Abstract
Isolation and analysis of circulating rare cells is a promising approach for early detection of cancer and other diseases and for prenatal diagnosis. Isolation of rare cells is usually difficult due to their heterogeneity as well as their low abundance in peripheral blood. We previously reported a two-stage ensemble-decision aliquot ranking platform (S-eDAR) for isolating circulating tumor cells from whole blood with high throughput, high recovery rate (>90%), and good purity (>70%), allowing detection of low surface antigen-expressing cancer cells linked to metastasis. However, due to the scarcity of these cells, large sample volumes and large quantities of antibodies were required to isolate sufficient cells for downstream analysis. Here, we drastically increased the number of nucleated cells analyzed by first concentrating peripheral blood mononuclear cells (PBMCs) from whole blood by density gradient centrifugation. The S-eDAR platform was capable of isolating rare cells from concentrated PBMCs (108/mL, equivalent to processing ~20 mL of whole blood in the 1 mL sample volume used by our instrument) at a high recovery rate (>85%). We then applied the S-eDAR platform for isolating rare fetal nucleated red blood cells (fNRBCs) from concentrated PBMCs spiked with umbilical cord blood cells and confirmed fNRBC recovery by immunostaining and fluorescence in situ hybridization, demonstrating the potential of the S-eDAR system for isolating rare fetal cells from maternal PBMCs to improve noninvasive prenatal diagnosis.
Graphical Abstract
Circulating rare cells are attractive for use in prognosis of cancer and viral infections and in prenatal diagnosis. (1) However, detection and isolation of circulating rare cells is difficult due to their scarcity (2–5) and heterogeneity in expression of surface markers. (6,7) Approaches for isolating rare cells fall into two categories: label-free methods and affinity or antibody-based methods. (8) Label-free isolation methods exploit differences in cell size, (9–11) density, (12) deformability, (13) and dielectric properties. (14–16) Because some rare cells such as some circulating tumor cells (CTCs) are larger than white blood cells (WBCs), they can be isolated by filtration. (17,18) However, the overlap in the sizes of rare cells and WBCs (19) can yield low cell purity following filtration. Label-free approaches to cell isolation also suffer from low recovery, clogging of filters, complicated integration of external force fields, and loss of cell viability, limiting their utility. (20)
Antibody-based methods can be employed to isolate rare cells based on expression of surface marker proteins. Three common antibody-based approaches that have been used are (1) immunomagnetic isolation, in which rare cells are captured by antibodies immobilized to magnetic beads that are separated by a magnetic field; (21,22) (2) microfluidics approaches, (22) in which rare cells are captured by antibodies immobilized on a microfluidic chip; (23,24) and (3) fluorescence-activated methods in which rare cells are incubated with fluorophore-labeled antibodies and the cells are detected and sorted based on sensitive laser-induced fluorescence. (25,26)
Fetal cells in maternal blood during pregnancy are potentially useful for noninvasive prenatal diagnosis because they contain the whole fetal genome, uncontaminated with maternal DNA. (27) Fetal cell types present in maternal blood during pregnancy include myeloid fetal progenitor cells, fetal lymphocytes, syncytiotrophoblasts, fNRBCs, and circulating extravillous trophoblasts (CTBs). (28–30) Fetal lymphocytes and myeloid fetal progenitor cells are poor candidates for prenatal diagnosis because they persist in maternal blood for years after pregnancies, causing contamination from previous pregnancies. (31) Syncytiotrophoblasts, large, multinucleated placental epithelial cells that shed into the maternal circulation during the first trimester, are poor targets for prenatal diagnosis because they become trapped in lung capillaries and are removed from circulation. (32) CTBs are a promising target for early fetal diagnosis because their short lifespan precludes contamination from previous pregnancies; however, they are extremely rare and methods to enrich them lacked consistency and repeatability. Furthermore, confined placental mosaicism is an obstacle to interpret the results obtained by analyzing CTBs. The fNRBCs are a promising cell type for noninvasive prenatal testing with a short lifespan (25–35 day half-life in adult circulation), preventing contamination from previous pregnancies. fNRBCs are abundant in fetal blood early in gestation, (33,34) but they are rare in maternal peripheral blood with a reported occurrence ranging from 3 to 26 cells per mL. (35–37) They have been successfully enriched from maternal blood by MACS, (38) FACS, (39) and microfluidics techniques. (40)
We previously developed an ensemble-decision aliquot ranking (eDAR) platform to isolate CTCs from whole blood, which showed a higher sensitivity than the FDA-approved CellSearch platform. (41) The eDAR platform includes an optical detection system, a microfluidic active cell sorting scheme, and an on-chip filter for cell enumeration. Fluorescently labeled cells in whole blood samples can be tracked in multiple color channels and enumerated from avalanche photodiode (APD) detection traces. (42) Upon detection, CTCs are diverted to a channel that leads to an on-chip filter, where they can be fixed, permeabilized, and labeled with confirmation antibodies prior to enumeration, or diverted to a collection well or tube for downstream analysis. A high CTC recovery rate (>90%) was achieved with the eDAR platform. The eDAR platform was further improved to allow sequential sorting (S-eDAR) by using a two-stage sorting technique, which increased the CTC purity from 1% to 70%. (25)
Here, we increased the number of nucleated cells analyzed with S-eDAR by first concentrating PBMCs from whole blood by density gradient centrifugation. We then used S-eDAR to isolate rare cells from concentrated PBMCs to characterize its performance in terms of rare cell recovery rate and signal-to-noise ratio (SNR). We also developed a process to analyze cells isolated by S-eDAR with fluorescence in situ hybridization (FISH). We then applied the S-eDAR platform for isolating fNRBCs from concentrated adult female PBMCs spiked with umbilical cord blood cells to demonstrate the potential of S-eDAR for isolating rare fetal cells from maternal blood for noninvasive prenatal diagnosis.
Experimental Section
Cell Culture
MCF-7 cells (ATCC, Manassas, VA) were used as rare cell surrogates and were spiked into concentrated PBMC samples. MCF-7 cells were cultured at 37 °C and 5% CO2 in EMEM media (ATCC) supplemented with 5% fetal bovine serum and 1% penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO).
Reagents and Materials
Whole blood samples from healthy donors were obtained from PlasmaLab International (Everett, WA). Umbilical cord blood samples from women delivering male fetuses were obtained from UW Medicine following regulations of the University of Washington Institutional Review Board. All patients provided informed consent, and the research was approved by the University of Washington Ethics Committee. All blood specimens were collected in anti-coagulant tubes and processed within 6 h.
