Abstract
The budding yeast Saccharomyces cerevisiae has been the principal organism used in experiments to examine genetic recombination in eukaryotes. Studies over the past decade have shown that meiotic recombination and probably most mitotic recombination arise from the repair of double-strand breaks (DSBs). There are multiple pathways by which such DSBs can be repaired, including several homologous recombination pathways and still other nonhomologous mechanisms. Our understanding has also been greatly enriched by the characterization of many proteins involved in recombination and by insights that link aspects of DNA repair to chromosome replication. New molecular models of DSB-induced gene conversion are presented. This review encompasses these different aspects of DSB-induced recombination in Saccharomyces and attempts to relate genetic, molecular biological, and biochemical studies of the processes of DNA repair and recombination.
The processes by which damaged DNA is repaired and the mechanisms of genetic recombination are intimately related. Much of what we know about these events has come from studies of the yeast Saccharomyces cerevisiae, for which the development of new molecular biological and genetic approaches has made it possible to appreciate the many different pathways used by eukaryotic cells. The study of these processes in a simple, unicellular eucaryote has the obvious advantages of the ease of manipulation of DNA sequences (all of which are now precisely known) and the possibility of studying specific repair and recombination events induced synchronously in a large proportion of cells. Equally important is the growing conviction that the processes that one can study with relative ease in yeast are identical in most respects to the ways in which human cells repair DNA damage and generate genetic diversity. The expanding list of human genetic diseases associated with defects in DNA metabolism makes it especially important in understanding how these processes occur. Moreover, defining these mechanisms has taken on added importance in the quest to develop more efficient mechanisms of gene targeting and gene replacement in mammalian cells.
Recombination can be initiated by several types of DNA damage. Single-strand DNA (ssDNA) lesions may result during DNA replication or during repair, after UV irradiation or the alkylation or cross-linking of DNA bases, or from intermediates of type I topoisomerases. Double-strand breaks (DSBs) can appear as a consequence of ionizing radiation, by mechanical stress, by endonucleases, or by replication of a single-stranded nicked chromosome. The repair of DNA resulting from nucleotide excision repair, base excision repair, and other types of damage affecting one strand of the DNA duplex has been well reviewed elsewhere (37, 136, 390). This review will concentrate on the types of recombination created by DSBs. DSBs are the sole instigators of recombination in meiotic cells and are a major factor in recombination in mitotic cells, although the origin of spontaneous mitotic recombination remains unknown. In addition to its relevance as a fundamental biological process, DSB-mediated recombination is the basis of gene modification in yeast and in other eukaryotes.
We classify DSB repair events into two major categories. Homologous recombination events of several types are characterized by the need for the damaged DNA strands to base pair with a homologous partner, where the extent of interaction generally involves hundreds of nearly perfectly matched base pairs. In contrast, illegitimate or nonhomologous repair events can seemingly join ends of DNA with no complementary base pairs at the junction, although in general it turns out that most of these events make use of a very small number of base pairs (microhomology). In yeast, nonhomologous repair events generally occur at significantly lower frequencies than homologous events, so that one could argue that some of the distinctions between homologous and nonhomologous repair are artificial, especially since homologous recombination can occur with surprisingly short homologous regions, albeit at low frequency. However, these types of events are distinctly different, because they have different genetic requirements.
For a complete overview of recombination and DSB repair in yeast and other organisms, we also direct the reader to several other reviews that have recently appeared (220, 235, 347, 377, 413, 461, 462). Also, the present review deals essentially with the budding yeast S. cerevisiae, but there are more and more available data about recombination in the fission yeast Schizosaccharomyces pombe, an organism that seem to behave more like higher eukaryotes. Several recent reviews have appeared that will give the reader a good overview (133, 264, 359).
Some Initial Thoughts about Homologous Recombination
Yeast, like mammals, has several ways to repair DSBs by homologous recombination mechanisms. These different pathways exist in a competitive hierarchy. Thus, in a wild-type cell, 90% of the repair events may proceed by a particular mechanism, but when that mechanism is eliminated, 90% of the cells do not fail to repair the broken chromosome (which is indeed a fatal condition). Instead, other apparently less efficient mechanisms will process and repair most of the DSBs. In some cases this can be demonstrated by a change in the kinetics of the repair process or by the appearance of a different product. Consequently, some mutations that profoundly affect the normally predominant repair pathway may not result in a severe phenotype.
Another problem to keep in mind is that the uncertainty principle enunciated for quantum physics applies to the measurement of recombination: attempts to know the exact location of an event may change its kinetics or its outcome. One often introduces heterologies into a region while attempting to monitor the extent of a repair tract, to find if there was crossing over, or to ensure that all the events were initiated at a specified location. In at least some cases, the introduction of these markers alters the spectrum of possible products.
Perhaps less philosophically daunting but no less problematical is the simple fact that different laboratories have used different assay systems to evaluate recombination and repair and that these systems do not all behave identically. Apparent differences in results may reflect the fact that alternative mechanisms of recombination are favored or excluded in these systems. In other cases the results appear to reflect significant differences in DNA sequence or chromatin structure. Moreover, unlike studies in Escherichia coli, where nearly all laboratories study descendants of a single progenitor strain, many different strains of S. cerevisiae are in circulation, some of which carry weak mutations in genes that influence recombination. Nevertheless, there is reasonably good agreement among different research groups studying the same types of events.
GENERAL STRATEGIES TO STUDY DNA RECOMBINATION AND REPAIR
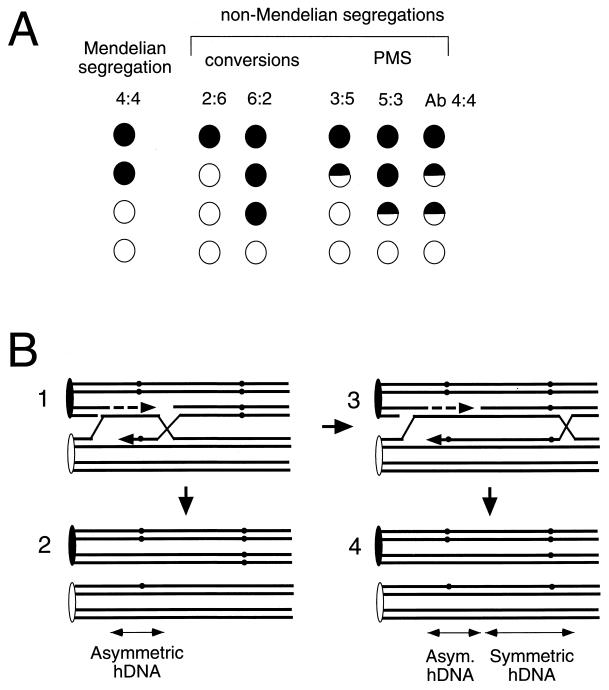
Although many of the fundamental ideas about recombination originated from studies of Drosophila, the analysis of fungi provided the opportunity to recover all four products of meiosis. This led to the discovery of non-Mendelian segregation of markers, both gene conversions and postmeiotic segregations, that provided the first insight into the molecular mechanisms of eukaryotic recombination. Although important observations were made with Neurospora crassa and Ascobolus immersus, it is Saccharomyces cerevisiae that has emerged as the model system of choice in studying both meiotic and mitotic recombination. The pioneering studies of Fogel and Mortimer with yeast (127–129, 193), as well as those of Rossignol et al. with Ascobolus (418) and Stadler with Neurospora (465), and insights from Hastings (176) and Whitehouse (549) established the basic framework by using naturally arising alleles in a variety of biosynthetic and pigment genes. However, it was the development of gene-targeting methods (183, 358, 421, 447) that allowed the creation of defined alterations of the genome and a refinement of these genetic approaches. The mechanism of gene targeting itself became the object of scrutiny, and much of our current thinking comes from the analysis of mitotic recombination of transformed DNA. More recently, two additional developments have provided new ways to investigate molecular events in greater detail. It is now possible to examine physical intermediates of recombination and thus to test the predictions of current recombination models. Moreover, in vitro biochemical studies of strand invasion, the central step of most recombination events, have provided direct tests of the role of proteins identified by genetic studies. The characterization of specific recombination proteins is discussed in a later section.
Genetic Assays
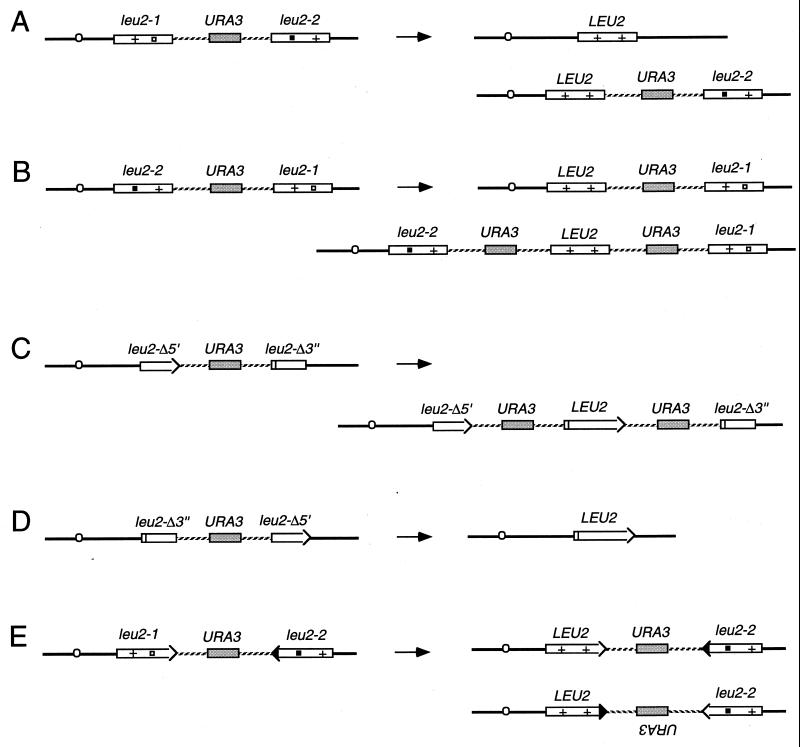
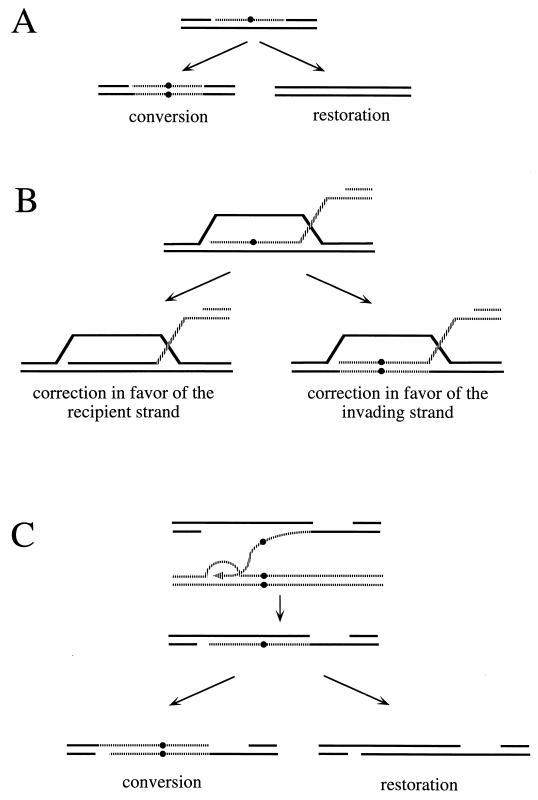
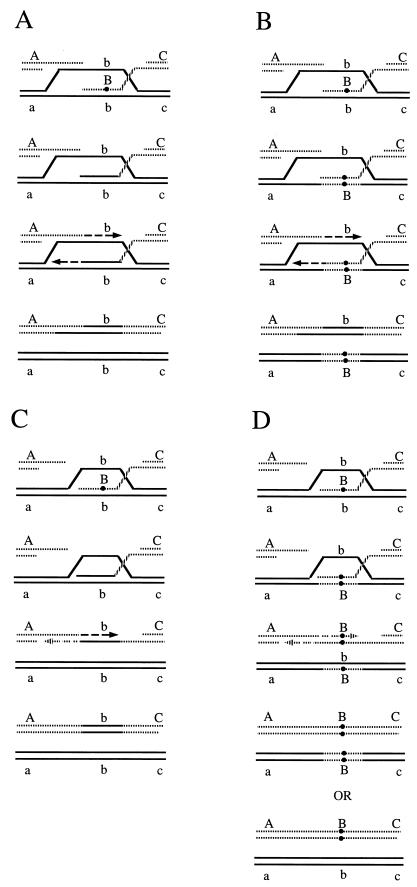
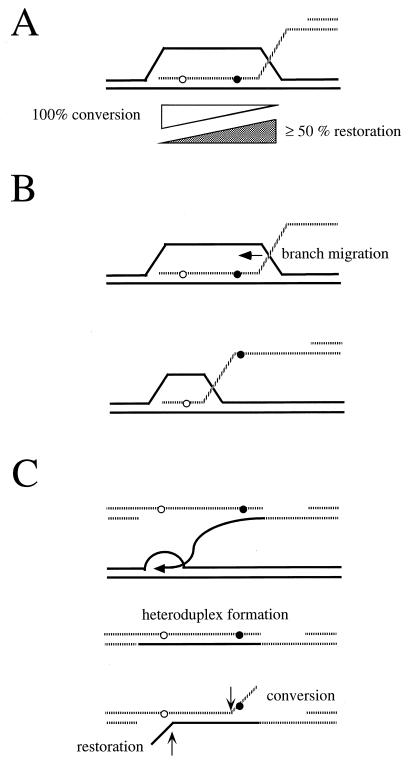
Recombination can be assessed genetically, for example by measuring gene conversion between heteroalleles of an easily scored nutritional marker (129, 448) (Fig. 1A and B). As initially defined in meiosis, a gene conversion is a nonreciprocal transfer of genetic information from one homologous chromosome to another. Although one might imagine that heteroallelic recombination could occur by a precise reciprocal exchange of DNA in the interval between the two alleles (Fig. 1C), studies of the fate of the alleles in diploids where a prototrophic cell has arisen show that more than 90% of the events are actually gene conversions, in which only one of the two participating alleles is unchanged (165, 268, 329). The literature is unfortunately replete with false distinctions between “gene conversions” (by which the authors mean gene conversions not associated with an exchange of flanking markers) and “reciprocal recombination” (by which the authors generally mean gene conversions associated with crossing over). Interchromosomal recombination can also be assessed by the loss of heterozygosity of nutritional markers. This can occur by gene conversion between two alleles of a scored marker (Fig. 1D) or by a reciprocal exchange anywhere between the marker and the centromere during the G2 stage of the cell cycle, followed by chromosome segregation (Fig. 1E).
FIG. 1.
Genetic assays for recombination. (A) Selection of heteroallelic recombination. Here, a functional LEU2 gene results from a conversion event not associated with crossing over. (B) Gene conversion associated with crossing over. (C) The LEU2 recombinant gene results from a reciprocal crossover event without any detectable gene conversion. (D) Assay for loss of heterozygosity. This results in Leu− cells. The event described here corresponds to a gene conversion with crossing over. (E) Loss of heterozygosity can also occur by reciprocal exchange between the centromere and the marker during the G2 stage, followed by segregation in the next cell division.
One can also assay specifically for crossovers. The simplest system involves a pair of alleles, distal to which are other markers that can be used to measure crossing over (Fig. 1E). Prototrophic recombinants can then be assessed for the arrangement of these flanking markers. If crossover occurs in G2, then—depending on the segregation of chromosomes—half of the prototrophic diploids should be homozygous for one or the other distal marker (Fig. 1E). If recombination occurs in G1, crossing over will be genetically silent, although the two heterozygous distal markers will have exchanged positions. It is also possible to detect such events if there are closely enough linked polymorphisms that can be analyzed on genomic blots or by inducing a chromosome loss event (to see which markers become coordinately lost) or sporulating the diploids and determining the genetic linkage of these markers to other markers on the chromosome.
Genetic exchange between (identical) sister chromatids cannot usually be detected; thus, to detect crossovers between sister chromatids, one must examine events involving tandem repeats that give rise to unequal sister chromatid exchange (USCE). The first such assays monitored the fate of an inserted gene into the repeated ribosomal DNA (rDNA) array (375, 495). One can score the appearance of sectored colonies, where one half is derived from a cell that lost the inserted marker. In some of these cases, the opposite sector has two copies of the marker and thus most probably arose from USCE. It should be noted, however, that in many instances only one copy of the marker is found in the opposite sector, which can be explained by intrachromosomal recombination or by gene conversion between misaligned rDNA repeats (144).
A second approach to look for USCE events is to examine prototrophic recombinants between tandem repeats of heteroalleles. Especially when the orientation of alleles is such that a simple “pop-out” event will not produce a prototrophic recombinant, a significant fraction of prototrophs prove to be triplications (Fig. 2B) resulting from USCE (199, 236). If the markers are arranged as shown in Fig. 2A, prototrophs containing a single copy of the repeat could be generated by USCE but could also arise from intrachromatid recombination. Another way to study USCE is to use partially overlapping truncated genes (119, 120). Here, two overlapping parts of a gene are inserted in an orientation that will not permit intrachromatid crossing over to produce a heritable recombined, complete gene. In contrast, USCE will yield a full-length gene (Fig. 2C).
FIG. 2.
Intrachromosomal recombination between direct or indirect repeats. (A) Recombination between two direct repeats. Here, Leu+ cells can arise by a deletion or a pop-out event that removes all the intervening sequences (top) or by a simple gene conversion of one of the two repeats (bottom). Both kinds of event involve only one chromatid. (B) Another case of recombination between direct repeats. Leu+ cells can arise by simple gene conversion (top). However, because of the orientation of the mutations, deletion is unlikely to result in a functional LEU2 gene, but a LEU2 gene can result from a USCE. Crossing over between one repeat from one chromatid and a second repeat from the other chromatid will result in a triplication (bottom). (C) Another case of USCE. The proximal leu2 copy is deleted at the 5′ end, and the distal one is deleted at the 3′ end. USCE can reconstitute a LEU2 copy. (D) Selection of deletion events between truncated direct repeats. (E) Selection of recombination between indirect repeats. Deletions cannot occur. Obtaining Leu+ cells depends on gene conversion events not associated or associated with crossing over (top and bottom, respectively).
Intrachromatid recombination can also be examined by using direct or inverted repeats, either at a chromosomal location or on a plasmid. Direct-repeat assays are commonly used. In one such assay, two copies of a gene, one truncated at the 3′ end and the other truncated at the 5′ end but with a homologous region, will recombine to restore gene function (452) (Fig. 2D). Alternatively, the deletion can also be scored by the loss of a marker in the interval between direct repeats (239, 503, 504) (Fig. 2D). Inverted repeats can also be used, either when recombination between heteroalleles will yield a functional gene (Fig. 2E) or when recombination will cause an inversion of the region in between the repeats to produce a complete gene (551) (Fig. 2E). However, recent results suggest that some apparently intrachromatid events might result from interchromatid gene conversion (71). In addition, recombination can be studied between sequences on two plasmids or between a plasmid and a chromosome.
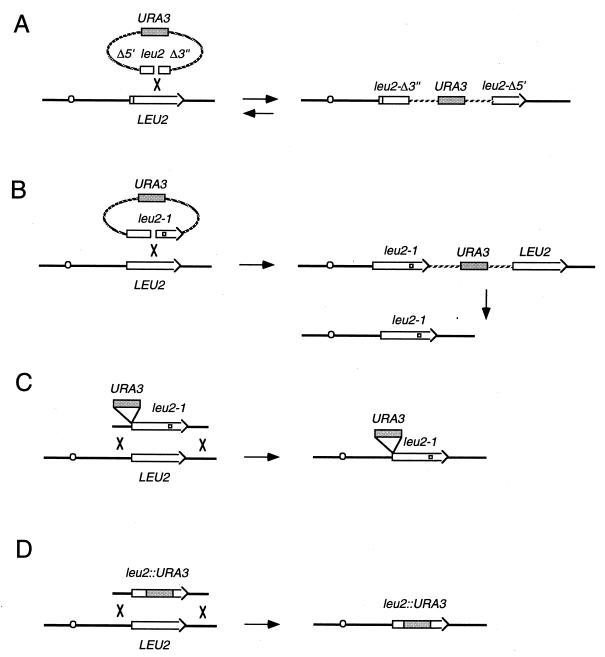
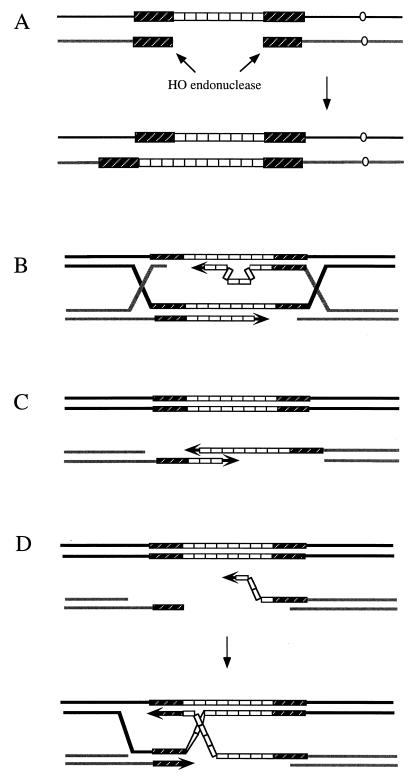
These approaches take advantage of the ease with which DNA sequences can be inserted into yeast by the introduction of a linearized fragment of transforming DNA. Two general methods are used to create new alleles on a chromosome. In the first (357, 358), a circular plasmid containing an in vitro-modified gene and a selectable marker such as URA3 is targeted by a DSB created by a restriction endonuclease within a region of homology (Fig. 3A and B). The integration creates a tandem duplication of nonidentical sequences. The process can effectively be reversed by screening for the loss of URA3, either randomly (447) or, more conveniently, by selecting such events on medium containing 5-fluoroorotic acid, whose presence is lethal to cells with a functional URA3 gene (43). Some of the pop-out events that eliminate the URA3 gene and other plasmid sequences will leave behind the modified DNA sequences in place of the original sequences (Fig. 3B). In this way it is possible to introduce known alleles into a specific strain, so that all derivatives are isogenic. Other genes, such as LYS2, can be selected for positively and negatively (69), but it is essentially the URA3 marker which has been used for this kind of procedure. Alternatively, one can introduce modified sequences in one step by transformation with a linearized fragment containing a selectable marker (421). This kind of recombination event is often called “ends-out” recombination, since the DSB ends point to opposite directions (Fig. 3C and D). Ends-out recombination is often used to disrupt or delete genes (Fig. 3D) or to insert other, adjacent unselected sequences into a novel location (Fig. 3C). Ends-out transformation with linearized, modified DNA is the basis of most knockout strategies in all organisms, about which more is discussed later.
FIG. 3.
Methods to create new alleles. (A) Gene disruption by recombination with a plasmid containing a leu2 copy deleted at both the 5′ and 3′ ends. This results in a duplication, where both copies are mutated. This duplication can be obtained by selecting with the URA3 marker. (B) The pop-in/pop-out method. This two-step method requires first the integration of a plasmid with a mutated copy of leu2 (pop-in, selected for with the URA3 marker) and then the excision of the plasmid (pop-out, selected for as loss of URA3) leaves only one copy of leu2, which can be the original one or the mutated one (the case shown here). (C) One-step gene replacement. Some of the Ura+ transformants have also integrated the mutation in leu2 and become Leu−. (D) One-step method of gene knockout. Most of the central part of the gene is replaced by the selectable URA3 marker.
Principles of Physical Assays
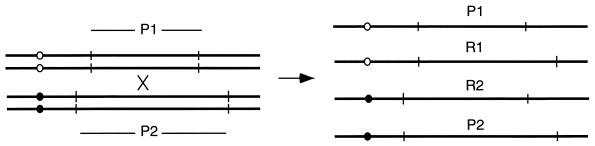
A powerful tool in studying mechanisms of recombination is the physical analysis of DNA to detect recombination in the absence of any easily scored genetic marker. For example, in a diploid in which the two homologous chromosomes have polymorphic restriction sites flanking a region of interest, it is possible to identify cases in which crossing over has occurred by the appearance of novel restriction fragments generated by reciprocal exchange (Fig. 4). This method also permits an analysis of the kinetics of recombination by isolating samples at intervals after the initiation of a recombination event (49, 50).
FIG. 4.
New restriction endonuclease fragments produced by reciprocal recombination. The appearance of these new restriction fragments can be monitored by Southern blotting at various time points of meiosis. Vertical lines stand for the restriction sites used in the diagnostic assay. P1 and P2, parental restriction fragments; R1 and R2, recombinant fragments resulting from crossing over.
An advantage of the physical assay is that it can give information not only about the products of recombination but also about intermediate steps. Knowledge of the structure of the recombinant molecule can help in constructing models, but these models will become tangible only when their individual steps can be physically monitored.
One may also examine the extent to which recombination can occur even under conditions where cells are unable to complete recombination or even to continue growing. For example, one could ask if recombination can be completed when cells are arrested at different stages of the cell cycle or after the elevation of the cells to the restrictive temperature of a conditional-lethal mutation. Thus, it is possible to carry out what we have termed in vivo biochemistry, i.e., to infer the biochemical roles of specific enzymes by determining which steps in recombination are affected by the inactivation of that enzyme and where the mutants become blocked (160).
Very recently, physical analysis of DNA has been dramatically applied to examine the position of meiotic crossovers along the entire genome in a single experiment. High-density oligonucleotide arrays, capable of detecting more than 3,700 allelic differences between two divergent yeast strains, were hybridized with the DNA from each of the four segregants of a meiotic tetrad, allowing Winzeler et al. (553) to map the position of every crossing over and many gene conversions not accompanied by reciprocal exchange.
Synchronous induction of DSBs.
Analysis of the kinetics of recombination, to discover the time of appearance of intermediates and products, is dependent upon the ability to initiate recombination synchronously in a large population of cells. This occurs naturally in meiotic cells, where DSBs arise at specific hot spots in a few percent of all chromatids. In mitotic cells, synchronous initiation of recombination can be accomplished by the induction of a site-specific endonuclease. Two such systems have been developed in yeast. The HO endonuclease recognizes a degenerate target of 22 bp (345) and normally cleaves only one site in the entire yeast genome: the mating-type (MAT) locus. Constructs in which the HO gene is fused to a galactose-inducible promoter have made it possible to express HO simply by adding galactose to cells grown on lactate, glycerol, or raffinose (203), three carbon sources that do not repress the galactose-inducible promoter. A second endonuclease is I-SceI, normally encoded and expressed only in yeast mitochondria to facilitate the movement of a mobile intron, ω (LSU.I) (82, 101, 102, 290). A synthetic version of this gene, replacing codons whose usage is different in mitochondria and the cytoplasm, was constructed, again under the control of a galactose-inducible promoter (381). A 45- to 90-min induction of either HO or I-SceI leads to the cleavage of a significant fraction (30% for I-SceI, 100% for HO) of target sites. HO endonuclease is also turned over rapidly, so that no activity remains 30 min after the end of the induction period (547). Once a DSB has been created, intermediate steps in recombination, along with the appearance of final products, can be identified.
Physical monitoring of HO-induced mitotic gene conversion: the example of MAT switching.
A paradigm for mitotic recombination is HO endonuclease-induced recombination, and more specifically, MAT switching. During switching (Fig. 5), the Ya- or Yα-specific sequences at MAT that specify the mating type are replaced by sequences copied from two unexpressed donor sequences, HMLα and HMRa (reviewed in references 161, 163, 164, and 471). The initiating event is a DSB catalyzed by the HO endonuclease at the Y/Z junction of the recipient MAT locus.
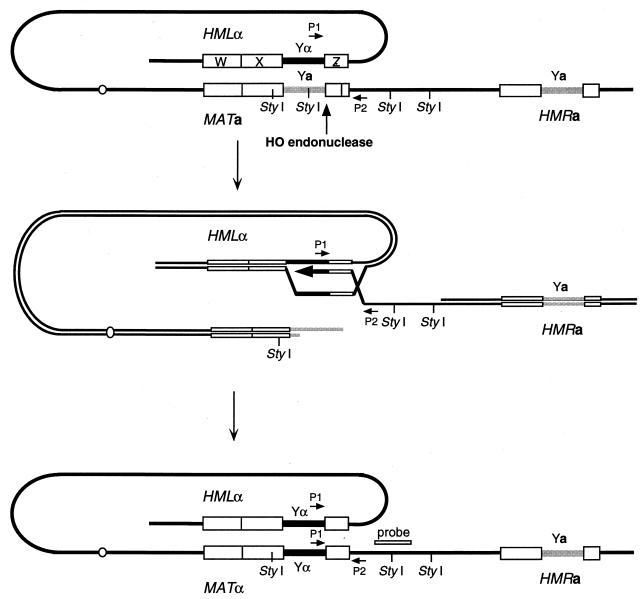
FIG. 5.
Mating-type switching in yeast. The MAT locus, which determines the a or α mating type, switches by gene conversion, using one of two silent cassettes, HMRa and HMLα, located on the same chromosome. The gene conversion event is initiated by the HO endonuclease, which creates a DSB at the border of the varying region (called Ya or Yα, according to the genotype). MAT, HMR, and HML share homology on both sides of the Y regions (W, X, and Z regions). Both strands of DNA are shown in the middle diagram. A PCR assay has been used to detect DNA synthesis during MAT switching. Using oligonucleotides P1 and P2, one cannot obtain any PCR products in MATa cells, but a PCR product appears when the cell switches to MATα, as soon as DNA synthesis initiated from the Z region of MAT proceeds to copy Yα from MATα.
The absence of a StyI site in Yα sequences and its presence in Ya sequences makes it easy to monitor MAT switching by the appearance of a novel StyI restriction fragment when MATa switches to MATα. Surprisingly, the time from the appearance of the HO-cut MATa locus until the appearance of the MATα is about 1 h, suggesting that there are a number of very slow steps in the process (547). Some of these steps may be slow because of a need for new protein synthesis (422). The kinetics of recombination appear to be cell cycle independent, since similar time courses were observed when synchronized cells were induced at different times in the cell cycle or in G1-arrested cells (85, 393). It remains possible that HO-induced events are different from the natural situation, where MAT switching occurs only in mother cells and only in the G1 phase of the cell cycle, because HO expression is tightly regulated. However, the same slowness of recombination is also seen in meiotic recombination (discussed below).
It is also possible to detect intermediates of recombination. In the time course of MAT switching, monitored on denaturing gels, one observes the transient appearance of one or more higher-molecular-weight DNA restriction fragments. These proved to be the result of extensive 5′-to-3′ degradation of the HO-cut end, so that one or more StyI sites were single stranded and could not be cut by the restriction endonuclease (547). Processing of DSB ends later proved to be a general feature of homologous DSB repair, with the resulting single-strand DNA being the pivotal intermediate in all homologous recombination pathways (see below). The extent of the resection is frequently more than 1 kb, far beyond the 320 bp of homology shared by MAT and its donor HML in the Z region. This process generates a long 3′-ended tail that can invade a homologous template. When later steps in recombination are prevented, for example when there is no homologous sequence with which MAT can recombine, 5′-to-3′ degradation appears to continue down the chromosome unabated (262). The rate of degradation can be estimated to be 1 to 2 nucleotides per s. The progress of 5′-to-3′ degradation can also be followed on dot blots by using strand-specific DNA probes (262, 474). Although the 5′-ended strand is extensively resected, there is little or no degradation of the 3′-ended strand (397, 474, 479, 547).
Strand invasion itself has not been assayed in vivo. No assay is yet available to detect the initial D-loop created by strand invasion, although by analogy to other processes such as the initiation of transcription, it should be possible to do so by reacting the displaced template strand in vivo with the single-strand-specific reagent KMnO4 (146). Once the invading strand assembles a DNA polymerase that begins to copy the template, it is possible to detect this early intermediate step by a sensitive PCR assay. Using one primer specific for the donor template (Yα) and one distal to the recipient (Fig. 5), it is possible to detect as little as 20 nucleotides of new DNA synthesis (547). This intermediate appears 15 to 30 min after HO cleavage but still 30 min prior to the completion of switching, which can be measured both on Southern blots and by a second PCR that detects the joining of Y donor sequences to those proximal to MAT. That 30 min elapses between the initial strand invasion-replication step and the completion of switching again argues that there are several slow steps in this recombination process.
MAT switching is perhaps unusually restrictive as a model for the study of DSB repair, because of the largely inaccessible chromatin structure of the donors. Consequently, additional studies have been carried out by inserting a 24- to 117-bp HO recognition site into other genes (245, 342, 344, 346, 371, 395, 422), allowing the characterization of recombination events occurring between chromosomal sequences (19, 124, 372, 394, 395, 422) and between chromosomal and plasmid sequences (342, 368, 487). One substrate that has received a great deal of attention in our laboratory is a centromere-containing plasmid carrying two copies of the E. coli lacZ gene, in either direct or inverted orientation (18, 123, 124, 195, 197, 366, 422). In general the results of MAT switching and these other HO-induced recombination systems have been similar. Analogous constructs involving I-SceI and an 18-bp recognition site have also been developed (117, 381).
DIFFERENT MECHANISMS OF HOMOLOGOUS DSB REPAIR
There are at least three different mechanisms of homologous recombination that can be used to repair a chromosomal DSB in mitotic yeast cells: gene conversion, single-strand annealing, and break-induced recombination. A fourth mechanism can account for the integration of foreign DNA into a homologous chromosomal locus. In addition, there are very probably two pathways of gene conversion.
Gene Conversion
Relationship between gene conversion and crossovers.
Gene conversion is defined as a nonreciprocal transfer of genetic information from one molecule to its homologue. Usually this occurs between two alleles of a gene (Fig. 1 and 2); however, gene conversions can embrace many contiguous genes, including the entire distal part of a chromosome arm. Gene conversions were initially defined in meiosis, where one could observe non-Mendelian segregation of alleles. The pioneering work of Mortimer and Fogel (329) established several key characteristics of gene conversions, including the idea that gene conversions exhibited polarity, whereby the probability that a nearby marker would be coconverted along with a specified gene-converted marker decreased with the distance between the markers. In meiosis, gene conversion tracts are on average 1 to 2 kb (46, 95, 293, 348, 478). In mitosis, some gene conversions cover very short distances (216, 305, 342) while others extend for hundreds of kilobases (see below).
A second key observation by Mortimer and Fogel (126, 127, 329) was that gene conversions were intimately associated with crossing over. Conversely, if one selects for crossovers, one will often find an associated gene conversion in meiosis (46, 492) as well as in mitosis (71, 551), and it is now taken for granted that most if not all crossovers arise from the same transfers of DNA strands that cause gene conversion. However, some crossovers will not be associated with a detectable gene conversion, either because the interval where crossing over occurs does not contain allelic differences between the homologous sequences or because intermediates that could give rise to a gene conversion can also be restored, with no detectable change in genotype. Willis and Klein (551) devised a system (similar to the one shown in Fig. 2E), in which mitotic intrachromosomal crossovers could be directly selected by inversion of a segment flanked by inverted repeats. Inversion led to a fortuitous increase in expression of a kanamycin resistance gene. By analyzing the pattern of gene conversion of markers in the repeats, Willis and Klein concluded that about 50% of crossovers had an associated, detectable gene conversion and that crossing over was more likely to occur when the gene conversion tract was long. As we noted above, crossovers without a detectable event are likely to have arisen by the same mechanism but with the original genotype restored.
The proportion of gene conversions that are accompanied by crossing over is much greater in meiosis than in mitosis. We will defer a discussion of the control of meiotic crossing over to a later section. The data suggest to us that meiotic recombination is fundamentally similar to mitotic recombination but modified in several ways, most notably in the proportion of gene conversions associated with crossing over. In mitosis, only a relatively small fraction of gene conversions are crossover associated, ranging from almost 0% to about 20%; however, in some special cases half of all gene conversions are associated with exchanges of flanking markers.
In transformation experiments where a plasmid, cut within a rDNA gene, was repaired by gene conversion with the chromosomal rDNA, Orr-Weaver and Szostak (357) observed that 50% of the repair events were associated with crossing over. This 50% ratio was also observed for recombination initiated by an HO-induced DSB in a centromeric plasmid containing two inverted repeats of the E. coli lacZ gene, one of which carried a recognition site for the site specific HO endonuclease (423). Similar results were found upon cleavage by another site-specific endonuclease, I-SceI (381). However, subsequent studies involving transformation or HO-induced DSB repair have generally found that the proportion of DSB repair events accompanied by crossing over was substantially less than 50% (380, 423). For example, when the same pair of inverted copies of the lacZ sequences are integrated into a chromosome, the proportion of HO-induced gene conversions accompanied by crossing over drops to 5% (423). In general, the frequency of crossover-associated events is low when one or both of the interacting molecules is a chromosomal locus, as opposed to plasmid-borne sequences. In this regard, the rDNA sequence first studied by Orr-Weaver et al. (357, 358) may be exceptional. In general, mitotic gene conversions, measured after selection for prototrophs between heteroalleles on homologous chromosomes, exhibit crossover frequencies of 10 to 20% (112, 165, 252).
Another major factor in the proportion of mitotic gene conversions associated with crossing over seems to be the nature of the recombining sequences. We mentioned above that Rudin et al. (423) observed 50% of crossovers among HO-induced recombination events between two inverted repeats of lacZ. When recombination occurs in a similar plasmid, but between two copies of the MAT sequence, crossovers are very rare (3%) (80, 475). Crossovers are also rare during normal MAT switching, when the chromosomal MAT locus is converted by one of the two silent cassettes, HMR and HML (234). Crossover events accompanying MAT switching would result in lethal chromosome deletions, and one can hypothesize that the MAT sequences might have evolved some intrinsic feature that inhibits crossovers.
Mechanisms of gene conversion.
Gene conversions can be explained by two different families of models, both of them supported by substantial experimental data. They are described below.
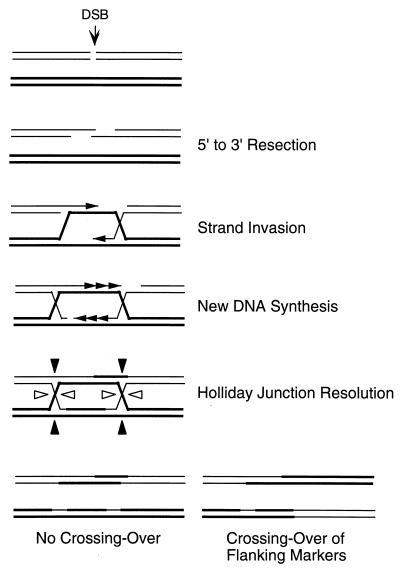
(i) DSB repair model of Szostak et al.
Because gene conversions are strongly associated with crossovers, molecular models were designed to account for this fact, culminating in the DSB repair model first suggested by Resnick and Martin (401) and later elaborated by Szostak and coworkers (479, 494) (Fig. 6). These models were based on earlier conceptions by Holliday (185) and by Meselson and Radding (312).
FIG. 6.
DSB repair model of Szostak et al. (494). DSB formation is followed by 5′-to-3′ resection of the ends. The resulting 3′ ends are recombinogenic and can invade a homologous template, to initiate new DNA synthesis. Two HJs are formed and are resolved independently by cutting the crossed (open arrowhead) or noncrossed (closed arrowhead) strands, resulting in crossover or noncrossover products.
Experimental support for recombination models initiated by a DSB came initially from transformation experiments carried out by Orr-Weaver et al. (357, 358), which established several important characteristics of DSB repair. First, a linearized plasmid, cut in a region of homology to a chromosome, could be repaired and integrated (by crossing over) into the chromosome. Second, a plasmid carrying a gap in the homologous region would be repaired by the filling in of all of the sequences present in the template but missing in the gapped broken molecule. Third, when the transforming cut plasmid contained a functional origin of DNA replication, so that noncrossover products (a recircularized plasmid) could also be recovered, approximately 50% of the repair events were accompanied by crossing over (leading to the integration of all the plasmid into the chromosome) and 50% yielded a repaired, autonomously replicated plasmid.
The initial version of the Szostak et al. model assumed that DSBs were resected on both strands to create large gaps flanked by rather short regions of single-stranded DNA that could invade a homologous template and initiate DNA repair. The version shown in Fig. 6 reflects current thinking and is based on observations from several laboratories that the 3′ ends of both meiotic and mitotic DSBs are not resected while the 5′ ends of the DNA can be chewed back for very long distances, often more than 1 kb (64, 262, 397, 479, 547). The 3′ ends are presumed to invade an intact homologous template in a manner similar to the way RecA-catalyzed strand exchange occurs in E. coli (105, 246, 461). The 3′ ends of the invading strands can then act as primers for the initiation of new DNA synthesis.
This process would lead to the formation of two Holliday junctions (HJs), four-stranded branched structures whose alternative resolution allows the formation of the crossover products. HJs theoretically can be cleaved by a resolvase by cutting either the two noncrossed strands or the two crossed strands (Fig. 6). If both HJs are cleaved in the same way, gene conversion will not be associated with crossing over, but if the noncrossover strands of one HJ are cleaved while the crossover strands on the second are cut, there will be an exchange of flanking markers. An equal number of crossovers and noncrossovers would be predicted if HJs were resolved randomly, which seems to be the case in the experiments of Orr-Weaver and Szostak (357). Lower frequencies of crossing over have often been explained by a bias in HJ resolution. In practice, it may be that an isomerization of the HJ is required, as proposed by Meselson and Radding (312), so that the crossed strands are always cleaved (32).
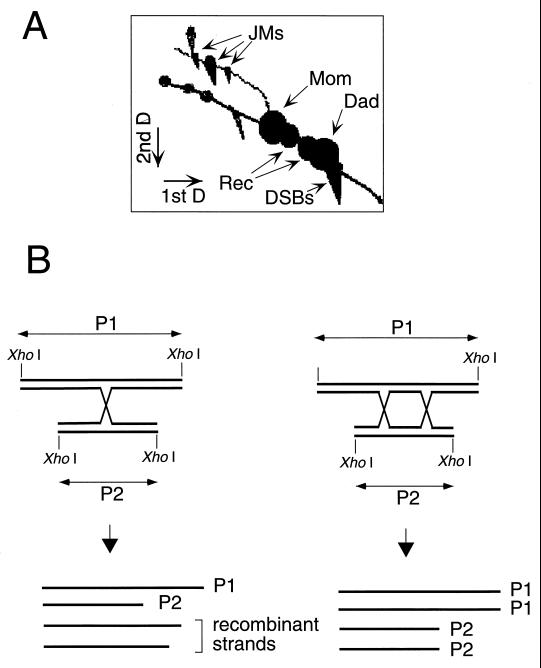
Recently, the existence of branched intermediates was physically demonstrated during meiotic DSB repair, using a two-dimensional (2D) gel electrophoresis procedure (83, 437–439). Schwacha and Kleckner could show that these structures indeed correspond to double Holliday junctions (437) (Fig. 7). Apparently, similar structures were also detected in the rDNA locus during vegetative growth (575).
FIG. 7.
