Abstract
Errors occurring during DNA replication can result in inaccurate, incomplete or re-replication, resulting in genome instability that can lead to diseases such as cancer or disorders such as autism. A great deal of progress has been made toward understanding the entire process of DNA replication in eukaryotes, including the mechanism of initiation and its control. This review focuses on the current understanding how the Origin Recognition Complex (ORC) contributes to determining the location of replication initiation in the multiple chromosomes within eukaryotic cells, as well as methods for mapping the location and temporal patterning of DNA replication. Origin specification and configuration varies substantially between eukaryotic species and in some cases co-evolved with gene silencing mechanisms. The possibility that centromeres and origins of DNA replication were originally derived from a common element and later separated during evolution is discussed.
eTOC for manuscript MOLECULAR-CELL-D-22-01584R1
The initiation of DNA replication occurs at multiple sites along eukaryotic cell chromosomes. The mechanism that defines the location of these replication origins varies considerably, ranging from DNA sequence-specific to epigenetically determined and inherited. It is possible that origins of DNA replication and centromeres evolved from a common ancestral element.
1. General DNA Replication Strategy
DNA replication is an essential process in all life forms for faithfully transmitting genetic information encoded within the nuclear and mitochondrial DNA to daughter cells during somatic cell division and to gametes for inheritance of “the chemistry of life” to the next generation. Bacteria generally have circular single or multiple chromosomes with a small genome size1. Typically, a single replication origin exists per bacterial chromosome2. Archaea have bacteria-like circular chromosomes3 and can have single or multiple clustered replication origins per archaeal chromosome4. In stark contrast, the polymer of DNA in each chromosome of a eukaryotic cell genome is much larger than bacterial and archaeal DNA5 and the entire genome is divided into multiple chromosomes (Table 1), creating a problem for ensuring that all the DNA molecules on separate chromosomes are replicated only once per cell division cycle and are then evenly segregated during mitosis. The size of eukaryotic genomes and the rate of DNA synthesis at each replication fork necessitates coordinated initiation of DNA replication from multiple origins in each chromosome so that the genome is duplicated in a timely manner during the cell division cycle. Furthermore, mechanisms exist to ensure that each replicon is duplicated once and only once per S phase.
Table 1 |. Origin recognition proteins in three domains of life.
Table shows the comparison of proteins that bind to origins of DNA replication in bacteria, archaea and eukaryotes.
| Domains of Life | Replication origin number | Chromosome number | Genome Size | Replication Initiator Proteins |
|---|---|---|---|---|
| Bacteria | Single origin per chromosome | Single or multiple circular chromosomes | ~0.6 to ~8.0 Mb | DnaA |
| Archaea | Single or multiple origins per chromosome | Single circular chromosome (typically) | ~0.5 to ~5.8 Mb | Orc1/Cdc6 |
| Eukaryote | Multiple origins per chromosome | Multiple linear chromosomes | ~10 to >100,000 Mb | ORC Cdc6 |
DNA replication is known to be bi-directional and temporally regulated in large domains (near-megabase-sized domains in mammals) (Figure 1a) with different parts of the genome replicating at different times (Figure 1b)6. Temporal patterning of DNA replication across the genome is influenced by different mechanisms, such as rate-limiting DNA replication factors, transcription, epigenetic control, chromosome structure and 3D chromatin organization in the nucleus7–10. Euchromatic regions tend to fill the nucleoplasm and replicate early during S phase, while the heterochromatic regions are more likely to be found at the nuclear and nucleolar peripheries and replicate late in S phase11. Chromatin is spatially folded into loops with the help of architectural proteins, such as CTCF and cohesins, and are further organized into large Topological Associated Domains (TADs) which serve as structural platforms for long-range chromatin interactions and cis-regulator contacts11. Remarkably TADs are highly correlated with DNA replication timing, with large megabase-sized A and B domains replicating at different times during S phase12–14. Replication timing is also dynamic and developmentally controlled15–17.
Figure 1 |. Replication Timing.
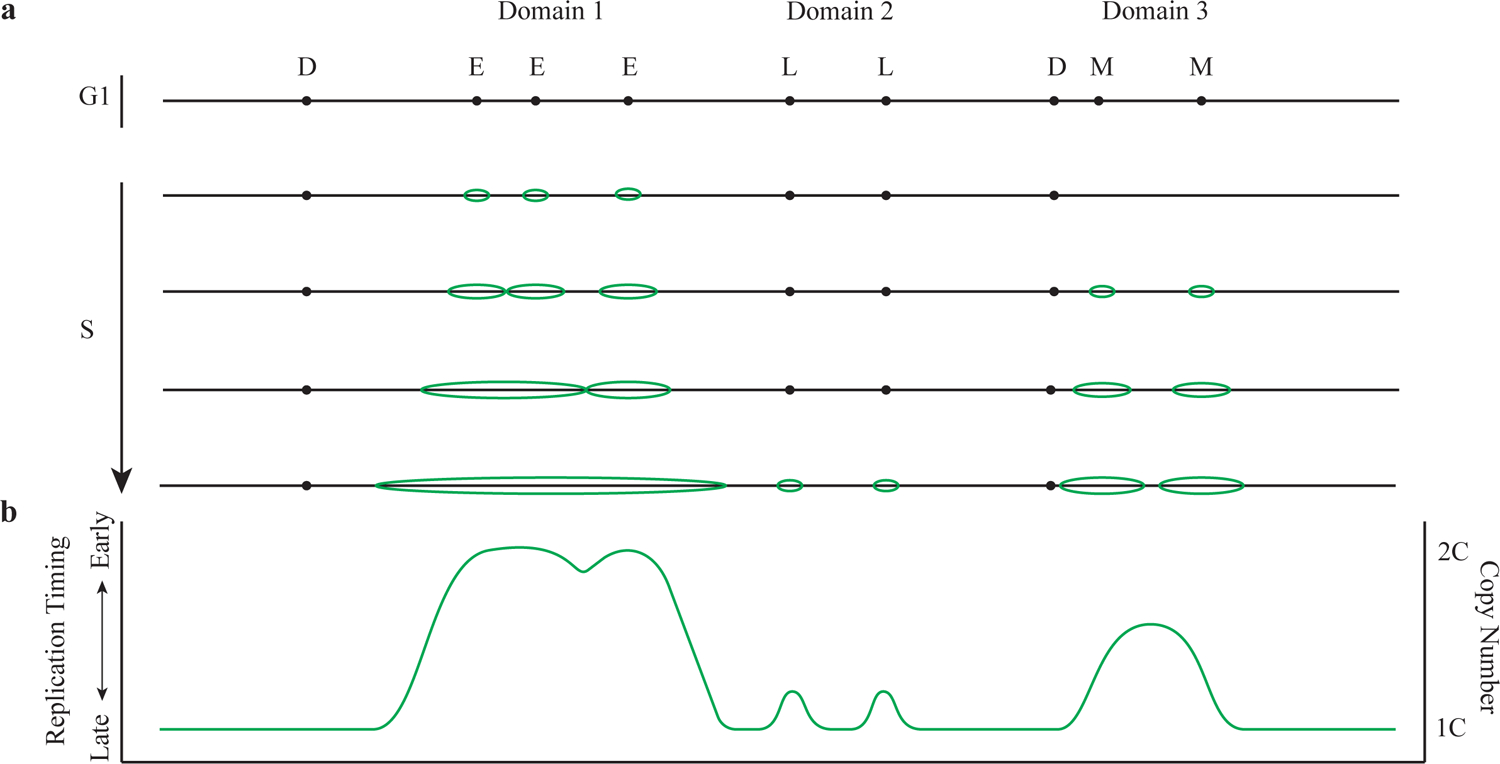
a shows the DNA replication process with various replication timing domains. Early-firing replication origins are indicated as E. Mid-firing replication origins are indicated as M. Late-firing replication origins are indicated as L. Dormant replication origins are indicated as D. Replication bubbles are indicated in green color. b shows the replication profile correspond to a measured in a population of cells with 1C and 2C genome copy number indicated. C equals to the genome copy number.
Early embryonic cells are highly proliferative and divide very rapidly and synchronously. DNA replication could occur in less than 4 mins in early-stage Drosophila embryos18, and less than 30 mins in early Xenopus cleavage embryos – 20-times faster than in somatic cells19. Replication origins are spaced much closer to each other during these rapid division stages, which can be 10-fold closer than the inter-origin distance in somatic cells and all origins become activated at roughly the same time. This density of replication origins declines when the embryo gets closer to the Mid Blastula Transition (MBT), during which zygotic transcription levels increase and cell division rates slow down15. In mammals, the replication fork rate is slower in the first cycle (2-cell-stage) with active replication origins closer to each other, compared to the second and the third cell division cycles (4- and 8-cell-stage) and replication fork rate increases as development proceeds. In addition, replication timing patterning, especially the early S-phase replication timing, is different between embryonic and 2-cell stage cells, which correlate with the transcription level of adjacent genes20. In Caenorhabditis elegans the temporal patterning of DNA replication during early embryogenesis precedes the onset of zygotic transcription suggesting that the temporal patterning of DNA replication is not dependent on cell type gene expression, but the mechanisms that determine replication timing may in turn influence developmental patterning of gene transcription21.
2. DNA Replication Initiation Mechanism
Because eukaryotic cells have multiple and often large chromosomes, it is necessary to establish starts sites for DNA replication across the entire genome with sufficient spacing so as to ensure complete replication within the S phase. Remarkably, this is achieved by the assembly of a large, multi-protein complex at every potential origin before any DNA synthesis occurs. Once the cell enters into S phase these complexes act as sites for assembly of the replisome that occurs in a sequential manner throughout S phase, creating the temporal patterns of DNA replication mentioned above. In the budding yeast Saccharomyces cerevisiae, the best characterized system to date, initiation of DNA replication occurs by assembly of a pre-Replicative Complex (pre-RC) at each potential origin of DNA replication by the sequential binding to DNA of the Origin Recognition Complex (ORC) and Cell Division Cycle 6 (Cdc6) that then recruit Chromatin Licensing and DNA Replication Factor 1 (Cdt1) that chaperones the DNA helicase Mini-chromosome Maintenance Proteins 2–7 (Mcm2-7), forming ORC/Cdc6/Cdt1/Mcm2-7 (OCCM) complex (Figure 2, a–e) [reviewed in22–24]. Complete assembly of the pre-RC involves the recruitment of a second Mcm2-7 hexamer to form head-to-head Mcm2-7 hexamers (Figure 2, f–h) that are destined to become the core components of the DNA helicases at the two replication forks that emanate from each origin25–28.
Figure 2 |. Pre-Replicative Complex Assembly Model.
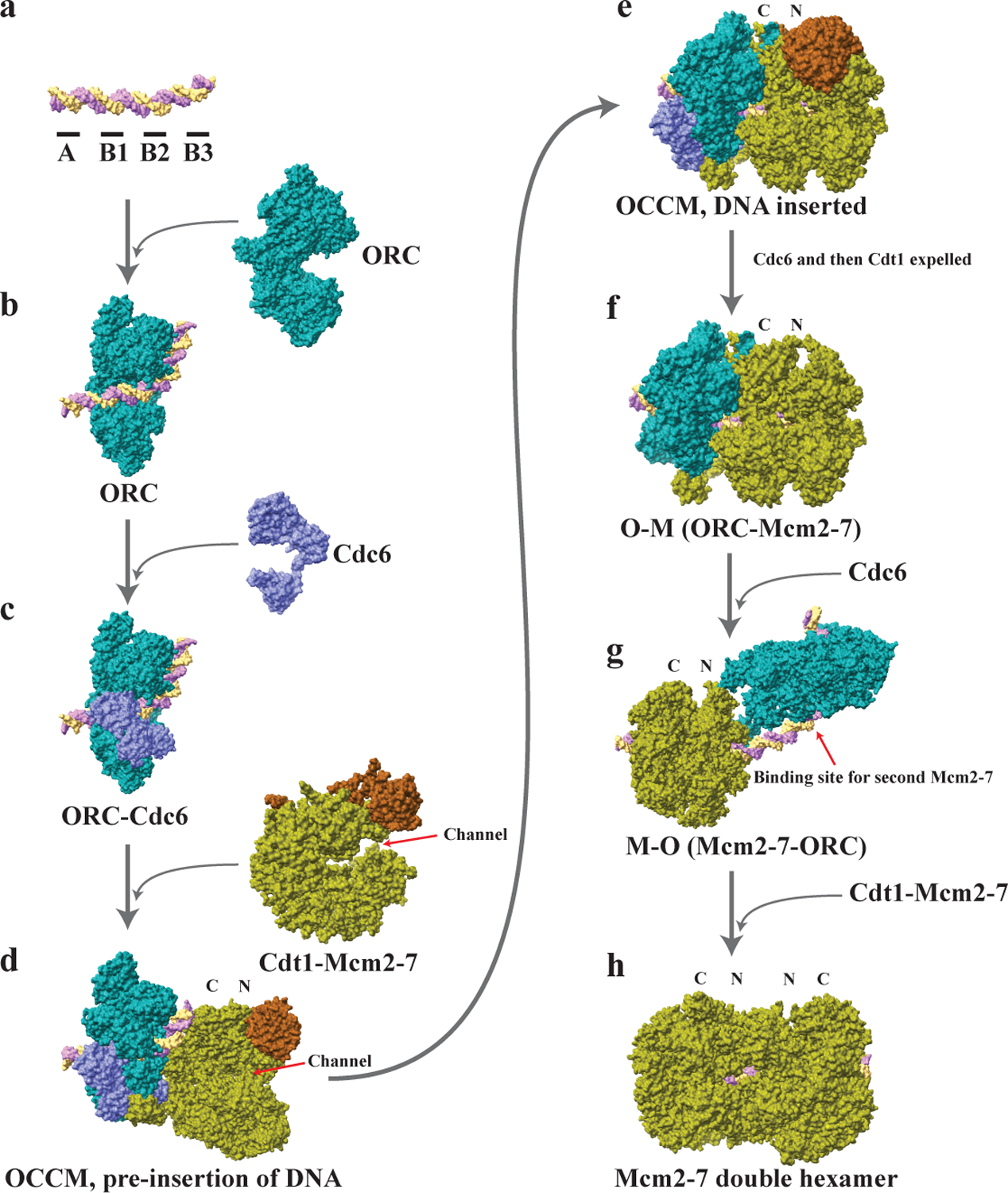
[Adapted from248 with image credit: molecular structures taken and adapted from the RCSB protein database, deposits 3JA8, 5ZR1139 5BK426, 5V8F249, 5XF8250, 6RQC251, 6F0L252, 6WGG and 6WGF253 and 7MCA254.) a shows the replication origin DNA (+ strand in cantaloupe color, - strand in lavender color), which in S. cerevisiae contains four elements (indicated as black segments) with A and B2 elements binding ORC in opposite orientations. b shows that ORC (in teal color) first binds to the A and B2 elements. c shows that ORC recruits Cdc6 (in orchid color). d shows that Cdt1-Mcm2-7 complex in open ring conformation (Cdt1 in Mocha color, Mcm2-7 in Asparagus color, a channel between Mcm2 and Mcm5 subunits is indicated) is recruited by ORC-Cdc6. DNA is aligned to the channel in the Mcm2-7 hexamer. The Mcm2-7 complex is oriented as the hexamer C-terminus binding to ORC-Cdc6. e shows the intermediate known as OCCM with the double stranded DNA inserted into the channel between Mcm2 and Mcm5 subunits in the Mcm2-7 hexamer. The hexamer is partially closed. f shows that the ATP hydrolysis by the Mcm2-7 expels the first Cdc6 and then Cdt1, creating the OM complex. g shows that ORC flips over to the N-terminus side of Mcm2-7 and presumably binds to the B2 element on DNA, creating the MO complex. The structure of the MO complex was modeled by real-space-refining docked coordinates of MCM (PDB 6EYC255), ORC (PDB 5ZR1139) and an N-terminal Orc6 homology134. h shows that ORC can now recruit a second Cdc6, creating a binding site for a second Cdt1-Mcm2-7 complex that can be loaded in an opposite orientation to the first Mcm2-7. The Mcm2-7 double hexamer, possibly with ORC still bound to the DNA, is then ready to be activated and can unwind the double stranded DNA when entering S phase.
