Abstract
This review summarizes a decade of research in which we have used molecular methods, in conjunction with more traditional approaches, to study hot spring cyanobacterial mats as models for understanding principles of microbial community ecology. Molecular methods reveal that the composition of these communities is grossly oversimplified by microscopic and cultivation methods. For example, none of 31 unique 16S rRNA sequences detected in the Octopus Spring mat, Yellowstone National Park, matches that of any prokaryote previously cultivated from geothermal systems; 11 are contributed by genetically diverse cyanobacteria, even though a single cyanobacterial species was suspected based on morphologic and culture analysis. By studying the basis for the incongruity between culture and molecular samplings of community composition, we are beginning to cultivate isolates whose 16S rRNA sequences are readily detected. By placing the genetic diversity detected in context with the well-defined natural environmental gradients typical of hot spring mat systems, the relationship between gene and species diversity is clarified and ecological patterns of species occurrence emerge. By combining these ecological patterns with the evolutionary patterns inherently revealed by phylogenetic analysis of gene sequence data, we find that it may be possible to understand microbial biodiversity within these systems by using principles similar to those developed by evolutionary ecologists to understand biodiversity of larger species. We hope that such an approach guides microbial ecologists to a more realistic and predictive understanding of microbial species occurrence and responsiveness in both natural and disturbed habitats.
This review focuses upon recent ecological studies of terrestrial hot spring microbial communities that are beginning to reshape our view of microbial biodiversity and of the composition, structure, and function of microbial communities. Other reviews provide general information about the types of microbial communities found in different terrestrial hot spring habitats (18, 23, 147, 148, 151, 160) and about the large numbers of new thermophilic bacterial isolates that have been obtained in recent years (136, 137), fueled by interest in biotechnology and the possible ancestral nature of hyperthermophily (96, 171; but see references 44 and 79). Here, we draw attention to how the application of molecular tools to the analysis of hot spring (and other) microbial communities is beginning to enable us to understand the organization of microbial communities as macroecologists understand the biodiversity of larger species, in terms of the specific populations that comprise higher-order structure within communities (Table 1). As suggested by an ecology text that we found to give an excellent introduction to the subject (11), “A first step is usually to search for patterns in community structure. … [R]ecognition of patterns leads in turn to the forming of hypotheses about the causes of these patterns.” These points were also recently emphasized by Lawton (69). We especially appreciate the natural view that Begon et al. (11) present in their text, i.e., that of the natural historian, who observes organisms in situ and considers organisms to be products of their evolution and ecology. Hence, we refer to this text to illustrate how such a view has helped us understand patterns that we have observed in our measurements of natural microbial populations.
TABLE 1.
Terms used by ecologists, evolutionary biologists, and bacteriologists to describe levels of biological organization above the level of individual organisms
| Term | Definition | Source |
|---|---|---|
| Population | “A group of individuals of one species in an area” | Begon et al. (11) |
| “A group of conspecific organisms that occupy a more or less well defined geographic region and exhibit reproductive continuity from generation to generation” | Futuyma (48) | |
| Ecotype | “A sub-set of individuals within a species with a characteristic ecology” | Begon et al. (11) |
| “A genetically determined phenotype of a species that is found as a local variant associated with certain ecological conditions” | Futuyma (48) | |
| Species | “The members in aggregate of a group of populations that interbreed or potentially interbreed with each other under natural conditions” | Futuyma (48) |
| “Strains with approximately 70% or greater DNA-DNA relatedness” | Wayne et al. (161) | |
| “The major biological meaning of reproductive isolation is that it provides protection for a genotype adapted for the utilization of a specific niche. … It is only where the criterion of reproductive isolation breaks down, as in the case of asexual clones, that one makes use of the criterion of niche occupation.” | Mayr (77) | |
| “A lineage …evolving separately from others and with its own unitary evolutionary role and tendencies. … Roles are definable by their equivalence to niches. | Simpson (130) | |
| Guild | “A group of species that exploit the same class of environmental resources in a similar way” | Begon et al. (11) |
| Community | “An assemblage of species populations which occur together in space and time” | Begon et al. (11) |
The recognition of patterns of occurrence of individual populations within microbial communities has been prevented by limitations inherent to the traditional methods of microbiology. One problem is the relatively small size and nondistinctive appearance of microorganisms, especially prokaryotes, which frustrates attempts to differentiate populations microscopically. The lack of morphological variation has led to another problem—the reliance upon cultivation of microorganisms for their identification. Many microbiologists, including Martinus Beijerinck and Sergei Winogradsky, who pioneered the use of culture methods to study the general biology of naturally occurring microorganisms, doubted that such an approach would provide an accurate description of microorganisms as they occur within natural habitats (see references 144 and 147 for citations and quotations). We certainly do not wish to condemn pure culture work; it is essential for characterizing microorganisms, and no one can deny the value of detailed investigation of pure cultures to the development of our understanding of microbial physiology and genetics. For example, pure culture studies have led to new molecular approaches (93) that are revolutionizing microbiology by providing an evolutionary framework and enabling new ecological approaches. However, the limitations of cultivation methods for objective analysis of populations within natural communities also cannot be denied (e.g., see references 19 and 147). We believe that the heavy emphasis on the pure culture approach during the past century has forced microbiologists to become overly comfortable with unnatural views of microbes in nature (146). Without an adequate means of detecting predominant native microbial populations, studies of microbial communities have mainly been ecophysiological studies of microbial process rates. These are, of course, essential for understanding those aspects of community function that it occurs to us to measure or which we have the capability to detect. Such measurements must, however, be considered assays of guild activities, since they potentially include the activities of many individual populations performing similar tasks (Table 1). Through the application of molecular methods, we appear to be entering an era in which it is possible to observe the patterns of occurrence of the individual populations upon which the structure of microbial communities is based.
The studies we review here mainly involve the use of 16S rRNA sequences (or the genes encoding them) as a means of avoiding the need to cultivate a microorganism to recognize its presence and measure its distribution in a community. Terrestrial hot spring microbial communities were among the first to be surveyed with this technology (134, 159) and thus were among the first in which the impressive diversity of uncultivated microbial populations in nature was revealed. Of course, this has been a typical finding in 16S rRNA gene surveys of microbial diversity in numerous habitats (13–16, 30, 47, 50, 56, 66, 71, 82, 91, 123, 132, 141). 16S rRNA studies of hyperthermal hot spring habitats have led to the discovery of novel uncultivated bacteria (e.g., Aquificales and Thermotogales relatives [98, 115] and archaea [e.g., Korarchaeota and others {4, 5, 97, 98}]) that are particularly interesting because they branch near the root of 16S rRNA-derived phylogenetic trees. Since such microorganisms may help us determine characteristics of the most ancestral cells, it is also quite exciting that some of them have recently been brought into culture (58, 59).
As ecologists, we wish to know why such diversity exists, how it is organized within a microbial community, and what value it may have to community structure and function. We are guided by the foundational thinking of macroecologists, who have reminded us of the intimate linkages among diversity, ecology, and evolution. Begon et al. (11), for example, in their introductory remarks extend T. H. Dobzhansky’s comment that “Nothing in biology makes sense, except in the light of evolution,” to emphasize that “very little in evolution makes sense, except in the light of ecology.” This is, of course, because the diversity of species that exists is a consequence of the interactions between organisms and environments—natural selection and genetic drift among ecologically or geographically isolated populations drive speciation (48, 117). The noted ecologist E. O. Wilson recently claimed in his autobiography (169) that “If I had it to do all over again and relive my vision in the twenty-first century, I would be a microbial ecologist. … Into that world I would go with the aid of modern microscopy and molecular analysis.” Perhaps this is because he was aware that there is potential to use molecular methods to understand microbial diversity in terms of evolutionary ecology, an approach that he and others developed to understand the diversity of larger species (e.g., see references 73 and 117). 16S rRNA sequence data, used so extensively to pioneer studies of microbial evolution (93, 98, 171, 172), not only permit culture-independent detection of native populations but also automatically provide a means of observing patterns of their evolution. The facility to probe 16S rRNA sequences (1, 84, 133) provides a means of observing the ecological patterns of occurrence of the populations detected. As microbial ecology enters a new era, in which molecular techniques permit improved detection of specific populations, it seems useful that microbial ecologists should consider whether patterns of occurrence of microbial species are governed by principles similar to those that explain the evolution and ecology of larger, more complex species.
TECHNIQUES AND TERMINOLOGY
Because we intend to emphasize results, we refer readers to several reviews describing what have become somewhat standard 16S rRNA-based cloning, sequencing, and oligonucleotide probe methods and their limitations (1, 92, 99, 133, 147), as well as to the specific studies we cite. We do, however, point out concerns about molecular methods as we review our own results. Most of the results we present here were obtained by denaturing gradient gel electrophoresis (DGGE) analysis (36, 84), which involves the separation of PCR-amplified 16S rRNA gene segments in an acrylamide gel containing increasing concentrations of denaturants. The separation is based on differences in melting characteristics of the double-stranded segments, which are in turn dependent upon sequence differences. The result is the simultaneous detection of many individual 16S rRNA molecules as a “profile” of bands, each of which can be reamplified and then sequenced.
In our opinion, because of possible PCR amplification and cloning biases (35, 108, 114, 139) and chimeric artifacts (65, 72) that can be difficult to detect (64, 116), these molecular methods should be viewed as providing a different way of sampling microbial biodiversity compared to cultivation and microscopy. Without further study, it cannot be presumed that methods such as these for detecting 16S rRNA sequences provide a complete and totally objective view of community composition and structure. Nevertheless, it is now possible to recognize evolutionary and ecological patterns of occurrence of those 16S rRNA sequences that are detected in natural habitats. It is essential to consider the meaning of such observations. Do 16S rRNA sequences measure gene diversity or organismal diversity, and, if the latter, at what biological level (population, species, above species) is diversity being measured? We will attempt to address these questions as data are presented below, employing terms which have been used by ecologists and evolutionary biologists, or by bacteriologists, to describe how genetic diversity is organized within communities (Table 1).
HOT SPRING CYANOBACTERIAL MATS AS MODELS FOR MICROBIAL COMMUNITY ECOLOGY
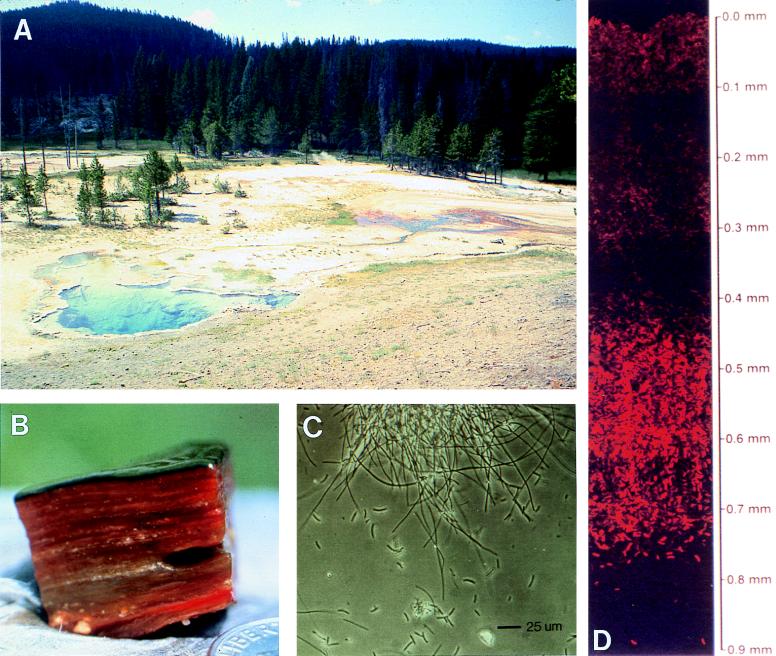
Since 1977, we have studied the cyanobacterial mat community inhabiting the effluent channels in Octopus Spring, Yellowstone National Park (Fig. 1), between ca. 74 and 42°C. These are the approximate upper temperature limits of the mat and of animals which graze upon the mats, respectively. Our rationale has been that the mat (Fig. 1B) is a relatively simple and stable community that can be used as a natural model system in which to investigate principles of microbial community ecology. The mat is typical of many other communities in the sense that organic matter formed through oxygenic photosynthesis in the uppermost 1 mm is almost completely recycled in deeper layers through aerobic and anaerobic decomposition (33, 149, 157). However, this thermal community is free of interactions involving plants and animals (and also eukaryotic microorganisms [156]); therefore, such investigations can be focused entirely on prokaryotes and their interactions. Hot spring mats will not model all microbial communities, especially those that are more heterogeneous and those in which greater microhabitat diversity may lead to greater microbial diversity (139a). However, studies of simpler and more stable systems make it easier to observe patterns that may influence the way we think about more complex systems.
FIG. 1.
Laminated cyanobacterial mat communities of alkaline siliceous hot springs in the Lower Geyser Basin, Yellowstone National Park, viewed at different scales. (A) Landscape showing green-orange mat at far edge of Octopus Spring and down effluent channel. (B) Cross section of ca. 50 to 55°C Octopus Spring cyanobacterial mat sample magnified at ca. × 1.8. (C) Phase-contrast microscopy image of homogenized 1-mm-thick upper green Octopus Spring mat layer showing the predominant cyanobacteria, sausage-shaped S. cf. lividus, embedded in a matrix of filaments, at least some of which are probably green nonsulfur bacteria such as C. aurantiacus (reprinted from reference 160). (D) Autofluorescence microscopy image of a vertical cryotome section through the upper 1-mm green layer of the 61°C Mushroom Spring mat showing banding of S. cf. lividus populations at different depths (111).
A distinct advantage of studying hot spring mats is that these communities were studied intensively for many years by Thomas D. Brock and his students (18), and also by many other microbiologists (summarized in reference 151), providing a solid base of knowledge from which to launch more detailed studies. In the past decade, we have in particular sought to understand the Octopus Spring mat community in terms of its component populations. At the outset, microscopic analysis (Fig. 1C and D) and selective enrichment culture led to the belief that a single unicellular cyanobacterium, morphologically resembling Synechococcus cf. lividus Copeland (23, 25) constructs such mats worldwide. (The modifier “cf.,” meaning “carried forward,” between the genus and species names indicates that the species in question resembles that described initially by Copeland on the basis of morphology.) Similar evidence suggested that a single filamentous green nonsulfur bacterium, Chloroflexus aurantiacus, co-occurs and provides physical cohesiveness to the mats. To borrow once again from the introductory remarks of Begon et al. (11), “the physicist Whitehead’s recipe for science is never more apposite than when applied to ecology: seek simplicity, but distrust it.”