Physical characterization of the double-HJ intermediate predicted by Szostak et al. (494). (A) Two-dimensional gel electrophoresis of DNA from meiotic cells at the pachytene stage. Restriction enzyme-digested DNA samples are run first slowly on a low-concentration (0.4%) agarose gel. The migration lane is then cut and inserted across a high-concentration (0.8 to 1.0%) agarose gel, for a second, quick migration. Various molecules or events can be identified by Southern blotting, such as parental molecules (Mom and Dad), recombinant molecules (Rec), DSBs, and JMs. There are three different JMs, corresponding to the Mom-Mom, Dad-Dad, and Mom-Dad (the bigger signal in the middle). D, dimension. This figure has been adapted from Fig. 1 of reference 439. (B) JMs are double Holliday junctions. The JMs corresponding to interchromosome recombination (Mom-Dad) can be extracted from the gel. These molecules are resolved into parental and recombinant molecules (437) by the RuvC resolvase, showing that they include HJs. The JMs can also be run on a denaturing electrophoresis gel, and the size and specific hybridization pattern of the strands will indicate if they are recombinant or nonrecombinant strands. The theoretical outcome expected for simple HJs is two parental strands and two recombinant strands (left). This was not observed (437, 438). Instead, only parental strands were observed, the expected outcome for double HJs (right).
The Szostak et al. model solved several problems that challenged the previous reigning model of Meselson and Radding (312). First, it accounted for the fact that the locus experiencing DNA damage (in this case a DSB) would generally be the recipient locus in a gene conversion. Second, the formation of two HJs allowed crossing over to occur both upstream and downstream of a site experiencing non-Mendelian segregation (128).
(ii) Synthesis-dependent strand annealing.
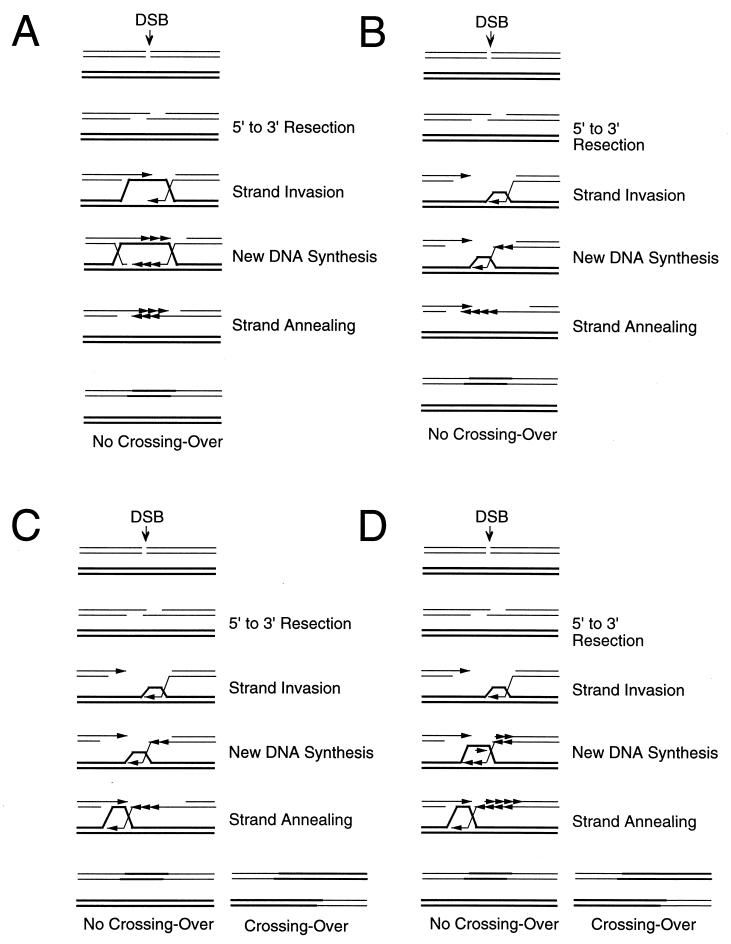
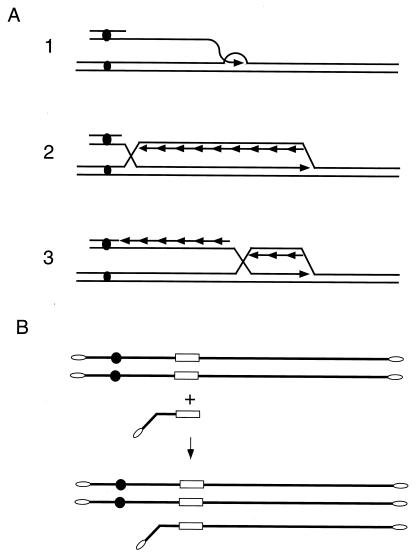
Because many mitotic gene conversions were infrequently associated with crossing over, a second family of gene conversion models emerged, beginning with those of Nasmyth (339) and Thaler and Stahl (499) and further elaborated by both Hastings (175) and McGill et al. (305). Similar alternative models appeared to explain results in other organism such as Drosophila (111, 150, 340), mammals (30), E. coli (250, 331), and Ustilago (121). The name we use for these kinds of mechanism, synthesis-dependent strand annealing (SDSA), was coined by Nassif et al. (340). The basic feature of these models is that the newly synthesized DNA strands are displaced from the template and returned to the broken molecule, allowing the two newly synthesized strands to anneal to each other. This could occur either because there are topoisomerases or helicases that actively dismantle the replication structure (305, 500) (Fig. 8A) or because the replication “bubble” remains small, with the newly synthesized strand being continuously unwound from its template (the bubble migration model) (130) (Fig. 8B). In both cases, DNA synthesis is conservative (all the newly synthesized sequences are on the same molecule) instead of semiconservative as in the Szostak et al. model. SDSA models were first designed to explain a lack of crossovers, but they received experimental backing from other observations that could best be explained by such a mechanism.
FIG. 8.
SDSA models. (A) Simple SDSA model. Both 3′ ends invade the template and initiate new DNA synthesis. (B) SDSA model with bubble migration. (C) SDSA model with crossing over. Following strand annealing of the second 3′ end with the displaced strand, a double HJ intermediate can occur. (D) Repair replication fork capture model. Strand invasion initiates both leading- and lagging-strand synthesis. Because of branch migration following DNA synthesis, this is an SDSA model.
First, in HO endonuclease-induced recombination between a donor and a recipient that contain polymorphic sites near the DSB, one can deduce the formation of heteroduplex DNA, with one strand from the donor and one from the recipient, by the appearance of a sectored colony derived from a single cell, each half of which is derived from one of the DNA strands of the heteroduplex-containing molecule. McGill et al. (305) showed, for mating-type (MAT) gene conversions, that such a heteroduplex was found only at the recipient locus. This was unexpected based on the predictions of the Szostak et al. model (494), where strand invasion should have left a region of heteroduplex DNA in the donor (Fig. 6). This result was more readily explained if the polymerase-extended invading strand was unwound to anneal with a second, newly synthesized strand that was copied from the donor sequence (Fig. 8A). Diagnostic events exhibiting post-repair segregation of markers are rare in wild-type cells, which are able to repair mismatched DNA. When mismatch repair is prevented by deleting the PMS1 gene, 85% of MAT conversions showed postswitching segregation of a marker only 8 bp from the end of the DSB (397). All the heteroduplex was found in the recipient locus. This observation also reinforces the conclusion that DSBs are rarely degraded on both strands to produce gaps, since a gap of 8 bp to the right of the DSB would have removed the possibility of detecting heteroduplex DNA.
SDSA models were also invoked to explain other observations not predicted by the model of Szostak et al. These also concern the location of heteroduplex regions. As noted above, the Szostak et al. model predicts the formation of two regions of heteroduplex that should always be on different chromatids. However, using alleles that form heteroduplexes that are poorly corrected by the mismatch repair proteins in meiosis, Porter et al. (384) and Gilbertson and Stahl (148) found very few such outcomes; instead, they found that both heteroduplex regions were found on a single chromatid, a result that can best be explained by SDSA models (Fig. 9). In addition, a number of DSB-induced gene conversions in both mitotic (342) and meiotic (148, 384) cells were found to be confined to markers on only one side of the DSB. These results are compatible with a strand invasion event where only one 3′ end of the DSB would form a heteroduplex with the template and then initiate DNA synthesis (Fig. 8B). Substantial heteroduplex formation would result from the annealing of the other 3′ end with the displaced strand (148, 384) or, as in an SDSA model, with the newly synthesized DNA (342) (Fig. 8B).
FIG. 9.
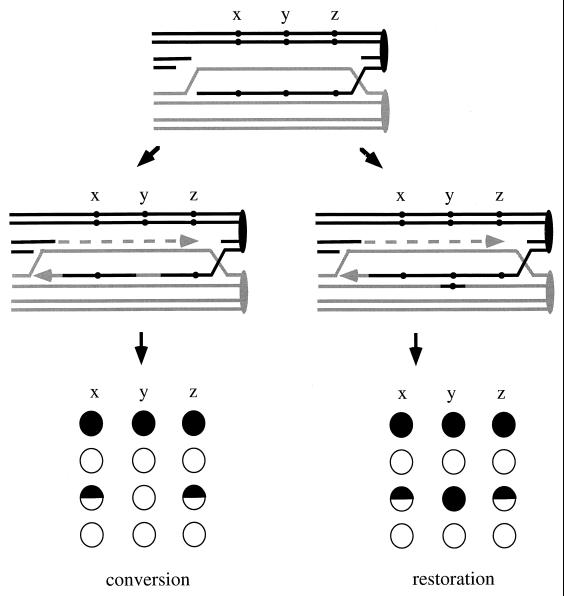
Some data in favor of SDSA models for meiotic recombination. Recombination can occur between two alleles differing by several mutations (A to C). These mutations can be on the same side of a meiotic DSB, as y and z, or on two different sides, as x and y or x and z. If these mutations result in high-PMS alleles, the revised DSB repair model of Szostak et al. (479, 494) predicts that y and z can be found in the same heteroduplex (D) and can be observed as two simultaneous PMS with the y and z alleles from one parent found in the same sector (F). In contrast, x will form a heteroduplex DNA on a different chromatid from y and z. In an SDSA model, y and z can also be found in the same heteroduplex (E). However, x can be found in another heteroduplex but on the same chromatid (E), resulting in simultaneous PMS with y and z, but now the x allele from one parent is found in the sector containing the y and z alleles from the other parent (G). This configuration, which is not predicted by the Szostak et al. model, has been observed by Porter et al. (384) and Gilbertson and Stahl (148).
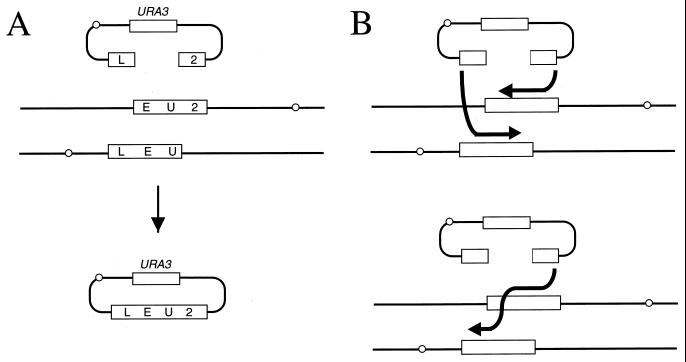
Further support for SDSA models is provided by studies showing that gene conversion can use two templates located on two different chromosomes. Silberman and Kupiec showed in a transformation experiment that a broken plasmid can acquire genetic information from two different loci on two different chromosomes (455). Pâques et al. (368) did a similar experiment, in which each end of a DSB on a plasmid was homologous to one of two overlapping truncated LEU2 genes on two different chromosomes (Fig. 10A). Gap repair to produce a complete LEU2 gene on the plasmid requires two separate strand invasion events and the subsequent annealing of DNA ends. The Leu+ recombinants resulting from this tripartite recombination occur with a frequency that is 2.5% of the frequency of regular gap repair (when the same HO-cut plasmid uses a single template for repair) (368). The absolute condition for this kind of repair event is that genetic information be copied from the two different templates and then assembled in the plasmid. This can occur only if newly synthesized sequences are unwound from their template and returned to the donor molecule. Two models are shown in Fig. 10B. One of these models is an avatar of the copy choice model, where genetic recombination is the consequence of template switching during DNA synthesis. In the other, each end of the DSB initiates DNA synthesis and then the newly synthesized strands are unwound from their templates and annealed.
FIG. 10.
Tripartite recombination. (A) A plasmid with a gapped copy of leu2 is gap repaired from two chromosomal templates. Genetic information must be recovered from two different chromosomes and assembled into the plasmid to create a complete LEU2 gene. (B) Two models to explain the production of a LEU2 gene. DNA synthesis can be initiated independently from both 3′ ends of the broken plasmid (top). Both newly synthesized strands are then unwound from the template and annealed. Alternatively, DNA synthesis can be initiated from one 3′ end, and template switching ensures the recovery of all the required LEU2 sequences (bottom). Then the newly synthesized strand anneals with the other 3′ end. In both cases, DNA synthesis is conservative: the newly synthesized DNA is unwound from its template.
Finally, several assays showed that sequence alterations occurring during DSB repair are found on the recipient molecule and not on the donor template. HO-induced recombination is accompanied by a high level of single-base-pair mutations (184, 472), which are almost always found in the recipient molecule. This distribution does not depend on the PMS1, MLH1, and MSH2 genes (304), which indicates that the absence of mutation in the donor template is not due to “restoration” by the mismatch repair machinery. HO-induced recombination also induces very frequent alterations in the copy number of a repeated locus (368) (Fig. 11). These alterations are nearly always found on the recipient molecule. These results also argue for a conservative mode of DNA synthesis during DSB repair, since both the single-base-pair substitution and the tandem repeat alterations associated with DSB repair are likely to be generated during the DNA synthesis step. Semiconservative DNA synthesis should let them appear on both donor and recipient molecules, but SDSA allows them to be confined to the recipient.
FIG. 11.
DSB-induced expansions and contractions of a tandem repeat. (A) To test directly if a DSB can induce rearrangements in tandem repeats in yeast, Pâques et al. (368) tested HO-induced gene conversions with a homologous donor sequence containing an intervening interval including 8 repeats of 375 bp. Perfect copying of the template should introduce the whole intervening repeated locus into the repaired molecule. However, only about half of the repaired chromosomes acquire an unmodified repeated array. The others have a variable number of repeats ranging from 1 to 13 copies. These frequent rearrangements are restricted to the repaired recipient molecule, with the donor template remaining unmodified. Similar result have been obtained with an artificial or real yeast 36-bp minisatellite locus (365) and with a microsatellite CTG locus (402). (B) The rearrangements may result from replication slippage occurring during the semiconservative kind of DNA synthesis predicted by Szostak et al. (494); however, the rearrangement would be expected to be found in the donor template as well as in the recipient. The clustering of the rearrangements in the recipient molecule can be better explained by an SDSA model. (C) In this SDSA model, both 3′ ends initiate DNA synthesis. The newly synthesized strands are then unwound from their template and annealed. Because of the redundant structure, there are many possibilities of annealing, resulting in expansions or contractions. (D) Another possibility is that the kind of DNA synthesis associated with SDSA (bubble migration for example) would easily generate slippage-like events. If DNA synthesis stops before the new strands overlap with the other 3′ end, reinvasion has to occur. Because of the redundant structure, there are many possibilities of reinvasion, which would be responsible for the expansions and contractions that are always found on the recipient molecule because the newly synthesized sequences return to the repaired molecule. Resolution by annealing can occur, but HJs also can be formed and lead to crossovers, as proposed in Fig. 8C. This last feature has the advantage of explaining why the infrequent crossover events found in this experiment were associated with tandem repeat rearrangements as often as were the noncrossover events.
(iii) Synthesis-dependent strand annealing with crossing over.
Gene conversion events not associated with crossing over can easily be explained by either the Szostak et al. model or the SDSA model. However, most formulations of SDSA do not allow for crossing over accompanying DSB repair; if this were the case, one might predict that outcomes characteristic of SDSA would not be found for gene conversions accompanied by crossing over. However, a version of SDSA that includes the possibility of crossing over has been suggested by Ferguson and Holloman (121). In this model, strand invasion is initiated by one end of the DSB and would proceed to copy across the template until one of two events occur. First, the newly synthesized strand may simply anneal with the second end, yielding a gene conversion without crossing over (Fig. 8B). Alternatively, the displaced D-loop created by the first strand may anneal with the second end, producing a single HJ that can be resolved with or without crossing over. We have subsequently proposed a similar model (368), wherein a double HJ would be formed instead of a single HJ (Fig. 8C). This idea of stabilizing the D-loop by annealing to the second end of the DSB was a feature of the mechanism proposed by Szostak et al., but there is a significant difference that might be used experimentally to distinguish between them: in the original DSB repair model the two HJs are found on either side of the DSB (494), while in our SDSA version, the two HJs are both on one side of the DSB (368). Crossovers could occur on either side of the DSB, depending on which end initiates DNA synthesis. We recognize that different positions of a double HJ could also result from branch migration of the HJs (437).
So far, only one experiment argues that SDSA associated with crossing over occurs in S. cerevisiae. We mentioned above that DSB repair induced frequent rearrangements in tandem repeats, nearly always in the recipient molecule (368). This accounts for noncrossover DSB repair events (where the donor and recipient molecules are clearly identifiable), which are the vast majority of the gene conversion events. However, some rare crossover events (about 5% of total) could also be found, and they were associated with tandem repeat rearrangements at the same frequency as noncrossover products were. Thus, SDSA may sometimes happen with crossing over. Figure 11D describes how SDSA could rearrange a tandem repeat and then allow crossover formation.
(iv) Repair replication fork capture.
In both the Szostak et al. DSB repair model and the standard version of SDSA, there seems to be a need only for leading-strand polymerization, primed by the two 3′ ends of the invading DNA strands. However, it would be possible for the invasion of one 3′-ended single strand to establish a modified replication fork, similar but not identical to the leading- and lagging-strand process of origin-dependent DNA replication (Fig. 8D).
This type of recombination-dependent, origin-independent DNA replication is also discussed in the next section to explain repair events that can copy all the way to a chromosome end. In the present context, we envision that this process is terminated when the repair-initiated replication fork is “captured” by the second end of the DSB. Data presented in a later section have led to the hypothesis that gene conversion requires both leading- and lagging-strand DNA synthesis (190). In this model, lagging-strand synthesis and conservative DNA synthesis are not incompatible: both newly synthesized strands can be returned to the recipient after synthesis has occurred. In the SDSA model proposed by McGill et al. (305), for example, this could be accomplished by a topoisomerase. An alternative view is that branch migration follows semiconservative DNA synthesis, as shown in Fig. 8D. This could be catalyzed by an enzyme complex similar to the RuvA and RuvB proteins in E. coli (546); it is noteworthy that ruvA and ruvB mutants have no noticeable recombination phenotype in the absence of UV radiation, except in recombination-dependent, origin-independent replication (241, 242).
Break-Induced Replication
As discussed above, a common view of gene conversion is that it involves short-patch events. However, this is not always the case, and several examples of very long conversion tracts have been reported in mitosis.
The first observation of such events was made by Esposito (112), who found gene conversion tracts that apparently extended from the TRP5 locus to the ADE5 locus on chromosome VII. Since the entire yeast genome has now been sequenced, we now know that ADE5 and TRP5 are 400 kb apart. Coconversion of the TRP5 and LEU1 loci, 25 kb apart on chromosome VII, was also observed at a frequency 1,200-fold higher than if those events were independent (152), with coconversion also of intervening markers (153). Such high levels of coconversions could be explained in two ways. First, the conversion tracts might be very large. Second, gene conversion could occur in a subset of cells that are especially prone to recombination, and these cells would convert any locus at a very high rate. This second hypothesis was ruled out by Golin and Tampe (154), who showed that only genetically linked loci were converted at high frequencies. These authors also showed that the coconversion frequency decreased with the distance between two loci, down to a certain distance (35 kb), where it no longer depended on distance. Thus, they defined two processes of coconversion, a distance-dependent one and (for very long distances) a distance-independent one. However, the authors did not propose any fundamentally different recombination model to explain the two kinds of events, perhaps because, at that time, recombination events occurring in yeast were explained in term of the Meselson and Radding or Szostak et al. models, involving the formation of heteroduplexes of extensive lengths.
Similar asymmetrical inheritance of distal markers was seen when recombination was initiated by the HOT1 sequence (528). Coconversions involving up to 70 kb accounted for 90% of the conversion events at one locus. Actually, the same study also showed that the rate of coconversion was 60% even for spontaneous recombination (not induced by HOT1). Although these events are gene conversion events (i.e., asymmetrical inheritance), Voelkel-Meiman and Roeder (528, 529) invoked a replicative model of DSB repair, analogous to the recombination-induced replication of phage T4 (130, 330) or of the E. coli chromosome (242). It is this kind of model that has since been favored to explain very long tracts of gene conversion. Three different versions are shown in Fig. 12A.
FIG. 12.
BIR. (A) Three models to explain BIR. In model 1, the 3′ end of the DNA fragment (or broken chromosome) invades the template and initiates synthesis of one DNA strand by bubble migration. The complementary strand has to be synthesized later. In model 2, another (more likely) possibility is that the 3′ end initiates both leading- and lagging-strand synthesis, in a true replication fork. Here, DNA synthesis is semiconservative. The branched structure has then to be resolved by an endonuclease. In model 3, the initiation of a true replication fork is compatible with conservative DNA synthesis provided that branch migration follows the progression of the replication fork (bottom). Semiconservative replication is constrained to a small bubble. This hybrid model corresponds to the gene conversion model proposed in Fig. 8D. (B) One example of BIR. A DNA fragment with subtelomeric sequences, a centromere, and a terminal sequence homologous to a chromosomal region is transformed into yeast. The subtelomeric sequence can recombine with a chromosomal subtelomeric region to result in a true telomere (ellipse). This step is not shown. The other end of the DNA fragment can acquire all the sequences distal to the chromosomal homologous region, up to the telomere.
DSB repair leading to long tracts of gene conversion has also been observed by Malkova et al. (291), who used MATa/MATα-inc diploids to study the repair of an HO-induced DSB. Nearly all of the repair of MATa occurs by recombination with the MATα-inc (noncleavable) homologous chromosome, since the cut chromosome lacks the donors HML and HMR. In a wild-type strain, nearly all the repair events are short-patch gene conversions, most of the time without crossovers. In rad52 diploids, the broken chromosome is almost always lost, but in rad51 diploids, the repair efficiency is 45% of the wild-type level. The repair of the DSB in rad51 diploids does not occur by classical gene conversion, however. All of the cells in which repair had occurred had become homozygous for MATα-inc and for all markers distal to MAT. This RAD52-dependent, RAD51-independent repair process was termed break-induced replication (BIR).
Another recent study supports the idea that one-ended strand invasion events do indeed result in extensive DNA synthesis involving whole chromosome arms. Morrow et al. (326) investigated the process of chromosome fragmentation developed by Vollrath et al. (530). Vollrath et al. transformed a linear fragment of DNA into yeast, with one end including Y′ subtelomeric sequences and therefore able to generate a new telomere by recombination with Y′ sequences on another chromosome end. The other end is homologous to a yeast chromosomal sequence far from a telomere. They recovered recombined chromosomes, including the transformed DNA linear fragment, with a new telomere on its Y′ side and, on the other side, all the sequences distal to the yeast homologous sequence (Fig. 12B). An obvious explanation was that this chromosome arose by a reciprocal exchange between the yeast gene present in the linear fragment and the chromosome. However, Morrow et al. showed that this was unlikely to be the case, because this new recombined chromosome was often found in addition to and not instead of the intact homologous yeast chromosome (Fig. 12B). Therefore, this kind of event had to involve extensive new DNA synthesis initiated from the non-Y′ end of the transformed fragment, adding a whole chromosome arm to this end. Here, also, the authors propose that a true replication fork would be initiated, leading to semiconservative replication, as during recombination-induced replication in E. coli or phage T4 (242, 330).
One theoretical feature of BIR is that after strand invasion of one 3′ end, there is no possible stabilization of the displaced strand by annealing with the second DSB end. Therefore, one has to envision two possibilities: either DNA synthesis occurs by bubble migration (130) (Fig. 12A, scheme 1), or BIR involves a true replication fork (Fig. 12A, scheme 2). However, with a bubble migration model, the synthesis of a complementary strand would be a secondary event. In contrast, recombination-induced replication forks have a rather well-characterized precedent in E. coli (242) and in bacteriophage T4 replication (330), a good reason to prefer this kind of model. We note that there is a satisfying unity of mechanism between BIR and the replication fork capture model described above. The third model (Fig. 12A, scheme 3) is directly derived from this fork capture model, with the progression of the replication fork closely followed by branch migration, resulting in conservative DNA synthesis. Once BIR starts, it can proceed to the chromosome end or be converted into gap repair if the second end of the DSB becomes involved.
BIR may also be a biologically very important repair pathway for the repair of chromosome ends. A chromosome that has lost a telomere has a single DSB end, and no second end can participate in a gene conversion repair event. One-ended events have been proposed for a long time to explain recombination at telomeres in wild-type cells (104, 534). Using HO-induced chromosome breaks in a diploid in which only one end of the broken chromosome has significant homology to its homologue, Bosco and Haber (51) found that repair was highly efficient; close to 70% of the broken chromosomes were repaired by apparently copying the 25 kb distal to the DSB from the homologous chromosome.
BIR accounts for the recombination-dependent maintenance of telomeres in cells in which telomerase, the enzyme that normally adds short TG1–3 sequences at the end of yeast chromosomes, is deleted. Although most of these telomerase-deficient cells die, a small proportion survive by apparently frequent recombination that regenerates and disperses sufficient TG1–3 at every chromosome end to keep cells alive (286). In addition, there are frequent rearrangements of subtelomeric sequences, including the proliferation to many ends of a subtelomeric Y′ element found normally at some chromosome ends. This whole process depends on the RAD52 gene and is affected by other recombination genes (260). It is not known if the cells that survive in this way have undergone some change that distinguishes them from the vast majority of cells that die without telomerase. Perhaps they have become hyperrecombination mutants, but genetic analysis has failed to reveal a single mutation to account for their survival (285).
A similar phenomenon has been observed in S. pombe. Cells with the trt1+ gene, encoding the catalytic unit of the telomerase, deleted can survive by two different processes: circularization of the chromosomes or apparent elongation of chromosomal ends without telomeres by a recombinational pathway, which might be BIR (337). The recombination process in trt1 mutant cells is enhanced by a mutation in the taz1+ gene, which encodes a telomeric DNA binding protein (337). This protein probably prevents the chromosome ends from entering the recombination process which is the fate of regular DNA ends.
Single-Strand Annealing
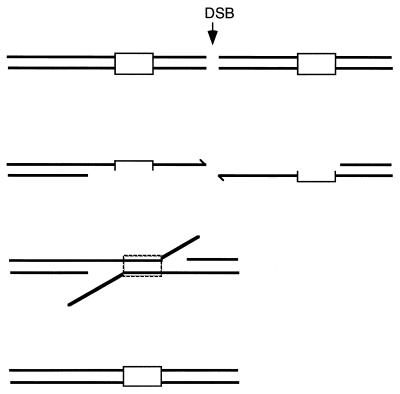
If a DSB occurs between two flanking homologous regions, repair of the broken chromosome is very efficient and results in a deletion containing a single copy of the repeated sequence. A mechanism that appears to account for these events is single-strand annealing (SSA) first suggested by Lin et al. for mammalian DNA repair (Fig. 13) (273, 274). SSA depends on the resection of the ends of the DSB by an exonuclease to produce long single-stranded tails in which complementary strands of the duplicated sequence are exposed and can reanneal. In yeast, SSA is nearly 100% efficient when homologous regions flanking the DSB are at least 400 bp, but 5% of the cells will survive a DSB when the repeats are only 60 bp (474). Repair is efficient even if the repeats are separated by as much as 15 kb.
FIG. 13.
SSA. SSA can occur when a DSB appears between (or within) two direct repeats. Resection of the DSB ends produces two complementary single strands that are annealed. After excision of the nonhomologous 3′ ends and new DNA synthesis, ligation restores two continuous strands.
SSA occurs in competition with other mechanisms of DSB repair. Fishman-Lobell et al. (124) used a plasmid containing direct repeats, one of which was cleaved by HO; thus either single-strand annealing or gene conversion could repair the DSB. The kinetics of the appearance of the noncrossover gene conversion product and of the deletion were different, arguing that these two outcomes arose predominantly by different mechanisms. Moreover, when 4 kb of additional DNA was inserted between the flanking repeats, the time to complete gene conversion remained the same while the time to produce the deletion increased by 60 min. This argued that a relatively slow exonuclease must traverse the entire intervening region before SSA could occur. Both in this plasmid assay and when substrates were integrated into the chromosome, deletions produced by SSA were three to four times more frequent than gene conversions, arguing that SSA is not a minor default pathway but a major repair pathway that may also explain the deletions of DNA between dispersed repeated sequences such as Alu in human DNA. Such deletions may be partially suppressed by the divergence of these dispersed sequences, given the observation in yeast that a 3% divergence between 205-bp repeats reduced SSA by a factor of 5 (476).
If a DSB is created within one of a pair of repeated sequences, deletions could happen in two ways: by SSA or by a gene conversion accompanied by crossing over. In the latter case, there would be a circular reciprocal product that would be retained only if it contained an origin of replication. Both genetic and physical experiments have failed to detect this reciprocal product except at very low levels, suggesting that SSA is the predominant route by which such a DSB is repaired (124, 395). SSA probably accounts for most of the spontaneous recombination events that are often called pop-out recombination.
SSA has provided a useful assay system to probe aspects of chromosome structure. To explore whether chromosomes lie in separate territories in the nucleus, Haber and Leung (166) created a strain in which two HO-induced DSBs on two different chromosomes could be repaired by competing SSA events: either by two intrachromosomal annealings (creating two deletions) or by two interchromosomal events (creating a pair of reciprocal translocations). Surprisingly, the interchromosomal events were as frequent as the intrachromosomal deletions. This argues that each DSB end could search the entire genome for a partner. SSA also has been useful as a default mechanism, in competition with gene conversion. In this way, it is possible to assess the efficiency with which an interchromosomal donor is used to repair a DSB, by comparing the frequency of gene conversion and a deletion event created by SSA between two repeated sequences flanking the DSB cut locus that is the target of gene conversion (371, 559).
Gene Targeting (Ends-Out Events)
Ends-out recombination events in eukaryotes is thought of essentially as an artificial event, useful for the researcher to knock out genes but not necessarily relevant to any biological pathway. In the one-step disruption method (421), a DNA fragment containing the gene one wishes to disrupt is modified in vitro, usually placing a selectable marker gene within, or instead of, the open reading frame. This disrupted gene is then transformed into yeast as a linear fragment. After selection for the marker gene, most of the transformants have integrated the DNA fragment at its homologous chromosomal counterpart (Fig. 3D). By placing sequences at each end of a selectable marker that were homologous to two very distant locations on a chromosome, Surosky and Tye (486) were able to create very large internal deletions of a chromosomal arm.
Current genome knockout strategies involve as little as 30 to 45 bp of DNA on either side of a KAN1 gene to knockout a target gene (531). In practice one needs two oligonucleotides each complementary to one end of the open reading frame and also overlapping the 5′ or 3′ end of the KAN1 gene. PCR amplification provides the transforming DNA. The success of this strategy is impressive—as many as 95% of the kanamycin-resistant transformants have knocked out the target gene. However, the results seem to vary with the strain and with the target sequences (213).
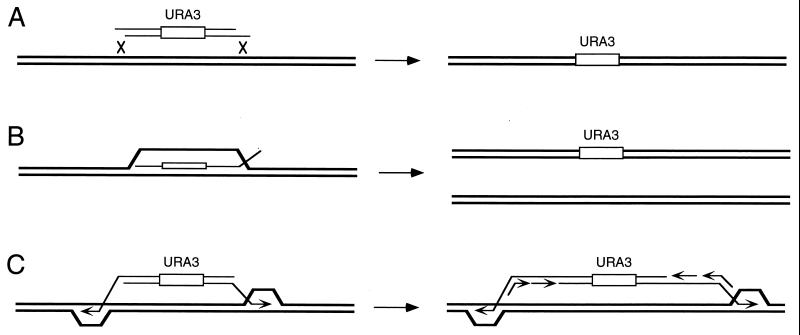
These ends-out recombination events have often been explained as the result of two crossovers at the ends of the transforming fragment, as illustrated in Fig. 14A. However, recent data have suggested another mechanism. Leung et al. (266) observed that such transformation events could result in sectored colonies, especially in strains mutated for the mismatch repair system. This outcome is not predicted by a double-crossover model, since both strands in the middle of the fragment should be integrated together. Instead, the authors proposed that ends-out events could involve an intermediate with a long heteroduplex created when one strand of the transforming fragment was assimilated into the homologous chromosomal sequence (Fig. 14B). This heteroduplex would often be aborted or corrected by the mismatch repair system, which appears to prefer to use the unbroken, resident strand as the template to correct the invading strand. Only about 5% of the time, correction occurs in favor of the transformed sequence. These data are supported by those of Negritto et al. (341), who also observed a 40-fold increase in gene replacement in a msh2 mismatch repair mutant when the linear fragment is perfectly homologous to the recipient site, except for the insertion of a selectable marker. However, these authors also observed that mismatches discouraged the integration of a linear fragment only when they were found at the edges of the fragment, suggesting that only the flanking parts of the fragment are engaged in heteroduplex formation. It is possible that the lack of effect of heterologies in the interior may be due to the fact that they were adjacent to a large, completely nonhomologous selectable marker.
FIG. 14.
Models for gene targeting (ends-out recombination). (A) A common view is that ends-out recombination is a double-crossover event. (B) Gene targeting could occur by the assimilation of one strand of the transformed DNA, which will thereafter convert the recipient by mismatch repair. (C) A third model, suggested by the work of Morrow et al. (326), envisions that the 3′ ends initiate BIR, resulting in a new chromosome that will replace or recombine with the original one.
A third model is strongly suggested by the results of Morrow et al. (326). After transformation of a DNA fragment whose terminal sequences were two inverted copies of a chromosomal locus, they screened for the formation of an isochromosome, in which the transformed fragment was now flanked by two copies of the same chromosome arm. These events most probably result from de novo synthesis of two chromosome arms, each initiated from the 3′ ends of the transformed fragment. This would correspond to two ends-out BIR events, with each end acting as a primer for extensive DNA synthesis. One striking feature of such unexpected events is that they occur with a frequency similar to that of the conventional knockout transformants. One can thus imagine that any ends-out event is initiated as follows. Strand invasion by the transforming fragment into the homologous locus would prime extensive DNA synthesis at each end of the fragment (Fig. 14C). Resolution with crossing over on each side would then integrate the transforming sequence. Alternatively, DNA synthesis might continue until the ends of the chromosome, resulting in chromosomal duplication.
During an “ends-in” integration, it may be sufficient that only one 3′ end invades the template, with the second end simply annealing to the D-loop or to the newly synthesized strand. If ends-out recombination events require a crossover at each end of the integrating fragment, they would also require two independent strand invasions. However, in some of the conceptions discussed above, only one end would have to invade. Hastings et al. (178) found that ends-out events are two- to ninefold less efficient than ends-in events, which seems to argue that there is an additional limiting step for ends-out events or that the processes are significantly different. Other studies (266, 326) found greater differences (up to 10- to 25-fold) between the two kind of events.
A combination of mechanisms may also account for the very efficient construction in vivo of recombinant plasmids by cotransforming a linearized, gapped autonomously replicating sequence (ARS)-containing plasmid molecule and a second linear fragment that is homologous to both sides of the DSB in the plasmid. Either by ends-out or ends-in recombination or even by SSA, a circular plasmid with newly introduced sequences is created (289).
PROTEINS INVOLVED IN MITOTIC RECOMBINATION
A complete characterization at the molecular level also includes the elucidation of the enzymatic machinery involved in the process. The combined power of genetics, molecular biology, and biochemistry has allowed researchers to assign specific roles to a number of proteins needed for the completion of each step during various recombination events. Biochemical studies are essentially attempts to reconstitute in vitro one or more steps in recombination, such as strand invasion, annealing, or 3′ nonhomologous-end removal.
Genes important for the repair of DSBs were identified primarily as mutations sensitive to X rays but not UV irradiation. These genes were classified as the RAD52 epistasis group. A mutation in rad52 was as radiation sensitive as a double-mutant of rad52 and one of the other rad mutations (141). Currently 10 genes fall into this group: RAD50, RAD51, RAD52, RAD53, RAD54, RAD55, RAD56, RAD57, MRE11, and XRS2; however, as discussed below, this classification masks a clear subdivision of these genes into at least four subgroups.
RAD52 stands alone as the one gene required for all homologous recombination events. RAD51, RAD54, RAD55, and RAD57 have common phenotypes, being required for some homologous recombination events but dispensable or less necessary for others. RAD50, MRE11, and XRS2 form another family of interacting proteins whose deletions have common phenotypes.
RAD53 has proven to be an essential gene that is not directly involved in DNA repair but is part of a complex network of checkpoint functions that arrest cell division and allow sufficient time for DNA repair to occur. The RAD56 gene has not yet been identified.
We also discuss several more recently identified genes involved in DSB repair: DMC1, RAD59, and TID1/RDH54. This list is certainly incomplete and does not include essential genes that are also required for DNA replication, for example. The roles of many of these replication genes have been learned by using conditional-lethal or hypomorphic mutations. In addition, we review surprising roles in recombination for a number of proteins initially implicated in nucleotide excision repair and mismatch repair.
Biochemical Properties of Recombination Proteins
The purification of eukaryotic recombination proteins and the subsequent characterization of their biochemical activities is a relatively recent field. So far, information has been obtained about eight proteins involved in general homologous recombination. These proteins can be classified in two families according to their biochemical properties: one contains Rad51p, Rad52p, Rad54p, Rad55p, and Rad57p, which participate in the strand transfer reaction, and the other contains Mre11p, Rad50p, and Xrs2p, which are involved in nuclease activity.
Rad51p, Rad52p, Rad54p, Rad55p, and Rad57p.
In vitro studies of strand exchange are supported by a number of experiments that demonstrate physical interactions among Rad51p, Rad52p, Rad54p, Rad55p, and Rad57p. This has led to the idea that these proteins form a recombinosome (179), but there is no direct evidence that they are all assembled into one complex. The complexities of eukaryotic recombination are also evident with the discovery of a meiosis-specific RAD51 homologue, named DMC1 (42), and of two other genes playing a role in mitotic homologous recombination: RAD59, a RAD52 homologue (18), and RDH54, a RAD54 homologue (100, 238, 451). Nothing is known so far about the biochemical activities of Rad59p and Rdh54p. We will reserve discussion of the role of Dmc1p until the section on meiotic recombination.
(i) Rad51p.
In E. coli and other bacteria, the pivotal protein involved in virtually all homologous recombination events is RecA. A large number of other rec genes are required, depending on the specific recombination event, but there appear to be two major RecA-dependent processes, known as the RecBCD and RecF pathways (105, 246, 461).
Rad51p is clearly a RecA homologue (1, 449), and a striking genetic demonstration of their relationship was shown by Chanet et al. (68), who isolated dominant recombination-defective RAD51 mutations and found that virtually all of them were in residues conserved among a wide variety of bacterial RecA proteins. One might expect that the yeast Rad51 protein would be indispensable for homologous recombination, but, surprisingly, RAD51 is not always required and often is less important than RAD52, even when the recombination events appear to be formally analogous to those in E. coli.
In many respects, the properties of RecA and the eukaryotic Rad51p protein from yeast or human are similar (for a review, see reference 38). Like E. coli RecA, Rad51p will catalyze an ATP-dependent strand exchange between a single-stranded circular molecule and a homologous linear duplex (338, 481, 484, 485), and, as with RecA, this reaction is enhanced by the presence of the yeast single-strand binding protein complex, RPA, a trimer of three different proteins with properties similar to E. coli SSB protein (27, 481, 484). Both Rad51p and RecA form filaments of oligomers on both ssDNA and double-stranded DNA (dsDNA) (26, 353) and extend the helical turn of dsDNA about 1.5 times relative to B-form dsDNA. It is argued that this extension also occurs when the RecA-coated ssDNA filament encounters its homologous dsDNA, making the base pairs more accessible for strand exchange to occur within the filament.
Using nuclear magnetic resonance spectroscopy techniques, Nishinaka et al. (349, 350) have further suggested that during the strand exchange process, the duplex DNA undergoes a rotational transition caused by a protein-mediated shift of the puckering of the deoxyribose moiety, so that the extended DNA goes from 18.6 bp per turn to 12.5 bp per turn but without a change in the extended distance between base pairs. This rotation is proposed to be the key step in the ATP-mediated exchange of strands (350). An identical change in DNA structure was found in E. coli RecA and S. cerevisiae Rad51p filaments (350).
However, some important differences have been found between RecA and Rad51p. Rad51p seems to bind dsDNA much more strongly than its bacterial counterpart does (26, 33, 481). Perhaps as a consequence of the increased affinity for dsDNA, strand exchange reactions catalyzed by Rad51p have a very narrow optimal ratio of protein to ssDNA (481, 484). Also, recent studies have provided evidence that the strand transfer reaction catalyzed by Rad51p occurs from 3′ to 5′ relative to the single strand (27, 484), since the RecA-catalyzed reaction usually displays the opposite polarity (for reviews, see references 28, 105, 246, and 461). However, other results argue that Rad51p can deal with either end (338). The direction that Rad51p chooses to assemble along ssDNA may be strongly influenced by the other proteins with which it interacts.
(ii) Rad52p.
By itself, Rad52p binds ssDNA and mediates DNA strand annealing (327) in a reaction that is stimulated by RPA (450, 477). Thus, it will promote the efficient annealing of two complementary single strands of DNA but will not catalyze the invasion of a single strand of DNA into a double-stranded molecule of the same sequence. Rad52p forms ring structures in vitro that interact with DNA (450).
However, interactions between Rad51p and Rad52p have been demonstrated both biochemically and genetically (99, 315, 433, 446, 449); therefore, the properties may be different in the presence of the complete set of recombination proteins. The importance of this interaction is shown by the discovery that expression of the Rad52p protein of another yeast, Kluyveromyces lactis, confers a dominant negative phenocopy of a rad52 mutation (315). This effect can be overcome by co-overexpression of the S. cerevisiae Rad51p, which suggests that the heterologous Rad52p interacts with S. cerevisiae Rad51p but the heterodimer cannot carry out some step in DNA repair (315).
One role of Rad52p in the Rad51p-catalyzed strand invasion seems to be to overcome an inhibitory effect of the RPA eukaryotic single-strand binding protein. In vitro, RPA is required for the strand transfer reaction, but it has to be added to the reaction mixture after the formation of the Rad51-ssDNA nucleoprotein filament. Incubation with RPA before this step actually inhibits strand transfer, but this inhibitory effect is overcome by the addition of Rad52p in both yeast and human protein studies (33a, 342a, 449a, 482). Consistent with the functional interactions of Rad52p and RPA in strand annealing and/or transfer, Rad52p has been found to physically interact with the middle subunit of RPA (450).
