CORENODE, a computational network decomposition algorithm, identifies an adaptive transcriptional mechanism of leukemia immune escape through combinatorial fluctuations of transcription factor expression.
Abstract
Relapse of acute myeloid leukemia (AML) after allogeneic bone marrow transplantation has been linked to immune evasion due to reduced expression of major histocompatibility complex class II (MHCII) genes through unknown mechanisms. In this work, we developed CORENODE, a computational algorithm for genome-wide transcription network decomposition that identified a transcription factor (TF) tetrad consisting of IRF8, MYB, MEF2C, and MEIS1, regulating MHCII expression in AML cells. We show that reduced MHCII expression at relapse is transcriptionally driven by combinatorial changes in the expression of these TFs, where MYB and IRF8 play major opposing roles, acting independently of the IFNγ/CIITA pathway. Beyond the MHCII genes, MYB and IRF8 antagonistically regulate a broad genetic program responsible for cytokine signaling and T-cell stimulation that displays reduced expression at relapse. A small number of cells with altered TF abundance and silenced MHCII expression are present at the time of initial leukemia diagnosis, likely contributing to eventual relapse.
Significance:
Our findings point to an adaptive transcriptional mechanism of AML evolution after allogeneic transplantation whereby combinatorial fluctuations of TF expression under immune pressure result in the selection of cells with a silenced T-cell stimulation program.
This article is highlighted in the In This Issue feature, p. 369
INTRODUCTION
Treatment of acute myeloid leukemia (AML) with curative intent often utilizes allogeneic hematopoietic stem cell transplantation (alloSCT), which relies on the graft-versus-leukemia effect (GvL), whereby donor-derived T cells eliminate residual leukemia cells (1). Although the immunologic mechanisms of GvL are complex, relapse of AML after allogeneic transplantation has been linked to the loss of major histocompatibility complex class II (MHCII) expression (2–4). Notably, treatment of AML blasts with IFNγ restored MHCII expression, and no new DNA coding mutations were detected in comparative analyses of pre- and post-alloSCT relapsed samples (3, 4). Thus, the reduced expression of MHCII at relapse appears to have a regulatory basis. Decreased expression of the MHCII transcriptional coactivator CIITA was observed in some cases (4), but loss of MHCII expression was also seen despite unchanged or elevated expression of CIITA (3), suggesting alternative mechanisms of immune escape.
RESULTS
Network Decomposition by Combinatorial Regression
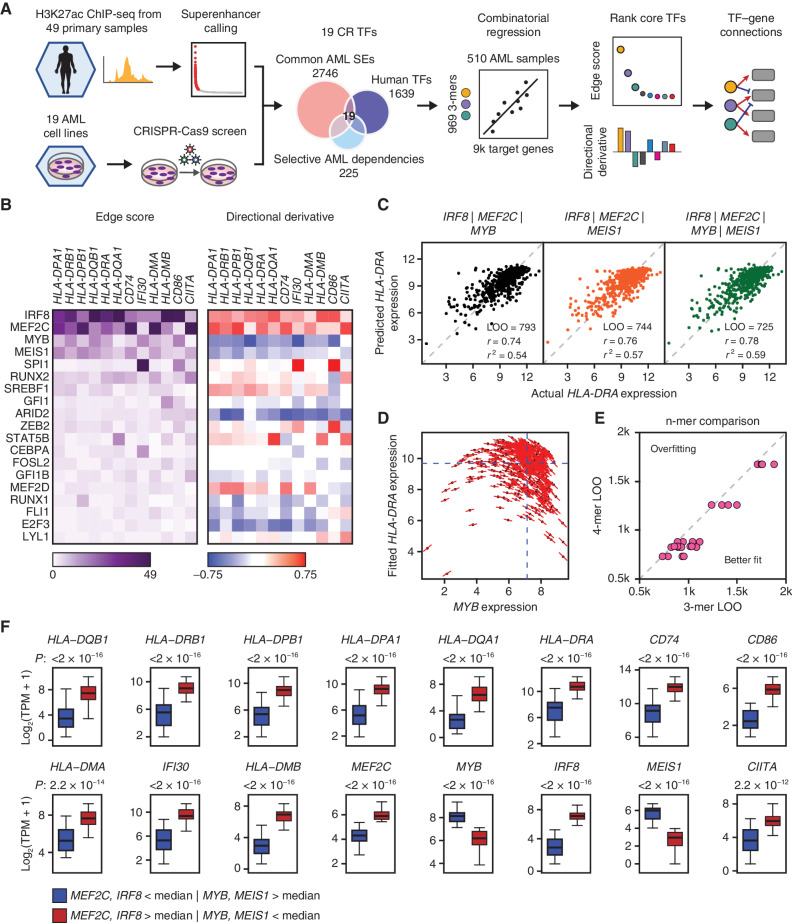
To identify the transcriptional regulators of MHCII in AML, we sought to reverse engineer a transcription regulatory network from public genome-scale data sets (Fig. 1A). In principle, the expression of a gene in a given context may be computationally inferred from the identities of its transcriptional regulators, their cellular abundance, and the gene-regulatory function that represents the quantitative relationship between a transcription factor (TF) and its target gene (5, 6). We began by defining a list of core regulatory (CR) TFs for AML lineage specification (7). Genes encoding CR TFs are typically marked by extended, closely spaced enhancers with markedly high histone acetylation and cofactor recruitment, termed superenhancers (SE; ref. 8). Among a chromatin immunoprecipitation sequencing (ChIP-seq) data set for the enhancer histone mark H3K27ac in a panel of 49 primary human AML samples (9), we identified 1,298 SEs present in at least 10 (∼20%) samples (Supplementary Fig. S1A–S1C). These common SEs were proximal to 2,748 genes, of which 220 encoded TFs. Because CR TFs are typically essential for lineage survival (10, 11), we focused this list further by asking which of the 220 SE-associated TFs were dependencies selective for AML versus other malignancies. We interrogated data from the Broad Cancer Dependency Map project, a collection of genome-scale CRISPR knockout screens of 18,119 genes in 769 cell lines, including 19 AML lines (12, 13). Applying a skewed-LRT test (14) to compare guide RNA drop out between AML and non-AML cell lines, we identified 40 TFs selectively essential in AML (Supplementary Fig. S1D). We then intersected the 40 essential TFs with the 220 SE-driven TFs, resulting in a list of 19 putative CR TFs (Fig. 1A).
Figure 1.
CORENODE identifies a tetrad of TFs regulating MHCII genes in AML. A, Study schematic. B, Heat maps of edge scores (ES) and directional derivatives (DD) representing computationally established edges between target genes (columns) and TFs (rows, sorted by average ES). Higher ES corresponds to higher confidence edges, and DD predicts amplitude and directionality (positive vs. negative) of TF-target regulation. C, 3- and 4-mer CORENODE fits for the gene HLA-DRA with indicated goodness-of-fit metrics. D, A graphic illustration of DD. MYB expression is plotted against the CORENODE-fitted HLA-DRA expression using an IRF8/MEF2C/MYB 3-mer. Red sticks indicate MYB slopes aggregated from the 4 MYB-containing terms (linear, quadratic, and two cross-terms) in each sample. Aggregation of all MYB slopes for all samples produces MYB DD. E, Leave-one-out (LOO) error improvement between 3-mers [4 per gene (4 combinations of 4 TFs taken 3 at a time)] and the 4-mer (1 per gene, combining all 4 TFs) for 6 MHCII genes. F, Gene expression in two populations of the Beat AML data set. The blue population (n = 46) represents patient samples with below-median expression of IRF8 and MEF2C and above-median expression of MYB and MEIS1, and the red population (n = 56) represents patient samples with the opposite pattern of TF expression. The elements of the box plots are as follows: center line, median; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range.
We sought next to define the gene-regulatory functions linking CR TFs to all expressed genes. We developed CORENODE (COmbinatorial REgression for NetwOrk DEcomposition), a coexpression-based network decomposition algorithm where TF–target gene connections, or edges, are inferred from naturally occurring variation in gene expression within a reasonably large set of samples. We hypothesized that the expression of each gene could be estimated from the expression of a small subset, possibly 3 to 5 TFs, of the entire CR TF set. Given that the intersection of dependency and SE profiling data nominated only 19 TFs, it was computationally feasible to design a comprehensive combinatorial regression approach to fit expression of each target gene against all possible combinations of the CR TFs (Fig. 1A; Supplementary Fig. S1A). Using the protein-coding transcriptomes of 510 primary AML samples from the Beat AML study (15), we regressed the mRNA expression values of all genes in the expressed genome against mRNA expression values of the 19 CR TFs taken 3 at a time, using linear, quadratic, and cross terms. Use of 3-mers provided the best balance between fit quality and overfitting risk genome-wide. Thus, for every target gene, we generated 969 gene-regulatory functions corresponding to all possible 3-mer combinations of the 19 TFs. We chose the top 5% of best-fitting 3-mers and calculated an edge score (ES) for each TF–target gene pair as the number of times the TF appeared in the top 5% of best fits. Finally, we estimated the direction and amplitude of positive or negative regulation by aggregating response slopes for each TF across all samples and 3-mers, which was termed directional derivative (DD). Details of the computational algorithm are found in the Supplementary Note and online data browser (https://corenode.shinyapps.io/corenode/). Genome-wide ES and DD outputs are found in Supplementary Data S1 and S2, respectively.