ORC was identified 30 years ago in budding yeast S. cerevisiae (herein referred to as ScORC) and is composed of six distinct subunits (Orc1-6) that binds in a sequence-specific manner to replication origins29. In some yeast, but not all, origins of DNA replication correspond to genetic elements called Autonomously Replicating Sequences (ARS) that confer upon a plasmid the ability to replicate as a mini chromosome once per cell division cycle like the rest of the genome29. ARSs may be DNA sequence specific, as in S. cerevisiae, but as discussed below, this is not a universal feature. Cdc6, which is related in amino acid sequence to the Orc1 subunit of ORC, was first identified in a screen for mutations that caused a cell division cycle arrest in S. cerevisiae30 and later found to bind to ORC and be essential for pre-RC assembly31,32. The Schizosaccharomyces pombe homolog Cdc18 was shown to be necessary for entry into DNA replication33. Cdt1 was named Cdc10-dependent transcript 1 since it was initially identified as a gene regulated by the Cdc10 transcription factor in fission yeast S. pombe34. Later, it was shown to be equivalent to the Replication Licensing Factor -B (RLF-B) that is required for loading of Mcm2-7 onto DNA in a Xenopus egg extract and thus for assembly of the pre-RC35–37 and has conserved functions throughout eukaryotes38. Thus, Cdt1 was renamed as chromatin licensing and DNA replication factor 1.
Genes encoding some of the six subunits of the Mcm2-7 helicase were originally identified in different genetic screens in S. cerevisiae, either as genes required for cell cycle progression39 or for selective mini-chromosome plasmid maintenance40. Subsequently, these Mini Chromosome Maintenance proteins were grouped together due to their sequence similarity41. Further studies indicated that they are involved in the initiation of DNA replication and for replication fork progression42–44. Factors that were identified and characterized in one species guided its homologue identification in other species. For example, Replication Licensing Factor-M that was required for licensing of DNA replication in Xenopus egg extracts consisted of Mcm2-7 proteins45–47. Likewise, the identification of ORC homologues in Xenopus and mammals was guided by ScORC48–50.
A key feature of DNA replication in eukaryotes is that the entire genome must be duplicated, but importantly, every chromosome or replicon must be copied only once per cell division cycle. A model for cell cycle-control of origin licensing and once per cell cycle replication was introduced by Blow and Laskey in which fully replicated DNA could not be re-replicated in a single cell cycle due to exclusion of origin licensing factors from the DNA by the nuclear envelope51. Although cell cycle-dependent exclusion of replication licensing factors does exist, this mechanism turned out not to control once per cell cycle replication. Instead, cell cycle-dependent Deoxyribonuclease I (DNase I) hypersensitive site changes at S. cerevisiae replication origins in vivo led to the idea that origin licensing occurred by pre-RC assembly at replication origins only during G1 phase of the cell cycle52. Furthermore, activation of pre-RCs during S phase also resulted in their destruction and hence, re-initiation could not occur on replicated DNA. Origin licensing is restricted to the G1 phase of the cell cycle when S phase cyclin-dependent kinase (CDK) activity is absent (Figure 3). Active CDK phosphorylates pre-RC components and inhibits pre-RC assembly and hence pre-RC assembly cannot occur once cells enter into S phase and can only re-occur after cells pass through mitosis when CDKs are destroyed53,54. Thus, low CDK activity is necessary for pre-RC assembly at replication origins55. A reduction of CDK activity56,57 or over-expression of a CDK inhibitor protein58 was found to lead to DNA re-replication. Phosphorylation of ORC, Cdc6 and Mcm2-7 protein by CDK control pre-RC assembly59. Paradoxically, the same CDK activity that prevents pre-RC assembly is activated just before S phase and is required for the initiation of DNA synthesis at each origin. Active CDK (S-CDK) phosphorylates two initiation proteins Sld2 and Sld3 to drive pre-Initiation Complex (pre-IC) formation, during which the active helicase is assembled prior to the start of actual DNA synthesis at the origin23,24,53,60–62. Another kinase essential for cell cycle regulation of DNA replication is Dbf4-dependent Cdc7 kinase (DDK), which phosphorylates Mcm2-7 and drives the recruitment of Cdc45, a component of the pre-Initiation Complex (pre-IC) to form an active helicase that contains the Cdc45-Mcm2-7-GINS tetramer complex, called the CMG23,24,63–65. Thus each pre-RC is destroyed when the Mcm2-7 double hexamer is converted into the two active helicases. Pre-IC formation is a subsequent stage following the pre-RC assembly, in which licensed origins initiate replication in a temporally regulated manner throughout S phase60,66. The pre-IC is formed by recruiting additional factors including Cdc45, GINS, Sld7, Sld3, Sld2, Dpb11, Pol ε, RPA and Mcm10 to the Mcm2-7 double hexamer23,24,67, thereby activating the 3’ to 5’ helicase activity22,23,68. Then DNA Polymerase α-primase is recruited to prime the initiation of replication and DNA polymerases drive the bi-directional replication forks, where Pol ε is dedicated to the replication of the leading strand, while Pol δ is dedicated to the replication of the lagging strand69–74. Pre-RC assembly with purified proteins was achieved first75,76. Later complete replication of DNA or chromatin templates was reconstituted with purified proteins that primarily were identified either from studies of Simian Virus 40 DNA replication in vitro or yeast biochemistry and genetics23,24,70–72,77,78.
Figure 3 |. Waves of cyclins and DNA replication proteins in human and budding yeast during cell cycle progression.
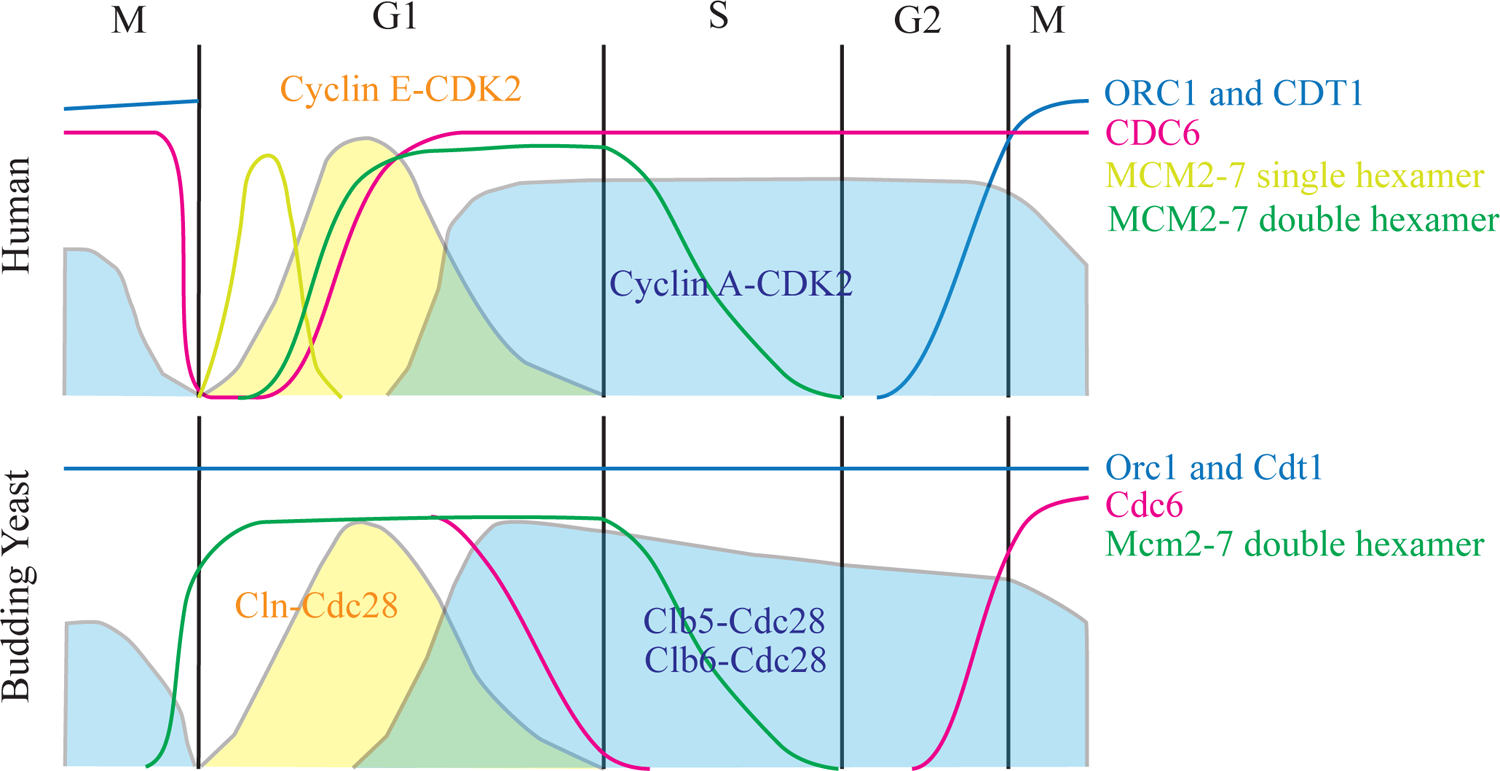
Human and budding yeast replication proteins ORC1/Orc1 and CDT1/Cdt1 (in blue) and CDC6/Cdc6 (in pink) proteins levels as well as Mcm2-7/Mcm2-7 single hexamer (in lime green) and Mcm2-7/Mcm2-7 double hexamer (in dark green) loading levels are shown as lines. Pre-RC assembly corresponds to Mcm2-7/Mcm2-7 double hexamer formation. Cyclin-dependent kinases activities are shown as solid areas with Cyclin E-CDK2 in human and Cln-Cdc28 in budding yeast are shown in yellow, while Cyclin A-CDK2 in human and Clb5-Cdc28 and Clb6-Cdc28 in budding yeast are shown in dark blue. G1, S, G2, M phases in cell cycle are indicated.
3. Origin Specification
4.1. Plasticity of Replication Origins
The location of origins of DNA replication can be as defined as 100~200 base pairs (bp) in budding yeast to as broad as ~150 kilo base pair (kb) initiation zones within megabase-sized replication domains in human chromosomes79–82 (Table 2). Despite the large diversity in configuration of replication origins, components of the pre-RC are conserved throughout eukaryotes83,84. However, the mechanism of how ORC recognizes and makes contacts with DNA varies considerably in cells across eukaryotic species.
Table 2 |. Replication origins configurations.
Table shows the comparison of three typical replication origins configurations.
| Species | Primary DNA Sequence | Sequence specificity | Replication origins defined by | Precession |
|---|---|---|---|---|
| Metazoan | GC-rich | Nonspecific | Epigenetically defined | ~1Mb replication domains with clustered initiation zones within. No obvious DNA sequence specificity. |
| S. pombe related fission yeast | AT-rich | Nonspecific | Orc4 AT-hook in ORC binds A.T rich DNA, not specific DNA sequences | ~500 to ~1500bp (can be as precise as ~100bp) |
| S. cerevisiae (most recently branching yeast) | AT-rich | Sequence specific, ACS identified | ORC subunits interact with defined ACS DNA sequence, both DNA sequence specific and non-specific | ~100 to ~200bp |
During DNA replication in Drosophila and Xenopus early embryos, plasmid or chromosomal DNA replication initiation was reported to be random or with greatly relaxed sequence specificity during the very rapid early cleavage stages85–87. The initiation of DNA replication in these early replication cycles occurs about every 3 kb along chromosomes with little temporal variation, but during the mid-blastula transition (MBT), when zygotic transcription begins, replication initiates from spaced out, specific sites in intergenic regions in a temporal order of replication throughout S phase88,89. The same pre-RC proteins are required for both the rapid embryonic cycles and the slower, temporally regulated somatic cell modes of DNA replication84,90. Thus, replication origin location has considerable plasticity during development. Such flexibility has also been apparent in yeasts where origins of replication can be DNA sequence-specific. In S. cerevisiae for example, when some or all the canonical origins on Chromosome III were deleted, the strains seem to grow relatively fine91,92 and DNA replication could initiate from non-canonical sites93. A similar phenomenon occurs in Archaea where Orc1/Cdc6 related proteins normally bind DNA to sequence-specific DNA replication origins, but deletion of all origins resulted in replication initiation at random sites94. Therefore, any stretch of DNA seems to have the potential for initiating replication and these observation raised the question as to whether DNA replication initiation required any sequence specificity at all51,86,95,96. To complicate mapping of origins even further, even in S. cerevisiae where ORC determines the location of pre-RC assembly at specific DNA sequences, the Mcm2-7 double hexamer can translocate along the chromosome driven by transcribing RNA polymerases so that CMG helicase activation and thus initiation of DNA replication could occur at a distance from the pre-RC assembly site97. Therefore, mapping DNA replication start sites (Table 3) may not be the equivalent of mapping the location of origin specification and pre-RC assembly.
Table 3. Origin mapping methods.
Table and figure motifs show many methods to map origins of DNA replication in cell populations and in single cells or molecules96.
| Method | Description | Cite | Comments | Illustration |
|---|---|---|---|---|
| 2D gel assays and fork direction analysis | Uses distinct gel migration patterns to characterize replication intermediates and detect origin firing efficiency | Brewer and Fangman117 Huberman et al., 118 | Pooled data using population cells |
|
| Density transfer | Uses density isotypes to distinguish newly synthesized DNA form parental DNA followed by DNA sequencing and quantitative copy number profiling to map general patterns of replication timing | Gilbert,84; Pope et al., 14; Rhind and Gilbert6. |
|
|
| ChlP-seq (chromatin immunoprecipitation followed by DNA sequencing) | Identify locations of ORC and MCM binding sites to predict the location of replication origins | Belsky et al., 256; Long et al., 214; Lucas and Raghuraman,257; MacAlpine et al190,258 |
|
|
| Bubble-seq | Trap replication bubbles and sequence DNA | Mesner et al., 259 |
|
|
| Ini-seq (initiation site sequencing) | Label nascent DNA with DNA analogs and either immuno-precipitate the labeled nascent DNA or separate the labeled nascent DNA from unreplicated DNA using density difference | Guilbaud et al., 107; Langley et al., 260. |
|
|
| OK-seq (Okazaki-fragment sequencing) | Isolate and sequence Okazaki fragments to map replication fork direction in asynchronous population of cells | Petryk et al., 82. |
|
|
| Repli-seq | Label nascent DNA with BrdU; look for the enrichment of BrdU-immunoprecipitated DNA in late S phase cells compared to early S phase cells | Zhao et al., 158. |
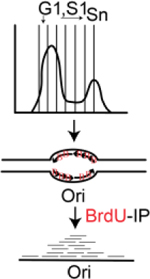
|
|
| SNS-seq (Short Nascent Strand-sequencing) | Identify nascent RNA-primed DNA synthesized at origins by the primase DNA polymerase a (Pol a) | Besnard et al., 261; Cadoret et al., 157; Cayrou et al., 262; Picard et al., 263 |
|
|
| S-jump based computational origin prediction | S-jump based method which computes the excess G over C and T over A along one DNA strand where abrupt changes correlate with the location of replication origins (N-domains and U-shaped domains of skew jumps), however the location of actual origins needs experimental validation | Brodie et al., 264. | Computational prediction data using population cells |
|
| Electron microscopy and fiber autoradiography | Visually observation of replication origins on radiolabeled DNA | Single molecule data using population cells |
|
|
| DNA combing | Fluorescently label nascent DNA. DNA molecules stretched and aligned along a slide and visualized by various fluorescence microscopy techniques | Bensimon et al., 265; Michalet et al., 266; Pasero et al.267. |
|
|
| DNAscent; FORK-seq etc. | Nanopore sequencing based methods that detect DNA analog labeled nascent DxwNA | Hennion et al., 268; Muller et al., 269; Georgieva et al., 270; Boemo271 |
|
|
| ORM (optical replication mapping) | Fluorescent nucleotide analog pulse-labeled nascent DNA in combination with the optical mapping method (Bionano Genomics) to map long individual DNA molecules | Wang et al., 109 |
|
|
| scRepli-seq | Uses a commercial whole genome amplification kit, SeqPlex from Sigma, to prepare sequencing libraries | Miura et al., 272; Takahashi et al., 273. | Single cell data |
|
| LIANTI (Linear Amplification via Transposon Insertion) | Combines the T7 in vitro transcription to the Tn5 transposition to avoid the intrinsic limitations of Tn5 transposition being symmetric and reduce biases and errors during the amplification steps | Chen et al., 274 |
|
|
| DLP (direct library preparation) | Uses nanoliter-volume reactions to adapt the conventional transposase-based library construction to single cells | Laks et al., 275; Zahn et al., 276. |
|
Further evidence for plasticity in the location of origins of DNA replication stems from the redundancy of initiation sites. Even in the S. cerevisiae genome, where origin location is sequence specific, there are ~12,000 potential matches to the core ARS consensus sequence (ACS), however there are only ~400 functional origins98. Within a given cell division cycle, not all of these ~400 origins become active, and within a single cell, the origin activity is known to be stochastic and vary extensively between cells99–104. These single molecule studies revealed more active origins than were found in bulk cell populations, likely due to some origins being used at a very low frequency that are not detected in the analysis of origin activity in a population of cells100,101. In metazoan cells and fission yeast, using single molecule methods (Table 3), DNA replication origin usage was also shown to be stochastic105–109. Even though different single cells could have different cohorts of active origins, the data from population studies show reproducible spatial, temporal and “non-random” replication timing profiles, indicating that origin redundancy provides the diversity and plasticity for a “controlled-stochastic” origin selection process. The existence of an excess of licensed origins over the actual number used to replicate the genome in any given cell cycle resulted in the idea of dormant origins that can become active if necessary to complete genome replication, particularly in situations when cells experience DNA damage or replication stress110–114.
