. 10.3390/antiox7020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Ramos M.; Vargas L. A.; Van Der Goes T. I. F.; Cervantes-Sandoval A.; Mendoza-Nú′nez V. M. Supplementation of Ascorbic Acid and α-Tocopherol Is Useful to Preventing Bone Loss Linked to Oxidative Stress in Elderly. J. Nutr. Health Aging 2010, 14 (6), 467–472. 10.1007/s12603-010-0099-5. [DOI] [PubMed] [Google Scholar]
- Balbi M. E.; Tonin F. S.; Mendes A. M.; Borba H. H.; Wiens A.; Fernandez-Llimos F.; Pontarolo R. Antioxidant Effects of Vitamins in Type 2 Diabetes: A Meta-Analysis of Randomized Controlled Trials. Diabetol. Metab. Syndr. 2018, 10 (1), 1–12. 10.1186/s13098-018-0318-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas E. R.; Ghosh R. Vitamin E: A Dark Horse at the Crossroad of Cancer Management. Biochem. Pharmacol. 2013, 86 (7), 845–852. 10.1016/j.bcp.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour-Gueddes S. B.; Saidana-Naija D. Vitamin E: Natural Antioxidant in the Mediterranean Diet. Vitam. E Health Dis. - Interact. Dis. Health Asp. 2021, 10.5772/intechopen.99705. [DOI] [Google Scholar]
- Retzlaff D.; Dörfler J.; Kutschan S.; Freuding M.; Büntzel J.; Hübner J. The Vitamin E Isoform α-Tocopherol Is Not Effective as a Complementary Treatment in Cancer Treatment: A Systematic Review. Nutr. Cancer 2022, 74, 2313. 10.1080/01635581.2021.2014905. [DOI] [PubMed] [Google Scholar]
- Galli F.; Bonomini M.; Bartolini D.; Zatini L.; Reboldi G.; Marcantonini G.; Gentile G.; Sirolli V.; Di Pietro N. Vitamin E (α-Tocopherol) Metabolism and Nutrition in Chronic Kidney Disease. Antioxidants 2022, 11 (5), 989–989. 10.3390/antiox11050989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamna F.; Spriano S. M. Vitamin E: A Review of Its Application and Methods of Detection When Combined with Implant Biomaterials. Materials 2021, 14 (13), 3691. 10.3390/ma14133691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salawi A.; Nazzal S. The Rheological and Textural Characterization of Soluplus/Vitamin E Composites. Int. J. Pharm. 2018, 546, 255–262. 10.1016/j.ijpharm.2018.05.049. [DOI] [PubMed] [Google Scholar]
- Lee C. H.; An D. S.; Lee S. C.; Park H. J.; Lee D. S. A Coating for Use as an Antimicrobial and Antioxidative Packaging Material Incorporating Nisin and α-Tocopherol. J. Food Eng. 2004, 62 (4), 323–329. 10.1016/S0260-8774(03)00246-2. [DOI] [Google Scholar]
- Saini R. K.; Keum Y. S. Tocopherols and Tocotrienols in Plants and Their Products: A Review on Methods of Extraction, Chromatographic Separation, and Detection. Food Res. Int. 2016, 82, 59–70. 10.1016/j.foodres.2016.01.025. [DOI] [Google Scholar]
- Cheng K.; Niu Y.; Zheng X. C.; Zhang H.; Chen Y.; Zhang M.; Huang X. X.; Zhang L. L.; Zhou Y. M.; Wang T. A Comparison of Natural (D-α-Tocopherol) and Synthetic (DL-α-Tocopherol Acetate) Vitamin E Supplementation on the Growth Performance, Meat Quality and Oxidative Status of Broilers. Asian-Australas. J. Anim. Sci. 2016, 29 (5), 681–688. 10.5713/ajas.15.0819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troesch B.; Hoeft B.; McBurney M. I.; Eggersdorfer M.; Weber P. Dietary Surveys Indicate Vitamin Intakes below Recommendations Are Common in Representative Western Countries. Br. J. Nutr. 2012, 108 (4), 692–698. 10.1017/S0007114512001808. [DOI] [PubMed] [Google Scholar]
- Péter S.; Friedel A.; Roos F. F.; Wyss A.; Eggersdorfer M.; Hoffmann K.; Weber P. A Systematic Review of Global α-Tocopherol Status as Assessed by Nutritional Intake Levels and Blood Serum Concentrations. Int. J. Vitam. Nutr. Res. 2015, 85, 261–281. 10.1024/0300-9831/a000281. [DOI] [PubMed] [Google Scholar]
- Takeda T.; Seki M.; Furusaki S.; Furuya T.. Effects of Rate Limiting Factors on Vitamin E Production Using Safflower Cells. In Biochemical Engineering for 2001; Furusaki S., Endo I., Matsuno R., Eds.; Springer: Tokyo, 1992; pp 283–285. [Google Scholar]
- Giménez T.; Mula D.; Gea-Botella S.; Martínez-Madrid M. C.; Martí N.; Valero M.; Saura D. Lipase Catalyzed Deacidification of Tocopherol-Rich Distillates Obtained from Natural Vitamin E Sources. Process Biochem. 2019, 77, 70–76. 10.1016/j.procbio.2018.11.008. [DOI] [Google Scholar]
- QYResearch. Global Vitamin E Market Insights and Forecast to 2028; QYRE-Auto-17134; Valuates Reports, 2022. https://reports.valuates.com/market-reports/QYRE-Auto-17134/global-vitamin-e/ (accessed July 15, 2022).
