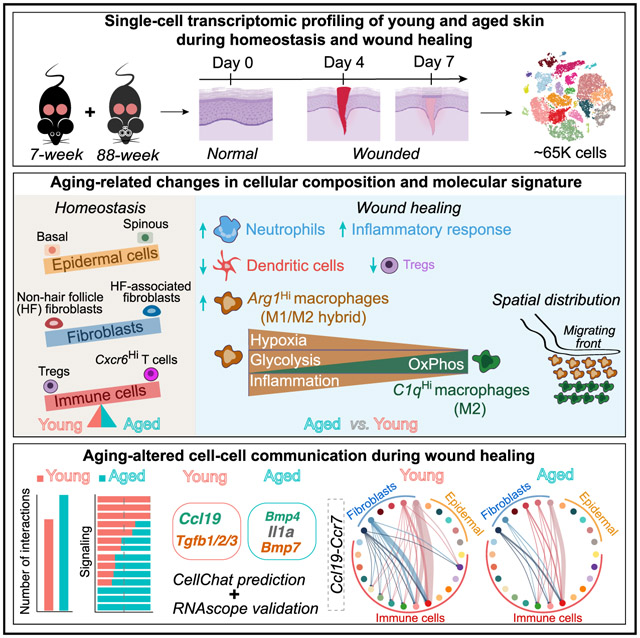
SUMMARY
Delayed and often impaired wound healing in the elderly presents major medical and socioeconomic challenges. A comprehensive understanding of the cellular/molecular changes that shape complex cell-cell communications in aged skin wounds is lacking. Here, we use single-cell RNA sequencing to define the epithelial, fibroblast, immune cell types, and encompassing heterogeneities in young and aged skin during homeostasis and identify major changes in cell compositions, kinetics, and molecular profiles during wound healing. Our comparative study uncovers a more pronounced inflammatory phenotype in aged skin wounds, featuring neutrophil persistence and higher abundance of an inflammatory/glycolytic Arg1Hi macrophage subset that is more likely to signal to fibroblasts via interleukin (IL)-1 than in young counterparts. We predict systems-level differences in the number, strength, route, and signaling mediators of putative cell-cell communications in young and aged skin wounds. Our study exposes numerous cellular/molecular targets for functional inter-rogation and provides a hypothesis-generating resource for future wound healing studies.
Graphical Abstract
In brief
A comprehensive understanding of cellular and molecular factors that underlie aging-induced decline in wound repair can expose targets for remedy. Vu et al. report single-cell and system-level analysis of young and aged skin during homeostasis and wound healing that reveals aging-related differences in cellular composition, molecular signature, and intercellular communication.
INTRODUCTION
Efficient and robust repair of tissue injury is crucial for survival, but this ability, along with tissue function itself, declines with age. Skin, body’s essential protective barrier, becomes thinner and drier, and cutaneous wound healing is delayed upon aging (Ding et al., 2021; Ge et al., 2020; Keyes et al., 2016; Wang and Dreesen, 2018). Healing of skin wounds is a concerted effort involving the actions of, and dynamic interactions among, a multitude of cell types in three overlapping phases: (1) inflammatory, characterized by blood clot formation and immune cell infiltration; (2) proliferative, characterized by proliferation and migration of keratinocytes and fibroblasts; and (3) resolution, where local inflammation subdues and extracellular matrix (ECM) is re-modeled (Eming et al., 2014; Minutti et al., 2017; Rognoni and Watt, 2018). Wound healing in aged skin associates with alterations in multiple stages, including prolonged inflammation and impaired immune cell function/signaling, increased cellular senescence, as well as delayed re-epithelialization, re-vascularization, granulation tissue formation, and wound closure (Bonham et al., 2020; Ding et al., 2021; Gosain and DiPietro, 2004; Sgonc and Gruber, 2013; Wang and Dreesen, 2018). However, the cellular and molecular mechanisms that coordinately contribute to such declined healing remain elusive. Particularly, a comprehensive picture of how cellular compositions and cell-cell communications in aged skin wounds differ from that in the young are still lacking. Single-cell RNA sequencing (scRNA-seq) presents a powerful approach to address these critical knowledge gaps.
We have recently developed a computational tool, CellChat (Jin et al., 2021), to analyze and compare intercellular communication networks. CellChat infers cell-cell communications from scRNA-seq data by computing the expression of signaling ligands, receptors, as well as soluble agonists, antagonists, and stimulatory or inhibitory membrane-bound co-receptors. However, the initial version of CellChat has a limited capability in systematically probing the underlying dysregulated intercellular communications that often lead to altered cellular composition and tissue phenotype.
Here we use scRNA-seq to dissect the microenvironmental cell community of unwounded (UW) and wounded skin of young and aged mice. Our analysis reveals altered abundance and molecular characteristics of multiple cell type/states as well as an overall more inflammatory profile in aged skin wounds featuring increased abundance of an Arg1Hi macrophage subset. We develop a comparative CellChat framework that enables the detection of signaling communication changes at different levels, including dysregulated cell populations, signaling pathways, and specific ligand-receptor pairs. Using this framework, we systematically identify aging-induced dysregulation in intercellular communications during wound healing, which are characterized by multi-compartmental alterations in growth factor, chemokine, and cytokine pathways.
RESULTS
Young and aged skin exhibit baseline differences across epithelial, fibroblast, and immune cell types during homeostasis
Previously, we sequenced single cells from UW and wounded back skin at day 4 post-wounding (4dpw) of young (6- to 8-week-old) mice (Haensel et al., 2020). To explore aging-associated changes in the heterogeneous wound microenvironment, we performed additional scRNA-seq experiments on young mice at 7dpw and aged (88-week-old) mice at comparable timepoints (UW, 4dpw, 7dpw) (Figure 1A). After quality control, we obtained sequences from 47,729 cells from three UW (14,549 cells), four 4dpw (19,465 cells), and three 7dpw (13,715 cells) samples for young mice, and a total of 17,106 cells from two UW (7,553 cells), two 4dpw (4,112 cells), two 7dpw (5,441 cells) samples for aged mice for downstream analysis (Figures S1A and S1B).
Figure 1. Heterogeneity of epidermal cells, fibroblasts, and immune cells in young and aged skin during homeostasis.
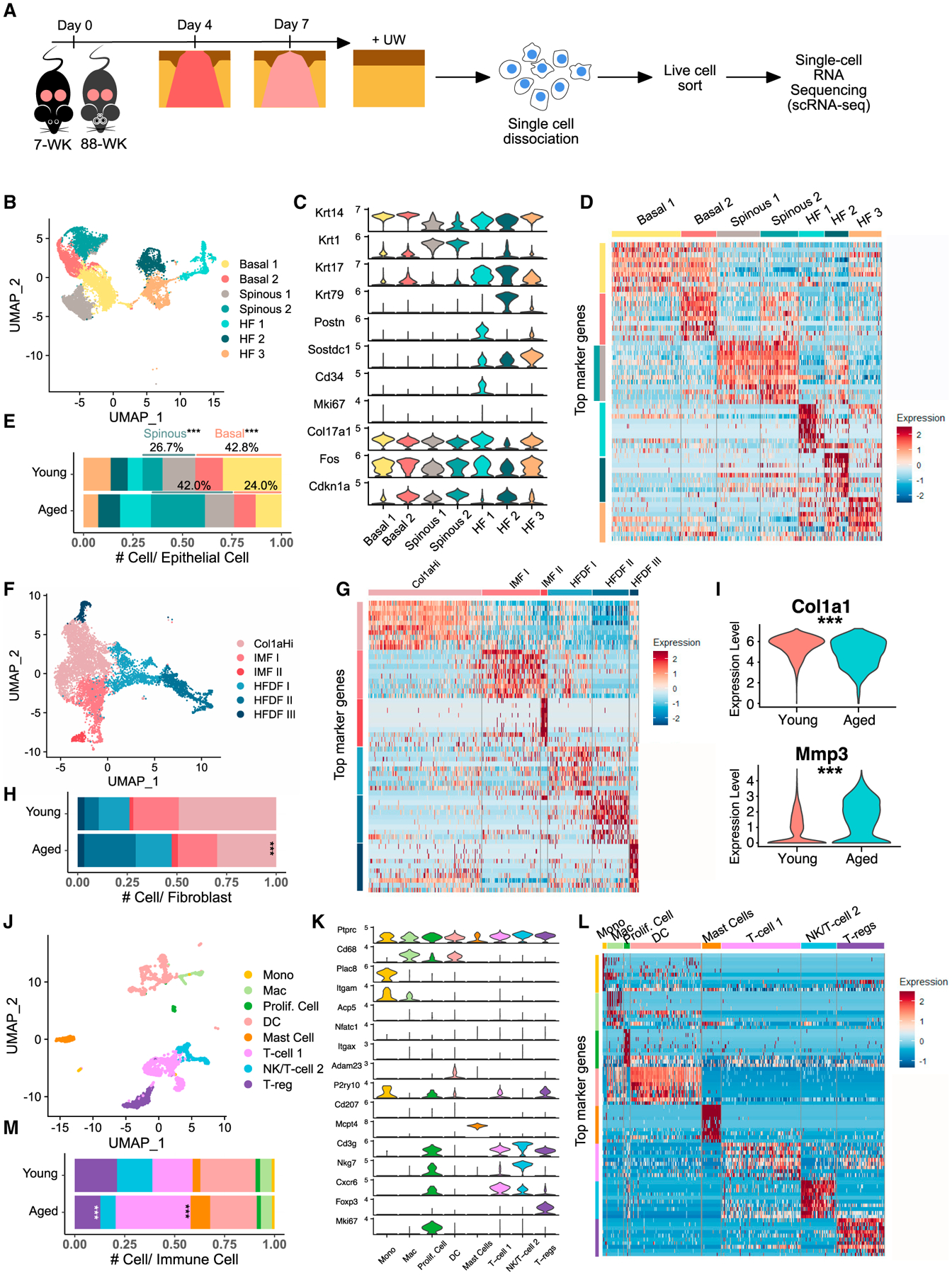
(A) Schematic diagram detailing single-cell isolation and collection.
(B) Uniform manifold approximation and projection (UMAP) of epithelial cells in young and aged UW skin.
(C) Expression of marker genes used for identification of major cell types in (B).
(D) Heatmap of top 10 differentially expressed marker genes of clusters in (B).
(E) Proportion of each epithelial subpopulation out of all epithelial cells in (B).
(F) UMAP of fibroblasts in young and aged UW skin.
(G) Heatmap of top 10 differentially expressed marker genes of clusters in (F).
(H) Proportion of each fibroblast subpopulation out of all fibroblasts in (F).
(I) Col1a1/Mmp3 expression in Col1aHi/IMF I/II fibroblasts from (F). See Table S1 for full list of differentially expressed genes.
(J) UMAP of immune cells in young and aged UW skin.
(K) Expression of marker genes used for identification of major cell types in (J).
(L) Heatmap of top 10 differentially expressed marker genes of clusters in (J).
(M) Proportion of each immune subpopulation out of all immune cells in (J). p values were calculated using prop.test function in R (E, H, and M) or Wilcoxon rank sum test (I). ***p < 0.001. Data presented in Figures 1–4 include both v2 and v3 samples unless specified otherwise.
To elucidate aging-associated changes in skin during homeostasis, we first compared young and aged UW samples. Specifically, we aggregated all UW skin cells in a single dataset and then computationally separated epithelial (Krt14+ or Krt1+), fibroblasts (Col1a2Hi and Rgs5−), and immune cells (Ptprc+) for analysis (Figures S1B–S1D).
The UW epithelial dataset contains 7,286 and 2,252 cells from young and aged skin, respectively, which were clustered into four interfollicular epidermal (IFE) (two basal [Krt14high] and two spinous [Krt1/10high]) and three hair follicle (HF)-associated (Krt17high or Krt79high) subpopulations (Figures 1B–1D; Table S1). Basal 1 cluster showed enriched expression of Col17a1, suggesting a Col17a1Hi stem cell-encompassing identity, whereas basal 2 cluster showed enriched expression of Fos and Cdkn1a (Figure 1C), suggesting an early response/growth arrest identity (Haensel et al., 2020; Liu et al., 2019). Spinous 2 cluster displayed overlapping marker genes with basal 2 cluster (Figure 1D), suggesting a transitional state identity. In aged skin, the relative abundance of basal keratinocytes decreased, whereas that of spinous keratinocytes (specifically spinous 2) increased (Figure 1E).
Since our data were collected using both v2 and v3 versions of Chromium Kits (Figure S1B; see STAR Methods), we also computationally isolated and analyzed UW epithelial cells that were analyzed by v3 experiments only to ensure consistency. Epithelial marker genes identified using v2 + v3 data (Figure 1D) were found to also mark the different epithelial cell subsets in v3-only data (Figure S2A). Furthermore, a similar trend of reduced basal-suprabasal ratio was observed in aged skin relative to young skin, although the difference was less drastic in v3-only than in v2 + v3 data (Figures 1E and S2B), likely due to differential sensitivity of v2 versus v3 chemistry as well as batch effects. Consistent with previous reports (Ge et al., 2020; Keyes et al., 2016), gene expression comparison between young and aged skin revealed only mild differences, with a small number of genes consistently (in both v2 + v3 and v3-only data) displaying more than 2-fold changes from young to aged skin in either basal or suprabasal keratinocytes (Table S1). For subsequent analyses, we used a similar strategy of comparing v2 + v3 with v3-only data and conclude only on robust differences that are supported by both.
Minor age-related alterations in fibroblasts were observed in UW skin. The UW fibroblast dataset contains 4,826 cells from young and 2,592 cells from aged skin, which were clustered into six subpopulations (Figures 1F and 1G). One cluster showed enriched expression of ECM genes (e.g., Col1a1, Col1a2) and was designed Col1aHi (Figures 1F and 1G; Table S1). Two clusters were enriched for immune-related genes (e.g., complements C3, Cxcl12, and Cxcl13) and designated immune-modulating fibroblast (IMF) I and II (Figures 1F and 1G; Table S1). Two clusters expressed known markers of dermal sheath (DS) or dermal papillae (DP) (e.g., Tagln, Mylk, and Crabp1) (Joost et al., 2020) and another cluster showed overlapping markers with one of these, and they were designed HF-associated dermal fibroblast (HFDF) I, II, and III (Figures 1F, 1G, and S2A; Table S1). Compared with young skin, aged skin showed increased frequency of HF-associated but decreased frequency of non-HF (especially Col1aHi) fibroblasts (Figures 1H and S2B). Furthermore, expression of ECM gene Col1a1 and ECM-regulatory gene Mmp3 was reduced and increased, respectively, in non-HF subsets of aged skin (Figures 1I and S2C; Table S1), consistent with previous reports of aging-associated ECM atrophy (Ding et al., 2021; Freitas-Rodríguez et al., 2017; Lago and Puzzi, 2019; Solé-Boldo et al., 2020; Varani et al., 2006).
