Abstract
The extremely large number of leaves produced by terrestrial and aquatic plants provide habitats for colonization by a diversity of microorganisms. This review focuses on the bacterial component of leaf microbial communities, with emphasis on Pseudomonas syringae—a species that participates in leaf ecosystems as a pathogen, ice nucleus, and epiphyte. Among the diversity of bacteria that colonize leaves, none has received wider attention than P. syringae, as it gained notoriety for being the first recombinant organism (Ice− P. syringae) to be deliberately introduced into the environment. We focus on P. syringae to illustrate the attractiveness and somewhat unique opportunities provided by leaf ecosystems for addressing fundamental questions of microbial population dynamics and mechanisms of plant-bacterium interactions. Leaf ecosystems are dynamic and ephemeral. The physical environment surrounding phyllosphere microbes changes continuously with daily cycles in temperature, radiation, relative humidity, wind velocity, and leaf wetness. Slightly longer-term changes occur as weather systems pass. Seasonal climatic changes impose still a longer cycle. The physical and physiological characteristics of leaves change as they expand, mature, and senesce and as host phenology changes. Many of these factors influence the development of populations of P. syringae upon populations of leaves. P. syringae was first studied for its ability to cause disease on plants. However, disease causation is but one aspect of its life strategy. The bacterium can be found in association with healthy leaves, growing and surviving for many generations on the surfaces of leaves as an epiphyte. A number of genes and traits have been identified that contribute to the fitness of P. syringae in the phyllosphere. While still in their infancy, such research efforts demonstrate that the P. syringae-leaf ecosystem is a particularly attractive system with which to bridge the gap between what is known about the molecular biology of genes linked to pathogenicity and the ecology and epidemiology of associated diseases as they occur in natural settings, the field.
Plants cover a significant proportion of the global land area. Each plant produces many leaves. Each leaf, in turn, may be inhabited by a qualitatively and quantitatively diverse assemblage of microorganisms, including fungi, yeasts, bacteria, and bacteriophages (cf. 158, 229, 249). Colonization of leaves by microbes is not limited to terrestrial plants but includes aquatic plants as well (90). When you view a tree (or any plant), think of it as a support system for 1,000 leaves. Then think of each of these leaves as a habitat for 1 to 10 million bacteria. Then multiply this by the almost infinite number of individual trees and other plants present on our planet, and the diversity of habitats that these leaves provide leads to an appreciation of the enormity and importance of leaf-microbe ecosystems.
In 1961, J. Ruinen published a paper entitled “The Phyllosphere—an Ecologically Neglected Milieu” (249). Up to that time, interest in leaves and their microbial inhabitants was largely centered on diseased leaves and the microbes that caused the various foliar diseases. Phytopathogenic microbes and the economic impact of diseases on crop production continue to provide a major motivation for research on the phyllosphere as an ecosystem. Interest in leaves as habitats for microorganisms, not merely the causal agents of disease, has grown since the midcentury. Indeed, Ruinen's research focused on the possible colonization of leaves of plants in the tropics (Indonesia and Surinam) by Beijerinckia and other nitrogen-fixing bacteria. Since then, a relatively small but devoted band of scientists has worked diligently to explore the microbiology of leaves. At 5-year intervals, beginning in 1970, international symposia have been organized to provide a forum for discussions on phyllosphere microbiology (1970, Newcastle-upon Tyne, England; 1975, Leeds, England; 1980, Aberdeen, Scotland; 1985, Wageningen, The Netherlands, 1990, Madison, Wisconsin; 1995, Bandol, France; 2000, Berkeley, California). The proceedings of these conferences summarize the progress made in understanding the nature of leaf habitats, the microbes that colonize leaves, and interactions among microbes and between microbes and plants (8, 26, 60, 78, 209, 229). In this review, we focus on the bacterial component of leaf ecosystems.
Although the phyllosphere may no longer be a completely neglected milieu, it continues to be a source of wonderful biological questions and undoubtedly will continue to hold many interesting surprises for future investigators. Not only are these communities of bacteria interesting in and of themselves, they are connected directly through their habitats to the reduction of most of the carbon upon which nearly all other organisms depend. Some of these bacteria are known pathogens, destroying or diminishing the photosynthetic output of their leaf habitats. Others are thought to be beneficial. The nature of the interactions, if any, of the majority of leaf-inhabiting bacteria with the leaves that they inhabit remains unknown, an indication of how little is really known about these bacteria.
Before one can integrate the parts of a puzzle into a whole, one must first find the parts and decide which puzzle they belong to. Our interest is the interactions of populations of bacteria with populations of leaves under “real world” field conditions. Factors that affect these interactions include plant and bacterial genetics, weather and other aspects of the physical environment, and plant phenology. Estimates of sizes of such populations are influenced by sampling and bacteriological and statistical methods. The synthesis summarized at the end of the review includes pieces drawn from these several areas. Although the synthesis is an attempt to create a unified picture of the entire puzzle, it still lacks much more than it contains.
We begin this review with a brief general description of the leaf as a habitat for bacteria and what is known about the types of bacteria that colonize leaves. In the remainder of the review we focus on Pseudomonas syringae, a species that participates in phyllosphere bacterial communities as a pathogen, ice nucleus, and epiphyte. Among the diversity of microbes that colonize leaves, none has received wider attention than P. syringae, as it gained notoriety for being the first recombinant organism (Ice− P. syringae) to be deliberately introduced into the environment. Our intent is to use this bacterium to illustrate what has been and can be learned about plant-bacterium interactions by integrating knowledge from population studies in the field and molecular biology studies conducted in the laboratory.
SETTING THE STAGE
General Characteristics of Leaf Habitats
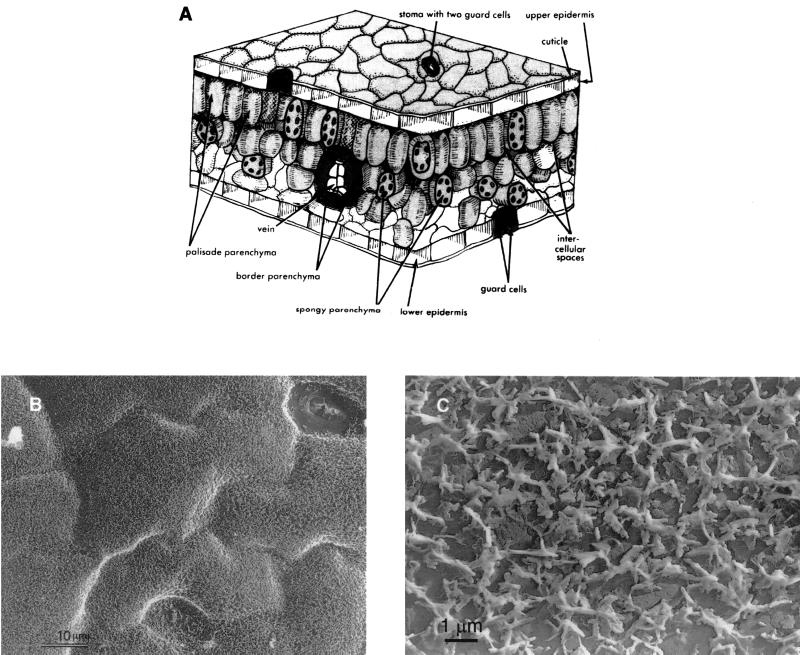
The first piece of the puzzle is the habitat itself. If we allow a rather gross anatomical simplification, leaves can be viewed as consisting of three general components—the epidermis, mesophyll, and veins or vascular bundles (Fig. 1A). The bacteria that are central to this review are found (or thought to be found—more about that later) on the surface of the epidermis and in the intercellular spaces (apoplast) of the mesophyll but not internal to plant cells. Bacteria pathogenic to leaves are thought to occupy the apoplast more frequently than commensal organisms, while both occupy the surface of the epidermis. Bacteria found within the vascular bundles are said to be endophytic and, regardless of their importance, are ignored in this review. Most of the research and interest in leaves as habitats for bacteria have focused on the nature of the leaf surface and its dwellers. This is reflected in terms such as phylloplane to refer specifically to the surface of leaves and epiphytes or epiphytic bacteria to set apart bacteria that are found on the leaf surface from those that are in the apoplast (Fig. 2). The term phyllosphere has been used to describe the environment of leaf surfaces. We interpret environment as including the zone below the surface as well (i.e., the entire leaf). Where appropriate, we use the term phyllosphere interchangeably with leaf associated to refer to bacterial populations in and on leaves.
FIG. 1.
(A) Three-dimensional schematic of a leaf. Phyllosphere bacteria colonize the surface of the epidermis and the intercellular spaces (apoplast). Reprinted from reference 299 with permission. (B) Low-magnification electron micrograph of the adaxial surface of a pea (Pisum sativum) leaf. Reprinted from reference 203 with permission. (C) Electron micrograph of a carbon replica of the adaxial surface of a corn (Zea mays) leaf. Note the waxy protuberances. Reprinted from reference 203 with permission.
FIG. 2.
(A) Scanning electron micrograph of bacteria on the surface of a corn (Z. mays) leaf. Photograph taken by J. Lindemann and M. Garment; reprinted from reference 312 with permission. (B) Imprint of a corn leaf. Segments of a field-grown corn leaf were gently pressed onto the surface of King's medium B (146), a nonselective medium.
Certain features of leaf surface topography set these habitats apart from others and evoke intriguing questions as to how bacteria have adapted to life at the interface between the leaf and the atmosphere. The cuticle layer coating the epidermis separates the leaf proper from the surrounding air and functions to protect and waterproof the plant surface (144). The layer, composed of cutin (mainly esterified hydroxy fatty acids) and waxes, forms a lipophilic barrier with low permeability. Dotting the surface landscape are stomates, trichomes, and other leaf surface appendages. All of these structures together with the surface waxes provide a topography that, scaled to the size of a bacterium, is far from smooth and uniform and may be imagined as a jumbled matrix of peaks, valleys, caves, and plains for bacterial colonization (Fig. 1B, 1C, and 2A).
In the past few decades there has been much interest in life in extreme environments, such as hydrothermal vents, acid seepage from mines, and similarly uninviting habitats. That bacteria colonize such environments is not at all surprising to us. After all, they flourish on leaf surfaces! This habitat as well has been frequently regarded as hostile to bacteria. Is the phyllosphere an extreme environment? Compared to a hot spring or hydrothermal vent, the 40 to 55°C sometimes found on leaf surfaces exposed to intense sunlight is relatively benign. Although desiccation of leaf surfaces may provide a stress for bacteria, that stress may be no greater than on exposed skin, very dry soil, or other such habitats. Similarly, the relatively cool 5 to 10°C temperatures and dilute (oligotrophic) substrate concentrations that may be typical of leaves that are wet with dew on a clear night may be relatively benign compared to the cold, dilute conditions of many streams or marine environments. Nor is the UV radiation any more intense on the surface of a leaf than in shallow, clear water. What is unique, or nearly so, to the leaf habitat is that a bacterium on a leaf may be exposed to all of these conditions during the course of a single day and is normally exposed to most of them every day. If the leaf surface is a harsh environment, then the property that makes it harsh is not the extremes to which it is exposed, but the frequent, repeated, rapid alteration between these very different conditions, any one of which may be considered stressful to at least some bacteria.
Thus, an important feature of leaf surfaces that sets the phyllosphere apart from most other types of microbial habitats is that epiphytic bacteria exist in a continuously fluctuating physical environment. Exposed to the atmosphere and the sun, leaf surfaces and, hence, their resident microbes are subjected to changes in aspects of microclimate such as temperature, relative humidity, wind speed, radiation, moisture, and others that occur on time scales of seconds to hours. As moisture levels change, for example, as might occur with the formation of dew during night and its subsequent evaporation during the day, so do substrate concentrations and osmolarity and other measures of water availability and activity. Intense rains may catastrophically perturb the system, as large numbers of bacteria (e.g., 105 CFU per bean leaflet in 15 min [171]) may be removed and deposited on the ground during rainfall.
Leaf habitats are intrinsically ephemeral in nature, surviving only a few weeks for many annual plants in the temperate zone to a few years for tropical or evergreen plants. For plants growing in temperate areas where climate changes are distinctively seasonal, leaves of annual plants emerge, develop, and senesce in a period of several weeks. During this period, the physical and biological nature of the habitat is continually changing as leaves expand, mature, and weather with age. With the onset of winter, these microbial habitats are destroyed. What happens to their resident bacterial communities?
The Players
Many pieces of the puzzle come in the forms of the kinds of bacteria that inhabit the phyllosphere. A diversity of bacterial species are known to colonize the phyllosphere (Fig. 2 and 3). The specific types and their relative abundances vary with a number of factors related to the plant (e.g., plant species, phenology, and age) and the environment in which the plants are grown (e.g., geographic area and weather conditions within a geographic area) (12, 13, 45, 59, 63, 68, 71, 86, 158, 159, 213, 248, 270, 271, 276, 285, 317). Additionally, what one finds depends on how one looks, and “looking” for phyllosphere bacteria has relied extensively on cultural methods. The extent to which reliance on cultural methods limits the study of bacterial populations and communities remains to be addressed for the phyllosphere (31). The only study to address this issue dealt with one species of bacteria on leaves of growth chamber-grown plants (308). Furthermore, the utility of various ribosomal DNA-based methods (reviewed in reference 6) for studies of bacterial communities in the phyllosphere remains unexplored.
FIG. 3.
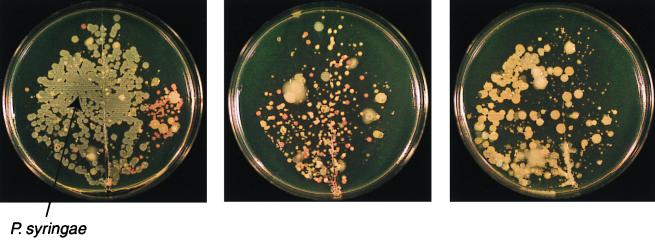
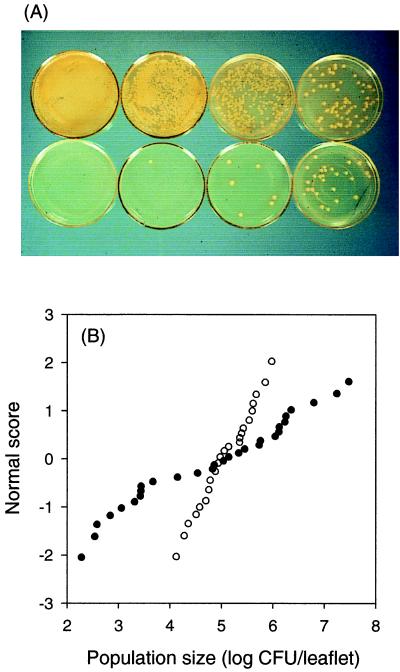
Qualitative variability in bacterial populations on individual leaflets of field-grown snap bean (Phaseolus vulgaris) plants. Leaflets, sampled on the same day, were gently pressed onto King's medium B (146).
One characteristic of communities of phyllosphere bacteria that has been frequently noted is the abundance (some have termed it preponderance) of pigmented forms (Fig. 3). Indeed, the abundance of chromogenic species common to the phyllosphere (e.g., Erwinia herbicola [renamed Pantoea agglomerans; however, we will refer to the bacterium by the more familiar name E. herbicola], xanthomonads, fluorescent pseudomonads, and pink-pigmented facultative methylotrophs) has led to the yet unresolved speculation that pigment production may confer a selective advantage to bacteria that colonize a habitat which is exposed to solar radiation daily (58, 182, 251, 272). At present there is some effort under way to determine if UV radiation affects the survival or fitness of bacteria in the phyllosphere (273). Once the importance of solar radiation to survival of bacteria adapted to the phyllosphere is better understood, the importance of pigmentation in this process may become better resolved.
Many phyllosphere bacteria are non-spore-forming, gram-negative or gram-positive heterotrophs (13, 68). Some of the bacterial species or individual strains that are well adapted to life on leaf surfaces may be distinct from those commonly found in the soil or rhizosphere (158, 270, 271).
Several studies have examined the distribution of specific groups or species of bacteria of interest across plant species (e.g., ice nucleation-active bacteria [189] and pink-pigmented facultative methylotrophs [45]). In general, such studies have found the same species or groups of bacteria present on a broad range of plants. The conclusion has been drawn that these groups are therefore well adapted to the phyllosphere. Note, however, that such studies do not separate the possibility that these species (or groups) are themselves highly variable and that very different individuals are each well adapted to a few plants from the possibility that most members of these groups are adapted to the phyllosphere of many plants.
There have been fewer attempts to determine bacterial community structure and diversity on a particular plant species (Table 1). Among these are bacteria from leaves of perennial rye (Lolium perenne) (59), olive (Olea europea) (68, 71), snap bean (Phaseolus vulgaris) (210), and sugar beet (Beta vulgaris) (276). Ercolani made two collections of bacteria from olive leaves, roughly 1,700 strains each, during each of two sampling periods spanning several years in two different decades. These collections, which included bacteria present on leaves of different age groups, were characterized phenetically (68–71) (Table 1). Differences in specific types were associated with leaf age and sampling time. Overall, the communities on olive leaves were dominated by strains of P. syringae (likely P. syringae pv. savastanoi, causal agent of olive knot disease). An interesting array of other gram-negative and gram-positive species were identified at lower frequencies. On leaves of perennial rye, the dominant components were identified as strains of Pseudomonas fluorescens and Xanthomonas campestris, with a number of other minor components (13). On bean, we have observed that the dominant species may change with plant age or with environmental conditions (118). Hence, while the bacterial species composition varies with plant species and other factors, the presence of a few dominant components with a number of species that occur at lower frequencies appears to be characteristic of the few phyllosphere bacterial communities characterized thus far. The distribution of bacterial species numbers among leaves thus fits a common pattern observed for other organisms (235).
TABLE 1.
| Plant and bacterial component or species | Relative abundancec (%) |
|---|---|
| Perennial rye | |
| Pseudomonas fluorescens | 20.12 |
| Xanthomonas campestris | 19.64 |
| “Coryneform bacteria” | 8.37 |
| Yellow chromogens | 4.83 |
| Flexibacter spp. | 4.66 |
| Listeria spp. | 4.02 |
| Pink chromogens | 3.86 |
| Staphylococcus saprophyticus | 1.77 |
| Other gram-negative rods | 1.61 |
| Klebsiella spp. | 0.96 |
| Acinetobacter spp. | 0.96 |
| Erwinia herbicola | 0.80 |
| Pseudomonas spp. | 0.64 |
| Staphylococcus spp. | 0.64 |
| Bacillus spp. | 0.32 |
| Micrococcus luteus | 0.32 |
| Orange chromogens | 0.32 |
| Unidentified isolates | 26.57 |
| Olive | |
| Pseudomonas syringae | 51.0 |
| Xanthomonas campestris | 6.7 |
| Erwinia herbicola | 6.0 |
| Acetobacter aceti | 4.7 |
| Gluconobacter oxydans | 4.3 |
| Pseudomonas fluorescens | 3.9 |
| Bacillus megaterium | 3.8 |
| Leuconostoc mesenteroides subsp. dextranicum | 3.1 |
| Lactobacillus plantarum | 2.8 |
| Curtobacterium plantarum | 2.2 |
| Micrococcus luteus | 2.2 |
| Arthrobacter globiformis | 1.4 |
| Klebsiella planticola | 1.2 |
| Streptococcus faecium | 1.2 |
| Clavibacter sp. | 0.98 |
| Micrococcus sp. | 0.82 |
| Serratia marcescens | 0.81 |
| Bacillus subtilis | 0.57 |
| Cellulomonas flavigena | 0.4 |
| Erwinia sp. | 0.37 |
| Zymomonas mobilis | 0.3 |
| Bacillus sp. | 0.29 |
| Alcaligenes faecalis | 0.27 |
| Erwinia carotovora | 0.08 |
| Pseudomonas aeruginosa | 0.04 |
What ecological roles do phyllosphere bacteria play? Do they have any effect on their habitat, or are they merely adapted to utilize resources otherwise “wasted” by the plant? What is their importance in the global scheme of things?
Bacteria that alter the leaf habitat in obvious ways.
We know most about those bacterial species that include strains that visibly modify their leaf habitats. Among these are some of the major groups of plant-pathogenic bacteria (140), pathovars of P. syringae and X. campestris and Erwinia spp., that to various degrees have been found as epiphytes on leaves of their susceptible host and nonhost plants (cf. 114, 161, 228). Bacterial ice nucleation alters the likelihood of frost injury at temperatures only slightly below 0°C. Some strains of several species that inhabit the phyllosphere are ice nucleation active (INA), including P. syringae, P. fluorescens, E. herbicola, and X. campestris pv. translucens (reviewed in references 121 and 175). Bacteria that produce plant hormones may also alter leaf and fruit habitats. For example, strains of E. herbicola that produce indoleacetic acid cause russetting of pear fruit (194). Habitat modification by pathogenic and ice nucleation-active P. syringae will be discussed further in subsequent sections.
Less is known about the ecological functions of the numerous other bacteria that may benefit their leaf habitats.
Nitrogen-fixing bacteria.
There are a number of reports on the isolation and identification of free-living nitrogen-fixing bacteria from the phyllosphere of various plants, most frequently from the tropics (2, 30, 139, 156, 214, 223, 248–252, 261, 262, 268, 287). The importance of these bacteria, their frequency or population sizes, and the overall role that they may play in the nitrogen cycle are difficult to assess. Most isolations have been by enrichment culture rather than direct plating, making quantitation difficult. Ruinen reported on the presence of large numbers of nitrogen-fixing bacteria on leaves of a range of plants grown in nitrogen-poor soils in Indonesia and Surinam (248–251). However, her quantitation was by direct microscopic observation of bacteria on leaf surfaces, a method that could not distinguish nitrogen-fixing bacteria from the many other genera present on leaves.
From the work of Jones (139), we do know that measurable amounts of 15N can be fixed on Douglas fir leaves in situ. Thus, there is no question that nitrogen fixation by foliar microorganisms does occur and may be of some importance for nitrogen cycling in nonfertilized systems. Identification of the actual microorganisms responsible for this nitrogen fixation is clouded, however, because Jones used enrichment culture to isolate such bacteria from his samples.
A large number of papers that report work done in India make it quite clear that nitrogen-fixing bacteria, most commonly Beijerinckia and Azotobacter spp., can be isolated from many crop plants (214, 222–224, 262–264, 287). Improved plant growth and/or yield of rice and wheat inoculated with strains of Azotobacter and Beijerinckia spp. has been reported. In addition, bacteria capable of nodulating aerial stems can be found as epiphytes. Adebayo et al. (2) found epiphytic populations of Azorhizobium caulinodans, a species that forms stem nodules on Sesbania rostrata, at densities of up to 5 × 105 cm−2 on leaves.
Thus, it is clear that nitrogen-fixing bacteria do occur on leaves, sometimes fairly frequently, and that measurable amounts of dinitrogen can be fixed in situ by at least one plant species, presumably because of the presence of nitrogen-fixing epiphytes. Many important issues such as the identities of the bacteria that are quantitatively important for the nitrogen fixation and the nature of the plant-bacterium interactions remain to be elucidated with regard to this potentially significant aspect of phyllosphere microbiology. Furthermore, there is sufficient data indicative of increases in yield and productivity in response to addition of nitrogen-fixing bacteria to the phyllosphere to suggest that important technological advances in agriculture may result from exploitation of nitrogen-fixing bacteria that inhabit the phyllosphere.
Almost nothing is known of the function in the phyllosphere of those bacteria for which there are no known detrimental or beneficial roles for their plant hosts.
Pink-pigmented facultative methylotrophs (PPFM).
