Abstract
Poly(3-hydroxyalkanoates) (PHAs) are a class of microbially produced polyesters that have potential applications as conventional plastics, specifically thermoplastic elastomers. A wealth of biological diversity in PHA formation exists, with at least 100 different PHA constituents and at least five different dedicated PHA biosynthetic pathways. This diversity, in combination with classical microbial physiology and modern molecular biology, has now opened up this area for genetic and metabolic engineering to develop optimal PHA-producing organisms. Commercial processes for PHA production were initially developed by W. R. Grace in the 1960s and later developed by Imperial Chemical Industries, Ltd., in the United Kingdom in the 1970s and 1980s. Since the early 1990s, Metabolix Inc. and Monsanto have been the driving forces behind the commercial exploitation of PHA polymers in the United States. The gram-negative bacterium Ralstonia eutropha, formerly known as Alcaligenes eutrophus, has generally been used as the production organism of choice, and intracellular accumulation of PHA of over 90% of the cell dry weight have been reported. The advent of molecular biological techniques and a developing environmental awareness initiated a renewed scientific interest in PHAs, and the biosynthetic machinery for PHA metabolism has been studied in great detail over the last two decades. Because the structure and monomeric composition of PHAs determine the applications for each type of polymer, a variety of polymers have been synthesized by cofeeding of various substrates or by metabolic engineering of the production organism. Classical microbiology and modern molecular bacterial physiology have been brought together to decipher the intricacies of PHA metabolism both for production purposes and for the unraveling of the natural role of PHAs. This review provides an overview of the different PHA biosynthetic systems and their genetic background, followed by a detailed summation of how this natural diversity is being used to develop commercially attractive, recombinant processes for the large-scale production of PHAs.
INTRODUCTION TO POLY(3-HYDROXYALKANOATES)
Storage Material
Poly(3-hydroxyalkanoates) (PHAs) are structurally simple macromolecules synthesized by many gram-positive and gram-negative bacteria. PHAs are accumulated as discrete granules to levels as high as 90% of the cell dry weight and are generally believed to play a role as sink for carbon and reducing equivalents. When nutrient supplies are imbalanced, it is advantageous for bacteria to store excess nutrients intracellularly, especially as their general fitness is not affected. By polymerizing soluble intermediates into insoluble molecules, the cell does not undergo alterations of its osmotic state and leakage of these valuable compounds out of the cell is prevented. Consequently, the nutrient stores will remain available at a relatively low maintenance cost and with a secured return on investment (36, 182, 239, 240, 286).
Once PHAs are extracted from the bacterial cell, however, these molecules show material properties that are similar to some common plastics such as polypropylene (20). The bacterial origin of the PHAs make these polyesters a natural material, and, indeed, many microorganisms have evolved the ability to degrade these macromolecules. Besides being biodegradable, PHAs are recyclable like the petrochemical thermoplasts. This review summarizes the chemical and physical properties of PHAs and the biochemical and genetic studies of the pathways involved in PHA metabolism. Within this framework, the scientific advances that have been made with the available pha genes for economic PHA production processes will be described.
Chemical Structure
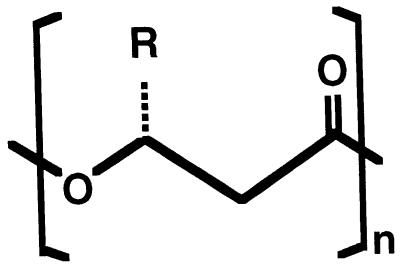
The many different PHAs that have been identified to date are primarily linear, head-to-tail polyesters composed of 3-hydroxy fatty acid monomers. In these polymers, the carboxyl group of one monomer forms an ester bond with the hydroxyl group of the neighboring monomer (Fig. 1). In all PHAs that have been characterized so far, the hydroxyl-substituted carbon atom is of the R configuration, except in some special cases where there is no chirality. At the same C-3 or β position, an alkyl group which can vary from methyl to tridecyl is positioned. However, this alkyl side chain is not necessarily saturated: aromatic, unsaturated, halogenated, epoxidized, and branched monomers have been reported as well (1, 25, 32, 44, 58–60, 85, 125, 126, 135, 247). Specialized, unnatural monomers such as 4-cyanophenylvalerate have been incorporated to obtain new polymers with special properties (124). As well as the variation in the alkyl substituent, the position of the hydroxyl group is somewhat variable, and 4-, 5- and 6-hydroxy acids have been incorporated (51, 131, 277–279). Substituents in the side chains of PHAs can be modified chemically, for instance by cross-linking of unsaturated bonds (39, 67, 68). This variation in the length and composition of the side chains and the ability to modify their reactive substituents is the basis for the diversity of the PHA polymer family and their vast array of potential applications that are described below.
FIG. 1.
Chemical structure of PHAs. PHAs are generally composed of (R)-β-hydroxy fatty acids, where the pendant group (R) varies from methyl (C1) to tridecyl (C13). Other fatty acids that have been incorporated have the hydroxy group at the γ, δ, or ɛ position, while the pendant group may be saturated or unsaturated or contain substituents. The best-known PHAs are P(3HB) (R = methyl), P(3HB-3HV) (R = methyl or ethyl), and P(3HO-3HH) (R = pentyl or propyl).
Historically, poly(3-hydroxybutyrate) [P(3HB)] has been studied most extensively and has triggered the commercial interest in this class of polymers. P(3HB) is the most common type of PHA, and the ability of bacteria to accumulate P(3HB) is often used as a taxonomic characteristic. Copolymers of P(3HB) can be formed by cofeeding of substrates and may result in the formation of polymers containing 3-hydroxyvalerate (3HV) or 4-hydroxybutyrate (4HB) monomers. Together, polymers containing such monomers form a class of PHAs typically referred to as short-side-chain PHAs (ssc-PHAs). In contrast, medium-side-chain PHAs (msc-PHAs) are composed of C6 to C16 3-hydroxy fatty acids. These PHAs are synthesized from fatty acids or other aliphatic carbon sources, and, typically, the composition of the resulting PHA depends on the growth substrate used (17, 105, 135). msc-PHAs are also synthesized from carbohydrates, but the composition of these PHAs is not related to the carbon source (84, 102, 270). The vast majority of microbes synthesize either ssc-PHAs containing primarily 3HB units or msc-PHAs containing 3-hydroxyoctanoate (3HO) and 3-hydroxydecanoate (3HD) as the major monomers (6, 142, 249, 252).
Physical Characteristics
The molecular mass of PHAs varies per PHA producer but is generally on the order of 50,000 to 1,000,000 Da. Although aliphatic polyesters have been studied extensively since the 1920s, their properties were not remarkable and did not initiate a great commercial interest at that time. This was primarily due to the use of relatively impure substrates at the time, which limited the molecular masses of these polymers to 20,000 to 30,000 Da (159). Bacterially produced P(3HB) and other PHAs, however, have a sufficiently high molecular mass to have polymer characteristics that are similar to conventional plastics such as polypropylene (Table 1).
TABLE 1.
Properties of PHAs and polypropylenea
| Parameter | Value forb:
|
||||
|---|---|---|---|---|---|
| P(3HB) | P(3HB-3HV) | P(3HB-4HB) | P(3HO-3HH) | PP | |
| Tm (°C)c | 177 | 145 | 150 | 61 | 176 |
| Tg (°C)d | 2 | −1 | −7 | −36 | −10 |
| Crystallinity (%) | 70 | 56 | 45 | 30 | 60 |
| Extension to break (%) | 5 | 50 | 444 | 300 | 400 |
Data from reference 42.
P(3HB) is poly(3-hydroxybutyrate), P(3HB-3HV) is poly(3-hydroxybutyrate-co-3-hydroxyvalerate) containing 20% 3HV, P(3HB-4HB) is poly(3-hydroxybutyrate-co-4-hydroxybutyrate) containing 16% 4HB, P(3HO-3HH) is poly(3-hydroxyoctanoate-co-3-hydroxyhexanoate) containing 11% 3HH, and PP is polypropylene.
Tm is melting temperature.
Tg is glass transition temperature.
Within the cell, P(3HB) exists in a fluid, amorphous state. However, after extraction from the cell with organic solvents, P(3HB) becomes highly crystalline (43) and in this state is a stiff but brittle material. Because of its brittleness, P(3HB) is not very stress resistant. Also, the relatively high melting temperature of P(3HB) (around 170°C) is close to the temperature where this polymer decomposes thermally and thus limits the ability to process the homopolymer. Initial biotechnological developments were therefore aimed at making PHAs that were easier to process. The incorporation of 3HV into the P(3HB) resulted in a poly(3-hydroxybutyrate-co-3-hydroxyvalerate) [P(3HB-3HV)] copolymer that is less stiff and brittle than P(3HB), that can be used to prepare films with excellent water and gas barrier properties reminiscent of polypropylene, and that can be processed at a lower temperature while retaining most of the other excellent mechanical properties of P(3HB) (159). In contrast to P(3HB) and P(3HB-3HV), msc-PHAs have a much lower level of crystallinity and are more elastic (73, 208). These msc-PHAs potentially have a different range of applications from the ssc-PHAs.
Biological Considerations
The diversity of different monomers that can be incorporated into PHAs, combined with a biological polymerization system that generates high-molecular weight materials, has resulted in a situation where an enormous range of new polymers are potentially available. The advent of genetic engineering combined with modern molecular microbiology now provides us with the exceptional framework for studying plastic properties as a function of genetic and metabolic blueprints. In fact, it presents an enormous challenge to our scientific discipline to fully explore this biology to ensure that environmentally friendly polyesters are available for generations to come.
Biodegradability.
Besides the typical polymeric properties described above, an important characteristic of PHAs is their biodegradability. In nature, a vast consortium of microorganisms is able to degrade PHAs by using secreted PHA hydrolases and PHA depolymerases (for a review of the microbiology and molecular genetics of PHA degradation, see reference 111). The activities of these enzymes may vary and depend on the composition of the polymer, its physical form (amorphous or crystalline), the dimensions of the sample, and, importantly, the environmental conditions. The degradation rate of a piece of P(3HB) is typically on the order of a few months (in anaerobic sewage [Fig. 2]) to years (in seawater) (111, 167–169).
FIG. 2.
Degradation of P(3HB-3HV) in aerobic sewage sludge. Bottles made of P(3HB-3HV) were incubated during the summer (average temperature, 20°C) in aerobic sewage sludge. The progress of degradation is demonstrated with bottles that have been subjected to this treatment for 0, 2, 4, 6, 8, and 10 weeks (from left to right). Photograph courtesy of Dieter Jendrossek, Georg-August-Universität, Göttingen, Germany.
Renewable nature.
As important as the biological characteristics and biodegradability of PHAs is the fact that their production is based on renewable resources. Fermentative production of PHAs is based on agricultural products such as sugars and fatty acids as carbon and energy sources. These agricultural feedstocks are derived from CO2 and water, and after their conversion to biodegradable PHA, the breakdown products are again CO2 and water. Thus, while for some applications the biodegradability is critical, PHAs receive general attention because they are based on renewable compounds instead of on our diminishing fossil fuel stockpiles (293).
Applications
PHAs are natural thermoplastic polyesters, and hence the majority of their applications are as replacements for petrochemical polymers currently in use for packaging and coating applications. The extensive range of physical properties of the PHA family of polymers and the broadened performance obtainable by compounding and blending provide a correspondingly broad range of potential end-use applications, as described in numerous patents.
Initial efforts focused on molding applications, in particular for consumer packaging items such as bottles, cosmetic containers, pens, and golf tees (9, 10, 287). U.S. patents 4,826,493 and 4,880,592 describe the manufacture of P(3HB) and P(3HB-3HV) films and their use as diaper backsheet (163, 164). These films can also be used to make laminates with other polymers such as polyvinyl alcohol (91). Diaper backsheet materials and other materials for manufacturing biodegradable or compostable personal hygiene articles from P(3HB) copolymers other than P(3HB-3HV) have been described (180, 181, 241). PHAs have also been processed into fibers which then were used to construct materials such as nonwoven fabrics (248). P(3HB) and P(3HB-3HV) have been described as hot-melt adhesives (118). PHAs with longer-side-chain hydroxyacids have been used in pressure-sensitive adhesive formulations (229). PHAs can also be used to replace petrochemical polymers in toner and developer compositions (65) or as ion-conducting polymers (221, 222). PHAs can be used as a latex, for instance for paper-coating applications (160), or can be used to produce dairy cream substitutes (298) or flavor delivery agents in foods (299).
In addition to its range of material properties and resulting applications, PHAs promise to be a new source of small molecules. PHA can be hydrolyzed chemically, and the monomers can be converted to commercially attractive molecules such as are β-hydroxy acids, 2-alkenoic acids, β-hydroxyalkanols, β-acyllactones, β-amino acids, and β-hydroxyacid esters (293). The last class of chemicals is currently receiving attention because of potential applications as biodegradable solvents.
PHA BIOSYNTHESIS IN NATURAL ISOLATES
Since 1987, the extensive body of information on P(3HB) metabolism, biochemistry, and physiology has been enriched by molecular genetic studies. Numerous genes encoding enzymes involved in PHA formation and degradation have been cloned and characterized from a variety of microorganisms. From these studies, it is becoming clear that nature has evolved several different pathways for PHA formation, each optimized for the ecological niche of the PHA-producing microorganism. Genetic studies have, furthermore, given insights into the regulation of PHA formation with respect to growth conditions. The cellular physiology of the cell and the important role of central metabolism have become apparent by studying PHA mutants with modifications in genes other than the phb genes. Not only do such studies provide a fundamental insight into microbial physiology, but also they provide the keys for designing and engineering recombinant organisms for PHA production. This section deals with the molecular details of the PHA enzymes and corresponding genes and how their activities blend with cellular metabolism to synthesize PHA only at times where their synthesis is useful.
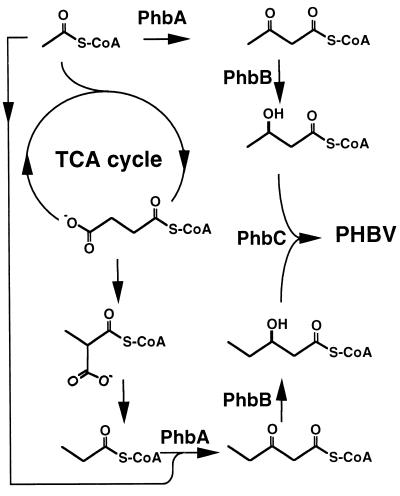
Of all the PHAs, P(3HB) is the most extensively characterized polymer, mainly because it was the first to be discovered, in 1926 by Lemoigne at the Institute Pasteur (152). The P(3HB) biosynthetic pathway consists of three enzymatic reactions catalyzed by three distinct enzymes (Fig. 3). The first reaction consists of the condensation of two acetyl coenzyme A (acetyl-CoA) molecules into acetoacetyl-CoA by β-ketoacyl-CoA thiolase (encoded by phbA). The second reaction is the reduction of acetoacetyl-CoA to (R)-3-hydroxybutyryl-CoA by an NADPH-dependent acetoacetyl-CoA dehydrogenase (encoded by phbB). Lastly, the (R)-3-hydroxybutyryl-CoA monomers are polymerized into poly(3-hydroxybutyrate) by P(3HB) polymerase (encoded by phbC). Although P(3HB) accumulation is a widely distributed prokaryotic phenotype, the biochemical investigations into the enzymatic mechanisms of β-ketoacyl-CoA thiolase, acetoacetyl-CoA reductase, and P(3HB) polymerase have focused on only two of the natural producers, Zoogloea ramigera and Ralstonia eutropha (formerly known as Alcaligenes eutrophus).
FIG. 3.
Biosynthetic pathway for P(3HB). P(3HB) is synthesized in a three-step pathway by the successive action of β-ketoacyl-CoA thiolase (PhbA), acetoacetyl-CoA reductase (PhbB), and P(3HB) polymerase (PhbC). The three enzymes are encoded by the genes of the phbCAB operon. A promoter upstream of phbC transcribes the complete operon.
Essential Genes for PHA Formation
The first phb gene to be isolated was from Z. ramigera (190), an interesting bacterium for biopolymer engineering since it produces both P(3HB) and extracellular polysaccharide (50). By using anti-thiolase antibodies the phbA gene was detected in Escherichia coli carrying a Z. ramigera gene library and was subsequently cloned (190). It was found that phbA and phbB form an operon, while phbC is located elsewhere on the chromosome of Z. ramigera (191). The cloning of phbA and phbB facilitated the purification of the encoded ketoacyl-CoA thiolase and acetoacetyl-CoA reductase for kinetic and mechanistic characterization of these enzymes as described in later sections.
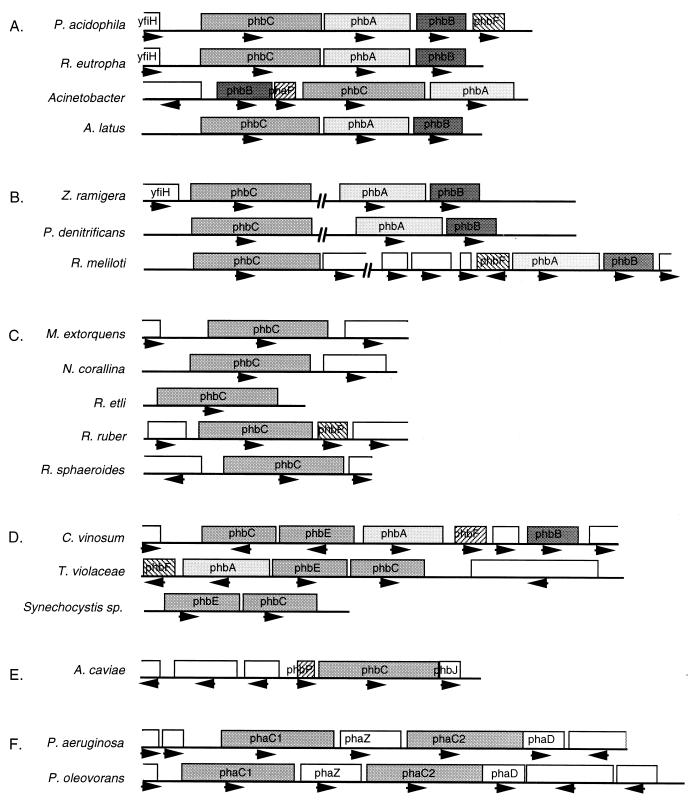
Since the original discovery of these phb genes, many genes encoding enzymes from the PHA pathway have been cloned from different organisms (Fig. 4). Given the diversity of P(3HB) biosynthetic pathways, it is not surprising that the pha loci have diverged considerably. In Acinetobacter spp., Alcaligenes latus, Pseudomonas acidophila, and R. eutropha, the phbCAB genes are in tandem on the chromosome although not necessarily in the same order (108, 192, 193, 232, 274). In Paracoccus denitrificans, Rhizobium meliloti, and Z. ramigera, the phbAB and phbC loci are unlinked (141, 191, 271, 273, 297). PHA polymerase in Chromatium vinosum, Thiocystis violacea, and Synechocystis is a two-subunit enzyme encoded by the phbE and phbC genes. In these organisms, phbAB and phbEC are in one locus but divergently oriented (87, 154, 155). The phb loci in C. vinosum, P. acidophila, R. eutropha, Rhizobium meliloti, and T. violacea all have an additional gene, phbF, that has a hitherto unknown function in PHA metabolism (202), while part of a gene encoding a protein homologous to the hypothetical E. coli protein YfiH is located upstream of the P. acidophila, R. eutropha, and Z. ramigera P(3HB) polymerase genes. In Methylobacterium extorquens, Nocardia corallina, Rhizobium etli, Rhodococcus ruber, and Rhodobacter sphaeroides, only the PHB polymerase-encoding gene has been identified thus far (23, 78, 109, 195, 280). The PHA polymerase gene in Aeromonas caviae is flanked by a unique PHA biosynthetic enzyme encoded by phaJ, which is discussed in further detail below (61). In msc-PHA-producing P. oleovorans and P. aeruginosa, the pha loci contain two phaC genes (107, 269) separated by phaZ, which encodes an intracellular PHA depolymerase (107). The two PHA polymerases are 50 to 60% identical in their primary structure and appear to have a very similar substrate specificity (102, 107).
FIG. 4.
pha and phb operons. The loci encoding the genes for PHA formation have been characterized from 18 different species. Genes specifying enzymes for ssc-PHA formation are designated phb, and those specifying enzymes for msc-PHA formation are designated pha. Not all pathways have completely been elucidated in these strains. The emerging picture is that pha and phb genes are not necessarily clustered and that the gene organization varies from species to species. Other genes possibly related to PHA metabolism may be linked to the essential pha and phb genes. (A) Complete phbCAB operons. (B) Interrupted phb loci. (C) Incomplete phb loci. (D) phb loci from organisms that encode two subunit P(3HB) polymerases. (E) The phbCJ locus of A. caviae involved in P(3HB-3HH) formation. (F) pha loci for msc-PHA formation in Pseudomonas.
Figure 4 provides grounds for some speculation on the evolution of PHA formation. When the first PHA-forming bacteria used this pathway, the purpose of the pathway was probably different from synthesis of a storage material (see also below). PHA formation was most probably a minor metabolic pathway in these organisms, perhaps resulting only from a side reaction. When PHA formation became beneficial for the microbe, evolution selected for improved PHA-accumulating strains under conditions of which we are unfortunately not aware. Knowledge of such conditions would be extremely helpful in the current efforts to optimize PHA production that employ recombinant PHA producers and are described in later sections. Over the course of evolution, phaC was sometimes combined with genes that supply monomer, such as phbAB or phaJ, or with genes involved in other aspects of PHA metabolism, such as phaZ. The selective pressures active at the time resulted in the clustering of pha genes in an operon in some organisms (as in P. acidophila, R. eutropha, Acinetobacter, Alcaligenes latus, and Aeromonas caviae) or as separate transcriptional units in others (as in Z. ramigera, P. denitrificans, Rhizobium meliloti, C. vinosum, T. violacea, P. oleovorans, P. putida, and perhaps other microorganisms for which no thiolase and reductase genes have been identified yet). A second evolutionary force must have worked on the pha genes since some but not all of these diversely structured loci contain phbF and phbP genes or homologs of yfiH. Whether the ancestral PHA polymerase was encoded by one (phaC) or two (phaEC) open reading frames is an open question. Since the two-subunit polymerase systems in C. vinosum and T. violaceae do have neighboring thiolase and reductase genes whereas phaEC in Synechocystis does not, fusion of phaEC or splicing of phaC may have preceded the rearrangements in the pha loci.
Although B. megaterium was the first strain from which P(3HB) was isolated and identified, its biosynthetic machinery has not yet been characterized. The recently isolated B. megaterium mutants impaired in P(3HB) formation (55) should allow the cloning and characterization of the phb genes from this historically relevant P(3HB) producer.
The Three-Step ssc-PHA Biosynthetic Pathway
β-Ketoacyl-CoA thiolase.
β-Ketoacyl-CoA thiolase catalyzes the first step in P(3HB) formation. The P(3HB) biosynthetic thiolase (acetyl-CoA:acetyl-CoA-acetyl transferase; EC 2.3.1.9) is a member of a family of enzymes involved in the thiolytic cleavage of substrates into acyl-CoA plus acetyl-CoA. These β-ketoacyl-CoA thiolases are found throughout nature from higher eukaryotes to yeasts to prokaryotes and are divided into two groups based on their substrate specificity. The first group consists of thiolases with a broad specificity for β-ketoacyl-CoAs ranging from C4 to C16. This class of enzymes is involved mainly in the degradation of fatty acids and is located in the cytoplasm of prokaryotes and in the mitochondria and peroxisomes of mammalian and plant cells. The second class of β-ketoacyl-CoA thiolases is considered biosynthetic and has a narrow range of chain length specificity, from C3 to C5. Throughout nature, these biosynthetic thiolases are specialized for a variety of roles such as ketone body formation, steroid and isoprenoid biosynthesis, and P(3HB) synthesis. The thiolase involved in P(3HB) formation is a biosynthetic thiolase with specificity primarily for acetoacetyl-CoA (166).
R. eutropha contains two β-ketothiolases (enzyme A and enzyme B) that are able to act in the biosynthetic pathway to P(3HB) synthesis. The major difference between these two enzymes is their substrate specificity. Enzyme A is a homotetramer of 44-kDa subunits and converts acetoacetyl-CoA and 3-ketopentanoyl-CoA (but only at 3% relative activity in comparison to acetoacetyl-CoA). In contrast, enzyme B, a homotetramer of 46-kDa subunits, has a broader substrate specificity and cleaves acetoacetyl-CoA as well as 3-ketopentanoyl-CoA, 3-ketohexanoyl-CoA, 3-ketoheptanoyl-CoA, 3-ketooctanoyl-CoA, and 3-ketodecanoyl-CoA (30, 17, 19, 10, and 12% activity relative to acetoacetyl-CoA, respectively). Originally it was thought that the major role of enzyme B is in fatty acid degradation while the primary role of enzyme A (PhbA) is in the biosynthesis of P(3HB) (81). Recently, however, it has been shown that enzyme B is the primary source of the 3HV monomer for P(3HB-3HV) formation (244).
The enzymatic mechanism of PhbA consists of two half-reactions that result in the condensation of two acetyl-CoA molecules into acetoacetyl-CoA. In the first half-reaction, an active-site cysteine attacks an acetyl-S-CoA molecule to form an acetyl-S-enzyme intermediate. In the second half-reaction, a second cysteine deprotonates another acetyl-CoA, resulting in an activated acetyl-CoA intermediate that is able to attack the acetyl-S-enzyme intermediate and form acetoacetyl-CoA (165). The involvement of a cysteine(s) in the active site of the P(3HB) thiolase was first hypothesized in 1953 because the thiolase was inhibited by sulfhydryl-blocking agents (156). In the late 1980s, the roles of cysteines in the active site of the P(3HB) thiolase were definitively determined, after the thiolase gene from Z. ramigera had been cloned and the enzyme had been overproduced and purified. The cysteine involved in the acetyl-S-enzyme intermediate was identified as Cys89 by peptide sequencing of the radioactive peptide after tryptic digestion of radiolabeled enzyme with [14C]iodoacetamide or [14C]acetyl-CoA (35, 267). A C89S thiolase mutant was also constructed and determined to be severely affected in catalysis but not substrate affinity (165, 267). The second cysteine in the active site of P(3HB) thiolase was determined by using affinity-labeled inactivators such as bromoacetyl-S-pantethene-11-pivalate. By using this inhibitor, Cys378 was identified as a potential residue for the second active-site cysteine that deprotonates the second acetyl-CoA molecule (34, 186) and the C378G mutant was virtually inactive (165, 186). So far, all P(3HB) thiolases contain these two active-site cysteines, and it is believed that all the P(3HB) thiolases use the same enzymatic mechanism to condense acetyl-CoA with either acetyl-CoA or acyl-CoA.
Acetoacetyl-CoA reductase.
Acetoacetyl-CoA reductase is an (R)-3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.36) and catalyzes the second step in the P(3HB) biosynthetic pathway by converting acetoacetyl-CoA into 3-hydroxybutyryl-CoA. The acetoacetyl-CoA reductase from Z. ramigera is a homotetramer of 25-kDa subunits and has been classified as an NADPH-dependent reductase (62, 198, 231). Although both NADPH- and NADH-dependent acetoacetyl-CoA reductase activities have been observed in cell extracts of R. eutropha, only the former is involved in P(3HB) synthesis (82). The only known NADH-dependent acetoacetyl-CoA reductase involved in P(3HB) formation to date was found in C. vinosum (155). Although the phbB gene product from Paracoccus denitrificans was initially ascribed to be NADH dependent (297), subsequent overexpression of this enzyme and characterization proved this reductase to be active only with NADPH (29).
