Abstract
The development of multicomponent crystal forms, such as cocrystals, represents a means to enhance the dissolution and absorption properties of poorly water-soluble drug compounds. However, the successful discovery of new pharmaceutical cocrystals remains a time- and resource-consuming process. This study proposes the use of a combined computational-experimental high-throughput approach as a tool to accelerate and improve the efficiency of cocrystal screening exemplified by posaconazole. First, we employed the COSMOquick software to preselect and rank cocrystal candidates (coformers). Second, high-throughput crystallization experiments (HTCS) were conducted on the selected coformers. The HTCS results were successfully reproduced by liquid-assisted grinding and reaction crystallization, ultimately leading to the synthesis of thirteen new posaconazole cocrystals (7 anhydrous, 5 hydrates, and 1 solvate). The posaconazole cocrystals were characterized by PXRD, 1H NMR, Fourier transform-Raman, thermogravimetry–Fourier transform infrared spectroscopy, and differential scanning calorimetry. In addition, the prediction performance of COSMOquick was compared to that of two alternative knowledge-based methods: molecular complementarity (MC) and hydrogen bond propensity (HBP). Although HBP does not perform better than random guessing for this case study, both MC and COSMOquick show good discriminatory ability, suggesting their use as a potential virtual tool to improve cocrystal screening.
Short abstract
We report the use of a combined cocrystal screening approach, employing computational tools and high-throughput experiments, in the discovery of new cocrystals of posaconazole.
Introduction
The molecular solid state strongly affects the physicochemical properties of organic compounds.1 To achieve solubility, dissolution rate, and absorption advantages, new solid forms of active pharmaceutical ingredients (APIs) are continuously developed, such as amorphous or coamorphous phases, polymorphs, and salts.2,3 In this regard, cocrystals have received particular attention because they are potentially very useful in overcoming several drug limitations (e.g., solubility, dissolution,4 stability,5 and hygroscopicity6).
Cocrystals are single-phase crystalline materials composed of two or more components, both of which are solid under ambient conditions. Typically, the components are present in a well-defined stoichiometric ratio and are held together by nonionic intermolecular interactions.7 This work refers to pharmaceutical cocrystals of an API and one or more cocrystal formers (coformers), chosen from the generally regarded as safe (GRAS)8 or everything added to food lists.9 In contrast to salts, cocrystal formation does not require the presence of acidic or basic functionalities with sufficient pKa differences between the interacting molecules. Intermolecular interactions such as hydrogen bonding, van der Waals forces, and π–π stacking are more dominant than proton transfer because the latter is incomplete or nonexistent.10
Searching for new cocrystals of a given API is a resource- and time-consuming process because hundreds of coformers may be tested experimentally by several methods.11 Hence, there is a need to streamline the approach to finding suitable cocrystals. A potential solution to the problem is represented by the efficient preselection of coformers that are expected to show a higher likelihood of cocrystal formation with the target molecule. For a long time, the supramolecular synthon approach12 has been employed for this purpose, but nowadays new computational tools are accessible.13
Sun et al. proposed a rough division of these virtual tools into three classes:14 (1) knowledge-based, (2) physics-based, and (3) machine learning approaches (ML). Knowledge-based methods15 rely on cocrystal structure information contained in the Cambridge Structural Database (CSD)16 to select coformers according to their shape [molecular complementarity (MC)],17,18 or probability of formation of intermolecular interactions [hydrogen bond propensity (HBP)].19 Physics-based methods describe the compound on a quantum chemical level and include the conductor-like screening model for real solvents (COSMO-RS),20,21 molecular electrostatic potential surfaces (MEPS),22,23 and crystal structure prediction (CSP).14,24 ML methods, such as artificial neural networks,25,26 employ large cocrystal databases to train predictive models for the ranking of promising coformers.27,28 More recently, several studies have reported on the combined use of multiple virtual approaches in the coformer selection for APIs.29−31
Despite the diverse methods proposed, most of them account only for the miscibility between the target molecule and the coformer, neglecting the contributions of the crystal structure.14 CSP is able to evaluate both terms but at the expense of power-demanding quantum chemical calculations that can last for days.24 Therefore, the choice of suitable virtual tools is a trade-off between accuracy and computational demands.
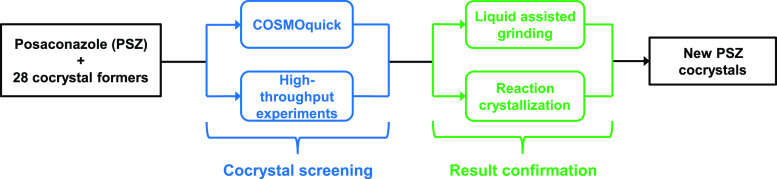
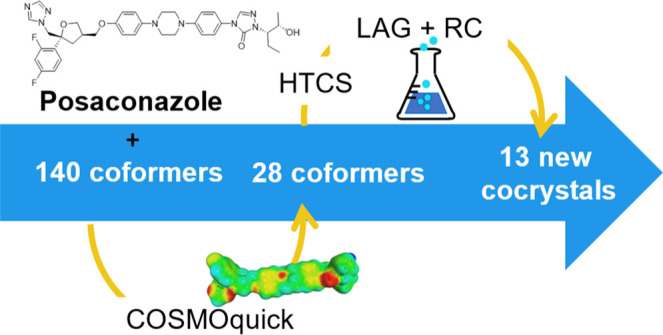
This paper aims to show how the combination of rapid computational tools and high-throughput experiments (Figure 1) can accelerate and improve the efficiency of the cocrystal screening process for a poorly water-soluble drug, posaconazole (Figure 2).
Figure 1.
Flowchart of the posaconazole cocrystal screening approach employed in the study.
Figure 2.
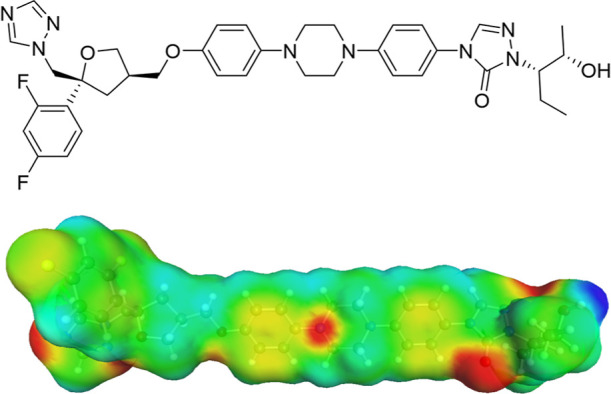
Top: Posaconazole molecular structure. Bottom: Posaconazole σ-surface reported from COSMOquick; areas with positive polarization charge density are highlighted in red (H-bond acceptor), while areas with negative polarization charge density area are highlighted in blue (H-bond donor).
Posaconazole (PSZ) is a triazole-based, wide-range antifungal drug.32 Posaconazole Form I33 appears to be the most stable polymorphic form and shows a low aqueous solubility of <1 μg/mL. The combination of low solubility with the high lipophilicity of log P 4.6 places PSZ in class II of the biopharmaceutical classification system (BCS).34,35 Although the functional groups of PSZ (pKa 3.6 and 4.6, piperazine and triazole respectively34) do not suggest salt formation with weak acids, they can still establish hydrogen bonds. As shown in Figure 2, PSZ interaction sites are represented by the triazole and triazolone rings, which possess both hydrogen bond donors (N=C–H) and acceptors (N and C=O), and by the amine and hydroxyl groups. To the best of the authors’ knowledge, one cocrystal of posaconazole with 4-aminobenzoic acid4 and two cocrystals with amino acids (l-glutamine and l-asparagine36), which displayed supersaturation and higher solubility compared to the free base, are reported in the literature.
The computational part of this work employed the COSMOquick software20 as a tool to preselect 28 coformers from a list of 140 compounds, according to their likelihood of cocrystal formation with posaconazole (Table 1). The classification is based on the COSMO-RS theory,21,37 which uses quantum mechanics and statistical thermodynamics to predict the miscibility of a drug molecule and a coformer in a supercooled liquid phase.20 The COSMOquick software avoids expensive quantum chemical calculation by using a database of precomputed polarization charge density surfaces (σ-surfaces).38 The experimental part involved the use of high-throughput experiments in order to test the preselected coformers rapidly and in parallel. This combined approach was able to discover new posaconazole cocrystals, which were reproduced by liquid-assisted grinding (LAG) and reaction crystallization (RC)39,40 and characterized through spectroscopic and thermal analysis. The study also provides a prospective head-to-head comparison of computations: the predictions of COSMOquick, MC,17 and HBP15,19 were directly compared to assess the performance of the methods.
