Abstract
There is ample evidence that cells of higher eukaryotes express double-stranded RNA molecules (dsRNAs) either naturally or as the result of viral infection or aberrant, bidirectional transcriptional readthrough. These duplex molecules can exist in either the cytoplasmic or nuclear compartments. Cells have evolved distinct ways of responding to dsRNAs, depending on the nature and location of the duplexes. Since dsRNA molecules are not thought to exist naturally within the cytoplasm, dsRNA in this compartment is most often associated with viral infections. Cells have evolved defensive strategies against such molecules, primarily involving the interferon response pathway. Nuclear dsRNA, however, does not induce interferons and may play an important posttranscriptional regulatory role. Nuclear dsRNA appears to be the substrate for enzymes which deaminate adenosine residues to inosine residues within the polynucleotide structure, resulting in partial or full unwinding. Extensively modified RNAs are either rapidly degraded or retained within the nucleus, whereas transcripts with few modifications may be transported to the cytoplasm, where they serve to produce altered proteins. This review summarizes our current knowledge about the function and fate of dsRNA in cells of higher eukaryotes and its potential manipulation as a research and therapeutic tool.
Numerous examples of naturally occurring antisense RNA-mediated regulation of gene expression in prokaryotes have been documented (140). In most cases the regulation occurs at the translational level (140, 297). The antisense transcript hybridizes to the sense transcript and blocks access of the translational machinery to the 5′ end of the sense transcript. This leads to reduced levels of protein synthesis. On the other hand, cellular compartmentalization in eukaryotes has allowed the development of more numerous and complex effects of antisense transcripts.
There might be some endogenous double-stranded RNA (dsRNA) within the nuclei of most or all eukaryotic cells. The first reports of dsRNA within cells came from studies of heterogeneous nuclear ribonucleoprotein (hnRNP) particles. The presence of highly repetitive sequences within the genome provides the potential for transcripts from different strands to yield RNAs that might anneal. Two groups reported that native RNP particles isolated from HeLa cells might contain 2 to 5% dsRNA, as judged both by nuclease resistance studies and by electron microscopy (42, 85). The majority of the sequences involved in these putative duplexes were reiterated, and the possible physiological significance of these structures was not clear. Since then, there have been many reports of naturally occurring antisense RNA with different modes of action and effects on the cell. The purpose of this review is to briefly summarize some of these reports and to discuss the different ways in which dsRNA can affect gene expression and the ways in which cells deal with the presence of dsRNA. In addition, a major cause of the presence of dsRNA in cells is viral infection. dsRNAs are formed in almost all viral infections and by all types of viruses. Since such molecules have profound effects on cellular physiology, they are reviewed here as well.
Naturally occurring antisense RNAs in higher eukaryotes may be grouped into molecules capable of forming short or imperfect RNA duplexes (less than 100 bp) and molecules capable of forming long, perfect duplexes. Also, examples may be grouped into duplexes in the cytoplasm and duplexes within the nucleus. Each of these scenarios may involve distinct fates for the molecules involved, as well as distinct consequences to the cell.
NATURALLY OCCURRING ANTISENSE RNA IN HIGHER EUKARYOTES
Short (or Imperfect) Duplex RNAs
Although there are a large number of RNA-RNA interactions within cells that are mediated by base pairing between stretches of less than 10 bp of complementary nucleotides, there are only a few examples of regulation of the expression of specific genes by short antisense molecules. These RNAs are generally shorter than 100 bp and are transcribed from a locus that is different from the locus of the sense RNA. Most short-RNA-RNA interactions, such as interactions between small nuclear RNP (snRNP) particle RNAs and pre-mRNA splicing substrates (reviewed in reference 170), as well as the involvement of small nucleolar RNAs in rRNA maturation (299), are beyond the scope of this review. The antisense regulation in each of the two systems described below presumably occurs in the cytoplasm, at the level of translational inhibition.
lin-4/lin-14 of C. elegans.
In Caenorhabditis elegans, antisense RNAs expressed from the lin-4 gene are complementary to the 3′ end of lin-14 mRNAs, which are expressed from a different genomic locus. There are two lin-4 transcripts, of approximately 22 and 61 nucleotides, and they are complementary to the 3′ untranslated region (UTR) of lin-14 mRNA. lin-4 transcripts are responsible for the temporal decrease in the levels of Lin-14 protein. In the absence of this regulation (in lin-4 null mutants) the levels of Lin-14 do not decrease temporally, and this causes a retarded phenotype, namely, the absence of adult structures and the failure to lay eggs (50). lin-14 transcript levels are constant throughout development, indicating that lin-14 is negatively regulated posttranscriptionally (330). The antisense RNAs are thought to cause downregulation of Lin-14 expression by interfering with its translation (9, 184, 330). lin-4 transcripts do not exhibit perfect complementarity to their sense mRNAs but, rather, contain unpaired regions that form bulges that have been shown to be necessary for antisense function (184, 330).
Chicken myosin tcRNA.
Another example of regulation by short antisense transcripts is seen in the regulation of the chicken myosin heavy-chain mRNAs by translation control RNAs (tcRNAs) (29, 121–123). tcRNAs, first isolated from embryonic chicken muscle, have complementarity to the 5′ end of chicken myosin heavy-chain mRNAs and inhibit its translation. McCarthy et al. (213) purified myosin heavy-chain mRNP particles from 13-day chicken embryonic skeletal muscle. These authors found a 102-nucleotide tcRNA associated with the mRNPs. tcRNA102 was capable of stoichiometrically inhibiting the translation of the mRNAs with which it associated. Under the same conditions, endogenous reticulocyte mRNA was not inhibited and tcRNA did not associate with rRNA or globin RNA. Like lin-4 transcripts, tcRNAs do not exhibit perfect complementarity to their target transcripts. As with lin-4, the mechanism of action of tcRNA is not known.
Long (or Perfect) Duplex RNAs
Long antisense transcripts are usually transcribed from the same locus as sense RNAs but in the opposite direction and overlapping the region of sense transcription. Williams and Fried (333) reported the first mammalian example of possible endogenous antisense RNA expression in the mouse surfeit locus (334). Processed mRNAs from two adjacent convergent transcription units, Surf-2 and Surf-4, showed an overlap of 133 bp at their 3′ untranslated ends. The potential interaction of these RNAs to confer regulation was discussed (333). Since that report was published, a number of other examples of convergent transcription units have been described. Some of these have been well studied and suggest antisense regulation, while others merely report the presence of antisense or convergent transcription. We shall first describe the best-studied examples and those that have been shown or are suspected to play regulatory roles. We only briefly mention some other examples of antisense RNA that have not been demonstrated to function in a regulatory manner. Nonmammalian systems are addressed after the mammalian ones. Table 1 provides a list of most reported instances of naturally occurring antisense transcripts in higher eukaryotes. Some of the systems described below have also been reviewed elsewhere (163, 168, 172, 297).
TABLE 1.
Some eukaryotic genes with reported complementary transcripts
| Gene | Organism | Primary reference(s) |
|---|---|---|
| surf-2/surf-4 | Mouse | 333, 334 |
| Human | 334 | |
| bFGF | Xenopus | 320 |
| Human | 169 | |
| 231 | ||
| Rat | 194 | |
| Chicken | 37 | |
| erbAα | Human, rat | 181 |
| eIF-2α | Human | 296 |
| p53 | Mouse | 73 |
| c-myc | Mouse | 71 |
| 233 | ||
| Human | 28 | |
| N-myc | Human | 173 |
| MBP (mld mice) | Mouse | 243 |
| Igf2r | Mouse | 338 |
| H19/Igf2 | Mouse | 226 |
| BCMA | Human | 177 |
| Gart | Drosophila | 118 |
| Ddc | Drosophila | 301 |
| PSV-A | Dictyostelium | 125 |
| GnRH | Rat | 6 |
| ERCC1 | Human | 317 |
| 4f-rnp | Drosophila | 254 |
| Dix-1, Dix-2 | Drosophila | 214 |
| IGF-II | Chicken | 308 |
| Mouse | 271 | |
| Hoxa 11 | Mouse | 130 |
| Hsp 70.2 | Mouse | 229 |
| EP1/PKN | Mouse | 19 |
| Ribosomal L27 | Mouse | 25 |
| c-myb | Mouse | 26 |
| WT1 | Human | 43 |
| 314 | ||
| GnRH | Rat | 6 |
| Collagen α1 (I) | Chicken | 84 |
bFGF.
Basic fibroblast growth factor (bFGF) is a highly conserved and ubiquitously distributed mitogen. It is overexpressed in glial tumor cells and promotes their unregulated proliferation. The bFGF gene locus is transcribed into a number of mRNA transcripts including an antisense mRNA derived from the opposite DNA strand. Expression of this natural antisense RNA has been implicated in regulation of the bFGF sense mRNA expression and turnover. While screening a Xenopus laevis cDNA library, Volk et al. isolated a clone representing a 1.5-kb polyadenylated transcript with an open reading frame coding for an unknown protein of 25 kDa (320). This putative mRNA spanned part of the bFGF exon 3, within the 3′ untranslated region but in the antisense orientation. The region of overlap and complementarity between the sense and antisense RNAs was greater than 900 bp. The sequence organization on the corresponding genomic fragments revealed that the antisense transcript is spliced. Sequence comparison with elongated transcripts from the bFGF gene in human cells revealed that the gene corresponding to the antisense mRNA is evolutionarily conserved. Recently, it has been shown that the long open reading frame within bFGF antisense RNA predicts a hypothetical protein with homology to the prokaryotic MutT family of nucleotide hydrolases (167, 195). Antibodies directed against the conserved MutT domain of the deduced human bFGF antisense protein revealed the expression of an immunoreactive 24-kDa protein in liver extracts from X. laevis and two proteins of 28 and 35 kDa in rat liver extracts. The bFGF antisense protein is expressed in a broad range of non-central nervous system (CNS) tissue in the postnatal period of the rat.
Kimelman and Kirschner (164) published data from Xenopus microinjection experiments suggesting that the antisense transcript can induce the modification of the mRNA encoding bFGF during maturation of the oocyte, converting almost half of the adenosine residues to inosine in the region of overlap between the sense and antisense transcripts. This suggested the action of dsRNA adenosine deaminase (ADAR) (which is discussed in greater detail below), but the results were most probably incorrect owing to the inadvertent microinjection of the modifying enzyme during the course of the experiments (276).
Borja et al. (37) studied the expression of sense and antisense bFGF transcripts during embryogenesis. The inversely proportional amounts of sense and antisense transcripts suggested a possible regulatory role of the antisense transcripts. Knee et al. (169) used reverse transcription-PCR and Northern hybridization to determine the presence of bFGF and its antisense RNA in unfertilized human oocytes and postnatal differentiated tissues. bFGF and the antisense transcripts were coexpressed in many tissues, with sense transcripts being more abundant than antisense transcripts in half of the tissues examined. Sense and antisense transcript expression was approximately equal in the kidneys and colon, whereas antisense transcripts predominated in the heart, liver, skeletal muscle, and testes.
Murphy and Knee (231) studied bFGF mRNA in human U87-MG glioma cells. They identified the human equivalent of the Xenopus antisense transcript and studied its role in bFGF mRNA stability. Analysis of the 3′ UTR of the human bFGF mRNA revealed two areas with greater than 75% homology to exons 3 and 4 of the Xenopus antisense transcript. A 1.5-kb antisense transcript was found in normal rat tissues and human T47D breast cancer cells, which contain very low levels of bFGF mRNA. In contrast, antisense transcript expression was undetectable in U87-MG cells, which overexpress the bFGF sense mRNA. The reciprocal relationship between bFGF sense and antisense mRNA expression suggested that antisense transcripts may regulate bFGF expression in mammalian cells and that disruption of normal sense/antisense mRNA ratios may lead to overexpression of bFGF in some tumors.
Li et al. (194) examined the developmental pattern of expression of the bFGF antisense transcript in fetal and postnatal rat tissues. Northern hybridization detected polyadenylated antisense RNA in all tissues examined. Both sense and antisense transcripts were detected in the developing brain, but the pattern of their expression was inversely related. These findings supported the possibility of a regulatory role for the antisense transcript in that tissue. However, there was no evidence for a reciprocal relationship between sense and antisense RNA expression in other tissues examined, indicating that the relationship between sense and antisense RNA expression may be tissue specific, at least for some genes.
Thyroid hormone receptor ErbAα/Rev-ErbAα.
The rat erbAα locus is associated with two alternatively spliced mRNAs, erbAα1 and erbAα2, which are identical except for their 3′-terminal exons. erbAα1 mRNA encodes a thyroid hormone receptor, while ErbAα2 encodes a similar protein with an altered ligand binding domain. Lazar et al. (181) isolated a rat cDNA encoding a novel member of the thyroid/steroid hormone receptor. This 56-kDa Rev-ErbAα protein is similar in structure to the erbAα product but does not bind thyroid hormone. Rev-ErbAα mRNA is present in many tissues. Interestingly, the mRNA overlaps the c-erbAα2 mRNA (but not the erbAα1 mRNA) by 269 bp but in the opposite orientation. The bidirectionally transcribed regions are conserved in the human genes, suggesting an important regulatory function (182). The ratio of erbAα1 to erbAα2 is highest in cells expressing high levels of Rev-ErbAα mRNA, leading to the hypothesis that base pairing with Rev-ErbAα blocks the splicing of erbAα2 mRNA, thereby favoring formation of the nonoverlapping erbAα1 (180). To test this model, Munroe and Lazar (228) used an in vitro splicing system and demonstrated that antisense transcripts spanning the erbAα2 3′ splice site could inhibit splicing, consistent with a mechanism in which base pairing with a complementary RNA regulates alternative processing of erbAα1 and erbAα2 mRNAs.
Finally, a recent report by Hastings et al. (116) examined the levels of erbAα mRNA in numerous B-cell lines representing different stages of differentiation. Expression of Rev-ErbAα was found to correlate strongly with an increase in the ratio of erbAα1 to erbAα2 mRNA. The correlation between Rev-ErbAα and erbAα mRNA is consistent with negative regulation of erbAα2 via antisense interactions with the complementary Rev-ErbAα mRNA.
eIF-2α.
Resting human peripheral blood T cells synthesize proteins at very low rates and contain very low levels of eukaryotic initiation factor eIF-2α mRNA. During mitogenic activation, the level of eIF-2α mRNA increases greater than 50-fold. This effect has been thought to result mainly from the intranuclear stabilization of the primary transcript (60). Analysis of sequences within the first intron (296) revealed a region with homology to the “initiator” (Inr) sequence first described by Smale and Baltimore (298). This Inr element is positioned about 450 bases downstream of the eIF2α promoter and is oriented in the antisense direction. Deletion or mutation of the Inr element resulted in higher expression from an eIF2α promoter-driven reporter gene. Noguchi et al. (237) used reverse transcription-PCR to demonstrate the presence of overlapping sense and antisense transcripts of the eIF-2α gene in resting and activated human T lymphocytes. These authors further characterized cis-acting elements that appear to regulate the antisense Inr. A model for the regulation of eIF-2α expression, which involves the rapid degradation of dsRNA generated by sense and antisense transcription, was presented.
p53.
In some cells, the expression of p53 might be regulated at the posttranscriptional level. Mouse F9 embryonal carcinoma stem cells differentiate after treatment with retinoic acid and dibutyryl cyclic AMP. This differentiation process is accompanied by the reduction of stable mRNA for p53, while the transcription rate of this gene is not altered (73). This type of regulation is conserved between chickens and mice and occurs during embryonal development (202). This control seemed to be the result of an induced RNA molecule (160).
Khochbin and Lawrence (159) localized the posttranscriptional regulation of p53 mRNA to the nuclear compartment of the cells. These workers identified a 1.3-kb polyadenylated nuclear RNA molecule and showed that it can anneal to the 5′ part of the first intron of the p53 gene but in the antisense orientation. This RNA was subsequently shown to be homologous to intron 1 as a result of a B1 repetitive element in the opposite orientation to one within the p53 first intron. More importantly, perhaps, a longer transcript residing in the nuclear compartment appears to represent an antisense transcript of the entire p53 gene and also appears to accumulate concomitantly with a decrease in sense-strand mRNAs (158).
c-myc.
In human, rodent and bovine cells, both strands of the c-myc gene are transcribed (28, 71, 165, 233). Sense transcription of the c-myc gene uses two major promoters, P1 and P2. In normal growing cells, transcripts initiated from P2 predominate over the ones initiated from P1.
Chang et al. (53) measured the effects of interleukin 3 (IL-3) on c-myc locus transcription in the IL-3-dependent pre-B-cell line Ba/F3. They found that IL-3 strongly influenced the relative use of the P1 and P2 promoters and that there was a rapid and reversible drop in the levels of c-myc mRNA after IL-3 deprivation, as well as a dramatic change in the relative use of P1 and P2. Interestingly, however, there was little change in the rate of initiation of c-myc pre-mRNA. Deprivation of IL-3 led to a large increase in antisense transcription. This correlation suggested a negative regulation of c-myc mRNA by antisense transcription, but stable antisense transcripts were not detected. Stable antisense RNAs were detected, however, in c-myc genes that were rearranged in murine plasmacytomas, where the oncogene was translocated to an immunoglobulin constant-region gene element (302). The opposite-strand RNAs are chimeric, containing c-myc antisense and immunoglobulin sense sequences. Spicer and Sonenshein (302) mapped the 5′ ends of the stable chimeric transcripts to a site within intron 2 of the c-myc gene and demonstrated that the antisense promoter is functional when linked to a reporter gene in transfection studies.
N-myc.
The human N-myc gene has bidirectional overlapping transcription units. There are multiple antisense transcripts initiating at various sites within the first intron of the N-myc pre-mRNA. Some of these are polyadenylated, and some are nonpolyadenylated (173). The nonpolyadenylated antisense transcripts have 5′ ends that are complementary to the 5′ ends of the N-myc sense mRNA. Interestingly, some of the nonpolyadenylated antisense transcripts were found in the cytoplasmic fraction, where most existed in RNA-RNA duplexes with approximately 5% of the sense N-myc mRNA. dsRNA formation appeared to occur only with some of the multiple forms of the N-myc mRNA. The transcriptional initiation site of the RNA appeared to play a role in determining this selectivity. The sense-antisense duplexes included sequences from both exon 1 and intron 1, suggesting that dsRNA formation might modulate RNA processing by inhibiting the splicing of intron 1.
Murine myelin basic protein.
Myelin-deficient (mld) mice are autosomal recessive mutants with hypomyelination of the CNS. Mutant mice express only about 2% of the myelin basic protein (MBP) and cytoplasmic mRNA concentrations present in normal mice. Okano et al. (243) demonstrated that in this mutant the MBP gene has undergone a tandem duplication coupled with an inversion. The upstream gene contains an inversion of exons 3 to 7 of the normal gene and therefore cannot give rise to mature mRNA and functional protein. However, the upstream gene does express an antisense transcript, which elongates through the inverted segment and past the transcriptional initiation site of the downstream gene. The suggestion was made that the antisense RNA interfered with downstream transcription initiation (243). Tosic et al. (314) reported that while the overall transcription rate of the wild-type MBP gene is normal in these mice (ruling out transcriptional interference as a cause of the defect), the rate of transcription of the inverted upstream gene is even higher. Further, the antisense transcript was shown to be nonpolyadenylated and retained within the nucleus. Thus, the high concentration of nuclear antisense RNA strongly suggested that posttranscriptional regulation occurs in mld mice through formation of dsRNA. Mikoshiba et al. (223) and Okano et al. (242) also showed that reduced MBP expression was the result of dsRNA formation within the nucleus and suggested that in this system naturally occurring antisense RNA induces the selective degradation of duplexes or inhibits the nucleocytoplasmic transport of MBP mRNA.
Imprinting.
