Abstract
The incidence of melioidosis cases caused by the gram-negative pathogen Burkholderia pseudomallei (BP) is seeing an increasing trend that has spread beyond its previously known endemic regions. Biofilms produced by BP have been associated with antimicrobial therapy limitation and relapse melioidosis, thus making it urgently necessary to understand the mechanisms of biofilm formation and their role in BP biology. Microbial cells aggregate and enclose within a self-produced matrix of extracellular polymeric substances (EPSs) to form biofilm. The transition mechanism of bacterial cells from planktonic state to initiate biofilm formation, which involves the formation of surface attachment microcolonies and the maturation of the biofilm matrix, is a dynamic and complex process. Despite the emerging findings on the biofilm formation process, systemic knowledge on the molecular mechanisms of biofilm formation in BP remains fractured. This review provides insights into the signaling systems, matrix composition, and the biosynthesis regulation of EPSs (exopolysaccharide, eDNA and proteins) that facilitate the formation of biofilms in order to present an overview of our current knowledge and the questions that remain regarding BP biofilms.
Keywords: Burkholderia pseudomallei, biofilm, exopolysaccharide, eDNA, cyclic-di-GMP, quorum sensing
Introduction
Burkholderia pseudomallei (BP) is a gram-negative, environmental saprophyte predominately found in the soil and surface groundwater of endemic tropical and subtropical regions worldwide [1, 2]. BP is the etiological agent of melioidosis, a life-threatening disease that accounts for approximately 89,000 deaths per year worldwide [2-6]. Diabetes mellitus remains a major risk factor for melioidosis; therefore, the rising global diabetes pandemic could further escalate the number of deaths attributed to melioidosis [2]. The virulence factors of BP include lipopolysaccharide (LPS), flagella, capsule, and type III secretory systems (TTSS), which have been identified to be involved in acute septicaemia and chronic melioidosis [1, 7]. These virulence factors enhance bacterial survival and persistence across a wide range of hosts and facilitate evasion of the host’s immune response [1, 7]. In addition, BP isolates that can form biofilm have been associated with the relapse of melioidosis, and BP within the biofilm community is more resistant to antibiotics [8].
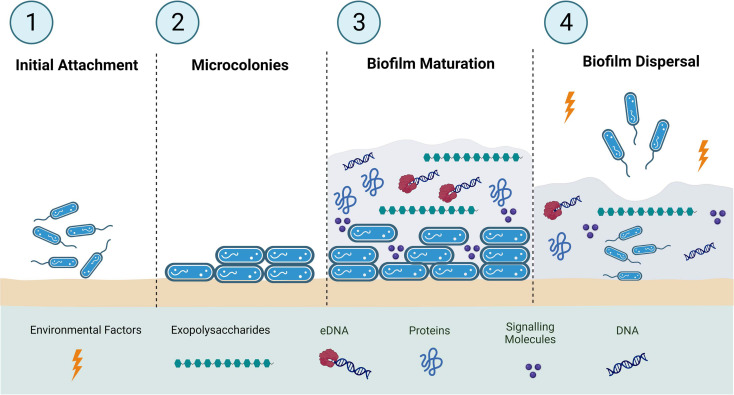
Many bacterial pathogens are known to form biofilm, which encloses the bacteria and facilitates cellular attachment and interaction [9]. In addition, the biofilm also renders the pathogen more tolerant to antibacterial agents and host immune molecules while aiding bacterial survival under nutrient-deficient conditions [10, 11]. Bacterial biofilms are composed of an aggregation of microbial cells on biotic or abiotic surfaces enclosed by a self-produced matrix of extracellular polymeric substances (EPSs) composed of proteins, polysaccharides, nucleic acids (DNA), lipopolysaccharides (LPS), and water [10-14]. A successful biofilm formation involves four main stages: (i) surface bacterial attachment, (ii) microcolony formation, (iii) maturation of biofilm architecture, and (iv) signals and environmental cues that trigger the dispersion of cells into the planktonic state [15-17]. The common biofilm formation processes in microbes are illustrated in Fig. 1.
Fig. 1. Schematic diagram representing four stages of biofilm formation (1) surface bacterial attachment, (2) microcolony formation, (3) maturation of biofilm architecture, and (4) dispersion of cells into the planktonic state (adapted from [15-17]).
In BP, biofilm formation is closely associated with its ability to adapt or survive in various environmental niches, as well as contributing to the bacteria’s pathogenicity [18, 19]. The emergence of resistance to antibiotics, including ceftazidime (CTZ), doxycycline (DOX), and imipenem, which are common drug treatments against melioidosis, is generally attributed to the presence of biofilm surrounding BP [20-22]. Despite the importance of biofilm formation features in BP that are linked to clinical pathogenicity and virulence, the detailed mechanism of biofilm formation in BP is yet to be elucidated. For the past decade, researchers have utilized genetics and ‘omics’ approaches targeting the biofilm biosynthesis pathway of BP in studies that have successfully identified genes and proteins that are crucial for BP biofilm formation. In addition, studies on other Burkholderia species that share high genome sequence similarity to BP have provided indirect evidence that further helps elucidate BP biofilm formation. Through this review, we aimed to provide current insights into EPS and biofilm formation in BP and highlight the potential genes, proteins, and pathways that warrant further investigation to develop effective therapeutics or successful vaccine candidates to treat melioidosis.
Signaling Systems That Promote BP Biofilm Formation
Environmental factors are known to trigger the formation or dispersal of biofilm in most bacteria [23, 24]. Environmental cues such as temperature, pH, nutrient deficiency, and glucose were reported to influence biofilm formation by BP [25-27]. These ecological factors are sensed by signaling molecules, which can influence gene expression in support of biofilm formation and facilitate the conversion of free-living planktonic cells into biofilm cells [25, 28]. Cyclic di-GMP (c-di-GMP), quorum-sensing (QS) molecules, and small RNAs (sRNAs) are known to be the major signaling molecules present in the biofilm community [29-31]. c-di-GMP signaling occurs during the early stages of biofilm formation to facilitate the conversion of free-living planktonic cells to biofilm cells, while QS signaling is involved during biofilm maturation and dispersion [29, 32-34]. sRNAs serve as regulatory molecules in several bacterial metabolic processes, including Burkholderia species for biofilm development [31, 35]; nonetheless, biofilm-associated sRNAs in BP have yet to be identified.
Cyclic-di-GMP Signaling
C-di-GMP is a bacterial universal intracellular secondary signaling molecule [36-38]. In bacterial biofilm formation, c-di-GMP is known to regulate genes responsible for synthesizing EPS components; extracellular polymeric exoenzymes, polysaccharides, and adhesins [39, 40]. In addition, c-di-GMP enhances bacterial adhesion and represses bacterial motility, further promoting biofilm production [32, 33, 41, 42]. Furthermore, depletion of c-di-GMP levels has been reported to trigger the dispersal of biofilms. For instance, inhibition of the final step of the denitrification pathway has been implicated in inducing biofilm dispersal [43]. Nitrate levels have been reported to significantly affect biofilm formation in BP, as they ultimately determine the fate of c-di-GMP [33]. The denitrification process, which involves the reduction of nitrate to nitrogen, is important in BP biofilms as it provides an alternative energy source under oxygen-limited conditions [33, 44]. The impact of inhibiting the denitrification pathway on biofilms was recently evaluated in B. thailandensis, a species closely related to BP [43]. Inhibiting the final step of denitrifying nitrous oxide to nitrogen catalyzed by nitrous oxide reductase leads to the accumulation of nitrous oxide, which in turn reduces c-di-GMP levels that ultimately trigger the dispersal process [43, 45]. As for BP, a recent transcriptome analysis between high and low BP biofilm-forming isolates revealed the overexpression of nitrous oxide reductase, bpsl1607, in the high biofilm-forming isolate. Furthermore, studies on BP 1026b isolate mutants involving a two-component, nitrate-sensing system in the form of narX-narL (equivalent to bpsl2313-bpsl2314) have further confirmed the regulation of the denitrification pathway in c-di-GMP production and biofilm formation [44].
The synthesis and breakdown of c-di-GMP in most bacteria are regulated by diguanylate cyclase (DGC) and phosphodiesterase (PDE), respectively. The activity of both proteins is affected by environmental cues, in agreement with the transition of bacteria from planktonic to biofilm state being regulated by c-di-GMP in response to changes in environmental stimuli [46-48]. DGC contains the conserved GGDEF domain, while PDE contains a conserved EAL or HD-GYP domain [11, 49]. DGC catalyzes the synthesis of c-di-GMP from the condensation of two GTP molecules, while PDE catalyzes the hydrolysis of c-di-GMP, resulting in two GMP molecules [50, 51]. Burkholderia cenocepacia is another Burkholderiaceae family member and closely related species to BP, and a number of genes encoding proteins that are homologous across the Burkholderia group responsible for the synthesis of c-di-GMP in B. cenocepacia have been identified and tabulated (Table 1) [51].
Table 1.
Proteins of B. pseudomallei that are involved in signaling system in the regulation of biofilms.
