Abstract
Following meiotic recombination, each pair of homologous chromosomes acquires at least one crossover, which ensures accurate chromosome segregation and allows reciprocal exchange of genetic information. Recombination failure often leads to meiotic arrest, impairing fertility, but the molecular basis of recombination remains elusive. Here, we report a homozygous M1AP splicing mutation (c.1074 + 2T > C) in patients with severe oligozoospermia owing to meiotic metaphase I arrest. The mutation abolishes M1AP foci on the chromosome axes, resulting in decreased recombination intermediates and crossovers in male mouse models. M1AP interacts with the mammalian ZZS (an acronym for yeast proteins Zip2‐Zip4‐Spo16) complex components, SHOC1, TEX11, and SPO16. M1AP localizes to chromosomal axes in a SPO16‐dependent manner and colocalizes with TEX11. Ablation of M1AP does not alter SHOC1 localization but reduces the recruitment of TEX11 to recombination intermediates. M1AP shows cytoplasmic localization in fetal oocytes and is dispensable for fertility and crossover formation in female mice. Our study provides the first evidence that M1AP acts as a copartner of the ZZS complex to promote crossover formation and meiotic progression in males.
Keywords: crossover, M1AP, meiotic recombination, severe oligospermia, ZZS complex
Subject Categories: Cell Cycle; DNA Replication, Recombination & Repair; Molecular Biology of Disease
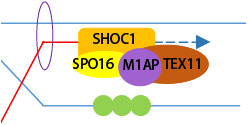
M1AP acts as a male‐specific copartner of the ZZS complex to stabilize recombination intermediates and promote crossover formation. Disruption of M1AP causes meiotic metaphase I arrest and impairs male fertility in humans and mice.
Introduction
Meiosis is a specialized cell division of sexual reproductive organisms, during which germ cells undergo a single round of DNA replication followed by two consecutive cell divisions to produce haploid gametes. Meiotic recombination plays a central role in this process. It occurs preferentially between nonsister chromatids of homologous chromosomes and permits the reciprocal exchange of genetic materials, assuring faithful segregation of chromosomes and creating genetic diversity in gametes (Baudat et al, 2013; Hunter, 2015; Zickler & Kleckner, 2015). Meiotic recombination is initiated by the formation of programmed DNA double‐strand breaks (DSBs) catalyzed by SPO11 and TOPOVIBL (Baudat et al, 2000; Romanienko & Camerini‐Otero, 2000; Robert et al, 2016; Vrielynck et al, 2016). DSBs are rapidly resected to form 3′ single‐stranded DNA (ssDNA) overhangs, which are coated with ssDNA‐binding proteins, replication protein A (RPA) complexes. Recombinases RAD51/DMC1 soon replace RPA and direct strand invasion into homologous chromosomes, leading to the formation of early recombination intermediates that are further processed to yield single‐end invasion (SEI) intermediates (Brown & Bishop, 2014). The intermediates that are designated for crossover formation are stabilized by a set of conserved proteins, known as “ZMM” proteins (an acronym for yeast proteins Zip1/Zip2/Zip3/Zip4, Msh4/Msh5, Mer3 and Spo16; Sym et al, 1993; Hollingsworth et al, 1995; Chua & Roeder, 1998; Nakagawa & Ogawa, 1999; Agarwal & Roeder, 2000; Novak et al, 2001; Börner et al, 2004; Snowden et al, 2004; Tsubouchi et al, 2006; Shinohara et al, 2008; Pyatnitskaya et al, 2019). Following the capture of the second DSB ends, double Holliday junctions (dHJs) are produced and subsequently resolved into crossovers catalyzed by endonuclease MLH1/3 (Baker et al, 1996; Edelmann et al, 1996; Wang et al, 1999; Lipkin et al, 2002; Nishant et al, 2008; Zakharyevich et al, 2012). Crossover formation is tightly regulated, such that only approximately 10% of DSBs finally become crossovers, and each homologous chromosome pair acquires at least one crossover (Baudat et al, 2013). Failures in meiotic recombination often cause meiotic arrest and infertility (Handel & Schimenti, 2010).
Three ZMM proteins, ZIP2, ZIP4, and SPO16, form a functionally conserved protein complex known as “ZZS,” which was initially described in budding yeast (Saccharomyces cerevisiae; Chua & Roeder, 1998; Tsubouchi et al, 2006; Adelman & Petrini, 2008; Macaisne et al, 2008; Shinohara et al, 2008; Yang et al, 2008b; De Muyt et al, 2018; Guiraldelli et al, 2018; Zhang et al, 2018, 2019b). During yeast meiosis, the ZZS complex localizes to early recombination intermediates shortly after strand invasion and stabilizes these early recombination intermediates to facilitate the formation of SEIs and subsequent crossovers (De Muyt et al, 2018; Arora & Corbett, 2019). The mammalian homologs of Zip2, Zip4, and Spo16 have been characterized as SHOC1, TEX11, and SPO16, respectively. In mice, all three ZZS proteins localize to chromosome axes as discrete foci and have similar foci kinetics in meiotic prophase I (Yang et al, 2008b; Guiraldelli et al, 2018; Zhang et al, 2018, 2019b; Liu et al, 2019). SHOC1 binds to early recombination intermediates after strand invasion (Zhang et al, 2018); SPO16 is then recruited by SHOC1 to stabilize its localization on the intermediates and further recruits TEX11 to facilitate the repair of DSBs through the dHJ pathway (Zhang et al, 2019b). Shoc1 −/− and Spo16 −/− mice showed severe defects in synapsis and meiotic arrest at a zygotene‐like or an early pachytene‐like stage without crossover formation (Zhang et al, 2018, 2019b), while knockout of Tex11 in mice caused a milder defect in synapsis and reduced crossover formation, leading to meiotic metaphase I (MMI) arrest (Adelman & Petrini, 2008; Yang et al, 2008b). However, how the ZZS complex regulates the processing of recombination intermediates is not fully understood in mice.
M1AP (meiosis 1 arresting protein) has been reported to be essential for male meiosis in mice, and its mutations have been found frequently in infertile men (Arango et al, 2013; Tu et al, 2020; Wyrwoll et al, 2020), but the molecular role of M1AP remains uncharacterized. In this study, we identified a homozygous M1AP splicing mutation (c.1074 + 2T > C) in a consanguineous Pakistani family with two cases of severe oligozoospermia owing to MMI arrest. Using mice carrying a splicing mutation equivalent to that of our patients, we showed that the mutation leads to abolished M1AP foci and reduced crossovers, resulting in MMI arrest. M1AP interacts with the ZZS complex and promotes the recruitment of TEX11 at the recombination intermediates. Our study demonstrates M1AP as a pathogenic gene for human male infertility using mouse models and unveils the molecular role of M1AP as a ZZS partner in male meiotic recombination.
Results
A homozygous M1AP splicing mutation was identified in patients with severe oligozoospermia and MMI arrest
We recruited a consanguineous Pakistani family with two brothers (V:2 and V:3) suffering from severe oligozoospermia (Fig 1A and Table EV1). According to the World Health Organization (WHO), the lower reference limit for sperm concentration is 15 × 106 spermatozoa per ml (WHO, 2010), but the mean sperm concentration for patient V:2 and V:3 was 0.8 ± 0.5 and 0.3 ± 0.3 million/ml, respectively (Table EV1). For patient V:3, no sperm were found in the last two semen analyses, so testicular biopsy was conducted. H&E staining of testicular sections revealed the presence of spermatogonia and spermatocytes in the seminiferous tubules of patient V:3, but we could hardly find spermatids or spermatozoa (Fig 1B). Noticeably, many meiotic metaphase cells with unaligned chromosomes were observed and confirmed by the staining of anti‐Histone H3 (phospho S10) in tubules of the patient (Fig 1B and C). These results indicate MMI arrest in the testis of our patient.
Figure 1. Homozygous M1AP splicing mutation was identified in a consanguineous Pakistani family with severe oligozoospermia and meiotic metaphase I arrest.
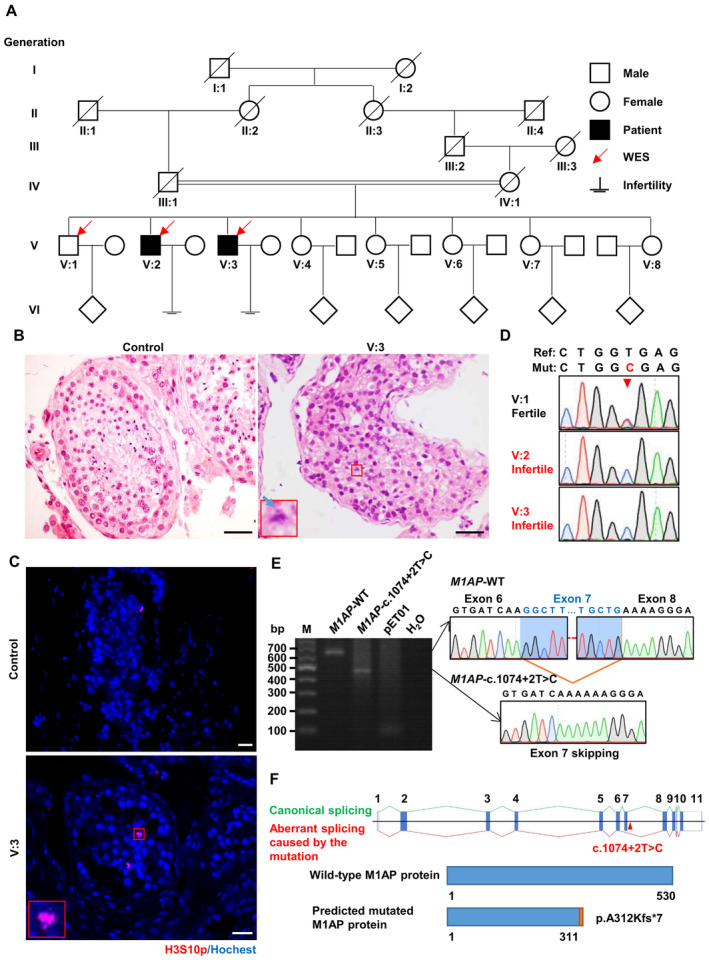
- The family pedigree. Arrows represent the individuals for whom whole‐exome sequencing was performed. Squares represent males, circles represent females, diamonds indicate offspring, and the slash symbols denote deceased family members. Solid squares indicate male patients suffering from severe oligozoospermia. Double horizontal lines indicate consanguineous marriage.
- Representative images of hematoxylin and eosin‐stained testicular sections from the patient (V:3) and a man diagnosed with obstructive azoospermia, serving as the control. The magnified view of the boxed area is shown in the lower left corner of the image from the patient. The blue arrow indicates unaligned chromosomes in the representative metaphase cells. Scale bars, 50 μm.
- Immunofluorescence staining of testicular sections from control and patient V:3 with antibodies against H3S10p (red), a marker of metaphase cells. The nuclei were stained with Hoechst 33342 (blue). The magnified view of the red box is shown in the lower left corner of the image from the patient. Scale bars, 50 μm.
- Sanger sequencing chromatograms showing the homozygous M1AP splicing mutation in patients and heterozygous mutation in their fertile brother. The arrowhead indicates the position of the mutation.
- Reverse transcription–PCR products of the pET01 empty vector and vectors expressing the wild‐type or c.1074 + 2T > C M1AP mini gene (containing genomic sequences spanning intron 5–6 and intron 8–9) in HEK‐293T cells. The splicing products of wild‐type and mutant mini genes were detected by Sanger sequencing. The wild‐type transcript is spliced canonically, while the mutant transcript is aberrantly spliced, leading to the skipping of exon 7. M, marker.
- Schematic representation of the aberrant splicing and the predicted protein truncation caused by the M1AP mutation. Vertical solid bars indicate exons of the human M1AP gene. The M1AP mutation (c.1074 + 2T > C) is located in intron 7–8 near exon 7 (Ensembl transcript ID ENST00000421985.2) and is indicated by the arrowhead. Green lines represent canonical splicing, and red lines represent aberrant splicing caused by the M1AP mutation.