- Ye Z.; Shi B.; Huang Y.; Ma T.; Xiang Z.; Hu B.; Kuang Z.; Huang M.; Lin X.; Tian Z.; Deng Z.; Shen K.; Liu T. Revolution of Vitamin E Production by Starting from Microbial Fermented Farnesene to Isophytol. Innovation 2022, 3, 100228–100228. 10.1016/j.xinn.2022.100228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiev V.; Slavov A.; Vasileva I.; Pavlov A. Plant Cell Culture as Emerging Technology for Production of Active Cosmetic Ingredients. Eng. Life Sci. 2018, 18 (11), 779–798. 10.1002/elsc.201800066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isah T.; Umar S.; Mujib A.; Sharma M. P.; Rajasekharan P. E.; Zafar N.; Frukh A. Secondary Metabolism of Pharmaceuticals in the Plant in Vitro Cultures: Strategies, Approaches, and Limitations to Achieving Higher Yield. Plant Cell Tissue Organ Cult. 2018, 132 (2), 239–265. 10.1007/s11240-017-1332-2. [DOI] [Google Scholar]
- Werner S.; Maschke R. W.; Eibl D.; Eibl R. Bioreactor Technology for Sustainable Production of Plant Cell-Derived Products 2018, 413–432. 10.1007/978-3-319-54600-1_6. [DOI] [Google Scholar]
- Marchev A. S.; Yordanova Z.; Georgiev M. I. Green (Cell) Factories for Advanced Production of Plant Secondary Metabolites. Crit. Rev. Biotechnol. 2020, 40 (4), 443–458. 10.1080/07388551.2020.1731414. [DOI] [PubMed] [Google Scholar]
- Chandran H.; Meena M.; Barupal T.; Sharma K. Plant Tissue Culture as a Perpetual Source for Production of Industrially Important Bioactive Compounds. Biotechnol. Rep. 2020, 26, e00450. 10.1016/j.btre.2020.e00450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J.; Zhang N. On the Way to Commercializing Plant Cell Culture Platform for Biopharmaceuticals: Present Status and Prospect. Pharm. Bioprocess. 2014, 2 (6), 499–518. 10.4155/pbp.14.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunanandaa B.; Qi Q.; Hao M.; Baszis S. R.; Jensen P. K.; Wong Y.-H. H.; Jiang J.; Venkatramesh M.; Gruys K. J.; Moshiri F.; Post-Beittenmiller D.; Weiss J. D.; Valentin H. E. Metabolically Engineered Oilseed Crops with Enhanced Seed Tocopherol. Metab. Eng. 2005, 7 (56), 384–400. 10.1016/j.ymben.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Liu F.; Xiang N.; Hu J. G.; Shijuan Y.; Xie L.; Brennan C. S.; Huang W.; Guo X. The Manipulation of Gene Expression and the Biosynthesis of Vitamin C, E and Folate in Light-and Dark-Germination of Sweet Corn Seeds. Sci. Rep. 2017, 7 (1), 7484–7484. 10.1038/s41598-017-07774-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye V. M.; Bhatia S. K. Metabolic Engineering for the Production of Clinically Important Molecules: Omega-3 Fatty Acids, Artemisinin, and Taxol. Biotechnol. J. 2012, 7 (1), 20–33. 10.1002/biot.201100289. [DOI] [PubMed] [Google Scholar]
- Bock R. Strategies for Metabolic Pathway Engineering with Multiple Transgenes. Plant Mol. Biol. 2013, 83 (1), 21–31. 10.1007/s11103-013-0045-0. [DOI] [PubMed] [Google Scholar]
- Dudareva N.; DellaPenna D. Plant Metabolic Engineering: Future Prospects and Challenges. Curr. Opin. Biotechnol. 2013, 24 (2), 226–228. 10.1016/j.copbio.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Dudareva N.; Klempien A.; Muhlemann J. K.; Kaplan I. Biosynthesis, Function and Metabolic Engineering of Plant Volatile Organic Compounds. New Phytol. 2013, 198 (1), 16–32. 10.1111/nph.12145. [DOI] [PubMed] [Google Scholar]
- Yoon J. M.; Zhao L.; Shanks J. V. Metabolic Engineering with Plants for a Sustainable Biobased Economy. Annu. Rev. Chem. Biomol. Eng. 2013, 4 (1), 211–237. 10.1146/annurev-chembioeng-061312-103320. [DOI] [PubMed] [Google Scholar]
- Moses T.; Pollier J.; Thevelein J. M.; Goossens A. Bioengineering of Plant (Tri)Terpenoids: From Metabolic Engineering of Plants to Synthetic Biology in Vivo and in Vitro. New Phytol. 2013, 200 (1), 27–43. 10.1111/nph.12325. [DOI] [PubMed] [Google Scholar]
- Staniek A.; Bouwmeester H. J.; Fraser P. D.; Kayser O.; Martens S.; Tissier A.; van der Krol S.; Wessjohann L. A.; Warzecha H. Natural Products - Modifying Metabolite Pathways in Plants. Biotechnol. J. 2013, 8 (10), 1159–1171. 10.1002/biot.201300224. [DOI] [PubMed] [Google Scholar]
- Farré G.; Blancquaert D.; Capell T.; Van Der Straeten D.; Christou P.; Zhu C. Engineering Complex Metabolic Pathways in Plants. Annu. Rev. Plant Biol. 2014, 65 (1), 187–223. 10.1146/annurev-arplant-050213-035825. [DOI] [PubMed] [Google Scholar]
- Lau W.; Fischbach M. A.; Osbourn A.; Sattely E. S. Key Applications of Plant Metabolic Engineering. PLOS Biol. 2014, 12 (6), e1001879. 10.1371/journal.pbio.1001879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ncube B.; Van Staden J. Tilting Plant Metabolism for Improved Metabolite Biosynthesis and Enhanced Human Benefit. Molecules 2015, 20 (7), 12698–12731. 10.3390/molecules200712698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J.; Chen Z.; Ban L.; Wu Y.; Huang J.; Chu J.; Fang S.; Wang Z.; Gao H.; Wang X. P-hydroxyphenylpyruvate dioxygenase from Medicago Sativa Is Involved in Vitamin E Biosynthesis and Abscisic Acid-Mediated Seed Germination. Sci. Rep. 2017, 7 (1), 40625–40625. 10.1038/srep40625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharti, Reetu; Kumar V.. Current Approaches and Key Applications of Plant Metabolic Engineering. In Recent Trends and Techniques in Plant Metabolic Engineering; Yadav S., Kumar V., Singh S., Eds.; Springer: Singapore, 2018; pp 47–61. [Google Scholar]
- Birchfield A. S.; McIntosh C. A. Metabolic Engineering and Synthetic Biology of Plant Natural Products – A Minireview. Curr. Plant Biol. 2020, 24, 100163. 10.1016/j.cpb.2020.100163. [DOI] [Google Scholar]
- Bailey J. E. Toward a Science of Metabolic Engineering. Science 1991, 252 (5013), 1668–1675. 10.1126/science.2047876. [DOI] [PubMed] [Google Scholar]