A Transcription Factor Tetrad Regulates MHCII Genes in AML
CORENODE prioritized four TFs (IRF8, MYB, MEF2C, and MEIS1) as candidate regulators of the MHCII and several additional genes involved in antigen presentation that have been associated with immune escape in AML (CD74, IFI30, HLA-DMA, HLA-DMB, and CD86; ref. 3; Fig. 1B). MYB and MEIS1 were predicted to negatively regulate the expression of these genes, whereas IRF8 and MEF2C were predicted to be positive regulators (Fig. 1B–D). In parallel work, we used additional data sets and criteria to define an extended AML core regulatory circuit of 31 members (16). For additional validation, we repeated CORENODE regressions with this extended set of TFs, as well as a 37-member set that included all selectively essential AML TFs above a minimal expression cutoff and regardless of the SE status. In all cases, the same 4 TFs were predicted to be the top regulators of MHCII expression (Supplementary Fig. S2A–S2D). Combining these TFs in a single 4-mer resulted in a modest improvement of the regression fits (compared with 3-mers) without overfitting (Fig. 1C and E; Supplementary Fig. S3A; Supplementary Data S3).
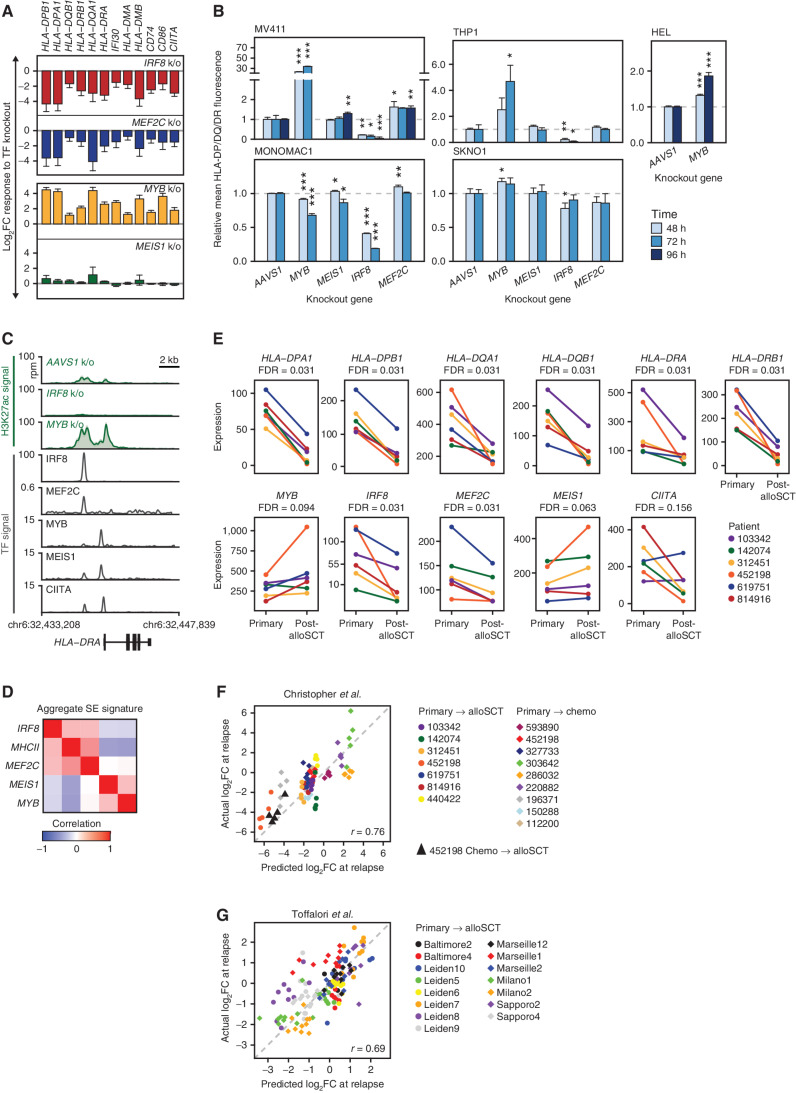
We examined how variation in the expression of the TF tetrad correlated with changes in MHCII expression in two independent data sets, Beat AML and TCGA, including 510 and 151 patients, respectively (15, 17). In both data sets, the expression of MHCII and other genes associated with immune escape in patients with high (above the median) IRF8/MEF2C and low (below the median) MYB/MEIS1 was up to 17-fold higher than in patients with the opposite pattern of TF expression (Fig. 1F; Supplementary Fig. S3B). To validate the impact of these TFs on MHCII regulation, we inactivated each TF with CRISPR/Cas9 editing followed by RNA-seq in an AML cell line (Supplementary Fig. S4A and S4B; Supplementary Data S4). Consistent with predictions of CORENODE, loss of IRF8 and MEF2C led to reduced expression of MHCII and other genes associated with immune escape, whereas loss of MYB and MEIS1 was associated with increased expression (Fig. 2A). We confirmed these findings at the protein level by measuring the cell-surface expression of MHCII molecules after TF knockout in a panel of AML cell lines (Fig. 2B). Overall, MYB and IRF8 had a major impact on MHCII expression, whereas the effects of MEIS1 and MEF2C were more subtle and variable between cell lines, suggesting context-specific roles of MEIS1 and MEF2C as fine-tuners of MHCII expression. One cell line with a particularly high baseline MHCII expression (MONOMAC1) demonstrated decreased, rather than increased, MHCII expression after MYB knockout (Fig. 2B; Supplementary Fig. S4B), but we found this effect to be epistatic with CIITA and reversed after a CIITA knockout (see below). Treatment of AML cells with the MYB inhibitor mebendazole (18) resulted in a modest induction of MHCII expression (Supplementary Fig. S4C), consistent with the effect of MYB knockout.
Figure 2.
IRF8, MEF2C, MYB, and MEIS1 regulate MHCII expression in AML. A, Transcriptional response of the indicated genes measured by mRNA-seq 72 hours after TF knockout. The experiment was performed in biological triplicates. Error bars represent standard error. MEF2C knockout changes were not significant after genome-wide adjustment for multiple hypothesis testing. Among MEIS1 knockout–induced changes only HLA-DRA and HLA-DQB1 had q-values <0.1. All shown MYB and IRF8 knockout–induced changes had q-values of <0.05. B, Changes in the surface expression of the MHCII molecules detected by immunostaining with a pan-specific anti–HLA-DP/DQ/DR antibody following TF knockout. An AAVS1 (“safe harbor”) targeting sgRNA was used as a control. The bar plots represent average values obtained from 3 replicates normalized to the AAVS1 control. Asterisks indicate P values calculated by a two-tailed t test comparing MHCII fluorescence between gene knockout and the AAVS1 control at the same time point. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. C, H3K27 acetylation and TF binding in the HLA-DRA locus. Green tracks are H3K27ac metatracks composed of overlaying semitransparent area plots representing two biological replicates of the indicated TF knockouts compared with AAVS1 control. The average profile is represented by a thick line. Gray tracks represent the binding of the indicated TFs. All of the shown ChIP-seq experiments were performed in MV411 except for CIITA, which was downloaded from ref. (38) and represents a lymphoid cell line. Refer to Supplementary Fig. S4 for a map of H3K27 acetylation and TF binding in the entire MHCII locus and genome-wide analysis of H3K27ac changes. D, Similarity matrix of SE scores associated with the TF tetrad and MHCII genes using data from ref. (9) E, mRNA expression in six paired primary and post-alloSCT samples from ref. (3). F and G, CORENODE accurately predicts MHCII expression changes at relapse. The plots depict predicted by CORENODE vs. observed log2 fold change in MHCII expression in paired samples from ref. (3). F, and ref. (4). G, between initial presentation and relapse. For each patient (color-coded as indicated) each data point reflects predicted and observed changes in the expression of one MHCII gene. The IRF8/MYB/MEIS1/MEF2C 4-mer was used in F, and CIITA was added as a fifth term in G resulting in a better fit.
The MHCII locus contains several SEs whose H3K27 acetylation correlates with the expression of MHCII genes in primary AML cells (Supplementary Fig. S5A). Using ChIP-seq, we confirmed binding of the TF tetrad at the MHCII SEs (Fig. 2C; Supplementary Fig. S5B). Spike-in controlled ChIP-seq analysis detected a significant increase in H3K27 acetylation at these SEs after MYB knockout, despite the expected global decrease of H3K27 acetylation elsewhere across the genome (Fig. 2C; Supplementary Fig. S5B and S5C). In contrast, IRF8 knockout was accompanied by the loss of H3K27 acetylation. Furthermore, the SEs associated with IRF8 and MEF2C in primary AML blast cells positively correlated with MHCII SEs, whereas SEs controlling MYB and MEIS1 negatively correlated (Fig. 2D; Supplementary Fig. S5D). These data provided further evidence of antagonistic functions of these TFs in MHCII regulation.
