Abstract
The human genome is arranged in the cell nucleus non-randomly, and phase separation has been proposed as an important driving force for genome organization. However, the cell nucleus is an active system, and the contribution of non-equilibrium activities to phase separation and genome structure and dynamics remains to be explored. We simulated the genome using an energy function parameterized with chromosome conformation capture (Hi-C) data with the presence of active, nondirectional forces that break the detailed balance. We found that active forces that may arise from transcription and chromatin remodeling can dramatically impact the spatial localization of heterochromatin. When applied to euchromatin, active forces can drive heterochromatin to the nuclear envelope and compete with passive interactions among heterochromatin that tend to pull them in opposite directions. Furthermore, active forces induce long-range spatial correlations among genomic loci beyond single chromosome territories. We further showed that the impact of active forces could be understood from the effective temperature defined as the fluctuation-dissipation ratio. Our study suggests that non-equilibrium activities can significantly impact genome structure and dynamics, producing unexpected collective phenomena.
Introduction
The eukaryotic genome is organized non-randomly in the three-dimensional space, and genomic regions with varying transcriptional activities occupy well-defined regions.1–9 Transcriptionally active chromosomes are frequently found near the nuclear interior,10 while heterochromatin tends to reside at the periphery close to the nuclear lamina.11 The lack of mixing among different chromatin types is evident from chromosome conformation capture (Hi-C)12,13 and single-cell imaging experiments.14,15 The contact maps produced from these experiments support that euchromatic regions share more interactions among themselves than with heterochromatin, giving rise to the so-called compartmentalization and the emergence of A/B compartments. Compartmentalization could arise from the microphase separation of polymeric systems, which has been proposed as a key mechanism for genome organization.16–26 Molecular mechanisms driving chromosome phase separation are beginning to emerge as well.
Euchromatin and heterochromatin are known to exhibit distinct chemical modifications27–30 and interact with different regulatory proteins28,29 and nuclear bodies.9,31–33 The chemical modifications themselves could drive chromatin demixing by altering nucleosome interactions. For example, acetylation could neutralize the positive charges on histone tails to weaken their binding with DNA,34–37 while methylation may lead to under-twisted DNA that strengthens its interactions with histones.38,39 It’s conceivable that nucleosomes with similar histone marks share more favorable interactions, driving their separation from other differentially modified ones. On the other hand, chromatin regulators associated with specific histone marks often contain intrinsically disordered regions that exhibit multivalent interactions to undergo liquid-liquid phase separation themselves.40–46 These proteins may further contribute to the compartmentalization of various chromatin types.
Besides passive interactions, phase separation can also arise due to the many non-equilibrium processes inside the cell nucleus.47–49 These processes, including transcription, chromatin remodeling, etc.,50,51 often consume ATP to drive conformational changes in molecular motors and exert forces on chromatin. Active forces break the detailed balance to produce out of equilibrium genome organizations beyond those dictated by passive interactions. Pioneering studies from the Menon group have shown that active processes could lead to non-random chromosome positions.47,52 However, the impact of non-equilibrium activities on the compartmentalization of different chromatin types has not been investigated. Since they are concentrated in euchromatic regions, active processes may produce a temperature gradient between euchromatin and heterochromatin,53 contributing to their phase separation.48,54 Active and passive mechanisms may both influence genome structure and dynamics, but their combined effect remains to be explored.
We carried out Brownian dynamics simulations to evaluate the role of passive interactions and active molecular motors on genome organization. A potential energy function derived from Hi-C data was used to account for effective interactions that fold individual chromosomes and drive their phase separation. Non-equilibrium activities were added to euchromatic regions by introducing nondirectional active forces. Computer simulations revealed a striking impact of active forces on genomic phase separation. While both passive interactions and active forces facilitate chromosome phase separation, they position B compartments in opposite directions. Competition between the two mechanisms produces genome organizations seen in normal and inverted nuclei. In addition to genome structures, the simulation framework naturally allowed the quantification of chromosome conformational dynamics. We observed that active forces with random orientations give rise to spatial correlations between genomic segments that are several micrometers apart on length scales comparable to those observed experimentally. Our study suggests that non-equilibrium activities could give rise to novel collective behaviors and provide new insights into genome organization.
Methods
Computational model of the human genome
We followed the simulation strategy outlined in Ref. 17 to study the diploid human genome. The model explicitly represents the 46 chromosomes as beads-on-spring polymers confined in a sphere depicting the nucleus envelope. Based on the contact maps of Hi-C data from GM12878 cells, we designated each one of the coarse-grained one MB size beads as compartment A, B, or centromeric C. This assignment results in 2424 beads as compartment A and 3130 as B.
We simplified the energy function introduced by Qi et al.17 to focus on the effect of non-equilibrium motors on genome organization. In particular, we enforced a common set of parameters for non-bonded interactions within the same and across different chromosomes. This setup provides a convenient system to explore molecular factors beyond simple passive interactions in positioning chromosomes inside the nucleus. Furthermore, we switched from a soft-core potential to the Lennard-Jones potential for non-specific interactions between genomic loci to avoid chain crossing. The soft-core potential was introduced originally to facilitate conformational sampling. However, the two potentials produce quantitatively similar trends for both dynamical and structural results (Figure S1). Detailed expressions of the energy function can be found in the Supporting Information.
We also adjusted the radius of the spherical confinement from 19.69 to 13.0 σ, where σ is the unit length and represents the diameter of a 1MB long genomic segment. The original confinement size was chosen to produce a DNA volume fraction of 0.1. Assuming a cell nucleus of 10 μm in diameter, we estimated σ as 254 nm using the original confinement size. However, for genomic regions of one MB in length, the average size determined via super-resolution imaging55 is larger than this value. Therefore, we rescaled the confinement size such that the estimated value for σ = 385 nm better matches experimental measurements.
Details of dynamical simulations
We implemented the genome model in the LAMMPS software56 to carry out Brownian dynamics simulations. Without active forces, the equation of motion for particle i is defined as
| (1) |
where ζ is the friction coefficient and UGenome(r) is the energy function of the genome defined in Eq. S1. The random force term ξi has zero means and satisfies the fluctuation-dissipation theorem as ⟨ξi(t)ξj(t′)⟩ = 6kBTζδijδ(t − t′).
Active forces were implemented by adding an additional noise term, fi, to the right hand side of Eq. 1. The active forces have zero mean, ⟨fi(t)⟩ = 0 and the correlation for forces at different times is
| (2) |
where ζ is the friction coefficient and kB is the Boltzmann constant. kBTa corresponds to the energy released by force generating events and τ accounts for the finite lifetime of these events. For τ = 0, the correlation function in Eq. 2 reduces to 6kBTaζδijδ(t−t′). Therefore, the two noise terms can be combined together to produce an effective equilibrium model with Teff = T + Ta. For non-zero τ, we treat the active force fi as dynamical variables that evolve with the following equation
| (3) |
where ηi(t) are Gaussian white noises that satisfy ⟨ηi(t) · ηi(t′)⟩ = 6δ(t − t′). Both Eq. 1 and 3 were integrated with the modified Euler-Maruyama algorithm.57
Dynamical simulations were initialized with an equilibrium structure obtained from a long-timescale trajectory performed in our previous study.17 We carried out additional 5×106 step-long equilibration simulations before switching to production runs that lasted for 2×107 steps. The second half of production trajectories were divided into five non-overlapping blocks to provide independent estimates for variables of interest. The standard deviations of these estimates were shown as error bars of the mean values.
