Abstract
Endothelial cell (EC) function declines with age and contributes to the development of many vascular-related disease processes. Currently, the effects of aging on the molecular regulatory mechanisms of liver ECs have not been fully elucidated. Here, we employed single-cell RNA sequencing to map the transcriptome of ECs and analyzed their relationship with aging. We identified 8 different EC subtypes, interestingly, 2 of which were specially expressed in aged mice ECs namely aged capillary ECs (Aged ECs) and pro-inflammation capillary ECs (Proinfla.ECs). Double immunostaining for an EC marker (Cd31) and a marker of these specialized EC phenotypes confirmed the single-cell RNA sequencing data. Gene ontology analysis revealed that Aged ECs and Proinfla.ECs were associated with inflammatory response. Then we found that liver proliferating capillary ECs (Prolife.ECs) were most affected by senescence. Single-cell transcript analysis suggests that Prolife.ECs and angiogenic capillary ECs may form a poor microenvironment that promotes angiogenesis and tumorigenesis. Pseudo-temporal trajectories revealed that Prolife.ECs have different differentiation pathways in young and aged mice. In aged mice, Prolife.ECs could specifically differentiate into an unstable state, which was mainly composed of angiogenic capillary ECs. Intercellular communication revealed inflammatory activation in old group. Overall, this work compared the single-cell RNA profiles of liver ECs in young and aged mice. These findings provide a new insight into liver aging and its molecular mechanisms, and further exploration of Aged ECs and Proinfla.ECs may help to elucidate the molecular mechanisms associated with senescence.
Hepatic vascular niche changes during aging. Aged capillary EC (Aged EC), pro-inflammation capillary EC (Proinfla.EC), proliferating capillary EC (Prolife.EC) and angiogenic capillary EC (Angio.EC) expand in the old niche, highlighted by the increase in senescence, inflammation, proliferation, and angiogenesis.
INTRODUCTION
Aging is an inevitable consequence of life. The aging process disrupts organismal function and homeostasis, leaving us vulnerable to chronic diseases and microbial infections.1,2 The optimal function of the biological organism depends on a rich network of blood vessels that provide a continuous supply of oxygenated blood to body tissues.3 It is now known that aging is associated with a deterioration of vascular function characterized by the occurrence of age-related endothelial cell (EC) dysfunction.4,5 ECs are located within the lumen of the blood and lymphatic vessels and are critical for nutrient and oxygen delivery, metabolite waste removal, and immune surveillance.6 Consistent with these critical roles, endothelial dysfunction is increasingly recognized as a major contributor to various age-associated diseases.7 Although previous studies have demonstrated the effects of aging on vascular and endothelial function, little is known about age-related molecular mechanistic changes in the liver vascular system, and therefore more research is greatly needed.
The liver is a central hub for metabolic regulation with a wide range of key functions, including regulation of lipid and glucose homeostasis, bile acid secretion, and protein synthesis.8 In addition, the liver vasculature displays unique anatomical and physiological features in that it receives both oxygen-rich blood from the hepatic artery and nutrient-rich blood from the portal vein. After merging, blood flow continues along the hepatic sinusoids, which are lined by discontinuous and fenestrated hepatic sinusoidal ECs.9 Liver ECs (LECs) have been extensively studied and are thought to play an important role in maintaining hepatic homeostasis, initiating immune responses, and progressive liver diseases such as fibrosis and carcinoma.10 Considering that LECs have these multifunctional properties, we aim to decipher age-related transcriptional changes in LECs and reveal potential therapeutic targets for age-related liver diseases.
The advent of single-cell RNA sequencing (scRNA-seq) technology has provided a powerful tool to study senescent cells and their molecular mechanisms at single-cell resolution.11,12 In our scRNA-seq analysis, we observed extensive differences in LECs composition, gene signatures, enriched pathways, transcriptional regulatory networks, differentiation programs, and intercellular communication between young and old mice. Overall, our work provides a comprehensive atlas of the effect of age on liver endothelium and expands our understanding of aging-driven changes in liver endothelium under homeostasis and pathological conditions.
MATERIALS AND METHODS
Mice and tissue collection
Experiments used wild-type 2-month-old and 2-year-old male C57BL6/J mice bred in-house, provided through the Animal Experimental Center of Zhongshan Ophthalmic Center. The mice adapted to the environment for 1 week. The method of ECs collection was approximately similar to a previously published protocol.13 After anesthesia with ketamine (0.9%)/xylazine (2%), mice were weighed before transcardial perfusion with ice-cold PBS, followed by perfusion with the digestion buffer containing DMEM, 1% penicillin/streptomycin, 2× Antibiotic-Antimycotic, 1 mM sodium pyruvate, 1× MEM nonessential amino acids solution and supplemented with 0.1% collagenase I, 0.1% collagenase IV, 2.5 U/mL Dispase II, and 7.5 mg/mL DNase I. Samples were incubated at 37°C in a water bath for 30 minutes, shaking every 5 minutes. At the end of the incubation time, PBS-based wash buffer was added and the cell suspension was filtered through a 100 mm cell strainer and centrifuged at 350g for 5 minutes. The obtained pellet was resuspended in a wash buffer and filtered through a 40 mm cell strainer. Samples were centrifuged at 350g for 5 minutes. The washing step was repeated once more. Next, we used CD31 MicroBeads (Miltenyi Biotec, Cat#130-097-418) to select ECs according to the manufacturer’s instructions. Finally, the cell suspension was stained with anti-CD45 antibodies, anti-CD31 antibodies, anti-CD11b antibodies, and DAPI (4',6-diamino-2-phenylindole). Viable CD11b– CD45– CD31+ ECs were sorted into collecting medium through flow cytometry. All procedures of this experiment were approved by the Animal Ethical Committee at Zhongshan Ophthalmic Center, Sun Yat-Sen University under the Permit ID, 2019-043.
RNAscope in situ hybridization and immunofluorescence
The livers were surgically dissected from wild-type 2-month-old and 2-year-old C57BL6/J mice. Four percent paraformaldehyde-fixed whole murine livers were sectioned and subjected to RNAscope in situ hybridization. Briefly, after deparaffinization and several washing steps, hybridization was performed with the RNAscope probes Ctss, Osm, Icam1, and Spn. RNAscope In Situ Hybridization service was provided by Guangzhou Exon biotechnology Co. Ltd, before immunofluorescence staining.
Library preparation and sequencing
Sorted single cells were washed and resuspended in PBS containing 0.04% BSA. RNA-seq libraries were prepared by using the Chromium Single Cell 3’ Reagent Kits v2 (10x Genomics). The libraries were sequenced on a HiSeq. 4000 instrument (Illumina) and the cellranger (10x Genomics, version 3.1.0) was used to generate matrixes from the FASTQ files.
Data processing, quality control, and integration
The raw sequencing data were preprocessed, counted by the cellranger and processed in R for visualization. (i) The first filtering step in Seurat (version 3.1.3) discarded cells with fewer than 200 and more than 6000 genes. (ii) Cells with the mitochondrial ratio of more than 5% were removed. We used “Harmony” to remove the batch effect for different samples.
Clustering and EC cluster identification
After data integration and scaling, principal component analysis was performed on the variable genes using the RunPCA function in Seurat and appropriate principal components were retained for downstream analysis. We used the “RunUMAP” function to reduce dimensionality. Besides these clusters were identified with the functions FindNeighbors and FindClusters. By setting p values <0.05 and |log2FC| > 0.25, we got the top 50 marker genes of each cluster with the “FindAllMarkers” function (Table S1, http://links.lww.com/HC9/A57). Canonical marker genes were used to identify cell types.
