Abstract
PAS domains are newly recognized signaling domains that are widely distributed in proteins from members of the Archaea and Bacteria and from fungi, plants, insects, and vertebrates. They function as input modules in proteins that sense oxygen, redox potential, light, and some other stimuli. Specificity in sensing arises, in part, from different cofactors that may be associated with the PAS fold. Transduction of redox signals may be a common mechanistic theme in many different PAS domains. PAS proteins are always located intracellularly but may monitor the external as well as the internal environment. One way in which prokaryotic PAS proteins sense the environment is by detecting changes in the electron transport system. This serves as an early warning system for any reduction in cellular energy levels. Human PAS proteins include hypoxia-inducible factors and voltage-sensitive ion channels; other PAS proteins are integral components of circadian clocks. Although PAS domains were only recently identified, the signaling functions with which they are associated have long been recognized as fundamental properties of living cells.
PAS domains are important signaling modules that monitor changes in light, redox potential, oxygen, small ligands, and overall energy level of a cell. Unlike most other sensor modules, PAS domains are located in the cytosol. There has been a long-standing search for a hypothetical sensor that measures the proton motive force or a similar parameter that reads the energy status inside the cell (18, 78, 221). The recent discovery of the Aer protein in Escherichia coli (24, 186) and progress in functional analysis of the NifL protein in Azotobacter vinelandii (97, 147, 207) resulted in a breakthrough in the search for internal energy sensors. These signal-transducing proteins have a PAS domain located inside the cell that senses redox changes in the electron transport system or overall cellular redox status. PAS domains can also sense environmental factors that cross the cell membrane and/or affect cell metabolism.
The advantage for cell survival of sensing oxygen, light, redox potential, and energy levels has been widely recognized. Oxygen is both a terminal acceptor for oxidative phosphorylation with its high ATP yield and a toxic agent that forms harmful reactive free radicals when partially reduced. Many microorganisms are adapted for living within a certain range of oxygen concentrations, as are cells in eukaryotic multicellular organisms. Sensing of light intensity and wavelength governs such cellular responses as phototropism in plants and phototaxis in bacteria. There is increasing evidence that depletion of cellular energy levels is first seen in a decreased electron transport and proton motive force that precede an observable change in ATP concentration. Monitoring electron transport or proton motive force can quickly alert a cell to energy loss. E. coli senses intracellular redox changes and migrates to a microenvironment with a preferred redox potential (23). The metabolic effects of oxygen, light, proton motive force, and redox potential are interrelated on the level of the flow of reducing equivalents through the electron transport system. As a result, it is sufficient for individual cells to sense any one of these parameters to monitor cell energy levels. Sensing of oxygen directly may be advantageous in cells that have enzyme reactions that are inactivated by oxygen. Sensing of proton motive force or redox potential may provide a more versatile measure of cellular energy. Recent studies suggest that PAS domains in various sensor proteins vary in the parameter that is sensed. That is, a PAS domain may sense oxygen, light, redox potential, or proton motive force as a way of monitoring energy changes in living cells.
PAS domains have been identified in proteins from all three kingdoms of life: Bacteria, Archaea, and Eucarya. These include histidine and serine/threonine kinases, chemoreceptors and photoreceptors for taxis and tropism, circadian clock proteins, voltage-activated ion channels, cyclic nucleotide phosphodiesterases, and regulators of responses to hypoxia and embryological development of the central nervous system. PAS domains are combined with a variety of regulatory modules in multidomain proteins. As a result, a spectrum of cell responses to changes in the environmental and intracellular conditions are controlled via PAS-containing receptors, transducers, and regulators.
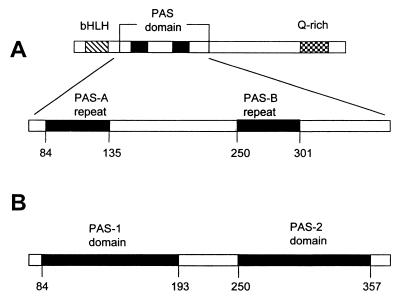
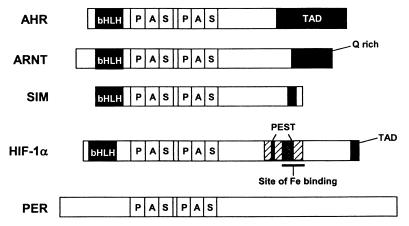
PAS is an acronym formed from the names of the proteins in which imperfect repeat sequences were first recognized: the Drosophila period clock protein (PER), vertebrate aryl hydrocarbon receptor nuclear translocator (ARNT), and Drosophila single-minded protein (SIM) (163). The earliest investigations identified the PAS domain in eukaryotes as a region of approximately 270 amino acid residues that contained two 50-residue conserved sequences termed PAS-A and PAS-B repeats (Fig. 1A) (40, 101). Recent studies suggest that a PAS domain comprises a region of approximately 100 to 120 amino acids. PAS-A and PAS-B repeats correspond to the N-terminal half of the respective PAS domains (Fig. 1B). It is typical to find PAS domains in pairs in eukaryotic transcriptional activators, such as SIM. Microbial proteins contain single, dual, or multiple (up to six) PAS domains.
FIG. 1.
Comparison of former (A) and present (B) definitions of PAS domains illustrated with the Drosophila SIM protein. (A) One PAS domain containing two PAS repeats as first described. (B) Two individual PAS domains have been identified in the SIM protein. Q-rich, glutamine-rich activation domain.
Several laboratories contributed to the current definition of a PAS domain. Lagarias et al. (131) identified a motif, similar to a PAS repeat, in an algal phytochrome and in 20 other proteins from both prokaryotes and eukaryotes. They also suggested that this 40-amino-acid motif represents a common fold that might be similar to the N terminus of the photoactive yellow protein, for which the crystallographic structure had been determined (26). Subsequently, it was recognized that this motif is the most highly conserved block (S1 box) of a larger PAS domain. The motif was extended in the carboxyl direction by defining the S2 box (or PAC motif), and complete PAS domains (including S1 and S2 boxes or PAS/PAC motifs) were identified in more than 200 proteins from different organisms throughout the phylogenetic tree (182, 266). More recently, the entire 125-residue photoactive yellow protein (PYP) (172), the heme domain of the FixL protein (82), and the N-terminal domain of the eukaryotic potassium channel HERG (160) were proposed as structural prototypes for the three-dimensional fold of the PAS domain superfamily. In this review, we present an alignment of the sequences from the PAS domain superfamily that supports this generalization.
The term “PAS domain” is used in this review to denote structures similar to the PYP, FixL, and HERG prototypes or the sequence that constitutes the PAS fold. To refer to regions of sequence similarity, we abandoned the use of S1/S2 (266) and PAS/PAC (182) in favor of referring to the PAS structural elements that the sequences specify (172). The recently described LOV domain (103) is a PAS domain by our definition, and we do not use the term “LOV.” When consulting the literature on the subject, readers should be aware of the progression in the meaning of “PAS domain.” Further confusion could be avoided by replacing the name “PAS” with a structural designation for the domain.
We have summarized the current knowledge of PAS domains with an emphasis on known and potential sensory and signaling roles in representative prokaryotic and eukaryotic systems. At the time this review was completed (August 1998), the number of identified PAS domains was growing rapidly. We have made no attempt to describe all proteins in which PAS domains are found but hope that our compilation will provide a broader picture of conservation and diversity in signal transduction pathways that involve these unique signaling modules.
PAS DOMAIN SUPERFAMILY
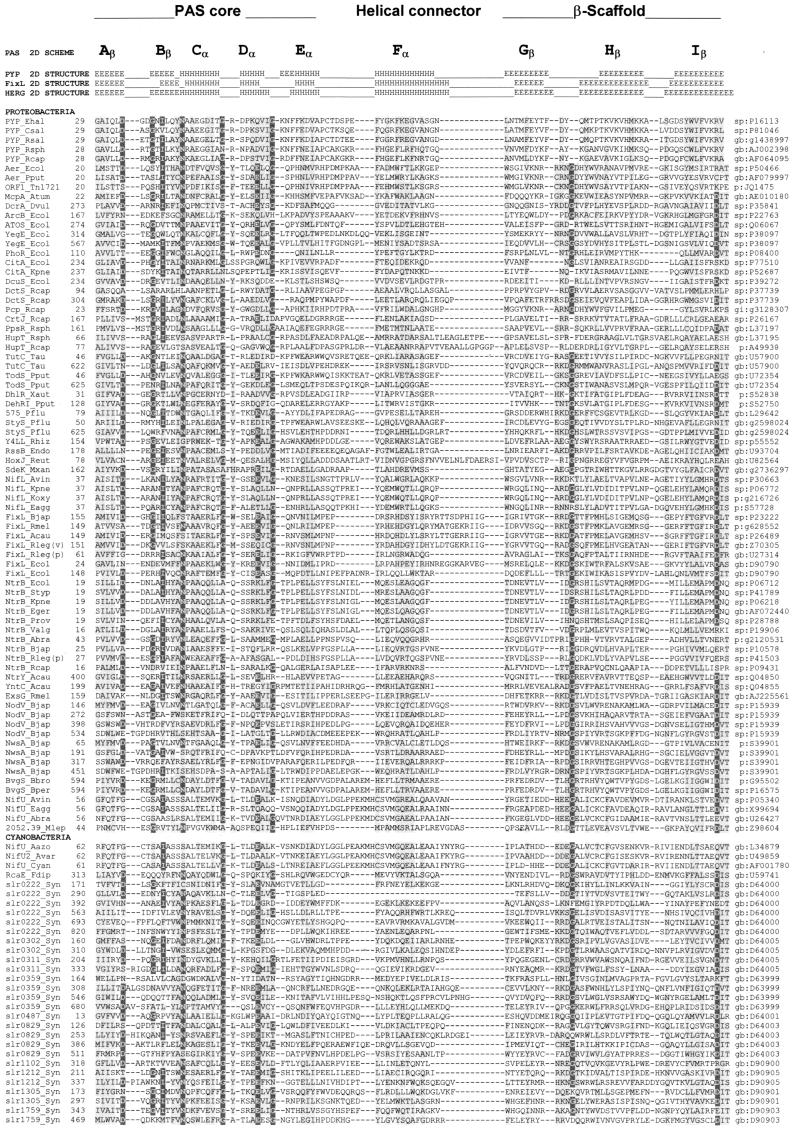
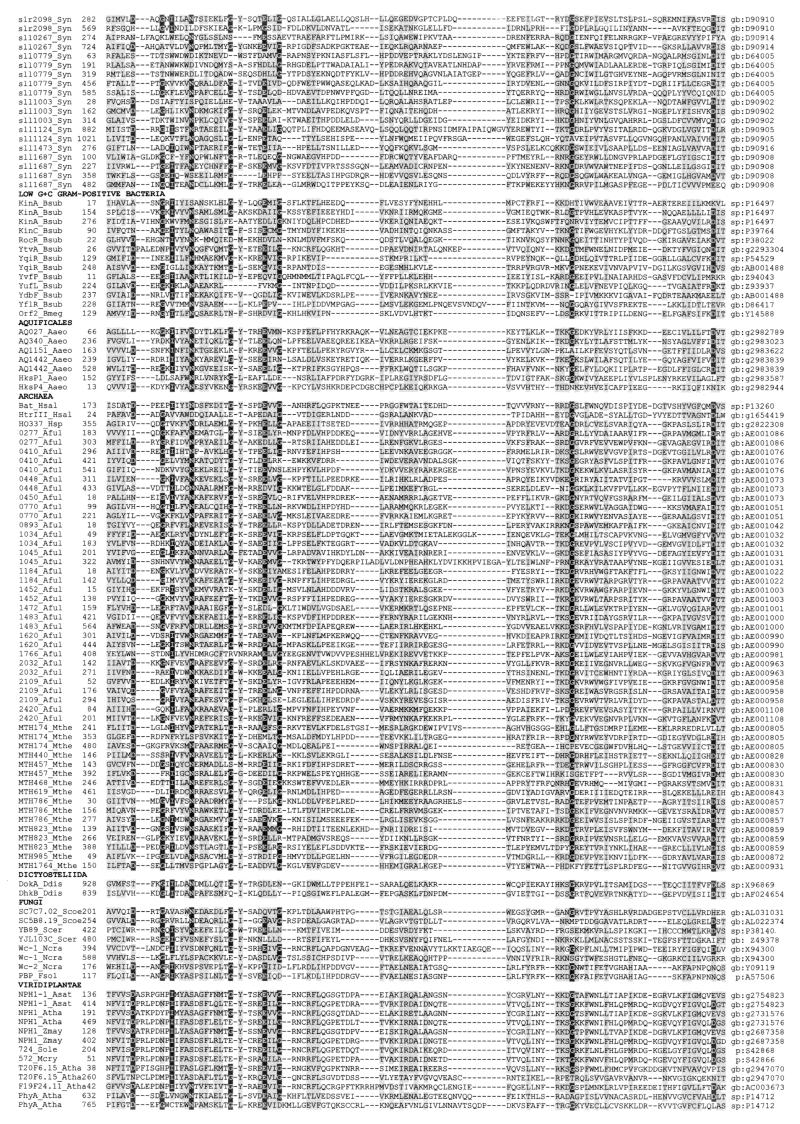
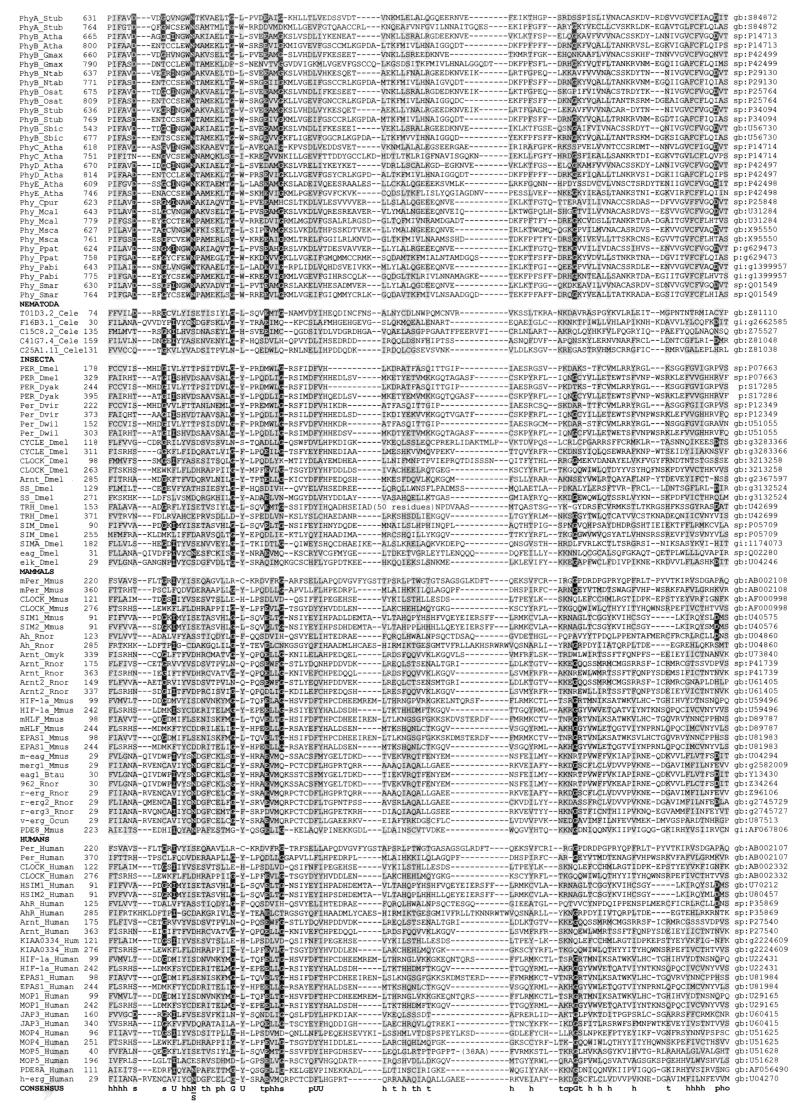
A multiple alignment of more than 300 PAS domains from more than 200 proteins is shown in Fig. 2, together with the secondary structures of PYP, FixL, and HERG determined by crystallographic analysis (82, 160, 172). Although a systematic study of the occurrence of PAS domains within the branches of the phylogenetic tree has not been performed, it is evident that PAS domains are not confined to specific phylogenetic groups. However, not all species have PAS domains. Analysis of completely sequenced bacterial and archaeal genomes revealed that within both Bacteria and Archaea some species contain no recognizable PAS domains whereas others have abundant PAS domains (182, 266). Table 1 illustrates the widespread distribution of PAS domains throughout the phylogenetic tree and the function of the corresponding proteins.
FIG. 2.
Multiple alignment of PAS domains. The alignment was constructed as described in “Search strategy” (see the text) and in reference 266 with modifications from reference 182. The secondary structures of PYP, FixL, and HERG were adapted from references 82, 160, and 172 and are numbered by using the convention of Gong et al. (82). Subdomains of the crystallographic structure of PYP (172) are shown above the secondary structure. The highly variable N-terminal cap segment is not included in the alignment. Identical amino acids that are conserved in at least 50% of sequences are in reverse contrast; similar residues conserved in at least 75% of PAS domains are shaded. Consensus sequences are shown below the alignment (threshold = 75%); c, charged (DEHKR), U, bulky hydrophobic (FILMVWY); h, hydrophobic (ACFGILMTVWY); o, hydroxy (S, T); p, polar (CDEHKNQRST); t, turn-like (ACDEGHNQRST); s, small (ACDGNPSTV). An updated version of the alignment is maintained at www.llu.edu/medicine/micro/PAS.
TABLE 1.
Representative PAS domain-containing proteins from the three kingdoms of life
| Protein or open reading frame and species | Description | Accession no.a |
|---|---|---|
| Bacteria | ||
| Proteobacteria | ||
| α-Subdivision | ||
| FixL (Sinorhizobium meliloti) | Sensor kinase, oxygen-dependent regulator of nitrogen fixation | P: S39984 |
| EsxG (Sinorhizobium meliloti) | Sensor kinase controlling succinoglycan synthesis | GB: AJ225561 |
| NtrY (Azorhizobium caulinodans) | Sensor kinase controlling nitrogen level | SP: Q04850 |
| NwsA (Bradyrhizobium japonicum) | Sensor kinase controlling nodulation response | P: S39901 |
| NodV (Bradyrhizobium japonicum) | Sensor kinase controlling nodulation response | SP: P15939 |
| NtrB (Azospirillum brasilense) | Sensor kinase controlling nitrogen assimilation | P: I39493 |
| PpsR (Rhodobacter sphaeroides) | Sensor kinase, redox-dependent regulator of photosynthesis | GB: L37197 |
| DctS (Rhodobacter capsulatus) | Sensor kinase controlling dicarboxylate transport | SP: P37739 |
| PleC (Caulobacter crescentus) | Sensor kinase controlling polar organelle development | P: S27533 |
| McpA (Agrobacterium tumefaciens) | Chemotaxis transducer (?) | GB: AF010180 |
| β-Subdivision | ||
| TutC (Thauera sp. strain T1) | Sensor kinase controlling toluene degradation | GB: U57900 |
| BvgS (Bordetella bronchiseptica) | Sensor kinase controlling virulence | P: S17944 |
| γ-Subdivision | ||
| Aer (Escherichia coli) | Oxygen (redox) taxis transducer | SP: P50466 |
| ArcB (Escherichia coli) | Sensor kinase, redox-dependent regulator of aerobic metabolism | SP: P22763 |
| PhoR (Escherichia coli) | Sensor kinase controlling phosphate regulon | SP: P08400 |
| ATOS (Escherichia coli) | Sensor kinase controlling ornithine decarboxylase antizyme | SP: Q06067 |
| TodS (Pseudomonas putida) | Sensor kinase controlling toluene degradation | GB: U72354 |
| StyS (Pseudomonas sp.) | Sensor kinase controlling styrene degradation | GB: AJ000330 |
| NifL (Azotobacter vinelandii) | Sensor, redox-dependent regulator of nitrogen fixation | SP: P30663 |
| δ-subdivision | ||
| SdeK (Myxococcus xanthus) | Sensor kinase controlling fruiting-body development | GB: AF031084 |
| DcrA (Desulfovibrio vulgaris) | Oxygen (redox) taxis transducer (?) | SP: P35841 |
| Cyanobacteria | ||
| RcaE (Fremyella diplosiphon) | Sensor kinase, phytochrome/ethylene receptor | GB: U59741 |
| Sll1003 (Synechocystis sp.) | Sensor kinase (?) | GB: D90902 |
| Slr1212 (Synechocystis sp.) | Sensor, ethylene response regulator (?) | GB: D90905 |
| Low G+C Gram-positive bacteria | ||
| KinA (Bacillus subtilis) | Sensor kinase controlling sporulation | SP: P16497 |
| KinC (Bacillus subtilis) | Sensor kinase controlling sporulation | SP: P39764 |
| Aquificales | ||
| HksP2 (Aquifex aeolicus) | Sensor kinase (?) | GB: AE000683 |
| Archaea | ||
| Euryarchaeota | ||
| Bat (Halobacterium salinarum) | Sensor, oxygen-dependent bacterio-opsin activator | SP: P13260 |
| AF0277 (Archaeoglobus fulgidis) | Sensor kinase (?) | GB: AE001086 |
| AF1034 (Archaeoglobus fulgidis) | Chemotaxis transducer (?) | GB: AE001032 |
| MTH174 (Methanobacterium ther-moautotrophicum) | Sensor kinase (?) | GB: AE000805 |
| Eucarya | ||
| Dictyosteliida | ||
| DokA (Dictyostelium discoideum) | Sensor kinase controlling osmotic response | GB: X96869 |
| Fungi | ||
| Hemiascomycetes | ||
| YB89 (Saccharomyces cerevisiae) | Transcriptional regulator (?) | SP: P38140 |
| Euascomycetes | ||
| Wc-1 (Neurospora crassa) | Transcriptional regulator of the blue-light response | GB: X94300 |
| Wc-2 (Neurospora crassa) | Phototransducer, clock component | GB: Y09119 |
| PBP (Fusarium solani) | Transcriptional regulator | P: A57506 |
| Viridiplantae | ||
| Charophyta | ||
| Phy1b (Mesotaenium caldariorum) | Phytochrome phototransducer | GB: U31284 |
| Phy (Mougeotia scalaris) | Phytochrome phototransducer | GB: X95550 |
| Embryophyta | ||
| PhyBb (Arabidopsis thaliana) | Phytochrome phototransducer | SP: P14713 |
| NPH1 (Arabidopsis thaliana) | Sensor kinase controlling phototropism | GB: AF030864 |
| Metazoa | ||
| Nematoda | ||
| T01D3.2. (Caenorhabditis elegans) | Single-minded protein (SIM) homolog | GB: Z81110 |
| Insecta | ||
| PER (Drosophila melanogaster) | Transcriptional regulator of circadian rhythms | SP: P07663 |
| ARNT (Drosophila melanogaster) | Aryl hydrocarbon receptor nuclear translocator (ARNT) | GB: AF016053 |
| SIM (Drosophila melanogaster) | Global transcriptional regulator | SP: P05709 |
| EAG (Drosophila melanogaster) | Voltage-sensitive potassium channel subunit | SP: Q02280 |
| Actinopterygii | ||
| ARNT (Oncrhynchus mykiss) | Aryl hydrocarbon receptor nuclear translocator (ARNT) | GB: U73840 |
| Mammalia | ||
| CLOCK (Mus musculus) | Transcriptional regulator of circadian rhythms | GB: AF000998 |
| ARNT (Mus musculus) | Aryl hydrocarbon receptor nuclear translocator (ARNT) | GB: U61405 |
| SIM1 (Mus musculus) | Single-minded protein (SIM) homolog | GB: U40575 |
| m-EAG (Mus musculus) | Voltage-sensitive potassium channel subunit | GB: U04294 |
| HIF-1α (Mus musculus) | Hypoxia-inducible factor 1α | GB: U59496 |
| Humans | ||
| ARNT (Homo sapiens) | Aryl hydrocarbon receptor nuclear translocator | GB: U61405 |
| h-ERG (Homo sapiens) | Voltage-sensitive potassium channel subunit | GB: U04270 |
| HIF-1α (Homo sapiens) | Hypoxia-inducible factor 1α | GB: U22431 |
| HIF-2β (Homo sapiens) | Hypoxia-inducible factor 2α | GB: U51626 |
The accession number for each sequence is for the SWISS-PROT (SP), GenBank (GB), or PIR (P) database.
