Abstract
Bacteriophage φ6 has a genome of three segments of double-stranded RNA. Each virus particle contains one each of the three segments. Packaging is effected by the acquisition, in a serially dependent manner, of the plus strands of the genomic segments into empty procapsids. The empty procapsids are compressed in shape and expand during packaging. The packaging program involves discrete steps that are determined by the amount of RNA inside the procapsid. The steps involve the exposure and concealment of binding sites on the outer surface of the procapsid for the plus strands of the three genomic segments. The plus strand of segment S can be packaged alone, while packaging of the plus strand of segment M depends upon prior packaging of S. Packaging of the plus strand of L depends upon the prior packaging of M. Minus-strand synthesis begins when the particle has a full complement of plus strands. Plus-strand synthesis commences upon the completion of minus-strand synthesis. All of the reactions of packaging, minus-strand synthesis, and plus-strand synthesis can be accomplished in vitro with isolated procapsids. Live-virus constructions that are in accord with the model have been prepared. Mutant virus with changes in the packaging program have been isolated and analyzed.
History and Structure
Bacteriophage φ6 is unusual in several ways. It has a polyhedral nucleocapsid that is covered by a lipid-containing membrane (82). Its genome consists of three segments of double-stranded RNA (dsRNA) (76). There are no other bacteriophages of similar known construction with the exception of a group of phages recently isolated by our laboratory that are very similar to φ6 in structural organization if not in sequence (unpublished data). In some ways, φ6 seems more closely related to members of the Reoviridae than it is to bacteriophages. The Reoviridae have genomes of 10, 11, or 12 segments of dsRNA (80). The genomic segments appear to be present in stoichiometric amounts, and the efficiency of plating of the viruses is high, indicating that most particles have accomplished precise packaging. The two groups also share a rare structural feature in that the inner core of the Reoviridae and φ6 contain 120 molecules of the major structural protein (13). In fact, it appears that even the fungal dsRNA virus-like particles, which package only one species of segment per particle, also have 120 molecules of the capsid protein (7). The question that is addressed in this review is how can the virus manage to package the three genomic segments in a precise manner. We have developed a model for the genomic packaging in φ6 that so far only applies to φ6 and involves unique mechanisms. The basic outline of the model is that a collapsed fully formed procapsid has binding sites on the outside for the plus strands of segment S. Upon successful packaging of S, the binding sites for S disappear and those for segment M appear. Upon successful packaging of M, the binding sites for M disappear and those for segment L appear. The changes in the availability of binding sites are determined by the amount of RNA inside the procapsid (69).
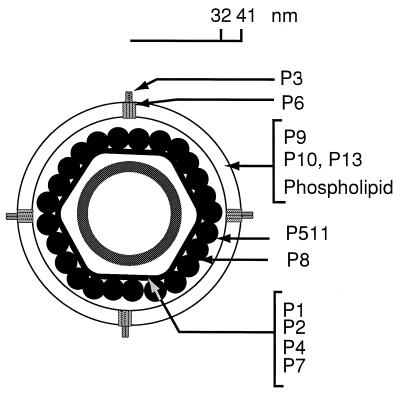
φ6 was first isolated from bean straw by Vidaver et al. (82). The phage could be propagated on many strains of Pseudomonas syringae (11). These investigators demonstrated that the virus had a genome consisting of three segments of dsRNA and that the nucleocapsid of the virion was enveloped in a lipid-containing membrane. The genomic RNA is located within a procapsid core that is composed of four proteins, P1, P2, P4, and P7 (44, 78) (Fig. 1). The procapsid is covered by a shell composed of a single protein, P8, to form the nucleocapsid (3, 53). The nucleocapsid is, in turn, covered by a lipid-containing membrane, which also contains proteins P9, P10, P6, P3, and P13 (53). Until recently, φ6 was the only bacteriophage isolate of its type; however, our laboratory has recently isolated about 10 relatives of φ6. The new isolates range from close relatives with 90% base sequence identity to distant ones that have essentially no base sequence or amino acid similarity but retain the general genomic organization of the family.
FIG. 1.
The φ6 virion is composed of a procapsid containing proteins P1, P2, P4, and P7, which is covered by a shell of protein P8 to form the nucleocapsid. The nucleocapsid is enveloped in a lipid-containing membrane composed of phospholipids and proteins P9, P10, P13, P3, and P6. P3 specifies the host range of the virus. P511 is a lytic endopeptidase that is associated with the surface of the nucleocapsid (4, 43).
Life Cycle
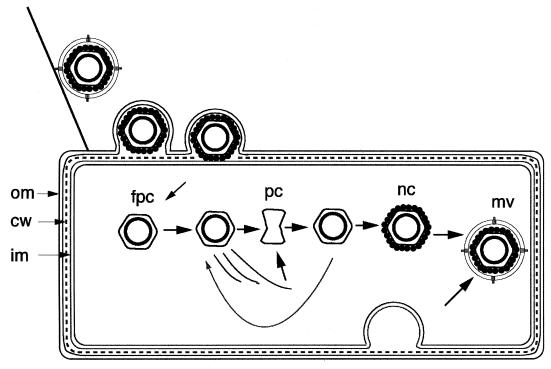
φ6 attaches to its host cell (Fig. 2), P. syringae, by adsorbing first to a type IV pilus (73, 74). The phage is brought into contact with the bacterial outer membrane by pilus retraction, and the membrane of the phage fuses with the outer membrane (1). This places the nucleocapsid in the periplasmic space. Protein P5 is located on the outside of the nucleocapsid and has endopeptidase activity (4, 45). The nucleocapsid then enters the host cell, whereupon it loses the shell of P8 and begins to transcribe copies of the three genomic segments. Isolated nucleocapsids have the ability to transcribe in vitro in the same manner if incubated with the four nucleoside triphosphates (NTPs) (16, 65). Transcription in φ6 involves the synthesis of plus strands on the dsRNA template and the displacement of the parental plus strand (14, 81). For the Reoviridae, transcription is conservative in that the new strand is extruded from the core (31).
FIG. 2.
Life cycle of φ6. The virion attaches to a pilus and is brought into contact with the outer membrane (om). The viral membrane fuses with the outer membrane to place the nucleocapsid in the periplasmic space. The murein (cw) is digested by viral protein P5, and the filled procapsid (fpc) penetrates the inner membrane (im) and enters the cell, leaving P8 behind. The procapsid transcriptase synthesizes complete copies of the three genomic segments. The L mRNA is translated to produce P1, P2, P4, and P7, which constitute the procapsid (pc). This is filled with dsRNA and continues transcription until it is covered by P8 to form nucleocapsids (nc). Membrane proteins are placed in the host membrane and then transferred to the virion (mv) along with host lipids. The membrane formation or translocation is dependent upon protein P12.
Several of the newly isolated relatives of φ6 do not adsorb to pili but instead attach directly to the rough lipopolysaccharide in the outer membrane of a wide range of gram-negative bacteria.
φ6 has a temporal program as part of its infection cycle. The proteins of the polymerase complex or procapsid are always synthesized early in infection (78). RNA synthesis during the early phase is restricted primarily to plus strands and these are synthesized in close to equimolar ratios. Later in infection, the plus strands of segments M and S are synthesized at levels 10 to 20 times that of the plus strand of segment L (10). During this period, the other proteins of the virion are synthesized and phage assembly takes place. Empty and filled procapsids are seen in infected cells, and they chase into nucleocapsids, and radioactivity in nucleocapsids chases into mature virions (15, 47). dsRNA is found only within procapsids; it is not found free in the cell. The membrane of the virion assembles within the cell through the mediation of nonstructural protein P12, and enveloped virions are seen inside the cell (36, 46). Ultimately, the cells lyse through the action of a membrane disorganizer, P10, and the endopeptidase P5, and about 300 virions are released into the medium (45).
