Abstract
Small molecule tools have enabled mechanistic investigations and therapeutic targeting of the protein kinase-like (PKL) superfamily. However, such tools are still lacking for many PKL members, including the highly conserved and disease-related UbiB family. Here, we sought to develop and characterize an inhibitor for the archetypal UbiB member, COQ8, whose function is essential for coenzyme Q (CoQ) biosynthesis. Guided by crystallography, activity assays, and cellular CoQ measurements, we repurposed the 4-anilinoquinoline scaffold to selectively inhibit human COQ8A in cells. Our chemical tool promises to lend mechanistic insights into the activities of these widespread and understudied proteins and to offer potential therapeutic strategies for human diseases connected to their dysfunction.
Introduction
The protein kinase-like (PKL) superfamily is an expansive set of more than 500 human proteins that perform critical roles in numerous biological processes1. Our understanding of these roles has been greatly advanced by the development of molecular tools capable of manipulating PKL activity2. Small molecule inhibition of PKL members has also emerged as a remarkably effective therapeutic strategy, with more than 70 new drugs approved in the past 20 years3,4. However, such tools and drugs are still lacking for many PKL members. Approximately 150 human PKL members are still considered to be understudied, including a large number of atypical kinases and pseudokinases5,6. Efforts to expand the repertoire of molecular tools for these proteins promise to further accelerate our understanding of their diverse functionality and expand the possibilities of manipulating their activities for therapeutic gain.
The UbiB proteins are a subset of atypical and understudied PKL family members. These proteins are conserved throughout all domains of life7 and, in eukaryotes, are found in cellular compartments responsible for the synthesis and distribution of prenylated metabolites8,9. The most well-studied UbiB members are the Saccharomyces cerevisiae protein Coq8p and its corresponding human homologs, COQ8A and COQ8B (collectively referred to here as COQ8). COQ8A and COQ8B each have well established connections to human disease, with inactivating mutations resulting in autosomal recessive cerebellar ataxia 210,11 and steroid-resistant nephrotic syndrome12, respectively. Early investigations identified COQ8 proteins as auxiliary factors required for the biosynthesis of coenzyme Q (CoQ)13,14, a redox-active prenyl lipid essential for biological processes including cellular respiration, pyrimidine biosynthesis, and cellular antioxidation15. COQ8 was initially proposed to be a protein kinase based on sequence homology, but structure/function studies of COQ8A revealed conserved features poised to prevent canonical protein kinase activity16. Instead, COQ8 may use its ATPase activity to maintain the integrity of a CoQ biosynthetic complex and to access hydrophobic CoQ intermediates embedded in the mitochondrial inner membrane17; however, this model requires further validation. Similar to other PKL members, the development of new molecular tools to manipulate COQ8 function could help elucidate its precise molecular function and establish it as a therapeutic target for human disease.
Previous efforts have set a foundation for the development of small molecular modulators of COQ818,19. We recently developed a covalent inhibitor against an engineered yeast Coq8p double mutant (M303C, V202C), but which lacked efficacy against the WT protein17. To our knowledge, no activators or inhibitors of WT COQ8 proteins have been validated in a biological context. Here, to address this limitation, we repurposed the 4-anilinoquinoline scaffold to create a custom endogenous inhibitor of COQ8A. This work expands the PKL small-molecule toolkit and offers resources to investigate the understudied UbiB family and its connection to human disease.
Results
4-Anilinoquin(az)olines bind and inhibit COQ8A in vitro
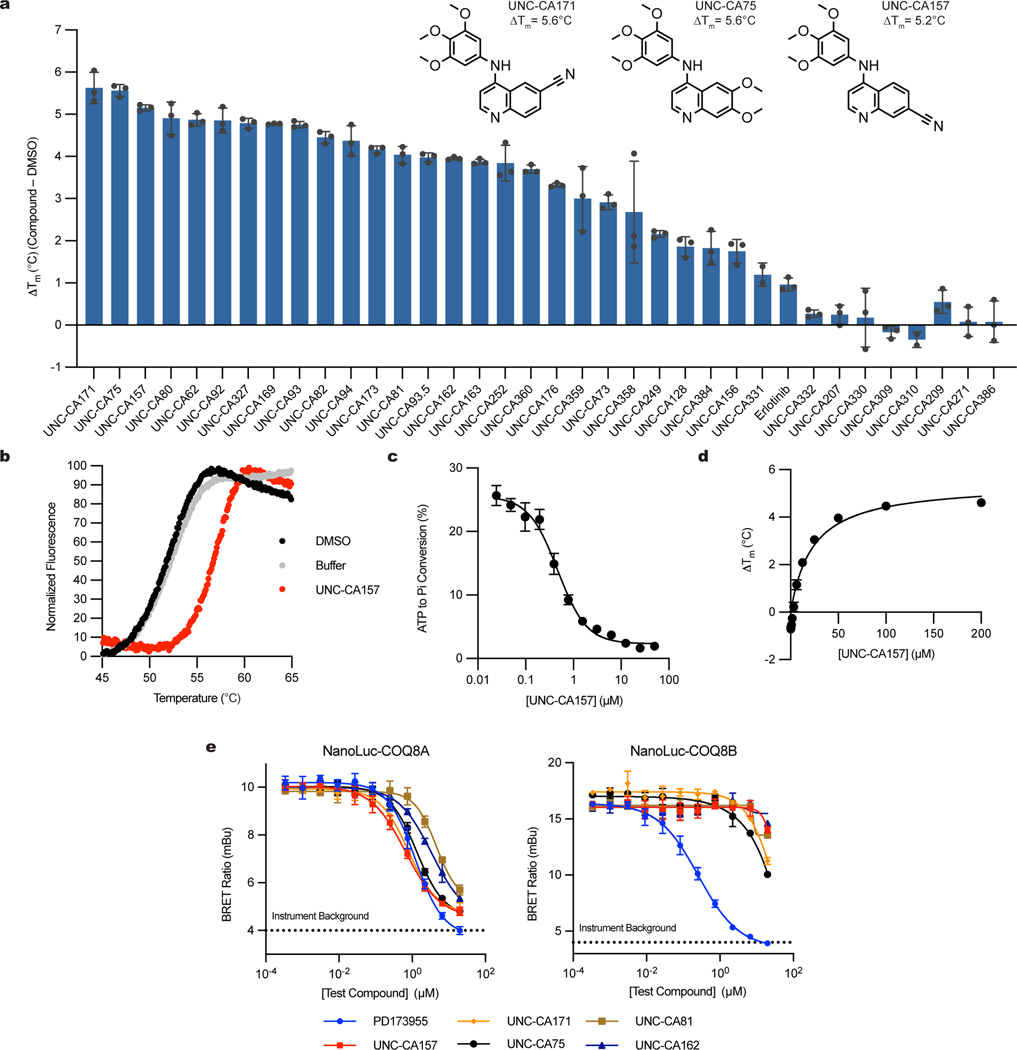
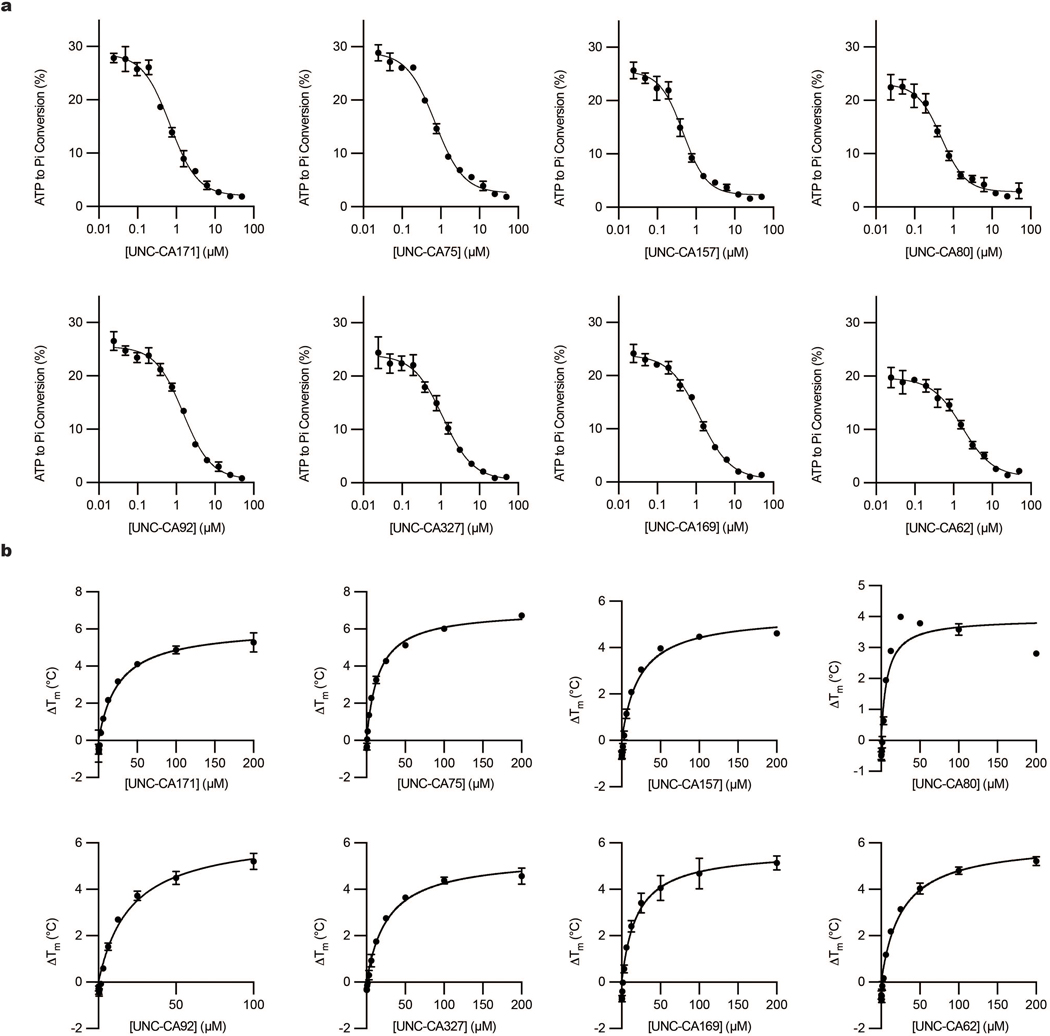
Functional investigations of COQ8 have been hindered by the lack of established inhibitors. To identify lead inhibitor compounds, we searched published kinase screening data and found COQ8A as a promising secondary target for 4-anilinoquinolines18,20,21. To further explore this scaffold, we tested the ability of a focused array of 35 distinct 4-anilinoquin(az)oline compounds to bind COQ8A by using differential scanning fluorimetry (DSF) (Fig. 1a, 1b). Twenty-five of these compounds increased COQ8A melting temperature significantly more than our weak-binding control, erlotinib. The strongest stabilizers featured methoxy groups on the aniline ring at the para- and both meta- positions and electronegative atoms at positions 6 or 7, suggesting important molecular features for COQ8A binding. We further tested the top eight compounds in an in vitro COQ8A ATPase activity assay in the presence of 2-PP and Triton X-100, compounds we previously found to enhance COQ8A activity17,19 (Fig. 1c, Extended Data Fig. 1a). All tested compounds inhibited COQ8ANΔ250 (which includes the full PKL domain and lacks only the mitochondrial targeting sequence and single-pass transmembrane domain)16 with IC50 values ranging from 0.47 to 1.7 μM. By DSF, these compounds possess Kd,app values ranging from 7.6 to 27 μM (Fig. 1d, Extended Data Fig. 1b).