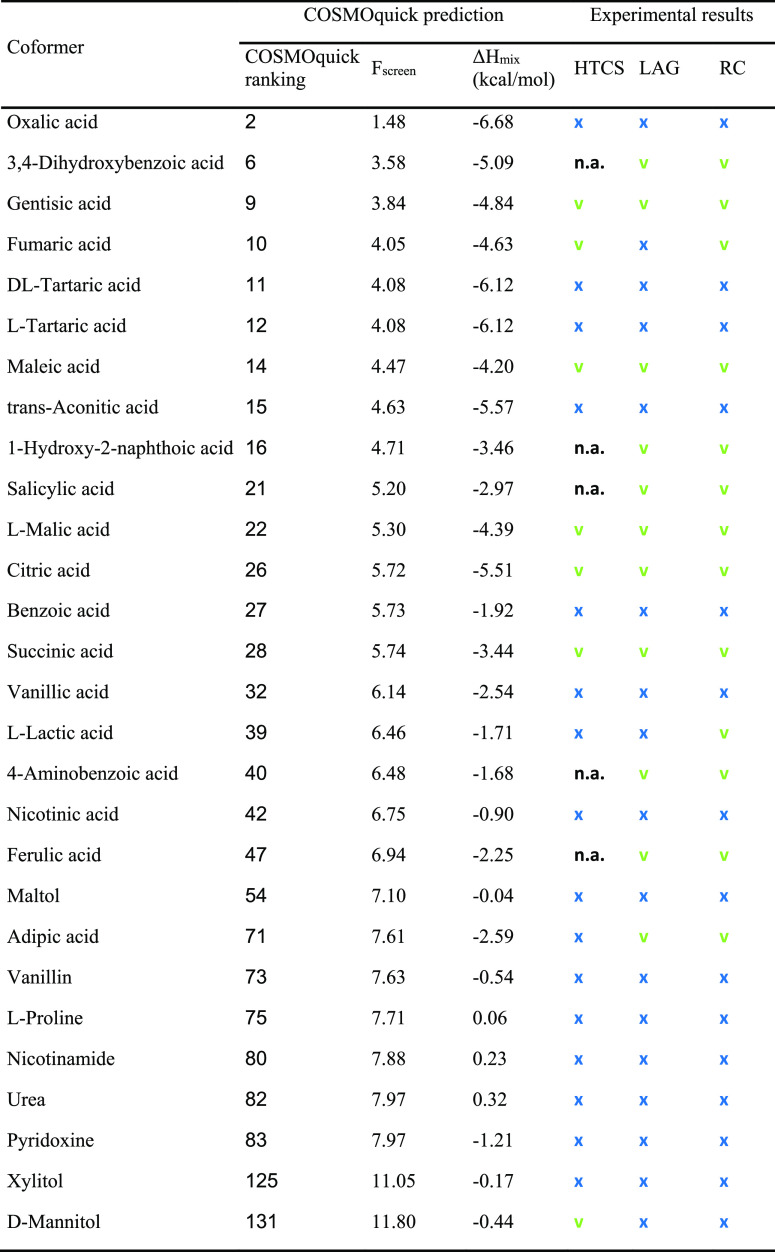
Table 1. COSMOquick Coformer Ranking Based on Penalty Function (Fscreen) Values, Calculated Excess Enthalpy (ΔHmix), and Experimental Results Obtained Using Different Methods: HTCS, LAG, and RCa.
Positive cocrystallization results are reported in green (v), while negative results are reported in blue (x).
Materials and Methods
Materials
Posaconazole (PSZ) was purchased from BOC Sciences (Shirley, NY) and used without further purification. Powder X-ray diffraction (PXRD) of the purchased PSZ showed that the sample was mainly amorphous with the presence of low-intensity peaks that are attributable to posaconazole Form I. The chemical identity of PSZ was confirmed by proton nuclear magnetic resonance (1H NMR) as shown in the Supporting Information (Figure S5). Coformers were purchased from Merck & Cie, (Schaffhausen, Switzerland) and used as received. A list of all the coformers used in the study is reported in the Supporting Information (Table S2).
Methods
Coformer Selection
A virtual cocrystal screening of posaconazole with a set of 140 coformers containing different functional groups (carboxylates, amides, and hydroxyl groups) was performed using COSMOquick (Supporting Information Table S1). The COSMOquick software is based on COSMO-RS theory,37 a statistical thermodynamic theory employed to predict equilibrium properties of molecules mostly in liquid systems. The approach employs a molecular polarization-charge surface, the σ-surface, to describe molecules embedded in a dielectric conductor. The normal COSMO-RS method employs time-consuming quantum chemical calculations for the σ-surface determination, whereas COSMOquick speeds up the calculation by combining surface fragments from a database of pre-computed molecules. The intermolecular interactions of the system (hydrogen bonding, van der Waals, electrostatic) are evaluated by statistical thermodynamic calculations of pairwise interactions of the σ-surface segments.41,42 The molecular interactions are employed to determine the chemical potential and other physicochemical properties of the molecule. Since COSMO-RS is a liquid-phase thermodynamic theory, it is assumed that the interactions in a crystal are similar to those of a supercooled liquid. The excess, or mixing, enthalpy (ΔHmix) of an API–coformer pair with given stoichiometry represents the strength of the interaction in the supercooled cocrystals. The mixing enthalpy is a rough approximation of the free energy of cocrystal formation (ΔGcc), as represented in eq 1.
| 1 |
In fact, the mixing entropy, ΔSmix, and the difference between the energy of fusion of cocrystal and reactants, ΔΔGfus, are assumed to be negligible: the former because both the reactants and the cocrystal are solid; the latter because the ΔΔGfus of reactants and cocrystal cancel each other.21
Finally, the tendency of an API–coformer pair to form a cocrystal is evaluated by the screening function Fscreen. This function accounts not only for the molecular interactions (ΔHmix) but also for the flexibility of the molecule in the form of the number of rotatable bonds. The screening function is reported in eq 2, where a is a fit parameter to be determined on a set of experimental results, and n is the total number of rotatable bonds of the drug and the coformer. The screening function, Fscreen, was introduced to consider floppy and highly flexible molecules, as it was observed experimentally that they penalize cocrystal formation.20,21
| 2 |
The virtual cocrystal screening of posaconazole was performed using COSMOquick software v.2020 (Dassault Systèmes Germany GmbH Biovia, Leverkusen, Germany) on the 140 compounds by selecting a 1:1 PSZ–coformer stoichiometry. A list of simplified molecular input line entries (SMILEs) of PSZ and the coformers was employed as input for the calculation. Approximate σ-profiles were generated by COSMOfrag, while the statistical thermodynamic calculations were carried out by COSMOtherm code. The output resulted in a list of coformers ranked according to the values of the penalty function (Fscreen) or excess enthalpy (ΔHmix) of the corresponding cocrystals with posaconazole. A set of 23 coformers was selected and analyzed by high-throughput cocrystal screening (HTCS). Then, five more compounds were added to the selection, and all 28 coformers were additionally tested by LAG and RC experiments. The candidates were selected in order to span the range of screening function values, with a preference for top-ranked compounds (Supporting Information Table S2). The selection of the coformer subset, which contains both top- and low-ranked compounds, allows for the testing of the predictive ability of the computational model.
High-Throughput Screening
The ability of 23 coformers to generate cocrystals with PSZ was experimentally tested by HTCS in a 96-well quartz microtiter plate. The screening experiments were performed in quadruplicate on samples of approximately 4 mg in total; each trial comprised one control well (free drug) and 23 test wells (one for each PSZ–coformer pair). The designed layout of the 96-well microtiter plate is available in the Supporting Information (Figure S1). The high-throughput method involved a slurry equilibration step using four different solvents, subsequently followed by an evaporation step; the final solid residues were analyzed by Raman microscopy (a detailed description of the procedure is available in Solvias’ patent US7892354B243). The slurry tests showed the presence of new features in the Raman spectra of some of the tested wells, potentially related to the presence of new solid forms.
LAG and RC Experiments
In order to verify the HTCS results, each of the selected coformer was also subjected to at least one LAG and one RC experiment.39 Additionally, five more coformers were selected and tested by the methods.
Liquid Assisted Grinding
Amounts of PSZ and coformer (approximately 100 mg in total) were placed into a 3 mL agate milling jar together with a small amount of solvent (usually acetonitrile) and two agate milling balls of 5 mm in diameter. If not stated otherwise, the mixture was agitated in an MM200 Retsch ball mill at the operating frequency of 30 Hz for 30 min. The resultant solid products were tested by Fourier transform (FT)-Raman spectroscopy.
Reaction Crystallization
The experiments were typically carried out in acetonitrile. PSZ (approximately 100 mg) was added to an almost saturated coformer solution and magnetically stirred for 2 days at RT.39,40 The suspension was filtered through a 0.2 μm PTFE filter, and the solid phase was characterized by FT-Raman spectroscopy and PXRD.
Posaconazole–4-Aminobenzoic Acid Cocrystal 2:3 (PSZ–ABA)
As a reference, the PSZ–ABA cocrystal was synthesized by the RC method following a similar procedure to the one reported by Kuminek et al.4 PSZ (100 mg, 0.14 mmol) was added to an almost saturated solution (1.5 mL) of ABA (60 mg, 0.44 mmol) in acetonitrile and magnetically stirred at RT for 2 days. A white powder was isolated after filtration. The stoichiometry was confirmed by NMR.
Posaconazole–l-Malic Acid Cocrystal 2:1 (PSZ–MLA)
PSZ–MLA cocrystal was obtained by the RC method. PSZ (100 mg, 0.14 mmol) was added to an equimolar solution (1.5 mL) of MLA (18.8 mg, 0.14 mmol) in acetonitrile and magnetically stirred at RT for 2 days. A white powder was isolated after filtration.
Posaconazole–Succinic Acid Cocrystal Hydrate 2:1:1 (PSZ–SUC–H2O)
PSZ–SUC–H2O cocrystal was synthesized by the RC method. PSZ (100 mg, 0.14 mmol) was added to an almost saturated solution (4 mL) of SUC (16.9 mg, 0.14 mmol) in acetonitrile and magnetically stirred at RT for 2 days. A white powder was isolated after filtration.