Genetic and Physical Organization
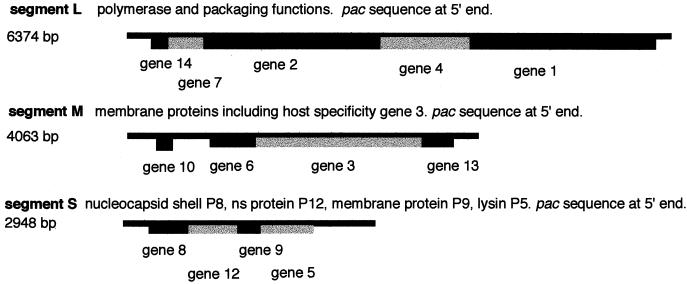
The genome of φ6 has been cloned as cDNA (23, 42, 48, 49, 72), and the genetic and physical maps of the three segments are shown in Fig. 3. The genome is organized such that all the polymerase functions are located on segment L, the genes for the host attachment proteins are on segment M, and the genes for the nucleocapsid shell (P8), the major membrane protein (P9), the lytic endopeptidase, and the membrane assembly nonstructural protein are on segment S. The cDNA copies are accurate in that plasmids carrying them are able to complement nonsense and temperature-sensitive mutants of the virus and ultimately to have their transcripts replace genomic segments of the virus. Plasmids containing the genes of segment L code for proteins P1, P2, P4, and P7. These four proteins comprise the procapsid of the virus. This is a dodecahedral structure that can be assembled in Escherichia coli strains that express the genes (25). The procapsids can be isolated from the cells and purified on sucrose gradients. Procapsids that have not packaged RNA are dodecahedral, but they are not expanded (Fig. 4). The five fold faces are inverted, giving the particle a compressed appearance (3). The procapsid contains 120 molecules of P1, which is the largest (85-kDa) procapsid protein and the major shape determinant; about 120 molecules of P4, which is a nonspecific nucleoside triphosphatase of 35 kDa; 60 molecules of P7 (21 kDa), which is an accessory protein involved in RNA synthesis and packaging; and, finally, 12 to 20 molecules of P2 (80 kDa), which has motifs of viral RNA polymerases. P1 alone can form dodecahedral structures (40, 55). The other three proteins bind to P1 independently of each other and can be added to particles from which they are missing (5, 38).
FIG. 3.
Physical and genetic maps of the genomic segments of φ6. Genes are identified below the segments (6, 43). The sequences necessary for packaging are located in the 5′ noncoding regions (left). The sequences necessary for minus-strand synthesis are at the 3′ ends.
FIG. 4.
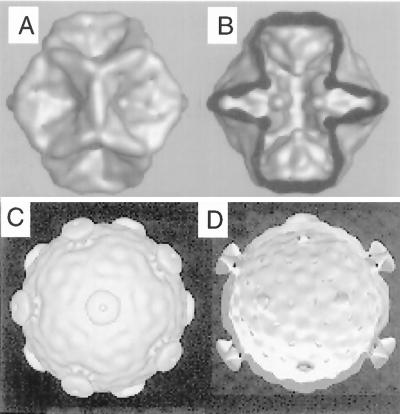
Reconstructions of the structures of empty procapsids and filled viral cores of φ6 determined from cryoelectron micrographs of procapsid particles containing proteins P1 and P4 (A and B) and cores visualized from viral nucleocapsids that have been stripped of the shell of protein P8 (C and D) (3). The structures in panels C and D are cut-open representations showing the inner surfaces of the structures viewed down a twofold axis of symmetry. The structures in panels A and B resemble particles that have not packaged RNA, while those in panels C and D resemble those that have packaged RNA and carried out minus-strand synthesis.
IN VITRO STUDIES
In Vitro Packaging of RNA by Isolated Procapsids
Procapsids isolated from E. coli will package φ6 plus-strand copies of the genomic segments (29). The packaging takes place in a rather simple buffer solution with PEG and salts and a source of NTP (26). There is no requirement for any bacterial proteins or for any other phage-encoded proteins besides the four components of the procapsid. Packaging is serially dependent in that the plus-strand copy of segment S can be packaged alone but packaging of M depends upon S and packaging of L depends upon S and M (67). The dependence in vitro is not absolute and varies from preparation to preparation (18, 20, 28); however, it appears that this is an important element in maintaining the high degree of precision in normal packaging. Segments M and L can be packaged independently (28), if inefficiently; however, given the normal range of plus-strand copies in an infected cell, the virus attains an efficiency of plating of 1, which implies that the packaging is close to perfect. In vitro packaging is generally measured by incubating radioactively labeled plus strands with procapsids and then treating the mixture with RNase I. The packaged RNA is protected and can be visualized on agarose gels (Fig. 5). The RNA molecules enter the procapsid from the 5′ end, and the portals through which they enter are small in that an RNA molecule with a strong hairpin stem (>20 G · C pairs) can be packaged from the 5′ end up to the hairpin stem (68) (Fig. 6). RNase I digests the hairpin and the 3′ extension. It was also observed that an S RNA with a hairpin was able to promote the packaging of a normal M molecule, indicating that at least two portals could function in a single procapsid. It is not known whether each segment has its assigned portal, but since the procapsid appears to be symmetrical, it is assumed that the portals are all equivalent. It is not known where the portals are located, but since both P4 and P7 seem to be involved in packaging, they both might comprise the portals. Packaging requires a source of NTP but is not very specific with respect to the base or the sugar (64). Protein P4 shows NTPase activities that are approximately the same as the base specificity for packaging (27). P4 molecules form multimers that may be precursors of their arrangement in the procapsids (39). There is an indication that the multimeric P4 complex might be placed at the center of the fivefold faces of the procapsid (13a). This would suggest that RNA uptake might occur at the center of the fivefold faces. This is an interesting possibility, since it is the same place where plus-strand transcripts of the genomic segments of Reoviridae are extruded from their cores (41).
FIG. 5.
In vitro packaging of radioactive plus strands of exact copies of genomic segments S and M and of a truncated segment L. Radioactive transcripts of plasmids pLM659 (s), pLM656 (m), and pLM1157 (l) were incubated with procapsids, treated with RNase I, and applied to a 2% agarose gel (67). An l strand of reduced size is used because its packaging is more efficient than that of normal-sized segment l. Note the dependence relationships in packaging.
FIG. 6.
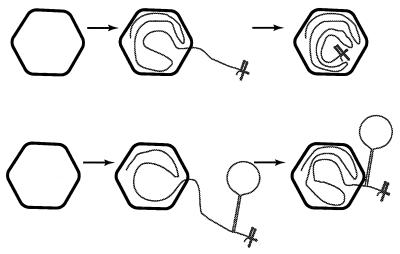
Diagram of the packaging of normal RNA and a hairpin structure by procapsids. The diagram is not to scale, and the small secondary structure is the normal 3′ end of the genomic segments. The upper pathway is that of a normal segment, and the lower pathway is that of a hairpin structure (68).
An important and useful adjunct to the in vitro packaging experiments was the development of a system for the construction of live virus from particles that had packaged either normal viral RNA or transcripts of cDNA plasmids (56). This was made possible by the finding that spheroplasts of the host cells could be infected by nucleocapsids of the virus in the absence of membrane and attachment proteins (54). Additionally, it was found that protein P8 could be removed from nucleocapsids to form filled cores that were noninfectious but could be reactivated by adding back P8 (57). This system made it possible to use P8 to coat procapsids that had undergone in vitro packaging and then to infect spheroplasts to obtain viable virus (56, 59).