- Chen F.; Dong W.; Zhang J.; Guo X.; Chen J.; Wang Z.; Lin Z.; Tang H.; Zhang L. The Sequenced Angiosperm Genomes and Genome Databases. Front. Plant Sci. 2018, 9, 418–418. 10.3389/fpls.2018.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H. R. A.; Doherty A.; Wu H. Review of Methodologies and a Protocol for the Agrobacterium -Mediated Transformation of Wheat. Plant Methods 2005, 1 (1), 5. 10.1186/1746-4811-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. M.; Jeng S. T.; To K. Y. In Vitro Regeneration, Agrobacterium-Mediated Transformation, and Genetic Assay of Chalcone Synthase in the Medicinal Plant Echinacea Pallida. Plant Cell Tissue Organ Cult. 2017, 130 (1), 117–130. 10.1007/s11240-017-1208-5. [DOI] [Google Scholar]
- Sundararajan S.; Sivaraman B.; Rajendran V.; Ramalingam S. Tissue Culture and Agrobacterium -Mediated Genetic Transformation Studies in Four Commercially Important Indica Rice Cultivars. J. Crop Sci. Biotechnol. 2017, 20 (3), 175–183. 10.1007/s12892-017-0045-0. [DOI] [Google Scholar]
- Stacey M. G.; Cahoon R. E.; Nguyen H. T.; Cui Y.; Sato S.; Nguyen C. T.; Phoka N.; Clark K. M.; Liang Y.; Forrester J.; Batek J.; Do T. P.; Sleper D. A.; Clemente T. E.; Cahoon E. B.; Stacey G. Identification of Homogentisate Dioxygenase as a Target for Vitamin E Biofortification in Oilseeds. Plant Physiol. 2016, 172 (3), 1506–1518. 10.1104/pp.16.00941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naqvi S.; Farré G.; Zhu C.; Sandmann G.; Capell T.; Christou P. Simultaneous Expression of Arabidopsis ρ-Hydroxyphenylpyruvate Dioxygenase and MPBQ Methyltransferase in Transgenic Corn Kernels Triples the Tocopherol Content. Transgenic Res. 2011, 20 (1), 177–181. 10.1007/s11248-010-9393-6. [DOI] [PubMed] [Google Scholar]
- Yabuta Y.; Tanaka H.; Yoshimura S.; Suzuki A.; Tamoi M.; Maruta T.; Shigeoka S. Improvement of Vitamin E Quality and Quantity in Tobacco and Lettuce by Chloroplast Genetic Engineering. Transgenic Res. 2013, 22 (2), 391–402. 10.1007/s11248-012-9656-5. [DOI] [PubMed] [Google Scholar]
- Lu Y.; Rijzaani H.; Karcher D.; Ruf S.; Bock R. Efficient Metabolic Pathway Engineering in Transgenic Tobacco and Tomato Plastids with Synthetic Multigene Operons. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (8), 2703–2704. 10.1073/pnas.1216898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Zhou Y.; Wang Z.; Sun X.; Tang K. Engineering Tocopherol Biosynthetic Pathway in Arabidopsis Leaves and Its Effect on Antioxidant Metabolism. Plant Sci. 2010, 178 (3), 312–320. 10.1016/j.plantsci.2010.01.004. [DOI] [Google Scholar]
- Singh R. K.; Chaurasia A. K.; Bari R.; Sane V. A. Tocopherol Levels in Different Mango Varieties Correlate with MiHPPD Expression and Its Over-Expression Elevates Tocopherols in Transgenic Arabidopsis and Tomato. 3 Biotech 2017, 7 (5), 352. 10.1007/s13205-017-0991-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowell E. F.; McGrath J. M.; Douches D. S. Accumulation of Vitamin E in Potato (Solanum Tuberosum) Tubers. Transgenic Res. 2008, 17 (2), 205–217. 10.1007/s11248-007-9091-1. [DOI] [PubMed] [Google Scholar]
- Ren W.; Zhao L.; Zhang L.; Wang Y.; Cui L.; Tang Y.; Sun X.; Tang K. Molecular Cloning and Characterization of 4-Hydroxyphenylpyruvate Dioxygenase Gene from Lactuca Sativa. J. Plant Physiol. 2011, 168 (10), 1076–1083. 10.1016/j.jplph.2010.12.017. [DOI] [PubMed] [Google Scholar]
- Raclaru M.; Gruber J.; Kumar R.; Sadre R.; Lühs W.; Zarhloul M. K.; Friedt W.; Frentzen M.; Weier D. Increase of the Tocochromanol Content in Transgenic Brassica Napus Seeds by Overexpression of Key Enzymes Involved in Prenylquinone Biosynthesis. Mol. Breed. 2006, 18 (2), 93–107. 10.1007/s11032-006-9014-5. [DOI] [Google Scholar]
- Seo Y. S.; Kim S. J.; Harn C. H.; Kim W. T. Ectopic Expression of Apple Fruit Homogentisate Phytyltransferase Gene (MdHPT1) Increases Tocopherol in Transgenic Tomato (Solanum Lycopersicum Cv. Micro-Tom) Leaves and Fruits. Phytochemistry 2011, 72 (4–5), 321–329. 10.1016/j.phytochem.2010.12.013. [DOI] [PubMed] [Google Scholar]
- Harish M. C.; Dachinamoorthy P.; Balamurugan S.; Murugan S.; Sathishkumar R. Overexpression of Homogentisate Phytyltransferase (HPT) and Tocopherol Cyclase (TC) Enhances α-Tocopherol Content in Transgenic Tobacco. Biol. Plant. 2013, 57 (2), 395–400. 10.1007/s10535-012-0298-5. [DOI] [Google Scholar]
- Collakova E.; DellaPenna D. Homogentisate Phytyltransferase Activity Is Limiting for Tocopherol Biosynthesis in Arabidopsis. Plant Physiol. 2003, 131 (2), 632–642. 10.1104/pp.015222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.; Rijzaani H.; Karcher D.; Ruf S.; Bock R. Efficient Metabolic Pathway Engineering in Transgenic Tobacco and Tomato Plastids with Synthetic Multigene Operons. Proc. Natl. Acad. Sci. U. S. A. 2013, 110 (8), E623–32. 10.1073/pnas.1216898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eenennaam A. L.; Lincoln K.; Durrett T. P.; Valentin H. E.; Shewmaker C. K.; Thorne G. M.; Jiang J.; Baszis S. R.; Levering C. K.; Aasen E. D.; Hao M.; Stein J. C.; Norris S. R.; Last R. L. Engineering Vitamin E Content: From Arabidopsis Mutant to Soy Oil. Plant Cell 2003, 15 (12), 3007–3019. 10.1105/tpc.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.; Lee S. M.; Park S.; Jung J.; Moon J.; Cheong J.; Kim M. Overexpression of Arabidopsis Homogentisate Phytyltransferase or Tocopherol Cyclase Elevates Vitamin E Content by Increasing γ -Tocopherol Level in Lettuce (Lactuca Sativa L.). Mol. Cells 2007, 24 (2), 301–306. [PubMed] [Google Scholar]
- Shintani D.; DellaPenna D. Elevating the Vitamin E Content of Plants through Metabolic Engineering. Science 1998, 282 (5396), 2098–2100. 10.1126/science.282.5396.2098. [DOI] [PubMed] [Google Scholar]