We next examined how changes in the MHCII gene expression after alloSCT correlated with expression of the TF tetrad. We accessed the published RNA-seq data from paired AML samples at initial presentation and relapse after alloSCT, where decreased MHCII expression was observed in 6 of the 7 examined patients (3). Despite a substantial variability of baseline expression, we observed significant downregulation of IRF8 and, to a lesser extent, MEF2C in all patients with reduced MHCII expression at relapse (Fig. 2E). In contrast, MYB and MEIS1 were upregulated in 5 of 6 patients at relapse, which was of borderline statistical significance. Each of these changes would be expected to lead to reduced expression of MHCII genes.
A Combinatorial Transcriptional Mechanism of Immune Escape
To this point, our computational predictions and knockout experiments demonstrated that perturbation of any one of the 4 candidate TFs was sufficient to effect a change in MHCII expression, and examination of AML relapses revealed changes in the abundance of these TFs in the direction consistent with reduced MHCII expression. However, the degree of observed TF change was often modest and, in the case of MYB and MEIS1, not evident in all post-alloSCT samples (Fig. 2E). This observation led us to hypothesize that immune escape may be enacted by combinatorial changes in the tetrad. To assess whether the observed changes in TF expression adequately accounted for the magnitude of MHCII downregulation at the time of relapse, we used the CORENODE gene-regulatory functions to predict the change in MHCII expression from the pre- and post-relapse expression values of the TF tetrad. For additional statistical robustness, we analyzed all paired samples from the two available studies (3, 4), including patients regardless of therapy (chemotherapy vs. alloSCT) or the direction of MHCII change. In both data sets, the predicted changes in MHCII expression closely correlated with the actual changes observed at the time of relapse (Fig. 2F and G). Given that the gene-regulatory functions were derived from an unrelated data set, these findings provided validation for the computational model and confirmed the combinatorial action of CR TFs in MHCII regulation.
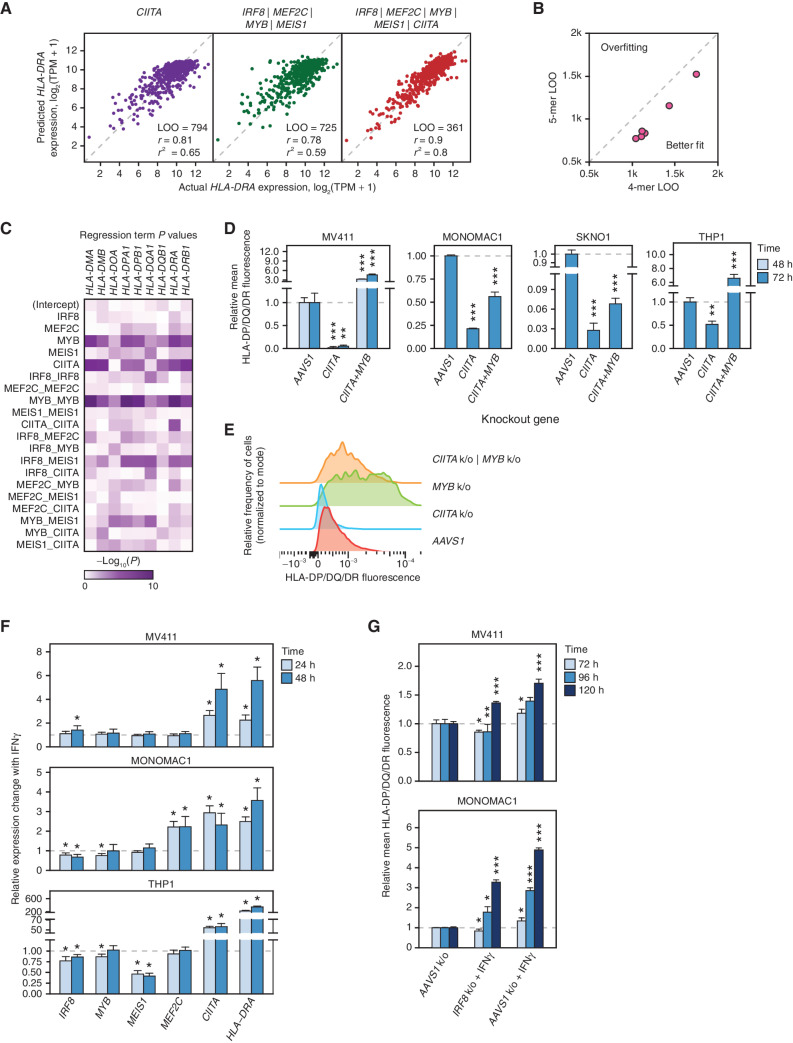
Immune Escape Is Regulated Independently of the IFNγ/CIITA Pathway
MHCII expression in myeloid cells is regulated by IFNγ, which acts via the Jak/STAT/IRF pathway to induce the expression of CIITA, a specific transcriptional coactivator of the MHCII genes (19–21). Accordingly, expression of MHCII genes correlates with CIITA expression (Fig. 3A; Supplementary Fig. S6A). Therefore, we asked if the TF tetrad regulates MHCII expression via the IFNγ/CIITA pathway. Indeed, CORENODE identified MEF2C, MEIS1, and IRF8 as candidate regulators of CIITA (Fig. 1B). Knockouts of IRF8 and MEF2C led to decreased CIITA expression, whereas a MYB knockout was associated with increased expression (Fig. 2A). Thus, at least three of the four TFs appear to participate in MHCII expression by modulating CIITA. However, post-alloSCT silencing of the MHCII genes was not consistently associated with reduced CIITA expression (Fig. 2E), pointing to an independent mechanism of MHCII regulation. In addition, ChIP-seq confirmed binding of the entire tetrad at the MHCII locus (Fig. 2C; Supplementary Fig. S5B), consistent with a possibility of direct action. These conflicting observations prompted us to investigate further CIITA-dependent and -independent actions of the tetrad. First, we repeated CORENODE regressions of MHCII genes using 4-mers and 5-mers where CIITA was used as an additional regression term. The inclusion of CIITA in the model resulted in a marked improvement of goodness-of-fit for all genes in the MHCII locus without overfitting (Fig. 3A and B). Furthermore, t and P values of individual regression terms, representing the probability of their independent contribution to the overall fit (see Supplementary Note), indicated that the TFs were independent variables, with the strongest contributions from MYB and CIITA (Fig. 3C; Supplementary Data S5 and S6). This analysis suggested that members of the TF tetrad—and particularly MYB—regulate MHCII in a CIITA-independent manner. To confirm this inference, we asked if the negatively acting TF MYB regulates MHCII expression in the absence of CIITA coactivation. We generated a panel of CIITA knockout cell lines (Supplementary Fig. S6B). As predicted, baseline expression of MHCII proteins was reduced in CIITA-deficient cells (Fig. 3D). Nonetheless, loss of MYB led to increased MHCII expression (Fig. 3D and E), consistent with a CIITA-independent mechanism of MHCII regulation. Notably, MYB knockout in wild-type MONOMAC1, a cell line with a very high baseline expression of CIITA and MHCII, decreased MHCII expression, whereas knockout of MYB in CIITA-deficient MONOMAC1 cells increased MHCII expression (Figs. 2B and 3D; Supplementary Fig. S4B). This result suggests context-specific epistasis between MYB and CIITA and confirms the role of MYB as a CIITA-independent repressor of MHCII in AML.
Figure 3.
CIITA-dependent and -independent regulation of MHCII genes by the TF tetrad. A, 1-mer CORENODE fit using CIITA (linear + quadratic term) as the only predictor of HLA-DRA expression, as well as 4- and 5-mer CORENODE HLA-DRA fits with indicated goodness-of-fit metrics. B, LOO improvement between TF 4-mers (6 fits, 1 per HLA gene) and 5-mers that include CIITA in addition to the 4 TFs. C, Heat map visualization of regression term P values representing the probability of the term's t-value being zero. Lower P values, visualized by denser color on the heat map, reflect a higher probability and magnitude of the term's independent contribution to the overall fit (see Supplementary Note for details). The statistics are calculated separately for each gene using the 5-mer composed of the TF tetrad and CIITA. D and E, Changes in the surface expression of HLA-DP/DQ/DR following CIITA ± MYB knockouts, detected by immunostaining with a pan-specific anti-HLA-DR/DP/DQ antibody. The bar plots (D) represent average fluorescence values obtained from 3 replicates with independent isotype controls and normalized to the AAVS1 control. Asterisks indicate P values calculated by a two-tailed t test comparing MHCII fluorescence between gene knockout and the AAVS1 control at the same time point. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. The FACS histogram (E) represents one of 3 replicate experiments performed in MV411. F, Changes in the expression of the tetrad TFs, CIITA and HLA-DRA induced by treatment of MV411 cells with 10 ng/mL IFNγ, measured by TaqMan ΔΔCt RT-PCR normalized to HPRT1 as internal control, in four biological replicates. G, Induction of MHCII expression (measured as in D) by 10 ng/mL IFNγ in wild type (AAVS1 control) and IRF8 knockout MV411 cells.