| Signaling Molecules | Annotation/Description | Species/isolate | Sequence identity to K96243 (% identity) | Burkholderia pseudomallei K96243 identifier code | Protein Description for Burkholderia pseudomallei K96243 | Reference |
|---|---|---|---|---|---|---|
| c-di-GMP | Bp1026b_II2523 DGC | Burkholderia pseudomallei 1026b | 99.89 | BPSS2342 | Hypothetical protein | [41,54] |
| Bp1026b_I2235 GGDEF domain | Burkholderia pseudomallei 1026b | 99.85 | BPSL1306 | Hypothetical protein | [41,54] | |
| Bp1026b_II0153 GGDEF domain | Burkholderia pseudomallei 1026b | 99.93 | BPSS0136 | Hypothetical protein | [41,54] | |
| Bp1026b_II1380 GGDEF domain | Burkholderia pseudomallei 1026b | 99.73 | BPSS1297 | Regulatory protein | [41,54] | |
| Bp1026b_II2115 GGDEF domain | Burkholderia pseudomallei 1026b | 99.87 | BPSS1971 | Two-component system fusion protein | [41,54] | |
| Bcam2836 putative DGC | Burkholderia cenocepacia J2315 | 85.70 | BPSS2342 | Hypothetical protein | [51,54] | |
| BTH_II2363 (pdcA) GGDEF domain | Burkholderia thailandensis E264 | 97.43 | BPSS2342 | Hypothetical protein | [53,54] | |
| BTH_II2364 (pdcB) CheC/CheX domain | Burkholderia thailandensis E264 | 98.52 | BPSS2343 | Hypothetical protein | [53,54] | |
| BTH_II2365 (pdcC) phosphate-accepting response regulator | Burkholderia thailandensis E264 | 96.72 | BPSS2344 | Hypothetical protein | [53,54] | |
| Bp1026b_I0571 EAL domain | Burkholderia pseudomallei 1026b | 99.88 | BPSL2744 | Hypothetical protein | [41,54] | |
| Bp1026b_I1579 EAL domain | Burkholderia pseudomallei 1026b | 100 | BPSL1635 | Hypothetical protein | [41,54] | |
| Bp1026b_I2260 EAL domain | Burkholderia pseudomallei 1026b | 99.38 | BPSL1286 | Hypothetical protein | [41,54] | |
| Bp1026b_I2659 EAL domain | Burkholderia pseudomallei 1026b | 99.53 | BPSL0887 | Hypothetical protein | [41,54] | |
| Bp1026b_I3148 EAL domain | Burkholderia pseudomallei 1026b | 99.84 | BPSL0358 | Hypothetical protein | [41,54] | |
| Bp1026b_II0879 EAL domain | Burkholderia pseudomallei 1026b | 99.48 | BPSS0799 | Hypothetical protein | [41,54] | |
| BCAL0652 EAL domain | Burkholderia cenocepacia J2315 | 30.17 | BPSL2744 | Hypothetical protein | [51,54] | |
| Bp1026b_I2284 (CdpA) GGDEF/EAL domain | Burkholderia pseudomallei 1026b | 99.95 | BPSL1263 | Hypothetical protein | [41,42,54] | |
| BCAL1069 (cdpA) GGDEF/EAL domain | Burkholderia cenocepacia J2315 | 85.52 | BPSL1263 | Hypothetical protein | [51,54,135] | |
| Bp1026b_I2456 GGDEF/EAL domain | Burkholderia pseudomallei 1026b | 99.79 | BPSL1080 | Hypothetical protein | [41,54] | |
| Bp1026b_I2928 GGDEF/EAL domain | Burkholderia pseudomallei 1026b | 99.40 | BPSL0602 | Hypothetical protein | [41,54] | |
| Bp1026b_II0885 GGDEF/EAL domain | Burkholderia pseudomallei 1026b | 99.71 | BPSS0805 | Hypothetical protein | [41,54] | |
| Bp1026b_II2498 GGDEF/EAL domain | Burkholderia pseudomallei 1026b | 99.83 | BPSS2318 | Hypothetical protein | [41,54] | |
| Bcam1160 putative c-di-GMP | Burkholderia cenocepacia | 86.75 | BPSL1080 | Hypothetical protein | [51,54] | |
| Bcam1349 CRP/FNR family transcriptional regulator | Burkholderia cenocepacia J2315 | 79.07 | BPSL0617 | Hypothetical protein | [45,54,96,98] | |
| CRP/FNR superfamily | Burkholderia pseudomallei K96243 | NA | BPSL0616 | Hypothetical Protein | [45] | |
| QS | BpsI autoinducer synthase | Burkholderia pseudomallei K96243, KHW, H11 | 100 | BPSS0885 BPSS1570 |
N-acyl-homoserine lactone synthase | [54,66] |
| BpsR autoinducer binding transcriptional regulator | Burkholderia pseudomallei K96243, KHW, H11 | 99.86 | BPSS0887 | N-acyl-homoserine lactone dependent regulatory protein | [54,66] | |
| PA0996 (pqsA) | Pseudomonas aeruginosa PAO1 | 30.36 | BPSS0481 | HhqA | [54,73-75,136] | |
| PA0997 (pqsB) | Pseudomonas aeruginosa PAO1 | 38.32 | BPSS0482 | HhqB | [54,73-75,136] | |
| PA0998 (pqsC) | Pseudomonas aeruginosa PAO1 | 38.59 | BPSS0483 | HhqC | [54,73-75,136] | |
| PA0999 (pqsD) | Pseudomonas aeruginosa PAO1 | 53.68 | BPSS0484 | HhqD | [54,73-75,136] | |
| PA1000 (pqsE) | Pseudomonas aeruginosa PAO1 | 30.36 | BPSS0485 | HhqE | [54,73-75,136] |
*NA- Not applicable
In BP, a putative DGC (bpss2342 or Bp1026b_II2523) that contains a conserved GGDEF domain was reported to influence the biofilm formation in a temperature-dependent manner in which increased biofilm formation was observed among mutant colonies grown at 37°C compared to 30°C [41, 52]. This observation highlights the correlation between c-di-GMP synthesis and environmental factors regulating BP's biofilm formation. Furthermore, the Bp1026b_II2523 (bpss2342) mutant was shown to affect various biological systems such as polysaccharide biosynthesis and several secretion systems (T3SS-3, T3SS-2, T6SS-3, and T6SS-6) and biosynthetic gene clusters (BGCs) that are involved in non-ribosomal peptide and polyketide synthesis and, predicted to encode small metabolites contributing to biofilm development [52]. Apart from that, the cdpA gene in BP KHW (corresponding to bpsl1263 in BP K96243) encoding phosphodiesterase proteins that contains a conserved EAL has been identified as PDE [42]. The cdpA deletion mutant was shown to exhibit high levels of c-di-GMP which favor biofilm production through increased exopolysaccharide and cellular aggregation [42]. Furthermore, higher expression of the cdpA gene was observed for BP exposed to exogenous sodium nitrate. This led to the upregulation of PDE activity which contributed to reduced c-di-GMP levels and, subsequently, poor biofilm formation [33]. Recently, a c-di-GMP signaling cascade mediated by a pdcABC operon that can regulate virulence, motility, and biofilm formation was reported for B. thailandensis [53]. pdcA encodes a DGC protein that produces c-di-GMP and is regulated by PdcC (phosphate-accepting response regulator). The phosphorylated PdcC inhibits PdcA by binding to its PAS domain. PdcB is a phosphatase that increases the activity of PdcA through dephosphorylation of PdcC [53]. Interestingly, an operon in BP shares high sequence identity with pdcABC (Table 1), suggesting that BP may share a similar pathway in modulating c-di-GMP levels.
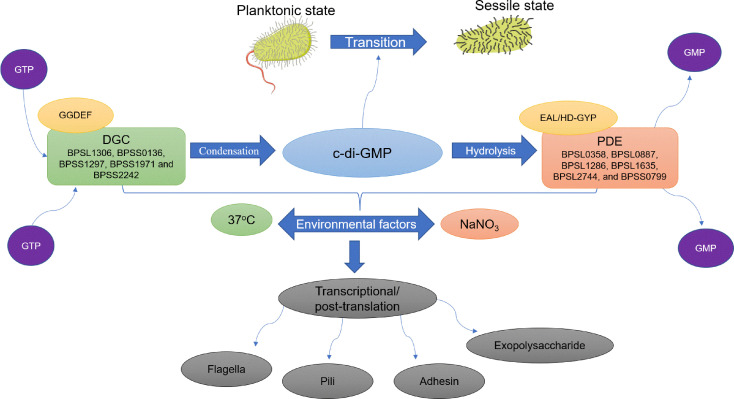
Several genes encoding proteins that contain the conserved GGDEF and EAL domains have been annotated in the BP genome (https://www.burkholderia.com/) and Plumley et al. [41, 54]. The proteins BPSL1306, BPSS0136, BPSS1297, BPSS1971, and BPSS2342 were predicted to carry the GGDEF domain with high sequence identity (Supplementary Fig. S1) while BPSL0358, BPSL0887, BPSL1286, BPSL1635, BPSL2744, and BPSS0799 have the EAL domain (Supplementary Fig. S2). Meanwhile, five other proteins (BPSL0602, BPSL1080, BPSL1263, BPSS0805, and BPSS2318) contain both the conserved GGDEF/EAL domains (Table 1) [41, 54]. By modulating the level of c-di-GMP, the GGDEF and EAL domains were reported to exert control at the transcriptional and post-translation level in regulating the expression of cell surface components (e.g., flagella, adhesins, pili, exopolysaccharides) (Fig. 2) [41]. Nitrate level and temperature have been proven to affect the DGC gene Bp1026b_II2523 (bpss2342) and cdpA, respectively. Nonetheless, the function of other predicted genes that encode DGC and PDE enzymes containing GGDEF/EAL domains in BP needs to be further characterized in terms of their correlation to other specific environmental cues. It is known that proteins containing GGDEF/EAL domain generally assemble an N-terminal sensory domain which may respond to specific environmental stimuli (oxygen, light, nitric oxide, etc.) in regulating the enzyme activity that may determine the production level of c-di-GMP [55-57]. Based on currently available reports, we proposed the c-di-GMP synthesis mechanism and its functional properties during BP biofilm formation and the factors that may influence the biosynthesis of c-di-GMP in enhancing the transition from free-living planktonic cells to sessile cells, as illustrated in Fig. 2.
Fig. 2. C-di-GMP synthesis mechanism and functional properties during BP biofilm formation.
The synthesis and breakdown of cyclic-di-GMP (c-di-GMP) are regulated by two enzymes, diguanylate cyclase (DGC) and phosphodiesterase (PDE), each containing a conserved GGDEF or EAL/HD-GYP domain respectively. Two guanosine-5’-triphosphate (GTP) molecules are utilized by DGC during the condensation reaction that results in the formation of c-di-GMP, which favors biofilm formation by enhancing the transition from free-living planktonic cells to sessile cells. PDE catalyzes the hydrolysis of c-di-GMP into two guanosine monophosphate (GMP) molecules. Both enzymes are influenced by environmental signals such as temperature and concentration of sodium nitrate (NaNO3) that ultimately determine the level of c-di-GMP. The phenotypic characteristics of the cells such as the presence of flagella, pili, adhesin, and exopolysaccharide may be regulated by these enzymes at the transcriptional and post-translation levels through determining the level of c-di-GMP [41].
Quorum Sensing (QS) Signaling
Quorum sensing is also a crucial signaling system involved in forming biofilms. Autoinducers produced by bacteria serve as chemical signal molecules and are released according to cell density [59, 60]. QS is utilized by both gram-positive and gram-negative bacteria [60]. In most Burkholderia spp., inhibition of this signaling system negatively affects biofilm formation, making the QS signaling system a suitable target for antimicrobials or anti-biofilm agents [61, 62].
N-acyl-homoserine lactones (AHLs) are the most common QS signaling molecule utilized by most gram-negative bacteria, including BP [63]. AHL signaling molecules are encoded by a class of genes that are homologous to the luxI and luxR of Vibrio fischeri and have been reported to mediate QS systems [65, 66]. luxI encodes AHL synthetases that are required for the synthesis of related signaling molecules, while LuxR family proteins serve as AHL molecular receptors [66, 67]. AHL autoinducers interact with the LuxR proteins to regulate the expression of genes that control relevant biological phenotypes, including biofilm formation [66, 67]. Similar QS systems in Pseudomonas aeruginosa, namely lasIR and rhlRI, are homologous to the LuxI-LuxR [68, 69]. In BP, the BpsI-BpsR QS system was reported as a homolog of LuxI-LuxR [70] and positively regulates biofilm formation.
