Significance
Although the genetic intricacies of acute myeloid leukemia have been unmasked, the transcriptomic landscape remains incompletely defined and poorly translated into clinical practice. In this study, we evaluated the transcriptome repertoire in a large multicenter AML cohort and established eight robust gene expression-based molecular subgroups (G1–G8) of AML, including two previously unidentified and three redefined subgroups. Each subgroup displayed characteristic clinical features, genetic lesions, and developmental hierarchies. This classification system reflects the complex interplay of regulatory circuitry in this disease and complements and enriches current, well-recognized genome-based categorization schemes, which may, thus, provide innovative insights into disease pathogenesis. Moreover, this transcriptomic classification demonstrated prognostic value and provided subgroup-specific drug sensitivity information, which may facilitate therapeutic decision-making for AML patients.
Keywords: acute myeloid leukemia, RNA-Seq, molecular classification, cell differentiation, drug sensitivity
Abstract
The current classification of acute myeloid leukemia (AML) relies largely on genomic alterations. Robust identification of clinically and biologically relevant molecular subtypes from nongenomic high-throughput sequencing data remains challenging. We established the largest multicenter AML cohort (n = 655) in China, with all patients subjected to RNA sequencing (RNA-Seq) and 619 (94.5%) to targeted or whole-exome sequencing (TES/WES). Based on an enhanced consensus clustering, eight stable gene expression subgroups (G1–G8) with unique clinical and biological significance were identified, including two unreported (G5 and G8) and three redefined ones (G4, G6, and G7). Apart from four well-known low-risk subgroups including PML::RARA (G1), CBFB::MYH11 (G2), RUNX1::RUNX1T1 (G3), biallelic CEBPA mutations or -like (G4), four meta-subgroups with poor outcomes were recognized. The G5 (myelodysplasia-related/-like) subgroup enriched clinical, cytogenetic and genetic features mimicking secondary AML, and hotspot mutations of IKZF1 (p.N159S) (n = 7). In contrast, most NPM1 mutations and KMT2A and NUP98 fusions clustered into G6–G8, showing high expression of HOXA/B genes and diverse differentiation stages, from hematopoietic stem/progenitor cell down to monocyte, namely HOX-primitive (G7), HOX-mixed (G8), and HOX-committed (G6). Through constructing prediction models, the eight gene expression subgroups could be reproduced in the Cancer Genome Atlas (TCGA) and Beat AML cohorts. Each subgroup was associated with distinct prognosis and drug sensitivities, supporting the clinical applicability of this transcriptome-based classification of AML. These molecular subgroups illuminate the complex molecular network of AML, which may promote systematic studies of disease pathogenesis and foster the screening of targeted agents based on omics.
Acute myeloid leukemia (AML) is a group of myeloid neoplasms characterized by high heterogeneity in clinical courses and responses to therapy (1). Leveraging advances in cytogenetics, molecular biology, and next-generation sequencing (NGS) technologies, an accumulating body of novel prognostic markers and therapeutic targets have been identified (2). The classification of AML has accordingly shifted from the French-American-British (FAB) morphological subtyping to the more refined World Health Organization (WHO) system (3). Additionally, the emergence of new targeted agents, as exemplified by FLT3, BCL2, and IDH1/2 inhibitors (4–7), has improved the long-term survival of a subset of AML patients.
Since 2010, several genomic and transcriptomic studies have been conducted in AML and other acute leukemia. The Cancer Genome Atlas (TCGA) program has dissected the genomic landscape of AML and proposed nine categories of mutated genes (8). Another landmark research has recommended 11 distinct AML classes based solely on genetic abnormalities. However, 4% of AML patients met the criteria of two or more classes, 11% remained unclassified, and 5% did not carry driver mutations (9). Hence, there is an urgent need to exploit and integrate more information beyond genomic alterations to further refine the classification and treatment strategies of the disease.
RNA-sequencing (RNA-Seq) provides comprehensive and multifaceted information, which can not only scrutinize gene fusions and mutations but also dissect the gene expression profiling (GEP), holding great potential for improving the classification framework of AML (10). Besides, recent works have reported that the cellular origin of AML can be inferred from bulk transcriptomes and have underscored the association between differentiation hierarchies and sensitivities to targeted inhibitors (11–14). Mer et al. (12) recently classified NPM1-mutated AML into primitive and committed subtypes based on the presence or absence of stemness. The primitive AML cells conferred stem cell signatures and poor prognosis, while the differentiated monocyte-like AML cells (committed) expressed immunomodulatory factors and suppressed T cells (11). Via mining large multicenter RNA-Seq data, we and others have successfully identified emerging molecular subtypes in B cell and T cell acute lymphoblastic leukemia (ALL) (15–17).
However, the robust classification of molecular subtypes based on integrative genomic and transcriptomic features in AML is still challenging. Inconsistent clustering results and complex cellular compositions have been observed in previous works (8, 18, 19). Small sample sizes, single hierarchical clustering without bias feature filtration or cross-validation, and limited biological/clinical interpretation have long hindered the stability and wide application of emerging classification systems in this disease. On the other hand, some published works have ignored dominant signals of specific genetic lesions or gene expression-like subtypes when analyzing the association between differentiation stages and molecular inhibitors. In fact, several genomic markers including t(8;21), inv(16)/t(16;16), and t(15;17) could be determinants of distinct gene expression clusters (20). Taken together, interpreting the complicated interplay and functional impact of leukemogenic events requires the establishment of stable molecular subtypes combining genetic abnormalities, gene expression landscapes, and cellular differentiation states in large AML cohorts.
To address these issues, we established the largest omics cohort of AML patients in China from three centers. Stable molecular subgroups (G1–G8) with characteristic clinical/biological signatures and cellular differentiation hierarchies were identified, which were reproduced in the TCGA LAML (8) and Beat AML (14, 19) cohorts.
Results
Genetic Mutations and Fusion Genes Identified in the Multicenter Cohort.
The study overview, flow diagram, and clinical characteristics of 655 newly diagnosed AML patients are provided (SI Appendix, Figs. S1 and S2 and Table S1 and Dataset S1). The frequency of recurrent genetic mutations was revealed by the combination of RNA-Seq and targeted or whole-exome sequencing (TES/WES). We identified at least one genetic lesion in 649 of 655 (99.1%) AML patients at diagnosis. A higher mutation rate of CEBPA gene was observed in this study, including 96 (14.7%) biallelic CEBPA (biCEBPA) and 24 (3.7%) monoallelic CEBPA (moCEBPA) mutations (SI Appendix, Fig. S3 A and B and Dataset S2). Consistently, Wilhelmson et al. (21) previously summarized that a higher incidence of biCEBPA mutations could be seen in Asian (6–15%; average 12%) as compared to Caucasian (2–6%; average 4%) populations, reflecting a possible difference in genetic backgrounds between the two counterparts. Several highly mutated genes in AML exhibited dysregulated expression levels, such as CEBPA, RUNX1, and FLT3 (SI Appendix, Fig. S4). Additionally, 38.0% (249/655) of newly diagnosed AML patients harbored at least one fusion, with entity-defining fusions, namely PML::RARA, CBFB::MYH11, RUNX1::RUNX1T1, and KMT2A translocations ranking the top (SI Appendix, Fig. S5), consistent with the TCGA and Beat reports (SI Appendix, Fig. S6). A total of 18 (2.7%) NUP98 fusions were detected. Among fusion-positive cases, 16 fusions were barely reported, including two NUP98 fusions (NUP98::HOXD12 and NUP98::TNRC18) (SI Appendix, Fig. S5 and Table S2). The RNA gene MIR99AHG was involved in RUNX1::MIR99AHG and NRIP1::MIR99AHG, and both of them exhibited significant upregulation of MIR99AHG (SI Appendix, Fig. S7).