Posaconazole–Fumaric Acid Cocrystal THF Solvate 1:1:1 (PSZ–FUM–THF)
PSZ–FUM–THF cocrystal was obtained by the RC method. PSZ (100 mg, 0.14 mmol) was added to an almost saturated solution (0.5 mL) of FUM (16.6 mg, 0.14 mmol) in THF and magnetically stirred at RT for 2 days. A yellow powder was isolated after filtration.
Posaconazole–Fumaric Acid Cocrystal Hydrate 1:1:0.5 (PSZ–FUM–H2O)
PSZ–FUM–H2O cocrystal was obtained by the RC method. PSZ (100 mg, 0.14 mmol) was added to an equimolar solution (4 mL) of FUM (16.6 mg, 0.14 mmol) in acetonitrile and magnetically stirred at RT for 2 days. A yellow powder was isolated after filtration.
Posaconazole–Ferulic Acid Cocrystal Hydrate 3:1:1 (PSZ–FER–H2O)
PSZ–FER–H2O cocrystal was synthesized by the RC method. PSZ (80 mg, 0.11 mmol) was added to an almost saturated solution (2 mL) of FER (22 mg, 0.11 mmol) in acetonitrile and magnetically stirred at RT for 2 days. A white powder was isolated after filtration.
Posaconazole–Maleic Acid Cocrystal 1:1 (PSZ–MLE)
PSZ–MLE cocrystal was synthesized by the RC method. PSZ (100 mg, 0.14 mmol) was added to an equimolar solution (2 mL) of MLE (16.6 mg, 0.14 mmol) in acetonitrile and magnetically stirred at RT for 2 days. A white powder was isolated after filtration.
Posaconazole–Citric Acid Cocrystal 4:1 (PSZ–CIT)
PSZ–CIT cocrystal was synthesized by the RC method. PSZ (100 mg, 0.14 mmol) was added to an equimolar solution (3 mL) of CIT (27.5 mg, 0.14 mmol) in acetonitrile and magnetically stirred at RT for 2 days. A white powder was isolated after filtration.
Posaconazole–1-Hydroxy-2-Naphthoic Acid Cocrystal 1:1 (PSZ–XIN)
PSZ–XIN cocrystal was synthesized by the RC method. PSZ (100 mg, 0.14 mmol) was added to a saturated solution (3 mL) of XIN (26.9 mg, 0.14 mmol) in acetonitrile and magnetically stirred at RT for 2 days. A white powder was isolated after filtration.
Posaconazole–Gentisic Acid Cocrystal 1:1 (PSZ–GEN)
PSZ–GEN cocrystal was obtained by adding PSZ (100 mg, 0.14 mmol) to a solution (2 mL) of GEN (44 mg, 0.29 mmol) in acetonitrile and magnetically stirred at RT for 2 days. A white powder was isolated after filtration.
Posaconazole–Salicylic Acid Cocrystal 1:1 (PSZ–SAL)
PSZ–SAL cocrystal was synthesized by the RC method. PSZ (100 mg, 0.14 mmol) was added to an almost saturated solution (1 mL) of SAL (60 mg, 0.43 mmol) in acetonitrile and magnetically stirred at RT for 2 days. A white powder was obtained after filtration.
Posaconazole–l-Lactic Acid Cocrystal 1:1 (PSZ–LLA)
PSZ–LLA cocrystal was synthesized by the RC method. PSZ (100 mg, 0.14 mmol) was added to an almost saturated solution (1.5 mL) of LLA (38 mg, 0.42 mmol) in ethyl acetate and magnetically stirred at RT for 2 days. A white powder was obtained after filtration.
Posaconazole–Adipic Acid Cocrystal Hydrate 2:1:1 (PSZ–ADI–H2O)
PSZ–ADI–H2O cocrystal was synthetized by the RC method. PSZ (90 mg, 0.13 mmol) was added to an almost saturated solution (4 mL) of ADI (20 mg, 0.14 mmol) in acetonitrile and magnetically stirred at RT for 2 days. A white powder was obtained after filtration.
Posaconazole–3,4-Dihydroxybenzoic Acid Cocrystal Hydrate 3:4:2 (PSZ–DHB–H2O)
PSZ–DHB–H2O was synthetized in an attempt of growing large crystals of the compound. PSZ (100 mg, 0.14 mmol) was added to an almost saturated solution (1 mL) of DHB (55 mg, 0.36 mmol) in acetonitrile and magnetically stirred for 2 days at RT. After filtration, the isolated white powder was dissolved in 4 mL of acetonitrile and the system was evaporated at RT. Transparent crystals appeared after two weeks.
Raman Microscopy
High-throughput experiments were analyzed using a Renishaw InVia Reflex Raman microscope (Renishaw, Wotton-under-Edge, United Kingdom) equipped with a 785 nm diode laser for excitation and a NIR-enhanced Peltier-cooled CCD camera as the detector. Measurements were carried out with a long working distance of 20× objective on a measurement range of 100–2000 cm–1.
FT-Raman Spectroscopy
FT-Raman spectra were recorded on a Bruker FT-Raman spectrometer MultiRAM (Bruker AG, Fällanden, Switzerland) with a near-infrared Nd:YAG laser operating at 1064 nm and a liquid-nitrogen-cooled germanium detector. A total number of 64 scans with a resolution of 2 cm–1 was accumulated in the range from 50 to 3500 cm–1 using a laser power of 100 mW. The samples were prepared by pressing the isolated powder into an aluminum sample holder.
Powder X-ray Diffraction
Diffractograms were collected using a Stoe Stadi P diffractometer (Stoe & Cie. GmbH, Darmstadt, Deutschland) equipped with a Mythen1K detector operating with Cu Kα1 radiation. The measurements were performed in transmission at a tube voltage of 40 kV and 40 mA tube power. A step size of 0.02° 2θ and a step time of 12 s over a 1.5–50.5° 2θ scanning range was applied. For a typical sample preparation about 10–20 mg of the sample was placed between two acetate foils and mounted into a Stoe transmission sample holder. The sample was rotated during the measurement. All sample preparation and measurement were done at RT in an ambient air atmosphere.
Thermogravimetry Coupled with Fourier Transform Infrared Spectroscopy
Thermogravimetry coupled with Fourier transform infrared spectroscopy (TG–FTIR) was performed on a Netzsch Thermo-Microbalance TG 209 (Netzsch, Selb, Germany), which is coupled with a Bruker FT-IR Spectrometer Vector 22. The aluminum crucibles used had a (micro) pinhole, and the measurements were carried out under a nitrogen atmosphere and at a heating rate of 10 °C/min over the range 25–250 °C.
Proton Nuclear Magnetic Resonance (1H NMR)
1H NMR analysis was carried out with a Bruker DPX300 spectrometer (Bruker AG, Fällanden, Switzerland) using a proton frequency of 300.13 MHz, a 30° excitation pulse, and a recycle delay of 1 s. Spectra were recorded by the accumulation of 16 scans in deuterated DMSO. The solvent peak was used for reference, and the chemical shifts were reported to the TMS scale.
Differential Scanning Calorimetry
Differential scanning calorimetry (DSC) analyses were carried out with a TA Instruments Q2000 or with a TA Instruments DSC 2500 (TA Instruments, New Castle, Delaware, USA). Approximately 2–3 mg of sample was heated at a rate of 10 or 20 K/min under nitrogen gas flow (50 mL/min). Standard aluminum sample pans that were either closed or perforated with a pinhole were used for the measurements.
Computational Cocrystal Screening
Molecular Complementarity
The method was developed by L. Fábián performing a statistical analysis on the cocrystals contained in the CSD.16 The investigation pointed out that molecules that cocrystallize tend to have similar or complementary molecular properties.17 The similarity is evaluated by comparing molecular descriptors. Two of them are related to the polarity of the molecule: the fraction of nitrogen and oxygen atoms (FNO) and the dipole moment. The remaining three are related to the shape and are defined considering the short (S), medium (M), and long (L) axes of a box enclosing the van der Waals molecular volume: S axis, M/L axis ratio, and S/L axis ratio. According to the cocrystal structures contained in the CSD, Fábián defined threshold values for the five molecular descriptors; any coformer with descriptor values that differ from the API beyond the thresholds would be unlikely to cocrystallize.17 The analysis is usually implemented as a PASS/FAIL test; it predicts cocrystal formation if, and only if, all five descriptors are within the cut-off values. A percentage hit rate is computed for each coformer and is averaged over the number of API conformations screened. The larger the hit rate, the higher the probability of cocrystal formation. Alternatively, coformers can be ranked by calculating a normalized complementarity score. As reported by Fábián and Frišcic,44 the complementarity score is calculated in eq 3 by dividing the difference between the API (XD,API) and the coformer descriptors (XD,cof) by the cut-off value (δD) and summing the results for each descriptor (D). The lower the complementarity score, the greater the similarity between the two molecules and their ability to cocrystallize.
| 3 |
The MC analysis was carried out with the CSD-material package supplied in Mercury 2020.3 (Cambridge, UK). A tutorial on how to perform the analysis is provided by the Cambridge Crystallographic Data Center (CCDC).45 Starting from the 2D molecular structure of posaconazole, ten likely conformations of the API were generated via the “Conformer generation” function in Mercury, while only one conformation was used for each one of the 28 coformers. The default settings using the five molecular descriptors were applied. Since the PASS/FAIL test did not show any discriminatory ability, the coformers were ranked according to their complementarity score. The complementarity score was calculated considering the five molecular descriptors for each PSZ–coformer pair and was averaged over the ten API conformations.
