Abstract
Introduction:
Sarcoidosis is a heterogeneous, granulomatous disease that can prove difficult to diagnose, with no accurate biomarkers of disease progression. Therefore, we profiled and integrated the DNA methylome, mRNAs, and microRNAs to identify molecular changes associated with sarcoidosis and disease progression that might illuminate underlying mechanisms of disease and potential genomic biomarkers.
Methods:
Bronchoalveolar lavage cells from 64 sarcoidosis subjects and 16 healthy controls were used. DNA methylation was profiled on Illumina HumanMethylationEPIC arrays, mRNA by RNA-sequencing, and miRNAs by small RNA-sequencing. Linear models were fit to test for effect of diagnosis and phenotype, adjusting for age, sex, and smoking. We built a supervised multi-omics model using a subset of features from each dataset.
Results:
We identified 46,812 CpGs, 1,842 mRNAs, and 5 miRNAs associated with sarcoidosis versus controls and 1 mRNA, SEPP1 - a protein that supplies selenium to cells, associated with disease progression. Our integrated model emphasized the prominence of the PI3K/AKT1 pathway in sarcoidosis, which is important in T cell and mTOR function. Novel immune related genes and miRNAs including LYST, RGS14, SLFN12L, and hsa-miR-199b-5p, distinguished sarcoidosis from controls. Our integrated model also demonstrated differential expression/methylation of IL20RB, ABCC11, SFSWAP, AGBL4, miR-146a-3p, and miR-378b between non-progressive and progressive sarcoidosis.
Conclusions:
Leveraging the DNA methylome, transcriptome, and miRNA-sequencing in sarcoidosis BAL cells, we detected widespread molecular changes associated with disease, many which are involved in immune response. These molecules may serve as diagnostic/prognostic biomarkers and/or drug targets, although future testing will be required for confirmation.
Introduction
Sarcoidosis is a heterogeneous disease characterized by non-caseating granulomatous inflammation that differentially impacts Black individuals and women1. The lungs are involved in over 90% of individuals2. Those with pulmonary sarcoidosis can be asymptomatic or demonstrate remission or resolution; however, progression can result in impairment and/or pulmonary fibrosis, the main cause of mortality3. The course of pulmonary sarcoidosis is unpredictable, with at least 25% of patients developing chronic or progressive disease requiring treatment4. Driven by a combination of genetic, environmental, and host immunologic factors, the underlying cause(s) of sarcoidosis are currently unknown. Multiple lines of evidence point towards an antigenic stimulus including HLA allele associations, environmental, seasonal, and regional patterns, and a Th1 predominate immune response in which CD4+ T cells secrete IFN-γ and TNF-α5. In addition, aberrant and dysfunctional immune responses are associated with sarcoidosis and supported by genome wide transcriptome studies6. While previous studies have elucidated many contributors to disease development and progression, many knowledge gaps remain.
Epigenetic mechanisms such as DNA methylation (DNAm) and microRNAs (miRNAs) mediate gene expression, are modified by exposures, and are dynamic and reversible, making them candidates for gene regulation in sarcoidosis as well as promising biomarkers and therapeutic targets. Epigenetic dysregulation has been identified in many lung diseases and likely drives sarcoidosis progression and manifestations. We have previously demonstrated changes in DNAm and mRNA gene expression in bronchoalveolar lavage (BAL) cells from chronic beryllium disease (a granulomatous lung disease caused by beryllium exposure) patients and a small sample of sarcoidosis patients7,8. No other studies have evaluated epigenome-wide DNAm in sarcoidosis although others have identified miRNAs associated with disease9,10. Additionally, BALF miRNAs such as miR-27b, miR-192, and miR-221 have been associated with pulmonary sarcoidosis progression11. These studies were limited by small sample sizes, targeted as opposed to genome-wide approaches, and a lack of integration of different omics modalities.
With a larger integrated approach utilizing several genomic data types, we hypothesize that epigenomic studies could link risk factors to disease pathobiology to better understand disease course, and subclassify patients based on molecular profiling; ultimately this would direct focused research on disease manifestations and treatment. In a first step, we conducted this study to profile genome-wide DNAm, mRNA and miRNA expression in sarcoidosis BAL cells, stratified by disease progression. Analyzing each dataset separately, we identified many molecular features associated with disease overall. By next constructing a sparse multi-omic model incorporating DNAm, mRNAs, and miRNAs, we identified features associated with sarcoidosis and pulmonary progression.
Methods
Study population
Sarcoidosis BAL cell samples were obtained from the Genomic Research in Alpha-1 Antitrypsin Deficiency and Sarcoidosis (GRADS) consortium12 (including National Jewish Health (NJH, n=17) and non-NJH GRADS cases (n=39)) and cases from the Granuloma Biorepository at NJH (n=8). Controls with no history of lung disease were obtained from the NJH Donor Lung Core (n=16). All sarcoidosis subjects met the ATS/ERS criteria13 for tissue biopsy confirmation of diagnosis of sarcoidosis. The non-progressive phenotype was defined as having either acute (i.e. consistent with Lofgren’s syndrome) or non-acute disease presentation, no new organ involvement, lung function testing with <10% decline in FVC or FEV1, <15% decline in DLCO, and stable chest imaging within 2 years after BAL. The progressive phenotype had a non-acute disease presentation; lung function testing with ≥10% decline in FVC or FEV1; or ≥15% decline in DLCO; worsening chest imaging; and/or if they required initiation of systemic immunosuppressive treatment any time up to 2 years after BAL. Non-NJH GRADS cases were phenotyped based on disease acuity, PFT, and treatment status only. See Supplemental Methods for more details.
