Abstract
As the home of the cell’s genetic information, the nucleus plays a critical role in determining the cell’s fate and function in response to various signals and stimuli. In addition to biochemical inputs, the nucleus is constantly exposed to intrinsic and extrinsic mechanical forces that trigger dynamic changes in nuclear structure and morphology. Emerging data suggest that the physical deformation of the nucleus modulates many cellular and nuclear functions, which have long been considered downstream of cytoplasmic signaling pathways and dictated by DNA genomic sequences. In this Review, we discuss an emerging perspective on the mechanoregulation of the genetic machinery that considers the physical connections from chromatin to nuclear lamina and cytoskeletal filaments as a single mechanical unit. We describe key mechanisms of the spatial and temporal coordination of nuclear deformations and provide a critical review of the structural and functional adaptive responses of the nucleus to deformations. We then consider the contribution of nuclear deformations to the regulation of important cellular functions, including muscle contraction, cell migration, and human disease pathogenesis. Collectively, these emerging insights shed new light on the dynamics of nuclear deformations and their roles in cellular mechanobiology.
Introduction
As the largest and stiffest organelle of eukaryotic cells1, the nucleus is constantly subjected to intrinsic and extrinsic forces that can lead to small and large scale nuclear deformations. Accumulating evidence suggests that the nucleus contributes to the cell’s perception of mechanical stimuli and the corresponding cellular response through dynamic changes of its structure and morphology2,3. The nucleus must therefore be considered not only as the primary site of gene replication and transcription but also as a fundamental mechanical component of the cell. Emerging views of the nucleus indicate a more dynamic organelle than anticipated, capable of mechanosensing and rapid remodeling, thereby orchestrating key cellular functions in response to mechanical stimulation, making the nucleus a key mechanoresponsive and mechanosensitive organelle.
Here, we define “mechanoresponsive” as responding to mechanical stimulation, either directly or downstream of other processes triggered by the mechanical stimulus. In contrast, “mechanosensitive” refers to the cellular elements or processes directly involved in the sensing of mechanical forces or deformations by converting the mechanical inputs into biochemical signals, a process known as “mechanotransduction”. Since ‘mechanotransduction’ is often found more broadly defined as also encompassing cellular responses downstream of the initial mechanosensing (or ‘transduction’) event, we will refer to ‘mechanosensing’ throughout the text to avoid any confusion.
The mechanoresponsive properties of the nucleus are now well recognized, including its ability to adapt to the cell’s physical microenvironment with changes in nuclear morphology or the expression of specific genes4,5. In contrast, the role of the nucleus as a mechanosensitive organelle has only recently begun to emerge. Nuclear deformation is a key component of the correct regulation of cell function in vivo and its importance has been highlighted by the observation of abnormally shaped nuclei6 and impaired mechanosensing mechanisms7 in many human diseases. Nuclear deformability plays crucial roles in activating and modulating cellular mechanoresponsive signaling. For example, several lines of evidence indicate that forces acting on the nucleus can induce sufficient nuclear deformations to modulate chromatin structure and trigger important protein conformational changes, thereby activating or repressing mechanoresponsive genes8,9.
In this Review, we provide an overview of the current understanding of the physical properties of the nucleus, describe the contribution of specific components, discuss the physical connections between the NE and the cytoskeleton, and shed light on the role of nuclear deformation in cellular mechanosensing and mechanoresponses.
Section 1 – Nuclear structure, organization, and connections to the cytoskeleton
Nuclear deformations are determined by the balance between the mechanical properties of the nucleus and the mechanical forces acting on it. Here we describe components constituting the nuclear structure from the inside out, as well as the physical connections between the nucleus and the cytoskeleton (Fig. 1). Importantly, extensive physical interactions between these components ensure that forces from the cytoskeleton are transmitted to and across the nuclear interior (see textbox “The interconnected nucleus”).
Figure 1 – The nuclear envelope and nucleo-skeletal interactions.
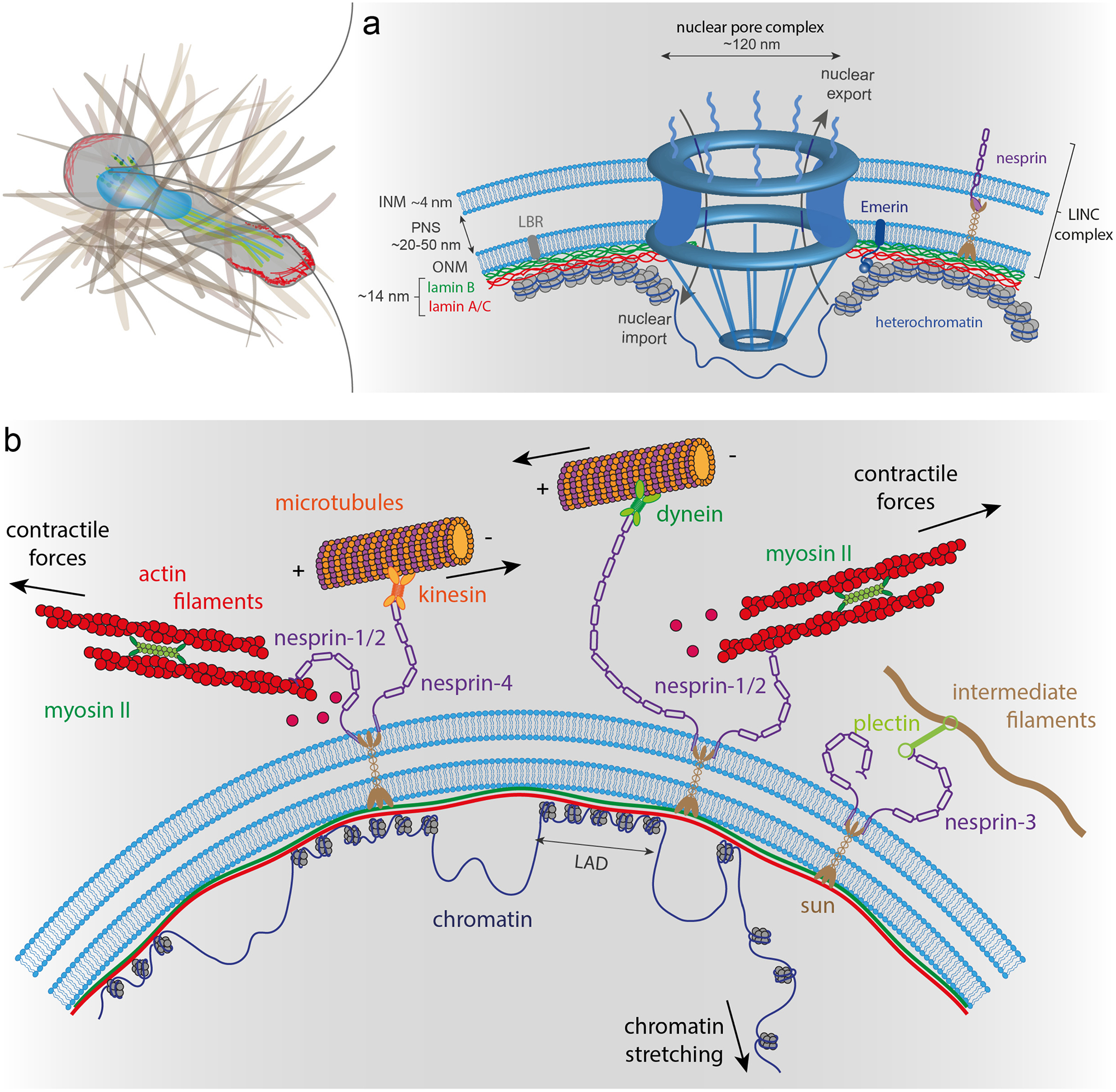
(a) The nuclear envelope (NE) is composed of the outer (ONM) and inner (INM) nuclear membranes, which form a double lipid bilayer. The nuclear lamina is attached to the INM and in close contact with condensed chromatin, while nuclear pore complexes (NPCs) are surrounded by less condensed chromatin. The genomic regions connected to the lamina are lamina-associated chromatin domains (LADs), which have low transcriptional activity. The nuclear interior is connected to cytoskeletal filaments by nesprins and SUN domain proteins. Nesprin-1 and nesprin-2 bind to actin filaments, whereas nesprin-3 interacts with intermediate filaments. Nesprins-1, −2, and −4 can interact with microtubules via kinesin and dynein molecular motors. (b) NPCs allow controlled nuclear import and export of large molecules. The nuclear lamina meshwork composed of A-type and B-type lamins binds to the INM. Lamins, along with other INM proteins, such as LBR and emerin, anchor chromatin to the NE. Nesprins, ONM, SUN domain proteins and INM form together the LINC complex.
Textbox 1 - The interconnected nucleus.
Chromatin binds to scaffolding at the nuclear periphery, particularly interacting with the nuclear lamina and NPCs. Heterochromatin interacts with the nuclear lamina via lamina-associated domains (LADs), while euchromatin associates with NPCs (Fig. 1a)223,224. Chromatin tethered to the nuclear periphery in LADs contains mostly silent or weakly expressed genes225. In contrast to LADs, genes associated with NPCs are often transcriptionally active226,227. LADS arise from many interactions, since lamins can bind non-specifically to histones and chromatin through bridging proteins such as LAP2α, lamin B receptors (LBR), Barrier-to-Autointegration Factor (BAF) and emerin (Fig. 1b)228. Specific NE proteins often interact with different DNA regions229, and the exact mechanisms that determine chromatin-nuclear lamina association remain to be elucidated230. The physical connections between chromatin and the NE provide not only control over gene expression, but also provide increase nuclear stiffness and stability, akin to the mechanical reinforcement used in composite materials or cross-linked polymer networks231–233.
Within the nuclear interior, chromatin from different chromosomes occupies distinct territories (Fig. 2). Inter-chromosome interactions are supported by mechanical crosslinking of neighboring chromatin at intervals of around 25 kb14,16–18. Heterochromatin protein 1 (HP1a) is a putative crosslinker that acts through either DNA, H3K9me3 heterochromatin, or phase separation236–238. Other candidates for linking the chromatin interior include proteins like CTCF and cohesin involved in chromatin looping and topologically associating domains (TADs)239 (Fig. 2), RNA scaffolding240, and nucleoli, whose periphery is covered with heterochromatin241. Obtaining a better understanding of the precise spatial organization of chromatin within the nuclear interior, and how this organization affects transcriptional regulation, remains a topic of extensive research 233,242.
The nuclear interior
The nuclear interior primarily consists of chromatin and nuclear bodies such as nucleoli, Cajal bodies, and promyelocytic leukemia (PML) bodies, which are membrane-less structures with specific signaling and processing functions10. Chromatin is composed of DNA and DNA-binding proteins, particularly histones (Fig. 2). Chromatin can be classified into two categories, depending on its level of compaction, transcriptional activity, and histone modifications. The loosely packed euchromatin is transcriptionally accessible and mostly localized in the nuclear interior and near nuclear pores. Densely packed heterochromatin is considered transcriptionally repressed and tends to be located at the nuclear periphery and around the nucleoli, with likely connections in between11.
Figure 2 – Chromatin organization and consequences of nuclear deformations in DNA organization.
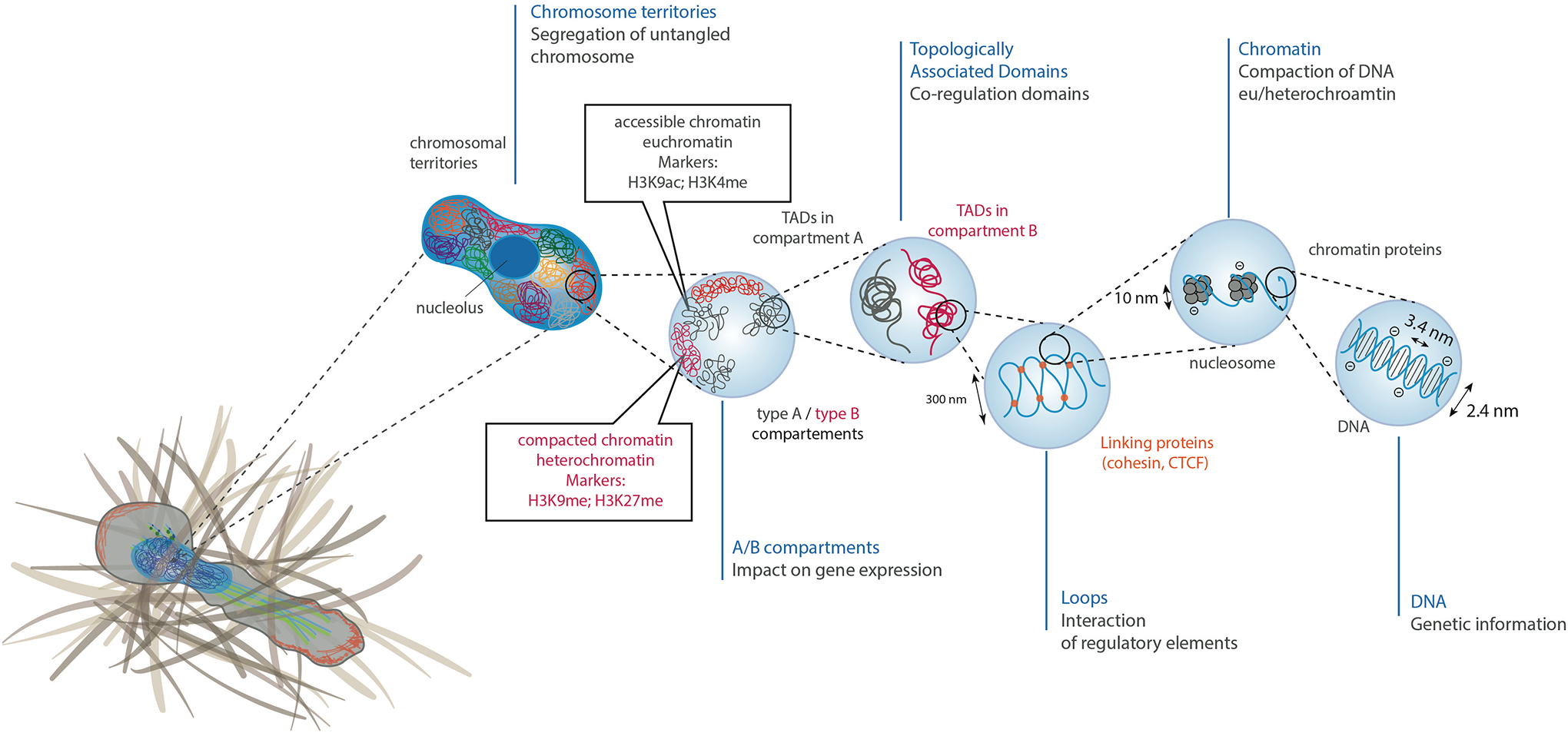
Chromosomal DNA is packaged inside the cell nucleus with the help of histones. At the simplest level, chromatin is a double-stranded helical structure of DNA. The negatively charged DNA double helix is complexed with histones, which are positively charged proteins, to form nucleosomes. Inside the interphase nucleus, chromosomes occupy distinct territories (highlighted by different colors). Within each chromosome territory, the chromatin is folded into multiple loops and segregated into two distinct compartments: compartment A clustered around nucleolus and nuclear bodies (permissive region, in grey), and compartment B (repressive region, in red) located at the nuclear periphery. Within compartments, chromatin is further partitioned into topologically associating domains (TADs), which have preferential intradomain interactions compared to interdomain interactions with the neighboring cis chromatin domains. Histone methylation, particularly at residues H3K9 and H3K27, are often associated with heterochromatin, whereas histone acetylation, particularly at residue H3K9 or histone methylation at residue H3K4 are typically associated with euchromatin.