Secondary Structure of the RNA pac Regions
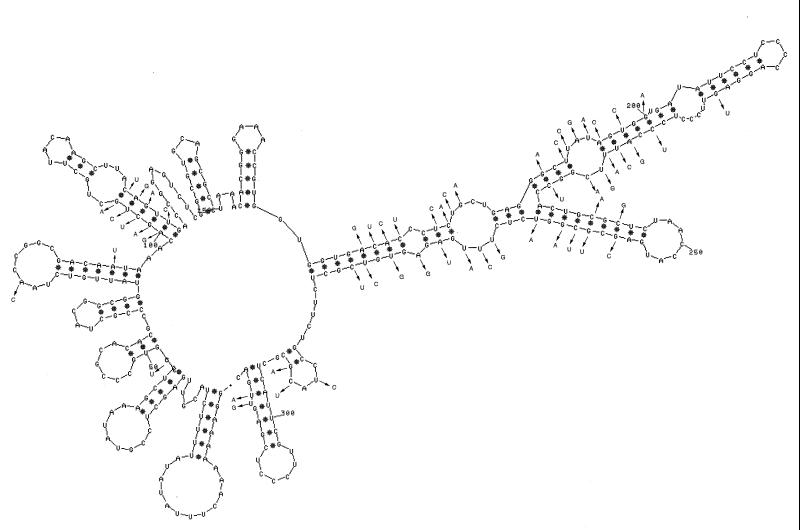
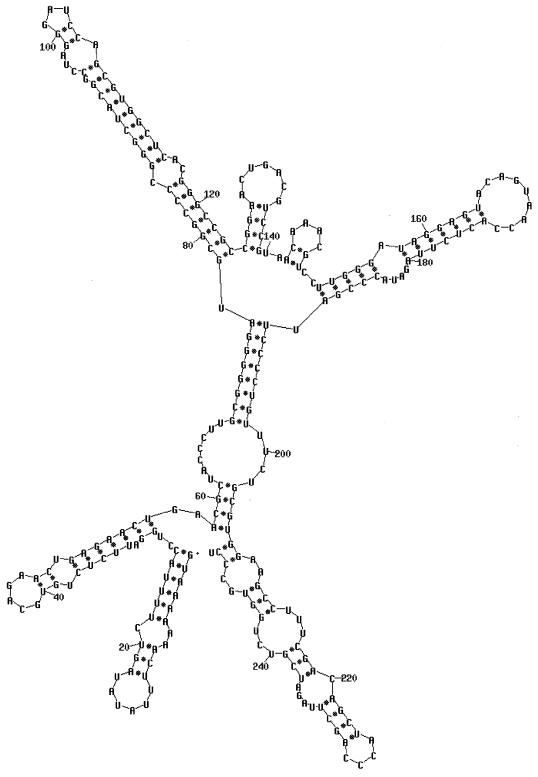
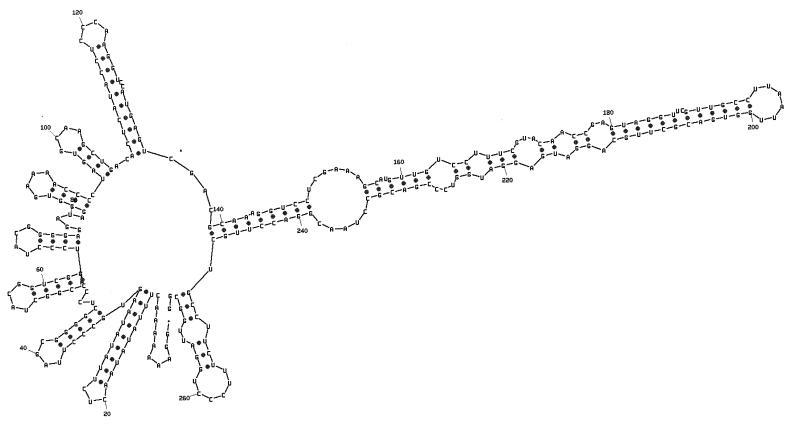
Packaging is specific to φ6 RNA and, as stated above, is specific with respect to each genomic segment. The RNA molecules are identified by two sequences. The first is an 18-base sequence that is essentially identical in all three segments and is located at the 5′ end (34). Its sequence is G(U/G)AAAAAAACUUUAUAUA. The second is a sequence of about 200 nucleotides that is unique for each segment and is located about 50 nucleotides from the 5′ sequence identity (24). The pac sequences have been defined by deletion studies involving in vitro packaging of φ6 procapsids. The 5′-terminal sequence and the segment-specific sequence are necessary and sufficient for packaging. Plus-strand copies of the first 300 nucleotides can be packaged by procapsids. Small changes within the pac sequences destroy packaging ability. The RNA within this region shows a high degree of secondary structure when analyzed with a program such as mfold (88) (Fig. 7 to 9). The relevance of the specific hairpin structures seen with mfold is not clear; however, comparison of the sequences of φ6, φ7, φ9, and φ10 shows changes in sequence that are compatible with the structures produced by mfold (88). In several of the hairpin stems, base changes have occurred in pairs so that base pairing is maintained. This is particularly striking in segment M near nucleotides 100 and 240 (Fig. 7). These newly isolated phages are able to interact genetically with φ6 at high frequency. This tends to support the notion that these hairpin stems do exist, are important, and probably play a role in packaging.
FIG. 7.
Secondary structure of the pac region of segment M of φ6. The first 305 nucleotides of segment M can be packaged by procapsids (24). The first 268 nucleotides cannot be packaged. Deletions of nucleotides 11 to 18, nucleotides 17 to 36, or nucleotides 23 to 43 does not prevent packaging, but deletion of nucleotides 11 to 43 does prevent packaging. Deletion of nucleotides 11 to 33 is not packaged but competes for packaging with normal segment M. The bases shown with arrows are all the changes in this region between φ6 and the related bacteriophage, φ7. It is notable that most of the changes found in proposed hairpin stems are compensated by changes that maintain base pairing.
FIG. 9.
Secondary structure of the pac region of segment L of φ6. The first 205 nucleotides are sufficient for packaging (24).
The three plus strands have a region of about 75 nucleotides at the 3′ end that has several predicted stem-loop structures. The sequences are similar but not identical between the three segments and can be exchanged by recombination with no apparent consequence (50, 51). The 3′ end is important for stability of the RNA and for polymerase recognition. However, most of the secondary structure can be deleted without destroying template function. A remnant as small as the terminal 12 nucleotides is enough to give template activity of about 30% of normal. A sequence of CCC is also sufficient to provide low template activity; however, most other sequences result in the total loss of template activity (50).
RNA Binding to the Outside of Procapsids
φ6 RNA binds to the outside of the procapsid (37). The binding is specific for φ6 RNA but does not seem to be segment specific. My group thought that there might be segment-specific binding that was masked by the general binding of RNA. We proposed that if there was segment-specific binding on the outside of the procapsids, we should be able to construct RNA molecules that would compete in packaging with normal segments but would not be packageable themselves. We prepared plus strands of segment S that had deletions of various sizes between the 5′ consensus sequence and the pac sequence (69) (Fig. 8). The deletions were of nucleotides 11 to 23, 11 to 32, and 11 to 43. We found that the smallest deletion had no great effect; deletion RNA competed with normal S RNA but was itself packaged. The largest deletion resulted in a loss of packaging and did not compete with normal RNA for packaging. The mid-sized deletion was just right. RNA containing this deletion competed for packaging but was not packaged. It also resulted in the lack of packaging of normal segments M and L. A similar set of deletions was prepared for segment M (Fig. 7). It was found that the proper deletion was not exactly the same as that for S but that it was possible to find a window of deletion that resulted in an RNA molecule that competed for packaging but was not packaged itself. RNA containing a deletion of nucleotides 17 to 43 was packaged normally, but RNA containing a deletion of 11 to 43 was not. This deletant (pLM794) competed for packaging with normal M. A satisfying correlate of the competition experiment was that the deletion fragment of segment M competed with M for packaging and also resulted in the lack of packaging of segment L; however, segment S packaging was unaffected. The working model for the binding on the outside of the procapsid is that the pac site, in cooperation with the 5′-terminal sequence, is responsible for specific binding to one of many sites on the surface of the procapsid. This binding places the 5′ end of the plus strand in close proximity to one of the entry portals, and the RNA is quickly taken up. The distance between the 5′ end of the molecule and the pac site is probably critical for the correct positioning.
FIG. 8.
Secondary structure of the pac region of segment S of φ6. The first 270 nucleotides are sufficient for packaging (24). The first 234 nucleotides cannot be packaged. Deletion of nucleotides 11 to 23 did not prevent packaging, but deletion of nucleotides 11 to 32 resulted in an RNA that competed with normal RNA for packaging but was not packaged itself. Deletion of nucleotides 11 to 43 resulted in RNA that neither competed nor packaged.
Minus-strand synthesis does not usually occur until all three plus strands have been packaged (19). The 3′ ends of the plus strands have a set of stem-loop structures that are similar to each other and can be interchanged with no loss of function (43). However, if the 3′ end is removed from the plus strand, the remaining plus strand usually cannot serve as a template for minus-strand synthesis. It can, however, count as a packaged segment to allow minus-strand synthesis on the other plus strands (19). It is even possible to package two plus strands that are lacking the normal 3′ ends, and these will serve to promote minus-strand synthesis on a third segment. The requirement for full genome packaging before the onset of minus-strand synthesis is another checkpoint that ensures that the composition of the virions will be precisely correct.
Packaging of Multiple Copies of Abnormally Small Segments
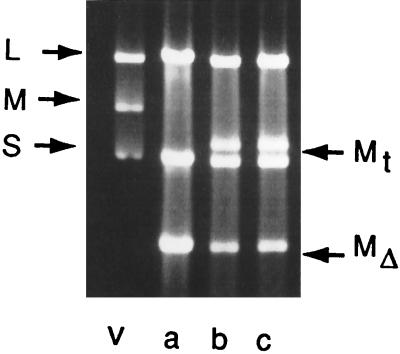
The question then arises as to the mechanism enabling the procapsid to keep track of the packaging progress. Another set of observations had to be rationalized. Viruses having large deletions in particular genomic segments were constructed (60). As an example, phage φ1980 has normal segments S and M but segment L is missing genes 7, 2, and 4. The size of segment L is normally 6.4 kbp, but in φ1980 it is 3.2 kbp. Upon examination of ethidium-stained gels of dsRNA of this phage, it was observed that the amount of L segment RNA in the virion was close to that of the normal L segment even though it contained a deletion reducing its size to half of normal. It appeared that two molecules of the shortened L segment were being packaged. We found the same result with a deletion in segment M (Fig. 10). In that case as well, we found that the amount of dsRNA in the virions indicated that two molecules of the segment must have been packaged or replicated. It did not appear as if the procapsid was simply accommodating more RNA to achieve a “headful” condition, since the additional RNA was always of the same class as the deletion segment. In the same set of experiments, it was found that the phages lacking viral genes that were expressed by plasmids could acquire the transcripts of the plasmids as fourth genomic segments (Fig. 10). This acquisition could occur at relatively high frequencies even though the transcripts had 5′ and 3′ extensions from the normal 5′ and 3′ ends of the genomic segments. The acquired transcripts were, however, found to have been tailored so that they matched the normal genomic segments exactly. A requirement for the acquisition of the transcript was that it contain the pac sequence of the deleted segment. We then tested whether the virion could acquire a transcript that had the pac sequence of another segment and found that, indeed, such transcripts could be picked up. However, sequence analysis showed that the acquired transcript underwent heterologous recombination with the deleted segment to change its 5′ end and pac sequence to that of the deleted segment (60). The conclusion appeared to be that the procapsid was able to determine the amount of RNA packaged in each class and that a selection pressure existed to yield virions with the correct amount of RNA for each segment class.