To explore the genetic cause of severe oligozoospermia in this family, whole‐exome sequencing (WES) was performed with genomic DNA from the patients and their fertile brother (V:1). Considering that the patients were born to consanguineous parents, variants following autosomal recessive inheritance were given priority. Taking the spermatogenic defects of our patients into account, we identified a homozygous splicing site mutation in M1AP (NM_001321739.2), c.1074 + 2T > C, as the candidate variant most likely pathogenic for the infertility of the two patients after variant filtration with a series of criteria (Appendix Fig S1A and B). Sanger sequencing of genomic DNA extracted from blood samples validated that the mutation is homozygous in both patients and heterozygous in the fertile brother (Fig 1D).
The M1AP mutation is located in the region of the splice donor site at the boundary between exon 7 and intron 7–8. To assess the impact of the mutation on M1AP splicing, minigene vectors containing the genomic sequence spanning intron 5–6 and intron 8–9 of the M1AP gene were transfected into HEK‐293T cells, followed by reverse transcription‐PCR (RT–PCR). The wild‐type M1AP minigene showed a canonical splicing pattern, producing a band corresponding to the junction of exons 6, 7, and 8, while the splicing mutation (c.1074 + 2T > C) in M1AP resulted in a band smaller than that of wild‐type (Fig 1E). Sanger sequencing showed that the splicing mutation caused exon 7 skipping, which is predicted to cause premature protein truncation (p. A312Kfs*7; Fig 1E and F). This result suggests that the splicing mutation in M1AP (c.1074 + 2T > C) induces aberrant mRNA splicing.
M1AP forms discrete foci on chromosome axes in spermatocytes
M1AP has been reported to be essential for male meiosis in mice (Arango et al, 2013), but the cellular localization and molecular role of M1AP in meiosis remain uncharacterized. To explore the expression and localization of M1AP, we generated an anti‐M1AP antibody that recognizes an epitope consisting of amino acids 415–529 of the mouse protein. M1AP protein was abundant in testes but was not detected in other adult tissues of mice (Fig 2A). M1AP protein was detected in mouse testes from 10 days of postpartum (dpp), when germ cells proceed to spermatocytes (Fig 2B). Immunofluorescence staining of spread spermatocytes revealed that M1AP forms discrete foci localizing on chromosome axes (Fig 2C). M1AP foci were detected from the leptotene stage, accumulated to the maximal level at the late zygotene stage (125.65 ± 4.59), decreased to 98.36 ± 1.95 at the early pachytene stage, and then declined sharply and disappeared in the mid/late pachytene stages (Fig 2D). Notably, most of the M1AP foci localize at synapsed regions. To investigate whether the M1AP foci are dependent on meiotic DSBs, we examined M1AP foci in spermatocytes from Spo11 −/− mice, in which meiotic programmed DSBs were not generated, and found that no M1AP foci were observed (Fig 2C), indicating that the localization of M1AP to the chromosome axes depends on programmed meiotic DSBs. The localization pattern of M1AP suggested a potential role in male meiotic recombination.
Figure 2. M1AP forms discrete foci on the chromosome axes in spermatocytes.
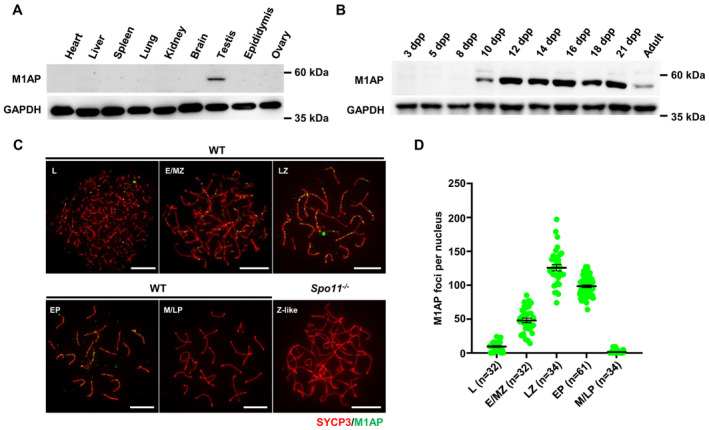
- Western blot analysis of M1AP expression in various tissues from adult wild‐type (WT) mice. GAPDH served as the loading control.
- Western blot analysis of M1AP expression in testes from mice at different days postpartum (dpp). GAPDH served as the loading control.
- Immunofluorescence staining of M1AP (green) and SYCP3 (red) on spermatocyte spreads of WT or Spo11 −/− mice. Scale bar, 10 μm.
- Quantification of M1AP foci in WT spermatocytes at the indicated stages. L, leptotene; E/MZ, early/mid zygotene; LZ, late zygotene; EP, early pachytene; M/LP, mid/late pachytene; Z‐like, zygotene‐like. Data are presented as the mean ± SEM. n shows the number of spermatocytes analyzed from three biological replicates.
M1ap KI /KI male mice recapitulate the meiotic defects of our patient
To assess the impacts of the M1AP splicing mutation on spermatogenesis, we generated M1ap mutant mice (M1ap KI/KI ) carrying the mutation equivalent to that of the patients using CRISPR/Cas9 technology (Fig EV1A).
Figure EV1. Generation of M1ap KI/KI mice.
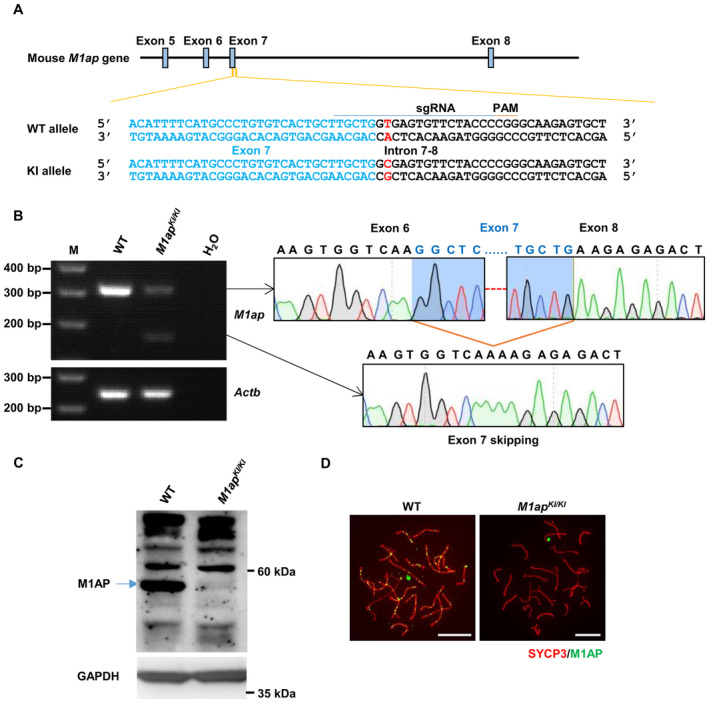
- The strategy to generate mice that carry an M1ap knockin (KI) mutation equivalent to that of our patients. The single guide RNA (sgRNA, underlined) was designed to target the splicing mutation site (written in red). In the KI allele, thymine was replaced by cytosine. Nucleotides from exon 7 are written in blue. WT, wild‐type. PAM, protospacer adjacent motif.
- Reverse transcription‐PCR analysis of WT and homozygous M1ap KI/KI mouse testes with primers spanning exons 6–8. Actb served as an internal control.
- Western blotting with testis lysates from WT and homozygous M1ap KI/KI mice using the anti‐M1AP antibody. GAPDH was used as the loading control. The band corresponding to WT full‐length M1AP protein is indicated by an arrow.
- Immunofluorescence staining of M1AP (green) and SYCP3 (red) on the spermatocyte spreads of WT and M1ap KI/KI mice. Scale bars, 10 μm.
Agarose gel electrophoresis of RT–PCR products with primers flanking exon 6 and exon 8 of M1ap cDNA detected one band corresponding to the canonical splicing product in the wild‐type testis. However, two bands, both much fainter than that in wild‐type testis, were observed in the M1ap KI/KI testes (Fig EV1B). Sanger sequencing showed that one band corresponds to the canonical splicing product, while the other band corresponds to the skipping of exon 7, which is predicted to result in protein truncation (Fig EV1B). Western blotting using the anti‐M1AP antibody revealed a very faint band just above the detection level at the size corresponding to the wild‐type M1AP proteins in the mutant mice (Fig EV1C). Notably, no M1AP foci were detected on chromosome axes in spread spermatocytes from M1ap KI/KI mice (Fig EV1D). Thus, in mice, this M1ap mutation causes an alteration of mRNA splicing, similar to the observation in patients, and abolished M1AP foci on chromosome axes.
M1ap KI/KI mice showed no obvious abnormalities and appeared healthy. However, the litter numbers and litter sizes obtained from mating of M1ap KI/KI male mice with wild‐type female mice were significantly reduced compared with those obtained from wild‐type males, suggesting that M1ap KI/KI male mice were subfertile (Table EV2). M1ap KI/KI mice displayed smaller testes and lower sperm numbers than their control littermates (Fig EV2A–D). Histological analyses of testes from the control mice revealed the presence of numerous spermatogenic cells of all developmental stages (Fig 3A). However, seminiferous tubules of M1ap KI/KI mice contained many MMI spermatocytes with unaligned chromosomes and showed an obvious reduction in the number of postmeiotic germ cells (Fig 3A), similar to the observations in our patient.
Figure EV2. The phenotype of M1ap KI/KI mice.
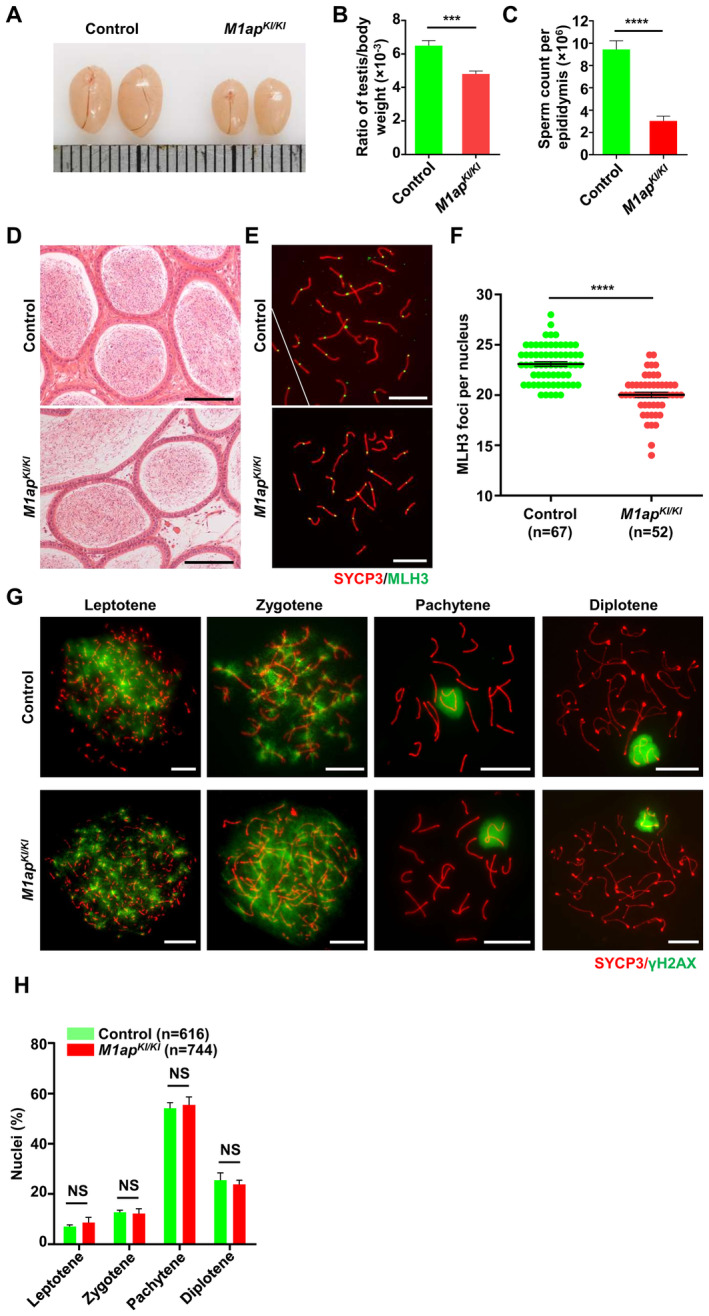
- Representative images of testes from 8‐week‐old control and M1ap KI/KI mice. Each grid represents 1 mm.