Antisense transcripts have been identified in several imprinted genes. The mouse insulin-like growth factor 2 receptor gene Igf2r is expressed only from the maternal chromosome (13). The second intron of Igf2r contains a 2-kb CpG island called region 2, which is thought to be an imprinting element (305). This region acquires a maternal imprinting pattern, and the maternal chromosome is methylated in diploid cells in the embryonic and adult stages. Paternal specific repression of Igf2r occurs, and in all these cases the presence of an antisense transcript has been observed (338). Unmethylated region 2 is required for the transcription of the antisense transcript but not for the production of the sense transcript. The maternal Igf2r makes only the sense transcript and cannot make the antisense RNA since region 2 is methylated in the maternal chromosome.
Multiple imprinted sense and antisense transcripts have also been found in a control region upstream of the imprinted Igf2 (insulin-like growth factor 2) gene in mice (226). In the mouse Igf2 gene, the maternal allele is silenced during fetal development. Numerous antisense transcripts, along with sense RNAs, are made from a region upstream of the Igf2 gene containing a tandem repeat. Imprinting depends upon production of both sense and antisense transcripts as well as parental specific methylation of regions flanking the tandem repeat. Both of these phenomena depend upon the presence of a functional H19 gene (226). Thus antisense transcripts may play an important role in the process of genomic imprinting.
Nonmammalian Systems
In addition to mammalian systems, several examples of antisense transcription have been reported in lower organisms. Of these, the best studied are the Dopa decarboxylase transcripts of Drosophila and the regulation of expression of the Dictyostelium PSV-A protein. Regulatory roles have been attributed to the antisense transcripts in both of these systems.
There is a region of overlap between the 3′ termini of a pair of convergent transcription units in the Dopa decarboxylase (Ddc) region of Drosophila melanogaster. This 88-bp genomic region is associated with the 3′ terminus of the mRNA for the Ddc enzyme and, in the opposite orientation, the 3′ terminus of the adjacent gene, whose function is unknown. An analysis of the temporal and spatial distribution of two transcripts within the organism revealed that levels of the two transcripts appear to be reciprocally regulated (301). Within the testes, where the 3′ transcript is maximally expressed, low levels of Ddc transcript were detected.
The Dictyostelium discoideum prespore gene product PSV-A is expressed in a highly regulated fashion during growth and development, although transcription of its gene is constitutive (125). The PSV-A mRNA accumulates only when cells form aggregates and establish the prespore-prestalk pattern. In early development and after stalk disaggregation, the mRNA is unstable. At these times, a 1.8-kb antisense transcript from the same locus is expressed. The antisense transcript does not encode a protein and is regulated by a promoter located within the open reading frame of the PSV-A gene. The reciprocal expression of sense and antisense transcripts suggests a possible antisense control of mRNA stability in this system (125).
A number of other nonmammalian systems have been reported to exhibit antisense transcription. Typical of such results are the Drosophila melanogaster 4f-rnp and Gart genes. The D. melanogaster 4f-rnp gene makes two alternatively spliced mRNAs, which are especially abundant in the CNS. The gene product contains an RNA recognition motif domain, implicated in RNA binding proteins (39). Several isolated cDNAs from the 4f-rnp region contained extensive A-to-G changes, suggesting the action of an ADAR on the original mRNA (254). Since the action of this enzyme is highly suggestive of the presence of dsRNA within the nucleus (see below), the implication is that the 4f-rnp gene is transcribed bidirectionally. This work did not show, however, whether the edited mRNAs are expressed into altered proteins in Drosophila, nor was the subcellular location of the edited mRNAs revealed.
The Gart gene of Drosophila melanogaster encodes three purine pathway enzymatic activities. Interestingly, a pupal cuticle protein gene was found within the first intron of this gene (118). The intronic gene is encoded on the opposite DNA strand from the purine pathway gene and contains an intron. The cuticle protein gene is expressed primarily over a 3-h period in the abdominal epidermal cells of prepupae that secrete the pupal cuticle, while the sense strand is expressed throughout development. It is not yet known how the expression of each gene might affect the expression of the other.
Conclusion
It is evident from the above discussion that although numerous examples of naturally occurring convergent transcription have been reported so far, a general role for antisense RNA in the regulation of gene expression is not yet firmly established. Future research is required not only to document the existence of more examples of antisense expression but also to carry out detailed mechanistic studies to learn how duplex formation can lead to the myriad of effects reported, including effects on transcription, pre-mRNA processing, mRNA transport, and RNA stability.
DOUBLE-STRANDED RNAS IN VIRUS-INFECTED CELLS
dsRNA formation may occur at some point during the life cycle of most viruses (21–24, 166, 183). Viruses that replicate within the cytoplasm (generally RNA viruses) are thought to generate dsRNA in that compartment. Viruses that express their RNAs in the nucleus (generally DNA viruses) could produce RNAs which form duplexes within the cytoplasm or the nucleus, and in most cases the site of dsRNA formation for these viruses has not been determined.
RNA Viruses
In single-stranded RNA (ssRNA) viruses, the replicative intermediates often consist of dsRNA. For example, by using antisera specific for dsRNA, such molecules were detected in the cytoplasm of cells infected with rubella virus and Semliki Forest virus (183). In dsRNA viruses, the genome itself is the source of dsRNA. However, due to the enormous physiological effects of dsRNAs on the cell, many dsRNA viruses have evolved replication strategies to prevent exposure of dsRNAs to the cytoplasm of cells. In reoviruses, for example, the dsRNA genome remains inside the capsid throughout the viral life cycle and only after the sense strand RNA for progeny viruses is packaged into subviral particles is the second genomic strand synthesized (284). The secondary structure of RNAs may also contribute to the double-stranded nature of RNAs, as seen from human immunodeficiency virus (HIV)- and human T-cell leukemia virus-infected cells (108, 109, 247, 295).
It has been suggested that there might be a role for antisense regulation in the life cycle of HIV-1. Most HIV-1 transcripts from integrated genomes have positive-strand polarity and initiate from the 5′ long terminal repeat. However, Michael et al. (222) reported that HIV-1 transcripts with negative-strand polarity could be isolated from acutely and chronically infected cell lines, as well as from peripheral blood mononuclear cell samples from 15 HIV-1-infected patients. Promoter elements critical for negative-strand synthesis were identified and shown to be regulated in a reciprocal fashion to the positive-strand promoter.
DNA Viruses
For DNA viruses, dsRNA arises most often as a result of converging bidirectional transcription. Such RNAs produced from overlapping regions give rise to complementary transcripts. dsRNAs have been found in cells infected by a number of different DNA viruses including adenoviruses (208, 255), herpes simplex virus (141), polyomavirus (4, 76, 106, 309, 316, 329), simian virus 40 (8), and vaccinia virus (36, 62).
The mouse polyomavirus has served as one of the best model systems to study antisense-induced regulation of gene expression. This virus is small and depends heavily on the host for its gene expression. The double-stranded, circular polyomavirus genome is divided into early and late transcription units that are expressed from opposite strands of the viral genome (Fig. 1A). The life cycle of this virus is divided into two phases: the early phase, which occurs immediately after infection and before DNA replication, and the late phase, which begins after the onset of DNA replication. During the early phase of productive infection, early-strand transcripts accumulate preferentially over late-strand transcripts (20, 59, 83, 88, 89, 133, 134, 151, 200, 257). At late times there is a dramatic change in the pattern of gene expression, and the late-strand transcripts are much more abundant than the early-strand transcripts (20, 59, 83, 88, 89, 133, 134, 151, 200, 257). Regulation of both early- and late-strand RNA levels is posttranscriptional (83, 133). During the late phase of infection, RNA polymerase II encircles the genome multiple times (1–3, 5, 32, 83, 134, 315) (Fig. 1B). These giant multigenomic transcripts are the precursors to most late viral mRNAs. Further, these multigenomic RNAs form sense-antisense hybrids with early-strand transcripts (Fig. 1B) and thereby downregulate early-strand mRNA levels (176). Importantly, the antisense portion of the late-strand transcripts is within an intron which is removed during pre-mRNA processing and which remains exclusively within the nucleus. The mechanism of antisense regulation in this system has been elucidated and is described in more detail below.
FIG. 1.
Temporal regulation of polyomavirus transcript levels. (A) During the early phase of viral infection, early-strand transcripts accumulate preferentially over late-strand transcripts. Late-strand transcripts are processed inefficiently and are relatively unstable. Before DNA replication, the ratio of late-strand to early-strand RNAs is less than 1:10. (B) During the late phase of infection, after the onset of DNA replication, late-strand transcripts are more abundant than early-strand transcripts. Transcription termination is inefficient during this period, allowing RNA polymerase II to encircle the genome multiple times. The resulting multigenomic transcripts contain sequences complementary to early-strand transcripts and act as natural antisense regulators within the nucleus (166). Hatched lines denote transcripts that are downregulated posttranscriptionally.
CELLULAR STRATEGIES TO DEAL WITH CYTOPLASMIC DOUBLE-STRANDED RNA
Double-Stranded RNA Can Induce Interferon
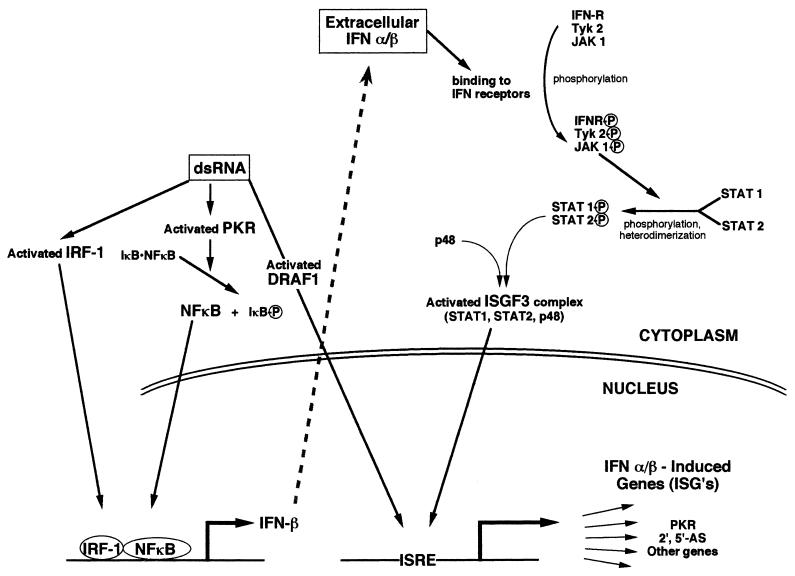
Duplex RNA molecules in the cytoplasm of cells can trigger a profound physiological reaction. As little as a single molecule of dsRNA is sufficient to induce the synthesis of interferon (IFN) (Fig. 2) (209). Since dsRNAs are formed in almost all viral infections, these can lead to the induction of IFN as well. Interferons are multifunctional cytokines that modulate host immunological functions and can inhibit tumor cell growth and virus multiplication. Most dsRNAs or virus infections induce type I IFNs, which include IFN-α and IFN-β. dsRNA [poly(rI-rC)] specifically induces IFN-β in mouse cells (155). In IFN-treated cells, dsRNA inhibits viral RNA and protein synthesis (reviewed in reference 192). Since the IFN pathways have been reviewed relatively recently (150), we only briefly discuss the most relevant aspects, which are also summarized in Fig. 2.
FIG. 2.
Signaling pathways of IFN-α, IFN-β, and cytoplasmic dsRNA. The major known pathways of signaling by cytoplasmic dsRNA and IFN-α/β are shown and are discussed in detail in the text. dsRNA directly activates PKR, IRF-1, and DRAF1. PKR phosphorylates IκB, which in turn leads to the nuclear localization of the transcription factor NF-κB. IRF-1 and NF-κB activate the IFN-β promoter, while DRAF1 can activate IFN-α- and IFN-β-induced genes (ISGs). IFN-β is secreted from cells (dashed arrow) and then binds to IFN receptors, leading to the activation of the signal transduction pathway shown, which itself stimulates the expression of ISGs.
Important cis-Acting Elements in the Alpha and Beta Interferon Promoters
A number of regulatory elements have been identified in the promoter of the IFN-β gene. There are four positive regulatory domains (PRD I to IV) (95, 96, 104, 105) and two negative regulatory domains (NRD I and II) (103, 104, 341). PRD I and III are binding sites for IFN regulatory factor I (IRF-1) (94, 212, 225, 268), PRD II is a binding site for NF-κB (190, 319), and PRD IV is recognized by activating transcription factor 2 (ATF-2/c-Jun) (74, 75). IRF-2 acts as a transcriptional repressor through the NRDs (114). Thanos and Maniatis (311) have suggested that virus induction of IFN-β gene expression in humans requires the assembly of a large complex (an enhanceosome) consisting of IRF-1, NF-κB, ATF-2, and the high-mobility group protein HMG I(Y). HMG I(Y) is not capable of transcriptional activation itself but is required for the binding as well as transcriptional activities of both NF-κB and ATF-2 (281, 311).
Although IFN-β and IFN-γ are produced from single-copy genes, IFN-α is expressed from a family of about 20 genes. All IFN-α genes have a virus-regulated element (VRE), which consists of two sets of repeats; the first contains multiple copies of the pentameric sequence CAGAA, and the other contains repeats of the octameric sequence A(A/T)GGAAAG. Both of these elements are important for virus inducibility of IFN-α (174, 275). The VRE of IFN-α contains a PRD I-like element whose exact sequence determines its ability to be induced by IRF-1 or viruses (206). No NF-κB binding sites are present.
Initiation of the Interferon Pathway by Double-Stranded RNA
A central player in cytoplasmic dsRNA activity is the dsRNA-activated protein kinase (PKR). Cells normally contain basal levels of PKR but in an unphosphorylated, inactive form. When dsRNA is introduced in the cells, PKR binds to and is activated by dsRNA, which induces autophosphorylation. Activated PKR can phosphorylate IκB, which in the unphosphorylated form is complexed with the transcription factor NF-κB and blocks its nuclear localization signal (175). Upon phosphorylation, IκB is released from NF-κB, which then translocates to the nucleus and activates transcription of genes having NF-κB binding sites. There is an NF-κB site in the PRD II element of the IFN-β gene promoter. dsRNA can also activate the expression of IFNs directly (265, 269, 325). It activates IRF-1, which can bind to the PRD I element present in the promoters of both IFN-α and IFN-β genes and may thus stimulate their expression (96, 256).
Following their synthesis, IFNs are secreted to neighboring cells, where they function as paracrine cytokines and induce a specific class of genes called IFN-stimulated genes (ISGs) (253). The activation of ISGs depends upon the presence of a promoter sequence called the IFN-stimulated response element (ISRE). The ISG factors (ISGFs) bind to this element and activate transcription from these genes.
Signal Transduction Pathway Activated by Interferons
Binding of IFN-α and IFN-β to their transmembrane receptors starts a signal transduction cascade that leads to the expression of ISGs (66, 68, 101, 137, 285). The pathway initiates with the receptor- and ligand-induced phosphorylation of Tyk and JAK kinases (68, 136, 332). Phosphorylated JAK kinases can subsequently phosphorylate signal transducers and activators of transcription (STATs), specifically STAT1 and STAT2 (68, 136). Activated STAT1 and STAT2 dimerize and together form the α subunit of ISGF3 (93, 286). The ISGF3α subunit then associates with the gamma subunit, p48, to form the active ISGF3 complex. This active ISGF3 complex translocates to the nucleus, binds to ISREs, and activates ISGs. A scheme for their mode of action is outlined in Fig. 2. The signal transduction cascades triggered by IFN-α and IFN-β are slightly different from that of IFN-γ, but they induce overlapping sets of genes.
Double-Stranded RNA Can Directly Induce Interferon-Stimulated and Other Genes
In addition to the IFN pathway of ISG induction, dsRNA can directly activate a number of genes, including ISGs (102, 112, 265, 325, 326). Daly and Reich (67) have reported the presence of two dsRNA-activated factors (DRAFs) that bind to the ISRE and flanking sequences and stimulate the expression of these genes directly, independent of the IFN pathway. One of these, DRAF1 (65, 67), is activated by dsRNA. DRAF1 can activate some but not all ISGs (67). Recently, Weaver et al. (328) further characterized the composition of DRAF1. DRAF1 includes IRF-3 and the transcriptional coactivators CREB binding protein and p300. This complex is dependent on dsRNA or viral induction. It has also been shown that both RNA viruses (Newcastle disease virus) and DNA viruses (adenovirus) can activate DRAF1, suggesting that this factor may be used widely for host defense against all types of viruses (65). It has also been reported that IRF-1, activated by dsRNA, can directly stimulate the expression of ISGs (11). Direct stimulation of ISRE-containing genes may be critical for the survival of virus-infected cells because it allows the antiviral effects to occur faster, without having to wait for the synthesis of IFN and the subsequent induction of the antiviral IFN pathway.
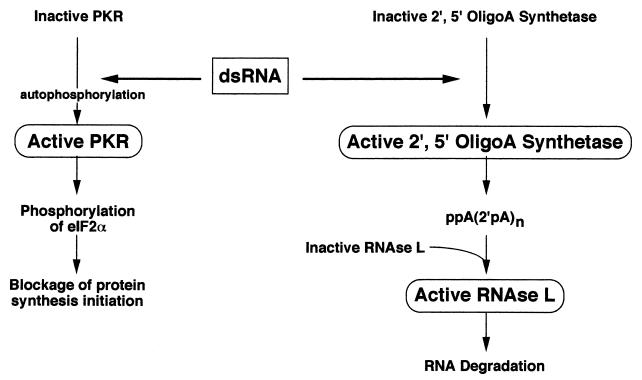
A large number of genes contain ISREs and can be activated by IFNs (150). Two of these are key players in pathways that are very important in the inhibition of cellular as well as viral growth. These are the RNA-dependent protein kinase (PKR) pathway and the 2′,5′-oligoadenylate synthetase (2′,5′-AS)/RNase L pathway (Fig. 3), and each of these is discussed in detail below.
FIG. 3.
Cytoplasmic effects of dsRNA. The PKR pathway and the 2′,5′-AS/RNase L pathway are directly activated by dsRNA, as described in the text. Activated PKR phosphorylates eIF-2α, which leads to the inhibition of protein synthesis initiation. Activated 2′,5′-AS generates oligoadenylates which activate RNase L, which can degrade viral and cellular RNAs.
RNA-Dependent Protein Kinase Pathway
A major pathway activated by IFNs and dsRNA is the PKR pathway (Fig. 2 and 3) (reviewed in references 263, 272, 279, 280, and 331). PKR is a serine/threonine kinase that is present in both the nucleus and the cytoplasm. About 20% of PKR exists in the nucleus (mainly in the nucleoli), and 80% is present in the cytoplasm (144, 145). A total of 80% of the cytoplasmic form of PKR is bound to ribosomes and is more phosphorylated than the nuclear form. PKR is normally present in an inactive or latent state (127, 191, 278) but can be activated by dsRNA.
Activated PKR can phosphorylate a number of substrates, including eukaryotic initiation factor 2α (eIF-2α) (97), IκB (175, 240), HIV Tat (215), and an unidentified cellular 90-kDa protein (270, 312). Phosphorylation of eIF2α has important consequences on cellular translation and can dramatically influence the cell’s response to viral infection. PKR phosphorylates eIF-2α at serine residue 51 (277). When phosphorylated, eIF-2α blocks the ability of eIF-2B to catalyze the guanine nucleotide exchange reaction required for protein synthesis initiation (57, 144, 279).