The enzymatic reactions involved in P(3HB) synthesis have been extensively analyzed by biochemical techniques and provide clues about the regulation of this pathway. The preferred reaction for the thiolase is thiolytic cleavage, which occurs in the direction opposite to the P(3HB) biosynthetic pathway. However, under P(3HB)-accumulating conditions the enzyme acts against its thermodynamically favored direction when the activities of acetoacetyl-CoA reductase and P(3HB) polymerase pull the condensation reaction (reviewed in reference 166). The availability of reducing equivalents in the form of NADPH is therefore considered to be the driving force for P(3HB) formation.
In the P(3HB) biosynthetic pathway, the reactions catalyzed by thiolase and reductase provide the monomer for PHA polymerization. The kinetic characteristics and substrate specificities of these two enzymes are therefore crucial in determining the range of products that can be expected to be synthesized in a thiolase, reductase, polymerase pathway, as depicted in Fig. 3. Table 2 shows a compilation of the kinetic characteristics of the best-studied thiolase and reductase enzymes, which provides insights in the use of these enzymes for the formation of P(3HB) copolymers. The concept of dividing PHA formation into monomer supply pathways and polymerization is important since in later sections it will be shown that monomers are not necessarily supplied by dedicated pathways. Some of the strategies currently used in fermentative production processes and also the new developments in metabolic engineering provide examples of the incorporation of monomers that are not supplied by thiolase and/or reductase mediated reactions.
TABLE 2.
Kinetic characteristics of P(3HB) biosynthetic enzymes
| Enzyme and species | Km (mM) | Substrate | Product | Reference |
|---|---|---|---|---|
| Thiolase (condensation) | ||||
| Z. ramigera | 0.33 | Acetyl-CoA | Acetoacetyl-CoA | 35 |
| Thiolase (thiolysis) | ||||
| Z. ramigera | 0.024 | Acetoacetyl-CoA | Acetyl-CoA | 35 |
| 0.46 | Acetoacetyl-pantheteine | Acetyl-CoA + acetyl-pantheteine | 35 | |
| 0.073 | Acetoacetyl-pantheteine-11-pivalate | Acetyl-CoA + acetyl-pantheteine-11-pivalate | 35 | |
| (50%)a | 3-Ketovaleryl-CoA | Acetyl-CoA + propionyl-CoA | 166 | |
| R. eutropha | 0.044 | Acetoacetyl-CoA | Acetyl-CoA | 252 |
| (3%)b | 3-Ketovaleryl-CoA | Acetyl-CoA + propionyl-CoA | 252 | |
| (0%)b | 3-Ketohexanoyl-CoA | Acetyl-CoA + butanoyl-CoA | 252 | |
| NADPH-dependent reductase | ||||
| Z. ramigera | 0.002 | Acetoacetyl-CoA | 3-Hydroxybutyryl-CoA | 198 |
| 0.002 | 3-Ketovaleryl-CoA | 3-Hydroxyvaleryl-CoA | 198 | |
| 0.010 | 3-Ketohexanoyl-CoA | 3-Hydroxyhexanoyl-CoA | 198 | |
| 0.99 | Acetoacetyl-pantheteine-11-pivalate | 3-Hydroxybutyryl-pantheteine-11-pivalate | 198 | |
| R. eutropha | 0.005 | Acetoacetyl-CoA | 3-Hydroxybutyryl-CoA | 252 |
| (18%)b | 3-Ketovaleryl-CoA | 3-Hydroxyvaleryl-CoA | 252 | |
| (3.6%)b | 3-Ketohexanoyl-CoA | 3-Hydroxyhexanoyl-CoA | 252 | |
| P(3HB) polymerase | ||||
| R. eutropha | 0.72 | 3-Hydroxybutyryl-CoA | P(3HB) | 252 |
| 1.63 | 3-Hydroxyvaleryl-CoA | PHV | 252 | |
| NDc | 3-Hydroxybutyryl-pantheteine-11-pivalate | None | 69 |
Vmax with respect to acetoacetyl-CoA.
Relative activity with respect to acetoacetyl-CoA and 3-hydroxybutyryl-CoA.
ND, not determined.
P(3HB) polymerase.
P(3HB) polymerase is the third enzyme in the biosynthetic pathway for P(3HB) production. The first phbC nucleotide sequence to be reported was from R. eutropha. This gene was isolated by complementation of R. eutropha P(3HB)-negative mutants (192), and the promoter that drives the expression of phbC (235) and the other genes in the phb operon (192, 193) was mapped. Expression of these three genes in E. coli resulted in the accumulation of P(3HB) up to levels exceeding 50% of the cell dry weight (192, 236, 245).
P(3HB) polymerase is just one member of the family of PHA polymerases. All of the polymerases have molecular masses of around 63,000 Da, except for the polymerases from C. vinosum (153), T. violacea (154), and Synechococcus spp. (87, 114), which are composed of two subunits with molecular masses of 40 and 45 kDa. Interestingly, there are only 15 fully conserved residues among the 26 known PHA polymerases, many of which lead only to ssc-PHA formation (Fig. 5). This is remarkable, since these 15 residues represent on average less than 3% of the total number of amino acids in these enzymes. Since PHA polymerase is found in both soluble (hydrophilic) and granule-bound (hydrophobic) states, it may be that evolution has selected for enzymes that are catalytically efficient while presenting few problems related to undesirable “protein–hydrophobic-surface” interactions. The broad variety of PHA-producing microbes would represent a vast spectrum of intracellular conditions to which these enzymes would have to be adapted. This could explain the low level of overall conserved sequence identity between the different PHA polymerases.
FIG. 5.
Sequence similarity of representatives of three types of PHA polymerases. R. eutropha ssc-PHA polymerase (PHB1), P. oleovorans msc-PHA polymerase (PHO), and the PhbC subunit of the two-subunit polymerase from Synechocystis sp. (PHB2) were aligned by using the program of Higgins (MacDNASIS; IntelliGenetics, Mountain View, Calif.). Residues conserved in all PHA polymerases identified to date are marked by an asterisk.
Early biochemical studies of PHB polymerase were hampered by the low activity of the protein purified from the natural PHB producers. These studies, however, indicated that the enzyme exists in both soluble and granule-bound forms (64, 83). It was proposed that two cysteine residues might be involved in catalysis, with one cysteine holding the growing PHA chain while the other cysteine holds the incoming monomer (72). To test this theory, two cysteines (Cys319 and Cys459) in the R. eutropha P(3HB) polymerase were mutated (70). Cys319 is conserved in all the synthases isolated to date (250), while Cys459 is conserved between only the R. eutropha and the P. oleovorans PHA polymerases. Cys319 was shown to be an active-site residue, because serine and alanine mutations rendered the enzyme inactive. In contrast, when the second cysteine (Cys459) was mutated to a serine, the enzyme retained 90% of the wild-type activity (70). By using the tritiated trimer (3HB)3-CoA, it was shown that the P(3HB) polymer is covalently bound to the P(3HB) polymerase through Cys319 (296).
To explain the ability of the enzyme to form ester bonds with only one cysteine residue, a second thiol was proposed to exist via posttranslational modification. Phosphopantetheine was proposed as a potential posttranslational modification moiety for P(3HB) polymerase (70). A phosphopantetheine posttranslational modification has been found in acyl-carrier protein and enzymes in enterobactin biosynthesis (110). By using a P(3HB) polymerase overexpression system, it was shown that the PhbC enzyme is radioactively labeled when β-[3H]alanine, a precursor of phosphopantetheine, is supplied to the culture. The most likely residue to be modified by phosphopantetheinylation is Ser260 (70), a residue conserved in all phaC genes characterized to date (Fig. 5) and part of a region that resembles similar sites in panthethenylated enzymes (70).
Given the function of the polymerases in forming ester bonds, it is not surprising to find the active-site cysteine residue of these enzymes in a lipase box, Gly-X-Cys319-X-Gly-Gly. The active site of a lipase generally consists of a nucleophile, either cysteine or serine, whose reactivity is enhanced by an aspartate residue and a histidine residue (16, 194, 295). Together, these three residues form a catalytic triad. Candidates for these aspartate and histidine residues are conserved in the polymerases, namely, aspartate residues at positions 351, 428, and 480 and histidine residues at positions 481 and 508 (Fig. 5). Given that PHA polymerase may have two active-site thiols, it is possible that two of the three conserved aspartate residues and both conserved histidines are part of a catalytic triad. The occurrence of the strictly conserved Trp425 in the proximity of Asp428 and the conserved dyads Asp480-His481 and Gly507-His508 underscores the likely importance of these residues in catalysis. Analogously, the strict conservation of Pro239, Asn248, Tyr251, and Asp254 in the direct vicinity of the critical Ser260 residues underscores the importance of this stretch of amino acids.
Model for PHA Granule Formation
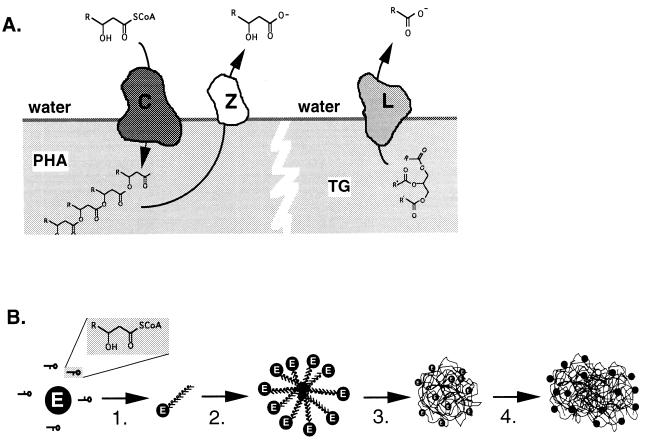
The resemblance of the active sites of PHA polymerases and lipases, as well as the preferred localization of these enzymes (Fig. 6A), suggests how the process of granule formation may proceed. Both enzymes act on ester bonds at the interface of a hydrophobic vesicle and water. The difference between these enzymes is in the direction of the reaction that they catalyze, either toward ester formation or towards ester hydrolysis. In the aqueous environment of the cytosol, the PHA polymerase is quite a remarkable enzyme since it performs an esterification reaction under typically unfavorable aqueous conditions.
FIG. 6.
(A) Similarities between PHA polymerase and lipase. PHA polymerase (C) acts at the surface of a PHA granule, where soluble precursors are polymerized and deposited in the hydrophobic environment of the granule. PHA depolymerase (Z) also acts at this surface and liberates the monomers from the polymer. Both enzymatic reactions are reminiscent of that of lipase (L), which cleaves ester bonds at triglyceride (TG)/water interfaces, yielding free acids and alkanols. (B) Proposed mechanism for the formation of PHA granules. Soluble enzyme converts monomer-CoA to oligomers, which remain enzyme bound (step 1). At a critical oligomer length and enzyme-oligomer concentration, the enzyme-oligomer complexes form micelles with the enzyme located at the interface, separating the PHA from the cytosol (step 2). Because of this compartmentalization, PHA polymerization is facilitated. Because the hydrophobic polymer can now be extruded into a hydrophobic environment instead of the aqueous phase, the reaction proceeds faster. The micelles are expanded and now appear as intracellular, granular structures visible with the phase-contrast microscope (step 3). As the number of granules increase, they may fuse and coalesce, giving rise to large aggregates of PHA (step 4).
Gerngross and Martin investigated P(3HB) granule formation in vitro and developed a model for P(3HB) granule formation (69). First, soluble P(3HB) polymerase interacts with increasing concentrations of 3-hydroxybutyryl-CoA in the cytoplasm, resulting in priming of the enzyme by an unknown mechanism. During an initial lag phase, HB oligomers are slowly formed and extruded from the enzyme. The HB oligomers then form micelles as the oligomers increase in length and hydrophobicity. Consequently, the micelle-like particles provide a two-phase boundary with the polymerase located at the interface. The enzyme then rapidly proceeds with P(3HB) synthesis, extruding more P(3HB) into the growing granule. Eventually the micelles are thought to coalesce into larger granules that can be visualized by microscopy (69) (Fig. 6B).
In vitro studies of the covalent linkage of the 3HB trimer support this model, since a shift in the conformation of the P(3HB) polymerase from monomer to dimer appeared to coincide with the binding of the trimer. Because the P(3HB) polymerase dimer was more active than the monomer and showed a greatly decreased lag time, it was suggested that the lag time in vitro is related to the initial acylation step. It is not yet clear whether this covalent catalysis in the polymerase-catalyzed reaction relates to in vivo priming (296). Physiologically this makes sense, however, since the formation of relatively few high-molecular-weight PHA molecules is expected to be favored over the formation of many low-molecular-weight PHA oligomers. As pointed out above, PHA is considered an osmotically inert macromolecule which depends on having a high molecular weight. Slow PHA polymerase activation in the priming process, combined with a rapid polymerization once activated enzyme forms micelle structures, appears to ensure the formation of high-molecular-weight materials.
The studies by Gerngross and Martin have, furthermore, established that the minimal requirements for P(3HB) synthesis are the (R)-3-hydroxybutyryl-CoA substrate and P(3HB) polymerase (69). P(3HB) polymerase is present both in soluble and granule-bound forms, but the soluble P(3HB) polymerase appears less active. Because of the higher activity when granule bound, optimal P(3HB) accumulation occurs when more enzyme is associated with the growing granule. Maintenance of the available surface is thus critical for efficient P(3HB) production. In subsequent studies, Martin and Gerngross observed that the size of in vitro-synthesized granules is related to the amount of protein added to the assay mixture, irrespective of whether this protein is PHB polymerase or an unrelated protein such as bovine serum albumin (161).
PhaP is a natural PHA-binding protein that determines the size of PHA granules. phaP was identified in genetic studies as a locus causing a P(3HB) leaky phenotype in R. eutropha. The phaP gene was cloned from a cosmid library and found to encode a 24-kDa protein that binds to the P(3HB) granule. Immunochemical analysis with anti-PhaP antibodies revealed that the protein is always granule bound and no free PhaP is present in the cytoplasm of the wild-type strain. Genetic studies have furthermore shown that the concentration of PhaP is inversely related to the size of the granule, since overexpression of PhaP resulted in the formation of many small P(3HB) granules while a phaP mutant contained only a single P(3HB) granule. The P(3HB) leaky phenotype in phaP mutants may therefore be the result of a decreased surface area available for P(3HB) synthesis and causes the observed low polymerase activity (289). This situation indicates an interesting regulatory phenomenon in which maximal activity is obtained by localization of the enzyme to a site which is created and maintained by a structural protein. PhaP is not essential in this regard, but in vivo this protein is likely to be involved in maintenance of the optimal intracellular environment for P(3HB) synthesis and utilization and as such provides guidance during the process of granule formation.
The characteristics of PhaP and related proteins are reminiscent of those of oleosins, proteins that associate exclusively with the oil bodies of oil-producing plants. For that reason, PhaP-like proteins are generally referred to as phasins. It appears that oleosins play a structural role in maintaining the integrity of individual oil bodies by preventing their coalescence (97). Such a role would be especially valuable upon germination of the seeds, when oil degradation is enhanced by a larger surface-to-volume ratio. PhaP and related proteins like GA14 from Rhodococcus ruber, GA14 and GA23 from Methylobacterium rhodesenium, GA13 from Acinetobacter, and the ORF1 gene product from Aeromonas caviae probably have such a function as well and are generally described as phasins (56, 57, 197, 234).
P(3HB)-negative and leaky mutants have been isolated from R. ruber, and subsequent immunochemical analysis showed that these phenotypes were both related to aberrant levels of a granule-associated protein, GA14. The absence of GA14 in P(3HB)-negative mutants is likely to be caused by the absolute requirement of the protein to bind P(3HB) granules, as was observed in R. eutropha, or by a polar effect on its expression by a phaC mutation (Fig. 4) (197). Two carboxy-terminal hydrophobic stretches were shown to be essential for the binding of PhaP to the P(3HB) granules, since PhaP derivatives that lack the two carboxy-terminal hydrophobic domains were unable to do so. This was further supported by the finding that when these carboxy-terminal hydrophobic regions were fused to acetaldehyde dehydrogenase II, the fusion protein localized to the surface of granules in vivo and in vitro rather than to the cytosol (196).
In vitro as well as in vivo studies revealed a role for PHA polymerase in the control of the molecular weight of P(3HB). Variation of the level of PHA polymerase suggested that the concentration of this enzyme is a critical factor in determining the molecular weight of in vitro-synthesized P(3HB). When decreasing amounts of enzyme were supplied to the assay mixture, a polymer was synthesized that had a higher molecular weight (69). New evidence from in vitro studies suggests that P(3HB) formation is a living polymerization in which no chain termination event takes place and that the molecular weight of the resulting polymer is simply dependent on the initial ratio of substrate to enzyme (257). By using an inducer-controlled system to vary PHA polymerase levels in a recombinant E. coli strain, the molecular weight of the formed P(3HB) could also be manipulated as a function of the inducer concentration in the culture medium (242).
Other Pathways for ssc-PHA Formation
P(3HB) is just one type of the many PHAs that are synthesized by thousands of different microorganisms, all originating from their own ecological niche and with their own evolutionary history. Not all these bacteria use the same biological pathways for PHA biosynthesis, since their metabolic blueprints undoubtedly vary. The three-step P(3HB) pathway involves the reactions catalyzed by thiolase, reductase, and polymerase, as exemplified by R. eutropha and Z. ramigera. However, some PHA producers use alternative pathways for PHA formation.
In the absence of a thiolase and reductase, Aeromonas caviae employs an enoyl-CoA hydratase for the formation of the (R)-3-hydroxy monomer from either crotonyl-CoA or hexenoyl-CoA. Other bacteria synthesize P(3HB-3HV) copolyesters from sugars by using a pathway in which 3-HV is derived from the methylmalonyl-CoA pathway. Two additional pathways are found in pseudomonads of rRNA homology group I, which involve either β-oxidation or fatty acid biosynthesis intermediates for msc-PHA production. The biosynthetic pathways for the two types of PHAs have therefore diverged at the level of monomer-CoA-supplying routes, while the polymerases evolved to accept either short- or medium-chain monomers. These pathways are discussed in more detail in this section.
PHA synthesis with an enoyl-CoA hydratase.
A. caviae produces a random copolymer of 3-hydroxybutyrate (3HB) and 3-hydroxyhexanoate (3HH) when growing on even-numbered fatty acids or olive oil as the sole carbon source. When grown on odd-numbered fatty acids, a PHA is produced that consists primarily of 3HV, but small amounts of 3HB are found as well (45). The crystallinity of a poly(3-hydroxybutyrate-3-hydroxyhexanoate) [P(3HB-3HH)] copolymer decreases from 60 to 18% with an increasing 3HH fraction. This property and its decreased melting temperature make P(3HB-3HH) an interesting polymer for several applications where a material that is more flexible than the P(3HB) homopolymer is desired.
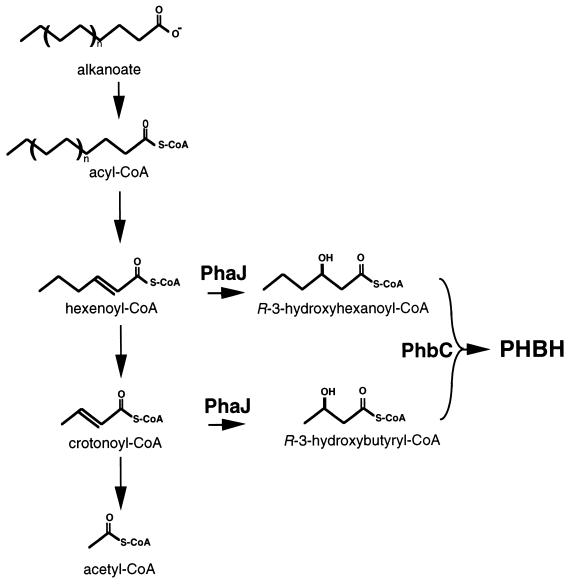
The pha locus from Aeromonas caviae has been cloned and characterized, shedding light on the metabolic pathway that results in P(3HB-3HH) formation (61, 63). It encodes PHA polymerase (encoded by phaC), enoyl-CoA hydratase (encoded by phaJ), and a phasin (encoded by ORF1 or phaP) and is sufficient for PHA formation in PHB-negative heterologous hosts (61, 63, 234). The identification of PhaJ as an (R)-specific enoyl-CoA hydratase suggested that the PHA biosynthetic pathway in A. caviae proceeds from enoyl-CoA derivatives of the fatty acid oxidation pathway (Fig. 7). Besides converting crotonyl-CoA to (R)-3-hydroxybutyryl-CoA, PhaJ converts pentenoyl-CoA and hexenoyl-CoA to PHA precursors, but it does not convert octenoyl-CoA. It was also shown that some PHA-negative mutants of A. caviae are complemented only by phaJ whereas others are complemented only by phaC. phaJ is therefore unique as the first ssc-PHA biosynthetic enzyme besides thiolase, reductase, and polymerase (61, 63).
FIG. 7.
Biosynthetic pathway for P(3HB-3HH). P(3HB-3HH) monomers are derived from fatty acid degradation by converting enoyl-CoA intermediates directly to (R)-3-hydroxyacyl-CoA precursors by an (R)-specific enoyl-CoA hydratase (PhaJ).
The molecular genetic data on P(3HB-3HH) formation in A. caviae provide a new perspective on the work of Moskowitz and Merrick from almost 30 years ago (171). In their work on Rhodospirillum rubrum, these authors proposed a pathway for P(3HB) synthesis that included two hydratases, one specific for the R enantiomer and the other specific for the S enantiomer (171). R. rubrum is able to synthesize PHAs from short- and medium-chain fatty acids up to 20% of the cell dry weight. The major monomers are the C4 and C5 fatty acids, depending on whether the carbon source has an even or odd number of carbons. Small amounts of C6 and C7 monomers were found in PHAs from R. rubrum as well (18). Although this pathway has not been paid much attention for many years, it may now see renewed interest in physiological studies on the formation of PHAs composed of both short- and medium-chain 3-hydroxy fatty acids.
Methylobacterium rhodesenium also uses the activities of two hydratases for P(3HB) synthesis (174). In addition to the two hydratases, this bacterium expresses two constitutive acetoacetyl-CoA reductases, one NADH dependent and one NADPH dependent (173). The combination of these four activities may allow for 3-hydroxybutyryl-CoA synthesis under a range of conditions in the absence of a significant transhydrogenase activity. The analysis of key cofactors in cellular metabolism demonstrated that the flux of acetyl-CoA to the tricarboxylic acid (TCA) cycle or to P(3HB) is determined primarily by the CoA levels (175). Interestingly, the growth substrate has a dramatic effect on the timing of the onset of P(3HB) formation in M. rhodesenium. During exponential growth on fructose, P(3HB) synthesis is used to prevent the formation of excess reducing equivalents. When methanol is the carbon source, reducing power is not excessive until growth is limited by deficiency of other nutrients and P(3HB) is not formed until the stationary phase (3, 172).
P(3HB-3HV) formation from sugars by the methylmalonyl-CoA pathway.
Rhodococcus ruber and Nocardia corallina accumulate PHAs containing 3HV even in the absence of typical HV precursors such as propionate or valerate in the feed (7, 275). Nuclear magnetic resonance spectroscopy (NMR) studies suggested that the 3HV monomer is derived from acetyl-CoA and propionyl-CoA, where the latter is a product of the methylmalonyl-CoA pathway (290). In this pathway, succinyl-CoA is converted to methylmalonyl-CoA, which is decarboxylated to propionyl-CoA (Fig. 8). A mutant strain of N. corallina was constructed in which the gene encoding the large subunit of methylmalonyl-CoA mutase was disrupted. The 3HV fraction in the PHAs formed by the resulting mutants was reduced from 70 to 4% compared to that in the wild-type strain. However, the mutants still accumulated P(3HB) on glucose and succinate and a P(3HB-3HV) copolyester on valerate (275). It appears that N. corallina derives PHA monomers from both the fatty acid degradation pathway and the traditional P(3HB) biosynthetic pathway, in contrast to A. caviae.
FIG. 8.
Biosynthetic pathway for P(3HB-3HV) from carbohydrates. Some microorganisms accumulate P(3HB-3HV) without supplementation of propionate, valerate, or other Codd fatty acids. Propionyl-CoA in these species is formed through the methylmalonyl-CoA pathway, which originates from succinyl-CoA in the TCA cycle. Propionyl-CoA and acetyl-CoA are converted to P(3HB-3HV) by the typical Phb enzymes.
Pathways for msc-PHA Formation
msc-PHAs from fatty acids.
msc-PHAs were not discovered until 1983, when Witholt and coworkers serendipitously found that P. oleovorans grown on 50% octane formed a material that was pliable under conditions where samples are prepared for freeze fracture electron microscopy. Because these materials left mushroom-like structures in the electron micrographs where P(3HB) formed spike structures, further characterization was warranted (41). By using chemically synthesized standards, the inclusions formed from n-octane were determined to be made of a copolyester consisting of 89% (R)-3-hydroxyoctanoate and 11% (R)-3-hydroxyhexanoate (135).
Subsequent studies showed that the composition of the PHAs formed by pseudomonads of the rRNA homology group I were directly related to the structure of the alkane, alkene, or fatty acid carbon source (17, 105, 135). When the carbon source consists of 6 to 12 carbon atoms, the monomers in the PHA are of the same length as the carbon source or have been shortened by 2, 4, or 6 carbon atoms. When the carbon source is a straight-chain C13 to C18 fatty acid, the composition of the polymer resembles that of the C11- and C12-grown bacteria (105). Use of mixtures of hydrocarbons or fatty acids as the carbon source results in the formation of PHAs in which the composition is a reflection of the ratio of the two carbon sources. For instance, when P. oleovorans is supplied with mixtures of octane and 1-octene, the ratio of monomers with an unsaturated bond ranged from 0 to 50% depending on the fraction of 1-octene in the substrate (135). By analogy, substituted 3-hydroxyalkanoates were introduced to different levels by supplying 7-methyloctanoate, 8-bromooctanoate, phenylundecanoate, or cyanophenoxyhexanoate as the cosubstrate (58–60, 85, 124, 126). Incorporation of the last of these substrates results in PHA with monomer constituents that are hyperpolarizable and may confer nonlinear optical properties to the polymer (124).
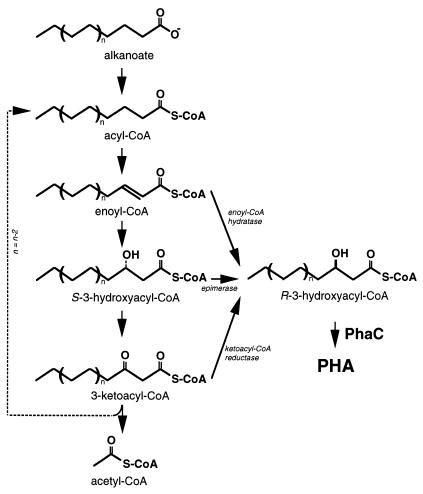
The composition of these PHAs and their direct relationship with the structure of the growth substrate suggested that the msc-PHA biosynthetic pathway is a direct branch of the fatty acid oxidation pathway (Fig. 9) (135). In this pathway, fatty acids are degraded by the removal of C2 units as acetyl-CoA. The remainder of the pathway oxidizes acyl-CoAs to 3-ketoacyl-CoAs via 3-hydroxyacyl-CoA intermediates. The substrate specificity of this msc-PHA polymerase ranges from C6 to C14 (R)-3-hydroxy-alkanoyl-CoAs, with a preference for the C8, C9, and C10 monomers (105). However, because the β-oxidation intermediate is (S)-3-hydroxyacyl-CoA, an additional biosynthetic step is required for synthesis of the (R)-3-hydroxyacyl-CoA monomer. Whether this PHA precursor is the product of a reaction catalyzed by a hydratase (as in A. caviae), by the epimerase activity of the β-oxidation complex, or by a specific 3-ketoacyl-CoA reductase is unknown.
FIG. 9.