- The ratio of testis/body weight of 8‐week‐old control and M1ap KI/KI mice.
- The sperm count per epididymis of 8‐week‐old control and M1ap KI/KI mice.
- Representative images of hematoxylin and eosin‐stained sections of cauda epididymides from 8‐week‐old wild‐type and M1ap KI/KI mice. Scale bars, 50 μm.
- Immunofluorescence staining of SYCP3 (red) and MLH3 (green) in spermatocyte spreads from 8‐week‐old control and M1ap KI/KI mice. Scale bars, 10 μm.
- Number of MLH3 foci per cell in control and M1ap KI/KI spermatocytes.
- Immunofluorescence staining of SYCP3 (red) and γH2AX (green) on spermatocyte spreads from 8‐week‐old control and M1ap KI/KI mice. Scale bars, 10 μm.
- The percentages of spermatocytes at each stage of meiotic prophase I in 8‐week‐old control and M1ap KI/KI mice.
Data information: (B and C), Data are shown as the mean ± SEM of three biological replicates. (F and H) Data are shown as the mean ± SEM. n shows the number of cells scored from three biological replicates. **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, not significant; two‐tailed Student's t‐test.
Figure 3. Meiotic arrest at metaphase I caused by decreased crossovers in M1ap KI/KI male mice.
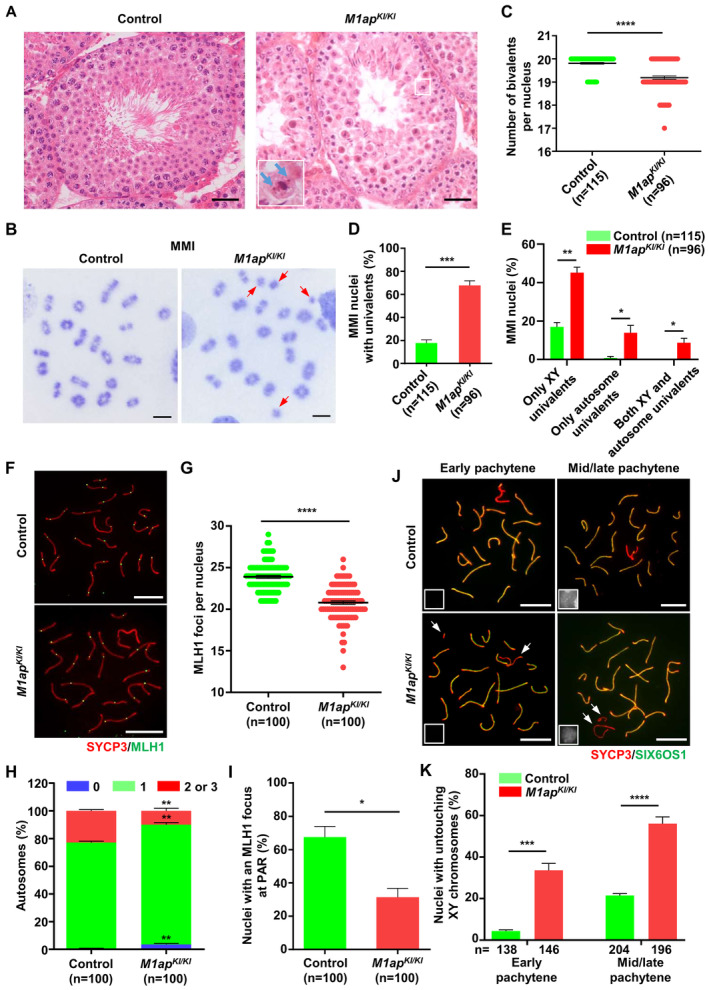
- Testicular histology from 8‐week‐old control and M1ap KI/KI mice. The magnified view of the boxed area is shown in the lower left corner of the image of M1ap KI/KI mice. The blue arrow indicates unaligned chromosomes in the representative metaphase cells. Scale bars, 50 μm.
- Meiotic metaphase I (MMI) spermatocytes stained with Giemsa. The arrows indicate univalent chromosomes. Scale bars, 10 μm.
- Number of bivalents per nucleus.
- Frequencies of nuclei with univalents.
- Frequencies of nuclei with only XY univalents, only autosome univalents, or both XY and autosome univalents.
- Representative spread spermatocytes stained for SYCP3 (green) and MLH1 (red). Scale bars, 10 μm.
- Number of MLH1 foci per nucleus.
- Frequencies of autosomal chromosomes with 0, 1, and 2 or 3 MLH1 foci.
- Frequencies of nuclei with an MLH1 focus at the pseudoautosomal region (PAR) of XY chromosomes.
- Representative spread spermatocytes stained for the lateral element (SYCP3, green) and the central element (SIX6OS1, red) in early and mid/late pachynema. Miniaturized H1t staining (white) images are shown in the left lower corner of the overlay images. The arrows indicate the untouching XY chromosomes. Scale bars, 10 μm.
- Quantification of nuclei with XY untouching in early and mid/late pachynema.
Data information: (C–E, G, and H–K), Data are presented as the mean ± SEM. n, the number of cells scored from three biological replicates. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001; two‐tailed Student's t‐test.
Increased prevalence of univalents in M1ap KI / KI MMI spermatocytes
The unaligned chromosomes in MMI spermatocytes may be a result of the absence of chiasmata between homologous chromosomes, which are usually present as univalents (Reynolds et al, 2013; Ma et al, 2022). Thus, we prepared metaphase chromosome spreads from control and M1ap KI/KI testes. Analysis of metaphase I chromosomes revealed that in control mice, 17.81% of MMI spermatocytes contained univalent chromosomes, among which 16.99% had only sex chromosome univalents and 0.82% had only autosome univalents (Fig 3B–E). However, univalent chromosomes were found in 67.81% of M1ap KI/KI MMI spermatocytes, with 45.19% having only sex chromosome univalents, 13.92% having only autosome univalents, and 8.70% having both autosome and sex chromosome univalents (Fig 3B–E). Hence, in M1ap KI/KI mice, autosomes and sex chromosomes are susceptible to precocious separation.
M1ap KI /KI spermatocytes displayed reduced crossover formation
Univalents in MMI spermatocytes could be caused by crossover formation failure. To evaluate whether crossover formation was impaired after M1ap mutated, we quantified the number of MLH1 and MLH3 foci, which mark sites of crossovers. The average number of MLH1 foci per nucleus was 23.89 ± 0.17 in control pachytene spermatocytes but 20.78 ± 0.21 in M1ap KI/KI mice (Fig 3F and G). Similarly, the average number of MLH3 foci per nucleus was also reduced (23.09 ± 0.23 in controls vs. 20.02 ± 0.28 in M1ap KI/KI mice; Fig EV2E and F). In control spermatocytes, autosomes without an MLH1 focus were seldom observed. In M1ap KI/KI mice, 3.64% of autosome pairs were detected without any MLH1 foci, in contrast to 0.62% in controls, and the ratio of autosome pairs with 2 or 3 MLH1 foci was significantly reduced compared with that in controls (Fig 3H). Consistent with the increased prevalence of XY univalent in MMI cells, the frequency of cells with MLH1 focus on the pseudoautosomal region (PAR) was significantly lower in M1ap KI/KI mice (31.37%) than in control mice (67.53%), and we also noticed a higher frequency of cells with XY untouching in M1ap KI/KI pachytene spermatocytes (Fig 3I–K). In addition, autosomal synapsis, γH2AX staining patterns, and meiotic prophase I progression in M1ap KI/KI mice were all comparable with those in controls (Fig EV2G and H). These observations together indicate that crossover formation is reduced on autosomes and particularly on the sex chromosomes in M1ap KI/KI mice.
M1AP ablation decreases recombination intermediates in males
To explore why crossover formation was deficient in M1ap KI/KI spermatocytes, we first quantified the number of MSH4 foci, which mark recombination intermediates that are committed to be converted into crossovers. In M1ap KI/KI spermatocytes, MSH4 foci were observed specifically at synapsed regions of homologous chromosomes in a manner indistinguishable from that in control cells; however, the average numbers of MSH4 foci per cell were decreased compared with those in control spermatocytes at all stages examined, including early/mid zygotene, late zygotene, and early pachytene (Fig 4A and B).
Figure 4. Decreased recombination intermediates in M1ap KI/KI male mice.
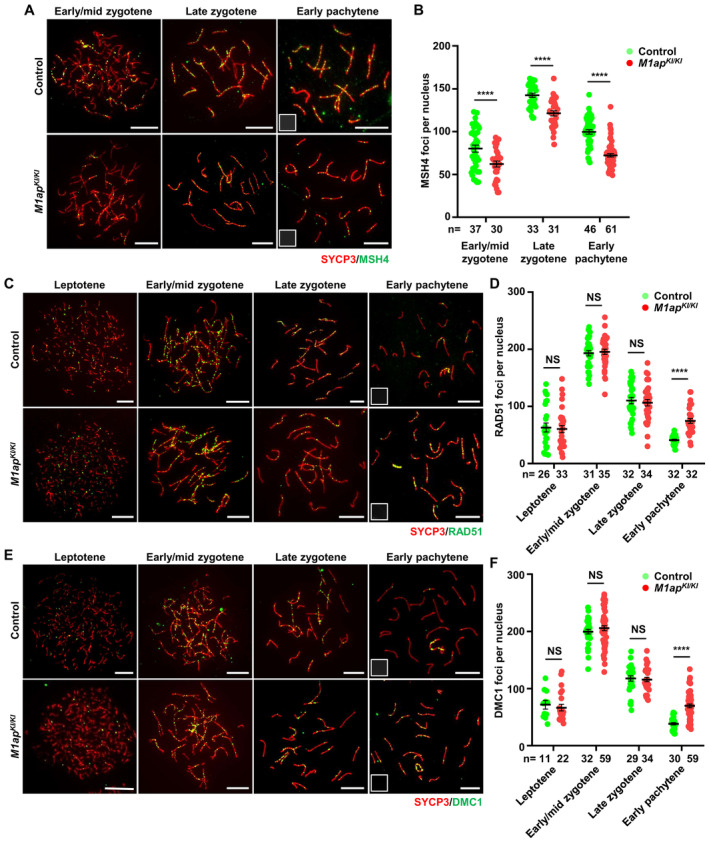
- Immunofluorescence staining with antibodies against SYCP3 (red) and MSH4 (green) on spermatocyte spreads. Scale bars, 10 μm.
- The mean number of MSH4 foci per nucleus in control and M1ap KI/KI spermatocytes at the indicated stages.
- Immunofluorescence staining with antibodies against SYCP3 (red) and RAD51 (green) on spermatocyte spreads. Scale bars, 10 μm.
- The mean number of RAD51 foci per nucleus in control and M1ap KI/KI spermatocytes at the indicated stages.
- Immunofluorescence staining with antibodies against SYCP3 (red) and DMC1 (green) on spermatocyte spreads. Scale bars, 10 μm.
- The mean number of DMC1 foci per nucleus in control and M1ap KI/KI spermatocytes at the indicated stages.
Data information: (B, D and F), Data are presented as the mean ± SEM. n, the number of cells scored from at least three biological replicates. ****P < 0.0001; NS, not significant; two‐tailed Student's t‐test.
The decreased recombination intermediates could be caused by impaired DSB formation, loading of recombination proteins, formation of recombination intermediates following strand invasion, or stabilization of recombination intermediates. To elucidate the defects in recombination, we quantified the kinetics of RAD51 and DMC1 foci at different substages of meiotic prophase. In control spermatocytes, RAD51 and DMC1 foci appear at leptotene; their numbers reach peaks at early/mid zygotene and gradually decline as DSB repair proceeds through pachytene. The numbers of RAD51 and DMC1 foci in M1ap KI/KI spermatocytes were comparable with those in control cells at leptotene, early/mid zygotene, and late zygotene stages (Fig 4C–F). However, we noticed significantly higher numbers of RAD51 and DMC1 foci at early pachytene in M1ap KI/KI spermatocytes than in control spermatocytes (Fig 4C–F). These findings lead us to conclude that there are no overt defects in meiotic DSB formation or loading of recombination proteins and strand invasion occurs, but recombination intermediates may not be formed successfully. Consistent with this, we also noticed that XY separation mainly occurs at early pachytene, as the frequency of cells with XY untouching at early pachytene was 33.5% in M1ap KI/KI mice, in contrast to 4.3% in control mice, and was increased to 56.1 and 21.5% in M1ap KI/KI and control mice, respectively (Fig 3J and K).