Age-related differential gene expression
With the threshold of adjusted p values <0.05 and |log2FC| > 0.25, we identified differentially expressed genes (DEGs) in each EC cluster between young and old samples with the “FindMarkers” function of Seurat (Table S2, http://links.lww.com/HC9/A57). And the gene sets related to liver diseases were obtained from DisGeNET (https://www.disgenet.org/home/).
Gene set score analysis
The senescence-associated secretory phenotype (SASP) genes that we acquired from a previously published study14 produced the aging score by the Seurat function “AddModuleScore.” The scores density-plot between young and old samples were visualized with the ggplot2 R package (version 3.3.5). All gene sets of this research are concluded in Table S3 (http://links.lww.com/HC9/A57).
Gene ontology (GO) term analysis
The GO analysis results were generated in R software. The “org.Mm.eg.db” R package can be used for gene ID conversion, and further we used the “clusterProfiler” (version 4.0.5) for enrichment analysis. With the threshold of p-value cutoff =0.05 and q value cutoff =0.05, the most significant genetic functions will be selected to represent the function of each cluster and DEGs.
Transcription factor (TF) activity inference
We infer TF activity in different ECs clusters using Dorothea (https://saezlab.github.io/dorothea/). The analysis of TF gene regulatory network was performed in the Metascape (http://metascape.org/gp/index.html). Only DEGs in proliferating capillary ECs (Prolife.ECs) were input as inferring for the transcriptional regulators in the age-related transcriptional regulatory network. Then the transcription regulatory network was visualized by Cytoscape (version 3.8.2) and igraph.
SCENIC
SCENIC (version 1.1.1-10) workflow was applied to identify the transcriptional regulatory network of aging-related DEGs with default parameters based on mm9 database from RcisTarget (version 1.6.0). Aging-related DEGs in Prolife.ECs were used to build a transcriptional regulator inferring network. The regulatory network was visualized by igraph (version 1.3.1).
Pseudo-time analysis
We used the Monocle2 R package to perform Pseudo-time analysis. Marker genes of different clusters were used as ordering genes with the threshold of adjusted p-value <0.05 and |log2FC| > 0.25. DDRTree dimensionality reduction method was applied to construct the trajectory that was plotted in 2-dimensional space. Time differentiation-related genes (TDGs) were obtained with a cutoff of q value <1×10−4.
Cell-cell communication analysis
The analysis of cell-cell communication was performed by using the and CellPhoneDB (version 2.0) and CellChat (version 1.1.3) package. We assessed the expression matrix of non-EC types in young and old liver from publicly available Tabula Muris Datasets (https://figshare.com/articles/dataset/Processed_files_to_use_with_scanpy_/8273102/2). The results of cell-cell communication can represent between any 2 cell types if their p-value <0.05, and the average expression of the interaction pair was >0.
RESULTS
Construction of a single-cell transcriptome atlas of young and aged liver
To map mice liver ECs and identify age-related transcriptional alterations, we collected liver tissues from 4 aged (2-y-old) and 3 young (2-mo-old) mice, efficiently isolated ECs by fluorescence-activated cell sorting (FACS) and performed scRNA-seq (Figure 1A). After stringent quality filtering and batch correction, a total of 10,543 young and 13,114 aged single cells were retained for downstream analysis. We performed data integration processing by harmony (Figure S1E, http://links.lww.com/HC9/A52) and unsupervised cell clustering methods and used the Uniform Manifold and Projection (UMAP) algorithm for visualization (Figure 1B).
FIGURE 1.
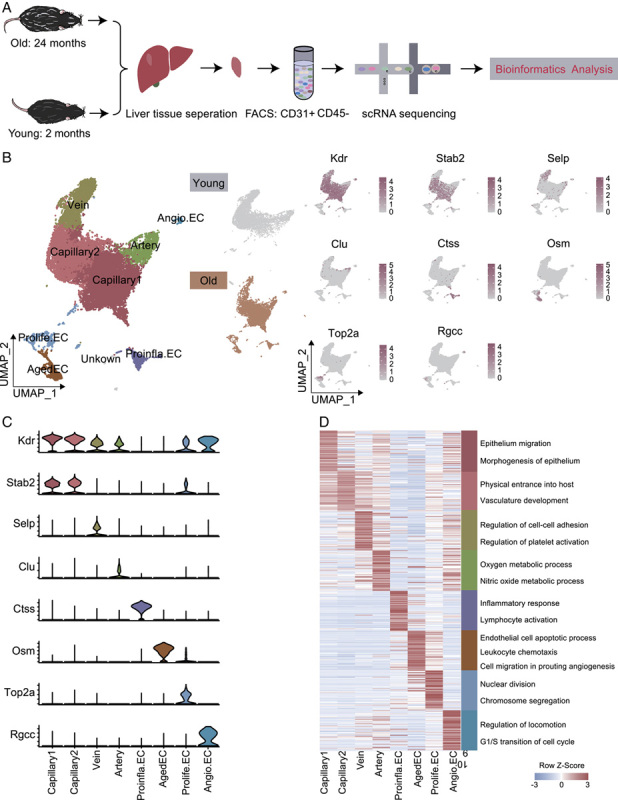
Construction of single-cell transcriptomic atlas of mouse liver endothelial cells (ECs). (A) Flow chart of scRNA-seq and bioinformatics analysis of mouse liver ECs. Young, n=3; old, n=4 mouses. (B) Left, UMAP plot showing the distribution of different cell types in mouse liver ECs. Middle, UMAP plots showing the distribution of different EC subpopulations in young (top) and old (bottom) mice. Right, UMAP plots showing the expression of representative marker genes for the corresponding cell types in mouse liver ECs. (C) Expressions of Kdr, Stab2, Selp, Clu, Ctss, Osm, Top2a, and Rgcc in each cluster. y axis represents log-normalized expression. (D) Heatmap of gene expression of the top 50 marker genes for each EC subcluster with their enriched functional annotations on the right. Abbreviations: Aged EC, aged capillary EC; Angio.EC, angiogenic capillary EC; Prolife.EC, proliferating capillary EC; Proinfla.EC, pro-inflammation capillary ECs.
We first established 9 cell clusters based on unique gene-expression profiles and successfully annotated 8 liver EC types by expression of established marker genes with reference to previous reports,13 which were not confounded by either a specific sample or condition (Figure S1G, http://links.lww.com/HC9/A52). These included capillary 1 ECs (Kdr + ), capillary 2 ECs (Stab2 + ), venous ECs (Selp + ), arterial ECs (Clu + ), Prolife.ECs (Top2a + ), and angiogenic capillary ECs (Angio.ECs) (Rgcc + ) (Figure 1B). Intriguingly, we detected pro-inflammation capillary ECs (Proinfla.ECs) with marker gene Ctss and aged capillary ECs (Aged ECs) with marker gene Osm in the liver of aged mice but not in the younger group. To confirm the scRNA-seq results, we labled marker gene Ctss and Osm identified above by performing double immunostaining, validating the liver Proinfla.ECs and Aged ECs in the old group (Figure S1H, http://links.lww.com/HC9/A52). By immunofluorescence and RNAscope experiments, we found Aged ECs in the old hepatic sinusoid region through coimmunostaining of Cd31 protein, p53 protein (the marker of senescent cells), and Osm mRNA transcript (Figure S1I, http://links.lww.com/HC9/A52). The immunofluorescence images and hierarchical clustering heatmap (Figure S1F, http://links.lww.com/HC9/A52) indicated that Aged ECs may originate from liver sinusoidal ECs.