Consensus Clustering Defines Stable Gene Expression Subgroups of AML.
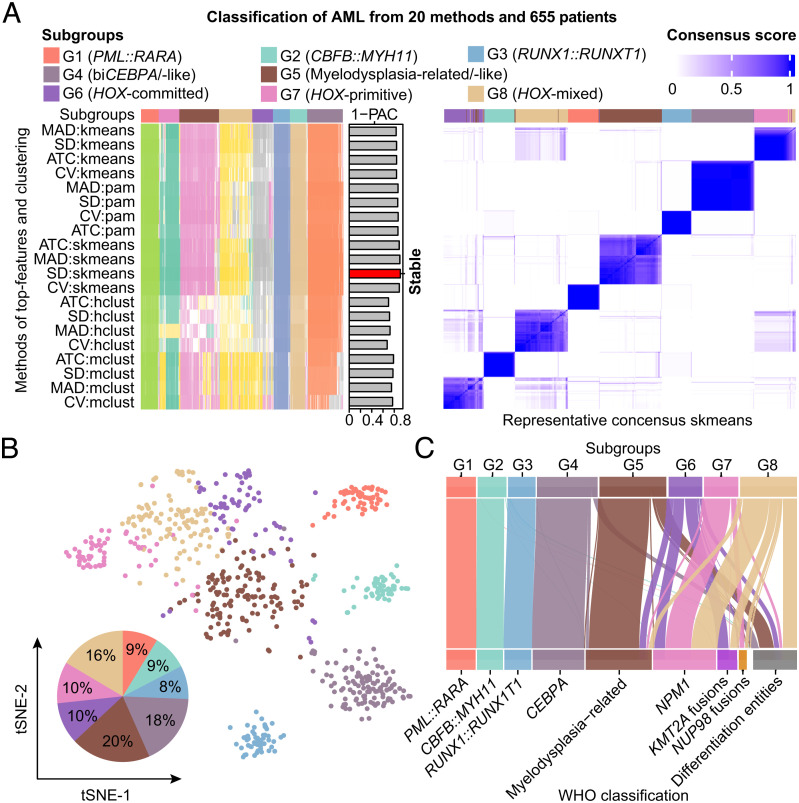
After adjusting the batch effect of three datasets from China (SI Appendix, Fig. S8), we identified eight stable subgroups (G1–G8) in AML (Fig. 1 A and B) based on consensus clustering (twenty methods and gradient sampling of rows/columns) of RNA-Seq data (n = 655) using 859 genes with the greatest variance (Datasets S3 and S4), which was more robust compared with unsupervised hierarchical clustering (SI Appendix, Fig. S9). The correlation between G1–G8 subgroups and AML entities defined by the latest WHO classification (3) was adopted to determine the cutoff of the top-feature selection (Fig. 1C). The former four subgroups (G1–G4) overlapped almost with the relatively favorable genetic lesions of the WHO classification. Transcription factor fusions PML::RARA, CBFB::MYH11, and RUNX1::RUNX1T1 exclusively clustered into the G1, G2, and G3 subgroups, respectively. The G4 subgroup harbored nearly all (95/96, 99.0%) biCEBPA mutations, eight (8/9, 88.9%) moCEBPA mutations with loss of heterozygosity (LOH) and 11 CEBPA wild-type (WT) cases, termed as biCEBPA/-like. Among moCEBPA mutations located in the basic leucine zipper (bZIP) region, except for two with LOH, the other three did not cluster with G4. By contrast, G5–G8 subgroups lacked a single strong subgroup-defining molecular event. The G5 subgroup was represented by AML, myelodysplasia-related (AML-MR), and those only defined by differentiation (FAB subtypes), hence this subgroup was nominated as myelodysplasia-related/-like (MR/-like). The G6–G8 subgroups encompassed NPM1 mutations, KMT2A and NUP98 fusions, and differentiation entities, indicating a relatively high heterogeneity in these gene expression-defined clusters (Fig. 1C).
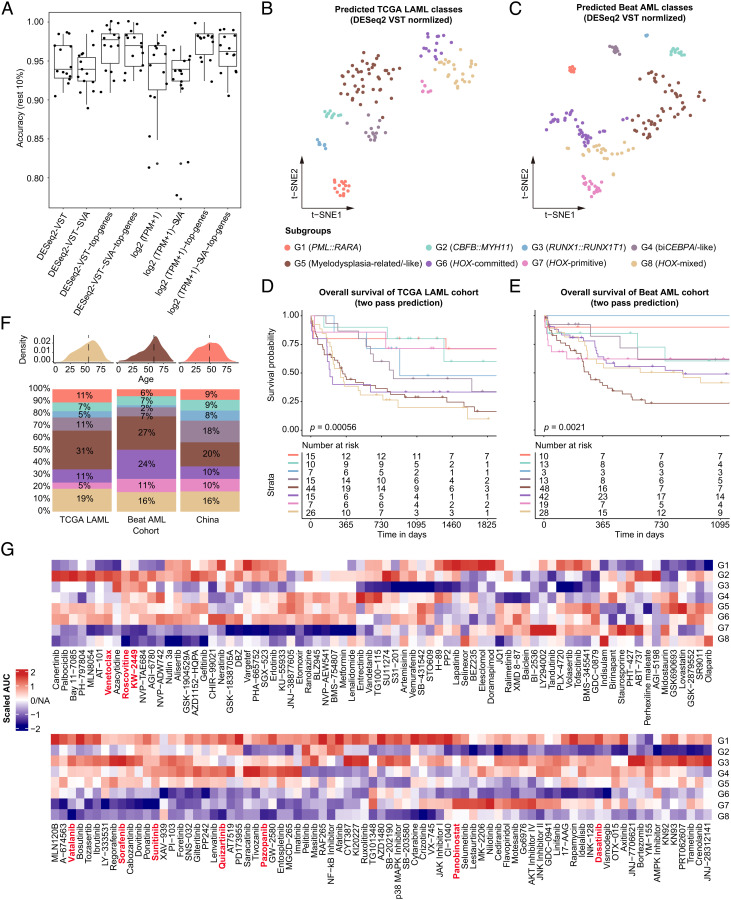
Fig. 1.
Characteristic AML subgroups defined by gene expression profiling. (A) Consensus clustering (membership heatmap) of gene expression profiling from 655 primary AML patients with the use of 20 different computational parameters. The PAC negatively indicates the stability of clustering results (Left). Heatmap displays representative consensus clustering method (skmeans) based on top variance genes, which shows the consistency of two samples in the same subgroup (Right). (B) Scatters of eight gene expression subgroups via tSNE visualization are shown. (C) Sankey plot indicates the relationship between the defined molecular subgroups and disease entities defined by WHO classification. PAC, proportion of ambiguous clustering; tSNE, t-distributed stochastic neighbor embedding; WHO, World Health Organization.