The human (221, 312), mouse (135, 306), and rat (248) PKR genes have been cloned. The PKR enzyme has two important functional domains: an N-terminal RNA binding domain and the C-terminal catalytic domain (279). The N-terminal half of PKR is necessary and sufficient for RNA binding. It binds dsRNA as well as structured ssRNA (87, 107, 154, 248). It does not require a specific sequence for activation and can be activated by very low concentrations (10 to 100 ng/ml) of perfectly duplexed dsRNAs but not by ssRNAs, DNA, RNA-DNA hybrids, or dsRNA duplexes containing modified bases. Short, highly structured viral ssRNAs may inhibit the activation of PKR. At high concentrations, long perfect dsRNAs also inhibit PKR. Activation of PKR by dsRNA most probably results from a conformational change induced by binding of dsRNA. Phosphorylation occurs at several sites within the N-terminal half (307).
Green and Mathews (107) proposed the hinge model for binding of PKR to dsRNA. This model would explain why PKR could bind dsRNAs with such a range of different sizes (22-mers to 80-mers or more). In this model, PKR has two dsRNA binding motifs (dsRBMs) separated by a hinge of about 20 amino acids. A single dsRBM is usually associated with a minimum of 11 bp of dsRNA (41, 157, 287). When dsRBMs bind RNA, the hinge can fold to different angles, to accommodate the binding of RNAs of different sizes. PKR is also known to require dimerization for activation (64, 179, 185, 249, 287, 313). Hunt and Ehrenfeld observed that at high concentrations of dsRNA, PKR is not activated (80, 132). In the dimer model, this would be explained if high concentrations of dsRNAs filled all dsRBMs, preventing two PKR molecules from dimerizing (272).
Some highly structured ssRNAs like the adenovirus VAI RNA can associate with PKR (55) yet can still inhibit its activity (see below). The signal for binding of dsRNA to PKR is the presence of two 2′-hydroxyl groups on the outside of a dsRNA. In highly structured ssRNAs, the two hydroxyl groups might be situated such that they mimic the 2′-hydroxyl groups in authentic dsRNAs (30).
In a recent report, a protein-protein interaction between STAT1 and PKR was shown (335). STAT1 is not a substrate for PKR phosphorylation, but in response to IFNs or dsRNA, the STAT1-PKR complex was dissociated concomitantly with an increase in the DNA binding activity of STAT1. This work suggests that PKR modulates the transcriptional activity of STAT1, which, if true, would further confirm the central role of PKR in both the dsRNA- and IFN-signaling pathways.
2′,5′-Oligoadenylate Synthetase/RNase L Pathway
Another pathway activated by dsRNA and IFNs is the 2′,5′-AS/RNase L pathway (Fig. 3). 2′,5′-AS is activated due to a conformational change that occurs on binding of dsRNA to the enzyme. Activated 2′,5′-AS is capable of polymerizing ATP and other nucleotides in novel 2′,5′ linkages (156). RNase L is activated by these 2′,5′-oligoadenylates. RNase L can cleave both cellular and viral RNAs and specifically cleaves ssRNAs at UpA, UpG, or UpU residues (10, 33, 90, 234, 289, 295, 336). RNase L can also exhibit antiviral effects by inducing apoptosis (45).
There are several distinct 2′,5′-AS activities, with different dsRNA requirements and subcellular locations (54, 138, 273, 282). Thus, they may act locally, to affect different cellular processes. About 70 bp of dsRNA is required for activation (224). The 110-kDa form requires low concentrations of dsRNA for activation and might be responsible for translational effects (128). The 67-kDa form is membrane associated and might affect signal transduction (54, 290). The 40- to 46-kDa nuclear form is associated with the nuclear matrix and activates an RNase that might cleave hnRNA (235).
Like PKR, 2′,5′-AS can bind to dsRNAs or ssRNAs with secondary structure. These substrates include adenovirus VAI RNA (72, 211), reovirus S1 mRNA (34), and HIV Tar RNA (78, 109, 207, 288, 292, 295). The consequences of binding of these RNAs are discussed below.
Other Effects of Double-Stranded RNA on Cells
dsRNA might inhibit protein synthesis in other ways. dsRNA can directly bind to and inactivate eIF-2, but this effect requires rather high intracellular concentrations of dsRNA (115, 149). In avian cells, dsRNA induces the secretion of a nuclease that degrades dsRNA (216). dsRNA sometimes inhibits cell growth in other ways. Low concentrations of dsRNA can inhibit the growth of some tumor cells (91, 318, 342), and this activity is sometimes independent of IFN (131, 196).
Cellular Proteins That Regulate RNA-Dependent Protein Kinase and 2′,5′-Oligoadenylate Synthetase Activities
A number of cellular factors interact with and regulate the activities of PKR and 2′,5′-AS. The most important of these are p58, p67, TAR RNA binding protein (TRBP), and the La autoantigen.
p58.
An inhibitor of PKR, p58 is found in human, monkey, bovine, and mouse cells (12, 186–189). p58 binds to PKR directly and inhibits autophosphorylation of PKR as well as phosphorylation of eIF-2α (262). This inhibitor is normally present in an inactive or latent state in the cell because it is complexed with an anti-inhibitor, p52. When p58 and PKR are coexpressed in yeast, p58 prevents PKR activation. However, when p52 was introduced into these strains, PKR activity as well as eIF-2α phosphorylation was restored (98). The anti-inhibitor and inhibitor can be separated by ammonium sulfate fractionation or during influenza virus infection (188). The p58 gene has been cloned and is a member of the tetratricopeptide family of proteins (12, 187). When p58 is overexpressed, cells in culture are transformed, perhaps as a result of an inhibition of PKR (12, 187).
p67.
The protein p67 is an eIF-2α-associated cellular protein (69) and can block eIF-2α phosphorylation by PKR (266). Serum starvation of cells leads to degradation of this inhibitor whereas subsequent mitogen stimulation induces its synthesis.
TRBP.
Human cellular TRBP (100) binds dsRNA (99, 247) and inhibits the activation of PKR in vitro (247). Overexpression of TRBP can reverse the effect of the vaccinia virus E3L deletion mutant (247). St. Johnston et al. (304) have identified a homologue of this protein in X. laevis. Using an infectious HIV-1 molecular clone, Benkirane et al. (27) have shown that overexpression of TRBP counteracts PKR-mediated inhibition of viral protein synthesis.
La autoantigen.
The La autoantigen can also regulate the activity of PKR. La is a 46.7-kDa cellular protein that is located in the nucleus as well as the cytoplasm of eukaryotic cells. It is an RNA binding protein and binds dsRNAs, snRNAs, and certain viral RNAs such as Epstein-Barr virus-encoded small RNAs (EBER) (56, 193) and VAI RNAs of human adenoviruses (92), which are discussed in greater detail below. The La autoantigen can also bind to the internal ribosome entry site of poliovirus transcripts and is required for efficient translation of the viral mRNAs (110, 217, 218). Xiao et al. (339) have demonstrated that the La autoantigen can inhibit dsRNA-dependent autophosphorylation of PKR as well as the ability of PKR to phosphorylate eIF-2α. Excess dsRNA can partially relieve this inhibition. These authors also reported that when the La autoantigen was incubated with synthetic or natural dsRNAs, it could unwind them and convert them into ssRNAs. It is therefore thought that the La autoantigen inhibits PKR activation by binding to dsRNA activators of PKR and converting them into ssRNAs that can no longer activate the kinase.
Other PKR inhibitors.
Oncogenic v-ras transformation of cells leads to the synthesis of a cellular PKR inhibitor. This 100-kDa inhibitor can act in trans to inhibit the auto phosphorylation and hence the activation of PKR (227). Little more is known about this factor. Also, a 15-kDa protein inhibitor of PKR is found in undifferentiated preadipocytes (147, 148). This protein is capable of blocking the interaction of PKR with dsRNA.
VIRAL STRATEGIES TO COUNTERACT HOST DEFENSE MECHANISMS
Many viruses have evolved ingenious strategies to counteract the antiviral effects of IFNs or of dsRNA expressed as a consequence of viral infection. There are a number of examples of this, deriving both from RNA viruses and DNA viruses and from viruses whose replication occurs in the cytoplasm or in the nucleus. Since a major cellular response to viral infections is an increase in the levels of cellular PKR, several viruses have devised strategies to downregulate and/or inactivate this kinase and thus prevent inhibition of protein synthesis. Most viruses use viral proteins or RNAs to downregulate PKR activity, either by direct interactions with PKR, thereby blocking its autophosphorylation and activation, or by sequestration of dsRNA activators of PKR. Other antiviral strategies may include activation of cellular proteins that may either degrade PKR, inhibit its activity, or inhibit the activity of the other major player in the antiviral response, namely, 2′,5′-AS.
DNA Viruses
Adenovirus.
VA RNA I is a small (160-nucleotide) viral RNA transcribed by RNA polymerase III and is expressed primarily at the late stages of infection, when it accumulates to very high concentrations (∼108 molecules/cell) in the cytoplasm (reviewed in reference 210). VA RNA II is a second RNA species, expressed to about ten-fold lower amounts than VA RNA I. Adenovirus mutant dl331, which does not synthesize VA RNA I, grows 10-fold less efficiently than the wild type (211). In this mutant, viral protein synthesis is inefficient late in infection. Host protein synthesis is also inhibited. Extracts of dl331-infected cells exhibit elevated levels of eIF-2α phosphorylation on serine 51 and reduced levels of guanine nucleotide exchange factor activity (211). In cells containing a serine 51-to-alanine mutation in eIF-2α, the effects of the dl331 mutation are suppressed. Therefore, it appears that VA RNA I may prevent the phosphorylation of eIF-2α kinase. VA RNA I acts by binding to PKR and inhibiting its activity. The viral molecule appears to bind PKR via an imperfectly duplexed stem loop structure (107). This binding apparently does not activate PKR because the duplex is too short and imperfect and may prevent the conformational change that occurs during activation when a long duplex RNA binds PKR.
EBV.
The human Epstein-Barr virus has two small RNAs, EBER-1 and EBER-2 (193). Like the adenovirus VA RNAs, these are small, are synthesized by RNA polymerase III, and exhibit extensi ve secondary structure (129). In vivo they interact with the La autoantigen (193). EBER-1 interacts with PKR in vitro and can inhibit its kinase activity (58, 81, 294).
Vaccinia virus.
Vaccinia virus uses several different mechanisms to inhibit the action of PKR. The viral E3L gene encodes two gene products that can bind dsRNA, sequester it away from PKR, and thus downregulate PKR activity (7, 52, 327, 340). If the E3L gene is deleted, there is a reversal of the kinase inhibitory effect, along with degradation of RNA, which occurs upon activation of the 2′,5′-AS/RNase L pathway (21). Binding of E3L to dsRNA appears to be necessary for vaccinia virus replication in human HeLa cells in culture, as seen in studies with E3L mutants (51). A second gene product of vaccinia virus, K3L, is also capable of inhibiting PKR activity. K3L has 28% identity to eIF-2α but lacks the important phosphorylation site which is found in native eIF-2α subunits (24). The fact that kinase regulation occurs in vivo in vaccinia virus-infected cells has been demonstrated with K3L-defective mutants (24). Therefore, it has been speculated that K3L regulates PKR by associating with PKR and inhibiting its autophosphorylation and activation, thereby preventing phosphorylation of the actual cellular eIF-2α subunit (24, 70, 142).
In addition, vaccinia virus expresses a 57-kDa protein which may inhibit the activity of 2′,5′-AS during viral infection (61, 244, 245). It is thought that p57 inhibits the 2′,5′-AS/RNase L pathway by sequestering its dsRNA activators.
Polyomavirus.
Whereas most DNA viruses express gene products (proteins or small RNAs) that interfere with PKR or 2′,5′-AS, the mouse polyomavirus uses a different strategy. Polyomavirus infection induces IFN (76, 106, 309, 316, 329). It has been reported recently that the polyomavirus large T antigen interferes with IFN-inducible gene expression (329). Cell lines derived from wild-type-virus-induced breast tumors are resistant to the growth-inhibitory action of IFN-β and IFN-γ. IFN-induced gene expression is blocked by wild-type virus but not by a mutant that lacks the pRB binding site of the viral large T antigen. The viral large T antigen inhibits IFN-inducible gene expression by binding to JAK-1 kinase and also inhibits the activation of ISGF3 (329). Overexpression of JAK-1 could reverse the IFN-inhibitory effect of the virus (329).
RNA Viruses
Poliovirus.
During poliovirus infection, cellular protein synthesis is inhibited at the stage of translation initiation due to proteolytic degradation of a 220-kDa component of eIF-4F (82, 171, 246, 310). eIF-4F is involved in the correct binding of capped mRNA to the 43S initiation complexes (220). Poliovirus mRNAs are uncapped and do not require p220 for its own mRNA translation, because initiation of translation occurs at an internal ribosome entry site (143). Furthermore, PKR was found to be degraded in poliovirus-infected cells (35). PKR is thought to be degraded by a cellular protease which is activated upon poliovirus infection, and the presence of dsRNA or structured ssRNA is necessary for kinase degradation (152).
Reovirus and influenza viruses.
Both reoviruses and influenza viruses use viral proteins to inactivate PKR. Reovirus uses the sigma 3 protein to inhibit PKR. The effect of this inhibition can be reversed by adding large amounts of dsRNA to in vitro reaction mixtures, suggesting that sigma 3 acts by sequestering dsRNA activators of PKR (139).
Influenza virus-infected cells exhibit a dramatic suppression of PKR activity and a decrease in the levels of eIF-2α phosphorylation (153). Moreover, it appears that viral protein synthesis and replication are required for kinase suppression (153). Influenza virus infection activates a 58-kDa, kinase-inhibitory activity, which is actually a cellular protein whose levels remain the same in mock- and virus-infected cells (189). In mock-infected cells, the 58-kDa suppressor protein is present in an inactive state because of its association with an inhibitor of p58 (I-p58). After virus infection, the inhibitor I-p58 may be released from p58. Active p58 is then able to block autophosphorylation of PKR as well as its kinase activity. In addition, the influenza virus NS1 protein can bind dsRNA (117) and PKR activity can be inhibited in vitro by NS1 (203, 264).
HIV.
HIV-1 has a stem-loop structure near the 5′ end of viral RNAs called the TAR element. This element can bind PKR (78, 108, 109, 207, 295, 340) and can inhibit its activation by dsRNA (108, 109). The interaction of TAR with PKR can be inhibited by the viral Tat protein (146). It has been reported recently that the viral Tat protein, which binds to the TAR element, not only is a substrate of PKR but also can inhibit it (38).
Conclusion
As should be evident from the above discussion, viruses have devised a wide variety of strategies to counteract host defense mechanisms against duplex RNA. As more viruses are studied in detail, yet more mechanisms or strategies may be revealed. Learning more about the molecular mechanisms by which viruses counteract the dsRNA and IFN systems may not only provide new insights into the underlying mechanisms of cellular responses to duplex RNA but also be of value in the design and implementation of more effective antiviral strategies.
CELLULAR STRATEGIES TO DEAL WITH NUCLEAR DOUBLE-STRANDED RNA
Whereas most viral dsRNAs are thought to be cytoplasmic, most naturally occurring antisense RNA is thought to act within the nucleus (63, 232). It is possible that many cRNAs are expressed within the nucleus, either by design (antisense regulation) or by unintended transcriptional readthrough. As far as is known, dsRNA within the nucleus does not trigger the PKR, IFN, or 2′,5′-AS pathways. What, then, is the fate of dsRNA in this compartment? As discussed above, claims have been made for a variety of effects of antisense RNA, including transcriptional regulation, inhibition of splicing, inhibition of mRNA transport, and induction of mRNA instability. Mechanistically, there are several distinct ways in which nuclear dsRNA molecules might be detected and resolved. They may be degraded by dsRNA-specific nucleases, unwound by dsRNA helicases, or edited by enzymes that modify dsRNAs.
Double-Stranded RNase
Are sense-antisense hybrids the targets of dsRNase activity within the nucleus? Cells might contain a nuclease(s) which is specific for dsRNA. There has been a recent report about the presence of a dsRNase in human cells (337). In this study, chimeric antisense oligonucleotides consisting of 2′-methoxy 5′- and 3′-flanking sequences on either side of an oligoribonucleotide gap were preincubated with sense RNAs and treated with cytosolic or nuclear extracts from T24 human bladder carcinoma cells, and the region of the RNA-RNA hybrid was cleaved. This cleavage was attributed to a dsRNase activity that has been partially purified from these cells. The significance and in vivo activity of such an enzyme is still unclear. However, it should be noted that when dsRNA was microinjected into Xenopus oocyte nuclei, it persisted for at least 16 h (16), suggesting that, at least in that system, dsRNAs are not rapidly destroyed within the nucleus.
Helicases
In theory, dsRNAs could be resolved within the cell by the action of helicases that unwind the two strands. A number of proteins which contain intrinsic RNA helicase activity have been identified (reviewed in references 303 and 324). However, there is no evidence that any of these factors acts on long duplexes within the nucleus. Rather, a primary function of such proteins is in the rearrangement of RNA-RNA interactions during RNA processing (303). Importantly, unwinding of duplexes might indeed be a common fate of most or all nuclear antisense RNA interactions. However, this fate might be achieved through the action of another family of enzymes, which are discussed below.
Adenosine Deaminases That Act on Double-Stranded RNA
Figure 4 summarizes what we believe are the consequences and fates of both short and long duplex RNAs formed within the nucleus. The most likely fate of duplex RNA in the nucleus is to be acted upon by a member of the class of enzymes, known as adenosine deaminases, that act on dsRNAs (DRADAs, dsRADs, or, more recently, ADARs [15]). In eukaryotes, a dsRNA unwinding and modifying activity was first discovered in the nucleus of X. laevis (16, 267). This enzyme, ADAR1, was subsequently found to be ubiquitous in the animal kingdom (323). ADAR activity is confined almost exclusively to the nucleus, although it was reported recently that a cytoplasmic form of this deaminase may be induced by IFN (198, 199, 250, 251). The first reports were that this enzyme might be a dsRNA-specific unwindase, but this proved not to be the case. ADARs catalyze the conversion of adenosines to inosines within dsRNA (17, 236, 322) by the mechanism of hydrolytic deamination (260). The resultant RNA contains I-U base pairs, which make the RNA duplex unstable and may lead to partial or complete unwinding (17). For ADAR1, the modified adenosines display a 5′-neighbor preference of U > A > C > G (258). The activity of this enzyme may require the presence of a metal ion but does not require any cofactors. In vitro studies suggest that the only substrates for this enzyme are perfectly or imperfectly duplexed RNAs. The activity cannot be competitively inhibited by ssRNA, dsDNA or ssDNA (321).
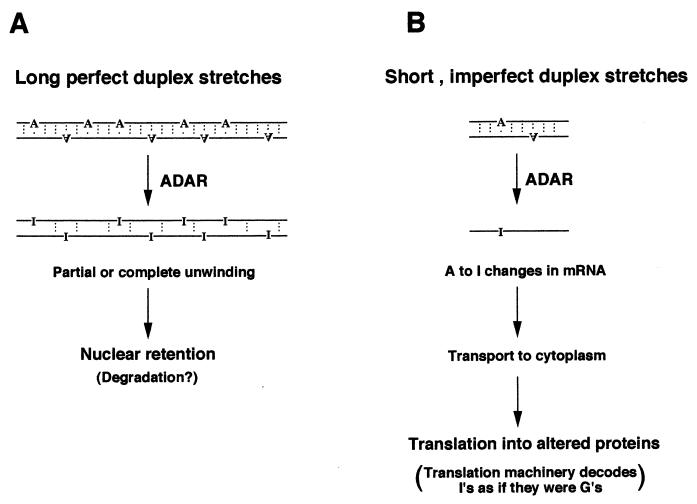
FIG. 4.
Nuclear effects of dsRNA. Antisense RNA within the nucleus most probably leads to adenosine modifications by a member of the ADAR family of dsRNA-dependent adenosine deaminases, as discussed in the text. (A) For long duplex stretches, extensive editing occurs. The two RNA strands are partially or fully unwound and are retained exclusively within the nucleus. (B) Short duplex stretches might lead to limited editing, with only one or a few adenosine-to-inosine modifications. Such edited mRNAs can be transported to the cytoplasm, where they are translated.