Fig. 1 |. Characterization of in vitro COQ8A inhibitors.
a, Differential scanning fluorimetry (DSF) screen of 4-anilinoquin(az)oline compounds (n=3 independent samples from one experiment, mean ± SD). Structures of all compounds are available in the supporting information b, DSF traces of COQ8ANΔ250 with UNC-CA157 compared to controls. c, Inhibition of COQ8ANΔ250 ATPase activity by UNC-CA157 (n=3 independent samples from one experiment, mean ± SD). d, UNC-CA157 binding curve, as determined by DSF (n=3 independent samples from one experiment, mean ± SD). e, Live cell target engagement for COQ8A (left) and COQ8B (right) assessed in HEK293 cells using a NanoBRET approach (n=3 independent samples, mean ± SD, representative of two independent experiments). PD173955 served as a control. COQ8 BRET assay development data are available in the supporting information.
To test the ability of these compounds to engage COQ8A and COQ8B in live HEK293 cells, we developed a bioluminescence resonance energy transfer (BRET) assay based on the NanoLuc luciferase technology22. The NanoBRET target engagement assay includes COQ8A or COQ8B fused to an N-terminal NanoLuc and a COQ8 BRET probe (formerly Transfer Probe 2)23. In the absence of inhibitor the COQ8 BRET probe and fusion protein generated a detectable BRET signal. Competitive displacement of the COQ8 BRET probe from the ATP binding site by our inhibitor series resulted in a dose-dependent loss of BRET signal from which we determined IC50 values (Fig. 1e). With the caveat that N-terminal tagging likely prevents COQ8 mitochondrial localization, 4-anilinoquinolines showed a clear preference for COQ8A over COQ8B, and UNC-CA157 (1) showed the lowest IC50 against COQ8A at 580 nM. From these data, we selected UNC-CA157 as the most promising lead candidate for further development.
UNC-CA157 slightly inhibits CoQ production in COQ8BKO cells
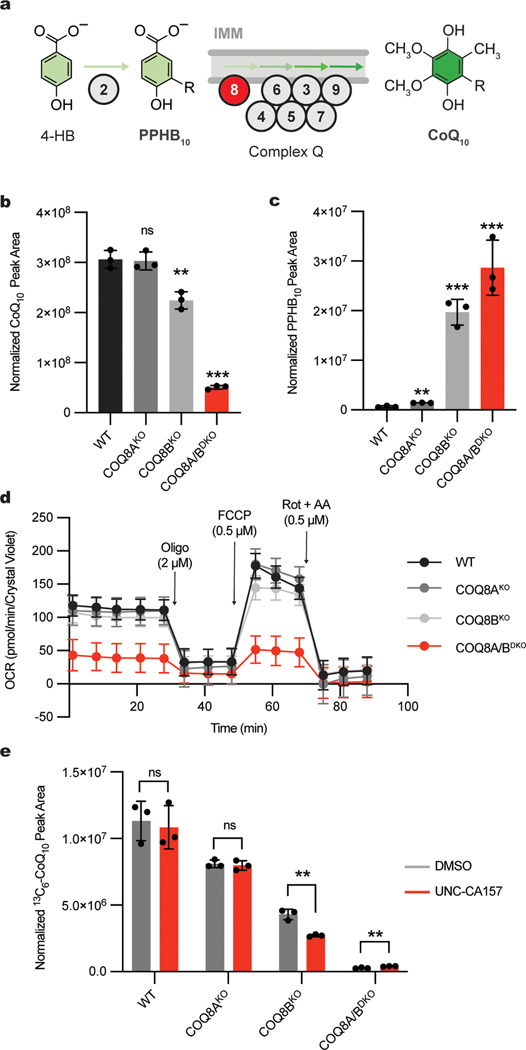
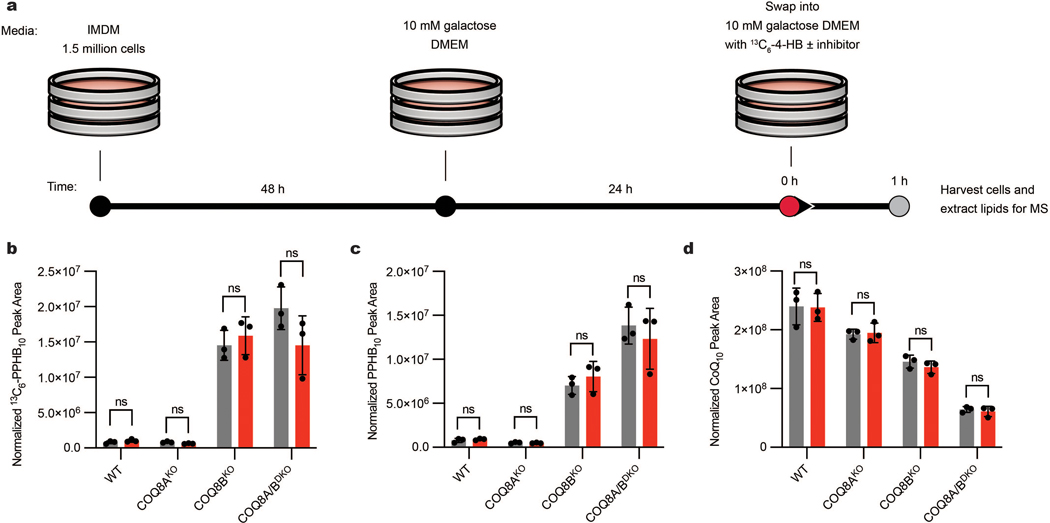
We next established a system to test the efficacy of our inhibitor in human HAP1 cells. In humans, COQ8A and COQ8B are paralogs that likely possess redundant functions in the CoQ biosynthesis pathway and that exhibit some tissue specificity12,24,25. In a recent study26, we determined that both paralogs are active in HAP1 cells, and that COQ8B is the more dominant version whose disruption causes the accumulation of the early CoQ precursor, polyprenyl hydroxybenzoate (PPHB10) (Fig. 2a). We performed mass spectrometry measurements and confirmed that the COQ8AKO HAP1 cells had no significant change in cellular CoQ10 levels, whereas the COQ8BKO cells had slight CoQ10 loss (Fig. 2b). Importantly, the double knockout line (COQ8A/BDKO) exhibited marked loss of CoQ10, demonstrating that each paralog can largely support the pathway in the absence of the other. The concomitant buildup of PPHB10 followed the same phenotypic pattern, with the largest increase seen in the COQ8A/BDKO cells (Fig. 2c). We further measured the impact of our COQ8 protein knockouts on cellular respiration by using Seahorse respirometry and again found that disrupting both COQ8 paralogs produced the strongest effect on basal and uncoupled oxygen consumption rates (OCR) (Fig. 2d). Together, these data demonstrate that both COQ8A and COQ8B are active in HAP1 cells, with COQ8B serving as the predominant paralog, thereby presenting a system to test the efficacy and specificity of our inhibitor.
Fig. 2 |. UNC-CA157 decreases de novo CoQ production in COQ8BKO HAP1 cells.
a, Abbreviated schematic of the CoQ biosynthesis pathway. COQ8 proteins are essential for converting PPHB10 to mature CoQ10. b, Total CoQ10 levels in WT, COQ8AKO, COQ8BKO, and COQ8A/BDKO HAP1 cells as determined by LC-MS (n=3 independent samples from one experiment, mean ± SD) (two sided students t-test, no adjustment for multiple comparisons, **p = 0.0048 WT vs. COQ8BKO, ***p = 0.00002 for WT vs. COQ8A/BDKO). c, Total PPHB10 levels in WT, COQ8AKO, COQ8BKO, and COQ8A/BDKO HAP1 cells as determined by LC-MS (n=3 independent samples from one experiment, mean ± SD) (two sided students t-test, no adjustment for multiple comparisons, **p = 0.0014 WT vs. COQ8AKO, ***p = 0.0002 WT vs. COQ8BKO, ***p = 0.0009 WT. vs. COQ8A/BDKO). d, Oxygen consumption rates from a Seahorse mitochondria stress test of WT, COQ8AKO, COQ8BKO, and COQ8A/BDKO HAP1 cells (n=11 (WT) or 12 (COQ8AKO, COQ8BKO, COQ8A/BDKO) independent wells of cells from one experiment, mean ± SD). Oligo, oligomycin; FCCP, carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone; Rot + AA, rotenone + antimycin A. e, 13C6-CoQ10 (de novo synthesized CoQ10) levels in WT, COQ8AKO, COQ8BKO, and COQ8A/BDKO HAP1 cells after treatment with 10 μM 13C6-4-HB and either DMSO or 20 μM UNC-CA157 (n=3 independent samples from one experiment, mean ± SD) (two sided students t-test, no adjustment for multiple comparisons, **p = 0.0024 COQ8BKO, **p = 0.0062 COQ8A/BDKO).
To begin, we measured the impact of UNC-CA157 on de novo CoQ production. We treated WT, COQ8AKO, COQ8BKO, and COQ8A/BDKO HAP1 cells with either 20 μM UNC-CA157 or matched DMSO and monitored the incorporation of the CoQ precursor 13C6-4-hydroxybenzoate (13C6-4-HB) into CoQ biosynthesis via liquid chromatography-mass spectrometry (LC-MS) (Extended Data Fig. 2a). Incorporation of the labeled 4-HB precursor followed the same phenotypic pattern observed in our cellular CoQ measurements, with single COQ8 knockouts showing decreased 13C6-CoQ10, and a further depletion when both paralogs were disrupted (Fig. 2e). Treatment with UNC-CA157 resulted in a significant decrease in 13C6-CoQ10 production only in the COQ8BKO, where cells are solely reliant on the COQ8A paralog for CoQ biosynthesis. No increase in 13C6-PPHB10 or unlabeled PPHB10 was seen, suggestive of a relatively weak inhibition (Extended Data Fig. 2b,c). Unsurprisingly, this short incubation with UNC-CA157 had no effect on unlabeled CoQ10 levels, given the long half-life of CoQ10 (Extended Data Fig. 2d). This directly demonstrated COQ8A inhibition in a biological context and set the foundation for further probe development around this scaffold.