Biosynthetic pathway for msc-PHA from hydrocarbons. Fluorescent pseudomonads of rRNA homology group I can derive monomers for PHA from fatty acid degradation. Intermediates from the β-oxidation cycle can be converted to (R)-3-hydroxyacyl-CoA by a hydratase (H), epimerase (E), or reductase (R) activity, whose nature is currently unknown. Any or all of these three enzymes and PHA polymerase determine the limits to the substrate specificity, which is from C6 to C16 3-hydroxy fatty acids.
Given the different biosynthetic pathways, it is not surprising that the pha loci in the msc-PHA-forming pseudomonads are very different from the pha loci in the ssc-PHA-forming bacteria (Fig. 4). Genes involved in msc-PHA formation have been characterized from P. oleovorans (107) and P. aeruginosa (269). In both species, two closely linked PHA polymerases were identified, and PHA polymerase genes are separated by one open reading frame. The two polymerases are approximately 50% identical in their primary structure and appear equally active in PHA synthesis from fatty acids (106, 107) or glucose (102). The open reading frame between phaC1 and phaC2 complements a mutation that prevents the utilization of accumulated PHA. The presence of a lipase box in the primary structure of the product of this gene, phaZ, and the homology of the gene product to other hydrolytic enzymes suggest that this gene encodes a PHA depolymerase (107). Downstream of phaC2 are three genes of unknown function, which may bind to the PHA granules (281).
In vivo experiments with P. putida showed that when either of the two PHA polymerase genes (phaC1 or phaC2) was introduced on a multicopy plasmid, the molecular weights of the PHAs decreased. These reductions were not caused by an increase in PHA depolymerase activity, since the molecular weight of PHA from a depolymerase mutant was not higher than that of PHA from the wild type (106). The latter observation prompted the hypothesis that the molecular weight of PHA is determined by the activity of the PHA polymerase. Based on in vitro analysis of the PHA polymerase from P. oleovorans, it has recently been suggested that the substrate is the limiting factor for PHA formation. Overall, these in vivo and in vitro experiments suggest that the substrate/enzyme ratio, and hence the substrate concentration and enzyme levels, determines the molecular weight of the resulting PHA (129, 130).
msc-PHAs from carbohydrates.
When fluorescent pseudomonads of rRNA homology group I are grown on sugars, a PHA that consists primarily of C10 and C8 monomers is formed (84, 102, 270). Evidence suggests that these monomers are derived from intermediates of fatty acid biosynthesis and that the composition of the PHAs is probably a reflection of the pool of fatty acid biosynthetic intermediates.
It is well known that temperature affects the fatty acid composition of bacterial membranes. Since this effect is due to enzyme activities in fatty acid biosynthesis, the PHA composition was studied in relation to the growth temperature. When P. putida was grown on decanoate, the PHA composition was almost identical irrespective of the growth temperature. In contrast, when glucose was the substrate, the fraction of unsaturated monomers increased from 10 to 20% and the fraction of monomers longer than C10 increased from 18 to 28% when the temperature was lowered from 30 to 15°C. Since the ratio of unsaturated to saturated monomers increases at lower temperature for both membrane lipids and PHA, a metabolic relationship between fatty acid biosynthesis and PHA formation from glucose was suggested (102).
Further corroboration of the involvement of fatty acid biosynthesis in PHA formation for glucose and β-oxidation from fatty acids was obtained by inhibition experiments. Nongrowing cultures of P. putida are able to synthesize PHA from either glucose or fatty acids when carbon sources are in excess. However when cerulenin (a fatty acid synthesis inhibitor) is added to such cell suspensions, no PHA is formed from glucose whereas PHA is still synthesized from fatty acids. Similarly, acrylic acid, a β-oxidation blocker, prevents the formation of PHA from octanoate but not from glucose (100).
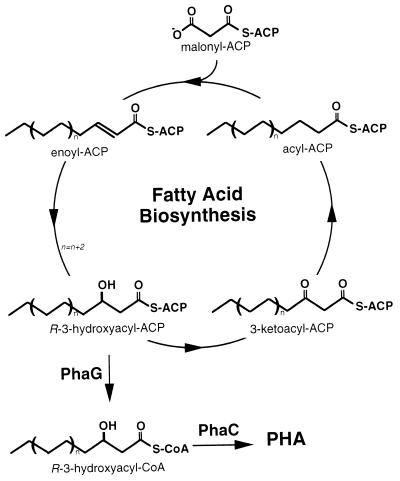
These experiments confirmed that PHA formation from glucose is linked to fatty acid biosynthesis (Fig. 10). Since fatty acid biosynthesis proceeds via (R)-3-hydroxyacyl-ACP, a new enzymatic activity was required that converts this intermediate to (R)-3-hydroxyacyl-CoA. Recently, Rehm et al. determined that the gene product of phaG is responsible for this conversion (214).
FIG. 10.
Biosynthetic pathway for msc-PHA from carbohydrates. Monomers for PHA are derived from the fatty acid biosynthesis pathway as (R)-3-hydroxyacyl-ACP intermediates and are converted to (R)-3-hydroxyacyl-CoA through an acyl-ACP:CoA transacylase encoded by the phaG gene.
Some Pseudomonas spp. can incorporate both ssc- and msc-PHA monomers in the same polymer chain. Typically, these PHAs are formed when these strains are grown on unrelated carbon source such as carbohydrates or 1,3-butanediol (2, 116, 139, 255). The PHA polymerases synthesizing these ssc- and msc-PHAs must therefore have a very broad substrate range. This type of mixed PHA is probably exceptional since it has been shown that physical constraints prevent the formation of mixed granules containing both P(3HB) and msc-PHA chains. This was concluded from experiments where a recombinant P. putida strain containing both the chromosomal phaC and a copy of the R. eutropha phbC on a plasmid was shown to accumulate individual granules composed of either P(3HB) or PHA (206, 268). The recent isolation of PHA polymerase genes from Pseudomonas sp. strain 61-3, which accumulates P(3HB) and P(3HB)-co-PHA granules from glucose (117), should provide further insights into the simultaneous metabolism of the two types of PHA.
Physiological and Genetic Regulation of PHA Production
The regulation of PHA production is quite complex, since it is exerted at the physiological level, through cofactor inhibition of the enzymes and availability of metabolites, and at the genetic level, through alternative ς-factors, two-component regulatory systems, and autoinducing molecules. Another level of regulation is discussed above and relates to granule size and molecular weight control by levels of PHA polymerase and phasins.
Several leaky mutants of R. eutropha that have a phenotype of reduced P(3HB) synthesis have been isolated. Mutations in phbHI alter the timing of P(3HB) synthesis, suggesting a regulatory role for the corresponding gene products. Whereas the wild-type strain synthesized P(3HB) to approximately 90%, phbHI mutants accumulated P(3HB) to 50% of their dry cell weight, although levels of the P(3HB) biosynthetic enzymes were similar in the wild-type and mutant strains. Upon continued incubation of the mutant strain, the polyester was degraded. This degradation of the polymer was not seen to an appreciable degree in the wild-type strain. The mutant also lacked the ability to transiently secrete 3HB (3 mM maximally), in contrast to the wild-type strain, and secreted pyruvate temporarily up to 8 mM instead (210).
Mapping and nucleotide sequencing of the Tn5 insertions indicated that the phbHI mutants resulted from the inactivation of genes encoding homologs of the E. coli phosphoenolpyruvate phosphotransferase system (PEP-PTS). PhbI has 39% identity to enzyme I of E. coli and Salmonella typhimurium, while phbH encodes a gene product with 35% identity to HPr from E. coli, S. typhimurium, and Staphylococcus aureus (210). The PEP-PTS is involved in the PEP-dependent uptake system of sugars in E. coli and S. typhimurium (201), but HPr has also been implicated in regulating chemotactic signaling in E. coli (74) and in regulating ς54-directed transcription (216). Pries et al. proposed that this “leaky” phenotype of phbHI mutants could actually be caused by aberrant regulation of the P(3HB) degradation pathway and suggested that the activity of the P(3HB)-degrading enzymes was controlled by phosphorylation through metabolic signaling that involves a PEP-PTS (210).
Mutants with mutations in phaL compose a second class of leaky mutants of R. eutropha. This gene encodes the lipoamide dehydrogenase component of the pyruvate dehydrogenase enzyme complex. The phaL mutation resulted in the accumulation of only one-third of the normal amount of P(3HB). Instead of funneling excess carbon into P(3HB) upon nitrogen limitation, this mutant secreted pyruvate up to 33 mM. After the complete consumption of the initial carbon source (fructose), pyruvate was utilized as the carbon source. Apparently the phaL mutation results in a decreased flux of carbon into acetyl-CoA and the TCA cycle. As a consequence, the cells do not efficiently metabolize pyruvate upon nitrogen exhaustion and secrete this intermediate. It is of interest that these mutants grow as well as the wild type, as it was expected that a decreased flux through the TCA cycle would affect the growth rate. Although the phaL mutation is a Tn5 insertion within the gene, the mutant still has residual lipoamide dehydrogenase activity. Indeed, it has been shown that R. eutropha has two enzymes that specify this activity. The regulation of these two genes and the role of the second lipoamide dehydrogenase remain to be determined (209).
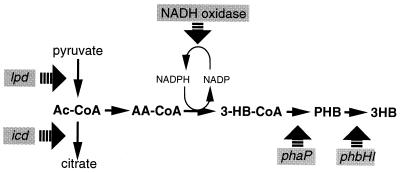
Azotobacter vinelandii UWD is a mutant strain that synthesizes P(3HB) during growth (184). This strain is impaired in NADH oxidase and uses the NADH-NADP transhydrogenase and P(3HB) synthesis to regenerate NAD during growth (158). The increased NADPH level that results from this mutation causes inhibition of citrate synthase and the TCA cycle. Consequently, acetyl-CoA accumulates and is converted to P(3HB) through the NADPH-dependent pathway. This branch point in acetyl-CoA metabolism to either the citric acid cycle or P(3HB) biosynthesis is also important in R. eutropha (89). Park et al. created an increased flux of acetyl-CoA to P(3HB) production by introducing a leaky mutation in the isocitrate dehydrogenase of R. eutropha (188). These findings indicate the importance of the redox balance in the cell in the control of PHB formation.
In Acinetobacter spp. P(3HB) synthesis is stimulated by low phosphate concentrations. A promoter that might be responsible for this regulation was identified by primer extension analysis and found to contain a sequence that is homologous to the pho box identified in E. coli. Whereas all three phb genes appear to be preceded by a promoter region, the phosphate-inducible promoter is only found upstream of the first gene, phaB. This could indicate that for efficient P(3HB) synthesis, the reductase enzyme is limiting and only under conditions of phosphate limitation is the P(3HB) biosynthetic pathway optimally induced (233).
Regulation of PHA synthesis in Pseudomonas has been studied to a limited extent. Many pseudomonads are able to synthesize PHAs by two different pathways: through fatty acid biosynthesis when grown on gluconate or through fatty acid degradation when grown on fatty acids. The two PHA polymerases that have been identified in P. putida are functional in either of the two biosynthetic pathways (102). In P. aeruginosa, the pathway from gluconate is strictly controlled by RpoN, the ς54 subunit of RNA polymerase, while the pathway from fatty acids is completely ς54 independent (269). In contrast to other msc-PHA producers, P. putida KT2442 synthesizes PHA during exponential growth when grown on fatty acids (106). Recently, the involvement of a two-component system homologous to the sensor kinase/response regulator couple LemA-GacA was found to regulate PHA synthesis in this strain (15). LemA, GacA, and their homologs can sense environmental conditions and relay these signals to control the expression of a diverse set of genes (30, 71, 95, 137, 228, 294). Given the potential role of PHAs in nature as a store of excess carbon and reducing equivalents, it is not unlikely that PHA formation is part of a regulon that is controlled by growth conditions.
The synthesis of P(3HB) in Vibrio harveyi is regulated by a 3-hydroxybutyryl-homoserine lactone (258), a signaling molecule that accumulates at high cell densities. A variety of microorganisms regulate the expression of genes at high cell density with such acyl-homoserine lactone derivatives (66). The possible involvement of such signals is consistent with the preferred production of PHAs in stationary phase. Since it was recently shown that GacA homologs and acyl-homoserine lactone derivatives may work through a common signaling pathway (137, 215), the regulatory circuits active on the PHA regulon become more complex. Further studies will clarify whether PHA accumulation is generally regulated by these signals and signal transducers and how environmental information is relayed to the PHA biosynthetic genes.
A hitherto unnoticed open reading frame (phaQ) is located on the opposite strand of all but two of the phaC genes (Table 3) (103). It is unknown whether this putative open reading frame is transcribed. Proteins possibly encoded by phaQ have no similarity to any other protein in the GenBank database. We can therefore only speculate on a function of this open reading frame, and a protein or RNA originating from this locus could be involved in regulating PHA metabolism.
TABLE 3.
Location of phaQ with respect to the endogenous PHA polymerase-encoding gene phaC
| Microorganism | Location ofa:
|
% Overlapb | |
|---|---|---|---|
| phaC | phaQ | ||
| A. caviae | 2640–4478 | 2657–4303 | 89.6 |
| Acinetobacter | 2351–4123 | None | |
| C. vinosum | 831–1898 | 907–1953 | 92.9 |
| M. extorquens | 1099–2736 | 591–2741 | 100 |
| N. corralina | 471–2156 | 551–2587 | 95.3 |
| P. aeruginosa 1 | 1266–2945 | 1472–2935 | 87.1 |
| P. aeruginosa 2 | 4259–5941 | 4687–6096 | 74.6 |
| P. denitrificans | 662–2536 | 205–1605 | 50.3 |
| P. oleovorans 1 | 552–2233 | 492–1908 | 80.6 |
| P. oleovorans 2 | 3217–4950 | 3093–5063 | 100 |
| R. eutropha | 842–2611 | 1075–2619 | 86.8 |
| R. etli | 121–2031 | 48–1400 | 67.0 |
| R. meliloti | 316–2049 | <1–1934 | 93.4 |
| R. sphaeroides | 1023–2828 | 918–2773 | 97.1 |
| R. ruber | 786–2462 | 119–2419 | 97.4 |
| Synechocystis | 2242–3378 | None | |
| T. violacea | 3028–4095 | 2028–4016 | 92.6 |
| Z. ramigera | 740–2470 | 733–2373 | 94.4 |
The location of the coding regions with respect to the reported pha sequences is indicated.
The percent overlap indicates the length of the phaC gene that has phaQ sequence on the complementary strand as part of the length of phaC. It is unknown whether phaQ represents coding information for an actual protein or RNA molecule.
Maintenance of Redox Balance in Nitrogen-Fixing Bacteria
PHA formation in Rhizobium spp. is not commonly studied for reasons of PHA production, but it provides an excellent example of the interplay between cellular metabolism and polyester formation. The symbiosis of Rhizobium species with their host plants provides the plant with a system to fix atmospheric nitrogen through the action of the bacterial nitrogenases in the bacteroid. The complex development of Rhizobium bacteria from free-living cells to bacteroids inside the plant vacuoles after infection of the plant root system is an important subject of study for the development of more efficient plant crops. Werner et al. have indicated that the activities of the enzymes acting on the amino acid pool of the bacteroid are directly related to the effectiveness of the nodules in nitrogen fixation (288). Bergersen et al. postulated that P(3HB) plays a role in the physiology of bacteroids in the nodule (11). The metabolic activity of the bacteroid is thus critical for the establishment of successful symbiosis.
Transposon mutants of Rhizobium meliloti with defects in P(3HB) formation were generated and examined for their effects in symbiosis. The phenotypes of four P(3HB)-negative mutants were similar to that of the wild-type strain with respect to induction of nodule formation on alfalfa (Medicago sativa). In addition, the ethylene-reducing activity, a measure of the nitrogenase activity, was also not affected in these phb mutants. Such results prompted the conclusion that efficient symbiosis between R. meliloti and alfalfa is not affected by alterations in the P(3HB) metabolic pathways (203). This finding is actually not surprising, given that R. meliloti bacteroids typically do not deposit P(3HB) (23).
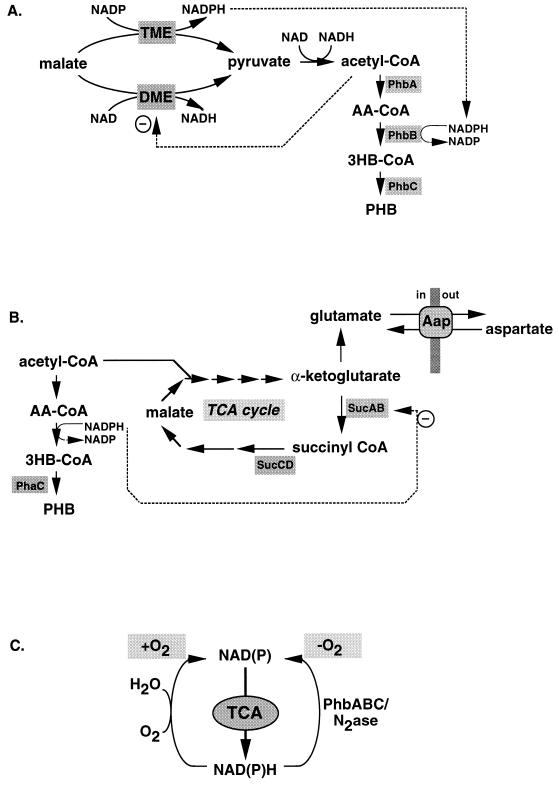
The inability of R. meliloti to form P(3HB) in the bacteroid may be due to low activity of the NADPH-dependent malic enzyme (49). Malate and other four-carbon dicarboxylic acids are provided by the plant and are the preferred carbon sources for the bacteroids (256). In fact, mutants with mutations in either the uptake system for these substrates or the malic enzymes are severely affected in nitrogen fixation. R. meliloti has two malic enzymes, one of which is NADH dependent (encoded by dme) and the other of which is NADPH dependent (encoded by tme). Whereas Dme and Tme are both expressed in the free-living state, Tme expression is repressed specifically in the bacteroid whereas Dme is inhibited by acetyl-CoA. As a consequence, P(3HB) formation is inhibited because too little substrate and too few reducing equivalents are present in the R. meliloti bacteroid to pull acetyl-CoA to 3-hydroxybutyryl-CoA (49) (Fig. 11A). Thus, metabolism in R. meliloti may have evolved so that P(3HB) is not formed in the bacteroid, since P(3HB) formation does not benefit the symbiosis.
FIG. 11.
P(3HB) metabolism and N2 fixation in Rhizobium. (A) In the bacteroid of R. meliloti in symbiosis with alfalfa, the Tme malic enzyme is not expressed while Dme is inhibited by excess acetyl-CoA. Consequently, the levels of NAD(P)H are too low to pull acetyl-CoA into the P(3HB) pathway. In the free-living state, however, both Tme and Dme are active and P(3HB) formation is initiated under the desired conditions. (B) A direct link in central metabolism between the TCA cycle, P(3HB) formation, and amino acid metabolism is apparent from studies of the R. leguminosarum amino acid permease. Mutants that are less sensitive to high levels of aspartate have an increased secretion of glutamate. This increased production of glutamate is caused by inhibition of the TCA cycle either by a mutation in one of the genes encoding a TCA cycle enzyme or by a mutation in the PHA polymerase gene. In the absence of P(3HB) synthesis, the TCA cycle cannot function optimally, since increased reducing equivalents inhibit α-ketoglutarate dehydrogenase. Both types of mutations cause accumulation of α-ketoglutarate, which is directly converted to glutamate. (C) Recycling of reducing equivalents in Rhizobium. The TCA cycle is the most important pathway for supplying precursors of amino acids. To keep the TCA cycle active in the anaerobic bacteroid, P(3HB) biosynthesis and nitrogenase oxidize reducing equivalents. Different Rhizobium spp. have evolved different means to regulate the three NAD(P)H-oxidizing pathways in the free-living or bacteroid state.
In contrast to R. meliloti, R. etli does form P(3HB) in both the free-living and bacteroid state. R. etli CE3 is auxotrophic for biotin and thiamin, cofactors for pyruvate dehydrogenase and α-ketoglutarate dehydrogenase, respectively, and in the absence of these vitamins P(3HB) was accumulated to high levels. As a result of these auxotrophies, the TCA cycle cannot function optimally even in the aerobic free-living state, and the role of the TCA cycle as an overflow mechanism for carbon and reducing equivalents appears to be taken over by P(3HB) formation (53). A P(3HB)-negative mutant of R. etli was constructed by insertion of an antibiotic resistance marker in the phaC gene. This mutant strain was growth impaired when glucose or pyruvate was the carbon source but not when succinate was the carbon source. On succinate the mutant excreted increased levels of organic acids and had a lower ratio of NAD to NADH compared to the parent strain (23). These data underscore the importance of P(3HB) formation for maintaining the redox balance and supporting a functional TCA cycle.
In contrast to the wild-type strain, nodules of the R. etli phaC mutant showed higher and prolonged nitrogenase activity, which fixes atmospheric nitrogen into ammonium ions. As a consequence, plants inoculated with the phaC mutants had a higher nitrogen content (23). It was proposed that the increase in reducing equivalents in the absence of P(3HB) formation is used by nitrogenase, similar to a Rhodobacter sphaeroides P(3HB)-negative mutant which uses the increased reductive power for hydrogen generation (109). The results obtained with the R. etli P(3HB)-negative mutant led to an explanation for the efficiency of alfalfa nodules in nitrogen fixation. These nodules are the result of a symbiosis with phenotypically P(3HB)-negative R. meliloti, which leads to an increased availability of reducing power for the nitrogenase enzyme (90). Apparently, nature has evolved the alfalfa-R. meliloti symbiosis to improve nitrogen fixation by preventing P(3HB) formation. Why the R. etli symbiosis with pea has not selected against P(3HB) formation is a mystery but prompts one to believe that P(3HB) plays another role in this relationship, possibly for survival of R. etli in the free-living state (23).
Hahn et al. studied the nif region of Bradyrhizobium japonicum and found that Tn5 mutants in the nitrogenase-encoding nifD, nifK, and nifH genes resulted in increased P(3HB) accumulation (76). Apparently, the absence of nitrogen fixation in these B. japonicum mutants also results in an energy status of the cell that supports increased P(3HB) synthesis. It seems that P(3HB) synthesis serves as an alternative pathway in these mutants for the regeneration of reducing equivalents.
Studies of amino acid uptake mutants in R. leguminosarum have also indicated a link between amino acid metabolism and P(3HB) formation (Fig. 11B). A general amino acid permease (Aap), which imports amino acids or exports glutamate, has been identified in this organism. However, when glutamate is secreted, no amino acids are taken up. Mutants with reduced activity of this transporter were isolated based on their resistance to aspartate, and the corresponding mutations were mapped in genes encoding the TCA cycle enzymes succinyl-CoA synthetase (sucCD) and 2-oxoglutarate dehydrogenase (sucAB). A second class of mutants had mutations in phaC, encoding P(3HB) polymerase. The increased secretion of glutamate due to mutations in either the TCA cycle or P(3HB) synthesis prevented aspartate uptake to confer the resistance phenotype. Glutamate therefore appears not to be important as a carbon and energy source; instead, the synthesis and secretion of glutamate is important to balance carbon and reducing equivalents, especially in the absence of a functional TCA cycle or PHB pathway. Because bacteroids are typically anaerobic, the TCA cycle requires cofactor regeneration by other means than oxidation with molecular oxygen. Apparently, both glutamate synthesis and P(3HB) synthesis play this role (283).
In the bacteroid stage, the nitrogen fixation apparatus is competing with P(3HB) formation for reducing equivalents. Rhizobium apparently evolved mechanisms to maintain a functional TCA cycle under anaerobic or microaerobic conditions (Fig. 11C). In the bacteroid, the reducing equivalents are used for nitrogen fixation to support symbiosis, but they can be used for P(3HB) formation as well. In the free-living state, nitrogenase is not expressed and P(3HB) plays a role as a sink for excess NAD(P)H when the TCA cycle is not completely active. By regulating the levels of the three different pathways to oxidize NAD(P)H, different Rhizobium spp. have evolved a variety of symbiotic conditions.
Conclusions
PHA biosynthesis proceeds through the action of only a few enzymes, which are specifically involved in PHA formation. The genes encoding these enzymes are essential for PHA formation. In addition, a range of other activities affects the amount of PHA that is accumulated, including enzymes that are involved in central metabolism, global metabolic regulation, or control and maintenance of the surface of PHA granules (Fig. 12). Taken together, these molecular genetic data provide a glimpse of the complexity of PHA metabolism. Since PHA formation is dependent on the fluxes in central metabolic pathways and the levels of precursors, a detailed knowledge of the molecular physiology of PHA metabolism is critical for successful implementation of transgenic PHA producers. Unlike the production of heterologous proteins, which relies mostly on sufficient gene expression, recombinant PHA production involves coordinated expression of heterologous enzymes over a prolonged period and with a concomitant redirection of the metabolism of the host. As a consequence of the metabolic changes introduced by expressing the pha and phb genes, the cell will induce its own responses, which are not necessarily favorable for PHA production. It is therefore critical to understand how bacteria normally regulate PHA formation and how undesired responses from a recombinant host can be prevented. Only then can recombinant processes be successfully developed and lead to what are expected to be the most efficient PHA production processes.
FIG. 12.
Ancillary genes encoding enzymes and proteins that affect PHA accumulation. Three enzymes encoded by three genes are essential for P(3HB) formation. Several other gene products, however, affect P(3HB) formation, and mutations in the corresponding genes may decrease P(3HB) levels. Such enzymes and proteins can act on different aspects of P(3HB) formation: monomer supply, cofactor regeneration, granule assembly, or polymer degradation.
PRODUCTION OF PHAS BY NATURAL ORGANISMS
The different examples provided in the previous section illustrate the diversity of the microbial community with respect to different metabolic pathways that are prominent in bacterial species isolated from different sources but that all lead to the formation of PHAs. It is this diversity of pathways that provides the bricks for the construction of an optimal recombinant PHA producer. Those optimal recombinant PHA producers can be evaluated only in the context of the wild-type organisms. Therefore, in this section the state of the art in PHA production by natural organisms is described to provide the background information needed to assess the merits and prospects of recombinant organisms.
P(3HB) was the only PHA known for almost 50 years until Wallen and Rohwedder (282) identified a number of additional 3-hydroxy fatty acids in active-sludge samples. The major force to commercialize PHAs was Imperial Chemical Industries, Ltd. (ICI), in the 1970s. Several bacterial species were evaluated as potential production organisms. The low cost of methanol and ICI’s experience with fermentations of methanol utilizers made methylotrophic bacteria the obvious first choice. However, the amount of polymer produced per cell was insufficient and its molecular weight was too low for the envisaged applications. The second organism of choice was Azotobacter, since it was microbiologically well understood and was recognized as a putative production organism. However, the studied strains were unstable and secreted polysaccharides. Obviously, the formation of any by-product is undesirable and should be kept to a minimum since it directly impacts the yield of product. The third organism of choice was R. eutropha, which produced high-molecular-weight P(3HB) on fructose. Accumulation of P(3HB) by R. eutropha proceeds preferentially under nitrogen- or phosphate-limiting conditions. The resulting production process with this bacterium was in 200,000-liter stirred fermentation vessels (20).
The first copolymer that was produced in fermentation systems also initiated the subsequent surge in interest in PHAs. A patent by Holmes described the controlled synthesis of P(3HB-3HV), in which the 3HV fraction in the polymer could be controlled by the concentration of propionate in the growth medium (92). After the discovery of polyhydroxyoctanoate [P(3HO-3HH)] (Fig. 1) in octane-grown P. oleovorans (41), the range of different constituents of PHAs expanded rapidly, and currently close to 100 different PHA monomers have been identified (254).
Comparison of PHA production by different organisms is generally not informative, due to the diversity of PHAs, production organisms, substrates, and growth conditions used by different laboratories. One should also consider that the rationale for the various studies may be different and that the different experimental details render the results not comparable. In sophisticated fermentation systems, higher cell densities can be obtained, which inherently lead to higher productivities per unit volume. In this section, we describe the different procedures that have been used to study the production of PHAs. The results are therefore generally presented in terms of “PHA accumulation as the percentage of the cell dry weight” and “monomer composition as the percentage of the polymer.” In general, these studies provide strategies and clues to increase productivities for industrial-scale operations. Production studies with the three most extensively studied organisms are described and are followed by a section on the use of raw but cheap carbon sources for PHA formation by other organisms.