ADAR1 cDNAs have been cloned from humans (162, 239), rats (239), frogs (126), and, recently, mice (339a), and the protein has been purified from frogs (126), chickens (120), and cows (162, 239). More dsRNA-dependent deaminases have been identified (219), and a family of deaminases is now thought to exist (15). ADAR has been postulated to be involved in antisense RNA regulation (14, 161), a view supported by recent results with polyomavirus (reference 176 and discussion below).
ADAR editing of dsRNA is sensitive to the length of the duplex. Duplexes smaller than 15 bp are not modified in vitro (236), and optimal activity is seen with dsRNAs of 100 bp or longer (17, 236). This could be an important property physiologically, and it might help the enzyme discriminate between sequences that are to be modified at a few specific sites and those that are to be extensively edited (such as in antisense RNA regulation). A number of in vivo substrates for ADARs have been described. Most of these have been viral, but several cellular targets are known. A final and interesting characteristic of A-to-I editing is that I’s are recognized as if they were G’s by the cellular translation machinery. Such a change can never create a translation stop codon, and so all A-to-I changes are missense in character.
Editing of short (or imperfect) duplex regions. (i) Glutamate receptor.
Several transcripts of the mammalian glutamate receptor subunits are edited, including gluR-B, gluR-C, gluR-5, and gluR-6 (291). Of these, the first-identified and best-characterized editing is the one found to occur in exon 11 of the gluR-B, gluR-5, and gluR-6 transcripts (124, 300). As a result of the editing, a glutamine codon (CAG) is converted to an arginine codon (CIG). This site is called the Q/R site, and this editing results in an ion channel with altered calcium permeability. Another site which is edited is the R/G site, where an arginine codon (AGA) is converted into a glycine codon (IGA), and this event leads to an ion channel with altered kinetic properties (201). Editing at these sites occurs due to the presence of a double-stranded secondary structure formed by base pairing between an exon and the downstream intron (79, 119, 201). ADAR1 can edit only the R/G site efficiently, and not the Q/R site (205). Another enzyme related to ADAR1, called RED1 or ADAR2, was identified (219). It was subsequently shown by in vitro studies that ADAR2 was capable of editing the Q/R site as well as the R/G site (238). However, the in vivo editing specificities of these enzymes remain to be elucidated. Therefore, there may exist a family of deaminases with each member having overlapping yet distinct specificities. Another member of this family, RED2, has been identified by homology to ADARs (219), and its expression appears to be confined only to the brain, unlike ADAR1 and ADAR2, which are expressed in many tissues.
(ii) Serotonin receptor.
Transcripts encoding the 2C subtype of the neurotransmitter serotonin receptor undergo RNA-editing events in which genomically encoded adenosine residues are converted to inosines (40). The serotonin C receptor RNA also appears to be modified by adenosine deamination. These receptors are G-protein-coupled receptors, and the A-to-I editing event in 2C leads to the synthesis of altered proteins in which the interaction between the receptors and the G-proteins is reduced by about 10- to 15-fold (40). Interestingly, in this system, as with the glutamate receptor, editing requires the interaction of exon sequences with downstream intron sequences.
Editing of the HDV antigenomic RNA.
RNA editing plays an important role in the hepatitis delta virus (HDV) life cycle. In HDV antigenomic RNA, an amber codon is converted into a codon for tryptophan, thus extending the open reading frame product by 19 amino acids (44, 178, 204). This editing event most probably occurs by adenosine deamination (259) and is crucial for the viral life cycle because it helps to generate two different protein products from a single viral RNA. Formation of both these proteins is essential for virus multiplication. The smaller form of the delta antigen is required for replication, while the larger protein, formed after RNA editing, represses replication and is required for packaging (178). It has been shown recently that editing of the amber/W site is suppressed by the hepatitis delta antigen (261).
Other examples of endogenous RNA editing.
As discussed above, the 4f-rnp gene in Drosophila has been reported to be subject to possible antisense regulation (254) . In this system, cDNAs that contained extensive A-to-I or A-to-G changes were isolated. Also in Drosophila, the mRNA for the Para sodium channel in the brain is edited in a fashion similar to that reported for gluR and the serotonin receptor (113).
A-to-I Editing in Long (or Perfect) Duplexes and in Viruses
Adenosine modifications have been seen in a number of RNA viruses. In a persistent measles virus infection, A-to-G as well as U-to-C transitions have been seen (31, 46–49). The cDNA from the matrix gene had about 50% of its U’s modified to C’s. These U-to-C changes were shown to be the result of adenosine deaminations of the opposite strand (18). Multiple modifications have also been observed in other negative-strand RNA viruses, namely, vesicular stomatitis virus (241), human parainfluenza virus (230), and respiratory syncytial virus (274). Certain retroviral genomes also exhibit modifications. The U3 region of an oncogene c-mil transducing retrovirus, IC4, has almost half the A’s of its parental Rous sarcoma virus U3 sequence modified to G’s (86). The genesis of these mutations is not known but is suggestive of ADAR action. There are two reports of editing in HIV-1. The HIV-1 TAR RNA element shows a single A-to-I mutation, and this change alters viral gene expression (293). Also, Hajjar and Linial (111) described a recombinant HIV-1 provirus generated during in vitro passage that contains a short region of A-to-G hypermutation. The edited region was restricted to complementary sequences present in the recombinant provirus.
In the polyomavirus model system, nuclear antisense RNA leads to both extensive editing and nuclear retention.
Work with the polyomavirus model system suggests a mechanism by which long, naturally occurring antisense RNA might function in mammalian cells (176). In polyomavirus-infected mouse NIH 3T3 fibroblasts, natural antisense RNA to viral early-strand transcripts is produced at late times in infection (Fig. 1). This antisense RNA is responsible for the downregulation of viral early-gene expression late in infection. Analysis of early-strand transcripts isolated late in infection revealed extensive base modifications. In many transcripts, about half of the adenosines were altered to inosines or guanosines. Probes that could detect only modified RNAs revealed that these molecules are actually relatively stable and accumulate within the nucleus. Since they are retained in the nucleus, they are inert for gene expression, even though they could theoretically encode mutant viral proteins. Antisense RNA-induced modifications could account for much of the observed regulation, with the lowered levels of early-strand RNAs commonly observed late in infection resulting from the fact that many transcripts are invisible to standard hybridization probes, owing to noncomplementarity. Recent work has shown that extensively edited polyomavirus early-strand RNAs can be polyadenylated and even spliced (176a).
Since polyomavirus is small, the mechanism of antisense RNA regulation observed in this system probably reflects a common mechanism used in all higher eukaryotes. This mechanism involves extensive modifications, leading to RNA “invisibility” to common hybridization probes (perhaps accounting for reports of antisense-induced RNA “decay”). Further, these data lead to the very important conclusion that nuclear antisense RNA leads to nuclear retention of target transcripts.
I-RNase
Scadden and Smith (283) have demonstrated the presence of an RNase in extracts from HeLa cells, sheep uterus, and pig brain that can specifically degrade ssRNAs containing inosines. This enzyme is a 3′-5′ exonuclease that produces 5′ nucleoside monophosphates. The authors suggest that this enzyme is capable of degrading synthetic dsRNAs edited in vitro by the enzyme ADAR2 (RED1). However, the dsRNA substrates edited by ADAR2 were heat denatured to allow the formation of ssRNAs prior to incubation with the enzyme. It has been demonstrated that synthetic dsRNA substrates of ADAR1 that have undergone almost 50% editing of A’s to I’s are only partially unwound but never completely separated into single strands (17, 322). Since I-RNase is specific for ssRNAs, it is unclear whether the ADAR substrates which never become completely single stranded are degraded by I-RNase. Data obtained Kumar and Carmichael (176) showed that in polyomavirus-infected cells, dsRNAs that have about 50% of the adenosines modified to inosines are maintained stably within the nucleus and can be detected readily even after 6 h of actinomycin D treatment. This suggests that the I-RNase does not act on ADAR substrates in the nuclei of mammalian cells. Moreover, certain substrates of ADARs that have been selectively deaminated at several residues are transported to the cytoplasm stably and are translated to form proteins with altered amino acids e.g., gluR-B. These must also escape degradation by I-RNase in the nucleus as well as the cytoplasm of cells. Further work is required to clarify the role of this interesting enzyme in the cellular response to dsRNA.
Nuclear Antisense RNA Might Generally Induce Adenosine Modifications
As we have seen, most naturally occurring antisense RNA probably acts within the nucleus (63, 232). This would avoid the multitude of physiological effects associated with cytoplasmic dsRNA. The fact that overlapping bidirectional transcription might occur frequently within the nuclei of eukaryotic cells, coupled with the likelihood that long stretches of dsRNA are recognized and extensively modified by ADAR, points to the fact that there may be a novel pool of nuclear RNAs containing inosines (176, 252). Many or most of these RNAs may have been undetected so far because standard hybridization probes will fail to hybridize to them. Inosines have been identified in poly(A)+ mRNAs from various tissues, and it has been found that the levels of inosines correlate with the levels of ADAR expression (252). These data suggest that modification of sense-antisense hybrid RNAs by adenosine deamination may play a very important role in regulating gene expression.
Antisense RNA-mediated gene regulation in eukaryotes has been demonstrated only in the polyomavirus system so far. As seen above, it occurs in the nucleus by ADAR-mediated deamination and nuclear retention of target transcripts. Other eukaryotic antisense systems must be studied in greater detail with the aim of elucidating the nature of antisense regulation and determining whether they also involve ADAR-mediated deamination or some other mechanism(s).
CONCLUSIONS AND FUTURE DIRECTIONS
As we have seen, dsRNA has a number of profound effects on cells, but these effects differ depending on the nature and location of the duplexes. Understanding and appreciating the various pathways of cellular responses to dsRNA will help not only in studies designed to combat viral infections but also in the design and implementation of effective antisense RNA technologies to regulate cellular gene expression.
Finally, is there a wider role for nuclear antisense RNA in the regulation of gene expression? As we have seen, there is increasing evidence for the presence of antisense transcripts associated with the complementary strand of a gene (Table 1). It has been reported that a large fraction of vertebrate mRNAs (perhaps more than 30%) have conserved regions in their 3′ and 5′ UTRs (77). These conserved regions comprise unique sequences in the genome and show sequence conservation only between corresponding regions of orthologous mRNAs in other species. Why is there such strong conservation of these noncoding sequences? Lipman has recently suggested the very interesting model that the long stretches of conserved regions in 3′ UTRs might actually be involved in regulation of RNA stability via the formation of long, perfectly matched sense-antisense duplexes with complementary RNAs (197). In this model, duplexes with perfect matches could be targets of cellular regulatory machinery designed to destabilize or modify the sense transcripts, as a novel mode of regulation. If this is true, antisense RNA regulation would be far more prevalent than has been believed to date.
ACKNOWLEDGMENTS
We thank Yingqun Huang, Jed Podoloff, Susan Young, and Kim Wimler for helpful comments on the manuscript.
This work was supported by grant CA45382 from the National Cancer Institute.
REFERENCES
- 1.Acheson N. Transcription during productive infection with polyoma virus and SV40. Cell. 1976;8:1–12. doi: 10.1016/0092-8674(76)90179-3. [DOI] [PubMed] [Google Scholar]
- 2.Acheson N. Efficiency of processing of viral RNA during the early and late phases of productive infection by polyoma virus. J Virol. 1981;37:628–635. doi: 10.1128/jvi.37.2.628-635.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Acheson N. Kinetics and efficiency of polyadenylation of late polyomavirus nuclear RNA: generation of oligomeric polyadenylated RNAs and their processing into mRNA. Mol Cell Biol. 1984;4:722–729. doi: 10.1128/mcb.4.4.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Acheson N, Buetti E, Scherrer K, Weil R. Transcription of the polyoma virus genome: synthesis and cleavage of giant late polyoma specific RNA. Proc Natl Acad Sci USA. 1971;68:2231–2235. doi: 10.1073/pnas.68.9.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Acheson N H. Polyoma giant RNAs contain tandem repeats of the nucleotide sequence of the entire viral genome. Proc Natl Acad Sci USA. 1978;75:4754–4758. doi: 10.1073/pnas.75.10.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adelman J P, Bond C T, Douglass J, Herbert E. Two mammalian genes transcribed from opposite strands of the same DNA locus. Science. 1987;235:1514–1517. doi: 10.1126/science.3547652. [DOI] [PubMed] [Google Scholar]
- 7.Akkaraju G R, Whitaker-Dowling P, Youngner J S, Jagus R. Vaccinia specific kinase inhibitory factor prevents translational inhibition by double-stranded RNA in rabbit reticulocyte lysate. J Biol Chem. 1989;264:10321–10325. [PubMed] [Google Scholar]
- 8.Aloni Y. Extensive symmetrical transcription of simian virus 40 DNA in virus-yielding cells. Proc Natl Acad Sci USA. 1972;69:2404–2409. doi: 10.1073/pnas.69.9.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambros V, Moss E G. Heterochronic genes and the temporal control of C. elegans development. Trends Genet. 1994;10:123–127. doi: 10.1016/0168-9525(94)90213-5. [DOI] [PubMed] [Google Scholar]
- 10.Baglioni C, Maroney P A, West D K. 2′5′Oligo(A) polymerase activity and inhibition of viral RNA synthesis in interferon-treated HeLa cells. Biochemistry. 1979;18:1765–1770. doi: 10.1021/bi00576a020. [DOI] [PubMed] [Google Scholar]
- 11.Bandyopadhyay S K, Leonard G T, Bandyopadhyay T, Stark G R, Sen G C. Transcriptional induction by double-stranded RNA is mediated by interferon-stimulated response elements without activation of interferon-stimulated gene factor 3. J Biol Chem. 1995;270:19624–19629. doi: 10.1074/jbc.270.33.19624. [DOI] [PubMed] [Google Scholar]
- 12.Barber G N, Thompson S, Lee T G, Strom T, Jagus R, Darveau A, Katze M G. The 58-kilodalton inhibitor of the interferon-induced double-stranded RNA-activated protein kinase is a tetratricopeptide repeat protein with oncogenic properties. Proc Natl Acad Sci USA. 1994;91:4278–4282. doi: 10.1073/pnas.91.10.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barlow D P, Stoger R, Herrmann B G, Saito K, Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349:84–87. doi: 10.1038/349084a0. [DOI] [PubMed] [Google Scholar]
- 14.Bass B L. The dsRNA unwinding/modifying activity: fact and fiction. Semin Dev Biol. 1992;3:425–433. [Google Scholar]
- 15.Bass B L, Nishikura K, Keller W, Seeburg P H, Emeson R B, O’Connell M A, Samuel C E, Herbert A. A standardized nomenclature for adenosine deaminases that act on RNA. RNA. 1997;3:947–949. [PMC free article] [PubMed] [Google Scholar]
- 16.Bass B L, Weintraub H. A developmentally regulated activity that unwinds RNA duplexes. Cell. 1987;48:607–613. doi: 10.1016/0092-8674(87)90239-x. [DOI] [PubMed] [Google Scholar]
- 17.Bass B L, Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 18.Bass B L, Weintraub H, Cattaneo R, Billeter M A. Biased hypermutation of viral RNA genomes could be due to unwinding/modification of double-stranded RNA. Cell. 1989;56:331. doi: 10.1016/0092-8674(89)90234-1. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 19.Batshake B, Sundelin J. The mouse genes for the EP1 prostanoid receptor and the PKN protein kinase overlap. Biochem Biophys Res Commun. 1996;227:70–76. doi: 10.1006/bbrc.1996.1469. [DOI] [PubMed] [Google Scholar]
- 20.Beard P, Acheson N H, Maxwell I H. Strand-specific transcription of polyoma virus DNA early in productive infection and in transformed cells. J Virol. 1976;17:20–26. doi: 10.1128/jvi.17.1.20-26.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beattie E, Denzler K L, Tartaglia J, Perkus M E, Paoletti E, Jacobs B L. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J Virol. 1995;69:499–505. doi: 10.1128/jvi.69.1.499-505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beattie E, Kauffman E B, Martinez H, Perkus M E, Jacobs B L, Paoletti E, Tartaglia J. Host-range restriction of vaccinia virus E3L-specific deletion mutants. Virus Genes. 1996;12:89–94. doi: 10.1007/BF00370005. [DOI] [PubMed] [Google Scholar]
- 23.Beattie E, Paoletti E, Tartaglia J. Distinct patterns of IFN sensitivity observed in cells infected with vaccinia K3L− and E3L− mutant viruses. Virology. 1995;210:254–263. doi: 10.1006/viro.1995.1342. [DOI] [PubMed] [Google Scholar]