DAPI (1 mg/mL) was purchased from Thermo Fisher Scientific (Waltham, MA). Antibodies used included phycoerythrin (PE)-anti-human EpCAM (BioLegend, Inc., San Diego, CA), Alexa Fluor 488 anti-human CD45 (BioLegend), PE-anti-human CD71 (Thermo Fisher Scientific, Waltham, MA), and APC-fetal hemoglobin monoclonal antibody (HbF; Thermo Fisher Scientific). Isoton II buffer (Beckman Coulter, Brea, CA) was used as a sheath flow for eDAR chips. A solution of 1% bovine serum albumin (BSA; Sigma-Aldrich)/0.05% Tween 20 (Sigma-Aldrich) in Isoton II buffer was used to pretreat glass-bottom multi-well plates (Cellvis, Mountain View, CA) and polytetrafluoroethylene tubing (SAI Infusion Technologies, Lake Villa, IL). Isoton II buffer with 0.1% BSA was used for labeling cells. Ficoll-Paque PLUS (Sigma-Aldrich) was used as the density gradient centrifugation medium for isolation of PBMCs. SRY FISH probes were purchased from Cytocell (Tarrytown, NY).
Microfluidic Chips
Silicon masters were created by using standard photolithographic techniques described previously. (43) SU-8 2050 photoresist (MicroChem, Westborough, MA) was used for spin coating. To fabricate microfluidic chips, polydimethylsiloxane (PDMS) with a 1:10 ratio of precursor to polymer base was cured and sealed to a glass substrate immediately after exposure to O2 plasma for 30 s. If not used immediately, chips were covered and stored for up to one month until use.
Isolating PBMCs from Healthy Donor Blood by Density Gradient Centrifugation
A 2 mL sample of whole blood was diluted with an equal volume of phosphate-buffered saline (PBS) and was carefully layered onto 3 mL of Ficoll-Paque PLUS. The sample was centrifuged at 400g for 30 min at RT, and the upper layer was removed, leaving the lymphocyte layer undisturbed at the interface. The lymphocyte layer was transferred to a clean centrifuge tube and was washed twice with PBS. Mononuclear cells from cord blood were isolated by the same procedure.
Cell Recovery Measurements
MCF-7 cells (1 × 106/mL) were labeled with PE-anti-EpCAM antibody at 0.5 μg/mL for 0.5 h. Cells were washed, counted with a hemocytometer, and serially diluted. A total of 50–500 cells were spiked into whole blood or into PBMCs at PBMC densities ranging from 2 × 106/mL to 1 × 108/mL. To establish stable sorting at two junctions before running cell samples, Isoton II buffer mixed with green food dye (COV Extract Company, Rockford, OH) was run on a sequential sorting chip at 30 μL/min, and stable sorting profiles were established by adjusting the sheath flow pressure with bright-field microscopy. After stable sorting events were observed, APD traces were recorded for the remaining samples containing PBMCs. When collecting sorted cells in a multi-well plate, fresh tubing treated with 1% BSA/0.05% Tween 20 was attached to the collection outlet and run into a pretreated well. To enumerate collected cells, the plate was spun at 450g for 10 min and PE-anti-EpCAM-positive cells were counted with an inverted fluorescence microscope and a 20× 0.75 NA objective (Nikon, Tokyo, Japan). The cell recovery rate was calculated as the number of counted MCF-7 cells divided by the number of MCF-7 cells spiked into the sample.
On-Chip and Single-Cell FISH
Female PBMCs were analyzed by S-eDAR with filters for on-chip FISH. Once female PBMCs were loaded into an eDAR chip, sorting events were triggered by switching two solenoids controlling sorting events at two sorting junctions, and sorted cells remained in the filters. Captured cells were fixed with 3:1 methanol:acetic acid solution for 10 min and were washed twice with 2× saline sodium citrate (SSC) buffer. The cells were then dehydrated in a series of 70%, 85%, and 100% ethanol solutions in water. A 10 μL volume of probe mixture was infused into the filters containing fixed, sorted cells. The sample and probe were denatured simultaneously by heating the chip on a hotplate at 75 °C for 2 min. The chip was then placed in a humid, opaque container at 37 °C overnight. The cells were washed with 0.4× SSC at 72 °C for 2 min, then with 2× SSC buffer, and 0.05% Tween-20 at room temperature for 30 s. DAPI antifade (10 μL) was infused into filters for 10 min to stain cell nuclei, and cells were imaged by fluorescence microscopy.
We also used single-cell off-chip FISH for analysis because of the difficulty of on-chip FISH due to bubble formation and high background. Male PBMCs were isolated by S-eDAR as above, and their identities were confirmed by single-cell off-chip FISH to demonstrate a typical workflow. Here, sorted cells were collected in a multi-well plate and a single cell was picked up with a micromanipulator and placed onto a photo-etched coverslip with grids. The coverslip was dried and immersed in fixation buffer (3:1 methanol:acetic acid) for 1 h without agitation before FISH analysis.
Identification on fNRBCs by Immunostaining and FISH
fNRBCs from cord blood were selected as a model for verifying the enrichment performance of the S-eDAR because of their low abundance in umbilical cord blood. Mononuclear cells were isolated from cord blood by density gradient centrifugation, labeled with PE-anti-CD71 antibody, and enriched by S-eDAR as described above. fNRBCs were alternatively isolated using the MiSelect R with the SelectChip Retrieval (MiCareo Inc., Taiwan). The collected cells were immunostained to confirm their identity as fNRBCs. Collected cells were fixed with 2% paraformaldehyde for 15 min, centrifuged, and washed twice. The cells were then permeabilized with 0.2% saponin and stained with Alexa Fluor 488 anti-CD45 antibody (10 μg/mL) for identification of WBCs and with APC anti-HbF antibody (5 μg/mL) for identification of fNRBCs. DAPI (1 μg/mL) was used to stain cell nuclei. Cells were then washed, resuspended in Isoton II buffer, and transferred to a glass-bottom multi-well plate for imaging. For single-cell FISH analysis, fNRBCs (CD71+, HbF+, CD45−, and DAPI+) were picked up with a micromanipulator and placed onto a photo-etched coverslip. The coverslip was dried and immersed in KCl solution (0.075 M, Sigma-Aldrich) for 30 min, and then single-cell FISH was performed as described above, and it exhibited one red dot indicating the SRY gene on the Y chromosome, and one cyan dot indicating the X chromosome.