UNC-CA157 is a type I kinase inhibitor
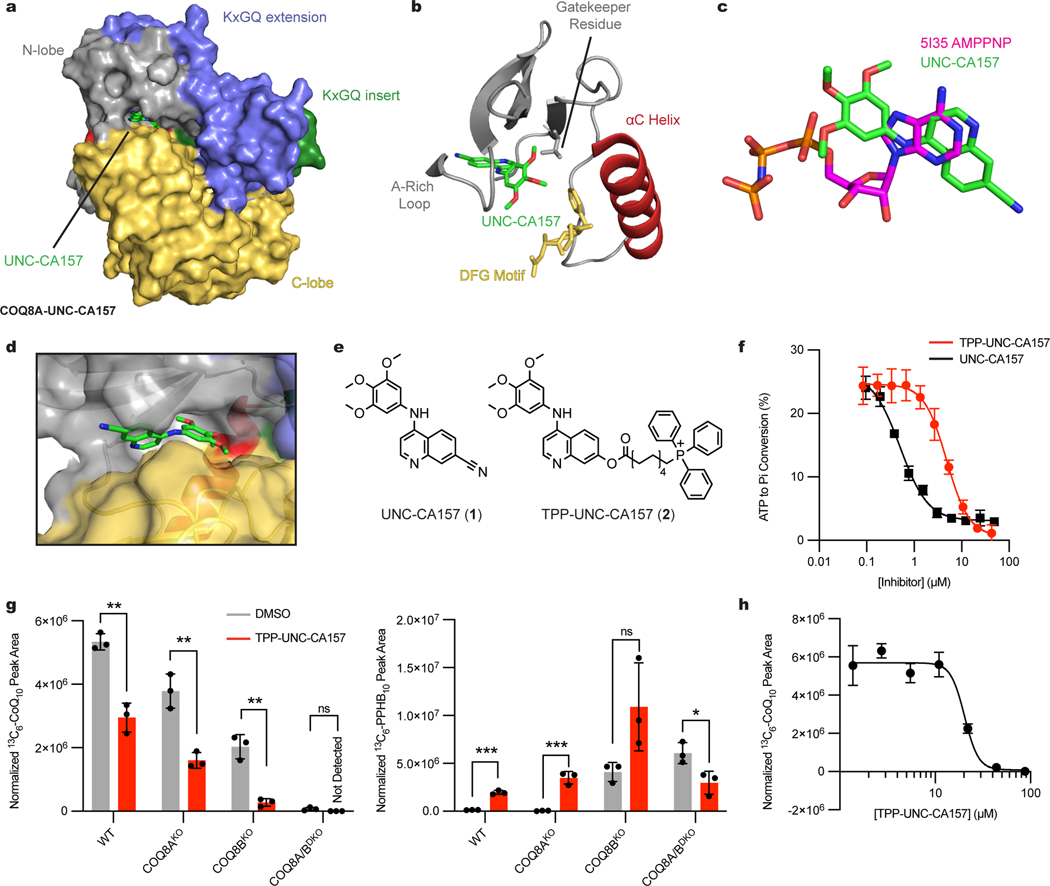
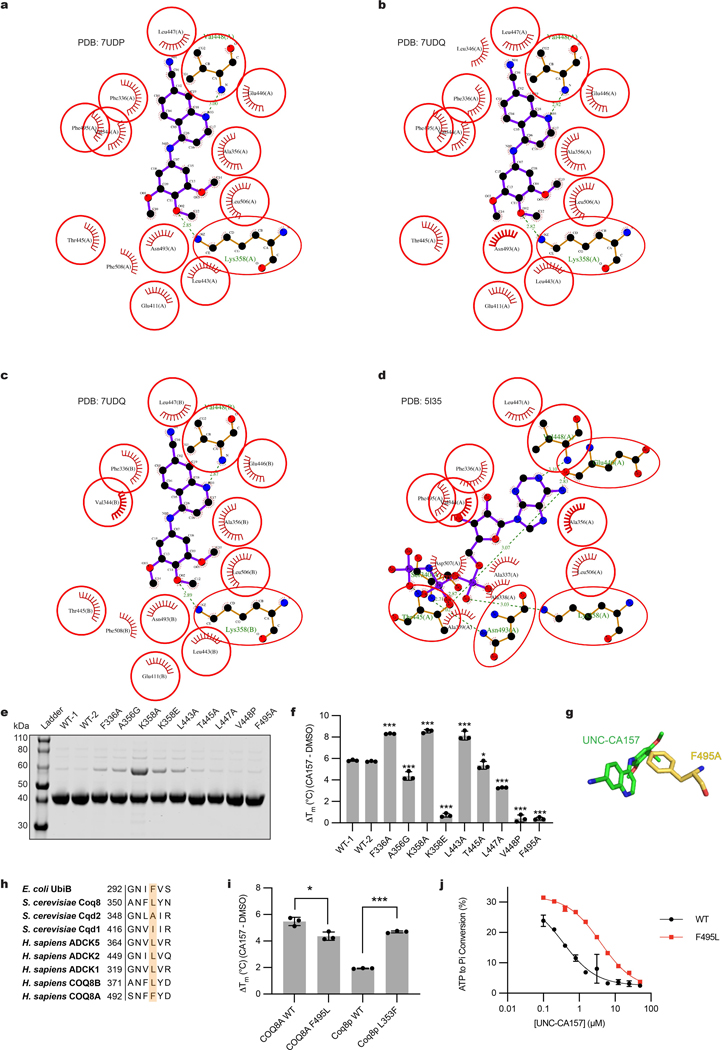
To further investigate the inhibitor-protein interaction, we purified COQ8ANΔ254, crystallized it in the presence of UNC-CA157, and solved two inhibitor-bound crystal structures (PDB=7UDP and 7UDQ) (Fig. 3a). The structures revealed that UNC-CA157 is a type I kinase inhibitor that binds to the COQ8A ATP pocket with the αC helix in the ‘in’ conformation and the DFG motif in an ‘intermediate’ conformation (Fig. 3b). Superimposing the UNC-CA157-bound structure with our previous nucleotide-bound structure (PDB=5I35)11 reveals that the quinoline ring from UNC-CA157 overlays with the adenosine ring from AMP-PNP (Fig. 3c). Additionally, the two ligands share common hydrogen bonding and hydrophobic interaction partners (Extended Data Fig. 3a–d), further suggesting direct competition with ATP as the mechanism of inhibition. The UNC-CA157-bound structures largely mimic the nucleotide-bound state of COQ8A, resulting in similar modifications to the KxGQ domain—the unique and invariant domain of the UbiB family—when compared to the apo form11.
Fig. 3 |. Mitochondrial targeting of TPP-UNC-CA157 increases cellular efficacy in HAP1 cells.
a, Surface representation of UNC-CA157-bound COQ8ANΔ254. b, UNC-CA157 bound to the protein active site reveals type I kinase inhibition. c, Overlay of AMPPNP (PDB=5I35) and UNC-CA157 in the COQ8A active site. d, UNC-CA157 positioning in the COQ8A active site revealed an aqueous-exposed cyano group. e, The addition of a triphenylphosphonium group to UNC-CA157 for mitochondrial targeting. f, Inhibition of in vitro COQ8ANΔ250 ATPase activity by UNC-CA157 and TPP-UNC-CA157 (n=3 independent samples from one experiment, mean ± SD). g, 13C6-CoQ10 (de novo synthesized CoQ10) and 13C6-PPHB10 (de novo synthesized PPHB10) levels in WT, COQ8AKO, COQ8BKO, and COQ8A/BDKO HAP1 cells after treatment with 10 μM 13C6-4-HB and either DMSO or 17.6 μM TPP-UNC-CA157 (n=3 independent samples from one experiment, mean ± SD) (two sided students t-test, no adjustment for multiple comparisons, **p = 0.0014 WT 13C6-CoQ10, **p = 0.0031 COQ8AKO 13C6-CoQ10, **p = 0.0016 COQ8BKO 13C6-CoQ10, ***p = 0.0001 WT 13C6-PPHB10, ***p = 0.0008 COQ8AKO 13C6-PPHB10, *p = 0.0290 COQ8A/BDKO 13C6-PPHB10). h, 13C6-CoQ10 levels in WT HAP1 cells after treatment with 10 μM 13C6-4-HB and 1.37–87.8 μM TPP-UNC-CA157 (n=3 independent samples from one experiment, mean ± SD).
COQ8A and COQ8B exhibit high sequence similarity; however, the kinase screening data from Asquith et al.20 and our cellular data above indicate that UNC-CA157 possesses specificity for COQ8A. To investigate what enables this specificity, we mutated select COQ8ANΔ250 active site residues that are in close proximity to UNC-CA157 in our crystal structures and purified the corresponding protein (Extended Data Fig. 3e). These mutations yielded variable effects, with some leading to slightly enhanced stabilization by UNC-CA157 and three causing near complete loss of stabilization as assessed by DSF (Extended Data Fig. 3f). Among these three, the F495A mutant stood out as particularly noteworthy given that this residue is not conserved among the human and yeast UbiB proteins, with all but COQ8A having leucine at this position (Extended Data Fig. 3g, 3h). We further tested this residue by purifying human COQ8ANΔ250 WT and F495L (converting phenylalanine to the typical residue found at this position within the family), along with S. cerevisiae Coq8pNΔ41 WT and L353F (introducing a phenylalanine into a homolog that lacks it). Substitution of this phenylalanine in the human protein again diminished binding and, notably, introduction of this residue into the yeast protein more than doubled inhibitor-induced thermal stability (Extended Data Fig. 3i). Additionally, we tested UNC-CA157 inhibition of human WT and F495L COQ8ANΔ250 ATPase activity and observed a greater than 10-fold increase in IC50 for the F495L mutant (Extended Data Fig. 3j). Overall, these results identify a COQ8A-specific phenylalanine required for UNC-CA157 potency.
TPP-UNC-CA157 inhibits CoQ biosynthesis in cells
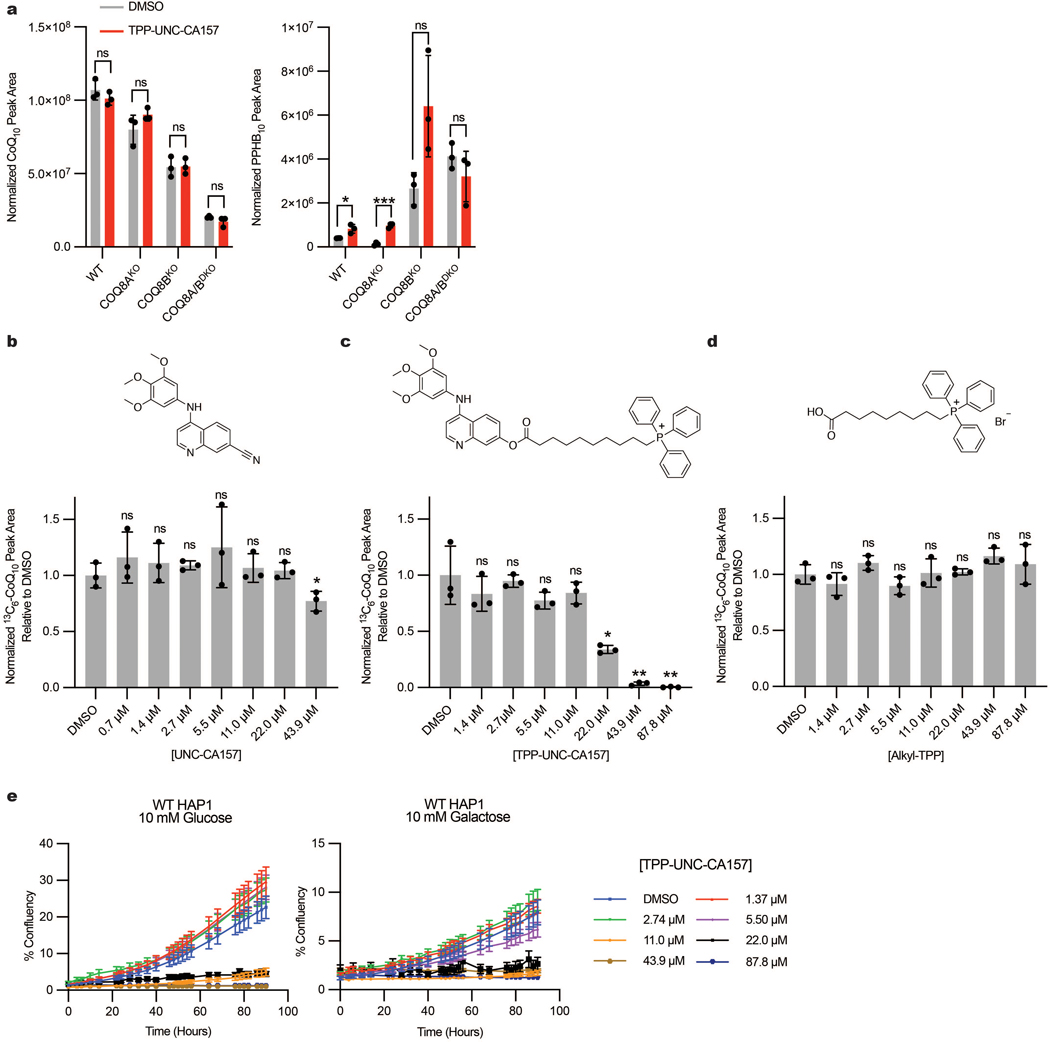
A further motivation for crystallizing UNC-CA157-bound COQ8A was to inform medicinal chemistry efforts toward improving inhibitor efficacy. COQ8A is the only known PKL protein that both resides in mitochondria and binds to 4-anilinoquin(az)olines20. Therefore, targeting UNC-CA157 to the mitochondrial matrix is a reasonable strategy to improve our compound specificity. The most common strategy for targeting a molecule to the mitochondrial matrix is to amend it with a lipophilic cation, such as triphenylphosphonium (TPP)27. Based on membrane potential and cellular conditions, greater than 1000-fold enrichments in mitochondria can be achieved using this strategy28. Our structure reveals that the cyano group of UNC-CA157 is positioned in the solvent-exposed portion of the ATP binding site and is, therefore, well positioned for further modification (Fig. 3d). We derivatized UNC-CA157 by replacing the cyano group with a carboxylate ester followed by a 9-carbon linker and TPP (Fig. 3e)29. This compound, TPP-UNC-CA157 (2), also inhibited COQ8ANΔ250 ATPase activity in vitro, albeit with a lower potency than UNC-CA157 alone (Fig. 3f). Despite this, we reasoned that the enrichment of this compound in the matrix should overcome the decreased potency and simultaneously improve its specificity by increasing its local cellular concentration.
