Abstract
One of the most salient features of Bacillus subtilis and related bacilli is their natural capacity to secrete a variety of proteins into their environment, frequently to high concentrations. This has led to the commercial exploitation of bacilli as major “cell factories” for secreted enzymes. The recent sequencing of the genome of B. subtilis has provided major new impulse for analysis of the molecular mechanisms underlying protein secretion by this organism. Most importantly, the genome sequence has allowed predictions about the composition of the secretome, which includes both the pathways for protein transport and the secreted proteins. The present survey of the secretome describes four distinct pathways for protein export from the cytoplasm and approximately 300 proteins with the potential to be exported. By far the largest number of exported proteins are predicted to follow the major “Sec” pathway for protein secretion. In contrast, the twin-arginine translocation “Tat” pathway, a type IV prepilin-like export pathway for competence development, and ATP-binding cassette transporters can be regarded as “special-purpose” pathways, through which only a few proteins are transported. The properties of distinct classes of amino-terminal signal peptides, directing proteins into the various protein transport pathways, as well as the major components of each pathway are discussed. The predictions and comparisons in this review pinpoint important differences as well as similarities between protein transport systems in B. subtilis and other well-studied organisms, such as Escherichia coli and the yeast Saccharomyces cerevisiae. Thus, they may serve as a lead for future research and applications.
GENERAL INTRODUCTION
A common feature in cells of prokaryotic and eukaryotic origin is the export of proteins from their site of synthesis, mostly the cytoplasm, to other destinations either inside or outside the cell. To achieve this, exported proteins are usually synthesized as precursors with an amino-terminal, transient “zip code” (signal peptide), which is recognized and deciphered by a cellular sorting and translocation machinery (318–320). Signal peptides consist of short stretches of amino acids which, after protein delivery to the correct subcellular compartment, are frequently removed by specialized signal peptidases. In general, a preprotein is first recognized by soluble targeting factors for its transport to the target membrane, where the protein becomes associated with a translocation machinery. Next, the polypeptide chain is transported through a proteinaceous channel. In most cases this transport process is driven by a translocation motor that binds and hydrolyzes nucleoside triphosphates. Finally, the signal peptide is removed, resulting in release of the mature protein from the translocase. If the protein is translocated in an unfolded conformation, the mature protein will fold into its native conformation shortly after release from the translocase. Notably, several integral membrane proteins retain their signal-like peptides and diffuse from the translocase laterally. These basic principles of protein transport across membranes apply to most eukaryotic and prokaryotic organisms (84, 216, 231, 249).
In eukaryotic cells, proteins can be transported to numerous destinations, such as the nucleus, the endoplasmic reticulum (ER), the Golgi apparatus, lysozomes, the plasma membrane, the cell wall, chloroplasts, mitochondria, peroxisomes, and the different membrane systems or compartments within the organelles mentioned. Furthermore, proteins can be secreted into the external environment of the cell. In contrast, in eubacterial and archaeal cells, protein sorting seems to be limited to a few compartments, such as the cytoplasmic membrane, the cell wall (gram-positive eubacteria and archaea), the periplasm, and the outer membrane (gram-negative eubacteria). In addition, eubacteria and archaea can secrete proteins directly into their growth medium. In order to do so, these unicellular organisms can exploit multiple pathways, such as the general secretory (Sec) pathway, the twin-arginine translocation (Tat) pathway, and ATP-binding cassette (ABC) transporters.
SCOPE OF THIS REVIEW—THE SECRETOME
In the following sections of this review, the known signal peptide-dependent protein transport pathways, as they are present in the gram-positive eubacterium Bacillus subtilis, will be discussed, with a strong focus on the Sec pathway. Notably, the protein export machineries of the gram-negative eubacterium Escherichia coli and certain eukarya, such as the yeast Saccharomyces cerevisiae, have in many cases been characterized in more detail than those of B. subtilis. For matters of comparison, these machineries will also be discussed where appropriate. Because of the differences in the cell envelope structure, differences in the machineries for protein export in B. subtilis and E. coli were anticipated more than a decade ago (205). As described in this review, such differences do indeed exist, especially at the early and late stages of protein export, making detailed characterization of the underlying molecular mechanisms a fascinating scientific challenge.
B. subtilis and related Bacillus species are well known for their industrial use in the production of secreted proteins. These eubacterial species are particularly attractive for this purpose because they have a high capacity to secrete proteins into the growth medium and because of their nonpathogenicity. Moreover, good fermentation technologies exist for various bacilli (36, 40, 41, 263). Many proteins can be secreted to very high levels by B. subtilis, such as the α-amylase AmyQ from Bacillus amyloliquefaciens (1 to 3 g/liter) (204), protein A from Staphylococcus aureus (>1 g/liter) (91), and human interleukin-3 (227). Although not precisely documented in the scientific literature, about 10-fold-higher secretion levels can be reached in optimized industrial fermentation systems using Bacillus amyloliquefaciens or Bacillus licheniformis strains. Unfortunately, the secretion of proteins of gram-negative eubacterial or eukaryotic origin by Bacillus species is often severely hampered due to several bottlenecks in the secretion pathway, such as poor targeting to the translocase, degradation of the secretory protein, and slow or incorrect folding. Therefore, it is not only of scientific but also of applied interest to define the so-called secretome of B. subtilis, which includes both the components of machineries for protein secretion and the native secreted proteins. In recent years, considerable progress has been made concerning the identification and characterization of host functions needed for protein secretion by B. subtilis. In particular, this progress was facilitated by the availability of the complete genome sequence of B. subtilis (149). Present research efforts on protein transport in B. subtilis are aimed first at obtaining a complete description of the secretome and second at identifying those secretome components that are limiting factors in secretion. This review will provide a first, largely genome-based survey of the secretome.
PROTEIN TRANSPORT IN B. SUBTILIS
At first sight, protein transport in B. subtilis appears to be a relatively simple process, as its cell structure is considerably less complicated than that of eukaryotic cells. The cytoplasm is surrounded by the cytoplasmic membrane, which is covered by a thick layer (10 to 50 nm) of peptidoglycan-containing anionic polymers, such as teichoic and teichuronic acid. All proteins of B. subtilis lacking transport signals will be retained in the cytoplasm and fold, with or without the aid of chaperones, such as GroEL-GroES and DnaK-DnaJ-GrpE, into their native conformation (16, 90, 111, 113, 193). Other proteins contain membrane-spanning domains that are required for their insertion into the cytoplasmic membrane.
Most proteins that are completely transported across the cytoplasmic membrane are synthesized with an amino-terminal signal peptide. As B. subtilis, like other gram-positive eubacteria, lacks an outer membrane, many of these proteins are secreted directly into the growth medium. In most cases, these secreted proteins are enzymes involved in the hydrolysis of natural polymers, such as proteases, lipases, carbohydrases, DNases, and RNases. Such degradative enzymes are frequently synthesized as part of an adaptive response to changes in the environment, allowing the cell to benefit optimally from the available resources (95, 177, 263). Subsequently, specialized uptake systems in the cytoplasmic membrane internalize (partially) degraded substrates (14, 104). A second well-described class of secreted proteins, consisting of seven relatively small proteins, denoted PhrA to PhrK, are used to sense the cell density of the population, thereby regulating the onset of post-exponential-phase processes, such as competence development and sporulation (152, 208). These Phr proteins are, after their secretion and processing into small peptides, reimported to fulfill their inhibitory action on certain cytoplasmic phosphatases (206, 207, 209, 271). In contrast to the degradative enzymes and Phr proteins, most other exported proteins, involved in processes such as cell wall turnover, substrate binding, or protein secretion (217, 281), have to be retained at the membrane-cell wall interface to fulfill their function. To prevent the loss of these proteins, they can contain signals for their attachment to the membrane (lipid modifications) or the cell wall. Alternatively, some exported proteins have the potential to form pilin-like structures at the membrane-cell wall interface.
Strikingly, under conditions of nutrient starvation, the formation of two genuine internal compartments, as encountered in organelles of the eukaryotic cell, is induced. These compartments, which ultimately develop into an endospore, are confined by two membranes, the forespore inner and outer membranes. The forespore inner membrane confines the cytosol of the forespore, while the forespore outer membrane forms the initial barrier between the forespore and the cytosol of the mother cell (89, 279) (for details, see the section on sporulation-specific protein transport). Recent data indicate that certain proteins are specifically sorted from the cytosol of the mother cell or the forespore to the intermembrane space (IMS) between the two forespore membranes. The process of subcellular compartmentalization during sporulation in particular underscores the fact that, though simple at first glance, complex mechanisms for protein sorting have evolved in B. subtilis.
In the following sections, the amino-terminal cleavable signal peptides which are involved in the transport of ribosomally synthesized proteins in B. subtilis will be discussed. Furthermore, the different protein export routes and retention mechanisms which prevent the loss of certain exported proteins in the environment will be described.
Amino-Terminal Signal Peptides
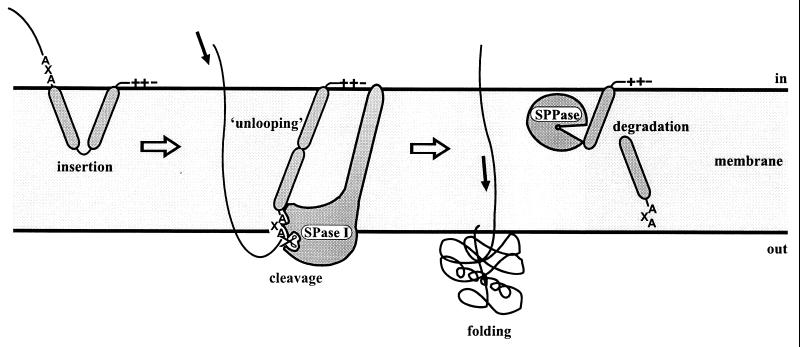
Although the primary structures of different amino-terminal signal peptides show little similarity, three distinct domains can nevertheless be recognized (225, 316–319, 321). The amino-terminal N-domain of signal peptides contains at least one arginine or lysine residue, although this positively charged residue does not seem to be strictly required for protein export (51, 101). The positively charged N-domain has been suggested to interact with the translocation machinery (1) and negatively charged phospholipids in the lipid bilayer of the membrane during translocation (71). The H-domain, following the N-domain, is formed by a stretch of hydrophobic residues that seem to adopt an α-helical conformation in the membrane (37). Helix-breaking glycine or proline residues are frequently present in the middle of this hydrophobic core. The latter residues might allow the signal peptide to form a hairpin-like structure that can insert into the membrane. In one model for signal peptide function, it was proposed that unlooping of this hairpin results in insertion of the complete signal peptide in the membrane (71) (Fig. 1). Helix-breaking residues found at the end of the H-domain, are thought to facilitate cleavage by a specific signal peptidase (SPase) (67, 202). The C-domain, following the H-domain, contains the cleavage site for SPase, which removes the signal peptide from the mature part of the secreted protein during or shortly after translocation. The mature part of the protein is thereby released from the membrane and can fold into its native conformation. Finally, the signal peptide is degraded by signal peptide peptidases (SPPases) and removed from the membrane (Fig. 1). Although different amino-terminal signal peptides tend to be quite similar in general structure, apparently small differences between individual signal peptides can cause cleavage by different SPases, export via different pathways, and transport to different destinations.
FIG. 1.
Model for signal peptide insertion into the cytoplasmic membrane and cleavage by SPase I. First, the positively charged N-domain of the signal peptide interacts with negatively charged phospholipids in the membrane, after which the H-domain integrates loopwise into the membrane. Next, the H-domain unloops, whereby the first part of the mature protein is pulled through the membrane. During or shortly after translocation by a translocation machinery (not shown), the signal peptide is cleaved by SPase I and subsequently degraded by SPPases. After its translocation across the membrane, the mature protein folds into its native conformation.
Signal Peptide Classification
At present, four major classes of amino-terminal signal peptides can be distinguished on the basis of the SPase recognition sequence. The first class is composed of “typical” signal peptides which are present in preproteins that are cleaved by one of the various type I SPases of B. subtilis (289, 290, 292). Although most proteins having such a signal seem to be secreted into the extracellular environment, some of them are retained in the cell wall or sorted specifically to the IMS of endospores after membrane translocation via the Sec pathway. Notably, a subgroup of these signal peptides contain a so called twin-arginine motif (RR-motif), which might direct proteins into a distinct translocation pathway known as the Tat pathway.
The second major class of signal peptides is present in prelipoproteins, which are cleaved by the lipoprotein-specific (type II) SPase of B. subtilis (Lsp) (223, 291, 293). The major difference between signal peptides of lipoproteins and secretory proteins is the presence of a well-conserved lipobox in lipoprotein precursors. This lipobox contains an invariable cysteine residue that is lipid modified by the diacylglyceryl transferase prior to precursor cleavage by SPase II. After translocation across the cytoplasmic membrane, exported lipid-modified proteins remain anchored to the membrane by their amino-terminal lipid-modified cysteine residue (see the section on lipoprotein signal peptides for details). Notably, some signal peptides of lipoproteins contain a typical RR-motif. Consequently, the possibility exists that certain lipoproteins are exported via the Tat pathway rather than the Sec pathway.
The third major class is formed by signal peptides of prepilin-like proteins, which, in B. subtilis, are cleaved by the prepilin-specific SPase ComC (53). The recognition sequence for the prepilin SPase is, in contrast to that of secretory and lipoproteins, localized between the N- and H-domains, leaving the H-domain attached to the mature pilin after cleavage (53, 54, 157, 225).
Finally, the fourth major class of signal peptides is found on ribosomally synthesized bacteriocins and pheromones that are exported by ABC transporters (11, 203, 335). These signal peptides lack a hydrophobic H-domain and are removed from the mature protein by a subunit of the ABC transporter that is responsible for the export of a particular bacteriocin or pheromone or by specific SPases.
HOW MANY PROTEINS ARE EXPORTED?
It is well established that B. subtilis can secrete certain proteins to high concentrations in the medium (184, 263). However, until recently it was difficult to estimate the number of exported proteins belonging to the secretome of B. subtilis. The completion of the B. subtilis genome sequencing project (149) and the availability of programs for the identification of signal peptides and transmembrane segments in large collections of protein sequences through worldwide web servers (194, 264) have now made it possible to predict the most likely location of all 4,107 annotated proteins (i.e., the proteome) of this organism. Computer-assisted studies have indicated that approximately 25% of the proteome of a given organism, such as B. subtilis, contains membrane sorting signals in the form of hydrophobic stretches of amino acids that can integrate in and span the membrane (35, 322; http://pedant.mips.biochem.mpg.de). Some of these putative membrane proteins contain amino-terminal signal peptides and may in fact be exported proteins, as indicated below.
Signal Peptide Predictions
To estimate the number of exported proteins, the amino termini of all annotated B. subtilis proteins in the SubtiList database (http://bioweb.pasteur.fr/GenoList/SubtiList) were used to predict amino-terminal signal peptides with the SignalP algorithm (194). This method incorporates a prediction of cleavage sites and a signal peptide/non-signal peptide prediction based on a combination of several artificial neural networks trained on the identification of signal peptides from gram-positive eubacteria. Next, all putative signal peptides were screened for the presence of a lipobox, RR-motif, or cleavage site for prepilin SPase. The numbers and features of each class of signal peptides are summarized in Fig. 2. It should be noted that polytopic membrane proteins, some of which can be cleaved by type I SPases (119, 288), were specifically excluded from the predictions, using the TopPred algorithm of Sipos and von Heijne (264). Furthermore, putative proteins with a single amino-terminal membrane-spanning domain, as encountered in certain type I SPases, might be falsely predicted to be secreted proteins. Finally, the neural networks of the SignalP algorithm, trained on data from gram-positive organisms, might not recognize some of the B. subtilis signal peptides with a more gram-negative or eukaryotic character.
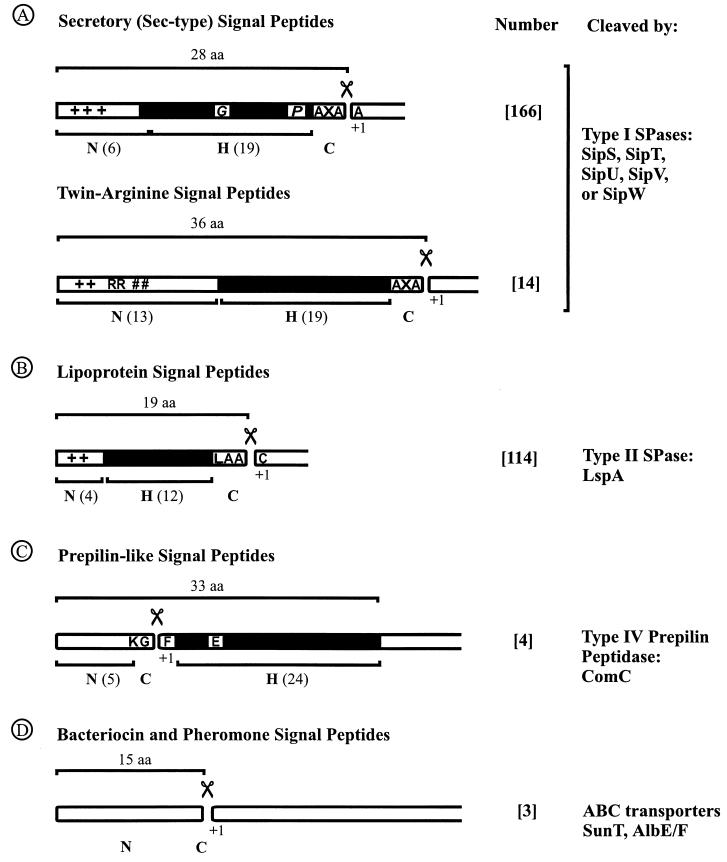
FIG. 2.
Numbers and features of predicted amino-terminal signal peptides found in (putative) exported proteins of B. subtilis. To estimate the number of exported proteins, the first 60 residues of all annotated proteins of B. subtilis in the SubtiList database (http://bioweb.pasteur.fr/GenoList/SubtiList) were used to predict amino-terminal signal peptides with the SignalP algorithm for the prediction of signal peptides of gram-positive eubacteria (194). Next, to distinguish between potential secretory proteins and multispanning membrane proteins, putative membrane-spanning segments in protein sequences with a putative signal peptide were predicted with the TopPred2 algorithm (61, 264). All proteins containing additional hydrophobic domains (upper cutoff, 1.0; lower cutoff, 0.6; window size top, 11; window size bottom, 21) were regarded as membrane proteins, and their amino termini were excluded from the primary set of signal peptides. Finally, all putative signal peptides were screened for the presence of a lipobox, twin-arginine motif, or a cleavage site for the prepilin SPase. On the basis of SPase cleavage sites, predicted signal peptides were divided into four distinct classes: A, secretory (Sec-type) signal peptides and twin-arginine signal peptides; B, lipoprotein signal peptides; C, prepilin-like signal peptides; and D, bacteriocin and pheromone signal peptides. The number of predicted B. subtilis signal peptides of each class and the SPases responsible for their cleavage are indicated. Most signal peptides have a tripartite structure: a positively charged N-domain (N), containing lysine and/or arginine residues (indicated with +); a hydrophobic H-domain (H, indicated by a black box); and a C-domain (C) which specifies the cleavage site for SPase, as indicated by the scissors symbol. The average lengths of the complete signal peptide, N-domain, H-domain, and consensus SPase recognition sequences are indicated. Furthermore, helix-breaking residues, mostly glycine or proline (G/P) in the H-domain of certain signal peptides, are indicated. These residues are thought to facilitate loopwise membrane insertion and cleavage by SPase I, respectively (Fig. 1) (202). Finally, where appropriate, the most frequently occurring first amino acid (aa) of the mature protein (+1) is indicated.
Secretory (Sec-type) signal peptides.
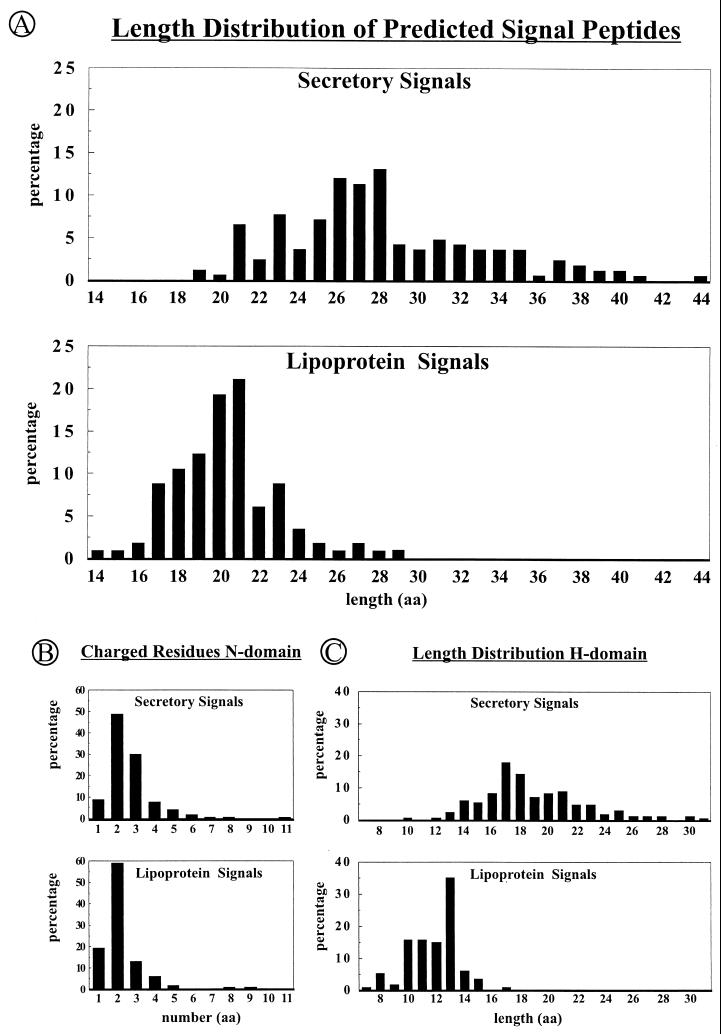
The signal peptide predictions resulted in the identification of 180 potential substrates for type I SPases. An RR-motif containing at least three residues of the R-R-X-#-# (where # is a hydrophobic residue) consensus sequence (18, 60) was found in 14 of these signals, suggesting that the corresponding preproteins are transported in a Sec-independent manner. The remaining 166 predicted “Sec-type” signal peptides (Table 1) had a length varying from 19 to 44 residues, with an average of 28 residues. These signal peptides contain on average two or three positively charged lysine (K) or arginine (R) residues in their N-domain, although some of the N-domains contain as many as 5 to 11 positively charged residues. The hydrophobic core (H-domain) has an average length of 19 residues, although a length of 17 or 18 residues seems to be preferred (Fig. 2 and 3). The C-domain of the predicted signal peptides carries a type I SPase cleavage site, with the consensus sequence A-X-A at position −3 to −1 relative to the SPase I cleavage site (Table 2). It is important to note that the C-domain must have an extended (β-sheeted) structure for efficient interaction with the active site of type I SPases. Based on the crystal structure of the type I SPase of E. coli, the side chains of residues at the −1 and −3 positions are thought to be bound in two shallow hydrophobic substrate-binding pockets (S1 and S3) of the active site, whereas the side chain of the residue at position −2 is pointing outwards from the enzyme (202). It is presumably for this reason that residues tolerated at positions −3 and −1 of the signal peptide are generally small and uncharged, while almost all residues (except cysteine and proline) seem to be allowed at position −2 (Table 2). Nevertheless, a preference for serine (18%) at position −2 of the signal peptide seems to exist in B. subtilis. According to the predictions, an alanine residue is most abundant (27%) at position +1 of the mature protein, but all other residues, with the exception of cysteine and proline, seem to be allowed at this position (Table 2). The absence of proline at the +1 position is consistent with the observation that the SPase I of E. coli was inhibited by recombinant preproteins with proline at this position (12, 196). Finally, approximately 60% of the predicted signal peptides contain a helix-breaking residue (mostly glycine) in the middle of the H-domain, and about 50% contain a helix-breaking residue (proline or glycine) at position −7 to −4 relative to the predicted processing site for SPase I.
TABLE 1.
Predicted secretory (Sec-type) signal peptides of B. subtilisa
Putative signal peptides were identified as described in the text under the heading Signal Peptide Predictions. Positively charged lysine (K) and arginine (R) residues in the N-domain are indicated in bold letters. The hydrophobic H-domain is indicated in gray shading. The residues at positions −3 to −1 relative to the predicted SPase I cleavage site are underlined, and the SPase cleavage site is indicated with a gap in the amino acid sequence. Proteins containing additional cell wall-binding repeats in the mature part of the protein are indicated with a superscript W.
FIG. 3.
Features of predicted secretory and lipoprotein signal peptides. (A) Length distribution of complete signal peptides (N-, H-, and C-domains). (B) Distribution of positively charged lysine or arginine residues in the N-domains of predicted signal peptides. (C) Length distribution of the hydrophobic H-domains in predicted signal peptides. Distributions are indicated as percentages of the total number of predicted secretory or lipoprotein signal peptides.
TABLE 2.