4.2. Replication Origin Configurations
The simplest eukaryotic replication origin configuration lies in a small clade of budding yeasts that include S. cerevisiae. Autonomously replicating sequences (ARSs) were identified based on the ability of some cloned, genomic fragments from the S. cerevisiae genome to confer high-frequency transformation of yeast cells with a circular plasmid DNA and thence stable inheritance of the resulting mini-chromosomes115,116. Later, 2D agarose gel electrophoretic analysis showed that DNA replication initiated at the ARS sequence and nowhere else on ARS containing plasmids117,118. DNA sequence analysis demonstrated that ARSs contained a short, 11-base pair conserved ACS DNA sequence (now called the A element) but this was not sufficient for origin activity119,120. Multiple elements within ARS1 (ARS416) were identified to be critical for origin activity121. The essential A element contains the ACS, and together with three important B elements, are conserved and provide binding sites for ORC and accessory proteins121–123. The B2 element was previously thought to be an instability region, called duplex unwinding element (DUE), where the initial unwinding of DNA occurs124. However, later studies found that B2 is unlikely to play this role but instead a role for pre-RC assembly66,125. Interestingly, the B2 element contains a partial, inverted ACS sequence motif to which ORC can bind29,125,126. Whether one127–130 or two126,131 ORCs are needed for loading the Mcm2-7 double hexamers has been investigated.133. Most studies to date have used the ARS1 (ARS416) origin that has the A and B1 elements as the primary ORC binding site and the B2 element as a weak, inverted ORC binding site with a short space between the two ORC binding sites122,126,132,133. Biochemical and structural studies of a pre-RC assembly intermediate Mcm2-7-ORC (MO) on ARS1 DNA and separate, single molecule analysis of pre-RC assembly show that ORC first binds to the A and B1 elements and loads the first Mcm2-7 hexamer by interacting with the Mcm2-7 C-terminal winged-helix domains and then flips to the amino-terminal side of the loaded Mcm2-7 to load a second Mcm2-7 hexamer (Figure 2, f–g), thereby creating the pre-RC with a single ORC134–137. It is possible that other origins with a different spacing between the A/B1 and B2 elements would employ two ORC molecules to load the two hexamers of Mcm2-7138, and this needs further investigation.
Precisely how ORC binds to origin DNA was not understood until structural studies of ORC bound to DNA suggested that an α-helix in the Orc4 subunit that binds to a major grove in the DNA and a loop in Orc2 subunit that contacts DNA contribute to DNA sequence specific ORC binding139,140. The Orc2 loop is essential and subtle mutations in the Orc4 α-helix, more specifically F485 and Y486 on the α-helix, bind to a different ACS recognition motif141. Furthermore, deletion of this α-helix within Orc4 has been shown to have more plasticity in its origin selection142. Strikingly, the Orc4 α-helix and the Orc2 loop are not found in ORC from most eukaryotes, including most fungi, plants and animals, raising the issue of how ORC from these species interacts with DNA and determines the location of origins of DNA replication. One possibility is that ORC from S. cerevisiae has lysine-rich sequence in an intrinsically disordered region (IDR) of the Orc1 subunit that binds to a minor grove in the origin DNA and this amino acid sequence is conserved in other eukaryotes and has been shown in ORC from human cells to bind DNA, possibly a short sequence the ORC1 IDR139,143.
In some fission yeasts, such as Schizosaccharomyces pombe (S. pombe), S. octosporus, and S. japonicus, replication origins lack consensus sequences144,145 but are found to be AT-rich, which are called AT islands and can be as short as ~100bp in length to the size of >500bp146,147. All these Schizosaccharomyces species contain an AT-hook domain at the N-terminus of the Orc4 protein that preferentially binds to AT-rich DNA sequences and is necessary for origin recognition148,149 (Table 2). There are nine AT-hook subdomains in S. pombe, while S. octosporus has four and S. japonicus has five146, and each subdomain is capable of binding 6–8 nucleotides of DNA150. Interestingly, bioinformatic analysis of some other fungi, such as Neurospora crassa and Aspergillus nidulans, suggest that AT-hooks at their Orc4 N-terminus might be used to specify replication origin location at AT-rich sequences in their genomes (N. Zali and B. Stillman, unpublished). But again, like the DNA sequence specific origins that bind the Orc4 α-helix, the A-T-hook mechanism of origin specification is restricted to a small fraction of eukaryotic species, raising the question of how the location of origins of DNA replication are specified in most eukaryotes.
In metazoan species, it has been more difficult to locate replication origins and independent studies with different methods do not show high concordance of origin features and locations90,151, likely due to the large genome sizes, the existence of large replication initiation zones, analysis of different cell types, movement of Mcm2-7 double hexamers and technical limitations. In the late 1960s, visualization of replication tracts in mammalian cells revealed that replication initiates from multiple sites that can be simultaneously active at adjacent sites in the chromosomes152. Following the release of the first draft of the human genome, techniques such as quantitative mapping of RNA-capped short nascent strands (SNS) and 2-dimensional gel electrophoresis were employed to map replication origins84,153–157. In the past few years, many methods have been developed to map replication origins in metazoan genomes (Table 3).
Data from these newer mapping approaches show that replication origins can be kilobase-sized zones in metazoans and were reported to fire stochastically with no sequence specificity82,109,158. They are organized into spatially and temporally regulated replication timing domains6. Consistently, metazoan ORC was shown to bind DNA with no apparent sequence specificity in vitro159–161. Nevertheless, a recent study of replication origins in mouse embryonic stem cells using a CRISPR-mediated deletion and inversion of TAD boundaries suggest that cis-acting DNA elements are required for DNA replication initiation162. How these cis-acting sequences contribute to DNA replication is not known. Despite the lack of sequence specificity, G-rich sequences which have the potential to form G-quadruplex secondary structure have been found to be associated with sites of efficient metazoan replication initiation and may play a role in defining replication origins, although this is not certain163–165. ORC from human cells was reported to preferentially bind to G4 motifs on single-stranded DNA while it binds to dsDNA randomly166,167, although how this relates to establishing the location of replication origins is not clear. The lack of defining DNA features at metazoan origins suggest that the dispersive initiation zones could be composed of multiple discrete replication origins that fire stochastically. Consistent with this line of reasoning, a recent modified Ini-seq method (Table 3), which shows precise mapping of 23,905 replication origins with a replication initiation efficiency score assigned to each origin indicates that origin firing within the dispersive initiation zones are not randomly distributed but are arranged hierarchically, with a set of highly efficient origins near defined zone boundaries107. Based on this study, the concept has emerged that dispersive replication initiation zones contain within them discrete, highly efficient replication origins. The question remains is how are these clustered sites and efficient origins determined?
For the bulk of eukaryotes, including animals, plants and most yeasts, the replication origins lack consensus DNA sequence motifs, with the notable exception sequence-specific ARSs in the small S. cerevisiae related clade of budding yeasts. Interestingly, ARSs are not strictly required for the initiation of DNA replication in vitro168,169 and can initiate from non-canonical sites in vivo when consensus sites are deleted from a chromosome93. Moreover, not all origins contain a clear consensus sequence170,171, and ORC can bind to non-origin DNA and load Mcm2-776. But ORC can direct sequence-specific replication in animal cells since fusion of a DNA binding protein such as Gal4 to ORC subunits in Drosophila can promote pre-RC assembly and initiation of DNA replication at a chromosomal Gal4 DNA binding site, even though replication in Drosophila is normally not sequence specific172. This raises the possibility that ORC may interact with sequence-specific DNA binding proteins to locate origins in the genome.
4.3. Local Environments Regulate Replication Origin Selection
Origin licensing and activation steps are critical and must be tightly regulated to ensure the accurate and complete duplication of the genome. In higher eukaryotes licensing in G1 phase is known to load an excess of Mcm2-7 protein onto the chromosomes over what is needed for genome replication, and some of this excess Mcm2-7 can be employed to activate dormant origins that protect the cells from replicative stress and rescue of stalled forks110–112. Even with random replication initiation, origins are spatially separated to ensure completion of a normal S phase173,174. What are the regulators that determine origin location and activity? Previous studies suggested that the chromatin-associated complexes may restrict pre-RC formation and the use of naked DNA for pre-RC assembly can bypass the restrictions imposed by chromatin168, implying the chromatin or local environments could play roles in regulating replication origin licensing and activation175,176. Multiple regulatory factors in local environments could play critical roles in specifying the location of origins of DNA replication, such as ORC associated proteins, epigenetic replication timing patterning, rate limiting replication factors, gene transcription, nucleosome occupancy, histone modifications, and chromatin structures and 3D organization of chromosomes.
For example, it is possible that within local chromatin environments, ORC-interacting proteins such as HMGA1177, the ORC-associated protein ORCA178–180 or noncoding RNAs181–183 could recruit ORC to specific chromatin sites and contribute to location-specific replication origins in metazoans. ORC, more specifically ORC1, has been shown to act like a pioneer factor by binding to the condensed chromosomes during mitosis, immediately after nuclear envelope breakdown184,185, similar to the transcription factor FoxA1 that also binds mitotic chromosomes and promotes site specific gene transcription in G1 phase186. When ORC1 is inherited into the newly formed daughter nuclei, it binds the other ORC subunits and forms spatiotemporal patterns throughout G1 phase that resemble the spatiotemporal patterns of DNA replication sites in S phase, suggesting an epigenetically inherited distribution of replication patterning that is determined in G1 phase by ORC187. How this temporal patterning of ORC1 in human cells occurs is not known. Since the activation of origins of DNA replication throughout S phase competes for rate limiting replication factors7,10,188, rate limiting factors that function in G1 phase may influence the nuclear patterns of ORC1. Alternatively, replication timing in S phase is known to be controlled by factors such as Rif1 that binds Protein Phosphatase 1189 and regulates DDK activity, so it is possible that these factors also play a role in G1 phase to influence ORC1 spatiotemporal nuclear localization.
The location of transcription start sites (TSSs)190 and CpG islands191 are known to correlate with the location of replication origins or pre-RC binding sites, suggesting that the mechanisms that specify TSSs may also determine the location of origins of replication. For example, the forkhead transcription factors (Fkh1 and Fkh2) were found to be necessary for replication from 30% of early-firing origins192. However, the relationship between transcription and origin localization is complicated because gene transcription can inhibit pre-RC assembly or restrict the location of where pre-RC assembly can occur, and even after the Mcm2-7 double-hexamers are loaded onto DNA, transcription can move them97,193–195. Furthermore, site-specific replication initiation can be artificially achieved by manipulation of the local chromatin environment such as DNA methylation196 or transcription factor targeting197, but clearly both methylation and transcription factors also affect transcription, raising cause and effect issues. Nevertheless, DNA methylation has been shown to be critical and required for maintaining replication timing and normal mammalian embryo development198,199. But whether this is a direct effect on the mechanism of origin licensing is not known.
Nucleosome depleted regions at replication origins have been shown to be critical for ORC binding and origin efficiency190,200–203, and ATP-dependent chromatin remodelers are required for establishing precisely positioned nucleosomes flanking an ACS on a template DNA in vitro77,204,205. Thus, at the very local level, chromatin structure can influence pre-RC assembly, perhaps because open chromatin would provide more accessibility to replication factors. But the chromatin structure over larger distances can influence the location or activation of origins of DNA replication. For example, the activation of one origin may increase the probability of firing of neighboring origins206–208, as long as the origins are not too close to each other, in which case suppression of origin firing can occur209,210. Chromatin context also influences the timing of initiation of DNA replication since ectopically positioning an early-firing origin to the late-replicating telomeric region led to the delayed firing of the translocated origin, and conversely, a late-firing origin can be replicated early when placed onto a small replicating plasmid211. Thus temporal patterning of DNA replication is not sequence specific, even in a cell where origin specification utilizes specific DNA sequences.
Replication efficiency in metazoan cells has been shown to correlate with DNA fragment size212. Furthermore, both cis-acting DNA elements and epigenetic factors can influence replication timing162,213. Histone modifications that define open chromatin have been linked to early replication origins, such as H2A.Z214, H3 and H4 acetylation, all three methylation states of H3K4 and all three methylation states of H4K20215–218. A very convincing correlation between replication origins and histone modifications has been reported in C. elegans21. In this study, the inter-origin distance was on average 40kb across the entire genome and histone modifications H3K4me2, H3K4me3 and H3K27Ac, all associated with active chromatin, co-localized with the location of origins in the genome. This is consistent with open chromatin influencing both transcription and replication start sites in the genome. In contrast, repressive chromatin modifications, such as H3K9me2/3 and H3K27me3, are linked to late replication origins in human cells219. Consistently, tethering a histone acetylase (HAT) or a histone deacetylase (HDAC) to the replication origin regions influenced replication timing9. Indeed, ORC can directly or indirectly interact with HAT or HDAC, such as HBO1(KAT7)220 and Sir2221 and directly with modified histones such as H4K20me2 and H3K9me3216,222. These enzymes could either recruit ORC or be recruited by ORC to specific genomic sites and have the potential to assemble pre-RCs and contribute to regulating replication timing.
However, the histone modifications correlated to certain groups of replication origins are not universal for all origins. There are also more modifications across the genome than replication origins and these modifications are involved in diverse processes, such as transcription and response to DNA damage, etc., indicating more factors are needed to determine replication origin locations. But the open chromatin idea raised another question: how does ORC localize to the heterochromatic regions of the genome? It is possible that the 3D chromosome organization and nuclear localization play a role in giving accessibility to replication factors in different spatiotemporal domains6. CTCFs and cohesins together with other proteins facilitate and shape loop extrusions in chromosomes223, and it has been proposed that the replication origins or initiation zones are colocalized with cohesins at the base of the loop extrusions and adjacent origins within these loops could have similar replication timing224–227. Pre-RC assembly occurs on euchromatin in early G1 phase and on heterochromatin in mid G1 guided by the ORC binding protein ORCA180 and this differential assembly may reflect the dynamic intranuclear changes in the localization of ORC1 observed during G1 phase187.
Taken together, all the regulatory factors display dynamic influences on individual origins and the integration of these multiple controls at each origin could determine differential origin activation probability and timing in single cells. These combinatorial factors could also contribute to the “non-random” optimal origin usage distribution profiles in cell populations, explaining the “controlled-stochastic” origin firing regulatory mechanism. Even so, replication timing is stochastic and can vary between cells and even between homologs, suggesting that origin activation can be independent of factors that determine global replication timing108.
In some special circumstances, DNA replication or replication origin firing is developmentally regulated by cis-acting factors that result in overriding the once-per-cycle replication rule. Egg shell genes in Drosophila nurse cells surrounding the egg are amplified by up to 16 to 32-fold by re-initiation of DNA replication228. ORC and the sequence-specific transcription factor E2F are required for re-amplifying the egg shell genes229. Interestingly, another exception occurs within stretched skin cells in zebrafish larvae where the cell can divide without DNA replication230. But these examples are in terminally differentiated cells, therefore, they no longer further differentiate and as a result, genome instability does not matter.