Multicomponent HBP
This tool is based on the key assumption that the strongest hydrogen bond, among all possible donor–acceptor pairs, directs the formation of a crystal structure.46 The HBP algorithm calculates the HBP index for the formation of hydrogen bonds for all possible combinations of donor and acceptor pairs. The propensity value, ranging from 0 to 1, represents the probability of the formation of hydrogen bonds in the specific chemical environment analyzed.19 First, the HBP approach creates a statistical model based on the 2D properties of a set of structures contained in the CSD. Second, the model is used to predict the propensity value for the target system. The discrimination level of the fitted model was estimated by the area under the curve (AUC); the higher the value, the more trustworthy the prediction.
The likelihood of cocrystal formation was assessed by comparing the propensity index for heteromeric (API–coformer) and homomeric (API–API or coformer–coformer) interactions. As shown in eq 4, the multicomponent score was calculated as the difference between the highest propensity in the cocrystal (interaction between PSZ and the coformer) and the highest propensity in the pure components (either between two PSZ molecules or two coformer molecules).19 Therefore, the multicomponent score will be greater than zero for pairs with a good chance to cocrystallize, and it will be lower than zero when pure forms are more likely. As a result, coformers were ranked according to their multicomponent score value.15
| 4 |
A tutorial on how to perform the HBP analysis is provided by the CCDC.45 Input molecular structures were provided for posaconazole and each coformer as .mol2 files. The HBP analysis generated a training set of crystal structures contained in the CSD depending on the functional groups present in each PSZ–coformer pair. The dataset was employed to fit a model by logistic regression that was subsequently evaluated by means of AUC. In the end, the model was used to assess the HBP of the posaconazole system under study, and coformers were ranked according to their multicomponent scores.
Results
Cocrystal Screening Results
The cocrystals with 4-aminobenzoic acid, l-glutamine, and l-asparagine are the only three examples of posaconazole cocrystals reported in the literature.4,36 However, several cocrystals of itraconazole, which is structurally similar to posaconazole, with dicarboxylic acids are known.47,48 This information suggested that a cocrystal screening for PSZ could reveal new multicomponent crystalline forms of the compound. To test this hypothesis, we screened potential coformers, including acidic and neutral compounds, by combining the mentioned computational tools with high-throughput screening experiments (Figure 2).
First, a computational screening using COSMOquick was performed to preselect coformers from a list of 140 compounds (Supporting Information Table S1). Instead of selecting the top-ranked candidates, we decided to pick a variety of top-, medium-, and low-ranked coformers in order to span the whole list of 140 compounds and test the predictive ability of COSMOquick. Second, a high-throughput PSZ cocrystal screening experiment was carried out on the selected subset of 23 coformers to identify the most promising screening hits. HTCS was conducted in a 96-well quartz microtiter plate by performing a slurry equilibration step, followed by additional evaporation to recover the product. The slurry residues produced lead to new solid phases with l-malic acid, fumaric acid, gentisic acid, citric acid, maleic acid, and mannitol. No positive screening hits were obtained from any of the other investigated coformers.
As a control, all 23 coformers from the HTCS experiment were also examined by the LAG and RC methods on an approximately twenty times larger scale (i.e., 50–100 mg). Furthermore, five top-ranked coformers that had not been included in the HTCS program were tested by the same methods, for a total of 28 candidates. In the end, thirteen new PSZ cocrystals were isolated by the two techniques and characterized in terms of their physicochemical properties. Table 1 compares the COSMOquick coformer ranking with the experimental results obtained by the HTCS, LAG, and RC methods.
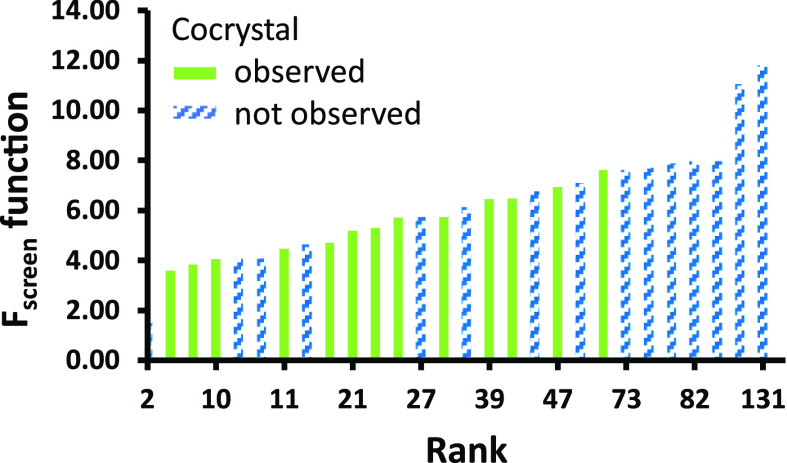
Table 1 underlines the good agreement between experimental and computational outcomes. Their ranking is illustrated in Figure 3, where, in general, cocrystals were successfully obtained with coformers characterized by lower Fscreen values, while no cocrystals were observed for compounds with the highest Fscreen values. It is worth noting that the COSMOquick prediction generally ranks neutral compounds at the bottom of the list, while all top-ranked compounds are organic acids. Experimentally, PSZ cocrystals were exclusively found with coformers containing acidic functionalities, and no cocrystals were discovered with neutral coformers. LAG or RC experiments validated the high-throughput screening results for 20 of the 23 (87%) coformers tested by the method, thus confirming the applicability of HTCS for the fast and reliable discovery of new cocrystals.
Figure 3.
Ranking of the 28 screened posaconazole coformers according to the corresponding Fscreen values. Green bars represent successful new cocrystals, and blue bars with line patterns represent unsuccessful cocrystallization.
Solid-State Characterization
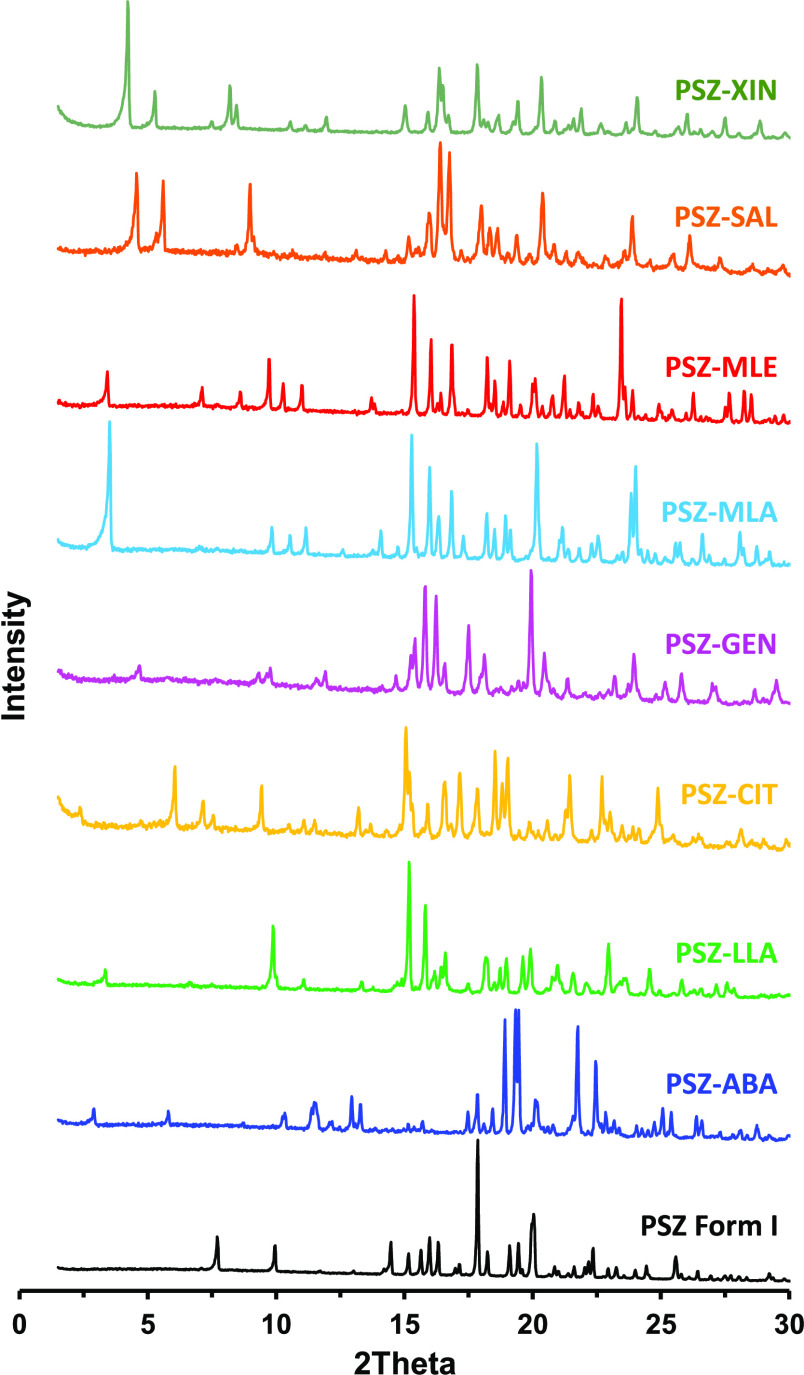
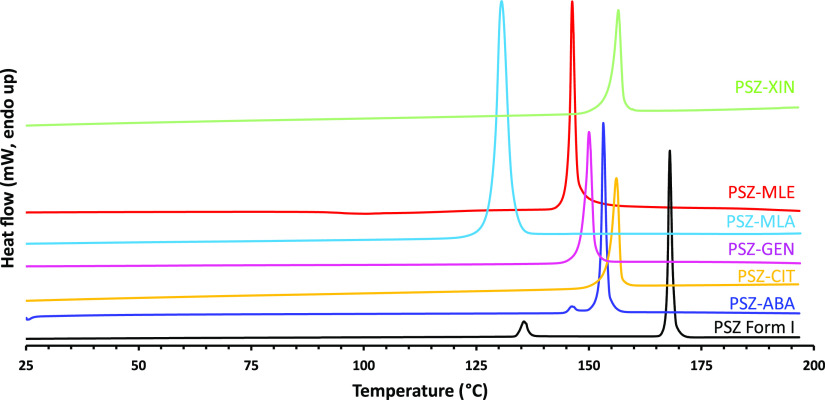
The experimental cocrystal screening of PSZ resulted in the isolation of seven new anhydrous cocrystals with maleic acid (PSZ–MLE), l-malic (PSZ–MLA), citric (PSZ–CIT), 1-hydroxy-2-naphthoic (PSZ–XIN), gentisic (PSZ–GEN), salicylic (PSZ–SAL), and l-lactic acid (PSZ–LLA), in addition to the already known cocrystal with 4-aminobenzoic acid (PSZ–ABA). Five cocrystal hydrates were also obtained with succinic (PSZ–SUC–H2O), fumaric (PSZ–FUM–H2O), ferulic (PSZ–FER–H2O), adipic (PSZ–ADI–H2O), and 3,4-dihydroxybenzoic acid (PSZ–DHB–H2O) and one solvate of a fumaric acid cocrystal (PSZ–FUM–THF). The formation of new crystalline solid phases was confirmed by PXRD; the multicomponent materials display unique diffraction patterns with no presence of posaconazole Form I or any other known posaconazole polymorph. The PXRD patterns of the nonsolvated cocrystals are depicted in Figure 4 and the remaining diffractograms are provided in the Supporting Information. The cocrystal stoichiometry was suggested by 1H NMR spectroscopy by comparing the integration of coformer and posaconazole characteristic proton signals. TG–FTIR, which couples thermogravimetry and infrared spectroscopy, was used to determine solvent contents or to demonstrate the absence of residual solvent in the crystal lattice. The melting points of selected nonsolvated cocrystals were determined by DSC, and the thermograms are reported in Figure 5. A summary of cocrystal physicochemical properties is reported in Table 2. Further information that established the existence of each cocrystal is provided in the Supporting Information.