Amino acid residues around (putative) SPase I cleavage sitesa
| Position and residue |
Frequency (% of total) |
|---|---|
| −3 | |
| A | 60 |
| V | 18 |
| I | 6 |
| S | 5 |
| G | 5 |
| T | 3 |
| L | 2 |
| F | 1 |
| Q | <1 |
| M | <1 |
| K | <1 |
| Y | <1 |
| W | <1 |
| −2 | |
| S | 18 |
| F | 11 |
| K | 10 |
| E | 8 |
| L | 8 |
| Q | 7 |
| A | 7 |
| H | 6 |
| I | 5 |
| D | 5 |
| T | 4 |
| G | 4 |
| N | 4 |
| V | 3 |
| M | 2 |
| R | 2 |
| Y | 2 |
| W | 1 |
| −1 | |
| A | 85 |
| S | 6 |
| G | 5 |
| V | 4 |
| L | 3 |
| I | 2 |
| E | 1 |
| N | <1 |
| +1 | |
| A | 27 |
| K | 10 |
| Q | 10 |
| S | 8 |
| V | 5 |
| F | 5 |
| D | 4 |
| N | 4 |
| Y | 3 |
| L | 4 |
| I | 4 |
| H | 3 |
| G | 2 |
| T | 2 |
| E | 1 |
| R | 1 |
| W | 1 |
| M | <1 |
The frequency of a particular amino acid at each position is given as the percentage of the total number of predicted signal peptides in which it appears.
Twin-arginine signal peptides.
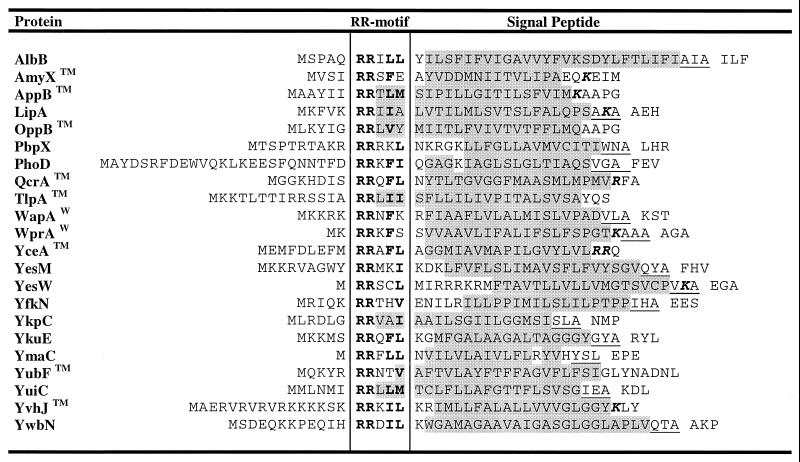
Proteins containing a signal peptide with the RR-motif (R-R-X-#-#, where # is a hydrophobic residue) may be transported via the Tat pathway. Through a database search for the presence of this motif in amino-terminal protein sequences, a total number of 27 putative RR-signal peptides were identified. Putative SPase I cleavage sites are present in 14 of these predicted signal peptides. These cleavage sites show no striking differences from those of the predicted Sec-type signal peptides (Fig. 2 and Table 3). Notably, the RR-motif was also found in the signal peptides of five putative lipoproteins, suggesting that these proteins might also be substrates for the Tat pathway (Table 4). Moreover, eight additional signal peptide-like sequences with the RR-motif but lacking cleavage sites for SPase I or SPase II were identified. The corresponding proteins have the potential to remain attached to the membrane with an amino-terminal transmembrane domain (Table 3). Interestingly, some of these proteins even contain additional transmembrane segments. Thus, the possibility exists that certain membrane proteins are translocated via the Tat pathway. Altogether, the N-domains of predicted RR-signal peptides of B. subtilis have an average length of 13 amino acid residues and are twice as long as the N-domains of the typical (Sec-type) signals. Strikingly, no significant differences are observed between the H-domains of predicted Sec-type and RR-signal peptides of B. subtilis. In contrast, it has been suggested that the H-domains of RR-signal peptides of E. coli are, on average, longer and less hydrophobic than those of Sec-type signal peptides of this gram-negative organism (60). These observations may suggest either that a difference in the H-domains of Sec-type and RR-signal peptides is not important for translocation via the Tat pathway in B. subtilis or that some of the predicted RR-signal peptides do not direct proteins into the Tat pathway. For example, it is conceivable that the latter possibility could apply to WapA and WprA, the secretion of which was impaired by Ffh or SecA depletion (119). Finally, positively charged residues (arginine or lysine) in the C-domain, which can function as a so-called Sec avoidance signal that prevents interactions with Sec pathway components (23), are present in 8 of the 27 putative twin-arginine signal peptides.
TABLE 3.
Predicted twin-arginine signal peptidesa
Amino-terminal signal peptides were identified as described in the text. Conserved residues of the RR-motif are indicated in bold letters. Only signals containing, in addition to the twin arginines, at least one other residue of the consensus sequence (R-R-X-#-#) (60) were included in this comparison. Positively charged residues in the C-domain that could function as a so-called Sec avoidance motif (23) are indicated in bold and italics. The hydrophobic H-domain is indicated in gray shading. In signal peptides with a predicted SPase I cleavage site, residues from positions −3 to −1 relative to the SPase I cleavage site are underlined, and this site is indicated with a gap in the amino acid sequence. Proteins containing additional cell wall retention signals in the mature part of the protein are indicated with a superscript W; proteins lacking a (putative) type I cleavage site, some of which contain additional transmembrane domains in their “mature” part, are indicated with a superscript TM.
TABLE 4.
Predicted lipoprotein signal peptidesa
Putative lipoprotein signal peptides were identified as described in the text. Positively charged lysine (K) and arginine (R) residues in the N-domain are indicated in bold letters. The hydrophobic H-domain is indicated in gray shading. The residues at positions −3 to +1, forming the lipobox, are underlined. The SPase II cleavage site is indicated with a gap in the amino acid sequence. Leucine residues at position −3 and strictly conserved cysteine residues at position +1 are indicated in bold letters. Lipoproteins containing (putative) transmembrane domains in the mature part of the protein are indicated with a superscript TM. Conserved residues of the RR-motif (R-R-X-#-#) (60) in the N-domains of the putative lipoproteins SpoIIIJ, YdeJ, YdhF, YdhK, and YmzC (indicated with a superscript RR) are enlarged. Based on theoretical considerations, the putative start sites of the potential lipoproteins YhaR and YhfQ (indicated with ∗) have been modified in a recent update of SubtiList. If the new annotation is correct, it is uncertain whether YhaR and YhfQ are lipoproteins.
Lipoprotein signal peptides.
As the signal peptides of lipoproteins are, in general, shorter than those of secretory proteins, not all lipoproteins are recognized by the SignalP algorithm (194; our unpublished observations). In addition, some lipoproteins, such as CtaC (17) (Table 4), contain multiple membrane-spanning segments that were excluded from predictions of the signal peptides of secretory proteins mentioned above. Therefore, additional putative lipoprotein signal peptides were identified through similarity searches in the SubtiList database with signal peptides of known lipoproteins, using the Blast algorithm (4). Putative lipoprotein sorting signals identified by the latter method were combined with those identified by SignalP, resulting in a total number of 114 (Table 4). Signal peptides from lipoproteins differ in several respects from those of secretory signals. First, the structural features of lipoprotein signal peptides are more conserved than those of secretory signal peptides. This suggests that less variation in these peptides is allowed by the components involved in lipid modification and processing of lipoproteins. The C-domain contains a so-called lipobox with the consensus sequence L-(A/S)-(A/G)-C (Table 5), of which the invariable cysteine residue is the target for lipid modification and becomes the first residue of the mature lipoprotein after cleavage by SPase II. Second, both the N-domain (average of four residues) and the H-domain (average of 12 residues) seem, on average, to be shorter than the corresponding domains of signal peptides of nonlipoproteins (Fig. 2 and 3). Finally, helix-breaking residues are less abundant (27%) in the H-domain of lipoprotein signal peptides than in the corresponding regions of nonlipoprotein signal peptides. As transmembrane helices seem to require at least 14 hydrophobic residues to span the membrane (34, 46), the latter findings suggest that not all lipoprotein signal peptides can span the membrane completely. This implies that the active site of SPase II may be embedded in the cytoplasmic membrane, as was suggested for SPase I (202, 309). Alternatively, the N-domain of the signal peptide may not stay fixed at the cytoplasmic surface of the membrane during translocation. Strikingly, aspartic acid was absent from the +2 position of predicted mature lipoproteins. In mature lipoproteins of gram-negative eubacteria, an aspartic acid residue at the +2 position specifically prevents the sorting of these proteins to the outer membrane (167, 168, 221). The fact that aspartic acid at the +2 position is absent from Bacillus lipoproteins suggests that this sorting signal has evolved exclusively in gram-negative eubacteria. Nevertheless, it has to be noted that glycine, phenylalanine, or tryptophan residues can be found at the +2 position of various lipoproteins of B. subtilis (Table 5). Like aspartic acid, these residues prevent lipoprotein sorting to the outer membrane of E. coli (260). Thus, even though B. subtilis lacks an outer membrane, residues with a potential sorting function can be found at the +2 position of mature lipoproteins of B. subtilis. It is therefore conceivable that such residues are involved in the targeting of lipoproteins to specific membrane locations in B. subtilis. Nevertheless, it has to be emphasized that presently no experimental data are available to support this idea.
TABLE 5.
Amino acid residues around (putative) SPase II cleavage sitesa
| Position and residue |
Frequency (% of total) |
|---|---|
| −3 | |
| L | 65 |
| V | 8 |
| F | 7 |
| T | 6 |
| I | 5 |
| M | 2 |
| G | 2 |
| S | <1 |
| W | <1 |
| −2 | |
| A | 36 |
| S | 24 |
| T | 11 |
| I | 6 |
| V | 6 |
| G | 6 |
| M | 2 |
| L | 2 |
| C | 2 |
| P | 2 |
| F | 2 |
| L | 2 |
| D | <1 |
| N | <1 |
| −1 | |
| A | 39 |
| G | 35 |
| L | 8 |
| I | 4 |
| S | 4 |
| V | 2 |
| T | 2 |
| F | 2 |
| P | 2 |
| M | <1 |
| C | <1 |
| E | <1 |
| +1 | |
| C | 100 |
| +2 | |
| G | 30 |
| S | 28 |
| A | 9 |
| I | 4 |
| T | 4 |
| F | 4 |
| W | 4 |
| K | 4 |
| L | 4 |
The frequency of a particular amino acid at each position is given as the percentage of the total number of predicted lipoprotein signal peptides in which it appears.
Type IV prepilin signal peptides.
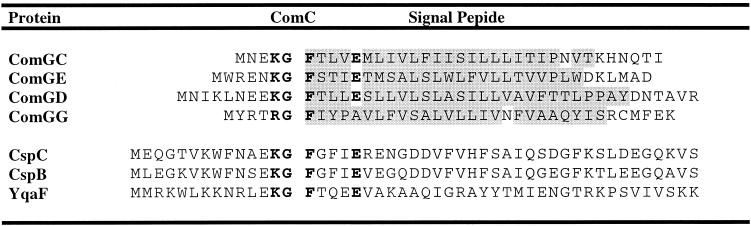
Only four proteins required for DNA binding and uptake during competence (ComGC, ComGE, ComGD, and ComGG), which are already known to contain type IV prepilin-like signal peptides (55), were identified in our SignalP and Blast homology searches for prepilin-like signal peptides in B. subtilis (Table 6). As the prepilin SPase (ComC) acts at the cytoplasmic side of the membrane (151, 157), the “C-domain” with the ComC cleavage site is localized between the N- and H-domains of the prepilin signal peptide (Fig. 2). After cleavage by ComC, the hydrophobic H-domain remains attached to the mature protein. Surprisingly, potential prepilin cleavage sites are also present in two known cold shock proteins, CspB and CspC (108), and one hypothetical protein, YqaF (Table 6), which have a (predicted) cytosolic localization as they lack an H-domain or transmembrane segments. Since the catalytic domain of ComC is located at the cytoplasmic side of the membrane, it is conceivable that cytoplasmic proteins containing a ComC cleavage site are substrates for this enzyme. However, it is presently not known whether CspB, CspC, or YqaF is processed.
TABLE 6.
Predicted type IV prepilin signal peptides and related sequencesa
Prepilin-like signal peptides were identified as described in the text. The hydrophobic H-domain is indicated in gray shading. The ComC recognition sequence in prepilin signal peptides and putative ComC recognition sequences in CspC, CspB, and YqaF are indicated in bold letters. Predicted ComC cleavage sites are indicated with a gap in the amino acid sequence.
Signal peptides of bacteriocins and pheromones.
Bacteriocins and pheromones (other than PhrA to PhrK) with a cleavable amino-terminal signal peptide form a distinct group of exported proteins that are exported via ABC transporters. Signal peptides of this class of secreted proteins cannot be predicted by the regular algorithms for signal peptide prediction, such as SignalP (see previous sections), as they consist only of N- and C-domains and completely lack a hydrophobic H-domain (Fig. 2). In B. subtilis 168, the three known signal peptides of this type direct the secretion of the bacteriocins sublancin 168 (203) and subtilosin (336) and the pheromone ComX (159). Thus far, no other signal peptides with a similar structure have been identified in the amino acid sequences of B. subtilis proteins. Obviously, this does not exclude the possibility that less related signal peptides of this type do exist, particularly in view of the fact that at least 77 (putative) ABC transporters have been identified in B. subtilis (149).
Protein Traffic
From the prediction of signal peptides and transmembrane regions, the percentage of the proteome that is transported from the cytoplasm to other cellular compartments can be estimated. The transport pathways followed by these (putative) preproteins will be discussed in more detail in the following paragraphs. Approximately 75% of the proteome of B. subtilis lacks an amino-terminal signal peptide or membrane anchor, and most of the corresponding proteins are likely to be localized in the cytoplasm. Proteins with (putative) amino-terminal signal peptides (∼7%) or transmembrane segments (∼18%) are likely to be targeted to the cytoplasmic membrane and (partially) translocated. A large portion (∼21%) of the B. subtilis proteins remain linked to the membrane as transmembrane proteins (∼18%), as lipid-modified proteins (∼2.5%) that remain linked to the extracytoplasmic surface of the membrane by their lipid moieties, or as pilin-like structures (<0.1%). A small portion of the putative exported proteins most likely remain specifically attached to the cell wall (∼0.5%; see the section on cell wall retention), whereas most of the remaining exported proteins (∼4%) have the potential to pass through the cell wall and be secreted into the environment.
Most proteins seem to be exported or inserted into the cytoplasmic membrane via the Sec pathway in B. subtilis. Nevertheless, several alternative export pathways seem to exist. First, the recently identified twin-arginine translocation (Tat) pathway seems to be present in B. subtilis, as judged from the identification of signal peptides with the RR-motif and conserved components of this pathway (see the section on Sec-independent protein transport). Protein secretion via this pathway was shown to be independent of Sec components in E. coli and plant chloroplasts. Possibly, this pathway has evolved specifically for the export of folded preproteins (65). Second, the assembly of extracellular prepilin-like structures depends on components which are, most likely, not involved in Sec-dependent protein secretion. Finally, at least three small prepeptides contain signal peptides lacking a hydrophobic domain. These prepeptides are transported across the membrane and cleaved by ABC transporters. The requirements for traveling via one of these export pathways are summarized in Fig. 4. In the following sections, each of these export pathways will be discussed in more detail.
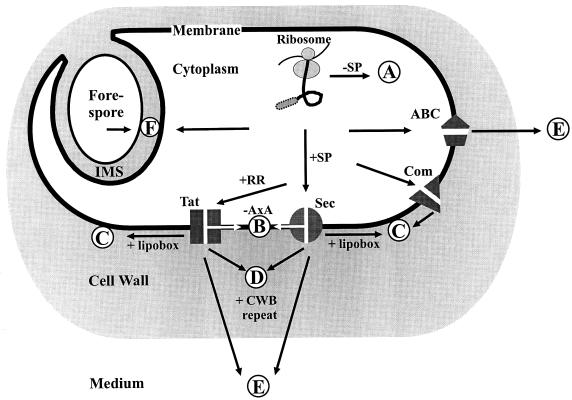
FIG. 4.
Predicted protein transport pathways in B. subtilis. Based on the predictions of signal peptides and various retention signals, it is hypothesized that at least four different protein transport pathways exist in B. subtilis that can direct proteins to at least five different subcellular destinations. Ribosomally synthesized proteins can be sorted to various destinations depending on the presence (+SP) or absence (−SP) of an amino-terminal signal peptide and specific retention signals such as lipid modification or cell wall-binding repeats (CWB). (A) Proteins devoid of a signal peptide remain in the cytoplasm. (B) Proteins with one or more membrane-spanning domains are inserted into the membrane either spontaneously (not shown), via the Sec pathway or, according to our predictions (Table 3), via the Tat pathway (+RR). (C) Proteins which have to be active at the extracytoplasmic side of the membrane can either be lipid-modified proteins (+lipobox) exported via the Sec or Tat pathways or prepilins exported by the Com system. (D) Proteins that need to be retained in the cell wall can be exported via the Sec or Tat pathway. In order to be retained, the mature part of these proteins contains cell wall-binding repeats (+CWB). (E) Proteins can be secreted into the medium via the Sec or Tat pathway or by ABC transporters. (F) Different mechanisms can be employed to transport proteins to the IMS of endospores.
THE SEC-DEPENDENT SECRETION MACHINERY
The various components of the Sec-dependent secretion machinery can be divided into six groups: cytosolic chaperones, the translocation motor (SecA), components of the translocation channel (SecYEG and SecDF-YajC), SPases, SPPases, and, finally, folding factors that function at the trans side of the membrane. The main components of the secretion machinery of B. subtilis are depicted in Fig. 5 and listed in Table 7. In addition, the homologous and/or analogous components from E. coli, the archaeon Methanococcus jannaschii, and the eukaryon S. cerevisiae are listed in Table 7. As protein secretion has not been studied experimentally in archaea, the identification of components of the secretion machinery of M. jannaschii is based entirely upon data deduced from the genome sequence (44).
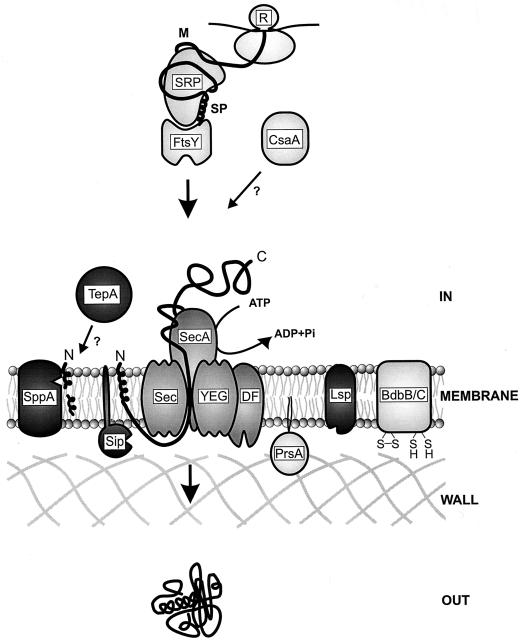
FIG. 5.
Main components of the Sec-dependent B. subtilis protein secretion machinery. The SRP complex consists of Ffh and the scRNA. See text for further details. C, carboxyl terminus; M, mature protein; N, amino terminus; R, ribosome; SP, signal peptide.
TABLE 7.
Components of Sec-dependent protein export machineries of B. subtilis, E. coli, M. jannaschii, and the ER of S. cerevisiaea
| Component | B. subtilis | E. coli | M. jannaschii | S. cerevisiae |
|---|---|---|---|---|
| Secretion-dedicated chaperones | Ffh, FtsY, scRNA | Ffh, FtsY, 4.5S RNA | Srp54/19, FtsY, 7S RNA | SRP complex,b DPα/β |
| SecB | ||||
| CsaA (?) | YgjH (?) | |||
| General chaperones | GroEL, GroES | GroEL, GroES | ||
| DnaK, DnaJ, GrpE | DnaK, DnaJ, GrpE | Hsp70, Ydj1 | ||
| Trigger factor | Trigger factor | |||
| Translocation motor | SecA | SecA | ||
| Ribosome | ||||
| Kar2 (Bipc) | ||||
| Translocation channel | SecY | SecY | SecY | Sec61, Ssh1 (Sec61αc) |
| SecG | SecG | Sec61β | Sbh1, Sbh2 (Sec61βc) | |
| SecE | SecE | SecE | Sss1 (Sec61γc) | |
| SecDF, YrbF | SecD/F, YajC | SecD/F | ||
| Sec62/63 | ||||
| Sec66/67 | ||||
| SPases | SipS/T/U/V/P | LepB | ||
| SipW | Sec11 | Sec11 | ||
| LspA | LspA | |||
| SPPases | SppA | SppA | SppA | |
| TepA | ||||
| OpdA | ||||
| Foldases (trans-acting) | PrsA | SurA, PpiD, RotA, FkpA | CPR2, CPR4, CPR5, CPR8, FPR2 | |
| BdbA/B/C | DsbA/B/C/D/E/G | PDI, Ero1, Eug1 | ||
| Kar2 (BiPc), Lhs1 (Hsp70c) |
Proteins with similar functions are placed in the same horizontal row.
The SRP complex consists of scR1 RNA, Srp72p, Srp68p, Srp54p, Sec65p, Srp21p, Srp14p, and Srp7p (338).
Synonymous names for mammalian homologues.
Cytosolic Chaperones
Most proteins that are destined for export can only be translocated across the membrane in a more or less unfolded conformation that allows them to pass through the translocation channel of the Sec pathway. To facilitate this, cytosolic factors aid in maintaining these preproteins in a so-called translocation-competent state. Such factors, called chaperones, bind to preproteins and prevent their folding and aggregation. Some of these chaperones are secretion dedicated and assist in protein targeting to the translocase.
Secretion-dedicated chaperones.
In B. subtilis, the only secretion-specific chaperone thus far identified is the Ffh protein (fifty-four homologue), a GTPase that is homologous to the 54-kDa subunit of the eukaryotic signal recognition particle (Srp54) (122). This protein forms a complex (denoted SRP) with the small cytoplasmic RNA (scRNA) that is functionally related to the eukaryotic 7S RNA (called scR1 RNA in S. cerevisiae) and the E. coli 4.5S RNA (186, 187). Recent data have shown that HBsu, a histone-like protein of B. subtilis, is also associated with the scRNA. Notably, HBsu was shown to bind to a region of scRNA that is not conserved in the 4.5S RNA of E. coli, suggesting that the E. coli SRP lacks an HBsu-like component (188, 333). The ternary ribonucleoprotein SRP complex of B. subtilis binds to the signal peptides of nascent chains emerging from the ribosome and is targeted to the membrane with the aid of the FtsY protein (also called Srb) (200). FtsY is a homologue of the eukaryotic SRP receptor α-subunit (DPα) that is essential for SRP-dependent protein secretion and cell viability, like the Ffh protein. In eukaryotic cells, SRP-dependent protein translocation occurs cotranslationally. The SRP of S. cerevisiae consists of a complex of seven subunits and the 7S RNA. Two of these subunits, Srp7 (Srp9 in mammalian cells) and Srp14, are responsible for a translation arrest as soon as the signal peptide emerges from the ribosome (262). The whole complex, consisting of the ribosome, nascent chain, and SRP, docks onto the SRP receptor (also termed docking protein), which consists of DPα and DPβ. Next, SRP is released and polypeptide translation by the ribosome is resumed. Protein synthesis is likely to provide the driving force for cotranslational protein translocation across membranes (249). A similar SRP-mediated translation arrest probably does not occur in eubacteria. First, it was shown that E. coli SRP and FtsY do not arrest translation in a eukaryotic in vitro translocation assay (222). Second, eubacteria lack the SRP components that are responsible for translation arrest in eukaryotes. Moreover, a specific translation arrest may not even be required for cotranslational translocation in eubacteria, because the traffic distances are short and the protein translocation rates are high compared to the translation rate (for E. coli, estimated at approximately 10-fold) (225, 302).
In E. coli, a second protein-targeting pathway utilizes the SecB protein that was shown to bind to the mature regions of a subset of preproteins (148) and the carboxyl terminus of SecA (92). Strikingly, recent results showed that the SecB-binding motif, consisting of a stretch of ∼9 amino acids enriched in aromatic and basic residues, occurs in SecB-dependent and -independent secretory proteins and in cytosolic proteins (141). Moreover, SecB appeared to bind these three classes of proteins with more or less similar affinities. This suggests that SecB may also be regarded as a general chaperone that promotes protein translocation by its specific binding to SecA, together with an associated preprotein (141). For E. coli, it was shown that the SRP route converges with the SecB-dependent targeting route at the translocase (302), and it was proposed that the SRP route acts primarily cotranslationally, while the SecB-dependent route acts mainly posttranslationally. Recent studies showed that SecA, SecB, SecE, and ATP are dispensable for the transfer of certain (nascent) membrane proteins to SecY and their subsequent insertion into the membrane, suggesting that SRP-FtsY and SecB-SecA constitute distinct targeting, or even translocation, routes (59, 69, 142, 256). The choice between these two routes seems to depend largely on the hydrophobicity of the targeting signal, as the more hydrophobic signal peptides target proteins primarily into the SRP pathway (300, 301). Altogether, these findings suggest that the SRP route of E. coli is mainly involved in the targeting of inner membrane proteins, whereas the SecA-SecB route is primarily involved in the targeting of periplasmic and outer membrane proteins (15, 256, 257, 299). As the signal peptides of B. subtilis are, on average, longer and more hydrophobic than those of E. coli (321), thereby closely resembling transmembrane segments of integral membrane proteins (see above), it seems likely that the majority of B. subtilis proteins are secreted in an SRP-dependent manner. This view seems to be supported by recent experiments of Hirose et al. (119), showing that the secretion of a large number of B. subtilis proteins was directly or indirectly Ffh dependent. Notably, the secretion of most of these proteins depended on SecA as well, suggesting that SRP-dependent protein secretion does not bypass SecA in B. subtilis.
