Abstract
There is a need to identify new biomarkers of radiation exposure for not only systemic total-body irradiation (TBI) but also to characterize partial-body irradiation and organ specific radiation injury. In the current study, we sought to develop novel biodosimetry models of radiation exposure using TBI and organ specific partial-body irradiation to only the brain, lung or gut using a multivariate proteomics approach. Subset panels of significantly altered proteins were selected to build predictive models of radiation exposure in a variety of sample cohort configurations relevant to practical field application of biodosimetry diagnostics during future radiological or nuclear event scenarios. Female C57BL/6 mice, 8–15 weeks old, received a single total-body or partial-body dose of 2 or 8 Gy TBI or 2 or 8 Gy to only the lung or gut, or 2, 8 or 16 Gy to only the brain using a Pantak X-ray source. Plasma was collected by cardiac puncture at days 1, 3 and 7 postirradiation for total-body exposures and only the lung and brain exposures, and at days 3, 7 and 14 postirradiation for gut exposures. Plasma was then screened using the aptamer-based SOMAscan proteomic assay technology, for changes in expression of 1,310 protein analytes. A subset panel of protein biomarkers which demonstrated significant changes (P < 0.01) in expression after irradiation were used to build predictive models of radiation exposure using different sample cohorts. Model 1 compared controls vs. all pooled irradiated samples, which included TBI and all organ specific partial irradiation. Model 2 compared controls vs. TBI vs. partial irradiation (with all organ specific partial exposure pooled within the partial-irradiated group), and model 3 compared controls vs. each individual organ specific partial-body exposure separately (brain, gut and lung). Detectable values were obtained for all 1,310 proteins included in the SOMAscan assay for all samples. Each model algorithm built using a unique sample cohort was validated with a training set of samples and tested with a separate new sample series. Overall predictive accuracies of 89%, 78% and 55% resulted for models 1–3, respectively, representing novel predictive panels of radiation responsive proteomic biomarkers. Though relatively high overall predictive accuracies were achieved for models 1 and 2, all three models showed limited accuracy at differentiating between the controls and partial-irradiated body samples. In our study we were able to identify novel panels of radiation responsive proteins useful for predicting radiation exposure and to create predictive models of partial-body exposure including organ specific radiation exposures. This proof-of-concept study also illustrates the inherent physiological limitations of distinguishing between small-body exposures and the unirradiated using proteomic biomarkers of radiation exposure. As use of biodosimetry diagnostics in future mass casualty settings will be complicated by the heterogeneity of partial-body exposure received in the field, further work remains in adapting these diagnostic tools for practical use.
INTRODUCTION
Research goals within the modern field of radiation biodosimetry have shifted over the last decade from focusing on development of diagnostics to quantify unknown received radiation dose to diagnostics which categorize the scale of severity of radiation injury and diagnostics which may determine organ specific partial-body exposure. These paradigm shifts reflect evolving Concept of Operations (CONOPS) at the federal level for mass casualty medical management of radiological or nuclear events from focusing on numerical received radiation dose towards more practical application of biodosimetry diagnostics for the inherently complex medical management of acute radiation injury. Clinical management of the Acute Radiation Syndrome (ARS) or acute partial-body exposure could be improved with more accurate assessments of the individual severity of radiation exposure and with determination of which organ systems are most affected, as measured by physiological markers of injury (1, 2). As nearly all radiation exposures in such scenarios will be heterogeneous, biodosimetry methodologies which may be used for both total-body irradiation (TBI) and partial-body irradiation are currently being developed.
Proof of concept partial-body biodosimetry models have been demonstrated using a variety of methodologies including use of genomic signatures, miRNA and mRNA expression profiles and cytogenetic approaches. These models have also been utilized across a range of animal model types including murine, non-human primate (NHP) and ex-vivo irradiated human blood samples (3–8). Development of proteomic biomarkers relevant for partial-body exposure have also been characterized in murine and NHP models (9–13). Yet, the number of studies characterizing partial-body biodosimetry models is far from robust. Our previous work using a multivariate proteomic approach to develop radiation exposure and dose prediction models demonstrated that models developed using TBI data exclusively have reduced performance when challenged with partial-body sample sets and that proteomic biomarker expression profiles are influenced not only by the percentage of body mass exposed to radiation but by the specific partial-body exposure profile relative to which organs are exposed. These previous studies also demonstrated the utility of using a multivariate approach for development of models of radiation exposure and that the prediction strength of an algorithm is determined by the number and diversity of the included biomarkers (10–12).
In the current study, we sought to develop novel biodosimetry models of exposure using TBI and organ specific partial-body exposure to only the brain, lung or gut. Using a multivariate proteomics approach, murine plasma samples were assayed using the SOMAlogic SOMAscan proteomics platform which included simultaneous analysis of over 1,300 proteomic targets. Our findings illustrate the utility of using a multivariate biomarker approach and the practical challenges of developing algorithms with simultaneous utility for prediction of both TBI and partial-body exposure.
METHODS
Animal Model
For this study female C57BL/6 mice, 8–15 weeks old received a single total-body or partial-body dose of 2 or 8 Gy for total-body, lung or gut exposures, or 2, 8 or 16 Gy to only the brain using a Pantak X-ray source. Animals were sourced from Charles River and housed under standard conditions with food and water ad libitum. All mice receiving total-body irradiation were confined using a standard pie jig preventing movement. Animals receiving radiation to the lung and gut were irradiated in custom-shielded jigs with restraints to prevent movement and each animal was individually aligned anatomically to ensure correct partial-body exposure. Animals receiving radiation to the brain were anesthetized with an IP injection of ketamine (100 mg/kg) and positioned in a custom-shielded jig for whole-brain irradiation. Murine TBI samples were utilized from our previous study which developed predictive models of radiation exposure and received dose using an exclusively TBI paradigm (14). Control samples were pooled from animals who received sham irradiation with restraint, sham irradiation with anesthesia and naïve animals. A separate statistical analysis yielded no significant changes in proteomic expression profiles between these respective control animals using the SOMAscan assay. (Data not shown.)