Differences in resident immune cells were also detected between young and aged skin. The UW immune cell dataset contains 1,273 cells from young and 1,213 cells from aged skin, and these were clustered into eight subpopulations designated monocytes (Plac8Hi) (Blecher-Gonen et al., 2019), macrophages (Cd68HiItgam+Plac8Low), dendritic cells (DCs) (Adam23+P2ry10+ Itgax+Itgam−; may also contain a small number of CD207+ Langerhans cells) (Tamoutounour et al., 2013), mast cells (Mcpt4+) (Groschwitz et al., 2009), T cells (Cd3g+), natural killer (NK) and NK T cells (Cd3g+Nkg7+) (Malarkannan, 2020), regulatory T cells (Tregs; Cd3g+Foxp3+), and proliferative cells (Mki67+) based on top markers (Figures 1J–1L and S2A; Table S1). Compared with young skin, aged skin showed a relative expansion of T cell 1 [Cxcr6+; Cxcr6 is required for localization of lung-resident memory CD8 T cells (Wein et al., 2019)] and a reduction of Tregs (Figures 1M and S2B).
Taken together, our data revealed baseline differences during homeostasis in young and aged skin across various epithelial, fibroblast, and immune cell populations.
Wound healing induces major alterations in epithelial, fibroblast, and immune cell populations in young and aged skin
Next, we examined the major cell types and changes in their pro-portions during wound healing by aggregating data for all three timepoints (UW, 4dpw, 7dpw) separately for young and aged samples (Figure 2A). In young skin, we observed 13 clusters assigned the following identities based on each cluster’s top markers and known marker genes (Haensel et al., 2020; Joost et al., 2016) (Figures 2B and 2C; Table S2): (1) two epithelial, a basal cluster with both IFE (Krt14+) and HF (Krt14+Krt17+) cells, and an epidermal spinous cluster (Krt1+); (2) two fibroblast, one present in both UW and wounded skin and termed homeostatic fibroblasts, and another enriched in wounded skin and termed wound-induced (WO) fibroblasts; (3) four immune, a cluster containing monocytes, macrophages (Cd68HiItgam+), and neutrophils (S100a8+Csf3r+) (Wilk et al., 2020), a DC (Itgax+) cluster, a T cell cluster, and a mast cell cluster; and (4) five additional cell-type populations, namely a pericyte (Rgs5Hi) cluster (He et al., 2020), a melanocyte (Pmel+) cluster (Solé-Boldo et al., 2020), an endothelial cell (Pecam1+) cluster (Avitabile et al., 2015; Wankell et al., 2001), a muscle cell (DesHi) cluster (Goldfarb and Dalakas, 2011), and a separate cluster of proliferative (Mki67+) cells.
Figure 2. Cellular changes during wound healing in young or aged skin.
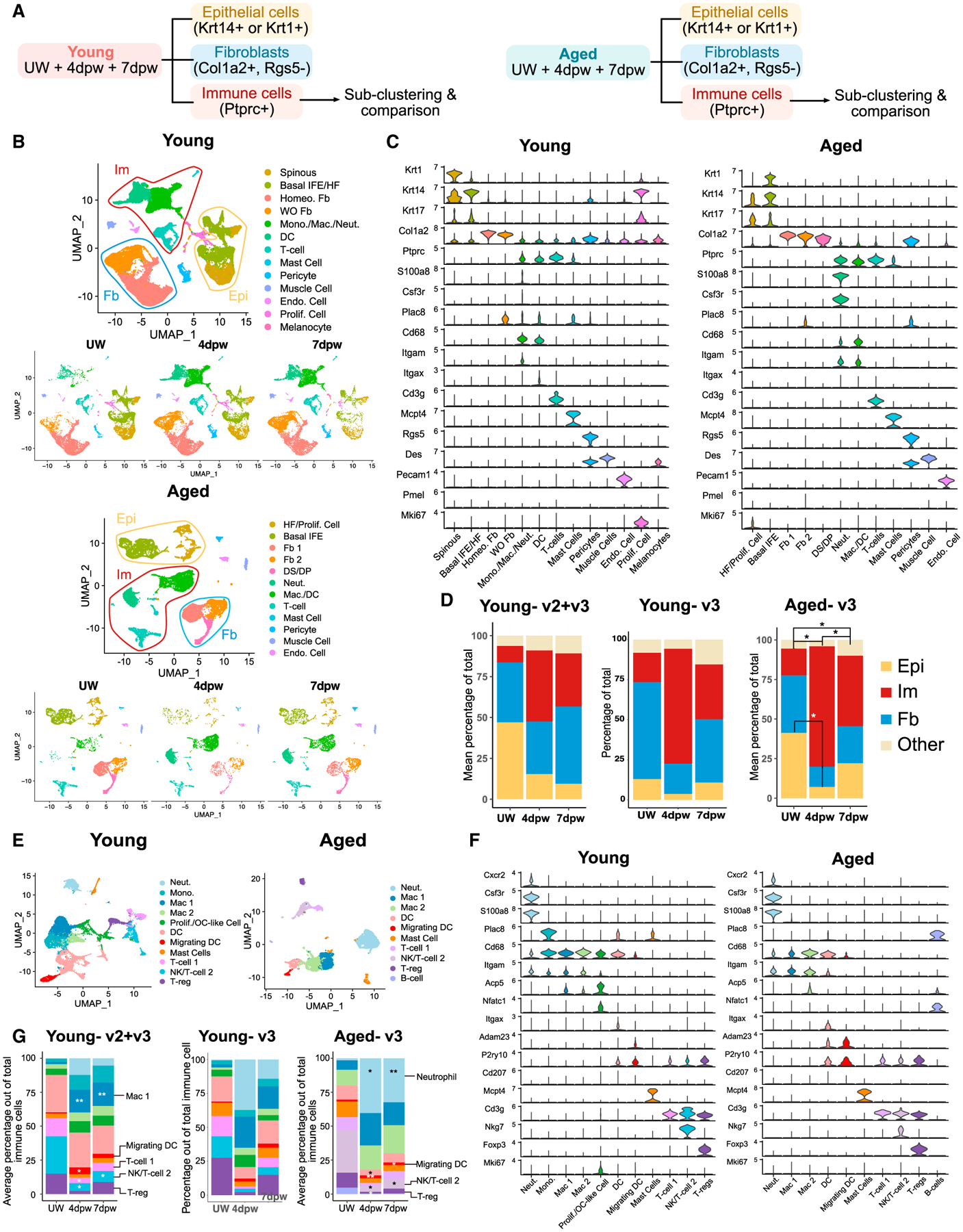
(A) Data analysis strategy. Young and aged samples are analyzed separately here.
(B) UMAPs of all cells in UW and wounded (4dpw and 7dpw) skin in young or aged samples.
(C) Expression of marker genes used for identification of cell populations in (B).
(D) Proportion of each major cell type out of all cells in (B) at the indicated timepoints. Epi, epithelial; Im, immune; Fb, fibroblast.
(E) UMAP of immune cells in young or aged samples.
(F) Expression of marker genes used for identification of cell populations in (E).
(G) Proportion of each immune population out of all immune cells in (E). p values were calculated using ANOVA and post hoc Tukey tests in R. **p < 0.01, *p < 0.05 compared with UW samples.
Similarly, we observed 12 clusters in aged skin: (1) two epithelial clusters, (2) three fibroblast clusters, (3) four immune cell clusters, and (4) three other cell-type clusters (Figures 2B and 2C; Table S2). Overall, epithelial, fibroblast, and immune cells were the predominant cell types detected in both young and aged skin/wounds (Figures 2B–2D). As expected, the relative abundance of epithelial cells and fibroblasts showed a trend of decrease during wound healing in both young and aged skin, evident especially at 4dpw (Figures 2D, S1C, and S1D). Conversely, the relative abundance of immune cells was increased during wound healing, especially at 4dpw. Fibroblast 1 and 2 clusters in aged skin showed the highest expression of young sample-derived markers of homeostatic and WO fibroblasts, respectively (Figure S1E; Tables S2 and S3). WO fibroblast subset in young skin expanded from UW skin to wounds, but fibroblast 2 subset in aged skin did not (Figure 2B), raising the possibility that aged skin fibroblasts may be compromised in their transcriptome response to wounding.
Immune cells play major roles in wound healing, including clearing pathogens and debris as well as facilitating keratinocyte/fibroblast proliferation and activation, but their apoptosis or exit from skin is also critical for resolution of inflammation and proper tissue repair (Boothby et al., 2020; Eming et al., 2014; Landén et al., 2016; Minutti et al., 2017). We next computationally isolated all-immune-cell clusters from the aggregated UW + 4dpw + 7dpw master dataset (Figure 2A) for finer analysis. Immune cells from young skin were further clustered into 11 subpopulations: neutrophils, monocytes, macrophages, proliferative and osteoclast-like (OC-like) (Acp5+Nfatc1+) (Humbert et al., 2021) cells, DCs (Ccr7−) and migrating DCs (Ccr7+) (Förster et al., 2008), mast cells, T cell 1, NK/T cell 2, and Tregs (Figures 2E and 2F; Table S2). From UW to wounded skin, percentage of myeloid populations (neutrophils, monocytes, and macrophages) increased, while that of T cell populations (Treg and NK/T cell 2) decreased, especially at 4dpw (Figure 2G). Immune cells from aged skin formed 10 subpopulations that contained all the cell types observed in young skin plus a small number of B cells (Figures 2E–2G). Wound-induced expansion in neutrophil/macrophage subpopulations at the cost of Treg/NK/T cell 2 subpopulations was also observed in aged skin (Figure 2G).
Taken together, our data provide an overview of the major tissue microenvironmental changes during wound healing and reveal a significant expansion of immune cells, but with seemingly differential dynamics and cell-type compositions in young versus aged skin wounds.
Aged skin wounds exhibit a more inflammatory profile than young counterparts, featuring neutrophil persistence, fewer DCs, and increased abundance of an Arg1Hi macrophage subset
To facilitate direct comparison of the immune microenvironment, we next aggregated data for young and aged samples at each post-wounding time point, and then computationally isolated immune cells for downstream subclustering analysis (Figures S1B, S1C, and S1E). At 4dpw, we identified 10 clusters, with neutrophils, macrophages, and DCs being the majority (87.2% total in young and 90.8% in aged) (Figures 3A–3C; Table S4). Neutrophil abundance showed a trend of increase per v2 + v3 but only mildly per v3-only data, whereas DC abundance was significantly decreased per both analyses, in aged skin wounds compared with young counterparts (Figures 3B and S3A). By 7dpw, neutrophil abundance was significantly higher in age skin wounds than in the young (Figures S3C–S3E; Table S4). Moreover, neutrophils in aged 4pdw and/or 7dpw wounds showed increased expression of inflammatory genes including Cxcl3, Ccl6, Spp1, Osm, and Il1a compared with young counterparts (Figures 3D, S3B, and S3F; Table S4). Reduction in DCs also persisted in aged 7dpw wounds (Figures S3C–S3E; Table S4). Additionally, although populational size was small, Tregs appeared to be fewer in aged skin wounds at 7dpw than in the young (Figures S3C–S3E).
Figure 3. Differential immune cell heterogeneities in young versus aged skin wounds.
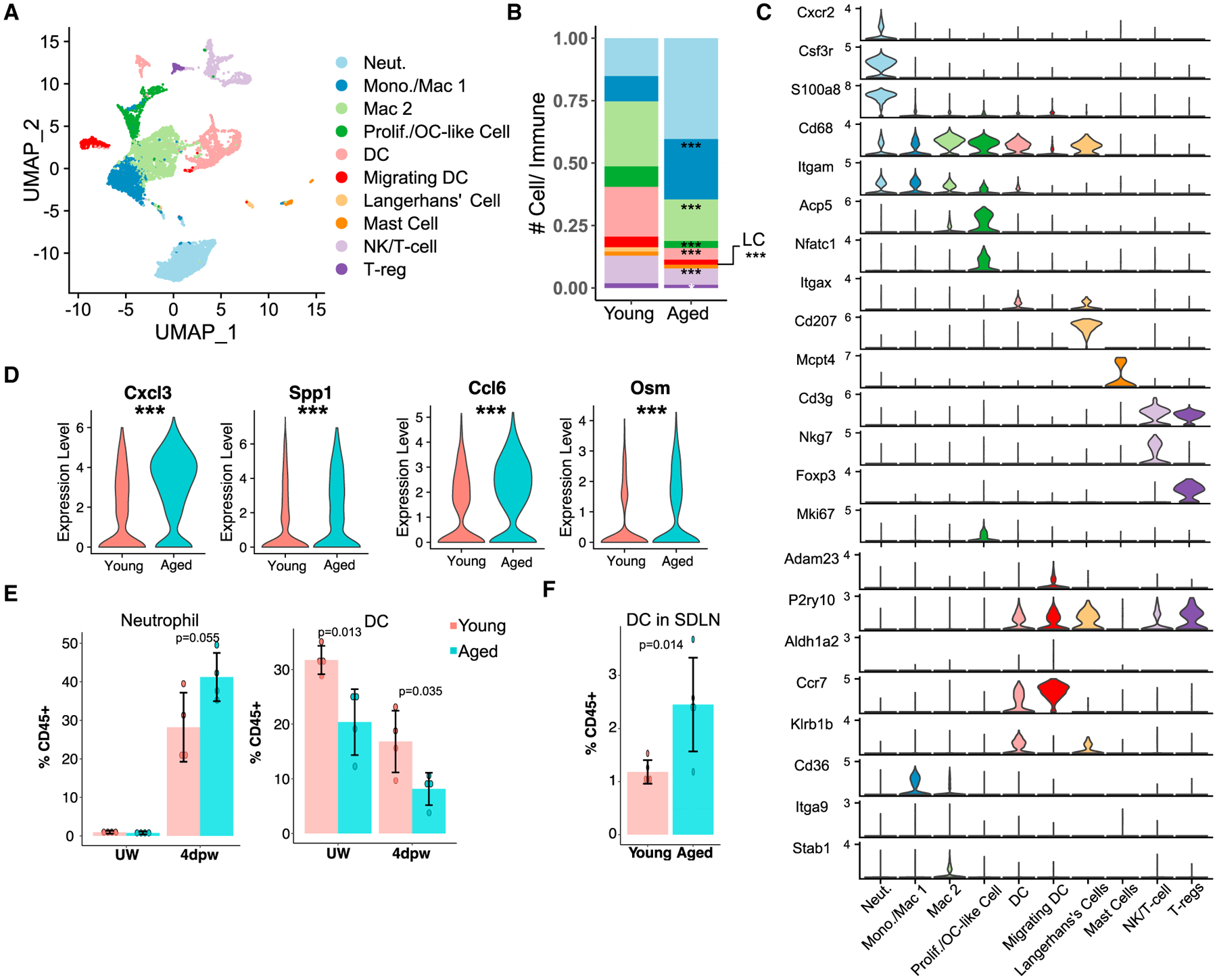
(A) UMAP of immune cells in young + aged 4dpw wounds.
(B) Proportion of each immune subpopulation out of all immune cells in (A).
(C) Expression of marker genes used for identification of cell populations in (A).
(D) Expression of select genes in neutrophils from (A). See Table S4 for full list of differentially expressed genes.
(E) Flow cytometry of neutrophils and DCs in UW and 4dpw skin. n = 4 each for young and aged samples.
(F) Flow cytometry of DCs in LNs at 4dpw. n = 5 each for young and aged samples. Bar graphs (E and F) represent mean ± SD. p values were calculated using prop.test function in R (B), Wilcoxon rank sum test (D), or unpaired two-tailed Student’s t test (E and F). ***p < 0.001.