Finally, we asked if the induction of MHCII expression by IFNγ is mediated by the tetrad TFs. We treated AML cells with IFNγ and measured the expression of the tetrad TFs, CIITA, and HLA-DRA by RT-qPCR. Although IFNγ dramatically induced the expression of HLA-DRA and CIITA, its effects on the tetrad TFs were modest (Fig. 3F). Furthermore, IFNγ induced expression of MHCII in AML cells with an IRF8 knockout (Fig. 3G). Collectively, our data indicate that the tetrad TFs regulate MHCII expression independently of the IFNγ/CIITA pathway.
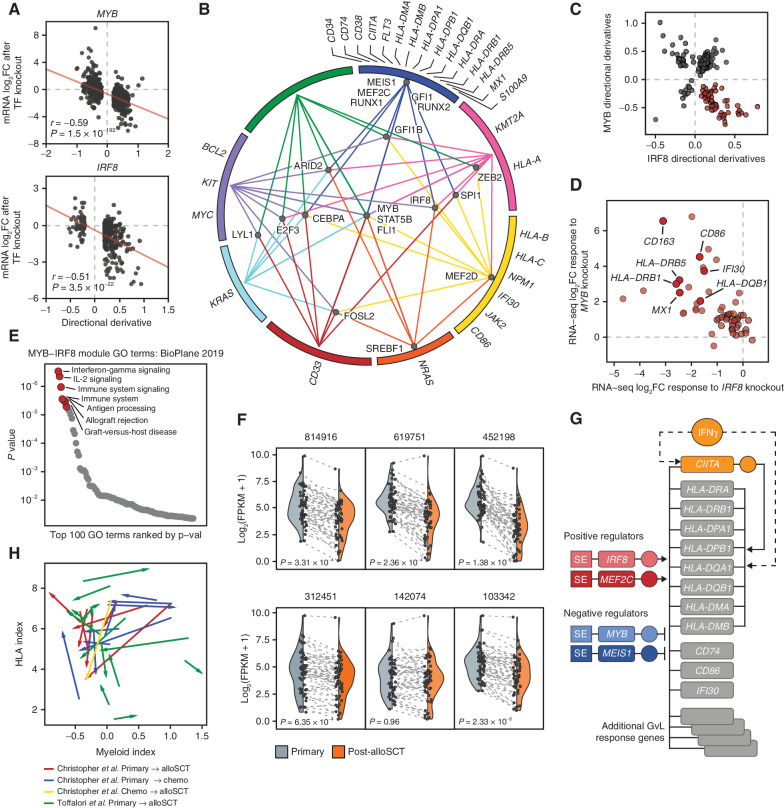
IRF8 and MYB Antagonistically Regulate a Graft-Versus-Leukemia Effect Response Module
We sought to place the MHCII genes in the context of genome-wide transcriptional regulation by the 19 CR TFs. We began by evaluating the global accuracy of CORENODE in predicting transcriptional regulation. To this end, we generated a list of high-confidence targets of each CR TF by applying a minimal ES cutoff of 15, and cross-plotted DDs for each target gene versus the actual response observed on RNA-seq after knockout of the corresponding TF in MV411 cells. A significant correlation was observed between predicted and actual changes in gene expression following TF loss (Fig. 4A; see Supplementary Note for a detailed discussion). Having confirmed CORENODE's precision in predicting genomic regulation, we built a map of high-confidence edges in the AML transcription regulatory network, revealing distinct functional modules of CR TFs and coregulated genes (Fig. 4B). The model highlighted several TFs (MYB, FLI1, STAT5B, CEBPA, IRF8, and SPI1) as genome-wide regulators, whereas other TFs (GFI1, MEIS1, RUNX1, RUNX2, MEF2C, and MEF2D) were projected to regulate more narrow transcriptional programs. In addition, the model predicted partially antagonistic functions for some TFs on a pairwise comparison (Supplementary Fig. S7A and S7B). We focused attention on the antagonistic actions of MYB and IRF8. CORENODE highlighted a module of 58 genes predicted to be upregulated by IRF8 and downregulated by MYB (Fig. 4C; Supplementary Data S7). The predicted outcome conformed closely to the actual responses following TF knockouts (Fig. 4D). The module was highly enriched for genes participating in multiple aspects of alloreactivity (Fig. 4E; Supplementary Fig. S8A), including response to IL2 and IFNγ (MAP2K1, OAS1, IFI30, MX1, and TNFRSF1B), T-cell stimulation (CD86, BTN3A1, TNFSF8, and SH2B3), adhesion (ITGB7 and TGFBI), inflammasome formation (NLRP1), and transcription regulation (NOD2, SPI1, MALT1, CBX7, and MAFB). Importantly, the module was downregulated in 5 of 6 patients with reduced MHCII expression after alloSCT, with the exception of the sole patient (142074) who displayed decreased MYB expression at relapse (Fig. 4F). In contrast, patients with relapse after chemotherapy displayed inconsistent changes in the expression of the module (Supplementary Fig. S8B). Thus, antagonistic actions of MYB and IRF8 facilitate immune evasion by silencing a broad transcriptional program that regulates the interaction between AML blasts and donor T cells (Fig. 4G).
Figure 4.
Global transcription network decomposition by CORENODE. A, CORENODE validation by TF knockout followed by mRNA-seq. DD values (predicted response) for high-confidence MYB and IRF8 edges (ES ≥ 15 and [DD] ≥ 0.2) are plotted against actual log2 fold change in mRNA expression measured by RNA-seq 72 hours after MYB knockout (observed response). Spike-in control was used to account for the global transcriptional collapse after MYB knockout (refer to Supplementary Note for a detailed discussion). B, A graphic representation of the AML transcription regulation network reconstructed from the CORENODE output. The circle represents clusters of target genes coregulated by the TFs, as illustrated by TF-cluster edges. The proximity of the TFs to the clusters reflects their specificity toward them (for example, MYB regulates all clusters, whereas MEIS1 regulates only one cluster). A geometrically optimal map with the shortest aggregate distance of the TF-cluster edges was produced by solving for the unweighted Weber problem (see Supplementary Note). C, CORENODE identifies a gene module predicted to be repressed by MYB and activated by IRF8. Visualized are DD scores of all genes with high confidence (ES ≥ 15) MYB and/or IRF8 edges; 58 of these genes (highlighted in red) are predicted to be negatively regulated by MYB and positively regulated by IRF8. D, Transcriptional responses of the 58 genes comprising the GvL module depicted in C to MYB and IRF8 knockouts measured by RNA-seq are cross-plotted, confirming the predicted directionality of regulation for the vast majority of genes. Spike-in normalization was not used for this plot (see Supplementary Note). E, Gene set enrichment analysis of the MYB-IRF8 (GvL) module genes in C using Enrichr (43). F, Decreased expression of the GvL module in paired primary and post-alloSCT patient samples from ref. (3) with reduced MHCII expression at relapse. P values were calculated by a paired two-tailed t test. Each data point represents one gene with the dotted line linking expression values at presentation and after alloSCT. The shaded areas reflect probability density of the data smoothed by a kernel density estimator. G, Schematic of the proposed regulation of the MHCII and other GvL genes in AML by the TF tetrad. H, MHCII expression at relapse does not correlate with the myeloid differentiation state. The index of myeloid differentiation is plotted against combined MHCII expression in each paired sample and the direction of change between initial presentation and relapse is marked by arrows.
Immune Escape Is Uncoupled from the Myeloid Differentiation State
Members of the TF tetrad have previously been implicated in the regulation of normal and malignant myelopoiesis: MYB and MEIS1 restrain myeloid differentiation, whereas IRF8 and MEF2C promote it (22–25). Additionally, AML cells form a functional hierarchy of differentiation states arising from a leukemia stem cell (LSC; ref. 26), and activation of immune pathways is a hallmark of monocytic development (27). Thus, we hypothesized that reduced expression of IRF8 and MEF2C and increased expression of MYB and MEIS1 at relapse reflect the evolution of leukemia into a less differentiated state. To test this hypothesis, we computed a myeloid differentiation index from the mRNA expression of 19 cell type–specific markers, which correctly reproduced both the normal myeloid trajectory and the functional AML hierarchy (Supplementary Fig. S9A and S9B). However, MHCII expression correlated poorly with myeloid differentiation in the Beat AML and TCGA data sets, and changes in the MHCII expression at relapse could not be predicted from the changes in the myeloid differentiation index (Fig. 4H; Supplementary Fig. S9C and S9D). We conclude that reduced MHCII expression at relapse is regulated independently from the myeloid differentiation state.