Figure 4.
Experimental diffraction patterns of PSZ Form I (black), PSZ–ABA cocrystal, and the new anhydrous cocrystals.
Figure 5.
DSC thermograms of PSZ Form I (black), PSZ–ABA cocrystal, and selected new anhydrous cocrystals.
Table 2. List of Obtained Posaconazole Cocrystals and Their Solid-State Properties.
| cocrystal | coformer | stoichiometry | coformer Tmelting (°C) | cocrystal Tmelting (°C) | TG–FTIR |
|---|---|---|---|---|---|
| PSZ–MLE | maleic acid | 1:1 | 130–135 | 146 | anhydrous |
| PSZ–MLA | l-malic acid | 2:1 | 101–103 | 131 | anhydrous |
| PSZ–CIT | citric acid | 4:1 | 153–159 | 156 | anhydrous |
| PSZ–XIN | 1-hydroxy-2-naphthoic acid | 1:1 | 195–200 | 157 | anhydrous |
| PSZ–GEN | gentisic acid | 1:1 | 182–184 | 150 | anhydrous |
| PSZ–SAL | salicylic acid | 1:1 | 158–161 | 125 | anhydrous |
| PSZ–ABA | 4-aminobenzoic acid | 2:3 | 187–189 | 153 | anhydrous |
| PSZ–LLA | l-lactic acid | 1:1 | 53–54 | 126 | anhydrous |
| PSZ–ADI–H2O | adipic acid | 2:1:1 | 151–154 | 1.3% H2O | |
| PSZ–FER–H2O | trans-ferulic acid | 3:1:1 | 168–172 | 0.9% H2O | |
| PSZ–SUC–H2O | succinic acid | 2:1:1 | 184–186 | 1% H2O | |
| PSZ–FUM–THF | fumaric acid | 1:1:1 | 298–300 | 10% THF | |
| PSZ–FUM–H2O | fumaric acid | 1:1:0.5 | 298–300 | 1% H2O | |
| PSZ–DHB–H2O | 3,4-dihydroxybenzoic acid | 3:4:2 | 197–200 | 1.4% H2O |
All synthesized cocrystals show unique FT-Raman spectra; the presence or disappearance of peaks indicates the formation of new intermolecular interactions in the solid state. It is worth noting that the FT-Raman spectrum of PSZ Form I is characterized by a peak at 798 cm–1, which is absent in all the new cocrystal forms found (Supporting Information Figure S18). Despite the synthesis in acetonitrile performed by the RC method, cocrystal hydrate formation was observed with five different coformers. The molar water content of the cocrystals was determined by the water release in the TG–FTIR; further analyses, such as dynamic vapor sorption (DVS), are required to establish if these new multicomponent crystals are stoichiometric or nonstoichiometric hydrates.
Regardless of numerous attempts, no anhydrous cocrystal with fumaric acid was observed, whereas a solvent-free cocrystal was obtained with its geometrical isomer, maleic acid. A PSZ–coformer stoichiometric ratio of 1 to 1 was suggested by 1H NMR for both PSZ–MLE and PSZ–FUM cocrystals, while the cocrystals with the aliphatic dicarboxylic acids, such as adipic and succinic acid, show a PSZ–coformer stoichiometry of 2 to 1.
The thermal behavior of the newly discovered anhydrous cocrystals and the PSZ–ABA cocrystal, characterized by one single endothermic peak corresponding to the melting point, is substantially different compared to that of the crystalline API. The DSC thermogram of PSZ Form I, as already described in the literature,4,49 shows a small endothermic peak at 135 °C and a melting peak at 168 °C (Figure 5). For comparison, the melting points of the coformers and the new cocrystals are summarized in Table 2. The melting points of PSZ–MLE (146 °C), PSZ–MLA (131 °C), and PSZ–LLA (126 °C) are in between the reported melting points of posaconazole and the corresponding coformer. For PSZ–CIT (156 °C), PSZ–XIN (157 °C), PSZ–SAL (125 °C), and PSZ–GEN (150 °C), the melting points are lower than both posaconazole and the coformer, as already reported by Kuminek et al. for the PSZ–ABA cocrystal.4
In addition, nine of the 14 synthetized cocrystals are characterized by a PSZ–coformer stoichiometric ratio that differs from 1 to 1. This highlights the necessity of carrying out RC experiments and not just LAG. Indeed, the latter employs API and coformer in a well-defined stoichiometric ratio, and it would require several experiments to find the exact relative ratio of the components, resulting in a pure cocrystal.
Discussion
Computational-Experimental Cocrystal Screening
The COSMOquick calculations for 140 potential coformers predicted that cocrystals with acidic compounds will generally have lower free energies (i.e., the lowest mixing enthalpy values) and will therefore more likely form cocrystals than compounds without carboxylate functional groups. Indeed, only 20% of the 50 top-ranked compounds are neutral coformers (Supporting Information Tables S1 and S2). For posaconazole, intermolecular interactions with carboxylic acids in the cocrystal appear to be stronger, and COSMOquick often predicts unfavorable interactions of PSZ with other compounds that lack a carboxylic acid group. These results are easily explained by the fact that posaconazole is a weakly dibasic molecule with pKa values of 3.6 and 4.6, containing basic functional groups that are able to interact with acidic functionalities present in the coformers. Thus, we preselected 28 representative coformers for an experimental screening, with the majority of them being carboxylic acids. The experimental screening results are in good agreement with the computational findings. However, it is worth mentioning that the COSMOquick’s high-ranking neutral compounds, myricetin (ranking no. 3) and methyl gallate (ranking no. 18), were not included in the experimental investigation.
The high throughput cocrystal screening (HTCS) enables the execution of many solution cocrystallization experiments in parallel using a small amount of material. For reasons of space in the 96-well plate, a subset of 23 coformers was examined with four different solvents, namely ethanol, acetonitrile, ethyl acetate, and a THF/H2O 50% v/v mixture. The formation of new solid phases was observed predominantly when the test solvents were acetonitrile or ethyl acetate; therefore, acetonitrile was the preferred solvent for the LAG and RC experiments. Acetonitrile was also favored because of the relative solubilities of PSZ and many of the coformers tested. Furthermore, acetonitrile had been used as the solvent to reproduce the already-known PSZ cocrystal with 4-aminobenzoic acid, and both techniques had been successful. We did not include l-glutamine and l-asparagine as a reference in the conducted cocrystal screening because their cocrystals with posaconazole were not reported in the literature at the time of this study. False negative HTCS leads were obtained for adipic acid and l-lactic acid as both cocrystals were produced during the follow-up LAG and RC experiments. This is not unusual because very small amounts of the substance were used. A false positive lead was obtained for mannitol; however, it is believed that the new Raman spectrum observed corresponds to another polymorphic form of mannitol that was not readily available as a reference.