To validate the scRNA-seq-revealed differences, we performed flow cytometry on both UW and wounded (4dpw) skin. Neutrophils (CD45+CD11b+Ly6G+) were not detectable in young or aged skin during homeostasis, whereas reduced presence of DCs (CD45+CD11c+MHCII+) in aged skin was already evident (Figure 3E). The results also confirmed a trending increase in neutrophils and a significant reduction in total DCs in aged skin wounds compared with the young (Figure 3E). To ask whether low DC abundance in aged skin might be due to their migration, we profiled DCs in skin-draining lymph nodes (LNs) of wounded mice using flow cytometry. Indeed, LNs of aged mice contained significantly more DCs than their young counterparts (Figure 3F). These findings suggest that DCs in aged mouse skin may be more likely to migrate to LNs, a feature reminiscent of DCs from aged human subjects that exhibit enhanced basal-level inflammatory responses (Agrawal et al., 2009; Panda et al., 2010).
Macrophages are critical orchestrators of skin wound healing through timely promotion and resolution of inflammation (Krzyszczyk et al., 2018; Minutti et al., 2017). We therefore zoomed in on their changes during wound healing in young versus aged skin by computationally isolating monocyte/macrophage clusters from all-immune-cell datasets at each wound time point for further subclustering (Figures S1B, 4A, and S4C). At 4dpw, we identified six monocytes/macrophage subpopulations in young plus aged skin wounds, and they were monocytes, Arg1Hi macrophages, C1qHi macrophages, OC-like, antigen-presenting macrophage 1 (APM 1; showing the highest expression of major histocompatibility complex (MHC) class II genes such as H2-Ab1 and H2-Eb1), and APM 2 (expressing both MHC class II markers and anti-inflammatory cytokines Il10 and Il1rn) (Figures 4A–4C, and S4A; Table S5). Interestingly, the abundance of Arg1Hi macrophages relative to other monocyte/macrophage subsets was significantly higher in aged skin wounds compared with the young (Figures 4C and S4B). At 7dpw, although only five monocyte/macrophage clusters were identified, four of them shared similar top markers with clusters observed at 4dpw (Arg1Hi/APM, C1qHi, OC-like, and monocytes), and one new cluster (enriched for Cd9 expression) emerged (Figures S4C and S4D; Table S4). The relative abundance of Arg1Hi/APM macrophages appeared slightly higher in aged 7dpw wounds than in young counterparts (Figure S4E).
Figure 4. Differential macrophage heterogeneities in young versus aged skin wounds.
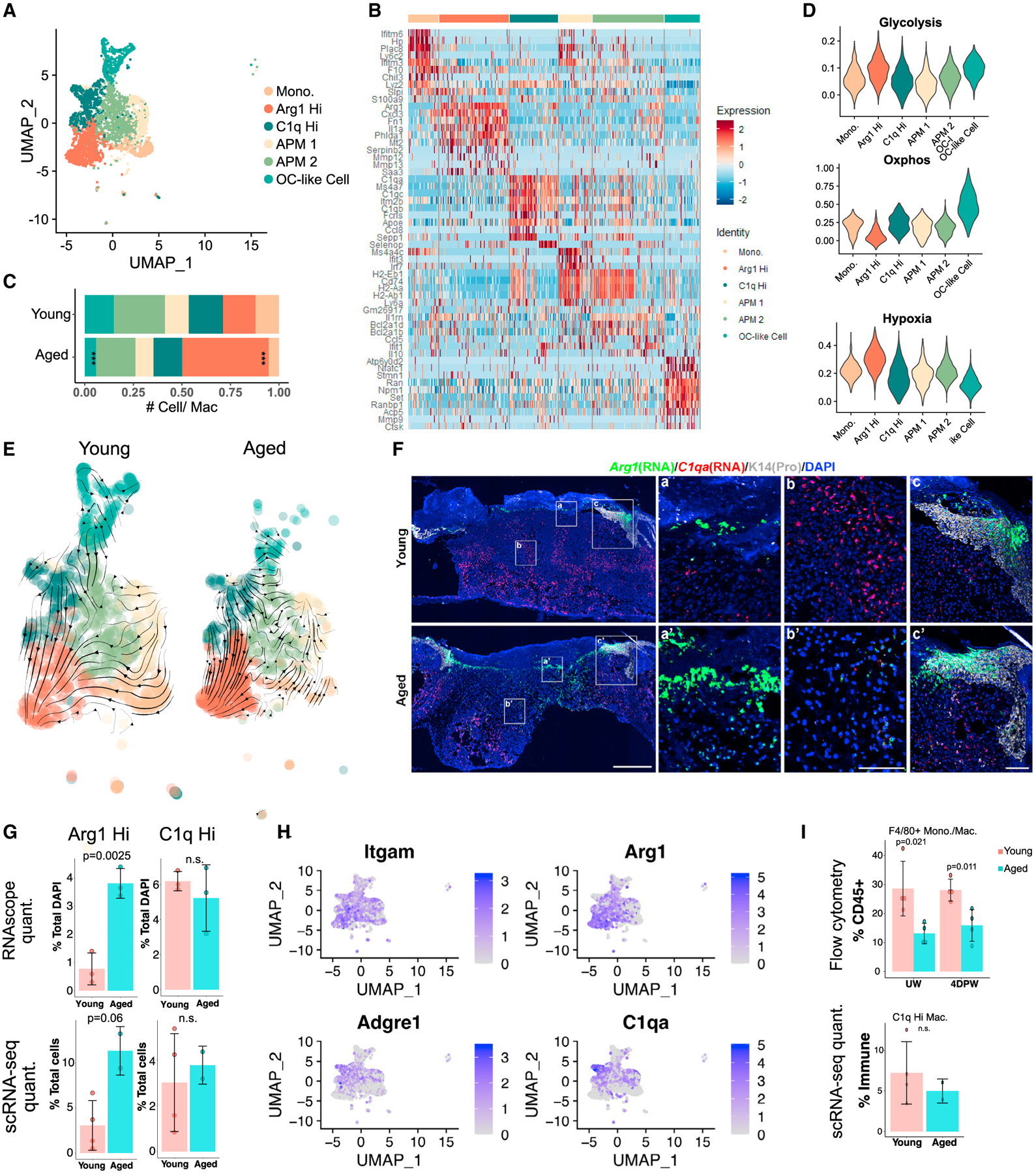
(A) UMAP of macrophage subpopulations in young + aged 4dpw wounds.
(B) Heatmap of top 10 differentially expressed marker genes of clusters in (A).
(C) Proportion of each macrophage subpopulation out of all monocyte/macrophages in (A).
(D) Gene scoring analysis of the macrophage subpopulations in (A).
(E) RNA velocity analysis of macrophages in (A).
(F) RNAScope showing spatial distribution of Arg1 and C1qa transcripts in young and aged 4dpw wounds (n = 3 each). DAPI stains the nuclei. Scale bars: 500 μm in low-magnification image (left), 100 μm in high-magnification images (middle, right).
(G) Quantification of percentage Arg1+ or C1qa+ cells per total DAPI-positive cells from RNAScope results in (F) (top) or percentage Arg1+ or C1q+ cells per total live cells from scRNA-seq experiments (bottom).
(H) Feature plots showing expression of the indicated genes in macrophage subpopulations from (A).
(I) Flow cytometry of F4/80+ macrophages in UW and 4dpw skin (top). n = 4 each for young and aged samples. Bottom, percentage C1q+ macrophages per total immune cells from scRNA-seq data. Bar graphs (G and I) represent mean ± SD. p values were calculated using prop.test function in R (C) or unpaired two-tailed Student’s t test (G and I). ***p < 0.001.
Next, we studied the molecular characteristics of the monocyte/macrophage subpopulations identified at 4dpw. C1qHi cluster showed enriched expression of known M2 macrophage marker Mrc1 (Martinez and Gordon, 2014; Orecchioni et al., 2019), and such cells were also present in UW skin (Tables S1, S2, S3, S4, and S5), suggesting a skin resident macrophage identity. Top markers of Arg1Hi cluster included pro-inflammatory chemokine/cytokines (Cxcl1/2/3, Il1a) and known M2 macrophage marker Arg1 (Figure 4C; Table S5), suggesting an inflammatory M1/M2 hybrid identity (Lawrence and Natoli, 2011). Arg1Hi cluster was among the highest-expressing cells for glycolysis-associated genes and scored lowest for oxidative phosphorylation (OxPhos)-associated genes (Figures 4D and S4F; Table S3). In contrast, C1qHi cluster scored low for glycolysis genes and high for OxPhos genes (Figures 4D and S4F; Table S3). Such gene scoring analysis also revealed higher and lower expression of a hypoxia-associated signature in Arg1Hi and C1qHi/OC-like clusters, respectively (Figures 4D and S4F). Similarly, at 7dpw, Arg1Hi/APM cluster showed higher hypoxia- and glycolysis-associated, but lower OxPhos-associated, gene expression than C1qHi cluster (Figure S4F; Table S3). These data highlight a metabolic preference of Arg1Hi cells for glycolysis over OxPhos that appears to be coupled to hypoxia response.
To explore potential lineage relationships among monocyte/macrophage subpopulations, we down-sampled their total cell number in young skin wounds to match that in their aged counterparts and performed RNA velocity analysis (Bergen et al., 2020; La Manno et al., 2018). In young 4dpw wounds, C1qaHi, APM 1 and 2, and monocyte clusters all showed a propensity to become Arg1Hi macrophages (Figure 4E; Table S5). In aged 4dpw wounds, however, only C1qaHi cluster showed this propensity, whereas monocyte and APM 2 clusters showed a propensity to become APM 1 (Figure 4E; Table S5). The situation appeared to be reversed at 7dpw, with many young cells transitioning away from the Arg1Hi state, whereas aged cells exhibited a more uniform trajectory toward Arg1Hi macrophages (Figure S4G; Table S5). These data suggest differential macrophage polarization kinetics in young versus aged skin wounds, and that multiple monocyte/macrophage subsets may have a potential to become Arg1Hi cells at some point during wound healing.
We next performed RNAScope experiments to examine the spatial expression of Arg1 and C1qa in young and aged skin wounds. Epithelial cells in wound hyperproliferative zone/leading edge were found to strongly express Arg1 (Figure 4F), consistent with a previous report (Kämpfer et al., 2003). Within the stroma, Arg1 expression was detected predominantly at the scab-wound bed interface and in upper wound bed, whereas C1qa expression was detected predominantly in lower wound bed and near the wound margin (Figures 4F and S4H). Supporting the scRNA-seq finding, there were more Arg1-expressing stromal cells in aged skin wound bed than in the young (Figures 4F, 4G, and S4H). These data suggest differential spatial locations of Arg1Hi and C1qHi macrophages within the 4dpw wound, and that Arg1Hi macrophages are more abundant in aged skin wounds.
F4/80 protein is a commonly used surface marker for monocyte/macrophages. Since Adgre1 (F4/80) mRNA expression fell within the C1qHi rather than Arg1Hi subset (Figures 4H and S4I), we wondered whether F4/80+ population was altered in aged skin wounds. Interestingly, flow cytometry detected lower relative abundance of CD45+CD11b+Ly6G−F4/80+ monocyte/macrophages in both UW skin and 4dpw wounds of aged mice compared with young mice, while quantification of scRNA-seq data did not show a significant difference in relative abundance of C1qHi macrophages between young and aged 4dpw wounds (Figure 4I). Additionally, immunostaining revealed a consistent reduction of F4/80+ cells in the dermis of aged 4dpw wounds compared with the young (Figure S4J). These results present an interesting discrepancy between mRNA (Adgre1) and protein (F4/80) expression, which was also noted previously (Haensel et al., 2020).
Taken together, our data reveal salient differences in immune cell compositions between young and aged skin wounds, featuring neutrophil persistence, DC mobilization, enrichment of Arg1Hi inflammatory macrophages at the cost of other monocyte/macrophages, and fewer immunosuppressive Tregs in the latter. As such, wound healing in aged skin is characterized by a stronger inflammatory immune microenvironment than in young skin.
Altered signaling interaction between Arg1Hi macrophages and fibroblasts in aged skin wounds
To gain insight into a signaling role of Arg1Hi macrophages, we examined their top marker genes and found Il1a (Table S5), which encodes a pro-inflammatory cytokine that is known to inhibit wound healing when present at excess (MacLeod and Mansbridge, 2016; Perrault et al., 2018). Arg1Hi macrophages are the predominant source of Il1a expression among all fibroblasts and immune cells in young skin wounds (Figure 5A). In contrast, Il1b is more broadly expressed in multiple immune cell types (Figure 5A). Fibroblasts are the major cell type expressing interleukin (IL)-1 receptor Il1r1, whereas expression of decoy receptor Il1r2 is more widespread but the highest in Col1a+ subset of all fibroblasts (Figures 5A, S2G, and S2H). Using RNAScope, we mapped the spatial locations of Il1a, Il1r1, and Il1r2 expression in young skin wounds. Il1a expression was weakly detected at the scab-wound bed interface, whereas Il1r1 expression was detected throughout the wound bed (Figure 5B). Il1r2 expression was detected prominently at the scab-wound bed interface and in the upper wound bed, areas where non-epithelial Arg1 expression was concentrated (Figures 5B, 4F, and S4H). Within the wound bed, Il1r1 and Il1r2 transcripts were detected in 22% and 12%, respectively, of the cells, but only <2% cells expressed both (Figure 5C). These data reveal interesting spatial patterning of IL-1 signaling ligands and receptors in the wound and implicate the existence of a negative regulatory mechanism (by decoy receptor) to restrict IL-1 signaling within the Arg1Hi macrophage-rich zone.
Figure 5. Differential Arg1Hi macrophage-associated signaling in young versus aged 4dpw wounds.