Analogous proteins are found in more than 20 species from this taxonomic group.
PAS domains are found predominantly in proteins that are involved directly or indirectly in signal transduction. Of the more than 200 proteins that contain PAS domains, most of those for which a function is either known or proposed are receptors, signal transducers, and transcriptional factors (266). Already the available information about PAS domains has enabled the assignment of putative function to many newly sequenced genes (Table 1). Some of the proteins in which PAS domains have now been identified, such as ArcB and NifL, have been extensively studied (110, 111, 125, 207). Identification of a PAS module in these proteins suggests new strategies for elucidating the signal transduction mechanism, which has proved elusive in the past. Investigation of signal transduction in other proteins lags because investigators have overlooked the presence of PAS domains. The oversight may reflect widespread confusion about what constitutes a PAS domain and the limited success of computerized searches for the PAS motif prior to the introduction of Gapped- and PSI-BLAST programs (7).
In members of the Bacteria and Archaea, PAS domains are found almost exclusively in sensors of two-component regulatory systems. This may be a universal rule since it is applicable to all six completely sequenced bacterial and archaeal genomes, where PAS domains have been identified (265). In members of the Eucarya, two major classes of PAS proteins have been recognized (266): transcriptional factors and voltage-sensitive ion channels. However, new classes of PAS proteins are emerging. These include proteins with kinase activity: both histidine and serine/threonine kinases have been found in members of the Eucarya. Histidine kinases first seemed to be limited to lower eukaryotes (3), such as Dictyostelium. Dictyostelium discoideum has an osmosensing histidine kinase DokA (202), with a PAS domain (182), and a novel histidine kinase that regulates spore germination (268), where we have identified a PAS domain (Fig. 2). Recently, histidine kinases have been found in plants (69, 119). Although PAS domains have not been identified in these proteins, they contain functional domains that are similar to PAS-containing cyanobacterial sequences. PAS domains have been identified in several plant serine/threonine protein kinases (182) (Fig. 2). The recent discovery of a PAS domain in a novel cyclic AMP-specific cyclic nucleotide phosphodiesterase in mammals (208) further extends the PAS domain superfamily. Phosphodiesterases regulate the intracellular level of cyclic nucleotides and are involved in the regulation of important physiological process such as visual and olfactory signal transduction (152, 254).
PAS-containing transcriptional factors have been found in fungi and metazoa, whereas PAS-containing ion channels have been found so far only in metazoa (Table 1). We and others (182, 266) have found PAS domains in representatives of a superfamily of voltage-activated potassium channels (243).
SENSING BY THE PAS DOMAIN
A Versatile Sensor Domain
Adaptation of the PAS domain structure to sense diverse stimuli such as oxygen, ligands, light, and redox potential is present in the simplest prokaryotes, and evidence is emerging that divergent PAS domains in a single protein may be functionally differentiated to sense different stimuli. PAS domains also determine the specificity of transcriptional factors in activating target genes. Chimeras constructed from the Trachealess (Trh) and Single-minded (Sim) proteins from Drosophila confirmed the specificity of the PAS domains in transcriptional activation. Replacement of the Trh PAS domain by the analogous region of Sim produced a chimera with the functional specificity of a Sim protein in gene activation (261).
In a signaling pathway, the receptor interacts with a stimulus and transduces a signal that can be processed by the cell. In some signaling pathways, the signal from the receptor is itself transduced into a different form of energy by a second protein. The second protein is termed a transducer to distinguish it from the receptor that detects the initial stimulus. This is a useful but arbitrary distinction because both proteins have a transduction function. Of the PAS proteins that sense light, PYP is a receptor in which blue light is captured by the 4-hydroxycinnamyl chromophore in the PAS domain (13). FixL is an oxygen receptor (77), in which oxygen binds directly to a heme that is coordinated to a histidine residue within a PAS domain (158). Other PAS proteins, such as Aer, are transducers that sense oxygen indirectly by sensing redox changes as the electron transport system responds to changes in oxygen concentration (132, 186, 224). At present, little is known about differences in transduction mechanisms between PAS receptors and transducers.
Input Modules of Two-Component Systems: Oxygen and Redox
Most PAS domains in prokaryotes are in histidine kinase sensor proteins (182, 265). The prototypical two-component regulatory system consists of a histidine kinase sensor and cognate response regulator (10, 84, 99, 167, 168). An N-terminal input module of the histidine kinase senses stimuli directly, or indirectly with an upstream receptor. A C-terminal transmitter module includes a conserved histidine that is the site of autophosphorylation. The phosphoryl moiety is transferred from the sensor histidine to a conserved aspartate in the receiver module of the response regulator, usually in the N terminus. As the result of phosphorylation, the output domain of the response regulator is activated and is capable of interacting with either DNA or another signaling protein. In most cases, the response regulator is a transcriptional activator.
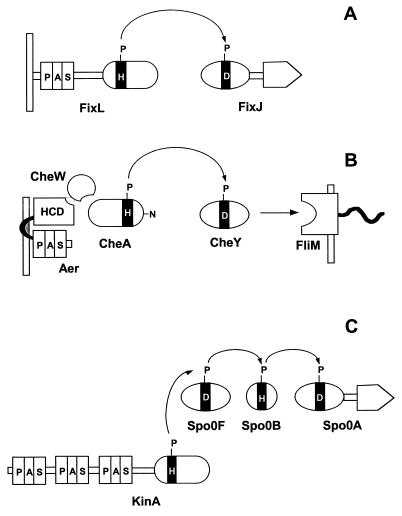
Figure 3 illustrates two-component signal transduction strategies in which PAS domains sense oxygen or redox potential. The FixL/FixJ pathway (Fig. 3A) in Sinorhizobium meliloti, Bradyrhizobium japonicum, and related bacteria is prototypical (46, 47). FixL is an oxygen sensor. Oxygen dissociation from the input PAS domain (22, 76, 77, 82, 158, 189) changes the conformation of the PAS domain, resulting in altered structure and increased autophosphorylation activity of the transmitter domain (76, 77, 82, 159). FixL catalyzes a His-Asp phosphoryl transfer to the receiver module of the response regulator FixJ. Phosphorylated FixJ acts as a transcriptional activator of the genes involved in nitrogen fixation.
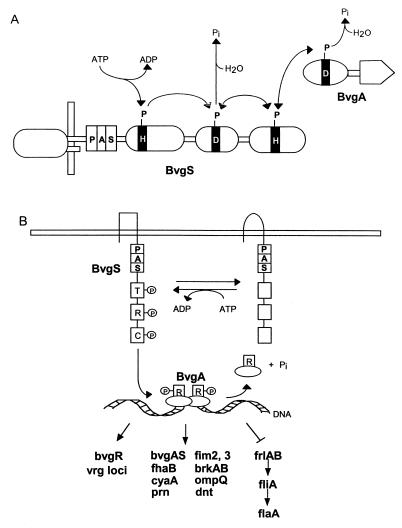
FIG. 3.
PAS domain modules in histidine kinase phosphorelay systems. (A) FixL-FixJ from Sinorhizobium meliloti. (B) Aerotaxis pathway from E. coli. (C) Pathway for initiation of sporulation in Bacillus subtilis.
In the signaling pathway for aerotaxis, the behavioral response of E. coli to oxygen, the output is not transcriptional regulation as in FixJ but direct protein-protein interaction with the flagellar motors. The PAS domain is upstream of the transmitter module (Fig. 3B). Oxygen binds to the terminal oxidases of the electron transport system. A PAS domain in the flavoprotein transducer Aer senses a change in redox potential in the electron transport system and inhibits a highly conserved domain (HCD) in the C-terminal segment of Aer (224). The HCD is a homolog of the chemotaxis-signaling domains that interact with the CheA histidine kinase and CheW protein, regulating the rate of autophosphorylation of CheA. The CheA sensor kinase has an unusual structural organization, with the transmitter being located centrally. The C-terminal segment functions as an input domain that couples CheA to CheW and the HCD (141, 167). The phosphorylated histidine in CheA is outside the transmitter module in a small N-terminal domain. CheY is a free-standing receiver domain that, when phosphorylated, binds to the FliM switch protein and reverses the direction of rotation of the flagellar motor (17, 246).
The modules of the two-component regulatory systems have been used in nature to construct more complex phosphorelay circuits. The four-step His-Asp-His-Asp phosphorelay that governs the initiation of sporulation in Bacillus subtilis illustrates the input role of PAS domains in a complex circuit (34, 85) (Fig. 3C). The KinA protein has three PAS domains in the N-terminal segment. A phosphoryl residue in KinA is transmitted from His to Asp to His to Asp, where it ultimately activates the Spo0A transcriptional regulator (Fig. 3C). KinA is a soluble cytoplasmic sensor kinase (Fig. 3C); the stimuli sensed by KinA have not been identified.
Photoreceptors, Phytochromes, and Clock Proteins: Light
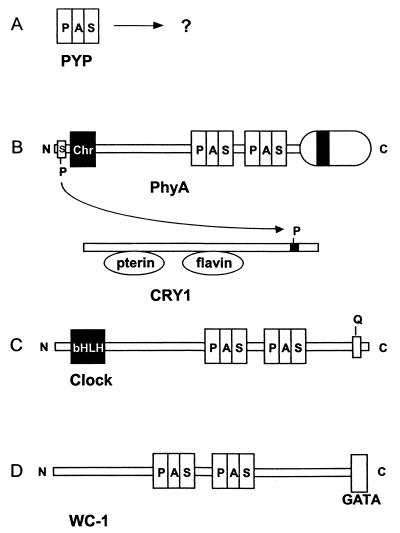
PAS domains are important modules in photoreceptors and in clock proteins, which are postulated to be derived from photoreceptors (Fig. 4). PYP is a bacterial photoreceptor postulated to govern a photophobic swimming response in Ectothiorhodospira halophila (211). The photoreceptor is an isolated PAS domain with a 4-hydroxycinnamyl chromophore attached (Fig. 4A). The structure and signal transduction mechanism of this photoreceptor are discussed below.
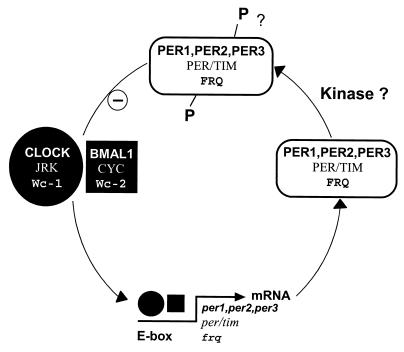
FIG. 4.
PAS domain modules in photoreceptor signaling pathways and clock proteins. (A) PYP. (B) PhyA-CRY1 phosphorelay in Arabidopsis for phytochrome regulation of cryptochrome. (C) Clock protein from mouse. (D) WC-1 clock-associated protein from Neurospora crassa. Abbreviations: chr, chromophore; GATA, GATA-like zinc finger domain.
Plant phytochromes have a photoreceptor domain with a linear tetrapyrrole chromophore. The receptor domain is separated from two PAS domains by a hinge region (185). A histidine kinase-like transmitter domain that has serine/threonine kinase activity (256) is C-terminal to the PAS domains (200) (Fig. 4B). There is no histidine phosphorylation in plant phytochromes, but a serine at the N terminus of oat phytochrome A is autophosphorylated (250) and the cryptochrome blue-light receptor CRY1 is regulated through phosphorylation by phytochrome A (1). It is conceivable that the PAS domain region transduces the light signal to regulate the kinase activity. In this regard, the cyanobacterial phytochrome Cph1 has a photosensory domain directly adjacent to a transmitter domain and has been shown to be a light-regulated histidine kinase (56, 257).
PAS domains have received widespread attention as a signature motif in circadian clocks (126, 197). In addition to being transcriptional regulators with DNA-binding modules, many clock proteins have PAS modules. For example, the mammalian CLOCK protein (9, 72, 127) has two PAS domains (Fig. 4C) and a typical basic helix-loop-helix (bHLH) DNA-binding domain (127) that is located N-terminal to the PAS domains, in contrast to PAS-containing histidine kinases, where the kinase domain is C-terminal to the PAS domain(s). White collar-1 (Wc-1) is a clock-associated protein that is essential for circadian blue-light responses in Neurospora crassa (14, 16, 41) (Fig. 4D). There are two PAS domains in Wc-1 (15, 182, 266) that are possible binding sites for the flavin chromophore in the protein (14). C-terminal to the PAS domains is a GATA-like zinc finger DNA-binding domain (16).
Voltage-Sensitive Ion Channels: Oxygen and/or Ion Motive Force?
PAS domains have been identified in a family of voltage-activated potassium channels that are related to the Drosophila Eag protein (243). The K+ channel is a homotetramer with a central pore. A single PAS domain has been identified in the N-terminal domain of the channel-forming subunit, which extends into the cytoplasm (182, 266). Mutations in the human homolog of Eag (HERG) are associated with cardiac arrhythmias (237, 241). The PAS domain regulates deactivation of the HERG channel (160), but the stimulus detected by the domain has not been determined. One possibility is that the primary role of the PAS domain is in sensing redox (oxygen) changes and regulating the channel activity accordingly. Oxygen-sensing potassium channels have been found previously in organisms ranging from bacteria to humans.
PAS as a Protein-Protein Interaction Domain
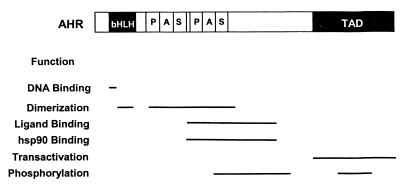
Protein-protein interactions mediate signal transduction by some PAS proteins, and the PAS core may determine the specificity of the interactions (105). PAS domains in PER proteins form homodimers in vitro, but PAS domains are usually involved in heterodimer formation. Where present in PAS proteins, such as the aryl hydrocarbon receptor (AHR) and aryl hydrocarbon receptor nuclear translocator (ARNT), the bHLH motif serves as an interface for heterodimerization. However, PAS domains add increased stability and specificity to the dimers (178). The ARNT protein forms heterodimers with AHR and mammalian hypoxia-inducible factor (HIF1α) in addition to forming homodimers. Heterodimers are also formed by CLOCK and CYCLE proteins and by PER and TIM proteins in circadian circuits (72, 197). The specificity of PAS transcriptional enhancers in binding to DNA response elements is determined by the composition of the dimer (193). A role of a PAS domain in dimerization of plant phytochromes has been suggested (55). As noted above, PAS domains are located in the N-terminal region of subunits that form tetramers of voltage-sensitive ion channels. Truncated subunits that have only the N-terminal PAS-containing region can form tetramers in solution (136). Photoactivation of the PYP photoreceptor and the resulting rearrangement of the chromophore in the PAS domain induces a conformational change that is transmitted to the surface of the protein, where it is proposed to alter the protein interaction site (74). However, the role of protein-protein interactions in signal transduction by the PAS domain may have been overemphasized. Crystallized PAS domains from FixL (82) and HERG (160) are monomeric.
Intracellular Location of PAS Domains
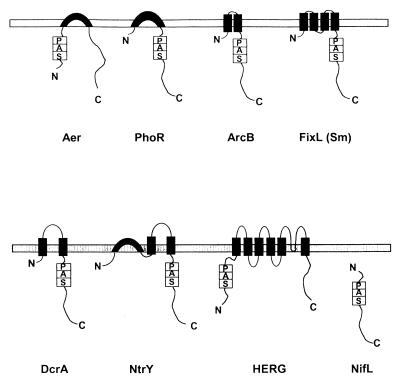
Figure 5 shows the topology of selected PAS domain-containing proteins in the cytoplasmic membrane. Regardless of the topology architecture, an intracellular location of single and multiple PAS domains was predicted in all analyzed proteins. Even in the PAS proteins with an extended periplasmic domain, such as DcrA, BvgS, and NtrY, PAS domains are located in the cytoplasm. PAS domains are also present in various soluble cytoplasmic sensors, such as NifL (49). The cytoplasmic location of PAS domains suggests that they sense changes in the intracellular environment. PAS domains can directly sense the environment outside the cell for stimuli that enter the cell, such as light, and can indirectly sense where the outside environment affects the intracellular environment.
FIG. 5.
Topology of selected PAS domain-containing proteins in the cytoplasmic membrane: Aer (Swiss-Prot accession no. P50466), PhoR (Swiss-Prot P08400), ArcB (Swiss-Prot P22761), FixL (PIR g628552), DcrA (Swiss-Prot P35841), NtrY (Swiss-Prot Q04850), HERG (GenBank U04270), and NifL (Swiss-Prot P30663). The putative transmembrane regions were predicted using the dense alignment surface (DAS) method (43) and comparing predictions to proteins with known topology.
The role of the membrane in signal transduction by PAS-containing proteins is unclear. The FixL protein of Sinorhizobium meliloti (Fig. 5) is an integral membrane protein with four putative transmembrane regions in its N terminus (143). We have predicted a similar topology for FixL from Azorhizobium caulinodans (120). However, FixL proteins from Bradyrhizobium japonicum (8), Rhizobium etli (48), and Rhizobium leguminosarum bv. viciae (169) do not appear to have any transmembrane region and apparently are soluble cytoplasmic proteins. Both oxygen-sensing and kinase activities appear to be similar in membrane-bound and soluble FixL proteins (75, 76). In all membrane-bound proteins, PAS domains are located adjacent to the transmembrane regions; therefore, it is possible that they interact with domains of other membrane-associated proteins.
IDENTIFICATION AND STRUCTURE OF THE PAS DOMAIN
Search Strategy
The discovery of PAS domains in a large variety of prokaryotic and eukaryotic sequences can be attributed to the overall improvement of computer-based macromolecule sequence analysis. Since PAS domains are usually present in proteins with multidomain architecture, it is necessary to reduce the noise during computerized searches for PAS domains. For example, in bacteria, most known PAS domain-containing proteins are protein kinases that contain the extremely widespread kinase domain. In a standard BLAST search (5), this domain always scores highest, returning a “noise” of histidine kinases that do not have a PAS domain. Similarly, the HCD (135) can mask PAS domains in searches with bacterial chemotaxis transducers.
A strategy for noise reduction is illustrated by the iterative process that we used to identify the PAS domain superfamily in the course of studying the newly discovered Aer transducer of E. coli (24, 186, 266). The initial standard BLAST search with a complete sequence of the Aer protein as a query revealed similarity between the N-terminal region of Aer, the NifL redox sensor, the Bat oxygen sensor, and the Wc-1 clock-associated protein (186). Further searches were performed after filtering the Aer sequence for known structural features, such as the HCD and a putative transmembrane region. That is, we restricted the queries to the first 166 N-terminal amino acid residues of Aer that were free of recognizable motifs and to homologous regions of NifL, Bat, and Wc-1. Eukaryotic PAS-containing proteins were found during these searches, and the similarity between the Aer N terminus and the PAS domains of ARNT and Sim indicated that Aer contains an authentic PAS domain. Multiple alignment of all returned hits that had similarity to the N terminus of Aer revealed that the PAS domain contains a variable (both in amino acid composition and in length) region between two more conserved motifs that we termed S-boxes (266). We then performed multiple reciprocal BLAST searches with, as queries, complete PAS motifs and individual S-boxes from all sequences found in the individual searches. A complete multiple alignment of generated sequences was constructed, and statistical analysis was used to verify that sequences included in the alignment had significant similarity (Z scores ranged from 3.7 to 14 [266]). The secondary structure of the region was predicted by using the PhD server (192). Similar results have been obtained independently by Ponting and Aravind (182). Important evidence for PAS as a functional domain came from a comparison of the predicted secondary structure with the known three-dimensional structures of PYP and of the FixL and HERG PAS domains (see “Structure of the PAS fold” below).
We recommend to investigators searching for PAS domains or for similar functional domains in their sequences of interest two guides that were published recently (6, 27). The strategy presented in the guides can improve the quality of searches in sequence databases, in-depth analysis of protein sequences, and prediction of functions from sequences. The gapped BLAST and position-specific iterative (PSI) BLAST programs (6, 7) have improved the accuracy of searches for PAS domains in newly sequenced proteins in both nonredundant (National Center for Biotechnology Information, National Institutes of Health, Bethesda, Md.) and some specialized (The Institute for Genomic Research, Rockville, Md.) databases. We recently analyzed the occurrence of PAS domains in completely sequenced microbial genomes by using gapped BLAST for exhaustive, iterative searches of nonredundant and specialized microbial databases (265) (see “PAS domains in microbial genomes” below).
Structure of the PAS Fold
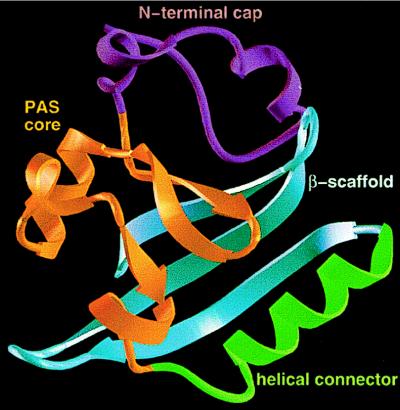
By mapping a typical PAS domain from the ARNT protein onto the crystallographic structure of the entire PYP, Getzoff and collaborators developed an argument for PYP as a prototypical PAS domain (172). This generalization is supported by the subsequent determination of the structure of PAS domains in the FixL protein from B. japonicum (82) and the human HERG protein (160). The Ectothiorhodospira halophila PYP is a self-contained, bacterial blue-light receptor with an unusual fold characterized by a central six-stranded β-sheet with N- and C-terminal β-strands (26) (Fig. 6). Four segments have been delineated in the overall PAS fold in PYP: (i) the N-terminal cap or lariat (residues 1 to 28), including the α1 and α2 helices; (ii) the PAS core with the first three β-strands of the central β-sheet (residues 26 to 69) and the α3 and α4 helices; (iii) the helical connector (residues 70 to 86) with the α5 helix, which diagonally crosses the β-sheet and connects two edge β-strands; and (iv) the β-scaffold, composed of β4, a connecting loop, and the β5-β6 hairpin that form the second three-stranded half of the central β-sheet (172) (Fig. 6). In PYP, there is a hydrophobic core on each side of the central β-sheet (26). The N-terminal cap encloses one side of the β-sheet to form the smaller hydrophobic core. The remaining helices and loops surround the other side of the central β-sheet to form the larger hydrophobic core, into which the 4-hydroxycinnamyl chromophore is inserted.
