Abstract
Accurate and precise identification of adeno-associated virus (AAV) vectors play an important role in dose-dependent gene therapy. Although solid-state nanopore techniques can potentially be used to characterize AAV vectors by capturing ionic current, the existing data analysis techniques fall short of identifying them from their ionic current profiles. Recently introduced machine learning methods such as deep convolutional neural network (CNN), developed for image identification tasks, can be applied for such classification. However, with smaller data set for the problem in hand, it is not possible to train a deep neural network from scratch for accurate classification of AAV vectors. To circumvent this, we applied a pre-trained deep CNN (GoogleNet) model to capture the basic features from ionic current signals and subsequently used fine-tuning-based transfer learning to classify AAV vectors. The proposed method is very generic as it requires minimal preprocessing and does not require any handcrafted features. Our results indicate that fine-tuning-based transfer learning can achieve an average classification accuracy between 90 and 99% in three realizations with a very small standard deviation. Results also indicate that the classification accuracy depends on the applied electric field (across nanopore) and the time frame used for data segmentation. We also found that the fine-tuning of the deep network outperforms feature extraction-based classification for the resistive pulse dataset. To expand the usefulness of the fine-tuning-based transfer learning, we have tested two other pre-trained deep networks (ResNet50 and InceptionV3) for the classification of AAVs. Overall, the fine-tuning-based transfer learning from pre-trained deep networks is very effective for classification, though deep networks such as ResNet50 and InceptionV3 take significantly longer training time than GoogleNet.
Keywords: adeno-associated virus, fine-tuning, feature extraction, deep learning, solid-state nanopore, signal processing, transfer learning
1. Introduction
Adeno associated virus (AAV) is a non-enveloped virus that can be engineered to deliver deoxyribonucleic acid (DNA) to target cells without any viral effects on the target cell [1]. As a result, AAV modulated gene therapy has been used to treat several diseases including hemophilia, cystic fibrosis [2], and cancer [3]. The ability to generate recombinant AAV particles containing DNA sequences of interest for various therapeutic applications has thus far proven to be one of the safest strategies for gene therapies [4]. However, after production, one considerable challenge is the difficulty of characterizing the AAV vectors based on their transgene packaging. Generally, enzyme-linked immunosorbent assay (ELISA) and polymerase chain reaction (PCR) are used for the characterization of capsid and genomic titers, respectively [5]. However, a blind study [6] revealed that ELISA or PCR results offer high variability, which is not acceptable for gene therapy since overdosing or under-dosing could trigger unexpected immune responses.
Recently, solid state nanopore based techniques have been employed to discriminate various AAVs based on their DNA loading [7]. In this type of sensor, nanopores are usually fabricated on thin (~50 nm) synthetic membranes that separate two chambers containing electrolytes. When an electric field is applied across the membrane via trans and cis electrodes, particles start to translocate through the pore from one chamber to another through the electrolyte solution. The electric field induced ionic current carries the information of analytes/particles that have been transported through the nanopore. For instance, when a virus particle passes through the pore, there is a leap or fall in base ionic current depending on the direction of the applied electric field and the properties of the particles and electrolyte. The resulting ionic current signal is known as the resistive pulse signal. In our previous work [8], we developed a silicon nitride (SixNy) based solid-state nanopore device to characterize three AAV vector types – empty (AAVEmpty), AAV with single-stranded DNA (AAVssDNA), and AAV with double-stranded DNA (AAVdsDNA). Furthermore, we used feature extraction based transfer learning where a deep network (ResNet50) was used to extract the feature map from the resistive-pulse signals; then, a support vector machine [9] was employed for the classification of AAVs and flagging the presence of empty capsids [8]. However, the feature extraction based transfer learning method was not very effective when it was applied to raw resistive-pulse signals.
Deep neural networks have recently been used in image processing/classifications, regression, computer vision, data clustering, scene labeling, action recognition, etc. because of their high accuracy. For example, Krizhevsky et al. developed a deep convolutional neural network (CNN) architecture, known as AlexNet [10], which demonstrated significant improvement in image classification tasks. Following the success of AlexNet, several other networks such as VGGNet [11], ZFNet [12], GoogleNet [13], and ResNet [14] were developed to improve image classification performance. Recently, robust temporal feature network (RTFN) [15–17] emerged as a very popular model for feature extraction in time series classification. The most striking ability of CNNs is the automatic discovery of essential features from the images (raw pixel intensity data) [18]. In contrast to the identification of features by hand, this automatic feature learning method has been shown to be easier, faster, and superior in efficacy [19–21]. Since, all of these high-performance networks are trained on the ImageNet dataset, direct use of these deep networks is only possible if the test/target data are also belong to one or multiple classes of the ImageNet dataset [22]. If a dataset (like AAV dataset) does not belong to ImageNet dataset, rebuilding a network from scratch could be a solution. However, rebuilding a new network requires a lot of labeled data. In fact, it is very expensive and sometimes impossible to collect enough training data by performing experiments to build a high-performance deep neural network on the domain of interest. Moreover, if sufficient labeled data are available, rebuilding a deep neural network from scratch and training the newly built model with the labeled data requires an enormous amount of computational time, effort, and power.
To address this problem, in recent years, transfer learning has emerged as a new learning framework [22–25]. Transfer learning is the idea of utilizing knowledge acquired from one task to solve correlated ones [26–28]. Although the transferability of features depends on the similarity between the base task and the target task [29], the initialization of a network with distant task features still performs better than initialization with random features [30]. Transfer learning is particularly useful because the initial layers of a deep network usually capture low-level features, like edges and blobs, which are commonly shared between different types of images.
Several transfer learning algorithms including joint training, feature extraction, and fine-tuning [31] have been developed so far. Although joint training works well on both old and new tasks, it is increasingly cumbersome in training as more tasks have to be learned and requires a great deal of storage [31]. A faster transfer learning method is based on feature extraction, in which the outputs (activations) of one or more layers are used as features. These features are then used to train another machine learning model, such as a support vector machine (SVM) [8, 32]. However, feature extraction is only useful if the dataset is comparatively small, and the new data is very similar to the original data. Moreover, loss of data interpretability can happen when feature extraction based transfer learning is used [33].
Transfer learning with fine-tuning [34] can eliminate several problems associated with feature extraction based transfer learning [31]. Fine-tuning is a process in which the task specific layers of an existing highly trained deep neural network are altered according to the requirements of the current task, and available data are used to train these layers. Since the pre-trained network has already learned a rich set of image features, fine-tuning a network is often faster and easier than constructing and training a new network. Fine-tuning reduces the training error and improves the classification performance of deep neural networks significantly in comparison to feature extraction-based networks [35, 36]. As a result, the transfer learning using fine-tuning has been widely used for various promising tasks such as speech recognition [37], plant recognition [38], biomedical image analysis [39], cellular morphological change analysis [40], text classification [41], brain tumor classification [42, 43] etc.
In this work, we have proposed a pre-trained deep CNN model to capture the essential features of ionic current signals and applied a fine-tuning-based transfer learning to discriminate different variants of AAV vectors based on their contents. In addition, we have compared the performances of fine-tuning-based transfer learning using three different pretrained networks: GoogleNet, ResNet50, and InceptionV3. Moreover, a support vector machine (SVM) has been used as feature extraction based approach to classify the different AAV vectors, where the input features for SVM were extracted using pretrained GoogleNet architecture. The performance of this feature extraction-based approach is compared with the corresponding performance of the fine-tuning-based transfer learning.
2. Method
2.1. GoogleNet architecture and its modification for transfer learning
For classification of AAVs using fine-tuning-based transfer learning, we first selected pretrained GoogleNet, a 22 layer deep (27 layers including pooling layers) architecture, as a deep network [13]. Figure 1a shows the zoomed-out structure of the full GoogleNet architecture, depicting the size of the feature maps in each layer. In addition to convolution (CNV) and maximum pooling (MP) operations, it introduces a module called “Inception”, which relies on the idea of running multiple operations (pooling, convolution) with several kernel sizes (1×1, 3×3, 5×5…) in parallel, to overcome the trade-off between the choices of pooling or convolutional operations. In total, there are 9 inception modules, and each inception layer consists of six convolution layers and one pooling layer in four parallel routes (Fig. 1b). These parallel routes are connected to a depth concatenation (DC) layer, which takes inputs of the same height and width and concatenates them along the depth dimension. Each inception module facilitates feature detection at different scales with different size filters. Moreover, it can minimize the computational cost of training a deep network significantly through dimensional reduction. The very last inception module (IN5b) is connected to a global average pooling (AP) layer. After that, GoogleNet utilizes a dropout (DO) layer which randomly selects 40% of the neurons and sets their weights to zero. Dropout offers the flexibility of a single model to imitate a large number of different network architectures by randomly dropping out nodes during training. Thus, dropout provides a computationally cheap and remarkably effective regularization method to reduce overfitting in deep neural networks [44]. Finally, classification is done through fully connected (FC) and softmax (SM) layers of 1000 nodes.
Fig. 1.