The nuclear envelope
The NE serves multiple pivotal functions: it controls access of cytoplasmic proteins to the genome, provides structural stability to the nucleus, and physically connects the nuclear interior and cytoskeleton (Fig. 1a). The NE is comprised of the nuclear membranes, the nuclear lamina, and nuclear pore complexes (NPCs). The inner and outer nuclear membranes (INM and ONM, respectively) are two concentric lipid bilayers, each ≈4 nm thick, separated by the ≈20–50 nm wide perinuclear space12 (Fig. 1b). The ONM is contiguous with the endoplasmic reticulum (ER) and provides an external reservoir of lipids to adapt nuclear surface area upon deformation, although membrane recruitment to the NE may be limited by resistance from the ER. Nuclear membrane wrinkling and folds present at low membrane tension provide an additional membrane reservoir for adjusting nuclear shape13. NPCs regulate the active nuclear transport of macromolecules larger than ≈50 kDa into and out of the nucleus14,15. NPCs are homogeneously distributed over the nuclear membrane surface by the underlying structure provided by lamins and nucleoporin ELYS15. NPCs expanding in response to mechanical stress can account for up to 10% of nuclear surface expansion during nuclear deformations16–18.The NE and ER additionally contain mechanosensitive ion channels such as Piezo119 and inositol triphosphate receptors (InsP3Rs)20 that can respond to nuclear membrane tension (see also textbox “Nuclear mechanoresponses and mechanosensing”). The nuclear lamina, a dense protein network underlying INM, is primarily comprised of lamins. Lamins assemble into 300–400 nm long and ≈3.5 nm thick non-polar filaments, and form a ≈14–30 nm thick meshwork21,22. Recent electron cryotomography imaging has revealed that the lamin meshwork organization in mammalian somatic cells is heterogenous21, substantially deviating from the regular meshwork structure reported for Xenopus oocytes.
Textbox 2 – Nuclear mechanoresponses and mechanosensing.
Although it is now well recognized that nuclear deformation has both rapid and long-lasting consequences on nuclear and cellular function, the precise mechanisms by which nuclear deformation is translated into biochemical signals, and to what degree the nucleus itself serves as a cellular mechanosensor, remain incompletely understood. As a note of caution, many nuclear changes cited as indicators of nuclear mechanosensing in response to external mechanical stimuli (e.g., altered nuclear shape, chromatin organization, gene expression), may reflect at least in part downstream effects of signaling pathways initiated in the cytoplasm or cell surface, rather than direct nuclear mechanosensing. In the following, we highlight recent findings and novel insights into established and proposed nuclear mechanosensing mechanisms. For a more detailed discussion, we refer the reader to some excellent recent reviews243–246.
Stretch-activated opening of channels in the nuclear membranes
NPCs allow passage of small molecules while excluding larger molecules that do not contain nuclear localization sequences or are transported by other proteins. Recent live cell imaging, electron microscopy, and cryoelectron-tomography studies found that NPCs are highly sensitive to nuclear membrane tension18,215,247, increasing their diameter in response to elevated nuclear membrane tension and thus facilitating nuclear import, including of the mechanoresponsive transcription factor YAP (Fig. 5c)248. The NE and ER contain various other stretch-sensitive ion channels, such as Piezo1 and InsP3R. Increased nuclear membrane tension, in response to cell compression, osmotic swelling, or stretching application, may trigger opening of these channels and release of calcium from the ER and perinuclear space, which can lead to changes in chromatin organization19 and increased cell contractility125,169. However, it remains unclear whether opening of these ion channels in response to cellular deformation occurs at the NE, ER, or the plasma membrane. One interesting hypothesis is that all three locations contribute to cellular mechanotransduction, and spatial coordination between ion channels in the these different membranes allow cells to distinguish between different sources of nuclear membrane strain, such as osmotic swelling and compression125,249.
Mechanosensing of the nuclear membranes and nuclear envelope proteins
Changes in the tension or curvature of the nuclear membranes alone can be sufficient to induce nuclear mechanoresponses by altering the packing and/or composition of nuclear membrane phospholipids, thereby facilitating binding of cytosolic phospholipase A2 (cPLA2) to the INM, which is further amplified by increased intranuclear calcium concentrations246,250,251. Recruitment of cPLA2 to the INM leads to the production of arachidonic acid and lysophosphatidic acid (LPA) production that can trigger increased cell contractility and other downstream responses (Fig. 5d)75,88,165,240.
Besides altering protein-interaction with the nuclear membranes, forces acting on the nucleus can also lead to local unfolding, conformational changes, and increased phosphorylation of lamins100,103,253–255 (Fig. 5e), although the functional relevance of these changes remain to be fully characterized. Furthermore, force application to the nucleus via nesprins leads to phosphorylation of emerin via Src kinases, resulting in the recruitment of lamins to the NE and nuclear stiffening256. Although it remains unclear whether the increased phosphorylation is due to mechanically induced activation of nuclear Src kinase or emerin becoming more accessible to the kinase, this study, which was conducted on isolated nuclei, provided some of the most direct evidence for nuclear mechanosensing.
Force-induced changes in chromatin organization
Several studies have demonstrated mechanically induced changes in chromatin organization that could affect gene expression, including in neutrophils that had migrated through tight constrictions257, macrophages under spatial confinement206, and a 3D chemomechanical model of the nuclear interior and its connections to the cytoskeleton258, but they did not completely address whether the effects were nucleus-intrinsic or mediated by cytoplasmic signals. Support for direct nuclear mechanosensing comes from two recent studies, which found that force application to the cell surface leads to near instantaneous chromatin deformation, visualized by tracking multiple GFP-LacI labeled genomic loci, and rapid (<15 sec) increase in transcription of the corresponding transgene and other genes203,204. The magnitude of the response was directly related to the extent of chromatin deformation and histone methylation status. Of note, the chromatin ‘stretching’ reported in these studies likely does not reflect stretching of the DNA itself, but rather partial unpacking of the chromatin, which may promote access to transcriptional regulators or polymerases204 (Fig. 5f). Depletion of lamins, emerin or LINC complex components abolished the force induced gene expression203, pointing to the importance of nucleo-cytoskeletal coupling. The effect of LINC complex disruption on the activation of mechanoresponsive genes contrasts with a previous study, in which LINC complex disruption did not alter expression of a several mechanoresponsive genes despite reducing nuclear deformation32, possibly reflecting differences in cell type, force application, or extent of nuclear deformation.
Another intriguing thought is that liquid-liquid phase separation (LLPS) of intrinsically disordered proteins within the nucleus could contribute to nuclear mechanosensing. LLPS inside the nucleus can exert significant mechanical forces that alter chromatin-organization and rearrangements84,259. Although the previous studies did not address whether these changes were associated with altered gene expression, one could speculate that externally applied forces and nuclear deformation could similarly affect LLPS events in the nucleus and thereby modulate nuclear function. In addition to LLPS, changes in molecular crowding resulting from mechanically induced alterations in nuclear volume13,162 could modulate various nuclear functions.
In mammalian somatic cells, the nuclear lamina is predominantly composed of four lamin isoforms: two A-type lamins (A and C), and two B-type lamins (B1 and B2)23. The LMNA gene encodes for lamin A and C and some rare isoforms, which arise from alternative splicing, and the LMNB1 and LMNB2 genes encode lamin B1 and lamin B2, respectively23. Each lamin isoform forms separate but interacting meshworks24,25. B-type lamins are modified by farnesylation and are thus primarily located at the nuclear membranes (Fig. 1b), whereas A-type lamins either lack (lamin C) or have their farnesylated C-terminus removed (lamin A) and can be localized both at the nuclear lamina and the nuclear interior26, with the intranuclear distribution of lamins mediated by LAP2α and other proteins27. Lamins interact with various binding partners, including NPC proteins, INM proteins, chromatin, and various transcriptional regulators23. Accordingly, the lamina has many structural and other functions, including contributing to nuclear shape, mechanical stability, nucleo-cytoskeletal coupling, nuclear positioning, genome organization, and mechanosensing28–30.
Physically connecting the nucleus and the cytoskeleton
Force transmission between the cytoskeleton and the nucleus is required for nuclear movement and positioning, for example, during cell migration, nucleokinesis, and muscle fiber regeneration31 (Fig. 3). Cytoskeletal connections to the large and rigid nucleus are also important for cytoskeletal organization, affecting stress fiber organization, focal adhesions, and cell-cell adhesion32,33. The physical coupling between the cytoskeleton and the nuclear interior is achieved by the linker of nucleoskeleton and cytoskeleton (LINC) complexes that span the NE31,32 (Fig. 1a), although additional mechanisms, such as molecular motors binding to NPCs34 or microtubules connecting to emerin and other nuclear envelope proteins35 may further contribute to nucleo-cytoskeletal coupling. LINC complexes are composed of nesprins (nuclear envelope spectrin repeat proteins) localized within the ONM that bind across the perinuclear space to SUN (Sad1p, UNC-84) domain containing proteins located on the INM via their C-terminal KASH (Klarsicht/ANC-1/Syne Homology) domain31,36,37. This interaction appears to be at least in part responsible for controlling the spacing between the INM and ONM31. On the cytoplasmic side, nesprin-1 and nesprin-2 bind to actin filaments38 and—via kinesins39 and dynein40 to microtubules. Nesprin-3 binds to intermediate filaments via plectin41. Nesprin-4, which is found in polarized epithelial cells, plays an important role in nuclear positioning via kinesin-142. KASH5 is a germ-cell specific KASH-domain protein required for proper meiosis43. On the nucleoplasmic side, SUN domain proteins bind to the nuclear lamina, nuclear pores, and chromatin. The current model considers that LINC complexes balance part of the cytoskeletal tensile force exerted on the ONM, with maximal stress values at nuclear poles44.
Figure 3 – Physiological sources of nuclear deformations.
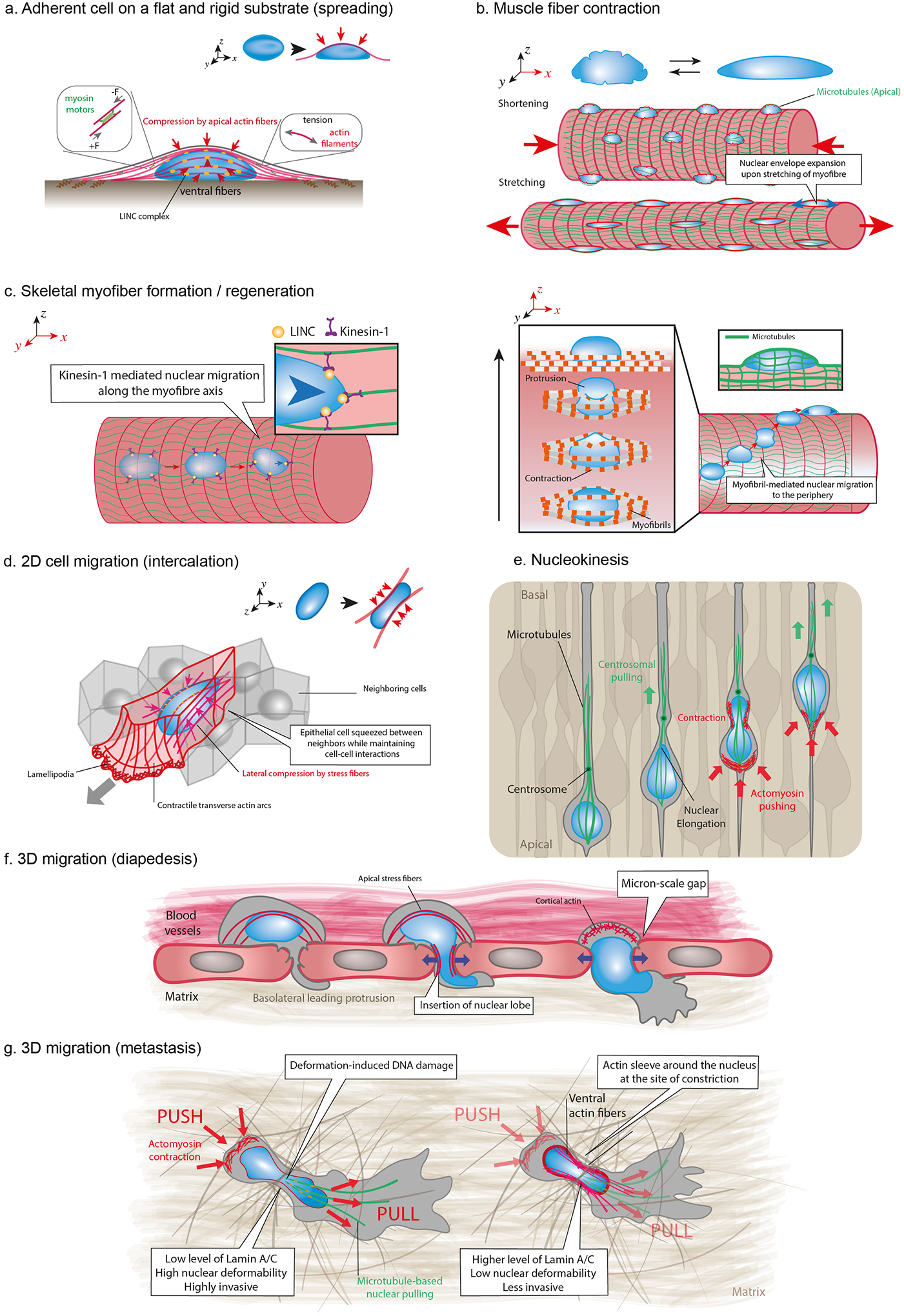
(a) Actomyosin contraction (in red) produces tension in actin fibers spanning the nucleus (in blue), which are connected to the NE via LINC complexes (in orange). Tension in apical actomyosin fibers generates vertical compressive forces that result in nuclear flattening. (b) Contraction and stretching of myofibers induce nuclear deformations, including NE wrinkling and expansion. Apical microtubules (in green) form cage-like structure around nuclei and exert compressive forces during myofibers elongation. (c) Formation and regeneration of skeletal myofibers require migration of nuclei along the myofiber axis through the interplay between LINC complex and microtubule associated motors such as kinesin-1. Myofibril contraction drives nuclear movement from the center to the periphery of the myofiber during muscle fiber maturation. This process requires myofibrils to exert contractile forces on the nucleus, resulting in large nuclear deformations. (d) Epithelial cell intercalation within dense tissues requires cellular elongation and nuclear deformation. Lateral compressive forces are exerted on both nuclear sides by ventral fibers, which are thick actomyosin bundles connected from their both ends to focal adhesions at the bottom of the cell. (e) Nucleokinesis events are observed during the development of the neuroepithelium of the central nervous system and is accompanied by considerable nuclear deformations. This mechanism occurs in densely packed tissues and involves pulling forces on the nucleus exerted by a microtubule cage towards the centrosome and pushing forces at the cell rear generated by actomyosin contraction, depending on the system. In mammals, microtubules exert pulling forces on the nuclear lamina through LINC complexes that move the nucleus towards the centrosome. (f) Immune cells and tumor cells can breach the endothelial barrier of blood vessels by inserting protrusion between or inside endothelial cells. Transendothelial migration through the small gaps (can be associated with a nuclear softening. Migration through the small openings (a few micrometers in diameter) is associated with large nuclear deformations. (g) Migrating cells translocate and deform their nucleus through narrow ECM pores or in between cells by using a combination of “push” and “pull” mechanisms. Nuclear deformations result from the balance between the amount and direction of the applied cytoskeletal force and the mechanical properties of the nucleus. Nuclear translocation requires both rear and front actomyosin contraction leading to pushing/pulling forces, respectively. At the front, microtubule motors are recruited to generate pulling forces. Together, the balance of forces results in the forward movement of the nucleus through the narrow constriction. High level of lamin A/C results in stiffer nuclei and highly invasive cells, whereas actin sleeve can be recruited at the site of the constriction to locally deform stiffer nuclei with higher level of lamin AC/ during the translocation of less invasive cells
LINC complex localization at the NE is associated with specific cellular functions. For example, LINC complex proteins are organized along apical stress fibers interacting with the cell nucleus45,46 and at the front of the nucleus as cells squeeze their nuclei through small pores47. Although our current understanding of how LINC complex localization and force transmission is regulated is still incomplete, recent findings indicate that disulfide bonds between the SUN and KASH domains can serve as a crucial modulator of nucleo-cytoskeletal coupling37,31. Several additional components have been identified that mediate LINC complex function and force transmission, including FHOD1 (Formin Homology 2 Domain Containing 1)48, torsinA49, Samp150 and lamins A/C51. Nesprins can also contribute to nucleo-cytoskeletal coupling independent of their actin and KASH-domains via their spectrin repeats52. Nonetheless, many questions remain regarding the precise regulation of LINC complex assembly and function.
Section 2 – Nuclear mechanics
Determining time and length scales of nuclear deformations is one of the key pieces of information for understanding how nuclear deformations mediate cellular functions. Here we analyze the nucleus from a mechanobiology perspective and discuss changes that influence nuclear mechanical properties and responses.