- 24.Beattie E, Tartaglia J, Paoletti E. Vaccinia virus-encoded eIF-2 alpha homolog abrogates the antiviral effect of interferon. Virology. 1991;183:419–422. doi: 10.1016/0042-6822(91)90158-8. [DOI] [PubMed] [Google Scholar]
- 25.Belhumeur P, Lussier M, Skup D. Expression of naturally occurring RNA molecules complementary to the murine L27 ribosomal protein mRNA. Gene. 1988;72:277–285. doi: 10.1016/0378-1119(88)90153-9. [DOI] [PubMed] [Google Scholar]
- 26.Bender T P, Thompson C B, Kuehl W M. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987;237:1473–1476. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- 27.Benkirane M, Neuveut C, Chun R F, Smith S M, Samuel C E, Gatignol A, Jeang K T. Oncogenic potential of TAR RNA binding protein TRBP and its regulatory interaction with RNA-dependent protein kinase PKR. EMBO J. 1997;16:611–624. doi: 10.1093/emboj/16.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bentley D L, Groudine M. A block to elongation is largely responsible for decreased transcription in c-myc in differentiated HL60 cells. Nature. 1986;312:702–706. doi: 10.1038/321702a0. [DOI] [PubMed] [Google Scholar]
- 29.Bester A J, Kennedy D S, Heywood S M. Two classes of translational control RNA: their role in the regulation of protein synthesis. Proc Natl Acad Sci USA. 1975;72:1523–1527. doi: 10.1073/pnas.72.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bevilacqua P C, Cech T R. Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry. 1996;35:9983–9994. doi: 10.1021/bi9607259. [DOI] [PubMed] [Google Scholar]
- 31.Billeter M A, Cattaneo R, Spielhofer P, Kaelin K, Huber M, Schmid A, Baczko K, ter Meulen V. Generation and properties of measles virus mutations typically associated with subacute sclerosing panencephalitis. Ann NY Acad Sci. 1994;724:367–377. doi: 10.1111/j.1749-6632.1994.tb38934.x. [DOI] [PubMed] [Google Scholar]
- 32.Birg F, Favaloro J, Kamen R. Analysis of polyoma viral nuclear RNA by miniblot hybridization. Proc Natl Acad Sci USA. 1977;74:3138–3142. doi: 10.1073/pnas.74.8.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bisbal C, Salehzada T, Lebleu B, Bayard B. Characterization of two murine (2′-5′)(A)n-dependent endonucleases of different molecular mass. Eur J Biochem. 1989;179:595–602. doi: 10.1111/j.1432-1033.1989.tb14588.x. [DOI] [PubMed] [Google Scholar]
- 34.Bischoff J R, Samuel C E. Mechanism of interferon action. Activation of the human P1/eIF-2 alpha protein kinase by individual reovirus s-class mRNAs: s1 mRNA is a potent activator relative to s4 mRNA. Virology. 1989;172:106–115. doi: 10.1016/0042-6822(89)90112-8. [DOI] [PubMed] [Google Scholar]
- 35.Black T L, Safer B, Hovanessian A, Katze M G. The cellular 68,000-Mr protein kinase is highly autophosphorylated and activated yet significantly degraded during poliovirus infection: implications for translational regulation. J Virol. 1989;63:2244–2251. doi: 10.1128/jvi.63.5.2244-2251.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boone R F, Parr R P, Moss B. Intermolecular duplexes formed from polyadenylylated vaccinia virus RNA. J Virol. 1979;30:365–374. doi: 10.1128/jvi.30.1.365-374.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borja A Z, Meijers C, Zeller R. Expression of alternatively spliced bFGF first coding exons and antisense mRNAs during chicken embryogenesis. Dev Biol. 1993;157:110–118. doi: 10.1006/dbio.1993.1116. [DOI] [PubMed] [Google Scholar]
- 38.Brand S R, Kobayashi R, Mathews M B. The Tat protein of human immunodeficiency virus type 1 is a substrate and inhibitor of the interferon-induced, virally activated protein kinase, PKR. J Biol Chem. 1997;272:8388–8395. doi: 10.1074/jbc.272.13.8388. [DOI] [PubMed] [Google Scholar]
- 39.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 40.Burns C M, Chu H, Rueter S M, Hutchinson L K, Canton H, Sanders-Bush E, Emeson R B. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 41.Bycroft M, Grunert S, Murzin A G, Proctor M, St. Johnston D. NMR solution structure of a dsRNA binding domain from Drosophila staufen protein reveals homology to the N-terminal domain of ribosomal protein S5. EMBO J. 1995;14:3563–3571. doi: 10.1002/j.1460-2075.1995.tb07362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Calvet J P, Pederson T. Secondary structure of heterogeneous nuclear RNA: two classes of double-stranded RNA in native ribonucleoprotein. Proc Natl Acad Sci USA. 1977;74:3705–3709. doi: 10.1073/pnas.74.9.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Campbell C E, Huang A, Gurney A L, Kessler P M, Hewitt J A, Williams B R. Antisense transcripts and protein binding motifs within the Wilms tumour (WT1) locus. Oncogene. 1994;9:583–595. [PubMed] [Google Scholar]
- 44.Casey J L, Gerin J L. Hepatitis D virus RNA editing: specific modification of adenosine in the antigenomic RNA. J Virol. 1995;69:7593–7600. doi: 10.1128/jvi.69.12.7593-7600.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Castelli J C, Hassel B A, Wood K A, Li X L, Amemiya K, Dalakas M C, Torrence P F, Youle R J. A study of the interferon antiviral mechanism: apoptosis activation by the 2-5A system. J Exp Med. 1997;186:967–972. doi: 10.1084/jem.186.6.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cattaneo R. Biased (A → I) hypermutation of animal RNA virus genomes. Curr Opin Genet Dev. 1994;4:895–900. doi: 10.1016/0959-437x(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 47.Cattaneo R. RNA editing. RNA duplexes guide base conversions. Curr Biol. 1994;4:134–136. doi: 10.1016/s0960-9822(94)00030-8. [DOI] [PubMed] [Google Scholar]
- 48.Cattaneo R, Billeter M A. Mutations and A/I hypermutations in measles virus persistent infections. Curr Top Microbiol Immunol. 1992;176:63–74. doi: 10.1007/978-3-642-77011-1_5. [DOI] [PubMed] [Google Scholar]
- 49.Cattaneo R, Schmid A, Eschle D, Baczko K, ter Meulen V, Billeter M A. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell. 1988;55:255–265. doi: 10.1016/0092-8674(88)90048-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chalfie M, Horvitz H R, Sulston J E. Mutations that lead to reiterations in the cell lineages of C. elegans. Cell. 1981;24:59–69. doi: 10.1016/0092-8674(81)90501-8. [DOI] [PubMed] [Google Scholar]
- 51.Chang H W, Uribe L H, Jacobs B L. Rescue of vaccinia virus lacking the E3L gene by mutants of E3L. J Virol. 1995;69:6605–6608. doi: 10.1128/jvi.69.10.6605-6608.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang H W, Watson J C, Jacobs B L. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc Natl Acad Sci USA. 1992;89:4825–4829. doi: 10.1073/pnas.89.11.4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang Y, Spicer D B, Sonenshein G E. Effects of IL-3 on promoter usage, attenuation and antisense transcription of the c-myc oncogene in the IL-3-dependent Ba/F3 early pre-B cell line. Oncogene. 1991;6:1979–1982. [PubMed] [Google Scholar]
- 54.Chebath J, Benech P, Hovanessian A, Galabru J, Revel M. Four different forms of interferon-induced 2′,5′-oligo(A) synthetase identified by immunoblotting in human cells. J Biol Chem. 1987;262:3852–3857. [PubMed] [Google Scholar]
- 55.Clarke P A, Mathews M B. Interactions between the double-stranded RNA binding motif and RNA: definition of the binding site for the interferon-induced protein kinase DAI (PKR) on adenovirus VA RNA. RNA. 1995;1:7–20. [PMC free article] [PubMed] [Google Scholar]
- 56.Clemens M J. The small RNAs of Epstein-Barr virus. Mol Biol Rep. 1993;17:81–92. doi: 10.1007/BF00996215. [DOI] [PubMed] [Google Scholar]
- 57.Clemens M J. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. Protein kinases that phosphorylate eIF-2 and eIF-2B, and their role in eukaryotic cell translational control; pp. 139–172. [Google Scholar]
- 58.Clemens M J, Laing K G, Jeffrey I W, Schofield A, Sharp T V, Elia A, Matys V, James M C, Tilleray V J. Regulation of the interferon-inducible eIF-2 alpha protein kinase by small RNAs. Biochimie. 1994;76:770–778. doi: 10.1016/0300-9084(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 59.Cogen B. Virus-specific early RNA in 3T6 cells infected by a tsA mutant of polyoma virus. Virology. 1978;85:222–230. doi: 10.1016/0042-6822(78)90426-9. [DOI] [PubMed] [Google Scholar]
- 60.Cohen R B, Boal T R, Safer B. Increased eIF-2 alpha expression in mitogen-activated primary T lymphocytes. EMBO J. 1990;9:3831–3837. doi: 10.1002/j.1460-2075.1990.tb07601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cohrs R J, Condit R C, Pacha R F, Thompson C L, Sharma O K. Modulation of ppp(A2′p)nA-dependent RNase by a temperature-sensitive mutant of vaccinia virus. J Virol. 1989;63:948–951. doi: 10.1128/jvi.63.2.948-951.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Colby C, Jurale C, Kates J R. Mechanism of synthesis of vaccinia virus double-stranded ribonucleic acid in vivo and in vitro. J Virol. 1971;7:71–76. doi: 10.1128/jvi.7.1.71-76.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cornelissen M. Nuclear and cytoplasmic sites for anti-sense control. Nucleic Acids Res. 1989;17:7203–7209. doi: 10.1093/nar/17.18.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cosentino G P, Venkatesan S, Serluca F C, Green S R, Mathews M B, Sonenberg N. Double-stranded-RNA-dependent protein kinase and TAR RNA-binding protein form homo- and heterodimers in vivo. Proc Natl Acad Sci USA. 1995;92:9445–9449. doi: 10.1073/pnas.92.21.9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Daly C, Reich N C. Double-stranded RNA activates novel factors that bind to the interferon-stimulated response element. Mol Cell Biol. 1993;13:3756–3764. doi: 10.1128/mcb.13.6.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Daly C, Reich N C. Receptor to nucleus signaling via tyrosine phosphorylation of the p91 transcription factor. Trends Endocrinol Metab. 1994;5:159–164. doi: 10.1016/1043-2760(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 67.Daly C, Reich N C. Characterization of specific DNA-binding factors activated by double-stranded RNA as positive regulators of interferon alpha/beta-stimulated genes. J Biol Chem. 1995;270:23739–23746. doi: 10.1074/jbc.270.40.23739. [DOI] [PubMed] [Google Scholar]
- 68.Darnell J E, Jr, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 69.Datta B, Ray M K, Chakrabarti D, Wylie D E, Gupta N K. Glycosylation of eukaryotic peptide chain initiation factor 2 (eIF-2)-associated 67-kDa polypeptide (p67) and its possible role in the inhibition of eIF-2 kinase-catalyzed phosphorylation of the eIF-2 alpha-subunit. J Biol Chem. 1989;264:20620–20624. [PubMed] [Google Scholar]
- 70.Davies M V, Elroystein O, Jagus R, Moss B, Kaufman R J. The vaccinia virus K3L-gene product potentiates translation by inhibiting double-stranded-RNA-activated protein kinase and phosphorylation of the alpha subunit of eukaryotic initiation factor-2. J Virol. 1992;66:1943–1950. doi: 10.1128/jvi.66.4.1943-1950.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dean M, Kent R B, Sonenshein G E. Transcriptional activation of immunoglobulin alpha heavy-chain genes by translocation of the c-myc oncogene. Nature. 1983;305:443–446. doi: 10.1038/305443a0. [DOI] [PubMed] [Google Scholar]
- 72.Desai S Y, Patel R C, Sen G C, Malhotra P, Ghadge G D, Thimmapaya B. Activation of interferon-inducible 2′-5′ oligoadenylate synthetase by adenoviral VAI RNA. J Biol Chem. 1995;270:3454–3461. doi: 10.1074/jbc.270.7.3454. [DOI] [PubMed] [Google Scholar]
- 73.Dony C, Kessel M, Gruss P. Post-transcriptional control of myc and p53 expression during differentiation of the embryonal carcinoma cell line F9. Nature. 1985;317:636–639. doi: 10.1038/317636a0. [DOI] [PubMed] [Google Scholar]
- 74.Du W, Maniatis T. An ATF/CREB binding site is required for virus induction of the human interferon beta gene. Proc Natl Acad Sci USA. 1992;89:2150–2154. doi: 10.1073/pnas.89.6.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Du W, Thanos D, Maniatis T. Mechanisms of transcriptional synergism between distinct virus-inducible enhancer elements. Cell. 1993;74:887–898. doi: 10.1016/0092-8674(93)90468-6. [DOI] [PubMed] [Google Scholar]
- 76.Dulbecco R, Johnson T. Interferon-sensitivity of the enhanced incorporation of thymidine into cellular DNA induced by polyoma virus. Virology. 1970;42:368–374. doi: 10.1016/0042-6822(70)90280-1. [DOI] [PubMed] [Google Scholar]
- 77.Duret L, Dorkeld F, Gautier C. Strong conservation of non-coding sequences during vertebrates evolution: potential involvement in post-transcriptional regulation of gene expression. Nucleic Acids Res. 1993;21:2315–2322. doi: 10.1093/nar/21.10.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Edery I, Petryshyn R, Sonenberg N. Activation of double-stranded RNA-dependent kinase (dsl) by the TAR region of HIV-1 mRNA: a novel translational control mechanism. Cell. 1989;56:303–312. doi: 10.1016/0092-8674(89)90904-5. [DOI] [PubMed] [Google Scholar]
- 79.Egebjerg J, Kukekov V, Heinemann S F. Intron sequence directs RNA editing of the glutamate receptor subunit GluR2 coding sequence. Proc Natl Acad Sci USA. 1994;91:10270–10274. doi: 10.1073/pnas.91.22.10270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ehrenfeld E, Hunt T. Double-stranded poliovirus RNA inhibits initiation of protein synthesis by reticulocyte lysates. Proc Natl Acad Sci USA. 1971;68:1075–1078. doi: 10.1073/pnas.68.5.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Elia A, Laing K G, Schofield A, Tilleray V J, Clemens M J. Regulation of the double-stranded RNA-dependent protein kinase PKR by RNAs encoded by a repeated sequence in the Epstein-Barr virus genome. Nucleic Acids Res. 1996;24:4471–4478. doi: 10.1093/nar/24.22.4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Etchison D, Milburn S C, Edery I, Sonenberg N, Hershey J W. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- 83.Farmerie W G, Folk W R. Regulation of polyoma-virus transcription by large tumor antigen. Proc Natl Acad Sci USA. 1984;81:6919–6923. doi: 10.1073/pnas.81.22.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Farrell C M, Lukens L N. Naturally occurring antisense transcripts are present in chick embryo chondrocytes simultaneously with the down-regulation of the alpha 1 (I) collagen gene. J Biol Chem. 1995;270:3400–3408. doi: 10.1074/jbc.270.7.3400. [DOI] [PubMed] [Google Scholar]
- 85.Fedoroff N, Wellauer P K, Wall R. Intermolecular duplexes in heterogeneous nuclear RNA from HeLa cells. Cell. 1977;10:597–610. doi: 10.1016/0092-8674(77)90092-7. [DOI] [PubMed] [Google Scholar]
- 86.Felder M P, Laugier D, Yatsula B, Dezelee P, Calothy G, Marx M. Functional and biological properties of an avian variant long terminal repeat containing multiple A-to-G conversions in the U3 sequence. J Virol. 1994;68:4759–4767. doi: 10.1128/jvi.68.8.4759-4767.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Feng G S, Chong K, Kumar A, Williams B R. Identification of double-stranded RNA-binding domains in the interferon-induced double-stranded RNA-activated p68 kinase. Proc Natl Acad Sci USA. 1992;89:5447–5451. doi: 10.1073/pnas.89.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fenton R G, Basilico C. Changes in the topography of early region transcription during polyomavirus lytic infection. Proc Natl Acad Sci USA. 1982;79:7142–7146. doi: 10.1073/pnas.79.23.7142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fenton R G, Basilico C. Regulation of polyoma virus early transcription in transformed cells by large T-antigen. Virology. 1982;121:384–392. doi: 10.1016/0042-6822(82)90176-3. [DOI] [PubMed] [Google Scholar]
- 90.Floyd-Smith G, Slattery E, Lengyel P. Interferon action: RNA cleavage pattern of a (2′-5′)oligoadenylate-dependent endonuclease. Science. 1981;212:1030–1032. doi: 10.1126/science.6165080. [DOI] [PubMed] [Google Scholar]
- 91.Forsberg K, Paulsson Y, Westermark B. Effect on platelet-derived growth factor-induced mitogenesis of double-stranded RNA: evidence for an autocrine growth inhibition mediated by interferon-beta. J Cell Physiol. 1988;136:266–272. doi: 10.1002/jcp.1041360208. [DOI] [PubMed] [Google Scholar]
- 92.Francoeur A M, Mathews M B. Interaction between VA RNA and the lupus antigen La: formation of a ribonucleoprotein particle in vitro. Proc Natl Acad Sci USA. 1982;79:6772–6776. doi: 10.1073/pnas.79.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fu X Y, Schindler C, Improta T, Aebersold R, Darnell J E., Jr The proteins of ISGF-3, the interferon alpha-induced transcriptional activator, define a gene family involved in signal transduction. Proc Natl Acad Sci USA. 1992;89:7840–7843. doi: 10.1073/pnas.89.16.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fujita T, Kimura Y, Miyamoto M, Barsoumian E L, Taniguchi T. Induction of endogenous IFN-alpha and IFN-beta genes by a regulatory transcription factor, IRF-1. Nature. 1989;337:270–272. doi: 10.1038/337270a0. [DOI] [PubMed] [Google Scholar]
- 95.Fujita T, Ohno S, Yasumitsu H, Taniguchi T. Delimitation and properties of DNA sequences required for the regulated expression of human interferon-beta gene. Cell. 1985;41:489–496. doi: 10.1016/s0092-8674(85)80022-2. [DOI] [PubMed] [Google Scholar]
- 96.Fujita T, Shibuya H, Hotta H, Yamanishi K, Taniguchi T. Interferon-beta gene regulation: tandemly repeated sequences of a synthetic 6 bp oligomer function as a virus-inducible enhancer. Cell. 1987;49:357–367. doi: 10.1016/0092-8674(87)90288-1. [DOI] [PubMed] [Google Scholar]
- 97.Galabru J, Hovanessian A. Autophosphorylation of the protein kinase dependent on double-stranded RNA. J Biol Chem. 1987;262:15538–15544. [PubMed] [Google Scholar]
- 98.Gale M, Jr, Blakely C M, Hopkins D A, Melville M W, Wambach M, Romano P R, Katze M G. Regulation of interferon-induced protein kinase PKR: modulation of P58IPK inhibitory function by a novel protein, P52rIPK. Mol Cell Biol. 1998;18:859–871. doi: 10.1128/mcb.18.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gatignol A, Buckler C, Jeang K T. Relatedness of an RNA-binding motif in human immunodeficiency virus type 1 TAR RNA-binding protein TRBP to human P1/dsI kinase and Drosophila staufen. Mol Cell Biol. 1993;13:2193–2202. doi: 10.1128/mcb.13.4.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gatignol A, Buckler-White A, Berkhout B, Jeang K T. Characterization of a human tar RNA-binding protein that activates the HIV-1 LTR. Science. 1991;251:1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- 101.Gilmour K C, Reich N C. Signal transduction and activation of gene transcription by interferons. Gene Expression. 1995;5:1–18. [PMC free article] [PubMed] [Google Scholar]