Isolating fNRBCs from Concentrated Adult Female PBMCs
fNRBCs from cord blood were first confirmed as positive controls, and then mononuclear cells from cord blood were mixed with PBMCs from healthy female donors, which were used as a mimic for fNRBCs in maternal PBMCs. The samples were labeled with PE-anti-CD71 antibody and were sorted by S-eDAR. Collected fNRBCs were identified by immunostaining and FISH analysis.
Results and Discussion
Workflow for Isolation and Confirmation of Rare Cells
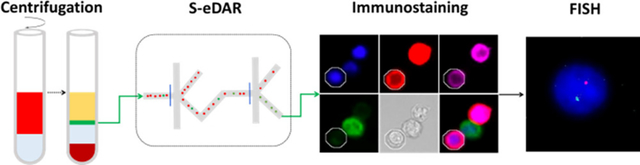
The workflow for rare cell isolation by S-eDAR and characterization by immunostaining and FISH is shown in Figure 1. PBMCs were isolated from whole blood by density gradient centrifugation (Figure 1a), labeled with PE-tagged antibodies, and loaded onto an S-eDAR chip for sorting. The identity of the sorted cells was confirmed by immunostaining and by FISH (Figure 1b), an especially useful technique for noninvasive prenatal diagnosis.
Figure 1.
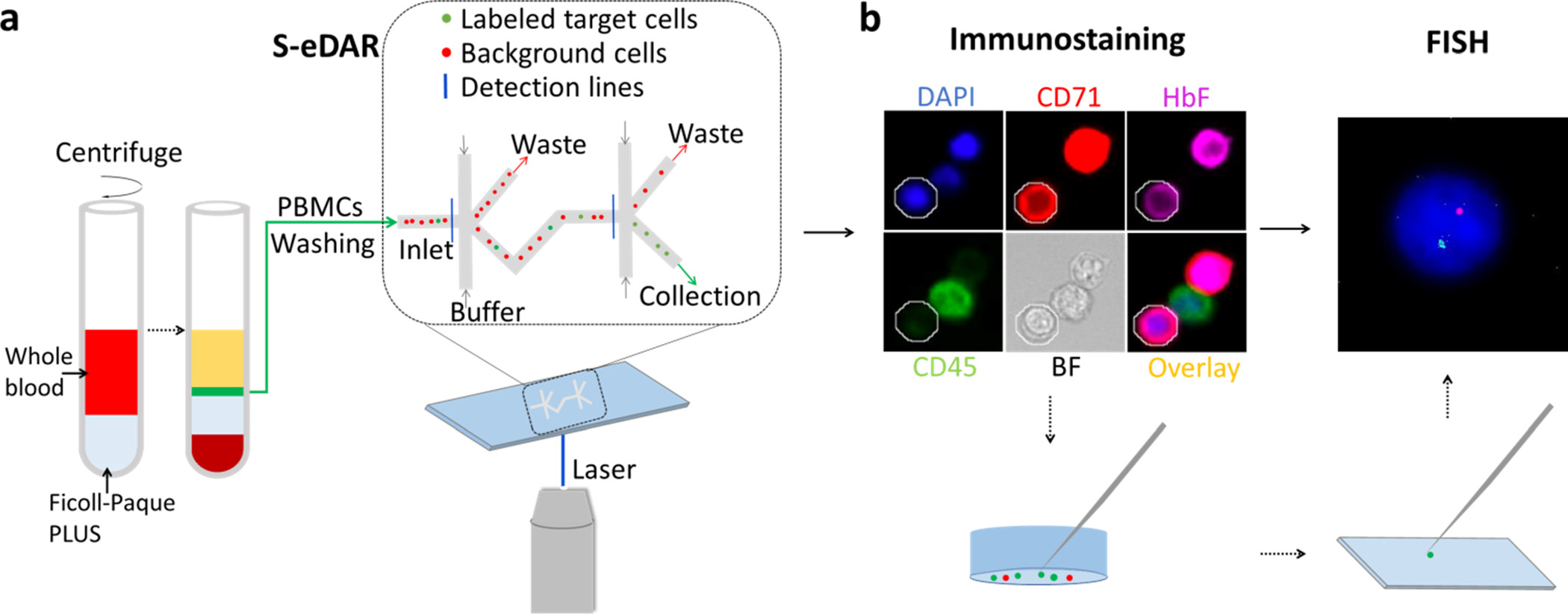
Workflow for isolation and confirmation of rare cells. (a) Diluted whole blood (red) was layered onto Ficoll-Paque PLUS (gray). After density gradient centrifugation, the thin middle layer (green) containing mononuclear cells between the plasma layer (yellow) and the Ficoll-Paque PLUS layer (light blue) was removed and washed twice and then loaded onto an S-eDAR chip. Labeled target cells were sorted twice at two sorting junctions, accompanied by background cells. (b) The identities of the sorted cells were confirmed by immunostaining and FISH analysis. Once a target cell was confirmed by immunostaining, the cell was picked up by a micromanipulator and transferred onto a slide for FISH analysis.
S-eDAR Performance on PBMCs and SNR
MCF-7 cells prelabeled with PE-anti-EpCAM antibody were spiked into concentrated PBMCs at different PBMC densities and cell sorting was performed using the S-eDAR system. Whole APD traces were recorded at two detection lines (Figure 2a,b). The SNRs at the first and second detection lines were 64.0 and 87.2, respectively, for a sample with 2.5 × 107 PBMCs/mL; 37.0 and 51.7 for 5.0 × 107 PBMCs/mL; and 34.7 and 58.1 for 1.0 × 108 PBMCs/mL (Figure 2c). As the PBMC density increased, SNR decreased but was still larger than 20, which was sufficiently sensitive for detection of MCF-7 cells, (39) allowing a recovery rate of over 85% even at the highest PBMC density (Table S1).
Figure 2.