All simulations were performed with reduced units. We estimated the length unit σ = 385nm, the energy unit ϵ = kBT, where T is the room temperature. The time unit τB = 0.125s was determined such that the diffusion constant of coarse grained particles match the experimentally determine values 10−2μm2/s.58–60
Results
Computational model for the motorized genome
We introduced a coarse-grained model for the diploid human genome to approximate the role of loop extrusion61,62 and phase separation16–19,21,22 on genome organization effectively. We modeled the genome at the one megabase (MB) resolution as a collection of 46 chromosomes inside spherical confinement (Figure 1). Each chromosome is represented as a string of one-megabase-size beads, which can be assigned as one of three types, A, B, or C. A and B correspond to the two-compartment types that contribute to the checkerboard patterns typically seen in Hi-C contact maps,12,63 and C marks centromeric regions. The compartment assignments provide chemical specificity for the coarse-grained beads to model specific interactions and capture microphase separation. In addition to the block co-polymer setup, we introduced intra-chromosome interactions that vary as a function of the sequence separation between two genomic segments. This “ideal” potential approximates the crosslinks produced by Cohesin molecules via their extrusion along chromatin, promoting chromosome territory formation.24,64,65 Detailed expressions of the energy function can be found in the Supporting Information. Interaction parameters in the ideal and compartment potential were tuned to reproduce various average contact probabilities determined from Hi-C experiments for GM12878 cells using the maximum entropy optimization algorithm.64,66–68
Figure 1:
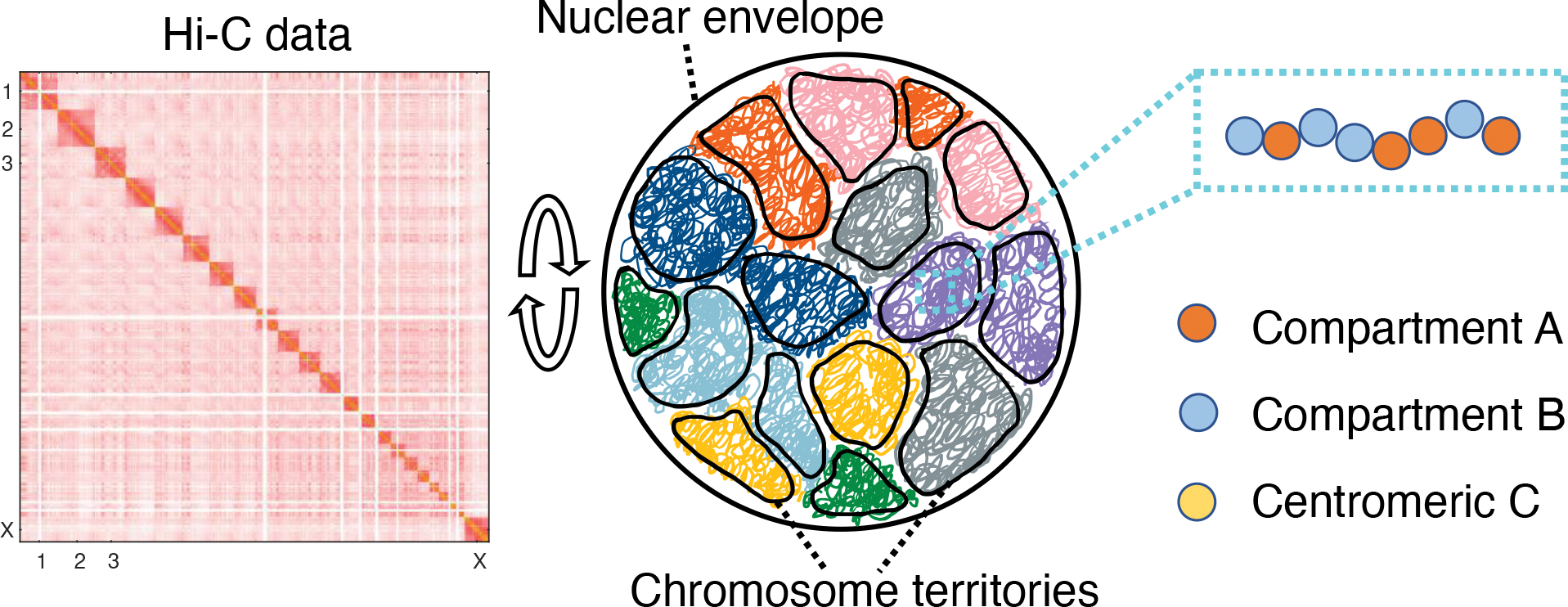
Illustration of the genome model parameterized with Hi-C data. The human genome consisting of 46 chromosomes was modeled in a spherical confinement of 10 μm in diameter. Each chromosome was represented as a string of coarse-grained particles that are one MB in size. We used Hi-C data to identify each bead as compartment A (orange), compartment B (blue), or centromeric region C (yellow) and to parameterize the interaction energy among them.
We carried out Brownian dynamics simulations of the coarse-grained model to produce an ensemble of genome structures (see Methods Section for simulation details). These structures support the formation of territories, and individual chromosomes exhibit collapsed conformations with minimal intermingling among them (Figure S2). We further determined an in-silico contact map by averaging over 3D structures to compare with Hi-C data. Given the genome model’s coarse resolution, the simulated contact map inevitably misses fine structural features such as chromatin loops63 and topologically associating domains.69,70 Nevertheless, it exhibits the typical plaid patterns that arise from the phase separation of A/B compartments (Figure S2).
An important feature of genome organization that’s amiss in the simulated structures is the peripheral localization of B compartments. As shown in the radial density profiles (Figure 2a), a significant amount of B compartments reside near the nuclear center. We note that since parameters were uniquely derived from Hi-C data, the observed mischaracterization of genome organization is due to the model’s inherent design, i.e., the specific functional form of the energy defined in Eq. S1, rather than a lack of parameter fine tuning. Indeed, as recognized in previous studies, explicit interactions with nuclear lamina,19,32,71,72 separate treatment of intra- and inter- chromosome interactions,17 or assuming repulsive interactions between the compartments,18 might be necessary to position B compartments towards the nuclear envelope.
Figure 2:
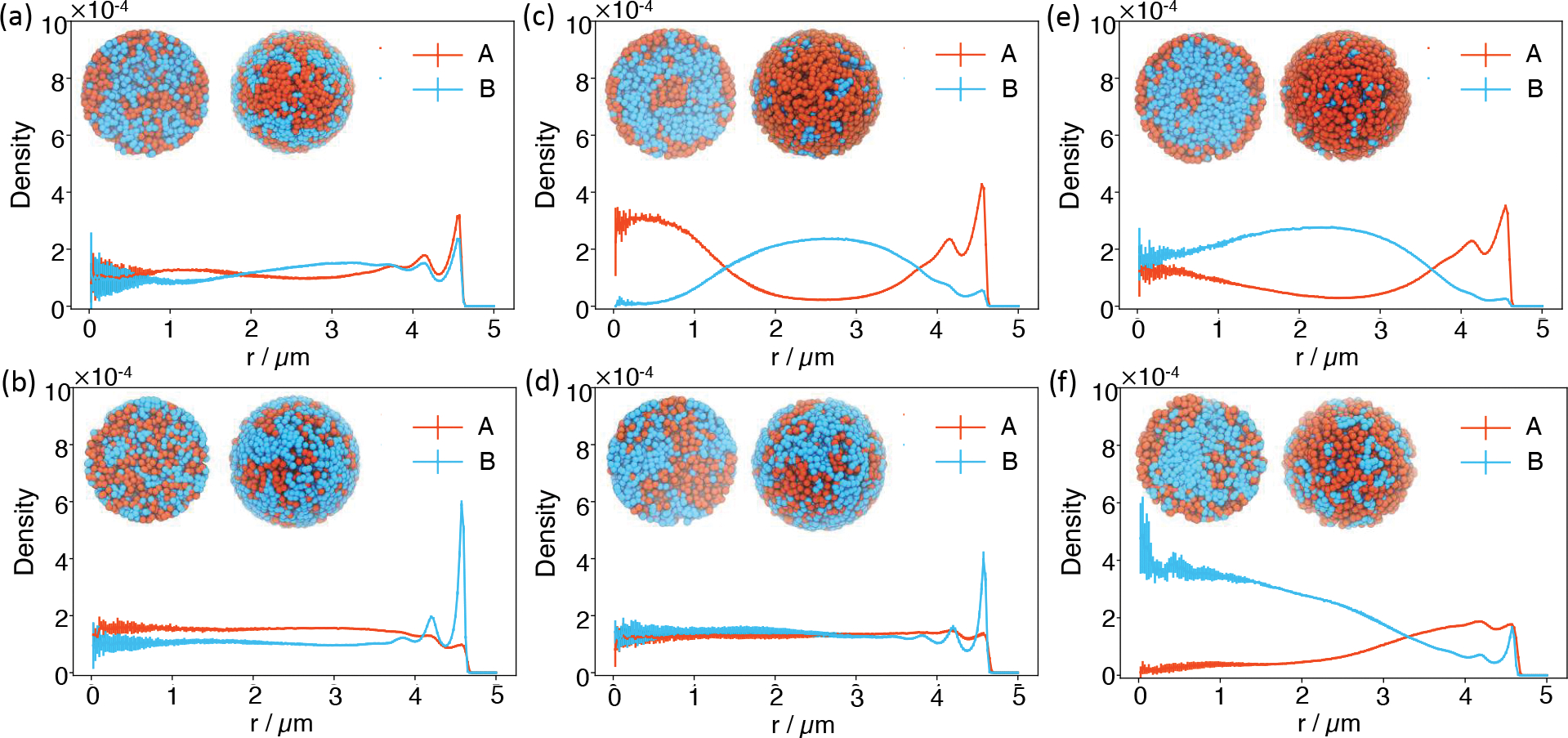
Radial distribution profiles of A/B compartments at different interaction strengths between B compartments without (a, c, and e) and with (b, d, and f) the presence of active forces. The interaction strength between B compartments was increased by two and three folds from a to c, and e, and from b to d, and f. The insets provide two different views of a representative configuration for each system. The left image corresponds to the front view of a cross-section of the genome sliced through the nuclear center, while the right image corresponds to the view from the nuclear envelope. In these images, A/B compartments are colored orange and blue, respectively.