- 102.Goldfeld A E, Maniatis T. Coordinate viral induction of tumor necrosis factor alpha and interferon beta in human B cells and monocytes. Proc Natl Acad Sci USA. 1989;86:1490–1494. doi: 10.1073/pnas.86.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Goodbourn S, Burstein H, Maniatis T. The human beta-interferon gene enhancer is under negative control. Cell. 1986;45:601–610. doi: 10.1016/0092-8674(86)90292-8. [DOI] [PubMed] [Google Scholar]
- 104.Goodbourn S, Maniatis T. Overlapping positive and negative regulatory domains of the human beta- interferon gene. Proc Natl Acad Sci USA. 1988;85:1447–1451. doi: 10.1073/pnas.85.5.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goodbourn S, Zinn K, Maniatis T. Human beta-interferon gene expression is regulated by an inducible enhancer element. Cell. 1985;41:509–520. doi: 10.1016/s0092-8674(85)80024-6. [DOI] [PubMed] [Google Scholar]
- 106.Gotlieb-Stematsky T, Rotem Z, Karby S. Production and susceptibility to interferon of polyoma virus variants of high and low oncogenic properties. J Natl Cancer Inst. 1966;37:99–103. [PubMed] [Google Scholar]
- 107.Green S R, Mathews M B. Two RNA-binding motifs in the double-stranded RNA-activated protein kinase, DAI. Genes Dev. 1992;6:2478–2490. doi: 10.1101/gad.6.12b.2478. [DOI] [PubMed] [Google Scholar]
- 108.Gunnery S, Green S R, Mathews M B. Tat-responsive region RNA of human immunodeficiency virus type 1 stimulates protein synthesis in vivo and in vitro: relationship between structure and function. Proc Natl Acad Sci USA. 1992;89:11557–11561. doi: 10.1073/pnas.89.23.11557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gunnery S, Rice A P, Robertson H D, Mathews M B. Tat-responsive region RNA of human immunodeficiency virus 1 can prevent activation of the double-stranded-RNA-activated protein kinase. Proc Natl Acad Sci USA. 1990;87:8687–8691. doi: 10.1073/pnas.87.22.8687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gutierrez-Escolano L, del Angel R M. Nuclear proteins bind to poliovirus 5′ untranslated region. Arch Med Res. 1996;27:413–419. [PubMed] [Google Scholar]
- 111.Hajjar A M, Linial M L. Modification of retroviral RNA by double-stranded RNA adenosine deaminase. J Virol. 1995;69:5878–5882. doi: 10.1128/jvi.69.9.5878-5882.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hall D J, Jones S D, Kaplan D R, Whitman M, Rollins B J, Stiles C D. Evidence for a novel signal transduction pathway activated by platelet-derived growth factor and by double-stranded RNA. Mol Cell Biol. 1989;9:1705–1713. doi: 10.1128/mcb.9.4.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hanrahan, C. J., M. J. Palladino, L. J. Bonneau, and R. A. Reenan. RNA editing of a Drosophila sodium channel. Ann. N. Y. Acad. Sci., in press. [DOI] [PubMed]
- 114.Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T, Taniguchi T. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- 115.Harary R, Kaempfer R. Distinct epitopes in eukaryotic initiation factor 2 for binding of mRNA and for ternary complex formation with methionyl-tRNA(f) and GTP. Biochim Biophys Acta. 1990;1050:129–133. doi: 10.1016/0167-4781(90)90153-s. [DOI] [PubMed] [Google Scholar]
- 116.Hastings M L, Milcarek C, Martincic K, Peterson M L, Munroe S H. Expression of the thyroid hormone receptor gene, erbAalpha, in B lymphocytes: alternative mRNA processing is independent of differentiation but correlates with antisense RNA levels. Nucleic Acids Res. 1997;25:4296–4300. doi: 10.1093/nar/25.21.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hatada E, Fukuda R. Binding of influenza A virus NS1 protein to dsRNA in vitro. J Gen Virol. 1992;73:3325–3329. doi: 10.1099/0022-1317-73-12-3325. [DOI] [PubMed] [Google Scholar]
- 118.Henikoff S, Keene M A, Fechtel K, Fristrom J W. Gene within a gene: nested Drosophila genes encode unrelated proteins on opposite DNA strands. Cell. 1986;44:33–42. doi: 10.1016/0092-8674(86)90482-4. [DOI] [PubMed] [Google Scholar]
- 119.Herb A, Higuchi M, Sprengel R, Seeburg P H. Q/R site editing in kainate receptor GluR5 and GluR6 pre-mRNAs requires distant intronic sequences. Proc Natl Acad Sci USA. 1996;93:1875–1880. doi: 10.1073/pnas.93.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Herbert A, Lowenhaupt K, Spitzner J, Rich A. Chicken double-stranded RNA adenosine deaminase has apparent specificity for Z-DNA. Proc Natl Acad Sci USA. 1995;92:7550–7554. doi: 10.1073/pnas.92.16.7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Heywood S M. tcRNA as a naturally occurring antisense RNA in eukaryotes. Nucleic Acids Res. 1986;14:6771–6772. doi: 10.1093/nar/14.16.6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Heywood S M, Kennedy D S. Purification of myosin translational control RNA and its interaction with myosin messenger RNA. Biochemistry. 1976;15:3314–3319. doi: 10.1021/bi00660a023. [DOI] [PubMed] [Google Scholar]
- 123.Heywood S M, Kennedy D S, Bester A J. Studies concerning the mechanism by which translational-control RNA regulates protein synthesis in embryonic muscle. Eur J Biochem. 1975;58:587–593. doi: 10.1111/j.1432-1033.1975.tb02409.x. [DOI] [PubMed] [Google Scholar]
- 124.Higuchi M, Single F N, Köhler M, Sommer B, Sprengel R, Seeburg P H. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 125.Hildebrandt M, Nellen W. Differential antisense transcription from the Dictyostelium EB4 gene locus: implications on antisense-mediated regulation of mRNA stability. Cell. 1992;69:197–204. doi: 10.1016/0092-8674(92)90130-5. [DOI] [PubMed] [Google Scholar]
- 126.Hough R F, Bass B L. Purification of the Xenopus laevis double-stranded RNA adenosine deaminase. J Biol Chem. 1994;269:9933–9939. [PubMed] [Google Scholar]
- 127.Hovanessian A G. The double stranded RNA-activated protein kinase induced by interferon: dsRNA-PK. J Interferon Res. 1989;9:641–647. doi: 10.1089/jir.1989.9.641. [DOI] [PubMed] [Google Scholar]
- 128.Hovanessian A G, Svab J, Marie I, Robert N, Chamaret S, Laurent A G. Characterization of 69- and 100-kDa forms of 2-5A-synthetase from interferon-treated human cells. J Biol Chem. 1988;263:4959–4959. [PubMed] [Google Scholar]
- 129.Howe J G, Shu M-D. Epstein-Barr virus small RNA (EBER) genes: unique transcription units that combine RNA polymerase II and III promoter elements. Cell. 1989;57:825–834. doi: 10.1016/0092-8674(89)90797-6. [DOI] [PubMed] [Google Scholar]
- 130.Hsieh-Li H M, Witte D P, Weinstein M, Branford W, Li H, Small K, Potter S S. Hoxa 11 structure, extensive antisense transcription, and function in male and female fertility. Development. 1995;121:1373–1385. doi: 10.1242/dev.121.5.1373. [DOI] [PubMed] [Google Scholar]
- 131.Hubbell H R. Synergistic antiproliferative effect of human interferons in combination with mismatched double-stranded RNA on human tumor cells. Int J Cancer. 1986;37:359–365. doi: 10.1002/ijc.2910370306. [DOI] [PubMed] [Google Scholar]
- 132.Hunt T, Ehrenfeld E. Cytoplasm from poliovirus-infected HeLa cells inhibits cell-free haemoglobin synthesis. Nat New Biol. 1971;230:91–94. doi: 10.1038/newbio230091a0. [DOI] [PubMed] [Google Scholar]
- 133.Hyde-DeRuyscher R, Carmichael G G. Polyomavirus early-late switch is not regulated at the level of transcription initiation and is associated with changes in RNA processing. Proc Natl Acad Sci USA. 1988;85:8993–8997. doi: 10.1073/pnas.85.23.8993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hyde-DeRuyscher R P, Carmichael G G. Polyomavirus late pre-mRNA processing: DNA-replication-associated changes in leader exon multiplicity suggest a role for leader-to-leader splicing in the early-late switch. J Virol. 1990;64:5823–5832. doi: 10.1128/jvi.64.12.5823-5832.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Icely P L, Gros P, Bergeron J J, Devault A, Afar D E, Bell J C. TIK, a novel serine/threonine kinase, is recognized by antibodies directed against phosphotyrosine. J Biol Chem. 1991;266:16073–16077. [PubMed] [Google Scholar]
- 136.Ihle J N. Signaling by the cytokine receptor superfamily in normal and transformed hematopoietic cells. Adv Cancer Res. 1996;68:23–65. doi: 10.1016/s0065-230x(08)60351-6. [DOI] [PubMed] [Google Scholar]
- 137.Ihle J N, Witthuhn B A, Quelle F W, Yamamoto K, Silvennoinen O. Signaling through the hematopoietic cytokine receptors. Annu Rev Immunol. 1995;13:369–398. doi: 10.1146/annurev.iy.13.040195.002101. [DOI] [PubMed] [Google Scholar]
- 138.Ilson D H, Torrence P F, Vilcek J. Two molecular weight forms of human 2′,5′-oligoadenylate synthetase have different activation requirements. J Interferon Res. 1986;6:5–12. doi: 10.1089/jir.1986.6.5. [DOI] [PubMed] [Google Scholar]
- 139.Imani F, Jacobs B L. Inhibitory activity for the interferon-induced protein kinase is associated with the reovirus serotype 1 sigma 3 protein. Proc Natl Acad Sci USA. 1988;85:7887–7891. doi: 10.1073/pnas.85.21.7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Inouye M. Antisense RNA: Its functions and applications in gene regulation. Gene. 1988;72:25–34. doi: 10.1016/0378-1119(88)90124-2. [DOI] [PubMed] [Google Scholar]
- 141.Jacquemont B, Roizman B. RNA synthesis in cells infected with herpes simplex virus. X. Properties of viral symmetric transcripts and of double-stranded RNA prepared from them. J Virol. 1975;15:707–713. doi: 10.1128/jvi.15.4.707-713.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Jagus R, Gray M M. Proteins that interact with PKR. Biochimie. 1994;76:779–791. doi: 10.1016/0300-9084(94)90082-5. [DOI] [PubMed] [Google Scholar]
- 143.Jang S K, Davies M V, Kaufman R J, Wimmer E. Initiation of protein synthesis by internal entry of ribosomes into the 5′ nontranslated region of encephalomyocarditis virus RNA in vivo. J Virol. 1989;63:1651–1660. doi: 10.1128/jvi.63.4.1651-1660.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jeffrey I W, Kadereit S, Meurs E F, Metzger T, Bachmann M, Schwemmle M, Hovanessian A G, Clemens M J. Nuclear localization of the interferon-inducible protein kinase PKR in human cells and transfected mouse cells. Exp Cell Res. 1995;218:17–27. doi: 10.1006/excr.1995.1126. [DOI] [PubMed] [Google Scholar]
- 145.Jimenez-Garcia L F, Green S R, Mathews M B, Spector D L. Organization of the double-stranded RNA-activated protein kinase DAI and virus-associated VA RNAI in adenovirus-2-infected HeLa cells. J Cell Sci. 1993;106:11–22. doi: 10.1242/jcs.106.1.11. [DOI] [PubMed] [Google Scholar]
- 146.Judware R, Li J, Petryshyn R. Inhibition of the dsRNA-dependent protein kinase by a peptide derived from the human immunodeficiency virus type 1 Tat protein. J Interferon Res. 1993;13:153–160. doi: 10.1089/jir.1993.13.153. [DOI] [PubMed] [Google Scholar]
- 147.Judware R, Petryshyn R. Partial characterization of a cellular factor that regulates the double-stranded RNA-dependent eIF-2 alpha kinase in 3T3-F442A fibroblasts. Mol Cell Biol. 1991;11:3259–3267. doi: 10.1128/mcb.11.6.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Judware R, Petryshyn R. Mechanism of action of a cellular inhibitor of the dsRNA-dependent protein kinase from 3T3-F442A cells. J Biol Chem. 1992;267:21685–21690. [PubMed] [Google Scholar]
- 149.Kaempfer R, Kaufman J. Inhibition of cellular protein synthesis by double-stranded RNA: inactivation of an initiation factor. Proc Natl Acad Sci USA. 1973;70:1222–1226. doi: 10.1073/pnas.70.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kalvakolanu D V R, Borden E C. An overview of the interferon system: signal transduction and mechanisms of action. Cancer Investig. 1996;14:25–53. doi: 10.3109/07357909609018435. [DOI] [PubMed] [Google Scholar]
- 151.Kamen R, Lindstrom D M, Shure H, Old R W. Virus-specific RNA in cells productively infected or transformed by polyoma virus. Cold Spring Harbor Symp Quant Biol. 1974;39:187–198. doi: 10.1101/sqb.1974.039.01.025. [DOI] [PubMed] [Google Scholar]
- 152.Katze M G. Games viruses play: a strategic initiative against the interferon-induced dsRNA-activated 68,000 Mr protein kinase. Semin Virol. 1993;4:259–268. [Google Scholar]
- 153.Katze M G, Tomita J, Black T, Krug R M, Safer B, Hovanessian A. Influenza virus regulates protein synthesis during infection by repressing autophosphorylation and activity of the cellular 68,000-Mr protein kinase. J Virol. 1988;62:3710–3717. doi: 10.1128/jvi.62.10.3710-3717.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Katze M G, Wambach M, Wong M L, Garfinkel M, Meurs E, Chong K, Williams B R, Hovanessian A G, Barber G N. Functional expression and RNA binding analysis of the interferon-induced, double-stranded RNA-activated, 68,000-Mr protein kinase in a cell-free system. Mol Cell Biol. 1991;11:5497–5505. doi: 10.1128/mcb.11.11.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kelley K A, Pitha P M. Differential effect of poly rI.rC and Newcastle disease virus on the expression of interferon and cellular genes in mouse cells. Virology. 1985;147:382–393. doi: 10.1016/0042-6822(85)90140-0. [DOI] [PubMed] [Google Scholar]
- 156.Kerr I M, Brown R E. pppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc Natl Acad Sci USA. 1978;75:256–260. doi: 10.1073/pnas.75.1.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kharrat A, Macias M J, Gibson T J, Nilges M, Pastore A. Structure of the dsRNA binding domain of E. coli RNase III. EMBO J. 1995;14:3572–3584. doi: 10.1002/j.1460-2075.1995.tb07363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Khochbin S, Brocard M P, Grunwald D, Lawrence J J. Antisense RNA and p53 regulation in induced murine cell differentiation. Ann N Y Acad Sci. 1992;660:77–87. doi: 10.1111/j.1749-6632.1992.tb21060.x. [DOI] [PubMed] [Google Scholar]
- 159.Khochbin S, Lawrence J J. An antisense RNA involved in p53 mRNA maturation in murine erythroleukemia cells induced to differentiate. EMBO J. 1989;8:4107–4114. doi: 10.1002/j.1460-2075.1989.tb08595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Khochbin S, Principaud E, Chabanas A, Lawrence J J. Early events in murine erythroleukemia cells induced to differentiate. Accumulation and gene expression of the transformation-associated cellular protein p53. J Mol Biol. 1988;200:55–64. doi: 10.1016/0022-2836(88)90333-6. [DOI] [PubMed] [Google Scholar]
- 161.Kim U, Nishikura K. Double-stranded RNA adenosine deaminase as a potential mammalian RNA editing factor. Semin Cell Biol. 1993;4:285–293. doi: 10.1006/scel.1993.1034. [DOI] [PubMed] [Google Scholar]
- 162.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kimelman D. Regulation of eukaryotic gene expression by natural antisense transcripts—the case of the modifying reaction. Gene Regul. 1992;1:1–10. [Google Scholar]
- 164.Kimelman D, Kirschner M W. An antisense mRNA directs the covalent modification of the transcript encoding fibroblast growth factor in Xenopus oocytes. Cell. 1989;59:687–96. doi: 10.1016/0092-8674(89)90015-9. [DOI] [PubMed] [Google Scholar]
- 165.Kindy M S, McCormack J E, Buckler A J, Levine R A, Sonenshein G E. Independent regulation of transcription of the two strands of the c-myc gene. Mol Cell Biol. 1987;7:2857–2862. doi: 10.1128/mcb.7.8.2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kitajewski J, Schneider R J, Safer B, Munemitsu S M, Samuel C E, Thimmappaya B, Shenk T. Adenovirus VAI RNA antagonizes the antiviral action of interferon by preventing activation of the interferon-induced eIF-2 alpha kinase. Cell. 1986;45:195–200. doi: 10.1016/0092-8674(86)90383-1. [DOI] [PubMed] [Google Scholar]
- 167.Knee R, Li A W, Murphy P R. Characterization and tissue-specific expression of the rat basic fibroblast growth factor antisense mRNA and protein. Proc Natl Acad Sci USA. 1997;94:4943–4947. doi: 10.1073/pnas.94.10.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Knee R, Murphy P R. Regulation of gene expression by natural antisense RNA transcripts. Neurochem Int. 1997;31:379–392. doi: 10.1016/s0197-0186(96)00108-8. [DOI] [PubMed] [Google Scholar]
- 169.Knee R S, Pitcher S E, Murphy P R. Basic fibroblast growth factor sense (FGF) and antisense (gfg) RNA transcripts are expressed in unfertilized human oocytes and in differentiated adult tissues. Biochem Biophys Res Commun. 1994;205:577–583. doi: 10.1006/bbrc.1994.2704. [DOI] [PubMed] [Google Scholar]
- 170.Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 171.Krausslich H G, Nicklin M J, Toyoda H, Etchison D, Wimmer E. Poliovirus proteinase 2A induces cleavage of eucaryotic initiation factor 4F polypeptide p220. J Virol. 1987;61:2711–2718. doi: 10.1128/jvi.61.9.2711-2718.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Krystal G W. Regulation of eukaryotic gene expression by naturally occurring antisense RNA. Gene Regul. 1992;1:11–20. [Google Scholar]
- 173.Krystal G W, Armstrong B C, Battey J F. N-myc mRNA forms an RNA-RNA duplex with endogenous antisense transcripts. Mol Cell Biol. 1990;10:4180–4191. doi: 10.1128/mcb.10.8.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kuhl D, de la Fuente J, Chaturvedi M, Parimoo S, Ryals J, Meyer F, Weissmann C. Reversible silencing of enhancers by sequences derived from the human IFN-alpha promoter. Cell. 1987;50:1057–1069. doi: 10.1016/0092-8674(87)90172-3. [DOI] [PubMed] [Google Scholar]
- 175.Kumar A, Haque J, Lacoste J, Hiscott J, Williams B R. Double-stranded RNA-dependent protein kinase activates transcription factor NF-kappa B by phosphorylating I kappa B. Proc Natl Acad Sci USA. 1994;91:6288–6292. doi: 10.1073/pnas.91.14.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kumar M, Carmichael G G. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc Natl Acad Sci USA. 1997;94:3542–3547. doi: 10.1073/pnas.94.8.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176a.Kumar, M., and G. G. Carmichael. Unpublished data.