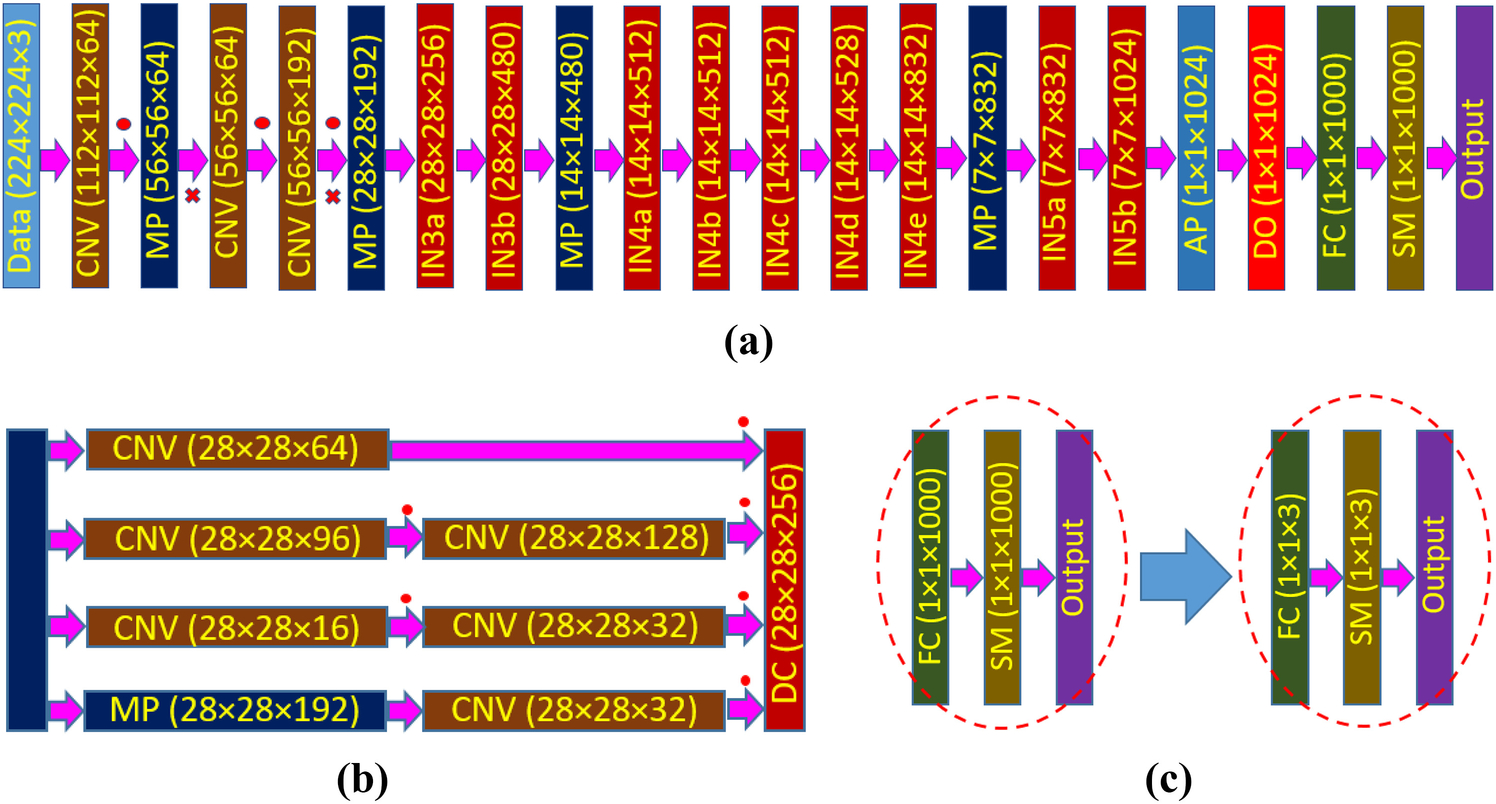
(a) Schematic of GoogleNet architecture, (b) detail of an inception (layer IN3a) block, (c) modification of last three layers of GoogleNet architecture to adopt the number of target classes for AAV classification problem. Here, CNV: convolution layer, MP: max pooling layer, IN: inception block, AP: average pooling layer, DO: dropout layer, FC: fully connected layer, SM: softmax layer, DC: depth concatenation layer. The red dot (∙) represents a ReLU operation and the red-cross (×) indicates a cross channel normalization operation.
The GoogleNet has already been trained on ImageNet’s data set (http://www.image-net.org/) that contains 1000 object categories including keyboard, mouse, pencil, and many animals. Thus, to use this network for the current problem of AAV classification, we need to modify the network according to our dataset. Therefore, in this work, we have replaced the last three layers: fully connected layer, softmax layer, and classification layer (output) of GoogleNet with three similar layers containing the number of outputs equal to the number of classes available in our current dataset (Fig. 1c). Since we are not using any labeled data of the source domain (ImageNet dataset) and we have labeled data in the target domain (AAV dataset), the method presented in this paper can be identified as inductive self-taught transfer learning [22]. Moreover, during the training (discussed later), we are freezing the set of shared parameters (all weights and biases from the input layer to dropout layer), while during fine-tuning only the task-specific parameters (weights and biases of fully connected layers) will be trained [31].
Since it is an application paper, we skip the details of the mathematical analysis of different layers. However, for the completeness of the paper, we have added cost function and parameter update methods. If ck denotes the known/true class label for an image, k, the cost function (CF) for a mini-batch m containing Nm examples can be formulated as [42, 45]
| (1) |
where is the predicted class label, xk is the input feature map for an image, k, respectively and γ2 is the variance which is assumed to be constant. The parameter vector (θ) includes weight matrix, W and bias vector, b. The cost function CFm is minimized by using stochastic gradient descent with momentum [45] over the mini-batches of size Nm and the training cost is approximated by the mini-batch cost. If θt is the parameters at iteration, t, one can update the weights and biases in the next iteration as follows [42]
| (2) |
| (3) |
where μ is the momentum that describes the influence of previously updated weights in the current iteration, η is the learning rate, and σt is the scheduling rate at iteration t.
2.2. Dataset
To test our modified deep network, we used a publicly available resistive pulse dataset from GitHub (https://github.com/mstfwsulab/AAV-classification), where electrical signals obtained from solid nanopore experiments are stored for three different types of AAVs: without any DNA (AAVEmpty), with single-stranded (AAVssDNA), and with double-stranded (AAVdsDNA) DNA [8]. Since pre-trained deep neural networks used for this work requires color (RGB) images as input, the time series resistive pulse data (current values across the nanopore with time) are plotted (2D plot) in MATLAB and saved as ‘jpg’ files. We have treated those 2D plots as RGB images and subsequently used them in the networks. However, for other types of datasets such as grayscale or CMYK images, the first convolutional layer can be altered by another one with appropriate dimensions that reflects the type of images in the dataset. To test the accuracy of the modified deep networks, images were created considering different time segments (1 sec, 2 sec, or 4 sec). Resistive pulse data from five different electric fields corresponding to the potential difference across the nanopore: −75 mV, −100 mV, −125 mV, −150 mV and −175 mV were used to demonstrate the usefulness of the approach. A representative segment (10 sec) of resistive pulse data is shown in Fig. 2 for different potential differences and AAV types.
Fig. 2.
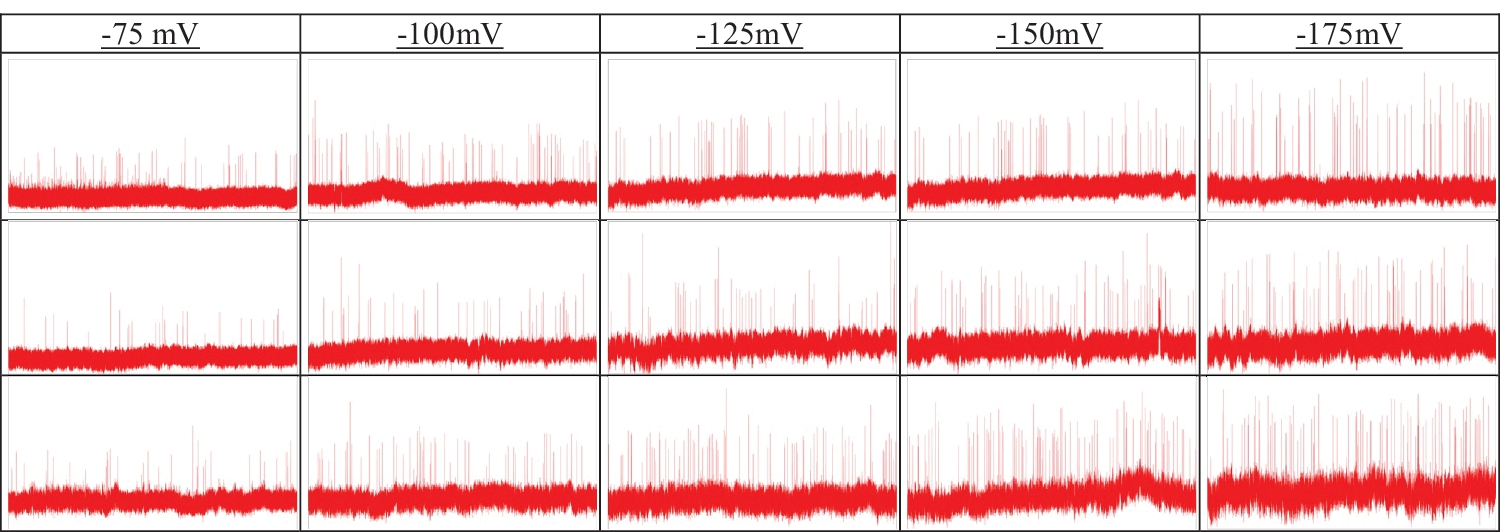
Representative current traces corresponding to the translocation of AAVs: AAVEmpty (top row), AAVssDNA (middle row), and AAVdsDNA (bottom row) for the different applied electric fields. The width and height of each image represent 10 sec and 2000 pA, respectively where pA stands for picoampere.
2.3. Image preparation for deep neural network
In this study, we have created two sets of data (images) to train and validate the model. The first set of images was plotted (aka raw data) without any constraints. For the second set of images (aka transformed data), the lower bound and upper bound of current values (vertical axis) were set as the global minimum and maximum value of current, respectively. For a particular potential difference across the nanopore, this bound was same for all AAV types. In other words, the transformed data were plotted by considering a fixed current change (in the vertical axis) for all three AAV classes. An example of raw and transformed images for all three AAV types is shown in Fig. 3 for an applied potential difference of −175 mV across the nanopore.
Fig. 3.
