Abstract
Pathogenic variants in the voltage-gated sodium channel gene family lead to early onset epilepsies, neurodevelopmental disorders, skeletal muscle channelopathies, peripheral neuropathies and cardiac arrhythmias. Disease-associated variants have diverse functional effects ranging from complete loss-of-function to marked gain-of-function. Therapeutic strategy is likely to depend on functional effect. Experimental studies offer important insights into channel function but are resource intensive and only performed in a minority of cases.
Given the evolutionarily conserved nature of the sodium channel genes, we investigated whether similarities in biophysical properties between different voltage-gated sodium channels can predict function and inform precision treatment across sodium channelopathies. We performed a systematic literature search identifying functionally assessed variants in any of the nine voltage-gated sodium channel genes until 28 April 2021. We included missense variants that had been electrophysiologically characterized in mammalian cells in whole-cell patch-clamp recordings. We performed an alignment of linear protein sequences of all sodium channel genes and correlated variants by their overall functional effect on biophysical properties.
Of 951 identified records, 437 sodium channel-variants met our inclusion criteria and were reviewed for functional properties. Of these, 141 variants were epilepsy-associated (SCN1/2/3/8A), 79 had a neuromuscular phenotype (SCN4/9/10/11A), 149 were associated with a cardiac phenotype (SCN5/10A) and 68 (16%) were considered benign. We detected 38 missense variant pairs with an identical disease-associated variant in a different sodium channel gene. Thirty-five out of 38 of those pairs resulted in similar functional consequences, indicating up to 92% biophysical agreement between corresponding sodium channel variants (odds ratio = 11.3; 95% confidence interval = 2.8 to 66.9; P < 0.001). Pathogenic missense variants were clustered in specific functional domains, whereas population variants were significantly more frequent across non-conserved domains (odds ratio = 18.6; 95% confidence interval = 10.9–34.4; P < 0.001). Pore-loop regions were frequently associated with loss-of-function variants, whereas inactivation sites were associated with gain-of-function (odds ratio = 42.1, 95% confidence interval = 14.5–122.4; P < 0.001), whilst variants occurring in voltage-sensing regions comprised a range of gain- and loss-of-function effects.
Our findings suggest that biophysical characterisation of variants in one SCN-gene can predict channel function across different SCN-genes where experimental data are not available. The collected data represent the first gain- versus loss-of-function topological map of SCN proteins indicating shared patterns of biophysical effects aiding variant analysis and guiding precision therapy. We integrated our findings into a free online webtool to facilitate functional sodium channel gene variant interpretation (http://SCN-viewer.broadinstitute.org).
Keywords: SCN1A, SCN2A, SCN4A, SCN5A, SCN8A
Brunklaus et al. report that biophysical characterization of variants in one voltage-gated sodium channel (SCN) gene can predict channel function across different SCN genes where experimental data are not available. Shared patterns of functional effects can aid variant interpretation and guide precision therapy.
See Mantegazza and Cestèle (https://doi.org/10.1093/brain/awac397) for a scientific commentary on this article.

For the podcast associated with this article, please visit https://academic.oup.com/brain/pages/podcast
See Mantegazza and Cestèle (https://doi.org/10.1093/brain/awac397) for a scientific commentary on this article.
Introduction
Voltage-gated sodium channel genes (SCN1–11A) encode a homologous family of nine functionally expressed sodium channels (SCN) from Nav1.1 to Nav1.9.1,2 They play a key role in initiating action potentials3 and are extensively distributed throughout the nervous system. Variants in SCN-genes are associated with early onset epilepsies, neurodevelopmental disorders, skeletal muscle channelopathies, peripheral neuropathies and cardiac conduction defects.4,5 All nine SCN-genes share common evolutionary origins and a conserved basic structure, consisting of four homologous domains (DI–IV), each containing six transmembrane segments (S1–6) with up to 85% amino acid sequence similarity between them.6,7 With the emergence of modern sequencing techniques facilitating genetic diagnosis, SCN-related disorders are promising candidates for precision therapies.8 However, predicting the impact of a variant on channel kinetics without prior functional characterization is challenging. Variants occurring within the same gene show remarkable phenotype variability depending on their location and effect on biophysical properties, while variants occurring in different SCN genes may result in similar phenotypes. Such genetic and clinical complexity hinders the establishment of genotype-phenotype correlations.
Genetic variants in SCN1/2/3/8A are responsible for a significant proportion of monogenic epilepsies and neurodevelopmental disorders. Loss-of-function (LoF) variants in SCN1A manifest variable phenotypes, ranging from milder presentations such as genetic epilepsy with febrile seizures plus to the severe developmental and epileptic encephalopathy Dravet syndrome, whereas gain-of-function (GoF) variants are associated with familial hemiplegic migraine type 3 (FHM3).9–14 Variants in SCN2/3/8A are clinically heterogenous, causing different forms of epilepsy ranging from self-limited infantile epilepsy to developmental and epileptic encephalopathies (DEEs) including early infantile DEE (Ohtahara syndrome) and infantile spasms syndrome.15–17 Patients with SCN5A variants showing GoF manifest long-QT syndrome (LQT3), while LoF variants cause Brugada syndrome.18,19 SCN4A variants are responsible for a significant proportion of skeletal muscle channelopathies,20,21 while SCN9/10/11A are primarily associated with peripheral neuropathies; both are predominantly caused by GoF variants and present with variable severity.22–24 Whilst the majority of SCN-related disorders are dominant conditions, newer SCN4A-linked phenotypes, including congenital myasthenia and congenital myopathy, have been identified to be caused by LoF variants inherited in a recessive manner.25,26
Sodium channelopathies display varying treatment responsiveness depending on the underlying functional effect. While many patients with SCN2/3/8A-related epilepsy remain treatment resistant, those caused by GoF variants, tend to respond to sodium channel blockers (SCBs), whereas those with LoF variants do not.27–30 In contrast, patients with Dravet syndrome due to LoF SCN1A-variants worsen with SCBs.31,32 Similarly, SCBs suppress pathological currents in patients with LQT3 caused by GoF SCN5A variants,33 whereas patients with Brugada syndrome caused by LoF are often treatment-resistant, relying on device implantation for arrhythmia control.34,35 As functional knowledge is a key determinant for optimal management, variant interpretation without requiring resource-intensive electrophysiological studies represents an unmet clinical need.
To improve our understanding of SCN-related disorders, our objective was to investigate relationships between variant type, location and biophysical properties across all SCN-subtypes. Applying evidence from functional studies we aimed to build a framework that informs clinical practice and guides precision therapy.
Materials and methods
Search strategy and selection criteria
In accordance with PRISMA guidance, we systematically searched PubMed to 28 April 2021 to identify studies published in English describing functional characteristics of missense variants using the terms ‘clamp AND SCN1A’. The same search was applied for SCN2-11A. To narrow search criteria for SCN5A, we added ‘Brugada’ or ‘QT’. In addition, we manually searched sodium channel mutation databases and bibliographies that were found through our systematic PubMed search. Any duplicate citations were removed, and all remaining studies were screened for relevance (Supplementary Fig. 1). Only missense variants electrophysiologically characterized by whole-cell patch clamp experiments were assessed for eligibility. To improve comparison of biophysical properties, only variants characterized in mammalian cells were included. We excluded variants if they had no evidence of a recognized human disease phenotype. We did not double count identical variants but did consider patch clamp data from different sources if available. Data were independently reviewed by three researchers (A.B., T.F. and S.S.).
Variant analysis
Variants were categorized as either GoF, LoF, mixed-function (mixed) or similar-to-wild-type function depending on their effect on biophysical properties (Supplementary material). We defined any biophysical change entailing an increase in the Na+ permeability as GoF, and the opposite for LoF. In some cases, variants demonstrated a paradoxical effect on channel properties, i.e. decreased peak current and increased persistent current. Where one effect was not clearly dominant, they were classified as ‘mixed’.36 Variants that exhibited wild-type-like function or lacked pronounced impact on channel function were classified as similar-to-wild-type and assumed benign. To detect analogous missense variants amongst SCN1-11A genes, we performed an alignment of linear protein sequences using the ‘PER viewer’ (pathogenic variant enriched regions) across genes and gene families (http://per.broadinstitute.org).37 Variants detected in the same alignment index position were investigated for similarities in their overall functional effects. Variants occurring in positions where the reference amino acid was different across SCN1-11A in the PER alignment were classified as ‘not conserved’, as this suggests that they were not conserved across evolution and thus less likely to be functionally significant.