Aged ECs were named by their elevated SASP score (Figure S1C, http://links.lww.com/HC9/A52). In addition, Osm (oncostain-M), a marker gene of Aged ECs, regulates cytokine production from ECs and promotes liver development and regeneration.15 Ctss (cathepsin S), marker gene of Proinfla.ECs was reported to positively regulate inflammatory response as the key thiol protease by MHC class II antigen presentation,16 and this cluster was referred to as Proinfla.ECs. Thus, Proinfla.ECs and Aged ECs were associated with inflammatory response, consistent with the notion that aging is accompanied by inflammation. We also identified top marker genes for LECs. Some genes were expressed only in a single cell type (Selp, Clu, Ctss, Top2a, and Rgcc, for vein, artery, Aged ECs, Prolife.ECs, and Angio.ECs, respectively), while other EC marker genes (Kdr, Stab2) were conserved across 2 or more cell types (Figure 1C).
GO analysis of the top 50 marker genes revealed distinct features corresponding to known biological processes and characteristics of ECs (Figure 1D). For example, the GO term “epithelium migration and morphogenesis” was enriched for the first 50 marker genes of capillary 1 ECs, “physical entrance into host” and “vasculature development” were enriched for capillary 2 ECs, while the GO terms “regulation of cell-cell adhesion and platelet activation” were specific to vein ECs. The GO terms “oxygen species metabolic process” and “nitric oxide metabolic process” correspond to molecular features of artery ECs. The GO terms “inflammatory response” and “lymphocyte activation” were specific to Proinfla.ECs, “leukocyte chemotaxis” and “cell migration involved in sprouting” were specific to Aged ECs, indicative of elevated inflammation with aging. Prolife.ECs increased the expression levels of genes involved in “nuclear division” and “chromosome segregation,” suggesting its essential roles in cell proliferation. Angio.ECs showed an enriched regulation of cell cycle-related signatures, indicative of its function in angiogenesis.
We next constructed transcriptional regulatory networks to reveal cell type-specific TF network (Figure S1D, http://links.lww.com/HC9/A52). For instance, Atf4 and Batf were shared by Proinfla.ECs and Aged ECs, suggesting their essential roles in liver aging. Atf4, cyclic AMP-dependent TF, binds the cAMP response element and promotes the transcription of genes resistant to oxidative stress. Batf (basic leucine zipper Atf-like transcriptional factor) was an AP-1 family TF that mediates the differentiation of lineage-specific cells in the immune system, especially in T-helper 17 cells (Th17).17 In addition, Cebpd (CCAAT/enhancer-binding protein delta), which acts as a transcription activator to regulate the expression of genes involved in immune and inflammatory responses and enhance Il6 transcription, is critical for the proper development of the liver Aged ECs. We also detected that E2f TFs (E2f1, E2f2, E2f3, and E2f4), regulating cell cycle and DNA damage repair by binding DNA in cooperation with DP proteins,18 were highly expressed in Prolife.ECs. Meanwhile, the prominent genes comprising the Prolife.ECs-specific TF network included Myc, Nr2c2, Rest, and Tfdp1. Myc activates transcription of genes involved in cell growth and sprouting angiogenesis, suggesting the involvement of Prolife.ECs in both liver repair and regeneration. For Angio.ECs, Esr1 with high TF-activity could remodel the blood vessel and accelerate endothelial healing.19
Since metabolism of ECs regulates vessel homeostasis and growth, we further explored the heterogeneity of metabolic gene-expression signatures of LECs in old group. Using all 1180 detectable metabolic genes in ECs and performing heatmap analysis of the expression levels of the top-15-ranking metabolic-gene transcripts, we observed that ECs from different cell types upregulated the expression of distinct sets of metabolic genes, especially the Aged ECs and Prolife.ECs (Figure S2A, http://links.lww.com/HC9/A53). Focusing on multiple genes within a single metabolic pathway, we observed that Prolife.ECs and Angio.ECs upregulated the expression of nucleotide metabolism involved in the cell proliferation (Figure S2B, http://links.lww.com/HC9/A53). Aged ECs and Proinfla.ECs upregulated the expression of genes involved in lipid metabolism, in line with reports that the liver regulates plasma cholesterol levels (Figure S2B, http://links.lww.com/HC9/A53). Further, capillary 2 and vein ECs (and to a lesser extent artery ECs) expressed higher transcript levels of genes involved in oxidative phosphorylation metabolism, supporting the notion that metabolic process of oxidative phosphorylation increases with aging (Figure S2B, http://links.lww.com/HC9/A53). Thus, ECs show heterogeneity in metabolic gene expression in different vascular beds in old liver.
Taken together, our work provides a single-cell transcriptome atlas of young and aged mouse LECs that can be used in our analyses to further elucidate the molecular changes induced by aging.
Characterization of the aging-associated cellular and molecular profiles of the LECs
To determine the age-related changes in the composition of EC types, we first compared the proportion of different ECs between the young and old livers. We found that capillary 1, Prolife.ECs and Angio.ECs were more abundant in the aged LECs compared with the young ones, but there was a decline in capillary 2 ECs with age (Figure S1A, B and G, http://links.lww.com/HC9/A52). Accordingly, we noticed that old liver ECs exhibited higher gene set score for SASP overall, especially for Aged ECs (Figure S1C, http://links.lww.com/HC9/A52).
To explore the involvement of ECs in aging, we next compared the gene expression profiles of individual LECs (Proinfla.ECs and Aged ECs not included). The DEGs included 2826 upregulated and 1883 downregulated genes (Figure 2A). The cell types most affected by aging included Prolife.ECs, vein ECs, and capillary 1 ECs. Further, through a GO enrichment analysis, we investigated molecular pathways most affected by aging in these cell types (Figure 2C). We noticed that decreased “regulation of angiogenesis” and “EC migration,” together with increased “viral process” in Prolife.ECs, pointed to angiogenesis and virus infection. Also contributing to age-related angiogenesis are Angio.ECs, as GO analysis of DEGs revealed increased expression of genes involved in “translation,” “cell growth,” and “cell cycle phase transition” implied for sprouting angiogenesis with age. Further, increased “phosphate metabolic process” and “apoptotic signaling” in capillary 1 and capillary 2 along with decreased “response to oxidative stress” in vein ECs, probably contributed to aging-related oxidative stress damage.
FIGURE 2.
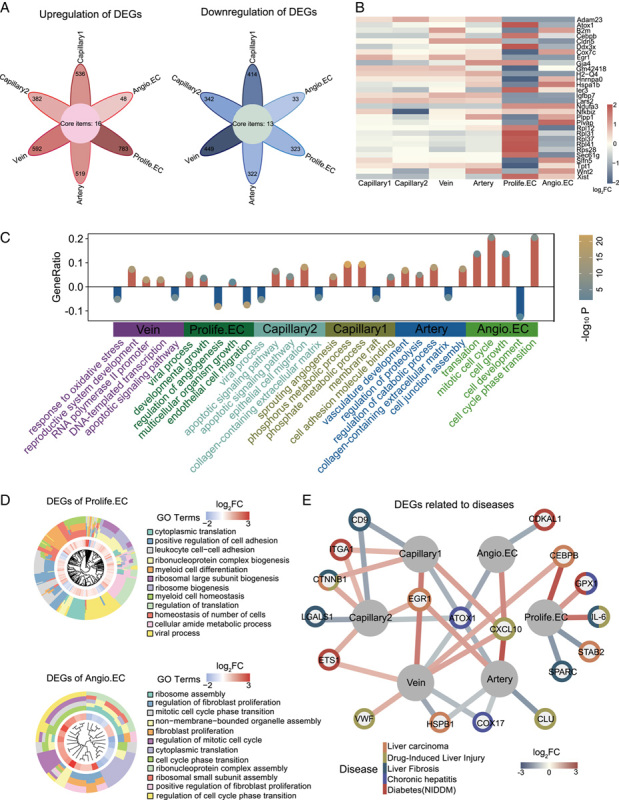
Cellular and molecular aging characteristics of aged mouse liver endothelial cells (ECs). (A) Flower plot showing aging-related up-regulation and down-regulation of DEGs in different cell types in mouse liver ECs (adjusted p-value <0.05, |log2FC| > 0.25). Core items represent commonly DEGs in 6 subclusters. (B) GO terms enriched for aging-related DEGs in different cell types from mouse liver ECs in a bar plot. The Y-axis represents the proportion of differentially expressed genes in the total DEGs in corresponding terms. (C) Heatmap showing DEGs expressed by at least two cell types in mouse liver ECs. (D) Circle graph showing various effects of DEGs on different cell types during aging. (E) Network plot showing DEGs associated with liver disease in different EC cell types in mouse liver. Abbreviations: Angio.EC, angiogenic capillary EC; DEGs, differentially expressed genes; GO, gene ontology; Prolife.EC, proliferating capillary EC.