Ralstonia eutropha
R. eutropha was the production organism of choice for ICI in the development of commercial production facilities for P(3HB-3HV) (20). This microorganism grows well in minimal medium at 30°C on a multitude of carbon sources but not on glucose. A glucose-utilizing mutant was therefore selected and used to produce P(3HB-3HV) at a scale of 300 tons per year (21). Chemie Linz GmbH, Linz, Austria, produced P(3HB) from sucrose at up to 1,000 kg per week by using Alcaligenes latus. A. latus is substantially different from R. eutropha and produces P(3HB) during exponential growth, whereas R. eutropha does not start PHA formation until stationary phase (79, 96).
The literature on PHA production by R. eutropha is somewhat confusing due to the different strains that have been used. The three strains that have been studied most extensively are the original P(3HB) producer H16 (ATCC 17699) and its glucose-utilizing mutant known as 11599 in the NCIMB collection. Other well-studied strains are ATCC 17697T, R. eutropha SH-69, and a natural isolate, Alcaligenes sp. strain AK201. R. eutropha has been studied intensively for potential copolymer formation to expand the properties range of ssc-PHAs. Two cultivation techniques have generally been used. In batch experiments, both cell growth and PHA formation are examined in the same medium. In nitrogen-free experiments, cells are pregrown in rich medium and then resuspended in a medium lacking a nitrogen source but with the carbon source of choice.
Feeding strategies for PHA copolymer production.
The first comonomer that was incorporated into P(3HB) in a defined growth medium was 3HV (92). 3HV can be formed by condensation of propionyl-CoA with acetyl-CoA by β-ketoacyl-CoA thiolase, followed by reduction to 3HV-CoA. By varying the ratio of acetate and propionate in the substrate, R. eutropha H16 accumulates P(3HB-3HV) up to 50% of the cell dry weight, with 3HV levels varying between 0 and 45% (46). By using 13C-labeled carbon sources, it was established that the P(3HB-3HV) biosynthetic pathway is through 3-ketoacyl-CoA thiolase, acetoacetyl-CoA reductase, and P(3HB) polymerase. When valerate was supplied as the carbon source to R. eutropha NCIMB 11599, the HV fraction in the polymer was 85%. When mixtures of 5-chlorovalerate and valerate were used, terpolyesters were formed containing 3HB, 3HV, and 5HV monomers up to 46% of the cell dry weight and with 52% 5HV monomer (47). R. eutropha H16 and R. eutropha NCIMB 11599 were directly compared in experiments where butyrate and valerate were used as the carbon source. NCIMB 11599 was able to direct more 3HV monomer to P(3HB-3HV) (90% 3HV) than was H16 (75%). Also, the molecular weight of the polymer produced by NCIMB 11599 was consistently higher. By using 13C-labeled carbon sources, it was established that these fatty acids were converted to P(3HB-3HV) without undergoing complete degradation to acetyl-CoA and propionyl-CoA. This means that either the (S)-3-hydroxyacyl-CoA or 3-ketoacyl-CoA is directly converted into monomer. Interestingly, this pathway operates in the presence of a nitrogen source, in contrast to the pathway from fructose (48). It is possible that inhibition of thiolase during active metabolism of carbohydrates prevents P(3HB) formation during growth whereas a pathway that involves only reductase and polymerase is insensitive to this inhibition.
R. eutropha H16 accumulates copolyesters of 3HB and 4-hydroxybutyrate (4HB) from mixtures of butyrate and 4HB (132) or mixtures with 4-chlorobutyrate, 1,4-butanediol, or γ-butyrolactone (131). With such mixtures of carbon sources, PHA levels reach 40% of the cell dry weight with 4HB levels up to 37%. As a result of the increased 4HB fraction, a lower melting temperature, a decreased crystallinity (132), and an enhanced rate of PHA degradation are obtained (131). Mixtures of butyrate, valerate, and 4HB led to the accumulation of a P(3HB-4HB-3HV) terpolymer with up to 45% 4HB and 23% 3HV (132). Even higher incorporation levels were achieved with mutants of R. eutropha H16 that cannot use valerate or 4HB as the carbon source. When such mutants are tested for copolymer formation, up to 96% 3 HV and 84% 4HB are incorporated (127). Although the total amount of accumulated PHA may be smaller in such mutants, they have great promise for further use in controlled fermentation systems where another carbon source is available to support growth.
Alcaligenes sp. strain AK201 has been studied for P(3HB-3HV) formation on C2 to C22 fatty acids. P(3HB) was formed up to 55% of the cell dry weight on Ceven fatty acids, whereas P(3HB-3HV) was formed on Codd substrates. As expected, the 3HV content of the polymer was higher on the shorter fatty acids. On plant oils and animal fats, P(3HB) levels were also around 50% of the cell dry weight. Interestingly, the molecular weight of the PHA formed was carbon source dependent and was maximal for C7–9 and C13–16 fatty acids (5). On dicarboxylic acids in the C4 to C9 range, P(3HB) homopolymer was accumulated to 50 to 60% of the cell dry weight (4). Further optimization of P(3HB) production on fatty substrates led to polymer levels over 60% of the cell dry weight in a palm oil fed fermentation. On the other hand, oleate, which is the main constituent of palm oil, supported P(3HB) formation to only 42% of the cell dry weight, and this polyester had a lower molecular weight (157). Apparently, palm oil and the free fatty acid that constitutes the oil have a sufficiently different effect on the cells, leading to variations in PHA productivities. Even though these two carbon sources are degraded by the same metabolic pathway, their nature (ionic/soluble or neutral/insoluble) affects PHA formation.
Copolymer production from central metabolites.
At high concentrations, short-chain fatty acids such as propionate and valerate are toxic for R. eutropha. Alternative means of introducing 3HV monomers have therefore been explored. Propionyl-CoA is an intermediate in the degradation pathway of threonine, valine, and isoleucine, and strains with mutations in these pathways were tested for P(3HB-3HV) production. R. eutropha R3 is a prototrophic revertant of an isoleucine auxotroph of R. eutropha H16 and accumulates P(3HB-3HV) with up to 7% 3HV on fructose, gluconate, succinate, acetate, and lactate. To compensate for a threonine dehydratase mutation, R. eutropha R3 overproduces acetolactate synthase and secretes valine and some leucine and isoleucine. Under nitrogen-deficient conditions, however, the precursors of these amino acids, 2-keto-3-isovalerate and 2-keto-3-methylvalerate, are overproduced and subsequently degraded through the propionyl-CoA intermediate (251) (Fig. 13).
FIG. 13.
Endogenous formation of propionyl-CoA in R. eutropha R3, which has altered metabolism of the branched-chain amino acids. This mutant overproduces the acetolactate synthase approximately 15-fold to compensate for a defective threonine dehydratase. The endogenous accumulation of propionyl-CoA under nitrogen-limiting conditions allows this strain to produce P(3HB-3HV) without the supplementation of the growth medium with propionate or other cofeeds.
Addition to threonine, isoleucine, and valine to cultures of R. eutropha SH-69 resulted in the incorporation of 53, 41, and 15% 3HV, respectively. Whereas threonine is toxic at high concentrations and consequently reduces biomass and PHA production, isoleucine and valine are not toxic up to concentrations of 50 mM. When the concentration of amino acid supplements exceeds 10 mM, the fraction of 3HV in the polymer is directly related to the concentration of the amino acid. In contrast, R. eutropha NCIMB 11599 does not incorporate 3HV from threonine and incorporates only up to 2% from isoleucine or valine (302). When R. eutropha H16 was resuspended in Na+- or O2-limiting medium with threonine as the sole carbon source, 6% PHA with 5% 3HV was accumulated (176).
These types of experiments prove that alternative, cell-derived substrates can be used for P(3HB-3HV) synthesis and that supplementation of carbon sources for alternative PHA monomers can be circumvented. Metabolic engineering of new PHA monomer biosynthetic pathways such as from the threonine pathway can thus lead to new P(3HB-3HV)-producing strains. The pathways involved in the biosynthesis of threonine, isoleucine, and valine are well characterized in E. coli and other amino acid producers, and engineered E. coli strains that produce 79 g of threonine per liter are commercially exploited (37). The combination of developments in metabolic engineering of amino acid and PHA pathways provides a tremendous benefit for the successful generation of economic P(3HB-3HV) producers. It is therefore to be expected that other biotechnological processes will aid in the production of some specific PHAs as well.
Fed-batch and continuous culture.
The preceding paragraphs show that the composition of ssc-PHAs is determined by multiple factors. The substrate for growth and PHA formation is an obvious parameter. More important is that central metabolism, especially amino acid metabolism, plays an important role. Recognition of such phenomena allows the metabolic engineer to design PHA-producing strains able to accumulate materials with a number of different compositions. The next paragraphs describe in more detail how R. eutropha is grown to obtain PHAs in large quantities from different carbon sources.
R. eutropha NCIMB 11599 has been studied intensively in high-cell-density fermentation studies. To reach cell densities of 100 g/liter, a fed-batch mode is the preferred way of operation. In fed-batch fermentations, the addition of nutrients is triggered by specific changes in the growth medium as a result of depletion of one of the required medium components. By using a pH-regulated system for glucose supplementation, P(3HB) was produced to 10 g/liter or 17% of the biomass at a productivity of 0.25 g/liter/h. Because the pH increase in response to carbon limitation is slow for this strain, improvements were sought by using the dissolved-oxygen value as the trigger for further glucose addition (DO-stat). When nitrogen was made limiting at a biomass of 70 g/liter and using a DO-stat, P(3HB) was produced to 121 g/liter, corresponding to 75% of the biomass, with a productivity of 2.42 g/liter/h. The yield of P(3HB) was 0.3 g/g of glucose (121). Since pH control under nitrogen-limiting conditions is achieved by the addition of NaOH, problems occur at high densities in large volumes because of the toxicity of highly concentrated hydroxide (230). In addition, it is very important to maintain phosphate and magnesium ion levels above 0.35 g/liter and 10 mg/liter, respectively (8). Ryu et al. therefore studied P(3HB) formation under phosphate-limiting conditions where the pH is controlled by ammonium hydroxide. Under these conditions, P(3HB) levels of 232 g/liter (80% of the cell dry weight) were obtained with a productivity of 3.14 g/liter/h (230). R. eutropha NCIMB 11599 was subsequently grown on tapioca hydrolysate (90% glucose) as a potential cheap carbon source, but unfortunately the presence of toxic compounds, possibly cyanate, in the substrate limited productivity to 1 g/liter/h for a 60-h fermentation (120).
Continuous-culture studies have shown that the theoretical maximal yield of P(3HB) on glucose (0.48 g/g) can be approached to within 5% at a growth rate of 0.05 h−1 (88). Such studies also indicated the importance of the growth rate on 3HV incorporation when a fructose-valerate mixture was used as the substrate (128). At dilution rates varying from 0.06 to 0.32, the 3HV content increased from 11 to 79%. Because the toxicity of propionate is pH dependent, P(3HB-3HV) copolymers with different 3HV contents can be produced by varying the pH of the culture as well (27). As described above, R. eutropha SH-69 accumulates P(3HB-3HV) from glucose as the only carbon source. For this strain, the 3HV fraction in the copolymer is strongly dependent on the glucose concentration in the medium. Maximal accumulation of P(3HB-3HV) occurs with 2 to 3% glucose and a dissolved oxygen concentration of at least 20%. Unfortunately, a 20% 3HV content is not obtained until 6% glucose is supplied (226). R. eutropha DSM 545 produces P(3HB-3HV) from glucose and propionate in fed-batch fermentations under conditions of nitrogen limitation and low dissolved oxygen concentrations. The yield of 3HB on glucose is independent of the dissolved-oxygen concentration, but the HV content is lower at high than at low dissolved-oxygen concentrations (21 and 29%, respectively) (151). The optimal conditions for 3HV incorporation appear to be determined by multiple parameters. As a consequence, the P(3HB-3HV) composition will be influenced to a large extent by the design and setup of the complete process.
Methylobacterium
Methanol is a relatively cheap carbon source and therefore is potentially useful as a substrate for PHA formation (204). Suzuki et al. demonstrated the feasibility of this concept in a series of experiments on P(3HB) formation by Protomonas extorquens sp. strain K (259–262). In a fully automated fed-batch culture, 136 g of P(3HB) per liter was formed in 175 h with a yield of 0.18 g of PHB per g of substrate. This polymer had a molecular mass of 300,000 Da. Improvement of the medium composition increased producing to 149 g/liter in 170 h (260, 261). The effect of physiological parameters such as temperature, pH, and methanol concentration were subsequently studied under the optimized conditions (259). When the growth temperature and pH were drastically different from the optimal conditions (30°C at pH 7.0), the molecular weight of the produced P(3HB) was significantly higher. However, such conditions also resulted in a dramatically reduced yield of P(3HB). The methanol concentration, on the other hand, proved to be a useful parameter for molecular weight control. At methanol concentrations of 0.05 to 2 g/liter, P(3HB) was deposited to 50 and 60% of the cell dry weight with molecular masses ranging from 70,000 to 600,000 Da. At higher methanol concentrations, the yield dropped to 30% and the molecular mass dropped to 30,000 Da (259). By using a slow methanol feed to prevent oxygen limitation in a fed-batch fermentation, P(3HB) was accumulated to 45% of the cell dry weight corresponding to 0.56 g/liter/h with a yield on methanol of 0.20 (14). As a result of the slow feed, a molecular mass of over 1,000,000 Da could be obtained.
By using a natural isolate of Methylobacterium extorquens, P(3HB-3HV) copolymers were produced from methanol-valerate mixtures. The optimal fermentation conditions consisted of a methanol concentration of 1.7 g/liter, and the addition of a complex nitrogen source. Under these conditions, P(3HB) was accumulated to 30% of the cell dry weight with a molecular mass of 250,000 Da (13). Still other isolates such as Methylobacterium sp. strain KCTC0048 have been studied for copolymer synthesis. This organism accumulates P(3HB-3HV), P(3HB-4HB), and poly(3-hydroxybutyrate-co-3-hydroxypropionate) (P[3HB-3HP]) to 30% of the cell dry weight with fractions of 3HV up to 0.7, 4HB up to 0.13, and 3HP up to 0.11 (115).
Whereas M. extorquens incorporates the methanol-derived formic acid into the serine pathway, another PHA producer, P. denitrificans, reduces formation to CO2, which is subsequently fixed by the ribulose bisphosphate pathway. Interestingly, these different pathways have clear effects on P(3HB-3HV) formation by these organisms (272). M. extorquens synthesizes 50% more PHA than P. denitrificans, while the latter incorporates twice as much 3HV on methanol-pentanol mixtures. The 3HV fraction in the PHA produced by P. denitrificans reaches 0.84 and is based on a relatively small amount of 3HB precursor. Under controlled growth conditions with pentanol as the only growth substrate, P. denitrificans accumulates PHV as a homopolymer up to 55% of its cell dry weight (300).
Pseudomonas
The PHA biosynthetic machinery of P. putida is most active toward monomers in the C8 to C10 range. Because long-side-chain fatty acids such as oleate (C18:1) need to be converted in multiple rounds of the β-oxidation pathway before the resulting C8 and C10 monomers can be incorporated, these substrates are less efficiently converted to PHA than is octanoate. Oleic acid, for instance, has to yield 4 acetyl-CoA molecules before a C10 monomer can be incorporated. This conversion yields 20 ATP equivalents in the reduction steps, which is unlikely to occur at a time when excess energy cannot be dissipated. In contrast, decanoic acid and octanoic acid yield 2 ATP equivalents before being incorporated into msc-PHA. As a consequence, the polymer yields per cell are often higher when medium-chain fatty acids are used. Unfortunately, medium-chain fatty acids are generally more expensive, and therefore a compromise between substrate price and conversion yield is being sought.
msc-PHA formation by Pseudomonas from fatty acids.
Inexpensive substrates have been tested for PHA production by Pseudomonas species. Tallow is an inexpensive fat that suffers a production overcapacity. Since it is a mixture of triglycerides with oleic, stearic, and palmitic acids as major fatty acid components, tallow represents an interesting substrate for PHA production. Although some of the better characterized Pseudomonas strains convert hydrolyzed tallow to PHAs at levels between 15 and 20% of their cell dry weight, these organisms do not secrete a lipase enzyme to facilitate tallow hydrolysis. P. resinovorans, however, provides both lipase activity and PHA biosynthetic capacity up to 15% of the cell dry weight (31). Whereas tallow is a widely available feedstock in the United States, other countries such as Malaysia have other carbon sources available for PHA production. Studies by Tan et al. (66) show that P. putida can convert saponified palm kernel oil to PHA. The major fatty acid constituents of palm oil are lauric and myristic acid (>55%). Whereas PHA from either lauric or myristic acid is semicrystalline, PHA from either oleate or saponified palm kernel oil is amorphous (266). Besides their lowest cost, long side-chain fatty acids offer an additional advantage, since they often contain functional groups that make the resulting PHA amenable to modification after isolation (52). The presence of double bonds in some fatty acids results in unsaturated monomers that provide sites for chemical modification of the PHA. When hydrolyzed linseed oil was used, PHA was accumulated up to 20% of the cell dry weight, with 51% of the monomers being polyunsaturated. The primary fatty acids in linseed oil are linolenic acid, oleic acid, and linoleic acid, and these substrates result in monomers with up to three unsaturated bonds. Interestingly, the initial PHA preparation was amorphous, but exposure to air for 3 days resulted in solidification of the material due to cross-linking of the polyunsaturated monomers (22).
Fed-batch and continuous culture.
The yield of PHA on glucose is relatively poor, and production of PHA by fermentation has therefore focused on using fatty acids and hydrocarbons. Initial fermentation studies of P. oleovorans on octane showed that cell growth is limited by the oxygen supply. When the growth rate was lowered by decreasing the growth temperature, a higher cell density was obtained (205). With the data from such batch experiments, fed-batch fermentations resulted in a final biomass of 37 g/liter, 33% of which is PHA with a productivity of 0.25 g of PHA/liter/h. Because octane is a flammable substrate, other production studies mostly involved the use of octanoic acid as the carbon source. By using pure oxygen, P. oleovorans was grown on octanoic acid to a cell density of 42 g/liter, accumulating 37% PHA with a productivity of 0.35 g/liter/h (145). In an experiment where cells were pregrown on a rich medium followed by resuspension in nitrogen-free minimal medium with octanoate, Hori et al. examined the effect of several physiological parameters on PHA production by P. putida (93). The rate of PHA formation is highest at 30°C with an octanoate concentration of 3.5 mM and a pH of 7.8. The molecular mass of the PHA is unchanged over the length of a fermentation process, but both lower temperature (15°C) and a lower octanoate concentration (1.5 mM) give a twofold-higher molecular mass (2.4 × 105 Da). Under these conditions in a two-stage fed-batch fermentation, the yield on octanoate was 0.3 and PHA was accumulated up to 50% of the cell dry weight (93).
Kim et al. studied the effects of the usage of separate carbon sources for growth and PHA production (123). With the use of octanoic acid throughout the fermentation, 25 g of PHA/liter was obtained at a yield of 0.28 g of PHA/g of octanoate. When glucose was used to obtain a biomass of 30 g/liter followed by the supplementation of octanoate for PHA production, the PHA concentration decreased to 18.6 g/liter although the yield was improved to 0.4. The simultaneous supply of both glucose and octanoic acid resulted in 35.9 g of PHA/liter (65% of the cell dry weight) with a high yield (0.4 g/g) and a productivity of 0.92 g/liter/h (123). From these experiments, it appears that mixtures of cheap growth substrates and more expensive substrates for product formation provide a valuable means of lowering PHA production costs. Because oleate is a cheaper substrate than octanoate, its use in a fed-batch production process was studied. Oleate supply was regulated by a DO-stat, and biomass was formed to 92 g/liter, of which 45% was PHA, in only 26 h. This resulted in the production of 1.6 g PHA/liter/h (100).
These studies show the tremendous impact of the growth conditions on PHA formation. Besides these fed-batch studies, optimization of PHA formation was also studied in continuous culture. Although continuous cultures are not industrially feasible and rarely reach the densities of fed-batch cultures, they often provide useful information for the scale-up of production processes.
At low biomass concentrations and a generation time of 0.1 generation/h, P. oleovorans produced PHA at a rate of 0.20 g/liter/h on either octane (207) or octanoate (213). Improvements in the medium composition led to a higher productivity (0.56 g/liter/h), primarily because of a higher biomass concentration (205). Similar studies by Huijberts and Eggink describe PHA production on oleate. The highest volumetric productivity obtained was 0.69 g/liter/h at a generation time of 0.1 h−1 (101). Although these productivities are lower than those obtained in fed-batch cultures, the data show the importance of the growth medium and give an indication of the optimal generation time during the later stages of growth.
PHA formation by Pseudomonas from carbohydrates.
Initially it was surprising when P. putida strains were found to be able to accumulate PHAs from glucose and other sugars. The first msc-PHA producer, P. oleovorans, was unable to do so, and it was expected that the msc-PHA pathway would be exclusively fatty acid based. However, several studies showed that P. putida and P. aeruginosa strains are able to convert acetyl-CoA to medium-chain-length monomers for PHA synthesis. In fact, it now turns out that rather than being the rule, P. oleovorans is an exception among the pseudomonads in being unable to synthesize PHAs from sugars. PHAs that are formed from gluconate or related sugars have a different composition from the PHAs from fatty acids. Whereas the latter PHAs have 3-hydroxyoctanoate as the main constituent, sugar-grown cells accumulate PHAs in which 3-hydroxydecanoate is the main monomer and small amounts of unsaturated monomer are present (84, 102, 270).
PHA Production by Other Microorganisms
PHA producers have been isolated from several waste stream treatment sites, since these facilities often provide a mixture of substrates that select for a variety of organisms. In addition, waste streams often contain high concentrations of organic molecules such as fatty acids, which are inexpensive substrates for PHA formation. Several investigators have studied PHA production by natural isolates from genera such as Sphaerotilus, Agrobacterium, Rhodobacter, Actinobacillus, and Azotobacter to convert organic waste into PHA.
Sphaerotilus natans is a typical inhabitant of activated sludge, where it is associated with the common problem of poor settling of the sludge. Wild-type isolates of this bacterium produce P(3HB) up to 30% of the cell dry weight, but mutants unable to form its encapsulating hydrophilic sheath overproduce P(3HB) up to 50% (265). The P(3HB)-overproducing mutant was found to be tolerant to 6 g of propionate per liter, which is at least sixfold higher than for R. eutropha. Consequently, S. natans is considered an excellent candidate for P(3HB-3HV) synthesis from glucose and propionate mixtures. The high concentration of propionate that can be supplied to the culture facilitates the fermentation process. The 3HV content and the final amount of PHA accumulated are pH dependent in this bacterium. The 3HV fraction varies from 15 to 43% between pH values of 7.3 and 5.9, establishing an additional means of controlling PHA composition besides substrate concentration. Under optimal conditions, PHA was accumulated to 67% of the cell dry weight (264).
Agrobacterium sp. strains SH-1 and GW-014 were isolated from activated sludge as organisms that accumulate P(3HB-3HV) from glucose. Depending on the carbon source, accumulation levels of 30 to 80% PHA with 3 to 11% 3HV were obtained. PHA yields of over 65% with 2 to 6% 3HV were obtained with hexoses such as glucose, fructose, mannitol, and sucrose. On the other hand, PHAs with 8 to 11% 3HV were accumulated when the pentose sugars arabinose and xylose were carbon sources, but only to 35% of the cell dry weight. The propionyl-CoA for 3HV formation is derived from succinate through the methylmalonyl-CoA pathway. It was shown that the specific production rate of the 3HV monomer was dependent on the concentration of Co2+ ions, which form part of the vitamin B12-dependent methylmalonyl-CoA mutase. Fed-batch cultivation on glucose-propionate resulted in PHA formation up to 75% of the dry cell weight with 50% 3HV monomer (140).
Rhodobacter sphaeroides has been studied for the formation of PHA from anaerobically treated palm oil mill effluent (POME). In Malaysia, POME is treated primarily such that the organic acids are converted into methane, which is released into the atmosphere. By combining processes in which POME is converted anaerobically to organic acids, followed by PHA production from these acids by a photosynthetic bacterium, carbon sources in the effluent can be converted to PHA (80).
Actinobacillus sp. strain EL-9 has been isolated from soil and accumulates PHA during the logarithmic growth phase. This strain was studied for the conversion of the reduced sugar components in alcoholic distillery wastewater to PHA. This waste stream is rich in sugar and nitrogeneous compounds, which have a high biological oxygen demand (BOD). Lowering of the BOD of this effluent by using it for PHA formation seems an environmentally sound solution for the treatment of this waste stream while simultaneously producing a useful material. Because Actinobacillus does not require nutrient-limiting conditions, P(3HB) can be formed continuously on the wastewater stream. Comparative studies of different carbon sources showed that enzyme-hydrolyzed alcoholic distillery wastewater gave the highest conversion of its components to biomass (4.8 g/liter), 47% of which is P(3HB) (246).
Azotobacter vinelandii was recognized early on for its ability to produce P(3HB) (20). A. vinelandii UWD was described as a strain that produces P(3HB) during growth, possibly as a result of a defective respiratory NADH oxidase (184). This strain was studied for P(3HB) formation on complex carbon sources such as corn syrup, cane molasses, beet molasses or malt extract (183), fatty acids (185) or swine waste liquor (24). With these different carbohydrates as growth substrates, similar levels and yields of P(3HB) were obtained. Perhaps the unrefined substrates have additional beneficial effects on the fermentation process, since they could promote growth (183). Swine waste liquor consists primarily of acetate, propionate, and butyrate and requires a high BOD. A. vinelandii UWD produces P(3HB-3HV) from twofold-diluted swine waste liquor, but the productivity can be remarkably increased by supplementation of additional carbohydrate sources (24).
Initially, the formation of polysaccharides by A. vinelandii was considered such a disadvantage that continuing exploration of this organism for commercial P(3HB) production was halted (20). In fact, it has been shown that the synthesis of alginate and P(3HB) are interrelated since they play a role in the response of the cell to growth conditions (19). The amounts of alginate and P(3HB) formed by A. vinelandii are dependent on the oxygenation, since a small amount of aeration promotes P(3HB) synthesis over alginate synthesis. The advent of genetic engineering since the initial efforts by ICI has provided mutants of A. vinelandii with diminished alginate formation. P(3HB) accumulation levels in these strains were increased from 46 to 75% of the cell dry weight, with a threefold higher yield on sucrose (162). This finding illustrates how modern molecular biological techniques can potentially have a direct impact on industrial P(3HB) production, as is discussed further in subsequent sections.
Conclusions
To discuss in great detail the vast number of organisms capable of producing PHAs would be beyond the scope of this review. The many PHA producers and the structures of the approximately 100 different monomers have been summarized previously (142, 254). It should be clear, however, that the study of the biosynthetic pathways of these diverse organisms provides insight into the processes necessary to engineer accumulation of a variety of PHA polymers in transgenic organisms. In addition, the study of mutants defective in PHA production will aid in identifying the genes required to efficiently express pha genes in heterologous organisms, such as E. coli and plants. Currently, molecular data on the PHA biosynthetic pathways from over 25 different bacterial species is available. These microorganisms, with their own unique metabolic versatility, provide the foundation from which engineered strains for the production of PHAs can be designed. Not only is this approach useful for recombinant bacterial strains, but also it will be indispensable for further development of a plant crop-based PHA production system.
PHA PRODUCTION BY RECOMBINANT BACTERIA
For the successful implementation of commercial PHA production systems, it is a prerequisite to optimize all facets of the fermentation conditions. The price of the PHA product will ultimately depend on parameters such as substrate cost, PHA yield on the substrate, and the efficiency of product formulation in the downstream processing. This means that high PHA levels as a percentage of the cell dry weight are desirable, as well as a high productivity in terms of gram of product per unit volume and time (38, 40).