BP owns three QS systems that produce AHL molecules, namely QS-1 (encoded by BpsI-BpsR), QS-2 (BpsI2-BpsR2), and QS-3 (BpsI3-BpsR3), which produce three types of AHLs, N-octanoylhomoserine lactone (C8HL), N-(3-hydroxy-octanoyl) homoserine lactone (OHC8HL), and N-(3-hydroxy-decanoyl) homoserine lactone [66, 71]. C8HL is synthesized by N-acyl-homoserine lactone synthase, encoded by bpss0885 (pmlI), which is also known as BpsI [54, 66]. The remaining two AHLs are mainly produced by BpsI2 (encoded by BPSS1180 in BP K96243) and BpsI3 (BPSS1570), respectively, which are paralogs to BpsI [54, 66]. The expression of the three BpsI enzymes is regulated by their corresponding AHL- dependent transcription regulators BpsR, BpsR2, and BpsR3, respectively. BP strains lacking the BpsI-BpsR system cannot form biofilm [34, 66] while individual bpsR and bpsI mutants have impaired biofilm formation [66, 72]. Biofilm formation is restored in the presence of exogenous C8HL. In contrast, the addition of exogenous OHC8HL further suppresses biofilm formation in the mutant strains, indicating that exogenous OHC8HL serves as an antagonist in suppressing the biofilm formed by BP [66, 72]. Moreover, it was reported that BpsR2 is not involved in biofilm formation while BpsR3 plays a partial role. Unlike BpsR, exogenous OHC8HL was not able to resume full biofilm formation of BpsR3 mutant. Taken together, only dedicated QS signaling systems (QS-1 and QS-3) in BP were shown to be involved in biofilm formation, suggesting the specificity of AHL-signaling molecules in regulating the biofilm formation mechanism.
Apart from the AHL molecules, BP is known for producing another type of QS molecule known as 4-hydroxy-3-methyl-2-alkylquinolines (HMAQs), which are similar to the Pseudomonas quinolone signal (PQS), 4-hydroxy-2-alkylquinolines (HAQs) that are found in P. aeruginosa [73]. The PQS molecule is synthesized by the pqsABCDE operon (pa0996-pa1000) which is homologous to the hhqABCDE (bpss0481-bpss0485) genes in BP [73-75]. In P. aeruginosa, anthranilic acid is the precursor molecule for the synthesis of HAQs and is supplied by three different pathways that includes anthranilate synthase encoded by phnAB and trpEG and the degradation of tryptophan through the kynurenine pathway [75]. Similarly, BP produces anthranilic acid via the TrpEG and kynurenine pathway [75, 76]. In addition, inhibition of the kynurenine pathway was reported to increase the production of biofilm and reduce motility in BP [76], suggesting the involvement of HMAQs in biofilm formation and as a virulence factor of the bacterium.
In 2008, another quorum-sensing signal, cis-2-dodecenoic acid, also known as Burkholderia diffusible signal factor (BDSF), was reported in B. cenocepacia [77]. The BDSF QS system was reported to exert control towards AHL signaling and biofilm formation and affects the virulency of B. cenocepacia [78-81]. A rpfF gene that encodes RpfFBC enzyme was found to be responsible for the synthesis of the BDSF, and the production of BDSF is regulated by the RqpSR two-component system [77, 82]. A neighboring gene of rpfF, namely rpfR, is a gene encoding protein containing a PAS-GGDEF-EAL domain associated with c-di-GMP synthesis. The deletion of rpfR resulted in increased intracellular c-di-GMP [80]. A further study shows that RpfR is a QS signal receptor that can interact with BDSF and a c-di-GMP phosphodiesterase that interacts with RpfF to inhibit BDSF production [83]. Moreover, RpfR can also act as a c-di-GMP sensor by interacting with the global regulator GtrR [83]. Interestingly, while homologs of RpfR, RqpSR two-component systems and GtrR were identified in BP, no RpfFBC homologs could be detected [84]. However, there have yet to be any reports on RpfR, RqpSR two-component systems, and GtrR in BP. Therefore, it is unknown if the BDSF QS system that regulates c-di-GMP signaling exists in BP. Hence, further studies are warranted for a better understanding of the BDSF QS system and c-di-GMP in regulating the biofilm formation of BP.
Regulation by Small RNAs (sRNAs)
sRNAs modulate protein expression by altering mRNA translation rates or via mRNA degradation [85]. Common metabolic processes regulated by sRNAs include QS, carbon metabolism, and iron homeostasis [86]. These metabolic processes were observed in a recent study on B. cenocepacia J3215 biofilm [85]. In addition, functional characterization of B. cenocepacia J3215 sRNAs through comparison between sRNA mutant and wild-type strains revealed high growth, cellular aggregation, and metabolic activity (upregulation of the tryptophan and phenylacetic acid degradation pathways) among the mutant strains [87]. A recent whole genome-level transcriptome study on B. cenocepacia J2315 biofilm and planktonic states highlighted the abundance of sRNAs in the biofilm transcriptome compared to bacteria in the planktonic state [85], thus suggesting that sRNAs may play a crucial role in the development of a successful biofilm. Fifteen of the identified sRNAs were highly conserved across Burkholderia spp. [85]. Nonetheless, to date, no biofilm-associated sRNAs have been described for BP. Therefore, further investigation to identify the presence and involvement of sRNAs is required to reach a better understanding of biofilm formation.
Biofilm Composition in BP
The EPS matrix forms a natural protection shield for many bacteria, where it enables the bacteria that have changed from the planktonic stage growth mode to live in biofilm in response to various environmental cues and stresses. The formation and degradation of the EPS matrix in the biofilm life cycle are highly regulated and specific mechanisms are involved in the synthesis and degeneration of each of the EPS matrix components. Several major EPS matrix components in BP, including exopolysaccharides, eDNA, and proteins, have been identified. This section provides an overview of the three major EPS components of BP.
Exopolysaccharide Biosynthesis
Exopolysaccharides are a major component of most bacterial biofilm matrices [40, 88, 89]. The exopolysaccharides have been categorized into various forms, such as capsular polysaccharides, free polysaccharides, and lipopolysaccharides (O-antigen) that have a key role in preventing the diffusion of antimicrobial agents within the biofilm community [89-91]. The exopolysaccharide in BP has been structurally classified to be acidic. It consists of a tetrasaccharide repeating unit composed of three galactose (with one bearing a 2-linked O-acetyl group) and a 3-deoxy-D-manno-2-octulosonic acid (KDO) residues ([→3)-β-D-Galp2Ac-(1→4)-A-D-Galp-(1→3)-β-D-Galp-(1→5)-β-Kdo-(2→]n) [92]. Later, glucose, mannose, and rhamnose were reported as the major type of monosaccharides predominantly found in BP biofilm exopolysaccharides [93]. While the chemical synthesis of the tetrasaccharide repeating unit of [→3)-β-D-Galp2Ac-(1→4)-A-D-Galp-(1→3)-β-D-Galp-(1→5)-β-Kdo-(2→] has been successfully carried out [94], the BP proteins that are responsible for the biosynthesis of KDO molecules remains unclear. A 3-deoxy-D-manno-octulosonate 8-phosphate phosphatase encoded by yrbI (bpsl0537) and responsible for hydrolysis of Kdo 8-phosphate to Kdo was found located in the operon bpsl0534-bpsl0538 [54, 95]. In this operon, bpsl0534 and bpsl0536 encode the lipopolysaccharide export system ATP-binding proteins (ABC transporter), while bpsl0535 and bpsl0536 were annotated to encode an Ost-A-like protein and an arabinose-5-phosphate isomerase, respectively [54]. The involvement of the operon in the BP biofilm exopolysaccharide synthesis is yet to be investigated.
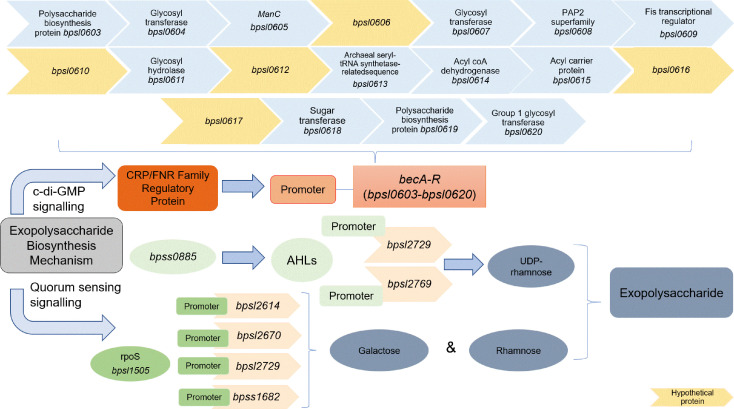
Recently, an exopolysaccharide gene cluster of 18 genes (becA-R) was identified. The becA-R is highly conserved within the Burkholderia spp. (B. pseudomallei, B. thailandensis, and B. mallei) [96]. The becA-R cluster encodes various enzymes such as glycosyl transferase, glycosyl hydrolase, capsular polysaccharide UDP-glucose lipid carrier transferase, and mannose-1-phosphate guanylyl transferase, which are required to synthesize exopolysaccharide components within the matrix [97, 98]. A transcriptome-level analysis of low and high BP biofilm producers revealed several genes within the becA-R gene cluster (bpsl0603, bpsl0605, bpsl0618, bpsl0619, and bpsl0620) were highly expressed in the high biofilm-producing strain [45]. Apart from the becA-R gene cluster, the wbiA gene cluster that consists of bpsl2670 and bpsl2671 was also identified to be involved in lipopolysaccharide biosynthesis. These genes encode UDP-glucose-4-epimerase and glycosyl transferase family protein, respectively [96]. Nonetheless, the detailed mechanism for exopolysaccharide biosynthesis has yet to be elucidated. Furthermore, several genes within the becA-R cluster encode hypothetical proteins, thus making elucidation of the exopolysaccharide synthesis mechanism more challenging.
Exopolysaccharide production in Burkholderia sp. biofilms is strongly influenced by c-di-GMP and QS signaling molecules [93, 97, 98]. The transcription regulation factors bpsI, ppk and rpoS were reported to influence the ratio of the monosaccharides glucose, galactose, mannose, and rhamnose of BP biofilm extracted exopolysaccharide [93]. In B. cenocepacia, c-di-GMP regulates exopolysaccharide biosynthesis at the post-translational level by promoting the binding between the CRP/FNR family transcriptional regulatory protein BCAM1349, (encoded by bcam1349) and the promoter region upstream of the becA-R gene cluster [97-99]. Two BP hypothetical proteins (BPSL0616 and BPSL0617) are reported to have a CRP/FNR superfamily domain, with BPSL0617 most likely an ortholog of BCAM1349 (Table 1) [45, 96].