Community Composition
Before we began molecular analysis of the community, numerous investigators had cultivated (or observed microscopically) many thermophiles (Table 2, column 2), including the cyanobacterium S. cf. lividus, the green nonsulfur bacterium C. aurantiacus, aerobic chemoorganotrophs like the planctomycete Isosphaera pallida, numerous gram-positive fermentative bacteria, a phylogenetically novel sulfate reducer, and a methanogen. In general, we doubted that microscopy and cultivation were adequate tools for fully understanding community composition and structure but we hoped that previous culture work would provide an internal means of verification of new approaches. For instance, we presumed that S. cf. lividus and C. aurantiacus would be readily observed in molecular analyses.
TABLE 2.
“Species list” for the Octopus Spring cyanobacterial mat community
| Phylogenetic typea | Previously cultivated organism | 16S rRNA sequence type | Recently enriched or cultivated strain | Physiological typeb | Highest relative population densityc (%) | GenBank accession no. | Reference(s) |
|---|---|---|---|---|---|---|---|
| Cyanobacteria | S. cf. lividusde | S. cf. lividus C1 | Oxygenic phototroph | L35345, L35479-80 | 40, 62 | ||
| A‴ | Oxygenic phototroph | U88530 | 37 | ||||
| A" | Oxygenic phototroph | U88069 | 41 | ||||
| A′ | Oxygenic phototroph | U42374 | 36 | ||||
| A | Oxygenic phototroph | X52544 | 159, 162 | ||||
| B′ | S. cf. lividus P3 | Oxygenic phototroph | 12.5 | U42375 | 36, 102, 111, 155 | ||
| B | S. cf. lividus P1,2 | Oxygenic phototroph | 12.5 | M62776 | 102, 155, 159, 162 | ||
| I | Oxygenic phototroph | L04709 | 162 | ||||
| J | Oxygenic phototroph | L04710 | 162 | ||||
| P | S. cf. lividus B10 | Oxygenic phototroph | 4 | L35331 | 40 | ||
| S. cf. lividus C9 | Oxygenic phototroph | 0.4 | L35481-3 | 40 | |||
| Green nonsulfur bacteria and relatives | C. aurantiacus Y-400-fle | Anoxygenic phototroph; aerobic chemoorganotroph | L04674 | 106, 162 | |||
| Thermomicrobium roseume,f | Aerobic chemoorganotroph | M34115 | 60, 95, 176, 177 | ||||
| C | M62775 | 159, 162 | |||||
| C′ | U42421 | 36 | |||||
| C" | U90433 | 41 | |||||
| OS-V-L-20 | L04703 | 162 | |||||
| env.OS_ace3 | Aerobic chemoorganotroph | 0.3 | L47199 | 121 | |||
| env.OS_ace4 | Aerobic chemoorganotroph | 0.3 | L47200 | 121 | |||
| env.OS_ace5 | Aerobic chemoorganotroph | 0.3 | L47201 | 121 | |||
| Green sulfur bacterium-like | E | X52548 | 147 | ||||
| E′ | U42419 | 36 | |||||
| E" | U42420 | 36 | |||||
| M | L04708 | 159 | |||||
| III-9 | L04705 | 147 | |||||
| Planctomycetes | I. pallidag | Oligotrophic aerobic chemoorganotroph | X64372 | 33, 52, 158 | |||
| Thermus/Deinococcush | T. aquaticus-like ac-1 | Aerobic chemoorganotroph | 0.000003 | L37520 | 22, 87, 121 | ||
| T. aquaticus-like ac-7 | Aerobic chemoorganotroph | 0.000004 | L37522 | 22, 87 | |||
| T. aquaticus-like ac-7′ | Aerobic chemoorganotroph | 0.0003 | L47202i | 121 | |||
| T. ruber-like ac-2 | Aerobic chemoorganotroph | 0.3 | L37521 | 87, 121 | |||
| T. ruber-like ac-17 | Aerobic chemoorganotroph | 0.001 | L37523 | 87 | |||
| Thermus spp. Ramaley-4 | Aerobic chemoorganotroph | X58344 | 6 | ||||
| Proteobacteria | |||||||
| α subdivision | O | L04706 | 147 | ||||
| env.OS_ace2 | Aerobic chemoorganotroph | 0.003 | L47198 | 121 | |||
| β subdivision | G | X52550 | 159 | ||||
| R | U46750 | 86 | |||||
| N | Isolate ac-15 | Aerobic chemoorganotroph | 0.03 | L04712, U46749 | 86 | ||
| Enriched population N′ | Aerobic chemoorganotroph | 0.003 | L47196 | 121 | |||
| Isolate ac-16 | Aerobic chemoorganotroph | 0.001 | U46748 | 86 | |||
| γ subdivision | Isolate env.OS_ace7 | Aerobic chemoorganotroph | 0.0000003 | L47204 | 86 | ||
| δ subdivision | env.OS_ace8j | Aerobic chemoorganotroph | 0.3 | L47205 | 121 | ||
| Spirochetes | H | X52551 | 159 | ||||
| OS-V-L-7 | L04704 | 162 | |||||
| Gram-positive bacteria | Heliobacterium modesticaldum | Anoxygenic phototroph | U14559 | 63, 138 | |||
| Thermoanaerobacter brockii HTD4k | Fermenter | L09165 | 8, 109, 175 | ||||
| Thermoanaerobacter ethanolicus JW200 and 39Ek | Fermenter | L09162, L09164 | 8, 109, 166, 167 | ||||
| Thermobacteroides acetoethylicus HTB2/W | Fermenter | L09163 | 8, 12, 109, 173 | ||||
| Thermoanaerobacterium thermosulfurigenes 4Bk | Fermenter | L09171 | 8, 109, 122 | ||||
| Moorella thermoautotrophica JW701k | Fermenter | X77849, L09168 | 8, 31, 109, 165 | ||||
| env.OS_ace1 | Aerobic chemoorganotroph | 3.3 | L47197 | 121 | |||
| env.OS_ace6 | Aerobic chemoorganotroph | 0.003 | L47203 | 121 | |||
| Isolate ac-18 | Aerobic chemoorganotroph | 0.01 | U46747 | 86 | |||
| Thermodesulfotobacterium | Thermodesulfotobacterium commune | Sulfate reducer | L10662 | 158, 171, 174 | |||
| Paraphyletic assemblage 2.5.1 Leptospirillum group | OP-I-2 | L22045 | 65 | ||||
| Paraphyletic assemblage 2.5.3 enviromental isolates | L | Enriched population 13 | Aerobic chemoorganotroph | 0.0000003 | L04707 | 121, 147 | |
| OS-I-25 | X67084-7 | 158 | |||||
| Paraphyletic assemblage 2.5.4 Nitrospina group | K | L04711 | 147, 162 | ||||
| Bacteria of uncertain lineage | D | X52547 | 159 | ||||
| F | X52549 | 159 | |||||
| Q | U42422 | 36 | |||||
| Archaea | Methanobacterium thermoautotrophicum | Methanogen | X68720 | 89, 120, 173 |
All sequences are placed as they occur in the Ribosomal Database Project (75), release 6.1. Some sequences were previously reported in different lineages (e.g., see references 38 and 153), based on comparison to earlier releases. We caution that two sequences which appear in this release (OS-VI-L-4 and OP-I-6) are not included in the table, as there is good evidence of these being chimeric artifacts (65, 116). A few sequences which do not appear yet in this database are placed according to our own phylogenetic analyses.
This may represent only a fraction of the population’s true physiological potential.
Expressed as a percentage of S. cf. lividus cells determined from direct microscopic counts. The estimate was made from the reciprocal of number of S. cf. lividus cells present in the diluted mat sample used to inoculate the enrichment that led to the observation of a DGGE band or to the cultivation of an isolate (× 100), assuming that at least one cell of the population led to its enrichment or isolation.
We used strain Y-7c-s, initially cultivated from a pH 5.5 spring at Clearwater Springs in Yellowstone National Park and provided by R. W. Castenholz, but several genetically similar strains are available (see reference 40).
Percent similarities between the 16S rRNA sequences of bacteria previously cultivated from the mat and their closest relative among phylogenetically similar sequences detected in the mat indicate how different the two samplings of diversity are: 93.1% between S. cf. lividus Y-7c-s and type I, 81.6% between C. aurantiacus and type C, and 75.4% between T. roseum and type C. The relationships among type A/B cyanobacterial sequences and type C sequences are quantified in Fig. 2. The green sulfur bacterium-like sequences exhibit 82.9 to 90.2% sequence variation; the E-like green sulfur bacterium-like sequences form a cluster which exhibits ca. 6.9% sequence variation. We caution, however, that percent sequence similarity is a function of which regions of the sequences are being compared (i.e., of the balance between conserved and variable regions being analyzed), so that these similarity values, which are based on portions of the 16S rRNA molecule, may not be accurate estimates of similarities of the entire 16S rRNA sequences.
Not cultivated from the Octopus Spring mat, but cells of this morphology were observed (33).
We could not find specific reference to prior cultivation of Thermus spp. from the Octopus Spring mat, though they have been cultivated from other nearby alkaline siliceous springs as well as from higher-temperature streamer communities in Octopus Spring.
Incorrectly reported in reference 121.
This designation is for the observation of a population as a DGGE band in an aerobic chemoorganotrophic enrichment culture (121); it is not the isolate cultivated from aerobic chemoorganotrophic enrichments and listed as ac-8 (86), whose 16S rRNA sequence is identical to that of type N.
Thermoanaerobacter brockii, Thermoanaerobacter ethanolicus, and Thermoanaerobacterium thermosulfurigenes are new names for Thermoanaerobium brockii, Clostridium thermohydrosulfuricum, and Clostridium thermosulfurogenes, respectively (70); Moorella thermoautotrophica is the new name for Clostridium thermoautotrophicum (31).
Table 2, column 3, summarizes what has been learned about the composition of this community through direct retrieval from the mat of 16S rRNA sequences by cloning methods or DGGE analysis of PCR-amplified 16S rRNA gene segments (36, 41). Cloning was based mainly on cDNA synthesis from 16S rRNA templates (163, 164) but to a lesser degree on PCR amplification of 16S rRNA genes (65). Many of the same 16S rRNA sequences were detected by more than one or all of these methods (152). The general relationships of 16S rRNA sequences detected in the mat to major phylogenetic lineages in the domains Bacteria and Archaea are given in Table 2 (column 1). Comparison of columns 2 and 3 in Table 2 reveals that none of the 31 unique 16S rRNA sequences observed is identical to the 16S rRNA sequence of any of the isolates previously cultivated from the mat. In fact, sequences obtained directly from the mat are at best 93% similar (see cautionary note in Table 2, footnote e) to their closest cultivated mat relatives and are also only distantly related to their closest cultivated or uncultivated relatives in the Ribosomal Database Project (75). No sequences were retrieved for many of the phylogenetic groups which are well represented in the mat culture collection (e.g., Thermus, Isosphaera, gram-positive and sulfate-reducing bacteria, and methanogenic archaea), and many sequences were retrieved from phylogenetic groups not represented in the mat culture collection (see below). It is obvious that the molecular approach samples the community in a totally different way than does the cultivation approach.
A note of caution is necessary regarding the possibility that cloned sequences might be chimeric artifacts formed from more than one 16S rRNA sequence during PCR. For instance, we do not include in Table 2 two sequences that we have detected, because we have evidence that they are chimeric Table 2, footnote a). Detection of chimeras is difficult (64, 116). In general, confidence that a sequence is real can be improved by obtaining more information about it. In our case, many of the sequences detected were observed by different methods of analysis (e.g., cloning from rRNA templates, cloning after PCR amplification from rDNA templates, PCR followed by DGGE analysis, oligonucleotide probing, and cultivation).
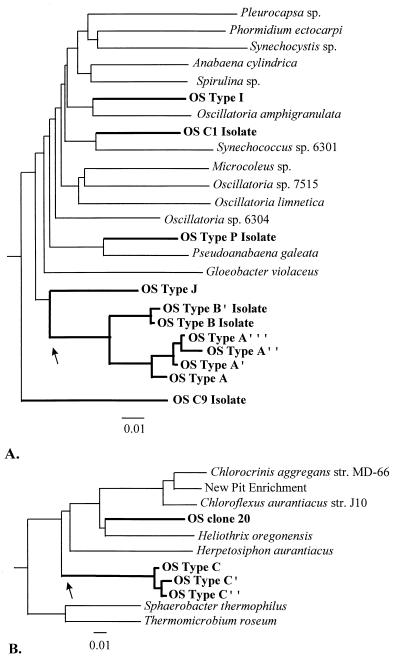
In addition to permitting inference of phylogenetic affiliations, 16S rRNA sequences sometimes permit inference of phenotypic properties for the organisms contributing them. Numerous sequences related to those of cyanobacteria were detected (Table 2 and Fig. 2A) but not the sequence of a Yellowstone cyanobacterial isolate of S. cf. lividus (strain Y-7c-s). It seems logical to infer an oxygenic photosynthetic phenotype for the populations contributing these sequences, since this is a metabolism common to all known members of the cyanobacterial lineage (including chloroplasts and prochlorophytes [53, 168]). In evolutionary terms, it is a shared, derived character defining oxygenic photosynthetic prokaryotes and chloroplasts as a monophyletic lineage. Although these sequences display wide variation in nucleotide composition, some are closely related (e.g., the type A/B cluster, including sequence types A, A′, A", A‴, B, and B′, which exhibit between 0.3 and 5.6% differences in 16S rRNA sequence). Very small sequence differences could be due to polymerase error (112), but again, further study may prove otherwise. For instance, in our earlier cloning work we were unwilling to define distinct 16S rRNA sequence types (e.g., type B) on the basis of a single nucleotide difference we felt might be due to polymerase error; hence, we described sequence type B allowing an ambiguous assignment for one nucleotide position (159, 162, 164). Later, however, DGGE analysis revealed that there were actually two distinct sequence types (types B and B′) (36).