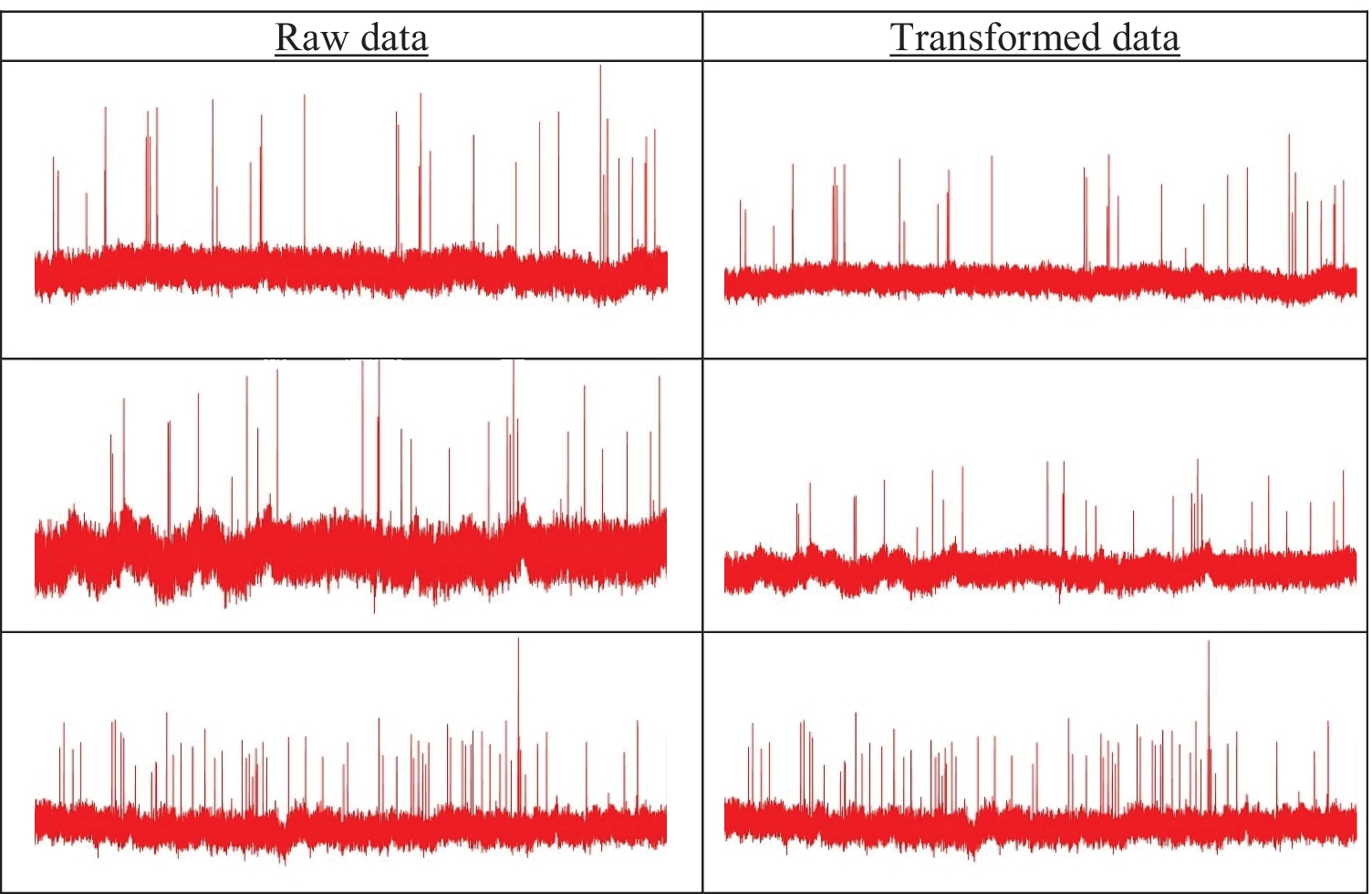
Concept of raw vs. transformed images: AAVEmpty (top row), AAVssDNA (middle row), and AAVdsDNA (bottom row). Since GoogleNet requires an image input size of 224 X 224, all images were resized to 224 X 224 ×3.
3. Results and Discussion
In this section, we examine the potential of the fine-tuning method applied to the resistive pulse images obtained from the nanopore translocation experiments. The fine-tuning-based transfer learning was conducted using MATLAB 2018b (academic use) and performed on an HP desktop computer with single GPU (Processor: Inter(R) Core(TM) i7–3770 CPU @ 3.4GHz, Installed RAM: 16.0 GB and GPU: Nvidia Geforce GT 640).
As stated earlier, we used 1, 2, and 4 sec time frames for segmenting the single resistive pulse experiment data into several images. Furthermore, we have considered five different resistive-pulse data sets corresponding to the applied potential differences of −75, −100, −125, −150, and −175 mV across the nanopore. Since all datasets are drawn following two styles: raw and transformed, there are 30 distinguishable datasets within three different classes. Table 1 shows the details of all datasets used in this study along with the number of available images. Fig. 4 illustrates the overall information processing used for fine-tuning-based transfer learning using a pre-trained deep neural network for AAV classifications.
Table 1.
Number of images in training and validation dataset for each AAV type.
| Applied potential difference (mV) | AAVs Type | Time Frame (sec) | |||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 4 | 1 | 2 | 4 | ||
| Number of Images | |||||||
| Training | Validation | ||||||
| −75 | Empty | 678 | 339 | 169 | 290 | 145 | 73 |
| ssDNA | 720 | 360 | 180 | 308 | 154 | 77 | |
| dsDNA | 501 | 251 | 125 | 215 | 107 | 54 | |
| −100 | Empty | 431 | 216 | 108 | 185 | 92 | 46 |
| ssDNA | 760 | 381 | 190 | 328 | 163 | 82 | |
| dsDNA | 720 | 360 | 180 | 308 | 154 | 77 | |
| −125 | Empty | 588 | 294 | 147 | 252 | 126 | 63 |
| ssDNA | 840 | 420 | 210 | 360 | 180 | 90 | |
| dsDNA | 459 | 230 | 115 | 197 | 98 | 49 | |
| −150 | Empty | 420 | 210 | 105 | 180 | 90 | 45 |
| ssDNA | 633 | 316 | 158 | 271 | 136 | 68 | |
| dsDNA | 420 | 210 | 105 | 180 | 90 | 45 | |
| −175 | Empty | 714 | 357 | 179 | 306 | 153 | 76 |
| ssDNA | 812 | 406 | 203 | 348 | 174 | 87 | |
| dsDNA | 384 | 192 | 96 | 164 | 82 | 41 | |
Fig. 4.
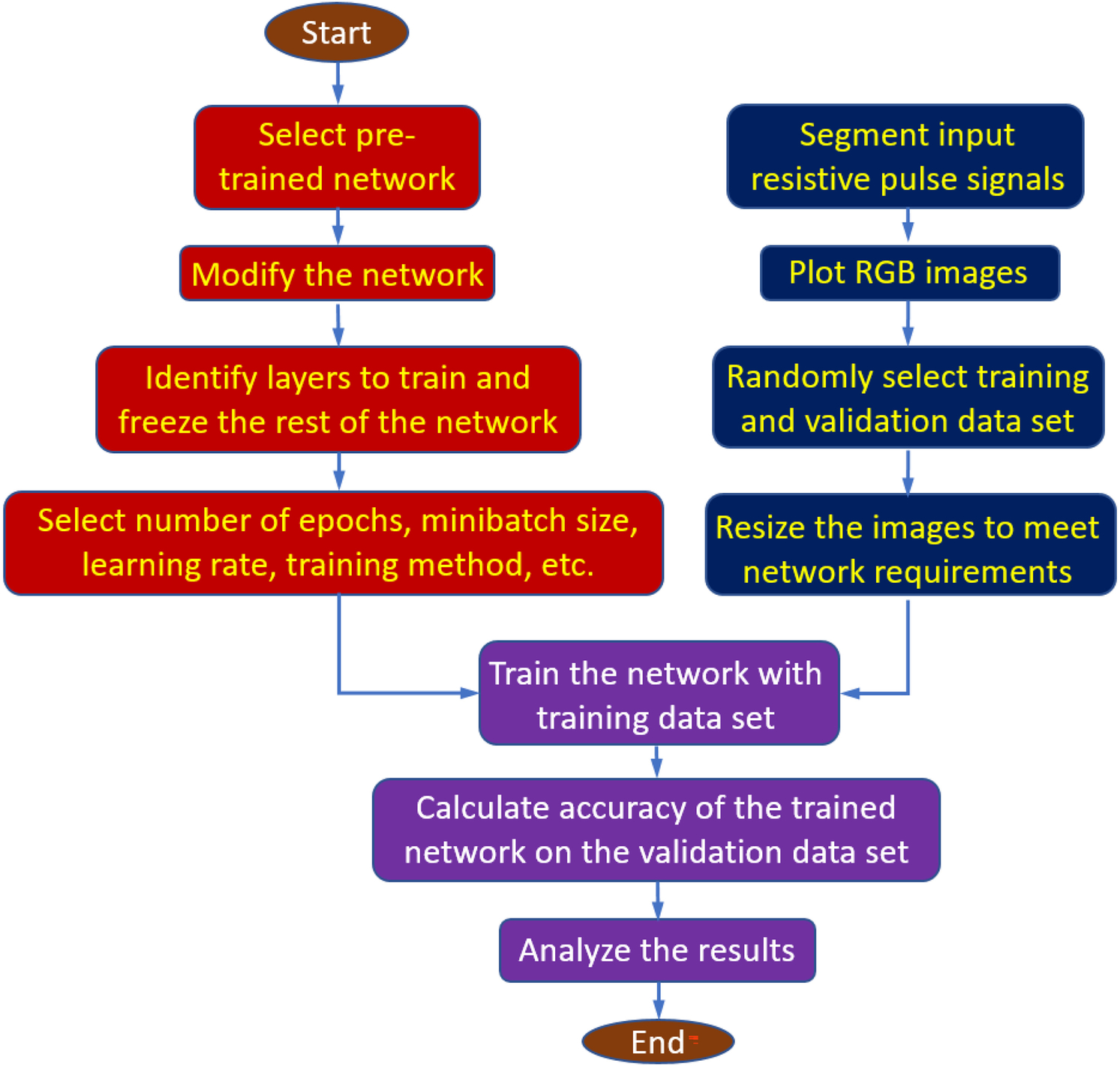
Flow chart of the fine-tuning-based deep learning method for AAV classification
3.1. Training
The training of the network starts from the first convolution layer and proceeds in the feedforward manner until the final classification layer by back-propagating the error from the classification layer towards the first convolutional layer. There are 4 million trainable parameters (weights) in the GoogleNet architecture, and thus a large dataset was necessary for the training and optimization of such a deep network. However, for a small dataset, like the problem in hand, it is very difficult to determine the appropriate global minima for the loss function, and the network will suffer from overfitting or underfitting. Therefore, we have used the data augmentation method to increase the number of samples to handle the overfitting or underfitting issue. In addition, we have initialized weights based on pre-trained GoogleNet, which was trained using the ImageNet dataset.