FIG. 6.
Proposed PAS three-dimensional fold illustrated on the PYP structure. The N-terminal cap (purple) includes residues 1 to 28; the PAS core (gold) includes residues 29 to 69, the helical connector (green) includes residues 70 to 87; and the β-scaffold (blue) includes residues 88 to 125. Courtesy of J. L. Pellequer and E. D. Getzoff. Reprinted from reference 172 with permission of the publisher.
The PAS core (Fig. 6), which has the highest density of conserved residues in PAS domains (172, 182, 266), corresponds approximately to the first reported PAS sequence motif (50 residues) (131, 182) and to the S1 box (266). This is the photosensing active site of PYP that has the Cys69 attachment site for the chromophore and forms most of the immediate environment of the chromophore, including all residues that hydrogen bond to the chromophore (172). The PAS core also contributes residues to a PAS protein-protein interaction site.
The β-scaffold in PYP constitutes a long platform with characteristic β-sheet twist that supports the PAS core and completes the central six-stranded β-sheet (172). The ω-loop between β4 and β5 closes a gap between the PAS core and the β-scaffold and completes the chromophore environment. The β-scaffold corresponds approximately to the PAC sequence motif (182) and the S2 box (266) introduced previously to designate segments of the PAS sequence homology. Similar crystallographic structures have been determined for the PAS domains from FixL (BjFixLH) and the human HERG potassium channel N terminus. The B. japonicum BjFixLH domain has only five β-strands; otherwise the domain structure closely resembles PYP (82). The N-terminal cap (corresponding to residues 1 to 25 in PYP) is disordered and therefore is not defined in the crystal lattice. This is also true for the HERG protein. A convenient nomenclature adopted for BjFixLH can be generalized for other PAS domains and is shown at the top of Fig. 2. The structural elements are designated Aβ, Bβ, Cα, Dα, Eα, Fα, Gβ, Hβ, and Iβ. Each loop is defined by the secondary structures that flank it (e.g., loop AB). A standardized residue-numbering system for the secondary structures is proposed (82). If universally adopted, this system would facilitate discussion of conserved residues in different PAS domains.
The largest differences between BjFixL and PYP pertain to enclosure of the cofactors, heme and hydroxycinnamic acid, respectively. The differences are localized around the central helix Fα (helical connector), to which the heme is coordinated, and the loops that flank it. This core region appears to be the critical regulatory region of the PAS domain family (82).
The HERG and PYP PAS domains also have highly similar three-dimensional structures (160). The major difference between the two proteins is again in the Fα helical connector. Of particular interest is a hydrophobic patch on the HERG PAS domain that is proposed to be a protein-protein interaction site by which the PAS domain adheres to the body of the potassium channel. Mutations that inactivate the HERG PAS domain are clustered at one boundary of the hydrophobic patch (160). Based on the structural predictions of Pellequer et al. (172), we reexamined the sequence alignment of more than 300 PAS domains (Fig. 2). The individual elements of the secondary structure of PYP are conserved throughout the alignment. We concluded that it is likely that all PAS domains have the PAS core, helical connector, and β-scaffold structural elements. In addition, there is an α-helix, which is attached to the C-terminal end of the PAS domain (82) and links the domain to another protein module.
The N-terminal cap is the least highly conserved segment of the PAS domain (172). Other structures that can protect the central β-sheet from solvent may replace the cap. Due to this variation, the N-terminal cap is not included in our compilation of PAS domain sequences (Fig. 2). The largest length variations in PAS sequences are in the FG loop joining the Fα helical connector region and Gβ strand (Fig. 2). The helical connector and β-scaffold also have a lower degree of sequence conservation than the PAS core. As predicted, PAS domains that have different cofactors also differ in the residues that surround and interact with the cofactors.
The functional importance of the shape of the PAS domain is clearly indicated by crystallographic analysis of profilin (155), and the Src homology 2 (SH2) domain (238). Profilin binds actin and is a signaling component in microfilament-based cell motility (198). SH2 domains bind phosphotyrosine and signal the phosphorylation state of regulatory proteins to the signal transduction pathway (129). Profilin, the SH2 domain, and PYP have strikingly similar three-dimensional structures but they do not share sequence homology (26). This suggests that the similar domain structures have independent origins. Further structure-function analysis of PAS, SH2 domains, and profilin are required to clarify what is so important about this structure.
Structure and Signal Transduction Mechanisms
In an elegant series of crystallographic analyses, Getzoff and collaborators have succeeded in monitoring, on a millisecond time scale, the excitation of the 4-hydroxycinnamyl chromophore and subsequent shift in protein residue alignment. They achieved this by trapping an early photocycle intermediate in a cryogenically cooled and then light-activated PYP crystal (73, 74). The chromophore thioester link to the protein undergoes rotation of the carbonyl group, and the protein rearranges slightly to accommodate the new chromophore configuration (73). Movement of Arg52 in the PAS core provides solvent access to the chromophore during the bleached signaling intermediate of the light cycle. Arg52 is in the putative protein interaction site and is proposed to participate in PYP interaction with a downstream signal transduction protein (172). The signal transduction mechanisms for FixL and HERG are discussed below.
The specificity of a PAS domain for detection of input signals is determined, in part, by the cofactor associated with the PAS domain. Known cofactors, in addition to 4-hydroxycinnamyl chromophore and heme, include flavin adenine dinucleotide (FAD) in NifL (97) and Aer (24) and putative 2Fe-2S centers in the NifU protein (67), where we have identified a PAS domain (Fig. 2). Where the site of attachment is known, each of these cofactors is attached to the PAS core “active site” of the PAS domain (Fig. 7). With minor modifications to each protein, the different cofactors might be accommodated in the major hydrophobic core of the PAS domain.
FIG. 7.
Site of attachment of prosthetic groups in selected PAS domains. The sites of attachment (asterisks) are shown for 4-hydroxycinnamyl chromophore in PYP (Swiss-Prot accession no. P16133), heme b in FixL (PIR g628552), and putative [2Fe-2S] centers in NuoE (Swiss-Prot P33601). The horizontal line indicates residues that constitute the PAS core. The NuoE protein is NADH dehydrogenase I subunit E, which does not have a complete PAS domain.
PAS DOMAINS IN MICROBIAL GENOMES
Single, Dual, and Multiple PAS Domains
The first proteins with PAS domains identified in eukaryotes (PER, ARNT, SIM, and phytochromes) have two PAS domains that may have diverged in origin. In contrast, many prokaryotic PAS domains are present as single domains (Table 2). Our recent analysis of PAS domains in the completely sequenced microbial genomes available through the public databases (265) provided a broader picture of the occurrence and possible role of PAS domains. Of 11 microbial genomes analyzed, 5 contain no PAS domains. The best-studied model microorganisms, E. coli and Bacillus subtilis, have 9 and 10 proteins with PAS domains, respectively. Interestingly, the functions of the proteins in the two species appear to be diverse: there is no single homolog between the proteins. Functions of some of these proteins are described in different sections of this review. Two of the nine E. coli proteins have two PAS domains each, whereas other proteins have a single PAS domain. Most PAS-containing proteins in B. subtilis have a single PAS domain. However, the KinA sensor kinase contains three PAS domains. Two microbial species, the cyanobacterium Synechocystis sp. strain PCC6803 (121–123) and the archaeon Archaeoglobus fulgidis (128) each have 17 proteins in which PAS domains have been identified. Comparison of the microbial genomes shows that in A. fulgidis most proteins have two PAS domains whereas in another archaeal species, Methanobacterium thermoautotrophicum (205), PAS domains are present more often as a single domain. Synechocystis sp. is the only species where more than four PAS domains have been identified in one protein. The only eukaryotic microbial genome sequenced at the time of this analysis (Saccharomyces cerevisiae) had only two PAS domains and few proteins involved in signal transduction; therefore, it is not known whether eukaryotes have sensor proteins with more than two PAS domains. Most of the eukaryotic transcriptional factors and clock proteins have two PAS domains whereas voltage-sensitive ion channels have a single PAS domain.
TABLE 2.
Single, dual, and multiple PAS domains in completely sequenced microbial genomes
| Species | No. of proteins containing the following no. of PAS domains:
|
Totala | |||||
|---|---|---|---|---|---|---|---|
| One | Two | Three | Four | Five | Six | ||
| E. coli | 7 | 2 | 9 (11) | ||||
| B. subtilis | 9 | 1 | 10 (12) | ||||
| A. aeolicus | 5 | 1 | 6 (7) | ||||
| M. thermoautotrophicum | 5 | 2 | 2 | 9 (15) | |||
| A. fulgidis | 5 | 9 | 3 | 17 (32) | |||
| Synechocystis sp. | 4 | 5 | 3 | 3 | 1 | 1 | 17 (47) |
| Total | 35 | 19 | 9 | 3 | 1 | 1 | 68 (124) |
Total number of proteins that contain a PAS domain, with the total number of PAS domains per genome in parentheses.
Origin of Diversity in PAS Domains
Considerable variation in PAS sequences is evident from even a casual search of available databases. We analyzed proteins with multiple PAS domains that were selected from the completely sequenced microbial genomes (265). In the Sll0779 protein from Synechocystis sp., three N-terminal PAS domains most probably originated from a simple duplication of one domain (265) (Fig. 8). Multiple copies of a similar domain may provide a selective advantage to the bacterium by amplifying the sensory signal. On the other hand, two C-terminal PAS domains in the same protein have different origins. All six PAS domains in another cyanobacterial protein, the Slr0222 sensor kinase, are unrelated. They are more similar to archaeal (such as the Bat oxygen sensor) and human (HIF1α) PAS domains than they are to each other (265). The observation that specific PAS domain sequences are conserved over long phylogenetic distances is an indication that PAS domains differentiated early in the phylogenetic tree. The fidelity with which differentiated PAS sequences have been maintained across kingdoms is best explained by a differentiated function for individual branches of the PAS domain lineage. Where different types of PAS domains are present, one sensor protein may respond to multiple input signals, each activating a specialized PAS domain.
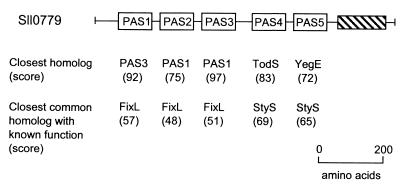
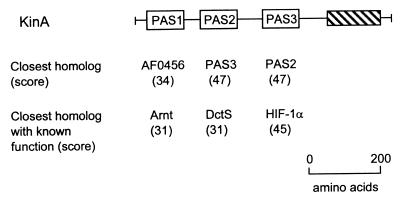
FIG. 8.
Domain structure of the Sll0779 protein from Synechocystis sp. strain PCC6803. The closest homolog and the closest homolog with a known function are shown for each PAS domain. Searches were performed individually with each PAS domain as a query, using the Gapped BLAST program (7). TodS, toluene sensor kinase from Pseudomonas putida (GenBank accession no. U72354); YegE, hypothetical sensor kinase from E. coli (Swiss-Prot P38097); StyS, styrene sensor kinase from Pseudomonas sp. (PID e1169869); FixL, oxygen sensor kinase from Bradyrhizobium japonicum (Swiss-Prot P23222). The hatched block represents a histidine kinase transmitter domain. Scores are given in bits. Reprinted from reference 265 with permission of the publisher.
Correlation of PAS Domains with Electron Transport Components
There is no correlation between the size of a bacterial genome and the total number of PAS domains present in the genome. However, we have found a correlation between the total number of PAS domains and the components of the respiratory and photosynthetic electron transport-associated proteins in completely sequenced microbial genomes (265). This is consistent with a hypothesis that the primary role of PAS domains is sensing oxygen, redox potential, and light (266). The species with the lowest incidence of electron transport proteins and the absence of PAS domains are animal parasites that live in an environment where they have little need for a complex electron transport system and redox sensing. The great number of electron transport-associated proteins in the hyperthermophilic archaeon Archaeoglobus fulgidis reflects multiple pathways for reduction of sulfate and alternative electron acceptors (128). The multiple PAS domains presumably provide A. fulgidis with enhanced flexibility in adapting to the complex redox environment. Another species with an abundance of PAS domains and multiple photosynthetic and respiratory electron transport pathways is the cyanobacterium Synechocystis sp. (Table 2), whose survival is aided by sensing light, oxygen, and redox potential.
REGULATION OF CELL FUNCTIONS IN PROKARYOTES
There is extensive knowledge of the role in the cell of signaling systems that have a PAS-containing component. Even where the participation of a PAS module is newly recognized, it is often possible to propose a role for the PAS domain based on known functions of PAS domains in similar signaling systems. In this section, we discuss known and putative regulatory roles of PAS-containing signaling systems in a wide range of biological systems. The emphasis is on the biological role of the PAS domain, and no attempt is made to provide a comprehensive review of each system. However, references that are cited can direct the reader to sources of more detailed information.
Bacterial Behavior
Motile bacteria are able to navigate rapidly to microenvironments where the concentration of oxygen is optimal for growth. This aerotaxis response has been most extensively studied in E. coli. The aerotaxis transducer Aer has a PAS domain in the N-terminal segment (186, 266). Evidence that the aerotaxis transducer in E. coli does not sense oxygen directly includes the following. (i) Aerotaxis requires a functional electron transport system (132). (ii) Alternative electron acceptors, such as nitrate, fumarate, and trimethylamine oxide, can mimic oxygen in eliciting a behavioral response, but only if they stimulate electron transport (223). (iii) At a constant oxygen concentration, perturbation of the electron transport system and proton motive force produces an aerotaxis-like behavioral response (23, 132). (iv) In anaerobic cells, Aer is a transducer for redox taxis that guides bacteria to the optimal redox potential (23).
The PAS domain in Aer has a noncovalently bound FAD as cofactor (24, 186). Current evidence suggests that Aer is representative of a class of PAS transducers that sense redox changes in the electron transport system or another component of the cell. Other transducers in this class include NifL (97, 207), ArcB (110, 111), and possibly the PpsR sensor from Rhodobacter sphaeroides, which also appears to be a redox transducer (81).
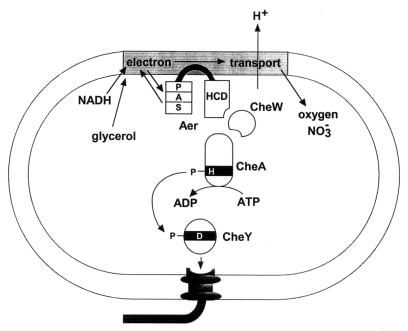
In the signal transduction pathway for aerotaxis (Fig. 9) Aer links the electron transport system to the CheA sensor kinase. The predicted structure of Aer provides clues to the transduction mechanism. A central hydrophobic sequence anchors two cytoplasmic domains to the membrane (186, 224). Aer forms a dimer in vivo (118). The C-terminal portion of Aer has an HCD that is found in all chemotaxis transducers (135). In the presence of CheW, the HCD serves as input domain for regulating the histidine kinase activity of CheA (Fig. 9). Phosphotransfer from CheA to the CheY response regulator activates CheY to bind to the FliM protein on the flagellar motors (17, 246). This reverses the direction of motor rotation from counterclockwise to clockwise and causes the bacteria to change the direction of swimming. Overexpression of Aer in an E. coli strain lacking all chemotaxis transducers imparts some clockwise rotation to the flagella (24), indicating that the carboxyl domain of Aer also interacts with CheA and CheW.
FIG. 9.
A proposed scheme for aerotaxis and redox (energy) sensing in E. coli.
The N-terminal portion of Aer consists of a PAS domain and a short linker to the transmembrane region (186). The FAD cofactor (24) is probably bound to the PAS domain (118), where oxidation and reduction of FAD generate the on and off signals for aerotaxis. Further research is required to identify how the PAS domain communicates first with the electron transport system and then with its C-terminal signaling (HCD) domain. Our model (Fig. 9) proposes that the PAS domain interacts with a component of the electron transport system. Interdomain communication between the PAS and the C-terminal domains of Aer may occur through direct contact of the domains, as proposed for HERG (160). Goudreau and Stock (84) have recently reviewed the importance of interdomain contact in signaling in two-component regulatory systems.
The importance of the PAS domain in signal transduction in aerotaxis has been confirmed by cysteine replacement mutagenesis of Aer. Serial mutation of 40 residues in the PAS domain, including the highly conserved amino acids, yielded mutants with various defective phenotypes (187). In addition to mutants with no aerotactic responses, the signaling in some mutants was locked in the signal-on (clockwise rotation) mode. One mutant had inverted responses to oxygen and redox stimuli; i.e., it reacted to attractants as repellents and vice versa. Many of the mutations that had a phenotype are located around the putative hydrophobic core.
Respiratory electron transport is limited by the availability of an electron acceptor, the supply of electron-donating substrates (usually carbon sources), or diversion of electrons from the system (224). Behavioral responses to environmental stimuli that act at each of these regulatory sites are signaled through the Aer transducer and are absent in an aer null mutant. This includes electron acceptor taxis, an aerotaxis-like response to alternative electron acceptors in anaerobic cells (221, 223), redox taxis to quinones (23), and glycerol taxis, an example of metabolism-dependent taxis to a carbon source in E. coli (264). We use the term “energy taxis” to include aerotaxis and these electron transport-dependent responses (224). This highlights the role of Aer in guiding E. coli away from microenvironments where respiration is impaired.
An Aer homolog identified recently in Pseudomonas putida (GenBank accession no. AF079997) has a PAS domain homologous to that of the E. coli protein, and the aer mutant has an impaired aerotactic response (96). The Aer-type redox-sensing transducers for bacterial behavior may be widespread. We have identified PAS domains in two putative chemotaxis transducers, AF1034 and AF1045, from the Archaeoglobus fulgidis genome (265) and in the putative chemotaxis transducer McpA from Agrobacterium tumefaciens (GenBank accession no. AF010180) (Fig. 2). We have also identified a PAS domain in the chemotaxis transducer HtpIII (HtpA) from Halobacterium salinarum (Fig. 2). Signal transduction in chemotaxis in H. salinarum is processed through 13 soluble and membrane-bound transducer proteins (194, 262). One of them, the membrane-bound HtrVIII transducer, governs the aerotactic response (31). The finding of a PAS domain in HtpIII suggests that this soluble transducer may be a second aerotaxis sensor in H. salinarum.
The DcrA protein from Desulfovibrio vulgaris Hildenborough (Fig. 2 and 5), an anaerobic, sulfate-reducing bacterium, is another candidate for a PAS-based redox sensor that regulates bacterial behavior. Early studies indicated homology of DcrA to the methyl-accepting chemotaxis transducers from enteric bacteria, and DcrA was proposed to serve as a receptor for negative aerotaxis (50, 66). The periplasmic N-terminal sensor domain was found to contain a putative heme-binding CHHCH motif, and a c-type heme was identified in DcrA. It was suggested that the protein was involved in redox sensing. Methyl labeling of DcrA decreased upon addition of oxygen and increased upon subsequent addition of the reducing agent dithionite, indicating possible chemotactic signaling by the sensor in response to oxygen concentration and/or redox potential (66). Subsequently, a PAS domain was identified in DcrA (182). Interestingly, it appears to be different from the proposed heme-binding periplasmic domain and is located in the predicted cytoplasmic portion of the protein followed by the C-terminal chemoreceptor-like signaling domain (Fig. 5). The exact attachment site for heme has not been established for DcrA, leaving the possibility open that heme is present in the PAS domain, not in the periplasmic portion of the protein. A potential heme-binding site (His300) is located within the PAS core of the DcrA PAS domain. Alternatively, two redox-sensing domains can be present in DcrA. Studies of the aerotactic response in the dcrA deletion strain showed that the aerotactic response is present in this mutant (222). Therefore, either DcrA is not an aerotaxis transducer or there is a second aerotaxis transducer in D. vulgaris, as in E. coli (186).
PYP has been proposed as a receptor for a photophobic swimming response in Ectothiorhodospira halophila (211). However, since little is known about motility in this species, the other components of the signal transduction pathway have not been identified. PYP was also detected in Rhodobacter sphaeroides (130), for which a great deal of information about motility and phototaxis is known (11, 12). It should be easier to establish the downstream elements of this photoresponse and the signal transduction pathways in R. sphaeroides than in E. halophila.
Global Regulation of Cell Metabolism and Development
Energy metabolism.
(i) ArcB.
The aerobic metabolism modulon in E. coli that is regulated by the ArcB-ArcA pathway includes regulons that encode enzymes for the tricarboxylic acid cycle, glyoxylate shunt, pathway for β-oxidation of fatty acids, cytochrome o and d complexes, and flavoprotein dehydrogenases (91, 109–111). Microaerobic control of cydAB (cytochrome d oxidase) gene expression involves ArcA-ArcB in conjunction with FNR (230).
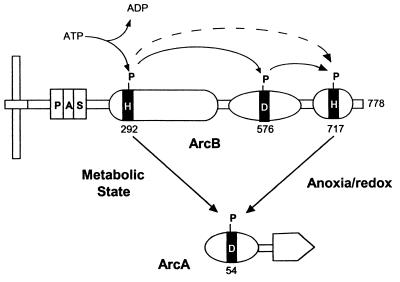
ArcB (Fig. 10) is a novel sensor kinase that has a PAS input domain, transmitter and receiver modules, and a histidine-containing phosphotransfer (HPt) domain (125, 231). The HPt domain is similar in prokaryotes and eukaryotes (125). His292 is autophosphorylated; Asp576 is a phosphoacceptor site, and His717 is an atypical phosphodonor site. The mechanism of oxygen sensing by ArcB is unknown, although redox-sensing rather than sensing of oxygen per se is indicated (108, 110). The PAS domain that we have identified between the transmembrane anchoring region and the N-terminal transmitter domain (266) is most probably an input domain for the redox signal. The ArcB protein is autophosphorylated at His292 in the transmitter domain. From there, the phosphoryl residue may be transferred first to Asp576 in the adjacent receiver domain and then to His717 in the HPt domain. The HPt domain has a characteristic four-helix bundle that is not autocatalytic or homologous to transmitter domains (125). Subsequently, the phosphoryl residue is transferred from the transmitter or HPt histidine to an aspartate residue in the receiver domain in the cognate response regulator ArcA (Fig. 10). Phosphorylated ArcA is a pleiotropic transcriptional factor that regulates the target genes. It was recently demonstrated that phosphorylation of ArcA by phospho-His717 is controlled by the anoxia/redox state and that direct phosphorylation of ArcA by phospho-His292 is controlled by a metabolic state of the bacteria (146, 151).
FIG. 10.
Schematic representation of communication modules in the ArcB-ArcA phosphorelay system. Transmembrane domains are shown in black. From the left, the modules in ArcB are PAS domain, transmitter domain, receiver domain, and HPt domain. Adapted from reference 151 with permission of the publisher.
PAS domains have been identified in several other sensors that are involved in controlling energy metabolism in bacteria (182, 266).
(ii) PpsR and CrtJ.