FIG. 2.
Distance matrix phylogenetic trees illustrating cyanobacterial (A) and green nonsulfur bacterium-like (B) 16S rRNA sequences detected directly in the Octopus Spring cyanobacterial mat or in isolates therefrom (thick lines) relative to representative major lines of descent in each lineage (thin lines). We used the OSC1 isolate sequence (identical to that of S. cf. lividus Y-7c-s) and the C. aurantiacus J10 sequence (98.2% similar to that of C. aurantiacus Y-400-fl) because more sequence data were available. The trees were constructed with the programs DNADIST and FITCH from the Phylogenetic Inference Package (PHYLIP), version 3.57c. Trees were inferred from nucleotides which align with Escherichia coli positions 332 to 452, 480 to 507, 712 to 892, and 1140 to 1364 (A) and 256 to 446, 485 to 598, 604 to 830, 856 to 922, 933 to 939, and 954 to 966 (B), except that for sequences shown to the right of arrows, fewer nucleotides were available for analysis. In these cases, smaller trees were inferred from available data and manually added to the appropriate branch. The tree in panel A was rooted by using the 16S rRNA sequences of Thermotoga maritima, Chlorobium vibrioforme, and E. coli, while the tree in panel B was rooted by using sequences from Methanobacterium formicicum, Thermodesulfobacterium commune, Aquifex pyrophilus, T. maritima, C. vibrioforme, Agrobacterium tumefaciens, Pseudomonas testosteroni, and E. coli. Evolutionary distance is indicated by horizontal lines; each bar corresponds to 0.01 fixed point mutations per sequence position.
As in the case of cyanobacteria, numerous sequences phylogenetically like those of green nonsulfur bacteria were detected (Table 2 and Figure 2B) but not that of an Octopus Spring isolate of C. aurantiacus (strain Y-400-fl). For the populations contributing these sequences, the phenotype cannot be confidently inferred since both anoxygenic photosynthetic and aerobic chemoorganotrophic bacteria are found in this lineage (95, 162). Like the cyanobacterial sequences, some green nonsulfur bacterium-like sequences exhibit large differences in nucleotide composition, while others are very closely related (e.g., type C-like sequences exhibit between 1 and 1.4% differences in 16S rRNA sequence) (Fig. 2B).
Sequences of numerous other lineages were detected, as summarized in Table 2 (column 3). The observation of several sequences possibly related to those of green sulfur bacteria is interesting. No cultures have been obtained from this mat, though a thermophilic Chlorobium species has been cultured from mats in New Zealand hot springs (28, 143). The inference of an anoxygenic photosynthetic phenotype for these populations is complicated by the fact that while Chlorobium species appear at present to be monophyletic (43), the related mat sequences appear to fall just outside of the Chlorobium group (94). Perhaps, like some of the Octopus Spring cyanobacterial sequences, they are contributed by deeply branching members of the green sulfur bacterial group, but proof that these sequences are from populations with this physiology awaits their cultivation and characterization.
Also detected were phylogenetic relatives of α- and β-subdivision proteobacteria, spirochetes, and members of paraphyletic groups which contain Leptospirillum or Nitrospina. Cultivated representatives of most of these lineages from the Octopus Spring mat have not been previously reported, though spirochetes have been previously observed microscopically (145) and have been cultivated from Oregon (103) and New Zealand hot springs (104). Some sequences cannot be ascribed to known phylogenetic groups and may represent novel lineages in the domain Bacteria. The Octopus Spring mat community is clearly an ensemble of bacteria with quite different evolutionary histories.
Why Are Culture and Molecular Samplings of Diversity Different?
Competitive exclusion during enrichment.
The incongruity between populations sampled by culture methods and those sampled by molecular methods prompted us to investigate culture methods in greater detail. By diluting inocula to extinction before enrichment (extincting dilution enrichment) we were able to recover cyanobacterial populations morphologically resembling S. cf. lividus, whose 16S rRNA sequences we had detected by cloning and/or DGGE analysis (types B, B′, and P) (40, 102, 155) (Table 2, column 4). This confirmed that common morphologies mask considerable genetic diversity among cyanobacterial mat inhabitants. If inocula were not diluted before enrichment, a single S. cf. lividus genotype (that of strain C1 [Table 2]) was recovered (40). In fact, all available culture collection strains of S. cf. lividus (including Yellowstone strain Y-7c-s) also exhibited this 16S rRNA sequence (40). The organism contributing it seems to be able to competitively exclude other more predominant Synechococcus populations in laboratory culture.
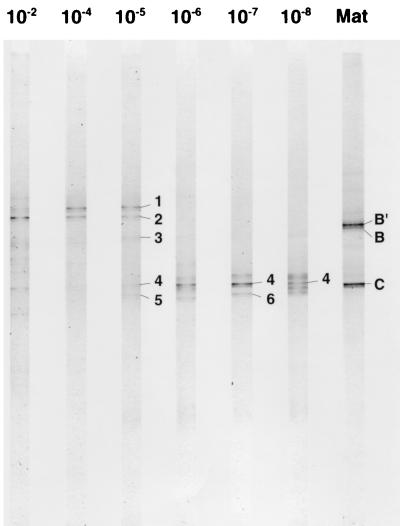
Perhaps the most clear illustration of competitive exclusion was observed by using DGGE to monitor liquid enrichments for aerobic chemoorganotrophic bacteria inhabiting the mat. Figure 3 exhibits the populations detected that grew in enrichments inoculated with increasingly diluted mat samples. Clearly, the populations that were detected in enrichments from low-dilution inocula (bands 1 to 3 in Fig. 3) were those that did not survive extreme dilution. Enrichments from highly diluted inocula selected for different and obviously more abundant organisms. In the example shown in Fig. 3, C. aurantiacus-like populations (bands 4 to 6, corresponding to populations env.OS_ace3 to -5 in Table 2, column 4) were 3 orders of magnitude more abundant than the populations that dominated low-dilution enrichments.
FIG. 3.
DGGE analysis of 16S rRNA gene segments of aerobic chemoorganotrophic populations enriched from 10-fold serially diluted samples of the Octopus Spring cyanobacterial mat and present in the mat itself. Bands labeled with numbers were characterized by purification and sequencing (see reference 121).
As in the case of cyanobacterial enrichments, it was possible to recover by plating from a high-dilution enrichment one aerobic chemoorganotrophic population whose 16S rRNA sequence had been observed by cloning (β-subdivision proteobacterial sequence type N [Table 2]). As mentioned above, the correspondence is usually too poor to permit inference of phenotypic properties of prokaryotes from their phylogenetic relationships. Thus, the recovery of isolates whose 16S rRNA sequences correspond to sequence types detected in natural samples helps link a possible physiology with a population otherwise defined only phylogenetically (Table 2, column 5). However, it is important to question whether the observed physiology reflects the true functional role of the population within the community. For example, while it is clear that the type N population is capable of aerobic chemoorganotrophy, it might also conduct numerous other metabolisms which characterize β-subdivision proteobacteria (e.g., anoxygenic photosynthesis, ammonia oxidation, denitrification, sulfide oxidation, iron oxidation, methanol oxidation) which we did not investigate. C. aurantiacus, which can grow as an aerobic chemoorganotroph but is also phototrophic, provides another good example of this problem. This raises an important general point about inferring functional roles from culture observations—the metabolisms observed in culture reflect only those which it has occurred to us to measure, they may constitute only a fraction of an organism’s metabolic potential, and they may not reflect in situ function at all.
Trophic structure.
S. cf. lividus populations whose sequences were frequently observed by cloning and DGGE analysis were cultivated from mat samples that had been diluted to near the limit of extinction of Synechococcus cells enumerated by direct microscopic count (i.e., diluted mat inocula contained only a few Synechococcus cells). This suggests that molecular methods may favor the detection of at least some of the most predominant populations within the community. Thus, the lack of congruence between culture and molecular samplings could reflect the rarity of some (many?) cultivated populations, which might be at low relative population densities. Numerical abundance estimates of some populations, derived from data on extincting dilution enrichment and isolation, are presented in Table 2 (column 6). Note, for instance, that S. cf. lividus populations, whose 16S rRNA sequences are frequently observed by cloning or DGGE analysis (e.g., types B and B′), appear to comprise a higher proportion of the direct S. cf. lividus microscopic count (12.5%) than do populations whose 16S rRNA sequences are rarely detected (e.g., type P, 4% of direct count) or have not yet been detected (e.g., strain C9, 0.4% of direct count). All of the aerobic chemoorganotrophic populations that we have enriched or isolated, including the C. aurantiacus-like populations mentioned above, exhibit estimated densities at or, more typically, far below 3.3% of the S. cf. lividus direct count. With two exceptions, their 16S rRNA sequences have never been detected. The exceptions include two populations whose sequence types have been detected in the mat and also in low-dilution enrichments (types N and L). Perhaps their 16S rRNA sequences are preferentially selected because of primer bias or unusual operon frequency. Most previously cultivated mat inhabitants (Table 2, column 2) were obtained in enrichment cultures from undiluted samples (i.e., without regard for their abundance or for the possibility that competitive exclusion might favor numerically inferior populations). The fact that their 16S rRNA sequences have not yet been detected is consistent with the possibility that they constitute low-density or possibly even zymogenous populations (i.e., those which persist but are inactive under present environmental conditions).
Mat trophic structure is also suggested by the relative abundances of pigment and lipid biomarkers (9, 32, 126, 127, 150, 154, 176, 177). The data suggest that cyanobacteria and green nonsulfur bacteria predominate over aerobic and anaerobic chemoorganotrophic bacteria, which in turn predominate over terminal members of anaerobic food chains, such as methanogenic archaea and sulfate-reducing bacteria (156). Thus, it is not surprising that primarily cyanobacterial and green nonsulfur bacterium-like 16S rRNA sequences have been recovered.
The hypothesis that trophic structure limits recovery of rare species can be tested by increased sampling. Ecologists can predict the total diversity within a system from the rate at which new populations are detected with sampling extent (rarefaction analysis) (140). We have resisted efforts to make such predictions, as the estimates depend on unbiased sampling, which cannot presently be assumed for either molecular or culture methods.
Different resources and conditions in situ than in culture.
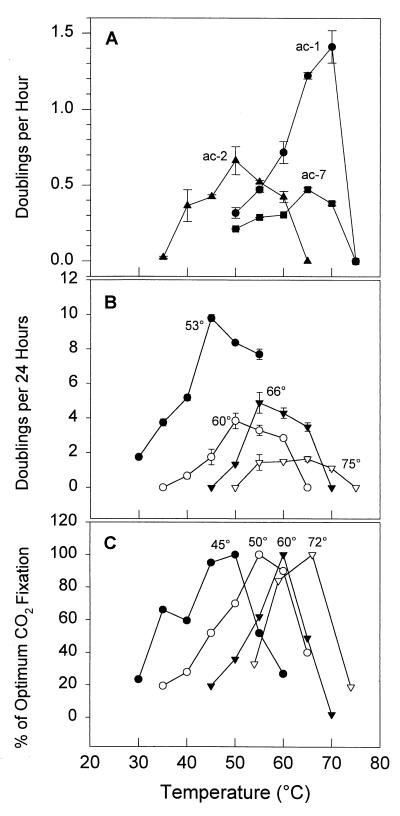
In addition to inoculum dilution, the outcome of enrichments for aerobic chemoorganotrophs was sensitive to substrate type, substrate concentration, and incubation temperature (121). A nice example of this was found with Thermus strains which co-occur in the ca. 50°C mat but exhibit different temperature optima (Fig. 4A). A high-temperature-adapted strain (ac-1) was selected when the enrichment was incubated at 70°C, even though strains adapted to lower temperatures were 2 (ac-7) to 4 (ac-2) orders of magnitude more abundant (87, 121). The most predominant low-temperature-adapted population was selected only when the incubation temperature matched the habitat temperature and the inoculum was diluted to extinction to prevent competitive exclusion. That is, the most abundant Thermus population was the one best suited for the environment in which it was found. Although enrichment at high temperature selected for the most fit population for the enrichment condition (i.e., the high-temperature-adapted strain), this was the least abundant and the least fit strain for the natural environment sampled. The types of aerobic chemoorganotrophic populations detected in liquid enrichments (121) differed from those recovered on solidified media of the same composition inoculated with such enrichments (86, 87). This led us to suspect that plating may also bias against recovery of some populations.
FIG. 4.
Evidence of the existence of temperature-adapted populations within hot spring cyanobacterial mats. (A) Effect of temperature on growth rate of Thermus strains ac-1, ac-2, and ac-7 isolated from a ca. 50°C Octopus Spring mat sample. Bars represent standard errors (modified from reference 87). (B) Effect of temperature on growth rate of S. cf. lividus strains isolated from the Hunter’s Springs, Oreg., mat. Bars represent ranges of replicate experiments (modified from reference 105). (C) Effect of temperature on bacterial photoautotrophy in samples of the Twin Butte Vista (Lower Geyser Basin, Yellowstone National Park) mat collected at various temperature-defined sites, as shown (redrawn from reference 9).
Incongruity between plate counts and direct microscopic counts (see reference 1) or between populations sampled by culture or molecular methods, has led some investigators to the opinion that many (most?) of the bacteria in natural environments are “uncultivatable.” We think the more likely explanation is our inability to understand and reproduce the real microenvironmental niches defined by physiochemical and also biotic features (e.g., in symbiotic relationships) that influence naturally occurring bacteria. Hence, we prefer to use the term “uncultivated,” which conveys the more optimistic view that microbiologists will eventually be able to more accurately reproduce the features of natural microenvironments in the laboratory. Our success in recovering numerically predominant Synechococcus populations provides an example that better correspondence between populations detected by culture and by molecular methods can be achieved.
Gene Diversity or Species Diversity?