While the model does not fully capture all features of genome organization, it stands out due to the simplicity of its design. The model serves as a valuable baseline to explore mechanisms in addition to loop extrusion and compartmentalization for genome organization.73
Active forces drive peripheral localization of heterochromatin
The cell nucleus is an active system enriched with ATP-driven processes. The ideal potential was specifically designed to account for the role of molecular motors like Cohesin that can compact individual chromosomes.61,62 Other active processes, including transcription and nucleosome remodeling,51,53,74–77 can also lead to non-conservative forces to perturb chromosome structures. The effect of these processes, together with passive interactions arising from epigenetic modifications and chromatin regulators, may produce the effective compartment specific potential inferred from Hi-C data.
To disentangle the role of passive and active forces on genome organization, following prior studies,47,78–80 we model the effect of transcription as self-propelled particles with random, nondirectional noises. For example, in addition to the typical random forces in the Brownian motion that satisfies the fluctuation-dissipation theorem, an extra term, fi(t) was added to the equation of motion for particle i at time t (see Methods for more details). Active forces were only applied to A compartments, i.e., euchromatin, which consists of approximately 40% of the genome.
We first studied genome structures with the presence of non-correlated active forces, i.e., white noises, in the limit of τ = 0. In this case, the correlation function becomes ⟨fi(t)fj(t′)⟩ = 6kBTaζδijδ(t − t′). We start with Ta = 10T, which is comparable to the amount of energy released from hydrolyzing a single ATP81 with T as the room temperature. Introducing active forces caused a dramatic change in the radial density profiles. There is a sharp peak for B compartments now at the nuclear periphery (Figure 2b), resembling the formation of lamina-associated domains (LADs).82 Correspondingly, the correlation between simulated and experimental contact frequency between chromosomes and the nuclear envelope improved (Figure S3). Accompanying the shift of B compartments is the increase of A compartments near the center, further driving the interior localization of chromosomes with more A compartments (Figure S4). As shown in Figure S5, these results are robust with respect to the strength of active forces. Restricting the activities to only the top half of most active A compartments also produced similar trends (Figure S6).
Balancing passive and active forces for heterochromatin localization
In our model, B compartments experience attractive interactions among themselves. Attractions could arise from direct contacts between histone proteins and the DNA44,83,84 or mediated by chromatin regulators, including HP1, PRC1, and PRC2,40,41,85–87 that promote multivalent interactions. To explore the combined role of passive and active forces on genome organization, we carried out additional simulations at different interaction strengths between B compartments with varying magnitudes of active forces.
As shown in Figures 2c and 2e, enhancing the interaction between B compartments by a factor of two or three causes the opposite effect on genome organization compared to adding active forces. The density profiles are in stark contrast to the one produced by the original interaction strength. Heterochromatin in the new simulations is closer to the nuclear interior, resulting in genome organizations that resemble the inverted nuclei. Strong interactions among heterochromatin have indeed been proposed to drive the formation of inverted nucleus observed in neuronal cells.88 Again, introducing active forces aligns B compartments towards the nuclear envelope (Figures 2d and 2f), though the effect becomes less dramatic at very strong passive interactions.
The competition between the two forces in positioning heterochromatin is summarized in an illustrative phase diagram (Figure 3a). We defined a collective variable that quantifies the radial distribution of B compartments using the ratio of the two average densities computed for regions close to and far away from the nuclear envelope. We separated the two regions with a distance cutoff of 3.85 μm. The variable increases for larger active forces and weaker passive interactions, supporting their counter effects on genome organization. We further classified systems with variable values larger than 0.65 and less than 0.2 as normal (blue) and inverted (red) nuclei, respectively based on the visual inspection of the simulated structures.
Figure 3:
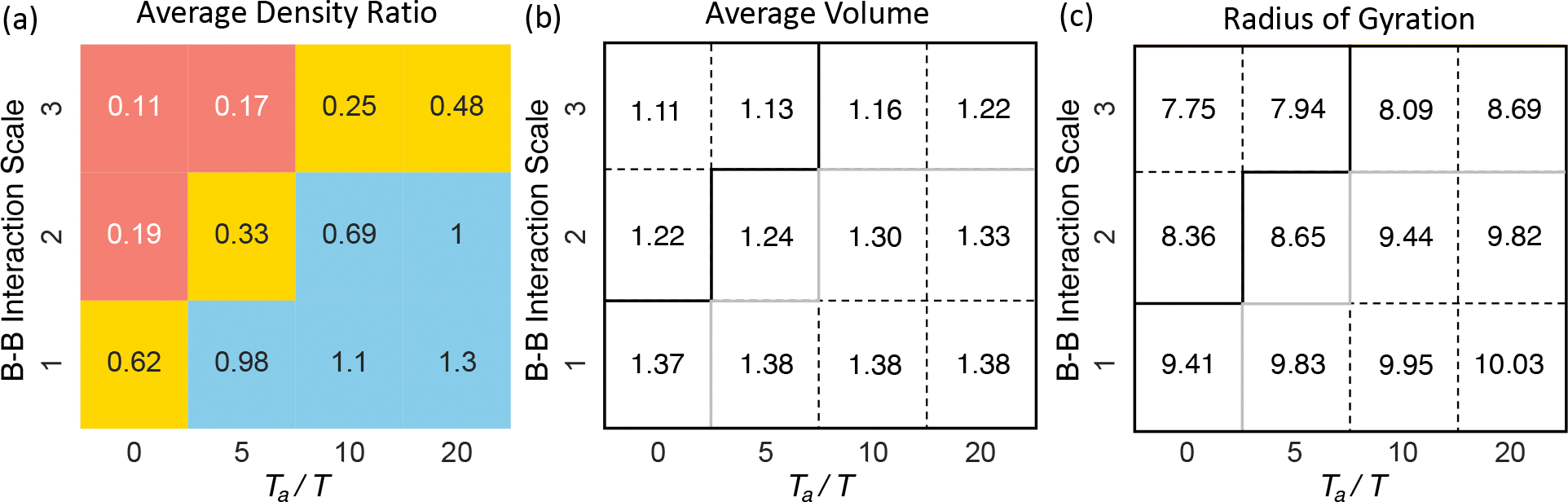
Illustrative phase diagram of genome organization. We identified three phases corresponding to the conventional (blue), intermediate (yellow), and inverted (red) nuclei using the average density ratio of the outer shell over the inner sphere. Representative configurations of the three phases are shown in Figure 2b (conventional), 2a (intermediate), and 2e (inverted). The x and y axes indicate the strength of active and passive forces. The number in each bin measures the average density ratio of the outer shell over the inner sphere (a), the average volume assigned to each heterochromatin region (b), and the radius of gyration for the largest cluster formed by B compartments (c). The solid lines in parts b and c correspond to the phase boundaries shown in part a. See also Figure S9 for the corresponding results for A compartments.