- Tavva V. S.; Kim Y. H.; Kagan I. A.; Dinkins R. D.; Kim K. H.; Collins G. B. Increased α-Tocopherol Content in Soybean Seed Overexpressing the Perilla Frutescens γ-Tocopherol Methyltransferase Gene. Plant Cell Rep. 2006, 26 (1), 61–70. 10.1007/s00299-006-0218-2. [DOI] [PubMed] [Google Scholar]
- Chen D. F.; Zhang M.; Wang Y. Q.; Chen X. W. Expression of γ-Tocopherol Methyltransferase Gene from Brassica Napus Increased α-Tocopherol Content in Soybean Seed. Biol. Plant. 2012, 56 (1), 131–134. 10.1007/s10535-012-0028-z. [DOI] [Google Scholar]
- Yusuf Mohd. A.; Sarin N. B. Antioxidant Value Addition in Human Diets: Genetic Transformation of Brassica Juncea with γ-TMT Gene for Increased α-Tocopherol Content. Transgenic Res. 2007, 16 (1), 109–113. 10.1007/s11248-006-9028-0. [DOI] [PubMed] [Google Scholar]
- Cho E. A.; Lee C. A.; Kim Y. S.; Baek S. H.; de los Reyes B. G.; Yun S. J. Expression of γ-Tocopherol Methyltransferase Transgene Improves Tocopherol Composition in Lettuce (Latuca Sativa L.). Mol. Cells 2005, 19 (1), 16–22. [PubMed] [Google Scholar]
- Ghimire B. K.; Seong E. S.; Lee C. O.; Lim J. D.; Lee J. G.; Yoo J. H.; Chung I.-M.; Kim N. Y.; Yu C. Y. Enhancement of α-Tocopherol Content in Transgenic Perilla Frutescens Containing the g-TMT Gene. Afr. J. Biotechnol. 2011, 10 (13), 2430–2439. 10.5897/AJB10.1541. [DOI] [Google Scholar]
- Lee B. K.; Kim S. L.; Kim K. H.; Yu S. H.; Lee S. C.; Zhang Z.; Kim M. S.; Park H. M.; Lee J. Y. Seed Specific Expression of Perilla γ-Tocopherol Methyltransferase Gene Increases α-Tocopherol Content in Transgenic Perilla (Perilla Frutescens). Plant Cell Tissue Organ Cult. 2007, 92 (1), 47–54. 10.1007/s11240-007-9301-9. [DOI] [Google Scholar]
- Thiele I.; Palsson B. O. A Protocol for Generating a High-Quality Genome-Scale Metabolic Reconstruction. Nat. Protoc. 2010, 5 (1), 93–121. 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson J. D.; Umayam L. A.; Dickinson T.; Hickey E. K.; White O. The Comprehensive Microbial Resource. Nucleic Acids Res. 2001, 29, 123–125. 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q.; Schlueter S. D.; Brendel V. PlantGDB, Plant Genome Database and Analysis Tools. Nucleic Acids Res. 2004, 32 (90001), D354–D359. 10.1093/nar/gkh046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C.; Jaiswal P.; Hebbard C.; Avraham S.; Buckler E. S.; Casstevens T. M.; Hurwitz B. L.; McCouch S. R.; Ni J.; Pujar A.; Ravenscroft D.; Ren L.; Spooner W.; Tecle I. Y.; Thomason J.; Tung C.-W.; Wei X.; Yap I.; Youens-Clark K.; Ware D.; Stein L. Gramene: A Growing Plant Comparative Genomics Resource. Nucleic Acids Res. 2007, 36, D947–D953. 10.1093/nar/gkm968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaver S. M. D.; Lerma-Ortiz C.; Conrad N.; Mikaili A.; Sreedasyam A.; Hanson A. D.; Henry C. S. PlantSEED Enables Automated Annotation and Reconstruction of Plant Primary Metabolism with Improved Compartmentalization and Comparative Consistency. Plant J. 2018, 95 (6), 1102–1113. 10.1111/tpj.14003. [DOI] [PubMed] [Google Scholar]
- Clark K.; Karsch-Mizrachi I.; Lipman D. J.; Ostell J.; Sayers E. W. GenBank. Nucleic Acids Res. 2016, 44, D67–D72. 10.1093/nar/gkv1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R.; Foerster H.; Fulcher C. A.; Kaipa P.; Krummenacker M.; Latendresse M.; Paley S.; Rhee S. Y.; Shearer A. G.; Tissier C.; Walk T. C.; Zhang P.; Karp P. D. The MetaCyc Database of Metabolic Pathways and Enzymes and the BioCyc Collection of Pathway/Genome Databases. Nucleic Acids Res. 2007, 36, D459–D471. 10.1093/nar/gkm900. [DOI] [Google Scholar]
- Kanehisa M.; Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28 (1), 27–30. 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomburg I.; Chang A.; Ebeling C.; Gremse M.; Heldt C.; Huhn G.; Schomburg D. BRENDA, the Enzyme Database: Updates and Major New Developments. Nucleic Acids Res. 2004, 32 (90001), 431–433. 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leinonen R.; Diez F. G.; Binns D.; Fleischmann W.; Lopez R.; Apweiler R. UniProt Archive. Bioinformatics 2004, 20 (17), 3236–3237. 10.1093/bioinformatics/bth191. [DOI] [PubMed] [Google Scholar]
- Ren Q.; Kang K. H.; Paulsen I. T. TransportDB: A Relational Database of Cellular Membrane Transport Systems. Nucleic Acids Res. 2004, 32 (90001), 284–288. 10.1093/nar/gkh016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller L. A.; Zhang P.; Rhee S. Y. AraCyc: A Biochemical Pathway Database for Arabidopsis. Plant Physiol. 2003, 132 (2), 453–460. 10.1104/pp.102.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P.; Wagg J.; Green M. L.; Kaiser D.; Krummenacker M.; Karp P. D. Computational Prediction of Human Metabolic Pathways from the Complete Human Genome. Genome Biol. 2004, 6 (1), R2. 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler I. M.; Collado-Vides J.; Gama-Castro S.; Ingraham J. L.; Paley S. M.; Paulsen I. T.; Peralta-Gil M.; Karp P. D. EcoCyc: A Comprehensive Database Resource for Escherichia Coli. Nucleic Acids Res. 2005, 33, 334–337. 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C. S; DeJongh M.; Best A. A; Frybarger P. M; Linsay B.; Stevens R. L High-Throughput Generation, Optimization and Analysis of Genome-Scale Metabolic Models. Nat. Biotechnol. 2010, 28 (9), 977–982. 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- Karp P. D.; Paley S. M.; Krummenacker M.; Latendresse M.; Dale J. M.; Lee T. J.; Kaipa P.; Gilham F.; Spaulding A.; Popescu L.; Altman T.; Paulsen I. T.; Keseler I. M.; Caspi R. Pathway Tools Version 13.0: Integrated Software for Pathway/Genome Informatics and Systems Biology. Brief. Bioinform. 2010, 11 (1), 40–79. 