After the weight transfer, we started the training process by sending the labeled data. Instead of training the whole dataset at one time, a minibatch (size 16) of images was applied to the network. The training was carried out by optimizing the cost function based on stochastic gradient descent (SGD) with momentum [45]. A small initial learning rate of η = 0.0001 was applied since a large change towards pretrained weights is not desirable during fine-tuning. To accelerate learning, we used a momentum of μ = 0.9 during the training process which describes the influence of previously updated weights in the current iteration. A scheduled rate of σt = 1 was used in training to facilitate a constant learning rate throughout the training process.
For supervised learning problems, a labeled dataset is usually split into two subsets: training and validation. The training set is used for preparing the model and the validation dataset is used for testing the generality. In our work, the ratio of the training to validation set was empirically set to 7:3 because this split option has shown the highest accuracy when compared to the other split options [46]. Xu and Goodacre [47] found that a good balance (50–70% for training) between the sizes of the training set and validation set is necessary to have a reliable estimation of model performance because having too many or too few samples in the training set had a negative effect on the estimated model performance. Since the validation prevents the network from overfitting and helps to monitor proper convergence, we validated the training process after every 3 training iterations. As stated earlier, a stochastic gradient decent method was used to update the parameters of the last three layers after each feed-forward training process. Though a validation run was executed after three training iterations, no parameters were updated during the validation process.
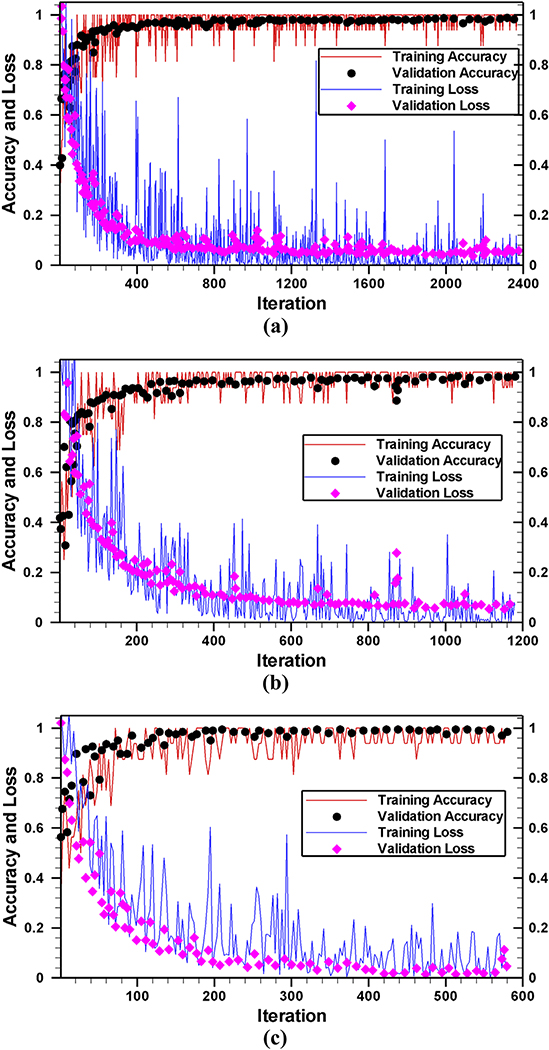
Fig. 5 shows the accuracy (as well as the loss) of the training and validation process on transformed images for the −175 mV case. As expected, both training and validation accuracies were quite low at the beginning of the training process. The training or validation accuracies as well as loss converged within 10 epochs (2380, 1180, and 580 iterations for 1 sec, 2 sec, and 4 sec time frames, respectively) in each run presented in this work. Therefore, we continued the training process up to 20 epochs ensuring the saturation of training and validation accuracies. As shown in Fig. 5, the overfitting is almost negligible since there is no observable difference between training and validation accuracies (as well as between training and validation loss). This indicates that despite the small dataset, fine-tuning of the last three layers does not hamper the generality of the network.
Fig. 5.
The accuracy of network on transformed images for time frame of (a) 1 sec, (b) 2 sec, and (c) 4 sec. After initializing the GoogleNet with known biases and weights, 16 images (minibatch) were passed through the network. Using the forward calculation, the class levels (predicted level) of those 16 images were found. The training or validation accuracies are calculated as the total number of correct predictions divided by the total number of images.
As stated earlier, we have replaced and trained the last three layers of GoogleNet to classify resistive pulse data sets and all other layers were kept frozen. Therefore, except those last three layers, all other layers do not contain any learnable parameters, and the number of FLOPs is the same as standard architecture. Thus, the training includes only 3,075 learnable parameters for the last three layers. These learnable parameters require total 6,155 FLOPs. The whole training and validation process (20 epoch) took approximately 296, 84, and 26 minutes for 1, 2, and 4 sec time frame images on a single GPU (Processor: Inter(R) Core (TM) i7–3770 CPU @ 3.4GHz) with a total of 2712, 1356, and 678 transformed images, respectively. A similar amount of processing time was required for raw data because the same number of training and validation images (Table 2) were employed. This result shows that as the number of images decreases, less and less amount of time is required for training without deteriorating the accuracy. For a linear drop in the total number of images, a quadratic reduction in processing time requirement has been observed. As shown in Fig. 5, in every case, the network reached the highest accuracy within 10 epochs; therefore, the proposed fine-tuned deep network is very fast. The processing time required for all different cases studied in this work is listed in Table 2.
Table 2.
Training time requirements for different datasets.
| Potential difference | Time frame (sec) | Training time (minutes) | |
|---|---|---|---|
| Raw data | Transformed data | ||
| −75 mV | 1 | 293 | 291 |
| 2 | 83 | 84 | |
| 4 | 26 | 26 | |
| −100 mV | 1 | 296 | 291 |
| 2 | 84 | 85 | |
| 4 | 26 | 27 | |
| −125 mV | 1 | 288 | 287 |
| 2 | 84 | 83 | |
| 4 | 26 | 26 | |
| −150 mV | 1 | 186 | 184 |
| 2 | 55 | 54 | |
| 4 | 18 | 18 | |
| −175 mV | 1 | 291 | 296 |
| 2 | 84 | 87 | |
| 4 | 26 | 26 | |
3.2. Classification accuracy
Classification accuracy obtained from the validation datasets is shown in Fig. 6 for 1 sec time frames. As shown in Fig. 6, the network performance is much better on transformed data than raw data. Since the transformed data set preserves the distinguishable features between classes, except in one case (see Fig. 6a: ssDNA), the mean accuracy for any class is always higher for transformed data than that of raw data. Similar results are observed for 2 and 4 sec time frames as shown in Supplementary Figs. S1 and S2, respectively. Because of the feature preserving nature of the transformed data, a higher overall mean accuracy has been observed for transformed data compared to the raw data, irrespective of class, potential difference, or time frame.
Fig. 6.
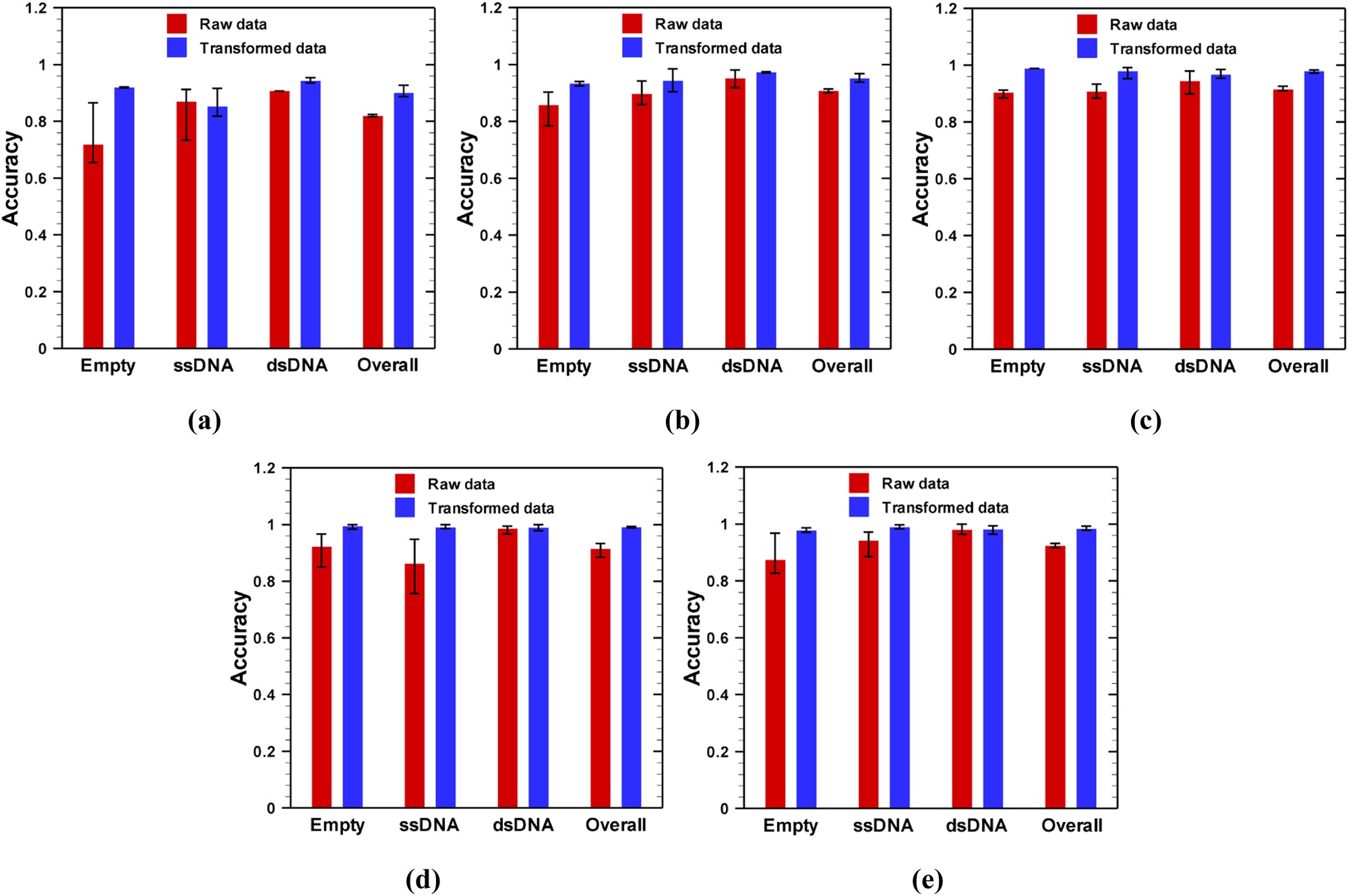
Efficacy of deep convolutional neural networks in classification of AAVs from nanopore experimental data obtained at different applied potential difference: (a) −75 mV, (b) −100 mV, (c) −125 mV, (d) −150 mV and (e) −175 mV for 1 sec time frame images. For a particular class, 70% of the data were randomly selected from all images of that class for training the network, while the rest of the images of that class were used for validation.