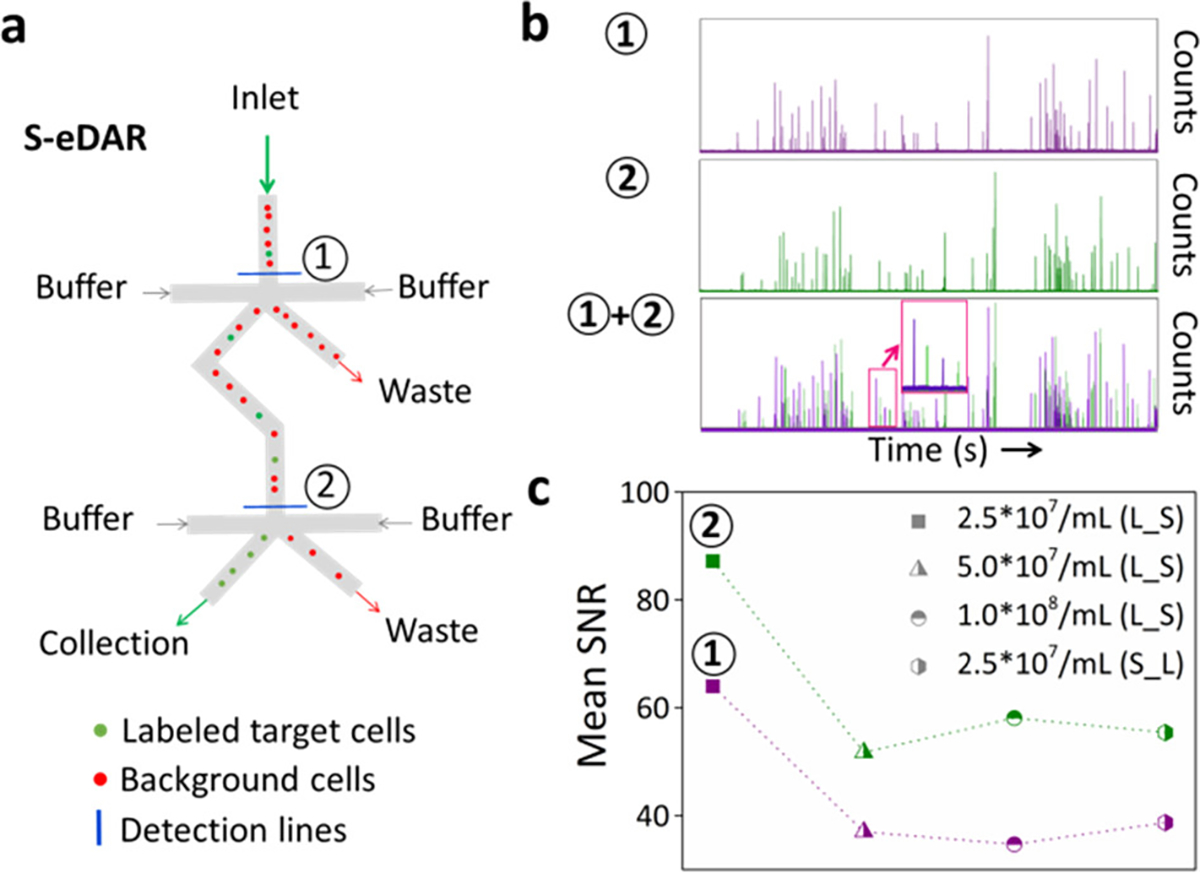
S-eDAR recovery rate and SNR. (a) Schematic showing how S-eDAR works. Labeled target cells are sorted at the two detection lines before collection. (b) Labeled MCF-7 cells were spiked into a suspension of PBMCs at 2.5 × 107 PBMCs/mL and then loaded onto an S-eDAR chip. APD traces at two detection lines and the overlay of two-channel traces were recorded. The inset in the overlay image shows that, after a signal from a labeled cell was recorded at the first detection line (purple), a signal from the same labeled cell is subsequently detected at the second detection line (green). (c) The mean SNR was calculated for traces from experiments with different PBMC densities (2.5 × 107, 5.0 × 107, and 1.0 × 108 PBMCs/mL). Since these three experiments used labeled MCF-7 cells spiked into PBMCs, they are referred to as “L_S”. Experiments in which unlabeled MCF-7 cells were spiked into PBMCs at 2.5 × 107 PBMCs/mL and then labeled are referred to as “S_L”.
We also tested an alternate method in which unlabeled rather than prelabeled MCF-7 cells were spiked into PBMCs at 2.5 × 107 PBMCs/mL, and the sample was then labeled with PE-anti-EpCAM antibody. With this method, the recovery rate was 87% and SNRs at the first and second detection lines were 38.7 and 55.4, respectively (Figure 2c). These SNRs were lower than when spiking prelabeled MCF-7 cells into 2.5 × 107 PBMCs/mL, likely due to free dyes in solution causing higher background.
On-Chip Filters and FISH Analysis
FISH analysis was performed to confirm the identity of the male fNRBCs following isolation from concentrated PBMC samples by S-eDAR. As a model system, spike-in MCF-7 cells were used as the rare cells in initial experiments. On-chip filters with a 5 μm slit size were used first since such filters retain all sorted cells (Figure 3a). MCF-7 cells emitting strong PE signals stayed in filters and could be easily counted and showed a high recovery rate (Figure 3b,c).
Figure 3.