10.1093/bib/bbp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchel F.; Rodriguez N.; Swainston N.; Wrzodek C.; Czauderna T.; Keller R.; Mittag F.; Schubert M.; Glont M.; Golebiewski M.; van Iersel M.; Keating S.; Rall M.; Wybrow M.; Hermjakob H.; Hucka M.; Kell D. B; Muller W.; Mendes P.; Zell A.; Chaouiya C.; Saez-Rodriguez J.; Schreiber F.; Laibe C.; Drager A.; Le Novere N. Path2Models: Large-Scale Generation of Computational Models from Biochemical Pathway Maps. BMC Syst. Biol. 2013, 7 (1), 116. 10.1186/1752-0509-7-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiele I.; Vlassis N.; Fleming R. M. T. FastGapFill: Efficient Gap Filling in Metabolic Networks. Bioinformatics 2014, 30 (17), 2529–2531. 10.1093/bioinformatics/btu321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker S.; Feist A.; Mo M. L.; Hannum G.; Palsson B.; Herrgard M. J. Quantitative Prediction of Cellular Metabolism with Constraint-Based Models: The COBRA Toolbox. Nat. Protoc. 2007, 2 (3), 727–738. 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- Klamt S.; Saez-Rodriguez J.; Gilles E. D. Structural and Functional Analysis of Cellular Networks with CellNetAnalyzer. BMC Syst. Biol. 2007, 1 (1), 2–2. 10.1186/1752-0509-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha I.; Maia P.; Evangelista P.; Vilaça P.; Soares S.; Pinto J. P.; Nielsen J.; Patil K. R.; Ferreira E. C.; Rocha M. OptFlux: An Open-Source Software Platform for in Silico Metabolic Engineering. BMC Syst. Biol. 2010, 4 (1), 45–45. 10.1186/1752-0509-4-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agren R.; Liu L.; Shoaie S.; Vongsangnak W.; Nookaew I.; Nielsen J. The RAVEN Toolbox and Its Use for Generating a Genome-Scale Metabolic Model for Penicillium Chrysogenum. PLOS Comput. Biol. 2013, 9 (3), e1002980. 10.1371/journal.pcbi.1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hucka M.; Finney A.; Sauro H. M.; Bolouri H.; Doyle J.; Kitano H.; Arkin A. P.; Bornstein B.; Bray D.; Cornish-Bowden A.; Cuellar A. A.; Dronov S.; Gilles E. D.; Ginkel M.; Gor V.; Goryanin I.; Hedley W. J.; Hodgman T. C.; Hofmeyr J.-H. S.; Hunter P. J.; Juty N. S.; Kasberger J. L.; Kremling A.; Kummer U.; Le Novère N.; Loew L. M.; Lucio D.; Mendes P.; Minch E.; Mjolsness E. D.; Nakayama Y.; Nelson M. R.; Nielsen P. F.; Sakurada T.; Schaff J. C.; Shapiro B. E.; Shimizu T. S.; Spence H. D.; Stelling J.; Takahashi K.; Tomita M.; Wagner J.; Wang J. The Systems Biology Markup Language (SBML): A Medium for Representation and Exchange of Biochemical Network Models. Bioinformatics 2003, 19 (4), 524–531. 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Olivier B. J.; Bergmann F. T. The Systems Biology Markup Language (SBML) Level 3 Package: Flux Balance Constraints. J. Integr. Bioinforma. 2015, 12 (2), 269. 10.1515/jib-2015-269. [DOI] [PubMed] [Google Scholar]
- Yun H.; Lee D. Y.; Jeong J.; Lee S.; Lee S. Y. MFAML: A Standard Data Structure for Representing and Exchanging Metabolic Flux Models. Bioinformatics 2005, 21 (15), 3329–3330. 10.1093/bioinformatics/bti502. [DOI] [PubMed] [Google Scholar]
- Demir E.; Cary M. P.; Paley S. M.; Fukuda K.; Lemer C.; Vastrik I.; Wu G.; D’Eustachio P.; Schaefer C. F.; Luciano J. S.; Schacherer F.; Martínez-Flores I.; Hu Z.; Jiménez-Jacinto V.; Joshi-Tope G.; Kandasamy K.; López-Fuentes A.; Mi H.; Pichler E.; Rodchenkov I.; Splendiani A.; Tkachev S.; Zucker J.; Gopinath G. R.; Rajasimha H.; Ramakrishnan R.; Shah I.; Syed M. H.; Anwar N.; Babur Ö.; Blinov M. L.; Brauner E.; Corwin D.; Donaldson S. L.; Gibbons F.; Goldberg R. N.; Hornbeck P.; Luna A.; Murray-Rust P.; Neumann E. K.; Reubenacker O.; Samwald M.; van Iersel M. P.; Wimalaratne S. M.; Allen K.; Braun B.; Whirl-Carrillo M.; Cheung K.-H.; Dahlquist K. D.; Finney A.; Gillespie M.; Glass E. M.; Gong L.; Haw R.; Honig M.; Hubaut O.; Kane D. W.; Krupa S.; Kutmon M.; Leonard J.; Marks D.; Merberg D.; Petri V.; Pico A. R.; Ravenscroft D.; Ren L.; Shah N. H.; Sunshine M.; Tang R.; Whaley R.; Letovksy S.; Buetow K. H.; Rzhetsky A.; Schächter V.; Sobral B. S.; Dogrusoz U.; McWeeney S. K.; Aladjem M. I.; Birney E.; Collado-Vides J.; Goto S.; Hucka M.; Le Novère N.; Maltsev N.; Pandey A.; Thomas P. D.; Wingender E.; Karp P. D.; Sander C.; Bader G. D. The BioPAX Community Standard for Pathway Data Sharing. Nat. Biotechnol. 2010, 28 (9), 935–942. 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glont M.; Nguyen T. V. N.; Graesslin M.; Hälke R.; Ali R.; Schramm J.; Wimalaratne S. M.; Kothamachu V. B.; Rodriguez N.; Swat M. J.; Eils J.; Eils R.; Laibe C.; Malik-Sheriff R. S.; Chelliah V.; Le Novère N.; Hermjakob H. BioModels: Expanding Horizons to Include More Modelling Approaches and Formats. Nucleic Acids Res. 2018, 46, D1248–D1253. 10.1093/nar/gkx1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkin A. P; Cottingham R. W; Henry C. S; Harris N. L; Stevens R. L; Maslov S.; Dehal P.; Ware D.; Perez F.; Canon S.; Sneddon M. W; Henderson M. L; Riehl W. J; Murphy-Olson D.; Chan S. Y; Kamimura R. T; Kumari S.; Drake M. M; Brettin T. S; Glass E. M; Chivian D.; Gunter D.; Weston D. J; Allen B. H; Baumohl J.; Best A. A; Bowen B.; Brenner S. E; Bun C. C; Chandonia J.-M.; Chia J.-M.; Colasanti R.; Conrad N.; Davis J. J; Davison B. H; DeJongh M.; Devoid S.; Dietrich E.; Dubchak I.; Edirisinghe J. N; Fang G.; Faria J. P; Frybarger P. M; Gerlach W.; Gerstein M.; Greiner A.; Gurtowski J.; Haun H. L; He F.; Jain R.; Joachimiak M. P; Keegan K. P; Kondo S.; Kumar V.; Land M. L; Meyer F.; Mills M.; Novichkov P. S; Oh T.; Olsen G. J; Olson R.; Parrello B.; Pasternak S.; Pearson E.; Poon S. S; Price G. A; Ramakrishnan S.; Ranjan P.; Ronald P. C; Schatz M. C; Seaver S. M D; Shukla M.; Sutormin R. A; Syed M. H; Thomason J.; Tintle N. L; Wang D.; Xia F.; Yoo H.; Yoo S.; Yu D. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36 (7), 566–569. 10.1038/nbt.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King Z. A.; Lu J. S.; Dräger A.; Miller P. C.; Federowicz S.; Lerman J. A.; Ebrahim A.; Palsson B. O.; Lewis N. E. BiGG Models: A Platform for Integrating, Standardizing and Sharing Genome-Scale Models. Nucleic Acids Res. 2016, 44, 515–522. 10.1093/nar/gkv1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondi M.; Liò P. Multi -Omics and Metabolic Modelling Pipelines: Challenges and Tools for Systems Microbiology. Microbiol. Res. 2015, 171, 52–64. 10.1016/j.micres.2015.01.003. [DOI] [PubMed] [Google Scholar]