As stated earlier, the training and validation sets are selected in a random manner; therefore, it should be tested how classification accuracy varies with randomness of the training dataset. To address this question, for each dataset, we trained the network for three times (which we called realization) with randomly selected training datasets. In each realization, the training started from the same point i.e., initialization of the network with pre-trained GoogleNet biases and weights, and the network was trained and validated using the method described before. The variation of classification accuracy with the randomness of the training dataset is shown by the error bar in Figs. 6, and supplementary results (Figs. S1 and S2). Our results show that the network trained on raw images is more sensitive to the randomness of the training data than that of transformed images as indicated by a larger error bar on results of raw data in Fig. 6. This is due to the fact that when resistive pulse data are drawn in a fixed geometrical frame (transformed), the difference between feature maps for training and validation data sets reduces. Similar results were obtained for 2 and 4 sec time frames as shown in supplementary Figs. S1 and S2.
Another interesting result is observed when the applied voltage magnitude changed from −75 mV to −175 mV. As shown in Fig. 6, as the absolute value of voltage increased, the network performed better as indicated by higher mean accuracy for both raw and transformed images. In addition to higher accuracy, at higher potential difference, the network is less vulnerable to the arbitrariness of the training dataset as indicated by smaller error bars for both raw and transformed data. At a smaller potential difference, a smaller number of particles are translocated through the nanopore as indicated by a lesser number of spikes in the resistive pulse signal (Fig. 2). Moreover, the height of the spikes is also very small for a low magnitude of applied electric field. Therefore, at a low electric field, the resistive pulse signal is mostly governed by the base (signal when there is no particle in the pore) current signal. Since the base signal does not depend on the particle types, it is very difficult for a network to discriminate between signals at low electric fields. Although similar behaviors were obtained with 2 and 4 sec time frame images as shown in Fig. S1 and S2, respectively, networks trained on 4 sec time frame images were more vulnerable to randomness of training data, especially at low electric field. Thus, a large potential difference (above 100 mV) is desirable for the characterization of AAVs through deep learning.
Although trained networks exhibit similar behavior for 1 sec (Fig. 6), 2 sec (supplementary Fig. S1), and 4 sec (supplementary Fig. S2) time frame images, the classification accuracy is slightly lower for 2 sec time frame case compared to 1 sec or 4 sec time frames for any applied potential difference. The reason for better performance with 1 sec or 4 sec images is as follows. First, although 1 sec time frame resistive-pulse images store only limited information, the number of training examples are much higher. On the other hand, 4 sec time frame images hold four times more information, though the number of training examples are small. Therefore, the slightly lower accuracy at 2 sec time frame is a combined effect of fewer number of training examples and less information stored by each training examples.
Nevertheless, the fine-tuning-based method is very effective in classifying AAVs from time series electric signals. Furthermore, it is important to note that our proposed breakdown of time sequence data clearly provides two benefits. First, it facilitates more images to train the network from a limited experimental dataset. For example, a single 10 min resistive-pulse signal obtained from a nanopore experiment can provide 600, 300 and 150 images for 1 sec, 2 sec and 4 sec segmentations, respectively. Second, proper prediction of a class is guaranteed from the time series resistive-pulse data, even though there are a few misclassifications. This is again because each input electrical signal being divided into many images, and the final prediction class of an electrical signal is based on the average performance of all images of that signal, not the individual flagging of each image.
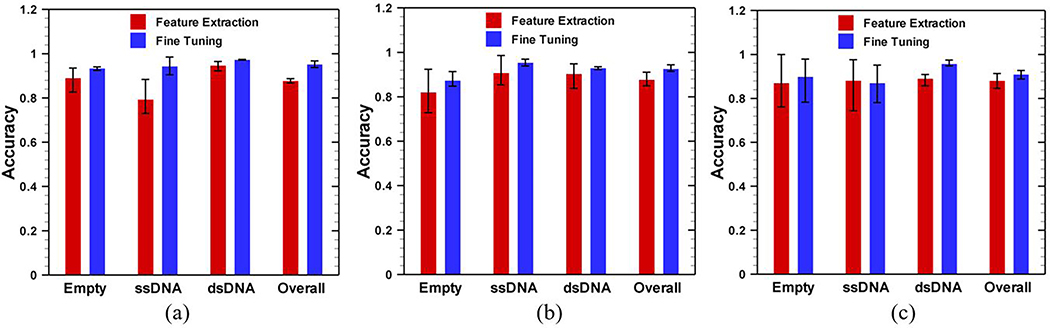
3.3. Comparison of performance between transfer learning methods
In this study, we have compared the classification performance of the fine-tuning method with feature extraction-based method. In case of feature extraction based transfer learning, we passed (just once) all available training examples through the pretrained GoogleNet and extracted the features from the training examples by using layer activations. In this work, activations of the fully connected layer (loss3-classifier) were taken as feature map; hence, the feature map has a dimension of 1×1×1000 for each image. From the extracted feature map, we have trained a multiclass support vector machine (SVM) using the one-versus-one method [48] for three different classes. After training, prediction of a new image was done through newly trained SVM classifier (for details please see [8]).
Figure 7 shows the prediction comparison between the fine-tuning and feature extraction based methods for different time frames (1 sec, 2 sec, and 4 sec) for −100 mV case. As shown in Fig. 7, the fine-tuning is always better than that of feature extraction. Although feature extraction uses a deep network to extract the feature, the final classification is done based on a shallow network such as support vector machine. On the other hand, in fine-tuning, the final classification is integrated in the deep network which allows the model to learn more. The reduced uncertainty in the prediction further results in a higher classification accuracy compared to feature extraction based method. The highest improvement in prediction accuracy is obtained with 1 sec time frame images (Fig. 7a) because there are enough training examples from which the network can learn through fine-tuning. However, when dataset is small (4 sec time frames for current example), the improvement is less significant since there is a smaller dataset to learn from (Fig. 7c). These results indicate that instead of using a pre-trained deep network as a feature extractor, a direct modification such as fine-tuning of the pre-trained deep network is preferred.
Fig. 7.
Comparison between feature extraction and fine-tuning methods for: a) 1 sec, b) 2 sec, and c) 4 sec time frames. Image data were obtained from solid state nanopore experiments for an applied potential difference of −100 mV.
For a multiclass problem, classification accuracy alone is not very reliable for a true prediction. In that case, a confusion matrix is a superior way to analyze results. Therefore, the overlapping results of a confusion matrix of 4 sec raw and the transformed data (only validation set) for different realizations (R1, R2, and R3) are listed in Table 3 and 4, respectively. The confusion matrix presents the number of correct and incorrect predictions with values broken down by each class. As shown in Table 3, the ternary classification tasks perform reasonably well on raw data with a high number of correct predictions and a few misclassifications. In addition, the behavior of the network is consistent with the random changes of training data, as number of correct predictions and misclassifications are similar among the three realizations. Moreover, as the magnitude of the applied voltage increases, the number of misclassifications decreases, resulting in a more accurate network. In comparison to the raw data (Table 3), the ternary classification tasks perform extremely well on the transformed data as shown in Table 4. For any applied electric field, the number of correct predictions increases for each class of AAVs, resulting in a reduced number of false negatives or false positives in the case of the transformed data. Nevertheless, even with some misclassifications, the fine-tuning-based transfer learning is quite promising for the classification of AAVs based on their transgene packaging. The confusion matrix for other cases, provided in supplementary materials (Table S1, S2, S3, and S4 for 1 sec raw, 1 sec transformed, 2 sec raw, and 2 sec transformed data, respectively), show the identical classification accuracy.
Table 3.