Population variants were collected for all SCN1A-11A genes where at least one pathogenic variant was previously identified from the Genome Aggregation Database (gnomAD, http://gnomad.broadinstitute.org), which provides access to germline variants from >140 000 exomes as well as 15 000 genomes from the general population. Assumed benign variants, which showed a wild-type like behaviour were added to the population variant cohort from gnomAD. Non-conserved variants that were also present in gnomAD were assumed benign. The term benign implies that these variants occur frequently without disease; however, they might still be associated with functional change.
In-silico prediction of functional variant effects
Heyne et al.38 recently developed an in-silico prediction tool (‘funNCion’; http://funNCion.broadinstitute.org) to estimate the functional consequences of voltage-gated sodium and calcium channels. The tool does not consider evidence from biophysical experiments but infers channel function from a large dataset of clinical disease phenotypes. We applied the ‘funNCion’ tool on our cohort of functionally characterized SCN variants to establish the tool’s accuracy and clinical utility compared with actual biophysical readouts.
Data analysis
A two-tailed Fisher’s Exact test with Bonferroni correction was performed to assess the burden of pathogenic variants across different regions of the SCN protein to compare pathogenic LoF and GoF variants and to determine categorical differences in functional effect and differences in phenotype or variant distribution across related SCN-subtypes. Similarly, Fisher’s Exact test was performed to test whether missense variant pairs with identical disease-associated variants across different SCN-genes more often have similar functional consequences than expected from random sampling. Significance for all tests was at the 5% level and analysis was performed using R version 4.0.3.
Data availability
All data used in this study are available in the manuscript, the Supplementary material and via the free online webtool http://SCN-viewer.broadinstitute.org.
Results
Literature search
From our systematic PubMed search, we found 951 records, and following elimination of 127 duplicate citations, screened 824 titles and abstracts. A total of 535 records were excluded, as they did not include a patch-clamp experiment on a missense SCN variant, and 289 records including 569 variants were subsequently assessed by full-text analysis. After additional manual searching and exclusion due to ineligibility, we identified 437 missense variants in the literature that were functionally characterized by whole-cell patch-clamp experiments. Of these, 369 (84%) variants were assessed as pathogenic, including 141 associated with epilepsy (SCN1/2/3/8A), 79 with a neuromuscular phenotype (SCN4/9/10/11A) and 149 with a cardiac phenotype (SCN5/10A). Sixty-eight (16%) variants were considered benign, as they either had properties similar to wild-type or were both not conserved and present in gnomAD (Supplementary Fig. 1 and Supplementary Table 1).
Similarities in variant function and distribution across related sodium channels
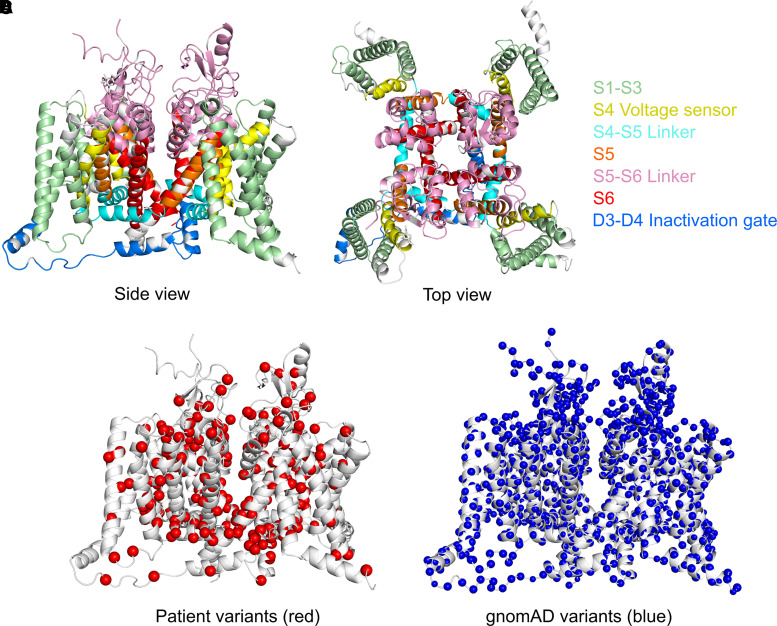
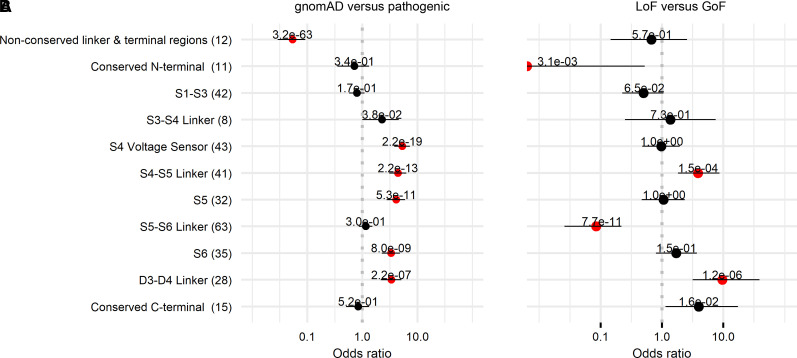
To illustrate the distribution of missense variants and similarities in functional consequences across related sodium channels, we plotted the position of 369 SCN1-11A pathogenic variants according to their corresponding SCN1A alignment index position and compared their position to the distribution of 3454 gnomAD variants (Fig. 1). Enrichment analysis comparing pathogenic versus gnomAD variants demonstrated that pathogenic missense variants are clustered in specific functional domains, whereas gnomAD variants are significantly more frequent across functionally less important and not conserved cytoplasmic domains [odds ratio (OR) = 18.6; 95% confidence interval (CI) = 10.9–34.4; P < 0.001; Fig. 2].
Figure 1.
SCN protein structure and position of disease-causing missense versus gnomAD variants. (A) and (B) SCN protein structure from side and top view. (C) Patient variants shown in red. (D) GnomAD variants shown in blue.
Figure 2.
Enrichment analysis of gnomAD versus pathogenic variants and LoF versus GoF variants. Enrichment analysis of (A) gnomAD versus pathogenic and (B) LoF versus GoF variants across different protein parts including four homologous domains (D1–D4), each consisting of six transmembrane segments (S1–S6) and large cytoplasmic loops.