4. The evolutionary transitions into sequence specific origins
Origins of DNA replication in bacteria are DNA sequence-specific and thus the finding of sequence-specific origins in S. cerevisiae was not a surprise, but these two sequence-specific origin systems are functionally very different from each other. Furthermore, the arrangement of S. cerevisiae sequence-specific origins that include a combination of essential and non-essential DNA sequence elements, reminiscent of gene promoters, was an unexpected and unprecedented organization of an origin of DNA replication121. Importantly, the presence of a conserved origin DNA sequence facilitated in the discovery of ORC that then accelerated both genetic and biochemical studies to understand pre-RC assembly and the initiation of DNA replication and its control. Thus, S. cerevisiae was a fortuitous choice to study the initiation of DNA replication because its origin sequence specificity gave confidence that ORC was the initiator, which then resulted in the identification of ORC in other eukaryotic cells. The existence of highly conserved ORC and other pre-RC proteins raises the question of why there is significant variation in mechanisms of origin structure and specification?
It is apparent that in most eukaryotes, origins of DNA replication lack DNA sequence-specificity and how ORC is localized to these chromosomes and how it determines the location of origins of DNA replication is not completely understood. Strict DNA sequence specific origins exist in a small clade of S. cerevisiae-like budding yeasts, whereas in some other fungi, like S. pombe, ORC localizes origins to AT-rich locations in the genome. Unlike other cis-acting elements, such as the centromeres and transcription factor binding sites, the loss of function of any one replication origin is unlikely to have a severe effect or apparent fitness loss for the cell231. These observations raise the question of why sequence-specific origins exist and are rare?
In the transition budding yeasts that are distant from the S. cerevisiae clade of yeasts and lack the Orc4 α-helix, such as Candida albicans and Pichia pastoris, ARS sequences that are independent of a centromere have been identified232,233. In the case of C. albicans, proposed origins (proORis) on chromosome arms were predicted based on the localization of ORC chromatin immunoprecipitation results and nucleosome free regions in the genome and a loose AC-rich DNA sequence was observed232. However multiple mutations in this sequence only reduced ARS activity by 3-fold. Some of these ARS sequences were shown by 2D gel analysis to have weak origin activity, whereas other proORis that functioned as ARSs lacked origin function. In the case of P. pastoris, analysis of ARS function on plasmids identified two separate ARS sequences, one class was a GC-rich sequence that when mutated eliminated ARS activity and a separate class with AT-rich sequences233. A representative of both classes was shown to colocalize with origin activity in the chromosomal context. In both the studies with C. albicans and P. pastoris, the biochemical role of the conserved sequences was not addressed. DNA sequences associated with ARS activity have been shown to either bind the initiator protein ORC, exclude nucleosomes or bind to other proteins that mediate ARS plasmid stability, such as proteins that tether plasmids to the nuclear periphery234. It is also possible that the GC-rich ARSs in P. pastoris overlap with gene promoters and these may contribute to origin activity. Thus, future studies of ARS activity in yeasts must address the biochemical function of any proposed conserved sequences to reveal how they contribute to ARS or origin activity, which may not be equivalent.
5.1. Co-evolutionary transitions of origin specificity, gene silencing mechanisms and centromeres
In species that lack DNA sequence-specific origins of replication, inherited transcriptional gene silencing can occur by RNA interference (RNAi)141. These species also have epigenetically defined centromeres (CEN)235. A group of intermediate branching yeasts seem to be in the transition of losing RNAi (e.g., C. albicans236; Figure 4), since they still carry some but not all genes that encode RNAi proteins (or non-canonical Dicer gene). The most recently branching budding yeasts, including S. cerevisiae (Figure 4), have completely lost RNAi and instead, they have gained ORC-Sir4 mediated gene silencing. This has been accompanied by the acquisition of the Orc4 α-helix, the Orc2 DNA interacting loop and sequence specific origins141 and a DNA sequence-specific, point CEN configuration235.
Figure 4 |. Co-evolution of gene silencing mechanisms, centromeres and replication origin sequence specificity.
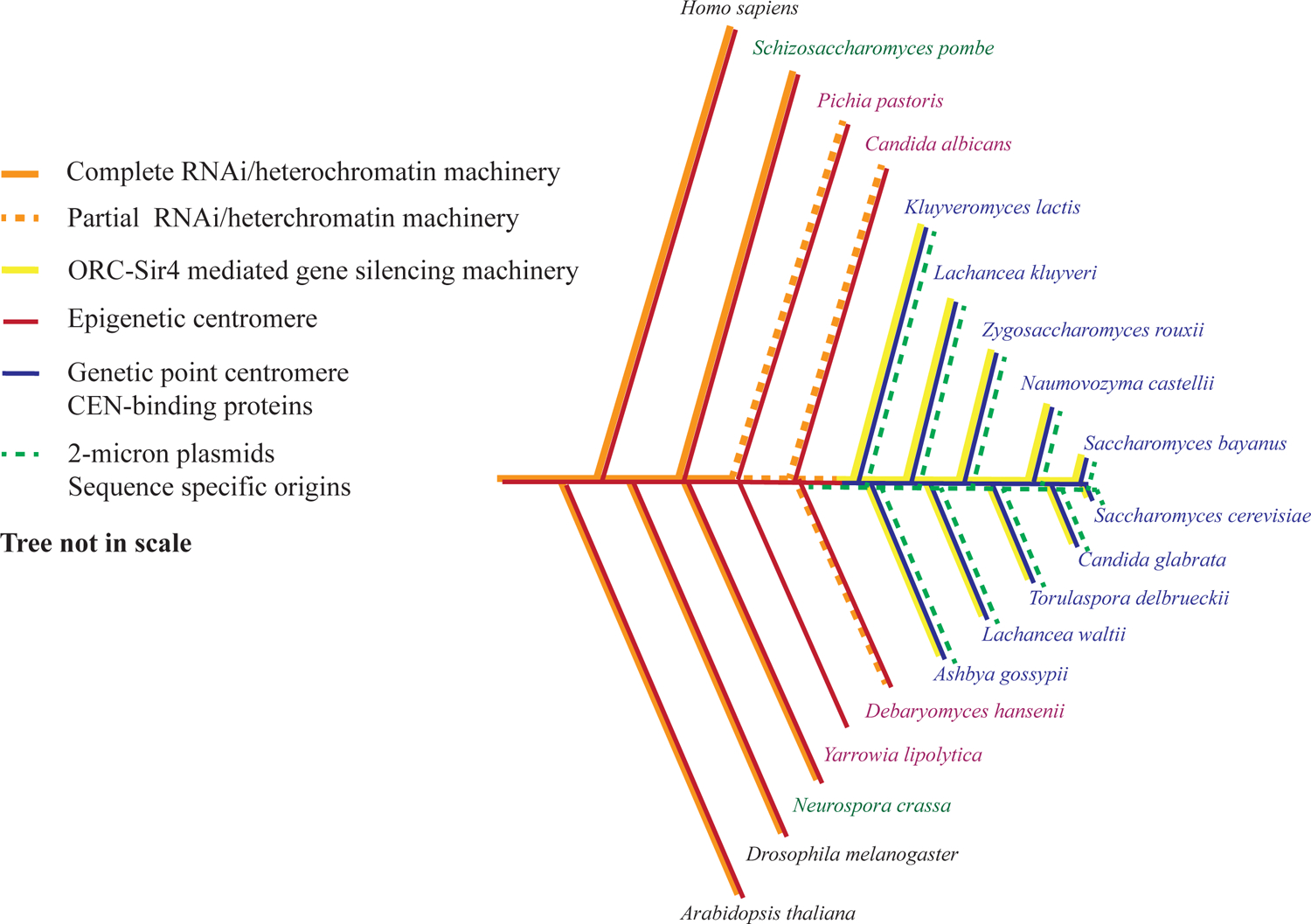
[Adapted from235 and141]. The phylogenetic tree is not drawn to scale. Most eukaryotes (font in black), including basal branching yeasts (font in green), have complete RNAi machinery or full complements of heterochromatin. Intermediate branching yeasts (font in purple) harbor partial components of RNAi or heterochromatin machinery, whereas the Saccharomycetaceae yeast family, the most recently branching yeasts, (font in blue) have completely lost RNAi/heterochromatin machinery and acquired ORC-Sir4 mediated gene silencing141. Meanwhile, the selfishly propagating 2-micron plasmids exist in Saccharomycetaceae lineage, where the strains have DNA sequence-defined point centromeres as well as sequence specific replication origins. Y. lipolytica lacks RNAi as well as the SIR proteins141, so it is not clear what gene silencing mechanism it uses.
5.2. Evolutionary driving forces
Previously, it has been suggested that acquiring and maintaining a beneficial killer virus could explain the loss of RNAi in budding yeast236. In addition, the loss of RNAi and the acquisition of sequence-specific point centromeres were suggested to co-evolve with maintenance of the circular 2-micron plasmid, which is known as a selfish DNA element that uses the host segregation apparatus components for plasmid stability235. It was proposed that DNA sequence-defined, point centromeres were acquired by integration of the selfish circular 2-micron plasmid into the genome, bringing with it DNA sequences that eventually functioned as centromeres235. RNAi contributes to the silencing and stability of repetitive DNA sequences like heterochromatin satellite repeated sequences at centromeres and remnants of transposable elements, as well as organization of rDNA repeats237. The S. cerevisiae and K. lactis (Figure 4) clade of budding yeasts have lost centromere-associated repeated sequences and have replaced RNAi with Silent Information Regulator (SIR) proteins that silence gene transcription in a DNA sequence-specific manner at the silent mating type loci, as well as maintaining rDNA and telomere repeats by preventing recombination238. Interestingly, the acquisition of the Sir4 protein that binds either directly to ORC in K. lactis or indirectly via Sir1 to ORC in S. cerevisiae, occurred with the acquisition of the Orc4 α-helix and the Orc2 loop, both of which contribute to the DNA sequence-specific DNA binding by ORC141. ORC binds in a DNA sequence-specific manner to the cis-acting silencer elements that flank the silent mating type loci, thereby establishing precise localization of transcriptionally silent regions in the genome by recruiting SIR proteins to these loci. The S. cerevisiae and K. lactis clade of budding yeasts have a dense genome with ~70 percent of the genome encoding protein and little gene spacing and repetitive DNA. To avoid conflicts between replication and transcription, it makes sense to place origins of DNA replication in intergenic regions and this may well be the reason why the acquisition of DNA sequence-specific origins of DNA replication in these yeasts evolved. But how were the sequence-specific origins acquired?
Interestingly, the naturally occurring replication origin from the 2-micron plasmid has similar properties to current S. cerevisiae chromosomal replication origins239,240. Like the previously proposed point centromere acquisition model235, it is appealing to propose that the DNA sequence specific replication origins may also have been acquired from the 2-micron plasmid. Unlike the centromere where one per chromosome is required, DNA sequence specific origins must have spread throughout the genome, particularly in intergenic regions. Hence, CEN and replication origin ARS sequence may originally have coincided with the same sequence that later evolved to become physically and functionally separated to ensure that the genetic information from the large sized eukaryotic genome can be faithfully replicated in time from multiple origins and the segregation into daughter cells during cell division to be well-coordinated by a single CEN. Indeed, one example of intermediate branching budding yeasts, Yarrowia lipolytica, has ARSs that contain physically separatable CEN and replication origin (ori) sequences241. But unlike S. cerevisiae ARS plasmids, Y. lipolytica ARSs require both CEN and ori to maintain replicating plasmids241. Only a few replication origin sequences that are associated with CEN sequences have been characterized in Y. lipolytica and they lack sequence similarity. Y. lipolytica also lacks RNAi, the SIR proteins and the Orc4 α-helix and the Orc2 loop141 (Figure 4). The nature of origins of DNA replication in Y. lipolytica remain enigmatic, but they may be specified by epigenetic means as proposed for animal cell replication.
Moreover, the Y. lipolytica genome is very GC-rich and is 1.6 times larger than the S. cerevisiae genome but has roughly the same number of protein coding genes which may provide a greater opportunity for DNA sequence-independent initiation in the larger intergenic regions, while still avoiding conflicts between DNA replication and transcription. Thus, it seems that CEN and replication origins may have started out as epigenetically defined and GC-rich and then evolved to become sequence specific in small clade of budding yeasts. One observation that supports this idea is that some of the basal branching yeasts, such as Pichia pastoris (Figure 4), have GC-rich replication origins, hence it was speculated that GC-rich is perhaps an ancestral trait of replication origins242.
In ancestral eukaryotes, CEN and ori elements may have been the same or tightly linked, but then evolved into separate functional elements. If this were the case, it may explain why ORC not only plays a role in the initiation of DNA replication, but why ORC subunits localize to centromeres in human cells and maintain the integrity of CEN associated α-satellite sequences243. Another intriguing potential link between origins of DNA replication and CEN sequences comes from the role of histone H3K4 methylation at origins and CEN sequences. As noted above, origins of DNA replication in C. elegans are associated with histone H4K4me2 and H3K4me3 modifications21. On the other hand, histone H3K4me2 modification at the α-satellite sequences of an artificial centromere is required to recruit the HJURP protein, the CEN-specific CENP-A histone244. C. elegans has holocentric centromeres which are multiple point centromeres located along the length of each of the chromosomes. Interestingly, CENP-A location in the genome has been determined245 and they co-localize with origins of DNA replication (Iestyn Whitehouse, personal communication). This observation lends strong support for the notion that CEN and ori have a common ancestor.
5.3. Perspectives for evolutionary driving forces
So, what advantages or disadvantages do these co-evolutionary transitions provide? We suggest that the loss of the RNAi system in the intermediate branching yeasts, by driving forces such as the beneficial killer virus infection236, would increase the transcription and replication conflicts and hence genome instability, thereby creating a higher mutation rate at fragile sites. A new gene silencing system was needed and meanwhile the possible integration of features from 2-micron plasmid into the genome could create the possibility of evolving to become new sequence-dependent systems for both CEN and chromosomal replication origins. Moreover, a paucity of replication origins could delay the chromosomal duplication completion and lead to the expression of fragile sites and elevate the rate of gross chromosomal rearrangements. Thus, the evolutionary transitions could be selected by limiting the fragile sites and decreasing genome instability by increasing the number of active and dormant, sequence-specific origins in the intergenic regions.
There are multiple essential questions that remain to be addressed: how does ORC localize to chromosomes in many different species? By which mechanisms does ORC contact DNA in different species? Independent studies suggested that any metazoan DNA sequence contained potential initiation sites and replication origins are epigenetically controlled in coordination with transcriptional activity. It raises the question whether metazoan origins have specific DNA elements and/or epigenetic markers or do not require such determinants. Whether or to what extent origins stochastically fire at spatially random sites or at multiple more discrete sites within the dispersive initiation zones remains a matter of debate and needs more precise metazoan replication origin mapping methods. On top of that, what role does transcription play in defining where replication initiates? In the species that have lost RNAi and have not yet gained sequence specificity, can ORC binding to DNA at random sites with lower affinity be removed by transcription, thereby placing origins of DNA replication in intergenic regions?
Intriguingly, species like Carpediemonas membranifera and Carpediemonas frisia seem to have lost canonical DNA replication proteins, such as ORC and Cdc6, and most structural kinetochore proteins, such as NDC80, during evolution246. What would be the mechanisms for replicating DNA in these species? Do they depend upon break induced replication (BIR) mechanisms247? How are their replication origins specified? Indeed, how ORC and CDC6 specify the location of origins of DNA replication remains an issue for most eukaryotic cells.
Acknowledgement:
This work was supported by grants from the National Institutes of Health (GM45436 and CA13106) and the Goldring Family Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests: The authors declare no competing interests.