(iii) Rad55p and Rad57p.
As with RecA, in vitro strand exchange can be enhanced by the addition of other recombination proteins. The bacterial proteins RecO and RecR help “load” RecA onto ssDNA when RecA is in competition with the single-strand binding protein (443, 520), and the yeast proteins Rad55p and Rad57p have been reported to play a similar role in vitro (482). Thus, while simultaneous addition of RPA and Rad51p inhibits strand exchange, the presence of Rad55p and Rad57p alleviates this problem, similarly to Rad52p. Rad55p and Rad57p form a heterodimer (482) and have some homology to Rad51p (222, 283), but they do not seem to participate directly in strand exchange. Both Rad55p and Rad57p contain Walker motifs suggestive of ATP binding. However, while a mutation in a conserved Walker type A lysine in Rad55p affects recombination, an analogous mutation in Rad57p has no effect (208).
Biochemical data have not yet revealed the difference between the respective roles of Rad52p and Rad55p-Rad57p. However, the genetic data clearly indicate that they play different roles. Rad52p is absolutely required for virtually all recombination events, whereas several genetic studies demonstrate that Rad55p and Rad57p play only a supporting role that is sometimes dispensable. First, deletions of these genes result in a cold-sensitive X-ray sensitivity. Cells are normal at 34°C but repair-defective at 18°C, when the formation of protein assemblies is theoretically more difficult (284). Second, overexpression of Rad51p compensates for the absence of Rad55p or Rad57p (179).
(iv) Rad54p.
Rad54p appears to be a member of a diverse family of chromatin-remodeling proteins including the transcription factors Swi2/Snf2p and Mot1p (107). These proteins carry the motifs also shared by many helicases, but no helicase activity has been demonstrated for any of these proteins. Rad54p is related to two other DNA repair proteins: Rad5p (6, 209), which is implicated in postreplication repair, and Rad16p (434), which is important in UV repair, especially when photodimers are found in heterochromatic locations, but is dispensable when UV damage is located in more accessible regions (524). This has led to the idea that these proteins help “open up” DNA for repair and recombination, an idea supported by an experiment with HO endonuclease-induced recombination, which is described below. Genetic studies have suggested that RAD5, RAD16, and RAD54 play partially overlapping roles in DNA repair (149). In vitro, Rad54p significantly increases Rad51p-mediated strand exchange (378). Like the other members of the Swi2/Snf2p family, Rad54p has a DNA-dependent ATPase activity (378). This activity essentially depends on dsDNA and is much stronger than the ssDNA-dependent ATPase activity of Rad51p (kcat > 1,000/min and kcat < 1/min, respectively), suggesting that it exerts its effect on homologous DNA pairing via an action on the incoming dsDNA.
Two-hybrid and coimmunoprecipitation experiments indicate an interaction between Rad51p and Rad54p (78, 204). Overexpression of Rad54p can suppress the methyl methanesulfonate (MMS) sensitivity of rad51 but not its defects in MAT switching or heteroallelic recombination (78). At present, we do not know the stoichiometry of these proteins in whatever complexes they may form.
Mre11, Rad50p, and Xrs2p.
MRE11, RAD50, and XRS2, three other members of the RAD52 epistasis group, also function as a complex, as indicated by two-hybrid and affinity purification experiments (214, 522). The strong association of Rad50p, Mre11p, and Xrs2p is substantiated by the identical phenotypes of their deletion mutations. These genes participate in a bewildering set of DNA repair and maintenance processes, including both homologous recombination, nonhomologous end joining (NHEJ), telomere maintenance, and the creation of Spo11p-mediated double-strand breaks in meiosis (reviewed in reference 162). These processes are discussed in detail in several sections of this review. These genes appear to play two major roles, one as a structural component and the other in controlling or directly processing DNA ends by a 5′-to-3′ exonuclease.
Mre11p and Rad50p are homologous to SbcD and SbcC, respectively, two interacting bacterial proteins. SbcD has double-strand exonuclease and single-strand endonuclease activity (444). Deletions of RAD50, XRS2, or MRE11 notably retard the rate of 5′-to-3′ exonuclease activity in vivo (198, 262, 474, 514). The mre11-58 mutation, with a mutation in a conserved phosphodiesterase motif of MRE11, markedly reduces 5′-to-3′ resection of an HO-induced DSB (514).
Surprisingly, a number of recent biochemical studies of human and yeast Mre11p have shown that instead of a 5′-to-3′ dsDNA exonuclease, Mre11 has a 3′-to-5′ ATP-independent exonuclease activity, at least with Mn2+ as the cofactor (no activity has been found with Mg2+) (138, 325, 374, 512, 522). The spectrum of activities seems to be the same whether the entire Mre11p complex (512) or the purified Mre11 protein (325, 374, 522) is assayed. An ssDNA endonuclease activity and an ssDNA 3′-to-5′ exonuclease activity have also been found for the yeast Mre11p (522). It is difficult to reconcile these findings with the fact that in vivo, Mre11p, Rad50p, and Xrs2p are apparently involved in the 5′-to-3′ resection of DSB ends. One way in which Mre11p, Xrs2p, or Rad50p could affect 5′-to-3′ degradation would be if its ssDNA endonuclease activity could clip off ssDNA unwound by an associated helicase, much as the RecBCD protein processes DSBs in E. coli (105, 246, 461). A possible role for a 3′ exonuclease activity would be to remove glycols from the end of X-ray-shattered DNA, which need not have 5′ phosphates and 3′ OH ends.
The nuclease activity(-ies) lies in the amino-terminal part of the protein, since mutations in this region eliminate the in vitro nuclease activity and/or reduce the 5′-to-3′ resection in vivo (138, 325, 514, 522). Also, mutations in the N-terminal part of Mre11 affect DSB repair after ionizing radiation (56). In contrast, mutations in the carboxy-terminal part of the protein do not affect the resection of DSB ends, although they affect the endonuclease activity of the meiotic Spo11 protein (138, 335), an issue we examine in more detail in the section on meiosis.
Recombination Proteins and Their Role In Vivo
Redefining the epistasis relationships of RAD52 group proteins.
A great deal of genetic and molecular biological evidence now supports the view that there are at least five major subdivisions of the RAD52 epistasis group, defined by their roles in mitotic recombination, which we call RAD52, the RAD51 family, the RAD59 family, the MRE11 family, and RAD53. RAD53 is not directly involved in DSB repair but is involved in cell cycle arrest, and we discuss its role in the checkpoint section. In addition, there is a newly discovered gene, RDH54, whose exact relationship to these subgroups remains unclear.
(i) RAD52.
As noted above, RAD52 is unique in being required for virtually all homologous recombination events. It is the only one of the X-ray sensitivity genes required for SSA (474), which is consistent with its role as a strand-annealing protein. The requirement for RAD52 in SSA diminishes as the length of the homologous regions flanking a DSB increases from 1 kb (where fewer than 1% of cells recombine) to 2 kb (about 10%) to >10 kb (nearly 100%) (360, 381, 422, 474).
There is also a residual (about 1% of the wild-type value) RAD52-independent pathway of recombination that is found in diploids, both in spontaneous recombination between heteroallelic markers (165) and when one of two homologues is cut by HO endonuclease (291). In both cases, nearly all of the recombinants are 2n − 1 diploids in which the retained chromosome has undergone a crossing over. Similar crossover-associated RAD52-independent events were noted by Jackson and Fink (199) for USCE. We suggest that such events may well arise by SSA, for example between the HO-cut chromosomes and a spontaneous DSB elsewhere on the homologous chromosome. Previous studies have shown that there is an elevated level of chromosome instability in rad52 diploids that could arise from DSBs created during DNA replication and that cannot be repaired by sister chromatid recombination (328).
The nonreciprocal half-crossovers that are the major type of outcome when gene conversions between heteroalleles are selected in a rad52 diploid may actually be a rare but significant pathway in recombination-proficient cells as well. Campbell and Fogel (62) noted that about 10% of Leu+ recombinants between two leu2 mutations in a haploid, disomic for chromosome III, were associated with chromosome loss. Conversely, both in wild-type strains (63) and in a chl1 mutant diploid, where chromosome loss is greatly increased (277), between 5 and 10% of the chromosome losses were associated with a detectable recombination event on the retained chromosome. Such events are consistent with some sort of one-ended strand invasion or with unrepaired lesions that are created during the resolution of recombination events.
In addition to its hyporecombination phenotype, a rad52 strain has a spontaneous mutator phenotype (511), with most of the events corresponding to deletions between short direct repeats. The way in which RAD52 participates in the avoidance of such events remains unclear. One possibility is that the absence of RAD52 prevents DSBs arising during replication from being repaired by homologous recombination with a sister chromatid; hence, cells resort to a RAD52-independent end-joining mechanism (described below).
(ii) RAD51 family.
In contrast to the strong effect of deleting RAD52, deletions of RAD51, RAD54, RAD55, and RAD57 sometimes have much less severe effects on mitotic homologous recombination. Spontaneous recombination between heteroalleles is reduced only 4- to 10-fold, compared to 100- to 1,000-fold for rad52 mutants (392). In fact, for spontaneous deletions between repeated sequences, mutations in these four genes give a modest hyperrecombination phenotype (303). RAD51, RAD54, RAD55, and RAD57 are also not required for HO-induced SSA (197). Similarly, the formation of HJs in the rDNA locus (575) depends on RAD52 but not on RAD51, RAD55, or RAD57.
Some insight into the roles of these proteins has been gained by studying HO-induced gene conversions. Mutations in any one of these genes completely prevent MAT switching using the silent, heterochromatic HML or HMR donors. Indeed, one cannot detect an early intermediate of new DNA synthesis after strand invasion. RAD51, RAD54, RAD55, and RAD57 are required for all gene conversion events induced by HO whenever the donor and recipient are located on bona fide chromosomes. However, HO-induced gene conversion between homologous sequences carried on a plasmid can be RAD51, RAD54, RAD55, and RAD57 independent, depending on the chromatin structure of the donor (475). If MAT and HMR donor sequences are on a plasmid, RAD51, RAD54, RAD55, and RAD57 are required to complete recombination when the HMR donor sequence is in its normal, inaccessible chromatin conformation but not when the donor sequences are unsilenced. Moreover, the kinetics and proportions of gene conversions associated with exchange are unaffected by the absence of Rad51p. One interpretation of these results is that the four proteins are not directly involved in the mechanics of strand exchange (despite the apparent similarity of RecAp and Rad51p); another, more likely possibility is that there are two independent recombination machines and that the second, Rad51p-independent pathway is unable to cope with donors that are not accessible. Consistent with this view, the homology between Rad54p and various chromatin-remodeling proteins would suggest that Rad54p is required to open up an otherwise inaccessible region and that Rad55p and Rad57p would be required to help Rad51p initiate strand exchange, as supported by in vitro studies (378, 482).
BIR also depends on Rad52p but not on Rad51p, Rad54p, Rad55p, and Rad57p. When an HO-induced DSB is created at the MATa locus on one chromosome, it can be repaired by gene conversion involving the MATα-inc region on a homologous chromosome (291). However, in rad51, rad54, rad55, and rad57 mutants, DSB repair is quite efficient (45%) but instead of gene conversion, repair occurs by BIR (291, 454), which appears to be initiated at some specific sites proximal to the DSB, which might have a more open conformation.
Another repair process that has been studied in yeast is the rescue of cells lacking essential components of yeast telomerase. In the absence of this essential RNA-dependent DNA polymerase, yeast telomeres gradually shorten until the cells die, apparently lacking the protection to chromosome ends afforded by telomeres. A very small fraction of cells survive by a RAD52-dependent recombination mechanism (which might be BIR) by maintaining chromosome ends with a minimal telomere segment (286, 458).
A recent study of cells lacking telomerase RNA (tlc1 mutants) has concluded that, as with many other DSB repair events, there is a less stringent requirement for RAD51, RAD54, RAD55, or RAD57 than for RAD52. Senescence is rapid, as with rad52 mutants, but survivors still appear when these genes are deleted. Moreover, their telomeres and subtelomeric sequence rearrangements are indistinguishable from those of the wild-type survivors lacking TLC1 (260). The absence of RAD50, XRS2, and MRE11 has no effect on the rate of cell death, but survivors appear later than those found in rad51 mutants. Significantly, a rad50 rad51 tlc1 mutant is similar to rad52 tlc1, since there are no survivors. This might suggest that there may be two RAD52-dependent ways to repair these ends, one pathway requiring RAD51 and one requiring RAD50. Alternatively, the two mutations may reduce but not eliminate the efficiency of sequential steps in a single repair process.
The question of what other proteins might substitute for Rad51p under the various conditions we have summarized is one of the most perplexing issues facing students of recombination. It seems clear that Rad51p mediates the most prevalent mechanism of homologous DSB repair and that other pathways become evident when Rad51p is absent. As noted above, HO-induced recombination between two inverted repeats of lacZ is nearly as efficient in rad51 mutants as in the wild type. However, this is in a situation where other efficient competitive pathways cannot operate. If one examines DSB repair in a nearly identical plasmid but with the two lacZ repeats in direct orientation, the effect of rad51 is strikingly different. In a wild-type cell, about 20% of the DSBs are repaired by gene conversion without crossing over whereas 80% undergo a deletion through SSA. However, in rad51 mutants, the proportion repaired by gene conversion is less than 1% (197). We conclude that Rad51p participates in the most efficient repair mechanism but that another, unknown pathway accomplishes much the same result: gene conversion both with and without crossing over. Here, too, rad51, rad54, rad55, and rad57 mutants form a distinctive subgroup.
(iii) RAD59.
The hypothesis that there is a Rad51-independent recombination pathway has been supported by the discovery of the RAD59 gene. As mentioned above, a rad51 mutation has little effect on spontaneous recombination in various assays. Rattray and Symington (391, 392) observed that inverted-repeat recombination is reduced only 4-fold in a rad51 mutant but more than 3,000-fold in a rad52 strain. A systematic search for mutants preventing rad51-independent events resulted in the isolation of the RAD59 gene (18). Rad59p has some homology to Rad52p, but RAD59-mediated events still require RAD52.
rad51 and rad59 single mutants display comparable modest defects in spontaneous recombination between chromosomal inverted repeats, but the rad51 rad59 double mutant is as defective as the rad52 mutant. However, this synergy is not observed for spontaneous or HO-induced recombination between plasmid-borne inverted repeats: rad51 has no effect, and the rad51 rad59 double mutant shows the same 10-fold decrease as rad59, indicating that RAD59 still does not define the only RAD51-independent pathway. For heteroallele recombination, it is the opposite: only RAD51 is required, and recombination is even increased sixfold in a rad59 mutant, which seems to indicate at least that RAD59 is not required for interchromosomal recombination. In addition, a rad59 deletion affects recombination between inverted repeats but not the ratio of associated crossover events, while these events are preferentially increased in a rad51 mutant (391, 392). Recent data also indicate that a rad59 deletion substantially reduces SSA (473).
It would be prudent to voice one word of caution about the notion that present data unequivocally place RAD51 and RAD59 in separate pathways. It is possible that each mutation very markedly reduces but does not eliminate the completion of a different step in one spontaneous recombination pathway.
(iv) TID1/RDH54.
Recently, additional complexities in homologous recombination have been revealed by the discovery of a RAD54 homologue also known as TID1 (100, 238, 451). This protein is another relative of the helicase-like Swi2/Snf2 protein family. Although this gene was identified by its interactions with a meiosis-specific RAD51 homologue, DMC1, tid1/rdh54 mutants have mitotic phenotypes. In haploid cells, a tid1/rdh54 mutation has no evident phenotype by itself but has a synergistic effect with rad54 for MMS sensitivity. However, the distinctive phenotype of tid1/rdh54 mutants is revealed in diploid cells: there is a marked decrease in interchromosomal recombination. Intrachromosomal ectopic recombination is not affected, although it should be pointed out that the intrachromosomal assay could predominantly result from SSA.
Exactly what process is blocked is confused by the fact that there are substantial differences in tid1/rdh54 phenotypes of two commonly used strains. In a W303 background, mitotic interchromosomal recombination is abolished in a tid1/rdh54/tid1/rdh54 homozygote (238). In an SK1 background, however, this effect on interchromosomal recombination is detected only in rad54/rad54 tid1/rdh54/tid1/rdh54 double mutants (451). Analogous defects in meiotic recombination (which is essentially interchromosomal) have been observed by these authors. We discuss this in the section on meiotic recombination (below), where we also discuss other discrepancies between similar strains.
TID1/RDH54 is also implicated in a complicated network of intersecting DNA repair pathways involving the 3′-to-5′ Srs2p helicase. SRS2 was initially identified as a suppressor of the UV-induced death of rad6 and rad18 mutations affecting postreplication repair (2). srs2 cells are UV sensitive, but the cell death is rescued by rad51 or rad52 mutations (1). It has been proposed that the SRS2 gene ensures that certain types of damage (possibly nicks or single-strand gaps) that are normally processed by postreplication repair do not enter the DSB repair pathway (1, 2).
srs2 is lethal in the absence of RAD54 (363), though why this should be the case is not understood. tid1/rdh54 also has a synergistic effect with srs2. An srs2 tid1/rdh54 haploid is viable, but an srs2/srs2 tid1/rdh54/tid1/rdh54 diploid is not (238). This cell death is suppressed by rad51. Possibly srs2/srs2 tid1/rdh54/tid1/rdh54 mutants are inviable because some spontaneous damage is improperly channeled into the DSB repair pathway, leading to a nonrepairable lethal intermediate. This would not be allowed in a rad51 background, which could imply either that TID1/RDH54 acts downstream of RAD51 and creates a lethal environment or that it acts before RAD51 in channeling damage toward the recombination pathway.
According to these results, there could be three recombinosomes, all of them including Rad52p. The best-characterized one also includes Rad51p, Rad54p, Rad55p, and Rad57p and is required in most situations involving chromosomes and when plasmid substrates have an inaccessible chromatin structure. A second recombinosome would require Rad59p and probably other proteins and would specialize in intrachromosomal events. Finally, a third recombinosome, involving Tid1/Rdh54p but probably largely overlapping with the Rad51p recombinosome, would specialize in interchromosomal recombination. The issue of a specific interchromosomal pathway is raised for meiotic recombination in a later section, when we address the existence of a fourth recombinosome, which includes Dmc1p, the meiosis-specific RecA homologue.
(v) MRE11 family.
In recombination between homologous chromosomes, the RAD50, XRS2, and MRE11 genes appear to play a surprisingly minor role despite the X-ray and MMS sensitivity resulting from deletions of these genes. In rad50, xrs2, or mre11 null mutants, HO-induced MAT switching is delayed about 1 h but recombination is nearly 100% successful (198, 514), in marked contrast to rad52 or rad51 mutations. When HO endonuclease is used to create a DSB on one homologous chromosome in a diploid strain, a 30% decrease in repair efficiency is observed in rad50 strains (292). Similarly, rad50 and xrs2 mutants have only a modest reduction in radiation-induced recombination (196).
Indeed, for spontaneous heteroallelic recombination, deletion of MRE11, RAD50, or XRS2 causes a 7- to 10-fold increase in recombination (156, 196, 295, 514). This increase is probably not attributable to the failure of these strains to religate DSBs (see “Mitotic nonhomologous recombination” below), which could increase the number of recombinogenic lesions, because other mutations, such as hdf1, that do not efficiently rejoin DSB ends are not hyperrecombination mutations (453). Hyperrecombination could reflect a decrease in the length of single-strand tails and hence a decrease in the proportion of gene conversions where both sites are coconverted (failing to yield a prototrophic recombinant). Alternatively, the hyperrecombination phenotype may result from a failure of these mutants to repair damage from a sister chromatid, thus increasing interchromosomal interactions.
There is strong evidence that these proteins play an especially important role in sister chromatid interactions. rad50 and xrs2 haploid cells in G2 are much more X-ray sensitive than are rad50 or xrs2 diploid cells in G1 (115, 196). Moreover, mre11/rad50/xrs2 mutants are as sensitive to low concentrations of hydroxyurea as are rad52 or rad51 mutants, where it is presumed that replication-associated DSBs are repaired by sister chromatid recombination (321).
MRE11, RAD50, and XRS2 play many other roles in DNA metabolism, some of which appear to be independent of any exo- or endonuclease activity. We discuss their participation in making and processing meiotic double-strand breaks in a later section. However, we should mention here several other phenotypes associated with mre11, rad50, and xrs2 deletions. First, these mutants have shortened telomeres (52, 232, 351). Moreover, mre11 and rad50 are lethal in combination with the telomere-associated protein mutation cdc13-1 and synthetically temperature hypersensitive in combination with a deletion of the yeast Ku homologues, HDF1 and HDF2 (351). One possibility is that Mre11p/Rad50p/Xrs2p signals to the RNA-dependent DNA polymerase, telomerase, that DNA replication has produced two sister chromatids and allows the coordinated addition of telomeres, but this cannot happen in cdc13-1 or hdf1 mutants. More generally, deletions of these three genes produce very slowly-growing colonies. The basis of the slow growth is not understood.
Yeast compared to mammals.
Whereas the biochemical studies on recombination proteins of yeast and mammals have given similar results for proteins such as Rad51p or Mre11p, the phenotypes of the corresponding mutants are quite different.
Rad51−/− mutant mice die early during embryonic development, and it is impossible to obtain Rad51−/− cell lines (272, 518). In chicken DT40 cells, when the RAD51 gene is under a repressible promoter, repression of RAD51 result in chromosome breaks, cell cycle arrest, and death (464). Interestingly, in mice, the embryonic death due to the Rad51 mutation is delayed by a mutation in the p53 tumor suppressor gene (272). Maybe the replication of the large genomes of higher eukaryotic is a tougher task than the replication of the yeast genome. There are also a surprisingly large number of other Rad51 homologues in mammals in addition to the meiotic DMC1 gene. Mutations in Rad51B, Rad51C, XRCC2, and XRCC3 all affect chicken cell recombination (496). Many replication fork collapses may result in DSBs, which would have to be repaired by a Rad51-dependent pathway.
However, Rad54−/− chicken cells and mice are perfectly viable although X-ray sensitive (36, 114). In addition, Rad52−/− knockout mutants are not only viable but also resistant to ionizing radiation (406, 564), although homologous recombination as measured by itself is defective in such mutants. It is possible that in the complex genome of the higher eukaryotes, there is also more than one RAD52 gene and more than one RAD54 gene. Another unidentified RAD52 homologue could be required for ionizing radiation resistance and for cell proliferation, together with RAD51 and an unidentified RAD54 homologue.
Another striking difference between yeast and mammalian cells is the requirement for Mre11p and Rad50p. Yeast mre11 and rad50 null mutant grow slowly, but an Mre11−/− or Rad50−/− homozygous mutation is lethal in mice (376, 560). Also, a mutation affecting the p95 gene, which seems to be the human equivalent of the yeast XRS2 gene in the Rad50p Mre11p Xrs2p trio, results in Nijmegen breakage syndrome (65, 523). This syndrome has many similarities to ataxia-telangiectasia, including sensitivity to ionizing radiation, predisposition to cancer, and failure to arrest in G1/S in response to DNA damage, suggesting a role in checkpoint arrest for Mre11p Rad50p p95 in humans. The yeast proteins have also been implicated in the adaptation from G2/M arrest in the presence of an unrepaired DSB (262).
Physical Monitoring of Recombination Intermediates in Various Mutant Backgrounds
Physical monitoring of DNA in HO-induced recombination events can be carried out in various mutant backgrounds, so that some of the roles of the wild-type proteins can be inferred for the steps that are affected. One liability of this approach is that generally it will allow one to see only the first step at which the protein acts; if the process is blocked by a mutation at an early step, one will not be able to see that the same protein might also be required at a much later point. We can identify at least four important steps in recombination characterized in this way: (i) resection of the DSB ends, (ii) strand invasion and initiation of new DNA synthesis, (iii) removal of nonhomologous DNA tails, and (iv) later steps in gene conversion. In addition, cytological approaches that provide important information on the roles of recombination proteins are beginning to be developed.
Resection of DNA ends.
During gene conversion and SSA, the first step to be observed after DSB formation is a resection of the ends, resulting in long 3′-ended tails. These tails can then invade a homologous template (gene conversion) or anneal (SSA). Whether the resection is by a 5′-to-3′ exonuclease or by an endonuclease associated with a helicase (as with RecBCD in E. coli) is not known, but deletions of RAD50, XRS2, and MRE11 all slow DNA degradation about twofold (198, 262, 474, 514). There must be several redundant activities that contribute to the overall rate of degradation. The Exo1p nuclease has a role in nucleotide excision repair and mismatch repair (122). Its function in the DSB response seems to be as a backup nuclease. A deletion of EXO1 has no effect on DNA degradation of DSBs by itself but eliminates much of the residual resection of DNA in an mre11 deletion (491, 513). However, even in an mre11 exo1 double mutant, there is still some 5′-to-3′ degradation and HO-induced recombination occurs at nearly wild-type levels (513).
Strand invasion and the initiation of new DNA synthesis.
In gap repair, the next apparent step is the invasion of the 3′ end into its homologous template to provide a primer for new DNA synthesis. No strand invasion is needed for SSA, where annealing provides the 3′ end necessary for synthesis initiation. A PCR assay designed to monitor the formation of newly synthesized strands during MAT switching (see “Physical monitoring of HO-induced mitotic gene conversion” above) was used to demonstrate that no DNA synthesis could proceed in a rad52, rad51, rad54, rad55, or rad57 mutant (396, 547). A similar result is found with conditional-lethal mutations of proliferating-cell nuclear antigen (PCNA) and the clamp loader RFC protein Cdc44 (190). It should be possible to carry out similar physical monitoring of equivalent steps with other gene conversion substrates where RAD51 or RAD54 do not seem to be required.
Removal of nonhomologous ends.
Strand invasion provides a primer for the initiation of new DNA synthesis. However, to provide a suitable primer, the invading 3′ end must be perfectly homologous to the template and any nonhomologous sequences must be removed from the 3′ end. During SSA also, the reannealed single strands are presumed to prime new DNA synthesis to fill the gaps resulting from 5′-to-3′ degradation, and the initiation of DNA synthesis would be equally impeded by 3′ nonhomologous sequences. The removal of such nonhomology must occur in the artificial substrates in which an HO cutting site is inserted into the recipient molecule, but it must also occur during MAT switching to replace the Y region (Fig. 5). The stability of the 3′ end when there is no recombination (262, 547) argues that removing nonhomology from the ends occurs in the context of pairing between an internal portion of the invading strand and the donor (gene conversion) or between two single strands (SSA).
The removal of these 3′ nonhomologous tails depends on the nucleotide excision repair genes RAD1 and RAD10 (123). Rad1p and Rad10p were shown to form an endonuclease that can cleave DNA with a “flap” of 3′-end ssDNA (22, 483, 509, 510). None of the other NER genes (RAD2, RAD3, RAD7, RAD14, RAD16, and RAD25) are required (195), but in vivo, the process depends on the MSH2 and MSH3 mismatch repair genes (476). Three other mismatch repair genes, MSH6, PMS1, and MLH1, are not needed. Thus, Msh2-Msh3 and Rad1-Rad10 proteins can participate in several different multiprotein complexes. However, MSH2-MSH3 and RAD1-RAD10 do not play precisely the same role in the removal of nonhomologous ends. During SSA, RAD1 and RAD10 are required independent of the length of the regions being annealed, but msh2 and msh3 mutations are needed only when the homologous region is less than 1 kb (476).
One interpretation of these results is that Msh2p and Msh3p stabilize the annealed intermediate structure by recognizing the unpaired single strands at the ends of the annealed region, allowing Rad1p and Rad10p to locate them and cleave off the 3′-ended tail. Msh2p and Msh3p could also recruit Rad1p and Rad10p and thus, promoting a quicker cleavage process, allow the processing of even unstable substrates. When the annealed regions during SSA are sufficiently long (e.g., 1 kb), the intermediate is apparently sufficiently stable without Msh2p and Msh3p. In contrast, during gene conversion, Msh2p and Msh3p are required to remove nonhomologous ends even when there are long homologous regions. In gene conversion, the invading strand can form only a side-by-side paranemic joint with its homologous sequence (554) before the DNA end is cut off, while during SSA two single strands of DNA can form interwound plectonemic molecules when the homologous segments are long enough. The SRS2 gene is also required for efficient removal of nonhomologous ends in gene conversion (366), and we hypothesized that the 5′-to-3′ Srs2p helicase also played a role in stabilizing the nascent joints.
There are alternative pathways for 3′ ends removal. Two different RAD1-independent pathways have been found. One can remove 3′ ends up to 20 bp and depends at least partly on the proofreading activity of DNA polymerase δ (366). Another pathway can remove a nonhomologous end on one side of the DSB but only after the other end has initiated strand invasion; thus, although Ya or Yα sequences must be excised during MAT switching, RAD1 is not required. A Southern blot analysis of DNA performed at intervals after HO cleavage suggests that Rad1p and Rad10p normally process the nonhomologous Y region, because the completion of MAT switching is delayed by about 60 min in a rad1 mutant (190). This is also true for other substrates containing an HO cleavage site (81). In the absence of Rad1p and Rad10p, there must be a second, slower but adequate system to remove this region and complete MAT gene conversion once the process has been initiated from a homologous end.
Enzymes required for DNA synthesis during gap repair.
DNA repair by homologous recombination requires new DNA synthesis. However, all the mutations of the RAD52 group, identified for the sensitivity of the mutants to X-rays or MMS, affect strand invasion or earlier steps. This a surprising result, because one might have assumed that among this collection of mutations, identified solely because their mutants could not complete DSB repair, some would be blocked at later stages of the repair process. That they all blocked so early suggests that genes needed for later steps have not been identified, possibly because they play indispensable roles in DNA replication.
Budd and Campbell used temperature-sensitive alleles of the three major DNA polymerases to demonstrate that repair of UV damage is prevented only when both polymerase δ (Polδ) and Polɛ are inactive (60), suggesting that either of the two polymerases can fill in ssDNA gaps created during excision repair. The primarily lagging-strand polymerase, Polα, appeared to have no effect. To investigate the role of these polymerases and other DNA replication functions in DSB repair, Holmes and Haber (190) analyzed MAT switching in temperature-sensitive mutants held at their restrictive temperatures and then induced for HO expression. Recombination was then monitored by Southern blotting and PCR. Conditional-lethal mutants of both POL30 (PCNA) and CDC44 (the largest subunit of the PCNA-loading complex) prevent mating-type switching but also fail to form the PCR-detectable intermediate indicative of new DNA synthesis after strand invasion. Temperature-sensitive mutants of both of PCNA-associated DNA Polδ and Polɛ are impaired in the kinetics and/or extent of MAT switching, with the Polɛ mutation having the more severe effect.
More surprising was the result obtained with a temperature-sensitive Polα mutant (pol1-17) and with a mutant with a temperature-sensitive mutation in DNA primase (pri2-1). Only about 15% of the cells were able to complete MAT switching at the restrictive temperature. This defect is not a reflection of the trapping of polymerases at stalled replication forks, because pri2-1 cells were defective in MAT switching even in cells arrested in G1, where DNA polymerases have not assembled at origins of replication. Furthermore, the absence of the nonessential Okazaki fragment-processing protein, Rad27p, also reduced and delayed switching. Most models of gap repair presume that DNA synthesis can be primed by the two 3′ ends of the DSB and thus should not require lagging-strand DNA synthesis enzymes. These results suggest that DSB repair involves both leading- and lagging-strand synthesis (Fig. 8D).
A viable mutation in the RFA1 gene (rfa1-t11), encoding the largest subunit of the essential single-stranded binding protein complex, RPA, allows cells to replicate their genome, but recombination is almost completely impaired (521). Therefore, these cells can successfully replicate 12,000,000 bp each cell cycle but cannot replace the 700 bp at MAT by recombination.
In addition, at least one nonessential polymerase gene plays a role in the DNA synthesis associated with DSB repair. Strathern et al. have found that DSB gap repair is error prone, so that the frequency of reversion of a mutant allele adjacent to the site of gap repair is elevated 1,000-fold over the background (472). REV3, encoding one subunit of Polζ, was shown to be responsible for most of this increase (184), at least for some sorts of mutations. Nevertheless, a deletion of REV3 has no detectable effect on the efficiency of repair of DSBs. This protein may be used redundantly with other polymerases, or the original gap repair event might leave behind lesions that are acted on by the Rev3 protein.
Mating-Type Regulation of Homologous Recombination Activities
MATa and MATα regulate many cell-type-specific genes through the action of the Mata1 and/or Matα1 and Matα2 regulator proteins (reviewed in references 164 and 180). In MATa/MATα diploids or in haploids expressing both mating-type genes, Mata1 and Matα2 form a repressor that turns off haploid cell-specific genes, including the HO endonuclease gene and some genes of the mating-pheromone signal transduction pathway, as well as negative regulators such as RME1, which represses the initiation of meiosis in haploid cells. There is at least one additional gene (probably several) that is controlled by mating type, which influences homologous recombination. MATa/MATα diploids are substantially more radiation resistant than MATa/MATa or MATαMATα diploids and have higher levels of spontaneous recombination (although this conceivably could result from a few cells entering meiosis) (119, 181). The expression of MATa and MATα in haploid cells suppresses the X-ray sensitivity of rad55 deletion mutants (284). Moreover, recent data suggest that HO-endonuclease-induced interchromosomal recombination is significantly more efficient in MATa-inc/MATα-inc diploids (with HO-insensitive MAT alleles) than in an isogenic strain where one or the other MAT gene is deleted (367). An inspection of the S. cerevisiae genome indicates at least 12 unknown open reading frames that have apparent consensus Mata1/Matα2 repressor binding sites in their upstream regions (103), which are candidates for mating-type-regulated genes that affect recombination.
There is also a fascinating difference in recombination seen between MATa and MATα cells. MATa selects the silent HML donor for MAT switching 80 to 95% of the time, whereas MATα has an equivalent preference for HMR. Donor preference is controlled by a small cis-acting recombination enhancer located 30 kb from the left end of chromosome III (538, 555, 558; for reviews, see references 161 and 164). In MATa cells, the recombination enhancer somehow activates the entire left arm of chromosome III to be “hot” in both HO-induced MAT switching and spontaneous recombination between leu2 alleles. In MATa, the left arm becomes unusually inaccessible for recombination, although there is no obvious difference in the level of transcription or the chromatin structure of the recombining sequences in the two mating types.
STIMULATION OF MITOTIC RECOMBINATION
Mitotic Hot Spots and Hyperrecombination Mutants: Connection to Transcription
It has been suggested that the activation of transcription in higher eukaryotes plays a critical role in the control of V(D)J and class-switching recombination in the immune system (158). There are several lines of evidence indicating that transcription can stimulate recombination in yeast, but in most of these cases it would appear that transcriptional initiation of recombination occurs when DNA sequences that are not normally transcribed experience the traversal of RNA polymerase.
The hpr1 mutation was initially characterized as a hyperrecombination mutation that specifically stimulates deletion between repeated sequences separated by plasmid or other sequences (3–5). The RAD1 and RAD52 dependence of hpr1-stimulated recombination suggests that the deletions arise from SSA. Recent studies by Aguilera’s laboratory have greatly clarified the effect of hpr1 (70, 385): the HPR1 gene codes for an RNA PolII elongation factor, and the stimulation of recombination by hpr1 depends on transcription through the region between the two repeated segments. Moreover, the 100-fold induction of recombination depends on the nature of the intervening sequences. Transcription through bacterial DNA stimulates recombination, while transcription through equivalently long yeast DNA does not. The difference appears to be that transcription is impeded when traversing bacterial DNA (which did not coevolve with histones or with the eukaryotic polymerase apparatus). The authors speculate that stalled transcription could result in an encounter with a converging DNA polymerase, causing recombinogenic damage. However, defects in the relaxation of transcription-induced supercoiling might also cause DNA damage.
These insights into the mechanism of hpr1-stimulated recombination may help to clarify another case of stimulation of recombination. The HOT1 sequence, which is the promoter region of rDNA, also stimulates recombination when it is inserted in a different DNA context, and the insertion of a terminator of rRNA transcription prevents HOT1 activity (227, 527–529). The traversal of RNA PolI across non-rDNA sequences could cause recombinogenic lesions, and, as with hpr1, recombination would be initiated by stalled transcription.
Actually, HOT1 could have the same effect on rDNA. We do not know whether it stimulates recombination in the rDNA locus by direct experiment, but a number of mutants isolated by Lin and Keil (276) decrease both HOT1-stimulated recombination outside the rDNA locus, and recombination in rDNA. Thus, transcription (or at least transcription from a PolI promoter) could be recombinogenic per se. There is a high level of spontaneous mitotic recombination in the ribosomal locus, as shown by genetic assays (75, 495). In addition, a physical assay designed to detect HJs found a very high level of this recombination intermediate in the rDNA locus (575). The level of HJs is clearly linked to replication, as shown by its sharp increase during S phase and by the increased abundance of the HJ signal in mutations affecting the Polα or Polδ DNA polymerases. As yet, however, the level of HJs has not been shown to correlate with rDNA recombination. As mentioned above, the encounter of a DNA polymerase with an RNA polymerase could result in collapse of the replication fork. Whether the rDNA recombination events detected here have any relationship to transcription is unknown.
A similar effect of transcription through regions that are not normally transcribed seems to account for the hyperrecombination activity in other assay systems. Thomas and Rothstein (503, 504) showed that induction of transcription in a tandem duplication of GAL1 genes stimulated recombination, but virtually all of the recombination events proved to be deletions between the repeated genes and not gene conversions without crossing over. Curiously, a recent paper suggests that the induction of recombination depends on which promoter sequences are found upstream (55).
Stimulation of Recombination by Mutations in Genes Involved in DNA Topology
It has been known for some time that mutations affecting DNA topoisomerase genes TOP1, TOP2, and TOP3 stimulate recombination (75, 143, 533). In top1 top2 double mutants, most of the rDNA genes are found as extrachromosomal circles, probably generated by intrachromatid crossovers within the rDNA array (228). The origin of this stimulation of recombination is unclear. Topological problems in the chromosome could generate DNA damage and induce recombination. Also, it is known that the TOP2 gene is required for untangling sister chromatids after replication (98, 188, 189, 415), with failure to do so resulting in chromosome nondisjunction and breakage that could initiate recombination. On the other hand, TOP1 is involved in transcription elongation (57, 74, 436), and the hyperrecombination phenotype of top1 mutants could be similar to that of hpr1.
The SGS1 gene, identified as a suppressor of the slow-growth phenotype of top3 mutants, also suppresses the hyperrecombination phenotype of top3 (143). However, sgs1 mutants display a modest hyperrecombination phenotype (143, 536) and frequent chromosomal nondisjunctions (537). Sgs1p is a putative helicase, homologous to the bacterial RecQ protein, and it interacts with the Top1 and Top2 proteins (143, 537). It has been proposed that a Sgs1p helicase activity would prepare a substrate for topoisomerase activity and that this substrate would be recombinogenic if not processed (143).
The yeast SGS1 gene has attracted a lot of attention since the human genes involved in the Bloom and Werner syndromes were shown to be SGS1 homologs (106, 568). Also, sgs1 mutants show rapid aging (457). Since the Werner syndrome is a premature-aging syndrome, the SGS1 yeast gene might provide a paradigm for studying the molecular basis of the disease. In yeast, the sgs1 mutation result in amplification of the rDNA in extrachromosomal circles, an event that has been shown to trigger nucleolar fragmentation and cell death (456, 457). However, the exact role of SGS1 in DNA topology and hyperrecombination is still unclear.
Stimulation of Recombination by Unusual DNA Structures
Recombination also can be stimulated by unusual structures. Deletions between direct repeats are stimulated by intervening inverted repeats that are long enough to form (at least theoretically) cruciform structures when DNA in that region is denatured. Gordenin et al. (155) have speculated that such regions become denatured at replication forks and permit annealing of inverted repeats in ssDNA, so that DNA polymerase can switch from one repeat template to the other, thus causing a deletion. Such a mechanism could also explain why long arrays of CAG repeats, which can form hairpin structures in vitro (72, 139, 317, 318, 567), cause DSBs in yeast and stimulate recombination between flanking homologous repeats (134). Another possibility is that inverted repeats are acted upon by endonucleases to create recombinogenic lesions, possibly similar to the induction of deletions in bacteria harboring such repeats (84). Such a model could also explain why palindromes induce DSBs in meiosis (332).
Stimulation of Recombination by Defects in DNA Replication
Among the hyperrecombination mutations identified by Klein (4, 237) were hpr1 and hpr5 (srs2), both previously discussed, but there were many other complementation groups, all of which turned out to be defects in various genes involved with DNA replication. A good example is a defect in DNA ligase I (CDC9) (116, 140, 211, 212), and another is the Okazaki fragment-processing enzyme, Rad27p (Rth1p) (399, 463).
MEIOTIC RECOMBINATION
During yeast meiosis, a diploid cell generates haploid spores, which do not differentiate into gametes but germinate and grow as haploid cells until they find an opportunity to mate. Following premeiotic replication, meiosis proceeds by two rounds of chromosome segregation, the reductional division meiosis I and the equational division meiosis II. The resulting chromosomes are a patchwork of alleles from the two homologues present in the diploid parent. Meiotic recombination between genetically linked genes remains the most popular example of genetic recombination. A clear link has been shown between gene conversion and crossing over (46, 126, 492, 551). However, as for mitotic recombination, the exact relation between crossing over and gene conversion is still a matter of active investigation.
Thorough reviews have been recently published about the entire meiotic process (235, 413, 574). The present review is limited to a discussion of the recombination process, which takes place during the prophase of meiosis I. The first feature that seems to make meiotic recombination different from mitotic recombination is that meiotic recombination affects the whole genome at very high frequencies. There are about 100 crossovers per yeast meiosis. It has been interpreted as a way to ensure the reassortment of all alleles or, in more fashionable terms, to generate diversity. In addition, crossovers play an essential mechanical role, establishing a physical link (chiasmata) between the homologous chromosomes to ensure their proper segregation during meiosis I. Chromosomes that fail to cross over exhibit a high frequency of meiosis I nondisjunction.
A second, very conspicuous difference between mitotic recombination and meiotic recombination is that in most organisms, meiotic recombination occurs in the context of a proteinaceous structure called the synaptonemal complex (SC), lying between synapsed homologues. In yeast, the formation of the SC depends on recombination and, in turn, some aspects of recombination are regulated by the SC. However, in Drosophila or in Caenorhabditis elegans, SC formation does not appear to require recombination (94, 309), and S. pombe gets by without an apparent SC (243). Mammals may be more Saccharomyces-like, in that mutations that prevent meiotic recombination do not show synapsis between homologous chromosomes (379, 566).
However, there is a major common feature between meiotic and mitotic recombination in S. cerevisiae. DSBs are clearly a major cause of recombination in mitosis. The evidence is even more compelling for meiosis. In S. cerevisiae, DSBs are probably the only cause of elevated recombination. If we currently know much more about meiosis in budding yeast than in any other organism, it is not only because it is a convenient microbe for the geneticist, the molecular biologist, and, to a certain degree, the cytologist, but also because the event initiating most if not all recombinations has been characterized: it is a DSB.