Overlapping results of confusion matrix obtained on 4 sec raw data (validation set) for different realizations (R1, R2, and r3) at different voltages (−75, −100, −125, −150, and −175 mV). In this table ‘ds’, ‘E’ and ‘ss’ stand for three types of AAVs: dsDNA, Empty, and ssDNA, respectively.
| −75 (mV) | −100 (mV) | −125 (mV) | −150 (mV) | −175 (mV) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ╲ | ds | E | ss | ds | E | ss | ds | E | ss | ds | E | ss | ds | E | ss | |
| R1 | ds | 40 | 12 | 2 | 67 | 6 | 4 | 42 | 3 | 4 | 44 | 0 | 1 | 41 | 0 | 0 |
| E | 5 | 63 | 5 | 2 | 40 | 4 | 1 | 57 | 5 | 2 | 41 | 2 | 0 | 66 | 10 | |
| ss | 3 | 7 | 67 | 8 | 3 | 71 | 2 | 0 | 88 | 7 | 0 | 61 | 1 | 2 | 84 | |
| R2 | ds | 40 | 11 | 3 | 69 | 0 | 8 | 42 | 3 | 4 | 43 | 0 | 2 | 41 | 0 | 0 |
| E | 10 | 58 | 5 | 4 | 37 | 5 | 4 | 57 | 2 | 0 | 43 | 2 | 0 | 71 | 5 | |
| ss | 5 | 5 | 67 | 1 | 1 | 80 | 4 | 1 | 85 | 2 | 2 | 64 | 0 | 9 | 78 | |
| R3 | ds | 45 | 5 | 4 | 74 | 0 | 3 | 42 | 3 | 4 | 40 | 0 | 5 | 40 | 0 | 1 |
| E | 13 | 55 | 5 | 2 | 43 | 1 | 1 | 57 | 5 | 0 | 37 | 8 | 0 | 64 | 12 | |
| ss | 4 | 2 | 71 | 11 | 5 | 66 | 2 | 0 | 88 | 0 | 0 | 68 | 1 | 3 | 83 | |
Table 4.
Overlapping results of confusion matrix obtained on 4 sec transformed data (validation set) for different realizations (R1, R2, and R3) at different voltages (−75, −100, −125, −150, and −175 mV). In this table ‘ds’, ‘E’ and ‘ss’ stand for three types of AAVs: dsDNA, Empty and ssDNA, respectively.
| −75 (mV) | −100 (mV) | −125 (mV) | −150 (mV) | −175 (mV) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ╲ | ds | E | ss | ds | E | ss | ds | E | ss | ds | E | ss | ds | E | ss | |
| R1 | ds | 43 | 5 | 6 | 75 | 0 | 2 | 48 | 1 | 0 | 44 | 0 | 1 | 41 | 0 | 0 |
| E | 1 | 68 | 4 | 3 | 43 | 0 | 2 | 61 | 0 | 0 | 43 | 2 | 1 | 74 | 1 | |
| ss | 1 | 3 | 73 | 9 | 1 | 72 | 5 | 0 | 85 | 0 | 0 | 68 | 0 | 0 | 87 | |
| R2 | ds | 53 | 1 | 0 | 73 | 3 | 1 | 42 | 0 | 7 | 43 | 0 | 2 | 41 | 0 | 0 |
| E | 5 | 57 | 11 | 0 | 45 | 1 | 2 | 61 | 0 | 1 | 44 | 0 | 2 | 73 | 1 | |
| ss | 1 | 2 | 74 | 12 | 6 | 64 | 3 | 0 | 87 | 1 | 0 | 67 | 0 | 0 | 87 | |
| R3 | ds | 50 | 0 | 4 | 73 | 0 | 4 | 41 | 0 | 8 | 44 | 0 | 1 | 41 | 0 | 0 |
| E | 5 | 63 | 5 | 4 | 36 | 6 | 0 | 63 | 0 | 0 | 44 | 1 | 1 | 74 | 1 | |
| ss | 1 | 0 | 76 | 4 | 0 | 78 | 1 | 0 | 89 | 0 | 0 | 68 | 0 | 0 | 87 | |
3.4. Comparison among deep networks
To assess the effectiveness of fine-tuning-based transfer learning, we have considered two other deep learning networks: InceptionV3 [49] and ResNet50 [14]. We have chosen InceptionV3 and ResNet50 because they have similar number of layers: 48 and 50, respectively, and both are widely used in numerous fields of science for image classification [50–53] due to their reputations. ResNet50 won the ImageNet large scale visual recognition challenge (ILSVRC) 2015, while InceptionV3 was the second runners up of ILSVRC 2015. For comparison of classification accuracy results with GoogleNet, we have changed the last three layers: fully connected, softmax and classification layers to accommodate our dataset. Fig. 8 shows the performance of each network on the 4 sec time frame validation datasets for an applied voltage of −175 mV. Here, the error bar is obtained by training the network with randomly selected training and validation dataset for three times. The performance of the networks on raw images is presented in Fig. 8a, while corresponding results for transformed data are shown in Fig. 8b. The performance (numeric value of validation accuracy) of different models: GoogleNet, InceptionV3 and ResNet50 are provided in Table S4 and Table S5 for 4 sec time frame raw data and transformed data, respectively. The comparison of performance of networks for other voltages (on 4 sec time frame data) is shown in Fig. S3 in supplementary materials. As shown in the Fig. 8a, on empty class of data, ResNet50 exhibits the highest average accuracy, while GoogleNet yields the lowest average accuracy. An opposite phenomenon is observed on the ssDNA class of data, while all the networks behave similarly on the dsDNA data. Thus, the overall accuracy for networks on the validation dataset are similar for all networks. As shown in Fig. 8b, for transformed images, all three networks perform in the same scale. For relatively higher applied voltage, the GoogleNet shows more consistency on the randomness of the training and validation dataset, as it has smaller error bars compare to ResNet50 or InceptionV3. However, at low magnitude of voltages (−75 mV or −100 mV), InceptionV3 and ResNet50 provide around 5% higher accuracy than GoogleNet (see Fig. S3 a–d). As the magnitude of the voltage increases, the accuracy of GoogleNet becomes comparable with InceptionV3 and ResNet50 (Fig. S3 e–h). Although the accuracy does not vary significantly with network architecture, the time consumption for training and validation process varies a lot with the network architecture because of different number of learnable parameters and corresponding number of FLOPs. For example, while training and validation of GoogleNet takes only 26 minutes on average for 3,075 (total 6,155 FLOPs) learnable parameters, the InceptionV3 and ResNet50 take ~ 182 and 155 minutes, respectively for 6,175 (total 12,299 FLOPs) learnable parameters. We also found that both InceptionV3 and ResNet50 start overfitting after 5 epochs while GoogleNet does not overfit at all (data is not shown). This may arise because InceptionV3 and ResNet50 has more layers and higher number of parameters than GoogleNet architecture. This result indicates that a comparatively less deep network works better for AAV classification because it offers faster training and validation with acceptable classification accuracy.
Fig. 8.
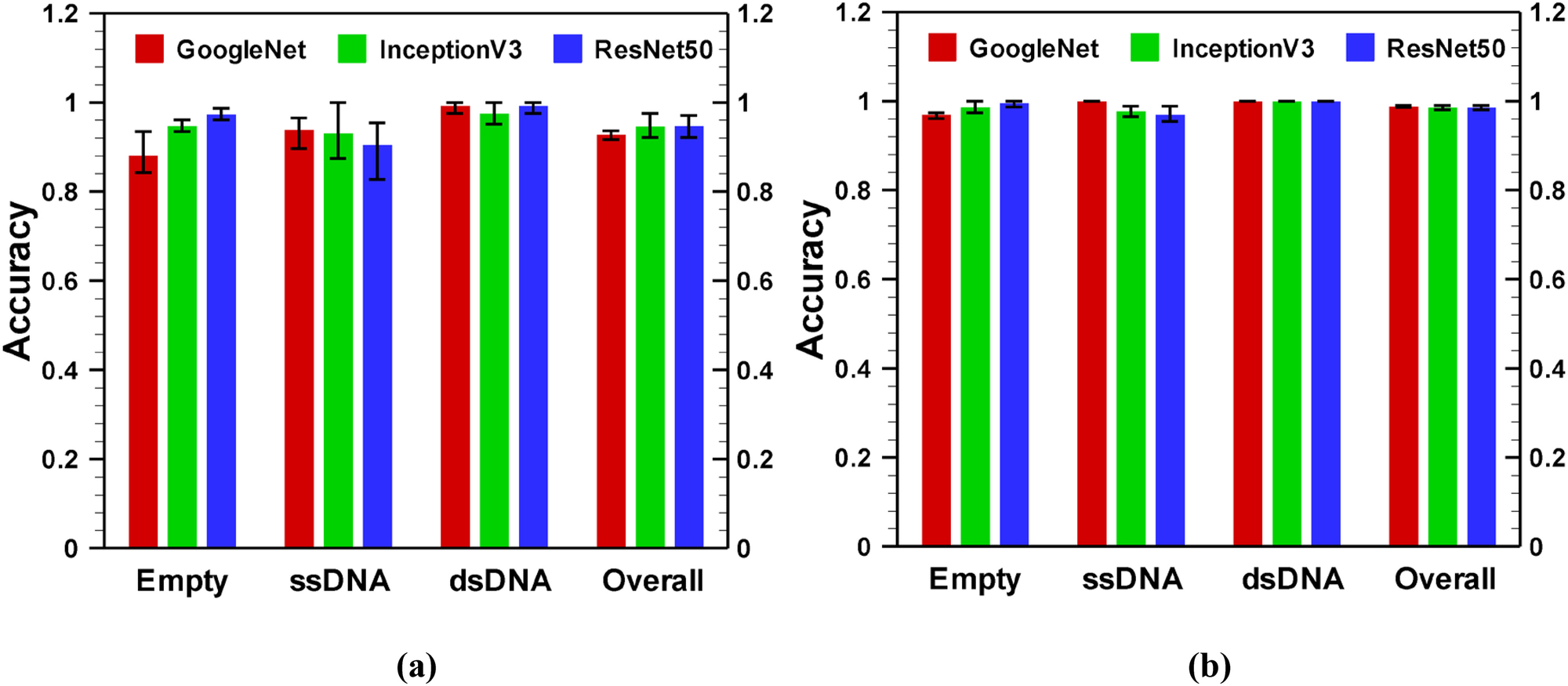
Performance comparison of GoogleNet, InceptionV3, and ResNet50 for (a) raw and (b) transformed images on 4 sec time frame data. Image data were obtained from solid state nanopore experiments for an applied voltage −175 mV.