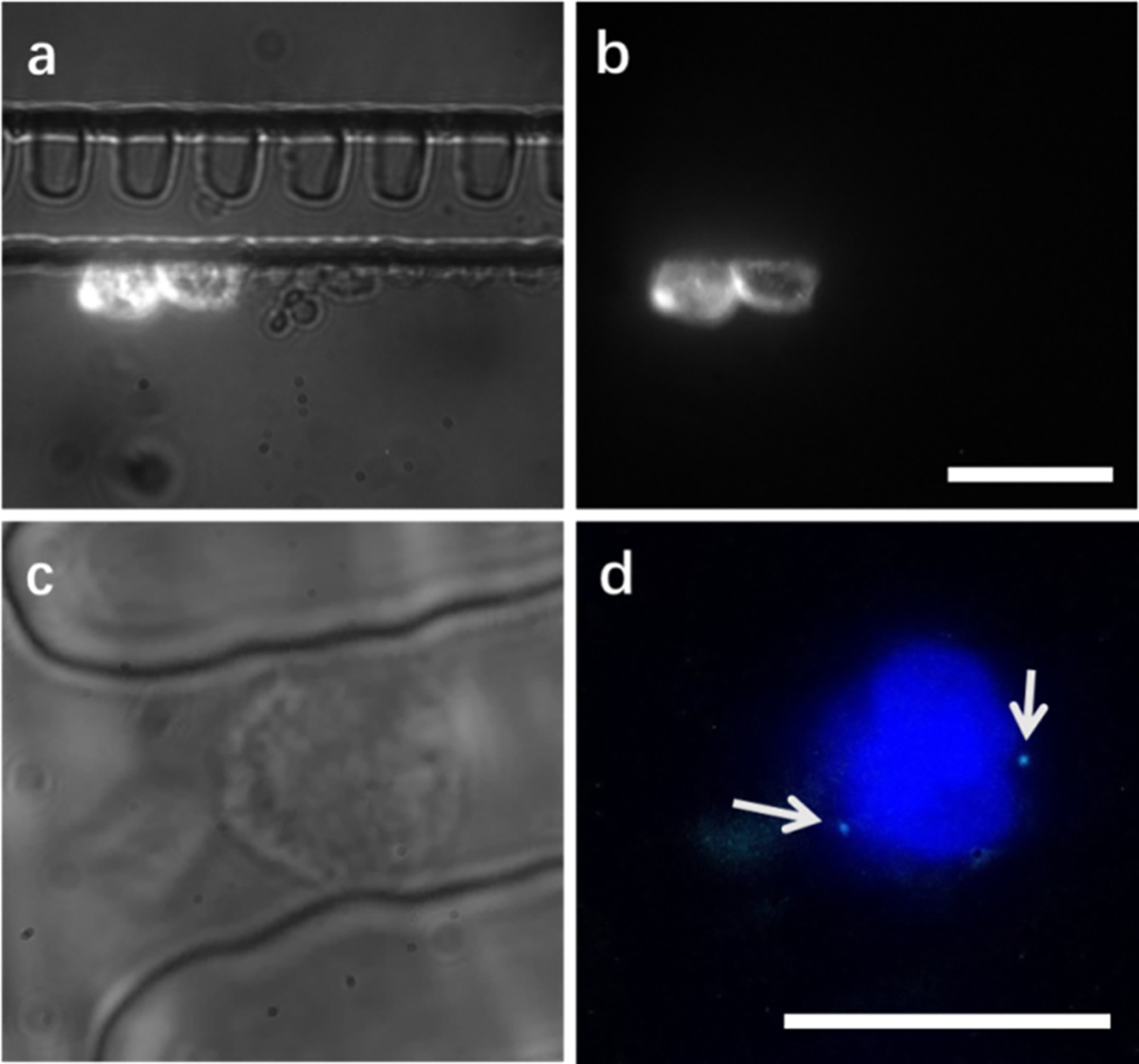
On-chip filters and FISH analysis. (a) Prelabeled MCF-7 cells were recovered by S-eDAR and remained in on-chip filters for enumeration by fluorescence microscopy. Overlay of fluorescence and bright-field images. (b) Corresponding fluorescence image. Scale bar, 20 μm. (c) Female PBMC retained in an on-chip filter for reaction with SRY FISH probe. (d) Corresponding fluorescence image showing the cell nucleus labeled by DAPI (blue) and two X chromosomes visible as two small cyan dots on opposite sides of the nucleus. Scale bar, 10 μm.
PBMCs from healthy male and female donors were also used as model cells to develop the FISH analysis protocol. The sex chromosomes of these PBMCs were labeled with an SRY FISH probe after these cells were isolated by S-eDAR. Female PBMCs exhibited two fluorescent dots for the two X chromosomes, and male PBMCs showed one fluorescent dot. On-chip FISH was performed on female PBMCs following S-eDAR. With manually triggered “positive events”, a small number of female PBMCs were sorted into filters where they were washed, fixed, dehydrated, denatured, and incubated overnight with SRY probes. One enriched cell is shown in a bright-field image in Figure 3d and in a fluorescence image in Figure 3e, showing the FISH results with two cyan dots indicating two X chromosomes.
Although on-chip filters allow a higher recovery rate, on-chip FISH posed several problems. First, micro-slits retained FISH probes, causing false-positive signals around cells. Second, bubbles entered the channels and caused high resistance, inhibiting subsequent infusion of buffer or reagents. Since we previously showed that off-chip collection with a multi-well plate can also achieve a high recovery rate (>85%), we used a micromanipulator to pick up single cells from a multi-well plate (Figure 4a) and place them on a coverslip (Figure 4b) for single-cell FISH. Figure 4
Figure 4.
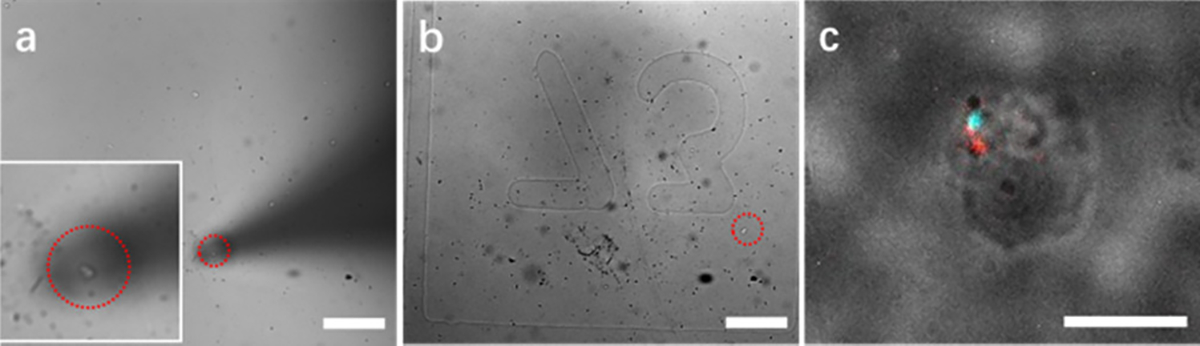
Single-cell FISH analysis. A single cell (inset) was picked up with a micromanipulator (a) and placed onto a coverslip with grids (b) for single-cell FISH analysis (c). Scale bars, 100 μm in (a) and (b) and 10 μm in (c).
Single-cell FISH images showed one red dot indicating the SRY gene on the Y chromosome and one cyan dot indicating the DXZ1 gene on the X chromosome (Figure 4c). To analyze the overall FISH success rate, five slides containing 261 total cells were tested. The success rate ranged from 64 to 80% for each slide (average, 71.6%) (Table S2). By adjusting the denaturation temperature to 78 °C, the overall success rate was improved to 85.7% (Table S3).
Isolation of fNRBCs from Adult Female PBMCs
We next applied S-eDAR to isolating male fNRBCs (from spike-in cord blood cells) from maternal PBMCs. As a positive control, we first isolated fNRBCs from cord blood by first labeling cord blood with PE-anti-CD71 antibody and then sorting by S-eDAR and immunostained with APC anti-HbF and Alexa Fluro 488 anti-CD45 antibodies (Figure 5a). SRY FISH probes were used to confirm the sorted fNRBCs. The fNRBCs (CD71+, HbF+, CD45−, and DAPI+) were characterized by single-cell FISH and exhibited one red dot indicating the SRY gene on the Y chromosome and one cyan dot indicating the X chromosome.