Transcriptional Plasticity Underlies Relapse
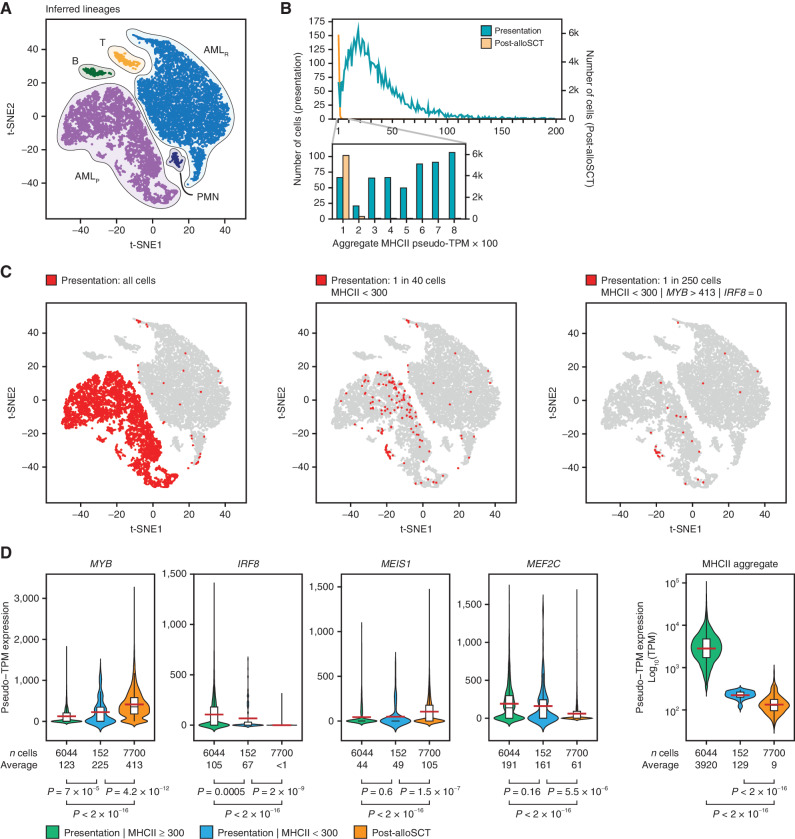
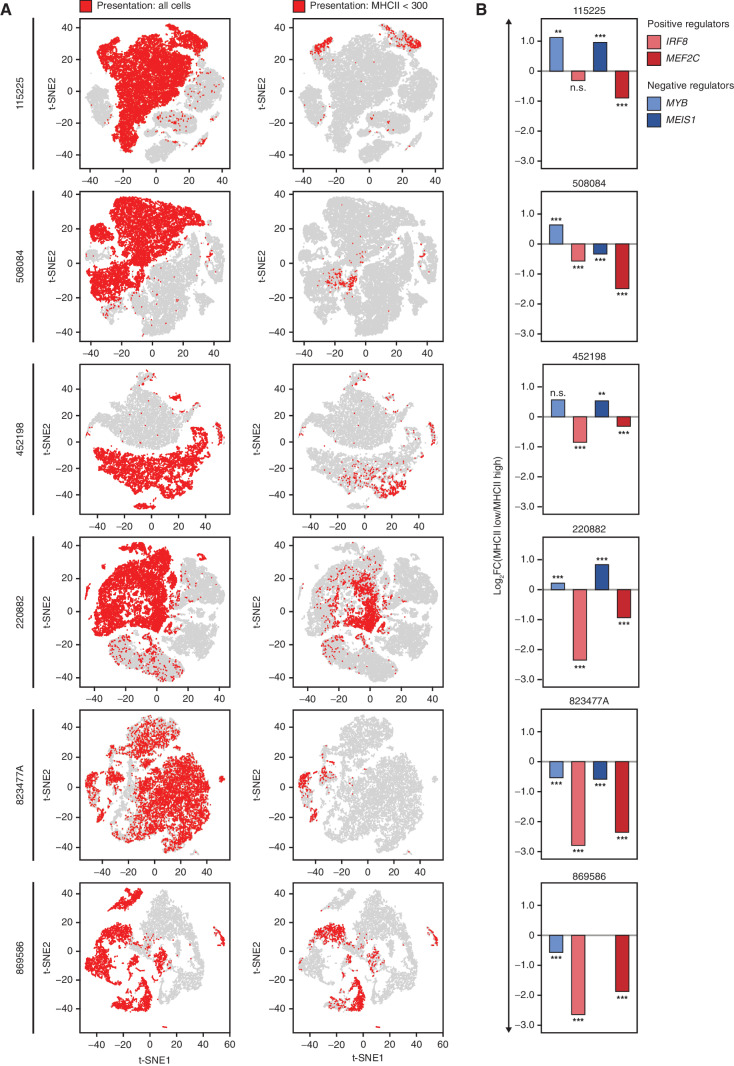
We examined the expression of MHCII genes in AML cells at the single-cell level. We analyzed single-cell transcriptomes obtained from patient 452198 at presentation and post-alloSCT relapse (3). Plotting all cells together using t-distributed stochastic neighbor embedding (t-SNE) revealed distinct clusters of AML cells corresponding to initial presentation and relapse (Fig. 5A). Although most AML cells displayed high MHCII expression at presentation, approximately 1 in 40 cells had low or undetectable MHCII similar to those seen at relapse (Fig. 5B and C). Some of these MHCII-low cells coclustered with the relapsed cell population, indicating similar global transcription patterns (Fig. 5C). The MHCII-low cells displayed significantly higher MYB and lower IRF8 compared with the cells expressing MHCII highly at presentation (Fig. 5D). The average MYB expression in the MHCII-low cell population at presentation was still lower, and IRF8 was still higher than the average of these TFs at relapse. However, up to 1 in 6 cells in the MHCII-low population (approximately 1 in 250 of all AML cells at presentation) expressed undetectable IRF8 and markedly elevated MYB that matched or exceeded the average MYB expression seen at relapse (Fig. 5C). Although expression of MEIS1 was increased and expression of MEF2C was decreased in most cells at relapse, we found no statistically significant change in their expression in the MHCII-low cells at presentation, indicating that stable changes in these TFs were acquired later in the course of leukemia evolution (Fig. 5D). We conclude that a minor population of cells with reduced MHCII expression driven by altered MYB and IRF8 expression was present in this patient at the time of initial presentation and likely contributed to the subsequent relapse. For additional validation, we examined scRNA-seq data from six diagnostic AML samples from another report (28). MHCII-low cells were universally present in all patients at the time of initial diagnosis (Fig. 6A). IRF8 and MEF2C were significantly lower in MHCII-low cells than in MHCII–high cells (Fig. 6B). MYB and MEIS1 were higher in MHCII-low cells in 4 of 6 patients (Fig. 6B). Importantly, in the 2 samples where MHCII-low cells displayed paradoxically lower expression of MYB and MEIS1, this was compensated by a much greater fractional decrease in IRF8 and MEF2C. Overall, these observations are consistent with the results of bulk RNA-seq of post-alloSCT samples (Fig. 2E) and lend further support to a combinatorial transcriptional mechanism of immune escape. Importantly, AML cells with transcriptionally driven low MHCII expression appear to be common at the time of initial diagnosis in all examined patients, suggesting a unifying potential mechanism of immune escape and eventual relapse.
Figure 5.
Single-cell RNA-seq reveals an adaptive mechanism of transcriptional evolution underlying relapse. A, scRNA-seq of AML cells obtained at initial presentation (AMLP) and post-alloSCT relapse (AMLR). Data from patient 452198 were downloaded from ref. (3) and clustered according to their genome-wide expression patterns using t-SNE. Cell lineages were inferred from the expression of lineage markers. B, Distribution of MHCII expression in AML cells. Individual cells are placed in bins according to their aggregate MHCII expression and cell frequency is plotted for each bin. C, t-SNE plots start with the same plot as in A but highlight cells from the initial presentation that fit the designated criteria. D, Single-cell expression of the TF tetrad in AML cells with low and high aggregate MHCII expression at presentation compared with relapse. P values were calculated by a two-tailed t test. The elements of the boxplots are as follows: black center line, median; red center line, mean; box limits, upper and lower quartiles; whiskers, 1.5× interquartile range. The shaded areas reflect the probability density of the data smoothed by a kernel density estimator.
Figure 6.
Single-cell RNA-seq reveals the universal presence of MHCII-low cells at diagnosis displaying an aberrant expression of the TF tetrad. A, scRNA-seq of AML cells obtained at initial presentation and relapse after alloSCT (patient 452198) or chemotherapy (all other patients). Data were downloaded from ref. (28) and clustered according to their genome-wide expression patterns using t-SNE. Cells from initial presentation and relapse are plotted together and highlighted according to the relapse status and MHCII expression, indicating the universal presence of MHCII-low cells (aggregate MHCII counts <300) in all patients at the time of initial presentation. B, Differences in the tetrad TF expression in the MHCII-low versus MHCII–high cells at the time of initial presentation. Asterisks indicate P values calculated by a two-tailed t test; **, P ≤ 0.01; ***, P ≤ 0.001; n.s., P > 0.05. MEIS1 was not expressed in sample 869586.