Blood samples were collected under anesthesia via cardiac puncture using a heparinized syringe at days 1, 3 and 7 postirradiation for total-body exposures and exposures to only the lung and brain, and at days 3, 7 and 14 postirradiation for the gut. Mice received 2.5–5.5% Isoflurane anesthesia during cardiac puncture for blood collection. A minimum of n = 6 animals were used for each exposure and collection time point in addition to n = 19 control animals which included animals both with and without anesthesia. In total n = 103 samples and n = 100 samples were used for the separate training and test sample cohorts, respectively, for model development.
Animal Model Dosimetry
Murine in vivo models utilized a Pantak X-ray source at a dose rate of 2.28 Gy/min. Dose rate was calibrated based upon the procedures described in American Association of Physicist in Medicine (AAPM) Task Group Report 61 (TG-61) with regard to the following conditions: X-ray tube potential was 300 kV, half value layer (HVL) is 0.9 mm copper (Cu), homogeneity coefficient (HC) is 0.33, source-to-surface distance (SSD) was 50 cm with a field size of 20 × 20 cm. Dose rate was measured at 2 cm depth in solid water phantom using a Preston-Tonks-Wallace (PTW) model: N23342 ion chamber and Inovision, model 35040 electrometer.
SOMAlogic SOMAscan assay
Approximately 160 μl of plasma per sample was used for the Somalogic SOMAscan Assay which uses a novel protein-capture aptamer-based technology (15). The SOMAscan platform provides a uniquely high-throughput proteomic screening analysis which allows characterization of dynamic changes in proteomic expression using a multiplex approach order of magnitude greater than other commercially available technologies. For this study the SOMAscan HTS Assay 1.3K was used and processed through the Center for Human Immunology at the National Institutes of Health. The assay included the measurement of 1,310 protein analytes.
Statistical Analysis
In brief, data was received in the form of Relative Fluorescent Units (RFU) for each of the 1,310 proteins in the SOMAscan assay after normalizing for intraplate and interplate variation. Each sample cohort was randomly split into two separate groups for model training and testing purposes with each sample representing a different animal. The RFU scores for each protein were log2 and z-score transformed. Statistical data analysis was performed using R (16). In this study, we investigated the effect of feature selection and prediction algorithms on the performance of prediction method. We considered the following feature selection and prediction methods implemented sequentially: Elastic net (Enet), recursive partitioning and regression trees (RPART), and linear discriminant analysis (LDA). We studied the effects of feature selection and the number of features on prediction for these methods (17–19). Supplementary Fig. S1 (https://doi.org/10.1667/RADE-22-00074.1.S1) contains a workflow of the relevant methodologies applied to this model building series.
Differential Expression Analysis
To remove invariant data from the analysis, we first performed t test or ANOVA analysis, respectively, depending on whether there were two groups (controls vs. radiation) or multiple groups (controls vs. partial exposure vs. TBI) or (controls vs. tissue type), respectively. Significance tests were used for filtering the features for further analysis. We used a threshold (P adjusted ≤ 0.01 for Anova tests), however, due to lack of enough variability between controls and pooled irradiated groups, we did not find significant P adjusted values and for this model and hence limited to (P ≤ 0.01).
Elastic-Net Analysis
Elastic-net (Enet) analysis was employed to select features for accurate classification and prediction (20). Ridge and LASSO penalties were employed to take advantage of both regularization methods (21). Enet provides shrinkage and automatic variable selection but given limitations in the number of exhaustive models it can handle, the input features needed to be moderately sized (30 covariates equal 1,073,741,824 based on the 2^n to calculate possible combinations). If the selected differential features were greater than 30, we implemented the generalized boosted regression (GBM) models machine learning algorithm to fine-tune variable importance and feature selection (22). Since Elastic-net feature selection results from random permutations, distinct sets of features are identified with each iteration. To identify a stable set of features for more comprehensive applications, we implemented 20 iterations of elastic-net computations resulting in 20 independent models. We then ranked the features by how often each feature was present in a maximum number of models and selected the top-ranked features (14).
Linear Discriminant Analysis
Linear discriminant analysis (LDA) was used to identify linear combinations of features that characterize or discriminate two or more classes and final feature selection and classification. A permutation test evaluated whether the specific classification of the individuals between groups is significantly better than random classification in any two arbitrary groups (23). Finally, we performed model performance evaluation with the new data for prediction accuracy. Histograms of the respective sample distributions and respective prediction matrices are shown in Supplementary Figs. S2–S4 (https://doi.org/10.1667/RADE-22-00074.1.S1).
RPART Tree
RPART is a binary recursive partitioning tree modeling technique that allows for the hierarchical modeling of interactions between variables of interest associated with sample classes (24). Variables were included at each possible split to evaluate whether they improved the node purity. Nodes were split using the best split values, maximizing the Gini index splitting criterion. After the initial tree growing from top to bottom, trees were pruned at the cost complexity value to minimize the Mean Square Error (MSE) for each split. Final trees were grown and validated using fivefold cross-validation. The left-most node on each tree, representing a control group of subjects, was used as the reference node to calculate ORs with 95% CIs.
Model Building for Partial-Body Exposure
Prediction analysis was conducted in a multi-step process. Feature selection was performed using exhaustive iterations the Enet option of the glmnet R package. An additional step using GBR was used to fine tune the feature selection as needed, resulting in generation of a smaller set of predictive features followed by supervised methods, RPART and LDA (18, 19). These methods were used for training and independent test data prediction corresponding to the comparison groups.