Figure 5.
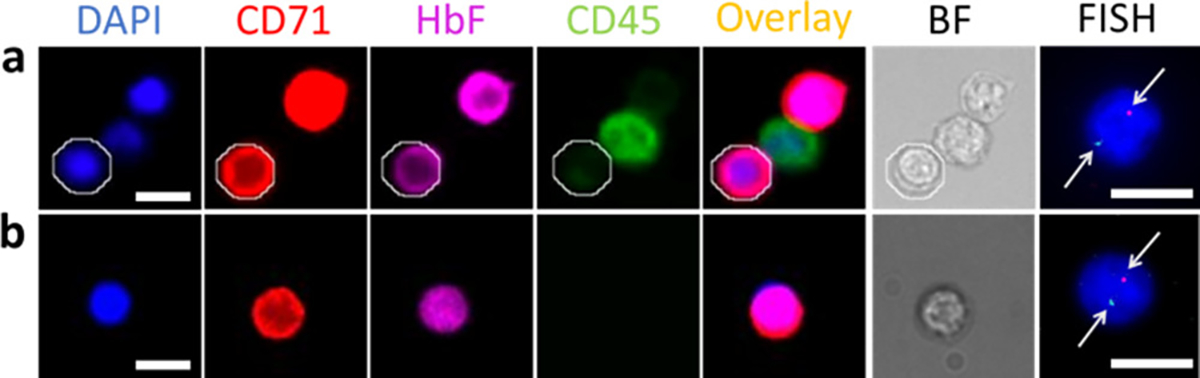
fNRBC isolation from maternal PBMCs. (a) fNRBCs from cord blood were labeled with PE-anti-CD71 antibody and sorted by S-eDAR. Enriched fNRBCs were identified by immunostaining with APC-anti-HbF and Alexa Fluor 488 anti-CD45 antibodies and with DAPI. fNRBCs were DAPI+, CD71+, HbF+, and CD45−, and white blood cells were DAPI+, CD71−, HbF−, and CD45+. One fNRBC is circled in white. In FISH analysis, male fNRBCs exhibited one red dot (arrow) indicating the SRY gene on the Y chromosome and one cyan dot (arrow) indicating the X chromosome. (b) Mononuclear cells from cord blood were mixed with PBMCs from healthy adult female donors. fNRBCs were enriched by S-eDAR and identified by immunostaining and FISH analysis. Scale bars, 10 μm.
Mononuclear cells from cord blood were then mixed with PBMCs from healthy adult female donors, which were used as a mimic for maternal PBMCs. The samples were labeled with PE-anti-CD71 antibody, enriched by S-eDAR, and immunostained. Immunostained fNRBCs were confirmed by FISH (Figure 5b). The positive SRY signal confirmed that fNRBCs were of fetal origin since adult female cells have no SRY gene.
Conclusions
100 × 106 PBMCs/mL at a recovery rate of over 85% and a high SNR, similar to the performance of S-eDAR when using whole blood samples. However, by using concentrated PBMCs obtained by density gradient centrifugation rather than whole blood, the required sample volume was reduced and the antibody cost was reduced 20-fold. More importantly, this method enables a dramatic increase in the number of nucleated cells analyzed: processing 1 mL of PBMCs at the demonstrated 108/mL is equivalent to screening at least 20 mL of whole blood since PBMCs yielded between 2 × 106 and 5 × 106 cells with density gradient separation from 1 mL whole blood, counted by using a hemocytometer. Rare cells isolated by S-eDAR remained viable for downstream analysis by immunostaining and FISH. Isolated cells were collected either in an on-chip filter or in a multi-well plate from which individual cells were picked up with a micromanipulator for single-cell FISH. We also applied S-eDAR to sorting rare fNRBCs from cord blood and from cord blood cells spiked into concentrated adult female PBMCs, demonstrating the potential of S-eDAR for isolating rare fetal cells from maternal blood for noninvasive prenatal diagnosis.
Supplementary Material
Acknowledgments
We thank Dr. Mengxia Zhao for his valuable advice and input on the project. We gratefully acknowledge support of this work by the NIH (R01HD089679) and the University of Washington.
Footnotes
The authors declare the following competing financial interest(s): DTC and PGS have financial interest in MiCareo, which has licensed the described technology from the University of Washington.
References
- 1.Chen Y; Li P; Huang P-H; Xie Y; Mai JD; Wang L; Nguyen N-T; Huang TJ Rare cell isolation and analysis in microfluidics. Lab Chip 2014, 14, 626–645, DOI: 10.1039/c3lc90136j [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Racila E; Euhus D; Weiss AJ; Rao C; McConnell J; Terstappen LW; Uhr JW Detection and characterization of carcinoma cells in the blood. Proc. Natl. Acad. Sci. U. S. A. 1998, 95, 4589–4594, DOI: 10.1073/pnas.95.8.4589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paterlini-Brechot P; Benali NL Circulating tumor cells (CTC) detection: clinical impact and future directions. Cancer Lett. 2007, 253, 180–204, DOI: 10.1016/j.canlet.2006.12.014 [DOI] [PubMed] [Google Scholar]
- 4.Dharmasiri U; Witek MA; Adams AA; Soper SA Microsystems for the capture of low-abundance cells. Annu. Rev. Anal. Chem. 2010, 3, 409–431, DOI: 10.1146/annurev.anchem.111808.073610 [DOI] [PubMed] [Google Scholar]
- 5.Alunni-Fabbroni M; Sandri MT Circulating tumour cells in clinical practice: Methods of detection and possible characterization. Methods 2010, 50, 289–297, DOI: 10.1016/j.ymeth.2010.01.027 [DOI] [PubMed] [Google Scholar]
- 6.Warkiani ME; Khoo BL; Wu L; Tay AKP; Bhagat AAS; Han J; Lim CT Ultra-fast, label-free isolation of circulating tumor cells from blood using spiral microfluidics. Nat. Protoc. 2016, 11, 134, DOI: 10.1038/nprot.2016.003 [DOI] [PubMed] [Google Scholar]
- 7.Powell AA; Talasaz AH; Zhang H; Coram MA; Reddy A; Deng G; Telli ML; Advani RH; Carlson RW; Mollick JA; Sheth S; Kurian AW; Ford JM; Stockdale FE; Quake SR; Pease RF; Mindrinos MN; Bhanot G; Dairkee SH; Davis RW; Jeffrey SS Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS One 2012, 7, e33788 DOI: 10.1371/journal.pone.0033788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bankó P; Lee SY; Nagygyörgy V; Zrínyi M; Chae CH; Cho DH; Telekes A Technologies for circulating tumor cell separation from whole blood. J. Hematol. Oncol. 2019, 12, 48, DOI: 10.1186/s13045-019-0735-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hyun K-A; Kwon K; Han H; Kim S-I; Jung H-I Microfluidic flow fractionation device for label-free isolation of circulating tumor cells (CTCs) from breast cancer patients. Biosens. Bioelectron. 2013, 40, 206–212, DOI: 10.1016/j.bios.2012.07.021 [DOI] [PubMed] [Google Scholar]