DISCUSSION
By developing a network decomposition approach that captures combinatorial TF interactions, we identified a tetrad of core TFs that regulate immune escape of AML cells after alloSCT, where IRF8 and MYB appear to act as major regulators of MHCII expression, acting antagonistically. Remarkably, the transcriptional activator MYB acts as a strong negative regulator of MHCII genes. Consistent with our observations, Myb-deficient mouse B cells exhibit elevated MHCII expression (29). Our data reinforce the status of MYB as an important therapeutic target in AML (30) and further indicate that even partial MYB inhibition may represent a viable strategy to prevent or reverse immune escape after alloSCT. Indeed, the tool compound mebendazole, which appears to inhibit MYB by targeting it for proteosomal degradation (18), increases MHCII expression, albeit modestly. However, a complete MYB knockout results in an extremely potent induction of MHCII expression, and it is likely that a potent direct MYB inhibitor would produce a similarly dramatic effect. Although IRF8 is an established effector of IFNγ and has been reported to control MHCII expression in myeloid cells (31–33), we demonstrate that both IRF8 and MYB regulate MHCII expression largely independently of the IFNγ/CIITA axis. Notably, various IRF paralogs (IRF1, IRF2, IRF4, and IRF8) have been implicated as transcriptional effectors of the IFNγ/Jak/STAT cascade, inducing MHCII expression both directly and via CIITA (19, 24, 31, 33, 34). We suspect that the IRF paralogs exhibit context-specific and partially redundant roles in MHCII regulation. Although binding of the tetrad TFs at the MHCII locus suggests that they may directly regulate the expression of MHCII genes, TF occupancy is a poor predictor of direct regulation (35, 36), and additional studies are needed to elucidate the mechanism by which the tetrad TFs regulate MHCII expression. Furthermore, the MHCII locus is a point of convergence of complex regulatory mechanisms (20), and it is likely that there are other important regulators of MHCII expression contributing to immune escape. Indeed, CORENODE is limited to a preselected set of regulators and is unable to capture posttranscriptional regulation. While this manuscript was in revision, a study by Gambacorta and colleagues reported epigenetic silencing by the polycomb repressive complex 2 (PRC2) as a mechanism of MHCII loss in post-alloSCT relapse (37). It would be interesting to examine whether altered expression of the tetrad TFs identified in our study contributes to the recruitment of PRC2 to the MHCII locus.
Beyond reducing MHCII expression, antagonistic actions of MYB and IRF8 facilitate immune escape by enacting a broader program of transcriptional changes resulting in reduced expression of multiple genes involved in antigen presentation, cytokine response, and T-cell costimulation. Notably, to the extent that these core TFs are essential for AML survival, post-SCT relapse appears to rely on relatively modest changes in their expression that cooperatively mediate immune evasion without affecting cell viability. A small population of such cells with reduced MHCII expression due to altered TF expression is present at the time of initial diagnosis and likely contributes to relapse under in vivo selection. Whether transcriptional heterogeneity is hard-wired (for example, by mutations in noncoding regulatory DNA elements) or driven by stochastic TF fluctuations remains to be elucidated. Nonetheless, we speculate that the immune pressure imposed by allogeneic transplantation selects AML cells with a pattern of CR TF expression that provides an optimal balance between immune escape and overall fitness. Such “transcriptional evolution” by the selection of cells with favorable transcriptional states mediated by altered combinatorial patterns of key TFs may drive the progression of other tumors and eventual relapse.
METHODS
External Data Sets
RNA-seq BAM files for 510 patients from the Beat AML project (15) were provided by Oregon Health & Science University and processed through the CCLE RNA processing pipeline (STAR/RSEM, described at https://github.com/broadinstitute/ccle_processing). Reads were normalized to transcripts per million (TPM) and filtered for protein-coding genes. The expression values were transformed to log2(TPM + 1). The genes were ranked by average expression across the samples and 9,000 top expressed genes were chosen for further analysis (Supplementary Fig. S1).
H3K27ac ChIP-seq data from primary AML samples (9) were downloaded from Sequence Read Archive (SRA) under accession number SRP103200 and processed using the AQUAS pipeline (https://github.com/kundajelab/chipseq_pipeline) with minor modifications and according to the ENCODE3 guidelines. Reads were aligned to the hg19 genome build using BWA-ALN and peaks were called using MACS2. For SE calling, each sample was processed using ROSE2 (https://github.com/linlabbcm/rose2), excluding 2,500 bp around TSSs (−t 2,500) and the hg19 Encode blacklisted regions. SE regions were then merged and ROSE2 was run again for each sample on the merged regions, producing the SE signal matrix, which was then normalized by a median signal for each sample.
CIITA ChIP-seq data (38) were downloaded from the GEO database (http://www.ncbi.nlm.nih.gov/geo/) under the accession number GSE52941.
TCGA mRNA (FPKM) data for 151 patients with AML was downloaded from the NCI Genomic Data Commons site using the GDCquery routine from the R package TCGAbiolinks (39); the procedure was last verified on September 18, 2019. TCGA mRNA gene information was converted from ENSG codes to hgnc symbols using the Bioconductor program biomaRt (40, 41). mRNA FPKM expression values were converted to TPM.
RNA-seq and associated metadata for normal hematopoietic progenitors and AML samples (42) for myeloid index analysis were downloaded from GEO GSE74246, converted to FPKM using transcript lengths acquired through the Bioconductor package “biomaRt” and then converted to TPM.
Paired processed RNA-seq data from the two available studies of patients with AML at the time of initial presentation and relapse (3, 4) were downloaded from https://www.nejm.org/doi/full/10.1056/NEJMoa1808777 and https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-7456/samples/.
Genetic dependency data are available for download at the Broad DepMap portal database (https://depmap.org/portal/download/). Data release 20q1 was used for this study. For the purpose of this study, we considered a gene to represent a selective AML dependency if it had a probability of dependency of ≥0.5 in 3 or more AML cell lines and not be considered a pan-lethal dependency based on the LRT score. This approach resulted in a total of 225 selective AML dependencies.
Gene set enrichment analysis was performed using the Enrichr database (43).
Cell Culture
AML cell lines were purchased from the American Type Culture Collection (https://www.atcc.org) within the past 10 years. The MV411 cell line was authenticated within the last year by confirming the presence of the t(4;11) translocation. Other cell lines were not independently authenticated. Cells were cultured in the RPMI-1640 media (Gibco 11875085) containing 10% FBS (Thermo Fisher A4766) and regularly tested to be free of Mycoplasma spp.
CRISPR-Cas9 Gene Knockouts
Synthetically modified sgRNA constructs were purchased from Synthego (refer to Supplementary Table S1 for sgRNA sequences). Ribonucleoprotein (RNP) assembly was performed by mixing 2 to 3 sgRNAs (a total of 120 pmol/L) with 8.5 μg recombinant Cas9 (Invitrogen A36499). The resulting RNP mix was electroporated into 0.3 × 106 cells using a Lonza 4D Nucleofector, program DJ-100, in 20 μL Nucleocuvette strips (Lonza V4XC-2032). Unless otherwise noted, cells were incubated in media for 72 hours postelectroporation before subsequent analyses. Knockout efficiency was confirmed by Western blotting (Supplementary Fig. S4A). The following antibodies were used: actin (Santa Cruz sc-47778), IRF8 (Cell Signaling 5628s), MEF2C (Cell Signaling 5030s), MYB (Abcam ab45150). A guide RNA targeting the AAVS1 “safe harbor” locus was used as a negative control (44).
Genome-Wide Occupancy Analysis
ChIP-seq was performed as previously described (45) and in accordance with the Encode guidelines (46). Briefly, 10 × 106 (for histone ChIPs) or 100 × 106 (for TF ChIPs) exponentially growing MV411 cells were fixed with 1% formaldehyde for 10 minutes at room temperature and then quenched with 125 mmol/L glycine for 5 minutes. Nuclei were isolated using the Nuclei EZ Kit (Sigma-Aldrich NUC101) and resuspended in 10 mmol/L Tris-HCl, pH 8.0, 1 mmol/L EDTA, 0.1% SDS with 1× HALT protease inhibitor (Thermo Fisher; 87786). Chromatin was fragmented by sonication on an E220 Covaris-focused sonication machine using the parameters of 140 mV, 5% duty factor, 200 cycles/burst for 14 minutes, and cleared by centrifugation. The following antibodies were used for ChIP: H3K27ac (Abcam ab4729), IRF8 (SantaCruz sc365042X), MYB (Abcam ab45150), MEIS1 (Abcam ab19867), and MEF2C (GeneTex GTX105433). ChIP-seq libraries were prepared using Swift S2 Acel reagents (Swift 21096) on a Beckman Coulter Biomek i7 liquid handling platform from approximately 1 ng of DNA according to the manufacturer's protocol and using 14 cycles of PCR amplification. Sequencing libraries were quantified by Qubit fluorometer and Agilent TapeStation 2200. Library pooling and indexing were evaluated by shallow sequencing on Illumina MiSeq. Subsequently, libraries were sequenced on NovaSeq targeting 40 million 100 bp read pairs by the Molecular Biology Core facilities at the Dana-Farber Cancer Institute. Sequencing reads were processed using the AQUAS pipeline (https://github.com/kundajelab/chipseq_pipeline) with minor modifications and according to the ENCODE3 guidelines. Reads were aligned to the hg38 genome build using BWA-ALN and peaks were called using MACS2. Quality control of the ChIP-seq data were performed using the nf-core pipeline (https://github.com/nf-core/chipseq), targeting FRiP scores of >0.3.