The physical properties of the nucleus
Insights from various experimental assays53 indicate that the nucleus behaves as a viscoelastic material, i.e., it exhibits both elastic and viscous behavior when subjected to external forces54. In this context, elastic deformations are defined as instantaneous, reversible deformations, like a spring that extends under an applied force and snaps back to its original length when the force is removed. In contrast, viscous, i.e., liquid-like materials, exhibit flow and thus a time-dependent, irreversible deformation when subjected to force.
Numerous assays have been developed to quantitatively capture the rheological properties of the nucleus, ranging from micropipette aspiration and microindentation to stretching intact cells or isolated nuclei55. A major challenge lies in the fact that the viscoelastic response of the nucleus reflects a complex coupling between chromatin, lamins, and other nuclear components, and thus the exact behavior can vary depending on the nature of the applied force/deformation and the molecular composition and organization of the cells being examined. Illustrating this challenge, some studies using micropipette aspiration found that the nucleus gradually deformed under an applied pressure before reaching a plateau, while in other cases the nucleus continued to deform under applied pressure, exhibiting a fluid-like behavior56,54,57–59. Stretching isolated nuclei at physiologically relevant strain rates revealed that for small deformations (<30% of the original length), the nuclear resistance is dominated by chromatin, whereas resistance to larger deformations is dominated by lamins A/C60. Furthermore, the nucleus undergoes strain stiffening, i.e., becomes more difficult to deform upon direct force application60,61.
After the removal of a mechanical strain, the elongated nucleus can relax with a nearly elastic response62–65 or with a delayed response and even residual plastic deformation, characteristic of viscoelastic material properties66,67. The elastic response requires the presence of lamin A/C, SUN-domain protein linkages and vimentin63. These differences of nuclear deformation and restoration dynamics may be explained by variations in nuclear lamina composition, chromatin organization, and cytoskeletal structure, composition, and remodeling.
Contribution of specific nuclear components to the mechanical properties of the nucleus
Although A- and B-type lamins share similar biochemical properties and filament structure, it is primarily the levels and assembly status of A-type lamins that determine nuclear stiffness and viscoelastic properties. Nonetheless, B-type lamins also contribute to nuclear stiffness and stability68,69, and loss of either lamin type results in abnormal nuclear shape and increased NE rupture62,70–73. Besides lamins, chromatin histone modification state and composition are major determinants of the mechanical properties of the nucleus, particularly for low nuclear deformation regimes74,60. Increasing the euchromatin content with histone deacetylase inhibitors, decreasing heterochromatin with histone methyltransferase inhibitors, or disrupting dynamics of the linker histone, all lead to softer nuclei and more nuclear blebbing events, without perturbing lamin levels74,60.
Although determining the physical state of condensed chromatin is critical for understanding mechanisms that modulate genome function, the mechanisms by which 10-nm chromatin fibers are packaged into higher-order heterochromatin and euchromatin domains and form phase-separated condensates at high cation concentrations75,76 have not been fully established.
The rheological behavior of the genome is highly complex and it is possible that the viscoelastic properties of chromatin are heterogeneous, locally tuned across the nucleus to accommodate different DNA-related biochemical processes such as transcription, replication, or DNA repair. Future experiments exploring the ATP dependence of DNA-related biochemical transactions as well as different time- and length-scales of chromatin organization both in vitro77, in vivo and in silico78 might provide further insights into the physical state of the genome. New evidence suggests that chromatin proteins such as HP1a, WDR5, BAF, and Numa also provide mechanical support to chromatin and regulate nuclear shape79–82. Interactions between chromatin and the NE further contribute to nuclear stiffness by forming an interconnected network. In addition, it is increasingly recognized that liquid-liquid phase separation (LLPS) of nucleoplasmic components, which leads to the formation of biomolecular condensates such as the nucleolus or heterochromatin83, may serve as a key principle governing nuclear organization84–87. The propensity to form liquid droplets is significantly enhanced in the vicinity of regions of low chromatin density because the higher mechanical energy required to deform the dense chromatin to create space for a growing protein droplet would generate an energetic penalty88. The growth of liquid droplets within the low chromatin density areas can lead to two distinct mechanical effects89. First, chromatin can be repelled as the drops growth by creating an effective repulsive interaction. A second effect can be driven by the tendency of the droplets to merge to minimize their surface energy. Indeed, regions of chromatin initially far apart and in separate droplets can be brought into close proximity when the droplets merge, creating an effective attractive interaction. The interplay between LLPS and chromatin is thus able to generate significant mechanical forces that can result in chromatin rearrangement90. Nonetheless, the relative contributions of LLPS versus other molecular mechanisms in determining the static and dynamic organization of chromatin within the nucleus remains to be fully elucidated.
Furthermore, the contribution of condensed chromatin to the mechanical integrity of the nucleus and its ability to respond to extranuclear forces are difficult to reconcile with a liquid state. Indeed, nuclear chromatin is mechanically responsive and can resist significant applied force91 which is a more consistent with a solid or gel state. Further studies that will consider chromatin fibers as viscoelastic filaments that can behave as both a viscoelastic solid and as a viscous liquid at different time- and length-scales may reconcile some of the apparently contradictory observations and ultimately provide a physical framework for the genome organization in space and time.
Determinants of nuclear volume and intranuclear pressure
Although the initial observation that the ratio between cellular and nuclear volumes is largely constant was made over 100 years ago92, and it is now well recognized that nuclear volume changes with chromatin organization and DNA content, the precise mechanisms underlying nuclear volume regulation remain incompletely understood. The nuclear volume is determined by the balance between outward pressures that originate from the nucleoplasm and tend to expand the nucleus, and inward pressures that originate from the cytoplasm and compress the nucleus. The outward pressure includes contribution from both the chromatin and the fluid inside the nucleus. Notable, despite the presence of NPCs that facilitate flow of fluid either into or out of the nucleus, cells are able to establish hydrostatic pressure differences between the nucleoplasm and cytoplasmic compartments93–95. To further understand, this mechanisms, biological factors implicated in nuclear size determination92 must be translated into quantifiable physical quantities to establish the force balance between the nucleus and cytoplasm that in mechanical—but not necessarily thermodynamic—equilibrium determines nuclear volume. Based on the concept that the interior of living cells is “crowded”, colloid osmotic pressure was introduced as a simple crowding metric to explain how mechanical works, such as inflating the nucleus, can be explained from protein aggregation and phase separation96. Very recent theoretical works suggest that the dominant pressure within the nucleus and cytoplasm originates from the osmotic pressure of the preferentially localized soluble molecules rather than the mechanical properties of large complexes such as the chromatin and cytoskeleton97,98. To go a step further, more sensitive subcellular osmometers97, such as genetically encoded biosensors, are needed to establish definitive physiological values of colloid osmotic pressure and to determine how crowding inside cells is regulated as a function of subcellular location and physiological inputs.
Adaptive changes in nuclear mechanics
Deformation of cells and the nucleus can lead to changes in chromatin organization and compaction, thus changing the mechanical properties of the nucleus and providing a mechanism to protect the nucleus from mechanical stress19,99 (see also textbox “Nuclear mechanoresponses and mechanosensing”). Furthermore, mechanical force application can lead to phosphorylation of emerin and subsequent recruitment of lamins to the NE, causing rapid stiffening of the nucleus, whereas reducing cytoskeletal tension can soften the nucleus by increasing lamin phosphorylation and turnover100, highlighting the importance of the interplay between the nucleus and the cytoskeleton. Of note, emerin is a recognized actin-binding protein that promotes actin polymerization101. Mechanically induced translocation of emerin from the INM to the ONM can thus lead to increased perinuclear actin polymerization102, which could alter nuclear deformability.
Differences in lamin expression between various tissues that affect the deformability and mechanical stability of nuclei may indicate tissue-specific adaptations to particular mechanical demands of the local microenvironment29,103–108 but may also reflect the role of lamins in tissue-specific gene expression104,105. For example, neutrophil nuclei have a particular lobulated morphology with characteristic low lamin A level and elevated condensed chromatin level109 that promote their perfusion and migration through tight spaces110, such as lung capillaries that are only few microns in diameter, or even smaller gaps between endothelial cells. However, whether individual cells can dynamically adapt their nuclear stiffness on short timescales to promote migration through tight spaces is still under debate. Confocal Brillouin microscopy revealed nuclear softening during transendothelial migration of breast cancer cells111. However, the origin and temporality of such nuclear softening remain poorly understood. Interestingly, metalloproteinase (MMP) inhibitor treatment leads to nuclear softening via lamin A/C phosphorylation, which is essential for migration through pores with subnuclear diameter (see also next section)112,113. This response requires an intact connection between the nucleus and the centrosome via the LINC complex protein nesprin-2 and the dynein adaptor Lis1112. Chromatin remodeling can further modulate nuclear stiffness and cell migration in 3D environments80. These findings suggest that dynamic chromatin modification and changes in lamin levels and organization can mediate nuclear mechanics and promote cell migration in confined 3D environments114,115, although reducing lamin A/C levels below a critical threshold may reduce cell survival under mechanical stress71,116–118.
Section 3 – Physiological sources of nuclear deformations
The nucleus is constantly exposed to forces from the surrounding cytoskeleton, including from active positioning of the nucleus during cell polarization119, migration119 or differentiation120. Recent advances in intravital imaging and modeling physiological microenvironments in vitro have documented large scale nuclear deformations in striated muscle121,122 and during ‘confined migration’, i.e., cells squeezing through three-dimensional spaces with pore sizes smaller than the size of the nucleus71,72,123, although similar nuclear deformations and functional consequences are expected to also occur during numerous other situations, such as embryonic development124,125 or nucleokinesis events126 (Fig. 3).
Nuclear deformation in cells adhering to flat and rigid substrates
Actin stress fibers and actomyosin contractility can impose vertical and lateral inward compressive forces on the nucleus. Lateral actin fibers can lead to nuclear deformation when cells migrate or are stretched127,128. Vertical compressive forces are exerted by apical actin stress fibers that form a dome-like structure across the nucleus and that are physically attached to the nuclear lamina through LINC complexes129. On flat rigid substrates, these forces flatten the nucleus during cell spreading (Fig. 3a) and can cause nuclear envelope rupture events130–132. In contrast, the nucleus remains more rounded in cells on soft substrates133, which are characterized by a lower amount of cytoskeletal tension and fewer actin stress fibers134, or when the actin cytoskeleton or LINC complex are disrupted130. Indeed, ventral actin fibers, which are thick actomyosin bundles connected from their both ends to focal adhesions at the bottom of the cell, can exert lateral compressive forces on both nuclear sides135. The high level of tension in ventral actin stress fibers can lead to nuclear indentations of the order of a few microns that are characterized by local enrichment of LINC complexes and segregated domains of condensed chromatin46,136. Collectively, these findings suggest that the amount of tension within the perinuclear actin fibers is an important parameter of nuclear deformation and nuclear mechanoresponses.
Nuclear deformation in skeletal and cardiac muscle
Actomyosin contractility also plays an important role in nuclear deformation in striated muscle cells. Large nuclear deformations were recently visualized in cardiac and skeletal muscle contraction in living fly larvae121. Increased expression of lamins A/C in muscle cells is essential to protect their nuclei from mechanical damage caused by muscle contraction137 (Fig. 3b). Another, more surprising mechanism responsible for mechanical stress on the nucleus are the cytoskeletal forces required to position muscle nuclei along the length of the muscle fiber and the nuclear periphery during muscle cell maturation and repair138,139. LINC complex proteins such as nesprin-1, together with the microtubule associated motors kinesin-1 and dynein, and other NE proteins such as emerin play a crucial role in moving myonuclei along the microtubule network to distribute them along the length of the muscle fiber138,140,141 (Fig. 3c). The physical stress during nuclear positioning results in nuclear rotation and nuclear deformation154,161,170. In lamin A/C-deficient or mutant cells, which have mechanically weaker nuclei, the kinesin-mediated forces can result in large-scale nuclear deformations and damage143. In later stages of muscle fiber maturation, myofibril contraction is needed to move skeletal muscle nuclei to the periphery of muscle fibers, incurring nuclear deformation in the process (Fig. 3e), particularly in lamin A/C-deficient cells144. Intriguingly, in lamin A/C-deficient and mutant mouse models that develop severe muscular dystrophy and dilated cardiomyopathy (see textbox “Human pathologies associated with nuclear deformations”), reducing the cytoskeletal forces acting on the fragile muscle cell nuclei by disrupting the LINC complex, prevents nuclear damage and results in improved muscle function and viability in vitro and in vivo118,145, pointing to promising new therapeutic approaches for these devastating diseases. However, given that mutations in nesprins and SUN proteins can lead to muscular dystrophy and heart disease146,further studies will need to evaluate the long-term risks and consequences of LINC complex disruption, using for example inducible LINC complex disruption models147.
Textbox 3 - Human pathologies associated with nuclear deformations.
Abnormalities in nuclear and chromatin organization, including defects in the lamina, are hallmarks of many diseases ranging from heart disease to premature aging and cancer260. For many years, aberrations in nuclear morphologies have been used by pathologist as a signature for disease such as cancer, where they can indicate metastatic potential261–263. Hundreds of mutations and variants have been found in genes encoding for NE components ranging from ONM proteins (e.g., nesprins) to INM proteins (e.g., emerin and SUN proteins) and lamins A/C. These mutations result in a set of dramatic diseases called nuclear envelopathies264. Mutations in the LMNA gene, which encodes Lamin A/C, are the second most frequent cause of congenital dilated cardiomyopathy (DCM), comprising 5–8% of cases265. So far, over 450 mutations associated with the LMNA gene have been identified266 and LMNA mutations are believed to cause over 13 human diseases. These diseases include various types of muscular dystrophy267, familial partial lipodystrophy268, and progeria269.
Muscular dystrophy
Several mutations in myopathic lamins associated with muscular dystrophy and dilated cardiomyopathy result in more deformable nuclei. Nuclear softening was exclusively associated with myopathic lamin mutations180. In skeletal muscle cells, lamin mutations were recently observed to cause extensive NE damages in skeletal muscle cells in vitro an in vivo, resulting from mechanical stress on the more fragile nuclei118. Lamin mutations associated with muscular dystrophy can also impair LINC complex function180,270,271 and other cellular processes. Abnormal YAP activity, known to be responsive to nuclear deformation and lamin A levels30,248, has been reported in muscular dystrophy and rhabdomyosarcoma272. In LMNA-related congenital muscular dystrophy, lamin mutations increase YAP nuclear localization via an increased nuclear import, implicating YAP as a potential pathogenic contributor in muscular dystrophies caused by NE defects273.
Hutchinson-Gilford Progeria Syndrome (HGPS)
HGPS is an exceptionally rare and dramatic premature aging disease caused by mutations in the LMNA gene. Most cases of HGPS result from a mutation that leads to alternative splicing, causing a truncated form of prelamin A (LAΔ50) that remains farnesylated. Cells from HGPS patients have irregular nuclear shapes274, increased nuclear stiffness, and increased sensitivity to mechanical stress275–278, which may be responsible for the progressive loss of vascular smooth muscle cells in HGPS. The formation of orientationally ordered microdomains of lamins in HGPS cells reduces the ability of the NE to dissipate mechanical stress278. Restoring the loss of heterochromatin alone in Hela cells expressing LAΔ50 and in patient HGPS cells is sufficient to restore normal nuclear shape, suggesting that heterochromatin loss may be responsible for many of the phenomena associated with HGPS279–281.
Neurodevelopmental defects
Deficiency of lamin B1 and lamin B2, but also increased expression of lamin B1, are associated with neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Altered nuclear lamin interferes with proper nucleokinesis, a nuclear translation process required during neuronal migration69. Defective migration of cortical neurons was observed in lamin B1- and lamin B2- deficient embryos, leading to neuronal layering abnormality in the cerebral cortex along with neonatal mortality282–284. The neuronal migration abnormality can be explained by a weakened nuclear lamina, as B-type lamin depletion was previously shown to affect nuclear mechanical properties285. Duplication of the gene encoding lamin B1 results in autosomal dominant leukodystrophy, which is characterized by widespread loss of myelin loss in the central nervous system286.