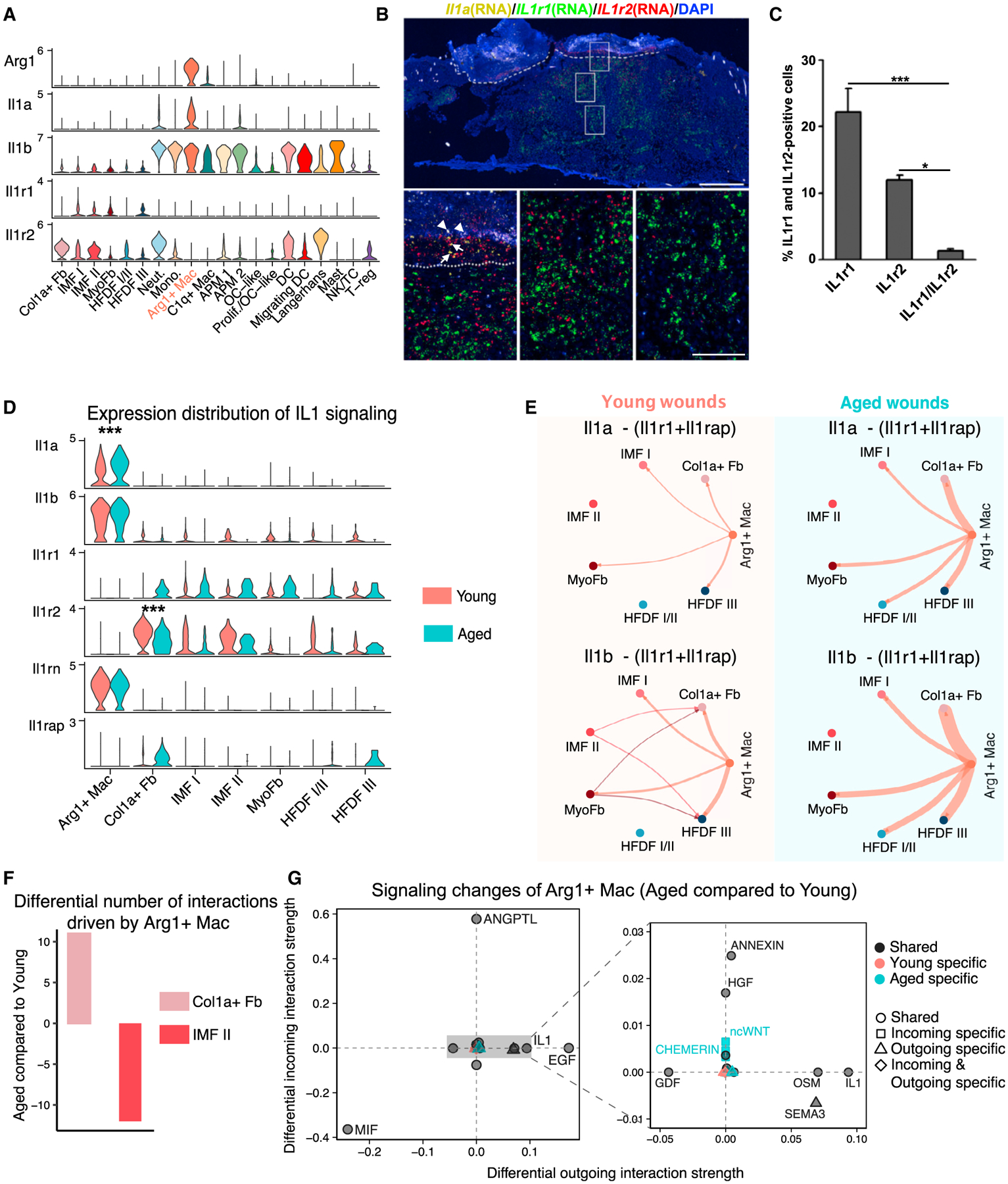
(A) Violin plots showing expression levels of the indicated genes in fibroblast and immune cell populations.
(B) RNAScope showing spatial distribution of Il1a, Il1r1, and Il1r2 transcripts in young 4dpw wound. Arrows and arrowheads indicate Il1a and background signals, respectively. Dashed line indicates scab-wound bed border. DAPI stains the nuclei. Scale bars: 500 μm in low-magnification image (top), 100 μm in high-magnification images (bottom).
(C) Quantitation of Il1r1-, Il1r2-, and Il1r1/Il1r2-positive cells in wound beds (B). n = 3.
(D) Comparison of expression levels of IL-1 signaling components in Arg1Hi macrophages and fibroblasts in young versus aged skin wounds.
(E) IL-1-mediated cell-cell communication networks between Arg1Hi macrophages and fibroblast populations in young versus aged skin wounds. Edge width is proportional to the inferred interaction strength. Edge color is consistent with the signaling source.
(F) Differential number of putative interactions driving Arg1Hi macrophage-to-fibroblast signaling in aged versus young skin wounds.
(G) Altered Arg1Hi macrophage-associated signaling pathways in young versus aged skin wounds. The x and y axes represent differential outgoing and incoming interaction strengths, respectively. Positive and negative values indicate increased and decreased signaling, respectively, in aged skin wounds compared with the young. Black, red, and green colors indicate whether a signaling pathway is shared between young and aged, or specific to either young or aged. Dot shapes indicate whether a signaling pathway is specific to either young or aged in its outgoing, incoming, or both outgoing and incoming signaling. Enlarged plot on the right shows the signaling pathways with relatively small alterations. Data presented in Figures 5–7 infer cell signaling from v2 + v3 samples unless specified otherwise. Cell-cell communication information that is qualitatively consistent between v2 + v3 and v3-only data analyses is shown in the main figures, whereas v3-only information is shown in supplemental figures. p values were calculated using unpaired two-tailed Student’s t test (C) or Wilcoxon rank sum test (D). ***p < 0.001, *p < 0.05.
We next compared the expression of IL-1 signaling components in Arg1Hi macrophages and fibroblasts of young and aged skin wounds. Compared with young counterparts, aged skin wounds exhibited decreased expression of Il1r2 in multiple fibroblast subsets at both 4dpw and 7dpw (Figures 5D, S2M, S2N, S5A, S5E, and S5H). RNAScope validated the reduced number of Il1r2-expressing cells within the wound stroma of aged skin compared with the young (Figure S4H). Il1a expression in Arg1Hi macrophages was consistently increased in aged skin wounds at 4dpw and 7dpw, whereas Il1b increase was more variable (Figures 5D, S5A, S5E, and S5H). Expression of IL-1R1 antagonist Il1rn in Arg1Hi macrophages was largely unchanged at 4dpw, whereas that of Il1rap, encoding an accessory protein essential for IL-1 signaling (Cullinan et al., 1998), was elevated in some fibroblast subsets of aged skin wounds at both 4dpw and 7dpw (Figures 5D, S5A, S5E, and S5H).
We then used CellChat to infer IL-1 signaling-mediated cell-cell communication between Arg1Hi macrophages and fibroblasts. We calculated communication probabilities between different cell populations by not only considering both Il1r1 and Il1rap at the signal-receiving end but also incorporating the inhibitory effect of Il1r2. At both 4dpw and 7dpw, total IL-1 signaling from Arg1Hi macrophages onto several fibroblast subsets (e.g., Col1aHi and myofibroblast) was consistently enhanced in aged skin wounds relative to the young (Figures 5E, S5B, S5F, and S5I). In addition to the spatial proximity between Il1r2-expressing cells and Arg1-expressing cells in young and aged skin wounds (Figure S4H), RNAScope experiments also showed a subset of Arg1-expressing cells to be adjacent to some Col1a1-expressing cells, particularly in aged skin wounds (Figure S5K). These findings support the possibility that Arg1Hi macrophages and Col1aHi fibroblasts can communicate with each other. Our data demonstrate that IL-1 signaling is elevated during wound healing in aged skin relative to young skin in a manner that cannot be simply explained by delayed healing.
To identify other putative molecular pathways that drive Arg1Hi macrophage-fibroblast signaling, we performed unbiased CellChat analysis using the CellChatDB database (Jin et al., 2021) of all known secreted ligands and associated signaling. We found that, at 4dpw, the number of signaling interactions of Arg1Hi macrophages-Col1aHi fibroblasts was prominently increased while that of Arg1Hi macrophages-IMF II was decreased in aged skin wounds compared with young counterparts (Figures 5F and S5C). To identify the specific signaling pathways altered in aged skin wounds, we compared the strength of outgoing and incoming interactions from and to Arg1Hi macrophages and then extracted information common to v2 + v3 and v3-only analyses (see STAR Methods). Some signaling pathways were present in both young and aged skin wounds (“shared”); of these, oncostatin M (OSM) and epidermal growth factor (EGF) showed enhanced outgoing (Arg1Hi macrophage-originated) signaling, whereas angiopoietin-like protein (ANGPTL) and (HGF) showed enhanced incoming (Arg1Hi macrophage-destined) signaling, in aged samples (Figures 5G and S5D). Macrophage migration inhibitory factor (MIF) showed remarkable decrease in both outgoing and incoming signaling in aged skin wounds (Figures 5G and S5D). Some of these signaling pathways (e.g., OSM, MIF) have previously been shown to regulate fibroblast functions (Ashcroft et al., 1997; Dewor et al., 2007; Ihn and Tamaki, 2000), testifying the overall validity of our findings. Age-dependent alterations were also observed at 7dpw for Arg1Hi macrophage/APM signaling interactions with fibroblasts (Figures S5G and S5J).
Collectively, our results reveal Arg1Hi macrophage-fibroblast communication as a target of altered cell-cell signaling during delayed wound healing in aged skin, presenting changes in multiple signaling pathways and featuring elevated pro-inflammatory IL-1 signaling due to altered expression of multiple signaling mediators and inhibitors.
Systems-level analysis reveals differences in cell-cell communication network structure and signaling strength across multiple cell populations in young versus aged skin wounds
To probe global differences in cell-cell communications between young and aged skin wounds, we employed comparative CellChat (see STAR Methods) to study signaling interactions among all major cell types at 4dpw and 7dpw. We combined the subclustering results of epithelial, fibroblast, and immune cells (Figures S2D, S2G, S2J, S2M, 3A, and S4C; Tables S5 and S6), leading to a total of 26 cell groups at 4dpw and 22 cell groups at 7dpw for analysis.
We first examined the total number of possible cell-cell communications and the prominently affected cell types. There was a higher number of possible interactions in aged skin wounds than in young skin wounds at both 4dpw and 7dpw (Figure 6A, S6A, S6H, and S6K). Putative signaling within and between the epithelial and fibroblast populations drastically increased, while signaling between the fibroblast and immune cell populations slightly increased, in aged skin wounds (Figures 6B, 6C, S6B, S6C, S6I, and S6L). Moreover, signaling from immune cells to epithelial cells was strengthened in aged skin wounds. These data show that, in general, aging increases both autocrine and paracrine signaling probabilities among all three major cell types during wound healing.
Figure 6. Alterations in network structure and signaling strength of putative cell-cell communications in young versus aged 4dpw wounds.
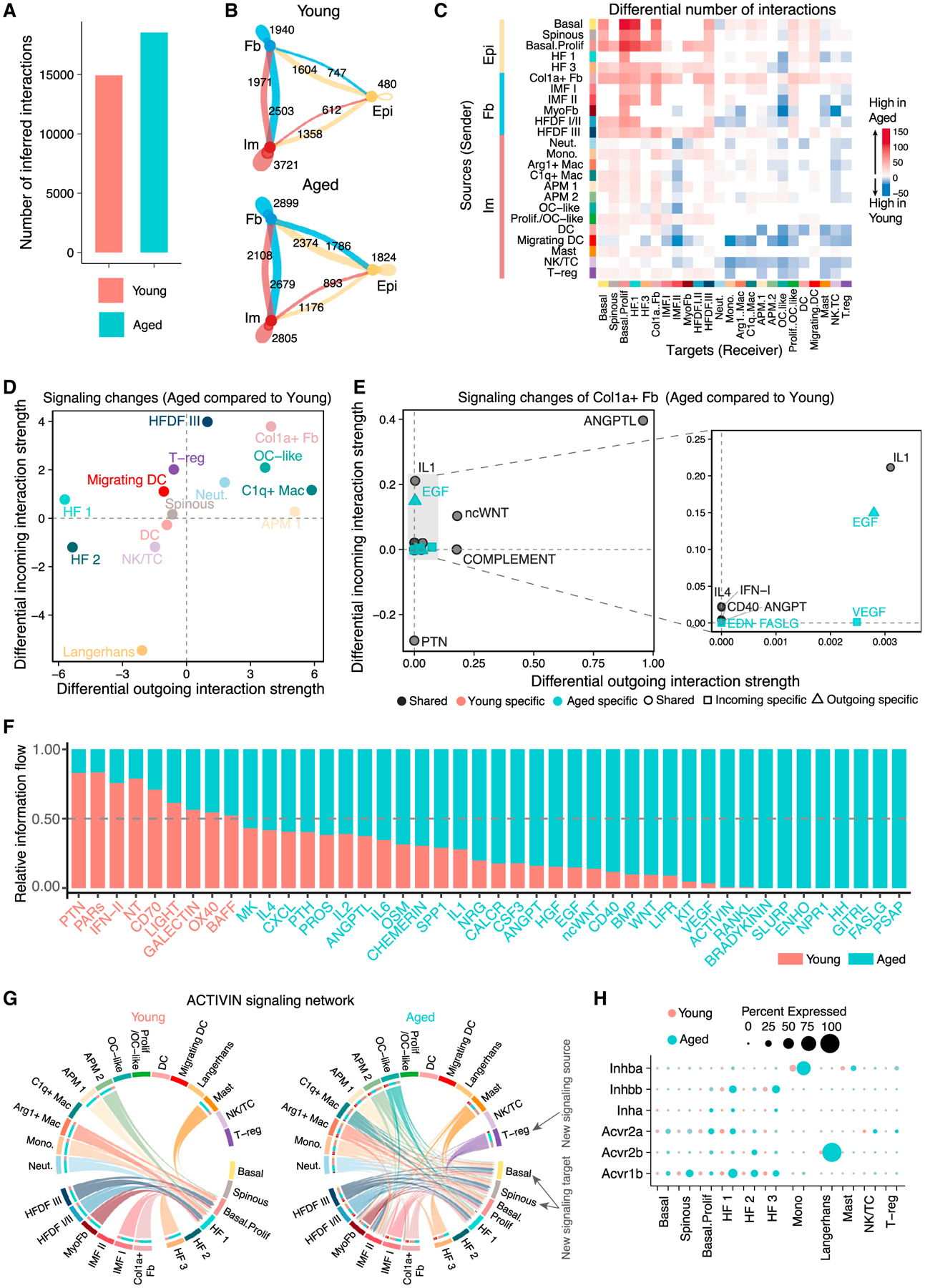
(A) Total number of possible interactions.
(B) Number of possible interactions between the three major cell types.
(C) Differential number of possible interactions between any two cell populations. Red (positive values) and blue (negative values) in the color bar indicate higher number of predicted interactions in aged and young skin wounds, respectively.
(D) Differential interaction analysis identifying prominently altered signaling sources and targets.
(E) Altered signaling pathways that associate with Col1aHi fibroblasts. See Figure 5G legends for more details.
(F) Significant signaling pathways were ranked based on their differences of overall information flow, calculated by summarizing all communication probabilities in a given inferred network. Those colored red and green are more enriched in young and aged skin wounds, respectively.
(G) Chord diagrams of inferred ACTIVIN signaling networks. Edge color is consistent with the signaling source. Segments with large arrows represent signaling targets and inner bars represent signaling sources in which the colors indicate signaling targets.
(H) Comparison of expression levels of ACTIVIN signaling components in epithelial and immune cells in young versus aged skin wounds.
We next studied how signaling patterns differ between young and aged skin wounds using network centrality analysis, which computes the outgoing and incoming interaction strength of each cell subpopulation to assess their likelihood as signaling sources and targets, respectively. Overall, fibroblasts (Col1aHi fibroblasts in particular) and immune cells (e.g., neutrophils, C1qHi macrophages) drastically increased their outgoing and/or incoming signaling in aged skin wounds (Figures 6D and S6D). Examination of age-associated changes in signaling between Col1aHi fibroblasts and all other cell types revealed a prominent increase in incoming and outgoing signals for ANGPTL, as well as in incoming signals for IL-1, in aged skin wounds (Figure 6E).
We next compared the information flow, defined by the sum of communication probability among all pairs of cell groups in the inferred network, for specific signaling pathways in young versus aged skin wounds. Certain pathways, such as Fas ligand (FASLG), ACTIVIN, IL-1, OSM, KIT proto-oncogene/receptor tyrosine kinase, and receptor activator of nuclear factor kappa-B ligandr (RANKL), prominently increased their information flow in aged skin wounds (Figures 6F and S6E). The key roles of ACTIVIN signaling in wound healing (Antsiferova and Werner, 2012; Wankell et al., 2001; Wietecha et al., 2020) prompted a closer examination of its overall network architecture. Compared with young skin wounds where proliferative basal and HF 1 cells were ACTIVIN targets, additional epithelial cell subsets in aged skin wounds gained ACTIVIN responsiveness (Figures 6G and S6F). Moreover, Tregs emerged as a new signaling source in aged skin. Interestingly, the expression levels of multiple ACTIVIN ligands and receptors were increased in multiple cell subpopulations in aged skin wounds compared with the young at both 4dpw and 7dpw (Figures 6H, S6G, S6J, and S6M), raising the possibility that inefficient healing in aged skin might be due to misdirected ACTIVIN signaling routes and/or deficiency in cell-type-specific downstream responses.