Bronchoscopy and nucleic acid processing
Bronchoscopy with BAL was performed as previously described.12,14 Cells were isolated and frozen at −80C in RLT buffer. DNA and RNA were extracted using the Qiagen AllPrep DNA/RNA extraction mini kit. Purified genomic DNA was bisulfite-converted with the Zymo EZ-96 DNA Methylation bisulfite conversion kit, followed by whole-genome amplification and enzymatic fragmentation. DNA was denatured and hybridized to Illumina Infinium HumanMethylationEPIC BeadChips, followed by single base extension. Hybridized BeadChips were stained, washed, and scanned using Illumina’s iScan System. mRNA libraries were prepared from 500 ng total RNA with TruSeq stranded mRNA library preparation kits (Illumina) and miRNA libraries were constructed using Lexogen Small RNA-Seq library preparation kits. RNA libraries were sequenced at an average depth of 80M 150bp paired-end reads on the Illumina NovaSeq 6000. RNA-Seq reads from an additional 28 samples were obtained from the GRADS consortium6. Additional details are outlined in the supplemental methods.
Data analysis
For each dataset, linear models were fit to each feature testing for an effect of sarcoidosis diagnosis or progressive vs. non-progressive disease while adjusting for age, sex, and smoking status. RNA-seq data was additionally adjusted for a surrogate variable capturing a batch effect of sequencing center15 (Supplementary Figure 1). Null test statistic distributions were derived for each analysis using bacon to reduce inflation16. P-values were adjusted to a 5% false discovery rate (FDR) to account for multiple testing using the Benjamini-Hochberg procedure17. Enrichment of significant results in the Gene Ontology (GO) resource18 and the Kyoto Encyclopedia of Genes and Genomes (KEGG)19 was performed using GOmeth20 for DNAm data and clusterProfiler21 for mRNA data. CpGs were annotated to CpG islands, shelves, and shores, and gene elements using the annotatr R package22. Experimentally confirmed miRNA target genes were obtained from MirTarBase23. For each miRNA, we retained target genes that had at least 2 sources of evidence (reporter assay, Western blot, qPCR, Microarray, NGS, pSILAC, CLIP-Seq, or other), including at least one source considered strong evidence (reporter assay, Western blot, or qPCR).
Data Integration Analysis for Biomarker discovery using Latent cOmponents (DIABLO)
Methylation M values and normalized mRNA and miRNA gene expression values were used as input for a DIABLO24 model implemented in the mixOmics R package25. Gene expression values were normalized to library size using DESeq226 and transformed to account for heteroscedasticity with a variance stabilizing transformation (VST)27. A subset of features from each dataset for 2 model components were selected for model input using LASSO regression and 5-fold cross validation repeated 10 times.
Results
Demographics of study population
We recruited 64 sarcoidosis cases, including 26 progressive and 38 non-progressive, and 16 healthy controls. Demographic information at time of BAL is displayed in Table 1. No significant differences were observed in race, ethnicity, or age. Non-progressive cases were more likely female than progressive cases. Progressive cases presented with significantly reduced FEV1 and FVC and more Stage 2 disease.
Table 1.
Demographic information of control, progressive sarcoidosis, and non-progressive sarcoidosis cases at time of bronchoscopy with lavage.
| Control | Non-Progressive | Progressive | p | |
|---|---|---|---|---|
| n | 16 | 38 | 26 | |
| Age (mean (SD)) | 55.62 (9.74) | 51.68 (9.88) | 51.19 (9.61) | 0.316* |
| Sex = Male (%) | 11 (68.8) | 11 (28.9) | 16 (61.5) | 0.006** |
| Race (%) | 0.242** | |||
| Asian | 1 (6.2) | 1 (2.6) | 0 (0.0) | |
| Black | 0 (0.0) | 6 (15.8) | 5 (19.2) | |
| White | 15 (93.8) | 31 (81.6) | 21 (80.8) | |
| Ethnicity = Non-Hispanic (%) | 15 (93.8) | 36 (94.7) | 25 (100.0) | 0.441** |
| Smoking Status = Former (%) | 0 | 12 (31.6) | 6 (23.1) | 0.0275** |
| FVC (mean (SD)) | 97.42 (11.25) | 85.92 (14.21) | 0.001# | |
| FEV1 (mean (SD)) | 101.03 (12.19) | 86.40 (15.04) | <0.001* | |
| DLCO (mean (SD)) | 87.71 (21.81) | 84.42 (12.64) | 0.491* | |
| Scadding Stage (%) | <0.001** | |||
| 0 | 13 (34.2) | 1 (3.8) | ||
| 1 | 20 (52.6) | 1 (3.8) | ||
| 2 | 4 (10.5) | 22 (84.6) | ||
| 3 | 1 (2.6) | 2 (7.7) | ||
One-way ANOVA
Fisher’s exact test
Mann-Whitney U test.
FVC: forced vital capacity. FEV1: forced expiratory volume over 1 second. DLCO: diffusing capacity of the lung for carbon monoxide.