RESULTS
This study sought to characterize organ specific proteomic biomarkers of radiation exposure and to build algorithms for the prediction of partial-body exposure to ionizing radiation. To this end, female C57BL6 mice received either TBI or organ specific partial-body irradiation to only the brain, gut or lung at doses of 2 or 8 Gy for TBI, lung or gut and 2, 8 or 16 Gy to the brain using a Pantak X-ray source. Blood samples were collected at days 1, 3 and 7 postirradiation for TBI and partial-body irradiation to the lung and brain and at days 3, 7 and 14 postirradiation for partial-body irradiation to the gut. Proteomic analysis of these samples was conducted using the Somalogic SOMAscan assay for 1,310 protein analytes (15). Radiation exposure prediction models were then constructed using these aggregated SOMAscan data.
For this series, models using three different treatment group combinations were explored. Model 1 compared controls vs. all pooled irradiated samples, including TBI and all organ specific partial irradiation. Model 2 compared controls vs. TBI vs. partial irradiation (with all organ specific partials pooled), and model 3 compared controls vs. each individual organ specific partial-body exposure separately (brain, gut and lung). These models and the corresponding proteomic panels of top ranked proteins for the prediction algorithm for each respective model are shown in Table 1. These expression profiles and the sample cohorts used to train and test each model included pooled samples for 2 and 8 Gy for TBI and partial exposure to only the gut and lung and for 2, 8 and 16 Gy for partial brain irradiation. These samples were collected at days 1, 3 and 7 for TBI and partial exposure to only the lung and brain and at days 3, 7 and 14 for partial exposure to only the gut.
TABLE 1.
Prediction Models for Radiation Exposure
| Model 1 |
Model 2 |
Model 3 |
|||
|---|---|---|---|---|---|
| Treatment group | Proteomic panel | Treatment group | Proteomic panel | Treatment group | Proteomic panel |
|
| |||||
| Controls | EDA2R | Controls | AMIGO2 | Controls | HAPLN1 |
| Irradiation (TBI and all partial-body irradiation pooled) | AK1 | TBI | INSR | Brain | IGF1 |
| MAPK12 | Partial-body irradiation (all partials pooled) | SSRP1 | Gut | PROS1 | |
| NCR1 | PIK3CA_PIK3R1 | Lung | PRKACA | ||
| MAPKAPK5 | SELE | MAPK14 | |||
| PTH | C3D | SPP1 | |||
| F7 | EDA2R | CYP3A4 | |||
| KPNBP1 | |||||
Notes. This includes the sample cohort distributions for each model and the top ranked selected proteins used to build each respective model. Protein selection was based on exhaustive GLM elastic net modeling with summarization of the top models.
For each model, predictive algorithms were generated using a RPART statistical methodology and separate murine sample cohorts were used for the training and testing of each model. Figure 1 highlights the expression distribution (panel A) and the relative ranking (panel B) as described in methods of the top ranked proteins used to build model 1 including ectodysplasin A2 receptor (EDA2R), adenylate kinase 1 (AK1), mitogen-activated protein kinase 12 (MAPK12), natural cytotoxicity triggering receptor 1 (NCR1), MAP kinase-activated protein kinase 5 (MAPKAPK5), parathyroid hormone (PTH) and coagulation factor VII (F7). Figure 1C illustrates the RPART tree used to differentiate between controls and all pooled irradiated samples. Most of the samples could be differentiated by an elevation in EDA2R, a member of the tumor necrosis factor receptor superfamily. The accuracy of this model using the training sample set was 99% and using a separate set of test samples was 89% (Table 2). Figure 2 highlights the expression distribution (panel A) and the relative ranking (panel B) of the top ranked proteins used to build model 2 including adhesion molecule with Ig like domain 2 (AMIGO2), insulin receptor (INSR), structure specific recognition protein 1 (SSRP1), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha/ phosphoinositide-3-kinase regulatory subunit 1 (PIK3CA/PIK3R1), selectin E (SELE), complement C3 (C3d), (EDA2R) and karyopherin subunit beta 1 (KPNB1). Figure 2C illustrates the RPART tree used to differentiate between samples grouped as controls, TBI and partial-body irradiation. This was a more complicated calculation and required 5 different proteins to achieve the maximal result. The accuracy of this model using the training sample set was 95% and using the separate test sample series was 78% (Table 2). Figure 3 highlights the expression distribution (panel A) and relative ranking (panel B) of the top ranked proteins used to build model 3 including hyaluronan and proteoglycan link protein 1 (HAPLN1), insulin like growth factor 1 (IGF1), protein S (PROS1), protein kinase CAMP-activated catalytic subunit alpha (PRKACA), mitogen-activated protein kinase 14 (MAPK14), secreted phosphoprotein 1 (SPP1), cytochrome P450 family 3 subfamily A member 4 (CYP3A4), with Fig. 3C depicting the RPART tree used to distinguish between controls and partial exposure to the brain, gut or lung samples. The overall respective accuracies of this model using the training sample set was 88% and using the separate test sample series was 55% (Table 2).
FIG. 1.