References
- 1.Koduru SK (2019). Recent Developments in Applied Microbiology and Biochemistry. 335–347. 10.1016/b978-0-12-816328-3.00024-6. [DOI] [Google Scholar]
- 2.Robinson NP, and Bell SD (2005). Origins of DNA replication in the three domains of life. Febs J 272, 3757–3766. 10.1111/j.1742-4658.2005.04768.x. [DOI] [PubMed] [Google Scholar]
- 3.Grogan DW (2013). Brenner’s Encyclopedia of Genetics (Second Edition). In Archaea, pp. 180–182. 10.1016/b978-0-12-374984-0.00092-9. [DOI] [Google Scholar]
- 4.Greci MD, and Bell SD (2020). Archaeal DNA Replication. Annu Rev Microbiol 74, 1–16. 10.1146/annurev-micro-020518-115443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sessions SK (2013). Brenner’s Encyclopedia of Genetics (Second Edition). In Genome Size, pp. 301–305. 10.1016/b978-0-12-374984-0.00639-2. [DOI] [Google Scholar]
- 6.Rhind N, and Gilbert DM (2013). DNA Replication Timing. CSH Perspect Biol 5, a010132. 10.1101/cshperspect.a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mantiero D, Mackenzie A, Donaldson A, and Zegerman P (2011). Limiting replication initiation factors execute the temporal programme of origin firing in budding yeast. Embo J 30, 4805–4814. 10.1038/emboj.2011.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogelauer M, Rubbi L, Lucas I, Brewer BJ, and Grunstein M (2002). Histone Acetylation Regulates the Time of Replication Origin Firing. Mol Cell 10, 1223–1233. 10.1016/s1097-2765(02)00702-5. [DOI] [PubMed] [Google Scholar]
- 9.Goren A, Tabib A, Hecht M, and Cedar H (2008). DNA replication timing of the human β-globin domain is controlled by histone modification at the origin. Gene Dev 22, 1319–1324. 10.1101/gad.468308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka S, Nakato R, Katou Y, Shirahige K, and Araki H (2011). Origin Association of Sld3, Sld7, and Cdc45 Proteins Is a Key Step for Determination of Origin-Firing Timing. Curr Biol 21, 2055–2063. 10.1016/j.cub.2011.11.038. [DOI] [PubMed] [Google Scholar]
- 11.Solovei I, Thanisch K, and Feodorova Y (2016). How to rule the nucleus: divide et impera. Curr Opin Cell Biol 40, 47–59. 10.1016/j.ceb.2016.02.014. [DOI] [PubMed] [Google Scholar]
- 12.Dileep V, Ay F, Sima J, Vera DL, Noble WS, and Gilbert DM (2015). Topologically associating domains and their long-range contacts are established during early G1 coincident with the establishment of the replication-timing program. Genome Res 25, 1104–1113. 10.1101/gr.183699.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilbert DM, Takebayashi S-I, Ryba T, Lu J, Pope BD, Wilson KA, and Hiratani I (2010). Space and Time in the Nucleus Developmental Control of Replication Timing and Chromosome Architecture. Cold Spring Harb Sym 75, 143–153. 10.1101/sqb.2010.75.011. [DOI] [PubMed] [Google Scholar]
- 14.Pope BD, Ryba T, Dileep V, Yue F, Wu W, Denas O, Vera DL, Wang Y, Hansen RS, Canfield TK, et al. (2014). Topologically associating domains are stable units of replication-timing regulation. Nature 515, 402–405. 10.1038/nature13986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kermi C, Furno EL, and Maiorano D (2017). Regulation of DNA Replication in Early Embryonic Cleavages. Genes-basel 8, 42. 10.3390/genes8010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siefert JC, Georgescu C, Wren JD, Koren A, and Sansam CL (2017). DNA replication timing during development anticipates transcriptional programs and parallels enhancer activation. Genome Res 27, 1406–1416. 10.1101/gr.218602.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nordman J, and Orr-Weaver TL (2012). Regulation of DNA replication during development. Development 139, 455–464. 10.1242/dev.061838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blumenthal AB, Kriegstein HJ, and Hogness DS (1974). The Units of DNA Replication in Drosophila melanogaster Chromosomes. Cold Spring Harb Sym 38, 205–223. 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- 19.Graham CF, and Morgan RW (1966). Changes in the cell cycle during early amphibian development. Dev Biol 14, 439–460. 10.1016/0012-1606(66)90024-8. [DOI] [Google Scholar]
- 20.Nakatani T, Lin J, Ji F, Ettinger A, Pontabry J, Tokoro M, Altamirano-Pacheco L, Fiorentino J, Mahammadov E, Hatano Y, et al. (2022). DNA replication fork speed underlies cell fate changes and promotes reprogramming. Nat Genet 54, 318–327. 10.1038/s41588-022-01023-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pourkarimi E, Bellush JM, and Whitehouse I (2016). Spatiotemporal coupling and decoupling of gene transcription with DNA replication origins during embryogenesis in C. elegans. Elife 5, e21728. 10.7554/elife.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Attali I, Botchan MR, and Berger JM (2021). Structural Mechanisms for Replicating DNA in Eukaryotes. Annu Rev Biochem 90, 1–30. 10.1146/annurev-biochem-090120-125407. [DOI] [PubMed] [Google Scholar]
- 23.Costa A, and Diffley JFX (2022). The Initiation of Eukaryotic DNA Replication. Annu Rev Biochem 91. 10.1146/annurev-biochem-072321-110228. [DOI] [PubMed] [Google Scholar]
- 24.Bell SP, and Labib K (2016). Chromosome Duplication in Saccharomyces cerevisiae. Genetics 203, 1027–1067. 10.1534/genetics.115.186452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewis JS, Gross MH, Sousa J, Henrikus SS, Greiwe JF, Nans A, Diffley JFX, and Costa A (2022). Mechanism of replication origin melting nucleated by CMG helicase assembly. Nature 606, 1007–1014. 10.1038/s41586-022-04829-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noguchi Y, Yuan Z, Bai L, Schneider S, Zhao G, Stillman B, Speck C, and Li H (2017). Cryo-EM structure of Mcm2-7 double hexamer on DNA suggests a lagging-strand DNA extrusion model. Proc National Acad Sci 114, E9529–E9538. 10.1073/pnas.1712537114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan Z, Georgescu R, Bai L, Zhang D, Li H, and O’Donnell ME (2020). DNA unwinding mechanism of a eukaryotic replicative CMG helicase. Nature Communications 11, 688–10. 10.1038/s41467-020-14577-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wasserman MR, Schauer GD, O’Donnell ME, and Liu S (2019). Replication Fork Activation Is Enabled by a Single- Stranded DNA Gate in CMG Helicase. Cell 178, 600611.e16. 10.1016/j.cell.2019.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bell SP, and Stillman B (1992). ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357, 128–134. 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- 30.Hartwell LH (1976). Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. Journal of Molecular Biology 104, 803–817. 10.1016/0022-2836(76)90183-2. [DOI] [PubMed] [Google Scholar]
- 31.Liang C, Weinreich M, and Stillman B (1995). ORC and Cdc6p interact and determine the frequency of initiation of DNA replication in the genome. Cell 81, 667–676. 10.1016/0092-8674(95)90528-6. [DOI] [PubMed] [Google Scholar]
- 32.Cocker JH, Piatti S, Santocanale C, Nasmyth K, and Diffley JF (1996). An essential role for the Cdc6 protein in forming the pre-replicative complexes of budding yeast. Nature 379, 180–182. 10.1038/379180a0. [DOI] [PubMed] [Google Scholar]
- 33.Kelly TJ, Martin GS, Forsburg SL, Stephen RJ, Russo A, and Nurse P (1993). The fission yeast cdc18+ gene product couples S phase to START and mitosis. Cell 74, 371–382. 10.1016/0092-8674(93)90427-r. [DOI] [PubMed] [Google Scholar]
- 34.Hofmann JF, and Beach D (1994). cdt1 is an essential target of the Cdc10/Sct1 transcription factor: requirement for DNA replication and inhibition of mitosis. Embo J 13, 425–434. 10.1002/j.1460-2075.1994.tb06277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishitani H, Lygerou Z, Nishimoto T, and Nurse P (2000). The Cdt1 protein is required to license DNA for replication in fission yeast. Nature 404, 625–628. 10.1038/35007110. [DOI] [PubMed] [Google Scholar]
- 36.Maiorano D, Moreau J, and Méchali M (2000). XCDT1 is required for the assembly of pre-replicative complexes in Xenopus laevis. Nature 404, 622–625. 10.1038/35007104. [DOI] [PubMed] [Google Scholar]
- 37.Gillespie PJ, Li A, and Blow JJ (2001). Reconstitution of licensed replication origins on Xenopus sperm nuclei using purified proteins. Bmc Biochem 2, 15. 10.1186/1471-2091-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pozo PN, and Cook JG (2016). Regulation and Function of Cdt1; A Key Factor in Cell Proliferation and Genome Stability. Genes-basel 8, 2. 10.3390/genes8010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moir D, Stewart SE, Osmond BC, and Botstein D (1982). COLD-SENSITIVE CELL-DIVISION-CYCLE MUTANTS OF YEAST: ISOLATION, PROPERTIES, AND PSEUDOREVERSION STUDIES. Genetics 100, 547–563. 10.1093/genetics/100.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maine GT, Sinha P, and Tye B-K (1984). MUTANTS OF S. CEREVISIAE DEFECTIVE IN THE MAINTENANCE OF MINICHROMOSOMES. Genetics 106, 365–385. 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chong J (1996). The role of MCM/P1 proteins in the licensing of DNA replication. Trends Biochem Sci 21, 102–107. 10.1016/0968-0004(96)10013-x. [DOI] [PubMed] [Google Scholar]
- 42.Aparicio OM, Weinstein DM, and Bell SP (1997). Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91, 59–69. 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 43.Labib K, Tercero JA, and Diffley JFX (2000). Uninterrupted Mcm2-7 Function Required for DNA Replication Fork Progression. Science 288, 1643–1647. 10.1126/science.288.5471.1643. [DOI] [PubMed] [Google Scholar]
- 44.Bochman ML, and Schwacha A (2008). The Mcm2-7 Complex Has In Vitro Helicase Activity. Mol Cell 31, 287–293. 10.1016/j.molcel.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 45.Thömmes P, Kubota Y, Takisawa H, and Blow JJ (1997). The RLF-M component of the replication licensing system forms complexes containing all six MCM/P1 polypeptides. Embo J 16, 3312–3319. 10.1093/emboj/16.11.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chong JP, Mahbubani HM, Khoo CY, and Blow JJ (1995). Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature 375, 418–421. 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- 47.Kubota Y, Mimura S, Nishimoto S, Masuda T, Nojima H, and Takisawa H (1997). Licensing of DNA replication by a multi-protein complex of MCM/P1 proteins in Xenopus eggs. Embo J 16, 3320–3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gavin KA, Hidaka M, and Stillman B (1995). Conserved Initiator Proteins in Eukaryotes. Science 270, 1667–1671. 10.1126/science.270.5242.1667. [DOI] [PubMed] [Google Scholar]
- 49.Takahara K, Bong M, Brevard R, Eddy RL, Haley LL, Sait SJ, Shows TB, Hoffman GG, and Greenspan DS (1996). Mouse and Human Homologues of the Yeast Origin of Replication Recognition Complex Subunit ORC2 and Chromosomal Localization of the Cognate Human Gene ORC2L. Genomics 31, 119–122. 10.1006/geno.1996.0018. [DOI] [PubMed] [Google Scholar]
- 50.Romanowski P, Madine MA, Rowles A, Blow JJ, and Laskey RA (1996). The Xenopus origin recognition complex is essential for DNA replication and MCM binding to chromatin. Current biology : CB 6, 1416–1425. [DOI] [PubMed] [Google Scholar]
- 51.Blow JJ, and Laskey RA (1988). A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature 332, 546–548. 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- 52.Diffley JFX, Cocker JH, Dowell SJ, and Rowley A (1994). Two steps in the assembly of complexes at yeast replication origins in vivo. Cell 78, 303–316. 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- 53.Tanaka S, Tak Y-S, and Araki H (2007). The role of CDK in the initiation step of DNA replication in eukaryotes. Cell Div 2, 16. 10.1186/1747-1028-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Diffley JF (1996). Once and only once upon a time: specifying and regulating origins of DNA replication in eukaryotic cells. Genes & development 10, 2819–2830. [DOI] [PubMed] [Google Scholar]
- 55.Pines J (1994). Protein kinases and cell cycle control. Semin Cell Biol 5, 399–408. 10.1006/scel.1994.1047. [DOI] [PubMed] [Google Scholar]
- 56.Broek D, Bartlett R, Crawford K, and Nurse P (1991). Involvement of p34cdc2 in establishing the dependency of S phase on mitosis. Nature 349, 388–393. 10.1038/349388a0. [DOI] [PubMed] [Google Scholar]
- 57.Dahmann C, Diffley JFX, and Nasmyth KA (1995). S-phase-promoting cyclin-dependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol 5, 1257–1269. 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- 58.Jallepalli PV, and Kelly TJ (1996). Rum1 and Cdc18 link inhibition of cyclin-dependent kinase to the initiation of DNA replication in Schizosaccharomyces pombe. Gene Dev 10, 541–552. 10.1101/gad.10.5.541. [DOI] [PubMed] [Google Scholar]
- 59.Nguyen VQ, Co C, and Li JJ (2001). Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature 411, 1068–1073. 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- 60.Zou L, and Stillman B (1998). Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science (New York, NY) 280, 593–596. [DOI] [PubMed] [Google Scholar]
- 61.Zegerman P, and Diffley JFX (2007). Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature 445, 281–285. 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 62.Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, and Araki H (2007). CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature 445, 328–332. 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 63.Moyer SE, Lewis PW, and Botchan MR (2006). Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proceedings of the National Academy of Sciences of the United States of America 103, 10236–10241. 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dolson A, Sauty SM, Shaban K, and Yankulov K (2021). Dbf4-Dependent Kinase: DDK-ated to post-initiation events in DNA replication. Cell Cycle, 1–13. 10.1080/15384101.2021.1986999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sheu Y-J, and Stillman B (2010). The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature 463, 113–117. 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zou L, and Stillman B (2000). Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Molecular and cellular biology 20, 3086–3096. DOI: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dhingra N, and Kaplan DL (2016). The Initiation of DNA Replication in Eukaryotes. In Introduction to Eukaryotic DNA Replication Initiation, pp. 1–21. 10.1007/978-3-319-24696-3_1. [DOI] [Google Scholar]
- 68.Dhingra N, and Kaplan DL (2016). The Initiation of DNA Replication in Eukaryotes. In Introduction to Eukaryotic DNA Replication Initiation, pp. 1–21. 10.1007/978-3-319-24696-3_1. [DOI] [Google Scholar]
- 69.Guilliam TA, and Yeeles JTP (2020). An updated perspective on the polymerase division of labor during eukaryotic DNA replication. Crit Rev Biochem Mol, 1–13. 10.1080/10409238.2020.1811630. [DOI] [PubMed] [Google Scholar]
- 70.Yeeles JTP, Janska A, Early A, and Diffley JFX (2017). How the Eukaryotic Replisome Achieves Rapid and Efficient DNA Replication. Molecular Cell 65, 105–116. 10.1016/j.molcel.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Devbhandari S, Jiang J, Kumar C, Whitehouse I, and Remus D (2017). Chromatin Constrains the Initiation and Elongation of DNA Replication. Molecular Cell 65, 131–141. 10.1016/j.molcel.2016.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yeeles JTP, Deegan TD, Janska A, Early A, and Diffley JFX (2015). Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 519, 431–435. 10.1038/nature14285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pursell ZF, Isoz I, Lundstrom E-B, Johansson E, and Kunkel TA (2007). Yeast DNA Polymerase Participates in Leading-Strand DNA Replication. Science 317, 127–130. 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.McElhinny SAN, Gordenin DA, Stith CM, Burgers PMJ, and Kunkel TA (2008). Division of Labor at the Eukaryotic Replication Fork. Molecular Cell 30, 137–144. 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, and Speck C (2009). A double-hexameric Mcm2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc. Natl. Acad. Sci USA 106, 20240–20245. 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, and Diffley JFX (2009). Concerted Loading of Mcm2-7 Double Hexamers around DNA during DNA Replication Origin Licensing. Cell 139, 719–730. 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kurat CF, Yeeles JTP, Patel H, Early A, and Diffley JFX (2017). Chromatin Controls DNA Replication Origin Selection, Lagging-Strand Synthesis, and Replication Fork Rates. Molecular Cell 65, 117–130. 10.1016/j.molcel.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waga S, and Stillman B (1998). The DNA replication fork in eukaryotic cells. Annual review of biochemistry 67, 721–751. 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 79.Ekundayo B, and Bleichert F (2019). Origins of DNA replication. PLoS genetics 15, e1008320. 10.1371/journal.pgen.1008320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vouzas AE, and Gilbert DM (2021). Mammalian DNA Replication Timing. CSH Perspect Biol, a040162. 10.1101/cshperspect.a040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Falaschi A, Biamonti G, and Riva S (2006). Encyclopedic Reference of Genomics and Proteomics in Molecular Medicine. 1635–1640. 10.1007/3-540-29623-9_2130. [DOI] [Google Scholar]