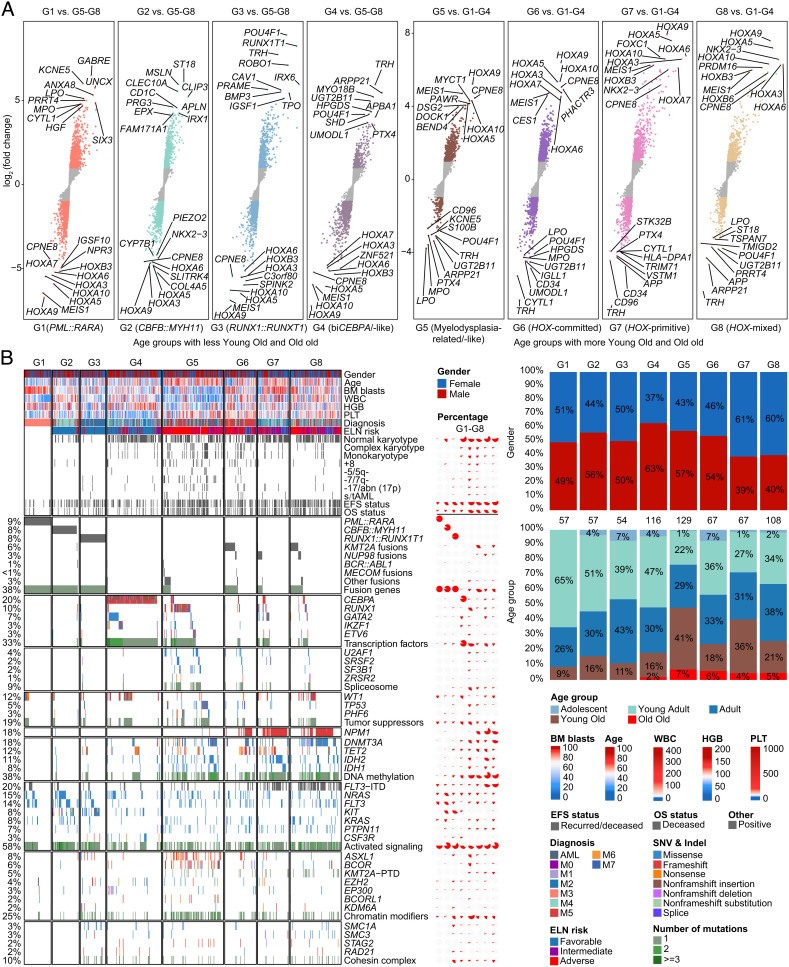
Compared with G1–G4, G5–G8 subgroups conferred higher expression levels of HOXA/B and MEIS1 and rarely reported calcium-dependent CPNE8 genes, representing the most striking discrepancies between the two counterparts (Fig. 2A and SI Appendix, Fig. S10 and Dataset S5). Genetic mutations involved in DNA methylation genes, chromatin modifiers, and spliceosomes were significantly enriched in G5–G8, which was concordant with the predominance of elderly patients, intermediate to high ELN risk, and more relapses and deaths in these subgroups (Fig. 2B and SI Appendix, Fig. S3 C and D and Table S3 and Dataset S6). The G5 subgroup mainly incorporated RUNX1, TP53, PHF6, and “secondary-type” mutations (ASXL1, BCOR, EZH2, STAG2, U2AF1, SRSF2, SF3B1, and ZRSR2), which were commonly enriched in myelodysplastic syndrome (MDS)-transformed AML (22). Consistently, patients in G5 carried more complex karyotype, monosomal karyotype, and abnormalities of chromosomes 5, 7, and 17, while they had lower bone marrow (BM) blasts and white blood cell count (WBC) at diagnosis (Fig. 2B). Meanwhile, rare IKZF1 N159S hotspot mutations (n = 7) uniformly clustered into the G5 subgroup, while the majority of other IKZF1 mutations cooccurred with biCEBPA and fell into the G4 subgroup. The last three subgroups (G6–G8) aggregated more NPM1 mutations, KMT2A and NUP98 fusions, FLT3-internal tandem duplication (FLT3-ITD), and KMT2A-partial tandem duplication (KMT2A-PTD). Notably, the frequency of DNMT3A/NPM1/FLT3-ITD triple-mutated AML was higher in the G8 subgroup, with a percentage of 4.5%, 6.0%, and 22.2% in G6, G7, and G8, respectively, whereas the concurrence of TET2 or IDH2 with NPM1/FLT3-ITD mutations was more common in G7, which accounted for 0, 28.4%, and 7.4% of G6, G7, and G8, respectively. Intriguingly, a female predominance could be observed in G7 (61%) and G8 (60%) in comparison with other subgroups though we have excluded the genes at X/Y chromosomes (Fig. 2B).
Fig. 2.
Clinical and molecular features of gene expression subgroups. (A) Differentially expressed genes between each subgroup in G1–G4 and G5–G8 as a whole (Left), and between each subgroup in G5–G8 and G1–G4 as a whole (Right). Each point represents a gene. All genes were ordered by the level of log2 (fold change) from low to high. (B) The Left panel shows clinical features, cytogenetic groups, outcomes, and recurrent gene fusions and mutations in AML, which are classified into diverse functional groups. Each column represents a patient, which is arranged according to the gene expression subgroup through G1–G8. The Middle panel depicts the percentage of clinical and molecular features in each subgroup. The Right panel presents the proportional distribution of gender (Upper) and age group (Lower).
Cell Differentiation Stage and Regulatory Characteristics of Gene Expression Subgroups.
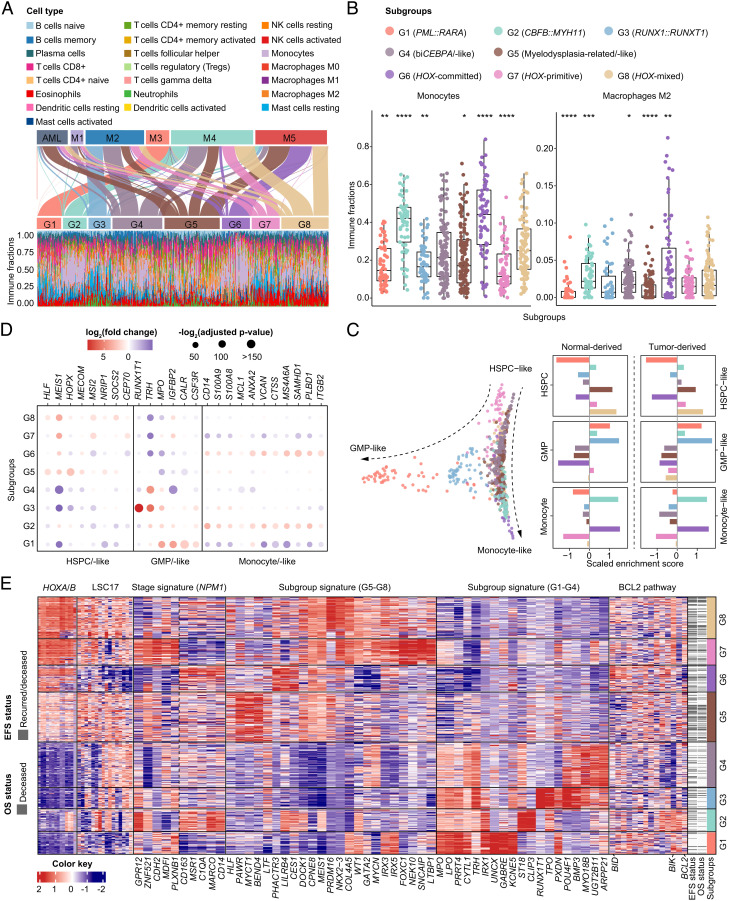
Next, we sought to decipher the intrinsic transcriptome deregulation of known or unreported subgroups. Considering cytomorphology is the traditional diagnostic approach showing the cell differentiation stage, we first compared gene expression subgroups with the FAB classification system. It was shown that G1 (PML::RARA), G2 (CBFB::MYH11), G3 (RUNX1::RUNX1T1), and G6, respectively, corresponded to M3, M4, and M2 with t(8;21) translocations and M5 in FAB nomenclature. Additionally, the G4 (biCEBPA/-like) subgroup was represented by M1/M2/M4, G5 (MR/-like) by AML/M2/M4/M5, G7 mainly by M2/M4, and G8 by M4/M5 (Fig. 3A). Moreover, molecular subtypes showed distinct immune cell abundances. G2 and G6 harbored more monocytes and macrophages (Fig. 3B), which was consistent with the FAB classification. Other immune fractions in defined molecular subgroups are provided in SI Appendix, Fig. S11.