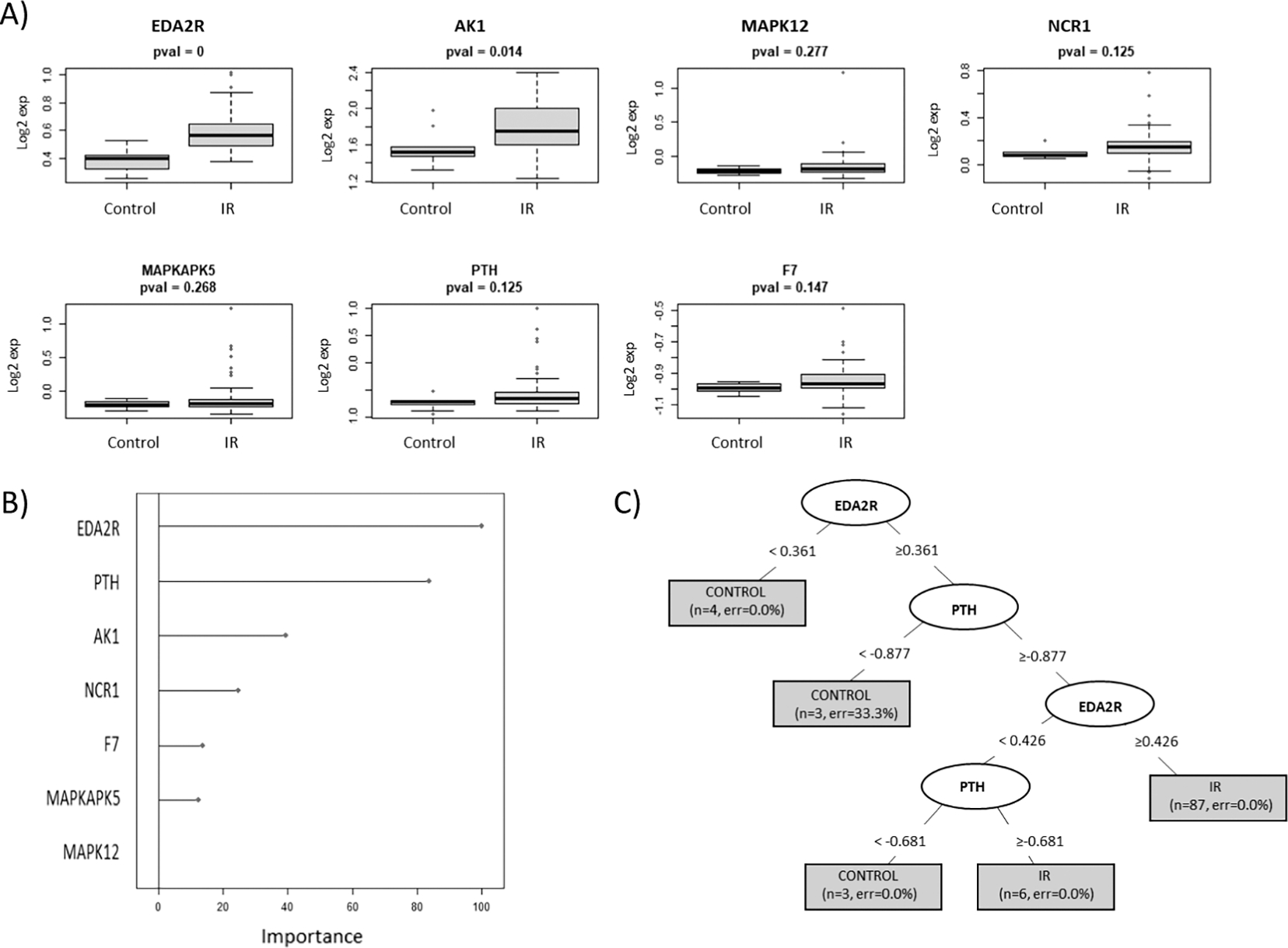
RPART model 1. This model series developed using recursive partitioning tree (RPART) methodology used a test and training sample cohort separated into controls vs. irradiated (all pooled irradiated samples to include TBI and partial-body irradiation to the brain, lung and gut). Panel A: Depicts the expression distribution of the top ranked selected proteins used to build model 1; panel B: the “Importance” value showing the variables with maximum influence on the classification of the cohort groups within the RPART tree; panel C: summary of the tree showing root, nodes and leaves. The tree is illustrated with the split in important variables to show classification (the shaded box included number of samples and class error rate).
TABLE 2.
Prediction Matrix Model Summary
| RPART model 1 prediction matrix | |||||||||
|
| |||||||||
| Training data set |
Test data set |
||||||||
| Controls | Irradiation | Controls | Irradiation | ||||||
|
| |||||||||
| Controls | 9 | 1 | Control | 5 | 6 | ||||
| Irradiation | 0 | 93 | Irradiation | 5 | 84 | ||||
| Accuracy | 0.99 | Accuracy | 0.89 | ||||||
| RPART model 2 prediction matrix | |||||||||
|
| |||||||||
| Training data set |
Test data set |
||||||||
| Controls | Partial | TBI | Controls | Partial | TBI | ||||
|
| |||||||||
| Controls | 7 | 2 | 0 | Controls | 5 | 2 | 0 | ||
| Partial | 2 | 73 | 0 | Partial | 5 | 63 | 8 | ||
| TBI | 0 | 1 | 18 | TBI | 0 | 7 | 10 | ||
| Accuracy | 0.95 | Accuracy | 0.78 | ||||||
| RPART model 3 prediction matrix | |||||||||
|
| |||||||||
| Training data set |
Test data set |
||||||||
| Controls | Brain | Gut | Lung | Controls | Brain | Gut | Lung | ||
|
| |||||||||
| Controls | 7 | 1 | 2 | 0 | Controls | 5 | 4 | 4 | 1 |
| Brain | 2 | 35 | 1 | 1 | Brain | 5 | 23 | 2 | 5 |
| Gut | 0 | 0 | 17 | 1 | Gut | 0 | 2 | 10 | 5 |
| Lung | 0 | 1 | 1 | 16 | Lung | 0 | 4 | 5 | 7 |
| Accuracy | 0.88 | Accuracy | 0.55 | ||||||
Notes. This includes summarized data of the overall prediction accuracies of each model and the specific sample cohort classifications in the associated confusion matrices. Numbers represent the actual number of samples analyzed (each taken from a different animal) in each type of radiation exposure cohort to power the study. Accuracy values state the percentage of correct sample classifications with the individual group accuracies representing the class error rate and the overall accuracy values representing the overall error rate. Total-body irradiated (TBI), ”Partial” includes all partial-body irradiation to the brain, lung and gut, and “Irradiation” refers to all pooled irradiated samples to include TBI and partial-body irradiation to the brain, lung and gut.
FIG. 2.