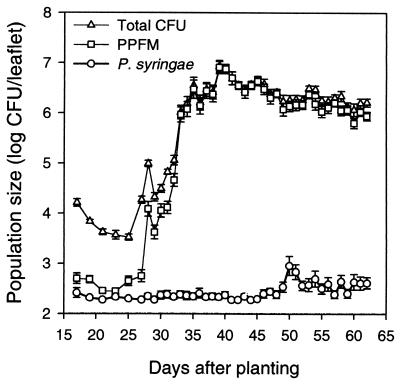
PPFMs in the genus Methylobacterium are a physiologically interesting group of bacteria that preferentially utilize substrates lacking carbon-carbon bonds (e.g., methanol and methyl amine) as sources of energy and carbon (92–94, 225). PPFMs (mainly Methylobacterium mesophilicum, formerly Pseudomonas mesophilica) have been found in large numbers from leaves of a diversity of plant species, including angiosperms (monocots and dicots), gymnosperms, and even lower plants (e.g., bryophytes and ferns) (12, 19, 43–45, 116, 118, 317). In the midwestern and northeastern United States, PPFMs may constitute a significant proportion of the bacterial community on leaves of field-grown plants, depending on plant species and a number of other factors. In other areas (or climates), these bacteria may be less abundant. PPFMs were not detected on leaves of olive plants in southern Italy (68, 71). Although they are not always present in large numbers, PPFMs frequently constituted up to 79% of the heterotrophic bacteria recovered from leaves of white clover (Trifolium repens) (45) or more than 90% of the bacteria culturable from snap bean (Phaseolus vulgaris) (118). We continue to be impressed with the consistent success (as reflected in their great abundance) of the PPFMs as leaf colonizers. Although they grow slowly in culture relative to, say, P. syringae or E. herbicola, by the time snap bean pods are ready for harvest PPFMs predictably establish large population sizes (>107 CFU per leaflet), regardless of weather conditions. (P. syringae, on the other hand, only does so under favorable weather conditions. More to be said about this in a later section.)
Corpe and Rheem (45) suggested that the success of the PPFMs may be due to their ability to uniquely utilize C1 compounds (such as methanol) which other, faster-growing heterotrophs are unable to metabolize. Methanol concentrations of leaves measured by Corpe and Rheem (45) and Nemecek-Marshall et al. (215) demonstrate the availability of this C1 compound on leaf surfaces. It has been suggested that methanol emission from leaves may be a major source of methanol found in the atmosphere (215). Hence, if PPFMs consume methanol as a source of carbon and energy, they may play an important ecological role in the carbon cycle in nature (see also reference 92). Whether this is the case remains to be tested experimentally.
There are several plausible reports that some sort of beneficial interaction with plants may exist for the PPFMs (19, 125, 126). Indeed, plant-PPFM associations were initially recognized when a PPFM was found to contaminate laboratory cultures of the bryophyte Scapania nemorosa (liverwort) (19). Because the PPFMs isolated from the liverwort stimulated growth and development of the liverwort and Streptocarpus prolixus (flowering plant) in tissue cultures, a positive commensal interaction was proposed (19, 44). That the effect may be due to production of vitamin B12 by the bacteria has been suggested by Basile et al. (18). The hypothesis is based on the associative findings that exogenous application of vitamin B12 to bryophyte cultures stimulated plant growth and a different strain of pink-pigmented methanol-utilizing bacterium produced vitamin B12 (277). Rigorous testing of the hypothesis and elimination of other alternatives remain to be accomplished. Effects of PPFMs on germinating soybean seeds have been suggested to be mediated by cytokinins or other plant growth substances (126). A possible connection between bacterial and plant nitrogen metabolism has also been suggested (125). Thus, findings from laboratory experiments have suggested interesting and unexpected interactions between PPFMs and plants. Additional research is needed to determine if such interactions occur in nature. At this time, what is eminently clear is that PPFMs are nearly ubiquitous on some kinds of plants in some climates. It is also likely that these bacteria may have some role in modifying their habitat (the plant) or the environment (e.g., via methanol degradation).
PSEUDOMONAS SYRINGAE
The Species: Pseudomonas syringae van Hall 1902
P. syringae was initially isolated from a diseased lilac (Syringa vulgaris L.) by M. W. Beijerinck in 1899 and subsequently characterized and named by C. J. J. van Hall (318). The species designation is thus linked to the diseased host where the bacterium was first found. Bacteria with similar characteristics have been isolated from diseased tissues of a very large number of other plant species. Although once numbering more than 40 species, all are now classed as the single species P. syringae (61). P. syringae is easily identified as a gram-negative strict aerobe in the γ subclass of the Proteobacteria which is rod-shaped, with polar flagella, with few exceptions produces fluorescent pigments, is oxidase and arginine dihydrolase negative (phenotypes that distinguish it from most of the other fluorescent pseudomonads), and does not rot potato (which distinguishes it from Pseudomonas viridiflava) (61).
The infrasubspecific epithet pathovar is used to distinguish among bacteria within the species that exhibit different pathogenic abilities (64, 319, 320). Nutritional, biochemical, physiological, and nucleic acid-based tests (e.g., DNA hybridization, restriction fragment length polymorphism, and repetitive DNA PCR-based genetic fingerprinting) have also been found useful in determining pathovars of P. syringae (49, 54–56, 66, 84, 104, 164, 198, 200, 244, 257, 259, 300, 318, 321). Findings from the various tests are generally in agreement with the groupings made on the basis of host range.
Strains within most of the pathovars exhibit rather narrow host ranges (29). The exception may be pathovar syringae, which includes the strain originally isolated from lilac (i.e., the type strain for the species). More than 80 plant species are listed as hosts for strains of P. syringae pv. syringae (29). From published data, it is not clear whether pv. syringae is a repository for strains that may in actuality have quite limited host ranges (36, 164, 198, 247, 253). A single plant species may serve as host for strains within two (or more) different pathovars, one of which is invariably pv. syringae. For example, P. syringae pv. tomato and P. syringae pv. syringae both cause disease on tomato, bacterial speck and fleck, respectively. Although the symptoms of the diseases are fairly similar (small necrotic lesions), strains within the pathovars are clearly different with respect to a number of phenotypes, including nutritional, biochemical, and serological parameters, phage sensitivity, DNA-based characteristics, and others (48, 54–56, 138).
Thus, the species P. syringae has generally evolved to interact with a wide range of plants in most regions of the world. However, within the species, there is a great deal of specialization with respect to plants with which individual strains are likely to interact. Bacteria that cause disease on a plant are usually much better colonists of that plant than those that do not. Further specialization is indicated by the observation that some strains differ with regard to the mechanisms of their interactions with a single plant, as evidenced by the fact that they cause different diseases of the same host.
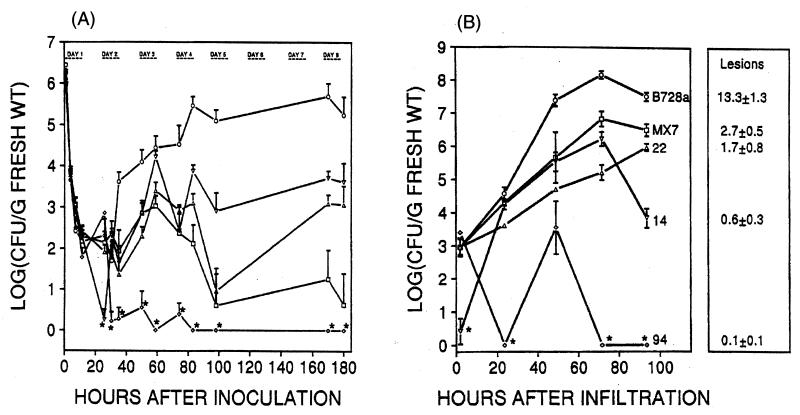
Must all strains of P. syringae be pathogenic? One finds what one looks for. Until the 1970s, plant pathologists were looking for pathogens in diseased tissue. Isolates unable to cause the symptoms of interest would have been discarded. Thus, all known strains of P. syringae were pathogens and pathogenicity was one criterion for belonging to the species P. syringae. When the association between frost injury to plants and ice-nucleating bacteria was discovered in the 1970s, searches began for bacteria that were active as ice nuclei (189). The bacteria most frequently found to be ice nucleation active, including the most efficient ice nuclei, had characteristics that placed them in the species P. syringae (Fig. 4). However, for some of these strains, plants could not be found on which they would cause disease. The question of whether it was possible that some strains of P. syringae might not cause disease at all became one of several important issues in the debate about the safety of deliberate environmental introduction of recombinant organisms. Among the many concerns surrounding the field tests of recombinant Ice− deletion mutants of P. syringae (the first deliberate release into the environment of a recombinant organism) was the pathogenic potential of these bacteria. However, the parent strains of P. syringae from which the mutants were derived were initially isolated from leaf surfaces of healthy potato (strain TLP2), citrus (Cit7) and strawberry (S203) (169, 177). Pathogenicity tests were conducted on 30 to 40 different plant species or cultivars, none of which could be identified as a susceptible host for either the parent or Ice− mutant strains. One could argue that plant species X was not tested. After all, is not P. syringae a plant pathogen? One could additionally argue that injection of plants grown in growth chambers or greenhouses may not always be a good indicator of the pathogenic potential of a strain, as plant reactions may not resemble those encountered in the field and hence be difficult to interpret. To add fuel to the fire, plant pathologists have long accepted the finding of Klement (153) that the rapid necrotic reaction that develops following inoculation of a nonhost (e.g., tobacco) with large doses of a strain of P. syringae, referred to as the hypersensitive reaction (HR), is indicative of its pathogenic potential. Hence, a strain that causes the HR on tobacco must be a pathogen of some other plant species. The parent Ice+ and mutant Ice− strains caused the HR when infiltrated into tobacco. On the other hand, if we were not so entrenched with the model that P. syringae is a plant pathogen and hence every strain must be pathogenic on some plant species, the results of Lindow (177) and Lindemann and Suslow (169) would not be so difficult to accept. Once again the dogma seemed in conflict with the data. Today many of us accept the concept that P. syringae has evolved to live in association with leaves, and incidentally, some (many?) strains cause lesions on some plants.
FIG. 4.
P. syringae—pathogen, ice nucleus, and epiphyte. (A) Foliar and (B) pod symptoms of bacterial brown spot disease of snap bean (P. vulgaris) caused by P. syringae pv. syringae. (C) An ice nucleation event occurred in the test tube on the right due to the large numbers of ice nucleation-active P. syringae present on the leaf. (D) Symptoms of frost injury to snap bean plants in the field. (E) Asymptomatic snap bean leaves—habitats for P. syringae. Figures B, C, and E are reprinted from reference 111 with the permission of the publisher.
P. SYRINGAE AND BACTERIAL ICE NUCLEATION
Discovery
Something associated with dead leaves—in one case derived from ground-up dry leaves, in the other from decaying leaf litter—became the common focal point for two groups, each pursuing completely different lines of investigation in disciplines as widely separated as atmospheric sciences and plant pathology (282). G. Vali, R. Schnell, and colleagues at the University of Wyoming searched for biogenic sources of ice nuclei that play a role in precipitation processes. S. E. Lindow, D. C. Arny, and C. D. Upper at the University of Wisconsin-Madison sought to understand how dried corn leaf powder affected the susceptibility of corn to frost injury. Their paths crossed with the isolation and identification of P. syringae from their respective leaf preparations and the discovery of its ability to nucleate supercooled water to form ice (9, 201). The story of the discovery of bacterial ice nucleation as narrated by Upper and Vali (282) is not only entertaining but informative and insightful in depicting the process of scientific inquiry—the delight in finding the unexpected; the frustration in overcoming accepted dogmas; and the skepticism toward accepting the unexpected. The discovery of biological ice nucleation opened new avenues of research ranging from such seemingly unrelated topics as winter survival of insects, snow making, and food processing (163). There are several excellent reviews on bacterial ice nucleation (175, 178, 294, 296, 297, 313, 314, 316). We include here only a few aspects deemed relevant to our overall goal of generating interest in the leaf ecosystem and its bacterial inhabitants.
Ice Nucleation-Active Bacteria, Genes, and Proteins
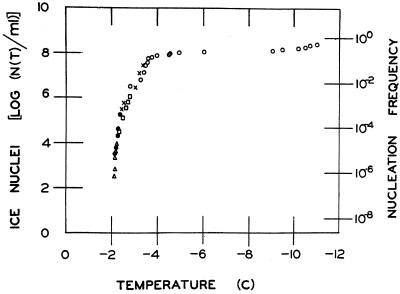
The ability of bacteria to nucleate supercooled water to form ice is uniquely limited to P. syringae and a handful of other bacterial species, many of which dwell in the phyllosphere. (Of course, ice-nucleating bacteria were first found associated with leaves, and this is where people have looked. Searches may not have been as extensive in other habitats.) Among these are strains of E. herbicola-like, Erwinia ananas, Erwinia uredovora, P. fluorescens, P. viridiflava, and X. campestris pv. translucens (9, 142, 145, 190, 201, 216, 217, 226). Not all strains within these species are ice nucleation active. Indeed, ice nucleation activity has been used as one (among several) trait to distinguish strains among some of the P. syringae pathovars (108, 138, 226). For example, strains within pv. syringae frequently exhibit the ice phenotype, while none of the strains tested thus far within pv. tomato or morsprunorum are ice nucleation active. Among strains that are active in ice nucleation, not every cell is active at a given time and temperature (Fig. 5) (9, 175, 195, 201). The fraction of cells within a population that are active as ice nuclei increases with decreasing temperatures below −1°C. That only a few bacterial species are ice nucleation active; that not all members within such species exhibit the phenotype; and the stochastic nature of the activity within strains that exhibit the phenotype have raised intriguing questions regarding the evolution and molecular basis for bacterial ice nucleation. Sequence analysis and biochemical characterization of ice genes and proteins have suggested some answers to these questions (292, 295, 314, 316).
FIG. 5.
Ice nucleation spectrum of a strain of P. syringae. Cells of P. syringae were suspended in phosphate buffer (0.1 M, pH 7.0) to ca. 108 CFU/ml. Tenfold serial dilutions were prepared from this suspension. Droplets from the original and each diluted suspension were placed on an aluminum block. The block was cooled, and the temperatures at which droplets froze were recorded. Each symbol represents determinations from one of the five suspensions. The concentration of ice nuclei N at temperature T [N(T)] in the suspension was calculated by the method of Vali (283). Nucleation frequency is the ratio of the number of ice nuclei to bacterial cell density.
Orser et al. (219, 220) were the first to demonstrate that the ice phenotype could be transferred to Escherichia coli by expression of DNA clones from the genome of P. syringae and E. herbicola. Since then, ice (or ina, for ice nucleation activity) genes have been cloned and sequenced from strains of P. syringae (96), P. fluorescens (291), E. herbicola (290), E. ananas (1), and X. campestris pv. translucens (325). In all cases, the genes contain a single open reading frame (ca. 3,600 to 4,000 bp) with a large central region consisting of a hierarchy of repeated motifs of 24, 48, and 144 nucleotides in length flanked by unique 5′ and 3′ sequences. The significance of the ordered internal repeats in the function of ice proteins as templates for ice formation has been noted (96, 291, 293). Results of a wide range of experimental and theoretical approaches suggest that ice proteins assemble to form aggregates of various sizes in association with the outer membrane of bacterial cells (32, 42, 52, 74, 91, 95, 141, 155, 196, 212, 267, 280, 290–294, 297, 298, 313–315). The number of protein monomers that are assembled in a proper conformation at any given time may account for the various temperatures at which individual cells are active in ice nucleation. The larger the aggregate, the more efficient the ice nucleus. For an excellent review on ice nucleation theory and measurement, see reference 283. For models of bacterial ice nuclei, see references 141 and 295.
It is curious that only a proportion of the strains within a bacterial species exhibit the ice phenotype. Edwards et al. (65) found a close correlation between the Ice+ phenotype and presence of an ice gene as detected by Southern hybridization using an 800-bp fragment from the internal repetitive region of the inaX gene from X. campestris pv. translucens. The probe hybridized to genomic digests from one of five E. herbicola strains, 9 of 26 P. syringae strains, one of two P. fluorescens strains, and 8 of 16 X. campestris strains. With the exception of two strains (one each of P. syringae and X. campestris), the strains that exhibited an Ice− phenotype did not hybridize to the probe. The two exceptions apparently contained the ice gene but did not express the Ice phenotype under the conditions tested. Thus, the gene was apparently missing, not just not expressed, in most strains that did not exhibit the Ice phenotype. Comparative analyses of the sequences of ice genes and proteins, including examination of codon usages, have led to the thought that ice genes present today diverged from a common ancestral gene. Wolber (316) suggested that the bacterial species known to contain members that are ice nucleation active acquired the gene via horizontal transfer, perhaps via conjugation between members of “epiphytic communities.” It might be interesting to compare the divergence of ice genes with those of other genes within ice-nucleating bacterial species.
Ice Nucleation-Active Bacteria and Frost Injury
Although we do not have clear answers as to what selective advantage(s) is conferred upon bacteria that express the Ice phenotype, it is clear that the presence of these bacteria on leaf surfaces can alter (i.e., destroy) leaf habitats at subzero temperatures (9, 174, 175, 188, 191, 195). Frost-sensitive plants are injured when ice forms within plant tissues. Without ice, such injury does not occur. In the absence of heterogeneous ice nuclei, water associated with leaves will supercool. Supercooling in the temperature range of 0 to roughly −5°C is primarily limited by the presence of INA bacteria. Below −5°C, other heterogeneous ice nuclei, including those produced by plants themselves, probably also limit supercooling. Thus, INA bacteria are responsible for ice formation, and hence injury to plants, mainly in the range from 0 to −5°C. Because most cells within a population of INA bacteria are not active at a given temperature and time, the larger the bacterial population, the greater the likelihood that one or more of the cells will be active at relatively warm temperatures (e.g., −2°C). Hence, the amount of frost injury sustained at a given temperature is a function of the population sizes of INA bacteria in the phyllosphere. Once the quantitative relationship between numbers of INA bacteria and frost injury was known, it was apparent that the hazard of frost injury to sensitive crops could be diminished by decreasing population sizes of INA bacteria (174–176).
S. E. Lindow and colleagues demonstrated that population sizes of INA bacteria could be reduced sufficiently to achieve measurable decreases in frost injury by application of naturally occurring and chemically induced Ice− strains of P. syringae, P. fluorescens, and E. herbicola (174, 176, 178, 184, 192, 193). Ice− strains were effective in preventing or minimizing colonization of plants by INA bacteria but not in eliminating established populations of the target microbes. Establishment of relatively large population sizes of the antagonists on leaf surfaces was required for effective exclusion of INA bacteria from leaf surfaces. Hence, manipulation of INA bacterial population sizes by application of non-INA strains is likely mediated by preemptive or competitive exclusion rather than displacement mechanisms (170, 178–180, 184). Competition for limiting resources on leaf surfaces (e.g., nutrients and space) was suggested as a possible mechanism by which established Ice− bacteria may prevent buildup of incoming Ice+ strains (309, 310). Because very similar strains of bacteria are involved in such exclusion, any density-dependent mechanism regulating population size is consistent with the available data. Thus, the term competition, in this context, appears to include a broad range of mechanisms, some of which may not normally be thought of as competition. Such a mechanism would imply that antagonist strains with ecological habitat requirements similar or identical to that of the target strains may be more effective in excluding the latter than antagonist strains that are not as closely related. This line of reasoning led to construction of recombinant Ice− strains of P. syringae and P. fluorescens by deleting a roughly 1- to 1.5-kb fragment (depending on construct) of the ice gene, followed by marker exchange of the mutated gene into the genome of the recipient strain (cf. 184, 296). These strains became the first recombinant microbes deliberately released into the environment (181, 184, 197, 296). With the exception of the Ice phenotype, the mutants were similar to their respective wild-type strains in all tests, including lack of pathogenic ability, relative ability to colonize and survive in association with plants in controlled environments, survival in soil, survival following freezing and thawing, and many other traits. Indeed, because of regulatory requirements for these first releases, no P. syringae strain has been as extensively and intensively characterized as the Ice− mutants and their respective wild-type strains (169, 181, 197).
After an arduous path through a maze of regulatory, political, and societal obstacles spanning more than 5 years, approval was granted to Lindow and researchers at DNA Plant Technology (formerly Advanced Genetic Sciences, Oakland, Calif.) to field test the Ice− mutants on potato and strawberry plants (181, 184). When the P. syringae Ice− mutants and their respective Ice+ parental strains were coinoculated onto potato plants in the field, population sizes of the parental strains were reduced over 300-fold (181, 184). Population sizes of naturally occurring (i.e., indigenous) Ice+ P. syringae were reduced about 50-fold on plants inoculated with the mutants compared to untreated controls. The larger reduction in population size of the Ice+ parental strains than of naturally occurring Ice+ P. syringae by the Ice− mutants is consistent with some degree of specificity in interactions among P. syringae strains on leaves. However, the efficacy of the Ice− mutants was similar to that found with some naturally occurring non-INA strains. The reduced population sizes of Ice+ bacteria treated with the Ice− strains correlated with reduced amounts of frost injury (ca. 70 to 80%) to potato plants during a natural radiative frost event.
None of the environmental disasters that were suggested prior to the deliberate release of recombinant Ice− bacteria have become reality. This is not surprising given that the experiments were soundly based on what was known about the ecology of P. syringae and phyllosphere bacterial population dynamics at the time. The monumental efforts of Lindow and scientists at DNA Plant Technology allowed other scientists, including us, to utilize the power of molecular tools to address fundamental ecological questions on interactions of bacteria with plants as they occur in the real world. In addition, a commercial preparation of a mixture of naturally occurring Ice− strains is now available for control of frost injury on some crops (184).
EPIPHYTIC POPULATIONS OF P. SYRINGAE
Discovery
For the first half of the century, P. syringae was isolated from diseased tissues of many plant species. Its role as a pathogen was firmly established. During this time, lesions were generally viewed as a primary source of inoculum for disease development. Bacteria were thought to move from lesions to susceptible tissue, grow in the intercellular spaces, and cause disease. Growth of the pathogen in other places was not considered important to the biology of the bacteria.
The “birth” of much of our current understanding of the epidemiology of foliar bacterial diseases and interest in leaves as habitats for bacteria can be traced to the seminal findings of J. E. Crosse (East Malling Research Station, U.K.) published in 1959 (46). Among several key findings was the revelation that populations of pathogenic P. syringae pv. morsprunorum, the causal agent of bacterial canker and leaf spot of stonefruit trees, could be recovered in large numbers from the surfaces of asymptomatic cherry leaves. Moreover, Crosse suggested that these surface or epiphytic populations of the pathogen and not populations in lesions on cherry leaves, as thought at the time, were the likely main source of inoculum for the infection of stems and branches leading to the development of canker infection. Crosse's pioneering work was subsequently generalized by C. Leben in the United States into a conceptual framework for the role of epiphytic populations in the epidemiology of diseases caused not only by pathovars of P. syringae but also by other phytopathogenic bacteria, most notably pathovars of X. campestris (161). The careful work of Crosse and advocacy of Leben gradually brought the possibility that these bacteria may not always reside in lesions to the consciousness of phytobacteriologists. Although the dogma that phytopathogenic bacteria are confined to lesions was gradually abandoned, the role of such bacteria growing in association with healthy tissues was not well understood. However, sufficient interest was generated in these so called epiphytic populations of phytopathogenic bacteria that much of the research in phytobacterial epidemiology during the 1960s and 1970s was centered on searching for and successfully finding pathogen populations on healthy leaves of host and nonhost plants (cf. 114). The role of P. syringae in the phyllosphere was extended to that of an epiphyte as well as pathogen. Because old ideas seldom die without a battle, however, there was substantial support for the idea that these bacteria resided in “latent lesions.” If the function of pathogens is to cause disease, they must be invading tissues and surviving or growing there until conditions that favor lesion development occur. Note the importance in the above argument of the assumption that the primary role of these bacteria is to cause disease. As we will describe below, we still do not fully understand the importance of the ability to cause disease to the overall fitness of P. syringae.
Quantitation of Epiphytic Bacterial Population Sizes
In his paper describing “epiphytic” populations of P. syringae pv. morsprunorum, Crosse described a method to quantitate leaf surface population sizes of the pathogen (46). Bacteria were removed from cherry leaves by vigorous shaking of leaf samples (2 min every half hour for up to 4 h) in sterile water (with or without a surfactant) followed by dilution plating of the washings. With some modifications, his basic leaf-washing method became a standard way of enumerating leaf surface bacterial population sizes. Others have since used leaf sonication to remove bacteria from leaves (reviewed in reference 136). Thus, measurement of epiphytic bacterial population sizes has been based on the ease with which culturable bacteria can be removed from leaves by either washing or sonication. While we conceptualize epiphytic bacteria as those that colonize the surfaces of leaves, what we actually measure are culturable bacteria that can be removed by methods such as leaf washing or sonication. The possibility that it may be more difficult to remove bacteria from some parts of the surface structures of leaves than from others and the possibility that either washing or sonication may remove some bacteria from the apoplast (i.e., intercellular spaces) demonstrate the practical difficulties encountered in enumerating “epiphytic” bacterial population sizes. Qualitatively, these bacteria clearly reside within and on leaves. This has been established microscopically. To this day, however, satisfactory methods have not been developed that allow facile quantitative separation of “epiphytic” from “apoplastic” populations of these bacteria. Thus, although it is likely that the bulk of bacteria in leaf washings or sonicates were on the “surface” (i.e., epiphytic), it is also possible that some bacteria in “protected sites” on the epidermal layer were not so readily removed or that some bacteria from the apoplast were. An alternative means to measure numbers of leaf-associated bacteria is to homogenize the leaf and plate from the homogenate. This method provides no information regarding where the bacteria were but does allow somewhat better recovery of bacteria from leaves. We have used homogenization as the method of choice for many years and refer to the bacterial populations enumerated as phyllosphere or leaf-associated bacteria.