Bacteria of the genus Rhodobacter are remarkably versatile in their growth capabilities. These anoxygenic phototrophic bacteria derive energy from aerobic respiration in the presence of oxygen. However, when the oxygen concentration drops below a threshold level (<1% dissolved oxygen), the cells differentiate and develop intracellular membranes that house the light-driven energy-generating photosystem (35). Oxygen and, to a lesser extent, light control the formation of the photosynthetic apparatus, partly by regulating several transcription factors that control the expression of photosynthesis genes (for reviews, see references 19, 51, and 260). A transcription factor termed PpsR (for “photopigment suppression”) in R. sphaeroides and CrtJ in R. capsulatus is an aerobic repressor of the light-harvesting antennae II (the puc operon), bacteriochlorophyll (bch), and carotenoid (crt) genes (80, 173, 181). A PAS domain has been identified in the N terminus of the PpsR/CrtJ protein (182). Surprisingly, the PAS domain in PpsR is not homologous to any known PAS domain from photosynthetic bacterial species but is similar (263) to a PAS domain in a putative sensor histidine kinase (MTH823) from the anaerobic archaeon Methanobacterium thermoautotrophicum (265).
The PpsR and CrtJ proteins contain a putative DNA-binding bHLH motif at the C terminus (174). The CrtJ protein binds to promoters of the controlled operons in a redox-dependent manner. Highly oxidizing conditions increase DNA binding; this is the opposite of the effect of DNA-binding redox-responding proteins, such as FNR, SoxR, and Fur (180). The repressor activity of PpsR requires other cellular factors to communicate to the PAS input domain the state of oxygen availability and changes in growth conditions (81). The PpsR protein is also required for normal regulation of the photosynthesis genes by light, but the mechanism of light control is less well understood (81). One hypothesis is that changes in light intensity generate changes in the cellular redox state that are sensed by PpsR through intervention of the AppA protein (81, 260). The DNA-binding affinity of the CrtJ protein in vitro is also modulated by redox potential (180). As in many other PAS-containing proteins, the PAS domain may confer dimerization specificity upon the CrtJ protein, which binds to DNA in vitro as a dimer (180). The CrtJ-type sensors provide bacteria with flexibility in utilizing available energy sources, such as light and carbon plus electron acceptors. Like ArcB in E. coli, PpsR and CrtJ in Rhodobacter species allow bacteria to switch on or off specific pathways for energy generation depending on the presence of energy sources in their environment.
(iii) TodS, TutC, and StyS.
The PAS-containing sensor, Aer, responds to changes in the concentration of a carbon source, such as glycerol, that donates reducing equivalents to the electron transport system (186, 264). PAS domains have also been identified in sensors for bacterial two-component systems that regulate the aerobic degradation of aromatic hydrocarbons. TutC of Thauera (37), TodS of toluene-degrading Pseudomonas (133), and StyS of styrene-degrading Pseudomonas (236) have extensive sequence homology. Lau et al. (133) reported the similarity of a domain in TodS to FixL oxygen sensors of rhizobia and suggested that TodS may respond to changes in oxygen concentration. Subsequently, we identified two PAS domains, separated by a sequence of similar length, in all three of the above-mentioned proteins. This suggests a common function (oxygen and/or redox sensing) for all three sensors. TodS has an unusual domain structure, expanding the domain repertoire known for histidine kinases. As in other histidine kinases, the PAS domains are located N-terminal to the histidine kinase domains. In addition, there is a basic region-leucine zipper dimerization motif at the N terminus of TodS (133). There is a remarkable similarity between the toluene response in Pseudomonas and responses triggered by dioxin in eukaryotes (133), and both the toluene response sensor TodS and the dioxin receptor AHR have two PAS domains. PAS domains in the bacterial sensors regulating aerobic utilization of aromatic hydrocarbons may play a role similar to that of other sensors involved in energy metabolism. Since degradation of the aromatics occurs under respiration conditions, the system may regulate the flow of reducing equivalents coming from the degradable hydrocarbons into the electron transport system.
(iv) PhoR.
Inorganic phosphate (Pi) is involved in a large number of cellular functions, including energy metabolism, and the transport and intracellular concentrations of Pi are regulated. The PhoR-PhoB (227, 242) two-component system of E. coli is a global regulator of phosphate metabolism in response to Pi starvation (228). The PhoR protein is a sensor kinase that phosphorylates the PhoB protein, a cognate response regulator (148, 252). PhoR acts as a negative regulator in the presence of excess phosphate and as a positive regulator when phosphate is limited. Upon phosphate starvation, phosphorylated PhoB is a positive regulator of at least 15 genes that constitute the phosphate regulon (among them, genes encoding alkaline phosphatase, porin E, transmembrane Pi channels, and glycerol-3-phosphate-binding and transport proteins). Phosphorylated PhoB binds to a specific region (the Pho box) upstream of each gene in the regulon and activates transcription (228).
The stimulus detected by the PhoR sensor is unknown. Analysis of a deduced amino acid sequence of PhoR suggested that this protein is anchored to the membrane and functional domains are located in the cytoplasm (149). A putative sensing domain of PhoR (C2) located between a transmembrane region and a histidine kinase domain (residues 52 to 220) was proposed to respond to an internal cellular stimulus. Mutants that had deletions in the C2 region were locked in the active kinase state, resulting in high-level expression of the pho regulon (201). Therefore, it was proposed that the C2 region is a signal-sensing domain that represses kinase activity. We identified a putative PAS domain in the PhoR protein of E. coli (Fig. 2 and 5) within the boundaries of the C2 region, supporting the concept that C2 is an internal sensor.
(v) DctS.
Malate, succinate, and fumarate are effective carbon sources for R. capsulatus under both aerobic conditions in the dark and anaerobic conditions in the light. Transport of these C4-dicarboxylates in this species is mediated by a high-affinity system, which belongs to a novel family of transporters, TRAP (for “tripartite ATP-independent periplasmic”) (64). Synthesis of this transport system is controlled by a two-component regulatory system, which consists of a sensor kinase, DctS, and a response regulator, DctR (92). Two transmembrane regions were identified in the N terminus of a deduced amino acid sequence of the sensor, with a histidine kinase domain being present in the C terminus (92). We and others (182, 266) identified a PAS domain in DctS, located in the cytoplasmic segment of the protein (residues 314 to 406) between the second transmembrane region and the linker which separates the PAS domain from the histidine kinase domain (92). The PAS domain in DctS could regulate autophosphorylation in response to changes in intracellular energy (redox) levels. Thus, PAS domain-containing two-component systems may control energy metabolism not only by regulating the expression of specific catabolic pathways but also by regulating the transport of carbon sources into a cell.
In Rhizobium leguminosarum and Sinorhizobium meliloti, there is a similar two-component system, DctB-DctD, that controls the transport of dicarboxylates (116, 244), but no PAS domain has been found in the DctB sensor of the rhizobial system. It seems likely that DctS responds to an intracellular signal, such as oxygen or/and redox potential, whereas DctB detects other signals, such as dicarboxylates (190, 191). Since Rhizobium and Rhodobacter are close relatives (alpha subdivision of the Proteobacteria), a difference in stimuli detection by homologous sensory systems may reflect the difference in their environments.
(vi) DcuS and CitA.
In E. coli, the genes encoding the anaerobic fumarate respiratory system are transcriptionally regulated by dicarboxylates. The DcuSR two-component system effects this regulation (79, 267). The DcuS sensor histidine kinase has a periplasmic domain, which is suggested to be involved in sensing of dicarboxylates (267), and a kinase domain. There is an “extra domain” between the periplasmic and kinase domains (267), which has been identified as a classical PAS domain (79). A similar domain organization, including the PAS domain (182), is found in the CitA sensor that regulates anaerobic citrate metabolism in Klebsiella pneumoniae (28, 29). The presence of a PAS domain in the DcuS sensor strongly suggests that it may respond to redox signals derived from dicarboxylate metabolism. It has been proposed previously that citrate, Na+, and oxygen are the stimuli that exert their regulatory effects on citrate catabolism by K. pneumoniae via the CitA sensor (28, 29). All the above stimuli may cause redox and/or energy changes that can be detected via the PAS domain.
Nitrogen fixation and nitrogen metabolism.
Biological nitrogen fixation carried out by various symbiotic and free-living bacteria is extremely sensitive to oxygen due to an oxygen-labile nitrogenase. Oxygen is the main environmental factor regulating the expression of nitrogen fixation genes in bacteria. Different diazotrophs use different strategies for oxygen sensing; however, they have something in common: all known oxygen sensors that control nitrogen fixation, as well as some putative sensors that are involved in regulating nitrogen fixation and nitrogen metabolism, contain PAS domains.
(i) FixL.
The expression of nitrogen fixation genes of a plant-symbiotic bacterium, Sinorhizobium meliloti, is induced under low oxygen concentrations by a two-component regulatory system consisting of the FixL and FixJ proteins. The FixL heme-containing protein kinase senses oxygen through a heme cofactor and transduces the signal by controlling the phosphorylation of FixJ (75) (Fig. 3A). The FixL-FixJ system is found in other rhizobial species, such as Azorhizobium caulinodans (120), B. japonicum (8), Rhizobium etli (48), and R. leguminosarum bv. viciae (169). Phosphorylated FixJ acts as a transcriptional activator of the nifA and fixK genes, which control the expression of nitrogen fixation genes and a high-affinity terminal oxidase complex, respectively (for reviews, see references 61 and 62). The PAS-containing ArcB-ArcA system in E. coli also regulates the synthesis of terminal oxidase complexes.
The FixL protein from B. japonicum is a true oxygen sensor, distinct from known heme-based oxygen carriers and electron transporters (77). The site of heme coordination is histidine residue 200, located on the Fα helix of the PAS core (82). Comparison of the three-dimensional structure of the BjFixLH (PAS) domain in the on (unliganded) and off (ligand-bound) conformations suggested a mechanism of signal transduction (82). On binding a strong-field ligand, the slightly puckered heme becomes more planar, causing a conformational change in the protein. The largest conformational shift is in the FG-loop region, which may accommodate an interaction of the kinase domain with the heme domain, inactivating the kinase activity. Since the FixL protein forms dimers in solution (77), the heme of one subunit is likely to interact with the kinase domain of the other subunit. The FG loop is one of the least conserved regions of PAS domains (Fig. 2) and may be adapted to the specific shape of the heme pocket. This mechanism is in contrast to PYP, where the regulatory conformational change is in the EF loop (73).
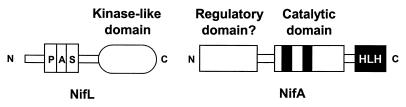
(ii) NifL.
Synthesis of nitrogenase in the free-living nitrogen-fixing bacterium Azotobacter vinelandii is controlled by an enhancer-binding protein, NifA, which activates transcription at ςN-dependent nif (nitrogen fixation) promoters, and a sensor protein, NifL, which antagonizes the NifA activity in response to external oxygen concentrations and fixed nitrogen (for a recent review, see reference 49). NifL has a C-terminal histidine kinase-like transmitter domain, and the N-terminal domain of the protein has significant homology to FixL proteins and the bat gene product of Halobacterium salinarum (88), indicating that this domain may be responsive to oxygen signals (25). A PAS motif was identified within the N-terminal domain (182, 266) (Fig. 2 and 11). Recently, Dixon and colleagues demonstrated that NifL is a flavoprotein with FAD as the prosthetic group (97) and identified the PAS domain as the flavin-binding redox-sensing domain distinct from the C-terminal nitrogen-responsive and nucleotide-binding domains (207). FAD has also been identified in the N terminus of the NifL protein from Klebsiella pneumonia (199). The high degree of identity between PAS domains of the NifL proteins from Azotobacter, Klebsiella, and Enterobacter agglomerans (203) suggests that these proteins share the structural and functional characteristics of the A. vinelandii NifL.
FIG. 11.
Domain structure of NifL and NifA from Azotobacter vinelandii. Adapted from reference 49 with permission of the publisher.
NifL is a homotetramer in vitro, with protein-protein interaction sites in both the PAS and C-terminal domains (207). A N-terminal fragment (residues 1 to 284) containing the PAS domain purifies as a tetramer, whereas a truncated NifL (residues 147 to 519) purifies as a dimer. The truncated protein, which lacks the PAS domain, does not respond to redox changes but does inhibit NifA in response to ADP in vitro and fixed nitrogen in vivo.
The redox-sensitive PAS domain acts as a switch for the regulatory activity of NifL. The NifL protein is inactive as an inhibitor of NifA when FAD is reduced and active when FAD is oxidized (147, 207). The redox potential of FAD in NifL is −226 mV at pH 8.0, so that it could be readily reduced in vitro by various electron donors and NAD(P)H-dependent enzymes (147). NifL from A. vinelandii responds to both oxygen and fixed nitrogen in E. coli (207). This indicates that A. vinelandii NifL can interact with appropriate electron donors in E. coli to maintain the protein in the inactive state under anaerobic conditions. Direct electron donation to NifL from the flavin of E. coli flavohemoprotein and spinach ferredoxin:NAD(P) oxidoreductase has been demonstrated with NADH as a reductant (207). NifL is rapidly reoxidized in the presence of air, raising the possibility that it senses intracellular oxygen.
The role of the C-terminal transmitter domain remains a puzzle (49, 207). Although there is homology to transmitter domains in sensor histidine kinases (Fig. 11), the conserved histidine that is phosphorylated in other kinases is not essential for the inhibitory activity of NifL (251). No autophosphorylation of NifL or phosphotransfer to NifA has been detected in purified proteins (49, 199). Furthermore, NifA does not have a receiver domain with the aspartate phosphoacceptor that is present in typical response regulators (52). Stoichiometric levels of NifL and NifA are required for bona fide regulation, suggesting that signal transduction involves protein-protein interactions in NifL-NifA instead of covalent modification.
(iii) NtrY.
Compared to other rhizobia, control of nitrogen fixation in Azorhizobium caulinodans involves another regulatory element in addition to the FixLJ system (62): the NtrY-NtrX two-component system (171). The NtrY protein is a putative sensor with a histidine kinase motif in its C terminus, and the NtrX protein is a putative cognate response regulator. The predicted topology of NtrY in the membrane suggested that NtrY was a sensor of the extracellular nitrogen concentration (171). The ntrY mutants had impaired growth on nitrate, but only on plates and in well-aerated cultures, suggesting that oxygen might be involved. Subsequently, a PAS domain was identified in the cytoplasmic segment of NtrY (182, 266) (Fig. 2 and 5) just upstream of the histidine kinase-like domain. This supports the experimental evidence for possible oxygen (redox) sensing in the regulation of nitrogen metabolism by this protein.
(iv) YntC.
A. caulinodans appears to use several PAS-containing proteins to regulate nitrogen fixation and nitrogen metabolism. We have identified a PAS domain in a predicted protein, YntC, from this species. The yntC gene is located between the ntrBC and ntrYX genes in this bacterium (171), and its function remains unknown. Interestingly, an open reading frame homologous to the yntC gene was found recently just upstream of the gene cluster that encodes respiratory nitrate reductase in R. sphaeroides, and a regulatory role in controlling nitrate reductase was suggested (188). Therefore, it is possible that YntC is involved in regulation of respiratory nitrate reductase in A. caulinodans. Oxygen or redox sensing obviously may be an important step in such regulation.
(v) NtrB.
The NtrB protein of a free-living diazotroph, Azospirillum brasilense, was shown not to be essential for nitrogen fixation as it is in other bacteria. However, the NtrB-NtrC system in this organism partially controls expression of the NifA protein (137). No FixL- or NifL-type protein has been detected in A. brasilense, and NtrB may contribute to the redox control of nitrogenase biosynthesis. We have identified a PAS domain in the N-terminal portion of the protein. NtrB proteins from other bacterial species also contain a putative PAS domain (Fig. 2). This raises a question whether the NtrB-NtrC system may be responsive to redox changes.
(vi) NifU.
The nifU gene is present in the nif gene clusters of many nitrogen-fixing bacteria (65, 94, 112, 153, 154, 162). Surprisingly, its exact function is not known in any of them. One of the suggested roles of NifU is the mobilization of iron and sulfur for nitrogenase-specific iron-sulfur cluster formation. NifU contains two identical [2Fe-2S] clusters (67). We have identified a PAS domain in the N-terminal portion of all known NifU proteins, and two of the four cysteinyl residues proposed to be involved in coordination of the iron-sulfur clusters are located within the PAS domain. It would be interesting to find whether the NifU protein utilizes the PAS domain as a fold to accommodate its redox cofactors without other signaling properties of PAS domains. Recent findings that the N-terminal domain of NifU is conserved in some proteins that are not related to nitrogen fixation or signal transduction but contain redox prosthetic groups (not only iron-sulfur centers but also FAD and heme) (106, 166) support this idea. The most recent studies on a nifU mutant of A. vinelandii also suggest a possible involvement of NifU in oxygen sensing and/or metabolism. Mutations in a cydR gene that encodes a transcriptional repressor of the cytochrome d operon (cydAB) suppressed nifU mutations (98).
Sporulation
Bacillus subtilis cells monitor their external and internal environments. When the external environment becomes hostile, they respond by forming dormant, heat-resistant endospores. Over 125 genes are involved in this process. The decision to sporulate is made by integrating diverse environmental and physiological signals, resulting in the activation of a key transcriptional factor, Spo0A (for a review, see reference 218). The signals arising from starvation, cell density, the tricarboxylic acid cycle, DNA synthesis, and DNA damage are funneled into a phosphorelay formed from homologs of modules of two-component regulatory systems (Fig. 3C) (215, 218). The phosphoryl residue is passed from his-asp-his-asp in a phosphorelay (34). The end product is phosphorylation and activation of Spo0A. The increase in phosphorylated Spo0A (Spo0A-P) production activates the transcription of at least seven genes that control entry into sporulation and the transition to a two-compartment sporangium in which gene transcription is regulated differentially (218).
The exact nature of the sporulation-inducing signals is still unknown, but the signals control the phosphorylation of two histidine kinases, KinA (175), and KinB (229), that initiate the phosphorelay, and aspartyl phosphatase, that regulates the flow of phosphate (165, 175).
KinA (Fig. 3C) is probably responsible for most of the phosphorylation entering the phosphorelay (215). Triple PAS domains with different origins were identified in KinA (182) (Fig. 2), a soluble kinase that senses the internal environment of the cell (Fig. 3C and 12). As an internal sensor, the PAS domain could respond to hypoxia, a decrease in the cellular energy, level or a change in redox potential (186, 224). Thus, unfavorable conditions for cell growth could contribute to the sporulation signal. KinB, which does not contain a PAS domain apparently responds to different signals than does KinA (134). Under some conditions, KinB may surpass KinA in contributing to the sporulation phosphorelay. The KinC sensor kinase has a PAS domain (182, 266) (Fig. 2), but there is no positive evidence that KinC is involved in the initiation of sporulation. KinA and KinB appear to have different functions, and they may be activated at different stages of cell growth and stationary phase (134). Although the primary control of phosphorylation of Spo0A is at the level of KinA-KinB, aspartyl protein phosphatases also play a critical role. These include the Spo0E phosphatase, which acts directly on Spo0A-P (89), and the Rap family of phosphatases, which dephosphorylate Spo0F and are controlled by the starvation-induced presporulation response (176, 215). A direct inhibitor of KinA autophosphorylation, KipI, has also been identified. KipI is also under complex environmental control (240).
FIG. 12.
Diversity of the PAS domains in the KinA protein. The closest homolog and closest homolog with known function are shown for each PAS domain. AF0456, signal transduction histidine kinase from A. fulgidis (GenBank accession no. AE001022); Arnt, aryl hydrocarbon receptor nuclear translocator from O. mykiss (GenBank U73840); DctS, dicarboxylate transport sensor kinase from R. capsulatus (Swiss-Prot P37739); HIF-1α, hypoxia-inducible factor from Mus musculus (GenBank AF045160). The methods used for this analysis are described in the legend to Fig. 8.
Animal-Microbe and Plant-Microbe Interactions
Virulence.
BvgS-BvgA, a two-component system, regulates virulence gene expression in the gram-negative Bordetella species that cause respiratory infections in a variety of hosts: Bordetella pertussis causes whooping cough in humans, B. bronchiseptica infects pigs and dogs, and B. avium causes coryza in birds. The BvgS-BvgA system regulates almost all of the known factors associated with the Bordetella pathogenesis (2). BvgS is a transmembrane sensor autokinase that detects environmental stimuli and phosphorylates a response regulator, BvgA, leading to activation and repression of various genes involved in virulence (Fig. 13). The BvgS-BvgA system mediates the expression of various phenotypic phases in response to the different environments encountered as the bacteria travel within and between mammalian hosts (2, 38). The BvgS-BvgA system acts as an activator of toxins, adhesins, fimbriae, and hemolysin (vir-activated) genes and controls negatively vir-repressed genes, such as flagellar genes in B. bronchioseptica and vrg loci in B. pertussis (2, 124, 269). BvgS is a 135-kDa integral cytoplasmic membrane protein (Fig. 13). The N-terminal periplasmic domain is linked by a membrane-spanning helix to cytoplasmic PAS (182), transmitter, receiver, and histidine phosphotransfer domains (232, 233). BvgA is a 23-kDa cytoplasmic response regulator that has receiver and C-terminal helix-turn-helix domains (216). In a complex phosphorelay, BvgS is autophosphorylated at His729 in the transmitter motif (Fig. 13). The phosphate is transferred to the receiver module (Asp1023), then to the C-terminal histidine phosphotransfer domain (His1172), and finally to Asp54 in the receiver domain of BvgA (38, 232, 233). As a result of phosphorylation, the affinity of BvgA is increased for Bvg-activated promoters (124, 214). The phosphoaspartate bond in the receiver domains of BvgS and BvgA is labile and can be hydrolyzed by water. As a result, the transmitter and C-terminal phosphorylation domain can be dephosphorylated. This may provide an additional control of the phosphorelay, akin to the action of the CheZ phosphatase in E. coli chemotaxis (233).
FIG. 13.
Communication modules and phosphorelay system for control of the BvgA-BvgS regulon in Bordetella. (A) Domain structure of the BvgS sensor kinase and BvgA response regulator showing histidine kinase, phosphotransfer, and phosphotase activities associated with each domain. (B) Regulation by BvgA of the BvgA-BvgS regulon. Adapted from references 2 and 38 with permission of the publishers.
The BvgS sensor is postulated to recognize various environmental signals (38); however, the exact stimuli sensed by the BvgS protein are unknown. Consequently, the signalling role of the PAS domain that has been identified in BvgS of Bordetella (182) has not been established. It is possible that the PAS domain senses oxygen or intracellular redox changes. Such changes may be indirect indicators of whether the bacterium is within or outside the host.
Nodulation.