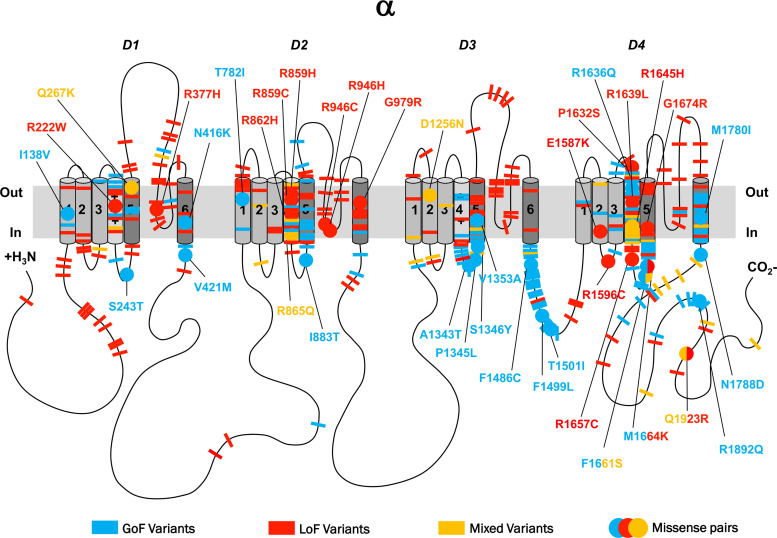
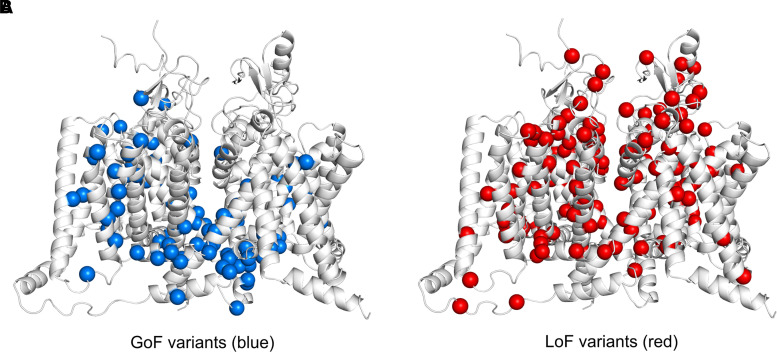
The majority of pathogenic variants were located in the homologous domains D1–4 (Fig. 3). Pathogenic variants distributed across all four S5–6 pore-loop regions appeared to be predominantly LoF (91%, 58/64), whereas variants occurring in voltage-sensing regions (including S3–4, S4 and S4–5) comprised a range of GoF (47%, 51/107), LoF (38%, 41/107) and mixed function (15%, 16/107) effects. In the fast inactivation gate (DIII–IV), 83% of variants were GoF (24/29), all occurring throughout the first half of the intracellular linker. Other sites implicated in inactivation gating, including the intracellular S4–5 regions in DIII and DIV as well as DIVS6, shared this pattern of harbouring predominantly GoF variants (69%, 34/49).39,40 Overall, pore-loop regions were frequently associated with LoF variants, whereas inactivation sites were associated with GoF (OR = 42.1, 95% CI = 14.5–122.4; P < 0.001). The C-terminus displayed a range of GoF (46%, 12/26), LoF (27%, 7/26) and mixed effects (27%, 7/26). Here, GoF and mixed variants appeared to cluster in the proximal region, whereas a minority of LoF variants were found distally. Overall, very few variants occurred in cytoplasmic regions (N-terminus, DI–II and DII–III linkers). A 3D illustration comparing GoF versus LoF locations across the SCN protein is detailed in Fig. 4.
Figure 3.
2D representation of pathogenic SCN variants with functional effects. 2D representation of the SCN protein. The alpha subunit consists of four homologous domains (D1–4) each formed of six transmembrane segments (S1–S6). Segment 4 represents the voltage sensor and S5–6 the pore region. Individual missense variants are displayed as different coloured bars. Blue denotes GoF, red LoF and yellow mixed function. Analogue missense pairs are displayed as circles with amino acid details.
Figure 4.
3D illustration comparing GoF with LoF locations across the SCN protein. (A) GoF variants are illustrated in blue. (B) LoF variants are illustrated in red.
Benign variants displayed a different distribution, comprising 40% of all variants identified in the N-terminus (8/20—mainly located in the initial segment), 56% in the large DI–II intracellular linker (10/18), 57% in the large DII–III intracellular linker (8/14) and 26% in the C-terminus (9/35—mainly located in the distal segment). In contrast, the fast inactivation gate was free of benign variants, and very few were found in other sites implicated in inactivation gating, including S4–5 of DIII and DIV, and DIVS6.
Corresponding variants in different SCN-genes have similar function
Among all functionally characterized SCN1-11A variants, we identified 38 random pairs with a corresponding analogous identical disease-associated variant in a different SCN-gene, including six previously reported pairs41 (Supplementary Table 2 and Fig. 3). The missense variants in each of these pairs have similar functional consequences in 35 out of the 38 pairs (92%) regardless of the type of voltage-gated sodium channel affected (OR = 11.3; 95% CI = 2.8–66.9; P < 0.001). Many of these pairs were found at conserved channel locations, including three LoF pairs between SCN1/2/5A at pore-loop regions, seven GoF pairs between SCN1-9A at sites implicated in channel inactivation and eight pairs between SCN1/4/5/8A at S4 showing a mixture of GoF, LoF and mixed-function effects (Fig. 3). The only three pairs with divergent function included the SCN1A F1661S/SCN4A F1473S42–44 variant pair and the SCN1A M1664K/SCN9A M1627K45–48 variant pair, both located in the D4 S4–5 linker region, as well as the SCN1A Q1923R/SCN5A Q1909R49–51 variant pair located in the distal C-terminus. The SCN4A variant was identified in a patient with paramyotonia congenita (PMC) due to GoF, the SCN9A variant in a patient with paroxysmal extreme pain disorder (PEPD), equally due to GoF, and the SCN5A variant in a patient with sudden infant death syndrome, associated with mixed function; the three corresponding SCN1A variants were identified in patients with genetic epilepsy with febrile seizures plus/Dravet syndrome and found to be LoF/mixed function. The SCN1A variants led to impaired channel trafficking and reduced cell surface expression, resulting in a reduction of peak current, not allowing for detailed biophysics to be recorded. In contrast, the trafficking of the SCN4A, SCN5A and SCN9A variants did not appear to be affected.
Comparison of reported biophysical effects versus predicted functional outcomes
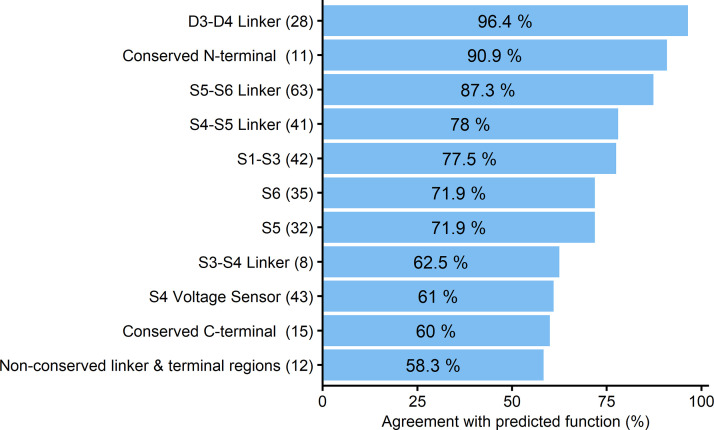
We applied the recently-developed in-silico prediction tool ‘funNCion’ to predict GoF versus LoF effects in the 369 biophysically characterized variants to evaluate the accuracy and usefulness of this type of tool. Compared with the gold standard whole-cell patch clamp experiment results, the in-silico tool achieved 78.5% agreement in the prediction of GoF properties and 75.0% agreement in the prediction of LoF properties. Agreement differed depending on location within the channel, with certain regions such as inactivation and pore loop sites achieving better agreement (78–96%) compared with others including the S4 region (<62%, Fig. 5).
Figure 5.
In-silico prediction versus reported biophysical SCN variant effects. Prediction agreement [as a percentage (%)] detailed according to different protein parts including four homologous domains (D1–D4), each consisting of six transmembrane segments (S1–S6) and large cytoplasmic loops. In silico prediction was performed according to ‘funNCion’ (http://funNCion.broadinstitute.org).