DSBs Initiate Most if Not All Meiotic Recombination Events
When Szostak et al. (494) proposed their DSB repair model, it was largely influenced by transformation experiments in mitotic cells (357, 358), but the authors proposed it as a model to account for meiotic recombination as well. The Szostak laboratory kept on this track until they could demonstrate that meiotic recombination at the ARG4 locus was due to an initiation event in the promoter of the gene (348) and that DSBs were created in this region at the beginning of the meiosis I prophase (478). In further experiments (479), they showed that the ends of this DSB were then resected, resulting not in a double-strand gap, as proposed in the original model (494), but in long single-stranded 3′ ends (up to 800 nucleotides), in accord with what had been shown to occur in mitosis at the MAT locus (547).
The discovery of a novel mutation allowed the characterization of meiotic DSBs in more detail. Whereas a deletion of RAD50 prevents the formation of DSBs, a separation-of-function mutation, rad50S, allowed the formation of DSBs but prevented their subsequent resection and recombination (11, 64). In such a mutant, meiotic DSBs persist, so that it is possible to precisely map their location. Analysis of chromosomes by pulsed-field gel electrophoresis allowed the localization of DSBs along an entire chromosome in rad50S strains, at a resolution level of a few kilobases (570). More recently, DSBs also have been mapped at a higher resolution (<50 bp) in a large chromosomal region (557) and even along the entire length of chromosome III (25). DSBs have been mapped at the nucleotide level at the ARG4 and CYC3 loci, at composite DSB hot spots resulting from a HIS4-bacterial DNA-LEU2 fusion and from an insertion of a telomeric sequence at the HIS4 locus (93, 278, 561, 562). These analyses showed that a hot spot spanned more than 100 bp and actually consisted of a number of sites where DNA cleavage occurred, yielding either 2-bp 5′ overhangs (278) or blunt ends (93), although the blunt ends may reflect filling in of the 5′ end during DNA isolation.
The conclusion of these studies is that DSBs are site specific but not sequence-specific. Hot spots appear to coincide with gene promoters that contain DNase I- or micrococcal nuclease-hypersensitive sites (118, 225, 356, 557); however, hot-spot activity does not depend on active transcription but presumably requires proper chromatin remodeling by transcription factors (118). At some hot spots at least, there are additional chromatin changes at the time when DSBs are created (356).
The mapping of all the DSBs hot spots of chromosome III allowed investigators to test the hypothesis that the prominent DSBs initiate all meiotic recombination events (25, 557). When the recombination frequencies (the genetic map) were compared with the DSB map, a good correlation was found for most of the chromosome. However, some discrepancies remained. For example, a 50-kb region representing 24 centimorgans (cM) accounts for only 2% of the DSBs mapped on the chromosome. Although this could be explained by non-site-specific DSBs (which could not be mapped because they would appear randomly), we cannot rule out that some recombination events could be initiated by a mysterious “something else,” although these events would also depend on the early genes, including SPO11.
Generation and Processing of DSBs
The initiation of meiotic recombination depends on 11 genes that have been shown by genetic or physical assays to be required for DSB formation, including RAD50, SPO11 (11, 64), MRE11 (214), XRS2 (196), MEI4 (310, 311), MER1 (108, 109, 467), MER2 (409), MRE2 (8, 336), REC102, REC104, and REC114 (61). In addition, the RED1, HOP1, and MEK1 genes, involved in the formation of the axial elements between sister chromatids (see below), appear to be required for full levels of meiotic DSBs (297, 563).
The meiotic endonuclease is almost certainly Spo11p.
In rad50S strains, where DSBs are not resected, a protein was found to be covalently linked to the 5′ ends (92, 278, 562), which probably explain why they are not resected. This discovery led to the hypothesis that this covalently attached protein was the endonuclease itself, stuck to the 5′ end after having catalyzed a transesterification step. Two recent and independent studies argue that this endonuclease is the product of the SPO11 gene. Keeney et al. (224) directly isolated the protein bound to the DSB 5′ end and showed that it was the product of the SPO11 gene. Bergerat et al. (34) found that an archaebacterial type II topoisomerase had homology to Spo11p. In most cleavage reactions catalyzed by a topoisomerase, a tyrosine residue attacks a phosphodiester bond, resulting in a transient covalent DNA-protein complex. When Bergerat et al. mutated a tyrosine conserved between Spo11p and its homologues, meiotic recombination was abolished. Sequences homologous to SPO11 have now been noted in S. pombe, C. elegans, and D. melanogaster. The S. pombe homologue, REC12, is required to initiate meiotic recombination (275, 460), but it has not yet been possible to visualize DSBs in fission yeast. A deletion of the C. elegans homologue also abolishes meiotic recombination (94), and the recombinationless mei-W68 mutation in Drosophila has now been shown to affect a SPO11 homologue (308). Therefore, this way of creating DSBs is highly conserved, and this conservation may extend to mammals. However, to date, Spo11p has not been demonstrated to have cleavage activity.
What is the role of other genes involved in DSB formation?
Although the type of reaction catalyzed by Spo11p seems to be understood, the mode of action of this endonuclease remains mysterious. Its activity requires the presence of many other genes, whose exact roles are unknown, with the exception of MER1 and MRE2, which regulate the splicing of the MER2 mRNA (108, 336) and probably also other transcripts, thereby affecting a number of early steps in DSB formation and metabolism.
(i) Chromosome pairing and chromatin remodeling.
Weiner and Kleckner (539) have documented early meiotic (pre-DSB) interactions between homologues by fluorescent in situ hybridization analysis. Such pairing is abolished or impaired in spo11 and rad50 mutants, which suggests that the complex involved in DSB formation begins to play a role even before cutting. The authors proposed that “an early meiotic pairing occurs by closely related paranemic DNA-DNA interactions subsequently converted directly to plectonemic recombination intermediates via DSBs” (539). However, it is clear that pairing between homologues is not a prerequisite for DSB formation, since DSBs are generated at nearly normal frequencies in haploid cells expressing both mating types, so that meiosis can be induced (92, 147, 532).
In normal diploid cells, a full level of DSB formation at a given hot spot depends on the homozygosity of the hot spot. When the two homologues differ over a few dozen or a few hundred base pairs in the hot-spot region, the level of DSBs is somewhat reduced on both chromosomes (408, 562). Further investigation showed that homozygosity favored an “open” configuration at the hot spot, as characterized by nuclease hypersensitivity, indicating a possible link between pairing and chromatin remodeling (408, 562). A recent study shows that Mre11p, Rad50p, Xrs2p, and Mre2p affect the micrococcal nuclease sensitivity at the ARG4 meiotic DSB hotspot, with mre11 and mre2 mutations decreasing the sensitivity and rad50 and xrs2 mutations increasing it (355). This is the first result that suggests separate functions for Mre11p versus Rad50p and Xrs2p. It also indicates that increased nuclease accessibility is insufficient to promote DSB formation.
(ii) Regulation of Spo11p activity.
The action of Spo11p is regulated in a complicated fashion. Just as there are associated proteins that regulate the bacterial gyrase, some of the “early” meiosis genes may encode such regulators. As mentioned above, Red1p and Hop1p, two components of the axial elements (the future lateral elements of the synaptonemal complex, see below), are required for full levels of DSBs. The same is true for Mek1p (563), which has been shown to be a protein kinase that phosphorylates Red1p (20, 91). However, in a rad50S mutant, where DSBs are not resected, red1 and mek1 mutations no longer affect the DSB level (563). Xu et al. (563) have suggested that cutting by Spo11p might be a reversible reaction (according to its topoisomerase-like nature), with Red1p and Mek1p affecting the reaction equilibrium in favor of the cut product, thus favoring stable cleavage. The effect of the mek1 and red1 mutations would not be seen in a mek1 rad50S or red1 rad50S double mutant, because even if irreversible cleavages arise at a lower rate, they are not turned over but, rather, accumulate in an unresected form.
(iii) Removal of Spo11p and resection of DSBs.
RAD50, MRE11, and XRS2 play multiple roles in DSB metabolism. Although null mutants are unable to initiate meiotic DSBs, separation-of-function mutations in both RAD50 and MRE11 allow SPO11-mediated DSBs to be formed but not resected. One of them is the rad50S allele, which we have already mentioned, and others are alleles of MRE11, mre11S (335), mre11-58 (73, 514), and mre11D16A (138). These mre11S, mre11-58, and mre11D16A mutations are in the N-terminal part of the protein, in or near conserved phosphoesterase motifs.
In a rad50S mutant, Spo11p catalyzes DSBs formation but remains covalently associated with the 5′ ends of the breaks. It seems logical that the covalently bound Spo11p has to be removed to allow further exonucleolytic resection. For example, in a rad50S mutant, DSB ends cannot be resected by λ exonuclease (278), an observation also made with a mre11-58 mutant (514). Therefore, one of the likely functions of Rad50p and Mre11p is to remove the Spo11p from the DSB ends in order to initiate DSB resection and allow meiotic recombination. It is generally assumed that Spo11p removal occurs by Mre11-mediated cleavage of the DNA near the protein, but it cannot yet be excluded that Mre11 and other proteins act allosterically to stimulate Spo11 to hydrolyze itself from the end.
Rad50p, Mre11p, and Xrs2p probably play a role in the 5′-to-3′ degradation itself during meiosis as they do in mitosis, where resection is reduced in rad50, mre11, and xrs2 null mutants and also in the mre11-58 mutant. Such an effect would be veiled by the strong meiosis-specific phenotypes of the corresponding mutations (no DSB formation in rad50, mre11, and xrs2 null mutants, and no resection at all in rad50S or mre11-58 mutants). The rad50S mutation is capable of degrading DSBs in mitosis (514), suggesting that in this mutant, it is probably only the ability to remove Spo11p from the DSBs ends that is affected.
Nairz and Klein (335) found an opposite set of separation-of-function alleles of MRE11. Transposon insertions in the C terminus of MRE11 abolish DSB formation. However, such alleles can be complemented by mre11-S in diploids where DSBs are both made and processed. Analogous C-terminal truncations have been created by other laboratories (138, 352). These C-terminal mutations are not radiation sensitive and may affect only the role of Mre11p in DSB formation. Two DNA binding motifs are found in the C-terminal part of Mre11p, and C-terminal truncations probably abolish the ability of Mre11p to take part in a DSB-creating complex that somehow interacts with Spo11p. Interestingly, immunoprecipitation of a glutathione S-transferase–C-terminal segment of Mre11p coprecipitates three additional meiosis-specific proteins, whose identity is unknown (352).
Finally, a meiosis-specific gene, SAE2/COM1, is also required for the removal of Spo11p from DSB ends, with the same subsequent phenotypes in meiosis as rad50S (226, 306, 387), but, unlike RAD50 and MRE11, this gene is not involved in DSB formation, since a null mutation affects only the resection process. sae2/com1 mutants are MMS sensitive in mitotic cells but have not been tested for other phenotypes shown by mre11Δ.
The conclusion from all these data is that there are several multiprotein complexes that perform five different functions early in meiosis I prophase, before the formation of any joint molecule: transient homologous sequence pairing, the remodeling of the chromatin structure, the initiation of DSBs, the excision of the Spo11p endonuclease, and DSB end resection. Only at this point would another set of proteins take over to initiate DNA strand transfer. However, additional data suggest a sixth precondition for recombination: the establishment of a marked preference for interhomologue recombination over interchromatid recombination. The relevant results are discussed in the next section.
Formation of Recombinant Products
Physical monitoring of meiotic recombination.
The ability to monitor the kinetics of the appearance of several meiotic intermediates, as well as the final products of recombination, has greatly aided our description of the sequence of molecular events.
(i) Kinetics of the appearance of gene conversion and crossover products.
Borts et al. (50) first constructed a chromosomal region containing restriction endonuclease differences on two homologous chromosomes that could be used to detect crossing over by the appearance of novel length restriction fragments on Southern blots. They showed that there was no crossing over in rad50 diploids but that both rad52 and rad57 cells did yield a significant level of exchange, despite producing only inviable spores (49). When it became possible to monitor the appearance of both DSBs and crossovers, Cao et al. (64) showed that there was at least a 1-h delay between DSB formation and the completion of recombination, an observation very similar to that seen in mitotic cells after HO cleavage of DNA. Subsequent studies have monitored the appearance of gene conversion events both with and without crossing over (467, 468).
(ii) Detection of Holliday junctions.
An intermediate predicted by various models of recombination is a double HJ (Fig. 6). HJ intermediates were first detected in 2μm plasmid DNA from meiotic cells by Bell and Byers (29), both on Southern blots and by electron microscopy. Currently such structures are identified by two-dimensional gel electrophoresis, similar to the technique used to capture replication intermediates (Fig. 7A). Branched DNA molecules appear shortly after DSB formation (83, 438), and subsequent analysis by Schwacha and Kleckner (437) confirmed that these are indeed double HJs: when these intermediates are denatured, only parental strands are recovered (Fig. 7B); but when the same intermediates are treated with the RuvC resolvase, which cleaves HJs (31, 32), both parental and recombinant strands are obtained. Double HJs disappear at the time that crossovers can be detected, at the same time that the SC disappears.
When the two parental chromosomes carried restriction site polymorphisms so that crossovers could be detected, three distinct joint molecule (JM) species are seen (Fig. 7A). The major spot is that predicted for recombination between the two homologues; the two minor spots are those expected for intersister recombination. The predominance of the biparental spot provides strong evidence that recombination between sisters is greatly suppressed, as inferred from previous genetic studies (165) and from physical analysis of whole chromosomes with chromosome-separating gels (142), in diploids carrying both linear and circular homologous chromosomes. Recent studies have used the ability to analyze intersister and biparental JMs to identify gene products that enforce the selection of nonsister DNA molecules for recombination (this is discussed further below).
(iii) Detection of recombinant strands.
Very recently, Bascom-Slack and Dawson (24) provided evidence that the formation of recombinant strands is an early event. They used an allele-specific PCR assay to detect the first appearance of recombinant strands carrying markers from the two different parent chromosomes and showed that they appeared at the time of commitment to recombination (which corresponds to the appearance of DSBs). The two markers are located on either side of the DSB hot spot, and the recombinant strands detected in this assay could be the result of HJ resolution, but we know that HJ resolution occurs later in meiosis (83, 438, 467, 468). Therefore, the recombinant strands are more likely to result from the DNA synthesis primed from the 3′ end of the invading strand.
(iv) Detection of heteroduplex DNA.
Genetic recombination is associated with heteroduplex DNA (hDNA) formation. Short restriction fragments containing mismatches can be detected on gradient thermal denaturing gels or by loss of restriction sites. In practice, mismatch repair is apparently too rapid to detect such intermediates, but they can be detected in DNA from cells in which a mismatch repair gene is deleted (269) or when the mismatch itself is poorly repaired (157, 333).
It is generally assumed that hDNA appears during the strand invasion step (see Fig. 16B). However, during meiosis, the appearance of hDNA seems far too late to reflect strand invasion; indeed, hDNA was not seen until the end of pachytene, at least 1 h after the time of appearance of recombinant strands and JMs and at about the same time that crossover products could be seen (157, 333, 438). It should be noted that the kinetics of hDNA and JM appearance were monitored in the same strain background but by different laboratories, monitoring different loci. It is possible that there is no conflict here but simply a difference in the timing of recombination at two different chromosome locations. Clearly these measurements need to be done for one set of markers in a single experiment. At the artificial HIS4::LEU2 locus, no hDNA has yet been found in JMs. In addition, heteroduplex regions smaller than 150 bp would have escaped detection in these studies, but hDNA is supposed to be formed over a much larger distance (95).
FIG. 16.
Formation and correction of hDNA. (A) In the model of Holliday (185), hDNA would be corrected after HJ resolution. Conversion would be the consequence of a correction in favor of the invading strand. Correction in favor of the recipient strand would yield a restoration event. (B) Some evidence suggest that hDNA formed during strand invasion is corrected immediately. In the Szostak et al. (494) model, conversion would result from correction in favor of the recipient strand (see Fig. 17A and B), which is the opposite of Holliday’s proposition. (C) hDNA can also result from annealing in SDSA models. Then it is impossible to define an invading and a recipient strand, and correction may be unbiased.
However, when Bascom-Slack and Dawson (24) showed that recombinant-strand formation was an early event, one of the diagnostic alleles they used was arg4-NspI, the same allele used by Goyon and Lichten (157), who detected hDNA only 1 h later, in the same strain. One explanation for the apparently late stage at which hDNA is detected would be that the detected hDNA does not reflect the initial strand invasion step. Bascom-Slack and Dawson propose that during meiotic recombination, strand invasion and the priming of new DNA synthesis involve only a short region of hDNA, similar to the bubble migration models discussed above and to a model proposed by Priebe et al. (386) for homeologous recombination. hDNA would be formed at a later step.
One possibility is that hDNA results from branch migration after strand invasion. One has to keep in mind that during DSB repair, two kinds of heteroduplex can appear: asymmetric ones resulting from strand invasion and symmetric ones resulting from branch migration (Fig. 15B). However, the genetic results are strongly at odds with the idea that branch migration would produce hDNA late in the process, at least as predicted by models such as that of Szostak et al. Branch migration of an HJ would produce hDNA symmetrically on both participating chromatids and would be expected to yield so-called aberrant 4:4 tetrads (Fig. 15A). In S. cerevisiae, aberrant 4:4 tetrads are quite rare (12, 128, 494). Such events can be found in the fungus Ascobolus immersus for markers distant from a hot spot, but only 5:3 and 3:5 segregations are found for high-postmeiotic segregation (PMS) alleles (Fig. 15B) closer to the hotspot (418, 419). The Meselson and Radding (312) model of recombination specifically accounted for this transition from an asymmetric heteroduplex next the site of initiation to symmetrical heteroduplex more distally (Fig. 15B).
FIG. 15.
hDNA formation and correction analyzed by meiotic tetrad analysis. (A) Tetrad analysis. Sporulation of a heterozygous diploid most of the time results in two spores with one allele and two spores with the other one. This is the normal Mendelian segregation (4:4). However, other patterns are also observed. Non-Mendelian segregations include conversions and PMS. In PMS, the two alleles will segregate only after the first cell division following meiosis, resulting in a sectored colony. Various patterns of conversions and PMS are shown here. Gene conversion are often referred to as 2:6 or 6:2 segregations, since PMS events are referred to as 3:5, 5:3, or aberrant 4:4 (to differentiate it from normal Mendelian 4:4). This nomenclature is derived from the one used with filamentous fungi, where meiosis is followed by an additional cell division, resulting in eight spores instead of four. Note that in yeast, the aberrant 4:4 pattern is very rarely observed. (B) Molecular interpretation of non-Mendelian segregation. Strand invasion results in an asymmetric hDNA (step 1). Asymetric hDNA is found on only one chromatid. The displaced strand is used as template for DNA synthesis and yields a homoduplex (step 2). If hDNA is not corrected, asymetric hDNA results in 5:3 or 3:5 segregation. If corrected, it results in 6:2 or 2:6 segregation, depending whether correction occurs in favor of the recipient strand or in favor of the invading strand. In addition to the initial asymmetric hDNA, symmetric hDNA could result from branch migration (step 3). Symmetric hDNA is found over the same portion of two different chromatids. If not corrected, it will result in aberrant 4:4 segregation (step 4). Such events are rare in yeast but are common in Ascobolus.
There is another possible explanation based on SDSA models. The displacement of a newly synthesized strand could yield the late appearance of asymmetric heteroduplex. hDNA would be the consequence not of strand invasion but of annealing of the newly synthesized strand with the other side of the DSB, as illustrated in Fig. 16C. The kind of hDNA tract shown in Fig. 16C would extend on only one side of the DSB and could account for the frequent unidirectional conversion tracts observed in meiosis (Fig. 9) (148, 383). Also, this kind of hDNA would arise late in the recombination process, which could explain why in meiosis, hDNA is detected after the appearance of joint molecules and recombinant strands and about at the same time as the final recombinant products.
In this view, most of the observed hDNA would be the consequence of an SDSA event without Holliday Junction formation (Fig. 8A and B). It would be the consequence of SDSA with crossing over (Fig. 8C and D), with hDNA formation only in a small region of annealing, possibly far from the DSB region. This kind of hDNA would appear at the same time as the HJs but could be undetected. This would explain why Schwacha and Kleckner (438) could not find hDNA in the joint molecules they examined. We return to this important issue below (see “Role of mismatch repair proteins in recombination”).
It is also possible that this discrepancy comes from technical differences. The detection of JM is substantially improved (more than 10-fold) when the DNA is cross-linked before extraction (438); without cross-linking, the level of observed JM is low (83), probably because of reverse branch migration. Reverse branch migration could dissociate the heteroduplex in the hDNA detection attempts mentioned above (done without cross-linking): the heteroduplexes and JM would be unstable until ligation occurs. This would imply that ligation occurs shortly before resolution.
Strand invasion proteins in meiosis.
It has been proposed that in mitosis, a recombinosome involving Rad52p, Rad51p, Rad54p, Rad55p, and Rad57p catalyzes the invasion of the unbroken molecule by the 3′ resected single strands of the broken one (179), although the discovery of Rad59p (18) and Rdh54/Tid1p (238, 451) shows that things are probably more complicated. In meiosis, recombination requires an additional RecA homologue, Dmc1p (42). DMC1 is expressed only in meiosis. In yeast dmc1 mutants, meiotic recombination is strongly impaired, and completion of meiosis is prevented in some strains (42, 100, 412, 439). The 5′ ends of meiotic DSBs become hyperresected, presumably because recombination is largely prevented. In mice, a dmc1 deletion completely prevents the completion of meiosis (379, 566).
(i) Distinct roles of Dmc1p and Rad51p.
The relationship between Dmc1p and Rad51p is unclear, with the main question being whether they act in one or two different pathways. Cytological testing, genetics, and molecular biology have been used to try to decipher their respective roles, and the answer is ambiguous. The cytological approach is the most controversial. During meiosis I prophase, Rad51p and Dmc1p are clustered in foci, which can be visualized by indirect immunofluorescence (40, 100). It has been proposed that these foci correspond to recombination nodules, the presumptive sites of recombination. Recombination nodules are structures revealed by electron microscopy in meiosis I prophase in metazoans and plants (66), and a distinction has been made between early nodules (seen in leptotene and zygotene) and late nodules (in pachytene). The number of late nodules fits well with the number of crossover events, and they have become good candidates for meiotic recombinosomes. Most of the immunofluorescence data on recombination nodules have been obtained with lilies, and a recent study gives good evidence that Rad51p and Dmc1p are indeed components of early nodules in these plants (14). In the yeast strain SK1, a large proportion of Dmc1p and Rad51p foci colocalize; Rad51p is necessary for the formation of most Dmc1p foci, and Dmc1p is necessary for the dissociation of the Rad51p foci (40). A few Dmc1p foci can be detected in rad51 mutants and persist longer than in wild-type strains (451). Another study, done in a different strain background, showed a lower correlation between the Rad51p and Dmc1p foci, which colocalized only in 21 to 53% of the cases (100).
Genetic and molecular approaches support these general conclusions, but, as with other meiotic phenomena, there are very important differences among strains that prevent us from having a fully coherent view. In strain SK1, a dmc1 mutant has a more severe phenotype than a rad51 mutant; it has a more pronounced effect on JM and crossover formation (100, 439, 451) and has a more severe arrest phenotype (dmc1 cells display a permanent arrest at the pachytene stage of meiosis, while a subset of the rad51 cells eventually bypass the arrest [563]). However, in the strain used by the Roeder laboratory, a dmc1 deletion does not cause pachytene arrest and the tetrads exhibit a 20% spore viability (411, 412). In contrast, rad51 abolishes sporulation. The genetic basis of this important difference has not yet been determined.
In strain SK1, a synergistic effect of rad51 and dmc1 has been observed on recombination (100, 451), but this has been assessed in return-to-growth experiments, where the respective roles of mitotic and meiotic recombination pathways are unclear. In such experiments, cells induced to initiate meiotic recombination (presumably to create meiotic DSBs) are removed from nitrogen starvation medium and returned to growth medium, where recombination is completed by what may be a more mitosis-like machinery. Recent studies have shown that SCs are quickly dismantled and that recombination proceeds more rapidly than in meiosis (571). Cells in a return-to-growth experiment tend to show the high proportion of crossovers characteristic of meiotic recombination (113).
The major difference between Rad51p and Dmc1p functions seems to be the specific role of DMC1 in interhomologue recombination, as illustrated by the important work of Schwacha and Kleckner (439). This study suggests that most DSBs are predetermined early in meiosis, in a Red1p-Hop1p-dependent fashion, to enter a Dmc1p-dependent pathway that results preferentially in interhomologue recombination. In meiosis, intersister recombination in SK1 occurs at lower level than interhomologue recombination, as demonstrated by two-dimensional gel experiments detecting intersister and interhomologue joint molecules (439). In a red1 mutant, the proportion of interhomologue JMs is greatly reduced. In dmc1 cells, both intersister and interhomologue recombination intermediates are undetectable. In contrast, JMs are late and less frequent in rad51 cells than in wild-type cells. However, in a red1 dmc1 double mutant, intersister JMs are restored, but the interhomologue joints are not.
red1 and hop1 alleles were identified in a screen for mutants defective for interhomologue reciprocal recombination (187). The results of Schwacha and Kleckner (439) show that for red1 and probably hop1 mutants, the decrease in interhomologue reciprocal recombination is probably due to an increase in intersister recombination. Interestingly, the screen that identified red1 and hop1 yielded mutations affecting various steps, since it also led to the discovery of a msh5 mutation, which plays a direct role in crossover control (see below).
(ii) Other components of the Dmc1 pathway.
An apparent partner for Dmc1 for interhomologue recombination is the Tid1/Rdh54 protein. In two-hybrid experiments, Tid1/Rdh54p interacts with Dmc1p (100). Moreover, Tid1/Rdh54p, which is not meiosis specific, specifically affects interhomologue recombination during mitosis (238). Whether it specifically affects interhomologue recombination during meiosis has not been tested, but the protein is necessary for sporulation and spore viability (although the phenotype is again strain dependent [238, 451]).
The strong lethality for spores observed in a spo13 tid1/rdh54 double mutant is alleviated in a spo13 tid1/rdh54 red1 triple mutant (in a spo13 background, the meiosis I reductional division does not occur; two viable diploid spores are produced, provided that the meiotic DSBs are repaired). This result is reminiscent of the interactions between RED1 and DMC1. TID1/RDH54 would be necessary for meiotic interhomologue recombination driven by RED1, and in a red1 tid1/rdh54 mutant, DSBs could be repaired by sister chromatid exchange.
The SAE3 gene could be a third component of this pathway, since a sae3 mutant has same phenotype as dmc1: hyperresection of DSBs, delayed SC formation, a recombination defect, and meiotic arrest in pachytene (307). A dmc1 sae3 double mutant has the same phenotype as either single mutant, at least for SC formation and meiotic arrest, indicating that DMC1 and SAE3 act in the same pathway. The phenotype of a sae3 red1 double mutant has not yet been reported.
Cytological Monitoring of Intermediate Steps in Recombination
The development of sensitive immunofluorescence methods has begun to make it possible to learn about the assembly of the recombination machinery. In meiosis, one can observe the appearance of foci formed by Rad51p and/or Dmc1p, presumably at sites of DSBs (see above) (40, 100, 451). Recently, Gasior et al. (145) have extended this type of analysis to investigate if such foci can be assembled in the absence of other proteins, both at normal meiotic DSBs and after X-ray irradiation. In meiosis, foci formation depends on the presence of Rad52p as well as Rad55p and Rad57p. RPA, the single-strand binding protein, colocalizes exactly with Rad52p (145). However, in mitotic cells, Rad51p foci appear after X-ray irradiation, even in the absence of Rad52p, Rad55p, and Rad57p (39). The ability to visualize proteins assemblies in this way will undoubtedly provide an important new way to monitor recombination.
We should also soon see the use of chromatin precipitation techniques in demonstrating the time at which recombination proteins become associated with donor and recipient DNA sequences, in much the same way as one monitors the progression of DNA replication (15).
Synaptonemal Complex
The milestones of the recombination process (DSB formation, JMs, and products) follow each other during the prophase of meiosis I. These stages have been correlated with the cytological appearance of chromosomes. The prominent structural change during this time is the formation of the SC.
Following premeiotic replication, two sister chromatids begin to be organized along a proteinaceous structure called the axial element during the leptotene stage of meiosis I. The homologous chromosomes start synapsis during the next stage, zygotene, as they form axial elements between sister chromatids. The homologous chromosomes are aligned, and the axial elements become the lateral elements of the SC. Synapsis is complete in pachytene, and the SC disassembles during diplotene, to allow diakinesis. DSBs appear during the leptotene (axial element assembly), double HJs appear at the beginning of the pachytene, and heteroduplexes and recombinant products appear at the end of the pachytene (157, 333, 362, 438, 467).
A few components of the SC have been identified. The Red1 and Hop1 proteins are two components of the axial-lateral element (186, 459). Zip1p is a component of the central element, and zip1 mutants fail to synapse the axial elements (488–490, 519). Zip1p is found along the whole central element and is thought to form a filament that zips the axial-lateral elements together. Another protein, Zip2p, is required for an early step of synapsis, since it is required for normal Zip1p distribution (77). Unlike Zip1p, which is assembled along the axis of the SC, Zip2p is found at only a few foci along the SC.
Another protein, Hop2p, is not a structural component of the SC but is necessary for proper synapsis: in a hop2 mutant, chromosomes synapse with nonhomologous partners (265). hop2 cells accumulate unrepaired DSBs, suggesting that they also fail to locate or to complete recombination with their homologous partners. It is possible that Hop2p plays a very early role in homologous recombination, perhaps in determining if a transient homology search had located a truly homologous template for DSB repair. This is discussed further in reference 159.
It is now clear that SC formation in yeast depends on recombination. Synapsis is totally absent from mutants that do not make meiotic DSBs (11, 409) and delayed in mutants that do not repair DSBs, such as rad51 and dmc1 mutants (42, 412, 449). In a zip1 mutant, defective for synapsis, homologous chromosomes are still paired at a few sites called axial associations (488). These connections disappear when the DSB repair machinery is also mutated (412), which suggests that axial associations are the sites of DSB formation and repair. Also, in a zip1 mutant, these pairing sites accumulate Zip2p (77), which indicates that they correspond to the preferential sites of the early steps of synapsis. Finally, in a rad50S mutant, where the DSBs are neither processed nor repaired, Zip2p colocalizes with Mre11p, providing further evidence that DSB sites are initiation sites for SC formation. A recent report demonstrates that Rad51p can localize in foci in a rad50S mutant (145), raising the possibility that Zip2, Mre11p, and Rad51p are all recruited to DSB sites, at least in the rad50S mutant. One might imagine that in a dmc1 mutant, where synapsis is delayed but not absent, there is still enough pairing activity in the cell (perhaps Rad51p dependent) to promote SC polymerization, and indeed in some strain backgrounds, dmc1 mutants give rise to about 20% viable spores (411, 412).
In mammals, as in yeast, synapsis depends on recombination, since SC formation is completely abolished in dmc1 mutants (379, 566). However, this is not a universally conserved feature, and SC formation in Drosophila or in C. elegans does not appear to require recombination (94, 309).
If recombination is necessary for SC assembly in yeast, the converse is not the case: zip1 and zip2 mutants do not assemble SC but are still competent to carry out recombination (468, 488, 489). However, the SC and/or its subcomponents, the axial-lateral elements, have a regulatory role at three levels of the recombination process: DSB formation, intersister versus interhomologues recombination, and crossover control.
However, the clearest role of the tripartite SC is in the regulation of the frequency, distribution, and timing of crossovers. This regulation is complex and deserves to be dealt with in a separate section.
Regulation of Crossover Events
Need for frequent meiotic crossover events.
Meiotic crossover events occur at much higher frequencies than in mitotic cells. This high frequency of crossovers could be simply a consequence of the generally higher frequency of recombination events, initiated by the Spo11p-catalyzed meiotic DSBs, but in fact, meiotic gene conversions are associated with crossovers much more frequently than are mitotic conversions. A systematic study of the proportion of meiotic gene conversions associated with crossing over was performed by Fogel et al. (126), who showed that the frequency of crossover-associated events was 35% on average. There has not been a single systematic study of many loci in one strain in mitotic cells; instead, there are a number of observations drawn from several strains, in which we do not know how or when in the cell cycle recombination was initiated. Taken together, these studies suggest that interhomologue gene conversion is associated with crossing over 10 to 20% of the time (112, 165, 252).
Mutations that reduce or eliminate crossovers cause a dramatic increase in chromosome nondisjunction, so that virtually all spores are aneuploid and inviable (295, 489). Thus, since the crossover is an essential step of meiotic mechanics and an evolutionarily desired outcome to generate diversity, one can imagine that the ideal career of a meiotic DSB is to become a crossover; however, too many crossovers may be as bad as too few. The fate of meiotic DSBs is controlled by a complex machinery whose purpose is to regulate the number of crossovers per chromosome.
A proper number of crossovers per chromosome.
Crossovers are regulated in overall frequency but also in their distribution along a chromosome. First, a crossover at one locus decreases the probability of another crossover in the vicinity. This phenomenon is known as crossover interference. Interference is abolished in zip1 mutants (489) but also in msh4 mutants (413), indicating that it does not involve only SC components.
The mechanism of interference remains elusive. A likely level of regulation by interference is the resolution of recombination intermediates, a hypothesis supported by the phenotype of zip1 mutants, in which crossover interference is suppressed and crossover frequencies are decreased but the levels of gene conversion are not (489). However, some of this regulation may also occur at the level of creating DSBs. Data are not yet available to rule out an interference mechanism that prevents nearby hot spots from being cleaved, but it is clear that a hot spot will change the frequency of DSBs at another neighboring hot spot. The ARG4 gene contains a prominent hot spot for DSBs in its promoter region (348, 478). However, when ARG4 is embedded in pBR322 sequences and integrated at other chromosomal locations, DSBs within the ARG4 promoter are surprisingly absent and all the DSBs are found in plasmid sequences. Deletion of the plasmid sequences restores cleavage at ARG4 (556).
It has been proposed that interference results from an inhibitory signal that is transduced from a crossover site to adjacent DSB repair sites (229). The role of Zip1p in interference argues that the SC is the transducing channel (489). Foss et al. (131) proposed a mechanism based on a “smart” resolvase that would move progressively down a chromosome, resolving two HJs as noncrossovers before resolving one as a crossover. One test of this model failed (132), but the authors suggest that this is a consequence of the biological uncertainty principle, i.e., that marking up a region to study it results in changing its behavior. A third model is based on the idea that crossovers relieve some sort of topological stress that builds up along the SC, with crossovers spaced similarly to the distribution of cracks in a polymer film coating a rigid, twisted beam (574).
A second essential feature of crossover regulation is that there is at least one crossover per chromosome, a phenomenon referred to as obligate chiasma. The total meiotic genetic map in yeast is about 5,000 cM, where a 50-cM distance is equivalent to one crossing over between two nonsister chromatids. Roughly speaking, there are about 100 crossovers distributed along 16 unequally sized chromosomes. Tetrad analysis of chromosomes marked from one end to the other indicates that almost all chromosomes experience at least one crossover whereas a Poisson distribution would predict a much higher proportion of small chromosomes lacking any crossovers (168, 219).
A third observation is that the crossover frequency is regulated by chromosome size. The number of crossovers per chromosome is not proportional to the chromosome size, as would be the case if crossovers occurred randomly; rather, the number of crossover per kilobase decreases as the size of the chromosome increases (215, 217, 219). The direct relationship between the size of the chromosome and the number of crossovers was definitively shown by Kaback et al. (218), who demonstrated that when a large chromosome was bisected, the two resulting small chromosomes underwent more crossing over per kilobase than did the large chromosome. The regulation by the size of the chromosome is likely to define a single phenomenon with crossover interference. This regulation does not seem to be at the level of DSB formation, because the frequency of total gene conversions at one locus does not change as the chromosome size is altered.
The three phenomena described above, i.e., crossover interference, obligate chiasma, and regulation by chromosome size, are probably three manifestations of a same mechanism of regulation at the chromosome level. However, other factors, possibly independent of this global regulation, affect the distribution of crossovers within the chromosome. While centromeric regions in higher eukaryotes are strongly suppressed for crossing over, there is only a very modest (fourfold) effect in yeast. This was shown by taking the audacious step of simultaneously deleting and inserting elsewhere the centromere of chromosome III and measuring crossing over in the same intervals (258, 259). The centromere does not form a barrier against gene conversion, since Symington and Petes (493) demonstrated that gene conversions identified at a locus 1 kb to one side of a centromere were frequently accompanied by coconversion of markers either within or on the other side of the centromere. There also is a modest repression of crossing over, but perhaps not of gene conversion, near telomeres. The frequency of crossing over in regions near telomeres appears to be reduced, per kilobase, compared to more internal regions, and this is reflected in an absence of meiotic DSBs in these regions (25, 240). However, ectopic gene conversion between genes inserted in subtelomeric regions is not notably reduced compared to similar events between sequences inserted at other locations (281).
Factors that modulate crossover frequency.
So far, six genes have been shown to be required for a full level of crossing over: MSH4 (416), MSH5 (187), MLH1 (21, 191), ZIP1 (468, 488, 489), ZIP2 (77), and MEI5 (319). An unknown gene whose splicing is controlled by MER1 and MRE2 (110, 336, 467) is also required for normal levels of crossing over.
msh4, msh5 and mlh1 mutations decrease the crossover frequency two- to threefold (187, 191, 416). Msh4p and Msh5p are two MutSp homologues with no role in mismatch repair. In contrast, Mlh1p, a MutLp homologue, was known to be a mismatch repair gene long before its role in crossover resolution was shown (388). The exact role of these genes is unknown. Msh4 and Msh5 proteins form a heterodimer (382), and Msh4 proteins are observed in foci during pachytene, with a distribution that could fit that of late recombination nodules (416). Msh4p and Msh5p may bind an HJ or other branched intermediate, just as Msh2p has been shown to do (10), and then recruit Mlh1p, but no biochemical data have yet been obtained to support these ideas.
Reciprocal recombination is decreased 1.4- to 4-fold in zip1 mutants, depending on the locus (468, 489), and in a comparable manner in zip2 mutants (77). We cannot really assign a specific role to these genes but, rather, conclude that the SC, in addition to its role in crossover interference, is required for a high level of crossovers. Thus, without the SC, the meiotic DSB repair more closely resembles mitotic recombination, with a low frequency of associated crossovers, randomly distributed.
The MER2 gene is required for DSB formation (409, 467), and its proper expression depends on the correct splicing of its mRNA, which is controlled by two genes, MER1 (110) and MRE2 (336). In mer1 and mre2 mutants, DSB formation can be rescued by overexpression of a MER2 gene or cDNA, but the recombination phenotype is not wild type: the crossover level remains low (108, 336, 467). The conclusion is that MER1 and MRE2 have another target that is involved in crossover regulation.
Finally, Modesti and Giroux (319) have identified the mei5 mutation that arrests in pachytene. A combination of DNA assays and return-to-growth experiments shows that mei5Δ allows a normal level of gene conversions without crossover but virtually eliminates crossovers. This new mutant appears to provide a new key to unlock the mysteries of crossover control.
In addition to these genes, the TAM1/NDJ1 gene (76, 86, 410) plays a role in crossover control, since a tam1/ndj1 mutation abolishes interference (76). However, in contrast to zip1, it has no effect on the crossover frequency at a given locus.
What is the mechanism of crossover regulation?
Although we have described several factors that directly modulate crossover frequency and distribution, the mechanism of regulation is not well understood. A common hypothesis is that the crossover regulation occurs at the level of the resolution of the HJ. No homologue of RuvC, the bacterial resolvase (32), has been isolated so far in eukaryotes, but an HJ is a good candidate to be the substrate of Msh4p and Msh5p. In wild-type cells, crossover and noncrossover products appear at the same time, at the end of pachytene (467). This seems to fit with the regulation at the HJ level, because it argues for a common mechanism for crossover and noncrossover recombination. However, Storlazzi et al. (468) showed that this synchrony is disrupted in a zip1 mutant. In such a mutant, noncrossover events represent most of the recombination events, and most of them appear at the same time as in the wild type. However, the crossover events and a small fraction of the noncrossover events appear later. This prompted the authors to suggest that Zip1p plays a role early in meiosis to determine which DSBs will be crossover associated. HJ formation (assayed by two-dimensional gel electrophoresis) is also decreased and delayed in a zip1 mutant (468), suggesting that a regulatory step in crossover formation could be the choice between a conversion pathway with HJs and a conversion pathway without, i.e., an SDSA pathway.
Recent data argue that SDSA could occur also in meiosis (148, 384) (Fig. 9). In this case, a key step of crossover regulation would take place before the formation of the double-HJ intermediate; by influencing whether this intermediate is to be formed, the cell would modulate the crossover formation. This could happen in one of two ways. First, the nature of the newly synthesized DNA could be regulated so that, in meiosis it would most often be inherently semiconservative, as predicted by Szostak et al., and then give rise to a double HJ, suitable for resolution as a crossover. In mitosis, such events could be rare because they proceed by SDSA. Alternatively, SDSA events themselves can undergo a transition to include a double HJ (Fig. 8C and D). This latter view is more consistent with the results of Porter et al. (384) and Gilbertson and Stahl (148) (discussed above). In this case, proteins such as Msh4p, Msh5p, Mlh1p, and Mei5p and SC components such as Zip1p and Zip2p would be involved in regulating this transition.
SEARCH FOR HOMOLOGOUS SEQUENCES: ECTOPIC RECOMBINATION
One of the most mysterious processes in recombination concerns the way a DSB end can find its homologous partner. Even mediated by a Rad51p-Dmc1p filament, how does the DNA inspect a double helix, with its base pairs largely inside the helix, to find homologous sequences? It is now evident from recent developments in gene-targeting strategies that as little as 35 bp at each end is enough to direct the integration of a transformed fragment at its homologous target site, often at least 25 to 50% of the time and sometimes up to 90% of the time (213, 531).
In mitotic recombination, the search for homology seems to be random; that is, the entire genome can be searched for a partner. This has been established in two ways. First, one can insert a leu2 or arg4 mutant sequence at various locations in the genome and then ask how often it recombines with another allele, at an ectopic location. These experiments have shown that all sites are roughly equivalent, suggesting that the entire genome can be searched (151, 270). A second approach has been to determine how the frequency of recombination varies as a function of the concentration of one of the two participating sequences. Wilson et al. (552) showed that successful recombination increased as the number of copies of the donor increased and argued that the search for homology must be one of the limiting steps in the process.
In meiotic recombination, similar ectopic recombination studies have been performed, leading to a somewhat different conclusion. In some cases, the frequency of ectopic recombination was as high as or even higher than allelic recombination (where the two heteroalleles were at the same chromosomal location) (151, 268). This was especially true when the allelic recombining sequences were inserted in a “cold” region of the genome and when the ectopic sequence was inserted in a “hot” region. On average, though, allelic recombination is favored about fivefold over ectopic recombination. The significance of this observation is still the topic of much discussion. In one view, this advantage reflects the fact that homologous chromosomes are already in a loose alignment, enforced by transient interactions between homologues that themselves do not lead to recombination (539). Such transient pairing may, however, simply be a reflection of a general orientation of chromosomes within the nucleus, with all the centromeres initially clustered together (205) and with the chromosome arms oriented toward the other end. Interestingly, genes inserted in telomeric regions are somehow isolated from ectopic interactions with sequences in the chromosome interior, but they engage in robust gene conversions with homologous sequences inserted near other telomeres (151, 281).