Rhizobial species have a unique ability to infect leguminous plants and establish a nitrogen-fixing symbiosis (for reviews, see references 63, 213, and 235). This process results in the formation of specific organs, root nodules (nodulation). In B. japonicum, there are more than 50 known nodulation genes that are organized in clusters, and their expression is regulated (213). Transcription of the bacterial nodulation genes is induced by flavonoids secreted by the plant roots (for a review, see reference 210). The nodD gene product is a constitutively expressed LysR-type transcriptional activator that responds to the flavonoid signals and is the central regulator of the expression of nodulation operons (63, 235). In addition to NodD, there is a two-component regulatory system, NodV-NodW, that is also involved in regulating the expression of nodulation genes (83). Like NodD, the NodV-NodW system positively regulates nodulation gene expression in response to flavonoid signals (142). The NodV protein appears to be a sensor kinase that phosphorylates the NodW protein in vitro, and this phosphorylation is induced by genistein, a plant isoflavonoid (142). It is not known whether phosphorylated or nonphosphorylated NodW interacts with nod gene promoters. A suppressor of NodW has been identified that appeared to be a part of another two-component regulatory system in B. japonicum, NwsA-NwsB (87). When overexpressed, the NwsB protein is able to suppress the Nod− phenotype in the nodW mutant. The NwsA-NwsB and NodV-NodW systems are highly homologous. A cross talk occurs between the two systems, and it appears that the activity of NwsB depends on either the NwsA or NodV sensor (86). The physiological importance of such coexistence of two similar systems is unclear.
Four PAS domains have been identified in the deduced amino acid sequence of both NodV and NwsA kinases (182). In NodV, the PAS domains are located between the expanded hydrophobic N-terminal sequence and the C-terminal histidine kinase domain (Fig. 14). NwsA has a similar architecture of PAS domains; however, this protein lacks a transmembrane region. Within an individual protein sequence, the PAS2 and PAS3 domains in both sensors are duplications whereas the PAS1 and PAS4 domains have different origins. Corresponding PAS domains in NodV and NwsA are highly homologous (Fig. 14), indicating gene duplication. As in KinA of B. subtilis, the multiple PAS domains in NodV and NwsA may provide the sensor with an ability to respond to various stimuli. Another interesting parallel between the sporulation and nodulation kinases is that in both cases one membrane-bound (KinA, NodV) and one soluble (KinC, NwsA) sensor communicate signals to a cognate response regulator. In all cases, PAS domains are located intracellularly.
FIG. 14.
Relatedness of PAS domains in NodV and NwsA nodulation proteins. The hatched block represents a histidine kinase transmitter domain. Black vertical bars represent transmembrane regions. The methods used for this analysis are described in the legend to Fig. 8.
REGULATION OF CELL FUNCTIONS IN EUKARYOTES
Light Sensing in Plants
Phytochromes form a family of soluble photoreceptors that monitor the light environment in plants and generate signals that regulate gene expression in seed germination, seedling deetiolation, shade avoidance, and flowering (184, 206). Phytochromes exist in two photointerconvertible forms: Pr, a red-light-absorbing form, and Pfr, a far-red-light-absorbing form. Pfr is believed to be the active form. Current research is centered on how the active form signals to the downstream regulatory pathways (30, 56). Phytochromes (Fig. 4B) from plants are homodimeric biliproteins. Each subunit possesses a photosensory domain containing a covalently linked linear tetrapyrrole separated from two PAS domains by a protease-sensitive hinge region (185). A histidine kinase-like module is found C-terminal to the PAS domains (200) (Fig. 4B). Since missense mutations that inactivate the regulatory activity of phytochrome cluster within the PAS domains (185), it is conceivable that this region transduces the light signal to regulate the kinase activity of the transmitter-related domain. In this regard, the cyanobacterial phytochrome Cph1 possesses a photosensory domain directly adjacent to a transmitter domain and is a light-regulated histidine kinase (257). By contrast, higher-plant phytochromes are not histidine kinases but appear to be light-regulated serine/threonine kinases. The PAS domain may represent a “transducer” domain, which mediates the signal from the input photosensory domain to the “transmitter-like” domain. In an alternative model for signal transduction by phytochromes, a phytochrome-interacting factor, PIF3, that is necessary for photo-induced signal transduction has been identified in Arabidopsis (164). PIF3, a bHLH protein, binds to the C-terminal domains of both phytochrome A and phytochrome B and localizes to the nucleus, where it may control gene expression. A PAS domain was reported for PIF3 (164) but our analysis indicates that there is no PAS motif in this protein.
Cryptochromes CRY1 and CRY2 are blue-light receptors in plants and animals and are the first photoreceptors that have been shown to entrain circadian rhythms (59, 209, 226). Phytochromes and cryptochromes are photoreceptors for alternative pathways in Arabidopsis for entrainment by light of the circadian clocks. Recently, phosphorylation of Arabidopsis cryptochromes CRY1 and CRY2 by phytochrome A was demonstrated (1). CRY1 is an 80-kDa protein with N-terminal homology (residues 1 to 506) to microbial type 1 photolyases and binding sites for both pterin and flavin chromophores (150). A unique C-terminal domain has protein-protein interaction and phosphorylation sites (Fig. 4B). The PAS domains of phytochrome A are required for interaction with CRY1 in a yeast two-hybrid assay, but since the PAS domains are required for phytochome A dimer formation, the role of PAS domains in interaction with CRY1 is not clear (1). Mutation of the first 10 serines in phytochrome A to alanines resulted in hyperactivity of the phytochrome with respect to the wild-type protein (217), suggesting that phosphorylation of an N-terminal serine down-regulates the activity of phytochrome (56). Plant phytochromes are not members of the serine/threonine/tyrosine kinase superfamily. They are more likely to be related to the mitochondrial protein kinase family (95) and the B. subtilis anti-sigma factor SpoIIAB, which phosphorylate at a serine but have a sequence that is similar to the transmitter domain of a prokaryote histidine kinase (156).
Phosphorylation of the CRY1 photoreceptor is increased by activation of the phytochrome with red light and represents an elegant mechanism of fine-tuning the cryptochrome light response by phytochrome A (1). PAS domains are essential for this coordination, but they may also be involved in signaling to PIF3.
The NPH1 protein is a PAS-containing photoreceptor in the pathway for blue-light phototropism (growth toward a light source) in Arabidopsis thaliana (103). The NPH1 homologs are found in other higher plants such as pea (138), spinach, and ice plant (20). NPH1 is a 120-kDa soluble protein that has two input PAS domains (also named LOV [103]), flavin mononucleotide, and a serine/threonine kinase output domain. Autophosphorylation of NPH1 is light dependent, suggesting that it may be a member of a class of eukaryotic two-component regulatory systems that have serine/threonine kinases, but the cognate response regulator has not been identified. The NPH1 protein is a soluble protein without membrane-spanning domains, but it is associated with the plasma membrane on isolation (103).
Circadian Clocks: from Fungi to Mammals
The circadian rhythms of biological processes in mammals are under the control of a master clock in the suprachiasmatic nuclei of the hypothalamus and are entrained by the light-dark cycle (127). A surprising development is the recent demonstration that Drosophila fruit flies have independent clocks throughout their bodies (177). The clock genes per and clock are active in many different tissues in the mouse, suggesting that mammals also have multiple clocks scattered throughout their bodies (127, 219, 225). The operation of circadian clocks in single cells was first discovered in prokaryotes (41, 126).
So far, PAS domains are the only motifs known to be conserved among widely diverse clock proteins. Clocks are made up of transcription factors that feed back and inhibit their own transcription (126). A clock molecule, by definition, must cycle. mRNA and protein are synthesized in increasing amounts during the daily cycle, eventually achieving concentrations at which they feed back and repress their own genes. If the clock molecule is artificially maintained at a constant level, the clock should stop. In addition, the molecule must respond rapidly to environmental signals that entrain the phase of the clock (126). Proteins that meet this strict test for a clock component include Frequency (Frq) and Wc-2 in Neurospora; Period (Per), Timeless (Tim), Clock (Clk), and Cycle (Cyc) in Drosophila; and BMAL1 and Clock in mice (126, 127). In addition, Wc-1 is a clock-associated protein that is essential for circadian blue-light responses in Neurospora (41, 140). Each of these proteins is a transcriptional regulator, and most of them have two PAS domains (126) (Fig. 4C and D). A connection has been made between PAS domains in photoreceptors (Wc-1 and PYP) and PAS domains in light-entrained clock components (PER, Clock, and Wc-2), with a hypothesis that PAS-containing clock proteins originated from photoreceptors (41, 126). With the current knowledge of the diverse role of PAS domains, the possibility must be considered that PAS domains in clock proteins transduce signals in addition to light, since various environmental signals can entrain circadian clocks. For example, short-duration heat pulses can reset circadian rhythms in Drosophila (204). The PAS-2 domain in Wc-1 from Neurospora is the only PAS domain that is known to be essential for environmental input to a clock-associated protein (15). An input role for PAS domains in other clock proteins is without experimental support at this time. Timeless is the only clock protein demonstrated to respond rapidly to light (220, 255), and it does not have a PAS domain.
In a generalized model for a circadian oscillator (Fig. 15), clock genes are expressed cyclically, increasing the level of clock RNAs and proteins (negative elements). The proteins then feed back after a lag to depress the level of their transcripts, by inhibiting positive elements that enhance the transcription of the clock genes (53). The concentration of clock molecules eventually falls below a threshold, and transcription of clock proteins restarts the cycle. In addition, there must be input into the cycle from an environmental signal that will advance or retard the cycle to maintain the clock in synchrony with the daily dark-light cycle.
FIG. 15.
A generalized circadian clock model showing the proposed role in negative feedback by proteins that contain a PAS domain. For each circadian clock component, proteins specific to mammals (top), Drosophila (middle), and Neurospora (bottom) are indicated. All proteins presented in the model, with the exception of FRQ, TIM, and the proposed kinase, contain a PAS domain. A question mark indicates hypothetical elements and events. JRK is also known as dCLOCK (4, 45).
The basic constituents of clocks in Neurospora, Drosophila, and mammals have been recently elucidated (Fig. 15). The bHLH-PAS CLOCK protein in mice (Fig. 4C) (9, 127) forms a heterodimer with another bHLH-PAS protein, BMAL1 (107) (also termed MOP3 (102). CLOCK has a glutamine-rich activation domain (Fig. 4C) (107, 127) and, when dimerized with BMAL1, binds to an E box upstream of the mouse per1 gene and activates transcription (72, 93, 102) (Fig. 15). Similarly, in Drosophila, the dCLOCK:CYCLE heterodimer, which is homologous to murine CLOCK:BMAL1, binds to the respective E boxes and activates transcription of the per and tim genes (Fig. 15) (4, 45, 195). Coexpression of PER and TIM blocked transactivation of the per and tim genes by dCLOCK, providing the negative feedback required for cycling of PER:TIM (45). The PAS domains provide specificity for bHLH-mediated dimerization. In Drosophila and Neurospora, posttranslational processing, including time-of-day-specific phosphorylation of PER and FRQ, may regulate clock protein turnover and contribute to entraining the clocks to external cycles (54, 70). Short pulses of light rapidly reset the Neurospora clock by induction of the frq gene (42). In constant light, negative autoregulation of frq is overcome by light induction. Of the two PAS domains in the N. crassa Wc-1 protein (Fig. 4D), the PAS1 domain is involved in the formation of homodimers and heterodimers with the Wc-2 protein. The PAS2 domain, which is similar to PAS domains in Bat (Halobacterium salinarum) and NPH1 (Arabidopsis thaliana), is capable of homodimerization only. Three blind Wc-1 strains have point mutations in the PAS2 domain (15).
Transcriptionally Regulated Hypoxia Response in Mammals
Prolonged hypoxia induces cellular responses that include a shift to glycolytic metabolism of carbohydrates, permanent restructuring of the blood supply to the cells, and stimulation of erythrocyte and hemoglobin production (90). Failure of the hypoxia response can contribute to diseases such as anemia, myocardial infarction, retinopathy, and tumor growth. The hypoxia response is under the control of a class of bHLH-PAS transcription factors, the best known of which is HIF1.
HIF1 is an α/β heterodimer, with each subunit containing a bHLH domain at the N terminus followed by two PAS domains (Fig. 16). The HIF1β subunit is identical to the ARNT protein and is absolutely required for HIF1 activity (71). Since HIF1β is also involved in the response to dioxins, HIF1α must be the hypoxia-specific component (90). In addition to the N-terminal bHLH and PAS domains, HIF1α contains in the C-terminal segment an oxygen-dependent degradation domain, which includes two PEST-like sequences (183) and one or two molecules of iron (212), and two transactivation domains (114, 183). In normoxic cells, the HIF1α subunit has a half-life of less than 5 min due to ubiquitination and degradation by the proteasome pathway (32, 104). In hypoxic cells, HIF1α is stabilized against ubiquitin-dependent proteolysis, forms heterodimers with ARNT, and transactivates HIF1-regulated genes. Additional hypoxia-inducible factors that are also bHLH-PAS proteins include hepatic nuclear factor 4 and HIF1-related factor (also known as HLF/E PAS-1) (58).
FIG. 16.
Schematic representation of the bHLH PAS-containing proteins AHR, ARNT, SIM, and HIF-1α. HIF-1α has an oxygen-dependent degradation domain that includes two PEST-like sequences, a transactivation domain (TAD), and a Fe center (104, 212). C-terminal transactivation (TAD) sites are shown in all proteins except PER. Redrawn from reference 193 with permission of the publisher.
The role of the PAS domains in HIF-1 has not been determined. The oxygen-dependent degradation domain (Fig. 16) is necessary and sufficient for oxygen-dependent degradation of HIF1α (104). Thus, the input domain for the hypoxia signal is distinct from the PAS domain. A redox reaction in the iron center may directly determine the survival of HIF1α under oxic conditions (212). The iron center is not a heme or [Fe-S] center and may be a hemerythrin-type di-iron center (212). The PAS domains are required for dimerization and probably determine the specificity of gene activation (113). A role of a PAS domain in hypoxia-enhanced HIF1 transactivation of the target genes has not been ruled out, but it is not the primary mechanism of HIF activation.
HIF1 is a global regulator of hypoxia gene expression. The best characterized of the genes regulated by hypoxia is the gene for erythropoietin, the growth factor that regulates erythrocyte production (32, 90). In hypoxia, HIF1 binds to the sequence 5′-ACGTGCT-3′ in an enhancer element at the 3′ end of the erythropoietin gene and up-regulates transcription (239). HIF1 is widely expressed in tissues, and HIF1-binding sites have been identified in genes for various glycolytic enzymes, vascular endothelial growth factor, nitric oxide synthase, heme oxygenase, and tyrosine hydroxylase (90). Thus, a single transcriptional factor, HIF1, controls many hypoxia responses including adaptation to anaerobic metabolism, erythropoiesis, angiogenesis, vasodilation, and possibly breathing (90). Hypoxia-like-factor appears to be responsible for regulating vascular endothelial growth factor under normoxic conditions and may be involved in the development of blood vessels and the tubular lining of the lungs (58).
Ion Channels: Fast Hypoxia Responses?
Voltage-activated ion channels are members of several conserved multigene families in organisms ranging from bacteria to humans. Oxygen-sensing ion channels participate in fast cellular responses to hypoxia. In contrast to hypoxia responses regulated by gene expression, the ion channel-mediated response occurs on a scale of seconds. One of the best-known examples of such responses in mammals is oxygen sensing by arterial chemoreceptors in the carotid bodies. The dopamine-containing glomus cells of the carotid body are electrically excitable and have oxygen-regulated potassium (K+) channels (145). Oxygen-sensitive ion channels are involved in several cardiocirculatory and respiratory responses to low oxygen levels in fetal and adult life (36, 258) and are associated with the pathophysiology of hypertension, cardiac arrhythmias, and ischemic neuronal damage (60, 115, 144, 245). When the oxygen concentration decreases, the potassium channels linearly decrease the probability of opening, leading to membrane potential depolarization and opening of voltage-sensitive calcium channels. A subsequent increase of the intracellular calcium level induces fast cellular responses, such as neurotransmitter release (234).
Although a variety of oxygen-sensitive ion channels have been described and their cellular role has been established, the mechanisms of oxygen sensing remain elusive. One recent hypothesis is that redox-based mechanisms are intermediates in oxygen sensing by ion channels in the carotid body and pulmonary smooth muscle. Reductants such as reduced glutathione or dithiothreitol can mimic the effect of hypoxia on the oxygen-regulated K+ channels (259), but the mechanism of redox sensing has not been established.
We and others (182, 266) have identified a PAS domain, a possible oxygen sensor, in the pore-forming subunit of voltage-activated K+ channels of the ether a go-go (eag) family (243). The PAS domain is located in the N terminus of the subunit and is followed by six putative transmembrane regions, the pore region, and a putative cyclic nucleotide-binding domain (cNBD), i.e., typical architecture for an ion channel (Fig. 5). One member of the eag family, human eag-related gene (HERG), is a focus of attention because it is associated with inherited and acquired cardiac arrhythmias (for reviews, see references 237 and 241). Mutations in HERG cause the LQT syndrome, a chromosome 7-linked inherited disorder causing sudden death from ventricular tachyarrhythmia (44). HERG, which is strongly expressed in the heart, encodes subunits of cardiac IKr channels. The rectification of the channel is a rapid (10-ms), voltage-dependent inactivation (196). Voltage-dependent gating is conferred by the arginine-rich fourth membrane-spanning segment. Deactivation is very slow and is controlled by a tight association of the PAS-containing N-terminal domain with the body of the potassium channel (160).
Ligand-Activated PAS Domains: AHR:ARNT
The dioxin (also named aryl hydrocarbon) receptor AHR in mammalian cells is a ligand-activated bHLH-PAS receptor that heterodimerizes with the ARNT bHLH-PAS protein (33, 57, 101, 247; for a review, see reference 193). The ligand-activated AHR:ARNT complex binds to an E-box motif in xenobiotic response elements present in upstream regions of dioxin-induced genes, such as the cytochrome P-450IA1 glutathione S-transferase Ya and NAD(P)H:quinone oxidoreductase (68, 170, 193). There is no known endogenous ligand for the dioxin receptor, but it is possible that the natural ligand or ligand precursor is found in the diet (193).
The domain structures of AHR and ARNT are similar in the N-terminal half of the proteins (Fig. 16). A basic region with adjacent positively charged residues and an HLH motif are present in the N-terminal domains. There are two PAS domains. Transactivation C-terminal domains are present in AHR and ARNT (193). The predicted structure of the ARNT PAS domain is similar to the structure of PYP (172).
In the cytosol, the unliganded AHR is found in a complex with a dimer of the heat shock protein hsp90 and a 43-kDa protein, p43 (248, 249). hsp90, which is a molecular chaperone, is apparently required for maintaining AHR in a nonactivated, ligand-binding conformation (179). After binding the ligand, AHR is translocated into the cell nucleus, where it dissociates from hsp90 and p43 and forms a heterodimer with ARNT (247, 249). The ligand-activated AHR:ARNT heterodimer binds specifically to the enhancer E-box DNA. Formation of human and mouse AHR:ARNT heterodimers requires phosphorylation of ARNT by protein kinase C, and DNA binding requires phosphorylation of ARNT and AHR (Fig. 16) (21).
The PAS2 but not the PAS1 domain (residues 230 to 421) is required for ligand or hsp90 binding to AHR (Fig. 17) (39). The chaperone role of hsp90 appears to prevent premature dimerization of the receptor to DNA-binding partners and assists in ligand binding to AHR by ensuring proper folding of the ligand-binding domain (39, 193). The role of the PAS domains in dimerization and DNA-binding specificity has been investigated extensively (105, 139, 178). In the presence of ARNT, an isolated bHLH domain from AHR bound specifically to the E box. However, it lacked specificity in dimerization and formed homodimers with AHR and heterodimers with the bHLH-leucine zipper USF. The addition of PAS1 to a bHLH domain from AHR restored absolute specificity for heterodimerization with ARNT (178). Thus, a functional differentiation has now been established between the role of PAS1 and PAS2 in AHR. PAS1 plays an important role in establishing the dimerization specificity of AHR, and PAS2 is required for binding ligand and hsp90. Recent studies of ARNT interaction with the PAS-bHLH proteins Trachealers (Trh) and Single-minded (Sim) in Drosophila indicated that the PAS domain also recruited factors in addition to ARNT that determined specificity in gene enhancement (261).
FIG. 17.
Structural and functional organization of AHR. TAD, transactivation domain. Redrawn from reference 193 with permission of the publisher.
Regulation of Central Nervous System Development
Development of the Drosophila central nervous system midline cells is dependent on the function of sim, which is a prototype PAS gene and a master regulator for central nervous system development (163). The proposed hierarchy of events underlying central nervous system development includes the following (163): sim is activated at a unique position in the blastoderm by maternal and zygotic genes that specify positional information along the dorsoventral axis; sim activates the transcription of specific midline genes, thereby initiating the developmental program of the central nervous system lineage; and sim positively autoregulates its expression to maintain this program, which is effected through expression of a large number of other midline-expressed genes that directly or indirectly require sim function. The ectopic expression of sim confirmed that Sim can direct cells of the lateral central nervous system to exhibit midline cell morphology and patterns of gene expression (163).
Sim is a bHLH-PAS protein that is similar to an ARNT protein with short truncations at the C and N termini (Fig. 16) (163). The C-terminal half of the protein has a series of transcriptional activation domains. The sensory role of the dual PAS domains has not been established, but they are likely to be critical elements in turning transcriptional activation by Sim on and off in response to cellular or extracellular signals.
Similar Sim homologs present in humans and mice also function as master regulators of central nervous system midline development (157, 253). The human sim gene is located on chromosome 21, in the Down’s syndrome-critical region, and its transcripts may contribute to the etiology and pathophysiology of Down’s syndrome (161, 253).
ARE PAS DOMAINS THE MISSING LINK IN PROTON MOTIVE FORCE AND ENERGY SENSING?
Over the last half century, there have been recurring proposals for a sensory pathway that monitors the energy state of cells (for reviews, see references 221 and 224). ATP was first proposed as the energy parameter that was monitored. Genetic experiments with bacterial strains deficient in the energy-transducing ATP synthase excluded ATP as the sensory signal. Subsequently, abundant evidence has been accumulated that bacteria and eukaryotic cells sense a change of energy level long before there is a drop in ATP concentrations (100, 117). That is not surprising if the role of the response is to protect the energy reserves of the cells.
A competing hypothesis, first proposed by Baryshev, Glagolev, and Shulachev (18, 78), is that cells sense the change in proton motive force. The name “protometer” was proposed for the link between the proton motive force and the sensory response. However, as evidence accumulated that cells do sense the proton motive force or a related parameter (Aer, ArcB, or HIF), the sensor link remained elusive until the PAS-containing Aer protein was shown to transduce the aerotactic response in E. coli (24, 186). Now it is evident that multiple proteins that have been identified as components of energy-sensing pathways have one or more PAS domains. These proteins include ArcB in E. coli, HIF1α, and possibly voltage-activated ion channels in eukaryotes. The evidence that PAS domains may sense redox changes in the electron transport system rather than the proton motive force does not exclude PAS domains as sensors of the proton motive force. Tight coupling of electron transport and the proton motive force enables cells to use either parameter to report changes in cellular energy levels.
Clearly, then, certain PAS domain-containing proteins should be considered a link between electron transport or proton motive force and the sensory response to a change in energy level. However, that is not the whole story. We still do not know how PAS domains interact with the electron transport system, so part of the link is still missing. Given the propensity of PAS domains for protein-protein interactions, it is possible that the still-missing protein has a cognate PAS domain and a domain that is part of, or interacts with, the electron transport system. Such a protein is the subject of an intensive search in our laboratories and in the laboratories of other investigators. The NuoE polypeptide, a chain of NADH dehydrogenase that contains a partial PAS domain (namely, the PAS core, a protein interaction site) and putative [2Fe-2S] centers (Fig. 7), is one of the candidates.