Detailed SCN1-11A variant analysis
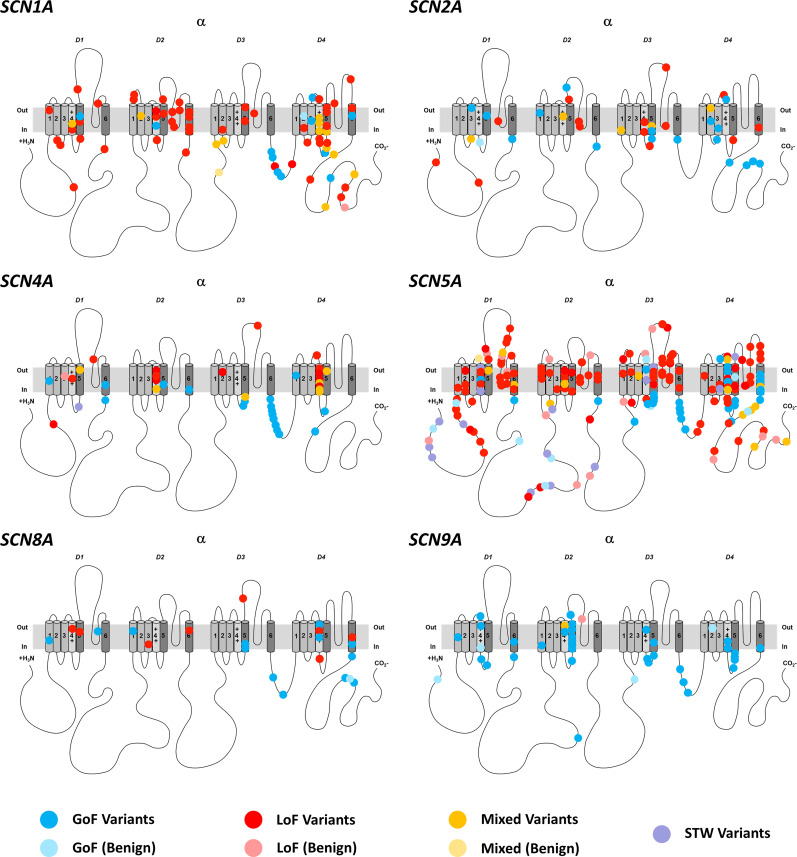
Each of the different SCN subtypes (SCN1–11A) presented with a specific distribution pattern of pathogenic versus benign variants according to channel function and location (Fig. 6). Supplementary Table 3 lists the channel specific clinical phenotypes and associated function.
Figure 6.
2D representation of SCN variants with functional effects according to single sodium channels. 2D representation of different SCN proteins. The alpha subunit consists of four homologous domains (D1–4) each formed of six transmembrane segments (S1–S6). Segment 4 represents the voltage sensor and segments S5–6 the pore region. Individual missense variants are displayed as different coloured circles. Blue denotes GoF, red LoF, yellow mixed function and purple similar-to-wild-type (STW). Benign variants are illustrated in pale colours.
Discussion
Comparing the distribution of disease-associated missense variants across all nine SCN-subtypes, we observed striking similarities in altered biophysical channel properties induced by missense variants at analogous position across SCN proteins. Almost all identified variant pairs in different SCN genes exhibited similar biophysical properties regardless of the SCN-subtype affected. Functional data of variants at analogous positions in SCN genes can predict variant effects in related sodium channel genes and may inform genotype-guided precision therapies in patients with neurological and cardiac sodium channelopathies.
Voltage-gated sodium channels contain a central pore composed of S5 and S6 segments from all four domains that line the inner cavity and form the intracellular exit. Intervening S5–6 pore-loops line the extracellular end of the pore, forming a large ion-selective filter.52 In keeping with previous work, we observed that variants occurring in the S5-6 pore-loop predominantly caused LoF effects.5,53 This region displayed clustering of epilepsy-associated LoF SCN1A variants, whereas very few epilepsy-associated SCN2/8A variants occurred here. The relationship between variant distribution and functional consequences was also observed in SCN5A. Similar to reported case series,54 the vast majority of Brugada syndrome-associated variants displayed LoF effects and clustered in S5–6 pore-loops. In contrast, neither LQT3-associated variants that predominantly caused GoF nor any GoF variants across other SCN-subtypes clustered in this region. This showed that, across different SCNs, variants occurring in S5–6 pore-loops often lead to LoF, causing detrimental effects on channel kinetics.
Sites implicated in inactivation gating harboured predominantly GoF variants. The short DIII–IV linker is responsible for fast inactivation. This loop folds into a hinged-lid structure that blocks the intracellular end of the pore during sustained depolarization and contains residues (IFM motif) that form a latch, holding the inactivation gate shut.52 Interference within this region leads to impaired inactivation and hyperexcitability that is in keeping with the underlying mechanism of SCN-related disorders.5 Respectively, SCN1A and SCN2/8A variants distributed across the DIII–IV linker were associated with FHM3 and epilepsy, while SCN4A variants were associated with myotonia. Similarly, a significant proportion of LQT3-associated SCN5A variants and painful neuropathy-associated SCN9A variants occurred across all regions implicated in inactivation gating. This suggested that variants occurring in inactivation sites frequently present GoF across different SCNs.
Voltage-sensing regions in each domain are composed of S4 and adjacent linkers. S4 serves as the voltage-sensor, containing a dense composition of highly-conserved arginine residues.7 In response to depolarization, this segment moves across the membrane and facilitates channel activation.52 We observed a mixture of biophysical effects in S4, constituting a cluster of painful neuropathy-associated SCN9/11A, hypokalaemic periodic paralysis-associated SCN4A and LQT3-associated SCN5A variants with a moderate distribution of epilepsy-associated SCN1/2/3/8A variants. Rather than a regional effect (as in the aforementioned S5–6 linker region), functional impact in S4 appears to be determined by individual variant changes that manifest a spectrum of disorders across all SCN-subtypes. The five conserved arginine residues R0–R4 serve as example for this observation: whilst pathogenic variants in R0 are associated with increased function, there appears to be a steady shift from nearly complete LoF in variants of R1/R2 to LoF/mixed in R3 and mainly mixed effects in R4, the deeper the progression into the S4 (Supplementary Fig. 2).
In contrast, the large non-conserved cytoplasmic regions and distal N- and C-terminal cytoplasmic regions are devoid of pathogenic variants across all SCN-subtypes.
Comparison of corresponding variants aids clinical prediction
Our alignment of all nine channel subtypes allowed a comparison of SCN-variants by their corresponding index position, revealing similar functional consequences occurring in specific regions. The clinical manifestation of the observed biophysical change depends on neuron type and neuronal network distribution. For example, while all variant pairs observed in the S5–6 pore-loop regions displayed the same LoF functional consequences, a LoF in SCN1A leads to epilepsy due to impaired expression of inhibitory interneurons,10 whilst the corresponding LoF SCN2A and SCN5A variants result in autism spectrum disorder and Brugada syndrome, respectively, due to high expression of these genes in excitatory brain-expressed SCN2A neurons and cardiac-expressed SCN5A channels.17,18 The same pattern was observed in sites of inactivation, where GoF variants in SCN1A cause FHM3, whilst analogous GoF variants in SCN2/8A cause epilepsy. Similarly, GoF variants at inactivation sites occurring in SCN4A cause myotonia due to hyperexcitability of the sarcolemma,20 while corresponding variants in SCN9/10/11A that display the same GoF properties cause painful peripheral neuropathy owing to expression in peripheral neurons.22 Importantly, these similarities in functional consequences across related SCNs apply in particular to regions that are conserved across evolution. For example, all variant pairs occurring in the S4 segment involved substitution of arginine residues which are highly-conserved across all related SCNs.55
In the three cases where there were discrepancies between how variants affected different channels (Supplementary Table 2), we noted that all three were cases where a variant produced LoF/mixed function (reduced or no peak current) of SCN1A but GoF or mixed effects in other channels. This discrepancy may be related to impaired channel trafficking by which SCN1A may be particularly affected. Where trafficking is suspected, pharmacological rescue is a possibility, first suggested when mexiletine partially-rescued trafficking for a mutation in SCN5A.56 Since then, trafficking mutations have been shown for SCN1A,51 and several of these can be rescued pharmacologically with anti-epileptic drugs such as phenytoin43 and other chaperonins.12,57 Indeed, SCN1A is so sensitive to trafficking that a mutation initially identified as LoF in cell lines due to trafficking becomes GoF simply by expressing in neurons.58 Thus, we propose that an additional use of our data may arise when classifying an SCN1A variant associated with LoF in functional studies but linked to a variant known to produce GoF or mixed effects in other channels when heterologously expressed. In this exceptional case, our data may support cautious interpretation of the functional data (which in almost every other case should be the gold standard). Our data suggest that in these cases it is particularly important to consider trafficking, as SCN1A may be more sensitive to trafficking deficits when expressed in non-native cells such as HEK cells. Where complete LoF may be seen in HEK cells, it is possible that functional channels—potentially with aberrant gating—are produced in native cells. As new genetic therapies may begin to distinguish between complete LoF and aberrant function (such as gating pore leaks), it will be particularly important to rule out the possibility of a mutant allele having gating changes or trafficking issues that could alter neuronal health if expression is increased. As treatments targeting trafficking become clinically available, these mutations may be prioritized for interrogation for these treatments: if in functional studies treatments that increase trafficking produce wild-type-like currents, then these mutations would represent a potentially valuable treatment via trafficking for children.