Treatment of HAP1 cells with TPP-UNC-CA157 resulted in a robust reduction in de novo CoQ production, with significant decreases observed in WT, COQ8AKO, and COQ8BKO cell lines (Fig. 3g). TPP-UNC-CA157 treatment also led to a reciprocal increase in 13C6-PPHB10 levels, confirming specific disruption of the headgroup modification portion of the CoQ biosynthetic pathway (Fig. 3g). This compound likewise appears to be more efficacious against COQ8A than COQ8B, with the strongest phenotype observed in the COQ8BKO cells. We again observed no changes in unlabeled CoQ10 but, owing to the low endogenous levels, a buildup of unlabeled PPHB10 was observed (Extended Data Fig. 4a). To determine the potency of our newfound inhibitor, we treated WT HAP1 cells with various concentrations of TPP-UNC-CA157 and were able to abolish completely de novo CoQ production in a dose-dependent manner (Fig. 3h). Both UNC-CA157 and an alkyl-TPP control showed lower efficacy compared to TPP-UNC-CA157 (Extended Data Fig. 4b–d). This compound is immediately useful as a tool for interrogating the CoQ biosynthesis pathway and, more broadly, demonstrates the promise of leveraging organellar targeting moieties for selective inhibition of mitochondrial processes. Nonetheless, owing to observed toxicity of TPP-UNC-CA157 at low μM concentrations, similar to that of alkyl-TPP alone30,31, further modifications will be needed before exploring the utility of this compound in higher organisms (Extended Data Fig. 4e).
Specificity of TPP-UNC-CA157 Across the Kinome
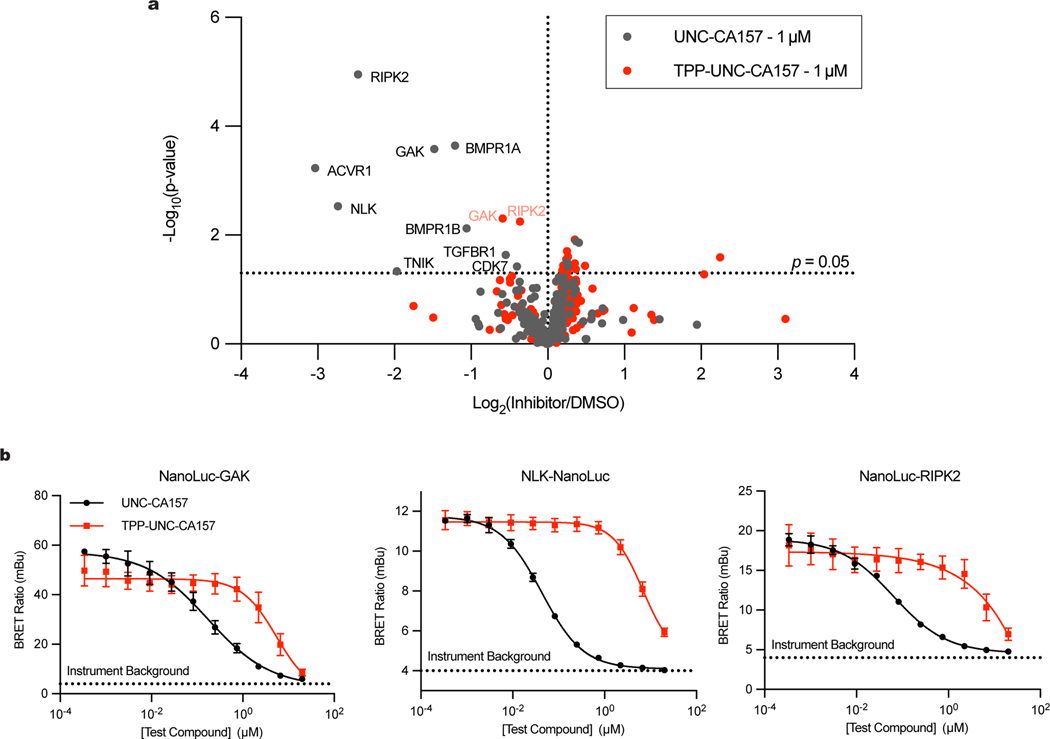
To determine the specificity of our inhibitor, we assessed the kinome profiles for UNC-CA157 and TPP-UNC-CA157 using a multiplexed kinase inhibitor bead set and quantitative mass spectrometry (MIBS-MS) (Fig. 4a)32–35. The MIBS-MS analysis captured 234 kinases from cell lysate. From among these, interactors were determined by the extent to which either UNC-CA157 or TPP-UNC-CA157 at 1 μM competitively displaced kinases from the beads. Both UNC-CA157 and TPP-UNC-CA157 showed narrow spectrum profiles across the kinome demonstrating that these compounds are relatively specific. COQ8A and COQ8B were not detected in this experiment. Potential off-targets that were detected in the MIBS-MS experiment include the cytosolic kinases GAK, RIPK2, and NLK, all of which have previously been shown to bind the 4-anilinoquinoline scaffold20,33–36.
Fig. 4 |. TPP-UNC-CA157 Kinome Specificity.
a, MIBS-MS kinome profiles for UNC-CA157 and TPP-UNC-CA157 (p-values determined using two sided students t-test, no adjustment for multiple comparisons). Full annotated kinome profiles are available in the supporting information. b, Representative NanoBRET assays for off-target cytosolic kinases GAK, NLK, and RIPK2 (n=3 independent samples, mean ± SD, representative of two independent experiments). Live cell target engagement summary table for COQ8A, COQ8B, ACVR1, GAK, NLK, PKN3, and RIPK2 using the NanoBRET approach with UNC-CA157 and TPP-UNC-CA157 is available in the supporting information.
Based on the MIBS-MS profiles, UNC-CA157 is more promiscuous in cell lysate than TPP-UNC-CA157. Additionally, mitochondrial targeting by TPP in intact cells should further alleviate cytosolic off-target effects. To validate this, we performed NanoBRET target engagement assays with UNC-CA157 and TPP-UNC-CA157 for COQ8A, COQ8B, and cytosolic kinases that were displaced in our MIBS-MS data (ACVR1, GAK, NLK, RIPK2) or that were previously shown to bind this scaffold (PKN3)35. COQ8B, ACVR1, and PKN3 all showed relatively weak affinity for our inhibitors in this experiment. Target engagement potency decreased for GAK, NLK, and RIPK2 upon addition of TPP, demonstrating that mitochondrial targeting alleviates cytosolic off-target effects (Fig. 4b). Decreased target engagement was also observed for NanoLuc-COQ8A with the addition of TPP, likely due to mislocalization of the protein to the cytosol by the addition of the N-terminal tag.
Discussion
In this work, we characterized a series of COQ8A inhibitors in vitro, solved inhibitor-bound crystal structures, specifically targeted our lead candidate to mitochondria, and demonstrated inhibition of endogenous COQ8 function in a biological context. This work sets a foundation for further chemical probe development against COQ8 proteins and the poorly characterized UbiB protein family at large. Small molecule tools have been instrumental in the biological characterization of PKL family members2,37. Development of specific inhibitors has informed kinase-mediated signaling and resulted in numerous therapeutically relevant molecules3. Kinase inhibition often does not phenocopy genetic knockouts, which has also enabled interrogation of protein functions independent of canonical phosphorylation or ATP binding5. Given the atypical nature of UbiB proteins and a lack of direct evidence for trans phosphorylation, development of chemical tools to modulate their activity in cells is sure to aid in elucidating their precise biological functions.
Although cells rely on CoQ biosynthesis for respiration and other metabolic processes15, there are potential benefits to inhibition18. Limitations to CoQ production through genetic or dietary intervention increase lifespan in C. elegans38 and mice39. CoQ is also used by the oxidoreductase FSP1 to help inhibit iron-mediated cell death in cancer40,41, suggesting that cell-specific suppression of CoQ biosynthesis may be an effective anti-cancer strategy. Furthermore, some established disease-related COQ8A mutations are likely to be gain-of-function variants10, suggesting that COQ8A inhibition could be therapeutically beneficial in select contexts. Last, there is still much to be learned about how the CoQ biosynthetic proteins interact and function, and specific inhibitory tools could aid in these investigations. Other inhibitors against CoQ biosynthesis were developed previously, including 4-nitrobenzoate (an inhibitor of COQ2)42, clioquinol (an inhibitor of COQ7)43, pyrazole derivatives (inhibitors of COQ7)44, oxazinoquinoline derivatives (inhibitors of COQ7 in Trypanosoma cruzi)45, and the osteoporosis drug zoledronic acid46. However, previously identified inhibitors are administered on the timescale of days, whereas TPP-UNC-CA157 presents a unique opportunity to perturb CoQ biosynthesis acutely and rapidly. This molecule has immediate use as a research tool to investigate the pathway, but toxicity issues at concentrations that are efficacious will need to be overcome before moving towards clinical application.
Our combined biochemical, genetic, and cellular data suggest that TPP-UNC-CA157 has high specificity. However, we have not yet established a drug-resistant COQ8 mutant. Given that UNC-CA157 and ATP share a binding pocket and many hydrogen bonding and hydrophobic interaction partners, generating this mutant will be difficult to do without negatively impacting the endogenous protein function. However, such a mutant may prove critical for ascribing new functionality to COQ8, and drug-resistant mutants have been generated for other members of the PKL family47. Our previously developed inhibitor against a genetically modified Coq8p (V202C, M303C) in S. cerevisiae has the benefit of lacking efficacy against the WT Coq8p protein, clearly demonstrating specificity. This specific inhibitor against a mutant yeast protein and the relatively specific inhibitor against the endogenous human homolog developed here form a useful set of initial tools for further investigating this protein family.