- 82.Petryk N, Kahli M, d’Aubenton-Carafa Y, Jaszczyszyn Y, Shen Y, Silvain M, Thermes C, Chen C-L, and Hyrien O (2016). Replication landscape of the human genome. Nat Commun 7, 10208. 10.1038/ncomms10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sun J, and Kong D (2010). DNA replication origins, ORC/DNA interaction, and assembly of pre-replication complex in eukaryotes. Acta biochimica et biophysica Sinica 42, 433–439. 10.1093/abbs/gmq048. [DOI] [PubMed] [Google Scholar]
- 84.Gilbert DM (2001). Making sense of eukaryotic DNA replication origins. Science 294, 96–100. 10.1126/science.1061724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hyrien O, and Méchali M (1993). Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopus early embryos. Embo J 12, 4511–4520. 10.1002/j.1460-2075.1993.tb06140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harland RM, and Laskey RA (1980). Regulated replication of DNA microinjected into eggs of Xenopus laevis. Cell 21, 761–771. 10.1016/0092-8674(80)90439-0. [DOI] [PubMed] [Google Scholar]
- 87.Shinomiya T, and Ina S (1991). Analysis of chromosomal replicons in early embryos of Drosophila melanogaster by two-dimensional gel electrophoresis. Nucleic Acids Res 19, 3935–3941. 10.1093/nar/19.14.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hyrien O, Maric C, and Méchali M (1995). Transition in Specification of Embryonic Metazoan DNA Replication Origins. Science 270, 994–997. 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- 89.Sasaki T, Sawado T, Yamaguchi M, and Shinomiya T (1999). Specification of Regions of DNA Replication Initiation during Embryogenesis in the 65-Kilobase DNApolα-dE2F Locus of Drosophila melanogaster. Mol Cell Biol 19, 547–555. 10.1128/mcb.19.1.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ganier O, Prorok P, Akerman I, and Méchali M (2019). Metazoan DNA replication origins. Curr Opin Cell Biol 58, 134–141. 10.1016/j.ceb.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 91.Dershowitz A, Snyder M, Sbia M, Skurnick JH, Ong LY, and Newlon CS (2007). Linear Derivatives of Saccharomyces cerevisiae Chromosome III Can Be Maintained in the Absence of Autonomously Replicating Sequence Elements ▿ †. Molecular and Cellular Biology 27. 10.1128/MCB.01246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dershowitz A, and Newlon CS (1993). The effect on chromosome stability of deleting replication origins. Mol Cell Biol 13, 391–398. 10.1128/mcb.13.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bogenschutz NL, Rodriguez J, and Tsukiyama T (2014). Initiation of DNA Replication from Non-Canonical Sites on an Origin-Depleted Chromosome. PLoS ONE 9, e114545. 10.1371/journal.pone.0114545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hawkins M, Malla S, Blythe MJ, Nieduszynski CA, and Allers T (2013). Accelerated growth in the absence of DNA replication origins. Nature 503, 544–547. 10.1038/nature12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Blow JJ, and Laskey RA (1986). Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell 47, 577–587. 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- 96.Méchali M, and Kearsey S (1984). Lack of specific sequence requirement for DNA replication in Xenopus eggs compared with high sequence specificity in yeast. Cell 38, 55–64. 10.1016/0092-8674(84)90526-9. [DOI] [PubMed] [Google Scholar]
- 97.Scherr MJ, Wahab SA, Remus D, and Duderstadt KE (2022). Mobile origin-licensing factors confer resistance to conflicts with RNA polymerase. Cell Reports 38, 110531. 10.1016/j.celrep.2022.110531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nieduszynski CA, Knox Y, and Donaldson AD (2006). Genome-wide identification of replication origins in yeast by comparative genomics. Gene Dev 20, 1874–1879. 10.1101/gad.385306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Czajkowsky DM, Liu J, Hamlin JL, and Shao Z (2008). DNA Combing Reveals Intrinsic Temporal Disorder in the Replication of Yeast Chromosome VI. J Mol Biol 375, 12–19. 10.1016/j.jmb.2007.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hennion M, Arbona J-M, Lacroix L, Cruaud C, Theulot B, Tallec BL, Proux F, Wu X, Novikova E, Engelen S, et al. (2020). FORK-seq: replication landscape of the Saccharomyces cerevisiae genome by nanopore sequencing. Genome Biol 21, 125. 10.1186/s13059-020-02013-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Müller CA, Boemo MA, Spingardi P, Kessler BM, Kriaucionis S, Simpson JT, and Nieduszynski CA (2019). Capturing the dynamics of genome replication on individual ultra-long nanopore sequence reads. Nat Methods 16, 429–436. 10.1038/s41592-019-0394-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bechhoefer J, and Rhind N (2012). Replication timing and its emergence from stochastic processes. Trends in Genetics 28, 374–381. 10.1016/j.tig.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Saner N, Karschau J, Natsume T, Gierliński M, Retkute R, Hawkins M, Nieduszynski CA, Blow JJ, Moura A.P.S. de, and Tanaka TU (2013). Stochastic association of neighboring replicons creates replication factories in budding yeast. J Cell Biology 202, 1001–1012. 10.1083/jcb.201306143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hawkins M, Retkute R, Müller CA, Saner N, Tanaka TU, de Moura APS, and Nieduszynski CA (2013). High-Resolution Replication Profiles Define the Stochastic Nature of Genome Replication Initiation and Termination. Cell Reports 5, 1132–1141. 10.1016/j.celrep.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kelly T, and Callegari AJ (2019). Dynamics of DNA replication in a eukaryotic cell. Proceedings of the National Academy of Sciences of the United States of America 116, 4973–4982. 10.1073/pnas.1818680116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kaykov A, and Nurse P (2015). The spatial and temporal organization of origin firing during the S-phase of fission yeast. Genome Research 25, 391–401. 10.1101/gr.180372.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guilbaud G, Murat P, Wilkes HS, Lerner LK, Sale JE, and Krude T (2022). Determination of human DNA replication origin position and efficiency reveals principles of initiation zone organisation. Nucleic Acids Res 50, 7436–7450. 10.1093/nar/gkac555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dileep V, and Gilbert DM (2018). Single-cell replication profiling to measure stochastic variation in mammalian replication timing. Nat Commun 9, 427. 10.1038/s41467-017-02800-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang W, Klein KN, Proesmans K, Yang H, Marchal C, Zhu X, Borrman T, Hastie A, Weng Z, Bechhoefer J, et al. (2021). Genome-wide mapping of human DNA replication by optical replication mapping supports a stochastic model of eukaryotic replication. Mol Cell 81, 2975–2988.e6. 10.1016/j.molcel.2021.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Woodward AM, Göhler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, and Blow JJ (2006). Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J Cell Biology 173, 673–683. 10.1083/jcb.200602108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ge XQ, Jackson DA, and Blow JJ (2007). Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Gene Dev 21, 3331–3341. 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ibarra A, Schwob E, and Méndez J (2008). Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc National Acad Sci 105, 8956–8961. 10.1073/pnas.0803978105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Saxena S, and Zou L (2022). Hallmarks of DNA replication stress. Mol Cell 82, 2298–2314. 10.1016/j.molcel.2022.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Alver RC, Chadha GS, and Blow JJ (2014). The contribution of dormant origins to genome stability: from cell biology to human genetics. DNA repair 19, 182–189. 10.1016/j.dnarep.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Stinchcomb DT, Struhl K, and Davis RW (1979). Isolation and characterisation of a yeast chromosomal replicator. Nature 282, 39–43. 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- 116.Hsiao C-L, and Carbon J (1979). High-frequency transformation of yeast by plasmids containing the cloned yeast ARG4 gene. Proc National Acad Sci 76, 3829–3833. 10.1073/pnas.76.8.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brewer BJ, and Fangman WL (1987). The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51, 463–471. 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 118.Huberman JA, Spotila LD, Nawotka KA, El-Assouli SM, and Davis LR (1987). The in vivo replication origin of the yeast 2μm plasmid. Cell 51, 473–481. 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- 119.Palzkill TG, and Newlon CS (1988). A yeast replication origin consists of multiple copies of a small conserved sequence. Cell 53, 441–450. 10.1016/0092-8674(88)90164-x. [DOI] [PubMed] [Google Scholar]
- 120.Broach JR, Li YY, Feldman J, Jayaram M, Abraham J, Nasmyth KA, and Hicks JB (1983). Localization and sequence analysis of yeast origins of DNA replication. Cold Spring Harb Sym 47 Pt 2, 1165–1173. 10.1101/sqb.1983.047.01.132. [DOI] [PubMed] [Google Scholar]
- 121.Marahrens Y, and Stillman B (1992). A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science 255, 817–823. 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 122.Rao H, Marahrens Y, and Stillman B (1994). Functional conservation of multiple elements in yeast chromosomal replicators. Molecular and cellular biology 14, 7643–7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Theis JF, and Newlon CS (1994). Domain B of ARS307 contains two functional elements and contributes to chromosomal replication origin function. Mol Cell Biol 14, 7652–7659. 10.1128/mcb.14.11.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Huang RY, and Kowalski D (1996). Multiple DNA elements in ARS305 determine replication origin activity in a yeast chromosome. Nucleic Acids Res 24, 816–823. 10.1093/nar/24.5.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wilmes GM, and Bell SP (2002). The B2 element of the Saccharomyces cerevisiae ARS1 origin of replication requires specific sequences to facilitate pre-RC formation. Proceedings of the National Academy of Sciences 99, 101–106. 10.1073/pnas.012578499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Coster G, and Diffley JFX (2017). Bidirectional eukaryotic DNA replication is established by quasi-symmetrical helicase loading. Science 357, 314–318. 10.1126/science.aan0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chistol G, and Walter JC (2015). Single-Molecule Visualization of Mcm2-7 DNA Loading: Seeing Is Believing. Cell 161, 429–430. 10.1016/j.cell.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 128.Duzdevich D, Warner MD, Ticau S, Ivica NA, Bell SP, and Greene EC (2015). The dynamics of eukaryotic replication initiation: origin specificity, licensing, and firing at the single-molecule level. Molecular Cell 58, 483–494. 10.1016/j.molcel.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sun J, Fernandez-Cid A, Riera A, Tognetti S, Yuan Z, Stillman B, Speck C, and Li H (2014). Structural and mechanistic insights into Mcm2-7 double-hexamer assembly and function. Gene Dev 28, 2291–2303. 10.1101/gad.242313.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ticau S, Friedman LJ, Ivica NA, Gelles J, and Bell SP (2015). Single-molecule studies of origin licensing reveal mechanisms ensuring bidirectional helicase loading. Cell 161, 513–525. 10.1016/j.cell.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yardimci H, and Walter JC (2014). Prereplication-complex formation: a molecular double take? Nat Struct Mol Biol 21, 20–25. 10.1038/nsmb.2738. [DOI] [PubMed] [Google Scholar]
- 132.Rowley A, Cocker JH, Harwood J, and Diffley JF (1995). Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. Embo J 14, 2631–2641. 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wilmes GM, and Bell SP (2002). The B2 element of the Saccharomyces cerevisiae ARS1 origin of replication requires specific sequences to facilitate pre-RC formation. Proceedings of the National Academy of Sciences 99, 101–106. 10.1073/pnas.012578499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Miller TCR, Locke J, Greiwe JF, Diffley JFX, and Costa A (2019). Mechanism of head-to-head MCM double-hexamer formation revealed by cryo-EM. Nature 575, 704–710. 10.1038/s41586-019-1768-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yuan Z, Schneider S, Dodd T, Riera A, Bai L, Yan C, Magdalou I, Ivanov I, Stillman B, Li H, et al. (2020). Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6. Proc. Natl. Acad. Sci. USA 117, 17747–17756. 10.1073/pnas.2006231117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Gupta S, Friedman LJ, Gelles J, and Bell SP (2021). A helicase-tethered ORC flip enables bidirectional helicase loading. Elife 10. 10.7554/elife.74282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Feng X, Noguchi Y, Barbon M, Stillman B, Speck C, and Li H (2021). The structure of ORC–Cdc6 on an origin DNA reveals the mechanism of ORC activation by the replication initiator Cdc6. Nat Commun 12, 3883. 10.1038/s41467-021-24199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Coster G, and Diffley JFX (2017). Bidirectional eukaryotic DNA replication is established by quasi-symmetrical helicase loading. Science 357, 314–318. 10.1126/science.aan0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li N, Lam WH, Zhai Y, Cheng J, Cheng E, Zhao Y, Gao N, and Tye B-K (2018). Structure of the origin recognition complex bound to DNA replication origin. Nature 559, 217–222. 10.1038/s41586-018-0293-x. [DOI] [PubMed] [Google Scholar]
- 140.Yuan Z, Riera A, Bai L, Sun J, Nandi S, Spanos C, Chen ZA, Barbon M, Rappsilber J, Stillman B, et al. (2017). Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1. Nat Struct Mol Biol 24, 316–324. 10.1038/nsmb.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hu Y, Tareen A, Sheu Y-J, Ireland WT, Speck C, Li H, Joshua-Tor L, Kinney JB, and Stillman B (2020). Evolution of DNA replication origin specification and gene silencing mechanisms. Nat Commun 11, 5175. 10.1038/s41467-020-18964-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lee CSK, Cheung MF, Li J, Zhao Y, Lam WH, Ho V, Rohs R, Zhai Y, Leung D, and Tye B-K (2021). Humanizing the yeast origin recognition complex. Nature Communications 12, 33. 10.1038/s41467-020-20277-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kawakami H, Ohashi E, Kanamoto S, Tsurimoto T, and Katayama T (2015). Specific binding of eukaryotic ORC to DNA replication origins depends on highly conserved basic residues. Scientific reports 5, 14929. 10.1038/srep14929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Xu J, Yanagisawa Y, Tsankov AM, Hart C, Aoki K, Kommajosyula N, Steinmann KE, Bochicchio J, Russ C, Regev A, et al. (2012). Genome-wide identification and characterization of replication origins by deep sequencing. Genome biology 13, R27. 10.1186/gb-2012-13-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dai J, Chuang R-Y, and Kelly TJ (2005). DNA replication origins in the Schizosaccharomyces pombe genome. Proceedings of the National Academy of Sciences 102, 337–342. 10.1073/pnas.0408811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Cotobal C, Segurado M, and Antequera F (2010). Structural diversity and dynamics of genomic replication origins in Schizosaccharomyces pombe. Embo J 29, 934–942. 10.1038/emboj.2009.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Antequera F (2004). Genomic specification and epigenetic regulation of eukaryotic DNA replication origins. Embo J 23, 4365–4370. 10.1038/sj.emboj.7600450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chuang RY, and Kelly TJ (1999). The fission yeast homologue of Orc4p binds to replication origin DNA via multiple AT-hooks. Proceedings of the National Academy of Sciences 96, 2656–2661. 10.1073/pnas.96.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee J-K, Moon K-Y, Jiang Y, and Hurwitz J (2001). The Schizosaccharomyces pombe origin recognition complex interacts with multiple AT-rich regions of the replication origin DNA by means of the AT-hook domains of the spOrc4 protein. Proc National Acad Sci 98, 13589–13594. 10.1073/pnas.251530398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Reeves R, and Nissen MS (1990). The A.T-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J Biol Chem 265, 8573–8582. 10.1016/s0021-9258(19)38926-4. [DOI] [PubMed] [Google Scholar]