These results collectively suggest that ablation of M1AP in male mice decreases recombination intermediates.
M1AP is dispensable for crossover formation and meiosis in oocytes
M1ap mRNA was detected in mouse fetal ovaries (Arango et al, 2006; Wang et al, 2020), but whether M1AP plays a role in female meiosis is unknown. We first examined the expression and localization of M1AP proteins in fetal oocytes. Western blotting confirmed the expression of M1AP protein in fetal ovaries at 16.5 days postcoitum (dpc), at a comparable level as in 14 dpp testes (Fig 5A). Intriguingly, immunofluorescence staining revealed that M1AP signals were barely detected in spread oocyte nuclei (Fig 5B). We speculated that M1AP protein may not localize in the nucleus in oocytes. We thus performed confocal laser scanning microscopy analyses of M1AP localization on spermatocyte and oocyte smears. As expected, M1AP signals were detected in the nuclei of zygotene/pachytene spermatocytes, forming discrete foci; by contrast, diffused M1AP signals were detected only in the cytoplasm, but not in the nucleus, of zygotene/pachytene oocytes (Fig 5C). In congruent with this, we did not observe any obvious difference in the number of MLH3 foci between M1ap KI/KI and control oocytes at 18.5 dpc (Fig 5D and E), nor in the numbers of total oocytes, primordial follicles, and primary follicles between control and the mutant mice at 5 dpp (Fig 5F and G), suggesting that the crossover formation and follicle development were not affected after M1AP ablation. Moreover, mating of M1ap KI/KI female mice with wild‐type male mice produced normal numbers of offspring (Table EV3).
Figure 5. M1AP shows cytoplasmic localization in fetal oocytes, and is dispensable for crossover formation in female mice.
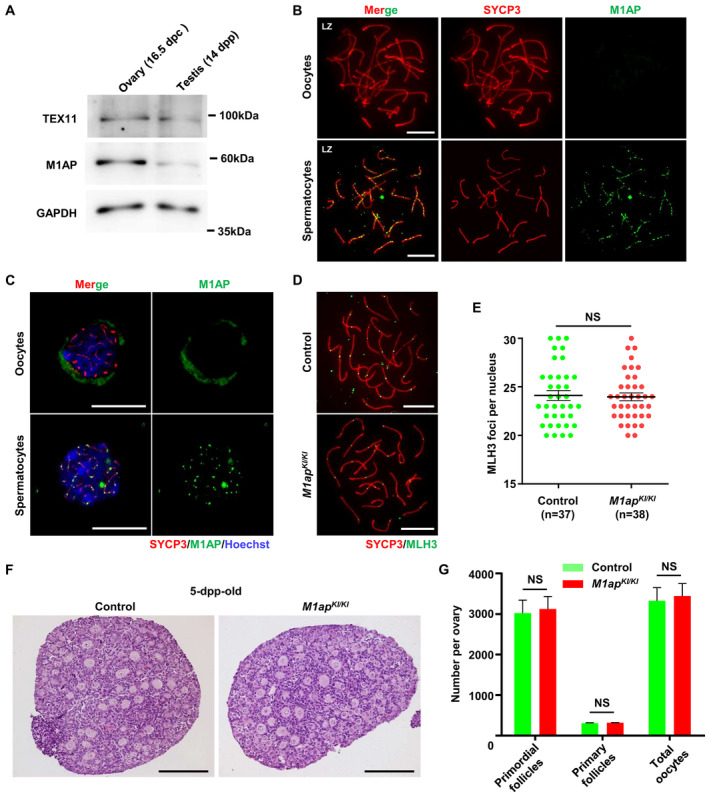
- Western blot analysis of M1AP and TEX11 expression in fetal ovaries (16.5 days postcoitum (dpc)) and testes (14 dpp) from WT mice. GAPDH served as the loading control.
- Immunofluorescence staining of M1AP (green) and SYCP3 (red) on meiotic spreads of fetal ovaries (16.5 dpc) and adult testes from WT mice. Scale bars, 10 μm.
- Confocal imaging of zygotene/pachytene meiocytes on cell smears of fetal ovaries (16.5 dpc) and adult testes from WT mice after immunofluorescence staining of M1AP (green) and SYCP3 (red). Scale bars, 10 μm.
- Immunofluorescence staining with antibodies against SYCP3 (red) and MLH3 (green) on oocyte spreads from control and M1ap KI/KI embryos (18.5 dpc). Scale bars, 10 μm.
- Number of MLH3 foci per cell in control and M1ap KI/KI oocytes.
- Representative haemotoxylin‐stained ovarian sections of 5‐dpp‐old control and M1ap KI/KI mice. Scale bars, 50 μm.
- Follicle counts and total oocyte counts per ovary at 5 dpp.
Data information: (E) Data are presented as the mean ± SEM. n shows the number of cells scored from two biological replicates. (G) Data are presented as the mean ± SEM of three biological replicates. NS, not significant; two‐tailed Student's t‐test.
Taken together, these results indicate that M1AP protein is expressed but shows cytoplasmic localization in fetal oocytes and is dispensable for female meiosis and fertility.
M1ap −/− mice resemble the meiotic defects of M1ap KI /KI mice
In our M1ap KI/KI males, crossover formation was reduced owing to decreased recombination intermediates, while autosomal pairing and synapsis were normal. However, M1ap lz/lz mice, carrying hypomorphic alleles in the M1ap gene, displayed Sertoli‐cell‐only seminiferous tubules, prolonged retention of γH2AX on chromosome axes, nonhomologous pairing, and synaptic defects on autosomes (Arango et al, 2013). The phenotypic differences raised the possibility that the trace amount of M1AP protein in M1ap KI/KI testes may retain some function. To test this possibility, we generated M1ap −/− mice using CRISPR/Cas9 technology (Fig EV3A). M1ap −/− mice carried a 10‐base‐pair deletion in exon 4, which introduced a premature stop codon (Fig EV3B). Western blotting and immunofluorescence staining of spermatocyte spreads confirmed the absence of M1AP proteins in M1ap −/− mice (Fig EV3C and D).
Figure EV3. Generation of M1ap −/− mice.
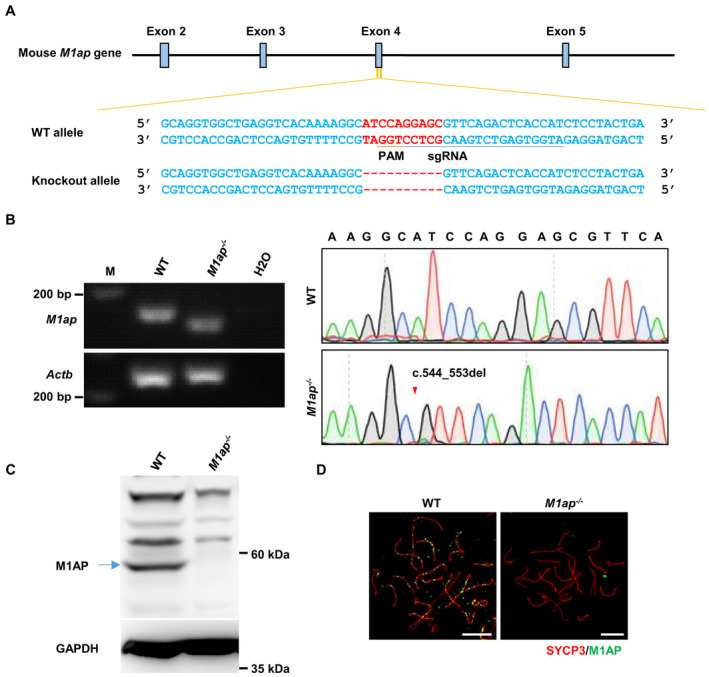
- The strategy to generate M1ap −/− mice. The single guide RNA (underlined) was designed to target exon 4 of the M1ap gene. A 10‐base pair deletion was detected in the M1ap knockout allele. WT, wild‐type. PAM, protospacer adjacent motif.
- Gel electrophoresis of the PCR products obtained from testis cDNA and subsequent Sanger sequencing showing the 10‐base pair deletion in the M1ap −/− mice. Actb served as an internal control. The arrowhead indicates the mutation site.
- Western blotting with testis lysates from WT and M1ap −/− mice using the anti‐M1AP antibody. GAPDH was used as the loading control. The band corresponding to full‐length WT M1AP protein is indicated by an arrow.
- Immunofluorescence staining of M1AP (green) and SYCP3 (red) on the spermatocyte spreads of WT and M1ap −/− mice. Scale bars, 10 μm.
Similar to the observations in M1ap KI/KI mice, M1ap −/− mice had smaller testes and lower sperm numbers than their control littermates (Fig EV4A–C). Histological analyses of M1ap −/− testes also revealed the presence of many MMI spermatocytes with obvious unaligned chromosomes and a reduction in the number of postmeiotic germ cells (Fig EV4D). Immunofluorescence staining of the spread spermatocytes from M1ap −/− mice revealed that meiotic prophase I progression was normal and autosomes were properly synapsed, while XY untouching was frequently observed (Fig EV4E–G). The average number of MLH3 foci per nucleus was decreased (20.86 ± 0.19 in M1ap −/− mice vs. 23.72 ± 0.16 in controls), leading to increased frequencies of univalent sex chromosomes and/or autosomes at the MMI in M1ap −/− mice (Fig EV4H–M). All these findings recapitulated the observations in M1ap KI/KI mice (Figs 3 and EV2) and indicated that M1ap −/− mice have the same meiotic defects as M1ap KI/KI mice.
Figure EV4. The phenotype of M1ap −/− mice.
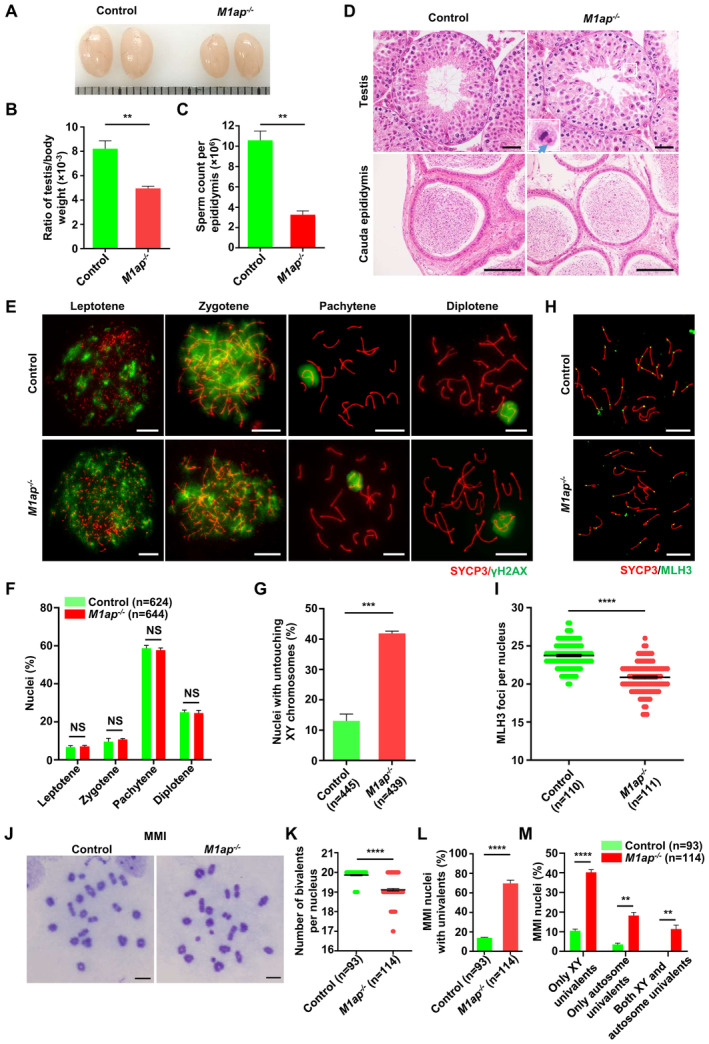
-
A–CRepresentative images of testes (A), the ratio of testis/body weight (B), and the sperm count per epididymis (C) from 8‐week‐old control and M1ap −/− mice. Each grid represents 1 mm in (A).