Tauopathies
Tauopathies refer to a class of neurodegenerative diseases involving the aggregation of Tau protein, a neuronal microtubule-associated protein into neurofibrillary or gliofibrillary tangles (NFTs) in the brain. Pathological accumulation of Tau is known to form Tau nuclear rods (TNRs) or Tau-positive nuclear indentations (TNIs)287. These structures have been identified in several neurodegenerative disorders ranging from Alzheimer’s disease, frontotemporal dementia to Huntington’s disease288,289. Several studies identify the Tau protein (MAPT) as central during pathogenesis associated to dementia290,291. However, the mechanism underlying Tau mediated pathogenesis is still unclear. Mutations in the Tau encoding gene MAPT result in Tau mislocalization to the cell bodies rather than neuronal axon. This leads to abnormal microtubule organization, which deform the NE via LINC complex based coupling292, causing large nuclear lamin invaginations and defects in nucleocytoplasmic trafficking291,293.
Why are laminopathies involved in a wide spectrum of diseases?
Although the pathological mechanisms underlying the diverse laminopathies are still not fully understood, various hypotheses have been put forward to explain the tissue-specific defects found in many laminopathies and the diverse phenotypes associated with specific LMNA mutations. The key role of lamins in establishing the mechanical properties of the nucleus suggests that defects in NE/lamina proteins can result in impaired nuclear stability and increased nuclear fragility, which can lead to mechanically induced nuclear damage and perturbations of mechanosensing pathways. This hypothesis is supported by numerous in vitro and in vivo observations of abnormalities in nuclear morphology (e.g., wrinkling, irregularities, blebs, and invaginations)180,65,294,295. Mechanically induced damage to the fragile laminopathic nuclei was associated with increased DNA damage, resulting from direct force transmission to the genome or NE rupture29,196,296. Besides their mechanical function, lamins have a key role in tethering and organizing chromatin, as well as in signaling involved in transcriptional regulation. In support of this, laminopathic nuclei often display alterations in the organization of chromatin, signaling, and broad alterations in gene expression297,298,280,299,300, which could contribute to the tissue specific phenotypes.
Nuclear deformation during development
In early Drosophila embryo, severe nuclear deformations occur when somatic nuclei at the periphery of the syncytial embryo move as the plasma membrane invaginates to form membranes around each nucleus, a process called ‘cellularization’. The nuclear deformations are caused by the polymerization of microtubules in bundles organized by dynein148. The nuclear deformations may be particularly pronounced because A-type lamin is not expressed in Drosophila during cellularization, leading to more deformable nuclei149. Nuclear movement during development also results in substantial nuclear deformations in the nematode Caernorhabditis elegans, which require cytoskeletal force transmission to the nucleus via the LINC complex150.
In epithelial systems, cellular intercalation is a common process occurring throughout development, where neighboring cells exchange their place to maintain epithelium integrity. Depending on the cell density, cellular intercalation can lead to transient cellular squeezing events and nuclear deformation (Fig. 3d), likely due to compression by neighboring cells and cytoskeletal remodeling151,152. Interkinetic nuclear migration is observed during the development of the neuroepithelium of the central nervous system and is accompanied by considerable nuclear deformations in the zebrafish embryos153 and within the mouse retinal tissue from P0 to P15 stages154, when A/C type lamins and Lamin B receptor (LBR) are not expressed. Interestingly, suppression of nuclear deformation in the mouse retina results in impairment of chromocenter clustering, suggesting that dynamic nuclear deformation could be an underlying driving force of nuclear architecture and spatiotemporal genomic reorganization155.
During brain development, nucleokinesis in neurons (Fig. 3e) is a beautiful example of how pulling the nucleus in densely packed tissues through the cytoskeletal leads to nuclear deformation156. Interkinetic nuclear migration is a nucleokinesis event occurring in progenitor cells that involves up and down movements of the nucleus in elongated cells, which are attached on their both ends. Both actin and microtubules have been involved in the process, depending on the system157. In mammals, microtubules exert pulling forces on the nuclear lamina through LINC complexes that move the nucleus towards the centrosome. Loss of either lamin B1 or lamin B2 cause both defective migration of cortical neurons in the developing brain and lead to severe nuclear defects (e.g. chromatin protrusions) during that phase, likely explaining the severe brain development defects and reduced neuronal survival69. It remains to be determined whether these defects are caused by disrupted transmission of force during saltatory nuclear movement or a more fragile nucleus unable to bear the stress generated during nucleokinesis. Besides nucleokinesis, live imaging studies have found remarkable nuclear deformation and rotation during the migration of cerebellar granule cells through narrow intercellular spaces in neural tissues158. During this process, microtubules steer the nucleus and drive its rotation and deformation through a dynamic interaction of nesprins with kinesin-1 and dynein. Given the apparent diversity of cytoskeletal organization in neuron species, further studies will be needed to obtain a better understanding of nuclear dynamics and nuclear shape regulatory mechanisms in neuronal cells.
Nuclear deformation during confined migration
Nuclear deformation is also a hallmark of important physiological and pathological situations involving cell migration. For instance, immune cells or invasive cancer cells must navigate through small interstitial spaces ranging from 1 to 20 μm in diameter159,160, which requires cells to deform their nucleus to squeeze through the available spaces (Fig. 3f). In the absence of matrix metalloproteinase (MMPs) proteolysis, the nucleus is often the main physical hindrance to cell migration through confined spaces162,173,174. Leucocytes can insert basolateral protrusion within (paracellular) or between (transcellular) endothelial cells to breach the endothelial barrier (Fig. 3f) and use actomyosin forces to push the nucleus through the pore, resulting in substantial nuclear deformation.
Tumor cells face similar challenges when invading tissues and intra- and extravasating blood vessels to metastasize to distant tissues161 (Fig. 3f). The primary sources of cytoskeletal forces to translocate and deform the nucleus are (i) actomyosin contractility that can cause both tension and compression of the nucleus by actin stress fibers pulling or pushing on the nucleus174,177,178 or by generating hydrostatic pressure within cellular compartments that acts on the nucleus162, and (ii) microtubule-associated motors, i.e., kinesins and dyneins163, which directly attach to the nucleus via nesprins and other NE proteins. Whether the nucleus is pulled and/or pushed is still debated164, although it is likely that cells can use multiple independent mechanism, depending on the particular context (Fig. 3f). For example, LINC complexes recruit dynein and kinesin-1 to pull nuclei towards the minus ends of polarized microtubule networks during C. elegans development 181, whereas evidence for nuclear pushing has been reported in breast cancer cells and in glioma invasion, where non-muscle myosin IIB (NMIIB) at the cell rear pushes the nucleus forward165–167. An additional mechanism has been observed in dendritic cells that uses Arp2/3 to generate lateral pushing forces that deform the nucleus to facilitate the migration process through narrow ECM pores168. In the context of cancer cells, fibroblasts and macrophages from the stromal microenvironment may further aid in the invasion process by physically pulling on the tumor cells or degrading the extracellular matrix.
Notably, nuclear deformation during confined migration may also involves dynamic or persistent changes in nuclear mechanical properties. For example, transient nuclear softening has been reported during transendothelial migration of cancer cells111, neutrophils develop highly lobulated and deformable nuclei during granulopoiesis that facilitates passages through tight spaces110, and highly invasive breast cancer cells are characterized by increased nuclear deformability and low lamin A/C levels113 (Fig. 3f).
Section 4 – Consequences of nuclear deformation
Given the central role of the cell nucleus in cellular function, it is easy to imagine how nuclear deformations can lead to various transient or persistent consequences, ranging from increased cell contractility, loss of NE integrity, DNA damage, and epigenetic modifications to altered cell differentiation. Notably, although these outcomes are now well established, the molecular mechanisms responsible, and whether the nuclear itself transduces mechanical signals into biological responses, often remains unresolved and a matter of active research (see textbox “Nuclear mechanoresponses and mechanosensing”).
Impact of NE tension on cellular proprioception and actomyosin contractility
Confinement of cells below a critical threshold, typically a fraction of the uncompressed nuclear height, results in nuclear flatting, an increase in nuclear membrane tension, and opening of nuclear membrane folds125,169. These events trigger the release of calcium from the ER and the recruitment of phospholipase cPLA2 to the INM, where it catalyzes the production of arachidonic acid (AA), an omega-6 polyunsaturated fatty acid. AA has been implicated to regulate myosin II activity both directly170 and indirectly via protein phosphorylation171. The resulting increase in cortical actomyosin contractility modulates cell morphology and promotes migration through narrow constrictions125,169. Unfolding of the NE under increasing membrane tension allows the nucleus to deform without exceeding critical membrane tension in the nuclear membranes172, but may also trigger downstream signaling events173. This nuclear mechanosensing of cellular confinement has been referred to as “cellular proprioception”. The physical properties of the large nucleus can also directly influence cellular processes. Recently, the microtubule mediated “frontward” positioning of the nucleus in amoeboid cell migration was shown to allow cells to use their nucleus as a mechanical gauge to determine the path of less resistance when encountering bifurcations of the path with pores of different size174, providing an example of how deformation of the nucleus aids cells in their ‘decision making’ during migration through confined environments.
Deformation-associated NE rupture and repair
NE rupture describes the (transient) loss of nuclear membrane integrity at localized sites, rather than global breakdown of the NE. Spontaneous NE rupture events, persisting typically from a few minutes to several dozens of minutes, were first observed in vitro in cells expressing the HIV protein VPR175, and subsequently in laminopathy patient fibroblasts176 and cancer cells177. Since then, it has become apparent that physical stress on the nucleus and the associated nuclear deformations can lead to transient NE ruptures, particularly during migration through confined environments, and that the probability of NE rupture increases with the degree of confinement71,72,178,179. NE ruptures have been documented in vitro and in vivo. The NE ruptures are often associated with loss of A-type or B-type lamins49,109,154,197, lamin mutations182–184, peripheral heterochromatin disruption91 or high-level of mechanical stress, resulting from tensile or compressive forces on the nucleus70–72,116,185–188. Based on super-resolution imaging and computational modeling, the NE rupture sites are estimated to be ≈100 nm in diameter69,70,206. A current hypothesis proposes that NE ruptures occur at pre-exiting gaps or defects in the nuclear lamina, particularly where the lamin B meshwork is weaker and thus cannot sufficiently support the nuclear membranes, causing the membrane to form a bleb that expands under continued mechanical stress and ultimately ruptures (Fig. 4)190,179. However, NE rupture and membrane blebs have also been observed in the absence of nuclear lamina gaps; they may thus generally arise when the nuclear membrane peels off the underlying nuclear lamina in response to increased nuclear pressure resulting from cytoskeletal forces71,191,192. A better understanding of the nucleation mechanism of nuclear ruptures will require to study the dynamics of the heterogenous lamina meshwork and its interaction with the nuclear membranes during nuclear deformations.
Figure 4 – Nuclear envelope rupture and repair.
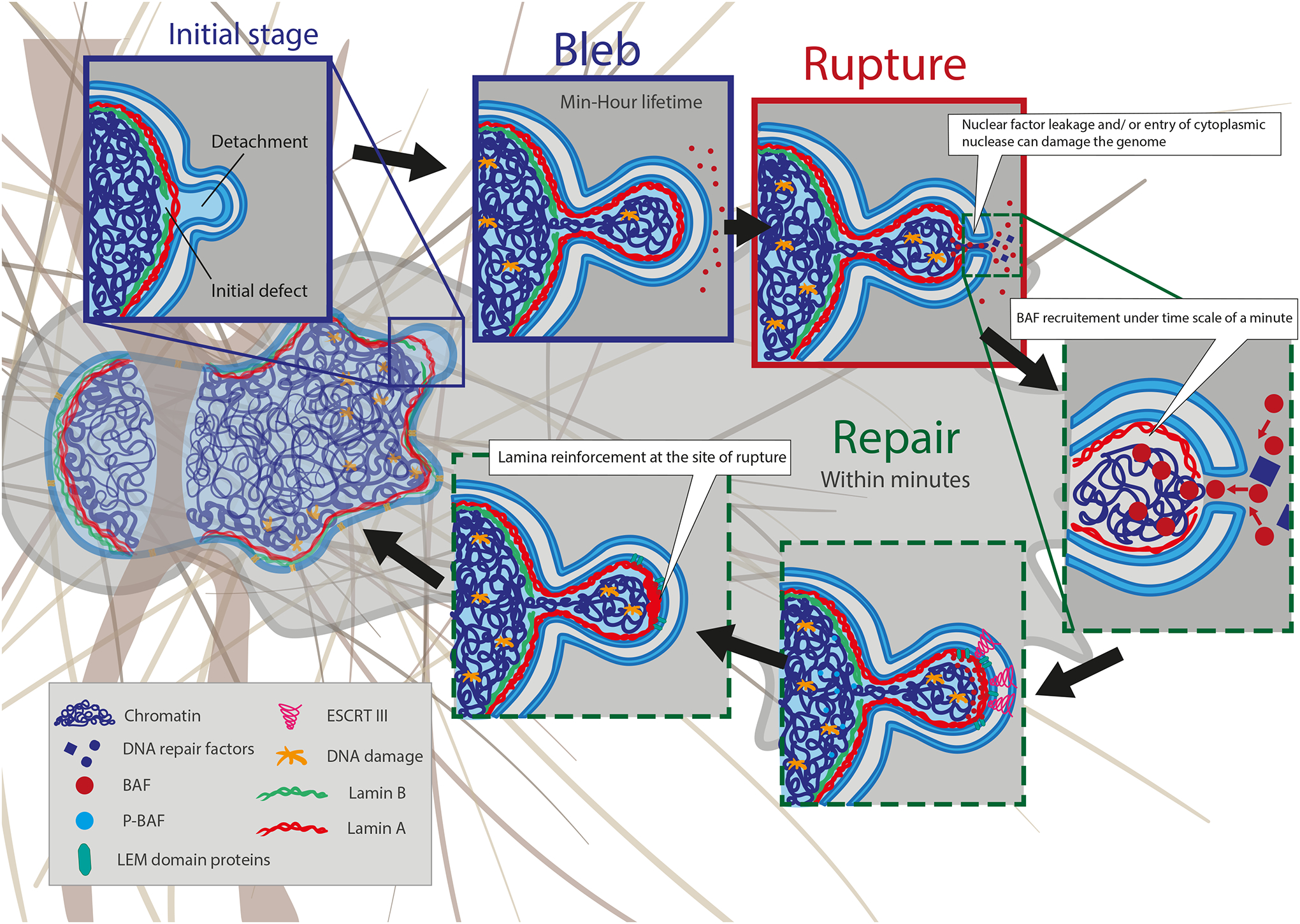
Migration through confined environments or external compression of cells can result in nuclear membrane bleb formation and NE rupture. The nuclear membrane rupture process is typically initiated by the formation of a nuclear membrane extrusion, or bleb. Blebs from at sites with high nuclear membrane curvature and where an initial defect in the nuclear lamina exists. Blebs are driven by increased hydrostatic pressure within the nucleus. Initially, only the nuclear membrane detaches from the lamina. Later, lamin A/C and chromatin can enter the bleb. The lifetime of blebs can be minutes to hours, but the rupture itself is usually quite short, on the orders of minutes. Blebs can have varying size and can contain chromatin or are just fluid filled. Nuclear blebs typically lack lamin B (green) and NPCs. Continued nuclear compression by confinement from the extracellular matrix, apical actin stress fibers, cell contractions, or external compression results in bleb expansion until the nuclear membranes in the bleb exceeds a critical threshold and ruptures, leading to the leakage of soluble proteins from the nucleoplasm into the cytoplasm and uncontrolled influx of cytoplasmic proteins into the nucleus. Following NE rupture, barrier-to-autointegration factor (BAF) is rapidly (< min) recruited to initiate NE repair. The recruitment of ESCRT III complexes further contributes to resealing the nuclear membranes. The process of repair/rescue is typically completed within 10–15 min and often associated with recruitment of lamin A/C to the site of rupture. Although the NE rupture is resealed, the bleb/protrusion often persists and is not fully resorbed.
As the transient nature of most NE ruptures documents, cells have robust mechanisms to repair their nuclear membrane during interphase, and even longer rupture events (few hours) can eventually be repaired193. The mechanisms involved in interphase nuclear membrane repair are largely shared with those during resealing of the NE post mitosis. The nuclear membrane repair mechanism is based on the recruitment of specific proteins to the sites of NE rupture, particularly BAF, LEM-domain proteins, the endosomal sorting complexes required for transport (ESCRT)-III remodeling complex, and CHMP7 72,181,186,193,194. Considering that the extent of rupture is correlated with the amount of cytoplasmic BAF accumulating at the rupture site72,193,194, a current model of nuclear membrane repair considers that the binding of cytosolic BAF to the exposed chromatin initializes recruitment of both new ER membranes to repair the membrane hole and the ESCRT-III complex to reseal the remaining gaps (Fig. 4). Interestingly, some nuclear processes such as transcription and DNA replication can be disturbed after NE rupture events, leading for instance to aneuploidy or extensive DNA damage such as double-stranded DNA breaks that cannot be repaired due to loss of DNA damage repair proteins132.