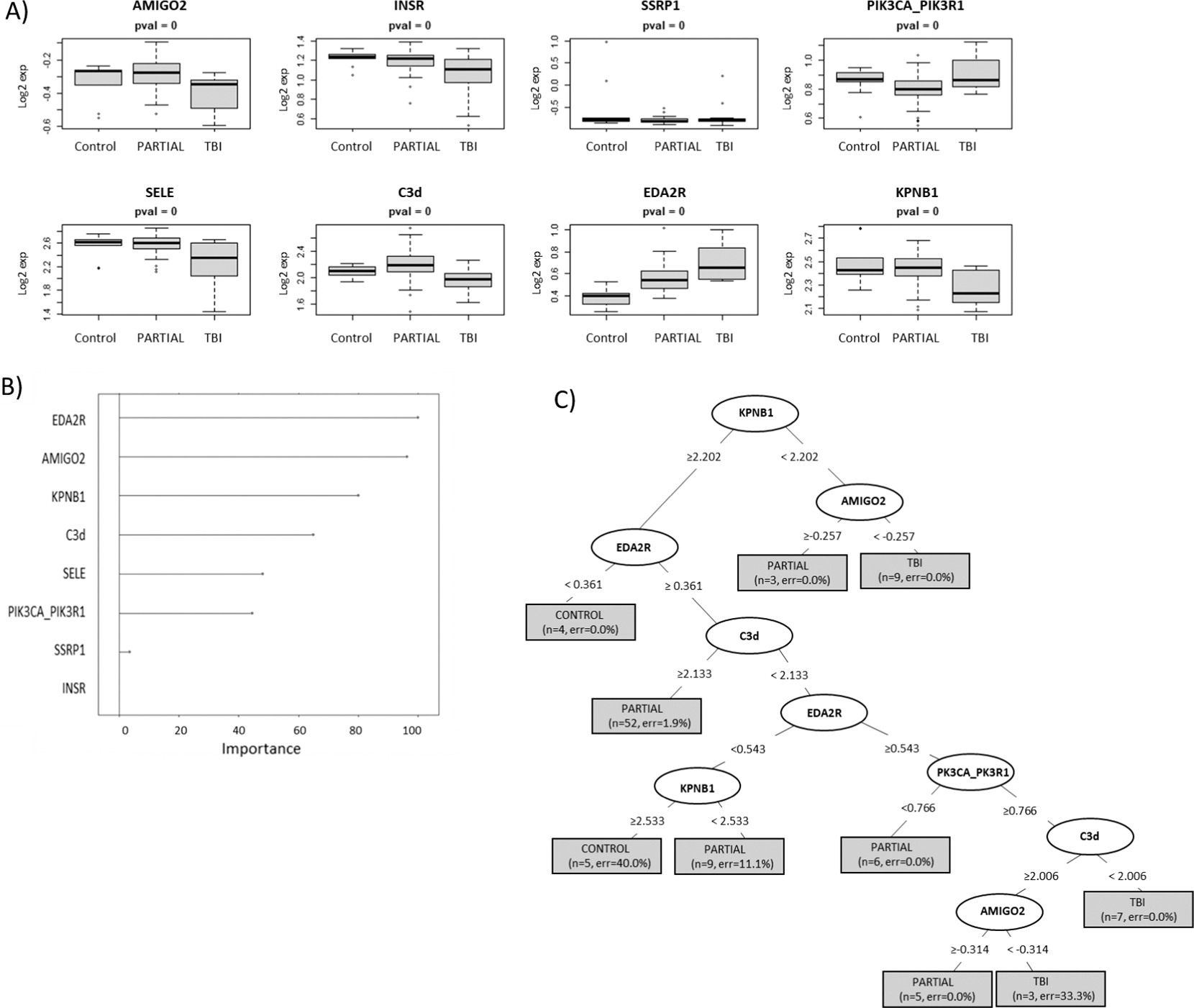
RPART model 2. This model series developed using recursive partitioning tree (RPART) methodology used a test and training sample cohort separated into controls vs. TBI vs. partial-body irradiation (with all organ specific partials pooled within the partials group). Panel A: Depicts the expression distribution of the top ranked selected proteins used to build model 1; panel B: the “Importance” value showing the variables with maximum influence on the classification of the cohort groups within the RPART tree; panel C: summary of the tree showing root, nodes and leaves. The tree is illustrated with the split in important variables to show classification (the shaded box included number of samples and class error rate).
FIG. 3.
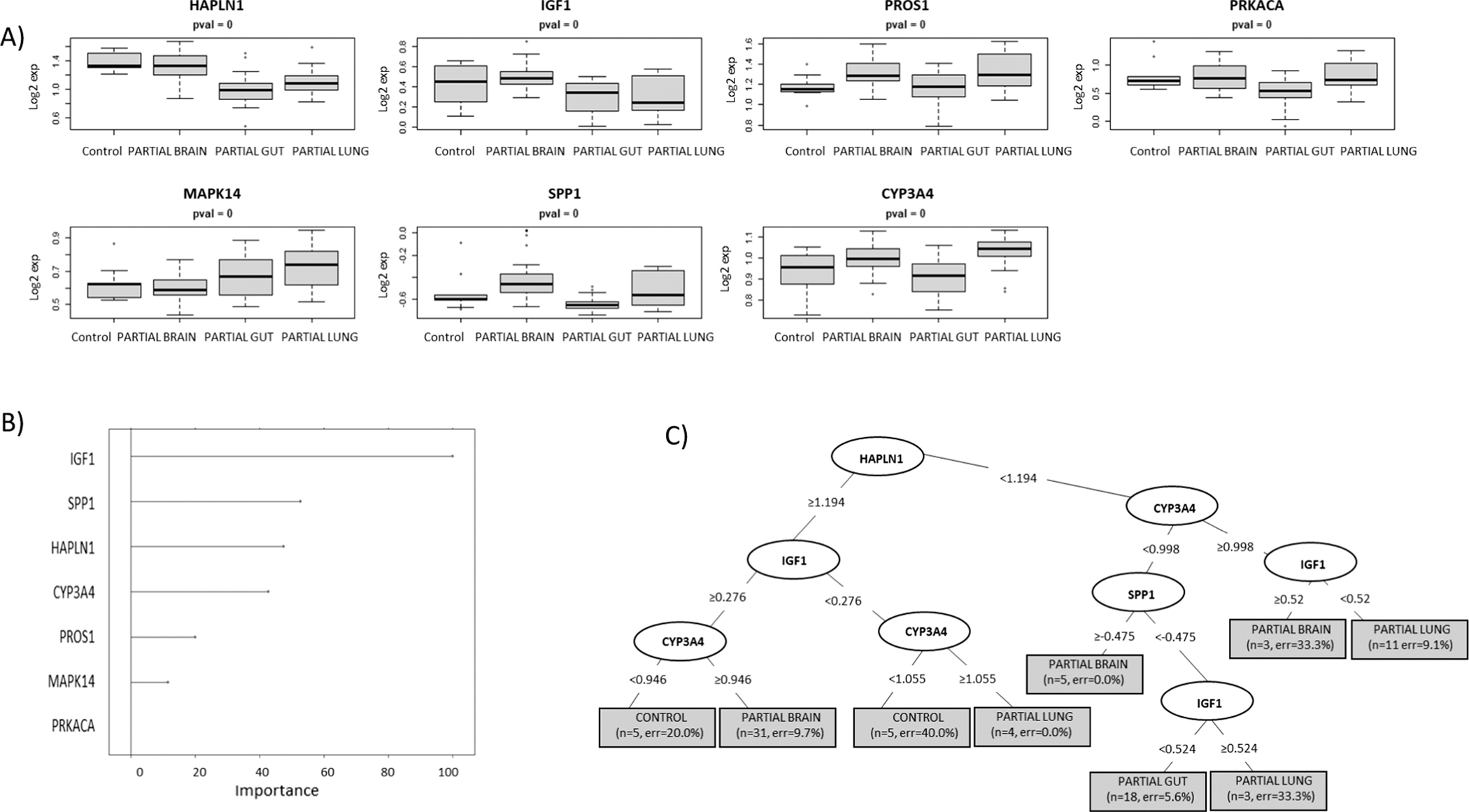
RPART model 3. This model series developed using recursive partitioning tree (RPART) methodology used a test and training sample cohort separated into controls vs. each individual organ specific partial-body irradiation separately (brain, gut and lung). Panel A: Depicts the expression distribution of the top ranked selected proteins used to build Model 1; panel B: the “Importance” value showing the variables with maximum influence on the classification of the cohort groups within the RPART tree; panel C: summary of the tree showing root, nodes and leaves. The tree is illustrated with the split in important variables to show classification (the shaded box included number of samples and class error rate).
Table 2 summarizes the overall prediction accuracies of each model and the specific sample cohort classifications in the associated confusion matrices. This data shows that though relatively high prediction accuracies of 89% and 78% were achieved for the test samples in models 1 and 2, respectively, both of these models’ algorithms misclassified 50% of the control samples into the irradiated cohort in the test group. For model 3, which sought to discriminate between controls and each of the partial exposure of brain, gut and lung, 50% of the control samples were again misclassified into one of the irradiated categories in the test group and the individual predictive accuracies were 70%, 48% and 39% for brain, gut and lung, respectively (Table 2). To improve on these models, other statistical methodologies for model building were also explored using a linear discriminant analysis (LDA) approach. The findings from these respective LDA model series 1–3 however, were comparable with the RPART approach and did not have significantly improved predictive accuracies. These findings are summarized in Supplementary Figs. S2–S4 (https://doi.org/10.1667/RADE-22-00074.1.S1).
DISCUSSION
This study sought to develop model algorithms specific for prediction of partial-body exposure of the brain, lung and gut in the event of radiological or nuclear incidents. As in such scenarios radiation exposure will be inherently heterogeneous, model building included uniform total-body exposure as well as partial-body exposure for comparison. Across models 1–3, each with a unique sample cohort distribution, the individual predictive accuracies within each sample subset were more illustrative than the overall prediction accuracies of the relative strength and weakness of each respective model. In model 1 and model 2 the overall predictive accuracies