- 10.Tan SJ; Lakshmi RL; Chen P; Lim W-T; Yobas L; Lim CT Versatile label free biochip for the detection of circulating tumor cells from peripheral blood in cancer patients. Biosens. Bioelectron. 2010, 26, 1701–1705, DOI: 10.1016/j.bios.2010.07.054 [DOI] [PubMed] [Google Scholar]
- 11.Warkiani ME; Guan G; Luan KB; Lee WC; Bhagat AAS; Chaudhuri PK; Tan DS-W; Lim WT; Lee SC; Chen PCY; Lim CT; Han J Slanted spiral microfluidics for the ultra-fast, label-free isolation of circulating tumor cells. Lab Chip 2014, 14, 128–137, DOI: 10.1039/C3LC50617G [DOI] [PubMed] [Google Scholar]
- 12.Gertler R; Rosenberg R; Fuehrer K; Dahm M; Nekarda H; Siewert JR Detection of circulating tumor cells in blood using an optimized density gradient centrifugation. In Molecular Staging of Cancer; Springer: 2003; pp. 149–155, DOI: 10.1007/978-3-642-59349-9_13. [DOI] [PubMed] [Google Scholar]
- 13.Hur SC; Henderson-MacLennan NK; McCabe ER; Di Carlo D Deformability-based cell classification and enrichment using inertial microfluidics. Lab Chip 2011, 11, 912–920, DOI: 10.1039/c0lc00595a [DOI] [PubMed] [Google Scholar]
- 14.Moon H-S; Kwon K; Kim S-I; Han H; Sohn J; Lee S; Jung H-I Continuous separation of breast cancer cells from blood samples using multi-orifice flow fractionation (MOFF) and dielectrophoresis (DEP). Lab Chip 2011, 11, 1118–1125, DOI: 10.1039/c0lc00345j [DOI] [PubMed] [Google Scholar]
- 15.Huang S-B; Wu M-H; Lin Y-H; Hsieh C-H; Yang C-L; Lin H-C; Tseng C-P; Lee G-B High-purity and label-free isolation of circulating tumor cells (CTCs) in a microfluidic platform by using optically-induced-dielectrophoretic (ODEP) force. Lab Chip 2013, 13, 1371–1383, DOI: 10.1039/c3lc41256c [DOI] [PubMed] [Google Scholar]
- 16.Li M; Anand RK High-throughput selective capture of single circulating tumor cells by dielectrophoresis at a wireless electrode array. J. Am. Chem. Soc. 2017, 139, 8950–8959, DOI: 10.1021/jacs.7b03288 [DOI] [PubMed] [Google Scholar]
- 17.Kahn HJ; Presta A; Yang L-Y; Blondal J; Trudeau M; Lickley L; Holloway C; McCready DR; Maclean D; Marks A Enumeration of circulating tumor cells in the blood of breast cancer patients after filtration enrichment: correlation with disease stage. Breast Cancer Res. Treat. 2004, 86, 237–247, DOI: 10.1023/B:BREA.0000036897.92513.72 [DOI] [PubMed] [Google Scholar]
- 18.Lin HK; Zheng S; Williams AJ; Balic M; Groshen S; Scher HI; Fleisher M; Stadler W; Datar RH; Tai Y-C; Cote RJ Portable filter-based microdevice for detection and characterization of circulating tumor cells. Clin. Cancer Res. 2010, 16, 5011–5018, DOI: 10.1158/1078-0432.CCR-10-1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coumans FA; van Dalum G; Beck M; Terstappen LW Filter characteristics influencing circulating tumor cell enrichment from whole blood. PLoS One 2013, 8, e61770 DOI: 10.1371/journal.pone.0061770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cima I; Wen Yee C; Iliescu FS; Min Phyo W; Hon Lim K; Iliescu C; Han Tan M Label-free isolation of circulating tumor cells in microfluidic devices: Current research and perspectives. Biomicrofluidics 2013, 7, 011810 DOI: 10.1063/1.4780062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Allard WJ; Matera J; Miller MC; Repollet M; Connelly MC; Rao C; Tibbe AG; Uhr JW; Terstappen LW Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904, DOI: 10.1158/1078-0432.CCR-04-0378 [DOI] [PubMed] [Google Scholar]
- 22.Xu H; Dong B; Xu S; Xu S; Sun X; Sun J; Yang Y; Xu L; Bai X; Zhang S; Yin Z; Song H High purity microfluidic sorting and in situ inactivation of circulating tumor cells based on multifunctional magnetic composites. Biomaterials 2017, 138, 69–79, DOI: 10.1016/j.biomaterials.2017.05.035 [DOI] [PubMed] [Google Scholar]
- 23.Dharmasiri U; Njoroge SK; Witek MA; Adebiyi MG; Kamande JW; Hupert ML; Barany F; Soper SA High-throughput selection, enumeration, electrokinetic manipulation, and molecular profiling of low-abundance circulating tumor cells using a microfluidic system. Anal. Chem. 2011, 83, 2301–2309, DOI: 10.1021/ac103172y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S; Liu K; Liu J; Yu ZT-F; Xu X; Zhao L; Lee T; Lee EK; Reiss J; Lee Y-K; Chung LWK; Huang J; Rettig M; Seligson D; Duraiswamy KN; Shen CK-F; Tseng H-R Highly efficient capture of circulating tumor cells by using nanostructured silicon substrates with integrated chaotic micromixers. Angew. Chem., Int. Ed. 2011, 123, 3140–3144, DOI: 10.1002/ange.201005853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson ES; Xu S; Yu H-M; Fang W-F; Qin Y; Wu L; Wang J; Zhao M; Schiro PG; Fujimoto B; Chen J-L; Chiu DT Isolating Rare Cells and Circulating Tumor Cells with High Purity by Sequential eDAR. Anal. Chem. 2019, 91, 14605–14610, DOI: 10.1021/acs.analchem.9b03690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson ES; Anand RK; Chiu DT Improved detection by ensemble-decision aliquot ranking of circulating tumor cells with low numbers of a targeted surface antigen. Anal. Chem. 2015, 87, 9389–9395, DOI: 10.1021/acs.analchem.5b02241 [DOI] [PubMed] [Google Scholar]