- 151.Hyrien O (2015). Peaks cloaked in the mist: The landscape of mammalian replication origins. J Cell Biol 208, 147–160. 10.1083/jcb.201407004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Huberman JA, and Riggs AD (1968). On the mechanism of DNA replication in mammalian chromosomes. J Mol Biol 32, 327–341. 10.1016/0022-2836(68)90013-2. [DOI] [PubMed] [Google Scholar]
- 153.Ladenburger E-M, Keller C, and Knippers R (2002). Identification of a Binding Region for Human Origin Recognition Complex Proteins 1 and 2 That Coincides with an Origin of DNA Replication. Mol Cell Biol 22, 1036–1048. 10.1128/mcb.22.4.1036-1048.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Altman AL, and Fanning E (2001). The Chinese Hamster Dihydrofolate Reductase Replication Origin Beta Is Active at Multiple Ectopic Chromosomal Locations and Requires Specific DNA Sequence Elements for Activity. Mol Cell Biol 21, 1098–1110. 10.1128/mcb.21.4.1098-1110.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dijkwel PA, and Hamlin JL (1999). Physical and genetic mapping of mammalian replication origins. Methods San Diego Calif 18, 418–431. 10.1006/meth.1999.0800. [DOI] [PubMed] [Google Scholar]
- 156.Kumar S, Giacca M, Norio P, Biamonti G, Riva S, and Falaschi A (1996). Utilization of the Same DNA Replication Origin by Human Cells of Different Derivation. Nucleic Acids Res 24, 3289–3294. 10.1093/nar/24.17.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cadoret J-C, Meisch F, Hassan-Zadeh V, Luyten I, Guillet C, Duret L, Quesneville H, and Prioleau M-N (2008). Genome-wide studies highlight indirect links between human replication origins and gene regulation. Proc National Acad Sci 105, 15837–15842. 10.1073/pnas.0805208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhao PA, Sasaki T, and Gilbert DM (2020). High-resolution Repli-Seq defines the temporal choreography of initiation, elongation and termination of replication in mammalian cells. Genome Biol 21, 76. 10.1186/s13059-020-01983-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Vashee S, Cvetic C, Lu W, Simancek P, Kelly TJ, and Walter JC (2003). Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Gene Dev 17, 1894–1908. 10.1101/gad.1084203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Remus D, Beall EL, and Botchan MR (2004). DNA topology, not DNA sequence, is a critical determinant for Drosophila ORC-DNA binding. The EMBO Journal 23, 897–907. 10.1038/sj.emboj.7600077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Schaarschmidt D, Baltin J, Stehle IM, Lipps HJ, and Knippers R (2004). An episomal mammalian replicon: sequence-independent binding of the origin recognition complex. Embo J 23, 191–201. 10.1038/sj.emboj.7600029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Sima J, Chakraborty A, Dileep V, Michalski M, Klein KN, Holcomb NP, Turner JL, Paulsen MT, Rivera-Mulia JC, Trevilla-Garcia C, et al. (2019). Identifying cis Elements for Spatiotemporal Control of Mammalian DNA Replication. Cell 176, 816–830.e18. 10.1016/j.cell.2018.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Valton A-L, and Prioleau M-N (2016). G-Quadruplexes in DNA Replication: A Problem or a Necessity? Trends Genet 32, 697–706. 10.1016/j.tig.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 164.Prioleau M-N (2018). DNA Replication, From Old Principles to New Discoveries. Adv Exp Med Biol 1042, 273–286. 10.1007/978-981-10-6955-0_13. [DOI] [PubMed] [Google Scholar]
- 165.Prorok P, Artufel M, Aze A, Coulombe P, Peiffer I, Lacroix L, Guédin A, Mergny J-L, Damaschke J, Schepers A, et al. (2019). Involvement of G-quadruplex regions in mammalian replication origin activity. Nat Commun 10, 3274. 10.1038/s41467-019-11104-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hoshina S, Yura K, Teranishi H, Kiyasu N, Tominaga A, Kadoma H, Nakatsuka A, Kunichika T, Obuse C, and Waga S (2013). Human Origin Recognition Complex Binds Preferentially to G-quadruplex-preferable RNA and Single-stranded DNA*. J Biol Chem 288, 30161–30171. 10.1074/jbc.m113.492504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Eladl A, Yamaoki Y, Hoshina S, Horinouchi H, Kondo K, Waga S, Nagata T, and Katahira M (2021). Investigation of the Interaction of Human Origin Recognition Complex Subunit 1 with G-Quadruplex DNAs of Human c-myc Promoter and Telomere Regions. Int J Mol Sci 22, 3481. 10.3390/ijms22073481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Gros J, Devbhandari S, and Remus D (2014). Origin plasticity during budding yeast DNA replication in vitro. Embo J 33, 621–636. 10.1002/embj.201387278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.On KF, Beuron F, Frith D, Snijders AP, Morris EP, and Diffley JFX (2014). Prereplicative complexes assembled in vitro support origin-dependent and independent DNA replication. Embo J 33, 605–620. 10.1002/embj.201387369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Theis JF, and Newlon CS (2001). Two Compound Replication Origins in Saccharomyces cerevisiae Contain Redundant Origin Recognition Complex Binding Sites. Mol Cell Biol 21, 2790–2801. 10.1128/mcb.21.8.2790-2801.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Theis JF, Yang C, Schaefer CB, and Newlon CS (1999). DNA Sequence and Functional Analysis of Homologous ARS Elements of Saccharomyces cerevisiae and S. carlsbergensis. Genetics 152, 943–952. 10.1093/genetics/152.3.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Crevel G, and Cotterill S (2012). Forced binding of the origin of replication complex to chromosomal sites in Drosophila S2 cells creates an origin of replication. J Cell Sci 125, 965–972. 10.1242/jcs.094409. [DOI] [PubMed] [Google Scholar]
- 173.Hyrien O, Marheineke K, and Goldar A (2003). Paradoxes of eukaryotic DNA replication: MCM proteins and the random completion problem. Bioessays 25, 116–125. 10.1002/bies.10208. [DOI] [PubMed] [Google Scholar]
- 174.Lucas I, Chevrier-Miller M, Sogo JM, and Hyrien O (2000). Mechanisms ensuring rapid and complete DNA replication despite random initiation in Xenopus early embryos. J Mol Biol 296, 769–786. 10.1006/jmbi.2000.3500. [DOI] [PubMed] [Google Scholar]
- 175.Ding Q, and MacAlpine DM (2011). Defining the replication program through the chromatin landscape. Crit Rev Biochem Mol 46, 165–179. 10.3109/10409238.2011.560139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Cayrou C, Ballester B, Peiffer I, Fenouil R, Coulombe P, Andrau JC, Helden JV, and Méchali M (2015). The chromatin environment shapes DNA replication origin organization and defines origin classes. Genome Research 25, 1873–1885. 10.1101/gr.192799.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Thomae AW, Pich D, Brocher J, Spindler M-P, Berens C, Hock R, Hammerschmidt W, and Schepers A (2008). Interaction between HMGA1a and the origin recognition complex creates site-specific replication origins. Proc National Acad Sci 105, 1692–1697. 10.1073/pnas.0707260105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Shen Z, Sathyan KM, Geng Y, Zheng R, Chakraborty A, Freeman B, Wang F, Prasanth KV, and Prasanth SG (2010). A WD-Repeat Protein Stabilizes ORC Binding to Chromatin. Mol Cell 40, 99–111. 10.1016/j.molcel.2010.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Shen Z, Chakraborty A, Jain A, Giri S, Ha T, Prasanth KV, and Prasanth SG (2012). Dynamic Association of ORCA with Prereplicative Complex Components Regulates DNA Replication Initiation. Mol Cell Biol 32, 3107–3120. 10.1128/mcb.00362-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mei L, Kedziora KM, Song E-A, Purvis JE, and Cook JG (2022). The consequences of differential origin licensing dynamics in distinct chromatin environments. Nucleic Acids Res, gkac003-. 10.1093/nar/gkac003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Norseen J, Thomae A, Sridharan V, Aiyar A, Schepers A, and Lieberman PM (2008). RNA-dependent recruitment of the origin recognition complex. Embo J 27, 3024–3035. 10.1038/emboj.2008.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Krude T, Christov CP, Hyrien O, and Marheineke K (2009). Y RNA functions at the initiation step of mammalian chromosomal DNA replication. J Cell Sci 122, 2836–2845. 10.1242/jcs.047563. [DOI] [PubMed] [Google Scholar]
- 183.Zhang AT, Langley AR, Christov CP, Kheir E, Shafee T, Gardiner TJ, and Krude T (2011). Dynamic interaction of Y RNAs with chromatin and initiation proteins during human DNA replication. J Cell Sci 124, 2058–2069. 10.1242/jcs.086561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kara N, Hossain M, Prasanth SG, and Stillman B (2015). Orc1 Binding to Mitotic Chromosomes Precedes Spatial Patterning during G1 Phase and Assembly of the Origin Recognition Complex in Human Cells*. J Biol Chem 290, 12355–12369. 10.1074/jbc.m114.625012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Okuno Y, McNairn AJ, den Elzen N, Pines J, and Gilbert DM (2001). Stability, chromatin association and functional activity of mammalian pre-replication complex proteins during the cell cycle. The EMBO journal 20, 4263–4277. 10.1093/emboj/20.15.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Caravaca JM, Donahue G, Becker JS, He X, Vinson C, and Zaret KS (2013). Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes & development 27, 251–260. 10.1101/gad.206458.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Kara N, Hossain M, Prasanth SG, and Stillman B (2015). Orc1 Binding to Mitotic Chromosomes Precedes Spatial Patterning during G1 Phase and Assembly of the Origin Recognition Complex in Human Cells*. J Biol Chem 290, 12355–12369. 10.1074/jbc.m114.625012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lynch KL, Alvino GM, Kwan EX, Brewer BJ, and Raghuraman MK (2019). The effects of manipulating levels of replication initiation factors on origin firing efficiency in yeast. Plos Genet 15, e1008430. 10.1371/journal.pgen.1008430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Richards L, Das S, and Nordman JT (2022). Rif1-Dependent Control of Replication Timing. Genes-basel 13, 550. 10.3390/genes13030550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.MacAlpine HK, Gordân R, Powell SK, Hartemink AJ, and MacAlpine DM (2010). Drosophila ORC localizes to open chromatin and marks sites of cohesin complex loading. Genome Res 20, 201–211. 10.1101/gr.097873.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Antequera F, and Bird A (1999). CpG islands as genomic footprints of promoters that are associated with replication origins. Curr Biol 9, R661–R667. 10.1016/s0960-9822(99)80418-7. [DOI] [PubMed] [Google Scholar]
- 192.Knott SRV, Peace JM, Ostrow AZ, Gan Y, Rex AE, Viggiani CJ, Tavaré S, and Aparicio OM (2012). Forkhead Transcription Factors Establish Origin Timing and Long-Range Clustering in S. cerevisiae. Cell 148, 99–111. 10.1016/j.cell.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Dimitrova DS (2006). Nuclear transcription is essential for specification of mammalian replication origins. Genes Cells 11, 829–844. 10.1111/j.1365-2443.2006.00981.x. [DOI] [PubMed] [Google Scholar]
- 194.Sasaki T, Ramanathan S, Okuno Y, Kumagai C, Shaikh SS, and Gilbert DM (2006). The Chinese Hamster Dihydrofolate Reductase Replication Origin Decision Point Follows Activation of Transcription and Suppresses Initiation of Replication within Transcription Units. Mol Cell Biol 26, 1051–1062. 10.1128/mcb.26.3.1051-1062.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Brison O, El-Hilali S, Azar D, Koundrioukoff S, Schmidt M, Nähse V, Jaszczyszyn Y, Lachages A-M, Dutrillaux B, Thermes C, et al. (2019). Transcription-mediated organization of the replication initiation program across large genes sets common fragile sites genome-wide. Nat Commun 10, 5693. 10.1038/s41467-019-13674-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Harvey KJ, and Newport J (2003). CpG Methylation of DNA Restricts Prereplication Complex Assembly in Xenopus Egg Extracts. Mol Cell Biol 23, 6769–6779. 10.1128/mcb.23.19.6769-6779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Cox LS, Hupp T, Midgley CA, and Lane DP (1995). A direct effect of activated human p53 on nuclear DNA replication. Embo J 14, 2099–2105. 10.1002/j.1460-2075.1995.tb07201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Du Q, Smith GC, Luu PL, Ferguson JM, Armstrong NJ, Caldon CE, Campbell EM, Nair SS, Zotenko E, Gould CM, et al. (2021). DNA methylation is required to maintain both DNA replication timing precision and 3D genome organization integrity. Cell Reports 36, 109722. 10.1016/j.celrep.2021.109722. [DOI] [PubMed] [Google Scholar]
- 199.Messerschmidt DM, Knowles BB, and Solter D (2014). DNA methylation dynamics during epigenetic reprogramming in the germline and preimplantation embryos. Gene Dev 28, 812–828. 10.1101/gad.234294.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, and Pugh BF (2008). A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18, 1073–1083. 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Shivaswamy S, Bhinge A, Zhao Y, Jones S, Hirst M, and Iyer VR (2008). Dynamic Remodeling of Individual Nucleosomes Across a Eukaryotic Genome in Response to Transcriptional Perturbation. Plos Biol 6, e65. 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Eaton ML, Galani K, Kang S, Bell SP, and MacAlpine DM (2010). Conserved nucleosome positioning defines replication origins. Gene Dev 24, 748–753. 10.1101/gad.1913210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Lipford JR, and Bell SP (2001). Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Molecular Cell 7, 21–30. [DOI] [PubMed] [Google Scholar]
- 204.Ito T, Bulger M, Pazin MJ, Kobayashi R, and Kadonaga JT (1997). ACF, an ISWI-Containing and ATP-Utilizing Chromatin Assembly and Remodeling Factor. Cell 90, 145–155. 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 205.Tsukiyama T, and Wu C (1995). Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell 83, 1011–1020. 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 206.Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, Laue ED, Tanay A, and Fraser P (2013). Single cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 502, 10.1038/nature12593. 10.1038/nature12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Guilbaud G, Rappailles A, Baker A, Chen C-L, Arneodo A, Goldar A, d’Aubenton-Carafa Y, Thermes C, Audit B, and Hyrien O (2011). Evidence for Sequential and Increasing Activation of Replication Origins along Replication Timing Gradients in the Human Genome. Plos Comput Biol 7, e1002322. 10.1371/journal.pcbi.1002322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Audit B, Zaghloul L, Vaillant C, Chevereau G, d’Aubenton-Carafa Y, Thermes C, and Arneodo A (2009). Open chromatin encoded in DNA sequence is the signature of ‘master’ replication origins in human cells. Nucleic Acids Res 37, 6064–6075. 10.1093/nar/gkp631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Marahrens Y, and Stillman B (1994). Replicator dominance in a eukaryotic chromosome. Embo J 13, 3395–3400. 10.1002/j.1460-2075.1994.tb06642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Brewer BJ, and Fangman WL (1993). Initiation at closely spaced replication origins in a yeast chromosome. Sci New York N Y 262, 1728–1731. 10.1126/science.8259517. [DOI] [PubMed] [Google Scholar]
- 211.Ferguson BM, and Fangman WL (1992). A position effect on the time of replication origin activation in yeast. Cell 68, 333–339. 10.1016/0092-8674(92)90474-q. [DOI] [PubMed] [Google Scholar]
- 212.Heinzel SS, Krysan PJ, Tran CT, and Calos MP (1991). Autonomous DNA replication in human cells is affected by the size and the source of the DNA. Mol Cell Biol 11, 2263–2272. 10.1128/mcb.11.4.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Klein KN, and Gilbert DM (2016). The Initiation of DNA Replication in Eukaryotes. In Epigenetic vs. Sequence-Dependent Control of Eukaryotic Replication Timing, pp. 39–63. 10.1007/978-3-319-24696-3_3. [DOI] [Google Scholar]