In addition, a total of 30 genes were differentially expressed in at least 2 cell types (Figure 2B). For example, Plvap, EC-specific membrane protein involved in the formation of endothelial fenestrae diaphragms and microvascular permeability,20 was downregulated in Prolife.ECs and upregulated in Angio.ECs. Igfbp7, which encodes an insulin-like growth factor binding protein and was previously reported to be involved in the suppression of HCC,21 was downregulated in both Prolife.ECs and Angio.ECs, indicative of highly likelihood of tumorigenesis with age. On the other hand, Nfkb upregulated in Prolife.ECs and Angio.ECs, was involved in the induction of inflammation and Th17 cells differentiation,22 suggesting the elevated proinflammatory response in the aged liver.
We next performed a comparative analysis of high-risk DEGs by annotating the hotspot genes associated with aging and various liver diseases such as liver carcinoma, drug-induced liver injury, liver cirrhosis, chronic hepatitis, and diabetes mellitus. These genes were derived from the DisGeNET (Figure 2E). Most of the high-risk DEGs were enriched in capillary 2 and vein ECs, suggesting that these 2 cell types were more susceptible to liver carcinoma, liver cirrhosis, and diabetes, as exemplified by the expression of Egr1, Cd9, Itga1. Meanwhile, some high-risk DEGs were associated with multiple diseases. For example, Ctnnb1 has been considered as a risk factor for HCC, cirrhosis, and drug-induced liver injury, as it encodes catenin, which plays a key role in the canonical Wnt signaling that regulates gene transcription and controls cell growth and differentiation, and its mutations are associated with various tumor types.23 Some of the high-risk genes (such as Egr1 and Atox1) were involved in more than 1 cell type.
Altogether, our findings identified aging-related molecular features of the mice LECs, demonstrating sprouting angiogenesis and elevated proinflammatory response as the most affected and potential hallmarks in the aged liver.
Aging-related molecular alterations along the differentiation trajectories
To gain insights into the molecular disruptions with age, we also applied pseudo-time analysis to infer the differentiation trajectories of LECs. The analysis predicted a trajectory of the vascular tree from the arteries, through the capillaries, and to the veins in the younger group (Figure 3A), which is consistent with the 1 for brain ECs in previous reports.13 We used previously identified marker genes to map the location of EC subpopulations on the pseudo-temporal trajectory. Expression of known hepatic arterial EC marker genes (Clu, Sdc1, Mgp) was detected at the arterial end of the pseudo-time trajectory, while expression of the known liver vein marker genes (Selp) was enriched at the venous end of the trajectory. Capillary marker genes (Kdr, Stab2) were most enriched in the middle part of the trajectory (Figure 3B). Compared with the younger group, aged mouse liver ECs showed a completely different cell type distribution along the trajectory (Figure S3A, http://links.lww.com/HC9/A54).
FIGURE 3.
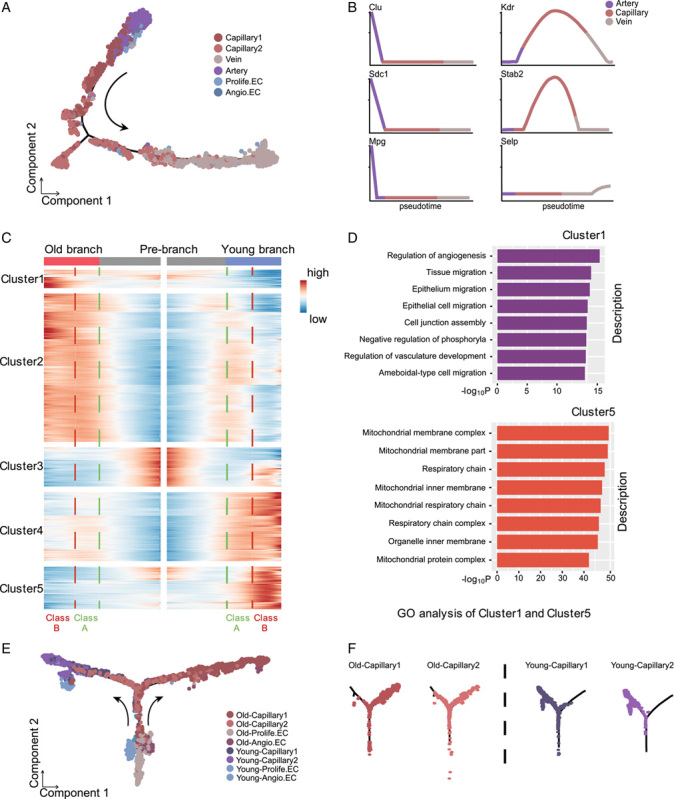
Pseudo-time trajectories of liver endothelial cells (ECs) phenotypes in young and old mice. (A) Pseudo-time trajectory of the phenotype of young liver ECs. (B) Gene expression dynamic of representative marker genes enriched in different EC phenotypes along the artery-capillary-vein axis. (C) Gene expression heatmap of 1025 top capillary DEGs (coded into 5 clusters, q value <1×10−4). Old and young liver ECs trajectories (including prebranch) are shown on the right and left, respectively. Class A and B indicate DEGs in the early and late stages after the prebranch, respectively. (D) GO analysis of class A DEGs in clusters 1 and 5, indicating the earliest transcriptomic events during aging. (E and F) Pseudo-time analysis of capillaries (including capillary 1 and capillary 2) in young and old mouse liver ECs. The dots are colored by cell types (left) and trajectory dot plots are divided by age and cell type (right). Abbreviations: Angio.EC, angiogenic capillary EC; DEGs, differentially expressed genes; GO, gene ontology; Prolife.EC, proliferating capillary EC.
We specifically examined the trajectories of capillary ECs, which exhibits a greater heterogeneity.13 We ordered the 4 capillary ECs (including capillary 1 and capillary 2, Prolife.ECs, and Angio.ECs) in a pseudo-temporal manner to deconstruct the population heterogeneity and reconstruct the developmental process. We found that Prolife.ECs progressed toward the capillary ECs state and bifurcated into 2 diverse branches (capillary 1 and capillary 2 ECs) in young and old, respectively, representing 2 major cell lineages in the late reprogramming stage (Figure 3E, F). To dissect the gene expression profiles that distinguish the 2 branches, we analyzed the expression profiles of the top 1025 DEGs of capillary 1 and capillary 2 ECs in pseudo-temporal order and divided cells after bifurcation into 2 classes: earlier stage (class A) and later stage (class B) (Figure 3C). In comparison with the young ones, old branch cells in class A expressed higher levels of genes enriched in “regulation of angiogenesis,” “negative regulation of phosphorylation,” and “regulation of vasculature development” (Figure 3D). While the DEGs in clusters 4 and 5 (downregulated genes) were strongly enriched for progression of energy metabolism, such as “respiratory chain” and “NADH dehydrogenase complex” (Figure 3D). Together, these unique transcriptional gene expression patterns of clusters 1, 4, and 5 characterize the earliest transcriptomic events of cell differentiation in capillary 1 and capillary 2 ECs.