- Varma A. H.; Palsson B. O. Metabolic Flux Balancing: Basic Concepts, Scientific and Practical Use. Nat. Biotechnol. 1994, 12 (10), 994–998. 10.1038/nbt1094-994. [DOI] [Google Scholar]
- Orth J. D.; Thiele I.; Palsson B. O. What Is Flux Balance Analysis. Nat. Biotechnol. 2010, 28 (3), 245–248. 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Dal’Molin C. G.; Nielsen L. K. Plant Genome-Scale Reconstruction: From Single Cell to Multi-Tissue Modelling and Omics Analyses. Curr. Opin. Biotechnol. 2018, 49, 42–48. 10.1016/j.copbio.2017.07.009. [DOI] [PubMed] [Google Scholar]
- Poolman M. G.; Miguet L.; Sweetlove L. J.; Fell D. A. A Genome-Scale Metabolic Model of Arabidopsis and Some of Its Properties. Plant Physiol. 2009, 151 (3), 1570–1581. 10.1104/pp.109.141267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Dal’Molin C. G.; Quek L.-E.; Palfreyman R. W.; Brumbley S. M.; Nielsen L. K. AraGEM, a Genome-Scale Reconstruction of the Primary Metabolic Network in Arabidopsis. Plant Physiol. 2010, 152 (2), 579–589. 10.1104/pp.109.148817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T. C. R.; Poolman M. G.; Howden A. J. M.; Schwarzlander M.; Fell D. a.; Ratcliffe R. G.; Sweetlove L. J. A Genome-Scale Metabolic Model Accurately Predicts Fluxes in Central Carbon Metabolism under Stress Conditions. Plant Physiol. 2010, 154 (1), 311–323. 10.1104/pp.110.158535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintz-Oron S.; Meir S.; Malitsky S.; Ruppin E.; Aharoni A.; Shlomi T. Reconstruction of Arabidopsis Metabolic Network Models Accounting for Subcellular Compartmentalization and Tissue-Specificity. Proc. Natl. Acad. Sci. U. S. A. 2012, 109 (1), 339–344. 10.1073/pnas.1100358109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C. Y. M.; Williams T. C. R.; Poolman M. G.; Fell D. A.; Ratcliffe R. G.; Sweetlove L. J. A Method for Accounting for Maintenance Costs in Flux Balance Analysis Improves the Prediction of Plant Cell Metabolic Phenotypes under Stress Conditions. Plant J. 2013, 75 (6), 1050–1061. 10.1111/tpj.12252. [DOI] [PubMed] [Google Scholar]
- Cheung C.Y. M.; Poolman M. G.; Fell D. A.; Ratcliffe R. G.; Sweetlove L. J. A Diel Flux Balance Model Captures Interactions between Light and Dark Metabolism during Day-Night Cycles in C3 and Crassulacean Acid Metabolism Leaves. Plant Physiol. 2014, 165 (2), 917–929. 10.1104/pp.113.234468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Dal’Molin C. G.; Orellana C. A.; Gebbie L.; Steen J. A.; Hodson M. P.; Chrysanthopoulos P.; Plan M. R.; McQualter R. B.; Palfreyman R. W.; Nielsen L. K. Metabolic Reconstruction of Setaria Italica: A Systems Biology Approach for Integrating Tissue-Specific Omics and Pathway Analysis of Bioenergy Grasses. Front. Plant Sci. 2016, 7 (1138), 1138–1138. 10.3389/fpls.2016.01138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold A.; Nikoloski Z. Bottom-up Metabolic Reconstruction of Arabidopsis and Its Application to Determining the Metabolic Costs of Enzyme Production. Plant Physiol. 2014, 165 (3), 1380–1391. 10.1104/pp.114.235358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan M.; Zhang Z.; Mohanty B.; Kwon J.-Y.; Choi H.-Y.; Nam H.-J.; Kim D.-I.; Lee D.-Y. Elucidating Rice Cell Metabolism under Flooding and Drought Stresses Using Flux-Based Modeling and Analysis. Plant Physiol. 2013, 162 (4), 2140–2150. 10.1104/pp.113.220178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmanan M.; Lim S.-H.; Mohanty B.; Kim J. K.; Ha S.-H.; Lee D.-Y. Unraveling the Light-Specific Metabolic and Regulatory Signatures of Rice through Combined in Silico Modeling and Multiomics Analysis. Plant Physiol. 2015, 169 (4), 3002–3020. 10.1104/pp.15.01379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poolman M. G.; Kundu S.; Shaw R.; Fell D. a. Responses to Light Intensity in a Genome-Scale Model of Rice Metabolism. Plant Physiol. 2013, 162 (2), 1060–1072. 10.1104/pp.113.216762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H.; Cheung C. Y. M.; Hilbers P. A. J.; van Riel N. A. W. Flux Balance Analysis of Plant Metabolism: The Effect of Biomass Composition and Model Structure on Model Predictions. Front. Plant Sci. 2016, 7, 537–537. 10.3389/fpls.2016.00537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R.; Suthers P. F.; Maranas C. D. Zea Mays Irs1563: A Comprehensive Genome-Scale Metabolic Reconstruction of Maize Metabolism. PLoS One 2011, 6 (7), e21784. 10.1371/journal.pone.0021784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira Dal’Molin C. G.; Quek L. E.; Palfreyman R. W.; Brumbley S. M.; Nielsen L. K. C4GEM, a Genome-Scale Metabolic Model to Study C4 Plant Metabolism. PLANT Physiol. 2010, 154 (4), 1871–1885. 10.1104/pp.110.166488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan A.; S V.; Raman K.; Srivastava S. Rational Metabolic Engineering for Enhanced α-Tocopherol Production in Helianthus Annuus Cell Culture. Biochem. Eng. J. 2019, 151, 107256. 10.1016/j.bej.2019.107256. [DOI] [Google Scholar]