Attempts have been made to estimate the proportion of bacteria that are internal, as opposed to on the surface, using various sterilants and UV irradiation of leaves (16, 143, 227, 258, 275, 307; unpublished data). In growth chamber assays, more than 99% of the bacteria (P. syringae and PPFMs) that were spray inoculated onto leaves immediately before treatment were killed when hydrogen peroxide was used as the sterilant (unpublished data). Almost none of the bacteria infiltrated into the apoplast before treatment were killed. Thus, this method appears to work quite well for highly artificial controls. When applied to bacteria that have grown in association with leaves in the growth chamber (307) or field (unpublished data), most of the P. syringae (75 to >90%) and >99% of the PPFMs were sensitive to the peroxide treatment. Results such as these are consistent with the view that the majority of P. syringae and virtually all of the PPFMs are “epiphytic” across a population of healthy leaves.
Enumeration of bacteria by cultural means is dependent on the assumption that the cells of interest (or at least a constant proportion thereof) are recoverable by plating. Wilson and Lindow (308) found that at least 25%, and usually more, of the viable P. syringae were recoverable by plating from leaves over a range of conditions in the growth chamber. Thus, estimates of bacterial population size based on culture methods should not be off by more than about a factor of 4. Although fourfold seems like a potentially large error, compared to the normal variability in the field (Fig. 6) and the five to six orders of magnitude across which such population sizes may fluctuate, it is of minor consequence. (The art of measuring population sizes of these bacteria from field samples is such that a factor of 4 is often close to the limit with which differences can be resolved.) Furthermore, the physical environment seemed to have the largest effect on the proportion of cells that were viable but not culturable. In field experiments, samples from all treatments (plots) are normally taken at the same time and hence under the same weather (physical) conditions. Thus, there is some reason to believe that differences in the proportion of viable but nonculturable cells across treatments should be small at any given sampling time. Nonetheless, the possibility that enumeration of phyllosphere bacterial population sizes from field experiments may be influenced by some variable proportion of cells that are viable but do not appear in culture remains an important caveat.
FIG. 6.
Quantitative variability in population sizes of P. syringae on individual leaves. (A) Each plate represents an equivalent dilution from washings of different individual rye leaves. (B) Lognormal distribution of population sizes of P. syringae on two sets of individual bean leaflets. The mean population size for both sets of leaflets is approximately 5.0 log CFU/leaflet. The population variances are 2.5 for set A (●) and 0.23 for set B (○). Reprinted from reference 111 with the permission of the publisher.
Crosse found great variability among population sizes of P. syringae pv. morsprunorum associated with individual leaves (46). Although he chose to combine large numbers of leaves that were washed and dilution plated to achieve some measure of the central tendency of population size, it has since been recognized that much of the information regarding interactions of bacteria with individual leaves resides in the variability about this central tendency (107, 109). Bacterial population sizes often follow (or, more correctly, can be modeled by) the lognormal distribution across populations of leaves (Fig. 6) (109). Less frequently, the related Weibull distribution provides a better “fit” to bacterial population data (134). These distributions are skewed. Estimation of means by bulking samples (several leaves combined to form a single sample) from such skewed distributions tends to overestimate the mean and underestimate the variability of epiphytic or leaf-associated bacterial population sizes. Variability in bacterial population sizes is present not only among individual leaves but also at other scales, including sites on and in a single leaf (149).
Relationship between Epiphytic Bacterial Population Sizes and Disease
Why are we so concerned about measuring population sizes of P. syringae in the phyllosphere? In his original paper, Crosse noted that larger numbers of P. syringae pv. morsprunorum were associated with a highly susceptible cultivar of cherry than with a more resistant one (46). Although it is generally accepted that epiphytic populations of P. syringae on asymptomatic leaves of susceptible and resistant host plants and on nonhost plants may serve as sources of inoculum for disease, there are few pathogen-host systems for which a quantitative relationship has been found between epiphytic population sizes and subsequent disease. The P. syringae pv. syringae-snap bean-bacterial brown spot disease is one such system (111, 112, 167, 246). The experimental approaches used by Lindemann et al. (167) and Rouse et al. (246) were similar in the sense that the relationship between epiphytic population sizes of P. syringae pv. syringae (measured by leaf washings) and amounts of brown spot disease were evaluated in what were essentially dose-response experiments conducted in the field. Different methods, however, were used to establish the necessary range of pathogen doses in experimental field plots. In both studies, pathogen population sizes on sets of individual bean leaflets collected at various times during the growing season were estimated, and information from the lognormal distribution of numbers of bacteria (population means and variances) was taken into account. Lindemann et al. (167) found that mean pathogen population sizes were not predictive of brown spot disease. However, the frequencies with which epiphytic population sizes of P. syringae pv. syringae were equal to or greater than approximately 104 CFU per leaflet on asymptomatic individual bean leaflets were predictive of disease a week later. The probit-lognormal model of Rouse et al. (246) generalized the threshold model by incorporating the notion that the probability of disease can be expressed in terms of the probability of a leaflet having a particular number of bacteria multiplied by the probability of disease occurring on that leaflet given that particular number of bacteria summed over populations of leaves. Fifty percent effective dose values of approximately 1 × 105 to 5 × 105 CFU per leaflet were obtained when the probit-lognormal model was applied to field data. Quantitative models relating bacterial population sizes on individual leaves have successfully predicted disease a short time later. Thus, a quantitative relationship was established between epiphytic population sizes of P. syringae pv. syringae and the amount of disease that followed. From this finding, in turn, comes the realization that the amount of disease will be determined by the development of large population sizes of the bacteria in association with leaves.
So we arrive at a rather large, paradoxical piece of the puzzle. P. syringae has two ways to destroy its own habitat, frost injury and lesion formation. To our knowledge, lesions may provide a place for the bacteria to survive during unfavorable weather conditions but little other advantage. Population sizes of the bacteria are highly variable in lesions but may remain quite large, even after several weeks of dry weather, when population sizes associated with asymptomatic leaves have declined markedly. Although several mechanisms have been proposed that may provide a selective advantage to the bacteria for causing frost injury, none has been demonstrated experimentally. Thus, there is no clear selection for destroying the leaf habitat. For most organisms, habitat destruction is regarded as highly unfavorable. Both of these events, lesion formation and frost injury, become highly likely only when the population sizes of the bacteria are relatively large, that is, when the bacteria have been particularly successful on or in their habitat. The bacteria reward themselves for success by destroying their habitat! Is this not paradoxical? To try to explain this phenomenon, we proposed a model in which lesion formation (and, we should add, frost injury) are unfortunate accidents of overpopulation that benefit neither the bacteria nor the plant (117). The real function of P. syringae is to live on healthy leaves. Perhaps it has means to communicate with some plants (we will call them hosts) in ways that improve the habitat for P. syringae in some way without causing (much) damage. Only when conditions become unusually favorable and population sizes of the bacteria become too large does the entire system crash, to the detriment of both host and bacteria.
When we first proposed this model a decade ago (117), we did so with tongue in cheek in an attempt to provoke discussion. To our amazement, nothing we have learned in the ensuing decade has provided strong evidence against this model. We reiterate it here to remind the reader that we tend to assign functions to organisms on the basis of those activities in which we are most interested, in this case causation of disease or frost injury. Perhaps we should look harder for those functions with real importance to the bacteria themselves.
Returning to our anthropomorphic interest, the realization that large bacterial population sizes are necessary for either disease or frost injury led to a search for those processes and factors that affect the relative abundance of P. syringae in the phyllosphere.
FACTORS THAT AFFECT RELATIVE ABUNDANCE
In this section, we explore what is known or what we think we know about the dynamics of population sizes of P. syringae across different time scales. In general, population sizes of P. syringae are frequently relatively small on young annual plants or emerging leaves during plant development and may increase to large numbers under suitable conditions (107, 174, 266). On perennials, population sizes of P. syringae are generally higher in spring than in summer (67, 68, 98, 174, 202, 243). The time scale over which such changes occur is remarkable. Increases occasionally occur in bursts lasting a day or more, with doubling times of a few hours (116). Infrequently, decreases can occur even more rapidly. For long periods, however, changes are relatively small (107).
At any given time, the number of P. syringae (and other bacterial components) present on a given leaf is the sum of immigration, emigration, growth, and death since the emergence of that leaf. Factors that affect the relative rates with which these population processes occur will, in turn, affect bacterial population sizes. For strains of P. syringae that are pathogenic and/or ice nucleation active, bacterial population sizes will, in turn, affect the probability of occurrence of disease or frost injury. The challenge is to determine the relative quantitative contributions of immigration, emigration, growth, and death to bacterial population sizes and identify those factors (biotic or abiotic) that influence these processes. This is a daunting task experimentally due to the complexity of interacting factors that operate in nature and difficulties associated with simultaneous measurements of rates at which each of the population processes occur in the field (cf. 147). However, some progress has been made, as described below.
Arrival (and Departure)
Bacteria may arrive on leaf surfaces in a number of ways (cf. 288). Many phytopathogenic bacteria, including several pathovars of P. syringae and X. campestris, may be seed borne (255). Indeed, production of (relatively) pathogen-free seed is an effective way to reduce the hazard from such diseases. Numerous other nonpathogenic phyllosphere bacteria, such as E. herbicola and the PPFMs, have been found associated with seeds (126, 286). P. syringae grows rapidly on preemergent seedlings and is available to colonize leaves as the plant emerges (80, 110). Presumably, other bacteria behave similarly. Thus, seed-borne pathovars of P. syringae are able to flourish in association with the plant below ground as well as above. Although the spermosphere is quite different from the rhizosphere, perhaps it is time to reexamine the issue of whether the separation between phyllosphere- and rhizosphere-associated bacteria is, indeed, as strong as it is thought to be. In field experiments, marked strains of P. syringae pv. syringae that were inoculated onto bean seeds or foliage were subsequently recovered from the rhizosphere of snap bean and wheat plants (119; unpublished data). The latter was planted as a fall-winter cover crop following a planting of snap beans that was inoculated with a marked strain of P. syringae (119). By growing in the spermosphere and in association with preemergent seedlings, P. syringae is able to make the transition from seed to leaves, its preferred habitat, and bridge the gap between generations of host plants. Although we have found P. syringae in the rhizosphere, we have no evidence that it is able to move from roots to aboveground plant parts. Emigration and immigration of bacteria from one aerial part of a plant to another (such as from buds to leaves) without physical departure of the microbes from plants is also expected but has been less critically demonstrated to occur in the field.
Leaf habitats are unusually open systems. Exposure to the atmosphere provides ample opportunity for aerial immigration and emigration of phyllosphere bacteria. Indeed, plant canopies were found to be a major source of bacteria in the troposphere, at least during the growing season in those parts of the temperate zone that support extensive plant growth (168). In a study on the effect of cropping patterns on population sizes of P. syringae pv. syringae, Lindemann et al. (166) found that numbers of the pathogen were larger on leaves of snap bean plants established in experimental plots within a major snap bean-growing area than in plots planted away from the commercial snap bean production area. The seed with which all of the plots were planted was naturally infested with the pathogen. Presumably, P. syringae that originated in the large commercial plantings of snap bean dispersed (probably aerially) to the experimental plots. These additional immigrants were sufficiently abundant to influence the population sizes in the experimental plots. Regardless of what the mechanism turns out to be, there is clearly a measurable effect of surrounding vegetation on population sizes of P. syringae (166; S. S. Hirano, J. A. Virata, K. A. Lindgren, and C. D. Upper, asbstr., Phytopathology 87:S42, 1997).
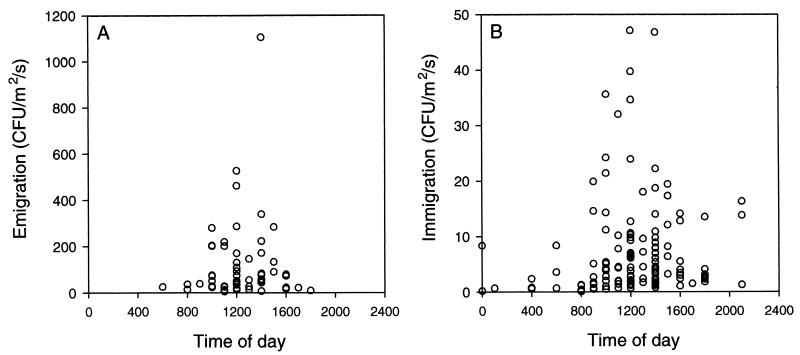
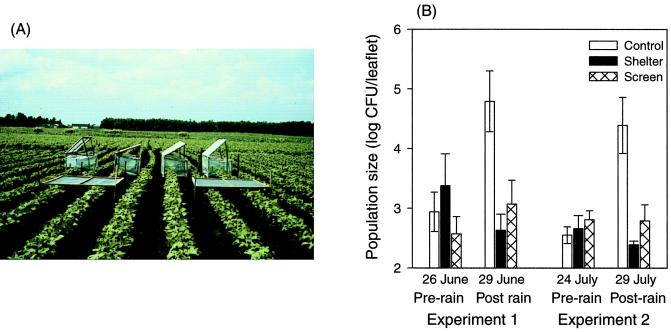
Lindemann and Upper (171) found that although viable bacteria were present in the air at all times of day, significant upward fluxes of bacteria (i.e., emigration) occurred only during the warmest part of sunny days, when leaves were dry and wind speeds exceeded about 1 m s−1. Emigration rates were highly variable and lognormally distributed, with a median value on the order of about 100 CFU m−2 s−1 (Fig. 7A). On average, this corresponds to a loss of roughly 103 CFU per h per bean leaflet with an area of 50 cm2. Immigration rates, as measured by the number of bacteria deposited onto the surface of solid medium in petri dishes exposed at canopy height, were also highly variable (Fig. 7B). Most of the time (67% of 130 measurements made over three growing seasons at various times during the day and night), deposition rates were between 1 and 10 CFU m−2 s−1. This corresponds to roughly 18 to 180 bacterial immigrants arriving per h on a bean leaflet 50 cm2 in area. From the estimates of Lindemann and Upper (171), we are left with the conclusion that at least during dry sunny weather, on average, there is a net loss of total culturable bacteria from bean canopies. The net loss was estimated to be over 104 CFU per bean leaflet per day during those periods when atmospheric conditions were conducive for dispersal.
FIG. 7.
Immigration and emigration of bacteria to snap bean canopies. (A) Emigration rates are based on measurements of concentrations of air-borne bacteria at canopy height and 1.5 m above the canopy using six-stage Andersen viable samplers (171). (B) Immigration was measured by the deposition of air-borne bacteria onto petri dishes filled to the rim with King's medium B (146). Measurements of immigration and emigration were made over canopies of snap beans during three growing seasons. Data from J. Lindemann; reprinted from reference 118 with the permission of the publisher.
On plants such as citrus where little growth of P. syringae occurs and population sizes of P. syringae are normally quite small, immigration appears to be an important contributor to the numbers of bacteria found on leaves (185, 187). Both bacterial population sizes and immigration rates, measured as rates of deposition on exposed petri dishes, were highest near the edge and decreased with distance away from the edge of a citrus grove (187). Epiphytic bacterial population sizes were only slightly larger than could be explained by total numbers of immigrants summed over several months. On this basis, it was suggested that there may have been no growth of bacteria on citrus leaves—that all of the bacteria on these leaves had grown on nearby plants, moved to the citrus leaves, and accumulated there by deposition (187).
The estimates of Lindemann and Upper (171) were for total culturable bacteria. Rates of immigration and emigration for specific bacterial components and species over snap bean canopies are expected to be smaller than those noted above. In field experiments in which numbers of P. syringae were estimated on a series of deposition plates exposed during a continuous 11- or 14-day period, the average number of P. syringae arriving on a bean leaf-sized area was about 35 CFU per day (S. S. Hirano, J. A. Virata, K. A. Lindgren, and C. D. Upper, abstr., Phytopathology 87:S42, 1997). Such numbers of immigrants are orders of magnitude too small to account for the population sizes of P. syringae frequently encountered on leaves of snap beans or other plants that carry relatively large numbers of bacteria in the field. So we are faced with another paradox. Immigration rates are too small to affect population sizes of P. syringae, yet strong circumstantial evidence points to the availability of immigrants as very important to relative population sizes of this bacterium on plants such as beans, where the bacteria grow to relatively large population sizes (166).
There is a plausible explanation for how small numbers of immigrants can have a relatively large effect on population sizes of P. syringae. The numbers of bacteria associated with very young leaves, as they first begin to expand, are very small. New leaves provide (nearly) empty habitats for bacterial colonization. If the immigration rate of P. syringae is low relative to that of other bacteria, then the probability that a given new leaf will receive even one immigrant of that species is very low. Immigrants of some other species will probably arrive first and begin the process of colonization of the leaf. Thus, by the time that a P. syringae immigrant arrives, its likelihood of success is smaller than if it had been the first bacterial cell to arrive on the leaf. When the numbers of P. syringae available to immigrate to a new leaf are large, then P. syringae is more likely to arrive early in the process of colonization of the leaf, when there is ample empty habitat. Hence, leaves subjected to large numbers of immigrants of P. syringae are likely to carry larger population sizes of that bacterium.
Immigration and emigration of phyllosphere bacteria also occur during rain. The classical explanation for the air-borne dissemination of foliar bacterial pathogens is that they are carried by the wind in aerosols or ballistic particles generated when raindrops strike diseased plant parts or plants supporting surface bacterial populations (33, 50, 72, 75–77, 288, 289). Thus, outbreaks or spread of bacterial foliar blights have been attributed to dissemination of the pathogen by rainsplash or windblown rain (50, 289) (more to be said about this later). Although bacteria may immigrate to leaves as they are dispersed by rainsplash, the contribution of these cells to bacterial population sizes is small. This is because during rain, the net flux of bacteria is downward (38, 118, 171). Most of the bacteria that are removed from leaves during rain move downward toward the soil. The large numbers of bacteria collected in rainwater below plant canopies suggest that emigration during rain could account for some of the large, rapid decreases in leaf surface bacterial populations associated with rain events (107, 111, 118, 171).
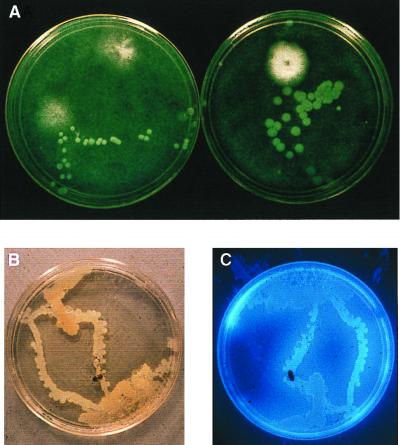
The role of insects in dissemination of phyllosphere bacteria is not totally clear. For some foliar pathogens, specific insect vectors are thought to be required for dispersal (cf. 288). From the studies of Lindemann et al. (171), we had concluded that in the absence of rain, dispersal occurred mainly during the day, when leaf surfaces are dry. Lindemann frequently observed insects visiting petri plates exposed to measure bacterial deposition. She marked such plates and examined them to determine any association between insect tracks and bacterial colonies. She found none. Due to the absence of bacterial colonies arising from the insect tracks, insects were not given much consideration as vectors of leaf surface bacteria. In more recent field experiments in which deposition plates were exposed “around the clock” for continuous periods of 11 to 14 days, larger numbers of bacteria were present on the plates exposed during the early morning hours, when leaves were wet with dew, than during midday (111; E. M. Groth, S. S. Hirano, L. S. Baker, and C. D. Upper, abstr., Phytopathology 85:1188, 1995; Hirano et al., abstr., 1997). The bacterial colonies that arose on the plates were not randomly distributed but rather followed “tracks” of faint footprints left by the insects (Fig. 8). It appears that when leaves are wet, insects traversing the surfaces become contaminated with epiphytic bacteria. Such bacteria, including P. syringae, emigrate from and immigrate to leaves as insects flit and fly about. The process appears to be independent of either insect species or bacterial species.
FIG. 8.
Insects as dispersal agents of P. syringae in the field. (A) Nonrandom distribution of P. syringae colonies on a semiselective medium (208) for P. syringae. The petri dishes, deployed at canopy height in a bean field, were exposed from 0600 to 0900 h, when leaves were wet with dew. (B and C) An insect (Glischrochilus quadrisignatus) was trapped in a sterile empty petri dish exposed in a bean canopy in the early morning. The trapped insect was transferred to a petri dish containing King's medium B (146) and allowed to walk over the surface of the medium. The white colonies in panel B are P. syringae. (C) The colonies fluoresced under UV light. Panel B is reprinted from reference 111 with the permission of the publisher.
To Grow or Not To Grow
The fate of a newly arrived P. syringae in its leaf habitat depends on a number of interacting factors related to the biological, physical, and chemical environments. A factor that plays a dominant role in the initial interaction is plant genotype. Plant species differ in the numbers of P. syringae and other bacteria that they normally support in the field. For example, plants such as potato and citrus generally harbor smaller numbers of bacteria than do plants such as almond and snap beans. Populations of bacteria on navel orange leaves appear to be largely accumulations of immigrants (187). It has been suggested that perhaps the thick waxy cuticle layer present on citrus leaves limits the diffusion of nutrients onto the leaf surface, thereby limiting bacterial growth (187). Alternative explanations, of course, remain to be ruled out. Large differences in population sizes of P. syringae are common even on leaves of plants of the same species. Breeding for disease-resistant plants has long been a practice to better manage plant diseases. In general, larger population sizes of pathogenic P. syringae are established on susceptible cultivars than on more resistant cultivars, even in the absence of disease (14, 47, 51, 206, 207, 269). Little work has addressed the mechanism(s) underlying the effect of host genotype on P. syringae population size despite the integral role that such growth plays within the larger question of the basis of disease resistance to P. syringae.
Some information regarding the overall role of climate on development of populations of P. syringae is available from a number of studies in which samples were taken weekly, monthly or even quarterly across one or more seasons (reviewed in references 115, 118, and 120). Generally speaking, net increases in bacterial population sizes were associated with relatively moist conditions (particularly rain and high relative humidity) and relatively cool temperatures. Declining or stable population sizes of P. syringae have similarly been associated with dry, hot weather conditions.
The physical conditions experienced by microorganisms on leaf habitats undergo extensive fluctuations each day. Thus, the more interesting question, one that may begin to explain differences between bacteria adapted to the phyllosphere and those better adapted to environments less likely to undergo extensive short-term fluctuations in the physical environment, is how does the weather affect the growth and death of P. syringae in the phyllosphere. Sets of individual leaves were sampled from field-grown snap bean plants either 5 or 7 days per week for 6 to 7 weeks (107, 111, 118). The bacterial population sizes that were measured were those of naturally occurring P. syringae (the plots were not artificially inoculated) on two cultivars that differ in susceptibility to bacterial brown spot disease. The data set that resulted included changes in bacterial population sizes from 11 plantings of snap beans spanning six different growing seasons. Examples are illustrated in Fig. 9. The effect of host genotype is illustrated by the generally larger population sizes of P. syringae on cultivar ‘Eagle’ than ‘Cascade’ (cv. ‘Eagle’ is more susceptible to brown spot disease than ‘Cascade’) (Fig. 9C). On a season-long basis, population sizes of P. syringae and the relative proportion of total culturable bacterial communities attributed to P. syringae may differ significantly from one planting to the next (Fig. 9C, D, and E). A comparison of daily changes in P. syringae population sizes and weather parameters across all plantings suggested a strong association between the occurrence of rain and the onset of periods of large increases in bacterial numbers. Estimates of immigration rates during either dry or rainy conditions were too small to account for the net increases in P. syringae population sizes. Hence, the net increases in numbers of P. syringae following rains were due to growth of the bacterium. In two of the plantings, weather conditions were dry and hot. Rainfall was minimal, and population sizes of P. syringae were generally below detection levels (Fig. 9E).
FIG. 9.