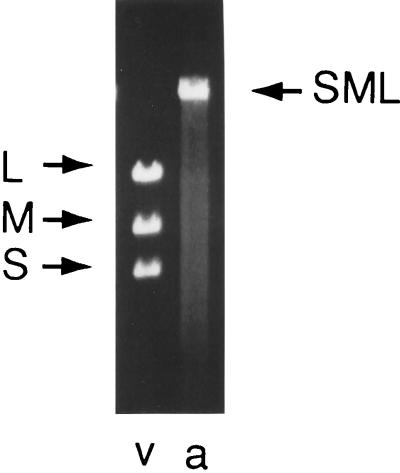
FIG. 10.
dsRNA extracted from virions with deletions in segment M. Lanes: v, wild-type virus, a, virus missing gene 3 of segment M; b and c, virus as in lane a that has acquired transcript RNA as a fourth segment. Mt is the segment formed from the transcript, and MΔ is the segment missing gene 3.
The question of packaging preferences within segment classes could be approached in another way. Labeled plus strands were prepared and packaging was studied in vitro with transcripts that had various sizes (52). It was found that small transcripts were packaged in numbers that approximated the normal amount of RNA carried by a genomic segment of a particular class. A plus strand of segment L that was only 1,200 bases was packaged in about five copies, so that its total amount was close to the normal size of segment L, 6,400 bases.
PACKAGING MODEL
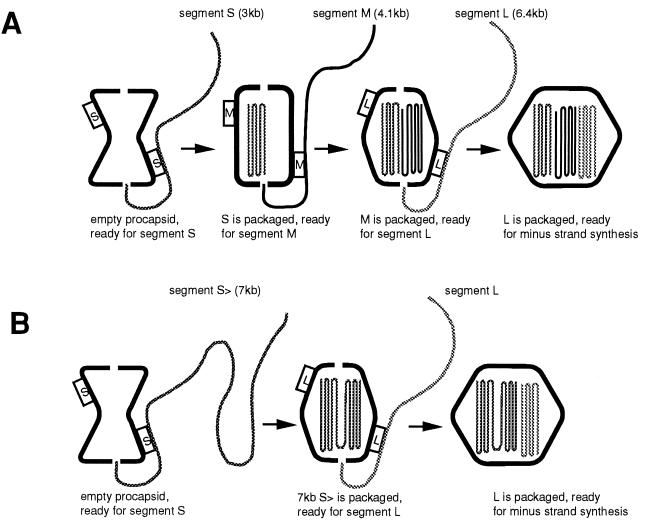
A model that rationalized the results of the packaging experiments and that made testable predictions was developed (69). The model proposes that a procapsid that has not packaged RNA has packaging and binding sites only for the plus strand of S on the outside and that there are no specific binding sites inside the procapsid. The binding sites are positioned so as to place the 5′ end of the plus strand at an entry portal. Upon completion of packaging of segment S, the particle expands and the conformation of the surface proteins changes so that the binding sites for S are lost and those for M appear (Fig. 11A). After the binding and packaging of segment M, the binding sites for M disappear and those for L appear. After the packaging of segment L, the binding sites for L disappear and the switch is activated for minus-strand synthesis. Once minus-strand synthesis is accomplished, the amount of RNA within the procapsid is doubled and the switch is activated to start transcription. Five different conformations of the procapsids are expected with this model.
FIG. 11.
(A) Packaging model (69). The procapsid shows only binding sites for segment S at the beginning. After a full-sized S is packaged, the S sites disappear and M sites appear. After a full-sized M is packaged, the M sites disappear and L sites appear. After a full-sized L is packaged, minus-strand synthesis commences. After minus-strand synthesis is completed, plus-strand synthesis commences. (B) If segment S is of a size equal to the sum of both S and M, the S sites will disappear after packaging of the 7-kb segment, the L sites will appear, and segment L will be packaged without segment M.
The model is consistent with the previous observations about packaging. It explains the packaging behavior of smaller-than-normal plus strands as well as the serial dependence of packaging. If the plus strand of segment S were half the normal size, we would expect that packaging one molecule would not be sufficient to expand the procapsid to the next stage of the packaging program. Upon packaging a second molecule, the particle would be able to lose the binding site for S and expose the site for M. Similarly, if the molecule were one-fifth the normal size, it would be necessary to package five molecules to achieve the proper extent of particle expansion before the binding sites were changed. A prediction of the model is that a molecule equal in size to the sum of segments S and M with the pac sequence of S would be packaged and would promote the packaging of segment L without the necessity for an independent segment M (Fig. 11B). This prediction was borne out (69). Another prediction was that an RNA molecule with the pac sequence of M and with a size equal to that of M and L would be packaged after segment S and would activate minus-strand synthesis even though it contained no sequence of segment L. This also was borne out by experiment. Finally, it was proposed that a single molecule the size of the entire genome of φ6, 14 kb, with the pac sequence of S could be packaged and would activate both minus- and plus-strand synthesis. This was also accomplished (69).
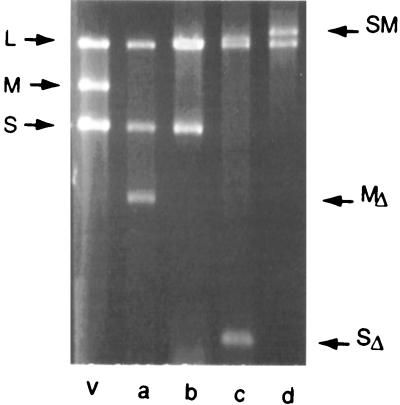
Just as the packaging experiments with smaller-than-normal plus strands were tested both in in vitro packaging and with the production of living virus, viral constructions that involved larger-than-normal segments were prepared (62). A chimeric molecule could be made that had segments S and M joined with the S portion at the 5′ end, therefore behaving as an S segment. This RNA could be acquired by virus either in vivo by transcript acquisition or in vitro by packaging of RNA into isolated procapsids. This RNA, along with a normal segment L, was able to produce a living virus that formed normal plaques and was genetically stable although it carried only two genomic segments (Fig. 12, lane d). If the chimeric molecule was constructed so that the M portion was at the 5′ end, a viable virus could be formed but it maintained a normal segment S as well (lane b). Segment S could be small enough that it carried no genes (798 bp), but the virus would not lose the third segment (lane c). This was consistent with the packaging program already described, in that segment S had to be packaged to prepare the procapsid for segment M. Additionally, it was found that a transcript of 14 kb containing all of the genes of φ6 with the pac sequence of S could form phages directly (Fig. 13). These findings support the notion that the program for the packaging and replication of the genome is not determined by an interaction of the pac sites of the genomic segments. This is of special interest because an argument has been made (20) that the packaging of segment L is the signal for the start of minus-strand synthesis and that the formation of dsRNA at the 5′ end of segment L is the signal for plus-strand synthesis. For the virus with one segment, there is only one 5′ end, and that is the pac region of segment S (62, 69). Minus- and plus-strand syntheses take place in the absence of the 5′ end of L.
FIG. 12.
dsRNA of virions with one or two genomic segments (62). Lanes: v, RNA from wild-type virus; a, RNA from a virus construct with a deletion in segment M; b, RNA from a virus containing normal segment S, normal segment L, and a chimera of M and S in which the pac sequence is that of M; c, RNA from a virus with normal L and the MS chimera and a segment S that has no genes; d, RNA from a two-segment virus in which there is normal segment L and a chimera of S and M with the pac sequence of S.
FIG. 13.
dsRNA from a virion that has the entire genome in one 14-kbp segment with the pac sequence of S. Lanes: v, RNA from wild type virus; a, RNA from the virus with the entire genome in one segment (62).