were higher than some the individual cohort subset predictive accuracies and in model 3 the overall predictive accuracy was lower than some of the individual cohort predictive accuracies. All the models performed poorly at differentiating control samples from partial-body exposure samples. This reflects the challenges of using biomarkers to differentiate between samples from unirradiated animals and samples from irradiated animals to relatively small percentages of body mass, as has been previously reported.
Further, the algorithms for model 3 which misclassified half of the control samples, classified them as partial brain exposure, representing the smallest percentage of body mass exposure within the partial brain, gut and lung exposures. This may correlate with overall physiological responses of injury to radiation exposure, as the partial brain exposure represent the smallest percentage of body mass exposed among the partial-body exposures in this study and may correlate with the least injury response as measured by plasma proteomic profiling. This illustrates the difficulty of differentiating changes in proteomic expression profiles between unirradiated and small body mass exposures, even with the benefit of a highly powered proteomic analysis with the use of ~1,300 proteomic targets. Misclassifications between partial lung and partial gut may be due to overlap in the exposure field due to the physiological proximity of these organs to each other in the body i.e., a partial gut exposure may include the lower edge of the lung and vice versa and represent real world scenarios where partial-body exposure will be heterogeneous and not confined to a specific organ.
The panels of proteins selected for each model type proved unique for each model except ectodysplasin A2 receptor (EDA2R), which was selected for both model 1 and model 2. This protein was also found to be significantly associated with TBI in our previously published controls vs. TBI model and was similarly associated with both model 1 and model 2 in the current study which included the same TBI sample cohort (14). Though different specific proteins were found to the best predictors of exposure in each model cohort, there was similarity in the functional physiological pathways which these proteins represent between the models. Model 1 and model 3 included members of the MAP kinase family which is involved with cellular stress and pro-inflammatory cytokine signaling and several proteins related to insulin regulation were found in model 1: INSR and PIK3CA/PIK3R1 and in model 3: IGF1. The various proteins in these panels represent a range of cellular processes and stress response pathways and may be useful predictors of radiation exposure. Though certain proteins may exhibit expression changes due to preexisting medical conditions or physical trauma, none of the candidate proteins are involved in the acute phase response, which is ideal for development of a mass screening tool for radiation exposure when combined injury may be present. Our previous work using multivariate approaches to development of biodosimetry algorithms has also demonstrated that inclusion of multiple biomarkers representing a range of physiological pathways strengthen dose prediction algorithms when challenged with more diverse sample cohorts and different radiation exposure profiles (12, 25).