Fig. 3.
Cellular hierarchies and regulatory pathways of gene expression subgroups. (A) Sankey plot shows reclassification from FAB subtypes to the defined molecular subgroups (Upper). Immune infiltration with specific cell type abundance in each subgroup as determined by CIBERSORTx (Lower). (B) Comparison of immune fractions of monocytes and macrophages M2 in G1–G8 subgroups. *P < .05; **P < .01; ***P < .005; ****P < .001. (C) Diffusion map for visualization of distinct cell differentiation stages of gene expression subgroups through dimensionality reduction, using HSPC-like, GMP-like, and monocyte-like cell signatures derived from scRNA-Seq data reported by Galen et al. (Left). Enrichment score of differentiation stage markers in each subgroup, using normal-derived and tumor-derived markers from the same scRNA-Seq data (Right). (D) Deregulation of representative molecular markers of each cell type in the eight subgroups. (E) Heatmap of gene expression signatures, including HOXA/B family genes, LSC17 score (Ng et al.), NPM1 stage signatures (Mer et al.), subgroup signatures representing the most significant differentially expressed genes in G5–G8 and G1–G4, and BCL2 family genes in each molecular subgroup. Columns indicate patients and are arranged according to the gene expression subgroup through G1–G8. The red and blue color represents relatively high and low gene expression, respectively. FAB, French-American-British; GMP, granulocyte-monocyte precursors; HSPC, hematopoietic stem/progenitor cells; scRNA-Seq, single-cell RNA sequencing.
In parallel, we referred to single-cell RNA-Seq (scRNA-Seq) data reported by Galen et al. (11) to pinpoint gene signatures of diverse differentiation states, including hematopoietic stem/progenitor cell-like (HSPC-like), granulocyte-monocyte precursor-like (GMP-like), and monocyte-like cells. Through diffusion map-based dimensionality reduction (23), each subgroup of G1–G8 enriched distinct cell-type signatures (Fig. 3C). Notably, G5, G7, and G8 subgroups displayed HSPC-like cell properties, although G5 and G8 involved a continuum of cell types projected along the HSPC to monocytic differentiation axis. G1 and G3 subgroups were characterized by GMP-like cell signatures, whereas differentiated monocytic features were more enriched in G2 and G6. The G4 subgroup lacked a prominent feature. Several molecular markers of each cell type showed diverse expression levels in G1–G8 (Fig. 3D). Single sample gene set enrichment analysis (ssGSEA) and hierarchical clustering of these cellular markers revealed similar hematopoietic cell differentiation hierarchies (SI Appendix, Figs. S12 and S13 and Dataset S7). In order to verify the inferred cell compositions from bulk RNA-Seq data, we randomly analyzed the immunophenotypes of 36 AML cases from G1–G8 by flow cytometry. The proportion of the CD34+CD38− component in total leukocytes was significantly higher in G5, G7, and G8 subgroups (SI Appendix, Fig. S14). In addition, samples from G1, G3, and G4 subgroups exhibited granulocytic differentiation immunophenotypes, while G2 and G6 showed typical monocytic differentiation (SI Appendix, Fig. S15).
Characteristic gene expression signatures of each subgroup were evaluated (Fig. 3E). HOXA/B family genes showed intermediate and high expression in G5 and G6–G8, respectively. Significantly, G5, G7, and G8 presented an upregulation of the previously well-established 17-gene leukemic stem cell (LSC17) signature (24). Using the reported differentially expressed genes that could classify NPM1-mutated AML into primitive and committed subtypes (12), G6 and G7 exhibited a more differentiated monocytic lineage feature and a stem cell signature, respectively, while G8 demonstrated a mixed feature. Accordingly, they were termed HOX-committed (G6, monocyte), HOX-primitive (G7, stem cell), and HOX-mixed (G8, with differentiation stage between stem cell and monocyte). Hierarchical clustering confirmed the intermediate status of the HOX-mixed subgroup (SI Appendix, Fig. S16). Apart from these well-defined gene sets, the expression levels of characteristic genes and BCL2 family genes in each subgroup were also delineated (Fig. 3E and SI Appendix, Fig. S17).
Oncogenic pathways were significantly upregulated mainly in the G5 subgroup, as exemplified by Rho GTPases, PI3K-AKT, JAK-STAT, and Calcium signaling pathways. The G2, G3, and G6 subgroups enriched more tumor microenvironment (TME)-related pathways, such as immunoregulatory, neutrophil degranulation, and IL-10 signaling. The platelet cytosolic Ca2+ pathway was upregulated in both G5 and G8 subgroups (SI Appendix, Fig. S18A). Multiple coexpression gene modules were associated with prognosis and G1–G8 subgroups (SI Appendix, Fig. S18B and Dataset S8). Selected HOX-, G5/G8-, and monocyte-related core networks are presented (SI Appendix, Figs. S19 and S20).
Prognostic Value of the established AML Subtypes.
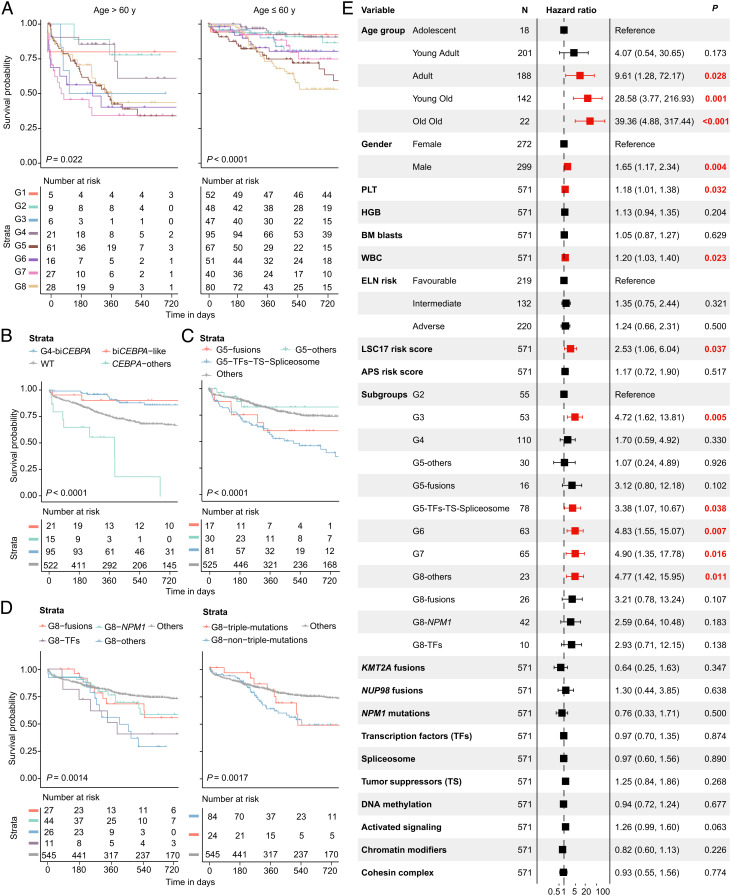
Clinical outcomes of the eight subgroups are displayed (SI Appendix, Fig. S21), with the survival of G1 (PML::RARA) as a reference. Despite G2 (CBFB::MYH11), G3 (RUNX1::RUNX1T1), and G4 (biCEBPA/-like) subgroups displaying a relatively long duration of overall survival (OS), a considerable proportion of these patients experienced disease recurrence. Patients in G5 (MR/-like) and G8 (HOX-mixed) subgroups had the poorest prognosis, in terms of both OS and event-free survival (EFS), while those in G6 (HOX-committed) and G7 (HOX-primitive) conferred slightly lower risk. Similar results were observed when only patients who received standard-of-care induction therapy were selected. Given that age significantly affects survival in AML, we stratified patients into two age groups. For patients older than 60 y, outcomes were uniformly dismal except for a few ELN low-risk subgroups. Nevertheless, both G5 and G8 could significantly predict an adverse prognosis in young (≤60 y) AML patients (Fig. 4A).