Apart from c-di-GMP, N-acyl-homoserine lactone synthase BpsI (AHL synthase or C8HL, BPSS0885), the regulatory protein polyphosphate kinase (PPK, BPSL1366) and an alternative sigma factor S (RpoS, BPSL1505) are known to regulate exopolysaccharide production. Polyphosphate kinase is essential in producing inorganic polyphosphate from ATP which is required in the activation of sugar precursors for exopolysaccharide production [93]. A bpsl1366 mutant showed increased susceptibility to antibiotics due to poor development of the exopolysaccharide framework [93]. AHL synthase and RpoS are crucial in regulating the expression of enzymes involved in the exopolysaccharide biosynthesis pathway to enhance the survival of biofilm cells under adverse conditions [93]. For example, UTP glucose-1-phosphate uridylyltransferase (BPSL2769) and GDP-mannose-4,6-dehydratase (WcbK) enzymes are involved in synthesizing UDP-rhamnose. The genes of these proteins are predicted to have a lux box promoter region that responds to BPSS0885 [93]. rpoS regulates a series of enzymes encoded by genes with an RpoS-dependent promoter region, such as glucokinase (BPSL2614), UDP-glucose 4-epimerase (BPSL2670), WcbK (BPSL2729), and UDP-glucose-1-phosphate uridylyltransferase (BPSS1682). These enzymes are involved in converting several monosaccharides into galactose and rhamnose [93]. The conversion of glucose into glucose-6-phosphate catalyzed by glucokinase is the first step in biofilm exopolysaccharide synthesis; this highlights the importance of rpoS in regulating exopolysaccharide synthesis in BP biofilms [93]. Furthermore, monosaccharides, particularly rhamnose, contribute to a robust biofilm matrix that significantly limits the diffusion of antibiotics [93]. Therefore, overexpression of bpss0885, bpsl1505, and bpsl1366 accompanied by the accumulation of c-di-GMP, may lead to the formation of a rigid biofilm [93, 97]. The mechanisms of c-di-GMP, QS signaling, and RpoS involved in exopolysaccharide biosynthesis for BP biofilm formation are proposed and illustrated in Fig. 3.
Fig. 3. Proposed BP exopolysaccharide biosynthesis regulation mechanism via c-di-GMP and QS signaling.
Signaling molecules, e.g. c-di-GMP, and QS molecules, e.g., RpoS and AHLs, regulate the development of the EPS components, particularly exopolysaccharides. c-di-GMP is reported to improve the binding between the regulatory protein and the promoter region of the becA-R gene cluster thereby triggering gene expression of the cluster to produce the enzymes that facilitate the synthesis of exopolysaccharides in the EPS.
Extracellular DNA (eDNA) in EPS
Extracellular DNA (eDNA) is a crucial component of EPS and biofilm development [100-102]. eDNA is proposed as a key component of many pathogenic bacteria that form biofilms where it contributes to shielding biofilm against antimicrobial agents, promoting adhesion, and strengthening the integrity of biofilms [101, 103, 104]. In some bacteria, eDNA is derived from chromosomal DNA that is released from the bacterial cells either by active secretion mediated by QS or through cell lysis [105-107]. These mechanisms of eDNA release have been widely described for Staphylococcus epidermidis and P. aeruginosa biofilms [108, 109] but are yet to be characterized for BP. However, there is some indication that eDNA production in BP occurs through the extrusion of DNA from living cells, which is controlled by the transcriptional regulator BPSL1887 [110]. Interestingly, recent studies aiming to determine and quantify the components of BP and B. thailandensis biofilms revealed that eDNA and other major components in the biofilms are synthesized by living cells [111]. In addition, strains lacking capsular polysaccharides (CPS I) compensate by producing high levels of eDNA to complete biofilm formation with the abundant eDNA contributing to the thickness of the biofilm matrix [111]. BP bpsl1036 and bpsl1037 mutants that lack the two-component signal transduction systems (TCSTS) implicated in virulence and drug resistance have increased eDNA levels which ultimately promotes biofilm formation in BP [112].
It was reported that eDNA is actively involved during the early stages of biofilm formation, facilitating initial attachment and bacterial aggregation under the planktonic and biofilm states [113, 114]. Deoxyribonucleases (DNAses) are able to completely inhibit eDNA activity which is reflected by a reduced biofilm mass. However, inhibition of eDNA activity beyond the initial biofilm formation step shows no significant changes in biofilm mass, due to limited access of DNAse towards eDNA in mature biofilm. Therefore, DNAse treatment could be an appropriate treatment strategy targeting eDNA during the early stages of biofilm infections [113]. The ability of eDNA to defend the biofilm community against antimicrobial agents arises from its chemical properties. The negatively charged eDNA binds to the positively charged ions on antibiotics such as aminoglycosides and antimicrobial peptides, thereby reducing the antimicrobial agents’ efficiency in eliminating biofilm-forming pathogens [100, 115]. When BP biofilm was subjected to DNase treatment, a drastic reduction in biofilm mass was observed which could not be restored following supplementation with exogenous DNA [113]. A similar observation was noted with Neisseria meningitidis [116], and taken together, implies the importance of BP eDNA in the formation of BP biofilms. More recently, the unraveling of the P. aeruginosa eDNA structure revealed that it was different from P. aeruginosa chromosomal DNA where purine-rich RNAs that were integrated into the eDNA framework enabled crosslinking of the extracellular matrix [117]. Moreover, the formation of G-quadruplexes occurs in eDNA due to the non-canonical Hoogsteen base pairing between thymine/uracil and guanine [118]. The presence of G-quadruplexes in the matrix of P. aeruginosa biofilms was verified by specific antibody binding while the loss of the G-quadruplexes resulted in a lack of eDNA fibers [118]. Hence, this marks the structural specificity of eDNA involved in biofilm formation.
eDNA also exists as a lattice structure stabilized by DNABII proteins [119]. The integration host factor (IHF) and histone-like protein (HU) are two common members of the DNABII protein family that contribute to the lattice structure of the eDNA, thereby increasing the structural stability of the biofilm [120-122]. The B. cenocepacia HU and IHF protein orthologs are present in BP [122] (Table 2), indicating similar structural integrity components among the Burkholderia biofilms. Targeting the DNABII proteins via anti-DNABII antibodies effectively reduced biofilm formation in P. aeruginosa 27853, B. cenocepacia K56, non-typeable Haemophilus influenzae 86-028NP, Moraxella catarrhalis 7169, and Staphylococcus aureus 29213 [123]. These findings highlight the significance of eDNA in the structural integrity of biofilms for most bacteria. Therefore, targeting the eDNA could be a therapeutic strategy to eradicate infections by biofilm-forming pathogens.
Table 2.
Genes/proteins involved in the contribution of extracellular polymeric matrix (EPS) components in B. pseudomallei biofilms.
| EPS components | Gene/gene cluster reported to be involved in EPS biosynthesis (Annotation/Description) | Species/isolate | Sequence identity to BP K96243 (% of identity) | Burkholderia pseudomallei K96243 identifier code | Protein Description of Burkholderia pseudomallei K96243 | Reference |
|---|---|---|---|---|---|---|
| Exopolysaccharide | Bcam1330 (putative exopolysaccharide export protein) | Burkholderia cenocepacia J2315 | 79.10 | BPSL2780 | Capsular polysaccharide transport protein | [54,97] |
| Bcam1331 (putative tyrosine kinase protein) | Burkholderia cenocepacia J2315 | - | - | - | [54,97] | |
| Bcam1332 (hypothetical protein) | Burkholderia cenocepacia J2315 | - | - | - | [54,97] | |
| Bcam1333 (putative exopolysaccharide acyltransferase) | Burkholderia cenocepacia J2315 | 73.68 | BPSL3087 | Acyltransferase | [54,97] | |
| Bcam1334 (hypothetical protein) | Burkholderia cenocepacia J2315 | 70.37 | BPSL0610 | Hypothetical protein | [54,97] | |
| Bcam1335 (glycosyltransferase) | Burkholderia cenocepacia J2315 | 71.77 | BPSL0604 | Glycosyltransferase | [54,97] | |
| Bcam1336 (putative exopolysaccharide transporter) | Burkholderia cenocepacia J2315 | 74.91 | BPSL0603 | polysaccharide biosynthesis protein | [54,97] | |
| Bcam1337 (glycosyltransferase) | Burkholderia cenocepacia J2315 | - | - | - | [54,97] | |
| Bcam1338 (glycosyltransferase) | Burkholderia cenocepacia J2315 | - | - | - | [54,97] | |
| Bcam1339 (hypothetical protein) | Burkholderia cenocepacia J2315 | 73.68 | BPSL1233 | Lipoprotein | [54,97] | |
| Bcam1340 (mannose-1-gyanylyltransferase) | Burkholderia cenocepacia J2315 | 83.60 | BPSL0605 | Mannose-1-phosphate guanylyltransferase (manC) | [45,54,97] | |
| Bcam1340 (mannose-1-gyanylyltransferase) | Burkholderia cenocepacia J2315 | 74.61 | BPSS1835 | LPS biosynthesis mannose-1-phosphate guanylyltransferase (BceA) | [54,97] | |
| Bcam1341 (hypothetical protein) | Burkholderia cenocepacia J2315 | 77.67 | BPSL0606 | Hypothetical protein | [54,97] | |
| - | Burkholderia pseudomallei K96243 | NA | BPSL0618 | putative sugar transferase | [45] | |
| - | Burkholderia pseudomallei K96243 | NA | BPSL0619 | putative polysaccharide biosynthesis/export protein | [45] | |
| - | Burkholderia pseudomallei K96243 | NA | BPSL0620 | glycosyl transferase group 1 protein | [45] | |
| - | Burkholderia pseudomallei K96243 | NA | BPSS1649 | sugar-binding protein | [45] | |
| - | Burkholderia pseudomallei K96243 | NA | BPSS1978 | EPS transport-related membrane protein kinase | [45] | |
| Bp 1026b_I2907-Bp1026b_I2927 becA-R | Burkholderia pseudomallei 1026b | NA | BPSL0603-BPSL0620 | Exopolysaccharide gene cluster | [96] | |
| Bp1026b-I0648 wbiA | Burkholderia pseudomallei 1026b | 99.1 | BPSL2671 | Glycosyltransferase family protein | [54,96] | |
| Bp1026b-I0649 wbiA | Burkholderia pseudomallei 1026b | 100 | BPSL2670 | UDP-glucose-4-epimerase | [54,96] | |
| bpsI | Burkholderia pseudomallei K96243 | NA | BPSS0885 | acyl homoserine lactone (AHL) | [93] | |
| rpoS | Burkholderia pseudomallei K96243 | NA | BPSL1505 | RNA polymerase sigma factor | [93] | |
| - | Burkholderia pseudomallei K96243 | NA | BPSL1366 | polyphosphate kinase | [93] | |
| wcbK | Burkholderia pseudomallei K96243 | NA | BPSL2729 | UTP glucose-1-phosphate | [93] | |
| eDNA | Bcal1585 (histone like protein) (hupb) | Burkholderia cenocepacia J2315 | 76.98 | BPSL0004 | DNA-binding protein HU-alpha | [54,122] |
| Bcal3530 (histone like protein) (hupA) | Burkholderia cenocepacia J2315 | 93.45 | BPSL0004 | DNA-binding protein HU-alpha | [54,122] | |
| Bcal1487 (integration host factor alpha) | Burkholderia cenocepacia J2315 | 88.04 | BPSL1939 | integration host factor alpha | [54,122] | |
| Bcal2949 (integration host factor beta) | Burkholderia cenocepacia J2315 | 89.56 | BPSL2514 | integration host factor beta | [54,122] | |
| BPSL1887 (transcriptional regulatory protein) | Burkholderia pseudomallei K96243 | NA | BPSL1887 | sigma-54 related transcriptional regulatory protein | [110] | |
| Proteins | - | Burkholderia pseudomallei K96243 | NA | BPSS0093 | outer membrane usher protein | [45] |
| - | Burkholderia pseudomallei K96243 | NA | BPSL1800 | outer membrane usher protein | [45] | |
| bceF | Burkholderia pseudomallei K96243 | NA | BPSS1830 | Tyrosine kinase | [54] | |
| AK34_RS27645 (Alginate lyase) | Burkholderia dolosa AU0158 | 85.42 | BPSL3363 | Hypothetical protein | [54] | |
| - | Burkholderia pseudomallei K96243 | NA | BPSL0782 | Type 4 Pili 1 | [130] | |
| - | Burkholderia pseudomallei K96243 | NA | BPSL1821 | Type 4 Pili 2 | [130] | |
| - | Burkholderia pseudomallei K96243 | NA | BPSL1899 | Type 4 Pili 3 | [130] | |
| - | Burkholderia pseudomallei K96243 | NA | BPSL2752 | Type 4 Pili 4 | [130] | |
| - | Burkholderia pseudomallei K96243 | NA | BPSL2756 | Type 4 Pili 4 | [130] | |
| - | Burkholderia pseudomallei K96243 | NA | BPSL3008 | Type 4 Pili 5 | [130] | |
| - | Burkholderia pseudomallei K96243 | NA | BPSL3170 | Type 4 Pili 6 | [130] | |
| - | Burkholderia pseudomallei K96243 | NA | BPSS1593 | Type 4 Pili 7 | [130] | |
| - | Burkholderia pseudomallei K96243 | NA | BPSS1595 | Type 4 Pili 7 | [130] | |
| - | Burkholderia pseudomallei K96243 | NA | BPSS2185 | Type 4 Pili 8 | [130] | |
| - | Burkholderia pseudomallei K96243 | NA | BPSS2186 | Type 4 Pili 8 | [130] |
*NA-Not applicable
Proteins in EPS
The abundance of proteins in EPSs has been examined recently in most bacteria capable of forming biofilms. The function of these proteins to achieve a successful biofilm are diverse [124]. Currently, proteins within EPSs are categorized as enzymes and structural proteins [125]. Numerous enzymes in EPSs are involved in the synthesis or degradation of matrix components. For instance, tyrosine kinase encoded by bceF has been implicated in favoring biofilm formation by mediating the synthesis of exopolysaccharides in B. cepacia IST408 [126]. On the contrary, enzymes that break down the EPS, such as alginate lyase, are involved in the breakdown of exopolysaccharides in P. aeruginosa biofilms [127]. The BP genes encoding tyrosine kinase and alginate lyase annotated in the Burkholderia Genome Database (https://www.burkholderia.com/) are shown in Table 2 [54]. Further investigation is required to assess the enzymatic activity of these enzymes towards the biofilm formed by BP.