- 177.Laabi Y, Gras M P, Brouet J C, Berger R, Larsen C J, Tsapis A. The BCMA gene, preferentially expressed during B lymphoid maturation, is bidirectionally transcribed. Nucleic Acids Res. 1994;22:1147–1154. doi: 10.1093/nar/22.7.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lai M M C. The molecular biology of hepatitis delta virus. Annu Rev Biochem. 1995;64:259–286. doi: 10.1146/annurev.bi.64.070195.001355. [DOI] [PubMed] [Google Scholar]
- 179.Langland J O, Jacobs B L. Cytosolic double-stranded RNA-dependent protein kinase is likely a dimer of partially phosphorylated Mr = 66,000 subunits. J Biol Chem. 1992;267:10729–10736. [PubMed] [Google Scholar]
- 180.Lazar M A, Hodin R A, Cardona G, Chin W W. Gene expression from the c-erbA alpha/Rev-ErbA alpha genomic locus. Potential regulation of alternative splicing by opposite strand transcription. J Biol Chem. 1990;265:12859–12863. [PubMed] [Google Scholar]
- 181.Lazar M A, Hodin R A, Darling D S, Chin W W. A novel member of the thyroid/steroid hormone receptor family is encoded by the opposite strand of the rat c-erbA alpha transcriptional unit. Mol Cell Biol. 1989;9:1128–1136. doi: 10.1128/mcb.9.3.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lazar M A, Jones K E, Chin W W. Isolation of a cDNA encoding human Rev-ErbA alpha: transcription from the noncoding DNA strand of a thyroid hormone receptor gene results in a related protein that does not bind thyroid hormone. DNA Cell Biol. 1990;9:77–83. doi: 10.1089/dna.1990.9.77. [DOI] [PubMed] [Google Scholar]
- 183.Lee J Y, Marshall J A, Bowden D S. Characterization of rubella virus replication complexes using antibodies to double-stranded RNA. Virology. 1994;200:307–312. doi: 10.1006/viro.1994.1192. [DOI] [PubMed] [Google Scholar]
- 184.Lee R C, Feinbaum R L, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 185.Lee S B, Green S R, Mathews M B, Esteban M. Activation of the double-stranded RNA (dsRNA)-activated human protein kinase in vivo in the absence of its dsRNA binding domain. Proc Natl Acad Sci USA. 1994;91:10551–10555. doi: 10.1073/pnas.91.22.10551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lee T G, Katze M G. Cellular inhibitors of the interferon-induced, dsRNA-activated protein kinase. Prog Mol Subcell Biol. 1994;14:48–65. doi: 10.1007/978-3-642-78549-8_4. [DOI] [PubMed] [Google Scholar]
- 187.Lee T G, Tang N, Thompson S, Miller J, Katze M G. The 58,000-dalton cellular inhibitor of the interferon-induced double-stranded RNA-activated protein kinase (PKR) is a member of the tetratricopeptide repeat family of proteins. Mol Cell Biol. 1994;14:2331–2342. doi: 10.1128/mcb.14.4.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lee T G, Tomita J, Hovanessian A G, Katze M G. Purification and partial characterization of a cellular inhibitor of the interferon-induced protein kinase of Mr 68,000 from influenza virus-infected cells. Proc Natl Acad Sci USA. 1990;87:6208–6212. doi: 10.1073/pnas.87.16.6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lee T G, Tomita J, Hovanessian A G, Katze M G. Characterization and regulation of the 58,000-dalton cellular inhibitor of the interferon-induced, dsRNA-activated protein kinase. J Biol Chem. 1992;267:14238–14243. [PubMed] [Google Scholar]
- 190.Lenardo M J, Fan C M, Maniatis T, Baltimore D. The involvement of NF-kappa B in beta-interferon gene regulation reveals its role as widely inducible mediator of signal transduction. Cell. 1989;57:287–294. doi: 10.1016/0092-8674(89)90966-5. [DOI] [PubMed] [Google Scholar]
- 191.Lengyel P. Biochemistry of interferons and their actions. Annu Rev Biochem. 1982;51:251–282. doi: 10.1146/annurev.bi.51.070182.001343. [DOI] [PubMed] [Google Scholar]
- 192.Lengyel P. Double-stranded RNA and interferon action. J Interferon Res. 1987;7:511–519. doi: 10.1089/jir.1987.7.511. [DOI] [PubMed] [Google Scholar]
- 193.Lerner M R, Andrews N C, Miller G, Steitz J A. Two small RNAs encoded by Epstein-Barr virus and complexed with protein are precipitated by antibodies from patients with systemic lupus erythematosus. Proc Natl Acad Sci USA. 1981;78:805–809. doi: 10.1073/pnas.78.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Li A W, Seyoum G, Shiu R P, Murphy P R. Expression of the rat BFGF antisense RNA transcript is tissue-specific and developmentally regulated. Mol Cell Endocrinol. 1996;118:113–123. doi: 10.1016/0303-7207(96)03772-0. [DOI] [PubMed] [Google Scholar]
- 195.Li A W, Too C K, Murphy P R. The basic fibroblast growth factor (FGF-2) antisense RNA (GFG) is translated into a MutT-related protein in vivo. Biochem Biophys Res Commun. 1996;223:19–23. doi: 10.1006/bbrc.1996.0839. [DOI] [PubMed] [Google Scholar]
- 196.Lin S L, Greene J J, Ts’o P O, Carter W A. Sensitivity and resistance of human tumor cells to interferon and rIn · rCn. Nature. 1982;297:417–419. doi: 10.1038/297417a0. [DOI] [PubMed] [Google Scholar]
- 197.Lipman D J. Making (anti)sense of non-coding sequence conservation. Nucleic Acids Res. 1997;25:3580–3583. doi: 10.1093/nar/25.18.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Liu Y, George C X, Patterson J B, Samuel C E. Functionally distinct double-stranded RNA-binding domains associated with alternative splice site variants of the interferon-inducible double-stranded RNA-specific adenosine deaminase. J Biol Chem. 1997;272:4419–4428. doi: 10.1074/jbc.272.7.4419. [DOI] [PubMed] [Google Scholar]
- 199.Liu Y, Samuel C E. Mechanism of interferon action: functionally distinct RNA-binding and catalytic domains in the interferon-inducible, double-stranded RNA-specific adenosine deaminase. J Virol. 1996;70:1961–1968. doi: 10.1128/jvi.70.3.1961-1968.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Liu Z, Carmichael G G. Polyoma virus early late switch—regulation of late RNA accumulation by DNA replication. Proc Natl Acad Sci USA. 1993;90:8494–8498. doi: 10.1073/pnas.90.18.8494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lomeli H, Mosbacher J, Melcher T, Hoger T, Geiger J R, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg P H. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 202.Louis J M, McFarland V W, May P, Mora P T. The phosphoprotein p53 is down-regulated post-transcriptionally during embryogenesis in vertebrates. Biochim Biophys Acta. 1988;950:395–402. doi: 10.1016/0167-4781(88)90136-4. [DOI] [PubMed] [Google Scholar]
- 203.Lu Y, Wambach M, Katze M G, Krug R M. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 204.Luo G X, Chao M, Hsieh S Y, Sureau C, Nishikura K, Taylor J. A specific base transition occurs on replicating hepatitis delta virus RNA. J Virol. 1990;64:1021–1027. doi: 10.1128/jvi.64.3.1021-1027.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Maas S, Melcher T, Herb A, Seeburg P H, Keller W, Krause S, Higuchi M, O’Connell M A. Structural requirements for RNA editing in glutamate receptor pre-mRNAs by recombinant double-stranded RNA adenosine deaminase. J Biol Chem. 1996;271:12221–12226. doi: 10.1074/jbc.271.21.12221. [DOI] [PubMed] [Google Scholar]
- 206.MacDonald N J, Kuhl D, Maguire D, Naf D, Gallant P, Goswamy A, Hug H, Bueler H, Chaturvedi M, de la Fuente J, et al. Different pathways mediate virus inducibility of the human IFN-alpha 1 and IFN-beta genes. Cell. 1990;60:767–779. doi: 10.1016/0092-8674(90)90091-r. [DOI] [PubMed] [Google Scholar]
- 207.Maitra R K, McMillan N A, Desai S, McSwiggen J, Hovanessian A G, Sen G, Williams B R, Silverman R H. HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzymes. Virology. 1994;204:823–827. doi: 10.1006/viro.1994.1601. [DOI] [PubMed] [Google Scholar]
- 208.Maran A, Mathews M B. Characterization of the double-stranded RNA implicated in the inhibition of protein synthesis in cells infected with a mutant adenovirus defective for VA RNA. Virology. 1988;164:106–113. doi: 10.1016/0042-6822(88)90625-3. [DOI] [PubMed] [Google Scholar]
- 209.Marcus P I. The interferon inducer moiety of viruses: a single molecule of dsRNA. Tex Rep Biol Med. 1981;41:70–75. [PubMed] [Google Scholar]
- 210.Mathews M B. Viral evasion of cellular defense mechanisms: regulation of the protein kinase DAI by RNA effectors. Semin Virol. 1993;4:247–257. [Google Scholar]
- 211.Mathews M B, Shenk T. Adenovirus virus-associated RNA and translation control. J Virol. 1991;65:5657–5662. doi: 10.1128/jvi.65.11.5657-5662.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Matsuyama T, Kimura T, Kitagawa M, Pfeffer K, Kawakami T, Watanabe N, Kundig T M, Amakawa R, Kishihara K, Wakeham A, et al. Targeted disruption of IRF-1 or IRF-2 results in abnormal type I IFN gene induction and aberrant lymphocyte development. Cell. 1993;75:83–97. [PubMed] [Google Scholar]
- 213.McCarthy T L, Siegel E, Mroczkowski B, Heywood S M. Characterization of translational-control ribonucleic acid isolated from embryonic chick muscle. Biochemistry. 1983;22:935–941. doi: 10.1021/bi00273a035. [DOI] [PubMed] [Google Scholar]
- 214.McGuinness T, Porteus M H, Smiga S, Bulfone A, Kingsley C, Qiu M, Liu J K, Long J E, Xu D, Rubenstein J L. Sequence, organization, and transcription of the Dlx-1 and Dlx-2 locus. Genomics. 1996;35:473–485. doi: 10.1006/geno.1996.0387. [DOI] [PubMed] [Google Scholar]
- 215.McMillan N A, Chun R F, Siderovski D P, Galabru J, Toone W M, Samuel C E, Mak T W, Hovanessian A G, Jeang K T, Williams B R. HIV-1 Tat directly interacts with the interferon-induced, double-stranded RNA-dependent kinase, PKR. Virology. 1995;213:413–424. doi: 10.1006/viro.1995.0014. [DOI] [PubMed] [Google Scholar]
- 216.Meegan J M, Marcus P I. Double-stranded ribonuclease coinduced with interferon. Science. 1989;244:1089–1091. doi: 10.1126/science.2471268. [DOI] [PubMed] [Google Scholar]
- 217.Meerovitch K, Pelletier J, Sonenberg N. A cellular protein that binds to the 5′-noncoding region of poliovirus RNA: implications for internal translation initiation. Genes Dev. 1989;3:1026–1034. doi: 10.1101/gad.3.7.1026. [DOI] [PubMed] [Google Scholar]
- 218.Meerovitch K, Svitkin Y V, Lee H S, Lejbkowicz F, Kenan D J, Chan E K, Agol V I, Keene J D, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Melcher T, Maas S, Herb A, Sprengel R, Seeburg P H, Higuchi M. A mammalian RNA editing enzyme. Nature. 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 220.Merrick W C. Mechanism and regulation of eukaryotic protein synthesis. Microbiol Rev. 1992;56:291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Meurs E, Chong K, Galabru J, Thomas N S, Kerr I M, Williams B R, Hovanessian A G. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 222.Michael N L, Vahey M T, d’Arcy L, Ehrenberg P K, Mosca J D, Rappaport J, Redfield R R. Negative-strand RNA transcripts are produced in human immunodeficiency virus type 1-infected cells and patients by a novel promoter downregulated by Tat. J Virol. 1994;68:979–987. doi: 10.1128/jvi.68.2.979-987.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Mikoshiba K, Okano H, Aruga J, Tamura T, Miura M, Ikenaka K, Nakagawa T. Chimeric and molecular genetic analysis of myelin-deficient (shiverer and mld) mutant mice. Ann NY Acad Sci. 1990;605:166–182. doi: 10.1111/j.1749-6632.1990.tb42391.x. [DOI] [PubMed] [Google Scholar]
- 224.Minks M A, West D K, Benvin S, Greene J J, Ts’o P O, Baglioni C. Activation of 2′,5′-oligo(A) polymerase and protein kinase of interferon-treated HeLa cells by 2′-O-methylated poly (inosinic acid) · poly(cytidylic acid): correlations with interferon-inducing activity. J Biol Chem. 1980;255:6403–6407. [PubMed] [Google Scholar]
- 225.Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-beta gene regulatory elements. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- 226.Moore T, Constancia M, Zubair M, Bailleul B, Feil R, Sasaki H, Reik W. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc Natl Acad Sci USA. 1997;94:12509–12514. doi: 10.1073/pnas.94.23.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Mundschau L J, Faller D V. Oncogenic ras induces an inhibitor of double-stranded RNA-dependent eukaryotic initiation factor 2 alpha-kinase activation. J Biol Chem. 1992;267:23092–23098. [PubMed] [Google Scholar]
- 228.Munroe S H, Lazar M A. Inhibition of c-erbA mRNA splicing by a naturally occurring antisense RNA. J Biol Chem. 1991;266:22083–22086. [PubMed] [Google Scholar]
- 229.Murashov A K, Wolgemuth D J. Sense and antisense transcripts of the developmentally regulated murine hsp70.2 gene are expressed in distinct and only partially overlapping areas in the adult brain. Brain Res Mol Brain Res. 1996;37:85–95. doi: 10.1016/0169-328x(95)00288-4. [DOI] [PubMed] [Google Scholar]
- 230.Murphy D G, Dimock K, Kang C Y. Numerous transitions in human parainfluenza virus 3 RNA recovered from persistently infected cells. Virology. 1991;181:760–763. doi: 10.1016/0042-6822(91)90913-v. [DOI] [PubMed] [Google Scholar]
- 231.Murphy P R, Knee R S. Identification and characterization of an antisense RNA transcript (gfg) from the human basic fibroblast growth factor gene. Mol Endocrinol. 1994;8:852–859. doi: 10.1210/mend.8.7.7984147. [DOI] [PubMed] [Google Scholar]
- 232.Murray J A H, Crockett N. Antisense techniques: an overview. In: Murray J A H, editor. in Modern cell biology. New York, N.Y: Wiley-Liss, Inc.; 1992. pp. 1–49. [Google Scholar]
- 233.Nepveu A, Marcu K B. Intragenic pausing and anti-sense transcription within the murine c-myc locus. EMBO J. 1986;5:2859–2865. doi: 10.1002/j.1460-2075.1986.tb04580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Nilsen T W, Maroney P A, Baglioni C. Synthesis of (2′-5′)oligoadenylate and activation of an endoribonuclease in interferon-treated HeLa cells infected with reovirus. J Virol. 1982;42:1039–1045. doi: 10.1128/jvi.42.3.1039-1045.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Nilsen T W, Wood D L, Baglioni C. Presence of 2′,5′-oligo(A) and of enzymes that synthesize, bind, and degrade 2′,5′-oligo(A) in HeLa cell nuclei. J Biol Chem. 1982;257:1602–1605. [PubMed] [Google Scholar]
- 236.Nishikura K. Modulation of double-stranded RNAs in vivo by RNA duplex unwindase. Ann N Y Acad Sci. 1992;660:240–250. doi: 10.1111/j.1749-6632.1992.tb21076.x. [DOI] [PubMed] [Google Scholar]
- 237.Noguchi M, Miyamoto S, Silverman T A, Safer B. Characterization of an antisense Inr element in the eIF-2 alpha gene. J Biol Chem. 1994;269:29161–29167. [PubMed] [Google Scholar]
- 238.O’Connell M A, Gerber A, Keller W. Purification of human double-stranded RNA-specific editase 1 (hRED1) involved in editing of brain glutamate receptor B pre-mRNA. J Biol Chem. 1997;272:473–478. doi: 10.1074/jbc.272.1.473. [DOI] [PubMed] [Google Scholar]
- 239.O’Connell M A, Krause S, Higuchi M, Hsuan J J, Totty N F, Jenny A, Keller W. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol Cell Biol. 1995;15:1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Offermann M K, Zimring J, Mellits K H, Hagan M K, Shaw R, Medford R M, Mathews M B, Goodbourn S, Jagus R. Activation of the double-stranded-RNA-activated protein kinase and induction of vascular cell adhesion molecule-1 by poly (I) · poly (C) in endothelial cells. Eur J Biochem. 1995;232:28–36. doi: 10.1111/j.1432-1033.1995.tb20777.x. [DOI] [PubMed] [Google Scholar]
- 241.O’Hara P J, Nichol S T, Horodyski F M, Holland J J. Vesicular stomatitis virus defective interfering particles can contain extensive genomic sequence rearrangements and base substitutions. Cell. 1984;36:915–924. doi: 10.1016/0092-8674(84)90041-2. [DOI] [PubMed] [Google Scholar]
- 242.Okano H, Aruga J, Nakagawa T, Shiota C, Mikoshiba K. Myelin basic protein gene and the function of antisense RNA in its repression in myelin-deficient mutant mouse. J Neurochem. 1991;56:560–567. doi: 10.1111/j.1471-4159.1991.tb08186.x. [DOI] [PubMed] [Google Scholar]
- 243.Okano H, Ikenaka K, Mikoshiba K. Recombination within the upstream gene of duplicated myelin basic protein genes of myelin deficient shimld mouse results in the production of antisense RNA. EMBO J. 1988;7:3407–3412. doi: 10.1002/j.1460-2075.1988.tb03214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Pacha R F, Condit R C. Characterization of a temperature-sensitive mutant of vaccinia virus reveals a novel function that prevents virus-induced breakdown of RNA. J Virol. 1985;56:395–403. doi: 10.1128/jvi.56.2.395-403.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Pacha R F, Meis R J, Condit R C. Structure and expression of the vaccinia virus gene which prevents virus-induced breakdown of RNA. J Virol. 1990;64:3853–3863. doi: 10.1128/jvi.64.8.3853-3863.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Palmenberg A C. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- 247.Park H, Davies M V, Langland J O, Chang H W, Nam Y S, Tartaglia J, Paoletti E, Jacobs B L, Kaufman R J, Venkatesan S. TAR RNA-binding protein is an inhibitor of the interferon-induced protein kinase PKR. Proc Natl Acad Sci USA. 1994;91:4713–4717. doi: 10.1073/pnas.91.11.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Patel R C, Sen G C. Identification of the double-stranded RNA-binding domain of the human interferon-inducible protein kinase. J Biol Chem. 1992;267:7671–7676. [PubMed] [Google Scholar]
- 249.Patel R C, Stanton P, McMillan N M, Williams B R, Sen G C. The interferon-inducible double-stranded RNA-activated protein kinase self-associates in vitro and in vivo. Proc Natl Acad Sci USA. 1995;92:8283–8287. doi: 10.1073/pnas.92.18.8283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Patterson J B, Samuel C E. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Patterson J B, Thomis D C, Hans S L, Samuel C E. Mechanism of interferon action: double-stranded RNA-specific adenosine deaminase from human cells is inducible by alpha and gamma interferons. Virology. 1995;210:508–511. doi: 10.1006/viro.1995.1370. [DOI] [PubMed] [Google Scholar]
- 252.Paul M S, Bass B L. Inosine exists in mRNA at tissue-specific levels and is most abundant in brain mRNA. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Pellegrini S, Schindler C. Early events in signalling by interferons. Trends Biochem Sci. 1993;18:338–342. doi: 10.1016/0968-0004(93)90070-4. [DOI] [PubMed] [Google Scholar]
- 254.Petschek J P, Mermer M J, Scheckelhoff M R, Simone A A, Vaughn J C. RNA editing in Drosophila 4f-rnp gene nuclear transcripts by multiple A-to-G conversions. J Mol Biol. 1996;259:885–890. doi: 10.1006/jmbi.1996.0365. [DOI] [PubMed] [Google Scholar]
- 255.Pettersson U, Philipson L. Synthesis of complementary RNA sequences during productive adenovirus infection. Proc Natl Acad Sci USA. 1974;71:4887–4891. doi: 10.1073/pnas.71.12.4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Pine R, Decker T, Kessler D S, Levy D E, Darnell J E., Jr Purification and cloning of interferon-stimulated gene factor 2 (ISGF2): ISGF2 (IRF-1) can bind to the promoters of both beta interferon- and interferon-stimulated genes but is not a primary transcriptional activator of either. Mol Cell Biol. 1990;10:2448–2457. doi: 10.1128/mcb.10.6.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Piper P. Polyoma virus transcription early during productive infection of mouse 3T6 cells. J Mol Biol. 1979;131:399–407. doi: 10.1016/0022-2836(79)90083-4. [DOI] [PubMed] [Google Scholar]
- 258.Polson A G, Bass B L. Preferential selection of adenosines for modification by double-stranded RNA adenosine deaminase. EMBO J. 1994;13:5701–5711. doi: 10.1002/j.1460-2075.1994.tb06908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Polson A G, Bass B L, Casey J L. RNA editing of hepatitis delta virus antigenome by dsRNA-adenosine deaminase. Nature. 1996;380:454–456. doi: 10.1038/380454a0. [DOI] [PubMed] [Google Scholar]