Gene expression representing immune pathways differs between cases and controls and to a lesser degree by disease progression phenotypes
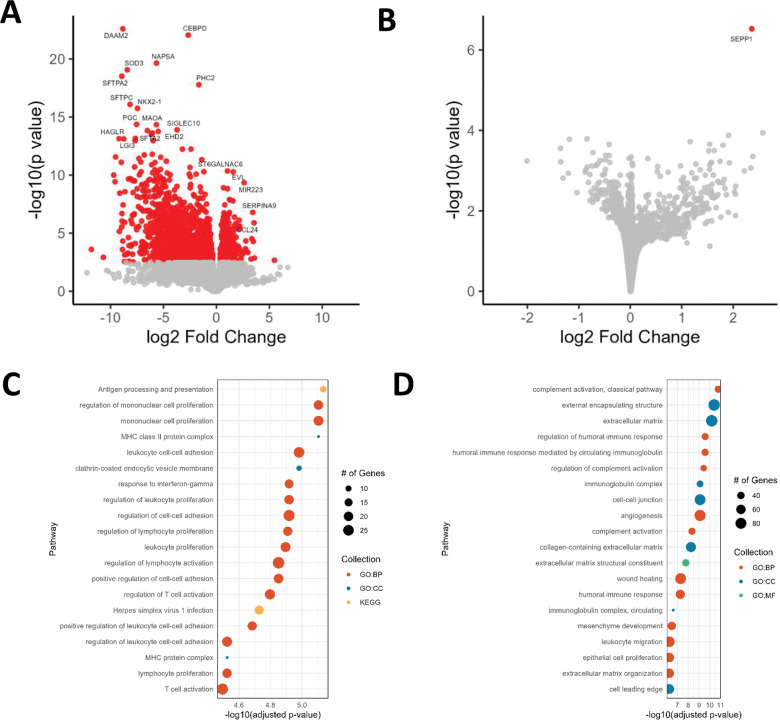
We analyzed gene expression data to assess differences between both cases and controls and progressive and non-progressive disease. In the sarcoidosis-control comparison, we detected 1,842 differentially expressed genes (DEGs), of which 379 (20.6%) were significantly increased in sarcoidosis (Figure 1A; Supplementary Table 1). Top significant upregulated genes involved many genes relating to immunity, cell migration, and cell adhesion including: EVL (Enah/Vasp-like) and SERPINA9 (serpin family A member 9). Most significant downregulated DEGs include SFTPA2 and SFTPC, surfactant genes and transcription factors such as CEBPD (CCAAT Enhancer Binding Protein Delta) and NKX2-1 (NK2 homeobox 1). A single DEG was found in progressive vs non-progressive cases: SEPP1 (Figure 1B).
Figure 1.
Differentially expressed genes in sarcoidosis. A) Differentially expressed genes in sarcoidosis vs. controls. B) Differentially expressed genes in progressive vs. non-progressive sarcoidosis. C) Pathway enrichment of upregulated mRNAs. D) Pathway enrichment of downregulated mRNAs.
We next tested whether sarcoidosis DEGs were overrepresented in GO18 and KEGG pathways using clusterProfiler21 (Figure 1C–D; Supplementary Table 2). Upregulated genes were enriched for 257 GO and 18 KEGG pathways. The most significantly enriched GO terms include MHC class II protein complex, regulation of mononuclear cell proliferation, leukocyte cell-cell adhesion, and response to interferon gamma. Top KEGG pathways include antigen processing and presentation, cell adhesion molecules, and multiple viral and autoimmune diseases including herpes simplex virus 1 infection, Epstein-Barr virus infection, viral myocarditis, Influenza A, tuberculosis, rheumatoid arthritis, and systemic lupus erythematosus. Downregulated genes were enriched for 512 GO and 0 KEGG pathways. The most significantly enriched GO terms include complement activation, classical pathway, extracellular matrix, regulation of humoral immune response, immunoglobulin complex, cell-cell junction, wound healing, mesenchyme development, and epithelial cell proliferation.
DNA methylation differs between cases and controls and overlaps with gene expression
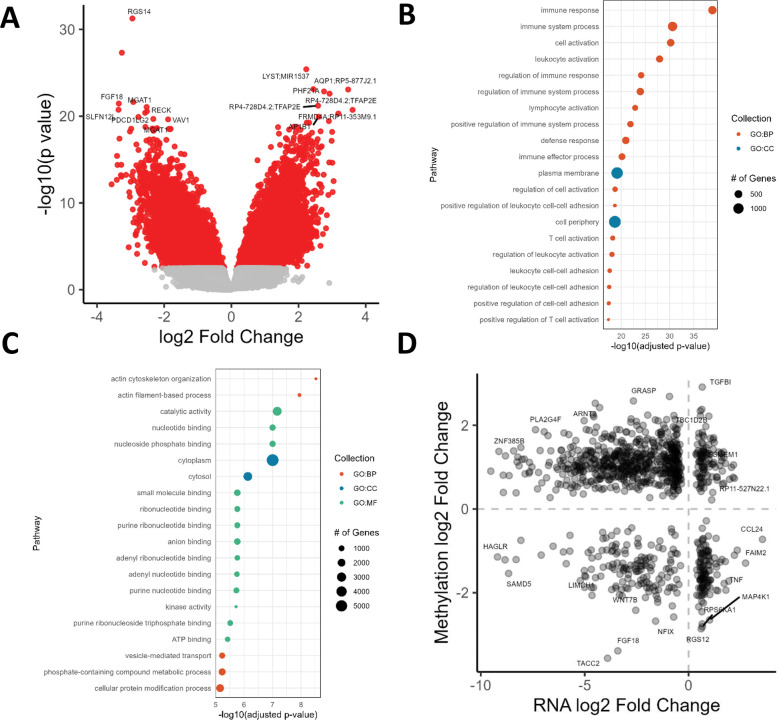
We next tested for differential methylation (DM) and identified 46,812 DNAm sites associated with sarcoidosis, of which 38,504 map to 15,208 unique genes (Figure 2A; Supplementary Table 3). We did not detect any DNAm sites significantly associated with disease progression. The majority of CpGs were hypermethylated in sarcoidosis (35,154; 75.1%). Significant probes were enriched for intronic regions (68.5% vs 60.8%; Fisher’s exact test p < 2.2 × 10−16) and FANTOM5 enhancers (9.17% vs 4.14%; Fisher’s exact test p < 2.2 × 10−16) relative to all tested DNAm sites. Hypomethylated DM sites were significantly enriched for 687 GO terms, with top hits including immune response, leukocyte activation, and defense response, and 88 KEGG pathways including viral protein interaction with cytokine and cytokine receptor, chemokine signaling pathway, Th17 cell differentiation, and T cell receptor signaling pathway (Figure 2B; Supplementary Table 4). Hypermethylated DM sites were significantly enriched for 146 GO terms, with top hits including actin cytoskeleton organization and 29 KEGG pathways including Rap1 signaling, Ras signaling, and phosphatidylinositol signaling system (Figure 2C). The most significant hypomethylated DM sites include CpGs that map to a predicted promoter region of RGS14, an intronic enhancer of FGF18, and MGAT1. Top hypermethylated DM sites mapped to genes such as LYST (lysosomal trafficking regulator), which regulates protein trafficking to lysosomes.