Overlapping boundaries between presumed benign and pathogenic variants
We observed that a significant proportion of SCN4/5/10A variants were not conserved or assumed benign, compared with very few SCN1/2/8A variants. This difference may be explained by the easily recognizable clinical presentation of SCN1/2/8A-related disease with difficult to treat epilepsy and severe intellectual disability.10 In comparison, SCN4/5/10A-related disorders appear to have less noticeable clinical features, often presenting with only one paroxysmal symptom with variable severity.59–61 Furthermore, there is significant evolutionary constraint among SCN genes, suggesting that whilst patients with SCN1/2/3/8A variants present with early onset severe dominant de novo disease and marked reduction in fecundity, variants associated with familial SCN disease such as SCN4/5/9/10/11A are better tolerated and might go unnoticed.36,62
The distribution pattern across SCN-subtypes for gnomAD/benign variants clearly contrasted that of pathogenic variants. Consistent with previous work, very few benign variants were found at sites of inactivation and pore-forming regions.63,64 A significant proportion of variants occurring at DI–II and DII–III linkers were assumed benign. Many did not share the same amino acid as their corresponding SCN1A position and were present in gnomAD, demonstrating how these regions are not conserved across evolution, and of lesser functional significance.65 The distribution of variants found in reference populations correlate strongly with the functional significance of these regions, and thus aids the interpretation of variant pathogenicity across different SCN-subtypes (http://per.broadinstitute.org).
Clinical implications on precision treatment and its limitations
Precision medicine aims to tailor individual treatment to reverse or modify an underlying disease pathophysiology.66,67 Knowledge of the functional impact of a SCN-variant can inform clinical management and therapeutic choice. Our approach will be particularly helpful for clinicians faced with early presentation in very young children with a novel mutation of unknown function, where biophysical estimation could guide treatment. Functional characterization remains the gold standard, but can lead to many months of delay, whilst our data will allow almost instantaneous information about the likely effect of a novel variant. However, we emphasize that this should not replace experimental functional analysis, which allows the actual identification of detailed effects and mechanisms. Evidence of successful precision treatment approaches have been reported across SCN-related disorders. GoF variant carriers are more likely to respond to SCBs in SCN2/8A-related epilepsies,27,68 SCN4A-related myotonias69 and SCN5A-mediated arrhythmias/LQT3.33,70 In contrast, use of SCBs in LoF cases is contraindicated. Inadvertently selecting incorrect treatment without knowledge of function can be detrimental to clinical symptoms.27,32 However, success with precision treatment is not guaranteed. A recent survey of precision medicine in genetic epilepsies demonstrated that many individuals with genetic epilepsy continue to have symptoms, with >50% seizure reduction observed in just 30% of patients.66 Limitations to precision medicine include the genetic background between patients, variation in gene expression, epigenetics and environmental factors. In vitro functional studies are not able to account for these modifying factors which limits their clinical utility.
We identified a number of SCN gene-specific clinical examples where knowledge of the underlying functional properties assists patient management. Early in disease presentation when phenotypes have not fully evolved, functional inference may guide early management and help predict the ultimate phenotype. In SCN1A-related disorders, this is particularly useful in those presenting <4 months of age as these can either be LoF, associated with Dravet syndrome, or GoF/mixed function, associated with early onset DEE.71,72 Our approach aids interpretation of novel SCN1A variants in FHM phenotypes, since these would be expected to be GoF. In SCN2A-related disorders it guides therapy for individuals with early onset DEE presenting between 2–4 months, since either GoF or LoF variant carriers might manifest around this age. Although many patients remain pharmacoresistant, GoF variant carriers tend to experience a reduction in seizure frequency with SCBs, in particular phenytoin in high doses, whereas SCBs lead to seizure aggravation in LoF variant carriers.27 Given that the number of reported variants and functional studies in SCN3A are very limited, our approach can be used to help determine pathogenicity of novel variants in this gene. In SCN8A-related epilepsies our approach guides therapy for those presenting between 9 months and 3 years of life that can harbour either GoF or LoF variants as recently shown in a large SCN8A cohort.30 Whilst many patients still have pharmacoresistant epilepsy, GoF variant carriers respond significantly better to SCBs than to other anti-seizure medications.30
In addition to conventional SCBs, more selective agents, including those that target mutations with specific effects on channel function73,74 and those that target specific types of sodium channels, are being developed.75,76 As these approaches rely on variant position and functional impact on the sodium channel, our method could facilitate identification of variants suitable for these treatments.
New disease modifying treatments are being developed for SCN1A, SCN2A and SCN8A-related disorders targeting gene specific LoF and GoF effects.77 Phase 1/2 clinical trials are ongoing/being developed for Dravet syndrome, aiming to increase SCN1A expression in LoF disease. This approach would likely be detrimental in GoF disease.77 Our method allows estimation of channel function that will be useful for variant interpretation in specific GoF- or Lof-directed disease modifying therapies. However, any decision of which therapeutic approach should be taken will be informed by multiple factors including functional estimation as well as clinical presentation.
In light of these developments, being able to map specific GoF and LoF regions across sodium channel paralogues allows clinicians to make the best use of extensive functional data to help inform both drug choice in the individual patient as well as allowing targeted drug compound development across different channels.76,78,79 Specific examples are discussed below, where LOF in SCN1A may be prioritized for new treatments improving trafficking, or where homology suggests a LOF might also induce toxic gating pore leaks, potentially precluding treatments aimed at increasing expression of both alleles. It has recently been shown how identifying key functional regions in one channel (Nav1.7/SCN9A) can be used to accelerate the design of next generation Nav modulators across other channels.80 Likewise, the increasing recognition that loss of S4 arginines may have a pathogenic mechanism of introducing an aberrant gating pore leak in multiple channels including SCN4A-related muscle channelopathies78 and SCN2A-related epilepsies81 means that drugs which specifically block gating pore leaks may be applicable in cases where gating pore leak is suspected and could apply across a range of channels, even where gating pore recordings are not possible. However, clinicians should consider functional variant information in the context of the clinical presentation of their patient, including seizure types, EEG signature, previous response to specific medications and medication side effects, all of which might offer important clues towards diagnosis and management.
Gold standard patch-clamp recordings in mammalian models are time and resource intensive, precluding their use in routine practice and the ability of functional variant interpretation without prior biophysical studies represents an unmet clinical need.82 However, our finding of up to 92% functional agreement between corresponding sodium channel variants illustrates that SCN paralogues can aid variant interpretation and guide precision treatment.
In comparison, agreement between the existing ‘funNCion’ prediction technique and gold standard patch-clamp recordings ranged between 50–96% (Fig. 5). This lack in precision of the ‘funNCion’ tool may in part be explained as the tool does not consider biophysical evidence but infers channel function from clinical datasets. We noticed that specific protein locations could be predicted with better accuracy than others. For example, functionally homogeneous GoF areas such as the D3–4 linker inactivation gate region and LoF areas such as the S5–6 linker pore region were predicted with high confidence. In contrast, mixed gain- and loss-of-function areas such as the S4 segments that include different arginine residues appear more challenging to predict using the ‘funNCion’ tool.