- Choi H. S.; Lee Y. J.; Kim T. Y.; Woo H. M. In Silico Identification of Gene Amplification Targets for Improvement of Lycopene Production. Appl. Environ. Microbiol. 2010, 76 (10), 3097–3105. 10.1128/AEM.00115-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mureşan L.; Clapa D.; Rusu T.; Wang T. T. Y.; Park J. B. Soybean Callus—A Potential Source of Tocopherols. Plants 2021, 10 (12), 2571. 10.3390/plants10122571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murthy H. N.; Lee E. J.; Paek K. Y. Production of Secondary Metabolites from Cell and Organ Cultures: Strategies and Approaches for Biomass Improvement and Metabolite Accumulation. Plant Cell Tissue Organ Cult. 2014, 118 (1), 1–16. 10.1007/s11240-014-0467-7. [DOI] [Google Scholar]
- Jamwal K.; Bhattacharya S.; Puri S. K. Plant Growth Regulator Mediated Consequences of Secondary Metabolites in Medicinal Plants. J. Appl. Res. Med. Aromat. Plants 2018, 9, 26–38. 10.1016/j.jarmap.2017.12.003. [DOI] [Google Scholar]
- Furuya T.; Yoshikawa T.; Kimura T.; Kaneko H. Production of Tocopherols by Cell Culture of Safflower. Phytochemistry 1987, 26 (10), 2741–2747. 10.1016/S0031-9422(00)83582-7. [DOI] [Google Scholar]
- Takeda T.; Seki M.; Furusaki S.; Furuya T. Factors Affecting Vitamin E Production Using Plant Cell Culture of Carthamus Tinctorius. J. Chem. Eng. Jpn. 1993, 26 (5), 470–474. 10.1252/jcej.26.470. [DOI] [Google Scholar]
- Furuya T.; Yoshikawa T.. Carthamus Tinctorius L. (Safflower): Production of Vitamin E in Cell Cultures. In Medicinal and Aromatic Plants III. Biotechnology in Agriculture and Forestry; Bajaj Y. P. S., Ed.; Springer: Berlin, Heidelberg, 1991; pp 142–155. [Google Scholar]
- Fachechi C.; Nisi R.; Gala R.; Leone A.; Caretto S. Tocopherol Biosynthesis Is Enhanced in Photomixotrophic Sunflower Cell Cultures. Plant Cell Rep. 2007, 26 (4), 525–530. 10.1007/s00299-006-0268-5. [DOI] [PubMed] [Google Scholar]
- Hüsemann W.; Barz W. Photoautotrophic Growth and Photosynthesis in Cell Suspension Cultures of Chenopodium Rubrum. Physiol. Plant. 1977, 40 (2), 77–81. 10.1111/j.1399-3054.1977.tb01498.x. [DOI] [PubMed] [Google Scholar]
- Geipel K.; Song X.; Socher M. L.; Kümmritz S.; Püschel J.; Bley T.; Ludwig-Müller J.; Steingroewer J. Induction of a Photomixotrophic Plant Cell Culture of Helianthus Annuus and Optimization of Culture Conditions for Improved α-Tocopherol Production. Appl. Microbiol. Biotechnol. 2014, 98 (5), 2029–2040. 10.1007/s00253-013-5431-7. [DOI] [PubMed] [Google Scholar]
- Caretto S.; Speth E. B.; Fachechi C.; Gala R.; Zacheo G.; Giovinazzo G. Enhancement of Vitamin E Production in Sunflower Cell Cultures. Plant Cell Rep. 2004, 23 (3), 174–179. 10.1007/s00299-004-0799-6. [DOI] [PubMed] [Google Scholar]
- Srikantan C.; Suraishkumar G. K.; Srivastava S. Effect of Light on the Kinetics and Equilibrium of the Textile Dye (Reactive Red 120) Adsorption by Helianthus Annuus Hairy Roots. Bioresour. Technol. 2018, 257, 84–91. 10.1016/j.biortech.2018.02.075. [DOI] [PubMed] [Google Scholar]
- Olgunsoy P.; Ulusoy S.; Akçay U. Ç. Metabolite Production and Antibacterial Activities of Callus Cultures from Rosa Damascena Mill. Petals. Marmara Pharm. J. 2017, 21 (3), 590–597. 10.12991/marupj.319331. [DOI] [Google Scholar]
- Jawahar G.; Punita D. L.; Rajasheker G.; Manoharachary C.; Venkatachalam P.; Kishor P. B. K. Feeding Elicitors and Precursors Enhance Colchicine Accumulation in Morphogenic Cultures of Gloriosa Superba L. Plant Cell Tissue Organ Cult. 2018, 135 (2), 235–245. 10.1007/s11240-018-1459-9. [DOI] [Google Scholar]
- Watcharatanon K.; Ingkaninan K.; Putalun W. Improved Triterpenoid Saponin Glycosides Accumulation in in Vitro Culture of Bacopa Monnieri (L.) Wettst with Precursor Feeding and LED Light Exposure. Ind. Crops Prod. 2019, 134, 303–308. 10.1016/j.indcrop.2019.04.011. [DOI] [Google Scholar]
- Srinivasan A.; Sundaram V.; Vidya Muthulakshmi M.; Srivastava S. Multi-Fold Enhancement in Vitamin E (α-Tocopherol) Production via Integration of Bioprocess Optimisation and Metabolic Engineering in Cell Suspension of Sunflower. J. Plant Biochem. Biotechnol. 2022, 31, 154–167. 10.1007/s13562-021-00671-3. [DOI] [Google Scholar]
- Rahimi S.; Kim Y. J.; Yang D. C. Production of Ginseng Saponins: Elicitation Strategy and Signal Transductions. Appl. Microbiol. Biotechnol. 2015, 99 (17), 6987–6996. 10.1007/s00253-015-6806-8. [DOI] [PubMed] [Google Scholar]
- Narayani M.; Srivastava S. Elicitation: A Stimulation of Stress in in Vitro Plant Cell/Tissue Cultures for Enhancement of Secondary Metabolite Production. Phytochem. Rev. 2017, 16 (6), 1227–1252. 10.1007/s11101-017-9534-0. [DOI] [Google Scholar]
- Antognoni F.; Faudale M.; Poli F.; Biondi S. Methyl Jasmonate Differentially Affects Tocopherol Content and Tyrosine Amino Transferase Activity in Cultured Cells of Amaranthus Caudatus and Chenopodium Quinoa. Plant Biol. 2009, 11 (2), 161–169. 10.1111/j.1438-8677.2008.00110.x. [DOI] [PubMed] [Google Scholar]
- Gala R.; Mita G.; Caretto S. Improving α-Tocopherol Production in Plant Cell Cultures. J. Plant Physiol. 2005, 162 (7), 782–784. 10.1016/j.jplph.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Nisi R.; Paradiso A.; De Gara L.; D’Amico L.; Caretto S. Cultivation of Arabidopsis Cell Cultures in a Stirred Bioreactor at Variable Oxygen Levels: Influence on Tocopherol Production. Plant Biosyst. 2010, 144 (3), 721–724. 10.1080/11263501003755572. [DOI] [Google Scholar]
- Miras-Moreno B.; Almagro L.; Pedreño M. A.; Sabater-Jara A. B. Enhanced Accumulation of Phytosterols and Phenolic Compounds in Cyclodextrin-Elicited Cell Suspension Culture of Daucus Carota. Plant Sci. 2016, 250, 154–164. 10.1016/j.plantsci.2016.06.008. [DOI] [PubMed] [Google Scholar]