Figure 2.
Differentially methylated sites in sarcoidosis. A) Differentially methylated sites in sarcoidosis vs. controls. B) Pathway enrichment of hypomethylated DNAm sites. C) Pathway enrichment of hypermethylated DNAm sites. D) Differentially expressed genes with DM DNAm sites.
We next overlapped DEGs and differentially methylated probes (DMPs) in sarcoidosis vs controls based on gene ID. 961 DEGs (52.2%) showed DM of 12,297 unique CpGs. 52.9% of associations (41,674/78,761) showed canonical inverse relationships: the directionality of DNAm was opposite that of gene expression (Figure 2D). Genes with canonical relationships and upregulated RNA include TGFB1. Canonical relationships with downregulated RNA include AQP1, FRMD4A, ARNT2, and BMP4. Genes with non-canonical relationships and downregulated RNA include ITGA9, an integrin component of receptor for VCAM1, LIMCH1, which positively regulates stress fiber assembly and stabilizes focal adhesions, and FGF18.
miRNA expression differs between cases and controls and targets DEGs
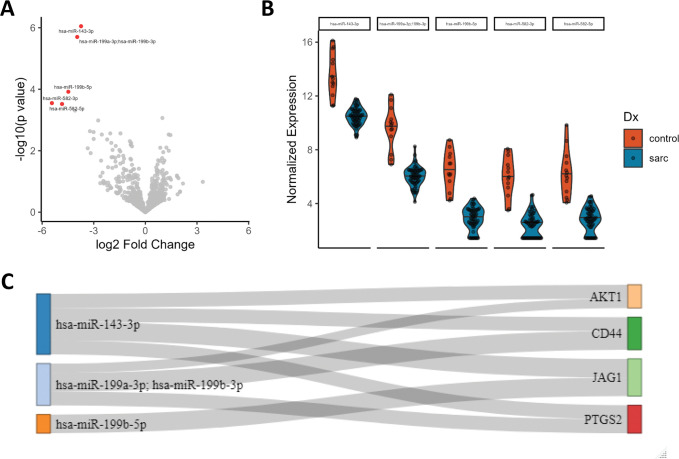
We next compared miRNA expression in sarcoidosis vs controls and identified 5 miRNAs (hsa-miR-143-3p, hsa-miR-199a-3p/hsa-miR-199b-3p, hsa-miR-199b-5p, hsa-miR-582-3p & hsa-miR-582-5p) downregulated in sarcoidosis (Figure 3A–B; Supplementary Table 5). No miRNAs were associated with progression. We derived experimentally validated target genes for each significant miRNA using MirTarBase23. We identified 67 target genes, of which 4 (AKT1, CD44, JAG1, PTGS2) are targeted by 2 DE miRNAs (Figure 3C), and a further 15 were DEGs in our mRNA analysis, including TNF.
Figure 3.
Differentially expressed microRNAs in sarcoidosis. A) Differentially expressed miRNAs in sarcoidosis vs. controls. B) Distribution of significant miRNAs’ expression in cases and controls. C) Sankey plot connecting DE miRNAs to target genes targeted by >1 DE miRNA. Connection width represents number of sources confirming relationship.
Integrated model reveals associations with sarcoidosis and with progression not found in individual analyses
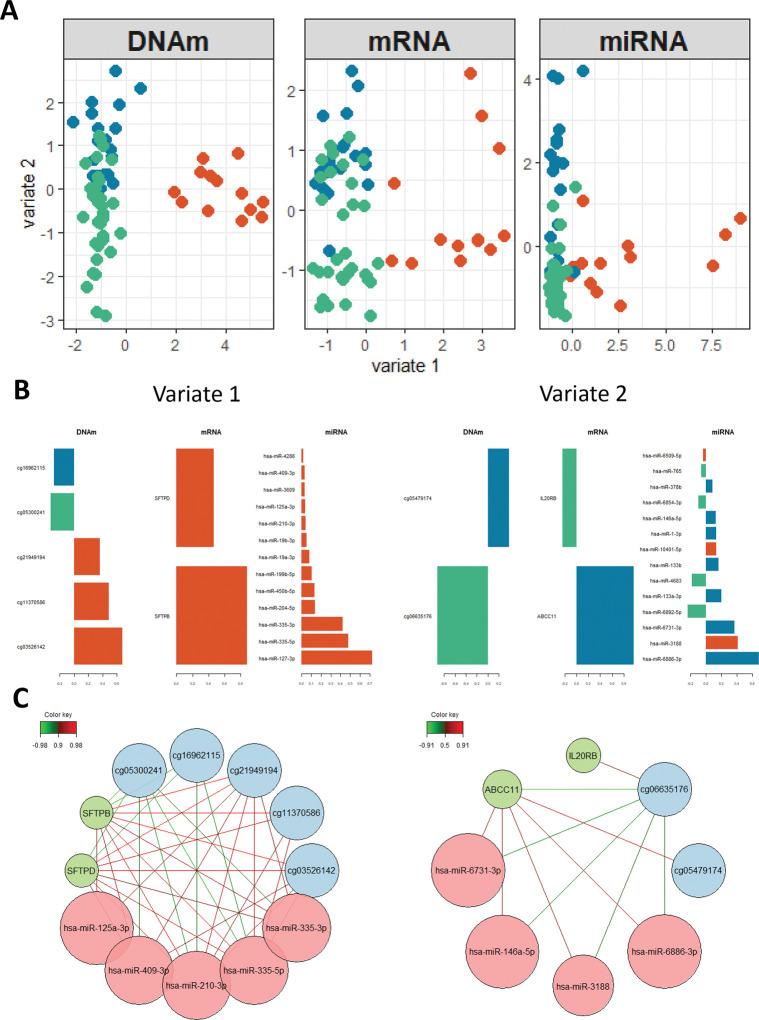
We used DIABLO to integrate the 3 datasets, including individuals present in all datasets after QC (n = 65; 13 controls, 19 progressive, 33 non-progressive sarcoidosis). With a goal to determine molecular features separating 3 groups (controls, progressive, and non-progressive sarcoidosis), we constructed a multi-omic model with 2 latent variables, which are linear combinations of input features. Using 5-fold cross validation repeated 10 times, we determined that selecting 5 DNAm sites, 2 mRNAs, and 13 miRNAs for latent variable 1 and 2 DNAm sites, 2 mRNAs, and 14 miRNAs for latent variable 2 maximized the prediction accuracy of the model and resulted in clustering based on diagnosis (Figure 4A; Supplementary Table 6). Latent variable 1 separates controls from sarcoidosis samples and latent variable 2 separates progressive from non-progressive sarcoidosis. Features included in each latent variable are shown in Figure 4B and Supplementary Table 7. We further constructed networks based on correlations of feature weights for each latent variable (Figure 4C). The two mRNAs contributing to latent variable 1 were SFTPB and SFTPD. DNAm sites contributing to latent variable 1 include DNAm sites hypermethylated in cases cg16962115, within an intron of LYST, and cg05300241 within an intron of PIK3CD. The remaining 3 DNAm sites were hypomethylated and included cg21949194 within an enhancer of SOS1 (SOS Ras/Rac guanine nucleotide exchange factor 1), cg11370586 within the predicted promoter of RGS14, and cg03526142 within an exon of SLFN12L. All DNAm sites contributing to latent variable 1 were DMPs in our previous methylation analysis. In addition to the DE miR hsa-miR-199b-5p, miRNA features on latent variable 1 include hsa-miR-204-5p. Features contributing to latent variable 2 include the DNAm sites cg05479174, within an exon of SFSWAP, and cg06635176 within an intron of AGBL4. IL20RB (interleukin 20 receptor subunit beta) mRNA was upregulated in non-progressive sarcoidosis, while ABCC11 (ATP binding cassette subfamily C member 11) expression was higher in progressive sarcoidosis. miRNAs contributing to latent variable 1 include hsa-miR-146a-3p and hsa-miR-378b.