Interestingly, no SecB homologue is present in B. subtilis, but it is conceivable that a SecB analogue does exist in this organism. For example, the SecB-binding domain of E. coli SecA is highly conserved in the SecA protein of B. subtilis (92). This binding domain might serve as a docking site for a SecB analogue, but obviously, other sites in the B. subtilis SecA protein could also serve this purpose. A candidate for a SecB analogue in B. subtilis is the CsaA protein, which was identified as a suppressor of an E. coli SecA(Ts) mutant (178). Recently, it was demonstrated that CsaA has chaperone-like activities (179) and affinity for SecA and preproteins (179a). These findings suggest that CsaA has an export-related function in B. subtilis. In conclusion, it is very well conceivable that B. subtilis, like E. coli, does contain two distinct routes for protein targeting to the translocase: first, a cotranslational targeting route via SRP, and second, a posttranslational targeting route in which CsaA might participate (Fig. 6). It has to be noted, however, that CsaA does not seem to bind to the conserved SecB-binding domain in SecA (179a). Thus, the precise role of CsaA in protein secretion in B. subtilis remains to be elucidated.
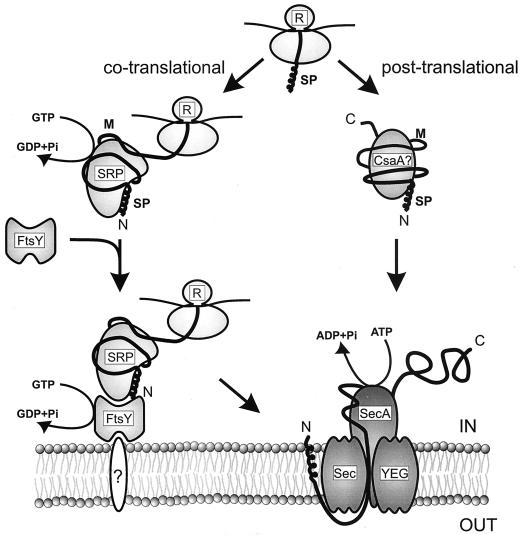
FIG. 6.
Model for protein targeting to the B. subtilis Sec translocase. Precursor proteins with the most hydrophobic targeting signals are bound cotranslationally by the SRP complex. The complex formed by the nascent chain, ribosome, and SRP docks the preprotein at an unidentified site at the membrane with the aid of FtsY. The subsequent release of the nascent chain and ribosome from the SRP-FtsY complex appears to be preceded or accompanied by GTP binding to the Ffh protein in SRP and FtsY. Hydrolysis of GTP bound to both Ffh and FtsY mediates the dissociation and recycling of these targeting components. A subset of precursor proteins containing relatively fewer hydrophobic targeting signals interact posttranslationally with an unidentified chaperone, possibly CsaA, which targets the complex to the translocase. After their binding to the Sec translocase, precursors are translocated across the membrane through cycles of ATP-dependent insertion and deinsertion of SecA into the translocation channel (see text for details). This model is largely based upon the model for the E. coli targeting routes proposed by Valent et al. (302). C, carboxyl terminus; M, mature protein; N, amino terminus; R, ribosome; SP, signal peptide.
General chaperones.
In addition to secretion-dedicated chaperones, chaperones with a general function in protein unfolding and folding might also function in protein translocation. Lill et al. (154) were the first to demonstrate that the so-called trigger factor (TF) is a cytosolic ribosome-bound protein that can maintain the translocation competence of the precursor form of the outer membrane protein OmpA in vitro. However, later studies indicated that depletion of TF did not block the export of proOmpA (109). Recently, it was shown that TF is a ribosome-bound peptidyl-prolyl cis/trans isomerase (PPIase, FK506 binding protein type [FKBP]) that interacts with both secretory and cytosolic proteins (117, 300, 301). Thus, the interaction with TF might represent a decision point for proteins to enter either the GroEL/ES folding pathway when these proteins are to remain cytosolic or to enter a targeting pathway to the translocase for secretory proteins. In addition, recent results indicate that secretory proteins are directed into the SecB-SecA-mediated posttranslational targeting pathway by means of their preferential recognition by TF (15). In B. subtilis, it was shown that TF, together with a second cytoplasmic PPIase (cyclophilin, also called PpiB), accounts for the entire PPIase activity in the cytoplasm (107). A direct involvement of TF or cyclophilin in protein secretion by B. subtilis has not been reported. However, it was recently shown that TF of the gram-positive eubacterium Streptococcus pyogenes is important for the secretion of the cysteine proteinase SCP (158). Interestingly, TF was required both for guiding SCP into the secretory pathway and for establishing an active conformation after translocation. This suggests that the cis-trans isomerization of certain peptidyl-prolyl bonds before translocation is important for the folding of the protein after translocation. However, an alternative explanation is that TF is involved in the secretion of an extracellular foldase that is required for folding of SCP at the trans side of the membrane.
In E. coli, it was shown that GroEL and GroES are important for the translocation of the SecB-independent precursor β-lactamase (21, 150). Furthermore, it was shown that GroEL interacts with SecA, and hence it was suggested that GroEL might be involved in the release of SecA from the membrane (22). Also, DnaK, DnaJ, and GrpE were shown to be involved in the export of a number of SecB-independent proteins, such as alkaline phosphatase, β-lactamase, and ribose-binding protein (327, 328). Similarly, DnaK and DnaJ homologues in S. cerevisiae, called Hsp70 and Ydj1, respectively, were shown to be involved in the posttranslational translocation of proteins across the ER membrane (338). In contrast, a role in protein secretion could not thus far be demonstrated for the corresponding heat shock chaperones in B. subtilis (T. Wiegert and W. Schumann, personal communication). Nevertheless, secretion of an antidigoxin single-chain antibody, which has the tendency to accumulate in inclusion bodies, was shown to be improved by about 60% through concerted overproduction of the GroEL-ES and DnaK-DnaJ-GrpE chaperone machineries (331). Even though the latter observation suggests that the effects of overproduction of these chaperone machineries are caused by the prevention of aggregation, the possibility that GroEL-ES and/or DnaK-DnaJ-GrpE are more directly involved in protein secretion by B. subtilis cannot presently be excluded.
The Translocase
The preprotein translocation machinery of the E. coli Sec pathway consists of at least seven proteins: SecA, which is the translocation motor, and the integral membrane proteins SecD, SecE, SecF, SecG, SecY, and YajC. Homologues of all of these components have been identified in B. subtilis. In the current model of preprotein translocation, which is based largely on results obtained in E. coli, several successive steps in the translocation of proteins are proposed (77, 79, 83, 304). First, SecA binds to acidic phospholipids and SecY (82, 110, 156, 161, 270) and is activated for recognition of SecB and the preprotein (110). Preprotein binding is followed by the binding and hydrolysis of ATP (155). The binding of ATP causes major conformational changes of SecA (72, 303), leading to a release of SecB (92, 93) and insertion of the carboxyl terminus of SecA into the membrane (85, 86, 94, 224). This membrane insertion, which occurs through the translocase complex (85, 87, 88, 224) promotes the translocation of a short fragment of the preprotein (251). Next, ATP is hydrolyzed by SecA, leading to release of the preprotein and deinsertion of SecA (85, 251). Once protein translocation is initiated by SecA, further translocation is driven by both repeated cycling of SecA through ATP binding and hydrolysis and the proton motive force (78, 100, 261).
The B. subtilis gene encoding SecA was initially identified as a gene called div, mutations in which affected cell division, sporulation, germination, protein secretion, autolysis, and the development of competence for DNA binding and uptake (236, 237). In fact, cloning and sequencing of the div gene, which is essential for cell viability, revealed that it encodes SecA (238). In addition, the B. subtilis secA gene was cloned independently, using hybridization with an E. coli secA probe (201). B. subtilis SecA can complement E. coli SecA mutants, provided that the protein is expressed at moderate levels (140). Notably, yeasts and archaea do not contain SecA homologues even though they contain a Sec-type protein-conducting channel (see below). In S. cerevisiae, the driving force for protein translocation across the ER membrane is generated either by the ribosome, in the case of cotranslational translocation, or by the ER-luminal Kar2 protein (an Hsp70 homologue), in the case of posttranslational translocation (249). The fact that SecA is absent from archaea, at least the ones for which the genome has been sequenced completely, suggests that these organisms use either another cytoplasmic ATPase, the SRP pathway, and/or protein synthesis as a force generator for protein translocation.
Notably, in addition to its role in preprotein translocation, SecA of B. subtilis could also fulfill the role of an export-specific chaperone, as suggested by Herbort et al. (116). Such a role for SecA of B. subtilis would be consistent with its recently documented low affinity for SecYEG (283). The latter observation implies that, at least in B. subtilis, the levels of cytosolic SecA are relatively high, which would facilitate early interactions with proteins destined for export.
A heterotrimeric complex of SecY, SecE, and SecG forms the main core of the translocation channel (42). This complex is found not only in eubacteria but also in eukaryotes (Sec61p complex) (106) and archaea (216). The SecY and SecE homologues in eukaryotes are called Sec61α and Sec61γ, respectively. SecG is not conserved, but this protein could be a functional analogue of the eukaryotic Sec61β. Based on sequence similarity and the observation that archaea probably contain a Sec61β homologue rather than SecG, it was suggested that the archael translocase is more related to the eukaryotic Sec61p complex (216). During the last decade, homologues of SecY, SecE, and SecG have been identified in B. subtilis, either by complementation studies using E. coli sec mutants or by DNA sequencing (131, 185, 280, 312). SecY, SecE, and SecG of B. subtilis are membrane proteins, with 10, 1, and 2 membrane-spanning domains, respectively (131, 185, 312). Notably, B. subtilis SecE is considerably smaller than E. coli SecE and has only one membrane-spanning domain, while E. coli SecE has three such domains (250). Nevertheless, SecE of B. subtilis was able to complement the cold-sensitive and export-defective phenotype of an E. coli SecE mutant, showing that it is a true SecE homologue (131). Based on data from posttranslational protein transport experiments in the S. cerevisiae ER membrane, Plath et al. (213) postulated that SecE and signal peptides bind to the same or overlapping regions in SecY and that SecE functions as a surrogate signal peptide when the SecY channel is in its closed form in the absence of translocating protein. Upon the arrival of a signal peptide, it would displace SecE and thus open the SecY channel for transport. In contrast to SecY and SecE, SecG is not strictly required for preprotein translocation and cell viability. Nevertheless, it is required for efficient translocation, possibly by facilitating the movement of preproteins through the translocation channel in concert with the insertion and deinsertion cycles of SecA (165). The absence of SecG from E. coli causes a cold-sensitive growth phenotype, as frequently encountered in E. coli strains in which protein secretion via the Sec pathway is compromised (215). Similarly, disruption of the B. subtilis secG (yvaL) gene caused secretion defects that resulted in cold-sensitive growth (312), confirming the earlier conclusion by Bolhuis et al. (27) that protein translocation in B. subtilis is intrinsically cold sensitive, as it is in E. coli. Furthermore, the cold sensitivity of the B. subtilis secG mutant was exacerbated by overproduction of secretory preproteins. Interestingly, the growth and secretion defects of the B. subtilis secG mutant could be complemented by the expression of the E. coli secG gene even when secretory preproteins were overproduced. Finally, consistent with the role of SecG in E. coli, B. subtilis SecG stimulated the ATP-dependent in vitro translocation of the precursor pre-PhoB by the B. subtilis SecA-SecYE complex (283).
In addition to the genes encoding the SecYEG core elements of the translocase, a gene encoding the SecDF protein was also identified in B. subtilis (27). In contrast to the secD and secF genes identified in most other organisms, B. subtilis was shown to contain a natural gene fusion between the equivalents of secD and secF. Consequently, SecDF of B. subtilis is a molecular Siamese twin, with 12 putative transmembrane domains. Notably, SecDF shows both sequence similarity and structural similarity to secondary solute transporters. It was demonstrated that B. subtilis SecDF, which is not essential for cell viability, is merely required to maintain a high capacity for protein secretion (27). Unlike the SecD subunit of E. coli (166), the B. subtilis SecDF protein does not seem to be required for the release of a mature secretory protein from the membrane. It has been suggested that SecD and SecF of E. coli modulate the cycling of SecA (83, 86, 137). However, it was also noted that archaea, which contain separate SecD and SecF proteins, do not contain a SecA homologue (216). Therefore, it is conceivable that SecD-SecF has another function in protein translocation, such as assembly of the translocase (216) or clearing of the translocation channel from signal peptides or misfolded proteins (27). The latter idea would be consistent with the observation that SecDF shows structural similarity to secondary solute transporters.
For E. coli, it was shown that SecD and SecF form a heterotrimeric subcomplex with a third protein (denoted YajC) and that this complex constitutes a large “holoenzyme” with the SecYEG complex (83). YajC is specified by the first gene of the SecDF operon. A gene encoding a homologue of the E. coli YajC protein, denoted yrbF, was also identified on the B. subtilis genome (53% identical plus conservative residues), but its involvement in protein secretion has not been documented so far. This yrbF gene is located in the same chromosomal region as the secDF gene but, in contrast to E. coli, it is not cotranscribed with secDF (27). Disruption of the yajC gene of E. coli did not have a clear effect on protein export, but it was shown that overproduction of YajC suppresses the dominant-negative phenotype of the secY-d1 mutation, an internal in-frame deletion in the secY gene (287).
Finally, the Sec translocon in the ER of eukaryotic cells contains, in addition to the Sec61 core components, a number of other membrane proteins (338). These include the components of the Sec62/63 and Sec66/67 (also called Sec71/72) complexes. In addition, translocation complexes in the ER of mammalian cells contain the TRAM protein. The function of these proteins is not fully clear, but they may have functions analogous to those proposed for the SecD/F proteins (216).
Type I SPases
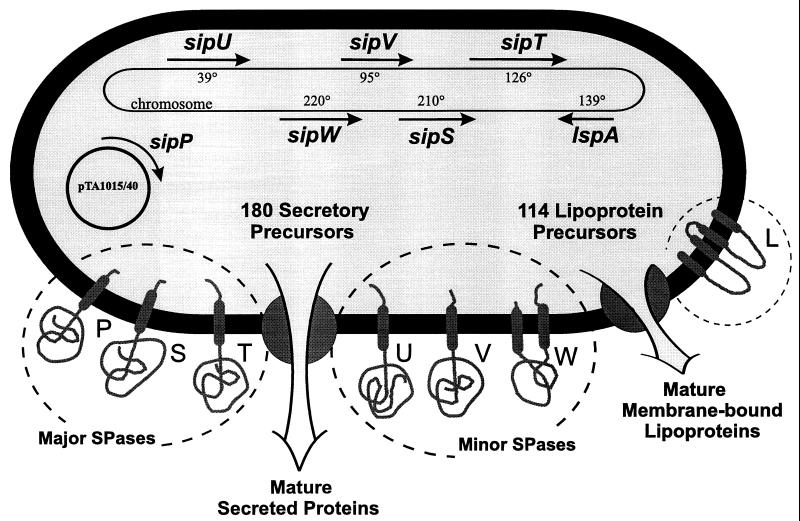
SPases remove signal peptides from secretory preproteins when their C-domain emerges at the extracytoplasmatic side of the membrane. This reaction is a prerequisite for the release of the mature secretory protein from the membrane (64, 67). One of the most remarkable features of the B. subtilis protein secretion machinery is the presence of multiple, paralogous type I SPases. This in contrast to the situation in many eubacteria, archaea, and yeasts, in which one type I SPase seems to be sufficient for the processing of secretory preproteins (44, 67, 105, 269). For most eukaryotic species, however, the presence of two paralogous SPases appears to be characteristic (67). The largest numbers of known paralogous SPases appear to be present in the archaeon Archaeoglobus fulgidus, which contains four genes for type I SPases (139), and B. subtilis, in which seven sip genes for type I SPases have been identified so far. Five of the sip genes of B. subtilis (sipS, sipT, sipU, sipV, and sipW) are located on the chromosome (26, 289, 290, 307, 308); two additional sip genes (sipP) are located on plasmids that were identified in natto-producing strains of B. subtilis (171, 172). As was shown for E. coli (66, 306) and S. cerevisiae (25), SPase I activity in B. subtilis is essential for cell viability. Although all five chromosomally encoded SPases can process secretory preproteins, only SipS and SipT are of major importance for preprotein processing and viability, whereas SipU, SipV, and SipW have a minor role in protein secretion (290). Notably, SipS and SipT can be functionally replaced by the plasmid-encoded SPase SipP (292). The latter three SPases are therefore considered to be the “major” SPases, which have substrate specificities that differ at least partly from those of SipV, SipU, and SipW, the “minor” SPases (Fig. 7). These findings indicate that the minor SPases are specifically required for the processing of a subset of the 180 predicted secretory preproteins. Indeed, SipW seems to be specifically involved in the processing of pre-TasA and pre-YqxM, two preproteins that are encoded by genes flanking the sipW gene (276, 277, 278). Surprisingly, SipW shows high degrees of sequence similarity not only to certain SPases found in sporulating gram-positive eubacteria, but also to the SPases of archaea and the eukaryotic ER membrane. Together these SipW-like SPases form the subfamily of ER-type SPases. In contrast, all other known B. subtilis SPases are of the prokaryotic type (P-type). Such P-type SPases have thus far been found exclusively in eubacteria, mitochondria, and chloroplasts (290). As demonstrated by site-directed mutagenesis of various P-type SPases, including SipS of B. subtilis (298, 305), and by X-ray crystallography of the E. coli SPase I (202), the P-type SPases make use of a serine-lysine catalytic dyad. In all known eubacterial P-type SPases, the active-site serine residue is predicted to be localized at the extracytoplasmic membrane surface. In SipS, SipT, SipU, SipV, and SipP of B. subtilis, this serine residue is kept in position by a unique amino-terminal membrane anchor. In contrast to the latter SPases, SipW appears to have a carboxyl-terminal membrane anchor in addition to an amino-terminal membrane anchor that precedes its active-site serine residue (290). However, the major difference between P- and ER-type SPases is that the catalytic lysine residue of the P-type SPases is replaced with a histidine residue in the ER-type SPases (67, 290, 307). Recent studies have shown that conserved serine, histidine, and aspartic acid residues are critical for the activity of SipW and the ER SPase Sec11 of S. cerevisiae, indicating that ER-type SPases employ a Ser-His-Asp catalytic triad or, alternatively, a Ser-His catalytic dyad (293a, 311).
FIG. 7.
Type I and II SPases of B. subtilis. The type I SPases, responsible for the processing of the 180 predicted secretory preproteins, can be divided into two groups: the major SPases SipS (S), SipT (T), and SipP (P), which are important for cell viability, and the minor SPases SipU (U), SipV (V), and SipW (W), which are not important for cell viability under laboratory conditions (290, 292). SipP is encoded by plasmid-borne genes on the plasmids pTA1015 and pTA1040, which are present in certain natto-producing B. subtilis strains. All other SPases are chromosomally encoded. The transcription of the genes for the major SPases increases during the postexponential growth phase in concert with the genes for most secretory proteins, whereas the minor SPases are transcribed at a low level during all growth phases. SipW is the only ER-type SPase, showing a high degree of similarity to eukaryotic and archaeal SPases. In contrast to the prokaryotic (P-)type SPases of B. subtilis, which have one amino-terminal membrane anchor, SipW appears to have an additional carboxyl-terminal membrane anchor. Finally, B. subtilis contains only one gene (lsp) for a type II SPase (L) with four membrane anchors, which is required for the processing of the 114 predicted lipoproteins (291) (see text for details).
Even though it is well established that various type I SPases of B. subtilis have different substrate specificities, the molecular basis for these differences with respect to the composition of the C-domain of signal peptides and the structure of the active sites of the different SPases is presently unknown.
Lipoprotein Processing by SPase II
In contrast to the type I SPases, B. subtilis contains only one gene for a type II SPase (lsp) (223), which is specifically required for the processing of lipid-modified preproteins. All known SPases of this type are integral membrane proteins with four (putative) membrane-spanning segments, the amino and carboxyl termini having a predicted cytosolic localization (183, 223, 293). The potential active site of SPase II, formed by two aspartic acid residues, is located in close proximity to the extracytoplasmic surface of the membrane, similar to the active-site serine residue of type I SPases (293).
Interestingly, cells lacking SPase II are viable under standard laboratory conditions. This indicates that processing of lipoproteins by SPase II in B. subtilis is not strictly required for lipoprotein function, as at least one lipoprotein, PrsA, is essential for viability (144, 291). Although certain lipoproteins are required for the development of genetic competence, sporulation, and germination, these developmental processes were not detectably affected in the absence of SPase II. Cells lacking SPase II accumulated lipid-modified precursor and, surprisingly, mature-like forms of the lipoprotein PrsA, which is involved in the folding of secreted proteins. These forms of PrsA appeared to be reduced in activity, as the secretion of the B. amyloliquefaciens α-amylase AmyQ, the folding of which is dependent on PrsA, was strongly impaired (291). It is presently not clear which proteases are responsible for the alternative processing of PrsA in the absence of SPase II. However, the involvement of type I SPases in this process appears to be highly unlikely (291). The cellular level of another lipoprotein, CtaC, is strongly reduced in the absence of SPase II, indicating that lipoprotein processing is important for the stability of certain proteins (17).
Diacylglyceryl modification of the cysteine residue at position +1 of the mature lipoprotein in lipoprotein precursors is catalyzed by the lipoprotein diacylglyceryl transferase (Lgt). This lipid modification is a prerequisite for processing of the lipoprotein precursor by SPase II (103, 147, 242, 243, 244, 294). Similar to the disruption of the lsp gene for SPase II, disruption of the lgt gene results in the accumulation of unprocessed (and unmodified) lipoproteins without affecting growth and cell viability (153, 291). Like SPase II, Lgt is required for the stability of several lipoproteins and the efficient secretion of AmyQ (17, 153). Interestingly, the processing rate of secreted nonlipoproteins was retarded in the absence of SPase II, which must be attributed to the malfunction of lipoproteins other than PrsA (291). Two candidate proteins that might be responsible for this effect are the putative lipoproteins SpoIIIJ and YqjG (Table 3), which show significant sequence similarity to the mitochondrial Oxa1 protein. The latter protein was shown to be required for export of the amino and carboxyl termini of the mitochondrially encoded precursor of cytochrome c oxidase subunit II (pre-CoxII) from the mitochondrial matrix to the intermembrane space (114, 115), proteolytic processing of pre-CoxII (13), and assembly of the cytochrome c oxidase and oligomycin-sensitive ATP synthase complexes (3, 33). As mitochondria lack Sec components, Oxa1p seems to represent a component of a specific protein export and/or assembly machinery that might be conserved in eukaryotic organelles and eubacteria (115, 235). Consistent with this hypothesis, it was recently shown that the E. coli homologue of SpoIIIJ/YqjG and Oxa1p, denoted YidC, is associated with the Sec translocase (256a).
Finally, processed lipoproteins of E. coli are further modified by aminoacylation of the diacylglyceryl-cysteine amino group (103, 147, 242, 243, 244, 294). The latter lipid modification step does not seem to be conserved in all eubacteria, as many organisms, including B. subtilis, lack an lnt gene for the lipoprotein N-acyltransferase (291).
SPPases
After being cleaved off from the mature protein, signal peptides are rapidly degraded. In E. coli, the signal peptide of the major lipoprotein (Lpp; also called Braun's lipoprotein) was shown to be degraded by the membrane-bound protease IV (also called signal peptide peptidase, encoded by the sppA gene) (123, 125, 126). Nevertheless, in the absence of protease IV, significant levels of signal peptide degradation were observed (282), suggesting that other proteases can replace protease IV in this process. In addition to the protease IV, the cytoplasmic oligopeptidase A (OpdA) was shown to be involved in the degradation of the Lpp signal peptide (197, 198). Current models imply that protease IV cleaves the signal peptide in the membrane in two fragments (Fig. 1), which are further degraded in the cytoplasm by OpdA (198). Interestingly, independent of its effect on natural signal peptides, OpdA was also identified in suppressor screens using secretory proteins with defective signal peptides. In such screens, it was noticed that the prlC (protein localization) mutation, which turned out to be a mutation in the opdA gene, could suppress the export defect of certain LamB signal sequence mutants (57). At present, it is not clear how mutations in OpdA can lead to the observed prlC phenotype.