In this work, we utilized a triphenylphosphonium moiety to target our small molecule inhibitor specifically to the mitochondrial matrix. This is a well-established strategy for mitochondrial targeting, most prominently for the mitochondrial antioxidant MitoQ48. Given the role of mitochondria as central hubs of metabolism, specific organellar targeting offers a wide range of potential applications to modulate cellular function. However, there is a growing awareness for the toxicity of TPP compounds30,31. Negative impacts of TPP compounds on bioenergetics were reported in the low μM range, including for MitoQ31. Efforts are underway to decrease this toxicity, including by developing modified lipophilic cations49, which could inform future efforts to improve our TPP-UNC-CA157 inhibitor.
UbiB proteins are found in all organisms and have established associations with prenyl lipid biology. In eukaryotes, the COQ8 proteins are likely ATPases required for CoQ biosynthesis; however, specifics regarding how this activity is coupled to CoQ production remain unknown17. More recently, the yeast intermembrane space UbiB proteins were connected to CoQ distribution throughout the cell, but their precise mechanism of action also remains to be determined50. Here, we demonstrate direct inhibition of an endogenous UbiB protein, expanding the molecular toolkit for the PKL superfamily and providing resources for establishing how auxiliary proteins assist the biosynthesis of CoQ. Beyond COQ8, we anticipate that these chemical tools will form the basis for selecting an even larger set of molecules that will enable mechanistic studies into the functions of the widespread UbiB protein family.
Methods
COQ8ANΔ250 General Expression and Purification
The general purification method has been documented previously16. COQ8ANΔ250 constructs were overexpressed in E. coli by autoinduction. Cells were isolated and frozen at −80 oC until further use. For protein purification, cells were thawed and resuspended in Lysis Buffer [20 mM HEPES (pH 7.2), 300 mM NaCl, 10% glycerol, 5 mM 2-mercaptoethanol (BME), 0.25 mM phenylmethylsulfonyl fluoride (PMSF)] (4 oC). The cells were lysed by sonication (4 oC, 75% amplitude, 20 s x 2). The lysate was clarified by centrifugation (15,000 g, 30 min, 4 oC). The cleared lysate was mixed with cobalt IMAC resin (Talon resin) and incubated (4 oC, 1 h). The resin was pelleted by centrifugation (700 g, 2 min, 4 oC) and washed four times with Wash Buffer [20 mM HEPES (pH 7.2), 300 mM NaCl, 10% glycerol, 5 mM BME, 0.25 mM PMSF, 10 mM imidazole]. His-tagged protein was eluted with Elution Buffer [20 mM HEPES (pH 7.2), 300 mM NaCl, 10% glycerol, 5 mM BME, 100 mM imidazole]. The eluted protein was concentrated with a MW-cutoff spin filter (50 kDa MWCO) and exchanged into Storage Buffer [20 mM HEPES (pH 7.2), 300 mM NaCl, 10% glycerol, 5 mM BME]. The concentration of 8His-MBP-[TEV]-COQ8ANΔ250 was determined by its absorbance at 280 nm (ε = 96,720 M−1cm−1)(MW=89.9 kDa). The fusion protein was incubated with Δ238TEV protease (1:50, TEV:fusion protein, mass:mass)(1 h, RT). The TEV protease reaction mixture was mixed with cobalt IMAC resin (Talon resin) and incubated (4 oC, 1 h). The resin was pelleted by centrifugation (700 g, 2 min, 4 oC). The unbound COQ8A was collected and concentrated with a MW-cutoff spin filter (30 kDa MWCO) and exchanged into storage buffer. The concentration of COQ8ANΔ250 was determined by Bio-Rad Protein Assay according to manufacturer protocol. The protein was aliquoted, frozen in N2 (l), and stored at −80 oC. Fractions from the protein preparation were analyzed by SDS-PAGE and imaged using a LI-COR Odyessey CLx with the Image Studio v5.2 software. Coq8pNΔ41 and mutants were also purified using this general method.
General 4-Anilinoquin(az)oline Synthesis
4-Chloroquin(az)oline derivative (150 mg, 0.67 mmol) and aniline (0.74 mmol) were suspended in ethanol (10 mL) and heated to reflux for 16 h. The crude mixture was purified by flash chromatography using EtOAc/hexane followed by 1–5% methanol in EtOAc (or by re-crystallization). After solvent removal under reduced pressure, the product was obtained. All compounds were >98% pure by 1H/13C NMR and LC-MS and melting points were consistent with previous reports. All compounds in the initial DSF screen have been previously described20,33,34,36,51,52. Full structures for all compounds in the DSF screen can be found in the supplemental information.
Differential Scanning Fluorimetry
The general DSF method has been documented previously17. For screening 4-anilinoquin(az)olines 20 μL reactions containing 10 μM COQ8ANΔ250, 100 μM inhibitor from 10 mM stock in DMSO, 5 mM MgCl2, 50 mM HEPES pH 7.5, 75 mM NaCl, and 4x Sypro Orange dye (Thermo S6651) were made in MicroAmp Optical 96-well reaction plates (Thermo N8010560), centrifuged (200 g, RT, 30 sec) and incubated at room temperature in the dark for 10 min. Fluorescence was then monitored with the ROX filter using a QuantStudio 6 Real-Time PCR system (QuantStudio Real-Time pCR v1.2 software) along a temperature gradient from 4–95 °C at a rate of 0.025 °C/s. Protein Thermal Shift software v1.3 (Applied Biosystems) was used to determine Tm values by fitting to a Boltzmann model. Melt curves flagged by the software were manually inspected. Two data points were omitted after inspection (UNC-CA331, UNC-CA310) resulting in only duplicate measurements reported for those compounds. Each inhibitor was run in triplicate and error bars represent SD. ΔTm values were determined by subtracting the average of matched vehicle controls from the same plate. UNC-CA157 binding to COQ8ANΔ250 and Coq8pNΔ41 WT and active site mutants were also analyzed using this method.
To determine the inhibitor Kd,app, 20 μL reactions contained 1 μM COQ8ANΔ250, 0–200 μM inhibitor with a final concentration of 2% DMSO, 4 mM MgCl2, 100 mM HEPES pH 7.5, 150 mM NaCl, and 4x Sypro Orange dye. Samples were run and analyzed as described above. ΔTm values were plotted against [inhibitor] in GraphPad Prism Version 8.4.3 and fit to nonlinear regression (one site specific binding) to determine Kd,app values. The experiments were performed in triplicate and error bars represent SD. Error values in Kd,app values represent 95% confidence intervals.
ATPase Assay
ATPase assays for COQ8ANΔ250 were performed essentially as described in Reidenbach et al17. The reaction was initiated by adding a mixture of ATP (Promega V703), 2-propylphenol (Sigma W352209), Triton X-100 (Sigma T9284), and indicated inhibitors from stocks in DMSO (Sigma D2650) in Reaction Buffer [100 mM HEPES pH 7.5, 4 mM MgCl2, 150 mM NaCl] to purified COQ8ANΔ250 also in reaction buffer. Final reaction conditions were 100 μM ATP, 1 mM 2-PP, 0.5 μM COQ8ANΔ250, 1 mM Triton X-100, and 0–50 μM indicated inhibitor with equivalent DMSO vehicle. The final reaction volume was 15 μL. Reactions were performed in clear 384 well plates (Fisher 12565506). Once the reactions were started and mixed, plates were sealed and incubated at 30°C for 45 min. Reactions were then quenched with 35 μL of Cytophos™ reagent (Cytoskeleton, Inc. BK054), incubated at RT for 10 min, and absorbance was read at 650 nm in a Biotek Cytation 3 plate reader with the Gen5 v3.02.2 software. Absorbance was converted to concentration of inorganic phosphate with a 0–50 μM standard curve run in parallel using the phosphate standard provided in the Cytophos™ kit diluted in reaction buffer. Experiments were performed in triplicate and error bars on the graph represent SD. To determine IC50 values the data were plotted in GraphPad Prism Version 8.4.3 and fit to a nonlinear regression ([inhibitor] vs. response, variable slope, four parameters). Indicated error in IC50 values represents 95% confidence intervals.
Cell Transfections and BRET Measurements
For cellular BRET measurements, N- or C-terminal NanoLuc/Kinase fusions were encoded in pFN31K expression vectors (Promega), including flexible Gly-Ser-Ser-Gly linkers between Nluc and each full-length kinase. For COQ8A and COQ8B open reading frames both corresponded to UNIPROT Isoform 1 (Q8NI60 and Q96D53, respectively). The N-terminal fusions to NanoLuc to COQ8A and COQ8B were found to be optimal for BRET and used for assay development and compound potency testing. For all other cellular kinase TE assays, the fusion vectors were obtained from Promega. HEK-293 cells were transfected with NanoLuc fusion constructs using FuGENE HD (Promega) according to the manufacturer’s protocol. Briefly, fusion constructs were diluted together into Transfection Carrier DNA (Promega) at a mass ratio of 1:9 (mass/mass/mass) in Opti-MEM (Gibco), after which FuGENE HD was added at a ratio of 1:3 (μg DNA: μL FuGENE HD). For example, for a 1 mL size transfection complex, 1μg of the NanoLuc fusion DNAs was combined with 9μg Transfection Carrier DNA in 1 mL of Opti-MEM. 1 part (vol) of FuGENE HD complexes thus formed were combined with 20 parts (vol) of HEK-293 cells suspended at a density of 2 × 105 per mL in Opti-MEM containing 1% (v/v) FBS. Briefly, transfected cell suspensions were seeded at 100 μL per well (20,000 cells/well) into white tissue culture-treated assay plates (Corning CAT# 3917) followed by incubation in a humidified, 37 °C/5% CO2 incubator for 18−24 h. The total concentrations for NanoLuc® fusion plasmids were 5 ng/well, and the total concentration of DNA was 50 ng/well. All chemical inhibitors were prepared as concentrated stock solutions in DMSO (Sigma-Aldrich) and diluted in Opti-MEM (unless otherwise noted) to prepare working stocks. Cells were equilibrated with BRET probe and test compound prior to BRET measurements, with an equilibration time of 2 h unless otherwise noted. BRET probes were prepared first at a stock concentration of 100X in DMSO, after which the 100X stock was diluted to a working concentration of 20X in BRET probe dilution buffer (12.5 mM HEPES, 31.25% PEG-400, pH 7.5). For COQ8 BRET probe dose response measurements, the COQ8 BRET probe (formerly Transfer Probe 2)23 was added to the cells in an 8 point, 2-fold dilution series starting at a final concentration of 1 μM. For target engagement analysis of COQ8A and COQ8B, the COQ8 BRET probe was added to the cells at a final concentration of 1 μM for COQ8A and 0.25 μM for COQ8B. For all other kinases, BRET probes (tracers) were added according to the manufacturer’s recommendations (GAK, Tracer K-10, 0.031 μM; RIPK2, Tracer K-4, 0.13 μM; NLK, Tracer K-10, 0.5 μM; ACVR1, Tracer K-11, 0.078 μM; PKN3, Tracer K-5, 2 μM). To measure BRET, NanoBRET Target Engagement Substrate (Promega) and Extracellular NanoLuc Inhibitor (Promega) were added according to the manufacturer’s recommended protocol, and filtered luminescence was measured on a GloMax Discover luminometer equipped with 450 nm BP filter (donor) and 600 nm LP filter (acceptor), using 0.5 s integration time. Raw BRET ratios were calculated by dividing the acceptor counts by the donor counts. Milli-BRET units (mBU) were calculated by multiplying the raw BRET values by 1000.
Apparent BRET probe affinity values (EC50) were determined using the sigmoidal dose-response (variable slope) equation available in GraphPad Prism (Equation 1);
For determination of test compound potency, competitive displacement data were plotted with GraphPad Prism software and data were fit to Equation 1 to determine the IC50 value.