-
DRepresentative images of hematoxylin and eosin‐stained sections of testes and cauda epididymides from 8‐week‐old control and M1ap −/− mice. A magnified view of the boxed representative metaphase cell is shown in the lower left corner of the image of M1ap −/− mice. The blue arrow indicates unaligned chromosomes. Scale bars, 50 μm.
-
EImmunofluorescence staining of SYCP3 (red) and γH2AX (green) on spermatocyte spreads from 8‐week‐old control and M1ap −/− mice. Scale bars, 10 μm.
-
FThe percentages of spermatocytes at each stage of meiotic prophase I in 8‐week‐old control and M1ap −/− mice.
-
GQuantification of nuclei with untouching XY chromosomes at pachytene.
-
HImmunofluorescence staining of SYCP3 (red) and MLH3 (green) in spermatocyte spreads from 8‐week‐old control and M1ap −/− mice. Scale bars, 10 μm.
-
INumber of MLH3 foci per cell.
-
JMeiotic metaphase I (MMI) spermatocytes stained with Giemsa. Scale bars, 10 μm.
-
KNumber of bivalents per nucleus.
-
LFrequencies of nuclei with univalents.
-
MFrequencies of nuclei with only XY univalents, only autosome univalents, or both XY and autosome univalents.
Data information: (B and C), Data are presented as the mean ± SEM of three biological replicates. (F, G, I, and K–M) Data are presented as mean ± SEM. n shows the number of cells scored from at least three biological replicates. **P < 0.01; ***P < 0.001; ****P < 0.0001; NS, not significant; two‐tailed Student's t‐test.
M1AP interacts with ZZS proteins
To explore the molecular mechanism by which M1AP promotes meiotic recombination, we performed co‐immunoprecipitation (IP) in lysates from 10‐week‐old wild‐type mouse testes using the anti‐M1AP antibody, followed by mass spectrometry (MS) analysis. Interestingly, in addition to M1AP, two ZZS proteins, TEX11 and SHOC1, were among the top three on the candidate interacting protein list (Fig 6A and Table EV4). Western blotting using anti‐TEX11 and anti‐SHOC1 antibodies validated that M1AP interacts with TEX11 and SHOC1 in mouse testes (Fig 6B). To further confirm the interaction between M1AP and the ZZS complex, we performed co‐IP in lysates of HEK‐293T cells co‐expressing M1AP with SHOC1, TEX11, or SPO16. The results showed that M1AP interacts with all three ZZS complex components (Fig 6C).
Figure 6. M1AP interacts with the ZZS complex.
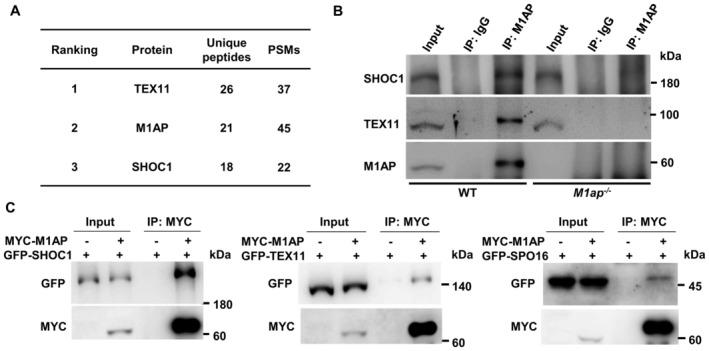
- Identification of proteins interacting with M1AP by coimmunoprecipitation and mass spectrometry (co‐IP/MS). The top three candidates on the ranking list sorted by the number of unique peptides are shown. PSMs, peptide‐spectrum matches.
- Co‐IP using anti‐M1AP antibody with whole‐testis lysates of WT mice and M1ap −/− mice, followed by western blot analyses of M1AP, SHOC1, and TEX11.
- Co‐IP detecting the interaction between M1AP (fused with an N‐terminal MYC tag) and each of the ZZS complex components (fused with an N‐terminal GFP tag) exogenously expressed in HEK‐293T cells.
The localization pattern of M1AP foci resembles that of ZZS proteins
ZZS proteins are known to localize at recombination intermediates during meiosis (Shinohara et al, 2008; Yang et al, 2008b; Zhang et al, 2018, 2019b). The kinetics of M1AP foci and the interaction between M1AP and the ZZS complex suggest that M1AP should have a cellular localization pattern similar to that of ZZS proteins. To test this hypothesis, we performed immunofluorescence staining for M1AP and TEX11, and found that they showed a high degree of colocalization on chromosome axes (Fig 7A). In late zygonema, the average fraction of M1AP foci colocalizing with TEX11 was 95.90 ± 0.53%, and the average fraction of TEX11 colocalizing with M1AP was 96.35 ± 0.71% (Fig 7B). Moreover, we found that M1AP foci seldom colocalized with DMC1 (14.04 ± 1.63%; Fig 7C and D), which marks resected 3′‐ssDNAs, but showed a high frequency of colocalization with RPA2 (88.10 ± 1.07%) at the early pachytene stage, which marks the sites of early recombination intermediates (Fig 7E and F). Intriguingly, similar to SPO16, M1AP foci were also detected on the asynapsed chromosome axes of Dmc1 −/− spermatocytes (Fig 7G), in which meiosis is arrested at a zygotene‐like stage without synapsis (Pittman et al, 1998), suggesting that M1AP foci may also localize at intersister chromatid joint molecules.
Figure 7. M1AP colocalizes with TEX11 and facilitates TEX11 recruitment at the recombination intermediates.
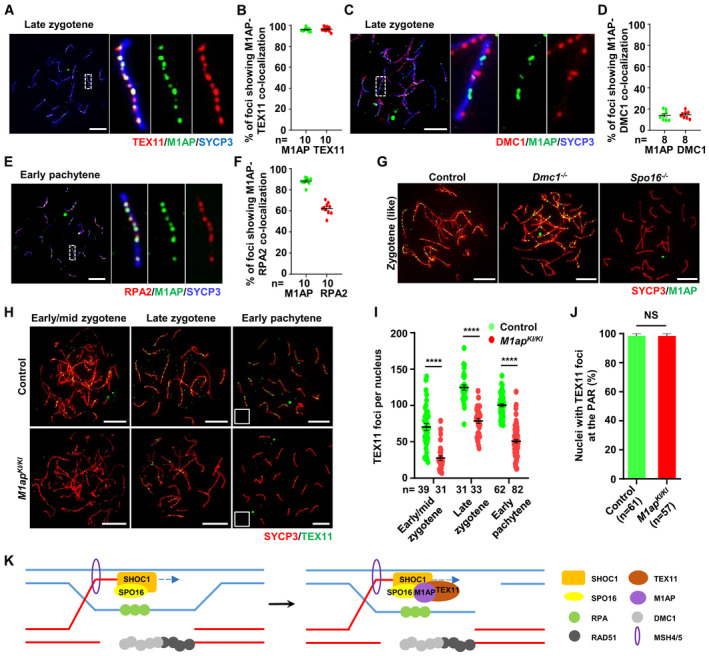
- Immunofluorescence staining of M1AP (green) and TEX11 (red) on the spermatocyte spreads of WT mice.
- Ratios of M1AP‐TEX11 co‐localizing foci to the M1AP total foci or TEX11 total foci at the indicated meiotic prophase substages. Each dot represents the ratio calculated from one nucleus.
- Immunofluorescence staining of M1AP (green) and DMC1 (red) on the spermatocyte spreads of WT mice.
- Ratios of M1AP‐DMC1 co‐localizing foci to the M1AP total foci or DMC1 total foci at the indicated meiotic prophase substages. Each dot represents the ratio calculated from one nucleus.
- Immunofluorescence staining of M1AP (green) and RPA2 (red) on the spermatocyte spreads of WT mice.
- Ratios of M1AP‐RPA2 co‐localizing foci to the M1AP total foci or RPA2 total foci at the indicated meiotic prophase substages. Each dot represents the ratio calculated from one nucleus.
- Immunofluorescence staining of M1AP (green) and SYCP3 (red) on spermatocyte spreads of control, Dmc1 −/− or Spo16 −/− mice.
- Immunofluorescence staining of TEX11 (green) and SYCP3 (red) on the spermatocyte spreads of control and M1ap KI/KI testes.
- The mean number of TEX11 foci per cell in control and M1ap KI/KI spermatocytes.
- Frequencies of nuclei with TEX11 foci detected at the PAR in spread early pachytene spermatocytes with touching XY chromosomes.
- Schematic diagram showing a proposed model in which M1AP acts as a partner of the ZZS complex in meiotic recombination in mouse meiotic prophase I. M1AP is recruited by SPO16 and facilitates TEX11 recruitment to stabilize recombination intermediates.
Data information: (A, C, E, G, and H) Scale bars, 10 μm. (B, D, F, I, and J) Data are presented as the mean ± SEM. n shows the number of cells scored from at least two biological replicates. (I and J) ****P < 0.0001; NS, not significant; two‐tailed Student's t‐test.
M1AP facilitates TEX11 recruitment
The interaction and similar localization pattern between M1AP and ZZS proteins indicate that they may have similar functions. To dissect the association between M1AP and the ZZS complex in meiotic recombination, we examined the localization of M1AP in spread spermatocytes from Spo16 −/− mice (Appendix Fig S2A and B), in which the assembly of MSH4/TEX11‐marked recombination intermediates was completely abolished (Zhang et al, 2019b). Immunofluorescence staining of spread spermatocytes showed the absence of M1AP foci in Spo16 −/− mice (Fig 7G), indicating that M1AP localization on chromosome axes depends on SPO16. To understand the impact of M1AP ablation on the ZZS complex, we performed immunofluorescence staining for SHOC1 and TEX11 in M1ap KI/KI spermatocytes. No obvious differences were observed in the number of SHOC1 foci between control and M1ap KI/KI spermatocytes (Fig EV5A and B), whereas the numbers of TEX11 foci were reduced in M1ap KI/KI spermatocytes compared with control cells from the early/mid zygotene to early pachytene stages (Fig 7H and I). Notably, in early pachytene cells with XY chromosomes touching, the frequency of nuclei with TEX11 foci at the PAR did not differ from that in controls (Fig 7J), which is in line with the observation that XY separation predominantly occurs at early pachytene (Fig 3K). Besides, the number of TEX11 foci in late zygotene or early pachytene oocytes showed no obvious difference between M1ap mutant and control embryos (Appendix Fig S3A and B). Taken together, we conclude that M1AP acts as a partner of the ZZS complex and facilitates the recruitment of TEX11, thereby stabilizing recombination intermediates in males (Fig 7K).
Figure EV5. No obvious differences were observed in the number of SHOC1 foci between control and M1ap KI/KI spermatocytes.
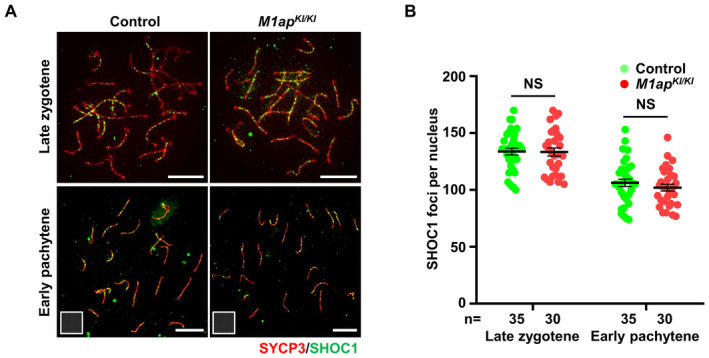
- Immunofluorescence staining with antibodies against SYCP3 (red) and SHOC1 (green) on spermatocyte spreads. Scale bars, 10 μm.