In macroecological terms, the information in Table 2 might be considered a species list were it not for uncertainties regarding the interpretation of individual 16S rRNA sequences as individual microbial populations and/or species. This uncertainty arises because some microorganisms are known to possess more than one rRNA operon with more than one unique 16S rRNA sequence (57, 78, 85, 90, 107). Two types of observations suggest that in the Octopus Spring mat system the unique 16S rRNA sequences we have detected correspond to unique organismal populations. First, analysis of all S. cf. lividus isolates (types B, B′, P, C9, and C1) suggested that each possesses only a single 16S rRNA sequence gene (40, 102). Second, multiple operons within a single microbial population should co-occur in distribution analyses based on amplification of genes. Thus, by noting that the natural distributions of these gene sequences are unique (see next section) we gain further confidence that each represents a unique microbial population (36, 121).
Whether the unique 16S rRNA-defined microbial populations in the Octopus Spring mat constitute unique species can be debated. The biological species concept, which is based on sexual isolation (Table 1, Futuyma definition of “species” [48]), does not readily apply to bacteria, which are mainly asexual. Bacteriologists have attempted to define “species” in terms of the degree of genetic difference (Table 1, Wayne et al. definition of species [161]). Applying this definition to the data of Fig. 2A, the six major cyanobacterial 16S rRNA lineages detected by cloning, DGGE, and cultivation (A/B cluster, type I, type J, type P, C9, and C1) would quite clearly be interpreted as being representative of cyanobacteria belonging to different “species,” in the sense that the 16S rRNA sequences defining these lineages exhibit much less than 97% similarity, corresponding to less than 70% similarity in DNA-DNA hybridization (131). Considering that the 16S rRNA sequence differences among the cyanobacterial populations in the mat rival those of all cultivated cyanobacteria so far investigated (Fig. 2A), the six divergent lineages of mat cyanobacteria probably differ at a considerably higher taxonomic level (family? order? phylum?). However, the A/B cyanobacterial cluster (Fig. 2A) and the type C green nonsulfur bacterium-like cluster (Fig. 2B) contain 16S rRNA sequences that differ by less than 3%. Assuming that such high 16S rRNA sequence similarity correlates with >70% DNA-DNA hybridization (not always true [131]), populations contributing these sequences would be considered by this quantitative definition to be within the same “species.” However, this approach to defining species seems very arbitrary (34, 76, 135). It contrasts sharply with the situation among macroorganismal species, which often exhibit much more than 70% similarity in DNA-DNA hybridization (and >97% rRNA similarity). For example, hybridization values of human DNA with chimpanzee, gorilla, and orangutan DNA are 98.4, 97.7, and 96.5% (rRNA similarities of 99.4, 99.2, and 98.1%), respectively (54, 128, 129). Note that these are differences among species belonging to different genera, not among species of a single genus.
The major problem with the biological species concept as applied to prokaryotes is the concept of reproductive isolation. Evolutionary biologists who have worked with animals and plants suggest that the origin of new species is driven by ecological forces, such as adaptation to environmental parameters and geographic isolation of gene pools accompanied by genetic drift (48, 117). Reproductive isolation itself may even be considered a reinforcing mechanism to prevent ecologically distinct sexual populations (i.e., new species) from undergoing frequent genetic exchange (Table 1, Mayr definition of species [77]), a force which would erode and perhaps eliminate the adaptive value gained by the ecologically based genetic divergence from an ancestral population. Macroorganismal species can be subdivided into distinct ecotypes (Table 1) under the biological species concept, since ecologically specialized populations can still interbreed. However, as pointed out by Mayr (77), asexual species may be defined by such ecological isolation (i.e., ecotypes are species). Simpson (130) preferred to define all species in ecological and evolutionary terms (Table 1). Some microbiologists and virologists are already comfortable with this more natural and more universal concept of species (26, 142, 146). Defining species in this way clearly requires that the populations in question be understood in terms of their ecology. In the following section we present evidence that suggests that even very closely related 16S rRNA sequences, such as those of the A/B cyanobacterial cluster and the type C-like green nonsulfur bacterium-like cluster, have unique ecological distributions and thus can be considered unique species.
Spatial Distribution of Populations within Individual Mats
A distinct advantage of hot spring microbial mat habitats is that the communities exist along well-defined environmental gradients against which population distributions can be measured. We first approached the study of population distribution by using oligonucleotide hybridization probes complementing specific regions of (i) cyanobacterial and green nonsulfur bacterium-like 16S rRNA sequences previously detected by cloning and (ii) S. cf. lividus Y-7c-s and C. aurantiacus Y-400-fl 16S rRNA sequences (118). However, DGGE analysis has allowed us to extend this work with greater ability to resolve individual populations (both in terms of detecting closely related sequences and distinguishing population from interoperon differences). DGGE also permits a more synecologic approach as it allows simultaneous evaluation of which populations are present, as opposed to an autecologic investigation of the presence of specifically targeted populations. Since DGGE depends on PCR, quantitative and qualitative biases are possible, but confidence in inferences is improved when similar observations are made by using more than one approach (152) (see below).
Temperature distribution.
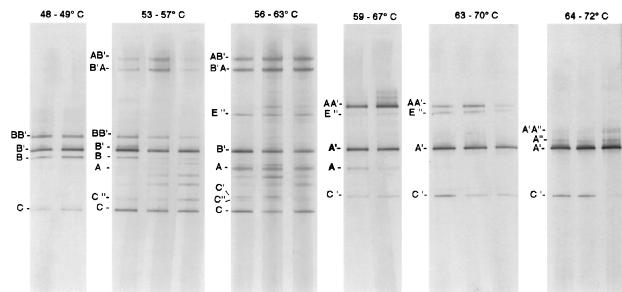
DGGE surveys have revealed that 16S rRNA gene distributions change along the thermal gradient which exists over many meters where the mat occurs in the Octopus Spring effluent channel (Fig. 1A and 5). The DGGE pattern cannot be taken directly as a profile of community composition because of the occurrence of heteroduplex bands (labeled with two letters in Fig. 5) formed as artifacts from reannealing of closely related single strands (i.e., there are more bands than actual populations) (41). For example, heteroduplex bands form from type A/B-like cyanobacterial sequences but not between more distantly related sequences. The presence of heteroduplex bands indicates that these PCR amplifications were inefficient, as template-template interactions are likely to have interfered with template-primer interactions (139). Thus, the correspondence between initial template concentrations and PCR products was not completely linear (41). We have also found it necessary to sequence bands to establish identity because bands of different sequence may migrate to similar or identical positions in the gradient (37, 46, 83).
FIG. 5.
Distribution of 16S rRNA gene segments of populations detected by DGGE in cyanobacterial mat samples collected from temperature-defined sites along the thermal gradient in the Octopus Spring effluent channel on 13 March 1995. Lanes within a single temperature interval are true replicate mat samples. Bands labeled with letters were characterized by purification and sequencing. Single letters denote homoduplex molecules from real populations, while double letters denote heteroduplex artifacts formed from the populations indicated by the two letters. Primes indicate sequences closely related to those of the same letter without primes (reprinted from reference 41).
Despite these limitations, DGGE provided a rapid means of detecting the 16S rRNA sequences of many mat inhabitants along the well-defined temperature gradient (41). The cyanobacterial sequences detected by DGGE appear to be from dominant populations, since the same sequences were recovered from Synechococcus isolates obtained from highly diluted inocula (see above). Five closely related yet distinct cyanobacterial 16S rRNA sequence types, A", A′, A, B′, and B, occur from high to low temperature along the Octopus Spring effluent channel, respectively. It is important to note that these are gene distributions. Hence, the lack of consistent co-occurrence of any two genes suggests that different organismal populations, as opposed to different operons within one organismal population, contribute each gene sequence. While it could be argued that these distributions arise as artifacts of skewed PCR amplification (139), our inference that each 16S rRNA sequence represents a unique organismal population is strengthened by the cultivation of Synechococcus isolates with distinct 16S rRNA sequences. Furthermore, probe studies, though less capable of resolving individual sequence type, confirmed the temperature distribution trends observed by DGGE analysis. Probe studies also suggested that these populations may be adapted to the temperatures at which they are found (118). Hence, the five populations may be interpreted as distinct ecotypes, and thus as species, according to the definitions of Mayr and Simpson (Table 1) (77, 130). This is particularly interesting considering the very high genetic similarities among the 16S rRNA sequences of these populations, in some cases >99% (Fig. 2A). Given their conserved nature, we must consider that each distinct 16S rRNA sequence might even represent more than one species (45, 101). Palys et al. (101) have recently demonstrated that ecologically distinct bacterial isolates with identical or nearly identical 16S rRNA sequences can be discerned through clustering of protein-encoding gene sequences, which evolve more rapidly. Since we are working with uncultivated populations defined by 16S rRNA sequences, we cannot use protein-encoding gene sequences and must use a more rapidly evolving genetic marker which is linked to the 16S rRNA gene to address this possibility. We have begun to discover genetic variants within specific Octopus Spring mat cyanobacterial 16S rRNA-defined populations, based on sequence variations in the intervening transcribed spacer (ITS) region separating the 16S and 23S rRNA genes (102). Pure cultures do not exhibit such variation, suggesting that natural ITS variants do not appear to be due to interoperon differences and may correspond to additional ecologically distinct cyanobacterial species with identical 16S rRNA sequences.
The cyanobacterial temperature distribution pattern we detected fits very nicely with that predicted by Castenholz (23), based on the cultivation of temperature-adapted strains of S. cf. lividus from an Oregon hot spring cyanobacterial mat (105) (Fig. 4B). These cultures were obviously genetically distinct Synechococcus populations, but the degree of genetic difference was not measured. The one surviving strain has a 16S rRNA sequence identical to that of other readily cultivated S. cf. lividus culture collection strains (e.g., strains C1 and Y-7c-s). Also, the numerical relevance of the isolates to the community was unknown, as they were obtained from low-dilution enrichments. Nevertheless, our results and those of Peary and Castenholz (105) suggest the existence of cyanobacterial populations adapted to several narrow temperature-defined niches along the ca. 30°C temperature range over which the mat occurs. The evolutionary and ecological patterns lead us to hypothesize that these cyanobacterial species formed through adaptive radiation from a recent common ancestor. We use the term adaptive radiation to describe the evolutionary divergence (via natural selection) of members of a single phylogenetic line into a variety of ecologically distinct populations (48, 117). The classic macrobiology analog is the radiation of Darwin’s Galapagos finches, which apparently evolved to exploit different food resources. Since our observations are from a single spring, we cannot say whether cyanobacterial evolutionary radiation occurred, as the finch radiation apparently did, because of niche partitioning in geographic isolation (e.g., within a single spring, geothermal basin, or region [see below]) without studies of broad-scale patterns of distribution.
Clusters of closely related 16S rRNA sequences have been commonly observed in molecular surveys of microbial diversity in numerous habitats (13–16, 30, 47, 50, 56, 66, 71, 82, 91, 100, 123, 132, 141). We wonder if adaptive radiations (i.e., evolution of ecologically specialized populations) are common in the microbial world. In this regard, it is interesting that closely related marine Prochlorococcus populations appear to exhibit different depth distributions (39) and that Prochlorococcus isolates with >97.3% similarity in 16S rRNA sequences exhibit unique light adaptations (81). It is also interesting to note that closely related marine proteobacteria appear to exhibit vertical stratification, suggesting that they may also be ecotypes (i.e., species) (42).
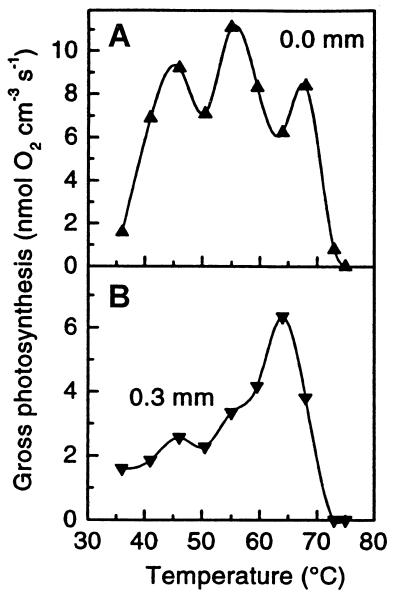
The existence of multiple cyanobacterial species fits the guild concept (Table 1). It is logical to predict that intraguild biodiversity has value to community structure, as it may help to stabilize guild processes. For example, in Octopus Spring, water flow surges cause temperatures to fluctuate as much as 10 to 15°C every 5 min (36). Photosynthesis, if conducted by a single temperature-adapted population, would be optimal only at a particular temperature. However, when shared among several populations adapted to different temperatures, the rate of photosynthesis might be more resistant to temperature fluctuation. Brock (17) pointed out many years ago that the optimal rate of photosynthesis in mat samples correlates with the temperature at the site of collection. Kühl et al. (68) have recently used microelectrodes to demonstrate that single samples of the Octopus Spring mat exhibit multiple temperature optima for oxygenic photosynthesis, consistent with the measured intraguild species diversity pattern (Fig. 6A). Except for small dips between optima, photosynthetic rates are relatively stable over a broad temperature range. This exemplifies how an emergent property of community structure can be a consequence of the evolution of biodiversity. It also helps us to appreciate the consequences of species extinction, since removal of one of the temperature-adapted cyanobacterial species would affect the temperature sensitivity of the guild process, oxygenic photosynthesis.
FIG. 6.
Rates of oxygenic photosynthesis determined by microelectrode analysis in a single ca. 60°C Octopus Spring mat sample incubated at different temperatures. (A) Upper mat surface. (B) 0.3 mm below the surface (modified from reference 68).
As with cyanobacteria, we found that the closely related green nonsulfur bacterium-like populations C, C′, and C" (Fig. 2B) are distributed at different temperatures (Fig. 5); following the reasoning developed above, we consider these to be distinct species (or possibly sets of species with identical 16S rRNA sequences). The similarity of the evolutionary patterns of the C-like and the A/B-like populations (Fig. 2) suggests the possibility that coevolutionary processes may have helped shape the development of the community. As mentioned above, we cannot confidently infer that the C-like populations conduct anoxygenic photosynthesis. Nevertheless, it is interesting that Bauld and Brock (9) demonstrated that anoxygenic photosynthetic activity in such mats was also optimized to habitat temperatures, suggesting the existence of temperature-adapted populations in this guild as well (Fig. 4C). Transfer of photoautotrophically fixed carbon from cyanobacteria to filamentous photoheterotrophs, such as Chloroflexus (2, 7, 10, 119, 157) is known to occur either via photoexcretion of glycolate (7) in the superoxic illuminated mat (113) or via fermentation of polyglucose (mainly to acetate) by cyanobacteria in a dark anoxic mat, and it may involve transfer of the majority of photoautotrophically fixed carbon from cyanobacteria to photoheterotrophs (88). It is interesting to speculate that a physiological linkage such as this (i.e., temperature-adapted cyanobacterial species cross-feeding temperature-adapted photoheterotrophic species) might explain coevolution between such phylogenetically disparate species.