- 214.Long H, Zhang L, Lv M, Wen Z, Zhang W, Chen X, Zhang P, Li T, Chang L, Jin C, et al. (2019). H2A.Z facilitates licensing and activation of early replication origins. Nature 577, 576–581. 10.1038/s41586-019-1877-9. [DOI] [PubMed] [Google Scholar]
- 215.Sherstyuk VV, Shevchenko AI, and Zakian SM (2014). Epigenetic landscape for initiation of DNA replication. Chromosoma 123, 183–199. 10.1007/s00412-013-0448-3. [DOI] [PubMed] [Google Scholar]
- 216.Song J, Ishibe-Murakami S, Chen JK, Patel DJ, and Gozani O (2012). The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier-Gorlin syndrome. Nature 484, 115–119. 10.1038/nature10956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Smith OK, Kim R, Fu H, Martin MM, Lin CM, Utani K, Zhang Y, Marks AB, Lalande M, Chamberlain S, et al. (2016). Distinct epigenetic features of differentiation-regulated replication origins. Epigenet Chromatin 9, 18. 10.1186/s13072-016-0067-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Hayashi-Takanaka Y, Hayashi Y, Hirano Y, Miyawaki-Kuwakado A, Ohkawa Y, Obuse C, Kimura H, Haraguchi T, and Hiraoka Y (2021). Chromatin loading of MCM hexamers is associated with di-/tri-methylation of histone H4K20 toward S phase entry. Nucleic Acids Res 49, 12152–12166. 10.1093/nar/gkab1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Du Q, Bert SA, Armstrong NJ, Caldon CE, Song JZ, Nair SS, Gould CM, Luu P-L, Peters T, Khoury A, et al. (2019). Replication timing and epigenome remodelling are associated with the nature of chromosomal rearrangements in cancer. Nat Commun 10, 416. 10.1038/s41467-019-08302-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Iizuka M, and Stillman B (1999). Histone acetyltransferase HBO1 interacts with the ORC1 subunit of the human initiator protein. The Journal of biological chemistry 274, 23027–23034. [DOI] [PubMed] [Google Scholar]
- 221.Hickman MA, and Rusche LN (2010). Transcriptional silencing functions of the yeast protein Orc1/Sir3 subfunctionalized after gene duplication. Proceedings of the National Academy of Sciences of the United States of America 107, 19384–19389. 10.1073/pnas.1006436107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Hossain M, and Stillman B (2016). Opposing roles for DNA replication initiator proteins ORC1 and CDC6 in control of Cyclin E gene transcription. eLife 5, 10.7554-eLife.12785. 10.7554/eLife.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Phillips-Cremins JE, Sauria MEG, Sanyal A, Gerasimova TI, Lajoie BR, Bell JSK, Ong C-T, Hookway TA, Guo C, Sun Y, et al. (2013). Architectural Protein Subclasses Shape 3D Organization of Genomes during Lineage Commitment. Cell 153, 1281–1295. 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Guillou E, Ibarra A, Coulon V, Casado-Vela J, Rico D, Casal I, Schwob E, Losada A, and Méndez J (2010). Cohesin organizes chromatin loops at DNA replication factories. Gene Dev 24, 2812–2822. 10.1101/gad.608210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Su QP, Zhao ZW, Meng L, Ding M, Zhang W, Li Y, Liu M, Li R, Gao Y-Q, Xie XS, et al. (2020). Superresolution imaging reveals spatiotemporal propagation of human replication foci mediated by CTCF-organized chromatin structures. Proc National Acad Sci 117, 15036–15046. 10.1073/pnas.2001521117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Costantino L, Hsieh T-HS, Lamothe R, Darzacq X, and Koshland D (2020). Cohesin residency determines chromatin loop patterns. Elife 9, e59889. 10.7554/elife.59889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Emerson DJ, Zhao PA, Cook AL, Barnett RJ, Klein KN, Saulebekova D, Ge C, Zhou L, Simandi Z, Minsk MK, et al. (2022). Cohesin-mediated loop anchors confine the locations of human replication origins. Nature, 1–8. 10.1038/s41586-022-04803-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Tower J (2004). DEVELOPMENTAL GENE AMPLIFICATION AND ORIGIN REGULATION. Genetics 38, 273–304. 10.1146/annurev.genet.37.110801.143851. [DOI] [PubMed] [Google Scholar]
- 229.Royzman I, Austin RJ, Bosco G, Bell SP, and Orr-Weaver TL (1999). ORC localization in Drosophila follicle cells and the effects of mutations in dE2F and dDP. Gene Dev 13, 827–840. 10.1101/gad.13.7.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Stubb A, and Wickström SA (2022). Stretched skin cells divide without DNA replication. Nature 605, 31–32. 10.1038/d41586-022-00790-4. [DOI] [PubMed] [Google Scholar]
- 231.Raghuraman MK, and Liachko I (2016). The Initiation of DNA Replication in Eukaryotes. In Sequence Determinants of Yeast Replication Origins The initiation of DNA replication in eukaryotes., Kaplan DL, ed. (Springer International Publishing; ), pp. 123–141. 10.1007/978-3-319-24696-3_7. [DOI] [Google Scholar]
- 232.Tsai H-J, Baller JA, Liachko I, Koren A, Burrack LS, Hickman MA, Thevandavakkam MA, Rusche LN, and Berman J (2014). Origin replication complex binding, nucleosome depletion patterns, and a primary sequence motif can predict origins of replication in a genome with epigenetic centromeres. Mbio 5, e01703–14. 10.1128/mbio.01703-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Liachko I, Youngblood RA, Tsui K, Bubb KL, Queitsch C, Raghuraman MK, Nislow C, Brewer BJ, and Dunham MJ (2014). GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast Pichia pastoris. Plos Genet 10, e1004169. 10.1371/journal.pgen.1004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Hoggard T, Liachko I, Burt C, Meikle T, Jiang K, Craciun G, Dunham MJ, and Fox CA (2016). High Throughput Analyses of Budding Yeast ARSs Reveal New DNA Elements Capable of Conferring Centromere-Independent Plasmid Propagation. G3 (Bethesda, Md.) 6, 993–1012. 10.1534/g3.116.027904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Malik HS, and Henikoff S (2009). Major Evolutionary Transitions in Centromere Complexity. Cell 138, 1067–1082. 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 236.Drinnenberg IA, Fink GR, and Bartel DP (2011). Compatibility with killer explains the rise of RNAi-deficient fungi. Science 333, 1592. 10.1126/science.1209575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Peng JC, and Karpen GH (2006). H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nature cell biology 9, 25–35. 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Rusche LN, and Hickman MA (2020). Sex in Fungi. 189–200. 10.1128/9781555815837.ch11. [DOI] [Google Scholar]
- 239.Murray JAH, Cesareni G, and Argos P (1988). Unexpected divergence and molecular coevolution in yeast plasmids. J Mol Biol 200, 601–607. 10.1016/0022-2836(88)90546-3. [DOI] [PubMed] [Google Scholar]
- 240.Futcher AB (1988). The 2 micron circle plasmid of Saccharomyces cerevisiae. Yeast Chichester Engl 4, 27–40. 10.1002/yea.320040104. [DOI] [PubMed] [Google Scholar]
- 241.Fournier P, Abbas A, Chasles M, Kudla B, Ogrydziak DM, Yaver D, Xuan JW, Peito A, Ribet AM, and Feynerol C (1993). Colocalization of centromeric and replicative functions on autonomously replicating sequences isolated from the yeast Yarrowia lipolytica. Proc National Acad Sci 90, 4912–4916. 10.1073/pnas.90.11.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Liachko I, Youngblood RA, Tsui K, Bubb KL, Queitsch C, Raghuraman MK, Nislow C, Brewer BJ, and Dunham MJ (2014). GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast Pichia pastoris. Plos Genet 10, e1004169. 10.1371/journal.pgen.1004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Prasanth SG, Shen Z, Prasanth KV, and Stillman B (2010). Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization. Proc National Acad Sci 107, 15093–15098. 10.1073/pnas.1009945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Bergmann JH, Rodríguez MG, Martins NMC, Kimura H, Kelly DA, Masumoto H, Larionov V, Jansen LET, and Earnshaw WC (2011). Epigenetic engineering shows H3K4me2 is required for HJURP targeting and CENP-A assembly on a synthetic human kinetochore. Embo J 30, 328–340. 10.1038/emboj.2010.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Steiner FA, and Henikoff S (2014). Holocentromeres are dispersed point centromeres localized at transcription factor hotspots. Elife 3, e02025. 10.7554/elife.02025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Salas-Leiva DE, Tromer EC, Curtis BA, Jerlström-Hultqvist J, Kolisko M, Yi Z, Salas-Leiva JS, Gallot-Lavallée L, Williams SK, Kops GJPL, et al. (2021). Genomic analysis finds no evidence of canonical eukaryotic DNA processing complexes in a free-living protist. Nat Commun 12, 6003. 10.1038/s41467-021-26077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Lydeard JR, Lipkin-Moore Z, Sheu Y-J, Stillman B, Burgers PM, and Haber JE (2010). Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genome research 24, 1133–1144. 10.1101/gad.1922610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Stillman B (2022). The remarkable gymnastics of ORC. Elife 11. 10.7554/elife.76475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Yuan Z, Riera A, Bai L, Sun J, Nandi S, Spanos C, Chen ZA, Barbon M, Rappsilber J, Stillman B, et al. (2017). Structural basis of Mcm2-7 replicative helicase loading by ORC–Cdc6 and Cdt1. Nat Struct Mol Biol 24, 316–324. 10.1038/nsmb.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Zhai Y, Cheng E, Wu H, Li N, Yung PYK, Gao N, and Tye BK (2017). Open-ringed structure of the Cdt1-Mcm2-7 complex as a precursor of the MCM double hexamer. Nature Structural & Molecular Biology 24, 300–308. 10.1038/nsmb.3374. [DOI] [PubMed] [Google Scholar]
- 251.Miller TCR, Locke J, Greiwe JF, Diffley JFX, and Costa A (2019). Mechanism of head-to-head MCM double-hexamer formation revealed by cryo-EM. Nature 575, 704–710. 10.1038/s41586-019-1768-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Ali FA, Douglas ME, Locke J, Pye VE, Nans A, Diffley JFX, and Costa A (2017). Cryo-EM structure of a licensed DNA replication origin. Nature Communications 8, 1–10. 10.1038/s41467-017-02389-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Yuan Z, Schneider S, Dodd T, Riera A, Bai L, Yan C, Magdalou I, Ivanov I, Stillman B, Li H, et al. (2020). Structural mechanism of helicase loading onto replication origin DNA by ORC-Cdc6. Proc. Natl. Acad. Sci. USA 117, 17747–17756. 10.1073/pnas.2006231117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Feng X, Noguchi Y, Barbon M, Stillman B, Speck C, and Li H (2021). The structure of ORC–Cdc6 on an origin DNA reveals the mechanism of ORC activation by the replication initiator Cdc6. Nat Commun 12, 3883. 10.1038/s41467-021-24199-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Croll TI (2018). ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr Sect D Struct Biology 74, 519–530. 10.1107/s2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Belsky JA, Macalpine HK, Lubelsky Y, Hartemink AJ, and MacAlpine DM (2015). Genome-wide chromatin footprinting reveals changes in replication origin architecture induced by pre-RC assembly. Genes & development 29, 212–224. 10.1101/gad.247924.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Lucas IA, and Raghuraman MK (2003). The Dynamics of Chromosome Replication in Yeast. Curr Top Dev Biol 55, 1–73. 10.1016/s0070-2153(03)01001-9. [DOI] [PubMed] [Google Scholar]
- 258.MacAlpine DM, Rodríguez HK, and Bell SP (2004). Coordination of replication and transcription along a Drosophila chromosome. Genes & development 18, 3094–3105. 10.1101/gad.1246404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Mesner LD, Valsakumar V, Cieślik M, Pickin R, Hamlin JL, and Bekiranov S (2013). Bubble-seq analysis of the human genome reveals distinct chromatin-mediated mechanisms for regulating early- and late-firing origins. Genome Res 23, 1774–1788. 10.1101/gr.155218.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Langley AR, Gräf S, Smith JC, and Krude T (2016). Genome-wide identification and characterisation of human DNA replication origins by initiation site sequencing (ini-seq). Nucleic Acids Research 44, 10230–10247. 10.1093/nar/gkw760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Besnard E, Babled A, Lapasset L, Milhavet O, Parrinello H, Dantec C, Marin J-M, and Lemaitre J-M (2012). Unraveling cell type–specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat Struct Mol Biol 19, 837–844. 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- 262.Cayrou C, Coulombe P, Vigneron A, Stanojcic S, Ganier O, Peiffer I, Rivals E, Puy A, Laurent-Chabalier S, Desprat R, et al. (2011). Genome-scale analysis of metazoan replication origins reveals their organization in specific but flexible sites defined by conserved features. Genome Res 21, 1438–1449. 10.1101/gr.121830.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Picard F, Cadoret J-C, Audit B, Arneodo A, Alberti A, Battail C, Duret L, and Prioleau M-N (2014). The Spatiotemporal Program of DNA Replication Is Associated with Specific Combinations of Chromatin Marks in Human Cells. Plos Genet 10, e1004282. 10.1371/journal.pgen.1004282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Brodie, E.B.B. of, Nicolay S, Touchon M, Audit B, d’Aubenton-Carafa Y, Thermes C, and Arneodo A (2005). From DNA Sequence Analysis to Modeling Replication in the Human Genome. Phys Rev Lett 94, 248103. 10.1103/physrevlett.94.248103. [DOI] [PubMed] [Google Scholar]
- 265.Bensimon A, Simon A, Chiffaudel A, Croquette V, Heslot F, and Bensimon D (1994). Alignment and Sensitive Detection of DNA by a Moving Interface. Science 265, 2096–2098. 10.1126/science.7522347. [DOI] [PubMed] [Google Scholar]
- 266.Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann JS, et al. (1997). Dynamic Molecular Combing: Stretching the Whole Human Genome for High-Resolution Studies. Science 277, 1518–1523. 10.1126/science.277.5331.1518. [DOI] [PubMed] [Google Scholar]
- 267.Pasero P, Bensimon A, and Schwob E (2002). Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes & development 16, 2479–2484. 10.1101/gad.232902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Hennion M, Arbona J-M, Lacroix L, Cruaud C, Theulot B, Tallec BL, Proux F, Wu X, Novikova E, Engelen S, et al. (2020). FORK-seq: replication landscape of the Saccharomyces cerevisiae genome by nanopore sequencing. Genome Biol 21, 125. 10.1186/s13059-020-02013-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Müller CA, Boemo MA, Spingardi P, Kessler BM, Kriaucionis S, Simpson JT, and Nieduszynski CA (2019). Capturing the dynamics of genome replication on individual ultra-long nanopore sequence reads. Nat Methods 16, 429–436. 10.1038/s41592-019-0394-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Georgieva D, Liu Q, Wang K, and Egli D (2020). Detection of base analogs incorporated during DNA replication by nanopore sequencing. Nucleic Acids Res 48, gkaa517-. 10.1093/nar/gkaa517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Boemo MA (2021). DNAscent v2: detecting replication forks in nanopore sequencing data with deep learning. Bmc Genomics 22, 430. 10.1186/s12864-021-07736-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Miura H, Takahashi S, Shibata T, Nagao K, Obuse C, Okumura K, Ogata M, Hiratani I, and Takebayashi S (2020). Mapping replication timing domains genome wide in single mammalian cells with single-cell DNA replication sequencing. Nature Protocols 15, 4058–4100. 10.1038/s41596-020-0378-5. [DOI] [PubMed] [Google Scholar]
- 273.Takahashi S, Miura H, Shibata T, Nagao K, Okumura K, Ogata M, Obuse C, Takebayashi S, and Hiratani I (2019). Genome-wide stability of the DNA replication program in single mammalian cells. Nature Genetics 51, 529–540. 10.1038/s41588-019-0347-5. [DOI] [PubMed] [Google Scholar]
- 274.Chen C, Xing D, Tan L, Li H, Zhou G, Huang L, and Xie XS (2017). Single-cell whole-genome analyses by Linear Amplification via Transposon Insertion (LIANTI). Science 356, 189–194. 10.1126/science.aak9787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Laks E, McPherson A, Zahn H, Lai D, Steif A, Brimhall J, Biele J, Wang B, Masud T, Ting J, et al. (2019). Clonal Decomposition and DNA Replication States Defined by Scaled Single-Cell Genome Sequencing. Cell 179, 1207–1221.e22. 10.1016/j.cell.2019.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Zahn H, Steif A, Laks E, Eirew P, VanInsberghe M, Shah SP, Aparicio S, and Hansen CL (2017). Scalable whole-genome single-cell library preparation without preamplification. Nat Methods 14, 167–173. 10.1038/nmeth.4140. [DOI] [PubMed] [Google Scholar]