Whereas natural PHA producers have become accustomed to accumulating PHA during evolution, they often have a long generation time and relatively low optimal growth temperature, are often hard to lyse, and contain pathways for PHA degradation. Bacteria such as E. coli do not have the capacity to synthesize or degrade PHAs; however, E. coli grows fast and at a higher temperature and is easy to lyse. The faster growth enables a shorter cycle time for the production process, while the higher growth temperature provides a cost saving associated with cooling of the fermentation vessel. The easier lysis of the cells provides cost savings during the purification of the PHA granules. This section gives an overview of the efforts to construct better PHA producers by applying the insights of genetic and metabolic engineering. The effects of altered expression levels of pha genes on PHA formation have been studied in natural PHA producers and are described first.
Recombinant Natural PHA Producers
Several studies report on the effects of additional copies of phb or pha genes on the formation of polymer by the wild-type organism. Although elevated levels of PHA were occasionally found, no dramatic effects of high-copy-number pha genes on PHA metabolism were observed. Such results are consistent with the multilayered regulation of PHA biosynthesis.
When the pha genes from P. oleovorans were introduced into itself or into P. putida, no increased PHA synthesis was observed. The only effect of additional copies of the PHA polymerase-expressing genes was a slight change in the composition of the polymer (107) and a decrease in its molecular weight (106). P(3HB) production in a Rhizobium meliloti P(3HB) mutant is also restored to only wild-type levels by a plasmid-encoded R. meliloti phbC gene (271), whereas an additional P. denitrificans phaC gene on a plasmid doubles the wild-type PHV levels in a pentanol-grown parent strain (273).
In recombinant R. eutropha strains that overexpress the phbCAB genes from a plasmid, the P(3HB) levels are increased from 33 to 40% of the cell dry weight (189). This small increase appears low in comparison to the 1.5- to 3-fold increase in the levels of the individual enzymes and suggests a major influence of the central metabolic pathways on P(3HB) formation. Subsequent studies with these strains in fed-batch cultures indicated that the use of recombinant R. eutropha strains could reduce the fermentation time by 20% while maintaining the same productivity (187). This reduction in fermentation time is significant for commercial production, since the overall productivity for a P(3HB) plant would be 20% higher.
The phbCAB operon from R. eutropha was also expressed in several Pseudomonas strains that normally do not accumulate P(3HB). The plasmid used in these studies expressed the genes successfully, since P(3HB) was deposited by P. aeruginosa, P. putida, P. oleovorans, P. syringae, and P. fluorescens. In contrast, the non-PHA producer P. stutzeri was unable to synthesize P(3HB) with the R. eutropha genes (253). Recently, PHB accumulation up to 25% of the cell dry weight was achieved in a recombinant Synechococcus sp. containing the phb genes from R. eutropha. PHB production was significantly enhanced under nitrogen-limiting conditions and with acetate as the carbon source, yielding a polymer with a molecular mass of 465,000 Da (263).
Recombinant E. coli as PHA Producer
The availability of a large number of PHA biosynthetic genes facilitates the construction of recombinant organisms for the production of P(3HB). Although R. eutropha is an excellent producer of P(3HB), this bacterium has certain limitations that prevent it from being useful for the commercial production of P(3HB). For instance, it grows slowly and is difficult to lyse. In addition, it is not well characterized genetically, which impedes its further manipulation for improved industrial performance. P(3HB) production with recombinant systems may be able to overcome these obstructions. Recombinant E. coli could potentially be used to address these problems, since it is genetically well characterized. P(3HB) production in E. coli must be engineered, because this organism does not naturally synthesize P(3HB) granules. Since the first phb genes were expressed in E. coli (192, 236, 245), a variety of other polymers, such as P(3HB-3HV), P(3HB-4HB), P4HB, and P(3HO-3HH), have been synthesized by E. coli following genetic and metabolic engineering.
P(3HB).
The first indication that P(3HB) could be synthesized in heterologous hosts was obtained when the phb genes from R. eutropha were cloned in E. coli and directed the formation of P(3HB) granules (192, 236, 245). Subsequent reports on cloning of phb genes from other prokaryotes often included similar heterologous expression studies. Even though recombinant E. coli is able to synthesize P(3HB) granules, these strains lack the ability to accumulate levels equivalent to the natural producers in defined media. The first P(3HB) production experiments in fed-batch cultures therefore were in Luria-Bertani (LB) broth, and P(3HB) levels of 90 g/liter were obtained in 42 h with a pH-stat controlled system (122).
In a comprehensive comparison of recombinant E. coli P(3HB)-producing strains, Lee et al. studied 10 different strains equipped with a parB-stabilized phbCAB plasmid (147). Among wild-type strains, E. coli B accumulated P(3HB) to 76% of the cell dry weight on 2% glucose–LB medium, while E. coli W, K-12, and EC3132 formed P(3HB) to only 15 to 33% of the cell dry weight. Typical cloning strains such as XL1-Blue, JM109, and HB101, on the other hand, accumulated P(3HB) to levels varying from 75 to 85% of the cell dry weight. By using stabilized plasmids derived from either medium- or high-copy-number plasmids, it was shown that only high-copy-number vectors support substantial P(3HB) accumulation in E. coli XL1-Blue (146). In a fed-batch fermentation on 2% glucose–LB medium, this strain produced 81% P(3HB) at a productivity of 2.1 g/liter/h (149). The P(3HB) productivity was reduced to 0.46 g/liter/h in minimal medium but could be recovered by the addition of complex nitrogen sources such as yeast extract, tryptone, Casamino Acids, and collagen hydrolysate (144). By supplementing different amino acids separately, it was apparent that P(3HB) formation in recombinant XL1-Blue is limited by available NADPH. Addition of either amino acids or oleate, both of which require substantial reducing equivalents for their synthesis, generally increased cellular P(3HB) levels (148).
Although recombinant E. coli XL1-Blue is able to synthesize substantial levels of P(3HB), growth is impaired by dramatic filamentation of the cells, especially in defined medium (143, 147, 285). By overexpression of FtsZ in this strain, biomass production was improved by 20% and P(3HB) levels were doubled (150). This recombinant strain produced 104 g of P(3HB) per liter in defined medium, corresponding to 70% of the cell dry weight. The volumetric productivity of 2 g/liter/h, however, is lower than achievable with R. eutropha (284).
One of the challenges of producing P(3HB) in recombinant organisms is the stable and constant expression of the phb genes during fermentation. P(3HB) production by recombinant organisms is often hampered by the loss of plasmid from the majority of the bacterial population. Such stability problems may be attributed to the metabolic load exerted by the need to replicate the plasmid and synthesize P(3HB), which diverts acetyl-CoA to P(3HB) rather than to biomass. In addition, plasmid copy numbers often decrease upon continued fermentation because only a few copies provide the required antibiotic resistance or prevent cell death by maintaining parB. For these reasons, Kidwell et al. designed a runaway plasmid to suppress the copy number of the plasmid at 30°C and induce plasmid replication by shifting the temperature to 38°C (119). By using this system, P(3HB) was produced to about 43% of the cell dry weight within 15 h after induction with a volumetric production of 1 g of P(3HB)/liter/h. Although this productivity is of the same order of magnitude as that of natural P(3HB) producers, strains harboring these parB-stabilized runaway replicons still lost the capacity to accumulate P(3HB) during prolonged fermentations.
Whereas the instability of the phb genes in high-cell-density fermentations affects the PHA cost by decreasing the cellular P(3HB) yields, another contributing factor to the comparatively high price of PHAs is the cost of the feedstock. The most common substrate used for P(3HB) production is glucose. Zhang et al. (303) examined E. coli and Klebsiella aerogenes strains for P(3HB) formation on molasses, which cost 33 to 50% less than glucose. The main carbon source in molasses is sucrose. Recombinant E. coli and K. aerogenes strains, carrying the phb locus on a plasmid, grown in minimal medium with 6% sugarcane molasses accumulated P(3HB) to approximately 3 g/liter, corresponding to 45% of the cell dry weight. When the K. aerogenes was grown in a fed-batch system in a 10-liter fermentor on molasses as the sole carbon source, P(3HB) was accumulated to 70% its cell dry weight, which corresponded to 24 g/liter. Although the phb plasmid in K. aerogenes was unstable, this strain shows promise as a P(3HB) producer on molasses, especially since fadR mutants incorporate 3HV up to 55% in the presence of propionate (303).
Morphologically, the number of granules in E. coli and R. eutropha and their size are not the same, even though they were synthesized by the same enzymes (170). By using differential scanning calorimetry, thermogravimetric analysis, and nuclear magnetic resonance, it was shown that the granules in E. coli are in a more crystalline form than the granules in R. eutropha (77). This may be because recombinant E. coli produces P(3HB) of higher molecular weight (133) or because of the absence of specific P(3HB)-binding proteins such as PhaP. The difference in crystallinity was thought to contribute to the differences in degradation of the polymer during purification (77). It was suggested that the increased crystallinity of this high-molecular-weight P(3HB) prevented the embrittlement seen for P(3HB) from natural sources such as R. eutropha (134), and recombinant P(3HB) may therefore have applications for which natural P(3HB) does not qualify.
As described above, the incorporation of other monomers in the growing P(3HB) chain results in polymers with drastically altered and improved mechanical properties. Therefore, recombinant production systems will have to be able to facilitate the production of a variety of copolymers.
P(3HB-3HV).
Engineering E. coli to produce P(3HB-3HV) involved altering the endogenous metabolism of E. coli rather than introducing a specialized set of genes. Supplementation with propionate had generally been used for P(3HB-3HV) formation in R. eutropha, and the initial strategy for recombinant P(3HB-3HV) was therefore similar. Because E. coli does not readily import propionic acid, cultures were adapted on acetate and then a glucose-propionate mixture was added (243). This system was improved by using E. coli strains that have constitutive expression of the ato operon and fad regulon to fully express fatty acid utilization enzymes (54, 243). The ato system transports acetoacetate into the cell, and this is initially activated to acetoacetyl-CoA by AtoAD. AtoAD is also able to transport propionate into the cell (28) (Fig. 14). The fad regulon encodes enzymes for complete degradation of fatty acids, including a broad-specificity thiolase (28). It was expected that the FadA thiolase was beneficial in the pathway for 3HV formation compared to PhbA. The 3HV fraction in the copolymer was dependent on the percentage of propionate used during the fermentation, but it never exceeded 40%. Because E. coli is resistant to 100 mM propionate (243) whereas 30 mM is already toxic for R. eutropha (212), it was suggested that P(3HB-3HV) fermentations may be more efficient with E. coli strains (243).
FIG. 14.
Propionate is an additional carbon source which is supplied as a cosubstrate for the synthesis of P(3HB-3HV) in recombinant E. coli. Several pathways have been shown to be involved in the uptake of propionate and are important in defining the optimal genotype for P(3HB-3HV) production strains. Both the acetoacetate degradation pathway (the Ato system) and the acetate secretion pathway (Ack/Pta) have been identified as contributing to propionate transport.
In subsequent studies, propionyl-CoA formation was studied in strains with mutations in ackA and pta or in strains that overexpress Ack. For efficient incorporation of 3HV into P(3HB-3HV), E. coli requires the Pta and Ack activities (Fig. 14), although the acetate-inducible acetyl-CoA synthase may also be involved (227). The prpE product is a recently discovered acetyl-CoA synthase homolog which actually may be even more specific to propionate (94). The recombinant production systems for P(3HB-3HV) exemplify the need to alter the metabolism of E. coli as well as to adjust feeding strategies in order to produce the desired copolymers. As in E. coli, the fadR mutation also enables Klebsiella oxytoca to produce P(3HB-3HV) when grown on glucose and propionate (303).
Yim et al. reported that these recombinant E. coli P(3HB-3HV) producers are unable to grow to a high density and therefore are unsuited for commercial processes (301). In an attempt to improve P(3HB-3HV) production in a recombinant strain, four E. coli strains (XL1-Blue, JM109, HB101, and DH5α) were tested. All four recombinant E. coli strains synthesized P(3HB-3HV) when grown on glucose and propionate with HV fractions of 7% (301). Unlike the strains studied previously (243), recombinant XL1-Blue incorporated less than 10% HV when the propionic acid concentration was varied between 0 and 80 mM. HV incorporation and PHA formation were increased by pregrowing cells on acetate followed by glucose-propionate addition at a cell density of around 108 cells per ml. Oleate supplementation also stimulated HV incorporation. This recombinant XL1-Blue strain, when pregrown on acetate and with oleate supplementation, reached a cell density of 8 g/liter, 75% of which was P(3HB-3HV), with an HV fraction of 0.16 (301).
P(3HB-4HB) and P(4HB).
P(4HB) is produced in E. coli by introducing genes from a metabolically unrelated pathway into a P(3HB) producer. The hbcT gene from Clostridium kluyveri encodes a 4-hydroxybutyric acid-CoA transferase (104). By engineering hbcT on the same plasmids as phbC from R. eutropha, recombinant E. coli produced 4HB-containing PHAs when grown in the presence of 4HB. Depending on the orientation of the phbC and hbcT genes in the vector and the growth conditions, up to 20% of the cell dry weight was made up of P(4HB) homopolymer. Interestingly, P(4HB) homopolymer was synthesized in the presence of glucose. In the absence of glucose, a P(3HB-4HB) copolymer was accumulated with up to 72% 3HB, even though phbA and phbB were absent. This suggests that E. coli contains an unknown pathway that allows the conversion of 4HB to 3HB (86).
Valentin and Dennis were able to produce P(3HB-4HB) directly from glucose (276). This was accomplished by introducing the succinate degradation pathway from C. kluyveri on a separate plasmid into an E. coli strain harboring a plasmid with the phb biosynthetic genes from R. eutropha. This copolymer is synthesized by redirecting succinyl-CoA from the TCA cycle to 4-hydroxybutyryl-CoA via succinic semialdehyde and 4HB (Fig. 15). P(3HB-4HB) was accumulated to 46% of the cell dry weight with a 1.5% 4HB (276).
FIG. 15.
Biosynthesis of P(3HB-4HB) in recombinant E. coli by using heterologous genes from Clostridium kluyveri. The 4HB monomer in the synthesis of P(3HB-4HB) is derived from succinate. Succinate is converted to 4HB-CoA by enzymes that are encoded by genes from the gram-positive, strictly anaerobic C. kluyveri microbe.
P(3HO-3HH).
E. coli has also been engineered to produce msc-PHAs by introducing the phaC1 and phaC2 gene from P. aeruginosa in a fadB::Kan mutant (136, 211). It was presumed that this mutant accumulated intermediates of the β-oxidation pathway that could be incorporated into PHA by the polymerases. The recombinant E. coli strain accumulated PHA up to 21% of the cell dry weight when grown in LB broth containing decanoate. The polymer contained primarily 3-hydroxydecanoate (73%) and 3-hydroxyoctanoate (19.0%) (136). Interestingly, the fadB mutation in this strain is an insertion mutation and not a point mutation and is noted to have undetectable FadAB activity. If FadAB is the only β-oxidation complex in E. coli, one would expect that this strain would not be capable of degrading fatty acids to PHA monomer.
The phaC1 gene from P. oleovorans also directs PHA formation in E. coli. Strains with a fadA or fadB mutation accumulated PHA up to 12% of the cell dry weight when grown on C8 to C18 fatty acids. By replacing the wild-type promoter of phaC1 with either the alk or lac promoter, polymerase levels were inducible, leading to 20 to 30% PHA formation with PHA polymerase 1 or 2. These experiments show that PHA polymerase is the only dedicated enzyme for PHA biosynthesis in Pseudomonas and that additional enzyme activities may be provided by ancillary enzymes (217).
Conclusions
With the identification of pha genes from multiple organisms, the possibilities of constructing recombinant PHA producers have emerged. History has repeated itself in that P3HB was again the first biological polyester, but now from a recombinant microorganism. The diversity of natural PHAs, however, was rapidly conferred to E. coli, and several ssc-PHAs and msc-PHAs have been synthesized in recombinant bacteria, albeit with various degrees of success. Significant progress must be made to produce a variety of PHAs in recombinant bacteria by cofeeding strategies, let alone from single-carbon sources. The optimization of fermentation systems for these recombinant organisms will also remain a challenge. Since PHAs are not natural products of E. coli, the responses by high-cell-density cultures to nutrient limitations that trigger subsequent feeds are unpredictable. New fermentation feeding strategies will therefore have to be developed.
METABOLIC ENGINEERING OF PHA BIOSYNTHETIC PATHWAYS IN HIGHER ORGANISMS
In an effort to reduce the cost of P(3HB) production, industrial interest has initiated programs for P(3HB) production systems in plant crops. Commercial oil-producing crops, such as Brassica, sunflower, or corn, have been bred to accumulate these oils to high levels. If one were able to replace the oil by PHAs and have the polymer be accumulated to 30% of the seed, PHA production per acre could be around 350 lb. Production of 1 billion lb of PHA would then require an area of 2.5 million acres (8% of the state of Iowa). The potential of an agricultural PHA production system is thus enormous (293). The prospects of producing P(3HB) in plant crops is encouraging now that several studies have reported the synthesis of PHAs in yeast, insect cells, and several plant species.
Saccharomyces cerevisiae
In contrast to E. coli, where the complete P(3HB) pathway had to be introduced for PHA formation to occur, P(3HB) was produced in yeast by expressing only part of the biosynthetic pathway. P(3HB) granules could be visualized in Saccharomyces cerevisiae cells when just the P(3HB) polymerase gene from R. eutropha was introduced into the cells. However, P(3HB) was accumulated to only 0.5% of the cell dry weight. This low level of P(3HB) may result from insufficient activity of the endogenous β-ketoacyl-CoA thiolase and acetoacetyl-CoA reductase enzymes. β-Ketoacyl-CoA thiolase (10 to 20 nmol/min/mg) and acetoacetyl-CoA reductase (150 to 200 nmol/min/mg) were detected and were thought to supply sufficient substrate for P(3HB) polymerase (138). Future improvements of this eukaryotic P(3HB) production system may require elevation of these activities.
Insect Cells
Expression of the R. eutropha phbC gene in insect cells was first achieved in Trichoplusia ni (cabbage looper) cells by using a baculovirus system. Expression of phbC was successful, since within 60 h after viral infection, 50% of the total protein was P(3HB) polymerase. In contrast to other recombinant systems, expression of phbC in insect cells allowed rapid purification of the soluble form of P(3HB) polymerase (291). This is surprising, since overexpression of PhbC in recombinant E. coli usually results in insoluble, inactive P(3HB) polymerase.
An elegant study with insect cells attempted to create a diverse set of PHA monomers endogenously by transfecting a mutant form of the rat fatty acid synthase into Spodoptera frugiperda (fall armyworm) cells by using a baculovirus (292). This previously characterized fatty acid synthase mutant does not extend fatty acids beyond 3HB (113), which was subsequently converted to P(3HB) by the cotransfected P(3HB) polymerase from R. eutropha. The presence of P(3HB) granules in the insect cells was visualized by immunofluorescence. Although P(3HB) production was achieved, only 1 mg of P(3HB) was isolated from 1 liter of cells, corresponding to 0.16% of the cell dry weight. These studies provide examples of the use of alternative, eukaryotic enzymes for the generation of P(3HB) intermediates and the ability to express the phb genes in heterologous hosts (292).
Plants
Recently, efforts have been made to produce P(3HB) in plants. Stable expression of the phb genes has been achieved and the P(3HB) produced is chemically identical to the bacterial products with respect to the thermal properties (Tm, Tg, ΔH), while the molecular weight distribution of the polymer was much broader. Still, a significant fraction of the plant P(3HB) had a molecular weight of 1,000,000, which indicated that plants can make P(3HB) of sufficient quality for industrial processing (200).
Since, in contrast to bacteria, eukaryotic cells are highly compartmentalized, there are a number of challenges in expressing phb genes in plants. phb genes must be targeted to the compartment of the plant cells where the concentration of acetyl-CoA is the highest but only in such a way that growth of the plant is not restricted.
Arabidopsis thaliana.
Although not a crop plant, Arabidopsis thaliana was the first plant of choice for transgenic P(3HB) studies since it is the model organism for heterologous expression studies in plants. The only enzyme of the P(3HB) synthesis pathway naturally found in A. thaliana is 3-ketoacyl-CoA thiolase. This cytoplasmic 3-ketoacyl-CoA thiolase produces mevalonate, the precursor of isoprenoids. Because of the presence of endogenous thiolase activity, only the phbB and phbC genes from R. eutropha were transfected, resulting in the accumulation of P(3HB) granules in the cytoplasm, vacuole, and nucleus. The expression of the phb genes had an adverse effect on growth which was possibly due to the depletion of acetyl-CoA from an essential biosynthetic pathway. Alternatively, P(3HB) accumulation in the nucleus could be detrimental (199). Similar growth defects and low P(3HB) yield were obtained with the commercial crop Brassica napus. These problems could not be surmounted by introducing phbA in the presence of phbB and phbC. This suggests that the endogenous thiolase activity may not have been the critical factor in the phenotypic problems associated to P(3HB) synthesis (178).
An improved plant production system was subsequently developed by expressing all three phb genes in the plastid of A. thaliana. The plastid was targeted for P(3HB) production because of the high level of acetyl-CoA in this organelle, which is the site for lipid biosynthesis. The P(3HB) content in the plastids gradually increased over time, and the maximum amount of P(3HB) in the leaves was 14% of the dry weight (179). In contrast to the broad molecular mass distribution of P(3HB) produced in the cytoplasm (200), P(3HB) isolated from the plastids had a uniform molecular mass of 500,000 Da (177).
Gossypium hirsutum (cotton).
Recently phb genes were engineered into cotton (Gossypium hirsutum) to determine whether P(3HB) formation could alter the characteristics of the cotton fiber. Constructs containing phbB and phbC were targeted to fiber cells. Expression of these constructs was switched on in the early fiber development stages (10 to 15 days postanthesis), under the control of the E6 promoter, or during the late fiber development stages (35 to 40 days postanthesis), when the genes were under the control of the FbL2A promoter. In the fibers of the transgenic plants, the endogenous thiolase activity varied between 0.01 and 0.03 μmol/min/mg and the reductase activity varied between 0.07 and 0.52 μmol/min/mg. Epifluorescence studies showed that P(3HB) granules had been deposited in the cytoplasm (112). Due to the presence of P(3HB) granules in the cotton fiber, the heat capacity of the purified cotton was increased and better insulation properties were obtained (26). Further improvement of P(3HB) and cotton fiber compositions is expected to improve cotton characteristics with respect to dyeability, warmth, and wrinklibility. Even though the maximum levels of P(3HB) amounted to only 3.4 mg/g of dry fiber, the incorporation of P(3HB) to this level already showed an effect.
Zea mays (corn).
The P(3HB) biosynthetic pathway from R. eutropha has also been expressed in Black Mexican sweet maize (Zea mays L.) cell cultures. Cell cultures were grown in a bioreactor for 2 years rather than in fully differentiated plants. The thiolase activity (0.140 U/mg) was constant, but the reductase activity was less stable and decreased from 0.64 to 0.12 U/mg. The phbC gene was initially detected, but after 1.5 years of cultivation it had been lost. In addition to the instability of the phbB and phbC genes, the transformed plant cells grew more slowly than the native cells did (75).
Conclusions
Although P(3HB) synthesis has been achieved in plants, the results obtained so far clearly indicate that a long road is still ahead. In contrast to microorganisms, metabolism in plants is mostly compartmentalized, which complicates the tasks at hand. Current and future developments in the molecular biology of plants will undoubtedly find rapid application in the pursuit of PHAs in plant crops. An intriguing development is the potential for transgenic P(3HB) to play a role in engineering new characteristics into existing materials such as cotton. Obviously, the limits of transgenic PHA production are unpredictable.
POTENTIAL ROLE FOR PHAS IN NATURE
Since bacteria did not evolve PHA production as a means of supplying plastics to mankind, the accumulation of PHAs by bacteria must have evolved out of an advantageous phenotype related to the deposition of these materials. Besides the discussed role as storage material for carbon and reducing equivalents, low-molecular-weight P(3HB) has been found to be part of bacterial Ca2+ channels and is also bound to protein and lipids in eukaryotic systems.
Voltage-Dependent Calcium Channel in Escherichia coli
An extensive body of knowledge was developed by Rosetta Reusch and coworkers at Michigan State University on the possible role and function of low-molecular-weight P(3HB) in microbial physiology (98, 99, 219, 223, 224). Recently it was established that P(3HB) in conjunction with polyphosphate can form a complex in E. coli that transports calcium ions. A model of such a complex is shown in Fig. 16. An alternative model has been based on the crystal structure of pure P(3HB) oligomers; however, that structure does not take the polyphosphate molecule into account (238).
FIG. 16.
Model of the P(3HB)-Ca2+-polyphosphate complex from E. coli. This P(3HB) complex forms a channel in the membrane to transport Ca2+ ions out of the cell. It is proposed that the channel is also involved in DNA uptake by competent E. coli cells. In this model, the Ca2+ ions (green) are localized between the inner polyphosphate molecule (yellow phosphorus atoms and red oxygen atoms) and a P(3HB) helix (red oxygen atoms, blue carbon atoms, and white hydrogen atoms). The methyl side groups of the P(3HB) helix face the outside of the channel and are in contact with the hydrophobic lipids of the membrane. The carbonyl oxygen atoms face the interior of the channel and ligand the Ca2+ ions. The phosphate groups play a similar role. Extrusion of Ca2+ ions may result from physical constraints on the structure or from enzymatic synthesis and degradation of the polyphosphate chain at the membrane/cytosol and membrane/periplasm interfaces.
Complexed P(3HB) (cPHB) is a low-molecular-mass P(3HB) (less than 15,000 Da) that has been found in low concentrations attached to cellular proteins (99) or complexed with calcium and polyphosphate in the form of a calcium channel in the cytoplasmic membrane (219, 224). It has been proposed that these latter structures aid the import of DNA after cells have been made genetically competent in procedures that use calcium ions. When cultures of A. vinelandii, Bacillus subtilis, Haemophilus influenzae, and E. coli are treated to make them genetically competent for DNA uptake, a specific change in the structure of the membrane of these cells is detected by fluorescence studies (223). Comparative studies indicated a close relationship between genetic competence, the appearance of this characteristic change in membrane structure, and the cPHB content of E. coli cells. In these studies, the transformation buffer that is generally used to make E. coli cells competent was varied such that instead of Ca2+ ions, a broad range of mono-, di-, and trivalent cations were examined for their capacity to make cells prone to take up DNA. From these studies, it was clear that only Ca2+ and Mg2+ ions can establish the competence state and that some ions support low efficiencies of transformation or even inhibit DNA uptake completely. For each metal ion, the transformation efficiency was closely related to the structure of the membrane as observed by fluorescence studies (98).
Because this type of P(3HB) is so different from the P(3HB) in the storage granules, new assays were developed to determine the amount of P(3HB) in biological samples. By using these techniques, it has been shown that competent E. coli cells contain cPHB in their cytoplasmic membranes and that the presence of cPHB was directly related to the transformability of the cells. The molar ratio of the components of the P(3HB)-polyphosphate-Ca2+ complex was determined from cPHB purified from genetically competent E. coli to be 1:1:0.5. These isolated cPHB complexes were able to form Ca2+ channels when introduced into liposomes (224) or voltage-activated Ca2+ channels in lipid bilayers. Identification of this channel as a calcium channel constitutes the first known biological nonproteinaceous Ca2+ channel (219). At present, no information is available for the genes and the corresponding gene products that are participating in cPHB biosynthesis. The elucidated genomic sequence of E. coli (12) does not show any significant homolog of a PHA polymerase-encoding gene.