Our findings show that both the network architecture and signaling strength of putative cell-cell communications are drastically changed in aged skin wounds compared with their young counterparts. Particularly, the potential of both self- and reciprocal interactions among the major cellular compartments is markedly increased in aged skin wounds.
Comparative analysis of molecular signaling identifies dysregulated growth factor, chemokine, and cytokine pathways during wound healing in aged skin
Having obtained an understanding of the general rules governing cell-cell communications in young versus aged skin wounds, we next investigated the molecular signaling mechanisms in detail to identify candidate pathways potentially underlying the altered cellular compositions and communications in aged skin. We identified dysregulated ligand-receptor pairs in aged skin wounds by combining differential expression analysis with cell-cell communication analysis. CellChat predicted significant upregulation of 24 ligand-receptor pairs from 11 signaling pathways in aged skin wounds at 4dpw compared with young counterparts. These include ligands Spp1, Areg, Bmp7, Bmp4, Tnf, and Cxcl12 (Figure 7A). Spp1 knockdown has been shown to accelerate wound healing and reduce scarring (Mori et al., 2008), whereas Bmp7 is known to interfere with transforming growth factor beta (TGF-β)-induced differentiation of fibroblasts into myofibroblasts and dermal papilla in vitro (Bin et al., 2013; Liu et al., 2020). Zooming in on fibroblasts, we found bone morphogenetic protein (BMP) 2/4/7 signaling to be among the many pathways with elevated potential to target aged skin wound fibroblasts compared with young counterparts (Figures 7B, 7C, and S7A; Table S7). Conversely, the potential of TGF-β1/2/3 signaling to fibroblasts was reduced in aged skin wounds (Figures 7B, S7A, and S7B). The known BMP-TGF-β antagonism in diverse contexts (Dituri et al., 2019; Hudnall et al., 2016; Keller et al., 2011; Ning et al., 2019; Oshimori and Fuchs, 2012; Sotiropoulos and Chitnis, 2020) together with the role of TGF-β signaling in promoting fibroblast proliferation and ECM production during wound healing (Cutroneo, 2007; Pakyari et al., 2013) suggest that an altered balance between BMP and TGF-β signaling might underlie compromised fibroblast activation in aged skin wounds. Implicating a possible consequence of such imbalance, aged skin wound fibroblasts showed reduced expression of ECM genes Col1a1, Col1a2, Mfap2, and Mfap4, and increased expression of Mmp3 and cell death-promoting gene Cyld (Lork et al., 2017), relative to the young counterparts (Figure S2P).
Figure 7. Dysregulated growth factor, chemokine, and cytokine signaling in aged 4dpw wounds.
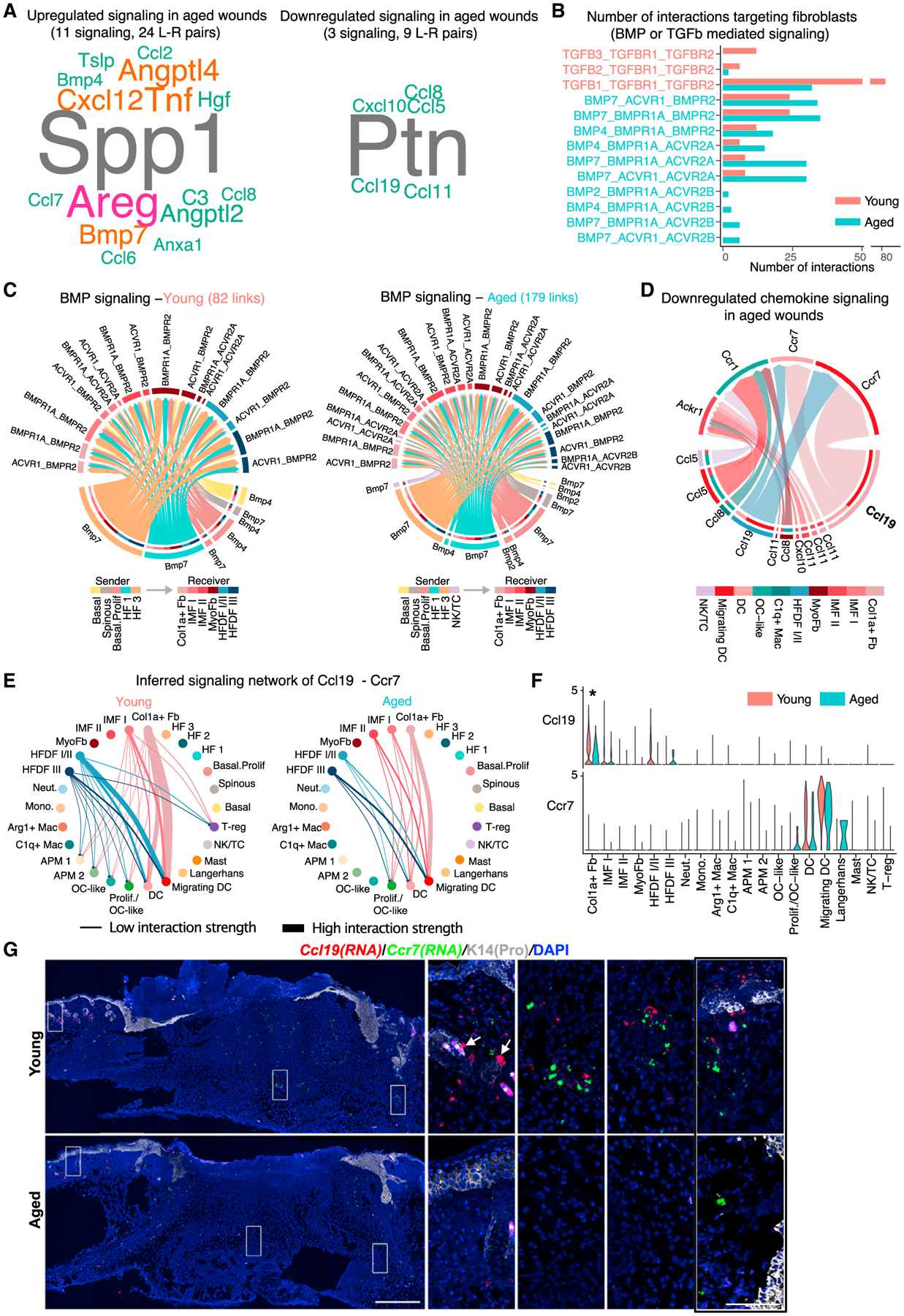
(A) Word cloud showing upregulated (left) and downregulated (right) signaling ligands in aged skin wounds compared with the young. Word size indicates the extent of enrichment in aged or young skin wounds.
(B) Number of possible interactions targeting wound fibroblasts via BMP or TGF-β signaling.
(C) Inferred BMP-mediated communications from non-fibroblast subsets to fibroblast subsets. The number of putative interactions (i.e., links in the diagram) is indicated on the top.
(D) Downregulated chemokine signaling in aged skin wounds compared with the young. Signaling sources and target are shown on the bottom and top, respectively. Colored segments indicate their cell identity information. Segment size is proportional to the total outgoing or incoming interaction strength associated with each ligand-receptor pair in the corresponding cell subpopulation.
(E) Circle plots of inferred Ccl19-Ccr7 signaling networks.
(F) Comparison of expression levels of Ccl19 and Ccr7 in epithelial, fibroblast, and immune cells in young versus aged skin wounds. *p < 0.05 (calculated using Wilcoxon rank sum test).
(G) RNAScope showing spatial distribution of Ccl19 and Ccr7 transcripts in young and aged skin wounds. DAPI stains the nuclei. Arrows point to Ccl19-positive cells that line HFs, and asterisk (*) indicates hair shaft autofluorescence. See Figure S7M for quantification (n = 3 pairs). Scale bars: 500 μm in low-magnification image (left), 100 μm in high-magnification images (right). The rightmost panel shows high-magnification image of an area near the wound margin from Figure S7L.
Only nine ligand-receptor pairs from three signaling pathways showed significant downregulation in aged skin wounds at 4dpw compared with the young, and these include Ptn, Ccl19, Ccl8/5/11, and Cxcl10 (Figure 7A). Ptn stimulates keratinocyte proliferation and has been implicated in angiogenesis (Florin et al., 2005; Perez-Pinera et al., 2007), so its downregulation might help explain the delayed re-epithelialization and reduced angiogenesis in aged skin wounds (Sgonc and Gruber, 2013). At 7dpw, similar upregulated (e.g., Spp1, Bmp7, Tnf, and Cxcl12) and downregulated (e.g., Ptn) ligand-receptor pairs were observed in aged skin wounds, but new changes also emerged, including downregulation of Th2 cytokine Il13 (Gieseck et al., 2018; Wynn, 2003) and macrophage/Langerhans cell-regulatory cytokine Il34 (Hieronymus et al., 2015; Wang and Colonna, 2014) (Figures S7F–S7I).
Chemokines are crucial modulators of wound healing through regulating leukocyte recruitment and integrating inflammatory events with tissue repair processes (Gillitzer and Goebeler, 2001). Altered chemokine signaling in aged skin wounds prompted us to examine and visualize the inferred chemokine signaling-mediated cell-cell communication network. Of the various ligand-receptor pairs, Ccl19 and its receptor Ccr7 displayed the strongest potential to signal from fibroblasts to DCs and migrating DCs (Figures 7D and S7C). Further analysis of the Ccl19-Ccr7 communication network showed that both the predicted strength of major communication routes (e.g., fibroblast-DC/migrating DC) and the total number of potential cell-cell interactions (e.g., disappearance of weak fibroblast-macrophage and fibroblast-Treg interactions) were decreased in aged skin wounds at 4dpw relative to young counterparts (Figures 7E and S7D). Decreased cell-cell communication strength was also seen at 7dpw, particularly for the fibroblast-DC/migrating DC and fibroblast-macrophage interaction pairs (Figures S7G–S7J). Reduced expression of Ccl19 by Col1aHi and HFDF I/II fibroblast subsets and slightly reduced expression of Ccr7 by DCs/migrating DCs are likely accountable for the reduction in potential signaling strength in aged skin wounds (Figures 7F, S7E, S7H, and S7K). RNAScope validated the reduced expression of both genes in aged skin wound dermis (Figures 7G, S7L, and S7M). In young skin wounds, both Ccl19 and Ccr7 mRNAs were detected in dermis of the unwounded region near the proliferative zone and within the wound bed, with Ccl19 expression seen in scattered dermal cells and what appear to be stromal cells lining the HFs. Ccl19- and Ccr7-positive cells tended to be in proximity of each other but were mutually exclusive (Figure 7G), suggesting a high likelihood of paracrine signaling interactions. In aged skin, Ccl19 and Ccr7 expression was decreased both within the wound bed and outside the wound margin (Figures 7G, S7L, and S7M). Taken together, these observations implicate the respective spatial locations of Col1aHi/HFDF I/II fibroblast subsets and DC/migrating DCs in wounded skin. More importantly, they identify reduced Ccl19 production by these fibroblast subsets as a possible mechanism for compromised retention and increased mobilization of DCs in aged skin and wounds.
DISCUSSION
Our work presents a comprehensive picture of the cellular compositions, cell-cell communications, and molecular mediators that underlie the wound healing processes in aged versus young skin. The resulting computational predictions and experimental findings not only recapitulate known biology to a substantial degree but also provide proof-of-principle examples for the utility of our scRNA-seq datasets as a useful resource to fuel future hypothesis-driven research that aims at identifying and manipulating critical cell populations and molecular pathways to help aged skin heal better.
Prolonged presence of macrophages in chronic wounds of humans suggests that dysfunctional macrophages likely contribute to impaired healing (Mast and Schultz, 1996). An M1-to-M2 macrophage switch has traditionally been viewed as the process that helps shift wound healing from inflammation to repair and regeneration (Landén et al., 2016), but recent studies have made it increasingly clear that macrophage states in vivo are far more complex (Orecchioni et al., 2019; Watanabe et al., 2019). We now provide evidence for an intermediate state of macrophages with both M1 and M2 characteristics in the healing wounds, namely Arg1Hi macrophages that both express inflammatory genes and exhibit a molecular profile indicative of high hypoxia and glycolysis but low OxPhos. In cancer, Arg1-expressing macrophages occupy the more hypoxic regions compared with macrophages that express Cd206 (Mrc1) (Carmona-Fontaine et al., 2017). We found Arg1Hi and C1qHi (Mrc1-expressing) macrophages to occupy distinct regions within the wound, suggesting skin wound may resemble cancer in its spatial demarcation of M1/M2 and M2 macrophages. Our RNA velocity data raise the intriguing possibility that other monocyte/macrophage subsets might have the potential to become Arg1Hi cells at some time point during wound healing. The increase of Arg1Hi and reduction of F4/80+ macrophages suggest that macrophages in aged skin wounds might be stuck at an Arg1Hi inflammatory M1/M2 hybrid stage.
Neutrophils are typically among the first inflammatory cells recruited to the wound to kill pathogens and they are later cleared by apoptosis and macrophage-mediated phagocytosis (Peiseler and Kubes, 2019; Silva, 2011; Wilgus et al., 2013). Alterations in neutrophil dynamics in aged versus young skin wounds remain controversial (Brubaker et al., 2013; Keyes et al., 2016; Mukai et al., 2019; Nishio et al., 2008; Swift et al., 2001), possibly due to differences in mouse lines used, wound size, and time point for analysis. Our study compares young and aged mice from the same source, alleviating potential complications from housing environments, microbiota compositions, and independently emerged mutations. We show that not only do neutrophils persist aberrantly in aged skin wounds but they also express higher levels of inflammatory genes such as neutrophil-attracting chemokine Cxcl3 (Rajarathnam et al., 2019) than their young counterparts. Interestingly, Arg1Hi macrophages express higher levels of Cxcl1, Cxcl2, and Cxcl3 than other monocyte/macrophage subsets. These findings suggest that chemotactic signaling from both neutrophils themselves and Arg1Hi macrophages might be responsible for elevated neutrophil infiltration and/or decreased neutrophil clearance, leading to unresolved inflammation that associates with delayed wound healing in aged skin.
Our analysis highlights elevated IL-1 signaling as a candidate pathway that drives aberrant Arg1Hi macrophage-fibroblast communication in aged skin wounds. Supporting the notion that high IL-1 signaling underlies delayed wound healing in aged skin, a previous study found inhibition of IL-1 signaling to improve healing in diabetic skin (Perrault et al., 2018). Our results also confirm, and significantly extend, previous findings (Bageghni et al., 2019; Suwara et al., 2014) to show that skin fibroblasts are a major target of Arg1Hi macrophage-originated IL-1 signaling in aged skin wounds, and that elevated signaling is achieved by both upregulation of the receptor and downregulation of the decoy inhibitor.