Finally, the results obtained for the tests with oxalic acid are noteworthy. Interestingly, COSMOquick sorted oxalic acid at the top of the ranking list, suggesting strong intermolecular interactions with the API. A new Raman spectrum was collected after LAG and RC tests, initially suggesting the presence of a new solid form. However, PXRD measurements performed on the very same samples revealed a poorly crystalline material identified by two broad peaks at 3.4 and 25.8° values of 2θ. The PXRD pattern of this material is presented in the Supporting Information (Figure S76). All experiments failed to isolate a truly crystalline material, and, therefore, the compound with PSZ and oxalic acid was not classified as a cocrystal. Nonetheless, there appears to be at least some degree of interaction between oxalic acid and PSZ, partially corroborated by the COSMOquick prediction.
The results of the experiments of PSZ with oxalic acid underline the importance of using multiple characterization techniques for the confirmation of cocrystal formation. This is particularly important for the performance assessment of computational cocrystal screening tools, for which reliable experimental results are required.
Computational Tools Comparison
COSMOquick was employed for the preselection of coformers in the combined cocrystal screening of posaconazole. However, other virtual tools are available and can be used for computational cocrystal screening. In this section, we compare the prediction performance of COSMOquick with that of two CSD-based approaches: MC and HBP. All these methods are described in the Materials and Methods section.
A pharmaceutical cocrystal is almost always more stable than the two pure components and, as a result, is characterized by a negative free energy of formation.50 This is also reflected by the three computational methods employed in the prediction, which select coformers able to generate a thermodynamically more stable crystal form with the API. The selection is done by COSMO-RS and HBP looking for stronger interactions in the cocrystal than in the starting compounds, while MC looks for a more efficient molecular packing.
COSMOquick and HBP have a different focus on the intermolecular interaction in the multicomponent system. COSMOquick computes electrostatic, hydrogen bonding, and van der Waals interactions in a supercooled liquid phase by statistical thermodynamic calculation of molecular σ-surface segments while HBP evaluates the strongest hydrogen bond in the molecular systems by training a model on cocrystal structures stored in the CSD. The performed HBP analysis associates a positive multicomponent score for almost all tested compounds, suggesting them as successful coformers (Supporting Information Table S3). The overestimation of positive results may be explained by the fact that HBP is trained on the CSD, which is a database containing only positive cocrystal examples.
In contrast, the MC approach neglects interactions but considers the similarity in shape and polarity of the API and the coformers by evaluating five molecular descriptors. Since the shape descriptors depend on the spatial arrangement of the molecules, different conformations must be considered for flexible molecules. These conformations should represent likely arrangements of the molecule in the crystal lattice. Despite previous studies mentioning the importance of choosing representative conformations of the target API (e.g., the molecular conformation present in polymorphs),18,30 many data about molecular crystal structures may not be available in the early stage of drug development. For this reason, we started from a two-dimensional chemical structure of posaconazole, as it was available from the PubChem library, to generate ten different low-energy conformations of the API. Only one conformation was employed for each coformer.
The MC results obtained using the default settings (5 molecular descriptors; all molecular descriptors below a threshold value to generate a “PASS”) appeared not to be sufficiently discriminatory: only three coformers (succinic, ferulic, and adipic acid) showed a hit rate that was different from 0% (Supporting Information Table S4); therefore, it was not possible to achieve a useful ranking of the coformers.51 This is explained by the fact that most of the coformers tested showed a lack of complementarity with posaconazole due to the shape, with the short axis (S) and the medium-to-long-axis ratio (M/L) being the main descriptors failing the analysis (Supporting Information Table S5).
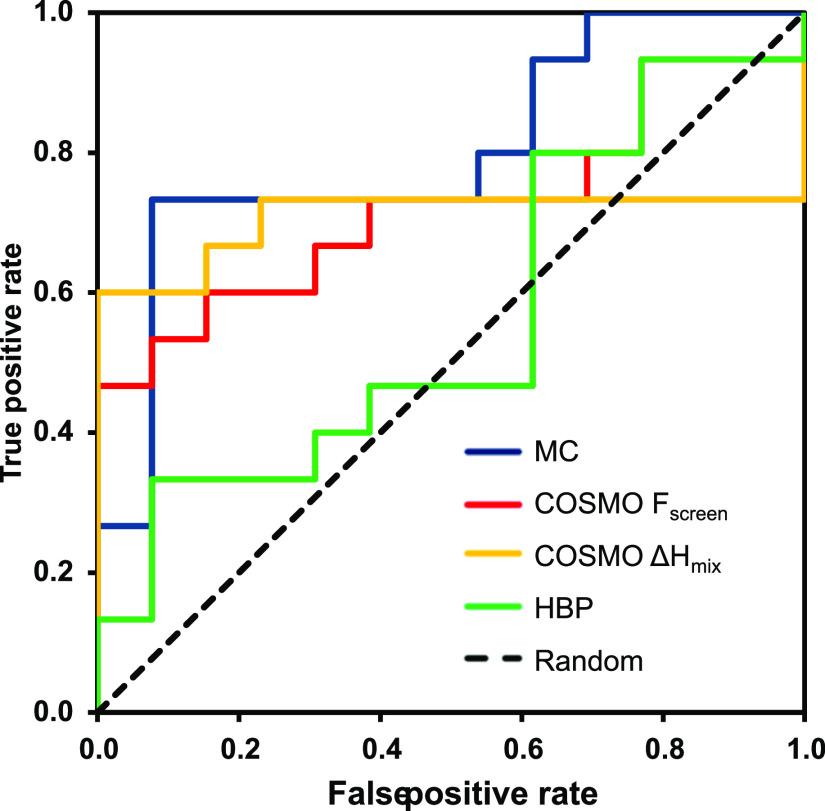
However, the values calculated by the MC tool for each descriptor and the established cut-off limits can be used to compute a complementarity score, which represents the degree of similarity between the coformer and target molecule. The complementarity score was averaged over the ten posaconazole conformations and then used to rank coformers in a list (Supporting Information Table S6). Having obtained a coformer ranking for each computational tool, a direct comparison between the prediction performance was made. The accuracy of the virtual tools was quantitatively assessed by building receiver operating characteristic (ROC) curves. A ROC curve plots the true-positive rate (number of true positive predictions/total number of positive observations) versus the false-positive rate (number of false-positive predictions/total number of negative observations) for a binary classifier system as its discrimination threshold (the screening function) is varied from small to higher values. The AUC measures the overall performance of the model. The higher the AUC, the better the predictive model. A random method, such as the flipping of a coin, has no discriminatory capacity and is characterized by an AUC value of 0.5, while a perfect prediction would have an AUC value of 1. Figure 6 depicts the ROC curves computed for the three virtual tools. The HBP approach does not perform better than a random method, as reflected by the AUC value of 0.56. In contrast, the MC and COSMOquick models performed well in the cocrystal screening of posaconazole, as shown by the AUC values of 0.80 and 0.72, respectively (Table 3).
Figure 6.
ROC plot of posaconazole cocrystal screening computed employing different approaches: COSMOquick using Fscreen and ΔHmix, HBP using the multicomponent score, and MC using the complementarity score.
Table 3. AUC Calculated for the Different Computational Cocrystal Screening toolsa.
| computational approach | ranking variable | AUC (I) | AUC (II) |
|---|---|---|---|
| COSMOquick | Fscreen | 0.72 | 0.42 |
| COSMOquick | ΔHmix | 0.71 | 0.37 |
| HBP | multicomponent score | 0.56 | 0.63 |
| MC | complementarity score | 0.80 | 0.79 |
AUC (I) is the AUC computed for posaconazole cocrystal screening on the 28 coformers; AUC (II) is computed for posaconazole cocrystal screening on the 20 acidic coformers.
Molecular flexibility is incorporated in the COSMOquick approach as the number of rotatable bonds. This parameter was introduced into the method because it improved the experimental screening function, and it seems to be related to crystallization kinetics.20 For the evaluation purpose, an additional ROC curve was plotted using ΔHmix as the screening function, and the AUC was compared to the one obtained with the Fscreen as the ranking variable. The results collected in Table 3 show that ranking compounds considering both intermolecular interactions and molecular flexibility results in a slightly better performance (AUC 0.72) compared with the simple use of ΔHmix (AUC 0.71).
The three virtual tools were also employed in ranking the subset of acidic coformers, excluding neutral compounds that do not form cocrystals with posaconazole. Although the ranking ability of the MC (AUC 0.79) and the HBP (AUC 0.63) methods was found to stay almost the same (Table 3), a large difference was observed for the COSMO-RS approach (AUC 0.42). The drop in ranking ability indicates that the model was no longer able to distinguish between positive and negative hits. The COSMO-RS approach successfully recognized carboxylic acids as the most suitable coformers for the formation of cocrystals with posaconazole but failed to provide their correct ranking order. One possible explanation for this shortcoming in sorting is that the COSMO-RS method does not consider the crystalline long-range order and the packing of molecules in the crystal; these factors, in parallel to intermolecular interactions, may drive the crystallization process. In addition, the difference between the energies of fusion of the two components, ΔΔGfus, may represent another source of deviation from experimental findings: if the energies of fusion of the two components do not compensate each other (equation 1), then ΔHmix cannot be used to assess the likelihood of cocrystal formation, because it is no longer a good approximation for the free energy of cocrystallization, ΔGcc.
These results suggest that the COSMO-RS screening seems to be most helpful in the preselection of coformers from a vast library of compounds with different chemical natures and functional groups.