Subsequent work proved that a channel with identical properties can be reconstituted from Ca2+ polyphosphate and synthetically prepared (R)-3-hydroxybutyrate oligomers (33). Recently, P(3HB) and polyphosphate have also been identified as components of purified Ca2+-ATPase from the human erythrocyte, a well-studied Ca2+ channel (220). Given the relative simplicity of the P(3HB)-polyphosphate complex in comparison with the proteinaceous Ca2+ channels, it is tempting to consider the possibility that these bacterial channels have a primordial origin.
Low-Molecular-Weight PHB in Eukaryotic Organisms
P(3HB) is not just an insoluble molecule made by bacteria but, rather, is a unique compound with a variety of roles and functions in nature. P(3HB) has also been found in a variety of plant and animal tissues (218). In human plasma, P(3HB) can be found associated with very-low-density lipoprotein and low-density lipoprotein, but not with high-density lipoprotein. In addition, a significant portion of P(3HB) is found associated with serum albumin. The lipid molecules and albumin are thought to be acting as transporters of P(3HB) through the blood, with albumin being the major carrier (225). If P(3HB) plays a physiological role in large eukaryotic organisms, the need for a P(3HB) carrier makes sense, since P(3HB) is highly insoluble in aqueous solutions.
Possible Evolutionary Precursors of PHB
Since PHB is such a high-molecular-weight molecule, it becomes an intriguing question to find which cellular function has driven its evolution. The direct involvement of DNA, RNA, and protein in sustaining life provides a simple clue for the presence of these macromolecules in the living cell. PHA, however, seems to be an inert molecule, and, as with polysaccharides, it is interesting to speculate about the roots of such molecules. Intracellular stores are obviously advantageous during prolonged periods of starvation, but what was the evolutionary, low-molecular-weight precursor? Why were 3-hydroxyacyl-CoAs found to be good substrates for deposition in intracellular granules, and could they have been abundant in the cell during starvation? Where did the enzymes that facilitate PHA synthesis come from? The most obvious hypothesis for its original biosynthetic pathway is suggested by similarities of its monomers to intermediates of fatty acid metabolism. 3-Hydroxy fatty acids are part of fatty acid biosynthesis and degradation, and these pathways do involve a β-ketoacyl-CoA thiolase and β-ketoacyl dehydrogenase. However, PHA polymerase, the enzyme involved in the unique step in PHA biosynthesis, does not have any significant homology to other proteins, and its evolutionary predecessor remains enigmatic.
By analogy, one can speculate about the origin of other ubiquitous storage materials such as starch, glycogen, or natural rubber. For these polymers, an evolutionary predecessor should also have a more essential function than being a storage molecule. Several oligosaccharides are essential for a bacterium. Trehalose is a dimer of glucose molecules and serves as an osmoprotectant for the cell. Lipopolysaccharides are oligosaccharides linked to diacylglycerol moieties and play a role in maintaining cell integrity and viability. Limited polymerization of glucose may have been an early evolutionary step in the eventual pathway to polysaccharides such as glycogen and starch. Other polysaccharides may have been synthesized by analogous pathways built on this scheme. In that context, oligomers of P(3HB) may have been, or may still be, important for life. Recently, oligomers of (R)-3-hydroxybutyrate were identified as pheromones in spiders (237). The P(3HB) component of Ca2+ channels and perhaps other transporters may be a subsequent low-molecular-weight predecessor of the high-molecular-weight material. Although unrelated to commercial PHA production, this evolutionary perspective suggests that cPHB may become a new paradigm in microbial physiology or even biology in general. As such, it may provide additional and unexpected clues to the future of biological polyesters.
CONCLUSIONS
An immense body of information is available presently to engineer organisms for the synthesis of almost any PHA. A polymerase-encoding gene for a specific composition can be chosen from a set of 18 identified genes. Depending on the pathway to be used for generating the desired monomers, phbAB, phaJ, or phaG genes are available. These can be chosen from a number of different organisms as well. In addition to these essential phb genes, other enzymes may be used to generate novel monomers. The opportunities seem limitless.
Recombinant production of molecules such as PHAs will undoubtedly thrive on the enormous biological diversity of nature, where novel protein activities can be obtained from exotic places, while gene cloning becomes less and less of a technological hurdle. In the future, bacterial fermentations will be able to support the production of a wide range of PHAs. For economic reasons, plant crops promise to be a more desired vehicle for PHA production. New procedures to introduce and express genes in plants are generated rapidly and will enable the timely expression of desired genes in the compartments of choice. Enzymes with all the desired characteristics will furthermore be obtained by new in vitro molecular breeding approaches as long as the screening tools are available. It is clear that at the start of the third millennium, transgenic PHA producers will be an important source of green plastics and chemicals to the world. With the advent of further developments in metabolic engineering, such biotechnologies will be the rule rather than the exception.
ACKNOWLEDGMENTS
We thank David Martin, Frank Skraly, Peter Yorgey, and Stacy Hernandez for helpful discussions and critical reading of the manuscript. We gratefully acknowledge Rosetta Reusch and Dieter Jendrossek for providing the photographs in Fig. 2 and 11.
REFERENCES
- 1.Abe C, Taima Y, Nakamura Y, Doi Y. New bacterial copolyesters of 3-hydroxyalkanoates and 3-hydroxy-ω-fluoroalkanoates produced by Pseudomonas oleovorans. Polym Commun. 1990;31:404–406. [Google Scholar]
- 2.Abe H, Doi Y, Fukushima T, Eya H. Biosynthesis from gluconate of a random copolyester consisting of 3-hydroxybutyrate and medium-chain-length 3-hydroxyalkanoates by Pseudomonas sp. 61-3. Int J Biol Macromol. 1994;16:115–119. doi: 10.1016/0141-8130(94)90036-1. [DOI] [PubMed] [Google Scholar]
- 3.Ackermann J-U, Babel W. Growth-associated synthesis of poly(hydroxybutyric acid) in Methylobacterium rhodesianum as an expression of an internal bottleneck. Appl Microbiol Biotechnol. 1997;47:144–149. [Google Scholar]
- 4.Akiyama M, Doi Y. Production of poly(3-hydroxyalkanoates) from α,ω-alkanedioic acids and hydroxylated fatty acids by Alcaligenes sp. Biotechnol Lett. 1993;15:163–168. [Google Scholar]
- 5.Akiyama M, Taima Y, Doi Y. Production of poly(3-hydroxyalkanoates) by a bacterium of the genus Alcaligenes utilizing long-chain fatty acids. Appl Microbiol Biotechnol. 1992;37:698–701. [Google Scholar]
- 6.Anderson A J, Dawes E A. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54:450–472. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson A J, Haywood G W, Williams D R, Dawes E A. The production of polyhydroxyalkanoates from unrelated carbon sources. In: Dawes E A, editor. Novel biodegradable microbial polymers. Vol. 186. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1990. pp. 119–129. [Google Scholar]
- 8.Asenjo J A, Schmidt A S, Anderson P R, Andrews B A. Effect of single nutrient limitation on poly-β-hydroxybutyrate molecular weight distribution in Alcaligenes eutrophus. Biotechnol Bioeng. 1995;46:497–502. doi: 10.1002/bit.260460514. [DOI] [PubMed] [Google Scholar]
- 9.Baptist. January 1963. U.S. patent 3,072,538.
- 10.Baptist. October 1963. U.S. patent 3,107,172.
- 11.Bergersen F J, Turner G L. Supply of O2 regulates O2 demand during utilization of reserves of poly-β-hydroxybutyrate in N2-fixing soybean bacteroids. Proc R Soc London Ser B. 1992;249:143–148. doi: 10.1098/rspb.1992.0096. [DOI] [PubMed] [Google Scholar]
- 12.Blattner F R, Plunkett G R, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 13.Bourque D, Ouellette B, André G, Groleau D. Production of poly-β-hydroxybutyrate from methanol: characterization of a new isolate of Methylobacterium extorquens. Appl Microbiol Biotechnol. 1992;37:7–12. [Google Scholar]
- 14.Bourque D, Pomerleau Y, Groleau D. High-cell-density production of poly-β-hydroxybutyrate (PHB) from methanol by Methylobacterium extorquens: production of high-molecular-mass PHB. Appl Microbiol Biotechnol. 1995;44:367–376. [Google Scholar]
- 15.Boynton, L., O. P. Peoples, and G. W. Huisman. 1998. Unpublished results.
- 16.Brady L, Brzozowski A M, Derewenda Z S, Dodson E, Dodson G, Tolley S, Turkenburg J P, Christiansen L, Huge-Jensen B, Norskov L, Thim L, Menge U. A serine protease triad forms the catalytic centre of a triglycerol lipase. Nature. 1990;343:767–770. doi: 10.1038/343767a0. [DOI] [PubMed] [Google Scholar]
- 17.Brandl H, Gross R A, Lenz R W, Fuller R C. Pseudomonas oleovorans as a source of poly(β-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl Environ Microbiol. 1988;54:1977–1982. doi: 10.1128/aem.54.8.1977-1982.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brandl H, Knee E J, Fuller R C. Ability of the phototrophic bacterium Rhodospirillum rubrum to produce various poly(β-hydroxyalkanoates): potential sources for biodegradable polyesters. Int J Biol Macromol. 1989;11:49–55. doi: 10.1016/0141-8130(89)90040-8. [DOI] [PubMed] [Google Scholar]
- 19.Brivonese A C, Sutherland I W. Polymer production by a mucoid stain of Azotobacter vinelandii in batch culture. Appl Microbiol Biotechnol. 1989;30:97–102. [Google Scholar]
- 20.Byrom D. Polymer synthesis by micro-organisms: technology and economics. Trends Biotechnol. 1987;5:246–250. [Google Scholar]
- 21.Byrom D. Production of poly-β-hydroxybutyrate:poly-β-hydroxyvalerate copolymers. FEMS Microbiol Rev. 1992;103:247–250. [Google Scholar]
- 22.Casini E, de Rijk T C, de Waard P, Eggink G. Synthesis of poly(hydroxyalkanoate) from hydrolyzed linseed oil. J Environ Polym Degr. 1997;5:153–158. [Google Scholar]
- 23.Cevallos M A, Encarnación S, Leija A, Mora Y, Mora J. Genetic and physiological characterization of a Rhizobium etli mutant strain unable to synthesize poly-β-hydroxybutyrate. J Bacteriol. 1996;178:1646–1654. doi: 10.1128/jb.178.6.1646-1654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho K-S, Ryu H W, Park C-H, Goodrich P R. Poly(hydroxybutyrate-co-hydroxyvalerate) from swine waste liquor by Azotobacter vinelandii UWD. Biotechnol Lett. 1997;19:7–10. [Google Scholar]
- 25.Choi M H, Yoon S C. Polyester biosynthesis characteristics of Pseudomonas citronellolis grown on various carbon sources, including 3-methyl-branched substrates. Appl Environ Microbiol. 1994;60:3245–3254. doi: 10.1128/aem.60.9.3245-3254.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chowdhury B, John M E. Thermal evaluation of transgenic cotton containing polyhydroxybutyrate. Thermochim Acta. 1998;313:43–53. [Google Scholar]
- 27.Chung Y J, Cha H J, Yeo J S, Yoo Y J. Production of poly(3-hydroxybutyric-co-3-hydroxyvaleric) acid using propionic acid by pH regulation. J Ferment Bioeng. 1997;83:492–495. [Google Scholar]
- 28.Clark D P, Cronan J E., Jr . Two-carbon compounds and fatty acids as carbon sources. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 343–357. [Google Scholar]
- 29.Clouart, J. D., and G. W. Huisman. 1997. Unpublished observations.
- 30.Corbell N, Loper J E. A global regulator of secondary metabolite production in Pseudomonas fluorescens Pf-5. J Bacteriol. 1995;177:6230–6236. doi: 10.1128/jb.177.21.6230-6236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cromwick A-M, Foglia T, Lenz R W. The microbial production of poly(hydroxyalkanoates) from tallow. Appl Microbiol Biotechnol. 1996;46:464–469. [Google Scholar]
- 32.Curley J M, Hazer B, Lenz R W, Fuller R C. Production of poly(3-hydroxyalkanoates) containing aromatic substituents by Pseudomonas oleovorans. Macromolecules. 1996;29:1762–1766. [Google Scholar]
- 33.Das S, Lengweiler U D, Seebach D, Reusch R N. Proof for a nonproteinaceous calcium-selective channel in Escherichia coli by total synthesis from (R)-3-hydroxybutanoic acid and inorganic polyphosphate. Proc Natl Acad Sci USA. 1997;94:9075–9079. doi: 10.1073/pnas.94.17.9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis J T, Chen H-H, Moore R, Nishitani Y, Masamune S, Sinskey A J, Walsh C T. Biosynthetic thiolase from Zoogloea ramigera. II. Inactivation with haloacetyl CoA analogs. J Biol Chem. 1987;262:90–96. [PubMed] [Google Scholar]
- 35.Davis J T, Moore R N, Imperiali B, Pratt A J, Kobayashi K, Masamune S, Sinskey A J, Walsh C T. Biosynthetic thiolase from Zoogloea ramigera. I. Preliminary characterization and analysis of proton transfer reaction. J Biol Chem. 1987;262:82–89. [PubMed] [Google Scholar]
- 36.Dawes E A, Senior P J. The role and regulation of energy reserve polymers in micro-organisms. Adv Microb Physiol. 1973;10:135–266. doi: 10.1016/s0065-2911(08)60088-0. [DOI] [PubMed] [Google Scholar]
- 37.Debabov, V. G., J. I. Kozlov, E. M. Khurges, V. A. Livshits, N. I. Zhdanova, M. M. Gusyatiner, A. K. Sokolov, T. A. Bachina, N. K. Yankovsky, J. D. Tsygankov, A. J. Chistoserdov, T. C. Plotnikova, I. C. Shakalis, A. V. Belareva, R. A. Arsatiants, A. F. Sholin, and T. M. Pozdnyakova. January 1998. U.S. patent 5,705,371.
- 38.de Koning G J M, Kellerhals M, van Meurs C, Witholt B. A process for the recovery of poly(3-hydroxyalkanoates) from pseudomonads. 2. Process development and economic evaluation. Bioprocess Eng. 1997;17:15–21. [Google Scholar]
- 39.de Koning G J M, van Bilsen H M M, Lemstra P J, Hazenberg W, Witholt B, Preusting H, van der Galiën J G, Schirmer A, Jendrossek D. A biodegradable rubber by crosslinking poly(hydroxyalkanoate) from Pseudomonas oleovorans. Polymer. 1994;35:2090–2097. [Google Scholar]
- 40.de Koning G J M, Witholt B. A process for the recovery of poly(3-hydroxyalkanoates) from pseudomonads. 1. Solubilization. Bioprocess Eng. 1997;17:7–13. [Google Scholar]
- 41.de Smet M J, Eggink G, Witholt B, Kingma J, Wynberg H. Characterization of intracellular inclusions formed by Pseudomonas oleovorans during growth on octane. J Bacteriol. 1983;154:870–878. doi: 10.1128/jb.154.2.870-878.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doi Y. Microbial polyesters. Yokohama, Japan: VCH Publishers, Inc.; 1990. [Google Scholar]
- 43.Doi Y. Microbial synthesis, physical properties, and biodegradability of polyhydroxyalkanoates. Macromol Symp. 1995;98:585–599. [Google Scholar]
- 44.Doi Y, Abe C. Biosynthesis and characterization of a new bacterial copolyester of 3-hydroxyalkanoates and 3-hydroxy-ω-chloroalkanoates. Macromolecules. 1990;23:3705–3707. [Google Scholar]
- 45.Doi Y, Kitamura S, Abe H. Microbial synthesis and characterization of poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) Macromolecules. 1995;28:4822–4828. [Google Scholar]
- 46.Doi Y, Kunioka M, Nakamura Y, Soga K. Biosynthesis of copolyesters in Alcaligenes eutrophus H16 from 13C-labeled acetate and propionate. Macromolecules. 1987;20:2988–2991. [Google Scholar]
- 47.Doi Y, Tamaki A, Kunioka M, Soga K. Biosynthesis of terpolyesters of 3-hydroxybutyrate, 3-hydroxyvalerate, and 5-hydroxyvalerate, in Alcaligenes eutrophus from 5-chloropentanoic and pentanoic acids. Makromol Chem Rapid Commun. 1987;8:631–635. [Google Scholar]
- 48.Doi Y, Tamaki A, Kunioka M, Soga K. Production of copolyesters of 3-hydroxybutyrate and 3-hydroxyvalerate by Alcaligenes eutrophus from butyric and pentanoic acids. Appl Microbiol Biotechnol. 1988;28:330–334. [Google Scholar]
- 49.Driscoll B T, Finan T M. Properties of NAD+- and NADP+-dependent malic enzymes of Rhizobium (Sinorhizobium) meliloti and differential expression of their genes in nitrogen-fixing bacteroids. Microbiology. 1997;143:489–498. doi: 10.1099/00221287-143-2-489. [DOI] [PubMed] [Google Scholar]
- 50.Easson D D, Jr, Sinskey A J, Peoples O P. Isolation of Zoogloea ramigera I-16-M exopolysaccharide biosynthetic genes and evidence for instability within this region. J Bacteriol. 1987;169:4518–4524. doi: 10.1128/jb.169.10.4518-4524.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eggink G, de Waard P, Huijberts G N M. Formation of novel poly(hydroxyalkanoates) from long-chain fatty acids. Can J Microbiol. 1995;41(Suppl. 1):14–21. doi: 10.1139/m95-163. [DOI] [PubMed] [Google Scholar]
- 52.Eggink G, van der Wal H, Huijberts G N M, de Waard P. Oleic acid as a substrate for poly-3-hydroxybutyrate formation in Alcaligenes eutrophus and Pseudomonas putida. Ind Crops Prod. 1993;1:157–163. [Google Scholar]
- 53.Encarnación S, Dunn M, Willms K, Mora J. Fermentative and aerobic metabolism of Rhizobium etli. J Bacteriol. 1995;177:3058–3066. doi: 10.1128/jb.177.11.3058-3066.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fidler S, Dennis D. Polyhydroxyalkanoate production in recombinant Escherichia coli. FEMS Microbiol Rev. 1992;103:231–236. doi: 10.1016/0378-1097(92)90314-e. [DOI] [PubMed] [Google Scholar]
- 55.Floccari M E, López N I, Méndez B S, Pieper-Fürst U, Steinbüchel A. Isolation and partial characterization of Bacillus megaterium mutants deficient in poly(3-hydroxybutyrate) synthesis. Can J Microbiol. 1995;41(Suppl. 1):77–79. [Google Scholar]
- 56.Föllner C G, Babel W, Steinbüchel A. Isolation and purification of granule-associated proteins relevant for poly(3-hydroxybutyric acid) biosynthesis from methylotrophic bacteria relying on the serine pathway. Can J Microbiol. 1995;41(Suppl. 1):124–130. [Google Scholar]
- 57.Föllner C G, Madkour M, Mayer F, Babel W, Steinbüchel A. Analysis of the PHA granule-associated proteins GA20 and GA11 in Methylobacterium extorquens and Methylobacterium rhodesianum. J Basic Microbiol. 1997;37:11–21. doi: 10.1002/jobm.3620370104. [DOI] [PubMed] [Google Scholar]
- 58.Fritzsche K, Lenz R W, Fuller R C. Bacterial polyesters containing branched poly(β-hydroxyalkanoate) units. Int J Biol Macromol. 1990;12:92–101. doi: 10.1016/0141-8130(90)90059-j. [DOI] [PubMed] [Google Scholar]
- 59.Fritzsche K, Lenz R W, Fuller R C. Production of unsaturated polyesters by Pseudomonas oleovorans. Int J Biol Macromol. 1990;12:85–91. doi: 10.1016/0141-8130(90)90058-i. [DOI] [PubMed] [Google Scholar]
- 60.Fritzsche K, Lenz R W, Fuller R C. An unusual bacterial polyester with a phenyl pendant group. Makromol Chem. 1990;191:1957–1965. [Google Scholar]
- 61.Fukui T, Doi Y. Cloning and analysis of the poly(3-hydroxybutyrate-co-3-hydroxyhexanoate) biosynthesis genes of Aeromonas caviae. J Bacteriol. 1997;179:4821–4830. doi: 10.1128/jb.179.15.4821-4830.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fukui T, Ito M, Saito S, Tomita K. Purification and characterization of NADP-linked acetoacetyl-CoA reductase from Zoogloea ramigera I-16-M. Biochim Biophys Acta. 1987;917:365–371. [PubMed] [Google Scholar]
- 63.Fukui T, Shiomi N, Doi Y. Expression and characterization of (R)-specific enoyl coenzyme A hydratase involved in polyhydroxyalkanoate biosynthesis by Aeromonas caviae. J Bacteriol. 1998;180:667–673. doi: 10.1128/jb.180.3.667-673.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fukui T, Yoshimoto A, Matsumoto M, Hosokawa S, Saito T, Nishikawa H, Tomita K. Enzymatic synthesis of poly-β-hydroxybutyrate in Zoogloea ramigera. Arch Microbiol. 1976;110:149–156. doi: 10.1007/BF00690222. [DOI] [PubMed] [Google Scholar]
- 65.Fuller, T. J., R. H. Marchessault, and T. L. Bluhm. April 1991. U.S. patent 5,004,664.
- 66.Fuqua W, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 67.Gagnon K D, Lenz R W, Farris R J, Fuller R C. Chemical modification of bacterial elastomers. 1. Peroxide crosslinking. Polymer. 1994;35:4358–4367. [Google Scholar]
- 68.Gagnon K D, Lenz R W, Farris R J, Fuller R C. Chemical modification of bacterial elastomers. 2. Sulfur vulcanization. Polymer. 1994;35:4368–4375. [Google Scholar]
- 69.Gerngross T U, Martin D P. Enzyme-catalyzed synthesis of poly[(R)-(−)-3-hydroxybutyrate]: formation of macroscopic granules in vitro. Proc Natl Acad Sci USA. 1995;92:6279–6283. doi: 10.1073/pnas.92.14.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gerngross T U, Snell K D, Peoples O P, Sinskey A J, Csuhai E, Masamune S, Stubbe J. Overexpression and purification of the soluble polyhydroxyalkanoate synthase from Alcaligenes eutrophus: evidence for a required posttranslational modification for catalytic activity. Biochemistry. 1994;33:9311–9320. doi: 10.1021/bi00197a035. [DOI] [PubMed] [Google Scholar]
- 71.Grewal S I S, Han B, Johnstone K. Identification and characterization of a locus which regulates multiple functions in Pseudomonas tolaasii, the cause of brown blotch disease of Agaricus bisporus. J Bacteriol. 1995;177:4658–4668. doi: 10.1128/jb.177.16.4658-4668.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Griebel R, Smith Z, Merrick J. Metabolism of poly-beta-hydroxybutyrate. I. Purification, composition, and properties of native poly-beta-hydroxybutyrate granules from Bacillus megaterium. Biochemistry. 1968;7:3676–3681. doi: 10.1021/bi00850a047. [DOI] [PubMed] [Google Scholar]
- 73.Gross R A, DeMello C, Lenz R W, Brandl H, Fuller R C. Biosynthesis and characterization of poly(β-hydroxyalkanoates) produced by Pseudomonas oleovorans. Macromolecules. 1989;22:1106–1115. [Google Scholar]
- 74.Grubl G, Vogler A P, Lengeler J W. Involvement of the histidine protein (HPr) of the phosphotransferase system in chemotactic signaling of Escherichia coli K-12. J Bacteriol. 1990;172:5871–5876. doi: 10.1128/jb.172.10.5871-5876.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hahn J J, Eschenlauer A C, Narrol M H, Somers D A, Srienc F. Growth kinetics, nutrient uptake, and expression of the Alcaligenes eutrophus poly(β-hydroxybutyrate) synthesis pathway in transgenic maize cell suspension cultures. Biotechnol Prog. 1997;13:347–354. doi: 10.1021/bp970033r. [DOI] [PubMed] [Google Scholar]
- 76.Hahn M, Meyer L, Studer D, Regensburger B, Hennecke H. Insertion and deletion mutations within the nif region of Rhizobium japonicum. Plant Mol Biol. 1984;3:159–168. doi: 10.1007/BF00016063. [DOI] [PubMed] [Google Scholar]
- 77.Hahn S K, Chang Y K, Lee S Y. Recovery and characterization of poly(3-hydroxybutyric acid) synthesized in Alcaligenes eutrophus and recombinant Escherichia coli. Appl Environ Microbiol. 1995;61:34–39. doi: 10.1128/aem.61.1.34-39.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hall B, Baldwin J, Rhie H G, Dennis D. Nocardia corallina polyhydroxyalkanoate synthase. 1998. GenBank accession no. AF019964. [DOI] [PubMed] [Google Scholar]
- 79.Hänggi U J. Pilot scale production of PHB with Alcaligenes latus. In: Dawes E A, editor. Novel biodegradable microbial polymers. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1990. pp. 65–70. [Google Scholar]
- 80.Hassan M A, Shirai Y, Kusubayashi N, Karim M I A, Nakanishi K, Hashimoto K. The production of polyhydroxyalkanoate from anaerobically treated palm oil mill effluent by Rhodobacter sphaeroides. J Ferment Bioeng. 1997;83:485–488. [Google Scholar]
- 81.Haywood G W, Anderson A J, Chu L, Dawes E A. Characterization of two 3-ketothiolases possessing differing substrate specificities in the polyhydroxyalkanoate synthesizing organism Alcaligenes eutrophus. FEMS Microbiol Lett. 1988;52:91–96. [Google Scholar]
- 82.Haywood G W, Anderson A J, Chu L, Dawes E A. The role of NADH- and NADPH-linked acetoacetyl-CoA reductases in the poly-3-hydroxybutyrate synthesizing organism Alcaligenes eutrophus. FEMS Microbiol Lett. 1988;52:259–264. [Google Scholar]
- 83.Haywood G W, Anderson A J, Dawes E A. The importance of PHB-synthase substrate specificity in polyhydroxyalkanoate synthesis by Alcaligenes eutrophus. FEMS Microbiol Lett. 1989;57:1–6. [Google Scholar]
- 84.Haywood G W, Anderson A J, Ewing D F, Dawes E A. Accumulation of a polyhydroxyalkanoate containing primarily 3-hydroxydecanoate from simple carbohydrate substrates by Pseudomonas sp. strain NCIMB 40135. Appl Environ Microbiol. 1990;56:3354–3359. doi: 10.1128/aem.56.11.3354-3359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hazer B, Lenz R W, Fuller R C. Biosynthesis of methyl-branched poly(β-hydroxyalkanoate)s by Pseudomonas oleovorans. Macromolecules. 1994;27:45–49. [Google Scholar]
- 86.Hein S, Söhling B, Gottschalk G, Steinbüchel A. Biosynthesis of poly(4-hydroxybutyric acid) by recombinant strains of Escherichia coli. FEMS Microbiol Lett. 1997;153:411–418. doi: 10.1111/j.1574-6968.1997.tb12604.x. [DOI] [PubMed] [Google Scholar]
- 87.Hein S, Tran H, Steinbuchel A. Synechocystis sp. PCC6803 possesses a two-component polyhydroxyalkanoic acid synthase similar to that of anoxygenic purple sulfur bacteria. Arch Microbiol. 1998;170:162–170. doi: 10.1007/s002030050629. [DOI] [PubMed] [Google Scholar]
- 88.Henderson R A, Jones C W. Physiology of poly-3-hydroxybutyrate (PHB) production by Alcaligenes eutrophus growing in continuous culture. Microbiology. 1997;143:2361–2371. doi: 10.1099/00221287-143-7-2361. [DOI] [PubMed] [Google Scholar]
- 89.Henderson R A, Jones C W. Poly-3-hydroxybutyrate production by washed cells of Alcaligenes eutrophus: purification, characterization and potential regulatory role of citrate synthase. Arch Microbiol. 1997;168:486–492. doi: 10.1007/s002030050526. [DOI] [PubMed] [Google Scholar]
- 90.Hirsch A M, Bang M, Ausubel F M. Ultrastructural analysis of ineffective alfalfa nodules formed by nif::Tn5 mutants of Rhizobium meliloti. J Bacteriol. 1983;155:367–380. doi: 10.1128/jb.155.1.367-380.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Holmes, P. A. November 1986. U.S. patent 4,620,999.