HAP1 Cell Growth and Lipid Extraction
WT, COQ8AKO, COQ8BKO, and COQ8A/BDKO HAP1 cells were obtained from Horizon Discovery and previously validated26. Three 10 cm plates were seeded with 1.4×106 cells for each cell line in IMDM (Thermo 12440053) supplemented with 10% heat inactivated FBS (HI FBS)(Atlanta Biologicals) and 100 U/mL Penicillin-Streptomycin (Pen/Strep)(Thermo 15140122). Cells were grown at 37 °C, 5% CO2 for three days to approximately 90% confluency. Each plate of cells was washed with 5 mL dPBS then isolated by trypsinization. Cells were pelleted (173 g, 5 min, RT) and media was removed. Cells were then resuspended in 5 mL media and counted using the Muse Count and Viability Assay (Luminex). 1.2×107 viable cells were collected per sample and pelleted by centrifugation (173 g, 5 min, RT). Media was removed and cells were resuspended in 5 mL dPBS. Cells were pelleted again by centrifugation and dPBS was removed. Cell pellets were resuspended in 600 μL cold methanol with 0.1 μM CoQ8 (Avanti Lipids 900151p-1mg) as the internal standard. Suspended cells were transferred to screw cap tubes and vortexed for 10 min at 4 °C to lyse. 400 μL cold petroleum ether was added to each tube. Samples were vortexed for 3 min at 4 °C then centrifuged at 800 g, 3 min, RT to separate the layers. The top petroleum ether layer was collected in a new tube and the extraction was repeated with an additional 400 μL petroleum ether. The two extractions were combined, and the ether was removed under argon gas. Once dried, the lipids were resuspended in 100 μL methanol, transferred to glass autosampler vials, and submitted for LC-MS lipid measurements.
Seahorse Mito Stress Test
WT, COQ8AKO, COQ8BKO, and COQ8A/BDKO HAP1 cells were split, counted via Muse Count and Viability Assay, and 40,000 viable cells per well were plated to a poly-D-lysine-coated XF96 Seahorse plate. Cells were allowed to adhere to the plate overnight at 37 °C, 5% CO2 in IMDM supplemented with 10% heat inactivated FBS and 100 U/mL penicillin-streptomycin. After 24 h, the medium was aspirated, cells were washed once with dPBS, and medium was replaced with 180 μL Seahorse XF Base Medium (Agilent 102353–100), pH 7.4, supplemented with 25 mM glucose and 2 mM glutamine. The plate was then incubated for 1 h at 37 °C in a non-CO2 incubator. Oxygen-consumption rates (OCR) and extracellular acidification rate were then monitored on a Seahorse XF96 Extracellular Flux Analyzer basally, and in the presence of a Seahorse XF Mito Stress Test (Agilent 103015–100). For the stress test, cells were treated with oligomycin (2 μM final concentration), FCCP (0.5 μM final concentration), and rotenone and antimycin A (0.5 μM final concentration). After the assay, cells were quantified by crystal violet staining as previously described26. Cells were fixed in 1% glutaraldehyde and incubated for 15 min. Cells were then washed by submersion in diH2O, air dried, and stained with 0.1% crystal violet for 20 min. Excess dye was removed by washing with diH2O and the plate was air dried. After release of the stain with 10% acetic acid, each well was read at an absorbance of 590 nm. OCR measurements for each well were normalized to relative crystal violet absorbance. OCR measurements were plotted and analyzed using GraphPad Prism Version 8.4.3 (n=11–12, error bars represent SD).
COQ8ANΔ254 Crystallography
COQ8ANΔ254 was expressed and purified as described for COQ8ANΔ250. After the final buffer exchange the protein was concentrated to less than 1 mL. The sample was centrifuged (20,000 g, 10 min, 4°C) and the soluble portion was filtered through a 0.22 μm filter. The protein was further purified using a HiLoad 16/600 Superdex 75 pg gel filtration column (GE) with an isocratic elution at 1 mL/min. The protein was exchanged into Crystallography Buffer [50 mM NaCl, 5 mM HEPES pH 7.5, 0.3 mM TCEP] during the FPLC run. Fractions were analyzed by SDS-PAGE. Pure fractions were pooled, concentrated with a MW-cutoff spin filter (30 kDa MWCO), aliquoted, frozen in N2 (l), and stored at −80°C for crystallization trials.
Crystallization screening and optimization was carried out in MRC SD2 plates using a TTP Labtech Mosquito using 50 microliters of reservoir solution. Mixtures of 0.39 mM COQ8ANΔ254 and 0.5 mM UNC-CA157 was incubated for 30 minutes at ambient temperature before crystallization setup.
Crystals were screened for optimum diffraction at sectors 21 and 23 of the Advanced Photon Source. Data sets for refinement were collected at GM/CA@APS on 2019–12-12 using JBluIce-EPICS V2019–353 beamline control software. Data was reduced using autoPROC V1.0.5(20190923)54 and XDS V2021032255. The structures were solved by molecular replacement using PDB 5I35 in Phaser V2.8.356. Geometric restraints for UNC-CA157 were prepared with phenix.elbow57. Structures were iteratively improved using model building in Coot V0.958 refinement in Phenix V1.20.1–448759, and sterochemical validation in MOLPROBITY V4.02–528. Final models were prepared for deposition using PDB_EXTRACT V3.27. SBGrid provided curated crystallographic software. Protein-ligand interaction diagrams were generated using LigPlot+ V2.2.560.
7UDP
Optimal crystals were obtained by mixing 200 nL COQ8A-UNC-CA157 with 250 nL of 0.65 M sodium succinate and 0.1 M HEPES buffer, pH 8. Data was collected at 100K on APS beamline 23ID-D with 1.0332 Å X-rays, on a Dectris Pilatus3–6M detector at 350 mm sample-detector distance, 1800 frames, 0.2 degree per frame.
7UDQ
Optimal crystals were obtained by mixing 200 nL COQ8A-UNC-CA157 with 250 nL of 0.65 M sodium succinate, pH 7. Data was collected at 100K on APS beamline 23ID-B with 1.0331 Å X-rays, using an Eiger 16M at 240 mm sample-detector distance, 1800 frames, 0.2 degree per frame.
Site Directed Mutagenesis
The COQ8ANΔ250, COQ8ANΔ254, and Coq8pNΔ41 in pVP68K vectors have been described previously11,16. Point mutations were introduced via PCR-based mutagenesis. Reactions were DpnI digested and transformed into DH5α competent E. coli cells. Plasmids were isolated from transformants and Sanger sequencing was used to identify those harboring the correct mutations. Oligonucleotides were purchased from IDT (Coralville, IA, USA) and sequences are included in the supporting information.
TPP-UNC-CA157 Synthesis
TPP-UNC-CA157 was synthesized in collaboration with the Washington University Center for Drug Discovery. Detailed synthesis methods can be found in the Supplemental Information. This general synthetic strategy for mitochondrial targeting has been described previously29.
De novo 13C-CoQ10 Production
This general strategy for tracking de novo CoQ production was used for yeast in Reidenbach et al17 and was optimized here for HAP1 cells. WT, COQ8AKO, COQ8BKO, and COQ8A/BDKO HAP1 cells were split, counted via Muse Count and Viability Assay, seeded in 10 cm plates at 1.5×106 cells per plate in IMDM with HI FBS and Pen/Strep, and allowed to grow at 37 °C, 5% CO2 for 48 h. Cells were then rinsed with 5 mL dPBS, swapped into 10 mL DMEM (no glucose) (Thermo 11966025) supplemented with HI FBS, Pen/Strep, and 10 mM galactose, and grown again at 37 °C, 5% CO2. After 24 h cells were swapped into the same media containing 10 μM 4-hydroxybenzoate-(phenyl-13C6) (Sigma 587869) and either indicated inhibitor or matched DMSO vehicle (Alkyl-TPP = Toronto Research Chemicals C178905). Cells were incubated for 1 h at 37 °C, 5% CO2. Cells were washed with dPBS then isolated by trypsinization. Cells were counted via Muse Count and Viability Assay and equivalent viable cells were collected in each experiment (UNC-CA157=2.92×106 cells; TPP-UNC-CA157=7.91×106 cells; TPP-UNC-CA157 dose response=4.53×106 cells; UNC-CA157 dose response=1.17×107 cells; Alkyl-TPP dose response=7.94×106 cells) via centrifugation (300 g, 5 min). Cells were washed with dPBS then again isolated by centrifugation (300 g, 5 min) and dPBS was removed. Cell pellets were frozen in N2 (l) and stored at −80 oC. On the day of analysis cell pellets were thawed on ice. Lipids were extracted and dried under Ar(g) as described above for untreated HAP1 cells. Dried lipids were resuspended in an equivalent volume of methanol per experiment (UNC-CA157=60 μL; TPP-UNC-CA157=100 μL; TPP-UNC-CA157 dose response=60 μL; UNC-CA157 dose response=100 μL; Alkyl-TPP dose response=100 μL), then submitted for LC-MS lipid measurements.
LC-MS Lipid Measurements
A Vanquish Horizon UHPLC system (Thermo Scientific) coupled to an Exploris 240 Orbitrap mass spectrometer (Thermo Scientific) was used for LC-MS analysis. A Waters Acquity CSH C18 column (100 mm × 2.1 mm, 1.7 μm) was held at 35 °C with the flow rate of 300 μL/min for the separation of the extracted lipids. A Vanquish binary pump system was employed, and mobile phase A consisted of 5 mM ammonium acetate in ACN/H2O (70/30, v/v) containing 125 μL/L acetic acid, while mobile phase B consisted of 5 mM ammonium acetate in IPA/ACN (90/10, v/v) with the same additive. The gradient was set as follows: B was held at 2% for 2 min and ramped up to 30% over the next 3 min, before further increased to 50% within 1 min and to 85% over 14 min, and then raised to 99% over 1 min and held for 4 min. The column was re-equilibrated for 5 min at 2% B before the injection of next sample. Samples were ionized by a heated ESI source with a vaporizer temperature of 350°C. Sheath gas was set to 50 units, auxiliary gas was set to 8 units, sweep gas was set to 1 unit. The ion transfer tube temperature was kept at 325 °C with 70% RF lens. Spray voltage was set to 3,500 V for positive mode and 2,500 V for negative mode. Each sample was injected twice to be analyzed in positive and negative modes separately. Full MS scans were acquired at 120,000 resolution from m/z 200 to 1,700 with in-source Easy-IC enabled. Normalized AGC target was set to 100% with the maximum ion injection time of 50 ms. Targeted quantitative analysis of CoQ10, PPHB10, 13C6-CoQ10, 13C6-PPHB10 and internal standard CoQ8 was processed using TraceFinder 5.1 (Thermo Scientific) with the mass accuracy of 5 ppm. The result of peak integration was manually examined. Peak areas were normalized to the CoQ8 internal standard. Experiments were performed in triplicate and error bars represent SD.
HAP1 Growth Assay
WT HAP1 cells were split and counted via Muse Count and Viability Assay. 2×104 cells were seeded per well in a 96 well plate (Greiner Bio-One 655090). Cells were allowed to adhere for 1 h at RT in the hood, then incubated at 37 °C, 5% CO2 overnight. The next day cells were washed once with dPBS then swapped into DMEM (no glucose) (Thermo 11966025) supplemented with HI FBS, Pen/Strep, either 10 mM glucose or galactose, and TPP-UNC-CA157 at indicated concentrations with equivalent DMSO vehicle. Cells were incubated at 37 °C, 5% CO2 for 90 h. Cell growth was monitored using a Sartorius IncuCyte S3 Live-Cell Analysis System at 10x magnification in the phase channel. Percent confluency and associated error were determined using the Incucyte S3 Software. Graphs were made using GraphPad Prism Version 8.4.3 (n=6, error represents SEM).