Alternatively, one would expect an increase in allelic over ectopic recombination if not all exchange events occurred at the same time. Once one recombination event has occurred anywhere along a pair of homologues, this will favor other interhomologue interactions and exclude ectopic exchanges. If DSB repair initiates synapsis in yeast, as suggested by many results, one allelic recombination event might be enough to cause the intimate association of the homologs along their total length. Hop2p could play a role in making sure that the first recombination events are indeed allelic events. In a hop2 mutant, synapsis occurs between nonhomologous chromosomes (265) and recombination is greatly reduced or absent. It can be restored in a return-to-growth experiment, but the preference for allelic recombination is then lost. The authors propose that Hop2p discourages ectopic recombination (or at least interaction) and thus suppresses inappropriate initiation of SC assembly.
It should also be noted that ectopic recombination, even between sequences smaller than 2 kb, is frequently accompanied by crossing over (151, 206, 268). Whether some of the mutants that affect the frequency of crossovers between homologues will also affect ectopic crossing over has not been examined. This might be a fruitful area of investigation, since it is unclear if one would expect SC formation between such short sequences.
ROLE OF MISMATCH REPAIR PROTEINS IN RECOMBINATION
The role of the mismatch repair proteins in both meiotic and mitotic recombination has been mentioned many times, but it is necessary to consolidate recent findings about the role of mismatch repair in DSB repair in a separate section.
Yeast Mismatch Repair System
The first mismatch repair mutant to be characterized, pms1, was isolated in a screen for mutants displaying high levels of PMS (550) and proved to be a homologue of the bacterial mismatch repair protein MutL (249). Other mismatch repair genes have been identified according to their homology to the bacterial mismatch repair proteins, including three homologues of the MutS protein, i.e., Msh2p, Msh3p, and Msh6p (298, 343, 400), three other homologues of MutLp, i.e., Mlh1p Mlh2p, and Mlh3p (125, 388), and the Exo1p nuclease (506). Three other MutS homologues have been identified in yeast, but they are not nuclear mismatch repair genes: Msh1p plays a role in mitochondrial mismatch repair (400), and Msh4p and Msh5p are involved in meiotic crossover control during meiosis.
The biochemistry of the yeast nuclear mismatch repair system is not fully elucidated. No homologue or equivalent of the bacterial MutH has been isolated, and thus we do not know what protein is responsible for endonucleolytic cleavage during mismatch repair, nor do we know how, during DNA replication, “old” and “new” strands are discriminated, since yeast DNA is not methylated. Most of the information we have concerns damage recognition. Mispairs other than C-C are recognized predominantly by a Msh2p-Msh6p heterodimer, while small heteroduplex loops (frameshift mutations) attract the Msh2p-Msh3p complex (9, 210, 298). Mismatch repair usually requires a Pms1p-Mlh1p heterodimer (169, 170, 389), but a Mlh1p-Mlh3p heterodimer has been implicated in repairing a fraction of the small heteroduplex loops (125).
An unexpected finding is a novel cooperative activity of components from both the mismatch repair machinery and the nucleotide excision repair machinery in processing structures other than mispairs and small insertions and deletions. We described above how MSH2, MSH3, RAD1, and RAD10, but not PMS1, MLH1, or MSH6, are involved together in mitotic recombination in the removal of 3′ nonhomologous sequences (476). Another study has shown that rad1 and msh2 mutations also produce a high level of PMS resulting from heteroduplexes between a 26-bp nonpalindromic insertion and wild-type sequences (231). Again, it argues that Msh2p-Msh3p and Rad1p-Rad10p are part of a novel complex that excises heterologies larger than a few nucleotides.
Role of Mismatch Repair in Gene Conversion
Heteroduplex correction.
The recombination model proposed by Holliday in 1964 (185) involved the formation of two symmetrical heteroduplexes. Conversion would result from the correction of hDNA. Occasional failures of hDNA correction would result in the PMS observed in yeast and other fungi. In yeast PMS, a spore gives rise to a sectored colony (Fig. 15), with the two sectors differing with respect to the alleles of one marker. In filamentous fungi, a replication step and an additional cell division immediately follow meiosis. The spores are formed only thereafter, so that meiosis results in eight spores and gene conversions are seen as 6:2 or 2:6 segregation of two alleles, as opposed to 4:4 segregation observed in most cases. Here, PMS can be identified as 5:3 segregations or aberrant 4:4 segregations that result from two symmetric PMS events. The nomenclature used for the genetics of filamentous fungi is also used for yeast (Fig. 15) since it is more convenient to describe a yeast PMS as a 5:3 event than, for example, a 2 1/2:1 1/2 event.
(i) hDNA correction in meiotic recombination.
The molecular nature of alleles that frequently produced PMS instead of gene conversion gave strong clues that gene conversion resulted from hDNA correction. Three high-PMS alleles were shown to be G-to-C mutations that would result in C-C mismatches (548). This sequence specificity was reminiscent of the bacterial mismatch repair system specificity, where C-C mismatches are poorly repaired, thus suggesting that a homologous eukaryotic system was responsible for gene conversion (548). Further genetic and physical studies supported the idea that C-C mismatches were not repaired whereas G-G mismatches (and other base-pair mismatches) were (41, 96, 247, 269). A variety of artificial palindromic insertions are also high-PMS alleles (334). In addition, although most insertions and deletions of more than a few base pairs are almost always gene converted, a special class of deletions, including the 38-bp deletion ade8-18 allele, yield high PMS. An analysis of several such high-PMS deletions led White et al. (548) to conclude that they all had a common sequence at the deletion junction that might bind a protein and prevent normal mismatch recognition.
The formation of hDNA during meiosis has been demonstrated directly by physical assays (157, 269, 333). Mismatch correction is sufficiently fast that one does not see G-G mismatches without using a strain defective in mismatch repair, while C-C mismatches are clearly visible. However, as we mentioned above, the timing hDNA can be detected only late in the meiotic process, about at the same time as the disappearance of the double HJs.
To various degrees, five mismatch repair proteins play a role in the correction of heteroduplex DNA in meiosis, i.e., those encoded by the PMS1, MLH1, MSH2, MSH3, and MSH6 genes (12, 192, 249, 343, 400, 550). In the context of such mutations, low-PMS alleles behave in roughly the same way as high-PMS alleles.
(ii) hDNA correction in mitotic recombination.
Evidence for hDNA formation and subsequent correction by the mismatch repair machinery was obtained for mitotic recombination in the case of MAT switching. Using XhoI linker insertions at several sites in the MAT locus, McGill et al. determined that the replacement of Ya sequences frequently involved coconversion of sites in the adjacent W and X regions (305). In some cases, the transfer of a single strand of the donor to form heteroduplex DNA with MAT could be directly inferred from the presence of sectored colonies in which one DNA strand gave rise to XhoI+ descendants and the other strand gave rise to XhoI− descendants. These events, sometimes referred as PSS (postswitching segregation [167]) are the mitotic equivalents of PMS.
Evidence for the formation and repair of heteroduplex DNA at MAT was also obtained for a single-base-pair change only 8 bp beyond the 3′ end of the HO-cleaved DSB (397). In this situation, mismatch repair is highly biased, since the heteroduplex was most often repaired in favor of the donor DNA sequence. A kinetic study, using PCR and DNA sequencing of the intermediate that arises after the MAT DNA invades the donor and begins new DNA synthesis, argued that mismatch correction was quite rapid (397).
We must add here that although hDNA formation has been extensively investigated during MAT switching, it appears to be a general feature of mitotic recombination and could be demonstrated genetically during recombination between direct repeats (304, 414) or transformation by a linear DNA fragment (266).
When is hDNA formed and repaired?
When Holliday formulated the hypothesis of hDNA correction (185), his idea was that hDNA would be corrected after there had been a stable transfer of DNA strand, to form regions of hDNA that would not subsequently unwind or anneal with other strands (Fig. 16A). In this view, hDNA correction occurs late in the process. In a more recent conception, hDNA is corrected immediately after its formation during strand invasion (Fig. 16B). Finally, hDNA might also appear after strand invasion, during the annealing step of a SDSA pathway (Fig. 16C), and we have described above how this may be the case for meiotic hDNA.
What about mitosis? Ray et al. (397) showed that during MAT switching, repair of the heteroduplex most probably occurs very soon after its formation. However, the only mismatch they actually monitored during the recombination process was 8 bp away from the break. Therefore, even a very short early hDNA would have included it. hDNA involving regions further away from the break (305) could be formed and corrected later. One must keep in mind that in an SDSA model, hDNA formation by strand invasion and hDNA formation by later annealing are not mutually exclusive, and there might be two steps of hDNA correction. For example, if an hDNA correction occurs during the initial invasion step, hDNA would appear in the donor template after the two strands of the donor molecule are reannealed (as shown in Fig. 17B). Data in favor of SDSA during mitotic recombination argue that such events actually happen.
FIG. 17.
Correction of hDNA resulting from strand invasion. (A) In the Szostak et al. (479, 494) model, correction in favor of the recipient strand leads to a gene conversion. (B) In the Szostak et al. (479, 494) model, correction in favor of the invading strand leads to a restoration at the B locus. Therefore, if one looks only at the B locus, nothing is detectable (normal Mendelian segregation). However, b is now associated with A and C, and B is associated with a and c (apparent double crossover). (C) In an SDSA model, correction in favor of the recipient strand results in a gene conversion. (D) In an SDSA model, correction in favor of the invading strand leads to a second round of mismatch repair that is initiated when the two strands of the template are reassociated. The final outcome can be gene conversion (genetic definition) or no detectable event (even no apparent double crossover). This outcome will be found with the SDSA models described in Fig. 8B, to D. In the SDSA model described in Fig. 8A, where a second strand is synthesized on the template, an additional mismatch will appear when the two newly synthesized strands are annealed (not shown). Four outcomes are then possible (no apparent event, apparent double crossover, gene conversion of B, gene conversion of b).
Directionality of hDNA correction in mitosis.
The directionality of hDNA repair observed at MAT strongly suggested that the invading DNA strand, with a free DNA end, provoked mismatch correction in favor of the resident, unbroken donor sequence. Neither Ray et al. (397) nor McGill et al. (305) found evidence of DNA transfer from MAT to the donor locus (“back conversion”). The nick could target the invading strand for correction, similarly to what has been shown in vitro for the bacterial mismatch repair system (320). This finding has changed our understanding of gene conversion.
When Holliday formulated the hypothesis of hDNA correction, his idea was that gene conversion resulted from correction in favor of the invading strand while correction in favor of the recipient, invaded strand would be a genetically silent restoration (Fig. 16A). In the context of the Szostak et al. model (Fig. 6), hDNA would form during strand invasion (Fig. 16B) and gene conversion would result from correction in favor of the recipient strand (Fig. 17A). Note that in an SDSA model also, if hDNA resulting from strand invasion is corrected in favor of the recipient strand, the genetic outcome is a gene conversion (Fig. 17C).
However, in an SDSA model, hDNA can be formed both at the time of strand invasion (Fig. 16B) and at the time of annealing (Fig. 16C). hDNA resulting from annealing would always be on the recipient molecule (Fig. 16C), and with this kind of hDNA, there is no need to refer to a nick-dependent directionality of mismatch repair to account for the lack of back conversion. Since we do not really know the amounts of hDNA resulting from strand invasion versus strand annealing, it is difficult to formally demonstrate the nick-directed mismatch repair hypothesis. Studies of mismatch correction during transformation (266) support the idea that there is a strong bias of correction in favor of the recipient, unbroken strand.
Restoration and conversion events in meiosis.
Nick-directed mismatch repair of hDNA has also been proposed to occur during meiosis (384). However, the data are very unclear, because, unlike HO-induced recombination, meiotic recombination does not allow the molecule which had been cut to be identified from among the recombinant products. In addition, the interpretation of the genetic data may depend on the recombination model.
An unappreciated prediction of the Szostak et al. model is that restoration would appear as a double crossover, as shown in Fig. 17B. If the invading DNA, forming heteroduplex B/b, is corrected in favor of the recipient strand, the final outcome would have 4:4 B:b segregation, but the linkage of markers has changed and there would appear to be a double crossover between flanking markers A and C. Extensive genetic data obtained by Mortimer and Fogel (329) clearly established that this was not observed.
If mismatch repair occurs during strand invasion (Fig. 16B) but recombination then follows an SDSA pathway (Fig. 17C and D), heteroduplex correction in favor of the invading strand would lead to a second round of hDNA correction when both strands of the donor reannealed (Fig. 17D). Here, the presence of a nick on both strands rules out the possibility of any systematic bias in correction. Some correction events would restore the former configuration (i.e., a restoration according to the genetic definition of no detectable event). The other possible correction would result in a gene conversion in the unexpected direction, that is, 1:3 B:b (back conversion) instead of the anticipated 3:1. There is no direct evidence for such an outcome.
However, restoration events probably exist. Using a combination of low-PMS alleles surrounded by high-PMS alleles, Hastings et al. (177) could isolate, in Ascobolus, PMS of the two flanking markers not associated with any apparent change in the central markers (Fig. 18). The PMS of the two flanking markers argues that the region in between was part of a heteroduplex, and the absence of non-Mendelian segregation for the central marker argues that this hDNA was corrected by a restoration event. In the same kind of experiment, Kirkpatrick et al. (230) could also find such events in yeast (Fig. 18). The frequency of these events rules out the simple alternative that there were two independent heteroduplexes including only the flanking high-PMS markers and not involving the central low-PMS allele.
FIG. 18.
Detection of restoration events in yeast. This experiment, developed for yeast by Kirkpatrick and Petes (230), is derived form an analogous experiment with Ascobolus by Hastings (174). A well corrected allele (y) is located between two high-PMS alleles (x and z). If the three alleles are found in the same heteroduplex, correction of the central allele might occur, even if the two flanking high-PMS alleles are not corrected. Correction in favor of the resident strand will result in gene conversion of y, while PMS will be observed for x and z. If correction occurs in favor of the invading strand (restoration event), there will be a 4:4 Mendelian segregation for y, even though simultaneous PMS will be observed for x and z.
However, the interpretation of these results is not easy. First, we are dealing with rare events, and it is difficult to infer from these data how much restoration there would be when a single mismatch is involved. Second, if hDNA results from annealing, as shown in Fig. 16C, restoration and conversion events could appear with equal frequency. Note that in this model, restoration would not appear as double-crossover events.
Also, if restoration events do exist, one prediction is that the total number of non-Mendelian segregations (gene conversions plus PMS) should increase in mismatch repair mutants, since one would expect that hDNA that is undetected among restoration events would now appear as PMS. However, this is not observed in pms1, msh2, msh3, or msh6 mutants (12, 191, 550). In contrast, mlh1 causes a 1.4- to 2.2-fold increase in the total number of non-Mendelian segregations (191). Whether Mlh1p is the only known mismatch repair protein involved in restoration events or whether it negatively regulates hDNA formation is unclear.
Conversion gradient.
Fogel and Mortimer (129) first noted that there was a polarity to gene conversion tracts. When a marker near a hot spot is converted, markers further away have a probability of being coconverted that decreases as a function of distance. With markers that are well repaired, it is rare that a marker far from a hot spot will be converted unless markers closer to the hot spot are also converted. Such gradients of conversions have been observed at many hot spots of meiotic recombination, such as ARG4 (126, 348), HIS4 (97), HIS1 (127), and HIS2 (293). Meiotic gene conversion tracts normally extend only 1 to 2 kb from a hot spot, in either direction (46, 435). At least at the HIS4 hot spot, although conversion occurs on either side of the DSB, the conversion tracts are frequently unidirectional (383).
Coconversion gradients are also observed in mitosis. The most extensive study has been done with an HO recognition site inserted into a plasmid-borne URA3 gene (342, 487). A donor URA3 gene carrying a number of heterologies across the gene was used to determine how often gene conversions that repaired the HO cut coconverted adjacent sites. Most of the conversion tracts extended only on one side of the DSB, consistent with what is observed in meiosis at the HIS4 hot spot. Coconversion tracts of at least 400 bp were found, but, surprisingly, 20% had conversion tracts of less than 53 bp. These results suggest that many repair events are surprisingly “local.” If this were true in meiosis, there could be a substantial number of meiotic repair events without a detectable gene conversion, and, indeed, not all selected crossovers have a detectable gene conversion (46, 492). However, it is possible that the presence of a large number of heterologies in the few base pairs surrounding the cut site resulted in the elimination of many attempted repair events through another action of the mismatch repair system, which is to discourage recombination between homeologous substrates (see below).
Models for the conversion gradient.
The initial DSB repair model of Szostak et al. explained the gradient in terms of the size of a gap that was resected from the site of the DSB (494). The realization that there was little 3′-to-5′ degradation but extensive 5′-to-3′ degradation (479, 547) restored the earlier idea that most gene conversions resulted from mismatch correction of hDNA that was generated after strand invasion (128, 185; see reference 466 for a more complete account). In this conception, the gradient of gene conversion would be explained by the extent of assimilation of the 3′ tail into hDNA. Subsequent results have shown that it is a more complicated process.
(i) Restoration-conversion model.
In the HIS4 locus, low-PMS markers close to the promoter region (the location of the DSB site) are converted about twice as often as markers at the 3′ end of the gene. When poorly repaired markers were used, both markers close to the hot spot and ones further away showed sectoring expected for PMS, but now the level of non-Mendelian segregation (gene conversions plus PMS events) was nearly equal at both ends of the gene (97). One interpretation of these results is that heteroduplex DNA is likely to extend across the entire gene but that some sort of bias in how mismatches are corrected accounts for the final gradient in conversion. This model has been referred to as the restoration-conversion model.
In a molecular view, hDNA would be formed by strand invasion, and the mismatches close to the DSB end would be preferentially corrected in favor of the allele on the unbroken molecule, since mismatch repair would be directed by the end of the invading DNA. As the distance between a mismatch and this end increases, the bias in repair could progressively disappear. At a certain distance, mismatches would be randomly corrected (Fig. 19A). Accordingly, Kirkpatrick et al. (230) detected more events that probably resulted from restoration (see above) for a marker far from the HIS4 DSB hot spot than for a proximal marker.
FIG. 19.
Three hypotheses to explain the gradient of meiotic gene conversion. (A) Restoration-conversion hypothesis. Next to the DSB, the mismatch is corrected preferentially in favor of the recipient molecule, resulting in a gene conversion. Further away, it will be corrected increasingly randomly, with the nick losing its ability to target the invading strand for the mismatch repair proteins. (B) Heteroduplex rejection model. Mismatches cause branch migration or unwinding of the heteroduplex, mediated by the mismatch repair proteins. Afterwards, the remaining hDNA is always corrected in favor of the recipient molecule. (C) Formation of a conversion gradient in an SDSA model. Both strands of the hDNA are processed by the mismatch repair machinery, which excises ssDNA tracts from each nick to the next mismatch. As a consequence, mismatches next to the DSB region result in conversion events whereas mismatches distal to the break more often result in restoration.
This model implies that correction in favor of the recipient allele would fall from 100% next to the DSB to 50% a few kilobases further away, resulting in a twofold gradient of gene conversion. This is indeed what is observed at the HIS4 locus. However, at other loci, such as ARG4 and CYC3, conversion near the hot spot is more than four times more frequent than at the 3′ end of the gene. In addition, as mentioned above, in a Szostak et al. model, such events would appear as double-crossover events (Fig. 17A), which are extremely rare and certainly cannot account for even a twofold gradient.
(ii) Heteroduplex rejection model.
Another hypothesis is that the mismatch repair machinery regulates branch migration (12) (Fig. 19B). It is sometimes referred to as the heteroduplex rejection model. In this model also, hDNA is formed during strand invasion. However, it could be unwound by the mismatch repair machinery. The hDNA region remote from the DSB would be dissociated first, and there would actually be a gradient of hDNA, but this gradient would occur only after processing by the mismatch repair system. The remaining hDNA would then be corrected by this mismatch repair system, thus involving it in two different steps of a complex process.
This hypothesis has the advantage of explaining steep gradients of gene conversion and fits better with the current models of recombination. It must also be pointed out that it does not necessarily contradict the observation of uniformly long heteroduplexes involving high-PMS alleles at the HIS4 locus. High-PMS alleles are noncorrected alleles and also could be unable to trigger mismatch repair-mediated reverse-branch migration.
Recently, Chen and Jinks-Robertson (71) found that some mitotic conversion tracts between diverged sequences were longer in mismatch repair-defective cells. In their experiments, the conversion tracts are supposed to result from segregation of uncorrected hDNA after replication. Thus, the hDNA length could be regulated by the mismatch repair system in mitosis also.
(iii) Conversion gradients in an SDSA model.
In the above two models, hDNA results from strand invasion. We now consider what happens when most of the hDNA results from annealing. hDNA will be formed between two nicked strands. The simplest hypothesis is that both of them are processed: the mismatch repair machinery removes excision tracts extending from a nick to the next mismatch (Fig. 19C). Thus markers proximal to the DSB would be preferentially converted, with distal markers being preferentially restored.
Inhibitory Effect of the Mismatch Repair Machinery on Homeologous Recombination
Another role of yeast mismatch repair proteins in recombination is their inhibitory effect on recombination between diverged sequences, during so-called homeologous recombination. It is known that the MutHLS system in bacteria establishes a recombination barrier during recombination of diverged sequences (445) and during interspecific conjugation of E. coli and the related species Salmonella typhimurium (299, 398). The PMS1 and MSH2 genes set up a similar genetic barrier between S. cerevisiae and its close relative Saccharomyces paradoxus. An interspecific hybrid between these yeasts gives rare viable spores with a high rate of aneuploidy, and the meiotic recombination level is very low. In msh2 and pms1 mutants, spore viability and meiotic recombination are increased as the rate of aneuploidy is decreased (192). Moreover, in an S. cerevisiae diploid in which one copy of chromosome III is derived from S. paradoxus, mismatch repair-dependent inhibition of recombination between the homeologous chromosomes is also observed, so that in an msh2 mutant, there is increased recombination between the two chromosomes III, accompanied by a reduction in nondisjunction and in spore inviability (67).
Even between two diverged genes, the mismatch repair machinery inhibits recombination; Selva et al. (442) showed that MSH2 and MSH3 discourage recombination between an S. cerevisiae gene and its S. pombe homologue, and in a different assay, Priebe et al. (386) found that PMS1 inhibits DSB-induced recombination between an S. cerevisiae gene and its diverged Saccharomyces carlbergensis counterpart. Even a single mismatch can discourage recombination, as shown in a mitotic assay of crossovers between two chromosomal inverted repeats (87, 88) or during transformation (266).
In a normal S. cerevisiae diploid, one detects another aspect of mismatch repair, termed mismatch repair-dependent recombination. Borts and Haber (47, 48) studied an artificial region in which the two homologous chromosomes had about 0.1% divergence. Compared with the same region without the mismatches, crossing over was reduced by 50% and there was a corresponding increase in aberrant events, which could be detected with the flanking markers. Recombination was restored to nearly normal by a pms1 mutation, leading the authors to propose that independent repair of these widely spaced mismatches, including extensive resection of hDNA, might lead to new double-strand breaks that could in turn stimulate a second round of (mismatch repair-dependent) recombination. These events were detected because of the presence of flanking repeated sequences and might not be easy to identify in other chromosomal regions (492).
ROLE OF DSB REPAIR IN TANDEM REPEAT INSTABILITY
A great deal of attention has been paid recently to the instability affecting single-sequence repeats, including micro- and minisatellites. Although the focus on this kind of genomic instability is the direct consequence of biomedical studies of humans, a number of laboratories have recently adopted S. cerevisiae as a model system to study simple repeat instability.
In microsatellites, the repeated motif is smaller than 12 nucleotides. Larger repeats, up to 100 nucleotides fall into the minisatellite category. Initially, these arbitrary classifications seemed to be justified by differences in their involvement in genetic disease. Indeed, microsatellites and, more specifically, trinucleotide repeats, e.g., (CTG)n, appeared as an especially at-risk motif, since expansions in such arrays were found to be involved in many genetic diseases, such as myotonic dystrophy, Huntington’s disease, fragile X syndrome, Friedreich’s ataxia, and others (for reviews, see references 404 and 535). The expansions can be relatively small, increasing the copy number less than 2-fold; but they can also be massive, multiplying the initial copy number by 3-, 10-, or even up to 80-fold. Expansions often seem to result from a two-step process. (i) A primary mutation (premutation) slightly increases the copy number, creating an unstable but functional allele. (ii) This unstable allele is then extremely prone to larger expansions (full mutation), which, in contrast, severely affect some aspect of the nearby or surrounding gene and result in a specific disease or in a fragile site. Such mutations have therefore been called dynamic mutations (404, 405).
However, recent studies have found that massive expansions are not specific to trinucleotides. Amplification of a dodecamer in the promoter of the EPM1 gene is responsible for certain types of epilepsy (256, 257, 526). Even larger repeats can be massively amplified: two cases of fragile chromosomes (yet not associated with disease), FRA10B and FRA16B, are the consequence of the expansion of a 42-mer and a 33-mer, respectively (182, 569). Thus, the distinction between micro- and minisatellites is not as clear any more. Part of this distinction also relies on the mechanisms that are thought to be involved in the instability of the micro- and minisatellites, and we discuss this issue below.
Despite the gloomy phenotypes associated with some expansions, instability of simple repeats is also a very useful tool for the scientist. Human minisatellite arrays are generally highly polymorphic and more or less stable (17, 45, 58, 59, 202, 207). Some loci can change in size in as much as 13% of the germ line transmissions (59). Relatively stable minisatellites can be used for positional cloning or haplotyping (for reviews, see references 17 and 200), and highly unstable ones can uniquely identify individuals, a property which can be used, for example, to identify a corpse (171).
Rearrangements in Minisatellites and Larger Sequences Are Probably the Consequence of DSB Repair
It is now assumed that minisatellite instability relies on recombinational mechanisms. First, a large number of human minisatellite rearrangements involve interallele transfer of information, i.e., a gene conversion (59, 202, 302). Second, a number of studies suggest that they are linked to meiotic recombination. Minisatellite instability occurs in the germ line (59, 202). At the MS32 minisatellite locus, the rearrangements are clustered on one side of the locus, suggesting an adjacent initiating sequence, as in meiotic recombination. Although most of the minisatellite rearrangements are not associated with crossing over, crossover mapping clearly indicate a meiotic recombination hot spot adjacent to (within 1 kb) of the unstable MS32 locus (201). Moreover, a variant allele of MS32 suppressed both minisatellite instability and the crossover hot spot (201), reinforcing the notion that meiotic recombination and minisatellite rearrangements are linked. Experiments with yeast also argue that meiotic recombination hot spots cause minisatellite instability: when the MS32 human minisatellite was inserted in yeast chromosome III next to the LEU2 hot spot, 10% of meiotic progeny exhibited a change in the array, whereas the insert was perfectly stable in mitosis (16).
Instability of larger repeats has also been observed in yeast and Drosophila and in both cases was alleged to result from genetic recombination. The yeast CUP1 repeated locus undergoes frequent rearrangement in meiosis (544, 545). In Drosophila, contractions and expansions of several different tandem repeats were observed when the repeated array was located within or near an P transposon (90, 253, 364, 369, 505). These rearrangements depends on expression of the transposase, which, when it excises the transposon, induces gene conversion, probably because the excision results in a DSB (111, 150). Thus, several results with various organisms argue that DSBs induce tandem repeats expansions and contractions.
We have recently directly tested this hypothesis in an experiment described in Fig. 11. When a DSB has to be repaired on a template that contains a repeated locus, up to 50% of the rearrangements are found in the repeated array incorporated into the recipient molecule (368). These rearrangements are definitively the consequence of DSB-induced gene conversion and can also be found associated with the rare crossover-associated gene conversion. This phenomenon does not depend on the nature of the repeated array, since DSB-induced rearrangements in yeast have been observed in a tandem repeat of 5S ribosomal genes (368), as well as in artificial or natural yeast minisatellites (365).
These results show that DSB can induce expansions and contractions in repeated arrays and, as explained in the legend of Fig. 11, support an SDSA model for gene conversion. However, the observed expansions were always small, not adding more than n − 1 units (if the initial size of the repeat was n). Thus, if DSB repair can easily explain the instability observed in many minisatellite locus, it is more difficult to invoke it to explain the massive expansion observed in EPM1, FRA10B, or FRA16B (182, 257, 526, 569). However, if the rearrangement results from slippage-like events (invasion, unwinding, and reinvasion of the template) during the DSB-associated DNA synthesis, as described in Fig. 11D, many slippage-like (reinvasion) steps might result in large amplification.
Is DSB Repair Involved in Trinucleotide Repeat Rearrangements?
If it is more or less assumed that minisatellite instability depends on DSB repair, the instability of smaller repeats, including trinucleotide repeats, is most often thought of as occurring during replication. This hypothesis is based mainly on two sets of results. First, mono- and dinucleotide repeats are destabilized in mismatch repair mutants in humans (194, 261, 373, 502) and in yeast (470). Second, a number of experiments with yeast (135, 301, 316) and E. coli (221) showed that the rate of rearrangement in a trinucleotide repeat in these organisms depends on the orientation of the repeat compared to an adjacent origin of replication. The interpretation of these results is based on the ability of at least some of the single-stranded trinucleotide repeats to form hairpins (72, 139, 317, 318, 567). Such hairpins could form on the lagging strand during replication, because it is single stranded over a short distance, but not in the leading strand. One strand of a trinucleotide repeat would be more prone to form hairpins than the other (for example, CTG more than CAG), and frequent rearrangements would therefore appear only when this strand happens to be the lagging strand during replication. Nevertheless, the events detected in these yeast systems were essentially confined to small variations in the copy number.
Recently, a rad27 mutation was shown to greatly increase microsatellite instability in yeast and to increase the rate of expansions versus contractions (134, 244, 440). rad27 also destabilizes an artificial minisatellite (20-bp repeat) in yeast (244). The Rad27 protein is a flap endonuclease involved in removing the 5′ RNA end of Okazaki fragments during replication, and a rad27 mutation causes duplications between very short homologies even within unique sequences (507).
However, a number of data argue for a role of recombination in trinucleotide repeat instability. One case of reversion at the MD (myotonic dystrophy) locus is associated with gene conversion (354). Also, trinucleotide rearrangements display some kind of polarity. At the FRM1 locus, the rearrangements affecting a CGG repeat seem to be confined to one side of the repeat array (251). Polarity has also been observed in yeast in the rearrangements affecting a CAG repeat (300). The experimental system described in Fig. 11 showed that DSBs can induce expansions and contractions in a CTG repeat as well as in minisatellites (402). A DSB-induced rearrangement could explain why trinucleotide repeats expansions occur about the time of meiosis, although it is still not clear if these expansions occur during meiosis or shortly after (137, 296). Nevertheless, it is difficult to conclude what mechanism is responsible for trinucleotide expansion in humans.
MITOTIC NONHOMOLOGOUS RECOMBINATION
Although repair of DSBs in Saccharomyces most often proceeds by homologous recombination, yeast also has the capacity to repair breaks by nonhomologous or illegitimate recombination. Indeed, there seem to be several pathways defined by differences in repair as a function of the cell cycle and also by various mutations.
Ligation of Complementary Ends
The simplest form of end joining is the ligation of complementary overhanging DNA ends produced by endonucleases. This type of repair has been assayed by transforming into yeast a linearized autonomously replicating plasmid cleaved by a restriction endonuclease, so that propagation of the plasmid requires the ligation of the ends. Repair is quite efficient; about 30 to 70% of cut plasmids survive compared to an uncut plasmid control (54, 313, 314). Alternatively, the HO endonuclease has been used to create DSBs in vivo in chromosomes that lack homologous donor sequences. Here, about 35% of cells survive HO cleavage (323). These events are independent of RAD1, RAD2, RAD51, RAD52, RAD54, RAD57, and, by inference, RAD55. Both types of ligation require a surprisingly large number of other gene products. End joining requires the second DNA ligase of yeast (427, 552a), termed LIG4 because of its closest resemblance to the fourth mammalian DNA ligase (498). Religation also requires RAD50, XRS2, and MRE11 (314, 323, 516) and the two subunits of the yeast homologues of the mammalian Ku70 and Ku80 proteins, known as HDF1 and HDF2, respectively (53, 54, 314). Three other genes, SIR2, SIR3, and SIR4, are also required for efficient religation of transformed plasmids with either 3′ or 5′ overhanging ends (517). However, the effect of sir mutants may be only to produce an a/α diploid-like state (by expressing HMLα and HMRa), and this only indirectly impedes ligation (263, 407). In addition, one gene, RAD5, appears to have a negative effect on end joining. A rad5 mutant shows a higher proportion of ligation of DSB ends that could also participate in gap repair (7); also, in an assay where only end joining can occur, a rad5 deletion increases plasmid end joining by about a factor of 5 (263).
It is likely that there are still other genes that play an important role. To date, there has been no systematic search for genes involved in this process. The RAD genes were surveyed on general principles, and the HDF genes were surveyed because of their apparently similar role in NHEJ in the mammalian immune system. The SIR genes were then implicated because of a search by two-hybrid analysis for genes interacting with Hdf1p, which turned up Sir4p (517).
Nonhomologous End Joining of Noncomplementary Ends
DNA end joining of noncomplementary ends has been studied in several ways. First, linearized plasmids with two different ends can be introduced. Second, chromosome breaks can be created by establishing a dicentric chromosome (248) or plasmid (517), where DNA breakage will promote deletions of one or the other centromere. Finally, in vivo HO-induced breaks can be used by subjecting cells to the continuous presence of the endonuclease, so that simple re-ligation will regenerate the site, which will be cut again (323); hence only cells that alter the HO cutting site will survive. In all of these cases there appear to be at least two distinctively different mechanisms of NHEJ, with quite different genetic requirements. Unlike the perfect ligation of overhanging complementary ends, NHEJ, producing deletions or small insertions, are rare in yeast; about 0.1% of DSBs are repaired in this way.
Misalignment and filling in of DNA ends.
When HO-induced DSBs are created at all stages of the cell cycle, more than 80% of the altered cleavage sites are created by misalignment and filling in of the ends (323) (Fig. 20). The terminal A of the top strand can pair with the terminal T of the bottom strand, and this intermediate, apparently stabilized by a single base pair, can then be filled in to produce a 3-bp insertion at the cleavage site. Interestingly, the much more prevalent product is a 2-bp fill-in, in which the terminal T of the bottom strand must have been excised. Why this more complex process should be more successful is unknown. It is possible that the HO endonuclease often remains associated with the terminal T of the bottom strand and prevents this base from pairing, but no data support the idea that HO has a covalent intermediate. These small additions to the DNA sequence depend on all of the same genes needed for ligation of complementary ends: RAD50, XRS2, MRE11, HDF1, and HDF2 (321, 323) (other genes have not been tested). It is not yet known what DNA polymerase is required to fill in the small gaps created by this process.
FIG. 20.
End joining of HO endonuclease-cleaved DNA. The 4-bp complementary ends can be ligated, which is an efficient process, or can be joined by NHEJ mechanisms that are inefficient. If the bases marked “f” pair, fill-in synthesis will generate a 3-bp insertion. If the bases marked “d” pair and the short tails are clipped off, a 3-bp deletion can result. Other, much larger deletions can occur in the same fashion, using bases exposed by the 5′-to-3′ resection of the DSB ends.
The efficiency of filling in of misaligned ends is surprisingly dependent on the stage in the cell cycle at which the DSB is generated. When HO endonuclease is induced only in the G1 stage, the efficiency of DNA end joining drops nearly 100-fold relative to that in cells where DSBs were induced over the entire cell cycle (323), and most of the remaining events are deletions.
Deletions.
Deletions range from a few base pairs to several kilobases. Sequencing of deletions reveals that most of them have junctions in which the two joined segments share 1 to 3 bp, although about 12% have no apparent base pairings at the exact junction (248). The distribution of base pairings at deletion junctions is remarkably similar to the pattern of deletions seen in mammalian cells (420). It should be noted that a few deletions share 5 bp or more at the junction point, yet these particular junctions are not frequently formed, suggesting that there is little advantage in having more than 1 or 2 bp stabilizing the intermediate that leads to a deletion. It is presumed that these deletions arise by resection and annealing followed by the removal of 3′ tails. Although Rad1p is essential in removing long 3′ tails during single-strand annealing, it is not responsible for processing intermediates leading to nonhomologous deletions (323).
Unlike complementary-end rejoining and noncomplementary end filling in, HO-induced deletions are not significantly diminished by mutations in RAD50, XRS2, or MRE11 (323), although the absence of HDF1 and HDF2 causes a 10- to 100-fold reduction in deletion formation (321, 515). There is, however, a notable shift in the junctions that are found among deletions in the absence of RAD50 or HDF1; deletions form only at those few sites where 5 bp or more of homology can be formed (321). These results are quite similar to those found in mouse cells for deletions formed in the absence of Ku86 (44, 172, 573). The use of these few sites with increased homology still does not require RAD52.
Nonhomologous Integration of Transformed DNA
There appears to be a close relationship between NHEJ and the integration of DNA fragments at nonhomologous sites. Schiestl and Petes first demonstrated that a linearized URA3 gene fragment would integrate at quasi-random sites in the genome if the homologous ura3 target site was completely deleted (430). Further investigation has revealed several very interesting features of this process. The first is that integration does not occur at random; many events occur at sites that have homology to the 4-bp overhanging ends of the restriction endonuclease-cleaved DNA. Another preferred site of integration appears to be cleavage sites for DNA topoisomerase I. The genetic requirements, insofar as they have been analyzed, seem to be very similar to those of NHEJ: the events are independent of RAD52 but dependent on RAD50 and HDF1 (428, 432). The second surprise was that these illegitimate insertion events were augmented by introducing the restriction enzyme that had cleaved the fragment (430). Restriction enzyme-mediated transformation specifically increases targeting at recognition sites throughout the genome. Restriction enzyme-mediated transformation has proven to be a boon to researchers studying other organisms such as Dictyostelium, where normal homologous integration is highly inefficient (254, 255, 280).
A surprising feature of some nonhomologous integrations was the inclusion of mitochondrial DNA (mtDNA) adjacent to the transformed URA3 fragment (429). How mtDNA could be liberated and captured is not understood, but another recent study has suggested that transformation itself may release mtDNA that can enter the nucleus and be captured. When the HO endonuclease gene was expressed just after it was introduced into yeast by transformation, to create DSBs that could be repaired only by NHEJ, 1 to 2% of the events had insertions of approximately 100-bp fragments of mtDNA at the site of the DSB (324). These events were not seen when the HO gene was already present in cells and was subsequently induced. Previously, Louis and Haber (282) had noted the presence of a 240-bp fragment of mtDNA between the X and Y’ subtelomeric elements. Since the sequence was perfectly homologous to mtDNA, it was not possible to determine if the sequence had been captured very recently, after transformation, or had been captured during the past eon.
The idea that exogenous DNA can be captured at the sites of DNA breaks with which it has a few base pairs of identity also received support from the discovery that about 1% of events that repair HO-induced DSBs at MAT contain insertions of unrelated DNA sequences. In otherwise wild-type cells, all of these insertions proved to be approximately 95 bp derived from the yeast retrotransposon, Ty1 (322). Further inspection of these inserts revealed that nearly all of them correspond to the so-called strong-stop cDNA fragment that is extended from a tRNA primer as the first step in converting Ty1 mRNA to cDNA. The inserted fragments were all in one orientation, in which there is some possible base pairing between one end of the inserted fragment and the Y region of the target MAT locus. The captured DNA contains a few bases that are part of the tRNA primer, raising the possibility of formation of an RNA-DNA hybrid during the process of integration and copying these sequences or the possibility that these sequences were first made in cDNA. As with nonhomologous integrations and NHEJ, these events are independent of RAD52 but reduced in rad50, mre11 and xrs2 mutants. In cells expressing both HO endonuclease and a Ty1::HIS3 mRNA that required pre-mRNA splicing before cDNA formation, similar but larger inserted fragments were found (497). The experiments of Teng et al. (497) established that these integrations depend on a functional reverse transcriptase but did not require Ty1 integrase. These unusual captures of cDNA fragments provide one possible explanation of how sequences such as Alu or pseudogenes could be inserted into the mammalian genome, at sites of random chromosome breakage.
Homologous-Nonhomologous Gene Targeting and Related Events
Current genome knockout strategies involve as few as 35 bp of DNA on either side of a KAN1 gene to replace a target gene (531). However, a careful inspection of the apparently properly targeted events reveals that at least 10% had apparently created a deletion at one end or the other. Such homologous-nonhomologous events have been well documented in mammalian transformations (30, 35, 403, 426).
DSB REPAIR CHECKPOINTS
DSBs are potentially lethal if they are not repaired, because they will result in chromosome loss. Therefore, it is important for the cell to optimize the repair conditions, and this is achieved by interrupting the cell cycle in response to unrepaired DSBs. It has been known for a long time that cell division can be arrested in response to various types of DNA damage (for reviews, see references 173 and 540). Weinert and Hartwell (542) showed that a G2/M arrest in response to ionizing radiation (supposedly inducing mostly DSBs) was suppressed in a rad9 mutant cell. The authors proposed that the role of this gene would be to monitor DNA damage and to interrupt the cell cycle in G2 in the presence of nonrepaired DNA to allow more time for repair to occur before mitosis creates aneuploid cells. This model, which assumes that cell cycle arrest is an active process under the control of a checkpoint, differs from the hypothesis that cell cycle arrest is a mere consequence of a structural defect caused by the damaging agent. Weinert and Hartwell (542) supported the checkpoint hypothesis by showing that the rad9 radiation and mutagen sensitivity can be rescued when the length of the G2 stage is artificially increased. When they irradiated cells blocked in G2 by a microtubule poison and released them from the arrest a few hours later, the rad9 cells recovered better, supposedly because their DNA repair machinery had been given enough time to accomplish its task.
A variety of different stimuli have been used to induce checkpoint arrest, but G2/M arrest will occur in response to even one unrepaired DSB induced by HO endonuclease expression (262, 294, 425, 508). A similar type of arrest occurs at the pachytene stage of meiosis I prophase in cells unable to repair DSBs (287). It is also with DSB-induced checkpoint arrest that a phenomenon called adaptation was characterized. The arrest induced by a single DSB is not irreversible but, rather, only imposes a delay in cell cycle progression. Sandell and Zakian (425) showed that a single DSB that could not be repaired was enough to cause a G2/M cell cycle arrest, but most of the cells resumed growth after transient arrest. Furthermore, the unrepaired chromosome could be propagated for up to 10 cell divisions, showing that the resumption of growth was not simply due to the loss of the signal (the broken molecule). Although the authors termed this phenomenon “recovery,” others preferred to call it “adaptation” (262, 508), a term which has been used to designate desensitization to a permanent stimulus in other biological processes.