HF fibroblast-derived CCL19 has been shown to accelerate wound closure in human experimental models (Topouzi et al., 2020). In this context, our finding of reduced Ccl19 expression in aged skin and wounds suggests that Ccl19 may play a similar role in mouse wound healing, and its reduction may contribute to delayed healing in aged skin. We found Col1aHi dermal fibroblasts and HF-associated (DS) fibroblasts to be the major sources of Ccl19 production and DCs, particularly migrating DCs, to be the major signaling targets. Ccl19 drives Ccr7 desensitization and internalization, and has been implicated in retaining skin DCs by suppressing their migration to draining LNs (Bardi et al., 2001; Bryce et al., 2016; Farnsworth et al., 2019; Kohout et al., 2004). Thus, reduction in fibroblast-derived Ccl19 signals in aged skin provides a possible molecular basis for precocious DC mobilization in aged skin wounds. Whether and how DC changes in aged skin functionally contributes to delayed wound healing merits further investigation.
Importantly, our results identify, with strong breadth and cellular resolution, myriad candidate cell-cell communication pairs and systems-level paths of signaling pathways that are affected in aging. Our discovery of increased total number of possible signaling interactions in aged skin wounds supports the idea that inefficient healing can be a consequence of overactive but misdirected signaling and/or lack of proper downstream response. This notion is in line with previous findings that intercellular communication can be detrimental in aging and related diseases (Fafián-Labora and O’Loghlen, 2020; López-Otín et al., 2013). Given that tissue aging is likely a net sum of varying degrees of cumulative changes in single cells (Todhunter et al., 2018), our single-cell/systems approach to skin aging and associated repair is applicable to understanding aging and regenerative decline of other tissues.
Limitations of the study
CellChat predictions of cell-cell communications that occur during wound healing in aged versus young skin have not yet incorporated downstream target gene expression and spatial information. Integrating signaling target genes into the computational pipeline faces the challenge that downstream responses to various extracellular signals are often context- and cell type specific. Furthermore, a comprehensive map of spatially resolved transcriptomes in young versus aged skin wounds awaits extensive future work. Experimental and computational studies combining improved scRNA-seq platforms with single-cell assay for transposase-accessible chromatin with sequencing and spatial transcriptomics are expected to uncover regulatory and spatial constraints that will improve prediction accuracy (Longo et al., 2021).
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to, and will be fulfilled by, the Lead Contact, Xing Dai (xdai@uci.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The scRNA-seq data reported in this study have been deposited in the GEO database under accession #GSE188432 (GEO: GSE188432).
CellChat (version 1.5) R package is publicly available at https://github.com/sqjin/CellChat.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Wild-type C57BL/6J mice are from the Jackson Laboratory (Stock # 000,664). Seven-week-old (young) and 88-week-old (aged) female mice were used for the studies. All maintenance, care, and experiments have been approved and abide by regulatory guidelines of the Institutional Animal Care and Use Committee of the University of California, Irvine.
METHOD DETAILS
Wounding
For single cell experiments, 7-week-old (p49, telogen) and 88-week-old C57BL/6J mice were anesthetized using isoflurane (Primal Healthcare; NDC-66794-017-25), their backs shaved, and then a 6-mm punch (Integra; 33–36) was used to generate a full-thickness wound on each side of the mouse. Wounds were collected 4 and 7 days later for analysis.
Single cell isolation for scRNA-Seq
For UW back skin, 7-week-old and 88-week-old C57BL/6J mice were shaved, back skin removed, fat scrapped off, and then skin was minced into pieces less than 1 mm in diameter. For wounded back skin, wound-encompassing skin was first removed, large pieces of fat attached to underside of the wound were carefully scraped off, and a 10-mm punch (Acuderm; 0413) was then used to capture the wound and a portion of un-wounded skin adjacent to the wound. The wounds were then minced into pieces less than 1 mm in diameter. The minced samples were placed in 15-mL conical tubes and digested with 10 mL of collagenase mix [0.25% collagenase (Sigma; C9891), 0.01M HEPES (Fisher; BP310), 0.001M sodium pyruvate (Fisher; BP356), and 0.1 mg/mL DNase (Sigma; DN25)]. Samples were incubated at 37°C for 2 h with rotation, and then filtered through 70-μm and 40-μm filters, spun down, and resuspended in 2% FBS. Cells were stained with SytoxBlue (Thermo Fisher; S34857) as per manufacturer’s instructions and live cells (SytoxBlue-negative) were sorted using BD FACSAria Fusion Sorter.
Flow cytometry
To obtain single cell suspension, minced samples were digested with 10 mL of a solution containing 0.15% collagenase (Sigma, C9891), 0.01 M HEPES (ThermoFisher, BP310), 0.001 M Sodium Pyruvate (ThermoFisher, BP356), and 0.1 mg/mL DNase (Sigma, DN25) at 37°C for 1 h with rotation, and then filtered through a 70-μm filter, spun down, and resuspended in 2% fetal bovine serum (FBS). Skin-draining LNs (Inguinal) were dissected and pressed on dish, then resuspended in 2% FBS and filtered through a 70-μm filter. Cells were stained by incubation for 30 min at room temperature with the following antibodies diluted in PBS/2% FBS: Alexa Fluor 488-conjugated anti-CD11b (Biolegend, 101,217), PE-conjugated anti-F4/80 (Biolegend, 123,110), APC-conjugated anti-CD45 (Tonbo biosciences, clone 30-F11, 20–0451), APC-Cy7-conjugated anti-Ly6G (Tonbo biosciences, clone 1A8, 25–1276), PE/Cyanine7 anti-CD11c (Biolegend, 117,317), FITC anti-MHC Class II (I-A/I-E) (Invitrogen, 11-5321-81).
Immunofluorescence and RNAScope
Immunofluorescence was performed as previously described (Dragan et al., 2022) using the following antibodies: anti-K14 antibody (rabbit, gift of Julie Segre, National Institutes of Health), anti-F4/80 monoclonal antibody (eBioSciences; Cat No. 14-4801-82).
RNAScope was performed as previously described9 using ACD Bio’s reagents. Briefly, wounds were freshly frozen in OCT (Fisher; 4585) and sectioned at 10 μm. Sectioned tissues were fixed for 1 h at room temperature with 4% PFA, and the RNAScope Multiplex Fluorescent v2 assay was run per manufacturer’s recommendations using the following probes: Arg1 (Cat No. 403431-C1), C1qa (Cat No. 441221-C2), IL1r1 (Cat No. 413211-C2), IL1r2 (Cat No. 539491-C3), Il1a (Cat No. 440391-C1), Ccl19 (Cat No. 432881-C3), Col1a2 (Cat No. 319371-C2), and Ccr7 (Cat No. 432871-C1) from ACD, along with immunofluorescence using anti-K14 antibody (rabbit or chicken). Images were obtained using a Zeiss LSM700 confocal microscope or Keyence BZ-X800 and analyzed using FIJI software.
Single cell library generation
FACS-sorted cells were washed in PBS containing 0.04% BSA and resuspended at a concentration of approximately 1,000 cell/μL. Library generation was performed following the Chromium Single Cell 3ʹ Reagents Kits v2 (two young wound samples collected at 7dpw) or v3 (other samples) (following the CG00052 Rev B. user guide) where we target 10,000 cells per sample for capture (see Figure S1B for a detail of the number of samples used for each kit chemistry). Additional reagents included: nuclease-free water (Thermo Fisher Scientific; AM9937), low TE buffer (Thermo Fisher Scientific; 12,090–015), ethanol (Millipore Sigma; E7023–500ML), SPRIselect Reagent Kit (Beckman Coulter; B23318), 10% Tween 20 (Bio-Rad; 1662404), glycerin (Ricca Chemical Company; 3290–32), Qiagen Buffer EB (Qiagen, 19086). Each library was sequenced on the Illumina HiSeq 4000 platform (Chromium Single Cell 3′ Reagents Kits v2 samples) or Illumina NovaSeq 6000 platform (Chromium Single Cell 3′ Reagents Kits v3 samples) to achieve an average of approximately 50,000 reads per cell.
Processing and quality control of scRNA-seq data
FASTQ files were aligned utilizing 10x Genomics Cell Ranger 2.1.0 (Chromium Single Cell 3′ Reagents Kits v2 samples) or 3.1.0 (Chromium Single Cell 3′ Reagents Kits v2 samples). Each library was aligned to an indexed mm10 genome using Cell Ranger Count. Cell Ranger Aggr function was used to normalize the number of mapped reads per cells across the libraries. Quality control parameters were used to filter cells with 200–5000 genes with a mitochondrial percentage under 15% for subsequent analysis. scRNA-seq data of young mice at UW and 4dpw from Haensel et al. (2020) were processed as previously described.
Clustering analysis of scRNA-seq data
Low quality and dead cells (nGenes <200 or >5,000 or percent mitochondria >15%) were excluded from analysis (Figure S1A). Further filtering, normalization, scaling, sample integration, principal component analysis, and cell clustering were performed using the Seurat R package version 3.1.0 (Stuart et al., 2019). Briefly, single cell data matrices were merged according to time point or age (Figures S1B and 2A) and then integrated following the standard workflow described in Seurat package vignettes. To identify cell clusters, the top 10 principal components with a resolution between 0.1 and 0.3 was used. For each v3-only analysis, cells from v3 runs were isolated from each particular aggregated and clustered dataset for plotting and differential gene expression analysis. FindAllMarker function with default parameters was used to find marker genes for each cluster. Major cell types of aggregated young and aged samples for each time point were isolated first and further subclustered. Gene scoring analysis was performed in AddModuleScore function using gene lists as previously described (Haensel et al., 2020).
RNA velocity analysis
RNA velocity was calculated based on the spliced and unspliced counts as previously reported (Bergen et al., 2020; La Manno et al., 2018). Loom files were generated using velocity package and used for downstream RNA velocity analysis using scVelo package (Bergen et al., 2020). One 7dpw sample (v2 kit) was not successfully mapped to the unspliced and spliced count and was removed and therefore eliminated from RNA velocity analysis.
Systematic inference of cell-cell communication
CellChat was used to systematically infer cell-cell communication by integrating scRNA-seq data with prior ligand-receptor interaction database CellChatDB (Jin et al., 2021). CellChat prediction of putative cell-cell communications occur in three steps. First, CellChat identifies differentially over-expressed ligands and receptors for each cell group. Second, CellChat quantifies the communication probability between two interacting cell groups based on the average expression values of a ligand by one cell group and that of a receptor by another cell group, as well as their cofactors. Third, CellChat infers significant interactions using permutation tests. CellChat outputs an intercellular communication network for each ligand-receptor pair, where the calculated communication probabilities are assigned as edge weights to quantify the interaction strength. An intercellular communication network of a signaling pathway is computed by summarizing the communication probabilities of all associated ligand-receptor pairs.
Quantitative comparison of cell-cell communication across conditions
To comprehensively identify signaling changes across conditions, we developed comparative CellChat, a computational framework by first focusing on the overall signaling changes on cell population levels and then drilling down to the dysregulated signaling pathways and ligand-receptor pairs. On the cell population level, we seek to identify the cell populations the interactions of which are significantly changed and the markedly altered signaling sources and targets from one condition to another. On the signaling pathway and ligand-receptor pair level, we seek to refine the overall signaling changes in a greater detail to identify significantly altered signaling pathways and ligand-receptor pairs in terms of network architecture and interaction strength. To facilitate intuitive user-guided data interpretation, we provide a variety of visualization outputs. We have implemented these new additions in our R package CellChat version 1.5, which is available at https://github.com/sqjin/CellChat. Below we briefly describe this framework.
Prediction of general principles of cell-cell communication
We start with the big picture to predict general principles of cell-cell communication. When comparing cell-cell communication across conditions, we seek to answer the following biological questions: (1) the interaction between which cell types is significantly changed; and (2) how the major sources and targets change from one condition to another.
To identify the interaction between which cell populations showing significant changes, we compare the number of interactions between any pair of cell populations across conditions. We compute the differential number of interactions in the cell-cell communication network across two conditions and visualize them using bar plot or heatmap (ComplexHeatmap R package (Gu et al., 2016)). In the heatmap, red and blue colors represent increased and decreased signaling in the second condition compared to the first one, respectively. To identify altered signaling sources and targets across conditions, we employed network centrality analysis by computing the outdegree as measurement of likelihood being signaling sources and indegree being signaling targets. The outdegree and indegree centralities are computed by summarizing each cell population-associated outgoing and incoming communication probabilities across all signaling pathways. We then compute the differential outdegree and indegree centralities of each cell population and visualize them in a 2-dimensional (2D) space, which allows ready identification of cell populations with significant changes in sending or receiving signals across conditions. Positive and negative values indicate increased and decreased signaling in the second condition compared to the first condition, respectively.
Identification of dysregulated signaling pathways
To identify signaling changes associated with one cell population, we compute the differential outgoing and incoming interaction strength of this cell population in each cell-cell communication network between two conditions, respectively. Positive and negative values indicate increased and decreased signaling in the second condition compared to the first condition.
Identification of upregulated and downregulated ligand-receptor pairs
To identify statistically significant upregulated and downregulated ligand-receptor pairs across conditions, we combine cell-cell communication analysis with differential gene expression analysis. Specifically, for each cell group, we perform Wilcoxon rank-sum test for gene expression of cells in the second condition vs. cells in the first condition. Signaling molecules are considered be upregulated in the second condition if (i) the p values are less than 0.05, (ii) the log fold-changes are higher than 0.25, and (iii) the percentage of cells with expression in the second condition is higher than 25%. Signaling molecules are considered be downregulated in the second condition if they are upregulated in the first condition. The ligand-receptor pairs are upregulated or downregulated if both ligands and receptors are upregulated or downregulated. The cell-cell communication mediated by these dysregulated ligand-receptor pairs is visualized by chord diagram using circlize R package (Gu et al., 2014).
To quantify the enrichment of ligand-receptor pairs in one condition, we define the following enrichment score (ES)
where logFCligand is the log fold change (FC) of ligand expression in the second condition compared to the first condition. Percent2ligand and Percent1ligand are the percent of ligand-expressed cells in the second and first conditions respectively. The enrichment of ligand-receptor pairs in one condition is visualized by word cloud plot using wordcloud R package (https://cran.r-project.org/web/packages/wordcloud/index.html). In the word cloud plot, the size of each word (representing each signaling pathway or ligand) indicates its enrichment in one condition compared to another.