PACKAGING WITHOUT GENOMIC SEGMENT S
φ6 can establish a true carrier state in infected cells (12). In this case, the virus replicates in equilibrium with the host cell and does not kill it. The cells form colonies that release phage and are somewhat resistant to superinfection. If the phage carries a reporter gene such as kanamycin resistance, the cells exhibit resistance to the drug (58). If the phage carries a gene for β-galactosidase, the colonies are Lac+. Since the genes on segment S are not necessary for the propagation of the carrier state, it is possible to lose them without losing the carrier state. Cultures of kanamycin-resistant carrier cells will show deletions in segment S and even the complete loss of the S segment in a strain called ERA12 (58). The finding of a stable φ6 replication system that is lacking segment S would seem to contradict the model of sequential dependence in packaging. A cDNA copy of segment L of the carrier-state cells was prepared and used to direct the synthesis of the procapsid proteins in E. coli. In vitro packaging was then investigated with normal procapsids as well as those derived from ERA12 (61). It was found that (i) segment S could be packaged by the ERA12 procapsids but that the packaging was subject to competition by segment M, (ii) segments M and L could be packaged without S, and (iii) minus-strand synthesis could be obtained with just segments M and L. If S was included in the incubation, it appeared as if it was excluded. The procapsids of ERA12, therefore, were behaving as if they had already packaged segment S (Fig. 14). A mutation that changed arginine to glycine (R14G) was localized in gene 1 of segment L. This mutation was sufficient to produce the packaging behavior seen with ERA12.
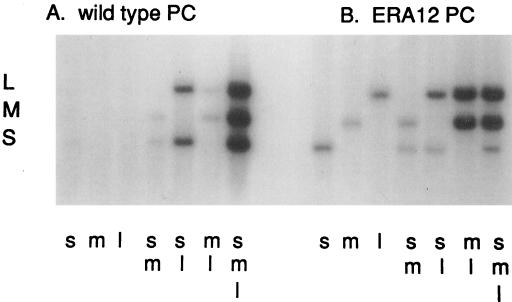
FIG. 14.
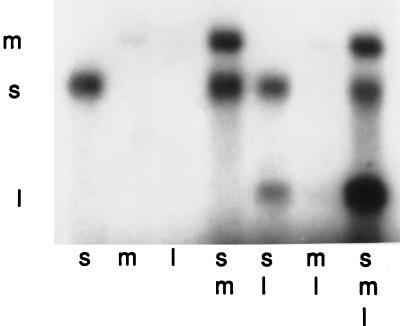
Minus-strand synthesis reactions of normal and mutant procapsids (61). Procapsids were prepared from cells producing proteins containing the mutation of ERA12 that results in a carrier-state infection where segment S is missing from the procapsids. Nonradioactive plus-strand RNA was added alone (s, m, l) or in combinations (sm, sl, ml, sml) to procapsids under conditions for minus-strand synthesis with radioactive nucleotides. Note that procapsids containing mutant P1 do not need segment S for minus-strand synthesis of segments M and L.
The findings with the mutant carrier-state virus in ERA12 and with the live-virus constructions involving chimeric segments or abnormally small segments are very important in the consideration of the packaging model. They show that the rules that obtain for in vitro packaging are also applicable to the live virus. During normal infection, the virus does not deal with segments of various sizes, and the packaging rules serve to maintain the precise stoichiometry of the genomic segments.
ROLES OF INDIVIDUAL PROTEINS IN PACKAGING
The φ6 procapsid has only four different proteins. It is possible to assemble procapsids without proteins P2 or P7 and to add these proteins back to deficient particles (5, 38). It is possible to prepare particles that are missing P4, but they are more difficult to isolate (63). Procapsids cannot be formed without P1, since this is the major structural protein of the particle. Procapsids missing P2 can still package RNA to a significant level, about 30% of normal. The stringency of the packaging program seems to be diminished somewhat in that segment M is able to package without segment S. It is not clear that P2 is involved in packaging specificity; the effect may be secondary. Procapsids that are missing P7 package RNA at about 5 to 10% of the normal level (38). It is interesting that the stringency of packaging remains high. Segment M still requires prior packaging of S (unpublished results). Packaging can be maintained in procapsids that are low in P4, but it is aberrant, and the extent of deficiency in the particles that are actually involved in packaging is not known, since the fraction of particles that are engaged is small. Particles that are completely missing P4 do not package RNA. Our working model is that protein P1 is the major determinant of packaging specificity, consistent with the mutation, but this may be in association with proteins P4 and P7. P7 does not seem to be absolutely necessary, but it plays a stimulatory role in packaging and in minus-strand and plus-strand synthesis. We assume that at least P4 is involved in the formation and function of the entry portals since the NTPase activity is necessary for packaging.
CAPSID EXPANSION IN GENOMIC PACKAGING OF DSDNA PHAGES
Bacteriophages that have genomes of dsDNA package their DNA into preformed capsid structures. The capsids usually go through an expansion that is coupled to packaging (79). It appears that the expansion occurs while the process is going on (35). The expansion of the T4 prohead involves a change in the arrangement of the proteins in the capsid and a change in the exposure of particular epitopes on the surface of the particle (79). Epitopes that are hidden in the prohead become exposed in the expanded particle, and some that are exposed at first become masked. No functional role for the change in exposure of epitopes has been proposed for the DNA phages, although changes in capsid stability often accompany maturation of virus particles. For φ6, we are proposing that the expansion of the procapsid plays an important role in determining the availability of binding sites for the particular genomic segments and ultimately for the activation of the minus-strand synthesis and plus-strand synthesis activities of the polymerase.
Another difference between genomic packaging in φ6 and dsDNA phages is that the specific DNA is recognized by an enzyme complex called terminase, which is not a permanent part of the capsid structure. It is usually a complex of two or more proteins that are responsible for recognizing the DNA, cutting it in some cases, and translocating it into the procapsid (2). The translocation is dependent upon the hydrolysis of ATP. In no case is the recognition of the DNA determined by the structural proteins of the capsid. For φ6, it appears that recognition of the viral RNA is determined by either P1 or P4 or both; these proteins are components of the procapsid structure. P4 has NTPase activity that has the same specificity as the packaging reaction. There are no serious models at present for the detailed mechanism of translocation of either DNA into proheads or RNA into the φ6 procapsid. Protein P4 of φ6, which is the likely motor for the translocation of the plus strands, has NTPase activity and seems to form hexameric rings (39). An interesting analog for the φ6 translocation system might be the RuvB protein of E. coli repair and recombination (84). The RuvB protein forms hexameric rings, has ATPase activity, and is thought to drive DNA through its structure as part of the branch migration process. DNA helicases also seem to have hexameric structures. In all of these cases, the protein is moving along the length of the nucleic acid. Although strong support has not been demonstrated, there are also models for the translocation of RNA helicases along the length of RNA (9). The suggestion has been made that the rotation of the gamma protein of F1 ATPase may have similarity to the movement of helicases in that the former has a clear demonstration of the alternating interaction of subunits in a rotary fashion (86). One can imagine a similar interaction between the nucleic acid backbone and the helicases or the packaging apparatus.
PACKAGING IN OTHER SYSTEMS WITH DSRNA GENOMES
In vitro packaging has not been accomplished yet in any of the other dsRNA systems. With the killer particles of yeast, it has been possible to achieve in vivo packaging of some constructions of the genomic segments (77, 85). It has also been possible to study the specificity of RNA binding to disrupted capsids. In this way, several stem loop structures that play important roles in packaging have been identified. A small stem-loop structure that contains a loop with the sequence GAUUC and a bulge of A in the stem was found to be necessary and sufficient for packaging in the L1/LA virus-like particle (21, 77, 85). Transcripts containing this structure, which can be as small as 34 bases, are competent for packaging or interference with normal packaging (21, 32). It seems that the plus-strand RNA is encapsidated by a process involving the nucleation of an array of subunits around the RNA and that the RNA does not interact with intact preformed empty procapsids. The capsid in the yeast virus-like particles is composed of 120 molecules of capsid protein with probably 2 molecules resulting from fusion, by frameshifting, of the capsid protein to a polymerase peptide. The polymerase portion of the protein is the determinant of specific packaging in this system. An interesting difference is seen in the packaging of small genomic segments by the yeast system as opposed to that of φ6. In φ6, a segment that is smaller than normal is packaged in multiples so as to approximate the weight of a normal segment. In the yeast particles, a single molecule is packaged and serves as template for minus-strand synthesis. The plus-strand transcripts that are subsequently produced are retained in the particle and serve as additional templates for minus-strand synthesis until the normal headful of RNA is accumulated (17). At that point, the transcripts are extruded from the particle.