- Almagro L.; Tudela L. R.; Sabater-Jara A. B.; Miras-Moreno B.; Pedreño M. A. Cyclodextrins Increase Phytosterol and Tocopherol Levels in Suspension Cultured Cells Obtained from Mung Beans and Safflower. Biotechnol. Prog. 2017, 33 (6), 1662–1665. 10.1002/btpr.2525. [DOI] [PubMed] [Google Scholar]
- Hegazi G.; Ibrahim W. M.; Hendawy M. H.; Salem H. M.; Ghareb H. Improving α-tocopherol accumulation in Argania spinosa suspension cultures by precursor and nano particles feeding. Plant Arch. 2020, 20, 2431–2437. [Google Scholar]
- Chavan S. P.; Lokhande V. H.; Nitnaware K. M.; Nikam T. D. Influence of Growth Regulators and Elicitors on Cell Growth and α-Tocopherol and Pigment Productions in Cell Cultures of Carthamus Tinctorius L. Appl. Microbiol. Biotechnol. 2011, 89 (6), 1701–1707. 10.1007/s00253-010-3014-4. [DOI] [PubMed] [Google Scholar]
- Naliwajski M. R.; Skłodowska M. The Oxidative Stress and Antioxidant Systems in Cucumber Cells during Acclimation to Salinity. Biol. Plant. 2014, 58 (1), 47–54. 10.1007/s10535-013-0378-1. [DOI] [Google Scholar]
- Çetin E. S.; Babalik Z.; Hallac-Turk F.; Gokturk-Baydar N. The Effects of Cadmium Chloride on Secondary Metabolite Production in Vitis Vinifera Cv. Cell Suspension Cultures. Biol. Res. 2014, 47 (1), 47–47. 10.1186/0717-6287-47-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namjooyan S.; Khavarinejad R.; Bernard F.; Namdjoyan S.; Piri H. The Effect of Cadmium on Growth and Antioxidant Responses in the Safflower (Carthamus Tinctorius L.) Callus. Turk. J. Agric. For. 2012, 36, 2. 10.3906/tar-1008-1175. [DOI] [Google Scholar]
- Cetin E. S. Induction of Secondary Metabolite Production by UV-C Radiation in Vitis Vinifera L. Öküzgözü Callus Cultures. Biol. Res. 2014, 47 (1), 37–37. 10.1186/0717-6287-47-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.; Ou S.; Peilin C. Optimization of Conditions for Safflower Cell Culture and Accumulation of Cellicolous Product Tocopherols. Chin. J. Biotechnol. 1999, 15 (4), 231. [PubMed] [Google Scholar]
- Werner S.; Greulich J.; Geipel K.; Steingroewer J.; Bley T.; Eibl D. Mass Propagation of Helianthus Annuus Suspension Cells in Orbitally Shaken Bioreactors : Improved Growth Rate in Single-Use Bag Bioreactors. Eng. Life Sci. 2014, 14 (6), 676–684. 10.1002/elsc.201400024. [DOI] [Google Scholar]
- Srikantan C.; Srivastava S.. Bioreactor Design and Analysis for Large-Scale Plant Cell and Hairy Root Cultivation. In Hairy Roots; Srivastava V., Mehrotra S., Mishra S., Eds.; Springer: Singapore, 2018; pp 147–182. [Google Scholar]
- Geipel K.; Socher M. L.; Haas C.; Bley T.; Steingroewer J. Growth Kinetics of a Helianthus Annuus and a Salvia Fruticosa Suspension Cell Line: Shake Flask Cultivations with Online Monitoring System. Eng. Life Sci. 2013, 13 (6), 593–602. 10.1002/elsc.201200148. [DOI] [Google Scholar]
- Scragg A. H. Large-Scale Cultivation of Helianthus Annuus Cell Suspensions. Enzyme Microb. Technol. 1990, 12 (2), 82–85. 10.1016/0141-0229(90)90077-4. [DOI] [Google Scholar]
- Hulst A. C.; Meyer M. M. T.; Breteler H.; Tramper J. Effect of Aggregate Size in Cell Cultures of Tagetes Patula on Thiophene Production and Cell Growth. Appl. Microbiol. Biotechnol. 1989, 30 (1), 18–25. 10.1007/BF00255991. [DOI] [Google Scholar]
- Liu Y.; Wang Z.; Zhang J.; Xia J.; Chu J.; Zhang S.; Zhuang Y. Quantitative Evaluation of the Shear Threshold on Carthamus Tinctorius L. Cell Growth with Computational Fluid Dynamics in Shaken Flask Bioreactors. Biochem. Eng. J. 2016, 113, 66. 10.1016/j.bej.2016.06.001. [DOI] [Google Scholar]
- Liu Y.; Wang Z. J.; Xia J.; Haringa C.; Liu Y.; Chu J.; Zhuang Y.; Zhang S. Application of Euler-Lagrange CFD for Quantitative Evaluating the Effect of Shear Force on Carthamus Tinctorius L. Cell in a Stirred Tank Bioreactor. Biochem. Eng. J. 2016, 114, 209–217. 10.1016/j.bej.2016.07.006. [DOI] [Google Scholar]
- Eibl R.; Eibl D. Design of Bioreactors Suitable for Plant Cell and Tissue Cultures. Phytochem. Rev. 2008, 7 (3), 593–598. 10.1007/s11101-007-9083-z. [DOI] [Google Scholar]
- Terrier B.; Courtois D.; Henault N.; Cuvier A. R. G.; Bastin M.; Aknin A.; Dubreuil J.; Petiard V. Two New Disposable Bioreactors for Plant Cell Culture: The Wave and Undertow Bioreactor and the Slug Bubble Bioreactor. Biotechnol. Bioeng. 2007, 96 (5), 914–923. 10.1002/bit.21187. [DOI] [PubMed] [Google Scholar]
- Haas C.; Weber J.; Ludwig-Müller J.; Deponte S.; Bley T.; Georgiev M. Flow Cytometry and Phytochemical Analysis of a Sunflower Cell Suspension Culture in a 5-L Bioreactor. Z. Für Naturforschung C 2008, 63, 699. 10.1515/znc-2008-9-1015. [DOI] [PubMed] [Google Scholar]
- Maschke R.; Geipel K.; Bley T. Modeling of Plant in Vitro Cultures: Overview and Estimation of Biotechnological Processes. Biotechnol. Bioeng. 2015, 112 (1), 1–12. 10.1002/bit.25346. [DOI] [PubMed] [Google Scholar]
- Takeda T.; Takeuchi Y.; Seki M.; Furusaki S.; Matsuoka H. Kinetic analysis of cell growth and vitamin e production in plant cell culture of Carthamus tinctorius using a structured model. Biochem. Eng. J. 1998, 1 (3), 233–242. 10.1016/S1369-703X(98)00007-2. [DOI] [Google Scholar]
- Borja-Martínez M.; Lozano-Sánchez J.; Borrás-Linares I.; Pedreño M. A.; Sabater-Jara A. B. Revalorization of Broccoli By-Products for Cosmetic Uses Using Supercritical Fluid Extraction. Antioxidants 2020, 9 (12), 1195. 10.3390/antiox9121195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafaei N.; Rempel C. B.; Scanlon M. G.; Jones P. J. H.; Eskin M. N. A. Application of Supercritical Fluid Extraction (SFE) of Tocopherols and Carotenoids (Hydrophobic Antioxidants) Compared to Non-SFE Methods. AppliedChem. 2022, 2 (2), 68–92. 10.3390/appliedchem2020005. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.