CONCLUSIONS
A PAS-domain superfamily of cytoplasmic sensory transduction elements has been identified as a result of improved techniques for computer-assisted homology searches. More than 300 conserved PAS domains (Fig. 2) have been found in diverse biological systems. Where a function is known, PAS domains mostly sense redox potential, oxygen, cellular energy, or light. Other proteins and predicted proteins that are proposed to have a redox potential- or energy-sensing role also have PAS domains. The 125-residue PYP, which is proposed as a prototype PAS domain, has a distinctive αβ fold with a central six-stranded β-sheet surrounded by the N-terminal cap and α-helices of the PAS core and a helical connector. The N-terminal cap is highly variable and does not show up as a conserved sequence in our compilation of PAS domains.
Analysis of 11 completely sequenced microbial genomes identified five species without a PAS domain. The other species had between 6 and 17 proteins with PAS domains, and there was a positive correlation between the number of PAS domains and the number of electron transport-associated proteins in the species. The number of PAS domains per protein varied from one to six. A comparison of conserved sequences indicated that PAS domains diverged early in the phylogenetic tree. The differentiated PAS domains may be highly conserved from bacteria to humans, consistent with different functions for different PAS domain homologs. The presence of homologous PAS domains in one protein suggests that there may be multiple sensory input.
In prokaryotes, PAS domains are mostly input domains for sensor kinases in two-component regulatory systems. The input domain may be in a separate protein. Some of the proteins with PAS input domains are global regulators of metabolism. In eukaryotes, it is more common for PAS domains to be in transcriptional activators and voltage-sensitive ion channels. Some, however, are in regulatory systems, in which serine or threonine is phosphorylated. Considerable variety and complexity exists in mixing and matching PAS domains with other modules to assemble multidomain proteins into signal transduction systems.
The mechanism of signaling by PAS domains to downstream components of the transduction pathway is not well understood. However, protein-protein interactions, particularly heterodimer formation with another PAS protein, may be a near-universal component of signaling. Recognition of this generalization led to recent progress in closing the loop for circadian clocks. However, even for clock proteins that have been extensively studied, a sensory role of PAS domains remains elusive. This is true for eukaryote PAS domains in general. We are at an early stage, then, in understanding the role of PAS domains, and the complete pathway for a PAS-dependent signaling system still awaits elucidation.
ACKNOWLEDGMENTS
We are grateful to Carl Bauer, Ray Dixon, Elizabeth Getzoff, Marie-Alda Gilles-Gonzalez, Rob Gunsalus, Jim Hoch, Steve Kay, Eugene Koonin, Clark Lagarias, Jeff F. Miller, and Karen Wager-Smith for helpful comments on various parts of the manuscript. We thank Simon Andrews, Michael Chan, Marie-Alda Gilles-Gonzalez, Caroline Harwood, Susan Hill, Samuel Kaplan, Christina Kennedy, Clark Lagarias, Nancy Nichols, and Giuseppe Macino for sending preprints and sharing unpublished results, and we thank Sean Bulloch for assistance with the artwork.
The work in our laboratories is supported by grants from the National Institute of General Medical Sciences (GM29481) and Loma Linda University (to B.L.T.) and from the U.S. Department of Agriculture/NRICGP (96-35305-3795) (to I.B.Z.).
REFERENCES
- 1.Ahmad M, Jarillo J A, Smirnova O, Cashmore A R. The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell. 1998;1:939–948. doi: 10.1016/s1097-2765(00)80094-5. [DOI] [PubMed] [Google Scholar]
- 2.Akerley B J, Miller J F. Understanding signal transduction during bacterial infection. Trends Microbiol. 1996;4:141–146. doi: 10.1016/0966-842x(96)10024-x. [DOI] [PubMed] [Google Scholar]
- 3.Alex L A, Simon M I. Protein histidine kinases and signal transduction in prokaryotes and eukaryotes. Trends Genet. 1994;10:133–138. doi: 10.1016/0168-9525(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 4.Allada R, White N E, So W V, Hall J C, Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. doi: 10.1016/s0092-8674(00)81440-3. [DOI] [PubMed] [Google Scholar]
- 5.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 6.Altschul S F, Koonin E V. Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem Sci. 1998;23:444–447. doi: 10.1016/s0968-0004(98)01298-5. [DOI] [PubMed] [Google Scholar]
- 7.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anthamatten D, Hennecke H. The regulatory status of the fixL- and fixJ-like genes in Bradyrhizobium japonicum may be different from that in Rhizobium meliloti. Mol Gen Genet. 1991;225:38–48. doi: 10.1007/BF00282640. [DOI] [PubMed] [Google Scholar]
- 9.Antoch M P, Song E J, Chang A M, Vitaterna M H, Zhao Y, Wilsbacher L D, Sangoram A M, King D P, Pinto L H, Takahashi J S. Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell. 1997;89:655–667. doi: 10.1016/s0092-8674(00)80246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Appleby J L, Parkinson J S, Bourret R B. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell. 1996;86:845–848. doi: 10.1016/s0092-8674(00)80158-0. [DOI] [PubMed] [Google Scholar]
- 11.Armitage J P. Behavioural responses of bacteria to light and oxygen. Arch Microbiol. 1997;168:249–261. doi: 10.1007/s002030050496. [DOI] [PubMed] [Google Scholar]
- 12.Armitage J P, Schmitt R. Bacterial chemotaxis: Rhodobacter sphaeroides and Sinorhizobium meliloti—variations on a theme? Microbiology. 1997;143:3671–3682. doi: 10.1099/00221287-143-12-3671. [DOI] [PubMed] [Google Scholar]
- 13.Baca M, Borgstahl G E, Boissinot M, Burke P M, Williams D R, Slater K A, Getzoff E D. Complete chemical structure of photoactive yellow protein: novel thioester-linked 4-hydroxycinnamyl chromophore and photocycle chemistry. Biochemistry. 1994;33:14369–14377. doi: 10.1021/bi00252a001. [DOI] [PubMed] [Google Scholar]
- 14.Ballario P, Macino G. White collar proteins: PASsing the light signal in Neurospora crassa. Trends Microbiol. 1997;5:458–462. doi: 10.1016/S0966-842X(97)01144-X. [DOI] [PubMed] [Google Scholar]
- 15.Ballario P, Talora C, Galli D, Linden H, Macino G. Roles in dimerization and blue light photoresponse of the PAS and LOV domains of Neurospora crassa white collar proteins. Mol Microbiol. 1998;29:719–729. doi: 10.1046/j.1365-2958.1998.00955.x. [DOI] [PubMed] [Google Scholar]
- 16.Ballario P, Vittorioso P, Magrelli A, Talora C, Cabibbo A, Macino G. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 1996;15:1650–1657. [PMC free article] [PubMed] [Google Scholar]
- 17.Barak R, Eisenbach M. Regulation of interaction between signaling protein CheY and flagellar motor during bacterial chemotaxis. Curr Top Cell Regul. 1996;34:137–158. doi: 10.1016/s0070-2137(96)80005-7. [DOI] [PubMed] [Google Scholar]
- 18.Baryshev V A, Glagolev A N, Skulachev V P. Sensing of ΔμH+ in phototaxis of Halobacterium halobium. Nature. 1981;292:338–340. [Google Scholar]
- 19.Bauer C E, Bird T H. Regulatory circuits controlling photosynthesis gene expression. Cell. 1996;85:5–8. doi: 10.1016/s0092-8674(00)81074-0. [DOI] [PubMed] [Google Scholar]
- 20.Baur B, Fischer K, Winter K, Dietz K J. cDNA sequence of a protein kinase from the inducible crassulacean acid metabolism plant Mesembryanthemum crystallinum L., encoding a SNF-1 homolog. Plant Physiol. 1994;106:1225–1226. doi: 10.1104/pp.106.3.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berghard A, Gradin K, Pongratz I, Whitelaw M, Poellinger L. Cross-coupling of signal transduction pathways: the dioxin receptor mediates induction of cytochrome P-450IA1 expression via a protein kinase C-dependent mechanism. Mol Cell Biol. 1993;13:677–689. doi: 10.1128/mcb.13.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertolucci C, Ming L J, Gonzalez G, Gilles-Gonzalez M A. Assignment of the hyperfine-shifted 1H-NMR signals of the heme in the oxygen sensor FixL from Rhizobium meliloti. Chem Biol. 1996;3:561–566. doi: 10.1016/s1074-5521(96)90147-7. [DOI] [PubMed] [Google Scholar]
- 23.Bespalov V A, Zhulin I B, Taylor B L. Behavioral responses of Escherichia coli to changes in redox potential. Proc Natl Acad Sci USA. 1996;93:10084–10089. doi: 10.1073/pnas.93.19.10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bibikov S I, Biran R, Rudd K E, Parkinson J S. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol. 1997;179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blanco G, Drummond M, Woodley P, Kennedy C. Sequence and molecular analysis of the nifL gene of Azotobacter vinelandii. Mol Microbiol. 1993;9:869–879. doi: 10.1111/j.1365-2958.1993.tb01745.x. [DOI] [PubMed] [Google Scholar]
- 26.Borgstahl G E, Williams D R, Getzoff E D. 1.4 A structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore. Biochemistry. 1995;34:6278–6287. doi: 10.1021/bi00019a004. [DOI] [PubMed] [Google Scholar]
- 27.Bork P, Koonin E V. Predicting functions from protein sequences—where are the bottlenecks? Nat Genet. 1998;18:313–318. doi: 10.1038/ng0498-313. [DOI] [PubMed] [Google Scholar]
- 28.Bott M. Anaerobic citrate metabolism and its regulation in enterobacteria. Arch Microbiol. 1997;167:78–88. [PubMed] [Google Scholar]
- 29.Bott M, Meyer M, Dimroth P. Regulation of anaerobic citrate metabolism in Klebsiella pneumoniae. Mol Microbiol. 1995;18:533–546. doi: 10.1111/j.1365-2958.1995.mmi_18030533.x. [DOI] [PubMed] [Google Scholar]
- 30.Bowler C, Frohnmeyer H, Schafer E, Neuhaus G, Chua N H. Phytochrome and UV signal transduction pathways. Acta Physiol Plant. 1997;19:475–483. [Google Scholar]
- 31.Brooun A, Bell J, Freitas T, Larsen R W, Alam M. An archaeal aerotaxis transducer combines subunit I core structures of eukaryotic cytochrome c oxidase and eubacterial methyl-accepting chemotaxis proteins. J Bacteriol. 1998;180:1642–1646. doi: 10.1128/jb.180.7.1642-1646.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bunn H, Gu J, Huang L, Park J W. Erythropoietin: a model system for studying oxygen-dependent gene regulation. J Exp Biol. 1998;201:1197–1201. doi: 10.1242/jeb.201.8.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burbach K M, Poland A, Bradfield C A. Cloning of the Ah-receptor cDNA reveals a distinctive ligand-activated transcription factor. Proc Natl Acad Sci USA. 1992;89:8185–8189. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burbulys D, Trach K A, Hoch J A. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991;64:545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- 35.Cohen-Bazire G, Sistrom W R, Stanier R Y. Kinetic studies of pigment synthesis by non-sulphur purple bacteria. J Cell Comp Physiol. 1957;49:25–68. doi: 10.1002/jcp.1030490104. [DOI] [PubMed] [Google Scholar]
- 36.Cornfield D N, Reeve H L, Tolarova S, Weir E K, Archer S. Oxygen causes fetal pulmonary vasodilation through activation of a calcium-dependent potassium channel. Proc Natl Acad Sci USA. 1996;93:8089–8094. doi: 10.1073/pnas.93.15.8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coschigano P W, Young L Y. Identification and sequence analysis of two regulatory genes involved in anaerobic toluene metabolism by strain T1. Appl Environ Microbiol. 1997;63:652–660. doi: 10.1128/aem.63.2.652-660.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cotter P A, Miller J F. A mutation in the Bordetella bronchiseptica bvgS gene results in reduced virulence and increased resistance to starvation, and identifies a new class of Bvg-regulated antigens. Mol Microbiol. 1997;24:671–685. doi: 10.1046/j.1365-2958.1997.3821741.x. [DOI] [PubMed] [Google Scholar]
- 39.Coumailleau P, Poellinger L, Gustafsson J A, Whitelaw M L. Definition of a minimal domain of the dioxin receptor that is associated with Hsp90 and maintains wild type ligand binding affinity and specificity. J Biol Chem. 1995;270:25291–25300. doi: 10.1074/jbc.270.42.25291. [DOI] [PubMed] [Google Scholar]
- 40.Crews S T, Thomas J B, Goodman C S. The Drosophila single-minded gene encodes a nuclear protein with sequence similarity to the per gene product. Cell. 1988;52:143–151. doi: 10.1016/0092-8674(88)90538-7. [DOI] [PubMed] [Google Scholar]
- 41.Crosthwaite S K, Dunlap J C, Loros J J. Neurospora wc-1 and wc-2: transcription, photoresponses, and the origins of circadian rhythmicity. Science. 1997;276:763–769. doi: 10.1126/science.276.5313.763. [DOI] [PubMed] [Google Scholar]
- 42.Crosthwaite S K, Loros J J, Dunlap J C. Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell. 1995;81:1003–1012. doi: 10.1016/s0092-8674(05)80005-4. [DOI] [PubMed] [Google Scholar]
- 43.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 44.Curran M E, Splawski I, Timothy K W, Vincent G M, Green E D, Keating M T. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 45.Darlington T K, Wager-Smith K, Ceriani M F, Staknis D, Gekakis N, Steeves T L, Weitz C J, Takahashi J S, Kay S A. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- 46.David M, Daveran M L, Batut J, Dedieu A, Domergue O, Ghai J, Hertig C, Boistard P, Kahn D. Cascade regulation of nif gene expression in Rhizobium meliloti. Cell. 1988;54:671–683. doi: 10.1016/s0092-8674(88)80012-6. [DOI] [PubMed] [Google Scholar]
- 47.de Philip P, Batut J, Boistard P. Rhizobium meliloti FixL is an oxygen sensor and regulates R. meliloti nifA and fixK genes differently in Escherichia coli. J Bacteriol. 1990;172:4255–4262. doi: 10.1128/jb.172.8.4255-4262.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.D’hooghe I, Michiels J, Vlassak K, Verreth C, Waelkens F, Vanderleyden J. Structural and functional analysis of the fixLJ genes of Rhizobium leguminosarum biovar phaseoli CNPAF512. Mol Gen Genet. 1995;249:117–126. doi: 10.1007/BF00290243. [DOI] [PubMed] [Google Scholar]
- 49.Dixon R. The oxygen-responsive NIFL-NIFA complex: a novel two-component regulatory system controlling nitrogenase synthesis in gamma-proteobacteria. Arch Microbiol. 1998;169:371–380. doi: 10.1007/s002030050585. [DOI] [PubMed] [Google Scholar]
- 50.Dolla A, Fu R, Brumlik M J, Voordouw G. Nucleotide sequence of dcrA, a Desulfovibrio vulgaris Hildenborough chemoreceptor gene, and its expression in Escherichia coli. J Bacteriol. 1992;174:1726–1733. doi: 10.1128/jb.174.6.1726-1733.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drews G. Adaptation of the bacterial photosynthetic apparatus to different light intensities. Trends Biochem Sci. 1986;6:255–257. [Google Scholar]
- 52.Drummond M, Whitty P, Wootton J. Sequence and domain relationships of ntrC and nifA from Klebsiella pneumoniae: homologies to other regulatory proteins. EMBO J. 1986;5:441–447. doi: 10.1002/j.1460-2075.1986.tb04230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dunlap J. Circadian rhythms. An end in the beginning. Science. 1998;280:1548–1549. doi: 10.1126/science.280.5369.1548. [DOI] [PubMed] [Google Scholar]
- 54.Edery I, Zwiebel L J, Dembinska M E, Rosbash M. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci USA. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgerton M D, Jones A M. Subunit interactions in the carboxy-terminal domain of phytochrome. Biochemistry. 1993;32:8239–8245. doi: 10.1021/bi00083a026. [DOI] [PubMed] [Google Scholar]
- 56.Elich T D, Chory J. Phytochrome: if it looks and smells like a histidine kinase, is it a histidine kinase? Cell. 1997;91:713–716. doi: 10.1016/s0092-8674(00)80458-4. [DOI] [PubMed] [Google Scholar]
- 57.Ema M, Sogawa K, Watanabe N, Chujoh Y, Matsushita N, Gotoh O, Funae Y, Fujii-Kuriyama Y. cDNA cloning and structure of mouse putative Ah receptor. Biochem Biophys Res Commun. 1992;184:246–253. doi: 10.1016/0006-291x(92)91185-s. [DOI] [PubMed] [Google Scholar]
- 58.Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii-Kuriyama Y. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc Natl Acad Sci USA. 1997;94:4273–4278. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emery P, So W V, Kaneko M, Hall J C, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 60.Fearon I M, Palmer A C, Balmforth A J, Ball S G, Mikala G, Schwartz A, Peers C. Hypoxia inhibits the recombinant alpha 1C subunit of the human cardiac L-type Ca2+ channel. J Physiol (London) 1997;500:551–556. doi: 10.1113/jphysiol.1997.sp022041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fischer H M. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev. 1994;58:352–386. doi: 10.1128/mr.58.3.352-386.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fischer H M. Environmental regulation of rhizobial symbiotic nitrogen fixation genes. Trends Microbiol. 1996;4:317–320. doi: 10.1016/0966-842x(96)10049-4. [DOI] [PubMed] [Google Scholar]
- 63.Fisher R F, Long S R. Rhizobium—plant signal exchange. Nature. 1992;357:655–660. doi: 10.1038/357655a0. [DOI] [PubMed] [Google Scholar]
- 64.Forward J A, Behrendt M C, Wyborn N R, Cross R, Kelly D J. TRAP transporters: a new family of periplasmic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse gram-negative bacteria. J Bacteriol. 1997;179:5482–5493. doi: 10.1128/jb.179.17.5482-5493.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Frazzon J, Schrank I S. Sequencing and complementation analysis of the nifUSV genes from Azospirillum brasilense. FEMS Microbiol Lett. 1998;159:151–158. doi: 10.1111/j.1574-6968.1998.tb12854.x. [DOI] [PubMed] [Google Scholar]
- 66.Fu R, Wall J D, Voordouw G. DcrA, a c-type heme-containing methyl-accepting protein from Desulfovibrio vulgaris Hildenborough, senses the oxygen concentration or redox potential of the environment. J Bacteriol. 1994;176:344–350. doi: 10.1128/jb.176.2.344-350.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fu W, Jack R F, Morgan T V, Dean D R, Johnson M K. nifU gene product from Azotobacter vinelandii is a homodimer that contains two identical [2Fe-2S] clusters. Biochemistry. 1994;33:13455–13463. doi: 10.1021/bi00249a034. [DOI] [PubMed] [Google Scholar]
- 68.Fujisawa-Sehara A, Sogawa K, Yamane M, Fujii-Kuriyama Y. Characterization of xenobiotic responsive elements upstream from the drug-metabolizing cytochrome P-450c gene: a similarity to glucocorticoid regulatory elements. Nucleic Acids Res. 1987;15:4179–4191. doi: 10.1093/nar/15.10.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gamble R L, Coonfield M L, Schaller G E. Histidine kinase activity of the ETR1 ethylene receptor from Arabidopsis. Proc Natl Acad Sci USA. 1998;95:7825–7829. doi: 10.1073/pnas.95.13.7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garceau N Y, Liu Y, Loros J J, Dunlap J C. Alternative initiation of translation and time-specific phosphorylation yield multiple forms of the essential clock protein FREQUENCY. Cell. 1997;89:469–476. doi: 10.1016/s0092-8674(00)80227-5. [DOI] [PubMed] [Google Scholar]
- 71.Gassmann M, Kvietikova I, Rolfs A, Wenger R H. Oxygen- and dioxin-regulated gene expression in mouse hepatoma cells. Kidney Int. 1997;51:567–574. doi: 10.1038/ki.1997.81. [DOI] [PubMed] [Google Scholar]
- 72.Gekakis N, Staknis D, Nguyen H B, Davis F C, Wilsbacher L D, King D P, Takahashi J S, Weitz C J. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 73.Genick U K, Borgstahl G E, Ng K, Ren Z, Pradervand C, Burke P M, Srajer V, Teng T Y, Schildkamp W, McRee D E, Moffat K, Getzoff E D. Structure of a protein photocycle intermediate by millisecond time-resolved crystallography. Science. 1997;275:1471–1475. doi: 10.1126/science.275.5305.1471. [DOI] [PubMed] [Google Scholar]
- 74.Genick U K, Soltis S M, Kuhn P, Canestrelli I L, Getzoff E D. Structure at 0.85 A resolution of an early protein photocycle intermediate. Nature. 1998;392:206–209. doi: 10.1038/32462. [DOI] [PubMed] [Google Scholar]
- 75.Gilles-Gonzalez M A, Ditta G S, Helinski D R. A haemoprotein with kinase activity encoded by the oxygen sensor of Rhizobium meliloti. Nature. 1991;350:170–172. doi: 10.1038/350170a0. [DOI] [PubMed] [Google Scholar]
- 76.Gilles-Gonzalez M A, Gonzalez G, Perutz M F. Kinase activity of oxygen sensor FixL depends on the spin state of its heme iron. Biochemistry. 1995;34:232–236. doi: 10.1021/bi00001a027. [DOI] [PubMed] [Google Scholar]
- 77.Gilles-Gonzalez M A, Gonzalez G, Perutz M F, Kiger L, Marden M C, Poyart C. Heme-based sensors, exemplified by the kinase FixL, are a new class of heme protein with distinctive ligand binding and autoxidation. Biochemistry. 1994;33:8067–8073. doi: 10.1021/bi00192a011. [DOI] [PubMed] [Google Scholar]
- 78.Glagolev A N. Reception of the energy level in bacterial taxis. J Theor Biol. 1980;82:171–185. doi: 10.1016/0022-5193(80)90097-1. [DOI] [PubMed] [Google Scholar]
- 79.Golby P, Davies S, Kelly D J, Guest J R, Andrews S C. Identification and characterization of a two-component sensor-kinase and response-regulator system (DcuS-DcuR) controlling gene expression in response to C4-dicarboxylates in Escherichia coli. J Bacteriol. 1999;181:1238–1248. doi: 10.1128/jb.181.4.1238-1248.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gomelsky M, Kaplan S. Genetic evidence that PpsR from Rhodobacter sphaeroides 2.4.1 functions as a repressor of puc and bchF expression. J Bacteriol. 1995;177:1634–1637. doi: 10.1128/jb.177.6.1634-1637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gomelsky M, Kaplan S. Molecular genetic analysis suggesting interactions between AppA and PpsR in regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J Bacteriol. 1997;179:128–134. doi: 10.1128/jb.179.1.128-134.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gong W, Hao B, Mansy S S, Gonzalez G, Gilles-Gonzalez M A, Chan M K. Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc Natl Acad Sci USA. 1998;96:15189–15193. doi: 10.1073/pnas.95.26.15177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gottfert M, Grob P, Hennecke H. Proposed regulatory pathway encoded by the nodV and nodW genes, determinants of host specificity in Bradyrhizobium japonicum. Proc Natl Acad Sci USA. 1990;87:2680–2684. doi: 10.1073/pnas.87.7.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goudreau P N, Stock A M. Signal transduction in bacteria: molecular mechanisms of stimulus-response coupling. Curr Opin Microbiol. 1998;1:160–169. doi: 10.1016/s1369-5274(98)80006-4. [DOI] [PubMed] [Google Scholar]
- 85.Grimshaw C E, Huang S, Hanstein C G, Strauch M A, Burbulys D, Wang L, Hoch J A, Whiteley J M. Synergistic kinetic interactions between components of the phosphorelay controlling sporulation in Bacillus subtilis. Biochemistry. 1998;37:1365–1375. doi: 10.1021/bi971917m. [DOI] [PubMed] [Google Scholar]
- 86.Grob P, Hennecke H, Gottfert M. Cross-talk between the two-component regulatory systems NodVW and NwsAB of Bradyrhizobium japonicum. FEMS Microbiol Lett. 1994;120:349–354. [Google Scholar]
- 87.Grob P, Michel P, Hennecke H, Gottfert M. A novel response-regulator is able to suppress the nodulation defect of a Bradyrhizobium japonicum nodW mutant. Mol Gen Genet. 1993;241:531–541. doi: 10.1007/BF00279895. [DOI] [PubMed] [Google Scholar]
- 88.Gropp F, Betlach M C. The bat gene of Halobacterium halobium encodes a trans-acting oxygen inducibility factor. Proc Natl Acad Sci USA. 1994;91:5475–5479. doi: 10.1073/pnas.91.12.5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Grossman A D, Losick R. Extracellular control of spore formation in Bacillus subtilis. Proc Natl Acad Sci USA. 1988;85:4369–4373. doi: 10.1073/pnas.85.12.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guillemin K, Krasnow M A. The hypoxic response: huffing and HIFing. Cell. 1997;89:9–12. doi: 10.1016/s0092-8674(00)80176-2. [DOI] [PubMed] [Google Scholar]
- 91.Gunsalus R P, Park S J. Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res Microbiol. 1994;145:437–450. doi: 10.1016/0923-2508(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 92.Hamblin M J, Shaw J G, Kelly D J. Sequence analysis and interposon mutagenesis of a sensor-kinase (DctS) and response-regulator (DctR) controlling synthesis of the high-affinity C4-dicarboxylate transport system in Rhodobacter capsulatus. Mol Gen Genet. 1993;237:215–224. doi: 10.1007/BF00282803. [DOI] [PubMed] [Google Scholar]
- 93.Hao H, Allen D L, Hardin P E. A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol Cell Biol. 1997;17:3687–3693. doi: 10.1128/mcb.17.7.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Harriott O T, Hosted T J, Benson D R. Sequences of nifX, nifW, nifZ, nifB and two ORF in the Frankia nitrogen fixation gene cluster. Gene. 1995;161:63–67. doi: 10.1016/0378-1119(95)00300-u. [DOI] [PubMed] [Google Scholar]
- 95.Harris R A, Popov K M, Zhao Y, Kedishvili N Y, Shimomura Y, Crabb D W. A new family of protein kinases—the mitochondrial protein kinases. Adv Enzyme Regul. 1995;35:147–162. doi: 10.1016/0065-2571(94)00020-4. [DOI] [PubMed] [Google Scholar]
- 96.Harwood, C., and N. Nichols. Personal communication.