Population dynamics of P. syringae and total culturable bacteria in association with snap bean leaves. (A and B) Plants at about 20 and 60 days after planting, respectively. (C and D) Population sizes of P. syringae and total CFU on bean cultivars that differ in susceptibility to brown spot disease. Cultivar ‘Eagle’ is more susceptible than cv. ‘Cascade’. Reprinted from reference 105 with the permission of the publisher. (E) Bacterial population dynamics on cv. ‘Cascade’ during a growing season with little rain and high temperatures. (C, D, and E) At each sampling time, 30 individual leaflets were collected from the top of the canopy at 0800 h. Each sample was processed individually by dilution plating of leaf homogenates. The data are the means and standard error (SE) for each set of 30 leaflets. Variability in population sizes among the individual leaflets within a set is exemplified by the data shown in Fig. 6B.
Such results only create a positive association between rain and relative abundance of P. syringae, which confirmed what others had previously found with less intensive samplings. Although the link between rains and increased population sizes was still just an association, we had added a few more pieces to the puzzle. Increases in population sizes of P. syringae associated with rains were due to growth, not immigration. In rain events that led to such bursts of growth, peak rainfall rates met or exceeded approximately 1 mm min−1 (105). Population sizes of P. syringae were usually lower if measurements were made very soon after rain, presumably due to washoff. Rains of lesser intensity did not trigger growth of P. syringae. Population sizes of P. syringae tended to remain the same or decline in the absence of intense rains, even though the leaves were wet with dew. Thus, there was no association between leaf wetness per se and bursts of bacterial growth. Furthermore, neither the temperature during the wet period nor the duration of time that leaves remained wet appeared to be an obvious factor in triggering the onset of growth of P. syringae on bean leaves.
Inert fiberglass screens were placed over rows of bean plants to absorb the momentum of raindrops but allow the natural volume of water to fall on the leaves without altering its chemical composition or quality (105). In each of three such experiments, population sizes of P. syringae increased, as expected, on plants that were exposed to the rains but not on plants under the screens or on plants shielded from the rains by plastic shelters (Fig. 10). The latter treatment provided direct evidence for the positive effect of rain on growth of P. syringae. The screen treatment further suggested that the momentum of raindrops plays an important role in triggering the onset of growth of P. syringae on bean leaves. The positive effect of rain on the relative abundance of P. syringae does not appear to apply to the PPFMs. Population sizes of the PPFMs were larger on plants that were shielded completely or screened from natural rains. Indeed, for the two plantings of snap beans in which P. syringae population sizes were at the limit of detection of our plating assay (Fig. 9E), the PPFMs constituted a significant proportion of total culturable bacterial population sizes (Fig. 11). Although both bacterial components are very successful colonists of leaves of snap bean plants grown in Wisconsin, as measured by their relative abundance, they have apparently evolved very different life strategies to achieve success.
FIG. 10.
Effect of raindrop momentum on population sizes of P. syringae. (A) Modification of the microclimate with polyethylene shelters and inert fiberglass screens. The shelters were used to shield bean plants from rain when rain was imminent; the screens were used to decrease the momentum of raindrops as they passed through the screens and dripped onto the plants. (B) Phyllosphere population sizes of P. syringae on bean plants exposed to natural rains (control), plants shielded from rains (shelter), and plants under screens. Each datum point represents the mean log CFU per sample and SE based on three replicate plots per treatment with 10 individual samples per plot. Reprinted from reference 105 with the permission of the publisher.
FIG. 11.
Population dynamics of PPFMs, P. syringae, and total culturable bacteria in association with leaves of field-grown snap bean plants (cultivar Cascade). Under the relatively hot and dry weather conditions that prevailed during the experiment, the PPFMs flourished while P. syringae did not. Each datum point represents the mean and SE of bacterial population sizes based on 30 samples, collected at 0800 h each morning, and processed individually by dilution plating of leaf homogenates.
The mechanism by which intense rain triggers growth of P. syringae remains unknown. Because P. syringae grows quite well following misting onto plants in the growth chamber, we lack the negative controls necessary to examine the triggering phenomenon under controlled conditions. This is but one example of how the behavior of P. syringae observed in the field differs from that in the growth chamber (281). Caution needs to be exercised in extrapolating findings from controlled environments to the field situation.
Intense rains play a significant role in the relative abundance of P. syringae in the phyllosphere. Such rain is not always required for growth of P. syringae in the phyllosphere, however. The bacterium does quite well in some climates and during some seasons when such rains are highly unlikely. Although we suspect that a combination of leaf wetness and extended periods of relatively low temperatures are the likely triggers that replace intense rain in these situations, we have no direct evidence that this is the case. In addition, other parameters of the physical environment (temperature, relative humidity, etc.) and other factors such as community composition and abundance and interactions among all of the biotic and abiotic factors likely influence the numbers of P. syringae at any given time and on any given leaf (70, 71, 85, 135, 148, 210, 218, 309, 310). The system is dynamic and only slightly understood.
The possibility has been raised that availability of nutrients has an effect on numbers of epiphytic bacteria (210, 309, 310). Nutrients in the form of carbohydrates, amino acids, and organic acids have been identified in leachates of leaves (57, 85, 278, 279). However, a consideration of the permeability of the cuticle has brought into question the routes by which such compounds may arrive on leaf surfaces (260). At some level, nutrients must be important for these bacteria to colonize leaves. After all, they cannot grow without a source of carbon, nitrogen, phosphorus, other minerals, and energy. The issue is what role, if any, nutrients play in determining or regulating population sizes of P. syringae and related bacteria on leaves. Available evidence suggests that P. syringae population sizes can be increased by addition of nutrients to the phyllosphere, either by spraying nutrients onto leaves (210, 309) or by supplying nutrients that might not otherwise be there by use of transgenic plants (311). Changes in bacterial population sizes in response to addition of nutrients to leaves were relatively small compared to normal variations in sizes of their populations in the field (210, 309).
Although the following experiments were done with a strain of P. fluorescens (A506) and not P. syringae, the findings provide some insight on the role of nutrients in regulating epiphytic bacterial population sizes while simultaneously raising a number of unresolved questions. To test the hypothesis that the availability of sugars on leaves determines population sizes of epiphytic bacteria, Mercier and Lindow examined the relationship between the abundance of sugars on leaves of greenhouse-grown snap bean plants and population sizes of P. fluorescens A506 during and after colonization of the leaves (205). Amounts of sugars that could be washed from individual leaves were lognormally distributed, as were population sizes of bacteria recovered from a different set of individual leaves. The extent of variation was similar for the two parameters. Amounts of mono- and disaccharides decreased to about 28 to 37% following bacterial inoculation, compared to amounts present at the time of inoculation. The authors interpreted this decrease as evidence that the bacteria utilized these carbohydrates. These and other arguments were presented as strong evidence that carbohydrates limit population sizes of epiphytic bacteria. However, it is curious that when sugars and bacterial population sizes were followed as a function of time, carbohydrate depletion stopped at about the time that the bacteria entered log phase (ca. 18 h postinoculation), before about 80% of the bacterial growth had occurred. Thus, it appears likely that the bacteria were not utilizing carbohydrates during log phase, in which case it is unlikely that their growth would be limited by depletion of carbohydrates. Alternatively, efflux of carbohydrates to leaf surfaces may have occurred during log-phase growth of Pf A506. (However, this seems an unlikely scenario, because amounts of carbohydrates were depleted following inoculation with the bacteria.) For carbohydrates to limit growth in this case, efflux would have had to stop at the end of exponential growth. Mercier and Lindow did note that carbohydrates were not exhausted from the leaves but decreased to a steady-state level 25 to 40% of initial amounts. They took this as evidence that the bacteria and this residual carbohydrate must be spatially separated on the leaves. Although this is a plausible explanation, so is the alternative explanation that the bacteria were neither growing on nor limited by carbohydrates on the leaves during mid- to late log phase.
Thus, when all of the work relating nutrient levels to population sizes of P. syringae and other bacteria in the phyllosphere is considered together, we must conclude that the relative importance of even this most intuitively obvious of the factors regulating population sizes of bacteria associated with leaves remains obscure.
GENES AND TRAITS THAT CONTRIBUTE TO FITNESS
A question of universal biological interest is what genes and traits (phenotypes) enable an organism to flourish in its habitat. Rhetorically, why do microbes live where they do, be it in an environment considered extreme (e.g., hydrothermal vents) or as commonplace as leaves? Efforts to identify genes and traits that confer fitness on P. syringae in the phyllosphere can be largely grouped into two not necessarily mutually exclusive efforts. What genes and traits contribute to the fitness of P. syringae as an epiphyte, i.e., to life on leaf surfaces? For strains of P. syringae that are pathogenic, to what extent do genes required for pathogenicity or virulence contribute to bacterial fitness in the phyllosphere? The sum of what has been learned from both efforts thus far tells us little more than that this is a necessary direction to follow if we are to fully understand the interactions of P. syringae with leaf habitats.
Traits and Genes That Confer Epiphytic Fitness
For a comprehensive treatment of this topic, we refer our readers to the reviews of Lindow (182) and Beattie and Lindow (20, 23, 24). The search for traits that confer epiphytic fitness stems largely from the perspective that the surfaces of leaves provide a rather hostile or harsh place for microbial life. Traits that have been suggested as contributing to the fitness of epiphytic bacteria include pigment production to tolerate UV irradiation, extracellular polysaccharide production to withstand desiccation, and a number of others. In no case has the actual role of these traits in epiphytic fitness been rigorously tested (reviewed in reference 182).
To identify traits that confer epiphytic fitness, Lindow et al. (183) used random transposon mutagenesis of P. syringae followed by a screen for mutants with impaired epiphytic growth or survival. It was thought that such an approach may reveal novel (as well as anticipated) traits that contribute to epiphytic fitness. Mutants of wild-type P. syringae pv. syringae strain B728a (causal agent of bacterial brown spot of snap bean) with altered epiphytic fitness were identified by their diminished population sizes on leaves of snap bean plants subjected to alternating wet-dry conditions. A tube nucleation test, which measures the frequency with which individual leaves carry ice nuclei active at or above −2.5°C, was the basis for a rapid assay to screen for diminished bacterial population sizes relative to the wild type. This is possible because the probability of ice nucleation events occurring at relatively warm temperatures (e.g., −2.5°C) increases with increasing numbers of INA bacteria. Of 5,300 mutants screened in this way, 109 were found that caused a smaller proportion of leaves to freeze compared to the parent strain (183, 186). The number was reduced to 82 when 27 were subsequently found to be reduced in ice nucleation activity but not population size, as measured by dilution plating.
The 82 mutants varied in the extent to which population sizes increased or decreased when they were subjected to the wet-dry-wet regimen (186). Nine of the mutants appeared to be similar to the parent strain in the magnitude of the decrease in population sizes during the dry period but differed from B728a in net growth during the wet periods. Most of the mutants, however, were impaired in their ability to survive the “stressful” (dry) conditions compared to the parent strain. Among this set of stress-intolerant mutants, net growth was either less than, equal to, or greater than that of B728a during the wet periods. Hence, characterization of the population dynamics of the large number of mutants relative to the wild type demonstrated the complex nature of a phenotype summarily described as epiphytic fitness.
Among the 82 epiphytic fitness mutants, only three were auxotrophs (methionine, tryptophan, and isoleucine/valine). Thus, the frequency of auxotrophs among epiphytic fitness mutants was only about four times that expected of auxotrophs among all mutants. This might suggest that auxotrophy per se is not a major impediment to successful colonization of bean leaves under laboratory conditions. The behavior of two of the auxotrophs (Met and Trp) and two other mutants was examined further in laboratory and field experiments (7, 21, 22) (Fig. 12A). The four mutants were selected as those that exhibited the greatest differences in epiphytic population sizes relative to the wild type in laboratory assays. The Trp auxotroph consistently exhibited the poorest ability to survive and grow on bean leaves under all conditions examined. In one field experiment in which population sizes of the mutants were monitored for 8 days following inoculation, the behavior of the mutants differed relative to each other and to the wild type, with the differences being greatest about 4 days after inoculation (Fig. 12A).
FIG. 12.
(A) Population dynamics of P. syringae pv. syringae B728a (○) and Tn5 mutant derivatives 14 (▿) and 22 (▵), methionine auxotroph MX7 (□), and tryptophan auxotroph 94 (◊) on bean plants under field conditions in June. Each point represents the mean and SE of 12 leaf samples. Periods of daylight are indicated by dashed lines. Because means could not be estimated for data sets with 10 or more leaves harboring undetectable populations, means for these data sets (∗) were calculated using log (CFU per gram [fresh weight]) values of 0 for leaves with no detectable population. Reprinted from reference 22 with permission. (B) Population dynamics of P. syringae pv. syringae B728a (○) and Tn5 mutant derivatives 14 (▿) and 22 (▵), methionine auxotroph MX7 (□), and tryptophan auxotroph 94 (◊) after vacuum infiltration into the intercellular spaces of leaves. Each point represents the mean and SE of eight samples, each composed of two 6.5-mm leaf disks. The values indicating the number of lesions induced by each strain are the mean and SE of seven plants, with the lesion number per plant represented by the mean number of lesions enumerated in three randomly chosen 1.2-cm2 leaf regions. Reprinted from reference 21 with permission.
The Met auxotroph has been characterized at the genetic level. Cloning and sequencing of the affected DNA locus yielded two consecutive open reading frames (ORFs). One (metX) encodes a protein with homology to several homoserine O-acetyltransferases that function in methionine biosynthesis in several different organisms. No similarity was found between the sequence of the second ORF (metW) to sequences in the databases. Although Tn5 had inserted into metX, the first of the two ORFs, it appears that both genes are required for wild-type epiphytic fitness (7). Exogenous methionine was applied to leaf surfaces to determine whether the reduced fitness of the Met auxotroph could be due to limiting methionine concentrations. Although small increases in population sizes of the mutant and B728a were found on leaves maintained under moist conditions, application of methionine did not increase the survival of the strains under dry (stressful) conditions. Hence, methionine prototrophy appears to be required for optimum fitness of B728a, but the underlying mechanism is not known at this time.
Perhaps as informative as what was found is what was not found in the search for mutants affected in their abilities to survive and grow on leaf surfaces (186). All of the mutants were similar to the parent strain in their ability to utilize the 31 different organic compounds tested. Pigment production has long been hypothesized as a trait that enables epiphytic bacteria to tolerate UV irradiation on leaf surfaces. However, all mutants produced a fluorescent pyoverdine siderophore, similar to the wild type. Indeed, no evidence could be found for in situ production of fluorescent pigments by P. syringae on bean leaf surfaces, as assessed in growth chamber experiments (199). The behaviors of a few mutants would appear to support the role of previously hypothesized fitness traits. Four of the mutants were reduced in motility, while two were nonmotile, as assessed on a semisolid medium. Findings from previous research with chemically induced nonmotile mutants suggested that motility contributed to the epiphytic fitness of P. syringae as assessed in laboratory experiments (100). Some of the mutants exhibited reduced osmotolerance, as measured by their ability to grow in medium containing high NaCl concentrations. A few mutants appeared to be reduced in production of extracellular polysaccharide (EPS), as assessed by colony morphology, but none were apparently devoid of EPS production. Alginate, an EPS produced by P. syringae, has been implicated as an epiphytic and virulence factor based on measured reductions in population sizes and disease severity of an alginate-deficient mutant as assessed under laboratory conditions (323).
Epiphytic fitness is undoubtedly influenced and affected by a number of traits and genes. With mutants that exhibit pleiotropic phenotypes, it may be difficult to ascribe causality to a phenotype. Cloning and sequencing of the affected loci of the large pool of epiphytic fitness mutants obtained from the Tn5 mutant hunt may reveal the nature of the genes affected that appear to contribute to epiphytic fitness.
A final point to be made is that phenotypic characterization of the four mutants that exhibited the greatest reduction in epiphytic fitness relative to the wild type revealed that all the mutants were indeed reduced in their ability to survive the conditions deemed stressful. However, in laboratory assays, the mutants were also reduced to different degrees in net growth on leaves kept moist (i.e., presumably nonstressful) and in their ability to grow when infiltrated into leaves (Fig. 12B). Hence, the mutants would appear to be generally affected in their ability to grow and survive in and on leaves. One is thus left with the conclusion that they are not strictly “epiphytic” mutants. This conclusion does not diminish the significance of the findings but rather provides additional information as to the possible effects that specific genes and traits may play in the interactions of P. syringae on the surface and in the apoplast of leaves. From the perspective of P. syringae, the leaf surface may not be as distinct a place to live compared to the apoplast as we may think. That some (many) of the epiphytic fitness genes may turn out to be what we tend to think of as “ordinary” housekeeping genes should not be surprising. Surely both the function and regulation of such housekeeping genes should be tuned to operate optimally in the organism's favored habitat—leaves.
The ability to attach or adhere to surfaces is a trait that is often mentioned in regard to bacterial fitness in a number of habitats. Although attachment per se is difficult to define in the context of the highly variable leaf habitat, mutants of P. syringae lacking pili have been shown to be more readily washed from leaves in short-term laboratory experiments (239–242, 274). In field experiments, mutants of P. syringae pv. tomato lacking type IV pili achieved slightly lower population sizes on leaves of tomato plants than those of the parental wild-type strain DC3000 (236). Thus, the presence of pili appears to affect the fitness of P. syringae in the phyllosphere.
Genes Associated with Pathogenicity
The substantial efforts to elucidate the molecular mechanisms of pathogenicity and virulence in P. syringae and other phytopathogenic bacteria have led to the identification of a large number of genes that in some way affect the ability of the bacteria to cause disease (reviewed in reference 4). Much of what is known about the phenotypes attributed to these genes is based on laboratory assays in which the bacteria are infiltrated into leaves of plants. Such methods circumvent a significant portion of the natural interactions that occur between bacteria and plants in the field. Yet even in these assays, symptom development (or lesion formation) is often not the only phenotype affected in many of the mutants that have been identified. For example, in many cases mutants that no longer cause a pathogenic reaction are also impaired in their ability to grow to wild-type levels in such assays.
The pleiotropic phenotypes of nonpathogenic mutants identified thus far precludes an assessment of the contribution of disease causation per se to overall fitness of P. syringae. However, the contribution of specific genes (not phenotypes) to fitness can be examined. Furthermore, we may be able to ask to what extent impaired plant-associated growth might contribute to a mutant's impaired ability to cause disease. From the dose-response experiments of Lindemann et al. (167) and Rouse et al. (246), the probability of brown spot disease occurring on snap bean plants in the field is a function of population sizes of P. syringae pv. syringae. Hence, any gene(s) that affects pathogen population sizes will also necessarily affect disease. For a system such as the P. syringae-leaf ecosystem, there is a nearly limitless supply of test subjects (i.e., plants and leaves) that can be manipulated in a natural setting—the field. Hence, the system has a distinct experimental advantage over, say, animal or human pathogens, in attempts to bridge the gap between what is known about the molecular biology of pathogenicity-associated genes and the ecology and epidemiology of the pathogen of interest.
In this section, we focus on two groups of pathogenicity-associated genes for which there is some information on the field behavior of mutants bearing defects in genes representative of the groups. We discuss how information from experiments conducted in the field can be used to better understand the functions of these genes in plant-bacterium interactions while simultaneously gaining insight into their impact on the field fitness of a bacterium for which disease causation is but one aspect of its life strategy.
Genes in the hrp regulon.
hrp (hypersensitive reaction and pathogenicity) genes were originally isolated from P. syringae pv. phaseolicola, the causal agent of halo blight on bean (173). The salient phenotypes of hrp mutants include an inability to elicit the HR on nonhost plants and resistant cultivars of susceptible hosts, an inability to cause disease on host plants, and an inability to grow to wild-type levels when infiltrated into leaves in laboratory assays. The HR, which is manifested by rapid collapse and death of plant cells, is thought to arrest pathogen growth and spread, leading to an incompatible reaction (87). Although the HR is believed to be an important component of disease resistance, whether this is indeed the case remains unclear at this time (322). Note that inability to cause the HR on a nonhost such as tobacco does not render an hrp mutant of P. syringae pathogenic to the nonhost.
Since the initial report by Lindgren et al. (173), hrp genes have been isolated and characterized from strains representative of nearly all major genera of gram-negative plant-pathogenic bacteria (reviewed in references 4, 5, 28, 88, 101, 102, 132, 133, 172, and 305). Genes with similarity to hrp genes have recently been found in a strain of P. fluorescens (230). This is highly significant because this bacterium is not a plant pathogen. Indeed, the specific strain of P. fluorescens examined is a colonist of the rhizosphere, with apparent plant growth-promoting activity.
hrp genes in plant-pathogenic bacteria are present as clusters spanning roughly 25 kb (Fig. 13). Putative functions of the proteins encoded by hrp genes include regulation of transcription, components of a secretion pathway, and proteins that are secreted via this pathway (reviewed in references 4, 5, 89, 102, 132, 133, and 172). The finding that a subset of hrp genes has similarity to genes that encode components of the novel type III secretion system present in animal pathogens such as Yersinia, Shigella, and Salmonella spp. has generated much interest in this group of genes (5, 15, 83, 102, 211). In animal systems, virulence effector molecules are exported via the type III secretory pathway, and in some cases, these molecules may pass directly into the cytoplasm of the target host cells in response to pathogen-host cell contact (reviewed in references 40, 41, 82, 83, 131, 162, and 204). Nine hrp genes that bear sequence similarity to genes that encode components of this pathway have been redesignated hrc genes (for HR conserved) to reflect the conserved nature of these genes among plant and animal pathogens (27). Under growth conditions that induce hrp gene expression, P. syringae pv. tomato strain DC3000 produces pili (237, 238). That the major structural proteins of these pili is coded by hrpA has led to the speculation that Hrp pili may serve as the conduit for direct delivery of effector proteins through the plant cell wall and into the cytoplasm.
FIG. 13.
Organization of the hrp gene cluster in P. syringae pv. syringae strain 61. The gene designation employs the unified nomenclature for widely conserved hrp genes (hrc) (27). Arrowheads indicate the direction of transcription for each operon. Genes encoding proteins predicted to be associated with the inner or outer membrane of the type III secretion system are stippled and hatched, respectively, but HrcJ may be associated with both membranes. The cluster of hrp genes from hrpK to hrpR is similar in strains 61 (weakly virulent) and B728a (highly virulent). Flanking regions are not as highly conserved in the two strains. Reprinted from reference 35 with permission.
Most of the molecular research on hrp genes has focused on their function in elicitation of the HR. Little is known about the ways in which the Hrp secretion system and its associated effector molecules contribute to pathogenicity. Although it has long been clear that hrp mutants do not grow well in leaves of susceptible hosts (173), the possibility that this inability to grow might explain the requirement for hrp genes in pathogenicity has been largely ignored.
What can be learned about the role(s) of hrp genes and the type III secretion system in pathogen-host interactions by examining these interactions across many bacterial generations as they occur in the “real world” with its fluctuating, variable, and dynamic environment? To address this question, we field-tested three hrp mutants of P. syringae pv. syringae strain B728a (ΔhrpZ::nptII, ΔhrcC::nptII, and hrpJ::ΩSpc) (106) (Fig. 14). hrpZ encodes a protein that is secreted via the Hrp type III secretion system (103). HrcC is thought to function as an outer membrane component of the Hrp secretion system (35, 53, 129, 130), similar to YscC in Yersinia spp. (154). The hrpJ mutant carried a polar mutation in hrpJ, the first of five genes in the hrpJ operon. One or more of the genes in the operon appears to function as an inner membrane component(s) of the secretion system (35).
FIG. 14.
Population dynamics of P. syringae pv. syringae B728a hrp mutants (ΔhrpZ::nptII, hrpJ::ΩSpc, and ΔhrcC::nptII) in association with field-grown snap bean plants. The bacterial strains were inoculated onto seeds immediately before planting. Samples were seeds or germinating seedlings collected on or before days 7 (A) and 9 (B) after planting; primary leaves were collected between 9 and 14 (A) and 12 and 20 (B) days after planting; and single leaflets from trifoliolate leaves were collected at all other times. Each datum point represents the mean log CFU per sample and SE based on three (A) or four (B) replicate plots with six or eight individual samples per plot. Inverted solid triangles indicate rain events with sustained rates of >1 mm min−1. Reprinted from reference 106 with the permission of the publisher.