To provide a more detailed characterization of the impact of active and passive forces on 3D genome organization, we performed Voronoi tessellation of the simulated structures (Figure S7). Through the tessellation, we identified genomic regions that are nearest neighbors in space and connected them to determine the largest connected component network for B compartments. We found that the networks include almost all B compartments in all setups (Figure S8), supporting the role of both active and passive forces in promoting phase separation. The average volume assigned to each B compartment is also relatively conserved with respect to parameter changes (Figure 3b). However, the radius of gyration of the network showed dramatic variations, indicating changes in the morphology and surface area of the phases (Figure 3c). Increasing the interactions among B compartments compacts the networks, while active forces applied to A compartments grow their branches.
Active forces enhance correlated motion among chromatin
In addition to affecting steady-state distributions, non-equilibrium activities are known to give rise to emergent dynamical behaviors absent in equilibrium systems.89 Chromosomes have indeed been reported to exhibit coherent motions across length scales beyond individual territories as a result of ATP-driven processes.90–93 Whether the so-called scalar noise, i.e., the ones studied here with no orientational preference, contributes to such coherent motion has been controversial. Several studies have suggested that scalar noises cannot significantly impact the dynamical correlation length.78,90 Instead, vector events that produce force dipoles have been proposed as essential for enhancing correlation, possibly via a nematic ordering of chromatin.78,90,94 However, these studies did not explicitly account for specific interactions and chromosome compartmentalization, which have been shown to produce dynamical correlations within individual chromosomes.80,95 We further studied how passive interactions and scalar forces impact whole-genome conformational dynamics.
In agreement with previous studies,80,95 we observed dynamical correlations among chromatin segments without the presence of active forces. Following Ref. 80, we characterized genome-wide correlated motions using the displacement correlation function defined as
| (4) |
where the angular brackets indicate ensemble averaging and i,j index over chromatin segments. Δri(Δt) corresponds to the displacement of particle i over the time interval Δt, · represents the dot-product of the two displacement vectors, and rij is the distance between particle i and j. As shown in Figure 4a, while the correlation function decays quickly as a function of the separation length r for small time intervals, it increases significantly for longer Δt. We further defined the correlation length, lc, using an exponential fit as to C(r,Δt). The dependence of lc on Δt is shown in Figure 4c.
Figure 4:
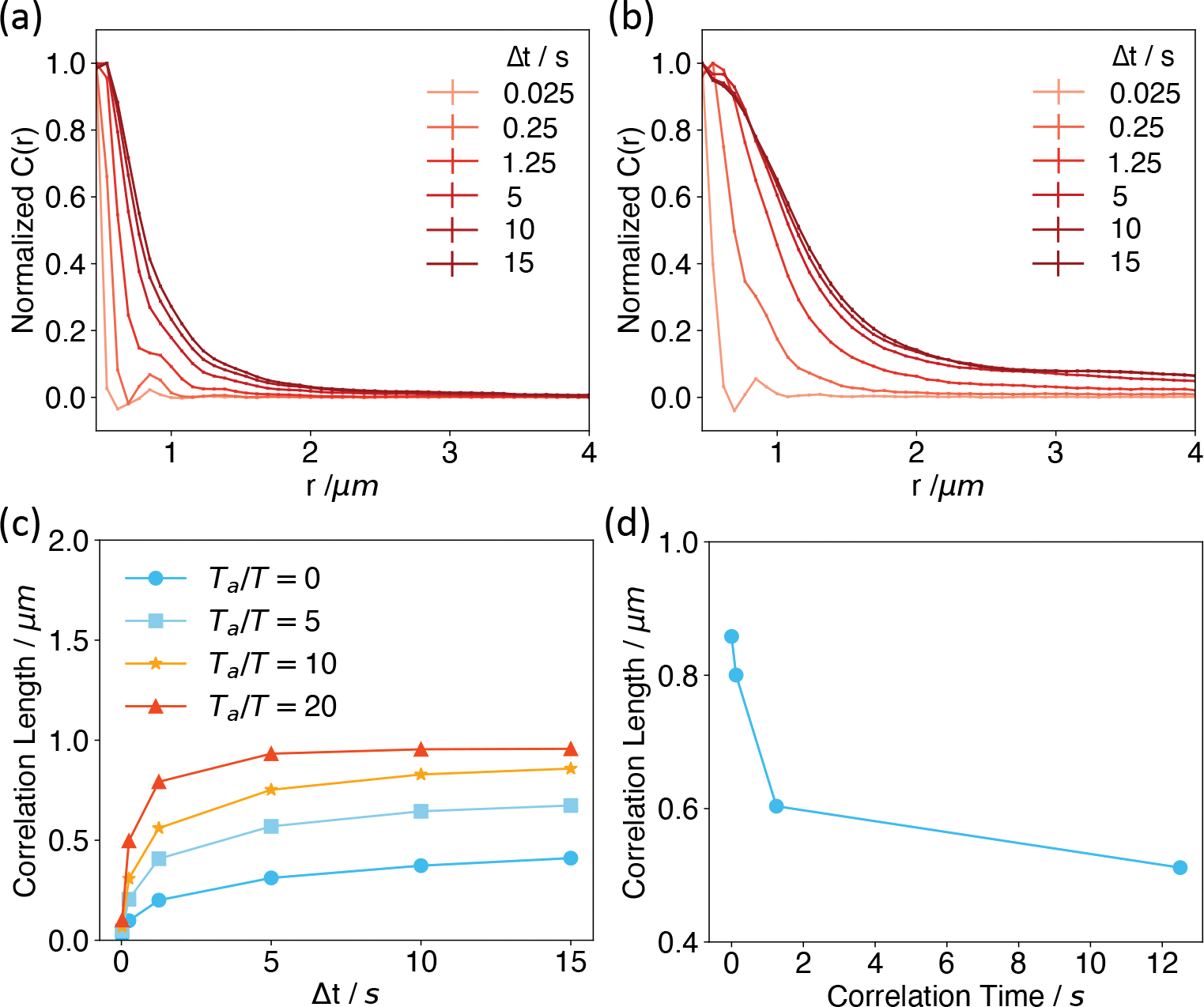
Active forces enhance long-range correlations among genomic loci. (a,b) Displacement correlation functions at various time separations computed without (a) and with (b) the presence of active forces. The correlation functions at different time intervals were normalized such that the maximum values are unity. See Figure S10 for results at different strengths of active forces. (c) Correlation lengths as a function of the time separation for different active forces. (d) Dependence of the spatial correlation length determined at Ta = 10T and Δt = 15s as a function of the correlation time of the active forces.
We next examined spatial chromosome correlation in the presence of active scalar forces with Ta = 10T. As shown in Figure 4b, a significant increase in the correlation length can be observed, especially for larger Δt. The precise dependence of lc on Δt at several values of Ta is provided in Figure 4c. These results support a significant role of molecular motors that produce nondirectional forces on chromosome dynamics.
To further break down the intra- and inter-chromosomal contributions, we limited the genomic pairs to those from the same or different chromosomes when computing C(r,Δt). As shown in Figure S11, active forces enhance correlations both within and across chromosomes. As expected, the correlation length within individual chromosomes cannot go beyond a certain value characteristic of the territory size. However, the inter-chromosomal correlations increase to much larger values.