Vertical distribution.
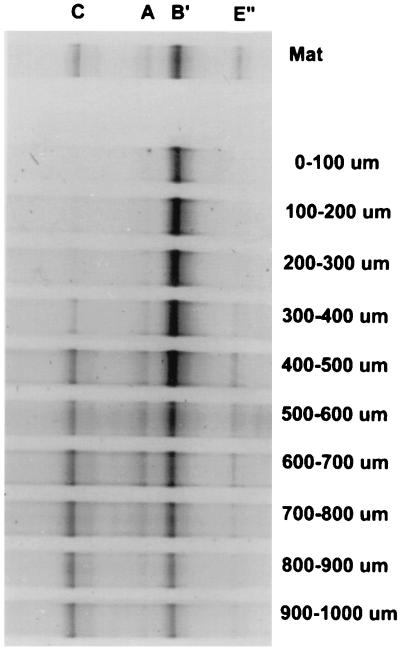
Microscopic analysis of vertical thin sections revealed that cyanobacterial populations in these mats are stratified within the upper approximately 1-mm-thick photic zone (Fig. 1D), consistent with the restriction of oxygenic photosynthesis to this zone, as measured by microelectrodes (113), and with the distribution of chlorophyll a (9) and cyanobacterial lipid biomarkers (177). By using a cryotome to section this interval into 100-μm-thick layers, we were able to observe vertical stratification of different populations detected by DGGE analysis (Fig. 7). The observations were made on 60°C Mushroom Spring mat samples because of hail damage to the Octopus Spring mat (see below). The populations detected, however, were the same as those found at comparable temperatures in the Octopus Spring mat (compare Fig. 5 and 7). The type B′ cyanobacterial population was most readily detected in the 0- to 500-μm interval, whereas the type A cyanobacterial population was detected only at depths corresponding to a well-defined layer of Synechococcus cells maximizing at ca. 600 μm below the mat surface (Fig. 1D). Several additional observations also suggest vertical stratification of cyanobacterial populations. Multiple maxima of oxygenic photosynthesis were not detected in microelectrode measurements in the vertical aspect of the undisturbed mat (113), possibly due to a minor contribution of deeper populations that receive less light. However, a bimodal vertical pattern of oxygenic photosynthesis was observed during recovery of the mat after disturbance when light penetration was greater (37), consistent with the existence of differentially adapted populations in the vertical orientation. The unique below-surface temperature optimum for photosynthesis also suggests that unique microbial populations are found below the mat surface (Fig. 6B). Synechococcus cells found deeper in the green layer exhibit higher autofluorescence per cell, possibly indicating higher chlorophyll a content, as would be expected of low-light-adapted cyanobacteria (Fig. 1D). It is interesting that only one 16S rRNA-defined cyanobacterial population (type B′) was detected at the Mushroom Spring mat surface by DGGE (Fig. 7) whereas three temperature optima for oxygenic photosynthesis were detected at the Octopus Spring mat surface by microelectrodes (Fig. 6A). Assuming that the two mats are similar, and that all cyanobacterial populations were detected by DGGE, the data suggest that temperature-adapted populations may have identical 16S rRNA sequences, perhaps corresponding to type B′ ITS variants.
FIG. 7.
Distribution of 16S rRNA gene segments of populations detected by DGGE through the upper 1-mm vertical interval of the 61°C Mushroom Spring cyanobacterial mat. Each lane represents DGGE analysis of a separate 100-μm-thick cryotome section. Bands are labeled as described in the legend to Fig. 5 (111).
The green sulfur bacterium-like population E" was detected at depths corresponding to the deeper cyanobacterial population. The green nonsulfur bacterium-like type C population was detected near the bottom of the green layer and probably also resides deeper in the orange mat underlayers. While type C-like populations cannot be assumed to be anoxygenic phototrophs (see above), it is interesting that their distribution correlates with the deeper distributions of bacteriochlorophyll (9) and lipid biomarkers (177) typical of C. aurantiacus. Recent results indicating an abundance maximum of green sulfur-like bacteria near the deep chlorophyll maximum, and of green nonsulfur-like bacteria at greater depths over the upper 250 m of marine water columns (51, 55), make us wonder if this is a typical pattern in aquatic habitats.
Many parameters vary across the upper few millimeters of these mats (e.g., light intensity and quality [151], ultraviolet light intensity [29, 49, 80], and nutritional resources supplied by diffusion from above or by recycling within the mat [e.g., fixed nitrogen, phosphate, carbon dioxide production in deeper mat layers, as inferred from pH profiles {113}]). Thus, we cannot yet identify the factors to which vertically stratified populations may have specialized. Differential population changes in response to light modification (118), distinct light-directed motility behaviors in different Synechococcus isolates (110), and high per-cell autofluorescence (see above) suggest that light adaptation is likely. These results indicate that the evolutionary radiation among closely related cyanobacterial populations of the type A/B cluster previously discussed resulted in species that are adapted to more parameters than just temperature. As we examine vertical distributions as a function of temperature and learn more about the associated adaptations, it may eventually be possible to correlate evolutionary patterns with specific ecological adaptations.
Temporal Distribution of Populations within the Octopus Spring Mat
One seeming advantage of the hot spring habitat is that many of the parameters subject to seasonal change in more temperate systems may not vary appreciably. Nevertheless, some parameters do vary predictably over diel, several-day, and seasonal time intervals, and stochastic disturbances can also be expected.
Light intensity is the most obvious predictably varying environmental parameter in this system. The observation that the mat becomes a darker green color within hours of experimental shading (18, 20, 21, 74) suggests either that mat cyanobacterial populations can physiologically acclimate to changes in light intensity or that motile, low-light-adapted (i.e., chlorophyll-rich) cyanobacterial populations might migrate upward, seeking optimal light intensity. Slow gliding motility in S. cf lividus isolates was recently quantified by image analysis techniques (110). Different S. cf. lividus isolates exhibited distinct motility behaviors relative to light intensity. Diel shifts in the pattern of vertical distribution, which might be anticipated for motile species, were not apparent in DGGE analysis (111), but it is not clear that DGGE could have detected the repositioning of a subdominant population. However, changes in cell orientation during a diel cycle were detected in this study.
Longer-term changes in light intensity, driven perhaps by a several-day period of extensive cloud cover, have the potential to cause physiological acclimation (74) or shifts in cyanobacterial populations (118). For instance, rRNA of the type J cyanobacterial population declined while that of the type B-like Synechococcus population remained stable during week-long reductions of light intensity of 93 and 100%. Sheridan (124) observed sun- and shade-adapted populations of the moderately thermophilic cyanobacterium Plectonema notatum, which he suggested might alternately predominate in the surface of temperate mats in summer and winter (125). The pattern of Synechococcus populations in the Octopus Spring mat revealed by DGGE analysis (Fiq. 5) was stable in all seasons despite an estimated 75% reduction of light intensity in winter compared to summer (41). One interesting, albeit speculative, explanation for why shade-adapted species might not dominate mats, even in low-light seasons, has to do with evidence that mat Synechococcus populations may be in an active but slowly growing or even nongrowing state (88). Perhaps, like trees in a climax forest community, they capture space and dominate biomass even when conditions are not optimal.
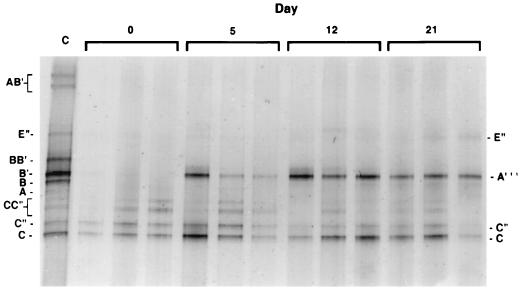
Macroecologists are well aware of the role stochastic disturbance plays in shaping community structure (e.g., the 1988 fires in Yellowstone National Park). Disturbances such as hailstorms or severe thunderstorms have been observed to disrupt the integrity of hot spring mat communities, which then become rapidly recolonized by S. cf. lividus-shaped cells (21). Recolonization by S. cf. lividus-shaped cells above experimental disturbances, such as silicon carbide layers spread atop the mat, has also been studied. However, microscopic analysis does not indicate which specific populations recolonize (33). We used DGGE to examine this in 55 to 62°C and 58 to 62°C sites within the Octopus Spring mat that were disrupted by the removal of the top green cyanobacterial layer (Fig. 8) (37). Scraping caused the removal of cyanobacterial (B, B′, and A) and green sulfur bacterium-like populations (E") but not the type C-like populations which reside deeper in such mats (Fig. 7). Interestingly, at the site shown in Fig. 8, a new cyanobacterial population (A‴) colonized as the upper layer of mat recovered, displacing the type B, B′, and A cyanobacterial populations which remained in adjacent undisturbed control sites. These observations suggest the possibility that some thermophilic cyanobacteria may be adapted for rapid growth after disturbance. This fits with the macroecological concept of r-selected species, which are effective at dispersal and adapted for rapid colonization, as opposed to k-selected species, which are better adapted for the highly competitive environment of climax communities (11). The results indicate that, as in macroorganismal communities, disturbance might have a stochastic effect on microbial community composition and structure and can be a force that increases species diversity within the community.
FIG. 8.
16S rRNA gene segments of populations detected by DGGE which recolonized the upper few millimeters of the Octopus Spring 55 to 62°C mat after removal of the upper green cyanobacterial layer. Triplicate mat samples were analyzed at each time point. Bands are labeled as described in the legend to Fig. 5. (reprinted from reference 37).
Distribution of Populations among Mats of Different Hot Springs
We have conducted a limited survey of mats in Yellowstone springs by using oligonucleotide hybridization probes specific for the 16S rRNAs of cultivated and uncultivated cyanobacterial and green nonsulfur bacterium-like populations described above (118). In later studies, we learned that these probes would not have allowed us to differentiate all closely related populations revealed by DGGE (36). These probes were used to demonstrate that type A-like and B-like cyanobacterial populations could be detected in springs up to ca. 100 km from Octopus Spring, as long as the temperature and pH in the springs were appropriate. Such evidence could be taken to suggest that at least some thermophiles are not geographically isolated among the springs of Yellowstone Park, though it should be kept in mind that the hybridization probes we used would not have distinguished sequences with slight differences within or outside the binding site. Type C-like and type J populations were detected in springs ranging from ca. 100 to 1000 m from Octopus Spring but not in more distant springs. Such evidence might indicate differences in dispersal among different populations. Clearly, there is a need for more robust experimentation on the issue of biodiversity across geographic gradients.
Springs with major chemical differences, such as those with lower pH values, exhibited different patterns of cyanobacterial species occurrence. For example, a pH 6.0 pool in the Clearwater Springs area was the only spring in which the S. cf. lividus Y-7c-s strain 16S rRNA sequence was detected. This is interesting, since the strain was almost certainly originally cultivated from this spring (61, 62). As mentioned above, cultures of S. cf. lividus with this 16S rRNA type have been recovered from Octopus Spring, and also Hunter’s Springs, Oreg., mats (both pH 8.0 to 8.4), suggesting that its geographical range may be broad. However, no evidence, such as the probe evidence for the pH 6.0 pool at Clearwater Springs or recovery of isolates from highly diluted inocula, exists to suggest that it is numerically abundant in these more alkaline springs. The same S. cf. lividus 16S rRNA sequence was also detected by probing RNA from mat samples collected in a more alkaline effluent in the Clearwater Springs group (pH 7.8) but only after it was incubated in the pH 6.0 spring for a week, suggesting that this population might be particularly competitive at lower pH even though it appears to be adapted to higher pH (61, 62). A yet-more-acidic pool in the Clearwater Springs group (pH 5.0) containing Synechococcus-shaped cells did not exhibit any probe responses. These results suggest the possible existence of Synechococcus species which are adapted to even lower pH. We presume that adaptation to pH would involve relatively large pH differences, since the pH within individual mats varies up to several pH units as light intensity changes alter photosynthetic CO2 consumption (113). Other major chemical differences among springs, such as sulfide concentration, might also select for different species, but this has not yet been investigated.
CONCLUSIONS AND FUTURE DIRECTIONS
The application of culture-independent 16S rRNA methods to the study of relatively simple and stable model microbial communities, in combination with an evolutionary ecology view (borrowed from theories developed to explain plant and animal ecology and evolution), forces us to appreciate the natural linkages among microbial biodiversity, ecology, and evolution. We are detecting at least some of the predominant species that have been masked by common morphology and have previously evaded cultivation. By better understanding the limitations of culture methods, we have learned how to lure some of these into pure culture, enabling us to combine culture and molecular approaches to more fully understand the ecology of the community. By studying the distributions of the 16S rRNA gene sequences we have detected, we are able to go beyond the detection of gene diversity to begin to understand how the gene sequences reflect the contributions of ecologically distinct community members. Such information helps us to appreciate that there may be a common ecological way to conceptualize all species.
Ecological patterns allow us to hypothesize that bacterial diversity, like diversity among plants and animals, is likely to have arisen, at least in part, from evolutionary radiations that resulted in ecologically distinct species. As in a forest community, different species in this “microbial forest” are distributed along environmental gradients (e.g., temperature rather than moisture and/or elevation) and, at given locations, shade-adapted species may live beneath canopy species. We hope that by mapping species distributions relative to environmental parameters on a microenvironmental scale, we will gain further insight into how to selectively cultivate the various species we have detected. With pure cultures in hand it will be possible to study adaptations and to test the hypothesis. As in forests, disturbance can create gaps, providing windows of opportunity for colonist species. A better appreciation for temporal as well as spatial variation in the environment of microorganisms will undoubtedly help us explain more of the diversity we detect.