Fig. 4.
Prognostic value of the established molecular subgroups. (A) Kaplan-Meier curves for overall survival of elderly (>60 y, Left) and young (≤60 y, Right) AML patients stratified by eight gene expression subgroups. (B–D) Kaplan-Meier curves for the probability of overall survival in G4 (B), G5 (C), and G8 (D) subgroups stratified by specific genetic lesions. (E) Multivariable Cox analysis for overall survival in non-M3 AML patients.
We then explored the within-subgroup heterogeneity in representative molecular subgroups. Patients with biCEBPA-like gene expression signatures achieved a similar good prognosis to those harboring biCEBPA mutations in the G4 subgroup. Nevertheless, CEBPA mutations in other subgroups yielded an extremely poor prognosis, almost all of which were moCEBPA (Fig. 4B). Within the G5 subgroup, patients who carried fusion genes (mainly rare and previously unreported ones), and mutations in transcription factors (TFs), tumor suppressors (TS), and spliceosome had more adverse survival (Fig. 4C). We explored the underlying pathogenesis of several previously unidentified fusions, among which, the injection of human CYB5A::DYM (G5) mRNA into zebrafish embryos led to an increased expression of myeloid markers lyz, mpx, and lcp1 (SI Appendix, Fig. S22). In the G8 subgroup, patients with TFs and other genetic lesions had a more dismal prognosis than those with fusion genes and NPM1 mutations, and those with and without DNMT3A/NPM1/FLT3-ITD triple-mutations showed similar poor outcomes (Fig. 4D). Moreover, these results facilitate the screen of prognostic genes from inter- and intrasubgroups (SI Appendix, Fig. S23).
To elucidate the independent prognostic value of these identified transcriptome-based subgroups, we conducted a multivariable Cox analysis in non-M3 AML patients (Fig. 4E). Age, male gender, platelet, WBC, and LSC17 risk score (24) were all unfavorable prognostic factors. Notably, as compared with the G2 (CBFB::MYH11) subgroup, the classification of G3, G5 with mutations in TFs, TS, and spliceosome and G6, G7, and G8 with other genetic lesions (except for fusions, NPM1, and TFs mutations) independently predicted an adverse OS after adjusting for established clinical and molecular prognostic parameters.
Prediction Models of Transcriptome-Based Classification Enable Individualized Risk-Adapted Therapy.
By utilizing automatic machine learning (AutoML)-based modeling algorithm and different preprocessing steps, the eight gene expression subgroups could be accurately predicted, with a median prediction accuracy of 0.95 (Fig. 5A). We selected newly diagnosed AML samples collected from BM with available data from the TCGA LAML (8) and Beat AML (14, 19) cohorts. Based on the established models, both cohorts could convincingly reproduce the G1–G8 subgroups, which showed the corresponding expression and differentiation signatures (Fig. 5 B and C and SI Appendix, Figs. S24–S26 and Dataset S9). Of note, patients in the predicted G5, G6, and G8 subgroups from the TCGA LAML cohort, and those in G5 from the Beat cohort conferred extremely adverse clinical outcomes (Fig. 5 D and E). Comparisons of clinical and molecular parameters of the three cohorts are provided (Fig. 5F and SI Appendix, Table S4 and Dataset S10).
Fig. 5.
Construction of prediction models and validation in the TCGA LAML and Beat AML cohort. (A) Construction of prediction models utilizing a customized modeling method with different data preprocessing, modeling methods, and random data sampling. Briefly, read counts were quantified by using Featurecounts, and the gene expression matrix was generated by DESeq2 or TPM normalization with or without batch effect adjustment. Then, 90% of all primary AML samples were used for model training by Autogluon with 10 times sampling. (B and C) tSNE visualization for scatters of eight gene expression subgroups predicted in the TCGA LAML (B) and Beat AML (C) cohort. (D and E) Kaplan-Meier curves for overall survival of eight gene expression subgroups in the TCGA LAML (D) and Beat AML (E) cohort. (F) Distribution of age (Upper) and gene expression subgroups (Lower) in the TCGA LAML, Beat AML, and our study cohort. (G) Heatmap shows treatment responses of gene expression subgroups to different small-molecule inhibitors, using the Beat AML ex vivo drug screen data. Drug responses were measured by the scaled AUC in each subgroup as compared with others, with blue and red color indicating sensitivity and resistance, respectively. AUC, area under the dose-response curve; tSNE, t-distributed stochastic neighbor embedding.
Based on ex vivo drug sensitivity data from the Beat AML cohort (14, 19), responses of gene expression subgroups to different types of drugs were predicted (Fig. 5G and Dataset S11). The G1, G4, and G5 subgroups showed resistance to multiple inhibitors of receptor tyrosine kinase (RTK) such as Sorafenib, Sunitinib, Quizartinib, Pazopanib, etc., whereas G6–G8 subgroups demonstrated high sensitivity to these agents. Notably, the G2, G5, and G6 subgroups were resistant to the BCL2 inhibitor Venetoclax. Of interest, we noticed an obvious sensitivity of the monocytic phenotype (G2 and G6) to the histone deacetylase (HDAC) inhibitor Panobinostat and RTK inhibitor Dasatinib, two drugs commonly used in hematological malignancies. Taken together, these data substantiated the clinical utility of defined gene expression subgroups in AML, which may facilitate more rational treatment, and lend support to the development of novel agents.
Discussion
Accurate molecular classification and risk assessment are indispensable for improving the prognosis of patients with AML (1). RNA-Seq has recently been proven to be a comprehensive NGS technique, allowing the detection of various genetic abnormalities in acute leukemia (25–28). Additionally, gene expression signatures provide valuable information on the molecular classification and differentiation hierarchies of AML, which may further advance our understanding of the disease (11–14).
To date, stable gene expression-based molecular subtypes have not been established and cross-validated in large-scale AML cohorts. Herein, we established the largest multicenter AML cohort from China including 655 RNA-Seq and 619 targeted/whole-exome sequencing data. Based on an enhanced consensus method and clustering algorithms with extra feature filtration steps, eight stable molecular subgroups (G1–G8) were identified in this study. Among them, several subtypes significantly overlap with the existing genomic classification of AML (3, 8, 9, 19). More importantly, the proposed G1–G8 subgroups further enrich the current classification system by integrating genetic anomalies, gene expression signatures, and putative differentiation trajectories in AML.