Mechanically induced DNA damage
Severe nuclear deformations, occurring for example during confined migration, external compression, or nuclear repositioning, can induce DNA damage upon NE rupture68,69,212,213 and even in the absence of NE rupture196. NE rupture can cause DNA damage by allowing access of the ER-associated exonuclease TREX1 into the nucleus172, or by loss of DNA damage repair factors from the nucleus195,197. Whereas NE rupture associated DNA damage occurs throughout all phases of the cell cycle, and more often in Ataxia Telangiectasia and Rad3-related protein (ATR)-defective cells198, the deformation-induced-DNA damage (i.e., DNA damage in the absence of NE rupture) occurs primarily in S/G2 phases, i.e., during active DNA replication, and is linked to increased replication stress, possibly due to torsional stress on DNA resulting from the nuclear deformation during confined migration or mechanically compression of cells196. Interestingly, different cell lines exhibit different propensities for these modes of DNA damage172,196, but the exact molecular reasons for these cell type specific differences remain to be elucidated.
What are the long-term consequences of DNA damage and NE rupture for cells and tissues homeostasis? Repeated migration through tight constrictions can lead to accumulation of DNA damage and changes in chromosome copy number195. Furthermore, TREX1-dependent DNA damage following NE rupture may favor tumor cell invasion by inducing a partial epithelial-to-mesenchymal transition (EMT) phenotype via ATM and SNAIL1 that leads to MMP dependant collagen degradation172. The precise mechanisms linking TREX1 and collagen degradation activity through MMP’s activity is still unknown, but is believed to be downstream of the ATM DNA damage response pathway199,200. NE rupture can also lead to activation of the cyclic GMP-AMP synthase-stimulator of interferon genes (cGAS-STING) DNA-sensing pathway, as it allows access of cytosolic cGAS to the genomic DNA at sites of NE rupture71,72,201. Intriguingly, a recent study found that increased cGAS-STING signaling can drive cancer metastasis in a mouse breast cancer model201, although in this case the cGAS-STING activation was primarily due to NE rupture of micronuclei, not primary nuclei.
Mechanically induced epigenetic modifications
Local cytoskeletal forces such as actin-based indentation acting on the nucleus can severely deform the NE and trigger reversible formation of heterochromatin at the NE periphery46,135,202. Local stresses applied to integrins can propagate to the LINC complex through the actin cytoskeleton and lead to chromatin un-packing203 and force-induced transcription that requires H3K9me3 demethylation204. Nuclear deformation during confined migration can also induce increased heterochromatin formation in a histone methylase and histone deacetylase dependent process, which promotes cell migration205. Furthermore, nuclear deformations associated with confined migration, cell compression, or cell stretching can lead to chromatin re-arrangements and the increased formation of heterochromatin that can last from hours to days205. In macrophages, spatial confinement can suppress late lipopolysaccharide (LPS)-activated inflammatory transcriptional programs (e.g., expression of IL-6, CXCL9, IL-1β, and iNOS) by modulating chromatin organization and epigenetic alterations216,206. In the context of cell migration, the increase in H3K9me3 and H3K27me3 heterochromatin marks promotes confined cell migration through yet to be defined mechanisms114,205. The molecular details by which mechanical deformation of the cell and nucleus result in increased heterochromatin formation remain incompletely understood, but two major contributors have emerged to date: an increase in intracellular cations (calcium and/or magnesium) by activation of stretch-activated ion channels, and remodeling of the nuclear and/or perinuclear actin network.
Repetitive stretching of mesenchymal stem cells activates mechanosensitive ion channels (Fig. 5a) such as Piezo1, leading to increased intracellular calcium levels and increased heterochromatin (H3K9me2,3) formation, and, ultimately promoting mesenchymal differentiation207,208. In epithelial cells, cyclic mechanical stretch triggers immediate nuclear deformation that leads to Piezo1-mediated calcium release from the ER, reducing lamina-associated heterochromatin (H3K9me3 marks) within a ≈30 minutes window, and resulting in nuclear softening that decreases stress and DNA damage in the stretched cells19. Long-term (8–12 hours) cyclic uniaxial stretch application causes transcriptional repression, increased heterochromatin (H3K27me3), and silencing of differentiation gene expression19. Intriguingly, activation of mechanosensitive ion channels by increasing extracellular multivalent ion concentrations, even in the absence of cell stretching or compression, is sufficient to trigger similar increase in heterochromatin91. The increased heterochromatin content mechanically strengthened the nucleus, rescued abnormal nuclear morphology in LMNA mutant and breast cancer cells, reduced NE ruptures, and prevented DNA damage91. Collectively, these findings demonstrate that mechanosensitive ion channels respond to mechanical stimuli, causing an increase in intracellular calcium that leads to chromatin modifications, which mechanically protect the nucleus and influence cell fate decisions. These stretch-sensitive ion channels can be found on the plasma membrane, the ER, and potentially the NE, with the contribution of specific channels and their locations likely depending on the particular cellular context. At least in some cases, influx of extracellular calcium, rather than release from intracellular stores, appears to be sufficient to trigger chromatin remodeling91,208.
Figure 5 – Schematic illustration of nuclear mechanoresponses and mechanosensing.
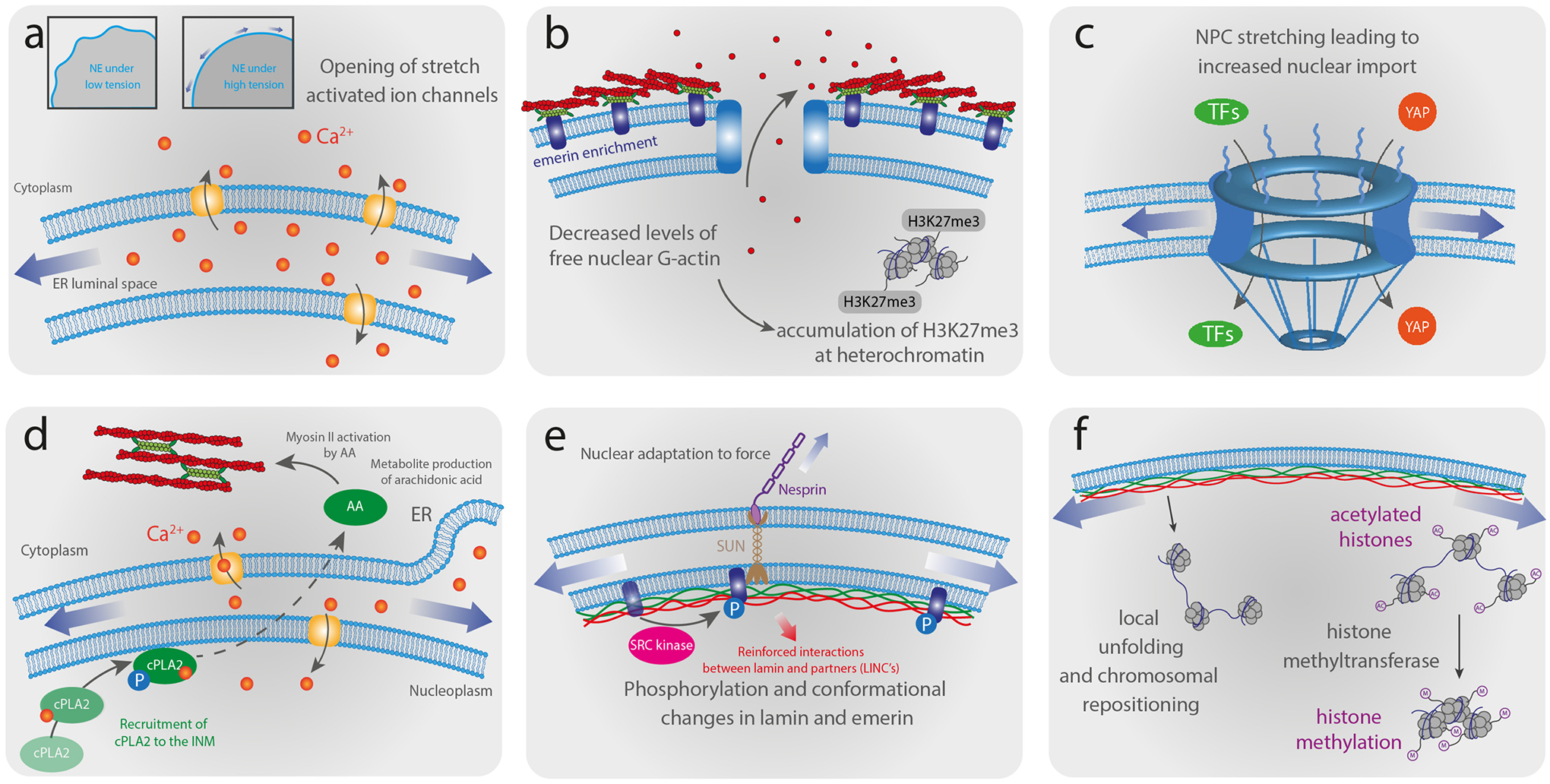
(a) High tension exerted on the NE during nuclear deformations induces unfolding of the wrinkled NE and the opening of stretch-activated ion channels. (b) Deformation of the nucleus induces enrichment of non-muscle myosin and emerin at the ONM. Relocalization of emerin to the ONM promotes perinuclear actin polymerization that leads to decreased levels of free nuclear monomeric actin, thereby reducing global transcriptional activity and increasing heterochromatin formation. The decrease of emerin at the INM leads to a loss of H3K9me2 and heterochromatin maintain their silenced state by recruiting H3K27me3. (c) Increased nuclear membrane tension stretch NPCs, leading to increased nuclear import of transcription factors (TFs) and mechanoresponsive transcriptional activators, such as YAP. (d) High NE tension resulting from nuclear deformation induces nuclear membrane unfolding, subsequent calcium release and the recruitment of cytosolic phospholipase A2 (cPLA2) activated by phosphorylation to the nuclear periphery, which promotes production of arachidonic acid (AA). The activation of the cPLA2-AA pathway leads to myosin II via AA-mediated Rhoa activation recruitment to the cell cortex, increasing actomyosin contractility. (e) Nuclear deformation induces phosphorylation of emerin and conformational changes in lamin A/C, which can alter the interaction with binding partners and induce further signaling events or recruit other proteins to the NE. (f) Forces acting on the nucleus may reposition or locally unfold chromatin domains, altering their transcriptional activity, and modulate the methylation level of histones by methyltransferases, regulating the transcriptional activity.
Changes in perinuclear actin polymerization, mediated by relocalization of emerin to the ONM (Fig. 5b), can result in increased facultative heterochromatin formation by depleting monomeric actin from the nucleus, reducing transcription, and activating the polycomb repressive complex (PRC2)102. Mechanically induced actin depolymerization can also lead to translocation of the histone deacetylase 3 (HDAC3) from the cytoplasm into the nucleus, resulting in increased heterochromatin formation209. Spatial confinement can similarly reduce actin polymerization, thereby reducing nuclear translocation of megakaryotic leukemia 1 (MKL1), a myocardin-related coactivator of the serum response factor that regulates many physiological processes, which can lead, for example, to altered inflammatory signaling in macrophages exposed to LPS206. Emerin-mediated actin polymerization can modulate nuclear translocation of the mechanoresponsive transcription factor MKL1, thereby altering expression of MKL1/SRF response genes such as Srf or vinculin210.
Nuclear deformations affect transcription factor translocation and cell differentiation
Nuclear deformations can also modulate the balance of nuclear and cytoplasmic pools of the mechanoresponsive transcriptional regulators YAP (Yes-associated protein) and TAZ (Transcriptional coactivator with PDZ-binding motif)211 (Fig. 5c), which play crucial roles in regulating a wide range of key biological processes212. For instance, the differentiation of myoblasts into myotubes which requires nuclear deformations was associated with the nuclear export of epigenetic regulator and transcription factors such as SMYD3 lysine methyltransferase and YAP213. Whereas activity and nuclear translocation of YAP/TAZ have been traditionally viewed as being controlled by cytoplasmic pathways214, recent findings point to nuclear deformation as an additional modulator. For example, compressive forces lead to nuclear flattening and NPC opening18, which increases YAP nuclear import215. More recently YAP nuclear export was associated with substrate curvature changes that impose nuclear deformation. Nuclei located on convex zones (i.e. crests) were flattened with YAP translocated to the nucleus and chromatin less condensed, whereas nuclei on concave zones (i.e. valleys) were highly elongated, contained more condensed chromatin, and YAP was predominantly cytoplasmic216. These findings support the notion of a control of YAP/TAZ by nuclear deformation and highlight the importance of mechanical and cytoskeletal regulation of the nuclear shape in YAP/TAZ regulation. Accumulative evidence has shown that similar effects can be observed by imposing nuclear shape changes with higher cell density217 or various external forces30,129,211,215, without changing the matrix properties. However, precisely how the intracellular localization of YAP is modulated by nuclear volume changes215 and how this observation relates to known regulators of YAP nuclear translocation remain to be elucidated.
Recent evidence suggests that the cytoskeleton can modify not only the physical state of the nucleus, but also the chromatin state. Sustained activity of MKL1, a key transcriptional co-activator of SRF that can be activated by mechanically induced actin polymerization and that regulates expression of many actin cytoskeletal genes, results in reduced nuclear volume and globally reduced chromatin accessibility218, consistent with a recent study that found that actin polymerization reduced pluripotency in induced pluripotent stem cells by decreasing chromatin accessibility. An additional example of how forces on the nucleus can modulate cell fate decisions comes from recent work on myofibroblasts, which found that the persistence of the myofibroblast phenotype relies on chromatin remodeling mediated by nuclear mechanosensing of cytoskeletal forces via LINC complexes219. Furthermore, human mesenchymal cells respond to matrix stiffening by increasing nuclear membrane tension and histone acetylation via deactivation of histone deacetylases (HDACs), leading to osteogenic fate determination220. On the other hand, LINC complex disruption, which presumably reduces nuclear membrane tension, leads to upregulation of HDACs and inhibits osteogenic differentiation220. In another example, migration of myoblasts through confined environments, which is associated with substantial nuclear deformations, led to delayed myoblast differentiation221. These findings may be particularly relevant to fibrotic tissues (e.g., in aging, muscular dystrophies), in which the regeneration ability of muscle stem cells is compromised222, and illustrate the impact of nuclear deformation on cell differentiation.
Conclusion and perspectives
Significant efforts in the recent years have started to shed light on the fascinating roles of nuclear deformations in cellular responses and human disease pathogenesis (see textbox “Human pathologies associated with nuclear deformations”). The disruption of nucleo-cytoskeletal coupling, alteration of nuclear mechanics, or defective mechanotransduction signaling can cause the emergence of various human diseases. Chromatin organization, compaction, stretching, and modification that arise from nuclear deformations control downstream expression of genes and cell fate decisions, although often the specific molecular mechanisms, and the precise site of where cellular mechanosensing occurs, remain to be explored in further detail. Altogether, these discoveries have revealed the remarkable mechanoresponsive nature of the nucleus and the key role of NE proteins in the cellular response to mechanical stimuli. However, many open questions remain, such as the mechanisms used by the nucleus to sense force and/or deformation, or how deformation of the nucleus may result in activation or epigenetic modification of specific genes. Although substantial progress has been made in the understanding of nucleo-cytoskeletal coupling, the precise mechanisms for the spatio-temporal regulation of force transmission across the LINC complex required for many cellular functions has yet to be fully elucidated. Connections between the nucleus, other organelles, and the plasma membrane have received far less attention and should be investigated in more detail. Inside the nucleus, a better understanding of the role nuclear F-actin and motor proteins, as well as LLPS processes, in the maintenance of the nuclear structure, genomic organization, and chromatin remodeling will require deeper investigation.