For the Reoviridae, it is not clear how genomic packaging works. It has been possible to effect minus-strand synthesis in vitro with disrupted core particles of rotavirus (8, 66, 83). The reaction is specific in that sequences at the 5′ and 3′ ends of the RNA are important in determining the extent of synthesis. The most dramatic effects are seen with changes at the 3′ ends. The sequences that are found at the ends of normal rotavirus plus strands are the most effective in promoting minus-strand synthesis. The reaction is dependent upon the major core protein VP2 and the polymerase VP1. At this point, it is not possible to differentiate the sequence requirements between those needed for polymerase recognition and those that might be needed for packaging. In φ6, the sequences at the 5′ end are necessary and sufficient for packaging while those at the 3′ end are necessary for polymerase recognition. Also for φ6, minus-strand synthesis can occur in the presence of manganese ions instead of magnesium ions. In this case, packaging is not necessary for minus-strand synthesis and the requirements for synthesis become much less stringent. The sequences at the 5′ ends are of little importance in these cases, while the sequences at the 3′ ends have a significant effect on the extent of minus-strand synthesis (unpublished results).
It is tempting to propose that the model for φ6 genomic packaging would also apply to the Reoviridae; however, there is no evidence either for or against such a proposition at present. There is no competing model of any substance, either. Several of the features of φ6 packaging have not (yet) been found in rotavirus, which is the most advanced system in the Reoviridae. Specifically, an analog of the NTPase P4 has not been identified. If such an analog existed, it could be a host protein or perhaps one of the nonstructural proteins, namely, NSP2 and NSP3 (22). The inner-core particles that have been assembled for rotavirus and blue tongue virus are in an expanded form (30). If a compressed form is a packaging intermediate, it has not been seen yet (but see reference 33). Although the stoichiometry of packaging in the Reoviridae is very good, it is not known how the various systems deal with segments that are missing or present in abnormally small sizes. So far, defective interfering segments have almost always been found in populations that also contain normal-sized segments. There is one case in which a cytoplasmic polyhedrosis virus, which is a member of the Reoviridae, has deleted much of its segment 2 to form a segment that is about one-sixth its normal size yet this smaller unit exists in a single copy in the virion (75). For wound tumor virus, there are several cases in which smaller segments have been formed and appear to be packaged in single copies (70, 71). There are also cases in which wound tumor virus has completely lost particular segments when growing in plants as opposed to insects (71). The possibility that this loss requires a mutation, similar to that observed for φ6, has not been tested. With the development of complementation systems (87), it should be possible to construct viral strains with novel sizes of segments and to facilitate the determination of the packaging rules.
ACKNOWLEDGMENTS
Much of the work described in this review was supported by grants GM34352 and GM31709 from the National Institutes of Health.
Discussions with D. H. Bamford and members of his group and with F. de la Cruz, David Dubnau, and Issar Smith were of great benefit to me in thinking about this subject.
REFERENCES
- 1.Bamford D H, Romantschuk M, Somerharju P J. Membrane fusion in procaryotes: bacteriophage φ6 membrane fuses with the Pseudomonas syringae outer membrane. EMBO J. 1987;6:1467–1473. doi: 10.1002/j.1460-2075.1987.tb02388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black L W. DNA packaging in dsDNA bacteriophages. In: Calendar R, editor. The bacteriophages. Vol. 2. New York, N.Y: Plenum Publishing Corp.; 1988. pp. 321–373. [Google Scholar]
- 3.Butcher S J, Dokland T, Ojala P M, Bamford D H, Fuller S D. Intermediates in the assembly pathway of the double-stranded RNA virus φ6. EMBO J. 1997;16:4477–4487. doi: 10.1093/emboj/16.14.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caldentey J, Bamford D H. The lytic enzyme of the Pseudomonas phage φ6. Purification and biochemical characterization. Biochim Biopys Acta. 1992;1159:44–50. doi: 10.1016/0167-4838(92)90073-m. [DOI] [PubMed] [Google Scholar]
- 5.Casini G, Qiao X, Mindich L. Reconstitution of active replicase in procapsids of the segmented dsRNA bacteriophage φ6. Virology. 1994;204:251–253. doi: 10.1006/viro.1994.1529. [DOI] [PubMed] [Google Scholar]
- 6.Casini G, Revel H R. A new small low-abundant nonstructural protein encoded by the L segment of the dsRNA bacteriophage φ6. Virology. 1994;203:221–228. doi: 10.1006/viro.1994.1479. [DOI] [PubMed] [Google Scholar]
- 7.Caston J R, Trus B L, Booy F P, Wickner R B, Wall J S, Steven A C. Structure of L-A virus: a specialized compartment for the transcription and replication of double-stranded RNA. J Cell Biol. 1997;138:975–985. doi: 10.1083/jcb.138.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen D, Zeng C Q-Y, Wentz M J, Gorziglia M, Estes M K, Ramig R F. Template-dependent, in vitro replication of rotavirus RNA. J Virol. 1994;68:7030–7039. doi: 10.1128/jvi.68.11.7030-7039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho H S, Ha N-C, Kang L-W, Chung K M, Back S H, Jang S K, Oh B-H. Crystal structure of RNA helicase from genotype 1b hepatitis C virus. J Biol Chem. 1998;273:15045–15052. doi: 10.1074/jbc.273.24.15045. [DOI] [PubMed] [Google Scholar]
- 10.Coplin D L, Van Etten J L, Vidaver A K. Synthesis of bacteriophage φ6 double-stranded ribonucleic acid. J Gen Virol. 1976;33:509–512. doi: 10.1099/0022-1317-33-3-509. [DOI] [PubMed] [Google Scholar]
- 11.Cuppels D A, Van Etten J L, Lambrecht P, Vidaver A K. Survey of phytopathogenic pseudomonads for a restriction and modification system active on the double-stranded ribonucleic acid phage φ6. Curr Microbiol. 1981;5:247–249. [Google Scholar]
- 12.Cuppels D A, Vidaver A K, Van Etten J L. Resistance to bacteriophage φ6 by Pseudomonas phaseolicola. J Gen Virol. 1979;44:493–504. [Google Scholar]
- 13.Day L A, Mindich L. The molecular weight of bacteriophage φ6 and its nucleocapsid. Virology. 1980;103:376–385. doi: 10.1016/0042-6822(80)90196-8. [DOI] [PubMed] [Google Scholar]
- 13a.de Haas, F., A. O. Paatero, L. Mindich, D. H. Bamford, and S. D. Fuller. Unpublished data.