Compared to the current gold standard, our approach is very effective and >90% accurate. We are not aware of any other techniques that would allow such immediate SCN functional prediction. Molecular dynamics can be useful for understanding specific channel properties including the movement of ions through the pore, but not for processes like inactivation and does not allow prediction of overall function.81 AlphaFold is a promising new approach to solving static protein structures that may be combined with molecular dynamics. However, for the present it may be envisaged that collated functional data from published electrophysiological studies will be needed to affirm predictions from molecular dynamics and AlphaFold in the future. At present, these approaches are too computationally expensive for prediction of the overall functional change caused by a variant.82
Based on our findings we created a freely accessible ‘SCN-Viewer’, allowing clinicians and scientists immediate access to published biophysical data across all voltage gated sodium channels (http://SCN-viewer.broadinstitute.org). The tool enables users to identify any SCN-paralogues detailing both functional and clinical characteristics associated with specific SCN1-11A variants.
In practice, when a new voltage gated sodium channel missense variant is identified and pathogenicity has been established by American College of Medical Genetics criteria, we propose the following key indicators to estimate the variants functional properties (Box 1). In the absence of gold standard functional data, these tools offer clinicians and scientists valuable insights when interpreting newly identified variants. Whilst this approach is not intended to replace clinical judgment, it will inform and complement clinical decision making based on objective and quantifiable data.
Box 1. Approach to functional SCN variant estimation.
Key indicators of variant characteristics
Identical missense variant in a paralogue sodium channel has been functionally characterized (http://SCN-viewer.broadinstitute.org): up to 92% likelihood that reported findings apply to new variant.
Use of in silico functional prediction tool (http://funNCion.broadinstitute.org): 58–96% likelihood that prediction applies to new variant (depending on variant location)’
Similar missense variant at equivalent position in a paralogue sodium channel has been reported in a related characteristic gain- or loss-of-function SCN disorder (http://per.broadinstitute.org): it is likely that the new variant is in keeping with the reported phenotypes.
There are several limitations to this study. We limited our literature search to peer reviewed studies reported in PubMed and sodium channel databases and did not include other sources. Whilst a number of reports will have been missed, this is unlikely to affect our main findings of functional similarity among different SCN-genes. Our simplified approach of variant categorization into gain-, loss- and mixed function often does not fully reflect the complex biophysical properties of SCNs; however, our data illustrate that there is value in this pragmatic approach of functional variant categorization.
Conclusion
Our findings suggest that biophysical characterization of variants in one SCN-gene can predict channel function across different SCN-genes where experimental data are not available. Shared patterns of functional effects aid variant interpretation and guide precision therapy.
Supplementary Material
Contributor Information
Andreas Brunklaus, The Paediatric Neurosciences Research Group, Royal Hospital for Children, Glasgow, UK; Institute of Health and Wellbeing, University of Glasgow, Glasgow, UK.
Tony Feng, The Paediatric Neurosciences Research Group, Royal Hospital for Children, Glasgow, UK; Institute of Health and Wellbeing, University of Glasgow, Glasgow, UK.
Tobias Brünger, Cologne Center for Genomics, University of Cologne, Cologne, Germany.
Eduardo Perez-Palma, Centro de Genética y Genómica, Facultad de Medicina Clínica Alemana, Universidad del Desarrollo, Santiago, Chile.
Henrike Heyne, Genomic and Personalized Medicine, Digital Health Center, Hasso Plattner Institute, Potsdam, Germany; Hasso Plattner Institute, Mount Sinai School of Medicine, New York, NY, USA; Institute for Molecular Medicine Finland: FIMM, Helsinki, Finland.
Emma Matthews, Atkinson Morley Neuromuscular Centre, St George’s University Hospitals NHS Foundation Trust, London, UK; Molecular and Clinical Sciences Research Institute, St George’s University of London, London, UK.
Christopher Semsarian, Agnes Ginges Centre for Molecular Cardiology at Centenary Institute, The University of Sydney, Sydney, Australia; Sydney Medical School Faculty of Medicine and Health, The University of Sydney, Sydney, Australia; Department of Cardiology, Royal Prince Alfred Hospital, Sydney, Australia.
Joseph D Symonds, The Paediatric Neurosciences Research Group, Royal Hospital for Children, Glasgow, UK; Institute of Health and Wellbeing, University of Glasgow, Glasgow, UK.
Sameer M Zuberi, The Paediatric Neurosciences Research Group, Royal Hospital for Children, Glasgow, UK; Institute of Health and Wellbeing, University of Glasgow, Glasgow, UK.
Dennis Lal, Epilepsy Center, Neurological Institute, Cleveland Clinic, Cleveland, USA; Stanley Center for Psychiatric Genetics, Broad Institute of MIT and Harvard, Cambridge, MA, USA.
Stephanie Schorge, Department of Neuroscience, Physiology and Pharmacology, UCL, London, UK.
Funding
D.L. and E.P. received a grant from the Dravet Syndrome Foundation (grant number, 272016) to support this work. E.P. is supported by the Agencia Nacional de Investigación y Desarrollo (ANID, PAI77200124) of Chile and the FamilieSCN2A foundation 2020 Action Potential Grant. A.B. and S.Z. received a grant from Dravet syndrome UK (grant number, 16GLW00) for the Glasgow SCN1A database. C.S. is the recipient of a National Health and Medical Research Council (NHMRC) Practitioner Fellowship (#1154992).
Competing interests
A.B. has received honoraria for presenting at educational events, advisory boards and consultancy work for Biocodex, GW Pharma, Encoded Therapeutics, Stoke Therapeutics, Nutricia and Zogenix. E.P. has received honoraria for consultancy work for the Friends of Faces foundation. S.M.Z. has received honoraria for presenting at educational events, advisory boards and consultancy work for GW Pharma, Zogenix, Biocodex, Encoded Therapeutics, Stoke Therapeutics and Nutricia. D.L. has received honoraria for advisory board work for Encoded Therapeutics. No other competing interests are reported.
Supplementary material
Supplementary material is available at Brain online.