Deciphering the complex mechanical interplay between chromatin, the NE, cytoskeletal filaments, and the cell surface in mechanobiology will benefit from interdisciplinary and integrative approaches, combining live-cell imaging with high spatial and temporal resolution, genetic manipulation, and precise mechanical manipulation. Much of our knowledge in nuclear mechanotransduction has come from innovative technologies. Addressing current challenges in this field will require further technological innovations, for instance to visualize gene expression in live cells while exerting sub-cellular deformations, ideally on a genome-wide scale, and yet with single cell resolution. In addition to these experimental breakthroughs, mechano-chemical models of the nucleus developed by theoretical modelling will be essential to explore how the cooperation between mechanical and biochemical parameters regulates feedback loops in nuclear signaling pathways. A better understanding of the molecular mechanisms governing nuclear mechanobiology would not only clarify how the various cellular mechanotransduction pathways are combined to determine downstream cellular function but may also guide the development of novel therapeutic strategies to treat human diseases that arise from impaired nuclear mechanotransduction signaling and disturbed nucleo-cytoskeletal force transmission.
Acknowledgements
We apologize to all authors whose work could not be included due to space constraints. A.D.S is supported by the Pathway to Independence Award (R00GM123195) and 4D Nucleome 2 center grant (1UM1HG011536). S.G. acknowledges funding from FEDER Prostem Research Project no. 1510614 (Wallonia DG06), the F.R.S.-FNRS Epiforce Project no. T.0092.21 and the Interreg MAT(T)ISSE project, which is financially supported by Interreg France-Wallonie-Vlaanderen (Fonds Européen de Développement Régional, FEDER-ERDF). Y.K. is financially supported by FRIA (F.R.S.-FNRS). J.L. is supported by awards from the National Institutes of Health (R01HL082792, R01GM137605, U54CA210184), the National Science Foundation (ID #2022048), and the VolkswagenStiftung (Az. 96733).
Glossary
- Mechanosensing
Active cellular process through which cells or cellular components detect changes in external forces or mechanical properties of their microenvironment
- Mechanotransduction
Molecular process in which mechanical stimuli are converted (or transduced) into biochemical signals. The mechanotransduction is the central process of cellular mechanosensing
- Chromatin compaction
Process that occurs predominantly at the mesoscale during nucleosome packing leading to chromatin fibers. Epigenetic modifications associated with heterochromatin lead to increased chromatin compaction
- Nuclear transport
Passive (cargo ≤ ≈50 kDa) or active (cargo ≥ ≈50 kDa) transport of molecules between the cytoplasm and nuclear interior through nuclear pore complexes
- Stress
Expression of the mechanical loading in terms of force applied per cross-sectional area of an object. Units of stress are N/m2 (or Pa)
- Farnesylation
post-translational modification of proteins catalyzed by the enzyme farnesyltransferase which adds a 15-carbon unsaturated hydrocarbon chain to a cysteine residue via a thioether linkage, thus anchoring the protein to a lipid membrane
- Focal adhesions
Integrin-mediated cell-substrate adhesion structure anchoring the ends of stress fibers. Focal adhesions mediate strong attachments to substrates and function as an integrin-signaling platform
- Stress fiber
Actin filaments assembly resulting from the interaction and merging of pre-existing radial fibers and transverse arcs (10–30 filaments). Stress fibers can reach a diameter of several hundreds of nanometers and are under constant prestress due to actomyosin contractility
- LINC complex
The Linker of Nucleoskeleton and Cytoskeleton complex consists of SUN-domain proteins at the INM and KASH domain proteins at the ONM and is crucial for force transmission across the NE. SUN-domain proteins are associated with both nuclear lamins and chromatin and cross the inner nuclear membrane. They interact with the KASH domain proteins in the perinuclear (lumen) space between the two membranes. The KASH domain proteins cross the outer nuclear membrane and interact with actin filaments, microtubule filaments (through dynein and kinesin motors), intermediate filaments (through plectin), centrosomes and cytoplasmic organelles
- Intermediate filaments
Large family of cytoskeletal filaments that includes keratins (types I and II), desmin and vimentin (type III), neurofilaments (type IV), and lamins (type V). Intermediate filaments form dimers that then assemble into larger non-polar filament structures that are characterized by their ability to extend substantially under mechanical stress
- Tensile force
Pulling force resulting in the extension of an object
- Viscoelastic or Viscoelasticity
Rheological behaviour of natural or synthetic materials that exhibit a combination of elastic and viscous properties
- Viscous or viscosity
Measure of the resistance of a fluid to deform under either shear or extensional stress, defined as the ratio of shear stress to shear flow. It is commonly perceived as the resistance of a liquid to flow, usually represented by a dashpot, and is related to the internal friction within the fluid. Units of viscosity are Pa/s
- Strain
Geometric measure of the amount of deformation in the direction of the applied force divided by the initial length of the object (unitless number)
- Strain stiffening
Mechanical material property corresponding to a sudden increase of the elastic modulus under strain, i.e., an increase in resistance to further deformation
- Plastic deformation or plasticity
Ability of a solid material to undergo permanent deformation (i.e. non-reversible change of shape) without rupture in response to applied forces
- BAF
Barrier-to-autointegration factor (BAF) is an essential 10 kDa chromatin-binding protein that is highly conserved in metazoan that helps DNA anchoring to the NE. BAF is involved in multiple pathways including NE reassembly (after mitosis and NE rupture), chromatin epigenetics, and DNA damage response. BAF’s function is controlled by phosphorylation/dephosphorylation waves that drive nuclear disassembly
- Colloid osmotic pressure
Pressure generated by solutions of macromolecules in contact with pores that are permeable to water and ions but not macromolecules. Colloid osmotic pressure generates depletion forces that push macromolecules together in crowded solutions and thus promotes aggregation and phase separation
- Confocal Brillouin microscopy
Optical technique combining Brillouin spectroscopy with confocal microscopy to provide a non-contact and direct readout of the mechanical properties of a material (i.e. stiffness, temperature or strain) at the micrometer scale. Spontaneous Brillouin light scattering arises from the interaction between photons and acoustic phonons (i.e., propagation of thermodynamic fluctuations) and permits to quantify intracellular longitudinal modulus without disturbing the cell
- Interkinetic nuclear migration
periodic movement of the nucleus between apical and basal surfaces of neuroepithelial progenitor cells as they progress through the cell cycle
- Epigenetic modification
Response and adaption of an organism to its environment by altering the (local) structure of chromatin, without changes in the genomic sequence, thereby altering the expression of genes. Epigenetic regulation is often achieved by acetylation or methylation of histones and/or DNA
- Laminopathy
Over 180 mutations have been reported in the genes of the nuclear lamina, in particular LMNA, causing diseases termed ‘laminopathies.’ The number of identified laminopathies has steadily increased in recent years, currently including 13 known conditions. heterogeneous genetic disorders that have been associated with mutations in LMNA and, most recently, LMNB1 and LMNB2
- LEM domain protein
The LAP2, Emerin and MAN1 (LEM)-domain is a ≈40-residue helix-loop-helix fold conserved both in eukaryotes and in prokaryotic DNA/RNA-binding proteins. Except Lap2 proteins, which have a second LEM-domain that binds DNA, the function of eukaryotic LEM-domains is to directly bind a conserved chromatin protein named barrier-to-autointegration factor (BAF)
- Aneuploidy
Deviation from the normal diploid karyotype of 46 chromosomes with the presence of one or more extra chromosomes or the absence of one or more chromosomes
- Epithelial-to-mesenchymal transition (EMT)
Transcriptionally governed process over which epithelial cells establish a front-rear polarity while acquiring a mesenchymal and motile phenotype
- TREX1
Three prime repair exonuclease 1 is major 3′ → 5′ DNA exonuclease in mammalian cells, metabolizes preferentially single-stranded DNA (ssDNA). It cleans the cytosol from DNA fragments coming from endogenous elements. Unless degraded, the accumulation of these DNA fragments can activate innate immune signaling
- cGAS-STING pathway
Cellular cytosolic double-stranded DNA (dsDNA) sensor, allowing innate immune response to infections, inflammation, and cancer
- Micronuclei
Small DNA-containing nuclear structures that are spatially isolated from the main nucleus. Micronuclei form from lagging chromosomes or chromosome fragments following mitotic errors or DNA damage, respectively
- Facultative heterochromatin
Condensed transcriptionally silent chromatin region that can decondense and adopt to allow transcription within temporal and spatial contexts. Facultative heterochromatin is not characterized by repetitive sequences, so at the DNA sequence level it is entirely different from constitutive heterochromatin
- Polycomb repressive complex (PRC2)
Major repressive chromatin complex formed by the polycom Group (PcG) proteins
- Serum response factor (SRF)
Transcription factor that plays a key role in the transduction of mechanical signals from cytoplasmic actin and extracellular matrix proteins to the nucleus. SRF is involved in various cellular processes, from cell proliferation to differentiation and devlopment
- Topologically associating domains (TADs)
Self-interacting megabase-scale genomic blocks in which DNA sequences exhibit significantly higher interaction frequencies with other DNA sequences within the domain than with those outside of the block
Bibliography
- 1.Lammerding J Mechanics of the Nucleus. Compr Physiol 1, 783–807 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szczesny SE & Mauck RL The Nuclear Option: Evidence Implicating the Cell Nucleus in Mechanotransduction. J. Biomechl Eng 139, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Long JT & Lammerding J Nuclear Deformation Lets Cells Gauge Their Physical Confinement. Dev. Cell 56, 156–158 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Thomas CH, Collier JH, Sfeir CS & Healy KE Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc. Natl Acad. Sci. U.S.A 99, 1972–1977 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skinner BM & Johnson EEP Nuclear morphologies: their diversity and functional relevance. Chromosoma 126, 195–212 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zink D, Fischer AH & Nickerson JA Nuclear structure in cancer cells. Nat. Rev. Cancer 4, 677–687 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Clippinger SR et al. Disrupted mechanobiology links the molecular and cellular phenotypes in familial dilated cardiomyopathy. Proc. Natl Acad. Sci. U.S.A 116, 17831–17840 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta S, Marcel N, Sarin A & Shivashankar GV Role of Actin Dependent Nuclear Deformation in Regulating Early Gene Expression. PLoS ONE 7, e53031 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miroshnikova YA, Nava MM & Wickström SA Emerging roles of mechanical forces in chromatin regulation. J. Cell. Sci 130, 2243–2250 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Zhu L & Brangwynne CP Nuclear bodies: The emerging biophysics of nucleoplasmic phases. Curr. Opin. Cell. Biol 34, 23–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buchwalter A, Kaneshiro JM & Hetzer MW Coaching from the sidelines: the nuclear periphery in genome regulation. Nat. Rev. Genet 20, 39–50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franke WW, Scheer U, Krohne G & Jarasch ED The nuclear envelope and the architecture of the nuclear periphery. J. Cell. Biol 91, 39s–50s (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim D-H et al. Volume regulation and shape bifurcation in the cell nucleus. J. Cell. Biol 128, 3375–3385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.García-González A et al. The Effect of Cell Morphology on the Permeability of the Nuclear Envelope to Diffusive Factors. Front. Physiol 9, 925 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jevtić P et al. The nucleoporin ELYS regulates nuclear size by controlling NPC number and nuclear import capacity. EMBO Rep 20, e47283 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donnaloja F, Jacchetti E, Soncini M & Raimondi MT Mechanosensing at the Nuclear Envelope by Nuclear Pore Complex Stretch Activation and Its Effect in Physiology and Pathology. Front. Physiol 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schuller AP, Wojtynek M, Mankus D et al. The cellular environment shapes the nuclear pore complex architecture. Nature 598, 667–671 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zimmerli CE et al. Nuclear pores dilate and constrict in cellulo. Science 374, 6573 (2021) [DOI] [PubMed] [Google Scholar]
- 19.Nava MM et al. Heterochromatin-Driven Nuclear Softening Protects the Genome against Mechanical Stress-Induced Damage. Cell 181, 800–817.e22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lomakin AJ et al. The nucleus acts as a ruler tailoring cell responses to spatial constraints. Science 370, eaba2894 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Turgay Y et al. The molecular architecture of lamins in somatic cells. Nature 543, 261–264 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tenga R & Medalia O Structure and unique mechanical aspects of nuclear lamin filaments. Curr. Opin. Struct. Biol 64, 152–159 (2020). [DOI] [PubMed] [Google Scholar]
- 23.de Leeuw R, Gruenbaum Y & Medalia O Nuclear Lamins: Thin Filaments with Major Functions. Trends in Cell Biol. 28, 34–45 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Shimi T et al. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol. Biol. Cell 26, 4075–4086 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolb T, Maass K, Hergt M, Aebi U & Herrmann H Lamin A and lamin C form homodimers and coexist in higher complex forms both in the nucleoplasmic fraction and in the lamina of cultured human cells. Nucleus 2, 425–433 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Nmezi B et al. Concentric organization of A- and B-type lamins predicts their distinct roles in the spatial organization and stability of the nuclear lamina. Proc. Natl Acad. Sci. U.S.A 116, 4307–4315 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naetar N et al. LAP2alpha maintains a mobile and low assembly state of A-type lamins in the nuclear interior. eLife 10, e63476 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pascual-Reguant L et al. Lamin B1 mapping reveals the existence of dynamic and functional euchromatin lamin B1 domains. Nat. Commun 9, 3420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho S et al. Mechanosensing by the Lamina Protects against Nuclear Rupture, DNA Damage, and Cell-Cycle Arrest. Dev. Cell 49, 920–935.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koushki N et al. Lamin A redistribution mediated by nuclear deformation determines dynamic localization of YAP http://biorxiv.org/lookup/doi/10.1101/2020.03.19.998708 (2020)
- 31.Jahed Z, Domkam N, Ornowski J, Yerima G & Mofrad MRK Molecular models of LINC complex assembly at the nuclear envelope. J. Cell. Sci 134 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Lombardi ML et al. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J. Biol. Chem 286, 26743–26753 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denis KB, Cabe JI, Danielsson BE, Tieu KV, Mayer CR, & Conway DE The LINC complex is required for endothelial cell adhesion and adaptation to shear stress and cyclic stretch. Mol. Biol. Cell 32, 1654–1663 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Splinter D et al. Bicaudal D2, Dynein, and Kinesin-1 Associate with Nuclear Pore Complexes and Regulate Centrosome and Nuclear Positioning during Mitotic Entry. PLoS Biol 8, e1000350 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salpingidou G, Smertenko A, Hausmanowa-Petrucewicz I, Hussey PJ & Hutchison CJ A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J. Cell. Biol 178, 897–904 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sosa BA, Rothballer A, Kutay U & Schwartz TU LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell 149, 1035–1047 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cruz VE, Esra Demircioglu F & Schwartz TU Structural Analysis of Different LINC Complexes Reveals Distinct Binding Modes. J. Mol. Biol 432, 6028–6041 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajgor D & Shanahan CM Nesprins: from the nuclear envelope and beyond. Expert Rev. Mol. Med 15, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson Meredith H, and Holzbaur Erika L F. Nesprins anchor kinesin-1 motors to the nucleus to drive nuclear distribution in muscle cells. Development 142, 218–228 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fridolfsson HN, Ly N, Meyerzon M & Starr DA UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Developmental Biology 338, 237–250 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilhelmsen K et al. Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J. Cell. Biol 171, 799–810 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roux KJ et al. Nesprin 4 is an outer nuclear membrane protein that can induce kinesin-mediated cell polarization. Proc. Natl Acad. Sci. U.S.A 106, 2194–2199 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horn HF et al. A mammalian KASH domain protein coupling meiotic chromosomes to the cytoskeleton. J. Cell. Biol 202, 1023–1039 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Agrawal A & Lele TP Mechanics of nuclear membranes. J. Cell. Sci 132, jcs229245 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoffman LM et al. Mechanical stress triggers nuclear remodeling and the formation of transmembrane actin nuclear lines with associated nuclear pore complexes. Mol. Biol. Cell 31, 1774–1787 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Versaevel M et al. Super-resolution microscopy reveals LINC complex recruitment at nuclear indentation sites. Sci. Rep 4, 7362 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davidson PM et al. Nesprin‐2 accumulates at the front of the nucleus during confined cell migration. EMBO Rep 21, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim SM, Cruz VE, Antoku S, Gundersen GG & Schwartz TU Structures of FHOD1-Nesprin1/2 complexes reveal alternate binding modes for the FH3 domain of formins. Structure 29, 540–552.e5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saunders CA et al. TorsinA controls TAN line assembly and the retrograde flow of dorsal perinuclear actin cables during rearward nuclear movement. J. Cell Biol 216, 657–674 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gudise S, Figueroa RA, Lindberg R, Larsson V & Hallberg E Samp1 is functionally associated with the LINC complex and A-type lamina networks. J. Cell Sci 124, 2077–2085 (2011). [DOI] [PubMed] [Google Scholar]