We next separately ordered the 4 capillary ECs of young and old liver tissues in a pseudo-temporal manner (Figure 4A, C). We assigned Prolife.ECs as the origin, considering its regenerative potential. We found that Prolife.ECs mainly differentiated into capillary 1 and capillary 2 ECs in young livers, but in old livers, they further differentiated into capillary 1, capillary 2 ECs and Angio.ECs (Figure 4A, C).
FIGURE 4.
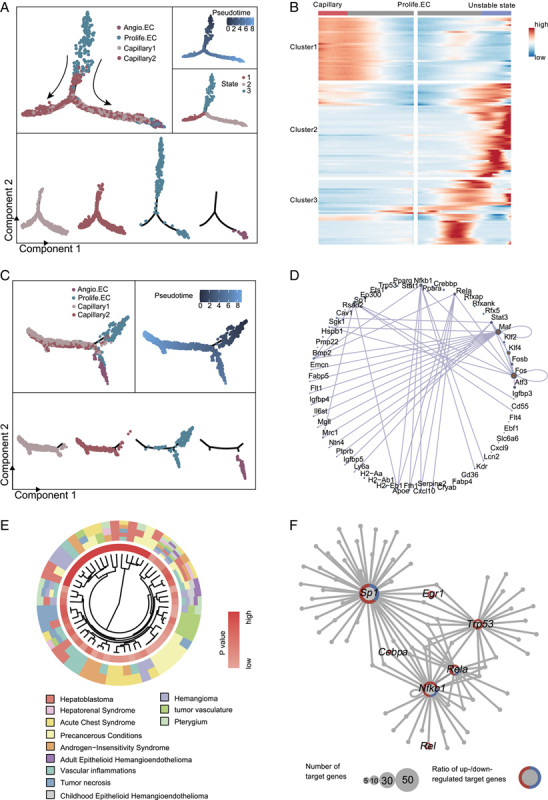
Pseudo-time trajectory analysis of capillaries in young and old liver. (A) Pseudo-time analysis of capillaries (including capillary 1, capillary 2, Prolife.ECs, and Angio.ECs) in young mouse liver endothelial cells (ECs). Cells are colored by cell types (top) and segmented by different cell types (bottom). (B) Heatmap showing the expression profiles of the top 130 TDGs (q value <1×10−4) that were significantly changed in pseudo-temporal order after the prebranch, and then grouped into 3 clusters by expression pattern. (C) Pseudo-time analysis of capillaries (including capillary 1, capillary 2, Prolife.ECs, and Angio.ECs) in old mouse liver ECs. Cells are colored by cell types (top) and segmented by different cell types (bottom). (D) Network diagram showing transcriptional regulators of the top 130 TDGs. (E) Circle plots showing the top 130 TDGs associated with liver carcinoma. (F) Network plot showing transcriptional regulators of aging-related DEGs in Prolife.ECs in mouse liver. Node size indicates the number of target genes. The outer circle of the node represents the proportion of upregulated (red) and downregulated (blue) target genes regulated by the corresponding transcriptional regulators. Abbreviations: Angio.EC, angiogenic capillary EC; DEGs, differentially expressed genes; Prolife.EC, proliferating capillary EC.
To elucidate the molecular dynamics that drive cell differentiation in old liver Prolife.ECs, we performed an expression heatmap of the top 130 TDGs in old liver capillary ECs (including capillary 1, capillary 2, Prolife.ECs, and Angio.ECs), which were divided into 3 distinct clusters (Figure 4B). DisGeNET enrichment analysis of TDGs of Prolife.ECs showed the enrichments of “hemangioma,” “precancerous conditions,” “hepatoblastoma,” and “tumor vasculature” indicating that the Prolife.ECs of old group may be responsible for tumor formation and metastasis in the liver (Figure 4E). The transcriptional regulatory network governing TDGs revealed a number of key TFs in capillary ECs (Figure 4D). For example, Maf is considered as a transcriptional factor to regulate cell differentiation and tumorigenesis.24 Kif4 is involved in both embryonic development and cell differentiation.25 Fos regulates cell proliferating and differentiation by binding to DNA sites through TGF-β-mediated signaling.26 Interestingly, some of the TFs in Prolife.ECs are closely related to senescence and tumorigenesis (Figure 4F). Notably, Sp1 (trans-acting TF 1) binds to GC-rich motifs and regulates the expression of genes related to cell growth and immune response.27,28 Rela (TF p65) and Nfkb1 are both pleiotropic TFs involved in a range of signal transduction events such as cell differentiation, cell growth and tumorigenesis.29 Trp53 (cellular tumor antigen p53) acts as a tumor suppressor to induce apoptosis and is involved in cell cycle regulation by controlling a set of genes.
Altogether, these findings indicated Prolife.ECs dysregulation along the trajectory constituted one of the major alterations in the aged liver and Prolife.ECs may facilitate liver tumorigenesis at an advanced age.
Intercellular communication networks support enhanced cell chemotaxis and inflammatory activation observed in an old liver
Although inflammation response with aging has been documented, the specific aging-induced cell-cell interactions in the hepatic vascular niche have not been investigated. To identify the cellular interactions affected by aging, we first explored cell-cell communication in the young and old liver using CellphoneDB and taking advantage of the public Tabula Muris data sets from the liver (Figure 5A, B). We compared the number of cellular interactions between groups and found that the number of predicted interactions was increased in the old group compared with the young group, consistent with the observations in other aged tissues.14 We then focused mainly on the aged cellular interactions in the following analysis.
FIGURE 5.
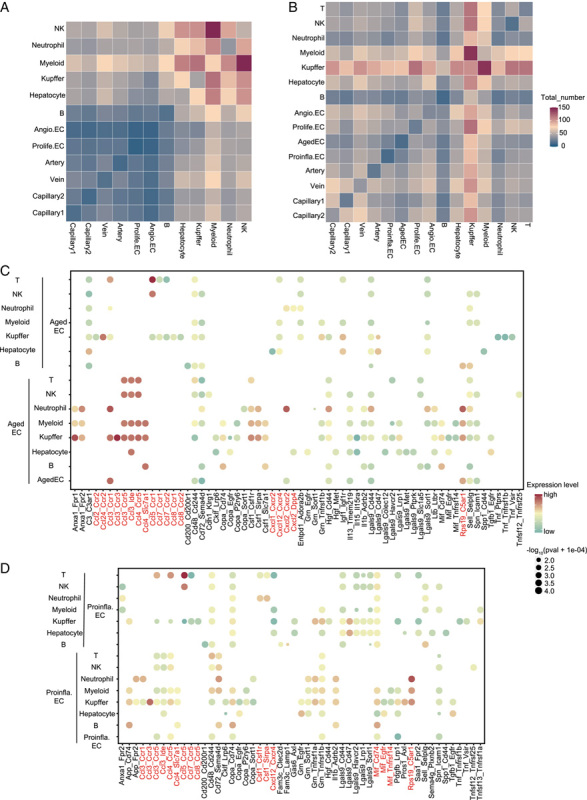
CellPhoneDB analysis of cell-cell interaction in young and old hepatic vascular niche. (A) Heatmap showing the total number of interactions between cell types in the young hepatic vascular niche. (B) Heatmap showing the total number of interactions between cell types in the old hepatic vascular niche. (C) Cell-cell interaction analysis between Aged EC and the indicated cell types in the old hepatic vascular niche. Ligand-receptor pairs are labeled on the x-axis. p Values are indicated by the circle sizes while different colors represent different expression levels of ligands and receptors. We carried out an mRNA assay to extrapolate protein interactions. (D) Cell-cell interaction analysis between Proinfla.EC and the indicated cell types in the old hepatic vascular niche. Ligand-receptor pairs are labeled on the x-axis. p Values are indicated by the circle sizes while different colors represent different expression levels of ligands and receptors. We carried out the mRNA assay to extrapolate protein interactions. Abbreviations: Aged EC, aged capillary EC; Angio.EC, angiogenic capillary EC; Prolife.EC, proliferating capillary EC; Proinfla.EC, pro-inflammation capillary ECs.