Well-known PML::RARA (G1), CBFB::MYH11 (G2), and RUNX1::RUNX1T1 (G3), respectively, defined a distinct gene expression subgroup and exhibited GMP-like (G1/G3) and monocyte-like (G2) gene signatures. In the G4 subgroup, the biCEBPA-like entity was identified, which incorporated moCEBPA mutations with LOH and several CEBPA WT cases, both exhibiting similar GEP to biCEBPA, such as high TRH and low HOXA/B gene expression (29). In contrast, apart from biCEBPA-positive cases, the recently proposed bZIP-CEBPA did not cluster closely at the gene expression level. Notably, the biCEBPA-like AML conferred an equally favorable prognosis to biCEBPA, whereas the non-G4 CEBPA mutations showed significantly poor outcomes.
Additionally, we observed four robust subgroups characterized by intermediate (G5) to high (G6–G8) expression of HOXA/B, MEIS1, and CPNE8 genes. The G5 (MR/-like) subgroup enriched more MDS-related changes, such as “secondary-type” mutations (22) and TP53 abnormalities/complex karyotype, suggesting that patients in this subgroup might originate from previously diagnosed or unrecognized MDS. It is worth noting that this subgroup, though being a previously undefined subtype at the transcriptome level, corresponds well to the redefined AML-MR in the fifth edition of the WHO classification, which has incorporated the eight “secondary-type” mutations (3). In light of these results, the transcriptome-based classification could serve as a potential surrogate for the diagnosis of AML-MR. Of interest, an unexpectedly high frequency of IKZF1 N159S mutation (1.1%, 7/655) was found in this study, which was reported as a germline and dominant negative mutation associated with T, B, and myeloid cell combined immunodeficiency and T-ALL (30, 31). All cases with IKZF1 N159S showed a distinct gene expression signature and clustered into the G5 subgroup, which has not been reported to date. By analogy, we previously defined a GEP-dependent subtype of IKZF1 N159Y mutation in B-progenitor acute lymphoblastic leukemia (15). Further studies concerning the functional mechanism of this hotspot mutation are warranted.
The G6 to G8 subgroups are represented by NPM1 mutations, KMT2A and NUP98 fusions, and FLT3-ITD and KMT2A-PTD. Leukemia cells in these three subgroups are inferred to be blocked at various differentiation stages. Consistent with this finding, a recent study has classified NPM1-mutated AML into primitive and committed subtypes based on stemness (12). Herein, we extend this classification to a larger AML population sharing similar GEP features, especially the significant overexpression of HOX family genes, even in the absence of NPM1 mutations. In comparison with the previous report (12), we redefined two subgroups with distinct differentiation properties, namely HOX-committed (G6) and HOX-primitive (G7), and identified an additional subgroup spanning a spectrum of cell types, referred to as HOX-mixed (G8). The HOX-committed (G6) subgroup was characterized by monocyte-like gene signatures, i.e., CD14, S100A8/9, and LILRB4 (32, 33), which was similar to CBFB::MYH11 (G2). In contrast, the G5 (MR/-like), HOX-primitive (G7), and HOX-mixed (G8) subgroup demonstrated HSPC-like signatures, which was in concordance with the well-recognized LSC17 score (24). Other genes upregulated in these subgroups, such as MYCT1, PAWR, HLF, and PRDM16, were reported to be associated with stem cell properties (34–36).
These GEP-dependent subgroups herald different clinical outcomes, with patients in G5 (MR/-like) and G8 (HOX-mixed) showing the worst prognosis, which may partly be attributed to the enrichment of high-risk genetic lesions in these subgroups. Besides, the G5 and G8 subgroups might bear both stemness and synergistic immunosuppressive properties that lead to an extremely adverse prognosis. Remarkably, the independent prognostic value of several gene expression subgroups and the within-subgroup heterogeneity were observed, as exemplified by different outcomes of specific genetic abnormalities in G5 and G8, indicating that the transcriptome-based molecular classification may lay the foundation for more accurate and efficient screening of prognostic indicators in AML.
Of note, the eight GEP-defined subgroups were successfully reproduced in both TCGA LAML and Beat AML cohorts using the established prediction models. More importantly, drug sensitivity data from the Beat cohort suggested that transcriptome-based molecular subgroups may guide therapeutic decisions for AML patients, as exemplified by the eventual sensitivity of G6–G8 subgroups to RTK inhibitors. Coinciding with recent reports (37, 38), a potential resistance to Venetoclax was observed in G2 and G6 possibly due to the monocytic phenotype in both subgroups. The HDAC inhibitor Panobinostat and RTK inhibitor Dasatinib might exert therapeutic efficacy for the two subgroups, which warrants further validation in preclinical and clinical studies. Besides, the G5 subgroup seemed resistant to Venetoclax and most kinase inhibitors, representing a putative treatment bottleneck in AML. In addition to intensive chemotherapy, potential treatment options for patients in G5 include CPX-351 (liposomal daunorubicin/cytarabine approved for AML-MR) (39), hypomethylation agents, targeted therapies, and hematopoietic stem cell transplantation. These results suggest that cellular compositions of AML correlate with different drug sensitivities. In this regard, it might be premature to ignore or phase out the FAB classification in AML diagnostics, as it can still reflect, to some extent, the stage of cell differentiation based on morphology.
To summarize, robust transcriptome-based molecular subgroups not only capture the clinically, morphologically, and genetically defined AML entities but also largely enrich the current widely used prognostic classification systems, which may constitute a paramount framework for understanding the cellular origin and genotype-phenotype associations of the disease. We envisage the widespread application of RNA-Seq and the established classification (G1–G8) in clinical routine will provide a prompt and comprehensive molecular landscape of AML and facilitate personally tailored disease management (Fig. 6).
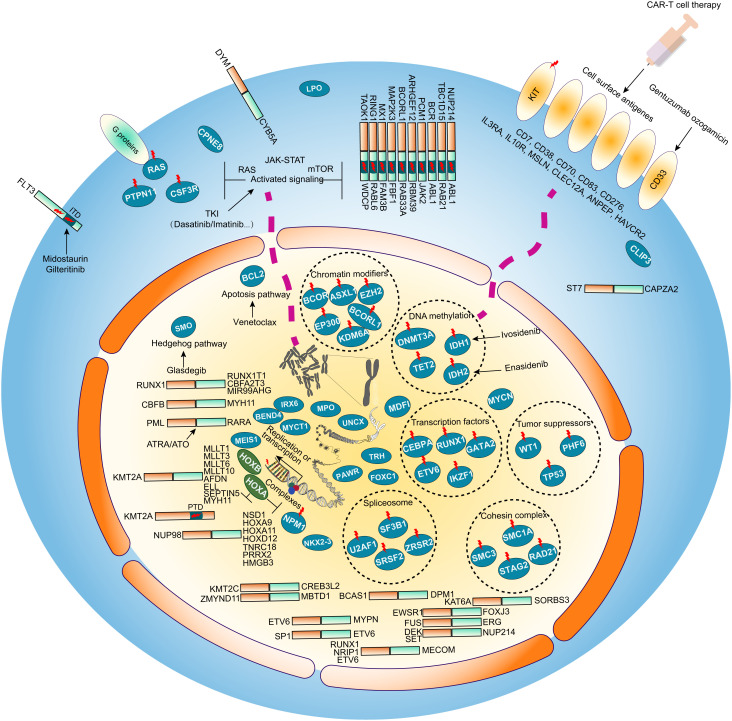
Fig. 6.