- 51.Zwerger M et al. Myopathic lamin mutations impair nuclear stability in cells and tissue and disrupt nucleo-cytoskeletal coupling. Hum. Mol. Genet 22, 2335–2349 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hao H et al. The Nesprin-1/−2 ortholog ANC-1 regulates organelle positioning in C. elegans independently from its KASH or actin-binding domains. eLife 10, e61069 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Versaevel M, Riaz M, Grevesse T & Gabriele S Cell confinement: Putting the squeeze on the nucleus. Soft Matter 9, 6665–6676 (2013). [Google Scholar]
- 54.Guilak F, Tedrow JR & Burgkart R Viscoelastic properties of the cell nucleus. Biochem. Biophys. Res. Commun 269, 781–786 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Hobson CM, Falvo MR & Superfine R A survey of physical methods for studying nuclear mechanics and mechanobiology. APL Bioeng 5, 041508 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ & Discher DE Physical plasticity of the nucleus in stem cell differentiation. Proc. Natl. Acad. Sci. U.S.A 104, 15619–15624 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Davidson PM et al. High-throughput microfluidic micropipette aspiration device to probe time-scale dependent nuclear mechanics in intact cells. Lab Chip 19, 3652–3663 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rowat AC, Foster LJ, Nielsen MM, Weiss M & Ipsen JH Characterization of the elastic properties of the nuclear envelope. J. R. Soc. Interface 2, 63–69 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dahl KN, Engler AJ, Pajerowski JD & Discher DE Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys. J 89, 2855–2864 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stephens AD, Banigan EJ, Adam SA, Goldman RD & Marko JF Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol. Biol. Cell 28, 1984–1996 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grevesse T, Dabiri BE, Parker KK & Gabriele S Opposite rheological properties of neuronal microcompartments predict axonal vulnerability in brain injury. Sci. Rep 5, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lammerding J et al. Lamins A and C but Not Lamin B1 Regulate Nuclear Mechanics. J. Biol. Chem 281, 25768–25780 (2006). [DOI] [PubMed] [Google Scholar]
- 63.Neelam S et al. Direct force probe reveals the mechanics of nuclear homeostasis in the mammalian cell. Proc. Natl Acad. Sci. U.S.A 112, 5720–5725 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lammerding J et al. Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J. Clin. Invest 113, 370–378 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lammerding J et al. Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J. Cell. Biol 170, 781–791 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cao X et al. A Chemomechanical Model for Nuclear Morphology and Stresses during Cell Transendothelial Migration. Biophys. J 111, 1541–1552 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Versaevel M et al. Probing cytoskeletal pre-stress and nuclear mechanics in endothelial cells with spatiotemporally controlled (de-)adhesion kinetics on micropatterned substrates. Cell. Adh. Migr 11, 98–109 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ferrera D et al. Lamin B1 overexpression increases nuclear rigidity in autosomal dominant leukodystrophy fibroblasts. FASEB J 28, 3906–3918 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen NY et al. An absence of lamin B1 in migrating neurons causes nuclear membrane ruptures and cell death. Proc. Natl Acad. Sci. U.S.A 116, 25870–25879 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hatch EM & Hetzer MW Nuclear envelope rupture is induced by actin-based nucleus confinement. J. Cell. Biol 215, 27–36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Raab M et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science 352, 359–362 (2016). [DOI] [PubMed] [Google Scholar]
- 72.Denais CM et al. Nuclear envelope rupture and repair during cancer cell migration. Science 352, 353–358 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Q et al. Local, transient tensile stress on the nuclear membrane causes membrane rupture. Mol. Cell. Biol 30, 899–906 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Furusawa T et al. Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat. Commun 6, 6138 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maeshima K, Tamura S, Hansen JC & Itoh Y Fluid-like chromatin: Toward understanding the real chromatin organization present in the cell. Curr. Opin. Cell Biol 64, 77–89 (2020). [DOI] [PubMed] [Google Scholar]
- 76.Hansen JC, Maeshima K & Hendzel MJ The solid and liquid states of chromatin. Epigenetics Chromatin 14, 50 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhuang X Spatially resolved single-cell genomics and transcriptomics by imaging. Nat. Methods 18, 18–22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fiorillo L et al. Comparison of the Hi-C, GAM and SPRITE methods using polymer models of chromatin. Nat. Methods 18, 482–490 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Samwer M, Schneider M, Hoefler R, Schmalhorst PS, Jude JG, Zuber J, & Gerlich DW DNA Cross-Bridging Shapes a Single Nucleus from a Set of Mitotic Chromosomes. Cell 170, 956–972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang P et al. WDR5 modulates cell motility and morphology and controls nuclear changes induced by a 3D environment. Proc. Natl Acad. Sci. U.S.A 115, 8581–8586 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Serra-Marques A, Houtekamer R, Hintzen D, Canty JT, Yildiz A, & Dumont S The mitotic protein NuMA plays a spindle-independent role in nuclear formation and mechanics. J. Cell. Biol 219, e202004202 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tamashunas AC, Tocco VJ, Matthews J, Zhang Q, Atanasova KR, Paschall L, Pathak S, Ratnayake R, Stephens AD, Luesch H, Licht JD, & Lele TP High-throughput gene screen reveals modulators of nuclear shape. Mol. Biol. Cell 31, 1392–1402 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Larson AG et al. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Welsh TJ, Shen Y, Levin A & Knowles TPJ Mechanobiology of Protein Droplets: Force Arises from Disorder. Cell 175, 1457–1459 (2018). [DOI] [PubMed] [Google Scholar]
- 85.Gibson BA et al. Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 179, 470–484 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zidovska A The rich inner life of the cell nucleus: dynamic organization, active flows, and emergent rheology. Biophys. Rev 12, 1093–1106 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Al Jord A et al. Cytoplasmic forces functionally reorganize nuclear condensates in oocytes https://www.biorxiv.org/content/10.1101/2021.03.15.434387v1 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bracha D et al. Mapping Local and Global Liquid Phase Behavior in Living Cells Using Photo-Oligomerizable Seeds. Cell 175, 1467–1480.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shin Y et al. Spatiotemporal control of intracellular phase transitions using light-activated optoDroplets. Cell 168, 159–171.e14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shin Y et al. Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome. Cell 175, 1481–1491.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Stephens Andrew D et al. Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. Mol. Biol. Cell 30, 2320–2330 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cantwell H & Nurse P Unravelling nuclear size control. Curr. Genet 65, 1281–1285 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Petrie RJ, Koo H & Yamada KM Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science 345, 1062–1065 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mistriotis P et al. Confinement hinders motility by inducing RhoA-mediated nuclear influx, volume expansion, and blebbing. J. Cell Biol 218, 4093–4111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Neelam S et al. Direct force probe reveals the mechanics of nuclear homeostasis in the mammalian cell. Proc. Natl Acad. Sci. U.S.A 112, 5720–5725 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mitchison TJ Colloid osmotic parameterization and measurement of subcellular crowding. Mol. Biol. Cell 30, 173–180 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lemière J, Real-Calderon P, Holt LJ, Fai TG & Chang F Control of nuclear size by osmotic forces in Schizosaccharomyces pombe https://www.biorxiv.org/content/10.1101/2021.12.05.471221v2 (2021) [DOI] [PMC free article] [PubMed]
- 98.Deviri D & Safran SA Balance of osmotic pressures determines the volume of the cell nucleus https://www.biorxiv.org/content/10.1101/2021.10.01.462771v1 (2021) [DOI] [PMC free article] [PubMed]
- 99.Takata H et al. Chromatin compaction protects genomic DNA from radiation damage. PloS One 8, e75622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Buxboim A et al. Matrix Elasticity Regulates Lamin-A,C Phosphorylation and Turnover with Feedback to Actomyosin. Curr. Biol 24, 1909–1917 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Holaska JM, Kowalski AK & Wilson KL Emerin Caps the Pointed End of Actin Filaments: Evidence for an Actin Cortical Network at the Nuclear Inner Membrane. PLoS Biol 2, e231 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Le HQ et al. Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat. Cell. Biol 18, 864–875 (2016). [DOI] [PubMed] [Google Scholar]
- 103.Swift J et al. Nuclear Lamin-A Scales with Tissue Stiffness and Enhances Matrix-Directed Differentiation. Science 341, 1240104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mattout A et al. An EDMD mutation in C. elegans lamin blocks muscle-specific gene relocation and compromises muscle integrity. Curr. Biol 21, 1603–1614 (2011). [DOI] [PubMed] [Google Scholar]
- 105.Solovei I et al. LBR and lamin A/C sequentially tether peripheral heterochromatin and inversely regulate differentiation. Cell 152, 584–598 (2013). [DOI] [PubMed] [Google Scholar]
- 106.Shin J-W et al. Lamins regulate cell trafficking and lineage maturation of adult human hematopoietic cells. Proc. Natl Acad. Sci. U.S.A 110, 18892–18897 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zuela N, Dorfman J & Gruenbaum Y Global transcriptional changes caused by an EDMD mutation correlate to tissue specific disease phenotypes in C. elegans. Nucleus 8, 60–69 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Iyer KV et al. Apico-basal cell compression regulates Lamin A/C levels in epithelial tissues. Nat. Commun 12, 1756 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Olins DE & Olins AL Granulocyte heterochromatin: defining the epigenome. BMC Cell Biol 6, 39 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rowat AC et al. Nuclear envelope composition determines the ability of neutrophil-type cells to passage through micron-scale constrictions. J Biol Chem 288, 8610–8618 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Roberts AB et al. Tumor cell nuclei soften during transendothelial migration. J. Biomech 121, 110400 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Infante E et al. LINC complex-Lis1 interplay controls MT1-MMP matrix digest-on-demand response for confined tumor cell migration. Nat. Commun 9, 2443 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bell ES et al. Low lamin A levels enhance confined cell migration and metastatic capacity in breast cancer https://www.biorxiv.org/content/10.1101/2021.07.12.451842v1 (2021). [DOI] [PMC free article] [PubMed]
- 114.Gerlitz G The Emerging Roles of Heterochromatin in Cell Migration. Front. Cell Dev. Biol 8, 394 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Krause M et al. Cell migration through three-dimensional confining pores: speed accelerations by deformation and recoil of the nucleus. Phil. Trans. R. Soc. B 374, 20180225 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Harada T et al. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J. Cell. Biol 204, 669–682 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mitchell MJ et al. Lamin A/C deficiency reduces circulating tumor cell resistance to fluid shear stress. Am. J. Physiol., Cell Physiol 309, C736–C746 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Earle AJ et al. Mutant lamins cause nuclear envelope rupture and DNA damage in skeletal muscle cells. Nat. Mater 19, 464–473 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gundersen GG & Worman HJ Nuclear Positioning. Cell 152, 1376–1389 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Roman W & Gomes ER Nuclear positioning in skeletal muscle. Semin. Cell Dev. Biol 82, 51–56 (2018). [DOI] [PubMed] [Google Scholar]
- 121.Lorber D, Rotkopf R & Volk T A minimal constraint device for imaging nuclei in live Drosophila contractile larval muscles reveals novel nuclear mechanical dynamics. Lab Chip 20, 2100–2112 (2020). [DOI] [PubMed] [Google Scholar]
- 122.Collins MA et al. Ensconsin-dependent changes in microtubule organization and LINC complex-dependent changes in nucleus-nucleus interactions result in quantitatively distinct myonuclear positioning defects. Mol. Biol. Cell 32 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Davidson PM, Denais C, Bakshi MC & Lammerding J Nuclear Deformability Constitutes a Rate-Limiting Step During Cell Migration in 3-D Environments. Cell. Mol. Bioeng 7, 293–306 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Smith ER et al. Nuclear envelope structural proteins facilitate nuclear shape changes accompanying embryonic differentiation and fidelity of gene expression. BMC Cell Biol 18, 8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Venturini V et al. The nucleus measures shape changes for cellular proprioception to control dynamic cell behavior. Science 370, eaba2644 (2020). [DOI] [PubMed] [Google Scholar]
- 126.Spear PC & Erickson CA Interkinetic nuclear migration: a mysterious process in search of a function. Dev. Growth Differ 54, 306–316 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Alam SG et al. The nucleus is an intracellular propagator of tensile forces in NIH 3T3 fibroblasts. J. Cell. Sci 128, 1901–1911 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tsukamoto S et al. Compressive forces driven by lateral actin fibers are a key to the nuclear deformation under uniaxial cell-substrate stretching. Biochem. Biophys. Res. Commun 597, 37–43 (2022). [DOI] [PubMed] [Google Scholar]
- 129.Aureille J et al. Nuclear envelope deformation controls cell cycle progression in response to mechanical force. EMBO Rep 20, e48084 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Khatau SB et al. A perinuclear actin cap regulates nuclear shape. Proc. Natl Acad. Sci. U.S.A 106, 19017–19022 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lammerding J & Wolf K Nuclear envelope rupture: Actin fibers are putting the squeeze on the nucleus. J. Cell. Biol 215, 5–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hatch EM Nuclear envelope rupture: little holes, big openings. Curr. Opin. Cell Biol 52, 66–72 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lovett DB, Shekhar N, Nickerson JA, Roux KJ & Lele TP Modulation of Nuclear Shape by Substrate Rigidity. Cell. Mol. Bioeng 6, 230–238 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gupta M et al. Adaptive rheology and ordering of cell cytoskeleton govern matrix rigidity sensing. Nat. Commun 6, 7525 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Versaevel M, Grevesse T & Gabriele S Spatial coordination between cell and nuclear shape within micropatterned endothelial cells. Nat. Commun 3, 671 (2012). [DOI] [PubMed] [Google Scholar]
- 136.Nagayama K, Yahiro Y & Matsumoto T Apical and Basal Stress Fibers have Different Roles in Mechanical Regulation of the Nucleus in Smooth Muscle Cells Cultured on a Substrate. Cell. Mol. Bioeng 6, 473–481 (2013). [Google Scholar]
- 137.Cho S et al. Mechanosensing by the Lamina Protects against Nuclear Rupture, DNA Damage, and Cell-Cycle Arrest. Dev. Cell 49, 920–935.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Azevedo M & Baylies MK Getting into Position: Nuclear Movement in Muscle Cells. Trends Cell Biol 30, 303–316 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Roman W et al. Muscle repair after physiological damage relies on nuclear migration for cellular reconstruction. Science 374, 355–359 (2021). [DOI] [PubMed] [Google Scholar]
- 140.Wilson MH, & Holzbaur EL Nesprins anchor kinesin-1 motors to the nucleus to drive nuclear distribution in muscle cells. Development 142, 218–228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gimpel P et al. Nesprin-1α-Dependent Microtubule Nucleation from the Nuclear Envelope via Akap450 Is Necessary for Nuclear Positioning in Muscle Cells. Curr. Biol 27, 2999–3009.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wilson MH & Holzbaur ELF Opposing microtubule motors drive robust nuclear dynamics in developing muscle cells. J. Cell. Sci 125, 4158–4169 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Earle AJ et al. Mutant lamins cause nuclear envelope rupture and DNA damage in skeletal muscle cells. Nat. Mater 19, 464–473 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Roman W et al. Myofibril contraction and crosslinking drive nuclear movement to the periphery of skeletal muscle. Nat. Cell. Biol 19, 1189–1201 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chai RJ et al. Disrupting the LINC complex by AAV mediated gene transduction prevents progression of Lamin induced cardiomyopathy. Nat. Commun 12, 4722 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Piccus R & Brayson D The nuclear envelope: LINCing tissue mechanics to genome regulation in cardiac and skeletal muscle. Biol. Lett 16, 20200302 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Razafsky D, Potter C & Hodzic D Validation of a Mouse Model to Disrupt LINC Complexes in a Cell-specific Manner. J. Vis. Exp 106, e53318 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hampoelz B et al. Microtubule-induced nuclear envelope fluctuations control chromatin dynamics in Drosophila embryos. Development 138, 3377–3386 (2011). [DOI] [PubMed] [Google Scholar]