Cell-cell communication analysis of young and aged wounds
We first applied CellChat to young and aged skin wound datasets separately to infer cell-cell communication at 4dpw or 7dpw. We then performed comparison analysis by merging the CellChat objects from young and aged wound data analyses. After analyzing v2+v3 samples and v3-only samples separately, we utilized our customed R codes to extract the cell-cell communication information that is common between v2+v3 and v3-only data analyses, such that we can highlight the qualitatively consistent results. Specifically, we combined the subclustering results of epithelial cells (Figure S2D), fibroblasts (Figure S2G), and immune cells (Figures 3A and 4A) at 4dpw, leading to a total of 26 cell groups for analysis, including 6 epithelial subsets (basal, spinous, proliferative basal, HF 1, HF 2, HF 3), 6 fibroblast subsets (Col1ahigh, IMF I, IMF II, myofibroblast, HFDF I/II, HFDF III), and 14 immune subsets (neutrophil, monocyte, Arg1high macrophage, C1qhigh macrophage, APM 1, APM 2, OC-like, proliferative and OC-like, DC, migrating DC, Langerhans cell, mast cell, NK and T cell, T-reg). Similarly, we combined the subclustering results of epithelial cells (Figure S2J), fibroblasts (Figure S2M), and immune cells (Figures S3C and S4C) at 7dpw, leading to a total of 22 cell groups for analysis. To infer cell-cell communication between interacting cell groups, we computed the average expression of signaling molecules per cell group using 10% truncated mean, i.e., the mean value after removing the top and bottom 10% cells based on their expression levels.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are presented as the mean ± standard deviation (SD), or as indicated. The sample sizes in each plot have been listed in the Results section and Figure Legends where appropriate. For data represented as violin plots, two-tailed Wilcoxon rank-sum test was performed using R (https://www.r-project.org/). For comparison of percentage changes, prop.test was performed using R. For differential gene expression analysis between cell clusters, Wilcoxon rank-sum test was performed using R. A significance threshold of p < 0.01 was used for defining marker genes of each cell cluster. For data presented in bar plot, unpaired two-tailed Student’s t-test was used.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit Anti-Mouse K14, Purified | Gift: Julie Segre, NIH | N/A |
| Chicken Anti-Mouse K14, Purified | Gift: Julie Segre, NIH | N/A |
| F4/80 Monoclonal Antibody | eBioSciences | Cat#: 14-4801-82; RRID:AB_467558 |
| Chemicals, peptides, and recombinant proteins | ||
| Collagenase | Sigma | Cat #: C9091 |
| HEPES | Fisher | Cat #: BP310 |
| Sodium Pyruvate | Fisher | Cat #: BP356 |
| DNase | Sigma | Cat #: DN25 |
| SytoxBlue | Thermo Fisher | Cat #: S34857 |
| Nuclease-free Water | Thermo Fisher | Cat #: AM9937 |
| Low TE Buffer | Thermo Fisher | Cat #: 12090–015 |
| Ethanol | Millipore Sigma | Cat #: E7023–500ML |
| 10% Tween 20 | Bio-Rad | Cat #: 1662404 |
| Glycerin | Ricca Chemical Company | Cat #: 3290–32 |
| DAPI | Thermo Fisher | Cat #: D1306 |
| DPBS | Corning Cellgro | Cat #: 21–031-CM |
| Arg1-Probe | ACD | Cat#: 403431-C1 |
| C1qa-Probe | ACD | Cat#: 441221-C2 |
| Ccr7-Probe | ACD | Cat#: 432871-C1 |
| Ccl19-Probe | ACD | Cat#: 432881-C3 |
| Il1a-Probe | ACD | Cat#: 440391-C1 |
| Il1r2-Probe | ACD | Cat#: 539491-C3 |
| Il1r1-Probe | ACD | Cat#: 413211-C2 |
| Col1a2-Probe | ACD | Cat#: 319371-C2 |
| Critical commercial assays | ||
| Chromium Single Cell 3’ Library & Gel Bead Kit v2 | 10x Genomics | Cat #: PN-120237 |
| Chromium Single Cell 3’ Library & Gel Bead Kit v3 | 10x Genomics | Cat #: PN-1000075 |
| Chromium Single Cell A Chip Kits | 10x Genomics | Cat #: PN-120236 |
| Chromium i7 Multiplex Kit | 10x Genomics | Cat #: PN-120262 |
| SPRIselect Reagent Kit | Beckman Coulter | Cat #: B23318 |
| Multiplex Fluorescent v2 system | ACD | Cat #: 323100 |
| Deposited data | ||
| Raw scRNA-seq data | This paper | GEO: GSE188432 |
| Experimental models: Organisms/strains | ||
| C57BL/6J | Jackson Laboratory | Stock #: JAX 000664 |
| Software and algorithm | ||
| Cell Ranger 2.1.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Cell Ranger 3.1.0 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| Seurat v3 | Stuart et al. (2019) | https://satijalab.org/seurat/articles/archive.html |
| scVelo v0.2.4 | Bergen et al. (2020) | https://scvelo.readthedocs.io/ |
| velocyto v0.17.17 | La Manno et al. (2018) | http://velocyto.org/ |
| UMAP | McInnes et al. (2018) | https://github.com/lmcinnes/umap |
| CellChatDB | Jin et al. (2021) | https://github.com/sqjin/CellChat |
| CellChat v1.5 | This paper | https://github.com/sqjin/CellChat |
| ComplexHeatmap v2.7.1 | Gu et al. (2016) | https://jokergoo.github.io/ComplexHeatmap-reference/book/ |
| circlize v0.4.12 | Gu et al. (2014) | https://github.com/jokergoo/circlize |
| wordcloud v2.6 | R package | https://cran.r-project.org/web/packages/wordcloud/index.html |
| R | R core | https://www.r-project.org/ |
| Python | Python Software Foundation | https://www.python.org/ |
Highlights.
Aging alters skin epithelial, fibroblast, and immune compositions during homeostasis
Aging alters inflammatory response/macrophage heterogeneity in skin wound healing
Aging restructures cell-cell communications at a system level during wound healing
ACKNOWLEDGMENTS
We thank the Genomics High Throughput Facility and the Institute for Immunology FACS Core Facility at University of California, Irvine for expert service. Skin images in the graphical abstract are adapted from BioRender. This work was supported by NIH grants U01-AR073159 (MPIs: Q.N., M.P., and X.D.), R01-AR068074, R35 GM145307, and R01GM123731 (X.D.); NSF grant DMS1763272; Simons Foundation grant 594598 (Q.N.); and NSFC grant 11801207 (S.J.). R.V. is supported by NSF predoctoral fellowship DGE-1839285.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111155.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Agrawal A, Tay J, Ton S, Agrawal S, and Gupta S (2009). Increased reactivity of dendritic cells from aged subjects to self-antigen, the human DNA. J. Immunol 182, 1138–1145. 10.4049/jimmunol.182.2.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antsiferova M, and Werner S (2012). The bright and the dark sides of activin in wound healing and cancer. J. Cell Sci 125, 3929–3937. 10.1242/jcs.094789. [DOI] [PubMed] [Google Scholar]
- Ashcroft GS, Horan MA, and Ferguson MW (1997). Aging is associated with reduced deposition of specific extracellular matrix components, an upregulation of angiogenesis, and an altered inflammatory response in a murine incisional wound healing model. J. Invest. Dermatol 108, 430–437. 10.1111/1523-1747.ep12289705. [DOI] [PubMed] [Google Scholar]
- Avitabile S, Odorisio T, Madonna S, Eyerich S, Guerra L, Eyerich K, Zambruno G, Cavani A, and Cianfarani F (2015). Interleukin-22 promotes wound repair in diabetes by improving keratinocyte pro-healing functions. J. Invest. Dermatol 135, 2862–2870. 10.1038/jid.2015.278. [DOI] [PubMed] [Google Scholar]
- Bageghni SA, Hemmings KE, Yuldasheva NY, Maqbool A, Gamboa-Esteves FO, Humphreys NE, Jackson MS, Denton CP, Francis S, Porter KE, et al. (2019). Fibroblast-specific deletion of IL-1 receptor-1 reduces adverse cardiac remodeling following myocardial infarction. JCI Insight 5, 125074. 10.1172/jci.insight.125074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardi G, Lipp M, Baggiolini M, and Loetscher P (2001). The T cell chemokine receptor CCR7 is internalized on stimulation with ELC, but not with SLC. Eur. J. Immunol 31, 3291–3297. . [DOI] [PubMed] [Google Scholar]
- Bergen V, Lange M, Peidli S, Wolf FA, and Theis FJ (2020). Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Bio-technol 38, 1408–1414. 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- Bin S, Li HD, Xu YB, Qi SH, Li TZ, Liu XS, Tang JM, and Xie JL (2013). BMP-7 attenuates TGF-β1-induced fibroblast-like differentiation of rat dermal papilla cells. Wound Repair Regen. 21, 275–281. 10.1111/wrr.12015. [DOI] [PubMed] [Google Scholar]
- Blecher-Gonen R, Bost P, Hilligan KL, David E, Salame TM, Roussel E, Connor LM, Mayer JU, Bahar Halpern K, Tóth B, et al. (2019). Single-cell analysis of diverse pathogen responses defines a molecular roadmap for generating antigen-specific immunity. Cell Syst. 8, 109–121.e6. 10.1016/j.cels.2019.01.001. [DOI] [PubMed] [Google Scholar]
- Bonham CA, Kuehlmann B, and Gurtner GC (2020). Impaired neovascularization in aging. Adv. Wound Care 9, 111–126. 10.1089/wound.2018.0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boothby IC, Cohen JN, and Rosenblum MD (2020). Regulatory T cells in skin injury : at the crossroads of tolerance and tissue repair. Sci. Immunol 5, 9631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brubaker AL, Rendon JL, Ramirez L, Choudhry MA, and Kovacs EJ (2013). Reduced neutrophil chemotaxis and infiltration contributes to delayed resolution of cutaneous wound infection with advanced age. J. Immunol 190, 1746–1757. 10.4049/jimmunol.1201213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryce SA, Wilson RAM, Tiplady EM, Asquith DL, Bromley SK, Luster AD, Graham GJ, and Nibbs RJB (2016). ACKR4 on stromal cells scavenges CCL19 to enable CCR7-dependent trafficking of APCs from inflamed skin to lymph nodes. J. Immunol 196, 3341–3353. 10.4049/jimmunol.1501542. [DOI] [PubMed] [Google Scholar]
- Carmona-Fontaine C, Deforet M, Akkari L, Thompson CB, Joyce JA, and Xavier JB (2017). Metabolic origins of spatial organization in the tumor microenvironment. Proc. Natl. Acad. Sci. USA 114, 2934–2939. 10.1073/pnas.1700600114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinan EB, Kwee L, Nunes P, Shuster DJ, Ju G, McIntyre KW, Chiz-zonite RA, and Labow MA (1998). IL-1 receptor accessory protein is an essential component of the IL-1 receptor. J. Immunol 161, 5614–5620. http://www.ncbi.nlm.nih.gov/pubmed/9820540. [PubMed] [Google Scholar]
- Cutroneo KR (2007). TGF-β–induced fibrosis and SMAD signaling: oligo decoys as natural therapeutics for inhibition of tissue fibrosis and scarring. Wound Repair Regen. 15, S54–S60. 10.1111/J.1524-475X.2007.00226.X. [DOI] [PubMed] [Google Scholar]
- Dewor M, Steffens G, Krohn R, Weber C, Baron J, and Bernhagen J (2007). Macrophage migration inhibitory factor (MIF) promotes fibroblast migration in scratch-wounded monolayers in vitro. FEBS Lett. 581, 4734–4742. 10.1016/j.febslet.2007.08.071. [DOI] [PubMed] [Google Scholar]
- Ding X, Kakanj P, Leptin M, and Eming SA (2021). Regulation of the wound healing response during aging. J. Invest. Dermatol 141, 1063–1070. 10.1016/j.jid.2020.11.014. [DOI] [PubMed] [Google Scholar]
- Dituri F, Cossu C, Mancarella S, and Giannelli G (2019). The interactivity between TGFβ and BMP signaling in organogenesis, fibrosis, and cancer. Cells 8, 1130. 10.3390/CELLS8101130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragan M, Sun P, Chen Z, Ma X, Vu R, Shi Y, Villalta SA, and Dai X (2022). Epidermis-intrinsic transcription factor Ovol1 coordinately regulates barrier maintenance and neutrophil accumulation in psoriasis-like inflammation. J. Invest. Dermatol 142, 583–593.e5. 10.1016/J.JID.2021.08.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eming SA, Martin P, and Tomic-canic M (2014). Wound repair and regeneration : mechanisms, signaling, and translation. Sci. Transl. Med 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fafián-Labora JA, and O’Loghlen A (2020). Classical and nonclassical intercellular communication in senescence and ageing. Trends Cell Biol. 30, 628–639. 10.1016/j.tcb.2020.05.003. [DOI] [PubMed] [Google Scholar]
- Farnsworth RH, Karnezis T, Maciburko SJ, Mueller SN, and Stacker SA (2019). The interplay between lymphatic vessels and chemokines. Front. Immunol 10, 518–614. 10.3389/fimmu.2019.00518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florin L, Maas-Szabowski N, Werner S, Szabowski A, and Angel P (2005). Increased keratinocyte proliferation by JUN-dependent expression of PTN and SDF-1 in fibroblasts. J. Cell Sci 118, 1981–1989. 10.1242/jcs.02303. [DOI] [PubMed] [Google Scholar]
- Förster R, Davalos-Misslitz AC, and Rot A (2008). CCR7 and its ligands: balancing immunity and tolerance. Nat. Rev. Immunol 8, 362–371. 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- Freitas-Rodríguez S, Folgueras AR, and López-Otín C (2017). The role of matrix metalloproteinases in aging: tissue remodeling and beyond. Biochim. Biophys. Acta. Mol. Cell Res 1864, 2015–2025. 10.1016/J.BBAMCR.2017.05.007. [DOI] [PubMed] [Google Scholar]
- Ge Y, Miao Y, Gur-Cohen S, Gomez N, Yang H, Nikolova M, Polak L, Hu Y, Verma A, Elemento O, et al. (2020). The aging skin microenvironment dictates stem cell behavior. Proc. Natl. Acad. Sci. USA 117, 5339–5350. 10.1073/PNAS.1901720117/SUPPL_FILE/PNAS.1901720117.SM05.MOV. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieseck RL, Wilson MS, and Wynn TA (2018). Type 2 immunity in tissue repair and fibrosis. Nat. Rev. Immunol 18, 62–76. 10.1038/nri.2017.90. [DOI] [PubMed] [Google Scholar]
- Gillitzer R, and Goebeler M (2001). Chemokines in cutaneous wound healing. J. Leukoc. Biol 69, 513–521. 10.1189/jlb.1106655. [DOI] [PubMed] [Google Scholar]
- Goldfarb LG, and Dalakas MC (2011). Tragedy in a heartbeat: malfunctioning desmin causes skeletal and cardiac muscle disease. J. Clin. Invest 121, 455. 10.1172/JCI45852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosain A, and DiPietro LA (2004). Aging and wound healing. World J. Surg 28, 321–326. 10.1007/s00268-003-7397-6. [DOI] [PubMed] [Google Scholar]
- Groschwitz KR, Ahrens R, Osterfeld H, Gurish MF, Han X,Åbrink M, Finkelman FD, Pejler G, and Hogan SP (2009). Mast cells regulate homeostatic intestinal epithelial migration and barrier function by a chymase/Mcpt4-dependent mechanism. Proc. Natl. Acad. Sci. USA 106, 22381–22386. 10.1073/pnas.0906372106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Eils R, and Schlesner M (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849. 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- Gu Z, Gu L, Eils R, Schlesner M, and Brors B (2014). Circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812. 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- Haensel D, Jin S, Sun P, Cinco R, Dragan M, Nguyen Q, Cang Z, Gong Y, Vu R, MacLean AL, et al. (2020). Defining epidermal basal cell states during skin homeostasis and wound healing using single-cell transcriptomics. Cell Rep. 30, 3932–3947.e6. 10.1016/j.celrep.2020.02.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Suryawanshi H, Morozov P, Gay-Mimbrera J, Del Duca E, Kim HJ, Kameyama N, Estrada Y, Der E, Krueger JG, et al. (2020). Single-cell transcriptome analysis of human skin identifies novel fibroblast subpopulation and enrichment of immune subsets in atopic dermatitis. J. Allergy Clin. Immunol 145, 1615–1628. 10.1016/j.jaci.2020.01.042. [DOI] [PubMed] [Google Scholar]