EPS proteins that contribute to structural stability include surface-associated proteins, such as pili and flagella, which mediate bacterial initial attachment and adhesion in H. influenzae biofilms and most other bacterial biofilms [124, 128]. Pili are known to facilitate bacterial adhesion, motility, DNA transfer, and biofilm formation [129]. BP is known to encode eight types of type IV pili (T4P) [130]. Recently, an uncharacterized type IV pili-associated protein (TFP8) encoded by bpss2185 was reported to be highly expressed during biofilm maturation and dispersal stages, highlighting that bacterial movement is crucial in stabilizing the structure of biofilm in BP [131]. Apart from that, proteomics analysis had discovered an abundance of outer membrane vesicle (OMV) proteins within the matrix of P. aeruginosa and B. multivorans biofilms [102]. The OMV proteins of gram-negative bacteria exist as spherical and bilayer membranes [132]. OMVs released during bacterial growth and and contain lipoproteins, lipopolysaccharides, and outer membrane proteins [133, 134]. OMVs are involved in several phenomena such as pathogenesis, bacterial communication, horizontal gene transfer, nutrient capture, bacterial-host interaction, and improvement of coaggregation during biofilm formation [102, 134]. Furthermore, OMVs can shield the biofilm community by releasing toxins that can target and affect the host defensive responses [133]. Meanwhile, bpss0093 and bpsl1800, two BP genes, were reported to be highly expressed during biofilm formation [45]. These genes are suggested to encode an outer membrane usher protein, presumably with a similar function to OMVs. Since the abundance of OMVs has been reported within the EPS, utilizing these OMVs to channel the antibacterial agents into the biofilm community serves as a strategy to eradicate biofilm infection [133].
Conclusion and Future Perspective
BP biofilms have been implicated as a virulence factor contributing to the pathogenesis of melioidosis during BP infections. This review systematically presents the genes and proteins that have been shown or predicted to be involved in the biosynthesis of essential B. pseudomallei EPS components. More than 60 genes and proteins representing 1.2% of the total annotated genes of BP have been identified as being involved in its biofilm formation. In this review, we have highlighted several knowledge gaps that require future investigation. These include: (i) the need to elucidate the roles of putative proteins that contain DGC and PDE domains for cyclic-di-GMP signaling; (ii) the determination of specific sRNAs that may have roles in regulating BP biofilm formation;(iii) the characterization of enzymes including hypothetical proteins in the becA-R gene cluster to decode the exopolysaccharide biosynthesis pathway; and (iv) to unravel the mechanistic role of eDNA in biofilm formation and its potential as a target for therapeutics. Furthermore, a systems biology approach could be adopted to characterize further the interrelationship between biofilm formation stages, signaling systems, regulation, and biosynthesis of EPS components.
Supplemental Materials
Supplementary data for this paper are available on-line only at http://jmb.or.kr.
Acknowledgments
This work is supported by research grants from the Ministry of Higher Education (MoHE) Malaysia (FRGS/1/ 2018/STG04/UKM/02/3) and Universiti Kebangsaan Malaysia (Geran Universiti Penyelidikan (GUP), GUP-2021-069). Graphical abstract and Figure 1 were created using BioRender.com. Part of Figure 2 was drawn by using pictures from Servier Medical Art, provided by Servier, licensed under a Creative Commons Attribution 3.0 unported license.
Footnotes
Conflict of Interest
The authors have no financial conflicts of interest to declare.
REFERENCES
- 1.Mariappan V, Vellasamy KM, Barathan M, Girija ASS, Shankar EM, Vadivelu J. Hijacking of the Host's Immune Surveillance Radars by Burkholderia pseudomallei. Front. Immunol. 2021;12:718719. doi: 10.3389/fimmu.2021.718719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiersinga WJ, Virk HS, Torres AG, et al. Melioidosis. Nat. Rev. Dis. Primers. 2018;4:17108. doi: 10.1038/nrdp.2017.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yip C-H, Ghazali A-K, Nathan S. Burkholderia pseudomallei pathogenesis and survival in different niches. Biochem. Soc. Trans. 2020;48:569–579. doi: 10.1042/BST20190836. [DOI] [PubMed] [Google Scholar]
- 4.Panomket P, Wongsana P, Wanram S, Wongratanacheewin S. Burkholderia pseudomallei biofilm plays a key role in chronic inflammation in C57BL/6 mice. Southeast Asian J. Trop. Med. Public Health. 2017;48:73–82. [PubMed] [Google Scholar]
- 5.Duangurai T, Indrawattana N, Pumirat P. Burkholderia pseudomallei adaptation for survival in stressful conditions. Biomed Res. Int. 2018;2018:3039106. doi: 10.1155/2018/3039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willcocks SJ, Cia F, Francisco AF, Wren BW. Revisiting aminocoumarins for the treatment of melioidosis. Int. J. Antimicrob. Agents. 2020;56:106002. doi: 10.1016/j.ijantimicag.2020.106002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarz S, Van Dijck P. Trehalose metabolism: A sweet spot for Burkholderia pseudomallei virulence. Virulence. 2017;8:5–7. doi: 10.1080/21505594.2016.1216295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Limmathurotsakul D, Paeyao A, Wongratanacheewin S, Saiprom N, Takpho N, Thaipadungpanit J, et al. Role of Burkholderia pseudomallei biofilm formation and lipopolysaccharide in relapse of melioidosis. Clin. Microbiol. Infect. 2014;20:O854–O856. doi: 10.1111/1469-0691.12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghosh R, Barman S, Mandal NC. Phosphate deficiency induced biofilm formation of Burkholderia on insoluble phosphate granules plays a pivotal role for maximum release of soluble phosphate. Sci. Rep. 2019;9:5477. doi: 10.1038/s41598-019-41726-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vestby LK, Grønseth T, Simm R, Nesse LL. Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics. 2020;9:59. doi: 10.3390/antibiotics9020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flemming H-C. EPS-Then and now. Microorganisms. 2016;4:41. doi: 10.3390/microorganisms4040041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neopane P, Nepal HP, Shrestha R, Uehara O, Abiko Y. In vitro biofilm formation by Staphylococcus aureus isolated from wounds of hospital-admitted patients and their association with antimicrobial resistance. Int. J. Gen. Med. 2018;11:25–32. doi: 10.2147/IJGM.S153268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan J, Bassler BL. Surviving as a community: Antibiotic tolerance and persistence in bacterial biofilms. Cell Host Microbe. 2019;26:15–21. doi: 10.1016/j.chom.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng N, Cai P, Mortimer M, Wu Y, Gao C, Huang Q. The exopolysaccharide-eDNA interaction modulates 3D architecture of Bacillus subtilis biofilm. BMC Microbiol. 2020;20:115. doi: 10.1186/s12866-020-01789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tolker-Nielsen T. Biofilm Development. Microbiol. Spectr. 2015;3:51–66. doi: 10.1128/9781555817466.ch3. [DOI] [PubMed] [Google Scholar]
- 16.Mcferrin A, Engineering C, Building JEB, Texas A. Insights on Escherichia coli biofilm formation and inhibition from wholetranscriptome profiling thomas. Environ. Microbiol. 2010;11:1–15. doi: 10.1111/j.1462-2920.2008.01768.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kostakioti M, Hadjifrangiskou M, Hultgren SJ. Bacterial biofilms: Development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 2013;3:a010306. doi: 10.1101/cshperspect.a010306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duangurai T, Indrawattana N, Pumirat P. Burkholderia pseudomallei adaptation for survival in stressful conditions. Biomed Res. Int. 2018;2018:3039106. doi: 10.1155/2018/3039106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunyanee C, Kamjumphol W, Taweechaisupapong S, Kanthawong S, Wongwajana S, Wongratanacheewin S, et al. Burkholderia pseudomallei biofilm promotes adhesion, internalization and stimulates proinflammatory cytokines in human epithelial A549 cells. PLoS One. 2016;11:e0160741. doi: 10.1371/journal.pone.0160741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castelo-Branco DSCM, Riello GB, Vasconcelos DC, Guedes GMM, Serpa R, Bandeira TJPG, et al. Farnesol increases the susceptibility of Burkholderia pseudomallei biofilm to antimicrobials used to treat melioidosis. J. Appl. Microbiol. 2016;120:600–606. doi: 10.1111/jam.13027. [DOI] [PubMed] [Google Scholar]
- 21.Sirijant N, Sermswan RW, Wongratanacheewin S. Burkholderia pseudomallei resistance to antibiotics in biofilm-induced conditions is related to efflux pumps. J. Med. Microbiol. 2016;65:1296–1306. doi: 10.1099/jmm.0.000358. [DOI] [PubMed] [Google Scholar]
- 22.Sawasdidoln C, Taweechaisupapong S, Sermswan RW, Tattawasart U, Tungpradabkul S, Wongratanacheewin S. Growing Burkholderia pseudomallei in biofilm stimulating conditions significantly induces antimicrobial resistance. PLoS One. 2010;5:e9196. doi: 10.1371/journal.pone.0009196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toyofuku M, Inaba T, Kiyokawa T, Obana N, Yawata Y, Nomura N. Environmental factors that shape biofilm formation. Biosci. Biotechnol. Biochem. 2016;80:7–12. doi: 10.1080/09168451.2015.1058701. [DOI] [PubMed] [Google Scholar]