Protease IV is highly conserved in eubacteria and archaea. In contrast, OpdA appears to be absent from gram-positive eubacteria and archaea. Like the protease IV of E. coli, its homologue in B. subtilis, denoted SppA (YteI), appears to be a membrane protein with three potential transmembrane segments. Disruption of the sppA gene of B. subtilis resulted in a decreased rate of processing of the α-amylase AmyQ but not of its translocation. This suggests that SPase I activity is negatively affected in the absence of SppA, for example, by the accumulation of certain signal peptides (30). Interestingly, B. subtilis contains a second gene (tepA or ymfB) for a potential protease with amino acid sequence similarity to SppA. In the absence of this so-called TepA protein (translocation-enhancing protein), the rate of translocation of a number of secretory proteins was strongly affected. Notably, the TepA protein, which probably has a cytoplasmic localization, also shows sequence similarity to the cytoplasmic protease ClpP (see below). Three possible roles of TepA in protein translocation have been suggested (30). First, TepA might be an analogue of E. coli OpdA, an idea which is based on the observation that signal peptides can inhibit protein translocation (62). Second, similar to what was observed for ClpP (176), TepA might have a regulatory function. Third, TepA might be a secretion-specific chaperone.
Extracytoplasmic Folding Catalysts
After leaving the translocation channel, secretory proteins have to fold into their native conformation at the trans side of the membrane. Since several proteases are present in this environment, rapid and correct folding is essential, in particular because (partly) unfolded proteins are very sensitive to proteases. This phenomenon is illustrated by the recent observation that an α-amylase from B. licheniformis (AmyL) was degraded when secreted by B. subtilis due to its relatively slow rate of folding. However, once folded into the native conformation, AmyL was stable in the growth medium (274). Folding at the trans side of the membrane is mediated by several extracellular folding catalysts, also termed foldases. In eubacteria, the foldases found thus far are PPIases, which catalyze the cis-trans isomerization of peptidyl-prolyl bonds, and thiol-disulfide oxidoreductases, which catalyze the formation and/or isomerization of disulfide bonds. In addition, other factors, such as certain cations, play a role in protein folding after translocation. Strictly speaking, the latter factors are not real components of the secretion machinery, but they can be regarded as such because they play an important role in the folding of secretory proteins of B. subtilis. Therefore, they are included in this overview.
PPIases.
One of the proteins involved in the folding of proteins after their translocation is PrsA, a lipoprotein that is anchored to the outer leaflet of the cytoplasmic membrane (144, 145). Based on sequence similarity, it was proposed that PrsA is a PPIase belonging to the parvulin family (228). PrsA is essential for viability, and strains containing mutant PrsA proteins were shown to secrete lower amounts of degradative enzymes, probably due to decreased stability, resulting in increased sensitivity to proteolysis of these exoproteins (130, 144, 145). E. coli contains four periplasmic PPIases (68, 174), denoted RotA (cyclophilin type), FkpA (FKBP type), and SurA and PpiD (parvulin type). Moreover, S. cerevisiae produces five PPIases (76), denoted CPR2, CPR4, CPR5, CPR8, (cyclophilin type), and FPR2 (FKBP type), which are either localized in the ER or secreted. In addition, S. cerevisiae contains eight other PPIases that are localized in the nucleus, the mitochondrion, or the cytoplasm (76). Interestingly, extracytoplasmic PPIases of the cyclophilin and FKBP types appear to be absent from B. subtilis.
Thiol-disulfide oxidoreductases.
Disulfide bonds are essential for the activity and stability of several proteins. Such bonds form spontaneously in vitro, but this process is much slower and less effective than the formation of disulfide bonds in vivo, which is catalyzed by thiol-disulfide oxidoreductases (210). In E. coli, the formation of disulfide bonds takes place in the periplasm. Six components involved in disulfide bond formation have been identified: DsbA, DsbB, DsbC, DsbD, DsbE, and DsbG (5). The Bdb (Bacillus disulfide bond) protein from Bacillus brevis was, until recently, the only known thiol-disulfide oxidoreductase from a gram-positive eubacterium (129). The bdb gene can complement a mutation in the E. coli dsbA gene, indicating that in the latter organism Bdb is translocated to the periplasm. It has been suggested that, in B. brevis, Bdb is also translocated across the membrane and localized at the membrane-cell wall interface.
Most likely, B. subtilis secretes only a limited number of proteins containing disulfide bonds (31). On the other hand, secreted proteins of eukaryotes often contain several disulfide bonds (133). Proteins containing several disulfide bonds, like the human serum albumin and the human pancreatic α-amylase, are secreted very poorly by B. subtilis (31, 248), which may be due to the formation of incorrect disulfide bonds or the lack of formation of such bonds. However, disulfide bonds in proteins secreted by B. subtilis can be formed properly. This is evident from the observation that human interleukin-3, containing one disulfide bond, can be secreted efficiently by B. subtilis (310). The same applies to an engineered neutral protease from B. subtilis that was stabilized to a great extent by the introduction of a disulfide bond (160), and the E. coli alkaline phosphatase (PhoA) protein, which contains two disulfide bonds (31). This suggests that, in B. subtilis, thiol-disulfide oxidoreductases are present at the trans side of the cytoplasmic membrane. Screening of the genome of B. subtilis revealed three genes for proteins with similarity to thiol-disulfide oxidoreductases (31). Disruption of two of these genes, called bdbB (yolK) and bdbC (yvgU), did indeed interfere with the efficient secretion of E. coli PhoA. These two genes encode putative membrane-bound thiol-disulfide oxidoreductases that are related to E. coli DsbB. Disruption of the bdbC gene had the strongest effect on PhoA secretion, which was reduced approximately 10-fold (as measured through PhoA activity). This gene was also shown to be involved in the folding of A13i-β-lactamase, a hybrid precursor that contains one disulfide bond. The third gene encoding a putative thiol-disulfide oxidoreductase in B. subtilis is bdbA (yolI). The deduced BdbA protein shows a high degree of sequence similarity to Bdb from B. brevis (129) and may be a secreted protein. Disruption of this gene did not affect the secretion of E. coli PhoA or A13i-β-lactamase (31).
Propeptides.
Propeptides, which are commonly present in secretory proteins from Bacillus species, are stretches of amino acids located between the signal peptide and the mature part of the protein. In B. subtilis, their lengths vary from 8 amino acids (α-amylase) (285) to 194 amino acids (neutral protease NprE) (334). Whereas no clear function has been assigned to short propeptides, long propeptides, which are mainly found in certain proteases, have two important functions. First, they prevent the activation of proteases prior to their translocation (323). Second, propeptides catalyze the folding and activation of the proteases once they have been translocated. It was shown that certain propeptides are functional not only in cis but also in trans, demonstrating their role as a molecular endochaperone (127, 128, 337). In general, the long propeptides in proteases are removed autocatalytically after translocation.
Other folding catalysts.
Some secretory proteins require cations, such as Fe3+ and Ca2+, for folding. The secretion of B. subtilis levansucrase is stimulated by growth of the cells in medium containing high concentrations of Fe3+, and in vitro refolding of B. subtilis levansucrase was greatly enhanced by this cation, even though Fe3+ is not bound to the mature protein (49). Furthermore, several extracellular proteins from B. subtilis, such as levansucrase, neutral protease, and α-amylase, are calcium-binding proteins, and they require Ca2+ for stability (212, 275, 313). Ca2+ is trapped in the B. subtilis cell wall, thereby creating a microenvironment that must play an important role in the late steps of secretion (211). Notably, the charge properties of the cell wall will also influence its interactions with secretory proteins, as a protein with overall positive charge can be retarded by the negatively charged polyanionic polymers in the wall (50). Indeed, in vitro folding assays with a derivative of an α-amylase from B. licheniformis having an increased net positive charge showed a decreased rate of folding in the presence of cell wall material which was not observed in the folding of wild-type α-amylase. In addition, the cell wall had the capacity to bind large amounts of the mutated α-amylase (275). The latter observations imply that the folding and subsequent secretion of heterologous proteins can, in principle, be improved by the removal of positively charged residues that are not required for their stability or biological function.
QUALITY CONTROL
As indicated in the first sections of this review, Bacillus species are prolific and commercially important producers of high-quality industrial enzymes and a few eukaryotic proteins, such as human interleukin-3. The high quality of these secreted proteins is, at least in part, due to the presence of cellular quality control systems that efficiently remove misfolded or incompletely synthesized proteins. Paradoxically, these quality control systems represent major bottlenecks for the production of many heterologous proteins at commercially significant concentrations, because their folding is usually inefficient (29).
One of the greatest problems is proteolytic degradation. Thus far, most attempts to find solutions were focused on the proteases that are secreted into the growth medium (263). Mutants lacking up to six of these extracellular proteases have been made (75, 134, 329, 331). In some cases, these mutant strains were able to secrete increased amounts of (heterologous) proteins (29). Nevertheless, the effects were only moderately positive, and some proteins, like OmpA of E. coli, were still degraded rapidly (170). This degradation is probably caused by membrane- or cell wall-associated proteases that are part of the quality control system for exported proteins. Indeed, one cell wall-bound protease, CWBP52 (encoded by the wprA gene) (163), was shown to be involved in the degradation of the α-amylase from B. licheniformis (274) and an unstable variant of the signal peptidase SipS (28). In addition to degradation by membrane- or wall-associated proteases, it is conceivable that the degradation of heterologous proteins occurs in the cytoplasm.
Several cytoplasmic, membrane-bound, and extracytoplasmic proteases of B. subtilis have been identified through genetic and biochemical analysis, but the availability of the complete genome sequence of B. subtilis (149) enabled a systematic evaluation of a large number of (putative) proteases (Table 8). Several cytoplasmic proteases, such as ClpP, ClpQ, and Lon, are probably mainly involved in general housekeeping functions, such as degradation of misfolded proteins. However, some of these may also have more specific functions in gene regulation through proteolysis, as it was recently shown that ClpP-mediated proteolysis is involved in controlling the levels of MecA, a negative regulator of competence (176). The MlpA protein, which is highly similar to mitochondrial processing peptidases, appeared to have a role in gene regulation through proteolytic processing. The latter conclusion is based on the observation that in a strain lacking a functional mlpA gene, expression of the subtilisin gene (aprE) was increased about fivefold (32). Furthermore, the intriguing possibility was recently put forward that some membrane proteins or even secretory proteins of B. subtilis could be degraded by cytosolic proteases after their translocation, which would require their retrograde transport to the cytoplasm (28). Even though there is no direct evidence for retrograde transport in B. subtilis, this phenomenon has been documented for the degradation of certain ER-luminal proteins (19, 146, 214, 326). The latter proteins were shown to be returned to the cytoplasm via the Sec61p complex and degraded by the proteasome, a multimeric complex that is responsible for much of the proteolysis within the cytoplasm of eukaryotic cells (118).
TABLE 8.
Putative proteases and peptidases of B. subtilis
| Proteasea | Description | Location | Reference or SubtiList accession no. |
|---|---|---|---|
| AlbE (YwhO) | Similar to Synechocystis sp. processing peptidase | Cytoplasm | 336 |
| AlbF (YwhN) | Similar to mitochondrial zinc-endoprotease | Cytoplasm | 336 |
| AmpS | Putative aminopeptidase; metalloprotease | Cytoplasm | BG10986 |
| AprX | Alkaline serine protease | Cytoplasm | BG12567 |
| ClpE | ATP-dependent Clp protease-like | Cytoplasm | BG12578 |
| ClpP | ATP-dependent Clp protease proteolytic subunit; requires ATP-binding subunit ClpA/ClpX | Cytoplasm | 315 |
| HslV (ClpQ, CodW) | Similar to β-subunit of the 20S proteasome; requires ATP-binding subunit HslU (ClpY) | Cytoplasm | 265 |
| IspA | Major intracellular serine protease | Cytoplasm | 143 |
| LonA | Heat shock ATP-dependent protease | Cytoplasm | 230 |
| LonB | Lon-like ATP-dependent protease | Cytoplasm | 230 |
| Map (AmpM) | Methionine aminopeptidase | Cytoplasm | 185 |
| MlpA (YmxG) | Similar to mitochondrial processing peptidase (metalloprotease) | Cytoplasm | 32 |
| Pcp | Pyrrolidone carboxyl peptidase | Cytoplasm | 7 |
| PepT | Tripeptidase | Cytoplasm | 255 |
| SpoVK | ATP-dependent FtsH-like protease | Cytoplasm | BG11039 |
| TepA | Similarity to both SPPase and ClpP-type proteases | Cytoplasm | 30 |
| YclE | Similar to prolyl aminopeptidase | Cytoplasm | BG12026 |
| YflG | Paralogue of methionine aminopeptidase (Map) | Cytoplasm | BG12942 |
| YjbG | Similar to oligoendopeptidase | Cytoplasm | BG13136 |
| YjoB | ATP-dependent FtsH-like protease | Cytoplasm | BG13216 |
| YkvY | Similar to Xaa-Pro dipeptidase | Cytoplasm | BG13326 |
| YlbL | Similar to Lon proteases | Cytoplasm | BG13364 |
| YmfG | Similar to processing peptidase, paralogue of AlbE | Cytoplasm | BG13427 |
| YmfH | Similar to mitochondrial processing peptidase (metalloprotease) | Cytoplasm | BG13428 |
| YpwA | Similar to carboxypeptidase; metalloprotease | Cytoplasm | BG11458 |
| YqgT | Similar to gamma-d-glutamyl-l-diamino acid endopeptidase I | Cytoplasm | BG11687 |
| YqhT | Similar to Xaa-Pro dipeptidase | Cytoplasm | BG11708 |
| YqjE | Similar to tripeptidase; paralogue of PepT | Cytoplasm | BG11734 |
| YrrN | Putative protease; similar to collagenase from Methanobacterium thermoautotrophicum | Cytoplasm | BG13795 |
| YrrO | Putative protease; paralogue of YrrN | Cytoplasm | BG13796 |
| YtjP | Similar to Xaa-His dipeptidase | Cytoplasm | BG13867 |
| YuiE | Similar to leucyl aminopeptidase | Cytoplasm | BG13970 |
| YusX | Similar to oligoendopeptidase | Cytoplasm | BG14036 |
| YwpE | Similar to surface protein sorting sulfhydryl protease (Staphylococcus aureus SrtA) | Cytoplasm | BG12499 |
| YuxL | Similar to acylaminoacyl-peptidase | Cytoplasm | BG10463 |
| ComC | Type IV prepilin-like SPase | Membrane | 53 |
| CtpA | Similar to carboxy-terminal processing protease (E. coli Tsp) | Membrane | 162 |
| FtsH | ATP-dependent zinc metallopeptidase | Membrane | 72 |
| HtrA (YkdA) | Similar to serine protease HtrA | Membrane | BG12608 |
| Lsp | Type II SPase | Membrane | 223 |
| SipS/T/U/V/W | Type I SPases | Membrane | 289, 290, 307 |
| SppA (YteI) | SPPase | Membrane | 30 |
| YhcS | Similar to surface protein sorting sulfhydryl protease (Staphylococcus aureus SrtA) | Membrane | BG11597 |
| YcdD | Similar to l-alanoyl-d-glutamate peptidase; putative lipoprotein | Membrane | BG12760 |
| YhfN (YzoA) | Similar to prenyl protein-specific endoprotease from S. cerevisiae; putative metalloprotease | Membrane | BG11029 |
| YvjB | Carboxy-terminal processing protease, similar to E. coli Tsp | Membrane | BG14110 |
| YvtA | Similar to serine protease HtrA from E. coli | Membrane | Noone and Devine, GenBank accession no. AAF03153 |
| YyxA (YycK) | Similar to serine protease HtrA from E. coli | Membrane | 45 |
| YodJ | Similar to d-alanyl-d-alanine carboxypeptidase | Cell wall | BG13538 |
| DacA | Penicillin-binding protein 5 (d-alanyl-d-alanine carboxypeptidase) | Cell wall | 324 |
| DacB | Penicillin-binding protein 5* (d-alanyl-d-alanine carboxypeptidase) | Cell wall | 43 |
| DacF | Penicillin-binding protein (d-alanyl-d-alanine carboxypeptidase) | Cell wall | 330 |
| LytE | Cell wall hydrolase; probable endopeptidase | Cell wall | 164 |
| WprA | Cell wall-associated protein precursor (serine protease) | Cell wall | 163 |
| AprE | Alkaline serine protease (subtilisin E) | Extracellular | 273 |
| Bpr (Bpf) | Bacillopeptidase F (serine protease) | Extracellular | 267 |
| Epr (Ipa-15r) | Minor serine protease | Extracellular | 266 |
| Mpr | “Metalloprotease,” but belongs to the family of serine proteases | Extracellular | 267 |
| NprB | Neutral protease B (metalloprotease) | Extracellular | 297 |
| NprE | Neutral protease E (metalloprotease) | Extracellular | 334 |
| Vpr (Ipa-45r) | Minor serine protease | Extracellular | 268 |
| YwaD (Ipa-8r) | Similar to aminopeptidase | Extracellular | BG10554 |
Synonymous names are in parentheses.
Accessible through http://bioweb.pasteur.fr/GenoList/SubtiList; BG codes are those used in SubtiList.
Three homologues of the HtrA protein from E. coli were identified in B. subtilis. HtrA of E. coli is a periplasmic heat shock protease involved in the removal of misfolded proteins from the periplasm (174). The B. subtilis HtrA homologues are encoded by the htrA, yvtA, and yyxA genes (for the correct sequence of YvtA, see D. Noone and K. M. Devine, GenBank accession no. AAF03153). These three genes are predicted to encode membrane-anchored proteins with their active sites located at the trans side of the membrane. Presently, the role of the HtrA homologues in B. subtilis is not known.
Several other proteases are present in the membrane that, most likely, have a quality control function for membrane-bound proteins. FtsH is a zinc-binding metalloprotease with its active site on the cytoplasmic side of the membrane. In B. subtilis, disruption of the ftsH gene resulted in a very pleiotropic phenotype (filamentous growth, sensitivity to heat and salt stress, and low levels of protein secretion) (71, 72). In E. coli, FtsH selectively degrades SecY when the latter protein is not in a complex with SecE, indicating that FtsH is indeed involved in quality control of proteins in the membrane (2). Notably, it was recently suggested that FtsH of E. coli could be involved in retrograde transport of membrane proteins and their degradation in the cytoplasm (136).
The type I SPases SipS, SipT, SipU, SipV, and SipW and the type II SPase are specifically involved in the cleavage of signal peptides from secretory preproteins. However, it cannot be excluded that these peptidases, in addition to preproteins, have other substrates. For example, it was demonstrated that the homologous Sec11p subunit of the SPase complex in the yeast ER membrane is involved in protein degradation (180).
CtpA and YvjB of B. subtilis are highly similar to the E. coli Tsp protein that is involved in the degradation of peptide-tagged proteins derived from truncated mRNAs. Keiler and coworkers (135) proposed a model in which truncated mRNAs that lack a stop codon are modified by the carboxyl-terminal addition of a peptide tag encoded by the SsrA RNA. This peptide tag is recognized by proteases, such as Tsp, that subsequently degrade the protein. Both CtpA and YjvB are predicted to remain attached to the membrane via an amino-terminal membrane anchor. The role of the two Tsp homologues and the SsrA RNA of B. subtilis in protein secretion or degradation has not yet been evaluated.
Finally, the YhfN protein, which is predicted to have seven membrane-spanning domains, shows sequence similarity to the Ste24 protease of S. cerevisiae, which is involved in the maturation of the α-factor, a small secreted pheromone (286). The function of YhfN is currently not known.
SEC-INDEPENDENT PROTEIN EXPORT
Although the Sec-dependent pathway seems to be responsible for the export of most proteins in B. subtilis, at least three alternative pathways seem to be present for the transport of small groups of specific proteins. These are a putative twin-arginine translocation (Tat) pathway, a prepilin-specific secretion and assembly pathway, and at least three ABC transporter-dependent secretion pathways. In the following sections we will discuss each of these three types of pathways.
A Twin-Arginine Translocation Pathway?
Like most other eubacteria, B. subtilis seems to contain a very recently identified Sec-independent secretion pathway, which is known as the Tat pathway. This pathway was first discovered in chloroplasts, in which it is involved in ΔpH-dependent protein import into the thylakoid lumen (47, 195, 232, 259). For the chloroplast system, it was shown that, in contrast to Sec-dependent translocation, proteins can be translocated in a folded conformation via this pathway (56, 58, 124, 233). Furthermore, it was demonstrated that two adjacent arginines combined with a hydrophobic determinant (preferably leucine) at position +2 or +3, relative to the twin arginines, were needed in the N-domain of signal peptides to direct precursors into this pathway (38, 39). In E. coli, precursors of several periplasmic cofactor (e.g., flavins, molybdopterins, and iron-sulfur clusters)-binding proteins have signal peptides carrying a similar RR-motif ([S/T]-R-R-x-F-L-K) immediately before their H-domain (18). Although the exact mechanism of protein export via the Tat pathway has yet to be unraveled, five components of the Tat pathway of E. coli have been identified. These are TatA (a putative membrane-bound receptor, homologous to the maize Hcf106 protein) (259), TatB (a TatA paralogue) (247), TatC (the putative translocase), TatD (a predicted soluble protein), and TatE (a TatA paralogue). These proteins are encoded by the tatABCD operon and the unlinked tatE gene (65, 246, 325). Disruption of the tatA to tatC and tatE genes affected the export of several preproteins with an RR-signal peptide, such as the trimethylamine N-oxide reductase TorA, which are transported in a Sec-independent manner (24, 245). The role of TatD, which is most likely a cytoplasmic protein, in the Tat pathway has not yet been established. Recent studies showed that a Sec-dependent periplasmic domain from the E. coli SPase I, also known as leader peptidase (lep), can be rerouted into the Tat pathway by the RR-signal peptide of TorA. In contrast, a full-length TorA-Lep fusion protein was not rerouted into the Tat pathway. Furthermore, it was shown that the TorA signal peptide could be converted into a Sec-targeting signal peptide by increasing the length and hydrophobicity of its H-domain (60). These findings indicate that the Sec and Tat pathways compete for preproteins, at least in E. coli, and that the overall hydrophobicity of the RR-signal peptide plays an important role in discrimination between these two pathways. Interestingly, B. subtilis contains three homologues of TatA/B/E (encoded by the ydiI, yczB, and ynzA genes), two homologues of TatC (encoded by the ydiJ and ycbT genes), and one TatD homologue (encoded by the yabD gene). Furthermore, at least one protein of B. subtilis with a putative RR-signal peptide is known to bind a cofactor. This is the QcrA protein, which contains an iron-sulfur cluster. It will be a major challenge for future research to determine the Tat-dependent exported fraction of the secretome.
Type IV Pilin Export
A second class of B. subtilis proteins that are exported in a Sec-independent manner consist of type IV pilin-like proteins encoded by the comGC, comGD, comGE, and comGG genes. The corresponding gene products are involved in the development of genetic competence. They resemble type IV pilins of various gram-negative eubacteria that are synthesized as precursors with cleavable signal peptides. Although prepilin signal peptides show certain similarities to signal peptides of secretory proteins and lipoproteins, the prepilin(-like) precursors are believed to bypass the Sec and Tat secretion pathways, as their translocation is dependent on a cleavage event at the cytoplasmic side of the membrane (54, 157, 199, 225, 226). ComC, the SPase that cleaves the comG products ComGF (an integral membrane protein) and ComGA (a putative ATPase located at the cytoplasmic side of the membrane), is known to be involved in the assembly of the pilin-like ComG proteins (54). The B. subtilis SPase ComC is an integral membrane protein with eight (putative) transmembrane regions, and this protein shows a high degree of similarity to prepilin peptidases of various other organisms (157). Processing of the comG products is required for the assembly and anchoring of the pilin-like structures to the membrane, which in turn is required for DNA binding during transformation in B. subtilis (55, 80). ComC-like SPases cleave the peptide bond between a glycine at position −1 and a phenylalanine at position +1 (Fig. 2). Notably, the amino acid at position +1 relative to the SPase cleavage site of these pilins is modified, like the +1 cysteine residue of mature lipoproteins. However, in the case of type IV pilins, the phenylalanine residue at the +1 position is amino-methylated (157). A second difference with lipoprotein processing is that ComC is bifunctional, being responsible for both prepilin processing and the subsequent methylation of the +1 phenylalanine residue. The cleavage site of prepilins is located amino-terminal to the hydrophobic H-domain (225). Consistent with the latter observation, the putative active site of prepilin SPases appears to be localized in the cytoplasm (151). Interestingly, even though prepilin SPases and type II SPases lack sequence similarity, the catalytic mechanism of prepilin SPases seems to be related to that of type II SPases, as the potential active sites of both SPases contain two catalytic aspartic acid residues. Furthermore, in both types of SPases, the potential active-site residues are predicted to reside in close proximity to the membrane surface. However, the putative catalytic aspartic acid residues of type IV prepilin SPases are located at the cytoplasmic side of the membrane, while those of type II SPases are located at the extracytoplasmic side (151, 293).