Overall, these findings show that proteomic profiling can be a useful tool for development of predictive algorithms of radiation exposure, but that allowances will need to be made for the inherent physiological limitations of distinguishing between minor body exposures vs. the unexposed. Ideally, injury specific biomarkers for individual organs could be used to characterize organ specific injury due to radiation exposure, but no robust organ specific singleplex biomarkers were identified in this ~1,300 panel. Use of the new Somalogic 7K plex panel may provide a more highly powered proteomic platform to further improve these radiation exposure diagnostic algorithms.
Though all exposures to radiation in the field will essentially be partial-body exposures, some will be more significant in percentage of body mass exposed, and total received dose than others. From an immediate field application perspective, exposure models based on TBI should be used for initial triage purposes to segregate individuals who have received significant radiation injury vs. the unexposed, or those receiving a partial-body exposure of less severity. Subsequently, other diagnostic algorithms could be applied to further differentiate between unexposed and those who have received less significant partial-body exposure. In both scenarios, diagnostics which could identify injury to specify organs would improve clinical decision making in medical management of radiation injury and further studies to identify organ specific biomarkers of radiation exposure are needed. Great strides have been made in the development of biodosimetry diagnostics for management of large-scale radiation casualties but challenges for practical application in the field still remain.
Supplementary Material
Fig. S1. Workflow of bioinformatics methods for multivariate model building normalized RFU values from training and test data were converted to z-score and log2 scale. For each model, filtering was performed by either student’s t test or Anova depending on the comparison groups. To prevent overfitting the prediction model, selective features were limited to 30. If the initial filter gave features greater than 30, a second robust feature selection was performed using the gradient boosting method followed by the elastic net method. The resulting features from each model were trained and independently validated with test data using 2 independent prediction methods, a generalized linear discriminant analysis (LDA) model and a tree-based model of recursive partitioning and regression tree (RPART). The number of selective features is shown following different filters in parenthesis. Generalized boosted regression (GBM) models function in R.
Fig. S2. LDA model 1. This model series developed using a linear discriminant analysis (LDA) approach used a test and training sample cohort separated into controls vs. irradiated [all pooled irradiated samples to include total-body irradiation (TBI) and partial-body irradiation to the brain, lung and gut]. Panel A: Depicts a histogram of the distribution of training (left) and test (right) sample sets showing the linear discriminant (LD) coefficients on dimension 1 that gave maximum separation and panel B summarizes the prediction accuracies of each sample set and the specific sample cohort classifications in the associated confusion matrices.
Fig. S3. LDA model 2. This model series using a linear discriminant analysis (LDA) approach used a test and training sample cohort separated into controls vs. total-body irradiation (TBI) vs. partial-body irradiation (with all organ specific partials pooled within the partials group). Panel A: Depicts a histogram of the distribution of training (left) and test (right) sample sets showing the linear discriminant (LD) coefficients on dimension 1 that gave maximum separation and panel B summarizes the prediction accuracies of each sample set and the specific sample cohort classifications in the associated confusion matrices.
Fig. S4. LDA model 3 This model series using a linear discriminant analysis (LDA) approach used a test and training sample cohort separated into controls vs. each individual organ specific partial-body exposure separately (brain, gut and lung). Panel A: Depicts a histogram of the distribution of training (left) and test (right) sample sets showing the linear discriminant (LD) coefficients on dimension 1 that gave maximum separation and panel B summarizes the prediction accuracies of each sample set and the specific sample cohort classifications in the associated confusion matrices.
ACKNOWLEDGMENTS
This research was supported in part by funding from the Radiation and Nuclear Countermeasures Program, #Y2-OD-0332-01 NIAID, by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, #ZIA SC 010373. The authors would also like to thank the Center for Human Immunology at the Clinical Center at the National Institutes of Health and Brian Sellers for their invaluable scientific and technical assistance with the Somalogic SOMAscan platform.