- The mean number of SHOC1 foci per cell in control and M1ap KI/KI spermatocytes at the indicated stages. Data are presented as the mean ± SEM. n shows the number of cells scored from three biological replicates. NS, not significant; two‐tailed Student's t‐test.
Discussion
Our study identified a homozygous M1AP splicing mutation (c.1074 + 2T > C) in a consanguineous Pakistani family with two brothers exhibiting MMI arrest and severe oligozoospermia. The pathogenicity of the mutation was validated using mouse models mimicking the M1AP mutation of our patients. Moreover, we found that M1AP interacts with the ZZS complex and facilitates TEX11 recruitment at recombination intermediates, promoting optimal crossover formation and meiotic progression in males.
Arango et al (2013) described the phenotype of mice carrying hypomorphic alleles in the M1ap gene (M1ap lz/lz ), in which a lacZ‐neomycin (β‐geo) cassette was introduced into intron 6 of the M1ap gene and led to variously reduced M1AP protein expression. They reported the presence of Sertoli‐cell‐only tubules in M1ap lz/lz testes and nonhomologous pairing of autosomes, XY asynapsis, retained γH2AX signals on the autosomes as late as diakinesis, and deficient crossover formation in M1ap lz/lz spermatocytes, which consequently resulted in severe oligozoospermia and male infertility (Arango et al, 2013). In our study, both lines of the M1ap mutant mice, M1ap KI/KI and M1ap −/− , showed very similar phenotypes that produced spermatozoa approximately one‐third of that in the control mice. Consistent with the observation in M1ap lz/lz mice, we also observed an increased frequency of XY untouching and reduced crossover formation, but we did not find any seminiferous tubules containing only Sertoli cells or abnormal synapses and persistent γH2AX signals on the autosomes. We believe that disruption of M1AP not resulting in Sertoli cell only seminiferous tubules is consistent with M1AP being first detected in testes at 10 dpp when leptotene spermatocytes appear, suggesting that M1ap ablation is not likely to affect early spermatogonia. Intriguingly, the meiotic defects in our M1ap KI/KI mice and M1ap −/− mice appeared much milder than those in the M1ap lz/lz mice. The possibility that the predicted truncated proteins in M1ap KI/KI mice and M1ap −/− mice (approximately two‐thirds and one‐third of the wild‐type protein length, respectively) may be produced and retain some function of M1AP gives a potential explanation. However, due to the lack of antibodies against the M1AP N‐terminus, we were unable to test this hypothesis. Future studies aimed at further dissecting the molecular function of M1AP in meiosis are warranted.
M1AP interacting with all three components of the ZZS complex and colocalizing with TEX11 suggest that M1AP likely functions as a copartner of the ZZS complex in meiotic recombination. Intriguingly, mice with any of the four genes knocked out have a different phenotype. Knockout of Shoc1 leads to the most severe phenotype—meiotic arrest at a zygotene stage with a complete absence of axial localization of the other two ZZS proteins (Zhang et al, 2018). Spo16 −/− mice exhibited meiotic arrest at a zygotene‐like or early pachytene‐like stage and the synapsis was less affected than in Shoc1 −/− mice (Zhang et al, 2019b). In Spo16 −/− spermatocytes, SHOC1 foci were reduced, and M1AP and TEX11 foci were completely abolished, suggesting that SPO16 likely stabilizes SHOC1 and recruits M1AP and TEX11 at the recombination intermediates. In Tex11 −/− mice, asynapsed chromosomes (two to 10 per nucleus) were seen in 38% of pachytene spermatocytes, crossovers could form but reduced, and meiosis arrested at the MMI (Adelman & Petrini, 2008; Yang et al, 2008b). Our present study revealed that M1AP disruption leads to a mild crossover reduction and a partial MMI arrest with no overt effect on autosomal synapsis. In M1ap‐deficient spermatocytes, the numbers of RAD51 and DMC1 foci were increased at early pachytene stages, suggesting that the recombination intermediates formed following strand invasion are likely unstable and subject to precocious dismantling, leading to delayed DSB repair, which are similar to the observations in Tex11 knockouts (Adelman & Petrini, 2008). Consistently, normal localization of SHOC1 but reduced number of TEX11 foci were observed. The number of TEX11 foci was reduced from the early/mid zygotene stage, and the reduction was maintained at the same level during the following stages, implying that the recruitment, but not the stabilization, of TEX11 on chromosome axes is affected in M1ap mutant mice. This was further supported by the finding that XY separation occurs predominantly at early pachytene in M1ap mutant mice and that the ratio of nuclei with TEX11 foci at the PAR in spread early pachytene spermatocytes with XY touching did not differ from that in controls. These findings lead us to propose that, following strand invasion, M1AP is recruited to recombination intermediates by SPO16 and stabilizes these early‐stage intermediates by promoting the recruitment of TEX11 but may be dispensable for stabilization and processing of intermediates at the later stages.
The disruption of M1AP seems to have a greater impact on crossover formation at the PAR of sex chromosomes than on autosomes. In M1ap mutant mice, the frequencies of cells with XY untouching and cells lacking an MLH1 focus at the PAR were approximately 30% higher than that in control mice, indicating that the number of early‐stage recombination intermediates was reduced by approximately one‐third. However, we did not observe any obvious defects in autosomal synapses, and the number of nucleus‐wide MLH1 foci was reduced by 13.02% in the mutants. The milder reduction in crossovers on autosomes could be explained by the crossover homeostasis (Cole et al, 2012; Wang et al, 2015). It is reasonable to postulate that when an early‐stage recombination intermediate is precociously dismantled, recombination intermediates nearby would have the opportunity to take over to generate an MLH1 crossover, thus buffering the perturbations on the ultimate number of crossovers. Given that the X and Y chromosomes have only a small region of homology (PAR), only one or two DSBs are produced in the tiny PAR (Kauppi et al, 2011). The compensation from adjacent intermediates through the homeostatic mechanism is limited and thus renders a higher susceptibility of the PAR to the decreased early‐stage recombination intermediates than autosomes.
The sexual dimorphisms in M1AP localization patterns and meiotic phenotypes of M1ap mutant mice indicate that M1AP, unlike the other ZZS proteins (Yang et al, 2008b; Zhang et al, 2018, 2019b), acts as a sex‐dependent regulator of meiotic recombination. The recruitment of TEX11 requires M1AP in spermatocytes but not in oocytes also underlines that early‐stage recombination intermediates are differentially regulated between males and females. Several meiotic recombination‐related proteins, despite expression in both spermatocytes and oocytes, have been found to exhibit sexually dimorphic functions in mammals, with a specific phenotype in males but not females when knocked‐out (Hua & Liu, 2021). For example, Tex15 −/− male mice were infertile and exhibited complete losses of RAD51 and DMC1 foci on chromosome axes, which, however, were not observed in the mutant females (Yang et al, 2008a). RAD51AP2 foci were detected in both spermatocytes and oocytes, but this protein is required for crossover formation only on XY chromosomes but not on XX and autosomes in mice (Ma et al, 2022). In addition, knockout of Meilb2 or Zcwpw1 also results in a much more severe meiotic DSB repair defects in males than in females (Li et al, 2019; Zhang et al, 2019a). Thus, our study further substantiates the awareness that the meiotic recombination in males may require additional factors, might owing to the longer meiotic DSB repair duration in males (7–8 days) than in females (4–5 days; Baudat et al, 2013) and the specific mechanism to ensure X‐Y recombination within the tiny homologous region (i.e., PAR), which is worthy of further investigations.
In conclusion, our study demonstrates that M1AP is a pathogenic gene for male infertility and that its mutation causes meiosis arrest at the metaphase I stage owing to unaligned chromosomes during spermatogenesis in humans and mice. Moreover, our study reveals the molecular role of M1AP in male meiotic recombination—cooperating with the ZZS complex in the stabilization of recombination intermediates. All these findings allow an in‐depth understanding of the role of M1AP and ZZS proteins in meiotic recombination, shed a new perspective for the differences between male and female meiosis, and provide a scientific basis for genetic counseling and diagnosis of male infertility.
Materials and Methods
Clinical samples
A consanguineous Pakistani family, having two brothers diagnosed with idiopathic male infertility, was enrolled in this study. Written informed consent was received from all participants prior to the onset of the study. For both patients, the reproductive hormones in the serum were examined in local laboratories; chromosome analysis revealed a normal karyotype, and no Y‐chromosome microdeletions were detected. Semen analyses were performed at least three times for each patient, following the guidelines of the World Health Organization (WHO, 2010). This study was approved by the institutional ethics committee of the University of Science and Technology of China (USTC approval number 2019‐KY‐168).
WES and variant filtration
Genomic DNA was extracted from blood samples for the two patients (V:2 and V:3) and their fertile brother (V:1) using a QIAamp Blood DNA Mini Kit (QIAGEN, 51106). WES and subsequent bioinformatics analyses were conducted for these three individuals as we previously described (Wu et al, 2021). Briefly, whole exomes were captured and sequenced, and the obtained reads were aligned to the human genome reference assembly (hg19). For each sequenced sample, the coverage of targeted exonic regions with ≥ 10 reads was 98%, and the average coverage depth after removing PCR duplicates was > 110×. After variant calling, the single‐nucleotide variants and small insertions/deletions within the coding regions and splicing sites were filtered following the pipeline described in Appendix Fig S1. The variant identified after filtration was validated by Sanger sequencing. The sequences of the primers are listed in Table EV5.
Immunofluorescence staining of human testicular sections
Human testicular tissues were fixed in formalin, embedded in paraffin, and then sectioned at 5 μm. For immunofluorescence staining, slides were blocked with 3% nonfat milk in PBS for 30 min and then incubated with primary antibodies overnight at 4°C. After three washes in PBS containing 0.05% Triton X‐100 (PBST), the slides were incubated with secondary antibodies for 1 h at 37°C. The slides were then washed three times in PBST and mounted with Vectashield medium (Vector Laboratories, H‐1000). Information on the antibodies is available in Table EV6.
Cell culture and transfection
HEK‐293T cells (ATCC, CRL‐3216) were cultured in high‐glucose Dulbecco's modified Eagle's medium (DMEM) with 10% FBS (Gibco, 16000‐044), 100 U/ml penicillin and 100 mg/ml streptomycin (Gibco, 15140‐122) and maintained at 5% CO2. Cells were tested negative for mycoplasma contamination. Cells were passaged 2–3 times after thawing and transfected at 70–80% confluency. Transfection of plasmids was performed using Lipofectamine 3000 (Invitrogen L3000015) according to the manufacturer's instructions.
RNA extraction and RT–PCR
Total RNA was extracted using TRIzol reagent (Takara, 9109), followed by the synthesis of complementary DNA (cDNA) using the PrimeScript RT reagent kit (Takara, RR047A) according to the manufacturer's instructions. EasyTaq DNA Polymerase (TransGen Biotech, AP111) was used for RT–PCR. The PCRs were performed using the following cycle conditions: 3 min at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at 60°C, and 30 s at 72°C. The sequences of the primers are listed in Table EV5.
Minigene splicing assay
The effect of the mutation (c.1074T > C) on the splicing of M1AP in humans was evaluated using the minigene splicing assay. Briefly, the M1AP minigene was amplified with genomic DNA from HEK‐293T cells (ATCC, CRL‐3216) by PCR, using primers flanking the sequences of intron 5–6 and intron 8–9 of the M1AP gene. Then, the PCR products were ligated with the backbone of pET01 vectors using the ClonExpress MultiS One Step Cloning Kit (Vazyme, C113) according to the manufacturer's instructions. The inserted sequences were verified by Sanger sequencing. After transfection of vectors expressing the wild‐type or mutant M1AP minigene, HEK‐293T cells were harvested for RNA extraction, followed by RT–PCR and Sanger sequencing. Sequences of primers for constructing the vectors and RT–PCR are listed in Table EV5.