4. Conclusions
We have presented a framework for fine-tuning-based transfer learning from pre-trained deep networks. Although there are numerous highly accurate deep neural networks for classification of images, we have selected GoogleNet, ResNet50 and InceptionV3 to distinguish adeno-associated virus vectors depending on their transgene packaging. The last three layers of those deep networks were modified to match the three object classes using a fine-tuning-based transfer learning method. Experimental data obtained from a nanopore translocation study was used to train and validate the modified neural networks. Our results suggest that the fine-tuning-based transfer learning can effectively classify different kind of AAVs from the resistive pulse signals/images. The comparative study of fine-tuning and feature extraction methods suggest that fine-tuning a network is a better choice to distinguish AAVs as it consistently yields a higher classification accuracy than the feature extraction. Additionally, for fine-tuning method, one can use any high performance pre-trained deep networks as our analysis shows a similar level of validation accuracy with GoogleNet, InceptionV3 or ResNet50. However, overfitting may be an issue if we choose a very deep network. For example, in contrast to InceptionV3 and ResNet50, GoogleNet does not show any sign of overfitting as the validation accuracy completely overlap with the training accuracy. Moreover, GoogleNet takes much shorter time for the overall training and validation process than those of InceptionV3 (one-seventh) and ResNet50 (one-sixth). Therefore, for an effective and fast classification of AAVs, a comparatively less deep network such as GoogleNet is preferable.
All the pretrained networks: GoogleNet, InceptionV3 and ResNet50 used in this work were trained with natural images (ImageNet dataset). Therefore, feeding the time series resistive pulse signal into the pre-trained networks was the main challenge of using fine-tuning-based transfer learning to classify AAV vectors. But, in this work, we have overcome this restriction by creating 2D plot from the resistive pulse signal with different time frames. Unlike classical machine learning, we used few experimental datasets for training (for a given applied electric field), but the segmentation of each resistive-pulse time series signal helped us to attain our desired goal in the data-driven classification. Furthermore, transformation of the data helped in better preservation of features between the training and validation data. For transformed data, the mean accuracy of the network was 95% or better for any class, when the applied potential difference across the nanopore was 100 V or higher. The classification accuracy can be further improved with increasing training data or by training more parameters in the deep learning architecture. In addition to very high classification accuracy, the fine-tuning-based method is very fast as overall training and validation process can be done in less than an hour. Therefore, the machine learning based method presented in this work can be integrated with nanopore or other experiments to detect viruses quickly and accurately in future.
Supplementary Material
Funding/Acknowledgements:
The research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number 1R21GM134544. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Biographies
 Aminul Islam Khan received his B.S. and M. S. from the Bangladesh University of Engineering and Technology where he also served as a Lecturer and Assistant Professor. Currently, Khan is a PhD candidate at Washington State University. He has been involved in multidisciplinary research including hands-on learning for STEM education, transport modeling in micro/nano scale device, and various inverse techniques including Bayesian inference, Monte Carlo methods, neural network, and deep/machine learning for adeno-associated virus and liposome characterization. In 2020, he was awarded the best Research Assistant award by the School of Mechanical and Materials Engineering at Washington State University. Khan plans to pursue a teaching career upon earning his PhD.
Aminul Islam Khan received his B.S. and M. S. from the Bangladesh University of Engineering and Technology where he also served as a Lecturer and Assistant Professor. Currently, Khan is a PhD candidate at Washington State University. He has been involved in multidisciplinary research including hands-on learning for STEM education, transport modeling in micro/nano scale device, and various inverse techniques including Bayesian inference, Monte Carlo methods, neural network, and deep/machine learning for adeno-associated virus and liposome characterization. In 2020, he was awarded the best Research Assistant award by the School of Mechanical and Materials Engineering at Washington State University. Khan plans to pursue a teaching career upon earning his PhD.
 MinJun Kim is presently the Robert C. Womack Endowed Chair Professor in the Department of Mechanical Engineering at Southern Methodist University. He received his B.S. and M.S. degrees in mechanical engineering from Yonsei University in Korea and Texas A&M University, respectively. Dr. Kim completed his Ph.D. degree in engineering at Brown University, where he held the prestigious Simon Ostrach Fellowship. Following his graduate studies, Dr. Kim was a postdoctoral research fellow in the Rowland Institute at Harvard University. He joined Drexel University in 2006 as an Assistant Professor and was later promoted to Professor of Mechanical Engineering and Mechanics. Since Aug. 2016, he has been the Director of Biological Actuation, Sensing and Transport Laboratory (BASTLab) at the Lyle School of Engineering. Dr. Kim has been exploring biological transport phenomena including cellular/molecular mechanics and engineering in novel nano/microscale architectures to produce new types of nanobiotechology, such as nanopore technology and nano/micro robotics.
MinJun Kim is presently the Robert C. Womack Endowed Chair Professor in the Department of Mechanical Engineering at Southern Methodist University. He received his B.S. and M.S. degrees in mechanical engineering from Yonsei University in Korea and Texas A&M University, respectively. Dr. Kim completed his Ph.D. degree in engineering at Brown University, where he held the prestigious Simon Ostrach Fellowship. Following his graduate studies, Dr. Kim was a postdoctoral research fellow in the Rowland Institute at Harvard University. He joined Drexel University in 2006 as an Assistant Professor and was later promoted to Professor of Mechanical Engineering and Mechanics. Since Aug. 2016, he has been the Director of Biological Actuation, Sensing and Transport Laboratory (BASTLab) at the Lyle School of Engineering. Dr. Kim has been exploring biological transport phenomena including cellular/molecular mechanics and engineering in novel nano/microscale architectures to produce new types of nanobiotechology, such as nanopore technology and nano/micro robotics.
 Prashanta Dutta received his PhD degree in Mechanical Engineering from Texas A&M University in 2001. He started his academic career as an Assistant Professor in the School of Mechanical and Materials Engineering of Washington State University right after his doctoral degree. He was promoted to the rank of Associate and Full Professor in 2007 and 2013, respectively. He served as a Visiting Professor at Konkuk University, Seoul, South Korea (2009–2010) and a Fulbright Faculty Fellow at the Technical University of Darmstadt, Germany (2017). Prof. Dutta is an elected Fellow of the American Society of Mechanical Engineers (ASME). He has authored and co-authored more than 175 peer-reviewed journal and conference papers and delivered more than 75 invited talks all over the world. Currently, he serves as a Deputy Editor for the Electrophoresis.
Prashanta Dutta received his PhD degree in Mechanical Engineering from Texas A&M University in 2001. He started his academic career as an Assistant Professor in the School of Mechanical and Materials Engineering of Washington State University right after his doctoral degree. He was promoted to the rank of Associate and Full Professor in 2007 and 2013, respectively. He served as a Visiting Professor at Konkuk University, Seoul, South Korea (2009–2010) and a Fulbright Faculty Fellow at the Technical University of Darmstadt, Germany (2017). Prof. Dutta is an elected Fellow of the American Society of Mechanical Engineers (ASME). He has authored and co-authored more than 175 peer-reviewed journal and conference papers and delivered more than 75 invited talks all over the world. Currently, he serves as a Deputy Editor for the Electrophoresis.
Footnotes
Conflicts of interest: The authors declare that they have no conflict of interest.