For quantitative ChIP-seq analysis of H3K27 acetylation, we used Drosophila chromatin/antibody spike-in control as previously described (47). Briefly, 4 μg of anti-H3K27ac antibody, 2 μg of spike-in antibody, and 20 ng of spike-in chromatin (Active Motif 61686 and 53083, respectively) were added to chromatin prepared from 2.5 × 106 MV411 cells 72 hours after RNP-mediated TF knockout. The rest of the ChIP-seq experiment was performed in the standard fashion. After ChIP-seq reads were mapped to the Drosophila genome and the hg38 human genome in parallel, human tag counts were normalized to Drosophila tag counts.
Global Transcriptome Analysis
For RNA-seq experiments, the total cellular RNA was extracted using the QuickRNA kit (Zymo Research R1054). Purified total RNA was mixed with the ERCC synthetic spike-in control (Invitrogen 4456740). RNA-seq libraries were prepared on a Beckman Coulter Biomek i7 liquid handling platform using Roche Kapa mRNA HyperPrep strand-specific sample preparation kits (Roche; 08098123702) from 200 ng of purified total RNA according to the manufacturer's protocol. Library quantification and Illumina sequencing were performed as described in the ChIP-seq section above.
RT-qPCR
The total cellular RNA was extracted using the RNeasy Plus Mini Kit (Qiagen; 74134). Target gene expression was quantified by a TaqMan ΔΔCt experiment using TaqPath 1-step master mix (Thermo Fisher A28525) and HPRT1 as an internal control. The following TaqMan assays were used: CIITA, Hs00172106_m1; MYB, Hs00920556_m1; MEIS1, Hs00180020_m1; MEF2C, Hs00231149_m1; IRF8, Hs00175238_m1; HPRT1, Hs99999909_m1; HLA-DRA, Hs00219575_m1).
Measurement of MHCII Expression by FACS
For MHCII analysis, 1 × 106 cells were first incubated for 10 minutes with 0.5 μg of Human Fc block (BD Biosciences; 564219). Cells were then split and stained for 20 minutes with either a pan-specific FITC-conjugated Mouse Anti-Human HLA-DR, DP, DQ antibody (BD Biosciences 555558, clone Tu39), or FITC-conjugated Mouse IgG2a, κ Isotype Control (BD Biosciences 555573). Data were analyzed in FlowJo (FlowJo LLC). Surface MHCII expression was calculated by subtracting the isotype control fluorescence from the pan–HLA-DR, DP, and DQ fluorescence.
Western Blotting
Whole-cell lysates were prepared in RIPA buffer (Boston Bio-Products BP-115-500) with a protease inhibitor cocktail (Thermo Fisher; 23225). Lysates were boiled in Laemmli buffer (Bio-Rad 1610737), separated by SDS-PAGE, and transferred and blocked using standard methodology. HRP-conjugated anti-mouse and anti-rabbit IgG secondary antibodies were used for imaging (Bio-Rad; 1706515 and 1706515) with an enhanced chemiluminescence substrate (PerkinElmer NEL104001EA) according to the manufacturers’ instructions. The following primary antibodies were used: anti-MYB (Abcam ab45150), anti-MEF2C (Cell Signaling; 5030S), anti-MEIS1 (Abcam; ab19867), anti-IRF8 (Cell Signaling; 5628S).
Statistical Analysis
Details of the statistical analyses pertaining to CORENODE are found in the Supplementary Note. All experiments were done in at least three replicates. Two-tailed Student t test was used to compare the mean fluorescence of antibody-stained cells following TF knockout. Changes in gene expression in patient samples between initial diagnosis and relapse were ascertained using a Wilcoxon signed-rank test. Statistical analysis of ChIP-seq and RNA-seq data were performed using the DESeq2 package (48) with genome-wide adjustment for multiple hypothesis testing.
Myeloid Differentiation Index
To identify markers of myeloid development, genome-wide mRNA expression values in “HSC” and “monocyte” samples from ref. (42) were processed to yield the mean and variance of expression by the gene. For each gene, the two variances were pooled [pooled variance = mean (HSC variance, monocyte variance)]. A separation index was then defined for each gene as the difference between the HSC and monocyte mean expressions divided by the square root of the pooled variance. Markers were chosen as the 19 genes with the highest separation indices. Lymphoid markers were identified with the same procedure, comparing HSC samples to the T-cell and B-cell samples. To compute the myeloid index, each sample's expression of the marker genes as determined above was converted to a z-score using the mean of all eight HSC and monocyte expressions and the pooled variance for that gene, and further normalized to a ± 1 scale by dividing by the maximum absolute value of all z-scores. For genes where the mean monocyte score was higher than the mean HSC score, normalized scores were multiplied by (−1). The normalized scores for the set of marker genes were summed to define the myeloid index for each sample. The same procedure was used for the lymphoid index, with appropriate cell-type substitution. Plotting normal and leukemia cell types according to the myeloid and lymphoid indices yielded the expected development vectors as shown in Supplementary Fig. S6. The same markers were then used to define a myeloid index for the samples from the other data sets (3, 4, 15, 17). The HLA index was defined as the mean expression (in log2[TPM + 1]) of the nine HLA-D genes.
Data and Materials Availability
The CORENODE code (written in R) is found at https://app.box.com/s/a3x7f4zwcgv3f4jgshphe4opsen13mwf. Genome-wide matrix of CR TF ESs and DDs can be found in Supplementary Data S1 and S2, respectively. Genome-wide CORENODE output, including n-mer fits and scores, is found at https://corenode.shinyapps.io/corenode/. Statistics on 5-mer CORENODE fits for the MHC type II genes is displayed in Supplementary Data S3. Gene-dependency scores from the CRISPR–Cas9 screen are available at https://depmap.org/portal/download/. ChIP-seq and RNA-seq data have been deposited to the SRA database under accession no. PRJNA751732. DESeq2 outputs from the RNA-seq experiments are found in Supplementary Data S4. CORENODE statistics for MHC type II 5-mer regressions are found in Supplementary Data S5. Plots of CORENODE n-mer regression fits for MHC type II genes are found in Supplementary Data S6. Expression of the GvL module in primary and post-allo-SCT relapse samples is found in Supplementary Data S7.
Supplementary Material
Acknowledgments
M. Pimkin was supported by a Damon Runyon–Sohn Pediatric Cancer Fellowship and a Young Investigator Award from the Alex's Lemonade Stand Foundation. This work was supported, in whole or in part, by research grants to M. Pimkin from the Curing Kids Cancer Foundation, When Everyone Survives Foundation, Pedals for Pediatrics, Children's Cancer Research Fund, Children's Leukemia Research Association, Hyundai Hope on Wheels, Kate Amato Foundation, and Boston Children's Hospital Office of Faculty Development. K. Stegmaier was supported by NIH 5R35 CA210030 and NIH P50 CA206963. N.V. Dharia was supported by the Julia's Legacy of Hope St. Baldrick's Foundation Fellowship. We thank Drs. L.S. Kean, G.C. Yuan and S. Avagyan for critical reading of this manuscript, and Drs. T. Ley and C. Miller for access to the scRNA-seq data.
Footnotes
Note: Supplementary data for this article are available at Blood Cancer Discovery Online (https://bloodcancerdiscov.aacrjournals.org/).