- 92.Holmes, P. A., S. H. Collins, and L. F. Wright. October 1984. U.S. patent 4,477,654.
- 93.Hori K, Soga K, Doi Y. Effects of culture conditions on molecular weights of poly(3-hydroxyalkanoates) produced by Pseudomonas putida from octanoate. Biotechnol Lett. 1994;16:709–714. [Google Scholar]
- 94.Horswill A R, Escalante-Semerena J C. Propionate catabolism in Salmonella typhimurium LT2: two divergently transcribed units comprise the prp locus at 8.5 centisomes, prpR encodes a member of the sigma-54 family of activators, and the prpBCDE genes constitute an operon. J Bacteriol. 1997;179:928–940. doi: 10.1128/jb.179.3.928-940.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hrabak E M, Willis D K. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J Bacteriol. 1992;174:3011–3020. doi: 10.1128/jb.174.9.3011-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hrabak O. Industrial production of poly-β-hydroxybutyrate. FEMS Microbiol Rev. 1992;103:251–256. [Google Scholar]
- 97.Huang A H C. Oil bodies and oleosins in seeds. Annu Rev Plant Physiol Plant Mol Biol. 1992;43:177–200. [Google Scholar]
- 98.Huang R, Reusch R N. Genetic competence in Escherichia coli requires poly-β-hydroxybutyrate/calcium polyphosphate membrane complexes and divalent cations. J Bacteriol. 1995;177:486–490. doi: 10.1128/jb.177.2.486-490.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang R, Reusch R N. Poly(3-hydroxybutyrate) is associated with specific proteins in the cytoplasm and membranes of Escherichia coli. J Biol Chem. 1996;271:22196–22202. doi: 10.1074/jbc.271.36.22196. [DOI] [PubMed] [Google Scholar]
- 100.Huijberts G N M. Ph.D. thesis. Groningen, The Netherlands: University of Groningen; 1996. [Google Scholar]
- 101.Huijberts G N M, Eggink G. Production of poly(3-hydroxyalkanoates) by Pseudomonas putida KT2442 in continuous cultures. Appl Microbiol Biotechnol. 1996;46:233–239. [Google Scholar]
- 102.Huijberts G N M, Eggink G, de Waard P, Huisman G W, Witholt B. Pseudomonas putida KT2442 cultivated on glucose accumulates poly(3-hydroxyalkanoates) consisting of saturated and unsaturated monomers. Appl Environ Microbiol. 1992;58:536–544. doi: 10.1128/aem.58.2.536-544.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huisman, G. W. 1998. Unpublished observations.
- 104.Huisman, G. W., and W. Buckel. 1998. Unpublished results.
- 105.Huisman G W, de Leeuw O, Eggink G, Witholt B. Synthesis of poly-3-hydroxyalkanoates is a common feature of fluorescent pseudomonads. Appl Environ Microbiol. 1989;55:1949–1954. doi: 10.1128/aem.55.8.1949-1954.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Huisman G W, Wonink E, de Koning G, Preusting H, Witholt B. Synthesis of poly(3-hydroxyalkanoates) by mutant and recombinant Pseudomonas strains. Appl Microbiol Biotechnol. 1992;38:1–5. [Google Scholar]
- 107.Huisman G W, Wonink E, Meima R, Kazemier B, Terpstra P, Witholt B. Metabolism of poly(3-hydroxyalkanoates) (PHAs) by Pseudomonas oleovorans. Identification and sequences of genes and function of the encoded proteins in the synthesis and degradation of PHA. J Biol Chem. 1991;266:2191–2198. [PubMed] [Google Scholar]
- 108.Huisman, G. W., L. P. Yap, and X. Z. Tu. 1998. Unpublished results.
- 109.Hustede E, Steinbüchel A. Characterization of the polyhydroxyalkanoate synthase gene locus of Rhodobacter sphaeroides. Biotechnol Lett. 1993;15:709–714. [Google Scholar]
- 110.Jackowski S. Biosynthesis of pantothenic acid and coenzyme A. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 687–694. [Google Scholar]
- 111.Jendrossek D, Schirmer A, Schlegel H G. Biodegradation of polyhydroxyalkanoic acids. Appl Microbiol Biotechnol. 1996;46:451–463. doi: 10.1007/s002530050844. [DOI] [PubMed] [Google Scholar]
- 112.John M E, Keller G. Metabolic pathway engineering in cotton: biosynthesis of polyhydroxybutyrate in fiber cells. Proc Natl Acad Sci USA. 1996;93:12768–12773. doi: 10.1073/pnas.93.23.12768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Joshi A K, Smith S. Construction, expression, and characterization of a mutated animal fatty acid synthase deficient in the dehydrase function. J Biol Chem. 1993;268:22508–22513. [PubMed] [Google Scholar]
- 114.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3(Suppl.):109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 115.Kang C K, Lee H S, Kim J H. Accumulation of PHA and its copolyesters by Methylobacterium sp. KCTC 0048. Biotechnol Lett. 1993;15:1017–1020. [Google Scholar]
- 116.Kato M, Bao H J, Kang C-K, Doi Y. Production of a novel copolyester of 3-hydroxybutyric acid and medium-chain-length 3-hydroxyalkanoic acids by Pseudomonas sp. 61-3 from sugars. Appl Microbiol Biotechnol. 1996;45:363–370. [Google Scholar]
- 117.Kato M, Fukui T, Iwata T, Doi Y. Genetic and morphological studies on biosynthesis of polyester blends of poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyalkanoates) in Pseudomonas sp. 61-3. In: Eggink G, Steinbüchel A, Poirier Y, Witholt B, editors. 1996 International Symposium on Bacterial Polyhydroxyalkanoates. Ottawa, Canada: NRC Research Press; 1996. pp. 173–180. [Google Scholar]
- 118.Kauffman, T., F. X. Brady, P. P. Puletti, and G. Raykovitz. December 1992. U.S. patent 5,169,889.
- 119.Kidwell J, Valentin H E, Dennis D. Regulated expression of the Alcaligenes eutrophus pha biosynthetic genes in Escherichia coli. Appl Environ Microbiol. 1995;61:1391–1398. doi: 10.1128/aem.61.4.1391-1398.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kim B S, Chang H N. Control of glucose feeding using exit gas data and its application to the production of PHB from tapioca hydrolysates by Alcaligenes eutrophus. Biotechnol Tech. 1995;9:311–314. [Google Scholar]
- 121.Kim B S, Lee S C, Lee S Y, Chang H N, Chang Y K, Woo S I. Production of poly(3-hydroxybutyric acid) by fed-batch culture of Alcaligenes eutrophus with glucose concentration control. Biotechnol Bioeng. 1994;43:892–898. doi: 10.1002/bit.260430908. [DOI] [PubMed] [Google Scholar]
- 122.Kim B S, Lee S Y, Chang H N. Production of poly-β-hydroxybutyrate by fed-batch culture of recombinant Escherichia coli. Biotechnol Lett. 1992;14:811–816. [Google Scholar]
- 123.Kim G J, Lee I Y, Yoon S C, Shin Y C, Prak Y H. Enhanced yield and a high production of medium-chain-length poly(3-hydroxyalkanoates) in a two-step fed-batch cultivation of Pseudomonas putida by combined use of glucose and octanoate. Enzyme Microb Technol. 1997;20:500–505. [Google Scholar]
- 124.Kim O, Gross R A, Rutherford D R. Bioengineering of poly(β-hydroxyalkanoates) for advanced material applications: incorporation of cyano and nitrophenoxy side chain substituents. Can J Microbiol. 1995;41(Suppl. 1):32–43. [Google Scholar]
- 125.Kim Y B, Lenz R W, Fuller R C. Poly(β-hydroxyalkanoate) copolymers containing brominated repeating units produced by Pseudomonas oleovorans. Macromolecules. 1992;25:1852–1857. [Google Scholar]
- 126.Kim Y B, Lenz R W, Fuller R C. Preparation and characterization of poly(β-hydroxyalkanoates) obtained from Pseudomonas oleovorans grown with mixtures of 5-phenylvaleric acid and n-alkanoic acids. Macromolecules. 1991;24:5256–5360. [Google Scholar]
- 127.Kitamura S, Doi Y. Production of copolyesters from organic acids by mutant strains of Alcaligenes eutrophus. In: Doi Y, Fukuda K, editors. Biodegradable plastics and polymers. Vol. 12. Amsterdam: Elsevier Biomedical Press; 1994. pp. 373–378. [Google Scholar]
- 128.Koyama N, Doi Y. Continuous production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) by Alcaligenes eutrophus. Biotechnol Lett. 1995;17:281–284. [Google Scholar]
- 129.Kraak M N, Kessler B, Witholt B. In vitro activities of granule-bound poly[(R)-3-hydroxyalkanoate) polymerase C1 of Pseudomonas oleovorans. Development of an activity test for medium-chain-length-poly(3-hydroxyalkanoate) polymerases. Eur J Biochem. 1997;250:432–439. doi: 10.1111/j.1432-1033.1997.0432a.x. [DOI] [PubMed] [Google Scholar]
- 130.Kraak M N, Smits T H M, Kessler B, Witholt B. Polymerase C1 levels and poly(R-3-hydroxyalkanoate) synthesis in wild-type and recombinant Pseudomonas strains. J Bacteriol. 1997;179:4985–4991. doi: 10.1128/jb.179.16.4985-4991.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kunioka M, Kawaguchi Y, Doi Y. Production of biodegradable copolyesters of 3-hydroxybutyrate and 4-hydroxybutyrate by Alcaligenes eutrophus. Appl Microbiol Biotechnol. 1989;30:569–573. [Google Scholar]
- 132.Kunioka M, Nakamura Y, Doi Y. New bacterial copolyesters produced in Alcaligenes eutrophus from organic acids. Polym Commun. 1989;29:174–176. [Google Scholar]
- 133.Kusaka S, Abe H, Lee S Y, Doi Y. Molecular mass of poly[(R)-3-hydroxybutyric acid] produced in a recombinant Escherichia coli. Appl Microbiol Biotechnol. 1997;47:140–143. doi: 10.1007/s002530050902. [DOI] [PubMed] [Google Scholar]
- 134.Kusaka S, Iwata T, Doi Y. Microbial synthesis and physical properties of ultra-high-molecular-weight poly[(R)-3-hydroxybutyrate] Pure Appl Chem. 1998;A35:319–335. [Google Scholar]
- 135.Lageveen R G, Huisman G W, Preusting H, Ketelaar P, Eggink G, Witholt B. Formation of polyesters by Pseudomonas oleovorans: effect of substrates on formation and composition of poly(R)-3-hydroxyalkanoates and poly(R)-3-hydroxyalkenoates. Appl Environ Microbiol. 1988;54:2924–2932. doi: 10.1128/aem.54.12.2924-2932.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Langenbach S, Rehm B H A, Steinbüchel A. Functional expression of the PHA synthase gene phaC1 from Pseudomonas aeruginosa in Escherichia coli results in poly(3-hydroxyalkanoate) synthesis. FEMS Microbiol Lett. 1997;150:303–309. doi: 10.1016/s0378-1097(97)00142-0. [DOI] [PubMed] [Google Scholar]
- 137.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarcical quorum-sensing cascase in Pseudomonas aeruginosa links the transcriptional activators LasR and RhlR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 138.Leaf T A, Peterson M S, Stoup S K, Somers D, Srienc F. Saccharomyces cerevisiae expressing bacterial polyhydroxybutyrate synthase produces poly-3-hydroxybutyrate. Microbiology. 1996;142:1169–1180. doi: 10.1099/13500872-142-5-1169. [DOI] [PubMed] [Google Scholar]
- 139.Lee E Y, Jendrossek D, Schirmer A, Choi C Y, Steinbüchel A. Biosynthesis of copolyesters consisting of 3-hydroxybutyric acid and medium-chain-length 3-hydroxyalkanoic acids from 1,3-butanediol or from 3-hydroxybutyric acid by Pseudomonas sp. A33. Appl Microbiol Biotechnol. 1995;42:901–909. [Google Scholar]
- 140.Lee E Y, Kang S H, Choi C Y. Biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) by newly isolated Agrobacterium sp. SH-1 and GW-014 from structurally unrelated single carbon substrates. J Ferment Bioeng. 1995;79:328–334. [Google Scholar]
- 141.Lee S P, Do V M, Huisman G W, Peoples O P. PHB polymerase from Zoogloea ramigera. 1996. GenBank accession no. U66242. [Google Scholar]
- 142.Lee S Y. Bacterial Polyhydroxyalkanoates. Biotechnol Bioeng. 1996;49:1–14. doi: 10.1002/(SICI)1097-0290(19960105)49:1<1::AID-BIT1>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 143.Lee S Y. Suppression of filamentation in recombinant Escherichia coli by amplified FtsZ activity. Biotechnol Lett. 1994;16:1247–1252. [Google Scholar]
- 144.Lee S Y, Chang H N. Production of poly(3-hydroxyalkanoates) Adv Biochem Eng Biotechnol. 1995;52:27–58. doi: 10.1007/BFb0102315. [DOI] [PubMed] [Google Scholar]
- 145.Lee S Y, Chang H N. Production of poly(hydroxyalkanoic acid) Adv Biochem Eng. 1995;52:27–58. doi: 10.1007/BFb0102315. [DOI] [PubMed] [Google Scholar]
- 146.Lee S Y, Chang H N, Chang Y K. Production of poly(β-hydroxybutyric) acid by recombinant Escherichia coli. Ann N Y Acad Sci. 1994;721:43–53. doi: 10.1111/j.1749-6632.1994.tb47375.x. [DOI] [PubMed] [Google Scholar]
- 147.Lee S Y, Lee K M, Chang H N, Steinbüchel A. Comparison of recombinant Escherichia coli strains for synthesis and accumulation of poly(3-hydroxybutyric acid) and morphological changes. Biotechnol Bioeng. 1994;44:1337–1347. doi: 10.1002/bit.260441110. [DOI] [PubMed] [Google Scholar]
- 148.Lee S Y, Lee Y K, Chang H N. Stimulatory effects of amino acids and oleic acid on poly(3-hydroxybutyric acid) synthesis by recombinant Escherichia coli. J Ferment Bioeng. 1995;79:177–180. [Google Scholar]
- 149.Lee S Y, Yim K S, Chang H N, Chang Y K. Construction of plasmids, estimation of plasmid stability, and use of stable plasmids for the production of poly(3-hydroxybutyric) acid by recombinant Escherichia coli. J Biotechnol. 1994;32:203–211. doi: 10.1016/0168-1656(94)90183-x. [DOI] [PubMed] [Google Scholar]
- 150.Lee Y, Lee S Y. Enhanced production of poly(3-hydroxybutyrate) by filamentation-suppressed recombinant Escherichia coli in a defined medium. J Environ Polymer Degrad. 1996;4:131–134. [Google Scholar]
- 151.Lefebvre G, Rocher M, Braunegg G. Effects of low dissolved-oxygen concentrations on poly-(3-hydroxybutyrate-co-3-hydroxyvalerate) production by Alcaligenes eutrophus. Appl Environ Microbiol. 1997;63:827–833. doi: 10.1128/aem.63.3.827-833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lemoigne M. Produits de déshydration et de polymerisation de l’acide b-oxobutyrique. Bull Soc Chem Biol (Paris) 1926;8:770–782. [Google Scholar]
- 153.Liebergesell M, Sonomoto K, Madkour M, Mayer F, Steinbüchel A. Purification and characterization of the poly(hydroxyalkanoic acid) synthase from Chromatium vinosum and localization of the enzyme at the surface of poly(hydroxyalkanoic acid) granules. Eur J Biochem. 1994;226:71–80. doi: 10.1111/j.1432-1033.1994.tb20027.x. [DOI] [PubMed] [Google Scholar]
- 154.Liebergesell M, Steinbüchel A. Cloning and molecular analysis of the poly(3-hydroxybutyric acid) biosynthetic genes of Thiocystis violacea. Appl Microbiol Biotechnol. 1993;38:493–501. doi: 10.1007/BF00242944. [DOI] [PubMed] [Google Scholar]
- 155.Liebergesell M, Steinbüchel A. Cloning and nucleotide sequences of genes relevant for biosynthesis of poly(3-hydroxybutyric acid) in Chromatium vinosum strain D. Eur J Biochem. 1992;209:135–150. doi: 10.1111/j.1432-1033.1992.tb17270.x. [DOI] [PubMed] [Google Scholar]
- 156.Lynen Functional group of coenzyme A and its metabolic relations, especially in the fatty acid cycle. Fed Proc. 1953;12:683–691. [PubMed] [Google Scholar]
- 157.Majid M I A, Hori K, Akiyama M, Doi Y. Production of poly(3-hydroxybutyrate) from plant oils by Alcaligenes sp. Stud Polym Sci. 1994;12:417–424. [Google Scholar]
- 158.Manchak J, Page W J. Control of polyhydroxyalkanoate synthesis in Azotobacter vinelandii strain UWD. Microbiology. 1994;140:953–963. [Google Scholar]
- 159.Marchessault R H. Tender morsels for bacteria: recent developments in microbial polyesters. Trends Polym Sci. 1996;4:163–168. [Google Scholar]
- 160.Marchessault, R. H., P. F. LePoutre, and P. E. Wrist. September 1995. U.S. patent 5451456.
- 161.Martin D P, Gerngross T U. Aspects of PHB granule formation in vitro. In: Eggink G, Steinbuuchel A, Poirier Y, Witholt B, editors. Abstracts of the 1996 International Symposium on Bacterial Polyhydroxyalkanoates. Davos, Switzerland: NRC Research Press; 1996. pp. 28–35. [Google Scholar]
- 162.Martínez P, Guzmán J, Espín G. A mutation impairing alginate production increased accumulation of poly-β-hydroxybutyrate in Azotobacter vinelandii. Biotechnol Lett. 1997;19:909–912. [Google Scholar]
- 163.Martini, F., L. Perazzo, and P. Vietto. May 1989. U.S. patent 4,826,493.
- 164.Martini, F., L. Perazzo, and P. Vietto. November 1989. U.S. patent 4,880,592.
- 165.Masamune S, Palmer M A J, Gamboni R, Thompson S, Davis J T, Williams S F, Peoples O P, Sinskey A J, Walsh C T. Bio-Claisen condensation catalyzed by thiolase from Zoogloea ramigera. Active site cysteine residues. J Am Chem Soc. 1989;111:1879–1881. [Google Scholar]
- 166.Masamune S, Walsh C T, Sinskey A J, Peoples O P. Poly-(R)-3-hydroxybutyrate (PHB) biosynthesis: mechanistic studies on the biological Claisen condensation catalyzed by β-ketoacyl thiolase. Pure Appl Chem. 1989;61:303–312. [Google Scholar]
- 167.Mergaert J, Anderson C, Wouters A, Swings J. Microbial degradation of poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in compost. J Environ Polym Degrad. 1994;2:177–183. doi: 10.1128/aem.59.10.3233-3238.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Mergaert J, Webb A, Anderson C, Wouters A, Swings J. Microbial degradation of poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in soils. Appl Environ Microbiol. 1993;59:3233–3238. doi: 10.1128/aem.59.10.3233-3238.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mergaert J, Wouters A, Anderson C, Swings J. In situ biodegradation of poly(3-hydroxybutyrate) and poly(3-hydroxybutyrate-co-3-hydroxyvalerate) in natural waters. Can J Microbiol. 1995;41(Suppl. 1):154–159. doi: 10.1139/m95-182. [DOI] [PubMed] [Google Scholar]
- 170.Middelberg A P J, Lee S Y, Martin J, Williams D R G, Chang H N. Size analysis of poly(3-hydroxybutyric acid) granules produced in recombinant Escherichia coli. Biotechnol Lett. 1995;17:205–210. [Google Scholar]
- 171.Moskowitz G J, Merrick J M. Metabolism of poly-β-hydroxybutyrate. II. Enzymatic syntesis of d-(−)-β-hydroxybutyryl coenzyme A by an enoyl hydrase from Rhodospirillum rubrum. Biochemistry. 1969;8:2748–2755. doi: 10.1021/bi00835a009. [DOI] [PubMed] [Google Scholar]
- 172.Mothes G, Ackermann J U, Babel W. Regulation of poly(3-hydroxybutyrate) synthesis in Methylobacterium rhodesianum MB 126 growing on methanol or fructose. Arch Microbiol. 1998;169:360–363. doi: 10.1007/s002030050583. [DOI] [PubMed] [Google Scholar]
- 173.Mothes G, Babel W. Methylobacterium rhodesianum MB 126 possesses two acetoacetyl-CoA reductases. Arch Microbiol. 1994;161:277–280. [Google Scholar]
- 174.Mothes G, Babel W. Methylobacterium rhodesianum MB 126 possesses two steriospecific crotonyl-CoA hydratases. Can J Microbiol. 1995;41(Suppl. 1):68–72. [Google Scholar]
- 175.Mothes G, Rivera I S, Babel W. Competition between β-ketothiolase and citrate synthase during poly(β-hydroxybutyrate) synthesis in Methylobacterium rhodesianum. Arch Microbiol. 1996;166:405–410. doi: 10.1007/BF01682987. [DOI] [PubMed] [Google Scholar]
- 176.Nakamura K, Gotô Y, Yoshie N, Inoue Y, Chûjô R. Biosynthesis of poly(3-hydroxyalkanoates) from threonine. Int J Biol Macromol. 1992;14:117–118. doi: 10.1016/0141-8130(92)90008-v. [DOI] [PubMed] [Google Scholar]
- 177.Nawrath C, Poirier Y. Review on polyhydroxyalkanoate formation in the model plant Arabidopsis thaliana. In: Eggink G, Steinbüchel A, Poirier Y, Witholt B, editors. Abstracts of the 1996 International Symposium on Bacterial Polyhydroxyalkanoates. Davos, Switzerland: NRC Research Press; 1996. pp. 119–126. [Google Scholar]
- 178.Nawrath C, Poirier Y, Somerville C. Plant polymers for biodegradeable plastics: cellulose, starch, and polyhydroxyalkanoates. Mol Breeding. 1995;1:105–122. [Google Scholar]
- 179.Nawrath C, Poirier Y, Somerville C. Targetting of the polyhydroxybutyrate biosynthetic pathway to the plastids of Arabidopsis thaliana results in high levels of polymer accumulation. Proc Natl Acad Sci USA. 1994;91:12760–12764. doi: 10.1073/pnas.91.26.12760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Noda, I. July 1996. U.S. patent 5,536,564.
- 181.Noda, I. February 1996. U.S. patent 5,489,470.
- 182.Oeding V, Schlegel H G. β-Ketothiolase from hydrogenomonas eutropha H16 and its significance in the regulation of poly-β-hydroxybutate metabolism. Biochem J. 1973;134:239–248. doi: 10.1042/bj1340239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Page W J. Production of poly-β-hydroxybutyrate by Azotobacter vinelandii strain UWD during growth on molasses and other complex carbon sources. Appl Microbiol Biotechnol. 1989;31:329–333. [Google Scholar]
- 184.Page W J, Knosp O. Hyperproduction of poly-β-hydroxybutyrate during exponential growth of Azotobacter vinelandii UWD. Appl Environ Microbiol. 1989;55:1334–1339. doi: 10.1128/aem.55.6.1334-1339.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Page W J, Manchak J, Rudy B. Formation of poly(hydroxybutyrate-co-hydroxyvalerate) by Azotobacter vinelandii UWD. Appl Environ Microbiol. 1992;58:2866–2873. doi: 10.1128/aem.58.9.2866-2873.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Palmer M A J, Differding E, Gamboni R, Williams S F, Peoples O P, Walsh C T, Sinskey A J, Masamune S. Biosynthetic thiolase from Zoogloea ramigera. Evidence for a mechanism involving Cys378 as the active site base. J Biol Chem. 1991;266:8369–8375. [PubMed] [Google Scholar]
- 187.Park J-S, Huh T-L, Lee Y H. Characteristics of cell growth and poly-β-hydroxybutyrate biosynthesis of Alcaligenes eutrophus transformants harboring cloned phbCAB genes. Enzyme Microb Technol. 1997;21:85–90. [Google Scholar]
- 188.Park J-S, Lee Y-H. Metabolic characteristics of isocitrate dehydrogenase leaky mutant of Alcaligenes eutrophus and its utilization for poly-β-hydroxybutyrate production. J Ferment Bioeng. 1996;81:197–205. [Google Scholar]
- 189.Park J-S, Park H-C, Huh T-L, Lee Y-H. Production of poly-β-hydroxybutyrate by Alcaligenes eutrophus transformants harbouring cloned phbCAB genes. Biotechnol Lett. 1995;17:735–740. [Google Scholar]
- 190.Peoples O P, Masamune S, Walsh C T, Sinskey A J. Biosynthetic thiolase from Zoogloea ramigera. III. Isolation and characterization of the structural gene. J Biol Chem. 1987;262:97–102. [PubMed] [Google Scholar]
- 191.Peoples O P, Sinskey A J. Fine structural analysis of the Zoogloea ramigera phbA-phbB locus encoding β-ketothiolase and acetoacetyl-CoA reductase: nucleotide sequence of phbB. Mol Microbiol. 1989;3:349–357. doi: 10.1111/j.1365-2958.1989.tb00180.x. [DOI] [PubMed] [Google Scholar]
- 192.Peoples O P, Sinskey A J. Poly-β-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC) J Biol Chem. 1989;264:15298–15303. [PubMed] [Google Scholar]
- 193.Peoples O P, Sinskey A J. Poly-β-hydroxybutyrate biosynthesis in Alcaligenes eutropha H16. Characterization of the genes encoding β-ketothiolase and acetoacetyl-CoA reductase. J Biol Chem. 1989;264:15293–15297. [PubMed] [Google Scholar]
- 194.Persson B, Bengtsson-Olivecrona G, Enerback S, Olivecrona T, Jornvall H. Structural features of lipoprotein lipase: lipase family relationships, binding interactions, non-equivalence of lipase cofactors, vitellogenin similarities and functional subdivision of lipoprotein lipase. Eur J Biochem. 1989;179:39–45. doi: 10.1111/j.1432-1033.1989.tb14518.x. [DOI] [PubMed] [Google Scholar]
- 195.Pieper U, Steinbüchel A. Identification, cloning and sequence analysis of the poly(3-hydroxyalkanoic acid) synthase gene of the gram-positive bacterium Rhodococcus ruber. FEMS Microbiol Lett. 1992;96:73–80. doi: 10.1016/0378-1097(92)90459-2. [DOI] [PubMed] [Google Scholar]
- 196.Pieper-Fürst U, Madkour M H, Mayer F, Steinbuchel A. Identification of the region of a 14-kilodalton protein of Rhodococcus ruber that is responsible for the binding of this phasin to polyhydroxyalkanoic acid granules. J Bacteriol. 1995;177:2513–2523. doi: 10.1128/jb.177.9.2513-2523.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Pieper-Fürst U, Madkour M H, Mayer F, Steinbüchel A. Purification and characterization of a 14 kilodalton protein that is bound to the surface of polyhydroxyalkanoic acid granules in Rhodococcus ruber. J Bacteriol. 1994;176:4328–4337. doi: 10.1128/jb.176.14.4328-4337.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ploux O, Masamune S, Walsh C T. The NADPH-linked acetoacetyl-CoA reductase from Zoogloea ramigera. Characterization and mechanistic studies of the cloned enzyme over-produced in Escherichia coli. Eur J Biochem. 1988;174:177–182. doi: 10.1111/j.1432-1033.1988.tb14079.x. [DOI] [PubMed] [Google Scholar]
- 199.Poirier Y, Dennis D E, Klomparens K, Somerville C. Polyhydroxybutyrate, a biodegradable thermoplastic, produced in transgenic plants. Science. 1992;256:520–523. doi: 10.1126/science.256.5056.520. [DOI] [PubMed] [Google Scholar]
- 200.Poirier Y, Somerville C, Schechtman L A, Satkowski M M, Noda I. Synthesis of high-molecular-weight poly([R]-(−)-3-hydroxybutyrate) in transgenic Arabidopsis thaliana plant cells. Int J Biol Macromol. 1995;17:7–12. doi: 10.1016/0141-8130(95)93511-u. [DOI] [PubMed] [Google Scholar]
- 201.Postma P W, Lengeler J W, Jacobson G R. Phosphoenoylpyruvate: carbohydrate phosphotransferase systems. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 1149–1174. [Google Scholar]
- 202.Povolo S, Casella S, Nuti M P. Abstracts of the 1996 International Symposium on Bacterial Polyhydroxyalkanoates, Davos, Switzerland. 1996. Involvement of an orf in the synthesis and/or degradation of polyhydroxyalkanoate (PHA) in Rhizobium meliloti; p. 4/01. [Google Scholar]
- 203.Povolo S, Tombolini R, Morea A, Anderson A J, Casella S, Nuti M P. Isolation and characterization of mutants of Rhizobium meliloti unable to synthesize poly-β-hydroxybutyrate. Can J Microbiol. 1994;40:823–829. [Google Scholar]
- 204.Powell, K. A., and B. A. Collinson. June 1982. U.S. patent 4,336,334.