By combining ecological and evolutionary patterns, we observe that ecological specialization can occur among closely related bacterial populations. This might offer an explanation for the widespread occurrence of clusters of highly related 16S rRNA sequences in all habitats so far examined. Our results also suggest that 16S rRNA sequences may be too conserved to detect all ecologically specialized populations and thus may underestimate microbial species diversity.
Geographic isolation is thought to be one of the most important forces driving speciation among plants and animals (48, 117). Many microbiologists assume that “everything is everywhere, but the milieu selects” (an often-cited quote from Baas Becking [3], who attributes the first half of the idea to Beijerinck). We hypothesize that geographic barriers to dispersal may exist for some microorganisms. Hot spring habitats constitute an extreme situation in which to observe the role of geographic isolation in prokaryote speciation, as geothermal “islands” are separated by a “sea” of low-temperature habitat through which high-temperature adapted microbial species must distribute. In this regard, it is interesting that Kristjansson et al. (67) have demonstrated differences in multilocus enzyme electrophoresis patterns among Thermus isolates from different Icelandic hot springs, indicating that geographic isolation and genetic drift are at play, even on a relatively local scale, for these thermophiles. Evaluation of the patterns of occurrence of predominant native thermophilic microbial species in geographically isolated hot springs, by the same methods described above for detailed analysis of individual model mats, should provide answers to interesting questions relating to microbial biogeography. Do geographic anomalies (e.g., the absence of Synechococcus in suitable habitats in Iceland and the absence of high-temperature forms of Synechococcus in New Zealand, Italy, and Japan [24, 27, 102]) indicate that limitations to dispersal exist? Are Synechococcus species of the type A/B cluster cosmopolitan, with low-temperature-adapted forms (e.g., B types) better at dispersal than high-temperature-adapted forms (e.g., A types)? Or did independent adaptive radiations occur in different geographic regions, in some cases, too recently for high-temperature-adapted forms to have evolved? Does the cultivation of S. cf. lividus strains of the same 16S rRNA genotype from hot springs in Yellowstone and Oregon imply that this species is easily dispersed and invasive? Or might it be zymogenous, a part of the “seed rain,” “watchful[ly]-waiting” (as Winogradsky [170] quoted Cohn) for better environmental conditions to develop? Another alternative is that 16S rRNA sequences may be too conserved for detection of geographically unique species.
As stated by Begon et al. (11) the objectives of ecology are “explanation, description, prediction and control.” Microbial ecologists are in a poor position to achieve these objectives when they are bound solely by the limitations of microscopic and culture methods. However, molecular methods are attractive new tools in the kit of the microbial ecologist, especially when used in concert with more traditional methods. They permit detection of the specific microorganisms that are important to microbial communities, recognition of their evolutionary history, and determination of how they are organized to accomplish community function. By adopting the view of macroecologists and macroevolutionary biologists as they interpret their findings, microbial ecologists can see if unifying principles control the diversity, ecology, and evolution of all organisms, large and small. Prokaryotes define two of life’s three primary evolutionary lineages; we know few of them, and yet their activities have a major influence on all ecosystems. Thus, we should be able to gain a more predictive knowledge of the intricacies of natural microbial communities upon which all life on Earth depends. Since the microorganisms associated with applied problems (including disease) must also obey the evolutionary and ecological rules which determine their existence, distribution, and fitness, we should also be able to gain a more predictive knowledge of how to control and exploit them to prevent or solve environmental problems. We should also learn how to better tap the vast biotechnological potential yet to be discovered among the multitude of uncultivated microorganisms.
ACKNOWLEDGMENTS
We appreciate the support provided by the U.S. National Park Service and in particular the personnel of Yellowstone National Park, who have permitted and facilitated our research there. We also appreciate the long-term support provided by the National Science Foundation Ecology Program (currently BSR-9708136) and the National Aeronautics and Space Administration Exobiology Program (currently NAGW5-3652) and, more recently, gifts from Stratagene Corporation that have funded our research.
We thank Jim Staley, Mike Madigan, Fred Cohan, Ed DeLong, and Anna Louise Reysenbach for thoughtful reviews of the manuscript and discussion of the topics therein.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson K L, Tayne T A, Ward D M. Formation and fate of fermentation products in hot spring cyanobacterial mats. Appl Environ Microbiol. 1987;53:2343–2352. doi: 10.1128/aem.53.10.2343-2352.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baas Becking L G M. Geobiologie of inleiding tot de milieukunde. The Hague, The Netherlands: W. P. van Stockum and Zoon N.V.; 1934. [Google Scholar]
- 4.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bateson M M, Thibault K J, Ward D M. Comparative analysis of 16S ribosomal RNA sequences of Thermus species. Syst Appl Microbiol. 1990;13:8–13. [Google Scholar]
- 7.Bateson M M, Ward D M. Photoexcretion and fate of glycolate in a hot spring cyanobacterial mat. Appl Environ Microbiol. 1988;54:1738–1743. doi: 10.1128/aem.54.7.1738-1743.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bateson M M, Wiegel J, Ward D M. Comparative analysis of 16S ribosomal RNA sequences of thermophilic fermentative bacteria isolated from hot spring cyanobacterial mats. Syst Appl Microbiol. 1989;12:1–7. [Google Scholar]
- 9.Bauld J, Brock T D. Ecological studies of Chloroflexis, a gliding photosynthetic bacterium. Arch Microbiol. 1973;92:267–284. [Google Scholar]
- 10.Bauld J, Brock T D. Algal excretion and bacterial assimilation in hot spring algal mats. J Phycol. 1974;10:101–106. [Google Scholar]
- 11.Begon M, Harper J L, Townsend C R. Ecology: individuals, populations and communities. Cambridge, Mass: Blackwell Scientific Publications; 1990. [Google Scholar]
- 12.Ben-Bassat A, Zeikus J G. Thermobacteroides acetoethylicus gen. nov. and spec. nov., a new chemoorganotrophic, anaerobic, thermophilic bacterium. Arch Microbiol. 1981;128:365–370. [Google Scholar]
- 13.Bintrim S B, Donohue T J, Handelsman J, Roberts G P, Goodman R M. Molecular phylogeny of Archaea from soil. Proc Natl Acad Sci USA. 1997;94:277–282. doi: 10.1073/pnas.94.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borneman J, Skrock P, O’Sullivan K M, Palus J A, Rumjanek N G, Jansen J L, Nienhuis J, Triplett E W. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl Environ Microbiol. 1996;62:1935–1943. doi: 10.1128/aem.62.6.1935-1943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Borneman J, Triplett E W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl Environ Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Britschgi T B, Giovannoni S J. Phylogenetic analysis of a natural marine bacterioplankton population by rRNA gene cloning and sequencing. Appl Environ Microbiol. 1991;57:1707–1713. doi: 10.1128/aem.57.6.1707-1713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brock T D. Micro-organisms adapted to high temperatures. Nature. 1967;214:882–885. doi: 10.1038/214882a0. [DOI] [PubMed] [Google Scholar]
- 18.Brock T D. Thermophilic microorganisms and life at high temperatures. New York, N.Y: Springer-Verlag; 1978. [Google Scholar]
- 19.Brock T D. The study of microorganisms in situ: progress and problems. Symp Soc Gen Microbiol. 1987;41:1–17. [Google Scholar]
- 20.Brock T D, Brock M L. Effect of light intensity on photosynthesis by thermal algae adapted to natural and reduced sunlight. Limnol Oceanogr. 1969;14:334–341. [Google Scholar]
- 21.Brock T D, Brock M L. Recovery of a hot spring community from a catastrophe. J Phycol. 1969;5:75–77. doi: 10.1111/j.1529-8817.1969.tb02580.x. [DOI] [PubMed] [Google Scholar]
- 22.Brock T D, Freeze H. Thermus aquaticus gen. n. and sp. n., a nonsporulating extreme thermophile. J Bacteriol. 1969;98:289–297. doi: 10.1128/jb.98.1.289-297.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castenholz R W. Ecology of blue-green algae in hot springs. In: Carr N G, Whitton B A, editors. The biology of blue-green algae. Los Angeles, Calif: University of California Press; 1973. pp. 379–414. [Google Scholar]
- 24.Castenholz R W. The biogeography of hot spring algae through enrichment cultures. Mitt Int Ver Limnol. 1978;21:296–315. [Google Scholar]
- 25.Castenholz R W. Isolation and cultivation of thermophilic cyanobacteria. In: Starr M P, Stolp H, Truper H G, Balows A, Schlegel H G, editors. The prokaryotes: a handbook in habitats, isolation, and identification of bacteria. Berlin, Germany: Springer-Verlag; 1981. pp. 236–246. [Google Scholar]
- 26.Castenholz R W. Species usage, concept, and evolution in the cyanobacteria (blue-green algae) J Phycol. 1992;28:737–745. [Google Scholar]
- 27.Castenholz R W. Endemism and biodiversity of thermophilic cyanobacteria. Nova Hedwigia. 1996;112:33–47. [Google Scholar]
- 28.Castenholz R W, Bauld J, Jørgensen B B. Anoxygenic microbial mats of hot springs: thermophilic Chlorobium sp. FEMS Microbiol Ecol. 1990;74:325–336. [Google Scholar]
- 29.Castenholz, R. W., and F. Garcia-Pichel. Cyanobacterial responses to UV radiation. In M. Potts and B. Whitton (ed.), Ecology of cyanobacteria, in press. Kluwer, Dordrecht, The Netherlands.
- 30.Choi B K, Paster B J, Dewhirst F E, Gobel U B. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect Immun. 1994;62:1889–1895. doi: 10.1128/iai.62.5.1889-1895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A E. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 32.Dobson G, Ward D M, Robinson N, Eglinton G. Biogeochemistry of hot spring environments: extractable lipids of a cyanobacterial mat. Chem Geol. 1988;68:155–179. [Google Scholar]
- 33.Doemel W N, Brock T D. Structure, growth, and decomposition of laminated algal-bacterial mats in alkaline hot springs. Appl Environ Microbiol. 1977;34:433–452. doi: 10.1128/aem.34.4.433-452.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Embley T M, Stackebrandt E. Species in practice: exploring uncultured prokaryote diversity in natural samples. In: Claridge M F, Dawah H A, Wilson M R, editors. Species: the units of biodiversity. London, England: Chapman & Hall; 1997. pp. 61–81. [Google Scholar]
- 35.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferris M J, Muyzer G, Ward D M. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol. 1996;62:340–346. doi: 10.1128/aem.62.2.340-346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferris M J, Nold S C, Revsbech N P, Ward D M. Population structure and physiological changes within a hot spring microbial mat community following disturbance. Appl Environ Microbiol. 1997;63:1367–1374. doi: 10.1128/aem.63.4.1367-1374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ferris, M. J., S. C. Nold, C. M. Santegoeds, and D. M. Ward. Examining bacterial population diversity within the Octopus Spring microbial mat community. In A.-L. Reysenbach (ed.), Thermophiles, in press. Plenum Publishing Corp., New York, N.Y.
- 39.Ferris, M. J., and B. P. Palenik. Cyanobacterial community structure changes with depth in an oligotrophic ocean environment. Nature, in press.
- 40.Ferris M J, Ruff-Roberts A L, Kopczynski E D, Bateson M M, Ward D M. Enrichment culture and microscopy conceal diverse thermophilic Synechococcus populations in a single hot spring microbial mat habitat. Appl Environ Microbiol. 1996;62:1045–1050. doi: 10.1128/aem.62.3.1045-1050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferris M J, Ward D M. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1997;63:1375–1381. doi: 10.1128/aem.63.4.1375-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Field K G, Gordon D, Wright T, Rappé M, Urbach E, Vergin K, Giovannoni S J. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol. 1997;63:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Figueras J B, Garcia-Gil L J, Abella C A. Phylogeny of the genus Chlorobium based on 16S rDNA sequence. FEMS Microbiol Lett. 1997;152:31–36. doi: 10.1111/j.1574-6968.1997.tb10405.x. [DOI] [PubMed] [Google Scholar]
- 44.Forterre P. Protein versus rRNA: problems in rooting the universal tree of life. ASM News. 1997;63:89–95. [Google Scholar]
- 45.Fox G E, Wisotzkey J D, Jurtshuk P., Jr How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int J Syst Bacteriol. 1992;42:166–170. doi: 10.1099/00207713-42-1-166. [DOI] [PubMed] [Google Scholar]
- 46.Friedrich, M., R. J. Grosser, E. A. Kern, W. P. Inskeep, and D. M. Ward. Characterization of 16S rRNA-defined phenanthrene-degrading microorganisms enriched in the presence of model organic phases by denaturing gradient gel electrophoresis. Unpublished data.