When inoculated onto bean seeds at the time of planting in the field, the hrp mutants behaved similarly to the wild-type B728a before plant emergence (Fig. 14) (106). All strains grew rapidly on germinating seeds and developing preemergent seedlings. However, phyllosphere population sizes of the hrp secretion mutants but not the hrpZ mutant were significantly reduced relative to that of B728a. The absence of substantial population increases during rainy periods favorable for growth of the wild-type strain suggested that the Hrp type III secretion system plays a critical role in enabling P. syringae to grow in the phyllosphere. Indeed, some hrp genes they may be considered fundamental phyllosphere growth (and possibly survival)-enabling genes that have a major impact on the field fitness of P. syringae. If the behavior of the secretion mutants is attributable primarily to defects in the Hrp type III secretion apparatus, then the requirement of the system for growth of P. syringae appears to be habitat and/or environment specific. The system is necessary for P. syringae to flourish in the phyllosphere but not in association with below-ground germinating seeds and developing preemergent seedlings.
The hrp secretion mutants caused brown spot lesions on primary bean leaves in the field (106). In the experiment shown in Fig. 14B, population sizes of the mutants and wild type were unusually large on below-ground plant parts. At the first sampling of primary leaves 12 days after planting, a few leaves harbored population sizes that were sufficiently large (>106 to 107 CFU per leaf) to be predictive of disease for strains capable of causing brown spot lesions. At this time, all leaves sampled were visibly asymptomatic. Two days later, brown spot lesions were detected in the plots (disease incidence of about 4% for the secretion mutants and 63% for the wild type). Characterization of bacteria isolated from the lesions confirmed that the hrcC and hrpJ mutants were the causal agents of the lesions detected in the field plots inoculated with these strains. The small amounts of disease caused by the secretion mutants are generally consistent with the dose-response experiments of Rouse et al. (246). The observation that hrp mutants are able to cause some disease in the field is also consistent with findings reported in the initial publication on hrp genes (174). Growth of various hrp mutants of P. syringae pv. phaseolicola as assessed in leaf infiltration assays was quantitatively different (174). The mutants grew to different extents relative to the wild type and in some cases caused correspondingly different amounts of disease (halo blight of bean), in some respects analogous to the behavior of the epiphytic fitness mutants (see Fig. 12B). Although this piece of the puzzle has been largely cast aside for over a decade, it is time that it be picked up once again and given careful reconsideration.
The role of the type III secretion systems in growth of a bacterial pathogen is not without precedent. Shea et al. (265) reported that the type III secretion system within Salmonella enterica serovar Typhimurium pathogenicity island 2 (SPI2) plays a role in growth of this pathogen. A mutation in ssaJ, a gene encoding a component of the SPI2 type III secretion system, affected growth but not survival of the pathogen in a mouse model system. How might the type III secretion system and its effector molecules affect growth of P. syringae in the phyllosphere? The answer is not yet within our grasp. However, a few pieces of information are worthy of mention. Atkinson and Baker (10, 11) previously reported that multiplication of P. syringae pv. syringae in the apoplast following infiltration of the bacterium into bean leaves was associated with the ability of the bacterium to induce a host plasma membrane K+/H+ exchange. According to their model, growth of the pathogen is due to leakage of sucrose from plant cells, which in turn is brought about by an increase in the pH of apoplast fluid (from about 5.5 to 7.5). Alkalization of the apoplast, as measured by monitoring the pH of solutions containing bean leaf disks inoculated with the pathogen, resulted in turn from the K+/H+ exchange. The impaired in planta growth ability of hrp mutants correlated with their inability to induce the ion exchange. Secretion of some proteins via the type III system in P. syringae (284), X. campestris (245), and serovar Typhimurium (25) has recently been reported to be pH dependent (i.e., stimulated at pH 5 to 6, depending on the system). Only time will tell whether these pieces of information will eventually fit into the puzzle of how the type III secretion system affects the growth of P. syringae in the phyllosphere.
Current mechanistic models for the function of hrp genes suggest that pathogenicity or virulence factors may traverse the Hrp type III pathway following intimate interactions of the pathogen with plant cells within the apoplast of leaves (4, 37, 211). However, to our knowledge, there is no case for which the effects of hrp genes on disease development have been demonstrated to be distinct from effects on growth in or on leaves. The possibility that the role of the type III secretion system in pathogenicity is mediated by its requirement for growth of P. syringae in the phyllosphere is a plausible alternative to current models. Furthermore, it appears likely that the Hrp secretion system is required for growth of P. syringae on leaf surfaces, not only in the apoplast. In its present form, the model contains elements that are difficult to reconcile with those early interactions between plant and bacterium which most likely occur at the plant surface rather than in the apoplast. Regardless of the underlying mechanism by which the Hrp type III secretion system affects interactions of P. syringae with leaves, the hrp regulon appears to play a fundamental and important role in the overall life strategy of the bacterium. Indeed, recent phylogenetic studies demonstrate that hrpL and hrpS (hrp genes with regulatory functions) appear to be as evolutionarily stable among pathovars of P. syringae as are gyrB and rpoD, two basic housekeeping genes (256).
If genes in the hrp regulon are not directly required for symptom development by P. syringae, what genes enable the bacterium to cause lesions?
Genes in the gacS regulon.
Willis et al. have identified a number of genes that are required for brown spot lesion formation in P. syringae pv. syringae strain B728a (127, 151, 152, 233, 234, 304, 306; D. K. Willis, J. J. Holmstadt, A. K. Savage, C. A. Hinckley, J. L. McEvoy, and T. G. Kinscherf, abstr., Phytopathology 89:S85, 1999). Unlike hrp genes, these genes are not required for elicitation of the HR. The first of these to be identified and extensively characterized is gacS (global activator sensor kinase). The gene was initially named lemA because of its requirement for lesion formation. However, with the finding that many other phenotypes are affected in gacS mutants, the gene was renamed to better reflect its requirement in P. syringae (151). GacS is similar to the transmembrane histidine kinase portion of a family of bacterial two-component regulatory systems (127). In these systems, the kinase is thought to serve as a sensor of environmental stimuli. The signal is then relayed by phosphorylation of the cytoplasmic response regulator, which in turn mediates changes in transcription (reviewed in references 124 and 221). The gacA gene has been identified genetically as encoding the cognate response regulator in P. syringae pv. syringae B728a (233, 306). The gacA gene was first described for P. fluorescens strain CHA0 as a global regulator of production of a number of secondary metabolites that contribute to the bacterium's biocontrol activity against a fungal root rot disease of tobacco (160).
Unlike hrp genes, the requirement for gacS in P. syringae appears to be pathovar specific (232). In P. syringae pv. syringae, the gene is required for lesion formation (brown spot disease of bean) and toxin production (syringomycin and syringopeptin). In P. syringae pv. coronafaciens, gacS is required for toxin production (tabtoxin) but not lesion formation (halo blight of oats) (17). In P. syringae pv. tomato (bacterial speck of tomato; the toxin is coronatine) and P. syringae pv. phaseolicola (halo blight of bean; the toxin is phaseolotoxin), mutations in gacS affect neither lesion formation nor toxin production (232). Homologs of the gacS and gacA genes have since been found in a number of other pseudomonads (e.g., P. fluorescens, P. viridiflava, and P. aeruginosa) (17, 34, 39, 81, 97, 160, 165, 231, 254, 301; S. T. Lam, T. D. Gaffney, R. A. Frazelle, K. Gates, J. Di Maio, N. Torkewitz, J. Ligon, S. Hill, S. Goodwin, and H. J. Kempf, abstr., Mol. Ecol. 3:620, 1994) and in Erwinia carotovora (73, 79), where they appear to affect bacterium-plant or bacterium-microbe interactions. In Salmonella serovar Typhimurium, the gacA homolog sirA has been shown to affect regulation of type III secretion systems present in this animal pathogen (3, 137). Similarly, the GacS and GacA counterparts BvgA and BvgS that control expression of virulence factors in Bordetella bronchiseptica appear to additionally regulate a type III secretion system in this animal pathogen (324). Thus, at least in these animal pathogens, a link between the equivalent hrc and gacA homologs has been made. Such a connection between the gacS/gacA and hrp regulons has been assumed not to occur in P. syringae because of the HR+ phenotype of gacA mutants. It will be interesting to learn if such a connection exists in P. syringae.
In addition to brown spot lesion formation and production of the toxins syringomycin and syringopeptin, gacS and gacA mutants of strain B728a are affected in production of extracellular proteases, acyl homoserine lactone, and alginate and in swarming behavior on soft agar medium (62, 128, 150, 151, 233; Willis et al., abstr., 1999) (Fig. 15). Unlike hrp mutants, gacS and gacA mutants are able to cause the HR in tobacco, and most significantly, they are fully able to grow as well as the wild type when infiltrated into leaves of bean plants (Fig. 16A). That gacS and gacA do not affect the in planta growth of P. syringae pv. syringae is unique among the many mutants of this pathogen that have been described as being nonpathogenic or reduced in virulence. Furthermore, that gacS and gacA mutants are indistinguishable from the wild type in their ability to multiply when infiltrated into leaves but are unable to cause disease demonstrated that bacterial growth in the apoplast is not sufficient for brown spot lesion formation (233, 304). The significance of this finding cannot be overemphasized.
FIG. 15.
gacS/gacA regulon in P. syringae pv. syringae B728a and phenotypes affected.
FIG. 16.
Population dynamics of a gacS mutant of P. syringae pv. syringae B728a. (A) Bacterial strains (106 CFU/ml) were infiltrated into primary leaves of growth chamber-grown bean plants. At each sampling time, leaf disks (6-mm diameter) were removed with a cork borer from each of five leaves. Each datum point represents the mean log CFU per sample and SE (n = 5). Unlike the hrpJ mutant, growth of the gacS mutant was indistinguishable from that of the wild type. (B) Population dynamics of a gacS mutant under field conditions. The bacterial strains were inoculated onto the foliage of 25-day-old snap bean plants. Population sizes were estimated by dilution plating of leaf homogenates. Each datum point represents the mean log CFU per sample and SE based on three replicate plots with eight individual leaf samples per plot. Reprinted from reference 110 with the permission of the publisher.
A number of genes that are regulated by GacS and GacA have been identified (151; Willis et al., abstr., 1999). Among these is salA (syringomycin and lesion formation) (151). Sequence analysis of salA suggests that it encodes a DNA-binding protein with little similarity to other proteins in the current databases. In laboratory assays, a salA mutant is impaired only in lesion formation and production of syringomycin. A salA mutant is similar to the wild type in production of extracellular proteases, acyl homoserine lactone, and alginate (151). Kitten et al. (151) have demonstrated that SalA positively regulates expression reporters inserted into the syringomycin biosynthesis gene syrB. A model for the gacS/gacA regulon places gacS/gacA at the top of a regulatory hierarchy in B728a. salA is regulated by GacS and GacA and in turn plays a regulatory role in activating the expression of a subset of genes in the regulon, including genes necessary for lesion formation and syringomycin production (Fig. 15).
In field experiments, population sizes of a gacS mutant were significantly reduced relative to B728a in association with bean leaves (Fig. 16B), but not with germinating seeds, similar to the hrp secretion mutants (110). Because a gacS mutant is able to grow when infiltrated into bean leaves in laboratory assays, it is tempting to speculate that a subset of genes regulated by gacS may be affecting the epiphytic fitness of B728a under field conditions. In this regard, it is interesting to note that gacS plays a role in regulating production of the exopolysaccharide alginate in P. syringae pv. syringae (Willis et al., abstr., 1999) and P. viridiflava (165). Alginate has been reported to contribute to the virulence and epiphytic fitness of P. syringae pv. syringae strain 3525, as measured in growth chamber experiments (323). The fitness of alginate-deficient mutants under field conditions remains to be examined.
gacS, salA, and syrB function at different tiers in the gacS regulon. GacS regulates expression of salA. SalA, in turn, regulates expression of the syringomycin biosynthesis gene syrB. In a field experiment in which the fitness of the mutants was compared to that of wild-type B728a and to each other, phyllosphere population sizes of the gacS mutant (gacS::Tn5) averaged roughly 125-fold less than wild-type population sizes (range, 7- to 1,250-fold over time from plant emergence to pod set), while population sizes of the salA mutant averaged only about 5- to 6-fold less than wild-type population sizes (range, 2- to 20-fold) (S. S. Hirano, D. K. Willis, and C. D. Upper, abstr., Phytopathology 89:S34, 1999). Hence, the overall difference in population sizes was greater between the salA and gacS mutants than between the salA mutant and the wild type. The data suggest that the subset of genes in the gacS regulon that are not regulated by salA play an important role in growth and survival of P. syringae pv. syringae in association with leaves in the field, in some respects similar to hrp genes. The salA mutant established population sizes that were sufficiently large to be predictive of disease but did not cause the expected amount of disease in the field (Hirano et al., abstr., 1999). Hence, the contribution of lesion formation to population sizes can be no more than the difference between the salA mutant and the wild type (average of 5- to 6-fold and range of 2- to 20-fold). Population sizes of the syrB mutant were roughly two- to threefold less than that of the wild type. The syrB mutant caused brown spot disease in the field; disease incidence was slightly reduced relative to the wild type. Hence, approximately half of the reduction in populations of the salA mutant relative to the wild type may be due to its inability to produce syringomycin. However, loss of syringomycin production did not explain the impaired ability of a salA mutant to cause brown spot lesions. Pursuit of the genes that are regulated by salA and may play a direct role in brown spot lesion is ongoing in the laboratory of D. K. Willis and colleagues.
The experiments described above have shown us that genes in the hrp and gacS regulons (and likely many others) play major roles in interactions of P. syringae with plants in the field. Characterization of these genes at the molecular level has progressed substantially in recent years, largely as two distinct regulons, each said to be required for pathogenicity in P. syringae. Both regulons affect the ability of the pathogen to grow in and on leaves in the field, an activity essential for disease epidemics to occur. Genes in the gacS regulon, in particular genes regulated by SalA, a key branch point in the regulon, appear to be directly required for lesion formation in the P. syringae pv. syringae-snap bean-brown spot system. It is useful to consider the function of genes in both regulons (and others as they are identified) in the integrative context of the interactions of P. syringae with plants in the field.
The field setting has provided useful information and new insight on the role(s) that pathogenicity-associated genes play in interactions of P. syringae and its susceptible host, bean plants. However, there remains an important caveat. Until a plant-associated phenotype (e.g., decreased population sizes) exhibited by a mutant can be restored to wild-type levels, we must be cautious in attributing the altered phenotype to the gene specifically mutated. The duration of our field experiments renders restoration in trans (i.e., wild-type copy on plasmid in mutant background) problematic due to possible plasmid instability over the many hundreds of bacterial generations that occur during these experiments. We have attempted to address this problem in several ways. For example, the experiments with the hrp secretion mutants involved two constructs in which the type III system was similarly affected, although different genes required for a functional system were mutated. We can more rigorously test the effects of mutations in gacS and other genes in the regulon by examining the behavior of constructs in which a specific gene has been mutated in different ways, thus demonstrating the reproducibility of observed phenotypes.
CONCLUDING REMARKS AND FUTURE DIRECTIONS
We have provided a brief tour of many, but surely not all, of the multitude of facets of the ecology of bacteria in the phyllosphere. Throughout we have used P. syringae as a model to provide examples of what is known of the various processes that influence the behaviors of bacteria associated with leaves. It is not yet clear whether the behavior of P. syringae in association with snap bean leaves is a model for plant-bacterium interactions or just a case study (120). This issue will only be resolved when the ecology of other bacterial species in the phyllosphere is understood as well as is the P. syringae-bean leaf system.
The diagram in Fig. 17 illustrates a number of the factors affecting the ecology of P. syringae on snap bean leaves, as discussed earlier in this review. These are pieces that are fairly well understood and that would seem to us to belong where they have been placed in this puzzle. Two points are not adequately made by Fig. 17, however. First, many pieces of the puzzle have been omitted because of our ignorance. Even if the P. syringae-bean leaf system is the best understood of phyllosphere-bacterium interactions at this time, there is undoubtedly far more to learn than we already know of how this puzzle fits together. Second, the model displays only two dimensions, whereas interactions in the field are more accurately represented in multidimensional space. By the time that plants are fully grown, there will be as many as 7 × 107 leaf habitats on each hectare planted to commercial snap beans. Each of the physical and biotic factors affecting bacterial population dynamics will vary across the field and within the bean canopy. Thus, the stochastic nature of the development of populations of bacteria on populations of leaves is not represented in the figure. Furthermore, regional effects on these processes are not represented. Bacteria emigrating from and immigrating to a particular field reflect what is happening in other fields. Those nearby will have a greater effect than more distant fields, yet an overall regional influence exists.
FIG. 17.
Interactions of P. syringae and leaf habitats—putting the puzzle together. An immigrant arrives on a leaf, either from growth on a germinating seed (bottom) or through the air. Conducive conditions such as a susceptible plant, intense rain, and early vegetative stage of plant development favor growth and establishment of large population sizes of P. syringae. Dry, hot conditions or an unfavorable plant will diminish growth and drive bacteria toward survival rather than growth. Some bacterial genes in the gac and hrp regulons and several housekeeping genes are necessary for sufficient growth to achieve large bacterial population sizes. Genes necessary to produce type IV pili may also enhance bacterial growth and survival. The larger the population size, the more bacteria are available to emigrate, either on insects when leaves are wet or as aerosols in dry sunny weather, or to be washed off by rain. As population sizes approach 106 cells per leaflet, disease becomes likely. Genes in the gac regulon (gacS/gacA and salA) are required for lesion formation. Lesions may provide a site for enhanced survival of P. syringae during unfavorable weather. These processes are repeated throughout the growing season on each of the 106 or more leaf habitats per hectare, with bacterial generation times on the order of 2 to 5 h. If the plants are allowed to mature, bacteria present on pods may move to seeds, where the bacteria can survive until the seeds are planted to begin the next plant generation. ↑ and ↓, positive and negative effects, respectively.
As illustrated in Fig. 17, lesion formation is only one of the many activities of P. syringae in association with bean leaves. Based on comparison of behaviors of various mutants in the field, the competitive advantage that lesion formation affords this bacterium is not as great as expected and may turn out to be quite small. The effects of bacterial genes required for growth in the phyllosphere and of physical and other biotic factors on the relative success of this bacterium are far greater than the effect of not being able to form a lesion. Thus, if we wish to understand the mechanisms associated with successful disease causation in the field, it is far more important to understand the processes that lead to development of large population sizes of this bacterium than to understand the mechanism by which lesions are formed per se.
We hope that we have left the reader with a clear idea of the importance of bacteria in the phyllosphere and the need to learn more about these fascinating, complex, highly interactive systems. There is so much yet to learn that there is ample room for greatly expanded efforts on these interesting problems from disciplines as diverse as genetics, ecology, and meteorology.
The powerful tools of molecular biology have greatly advanced our understanding of plant-microbe interactions at the molecular level. Such studies, of course, are no stronger than the limitations provided by the biological assays upon which they depend. Regardless of the sophistication of the molecular mechanisms proposed, they are of little value until a relationship is tightly established between the mechanisms and the phenomena that they are thought to explain. At the moment many proposed mechanisms are available for bacterium-plant interactions in the phyllosphere, but few, if any, have been clearly shown to explain (or even be related to) the phenomena which they are supposed to explicate. For examples, the role of pigmentation in fitness of phyllosphere bacteria, the roles of specific genes thought to affect plant-bacterium interactions, the role of nutrients or other mechanisms for regulating bacterial population sizes associated with leaves, the importance of immigration in a growth-driven system, and many other issues as discussed above clearly deserve careful examination. We believe it is time to devote greater effort to the biological side of these systems, to do the definitive experiments that will “close the loop” and create the necessary links between mechanisms and phenomena. Unfortunately, at least some of this work will necessarily have to be done in that most difficult of all laboratories—the field.
ACKNOWLEDGMENTS
We acknowledge the Agricultural Research Service of the U.S. Department of Agriculture and the USDA CSREES National Research Initiative Competitive Grants Program for their support of our research efforts during the past decades.
We thank M. Barbour, G. Beattie, A. Charkowski, B. Juniper, and J. Lindemann for kindly allowing use of their photographs or figures from previously published materials. Special thanks are extended to B. Juniper (Oxford University) for locating original electron micrographs of leaf surfaces.