- 205.Preusting H, Hazenberg W, Witholt B. Continuous production of poly(3-hydroxyalkanoates) by Pseudomonas oleovorans in a high cell density two-liquid phase chemostat. Enzyme Microb Technol. 1993;15:311–316. [Google Scholar]
- 206.Preusting H, Kingma J, Huisman G, Steinbüchel A, Witholt B. Formation of polyester blends by a recombinant strain of Pseudomonas oleovorans: different poly(3-hydroxyalkanoates) are stored in separate granules. J Environ Polym Degrad. 1993;1:11–21. [Google Scholar]
- 207.Preusting H, Kingma J, Witholt B. Physiology and polyester formation of Pseudomonas oleovorans in continuous two-liquid phase cultures. Enzyme Microb Technol. 1991;13:770–780. [Google Scholar]
- 208.Preusting H, Nijenhuis A, Witholt B. Physical characteristics of poly(3-hydroxyalkanoates) and poly(3-hydroxyalkenoates) produced by Pseudomonas oleovorans grown on aliphatic hydrocarbons. Macromolecules. 1990;23:4220–4224. [Google Scholar]
- 209.Pries A, Hein S, Steinbüchel A. Identification of lipoamide dehydrogenase as second locus affected in poly(3-hydroxybutyric acid)-leaky mutants of Alcaligenes eutrophus. FEMS Microbiol. 1992;97:227–234. doi: 10.1016/0378-1097(92)90341-k. [DOI] [PubMed] [Google Scholar]
- 210.Pries A, Priefert H, Krüger N, Steinbüchel A. Identification and characterization of two Alcaligenes eutrophus gene loci relevant to the poly(β-hydroxybutyric acid)-leaky phenotype which exhibit homology to ptsH and ptsI of Escherichia coli. J Bacteriol. 1991;173:5843–5853. doi: 10.1128/jb.173.18.5843-5853.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Qi Q, Rehm B H A, Steinbuchel A. Synthesis of poly(3-hydroxyalkanoates) in Escherichia coli expressing the PHA synthase gene phaC2 from Pseudomonas aeruginosa: comparison of PhaC1 and PhaC2. FEMS Microbiol Lett. 1997;157:155–162. doi: 10.1111/j.1574-6968.1997.tb12767.x. [DOI] [PubMed] [Google Scholar]
- 212.Ramsay B A, Lomaliza K, Chavarie C, Dube B, Bataille P, Ramsay J A. Production of poly-(beta-hydroxybutyric-co-beta-hydroxyvaleric) acids. Appl Environ Microbiol. 1990;56:2093–2098. doi: 10.1128/aem.56.7.2093-2098.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Ramsay B A, Saracovan I, Ramsay J A, Marchessault R H. Continuous production of long-side-chain poly-β-hydroxyalkanoates by Pseudomonas oleovorans. Appl Environ Microbiol. 1991;57:625–629. doi: 10.1128/aem.57.3.625-629.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Rehm B H A, Krüger N, Steinbüchel A. A new metabolic link between fatty acid de novo synthesis and polyhydroxyalkanoic acid synthesis. J Biol Chem. 1998;273:24044–24051. doi: 10.1074/jbc.273.37.24044. [DOI] [PubMed] [Google Scholar]
- 215.Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Haas D. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide and lipase. Mol Microbiol. 1997;24:309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- 216.Reizer J, Reizer A, Saier M H, Jacobson G R. A proposed link between nitrogen and carbon metabolism involving protein phosphorylation in bacteria. Protein Sci. 1992;1:722–726. doi: 10.1002/pro.5560010604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ren Q, Kessler B, Witholt B. Abstracts of the 1996 International Symposium on Bacterial Polyhydroxyalkanoates. 1996. Production of medium chain length poly-3-hydroxyalkanoates: from Pseudomonas to Escherichia coli; p. 2/03. Davos, Switzerland. [Google Scholar]
- 218.Reusch R N. Poly-beta-hydroxybutyrate/calcium polyphosphate complexes in eukaryotic membranes. Proc Soc Exp Biol Med. 1989;191:377–381. doi: 10.3181/00379727-191-42936. [DOI] [PubMed] [Google Scholar]
- 219.Reusch R N, Huang R, Bramble L L. Poly-3-hydroxybutyrate/polyphosphate complexes form voltage-activated Ca+2 channels in the plasma membrane of Escherichia coli. Biophys J. 1995;69:754–766. doi: 10.1016/S0006-3495(95)79958-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Reusch R N, Huang R, Kosk-Kosicka D. Novel components and enzymatic activities of the human erythrocyte plasma membrane calcium pump. FEBS Lett. 1997;412:592–596. doi: 10.1016/s0014-5793(97)00863-6. [DOI] [PubMed] [Google Scholar]
- 221.Reusch, R. N., and W. H. Reusch. November 1993. U.S. patent 5,266,422.
- 222.Reusch, R. N., and W. H. Reusch. May 1996. U.S. patent RE035257.
- 223.Reusch R N, Sadoff H L. d-(−)-Poly-β-hydroxybutyrate in membranes of genetically competent bacteria. J Bacteriol. 1983;156:778–788. doi: 10.1128/jb.156.2.778-788.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Reusch R N, Sadoff H L. Putative structure and functions of a poly-β-hydroxybutyrate/calcium polyphosphate channel in bacterial plasma membranes. Proc Natl Acad Sci USA. 1988;85:4176–4180. doi: 10.1073/pnas.85.12.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Reusch R N, Sparrow A W, Gardiner J. Transport of poly-β-hydroxybutyrate in human plasma. Biochim Biophys Acta. 1992;1123:33–40. doi: 10.1016/0005-2760(92)90168-u. [DOI] [PubMed] [Google Scholar]
- 226.Rhee Y H, Jang J-H, Rogers P L. Production of copolymer consisting of 3-hydroxybutyrate and 3-hydroxyvalerate by fed-batch culture of Alcaligenes sp. SH-69. Biotechnol Lett. 1993;15:377–382. [Google Scholar]
- 227.Rhie H G, Dennis D. The function of ackA and pta genes is necessary for poly(3-hydroxybutyrate-co-3-hydroxyvalerate) synthesis in recombinant pha+ Escherichia coli. Can J Microbiol. 1995;41(Suppl. 1):200–206. doi: 10.1139/m95-188. [DOI] [PubMed] [Google Scholar]
- 228.Rich J J, Kinscherf T G, Kitten T, Willis D K. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J Bacteriol. 1994;176:7468–7475. doi: 10.1128/jb.176.24.7468-7475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Rutherford, D. R., W. J. Hammar, and G. N. Babu. March 1997. U.S. patent 5,614,576.
- 230.Ryu H W, Hahn S K, Chang Y K, Chang H N. Production of poly(3-hydroxybutyrate) by high cell density fed-batch culture of Alcaligenes eutrophus with phosphate limitation. Biotechnol Bioeng. 1997;55:28–32. doi: 10.1002/(SICI)1097-0290(19970705)55:1<28::AID-BIT4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 231.Saito T, Fukui T, Ikeda F, Tanaka Y, Tomita K. An NADP-linked acetoacetyl CoA reductase from Zoogloea ramigera. Arch Microbiol. 1977;114:211–217. doi: 10.1007/BF00446864. [DOI] [PubMed] [Google Scholar]
- 232.Schembri M A, Bayly R C, Davies J K. Cloning and analysis of the polyhydroxyalkanoic acid synthase gene from an Acinetobacter sp.: evidence that the gene is both plasmid and chromosomally located. FEMS Microbiol Lett. 1994;118:145–152. doi: 10.1111/j.1574-6968.1994.tb06817.x. [DOI] [PubMed] [Google Scholar]
- 233.Schembri M A, Bayly R C, Davies J K. Phosphate concentration regulates transcription of the Acinetobacter polyhydroxyalkanoic acid biosynthetic genes. J Bacteriol. 1995;177:4501–4507. doi: 10.1128/jb.177.15.4501-4507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Schembri M A, Woods A A, Bayly R C, Davies J K. Identification of a 13-kDa protein associated with the polyhydroxyalkanoic acid granules from Acinetobacter spp. FEMS Microbiol Lett. 1995;133:277–283. doi: 10.1111/j.1574-6968.1995.tb07897.x. [DOI] [PubMed] [Google Scholar]
- 235.Schubert P, Kruger N, Steinbuchel A. Molecular analysis of the Alcaligenes eutrophus poly(3-hydroxybutyrate) biosynthesis operon: identification of the N terminus of poly(3-hydroxybutyrate) synthase and the identification of the promoter. J Bacteriol. 1991;173:168–175. doi: 10.1128/jb.173.1.168-175.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Schubert P, Steinbüchel A, Schlegel H G. Cloning of the Alcaligenes eutrophus genes for the synthesis of poly-β-hydroxybutyrate (PHB) and synthesis of PHB in Escherichia coli. J Bacteriol. 1988;170:5837–5847. doi: 10.1128/jb.170.12.5837-5847.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Schulz S, Toft S. Identification of a sex pheromone from a spider. Science. 1993;260:1635–1637. doi: 10.1126/science.260.5114.1635. [DOI] [PubMed] [Google Scholar]
- 238.Seebach D, Brunner A, Bürger H M, Reusch R N, Bramble L L. Channel-forming activity of 3-hydroxybutanoic-acid oligomers in planar lipid bilayers. Helv Chim Acta. 1996;79:507–517. [Google Scholar]
- 239.Senior P J, Beech G A, Ritchie G A F, Dawes E A. The role of oxygen limitation in the formation of poly-β-hydroxybutyrate during batch and continuous culture of Azotobacter beijerinckii. Biochem J. 1972;128:1193–1201. doi: 10.1042/bj1281193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Senior P J, Dawes E A. The regulation of poly-β-hydroxybutyrate metabolism in Azotobacter beijerinckii. Biochem J. 1973;134:225–238. doi: 10.1042/bj1340225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Shiotani, T., and G. Kobayashi. March 1994. U.S. patent 5,292,860.
- 242.Sim S J, Snell K D, Hogan S A, Stubbe J, Rha C, Sinskey A J. PHA synthase activity controls the molecular weight and polydispersity of polyhydroxybutyrate in vivo. Nat Biotechnol. 1997;15:63–67. doi: 10.1038/nbt0197-63. [DOI] [PubMed] [Google Scholar]
- 243.Slater S, Gallaher T, Dennis D. Production of poly-(3-hydroxybutyrate-co-3-hydroxyvalerate) in a recombinant Escherichia coli strain. Appl Environ Microbiol. 1992;58:1089–1094. doi: 10.1128/aem.58.4.1089-1094.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Slater S, Houmiel K L, Tran M, Mitsky T A, Taylor N B, Padgette S R, Gruys K J. Multiple β-ketothiolases mediate poly(β-hydroxyalkanoate) copolymer synthesis in Ralstonia eutropha. J Bacteriol. 1998;180:1979–1987. doi: 10.1128/jb.180.8.1979-1987.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Slater S C, Voige W H, Dennis D E. Cloning and expression in Escherichia coli of the Alcaligenes eutrophus H16 poly-β-hydroxybutyrate biosynthetic pathway. J Bacteriol. 1988;170:4431–4436. doi: 10.1128/jb.170.10.4431-4436.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Son H, Park G, Lee S. Growth-associated production of poly-β-hydroxybutyrate from glucose or alcoholic distillery wastewater by Actinobacillus sp. EL-9. Biotechnol Lett. 1996;18:1229–1234. [Google Scholar]
- 247.Song J J, Yoon S C. Biosynthesis of novel aromatic copolyesters from insoluble 11-phenoxyundecanoic acid by Pseudomonas putida BMO1. Appl Environ Microbiol. 1996;62:536–544. doi: 10.1128/aem.62.2.536-544.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Steel, M. L., and P. Norton-Berry. July 1986. U.S. patent 4,603,070.
- 249.Steinbüchel A. Polyhydroxyalkanoaic acids. In: Byrom D, editor. Biomaterials: novel materials from biological sources. New York, N.Y: Stockton Press; 1991. pp. 124–213. [Google Scholar]
- 250.Steinbüchel A, Hustede E, Liebergesell M, Pieper U, Timm A, Valentin H. Molecular basis for biosynthesis and accumulation of polyhydroxyalkanoic acids in bacteria. FEMS Microbiol Rev. 1992;103:217–230. doi: 10.1111/j.1574-6968.1992.tb05841.x. [DOI] [PubMed] [Google Scholar]
- 251.Steinbüchel A, Pieper U. Production of a copolyester of 3-hydroxybutyric acid and 3-hydroxyvaleric acid from single unrelated carbon sources by a mutant of Alcaligenes eutrophus. Appl Microbiol Biotechnol. 1992;37:1–6. [Google Scholar]
- 252.Steinbüchel A, Schlegel H G. Physiology and molecular genetics of poly(β-hydroxyalkanoic acid) synthesis in Alcaligenes eutrophus. Mol Microbiol. 1991;5:523–542. doi: 10.1111/j.1365-2958.1991.tb00725.x. [DOI] [PubMed] [Google Scholar]
- 253.Steinbüchel A, Schubert P. Expression of the Alcaligenes eutrophus poly(β-hydroxybutyric acid)-synthetic pathway in Pseudomonas sp. Arch Microbiol. 1989;153:101–104. [Google Scholar]
- 254.Steinbüchel A, Valentin H E. Diversity of bacterial polyhydroxyalkanoic acids. FEMS Microbiol Lett. 1995;128:219–228. [Google Scholar]
- 255.Steinbüchel A, Wiese S. A Pseudomonas strain accumulating polyesters of 3-hydroxybutyric acid and medium-chain-length 3-hydroxyalkanoic acids. Appl Microbiol Biotechnol. 1992;37:691–697. [Google Scholar]
- 256.Streeter J G. Transport and metabolism of carbon and nitrogen in legume nodules. Adv Bot Res. 1991;18:129–187. [Google Scholar]
- 257.Su, L., R. W. Lenz, and D. P. Martin. Enzymatic polymerization of (R)-3-hydroxyalkanoates by a bacterial polymerase. Macromolecules, in press.
- 258.Sun W, Cao J-G, Teng K, Meighen E A. Biosynthesis of poly-3-hydroxybutyrate in the luminescent bacterium, Vibrio harveyi, and regulation by the lux autoinducer, N-(3-hydroxybutanoyl)homoserine lactone. J Biol Chem. 1994;269:20785–20790. [PubMed] [Google Scholar]
- 259.Suzuki T, Deguchi H, Yamane T, Shimizu S, Gekko K. Control of molecular weight of poly-β-hydroxybutyric acid produced in fed-batch culture of Protomonas extorquens. Appl Microbiol Biotechnol. 1988;27:487–491. [Google Scholar]
- 260.Suzuki T, Yamane T, Shimizu S. Kinetics and effect of nitrogen source feeding on production of poly-β-hydroxybutyric acid by fed-batch culture. Appl Microbiol Biotechnol. 1986;24:366–369. [Google Scholar]
- 261.Suzuki T, Yamane T, Shimizu S. Mass production of poly-β-hydroxybutyric acid by fed-batch culture with controlled carbon/nitrogen feeding. Appl Microbiol Biotechnol. 1986;24:370–374. [Google Scholar]
- 262.Suzuki T, Yamane T, Shimizu S. Mass production of poly-β-hydroxybutyric acid by fully automatic fed-batch culture of a methylotroph. Appl Microbiol Biotechnol. 1986;23:322–329. [Google Scholar]
- 263.Takahashi H, Miyake M, Tokiwa Y, Asada Y. Improved accumulation of poly-3-hydroxybutyrate by a recombinant cyanobacterium. Biotechnol Lett. 1998;20:183–186. [Google Scholar]
- 264.Takeda M, Matsuoka H, Ban H, Ohashi Y, Hikuma M, Koizumi J-I. Biosynthesis of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) by a mutant of Sphaerotilus natans. Appl Microbiol Biotechnol. 1995;44:37–42. [Google Scholar]
- 265.Takeda M, Matsuoka H, Hamana H, Hikuma M. Biosynthesis of poly-3-hydroxybutyrate by Sphaerotilus natans. Appl Microbiol Biotechnol. 1995;43:31–34. [Google Scholar]
- 266.Tan I K P, Kumar K S, Theanmalar M, Gan S N, Gordon B., III Saponified palm kernel oil and its major free fatty acids as carbon substrates for the production of polyhydroxyalkanoates in Pseudomonas putida PGA1. Appl Microbiol Biotechnol. 1997;47:207–211. [Google Scholar]
- 267.Thompson S, Mayer F, Peoples O P, Masamune S, Sinskey A J, Walsh C T. Mechanistic studies on β-ketoacyl thiolase from Zoogloea ramigera: identification of the active site nucleophile as Cys89, its mutation to Ser89, and kinetic and thermodynamic characterization of wildtype and mutant enzymes. Biochemistry. 1989;28:5735–5742. doi: 10.1021/bi00440a006. [DOI] [PubMed] [Google Scholar]
- 268.Timm A, Byrom D, Steinbüchel A. Formation of blends of various poly(3-hydroxyalkanoic acids) by a recombinant strain of Pseudomonas oleovorans. Appl Microbiol Biotechnol. 1990;33:296–301. [Google Scholar]
- 269.Timm A, Steinbüchel A. Cloning and molecular analysis of the poly(3-hydroxyalkanoic acid) gene locus of Pseudomona aeroginosa PAO1. Eur J Biochem. 1992;209:15–30. doi: 10.1111/j.1432-1033.1992.tb17256.x. [DOI] [PubMed] [Google Scholar]
- 270.Timm A, Steinbüchel A. Formation of polyesters consisting of medium-chain-length 3-hydroxyalkanoic acids from gluconate by Pseudomonas aeruginosa and other fluorescent pseudomonads. Appl Environ Microbiol. 1990;56:3360–3367. doi: 10.1128/aem.56.11.3360-3367.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Tombolini R, Povolo S, Buson A, Squartini A, Nuti M P. Poly-β-hydroxybutyrate (PHB) biosynthetic genes in Rhizobium meliloti 41. Microbiology. 1995;141:2553–2559. doi: 10.1099/13500872-141-10-2553. [DOI] [PubMed] [Google Scholar]
- 272.Ueda S, Matsumoto S, Takagi A, Yamane T. Synthesis of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) from methanol and n-amyl alcohol by the methylotrophic bacteria Paracoccus denitrificans and Methylobacterium extorquens. Appl Environ Microbiol. 1992;58:3574–3579. doi: 10.1128/aem.58.11.3574-3579.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Ueda S, Yabutani T, Maehara A, Yamane T. Molecular analysis of the poly(3-hydroxyalkanoate) synthase gene from a methylotrophic bacterium, Paracoccus denitrificans. J Bacteriol. 1996;178:774–779. doi: 10.1128/jb.178.3.774-779.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Umeda F, Kitano Y, Murakami Y, Yagi K, Miura Y, Mizoguchi T. Cloning and sequence analysis of the poly(3-hydroxyalkanoic acid)-synthesis genes of Pseudomonas acidophila. Appl Biochem Biotechnol. 1998;70–72:341–352. doi: 10.1007/BF02920150. [DOI] [PubMed] [Google Scholar]
- 275.Valentin H E, Dennis D. Metabolic pathway for poly(3-hydroxybutyrate-co-3-hydroxyvalerate) formation in Nocardia corallina: Inactivation of mutB by chromosomal integration of a kanamycin resistance gene. Appl Environ Microbiol. 1996;62:372–379. doi: 10.1128/aem.62.2.372-379.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Valentin H E, Dennis D. Production of poly(3-hydroxybutyrate-co-4-hydroxybutyrate) in recombinant Escherichia coli grown on glucose. J Biotechnol. 1997;58:33–38. doi: 10.1016/s0168-1656(97)00127-2. [DOI] [PubMed] [Google Scholar]
- 277.Valentin H E, Lee E Y, Choi C Y, Steinbüchel A. Identification of 4-hydroxyhexanoic acid as a new constituent of biosynthetic polyhydroxyalkanoic acids from bacteria. Appl Microbiol Biotechnol. 1994;40:710–716. [Google Scholar]
- 278.Valentin H E, Schönebaum A, Steinbüchel A. Identification of 4-hydroxyvaleric acid as a constituent in biosynthetic polyhydroxyalkanoic acids from bacteria. Appl Microbiol Biotechnol. 1992;36:507–514. [Google Scholar]
- 279.Valentin H E, Schönebaum A, Steinbüchel A. Identification of 5-hydroxyhexanoic acid, 4-hydroxyheptanoic acid and 4-hydroxyoctanoic acid as new constituents in bacterial polyhydroxyalkanoic acids. Appl Microbiol Biotechnol. 1996;46:261–267. [Google Scholar]
- 280.Valentin H E, Steinbuchel A. Cloning and characterization of the Methylobacterium extorquens polyhydroxyalkanoic-acid-synthase structural gene. Appl Microbiol Lett. 1993;39:309–317. doi: 10.1007/BF00192084. [DOI] [PubMed] [Google Scholar]
- 281.Valentin H E, Stuart E S, Fuller R C, Lenz R W, Dennis D. Investigation of the function of proteins associated to polyhydroxyalkanoate inclusions in Pseudomonas putida BAO1. J Biotechnol. 1998;64:145–157. doi: 10.1016/s0168-1656(98)00097-2. [DOI] [PubMed] [Google Scholar]
- 282.Wallen L L, Rohwedder W K. Poly-β-hydroxyalkanoate from activated sludge. Environ Science Technol. 1974;8:576–579. [Google Scholar]
- 283.Walshaw D L, Wilkinson A, Mundy M, Smith M, Poole P S. Regulation of the TCA cycle and the general amino acid permease by overflow metabolism in Rhizobium leguminosarum. Microbiology. 1997;143:2209–2221. doi: 10.1099/00221287-143-7-2209. [DOI] [PubMed] [Google Scholar]
- 284.Wang F, Lee S Y. High cell density culture of metabolically engineered Escherichia coli for the production of poly(3-hydroxybutyrate) in a defined medium. Biotechnol Bioeng. 1998;58:325–328. [PubMed] [Google Scholar]
- 285.Wang F, Lee S Y. Production of poly(3-hydroxybutyrate) by fed-batch culture of filamentation-suppressed recombinant Escherichia coli. Appl Environ Microbiol. 1997;63:4765–4769. doi: 10.1128/aem.63.12.4765-4769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Wang J G, Bakken L R. Screening of soil bacteria for poly-β-hydroxybutyric acid production and its role in the survival of starvation. Microbiol Ecol. 1998;35:94–101. doi: 10.1007/s002489900063. [DOI] [PubMed] [Google Scholar]
- 287.Webb, A. February 1990. U.S. patent 4900299.
- 288.Werner D, Mörschel E, Stripf R, Winchenbach B. Development of nodules of glycine max infected with an ineffective strain of Rhizobium japonicum. Planta. 1980;147:320–329. doi: 10.1007/BF00379840. [DOI] [PubMed] [Google Scholar]
- 289.Wieczorek R, Pries A, Steinbuchel A, Mayer F. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J Bacteriol. 1995;177:2425–2435. doi: 10.1128/jb.177.9.2425-2435.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Williams D R, Anderson A J, Dawes E A, Ewing D F. Production of a co-polyester of 3-hydroxybutyric acid and 3-hydroxyvaleric acid from succinic acid by Rhodococcus ruber: biosynthetic considerations. Appl Microbiol Biotechnol. 1994;40:717–723. [Google Scholar]
- 291.Williams M D, Fieno A M, Grant R A, Sherman D H. Expression and analysis of a bacterial poly(hydroxyalkanoate) synthase in insect cells using baculovirus system. Protein Expression Purif. 1996;7:203–211. doi: 10.1006/prep.1996.0028. [DOI] [PubMed] [Google Scholar]
- 292.Williams M D, Rahn J A, Sherman D H. Production of a polyhydroxyalkanoate biopolymer in insect cells with a modified eukaryotic fatty acid synthase. Appl Environ Microbiol. 1996;62:2540–2546. doi: 10.1128/aem.62.7.2540-2546.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Williams S F, Peoples O P. Biodegradable plastics from plants. Chemtech. 1996;26:38–44. [Google Scholar]
- 294.Willis D K, Hrabak E M, Rich J J, Barta M, Lindow S E, Panopoulos N J. Isolation and characterization of a Pseudomonas syringae pv. syringae mutant deficient in lesion-forming ability on bean. Mol Plant-Microbe Interact. 1990;3:149–156. [Google Scholar]
- 295.Winkler F K, D’Arcy A, Hunziker W. Structure of human pancreatic lipase. Nature. 1990;343:771–774. doi: 10.1038/343771a0. [DOI] [PubMed] [Google Scholar]
- 296.Wodzinska J, Snell K D, Rhomberg A, Sinskey A J, Biemann K, Stubbe J. Polyhydroxybutyrate synthase: evidence for covalent catalysis. J Am Chem Soc. 1996;118:6319–6320. [Google Scholar]
- 297.Yabutani T, Maehara A, Ueda S, Yamane T. Analysis of β-ketothiolase and acetoacetyl-CoA reductase genes of a methylotrophic bacterium, Parracoccus denitrificans, and their expression in Escherichia coli. FEMS Microbiol Lett. 1995;133:85–90. doi: 10.1111/j.1574-6968.1995.tb07865.x. [DOI] [PubMed] [Google Scholar]
- 298.Yalpani, M. July 1993. U.S. patent 5,229,158.
- 299.Yalpani, M. July 1993. U.S. patent 5,225,227.
- 300.Yamane T, Chen X-F, Ueda S. Growth-associated production of poly(3-hydroxyvalerate) from n-pentanol by a methylotrophic bacterium, Paracoccus denitrificans. Appl Environ Microbiol. 1996;62:380–384. doi: 10.1128/aem.62.2.380-384.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Yim K S, Lee S Y, Chang H N. Synthesis of poly-(3-hydroxybutyrate-co-3-hydroxyvalerate) by recombinant Escherichia coli. Biotechnol Bioeng. 1996;49:495–503. doi: 10.1002/(SICI)1097-0290(19960305)49:5<495::AID-BIT2>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 302.Yoon J S, Kim J Y, Rhee Y H. Effects of amino acid addition on molar fraction of 3-hydroxyvalerate in copolyester of 3-hydroxybutyrate and 3-hydroxyvalerate synthesized by Alcaligenes sp. SH-69. J Ferment Bioeng. 1995;80:350–354. [Google Scholar]
- 303.Zhang H, Obias V, Gonyer K, Dennis D. Production of polyhydroxyalkanoates in sucrose-utilizing recombinant Escherichia coli and Klebsiella strains. Appl Environ Microbiol. 1994;60:1198–1205. doi: 10.1128/aem.60.4.1198-1205.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]