- 24.Lin Chua S, Liu Y, Li Y, Ting HJ, Kohli GS, Cai Z, et al. Reduced intracellular c-di-GMP content increases expression of quorum sensing-regulated genes in Pseudomonas aeruginosa. Front. Cell. Infect. Microbiol. 2017;7:451. doi: 10.3389/fcimb.2017.00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramli NSK, Eng Guan C, Nathan S, Vadivelu J. The effect of environmental conditions on biofilm formation of Burkholderia pseudomallei clinical isolates. PLoS One. 2012;7:e44104. doi: 10.1371/journal.pone.0044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paksanont S, Sintiprungrat K, Yimthin T, Pumirat P, Peacock SJ, Chantratita N. Effect of temperature on Burkholderia pseudomallei growth, proteomic changes, motility and resistance to stress environments. Sci. Rep. 2018;8:9196. doi: 10.1038/s41598-018-27356-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anutrakunchai C, Bolscher JGM, Krom BP, Kanthawong S, Chareonsudjai S, Taweechaisupapong S. Impact of nutritional stress on drug susceptibility and biofilm structures of Burkholderia pseudomallei and Burkholderia thailandensis grown in static and microfluidic systems. PLoS One. 2018;13:e0194946. doi: 10.1371/journal.pone.0194946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou G, Yuan J, Gao H. Regulation of biofilm formation by BpfA, BpfD, and BpfG in Shewanella oneidensis. Front. Microbiol. 2015;6:790. doi: 10.3389/fmicb.2015.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim B, Park J-S, Choi H-Y, Yoon SS, Kim W-G. Terrein is an inhibitor of quorum sensing and c-di-GMP in Pseudomonas aeruginosa: a connection between quorum sensing and c-di-GMP. Sci. Rep. 2018;8:8617. doi: 10.1038/s41598-018-26974-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Opoku-Temeng C, Sintim HO. Targeting c-di-GMP Signaling, Biofilm Formation, and Bacterial Motility with Small Molecules. In: Sauer K, editor. c-di-GMP Signaling: Methods and Protocols. Springer New York; New York, NY: 2017. pp. 419–430 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Fazli M, Almblad H, Rybtke ML, Givskov M, Eberl L, Tolker-Nielsen T. Regulation of biofilm formation in Pseudomonas and Burkholderia species. Environ. Microbiol. 2014;16:1961–1981. doi: 10.1111/1462-2920.12448. [DOI] [PubMed] [Google Scholar]
- 32.Jones CJ, Utada A, Davis KR, et al. c-di-GMP Regulates motile to sessile transition by modulating MshA Pili Biogenesis and near-surface motility behavior in Vibrio cholerae. PLoS Pathog. 2015;11:e1005068. doi: 10.1371/journal.ppat.1005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mangalea MR, Plumley BA, Borlee BR. Nitrate sensing and metabolism inhibit biofilm formation in the opportunistic pathogen Burkholderia pseudomallei by reducing the intracellular concentration of c-di-GMP. Front. Microbiol. 2017;8:1353. doi: 10.3389/fmicb.2017.01353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tseng BS, Majerczyk CD, Passos da Silva D, Chandler JR, Peter Greenberg E, Parsek MR. Quorum sensing influences Burkholderia thailandensis biofilm development and matrix production. J. Bacteriol. 2016;198:2643–2650. doi: 10.1128/JB.00047-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Acker H, Crabbé A, Jurėnas D, Ostyn L, Sass A, Daled S, et al. The role of small proteins in Burkholderia cenocepacia J2315 biofilm formation, persistence and intracellular growth. Biofilm. 2019;1:100001. doi: 10.1016/j.bioflm.2019.100001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen L-H, Köseoğlu VK, Güvener ZT, Myers-Morales T, Reed JM, D'Orazio SEF, et al. Cyclic di-GMP-dependent Signaling pathways in the pathogenic firmicute Listeria monocytogenes. PLoS Pathog. 2014;10:e1004301. doi: 10.1371/journal.ppat.1004301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jenal U, Reinders A, Lori C. Cyclic di-GMP: Second messenger extraordinaire. Nat. Rev. Microbiol. 2017;15:271–284. doi: 10.1038/nrmicro.2016.190. [DOI] [PubMed] [Google Scholar]
- 38.Papenfort K, Bassler BL. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016;14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang Y, Geng M, Bai L. Targeting biofilms therapy: Current research strategies and development hurdles. Microorganisms. 2020;8:1222. doi: 10.3390/microorganisms8081222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bellich B, Jou IA, Caterino M, Rizzo R, Ravenscroft N, Fazli M, et al. Burkholderia cenocepacia H111 produces a waterinsoluble exopolysaccharide in biofilm: Structural determination and molecular modelling. Int. J. Mol. Sci. 2020;21:1702. doi: 10.3390/ijms21051702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plumley BA, Martin KH, Borlee GI, Marlenee NL, Burtnick MN, Brett PJ, et al. Thermoregulation of biofilm formation in Burkholderia pseudomallei is disrupted by mutation of a putative diguanylate cyclase. J. Bacteriol. 2017;199:e00780–16. doi: 10.1128/JB.00780-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee HS, Gu F, Ching SM, Lam Y, Chua KL. CdpA is a Burkholderia pseudomallei cyclic di-GMP phosphodiesterase involved in autoaggregation, flagellum synthesis, motility, biofilm formation, cell invasion, and cytotoxicity. Infect. Immun. 2010;78:1832–1840. doi: 10.1128/IAI.00446-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vitale A, Paszti S, Takahashi K, Toyofuku M, Pessi G, Eberl L. Mapping of the denitrification pathway in Burkholderia thailandensis by genome-wide mutant profiling. J. Bacteriol. 2020;202:e00304–20. doi: 10.1128/JB.00304-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mangalea MR, Borlee BR. The NarX-NarL two-component system regulates biofilm formation, natural product biosynthesis, and host-associated survival in Burkholderia pseudomallei. Sci. Rep. 2022;12:203. doi: 10.1038/s41598-021-04053-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chin C-Y, Hara Y, Ghazali A-K, Yap S-J, Kong C, Wong Y-C, et al. Global transcriptional analysis of Burkholderia pseudomallei high and low biofilm producers reveals insights into biofilm production and virulence. BMC Genomics. 2015;16:471. doi: 10.1186/s12864-015-1692-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Guo J, Zhang N, Yuan W, Lin Z, Huang W. Characterization and analysis of a novel diguanylate cyclase PA0847 from Pseudomonas aeruginosa PAO1. Infect. Drug Resist. 2019;12:655–665. doi: 10.2147/IDR.S194462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Römling U, Galperin MY, Gomelsky M. Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 2013;77:1–52. doi: 10.1128/MMBR.00043-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orr MW, Donaldson GP, Severin GB, et al. Oligoribonuclease is the primary degradative enzyme for pGpG in Pseudomonas aeruginosa that is required for cyclic-di-GMP turnover. Proc. Natl. Acad. Sci. USA. 2015;112:E5048–E5057. doi: 10.1073/pnas.1507245112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ha D-G, O'Toole GA. c-di-GMP and its effects on biofilm formation and dispersion: a Pseudomonas aeruginosa review. Microbiol. Spectr. 2015;3:MB-0003-2014. doi: 10.1128/microbiolspec.MB-0003-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicastro GG, Kaihami GH, Pulschen AA, et al. c-di-GMP-related phenotypes are modulated by the interaction between a diguanylate cyclase and a polar hub protein. Sci. Rep. 2020;10:3077. doi: 10.1038/s41598-020-59536-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Richter AM, Fazli M, Schmid N, Shilling R, Suppiger A, Givskov M, et al. Key players and individualists of Cyclic-di-GMP signaling in Burkholderia cenocepacia. Front. Microbiol. 2019;10:3286. doi: 10.3389/fmicb.2018.03286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Borlee GI, Mangalea MR, Martin KH, Plumley BA, Golon SJ, Borlee BR. Disruption of c-di-GMP signaling networks unlocks cryptic expression of secondary metabolites during biofilm growth in Burkholderia pseudomallei. Appl. Environ. Microbiol. 2022;88:e02431–21. doi: 10.1128/aem.02431-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Z, Xie X, Shang D, Xie L, Hua Y, Song L, et al. A c-di-GMP signaling cascade controls motility, biofilm formation, and virulence in Burkholderia thailandensis. Appl. Environ. Microbiol. 2022;88:e02529–21. doi: 10.1128/aem.02529-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winsor GL, Khaira B, Van Rossum T, Lo R, Whiteside MD, Brinkman FSL. The Burkholderia genome database: facilitating flexible queries and comparative analyses. Bioinformatics. 2008;24:2803–2804. doi: 10.1093/bioinformatics/btn524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Galperin MY, Nikolskaya AN, Koonin EV. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 2001;203:11–21. doi: 10.1111/j.1574-6968.2001.tb10814.x. [DOI] [PubMed] [Google Scholar]
- 56.Whiteley CG, Lee D-J. Bacterial diguanylate cyclases: structure, function and mechanism in exopolysaccharide biofilm development. Biotechnol. Adv. 2015;33:124–141. doi: 10.1016/j.biotechadv.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 57.Hengge R, Galperin MY, Ghigo J-M, Gomelsky M, Green J, Hughes KT, et al. Systematic nomenclature for GGDEF and EAL domain-containing cyclic di-GMP turnover proteins of Escherichia coli. J. Bacteriol. 2016;198:7–11. doi: 10.1128/JB.00424-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rutherford ST, Bassler BL. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012;2:1–25. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reen FJ, Gutiérrez-Barranquero JA, Parages ML. Coumarin: a novel player in microbial quorum sensing and biofilm formation inhibition. Appl. Microbiol. Biotechnol. 2018;102:2063–2073. doi: 10.1007/s00253-018-8787-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao X, Yu Z, Ding T. Quorum-sensing regulation of antimicrobial resistance in bacteria. Microorganisms. 2020;8:425. doi: 10.3390/microorganisms8030425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brackman G, Hillaert U, Van Calenbergh S, Nelis HJ, Coenye T. Use of quorum sensing inhibitors to interfere with biofilm formation and development in Burkholderia multivorans and Burkholderia cenocepacia. Res. Microbiol. 2009;160:144–151. doi: 10.1016/j.resmic.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 62.Brackman G, Coenye T. Quorum sensing inhibitors as anti-biofilm agents. Curr. Pharm. Des. 2015;21:5–11. doi: 10.2174/1381612820666140905114627. [DOI] [PubMed] [Google Scholar]