The results presented so far are for active forces with zero correlation time, i.e., τ = 0 as defined in Eq. 2. However, forces at different time intervals could be correlated due to the finite lifetime of chemical processes, producing nonzero values for τ. Since precise values for τ are not available, we consider a range of values from 0.125 to 12.5 s that roughly matches the rate for chromatin remodeling.96 As shown in Figure 4d, the correlation length for Δt = 15s decreases significantly as τ increases. In addition, we found that nonzero values for τ also reduced the impact of active forces on the peripheral localization of B compartments (Figure S12). A physical explanation of the dramatic impact of τ on genome structure and dynamics is provided in the next section.
Effective temperature model for motors
We further resorted to the theory of effective temperature97 to provide intuitions for the role of active forces on genome organization. Effective temperature has proven useful for studying non-equilibrium systems and glasses.98–100 We quantify the effective temperature with the fluctuation-dissipation ratio53,98–101 defined as
| (5) |
C(t) represents the correlation function for the single particle density fluctuation with observables defined as and , and is the corresponding response function. More detailed expressions for computing the effective temperature are provided in the Supporting Information.
We first analyzed systems with non-correlated active forces at τ = 0. In such cases, the equation of motion for A compartments is identical to the one without active forces but at an elevated thermal temperature of Ta + T. We found that the susceptibility-correlation plots exhibit a single linear regime across all timescales (Figure 5a). The effective temperatures defined from the negative slope of such plots indeed agree with Ta+T (Figure 5b). Therefore, our analyses suggest that the overall system is equivalent to A/B compartments at equilibrium with two separate thermostats of different temperatures.47,53,102 Phase separation of liquid mixtures with differential temperature has indeed been well-documented,48,79,103,104 which may explain the changes seen in Figure 2. The higher temperature of A compartments makes them entropically favorable to stay in the interior with more conformational space.
Figure 5:
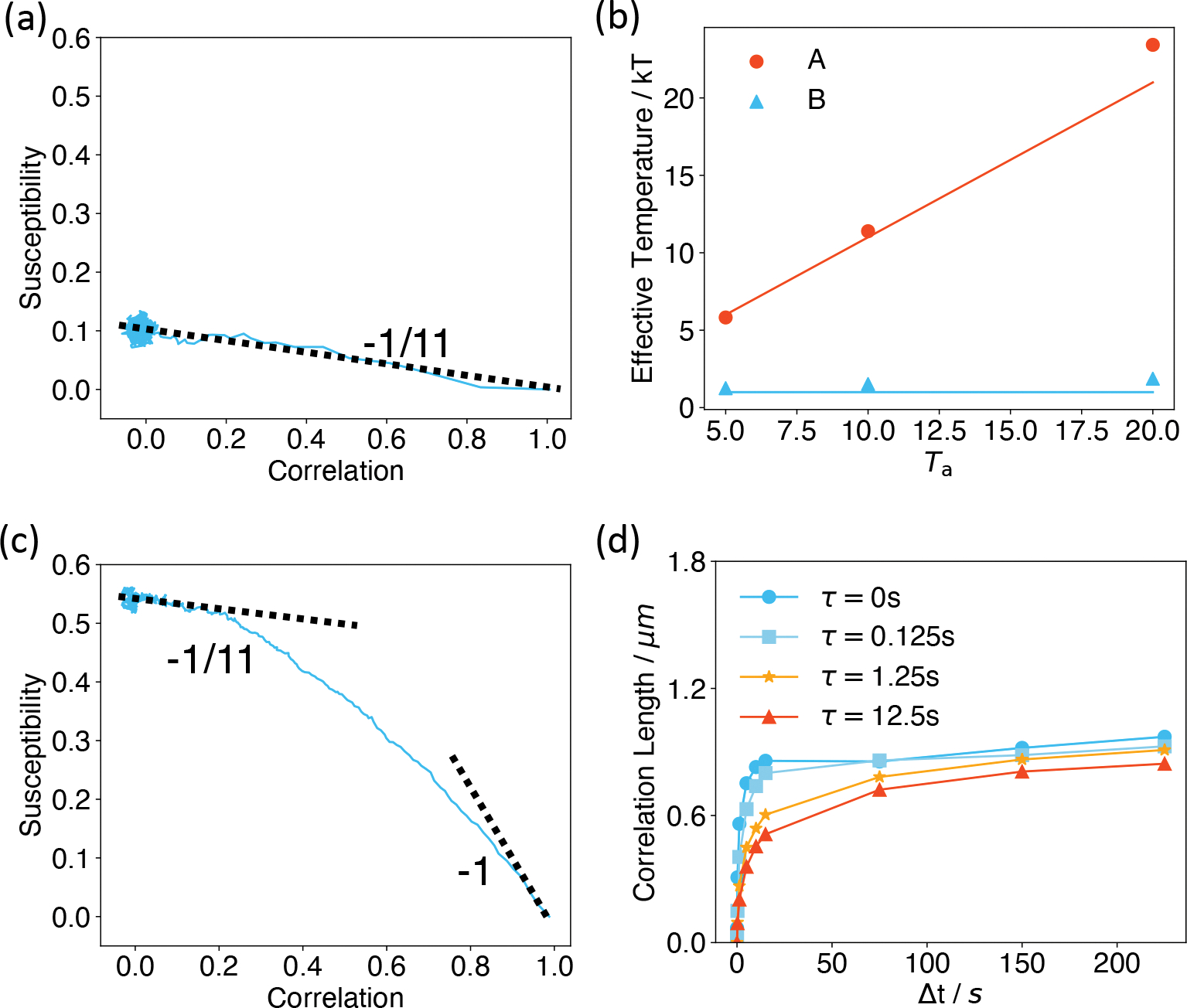
The effective temperature of active systems computed using the fluctuation-dissipation ratio. (a,c) Susceptibility as a function of correlation for the system with Ta = 10T, τ = 0 s (a) and τ = 1.25 s (c). The dashed lines correspond to the thermal temperature −1/T and the theoretical value −1/(Ta + T) for non-correlated active forces. (b) Effective temperatures of A/B compartments computed using the fluctuation-dissipation ratio for Ta = 5, 10, and 20T at τ = 0 s are shown as dots. The red line corresponds to Ta + T, while the blue line represents the constant T. (d) Correlation lengths as functions of time interval Δt for different correlation time τ of the colored noise.
For correlated noises with nonzero τ, we found that the systems exhibit two timescales (Figure 5c and S13). At smaller timescales, the susceptibility-correlation plot for A compartments is approximately linear with a slope close to −1/T, indicating that the system is in thermal equilibrium. At much longer timescales, the effect of non-equilibrium motors dominates, and the plot again becomes linear but with a different slope that approaches −1/(Ta + T). Furthermore, the transition regime between the two timescales increases with τ (Figure S13), suggesting that the system stays in the thermal temperature longer at larger τ. Therefore, for the same Δt, larger τ leads to a lower effective temperature and the decreased correlation length seen in Figure 4d. The appearance of a higher effective temperature at longer timescales has indeed been predicted in prior simulation and theoretical studies.100,105,106 As in the non-correlated noise, we anticipate that the higher effective temperature might lead to an increase in the correlation length at longer timescales. Indeed, we found that while lc(Δt) has plateaued after around 15s for τ = 0, it continues to rise for larger τ. The value lc(Δt) at different τ almost converges to the same value for Δt = 225s (Figure 5d and S14).
Conclusions and Discussion
We studied the impact of non-equilibrium activities on the structure and dynamics of the human genome. We found that activities such as transcription that are restricted to A compartments significantly influence the phase separation of euchromatin and heterochromatin. Heterochromatin is driven more towards the nuclear envelope, leaving more room in the interior for euchromatin. This effect can be largely understood from the presence of a temperature gradient. The impact on phase separation differs from those conferred by passive interactions, which stabilize the opposite trend by moving heterochromatin towards the nuclear interior, giving rise to the inverted nucleus.