REFERENCES
- 1.Abe K, Watabe S, Emori Y, Watanabe M, Arai S. An ice nucleation active gene of Erwinia ananas: sequence similarity to those of Pseudomonas species and regions required for ice nucleation activity. FEBS Lett. 1989;258:297–300. doi: 10.1016/0014-5793(89)81678-3. [DOI] [PubMed] [Google Scholar]
- 2.Adebayo A, Watanabe I, Ladha J K. Epiphytic occurrence of Azorhizobium caulinodans and other rhizobia on host and nonhost legumes. Appl Environ Microbiol. 1989;55:2407–2409. doi: 10.1128/aem.55.9.2407-2409.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmer B M M, van Reeuwijk J, Watson P R, Wallis T S, Heffron F. Salmonella SirA is a global regulator of genes mediating enteropathogenesis. Mol Microbiol. 1999;31:971–982. doi: 10.1046/j.1365-2958.1999.01244.x. [DOI] [PubMed] [Google Scholar]
- 4.Alfano J R, Collmer A. Bacterial pathogens in plants: life up against the wall. Plant Cell. 1996;8:1683–1698. doi: 10.1105/tpc.8.10.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alfano J R, Collmer A. The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J Bacteriol. 1997;179:5655–5662. doi: 10.1128/jb.179.18.5655-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersen G L, Beattie G A, Lindow S E. Molecular characterization and sequence of a methionine biosynthetic locus from Pseudomonas syringae. J Bacteriol. 1998;180:4497–4507. doi: 10.1128/jb.180.17.4497-4507.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrews J H, Hirano S S, editors. Microbial ecology of leaves. New York, N.Y: Springer-Verlag; 1991. [Google Scholar]
- 9.Arny D C, Lindow S E, Upper C D. Frost sensitivity of Zea mays increased by application of Pseudomonas syringae. Nature. 1976;262:282–284. [Google Scholar]
- 10.Atkinson M M, Baker C J. Association of host plasma membrane K+/H+ exchange with multiplication of Pseudomonas syringae pv. syringae in Phaseolus vulgaris. Phytopathology. 1987;77:1273–1279. [Google Scholar]
- 11.Atkinson M M, Baker C J. Alteration of plasmalemma sucrose transport in Phaseolus vulgaris by Pseudomonas syringae pv. syringae and its association with K+/H+ exchange. Phytopathology. 1987;77:1573–1578. [Google Scholar]
- 12.Austin B, Goodfellow M. Pseudomonas mesophilica, a new species of pink bacteria isolated from leaf surfaces. Int J Syst Bacteriol. 1979;29:373–378. [Google Scholar]
- 13.Austin B, Goodfellow M, Dickinson C H. Numerical taxonomy of phylloplane bacteria isolated from Lolium perenne. J Gen Microbiol. 1978;104:139–155. doi: 10.1099/00221287-97-2-219. [DOI] [PubMed] [Google Scholar]
- 14.Babelegoto N M, Varvaro L, Cirulli M. Epiphytic and endophytic multiplication of Pseudomonas syringae pv. tomato (Okabe) Young et al. in susceptible and resistant tomato leaves. Phytopathol Mediterr. 1988;27:138–144. [Google Scholar]
- 15.Barinaga M. A shared strategy for virulence. Science. 1996;272:1261–1263. doi: 10.1126/science.272.5266.1261. [DOI] [PubMed] [Google Scholar]
- 16.Barnes E H. Bacteria on leaf surfaces and in intercellular leaf spaces. Science. 1965;147:1151–1152. doi: 10.1126/science.147.3662.1151. [DOI] [PubMed] [Google Scholar]
- 17.Barta T M, Kinscherf T G, Willis D K. Regulation of tabtoxin production by the lemA gene in Pseudomonas syringae. J Bacteriol. 1992;174:3021–3029. doi: 10.1128/jb.174.9.3021-3029.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Basile D V, Basile M R, Li Q-Y, Corpe W A. Vitamin B12-stimulated growth and development of Jungermannia leiantha Grolle and Gymnocolea inflata (Huds.) Dum. (Hepaticae) Bryologist. 1985;88:77–81. [Google Scholar]
- 19.Basile D V, Slade L L, Corpe W A. An association between a bacterium and a liverwort, Scapania nemorosa. Bull Torrey Bot Club. 1969;96:711–714. [Google Scholar]
- 20.Beattie G A, Lindow S E. Epiphytic fitness of phytopathogenic bacteria: physiological adaptations for growth and survival. In: Dangl J L, editor. Bacterial pathogenesis of plants and animals: molecular and cellular mechanisms. New York, N.Y: Springer-Verlag; 1994. pp. 1–27. [DOI] [PubMed] [Google Scholar]
- 21.Beattie G A, Lindow S E. Survival, growth, and localization of epiphytic fitness mutants of Pseudomonas syringae on leaves. Appl Environ Microbiol. 1994;60:3790–3798. doi: 10.1128/aem.60.10.3790-3798.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beattie G A, Lindow S E. Comparison of the behavior of epiphytic fitness mutants of Pseudomonas syringae under controlled and field conditions. Appl Environ Microbiol. 1994;60:3799–3808. doi: 10.1128/aem.60.10.3799-3808.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beattie G A, Lindow S E. The secret life of foliar bacterial pathogens on leaves. Annu Rev Phytopathol. 1995;33:145–172. doi: 10.1146/annurev.py.33.090195.001045. [DOI] [PubMed] [Google Scholar]
- 24.Beattie G A, Lindow S E. Bacterial colonization of leaves: a spectrum of strategies. Phytopathology. 1999;89:353–359. doi: 10.1094/PHYTO.1999.89.5.353. [DOI] [PubMed] [Google Scholar]
- 25.Beuzón C R, Banks G, Deiwick J, Hensel M, Holden D W. pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol Microbiol. 1999;33:806–816. doi: 10.1046/j.1365-2958.1999.01527.x. [DOI] [PubMed] [Google Scholar]
- 26.Blakeman J P, editor. Microbial ecology of the phylloplane. New York, N.Y: Academic Press; 1981. [Google Scholar]
- 27.Bogdanove A J, Beer S V, Bonas U, Boucher C A, Collmer A, Coplin D L, Cornelis G R, Huang H-C, Hutcheson S W, Panopoulos N J, Van Gijsegem F. Unified nomenclature for broadly conserved hrp genes of phytopathogenic bacteria. Mol Microbiol. 1996;20:681–683. doi: 10.1046/j.1365-2958.1996.5731077.x. [DOI] [PubMed] [Google Scholar]
- 28.Bonas U. hrp genes of phytopathogenic bacteria. In: Dangl J L, editor. Bacterial pathogenesis of plants and animals: molecular and cellular mechanisms. New York, N.Y: Springer-Verlag; 1994. pp. 79–98. [Google Scholar]
- 29.Bradbury J F. Guide to plant pathogenic bacteria. Farnham Royal, Great Britain: CAB International; 1986. [Google Scholar]
- 30.Brighigna L, Montaini P, Favilli F, Carabez Trejo A. Role of the nitrogen-fixing bacterial microflora in the epiphytism of Tillandsia (Bromeliaceae) Am J Bot. 1992;79:723–727. [Google Scholar]
- 31.Brock T D. The study of microorganisms in situ: progress and problems. Symp Soc Gen Microbiol. 1987;41:1–17. [Google Scholar]
- 32.Burke M J, Lindow S E. Surface properties and size of the ice nucleation site in ice nucleation active bacteria: theoretical considerations. Cryobiology. 1990;27:80–84. [Google Scholar]
- 33.Butterworth J, McCartney H A. The dispersal of bacteria from leaf surfaces by water splash. J Appl Bacteriol. 1991;71:484–496. [Google Scholar]
- 34.Chancey S T, Wood D W, Pierson L S., III Two-component transcriptional regulation of N-acyl-homoserine lactone production in Pseudomonas aureofaciens. Appl Environ Microbiol. 1999;65:2294–2299. doi: 10.1128/aem.65.6.2294-2299.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Charkowski A O, Huang H-C, Collmer A. Altered localization of HrpZ in Pseudomonas syringae pv. syringae hrp mutants suggests that different components of the type III secretion pathway control protein translocation across the inner and outer membranes of gram-negative bacteria. J Bacteriol. 1997;179:3866–3874. doi: 10.1128/jb.179.12.3866-3874.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng G Y, Legard D E, Hunter J E, Burr T J. Modified bean pod assay to detect strains of Pseudomonas syringae pv. syringae that cause bacterial brown spot of snap bean. Plant Dis. 1989;73:419–423. [Google Scholar]
- 37.Collmer A. Determinants of pathogenicity and avirulence in plant pathogenic bacteria. Curr Opin Plant Biol. 1998;1:329–335. doi: 10.1016/1369-5266(88)80055-4. [DOI] [PubMed] [Google Scholar]
- 38.Constantinidou H A, Hirano S S, Baker L S, Upper C D. Atmospheric dispersal of ice nucleation-active bacteria: the role of rain. Phytopathology. 1990;80:934–937. [Google Scholar]
- 39.Corbell N, Loper J E. A global regulator of secondary metabolite production in Pseudomonas fluorescens Pf-5. J Bacteriol. 1995;177:6230–6236. doi: 10.1128/jb.177.21.6230-6236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cornelis G R. The Yersinia deadly kiss. J Bacteriol. 1998;180:5495–5504. doi: 10.1128/jb.180.21.5495-5504.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cornelis G R, Boland A, Boyd A P, Geuijen C, Iriarte M, Neyt C, Sory M-P, Stainier I. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Corotto L V, Wolber P K, Warren G J. Ice nucleation activity of Pseudomonas fluorescens: mutagenesis, complementation analysis and identification of a gene product. EMBO J. 1986;5:231–236. doi: 10.1002/j.1460-2075.1986.tb04203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corpe W A. A method for detecting methylotrophic bacteria on solid surfaces. J Microbiol Methods. 1985;3:215–221. [Google Scholar]
- 44.Corpe W A, Basile D V. Methanol-utilizing bacteria associated with green plants. Dev Ind Microbiol. 1982;23:483–493. [Google Scholar]
- 45.Corpe W A, Rheem S. Ecology of the methylotrophic bacteria on living leaf surfaces. FEMS Microbiol Ecol. 1989;62:243–250. [Google Scholar]
- 46.Crosse J E. Bacterial canker of stone-fruits. IV. Investigation of a method for measuring the inoculum potential of cherry trees. Ann Appl Biol. 1959;47:306–317. [Google Scholar]
- 47.Crosse J E. Bacterial canker of stone-fruits. V. A comparison of leaf-surface populations of Pseudomonas mors-prunorum in autumn on two cherry varieties. Ann Appl Biol. 1963;52:97–104. [Google Scholar]
- 48.Cuppels D A. The use of pathovar-indicative bacteriophages for rapidly detecting Pseudomonas syringae pv. tomato in tomato leaf and fruit lesions. Phytopathology. 1984;74:891–894. [Google Scholar]
- 49.Cuppels D A, Ainsworth T. Molecular and physiological characterization of Pseudomonas syringae pv. tomato and Pseudomonas syringae pv. maculicola strains that produce the phytotoxin coronatine. Appl Environ Microbiol. 1995;61:3530–3536. doi: 10.1128/aem.61.10.3530-3536.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daft G C, Leben C. Bacterial blight of soybeans: epidemiology of blight outbreaks. Phytopathology. 1972;62:57–62. [Google Scholar]
- 51.Daub M E, Hagedorn D J. Epiphytic populations of Pseudomonas syringae on susceptible and resistant bean lines. Phytopathology. 1981;71:547–550. [Google Scholar]
- 52.Deininger C A, Mueller G M, Wolber P K. Immunological characterization of ice nucleation proteins from Pseudomonas syringae, Pseudomonas fluorescens, and Erwinia herbicola. J Bacteriol. 1988;170:669–675. doi: 10.1128/jb.170.2.669-675.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng W-L, Preston G, Collmer A, Chang C-J, Huang H-C. Characterization of the hrpC and hrpRS operons of Pseudomonas syringae pathovars syringae, tomato, and glycinea and analysis of the ability of hrpF, hrpG, hrcC, hrpT, and hrpV mutants to elicit the hypersensitive response and disease in plants. J Bacteriol. 1998;180:4523–4531. doi: 10.1128/jb.180.17.4523-4531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Denny T P. Differentiation of Pseudomonas syringae pv. tomato from P. s. syringae with a DNA hybridization probe. Phytopathology. 1988;78:1186–1193. [Google Scholar]
- 55.Denny T P. Phenotypic diversity in Pseudomonas syringae pv. tomato. J Gen Microbiol. 1988;134:1939–1948. doi: 10.1099/00221287-134-7-1949. [DOI] [PubMed] [Google Scholar]
- 56.Denny T P, Gilmour M N, Selander R K. Genetic diversity and relationships of two pathovars of Pseudomonas syringae. J Gen Microbiol. 1988;134:1949–1960. doi: 10.1099/00221287-134-7-1949. [DOI] [PubMed] [Google Scholar]
- 57.Derridj S. Nutrients on the leaf surface. In: Morris C E, Nicot P C, Nguyen-The C, editors. Aerial plant surface microbiology. New York, N.Y: Plenum Press; 1996. pp. 25–42. [Google Scholar]
- 58.Dickinson C H. Adaptations of micro-organisms to climatic conditions affecting aerial plant surfaces. In: Fokkema N J, van den Heuvel J, editors. Microbiology of the phyllosphere. Cambridge, U.K: Cambridge University Press; 1986. pp. 77–100. [Google Scholar]
- 59.Dickinson C H, Austin B, Goodfellow M. Quantitative and qualitative studies of phylloplane bacteria from Lolium perenne. J Gen Microbiol. 1975;91:157–166. [Google Scholar]
- 60.Dickinson C H, Preece T F, editors. Microbiology of aerial plant surfaces. London, U.K: Academic Press; 1976. [Google Scholar]
- 61.Doudoroff M, Palleroni N J. Genus I. Pseudomonas Migula. In: Buchanan R E, Gibbons N E, editors. Bergey's manual of determinative bacteriology. 8th ed. Baltimore, Md: Williams & Wilkins; 1974. pp. 217–243. [Google Scholar]
- 62.Dumenyo C K, Mukherjee A, Chun W, Chatterjee A K. Genetic and physiological evidence for the production of N-acyl homoserine lactones by Pseudomonas syringae pv. syringae and other fluorescent plant pathogenic Pseudomonas species. Eur J Plant Pathol. 1998;104:569–582. [Google Scholar]
- 63.Dunleavy J M. Curtobacterium plantarum sp. nov. is ubiquitous in plant leaves and is seed transmitted in soybean and corn. Int J Syst Bacteriol. 1989;39:240–249. [Google Scholar]
- 64.Dye D W, Bradbury J F, Goto M, Hayward A C, Lelliott R A, Schroth M N. International standards for naming pathovars of phytopathogenic bacteria and a list of pathovar names and pathotype strains. Rev Plant Pathol. 1980;59:153–168. [Google Scholar]
- 65.Edwards A R, Van den Bussche R A, Wichman H A, Orser C S. Unusual pattern of bacterial ice nucleation gene evolution. Mol Biol Evol. 1994;11:911–920. doi: 10.1093/oxfordjournals.molbev.a040172. [DOI] [PubMed] [Google Scholar]
- 66.Endert E, Ritchie D F. Detection of pathogenicity, measurement of virulence, and determination of strain variation in Pseudomonas syringae pv. syringae. Plant Dis. 1984;68:677–680. [Google Scholar]
- 67.Ercolani G L. Occurrence of Pseudomonas savastanoi (E. F. Smith) Stevens as an epiphyte of olive trees, in Apulia. Phytopathol Mediterr. 1971;10:130–132. [Google Scholar]
- 68.Ercolani G L. Pseudomonas savastanoi and other bacteria colonizing the surface of olive leaves in the field. J Gen Microbiol. 1978;109:245–257. [Google Scholar]
- 69.Ercolani G L. Variability among isolates of Pseudomonas syringae pv. savastanoi from the phylloplane of the olive. J Gen Microbiol. 1983;129:901–916. doi: 10.1099/00221287-129-4-901. [DOI] [PubMed] [Google Scholar]
- 70.Ercolani G L. Factor analysis of fluctuation in populations of Pseudomonas syringae pv. savastanoi on the phylloplane of the olive. Microb Ecol. 1985;11:41–49. doi: 10.1007/BF02015107. [DOI] [PubMed] [Google Scholar]
- 71.Ercolani G L. Distribution of epiphytic bacteria on olive leaves and the influence of leaf age and sampling time. Microb Ecol. 1991;21:35–48. doi: 10.1007/BF02539143. [DOI] [PubMed] [Google Scholar]
- 72.Ercolani G L, Hagedorn D J, Kelman A, Rand R E. Epiphytic survival of Pseudomonas syringae on hairy vetch in relation to epidemiology of bacterial brown spot of bean in Wisconsin. Phytopathology. 1974;64:1330–1339. [Google Scholar]
- 73.Eriksson A R B, Andersson R A, Pirhonen M, Palva E T. Two-component regulators involved in the global control of virulence in Erwinia carotovora subsp. carotovora. Mol Plant-Microbe Interact. 1998;11:743–752. doi: 10.1094/MPMI.1998.11.8.743. [DOI] [PubMed] [Google Scholar]
- 74.Fall R, Wolber P K. Biochemistry of bacterial ice nuclei. In: Lee R E Jr, Warren G J, Gusta L V, editors. Biological ice nucleation and its applications. St. Paul, Minn: American Phytopathological Society; 1995. pp. 63–83. [Google Scholar]
- 75.Faulwetter R C. Dissemination of the angular leafspot of cotton. J Agric Res. 1917;8:457–475. [Google Scholar]
- 76.Faulwetter R C. Wind-blown rain, a factor in disease dissemination. J Agric Res. 1917;10:639–648. [Google Scholar]
- 77.Fitt B D L, McCartney H A, Walklate P J. The role of rain in dispersal of pathogen inoculum. Annu Rev Phytopathol. 1989;27:241–270. [Google Scholar]
- 78.Fokkema N J, van den Heuvel J, editors. Microbiology of the phyllosphere. Cambridge, U.K: Cambridge University Press; 1986. [Google Scholar]
- 79.Frederick R D, Chiu J, Bennetzen J L, Handa A K. Identification of a pathogenicity locus, rpfA, in Erwinia carotovora subsp. carotovora that encodes a two-component sensor-regulator protein. Mol Plant-Microbe Interact. 1997;10:407–415. doi: 10.1094/MPMI.1997.10.3.407. [DOI] [PubMed] [Google Scholar]
- 80.Fryda S J, Otta J D. Epiphytic movement and survival of Pseudomonas syringae on spring wheat. Phytopathology. 1978;68:1064–1067. [Google Scholar]
- 81.Gaffney T D, Lam S T, Ligon J, Gates K, Frazelle A, Di Maio J, Hill S, Goodwin S, Torkewitz N, Allshouse A M, Kempf H-J, Becker J O. Global regulation of expression of antifungal factors by a Pseudomonas fluorescens biological control strain. Mol Plant-Microbe Interact. 1994;7:455–463. doi: 10.1094/mpmi-7-0455. [DOI] [PubMed] [Google Scholar]
- 82.Galán J E, Bliska J B. Cross-talk between bacterial pathogens and their host cells. Annu Rev Cell Dev Biol. 1996;12:221–255. doi: 10.1146/annurev.cellbio.12.1.221. [DOI] [PubMed] [Google Scholar]
- 83.Galán J E, Collmer A. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 1999;284:1322–1328. doi: 10.1126/science.284.5418.1322. [DOI] [PubMed] [Google Scholar]
- 84.Gardan L, Shafik H, Belouin S, Broch R, Grimont F, Grimont P A D. DNA relatedness among the pathovars of Pseudomonas syringae and description of Pseudomonas tremae sp. nov. and Pseudomonas cannabina sp. nov. (ex Sutic and Dowson 1959) Int J Syst Bacteriol. 1999;49:469–478. doi: 10.1099/00207713-49-2-469. [DOI] [PubMed] [Google Scholar]
- 85.Godfrey B E S. Leachates from aerial parts of plants and their relation to plant surface microbial populations. In: Dickinson C H, Preece T F, editors. Microbiology of aerial plant surfaces. London, U.K: Academic Press; 1976. pp. 433–439. [Google Scholar]
- 86.Goodfellow M, Austin B, Dickinson C H. Numerical taxonomy of some yellow-pigmented bacteria isolated from plants. J Gen Microbiol. 1976;97:219–233. doi: 10.1099/00221287-97-2-219. [DOI] [PubMed] [Google Scholar]
- 87.Goodman R N, Novacky A J. The hypersensitive reaction in plants to pathogens—a resistance phenomenon. St. Paul, Minn: American Phytopathological Society; 1994. [Google Scholar]
- 88.Gopalan S, He S Y. Bacterial genes involved in the elicitation of hypersensitive response and pathogenesis. Plant Dis. 1996;80:604–610. [Google Scholar]
- 89.Gopalan S, Wei W, Hei S Y. hrp gene-dependent induction of hin1: a plant gene activated rapidly by both harpins and the avrPto gene-mediated signal. Plant J. 1996;10:591–600. doi: 10.1046/j.1365-313x.1996.10040591.x. [DOI] [PubMed] [Google Scholar]
- 90.Goulder R, Baker J H. Submerged leaf surfaces as a microbial habitat. In: Andrews J H, Hirano S S, editors. Microbial ecology of leaves. New York, N.Y: Springer-Verlag; 1991. pp. 60–86. [Google Scholar]
- 91.Govindarajan A G, Lindow S E. Size of bacterial ice-nucleation sites measured in situ by radiation inactivation analysis. Proc Natl Acad Sci USA. 1988;85:1334–1338. doi: 10.1073/pnas.85.5.1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Green P N. The genus Methylobacterium. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. III. New York, N.Y: Springer-Verlag; 1992. pp. 2342–2349. [Google Scholar]
- 93.Green P N, Bousfield I J. A taxonomic study of some gram-negative facultatively methylotrophic bacteria. J Gen Microbiol. 1982;128:623–638. [Google Scholar]
- 94.Green P N, Bousfield I J. Emendation of Methylobacterium Patt, Cole, and Hanson 1976; Methylobacterium rhodinum (Heumann 1962) comb. nov. corrig.; Methylobacterium radiotolerans (Ito and Iizuka 1971) comb. nov. corrig.; and Methylobacterium mesophilicum (Austin and Goodfellow 1979) comb. nov. Int J Syst Bacteriol. 1983;33:875–877. [Google Scholar]
- 95.Green R L, Corotto L V, Warren G J. Deletion mutagenesis of the ice nucleation gene from Pseudomonas syringae S203. Mol Gen Genet. 1988;215:165–172. doi: 10.1007/BF00331320. [DOI] [PubMed] [Google Scholar]
- 96.Green R L, Warren G J. Physical and functional repetition in a bacterial ice nucleation gene. Nature. 1985;317:645–648. [Google Scholar]
- 97.Grewal S I S, Han B, Johnstone K. Identification and characterization of a locus which regulates multiple functions in Pseudomonas tolaasii, the cause of brown blotch disease of Agaricus bisporus. J Bacteriol. 1995;177:4658–4668. doi: 10.1128/jb.177.16.4658-4668.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gross D C, Cody Y S, Proebsting E L, Jr, Radamaker G K, Spotts R A. Distribution, population dynamics, and characteristics of ice nucleation-active bacteria in deciduous fruit tree orchards. Appl Environ Microbiol. 1983;46:1370–1379. doi: 10.1128/aem.46.6.1370-1379.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Reference deleted.
- 100.Haefele D M, Lindow S E. Flagellar motility confers epiphytic fitness advantages upon Pseudomonas syringae. Appl Environ Microbiol. 1987;53:2528–2533. doi: 10.1128/aem.53.10.2528-2533.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.He S Y. Elicitation of plant hypersensitive response by bacteria. Plant Physiol. 1996;112:865–869. doi: 10.1104/pp.112.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.He S Y. Type III protein secretion systems in plant and animal pathogenic bacteria. Annu Rev Phytopathol. 1998;36:363–392. doi: 10.1146/annurev.phyto.36.1.363. [DOI] [PubMed] [Google Scholar]
- 103.He S Y, Huang H-C, Collmer A. Pseudomonas syringae pv. syringae harpinPss: a protein that is secreted via the hrp pathway and elicits the hypersensitive response in plants. Cell. 1993;73:1255–1266. doi: 10.1016/0092-8674(93)90354-s. [DOI] [PubMed] [Google Scholar]
- 104.Hildebrand D C, Schroth M N, Huisman O C. The DNA homology matrix and non-random variation concepts as the basis for the taxonomic treatment of plant pathogenic and other bacteria. Annu Rev Phytopathol. 1982;20:235–256. [Google Scholar]
- 105.Hirano S S, Baker L S, Upper C D. Raindrop momentum triggers growth of leaf-associated populations of Pseudomonas syringae on field-grown snap bean plants. Appl Environ Microbiol. 1996;62:2560–2566. doi: 10.1128/aem.62.7.2560-2566.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hirano S S, Charkowski A O, Collmer A, Willis D K, Upper C D. Role of the Hrp type III protein secretion system in growth of Pseudomonas syringae pv. syringae B728a on host plants in the field. Proc Natl Acad Sci USA. 1999;96:9851–9856. doi: 10.1073/pnas.96.17.9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hirano S S, Clayton M K, Upper C D. Estimation of and temporal changes in means and variances of populations of Pseudomonas syringae on snap bean leaflets. Phytopathology. 1994;84:934–940. [Google Scholar]
- 108.Hirano S S, Maher E A, Kelman A, Upper C D. Proceedings of the 4th International Conference on Plant Pathogenic Bacteria. Beaucozé, France: Station de Pathologie Végétale et Phytobactériologie; 1978. Ice nucleation activity of fluorescent plant pathogenic pseudomonads; pp. 717–724. [Google Scholar]
- 109.Hirano S S, Nordheim E V, Arny D C, Upper C D. Lognormal distribution of epiphytic bacterial populations on leaf surfaces. Appl Environ Microbiol. 1982;44:695–700. doi: 10.1128/aem.44.3.695-700.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hirano S S, Ostertag E M, Savage S A, Baker L S, Willis D K, Upper C D. Contribution of the regulatory gene lemA to field fitness of Pseudomonas syringae pv. syringae. Appl Environ Microbiol. 1997;63:4304–4312. doi: 10.1128/aem.63.11.4304-4312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hirano S S, Rouse D I, Clayton M K, Upper C D. Pseudomonas syringae pv. syringae and bacterial brown spot of snap bean: a study of epiphytic phytopathogenic bacteria and associated disease. Plant Dis. 1995;79:1085–1093. [Google Scholar]
- 112.Hirano S S, Rouse D I, Upper C D. Bacterial ice nucleation as a predictor of bacterial brown spot disease on snap beans. Phytopathology. 1987;77:1078–1084. [Google Scholar]
- 113.Reference deleted.
- 114.Hirano S S, Upper C D. Ecology and epidemiology of foliar bacterial plant pathogens. Annu Rev Phytopathol. 1983;21:243–269. [Google Scholar]
- 115.Hirano S S, Upper C D. Temporal, spatial, and genetic variability of leaf-associated bacterial populations. In: Fokkema N J, van den Heuvel J, editors. Microbiology of the phyllosphere. Cambridge, U.K: Cambridge University Press; 1986. pp. 235–251. [Google Scholar]
- 116.Hirano S S, Upper C D. Diel variation in population size and ice nucleation activity of Pseudomonas syringae on snap bean leaflets. Appl Environ Microbiol. 1989;55:623–630. doi: 10.1128/aem.55.3.623-630.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hirano S S, Upper C D. Population biology and epidemiology of Pseudomonas syringae. Annu Rev Phytopathol. 1990;28:155–177. [Google Scholar]
- 118.Hirano S S, Upper C D. Bacterial community dynamics. In: Andrews J H, Hirano S S, editors. Microbial ecology of leaves. New York, N.Y: Springer-Verlag; 1991. pp. 271–294. [Google Scholar]
- 119.Hirano S S, Upper C D. Dynamics, spread, and persistence of a single genotype of Pseudomonas syringae relative to those of its conspecifics on populations of snap bean leaflets. Appl Environ Microbiol. 1993;59:1082–1091. doi: 10.1128/aem.59.4.1082-1091.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hirano S S, Upper C D. Autecology of foliar pseudomonads. In: Blakeman J P, Williamson B, editors. Ecology of plant pathogens. Wallingford, U.K: CAB International; 1994. pp. 227–243. [Google Scholar]
- 121.Hirano S S, Upper C D. Ecology of ice nucleation-active bacteria. In: Lee R E Jr, Warren G J, Gusta L V, editors. Biological ice nucleation and its applications. St. Paul, Minn: American Phytopathological Society; 1995. pp. 41–61. [Google Scholar]
- 122.Reference deleted.