MIBS-MS
The multiplexed inhibitor bead coupled with mass spectrometry (MIBS/MS) method of screening for kinase inhibitor selectivity has been described previously32–35. Briefly, SUM159PT cells were grown to 80% confluency in a 1:1 mixture of DMEM and Nutrient Mixture F-12 medium supplemented with 5% fetal bovine serum, 1% penn/strep, 5 mg/mL insulin, and 1 mg/mL hydrocortisone at 37 °C in a humidified 5% CO2 atmosphere. Cells were then washed three times with ice-cold PBS and harvested by scraping. Cell pellets were resuspended in lysis buffer (50 mM HEPES, pH 7.5, 150 mM NaCl, 0.5% Triton X-100, 1 mM EDTA, 1 mM EGTA, 10 mM NaF, 2.5 mM Na3VO4, complete protease inhibitor cocktail (Roche), and phosphatase inhibitor cocktails 2 and 3 (Sigma)) and incubated on ice for 10 minutes. Lysates were then sonicated using a microtip sonicator in 10 s bursts at 35% amplitude three times. Lysates were clarified by centrifugration at 14,000 × g for 15 minutes at 4°C then filtered through a 0.2 μm syringe filter, flash frozen, and stored at −80 °C until used. Protein concentration was quantified using a Bradford assay the day of the experiment. Aliquots of lysate containing 4 mg of total protein in 1 mL of lysis buffer was incubated with 1 μM of the indicated compound or an equivalent amount of DMSO on ice for 30 minutes. Lysates were then diluted to a final volume of 3 mL with lysis buffer containing 1 μM of the indicated inhibitor or DMSO then 5 M NaCl added to a final concentration to 1 M. A column consisting of a 175 μL bed volume of settled MIBs: 25 μL each of purvalanol B, PP58, UNC21474A, and UNC8088A and 37.5 μL of VI-16832 and CTx-0294885 was equilibrated in high-salt buffer (50 mM HEPES pH 7.5, 1 M NaCl, 0.5% Triton X-100, 1 mM EDTA, 1 mM EGTA). Lysates were applied to MIBs columns by gravity flow followed by sequential washes with high-salt buffer, low-salt buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 0.5% Triton X-100, 1 mM EDTA, and 1 mM EGTA), and SDS buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 0.5% Triton X-100, 1 mM EDTA, 1 mM EGTA, and 0.1% SDS). Bound proteins were eluted by boiling samples in elution buffer (100 mM Tris-HCl pH 6.8, 0.5% SDS, and 1% 2-mercaptoethanol) twice for 15 min each. Eluted proteins were reduced by addition of dithiothreitol (DTT) to a final concentration of 5 mM and incubation at 60 °C for 25 min. Samples were allowed to cool to room temperature on ice prior to addition of of iodacetamide to a final concentration of 20 mM and incubation for 30 minutes in the dark. Samples were then concentrated to a final volume of ~100 μL in 10 K Amicon Ultra centrifugal concentrators (Millipore) followed by methanol/chloroform precipitation of proteins. Precipitated protein was re-suspended in 50 mM HEPES pH 8.0 and digested with trypsin at 37 °C overnight. Residual detergent was removed by three sequential ethyl acetate extractions. Samples were desalted using BioPureSPN MINI desalting columns (NEST) according to the manufacturer’s protocol.
Each sample was analyzed by LC-MS/MS using an Easy-nLC 1200 coupled to a QExactive HF mass spectrometer equipped with an Easy Spray source (Thermo Scientific). Dried peptide samples were reconstituted in 2% MeCN, 0.1% formic acid and loaded onto an EASY-Spray HPLC column (75 μm id x 25 cm, 2 μm particle size) (Thermo Scientific). Peptide separation was achieved over a 120 min gradient consisting of 5–45% mobile phase B at a 250 nL/min flow rate, where mobile phase A was 0.1% formic acid in water and mobile phase B consisted of 0.1% formic acid in 80% MeCN. The QExactive HF was operated in data-dependent mode where the top 15 precursor ions were selected for subsequent fragmentation and MS2 analysis. For all runs, ESI parameters: 3 × 106 AGC MS1, 100 ms MS1 max inject time, 1 × 105 AGC MS2, 60 ms MS2 max inject time, 1.6 m/z isolation window, 30 s dynamic exclusion. Spectra were searched against the human Uniprot/Swiss-Prot proteome using the integrated MaxQuant search engine, ANDROMEDA. Label-free quantification was used to calculate kinase abundance using the MaxQuant software package (ver. 1.6.2.10) with a false discovery rate of 5%. The following parameters were used to identify tryptic peptides for protein identification: up to two missed trypsin cleavage sites; carbamidomethylation (C) was set as a fixed modification; and oxidation (M), and acetyl (protein N-term) were set as variable modifications. The ratios of label-free quantification values of inhibitor treated samples over DMSO treated samples were log2 transformed and reported.
Statistical Analysis
In all cases, “mean” refers to the arithmetic mean, “SD” refers to standard deviation, “CI” refers to confidence interval, and “SEM” refers to standard error of the mean. Statistical analyses were performed using GraphPad Prism Version 8.4.3 unless otherwise noted. For significance claims p-values were calculated using an unpaired, two-tailed Student’s t-test and p < 0.05 was considered significant. “ns” stands for not significant. No adjustments were made for multiple comparisons. In all cases, n represents independent replicates of an experiment.
Data Availability
LC-MS lipid measurement data that support the findings of this study were deposited into the MassIVE data repository under accession #MSV000090082. Structural data generated in this study have been deposited in the PDB with accession codes 7UDP and 7UDQ. Structural data that were used to support the findings in this study are available in the PDB with accession codes 5I35 and 4PED.
Material Availability
All unique biological materials are available from the authors upon request.
Extended Data
Extended Data Fig. 1. COQ8A inhibition in vitro.
a, Inhibition of COQ8ANΔ250 ATPase activity by the top candidate inhibitors (n=3 independent samples from one experiment, mean ± SD) (IC50 data are available in the supporting information). b, Top candidate inhibitor binding curves, as determined by DSF (n=3 independent samples from one experiment, mean ± SD) (Kd,app data are available in the supporting information).
Extended Data Fig. 2. De novo CoQ10 biosynthesis measurements.
a, Schematic outlining the experimental plan to measure de novo CoQ production. b, De novo production of 13C6-PPHB10 in WT, COQ8AKO, COQ8BKO, and COQ8A/BDKO HAP1 cells after treatment with 10 μM 13C6-4-HB and either DMSO or 20 μM UNC-CA157 (grey=DMSO, red=UNC-CA157, n=3 independent samples from one experiment, mean ± SD) (two sided students t-test, no adjustment for multiple comparisons). c, Unlabeled PPHB10 levels in WT, COQ8AKO, COQ8BKO, and COQ8A/BDKO HAP1 cells after treatment with 10 μM 13C6-4-HB and either DMSO or 20 μM UNC-CA157 (grey=DMSO, red=UNC-CA157, n=3 independent samples from one experiment, mean ± SD) (two sided students t-test, no adjustment for multiple comparisons). d, Unlabeled CoQ10 levels in WT, COQ8AKO, COQ8BKO, and COQ8A/BDKO HAP1 cells after treatment with 10 μM 13C6-4-HB and either DMSO or 20 μM UNC-CA157 (grey=DMSO, red=UNC-CA157, n=3 independent samples from one experiment, mean ± SD) (two sided students t-test, no adjustment for multiple comparisons).
Extended Data Fig. 3. F495 is required for potent inhibition.
a-c, Protein-ligand interaction networks for three independent complexes of COQ8ANΔ254 and UNC-CA157 generated in LigPlot+ (PDB: 7UDQ and 7UDP) (hydrogen bond = dashed line; hydrophobic contact = arc with spokes). d, Protein-ligand interaction network for COQ8ANΔ254 R611K and AMPPNP generated in LigPlot+ (PDB: 5I35) (hydrogen bond = dashed line; hydrophobic contact = arc with spokes). e, SDS-PAGE analysis of COQ8ANΔ250 purifications. f, COQ8ANΔ250 WT and binding pocket mutant thermal stabilization by UNC-CA157, as determined by DSF (n=3 independent samples from one experiment, mean ± SD) (two sided students t-test, no adjustment for multiple comparisons, ***p = 1.2×10−10 F336A, ***p = 4.5×10−5 A356G, ***p = 2.4×10−9 K358A, ***p = 4.7×10−11 K358E, ***p = 1.7×10−6 L443A, *p = 0.0264 T445A, ***p = 1.8×10−10 L447A, ***p = 1.4×10−9 V448P, ***p = 9.5×10−12 F495A; all six WT-1 and WT-2 data points were used for statistical analysis). g, Stick representation of UNC-CA157 and adjacent F495 residue. h, Conservation of F495 across the UbiB protein family. i, Thermal stabilization of COQ8ANΔ250 WT and F495L as well as Coq8pNΔ41 WT and L353F by UNC-CA157 (n=3 independent samples from one experiment, mean ± SD) (two sided students t-test, no adjustment for multiple comparisons, *p = 0.0126 COQ8A WT vs. F495L, ***p = 5.6×10−7 Coq8p WT vs. L353F). j, Inhibition of COQ8ANΔ250 WT and F495L ATPase activity by UNC-CA157 (n=3 independent samples from one experiment, mean ± SD).
Extended Data Fig. 4. Results from de novo CoQ production with TPP-UNC-CA157 and inhibitor toxicity analysis.
a, Unlabeled CoQ10 and PPHB10 levels in WT, COQ8AKO, COQ8BKO, and COQ8A/BDKO HAP1 cells after treatment with 10 μM 13C6-4-HB and either DMSO or 17.6 μM TPP-UNC-CA157 (n=3 independent samples from one experiment, mean ± SD) (two sided students t-test, no adjustment for multiple comparisons, *p = 0.0228 WT PPHB10, ***p = 0.0003 COQ8AKO PPHB10). b-d, 13C6-CoQ10 levels in WT HAP1 cells after treatment with 10 μM 13C6-4-HB and indicated concentrations of UNC-CA157, TPP-UNC-CA157 or Alkyl-TPP (n=3 independent samples from one experiment, mean ± SD) (two sided students t-test, no adjustment for multiple comparisons, *p =0.0482 DMSO vs. 43.9 μM UNC-CA157, *p = 0.0118 DMSO vs. 22.0 μM TPP-UNC-CA157, **p = 0.0030 DMSO vs. 43.9 μM TPP-UNC-CA157, **p = 0.0026 DMSO vs. 87.8 μM TPP-UNC-CA157). e, Growth analysis of WT HAP1 cells in either 10 mM glucose or 10 mM galactose with indicated concentrations of TPP-UNC-CA157 (n=3 biologically independent wells of cells in one experiment, mean ± SEM).