Export via ABC Transporters
Several Bacillus species produce peptide antibiotics which are synthesized through either a ribosomal or nonribosomal mechanism (81, 99, 189). Some of the ribosomally synthesized antimicrobial peptides contain signal peptides for their translocation across the membrane by dedicated ABC transporters (70, 112, 239). In B. subtilis 168, the sunS-sunT operon has recently been shown to encode, respectively, the lantibiotic sublancin 168 and the ABC transporter SunT, which is required for sublancin production (203). Mature sublancin 168 contains one β-methyllanthionine bond and two disulfide bonds. Like signal peptides of other lantibiotics (48, 192), the sublancin signal peptide contains a double glycine motif amino-terminal to the processing site. Interestingly, ABC transporters such as SunT have a dual role in secretion, as they are responsible both for removal of the signal peptide and for translocation of the mature lantibiotic across the cytoplasmic membrane. The protease domain of these so-called dual-function transporters is localized in their conserved amino terminus, which contains cysteine and histidine residues involved in precursor cleavage (98, 112, 314). A second lantibiotic known as subtilin is secreted by the B. subtilis strain ATCC 6633. This lantibiotic contains a signal peptide that is unrelated to those of sublancin and the bacteriocin subtilosin (see below). Nevertheless, subtilin is secreted by a dedicated ABC transporter, like sublancin and subtilosin (11, 52, 138).
Similar to sublancin and subtilin, the antilisterial bacteriocin subtilosin is ribosomally synthesized as a precursor; it then matures and is exported as a cyclic peptide containing several modifications, including one disulfide bridge. Notably, presubtilosin lacks the sequence motifs known to be required for prelantibiotic processing, as found in sublancin (336). The downstream operon of the sbo gene coding for subtilosin contains seven genes, albA to albG (ywiA to ywhM). This operon was shown to be essential for subtilosin production. Genes in the alb operon encode proteins, such as an ABC transporter (AlbC) and two peptidases (AlbE and AlbF), which are likely to be required for the processing and export of subtilosin (336). Interestingly, the albA gene product is homologous to proteins involved in cofactor synthesis (e.g., molybdenum cofactors) (336), while the albB (ywhR) gene encodes a putative protein with an RR-signal peptide (Table 3). Thus, it is conceivable that AlbB is exported via the Tat pathway, which in gram-negative eubacteria, is known to be required for the export of cofactor-binding proteins (18, 245).
Finally, the extracellular pheromone ComX, which is involved in cell density-controlled onset of the transcription of competence genes, is also ribosomally synthesized as a precursor and modified prior to secretion (159, 272). Although not documented, it is conceivable that a dedicated ABC transporter is responsible for the processing and secretion of this pheromone.
CELL WALL RETENTION
The cell wall of B. subtilis defines a cellular compartment containing approximately 9% of the total cellular protein content, analogous to the gram-negative periplasm (217). In B. subtilis, proteins retained in the cell wall include DNases, RNases (173), proteases (10, 163, 274), enzymes involved in the synthesis of peptidoglycan (penicillin-binding proteins), and cell wall hydrolases (20, 96, 97) that are involved in cell wall turnover during cell growth, cell division, sporulation, and germination (181, 182, 219, 335).
Cell Wall Retention Signals
Several B. subtilis enzymes involved in cell wall turnover contain a variable number of repeated domains which have affinity for components of the cell wall (102, 164, 229). These repeats are thought to direct enzymes for cell wall assembly and turnover to specific sites where cell wall synthesis and/or hydrolysis takes place, as was shown for Staphylococcus aureus (8, 9). Most likely, this specific targeting is promoted by certain components of the cell wall, such as choline, which was shown to be a receptor for several cell wall proteins of Streptococcus pneumoniae (234, 240, 241). Like the enzymes involved in cell wall turnover, other B. subtilis proteins also retained in the cell wall, such as WprA and WapA, contain repeated motifs for cell wall binding. At present it is not known which receptors are involved in directed targeting of cell wall proteins in B. subtilis. The mature parts of 16 of the proteins with cleavable signal peptides (Tables 1 and 3) contain (putative) cell wall-binding repeats, indicating that they may be retained in the cell wall. Surprisingly, potential cell wall-binding regions also might be present in some (predicted) membrane proteins, such as HtrA, YclI, and YxcE, suggesting that these proteins are active at sites of intimate contact between the membrane and the cell wall.
Covalent Attachment to the Cell Wall?
A second group of surface proteins of gram-positive organisms are covalently anchored to the cell wall via the carboxyl terminus. The amino-terminal domains of these molecules are displayed on the eubacterial surface (252, 253). Cell wall anchoring of surface proteins in S. aureus requires, in addition to the amino-terminal signal peptide, a carboxyl-terminal cell wall sorting signal consisting of the so-called LPxTG motif, a carboxyl-terminal hydrophobic domain, and a positively charged tail (190, 191, 254). During the export of surface proteins, the cell wall sorting signal is cleaved between the Thr and Gly residues of the LPxTG motif by the so-called sortase (SrtA) (169). Simultaneously, the carboxyl group of the Thr residue is linked to the cell wall by a branched anchor peptide (295).
In contrast to S. aureus, none of the putative exported B. subtilis proteins contains a carboxyl-terminal LPxTG retention signal for covalent attachment to the cell wall. Nevertheless, genes for two SrtA homologues could be identified in the B. subtilis genome. First, the yhcS gene codes for a putative exported protein of 198 amino acids with an amino-terminal membrane anchor showing 22% sequence identity to SrtA of S. aureus. This protein contains the carboxyl-terminal LxTC motif, of which the cysteine is essential for SrtA activity (296). Second, a small open reading frame called ywpE codes for a protein of 102 amino acids that also contains the LxTC motif, showing 23% sequence identity with the carboxyl terminus of SrtA. Notably, the latter protein is predicted to be a cytoplasmic protein. Taken together, these findings suggest that at least one sortase-like enzyme for the cleavage and linkage of surface proteins is present in B. subtilis. If so, this enzymes recognizes amino acid sequences that are different from the LPxTG motif of S. aureus surface proteins. Alternatively, the sortase-like proteins of B. subtilis may be involved in completely different, as yet unidentified, processing events.
SPORULATION-SPECIFIC PROTEIN TRANSPORT
In response to nutrient starvation, B. subtilis develops endospores to resist a variety of harsh conditions, such as extreme cold or heat (279). At the beginning of the sporulation process, an asymmetrically positioned septum is formed that divides the cell into two unequally sized compartments. Subsequently, the larger compartment (the mother cell) engulfs the smaller compartment (the forespore), which ultimately becomes the spore. Thus, the forespore is surrounded by two membranes. The IMS between these two membranes is the assembly site of two layers of specialized peptidoglycan, called the germ cell wall and the cortex. Consequently, proteins residing in the germ cell wall or cortex must be sorted to the IMS between the forespore and the mother cell. This subcellular compartmentalization imposes a requirement for intracellular protein sorting on the cells, because protein synthesis is limited to the cytoplasm of the mother cell and the forespore.
Spore Protein Traffic
One of the processes that requires protein transport during sporulation is the communication between the mother cell and the forespore. Two proteins were shown to be exported by the forespore and to interact with membrane proteins of the mother cell. First, the SpoIIR protein, synthesized in the forespore prior to engulfment, contains a functional signal peptide which can drive the export of the mature part of the protein. The mature SpoIIR protein is thought to activate, directly or indirectly, the receptor/protease SpoIIGA, which is required for pro-ςE processing (120, 121). Second, the SpoIVB protein is synthesized in the forespore and transported across the forespore inner membrane after engulfment has taken place. SpoIVB probably remains anchored to the latter membrane, but a smaller form seems to be released into the IMS of the spore, allowing the activation of receptors and proteases in the outer forespore membrane that are responsible for pro-ςK processing (63). As the amino terminus of SpoIVB contains a putative signal peptide but lacks a putative SPase I cleavage site, it is presently unclear which protease is responsible for the processing and subsequent release of SpoIVB in the IMS of the forespore. Other processes in sporulation which require transport of proteins are the biogenesis of the germ wall and spore-cortex in the IMS of the forespore and the degradation of the spore peptidoglycan during germination. CwlD and DacB (also known as PBP5*) (218, 220) are the only proteins with a putative signal peptide that were reported to be involved in cortex synthesis. However, the precise subcellular localization of DacB and CwlD has not yet been documented. The germination-specific amidase SleB was found to be localized on the exterior side of the cortex in spores, while its synthesis is forespore specific (175). The fact that pre-SleB has to be transported across the forespore inner membrane and processed into its mature form to reach the IMS implies that a functional protein translocation machinery and at least one of the type I SPases are present in the forespore inner membrane. Other proteins involved in spore-cortex synthesis, such as SpoVB and SpoVE (279), are predicted to be transmembrane proteins with loops exposed in the IMS of the forespore. Finally, the recent finding that TasA (for translocated antibacterial spore-associated protein), a protein with a broad spectrum of antibacterial activity, is transported to B. subtilis endospores provides another example of spore-specific protein sorting. TasA is thought to confer a competitive advantage to the spore during the onset of sporulation and later, during germination, by inhibiting the growth of other organisms (276). In addition, TasA has been suggested to be required for proper spore coat assembly (258).
Factors Involved in Spore Protein Traffic
Although not much is known about the factors involved in protein transport from one sporulation-specific compartment to another, it is conceivable that certain factors involved in the secretion of proteins by vegetative cells are also involved in the sorting of proteins, such as TasA and SleB, to the IMS of forespores. Three lines of evidence indicate that this is indeed the case. First, SecA is likely to be involved, directly or indirectly via the secretion of certain Phr peptides, in some of these sorting events, as divA mutant strains are defective in sporulation (6, 238, 284). Second, a requirement for the type I SPases SipT and SipV for sporulation was recently reported (132). Finally, recent studies to identify determinants of the subcellular sorting of TasA showed that SipW is required for this process (258, 276, 293a).
PERSPECTIVES
As described in this review, important progress has been made in characterization of the protein transport machineries of B. subtilis, the Sec machinery in particular, since the complete genome sequence of this versatile and amenable organism was published by Kunst et al. in 1997 (149). The present “genome-based” predictions concerning the various types of signal peptides that direct protein transport in B. subtilis have fruitfully built upon these sequencing data and will, hopefully, form a solid basis for further studies leading to a full definition of the secretome. Major challenges for future research are provided by the recent identification of alternative Sec-independent pathways for protein transport, such as the Tat pathway. First, it will be very important to identify the critical components of these novel pathways. This is of particular relevance in the case of the protein transport pathways facilitating spore protein traffic, of which close to nothing is presently known. Second, and perhaps even more interesting, the factors that direct different exported proteins into distinct pathways for protein transport in B. subtilis, for example the Sec or the Tat pathway, need to be defined. Finally, from the applied point of view, it will be of particular importance to unravel the mechanisms that the Bacillus cell uses for optimization of its specificity and capacity for protein secretion, the folding of secreted proteins, and their quality control. The fact that B. subtilis can modulate its capacity and specificity for protein secretion through temporally controlled expression of the gene (sipS) for a secretion pathway component was first documented by Bolhuis et al. in 1996 (26). Strikingly, those studies showed that the sipS gene was expressed in concert with the genes for degradative enzymes, its transcription being under the control of the DegS-DegU two-component regulatory system, which is one of the major regulatory systems involved in the synthesis of degradative enzymes in the postexponential growth phase (95). In the meantime, it has been shown that the expression of genes for other secretion pathway components, such as SecA (116), SecDF (27), SipP (292), SipT (289, 290), SipW (276, 277), SPase II (291), SppA (30), BdbA, and BdbB (31), is regulated in a growth phase-dependent manner and/or in response to changes in the environment. In contrast, the genes for other components, such as TepA (30), SipU, SipV (289, 290), and BdbC (31) appeared to be expressed constitutively. Strikingly, the expression patterns for many of these genes show considerable differences (31). It seems likely that the different patterns reflect different responses of the cell to prevent potentially detrimental situations that can be caused by high-level protein secretion, but the molecular basis for such responses is presently not clearly understood. The implementation of advanced technologies, including DNA array and proteome-secretome analyses, will be required to evaluate the biological relevance of the latter observations and, ultimately, to exploit the natural protection mechanisms against “secretion stress” for the high-level production of an extended range of proteins of commercial value by the B. subtilis cell factory. These are major aims of our ongoing research that is carried out in close collaboration with partners in the so-called European Bacillus Secretion Group.
ACKNOWLEDGMENTS
We thank A. de Jong for technical assistance with signal peptide predictions and G. Venema and members of the European Bacillus Secretion Group (http://www.ncl.ac.uk/ebsg) for stimulating discussions.
H.T. was supported by Genencor International (Rijswijk, The Netherlands) and Gist-brocades B.V. (Delft, The Netherlands); A.B., S.B., and J.M.V.D. were supported by Biotechnology grants Bio2-CT93-0254, Bio4-CT95-0278, and Bio4-CT96-0097 from the European Union. J.D.H.J. was supported by a grant (805-33.605) from SLW (Stichting Levenswetenschappen). In addition, S.B. and J.M.V.D. were supported by “Quality of Life and Management of Living Resources” grants QLK3-CT-1999-00415 and QLK3-CT-1999-00917 from the European Union (Framework V Programme).
The first two authors contributed equally to this work.
REFERENCES
- 1.Akita M, Sasaki S, Matsuyama S, Mizushima S. SecA interacts with secretory proteins by recognizing the positive charge at the amino terminus of the signal peptide in Escherichia coli. J Biol Chem. 1990;265:8162–8169. [PubMed] [Google Scholar]
- 2.Akiyama Y, Kihara A, Tokuda H, Ito K. FtsH (HflB) is an ATP-dependent protease selectively acting on SecY and some other membrane proteins. J Biol Chem. 1996;271:31196–31201. doi: 10.1074/jbc.271.49.31196. [DOI] [PubMed] [Google Scholar]
- 3.Altamura N, Capitanio N, Bonnefoy N, Papa S, Dujardin G. The Saccharomyces cerevisiae OXA1 gene is required for the correct assembly of cytochrome c oxidase and oligomycin-sensitive ATP synthase. FEBS Lett. 1996;382:111–115. doi: 10.1016/0014-5793(96)00165-2. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersen C L, Matthey-Dupraz A, Missiakas D, Raina S. A new Escherichia coli gene, dsbG, encodes a periplasmic protein involved in disulfide bond formation, required for recycling DsbA/DsbB and DsbC redox proteins. Mol Microbiol. 1997;26:121–132. doi: 10.1046/j.1365-2958.1997.5581925.x. [DOI] [PubMed] [Google Scholar]
- 6.Asai K, Kawamura F, Sadaie Y, Takahashi H J. Isolation and characterization of a sporulation initiation mutation in the Bacillus subtilis secA gene. J Bacteriol. 1997;179:544–547. doi: 10.1128/jb.179.2.544-547.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Awade A, Cleuziat P, Gonzales T, Robert-Baudouy J. Characterization of the pcp gene encoding the pyrrolidone carboxyl peptidase of Bacillus subtilis. FEBS Lett. 1992;305:67–73. doi: 10.1016/0014-5793(92)80656-2. [DOI] [PubMed] [Google Scholar]
- 8.Baba T, Schneewind O. Target cell specificity of a bacteriocin molecule: a C-terminal signal directs lysostaphin to the cell wall of Staphylococcus aureus. EMBO J. 1996;15:4789–4797. [PMC free article] [PubMed] [Google Scholar]
- 9.Baba T, Schneewind O. Targeting of muralytic enzymes to the cell division site of Gram-positive bacteria: repeat domains direct autolysin to the equatorial surface ring of Staphylococcus aureus. EMBO J. 1998;17:4639–4646. doi: 10.1093/emboj/17.16.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babe L M, Schmidt B. Purification and biochemical analysis of WprA, a 52-kDa serine protease secreted by Bacillus subtilis as an active complex with its 23-kDa propeptide. Biochim Biophys Acta. 1998;1386:211–219. doi: 10.1016/s0167-4838(98)00110-1. [DOI] [PubMed] [Google Scholar]
- 11.Banerjee S, Hansen J N. Structure and expression of a gene encoding the precursor of subtilin, a small protein antibiotic. J Biol Chem. 1988;263:9508–9514. [PubMed] [Google Scholar]
- 12.Barkocy-Gallagher G A, Bassford P J. Synthesis of precursor maltose-binding protein with proline in the +1 position of the cleavage site interferes with the activity of Escherichia coli signal peptidase I in vivo. J Biol Chem. 1992;267:1231–1238. [PubMed] [Google Scholar]
- 13.Bauer M, Behrens M, Esser K, Michaelis G, Pratje E. PET1402, a nuclear gene required for proteolytic processing of cytochrome oxidase subunit 2 in yeast. Mol Gen Genet. 1994;245:272–278. doi: 10.1007/BF00290106. [DOI] [PubMed] [Google Scholar]
- 14.Beaman T C, Hitchins A D, Ochi K, Vasantha N, Endo T, Freese E L. Specificity and control of uptake of purines and other compounds in Bacillus subtilis. J Bacteriol. 1983;156:1107–1117. doi: 10.1128/jb.156.3.1107-1117.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beck K, Wu L F, Brunner J, Muller M. Discrimination between SRP- and SecA/SecB-dependent substrates involves selective recognition of nascent chains by SRP and trigger factor. EMBO J. 2000;19:134–143. doi: 10.1093/emboj/19.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beissinger M, Buchner J. How chaperones fold proteins. J Biol Chem. 1998;379:245–259. [PubMed] [Google Scholar]
- 17.Bengtsson J, Tjalsma H, Rivolta C, Hederstedt L. Subunit II of Bacillus subtilis cytochrome c oxidase is a lipoprotein. J Bacteriol. 1999;181:685–688. doi: 10.1128/jb.181.2.685-688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berks B C. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol. 1996;22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 19.Biederer T, Volkwein C, Sommer T. Role of Cue1p in ubiquitination and degradation at the ER surface. Science. 1997;278:1806–1809. doi: 10.1126/science.278.5344.1806. [DOI] [PubMed] [Google Scholar]
- 20.Blackman S A, Smith T J, Foster S J. The role of autolysins during vegetative growth of Bacillus subtilis 168. Microbiol. 1998;144:73–82. doi: 10.1099/00221287-144-1-73. [DOI] [PubMed] [Google Scholar]
- 21.Bochkareva E S, Lissin N M, Girshovich A S. Transient association of newly synthesized unfolded proteins with the heat-shock GroEL protein. Nature. 1988;336:254–257. doi: 10.1038/336254a0. [DOI] [PubMed] [Google Scholar]
- 22.Bochkareva E S, Solovieva M E, Girshovich A S. Targeting of GroEL to SecA on the cytoplasmic membrane of Escherichia coli. Proc Natl Acad Sci USA. 1998;95:478–483. doi: 10.1073/pnas.95.2.478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bogsch E, Brink S, Robinson C. Pathway specificity for a delta pH-dependent precursor thylakoid lumen protein is governed by a ‘Sec-avoidance’ motif in the transfer peptide and a ‘Sec-incompatible’ mature protein. EMBO J. 1997;16:3851–3859. doi: 10.1093/emboj/16.13.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bogsch E, Sargent F, Stanley N R, Berks B C, Robinson C, Palmer T. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J Biol Chem. 1998;273:18003–18006. doi: 10.1074/jbc.273.29.18003. [DOI] [PubMed] [Google Scholar]
- 25.Böhni P C, Deshaies R J, Schekman R W. Sec11 is required for signal peptide processing and yeast cell growth. J Cell Biol. 1988;106:1035–1042. doi: 10.1083/jcb.106.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolhuis A, Sorokin A, Azevedo V, Ehrlich S D, Braun P G, de Jong A, Venema G, Bron S, van Dijl J M. Bacillus subtilis can modulate its capacity and specificity for protein secretion by temporally controlled expression of the sipS gene for signal peptidase I. Mol Microbiol. 1996;22:605–618. doi: 10.1046/j.1365-2958.1996.d01-4676.x. [DOI] [PubMed] [Google Scholar]
- 27.Bolhuis A, Broekhuizen C P, Sorokin A, van Roosmalen M L, Venema G, Bron S, Quax W J, van Dijl J M. SecDF of Bacillus subtilis, a molecular Siamese twin required for the efficient secretion of proteins. J Biol Chem. 1998;273:21217–21224. doi: 10.1074/jbc.273.33.21217. [DOI] [PubMed] [Google Scholar]
- 28.Bolhuis A, Tjalsma H, Stephenson K, Harwood C R, Venema G, Bron S, van Dijl J M. Different mechanisms for thermal inactivation of Bacillus subtilis signal peptidase mutants. J Biol Chem. 1999;274:15865–15868. doi: 10.1074/jbc.274.22.15865. [DOI] [PubMed] [Google Scholar]
- 29.Bolhuis A, Tjalsma H, Smith H E, de Jong A, Meima R, Venema G, Bron S, van Dijl J M. Evaluation of bottlenecks in the late stages of protein secretion in Bacillus subtilis. Appl Environ Microbiol. 1999;65:2934–2941. doi: 10.1128/aem.65.7.2934-2941.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bolhuis A, Matzen A, Hyyrylaïnen H L, Kontinen V P, Meima R, Chapuis J, Venema G, Bron S, Freudl R, van Dijl J M. Signal peptide peptidase- and ClpP-like proteins of Bacillus subtilis are required for efficient translocation and processing of secretory proteins. J Biol Chem. 1999;274:24585–24592. doi: 10.1074/jbc.274.35.24585. [DOI] [PubMed] [Google Scholar]
- 31.Bolhuis A, Venema G, Quax W J, Bron S, van Dijl R J M. Functional analysis of paralogous thiol-disulfide oxidoreductases in Bacillus subtilis. J Biol Chem. 1999;274:24531–24538. doi: 10.1074/jbc.274.35.24531. [DOI] [PubMed] [Google Scholar]
- 32.Bolhuis A, Koetje E, Dubois J-Y, Vehmaanperä J, Venema G, Bron S, van Dijl J M. Did the mitochondrial processing peptidase evolve from a eubacterial regulator of gene expression? Mol Biol Evol. 2000;17:198–201. doi: 10.1093/oxfordjournals.molbev.a026232. [DOI] [PubMed] [Google Scholar]
- 33.Bonnefoy N, Kermorgan M, Groudinsky O, Minet M, Slonimski P P, Dujardin G. Cloning of a human gene involved in cytochrome oxidase assembly by functional complementation of an oxa1-mutation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1994;91:11978–11982. doi: 10.1073/pnas.91.25.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowie J U. Helix packing in membrane proteins. J Mol Biol. 1997;272:780–789. doi: 10.1006/jmbi.1997.1279. [DOI] [PubMed] [Google Scholar]