- 63.Ulrich RL, Ulrich RL. Quorum quenching: Enzymatic disruption of N-acylhomoserine lactone-mediated bacterial communication in Burkholderia thailandensis. Appl. Environ. Microbiol. 2004;70:6173–6180. doi: 10.1128/AEM.70.10.6173-6180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tan W-S, Law JW-F, Letchumanan V, Chan K-G. Decoding the mystery of how bacteria "talk": Among Gram-negative microorganisms. Prog. Microbes Mol. Biol. 2019;2 doi: 10.36877/pmmb.a0000038. DOI: https://doi.org/10.36877/pmmb.a0000038 . [DOI] [Google Scholar]
- 65.Klaus JR, Deay J, Neuenswander B, et al. Malleilactone is a Burkholderia pseudomallei virulence factor regulated by antibiotics and quorum sensing. J. Bacteriol. 2018;200:e00008–18. doi: 10.1128/JB.00008-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gamage AM, Shui G, Wenk MR, Chua KL. N-Octanoylhomoserine lactone signalling mediated by the BpsI-BpsR quorum sensing system plays a major role in biofilm formation of Burkholderia pseudomallei. Microbiology. 2011;157:1176–1186. doi: 10.1099/mic.0.046540-0. [DOI] [PubMed] [Google Scholar]
- 67.Kumari A, Pasini P, Daunert S. Detection of bacterial quorum sensing N-acyl homoserine lactones in clinical samples. Anal. Bioanal. Chem. 2008;391:1619–1627. doi: 10.1007/s00216-008-2002-3. [DOI] [PubMed] [Google Scholar]
- 68.Mangwani N, Kumari S, Das S. Involvement of quorum sensing genes in biofilm development and degradation of polycyclic aromatic hydrocarbons by a marine bacterium Pseudomonas aeruginosa N6P6. Appl. Microbiol. Biotechnol. 2015;99:10283–10297. doi: 10.1007/s00253-015-6868-7. [DOI] [PubMed] [Google Scholar]
- 69.Mukherjee S, Moustafa D, Smith CD, Goldberg JB, Bassler BL. The RhlR quorum-sensing receptor controls Pseudomonas aeruginosa pathogenesis and biofilm development independently of its canonical homoserine lactone autoinducer. PLoS Pathog. 2017;13:1–25. doi: 10.1371/journal.ppat.1006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Song Y, Xie C, Ong Y-M, Gan Y-H, Chua K-L. The BpsIR quorum-sensing system of Burkholderia pseudomallei. J. Bacteriol. 2005;187:785–790. doi: 10.1128/JB.187.2.785-790.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mott T, Panchal RG, Rajamani S. Quorum sensing in Burkholderia pseudomallei and other Burkholderia species. Curr. Trop. Med. Rep. 2017;4:199–207. doi: 10.1007/s40475-017-0127-1. [DOI] [Google Scholar]
- 72.Lazar Adler NR, Dean RE, Saint RJ, Stevens MP, Prior JL, Atkins TP, et al. Identification of a predicted trimeric autotransporter adhesin required for biofilm formation of Burkholderia pseudomallei. PLoS One. 2013;8:e79461. doi: 10.1371/journal.pone.0079461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Coulon PML, Zlosnik JEA, Déziel E. Presence of the Hmq system and production of 4-Hydroxy-3-Methyl-2-Alkylquinolines are heterogeneously distributed between Burkholderia cepacia complex species and more prevalent among environmental than clinical isolates. Microbiol. Spectr. 2021;9:e00127–21. doi: 10.1128/Spectrum.00127-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heeb S, Fletcher MP, Chhabra SR, Diggle SP, Williams P, Cámara M. Quinolones: from antibiotics to autoinducers. FEMS Microbiol. Rev. 2011;35:247–274. doi: 10.1111/j.1574-6976.2010.00247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vial L, Lépine F, Milot S, Groleau M-C, Dekimpe V, Woods DE, et al. Burkholderia pseudomallei, B. thailandensis, and B. ambifaria produce 4-Hydroxy-2-alkylquinoline analogues with a methyl group at the 3 position that is required for quorum-sensing regulation. J. Bacteriol. 2008;190:5339–5352. doi: 10.1128/JB.00400-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Butt A, Halliday N, Williams P, Atkins HS, Bancroft GJ, Titball RW. Burkholderia pseudomallei kynB plays a role in AQ production, biofilm formation, bacterial swarming and persistence. Res. Microbiol. 2016;167:159–167. doi: 10.1016/j.resmic.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 77.Boon C, Deng Y, Wang L-H, He Y, Xu J-L, Fan Y, et al. A novel DSF-like signal from Burkholderia cenocepacia interferes with Candida albicans morphological transition. ISME J. 2008;2:27–36. doi: 10.1038/ismej.2007.76. [DOI] [PubMed] [Google Scholar]
- 78.Deng Y, Boon C, Eberl L, Zhang L-H. Differential modulation of Burkholderia cenocepacia virulence and energy metabolism by the quorum-sensing signal BDSF and its synthase. J. Bacteriol. 2009;191:7270–7278. doi: 10.1128/JB.00681-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang C, Cui C, Ye Q, Kan J, Fu S, Song S, et al. Burkholderia cenocepacia integrates cis-2-dodecenoic acid and cyclic dimeric guanosine monophosphate signals to control virulence. Proc. Natl. Acad. Sci. USA. 2017;114:13006–13011. doi: 10.1073/pnas.1709048114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Deng Y, Schmid N, Wang C, Wang J, Pessi G, Wu D, et al. Cis-2-dodecenoic acid receptor RpfR links quorum-sensing signal perception with regulation of virulence through cyclic dimeric guanosine monophosphate turnover. Proc. Natl. Acad. Sci. USA. 2012;109:15479–15484. doi: 10.1073/pnas.1205037109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Deng Y, Lim A, Wang J, Zhou T, Chen S, Lee J, et al. Cis-2-dodecenoic acid quorum sensing system modulates N-acyl homoserine lactone production through RpfR and cyclic di-GMP turnover in Burkholderia cenocepacia. BMC Microbiol. 2013;13:148. doi: 10.1186/1471-2180-13-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cui C, Yang C, Song S, et al. A novel two-component system modulates quorum sensing and pathogenicity in Burkholderia cenocepacia. Mol. Microbiol. 2018;108:32–44. doi: 10.1111/mmi.13915. [DOI] [PubMed] [Google Scholar]
- 83.Waldron EJ, Snyder D, Fernandez NL, Sileo E, Inoyama D, Freundlich JS, et al. Structural basis of DSF recognition by its receptor RpfR and its regulatory interaction with the DSF synthase RpfF. PLoS Biol. 2019;17:e3000123. doi: 10.1371/journal.pbio.3000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang M, Li X, Song S, Cui C, Zhang L-H, Deng Y. The cis-2-dodecenoic acid (BDSF) quorum sensing system in Burkholderia cenocepacia. Appl. Environ. Microbiol. 2022;88:e02342–21. doi: 10.1128/aem.02342-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sass A, Kiekens S, Coenye T. Identification of small RNAs abundant in Burkholderia cenocepacia biofilms reveal putative regulators with a potential role in carbon and iron metabolism. Sci. Rep. 2017;7:15665. doi: 10.1038/s41598-017-15818-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pita T, Feliciano JR, Leitão JH. Small noncoding regulatory RNAs from Pseudomonas aeruginosa and Burkholderia cepacia complex. Int. J. Mol. Sci. 2018;19:3759. doi: 10.3390/ijms19123759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kiekens S, Sass A, Van Nieuwerburgh F, Deforce D, Coenye T. The small RNA ncS35 regulates growth in Burkholderia cenocepacia J2315. mSphere. 2018;3:e00579–17. doi: 10.1128/mSphere.00579-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Rossi F, De Philippis R. Role of cyanobacterial exopolysaccharides in phototrophic biofilms and in complex microbial mats. Life. 2015;5:1218–1238. doi: 10.3390/life5021218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cuzzi B, Herasimenka Y, Silipo A, Lanzetta R, Liut G, Rizzo R, et al. Versatility of the Burkholderia cepacia complex for the biosynthesis of exopolysaccharides: A comparative structural investigation. PLoS One. 2014;9:e94372. doi: 10.1371/journal.pone.0094372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ciofu O, Tolker-Nielsen T. Tolerance and resistance of Pseudomonas aeruginosa biofilms to antimicrobial agents-how P. aeruginosa can escape antibiotics. Front. Microbiol. 2019;10:913. doi: 10.3389/fmicb.2019.00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mangalea MR, Borlee GI, Borlee BR. The current status of extracellular polymeric substances produced by Burkholderia pseudomallei. Curr. Trop. Med. Reports. 2017;4:117–126. doi: 10.1007/s40475-017-0118-2. [DOI] [Google Scholar]
- 92.Nimtz M, Wray V, Domke T, Brenneke B, Häussler S, Steinmetz I. Structure of an acidic exopolysaccharide of Burkholderia pseudomallei. Eur. J. Biochem. 1997;250:608–616. doi: 10.1111/j.1432-1033.1997.0608a.x. [DOI] [PubMed] [Google Scholar]
- 93.Mongkolrob R, Taweechaisupapong S, Tungpradabkul S. Correlation between biofilm production, antibiotic susceptibility and exopolysaccharide composition in Burkholderia pseudomallei bpsI, ppk, and rpoS mutant strains. Microbiol. Immunol. 2015;59:653–663. doi: 10.1111/1348-0421.12331. [DOI] [PubMed] [Google Scholar]
- 94.Laroussarie A, Barycza B, Andriamboavonjy H, Tamigney Kenfack M, Blériot Y, Gauthier C. Synthesis of the tetrasaccharide repeating unit of the β-Kdo-containing exopolysaccharide from Burkholderia pseudomallei and B. cepacia complex. J. Org. Chem. 2015;80:10386–10396. doi: 10.1021/acs.joc.5b01823. [DOI] [PubMed] [Google Scholar]
- 95.Park J, Lee D, Kim M-S, Kim DY, Shin DH. A preliminary X-ray study of 3-deoxy-d-manno-oct-2-ulosonic acid 8-phosphate phosphatase (YrbI) from Burkholderia pseudomallei. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2015;71:790–793. doi: 10.1107/S2053230X15006135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Borlee GI, Plumley BA, Martin KH, Somprasong N, Mangalea MR, Islam MN, et al. Genome-scale analysis of the genes that contribute to Burkholderia pseudomallei biofilm formation identifies a crucial exopolysaccharide biosynthesis gene cluster. PLoS Negl. Trop. Dis. 2017;11:e0005689. doi: 10.1371/journal.pntd.0005689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fazli M, McCarthy Y, Givskov M, Ryan RP, Tolker‐Nielsen T. The exopolysaccharide gene cluster Bcam1330-Bcam1341 is involved in Burkholderia cenocepacia biofilm formation, and its expression is regulated by c-di-GMP and Bcam1349. Microbiologyopen. 2013;2:105–122. doi: 10.1002/mbo3.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fazli M, O'Connell A, Nilsson M, Niehaus K, Dow JM, Givskov M, et al. The CRP/FNR family protein Bcam1349 is a c-di-GMP effector that regulates biofilm formation in the respiratory pathogen Burkholderia cenocepacia. Mol. Microbiol. 2011;82:327–341. doi: 10.1111/j.1365-2958.2011.07814.x. [DOI] [PubMed] [Google Scholar]