- 27.Hou S; Chen JF; Song M; Zhu Y; Jan YJ; Chen SH; Weng TH; Ling DA; Chen SF; Ro T; Liang AJ; Lee T; Jin H; Li M; Liu L; Hsiao YS; Chen P; Yu HH; Tsai MS; Pisarska MD; Chen A; Chen LC; Tseng HR Imprinted NanoVelcro Microchips for Isolation and Characterization of Circulating Fetal Trophoblasts: Toward Noninvasive Prenatal Diagnostics. ACS Nano 2017, 11, 8167–8177, DOI: 10.1021/acsnano.7b03073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tjoa ML; Delli-Bovi L; Johnson KL; Bianchi DW Antibodies to trophoblast antigens HLA-G, placenta growth factor, and neuroD2 do not improve detection of circulating trophoblast cells in maternal blood. Fetal Diagn. Ther. 2007, 22, 85–89, DOI: 10.1159/000097102 [DOI] [PubMed] [Google Scholar]
- 29.Zhang L; Wang Y; Liao AH Quantitative abnormalities of fetal trophoblast cells in maternal circulation in preeclampsia. Prenatal Diagn. 2008, 28, 1160–1166, DOI: 10.1002/pd.2135 [DOI] [PubMed] [Google Scholar]
- 30.Bianchi DW; Zickwolf GK; Weil GJ; Sylvester S; DeMaria MA Male fetal progenitor cells persist in maternal blood for as long as 27 years postpartum. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 705–708, DOI: 10.1073/pnas.93.2.705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pertl B; Bianchi DW First trimester prenatal diagnosis: Fetal cells in the maternal circulation. Semin. Perinatol. 1999, 23, 393–402, DOI: 10.1016/S0146-0005(99)80005-6 [DOI] [PubMed] [Google Scholar]
- 32.Oudejans CBM; Tjoa ML; Westerman BA; Mulders MAM; Van Wijk IJ; Van Vugt JMG Circulating trophoblast in maternal blood. Prenatal Diagn. 2003, 23, 111–116, DOI: 10.1002/pd.539 [DOI] [PubMed] [Google Scholar]
- 33.Thomas DB; Yoffey JM Human foetal haemopoiesis I. The cellular composition of foetal blood. Br. J. Haematol. 1962, 8, 290–295, DOI: 10.1111/j.1365-2141.1962.tb06523.x [DOI] [PubMed] [Google Scholar]
- 34.Millar DS; Davis LR; Rodeck CH; Nicolaides KH; Mibashan RS Normal blood cell values in the early mid-trimester fetus. Prenatal Diagn. 1985, 5, 367–373, DOI: 10.1002/pd.1970050602 [DOI] [PubMed] [Google Scholar]
- 35.Simpson JL; Elias S Isolating fetal cells from maternal blood: advances in prenatal diagnosis through molecular technology. JAMA 1993, 270, 2357–2361, DOI: 10.1001/jama.1993.03510190113036 [DOI] [PubMed] [Google Scholar]
- 36.Wei X; Cai B; Chen K; Cheng L; Zhu Y; Wang Z; Sun Y; Liu W; Guo S-S; Zhang Y; Zhao X-Z Enhanced isolation and release of fetal nucleated red blood cells using multifunctional nanoparticle-based microfluidic device for non-invasive prenatal diagnostics. Sens. Actuators, B 2019, 281, 131–138, DOI: 10.1016/j.snb.2018.10.027 [DOI] [Google Scholar]
- 37.Sun Y; Li N; Cai B; Wei X; Wang Z; Cui H; Zhao D; Zhang Y; Zhao XZ A Biocompatible Nanofibers-Based Microchip for Isolation and Nondestructive Release of Fetal Nucleated Red Blood Cells. Adv. Mater. Interfaces 2020, 7, 2001028, DOI: 10.1002/admi.202001028 [DOI] [Google Scholar]
- 38.Zheng YL; Carter NP; Price CM; Colman SM; Milton PJ; Hackett GA; Greaves MF; Ferguson-Smith MA Prenatal diagnosis from maternal blood: simultaneous immunophenotyping and FISH of fetal nucleated erythrocytes isolated by negative magnetic cell sorting. J. Med. Genet. 1993, 30, 1051–1056, DOI: 10.1136/jmg.30.12.1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bianchi DW; Flint AF; Pizzimenti MF; Knoll JH; Latt SA Isolation of fetal DNA from nucleated erythrocytes in maternal blood. Proc. Natl. Acad. Sci. U. S. A. 1990, 87, 3279–3283, DOI: 10.1073/pnas.87.9.3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.He Z; Guo F; Feng C; Cai B; Lata JP; He R; Huang Q; Yu X; Rao L; Liu H; Guo S; Liu W; Zhang Y; Huang TJ; Zhao X Fetal nucleated red blood cell analysis for non-invasive prenatal diagnostics using a nanostructure microchip. J. Mater. Chem. B 2017, 5, 226–235, DOI: 10.1039/C6TB02558G [DOI] [PubMed] [Google Scholar]
- 41.Schiro PG; Zhao M; Kuo JS; Koehler KM; Sabath DE; Chiu DT Sensitive and High-Throughput Isolation of Rare Cells from Peripheral Blood with Ensemble-Decision Aliquot Ranking. Angew. Chem., Int. Ed. 2012, 51, 4618–4622, DOI: 10.1002/anie.201108695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao M; Nelson WC; Wei B; Schiro PG; Hakimi BM; Johnson ES; Anand RK; Gyurkey GS; White LM; Whiting SH; Coveler AL; Chiu DT New generation of ensemble-decision aliquot ranking based on simplified microfluidic components for large-capacity trapping of circulating tumor cells. Anal. Chem. 2013, 85, 9671–9677, DOI: 10.1021/ac401985r [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qin Y; Wu L; Schneider T; Yen GS; Wang J; Xu S; Li M; Paguirigan AL; Smith JL; Radich JP; Anand RK; Chiu DT A Self-Digitization Dielectrophoretic (SD-DEP) Chip for High-Efficiency Single-Cell Capture, On-Demand Compartmentalization, and Downstream Nucleic Acid Analysis. Angew. Chem., Int. Ed. 2018, 130, 11548–11553, DOI: 10.1002/ange.201807314 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