We detected that aging induced several interactions which were mainly involved in recruiting immune cells to hepatic vascular niche, including Ccl-related and Cxcl-related pathways (Figure 5C; Figure S4A, C, http://links.lww.com/HC9/A55).30 Specifically, the old group showed increased enrichment of intercellular communications, including Ccl3-Ccr3 between Aged ECs and KC; Rps19-C5ar1 between Aged ECs and neutrophils. Rps19-C5ar1 was also the most frequent ligand-receptor pair between old Proinfla.ECs and neutrophils (Figure 5D), which play important roles in antitumor immune responses.31 Hepatocytes also had broad interactions with capillary ECs (Figure S4B, http://links.lww.com/HC9/A55). For instance, upregulated interaction Igf1-Igf1r was observed between old liver ECs (Angio.ECs, Prolife.ECs, Aged ECs, capillary 1, and capillary 2 ECs) and hepatocytes, suggesting the association between liver ECs activation and liver tumorigenesis.32 Next, upregulated signaling events, between common EC clusters and old hepatic vascular niches, were disclosed (Figure S4D-I, http://links.lww.com/HC9/A55). We found that upregulated cell-cell interactions were concentrated on Prolife.ECs and Angio.ECs involved signalings, including Mif-Cd74 interaction participated in the recruitment of immune cells33; Lgals9-related pathway and other immune checkpoint receptor interactions (eg, Havcr2 and Cd200r)34,35 which signified a protumorigenic milieu. Upregulated Ccl-related and Cxcl-related pathways were concentrated on capillary 1, capillary 2, and vein. Notably, we identified increased Ackr1-related and Ackr2-related pathways which facilitated angiogenesis and the recruitment of immune cells towards the inflammatory niche.36
To further examine the key signaling events with age, we performed cell-cell interaction analysis in liver EC types using Cellchat. There was an overall increase in cell-cell interactions in aged mouse liver ECs (Figure S5A, http://links.lww.com/HC9/A56). We compared the information flow for each signaling pathway between young and old liver (Figure S5B, http://links.lww.com/HC9/A56), and found that both Jam and Edn pathways maintain similar flow. It may be that the 2 pathways represent core signaling pathways necessary for liver function, independent of the specific point in the biological time scale (ie, young vs. old). However, some important pathways (such as Notch, Collagen, Cdh5, and Vegf) significantly increased their information flow in the old livers as compared to the young ones. We identified 2 pathways that were specifically activated in young liver ECs, including cell adhesion signaling Esam (EC-selective adhesion molecule) and Mpz (Myelin protein zero). Some other pathways were active only in the old liver endothelium (Figure S5B, http://links.lww.com/HC9/A56), including Icam pathway (Figure S5C, http://links.lww.com/HC9/A56), the one for leukocyte-EC adhesion. Icam1 is a ligand for the leukocyte adhesion protein regulating inflammatory cells adhesion to vascular EC and T-cell activation.37 In addition, RNAscope and immunofluorescence staining indicated that molecules involved in Icam1-Spn signaling were expressed in Aged ECs (Figure S5D, E, http://links.lww.com/HC9/A56). Further studies are needed to explore the immuno-regulation and proinflammatory role of Icam1-Spn signaling between Aged ECs and niche immune cells in the hepatic vascular niche. Collectively, these cellular interaction findings indicated inflammatory activation and underlying molecular features during liver aging, which may exacerbate age-related liver diseases.
DISCUSSION
In the present study, we established the first mice single-cell transcriptomic atlas of LECs aging. According to unique transcriptional signatures, we identified 8 distinct cell types, comprised of the capillary lineage (including capillary 1, capillary 2, Prolife.ECs, Angio.ECs, Proinfla.ECs, and Aged ECs), arterial and venous ECs. Consistently, we captured a broad spectrum of cell types in the liver that had been previously reported by Kalucka et al.13 who performed a single-cell RNA study on ECs of 11 mouse tissues.
Firstly, we detected Aged ECs, especially expressed in the old liver with highest SASP score and related with inflammatory response indicated by marker gene Osm,15 in line with previous studies that aging was associated with chronic, low-grade inflammation (Figure 1C).38 Among the 8 identified cell types, capillary ECs were most affected by aging in line with previous reports.39,40 According to the vascular theory of aging41 and our study, we speculated that Aged ECs might originate from liver sinusoidal ECs which may possessed increased proliferating capability under the influence of complex hepatic niche. In addition, the interactions between Aged ECs and other cells were highly enriched in Ccl3-Ccr1, Ccl3-Ccr5, Ccl3-Ide and other chemokines in old hepatic vascular niche. Ccl3-Ccr1 was reported to recruit circulating neutrophils to the inflamed region, which could mediate pathogen defenses and self-protecting mechanism with aging.42 Moreover, Ccl3-Ccr5 has pro-tumorigenic effects by inflammatory and angiogenic pathways.43 Our study suggested that interactions between Aged ECs and other niche cells, including Ccl-related pathway and Icam1-Spn signaling, may underlie chronic inflammatory liver diseases with aging.
Secondly, we found Proinfla.ECs in old group associated with promoting inflammation. The interactions between Proinfla.ECs and liver immune cells, and other ECs were highly enriched in Ccl3-Ccr5, Ccl3-Ccr3, Csf1-Csf1r and other chemokines, indicating an important role in inflammatory processes and elevated pro-inflammatory chemokines levels. In addition, this Proinfla.ECs phenotype has also been demonstrated in older healthy humans,38,44 and may link to endothelial dysfunction with advancing age.
Thirdly, aging-related DEG analysis unraveled that Prolife.ECs were the most susceptible cell types to aging as they manifested the most DEGs, including genes annotated as high-risk genes for liver diseases. GO analysis of DEGs in Prolife.ECs implied age-related angiogenesis and liver tumorigenesis with age (Figure 2B, C). Moreover, in-depth analysis of the dynamic gene expression signatures of capillary development trajectory revealed in aged livers, Prolife.ECs could differentiate into Angio.ECs, of which TDGs indicated tumor formation and metastasis in the liver (Figure 4E). Cellular interaction analysis demonstrated Rps19-C5ar1 was highly enriched in Proinfla.ECs, Aged ECs, and Prolife.ECs, which play important roles in antitumor immune response and was found to contribute to increased risk of tumor metastasis with aging, corresponding with pioneering studies that age-associated alterations increase the metastatic capacity of tumor cells.31
Lastly, Angio.ECs were analyzed to be related with hepatic angiogenesis, which plays a major role in wound healing and scar formation.45 Previous studies revealed that ECs contribute to liver angiogenesis,46 and angiogenesis significantly linked to progression of fibrosis to cirrhosis in patients with chronic liver diseases.47,48 In addition, Rgcc, upregulated gene in old group angiogenic ECs was reported to regulate the cell cycle and contributed to the pathogenesis of fibrosis.49 Therefore, Angio.ECs and Prolife.ECs of aged liver may result in a highly vascularized tumor and promote tumorigenesis and the development of metastasis. Future studies are needed to explore whether targeting Prolife.ECs and Angio.ECs can effectively inhibit angiogenesis in cancer or age-related diseases.