Mice
M1ap KI/KI mice, M1ap −/− mice and Spo16 −/− mice were generated using CRISPR/Cas9 technology as we described previously (Jiang et al, 2017; Li et al, 2021). Briefly, to generate M1ap KI/KI mice, single‐strand oligodeoxynucleotides (ssODNs) harboring a mutation equivalent to the M1AP splicing mutation identified in our patients were synthesized by Sangon Biotech (Shanghai). The sgRNA, Cas9 mRNAs, and ssODNs (for generating M1ap KI/KI mice) were microinjected into zygotes of C57BL/6 mice, followed by transfer of the microinjected zygotes into the oviducts of pseudopregnant ICR female mice. Sanger sequencing with genomic DNA extracted from mouse toes was performed to detect the mutations in the F0 generation mice. The founder mice carrying the mutation of interest were backcrossed with wild‐type C57BL/6 mice for at least one generation. The obtained heterozygous mutant mice were intercrossed to produce homozygous mutant mice. All mouse studies were conducted in accordance with the guidelines approved by the Institutional Animal Care Committee of USTC. The sequences of sgRNA, ssODNs, and primers for mouse genotyping are listed in Table EV5.
Hematoxylin and eosin staining
Mouse testicular and epididymal tissues were harvested and fixed in Bouin's solution. Human testicular tissues were fixed in formalin. After embedding in paraffin, the testicular tissue was sectioned at 5 μm. Hematoxylin and eosin (H&E) staining was performed as we described previously (Jiang et al, 2015). Mouse ovaries were harvested and fixed in modified Davidson's fluid overnight. Paraffin‐embedded ovaries were serially sectioned and stained with hematoxylin for counting the numbers of follicles as we described previously (Jiao et al, 2020).
Spermatocyte chromosome spreads and immunofluorescence staining
Spermatocyte chromosome spreads and subsequent immunofluorescence staining were performed as previously reported (Jiang et al, 2014; Ma et al, 2022). Information on the antibodies is available in Table EV6.
Diakinesis/metaphase I chromosome spreading
Chromosome spreads of meiotic diakinesis/metaphase cells were prepared as previously described and stained with Giemsa (Jiang et al, 2017).
Oocyte chromosome spreads
Fetal ovaries were collected from embryos at 16.5 dpc or 18.5 dpc and spread as previously described (Hwang et al, 2018; Ma et al, 2022). Briefly, ovaries were placed in hypo‐extraction buffer for 15 min, transferred into 100 mM sucrose drop, and teased apart with two needles to release cells. Twenty microliters of the cell suspension was added onto slides mounted with 100 μl of 1% paraformaldehyde containing 0.15% Triton X‐100, evenly spread on the slides, and then incubated in a closed humid chamber for 2 h at room temperature. The slides were then air‐dried and subsequent immunofluorescence staining was performed as described above for spermatocyte spreads.
Cell smear
Testes from adult mice and ovaries from embryos at 16.5 dpc were harvested. The tissues ware placed in PBS supplemented with 10% FBS, teased apart to release cells. A drop of cell suspension was added to one side of the glass slide and spread gently using a glass capillary tube. After air‐dried, the slide was fixed in 4% PFA for 10 min and permeabilized in 0.2% PBST for 30 min, followed by immunofluorescence staining as described for spermatocyte spreads.
Coimmunoprecipitation
The mouse testes were lysed in immunoprecipitation (IP) buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5% Triton X‐100, and 2.5 mM EDTA) supplemented with 1 mM phenylmethanesulfonyl fluoride (PMSF, Thermo Fisher Scientific, 36978). The lysates were sonicated for 12 cycles (2 s on/off) with 10% pulses and centrifuged at 15,000 g at 4°C for 15 min. The supernatant was divided into two aliquots and incubated with precleared Protein A/G agarose beads (Santa Cruz, sc‐2003) and 2 μg anti‐M1AP C‐terminal antibody (epitope: residues 415–529 of mouse protein; customizedly produced by ABclonal) or rabbit IgG antibody (ABclonal, AC005). After incubation at 4°C for 6 h, the agarose beads were washed in IP buffer five times and stored at −80°C. Western blotting using the anti‐M1AP antibody, as well as silver staining, was performed to assess the quality of Co‐IP. The IPed samples (including one sample obtained using anti‐M1AP antibody and one sample obtained using anti‐rabbit IgG antibody) were subjected to MS at the National Center for Protein Science Shanghai.
HPLC‐tandem MS (MS/MS) analysis
For protein interaction analysis, on beads digestion was performed. Briefly, beads were washed and denatured using 8 M urea with 100 mM Tris‐Cl (pH 8.5), reduced by incubation with 10 mM Tris(2‐Carboxyethyl)‐Phosphine HCl (TCEP, Thermo Scientific, T2556), and then alkylated with 15 mM iodoacetamide (Sigma, 16125) for 30 min at room temperature in the dark. The samples were digested with Trypsin at 1:50 (w/w Promega) overnight and stopped by 5% Formic Acid (FA, Thermo Scientific, 28905). The peptide mixture was then desalted by MonoSpinTM C18 column (GL Science, 5010‐21671). Desalted mixture was dried with a SpeedVac and resuspended in 0.1% FA for MS analysis.
The peptide mixture was analyzed using a homemade 30 cm‐long pulled‐tip analytical column (75 μm ID packed with ReproSil‐Pur C18‐AQ 1.9 μm resin, Dr. Maisch GmbH), and the column was then placed in‐line with an Easy‐nLC 1200 nano HPLC (Thermo Scientific) for MS analysis. The analytical column temperature was set at 55°C during the experiments. The mobile phase and elution gradient used for peptide separation were as follows: 0.1% formic acid in water as buffer A and 0.1% formic acid in 80% acetonitrile as buffer B, 0–1 min, 5–10% B; 1–96 min, 10–40% B; 96–104 min, 40–60% B, 104–105 min, 60–100% B, 105–120 min, 100% B. The flow rate was set as 300 nL/min.
Data‐dependent MS/MS analysis was performed with a Q Exactive Orbitrap mass spectrometer (Thermo Scientific). Peptides eluted from the LC column were directly electrosprayed into the mass spectrometer with the application of a distal 2.5‐kV spray voltage. A cycle of one full‐scan MS spectrum (m/z 300–1,800) was acquired followed by top 20 MS/MS events, sequentially generated on the first to the twentieth most intense ions selected from the full MS spectrum at a 30% normalized collision energy. Full scan resolution was set to 70,000 with automated gain control (AGC) target of 3e6. MS/MS scan resolution was set to 17,500 with isolation window of 1.8 m/z and AGC target of 1e5. The number of microscans was one for both MS and MS/MS scans and the maximum ion injection time was 50 and 100 ms, respectively. The dynamic exclusion settings used were as follows: charge exclusion, 1 and > 8; exclude isotopes, on; and exclusion duration, 30 s. MS scan functions and LC solvent gradients were controlled by the Xcalibur data system (Thermo Scientific).
For protein interaction analysis, the acquired MS/MS data were analyzed against a UniProtKB Mus musculus (database released on Jan. 28, 2021) using Proteome Discoverer 2.4 (Thermo Scientific). Mass tolerances for precursor ions were set at 20 ppm and for fragment were set at 0.1 Da. Trypsin was defined as cleavage enzyme; cysteine alkylation by iodoacetamide was specified as fixed modification with mass shift 57.02146; methionine oxidation was set as dynamic modification with mass shift 15.9949. In order to accurately estimate peptide probabilities and false discovery rates (FDR), a decoy database containing the reversed sequences of all the proteins was appended to the target database to accurately estimate peptide probabilities and FDR, and the FDR was set at 0.01.
The list of all the peptides identified by MS in M1AP‐IPed or IgG‐IPed lysates are available in Dataset EV1. Proteins, identified in M1AP‐IPed lysates (the number of unique peptides greater than 2) but not the IgG‐IPed lysates, were considered as the candidate interacting proteins of M1AP (Table EV4).
Co‐IP in cultured cells
To construct vectors expressing MYC‐tagged mouse M1AP protein and vectors expressing GFP‐tagged mouse SHOC1, TEX11, or SPO16 proteins, the coding sequences of M1ap, Shoc1, Tex11, and Spo16 were amplified from mouse testis cDNA by PCR and ligated with the backbone from pEGFP‐N1 vectors. The inserted sequences were verified by Sanger sequencing. Sequences of primers for constructing the vectors are listed in Table EV5.
To investigate whether M1AP interacts with the ZZS proteins directly, we cotransfected vectors expressing N‐terminal MYC‐tagged M1AP and vectors expressing N‐terminal GFP‐tagged SHOC1, TEX11, or SPO16 into HEK‐293T cells. Forty‐eight hours after transfection, cells were harvested for protein extraction and lysed in IP buffer supplemented with 1 mM PMSF. The obtained protein lysates were incubated with Pierce anti‐c‐Myc magnetic beads (Thermo Fisher Scientific, 88842) under gentle rotation at 4°C for 6 h. After washing with IP buffer five times, the beads were boiled in 1× SDS sample buffer (100 mM Tris–HCl pH 7.4, 2% SDS, 15% glycerol, 0.1% bromophenol blue and 5 mM dithiothreitol [DTT]) for 10 min, followed by Western blotting analyses.
Statistical analysis
Data are presented as the mean ± SEM. Two‐tailed Student's unpaired t‐test was applied for all statistical analyses using the GraphPad Prism software. The difference was considered significant when the P value was < 0.05. The investigators were not blinded to all experiments.
Author contributions
Yang Li: Data curation; formal analysis; investigation; methodology; writing – original draft. Yufan Wu: Data curation; formal analysis; validation; investigation; methodology. Ihsan Khan: Resources; data curation; investigation; methodology. Jianteng Zhou: Formal analysis; methodology. Yue Lu: Investigation; methodology. Jingwei Ye: Investigation; methodology. Junyan Liu: Investigation; methodology. Xuefeng Xie: Investigation; methodology. Congyuan Hu: Investigation; methodology. Hanwei Jiang: Funding acquisition. Suixing Fan: Formal analysis. Huan Zhang: Investigation; project administration. Yuanwei Zhang: Funding acquisition; methodology; project administration. Xiaohua Jiang: Methodology; project administration. Bo Xu: Methodology; project administration. Hui Ma: Formal analysis; supervision; funding acquisition; writing – original draft; project administration; writing – review and editing. Qinghua Shi: Formal analysis; supervision; funding acquisition; project administration; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Table EV4
Table EV5
Table EV6
Dataset EV1
PDF+
Acknowledgments
We thank all the participants for their cooperation. We thank Dr. Mengcheng Luo (Wuhan University, China) for generously gifting us the anti‐MLH3 antibodies and Dr. Chao Yu (Zhejiang University) for the anti‐SHOC1 and the goat anti‐TEX11 antibodies. We thank Dr. Jingwen Li and Dr. Chao Peng from the Mass Spectrometry System at the National Facility for Protein Science in Shanghai (NFPS), Shanghai Advanced Research Institute, Chinese Academy of Science, Shanghai 201210, China for MS sample preparation, data collection and data analysis. We also thank the Bioinformatics Center of the University of Science and Technology of China, School of Life Sciences, for providing supercomputing resources. This work was supported by the National Natural Science Foundation of China (31890780 to QS, U21A20204 to QS, 82071709 to XJ and 32061143006 to QS) and the National Key Research and Developmental Program of China (2021YFC2700200 to QS, 2021YFC2700100 to YZ, and 2019YFA0802600 to HM).
EMBO reports (2023) 24: e55778
Contributor Information
Hui Ma, Email: clsmh@ustc.edu.cn.
Qinghua Shi, Email: qshi@ustc.edu.cn.
Data availability
This study includes no data deposited in public databases.