- 14.Emori Y, Iba H, Okada Y. Semi-conservative transcription of double-stranded RNA catalyzed by bacteriophage φ6 RNA polymerase. J Biochem. 1980;88:1569–1575. doi: 10.1093/oxfordjournals.jbchem.a133131. [DOI] [PubMed] [Google Scholar]
- 15.Emori Y, Iba H, Okada Y. Morphogenetic pathway of bacteriophage φ6. A flow analysis of subviral and viral particles in infected cells. J Mol Biol. 1982;154:287–310. doi: 10.1016/0022-2836(82)90065-1. [DOI] [PubMed] [Google Scholar]
- 16.Emori Y, Iba H, Okada Y. Transcriptional regulation of three double-stranded RNA segments of bacteriophage φ6 in vitro. J Virol. 1983;46:196–203. doi: 10.1128/jvi.46.1.196-203.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esteban R, Wickner R B. A deletion mutant of L-A double-stranded RNA replicates like M1 double-stranded RNA. J Virol. 1988;62:1278–1285. doi: 10.1128/jvi.62.4.1278-1285.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frilander M, Bamford D H. In vitro packaging of the single-stranded RNA genomic precursors of the segmented double-stranded RNA bacteriophage φ6: the three segments modulate each other’s packaging efficiency. J Mol Biol. 1995;246:418–428. doi: 10.1006/jmbi.1994.0096. [DOI] [PubMed] [Google Scholar]
- 19.Frilander M, Gottlieb P, Strassman J, Bamford D H, Mindich L. Dependence of minus-strand synthesis upon complete genomic packaging in the double-stranded RNA bacteriophage φ6. J Virol. 1992;66:5013–5017. doi: 10.1128/jvi.66.8.5013-5017.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frilander M, Poranen M, Bamford D H. The large genome segment of dsRNA bacteriophage φ6 is the key regulator in the in vitro minus and plus strand synthesis. RNA. 1995;1:510–518. [PMC free article] [PubMed] [Google Scholar]
- 21.Fujimura T, Esteban R, Esteban L M, Wickner R B. Portable encapsidation signal of the L-A double-stranded RNA virus of S. cerevisiae. Cell. 1990;62:819–828. doi: 10.1016/0092-8674(90)90125-x. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez R A, Torres-Vega M A, Lopez S, Arias C F. In vivo interactions among rotavirus nonstructural proteins. Arch Virol. 1998;143:981–986. doi: 10.1007/s007050050347. [DOI] [PubMed] [Google Scholar]
- 23.Gottlieb P, Metzger S, Romantschuk M, Carton J, Strassman J, Bamford D H, Kalkkinen N, Mindich L. Nucleotide sequence of the middle dsRNA segment of bacteriophage φ6: placement of the genes of membrane-associated proteins. Virology. 1988;163:183–190. doi: 10.1016/0042-6822(88)90245-0. [DOI] [PubMed] [Google Scholar]
- 24.Gottlieb P, Qiao X, Strassman J, Frilander M, Mindich L. Identification of the packaging regions within the genomic RNA segments of bacteriophage φ6. Virology. 1994;200:42–47. doi: 10.1006/viro.1994.1160. [DOI] [PubMed] [Google Scholar]
- 25.Gottlieb P, Strassman J, Bamford D H, Mindich L. Production of a polyhedral particle in Escherichia coli from a cDNA copy of the large genomic segment of bacteriophage φ6. J Virol. 1988;62:181–187. doi: 10.1128/jvi.62.1.181-187.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gottlieb P, Strassman J, Frucht A, Qiao X, Mindich L. In vitro packaging of the bacteriophage φ6 ssRNA genomic precursors. Virology. 1991;181:589–594. doi: 10.1016/0042-6822(91)90892-f. [DOI] [PubMed] [Google Scholar]
- 27.Gottlieb P, Strassman J, Mindich L. Protein P4 of the bacteriophage φ6 procapsid has a nucleoside triphosphate-binding site with associated nucleoside triphosphate phosphohydrolase activity. J Virol. 1992;66:6220–6222. doi: 10.1128/jvi.66.10.6220-6222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gottlieb P, Strassman J, Qiao X, Frilander M, Frucht A, Mindich L. In vitro packaging and replication of individual genomic segments of bacteriophage φ6 RNA. J Virol. 1992;66:2611–2616. doi: 10.1128/jvi.66.5.2611-2616.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottlieb P, Strassman J, Qiao X, Frucht A, Mindich L. In vitro replication, packaging and transcription of the segmented double-stranded RNA genome of bacteriophage φ6: studies with procapsids assembled from plasmid encoded proteins. J Bacteriol. 1990;172:5774–5782. doi: 10.1128/jb.172.10.5774-5782.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grimes J M, Burroughs J N, Gouet P, Diprose J M, Malby R, Zientara S, Mertens P P C, Stuart D I. The atomic structure of the bluetongue virus core. Nature. 1998;395:470–478. doi: 10.1038/26694. [DOI] [PubMed] [Google Scholar]
- 31.Hay A J, Joklik W K. Demonstration that the same strand of reovirus genome RNA is transcribed in vitro and in vivo. Virology. 1971;44:450–453. doi: 10.1016/0042-6822(71)90276-5. [DOI] [PubMed] [Google Scholar]
- 32.Huan B, Shen Y, Bruenn J A. In vivo mapping of a sequence required for interference with the yeast killer virus. Proc Natl Acad Sci USA. 1991;88:1271–1275. doi: 10.1073/pnas.88.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hulsmans H, van Dijk A A, Els H J. Uncoating of parental bluetongue virus to core and subcore particles in infected L cells. Virology. 1987;157:180–188. doi: 10.1016/0042-6822(87)90327-8. [DOI] [PubMed] [Google Scholar]
- 34.Iba H, Watanabe T, Emori Y, Okada Y. Three double-stranded RNA genome segments of bacteriophage φ6 have homologous terminal sequences. FEBS Lett. 1982;141:111–115. doi: 10.1016/0014-5793(82)80027-6. [DOI] [PubMed] [Google Scholar]
- 35.Jardine P J, Coombs D H. Capsid expansion follows the initiation of DNA packaging in bacteriophage T4. J Mol Biol. 1998;284:661–672. doi: 10.1006/jmbi.1998.2179. [DOI] [PubMed] [Google Scholar]
- 36.Johnson M D, III, Mindich L. Plasmid directed assembly of the lipid-containing membrane of bacteriophage φ6. J Bacteriol. 1994;176:4124–4132. doi: 10.1128/jb.176.13.4124-4132.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juuti J J, Bamford D H. RNA binding, packaging and polymerase activities of the different incomplete polymerase complex particles of dsRNA bacteriophage φ6. J Mol Biol. 1995;249:545–554. doi: 10.1006/jmbi.1995.0317. [DOI] [PubMed] [Google Scholar]
- 38.Juuti J T, Bamford D H. Protein P7 of phage φ6 RNA polymerase complex, acquiring of RNA packaging activity by in vitro assembly of the purified protein onto deficient particles. J Mol Biol. 1997;266:891–900. doi: 10.1006/jmbi.1996.0817. [DOI] [PubMed] [Google Scholar]
- 39.Juuti J T, Bamford D H, Tuma R, Thomas G J., Jr Structure and NTPase activity of the RNA-translocating protein (P4) of bacteriophage φ6. J Mol Biol. 1998;279:347–359. doi: 10.1006/jmbi.1998.1772. [DOI] [PubMed] [Google Scholar]
- 40.Ktistakis N T, Lang D. The dodecahedral framework of the bacteriophage φ6 nucleocapsid is composed of protein P1. J Virol. 1987;61:2621–2623. doi: 10.1128/jvi.61.8.2621-2623.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawton J A, Estes M K, Prasad B V. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat Struct Biol. 1997;4:118–121. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- 42.McGraw T, Mindich L, Frangione B. Nucleotide sequence of the small double-stranded RNA segment of bacteriophage φ6: Novel mechanism of natural translational control. J Virol. 1986;58:142–151. doi: 10.1128/jvi.58.1.142-151.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mindich L. Bacteriophage φ6: a unique virus having a lipid-containing membrane and a genome composed of three dsRNA segments. Adv Virus Res. 1988;35:137–176. doi: 10.1016/s0065-3527(08)60710-1. [DOI] [PubMed] [Google Scholar]
- 44.Mindich L, Abelson R D. The characterization of a 120 S particle formed during φ6 infection. Virology. 1980;103:386–391. doi: 10.1016/0042-6822(80)90197-x. [DOI] [PubMed] [Google Scholar]
- 45.Mindich L, Lehman J. Cell wall lysine as a component of the bacteriophage φ6 virion. J Virol. 1979;30:489–496. doi: 10.1128/jvi.30.2.489-496.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mindich L, Lehman J. Characterization of φ6 mutants that are temperature sensitive in the morphogenetic protein P12. Virology. 1983;127:438–445. doi: 10.1016/0042-6822(83)90156-3. [DOI] [PubMed] [Google Scholar]
- 47.Mindich L, Lehman J, Sinclair J F. Biogenesis of φ6, a bacteriophage that contains lipid. In: Schlessinger D, editor. Microbiology—1979. Washington, D.C: American Society for Microbiology; 1979. pp. 38–41. [Google Scholar]
- 48.Mindich L, MacKenzie G, Strassman J, McGraw T, Metzger S, Romantschuk M, Bamford D. cDNA cloning of portions of the bacteriophage φ6 genome. J Bacteriol. 1985;162:992–999. doi: 10.1128/jb.162.3.992-999.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mindich L, Nemhauser I, Gottlieb P, Romantschuk M, Carton J, Frucht S, Strassman J, Bamford D H, Kalkkinen N. Nucleotide sequence of the large double-stranded RNA segment of bacteriophage φ6: the genes specifying the viral replicase and transcriptase. J Virol. 1988;62:1180–1185. doi: 10.1128/jvi.62.4.1180-1185.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mindich L, Qiao X, Onodera S, Gottlieb P, Frilander M. RNA structural requirements for stability and minus strand synthesis in the dsRNA bacteriophage φ6. Virology. 1994;202:258–263. doi: 10.1006/viro.1994.1341. [DOI] [PubMed] [Google Scholar]