Supplementary Material
Acknowledgements
We thank members of the Pagliarini lab for helpful discussions throughout this project. This work was supported by NIH awards R35GM131795 (D.J.P.) and T32GM008505 (N.H.M); funds from the BJC Investigator Program (D.J.P.); and NSF DGE-1747503 (N.H.M.). We would like to thank Katherine Overmyer, Brett Paulson, Edna Trujillo, and Joshua Coon for assistance with LC-MS method development. We thank Galen Hicks for graphic design assistance. This study made use of the Washington University in St. Louis Genome Engineering and iPSC Center for instrument use and the Washington University in St. Louis Center for Drug Discovery for synthesis services. The SGC is a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada, Innovative Medicines Initiative (EU/EFPIA) [ULTRA-DD grant no. 115766], Janssen, Merck KGaA Darmstadt Germany, MSD, Novartis Pharma AG, Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and Wellcome [106169/ZZ14/Z]. The NIH is acknowledged for support (1U24DK11604–01). We thank Biocenter Finland/DDCB for financial support and the CSC-IT Center for Science Ltd. (Finland) for allocation of computational resources. We also thank Dr. Brandie Ehrmann and Ms. Diane E. Weatherspoon for for LC-MS/HRMS support provided by in the Mass Spectrometry Core Laboratory at the University of North Carolina at Chapel Hill. The core is supported by the National Science Foundation under Grant No. (CHE-1726291). Dr. Tatu Pantsar (University of Eastern Finland) is thanked for useful discussions. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02–06CH11357. GM/CA@APS has been funded by the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006, P30GM138396). The Eiger 16M detector at GM/CA-XSD was funded by NIH grant S10 OD012289. With thank Craig Ogata for beamline support. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). The Collaborative Crystallography Core in the Department of Biochemistry, UW-Madison received support from the Department of Biochemistry endowment.
Footnotes
Competing interest declaration
J.D.V., C.R.C., C.A.Z., and M.B.R. are employed by Promega Corporation. The remaining authors declare no competing interests.
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T. & Sudarsanam S. The protein kinase complement of the human genome. Science 298, 1912–34 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Dar AC & Shokat KM The evolution of protein kinase inhibitors from antagonists to agonists of cellular signaling. Annu Rev Biochem 80, 769–95 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Cohen P, Cross D. & Jänne PA Kinase drug discovery 20 years after imatinib: progress and future directions. Nat Rev Drug Discov 20, 551–569 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attwood MM, Fabbro D, Sokolov AV, Knapp S. & Schiöth HB Trends in kinase drug discovery: targets, indications and inhibitor design. Nat Rev Drug Discov 20, 839–861 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Kung JE & Jura N. Prospects for pharmacological targeting of pseudokinases. Nat Rev Drug Discov 18, 501–526 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oprea TI et al. Unexplored therapeutic opportunities in the human genome. Nat Rev Drug Discov 17, 317–332 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leonard CJ, Aravind L. & Koonin EV Novel families of putative protein kinases in bacteria and archaea: evolution of the “eukaryotic” protein kinase superfamily. Genome Res 8, 1038–47 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Pagliarini DJ et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–23 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lundquist PK, Davis JI & Wijk KJ van. ABC1K atypical kinases in plants: filling the organellar kinase void. Trends Plant Sci 17, 546–55 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Traschütz A. et al. Clinico-Genetic, Imaging and Molecular Delineation of COQ8A-Ataxia: A Multicenter Study of 59 Patients. Ann Neurol 88, 251–263 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefely JA et al. Cerebellar Ataxia and Coenzyme Q Deficiency through Loss of Unorthodox Kinase Activity. Mol Cell 63, 608–620 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashraf S. et al. ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J Clin Invest 123, 5179–89 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poon WW et al. Identification of Escherichia coli ubiB, a gene required for the first monooxygenase step in ubiquinone biosynthesis. J Bacteriol 182, 5139–46 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Do TQ, Hsu AY, Jonassen T, Lee PT & Clarke CF A defect in coenzyme Q biosynthesis is responsible for the respiratory deficiency in Saccharomyces cerevisiae abc1 mutants. J Biol Chem 276, 18161–8 (2001). [DOI] [PubMed] [Google Scholar]
- 15.Stefely JA & Pagliarini DJ Biochemistry of Mitochondrial Coenzyme Q Biosynthesis. Trends Biochem Sci 42, 824–843 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stefely JA et al. Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthesis. Mol Cell 57, 83–94 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reidenbach AG et al. Conserved Lipid and Small-Molecule Modulation of COQ8 Reveals Regulation of the Ancient Kinase-like UbiB Family. Cell Chem Biol 25, 154–165.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asquith CRM, Murray NH & Pagliarini DJ ADCK3/COQ8A: the choice target of the UbiB protein kinase-like family. Nat Rev Drug Discov 18, 815 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray NH et al. 2-Propylphenol Allosterically Modulates COQ8A to Enhance ATPase Activity. Acs Chem Biol (2022) doi: 10.1021/acschembio.2c00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asquith CRM et al. SGC-GAK-1: A Chemical Probe for Cyclin G Associated Kinase (GAK). J Med Chem 62, 2830–2836 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wells CI et al. The Kinase Chemogenomic Set (KCGS): An Open Science Resource for Kinase Vulnerability Identification. Int J Mol Sci 22, 566 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robers MB et al. Target engagement and drug residence time can be observed in living cells with BRET. Nat Commun 6, 10091 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vasta JD et al. Quantitative, Wide-Spectrum Kinase Profiling in Live Cells for Assessing the Effect of Cellular ATP on Target Engagement. Cell Chem Biol 25, 206–214.e11 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lagier-Tourenne C. et al. ADCK3, an ancestral kinase, is mutated in a form of recessive ataxia associated with coenzyme Q10 deficiency. Am J Hum Genetics 82, 661–72 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fonseca LV et al. Mutations in COQ8B (ADCK4) found in patients with steroid-resistant nephrotic syndrome alter COQ8B function. Hum Mutat 39, 406–414 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rensvold JW et al. Defining mitochondrial protein functions through deep multiomic profiling. Nature 606, 382–388 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zielonka J. et al. Mitochondria-Targeted Triphenylphosphonium-Based Compounds: Syntheses, Mechanisms of Action, and Therapeutic and Diagnostic Applications. Chem Rev 117, 10043–10120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith RAJ, Porteous CM, Gane AM & Murphy MP Delivery of bioactive molecules to mitochondria in vivo. Proc National Acad Sci 100, 5407–5412 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trionnaire SL et al. The synthesis and functional evaluation of a mitochondria-targeted hydrogen sulfide donor, (10-oxo-10-(4-(3-thioxo-3 H −1,2-dithiol-5-yl)phenoxy)decyl)triphenylphosphonium bromide (AP39). Medchemcomm 5, 728–736 (2014). [Google Scholar]
- 30.Trnka J, Elkalaf M. & Anděl M. Lipophilic Triphenylphosphonium Cations Inhibit Mitochondrial Electron Transport Chain and Induce Mitochondrial Proton Leak. Plos One 10, e0121837 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reily C. et al. Mitochondrially targeted compounds and their impact on cellular bioenergetics. Redox Biol 1, 86–93 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duncan JS et al. Dynamic Reprogramming of the Kinome in Response to Targeted MEK Inhibition in Triple-Negative Breast Cancer. Cell 149, 307–321 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asquith CRM et al. Identification and Optimization of 4-Anilinoquinolines as Inhibitors of Cyclin G Associated Kinase. Chemmedchem 13, 48–66 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Asquith CRM et al. Design of a Cyclin G Associated Kinase (GAK)/Epidermal Growth Factor Receptor (EGFR) Inhibitor Set to Interrogate the Relationship of EGFR and GAK in Chordoma. J Med Chem 62, 4772–4778 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Asquith CRM et al. Identification of 4-Anilinoquin(az)oline as a Cell-Active Protein Kinase Novel 3 (PKN3) Inhibitor Chemotype**. Chemmedchem e202200161 (2022) doi: 10.1002/cmdc.202200161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asquith CRM et al. Design and Analysis of the 4-Anilinoquin(az)oline Kinase Inhibition Profiles of GAK/SLK/STK10 Using Quantitative Structure-Activity Relationships. Chemmedchem 15, 26–49 (2020). [DOI] [PubMed] [Google Scholar]
- 37.Serafim RAM, Elkins JM, Zuercher WJ, Laufer SA & Gehringer M. Chemical Probes for Understudied Kinases: Challenges and Opportunities. J Med Chem 65, 1132–1170 (2022). [DOI] [PubMed] [Google Scholar]
- 38.Larsen PL & Clarke CF Extension of life-span in Caenorhabditis elegans by a diet lacking coenzyme Q. Science 295, 120–3 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Liu X. et al. Evolutionary conservation of the clk-1-dependent mechanism of longevity: loss of mclk1 increases cellular fitness and lifespan in mice. Gene Dev 19, 2424–2434 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Doll S. et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575, 693–698 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Bersuker K. et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 575, 688–692 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forsman U, Sjöberg M, Turunen M. & Sindelar PJ 4-Nitrobenzoate inhibits coenzyme Q biosynthesis in mammalian cell cultures. Nat Chem Biol 6, 515–517 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Wang Y. et al. The Anti-neurodegeneration Drug Clioquinol Inhibits the Aging-associated Protein CLK-1*. J Biol Chem 284, 314–323 (2009). [DOI] [PubMed] [Google Scholar]
- 44.Tsuganezawa K. et al. Identification of small molecule inhibitors of human COQ7. Bioorgan Med Chem 28, 115182 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Nara T. et al. The ubiquinone synthesis pathway is a promising drug target for Chagas disease. Plos One 16, e0243855 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernández-del-Río L. et al. Regulation of hepatic coenzyme Q biosynthesis by dietary omega-3 polyunsaturated fatty acids. Redox Biol 102061 (2021) doi: 10.1016/j.redox.2021.102061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ward RA et al. Challenges and Opportunities in Cancer Drug Resistance. Chem Rev 121, 3297–3351 (2021). [DOI] [PubMed] [Google Scholar]
- 48.Kelso GF et al. Selective Targeting of a Redox-active Ubiquinone to Mitochondria within Cells. J Biol Chem 276, 4588–4596 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Kulkarni CA et al. A Novel Triphenylphosphonium Carrier to Target Mitochondria without Uncoupling Oxidative Phosphorylation. J Med Chem 64, 662–676 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kemmerer ZA et al. UbiB proteins regulate cellular CoQ distribution in Saccharomyces cerevisiae. Nat Commun 12, 4769 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Asquith CRM et al. Targeting an EGFR Water Network with 4-Anilinoquin(az)oline Inhibitors for Chordoma. Chemmedchem 14, 1693–1700 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Asquith CRM, Treiber DK & Zuercher WJ Utilizing comprehensive and mini-kinome panels to optimize the selectivity of quinoline inhibitors for cyclin G associated kinase (GAK). Bioorg Med Chem Lett 29, 1727–1731 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Stepanov S. et al. JBluIce-EPICS control system for macromolecular crystallography. Acta Crystallogr Sect D Biological Crystallogr 67, 176–88 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vonrhein C. et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr Sect D Biological Crystallogr 67, 293–302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kabsch W. XDS. Acta Crystallogr Sect D Biological Crystallogr 66, 125–32 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCoy AJ et al. Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moriarty NW, Grosse-Kunstleve RW & Adams PD electronic Ligand Builder and Optimization Workbench (eLBOW): a tool for ligand coordinate and restraint generation. Acta Crystallogr Sect D Biological Crystallogr 65, 1074–80 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emsley P, Lohkamp B, Scott WG & Cowtan K. Features and development of Coot. Acta Crystallogr Sect D Biological Crystallogr 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Afonine PV et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr Sect D Biological Crystallogr 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laskowski RA & Swindells MB LigPlot+: Multiple Ligand–Protein Interaction Diagrams for Drug Discovery. J Chem Inf Model 51, 2778–2786 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
LC-MS lipid measurement data that support the findings of this study were deposited into the MassIVE data repository under accession #MSV000090082. Structural data generated in this study have been deposited in the PDB with accession codes 7UDP and 7UDQ. Structural data that were used to support the findings in this study are available in the PDB with accession codes 5I35 and 4PED.