References
- 1.Goldin AL, Barchi RL, Caldwell JH, et al. . Nomenclature of voltage-gated sodium channels. Neuron. 2000;28(2):365–368. [DOI] [PubMed] [Google Scholar]
- 2.Yu FH, Catterall WA. Overview of the voltage-gated sodium channel family. Genome Biol. 2003;4(3):207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117(4):500–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spillane J, Kullmann DM, Hanna MG. Genetic neurological channelopathies: molecular genetics and clinical phenotypes. J Neurol Neurosurg Psychiatry. 2016;87(1):37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunklaus A, Ellis R, Reavey E, Semsarian C, Zuberi SM. Genotype phenotype associations across the voltage-gated sodium channel family. J Med Genet. 2014;51(10):650–658. [DOI] [PubMed] [Google Scholar]
- 6.Catterall WA, Goldin AL, Waxman SG. International union of pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol Rev. 2005;57(4):397–409. [DOI] [PubMed] [Google Scholar]
- 7.Yu FH, Catterall WA. The VGL-chanome: a protein superfamily specialized for electrical signaling and ionic homeostasis. Sci STKE. 2004;2004(253):re15. [DOI] [PubMed] [Google Scholar]
- 8.Brunklaus A. Precision medicine in sodium channelopathies - Moving beyond seizure control towards disease modification. Eur J Paediatr Neurol. 2020;24:7. [DOI] [PubMed] [Google Scholar]
- 9.Yu FH, Mantegazza M, Westenbroek RE, et al. . Reduced sodium current in GABAergic interneurons in a mouse model of severe myoclonic epilepsy in infancy. Nat Neurosci. 2006;9(9):1142–1149. [DOI] [PubMed] [Google Scholar]
- 10.Mantegazza M, Broccoli V. SCN1A/NaV1.1 channelopathies: Mechanisms in expression systems, animal models, and human iPSC models. Epilepsia. 2019;60(S3):S25–S38. [DOI] [PubMed] [Google Scholar]
- 11.Cestèle S, Scalmani P, Rusconi R, Terragni B, Franceschetti S, Mantegazza M. Self-limited hyperexcitability: Functional effect of a familial hemiplegic migraine mutation of the Nav1.1 (SCN1A) Na+ channel. J Neurosci. 2008;28(29):7273–7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cestèle S, Schiavon E, Rusconi R, Franceschetti S, Mantegazza M. Nonfunctional NaV1.1 familial hemiplegic migraine mutant transformed into gain of function by partial rescue of folding defects. Proc Natl Acad Sci USA. 2013;110(43):17546–17551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kahlig KM, Rhodes TH, Pusch M, et al. . Divergent sodium channel defects in familial hemiplegic migraine. Proc Natl Acad Sci USA. 2008;105(28):9799–9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barbieri R, Bertelli S, Pusch M, Gavazzo P. Late sodium current blocker GS967 inhibits persistent currents induced by familial hemiplegic migraine type 3 mutations of the SCN1A gene. J Headache Pain. 2019;20(1):107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berkovic SF, Heron SE, Giordano L, et al. . Benign familial neonatal-infantile seizures: characterization of a new sodium channelopathy. Ann Neurol. 2004;55(4):550–557. [DOI] [PubMed] [Google Scholar]
- 16.Nakamura K, Kato M, Osaka H, et al. . Clinical spectrum of SCN2A mutations expanding to Ohtahara syndrome. Neurology. 2013;81(11):992–998. [DOI] [PubMed] [Google Scholar]
- 17.Shi X, Yasumoto S, Kurahashi H, et al. . Clinical spectrum of SCN2A mutations. Brain Dev. 2012;34(7):541–545. [DOI] [PubMed] [Google Scholar]
- 18.Bennett PB, Yazawa K, Makita N, George AL, Jr. Molecular mechanism for an inherited cardiac arrhythmia. Nature. 1995;376(6542):683–685. [DOI] [PubMed] [Google Scholar]
- 19.Fonseca DJ, Vaz da Silva MJ. Cardiac channelopathies: The role of sodium channel mutations. Rev Port Cardiol. 2018;37(2):179–199. [DOI] [PubMed] [Google Scholar]
- 20.Jurkat-Rott K, Holzherr B, Fauler M, Lehmann-Horn F. Sodium channelopathies of skeletal muscle result from gain or loss of function. Pflugers Arch. 2010;460(2):239–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SC, Kim HS, Park YE, Choi YC, Park KH, Kim DS. Clinical diversity of SCN4A-mutation-associated skeletal muscle sodium channelopathy. J Clin Neurol. 2009;5(4):186–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waxman SG. Painful Na-channelopathies: an expanding universe. Trends Mol Med. 2013;19(7):406–409. [DOI] [PubMed] [Google Scholar]
- 23.Faber CG, Lauria G, Merkies ISJ, et al. . Gain-of-function Nav1.8 mutations in painful neuropathy. Proc Natl Acad Sci USA. 2012;109(47):19444–19449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J, Han C, Estacion M, et al. . Gain-of-function mutations in sodium channel Na(v)1.9 in painful neuropathy. Brain. 2014;137(Pt 6):1627–1642. [DOI] [PubMed] [Google Scholar]
- 25.Elia N, Palmio J, Castañeda MS, et al. . Myasthenic congenital myopathy from recessive mutations at a single residue in Na(V)1.4. Neurology. 2019;92(13):e1405–e1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Habbout K, Poulin H, Rivier F, et al. . A recessive Nav1.4 mutation underlies congenital myasthenic syndrome with periodic paralysis. Neurology. 2016;86(2):161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolff M, Johannesen KM, Hedrich UBS, et al. . Genetic and phenotypic heterogeneity suggest therapeutic implications in SCN2A-related disorders. Brain. 2017;140(5):1316–1336. [DOI] [PubMed] [Google Scholar]
- 28.Sanders SJ, Campbell AJ, Cottrell JR, et al. . Progress in understanding and treating SCN2A-mediated disorders. Trends Neurosci. 2018;41(7):442–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Møller RS, Johannesen KM. Precision medicine: SCN8A encephalopathy treated with sodium channel blockers. Neurotherapeutics. 2016;13(1):190–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johannesen KM, Liu Y, Koko M, et al. . Genotype-phenotype correlations in SCN8A-related disorders reveal prognostic and therapeutic implications. Brain. 2022;145:2991–3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunklaus A, Ellis R, Reavey E, Forbes GH, Zuberi SM. Prognostic, clinical and demographic features in SCN1A mutation-positive Dravet syndrome. Brain. 2012;135(Pt 8):2329–2336. [DOI] [PubMed] [Google Scholar]
- 32.Guerrini R, Dravet C, Genton P, Belmonte A, Kaminska A, Dulac O. Lamotrigine and seizure aggravation in severe myoclonic epilepsy. Epilepsia. 1998;39(5):508–512. [DOI] [PubMed] [Google Scholar]
- 33.Mazzanti A, Maragna R, Faragli A, et al. . Gene-specific therapy with mexiletine reduces arrhythmic events in patients with long QT syndrome type 3. J Am Coll Cardiol. 2016;67(9):1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kambouris NG, Nuss HB, Johns DC, Marbán E, Tomaselli GF, Balser JR. A revised view of cardiac sodium channel “blockade” in the long-QT syndrome. J Clin Invest. 2000;105(8):1133–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Priori SG, Wilde AA, Horie M, et al. . HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes: document endorsed by HRS, EHRA, and APHRS in May 2013 and by ACCF, AHA, PACES, and AEPC in June 2013. Heart Rhythm. 2013;10(12):1932–1963. [DOI] [PubMed] [Google Scholar]
- 36.Brunklaus A, Schorge S, Smith AD, et al. . SCN1A variants from bench to bedside-improved clinical prediction from functional characterization. Hum Mutat. 2020;41(2):363–374. [DOI] [PubMed] [Google Scholar]
- 37.Pérez-Palma E, May P, Iqbal S, et al. . Identification of pathogenic variant enriched regions across genes and gene families. Genome Res. 2020;30(1):62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heyne HO, Baez-Nieto D, Iqbal S, et al. . Predicting functional effects of missense variants in voltage-gated sodium and calcium channels. Sci Transl Med. 2020;12(556):eaay6848. [DOI] [PubMed] [Google Scholar]
- 39.Meisler MH, Hill SF, Yu W. Sodium channelopathies in neurodevelopmental disorders. Nat Rev Neurosci. 2021;22(3):152–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mantegazza M, Cestèle S, Catterall WA. Sodium channelopathies of skeletal muscle and brain. Physiol Rev. 2021;101(4):1633–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brunklaus A, Du J, Steckler F, et al. . Biological concepts in human sodium channel epilepsies and their relevance in clinical practice. Epilepsia. 2020;61(3):387–399. [DOI] [PubMed] [Google Scholar]
- 42.Rhodes TH, Lossin C, Vanoye CG, Wang DW, George AL Jr. Noninactivating voltage-gated sodium channels in severe myoclonic epilepsy of infancy. Proc Natl Acad Sci USA. 2004;101(30):11147–11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson CH, Porter JC, Kahlig KM, Daniels MA, George AL Jr. Nontruncating SCN1A mutations associated with severe myoclonic epilepsy of infancy impair cell surface expression. J Biol Chem. 2012;287(50):42001–42008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fleischhauer R, Mitrovic N, Deymeer F, Lehmann-Horn F, Lerche H. Effects of temperature and mexiletine on the F1473S Na+ channel mutation causing paramyotonia congenita. Pflugers Arch. 1998;436(5):757–765. [DOI] [PubMed] [Google Scholar]