References
- Adelman CA, Petrini JH (2008) ZIP4H (TEX11) deficiency in the mouse impairs meiotic double strand break repair and the regulation of crossing over. PLoS Genet 4: e1000042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S, Roeder GS (2000) Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102: 245–255 [DOI] [PubMed] [Google Scholar]
- Arango NA, Huang TT, Fujino A, Pieretti‐Vanmarcke R, Donahoe PK (2006) Expression analysis and evolutionary conservation of the mouse germ cell‐specific D6Mm5e gene. Dev Dyn 235: 2613–2619 [DOI] [PubMed] [Google Scholar]
- Arango NA, Li L, Dabir D, Nicolau F, Pieretti‐Vanmarcke R, Koehler C, McCarrey JR, Lu N, Donahoe PK (2013) Meiosis I arrest abnormalities lead to severe oligozoospermia in meiosis 1 arresting protein (M1ap)‐deficient mice. Biol Reprod 88: 76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora K, Corbett KD (2019) The conserved XPF:ERCC1‐like Zip2:Spo16 complex controls meiotic crossover formation through structure‐specific DNA binding. Nucleic Acids Res 47: 2365–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC, Yao X, Christie DM, Monell C, Arnheim N, Bradley A et al (1996) Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet 13: 336–342 [DOI] [PubMed] [Google Scholar]
- Baudat F, Manova K, Yuen JP, Jasin M, Keeney S (2000) Chromosome synapsis defects and sexually dimorphic meiotic progression in mice lacking Spo11. Mol Cell 6: 989–998 [DOI] [PubMed] [Google Scholar]
- Baudat F, Imai Y, de Massy B (2013) Meiotic recombination in mammals: localization and regulation. Nat Rev Genet 14: 794–806 [DOI] [PubMed] [Google Scholar]
- Börner GV, Kleckner N, Hunter N (2004) Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45 [DOI] [PubMed] [Google Scholar]
- Brown MS, Bishop DK (2014) DNA strand exchange and RecA homologs in meiosis. Cold Spring Harb Perspect Biol 7: a016659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua PR, Roeder GS (1998) Zip2, a meiosis‐specific protein required for the initiation of chromosome synapsis. Cell 93: 349–359 [DOI] [PubMed] [Google Scholar]
- Cole F, Kauppi L, Lange J, Roig I, Wang R, Keeney S, Jasin M (2012) Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat Cell Biol 14: 424–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Pyatnitskaya A, Andréani J, Ranjha L, Ramus C, Laureau R, Fernandez‐Vega A, Holoch D, Girard E, Govin J et al (2018) A meiotic XPF‐ERCC1‐like complex recognizes joint molecule recombination intermediates to promote crossover formation. Genes Dev 32: 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelmann W, Cohen PE, Kane M, Lau K, Morrow B, Bennett S, Umar A, Kunkel T, Cattoretti G, Chaganti R et al (1996) Meiotic pachytene arrest in MLH1‐deficient mice. Cell 85: 1125–1134 [DOI] [PubMed] [Google Scholar]
- Guiraldelli MF, Felberg A, Almeida LP, Parikh A, de Castro RO, Pezza RJ (2018) SHOC1 is a ERCC4‐(HhH)2‐like protein, integral to the formation of crossover recombination intermediates during mammalian meiosis. PLoS Genet 14: e1007381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handel MA, Schimenti JC (2010) Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 11: 124–136 [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Ponte L, Halsey C (1995) MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev 9: 1728–1739 [DOI] [PubMed] [Google Scholar]
- Hua R, Liu M (2021) Sexual dimorphism in mouse meiosis. Front Cell Dev Biol 9: 670599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N (2015) Meiotic recombination: the essence of heredity. Cold Spring Harb Perspect Biol 7: a016618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang GH, Hopkins JL, Jordan PW (2018) Chromatin spread preparations for the analysis of mouse oocyte progression from prophase to metaphase II. J Vis Exp 132: 56736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wang L, Cui Y, Xu Z, Guo T, Cheng D, Xu P, Yu W, Shi Q (2014) Meiotic chromosome behavior in a human male t (8; 15) carrier. J Genet Genomics 41: 177–185 [DOI] [PubMed] [Google Scholar]
- Jiang X, Ma T, Zhang Y, Zhang H, Yin S, Zheng W, Wang L, Wang Z, Khan M, Sheikh SW et al (2015) Specific deletion of Cdh2 in Sertoli cells leads to altered meiotic progression and subfertility of mice. Biol Reprod 92: 79 [DOI] [PubMed] [Google Scholar]
- Jiang L, Li T, Zhang X, Zhang B, Yu C, Li Y, Fan S, Jiang X, Khan T, Hao Q et al (2017) RPL10L is required for male meiotic division by compensating for RPL10 during meiotic sex chromosome inactivation in mice. Curr Biol 27: 1498–1505 [DOI] [PubMed] [Google Scholar]
- Jiao Y, Fan S, Jabeen N, Zhang H, Khan R, Murtaza G, Jiang H, Ali A, Li Y, Bao J et al (2020) A TOP6BL mutation abolishes meiotic DNA double‐strand break formation and causes human infertility. Sci Bull 65: 2120–2129 [DOI] [PubMed] [Google Scholar]
- Kauppi L, Barchi M, Baudat F, Romanienko PJ, Keeney S, Jasin M (2011) Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 331: 916–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Huang T, Li MJ, Zhang CX, Yu XC, Yin YY, Liu C, Wang X, Feng HW, Zhang T et al (2019) The histone modification reader ZCWPW1 is required for meiosis prophase I in male but not in female mice. Sci Adv 5: eaax1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wu Y, Zhou J, Zhang H, Zhang Y, Ma H, Jiang X, Shi Q (2021) A recurrent ZSWIM7 mutation causes male infertility resulting from decreased meiotic recombination. Hum Reprod 36: 1436–1445 [DOI] [PubMed] [Google Scholar]
- Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, Gilgeous A, Thomas J, Cheng J, Touchman JW, Green ED et al (2002) Meiotic arrest and aneuploidy in MLH3‐deficient mice. Nat Genet 31: 385–390 [DOI] [PubMed] [Google Scholar]
- Liu H, Huang T, Li M, Li M, Zhang C, Jiang J, Yu X, Yin Y, Zhang F, Lu G et al (2019) SCRE serves as a unique synaptonemal complex fastener and is essential for progression of meiosis prophase I in mice. Nucleic Acids Res 47: 5670–5683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Li T, Xie X, Jiang L, Ye J, Gong C, Jiang H, Fan S, Zhang H, Shi B et al (2022) RAD51AP2 is required for efficient meiotic recombination between X and Y chromosomes. Sci Adv 8: eabk1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaisne N, Novatchkova M, Peirera L, Vezon D, Jolivet S, Froger N, Chelysheva L, Grelon M, Mercier R (2008) SHOC1, an XPF endonuclease‐related protein, is essential for the formation of class I meiotic crossovers. Curr Biol 18: 1432–1437 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Ogawa H (1999) The Saccharomyces cerevisiae MER3 gene, encoding a novel helicase‐like protein, is required for crossover control in meiosis. EMBO J 18: 5714–5723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishant KT, Plys AJ, Alani E (2008) A mutation in the putative MLH3 endonuclease domain confers a defect in both mismatch repair and meiosis in Saccharomyces cerevisiae . Genetics 179: 747–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak JE, Ross‐Macdonald PB, Roeder GS (2001) The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics 158: 1013–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittman DL, Cobb J, Schimenti KJ, Wilson LA, Cooper DM, Brignull E, Handel MA, Schimenti JC (1998) Meiotic prophase arrest with failure of chromosome synapsis in mice deficient for Dmc1, a germline‐specific RecA homolog. Mol Cell 1: 697–705 [DOI] [PubMed] [Google Scholar]
- Pyatnitskaya A, Borde V, De Muyt A (2019) Crossing and zipping: molecular duties of the ZMM proteins in meiosis. Chromosoma 128: 181–198 [DOI] [PubMed] [Google Scholar]
- Reynolds A, Qiao H, Yang Y, Chen JK, Jackson N, Biswas K, Holloway JK, Baudat F, de Massy B, Wang J et al (2013) RNF212 is a dosage‐sensitive regulator of crossing‐over during mammalian meiosis. Nat Genet 45: 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert T, Nore A, Brun C, Maffre C, Crimi B, Bourbon HM, de Massy B (2016) The TopoVIB‐like protein family is required for meiotic DNA double‐strand break formation. Science 351: 943–949 [DOI] [PubMed] [Google Scholar]
- Romanienko PJ, Camerini‐Otero RD (2000) The mouse Spo11 gene is required for meiotic chromosome synapsis. Mol Cell 6: 975–987 [DOI] [PubMed] [Google Scholar]
- Shinohara M, Oh SD, Hunter N, Shinohara A (2008) Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat Genet 40: 299–309 [DOI] [PubMed] [Google Scholar]
- Snowden T, Acharya S, Butz C, Berardini M, Fishel R (2004) hMSH4‐hMSH5 recognizes Holliday junctions and forms a meiosis‐specific sliding clamp that embraces homologous chromosomes. Mol Cell 15: 437–451 [DOI] [PubMed] [Google Scholar]
- Sym M, Engebrecht JA, Roeder GS (1993) ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72: 365–378 [DOI] [PubMed] [Google Scholar]
- Tsubouchi T, Zhao H, Roeder GS (2006) The meiosis‐specific zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with zip2. Dev Cell 10: 809–819 [DOI] [PubMed] [Google Scholar]
- Tu C, Wang Y, Nie H, Meng L, Wang W, Li Y, Li D, Zhang H, Lu G, Lin G et al (2020) An M1AP homozygous splice‐site mutation associated with severe oligozoospermia in a consanguineous family. Clin Genet 97: 741–746 [DOI] [PubMed] [Google Scholar]
- Vrielynck N, Chambon A, Vezon D, Pereira L, Chelysheva L, De Muyt A, Mézard C, Mayer C, Grelon M (2016) A DNA topoisomerase VI‐like complex initiates meiotic recombination. Science 351: 939–943 [DOI] [PubMed] [Google Scholar]
- Wang TF, Kleckner N, Hunter N (1999) Functional specificity of MutL homologs in yeast: Evidence for three Mlh1‐based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc Natl Acad Sci U S A 96: 13914–13919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zickler D, Kleckner N, Zhang L (2015) Meiotic crossover patterns: obligatory crossover, interference and homeostasis in a single process. Cell Cycle 14: 305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JJ, Ge W, Zhai QY, Liu JC, Sun XW, Liu WX, Li L, Lei CZ, Dyce PW, De Felici M et al (2020) Single‐cell transcriptome landscape of ovarian cells during primordial follicle assembly in mice. PLoS Biol 18: e3001025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO (2010) WHO laboratory manual for the examination and processing of human semen, 5th edn. Geneva: WHO Press; [Google Scholar]
- Wu Y, Li Y, Murtaza G, Zhou J, Jiao Y, Gong C, Hu C, Han Q, Zhang H, Zhang Y et al (2021) Whole‐exome sequencing of consanguineous families with infertile men and women identifies homologous mutations in SPATA22 and MEIOB. Hum Reprod 36: 2793–2804 [DOI] [PubMed] [Google Scholar]
- Wyrwoll MJ, Temel ŞG, Nagirnaja L, Oud MS, Lopes AM, van der Heijden GW, Heald JS, Rotte N, Wistuba J, Wöste M et al (2020) Bi‐allelic mutations in M1AP are a frequent cause of meiotic arrest and severely impaired spermatogenesis leading to male infertility. Am J Hum Genet 107: 342–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Eckardt S, Leu NA, McLaughlin KJ, Wang PJ (2008a) Mouse TEX15 is essential for DNA double‐strand break repair and chromosomal synapsis during male meiosis. J Cell Biol 180: 673–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Gell K, van der Heijden GW, Eckardt S, Leu NA, Page DC, Benavente R, Her C, Höög C, McLaughlin KJ et al (2008b) Meiotic failure in male mice lacking an X‐linked factor. Genes Dev 22: 682–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Tang S, Ma Y, Hunter N (2012) Delineation of joint molecule resolution pathways in meiosis identifies a crossover‐specific resolvase. Cell 149: 334–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Shao J, Fan HY, Yu C (2018) Evolutionarily‐conserved MZIP2 is essential for crossover formation in mammalian meiosis. Commun Biol 1: 147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Fujiwara Y, Yamamoto S, Shibuya H (2019a) A meiosis‐specific BRCA2 binding protein recruits recombinases to DNA double‐strand breaks to ensure homologous recombination. Nat Commun 10: 722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Ji SY, Busayavalasa K, Yu C (2019b) SPO16 binds SHOC1 to promote homologous recombination and crossing‐over in meiotic prophase I. Sci Adv 5: eaau9780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N (2015) Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol 7: a016626 [DOI] [PMC free article] [PubMed] [Google Scholar]