Figure 4.
A sparse multi-omic model of sarcoidosis and progression. A) Projection of samples based on selected feature weights. B) Weights of selected features for latent variables. C) Network constructed from feature correlations on latent variables.
Discussion
In this study, we present the first application of multiomic integration in sarcoidosis, leveraging coding and miRNA expression with DNA methylation data to construct a multiomic network. Through initial genome-wide profiling of DNA methylation, mRNA expression, and miRNA expression in sarcoidosis BAL, we identified 1,846 DEGs, 46,812 DMPs, and 5 DE miRNAs in sarcoidosis relative to healthy controls, as well as 1 DEG associated with sarcoidosis progression. By integrating omic datasets, we define pathogenic molecules/genes in sarcoidosis not identified using conventional modeling methods in single-omics datasets. While our single-omics approach only demonstrated 1 DEG for progression, the multiomic model identified several progression-associated methylation, mRNA, and miRNA features. We identify previously reported molecules and pathways associated with disease as well as implicate novel molecular features as potential drivers and modifiers of sarcoidosis, thus demonstrating the potential of integrative approaches.
Our single-omic analyses implicate multiple genes in the pathogenesis of sarcoidosis, with many of these genes involved in processes relating to inflammation and immunity (potentially associated with those involved in viral/atypical bacterial infections and autoimmunity) and extracellular matrix signaling. While we detected many associations with case status, we detected only a single omic association with progression: SEPP1 mRNA levels. Results from the GRADS study demonstrated that SEPP1 levels were inversely correlated with DLCO (%) and FVC (% pred) in sarcoidosis individuals6. These finding support a progressive phenotype, including lung function changes that we and others have used in the definition of progressive pulmonary disease.
Our study also demonstrated 5 DE miRNAs downregulated in sarcoidosis cases in the first study of genome-wide miRNAs in sarcoidosis. While these associations are novel, multiple target genes of these miRNAs have been implicated in sarcoidosis previously, including the genes AKT1, CD44, JAG1, and PTGS2. For example, CD44 has been found in areas of granuloma formation and fibrosis28 and is differentially expressed between Lofgren syndrome versus non-Lofgren syndrome subjects29. Through integrating genes with evidence of both DE and DM, we identified biologically relevant genes in sarcoidosis versus controls, including HIF1A and TGFB1. A previous study showed that HIF1A protein and mRNA levels were decreased in sarcoidosis granulomas30. We and others have found alterations in TGFB1, which encodes TGF-β, in pulmonary sarcoidosis31,32. Additionally, TGF-β genotypes have been associated with sarcoidosis severity31.
Our integrated model revealed several novel genes and miRNAs between sarcoidosis and controls, including LYST, RGS14, SLFN12L, and hsa-miR-199b-5p. The first 3 genes appear to have important roles in immune function. Specifically, LYST, a regulator of endosome/lysosome trafficking, can regulate TLR3 and TLR4 mediated pathways33, genes involved in the innate immune system which have been implicated in sarcoidosis6,34,35. RGS14 is expressed in lymphocytes and regulates chemokine receptors to control immune responses to exogenous agents36. Finally, SLFN21L regulates thymocyte development and is downregulated in T-cell activation, suggesting a role as an immune response regulator37.