- 97.Hill S, Austin S, Eydmann T, Jones T, Dixon R. Azotobacter vinelandii NIFL is a flavoprotein that modulates transcriptional activation of nitrogen-fixation genes via a redox-sensitive switch. Proc Natl Acad Sci USA. 1996;93:2143–2148. doi: 10.1073/pnas.93.5.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hill, S., L. He, and C. Kennedy. Characterization of an Azotobacter vinelandii nifU mutant and its spontaneous revertants that over-produce cytochrome bd. Submitted for publication.
- 99.Hoch J A, Silhavy T J. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. [Google Scholar]
- 100.Hochachka P W, Buck L T, Doll C J, Land S C. Unifying theory of hypoxia tolerance: molecular/metabolic defense and rescue mechanisms for surviving oxygen lack. Proc Natl Acad Sci USA. 1996;93:9493–9498. doi: 10.1073/pnas.93.18.9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hoffman E C, Reyes H, Chu F F, Sander F, Conley L H, Brooks B A, Hankinson O. Cloning of a factor required for activity of the Ah (dioxin) receptor. Science. 1991;252:954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- 102.Hogenesch J B, Gu Y Z, Jain S, Bradfield C A. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci USA. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huala E, Oeller P W, Liscum E, Han I S, Larsen E, Briggs W R. Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science. 1997;278:2120–2123. doi: 10.1126/science.278.5346.2120. [DOI] [PubMed] [Google Scholar]
- 104.Huang L E, Gu J, Schau M, Bunn H F. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Huang Z J, Edery I, Rosbash M. PAS is a dimerization domain common to Drosophila period and several transcription factors. Nature. 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- 106.Hwang D M, Dempsey A, Tan K T, Liew C C. A modular domain of NifU, a nitrogen fixation cluster protein, is highly conserved in evolution. J Mol Evol. 1996;43:536–540. doi: 10.1007/BF02337525. [DOI] [PubMed] [Google Scholar]
- 107.Ikeda M, Nomura M. cDNA cloning and tissue-specific expression of a novel basic helix-loop-helix/PAS protein (BMAL1) and identification of alternatively spliced variants with alternative translation initiation site usage. Biochem Biophys Res Commun. 1997;233:258–264. doi: 10.1006/bbrc.1997.6371. [DOI] [PubMed] [Google Scholar]
- 108.Iuchi S, Chepuri V, Fu H A, Gennis R B, Lin E C. Requirement for terminal cytochromes in generation of the aerobic signal for the arc regulatory system in Escherichia coli: study utilizing deletions and lac fusions of cyo and cyd. J Bacteriol. 1990;172:6020–6025. doi: 10.1128/jb.172.10.6020-6025.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Iuchi S, Lin E C. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci USA. 1988;85:1888–1892. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Iuchi S, Lin E C. Adaptation of Escherichia coli to redox environments by gene expression. Mol Microbiol. 1993;9:9–15. doi: 10.1111/j.1365-2958.1993.tb01664.x. [DOI] [PubMed] [Google Scholar]
- 111.Iuchi S, Matsuda Z, Fujiwara T, Lin E C. The arcB gene of Escherichia coli encodes a sensor-regulator protein for anaerobic repression of the arc modulon. Mol Microbiol. 1990;4:715–727. doi: 10.1111/j.1365-2958.1990.tb00642.x. [DOI] [PubMed] [Google Scholar]
- 112.Jacobson M R, Brigle K E, Bennett L T, Setterquist R A, Wilson M S, Cash V L, Beynon J, Newton W E, Dean D R. Physical and genetic map of the major nif gene cluster from Azotobacter vinelandii. J Bacteriol. 1989;171:1017–1027. doi: 10.1128/jb.171.2.1017-1027.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jiang B H, Rue E, Wang G L, Roe R, Semenza G L. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J Biol Chem. 1996;271:17771–17778. doi: 10.1074/jbc.271.30.17771. [DOI] [PubMed] [Google Scholar]
- 114.Jiang B H, Zheng J Z, Leung S W, Roe R, Semenza G L. Transactivation and inhibitory domains of hypoxia-inducible factor 1alpha. Modulation of transcriptional activity by oxygen tension. J Biol Chem. 1997;272:19253–19260. doi: 10.1074/jbc.272.31.19253. [DOI] [PubMed] [Google Scholar]
- 115.Jiang C, Haddad G G. A direct mechanism for sensing low oxygen levels by central neurons. Proc Natl Acad Sci USA. 1994;91:7198–7201. doi: 10.1073/pnas.91.15.7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jiang J, Gu B H, Albright L M, Nixon B T. Conservation between coding and regulatory elements of Rhizobium meliloti and Rhizobium leguminosarum dct genes. J Bacteriol. 1989;171:5244–5253. doi: 10.1128/jb.171.10.5244-5253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Johnson M S, Taylor B L. Comparison of methods for specific depletion of ATP in Salmonella typhimurium. Appl Environ Microbiol. 1993;59:3509–3512. doi: 10.1128/aem.59.10.3509-3512.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Johnson, M. S., and B. L. Taylor. Unpublished data.
- 119.Kakimoto T. CKI1, a histidine kinase homolog implicated in cytokinin signal transduction. Science. 1996;274:982–985. doi: 10.1126/science.274.5289.982. [DOI] [PubMed] [Google Scholar]
- 120.Kaminski P A, Elmerich C. Involvement of fixLJ in the regulation of nitrogen fixation in Azorhizobium caulinodans. Mol Microbiol. 1991;5:665–673. doi: 10.1111/j.1365-2958.1991.tb00738.x. [DOI] [PubMed] [Google Scholar]
- 121.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 122.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions (supplement) DNA Res. 1996;3:185–209. doi: 10.1093/dnares/3.3.185. [DOI] [PubMed] [Google Scholar]
- 123.Kaneko T, Tabata S. Complete genome structure of the unicellular cyanobacterium Synechocystis sp. PCC6803. Plant Cell Physiol. 1997;38:1171–1176. doi: 10.1093/oxfordjournals.pcp.a029103. [DOI] [PubMed] [Google Scholar]
- 124.Karimova G, Bellalou J, Ullmann A. Phosphorylation-dependent binding of BvgA to the upstream region of the cyaA gene of Bordetella pertussis. Mol Microbiol. 1996;20:489–496. doi: 10.1046/j.1365-2958.1996.5231057.x. [DOI] [PubMed] [Google Scholar]
- 125.Kato M, Mizuno T, Shimizu T, Hakoshima T. Insights into multistep phosphorelay from the crystal structure of the C-terminal HPt domain of ArcB. Cell. 1997;88:717–723. doi: 10.1016/s0092-8674(00)81914-5. [DOI] [PubMed] [Google Scholar]
- 126.Kay S A. PAS, present, and future: clues to the origins of circadian clocks. Science. 1997;276:753–754. doi: 10.1126/science.276.5313.753. [DOI] [PubMed] [Google Scholar]
- 127.King D P, Zhao Y, Sangoram A M, Wilsbacher L D, Tanaka M, Antoch M P, Steeves T D, Vitaterna M H, Kornhauser J M, Lowrey P L, Turek F W, Takahashi J S. Positional cloning of the mouse circadian clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B, Venter J C. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 129.Koch C A, Anderson D, Moran M F, Ellis C, Pawson T. SH2 and SH3 domains: elements that control interactions of cytoplasmic signaling proteins. Science. 1991;252:668–674. doi: 10.1126/science.1708916. [DOI] [PubMed] [Google Scholar]
- 130.Kort R, Phillips-Jones M K, van Aalten D F, Haker A, Hoffer S M, Hellingwerf K J, Crielaard W. Sequence, chromophore extraction and 3-D model of the photoactive yellow protein from Rhodobacter sphaeroides. Biochim Biophys Acta. 1998;1385:1–6. doi: 10.1016/s0167-4838(98)00050-8. [DOI] [PubMed] [Google Scholar]
- 131.Lagarias D M, Wu S H, Lagarias J C. Atypical phytochrome gene structure in the green alga Mesotaenium caldariorum. Plant Mol Biol. 1995;29:1127–1142. doi: 10.1007/BF00020457. [DOI] [PubMed] [Google Scholar]
- 132.Laszlo D J, Taylor B L. Aerotaxis in Salmonella typhimurium: role of electron transport. J Bacteriol. 1981;145:990–1001. doi: 10.1128/jb.145.2.990-1001.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lau P C, Wang Y, Patel A, Labbe D, Bergeron H, Brousseau R, Konishi Y, Rawlings M. A bacterial basic region leucine zipper histidine kinase regulating toluene degradation. Proc Natl Acad Sci USA. 1997;94:1453–1458. doi: 10.1073/pnas.94.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.LeDeaux J R, Yu N, Grossman A D. Different roles for KinA, KinB, and KinC in the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1995;177:861–863. doi: 10.1128/jb.177.3.861-863.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Le Moual H, Koshland D E J. Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J Mol Biol. 1996;261:568–585. doi: 10.1006/jmbi.1996.0483. [DOI] [PubMed] [Google Scholar]
- 136.Li X, Xu J, Li M. The human delta1261 mutation of the HERG potassium channel results in a truncated protein that contains a subunit interaction domain and decreases the channel expression. J Biol Chem. 1997;272:705–708. doi: 10.1074/jbc.272.2.705. [DOI] [PubMed] [Google Scholar]
- 137.Liang Y Y, Arsene F, Elmerich C. Characterization of the ntrBC genes of Azospirillum brasilense Sp7: their involvement in the regulation of nitrogenase synthesis and activity. Mol Gen Genet. 1993;240:188–196. doi: 10.1007/BF00277056. [DOI] [PubMed] [Google Scholar]
- 138.Lin X, Feng X H, Watson J C. Differential accumulation of transcripts encoding protein kinase homologs in greening pea seedlings. Proc Natl Acad Sci USA. 1991;88:6951–6955. doi: 10.1073/pnas.88.16.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lindebro M C, Poellinger L, Whitelaw M L. Protein-protein interaction via PAS domains: role of the PAS domain in positive and negative regulation of the bHLH/PAS dioxin receptor-Arnt transcription factor complex. EMBO J. 1995;14:3528–3539. doi: 10.1002/j.1460-2075.1995.tb07359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Linden H, Ballario P, Macino G. Blue light regulation in Neurospora crassa. Fungal Genet Biol. 1997;22:141–150. doi: 10.1006/fgbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- 141.Liu J D, Parkinson J S. Genetic evidence for interaction between the CheW and Tsr proteins during chemoreceptor signaling by Escherichia coli. J Bacteriol. 1991;173:4941–4951. doi: 10.1128/jb.173.16.4941-4951.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Loh J, Garcia M, Stacey G. NodV and NodW, a second flavonoid recognition system regulating nod gene expression in Bradyrhizobium japonicum. J Bacteriol. 1997;179:3013–3020. doi: 10.1128/jb.179.9.3013-3020.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Lois A F, Ditta G S, Helinski D R. The oxygen sensor FixL of Rhizobium meliloti is a membrane protein containing four possible transmembrane segments. J Bacteriol. 1993;175:1103–1109. doi: 10.1128/jb.175.4.1103-1109.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lopez-Barneo J. Oxygen-sensing by ion channels and the regulation of cellular functions. Trends Neurosci. 1996;19:435–440. doi: 10.1016/0166-2236(96)10050-3. [DOI] [PubMed] [Google Scholar]
- 145.Lopez-Barneo J, Lopez-Lopez J R, Urena J, Gonzalez C. Chemotransduction in the carotid body: K+ current modulated by PO2 in type I chemoreceptor cells. Science. 1988;241:580–582. doi: 10.1126/science.2456613. [DOI] [PubMed] [Google Scholar]
- 146.Lynch A S, Lin E C C. Responses to molecular oxygen. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 1526–1538. [Google Scholar]
- 147.Macheroux P, Hill S, Austin S, Eydmann T, Jones T, Kim S O, Poole R, Dixon R. Electron donation to the flavoprotein NifL, a redox-sensing transcriptional regulator. Biochem J. 1998;332:413–419. doi: 10.1042/bj3320413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Makino K, Shinagawa H, Amemura M, Kawamoto T, Yamada M, Nakata A. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J Mol Biol. 1989;210:551–559. doi: 10.1016/0022-2836(89)90131-9. [DOI] [PubMed] [Google Scholar]
- 149.Makino K, Shinagawa H, Amemura M, Nakata A. Nucleotide sequence of the phoR gene, a regulatory gene for the phosphate regulon of Escherichia coli. J Mol Biol. 1986;192:549–556. doi: 10.1016/0022-2836(86)90275-5. [DOI] [PubMed] [Google Scholar]
- 150.Malhotra K, Kim S T, Batschauer A, Dawut L, Sancar A. Putative blue-light photoreceptors from Arabidopsis thaliana and Sinapis alba with a high degree of sequence homology to DNA photolyase contain the two photolyase cofactors but lack DNA repair activity. Biochemistry. 1995;34:6892–6899. doi: 10.1021/bi00020a037. [DOI] [PubMed] [Google Scholar]
- 151.Matsushika A, Mizuno T. A dual-signaling mechanism mediated by the ArcB hybrid sensor kinase containing the histidine-containing phosphotransfer domain in Escherichia coli. J Bacteriol. 1998;180:3973–3977. doi: 10.1128/jb.180.15.3973-3977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.McLaughlin M E, Sandberg M A, Berson E L, Dryja T P. Recessive mutations in the gene encoding the beta-subunit of rod phosphodiesterase in patients with retinitis pigmentosa. Nat Genet. 1993;4:130–134. doi: 10.1038/ng0693-130. [DOI] [PubMed] [Google Scholar]
- 153.Meijer W G, Tabita F R. Isolation and characterization of the nifUSVW-rpoN gene cluster from Rhodobacter sphaeroides. J Bacteriol. 1992;174:3855–3866. doi: 10.1128/jb.174.12.3855-3866.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Merrick M, Filser M, Dixon R, Elmerich C, Sibold L, Houmard J. The use of translocatable genetic elements to construct a fine-structure map of the Klebsiella pneumoniae nitrogen fixation (nif) gene cluster. J Gen Microbiol. 1980;117:509–520. doi: 10.1099/00221287-117-2-509. [DOI] [PubMed] [Google Scholar]
- 155.Metzler W J, Farmer B T, Constantine K L, Friedrichs M S, Lavoie T, Mueller L. Refined solution structure of human profilin I. Protein Sci. 1995;4:450–459. doi: 10.1002/pro.5560040312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Min K T, Hilditch C M, Diederich B, Errington J, Yudkin M D. Sigma F, the first compartment-specific transcription factor of B. subtilis, is regulated by an anti-sigma factor that is also a protein kinase. Cell. 1993;74:735–742. doi: 10.1016/0092-8674(93)90520-z. [DOI] [PubMed] [Google Scholar]
- 157.Moffett P, Dayo M, Reece M, McCormick M K, Pelletier J. Characterization of msim, a murine homologue of the Drosophila sim transcriptional factor. Genomics. 1996;35:144–155. doi: 10.1006/geno.1996.0333. [DOI] [PubMed] [Google Scholar]
- 158.Monson E K, Ditta G S, Helinski D R. The oxygen sensor protein, FixL, of Rhizobium meliloti. Role of histidine residues in heme binding, phosphorylation, and signal transduction. J Biol Chem. 1995;270:5243–5250. doi: 10.1074/jbc.270.10.5243. [DOI] [PubMed] [Google Scholar]
- 159.Monson E K, Weinstein M, Ditta G S, Helinski D R. The FixL protein of Rhizobium meliloti can be separated into a heme-binding oxygen-sensing domain and a functional C-terminal kinase domain. Proc Natl Acad Sci USA. 1992;89:4280–4284. doi: 10.1073/pnas.89.10.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Morais Cabral J H, Lee A, Cohen S L, Chait B T, Li M, Mackinnon R. Crystal structure and functional analysis of the HERG potassium channel N terminus: a eukaryotic PAS domain. Cell. 1998;95:649–655. doi: 10.1016/s0092-8674(00)81635-9. [DOI] [PubMed] [Google Scholar]
- 161.Muenke M, Bone L J, Mitchell H F, Hart I, Walton K, Hall-Johnson K, Ippel E F, Dietz-Band J, Kvaloy K, Fan C M. Physical mapping of the holoprosencephaly critical region in 21q22.3, exclusion of SIM2 as a candidate gene for holoprosencephaly, and mapping of SIM2 to a region of chromosome 21 important for Down syndrome. Am J Hum Genet. 1995;57:1074–1079. [PMC free article] [PubMed] [Google Scholar]
- 162.Mulligan M E, Haselkorn R. Nitrogen fixation (nif) genes of the cyanobacterium Anabaena species strain PCC 7120. The nifB-fdxN-nifS-nifU operon. J Biol Chem. 1989;264:19200–19207. [PubMed] [Google Scholar]
- 163.Nambu J R, Lewis J O, Wharton K A J, Crews S T. The Drosophila single-minded gene encodes a helix-loop-helix protein that acts as a master regulator of CNS midline development. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- 164.Ni M, Tepperman J M, Quail P H. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- 165.Ohlsen K L, Grimsley J K, Hoch J A. Deactivation of the sporulation transcription factor Spo0A by the Spo0E protein phosphatase. Proc Natl Acad Sci USA. 1994;91:1756–1760. doi: 10.1073/pnas.91.5.1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ouzounis C, Bork P, Sander C. The modular structure of NifU proteins. Trends Biochem Sci. 1994;19:199–200. doi: 10.1016/0968-0004(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 167.Parkinson J S. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 168.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 169.Patschkowski T, Schluter A, Priefer U B. Rhizobium leguminosarum bv. viciae contains a second fnr/fixK-like gene and an unusual fixL homologue. Mol Microbiol. 1996;21:267–280. doi: 10.1046/j.1365-2958.1996.6321348.x. [DOI] [PubMed] [Google Scholar]
- 170.Paulson K E, Darnell J E J, Rushmore T, Pickett C B. Analysis of the upstream elements of the xenobiotic compound-inducible and positionally regulated glutathione S-transferase Ya gene. Mol Cell Biol. 1990;10:1841–1852. doi: 10.1128/mcb.10.5.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pawlowski K, Klosse U, de Bruijn F J. Characterization of a novel Azorhizobium caulinodans ORS571 two-component regulatory system, NtrY/NtrX, involved in nitrogen fixation and metabolism. Mol Gen Genet. 1991;231:124–138. doi: 10.1007/BF00293830. [DOI] [PubMed] [Google Scholar]
- 172.Pellequer J L, Wager-Smith K A, Kay S A, Getzoff E D. Photoactive yellow protein: a structural prototype for the three-dimensional fold of the PAS domain superfamily. Proc Natl Acad Sci USA. 1998;95:5884–5890. doi: 10.1073/pnas.95.11.5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Penfold R J, Pemberton J M. A gene from the photosynthetic gene cluster of Rhodobacter sphaeroides induces in trans suppression of bacteriochlorophyll and carotenoid levels in R. sphaeroides and R. capsulata. Curr Microbiol. 1991;23:259–263. [Google Scholar]
- 174.Penfold R J, Pemberton J M. Sequencing, chromosomal inactivation, and functional expression in Escherichia coli of ppsR, a gene which represses carotenoid and bacteriochlorophyll synthesis in Rhodobacter sphaeroides. J Bacteriol. 1994;176:2869–2876. doi: 10.1128/jb.176.10.2869-2876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Perego M, Cole S P, Burbulys D, Trach K, Hoch J A. Characterization of the gene for a protein kinase which phosphorylates the sporulation-regulatory proteins Spo0A and Spo0F of Bacillus subtilis. J Bacteriol. 1989;171:6187–6196. doi: 10.1128/jb.171.11.6187-6196.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Perego M, Hanstein C, Welsh K M, Djavakhishvili T, Glaser P, Hoch J A. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis. Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 177.Plautz J D, Kaneko M, Hall J C, Kay S A. Independent photoreceptive circadian clocks throughout Drosophila. Science. 1997;278:1632–1635. doi: 10.1126/science.278.5343.1632. [DOI] [PubMed] [Google Scholar]
- 178.Pongratz I, Antonsson C, Whitelaw M L, Poellinger L. Role of the PAS domain in regulation of dimerization and DNA binding specificity of the dioxin receptor. Mol Cell Biol. 1998;18:4079–4088. doi: 10.1128/mcb.18.7.4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Pongratz I, Mason G G, Poellinger L. Dual roles of the 90-kDa heat shock protein hsp90 in modulating functional activities of the dioxin receptor. Evidence that the dioxin receptor functionally belongs to a subclass of nuclear receptors which require hsp90 both for ligand binding activity and repression of intrinsic DNA binding activity. J Biol Chem. 1992;267:13728–13734. [PubMed] [Google Scholar]
- 180.Ponnampalam S N, Bauer C E. DNA binding characteristics of CrtJ. A redox-responding repressor of bacteriochlorophyll, carotenoid, and light harvesting-II gene expression in Rhodobacter capsulatus. J Biol Chem. 1997;272:18391–18396. doi: 10.1074/jbc.272.29.18391. [DOI] [PubMed] [Google Scholar]
- 181.Ponnampalam S N, Buggy J J, Bauer C E. Characterization of an aerobic repressor that coordinately regulates bacteriochlorophyll, carotenoid, and light harvesting-II expression in Rhodobacter capsulatus. J Bacteriol. 1995;177:2990–2997. doi: 10.1128/jb.177.11.2990-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ponting C P, Aravind L. PAS: a multifunctional domain family comes to light. Curr Biol. 1997;7:R674–R677. doi: 10.1016/s0960-9822(06)00352-6. [DOI] [PubMed] [Google Scholar]
- 183.Pugh C W, O’Rourke J F, Nagao M, Gleadle J M, Ratcliffe P J. Activation of hypoxia-inducible factor-1: definition of regulatory domains within the alpha subunit. J Biol Chem. 1997;272:11205–11214. doi: 10.1074/jbc.272.17.11205. [DOI] [PubMed] [Google Scholar]
- 184.Quail P H. The phytochromes: a biochemical mechanism of signaling in sight? Bioessays. 1997;19:571–579. doi: 10.1002/bies.950190708. [DOI] [PubMed] [Google Scholar]
- 185.Quail P H, Boylan M T, Parks B M, Short T W, Xu Y, Wagner D. Phytochromes: photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- 186.Rebbapragada A, Johnson M S, Harding G P, Zuccarelli A J, Fletcher H M, Zhulin I B, Taylor B L. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci USA. 1997;94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Repik, A. V., A. Rebbapragada, and B. L. Taylor. Unpublished data.