- 45.Bechi G, Rusconi R, Cestèle S, Striano P, Franceschetti S, Mantegazza M. Rescuable folding defective NaV1.1 (SCN1A) mutants in epilepsy: properties, occurrence, and novel rescuing strategy with peptides targeted to the endoplasmic reticulum. Neurobiol Dis. 2015;75:100–114. [DOI] [PubMed] [Google Scholar]
- 46.Fertleman CR, Baker MD, Parker KA, et al. . SCN9A mutations in paroxysmal extreme pain disorder: allelic variants underlie distinct channel defects and phenotypes. Neuron. 2006;52(5):767–774. [DOI] [PubMed] [Google Scholar]
- 47.Dib-Hajj SD, Estacion M, Jarecki BW, et al. . Paroxysmal extreme pain disorder M1627K mutation in human Nav1.7 renders DRG neurons hyperexcitable. Mol Pain. 2008;4:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Theile JW, Jarecki BW, Piekarz AD, Cummins TR. Nav1.7 mutations associated with paroxysmal extreme pain disorder, but not erythromelalgia, enhance Navbeta4 peptide-mediated resurgent sodium currents. J Physiol. 2011;589(Pt 3):597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winkel BG, Yuan L, Olesen MS, et al. . The role of the sodium current complex in a nonreferred nationwide cohort of sudden infant death syndrome. Heart Rhythm. 2015;12(6):1241–1249. [DOI] [PubMed] [Google Scholar]
- 50.Abdelsayed M, Baruteau AE, Gibbs K, et al. . Differential calcium sensitivity in NaV 1.5 mixed syndrome mutants. J Physiol. 2017;595(18):6165–6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rusconi R, Scalmani P, Cassulini RR, et al. . Modulatory proteins can rescue a trafficking defective epileptogenic Nav1.1 Na+ channel mutant. J Neurosci. 2007;27(41):11037–11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Catterall WA. Structure and function of voltage-gated sodium channels at atomic resolution. Exp Physiol. 2014;99(1):35–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zuberi SM, Brunklaus A, Birch R, Reavey E, Duncan J, Forbes GH. Genotype-phenotype associations in SCN1A-related epilepsies. Neurology. 2011;76(7):594–600. [DOI] [PubMed] [Google Scholar]
- 54.Tarradas A, Selga E, Beltran-Alvarez P, et al. . A novel missense mutation, I890T, in the pore region of cardiac sodium channel causes Brugada syndrome. PLoS One. 2013;8(1):e53220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liebeskind BJ, Hillis DM, Zakon HH. Evolution of sodium channels predates the origin of nervous systems in animals. Proc Natl Acad Sci USA. 2011;108(22):9154–9159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Valdivia CR, Ackerman MJ, Tester DJ, et al. . A novel SCN5A arrhythmia mutation, M1766L, with expression defect rescued by mexiletine. Cardiovasc Res. 2002;55(2):279–289. [DOI] [PubMed] [Google Scholar]
- 57.Bechi G, Rusconi R, Cestele S, Striano P, Franceschetti S, Mantegazza M. Rescuable folding defective NaV1.1 (SCN1A) mutants in epilepsy: properties, occurrence, and novel rescuing strategy with peptides targeted to the endoplasmic reticulum. Neurobiol Dis. 2015;75:100–114. [DOI] [PubMed] [Google Scholar]
- 58.Dhifallah S, Lancaster E, Merrill S, Leroudier N, Mantegazza M, Cesteèle S. Gain of function for the SCN1A/hNav1.1-L1670W mutation responsible for familial hemiplegic migraine. Front Mol Neurosci. 2018;11:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Horga A, Raja Rayan DL, Matthews E, et al. . Prevalence study of genetically defined skeletal muscle channelopathies in England. Neurology. 2013;80(16):1472–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loussouarn G, Sternberg D, Nicole S, et al. . Physiological and pathophysiological insights of Nav1.4 and Nav1.5 comparison. Front Pharmacol. 2015;6:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Antzelevitch C, Brugada P, Borggrefe M, et al. . Brugada syndrome: report of the second consensus conference. Heart Rhythm. 2005;2(4):429–440. [DOI] [PubMed] [Google Scholar]
- 62.Ku CS, Polychronakos C, Tan EK, et al. . A new paradigm emerges from the study of de novo mutations in the context of neurodevelopmental disease. Mol Psychiatry. 2013;18(2):141–153. [DOI] [PubMed] [Google Scholar]
- 63.Holland KD, Bouley TM, Horn PS. Location: A surrogate for personalized treatment of sodium channelopathies. Ann Neurol. 2018;84(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yoshinaga H, Sakoda S, Shibata T, et al. . Phenotypic variability in childhood of skeletal muscle sodium channelopathies. Pediatr Neurol. 2015;52(5):504–508. [DOI] [PubMed] [Google Scholar]
- 65.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15(7):901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Balestrini S, Chiarello D, Gogou M, et al. . Real-life survey of pitfalls and successes of precision medicine in genetic epilepsies. J Neurol Neurosurg Psychiatry. 2021;92(10):1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.National Research Council. Toward precision medicine: building a knowledge network for biomedical research and a new taxonomy of disease. The National Academies Press; 2011:142. [PubMed] [Google Scholar]
- 68.Gardella E, Møller RS. Phenotypic and genetic spectrum of SCN8A-related disorders, treatment options, and outcomes. Epilepsia. 2019;60(Suppl 3):S77–S85. [DOI] [PubMed] [Google Scholar]
- 69.Suetterlin KJ, Bugiardini E, Kaski JP, et al. . Long-term safety and efficacy of mexiletine for patients with skeletal muscle channelopathies. JAMA Neurol. 2015;72(12):1531–1553. [DOI] [PubMed] [Google Scholar]
- 70.Mann SA, Castro ML, Ohanian M, et al. . R222Q SCN5A mutation is associated with reversible ventricular ectopy and dilated cardiomyopathy. J Am Coll Cardiol. 2012;60(16):1566–1573. [DOI] [PubMed] [Google Scholar]
- 71.Sadleir LG, Mountier EI, Gill D, et al. . Not all SCN1A epileptic encephalopathies are Dravet syndrome: Early profound Thr226Met phenotype. Neurology. 2017;89(10):1035–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Berecki G, Bryson A, Terhag J, et al. . SCN1A gain of function in early infantile encephalopathy. Ann Neurol. 2019;85(4):514–525. [DOI] [PubMed] [Google Scholar]
- 73.Anderson LL, Thompson CH, Hawkins NA, et al. . Antiepileptic activity of preferential inhibitors of persistent sodium current. Epilepsia. 2014;55(8):1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Männikkö R, Shenkarev ZO, Thor MG, et al. . Spider toxin inhibits gating pore currents underlying periodic paralysis. Proc Natl Acad Sci USA. 2018;115(17):4495–4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weuring WJ, Singh S, Volkers L, et al. . NaV1.1 and NaV1.6 selective compounds reduce the behavior phenotype and epileptiform activity in a novel zebrafish model for Dravet Syndrome. PLoS One. 2020;15(3):e0219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xu H, Li T, Rohou A, et al. . Structural basis of Nav1.7 inhibition by a gating-modifier spider toxin. Cell. 2019;176(4):702–715. [DOI] [PubMed] [Google Scholar]
- 77.Han Z, Chen C, Christiansen A, et al. . Antisense oligonucleotides increase Scn1a expression and reduce seizures and SUDEP incidence in a mouse model of Dravet syndrome. Sci Transl Med. 2020;12(558):eaaz6100. [DOI] [PubMed] [Google Scholar]
- 78.Matthews E, Labrum R, Sweeney MG, et al. . Voltage sensor charge loss accounts for most cases of hypokalemic periodic paralysis. Neurology. 2009;72(18):1544–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Castañeda MS, Zanoteli E, Scalco RS, et al. . A novel ATP1A2 mutation in a patient with hypokalaemic periodic paralysis and CNS symptoms. Brain. 2018;141(12):3308–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Xu H, Li T, Rohou A, et al. . Structural basis of Nav1.7 inhibition by a gating-modifier spider toxin. Cell. 2019;176(4):702–715.e14. [DOI] [PubMed] [Google Scholar]
- 81.Mason ER, Wu F, Patel RR, Xiao Y, Cannon SC, Cummins TR. Resurgent and gating pore currents induced by De Novo SCN2A epilepsy mutations. eNeuro. 2019;6(5):ENEURO.0141-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dunlop J, Bowlby M, Peri R, Vasilyev D, Arias R. High-throughput electrophysiology: an emerging paradigm for ion-channel screening and physiology. Nat Rev Drug Discov. 2008;7(4):358–368. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data used in this study are available in the manuscript, the Supplementary material and via the free online webtool http://SCN-viewer.broadinstitute.org.