- 35.Boyd D, Schierle C, Beckwith J. How many membrane proteins are there? Protein Sci. 1998;7:201–205. doi: 10.1002/pro.5560070121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braun P, Gerritse G, van Dijl J M, Quax W J. Improving protein secretion by engineering components of the bacterial translocation machinery. Curr Opin Biotechnol. 1999;10:376–381. doi: 10.1016/s0958-1669(99)80068-8. [DOI] [PubMed] [Google Scholar]
- 37.Briggs M S, Cornell D G, Dluhy R A, Gierasch L M. Conformations of signal peptides induced by lipids suggest initial steps in protein export. Science. 1986;233:206–208. doi: 10.1126/science.2941862. [DOI] [PubMed] [Google Scholar]
- 38.Brink S, Bogsch E G, Edwards W R, Hynds P J, Robinson C. Targeting of thylakoid proteins by the delta pH-driven twin-arginine translocation pathway requires a specific signal in the hydrophobic domain in conjunction with the twin-arginine motif. FEBS Lett. 1998;434:425–430. doi: 10.1016/s0014-5793(98)01028-x. [DOI] [PubMed] [Google Scholar]
- 39.Brink S, Bogsch E G, Mant A, Robinson C. Unusual characteristics of amino-terminal and hydrophobic domains in nuclear-encoded thylakoid signal peptides. Eur J Biochem. 1997;245:340–348. doi: 10.1111/j.1432-1033.1997.00340.x. [DOI] [PubMed] [Google Scholar]
- 40.Bron S, Bolhuis A, Tjalsma H, Holsappel S, Venema G, van Dijl J M. Protein secretion and possible roles for multiple signal peptidases for precursor processing in bacilli. J Biotechnol. 1998;64:3–13. doi: 10.1016/s0168-1656(98)00099-6. [DOI] [PubMed] [Google Scholar]
- 41.Bron S, Meima R, van Dijl J M, Wipat A, Harwood C. Molecular biology and genetics of Bacillus spp., p 392–416. In: Demain A L, Davies E, editors. Manual of industrial microbiology and biotechnology. 2nd ed. Washington, D.C.: ASM Press; 1999. [Google Scholar]
- 42.Brundage L, Hendrick J P, Schiebel E, Driessen A J M, Wickner W. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- 43.Buchanan C E, Ling M L. Isolation and sequence analysis of dacB, which encodes a sporulation-specific penicillin-binding protein in Bacillus subtilis. J Bacteriol. 1992;174:1717–1725. doi: 10.1128/jb.174.6.1717-1725.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, et al. Complete genome sequence of the methanogenic archaeon Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 45.Calogero S, Gardan R, Glaser P, Schweizer J, Rapoport G, Debarbouille M. RocR, a novel regulatory protein controlling arginine utilization in Bacillus subtilis, belongs to the NtrC/NifA family of transcriptional activators. J Bacteriol. 1994;176:1234–1241. doi: 10.1128/jb.176.5.1234-1241.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casadio R, Fariselli P, Taroni C, Compiani M. A predictor of transmembrane alpha-helix domains of proteins based on neural networks. Eur Biophys J. 1996;24:165–178. doi: 10.1007/BF00180274. [DOI] [PubMed] [Google Scholar]
- 47.Chaddock A M, Mant A, Karnauchov I, Brink S, Herrmann R G, Klösgen R B, Robinson C. A new type of signal peptide: central role of a twin-arginine motif in transfer signals for the delta pH-dependent thylakoidal protein translocase. EMBO J. 1995;14:2715–2722. doi: 10.1002/j.1460-2075.1995.tb07272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chakicherla A, Hansen J N. Role of the leader and structural regions of prelantibiotic peptides as assessed by expressing nisin-subtilin chimeras in Bacillus subtilis 168, and characterization of their physical, chemical, and antimicrobial properties. J Biol Chem. 1995;270:23533–23539. doi: 10.1074/jbc.270.40.23533. [DOI] [PubMed] [Google Scholar]
- 49.Chambert R, Benyahia F, Petit-Glatron M F. Secretion of Bacillus subtilis levansucrase. Fe(III) could act as a cofactor in an efficient coupling of the folding and translocation processes. Biochem J. 1990;265:375–382. doi: 10.1042/bj2650375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chambert R, Petit-Glatron M F. Anionic polymers of Bacillus subtilis cell wall modulate the folding rate of secreted proteins. FEMS Microbiol Lett. 1999;179:43–47. doi: 10.1111/j.1574-6968.1999.tb08705.x. [DOI] [PubMed] [Google Scholar]
- 51.Chen M, Nagarajan V. Effect of alteration of charged residues at the N termini of signal peptides on protein export in Bacillus subtilis. J Bacteriol. 1994;176:5796–5801. doi: 10.1128/jb.176.18.5796-5801.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chung Y J, Steen M T, Hansen J N. The subtilin gene of Bacillus subtilis ATCC 6633 is encoded in an operon that contains a homolog of the hemolysin B transport protein. J Bacteriol. 1992;174:1417–1422. doi: 10.1128/jb.174.4.1417-1422.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chung Y S, Dubnau D. ComC is required for the processing and translocation of ComGC, a pilin-like competence protein of Bacillus subtilis. Mol Microbiol. 1995;15:543–551. doi: 10.1111/j.1365-2958.1995.tb02267.x. [DOI] [PubMed] [Google Scholar]
- 54.Chung Y S, Dubnau D. All seven comG open reading frames are required for DNA binding during transformation of competent Bacillus subtilis. J Bacteriol. 1998;180:41–45. doi: 10.1128/jb.180.1.41-45.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chung Y S, Breidt F, Dubnau D. Cell surface localization and processing of the ComG proteins, required for DNA binding during transformation of Bacillus subtilis. Mol Microbiol. 1998;29:905–913. doi: 10.1046/j.1365-2958.1998.00989.x. [DOI] [PubMed] [Google Scholar]
- 56.Clark S A, Theg S M. A folded protein can be transported across the chloroplast envelope and thylakoid membranes. Mol Biol Cell. 1997;8:923–934. doi: 10.1091/mbc.8.5.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Conlin C A, Trun N J, Silhavy T J, Miller C G. Escherichia coli prlC encodes an endopeptidase and is homologous to the Salmonella typhimurium opdA gene. J Bacteriol. 1992;174:5881–5887. doi: 10.1128/jb.174.18.5881-5887.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Creighton A M, Hulford A, Mant A, Robinson D, Robinson C. A monomeric, tightly folded stromal intermediate on the ΔpH-dependent thylakoidal protein transport pathway. J Biol Chem. 1995;270:1663–1669. doi: 10.1074/jbc.270.4.1663. [DOI] [PubMed] [Google Scholar]
- 59.Cristóbal S, Scotti P, Luirink J, von Heijne G, de Gier J W. The signal recognition particle-targeting pathway does not necessarily deliver proteins to the Sec-translocase in Escherichia coli. J Biol Chem. 1999;274:20068–20070. doi: 10.1074/jbc.274.29.20068. [DOI] [PubMed] [Google Scholar]
- 60.Cristóbal S, de Gier J W, Nielsen H, von Heijne G. Competition between Sec- and Tat-dependent protein translocation in Escherichia coli. EMBO J. 1999;18:2982–2990. doi: 10.1093/emboj/18.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;6:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 62.Cunningham K, Wickner W. Specific recognition of the leader region of precursor proteins is required for the activation of translocation ATPase of Escherichia coli. Proc Natl Acad Sci USA. 1989;86:8630–8634. doi: 10.1073/pnas.86.22.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cutting S, Driks A, Schmidt R, Kunkel B, Losick R. Forespore-specific transcription of a gene in the signal transduction pathway that governs pro-sigma K processing in Bacillus subtilis. Genes Dev. 1991;5:456–466. doi: 10.1101/gad.5.3.456. [DOI] [PubMed] [Google Scholar]
- 64.Dalbey R E, von Heijne G. Signal peptidases in prokaryotes and eukaryotes: a new protease family. Trends Biochem Sci. 1992;17:474–478. doi: 10.1016/0968-0004(92)90492-r. [DOI] [PubMed] [Google Scholar]
- 65.Dalbey R E, Robinson C. Protein translocation into and across the bacterial plasma membrane and the plant thylaloid membrane. Trends Biochem Sci. 1999;24:17–22. doi: 10.1016/s0968-0004(98)01333-4. [DOI] [PubMed] [Google Scholar]
- 66.Dalbey R E, Wickner W. Leader peptidase catalyzes the release of exported proteins from the outher surface of the Escherichia coli plasma membrane. J Biol Chem. 1985;260:15925–15931. [PubMed] [Google Scholar]
- 67.Dalbey R E, Lively M O, Bron S, van Dijl J M. The chemistry and enzymology of the type I signal peptidases. Protein Sci. 1997;6:1129–1138. doi: 10.1002/pro.5560060601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dartigalongue C, Raina S. A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J. 1998;17:3968–3980. doi: 10.1093/emboj/17.14.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.de Gier J W, Scotti P A, Saaf A, Valent Q A, Kuhn A, Luirink J, von Heijne G. Differential use of the signal recognition particle translocase targeting pathway for inner membrane protein assembly in Escherichia coli. Proc Natl Acad Sci USA. 1998;95:14646–146451. doi: 10.1073/pnas.95.25.14646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.den Blaauwen T, Fekkes P, de Wit J G, Kuiper W, Driessen A J M. Domain interactions of the peripheral pre-protein translocase subunit SecA. Biochemistry. 1996;35:11194–12004. doi: 10.1021/bi9605088. [DOI] [PubMed] [Google Scholar]
- 71.Deuerling E, Mogk A, Richter C, Purucker M, Schumann W. The ftsH gene of Bacillus subtilis is involved in major cellular processes such as sporulation, stress adaptation and secretion. Mol Microbiol. 1997;23:921–933. doi: 10.1046/j.1365-2958.1997.2721636.x. [DOI] [PubMed] [Google Scholar]
- 72.Deuerling E, Paeslack B, Schumann W. The ftsH gene of Bacillus subtilis is transiently induced after osmotic and temperature upshift. J Bacteriol. 1995;177:4105–4112. doi: 10.1128/jb.177.14.4105-4112.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.de Vos W M, Kuipers O P, van der Meer J R, Siezen R J. Maturation pathway of nisin and other lantibiotics: post-translationally modified antimicrobial peptides exported by Gram-positive bacteria. Mol Microbiol. 1995;17:427–437. doi: 10.1111/j.1365-2958.1995.mmi_17030427.x. [DOI] [PubMed] [Google Scholar]
- 74.de Vrije T, Batenburg A M, Killian J A, De Kruijff B. Lipid involvement in protein translocation in Escherichia coli. Mol Microbiol. 1990;4:143–150. doi: 10.1111/j.1365-2958.1990.tb02024.x. [DOI] [PubMed] [Google Scholar]
- 75.Doi R H. Proteolytic activities in Bacillus. Curr Opin Biotechnol. 1991;2:682–684. doi: 10.1016/0958-1669(91)90034-3. [DOI] [PubMed] [Google Scholar]
- 76.Dolinski K, Muir S, Cardenas M, Heitman J. All cyclophilins and FK506 binding proteins are, individually and collectively, dispensable for viability in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:13093–13098. doi: 10.1073/pnas.94.24.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Driessen A J M, Fekkes P, van der Wolk J P. The Sec system. Curr Opin Microbiol. 1998;1:216–222. doi: 10.1016/s1369-5274(98)80014-3. [DOI] [PubMed] [Google Scholar]
- 78.Driessen A J M. Bacterial protein translocation: kinetic and thermodynamic role of ATP and the protonmotive force. Trends Biochem Sci. 1992;17:219–223. doi: 10.1016/0968-0004(92)90381-i. [DOI] [PubMed] [Google Scholar]
- 79.Driessen A J M. How proteins cross the bacterial cytoplasmic membrane. J Membr Biol. 1994;142:145–159. doi: 10.1007/BF00234937. [DOI] [PubMed] [Google Scholar]
- 80.Dubnau D. Binding and transport of transforming DNA by Bacillus subtilis: the role of type-IV pilin-like proteins—a review. Gene. 1997;192:191–198. doi: 10.1016/s0378-1119(96)00804-9. [DOI] [PubMed] [Google Scholar]
- 81.Duitman E H, Hamoen L W, Rembold W, Venema G, Seitz H, Saenger W, Bernhard F, Reinhardt R, Schmidt M, Ullrich C, Stein T, Leenders F, Vater J. The mycosubtilin synthetase of Bacillus subtilis ATCC 6633: a multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc Natl Acad Sci USA. 1999;96:13294–13299. doi: 10.1073/pnas.96.23.13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Duong F, Wickner W. Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of pre-protein translocase holoenzyme. EMBO J. 1997;16:2756–2768. doi: 10.1093/emboj/16.10.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Duong F, Eichler J, Price A, Leonard M R, Wickner W. Biogenesis of the Gram-negative bacterial envelope. Cell. 1997;91:567–573. doi: 10.1016/s0092-8674(00)80444-4. [DOI] [PubMed] [Google Scholar]
- 84.Economou A. Bacterial pre-protein translocase: mechanism and conformational dynamics of a processive enzyme. Mol Microbiol. 1998;27:511–518. doi: 10.1046/j.1365-2958.1998.00713.x. [DOI] [PubMed] [Google Scholar]
- 85.Economou A, Wickner W. SecA promotes pre-protein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 86.Economou A, Pogliano J A, Beckwith J, Oliver D B, Wickner W. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell. 1995;83:1171–1181. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 87.Eichler J, Brunner J, Wickner W. The protease-protected 30 kDa domain of SecA is largely inaccessible to the membrane lipid phase. EMBO J. 1997;16:2188–2196. doi: 10.1093/emboj/16.9.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Eichler J, Wickner W. Both an N-terminal 65-kDa domain and a C-terminal 30-kDa domain of SecA cycle into the membrane at SecYEG during translocation. Proc Natl Acad Sci USA. 1997;94:5574–5578. doi: 10.1073/pnas.94.11.5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ewalt K L, Hendrick J P, Houry W A, Hartl F U. In vivo observation of polypeptide flux through the bacterial chaperonin system. Cell. 1997;90:491–500. doi: 10.1016/s0092-8674(00)80509-7. [DOI] [PubMed] [Google Scholar]
- 91.Fahnestock S R, Fisher K E. Expression of the staphylococcal protein A gene in Bacillus subtilis by gene fusions utilizing the promoter from a Bacillus amyloliquefaciens α-amylase gene. J Bacteriol. 1986;165:796–804. doi: 10.1128/jb.165.3.796-804.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fekkes P, van der Does C, Driessen A J M. The molecular chaperone SecB is released from the carboxyl terminus of SecA during initiation of precursor potein translocation. EMBO J. 1997;16:6105–6113. doi: 10.1093/emboj/16.20.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fekkes P, de Wit J G, van der Wolk J P, Kimsey H H, Kumamoto C A, Driessen A J M. Pre-protein transfer to the Escherichia coli translocase requires the co-operative binding of SecB and the signal sequence to SecA. Mol Microbiol. 1998;29:1179–1190. doi: 10.1046/j.1365-2958.1998.00997.x. [DOI] [PubMed] [Google Scholar]
- 94.Fekkes P, Driessen A J M. Protein targeting to the bacterial cytoplasmic membrane. Microbiol Mol Biol Rev. 1999;63:161–173. doi: 10.1128/mmbr.63.1.161-173.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ferrari E, Jarnagin A S, Schmidt B F. Commercial production of extracellular enzymes. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 917–937. [Google Scholar]
- 96.Foster S J. Analysis of the autolysins of Bacillus subtilis 168 during vegetative growth and differentiation by using renaturing polyacrylamide gel electrophoresis. J Bacteriol. 1992;174:464–470. doi: 10.1128/jb.174.2.464-470.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Foster S J. Molecular analysis of three major wall-associated proteins of Bacillus subtilis 168: evidence for processing of the product of a gene encoding a 258 kDa precursor two-domain ligand-binding protein. Mol Microbiol. 1993;8:299–310. doi: 10.1111/j.1365-2958.1993.tb01574.x. [DOI] [PubMed] [Google Scholar]
- 98.Franke C M, Tiemersma J, Venema G, Kok J. Membrane topology of the lactococcal bacteriocin ATP-binding cassette transporter protein LcnC. Involvement of LcnC in lactococcin a maturation. J Biol Chem. 1999;274:8484–8490. doi: 10.1074/jbc.274.13.8484. [DOI] [PubMed] [Google Scholar]
- 99.Galli G, Rodriguez F, Cosmina P, Pratesi C, Nogarotto R, de Ferra F, Grandi G. Characterization of the surfactin synthetase multi-enzyme complex. Biochim Biophys Acta. 1994;1205:19–28. doi: 10.1016/0167-4838(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 100.Geller B L, Movva N R, Wickner W. Both ATP and the electrochemical potential are required for optimal assembly of pro-OmpA into Escherichia coli inner membrane vesicles. Proc Natl Acad Sci USA. 1986;83:4219–4222. doi: 10.1073/pnas.83.12.4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gennity J, Goldstein J, Inouye M. Signal peptide mutants of Escherichia coli. J Bioenerg Biomembr. 1990;22:233–269. doi: 10.1007/BF00763167. [DOI] [PubMed] [Google Scholar]
- 102.Ghuysen J M, Lamotte-Brasseur J, Joris B, Shockman G D. Binding site-shaped repeated sequences of bacterial wall peptidoglycan hydrolases. FEBS Lett. 1994;342:23–28. doi: 10.1016/0014-5793(94)80577-6. [DOI] [PubMed] [Google Scholar]
- 103.Giam C Z, Chai T, Hayashi S, Wu H C. Prolipoprotein modification and processing in Escherichia coli: a unique secondary structure in prolipoprotein signal sequence for the recognition by glyceryl transferase. Eur J Biochem. 1984;141:331–337. doi: 10.1111/j.1432-1033.1984.tb08196.x. [DOI] [PubMed] [Google Scholar]
- 104.Glover G I, D'Ambrosio S M, Jensen R A. Versatile properties of a nonsaturable, homogeneous transport system in Bacillus subtilis: genetic, kinetic, and affinity labeling studies. Proc Natl Acad Sci USA. 1975;72:14–18. doi: 10.1073/pnas.72.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goffeau A, Aert R, Agostini-Carbone M L, Ahmed A, Aigle M, Alberghina L, Albermann K, Albers M, Aldea M, Alexandraki D, et al. The yeast genome directory. Nature. 1997;387:5–105. [Google Scholar]
- 106.Görlich D, Rapoport T A. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- 107.Gothel S F, Scholz C, Schmid F X, Marahiel M A. Cyclophilin and trigger factor from Bacillus subtilis catalyze in vitro protein folding and are necessary for viability under starvation conditions. Biochemistry. 1998;37:13392–13399. doi: 10.1021/bi981253w. [DOI] [PubMed] [Google Scholar]
- 108.Graumann P, Wendrich T M, Weber M H, Schroder K, Marahiel M A. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol Microbiol. 1997;25:741–756. doi: 10.1046/j.1365-2958.1997.5121878.x. [DOI] [PubMed] [Google Scholar]
- 109.Guthrie B, Wickner W. Trigger factor depletion or overproduction causes defective cell division but does not block protein export. J Bacteriol. 1990;172:5555–5562. doi: 10.1128/jb.172.10.5555-5562.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hartl F U, Lecker S, Schiebel E, Hendrick J P, Wickner W. The binding cascade of SecB to SecA to SecY/E mediates pre-protein targeting to the E. coli plasma membrane. Cell. 1990;63:269–279. doi: 10.1016/0092-8674(90)90160-g. [DOI] [PubMed] [Google Scholar]
- 111.Hartl F U. Highlight: protein folding in vivo. J Biol Chem. 1998;379:235. [PubMed] [Google Scholar]
- 112.Havarstein L S, Diep D B, Nes I F. A family of bacteriocin ABC transporters carry out proteolytic processing of their substrates concomitant with export. Mol Microbiol. 1995;16:229–240. doi: 10.1111/j.1365-2958.1995.tb02295.x. [DOI] [PubMed] [Google Scholar]
- 113.Hecker M, Schumann W, Volker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 114.Hell K, Herrmann J M, Pratje E, Neupert W, Stuart R A. Oxa1p mediates the export of the N- and C-termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett. 1997;418:367–370. doi: 10.1016/s0014-5793(97)01412-9. [DOI] [PubMed] [Google Scholar]
- 115.Hell K, Herrmann J M, Pratje E, Neupert W, Stuart R A. Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc Natl Acad Sci USA. 1998;95:2250–2255. doi: 10.1073/pnas.95.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Herbort M, Klein M, Manting E H, Driessen A J M, Freudl R J. Temporal expression of the Bacillus subtilis secA gene, encoding a central component of the preprotein translocase. J Bacteriol. 1999;181:493–500. doi: 10.1128/jb.181.2.493-500.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hesterkamp T, Hauser S, Lutcke H, Bukau B. Escherichia coli trigger factor is a prolyl isomerase that associates with nascent polypeptide chains. Proc Natl Acad Sci USA. 1996;93:4437–4444. doi: 10.1073/pnas.93.9.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hilt W, Wolf D H. Proteasomes: destruction as a programme. Trends Biochem Sci. 1996;21:96–102. [PubMed] [Google Scholar]
- 119.Hirose I, Sano K, Shiosa I, Kumano M, Nakamura K, Yamane K. Proteome analysis of Bacillus subtilis extracellular proteins: a two-dimensional protein electrophoretic study. Microbiology. 1999;146:65–75. doi: 10.1099/00221287-146-1-65. [DOI] [PubMed] [Google Scholar]
- 120.Hofmeister A. Activation of the proprotein transcription factor pro-sigma E is associated with its progression through three patterns of subcellular localization during sporulation in Bacillus subtilis. J Bacteriol. 1998;180:2426–2433. doi: 10.1128/jb.180.9.2426-2433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hofmeister A, Londono-Vallejo A, Harry E, Stragier P, Losick R. Extracellular signal protein triggering the proteolytic activation of a developmental transcription factor in Bacillus subtilis. Cell. 1995;83:219–226. doi: 10.1016/0092-8674(95)90163-9. [DOI] [PubMed] [Google Scholar]
- 122.Honda K, Nakamura K, Nishiguchi M, Yamane K. Cloning and characterization of a Bacillus subtilis gene encoding a homolog of the 54-kilodalton subunit of mammalian signal recognition particle and Escherichia coli Ffh. J Bacteriol. 1993;175:4885–4894. doi: 10.1128/jb.175.15.4885-4894.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hussain M, Ozawa Y, Ichihara S, Mizushima S. Signal peptide digestion in Escherichia coli: effect of protease inhibitors on hydrolysis of the cleaved signal peptide of the major outer-membrane lipoprotein. Eur J Biochem. 1982;129:233–239. doi: 10.1111/j.1432-1033.1982.tb07044.x. [DOI] [PubMed] [Google Scholar]
- 124.Hynds P J, Robinson D, Robinson C. The sec-independent twin-arginine translocation system can transport both tightly folded and malfolded proteins across the thylakoid membrane. J Biol Chem. 1998;273:34868–34874. doi: 10.1074/jbc.273.52.34868. [DOI] [PubMed] [Google Scholar]
- 125.Ichihara S, Beppu N, Mizushima S. Protease IV, a cytoplasmic membrane protein of Escherichia coli, has signal peptide peptidase activity. J Biol Chem. 1984;259:9853–9857. [PubMed] [Google Scholar]
- 126.Ichihara S, Suzuki T, Suzuki M, Mizushima S. Molecular cloning and sequencing of the sppA gene and characterization of the encoded protease IV, a signal peptide peptidase. J Biol Chem. 1986;261:9405–9411. [PubMed] [Google Scholar]
- 127.Ikemura H, Inouye H. In vitro processing of pro-subtilisin produced in Escherichia coli. J Biol Chem. 1988;263:12959–12963. [PubMed] [Google Scholar]
- 128.Ikemura H, Takagi H, Inouye H. Requirement of pro-sequence for the production of active subtilsin E in Escherichia coli. J Biol Chem. 1987;262:7859–7864. [PubMed] [Google Scholar]
- 129.Ishihara T, Tomita H, Hasegawa Y, Tsukagoshi N, Yamagata H, Udaka S. Cloning and characterization of the gene for a protein thiol-disulfide oxidoreductase in Bacillus brevis. J Bacteriol. 1995;177:745–749. doi: 10.1128/jb.177.3.745-749.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jacobs M, Andersen J B, Kontinen V P, Sarvas M. Bacillus subtilis PrsA is required in vivo as an extracytoplasmic chaperone for secretion of active enzymes synthesized either with or without pro-sequences. Mol Microbiol. 1993;8:957–966. doi: 10.1111/j.1365-2958.1993.tb01640.x. [DOI] [PubMed] [Google Scholar]
- 131.Jeong S M, Yoshikawa H, Takahashi H. Isolation and characterization of the secE homologue gene of Bacillus subtilis. Mol Microbiol. 1993;10:133–142. doi: 10.1111/j.1365-2958.1993.tb00910.x. [DOI] [PubMed] [Google Scholar]
- 132.Jiang M, Grau R, Perego M. Differential processing of propeptide inhibitors of Rap phosphatases in Bacillus subtilis. J Bacteriol. 2000;182:303–310. doi: 10.1128/jb.182.2.303-310.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Joly J C, Swartz J R. Protein folding activities of Escherichia coli protein disulfide isomerase. Biochemistry. 1994;33:4231–4236. doi: 10.1021/bi00180a017. [DOI] [PubMed] [Google Scholar]
- 134.Kawamura F, Doi R H. Construction of a Bacillus subtilis double mutant deficient in extracellular alkaline and neutral proteases. J Bacteriol. 1984;160:442–444. doi: 10.1128/jb.160.1.442-444.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Keiler K C, Waller P R, Sauer R T. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- 136.Kihara A, Akiyama Y, Ito K. Dislocation of membrane proteins in FtsH-mediated proteolysis. EMBO J. 1999;18:2970–2981. doi: 10.1093/emboj/18.11.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kim Y J, Rajapandi T, Oliver D. SecA protein is exposed to the periplasmic surface of the Escherichia coli inner membrane in its active state. Cell. 1994;78:845–853. doi: 10.1016/s0092-8674(94)90602-5. [DOI] [PubMed] [Google Scholar]