- 99.Poulin MB, Kuperman LL. Regulation of biofilm exopolysaccharide production by cyclic di-guanosine monophosphate. Front. Microbiol. 2021;12:2506. doi: 10.3389/fmicb.2021.730980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chiang W-C, Nilsson M, Jensen PØ, Høiby N, Nielsen TE, Givskov M, et al. Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 2013;57:2352–2361. doi: 10.1128/AAC.00001-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kavanaugh JS, Flack CE, Lister J, Ricker EB, Ibberson CB, Jenul C, et al. Identification of extracellular DNA-binding proteins in the biofilm matrix. mBio. 2019;10:e01137–19. doi: 10.1128/mBio.01137-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Terán LC, Distefano M, Bellich B, Petrosino S, Bertoncin P, Cescutti P, et al. Proteomic studies of the biofilm matrix including outer membrane vesicles of Burkholderia multivorans c1576, a strain of clinical importance for cystic fibrosis. Microorganisms. 2020;8:1826. doi: 10.3390/microorganisms8111826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Panlilio H, Rice CV. The role of extracellular DNA in the formation, architecture, stability, and treatment of bacterial biofilms. Biotechnol. Bioeng. 2021;118:2129–2141. doi: 10.1002/bit.27760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wongkaewkhiaw S, Kanthawong S, Bolscher JGM, Nazmi K, Taweechaisupapong S, Krom BP. DNase-mediated eDNA removal enhances D-LL-31 activity against biofilms of bacteria isolated from chronic rhinosinusitis patients. Biofouling. 2020;36:1117–1128. doi: 10.1080/08927014.2020.1857741. [DOI] [PubMed] [Google Scholar]
- 105.Ibáñez de Aldecoa AL, Zafra O, González-Pastor JE. Mechanisms and regulation of extracellular DNA release and its biological roles in microbial communities. Front. Microbiol. 2017;8:1390. doi: 10.3389/fmicb.2017.01390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jakubovics NS, Shields RC, Rajarajan N, Burgess JG. Life after death: The critical role of extracellular DNA in microbial biofilms. Lett. Appl. Microbiol. 2013;57:467–475. doi: 10.1111/lam.12134. [DOI] [PubMed] [Google Scholar]
- 107.Das T, Sehar S, Manefield M. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ. Microbiol. Rep. 2013;5:778–786. doi: 10.1111/1758-2229.12085. [DOI] [PubMed] [Google Scholar]
- 108.Qin Z, Ou Y, Yang L, Zhu Y, Tolker-Nielsen T, Molin S, et al. Role of autolysin-mediated DNA release in biofilm formation of Staphylococcus epidermidis. Microbiology. 2007;153:2083–2092. doi: 10.1099/mic.0.2007/006031-0. [DOI] [PubMed] [Google Scholar]
- 109.Petrova OE, Schurr JR, Schurr MJ, Sauer K. The novel Pseudomonas aeruginosa two-component regulator BfmR controls bacteriophage-mediated lysis and DNA release during biofilm development through PhdA. Mol. Microbiol. 2011;81:767–783. doi: 10.1111/j.1365-2958.2011.07733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Austin CR, Goodyear AW, Bartek IL, Stewart A, Sutherland MD, Silva EB, et al. A Burkholderia pseudomallei colony variant necessary for gastric colonization. mBio. 2015;6:e02462--14. doi: 10.1128/mBio.02462-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Okaro U, Mou S, DeShazer D. Production and molecular composition of Burkholderia pseudomallei and Burkholderia thailandensis biofilms. Authorea. 2021. [Google Scholar]
- 112.Alwis PA, Treerat P, Gong L, Lucas DD, Allwood EM, Prescott M, et al. Disruption of the Burkholderia pseudomallei twocomponent signal transduction system BbeR-BbeS leads to increased extracellular DNA secretion and altered biofilm formation. Vet. Microbiol. 2020;242:108603. doi: 10.1016/j.vetmic.2020.108603. [DOI] [PubMed] [Google Scholar]
- 113.Pakkulnan R, Anutrakunchai C, Kanthawong S, Taweechaisupapong S, Chareonsudjai P, Chareonsudjai S. Extracellular DNA facilitates bacterial adhesion during Burkholderia pseudomallei biofilm formation. PLoS One. 2019;14:e0213288. doi: 10.1371/journal.pone.0213288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sena-Vélez M, Redondo C, Graham JH, Cubero J. Presence of extracellular DNA during biofilm formation by Xnthomonas citri subsp. citri strains with different host range. PLoS One. 2016;11:e0156695. doi: 10.1371/journal.pone.0156695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Okshevsky M, Meyer RL. The role of extracellular DNA in the establishment, maintenance and perpetuation of bacterial biofilms. Crit. Rev. Microbiol. 2015;41:341–352. doi: 10.3109/1040841X.2013.841639. [DOI] [PubMed] [Google Scholar]
- 116.Lappann M, Claus H, Van Alen T, Harmsen M, Elias J, Molin S, et al. A dual role of extracellular DNA during biofilm formation of Neisseria meningitidis. Mol. Microbiol. 2010;75:1355–1371. doi: 10.1111/j.1365-2958.2010.07054.x. [DOI] [PubMed] [Google Scholar]
- 117.Seviour T, Winnerdy FR, Wong LL, Shi X, Mugunthan S, et al. The biofilm matrix scaffold of Pseudomonas species contains non-canonically base paired extracellular DNA and RNA. bioRxiv. 2020 doi: 10.1101/527267. doi: https://doi.org/10.1101/527267 . [DOI] [Google Scholar]
- 118.Seviour T, Winnerdy FR, Wong LL, Shi X, Mugunthan S, Foo YH, et al. The biofilm matrix scaffold of Peudomonas aeruginosa contains G-quadruplex extracellular DNA structures. NPJ Biofilms Microbiomes. 2021;7:27. doi: 10.1038/s41522-021-00197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Goodman SD, Obergfell KP, Jurcisek JA, Novotny LA, Downey JS, Ayala EA, et al. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol. 2011;4:625–637. doi: 10.1038/mi.2011.27. [DOI] [PubMed] [Google Scholar]
- 120.Devaraj A, Buzzo JR, Mashburn-Warren L, et al. The extracellular DNA lattice of bacterial biofilms is structurally related to Holliday junction recombination intermediates. Proc. Natl. Acad. Sci. USA. 2019;116:25068 LP-25077. doi: 10.1073/pnas.1909017116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jurcisek JA, Brockman KL, Novotny LA, Goodman SD, Bakaletz LO. Nontypeable haemophilus influenzae releases DNA and DNABII proteins via a T4SS-like complex and ComE of the type IV pilus machinery. Proc. Natl. Acad. Sci. USA. 2017;114:E6632–E6641. doi: 10.1073/pnas.1705508114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Novotny LA, Amer AO, Brockson ME, Goodman SD, Bakaletz LO. Structural stability of Burkholderia cenocepacia biofilms is reliant on eDNA structure and presence of a bacterial nucleic acid binding protein. PLoS One. 2013;8:e67629. doi: 10.1371/journal.pone.0067629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Novotny LA, Goodman SD, Bakaletz LO. Targeting a bacterial DNABII protein with a chimeric peptide immunogen or humanised monoclonal antibody to prevent or treat recalcitrant biofilm-mediated infections. EBioMedicine. 2020;59:102867. doi: 10.1016/j.ebiom.2020.102867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fong JNC, Yildiz FH. Biofilm matrix proteins. Microbiol. Spectr. 2015;3:10.1128/microbiolspec.MB-0004-2014. doi: 10.1128/microbiolspec.MB-0004-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Flemming H-CC, Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 126.Ferreira AS, Silva IN, Oliveira VH, Becker JD, Givskov M, Ryan RP, et al. Comparative transcriptomic analysis of the Burkholderia cepacia tyrosine kinase bceF mutant reveals a role in tolerance to stress, biofilm formation, and virulence. Appl. Environ. Microbiol. 2013;79:3009–3020. doi: 10.1128/AEM.00222-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Blanco-Cabra N, Paetzold B, Ferrar T, Mazzolini R, Torrents E, Serrano L, et al. Characterization of different alginate lyases for dissolving Pseudomonas aeruginosa biofilms. Sci. Rep. 2020;10:9390. doi: 10.1038/s41598-020-66293-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wu S, Baum MM, Kerwin J, Guerrero D, Webster S, Schaudinn C, et al. Biofilm-specific extracellular matrix proteins of nontypeable Haemophilus influenzae. Pathog. Dis. 2014;72:143–160. doi: 10.1111/2049-632X.12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Berne C, Ducret A, Hardy GG, Brun YV. Microbial Biofilms. John Wiley & Sons, Ltd; 2015. Adhesins involved in attachment to abiotic surfaces by Gram-negative bacteria [Internet] pp. 163–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Essex-Lopresti AE, Boddey JA, Thomas R, et al. A type IV Pilin, PilA, contributes to Adherence of Burkholderia pseudomallei and virulence in vivo. Infect. Immun. 2005;73:1260–1264. doi: 10.1128/IAI.73.2.1260-1264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Okaro U, Mou S, Lenkoue G, Williams JA, Bonagofski A, Larson P, et al. A type IVB pilin influences twitching motility and in vitro adhesion to epithelial cells in Burkholderia pseudomallei. Microbiology. 2022;168:001150. doi: 10.1099/mic.0.001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jan AT. Outer Membrane Vesicles (OMVs) of Gram-negative bacteria: A perspective update. Front. Microbiol. 2017;8:1053. doi: 10.3389/fmicb.2017.01053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang W, Chanda W, Zhong M. The relationship between biofilm and outer membrane vesicles: a novel therapy overview. FEMS Microbiol. Lett. 2015;362:fnv117. doi: 10.1093/femsle/fnv117. [DOI] [PubMed] [Google Scholar]
- 134.Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kumar B, Sorensen JL, Cardona ST. A c-di-GMP-modulating protein regulates swimming motility of Burkholderia cenocepacia in response to arginine and glutamate. Front. Cell. Infect. Microbiol. 2018;8:56. doi: 10.3389/fcimb.2018.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FSL. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44:D646–53. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Madeira F, Pearce M, Tivey ARN, Basutkar P, Lee J, Edbali O, et al. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022;50:W276–W279. doi: 10.1093/nar/gkac240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data for this paper are available on-line only at http://jmb.or.kr.