Schematic of molecular alterations and potential therapeutic targets in AML. Accumulation of diverse genomic and transcriptomic aberrations is related to the risk and prognosis of AML. Potential AML-related abnormalities identified in this work are displayed in the cell diagram (cell membrane, cytoplasm, nucleus) including fusion transcripts, genetic mutations, prognostic gene expression and alternative splicing events. Genetic mutations involving activated signaling molecules are common in the cytoplasm and cell membrane. Nuclear regulatory factors may contribute to the instability of the genome resulting in specific genetic mutations and gene expression profiles. Genes with sequence variations are marked with red lightning marks. The known and emerging target therapy agents are also labeled accompanying the target gene or pathway. Venetoclax and Glasdegib are respectively targeting the de-regulated apoptosis pathway gene BCL2 and the Hedgehog pathway SMO. Inhibitors Midostaurin and Gilteritinib can be used to treat FLT3 mutant AMLs. Cellular immunotherapy is another treatment option for AML patients with specific cell surface markers including the Gentuzumab ozogamicin targeting CD33.
Materials and Methods
Patients.
A total of 655 primary AML patients were enrolled in this study, among them, 442 were from Shanghai Institute of Hematology (SIH), 110 were from Jiangsu Institute of Hematology (JIH), and 103 were from Zhejiang Institute of Hematology (ZIH). All BM samples from 655 AML patients were subjected to RNA-Seq, while TES and WES were performed in 576 and 43 patients, respectively. Treatment protocols are provided in SI Appendix.
This study was approved by the Ethics Committee of Ruijin Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, the First Affiliated Hospital of Soochow University, and the First Affiliated Hospital of Zhejiang University College of Medicine. All patients had given informed consent for both treatment and cryopreservation of BM and peripheral blood samples according to the Declaration of Helsinki.
Gene Expression Profiling-Dependent Subgroups.
Raw RNA-Seq reads counts were extracted by both genome alignment-based Featurecounts v2.0.1 (40) and Htseq v0.11.3 (41) and alignment-free methods salmon v1.2.1 (42) and Kallisto v0.46.2 (43). Normalization of the counts matrix was simultaneously computed based on the R DESeq2 (v1.28.0) (44) transformation and the Transcripts Per Kilobase Million (TPM) value, which were used as the gene expression matrix for downstream analysis. ComBat function in the R sva package (v3.40.0) (45) was used to adjust the batch effect. Unsupervised clustering of top variance genes was conducted in R using the ComplexHeatmap (46) and a modified consensus clustering workflow. Autogluon (v0.2.0) (https://github.com/awslabs/autogluon) in Python was applied in the training and assessment of predictive models of GEP-defined subgroups. Detailed information and parameters related to the gene expression analysis are provided in SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Dataset S09 (XLSX)
Dataset S10 (XLSX)
Dataset S11 (XLSX)
Acknowledgments
We thank Weng Xiang-Qin for performing flow cytometric analysis. This work was supported by the State Key Laboratory of Medical Genomics, the Double First-Class Project (WF510162602) from the Ministry of Education, the Shanghai Collaborative Innovation Program on Regenerative Medicine and Stem Cell Research (2019CXJQ01), the Overseas Expertise Introduction Project for Discipline Innovation (111 Project; B17029), the National Natural Science Foundation of China (NSFC 81861148030, 82230006, and 82270166), the Shanghai Clinical Research Center for Hematological disease (19MC1910700), the Shanghai Shenkang Hospital Development Center (SHDC2020CR5002), the Shanghai Major Project for Clinical Medicine (2017ZZ01002), the Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20161406), the Innovative Research Team of High-level Local Universities in Shanghai, and the Shanghai Guangci Translational Medical Research Development Foundation.
Author contributions
Z.C., J.J., D.-P.W., Y.S., and S.-J.C. designed research; W.-Y.C., J.-F.L., Y.-M.Z., Xiang-Jie L., L.-J.W., F.Z., Xiao-Jing L., N.Q., W.Y., J.-N.Z., Y.X., T.-T.Z., S.-N.C., and H.-H.Z. performed research; H.F., S.-Y.W., L.J., and X.-J.S. contributed new reagents/analytic tools; W.-Y.C., J.-F.L., Y.-L.Z., M.Z., and Y.-T.D. analyzed data; W.-Y.C. and Y.-M.Z. performed validation experiments; Xiang-Jie L., L.-J.W., W.Y., J.-N.Z., Y.X., T.-T.Z., S.-N.C., and H.-H.Z. collected clinical data; F.Z. performed zebrafish experiments; S.-Y.W. and L.J. performed RNA sequencing; and W.-Y.C. and J.-F.L. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: R.E.G., Albert Einstein College of Medicine; and G.M., City of Hope National Medical Center.
Contributor Information
Zhu Chen, Email: zchen@stn.sh.cn.
Jie Jin, Email: jiej0503@zju.edu.cn.
De-Pei Wu, Email: wudepei@suda.edu.cn.
Yang Shen, Email: yang_shen@sjtu.edu.cn.
Sai-Juan Chen, Email: sjchen@stn.sh.cn.
Data, Materials, and Software Availability
Available scripts and programs of this study were deployed in https://github.com/clindet and Hiplot website (47). Anonymized [RNA sequencing] data have been deposited in [The Genome Sequence Archive for Human (GSA-Human, https://ngdc.cncb.ac.cn/gsa-human)](HRA002693). All study data are included in the article and/or SI Appendix. Previously published data were used for this work (1. X. Lin et al. Integration of Genomic and Transcriptomic Markers Improves the Prognosis Prediction of Acute Promyelocytic Leukemia. Clin. Cancer. Res. 27, 3683-3694 (2021). 2. P. Jin et al. Large-scale in vitro and in vivo CRISPR-Cas9 knockout screens identify a 16-gene fitness score for improved risk assessment in AML. Clin. Cancer. Res. 10.1158/1078-0432.Ccr-22-1618 (2022). 3. T. J. Ley et al. Genomic and epigenomic landscapes of adult de novo AML. N. Engl. J. Med. 368, 2059-2074 (2013). 4. J. W. Tyner et al., Functional genomic landscape of AML. Nature 562, 526-531 (2018). 5. D. Bottomly et al. Integrative analysis of drug response and clinical outcome in acute myeloid leukemia. Cancer Cell 40, 850-864.e859 (2022)).