- 47.Fuhrman J A, McCallum K, Davis A A. Phylogenetic diversity of subsurface marine microbial communities from the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1993;59:1294–1302. doi: 10.1128/aem.59.5.1294-1302.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Futuyma D J. Evolutionary biology. Sunderland, Mass: Sinauer Associates; 1986. [Google Scholar]
- 49.Garcia-Pichel F, Castenholz R W. On the significance of solar ultraviolet radiation for the ecology of microbial mats. In: Stal L, Caumette P, editors. Microbial mats. Heidelberg, Germany: Springer-Verlag; 1994. pp. 77–84. [Google Scholar]
- 50.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;344:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 51.Giovannoni S J, Rappe M S, Vergin K L, Adair N L. 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the green non-sulfur bacteria. Proc Natl Acad Sci USA. 1996;93:7979–7984. doi: 10.1073/pnas.93.15.7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giovannoni S J, Schabtach E, Castenholz R W. Isosphaera pallida, gen. and comb. nov., a gliding, budding eubacterium from hot springs. Arch Microbiol. 1987;147:276–284. doi: 10.1128/jb.169.6.2702-2707.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giovannoni S J, Turner S, Olsen G J, Barns S, Lane D J, Pace N R. Evolutionary relationships among cyanobacteria and green chloroplasts. J Bacteriol. 1988;170:3584–3592. doi: 10.1128/jb.170.8.3584-3592.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gonzalez I L, Sylvester J E, Smith T F, Stambolian D, Schmickel R D. Ribosomal RNA gene sequences and hominoid phylogeny. Mol Biol Evol. 1990;7:203–219. doi: 10.1093/oxfordjournals.molbev.a040600. [DOI] [PubMed] [Google Scholar]
- 55.Gordon D A, Giovannoni S J. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific Oceans. Appl Environ Microbiol. 1996;62:1171–1177. doi: 10.1128/aem.62.4.1171-1177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gray J P, Herwig R P. Phylogenetic analysis of the bacterial communities in marine sediments. Appl Environ Microbiol. 1996;62:4049–4059. doi: 10.1128/aem.62.11.4049-4059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gunderson J H, Sogin M L, Wollett G, Hollingdale M, de la Cruz V F, Waters A P, McCutchan T F. Structurally distinct, stage-specific ribosomes occur in Plasmodium. Science. 1987;238:933–937. doi: 10.1126/science.3672135. [DOI] [PubMed] [Google Scholar]
- 58.Huber R, Burggraf S, Mayer T, Barns S M, Rossnagel P, Stetter K O. Isolation of a hyperthermophilic archaeum predicted by in situ RNA analysis. Nature. 1995;376:57–58. doi: 10.1038/376057a0. [DOI] [PubMed] [Google Scholar]
- 59.Huber R, Eder W, Heldwein S, Wanner G, Huber H, Rachel R, Stetter K O. Thermocrinis ruber gen. nov., sp. nov., a pink-filament-forming hyperthermophilic bacterium isolated from Yellowstone National Park. Appl Environ Microbiol. 1998;64:3576–3583. doi: 10.1128/aem.64.10.3576-3583.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jackson T J, Ramaley R F, Meinschein W G. Thermomicrobium, a new genus of extremely thermophilic bacteria. Int J Syst Bacteriol. 1973;23:28–36. [Google Scholar]
- 61.Kallas T, Castenholz R W. Internal pH and ATP-ADP pools in the cyanobacterium Synechococcus sp. during exposure to growth-inhibiting low pH. J Bacteriol. 1982;149:229–236. doi: 10.1128/jb.149.1.229-236.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kallas T, Castenholz R W. Rapid transient growth at low pH in the cyanobacterium Synechococcus sp. J Bacteriol. 1982;149:237–246. doi: 10.1128/jb.149.1.237-246.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kimble L K, Mandelco L, Woese C R, Madigan M T. Heliobacterium modesticaldum, sp. nov., a thermophilic heliobacterium of hot springs and volcanic soils. Arch Microbiol. 1995;163:259–267. [Google Scholar]
- 64.Komatsoulis G A, Waterman M S. A new computational method for detection of chimeric 16S rRNA artifacts generated by PCR amplification from mixed bacterial populations. Appl Environ Microbiol. 1997;63:2338–2346. doi: 10.1128/aem.63.6.2338-2346.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kopczynski E D, Bateson M M, Ward D M. Recognition of chimeric small-subunit ribosomal DNAs composed of genes from uncultivated microorganisms. Appl Environ Microbiol. 1994;60:746–748. doi: 10.1128/aem.60.2.746-748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kowalchuk G A, Stephen J R, De Boer W, Prosser J I, Embley T M, Woldendorp J W. Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl Environ Microbiol. 1997;63:1489–1497. doi: 10.1128/aem.63.4.1489-1497.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kristjansson J K, Hreggvidsson G O, Petursdottir S K, Konradsdottir M, Helgason E, Gislason J, Mathur E J. Thermophiles ’96 International Conference on “The Biology, Ecology, and Biotechnology of Thermophilic Microorganisms” abstracts. 1996. Population structure of aerobic, nonsporeforming, thermophilic eubacteria; p. 8. [Google Scholar]
- 68.Kühl, M., F. Garcia-Pichel, and R. W. Castenholz. Different temperature adaptation of photosynthetic populations within a single hot spring microbial mat habitat as measured with oxygen microelectrodes. Unpublished data.
- 69.Lawton J H. Patterns in ecology. Oikos. 1996;75:145–147. [Google Scholar]
- 70.Lee Y, Jain M K, Lee C, Lowe C E, Zeikus J G. Taxonomic distinction of saccharolytic thermophilic anaerobes: description of Thermoanaerobacterium xylanolyticum gen. nov., sp. nov., and Thermoanaerobacterium saccharolyticum gen. nov., sp. nov.; reclassification of Thermoanaerobium brockii, Clostridium thermosulfurogenes, and C. thermohydrosulfuricum E11-69 as Thermoanaerobacter brockii comb. nov., Thermoanaerobacterium thermosulfurigenes comb. nov., and Thermoanaerobacter thermohydrosulfuricus comb. nov., respectively; and transfer of Clostridium thermohydrosulfuricum 39E to Thermoanaerobacter ethanolicus. Int J Syst Bacteriol. 1993;43:41–51. [Google Scholar]
- 71.Liesack W, Stackebrandt E. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J Bacteriol. 1992;174:5072–5078. doi: 10.1128/jb.174.15.5072-5078.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liesack W, Weyland H, Stackebrandt E. Potential risks of gene amplification by PCR as determined by 16S rDNA analysis of a mixed-culture of strict barophilic bacteria. Microb Ecol. 1991;21:191–198. doi: 10.1007/BF02539153. [DOI] [PubMed] [Google Scholar]
- 73.MacArthur R H, Wilson E O. The theory of island biogeography. Princeton, N.J: Princeton University Press; 1967. [Google Scholar]
- 74.Madigan M T, Brock T D. Adaptation by hot spring phototrophs to reduced light intensities. Arch Microbiol. 1977;113:111–120. doi: 10.1007/BF00428590. [DOI] [PubMed] [Google Scholar]
- 75.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maynard-Smith J. Do bacteria have population genetics? Symp Soc Gen Microbiol. 1995;52:1–12. [Google Scholar]
- 77.Mayr E. The growth of biological thought: diversity, evolution, and inheritance. Cambridge, Mass: Harvard University Press; 1982. [Google Scholar]
- 78.McCutchan T F, de la Cruz V F, Lal A A, Gunderson J H, Elwood H J, Sogin M L. Primary sequences of two small subunit ribosomal RNA genes from Plasmodium falciparum. Mol Biochem Parasitol. 1988;28:63–68. doi: 10.1016/0166-6851(88)90181-8. [DOI] [PubMed] [Google Scholar]
- 79.Miller S L, Lazcano A. The origin of life: did it occur at high temperatures? J Mol Evol. 1995;41:689–692. doi: 10.1007/BF00173146. [DOI] [PubMed] [Google Scholar]
- 80.Miller S R, Wingard C E, Castenholz R W. Effects of visible light and UV radiation on photosynthesis in a population of a hot spring cyanobacterium, a Synechococcus sp., subjected to high-temperature stress. Appl Environ Microbiol. 1998;64:3893–3899. doi: 10.1128/aem.64.10.3893-3899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moore L R, Rocap G, Chisholm S W. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 82.Mullins T D, Britschgi T B, Krest R L, Giovannoni S J. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol Oceanogr. 1995;40:148–158. [Google Scholar]
- 83.Murray A E, Hollibaugh J T, Orrego C. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl Environ Microbiol. 1996;62:2676–2680. doi: 10.1128/aem.62.7.2676-2680.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Muyzer G, De Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mylvaganam S, Dennis P P. Sequence heterogeneity between the two genes encoding 16S rRNA from the halophilic archaebacterium Haloarcula marismortui. Genetics. 1992;130:399–410. doi: 10.1093/genetics/130.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nold S C, Kopczynski E D, Ward D M. Cultivation of aerobic chemoorganotrophic proteobacteria and gram-positive bacteria from a hot spring microbial mat. Appl Environ Microbiol. 1996;62:3917–3921. doi: 10.1128/aem.62.11.3917-3921.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nold S C, Ward D M. Diverse Thermus species inhabit a single hot spring microbial mat. Syst Appl Microbiol. 1995;18:274–278. doi: 10.1016/s0723-2020(11)80398-x. [DOI] [PubMed] [Google Scholar]
- 88.Nold S C, Ward D M. Photosynthate partitioning and fermentation in hot spring microbial mat communities. Appl Environ Microbiol. 1996;62:4598–4607. doi: 10.1128/aem.62.12.4598-4607.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nölling J, Hahn D, Ludwig W, De Vos W M. Phylogenetic analysis of thermophilic Methanobacterium sp.: evidence for a formate-utilizing ancestor. Syst Appl Microbiol. 1993;16:208–215. [Google Scholar]
- 90.Nübel U, Engelen B, Felske A, Snaidr J, Wieshuber A, Amann R I, Ludwig W, Backhaus H. Sequence heterogeneities of genes encoding 16S rRNAs in Paenibacillus polymyxa detected by temperature gradient gel electrophoresis. J Bacteriol. 1996;178:5636–5643. doi: 10.1128/jb.178.19.5636-5643.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ohkuma M, Kudo T. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl Environ Microbiol. 1996;62:461–468. doi: 10.1128/aem.62.2.461-468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Olsen G J, Lane D J, Giovannoni S J, Pace N R. Microbial ecology and evolution: a ribosomal RNA approach. Annu Rev Microbiol. 1986;40:337–365. doi: 10.1146/annurev.mi.40.100186.002005. [DOI] [PubMed] [Google Scholar]
- 93.Olsen G J, Woese C R, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Overmann J, Tuschak C. Phylogeny and molecular fingerprinting of green sulfur bacteria. Arch Microbiol. 1997;167:302–309. doi: 10.1007/s002030050448. [DOI] [PubMed] [Google Scholar]
- 95.Oyaizu H, Debrunner-Vossbrinck B, Mandelco L, Studier J A, Woese C R. The green non-sulfur bacteria: a deep branching in the eubacterial line of descent. Syst Appl Microbiol. 1987;9:47–53. doi: 10.1016/s0723-2020(87)80055-3. [DOI] [PubMed] [Google Scholar]
- 96.Pace N R. Origin of life—facing up to the physical setting. Cell. 1991;65:531–533. doi: 10.1016/0092-8674(91)90082-a. [DOI] [PubMed] [Google Scholar]
- 97.Pace N R. New perspective on the natural microbial world: molecular microbial ecology. ASM News. 1996;62:463–470. [Google Scholar]
- 98.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 99.Pace N R, Stahl D A, Lane D J, Olsen G J. The analysis of natural microbial populations by ribosomal RNA sequences. Adv Microb Ecol. 1986;9:1–55. [Google Scholar]
- 100.Palenik B. Cyanobacterial community structure as seen from RNA polymerase gene sequence analysis. Appl Environ Microbiol. 1994;60:3212–3219. doi: 10.1128/aem.60.9.3212-3219.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Palys T, Nakamura L K, Cohan F M. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int J Syst Bacteriol. 1997;47:1145–1156. doi: 10.1099/00207713-47-4-1145. [DOI] [PubMed] [Google Scholar]
- 102.Papke, R. T., N. B. Ramsing, and D. M. Ward. Unpublished data.