Declarations
Code availability: All codes are available upon request
Ethics approval: Not applicable
Availability of data and material:
All data associated with this paper is publicly available at https://github.com/mstfwsulab/AAV-classification
References
- [1].Li C, and Samulski RJ (2020). Engineering adeno-associated virus vectors for gene therapy. Nature Reviews Genetics, 21(4), 255–272. [DOI] [PubMed] [Google Scholar]
- [2].F Flotte TR, Afione SA, Conrad C, McGrath SA, Solow R, Oka H, Zeitlin PL, Guggino WB, and Carter BJ (1993). Stable in vivo expression of the cystic fibrosis transmembrane conductance regulator with an adeno-associated virus vector. Proceedings of the National Academy of Sciences, 90(22), 10613–10617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Li C, Bowles DE, van Dyke T, and Samulski RJ (2005). Adeno-associated virus vectors: potential applications for cancer gene therapy. Cancer Gene Therapy, 12(12), 913–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Naso MF, Tomkowicz B, Perry WL, and Strohl WR (2017). Adeno-Associated Virus (AAV) as a vector for gene therapy. Biodrugs, 31(4), 317–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Gimpel AL, Katsikis G, Sha S, Maloney AJ, Hong MS, Nguyen TNT, Wolfrum J, Springs SL, Sinskey AJ, Manalis SR, Barone PW, and Braatz RD (2021). Analytical methods for process and product characterization of recombinant adeno-associated virus-based gene therapies. Molecular Therapy-Methods & Clinical Development, 20, 740–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lock M, McGorray S, Auricchio A, Ayuso E, Beecham EJ, Blouin-Tavel V, Bosch F, Bose M, Byrne BJ, Caton T, Chiorini JA, Chtarto A, Clark KR, Conlon T, Darmon C, Doria M, Douar A, Flotte TR, Francis JD, Francois A, Giacca M, Korn MT, Korytov I, Leon X, Leuchs B, Lux G, Melas C, Mizukami H, Moullier P, Muller M, Ozawa K, Philipsberg T, Poulard K, Raupp C, Riviere C, Roosendaal SD, Samulski RJ, Soltys SM, Surosky R, Tenenbau L, Thomas DL, van Montfort B, Veres G, Wright JF, Xu YL, Zelenaia O, Zentilin L, and Snyder RO (2010). Characterization of a recombinant adeno-associated virus type 2 reference standard material. Human Gene Therapy, 21(10), 1273–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fried JP, Swett JL, Nadappuram BP, Mol JA, Edel JB, Ivanov AP, and Yates JR (2021). In situ solid-state nanopore fabrication. Chemical Society Reviews, 50(8), 4974–4992. [DOI] [PubMed] [Google Scholar]
- [8].Karawdeniya BI, Bandara Y, Khan AI, Chen WT, Vu HA, Morshed A, Suh J, Dutta P, and Kim MJ (2020). Adeno-associated virus characterization for cargo discrimination through nanopore responsiveness. Nanoscale, 12(46), 23721–23731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Marques AD, Kummer M, Kondratov O, Banerjee A, Moskalenko O, and Zolotukhin S (2021). Applying machine learning to predict viral assembly for adeno-associated virus capsid libraries. Molecular Therapy-Methods & Clinical Development, 20, 276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Krizhevsky A, Sutskever I, and Hinton GE (2012). Imagenet classification with deep convolutional neural networks. Proc. Advances in Neural Information Processing Systems 25, 1097–1105. [Google Scholar]
- [11].Simonyan K, and Zisserman A (2014). Very deep convolutional networks for large-scale image recognition, arXiv preprint arXiv:1409.1556. [Google Scholar]
- [12].Zeiler MD, Fergus R (2014). Visualizing and understanding convolutional networks. In: Fleet D, Pajdla T, Schiele B, Tuytelaars T (eds) Computer Vision – ECCV 2014. ECCV 2014. Lecture Notes in Computer Science, vol 8689. Springer, Cham. 10.1007/978-3-319-10590-1_53 [DOI] [Google Scholar]
- [13].Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, and Rabinovich A (2015). Going deeper with convolutions. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 1–9. [Google Scholar]
- [14].He KM, Zhang XY, Ren SQ, Sun J, and IEEE. (2016) Deep residual learning for image recognition, Proc. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 770–778. [Google Scholar]
- [15].Xiao ZW, Xu X, Xing HL, Qu R, Song FH, Zhao BW, and IEEE. (2021). RNTS: Robust neural temporal search for time series classification. Proceedings of International Joint Conference on Neural Networks (IJCNN), 1–8. [Google Scholar]
- [16].Xiao ZW, Xu X, Zhang HX, and Szczerbicki E (2021), A new multi-process collaborative architecture for time series classification. Knowledge-Based Systems, 220, 106934. [Google Scholar]
- [17].Xiao ZW, Xu X, Xing HL, Luo SX, Dai PL, and Zhan DW (2021), RTFN: A robust temporal feature network for time series classification,” Information Sciences, 571, pp. 65–86. [Google Scholar]
- [18].LeCun Y, Bengio Y, and Hinton G (2015). Deep learning. Nature, 521(7553), 436–444. [DOI] [PubMed] [Google Scholar]
- [19].Xu Y, Mo T, Feng Q, Zhong P, Lai M, Eric I, and Chang C (2014). Deep learning of feature representation with multiple instance learning for medical image analysis. Proc. 2014 IEEE international conference on acoustics, speech and signal processing (ICASSP), IEEE, 1626–1630. [Google Scholar]
- [20].Pärnamaa T, and Parts L (2017). Accurate classification of protein subcellular localization from high-throughput microscopy images using deep learning. G3: Genes, Genomes, Genetics, 7(5), 1385–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nanni L, Ghidoni S, and Brahnam S (2017). Handcrafted vs. non-handcrafted features for computer vision classification, Pattern Recognition, 71, 158–172. [Google Scholar]
- [22].Pan SJ, and Yang QA (2010). A survey on transfer learning. IEEE Transactions on Knowledge and Data Engineering, 22(10), 1345–1359. [Google Scholar]
- [23].Weiss K, Khoshgoftaar TM, and Wang D (2016). A survey of transfer learning. Journal of Big data, 3(1), 9. [Google Scholar]
- [24].Tan CQ, Sun FC, Kong T, Zhang WC, Yang C, and Liu CF (2018). A survey on deep transfer learning. Artificial Neural Networks and Machine Learning - ICANN 2018, Pt Iii, 11141, 270–279. [Google Scholar]
- [25].Mabu S, Atsumo A, Kido S, Kuremoto T, and Hirano Y (2020). Investigating the effects of transfer learning on ROI-based classification of chest CT scan images: A case study on diffuse lung diseases. Journal of Signal Processing Systems 92(3), 307–313. [Google Scholar]
- [26].Sharif Razavian A, Azizpour H, Sullivan J, and Carlsson S (2014). CNN features off-the-shelf: an astounding baseline for recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, 806–813. [Google Scholar]
- [27].Donahue J, Jia Y, Vinyals O, Hoffman J, Zhang N, Tzeng E, and Darrell T (2014). DeCAF: A deep convolutional activation feature for generic visual recognition. Proc. International Conference on Machine Learning, 647–655. [Google Scholar]
- [28].Hur C, and Kang S (2020). On-device partial learning technique of convolutional neural network for new classes. Journal of Signal Processing Systems. 10.1007/s11265-020-01520-7 [DOI] [Google Scholar]
- [29].Yosinski J, Clune J, Bengio Y, and Lipson H (2014). How transferable are features in deep neural networks?. Proc. Advances in Neural Information Processing Systems, 3320–3328. [Google Scholar]
- [30].Bayramoglu N, and Heikkilä J (2014). Transfer learning for cell nuclei classification in histopathology images. Proc. European Conference on Computer Vision, 532–539. [Google Scholar]
- [31].Li ZZ, and Hoiem D (2018). Learning without forgetting. IEEE Transactions on Pattern Analysis and Machine Intelligence, 40(12), 2935–2947. [DOI] [PubMed] [Google Scholar]
- [32].Shia WC, and Chen DR (2021). Classification of malignant tumors in breast ultrasound using a pretrained deep residual network model and support vector machine. Computerized Medical Imaging and Graphics, 87, 101829. [DOI] [PubMed] [Google Scholar]
- [33].Hira ZM, and Gillies DF (2015). A review of feature selection and feature extraction methods applied on microarray data. Advances in Bioinformatics, 2015, 198363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Guo Y, Shi H, Kumar A, Grauman K, Rosing T, and Feris R (2018). SpotTune: transfer learning through adaptive fine-tuning. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 4805–4814. [Google Scholar]
- [35].Ali M, Son DH, Kang SH, and Nam SR (2017). An accurate CT saturation classification using a deep learning approach based on unsupervised feature extraction and supervised fine-tuning strategy. Energies, 10(11), 1830. [Google Scholar]
- [36].Boyd A, Czajka A, and Bowyer K (2019). Deep learning-based feature extraction in iris recognition: Use existing models, fine-tune or train from scratch? Proc. 2019 IEEE 10th International Conference on Biometrics Theory, Applications and Systems (BTAS), IEEE, 1–9. [Google Scholar]
- [37].Bai Y, Yi JY, Tao JH, Wen ZQ, and Fan CH (2020). A public Chinese dataset for language model adaptation. Journal of Signal Processing Systems, 92(8), 839–851. [Google Scholar]
- [38].Reyes AK, Caicedo JC, and Camargo JE (2015). Fine-tuning deep convolutional networks for plant recognition. CLEF (Working Notes), 1391, 467–475. [Google Scholar]
- [39].Zhou Z, Shin J, Zhang L, Gurudu S, Gotway M, and Liang J (2017). Fine-tuning convolutional neural networks for biomedical image analysis: actively and incrementally. Proceedings of the IEEE Conference on Computer Vision And Pattern Recognition, 7340–7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kensert A, Harrison PJ, and Spjuth O (2019). Transfer learning with deep convolutional neural networks for classifying cellular morphological changes. SLAS DISCOVERY: Advancing Life Sciences R&D, 24(4), 466–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sun C, Qiu X, Xu Y, and Huang X (2019). How to fine-tune BERT for text classification? Proc. China National Conference on Chinese Computational Linguistics, Springer, 194–206. [Google Scholar]
- [42].Swati ZNK, Zhao Q, Kabir M, Ali F, Ali Z, Ahmed S, and Lu J (2019). Brain tumor classification for MR images using transfer learning and fine-tuning. Computerized Medical Imaging and Graphics, 75, 34–46. [DOI] [PubMed] [Google Scholar]
- [43].Nazir M, Shakil S, and Khurshid K (2021). Role of deep learning in brain tumor detection and classification (2015 to 2020): A review. Computerized Medical Imaging and Graphics, 101940. [DOI] [PubMed] [Google Scholar]
- [44].Srivastava N, Hinton G, Krizhevsky A, Sutskever I, and Salakhutdinov R (2014). Dropout: A simple way to prevent neural networks from overfitting. Journal of Machine Learning Research, 15, 1929–1958. [Google Scholar]
- [45].Goodfellow I, Bengio Y, and Courville A (2016). Deep Learning, MIT press Cambridge. [Google Scholar]
- [46].Adelabu S, Mutanga O, and Adam E (2015). Testing the reliability and stability of the internal accuracy assessment of random forest for classifying tree defoliation levels using different validati on methods. Geocarto International, 30(7), 810–821. [Google Scholar]
- [47].Xu Y, and Goodacre R (2018). On splitting training and validation set: a comparative study of cross-validation, bootstrap and systematic sampling for estimating the generalization performance of supervised learning. Journal of Analysis and Testing, 2(3), 249–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hsu CW, and Lin CJ (2002). A comparison of methods for multiclass support vector machines. IEEE Transactions on Neural Networks, 13(2), 415–425. [DOI] [PubMed] [Google Scholar]
- [49].Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z, and IEEE. (2016). Rethinking the Inception Architecture for Computer Vision. Proceddings of the IEEE Conference on Computer Vision and Pattern Recognition, 2818–2826. [Google Scholar]
- [50].Dong N, Zhao L, Wu CH, and Chang JF (2020). Inception v3 based cervical cell classification combined with artificially extracted features. Applied Soft Computing, 93, 106311. [Google Scholar]
- [51].Xia XL, Xu C, Nan B, and IEEE. (2017). Inception-v3 for Flower Classification. 2nd International Conference on Image, Vision and Computing, 783–787. [Google Scholar]
- [52].Tian X, Chen C, and IEEE. (2019). Modulation Pattern Recognition Based on Resnet50 Neural Network. 2nd IEEE International Conference on Information Communication and Signal Processing, 34–38. [Google Scholar]
- [53].Wang C, Chen DL, Hao L, Liu XB, Zeng Y, Chen JW, and Zhang GK (2019). Pulmonary image classification based on inception-v3 transfer learning model. IEEE Access, 7, 146533–146541. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this paper is publicly available at https://github.com/mstfwsulab/AAV-classification