- 138.Klein C, Entian K D. Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC 6633. Appl Environ Microbiol. 1994;60:2793–2801. doi: 10.1128/aem.60.8.2793-2801.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 140.Klose M, Schimz K L, van der Wolk J P, Driessen A J M, Freudl R. Lysine 106 of the putative catalytic ATP-binding site of the Bacillus subtilis SecA protein is required for functional complementation of Escherichia coli secA mutants in vivo. J Biol Chem. 1993;268:4504–4510. [PubMed] [Google Scholar]
- 141.Knoblauch N T, Rudiger S, Schonfeld H J, Driessen A J M, Schneider-Mergener J, Bukau B. Substrate specificity of the SecB chaperone. J Biol Chem. 1999;274:34219–34225. doi: 10.1074/jbc.274.48.34219. [DOI] [PubMed] [Google Scholar]
- 142.Koch H G, Hengelage T, Neumann-Haefelin C, MacFarlane J, Hoffschulte H K, Schimz K L, Mechler B, Muller M. In vitro studies with purified components reveal signal recognition particle (SRP) and SecA/SecB as constituents of two independent protein-targeting pathways of Escherichia coli. Mol Biol Cell. 1999;10:2163–2173. doi: 10.1091/mbc.10.7.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Koide, Y., A. Nakamura, T. Uozumi, and T. Beppu. 1986. Cloning and sequencing of the major intracellular serine protease gene of Bacillus subtilis. 167:110–116. [DOI] [PMC free article] [PubMed]
- 144.Kontinen V P, Sarvas M. The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol Microbiol. 1993;8:727–737. doi: 10.1111/j.1365-2958.1993.tb01616.x. [DOI] [PubMed] [Google Scholar]
- 145.Kontinen V P, Saris P, Sarvas M. A gene (prsA) of Bacillus subtilis involved in a novel, late stage of protein export. Mol Microbiol. 1991;5:1273–1283. doi: 10.1111/j.1365-2958.1991.tb01901.x. [DOI] [PubMed] [Google Scholar]
- 146.Kopito R R. ER quality control: the cytoplasmic connection. Cell. 1997;88:427–430. doi: 10.1016/s0092-8674(00)81881-4. [DOI] [PubMed] [Google Scholar]
- 147.Kosic N, Sugai M, Fan C K, Wu H C. Processing of lipid-modified prolipoprotein requires energy and sec gene products in vivo. J Bacteriol. 1993;175:6113–6117. doi: 10.1128/jb.175.19.6113-6117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kumamoto C A, Francetic O. Highly selective binding of nascent polypeptides by an Escherichia coli chaperone protein in vivo. J Bacteriol. 1993;175:2184–2188. doi: 10.1128/jb.175.8.2184-2188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, et al. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 150.Kusukawa N, Yura T, Ueguchi C, Akiyama Y, Ito K. Effects of mutations in heat-shock genes groES and groEL on protein export in Escherichia coli. EMBO J. 1989;8:3517–3512. doi: 10.1002/j.1460-2075.1989.tb08517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.LaPointe C F, Taylor R K. The type IV prepilin peptidases comprise a novel family of aspartic acid proteases. J Biol Chem. 2000;275:1502–1510. doi: 10.1074/jbc.275.2.1502. [DOI] [PubMed] [Google Scholar]
- 152.Lazazzera B A, Solomon J M, Grossman A D. An exported peptide functions intracellularly to contribute to cell density signaling in Bacillus subtilis. Cell. 1997;13:917–925. doi: 10.1016/s0092-8674(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 153.Leskelä S, Wahlström E, Kontinen V P, Sarvas M. Lipid modification of prelipoproteins is dispensable for growth but essential for efficient protein secretion in Bacillus subtilis: characterization of the lgt gene. Mol Microbiol. 1999;31:1075–1085. doi: 10.1046/j.1365-2958.1999.01247.x. [DOI] [PubMed] [Google Scholar]
- 154.Lill R, Crooke E, Guthrie B, Wickner W. The “trigger factor cycle” includes ribosomes, presecretory proteins and the plasma membrane. Cell. 1988;54:1013–1018. doi: 10.1016/0092-8674(88)90116-x. [DOI] [PubMed] [Google Scholar]
- 155.Lill R, Cinningham K, Brundage L A, Ito K, Oliver D, Wickner W. SecA protein hydrolyses ATP and is an essential component of the protein translocation ATPase of Escherichia coli. EMBO J. 1989;8:961–966. doi: 10.1002/j.1460-2075.1989.tb03458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 157.Lory S. Leader peptidase of type IV prepilins and related proteins. In: von Heijne G, editor. Signal peptidases. R. G. Austin, Tex: Landes Company; 1994. pp. 17–29. [Google Scholar]
- 158.Lyon R L, Gibson C M, Caparon M G. A role for trigger factor and an Rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Magnuson R, Solomon J, Grossman A D. Biochemical and genetic characterization of a competence pheromone from Bacillus subtilis. Cell. 1994;77:207–216. doi: 10.1016/0092-8674(94)90313-1. [DOI] [PubMed] [Google Scholar]
- 160.Mansfeld J, Vriend G, Dijkstra B W, Veltman O R, van den Burg B, Venema G, Ulbrich-Hofmann R, Eijsink V G. Extreme stabilization of a thermolysin-like protease by an engineered disulfide bond. J Biol Chem. 1997;272:11152–11156. doi: 10.1074/jbc.272.17.11152. [DOI] [PubMed] [Google Scholar]
- 161.Manting E H, van der Does C, Driessen A J M. In vivo cross-linking of the SecA and SecY subunits of the Escherichia coli preprotein translocase. J Bacteriol. 1997;179:5699–5704. doi: 10.1128/jb.179.18.5699-5704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Marasco R, Varcamonti M, Ricca E, Sacco M. A new Bacillus subtilis gene with homology to Escherichia coli prc. Gene. 1996;183:149–152. doi: 10.1016/s0378-1119(96)00543-4. [DOI] [PubMed] [Google Scholar]
- 163.Margot P, Karamata D. The wprA gene of Bacillus subtilis 168, expressed during exponential growth, encodes a cell-wall-associated protease. Microbiology. 1996;142:3437–3444. doi: 10.1099/13500872-142-12-3437. [DOI] [PubMed] [Google Scholar]
- 164.Margot P, Wahlen M, Gholamhoseinian A, Piggot P, Karamata D. The lytE gene of Bacillus subtilis 168 encodes a cell wall hydrolase. J Bacteriol. 1998;180:749–752. doi: 10.1128/jb.180.3.749-752.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Matsumoto G, Mori H, Ito K. Roles of SecG in ATP- and SecA-dependent protein translocation. Proc Natl Acad Sci USA. 1998;95:13567–13572. doi: 10.1073/pnas.95.23.13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Matsuyama S, Fujita Y, Mizushima S. SecD is involved in the release of translocated secretory proteins from the cytoplasmic membrane of Escherichia coli. EMBO J. 1993;12:265–270. doi: 10.1002/j.1460-2075.1993.tb05652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Matsuyama S, Tajima T, Tokuda H. A novel periplasmic carrier protein involved in the sorting and transport of Escherichia coli lipoproteins destined for the outer membrane. EMBO J. 1995;14:3365–3372. doi: 10.1002/j.1460-2075.1995.tb07342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Matsuyama S, Yokota N, Tokuda H. A novel outer membrane lipoprotein, LolB (HemM), involved in the LolA (p20)-dependent localization of lipoproteins to the outer membrane of Escherichia coli. EMBO J. 1997;16:6947–6955. doi: 10.1093/emboj/16.23.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mazmanian S K, Liu G, Ton-That H, Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 170.Meens J, Herbort M, Klein M, Freudl R. Use of the pre-pro part of Staphylococcus hyicus lipase as a carrier for secretion of Escherichia coli outer membrane protein A (OmpA) prevents proteolytic degradation of OmpA by cell-associated protease(s) in two different gram-positive bacteria. Appl Environ Microbiol. 1997;63:2814–2820. doi: 10.1128/aem.63.7.2814-2820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Meijer W J J, de Jong A, Wisman G B A, Tjalsma H, Venema G, Bron S, van Dijl J M. The endogenous Bacillus subtilis (natto) plasmids pTA1015 and pTA1040 contain signal peptidase-encoding genes: identification of a new structural module on cryptic plasmids. Mol Microbiol. 1995;17:621–631. doi: 10.1111/j.1365-2958.1995.mmi_17040621.x. [DOI] [PubMed] [Google Scholar]
- 172.Meijer W J J, Wisman G B, Terpstra P, Thorsted P B, Thomas C M, Holsappel S, Venema G, Bron S. Rolling-circle plasmids from Bacillus subtilis: complete nucleotide sequences and analyses of genes of pTA1015, pTA1040, pTA1050 and pTA1060, and comparisons with related plasmids from Gram-positive bacteria. FEMS Microbiol Rev. 1998;21:337–368. doi: 10.1111/j.1574-6976.1998.tb00357.x. [DOI] [PubMed] [Google Scholar]
- 173.Merchante R, Pooley H M, Karamata D. A periplasm in Bacillus subtilis. J Bacteriol. 1995;177:6176–6183. doi: 10.1128/jb.177.21.6176-6183.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Missiakas D, Raina S. Protein misfolding in the cell envelope of Escherichia coli: new signaling pathways. Trends Biochem Sci. 1997;22:59–63. doi: 10.1016/s0968-0004(96)10072-4. [DOI] [PubMed] [Google Scholar]
- 175.Moriyama R, Fukuoka H, Miyata S, Kudoh S, Hattori A, Kozuka S, Yasuda Y, Tochikubo K, Makino S. Expression of a germination-specific amidase, SleB, of bacilli in the forespore compartment of sporulating cells and its localization on the exterior side of the cortex in dormant spores. J Bacteriol. 1999;181:2373–2378. doi: 10.1128/jb.181.8.2373-2378.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Msadek T, Dartois V, Kunst F, Herbaud M L, Denizot F, Rapoport G. ClpP of Bacillus subtilis is required for competence development, motility, degradative enzyme synthesis, growth at high temperature and sporulation. Mol Microbiol. 1998;27:899–914. doi: 10.1046/j.1365-2958.1998.00735.x. [DOI] [PubMed] [Google Scholar]
- 177.Msadek T, Kunst F, Rapoport G. Two-component regulatory systems. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 729–745. [Google Scholar]
- 178.Müller J, Walter F, van Dijl J M, Behnke D. Suppression of the growth and export defects of an Escherichia coli secA(Ts) mutant by a gene cloned from Bacillus subtilis. Mol Gen Genet. 1992;235:89–96. doi: 10.1007/BF00286185. [DOI] [PubMed] [Google Scholar]
- 179.Müller J, Bron S, Venema G, van Dijl J M. Chaperone-like activities of the CsaA protein of Bacillus subtilis. Microbiology. 2000;146:77–88. doi: 10.1099/00221287-146-1-77. [DOI] [PubMed] [Google Scholar]
- 179a.Müller J, Ozegowski J, Vettermann S, Swaving J, van Wely K H, Driessen A J M. Interaction of Bacillus subtilis CsaA with SecA and precursor proteins. Biochem J. 2000;348:367–373. [PMC free article] [PubMed] [Google Scholar]
- 180.Mullins C, Lu Y, Campbell A, Fang H, Green N. A mutation affecting signal peptidase inhibits degradation of an abnormal membrane protein in Saccharomyces cerevisiae. J Biol Chem. 1995;270:7139–7147. doi: 10.1074/jbc.270.29.17139. [DOI] [PubMed] [Google Scholar]
- 181.Murray T, Popham D L, Setlow P. Identification and characterization of pbpC, the gene encoding Bacillus subtilis penicillin-binding protein 3. J Bacteriol. 1996;178:6001–6005. doi: 10.1128/jb.178.20.6001-6005.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Murray T, Popham D L, Setlow P. Identification and characterization of pbpA, encoding Bacillus subtilis penicillin-binding protein 2A. J Bacteriol. 1997;179:3021–3029. doi: 10.1128/jb.179.9.3021-3029.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Muñoa F J, Miller K W, Beers R, Graham M, Wu H C. Membrane topology of Escherichia coli prolipoprotein signal peptidase (signal peptidase II) J Biol Chem. 1991;266:17667–17672. [PubMed] [Google Scholar]
- 184.Nagarajan V. Protein secretion. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 713–7726. [Google Scholar]
- 185.Nakamura K, Nakamura A, Takamatsu H, Yoshikawa H, Yamane K. Cloning and characterization of a Bacillus subtilis gene homologous to E. coli secY. J Biochem. 1990;107:603–607. doi: 10.1093/oxfordjournals.jbchem.a123093. [DOI] [PubMed] [Google Scholar]
- 186.Nakamura K, Imai Y, Nakamura A, Yamane K. Small cytoplasmic RNA of Bacillus subtilis: functional relationship with human signal recognition particle 7S RNA and Escherichia coli 4.5S RNA. J Bacteriol. 1992;174:2185–2192. doi: 10.1128/jb.174.7.2185-2192.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Nakamura K, Nishiguchi M, Honda K, Yamane K. The Bacillus subtilis SRP54 homologue, Ffh, has an intrinsic GTPase activity and forms a ribonucleoprotein complex with small cytoplasmic RNA in vivo. Biochem Biophys Res Commun. 1994;199:1394–1399. doi: 10.1006/bbrc.1994.1385. [DOI] [PubMed] [Google Scholar]
- 188.Nakamura K, Yahagi S, Yamazaki T, Yamane K. Bacillus subtilis histone-like protein, HBsu, is an integral component of a SRP-like particle that can bind the Alu domain of small cytoplasmic RNA. J Biol Chem. 1999;274:13569–13576. doi: 10.1074/jbc.274.19.13569. [DOI] [PubMed] [Google Scholar]
- 189.Nakano M M, Zuber P. Molecular biology of antibiotic production in Bacillus. Crit Rev Biotechnol. 1990;10:223–240. doi: 10.3109/07388559009038209. [DOI] [PubMed] [Google Scholar]
- 190.Navarre W W, Schneewind O. Proteolytic cleavage and cell wall anchoring at the LPXTG motif of surface proteins in Gram-positive bacteria. Mol Microbiol. 1994;14:115–121. doi: 10.1111/j.1365-2958.1994.tb01271.x. [DOI] [PubMed] [Google Scholar]
- 191.Navarre W W, Daefler S, Schneewind O. Cell wall sorting of lipoproteins in Staphylococcus aureus. J Bacteriol. 1996;178:441–446. doi: 10.1128/jb.178.2.441-446.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Nes I F, Tagg J R. Novel lantibiotics and their pre-peptides. Antonie Leeuwenhoek. 1996;69:89–97. doi: 10.1007/BF00399414. [DOI] [PubMed] [Google Scholar]
- 193.Netzer W J, Hartl F U. Protein folding in the cytosol: chaperonin-dependent and -independent mechanisms. Trends Biochem Sci. 1998;23:68–73. doi: 10.1016/s0968-0004(97)01171-7. [DOI] [PubMed] [Google Scholar]
- 194.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 195.Nielsen V S, Mant A, Knoetzel J, Moller B L, Robinson C. Import of barley photosystem I subunit N into the thylakoid lumen is mediated by a bipartite presequence lacking an intermediate processing site: role of the delta pH in translocation across the thylakoid membrane. J Biol Chem. 1994;269:3762–3766. [PubMed] [Google Scholar]
- 196.Nilsson I, von Heijne G. A signal peptide with a proline next to the cleavage site inhibits leader peptidase when present in a Sec-independent protein. FEBS Lett. 1992;299:243–246. doi: 10.1016/0014-5793(92)80124-y. [DOI] [PubMed] [Google Scholar]
- 197.Novak P, Ray P H, Dev I K. Localization and purification of two enzymes from Escherichia coli capable of hydrolyzing a signal peptide. J Biol Chem. 1986;261:420–427. [PubMed] [Google Scholar]
- 198.Novak P, Dev I K. Degradation of a signal peptide by protease IV and oligopeptidase A. J Bacteriol. 1988;170:5067–5075. doi: 10.1128/jb.170.11.5067-5075.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Nunn D N, Lory S. Product of the Pseudomonas aeruginosa gene pilD is a prepilin leader peptidase. Proc Natl Acad Sci USA. 1991;15:3281–3285. doi: 10.1073/pnas.88.8.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ogura A, Kakeshita H, Honda K, Takamatsu H, Nakamura K, Yamane K. srb: a Bacillus subtilis gene encoding a homologue of the alpha-subunit of the mammalian signal recognition particle receptor. DNA Res. 1995;2:95–100. doi: 10.1093/dnares/2.2.95. [DOI] [PubMed] [Google Scholar]
- 201.Overhoff B, Klein M, Spies M, Freudl R. Identification of a gene fragment which codes for the 364 amino-terminal amino acid residues of a SecA homologue from Bacillus subtilis: further evidence for the conservation of the protein export apparatus in Gram-positive and Gram-negative bacteria. Mol Gen Genet. 1991;228:417–423. doi: 10.1007/BF00260635. [DOI] [PubMed] [Google Scholar]
- 202.Paetzel M, Dalbey R E, Strynadka N C. Crystal structure of a bacterial signal peptidase in complex with a beta-lactam inhibitor. Nature. 1998;396:186–190. doi: 10.1038/24196. [DOI] [PubMed] [Google Scholar]
- 203.Paik S H, Chakicherla A, Hansen J N. Identification and characterization of the structural and transporter genes for, and the chemical and biological properties of, sublancin 168, a novel lantibiotic produced by Bacillus subtilis 168. J Biol Chem. 1998;273:23134–23142. doi: 10.1074/jbc.273.36.23134. [DOI] [PubMed] [Google Scholar]
- 204.Palva I. Molecular cloning of α-amylase gene from Bacillus amyloliquefaciens and its expression in Bacillus subtilis. Gene. 1982;19:81–87. doi: 10.1016/0378-1119(82)90191-3. [DOI] [PubMed] [Google Scholar]
- 205.Palva I. Engineering for secretion of proteins by bacteria. In: Baumberg S, Hunter I, Rhodes M, editors. Microbial products: new approaches. Cambridge, United Kingdom: Cambridge University Press; 1989. pp. 255–269. [Google Scholar]
- 206.Perego M, Hanstein C, Welsh K M, Djavakhishvili T, Glaser P, Hoch J A. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in Bacillus subtilis. Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 207.Perego M, Glaser P, Hoch J A. Aspartyl-phosphate phosphatases deactivate the response regulator components of the sporulation signal transduction system in Bacillus subtilis. Mol Microbiol. 1996;19:1151–1157. doi: 10.1111/j.1365-2958.1996.tb02460.x. [DOI] [PubMed] [Google Scholar]
- 208.Perego M, Hoch J A. Cell-cell communication regulates the effects of protein aspartate phosphatases on the phosphorelay controlling development in Bacillus subtilis. Proc Natl Acad Sci USA. 1996;93:1549–1553. doi: 10.1073/pnas.93.4.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Perego M. A peptide export-import control circuit modulating bacterial development regulates protein phosphatases of the phosphorelay. Proc Natl Acad Sci USA. 1997;94:8612–8617. doi: 10.1073/pnas.94.16.8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Peters T, Davidson L K. The biosynthesis of rat serum albumin: in vivo studies on the formation of the disulfide bonds. J Biol Chem. 1982;257:8847–8853. [PubMed] [Google Scholar]
- 211.Petit-Glatron M F, Grajcar L, Munz A, Chambert R. The contribution of the cell wall to a transmembrane calcium gradient could play a key role in Bacillus subtilis protein secretion. Mol Microbiol. 1993;9:1097–1106. doi: 10.1111/j.1365-2958.1993.tb01239.x. [DOI] [PubMed] [Google Scholar]
- 212.Petit-Glatron M F, Monteil I, Benyahia F, Chambert R. Bacillus subtilis levansucrase: amino acid substitutions at one site affect secretion efficiency and refolding kinetics mediated by metals. Mol Microbiol. 1990;4:2063–2070. doi: 10.1111/j.1365-2958.1990.tb00566.x. [DOI] [PubMed] [Google Scholar]
- 213.Plath K, Mothes W, Wilkinson B M, Stirling C J, Rapoport T A. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- 214.Plemper R K, Bohmler S, Bordallo J, Sommer T, Wolf D H. Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature. 1997;388:891–895. doi: 10.1038/42276. [DOI] [PubMed] [Google Scholar]
- 215.Pogliano K J, Beckwith J. The Cs sec mutants of Escherichia coli reflect the cold sensitivity of protein export itself. Genetics. 1993;133:763–773. doi: 10.1093/genetics/133.4.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Pohlschröder M, Prinz W A, Hartmann E, Beckwith J. Protein translocation in the three domains of life: variations on a theme. Cell. 1997;91:563–566. doi: 10.1016/s0092-8674(00)80443-2. [DOI] [PubMed] [Google Scholar]
- 217.Pooley H M, Merchante R, Karamata D. Overall protein content and induced enzyme components of the periplasm of Bacillus subtilis. Microb Drug Resist. 1996;2:9–15. doi: 10.1089/mdr.1996.2.9. [DOI] [PubMed] [Google Scholar]
- 218.Popham D L, Illades-Aguiar B, Setlow P. The Bacillus subtilis dacB gene, encoding penicillin-binding protein 5*, is part of a three-gene operon required for proper spore cortex synthesis and spore core dehydration. J Bacteriol. 1995;177:4721–4729. doi: 10.1128/jb.177.16.4721-4729.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Popham D L, Helin J, Costello C E, Setlow P. Analysis of the peptidoglycan structure of Bacillus subtilis endospores. J Bacteriol. 1996;178:6451–6458. doi: 10.1128/jb.178.22.6451-6458.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Popham D L, Meador-Parton J, Costello C E, Setlow P. Spore peptidoglycan structure in a cwlD dacB double mutant of Bacillus subtilis. J Bacteriol. 1999;181:6205–6209. doi: 10.1128/jb.181.19.6205-6209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Poquet I, Kornacker M G, Pugsley A P. The role of the lipoprotein sorting signal (aspartate +2) in pullulanase secretion. Mol Microbiol. 1993;9:1061–1069. doi: 10.1111/j.1365-2958.1993.tb01235.x. [DOI] [PubMed] [Google Scholar]
- 222.Powers T, Walter P. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 1997;16:4880–4886. doi: 10.1093/emboj/16.16.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Prágai Z, Tjalsma H, Bolhuis A, van Dijl J M, Venema G, Bron S. The signal peptidase II (lsp) gene of Bacillus subtilis. Microbiology. 1997;143:1327–1333. doi: 10.1099/00221287-143-4-1327. [DOI] [PubMed] [Google Scholar]
- 224.Price A, Economou A, Duong F, Wickner W. Separable ATPases and membrane insertion domains of the secA subunit of pre-protein translocase. J Biol Chem. 1996;271:31580–31584. doi: 10.1074/jbc.271.49.31580. [DOI] [PubMed] [Google Scholar]
- 225.Pugsley A P. The complete general secrotory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Pugsley A P, Kornacker M G, Poquet I. The general protein-export pathway is directly required for extracellular pullulanase secretion in Escherichia coli K12. Mol Microbiol. 1991;5:343–352. doi: 10.1111/j.1365-2958.1991.tb02115.x. [DOI] [PubMed] [Google Scholar]
- 227.Quax W J. Merits of secretion of heterologous proteins from industrial micro-organisms. Folia Microbiol (Prague) 1997;42:99–103. doi: 10.1007/BF02898715. [DOI] [PubMed] [Google Scholar]
- 228.Rahfeld J U, Rucknagel K P, Schelbert B, Ludwig B, Hacker J, Mann K, Fischer G. Confirmation of the existence of a third family among peptidyl-prolyl cis/trans isomerases: amino acid sequence and recombinant production of parvulin. FEBS Lett. 1994;352:180–184. doi: 10.1016/0014-5793(94)00932-5. [DOI] [PubMed] [Google Scholar]
- 229.Rashid M H, Mori M, Sekiguchi J. Glucosaminidase of Bacillus subtilis: cloning, regulation, primary structure and biochemical characterization. Microbiology. 1995;141:2391–2404. doi: 10.1099/13500872-141-10-2391. [DOI] [PubMed] [Google Scholar]
- 230.Riethdorf S, Volker U, Gerth U, Winkler A, Engelmann S, Hecker M. Cloning, nucleotide sequence, and expression of the Bacillus subtilis lon gene. J Bacteriol. 1994;76:6518–6527. doi: 10.1128/jb.176.21.6518-6527.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Riezman H. The ins and outs of protein translocation. Science. 1997;278:1728–1729. doi: 10.1126/science.278.5344.1728. [DOI] [PubMed] [Google Scholar]
- 232.Robinson C, Cai D, Hulford A, Brock I W, Michl D, Hazell L, Schmidt I, Herrmann R G, Klösgen R B. The presequence of a chimeric construct dictates which of two mechanisms are utilized for translocation across the thylakoid membrane: evidence for the existence of two distinct translocation systems. EMBO J. 1994;13:279–285. doi: 10.1002/j.1460-2075.1994.tb06260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Robinson C, Mant A. Targeting of proteins into and across the thylakoid membrane. Trends Plant Sci. 1997;2:431–437. [Google Scholar]
- 234.Rosenow C, Ryan P, Weiser J N, Johnson S, Fontan P, Ortqvist A, Masure H R. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol Microbiol. 1997;25:819–829. doi: 10.1111/j.1365-2958.1997.mmi494.x. [DOI] [PubMed] [Google Scholar]
- 235.Saaf A, Monne M, de Gier J W, von Heijne G. Membrane topology of the 60-kDa Oxa1p homologue from Escherichia coli. J Biol Chem. 1998;273:30415–30418. doi: 10.1074/jbc.273.46.30415. [DOI] [PubMed] [Google Scholar]
- 236.Sadaie Y, Kada T. Effect of septum initiation mutation on sporulation and competent cell formation in Bacillus subtilis. Mol Gen Genet. 1983;190:176–178. [Google Scholar]
- 237.Sadaie Y, Kada T. Bacillus subtilis gene involved in cell division, sporulation, and exoenzyme scretion. J Bacteriol. 1985;163:648–653. doi: 10.1128/jb.163.2.648-653.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Sadaie Y, Takamatsu H, Nakamura K, Yamane K. Sequencing reveals similarity of the wild-type div+ gene of Bacillus subtilis to the Escherichia coli secA gene. Gene. 1991;98:101–105. doi: 10.1016/0378-1119(91)90110-w. [DOI] [PubMed] [Google Scholar]