It should be noted that the EC clusters identified in young mice in our study and those published by Kalucka et al.13 are similar. Specifically, both studies identified capillary, arterial, venous, and proliferating ECs. Nevertheless, our study additionally identified a very small group of angiogenic ECs (0.31%), whereas the Kalucka et al.13 study additionally identified a small group of lymphatic ECs (0.8%). The differences between the 2 studies may be attributed to the possibility that during dissociation of liver tissues in different enzymatic dissociation medium and FACS-based enrichment of ECs, different rare cell types could have been captured or lost.
The present study has some limitations. Firstly, we acknowledge that protein analysis and functional experiments are both needed to better confirm the role of each vascular endothelial phenotype. Secondly, the presumed differentiation tree localization of the vascular endothelium requires spatial transcriptomic validation. Thirdly, we cannot exclude that some EC types were lost during tissue isolation and were not detected.
Nevertheless, our work is the first to describe multiple age differences in liver ECs, including cell composition, DEGs, enrichment pathways, differentiation trajectories, and cell-cell communication. Further studies of Proinfla.ECs, Aged ECs, Angio.ECs, and Prolife.ECs in the future may elucidate the underlying mechanisms of liver aging-related diseases.
Supplementary Material
ACKNOWLEDGMENTS
This study was funded by the National Natural Science Foundation of China (81721003); the State Key Laboratory of Ophthalmology, Zhongshan Ophthalmic Center, Sun Yat-sen University. The funding body had no role in study design, collection, management, analysis and interpretation of data, writing of the manuscript, and the decision to submit the manuscript for publication.
CONFLICT OF INTEREST
Nothing to report.
DATA AVAILABILITY STATEMENT
The scRNA-seq data analyzed in this study was deposited in the Genome Sequence Archive (GSA) under project number (PRJCA014447).
Footnotes
Funding information This study was funded by the National Natural Science Foundation of China (81721003); the State Key Laboratory of Ophthalmology, Zhongshan Ophthalmic Center, Sun Yat-sen University. The funding body had no role in the study design, collection, management, analysis and interpretation of data, writing of the manuscript, and the decision to submit the manuscript for publication.
Abbreviations: EC, endothelial cells; scRNA-seq, single-cell RNA sequencing; Aged ECs, aged capillary ECs; Proinfla.ECs, pro-inflammation capillary ECs; Prolife.ECs, proliferating capillary ECs; Angio.ECs, angiogenic capillary ECs; LECs, liver ECs; DEGs, differentially expressed genes; SASP, senescence-associated secretory phenotype; TDG, time differentiation-related gene; TF, transcription factor; GO, gene ontology, GO.
D.W., M.L., and J.L. contributed equally to this work.
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website, www.hepcommjournal.com.
Contributor Information
Dongliang Wang, Email: wangdliang3@mail2.sysu.edu.cn.
Mengke Li, Email: limengke@gzzoc.com.
Jie Ling, Email: lingjie2018@163.com.
Shuxia Chen, Email: chenshuxia@gzzoc.com.
Qikai Zhang, Email: 1592789885@qq.com.
Zhong Liu, Email: liu18390988308@163.com.
Yanjing Huang, Email: olivehuang17@gmail.com.
Caineng Pan, Email: pancaineng@gzzoc.com.
Yuheng Lin, Email: linyuheng@gzzoc.com.
Ping Zhang, Email: zhangping@gzzoc.com.
Yingfeng Zheng, Email: zhyfeng@mail.sysu.edu.cn.
REFERENCES
- 1.Kirkland JL. Translating the science of aging into therapeutic interventions. Cold Spring Harb Perspect Med. 2016;6:a025908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J, Litvinova M, Liang Y, Wang Y, Wang W, Zhao S, et al. Changes in contact patterns shape the dynamics of the COVID-19 outbreak in China. Science. 2020;368:1481–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cines DB, Pollak ES, Buck CA, Loscalzo J, Zimmerman GA, McEver RP, et al. Endothelial cells in physiology and in the pathophysiology of vascular disorders. Blood. 1998;91:3527–3561. [PubMed] [Google Scholar]
- 4.Di Micco R, Krizhanovsky V, Baker D, d’Adda di Fagagna F. Cellular senescence in ageing: from mechanisms to therapeutic opportunities. Nat Rev Mol Cell Biol. 2021;22:75–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grosse L, Wagner N, Emelyanov A, Molina C, Lacas-Gervais S, Wagner KD, et al. Defined p16(high) senescent cell types are indispensable for mouse healthspan. Cell Metab. 2020;32:87–99 e86. [DOI] [PubMed] [Google Scholar]
- 6.Potente M, Makinen T. Vascular heterogeneity and specialization in development and disease. Nat Rev Mol Cell Biol. 2017;18:477–494. [DOI] [PubMed] [Google Scholar]
- 7.Bloom SI, Islam MT, Lesniewski LA, Donato AJ. Mechanisms and consequences of endothelial cell senescence. Nat Rev Cardiol. 2023;20:38–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rui L. Energy metabolism in the liver. Compr Physiol. 2014;4:177–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koch PS, Lee KH, Goerdt S, Augustin HG. Angiodiversity and organotypic functions of sinusoidal endothelial cells. Angiogenesis. 2021;24:289–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shetty S, Lalor PF, Adams DH. Liver sinusoidal endothelial cells—gatekeepers of hepatic immunity. Nat Rev Gastroenterol Hepatol. 2018;15:555–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grun D, van Oudenaarden A. Design and analysis of single-cell sequencing experiments. Cell. 2015;163:799–810. [DOI] [PubMed] [Google Scholar]
- 12.He X, Memczak S, Qu J, Belmonte JCI, Liu GH. Single-cell omics in ageing: a young and growing field. Nat Metab. 2020;2:293–302. [DOI] [PubMed] [Google Scholar]
- 13.Kalucka J, de Rooij L, Goveia J, Rohlenova K, Dumas SJ, Meta E, et al. Single-cell transcriptome atlas of murine endothelial cells. Cell. 2020;180:764–779 e720. [DOI] [PubMed] [Google Scholar]
- 14.Ma S, Sun S, Geng L, Song M, Wang W, Ye Y, et al. Caloric restriction reprograms the single-cell transcriptional landscape of Rattus Norvegicus. Aging Cell. 2020;180:984–1001 e1022. [DOI] [PubMed] [Google Scholar]
- 15.Richards CD. The enigmatic cytokine oncostatin m and roles in disease. ISRN Inflamm. 2013;2013:512103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng K, Liu H, Yan B, Meng XW, Song SY, Ji FH, et al. Inhibition of cathepsin S attenuates myocardial ischemia/reperfusion injury by suppressing inflammation and apoptosis. J Cell Physiol. 2021;236:1309–20. [DOI] [PubMed] [Google Scholar]
- 17.Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U, et al. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nat Immunol. 2011;12:536–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Song X, Herrup K. Context-dependent functions of E2F1: cell cycle, cell death, and DNA damage repair in cortical neurons. Mol Neurobiol. 2020;57:2377–90. [DOI] [PubMed] [Google Scholar]