REFERENCES
- 1.Prasanna PG, Blakely WF, Bertho JM, Chute JP, Cohen EP, Goans RE, et al. Synopsis of partial-body radiation diagnostic biomarkers and medical management of radiation injury workshop. Radiat Res. 2010; 173(2):245–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sullivan JM, Prasanna PGS, Grace MB, Wathen LK, Wallace RL, Koerner JF, et al. Assessment of biodosimetry methods for a mass-casualty radiological incident: medical response and management considerations. Health Phys. 2013; 105(6):540–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostheim P, Haupt J, Herodin F, Valente M, Drouet M, Majewski M, et al. miRNA expression patterns differ by total- or partial-body radiation exposure in baboons. Radiat Res. 2019; 192(6):579–88. [DOI] [PubMed] [Google Scholar]
- 4.Ostheim P, Haupt J, Schüle S, Herodin F, Valente M, Drouet M, et al. Differentiating total- or partial-body irradiation in baboons using mRNA expression patterns: A proof of concept. Radiat Res. 2020; 194(5):476–84. [DOI] [PubMed] [Google Scholar]
- 5.Shirley BC, Knoll JHM, Moquet J, Ainsbury E, Pham N-D, Norton F, et al. Estimating partial-body ionizing radiation exposure by automated cytogenetic biodosimetry. Int J Radiat Biol. 2020; 96(11):1492–503. [DOI] [PubMed] [Google Scholar]
- 6.Meadows SK, Dressman HK, Daher P, Himburg H, Russell JL, Doan P, et al. Diagnosis of partial body radiation exposure in mice using peripheral blood gene expression profiles. PLoS One. 2010; 5(7):e11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horn S, Barnard S, Rothkamm K. Gamma-H2AX-based dose estimation for whole and partial body radiation exposure. PLoS One. 2011; 6(9):e25113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shuryak I, Turner HC, Perrier JR, Cunha L, Canadell MP, Durrani MH, et al. A high throughput approach to reconstruct partial-body and neutron radiation exposures on an individual basis. Sci Rep. 2020; 10(1):2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blakely WF, Sandgren DJ, Nagy V, Kim S-Y, Sigal GB, Ossetrova NI. Further biodosimetry investigations using murine partial-body irradiation model. Radiat Prot Dosimet. 2014; 159(1–4):46–51. [DOI] [PubMed] [Google Scholar]
- 10.Sproull M, Kramp T, Tandle A, Shankavaram U, Camphausen K. Serum amyloid A as a biomarker for radiation exposure. Radiat Res. 2015; 184(1):14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sproull M, Avondoglio D, Kramp T, Shankavaram U, Camphausen K. Correlation of plasma FL expression with bone marrow irradiation dose. PLoS One. 2013; 8(3):e58558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sproull M, Kramp T, Tandle A, Shankavaram U, Camphausen K. Multivariate analysis of radiation responsive proteins to predict radiation exposure in total-body irradiation and partial-body irradiation models. Radiat Res. 2017; 187:251–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blakely WF, Bolduc DL, Debad J, Sigal G, Port M, Abend M, et al. Use of proteomic and hematology biomarkers for prediction of hematopoietic acute radiation syndrome severity in baboon radiation models. Health Phys. 2018; 115(1). [DOI] [PubMed] [Google Scholar]
- 14.Sproull M, Shankavaram U, Camphausen K. Novel Murine Biomarkers of radiation exposure using an aptamer-based proteomic technology. Front Pharmacol. 2021; 12:633131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohloff JC, Gelinas AD, Jarvis TC, Ochsner UA, Schneider DJ, Gold L, et al. Nucleic acid ligands with protein-like side chains: modified aptamers and their use as diagnostic and therapeutic agents. Mol Ther Nucleic Acids. 2014; 3(10):e201–e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Team RC. R: A language and environment for statistical computing. Vienna, Austria: RFoundation for Statistical Computing; 2015. [Google Scholar]
- 17.Fisher RA. The use of multiple measurements in taxonomic problems. Ann Eugen. 1936; 7(2):179–88. [Google Scholar]
- 18.Breiman L, Friedman JH, Olshen RA, Stone CJ. Classification and regression trees. Belmont, CA: Wadsworth; 1983. [Google Scholar]
- 19.Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010; 33(1):1–22. [PMC free article] [PubMed] [Google Scholar]
- 20.Algamal ZY, Lee MH. Regularized logistic regression with adjusted adaptive elastic net for gene selection in high dimensional cancer classification. Comput Biol Med. 2015; 67:136–45. [DOI] [PubMed] [Google Scholar]
- 21.Zou H, Hastie T. Regularization and variable selection via the elastic net. J Royal Stat Soc: Series B (Statistical Methodology). 2005; 67(2):301–20. [Google Scholar]
- 22.Friedman JH. Greedy Function approximation: A gradient boosting machine. Ann Stat. 2001; 29(5):1189–232. [Google Scholar]
- 23.Bylesjö M, Rantalainen M, Cloarec O, Nicholson JK, Holmes E, Trygg J. OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J Chemomet. 2006; 20(8–10):341–51. [Google Scholar]
- 24.Therneau TM, Atkinson EJ, editors. An Introduction to Recursive Partitioning Using the RPART Routines. 2015.
- 25.Sproull M, Shankavaram U, Camphausen K. Comparison of Proteomic biodosimetry biomarkers across five different murine strains. Radiat Res. 2019; 192(6):640–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Workflow of bioinformatics methods for multivariate model building normalized RFU values from training and test data were converted to z-score and log2 scale. For each model, filtering was performed by either student’s t test or Anova depending on the comparison groups. To prevent overfitting the prediction model, selective features were limited to 30. If the initial filter gave features greater than 30, a second robust feature selection was performed using the gradient boosting method followed by the elastic net method. The resulting features from each model were trained and independently validated with test data using 2 independent prediction methods, a generalized linear discriminant analysis (LDA) model and a tree-based model of recursive partitioning and regression tree (RPART). The number of selective features is shown following different filters in parenthesis. Generalized boosted regression (GBM) models function in R.
Fig. S2. LDA model 1. This model series developed using a linear discriminant analysis (LDA) approach used a test and training sample cohort separated into controls vs. irradiated [all pooled irradiated samples to include total-body irradiation (TBI) and partial-body irradiation to the brain, lung and gut]. Panel A: Depicts a histogram of the distribution of training (left) and test (right) sample sets showing the linear discriminant (LD) coefficients on dimension 1 that gave maximum separation and panel B summarizes the prediction accuracies of each sample set and the specific sample cohort classifications in the associated confusion matrices.
Fig. S3. LDA model 2. This model series using a linear discriminant analysis (LDA) approach used a test and training sample cohort separated into controls vs. total-body irradiation (TBI) vs. partial-body irradiation (with all organ specific partials pooled within the partials group). Panel A: Depicts a histogram of the distribution of training (left) and test (right) sample sets showing the linear discriminant (LD) coefficients on dimension 1 that gave maximum separation and panel B summarizes the prediction accuracies of each sample set and the specific sample cohort classifications in the associated confusion matrices.
Fig. S4. LDA model 3 This model series using a linear discriminant analysis (LDA) approach used a test and training sample cohort separated into controls vs. each individual organ specific partial-body exposure separately (brain, gut and lung). Panel A: Depicts a histogram of the distribution of training (left) and test (right) sample sets showing the linear discriminant (LD) coefficients on dimension 1 that gave maximum separation and panel B summarizes the prediction accuracies of each sample set and the specific sample cohort classifications in the associated confusion matrices.