- 239.Sahl H G, Jack R W, Bierbaum G. Biosynthesis and biological activities of lantibiotics with unique post-translational modifications. Eur J Biochem. 1995;230:827–853. doi: 10.1111/j.1432-1033.1995.tb20627.x. [DOI] [PubMed] [Google Scholar]
- 240.Sánchez-Puelles J M, Ronda C, García J L, Lopez R, García E. searching for autolysin functions: characterization of a pneumococcal mutant deleted in the lytA gene. Eur J Biochem. 1986;158:289–293. doi: 10.1111/j.1432-1033.1986.tb09749.x. [DOI] [PubMed] [Google Scholar]
- 241.Sánchez-Puelles J M, Sanz J M, García J L, García E. Cloning and expression of gene fragments encoding the choline-binding domain of pneumococcal murein hydrolases. Gene. 1990;89:69–75. doi: 10.1016/0378-1119(90)90207-8. [DOI] [PubMed] [Google Scholar]
- 242.Sankaran K, Wu H C. Lipid modification of bacterial prolipoprotein; transfer of diacylglyceryl moiety from phosphatidylglycerol. J Biol Chem. 1994;269:19701–19706. [PubMed] [Google Scholar]
- 243.Sankaran K, Gupta S D, Wu H C. Modification of bacterial lipoproteins. Methods Enzymol. 1995;250:683–697. doi: 10.1016/0076-6879(95)50105-3. [DOI] [PubMed] [Google Scholar]
- 244.Sankaran K, Gan K, Rash B, Qi H Y, Wu H C, Rick P D. Roles of histidine-103 and tyrosine-235 in the function of the prolipoprotein diacylglyceryl transferase of Escherichia coli. J Bacteriol. 1997;179:2944–2948. doi: 10.1128/jb.179.9.2944-2948.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Santini C L, Ize B, Chanal A, Muller M, Giordano G, Wu L F. A novel Sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 1998;17:101–112. doi: 10.1093/emboj/17.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Sargent F, Bogsch E G, Stanley N R, Wexler M, Robinson C, Berks B C, Palmer T. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 1998;17:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Sargent F, Stanley N R, Berks B C, Palmer T. Sec-independent protein translocation in Escherichia coli: a distinct and pivotal role for the TatB protein. J Biol Chem. 1999;274:36073–36082. doi: 10.1074/jbc.274.51.36073. [DOI] [PubMed] [Google Scholar]
- 248.Saunders C W, Schmidt B J, Mallonee R L, Guyer M S. Secretion of human serum albumin from Bacillus subtilis. J Bacteriol. 1985;169:2917–2925. doi: 10.1128/jb.169.7.2917-2925.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 250.Schatz P J, Riggs P D, Jacq A, Fath M J, Beckwith J. The secE gene encodes an integral membrane protein required for protein export in Escherichia coli. Genes Dev. 1989;3:1035–1044. doi: 10.1101/gad.3.7.1035. [DOI] [PubMed] [Google Scholar]
- 251.Schiebel E, Driessen A J M, Hartl F U, Wickner W. ΔμH+ and ATP function at different steps of the catalytic cycle of pre-protein translocase. Cell. 1991;64:927–939. doi: 10.1016/0092-8674(91)90317-r. [DOI] [PubMed] [Google Scholar]
- 252.Schneewind O, Fowler A, Faull K F. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science. 1995;268:103–106. doi: 10.1126/science.7701329. [DOI] [PubMed] [Google Scholar]
- 253.Schneewind O, Mihaylova-Petkov D, Model P. Cell wall sorting signals in surface proteins of Gram-positive bacteria. EMBO J. 1993;12:4803–4811. doi: 10.1002/j.1460-2075.1993.tb06169.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Schneewind O, Model P, Fischetti V A. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 255.Schrögel O, Krispin O, Allmansberger R. Expression of a pepT homologue from Bacillus subtilis. FEMS Microbiol Lett. 1996;145:341–348. doi: 10.1111/j.1574-6968.1996.tb08598.x. [DOI] [PubMed] [Google Scholar]
- 256.Scotti P A, Valent Q A, Manting E H, Urbanus M L, Driessen A J M, Oudega B, Luirink J. SecA is not required for signal recognition particle-mediated targeting and initial membrane insertion of a nascent inner membrane protein. J Biol Chem. 1999;274:29883–29888. doi: 10.1074/jbc.274.42.29883. [DOI] [PubMed] [Google Scholar]
- 256a.Scotti P A, Urbanus M L, Brunner J, de Gier J W, von Heijne G, van der Does C, Driessen A J M, Oudega B, Luirink J. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 2000;19:542–549. doi: 10.1093/emboj/19.4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Seluanov A, Bibi E. FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J Biol Chem. 1997;272:2053–2055. doi: 10.1074/jbc.272.4.2053. [DOI] [PubMed] [Google Scholar]
- 258.Serrano M, Zilhao R, Ricca E, Ozin A J, Moran Jr C P, Henriques A O. A Bacillus subtilis secreted protein with a role in endospore coat assembly and function. J Bacteriol. 1999;181:3632–3643. doi: 10.1128/jb.181.12.3632-3643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Settles A M, Yonetani A, Baron A, Bush D R, Cline K, Martienssen R. Sec-independent protein translocation by the maize Hcf106 protein. Science. 1997;278:1467–1470. doi: 10.1126/science.278.5342.1467. [DOI] [PubMed] [Google Scholar]
- 260.Seydel A, Gounon P, Pugsley A P. Testing the ‘+2 rule’ for lipoprotein sorting in the Escherichia coli cell envelope with a new genetic selection. Mol Microbiol. 1999;34:810–821. doi: 10.1046/j.1365-2958.1999.01647.x. [DOI] [PubMed] [Google Scholar]
- 261.Shiozuka K, Tani K, Mizushima S, Tokuda H. The proton motive force lowers the level of ATP required for the in vitro translocation of a secretory protein in Escherichia coli. J Biol Chem. 1990;265:18843–18847. [PubMed] [Google Scholar]
- 262.Siegel V, Walter P. Each of the activities of signal recognition particle (SRP) is contained within a distinct domain: analysis of biochemical mutants of SRP. Cell. 1988;52:39–49. doi: 10.1016/0092-8674(88)90529-6. [DOI] [PubMed] [Google Scholar]
- 263.Simonen M, Palva I. Protein secretion in Bacillus species. Microbiol Rev. 1993;57:109–137. doi: 10.1128/mr.57.1.109-137.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Sipos L, von Heijne G. Predicting the topology of eukaryotic membrane proteins. Eur J Biochem. 1993;213:1333–1340. doi: 10.1111/j.1432-1033.1993.tb17885.x. [DOI] [PubMed] [Google Scholar]
- 265.Slack F J, Serror P, Joyce E, Sonenshein A L. A gene required for nutritional repression of the Bacillus subtilis dipeptide permease operon. Mol Microbiol. 1995;15:689–702. doi: 10.1111/j.1365-2958.1995.tb02378.x. [DOI] [PubMed] [Google Scholar]
- 266.Sloma A, Ally A, Ally D, Pero J. Gene encoding a minor extracellular protease in Bacillus subtilis. J Bacteriol. 1988;170:5557–5563. doi: 10.1128/jb.170.12.5557-5563.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Sloma A, Rufo G A, Jr, Rudolph C F, Sullivan B J, Theriault K A, Pero J. Bacillopeptidase F of Bacillus subtilis: purification of the protein and cloning of the gene. J Bacteriol. 1990;172:1470–1477. doi: 10.1128/jb.172.3.1470-1477.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Sloma A, Rufo G A, Jr, Theriault K A, Dwyer M, Wilson S W, Pero J. Cloning and characterization of the gene for an additional extracellular serine protease of Bacillus subtilis. J Bacteriol. 1991;173:6889–6895. doi: 10.1128/jb.173.21.6889-6895.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Snyders S, Ramamurthy V, Oliver D. Identification of a region of interaction between Escherichia coli SecA and SecY proteins. J Biol Chem. 1997;272:11302–11306. doi: 10.1074/jbc.272.17.11302. [DOI] [PubMed] [Google Scholar]
- 271.Solomon J M, Magnuson R, Srivastava A, Grossman A D. Convergent sensing pathways mediate response to two extracellular competence factors in Bacillus subtilis. Genes Dev. 1995;9:547–558. doi: 10.1101/gad.9.5.547. [DOI] [PubMed] [Google Scholar]
- 272.Solomon J M, Lazazzera B A, Grossman A D. Purification and characterization of an extracellular peptide factor that affects two different developmental pathways in Bacillus subtilis. Genes Dev. 1996;10:2014–2024. doi: 10.1101/gad.10.16.2014. [DOI] [PubMed] [Google Scholar]
- 273.Stahl M L, Ferrari E. Replacement of the Bacillus subtilis subtilisin structural gene with an in vitro-derived deletion mutation. J Bacteriol. 1984;158:411–418. doi: 10.1128/jb.158.2.411-418.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Stephenson K, Harwood C R. Influence of a cell wall-associated protease on production of alpha-amylase by Bacillus subtilis. Appl Environ Microbiol. 1998;64:2875–2881. doi: 10.1128/aem.64.8.2875-2881.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Stephenson K, Carter N M, Harwood C R, Petit-Glatron M F, Chambert R. The influence of protein folding on late stages of the secretion of α-amylases from Bacillus subtilis. FEBS Lett. 1998;430:385–389. doi: 10.1016/s0014-5793(98)00698-x. [DOI] [PubMed] [Google Scholar]
- 276.Stöver A G, Driks A. Secretion, localization, and antibacterial activity of TasA, a Bacillus subtilis spore-associated protein. J Bacteriol. 1999;181:1664–1672. doi: 10.1128/jb.181.5.1664-1672.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Stöver A G, Driks A. Regulation of synthesis of the Bacillus subtilis transition-phase, spore-associated antibacterial protein TasA. J Bacteriol. 1999;181:5476–5481. doi: 10.1128/jb.181.17.5476-5481.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Stöver A G, Driks A. Control of synthesis and secretion of the Bacillus subtilis protein YqxM. J Bacteriol. 1999;181:7065–7069. doi: 10.1128/jb.181.22.7065-7069.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 280.Suh J W, Boylan S A, Thomas S M, Dolan K M, Oliver D B, Price C W. Isolation of a secY homologue from Bacillus subtilis: evidence for a common protein export pathway in eubacteria. Mol Microbiol. 1990;4:305–314. doi: 10.1111/j.1365-2958.1990.tb00597.x. [DOI] [PubMed] [Google Scholar]
- 281.Sutcliffe I C, Russell R R B. Lipoproteins of gram-positive bacteria. J Bacteriol. 1995;177:1123–1128. doi: 10.1128/jb.177.5.1123-1128.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Suzuki T, Itoh A, Ichihara S, Mizushima S. Characterization of the sppA gene encoding protease IV, a signal peptide peptidase of Escherichia coli. J Bacteriol. 1987;169:2523–2528. doi: 10.1128/jb.169.6.2523-2528.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Swaving J, van Wely K H, Driessen A J M. Pre-protein translocation by a hybrid translocase composed of Escherichia coli and Bacillus subtilis subunits. J Bacteriol. 1999;181:7021–7027. doi: 10.1128/jb.181.22.7021-7027.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Takamatsu H, Fuma S, Nakamura K, Sadaie Y, Shinkai A, Matsuyama S, Mizushima S, Yamane K. In vivo and in vitro characterization of the secA gene product of Bacillus subtilis. J Bacteriol. 1992;174:4308–4316. doi: 10.1128/jb.174.13.4308-4316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Takase K, Mizuno H, Yamane K. NH2-terminal processing of Bacillus subtilis α-amylase. J Biol Chem. 1988;263:11548–11553. [PubMed] [Google Scholar]
- 286.Tam A, Nouvet F J, Fujimura-Kamada K, Slunt H, Sisodia S S, Michaelis S. Dual roles for Ste24p in yeast α-factor maturation:NH2-terminal proteolysis and COOH-terminal CAAX processing. J Cell Biol. 1998;142:635–649. doi: 10.1083/jcb.142.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Taura T, Akiyama Y, Ito K. Genetic analysis of SecY: additional export-defective mutations and factors affecting their phenotypes. Mol Gen Genet. 1994;243:261–269. doi: 10.1007/BF00301061. [DOI] [PubMed] [Google Scholar]
- 288.Thony-Meyer L, James P, Hennecke H. From one gene to two proteins: the biogenesis of cytochromes b and c1 in Bradyrhizobium japonicum. Proc Natl Acad Sci USA. 1991;88:5001–5005. doi: 10.1073/pnas.88.11.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Tjalsma H, Noback M A, Bron S, Venema G, Yamane K, van Dijl J M. Bacillus subtilis contains four closely related type I signal peptidases with overlapping substrate specificities: constitutive and temporally controlled expression of different sip genes. J Biol Chem. 1997;272:25983–25992. doi: 10.1074/jbc.272.41.25983. [DOI] [PubMed] [Google Scholar]
- 290.Tjalsma H, Bolhuis A, van Roosmalen M L, Wiegert T, Schumann W, Broekhuizen C P, Quax W, Venema G, Bron S, van Dijl J M. Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes Dev. 1998;12:2318–2331. doi: 10.1101/gad.12.15.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Tjalsma H, Kontinen V P, Prágai Z, Wu H, Meima R, Venema G, Bron S, Sarvas M, van Dijl J M. The role of lipoprotein processing by signal peptidase II in the Gram-positive eubacterium Bacillus subtilis: signal peptidase II is required for the efficient secretion of α-amylase, a non-lipoprotein. J Biol Chem. 1999;274:1698–1707. doi: 10.1074/jbc.274.3.1698. [DOI] [PubMed] [Google Scholar]
- 292.Tjalsma H, van den Dolder J, Meijer W J J, Venema G, Bron S, van Dijl J M. The plasmid-encoded type I signal peptidase SipP can functionally replace the major signal peptidases SipS and SipT of Bacillus subtilis. J Bacteriol. 1999;181:2448–2454. doi: 10.1128/jb.181.8.2448-2454.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Tjalsma H, Zanen G, Venema G, Bron S, van Dijl J M. The potential active site of the lipoprotein-specific (type II) signal peptidase of Bacillus subtilis. J Biol Chem. 1999;274:28191–28197. doi: 10.1074/jbc.274.40.28191. [DOI] [PubMed] [Google Scholar]
- 293a.Tjalsma H, Stöver A G, Driks A, Venema G, Bron S, van Dijl J M. Conserved serine and histidine residues are critical for activity of the ER-type signal peptidase SipW of Bacillus subtilis. J Biol Chem. 2000;275:25102–25108. doi: 10.1074/jbc.M002676200. [DOI] [PubMed] [Google Scholar]
- 294.Tokunaga M, Tokunaga H, Wu H C. Post-translational modification and processing of Escherichia coli prolipoprotein in vitro. Proc Natl Acad Sci USA. 1982;79:2253–2259. doi: 10.1073/pnas.79.7.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Ton-That H, Faull K F, Schneewind O. Anchor structure of staphylococcal surface proteins. A branched peptide that links the carboxyl terminus of proteins to the cell wall. J Biol Chem. 1997;272:22285–22292. doi: 10.1074/jbc.272.35.22285. [DOI] [PubMed] [Google Scholar]
- 296.Ton-That H, Liu G, Mazmanian S K, Faull K F, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Tran L, Wu X C, Wong S L. Cloning and expression of a novel protease gene encoding an extracellular neutral protease from Bacillus subtilis. J Bacteriol. 1991;173:6364–6372. doi: 10.1128/jb.173.20.6364-6372.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Tschantz W R, Sung M, Delgado-Partin V M, Dalbey R E. A serine and a lysine residue implicated in the catalytic mechanism of the Escherichia coli leader peptidase. J Biol Chem. 1993;268:27349–27354. [PubMed] [Google Scholar]
- 299.Ulbrandt N D, Newitt J A, Bernstein H D. The Escherichia coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell. 1997;88:187–196. doi: 10.1016/s0092-8674(00)81839-5. [DOI] [PubMed] [Google Scholar]
- 300.Valent Q A, Kendall D A, High S, Kusters R, Oudega B, Luirink J. Early events in pre-protein recognition in E. coli: interaction of SRP and trigger factor with nascent polypeptides. EMBO J. 1995;14:5494–5505. doi: 10.1002/j.1460-2075.1995.tb00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Valent Q A, de Gier J W, von Heijne G, Kendall D A, ten Hagen-Jongman C M, Oudega B, Luirink J. Nascent membrane and presecretory proteins synthesized in Escherichia coli associate with signal recognition particle and trigger factor. Mol Microbiol. 1997;25:53–64. doi: 10.1046/j.1365-2958.1997.4431808.x. [DOI] [PubMed] [Google Scholar]
- 302.Valent Q A, Scotti P A, High S, de Gier J W, von Heijne G, Lentzen G, Wintermeyer W, Oudega B, Luirink J. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 1998;17:2504–2512. doi: 10.1093/emboj/17.9.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.van der Does C, Manting E H, Kaufmann A, Lutz M, Driessen A J M. Interaction between SecA and SecYEG in micellar solution and formation of the membrane-inserted state. Biochemistry. 1998;37:201–210. doi: 10.1021/bi972105t. [DOI] [PubMed] [Google Scholar]
- 304.van der Wolk J P, de Wit J G, Driessen A J M. The catalytic cycle of the Escherichia coli SecA ATPase comprises two distinct pre-protein translocation events. EMBO J. 1997;16:7297–7304. doi: 10.1093/emboj/16.24.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.van Dijl J M, de Jong A, Venema G, Bron S. Identification of the potential active site of the signal peptidase SipS of Bacillus subtilis: structural and functional similarities with LexA-like proteases. J Biol Chem. 1995;270:3611–3618. doi: 10.1074/jbc.270.8.3611. [DOI] [PubMed] [Google Scholar]
- 306.van Dijl J M, de Jong A, Smith H, Bron S, Venema G. Synthesis and processing of Escherichia coli TEM-β-lactamase and Bacillus licheniformis α-amylase in Escherichia coli: the role of signal peptidase I. Mol Gen Genet. 1988;214:55–61. doi: 10.1007/BF00340179. [DOI] [PubMed] [Google Scholar]
- 307.van Dijl J M, de Jong A, Vehmaanperä J, Venema G, Bron S. Signal peptidase I of Bacillus subtilis: patterns of conserved amino acids in prokaryotic and eukaryotic type I signal peptidases. EMBO J. 1992;11:2819–2828. doi: 10.1002/j.1460-2075.1992.tb05349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.van Dijl J M, Bolhuis A, Tjalsma H, Venema G, Bron S. The signal peptidase SipS. In: Barret A J, Rawlings N D, Woesner J F Jr, editors. The handbook of proteolytic enzymes. London, England: Academic Press, Ltd.; 1998. pp. 451–452. [Google Scholar]
- 309.van Klompenburg W, Ridder A N J A, van Raalte A L, Killian A J, von Heijne G, de Kruijff B. In vitro membrane integration of leader peptidase depends on the Sec machinery and anionic phospholipids and can occur post-translationally. FEBS Lett. 1997;413:109–114. doi: 10.1016/s0014-5793(97)00888-0. [DOI] [PubMed] [Google Scholar]
- 310.van Leen R W, Bakhuis J G, van Beckhoven F W C, Burger H, Dorssers J, Hommes R W J, Lemson P J, Noordam B, Persoon N L M, Wagemaker G. Production of human interleukin-3 using industrial micro-organisms. Bio/Technology. 1991;9:47–52. doi: 10.1038/nbt0191-47. [DOI] [PubMed] [Google Scholar]
- 311.VanValkenburgh C, Chen X, Mullins C, Fang H, Green N. The catalytic mechanism of endoplasmic reticulum signal peptidase appears to be distinct from most eubacterial signal peptidases. J Biol Chem. 1999;274:11519–11525. doi: 10.1074/jbc.274.17.11519. [DOI] [PubMed] [Google Scholar]
- 312.van Wely K H M, Swaving J, Broekhuizen C P, Rose M, Quax W J, Driessen A J M. Functional identification of the product of the Bacillus subtilis yvaL gene as a SecG homologue. J Bacteriol. 1999;181:1786–1792. doi: 10.1128/jb.181.6.1786-1792.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Veltman O R, Vriend G, Hardy F, Mansfeld J, van den Burg B, Venema G, Eijsink V G H. Analysis of a calcium binding surface loop critical for the stability of the thermolysin-like protease of Bacillus stearothermophilus. Eur J Biochem. 1997;248:433–440. doi: 10.1111/j.1432-1033.1997.00433.x. [DOI] [PubMed] [Google Scholar]
- 314.Venema K, Dost M H, Beun P A, Haandrikman A J, Venema G, Kok J. The genes for secretion and maturation of lactococcins are located on the chromosome of Lactococcus lactis IL1403. J Appl Environ Microbiol. 1996;62:1689–1692. doi: 10.1128/aem.62.5.1689-1692.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Volker U, Engelmann S, Maul B, Riethdorf S, Volker A, Schmid R, Mach H, Hecker M. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology. 1994;140:741–752. doi: 10.1099/00221287-140-4-741. [DOI] [PubMed] [Google Scholar]
- 316.von Heijne G. How signal sequences maintain cleavage specific. J Mol Biol. 1984;173:243–251. doi: 10.1016/0022-2836(84)90192-x. [DOI] [PubMed] [Google Scholar]
- 317.von Heijne G. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 1989;2:531–534. doi: 10.1093/protein/2.7.531. [DOI] [PubMed] [Google Scholar]
- 318.von Heijne G. Protein targeting signals. Curr Opin Cell Biol. 1990;2:604–608. doi: 10.1016/0955-0674(90)90100-s. [DOI] [PubMed] [Google Scholar]
- 319.von Heijne G. The signal peptide. J Membr Biol. 1990;115:195–201. doi: 10.1007/BF01868635. [DOI] [PubMed] [Google Scholar]
- 320.von Heijne G. Life and death of a signal peptide. Nature. 1998;396:111. doi: 10.1038/24036. , 113. [DOI] [PubMed] [Google Scholar]
- 321.von Heijne G, Abrahmsén L. Species-specific variation in signal peptide design. Implications for protein secretion in foreign hosts. FEBS Lett. 1989;27:439–446. doi: 10.1016/0014-5793(89)80579-4. [DOI] [PubMed] [Google Scholar]
- 322.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1997;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Wandersman C. Secretion, processing and activation of bacterial extracellular proteases. Mol Microbiol. 1989;3:1825–1831. doi: 10.1111/j.1365-2958.1989.tb00169.x. [DOI] [PubMed] [Google Scholar]
- 324.Waxman D J, Strominger J L. Sequence of active site peptides from the penicillin-sensitive d-alanine carboxypeptidase of Bacillus subtilis: mechanism of penicillin action and sequence homology to beta-lactamases. J Biol Chem. 1980;255:3964–3976. [PubMed] [Google Scholar]
- 325.Weiner J H, Bilous P T, Shaw G M, Lubitz S P, Frost L, Thomas G H, Cole J A, Turner R J. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 326.Wiertz E J, Tortorella D, Bogyo M, Yu J, Mothes W, Jones T R, Rapoport T A, Ploegh H L. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 327.Wild J, Altman E, Yura T, Gross C A. DnaK and DnaJ heat shock proteins participate in protein export in Escherichia coli. Genes Dev. 1992;6:1165–1172. doi: 10.1101/gad.6.7.1165. [DOI] [PubMed] [Google Scholar]
- 328.Wild J, Rossmeissl P, Walter W A, Gross C A. Involvement of the DnaK-DnaJ-GrpE chaperone team in protein secretion in Escherichia coli. J Bacteriol. 1996;178:3608–3613. doi: 10.1128/jb.178.12.3608-3613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Wong S L. Advances in the use of Bacillus subtilis for the expression and secretion of heterologous proteins. Curr Opin Biotechnol. 1995;6:517–522. doi: 10.1016/0958-1669(95)80085-9. [DOI] [PubMed] [Google Scholar]
- 330.Wu J J, Schuch R, Piggot P J. Characterization of a Bacillus subtilis sporulation operon that includes genes for an RNA polymerase sigma factor and for a putative dd-carboxypeptidase. J Bacteriol. 1992;174:4885–4892. doi: 10.1128/jb.174.15.4885-4892.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Wu S C, Ye R, Wu X C, Ng S C, Wong S L. Enhanced secretory production of a single-chain antibody fragment from Bacillus subtilis by coproduction of molecular chaperones. J Bacteriol. 1998;180:2830–2835. doi: 10.1128/jb.180.11.2830-2835.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Wu X C, Lee W, Tran L, Wong S L. Engineering a Bacillus subtilis expression-secretion system with a strain deficient in six extracellular proteases. J Bacteriol. 1991;173:4952–4958. doi: 10.1128/jb.173.16.4952-4958.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Yamazaki T, Yahagi S, Nakamura K, Yamane K. Depletion of Bacillus subtilis histone-like protein, HBsu, causes defective protein translocation and induces upregulation of small cytoplasmic RNA. Biochem Biophys Res Commun. 1999;258:211–214. doi: 10.1006/bbrc.1999.0615. [DOI] [PubMed] [Google Scholar]
- 334.Yang M Y, Ferrari E, Henner D J. Cloning of the neutral protease gene of Bacillus subtilis and the use of the cloned gene to create an in vitro-derived deletion mutation. J Bacteriol. 1984;160:15–21. doi: 10.1128/jb.160.1.15-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Yanouri A, Daniel R A, Errington J, Buchanan C E. Cloning and sequencing of the cell division gene pbpB, which encodes penicillin-binding protein 2B in Bacillus subtilis. J Bacteriol. 1993;175:7604–7616. doi: 10.1128/jb.175.23.7604-7616.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Zheng G, Yan L Z, Vederas J C, Zuber P J. Genes of the sbo-alb locus of Bacillus subtilis are required for production of the antilisterial bacteriocin subtilosin. J Bacteriol. 1999;181:7346–7355. doi: 10.1128/jb.181.23.7346-7355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Zhu X L, Ohta Y, Jordan F, Inouye M. Pro-sequence of subtilisin can guide the refolding of denatured subtilisin in an intermolecular process. Nature. 1989;339:483–484. doi: 10.1038/339483a0. [DOI] [PubMed] [Google Scholar]
- 338.Zimmermann R. The role of molecular chaperones in protein transport into mammalian endoplasmic reticulum. Biol Chem. 1998;379:275–282. [PubMed] [Google Scholar]