DNA Damage Checkpoints
Since the discovery of the G2/M DNA damage checkpoint, other checkpoints have been identified. For example, microtubule assembly is also subject to checkpoint control (for a review, see reference 469). A number of genes have been implicated in five different checkpoints for DNA damage in S. cerevisiae: G2/M, G1/M, S/M, intra-S, and meiotic checkpoints (for a review, see reference 540). These checkpoint genes respond to various damages, since the corresponding mutants are sensitive to ionizing radiation or MMS (supposed to induce mainly DSBs) and UV light (inducing mainly photodimers repaired by the nucleotide excision repair pathway).
As mentioned above, the G2/M checkpoint can be activated by single unrepaired DSB (262, 294, 425, 508). It is under the control of at least eight genes: RAD9, RAD17, RAD24, RAD53, MEC1, MEC3, PDS1, and DDC1 (13, 79, 223, 279, 361, 431, 525, 540–543, 565, 572).
Another checkpoint of particular interest for people studying DSB repair is the meiotic checkpoint, which subordinates the exit of meiosis I prophase to the repair of the Spo11p-induced DSBs. Some mutants in which meiotic recombination is affected, such as rad51, dmc1, zip1, rad50S, com1/sae2, and sae3 mutants, are also arrested in late meiosis I prophase (42, 306, 307, 387, 449, 488, 563). The exact nature of the arrest (transient or permanent, and what proportion of the cells are affected) can depend largely on the strain background. zip1 mutants, for example, display a total permanent arrest in a strain background used in the Roeder laboratory (488), but this arrest is largely alleviated in the SK1 background (488, 563). On the other hand, mutations that impair DSB formation, such as rad50, do not affect cell cycle progression through meiosis (64, 563). Therefore, the meiotic checkpoint arrest clearly seems to be a response to unrepaired DSBs (or improperly repaired DSBs in the case of zip1).
The discovery that rad17, rad24, and mec3 mutants have a very low level of viable spores (288) made the corresponding genes candidates for meiotic checkpoint genes. In contrast, a rad9 mutant has a nearly wild-type level of viable spores. Lydall et al. (287) showed in a SK1 background that the total and permanent arrest observed in a dmc1 mutant was alleviated by a rad17, rad24, or mec1 mutation but not by a rad9 mutation. Thus, RAD17, RAD24, and MEC1 (and probably MEC3 and DDC1) take part in a meiotic checkpoint. This checkpoint differs from the G2/M checkpoint because of the absence of any requirement for RAD9; however, dmc1 mutants still have a functional RAD51 gene, and we do not know to what extent DSBs may be committed to an abortive repair pathway that the Rad9 protein could not monitor. In this regard, it must be pointed out that in the cells mutated for dmc1 and rad24, Rad51p foci are retained at least until the first meiotic division (287).
Recently, Xu et al. (563) reported that the RED1 and MEK1 genes (involved in axial-element formation) also play a role in the meiotic checkpoint. The arrest or sporulation delay observed in rad51, dmc1, zip1, or rad50S mutants is alleviated by a red1 or mek1 deletion. Since the red1 and mek1 mutations also severely affects the meiotic DSB level (297, 563) they might simply reduce the signal for checkpoint arrest. However, in rad50S mutants, the DSB level is not affected by red1 and mek1, yet these two mutations still suppress the meiotic delay (563). At least in this case, it is the checkpoint itself, and not the checkpoint signal, which seems to be alleviated.
Meanwhile, other results have made clear that meiotic DSBs are not processed in the same way in the presence or absence of Red1p. As described above, DMC1 is not required for meiotic DSB repair in a red1 strain (439). It is possible that without Red1p, DSBs enter a pathway that excludes them from checkpoint control.
What Initiates Checkpoint Arrest?
As clearly enunciated in the review of Lydall and Weinert (288), a checkpoint can be viewed as a process going from a sensor (which recognizes the damage) to an effector (which arrests the cell cycle) through a transduction cascade. Because Mec1p and Rad53p protein kinases are involved in all the DNA damage checkpoint pathways, they are decent candidates for a transduction cascade. Mec1p appears to phosphorylate Rad53p in response to DNA damage and would thus be upstream of Rad53p in the activation cascade (424, 480). However, things are still very unclear when it comes to the effector and the sensor of the G2/M and meiotic checkpoints. For the effectors, we prefer to direct the reader to a relevant review (540) and simply mention that there are contradictory data on a possible role of the yeast p34cdc28 protein in G2/M arrest and that we have no information about meiotic effectors. In the context of this review on recombination, we are much more interested in the connection between the DSBs and the checkpoint arrest, i.e., the exact nature of the signal for arrest, and of the sensor that responds to this signal.
Upstream of the Mec1p-Rad53p cascade, there are two different pathways, the RAD17-RAD24-MEC3-DDC1 pathway, which plays a role in most of the checkpoints, including the G2/M and meiotic ones (279, 287, 361, 543), and the RAD9 pathway, which, except in meiosis, acts with the RAD17 pathway. These two different pathways were revealed by the synergistic effect of rad9 mutations with rad17, rad24, and mec3 mutations for MMS and UV sensitivity (288) and for suppression of UV-induced cell cycle arrest (89).
Lydall and Weinert (288) observed that a rad24 and rad17 mutations slowed the formation of ssDNA at the end of the chromosome in a thermosensitive cdc13-1 mutant (at the nonpermissive temperature, cdc13-1 generates DNA damage in the vicinity of the telomere, characterized by the formation of ssDNA). The authors inferred that these genes may have a direct effect on the processing of the DNA damage appearing in a cdc13 strain. Rad17p is homologous to the Rec1 protein of Ustilago maydis and to the human Rad1p, with both of them having a 3′-to-5′ exonuclease activity in vitro (370, 501). Thus, Rad17p acting with Rad24p and Mec3p could process subtelomeric nicks occurring in the cdc13 mutant, and this processing would activate the checkpoint. In contrast, single-strand formation was enhanced in a rad9 mutant, and obviously the role of Rad9p must be different, maybe competing with Rad17p and thus inhibiting the processing and sending an independent signal to the Mec1p-Rad53p cascade.
The kind of damage appearing in a cdc13 strain derives from a failure of telomere maintenance, but the mechanism proposed above may also act for internal chromosomal DSBs. Lydall et al. (287) pointed out that the Rad17p meiotic checkpoint arrest was activated in cells defective for dmc1, rad51, or rad52, where the DSB is resected but cannot be repaired, since it is not activated in a rad50S mutant, where the Spo11p-blocked 5′ end prevents exonucleolytic resection of DSBs. They interpreted this as a further evidence that a complex involving ssDNA and the gene products of the RAD24 pathway might act as a signal for DSB-induced checkpoint arrest. However, they also observed that the checkpoint genes do not affect the amount of ssDNA, which also appears to be true for mitotic DSBs (262). Thus, interaction of the ssDNA with Rad17p and/or other proteins could be sufficient for checkpoint activation. The conclusion is that an interaction between ssDNA and Rad17p-Rad24p-Mec3p-Ddc1p or Rad9p is so far the best candidate we have for an initiator of checkpoint activation.
Mechanism of Adaptation
Confronted with a single unrepairable DSB, a yeast cell remains arrested for several hours but eventually resumes growth until death results from the loss of the broken chromosome. Recently, three adaptation-defective mutants have been isolated by Toczyski et al. (508). In these mutants, a single DSB that cannot be repaired causes a permanent RAD9-dependent cell cycle arrest. The mutation with the strongest effect resides in the CDC5 gene. CDC5 codes for an essential kinase that regulates mitosis (233). The two other genes found to be involved in adaptation are CBK1 and CBK2, coding for the two subunits of the nonessential casein kinase II (CKII).
If CDC5 and CKII act positively on the adaptation process, a recent study has revealed that a basic by-product of DNA damage, ssDNA, acts negatively. To study DSB induced arrest, Sandell and Zakian (425) and Toczyski et al. (508) used a rad52 disomic strain, where the DSB could not be repaired by homologous recombination but the broken chromosome could nevertheless be lost without affecting the viability of the cell. Lee et al. (262) used a slightly different experimental system: they induced a DSB at various sites in a haploid strain. A single unrepairable DSB at either MAT or URA3 resulted in a transient arrest, controlled by RAD17 and RAD9. However, two DSBs induced a permanent arrest. In addition, proteins that affect the resection of the DSBs also affect the arrest: in a hdf1 mutant, lacking the yeast Ku70p, exonucleolytic resection of the DSBs is increased twofold. Then, a single DSB at MAT or URA3 is enough to induce permanent arrest (262). Barnes and Rio (23) and Lewis et al. (267) also observed a permanent arrest in hdf1 yeast in the presence of multiple cuts (induced by a transgenic restriction enzyme). In contrast, an mre11 mutation, which reduces exonucleolytic resection about twofold, suppresses the permanent arrest seen in both hdf1 cells with one DSB or HDF1 cells with two breaks (262). Thus, the permanent arrest seems to depend on the amount of ssDNA in the cell.
If ssDNA is a signal that inhibits the growth resumption, a single-strand binding protein could be the sensor. Indeed, the permanent arrest resulting from two DSBs or from a single DSB in a hdf1 background is suppressed by a rfa1-t11 mutation (262). This mutation affects the largest subunit of RPA (the essential single-strand binding protein involved in replication) and impairs DSB repair in yeast without preventing replication (521). We must point out that what has been shown here is that the amount of ssDNA monitored by RPA is the signal for the prolongation of the checkpoint arrest, not for the initiation of this arrest. Neither rfa1-t11 nor mre11 affects the initial arrest. If ssDNA also plays a role in the initiation, as proposed by Lydall et al. (288), the amount required to act as a signal for arrest would be much smaller than that required to maintain arrest. One can then imagine a single mechanism, where a small amount of ssDNA would be enough to trigger the arrest but the maintenance of the arrest would depend on the constant formation of more and more ssDNA.
FUTURE DIRECTIONS
One cannot help but be impressed with how much information has been acquired in the past decade, with the flourishing of recombinant DNA-based analysis of recombination and repair. However, there remain dozens of fundamental questions for which we have no answers and not even very good guesses. Among the most relevant questions are the following.
How Many Pathways Are There?
The closer one looks, the more pathways to repair DSBs seem to appear. Are they all “real,” or are some of them jerry-rigged out of available components when normal recombination is disrupted? For example, there is a residual recombination process in the absence of RAD52. However, it is not known if it is RAD52 independent or rad52 defective (i.e., an event that depends on an aberrant intermediate that appears only in the absence of RAD52). The number of apparent mechanisms we have mentioned is already large and is certain to grow.
What Are the Precise Roles of the Recombination Proteins?
More and more data have been obtained recently on the biochemical activities of proteins involved in recombination, such as Rad51p, Rad52p, Rad54p, Rad55p, Rad57p, and Mre11p. However, we are still far from understanding their exact role in vivo, especially when we consider the different recombination pathways. For example, why is a Rad52p homologue such as Rad59p required for some kind of Rad51-independent recombination events? What makes Tid1/Rdh54p different from Rad54p? How do the specific components of the meiotic recombination machinery act to confer on meiotic recombination its specific features (preferential interhomologue recombination and high crossover proficiency)?
How is a DSB “Channeled” into Different Repair Pathways?
Although misalignment/fill-in and deletion repair of a DSB are rare events (1 in 1,000 cells), perfect religation of 3′ overhanging ends is competitive with the efficiency of homologous recombination. We imagine that the early steps of homologous recombination involve 5′-to-3′ resection of the DNA ends and most probably the formation of a Rad51p filament that would engage in a search for a homologous partner. In contrast, end ligation appears to involve the interaction of DNA ends with a number of DNA binding proteins, at least Ku70p-Ku80p and Mre11p-Rad50p-Xrs2p. How are ends directed to one pathway or another? The Rad5 protein seems to affect the ratio of homologous to nonhomologous repair but does so by a totally unknown mechanism (7). Although the Srs2 helicase has been implicated in shuttling DNA damage from a RAD6-, RAD18-mediated postreplicative pathway to a RAD52-mediated homologous repair pathway (1, 2), we are just beginning to realize the importance of this type of control.
What Regulates the Transition from Break-Induced Replication to Gene Conversion?
If recombination is initiated by the invasion of one end of the DSB, it may set up a (modified) replication fork that, at least under some circumstances, seems capable of proceeding all the way to the end of the chromosome. However, most of the time, the DSB is repaired by gene conversion, in which we imagine that the second end of the DSB becomes engaged. How often does BIR occur relative to gene conversion, and how is the second end recruited? What proteins are involved at this step?
How Do the Ends of a DSB Find Their Homologous Partners?
Once the DSB is created, where do the two ends go? Are they often held in the same region of the nucleus, or do they just sweep out the available space, searching for a homologous partner? The ability to search the entire genome for a relatively small homologous partner is well documented in fungi. In the fungi Neurospora and Ascobolus, virtually all the cytosines in a duplicated sequence, be it endogenous or artificial, can be methylated and/or mutated (processes known as MIP and RIP) (417, 441). This occurs at a premeiotic stage in a haploid nucleus and can involve homologous segments of less than 1 kb. It occurs without evident genetic exchange (although the relationship of this process to recombination by using recombination-defective mutations has not yet been explored). Saccharomyces can carry out recombination between ectopically located homologous sequences (151, 207, 269–271), but we still have no idea of the basic mechanism of homology searching throughout the genome.
How Does New DNA Synthesis Occur during Recombination?
Is DNA replication during gene conversion conservative? Although many genetic results support this idea, it remains to be demonstrated by physical means. In addition, it suggests that a lot of genes involved in DNA repair remain to be identified: genes involved in DNA synthesis during DNA repair but not during replication, genes regulating the association of the newly synthesized strands with their template, genes responsible for branch migration, and these three probably overlapping categories are not exclusive of other ones. We shall be employed for many years to come.
ACKNOWLEDGMENTS
We are grateful for discussions over several years with many of the authors cited in this work, particularly to Sue Jinks-Robertson, Michael Lichten, Patrick Sung, Stephen West, and three anonymous reviewers for their critical reading of the manuscript. Members of the Haber laboratory contributed their ideas, criticisms, and support.
Research done in this laboratory that is cited in this review was supported by grants from the National Institutes of Health, National Science Foundation, and Department of Energy. F.P. was supported by fellowships from the Association pour la Recherche contre le cancer, the Jane Coffin Memorial Fund for Medical Research, and the American Cancer Society.
REFERENCES
- 1.Aboussekhra A, Chanet R, Adjiri A, Fabre F. Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol Cell Biol. 1992;12:3224–3234. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aboussekhra A, Chanet R, Zgaga Z, Cassier-Chauvat C, Heude M, Fabre F. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 1989;17:7211–7220. doi: 10.1093/nar/17.18.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aguilera A, Klein H. Genetic and molecular analysis of recombination events in Saccharomyces cerevisiae occurring in the presence of the hyper-recombination mutation hpr1. Genetics. 1989;122:503–517. doi: 10.1093/genetics/122.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aguilera A, Klein H L. Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae. I. Isolation and genetic characterization of hyper-recombination mutations. Genetics. 1988;119:779–790. doi: 10.1093/genetics/119.4.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aguilera A, Klein H L. HPR1, a novel yeast gene that prevents intrachromosomal excision recombination, shows carboxy-terminal homology to the Saccharomyces cerevisiae TOP1 gene. Mol Cell Biol. 1990;10:1439–1451. doi: 10.1128/mcb.10.4.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahne F, Baur M, Eckardt-Schupp F. The REV2 gene of Saccharomyces cerevisiae: cloning and DNA sequence. Curr Genet. 1992;22:277–282. doi: 10.1007/BF00317921. [DOI] [PubMed] [Google Scholar]
- 7.Ahne F, Jha B, Eckardt-Schupp F. The RAD5 gene product is involved in the avoidance of non-homologous end-joining of DNA double strand breaks in the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25:743–749. doi: 10.1093/nar/25.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ajimura M, Leem S H, Ogawa H. Identification of new genes required for meiotic recombination in Saccharomyces cerevisiae. Genetics. 1993;133:51–66. doi: 10.1093/genetics/133.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alani E, Chi N W, Kolodner R. The Saccharomyces cerevisiae Msh2 protein specifically binds to duplex oligonucleotides containing mismatched DNA base pairs and insertions. Genes Dev. 1995;9:234–247. doi: 10.1101/gad.9.2.234. [DOI] [PubMed] [Google Scholar]
- 10.Alani E, Lee S, Kane M F, Griffith J, Kolodner R D. Saccharomyces cerevisiae Msh2p, a mispaired base recognition protein, also recognizes Holliday junctions in DNA. J Mol Biol. 1997;265:289–301. doi: 10.1006/jmbi.1996.0743. [DOI] [PubMed] [Google Scholar]
- 11.Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- 12.Alani E, Reenan R A, Kolodner R D. Interaction between mismatch repair and genetic recombination in Saccharomyces cerevisiae. Genetics. 1994;137:19–39. doi: 10.1093/genetics/137.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allen J B, Zhou Z, Siede W, Friedberg E C, Elledge S J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage induced transcription in yeast. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 14.Anderson L K, Offenberg H H, Verkuijlen W M H C, Heyting C. RecA-like proteins are components of early meiotic nodules in lily. Proc Natl Acad Sci USA. 1997;94:6868–6873. doi: 10.1073/pnas.94.13.6868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 16.Appelgren H, Cederberg H, Rannug U. Mutations at the human minisatellite MS32 integrated in yeast occur with high frequency in meiosis and involve complex recombination events. Mol Gen Genet. 1997;256:7–17. doi: 10.1007/s004380050540. [DOI] [PubMed] [Google Scholar]
- 17.Armour J A, Jeffreys A J. Biology and applications of human minisatellite loci. Curr Opin Genet Dev. 1992;2:850–856. doi: 10.1016/s0959-437x(05)80106-6. [DOI] [PubMed] [Google Scholar]
- 18.Bai Y, Symington L S. A RAD52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- 19.Bailis A M, Maines S, Negritto M T. The essential helicase gene RAD3 suppresses short-sequence recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:3998–4008. doi: 10.1128/mcb.15.8.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailis J M, Roeder G S. Synaptonemal complex morphogenesis and sister-chromatid cohesion require Mek1-dependent phosphorylation of a meiotic chromosomal protein. Genes Dev. 1998;12:3551–3563. doi: 10.1101/gad.12.22.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baker S M, Plug A W, Prolla T A, Bronner C E, Harris A C, Yao X, Christie D M, Monell C, Arnheim N, Bradley A, Ashley T, Liskay R M. Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 22.Bardwell A J, Bardwell L, Tomkinson A E, Friedberg E C. Specific cleavage of model recombination and repair intermediates by the yeast Rad1-Rad10 DNA endonuclease. Science. 1994;265:2082–2085. doi: 10.1126/science.8091230. [DOI] [PubMed] [Google Scholar]
- 23.Barnes G, Rio D. DNA double-strand-break sensitivity, DNA replication, and cell cycle arrest phenotypes of Ku-deficient Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:867–872. doi: 10.1073/pnas.94.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bascom-Slack C A, Dawson D. A physical assay for detection of early meiotic recombination intermediates in Saccharomyces cerevisiae. Mol Gen Genet. 1998;258:512–520. doi: 10.1007/s004380050762. [DOI] [PubMed] [Google Scholar]
- 25.Baudat F, Nicolas A. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc Natl Acad Sci USA. 1997;94:5213–5218. doi: 10.1073/pnas.94.10.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baumann P, Benson F E, West S C. Human Rad51 protein promotes ATP-dependent homologous pairing and strand transfer reactions in vitro. Cell. 1996;87:757–766. doi: 10.1016/s0092-8674(00)81394-x. [DOI] [PubMed] [Google Scholar]
- 27.Baumann P, West S C. The human Rad51 protein: polarity of strand transfer and stimulation by hRP-A. EMBO J. 1997;16:5198–5206. doi: 10.1093/emboj/16.17.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baumann P, West S C. Role of the human Rad51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci. 1998;23:247–251. doi: 10.1016/s0968-0004(98)01232-8. [DOI] [PubMed] [Google Scholar]
- 29.Bell L, Byers B. Occurrence of crossed strand-exchange forms in yeast DNA during meiosis. Proc Natl Acad Sci USA. 1979;76:3445–3449. doi: 10.1073/pnas.76.7.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Belmaaza A, Milot E, Villemure J F, Chartrand P. Interference of DNA sequence divergence with precise recombinational DNA repair in mammalian cells. EMBO J. 1994;13:5355–5360. doi: 10.1002/j.1460-2075.1994.tb06870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennett R J, Dunderdale H J, West S C. Resolution of Holliday junctions by RuvC resolvase: cleavage specificity and DNA distortion. Cell. 1993;74:1021–1031. doi: 10.1016/0092-8674(93)90724-5. [DOI] [PubMed] [Google Scholar]
- 32.Bennett R J, West S C. RuvC protein resolves Holliday junctions via cleavage of the continuous (noncrossover) strands. Proc Natl Acad Sci USA. 1995;92:5635–5639. doi: 10.1073/pnas.92.12.5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benson F E, Stasiak A, West S C. Purification and characterization of the human Rad51 protein, an analogue of E. coli RecA. EMBO J. 1994;13:5764–5771. doi: 10.1002/j.1460-2075.1994.tb06914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Benson F E, Baumann P, West S C. Synergistic actions of Rad51 and Rad52 in recombination and DNA repair. Nature. 1998;391:401–404. doi: 10.1038/34937. [DOI] [PubMed] [Google Scholar]
- 34.Bergerat A, de Massy B, Gadelle D, Varoutas P C, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 35.Berinstein N, Pennell N, Ottaway C A, Shulman M J. Gene replacement with one-sided homologous recombination. Mol Cell Biol. 1992;12:360–367. doi: 10.1128/mcb.12.1.360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bezzubova O, Silbergleit A, Yamaguchi I Y, Takeda S, Buerstedde J M. Reduced X-ray resistance and homologous recombination frequencies in a RAD54−/− mutant of the chicken DT40 cell line. Cell. 1997;89:185–193. doi: 10.1016/s0092-8674(00)80198-1. [DOI] [PubMed] [Google Scholar]
- 37.Bhatia P K, Wang Z, Friedberg E C. DNA repair and transcription. Curr Opin Genet Dev. 1996;6:146–150. doi: 10.1016/s0959-437x(96)80043-8. [DOI] [PubMed] [Google Scholar]
- 38.Bianco P R, Tracy R B, Kowalczykowski S C. DNA strand exchange proteins: a biochemical and physical comparison. Front Biosci. 1998;3:570–603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- 39.Bishop, D. K. Personal communication.
- 40.Bishop D K. RecA homologs Dmc1 and Rad51 interact to form multiple nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 41.Bishop D K, Anderson J, Kolodner R D. Specificity of mismatch repair following transformation of Saccharomyces cerevisiae with heteroduplex plasmid DNA. Proc Natl Acad Sci USA. 1989;86:3713–3717. doi: 10.1073/pnas.86.10.3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bishop D K, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 43.Boeke J D, Trueheart J, Natsoulis G, Fink G R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 44.Bogue M A, Wang C, Zhu C, Roth D B. V(D)J recombination in Ku86-deficient mice: distinct effects on coding, signal, and hybrid joint formation. Immunity. 1997;7:37–47. doi: 10.1016/s1074-7613(00)80508-7. [DOI] [PubMed] [Google Scholar]
- 45.Bois P, Williamson J, Brown J, Dubrova Y E, Jeffreys A J. A novel unstable mouse VNTR family expanded from SINE B1 elements. Genomics. 1998;49:122–128. doi: 10.1006/geno.1998.5228. [DOI] [PubMed] [Google Scholar]
- 46.Borts R H, Haber J E. Length and distribution of meiotic gene conversion tracts and crossovers in Saccharomyces cerevisiae. Genetics. 1989;123:69–80. doi: 10.1093/genetics/123.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borts R H, Haber J E. Meiotic recombination in yeast: alteration by multiple heterozygosities. Science. 1987;237:1459–1465. doi: 10.1126/science.2820060. [DOI] [PubMed] [Google Scholar]
- 48.Borts R H, Leung W Y, Kramer W, Kramer B, Williamson M, Fogel S, Haber J E. Mismatch repair-induced meiotic recombination requires the pms1 gene product. Genetics. 1990;124:573–584. doi: 10.1093/genetics/124.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Borts R H, Lichten M, Haber J E. Analysis of meiosis-defective mutations in yeast by physical monitoring of recombination. Genetics. 1986;113:551–567. doi: 10.1093/genetics/113.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borts R H, Lichten M, Hearn M, Davidow L S, Haber J E. Physical monitoring of meiotic recombination in Saccharomyces cerevisiae. Cold Spring Harbor Symp Quant Biol. 1984;49:67–76. doi: 10.1101/sqb.1984.049.01.010. [DOI] [PubMed] [Google Scholar]
- 51.Bosco G, Haber J E. Chromosome break-induced DNA replication leads to non-reciprocal translocations and telomere capture. Genetics. 1998;150:1037–1047. doi: 10.1093/genetics/150.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boulton S J, Jackson S P. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boulton S J, Jackson S P. Identification of a Saccharomyces cerevisiae Ku80 homologue: roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boulton S J, Jackson S P. Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 55.Bratty J, Ferbeyre G, Molinaro C, Cedergren R. Stimulation of mitotic recombination upon transcription from the yeast GAL1 promoter but not from other RNA polymerase I, II and III promoters. Curr Genet. 1996;30:381–388. doi: 10.1007/s002940050146. [DOI] [PubMed] [Google Scholar]
- 56.Bressan D A, Olivares H A, Nelms B E, Petrini J H. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae Mre11. Genetics. 1998;150:591–600. doi: 10.1093/genetics/150.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brill S J, DiNardo S, Voelkel-Meiman K, Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 1987;326:414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- 58.Buard J, Jeffreys A J. Big, bad minisatellites. Nat Genet. 1997;15:327–328. doi: 10.1038/ng0497-327. [DOI] [PubMed] [Google Scholar]
- 59.Buard J, Vergnaud G. Complex recombination events at the hypermutable minisatellite CEB1 (D2S90) EMBO J. 1994;13:3203–3210. doi: 10.1002/j.1460-2075.1994.tb06619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Budd M E, Campbell J L. DNA polymerases required for repair of UV-induced damage in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:2173–2179. doi: 10.1128/mcb.15.4.2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bullard S A, Kim S, Galbraith A M, Malone R E. Double strand breaks at the HIS2 recombination hot spot in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1996;93:13054–13059. doi: 10.1073/pnas.93.23.13054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campbell D A, Fogel S. Association of chromosome loss with centromere-adjacent mitotic recombination in yeast disomic haploid. Genetics. 1977;85:573–585. doi: 10.1093/genetics/85.4.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Campbell D A, Fogel S, Lusnak K. Mitotic chromosome loss in a disomic haploid of Saccharomyces cerevisiae. Genetics. 1975;79:383–396. doi: 10.1093/genetics/79.3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- 65.Carney J P, Maser R S, Olivares H, Davis E M, Le Beau M, Yates III J R, Hays L, Morgan W F, Petrini J H. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- 66.Carpenter A T C. Thoughts on recombination nodules, meiotic recombination and chiasmata. In: Kucherlapati R, Smith G R, editors. Genetic recombination. Washington, D.C: American Society for Microbiology; 1988. pp. 529–548. [Google Scholar]
- 67.Chambers S R, Hunter N, Louis E J, Borts R H. The mismatch repair system reduces meiotic homeologous recombination and stimulates recombination-dependent chromosome loss. Mol Cell Biol. 1996;16:6110–6120. doi: 10.1128/mcb.16.11.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chanet R, Heude M, Adjiri A, Maloisel L, Fabre F. Semidominant mutations in the yeast Rad51 protein and their relationships with the Srs2 helicase. Mol Cell Biol. 1996;16:4782–4789. doi: 10.1128/mcb.16.9.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chattoo B B, Sherman F, Atubalis D A, Fiellstedt T A, Mehnert D, et al. Selection of lys2 mutants on the yeast Saccharomyces cerevisiae by the utilization of α-aminoadipate. Genetics. 1979;93:51–65. doi: 10.1093/genetics/93.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chavez S, Aguilera A. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 1997;11:3459–3470. doi: 10.1101/gad.11.24.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen W, Jinks-Robertson S. Mismatch repair proteins regulate heteroduplex formation during mitotic recombination in yeast. Mol Cell Biol. 1998;18:6525–6537. doi: 10.1128/mcb.18.11.6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen X, Mariappan S V, Catasti P, Ratliff R, Moyzis R K, Laayoun A, Smith S S, Bradbury E M, Gupta G. Hairpins are formed by the single DNA strands of the fragile X triplet repeats: structure and biological implications. Proc Natl Acad Sci USA. 1995;92:5199–5203. doi: 10.1073/pnas.92.11.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chepurnaya O V, Kozhin S A, Peshekhonov V T, Korolev V G. RAD58 (XRS4), a new gene in the RAD52 epistasis group. Curr Genet. 1995;28:274–279. doi: 10.1007/BF00309787. [DOI] [PubMed] [Google Scholar]
- 74.Choder M. A general topoisomerase I-dependent transcriptional repression in the stationary phase in yeast. Genes Dev. 1991;5:2315–2326. doi: 10.1101/gad.5.12a.2315. [DOI] [PubMed] [Google Scholar]
- 75.Christman M F, Dietrich F S, Fink G R. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell. 1988;55:413–425. doi: 10.1016/0092-8674(88)90027-x. [DOI] [PubMed] [Google Scholar]
- 76.Chua P R, Roeder G S. Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes Dev. 1997;11:1786–1800. doi: 10.1101/gad.11.14.1786. [DOI] [PubMed] [Google Scholar]
- 77.Chua P R, Roeder G S. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell. 1998;93:349–359. doi: 10.1016/s0092-8674(00)81164-2. [DOI] [PubMed] [Google Scholar]
- 78.Clever B, Interthal H, Schmuckli M J, King J, Sigrist M, Heyer W D. Recombinational repair in yeast: functional interactions between Rad51 and Rad54 proteins. EMBO J. 1997;16:2535–2544. doi: 10.1093/emboj/16.9.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cohen-Fix O, Koshland D. The anaphase inhibitor of Saccharomyces cerevisiae Pds1p is a target of the DNA damage checkpoint pathway. Proc Natl Acad Sci USA. 1997;94:14361–14366. doi: 10.1073/pnas.94.26.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Colaiácovo, M. P., and J. E. Haber. Unpublished results.
- 81.Colaiácovo, M. P., F. Pâques, and J. E. Haber. Removal of nonhomologous DNA ends during gene conversion by a RAD1-, MSH2-independent pathway. Genetics, in press. [DOI] [PMC free article] [PubMed]
- 82.Colleaux L, d’Auriol L, Betermier M, Cottarel G, Jacquier A, Galibert F, Dujon B. Universal code equivalent of a yeast mitochondrial intron reading frame is expressed into E. coli as a specific double strand endonuclease. Cell. 1986;44:521–533. doi: 10.1016/0092-8674(86)90262-x. [DOI] [PubMed] [Google Scholar]
- 83.Collins I, Newlon C S. Meiosis-specific formation of joint DNA molecules containing sequences from homologous chromosomes. Cell. 1994;76:65–75. doi: 10.1016/0092-8674(94)90173-2. [DOI] [PubMed] [Google Scholar]
- 84.Connelly J C, Kirkham L A, Leach D R F. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc Natl Acad Sci USA. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Connolly B, White C I, Haber J E. Physical monitoring of mating type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2342–2349. doi: 10.1128/mcb.8.6.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Conrad M N, Dominguez A M, Dresser M E. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science. 1997;276:1252–1255. doi: 10.1126/science.276.5316.1252. [DOI] [PubMed] [Google Scholar]
- 87.Datta A, Adjiri A, New L, Crouse G F, Jinks R S. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1085–1093. doi: 10.1128/mcb.16.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Datta A, Hendrix M, Lipsitch M, Jinks R S. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc Natl Acad Sci USA. 1997;94:9757–9762. doi: 10.1073/pnas.94.18.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.de la Torre-Ruiz M A, Green C M, Lowndes N F. RAD9 and RAD24 define two additive, interacting branches of the DNA damage checkpoint pathway in budding yeast normally required for Rad53 modification and activation. EMBO J. 1998;17:2687–2698. doi: 10.1093/emboj/17.9.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Delattre M, Anxolabehere D, Coen D. Prevalence of localized rearrangements vs. transpositions among events induced by Drosophila P element transposase on a P transgene. Genetics. 1995;141:1407–1424. doi: 10.1093/genetics/141.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.de los Santos T, Hollingsworth N M. Red1p: a MEK1-dependent phosphoprotein that physically interacts with Hop1p during meiosis in yeast. J Biol Chem. 1999;274:1783–1790. doi: 10.1074/jbc.274.3.1783. [DOI] [PubMed] [Google Scholar]
- 92.de Massy B, Baudat F, Nicolas A. Initiation of recombination in Saccharomyces cerevisiae haploid meiosis. Proc Natl Acad Sci USA. 1994;91:11929–11933. doi: 10.1073/pnas.91.25.11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.de Massy B, Rocco V, Nicolas A. The nucleotide mapping of DNA double-strand breaks at the CYS3 initiation site of meiotic recombination in Saccharomyces cerevisiae. EMBO J. 1995;14:4589–4598. doi: 10.1002/j.1460-2075.1995.tb00138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dernburg A F, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve A M. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- 95.Detloff P, Petes T D. Measurements of excision repair tracts formed during meiotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1805–1814. doi: 10.1128/mcb.12.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Detloff P, Sieber J, Petes T D. Repair of specific base pair mismatches formed during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:737–745. doi: 10.1128/mcb.11.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Detloff P, White M A, Petes T D. Analysis of a gene conversion gradient at the HIS4 locus in Saccharomyces cerevisiae. Genetics. 1992;132:113–123. doi: 10.1093/genetics/132.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.DiNardo S, Voelkel K, Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc Natl Acad Sci USA. 1984;81:2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Donovan J W, Milne G T, Weaver D T. Homotypic and heterotypic protein associations control Rad51 function in double-strand break repair. Genes Dev. 1994;8:2552–2562. doi: 10.1101/gad.8.21.2552. [DOI] [PubMed] [Google Scholar]
- 100.Dresser M E, Ewing D J, Conrad M N, Dominguez A M, Barstead R, Jiang H, Kodadek T. DMC1 functions in a Saccharomyces cerevisiae meiotic pathway that is largely independent of the RAD51 pathway. Genetics. 1997;147:533–544. doi: 10.1093/genetics/147.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dujon B. Group I introns as mobile genetic elements: facts and mechanistic speculations—a review. Gene. 1989;82:91–114. doi: 10.1016/0378-1119(89)90034-6. [DOI] [PubMed] [Google Scholar]
- 102.Dujon B, Belfort M, Butow R A, Jacq C, Lemieux C, Perlman P S, Vogt V M. Mobile introns: definition of terms and recommended nomenclature. Gene. 1989;82:115–118. doi: 10.1016/0378-1119(89)90035-8. [DOI] [PubMed] [Google Scholar]
- 103.Dunn, B. Personal communication.