- 103.Paster B J, Dewhirst F E, Weisburg W G, Tordoff L A, Fraser G J, Hespell R B, Stanton T B, Zablen L, Mandelco L, Woese C R. Phylogenetic analysis of the spirochetes. J Bacteriol. 1991;173:6101–6109. doi: 10.1128/jb.173.19.6101-6109.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Patel B K C, Morgan H W, Daniel R M. Thermophilic anaerobic spirochetes in New Zealand hot springs. FEMS Microbiol Lett. 1985;26:101–106. [Google Scholar]
- 105.Peary J A, Castenholz R W. Temperature strains of a thermophilic blue-green alga. Nature. 1964;202:720–721. [Google Scholar]
- 106.Pierson B K, Castenholz R W. A phototrophic gliding filamentous bacterium of hot springs, Chloroflexus auranticus, gen. and sp. nov. Arch Microbiol. 1974;100:5–24. doi: 10.1007/BF00446302. [DOI] [PubMed] [Google Scholar]
- 107.Rainey F A, Ward-Rainey N L, Janssen P H, Hippe H, Stackebrandt E. Clostridium paradoxum DSM 7308T contains multiple 16S rRNA genes with heterogeneous intervening sequences. Microbiology. 1996;142:2087–2095. doi: 10.1099/13500872-142-8-2087. [DOI] [PubMed] [Google Scholar]
- 108.Rainey F A, Ward N, Sly L I, Stackebrandt E. Dependence on the taxon composition of clone libraries for PCR amplified, naturally occurring 16S rDNA, on the primer pair and the cloning system used. Experientia. 1994;50:796–797. [Google Scholar]
- 109.Rainey F A, Ward N L, Morgan H W, Toalster R, Stackebrandt E. Phylogenetic analysis of anaerobic thermophilic bacteria: aid for their reclassification. J Bacteriol. 1993;175:4772–4779. doi: 10.1128/jb.175.15.4772-4779.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ramsing N B, Ferris M J, Ward D M. Light-induced motility of thermophilic Synechococcus isolates from Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1997;63:2347–2354. doi: 10.1128/aem.63.6.2347-2354.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ramsing, N. B., M. J. Ferris, and D. M. Ward. Highly ordered vertical structure of Synechococcus populations within the one-millimeter thick photic zone of a hot spring cyanobacterial mat. Unpublished data. [DOI] [PMC free article] [PubMed]
- 112.Relman D A, Loutit J S, Schmidt T M, Falkow S, Tompkins L S. The agent of bacillary angiomatosis: an approach to the identification of uncultured pathogens. N Engl J Med. 1990;323:1573–1580. doi: 10.1056/NEJM199012063232301. [DOI] [PubMed] [Google Scholar]
- 113.Revsbech N P, Ward D M. Microelectrode studies of interstitial water chemistry and photosynthetic activity in a hot spring microbial mat. Appl Environ Microbiol. 1984;48:270–275. doi: 10.1128/aem.48.2.270-275.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Reysenbach A-L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Reysenbach A-L, Wickham G S, Pace N R. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring, Yellowstone National Park. Appl Environ Microbiol. 1994;60:2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Robison-Cox J F, Bateson M M, Ward D M. Evaluation of nearest-neighbor methods for detection of chimeric small-subunit rRNA sequences. Appl Environ Microbiol. 1995;61:1240–1245. doi: 10.1128/aem.61.4.1240-1245.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rosenzweig M L. Species diversity in space and time. Cambridge, England: University Press; 1995. [Google Scholar]
- 118.Ruff-Roberts A L, Kuenen J G, Ward D M. Distribution of cultivated and uncultivated cyanobacteria and Chloroflexus-like bacteria in hot spring microbial mats. Appl Environ Microbiol. 1994;60:697–704. doi: 10.1128/aem.60.2.697-704.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sandbeck K A, Ward D M. Fate of immediate methane precursors in low-sulfate, hot-spring algal-bacterial mats. Appl Environ Microbiol. 1981;41:775–782. doi: 10.1128/aem.41.3.775-782.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sandbeck K A, Ward D M. Temperature adaptations in the terminal processes of anaerobic decomposition of Yellowstone National Park and Icelandic hot spring microbial mats. Appl Environ Microbiol. 1982;44:844–851. doi: 10.1128/aem.44.4.844-851.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Santegoeds C M, Nold S C, Ward D M. Denaturing gradient gel electrophoresis used to monitor the enrichment culture of aerobic chemoorganotrophic bacteria from a hot spring cyanobacterial mat. Appl Environ Microbiol. 1996;62:3922–3928. doi: 10.1128/aem.62.11.3922-3928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schink B, Zeikus J G. Clostridium thermosulfurogenes sp. nov., a new thermophile that produces elemental sulfur from thiosulphate. J Gen Microbiol. 1983;129:1149–1158. [Google Scholar]
- 123.Schmidt T M, DeLong E F, Pace N R. Analysis of a marine picoplankton community using 16S rRNA gene cloning and sequencing. J Bacteriol. 1991;173:4371–4378. doi: 10.1128/jb.173.14.4371-4378.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sheridan R P. Sun-shade ecotypes of a bluegreen alga in a hot spring. J Phycol. 1976;12:279–285. [Google Scholar]
- 125.Sheridan R P. Seasonal variation in sun-shade ecotypes of Plectonema notatum (cyanophyta) J Phycol. 1979;15:223–226. [Google Scholar]
- 126.Shiea J, Brassell S C, Ward D M. Mid-chain branched mono- and dimethyl alkanes in hot spring cyanobacterial mats: a direct biogenic source for branched alkanes in ancient sediments? Org Geochem. 1990;15:223–231. [Google Scholar]
- 127.Shiea J, Brassell S C, Ward D M. Comparative analysis of extractable lipids in hot spring microbial mats and their component photosynthetic bacteria. Org Geochem. 1991;17:309–319. [Google Scholar]
- 128.Sibley C G, Ahlquist J E. DNA hybridization evidence of hominoid phylogeny: results from an expanded data set. J Mol Evol. 1987;26:99–121. doi: 10.1007/BF02111285. [DOI] [PubMed] [Google Scholar]
- 129.Sibley C G, Comstock J A, Ahlquist J E. DNA hybridization evidence of hominoid phylogeny: a reanalysis of the data. J Mol Evol. 1990;30:202–236. doi: 10.1007/BF02099992. [DOI] [PubMed] [Google Scholar]
- 130.Simpson G G. Principles of animal taxonomy. New York, N.Y: Columbia University Press; 1961. [DOI] [PubMed] [Google Scholar]
- 131.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. [Google Scholar]
- 132.Stackebrandt E, Liesack W, Goebel B M. Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rDNA analysis. FASEB J. 1993;7:232–236. doi: 10.1096/fasebj.7.1.8422969. [DOI] [PubMed] [Google Scholar]
- 133.Stahl D A, Amann R. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, England: John Wiley & Sons, Ltd.; 1991. pp. 205–248. [Google Scholar]
- 134.Stahl D A, Lane D J, Olsen G J, Pace N R. Characterization of a Yellowstone hot spring microbial community by 5S rRNA sequences. Appl Environ Microbiol. 1985;49:1379–1384. doi: 10.1128/aem.49.6.1379-1384.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Staley J T. Biodiversity: are microbial species threatened? Curr Opin Biotechnol. 1997;8:340–345. doi: 10.1016/s0958-1669(97)80014-6. [DOI] [PubMed] [Google Scholar]
- 136.Stetter K O. Hyperthermophilic procaryotes. FEMS Microbiol Rev. 1996;18:149–158. [Google Scholar]
- 137.Stetter K O, Fiala G, Huber G, Huber R, Segerer A. Hyperthermophilic microorganisms. FEMS Microbiol Rev. 1990;75:117–124. [Google Scholar]
- 138.Stevenson A K, Kimble L K, Woese C R, Madigan M T. Characterization of new heliobacteria and their habitats. Photosynth Res. 1997;53:1–12. [Google Scholar]
- 139.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139a.Tiedje J M, Zhou J-Z, Nüsslein K, Moyer C L, Fulthorpe R R. Extent and patterns of soil microbial diversity. In: Martins M T, Sato M I Z, Tiedje J M, Hagler L C N, Dobereiner J, Sanchez P S, editors. Progress in microbial ecology. Proceedings of the Seventh International Symposium on Microbial Ecology. Brazilian Society for Microbiology/International; 1997. pp. 35–41. Committee on Microbial Ecology, Sao Paulo, Brazil. [Google Scholar]
- 140.Tipper J C. Rarefaction and rarefiction—the use and abuse of a method in paleoecology. Paleobiology. 1979;5:423–434. [Google Scholar]
- 141.Ueda T, Suga Y, Matsuguchi T. Molecular phylogenetic analysis of a soil microbial community in a soybean field. Eur J Soil Sci. 1995;46:415–421. [Google Scholar]
- 142.Van Regenmortel M H V. Viral species. In: Claridge M F, Dawah H A, Wilson M R, editors. Species: the units of biodiversity. London, England: Chapman & Hall; 1997. pp. 17–24. [Google Scholar]
- 143.Wahlund T M, Woese C R, Castenholz R W, Madigan M T. A thermophilic green sulfur bacterium from New Zealand hot springs, Chlorobium tepidum sp. nov. Arch Microbiol. 1991;156:81–90. [Google Scholar]
- 144.Ward D M. Molecular probes for analysis of microbial communities. In: Characklis W G, Wilderer P A, editors. Structure and function of biofilms. Chichester, England: John Wiley & Sons, Ltd.; 1989. pp. 145–163. [Google Scholar]
- 145.Ward, D. M. Unpublished observations.
- 146.Ward D M. A natural species concept for prokaryotes. Curr Opin Microbiol. 1998;1:271–277. doi: 10.1016/s1369-5274(98)80029-5. [DOI] [PubMed] [Google Scholar]
- 147.Ward D M, Bateson M M, Weller R, Ruff-Roberts A L. Ribosomal RNA analysis of microorganisms as they occur in nature. Adv Microb Ecol. 1992;12:219–286. [Google Scholar]
- 148.Ward D M, Bauld J, Castenholz R W, Pierson B K. Modern phototrophic microbial mats: anoxygenic, intermittently oxygenic/anoxygenic, thermal, eukaryotic, and terrestrial. In: Schopf J W, Klein C, editors. The proterozoic biosphere—a multidisciplinary study. Cambridge, England: Cambridge University Press; 1992. pp. 309–324. [Google Scholar]
- 149.Ward D M, Beck E, Revsbech N P, Sandbeck K A, Winfrey M R. Decomposition of hot spring microbial mats. In: Cohen Y, Castenholz R W, Halvorson H O, editors. Microbial mats: stromatolites. New York, N.Y: Alan R. Liss; 1984. pp. 191–214. [Google Scholar]
- 150.Ward D M, Brassell S C, Eglinton G. Archaebacterial lipids in hot-spring microbial mats. Nature. 1985;318:656–659. [Google Scholar]
- 151.Ward, D. M., and R. W. Castenholz. Cyanobacteria in geothermal habitats. In M. Potts and B. Whitton (ed.), Ecology of cyanobacteria, in press. Kluwer, Dordrecht, The Netherlands.
- 152.Ward D M, Ferris M J, Bateson M M. Organization of native populations within hot spring microbial mat communities: need for a more ecological approach. In: Martins M T, Sato M I Z, Tiedje J M, Hagler L C N, Dobereiner J, Sanchez P S, editors. Progress in microbial ecology. Proceedings of the Seventh International Symposium on Microbial Ecology. Brazilian Society for Microbiology/International; 1997. pp. 147–153. Committee on Microbial Ecology, Sao Paulo, Brazil. [Google Scholar]
- 153.Ward D M, Ferris M J, Nold S C, Bateson M M, Kopczynski E D, Ruff-Roberts A L. Species diversity in hot spring microbial mats as revealed by both molecular and enrichment culture approaches—relationship between biodiversity and community structure. NATO ASI Ser. 1994;G35:33–44. [Google Scholar]
- 154.Ward D M, Panke S, Klöppel K-D, Christ R, Fredrickson H. Complex polar lipids of a hot spring cyanobacterial mat and its cultivated inhabitants. Appl Environ Microbiol. 1994;60:3358–3367. doi: 10.1128/aem.60.9.3358-3367.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ward D M, Santegoeds C M, Nold S C, Ramsing N B, Ferris M J, Bateson M M. Biodiversity within hot spring microbial mat communities: molecular monitoring of enrichment cultures. Antonie Leeuwenhoek. 1997;71:143–150. doi: 10.1023/a:1000131426164. [DOI] [PubMed] [Google Scholar]
- 156.Ward D M, Shiea J, Zeng Y B, Dobson G, Brassell S, Eglinton G. Lipid biochemical markers and the composition of microbial mats. In: Cohen Y, Rosenberg E, editors. Microbial mats—physiological ecology of benthic microbial communities. Washington, D.C: American Society for Microbiology; 1989. pp. 439–454. [Google Scholar]
- 157.Ward D M, Tayne T A, Anderson K L, Bateson M M. Community structure and interactions among community members in hot spring cyanobacterial mats. Symp Soc Gen Microbiol. 1987;41:179–210. [Google Scholar]
- 158.Ward D M, Weller D, Bateson M M. 16S rRNA sequences reveal uncultured inhabitants of a well-studied thermal community. FEMS Microbiol Rev. 1990;75:105–116. doi: 10.1111/j.1574-6968.1990.tb04088.x. [DOI] [PubMed] [Google Scholar]
- 159.Ward D M, Weller R, Bateson M M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990;344:63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- 160.Ward D M, Weller R, Shiea J, Castenholz R W, Cohen Y. Hot spring microbial mats: anoxygenic and oxygenic mats of possible evolutionary significance. In: Cohen Y, Rosenberg E, editors. Microbial mats—physiological ecology of benthic microbial communities. Washington, D.C: American Society for Microbiology; 1989. pp. 3–15. [Google Scholar]
- 161.Wayne L G, Brenner D J, Colwell R R, Grimont P A D, Kandler O, Krichevsky M I, Moore L H, Moore W E C, Murray R G E, Stackebrandt E, Starr M P, Trüper H G. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol. 1987;37:463–464. [Google Scholar]
- 162.Weller R, Bateson M M, Heimbuch B K, Kopczynski E D, Ward D M. Uncultivated cyanobacteria, Chloroflexus-like and spirochete-like inhabitants of a hot spring microbial mat. Appl Environ Microbiol. 1992;58:3964–3969. doi: 10.1128/aem.58.12.3964-3969.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Weller R, Ward D M. Selective recovery of 16S rRNA sequences from natural microbial communities in the form of cDNA. Appl Environ Microbiol. 1989;55:1818–1822. doi: 10.1128/aem.55.7.1818-1822.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Weller R, Weller J W, Ward D M. 16S rRNA sequences of uncultivated hot spring cyanobacterial mat inhabitants retrieved as randomly primed cDNA. Appl Environ Microbiol. 1991;57:1146–1151. doi: 10.1128/aem.57.4.1146-1151.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wiegel J, Braun M, Gottschalk G. Clostridium thermoautotrophicum species novum, a thermophile producing acetate from molecular hydrogen and carbon dioxide. Curr Microbiol. 1981;5:255–260. [Google Scholar]
- 166.Wiegel J, Ljungdahl L G. Thermoanaerobacter ethanolicus gen. nov., spec. nov., a new, extreme thermophilic, anaerobic bacterium. Arch Microbiol. 1981;128:343–348. [Google Scholar]
- 167.Wiegel J, Ljungdahl L G, Rawson J R. Isolation from soil and properties of the extreme thermophile Clostridium thermohydrosulfuricum. J Bacteriol. 1979;139:800–810. doi: 10.1128/jb.139.3.800-810.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wilmotte A. Molecular evolution and taxonomy of the cyanobacteria. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 1–25. [Google Scholar]
- 169.Wilson E O. Naturalist. New York, N.Y: Warner Books; 1994. [Google Scholar]
- 170.Winogradsky S. Microbiologie du sol, problèmes et méthodes. Paris, France: Masson et Cie; 1949. [Google Scholar]
- 171.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Woese C R. There must be a prokaryote somewhere: microbiology’s search for itself. Microbiol Rev. 1994;58:1–9. doi: 10.1128/mr.58.1.1-9.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zeikus J G, Ben-Bassat A, Hegge P W. Microbiology of methanogenesis in thermal, volcanic environments. J Bacteriol. 1980;143:432–440. doi: 10.1128/jb.143.1.432-440.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zeikus J G, Dawson M A, Thompson T E, Ingvorsen K, Hatchikian E C. Microbial ecology of volcanic sulphidogenesis: isolation and characterization of Thermodesulfobacterium commune gen. nov. and sp. nov. J Gen Microbiol. 1983;129:1159–1169. [Google Scholar]
- 175.Zeikus J G, Hegge P W, Anderson M A. Thermoanaerobium brockii gen. nov. and sp. nov., a new chemoorganotrophic, caldoactive, anaerobic bacterium. Arch Microbiol. 1979;122:41–48. [Google Scholar]
- 176.Zeng Y B, Ward D M, Brassell S C, Eglinton G. Biogeochemistry of hot spring environments. 2. Lipid compositions of Yellowstone (Wyoming, U.S.A.) cyanobacterial and Chloroflexus mats. Chem Geol. 1992;95:327–345. [Google Scholar]
- 177.Zeng Y B, Ward D M, Brassell S C, Eglinton G. Biogeochemistry of hot spring environments. 3. Apolar and polar lipids in the biologically active layers of a cyanobacterial mat. Chem Geol. 1992;95:347–360. [Google Scholar]