Supporting Information
References
- 1.Newell L. F., Cook R. J., Advances in acute myeloid leukemia. BMJ 375, n2026 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Charrot S., Armes H., Rio-Machin A., Fitzgibbon J., AML through the prism of molecular genetics. Br. J. Haematol. 188, 49–62 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Khoury J. D., The 5th edition of the World Health Organization classification of haematolymphoid tumours: Myeloid and histiocytic/dendritic neoplasms. Leukemia 36, 1703–1719 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perl A. E., et al. , Gilteritinib or chemotherapy for relapsed or refractory FLT3-mutated AML. N. Engl. J. Med. 381, 1728–1740 (2019). [DOI] [PubMed] [Google Scholar]
- 5.DiNardo C. D., et al. , Azacitidine and venetoclax in previously untreated acute myeloid leukemia. N. Engl. J. Med. 383, 617–629 (2020). [DOI] [PubMed] [Google Scholar]
- 6.DiNardo C. D., et al. , Durable remissions with ivosidenib in IDH1-mutated relapsed or refractory AML. N. Engl. J. Med. 378, 2386–2398 (2018). [DOI] [PubMed] [Google Scholar]
- 7.Stein E. M., et al. , Enasidenib in mutant IDH2 relapsed or refractory acute myeloid leukemia. Blood 130, 722–731 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ley T. J., et al. , Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 368, 2059–2074 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papaemmanuil E., et al. , Genomic classification and prognosis in acute myeloid leukemia. N. Engl. J. Med. 374, 2209–2221 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Handschuh L., Not only mutations matter: Molecular picture of acute myeloid leukemia emerging from transcriptome studies. J. Oncol. 2019, 7239206 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Galen P., et al. , Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell 176, 1265–1281.e1224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mer A. S. et al. , Biological and therapeutic implications of a unique subtype of NPM1 mutated AML. Nat. Commun. 12, 1054 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zeng A. G. X., et al. , A cellular hierarchy framework for understanding heterogeneity and predicting drug response in acute myeloid leukemia. Nat. Med. 28, 1212–1223 (2022), 10.1038/s41591-022-01819-x. [DOI] [PubMed] [Google Scholar]
- 14.Bottomly D., et al. , Integrative analysis of drug response and clinical outcome in acute myeloid leukemia. Cancer Cell 40, 850–864.e859 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J. F., et al. , Transcriptional landscape of B cell precursor acute lymphoblastic leukemia based on an international study of 1,223 cases. Proc. Natl. Acad. Sci. U.S.A. 115, E11711–E11720 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gu Z., et al. , PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat. Genet. 51, 296–307 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai Y. T., et al. , Transcriptome-wide subtyping of pediatric and adult T cell acute lymphoblastic leukemia in an international study of 707 cases. Proc. Natl. Acad. Sci. U.S.A. 119, e2120787119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valk P. J., et al. , Prognostically useful gene-expression profiles in acute myeloid leukemia. N. Engl. J. Med. 350, 1617–1628 (2004). [DOI] [PubMed] [Google Scholar]
- 19.Tyner J. W., et al. , Functional genomic landscape of acute myeloid leukaemia. Nature 562, 526–531 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lilljebjörn H., Orsmark-Pietras C., Mitelman F., Hagström-Andersson A., Fioretos T., Transcriptomics paving the way for improved diagnostics and precision medicine of acute leukemia. Semin. Cancer Biol. 84, 40–49 (2022). [DOI] [PubMed] [Google Scholar]
- 21.Wilhelmson A. S., Porse B. T., CCAAT enhancer binding protein alpha (CEBPA) biallelic acute myeloid leukaemia: Cooperating lesions, molecular mechanisms and clinical relevance. Br. J. Haematol. 190, 495–507 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindsley R. C., et al. , Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood 125, 1367–1376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Angerer P., et al. , Destiny: Diffusion maps for large-scale single-cell data in R. Bioinformatics 32, 1241–1243 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Ng S. W., et al. , A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 540, 433–437 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Gu M., et al. , RNAmut: Robust identification of somatic mutations in acute myeloid leukemia using RNA-seq. Haematologica 105, e290–e293 (2019), 10.3324/haematol.2019.230821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arindrarto W., et al. , Comprehensive diagnostics of acute myeloid leukemia by whole transcriptome RNA sequencing. Leukemia 35, 47–61 (2020), 10.1038/s41375-020-0762-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Docking T. R., et al. , A clinical transcriptome approach to patient stratification and therapy selection in acute myeloid leukemia. Nat. Commun. 12, 2474 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anonymous, Brown L. M., et al. , The application of RNA sequencing for the diagnosis and genomic classification of pediatric acute lymphoblastic leukemia. Blood Adv. 4, 930–942 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wouters B. J., et al. , Double CEBPA mutations, but not single CEBPA mutations, define a subgroup of acute myeloid leukemia with a distinctive gene expression profile that is uniquely associated with a favorable outcome. Blood 113, 3088–3091 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida N., et al. , Germline IKAROS mutation associated with primary immunodeficiency that progressed to T-cell acute lymphoblastic leukemia. Leukemia 31, 1221–1223 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Boutboul D., et al. , Dominant-negative IKZF1 mutations cause a T, B, and myeloid cell combined immunodeficiency. J. Clin. Invest. 128, 3071–3087 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laouedj M., et al. , S100A9 induces differentiation of acute myeloid leukemia cells through TLR4. Blood 129, 1980–1990 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Deng M., et al. , LILRB4 signalling in leukaemia cells mediates T cell suppression and tumour infiltration. Nature 562, 605–609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lavallee V. P., et al. , EVI1-rearranged acute myeloid leukemias are characterized by distinct molecular alterations. Blood 125, 140–143 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garg S., et al. , Hepatic leukemia factor is a novel leukemic stem cell regulator in DNMT3A, NPM1, and FLT3-ITD triple-mutated AML. Blood 134, 263–276 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Shiba N., et al. , High PRDM16 expression identifies a prognostic subgroup of pediatric acute myeloid leukaemia correlated to FLT3-ITD, KMT2A-PTD, and NUP98-NSD1: The results of the japanese paediatric leukaemia/lymphoma study group AML-05 trial. Br. J. Haematol. 172, 581–591 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Pei S., et al. , Monocytic subclones confer resistance to venetoclax-based therapy in patients with acute myeloid leukemia. Cancer Discov. 10, 536–551 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kurtz S. E., et al. , Associating drug sensitivity with differentiation status identifies effective combinations for acute myeloid leukemia. Blood Adv. 6, 3062–3067 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lancet J. E., et al. , CPX-351 (cytarabine and daunorubicin) liposome for injection versus conventional cytarabine plus daunorubicin in older patients with newly diagnosed secondary acute myeloid leukemia. J. Clin. Oncol. 36, 2684–2692 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao Y., Smyth G. K., Shi W., Featurecounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Anders S., Pyl P. T., Huber W., HTSeq–a python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bray N. L., Pimentel H., Melsted P., Pachter L., Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leek J. T., Johnson W. E., Parker H. S., Jaffe A. E., Storey J. D., The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu Z., Eils R., Schlesner M., Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Li J., et al. , Hiplot: A comprehensive and easy-to-use web service for boosting publication-ready biomedical data visualization. Brief. Bioinform. 23, bbac261 (2022). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Dataset S08 (XLSX)
Dataset S09 (XLSX)
Dataset S10 (XLSX)
Dataset S11 (XLSX)
Data Availability Statement
Available scripts and programs of this study were deployed in https://github.com/clindet and Hiplot website (47). Anonymized [RNA sequencing] data have been deposited in [The Genome Sequence Archive for Human (GSA-Human, https://ngdc.cncb.ac.cn/gsa-human)](HRA002693). All study data are included in the article and/or SI Appendix. Previously published data were used for this work (1. X. Lin et al. Integration of Genomic and Transcriptomic Markers Improves the Prognosis Prediction of Acute Promyelocytic Leukemia. Clin. Cancer. Res. 27, 3683-3694 (2021). 2. P. Jin et al. Large-scale in vitro and in vivo CRISPR-Cas9 knockout screens identify a 16-gene fitness score for improved risk assessment in AML. Clin. Cancer. Res. 10.1158/1078-0432.Ccr-22-1618 (2022). 3. T. J. Ley et al. Genomic and epigenomic landscapes of adult de novo AML. N. Engl. J. Med. 368, 2059-2074 (2013). 4. J. W. Tyner et al., Functional genomic landscape of AML. Nature 562, 526-531 (2018). 5. D. Bottomly et al. Integrative analysis of drug response and clinical outcome in acute myeloid leukemia. Cancer Cell 40, 850-864.e859 (2022)).