Conclusions
This study demonstrated the successful application of a combined computational-experimental high-throughput approach in the cocrystal screening of posaconazole with 28 coformers. COSMOquick was employed to narrow down a list of approximately 140 potential coformers to a selection of 28 candidates. In addition, the computational step successfully ranked acidic coformers at the top of the test list. The experimental high-throughput slurry approach highlighted the formation of new cocrystals. The method, combined with scale-up experiments, ultimately led to the discovery of thirteen new cocrystals that were confirmed and characterized by spectroscopic and thermal analysis.
Based on these experimental results, the present work retrospectively compared three computational approaches, namely, MC, HBP, and COSMOquick, according to their ability to successfully select coformers of posaconazole. HBP did not perform better than a random method in the virtual cocrystal screening of posaconazole (AUC of 0.56) on this subset of coformers. However, MC and COSMOquick resulted in an enhanced likelihood prediction of the best coformers at the top of the ranking list, as demonstrated by the AUC values of 0.80 and 0.72, respectively. The COSMO-RS approach exhibited some limitations in ranking the best acidic coformers.
Overall, MC and COSMOquick showed promising predictive ability for the posaconazole case, and it would be interesting to test them on different molecular systems in order to assess their performance in a wider chemical space.
Acknowledgments
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement no. 955756. The authors thank Dr. Susan De Paul for critically reviewing the manuscript and the teams from the Solvias PXRD and thermal analysis laboratories for their support.
Glossary
Abbreviations
- API
active pharmaceutical ingredient
- AUC
area under the curve
- BCS
biopharmaceutical classification system
- CCDC
Cambridge crystallographic data center
- COSMO-RS
conductor-like screening model for real solvent
- CSD
Cambridge structural database
- CSP
crystal structure prediction
- DSC
differential scanning calorimetry
- FNO
fraction of nitrogen and oxygen atoms
- GRAS
generally regarded as safe
- HBP
hydrogen bond propensity
- HTCS
high-throughput cocrystal screening
- LAG
liquid assisted grinding
- MC
molecular complementarity
- MEPS
molecular electrostatic potential surfaces
- ML
machine learning
- PSZ
posaconazole
- PXRD
powder x-ray diffraction
- RC
reaction crystallization
- ROC
receiver operating characteristic
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.cgd.2c01072.
List of coformers employed in the cocrystal screening; experimental details; PXRD, FT-Raman, TG-FTIR, DSC, and solution 1H NMR spectra for the posaconazole starting material and the new cocrystals obtained; and COSMOquick, MC, and HBP calculation results (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- von Raumer M.; Hilfiker R.. Solid State and Polymorphism of the Drug Substance in the Context of Quality by Design and ICH Guidelines Q8-Q12. Polymorphism in the Pharmaceutical Industry; Wiley-VCH Verlag GmbH & Co. KGaA, 2018, pp 1–30. [Google Scholar]
- Kalepu S.; Nekkanti V. Insoluble Drug Delivery Strategies: Review of Recent Advances and Business Prospects. Acta Pharm. Sin. B 2015, 5, 442–453. 10.1016/j.apsb.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aaltonen J.; Alleso M.; Mirza S.; Koradia V.; Gordon K.; Rantanen J. Solid form screening—A review. Eur. J. Pharm. Biopharm. 2009, 71, 23–37. 10.1016/j.ejpb.2008.07.014. [DOI] [PubMed] [Google Scholar]
- Kuminek G.; Cavanagh K. L.; da Piedade M. F. M.; Rodríguez-Hornedo N. Posaconazole Cocrystal with Superior Solubility and Dissolution Behavior. Cryst. Growth Des. 2019, 19, 6592–6602. 10.1021/acs.cgd.9b01026. [DOI] [Google Scholar]
- Wong S. N.; Chen Y. C. S.; Xuan B.; Sun C. C.; Chow S. F. Cocrystal Engineering of Pharmaceutical Solids: Therapeutic Potential and Challenges. CrystEngComm 2021, 23, 7005–7038. 10.1039/D1CE00825K. [DOI] [Google Scholar]
- Viertelhaus M.; Hilfiker R.; Blatter F.; Neuburger M. Piracetam Co-Crystals with OH-Group Functionalized Carboxylic Acids. Cryst. Growth Des. 2009, 9, 2220–2228. 10.1021/cg800942n. [DOI] [Google Scholar]
- Aitipamula S.; Banerjee R.; Bansal A. K.; Biradha K.; Cheney M. L.; Choudhury A. R.; Desiraju G. R.; Dikundwar A. G.; Dubey R.; Duggirala N.; Ghogale P. P.; Ghosh S.; Goswami P. K.; Goud N. R.; Jetti R. R. K. R.; Karpinski P.; Kaushik P.; Kumar D.; Kumar V.; Moulton B.; Mukherjee A.; Mukherjee G.; Myerson A. S.; Puri V.; Ramanan A.; Rajamannar T.; Reddy C. M.; Rodriguez-Hornedo N.; Rogers R. D.; Row T. N. G.; Sanphui P.; Shan N.; Shete G.; Singh A.; Sun C. C.; Swift J. A.; Thaimattam R.; Thakur T. S.; Kumar Thaper R.; Thomas S. P.; Tothadi S.; Vangala V. R.; Variankaval N.; Vishweshwar P.; Weyna D. R.; Zaworotko M. J. Polymorphs, Salts, and Cocrystals: What’s in a Name?. Cryst. Growth Des. 2012, 12, 2147–2152. 10.1021/cg3002948. [DOI] [Google Scholar]
- GRAS Substances (SCOGS) Database; FDA. https://www.fda.gov/food/generally-recognized-safe-gras/gras-substances-scogs-database (accessed July 18, 2022).
- Substances Added to Food (Formerly EAFUS); FDA. https://www.fda.gov/food/food-additives-petitions/substances-added-food-formerly-eafus (accessed July 18, 2022).
- Childs S. L.; Stahly G. P.; Park A. The Salt–Cocrystal Continuum: The Influence of Crystal Structure on Ionization State. Mol. Pharm. 2007, 4, 323–338. 10.1021/mp0601345. [DOI] [PubMed] [Google Scholar]
- Karimi-Jafari M.; Padrela L.; Walker G. M.; Croker D. M. Creating Cocrystals: A Review of Pharmaceutical Cocrystal Preparation Routes and Applications. Cryst. Growth Des. 2018, 18, 6370–6387. 10.1021/acs.cgd.8b00933. [DOI] [Google Scholar]
- Desiraju G. R. Supramolecular Synthons in Crystal Engineering-A New Organic Synthesis. Angew. Chem., Int. Ed. Engl. 1995, 34, 2311–2327. 10.1002/anie.199523111. [DOI] [Google Scholar]
- Kumar A.; Nanda A. In-Silico Methods of Cocrystal Screening: A Review on Tools for Rational Design of Pharmaceutical Cocrystals. J. Drug Deliv. Sci. Technol. 2021, 63, 102527. 10.1016/j.jddst.2021.102527. [DOI] [Google Scholar]
- Sun G.; Jin Y.; Li S.; Yang Z.; Shi B.; Chang C.; Abramov Y. A. Virtual Coformer Screening by Crystal Structure Predictions: Crucial Role of Crystallinity in Pharmaceutical Cocrystallization. J. Phys. Chem. Lett. 2020, 11, 8832–8838. 10.1021/acs.jpclett.0c02371. [DOI] [PubMed] [Google Scholar]
- Wood P. A.; Feeder N.; Furlow M.; Galek P. T. A.; Groom C. R.; Pidcock E. Knowledge-Based Approaches to Co-Crystal Design. CrystEngComm 2014, 16, 5839. 10.1039/c4ce00316k. [DOI] [Google Scholar]
- Groom C. R.; Bruno I. J.; Lightfoot M. P.; Ward S. C. The Cambridge Structural Database. Acta Crystallogr., Sect. B: Struct. Sci., Cryst. Eng. Mater. 2016, 72, 171–179. 10.1107/S2052520616003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fábián L. Cambridge Structural Database Analysis of Molecular Complementarity in Cocrystals. Cryst. Growth Des. 2009, 9, 1436–1443. 10.1021/cg800861m. [DOI] [Google Scholar]
- Cappuccino C.; Cusack D.; Flanagan J.; Harrison C.; Holohan C.; Lestari M.; Walsh G.; Lusi M. How Many Cocrystals Are We Missing? Assessing Two Crystal Engineering Approaches to Pharmaceutical Cocrystal Screening. Cryst. Growth Des. 2022, 22, 1390–1397. 10.1021/acs.cgd.1c01342. [DOI] [Google Scholar]
- Galek P. T. A.; Fábián L.; Motherwell W. D. S.; Allen F. H.; Feeder N. Knowledge-Based Model of Hydrogen-Bonding Propensity in Organic Crystals. Acta Crystallogr., Sect. B: Struct. Sci. 2007, 63, 768–782. 10.1107/S0108768107030996. [DOI] [PubMed] [Google Scholar]