- 104.Dunn B, Szauter P, Pardue M L, Szostak J W. Transfer of yeast telomeres to linear plasmids by recombination. Cell. 1984;39:191–201. doi: 10.1016/0092-8674(84)90205-8. [DOI] [PubMed] [Google Scholar]
- 105.Eggleston A K, West S C. Exchanging partners: recombination in E. coli. Trends Genet. 1996;12:20–26. doi: 10.1016/0168-9525(96)81384-9. [DOI] [PubMed] [Google Scholar]
- 106.Ellis N A, Groden J, Ye T Z, Straughen J, Lennon D J, Ciocci S, Proytcheva M, German J. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 107.Emery H S, Schild D, Kellogg D E, Mortimer R K. Sequence of RAD54, a Saccharomyces cerevisiae gene involved in recombination and repair. Gene. 1991;104:103–106. doi: 10.1016/0378-1119(91)90473-o. [DOI] [PubMed] [Google Scholar]
- 108.Engebrecht J, Hirsch J, Roeder G S. Meiotic gene conversion and crossing over: their relationship to each other and to chromosome synapsis and segregation. Cell. 1990;62:927–937. doi: 10.1016/0092-8674(90)90267-i. [DOI] [PubMed] [Google Scholar]
- 109.Engebrecht J, Roeder G S. Yeast mer1 mutants display reduced levels of meiotic recombination. Genetics. 1989;121:237–247. doi: 10.1093/genetics/121.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Engebrecht J A, Voelkel-Meiman K, Roeder G S. Meiosis-specific RNA splicing in yeast. Cell. 1991;66:1257–1268. doi: 10.1016/0092-8674(91)90047-3. [DOI] [PubMed] [Google Scholar]
- 111.Engels W R, Johnson-Schlitz D M, Eggleston W B, Sved J. High-frequency P element loss in Drosophila is homolog dependent. Cell. 1990;62:515–525. doi: 10.1016/0092-8674(90)90016-8. [DOI] [PubMed] [Google Scholar]
- 112.Esposito M S. Evidence that spontaneous mitotic recombination occurs at the two- strand stage. Proc Natl Acad Sci USA. 1978;75:4436–4440. doi: 10.1073/pnas.75.9.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Esposito R E, Esposito M S. Genetic recombination and commitment to meiosis in Saccharomyces. Proc Natl Acad Sci USA. 1974;71:3172–3176. doi: 10.1073/pnas.71.8.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Essers J, Hendriks R W, Swagemakers S M, Troelstra C, de Wit J, Bootsma D, Hoeijmakers J H, Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195–204. doi: 10.1016/s0092-8674(00)80199-3. [DOI] [PubMed] [Google Scholar]
- 115.Fabre F, Boulet A, Roman H. Gene conversion at different points in the mitotic cycle of Saccharomyces cerevisiae. Mol Gen Genet. 1984;195:139–143. doi: 10.1007/BF00332736. [DOI] [PubMed] [Google Scholar]
- 116.Fabre F, Roman H. Evidence that a single DNA ligase is involved in replication and recombination in yeast. Proc Natl Acad Sci USA. 1979;76:4586–4588. doi: 10.1073/pnas.76.9.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Fairhead C, Dujon B. Consequences of unique double-stranded breaks in yeast chromosomes: death or homozygosis. Mol Gen Genet. 1993;240:170–178. doi: 10.1007/BF00277054. [DOI] [PubMed] [Google Scholar]
- 118.Fan Q Q, Petes T D. Relationship between nuclease-hypersensitive sites and meiotic recombination hot spot activity at the HIS4 locus of Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2037–2043. doi: 10.1128/mcb.16.5.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fasullo M, Dave P. Mating type regulates the radiation-associated stimulation of reciprocal translocation events in Saccharomyces cerevisiae. Mol Gen Genet. 1994;243:63–70. doi: 10.1007/BF00283877. [DOI] [PubMed] [Google Scholar]
- 120.Fasullo M T, Davis R W. Direction of chromosome rearrangements in Saccharomyces cerevisiae by use of his3 recombinational substrates. Mol Cell Biol. 1988;8:4370–4380. doi: 10.1128/mcb.8.10.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ferguson D O, Holloman W K. Recombinational repair of gaps in DNA is asymmetric in Ustilago maydis and can be explained by a migrating D-loop model. Proc Natl Acad Sci USA. 1996;93:5419–5424. doi: 10.1073/pnas.93.11.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fiorentini P, Huang K N, Tishkoff D X, Kolodner R D, Symington L S. Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol Cell Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fishman-Lobell J, Haber J E. Removal of nonhomologous DNA ends in double-strand break recombination: the role of the yeast ultraviolet repair gene RAD1. Science. 1992;258:480–484. doi: 10.1126/science.1411547. [DOI] [PubMed] [Google Scholar]
- 124.Fishman-Lobell J, Rudin N, Haber J E. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol Cell Biol. 1992;12:1292–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Flores-Rozas H, Kolodner R D. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc Natl Acad Sci USA. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fogel S, Mortimer R K, Lusnak K. Mechanisms of meiotic gene conversion, or “wandering on a foreign strand,”. In: Strathern J N, Jones E W, Broach J R, editors. The molecular analysis of the yeast Saccharomyces cerevisiae: life cycle and inheritance. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1981. pp. 289–339. [Google Scholar]
- 127.Fogel S, Hurst D D. Meiotic gene conversion in yeast tetrads and the theory of recombination. Genetics. 1967;57:455–481. doi: 10.1093/genetics/57.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fogel S, Mortimer R, Lusnak K, Tavares F. Meiotic gene conversion: a signal of the basic recombination event in yeast. Cold Spring Harbor Symp Quant Biol. 1979;43:1325–1341. doi: 10.1101/sqb.1979.043.01.152. [DOI] [PubMed] [Google Scholar]
- 129.Fogel S, Mortimer R K. Informational transfer in meiotic gene conversion. Proc Natl Acad Sci USA. 1969;62:96–103. doi: 10.1073/pnas.62.1.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Formosa T, Alberts B M. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell. 1986;47:793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- 131.Foss E, Lande R, Stahl F W, Steinberg C M. Chiasma interference as a function of genetic distance. Genetics. 1993;133:681–691. doi: 10.1093/genetics/133.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Foss E J, Stahl F W. A test of a counting model for chiasma interference. Genetics. 1995;139:1201–1209. doi: 10.1093/genetics/139.3.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fox M E, Smith G R. Control of meiotic recombination in Schizosaccharomyces pombe. Prog Nucleic Acid Res Mol Biol. 1998;61:345–378. doi: 10.1016/s0079-6603(08)60831-4. [DOI] [PubMed] [Google Scholar]
- 134.Freudenreich C H, Kantrow S M, Zakian V A. Expansion and length-dependent fragility of CTG repeats in yeast. Science. 1998;279:853–856. doi: 10.1126/science.279.5352.853. [DOI] [PubMed] [Google Scholar]
- 135.Freudenreich C H, Stavenhagen J B, Zakian V A. Stability of a CTG/CAG trinucleotide repeat in yeast is dependent on its orientation in the genome. Mol Cell Biol. 1997;17:2090–2098. doi: 10.1128/mcb.17.4.2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Friedberg E C, Bardwell A J, Bardwell L, Feaver W J, Kornberg R D, Svejstrup J Q, Tomkinson A E, Wang Z. Nucleotide excision repair in the yeast Saccharomyces cerevisiae: its relationship to specialized mitotic recombination and RNA polymerase II basal transcription. Philos Trans R Soc London Ser B. 1995;347:63–68. doi: 10.1098/rstb.1995.0010. [DOI] [PubMed] [Google Scholar]
- 137.Fu Y H, Kuhl D P, Pizzuti A, Pieretti M, Sutcliffe J S, Richards S, Verkerk A J, Holden J J, Fenwick R G, Jr, Warren S T, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 138.Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K. Distinct roles of two separable activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gacy A M, Goellner G, Juranic N, Macura S, McMurray C T. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 140.Game J C, Johnston L H, von Borstel R C. Enhanced mitotic recombination in a ligase-defective mutant of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1979;76:4589–4592. doi: 10.1073/pnas.76.9.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Game J C, Mortimer R K. A genetic study of X-ray sensitive mutants in yeast. Mutat Res. 1974;24:281–892. doi: 10.1016/0027-5107(74)90176-6. [DOI] [PubMed] [Google Scholar]
- 142.Game J C, Sitney K C, Cook V E, Mortimer R K. Use of a ring chromosome and pulsed-field gels to study interhomolog recombination, double-strand DNA breaks and sister-chromatid exchange in yeast. Genetics. 1989;123:695–713. doi: 10.1093/genetics/123.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gangloff S, McDonald J P, Bendixen C, Arthur L, Rothstein R. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: a potential eukaryotic reverse gyrase. Mol Cell Biol. 1994;14:8391–8398. doi: 10.1128/mcb.14.12.8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gangloff S, Zou H, Rothstein R. Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J. 1996;15:1715–1725. [PMC free article] [PubMed] [Google Scholar]
- 145.Gasior S L, Wong A K, Kora Y, Shinohara A, Bishop D K. Rad52 associates with RPA and functions with Rad55 and Rad57 to assemble meiotic recombination complexes. Genes Dev. 1998;12:2208–2221. doi: 10.1101/gad.12.14.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Giardina C, Lis J T. DNA melting on yeast RNA polymerase II promoters. Science. 1993;261:759–762. doi: 10.1126/science.8342041. [DOI] [PubMed] [Google Scholar]
- 147.Gilbertson L A, Stahl F W. Initiation of meiotic recombination is independent of interhomologue interactions. Proc Natl Acad Sci USA. 1994;91:11934–11937. doi: 10.1073/pnas.91.25.11934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Gilbertson L A, Stahl F W. A test of the double-strand break repair model for meiotic recombination in Saccharomyces cerevisiae. Genetics. 1996;144:27–41. doi: 10.1093/genetics/144.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Glassner B J, Mortimer R K. Synergistic interactions between RAD5, RAD16 and RAD54, three partially homologous yeast DNA repair genes each in a different repair pathway. Radiat Res. 1994;139:24–33. [PubMed] [Google Scholar]
- 150.Gloor G B, Nassif N A, Johnson-Schlitz D M, Preston C R, Engels W R. Targeted gene replacement in Drosophila via P element-induced gap repair. Science. 1991;253:1110–1117. doi: 10.1126/science.1653452. [DOI] [PubMed] [Google Scholar]
- 151.Goldman A S, Lichten M. The efficiency of meiotic recombination between dispersed sequences in Saccharomyces cerevisiae depends upon their chromosomal location. Genetics. 1996;144:43–55. doi: 10.1093/genetics/144.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Golin J E, Esposito M S. Coincident gene conversion during mitosis in Saccharomyces. Genetics. 1984;107:355–365. doi: 10.1093/genetics/107.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Golin J E, Falco S C. The behavior of insertions near a site of mitotic gene conversion in yeast. Genetics. 1988;119:535–540. doi: 10.1093/genetics/119.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Golin J E, Tampe H. Coincident recombination during mitosis in Saccharomyces: distance-dependent and -independent components. Genetics. 1988;119:541–547. doi: 10.1093/genetics/119.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gordenin D A, Lobachev K S, Degtyareva N P, Malkova A L, Perkins E, Resnick M A. Inverted DNA repeats: a source of eukaryotic genomic instability. Mol Cell Biol. 1993;13:5315–5322. doi: 10.1128/mcb.13.9.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Gottlieb S, Wagstaff J, Esposito R E. Evidence for two pathways of meiotic intrachromosomal recombination in yeast. Proc Natl Acad Sci USA. 1989;86:7072–7076. doi: 10.1073/pnas.86.18.7072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Goyon C, Lichten M. Timing of molecular events in meiosis in Saccharomyces cerevisiae: stable heteroduplex DNA is formed late in meiotic prophase. Mol Cell Biol. 1993;13:373–382. doi: 10.1128/mcb.13.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Grawunder U, West R B, Lieber M R. Antigen receptor gene rearrangement. Curr Opin Immunol. 1998;10:172–180. doi: 10.1016/s0952-7915(98)80246-x. [DOI] [PubMed] [Google Scholar]
- 159.Haber J E. Avoiding inappropriate relationships. Curr Biol. 1998;8:832–835. doi: 10.1016/s0960-9822(07)00524-6. [DOI] [PubMed] [Google Scholar]
- 160.Haber J E. In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. Bioessays. 1995;17:609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- 161.Haber J E. A locus control region in yeast that regulates recombination. Trends Genet. 1998;14:317–321. doi: 10.1016/s0168-9525(98)01501-7. [DOI] [PubMed] [Google Scholar]
- 162.Haber J E. The many interfaces of Mre11. Cell. 1998;95:583–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- 163.Haber J E. Mating-type gene switching in Saccharomyces cerevisiae. Trends Genet. 1992;8:446–452. doi: 10.1016/0168-9525(92)90329-3. [DOI] [PubMed] [Google Scholar]
- 164.Haber J E. Mating-type gene switching in Saccharomyces cerevisiae. Annu Rev Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 165.Haber J E, Hearn M. RAD52-independent mitotic gene conversion in Saccharomyces cerevisiae frequently results in chromosomal loss. Genetics. 1985;111:7–22. doi: 10.1093/genetics/111.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Haber J E, Leung W Y. Lack of chromosome territoriality in yeast: promiscuous rejoining of broken chromosome ends. Proc Natl Acad Sci USA. 1996;93:13949–13954. doi: 10.1073/pnas.93.24.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Haber J E, Ray B L, Kolb J M, White C I. Rapid kinetics of mismatch repair of heteroduplex DNA that is formed during recombination in yeast. Proc Natl Acad Sci USA. 1993;90:3363–3367. doi: 10.1073/pnas.90.8.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Haber J E, Thorburn P C, Rogers D. Meiotic and mitotic behavior of dicentric chromosomes in Saccharomyces cerevisiae. Genetics. 1984;106:185–205. doi: 10.1093/genetics/106.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Habraken Y, Sung P, Prakash L, Prakash S. ATP-dependent assembly of a ternary complex consisting of a DNA mismatch and the yeast Msh2-Msh6 and Mlh1-Pms1 protein complexes. J Biol Chem. 1998;273:9837–9841. doi: 10.1074/jbc.273.16.9837. [DOI] [PubMed] [Google Scholar]
- 170.Habraken Y, Sung P, Prakash L, Prakash S. Enhancement of Msh2-Msh3-mediated mismatch recognition by the yeast Mlh1-Pms1 complex. Curr Biol. 1997;7:790–793. doi: 10.1016/s0960-9822(06)00337-x. [DOI] [PubMed] [Google Scholar]
- 171.Hagelberg E, Gray I C, Jeffreys A J. Identification of the skeletal remains of a murder victim by DNA analysis. Nature. 1991;352:427–429. doi: 10.1038/352427a0. [DOI] [PubMed] [Google Scholar]
- 172.Han J O, Steen S B, Roth D B. Ku86 is not required for protection of signal ends or for formation of nonstandard V(D)J recombination products. Mol Cell Biol. 1997;17:2226–2234. doi: 10.1128/mcb.17.4.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hartwell L H, Weinert T A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- 174.Hastings P J. Measurement of restoration and conversion: its meaning for the mismatch repair hypothesis of conversion. Cold Spring Harbor Symp Quant Biol. 1984;49:49–53. doi: 10.1101/sqb.1984.049.01.008. [DOI] [PubMed] [Google Scholar]
- 175.Hastings P J. Recombination in the eukaryotic nucleus. Bioessays. 1988;9:61–64. doi: 10.1002/bies.950090206. [DOI] [PubMed] [Google Scholar]
- 176.Hastings P J. Some aspects of recombination in eukaryotic organisms. Annu Rev Genet. 1975;9:129–144. doi: 10.1146/annurev.ge.09.120175.001021. [DOI] [PubMed] [Google Scholar]
- 177.Hastings P J, Kalogeropoulos A, Rossignol J-L. Restoration to the parental genotype of mismatches formed in recombinant DNA heteroduplex. Curr Genet. 1980;2:169–174. doi: 10.1007/BF00420629. [DOI] [PubMed] [Google Scholar]
- 178.Hastings P J, McGill C, Shafer B, Strathern J N. Ends-in vs. ends-out recombination in yeast. Genetics. 1993;135:973–980. doi: 10.1093/genetics/135.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hays S L, Firmenich A A, Berg P. Complex formation in yeast double-strand break repair: participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc Natl Acad Sci USA. 1995;92:6925–6929. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Herskowitz I. A regulatory hierarchy for cell specialization in yeast. Nature. 1989;342:749–757. doi: 10.1038/342749a0. [DOI] [PubMed] [Google Scholar]
- 181.Heude M, Fabre F. a/alpha-control of DNA repair in the yeast Saccharomyces cerevisiae: genetic and physiological aspects. Genetics. 1993;133:489–498. doi: 10.1093/genetics/133.3.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Hewett D R, Handt O, Hobson L, Mangelsdorf M, Eyre H J, Baker E, Sutherland G R, Schuffenhauer S, Mao J I, Richards R I. FRA10B structure reveals common elements in repeat expansion and chromosomal fragile site genesis. Mol Cell. 1998;1:773–781. doi: 10.1016/s1097-2765(00)80077-5. [DOI] [PubMed] [Google Scholar]
- 183.Hicks J B, Hinnen A, Fink G R. Properties of yeast transformation. Cold Spring Harbor Symp Quant Biol. 1979;43:1305–1313. doi: 10.1101/sqb.1979.043.01.149. [DOI] [PubMed] [Google Scholar]
- 184.Holbeck S L, Strathern J N. A role for REV3 in mutagenesis during double-strand break repair in Saccharomyces cerevisiae. Genetics. 1997;147:1017–1024. doi: 10.1093/genetics/147.3.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 186.Hollingsworth N M, Goetsch L, Byers B. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell. 1990;61:73–84. doi: 10.1016/0092-8674(90)90216-2. [DOI] [PubMed] [Google Scholar]
- 187.Hollingsworth N M, Ponte L, Halsey C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 1995;9:1728–1739. doi: 10.1101/gad.9.14.1728. [DOI] [PubMed] [Google Scholar]
- 188.Holm C, Goto T, Wang J C, Botstein D. DNA topoisomerase II is required at the time of mitosis in yeast. Cell. 1985;41:553–563. doi: 10.1016/s0092-8674(85)80028-3. [DOI] [PubMed] [Google Scholar]
- 189.Holm C, Stearns T, Botstein D. DNA topoisomerase II must act at mitosis to prevent nondisjunction and chromosome breakage. Mol Cell Biol. 1989;9:159–168. doi: 10.1128/mcb.9.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Holmes A, Haber J E. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- 191.Hunter N, Borts R H. Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 1997;11:1573–1582. doi: 10.1101/gad.11.12.1573. [DOI] [PubMed] [Google Scholar]
- 192.Hunter N, Chambers S R, Louis E J, Borts R H. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 1996;15:1726–1733. [PMC free article] [PubMed] [Google Scholar]
- 193.Hurst D D, Fogel S, Mortimer R K. Conversion-associated recombination in yeast (hybrids-meiosis-tetrads-marker loci-models) Proc Natl Acad Sci USA. 1972;69:101–105. doi: 10.1073/pnas.69.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ionov Y, Peinado M A, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 195.Ivanov E L, Haber J E. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:2245–2251. doi: 10.1128/mcb.15.4.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ivanov E L, Korolev V G, Fabre F. XRS2, a DNA repair gene of Saccharomyces cerevisiae, is needed for meiotic recombination. Genetics. 1992;132:651–664. doi: 10.1093/genetics/132.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ivanov E L, Sugawara N, Fishman L J, Haber J E. Genetic requirements for the single-strand annealing pathway of double- strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ivanov E L, Sugawara N, White C I, Fabre F, Haber J E. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jackson J A, Fink G R. Gene conversion between duplicated genetic elements in yeast. Nature. 1981;292:306–311. doi: 10.1038/292306a0. [DOI] [PubMed] [Google Scholar]
- 200.Jeffreys A J. DNA typing: approaches and applications. J Forensic Sci Soc. 1993;33:204–211. doi: 10.1016/s0015-7368(93)73016-9. [DOI] [PubMed] [Google Scholar]
- 201.Jeffreys A J, Murray J, Neumann R. High-resolution mapping of crossovers in human sperm defines a minisatellite-associated recombination hotspot. Mol Cell. 1998;2:267–273. doi: 10.1016/s1097-2765(00)80138-0. [DOI] [PubMed] [Google Scholar]
- 202.Jeffreys A J, Tamaki K, MacLeod A, Monckton D G, Neil D L, Armour J A. Complex gene conversion events in germline mutation at human minisatellites. Nat Genet. 1994;6:136–145. doi: 10.1038/ng0294-136. [DOI] [PubMed] [Google Scholar]
- 203.Jensen R E, Herskowitz I. Directionality and regulation of cassette substitution in yeast. Cold Spring Harbor Symp Quant Biol. 1984;49:97–104. doi: 10.1101/sqb.1984.049.01.013. [DOI] [PubMed] [Google Scholar]
- 204.Jiang H, Xie Y, Houston P, Stemke-Hale K, Mortensen U H, Rothstein R, Kodadek T. Direct association between the yeast Rad51 and Rad54 recombination proteins. J Biol Chem. 1996;271:33181–33186. doi: 10.1074/jbc.271.52.33181. [DOI] [PubMed] [Google Scholar]
- 205.Jin Q, Trelles-Sticken E, Scherthan H, Loidl J. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J Cell Biol. 1998;141:21–29. doi: 10.1083/jcb.141.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Jinks-Robertson S, Petes T D. Chromosomal translocations generated by high-frequency meiotic recombination between repeated yeast genes. Genetics. 1986;114:731–752. doi: 10.1093/genetics/114.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Jinks-Robertson S, Petes T D. High-frequency meiotic gene conversion between repeated genes on nonhomologous chromosomes in yeast. Proc Natl Acad Sci USA. 1985;82:3350–3354. doi: 10.1073/pnas.82.10.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Johnson R D, Symington L S. Functional differences and interactions among the putative RecA homologs Rad51, Rad55, and Rad57. Mol Cell Biol. 1995;15:4843–4850. doi: 10.1128/mcb.15.9.4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Johnson R E, Henderson S T, Petes T D, Prakash S, Bankmann M, Prakash L. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol Cell Biol. 1992;12:3807–3818. doi: 10.1128/mcb.12.9.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Johnson R E, Kovvali G K, Prakash L, Prakash S. Requirement of the yeast MSH3 and MSH6 genes for MSH2-dependent genomic stability. J Biol Chem. 1996;271:7285–7288. doi: 10.1074/jbc.271.13.7285. [DOI] [PubMed] [Google Scholar]
- 211.Johnston L H. The DNA repair capability of cdc9, the Saccharomyces cerevisiae mutant defective in DNA ligase. Mol Gen Genet. 1979;170:89–92. doi: 10.1007/BF00268583. [DOI] [PubMed] [Google Scholar]
- 212.Johnston L H, Nasmyth K A. Saccharomyces cerevisiae cell cycle mutant cdc9 is defective in DNA ligase. Nature. 1978;274:891–893. doi: 10.1038/274891a0. [DOI] [PubMed] [Google Scholar]
- 213.Johnston, M. Personal communication.
- 214.Johzuka K, Ogawa H. Interaction of Mre11 and Rad50: two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics. 1995;139:1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Jones G H. The control of chiasma distribution. Symp Soc Exp Biol. 1984;38:293–320. [PubMed] [Google Scholar]
- 216.Judd S R, Petes T D. Physical lengths of meiotic and mitotic gene conversion tracts in Saccharomyces cerevisiae. Genetics. 1988;118:401–410. doi: 10.1093/genetics/118.3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Kaback D B. Chromosome-size dependent control of meiotic recombination in humans. Nat Genet. 1996;13:20–21. doi: 10.1038/ng0596-20. [DOI] [PubMed] [Google Scholar]
- 218.Kaback D B, Guacci V, Barber D, Mahon J W. Chromosome size-dependent control of meiotic recombination. Science. 1992;256:228–232. doi: 10.1126/science.1566070. [DOI] [PubMed] [Google Scholar]
- 219.Kaback D B, Steensma H Y, de Jonge P. Enhanced meiotic recombination on the smallest chromosome of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1989;86:3694–3698. doi: 10.1073/pnas.86.10.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Kanaar R, Hoeijmakers J H. Recombination and joining: different means to the same ends. Genes Funct. 1997;1:165–174. doi: 10.1046/j.1365-4624.1997.00016.x. [DOI] [PubMed] [Google Scholar]
- 221.Kang S, Jaworski A, Ohshima K, Wells R D. Expansion and deletion of CTG repeats from human disease genes are determined by the direction of replication in E. coli. Nat Genet. 1995;10:213–218. doi: 10.1038/ng0695-213. [DOI] [PubMed] [Google Scholar]
- 222.Kans J A, Mortimer R K. Nucleotide sequence of the RAD57 gene of Saccharomyces cerevisiae. Gene. 1991;105:139–140. doi: 10.1016/0378-1119(91)90527-i. [DOI] [PubMed] [Google Scholar]
- 223.Kato R, Ogawa H. An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1994;22:3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Keeney S, Giroux C N, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 225.Keeney S, Kleckner N. Communication between homologous chromosomes: genetic alterations at a nuclease-hypersensitive site can alter mitotic chromatin structure at that site both in cis and in trans. Genes Cells. 1996;1:475–489. doi: 10.1046/j.1365-2443.1996.d01-257.x. [DOI] [PubMed] [Google Scholar]
- 226.Keeney S, Kleckner N. Covalent protein-DNA complexes at the 5′ strand termini of meiosis-specific double-strand breaks in yeast. Proc Natl Acad Sci USA. 1995;92:11274–11278. doi: 10.1073/pnas.92.24.11274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Keil R L, Roeder G S. Cis-acting, recombination-stimulating activity in a fragment of the ribosomal DNA of S. cerevisiae. Cell. 1984;39:377–386. doi: 10.1016/0092-8674(84)90016-3. [DOI] [PubMed] [Google Scholar]
- 228.Kim R A, Wang J C. A subthreshold level of DNA topoisomerases leads to the excision of yeast rDNA as extrachromosomal rings. Cell. 1989;57:975–985. doi: 10.1016/0092-8674(89)90336-x. [DOI] [PubMed] [Google Scholar]
- 229.King J S, Mortimer R K. A polymerization model of chiasma interference and corresponding computer simulation. Genetics. 1990;126:1127–1138. doi: 10.1093/genetics/126.4.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Kirkpatrick D T, Dominska M, Petes T D. Conversion-type and restoration-type repair of DNA mismatches formed during meiotic recombination in Saccharomyces cerevisiae. Genetics. 1998;149:1693–1705. doi: 10.1093/genetics/149.4.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Kirkpatrick D T, Petes T D. Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature. 1997;387:929–931. doi: 10.1038/43225. [DOI] [PubMed] [Google Scholar]
- 232.Kironmai K M, Muniyappa K. Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells. 1997;2:443–455. doi: 10.1046/j.1365-2443.1997.1330331.x. [DOI] [PubMed] [Google Scholar]
- 233.Kitada K, Johnson A L, Johnston L H, Sugino A. A multicopy suppressor gene of the Saccharomyces cerevisiae G1 cell cycle mutant gene dbf4 encodes a protein kinase and is identified as CDC5. Mol Cell Biol. 1993;13:4445–4457. doi: 10.1128/mcb.13.7.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Klar A J, Strathern J N. Resolution of recombination intermediates generated during yeast mating type switching. Nature. 1984;310:744–788. doi: 10.1038/310744a0. [DOI] [PubMed] [Google Scholar]
- 235.Kleckner N. Meiosis: how could it work? Proc Natl Acad Sci USA. 1996;93:8167–8174. doi: 10.1073/pnas.93.16.8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Klein H L. Different types of recombination events are controlled by the RAD1 and RAD52 genes of Saccharomyces cerevisiae. Genetics. 1988;120:367–377. doi: 10.1093/genetics/120.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Klein H L. Examination of mitotic recombination by means of hyper-recombination mutants in Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol. 1995;51:271–303. doi: 10.1016/s0079-6603(08)60881-8. [DOI] [PubMed] [Google Scholar]
- 238.Klein H L. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics. 1997;147:1533–1543. doi: 10.1093/genetics/147.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Klein H L, Petes T D. Intrachromosomal gene conversion in yeast. Nature. 1981;289:144–148. doi: 10.1038/289144a0. [DOI] [PubMed] [Google Scholar]
- 240.Klein S, Zenvirth D, Dror V, Barton A B, Kaback D B, Simchen G. Patterns of meiotic double-strand breakage on native and artificial yeast chromosomes. Chromosoma. 1996;105:276–284. doi: 10.1007/BF02524645. [DOI] [PubMed] [Google Scholar]
- 241.Kogoma T. Recombination by replication. Cell. 1996;85:625–627. doi: 10.1016/s0092-8674(00)81229-5. [DOI] [PubMed] [Google Scholar]
- 242.Kogoma T. Stable DNA replication: interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev. 1997;61:212–238. doi: 10.1128/mmbr.61.2.212-238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Kohli J, Bahler J. Homologous recombination in fission yeast: absence of crossover interference and synaptonemal complex. Experientia. 1994;50:295–306. doi: 10.1007/BF01924013. [DOI] [PubMed] [Google Scholar]
- 244.Kokoska R J, Stefanovic L, Tran H T, Resnick M A, Gordenin D A, Petes T D. Destabilization of yeast micro- and minisatellite DNA sequences by mutations affecting a nuclease involved in Okazaki fragment processing (rad27) and DNA polymerase delta (pol3-t) Mol Cell Biol. 1998;18:2779–2788. doi: 10.1128/mcb.18.5.2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Kolodkin A L, Klar A J, Stahl F W. Double-strand breaks can initiate meiotic recombination in S. cerevisiae. Cell. 1986;46:733–740. doi: 10.1016/0092-8674(86)90349-1. [DOI] [PubMed] [Google Scholar]
- 246.Kowalczykowski S C, Dixon D A, Eggleston A K, Lauder S D, Rehrauer W M. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Kramer B, Kramer W, Williamson M S, Fogel S. Heteroduplex DNA correction in Saccharomyces cerevisiae is mismatch specific and requires functional PMS genes. Mol Cell Biol. 1989;9:4432–4440. doi: 10.1128/mcb.9.10.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Kramer K M, Brock J A, Bloom K, Moore J K, Haber J E. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol Cell Biol. 1994;14:1293–1301. doi: 10.1128/mcb.14.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Kramer W, Kramer B, Williamson M S, Fogel S. Cloning and nucleotide sequence of DNA mismatch repair gene PMS1 from Saccharomyces cerevisiae: homology of PMS1 to procaryotic MutL and HexB. J Bacteriol. 1989;171:5339–5346. doi: 10.1128/jb.171.10.5339-5346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Kreuzer K N, Saunders M, Weislo L J, Kreuzer H W. Recombination-dependent DNA replication stimulated by double-strand breaks in bacteriophage T4. J Bacteriol. 1995;177:6844–6853. doi: 10.1128/jb.177.23.6844-6853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Kunst C B, Warren S T. Cryptic and polar variation of the fragile X repeat could result in predisposing normal alleles. Cell. 1994;77:853–861. doi: 10.1016/0092-8674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- 252.Kupiec M, Petes T D. Allelic and ectopic recombination between Ty elements in yeast. Genetics. 1988;119:549–559. doi: 10.1093/genetics/119.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Kurkulos M, Weinberg J M, Roy D, Mount S M. P element-mediated in vivo deletion analysis of white-apricot: deletions between direct repeats are strongly favored. Genetics. 1994;136:1001–1011. doi: 10.1093/genetics/136.3.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Kuspa A, Loomis W F. REMI-RFLP mapping in the Dictyostelium genome. Genetics. 1994;138:665–674. doi: 10.1093/genetics/138.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Kuspa A, Loomis W F. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc Natl Acad Sci USA. 1992;89:8803–8807. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Lalioti M D, Scott H S, Buresi C, Rossier C, Bottani A, Morris M A, Malafosse A, Antonarakis S E. Dodecamer repeat expansion in cystatin B gene in progressive myoclonus epilepsy. Nature. 1997;386:847–851. doi: 10.1038/386847a0. [DOI] [PubMed] [Google Scholar]
- 257.Lalioti M D, Scott H S, Genton P, Grid D, Ouazzani R, M’Rabet A, Ibrahim S, Gouider R, Dravet C, Chkili T, Bottani A, Buresi C, Malafosse A, Antonarakis S E. A PCR amplification method reveals instability of the dodecamer repeat in progressive myoclonus epilepsy (EPM1) and no correlation between the size of the repeat and age at onset. Am J Hum Genet. 1998;62:842–847. doi: 10.1086/301798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Lambie E J, Roeder G S. Repression of meiotic crossing over by a centromere (CEN3) in Saccharomyces cerevisiae. Genetics. 1986;114:769–789. doi: 10.1093/genetics/114.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Lambie E J, Roeder G S. A yeast centromere acts in cis to inhibit meiotic gene conversion of adjacent sequences. Cell. 1988;52:863–873. doi: 10.1016/0092-8674(88)90428-x. [DOI] [PubMed] [Google Scholar]
- 260.Le, S., J. K. Moore, J. E. Haber, and C. Greider. Different roles of recombination proteins in the formation of cells able to grow in the absence of telomerase. Genetics, in press.
- 261.Leach F S, Nicolaides N C, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen L A, Nystrom-Lahti M, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 262.Lee S E, Moore J K, Holmes A, Umezu K, Kolodner R, Haber J E. Saccharomyces Ku70, Mre11/Rad50 and RPA proteins regulate adaptation to G2/M arrest DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 263.Lee, S. E., and J. E. Haber. Unpublished results.
- 264.Lehmann A R. Molecular biology of DNA repair in the fission yeast Schizosaccharomyces pombe. Mutat Res. 1996;363:147–161. doi: 10.1016/0921-8777(96)00017-1. [DOI] [PubMed] [Google Scholar]
- 265.Leu J-Y, Chua P R, Roeder G S. The meiosis-specific Hop2 protein of S. cerevisiae ensures synapsis between homologous chromosomes. Cell. 1998;94:375–386. doi: 10.1016/s0092-8674(00)81480-4. [DOI] [PubMed] [Google Scholar]
- 266.Leung W, Malkova A, Haber J E. Gene targeting by linear duplex DNA frequently occurs by assimilation of a single strand that is subject to preferential mismatch correction. Proc Natl Acad Sci USA. 1997;94:6851–6856. doi: 10.1073/pnas.94.13.6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Lewis L K, Kirchner J M, Resnick M A. Requirement for end-joining and checkpoint functions, but not RAD52-mediated recombination, after EcoRI endonuclease cleavage of Saccharomyces cerevisiae DNA. Mol Cell Biol. 1998;18:1891–1902. doi: 10.1128/mcb.18.4.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Lichten M, Borts R H, Haber J E. Meiotic gene conversion and crossing over between dispersed homologous sequences occurs frequently in Saccharomyces cerevisiae. Genetics. 1987;115:233–246. doi: 10.1093/genetics/115.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Lichten M, Goyon C, Schultes N P, Treco D, Szostak J W, Haber J E, Nicolas A. Detection of heteroduplex DNA molecules among the products of Saccharomyces cerevisiae meiosis. Proc Natl Acad Sci USA. 1990;87:7653–7657. doi: 10.1073/pnas.87.19.7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Lichten M, Haber J E. Position effects in ectopic and allelic mitotic recombination in Saccharomyces cerevisiae. Genetics. 1989;123:261–268. doi: 10.1093/genetics/123.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Liefshitz B, Parket A, Maya R, Kupiec M. The role of DNA repair genes in recombination between repeated sequences in yeast. Genetics. 1995;140:1199–1211. doi: 10.1093/genetics/140.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Lim D S, Hasty P. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol Cell Biol. 1996;16:7133–7143. doi: 10.1128/mcb.16.12.7133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Lin F-L, Sperle K, Sternberg N. Recombination in mouse L cells between DNA introduced into cells and homologous chromosomal sequences. Proc Natl Acad Sci USA. 1985;82:1391–1395. doi: 10.1073/pnas.82.5.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Lin F L, Sperle K, Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984;4:1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Lin Y, Smith G R. Transient, meiosis-induced expression of the rec6 and rec12 genes of Schizosaccharomyces pombe. Genetics. 1994;136:769–779. doi: 10.1093/genetics/136.3.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Lin Y H, Keil R L. Mutations affecting RNA polymerase I-stimulated exchange and rDNA recombination in yeast. Genetics. 1991;127:31–38. doi: 10.1093/genetics/127.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Liras P, McCusker J, Mascioli S, Haber J E. Characterization of a mutation in yeast causing nonrandom chromosome loss during mitosis. Genetics. 1978;88:651–671. [PMC free article] [PubMed] [Google Scholar]
- 278.Liu J, Wu T C, Lichten M. The location and structure of double-strand DNA breaks induced during yeast meiosis: evidence for a covalently linked DNA-protein intermediate. EMBO J. 1995;14:4599–4608. doi: 10.1002/j.1460-2075.1995.tb00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Longhese M P, Paciotti V, Fraschini R, Zaccarini R, Plevani P, Lucchini G. The novel DNA damage checkpoint protein ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J. 1997;16:5216–5226. doi: 10.1093/emboj/16.17.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Loomis W F, Welker D, Hughes J, Maghakian D, Kuspa A. Integrated maps of the chromosomes in Dictyostelium discoideum. Genetics. 1995;141:147–157. doi: 10.1093/genetics/141.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Louis, E. Personal communication.
- 282.Louis E J, Haber J E. Evolutionarily recent transfer of a group I mitochondrial intron to telomere regions in Saccharomyces cerevisiae. Curr Genet. 1991;20:411–415. doi: 10.1007/BF00317070. [DOI] [PubMed] [Google Scholar]
- 283.Lovett S T. Sequence of the RAD55 gene of Saccharomyces cerevisiae: similarity of Rad55 to prokaryotic RecA and other RecA-like proteins. Gene. 1994;142:103–106. doi: 10.1016/0378-1119(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 284.Lovett S T, Mortimer R K. Characterization of null mutants of the RAD55 gene of Saccharomyces cerevisiae: effects of temperature, osmotic strength and mating type. Genetics. 1987;116:547–553. doi: 10.1093/genetics/116.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Lundblad, V. Personal communication.
- 286.Lundblad V, Blackburn E H. An alternative pathway for yeast telomere maintenance rescues est1− senescence. Cell. 1993;73:347–360. doi: 10.1016/0092-8674(93)90234-h. [DOI] [PubMed] [Google Scholar]
- 287.Lydall D, Nikolsky Y, Bishop D K, Weinert T. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature. 1996;383:840–843. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- 288.Lydall D, Weinert T. Yeast checkpoint genes in DNA damage processing: implications for repair and arrest. Science. 1995;270:1488–1491. doi: 10.1126/science.270.5241.1488. [DOI] [PubMed] [Google Scholar]
- 289.Ma H, Kunes S, Schatz P J, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- 290.Macreadie I G, Scott R M, Zinn A R, Butow R A. Transposition of an intron in yeast mitochondria requires a protein encoded by that intron. Cell. 1985;41:395–402. doi: 10.1016/s0092-8674(85)80012-x. [DOI] [PubMed] [Google Scholar]
- 291.Malkova A, Ivanov E L, Haber J E. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc Natl Acad Sci USA. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Malkova A, Ross L, Dawson D, Hoekstra M F, Haber J E. Meiotic recombination initiated by a double-strand break in rad50 delta yeast cells otherwise unable to initiate meiotic recombination. Genetics. 1996;143:741–754. doi: 10.1093/genetics/143.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Malone R E, Bullard S, Lundquist S, Kim S, Tarkowski T. A meiotic gene conversion gradient opposite to the direction of transcription. Nature. 1992;359:154–155. doi: 10.1038/359154a0. [DOI] [PubMed] [Google Scholar]
- 294.Malone R E, Esposito R E. The RAD52 gene is required for homothallic interconversion of mating types and spontaneous mitotic recombination in yeast. Proc Natl Acad Sci USA. 1980;77:503–507. doi: 10.1073/pnas.77.1.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Malone R E, Esposito R E. Recombinationless meiosis in Saccharomyces cerevisiae. Mol Cell Biol. 1981;1:891–901. doi: 10.1128/mcb.1.10.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Malter H E, Iber J C, Willemsen R, de Graaff E, Tarleton J C, Leisti J, Warren S T, Oostra B A. Characterization of the full fragile X syndrome mutation in fetal gametes. Nat Genet. 1997;15:165–169. doi: 10.1038/ng0297-165. [DOI] [PubMed] [Google Scholar]
- 297.Mao-Draayer Y, Galbraith A M, Pittman D L, Cool M, Malone R E. Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. Genetics. 1996;144:71–86. doi: 10.1093/genetics/144.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Marsischky G T, Filosi N, Kane M F, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 299.Matic I, Rayssiguier C, Radman M. Interspecies gene exchange in bacteria: the role of SOS and mismatch repair systems in evolution of species. Cell. 1995;80:507–515. doi: 10.1016/0092-8674(95)90501-4. [DOI] [PubMed] [Google Scholar]
- 300.Maurer D J, O’Callaghan B L, Livingston D M. Mapping the polarity of changes that occur in interrupted CAG repeat tracts in yeast. Mol Cell Biol. 1998;18:4597–4604. doi: 10.1128/mcb.18.8.4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Maurer D J, O’Callaghan B L, Livingston D M. Orientation dependence of trinucleotide CAG repeat instability in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:6617–6622. doi: 10.1128/mcb.16.12.6617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.May C A, Jeffreys A J, Armour J A. Mutation rate heterogeneity and the generation of allele diversity at the human minisatellite MS205 (D16S309) Hum Mol Genet. 1996;5:1823–1833. doi: 10.1093/hmg/5.11.1823. [DOI] [PubMed] [Google Scholar]
- 303.McDonald J P, Rothstein R. Unrepaired heteroduplex DNA in Saccharomyces cerevisiae is decreased in RAD1 RAD52-independent recombination. Genetics. 1994;137:393–405. doi: 10.1093/genetics/137.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.McGill C B, Holbeck S L, Strathern J N. The chromosome bias of misincorporations during double-strand break-repair is not altered in mismatch repair-defective strains of Saccharomyces cerevisiae. Genetics. 1998;148:1525–1533. doi: 10.1093/genetics/148.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.McGill C, Shafer B, Strathern J. Coconversion of flanking sequences with homothallic switching. Cell. 1989;57:459–467. doi: 10.1016/0092-8674(89)90921-5. [DOI] [PubMed] [Google Scholar]
- 306.McKee A H, Kleckner N. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics. 1997;146:797–816. doi: 10.1093/genetics/146.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.McKee A H, Kleckner N. Mutations in Saccharomyces cerevisiae that block meiotic prophase chromosome metabolism and confer cell cycle arrest at pachytene identify two new meiosis-specific genes SAE1 and SAE3. Genetics. 1997;146:817–834. doi: 10.1093/genetics/146.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.McKim, K. Personal communication.
- 309.McKim K S, Green-Marroquin B L, Sekelsky J J, Chin G, Steinberg C, Khodosh R, Hawley R S. Meiotic synapsis in the absence of recombination. Science. 1998;279:876–878. doi: 10.1126/science.279.5352.876. [DOI] [PubMed] [Google Scholar]
- 310.Menees T M, Roeder G S. MEI4, a yeast gene required for meiotic recombination. Genetics. 1989;123:675–682. doi: 10.1093/genetics/123.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Menees T M, Ross-MacDonald P B, Roeder G S. MEI4, a meiosis-specific yeast gene required for chromosome synapsis. Mol Cell Biol. 1992;12:1340–1351. doi: 10.1128/mcb.12.3.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Meselson M M, Radding C M. A general model for genetic recombination. Proc Natl Acad Sci USA. 1975;72:358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Mezard C, Nicolas A. Homologous, homeologous, and illegitimate repair of double-strand breaks during transformation of a wild-type strain and a rad52 mutant strain of Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:1278–1292. doi: 10.1128/mcb.14.2.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Milne G T, Jin S, Shannon K B, Weaver D T. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4189–4198. doi: 10.1128/mcb.16.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Milne G T, Weaver D T. Dominant negative alleles of RAD52 reveal a DNA repair/recombination complex including Rad51 and Rad52. Genes Dev. 1993;7:1755–1765. doi: 10.1101/gad.7.9.1755. [DOI] [PubMed] [Google Scholar]
- 316.Miret J J, Pessoa-Brandao L, Lahue R S. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:12438–12443. doi: 10.1073/pnas.95.21.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Mitas M, Yu A, Dill J, Haworth I S. The trinucleotide repeat sequence d(CGG)15 forms a heat-stable hairpin containing Gsyn.Ganti base pairs. Biochemistry. 1995;34:12803–12811. doi: 10.1021/bi00039a041. [DOI] [PubMed] [Google Scholar]
- 318.Mitas M, Yu A, Dill J, Kamp T J, Chambers E J, Haworth I S. Hairpin properties of single-stranded DNA containing a GC-rich triplet repeat: (CTG)15. Nucleic Acids Res. 1995;23:1050–1059. doi: 10.1093/nar/23.6.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Modesti, M., and C. N. Giroux. Personal communication.
- 320.Modrich P. Mechanisms and biological effects of mismatch repair. Annu Rev Genet. 1991;25:229–253. doi: 10.1146/annurev.ge.25.120191.001305. [DOI] [PubMed] [Google Scholar]
- 321.Moore, J. K., and J. E. Haber. Unpublished results.
- 322.Moore J K, Haber J E. Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature. 1996;383:644–646. doi: 10.1038/383644a0. [DOI] [PubMed] [Google Scholar]
- 323.Moore J K, Haber J E. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Moore, J. K., L. Sapoghnikova, and J. E. Haber. Unpublished results.
- 325.Moreau S, Ferguson H R, Symington L S. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Morrow D M, Connelly C, Hieter P. “Break copy” duplication: a model for chromosome fragment formation in Saccharomyces cerevisiae. Genetics. 1997;147:371–382. doi: 10.1093/genetics/147.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Mortensen U H, Bendixen C, Sunjevaric I, Rothstein R. DNA strand annealing is promoted by the yeast Rad52 protein. Proc Natl Acad Sci USA. 1996;93:10729–10734. doi: 10.1073/pnas.93.20.10729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Mortimer R K, Contopoulou R, Schild D. Mitotic chromosome loss in a radiation-sensitive strain of the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1981;78:5778–5782. doi: 10.1073/pnas.78.9.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Mortimer R K, Fogel S. Genetical interference and gene conversion. In: Grell R F, editor. Mechanisms in recombination. New York, N.Y: Plenum Press; 1969. pp. 263–275. [Google Scholar]
- 330.Mosig G. The essential role of recombination in phage T4 growth. Annu Rev Genet. 1987;21:347–371. doi: 10.1146/annurev.ge.21.120187.002023. [DOI] [PubMed] [Google Scholar]
- 331.Mueller J E, Clyman J, Huang Y J, Parker M M, Belfort M. Intron mobility in phage T4 occurs in the context of recombination-dependent DNA replication by way of multiple pathways. Genes Dev. 1996;10:351–364. doi: 10.1101/gad.10.3.351. [DOI] [PubMed] [Google Scholar]
- 332.Nag D K, Kurst A. A 140-bp-long palindromic sequence induces double-strand breaks during meiosis in the yeast Saccharomyces cerevisiae. Genetics. 1997;146:835–847. doi: 10.1093/genetics/146.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Nag D K, Petes T D. Physical detection of heteroduplexes during meiotic recombination in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2324–2331. doi: 10.1128/mcb.13.4.2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Nag D K, White M A, Petes T D. Palindromic sequences in heteroduplex DNA inhibit mismatch repair in yeast. Nature. 1989;340:318–320. doi: 10.1038/340318a0. [DOI] [PubMed] [Google Scholar]
- 335.Nairz K, Klein F. mre11S, a yeast mutation that blocks double-strand-break processing and permits nonhomologous synapsis in meiosis. Genes Dev. 1997;11:2272–2290. doi: 10.1101/gad.11.17.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Nakagawa T, Ogawa H. Involvement of the MRE2 gene of yeast in formation of meiosis-specific double-strand breaks and crossover recombination through RNA splicing. Genes Cells. 1997;2:65–79. doi: 10.1046/j.1365-2443.1997.d01-283.x. [DOI] [PubMed] [Google Scholar]
- 337.Nakamura T M, Cooper J P, Cech T R. Two modes of survival of fission yeast without telomerase. Science. 1998;282:493–496. doi: 10.1126/science.282.5388.493. [DOI] [PubMed] [Google Scholar]
- 338.Namsaraev E, Berg P. Characterization of strand exchange activity of yeast Rad51 protein. Mol Cell Biol. 1997;17:5359–5368. doi: 10.1128/mcb.17.9.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Nasmyth K A. Molecular genetics of yeast mating type. Annu Rev Genet. 1982;16:439–500. doi: 10.1146/annurev.ge.16.120182.002255. [DOI] [PubMed] [Google Scholar]
- 340.Nassif N, Penney J, Pal S, Engels W R, Gloor G B. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Negritto M T, Wu X, Kuo T, Chu S, Bailis A M. Influence of DNA sequence identity on efficiency of targeted gene replacement. Mol Cell Biol. 1997;17:278–286. doi: 10.1128/mcb.17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Nelson H H, Sweetser D B, Nickoloff J A. Effects of terminal nonhomology and homeology on double-strand-break-induced gene conversion tract directionality. Mol Cell Biol. 1996;16:2951–2957. doi: 10.1128/mcb.16.6.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342a.New J H, Sugiyama T, Zaitseva E, Kowalczykowski S C. Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature. 1998;391:407–410. doi: 10.1038/34950. [DOI] [PubMed] [Google Scholar]
- 343.New L, Liu K, Crouse G F. The yeast gene MSH3 defines a new class of eukaryotic MutS homologues. Mol Gen Genet. 1993;239:97–108. doi: 10.1007/BF00281607. [DOI] [PubMed] [Google Scholar]
- 344.Nickoloff J A, Chen E Y, Heffron F. A 24-base-pair DNA sequence from the MAT locus stimulates intergenic recombination in yeast. Proc Natl Acad Sci USA. 1986;83:7831–7835. doi: 10.1073/pnas.83.20.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Nickoloff J A, Singer J D, Heffron F. In vivo analysis of the Saccharomyces cerevisiae HO nuclease recognition site by site-directed mutagenesis. Mol Cell Biol. 1990;10:1174–1179. doi: 10.1128/mcb.10.3.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Nickoloff J A, Singer J D, Hoekstra M F, Heffron F. Double-strand breaks stimulate alternative mechanisms of recombination repair. J Mol Biol. 1989;207:527–541. doi: 10.1016/0022-2836(89)90462-2. [DOI] [PubMed] [Google Scholar]
- 347.Nickoloff J A, Hoekstra M F. Double-strand break and recombinational repair in Saccharomyces cerevisiae. In: Nickoloff J A, Hoekstra M F, editors. DNA damage and repair. 1. DNA repair in prokaryotes and lower eukaryotes. Totowa, N.J: Humana Press Inc.; 1998. pp. 335–362. [Google Scholar]
- 348.Nicolas A, Treco D, Schultes N P, Szostak J W. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature. 1989;338:35–39. doi: 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]
- 349.Nishinaka T, Ito Y, Yokoyama S, Shibata T. An extended DNA structure through deoxyribose-base stacking induced by RecA protein. Proc Natl Acad Sci USA. 1997;94:6623–6228. doi: 10.1073/pnas.94.13.6623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Nishinaka T, Shinohara A, Ito Y, Yokoyama S, Shibata T. Base pair switching by interconversion of sugar puckers in DNA extended by proteins of RecA-family: a model for homology search in homologous genetic recombination. Proc Natl Acad Sci USA. 1998;95:11071–11076. doi: 10.1073/pnas.95.19.11071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Nugent C I, Bosco G, Ross L O, Evans S K, Salinger A P, Moore J K, Haber J E, Lundblad V. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- 352.Ogawa, T. Personal communication.