Non-equilibrium phase separation between active and inactive particles has been shown to be sensitive to density.48,107 We repeated the active force simulations in larger nuclei to examine the impact of density on genome organization. Figure S15 shows that the preferential localization of B compartments towards the nuclear envelope becomes less apparent in these simulations. The dependence of activity-driven phase separation on nuclear volume may help rationalize conflicting observations of changes in LAD localization upon depleting lamin proteins. Several papers have reported a minimal change in Lamin B DamID profiles upon removing all lamin proteins.108,109 It is possible that passive interactions between chromatin and the nuclear envelope persist as a result of unknown mechanisms.110 On the other hand, the activity-driven phase separation could contribute to the residual association of LADs with the nuclear envelope observed in these studies. In cells where significant LAD dissociation from the nuclear envelope was observed,111–113 the nuclei volume is often larger than expected. The change in LAD distribution could arise from both a disruption in lamin expression and a weakened effect of the activity-driven phase separation.
We note that the model presented here is still an approximation of the cell nucleus. Additional factors not accounted for in our model, including hydrodynamic effects,90 vectorial forces,78 and deformability of the nucleus envelope,114 could further enhance the spatial correlation length. Including these factors will be interesting in the future for building more quantitative models of the genome.
Supplementary Material
Acknowledgement
This work was supported by the National Institutes of Health grant R35GM133580.
Footnotes
- Details of the diploid Human genome model parameterized with Hi-C data, simulation details, and numerical analysis details of effective temperature calculations, radial density profiles and chromosome clustering with Voronoi tessellation, (Figure S1) demonstration of model robustness with a soft-core potential, (Figure S2) a representative chromosome configuration and a comparison between simulated and experimental contact maps, (Figure S3) the correlation between simulated and experimental contact frequency between chromosomes and the nuclear envelope, (Figure S4) comparison between experimental and simulated chromosome radial positions, (Figure S5) radial density profiles of A/B compartments determined from simulations performed at different parameter sets, (Figure S6) radial density profiles determined from simulations in which the active forces were limited to the top half of most active A compartments, (Figure S7) two-dimensional illustration of the Voronoi tesselation method for identifying the largest cluster, (Figure S8) the average size of the largest connected components for A and B compartments at different parameter sets, (Figure S9) illustrative phase diagram of genome organization for A compartments, (Figure S10) chromosome spatial correlation with different active forces, (Figure S11) contributions from intra and inter-chromosome genomic pairs to chromosome spatial correlation, (Figure S12) radial density profiles calculated from simulations with active forces at Ta = 10T for various correlation time, (Figure S13) susceptibility-correlation curves for A and B compartments obtained from simulations with correlated active forces at Ta = 10T, (Figure S14) normalized displacement correlation plots determined from simulations with active forces of Ta = 10T and different correlation time, (Figure S15) radial density profiles calculated from simulations with spherical confinements of radius 5.4 μm and 5.8 μm.
References
- (1).Lin X; Qi Y; Latham AP; Zhang B Multiscale modeling of genome organization with maximum entropy optimization. J. Chem. Phys. 2021, 155, 010901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Bickmore WA The Spatial Organization of the Human Genome. Annu. Rev. Genomics Hum. Genet. 2013, 14, 67–84. [DOI] [PubMed] [Google Scholar]
- (3).Gorkin DU; Leung D; Ren B The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell 2014, 14, 762–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Cremer T; Cremer C Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Genet. 2001, 2, 292–301. [DOI] [PubMed] [Google Scholar]
- (5).van Steensel B; Belmont AS Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell 2017, 169, 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Dekker J; Mirny L The 3D Genome as Moderator of Chromosomal Communication. Cell 2016, 164, 1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Bonev B; Cavalli G Organization and function of the 3D genome. Nat. Rev. Genet. 2016, 17, 661–678. [DOI] [PubMed] [Google Scholar]
- (8).Finn EH; Misteli T Molecular basis and biological function of variability in spatial genome organization. Science 2019, 365, eaaw9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Bhat P; Honson D; Guttman M Nuclear compartmentalization as a mechanism of quantitative control of gene expression. Nat. Rev. Mol. Cell Biol. 2021, 0123456789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Boyle S; Gilchrist S; Bridger JM; Mahy NL; Ellis J; Bickmore WA The spatial organization of human chromosomes within the nuclei of normal and emerinmutant cells. Hum. Mol. Genet. 2001, 10, 211–219. [DOI] [PubMed] [Google Scholar]
- (11).Lemaˆıtre C; Bickmore WA Chromatin at the nuclear periphery and the regulation of genome functions. Histochem. Cell Biol. 2015, 144, 111–122. [DOI] [PubMed] [Google Scholar]
- (12).Lieberman-Aiden E; Van Berkum NL; Williams L; Imakaev M; Ragoczy T; Telling A; Amit I; Lajoie BR; Sabo PJ; Dorschner MO et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Dekker J; Marti-Renom MA; Mirny LA Exploring the three-dimensional organization of genomes: interpreting chromatin interaction data. Nat. Rev. Genet. 2013, 14, 390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Su JH; Zheng P; Kinrot SS; Bintu B; Zhuang X Genome-Scale Imaging of the 3D Organization and Transcriptional Activity of Chromatin. Cell 2020, 182, 1641–1659.e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Takei Y; Yun J; Zheng S; Ollikainen N; Pierson N; White J; Shah S; Thomassie J; Suo S; Eng C-HL et al. Integrated spatial genomics reveals global architecture of single nuclei. Nature 2021, 590, 344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Falk M; Feodorova Y; Naumova N; Imakaev M; Lajoie BR; Leonhardt H; Joffe B; Dekker J; Fudenberg G; Solovei I et al. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 2019 570:7761 2019, 570, 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Qi Y; Reyes A; Johnstone SE; Aryee MJ; Bernstein BE; Zhang B Data-Driven Polymer Model for Mechanistic Exploration of Diploid Genome Organization. Biophys. J. 2020, 119, 1905–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Fujishiro S; Sasai M Generation of dynamic three-dimensional genome structure through phase separation of chromatin. bioRxiv 2021, [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Laghmach R; Di Pierro M; Potoyan DA The interplay of chromatin phase separation and lamina interactions in nuclear organization. Biophys. J. 2021, 120, 5005–5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Xie WJ; Qi Y; Zhang B Characterizing chromatin folding coordinate and landscape with deep learning. PLoS computational biology 2020, 16, e1008262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Liu S; Zhang L; Quan H; Tian H; Meng L; Yang L; Feng H; Gao YQ From 1D sequence to 3D chromatin dynamics and cellular functions: a phase separation perspective. Nucleic Acids Res. 2018, 46, 9367–9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Shi G; Liu L; Hyeon C; Thirumalai D Interphase human chromosome exhibits out of equilibrium glassy dynamics. Nat. Commun. 2018, 9, 3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Jost D; Carrivain P; Cavalli G; Vaillant C Modeling epigenome folding: Formation and dynamics of topologically associated chromatin domains. Nucleic Acids Res. 2014, 42, 9553–9561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Di Pierro M; Zhang B; Aiden EL; Wolynes PG; Onuchic JN Transferable model for chromosome architecture. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 12168–12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Barbieri M; Chotalia M; Fraser J; Lavitas L-M; Dostie J; Pombo A; Nicodemi M Complexity of chromatin folding is captured by the strings and binders switch model. Proc. Natl. Acad. Sci. 2012, 109, 16173–16178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Erdel F; Rippe K Formation of Chromatin Subcompartments by Phase Separation. Biophys. J. 2018, 114, 2262–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Xie WJ; Zhang B Learning the formation mechanism of domain-level chromatin states with epigenomics data. Biophys. J. 2019, 116, 2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Morrison O; Thakur J Molecular Sciences Molecular Complexes at Euchromatin, Heterochromatin and Centromeric Chromatin. Int. J. Mol. Sci. 2021, 22, 6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Allshire RC; Madhani HD Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 2018, 19, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Bannister AJ; Kouzarides T Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Belmont AS Nuclear Compartments: An Incomplete Primer to Nuclear Compartments, Bodies, and Genome Organization Relative to Nuclear Architecture. Cold Spring Harb. Perspect. Biol. 2021, a041268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Kamat K; Qi Y; Wang Y; Ma J; Zhang B Genome Compartmentalization with Nuclear Landmarks: Random yet Precise. bioRxiv 2021, [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Hildebrand EM; Dekker J Mechanisms and functions of chromosome compartmentalization. Trends Biochem. Sci. 2020, 45, 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Potoyan DA; Papoian GA Regulation of the H4 tail binding and folding landscapes via Lys-16 acetylation. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 17857–17862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Parsons T; Zhang B Critical role of histone tail entropy in nucleosome unwinding. J. Chem. Phys. 2019, 150, 185103. [DOI] [PubMed] [Google Scholar]
- (36).Collepardo-Guevara R; Portella G; Vendruscolo M; Frenkel D; Schlick T; Orozco M Chromatin unfolding by epigenetic modifications explained by dramatic impairment of internucleosome interactions: A multiscale computational study. J. Am. Chem. Soc. 2015, 137, 10205–10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Arya G; Schlick T Role of histone tails in chromatin folding revealed by a mesoscopic oligonucleosome model. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 16236–16241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Kouzarides T Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 2002, 12, 198–209. [DOI] [PubMed] [Google Scholar]
- (39).Li S; Peng Y; Landsman D; Panchenko AR DNA methylation cues in nucleosome geometry, stability and unwrapping. Nucleic Acids Res. 2022, 50, 1864–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Larson AG; Elnatan D; Keenen MM; Trnka MJ; Johnston JB; Burlingame AL; Agard DA; Redding S; Narlikar GJ Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Strom AR; Emelyanov AV; Mir M; Fyodorov DV; Darzacq X; Karpen GH Phase separation drives heterochromatin domain formation. Nature 2017, 547, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Latham AP; Zhang B Consistent Force Field Captures Homologue-Resolved HP1 Phase Separation. J. Chem. Theory Comput. 2021, 17, 3134–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Sabari BR; Dall’Agnese A; Young RA Biomolecular Condensates in the Nucleus. Trends Biochem. Sci. 2020, 45, 961–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Gibson BA; Doolittle LK; Schneider MW; Jensen LE; Gamarra N; Henry L; Gerlich DW; Redding S; Rosen MK Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 2019, 179, 470–484.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Strom AR; Brangwynne CP The liquid nucleome - phase transitions in the nucleus at a glance. J. Cell Sci. 2019, 132, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Xie L; Dong P; Qi Y; Hsieh T-HS; English BP; Jung S; Chen X; De Marzio M; Casellas R; Chang HY et al. BRD2 compartmentalizes the accessible genome. Nature Genetics 2022, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Ganai N; Sengupta S; Menon GI Chromosome positioning from activity-based segregation. Nucleic Acids Res. 2014, 42, 4145–4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Grosberg AY; Joanny JF Nonequilibrium statistical mechanics of mixtures of particles in contact with different thermostats. Phys. Rev. E 2015, 92, 1–10. [DOI] [PubMed] [Google Scholar]
- (49).Binder K; Virnau P Phase transitions and phase coexistence: equilibrium systems versus externally driven or active systems - Some perspectives. Soft Materials 2021, 19, 267–285. [Google Scholar]
- (50).van Steensel B; Furlong EE The role of transcription in shaping the spatial organization of the genome. Nat. Rev. Mol. Cell Biol. 2019, 20, 327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Struhl K; Segal E Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 2013, 20, 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Agrawal A; Ganai N; Sengupta S; Menon GI Nonequilibrium Biophysical Processes Influence the Large-Scale Architecture of the Cell Nucleus. Biophys. J. 2020, 118, 2229–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Jiang Z; Zhang B Theory of Active Chromatin Remodeling. Phys. Rev. Lett. 2019, 123, 208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Jiang H; Dou N; Fan G; Yang Z; Zhang X Effect of temperature gradient on liquid-liquid phase separation in a polyolefin blend. J. Chem. Phys 2013, 139, 124903. [DOI] [PubMed] [Google Scholar]
- (55).Su JH; Zheng P; Kinrot SS; Bintu B; Zhuang X Genome-Scale Imaging of the 3D Organization and Transcriptional Activity of Chromatin. Cell 2020, 182, 1641–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Thompson AP; Aktulga HM; Berger R; Bolintineanu DS; Brown WM; Crozier PS; in ‘t Veld PJ; Kohlmeyer A; Moore SG; Nguyen TD et al. LAMMPS - a flexible simulation tool for particle-based materials modeling at the atomic, meso, and continuum scales. Comp. Phys. Comm. 2022, 271, 108171. [Google Scholar]
- (57).Leimkuhler B; Matthews C Rational construction of stochastic numerical methods for molecular sampling. Appl. Math. Res. Express 2013, 2013, 34–56. [Google Scholar]
- (58).Chen B; Gilbert L; Cimini B; Schnitzbauer J; Zhang W; Li G-W; Park J; Blackburn E; Weissman J; Qi L et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell 2013, 155, 1479–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Qin P; Parlak M; Kuscu C; Bandaria J; Mir M; Szlachta K; Singh R; Darzacq X; Yildiz A; Adli M ARTICLE Live cell imaging of low-and non-repetitive chromosome loci using CRISPR-Cas9. Nature Communications 2017, [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Chubb JR; Boyle S; Perry P; Bickmore WA Chromatin Motion Is Constrained by Association with Nuclear Compartments in Human Cells. Current Biology 2002, 12, 439–445. [DOI] [PubMed] [Google Scholar]
- (61).Sanborn AL; Rao SSP; Huang S-CC; Durand NC; Huntley MH; Jewett AI; Bochkov ID; Chinnappan D; Cutkosky A; Li J et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, E6456–6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Fudenberg G; Imakaev M; Lu C; Goloborodko A; Abdennur N; Mirny LALA Formation of Chromosomal Domains by Loop Extrusion. Cell Rep 2016, 15, 2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Rao SS; Huntley MH; Durand NC; Stamenova EK; Bochkov ID; Robinson JT; Sanborn AL; Machol I; Omer AD; Lander ES et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014, 159, 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Qi Y; Zhang B Predicting three-dimensional genome organization with chromatin states. PLOS Comput. Biol. 2019, 15, e1007024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Brahmachari S; Contessoto VG; Di Pierro M; Onuchic JN Shaping the genome via lengthwise compaction, phase separation, and lamina adhesion. Nucleic Acids Res. 2022, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Latham APA; Zhang B Improving Coarse-Grained Protein Force Fields with Small-Angle X-ray Scattering Data. J. Phys. Chem. B 2019, 123, 1026–1034. [DOI] [PubMed] [Google Scholar]
- (67).Chu X; Wang J PLoS Comput. Biol; 2021; Vol. 17; pp 1–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Shi G; Thirumalai D From Hi-C Contact Map to Three-Dimensional Organization of Interphase Human Chromosomes. Phys. Rev. X 2021, 11, 11051. [Google Scholar]