- 19.Favre J, Vessieres E, Guihot AL, Proux C, Grimaud L, Rivron J, et al. Membrane estrogen receptor alpha (ERalpha) participates in flow-mediated dilation in a ligand-independent manner. Elife. 2021;10:e68695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stan RV, Kubitza M, Palade GE. PV-1 is a component of the fenestral and stomatal diaphragms in fenestrated endothelia. Proc Natl Acad Sci U S A. 1999;96:13203–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Akiel M, Guo C, Li X, Rajasekaran D, Mendoza RG, Robertson CL, et al. IGFBP7 deletion promotes hepatocellular carcinoma. Cancer Res. 2017;77:4014–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeltsch KM, Hu D, Brenner S, Zoller J, Heinz GA, Nagel D, et al. Cleavage of roquin and regnase-1 by the paracaspase MALT1 releases their cooperatively repressed targets to promote T(H)17 differentiation. Nat Immunol. 2014;15:1079–89. [DOI] [PubMed] [Google Scholar]
- 23.Mao L, Yang J, Yue J, Chen Y, Zhou H, Fan D, et al. Decorin deficiency promotes epithelial-mesenchymal transition and colon cancer metastasis. Matrix Biol. 2021;95:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blank V, Andrews NC. The Maf transcription factors: regulators of differentiation. Trends Biochem Sci. 1997;22:437–41. [DOI] [PubMed] [Google Scholar]
- 25.Ye B, Liu B, Hao L, Zhu X, Yang L, Wang S, et al. Klf4 glutamylation is required for cell reprogramming and early embryonic development in mice. Nat Commun. 2018;9:1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J. Molecular interpretation of ERK signal duration by immediate early gene products. Nat Cell Biol. 2002;4:556–64. [DOI] [PubMed] [Google Scholar]
- 27.Yu J, Wei M, Boyd Z, Lehmann EB, Trotta R, Mao H, et al. Transcriptional control of human T-BET expression: the role of Sp1. Eur J Immunol. 2007;37:2549–61. [DOI] [PubMed] [Google Scholar]
- 28.Beishline K, Azizkhan-Clifford J. Sp1 and the “hallmarks of cancer”. FEBS J. 2015;282:224–58. [DOI] [PubMed] [Google Scholar]
- 29.Levy D, Kuo AJ, Chang Y, Schaefer U, Kitson C, Cheung P, et al. Lysine methylation of the NF-kappaB subunit RelA by SETD6 couples activity of the histone methyltransferase GLP at chromatin to tonic repression of NF-kappaB signaling. Nat Immunol. 2011;12:29–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menten P, Wuyts A, Van Damme J. Macrophage inflammatory protein-1. Cytokine Growth Factor Rev. 2002;13:455–81. [DOI] [PubMed] [Google Scholar]
- 31.Markiewski MM, Vadrevu SK, Sharma SK, Chintala NK, Ghouse S, Cho JH, et al. The ribosomal protein S19 suppresses antitumor immune responses via the complement C5a receptor 1. J Immunol. 2017;198:2989–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kerr A, Baxter RC. Noncoding RNA actions through IGFs and IGF binding proteins in cancer. Oncogene. 2022;41:3385–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bernhagen J, Krohn R, Lue H, Gregory JL, Zernecke A, Koenen RR, et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med. 2007;13:587–96. [DOI] [PubMed] [Google Scholar]
- 34.Yang R, Sun L, Li CF, Wang YH, Yao J, Li H, et al. Galectin-9 interacts with PD-1 and TIM-3 to regulate T cell death and is a target for cancer immunotherapy. Nat Commun. 2021;12:832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinjab A, Han G, Treekitkarnmongkol W, Hara K, Brennan PM, Dang M, et al. Resolving the spatial and cellular architecture of lung adenocarcinoma by multiregion single-cell sequencing. Cancer Discov. 2021;11:2506–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nibbs RJ, Graham GJ. Immune regulation by atypical chemokine receptors. Nat Rev Immunol. 2013;13:815–29. [DOI] [PubMed] [Google Scholar]
- 37.Hogg N, Patzak I, Willenbrock F. The insider’s guide to leukocyte integrin signalling and function. Nat Rev Immunol. 2011;11:416–26. [DOI] [PubMed] [Google Scholar]
- 38.Donato AJ, Black AD, Jablonski KL, Gano LB, Seals DR. Aging is associated with greater nuclear NF kappa B, reduced I kappa B alpha, and increased expression of proinflammatory cytokines in vascular endothelial cells of healthy humans. Aging Cell. 2008;7:805–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hilmer SN, Cogger VC, Fraser R, McLean AJ, Sullivan D, Le Couteur DG. Age-related changes in the hepatic sinusoidal endothelium impede lipoprotein transfer in the rat. Hepatology. 2005;42:1349–54. [DOI] [PubMed] [Google Scholar]
- 40.Le Couteur DG, Cogger VC, Markus AM, Harvey PJ, Yin ZL, Ansselin AD, et al. Pseudocapillarization and associated energy limitation in the aged rat liver. Hepatology. 2001;33:537–43. [DOI] [PubMed] [Google Scholar]
- 41.Le Couteur DG, Lakatta EG. A vascular theory of aging. J Gerontol A Biol Sci Med Sci. 2010;65:1025–7. [DOI] [PubMed] [Google Scholar]
- 42.Metzemaekers M, Gouwy M, Proost P. Neutrophil chemoattractant receptors in health and disease: double-edged swords. Cell Mol Immunol. 2020;17:433–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kodama T, Koma YI, Arai N, Kido A, Urakawa N, Nishio M, et al. CCL3-CCR5 axis contributes to progression of esophageal squamous cell carcinoma by promoting cell migration and invasion via Akt and ERK pathways. Lab Invest. 2020;100:1140–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Donato AJ, Eskurza I, Silver AE, Levy AS, Pierce GL, Gates PE, et al. Direct evidence of endothelial oxidative stress with aging in humans: relation to impaired endothelium-dependent dilation and upregulation of nuclear factor-kappaB. Circ Res. 2007;100:1659–66. [DOI] [PubMed] [Google Scholar]
- 45.Carmeliet P. Mechanisms of angiogenesis and arteriogenesis. Nat Med. 2000;6:389–95. [DOI] [PubMed] [Google Scholar]
- 46.Ankoma-Sey V, Wang Y, Dai Z. Hypoxic stimulation of vascular endothelial growth factor expression in activated rat hepatic stellate cells. Hepatology. 2000;31:141–8. [DOI] [PubMed] [Google Scholar]
- 47.Ehling J, Bartneck M, Wei X, Gremse F, Fech V, Mockel D, et al. CCL2-dependent infiltrating macrophages promote angiogenesis in progressive liver fibrosis. Gut. 2014;63:1960–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Medina J, Sanz-Cameno P, Garcia-Buey L, Martin-Vilchez S, Lopez-Cabrera M, Moreno-Otero R. Evidence of angiogenesis in primary biliary cirrhosis: an immunohistochemical descriptive study. J Hepatol. 2005;42:124–31. [DOI] [PubMed] [Google Scholar]
- 49.Li Z, Xie WB, Escano CS, Asico LD, Xie Q, Jose PA, et al. Response gene to complement 32 is essential for fibroblast activation in renal fibrosis. J Biol Chem. 2011;286:41323–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The scRNA-seq data analyzed in this study was deposited in the Genome Sequence Archive (GSA) under project number (PRJCA014447).