Our integrated analyses demonstrated miRNAs DE between progressive/non-progressive sarcoidosis including those previously described. Specifically, miR-146a-3p upregulated in progressive sarcoidosis is an indicator of inflammation and oxidative stress that may target TLR4 and was previously found elevated in sarcoidosis BALF, as well as serum38,39. miR-378b, which was upregulated in progressive cases, was previously found associated with sarcoidosis40. We also identified novel genes including IL20RB, ABCC11, SFSWAP and AGBL4. Interestingly, IL20RB expression was increased in non-progressive versus progressive sarcoidosis in our study, suggesting that the activation of this inflammatory pathway is increased in the non-progressive phenotype. ABCC11 is a gene that influences macrophage differentiation and induces TNF-α and IL17 through TLR4 signaling41; these results as well as the novel findings above support the importance of innate immune response genes in sarcoidosis.
Both our individual and integrative analyses identify molecules involved in PI3K/AK1 signaling, a pathway already recognized in sarcoidosis pathogenesis. For example, we observe hypermethylation of PI3KCD (phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta), which encodes a component of PI3K, and complexes with AKT1 to impact T cell differentiation and function42. Our miRNA results demonstrate AKT1 as a target of downregulated miRNAs (miR-143-3p and miR-199a/miR-199b) in sarcoidosis. Both downregulation and upregulation of the PI3K/AKT signaling pathways have been associated with sarcoidosis42,43. Previous studies demonstrated both reduced proliferative response and exhaustion of T cells in progressive sarcoidosis is thought to be driven in part by inhibition of PI3K/AKT1 signaling42. The PI3K/AKT1 pathway has also been implicated in activation of mTOR43, which has been associated with granuloma formation44. Interestingly, in the recent GRADS study, PI3K activation was associated with an endotype of sarcoidosis characterized by hilar lymphadenopathy, pulmonary reticulation, less multiorgan involvement, and more environmental associations6.
While our sample size is larger than most previous sarcoidosis omic studies, power in our analyses was limited, especially in our phenotype analyses. In future studies, it will be important to both increase sample sizes and investigate associations with better-powered quantitative measures related to disease such as pulmonary function testing variables. We were unable to correct for cell proportions in sarcoidosis subjects relative to controls as our controls lacked cell differentials. Altered cell proportions may explain the downregulated epithelial genes in sarcoidosis BAL (e.g. decreased surfactant proteins in sarcoidosis) and some of the DNAm sites and DEGs detected in sarcoidosis versus control comparisons, although in general epithelial cells are not a large proportion of BAL. Finally, our multiomic models did not take into account demographic information. Regressing out demographic information such as age and sex may lead to more predictive networks. Despite these shortcomings, we detected widespread molecular changes and constructed multiomic networks associated with sarcoidosis as well as progression. Molecules discovered in these analyses shed light on disease pathogenesis and may also be leveraged therapeutically or as biomarkers after replication/validation in additional populations.
Supplementary Material
Supplementary Figure 1. Accounting for RNA-Sequencing batch effect. A) Principal component analysis of mRNA data, colored by sequencing location. B) Surrogate variable analysis of mRNA data, colored by sequencing location. C) Principal component analysis of mRNA data after regressing out SV1.
Supplementary Table 1. Differentially expressed genes associated with sarcoidosis.
Supplementary Table 2. Pathway over-representation analysis of DEGs.
Supplementary Table 3. Differentially methylated probes associated with sarcoidosis.
Supplementary Table 4. Pathway over-representation analysis of DMPs.
Supplementary Table 5. Differentially expressed miRNAs associated with sarcoidosis.
Supplementary Table 6. DIABLO cross-validation balanced error rates.
Supplementary Table 7. DIABLO loadings.
Acknowledgments
Supported by NIH awards R01HL140357, TL1TR0025331 and UL1TR001082; FSR grant 22-505-RFP.
References
- 1.Baughman R.P., Field S., Costabel U., Crystal R.G., Culver D.A., Drent M., Judson M.A., and Wolff G. (2016). Sarcoidosis in America. Analysis Based on Health Care Use. Annals of the American Thoracic Society 13, 1244–1252. 10.1513/AnnalsATS.201511-760OC. [DOI] [PubMed] [Google Scholar]
- 2.Baughman R.P., Teirstein A.S., Judson M.A., Rossman M.D., Yeager H., Bresnitz E.A., DePalo L., Hunninghake G., Iannuzzi M.C., Johns C.J., et al. (2001). Clinical Characteristics of Patients in a Case Control Study of Sarcoidosis. American Journal of Respiratory and Critical Care Medicine 164, 1885–1889. 10.1164/ajrccm.164.10.2104046. [DOI] [PubMed] [Google Scholar]
- 3.Swigris J.J., Olson A.L., Huie T.J., Fernandez-Perez E.R., Solomon J., Sprunger D., and Brown K.K. (2011). Sarcoidosis-related Mortality in the United States from 1988 to 2007. American Journal of Respiratory and Critical Care Medicine 183, 1524–1530. 10.1164/rccm.201010-1679OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerke A.K., Judson M.A., Cozier Y.C., Culver D.A., and Koth L.L. (2017). Disease Burden and Variability in Sarcoidosis. Annals of the American Thoracic Society 14, S421–S428. 10.1513/AnnalsATS.201707-564OT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asano M., Minagawa T., Ohmichi M., and Hiraga Y. (1991). Detection of endogenous cytokines in sera or in lymph nodes obtained from patients with sarcoidosis. Clin Exp Immunol 84, 92–96. 