- 123.Reference deleted.
- 124.Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C.: American Society for Microbiology; 1995. [Google Scholar]
- 125.Holland M A, Polacco J C. Urease-null and hydrogenase-null phenotypes of a phylloplane bacterium reveal altered nickel metabolism in two soybean mutants. Plant Physiol. 1992;98:942–948. doi: 10.1104/pp.98.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Holland M A, Polacco J C. PPFMs and other covert contaminants: is there more to plant physiology than just plant? Annu Rev Plant Physiol Plant Mol Biol. 1994;45:197–209. [Google Scholar]
- 127.Hrabak E M, Willis D K. The lemA gene required for pathogenicity of Pseudomonas syringae pv. syringae on bean is a member of a family of two-component regulators. J Bacteriol. 1992;174:3011–3020. doi: 10.1128/jb.174.9.3011-3020.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hrabak E M, Willis D K. Involvement of the lemA gene in production of syringomycin and protease by Pseudomonas syringae pv. syringae. Mol Plant-Microbe Interact. 1993;6:368–375. [Google Scholar]
- 129.Huang H-C, He S Y, Bauer D W, Collmer A. The Pseudomonas syringae pv. syringae 61 hrpH product, an envelope protein required for elicitation of the hypersensitive response in plants. J Bacteriol. 1992;174:6878–6885. doi: 10.1128/jb.174.21.6878-6885.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Huang H-C, Lin R-H, Chang C-J, Collmer A, Deng W-L. The complete hrp gene cluster of Pseudomonas syringae pv. syringae 61 includes two blocks of genes required for harpinPss secretion that are arranged colinearly with Yersinia ysc homologs. Mol Plant-Microbe Interact. 1995;8:733–746. doi: 10.1094/mpmi-8-0733. [DOI] [PubMed] [Google Scholar]
- 131.Hueck C J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hutcheson S W, Heu S, Jin S, Lidell M C, Pirhonen M U, Rowley D L. Function and regulation of Pseudomonas syringae hrp genes. In: Nakazawa T, Furukawa K, Harayama S, Silvers S, editors. Pseudomonas: molecular biology and biotechnology. Washington, D.C.: American Society for Microbiology; 1995. pp. 512–521. [Google Scholar]
- 133.Hutcheson S W, Jin S, Lidell M C, Fu Z. Pseudomonas syringae hrp genes: regulation and role in avirulence phenotypes. In: Stacey G, Mullin B, Gresshoff P M, editors. Biology of plant-microbe interactions. St. Paul, Minn: International Society for Molecular Plant-Microbe Interactions; 1996. pp. 153–158. [Google Scholar]
- 134.Ishimaru C, Eskridge K M, Vidaver A K. Distribution analyses of naturally occurring epiphytic populations of Xanthomonas campestris pv. phaseoli on dry beans. Phytopathology. 1991;81:262–268. [Google Scholar]
- 135.Jacques M-A. The effect of leaf age and position on the dynamics of microbial populations on aerial plant surfaces. In: Morris C E, Nicot P C, Nguyen-The C, editors. Aerial plant surface microbiology. New York, N.Y: Plenum Press; 1996. pp. 233–248. [Google Scholar]
- 136.Jacques M-A, Morris C E. A review of issues related to the quantification of bacteria from the phyllosphere. FEMS Microbiol Ecol. 1995;18:1–14. [Google Scholar]
- 137.Johnston C, Pegues D A, Hueck C J, Lee C A, Miller S I. Transcriptional activation of Salmonella typhimurium invasion genes by a member of the phosphorylated response-regulator superfamily. Mol Microbiol. 1996;22:715–727. doi: 10.1046/j.1365-2958.1996.d01-1719.x. [DOI] [PubMed] [Google Scholar]
- 138.Jones J B, Gitaitis R D, McCarter S M. Evaluation of indirect immunofluorescence and ice nucleation activity as rapid tests for identifying foliar diseases of tomato transplants incited by fluorescent pseudomonads. Plant Dis. 1983;67:684–687. [Google Scholar]
- 139.Jones K. Nitrogen fixation in the phyllosphere of the Douglas fir, Pseudotsuga douglasii. Ann Bot. 1970;34:239–244. [Google Scholar]
- 140.Kado C I. Plant pathogenic bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. I. New York, N.Y: Springer-Verlag; 1992. pp. 659–674. [Google Scholar]
- 141.Kajava A V. Molecular modeling of the three-dimensional structure of bacterial Ina proteins. In: Lee R E Jr, Warren G J, Gusta L V, editors. Biological ice nucleation and its applications. St. Paul, Minn: American Phytopathological Society; 1995. pp. 101–114. [Google Scholar]
- 142.Kaneda T. Seasonal population changes and characterization of ice-nucleating bacteria in farm fields of central Alberta. Appl Environ Microbiol. 1986;52:173–178. doi: 10.1128/aem.52.1.173-178.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kennedy B W, Ercolani G L. Soybean primary leaves as a site for epiphytic multiplication of Pseudomonas glycinea. Phytopathology. 1978;68:1196–1201. [Google Scholar]
- 144.Kerstiens G, editor. Plant cuticles: an integrated functional approach. Oxford, U.K: BIOS Scientific Publishers; 1996. [Google Scholar]
- 145.Kim H K, Orser C, Lindow S E, Sands D C. Xanthomonas campestris pv. translucens strains active in ice nucleation. Plant Dis. 1987;71:994–997. [Google Scholar]
- 146.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 147.Kinkel L L. Microbial population dynamics on leaves. Annu Rev Phytopathol. 1997;35:327–347. doi: 10.1146/annurev.phyto.35.1.327. [DOI] [PubMed] [Google Scholar]
- 148.Kinkel L L, Lindow S E. Invasion and exclusion among coexisting Pseudomonas syringae strains on leaves. Appl Environ Microbiol. 1993;59:3447–3454. doi: 10.1128/aem.59.10.3447-3454.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kinkel L L, Wilson M, Lindow S E. Effect of sampling scale on the assessment of epiphytic bacterial populations. Microb Ecol. 1995;29:283–297. doi: 10.1007/BF00164891. [DOI] [PubMed] [Google Scholar]
- 150.Kinscherf T G, Willis D K. Swarming by Pseudomonas syringae B728a requires gacS (lemA) and gacA but not the acyl-homoserine lactone biosynthetic gene ahlI. J Bacteriol. 1999;181:4133–4136. doi: 10.1128/jb.181.13.4133-4136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kitten T, Kinscherf T G, McEvoy J L, Willis D K. A newly-identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol Microbiol. 1998;28:917–929. doi: 10.1046/j.1365-2958.1998.00842.x. [DOI] [PubMed] [Google Scholar]
- 152.Kitten T, Willis D K. Suppression of a sensor kinase-dependent phenotype in Pseudomonas syringae by ribosomal proteins L35 and L20. J Bacteriol. 1996;178:1548–1555. doi: 10.1128/jb.178.6.1548-1555.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Klement Z. Rapid detection of the pathogenicity of phytopathogenic pseudomonads. Nature. 1963;199:299–300. doi: 10.1038/199299b0. [DOI] [PubMed] [Google Scholar]
- 154.Koster M, Bitter W, de Cock H, Allaoui A, Cornelis G R, Tommassen J. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocolitica forms a ring-shaped multimeric complex. Mol Microbiol. 1997;26:789–797. doi: 10.1046/j.1365-2958.1997.6141981.x. [DOI] [PubMed] [Google Scholar]
- 155.Kozloff L M, Turner M A, Arellano F. Formation of bacterial membrane ice-nucleating lipoglycoprotein complexes. J Bacteriol. 1991;173:6528–6536. doi: 10.1128/jb.173.20.6528-6536.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ladha J K, Barraquio W L, Watanabe I. Isolation and identification of nitrogen-fixing Enterobacter cloacae and Klebsiella planticola associated with rice plants. Can J Microbiol. 1983;29:1301–1308. doi: 10.1139/m83-141. [DOI] [PubMed] [Google Scholar]
- 157.Reference deleted.
- 158.Last F T, Deighton F C. The non-parasitic microflora on the surfaces of living leaves. Trans Br Mycol Soc. 1965;48:83–99. [Google Scholar]
- 159.Last F T, Warren R C. Non-parasitic microbes colonizing green leaves—their form and functions. Endeavour. 1972;31:143–150. [Google Scholar]
- 160.Laville J, Voisard C, Keel C, Maurhofer M, Défago G, Haas D. Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco. Proc Natl Acad Sci USA. 1992;89:1562–1566. doi: 10.1073/pnas.89.5.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Leben C. Epiphytic microorganisms in relation to plant disease. Annu Rev Phytopathol. 1965;3:209–230. [Google Scholar]
- 162.Lee C A. Type III secretion systems: machines to deliver bacterial proteins into eukaryotic cells? Trends Microbiol. 1997;5:148–156. doi: 10.1016/S0966-842X(97)01029-9. [DOI] [PubMed] [Google Scholar]
- 163.Lee R E Jr, Warren G J, Gusta L V, editors. Biological ice nucleation and its applications. St. Paul, Minn: American Phytopathological Society; 1995. [Google Scholar]
- 164.Legard D E, Aquadro C F, Hunter J E. DNA sequence variation and phylogenetic relationships among strains of Pseudomonas syringae pv. syringae inferred from restriction site maps and restriction fragment length polymorphism. Appl Environ Microbiol. 1993;59:4180–4188. doi: 10.1128/aem.59.12.4180-4188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Liao C-H, McCallus D E, Fett W F. Molecular characterization of two gene loci required for production of the key pathogenicity factor pectate lyase in Pseudomonas viridiflava. Mol Plant-Microbe Interact. 1994;7:391–400. doi: 10.1094/mpmi-7-0391. [DOI] [PubMed] [Google Scholar]
- 166.Lindemann J, Arny D C, Upper C D. Epiphytic populations of Pseudomonas syringae pv. syringae on snap bean and nonhost plants and the incidence of bacterial brown spot disease in relation to cropping patterns. Phytopathology. 1984;74:1329–1333. [Google Scholar]
- 167.Lindemann J, Arny D C, Upper C D. Use of an apparent infection threshold population of Pseudomonas syringae to predict incidence and severity of brown spot of bean. Phytopathology. 1984;74:1334–1339. [Google Scholar]
- 168.Lindemann J, Constantinidou H A, Barchet W R, Upper C D. Plants as sources of airborne bacteria, including ice nucleation-active bacteria. Appl Environ Microbiol. 1982;44:1059–1063. doi: 10.1128/aem.44.5.1059-1063.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lindemann J, Suslow T V. Characteristics relevant to the question of environmental fate of genetically engineered INA− deletion mutant strains of Pseudomonas. In: Civerolo E L, Collmer A, Davis R E, Gillaspie A G, editors. Plant pathogenic bacteria. Dordrecht, The Netherlands: Martinus Nijhoff Publishers; 1987. pp. 1005–1012. [Google Scholar]
- 170.Lindemann J, Suslow T V. Competition between ice nucleation-active wild type and ice nucleation-deficient deletion mutant strains of Pseudomonas syringae and P. fluorescens biovar I and biological control of frost injury on strawberry blossoms. Phytopathology. 1987;77:882–886. [Google Scholar]
- 171.Lindemann J, Upper C D. Aerial dispersal of epiphytic bacteria over bean plants. Appl Environ Microbiol. 1985;50:1229–1232. doi: 10.1128/aem.50.5.1229-1232.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lindgren P B. The role of hrp genes during plant-bacterial interactions. Annu Rev Phytopathol. 1997;35:129–152. doi: 10.1146/annurev.phyto.35.1.129. [DOI] [PubMed] [Google Scholar]
- 173.Lindgren P B, Peet R C, Panopoulos N J. Gene cluster of Pseudomonas syringae pv. “phaseolicola” controls pathogenicity on bean plants and hypersensitivity on nonhost plants. J Bacteriol. 1986;168:512–522. doi: 10.1128/jb.168.2.512-522.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lindow S E. Population dynamics of epiphytic ice nucleation active bacteria on frost sensitive plants and frost control by means of antagonistic bacteria. In: Li P H, Sakai A, editors. Plant cold hardiness and freezing stress—mechanisms and crop implications. Vol. 2. New York, N.Y: Academic Press; 1982. pp. 395–416. [Google Scholar]
- 175.Lindow S E. The role of bacterial ice nucleation in frost injury to plants. Annu Rev Phytopathol. 1983;21:363–384. [Google Scholar]
- 176.Lindow S E. Methods of preventing frost injury caused by epiphytic ice-nucleation-active bacteria. Plant Dis. 1983;67:327–333. [Google Scholar]
- 177.Lindow S E. Ecology of Pseudomonas syringae relevant to the field use of Ice− deletion mutants constructed in vitro for plant frost control. In: Halvorson H O, Pramer D, Rogul M, editors. Engineered organisms in the environment: scientific issues. Washington, D.C.: American Society for Microbiology; 1985. pp. 23–35. [Google Scholar]
- 178.Lindow S E. Strategies and practice of biological control of ice nucleation-active bacteria on plants. In: Fokkema N J, van den Heuvel J, editors. Microbiology of the phyllosphere. Cambridge, U.K: Cambridge University Press; 1986. pp. 293–311. [Google Scholar]
- 179.Lindow S E. Construction of isogenic Ice− strains of Pseudomonas syringae for evaluation of specificity of competition on leaf surfaces. In: Megusar F, Gantar M, editors. Perspectives in microbial ecology. Ljubljana, Yugoslavia: Slovene Society for Microbiology; 1986. pp. 509–515. [Google Scholar]
- 180.Lindow S E. Competitive exclusion of epiphytic bacteria by Ice−Pseudomonas syringae mutants. Appl Environ Microbiol. 1987;53:2520–2527. doi: 10.1128/aem.53.10.2520-2527.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lindow S E. Design and results of field tests of recombinant Ice−Pseudomonas syringae strains. In: Marois J J, Bruening G, editors. Risk assessment in agricultural biotechnology: proceedings of the international conference. Calif: University of California, Davis; 1990. pp. 61–69. [Google Scholar]
- 182.Lindow S E. Determinants of epiphytic fitness in bacteria. In: Andrews J H, Hirano S S, editors. Microbial ecology of leaves. New York, N.Y: Springer-Verlag; 1991. pp. 295–314. [Google Scholar]
- 183.Lindow S E. Novel method for identifying bacterial mutants with reduced epiphytic fitness. Appl Environ Microbiol. 1993;59:1586–1592. doi: 10.1128/aem.59.5.1586-1592.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lindow S E. Control of epiphytic ice nucleation-active bacteria for management of plant frost injury. In: Lee R E Jr, Warren G J, Gusta L V, editors. Biological ice nucleation and its applications. St. Paul, Minn: American Phytopathological Society; 1995. pp. 239–256. [Google Scholar]
- 185.Lindow S E. Role of immigration and other processes in determining epiphytic bacterial populations. In: Morris C E, Nicot P C, Nguyen-The C, editors. Aerial plant surface microbiology. New York, N.Y: Plenum Press; 1996. pp. 155–168. [Google Scholar]
- 186.Lindow S E, Andersen G, Beattie G A. Characteristics of insertional mutants of Pseudomonas syringae with reduced epiphytic fitness. Appl Environ Microbiol. 1993;59:1593–1601. doi: 10.1128/aem.59.5.1593-1601.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lindow S E, Andersen G L. Influence of immigration on epiphytic bacterial populations on navel orange leaves. Appl Environ Microbiol. 1996;62:2978–2987. doi: 10.1128/aem.62.8.2978-2987.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Lindow S E, Arny D C, Barchet W R, Upper C D. The role of bacterial ice nuclei in frost injury to sensitive plants. In: Li P H, Sakai A, editors. Plant cold hardiness and freezing stress—mechanisms and crop implications. New York, N.Y: Academic Press; 1978. pp. 249–263. [Google Scholar]
- 189.Lindow S E, Arny D C, Upper C D. Distribution of ice nucleation-active bacteria on plants in nature. Appl Environ Microbiol. 1978;36:831–838. doi: 10.1128/aem.36.6.831-838.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lindow S E, Arny D C, Upper C D. Erwinia herbicola: a bacterial ice nucleus active in increasing frost injury to corn. Phytopathology. 1978;68:523–527. [Google Scholar]
- 191.Lindow S E, Arny D C, Upper C D. Bacterial ice nucleation: a factor in frost injury to plants. Plant Physiol. 1982;70:1084–1089. doi: 10.1104/pp.70.4.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lindow S E, Arny D C, Upper C D. Biological control of frost injury: an isolate of Erwinia herbicola antagonistic to ice nucleation active bacteria. Phytopathology. 1983;73:1097–1102. [Google Scholar]
- 193.Lindow S E, Connell J H. Reduction of frost injury to almond by control of ice nucleation active bacteria. J Am Soc Hort Sci. 1984;109:48–53. [Google Scholar]
- 194.Lindow S E, Desurmont C, Elkins R, McGourty G, Clark E, Brandl M T. Occurrence of indole-3-acetic acid-producing bacteria on pear trees and their association with fruit russet. Phytopathology. 1998;88:1149–1157. doi: 10.1094/PHYTO.1998.88.11.1149. [DOI] [PubMed] [Google Scholar]
- 195.Lindow S E, Hirano S S, Barchet W R, Arny D C, Upper C D. Relationship between ice nucleation frequency of bacteria and frost injury. Plant Physiol. 1982;70:1090–1093. doi: 10.1104/pp.70.4.1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Lindow S E, Lahue E, Govindarajan A G, Panopoulos N J, Gies D. Localization of ice nucleation activity and the iceC gene product in Pseudomonas syringae and Escherichia coli. Mol Plant-Microbe Interact. 1989;2:262–272. doi: 10.1094/mpmi-2-262. [DOI] [PubMed] [Google Scholar]
- 197.Lindow S E, Panopoulos N J. Field tests of recombinant Ice−Pseudomonas syringae for biological frost control in potato. In: Sussman M, Collins C H, Skinner F A, Stewart-Tull D E, editors. Release of genetically-engineered micro-organisms. New York, N.Y: Academic Press; 1988. pp. 121–138. [Google Scholar]
- 198.Little E L, Bostock R M, Kirkpatrick B C. Genetic characterization of Pseudomonas syringae pv. syringae strains from stone fruits in California. Appl Environ Microbiol. 1998;64:3818–3823. doi: 10.1128/aem.64.10.3818-3823.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Loper J E, Lindow S E. Lack of evidence for in situ fluorescent pigment production by Pseudomonas syringae pv. syringae on bean leaf surfaces. Phytopathology. 1987;77:1449–1454. [Google Scholar]
- 200.Louws F J, Fulbright D W, Stephens C T, de Bruijn F J. Specific genomic fingerprints of phytopathogenic Xanthomonas and Pseudomonas pathovars and strains generated with repetitive sequences and PCR. Appl Environ Microbiol. 1994;60:2286–2295. doi: 10.1128/aem.60.7.2286-2295.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Maki L R, Galyan E L, Chang-Chien M-M, Caldwell D R. Ice nucleation induced by Pseudomonas syringae. Appl Microbiol. 1974;28:456–459. doi: 10.1128/am.28.3.456-459.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Martin J M S. Characteristics and population densities of fluorescent pseudomonads from cherry and apricot leaf surfaces in Portugal. Garcia De Orta Ser Estud Agron. 1982;9:249–254. [Google Scholar]
- 203.Martin J T, Juniper B E. The cuticles of plants. New York, N.Y: St. Martin's Press; 1970. [Google Scholar]
- 204.Mecsas J, Strauss E J. Molecular mechanisms of bacterial virulence: type III secretion and pathogenicity islands. Emerg Infect Dis. 1996;2:271–288. doi: 10.3201/eid0204.960403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Mercier J, Lindow S E. Role of leaf surface sugars in colonization of plants by bacterial epiphytes. Appl Environ Microbiol. 2000;66:369–374. doi: 10.1128/aem.66.1.369-374.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Mew T W, Kennedy B W. Growth of Pseudomonas glycinea on the surface of soybean leaves. Phytopathology. 1971;61:715–716. [Google Scholar]
- 207.Mew T W, Vera Cruz C M. Epiphytic colonization of host and non-host plants by phytopathogenic bacteria. In: Fokkema N J, van den Heuvel J, editors. Microbiology of the phyllosphere. Cambridge, U.K: Cambridge University Press; 1986. pp. 269–282. [Google Scholar]
- 208.Mohan S K, Schaad N W. An improved agar plating assay for detecting Pseudomonas syringae pv. syringae and P. s. pv. phaseolicola in contaminated bean seed. Phytopathology. 1987;77:1390–1395. [Google Scholar]
- 209.Morris C E, Nicot P C, Nguyen-The C, editors. Aerial plant surface microbiology. New York, N.Y: Plenum Press; 1996. [Google Scholar]
- 210.Morris C E, Rouse D I. Role of nutrients in regulating epiphytic bacterial populations. In: Windels C E, Lindow S E, editors. Biological control on the phylloplane. St. Paul, Minn: American Phytopathological Society; 1985. pp. 63–82. [Google Scholar]
- 211.Mudgett M B, Staskawicz B J. Protein signaling via type III secretion pathways in phytopathogenic bacteria. Curr Opin Microbiol. 1998;1:109–115. doi: 10.1016/s1369-5274(98)80150-1. [DOI] [PubMed] [Google Scholar]
- 212.Mueller G M, Wolber P K, Warren G J. Clustering of ice nucleation protein correlates with ice nucleation activity. Crybiology. 1990;27:416–422. doi: 10.1016/0011-2240(90)90018-y. [DOI] [PubMed] [Google Scholar]
- 213.Mullins J S, Davis D G. Studies on corn leaf surface bacterial flora. Proc ND Acad Sci. 1975;27:120–127. [Google Scholar]
- 214.Murty M G. Phyllosphere of cotton as a habitat for diazotrophic microorganisms. Appl Environ Microbiol. 1984;48:713–718. doi: 10.1128/aem.48.4.713-718.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Nemecek-Marshall M, MacDonald R C, Franzen J J, Wojciechowski C L, Fall R. Methanol emission from leaves. Plant Physiol. 1995;108:1359–1368. doi: 10.1104/pp.108.4.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Newton D, Hayward A C. Ice nucleation activity of some reference cultures of Pseudomonas syringae and field isolates of bacteria from wheat and barley in Queensland. Australas Plant Pathol. 1986;15:71–73. [Google Scholar]
- 217.Obata H, Takinami K, Tanishita J, Hasegawa Y, Kawate S, Tokuyama T, Ueno T. Identification of a new ice nucleating bacterium and its ice nucleation properties. Agric Biol Chem. 1990;54:725–730. [Google Scholar]
- 218.O'Brien R D, Lindow S E. Effect of plant species and environmental conditions on epiphytic population sizes of Pseudomonas syringae and other bacteria. Phytopathology. 1989;79:619–627. [Google Scholar]
- 219.Orser C, Staskawicz B J, Loper J, Panopoulos N J, Dahlbeck D, Lindow S E, Schroth M N. Cloning of genes involved in bacterial ice nucleation and fluorescent pigment/siderophore production. In: Pühler A, editor. Molecular genetics of the bacteria-plant interaction. Berlin, Germany: Springer-Verlag; 1983. pp. 353–361. [Google Scholar]
- 220.Orser C, Staskawicz B J, Panopoulos N J, Dahlbeck D, Lindow S E. Cloning and expression of bacterial ice nucleation genes in Escherichia coli. J Bacteriol. 1985;164:359–366. doi: 10.1128/jb.164.1.359-366.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 222.Pati B R. Effect of spraying nitrogen fixing phyllospheric bacterial isolates on rice plants. Zentralbl Mikrobiol. 1992;147:441–446. [Google Scholar]