- 51.Mindich L, Qiao X, Onodera S, Gottlieb P, Strassman J. Heterologous recombination in the dsRNA bacteriophage φ6. J Virol. 1992;66:2605–2610. doi: 10.1128/jvi.66.5.2605-2610.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mindich L, Qiao X, Qiao J. Packaging of multiple copies of small-sized genomic segments by bacteriophage φ6. Virology. 1995;212:213–217. doi: 10.1006/viro.1995.1470. [DOI] [PubMed] [Google Scholar]
- 53.Mindich L, Sinclair J F, Cohen J. The morphogenesis of bacteriophage φ6: particles formed by nonsense mutants. Virology. 1976;75:224–231. doi: 10.1016/0042-6822(76)90021-0. [DOI] [PubMed] [Google Scholar]
- 54.Ojala P M, Romantschuk M, Bamford D H. Purified φ6 nucleocapsids are capable of productive infection of host cells with partially disrupted outer membrane. Virology. 1990;178:364–372. doi: 10.1016/0042-6822(90)90333-m. [DOI] [PubMed] [Google Scholar]
- 55.Olkkonen V M, Bamford D H. The nucleocapsid of the lipid-containing double-stranded RNA bacteriophage φ6 contains a protein skeleton consisting of a single polypeptide species. J Virol. 1987;61:2362–2367. doi: 10.1128/jvi.61.8.2362-2367.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olkkonen V M, Gottlieb P, Strassman J, Qiao X, Bamford D H, Mindich L. In vitro assembly of infectious nucleocapsids of bacteriophage φ6: formation of a recombinant double-stranded RNA virus. Proc Natl Acad Sci USA. 1990;87:9173–9177. doi: 10.1073/pnas.87.23.9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olkkonen V M, Ojala P, Bamford D H. Generation of infectious nucleocapsids by in vitro assembly of the shell protein onto the polymerase complex of the dsRNA bacteriophage φ6. J Mol Biol. 1991;218:569–581. doi: 10.1016/0022-2836(91)90702-8. [DOI] [PubMed] [Google Scholar]
- 58.Onodera S, Olkkonen V M, Gottlieb P, Strassman J, Qiao X, Bamford D H, Mindich L. Construction of a transducing virus from double-stranded RNA bacteriophage φ6: establishment of carrier states in host cells. J Virol. 1992;66:190–196. doi: 10.1128/jvi.66.1.190-196.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Onodera S, Qiao X, Gottlieb P, Strassman J, Frilander M, Mindich L. RNA structure and heterologous recombination in the double-stranded RNA bacteriophage φ6. J Virol. 1993;67:4914–4922. doi: 10.1128/jvi.67.8.4914-4922.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Onodera S, Qiao X, Qiao J, Mindich L. Acquisition of a fourth genomic segment in bacteriophage φ6: a bacteriophage with a genome of three segments of dsRNA. Virology. 1995;212:204–212. doi: 10.1006/viro.1995.1469. [DOI] [PubMed] [Google Scholar]
- 61.Onodera S, Qiao X, Qiao J, Mindich L. Isolation of a mutant that changes genomic packaging specificity in φ6. Virology. 1998;252:438–442. doi: 10.1006/viro.1998.9479. [DOI] [PubMed] [Google Scholar]
- 62.Onodera S, Qiao X, Qiao J, Mindich L. Directed changes in the number of dsRNA genomic segments in bacteriophage φ6. Proc Natl Acad Sci USA. 1998;95:3920–3924. doi: 10.1073/pnas.95.7.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paatero A, Mindich L, Bamford D H. Mutational analysis of the role of the NTPase P4 in the assembly of the RNA polymerase complex of bacteriophage φ6. J Virol. 1998;72:10058–10065. doi: 10.1128/jvi.72.12.10058-10065.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Paatero A O, Syvaoja J E, Bamford D H. Double-stranded RNA bacteriophage φ6 protein P4 is an unspecific nucleoside triphosphatase activated by calcium ions. J Virol. 1995;69:6729–6734. doi: 10.1128/jvi.69.11.6729-6734.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Partridge J E, Van Etten J L, Burbank D E, Vidaver A K. RNA polymerase activity associated with bacteriophage φ6 nucleocapsid. J Gen Virol. 1979;43:299–307. [Google Scholar]
- 66.Patton J T, Wentz M, Xiaobo J, Ramig R F. cis-acting signals that promote genome replication in rotavirus mRNAs. J Virol. 1996;70:3961–3971. doi: 10.1128/jvi.70.6.3961-3971.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qiao X, Casini G, Qiao J, Mindich L. In vitro packaging of individual genomic segments of bacteriophage φ6 RNA: serial dependence relationships. J Virol. 1995;69:2926–2931. doi: 10.1128/jvi.69.5.2926-2931.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qiao X, Qiao J, Mindich L. Interference of bacteriophage φ6 genomic RNA packaging by hairpin structures. J Virol. 1995;69:5502–5505. doi: 10.1128/jvi.69.9.5502-5505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qiao X, Qiao J, Mindich L. Stoichiometric packaging of the three genomic segments of dsRNA bacteriophage φ6. Proc Natl Acad Sci USA. 1997;94:4074–4079. doi: 10.1073/pnas.94.8.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reddy D V R, Black L M. Deletion mutations of the genome segments of wound tumor virus. Virology. 1974;61:458–473. doi: 10.1016/0042-6822(74)90282-7. [DOI] [PubMed] [Google Scholar]
- 71.Reddy D V R, Black L M. Isolation and replication of mutant populations of wound tumor virions lacking certain genome segments. Virology. 1977;80:336–346. doi: 10.1016/s0042-6822(77)80009-3. [DOI] [PubMed] [Google Scholar]
- 72.Revel H R, Ewen M E, Busslan J, Pagratis N. Generation of cDNA clones of the bacteriophage φ6 segmented dsRNA genome: characterization and expression of L segment clones. Virology. 1986;155:402–417. doi: 10.1016/0042-6822(86)90203-5. [DOI] [PubMed] [Google Scholar]
- 73.Roine E, Nunn D N, Paulin L, Romantschuk M. Characterization of genes required for pilus expression in Pseudomonas syringae pathovar phaseolicola. J Bacteriol. 1996;178:410–417. doi: 10.1128/jb.178.2.410-417.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Romantschuk M, Bamford D H. Function of pili in bacteriophage φ6 penetration. J Gen Virol. 1985;66:2461–2469. doi: 10.1099/0022-1317-66-11-2461. [DOI] [PubMed] [Google Scholar]
- 75.Rubinstein R, Harley E H. Reproducible alteration of cytoplasmic polyhedrosis virus double-stranded RNA genome patterns on laboratory passage. Virology. 1978;84:195–198. doi: 10.1016/0042-6822(78)90232-5. [DOI] [PubMed] [Google Scholar]
- 76.Semancik J S, Vidaver A K, Van Etten J L. Characterization of a segmented double-helical RNA from bacteriophage φ6. J Mol Biol. 1973;78:617–625. doi: 10.1016/0022-2836(73)90283-0. [DOI] [PubMed] [Google Scholar]
- 77.Shen Y, Bruenn J A. RNA structural requirements for RNA binding, replication, and packaging in the yeast double-stranded virus. Virology. 1993;195:481–491. doi: 10.1006/viro.1993.1399. [DOI] [PubMed] [Google Scholar]
- 78.Sinclair J F, Tzagoloff A, Levine D, Mindich L. Proteins of bacteriophage φ6. J Virol. 1975;16:685–695. doi: 10.1128/jvi.16.3.685-695.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Steven A C, Bauer A C, Bisher M E, Robey F A, Black L W. The maturation-dependent conformational change of phage T4 capsid involves the translocation of specific epitopes between the inner and the outer capsid surfaces. J Struct Biol. 1991;106:221–236. doi: 10.1016/1047-8477(91)90072-5. [DOI] [PubMed] [Google Scholar]
- 80.Tyler K L, Fields B N. Reoviridae: a brief introduction. In: Fields B N, Knipe D M, editors. Virology. New York, N.Y: Raven Press; 1990. pp. 1271–1273. [Google Scholar]
- 81.Usala S J, Brownstein B H, Haselkorn R. Displacement of parental RNA strands during in vitro transcription by bacteriophage φ6 nucleocapsids. Cell. 1980;19:855–862. doi: 10.1016/0092-8674(80)90076-8. [DOI] [PubMed] [Google Scholar]
- 82.Vidaver A K, Koski R K, Van Etten J L. Bacteriophage φ6: a lipid-containing virus of Pseudomonas phaseolicola. J Virol. 1973;11:799–805. doi: 10.1128/jvi.11.5.799-805.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wentz M J, Patton J T, Ramig R F. The 3′-terminal consensus sequence of rotavirus mRNA is the minimal promoter of negative-strand RNA synthesis. J Virol. 1996;70:7833–7841. doi: 10.1128/jvi.70.11.7833-7841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.West S C. Processing of recombination intermediates by the RuvABC proteins. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 85.Wickner R B. Double-stranded RNA virus replication and packaging. J Biol Chem. 1993;268:3797–3800. [PubMed] [Google Scholar]
- 86.Zhou Y, Duncan T M, Cross R L. Subunit rotation in Escherichia coli FoF1-ATP synthase during oxidative phosphorylation. Proc Natl Acad Sci USA. 1997;94:10583–10587. doi: 10.1073/pnas.94.20.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zou S, Brown E G. Stable expression of the reovirus mu2 protein in mouse L cells complements the growth of a reovirus ts mutant with a defect in its M1 gene. Virology. 1996;217:42–48. doi: 10.1006/viro.1996.0091. [DOI] [PubMed] [Google Scholar]
- 88.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]