- Loschen C.; Klamt A. Solubility Prediction, Solvate and Cocrystal Screening as Tools for Rational Crystal Engineering. J. Pharm. Pharmacol. 2015, 67, 803–811. 10.1111/jphp.12376. [DOI] [PubMed] [Google Scholar]
- Loschen C.; Klamt A.; Abramov Y. A.. New Developments in Prediction of Solid-State Solubility and Cocrystallization Using COSMO-RS Theory. Computational Pharmaceutical Solid State Chemistry; John Wiley & Sons, Inc: Hoboken, NJ, 2016, pp 211–233. [Google Scholar]
- Musumeci D.; Hunter C. A.; Prohens R.; Scuderi S.; McCabe J. F. Virtual Cocrystal Screening. Chem. Sci. 2011, 2, 883. 10.1039/c0sc00555j. [DOI] [Google Scholar]
- Grecu T.; Hunter C. A.; Gardiner E. J.; McCabe J. F. Validation of a Computational Cocrystal Prediction Tool: Comparison of Virtual and Experimental Cocrystal Screening Results. Cryst. Growth Des. 2014, 14, 165–171. 10.1021/cg401339v. [DOI] [Google Scholar]
- Sugden I. J.; Braun D. E.; Bowskill D. H.; Adjiman C. S.; Pantelides C. C. Efficient Screening of Coformers for Active Pharmaceutical Ingredient Cocrystallization. Cryst. Growth Des. 2022, 22, 4513–4527. 10.1021/acs.cgd.2c00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devogelaer J.; Meekes H.; Tinnemans P.; Vlieg E.; Gelder R. Co-crystal Prediction by Artificial Neural Networks**. Angew. Chem., Int. Ed. 2020, 59, 21711–21718. 10.1002/anie.202009467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devogelaer J.-J.; Charpentier M. D.; Tijink A.; Dupray V.; Coquerel G.; Johnston K.; Meekes H.; Tinnemans P.; Vlieg E.; ter Horst J. H.; de Gelder R. Cocrystals of Praziquantel: Discovery by Network-Based Link Prediction. Cryst. Growth Des. 2021, 21, 3428–3437. 10.1021/acs.cgd.1c00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker J. G. P.; Crowley L. M.; Robshaw O.; Little E. J.; Stokes S. P.; Cooper R. I.; Lawrence S. E. Will They Co-Crystallize?. CrystEngComm 2017, 19, 5336–5340. 10.1039/C7CE00587C. [DOI] [Google Scholar]
- Wang D.; Yang Z.; Zhu B.; Mei X.; Luo X. Machine-Learning-Guided Cocrystal Prediction Based on Large Data Base. Cryst. Growth Des. 2020, 20, 6610–6621. 10.1021/acs.cgd.0c00767. [DOI] [Google Scholar]
- Yuan J.; Liu X.; Wang S.; Chang C.; Zeng Q.; Song Z.; Jin Y.; Zeng Q.; Sun G.; Ruan S.; Greenwell C.; Abramov Y. A. Virtual Coformer Screening by a Combined Machine Learning and Physics-Based Approach. CrystEngComm 2021, 23, 6039–6044. 10.1039/D1CE00587A. [DOI] [Google Scholar]
- Khalaji M.; Potrzebowski M. J.; Dudek M. K. Virtual Cocrystal Screening Methods as Tools to Understand the Formation of Pharmaceutical Cocrystals-A Case Study of Linezolid, a Wide-Range Antibacterial Drug. Cryst. Growth Des. 2021, 21, 2301–2314. 10.1021/acs.cgd.0c01707. [DOI] [Google Scholar]
- Nunes Costa R.; Choquesillo-Lazarte D.; Cuffini S. L.; Pidcock E.; Infantes L. Optimization and Comparison of Statistical Tools for the Prediction of Multicomponent Forms of a Molecule: The Antiretroviral Nevirapine as a Case Study. CrystEngComm 2020, 22, 7460–7474. 10.1039/D0CE00948B. [DOI] [Google Scholar]
- Schiller D.; Fung H. P. Posaconazole: An extended-spectrum triazole antifungal agent. Clin. Therapeut. 2007, 29, 1862–1886. 10.1016/j.clinthera.2007.09.015. [DOI] [PubMed] [Google Scholar]
- McQuiston D. K.; Mucalo M. R.; Saunders G. C. The Structure of Posaconazole and Its Solvates with Methanol, and Dioxane and Water: Difluorophenyl as a Hydrogen Bond Donor. J. Mol. Struct. 2019, 1179, 477–486. 10.1016/j.molstruc.2018.11.031. [DOI] [Google Scholar]
- Courtney R.; Wexler D.; Radwanski E.; Lim J.; Laughlin M. Effect of food on the relative bioavailability of two oral formulations of posaconazole in healthy adults. Br. J. Clin. Pharmacol. 2003, 57, 218–222. 10.1046/j.1365-2125.2003.01977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkhabaz A.; Moseson D. E.; Brouwers J.; Augustijns P.; Taylor L. S. Interplay of Supersaturation and Solubilization: Lack of Correlation between Concentration-Based Supersaturation Measurements and Membrane Transport Rates in Simulated and Aspirated Human Fluids. Mol. Pharm. 2019, 16, 5042–5053. 10.1021/acs.molpharmaceut.9b00956. [DOI] [PubMed] [Google Scholar]
- Vemuri V. D.; Lankalapalli S.; Chandra Reddy Guntaka P. Posaconazole-Amino Acid Cocrystals for Improving Solubility and Oral Bioavailability While Maintaining Antifungal Activity and Low In Vivo Toxicity. J. Drug Deliv. Sci. Technol. 2022, 74, 103491. 10.1016/j.jddst.2022.103491. [DOI] [Google Scholar]
- Klamt A. Conductor-like Screening Model for Real Solvents: A New Approach to the Quantitative Calculation of Solvation Phenomena. J. Phys. Chem. 1995, 99, 2224–2235. 10.1021/j100007a062. [DOI] [Google Scholar]
- Hornig M.; Klamt A. COSMOfrag: A Novel Tool for High-Throughput ADME Property Prediction and Similarity Screening Based on Quantum Chemistry. J. Chem. Inf. Model. 2005, 45, 1169–1177. 10.1021/ci0501948. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Hornedo N.; Nehm S. J.; Seefeldt K. F.; Pagán-Torres Y.; Falkiewicz C. J. Reaction Crystallization of Pharmaceutical Molecular Complexes. Mol. Pharm. 2006, 3, 362–367. 10.1021/mp050099m. [DOI] [PubMed] [Google Scholar]
- Rager T.; Hilfiker R. Stability Domains of Multi-Component Crystals in Ternary Phase Diagrams. Z. Phys. Chem. 2009, 223, 793–813. 10.1524/zpch.2009.5454. [DOI] [Google Scholar]
- Klamt A. The COSMO and COSMO-RS solvation models. Wiley Interdiscip. Rev.: Comput. Mol. Sci. 2011, 1, 699–709. 10.1002/wcms.56. [DOI] [Google Scholar]
- Klamt A.; Eckert F. RS. COSMO-RS: a novel and efficient method for the a priori prediction of thermophysical data of liquids. Fluid Phase Equilib. 2000, 172, 43–72. 10.1016/S0378-3812(00)00357-5. [DOI] [Google Scholar]
- Blatter F.; Szelagiewicz M.; von Raumer M.. Process for the Parallel Detection of Crystalline Forms of Molecular Solids. U.S. Patent 7,892,354 B2, February 22, 2011.
- Fábián L.; Frišcic T.; Quéré L.. Chapter 5. Shape and Polarity in Co-Crystal Formation: Database Analysis and Experimental Validation. In Pharmaceutical Salts and Co-crystals; Wouters J., Quere L., Eds.; Drug Discovery; Royal Society of Chemistry: Cambridge, 2011, pp 89–109. [Google Scholar]
- CSD-Materials Workshops—The Cambridge Crystallographic Data Centre (CCDC). https://www.ccdc.cam.ac.uk/Community/educationalresources/workshop-materials/csd-materials-workshops/ (accessed July 24, 2022).
- Etter M. C. Encoding and Decoding Hydrogen-Bond Patterns of Organic Compounds. Acc. Chem. Res. 1990, 23, 120–126. 10.1021/ar00172a005. [DOI] [Google Scholar]
- Remenar J. F.; Morissette S. L.; Peterson M. L.; Moulton B.; MacPhee J. M.; Guzmán H. R.; Almarsson Ö. Crystal Engineering of Novel Cocrystals of a Triazole Drug with 1,4-Dicarboxylic Acids. J. Am. Chem. Soc. 2003, 125, 8456–8457. 10.1021/ja035776p. [DOI] [PubMed] [Google Scholar]
- Shevchenko A.; Miroshnyk I.; Pietilä L.-O.; Haarala J.; Salmia J.; Sinervo K.; Mirza S.; van Veen B.; Kolehmainen E.; Nonappa; Yliruusi J. Diversity in Itraconazole Cocrystals with Aliphatic Dicarboxylic Acids of Varying Chain Length. Cryst. Growth Des. 2013, 13, 4877–4884. 10.1021/cg401061t. [DOI] [Google Scholar]
- Adrjanowicz K.; Kaminski K.; Wlodarczyk P.; Grzybowska K.; Tarnacka M.; Zakowiecki D.; Garbacz G.; Paluch M.; Jurga S. Molecular Dynamics of the Supercooled Pharmaceutical Agent Posaconazole Studied via Differential Scanning Calorimetry and Dielectric and Mechanical Spectroscopies. Mol. Pharm. 2013, 10, 3934–3945. 10.1021/mp4003915. [DOI] [PubMed] [Google Scholar]
- Taylor C. R.; Day G. M. Evaluating the Energetic Driving Force for Cocrystal Formation. Cryst. Growth Des. 2018, 18, 892–904. 10.1021/acs.cgd.7b01375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- It should be noted that all three of these coformers did indeed form experimental cocrystals with posaconazole.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.