- 223.Pati B R, Chandra A K. Effect of spraying nitrogen-fixing phyllospheric bacterial isolates on wheat plants. Plant Soil. 1981;61:419–427. [Google Scholar]
- 224.Pati B R, Chandra A K. Nitrogen fixing potentialities of the phyllospheric bacteria in relation to concentration of sucrose in the medium. Zentralbl Mikrobiol. 1992;147:435–440. [Google Scholar]
- 225.Patt T E, Cole G C, Hanson R S. Methylobacterium, a new genus of facultatively methylotrophic bacteria. Int J Syst Bacteriol. 1976;26:226–229. [Google Scholar]
- 226.Paulin J-P, Luisetti J. Proceedings of the 4th International Conference on Plant Pathogenic Bacteria. Beaucozé, France: Station de Pathologie Végétale et Phytobactériologie; 1978. Ice nucleation activity among phytopathogenic bacteria; pp. 725–731. [Google Scholar]
- 227.Pennycook S R, Newhook F J. Ultraviolet sterilization in phylloplane studies. Trans Br Mycol Soc. 1982;78:360–361. [Google Scholar]
- 228.Perombelon M C. The ecology of erwinias on aerial plant surfaces. In: Blakeman J P, editor. Microbial ecology of the phylloplane. New York, N.Y: Academic Press; 1981. pp. 411–431. [Google Scholar]
- 229.Preece T F, Dickinson C H, editors. Ecology of leaf surface micro-organisms. London, U.K: Academic Press; 1971. [Google Scholar]
- 230.Preston G M, Haubold B, Rainey P B. Bacterial genomics and adaptation to life on plants: implications for the evolution of pathogenicity and symbiosis. Curr Opin Microbiol. 1998;1:589–597. doi: 10.1016/s1369-5274(98)80094-5. [DOI] [PubMed] [Google Scholar]
- 231.Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A, Haas D. The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol. 1997;24:309–319. doi: 10.1046/j.1365-2958.1997.3291701.x. [DOI] [PubMed] [Google Scholar]
- 232.Rich J J, Hirano S S, Willis D K. Pathovar-specific requirement for the Pseudomonas syringae lemA gene in disease lesion formation. Appl Environ Microbiol. 1992;58:1440–1446. doi: 10.1128/aem.58.5.1440-1446.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Rich J J, Kinscherf T G, Kitten T, Willis D K. Genetic evidence that the gacA gene encodes the cognate response regulator for the lemA sensor in Pseudomonas syringae. J Bacteriol. 1994;176:7468–7475. doi: 10.1128/jb.176.24.7468-7475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Rich J J, Willis D K. Multiple loci of Pseudomonas syringae pv. syringae are involved in pathogenicity on bean: restoration of one lesion-deficient mutant requires two tRNA genes. J Bacteriol. 1997;179:2247–2258. doi: 10.1128/jb.179.7.2247-2258.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Ricklefs R E, Miller G L. Ecology. 4th ed. New York, N.Y: W. H. Freeman; 2000. [Google Scholar]
- 236.Roine E, Raineri D M, Romantschuk M, Wilson M, Nunn D N. Characterization of type IV pilus genes in Pseudomonas syringae pv. tomato DC3000. Mol Plant-Microbe Interact. 1998;11:1048–1056. doi: 10.1094/MPMI.1998.11.11.1048. [DOI] [PubMed] [Google Scholar]
- 237.Roine E, Saarinen J, Kalkkinen N, Romantschuk M. Purified HrpA of Pseudomonas syringae pv. tomato DC3000 reassembles into pili. FEBS Lett. 1997;417:168–172. doi: 10.1016/s0014-5793(97)01276-3. [DOI] [PubMed] [Google Scholar]
- 238.Roine E, Wei W, Yuan J, Nurmiaho-Lassila E-L, Kalkkinen N, Romantschuk M, He S Y. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Romantschuk M. Attachment of plant pathogenic bacteria to plant surfaces. Annu Rev Phytopathol. 1992;30:225–243. doi: 10.1146/annurev.py.30.090192.001301. [DOI] [PubMed] [Google Scholar]
- 240.Romantschuk M, Bamford D H. The causal agent of halo blight in bean, Pseudomonas syringae pv. phaseolicola, attaches to stomata via its pili. Microb Pathog. 1986;1:139–148. doi: 10.1016/0882-4010(86)90016-1. [DOI] [PubMed] [Google Scholar]
- 241.Romantschuk M, Nurmiaho-Lassila E-L, Roine E, Suoniemi A. Pilus-mediated adsorption of Pseudomonas syringae to the surface of host and non-host plant leaves. J Gen Microbiol. 1993;139:2251–2260. [Google Scholar]
- 242.Romantschuk M, Roine E, Björklöf K, Ojanen T, Nurmiaho-Lassila E-L, Haahtela K. Microbial attachment to plant aerial surfaces. In: Morris C E, Nicot P C, Nguyen-The C, editors. Aerial plant surface microbiology. New York, N.Y: Plenum Press; 1996. pp. 43–57. [Google Scholar]
- 243.Roos I M M, Hattingh M J. Resident populations of Pseudomonas syringae on stone fruit tree leaves in South Africa. Phytophylactica. 1986;18:55–58. [Google Scholar]
- 244.Roos I M M, Hattingh M J. Pathogenicity and numerical analysis of phenotypic features of Pseudomonas syringae strains isolated from deciduous fruit trees. Phytopathology. 1987;77:900–908. [Google Scholar]
- 245.Rossier O, Wengelnik K, Hahn K, Bonas U. The Xanthomonas Hrp type III system secretes proteins from plant and mammalian bacterial pathogens. Proc Natl Acad Sci USA. 1999;96:9368–9373. doi: 10.1073/pnas.96.16.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Rouse D I, Nordheim E V, Hirano S S, Upper C D. A model relating the probability of foliar disease incidence to the population frequencies of bacterial plant pathogens. Phytopathology. 1985;75:505–509. [Google Scholar]
- 247.Rudolph K. Bacterial brown spot disease of bush bean (Phaseolus vulgaris L.) in Germany, incited by Pseudomonas syringae van Hall s. s. pathovar phaseoli. Z Pflanzenkr Pflanzenschutz. 1979;86:75–85. [Google Scholar]
- 248.Ruinen J. Occurrence of Beijerinckia species in the ‘phyllosphere.’. Nature. 1956;177:220–221. [Google Scholar]
- 249.Ruinen J. The phyllosphere. I. An ecologically neglected milieu. Plant Soil. 1961;15:81–109. [Google Scholar]
- 250.Ruinen J. The phyllosphere. III. Nitrogen fixation in the phyllosphere. Plant Soil. 1965;22:375–394. [Google Scholar]
- 251.Ruinen J. Nitrogen fixation in the phyllosphere. In: Quispel A, editor. The biology of nitrogen fixation. Amsterdam, The Netherlands: North-Holland Publishing Co.; 1974. pp. 121–167. [Google Scholar]
- 252.Ruinen J. Nitrogen fixation in the phyllosphere. In: Stewart W D P, editor. Nitrogen fixation by free-living micro-organisms. Cambridge, U.K: Cambridge University Press; 1975. pp. 85–100. [Google Scholar]
- 253.Saad S M, Hagedorn D J. Relationship of isolate source to virulence of Pseudomonas syringae on Phaseolus vulgaris. Phytopathology. 1972;62:678–680. [Google Scholar]
- 254.Sacherer P, Défago G, Haas D. Extracellular protease and phospholipase C are controlled by the global regulatory gene gacA in the biocontrol strain Pseudomonas fluorescens CHAO. FEMS Microbiol Lett. 1994;116:155–160. doi: 10.1111/j.1574-6968.1994.tb06694.x. [DOI] [PubMed] [Google Scholar]
- 255.Saettler A W, Schaad N W, Roth D A, editors. Detection of bacteria in seed and other planting material. St. Paul, Minn: American Phytopathological Society; 1989. [Google Scholar]
- 256.Sawada H, Suzuki F, Matsuda I, Saitou N. Phylogenetic analysis of Pseudomonas syringae pathovars suggests the horizontal gene transfer of argK and the evolutionary stability of hrp gene cluster. J Mol Evol. 1999;49:627–644. doi: 10.1007/pl00006584. [DOI] [PubMed] [Google Scholar]
- 257.Scheck H J, Canfield M L, Pscheidt J W, Moore L W. Rapid evaluation of pathogenicity in Pseudomonas syringae pv. syringae with a lilac tissue culture bioassay and syringomycin DNA probes. Plant Dis. 1997;81:905–910. doi: 10.1094/PDIS.1997.81.8.905. [DOI] [PubMed] [Google Scholar]
- 258.Schneider R W, Grogan R G. Tomato leaf trichomes, a habitat for resident populations of Pseudomonas tomato. Phytopathology. 1977;67:898–902. [Google Scholar]
- 259.Scholz B K, Jakobek J L, Lindgren P B. Restriction fragment length polymorphism evidence for genetic homology within a pathovar of Pseudomonas syringae. Appl Environ Microbiol. 1994;60:1093–1100. doi: 10.1128/aem.60.4.1093-1100.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Schönherr J, Baur P. Cuticle permeability studies: a model for estimating leaching of plant metabolites to leaf surfaces. In: Morris C E, Nicot P C, Nguyen-The C, editors. Aerial plant surface microbiology. New York, N.Y: Plenum Press; 1996. pp. 1–23. [Google Scholar]
- 261.Sebastian J, Chandra A K, Kolattukudy P E. Discovery of a cutinase-producing Pseudomonas sp. cohabiting with an apparently nitrogen-fixing Corynebacterium sp. in the phyllosphere. J Bacteriol. 1987;169:131–136. doi: 10.1128/jb.169.1.131-136.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Sen Gupta B, Nandi A S, Samanta R K, Pal D, Sengupta D N, Sen S P. Nitrogen fixation in the phyllosphere of tropical plants: occurrence of phyllosphere nitrogen-fixing micro-organisms in eastern India and their utility for the growth and nitrogen nutrition of host plants. Ann Bot. 1981;48:705–716. [Google Scholar]
- 263.Sen Gupta B, Nandi A S, Sen S P. Utility of phyllosphere N2-fixing micro-organisms in the improvement of crop growth. I. Rice. Plant Soil. 1982;68:55–67. [Google Scholar]
- 264.Sen Gupta B, Sen S P. Utility of phyllosphere N2-fixing micro-organisms in the improvement of crop growth. II. Wheat. Plant Soil. 1982;68:69–74. [Google Scholar]
- 265.Shea J E, Beuzón C R, Gleeson C, Mundy R, Holden D W. Influence of the Salmonella typhimurium pathogenicity island 2 type III secretion system on bacterial growth in the mouse. Infect Immun. 1999;67:213–219. doi: 10.1128/iai.67.1.213-219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Smitley D R, McCarter S M. Spread of Pseudomonas syringae pv. tomato and role of epiphytic populations and environmental conditions in disease development. Plant Dis. 1982;66:713–717. [Google Scholar]
- 267.Southworth M W, Wolber P K, Warren G J. Nonlinear relationship between concentration and activity of a bacterial ice nucleation protein. J Biol Chem. 1988;263:15211–15216. [PubMed] [Google Scholar]
- 268.Sprent J I, Sprent P. Nitrogen fixing organisms. London, U.K: Chapman and Hall; 1990. [Google Scholar]
- 269.Stadt S J, Saettler A W. Effect of host genotype on multiplication of Pseudomonas phaseolicola. Phytopathology. 1981;71:1307–1310. [Google Scholar]
- 270.Stout J D. Biological studies of some tussock-grassland soils. XV. Bacteria of two cultivated soils. N Z J Agric Res. 1960;3:214–223. [Google Scholar]
- 271.Stout J D. Bacteria of soil and pasture leaves at Claudelands Showgrounds. N Z J Agric Res. 1960;3:413–430. [Google Scholar]
- 272.Sundin G W, Jacobs J L. Ultraviolet radiation (UVR) sensitivity analysis and UVR survival strategies of a bacterial community from the phyllosphere of field-grown peanut (Arachis hypogeae L.) Microb Ecol. 1999;38:27–38. doi: 10.1007/s002489900152. [DOI] [PubMed] [Google Scholar]
- 273.Sundin G W, Kidambi S P, Ullrich M, Bender C L. Resistance to ultraviolet light in Pseudomonas syringae: sequence and functional analysis of the plasmid-encoded rulAB genes. Gene. 1996;177:77–81. doi: 10.1016/0378-1119(96)00273-9. [DOI] [PubMed] [Google Scholar]
- 274.Suoniemi A, Björklöf K, Haahtela K, Romantschuk M. Pili of Pseudomonas syringae pathovar syringae enhance initiation of bacterial epiphytic colonization of bean. Microbiology. 1995;141:497–503. doi: 10.1099/13500872-141-10-2719. [DOI] [PubMed] [Google Scholar]
- 275.Sztejnberg A, Blakeman J P. Ultraviolet-induced changes in populations of epiphytic bacteria on beetroot leaves and their effect on germination of Botrytis cinerea spores. Physiol Plant Pathol. 1973;3:443–451. [Google Scholar]
- 276.Thompson I P, Bailey M J, Fenlon J S, Fermor T R, Lilley A K, Lynch J M, McCormack P J, McQuilken M P, Purdy K J, Rainey P B, Whipps J M. Quantitative and qualitative seasonal changes in the microbial community from the phyllosphere of sugar beet (Beta vulgaris) Plant Soil. 1993;150:177–191. [Google Scholar]
- 277.Toraya T, Yongsmith B, Tanaka A, Fukui S. Vitamin B12 production by a methanol-utilizing bacterium. Appl Microbiol. 1975;30:477–479. doi: 10.1128/am.30.3.477-479.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Tukey H B., Jr The leaching of substances from plants. Annu Rev Plant Physiol. 1970;21:305–324. [Google Scholar]
- 279.Tukey H B., Jr . Leaching of substances from plants. In: Preece T F, Dickinson C H, editors. Ecology of leaf surface micro-organisms. London, U.K: Academic Press; 1971. pp. 67–80. [Google Scholar]
- 280.Turner M A, Arellano F, Kozloff L M. Components of ice nucleation structures of bacteria. J Bacteriol. 1991;173:6515–6527. doi: 10.1128/jb.173.20.6515-6527.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Upper C D, Hirano S S. Predicting behavior of phyllosphere bacteria in the growth chamber from field studies. In: Morris C E, Nicot P C, Nguyen-The C, editors. Aerial plant surface microbiology. New York, N.Y: Plenum Press; 1996. pp. 277–284. [Google Scholar]
- 282.Upper C D, Vali G. The discovery of bacterial ice nucleation and its role in the injury of plants by frost. In: Lee R E Jr, Warren G J, Gusta L V, editors. Biological ice nucleation and its applications. St. Paul, Minn: American Phytopathological Society; 1995. pp. 29–39. [Google Scholar]
- 283.Vali G. Principles of ice nucleation. In: Lee R E Jr, Warren G J, Gusta L V, editors. Biological ice nucleation and its applications. St. Paul, Minn: American Phytopathological Society; 1995. pp. 1–28. [Google Scholar]
- 284.van Dijk K, Fouts D E, Rehm A H, Hill A R, Collmer A, Alfano J R. The Avr (effector) proteins HrmA (HopPsyA) and AvrPto are secreted in culture from Pseudomonas syringae pathovars via the Hrp (type III) protein secretion system in a temperature- and pH-sensitive manner. J Bacteriol. 1999;181:4790–4797. doi: 10.1128/jb.181.16.4790-4797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Van Outryve M F, Gosselé F, Swings J. The bacterial microflora of witloof chicory (Cichorium intybus L. var. foliosum Hegi) leaves. Microb Ecol. 1989;18:175–186. doi: 10.1007/BF02030125. [DOI] [PubMed] [Google Scholar]
- 286.Van Outryve M F, Gosselé V, Gosselé F, Swings J. Composition of the microflora of witloof chicory seeds. Microb Ecol. 1988;16:339–348. doi: 10.1007/BF02011705. [DOI] [PubMed] [Google Scholar]
- 287.Vasantharajan V N, Bhat J V. Interrelations of micro-organisms and mulberry. II. Phyllosphere microflora and nitrogen fixation in leaf and root surfaces. Plant Soil. 1968;28:258–267. [Google Scholar]
- 288.Venette J R. How bacteria find their hosts. In: Mount M S, Lacy G H, editors. Phytopathogenic prokaryotes. Vol. 2. New York, N.Y: Academic Press; 1982. pp. 3–30. [Google Scholar]
- 289.Walker J C, Patel P N. Splash dispersal and wind as factors in epidemiology of halo blight of bean. Phytopathology. 1964;54:140–141. [Google Scholar]
- 290.Warren G, Corotto L. The consensus sequence of ice nucleation proteins from Erwinia herbicola, Pseudomonas fluorescens and Pseudomonas syringae. Gene. 1989;85:239–242. doi: 10.1016/0378-1119(89)90488-5. [DOI] [PubMed] [Google Scholar]
- 291.Warren G, Corotto L, Wolber P. Conserved repeats in diverged ice nucleation structural genes from two species of Pseudomonas. Nucleic Acids Res. 1986;14:8047–8060. doi: 10.1093/nar/14.20.8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Warren G, Wolber P. Molecular aspects of microbial ice nucleation. Mol Microbiol. 1991;5:239–243. doi: 10.1111/j.1365-2958.1991.tb02104.x. [DOI] [PubMed] [Google Scholar]
- 293.Warren G, Wolber P, Green R. Functional significance of oligonucleotide repeats in a bacterial ice nucleation gene. In: Civerolo E L, Collmer A, Davis R E, Gillaspie A G, editors. Plant pathogenic bacteria. Dordrecht, The Netherlands: Martinus Nijhoff Publishers; 1987. pp. 1013–1017. [Google Scholar]
- 294.Warren G J. Bacterial ice nucleation: molecular biology and applications. Biotechnol Genet Eng Rev. 1987;5:107–135. [Google Scholar]
- 295.Warren G J. Identification and analysis of ina genes and proteins. In: Lee R E Jr, Warren G J, Gusta L V, editors. Biological ice nucleation and its applications. St. Paul, Minn: American Phytopathological Society; 1995. pp. 85–99. [Google Scholar]
- 296.Warren G J, Lindemann J, Suslow T V, Green R L. Ice nucleation-deficient bacteria as frost protection agents. In: LeBaron H M, Mumma R O, Honeycutt R C, Duesing J H, editors. Biotechnology in agricultural chemistry. Washington, D.C.: American Chemical Society; 1987. pp. 215–227. [Google Scholar]
- 297.Warren G J, Wolber P K. Heterogeneous ice nucleation by bacteria. Cryo Lett. 1987;8:204–215. [Google Scholar]
- 298.Watanabe N M, Southworth M W, Warren G J, Wolber P K. Rates of assembly and degradation of bacterial ice nuclei. Mol Microbiol. 1990;4:1871–1879. doi: 10.1111/j.1365-2958.1990.tb02036.x. [DOI] [PubMed] [Google Scholar]
- 299.Weier T E, Stocking C R, Barbour M G. Botany—an introduction to plant biology. 5th ed. New York, N.Y: John Wiley & Sons, Inc.; 1974. [Google Scholar]
- 300.Weingart H, Völksch B. Genetic fingerprinting of Pseudomonas syringae pathovars using ERIC-, REP-, and IS50-PCR. J Phytopathol. 1997;145:339–345. [Google Scholar]
- 301.Whistler C A, Cörbell N A, Sarniguet A, Ream W, Loper J E. The two-component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor ςS and the stress response in Pseudomonas fluorescens Pf-5. J Bacteriol. 1998;180:6635–6641. doi: 10.1128/jb.180.24.6635-6641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Reference deleted.
- 303.Reference deleted.
- 304.Willis D K, Hrabak E M, Rich J J, Barta T M, Lindow S E, Panopoulos N J. Isolation and characterization of a Pseudomonas syringae pv. syringae mutant deficient in lesion formation on bean. Mol Plant-Microbe Interact. 1990;3:149–156. [Google Scholar]
- 305.Willis D K, Rich J J, Hrabak E M. hrp genes of phytopathogenic bacteria. Mol Plant-Microbe Interact. 1991;4:132–138. [Google Scholar]
- 306.Willis D K, Rich J J, Kinscherf T G, Kitten T. Genetic regulation in plant pathogenic pseudomonads. In: Setlow J K, editor. Genetic engineering: principles and methods. Vol. 16. New York, N.Y: Plenum Press; 1994. pp. 167–193. [PubMed] [Google Scholar]
- 307.Wilson M, Hirano S S, Lindow S E. Location and survival of leaf-associated bacteria in relation to pathogenicity and potential for growth within the leaf. Appl Environ Microbiol. 1999;65:1435–1443. doi: 10.1128/aem.65.4.1435-1443.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Wilson M, Lindow S E. Relationship of total viable and culturable cells in epiphytic populations of Pseudomonas syringae. Appl Environ Microbiol. 1992;58:3908–3913. doi: 10.1128/aem.58.12.3908-3913.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Wilson M, Lindow S E. Ecological similarity and coexistence of epiphytic ice-nucleation (Ice+) Pseudomonas syringae strains and a non-ice-nucleating (Ice−) biological control agent. Appl Environ Microbiol. 1994;60:3128–3137. doi: 10.1128/aem.60.9.3128-3137.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Wilson M, Lindow S E. Coexistence among epiphytic bacterial populations mediated through nutritional resource partitioning. Appl Environ Microbiol. 1994;60:4468–4477. doi: 10.1128/aem.60.12.4468-4477.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Wilson M, Savka M A, Hwang I, Farrand S K, Lindow S E. Altered epiphytic colonization of mannityl opine-producing transgenic tobacco plants by a mannityl opine-catabolizing strain of Pseudomonas syringae. Appl Environ Microbiol. 1995;61:2151–2158. doi: 10.1128/aem.61.6.2151-2158.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Windels C E, Lindow S E, editors. Biological control on the phylloplane. St. Paul, Minn: American Phytopathological Society; 1985. [Google Scholar]
- 313.Wolber P, Warren G. Bacterial ice-nucleation proteins. TIBS Trends Biochem Sci. 1989;14:179–182. doi: 10.1016/0968-0004(89)90270-3. [DOI] [PubMed] [Google Scholar]
- 314.Wolber P K. Bacterial ice nucleation. Adv Microb Physiol. 1993;34:203–237. doi: 10.1016/s0065-2911(08)60030-2. [DOI] [PubMed] [Google Scholar]
- 315.Wolber P K, Deininger C A, Southwort M W, Vandekerckhove J, van Montagu M, Warren G J. Identification and purification of a bacterial ice-nucleation protein. Proc Natl Acad Sci USA. 1986;83:7256–7260. doi: 10.1073/pnas.83.19.7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Wolber P K, Warren G J. Evolutionary perspective on the ice nucleation gene-encoded membrane protein. In: Andrews J H, Hirano S S, editors. Microbial ecology of leaves. New York, N.Y: Springer-Verlag; 1991. pp. 315–330. [Google Scholar]
- 317.Yoshimura F. Phylloplane bacteria in a pine forest. Can J Microbiol. 1982;28:580–592. [Google Scholar]
- 318.Young J M. Pathogenicity and identification of the lilac pathogen, Pseudomonas syringae pv. syringae van Hall 1902. Ann Appl Biol. 1991;118:283–298. [Google Scholar]
- 319.Young J M, Bradbury J F, Davis R E, Dickey R S, Ercolani G L, Hayward A C, Vidaver A K. Nomenclatural revisions of plant pathogenic bacteria and list of names 1980–1988. Rev Plant Pathol. 1991;70:211–221. [Google Scholar]
- 320.Young J M, Dye D W, Bradbury J F, Panagopoulos C G, Robbs C F. A proposed nomenclature and classification for plant pathogenic bacteria. N Z J Agric Res. 1978;21:153–177. [Google Scholar]
- 321.Young J M, Triggs C M. Evaluation of determinative tests for pathovars of Pseudomonas syringae van Hall 1902. J Appl Bacteriol. 1994;77:195–207. doi: 10.1111/j.1365-2672.1994.tb03064.x. [DOI] [PubMed] [Google Scholar]
- 322.Yu I-C, Parker J, Bent A F. Gene-for-gene disease resistance without the hypersensitive response in Arabidopsis dnd1 mutant. Proc Natl Acad Sci USA. 1998;95:7819–7824. doi: 10.1073/pnas.95.13.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Yu J, Peñaloza-Vázquez A, Chakrabarty A M, Bender C L. Involvement of the exopolysaccharide alginate in the virulence and epiphytic fitness of Pseudomonas syringae pv. syringae. Mol Microbiol. 1999;33:712–720. doi: 10.1046/j.1365-2958.1999.01516.x. [DOI] [PubMed] [Google Scholar]
- 324.Yuk M H, Harvill E T, Miller J F. The BvgAS virulence control system regulates type III secretion in Bordetella bronchiseptica. Mol Microbiol. 1998;28:945–959. doi: 10.1046/j.1365-2958.1998.00850.x. [DOI] [PubMed] [Google Scholar]
- 325.Zhao J-L, Orser C S. Conserved repetition in the ice nucleation gene inaX from Xanthomonas campestris pv. translucens. Mol Gen Genet. 1990;223:163–166. doi: 10.1007/BF00315811. [DOI] [PubMed] [Google Scholar]