- 188.Reyes F, Gavira M, Castillo F, Moreno V. Periplasmic nitrate-reducing system of the phototrophic bacterium Rhodobacter sphaeroides DSM 158: transcriptional and mutational analysis of the napKEFDABC gene cluster. Biochem J. 1998;331:897–904. doi: 10.1042/bj3310897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Rodgers K R, Lukat-Rodgers G S, Barron J A. Structural basis for ligand discrimination and response initiation in the heme-based oxygen sensor FixL. Biochemistry. 1996;35:9539–9548. doi: 10.1021/bi9530853. [DOI] [PubMed] [Google Scholar]
- 190.Ronson C W, Astwood P M, Nixon B T, Ausubel F M. Deduced products of C4-dicarboxylate transport regulatory genes of Rhizobium leguminosarum are homologous to nitrogen regulatory gene products. Nucleic Acids Res. 1987;15:7921–7934. doi: 10.1093/nar/15.19.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ronson C W, Nixon B T, Ausubel F M. Conserved domains in bacterial regulatory proteins that respond to environmental stimuli. Cell. 1987;49:579–581. doi: 10.1016/0092-8674(87)90530-7. [DOI] [PubMed] [Google Scholar]
- 192.Rost B, Sander C, Schneider R. PHD—an automatic mail server for protein secondary structure prediction. Comput Appl Biosci. 1994;10:53–60. doi: 10.1093/bioinformatics/10.1.53. [DOI] [PubMed] [Google Scholar]
- 193.Rowlands J C, Gustafsson J A. Aryl hydrocarbon receptor-mediated signal transduction. Crit Rev Toxicol. 1997;27:109–134. doi: 10.3109/10408449709021615. [DOI] [PubMed] [Google Scholar]
- 194.Rudolph J, Nordmann B, Storch K F, Gruenberg H, Rodewald K, Oesterhelt D. A family of halobacterial transducer proteins. FEMS Microbiol Lett. 1996;139:161–168. doi: 10.1111/j.1574-6968.1996.tb08197.x. [DOI] [PubMed] [Google Scholar]
- 195.Rutila J E, Suri V, Le M, So W V, Rosbash M, Hall J C. CYCLE is a second bHLH-PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell. 1998;93:805–814. doi: 10.1016/s0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- 196.Sanguinetti M C, Jiang C, Curran M E, Keating M T. A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell. 1995;81:299–307. doi: 10.1016/0092-8674(95)90340-2. [DOI] [PubMed] [Google Scholar]
- 197.Schibler U. New cogwheels in the clockworks. Nature. 1998;393:620–621. doi: 10.1038/31337. [DOI] [PubMed] [Google Scholar]
- 198.Schluter K, Jockusch B M, Rothkegel M. Profilins as regulators of actin dynamics. Biochim Biophys Acta. 1997;1359:97–109. doi: 10.1016/s0167-4889(97)00100-6. [DOI] [PubMed] [Google Scholar]
- 199.Schmitz R A. NifL of Klebsiella pneumoniae carries an N-terminally bound FAD cofactor, which is not directly required for the inhibitory function of NifL. FEMS Microbiol Lett. 1997;157:313–318. doi: 10.1111/j.1574-6968.1997.tb12791.x. [DOI] [PubMed] [Google Scholar]
- 200.Schneider-Poetsch H A, Braun M, Marx S, Schaumburg A. Phytochromes and bacterial sensor proteins are related by structural and functional homologies. Hypothesis on phytochrome-mediated signal-transduction. FEBS Lett. 1991;281:245–249. doi: 10.1016/0014-5793(91)80403-p. [DOI] [PubMed] [Google Scholar]
- 201.Scholten M, Tommassen J. Topology of the PhoR protein of Escherichia coli and functional analysis of internal deletion mutants. Mol Microbiol. 1993;8:269–275. doi: 10.1111/j.1365-2958.1993.tb01571.x. [DOI] [PubMed] [Google Scholar]
- 202.Schuster S S, Noegel A A, Oehme F, Gerisch G, Simon M I. The hybrid histidine kinase DokA is part of the osmotic response system of Dictyostelium. EMBO J. 1996;15:3880–3889. [PMC free article] [PubMed] [Google Scholar]
- 203.Siddavattam D, Steibl H D, Kreutzer R, Klingmuller W. Regulation of nif gene expression in Enterobacter agglomerans: nucleotide sequence of the nifLA operon and influence of temperature and ammonium on its transcription. Mol Gen Genet. 1995;249:629–636. doi: 10.1007/BF00418032. [DOI] [PubMed] [Google Scholar]
- 204.Sidote D, Majercak J, Parikh V, Edery I. Differential effects of light and heat on the Drosophila circadian clock proteins PER and TIM. Mol Cell Biol. 1998;18:2004–2013. doi: 10.1128/mcb.18.4.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Smith H. Physiological and ecological function within the phytochrome family. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:289–315. [Google Scholar]
- 207.Soderback E, Reyes-Ramirez F, Eydmann T, Austin S, Hill S, Dixon R. The redox- and fixed nitrogen-responsive regulatory protein NIFL from Azotobacter vinelandii comprises discrete flavin and nucleotide-binding domains. Mol Microbiol. 1998;28:179–192. doi: 10.1046/j.1365-2958.1998.00788.x. [DOI] [PubMed] [Google Scholar]
- 208.Soderling S H, Bayuga S J, Beavo J A. Cloning and characterization of a cAMP-specific cyclic nucleotide phosphodiesterase. Proc Natl Acad Sci USA. 1998;95:8991–8996. doi: 10.1073/pnas.95.15.8991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Somers D E, Devlin P F, Kay S A. Phytochromes and cryptochromes in the entrainment of the arabidopsis circadian clock. Science. 1998;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 210.Spaink H P. The molecular basis of the host specificity of the Rhizobium bacteria. Antonie Leeuwenhoek. 1994;65:81–98. doi: 10.1007/BF00871750. [DOI] [PubMed] [Google Scholar]
- 211.Sprenger W W, Hoff W D, Armitage J P, Hellingwerf K J. The eubacterium Ectothiorhodospira halophila is negatively phototactic, with a wavelength dependence that fits the absorption spectrum of the photoactive yellow protein. J Bacteriol. 1993;175:3096–3104. doi: 10.1128/jb.175.10.3096-3104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Srinivas V, Zhu X, Salceda S, Nakamura R, Caro J. Hypoxia-inducible factor 1α (HIF-1α) is a non-heme iron protein: implications for oxygen sensing. J Biol Chem. 1998;273:18019–18022. doi: 10.1074/jbc.273.29.18019. [DOI] [PubMed] [Google Scholar]
- 213.Stacey G. Bradyrhizobium japonicum nodulation genetics. FEMS Microbiol Lett. 1995;127:1–9. doi: 10.1111/j.1574-6968.1995.tb07441.x. [DOI] [PubMed] [Google Scholar]
- 214.Steffen P, Goyard S, Ullmann A. Phosphorylated BvgA is sufficient for transcriptional activation of virulence-regulated genes in Bordetella pertussis. EMBO J. 1996;15:102–109. [PMC free article] [PubMed] [Google Scholar]
- 215.Stephens C. Bacterial sporulation: a question of commitment? Curr Biol. 1998;8:R45–R48. doi: 10.1016/s0960-9822(98)70031-4. [DOI] [PubMed] [Google Scholar]
- 216.Stibitz S, Yang M S. Subcellular localization and immunological detection of proteins encoded by the vir locus of Bordetella pertussis. J Bacteriol. 1991;173:4288–4296. doi: 10.1128/jb.173.14.4288-4296.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Stockhaus J, Nagatani A, Halfter U, Kay S, Furuya M, Chua N H. Serine-to-alanine substitutions at the amino-terminal region of phytochrome A result in an increase in biological activity. Genes Dev. 1992;6:2364–2372. doi: 10.1101/gad.6.12a.2364. [DOI] [PubMed] [Google Scholar]
- 218.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 219.Sun Z S, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee C C. RIGUI, a putative mammalan ortholog of the Drosophila period gene. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 220.Suri V, Qian Z, Hall J C, Rosbash M. Evidence that the TIM light response is relevant to light-induced phase shifts in Drosophila melanogaster. Neuron. 1998;21:225–234. doi: 10.1016/s0896-6273(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 221.Taylor B L. Role of proton motive force in sensory transduction in bacteria. Annu Rev Microbiol. 1983;37:551–573. doi: 10.1146/annurev.mi.37.100183.003003. [DOI] [PubMed] [Google Scholar]
- 222.Taylor, B. L., M. S. Johnson, I. B. Zhulin, R. Fu, and G. Voordouw. Unpublished data.
- 223.Taylor B L, Miller J B, Warrick H M, Koshland D E J. Electron acceptor taxis and blue light effect on bacterial chemotaxis. J Bacteriol. 1979;140:567–573. doi: 10.1128/jb.140.2.567-573.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Taylor B L, Zhulin I B. In search of higher energy: metabolism-dependent behaviour in bacteria. Mol Microbiol. 1998;28:683–690. doi: 10.1046/j.1365-2958.1998.00835.x. [DOI] [PubMed] [Google Scholar]
- 225.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature. 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 226.Thresher R J, Vitaterna M H, Miyamoto Y, Kazantsev A, Hsu D S, Petit C, Selby C P, Dawut L, Smithies O, Takahashi J S, Sancar A. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282:1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 227.Tommassen J, de Geus P, Lugtenberg B, Hackett J, Reeves P. Regulation of the pho regulon of Escherichia coli K-12. Cloning of the regulatory genes phoB and phoR and identification of their gene products. J Mol Biol. 1982;157:265–274. doi: 10.1016/0022-2836(82)90233-9. [DOI] [PubMed] [Google Scholar]
- 228.Torriani A. From cell membrane to nucleotides: the phosphate regulon in Escherichia coli. Bioessays. 1990;12:371–376. doi: 10.1002/bies.950120804. [DOI] [PubMed] [Google Scholar]
- 229.Trach K A, Hoch J A. Multisensory activation of the phosphorelay initiating sporulation in Bacillus subtilis: identification and sequence of the protein kinase of the alternate pathway. Mol Microbiol. 1993;8:69–79. doi: 10.1111/j.1365-2958.1993.tb01204.x. [DOI] [PubMed] [Google Scholar]
- 230.Tseng C P, Albrecht J, Gunsalus R P. Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J Bacteriol. 1996;178:1094–1098. doi: 10.1128/jb.178.4.1094-1098.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Tsuzuki M, Ishige K, Mizuno T. Phosphotransfer circuitry of the putative multi-signal transducer, ArcB, of Escherichia coli: in vitro studies with mutants. Mol Microbiol. 1995;18:953–962. doi: 10.1111/j.1365-2958.1995.18050953.x. [DOI] [PubMed] [Google Scholar]
- 232.Uhl M A, Miller J F. Central role of the BvgS receiver as a phosphorylated intermediate in a complex two-component phosphorelay. J Biol Chem. 1996;271:33176–33180. doi: 10.1074/jbc.271.52.33176. [DOI] [PubMed] [Google Scholar]
- 233.Uhl M A, Miller J F. Integration of multiple domains in a two-component sensor protein: the Bordetella pertussis BvgAS phosphorelay. EMBO J. 1996;15:1028–1036. [PMC free article] [PubMed] [Google Scholar]
- 234.Urena J, Fernandez-Chacon R, Benot A R, Alvarez de Toledo G A, Lopez-Barneo J. Hypoxia induces voltage-dependent Ca2+ entry and quantal dopamine secretion in carotid body glomus cells. Proc Natl Acad Sci USA. 1994;91:10208–10211. doi: 10.1073/pnas.91.21.10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.van Rhijn P, Vanderleyden J. The Rhizobium-plant symbiosis. Microbiol Rev. 1995;59:124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Velasco A, Alonso S, Garcia J L, Perera J, Diaz E. Genetic and functional analysis of the styrene catabolic cluster of Pseudomonas sp. strain Y2. J Bacteriol. 1998;180:1063–1071. doi: 10.1128/jb.180.5.1063-1071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Vincent G M. The molecular genetics of the long QT syndrome: genes causing fainting and sudden death. Annu Rev Med. 1998;49:263–274. doi: 10.1146/annurev.med.49.1.263. [DOI] [PubMed] [Google Scholar]
- 238.Waksman G, Kominos D, Robertson S C, Pant N, Baltimore D, Birge R B, Cowburn D, Hanafusa H, Mayer B J, Overduin M. Crystal structure of the phosphotyrosine recognition domain SH2 of v-src complexed with tyrosine-phosphorylated peptides. Nature. 1992;358:646–653. doi: 10.1038/358646a0. [DOI] [PubMed] [Google Scholar]
- 239.Wang G L, Semenza G L. Molecular basis of hypoxia-induced erythropoietin expression. Curr Opin Hematol. 1996;3:156–162. doi: 10.1097/00062752-199603020-00009. [DOI] [PubMed] [Google Scholar]
- 240.Wang L, Grau R, Perego M, Hoch J A. A novel histidine kinase inhibitor regulating development in Bacillus subtilis. Genes Dev. 1997;11:2569–2579. doi: 10.1101/gad.11.19.2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Wang Q, Chen Q, Towbin J A. Genetics, molecular mechanisms and management of long QT syndrome. Ann Med. 1998;30:58–65. doi: 10.3109/07853899808999385. [DOI] [PubMed] [Google Scholar]
- 242.Wanner B L, Chang B D. The phoBR operon in Escherichia coli K-12. J Bacteriol. 1987;169:5569–5574. doi: 10.1128/jb.169.12.5569-5574.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Warmke J W, Ganetzky B. A family of potassium channel genes related to eag in Drosophila and mammals. Proc Natl Acad Sci USA. 1994;91:3438–3442. doi: 10.1073/pnas.91.8.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Watson R J. Analysis of the C4-dicarboxylate transport genes of Rhizobium meliloti: nucleotide sequence and deduced products of dctA, dctB and dctD. Mol Plant-Microbe Interact. 1990;3:174–181. doi: 10.1094/mpmi-3-174. [DOI] [PubMed] [Google Scholar]
- 245.Weir E K, Archer S L. The mechanism of acute hypoxic pulmonary vasoconstriction: the tale of two channels. FASEB J. 1995;9:183–189. doi: 10.1096/fasebj.9.2.7781921. [DOI] [PubMed] [Google Scholar]
- 246.Welch M, Oosawa K, Aizawa S, Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci USA. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Whitelaw M, Pongratz I, Wilhelmsson A, Gustafsson J A, Poellinger L. Ligand-dependent recruitment of the Arnt coregulator determines DNA recognition by the dioxin receptor. Mol Cell Biol. 1993;13:2504–2514. doi: 10.1128/mcb.13.4.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Whitelaw M L, McGuire J, Picard D, Gustafsson J A, Poellinger L. Heat shock protein hsp90 regulates dioxin receptor function in vivo. Proc Natl Acad Sci USA. 1995;92:4437–4441. doi: 10.1073/pnas.92.10.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Wilhelmsson A, Cuthill S, Denis M, Wikstrom A C, Gustafsson J A, Poellinger L. The specific DNA binding activity of the dioxin receptor is modulated by the 90 kd heat shock protein. EMBO J. 1990;9:69–76. doi: 10.1002/j.1460-2075.1990.tb08081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Wong Y S, Cheng H C, Walsh D A, Lagarias J C. Phosphorylation of Avena phytochrome in vitro as a probe of light-induced conformational changes. J Biol Chem. 1986;261:12089–12097. [PubMed] [Google Scholar]
- 251.Woodley P, Drummond M. Redundancy of the conserved His residue in Azotobacter vinelandii NifL, a histidine autokinase homologue which regulates transcription of nitrogen fixation genes. Mol Microbiol. 1994;13:619–626. doi: 10.1111/j.1365-2958.1994.tb00456.x. [DOI] [PubMed] [Google Scholar]
- 252.Yamada M, Makino K, Shinagawa H, Nakata A. Regulation of the phosphate regulon of Escherichia coli: properties of phoR deletion mutants and subcellular localization of PhoR protein. Mol Gen Genet. 1990;220:366–372. doi: 10.1007/BF00391740. [DOI] [PubMed] [Google Scholar]
- 253.Yamaki A, Noda S, Kudoh J, Shindoh N, Maeda H, Minoshima S, Kawasaki K, Shimizu Y, Shimizu N. The mammalian single-minded (SIM) gene: mouse cDNA structure and diencephalic expression indicate a candidate gene for Down syndrome. Genomics. 1996;35:136–143. doi: 10.1006/geno.1996.0332. [DOI] [PubMed] [Google Scholar]
- 254.Yan C, Zhao A Z, Bentley J K, Loughney K, Ferguson K, Beavo J A. Molecular cloning and characterization of a calmodulin-dependent phosphodiesterase enriched in olfactory sensory neurons. Proc Natl Acad Sci USA. 1995;92:9677–9681. doi: 10.1073/pnas.92.21.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Yang Z, Emerson M, Su H S, Sehgal A. Response of the timeless protein to light correlates with behavioral entrainment and suggests a nonvisual pathway for circadian photoreception. Neuron. 1998;21:215–223. doi: 10.1016/s0896-6273(00)80528-0. [DOI] [PubMed] [Google Scholar]
- 256.Yeh K C, Lagarias J C. Eukaryotic phytochromes: light-regulated serine/threonine protein kinases with histidine kinase ancestry. Proc Natl Acad Sci USA. 1998;95:13976–13981. doi: 10.1073/pnas.95.23.13976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Yeh K C, Wu S H, Murphy J T, Lagarias J C. A cyanobacterial phytochrome two-component light sensory system. Science. 1997;277:1505–1508. doi: 10.1126/science.277.5331.1505. [DOI] [PubMed] [Google Scholar]
- 258.Youngson C, Nurse C, Yeger H, Cutz E. Oxygen sensing in airway chemoreceptors. Nature. 1993;365:153–155. doi: 10.1038/365153a0. [DOI] [PubMed] [Google Scholar]
- 259.Yuan X J, Tod M L, Rubin L J, Blaustein M P. Deoxyglucose and reduced glutathione mimic effects of hypoxia on K+ and Ca2+ conductances in pulmonary artery cells. Am J Physiol. 1994;267:L52–L63. doi: 10.1152/ajplung.1994.267.1.L52. [DOI] [PubMed] [Google Scholar]
- 260.Zeilstra-Ryalls J, Gomelsky M, Eraso J M, Yeliseev A, O’Gara J, Kaplan S. Control of photosystem formation in Rhodobacter sphaeroides. J Bacteriol. 1998;180:2801–2809. doi: 10.1128/jb.180.11.2801-2809.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Zelzer E, Wappner P, Shilo B Z. The PAS domain confers target gene specificity of Drosophila bHLH/PAS proteins. Genes Dev. 1997;11:2079–2089. doi: 10.1101/gad.11.16.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Zhang W, Brooun A, McCandless J, Banda P, Alam M. Signal transduction in the archaeon Halobacterium salinarium is processed through three subfamilies of 13 soluble and membrane-bound transducer proteins. Proc Natl Acad Sci USA. 1996;93:4649–4654. doi: 10.1073/pnas.93.10.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Zhulin, I. B. Unpublished data.
- 264.Zhulin I B, Rowsell E H, Johnson M S, Taylor B L. Glycerol elicits energy taxis of Escherichia coli and Salmonella typhimurium. J Bacteriol. 1997;179:3196–3201. doi: 10.1128/jb.179.10.3196-3201.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Zhulin I B, Taylor B L. Correlation of PAS domains with electron transport-associated proteins in completely sequenced microbial genomes. Mol Microbiol. 1998;29:1522–1523. . (Erratum, 31:741, 1999.) [PubMed] [Google Scholar]
- 266.Zhulin I B, Taylor B L, Dixon R. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem Sci. 1997;22:331–333. doi: 10.1016/s0968-0004(97)01110-9. [DOI] [PubMed] [Google Scholar]
- 267.Zientz E, Bongaerts J, Unden G. Fumarate regulation of gene expression in Escherichia coli by the DcuSR (dcuSR genes) two-component regulatory system. J Bacteriol. 1998;180:5421–5425. doi: 10.1128/jb.180.20.5421-5425.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Zinda M J, Singleton C K. The hybride histidine kinase dhkB regulates spore germination in Dictyostelium discoideum. Dev Biol. 1998;196:171–183. doi: 10.1006/dbio.1998.8854. [DOI] [PubMed] [Google Scholar]
- 269.Zu T, Manetti R, Rappuoli R, Scarlato V. Differential binding of BvgA to two classes of virulence genes of Bordetella pertussis directs promoter selectivity by RNA polymerase. Mol Microbiol. 1996;21:557–565. doi: 10.1111/j.1365-2958.1996.tb02564.x. [DOI] [PubMed] [Google Scholar]