- 260.Polson A G, Crain P F, Pomerantz S C, McCloskey J A, Bass B L. The mechanism of adenosine to inosine conversion by the double-stranded RNA unwinding/modifying activity: a high-performance liquid chromatography-mass spectrometry analysis. Biochemistry. 1991;30:11507–11514. doi: 10.1021/bi00113a004. [DOI] [PubMed] [Google Scholar]
- 261.Polson A G, Ley H L, Bass B L, Casey J L. Hepatitis delta virus RNA editing is highly specific for the amber/W site and is suppressed by hepatitis delta antigen. Mol Cell Biol. 1998;18:1919–1926. doi: 10.1128/mcb.18.4.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Polyak S J, Tang N, Wambach M, Barber G N, Katze M G. The P58 cellular inhibitor complexes with the interferon-induced, double-stranded RNA-dependent protein kinase, PKR, to regulate its autophosphorylation and activity. J Biol Chem. 1996;271:1702–1707. doi: 10.1074/jbc.271.3.1702. [DOI] [PubMed] [Google Scholar]
- 263.Proud C G. PKR: a new name and new roles. Trends Biochem Sci. 1995;20:241–246. doi: 10.1016/s0968-0004(00)89025-8. [DOI] [PubMed] [Google Scholar]
- 264.Qian X Y, Chien C Y, Lu Y, Montelione G T, Krug R M. An amino-terminal polypeptide fragment of the influenza virus NS1 protein possesses specific RNA-binding activity and largely helical backbone structure. RNA. 1995;1:948–956. [PMC free article] [PubMed] [Google Scholar]
- 265.Ray A, Tatter S B, May L T, Sehgal P B. Activation of the human “beta 2-interferon/hepatocyte-stimulating factor/interleukin 6” promoter by cytokines, viruses, and second messenger agonists. Proc Natl Acad Sci USA. 1988;85:6701–6705. doi: 10.1073/pnas.85.18.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Ray M K, Datta B, Chakraborty A, Chattopadhyay A, Meza-Keuthen S, Gupta N K. The eukaryotic initiation factor 2-associated 67-kDa polypeptide (p67) plays a critical role in regulation of protein synthesis initiation in animal cells. Proc Natl Acad Sci USA. 1992;89:539–543. doi: 10.1073/pnas.89.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Rebagliati M R, Melton D A. Antisense RNA injections in fertilized frog eggs reveal an RNA duplex unwinding activity. Cell. 1987;48:599–605. doi: 10.1016/0092-8674(87)90238-8. [DOI] [PubMed] [Google Scholar]
- 268.Reis L F, Harada H, Wolchok J D, Taniguchi T, Vilcek J. Critical role of a common transcription factor, IRF-1, in the regulation of IFN-beta and IFN-inducible genes. EMBO J. 1992;11:185–193. doi: 10.1002/j.1460-2075.1992.tb05041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Reis L F, Lee T H, Vilcek J. Tumor necrosis factor acts synergistically with autocrine interferon-beta and increases interferon-beta mRNA levels in human fibroblasts. J Biol Chem. 1989;264:16351–16354. [PubMed] [Google Scholar]
- 270.Rice A P, Kostura M, Mathews M B. Identification of a 90-kDa polypeptide which associates with adenovirus VA RNAI and is phosphorylated by the double-stranded RNA-dependent protein kinase. J Biol Chem. 1989;264:20632–20637. [PubMed] [Google Scholar]
- 271.Rivkin M, Rosen K M, Villa-Komaroff L. Identification of an antisense transcript from the IGF-II locus in mouse. Mol Reprod Dev. 1993;35:394–397. doi: 10.1002/mrd.1080350413. [DOI] [PubMed] [Google Scholar]
- 272.Robertson H D, Mathews M B. The regulation of the protein kinase PKR by RNA. Biochimie. 1996;78:909–914. doi: 10.1016/s0300-9084(97)86712-0. [DOI] [PubMed] [Google Scholar]
- 273.Rosenblum M G, Cheung L, Kessler D. Differential activity of the 30-kD and the 100-kD forms of 2′-5′An synthetase induced by recombinant human interferon-alpha and interferon-gamma. J Interferon Res. 1988;8:275–282. doi: 10.1089/jir.1988.8.275. [DOI] [PubMed] [Google Scholar]
- 274.Rueda P, Garcia-Barreno B, Melero J A. Loss of conserved cysteine residues in the attachment (G) glycoprotein of two human respiratory syncytial virus escape mutants that contain multiple A-G substitutions (hypermutations) Virology. 1994;198:653–662. doi: 10.1006/viro.1994.1077. [DOI] [PubMed] [Google Scholar]
- 275.Ryals J, Dierks P, Ragg H, Weissmann C. A 46-nucleotide promoter segment from an IFN-alpha gene renders an unrelated promoter inducible by virus. Cell. 1985;41:497–507. doi: 10.1016/s0092-8674(85)80023-4. [DOI] [PubMed] [Google Scholar]
- 276.Saccomanno L, Bass B L. The cytoplasm of Xenopus oocytes contains a factor that protects double-stranded RNA from adenosine-to-inosine modification. Mol Cell Biol. 1994;14:5425–5432. doi: 10.1128/mcb.14.8.5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Samuel C E. Mechanism of interferon action: phosphorylation of protein synthesis initiation factor eIF-2 in interferon-treated human cells by a ribosome-associated kinase processing site specificity similar to hemin- regulated rabbit reticulocyte kinase. Proc Natl Acad Sci USA. 1979;76:600–604. doi: 10.1073/pnas.76.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Samuel C E. Antiviral actions of interferon—interferon-regulated cellular proteins and their surprisingly selective antiviral activities. Virology. 1991;183:1–11. doi: 10.1016/0042-6822(91)90112-o. [DOI] [PubMed] [Google Scholar]
- 279.Samuel C E. The eIF-2-alpha protein kinases, regulators of translation in eukaryotes from yeasts to humans. J Biol Chem. 1993;268:7603–7606. [PubMed] [Google Scholar]
- 280.Samuel C E, Kuhen K L, George C X, Ortega L G, Rende-Fournier R, Tanaka H. The PKR protein kinase--an interferon-inducible regulator of cell growth and differentiation. Int J Hematol. 1997;65:227–237. doi: 10.1016/s0925-5710(96)00544-0. [DOI] [PubMed] [Google Scholar]
- 281.Sato M, Tanaka N, Hata N, Oda E, Taniguchi T. Involvement of the IRF family transcription factor IRF-3 in virus-induced activation of the IFN-beta gene. FEBS Lett. 1998;425:112–116. doi: 10.1016/s0014-5793(98)00210-5. [DOI] [PubMed] [Google Scholar]
- 282.Saunders M E, Gewert D R, Tugwell M E, McMahon M, Williams B R G. Human 2-5A synthetase: characterization of a novel cDNA and corresponding gene structure. EMBO J. 1985;4:1761–1768. doi: 10.1002/j.1460-2075.1985.tb03848.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Scadden A D, Smith C W. A ribonuclease specific for inosine-containing RNA: a potential role in antiviral defence? EMBO J. 1997;16:2140–2149. doi: 10.1093/emboj/16.8.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Schiff L A, Fields B N. Reoviruses and their replication. In: Fields B N, Knipe D M, editors. Virology. New York, N.Y: Raven Press; 1990. pp. 1275–1306. [Google Scholar]
- 285.Schindler C, Darnell J E., Jr Transcriptional responses to polypeptide ligands: the JAK-STAT pathway. Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 286.Schindler C, Fu X Y, Improta T, Aebersold R, Darnell J E., Jr Proteins of transcription factor ISGF-3: one gene encodes the 91- and 84-kDa ISGF-3 proteins that are activated by interferon alpha. Proc Natl Acad Sci USA. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Schmedt C, Green S R, Manche L, Taylor D R, Ma Y, Mathews M B. Functional characterization of the RNA-binding domain and motif of the double-stranded RNA-dependent protein kinase DAI (PKR) J Mol Biol. 1995;249:29–44. doi: 10.1006/jmbi.1995.0278. [DOI] [PubMed] [Google Scholar]
- 288.Schröder H C, Ugarkovic D, Wenger R, Reuter P, Okamoto T, Muller W E. Binding of Tat protein to TAR region of human immunodeficiency virus type 1 blocks TAR-mediated activation of (2′-5′)oligoadenylate synthetase. AIDS Res Hum Retroviruses. 1990;6:659–672. doi: 10.1089/aid.1990.6.659. [DOI] [PubMed] [Google Scholar]
- 289.Schröder H C, Wenger R, Kuchino Y, Muller W E G. Modulation of nuclear matrix-associated (2′-5′) oligoadenylate metabolism and ribonuclease L activity in H9 cells by human immunodeficiency virus. J Biol Chem. 1989;264:5669–5673. [PubMed] [Google Scholar]
- 290.Schröder H C, Wenger R, Rottmann M, Muller W E. Alteration of nuclear (2′-5′)oligoriboadenylate synthetase and nuclease activities preceding replication of human immunodeficiency virus in H9 cells. Biol Chem Hoppe-Seyler. 1988;369:985–995. doi: 10.1515/bchm3.1988.369.2.985. [DOI] [PubMed] [Google Scholar]
- 291.Seeburg P H. The role of RNA editing in controlling glutamate receptor channel properties. J Neurochem. 1996;66:1–5. doi: 10.1046/j.1471-4159.1996.66010001.x. [DOI] [PubMed] [Google Scholar]
- 292.SenGupta D N, Silverman R H. Activation of interferon-regulated, dsRNA-dependent enzymes by human immunodeficiency virus-1 leader RNA. Nucleic Acids Res. 1989;17:969–978. doi: 10.1093/nar/17.3.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Sharmeen L, Bass B, Sonenberg N, Weintraub H, Groudine M. Tat-dependent adenosine-to-inosine modification of wild-type transactivation response RNA. Proc Natl Acad Sci USA. 1991;88:8096–100. doi: 10.1073/pnas.88.18.8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Sharp T V, Schwemmle M, Jeffrey I, Laing K, Mellor H, Proud C G, Hilse K, Clemens M J. Comparative analysis of the regulation of the interferon-inducible protein kinase PKR by Epstein-Barr virus RNAs EBER-1 and EBER-2 and adenovirus VAI RNA. Nucleic Acids Res. 1993;21:4483–4490. doi: 10.1093/nar/21.19.4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Silverman R H, Sengupta D N. Translational regulation by HIV leader RNA, TAT, and interferon-inducible enzymes. J Exp Pathol. 1990;5:69–77. [PubMed] [Google Scholar]
- 296.Silverman T A, Noguchi M, Safer B. Role of sequences within the first intron in the regulation of expression of eukaryotic initiation factor 2 alpha. J Biol Chem. 1992;267:9738–9742. [PubMed] [Google Scholar]
- 297.Simons R W. Naturally occurring antisense RNA control—a brief review. Gene. 1988;72:35–44. doi: 10.1016/0378-1119(88)90125-4. [DOI] [PubMed] [Google Scholar]
- 298.Smale S T, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 299.Smith C M, Steitz J A. Sno storm in the nucleolus: new roles for myriad small RNPs. Cell. 1997;89:669–672. doi: 10.1016/s0092-8674(00)80247-0. [DOI] [PubMed] [Google Scholar]
- 300.Sommer B, Kohler M, Sprengel R, Seeburg P H. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 301.Spencer C A, Gietz R D, Hodgetts R B. Overlapping transcription units in the Dopa decarboxylase region of Drosophila. Nature. 1986;322:279–281. doi: 10.1038/322279a0. [DOI] [PubMed] [Google Scholar]
- 302.Spicer D B, Sonenshein G E. An antisense promoter of the murine c-myc gene is localized within intron 2. Mol Cell Biol. 1992;12:1324–1329. doi: 10.1128/mcb.12.3.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Staley J P, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 304.St. Johnston D, Brown N H, Gall J G, Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci USA. 1992;89:10979–10983. doi: 10.1073/pnas.89.22.10979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Stoger R, Kubicka P, Liu C G, Kafri T, Razin A, Cedar H, Barlow D P. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- 306.Tanaka H, Samuel C E. Mechanism of interferon action: structure of the mouse PKR gene encoding the interferon-inducible RNA-dependent protein kinase. Proc Natl Acad Sci USA. 1994;91:7995–7999. doi: 10.1073/pnas.91.17.7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Taylor D R, Lee S B, Romano P R, Marshak D R, Hinnebusch A G, Esteban M, Mathews M B. Autophosphorylation sites participate in the activation of the double-stranded-RNA-activated protein kinase PKR. Mol Cell Biol. 1996;16:6295–6302. doi: 10.1128/mcb.16.11.6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Taylor E R, Seleiro E A, Brickell P M. Identification of antisense transcripts of the chicken insulin-like growth factor-II gene. J Mol Endocrinol. 1991;7:145–154. doi: 10.1677/jme.0.0070145. [DOI] [PubMed] [Google Scholar]
- 309.Taylor-Papadimitriou J, Stoker M. Effect of interferon on some aspects of transformation by polyoma virus. Nat New Biol. 1971;230:114–117. doi: 10.1038/newbio230114a0. [DOI] [PubMed] [Google Scholar]
- 310.Thach R E. Cap recap: the involvement of eIF-4F in regulating gene expression. Cell. 1992;68:177–180. doi: 10.1016/0092-8674(92)90461-k. [DOI] [PubMed] [Google Scholar]
- 311.Thanos D, Maniatis T. Virus induction of human IFN beta gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 312.Thomis D C, Doohan J P, Samuel C E. Mechanisms of interferon action: cDNA structure, expression, and regulation of the interferon-induced, RNA-dependent P1/eIF-2 alpha protein kinase from human cells. Virology. 1992;188:33–46. doi: 10.1016/0042-6822(92)90732-5. [DOI] [PubMed] [Google Scholar]
- 313.Thomis D C, Samuel C E. Mechanism of interferon action: characterization of the intermolecular autophosphorylation of PKR, the interferon-inducible, RNA-dependent protein kinase. J Virol. 1995;69:5195–5198. doi: 10.1128/jvi.69.8.5195-5198.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Tosic M, Roach A, de Rivaz J C, Dolivo M, Matthieu J-M. Post-transcriptional events are responsible for low expression of myelin basic protein in myelin deficient mice: role of natural antisense RNA. EMBO J. 1990;9:401–406. doi: 10.1002/j.1460-2075.1990.tb08124.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Treisman R, Kamen R. Structure of polyoma virus late nuclear RNA. J Mol Biol. 1981;148:273–301. doi: 10.1016/0022-2836(81)90539-8. [DOI] [PubMed] [Google Scholar]
- 316.Ustacelebi S, Williams J F. Induction of interferon in chick cells by polyoma virus. J Gen Virol. 1973;21:163–168. doi: 10.1099/0022-1317-21-1-163. [DOI] [PubMed] [Google Scholar]
- 317.van Duin M, van Den Tol J, Hoeijmakers J H, Bootsma D, Rupp I P, Reynolds P, Prakash L, Prakash S. Conserved pattern of antisense overlapping transcription in the homologous human ERCC-1 and yeast RAD10 DNA repair gene regions. Mol Cell Biol. 1989;9:1794–1798. doi: 10.1128/mcb.9.4.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Vilcek J, Kohase M, Henriksen-DeStefano D. Mitogenic effect of double-stranded RNA in human fibroblasts: role of autogenous interferon. J Cell Physiol. 1987;130:37–43. doi: 10.1002/jcp.1041300107. [DOI] [PubMed] [Google Scholar]
- 319.Visvanathan K V, Goodbourn S. Double-stranded RNA activates binding of NF-kappa B to an inducible element in the human beta-interferon promoter. EMBO J. 1989;8:1129–1138. doi: 10.1002/j.1460-2075.1989.tb03483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Volk R, Koster M, Poting A, Hartmann L, Knochel W. An antisense transcript from the Xenopus laevis bFGF gene coding for an evolutionarily conserved 24 kd protein. EMBO J. 1989;8:2983–2988. doi: 10.1002/j.1460-2075.1989.tb08448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Wagner R W, Nishikura K. Cell cycle expression of RNA duplex unwindase activity in mammalian cells. Mol Cell Biol. 1988;8:770–777. doi: 10.1128/mcb.8.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Wagner R W, Smith J E, Cooperman B S, Nishikura K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc Natl Acad Sci USA. 1989;86:2647–51. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Wagner R W, Yoo C, Wrabetz L, Kamholz J, Buchhalter J, Hassan N F, Khalili K, Kim S U, Perussia B, McMorrin F A, et al. Double-stranded RNA unwinding and modifying activity is detected ubiquitously in primary tissues and cell lines. Mol Cell Biol. 1990;10:5586–5590. doi: 10.1128/mcb.10.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Wassarman D A, Steitz J A. RNA splicing-alive with DEAD proteins. Nature. 1991;349:463–464. doi: 10.1038/349463a0. [DOI] [PubMed] [Google Scholar]
- 325.Wathelet M G, Clauss I M, Content J, Huez G A. Regulation of two interferon-inducible human genes by interferon, poly(rI) · poly(rC) and viruses. Eur J Biochem. 1988;174:323–329. doi: 10.1111/j.1432-1033.1988.tb14101.x. [DOI] [PubMed] [Google Scholar]
- 326.Wathelet M G, Clauss I M, Nols C B, Content J, Huez G A. New inducers revealed by the promoter sequence analysis of two interferon-activated human genes. Eur J Biochem. 1987;169:313–321. doi: 10.1111/j.1432-1033.1987.tb13614.x. [DOI] [PubMed] [Google Scholar]
- 327.Watson J C, Chang H W, Jacobs B L. Characterization of a vaccinia virus-encoded double-stranded RNA-binding protein that may be involved in inhibition of the double-stranded RNA-dependent protein kinase. Virology. 1991;185:206–216. doi: 10.1016/0042-6822(91)90768-7. [DOI] [PubMed] [Google Scholar]
- 328.Weaver B K, Kumar K P, Reich N C. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Weihua X, Ramanujam S, Lindner D J, Kudaravalli R D, Freund R, Kalvakolanu D V. The polyoma virus T antigen interferes with interferon-inducible gene expression. Proc Natl Acad Sci USA. 1998;95:1085–1090. doi: 10.1073/pnas.95.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 331.Williams B R. Role of the double-stranded RNA-activated protein kinase (PKR) in cell regulation. Biochem Soc Trans. 1997;25:509–513. doi: 10.1042/bst0250509. [DOI] [PubMed] [Google Scholar]
- 332.Williams B R, Haque S J. Interacting pathways of interferon signaling. Semin Oncol. 1997;24:S970–S977. [PubMed] [Google Scholar]
- 333.Williams T, Fried M. A mouse locus at which transcription from both DNA strands produces mRNAs complementary at their 3′ ends. Nature. 1986;322:275–279. doi: 10.1038/322275a0. [DOI] [PubMed] [Google Scholar]
- 334.Williams T, Yon J, Huxley C, Fried M. The mouse surfeit locus contains a very tight cluster of four “housekeeping” genes that is conserved through evolution. Proc Natl Acad Sci USA. 1988;85:3527–3530. doi: 10.1073/pnas.85.10.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Wong A H, Tam N W, Yang Y L, Cuddihy A R, Li S, Kirchhoff S, Hauser H, Decker T, Koromilas A E. Physical association between STAT1 and the interferon-inducible protein kinase PKR and implications for interferon and double-stranded RNA signaling pathways. EMBO J. 1997;16:1291–1304. doi: 10.1093/emboj/16.6.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Wreschner D H, McCauley J W, Skehel J J, Kerr I M. Interferon action-sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature. 1981;289:414–417. doi: 10.1038/289414a0. [DOI] [PubMed] [Google Scholar]
- 337.Wu H, MacLeod A R, Lima W F, Crooke S T. Identification and partial purification of human double strand RNase activity. A novel terminating mechanism for oligoribonucleotide antisense drugs. J Biol Chem. 1998;273:2532–2542. doi: 10.1074/jbc.273.5.2532. [DOI] [PubMed] [Google Scholar]
- 338.Wutz A, Smrzka O W, Schweifer N, Schellander K, Wagner E F, Barlow D P. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature. 1997;389:745–749. doi: 10.1038/39631. [DOI] [PubMed] [Google Scholar]
- 339.Xiao Q, Sharp T V, Jeffrey I W, James M C, Pruijn G J, van Venrooij W J, Clemens M J. The La antigen inhibits the activation of the interferon-inducible protein kinase PKR by sequestering and unwinding double-stranded RNA. Nucleic Acids Res. 1994;22:2512–2518. doi: 10.1093/nar/22.13.2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339a.Young, S. B., and G. G. Carmichael. Unpublished data.
- 340.Yuwen H, Cox J H, Yewdell J W, Bennink J R, Moss B. Nuclear localization of a double-stranded RNA-binding protein encoded by the vaccinia virus E3L gene. Virology. 1993;195:732–744. doi: 10.1006/viro.1993.1424. [DOI] [PubMed] [Google Scholar]
- 341.Zinn K, DiMaio D, Maniatis T. Identification of two distinct regulatory regions adjacent to the human beta-interferon gene. Cell. 1983;34:865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]
- 342.Zullo J N, Cochran B H, Huang A S, Stiles C D. Platelet-derived growth factor and double-stranded ribonucleic acids stimulate expression of the same genes in 3T3 cells. Cell. 1985;43:793–800. doi: 10.1016/0092-8674(85)90252-1. [DOI] [PubMed] [Google Scholar]