10.1111/j.1365-2249.1991.tb08129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vukmirovic M., Yan X., Gibson K.F., Gulati M., Schupp J.C., DeIuliis G., Adams T.S., Hu B., Mihaljinec A., Woolard T.N., et al. (2021). Transcriptomics of bronchoalveolar lavage cells identifies new molecular endotypes of sarcoidosis. Eur Respir J 58. 10.1183/13993003.02950-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang I.V., Konigsberg I., MacPhail K., Li L., Davidson E.J., Mroz P.M., Hamzeh N., Gillespie M., Silveira L.J., Fingerlin T.E., and Maier L.A. (2019). DNA Methylation Changes in Lung Immune Cells Are Associated with Granulomatous Lung Disease. American Journal of Respiratory Cell and Molecular Biology 60, 96–105. 10.1165/rcmb.2018-0177OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li L., Konigsberg I.R., Bhargava M., Liu S., MacPhail K., Mayer A., Davidson E.J., Liao S.Y., Lei Z., Mroz P.M., et al. (2022). Multiomic Signatures of Chronic Beryllium Disease Bronchoalveolar Lavage Cells Relate to T-Cell Function and Innate Immunity. Am J Respir Cell Mol Biol 67, 632–640. 10.1165/rcmb.2022-0077OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ascoli C., Huang Y., Schott C., Turturice B.A., Metwally A., Perkins D.L., Finn P.W., and Group A.R. (2018). A Circulating MicroRNA Signature Serves as a Diagnostic and Prognostic Indicator in Sarcoidosis. Am J Respir Cell Mol Biol 58, 40–54. 10.1165/rcmb.2017-0207OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou T., Casanova N., Pouladi N., Wang T., Lussier Y., Knox K.S., and Garcia J.G.N. (2017). Identification of Jak-STAT signaling involvement in sarcoidosis severity via a novel microRNA-regulated peripheral blood mononuclear cell gene signature. Sci Rep 7, 4237. 10.1038/s41598-017-04109-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kiszalkiewicz J., Piotrowski W.J., Pastuszak-Lewandoska D., Gorski P., Antczak A., Gorski W., Domanska-Senderowska D., Migdalska-Sek M., Czarnecka K.H., Nawrot E., and Brzezianska-Lasota E. (2016). Altered miRNA expression in pulmonary sarcoidosis. BMC Med Genet 17, 2. 10.1186/s12881-016-0266-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moller D.R., Koth L.L., Maier L.A., Morris A., Drake W., Rossman M., Leader J.K., Collman R.G., Hamzeh N., Sweiss N.J., et al. (2015). Rationale and Design of the Genomic Research in Alpha-1 Antitrypsin Deficiency and Sarcoidosis (GRADS) Study. Sarcoidosis Protocol. Ann Am Thorac Soc 12, 1561–1571. 10.1513/AnnalsATS.201503-172OT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crouser E.D., Maier L.A., Wilson K.C., Bonham C.A., Morgenthau A.S., Patterson K.C., Abston E., Bernstein R.C., Blankstein R., Chen E.S., et al. (2020). Diagnosis and Detection of Sarcoidosis. An Official American Thoracic Society Clinical Practice Guideline. American Journal of Respiratory and Critical Care Medicine 201, e26–e51. 10.1164/rccm.202002-0251ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maier L.A., Sawyer R.T., Tinkle S.S., Kittle L.A., Barker E.A., Balkissoon R., Rose C., and Newman L.S. (2001). IL-4 fails to regulate in vitro beryllium-induced cytokines in berylliosis. Eur Respir J 17, 403–415. 10.1183/09031936.01.17304030. [DOI] [PubMed] [Google Scholar]
- 15.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., and Storey J.D. (2012). The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883. 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Iterson M., van Zwet E.W., and Heijmans B.T. (2017). Controlling bias and inflation in epigenome- and transcriptome-wide association studies using the empirical null distribution. Genome Biology 18. 10.1186/s13059-016-1131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benjamini Y., and Hochberg Y. (1995). Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 57, 289–300. [Google Scholar]
- 18.Carbon S., Douglass E., Good B.M., Unni D.R., Harris N.L., Mungall C.J., Basu S., Chisholm R.L., Dodson R.J., Hartline E., et al. (2021). The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Research 49, D325–D334. 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M., Furumichi M., Sato Y., Ishiguro-Watanabe M., and Tanabe M. (2021). KEGG: integrating viruses and cellular organisms. Nucleic Acids Research 49, D545–D551. 10.1093/nar/gkaa970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maksimovic J., Oshlack A., and Phipson B. (2021). Gene set enrichment analysis for genome-wide DNA methylation data. Genome Biology 22. 10.1186/s13059-021-02388-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. (2021). clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. The Innovation 2. 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cavalcante R.G., and Sartor M.A. (2017). annotatr: genomic regions in context. Bioinformatics 33, 2381–2383. 10.1093/bioinformatics/btx183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang H.Y., Lin Y.C., Cui S., Huang Y., Tang Y., Xu J., Bao J., Li Y., Wen J., Zuo H., et al. (2021). miRTarBase update 2022: an informative resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 10.1093/nar/gkab1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh A., Shannon C.P., Gautier B., Rohart F., Vacher M., Tebbutt S.J., and Le Cao K.A. (2019). DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics 35, 3055–3062. 10.1093/bioinformatics/bty1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneidman D., Rohart F., Gautier B., Singh A., and Lê Cao K.-A. (2017). mixOmics: An R package for ‘omics feature selection and multiple data integration. PLOS Computational Biology 13. 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love M.I., Huber W., and Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anders S., and Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol 11, R106. 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aleksoniene R., Besusparis J., Gruslys V., Jurgauskiene L., Laurinaviciene A., Laurinavicius A., Malickaite R., Norkuniene J., Zablockis R., Zurauskas E., and Danila E. (2021). CD31(+), CD38(+), CD44(+), and CD103(+) lymphocytes in peripheral blood, bronchoalveolar lavage fluid and lung biopsy tissue in sarcoid patients and controls. J Thorac Dis 13, 2300–2318. 