Authors’ Disclosures
K. Eagle reports personal fees from Flare Therapeutics and Third Rock Ventures outside the submitted work; in addition, K. Eagle has a patent for Method of Inducing Therapeutic Selenium Deficiency pending. C.Y. Lin reports other support from Kronos Bio outside the submitted work. N.V. Dharia reports grants from Julia's Legacy of Hope St. Baldrick's Foundation Fellowship during the conduct of the study; other support from Genentech, Inc., a member of the Roche Group outside the submitted work. K. Stegmaier reports grants from NCI during the conduct of the study; personal fees from AstraZeneca, grants from KronosBio, Novartis, and other support from Auron Therapeutics outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
K. Eagle: Conceptualization, data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. T. Harada: Validation, investigation, visualization, methodology, writing–review and editing. J. Kalfon: Data curation, software, formal analysis, investigation, visualization, methodology. M.W. Perez: Data curation, software, formal analysis, investigation, visualization. Y. Heshmati: Formal analysis, validation, investigation, methodology, writing–review and editing. J. Ewers: Investigation. J. Vrabič Koren: Data curation, formal analysis, methodology. J.M. Dempster: Data curation. G. Kugener: Data curation. V.R. Paralkar: Conceptualization, investigation, writing–review and editing. C.Y. Lin: Conceptualization, supervision, investigation, writing–review and editing. N.V. Dharia: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation, project administration, writing–review and editing. K. Stegmaier: Resources, supervision, funding acquisition, investigation, project administration, writing–review and editing. S.H. Orkin: Conceptualization, resources, supervision, funding acquisition, investigation, writing–original draft, project administration, writing–review and editing. M. Pimkin: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Sweeney C, Vyas P. The graft-versus-leukemia effect in AML. Front Oncol 2019;9:1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dermime S, Mavroudis D, Jiang YZ, Hensel N, Molldrem J, Barrett AJ. Immune escape from a graft-versus-leukemia effect may play a role in the relapse of myeloid leukemias following allogeneic bone marrow transplantation. Bone Marrow Transplant 1997;19:989–99. [DOI] [PubMed] [Google Scholar]
- 3. Christopher MJ, Petti AA, Rettig MP, Miller CA, Chendamarai E, Duncavage EJ, et al. Immune escape of relapsed AML cells after allogeneic transplantation. N Engl J Med 2018;379:2330–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Toffalori C, Zito L, Gambacorta V, Riba M, Oliveira G, Bucci G, et al. Immune signature drives leukemia escape and relapse after hematopoietic cell transplantation. Nat Med 2019;25:603–11. [DOI] [PubMed] [Google Scholar]
- 5. Kim HD, Shay T, O'Shea EK, Regev A. Transcriptional regulatory circuits: predicting numbers from alphabets. Science 2009;325:429–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. He B, Tan K. Understanding transcriptional regulatory networks using computational models. Curr Opin Genet Dev 2016;37:101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Saint-André V, Federation AJ, Lin CY, Abraham BJ, Reddy J, Lee TI, et al. Models of human core transcriptional regulatory circuitries. Genome Res 2016;26:385–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013;153:307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McKeown MR, Corces MR, Eaton ML, Fiore C, Lee E, Lopez JT, et al. Superenhancer analysis defines novel epigenomic subtypes of non-APL AML, including an RARα dependency targetable by SY-1425, a potent and selective RARα agonist. Cancer Discov 2017;7:1136–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mack SC, Singh I, Wang X, Hirsch R, Wu Q, Villagomez R, et al. Chromatin landscapes reveal developmentally encoded transcriptional states that define human glioblastoma. J Exp Med 2019;216:1071–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Durbin AD, Zimmerman MW, Dharia NV, Abraham BJ, Iniguez AB, Weichert-Leahey N, et al. Selective gene dependencies in MYCN-amplified neuroblastoma include the core transcriptional regulatory circuitry. Nat Genet 2018;50:1240–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dempster JM, Rossen J, Kazachkova M, Pan J, Kugener G, Root DE, et al. Extracting biological insights from the project achilles genome-scale CRISPR screens in cancer cell lines. bioRxiv 2019;20:720243. [Google Scholar]
- 13. Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H, et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat Genet 2017;49:1779–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McDonald ER, de WA, Schlabach MR, Billy E, Mavrakis KJ, Hoffman GR, et al. Project DRIVE: a compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell 2017;170:577–92. [DOI] [PubMed] [Google Scholar]
- 15. Tyner JW, Tognon CE, Bottomly D, Wilmot B, Kurtz SE, Savage SL, et al. Functional genomic landscape of acute myeloid leukaemia. Nature 2018;562:526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Harada T, Heshmati Y, Kalfon J, Ferrucio JX, Perez M, Ewers J, et al. A distinct core regulatory module enforces oncogene expression in KMT2A-rearranged leukemia. bioRxiv 2021;2021.08.03.454902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Network CGAR. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med 2013;368:2059–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Walf-Vorderwülbecke V, Pearce K, Brooks T, Hubank M, H-EMMv den, Zwaan CM, et al. Targeting acute myeloid leukemia by drug-induced c-MYB degradation. Leukemia 2018;32:882–9. [DOI] [PubMed] [Google Scholar]
- 19. Ting JP-Y, Trowsdale J. Genetic control of MHC class II expression. Cell 2002;109Suppl:S21–33. [DOI] [PubMed] [Google Scholar]
- 20. Wright KL, Ting JP-Y. Epigenetic regulation of MHC-II and CIITA genes. Trends Immunol 2006;27:405–12. [DOI] [PubMed] [Google Scholar]
- 21. Raval A, Howcroft TK, Weissman JD, Kirshner S, Zhu XS, Yokoyama K, et al. Transcriptional coactivator, CIITA, is an acetyltransferase that bypasses a promoter requirement for TAF(II)250. Mol Cell 2001;7:105–15. [DOI] [PubMed] [Google Scholar]
- 22. Schüler A, Schwieger M, Engelmann A, Weber K, Horn S, Müller U, et al. The MADS transcription factor Mef2c is a pivotal modulator of myeloid cell fate. Blood 2008;111:4532–41. [DOI] [PubMed] [Google Scholar]
- 23. Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer 2008;8:523–34. [DOI] [PubMed] [Google Scholar]
- 24. Tamura T, Kurotaki D, Koizumi S. Regulation of myelopoiesis by the transcription factor IRF8. Int J Hematol 2015;101:342–51. [DOI] [PubMed] [Google Scholar]
- 25. Collins CT, Hess JL. Deregulation of the HOXA9/MEIS1 axis in acute leukemia. Curr Opin Hematol 2016;23:354–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vetrie D, Helgason GV, Copland M. The leukaemia stem cell: similarities, differences and clinical prospects in CML and AML. Nat Rev Cancer 2020;20:158–73. [DOI] [PubMed] [Google Scholar]
- 27. Coillard A, Segura E. In vivo differentiation of human monocytes. Front Immunol 2019;10:1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Petti AA, Khan SM, Xu Z, Helton N, Fronick CC, Fulton R, et al. Genetic and transcriptional contributions to relapse in normal karyotype acute myeloid leukemiagenetic and transcriptional contributions to relapse in AML. Blood Cancer Discov 2021;3:32–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thomas MD, Kremer CS, Ravichandran KS, Rajewsky K, Bender TP. c-Myb is critical for B cell development and maintenance of follicular B cells. Immunity 2005;23:275–86. [DOI] [PubMed] [Google Scholar]
- 30. Henley MJ, Koehler AN. Advances in targeting ‘undruggable’ transcription factors with small molecules. Nat Rev Drug Discov 2021;20:669–88. [DOI] [PubMed] [Google Scholar]
- 31. Marquis J-F, Kapoustina O, Langlais D, Ruddy R, Dufour CR, Kim B-H, et al. Interferon regulatory factor 8 regulates pathways for antigen presentation in myeloid cells and during tuberculosis. PLos Genet 2011;7:e1002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sharma A, Yun H, Jyotsana N, Chaturvedi A, Schwarzer A, Yung E, et al. Constitutive IRF8 expression inhibits AML by activation of repressed immune response signaling. Leukemia 2015;29:157–68. [DOI] [PubMed] [Google Scholar]
- 33. Kurotaki D, Osato N, Nishiyama A, Yamamoto M, Ban T, Sato H, et al. Essential role of the IRF8-KLF4 transcription factor cascade in murine monocyte differentiation. Blood 2013;121:1839–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reith W, LeibundGut-Landmann S, Waldburger J-M. Regulation of MHC class II gene expression by the class II transactivator. Nat Rev Immunol 2005;5:793–806. [DOI] [PubMed] [Google Scholar]
- 35. Muhar M, Ebert A, Neumann T, Umkehrer C, Jude J, Wieshofer C, et al. SLAM-seq defines direct gene-regulatory functions of the BRD4-MYC axis. Science 2018;360:800–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Harada T, Heshmati Y, Kalfon J, Perez MW, Ferrucio JX, Ewers J, et al. A distinct core regulatory module enforces oncogene expression in KMT2A-rearranged leukemia. Gene Dev 2022;36:368–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. gambacorta V, Beretta S, Ciccimarra M, Zito L, Giannetti K, Andrisani A, et al. Integrated multiomic profiling identifies the epigenetic regulator PRC2 as a therapeutic target to counteract leukemia immune escape and relapse epigenetic control of immune evasion and relapse after HCT. Cancer Discov 2022;OF1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Scharer CD, Choi NM, Barwick BG, Majumder P, Lohsen S, Boss JM. Genome-wide CIITA-binding profile identifies sequence preferences that dictate function versus recruitment. Nucleic Acids Res 2015;43:3128–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 2016;44:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods 2015;12:115–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Durinck S, Spellman PT, Birney E, Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat Protoc 2009;4:1184–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet 2016;48:1193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Demirci S, Zeng J, Wu Y, Uchida N, Shen AH, Pellin D, et al. BCL11A enhancer–edited hematopoietic stem cells persist in rhesus monkeys without toxicity. J Clin Invest 2020;130:6677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chapuy B, McKeown MR, Lin CY, Monti S, Roemer MGM, Qi J, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell 2013;24:777–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res 2012;22:1813–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Egan B, Yuan C-C, Craske ML, Labhart P, Guler GD, Arnott D, et al. An alternative approach to ChIP-seq normalization enables detection of genome-wide changes in histone H3 Lysine 27 trimethylation upon EZH2 inhibition. PLoS One 2016;11:e0166438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.