- Hieronymus T, Zenke M, Baek JH, and Seré K (2015). The clash of Langerhans cell homeostasis in skin: should I stay or should I go? Semin. Cell Dev. Biol 41, 30–38. 10.1016/j.semcdb.2014.02.009. [DOI] [PubMed] [Google Scholar]
- Hudnall AM, Arthur JW, and Lowery JW (2016). Clinical relevance and mechanisms of antagonism between the BMP and activin/TGF-β signaling pathways. J. Am. Osteopath. Assoc 116, 452–461. 10.7556/JAOA.2016.089/ASSET/GRAPHIC/J_JAOA.2016.089_FIG_001.JPG. [DOI] [PubMed] [Google Scholar]
- Humbert P, Brennan M, De Lima J, Brion R, Adrait A, Charrier C, Brulin B, Trichet V, Couté Y, Blanchard F, and Layrolle P (2021). Apoptotic mesenchymal stromal cells support osteoclastogenesis while inhibiting multi-nucleated giant cells formation in vitro. Sci. Rep 11, 12144–12214. 10.1038/s41598-021-91258-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihn H, and Tamaki K (2000). Oncostatin M stimulates the growth of dermal fibroblasts via a mitogen-activated protein kinase-dependent pathway. J. Immunol 165, 2149–2155. 10.4049/jimmunol.165.4.2149. [DOI] [PubMed] [Google Scholar]
- Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, and Nie Q (2021). Inference and analysis of cell-cell communication using CellChat. Nat. Commun 12, 1088–1120. 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joost S, Annusver K, Jacob T, Sun X, Dalessandri T, Sivan U, Sequeira I, Sandberg R, and Kasper M (2020). The molecular anatomy of mouse skin during hair growth and rest. Cell Stem Cell 26, 441–457.e7. 10.1016/j.stem.2020.01.012. [DOI] [PubMed] [Google Scholar]
- Joost S, Zeisel A, Jacob T, Sun X, La Manno G, Lönnerberg P, Linnarsson S, and Kasper M (2016). Single-cell transcriptomics reveals that differentiation and spatial signatures shape epidermal and hair follicle heterogeneity. Cell Syst. 3, 221–237.e9. 10.1016/j.cels.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kämpfer H, Pfeilschifter J, and Frank S (2003). Expression and activity of arginase isoenzymes during normal and diabetes-impaired skin repair. J. Invest. Dermatol 121, 1544–1551. 10.1046/j.1523-1747.2003.12610.x. [DOI] [PubMed] [Google Scholar]
- Keller B, Yang T, Chen Y, Munivez E, Bertin T, Zabel B, and Lee B (2011). Interaction of TGFβ and BMP signaling pathways during chondrogenesis. PLoS One 6, e16421. 10.1371/JOURNAL.PONE.0016421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyes BE, Liu S, Asare A, Naik S, Levorse J, Polak L, Lu CP, Nikolova M, Pasolli HA, and Fuchs E (2016). Impaired epidermal to dendritic T cell signaling slows wound repair in aged skin. Cell 167, 1323–1338.e14. 10.1016/j.cell.2016.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohout TA, Nicholas SL, Perry SJ, Reinhart G, Junger S, and Struthers RS (2004). Differential desensitization, receptor phosphorylation, β-arrestin recruitment, and ERK1/2 activation by the two endogenous ligands for the CC chemokine receptor 7. J. Biol. Chem 279, 23214–23222. 10.1074/jbc.M402125200. [DOI] [PubMed] [Google Scholar]
- Krzyszczyk P, Schloss R, Palmer A, and Berthiaume F (2018). The role of macrophages in acute and chronic wound healing and interventions to promote pro-wound healing phenotypes. Front. Physiol 9, 419–422. 10.3389/fphys.2018.00419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lago JC, and Puzzi MB (2019). The effect of aging in primary human dermal fibroblasts. PLoS One 14. 10.1371/journal.pone.0219165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landén NX, Li D, and Ståhle M (2016). Transition from inflammation to proliferation: a critical step during wound healing. Cell. Mol. Life Sci 73, 3861–3885. 10.1007/s00018-016-2268-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence T, and Natoli G (2011). Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nat. Rev. Immunol 11, 750–761. 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- Liu J, Zhao B, Zhu H, Pan Q, Cai M, Bai X, Li X, Hu X, Zhang M, Shi J, et al. (2020). Wnt4 negatively regulates the TGF-β1-induced human dermal fibroblast-to-myofibroblast transition via targeting Smad3 and ERK. Cell Tissue Res. 379, 537–548. 10.1007/s00441-019-03110-x. [DOI] [PubMed] [Google Scholar]
- Liu N, Matsumura H, Kato T, Ichinose S, Takada A, Namiki T, Asakawa K, Morinaga H, Mohri Y, De Arcangelis A, et al. (2019). Stem cell competition orchestrates skin homeostasis and ageing. Nature 568, 344–350. 10.1038/s41586-019-1085-7. [DOI] [PubMed] [Google Scholar]
- Longo SK, Guo MG, Ji AL, and Khavari PA (2021). Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat. Rev. Genet 22, 627–644. 10.1038/s41576-021-00370-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Otín C, Blasco MA, Partridge L, Serrano M, and Kroemer G (2013). The hallmarks of aging. Cell 153, 1194–1217. 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lork M, Verhelst K, and Beyaert R (2017). CYLD, A20 and OTULIN deubiquitinases in NF-κB signaling and cell death: so similar, yet so different. Cell Death Differ. 24, 1172–1183. 10.1038/cdd.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLeod AS, and Mansbridge JN (2016). The innate immune system in acute and chronic wounds. Adv. Wound Care 5, 65–78. 10.1089/wound.2014.0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malarkannan S (2020). NKG7 makes a better killer. Nat. Immunol 21, 1137–1139. 10.1038/s41590-020-0775-5. [DOI] [PubMed] [Google Scholar]
- Martinez FO, and Gordon S (2014). The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 6, 13. 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mast BA, and Schultz GS (1996). Interactions of cytokines, growth factors, and proteases in acute and chronic wounds. Wound Repair Regen. 4, 411–420. 10.1046/j.1524-475X.1996.40404.x. [DOI] [PubMed] [Google Scholar]
- McInnes L, Healy J, Saul N, and Großberger L (2018). UMAP: uniform manifold approximation and projection. J. Open Source Softw 3, 861. 10.21105/joss.00861. [DOI] [Google Scholar]
- Minutti CM, Knipper JA, Allen JE, and Zaiss DMW (2017). Tissue-specific contribution of macrophages to wound healing. Semin. Cell Dev. Biol 61, 3–11. 10.1016/j.semcdb.2016.08.006. [DOI] [PubMed] [Google Scholar]
- Mori R, Shaw TJ, and Martin P (2008). Molecular mechanisms linking wound inflammation and fibrosis: knockdown of osteopontin leads to rapid repair and reduced scarring. J. Exp. Med 205, 43–51. 10.1084/jem.20071412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai K, Nakajima Y, Asano K, and Nakatani T (2019). Topical estrogen application to wounds promotes delayed cutaneous wound healing in 80-week-old female mice. PLoS One 14, e0225880–14. 10.1371/journal.pone.0225880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning J, Zhao Y, Ye Y, and Yu J (2019). Opposing roles and potential antagonistic mechanism between TGF-β and BMP pathways: implications for cancer progression. EBioMedicine 41, 702–710. 10.1016/J.EBIOM.2019.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio N, Okawa Y, Sakurai H, and Isobe KI (2008). Neutrophil depletion delays wound repair in aged mice. Age 30, 11–19. 10.1007/s11357-007-9043-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orecchioni M, Ghosheh Y, Pramod AB, and Ley K (2019). Macrophage polarization: different gene signatures in M1(Lps+) vs. Classically and M2(LPS−) vs. Alternatively activated macrophages. Front. Immunol 10, 1084–1114. 10.3389/fimmu.2019.01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori N, and Fuchs E (2012). Paracrine TGF-β signaling counterbalances BMP-mediated repression in hair follicle stem cell activation. Cell Stem Cell 10, 63–75. 10.1016/J.STEM.2011.11.005/ATTACH-MENT/3CFB81A3-8462-4227-8FBC-5E8F1EB1EBE3/MMC1.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakyari M, Farrokhi A, Maharlooei MK, and Ghahary A (2013). Critical role of transforming growth factor beta in different phases of wound healing. Adv. Wound Care 2, 215–224. 10.1089/wound.2012.0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda A, Qian F, Mohanty S, van Duin D, Newman FK, Zhang L, Chen S, Towle V, Belshe RB, Fikrig E, et al. (2010). Age-associated decrease in TLR function in primary human dendritic cells predicts influenza vaccine response. J. Immunol 184, 2518–2527. 10.4049/jimmunol.0901022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiseler M, and Kubes P (2019). More friend than foe: the emerging role of neutrophils in tissue repair. J. Clin. Invest 129, 2629–2639. 10.1172/JCI124616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Chang Y, and Deuel TF (2007). Pleiotrophin, a multifunctional tumor promoter through induction of tumor angiogenesis, remodeling of the tumor microenvironment and activation of stromal fibroblasts. Cell Cycle 6, 2877–2883. 10.4161/cc.6.23.5090. [DOI] [PubMed] [Google Scholar]
- Perrault DP, Bramos A, Xu X, Shi S, and Wong AK (2018). Local administration of interleukin-1 receptor antagonist improves diabetic wound healing. Ann. Plast. Surg 80 (Suppl 5), S317–S321. 10.1097/SAP.0000000000001417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajarathnam K, Schnoor M, Richardson RM, and Rajagopal S (2019). How do chemokines navigate neutrophils to the target site: dissecting the structural mechanisms and signaling pathways. Cell. Signal 54, 69–80. 10.1016/j.cellsig.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognoni E, and Watt FM (2018). Skin cell heterogeneity in development, wound healing, and cancer. Trends Cell Biol. 28, 709–722. 10.1016/j.tcb.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgonc R, and Gruber J (2013). Age-related aspects of cutaneous wound healing: a mini-review. Gerontology 59, 159–164. 10.1159/000342344. [DOI] [PubMed] [Google Scholar]
- Silva MT (2011). Macrophage phagocytosis of neutrophils at inflammatory/infectious foci: a cooperative mechanism in the control of infection and infectious inflammation. J. Leukoc. Biol 89, 675–683. 10.1189/jlb.0910536. [DOI] [PubMed] [Google Scholar]
- Solé-Boldo L, Raddatz G, Schütz S, Mallm JP, Rippe K, Lonsdorf AS, Rodríguez-Paredes M, and Lyko F (2020). Single-cell transcriptomes of the human skin reveal age-related loss of fibroblast priming. Commun. Biol 3, 188–212. 10.1038/s42003-020-0922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulos MG, and Chitnis T (2020). Opposing and potentially antagonistic effects of BMP and TGF-β in multiple sclerosis: the “Yin and Yang” of neuro-immune Signaling. J. Neuroimmunol 347, 577358. 10.1016/J.JNEUROIM.2020.577358. [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwara MI, Green NJ, Borthwick LA, Mann J, Mayer-Barber KD, Barron L, Corris PA, Farrow SN, Wynn TA, Fisher AJ, and Mann DA (2014). IL-1a released from damaged epithelial cells is sufficient and essential to trigger inflammatory responses in human lung fibroblasts. Mucosal Immunol. 7, 684–693. 10.1038/mi.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift ME, Burns AL, Gray KL, and DiPietro LA (2001). Age-related alterations in the inflammatory response to dermal injury. J. Invest. Dermatol 117, 1027–1035. 10.1046/j.0022-202x.2001.01539.x. [DOI] [PubMed] [Google Scholar]
- Tamoutounour S, Guilliams M, MontananaSanchis F, Liu H, Terhorst D, Malosse C, Pollet E, Ardouin L, Luche H, Sanchez C, et al. (2013). Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity 39, 925–938. 10.1016/j.immuni.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Todhunter ME, Sayaman RW, Miyano M, and LaBarge MA (2018). Tissue aging: the integration of collective and variant responses of cells to entropic forces over time. Curr. Opin. Cell Biol 54, 121–129. 10.1016/J.CEB.2018.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topouzi H, Boyle CJ, Williams G, and Higgins CA (2020). Harnessing the secretome of hair follicle fibroblasts to accelerate ex vivo healing of human skin wounds. J. Invest. Dermatol 140, 1075–1084.e11. 10.1016/j.jid.2019.09.019. [DOI] [PubMed] [Google Scholar]
- Varani J, Dame MK, Rittie L, Fligiel SEG, Kang S, Fisher GJ, and Voorhees JJ (2006). Decreased collagen production in chronologically aged skin: roles of age-dependent alteration in fibroblast function and defective mechanical stimulation. Am. J. Pathol 168, 1861–1868. 10.2353/ajpath.2006.051302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang AS, and Dreesen O (2018). Biomarkers of cellular senescence and skin aging. Front. Genet 9, 247–314. 10.3389/fgene.2018.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, and Colonna M (2014). Interkeukin-34, a cytokine crucial for the differentiation and maintenance of tissue resident macrophages and Langerhans cells. Eur. J. Immunol 44, 1575–1581. 10.1002/eji.201344365.Interkeukin-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wankell M, Munz B, Hübner G, Hans W, Wolf E, Goppelt A, and Werner S (2001). Impaired wound healing in transgenic mice overexpressing the activin antagonist follistatin in the epidermis. EMBO J. 20, 5361–5372. 10.1093/emboj/20.19.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Alexander M, Misharin AV, and Budinger GRS (2019). The role of macrophages in the resolution of inflammation. J. Clin. Invest 129, 2619–2628. 10.1172/JCI124615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wein AN, McMaster SR, Takamura S, Dunbar PR, Cartwright EK, Hayward SL, McManus DT, Shimaoka T, Ueha S, Tsukui T, et al. (2019). CXCR6 regulates localization of tissue-resident memory CD8 T cells to the airways. J. Exp. Med 216, 2748–2762. 10.1084/JEM.20181308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wietecha MS, Pensalfini M, Cangkrama M, Müller B, Jin J, Brinckmann J, Mazza E, and Werner S (2020). Activin-mediated alterations of the fibroblast transcriptome and matrisome control the biomechanical properties of skin wounds. Nat. Commun 11, 2604. 10.1038/s41467-020-16409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilgus TA, Roy S, and McDaniel JC (2013). Neutrophils and wound repair: positive actions and negative reactions. Adv. Wound Care 2, 379–388. 10.1089/wound.2012.0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk AJ, Rustagi A, Zhao NQ, Roque J, Martínez-Colón GJ, McKechnie JL, Ivison GT, Ranganath T, Vergara R, Hollis T, et al. (2020). A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med 26, 1070–1076. 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA (2003). IL-13 effector functions. Annu. Rev. Immunol 21, 425–456. 10.1146/annurev.immunol.21.120601.141142. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq data reported in this study have been deposited in the GEO database under accession #GSE188432 (GEO: GSE188432).
CellChat (version 1.5) R package is publicly available at https://github.com/sqjin/CellChat.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