- 353.Ogawa T, Yu X, Shinohara A, Egelman E H. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- 354.O’Hoy K L, Tsilfidis C, Mahadevan M S, Neville C E, Barcelo J, Hunter A G, Korneluk R G. Reduction in size of the myotonic dystrophy trinucleotide repeat mutation during transmission. Science. 1993;259:809–812. doi: 10.1126/science.8094260. [DOI] [PubMed] [Google Scholar]
- 355.Ohta K, Nicolas A, Furuse M, Nabetani A, Ogawa H, Shibata T. Mutations in the MRE11, RAD50, XRS2, and MRE2 genes alter chromatin configuration at meiotic DNA double-stranded break sites in premeiotic and meiotic cells. Proc Natl Acad Sci USA. 1998;95:646–651. doi: 10.1073/pnas.95.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Ohta K, Shibata T, Nicolas A. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 1994;13:5754–5763. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Orr-Weaver T L, Szostak J W. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc Natl Acad Sci USA. 1983;80:4417–4421. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Orr-Weaver T L, Szostak J W, Rothstein R J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci USA. 1981;78:6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Osman F, Subramani S. Double-strand break-induced recombination in eukaryotes. Prog Nucleic Acid Res Mol Biol. 1998;58:263–299. doi: 10.1016/s0079-6603(08)60039-2. [DOI] [PubMed] [Google Scholar]
- 360.Ozenberger B A, Roeder G S. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol Cell Biol. 1991;11:1222–1231. doi: 10.1128/mcb.11.3.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Paciotti V, Lucchini G, Plevani P, Longhese M P. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 1998;17:4199–4209. doi: 10.1093/emboj/17.14.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Padmore R, Cao L, Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- 363.Palladino F, Klein H L. Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics. 1992;132:23–37. doi: 10.1093/genetics/132.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Pâques F, Bucheton B, Wegnez M. Rearrangements involving repeated sequences within a P element preferentially occur between units close to the transposon extremities. Genetics. 1996;142:459–470. doi: 10.1093/genetics/142.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Pâques, F., and J. E. Haber. Unpublished results.
- 366.Pâques F, Haber J E. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Pâques, F., W.-Y. Leung, J. Sylvan, and J. E. Haber. Unpublished results.
- 368.Pâques F, Leung W Y, Haber J E. Expansions and contractions in a tandem repeat induced by double-strand break repair. Mol Cell Biol. 1998;18:2045–2054. doi: 10.1128/mcb.18.4.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Pâques F, Wegnez M. Deletions and amplifications of tandemly arranged ribosomal 5S genes internal to a P element occur at a high rate in a dysgenic context. Genetics. 1993;135:469–476. doi: 10.1093/genetics/135.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Parker A E, Van de Weyer I, Laus M C, Oostveen I, Yon J, Verhasselt P, Luyten W H. A human homologue of the Schizosaccharomyces pombe rad1+ checkpoint gene encodes an exonuclease. J Biol Chem. 1998;273:18332–18339. doi: 10.1074/jbc.273.29.18332. [DOI] [PubMed] [Google Scholar]
- 371.Parket A, Inbar O, Kupiec M. Recombination of Ty elements in yeast can be induced by a double-strand break. Genetics. 1995;140:67–77. doi: 10.1093/genetics/140.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Parket A, Kupiec M. Ectopic recombination between Ty elements in Saccharomyces cerevisiae is not induced by DNA damage. Mol Cell Biol. 1992;12:4441–4448. doi: 10.1128/mcb.12.10.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Parsons R, Li G M, Longley M J, Fang W H, Papadopoulos N, Jen J, de la Chapelle A, Kinzler K W, Vogelstein B, Modrich P. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- 374.Paull T T, Gellert M. The 3′ to 5′ exonuclease activity of Mre11 facilitates repair of DNA double-strands breaks. Mol Cell. 1998;1:969–980. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- 375.Petes T D. Unequal meiotic recombination within tandem arrays of yeast ribosomal DNA genes. Cell. 1980;19:765–774. doi: 10.1016/s0092-8674(80)80052-3. [DOI] [PubMed] [Google Scholar]
- 376.Petrini, J. H. Personal communication.
- 377.Petrini J H, Bressan D A, Yao M S. The RAD52 epistasis group in mammalian double strand break repair. Semin Immunol. 1997;9:181–188. doi: 10.1006/smim.1997.0067. [DOI] [PubMed] [Google Scholar]
- 378.Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 379.Pittman D L, Cobb J, Schimenti K J, Wilson L A, Cooper D M, Brignull E, Handel M A, Schimenti J C. Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline-specific RecA homolog. Mol Cell. 1998;1:697–705. doi: 10.1016/s1097-2765(00)80069-6. [DOI] [PubMed] [Google Scholar]
- 380.Plessis A, Dujon B. Multiple tandem integrations of transforming DNA sequences in yeast chromosomes suggest a mechanism for integrative transformation by homologous recombination. Gene. 1993;134:41–50. doi: 10.1016/0378-1119(93)90172-y. [DOI] [PubMed] [Google Scholar]
- 381.Plessis A, Perrin A, Haber J E, Dujon B. Site-specific recombination determined by I-SceI, a mitochondrial group I intron-encoded endonuclease expressed in the yeast nucleus. Genetics. 1992;130:451–460. doi: 10.1093/genetics/130.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Pochart P, Woltering D, Hollingsworth N M. Conserved properties between functionally distinct MutS homologs in yeast. J Biol Chem. 1997;272:30345–30349. doi: 10.1074/jbc.272.48.30345. [DOI] [PubMed] [Google Scholar]
- 383.Porter G, Westmoreland J, Priebe S, Resnick M A. Homologous and homeologous intermolecular gene conversion are not differentially affected by mutations in the DNA damage or the mismatch repair genes RAD1, RAD50, RAD51, RAD52, RAD54, PMS1 and MSH2. Genetics. 1996;143:755–767. doi: 10.1093/genetics/143.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Porter S E, White M A, Petes T D. Genetic evidence that the meiotic recombination hotspot at the HIS4 locus of Saccharomyces cerevisiae does not represent a site for a symmetrically processed double-strand break. Genetics. 1993;134:5–19. doi: 10.1093/genetics/134.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Prado F, Piruat J I, Aguilera A. Recombination between DNA repeats in yeast hpr1 delta cells is linked to transcription elongation. EMBO J. 1997;16:2826–2835. doi: 10.1093/emboj/16.10.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Priebe S D, Westmoreland J, Nilsson T T, Resnick M A. Induction of recombination between homologous and diverged DNAs by double-strand gaps and breaks and role of mismatch repair. Mol Cell Biol. 1994;14:4802–4814. doi: 10.1128/mcb.14.7.4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Prinz S, Amon A, Klein F. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics. 1997;146:781–795. doi: 10.1093/genetics/146.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Prolla T A, Christie D M, Liskay R M. Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol Cell Biol. 1994;14:407–415. doi: 10.1128/mcb.14.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Prolla T A, Pang Q, Alani E, Kolodner R D, Liskay R M. MLH1, PMS1, and MSH2 interactions during the initiation of DNA mismatch repair in yeast. Science. 1994;265:1091–1093. doi: 10.1126/science.8066446. [DOI] [PubMed] [Google Scholar]
- 390.Ramotar D, Masson J Y. Saccharomyces cerevisiae DNA repair processes: an update. Mol Cell Biochem. 1996;158:65–75. doi: 10.1007/BF00225884. [DOI] [PubMed] [Google Scholar]
- 391.Rattray A J, Symington L S. Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:45–56. doi: 10.1093/genetics/139.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Rattray A J, Symington L S. Use of a chromosomal inverted repeat to demonstrate that the RAD51 and RAD52 genes of Saccharomyces cerevisiae have different roles in mitotic recombination. Genetics. 1994;138:587–595. doi: 10.1093/genetics/138.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Raveh D, Hughes S H, Shafer B K, Strathern J N. Analysis of the HO-cleaved MAT DNA intermediate generated during the mating type switch in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1989;220:33–42. [PubMed] [Google Scholar]
- 394.Ray A, Machin N, Stahl F W. A DNA double chain break stimulates triparental recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1989;86:6225–6229. doi: 10.1073/pnas.86.16.6225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Ray A, Siddiqi I, Kolodkin A L, Stahl F W. Intra-chromosomal gene conversion induced by a DNA double-strand break in Saccharomyces cerevisiae. J Mol Biol. 1988;201:247–260. doi: 10.1016/0022-2836(88)90136-2. [DOI] [PubMed] [Google Scholar]
- 396.Ray, B., and J. E. Haber. Unpublished results.
- 397.Ray B L, White C I, Haber J E. Heteroduplex formation and mismatch repair of the “stuck” mutation during mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5372–5380. doi: 10.1128/mcb.11.10.5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Rayssiguier C, Thaler D S, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- 399.Reagan M S, Pittenger C, Siede W, Friedberg E C. Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J Bacteriol. 1995;177:364–371. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Reenan R A, Kolodner R D. Characterization of insertion mutations in the Saccharomyces cerevisiae MSH1 and MSH2 genes: evidence for separate mitochondrial and nuclear functions. Genetics. 1992;132:975–985. doi: 10.1093/genetics/132.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Resnick M A, Martin P. The repair of double-stranded breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol Gen Genet. 1976;143:119–145. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- 402.Richard, G.-F., and J. E. Haber. Unpublished results.
- 403.Richard M, Belmaaza A, Gusew N, Wallenburg J C, Chartrand P. Integration of a vector containing a repetitive LINE-1 element in the human genome. Mol Cell Biol. 1994;14:6689–6695. doi: 10.1128/mcb.14.10.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Richards R I, Sutherland G R. Dynamic mutation: possible mechanisms and significance in human disease. Trends Biochem Sci. 1997;22:432–436. doi: 10.1016/s0968-0004(97)01108-0. [DOI] [PubMed] [Google Scholar]
- 405.Richards R I, Sutherland G R. Dynamic mutations: a new class of mutations causing human disease. Cell. 1992;70:709–712. doi: 10.1016/0092-8674(92)90302-s. [DOI] [PubMed] [Google Scholar]
- 406.Rijkers T, Van Den Ouweland J, Morolli B, Rolink A G, Baarends W M, Van Sloun P P H, Lohman P H M, Pastink A. Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol Cell Biol. 1998;18:6423–6429. doi: 10.1128/mcb.18.11.6423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Rine, J. Personal communication.
- 408.Rocco V, Nicolas A. Sensing of DNA non-homology lowers the initiation of meiotic recombination in yeast. Genes Cells. 1996;1:645–661. doi: 10.1046/j.1365-2443.1996.00256.x. [DOI] [PubMed] [Google Scholar]
- 409.Rockmill B, Engebrecht J A, Scherthan H, Loidl J, Roeder G S. The yeast MER2 gene is required for chromosome synapsis and the initiation of meiotic recombination. Genetics. 1995;141:49–59. doi: 10.1093/genetics/141.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Rockmill B, Roeder G S. Telomere-mediated chromosome pairing during meiosis in budding yeast. Genes Dev. 1998;12:2574–2586. doi: 10.1101/gad.12.16.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Rockmill B, Roeder G S. The yeast med1 mutant undergoes both meiotic homolog nondisjunction and precocious separation of sister chromatids. Genetics. 1994;136:65–74. doi: 10.1093/genetics/136.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Rockmill B, Sym M, Scherthan H, Roeder G S. Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev. 1995;9:2684–2695. doi: 10.1101/gad.9.21.2684. [DOI] [PubMed] [Google Scholar]
- 413.Roeder G S. Meiotic chromosomes: it takes two to tango. Genes Dev. 1997;11:2600–2621. doi: 10.1101/gad.11.20.2600. [DOI] [PubMed] [Google Scholar]
- 414.Ronne H, Rothstein R. Mitotic sectored colonies: evidence of heteroduplex DNA formation during direct repeat recombination. Proc Natl Acad Sci USA. 1988;85:2696–2700. doi: 10.1073/pnas.85.8.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Rose D, Thomas W, Holm C. Segregation of recombined chromosomes in meiosis I requires DNA topoisomerase II. Cell. 1990;60:1009–1017. doi: 10.1016/0092-8674(90)90349-j. [DOI] [PubMed] [Google Scholar]
- 416.Ross M P, Roeder G S. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell. 1994;79:1069–1080. doi: 10.1016/0092-8674(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 417.Rossignol J L, Faugeron G. MIP: an epigenetic gene silencing process in Ascobolus immersus. Curr Top Microbiol Immunol. 1995;197:179–191. doi: 10.1007/978-3-642-79145-1_12. [DOI] [PubMed] [Google Scholar]
- 418.Rossignol J L, Nicolas A, Hamza H, Langin T. Origins of gene conversion and reciprocal exchange in Ascobolus. Cold Spring Harbor Symp Quant Biol. 1984;49:13–21. doi: 10.1101/sqb.1984.049.01.004. [DOI] [PubMed] [Google Scholar]
- 419.Rossignol J L, Paquette N, Nicolas A. Aberrant 4:4 asci, disparity in the direction of conversion, and frequencies of conversion in Ascobolus immersus. Cold Spring Harbor Symp Quant Biol. 1979;43:1343–1352. doi: 10.1101/sqb.1979.043.01.153. [DOI] [PubMed] [Google Scholar]
- 420.Roth D B, Wilson J H. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol. 1986;6:4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Rothstein R J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 422.Rudin N, Haber J E. Efficient repair of HO-induced chromosomal breaks in Saccharomyces cerevisiae by recombination between flanking homologous sequences. Mol Cell Biol. 1988;8:3918–3928. doi: 10.1128/mcb.8.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Rudin N, Sugarman E, Haber J E. Genetic and physical analysis of double-strand break repair and recombination in Saccharomyces cerevisiae. Genetics. 1989;122:519–534. doi: 10.1093/genetics/122.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Sanchez Y, Desany B A, Jones W J, Liu Q, Wang B, Elledge S J. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 425.Sandell L L, Zakian V A. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- 426.Sargent R G, Rolig R L, Kilburn A E, Adair G M, Wilson J H, Nairn R S. Recombination-dependent deletion formation in mammalian cells deficient in the nucleotide excision repair gene ERCC1. Proc Natl Acad Sci USA. 1997;94:13122–13127. doi: 10.1073/pnas.94.24.13122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Schar P, Herrmann G, Daly G, Lindahl T. A newly identified DNA ligase of Saccharomyces cerevisiae involved in RAD52-independent repair of DNA double-strand breaks. Genes Dev. 1997;11:1912–1924. doi: 10.1101/gad.11.15.1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Schiestl, R. Personal communication.
- 429.Schiestl R H, Dominska M, Petes T D. Transformation of Saccharomyces cerevisiae with nonhomologous DNA: illegitimate integration of transforming DNA into yeast chromosomes and in vivo ligation of transforming DNA to mitochondrial DNA sequences. Mol Cell Biol. 1993;13:2697–2705. doi: 10.1128/mcb.13.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Schiestl R H, Petes T D. Integration of DNA fragments by illegitimate recombination in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1991;88:7585–7589. doi: 10.1073/pnas.88.17.7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Schiestl R H, Reynolds P, Prakash S, Prakash L. Cloning and sequence analysis of the Saccharomyces cerevisiae RAD9 gene and further evidence that its product is required for cell cycle arrest induced by DNA damage. Mol Cell Biol. 1989;9:1882–1896. doi: 10.1128/mcb.9.5.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Schiestl R H, Zhu J, Petes T D. Effect of mutations in genes affecting homologous recombination on restriction enzyme-mediated and illegitimate recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:4493–4500. doi: 10.1128/mcb.14.7.4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Schild D. Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics. 1995;140:115–127. doi: 10.1093/genetics/140.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Schild D, Glassner B J, Mortimer R K, Carlson M, Laurent B C. Identification of RAD16, a yeast excision repair gene homologous to the recombinational repair gene RAD54 and to the SNF2 gene involved in transcriptional activation. Yeast. 1992;8:385–395. doi: 10.1002/yea.320080506. [DOI] [PubMed] [Google Scholar]
- 435.Schultes N P, Szostak J W. Decreasing gradients of gene conversion on both sides of the initiation site for meiotic recombination at the ARG4 locus in yeast. Genetics. 1990;126:813–822. doi: 10.1093/genetics/126.4.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Schultz M C, Brill S J, Ju Q, Sternglanz R, Reeder R H. Topoisomerases and yeast rRNA transcription: negative supercoiling stimulates initiation and topoisomerase activity is required for elongation. Genes Dev. 1992;6:1332–1341. doi: 10.1101/gad.6.7.1332. [DOI] [PubMed] [Google Scholar]
- 437.Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- 438.Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- 439.Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- 440.Schweitzer J K, Livingston D M. Expansions of CAG repeat tracts are frequent in a yeast mutant defective in Okazaki fragment maturation. Hum Mol Genet. 1998;7:69–74. doi: 10.1093/hmg/7.1.69. [DOI] [PubMed] [Google Scholar]
- 441.Selker E U. Epigenetic phenomena in filamentous fungi: useful paradigms or repeat-induced confusion? Trends Genet. 1997;13:296–301. doi: 10.1016/s0168-9525(97)01201-8. [DOI] [PubMed] [Google Scholar]
- 442.Selva E M, New L, Crouse G F, Lahue R S. Mismatch correction acts as a barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:1175–1188. doi: 10.1093/genetics/139.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Shan Q, Bork J M, Webb B L, Inman R B, Cox M M. RecA protein filaments: end-dependent dissociation from ssDNA and stabilization by RecO and RecR proteins. J Mol Biol. 1997;265:519–540. doi: 10.1006/jmbi.1996.0748. [DOI] [PubMed] [Google Scholar]
- 444.Sharples G J, Leach D R. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the Rad50 and Mre11 (Rad32) recombination and repair proteins of yeast. Mol Microbiol. 1995;17:1215–1217. doi: 10.1111/j.1365-2958.1995.mmi_17061215_1.x. [DOI] [PubMed] [Google Scholar]
- 445.Shen P, Huang H V. Effect of base mismatches on recombination via the RecBCD pathway. Mol Gen Genet. 1989;218:359–360. doi: 10.1007/BF00331291. [DOI] [PubMed] [Google Scholar]
- 446.Shen Z, Cloud K G, Chen D J, Park M S. Specific interactions between the human RAD51 and RAD52 proteins. J Biol Chem. 1996;271:148–152. doi: 10.1074/jbc.271.1.148. [DOI] [PubMed] [Google Scholar]
- 447.Sherer S, Davis R W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci USA. 1979;76:4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Sherman F, Roman H. Evidence for two types of allelic recombination in yeast. Genetics. 1963;48:253–271. doi: 10.1093/genetics/48.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 449a.Shinohara A, Ogawa T. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature. 1998;391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 450.Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells. 1998;3:145–156. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- 451.Shinohara M, Shita Y E, Buerstedde J M, Shinagawa H, Ogawa H, Shinohara A. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics. 1997;147:1545–1456. doi: 10.1093/genetics/147.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Shortle D, Haber J E, Botstein D. Lethal disruption of the yeast actin gene by integrative DNA transformation. Science. 1982;217:371–373. doi: 10.1126/science.7046050. [DOI] [PubMed] [Google Scholar]
- 453.Siede W, Friedl A A, Dianova I, Eckardt S F, Friedberg E C. The Saccharomyces cerevisiae Ku autoantigen homologue affects radiosensitivity only in the absence of homologous recombination. Genetics. 1996;142:91–102. doi: 10.1093/genetics/142.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Signon, L., and J. E. Haber. Unpublished results.
- 455.Silberman R, Kupiec M. Plasmid-mediated induction of recombination in yeast. Genetics. 1994;137:414–418. doi: 10.1093/genetics/137.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Sinclair D A, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 457.Sinclair D A, Mills K, Guarente L. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- 458.Singer M S, Gottschling D E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 459.Smith A V, Roeder G S. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J Cell Biol. 1997;136:957–967. doi: 10.1083/jcb.136.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Smith, G. R. Personal communication.
- 461.Smith G R. DNA double-strand break repair and recombination in Escherichia coli. In: Nickoloff J A, Hoekstra M F, editors. DNA damage and repair. 1. DNA repair in prokaryotes and lower eukaryotes. Totowa, N.J: Humana Press Inc.; 1998. pp. 135–162. [Google Scholar]
- 462.Smith K N, Nicolas A. Recombination at work for meiosis. Curr Opin Genet Dev. 1998;8:200–211. doi: 10.1016/s0959-437x(98)80142-1. [DOI] [PubMed] [Google Scholar]
- 463.Sommers C H, Miller E J, Dujon B, Prakash S, Prakash L. Conditional lethality of null mutations in RTH1 that encodes the yeast counterpart of a mammalian 5′- to 3′-exonuclease required for lagging strand DNA synthesis in reconstituted systems. J Biol Chem. 1995;270:4193–4196. doi: 10.1074/jbc.270.9.4193. [DOI] [PubMed] [Google Scholar]
- 464.Sonoda E, Sasaki M S, Buerstedde J M, Bezzubova O, Shinohara A, Ogawa H, Takata M, Yamaguchi-Iwai Y, Takeda S. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Stadler D R. The mechanism of intragenic recombination. Annu Rev Genet. 1973;7:113–127. doi: 10.1146/annurev.ge.07.120173.000553. [DOI] [PubMed] [Google Scholar]
- 466.Stahl F W. The Holliday junction on its thirtieth anniversary. Genetics. 1994;138:241–246. doi: 10.1093/genetics/138.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 467.Storlazzi A, Xu L, Cao L, Kleckner N. Crossover and noncrossover recombination during meiosis: timing and pathway relationships. Proc Natl Acad Sci USA. 1995;92:8512–8516. doi: 10.1073/pnas.92.18.8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 468.Storlazzi A, Xu L, Schwacha A, Kleckner N. Synaptonemal complex (SC) component Zip1 plays a role in meiotic recombination independent of SC polymerization along the chromosomes. Proc Natl Acad Sci USA. 1996;93:9043–9048. doi: 10.1073/pnas.93.17.9043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Straight A F, Murray A W. The spindle assembly checkpoint in budding yeast. Methods Enzymol. 1997;283:425–440. doi: 10.1016/s0076-6879(97)83035-2. [DOI] [PubMed] [Google Scholar]
- 470.Strand M, Prolla T A, Liskay R M, Petes T D. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–276. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- 471.Strathern J N. Control and execution of mating type switching in Saccharomyces cerevisiae. In: Kucherlapati R, Smith G R, editors. Genetic recombination. Washington, D.C: American Society for Microbiology; 1988. pp. 445–464. [Google Scholar]
- 472.Strathern J N, Shafer B K, McGill C B. DNA synthesis errors associated with double-strand-break repair. Genetics. 1995;140:965–972. doi: 10.1093/genetics/140.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Sugawara, N., and J. E. Haber. Unpublished results.
- 474.Sugawara N, Haber J E. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Sugawara N, Ivanov E L, Fishman L J, Ray B L, Wu X, Haber J E. DNA structure-dependent requirements for yeast RAD genes in gene conversion. Nature. 1995;373:84–86. doi: 10.1038/373084a0. [DOI] [PubMed] [Google Scholar]
- 476.Sugawara N, Pâques F, Colaiácovo M, Haber J E. Role of Saccharomyces cerevisiae Msh2 and Msh3 repair proteins in double-strand break-induced recombination. Proc Natl Acad Sci USA. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Sugiyama T, New J H, Kowalczykowski S C. DNA annealing by Rad52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc Natl Acad Sci USA. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Sun H, Treco D, Schultes N P, Szostak J W. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 479.Sun H, Treco D, Szostak J W. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- 480.Sun Z, Fay D S, Marini F, Foiani M, Stern D F. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- 481.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast Rad51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 482.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 483.Sung P, Reynolds P, Prakash L, Prakash S. Purification and characterization of the Saccharomyces cerevisiae Rad1/Rad10 endonuclease. J Biol Chem. 1993;268:26391–26399. [PubMed] [Google Scholar]
- 484.Sung P, Robberson D L. DNA strand exchange mediated by a Rad51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- 485.Sung P, Stratton S A. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J Biol Chem. 1996;271:27983–27986. doi: 10.1074/jbc.271.45.27983. [DOI] [PubMed] [Google Scholar]
- 486.Surosky R T, Tye B K. Construction of telocentric chromosomes in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1985;82:2106–2110. doi: 10.1073/pnas.82.7.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Sweetser D B, Hough H, Whelden J F, Arbuckle M, Nickoloff J A. Fine-resolution mapping of spontaneous and double-strand break-induced gene conversion tracts in Saccharomyces cerevisiae reveals reversible mitotic conversion polarity. Mol Cell Biol. 1994;14:3863–3875. doi: 10.1128/mcb.14.6.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Sym M, Engebrecht J A, Roeder G S. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- 489.Sym M, Roeder G S. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 1994;79:283–292. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- 490.Sym M, Roeder G S. Zip1-induced changes in synaptonemal complex structure and polycomplex assembly. J Cell Biol. 1995;128:455–466. doi: 10.1083/jcb.128.4.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Symington, L. Personal communication.
- 492.Symington L S, Petes T D. Expansions and contractions of the genetic map relative to the physical map of yeast chromosome III. Mol Cell Biol. 1988;8:595–604. doi: 10.1128/mcb.8.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Symington L S, Petes T D. Meiotic recombination within the centromere of a yeast chromosome. Cell. 1988;52:237–240. doi: 10.1016/0092-8674(88)90512-0. [DOI] [PubMed] [Google Scholar]
- 494.Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 495.Szostak J W, Wu R. Unequal crossing-over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 1980;284:426–430. doi: 10.1038/284426a0. [DOI] [PubMed] [Google Scholar]
- 496.Takeda, S. Personal communication.
- 497.Teng S C, Kim B, Gabriel A. Retrotransposon reverse-transcriptase-mediated repair of chromosomal breaks. Nature. 1996;383:641–644. doi: 10.1038/383641a0. [DOI] [PubMed] [Google Scholar]
- 498.Teo S H, Jackson S P. Identification of Saccharomyces cerevisiae DNA ligase IV: involvement in DNA double-strand break repair. EMBO J. 1997;16:4788–4795. doi: 10.1093/emboj/16.15.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 499.Thaler D S, Stahl F W. DNA double-chain breaks in recombination of phage lambda and of yeast. Annu Rev Genet. 1988;22:169–197. doi: 10.1146/annurev.ge.22.120188.001125. [DOI] [PubMed] [Google Scholar]
- 500.Thaler D S, Stahl M M, Stahl F W. Tests of the double-strand-break repair model for red-mediated recombination of phage lambda and plasmid lambda dv. Genetics. 1987;116:501–511. doi: 10.1093/genetics/116.4.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 501.Thelen M P, Onel K, Holloman W K. The REC1 gene of Ustilago maydis involved in the cellular response to DNA damage encodes an exonuclease. J Biol Chem. 1994;269:747–754. [PubMed] [Google Scholar]
- 502.Thibodeau S N, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 503.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 504.Thomas B J, Rothstein R. The genetic control of direct-repeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics. 1989;123:725–738. doi: 10.1093/genetics/123.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Thompson-Stewart D, Karpen G H, Spradling A C. A transposable element can drive the concerted evolution of tandemly repetitious DNA. Proc Natl Acad Sci USA. 1994;91:9042–9046. doi: 10.1073/pnas.91.19.9042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Tishkoff D X, Boerger A L, Bertrand P, Filosi N, Gaida G M, Kane M F, Kolodner R D. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci USA. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 507.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 508.Toczyski D P, Galgoczy D J, Hartwell L H. CDC5 and CKII control adaptation to the yeast DNA damage checkpoint. Cell. 1997;90:1097–1106. doi: 10.1016/s0092-8674(00)80375-x. [DOI] [PubMed] [Google Scholar]
- 509.Tomkinson A E, Bardwell A J, Bardwell L, Tappe N J, Friedberg E C. Yeast DNA repair and recombination proteins Rad1 and Rad10 constitute a single-stranded-DNA endonuclease. Nature. 1993;362:860–862. doi: 10.1038/362860a0. [DOI] [PubMed] [Google Scholar]
- 510.Tomkinson A E, Bardwell A J, Tappe N, Ramos W, Friedberg E C. Purification of Rad1 protein from Saccharomyces cerevisiae and further characterization of the Rad1/Rad10 endonuclease complex. Biochemistry. 1994;33:5305–5311. doi: 10.1021/bi00183a038. [DOI] [PubMed] [Google Scholar]
- 511.Tran H T, Degtyareva N P, Koloteva N N, Sugino A, Masumoto H, Gordenin D A, Resnick M A. Replication slippage between distant short repeats in Saccharomyces cerevisiae depends on the direction of replication and the RAD50 and RAD52 genes. Mol Cell Biol. 1995;15:5607–5617. doi: 10.1128/mcb.15.10.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Trujillo K M, Yuan S S, Lee E-Y, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 513.Tsubouchi, H., and H. Ogawa. Personal communication.
- 514.Tsubouchi H, Ogawa H. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol Cell Biol. 1998;18:260–268. doi: 10.1128/mcb.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Tsukamoto Y, Kato J, Ikeda H. Budding yeast Rad50, Mre11, Xrs2, and Hdf1, but not Rad52, are involved in the formation of deletions on a dicentric plasmid. Mol Gen Genet. 1997;255:543–547. doi: 10.1007/s004380050527. [DOI] [PubMed] [Google Scholar]
- 516.Tsukamoto Y, Kato J, Ikeda H. Effects of mutations of RAD50, RAD51, RAD52, and related genes on illegitimate recombination in Saccharomyces cerevisiae. Genetics. 1996;142:383–391. doi: 10.1093/genetics/142.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Tsukamoto Y, Kato J, Ikeda H. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature. 1997;388:900–903. doi: 10.1038/42288. [DOI] [PubMed] [Google Scholar]
- 518.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci USA. 1996;93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Tung K S, Roeder G S. Meiotic chromosome morphology and behavior in zip1 mutants of Saccharomyces cerevisiae. Genetics. 1998;149:817–832. doi: 10.1093/genetics/149.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Umezu K, Kolodner R D. Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J Biol Chem. 1994;269:30005–30013. [PubMed] [Google Scholar]
- 521.Umezu K, Sugawara N, Chen C, Haber J E, Kolodner R D. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics. 1998;148:989–1005. doi: 10.1093/genetics/148.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Usui T, Ohta T, Oshumi H, Tsubouchi H, Tomizawa J-I, Ogawa H, Ogawa T. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- 523.Varon R, Vissinga C, Platzer M, Cerosaletti K M, Chrzanowska K H, Saar K, Beckmann G, Seemanova E, Cooper P R, Nowak N J, Stumm M, Weemaes C M, Gatti R A, Wilson R K, Digweed M, Rosenthal A, Sperling K, Concannon P, Reis A. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- 524.Verhage R, Zeeman A M, de Groot N, Gleig F, Bang D D, van de Putte P, Brouwer J. The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6135–6142. doi: 10.1128/mcb.14.9.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Vialard J E, Gilbert C S, Green C M, Lowndes N F. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 1998;17:5679–5688. doi: 10.1093/emboj/17.19.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 526.Virtaneva K, D’Amato E, Miao J, Koskiniemi M, Norio R, Avanzini G, Franceschetti S, Michelucci R, Tassinari C A, Omer S, Pennacchio L A, Myers R M, Dieguez-Lucena J L, Krahe R, de la Chapelle A, Lehesjoki A E. Unstable minisatellite expansion causing recessively inherited myoclonus epilepsy, EPM1. Nat Genet. 1997;15:393–396. doi: 10.1038/ng0497-393. [DOI] [PubMed] [Google Scholar]
- 527.Voelkel-Meiman K, Keil R L, Roeder G S. Recombination-stimulating sequences in yeast ribosomal DNA correspond to sequences regulating transcription by RNA polymerase I. Cell. 1987;48:1071–1079. doi: 10.1016/0092-8674(87)90714-8. [DOI] [PubMed] [Google Scholar]
- 528.Voelkel-Meiman K, Roeder G S. A chromosome containing HOT1 preferentially receives information during mitotic interchromosomal gene conversion. Genetics. 1990;124:561–572. doi: 10.1093/genetics/124.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Voelkel-Meiman K, Roeder G S. Gene conversion tracts stimulated by HOT1-promoted transcription are long and continuous. Genetics. 1990;126:851–867. doi: 10.1093/genetics/126.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Vollrath D, Davis R W, Connelly C, Hieter P. Physical mapping of large DNA by chromosome fragmentation. Proc Natl Acad Sci USA. 1988;85:6027–6031. doi: 10.1073/pnas.85.16.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 531.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 532.Wagstaff J E, Klapholz S, Esposito R E. Meiosis in haploid yeast. Proc Natl Acad Sci USA. 1982;79:2986–2990. doi: 10.1073/pnas.79.9.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Wallis J W, Chrebet G, Brodsky G, Rolfe M, Rothstein R. A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell. 1989;58:409–419. doi: 10.1016/0092-8674(89)90855-6. [DOI] [PubMed] [Google Scholar]
- 534.Walmsley R W, Chan C S, Tye B K, Petes T D. Unusual DNA sequences associated with the ends of yeast chromosomes. Nature. 1984;310:157–160. doi: 10.1038/310157a0. [DOI] [PubMed] [Google Scholar]
- 535.Warren S T. The expanding world of trinucleotide repeats. Science. 1996;271:1374–1375. doi: 10.1126/science.271.5254.1374. [DOI] [PubMed] [Google Scholar]
- 536.Watt P M, Hickson I D, Borts R H, Louis E J. SGS1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics. 1996;144:935–945. doi: 10.1093/genetics/144.3.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Watt P M, Louis E J, Borts R H, Hickson I D. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 538.Weiler K S, Szeto L, Broach J R. Mutations affecting donor preference during mating type interconversion in Saccharomyces cerevisiae. Genetics. 1995;139:1495–1510. doi: 10.1093/genetics/139.4.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Weiner B M, Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994;77:977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- 540.Weinert T. DNA damage checkpoints update: getting molecular. Curr Opin Genet Dev. 1998;8:185–193. doi: 10.1016/s0959-437x(98)80140-8. [DOI] [PubMed] [Google Scholar]
- 541.Weinert T A, Hartwell L H. Characterization of RAD9 of Saccharomyces cerevisiae and evidence that its function acts posttranslationally in cell cycle arrest after DNA damage. Mol Cell Biol. 1990;10:6554–6564. doi: 10.1128/mcb.10.12.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 542.Weinert T A, Hartwell L H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 543.Weinert T A, Kiser G L, Hartwell L H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 544.Welch J W, Maloney D H, Fogel S. Gene conversions within the Cup1r region from heterologous crosses in Saccharomyces cerevisiae. Mol Gen Genet. 1991;229:261–266. doi: 10.1007/BF00272164. [DOI] [PubMed] [Google Scholar]
- 545.Welch J W, Maloney D H, Fogel S. Unequal crossing-over and gene conversion at the amplified CUP1 locus of yeast. Mol Gen Genet. 1990;222:304–310. doi: 10.1007/BF00633833. [DOI] [PubMed] [Google Scholar]
- 546.West S C. Processing of recombination intermediates by the RuvABC proteins. Annu Rev Genet. 1997;31:213–244. doi: 10.1146/annurev.genet.31.1.213. [DOI] [PubMed] [Google Scholar]
- 547.White C I, Haber J E. Intermediates of recombination during mating type switching in Saccharomyces cerevisiae. EMBO J. 1990;9:663–673. doi: 10.1002/j.1460-2075.1990.tb08158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.White J H, Lusnak K, Fogel S. Mismatch-specific post-meiotic segregation frequency in yeast suggests a heteroduplex recombination intermediate. Nature. 1985;315:350–352. doi: 10.1038/315350a0. [DOI] [PubMed] [Google Scholar]
- 549.Whitehouse H L. The mechanism of genetic recombination. Biol Rev Camb Philos Soc. 1970;45:265–315. doi: 10.1111/j.1469-185x.1970.tb01633.x. [DOI] [PubMed] [Google Scholar]
- 550.Williamson M S, Game J C, Fogel S. Meiotic gene conversion mutants in Saccharomyces cerevisiae. I. Isolation and characterization of pms1-1 and pms1-2. Genetics. 1985;110:609–646. doi: 10.1093/genetics/110.4.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 551.Willis K K, Klein H L. Intrachromosomal recombination in Saccharomyces cerevisiae: reciprocal exchange in an inverted repeat and associated gene conversion. Genetics. 1987;117:633–643. doi: 10.1093/genetics/117.4.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552.Wilson J H, Leung W Y, Bosco G, Dieu D, Haber J E. The frequency of gene targeting in yeast depends on the number of target copies. Proc Natl Acad Sci USA. 1994;91:177–181. doi: 10.1073/pnas.91.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 552a.Wilson T, Grawunder U, Lieber M R. S. cerevisiae DNA ligase IV mediates DNA end-joining. Nature. 1997;338:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 553.Winzeler E A, Richards D R, Conway A R, Goldstein A L, Kalman S, McCullough M J, McCusker J H, Stevens D A, Wodicka L, Lockhart D J, Davis R W. Direct allelic variation scanning of the yeast genome. Science. 1998;281:1194–1197. doi: 10.1126/science.281.5380.1194. [DOI] [PubMed] [Google Scholar]
- 554.Wu A M, Kahn R, DasGupta C, Radding C M. Formation of nascent heteroduplex structures by RecA protein and DNA. Cell. 1982;30:37–44. doi: 10.1016/0092-8674(82)90009-5. [DOI] [PubMed] [Google Scholar]
- 555.Wu C, Weiss K, Yang C, Harris M A, Tye B K, Newlon C S, Simpson R T, Haber J E. Mcm1 regulates donor preference controlled by the recombination enhancer in Saccharomyces mating-type switching. Genes Dev. 1998;12:1726–1737. doi: 10.1101/gad.12.11.1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Wu T C, Lichten M. Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics. 1995;140:55–66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 557.Wu T C, Lichten M. Meiosis-induced double-strand break sites determined by yeast chromatin structure. Science. 1994;263:515–518. doi: 10.1126/science.8290959. [DOI] [PubMed] [Google Scholar]
- 558.Wu X, Haber J E. A 700 bp cis-acting region controls mating-type dependent recombination along the entire left arm of yeast chromosome III. Cell. 1996;87:277–285. doi: 10.1016/s0092-8674(00)81345-8. [DOI] [PubMed] [Google Scholar]
- 559.Wu X, Wu C, Haber J E. Rules of donor preference in Saccharomyces mating-type gene switching revealed by a competition assay involving two types of recombination. Genetics. 1997;147:399–407. doi: 10.1093/genetics/147.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Xiao Y, Weaver D T. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 561.Xu F, Petes T D. Fine-structure mapping of meiosis-specific double-strand DNA breaks at a recombination hotspot associated with an insertion of telomeric sequences upstream of the HIS4 locus in yeast. Genetics. 1996;143:1115–1125. doi: 10.1093/genetics/143.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 562.Xu L, Kleckner N. Sequence non-specific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hot spot in yeast. EMBO J. 1995;14:5115–5128. doi: 10.1002/j.1460-2075.1995.tb00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Xu L, Weiner B M, Kleckner N. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 1997;11:106–118. doi: 10.1101/gad.11.1.106. [DOI] [PubMed] [Google Scholar]
- 564.Yamaguchi-Iwai Y, Sonoda E, Buerstedde J M, Bezzubova O, Morrison C, Takata M, Shinohara A, Takeda S. Homologous recombination, but not DNA repair, is reduced in vertebrate cells deficient in RAD52. Mol Cell Biol. 1998;18:6430–6435. doi: 10.1128/mcb.18.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 565.Yamamoto A, Guacci V, Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.Yoshida K, Kondoh G, Matsuda Y, Habu T, Nishimune Y, Morita T. The mouse RecA-like gene Dmc1 is required for homologous chromosome synapsis during meiosis. Mol Cell. 1998;1:707–718. doi: 10.1016/s1097-2765(00)80070-2. [DOI] [PubMed] [Google Scholar]
- 567.Yu A, Dill J, Wirth S S, Huang G, Lee V H, Haworth I S, Mitas M. The trinucleotide repeat sequence d(GTC)15 adopts a hairpin conformation. Nucleic Acids Res. 1995;23:2706–2714. doi: 10.1093/nar/23.14.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 568.Yu C E, Oshima J, Fu Y H, Wijsman E M, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin G M, Mulligan J, Schellenberg G D. Positional cloning of the Werner’s syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 569.Yu S, Mangelsdorf M, Hewett D, Hobson L, Baker E, Eyre H J, Lapsys N, Le Paslier D, Doggett N A, Sutherland G R, Richards R I. Human chromosomal fragile site FRA16B is an amplified AT-rich minisatellite repeat. Cell. 1997;88:367–374. doi: 10.1016/s0092-8674(00)81875-9. [DOI] [PubMed] [Google Scholar]
- 570.Zenvirth D, Arbel T, Sherman A, Goldway M, Klein S, Simchen G. Multiple sites for double-strand breaks in whole meiotic chromosomes of Saccharomyces cerevisiae. EMBO J. 1992;11:3441–3447. doi: 10.1002/j.1460-2075.1992.tb05423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Zenvirth D, Loidl J, Klein S, Arbel A, Shemesh R, Simchen G. Switching yeast from meiosis to mitosis: double-strand break repair, recombination and synaptonemal complex. Genes Cells. 1997;2:487–498. doi: 10.1046/j.1365-2443.1997.1370335.x. [DOI] [PubMed] [Google Scholar]
- 572.Zheng P, Fay D S, Burton J, Xiao H, Pinkham J L, Stern D F. SPK1 is an essential S-phase-specific gene of Saccharomyces cerevisiae that encodes a nuclear serine/threonine/tyrosine kinase. Mol Cell Biol. 1993;13:5829–5842. doi: 10.1128/mcb.13.9.5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Zhu C, Bogue M A, Lim D S, Hasty P, Roth D B. Ku86-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86:379–389. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]
- 574.Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu Rev Genet. 1998;32:619–697. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- 575.Zou H, Rothstein R. Holliday junctions accumulate in replication mutants via a RecA homolog- independent mechanism. Cell. 1997;90:87–96. doi: 10.1016/s0092-8674(00)80316-5. [DOI] [PubMed] [Google Scholar]