10.21037/jtd-20-2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaiser Y., Lakshmikanth T., Chen Y., Mikes J., Eklund A., Brodin P., Achour A., and Grunewald J. (2017). Mass Cytometry Identifies Distinct Lung CD4(+) T Cell Patterns in Lofgren’s Syndrome and Non-Lofgren’s Syndrome Sarcoidosis. Front Immunol 8, 1130. 10.3389/fimmu.2017.01130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tzouvelekis A., Ntolios P., Karameris A., Koutsopoulos A., Boglou P., Koulelidis A., Archontogeorgis K., Zacharis G., Drakopanagiotakis F., Steiropoulos P., et al. (2012). Expression of hypoxia-inducible factor (HIF)-1a-vascular endothelial growth factor (VEGF)-inhibitory growth factor (ING)-4- axis in sarcoidosis patients. BMC Res Notes 5, 654. 10.1186/1756-0500-5-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonth A.C., Silveira L., Fingerlin T.E., Sato H., Luby J.C., Welsh K.I., Rose C.S., Newman L.S., du Bois R.M., Maier L.A., and Group A. (2007). TGF-beta 1 variants in chronic beryllium disease and sarcoidosis. J Immunol 179, 4255–4262. 10.4049/jimmunol.179.6.4255. [DOI] [PubMed] [Google Scholar]
- 32.Kobak S., Akyildiz M., Gokduman A., Atabay T., and Vural H. (2021). Serum galectin-3 and TGF-beta levels in patients with sarcoidosis. Reumatol Clin (Engl Ed) 17, 562–565. 10.1016/j.reumae.2020.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Westphal A., Cheng W., Yu J., Grassl G., Krautkramer M., Holst O., Foger N., and Lee K.H. (2017). Lysosomal trafficking regulator Lyst links membrane trafficking to toll-like receptor-mediated inflammatory responses. J Exp Med 214, 227–244. 10.1084/jem.20141461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiken M., Grunewald J., Eklund A., and Wahlstrom J. (2009). Higher monocyte expression of TLR2 and TLR4, and enhanced pro-inflammatory synergy of TLR2 with NOD2 stimulation in sarcoidosis. J Clin Immunol 29, 78–89. 10.1007/s10875-008-9225-0. [DOI] [PubMed] [Google Scholar]
- 35.Schurmann M., Kwiatkowski R., Albrecht M., Fischer A., Hampe J., Muller-Quernheim J., Schwinger E., and Schreiber S. (2008). Study of Toll-like receptor gene loci in sarcoidosis. Clin Exp Immunol 152, 423–431. 10.1111/j.1365-2249.2008.03621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moratz C., Harrison K., and Kehrl J.H. (2004). Regulation of chemokine-induced lymphocyte migration by RGS proteins. Methods Enzymol 389, 15–32. 10.1016/S0076-6879(04)89002-5. [DOI] [PubMed] [Google Scholar]
- 37.Puck A., Aigner R., Modak M., Cejka P., Blaas D., and Stockl J. (2015). Expression and regulation of Schlafen (SLFN) family members in primary human monocytes, monocyte-derived dendritic cells and T cells. Results Immunol 5, 23–32. 10.1016/j.rinim.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dyskova T., Fillerova R., Novosad T., Kudelka M., Zurkova M., Gajdos P., Kolek V., and Kriegova E. (2015). Correlation Network Analysis Reveals Relationships between MicroRNAs, Transcription Factor T-bet, and Deregulated Cytokine/Chemokine-Receptor Network in Pulmonary Sarcoidosis. Mediators Inflamm 2015, 121378. 10.1155/2015/121378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novosadova E., Chabronova A., Kolek V., Petrek M., and Navratilova Z. (2016). The Serum Expression of Selected miRNAs in Pulmonary Sarcoidosis with/without Lofgren’s Syndrome. Mediators Inflamm 2016, 1246129. 10.1155/2016/1246129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jazwa A., Kasper L., Bak M., Sobczak M., Szade K., Jozkowicz A., Sladek K., and Dulak J. (2015). Differential inflammatory microRNA and cytokine expression in pulmonary sarcoidosis. Arch Immunol Ther Exp (Warsz) 63, 139–146. 10.1007/s00005-014-0315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogl T., Tenbrock K., Ludwig S., Leukert N., Ehrhardt C., van Zoelen M.A., Nacken W., Foell D., van der Poll T., Sorg C., and Roth J. (2007). Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat Med 13, 1042–1049. 10.1038/nm1638. [DOI] [PubMed] [Google Scholar]
- 42.Celada L.J., Rotsinger J.E., Young A., Shaginurova G., Shelton D., Hawkins C., and Drake W.P. (2017). Programmed Death-1 Inhibition of Phosphatidylinositol 3-Kinase/AKT/Mechanistic Target of Rapamycin Signaling Impairs Sarcoidosis CD4(+) T Cell Proliferation. Am J Respir Cell Mol Biol 56, 74–82. 10.1165/rcmb.2016-0037OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhargava M., Viken K.J., Barkes B., Griffin T.J., Gillespie M., Jagtap P.D., Sajulga R., Peterson E.J., Dincer H.E., Li L., et al. (2020). Novel protein pathways in development and progression of pulmonary sarcoidosis. Sci Rep 10, 13282. 10.1038/s41598-020-69281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Linke M., Pham H.T., Katholnig K., Schnoller T., Miller A., Demel F., Schutz B., Rosner M., Kovacic B., Sukhbaatar N., et al. (2017). Chronic signaling via the metabolic checkpoint kinase mTORC1 induces macrophage granuloma formation and marks sarcoidosis progression. Nat Immunol 18, 293–302. 10.1038/ni.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Accounting for RNA-Sequencing batch effect. A) Principal component analysis of mRNA data, colored by sequencing location. B) Surrogate variable analysis of mRNA data, colored by sequencing location. C) Principal component analysis of mRNA data after regressing out SV1.
Supplementary Table 1. Differentially expressed genes associated with sarcoidosis.
Supplementary Table 2. Pathway over-representation analysis of DEGs.
Supplementary Table 3. Differentially methylated probes associated with sarcoidosis.
Supplementary Table 4. Pathway over-representation analysis of DMPs.
Supplementary Table 5. Differentially expressed miRNAs associated with sarcoidosis.
Supplementary Table 6. DIABLO cross-validation balanced error rates.
Supplementary Table 7. DIABLO loadings.