- (69).Pope BD; Ryba T; Dileep V; Yue F; Wu W; Denas O; Vera DL; Wang Y; Hansen RS; Canfield TK et al. Topologically associating domains are stable units of replication-timing regulation. Nature 2014, 515, 402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Dekker J; Heard E Structural and functional diversity of topologically associating domains. FEBS letters 2015, 589, 2877–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Chiang M; Michieletto D; Brackley CA; Rattanavirotkul N; Mohammed H; Marenduzzo D; Chandra T Polymer Modeling Predicts Chromosome Reorganization in Senescence. Cell Rep. 2019, 28, 3212–3223.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Amiad-Pavlov D; Lorber D; Bajpai G; Reuveny A; Roncato F; Alon R; Safran S; Volk T Live imaging of chromatin distribution reveals novel principles of nuclear architecture and chromatin compartmentalization. Sci. Adv. 2021, 7, eabf6251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Qi Y; Zhang B Chromatin network retards nucleoli coalescence. Nat Commun 2021, 12, 6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Jiang Z; Zhang B On the role of transcription in positioning nucleosomes. PLOS Comput. Biol. 2021, 17, e1008556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Fisher ME; Kolomeisky AB The force exerted by a molecular motor. Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 6597–6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 2013, 154, 490–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Shin S; Cho HW; Shi G; Thirumalai D Transcription-induced active forces suppress chromatin motion by inducing a transient disorder-to-order transition. bioxiv 2022, [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Bruinsma R; Grosberg AY; Rabin Y; Zidovska A Chromatin hydrodynamics. Biophys. J. 2014, 106, 1871–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Smrek J; Kremer K Small Activity Differences Drive Phase Separation in Active-Passive Polymer Mixtures. Phys. Rev. Lett. 2017, 118, 1–5. [DOI] [PubMed] [Google Scholar]
- (80).Liu L; Shi G; Thirumalai D; Hyeon C Chain organization of human interphase chromosome determines the spatiotemporal dynamics of chromatin loci. PLOS Comput. Biol. 2018, 14, e1006617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Lodish H; Lodish UH; Berk A; Fund RECM; Matsudaira UP; Kaiser CA; Matsudaira P; Kaiser UCA; Krieger M; Darnell J et al. Molecular Cell Biology; W. H. Freeman, 2004. [Google Scholar]
- (82).Briand N; Collas P Lamina-associated domains: Peripheral matters and internal affairs. Genome Biol. 2020, 21, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Hansen JC; Maeshima K; Hendzel MJ The solid and liquid states of chromatin. Epigenetics Chromatin 2021, 14, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Liu S; Lin X; Zhang B Chromatin fiber breaks into clutches under tension and crowding. bioRxiv 2021, [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Plys AJ; Davis CP; Kim J; Rizki G; Keenen MM; Marr SK; Kingston RE Phase separation of polycomb-repressive complex 1 is governed by a charged disordered region of CBX2. Genes Dev. 2019, 33, 799–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Lin X; Leicher R; Liu S; Zhang B Cooperative DNA looping by PRC2 complexes. Nucleic Acids Res. 2021, 49, 6238–6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Kent S; Brown K; hsun Yang C; Alsaihati N; Tian C; Wang H; Ren X Phase-Separated Transcriptional Condensates Accelerate Target-Search Process Revealed by Live-Cell Single-Molecule Imaging. Cell Rep. 2020, 33, 108248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Falk M; Feodorova Y; Naumova N; Imakaev M; Lajoie BR; Leonhardt H; Joffe B; Dekker J; Fudenberg G; Solovei I et al. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 2019, 570, 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Gnesotto FS; Mura F; Gladrow J; Broedersz CP Broken detailed balance and non-equilibrium dynamics in living systems: a review. Reports Prog. Phys. 2018, 81, 066601. [DOI] [PubMed] [Google Scholar]
- (90).Saintillan D; Shelley MJ; Zidovska A Extensile motor activity drives coherent motions in a model of interphase chromatin. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, 11442–11447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Zidovska A; Weitz DA; Mitchison TJ Micron-scale coherence in interphase chromatin dynamics. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 15555–15560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Shaban HA; Barth R; Bystricky K Formation of correlated chromatin domains at nanoscale dynamic resolution during transcription. Nucleic Acids Res. 2018, 46, e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Barth R; Bystricky K; Shaban HA Coupling chromatin structure and dynamics by live super-resolution imaging. Sci. Adv. 2020, 6, eaaz2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Put S; Sakaue T; Vanderzande C Active dynamics and spatially coherent motion in chromosomes subject to enzymatic force dipoles. Phys. Rev. E 2019, 99, 5–10. [DOI] [PubMed] [Google Scholar]
- (95).Pierro MD; Potoyan DA; Wolynes PG; Onuchic JN Anomalous diffusion, spatial coherence, and viscoelasticity from the energy landscape of human chromosomes. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, 7753–7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Deindl S; Hwang WL; Hota SK; Blosser TR; Prasad P; Bartholomew B; Zhuang X ISWI remodelers slide nucleosomes with coordinated multi-base-pair entry steps and single-base-pair exit steps. Cell 2013, 152, 442–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Cugliandolo LF; Kurchan J; Peliti L Energy flow, partial equilibration, and effective temperatures in systems with slow dynamics. Phys. Rev. E 1997, 55, 3898–3914. [Google Scholar]
- (98).Berthier L; Barrat JL Nonequilibrium dynamics and fluctuation-dissipation relation in a sheared fluid. J. Chem. Phys. 2002, 116, 6228–6242. [DOI] [PubMed] [Google Scholar]
- (99).Berthier L; Barrat JL Shearing a Glassy Material: Numerical Tests of Nonequilibrium Mode-Coupling Approaches and Experimental Proposals. Phys. Rev. Lett. 2002, 89, 1–4. [DOI] [PubMed] [Google Scholar]
- (100).Loi D; Mossa S; Cugliandolo LF Non-conservative forces and effective temperatures in active polymers. Soft Matter 2011, 7, 10193–10209. [Google Scholar]
- (101).Barrat JL; Berthier L Fluctuation-dissipation relation in a sheared fluid. Phys. Rev. E 2001, 63, 0125031–0125034. [DOI] [PubMed] [Google Scholar]
- (102).Wang S; Wolynes PG Communication: Effective temperature and glassy dynamics of active matter. J. Chem. Phys 2011, 135, 051101. [DOI] [PubMed] [Google Scholar]
- (103).Stenhammar J; Wittkowski R; Marenduzzo D; Cates ME Activity-Induced Phase Separation and Self-Assembly in Mixtures of Active and Passive Particles. Phys. Rev. Lett. 2015, 114, 018301. [DOI] [PubMed] [Google Scholar]
- (104).Weber SN; Weber CA; Frey E Binary Mixtures of Particles with Different Diffusivities Demix. Phys. Rev. Lett. 2016, 116, 1–5. [DOI] [PubMed] [Google Scholar]
- (105).Morozov KI; Pismen LM Motor-driven effective temperature and viscoelastic response of active matter. Phys. Rev. E 2010, 81, 1–8. [DOI] [PubMed] [Google Scholar]
- (106).Joo S; Durang X; Lee O.-c.; Jeon J-H Anomalous diffusion for active Brownian particles cross-linked to a networked polymer: Langevin dynamics simulation and theory. Soft Matter 2020, [DOI] [PubMed] [Google Scholar]
- (107).Chari SSN; Dasgupta C; Maiti PK Scalar activity induced phase separation and liquid-solid transition in a Lennard-Jones system. Soft Matter 2019, 15, 7275–7285. [DOI] [PubMed] [Google Scholar]
- (108).Amendola M; Steensel B Nuclear lamins are not required for lamina-associated domain organization in mouse embryonic stem cells. EMBO Rep. 2015, 16, 610–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).Zheng X; Hu J; Yue S; Kristiani L; Kim M; Sauria M; Taylor J; Kim Y; Zheng Y Lamins Organize the Global Three-Dimensional Genome from the Nuclear Periphery. Mol. Cell 2018, 71, 802–815.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (110).Manzo SG; Dauban L; van Steensel B Lamina-associated domains: Tethers and looseners. Curr. Opin. Cell Biol. 2022, 74, 80–87. [DOI] [PubMed] [Google Scholar]
- (111).Ugarte F; Sousae R; Cinquin B; Martin EW; Krietsch J; Sanchez G; Inman M; Tsang H; Warr M; Passegué E et al. Progressive chromatin condensation and H3K9 methylation regulate the differentiation of embryonic and hematopoietic stem cells. Stem Cell Rep. 2015, 5, 728–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Johnstone SE; Reyes A; Qi Y; Adriaens C; Hegazi E; Pelka K; Chen JH; Zou LS; Drier Y; Hecht V et al. Large-Scale Topological Changes Restrain Malignant Progression in Colorectal Cancer. Cell 2020, 182, 1474–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (113).Chang L; Li M; Shao S; Li C; Ai S; Xue B; Hou Y; Zhang Y; Li R; Fan X et al. Nuclear peripheral chromatin-lamin B1 interaction is required for global integrity of chromatin architecture and dynamics in human cells. Protein Cell 2022, 13, 258–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Liu K; Patteson AE; Banigan EJ; Schwarz JM Dynamic Nuclear Structure Emerges from Chromatin Cross-Links and Motors. Phys. Rev. Lett. 2021, 126, 158101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