- 149.Schulze SR et al. A Comparative Study of Drosophila and Human A-Type Lamins. PLoS ONE 4, e7564 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Starr DA A network of nuclear envelope proteins and cytoskeletal force generators mediates movements of and within nuclei throughout Caenorhabditis elegans development. Exp. Biol. Med 244, 1323–1332 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Driver EC, Northrop A & Kelley MW Cell migration, intercalation and growth regulate mammalian cochlear extension. Development 144, 3766–3776 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mohammed D et al. Substrate area confinement is a key determinant of cell velocity in collective migration. Nat. Phys 15, 858–866 (2019). [Google Scholar]
- 153.Norden C, Young S, Link BA & Harris WA Actomyosin Is the Main Driver of Interkinetic Nuclear Migration in the Retina. Cell 138, 1195–1208 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Nickerson PE et al. Live imaging and analysis of postnatal mouse retinal development. BMC Dev. Biol 13, 24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Seirin-Lee S et al. Role of dynamic nuclear deformation on genomic architecture reorganization. PLoS Comput. Biol 15, e1007289 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tsai L-H & Gleeson JG Nucleokinesis in neuronal migration. Neuron 46, 383–388 (2005). [DOI] [PubMed] [Google Scholar]
- 157.Cooper JA Mechanisms of cell migration in the nervous system. J. Cell Biol 202, 725–734 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wu YK, Umeshima H, Kurisu J & Kengaku M Nesprins and opposing microtubule motors generate a point force that drives directional nuclear motion in migrating neurons. Development 145, (2018). [DOI] [PubMed] [Google Scholar]
- 159.Wolf K et al. Collagen-based cell migration models in vitro and in vivo. Sem. Cell Dev. Biol 20, 931–941 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yamada KM & Sixt M Mechanisms of 3D cell migration. Nat. Rev. Mol. Cell Biol 20, 738–752 (2019). [DOI] [PubMed] [Google Scholar]
- 161.Roberts AB et al. Tumor cell nuclei soften during transendothelial migration. J. Biomech 121, 110400 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mistriotis P et al. Confinement hinders motility by inducing RhoA-mediated nuclear influx, volume expansion, and blebbing. J. Cell Biol 218, 4093–4111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fridolfsson HN & Starr DA Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J. Cell Biol 191, 115–128 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Marks PC & Petrie RJ Push or pull: how cytoskeletal crosstalk facilitates nuclear movement through 3D environments. Phys. Biol 19, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ivkovic S et al. Direct inhibition of myosin II effectively blocks glioma invasion in the presence of multiple motogens. Mol. Biol. Cell 23, 533–542 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Thomas DG et al. Non-muscle myosin IIB is critical for nuclear translocation during 3D invasion. J. Cell. Biol 210, 583–594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Picariello HS et al. Myosin IIA suppresses glioblastoma development in a mechanically sensitive manner. Proc. Natl Acad. Sci. U.S.A 116, 15550–15559 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Thiam HR, Vargas P, Carpi N et al. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat. Commun 7, 10997 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Lomakin AJ et al. The nucleus acts as a ruler tailoring cell responses to spatial constraints. Science 370, eaba2894 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Katayama T et al. Stimulatory effects of arachidonic acid on myosin ATPase activity and contraction of smooth muscle via myosin motor domain. Am. J. Physiol. Heart Circ. Physiol 298, H505–514 (2010). [DOI] [PubMed] [Google Scholar]
- 171.Brown M, Roulson J-A, Hart CA, Tawadros T & Clarke NW Arachidonic acid induction of Rho-mediated transendothelial migration in prostate cancer. Br. J. Cancer 110, 2099–2108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.de F. Nader GP et al. Compromised nuclear envelope integrity drives TREX1-dependent DNA damage and tumor cell invasion. Cell 184, 5230–5246.e22 (2021). [DOI] [PubMed] [Google Scholar]
- 173.Cosgrove BD et al. Nuclear envelope wrinkling predicts mesenchymal progenitor cell mechano-response in 2D and 3D microenvironments. Biomaterials 270, 120662 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Renkawitz J et al. Nuclear positioning facilitates amoeboid migration along the path of least resistance. Nature 568, 546–550 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.de Noronha CM, Sherman MP, Lin HW, Cavrois MV, Moir RD, Goldman RD, & Greene WC Dynamic disruptions in nuclear envelope architecture and integrity induced by HIV-1 Vpr. Science 294, 1105–1108 (2001). [DOI] [PubMed] [Google Scholar]
- 176.De Vos WH et al. Repetitive disruptions of the nuclear envelope invoke temporary loss of cellular compartmentalization in laminopathies. Hum. Mol. Genet 20, 4175–4186 (2011). [DOI] [PubMed] [Google Scholar]
- 177.Vargas JD, Hatch EM, Anderson DJ & Hetzer MW Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus 3, 88–100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Maciejowski J & Hatch EM Nuclear Membrane Rupture and Its Consequences. Annu Rev. Cell Dev. Biol 36, 85–114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Srivastava N et al. Nuclear fragility, blaming the blebs. Curr. Opin. Cell Biol 70, 100–108 (2021). [DOI] [PubMed] [Google Scholar]
- 180.Zwerger M et al. Myopathic lamin mutations impair nuclear stability in cells and tissue and disrupt nucleo-cytoskeletal coupling. Hum. Mol. Genet 22, 2335–2349 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Penfield L et al. Dynein pulling forces counteract lamin-mediated nuclear stability during nuclear envelope repair. Mol. Biol. Cell 29, 852–868 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Goldman RD et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl Acad. Sci. U.S.A 101, 8963–8968 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Muchir A et al. Nuclear envelope alterations in fibroblasts from patients with muscular dystrophy, cardiomyopathy, and partial lipodystrophy carrying lamin A/C gene mutations. Muscle Nerve 30, 444–450 (2004). [DOI] [PubMed] [Google Scholar]
- 184.Karoutas A, Szymanski W, Rausch T et al. The NSL complex maintains nuclear architecture stability via lamin A/C acetylation. Nat. Cell. Biol 21, 1248–1260 (2019). [DOI] [PubMed] [Google Scholar]
- 185.Rowat AC, Lammerding J & Ipsen JH Mechanical Properties of the Cell Nucleus and the Effect of Emerin Deficiency. Biophys. J 91, 4649–4664 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Le Berre M, Aubertin J & Piel M Fine control of nuclear confinement identifies a threshold deformation leading to lamina rupture and induction of specific genes. Integr. Biol 4, 1406–1414 (2012). [DOI] [PubMed] [Google Scholar]
- 187.Thiam HR, Vargas P, Carpi N et al. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat. Commun 7, 10997 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Takaki T et al. Actomyosin drives cancer cell nuclear dysmorphia and threatens genome stability. Nat. Commun 8, 16013 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Deviri D, Discher DE & Safran SA Rupture Dynamics and Chromatin Herniation in Deformed Nuclei. Biophys. J 113, 1060–1071 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Isermann P & Lammerding J Consequences of a tight squeeze: Nuclear envelope rupture and repair. Nucleus 8, 268–274 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Deviri D et al. Scaling laws indicate distinct nucleation mechanisms of holes in the nuclear lamina. Nat. Phys 15, 823–829 (2019). [Google Scholar]
- 192.Xia Y et al. Nuclear rupture at sites of high curvature compromises retention of DNA repair factors. J. Cell Biol 217, 3796–3808 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Halfmann CT et al. Repair of nuclear ruptures requires barrier-to-autointegration factor. J. Cell Biol 218, 2136–2149 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Young AM, Gunn AL & Hatch EM BAF facilitates interphase nuclear membrane repair through recruitment of nuclear transmembrane proteins. Mol. Biol. Cell 31, 1551–1560 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Irianto J et al. DNA Damage Follows Repair Factor Depletion and Portends Genome Variation in Cancer Cells after Pore Migration. Curr. Biol 27, 210–223 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Shah P et al. Nuclear Deformation Causes DNA Damage by Increasing Replication Stress. Curr. Biol 31, 753–765.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Pfeifer CR, Vashisth M, Xia Y & Discher DE Nuclear failure, DNA damage, and cell cycle disruption after migration through small pores: a brief review. Essays Biochem 63, 569–577 (2019). [DOI] [PubMed] [Google Scholar]
- 198.Kidiyoor GR et al. ATR is essential for preservation of cell mechanics and nuclear integrity during interstitial migration. Nat. Commun 11, 4828 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jiang YN et al. Interleukin 6-triggered ataxia-telangiectasia mutated kinase activation facilitates epithelial-to-mesenchymal transition in lung cancer by upregulating vimentin expression. Exp. Cell. Res 381, 165–171 (2019). [DOI] [PubMed] [Google Scholar]
- 200.Peng B, Ortega J, Gu L, Chang Z & Li G-M Phosphorylation of proliferating cell nuclear antigen promotes cancer progression by activating the ATM/Akt/GSK3β/Snail signaling pathway. J. Biol. Chem 294, 7037–7045 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Bakhoum SF et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Almonacid M et al. Active Fluctuations of the Nuclear Envelope Shape the Transcriptional Dynamics in Oocytes. Dev. Cell 51, 145–157.e10 (2019). [DOI] [PubMed] [Google Scholar]
- 203.Tajik A et al. Transcription upregulation via force-induced direct stretching of chromatin. Nat. Mater 15, 1287–1296 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Sun J, Chen J, Mohagheghian E & Wang N Force-induced gene up-regulation does not follow the weak power law but depends on H3K9 demethylation. Sci. Adv 6, eaay9095 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hsia C-R et al. Confined Migration Induces Heterochromatin Formation and Alters Chromatin Accessibility 2021.09.22.461293 https://www.biorxiv.org/content/10.1101/2021.09.22.461293v1 (2021). [DOI] [PMC free article] [PubMed]
- 206.Jain N & Vogel V Spatial confinement downsizes the inflammatory response of macrophages. Nat. Mater 17, 1134–1144 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Heo S-J et al. Mechanically Induced Chromatin Condensation Requires Cellular Contractility in Mesenchymal Stem Cells. Biophys. J 111, 864–874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Heo S-J et al. Biophysical Regulation of Chromatin Architecture Instills a Mechanical Memory in Mesenchymal Stem Cells. Sci. Rep 5, 16895 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Damodaran K et al. Compressive force induces reversible chromatin condensation and cell geometry-dependent transcriptional response. Mol. Biol. Cell 29, 3039–3051 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ho CY, Jaalouk DE, Vartiainen MK & Lammerding J Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature 497, 507–511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Driscoll TP, Cosgrove BD, Heo S-J, Shurden ZE & Mauck RL Cytoskeletal to Nuclear Strain Transfer Regulates YAP Signaling in Mesenchymal Stem Cells. Biophys. J 108, 2783–2793 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Moya IM & Halder G Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol 20, 211–226 (2019). [DOI] [PubMed] [Google Scholar]
- 213.Bruyère C et al. Actomyosin contractility scales with myoblast elongation and enhances differentiation through YAP nuclear export. Sci. Rep 9, 15565 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Dupont S Regulation of YAP/TAZ Activity by Mechanical Cues: An Experimental Overview. Methods Mol. Biol 1893, 183–202 (2019). [DOI] [PubMed] [Google Scholar]
- 215.Elosegui-Artola A et al. Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell 171, 1397–1410.e14 (2017). [DOI] [PubMed] [Google Scholar]
- 216.Luciano M et al. Cell monolayers sense curvature by exploiting active mechanics and nuclear mechanoadaptation. Nat. Phys 17, 1382–1390 (2021) [Google Scholar]
- 217.Aragona M et al. A Mechanical Checkpoint Controls Multicellular Growth through YAP/TAZ Regulation by Actin-Processing Factors. Cell 154, 1047–1059 (2013). [DOI] [PubMed] [Google Scholar]
- 218.Hoffman LM et al. Mechanical stress triggers nuclear remodeling and the formation of transmembrane actin nuclear lines with associated nuclear pore complexes. Mol. Biol. Cell 31, 1774–1787 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Walker CJ et al. Nuclear mechanosensing drives chromatin remodelling in persistently activated fibroblasts. Nat. Biomed. Eng 5,1485–1499 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Killaars AR, Walker CJ & Anseth KS Nuclear mechanosensing controls MSC osteogenic potential through HDAC epigenetic remodeling. Proc. Natl Acad. Sci. U.S.A 117, 21258–21266 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Smith LR, Irianto J, Xia Y, Pfeifer CR & Discher DE Constricted migration modulates stem cell differentiation. Mol. Biol. Cell 30, 1985–1999 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Smith LR & Barton ER Regulation of fibrosis in muscular dystrophy. Matrix Biol 68–69, 602–615 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Schermelleh L et al. Subdiffraction Multicolor Imaging of the Nuclear Periphery with 3D Structured Illumination Microscopy. Science 320, 1332–1336 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Sabinina VJ et al. Three-dimensional superresolution fluorescence microscopy maps the variable molecular architecture of the nuclear pore complex. Mol. Biol. Cell 32, 1523–1533 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Kind J & van Steensel B Genome-nuclear lamina interactions and gene regulation. Curr. Opin. Cell Biol 22, 320–325 (2010). [DOI] [PubMed] [Google Scholar]
- 226.Pascual-Garcia P & Capelson M Nuclear pores in genome architecture and enhancer function. Curr. Opin. Cell Biol 58, 126–133 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Kalverda B, Pickersgill H, Shloma VV, & Fornerod M Nucleoporins Directly Stimulate Expression of Developmental and Cell-Cycle Genes Inside the Nucleoplasm. Cell 140, 360–371 (2010). [DOI] [PubMed] [Google Scholar]
- 228.Briand N & Collas P Lamina-associated domains: peripheral matters and internal affairs. Genome Biol 21, 85 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Gesson K et al. A-type lamins bind both hetero- and euchromatin, the latter being regulated by lamina-associated polypeptide 2 alpha. Genome Res 26, 462–473 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Rullens PMJ & Kind J Attach and stretch: Emerging roles for genome–lamina contacts in shaping the 3D genome. Curr. Opin. Cell Biol 70, 51–57 (2021). [DOI] [PubMed] [Google Scholar]
- 231.Briand N & Collas P Laminopathy-causing lamin A mutations reconfigure lamina-associated domains and local spatial chromatin conformation. Nucleus 9, 216–226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Lionetti MC et al. Chromatin and Cytoskeletal Tethering Determine Nuclear Morphology in Progerin-Expressing Cells. Biophys. J 118, 2319–2332 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Schreiner SM, Koo PK, Zhao Y, Mochrie SGJ & King MC The tethering of chromatin to the nuclear envelope supports nuclear mechanics. Nat. Commun 6, 7159 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Tavares-Cadete F, Norouzi D, Dekker B, Liu Y & Dekker J Multi-contact 3C reveals that the human genome during interphase is largely not entangled. Nat. Struct. Mol. Biol 27, 1105–1114 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Belaghzal H et al. Liquid chromatin Hi-C characterizes compartment-dependent chromatin interaction dynamics. Nat. Genet 53, 367–378 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Williams JF et al. Phase separation enables heterochromatin domains to do mechanical work https://www.biorxiv.org/content/10.1101/2020.07.02.184127v1 (2020)
- 237.Strom AR et al. HP1α is a chromatin crosslinker that controls nuclear and mitotic chromosome mechanics. eLife 10, e63972 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Keenen MM et al. HP1 proteins compact DNA into mechanically and positionally stable phase separated domains. eLife 10, e64563 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Cattoglio C, Pustova I, Walther N, Ho JJ, Hantsche-Grininger M, Inouye CJ, Hossain MJ, Dailey GM, Ellenberg J, Darzacq X, Tjian R, & Hansen AS Determining cellular CTCF and cohesin abundances to constrain 3D genome models. eLife 8, e40164 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Li X & Fu X-D Chromatin-associated RNAs as facilitators of functional genomic interactions. Nat. Rev. Genet 20, 503–519 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Bizhanova A & Kaufman P Close to the edge: Heterochromatin at the nucleolar and nuclear peripheries. Biochim Biophys Acta Gene Regul Mech 1864, 194666 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Miron E et al. Chromatin arranges in chains of mesoscale domains with nanoscale functional topography independent of cohesin. Sci. Adv 6, eaba8811 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Cho S, Irianto J & Discher DE Mechanosensing by the nucleus: From pathways to scaling relationships. J. Cell Biol 216, 305–315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Kirby TJ & Lammerding J Emerging views of the nucleus as a cellular mechanosensor. Nat. Cell. Biol 20, 373–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Miroshnikova YA & Wickström SA Mechanical Forces in Nuclear Organization. Cold Spring Harb. Perspect. Biol 14, a039685 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Niethammer P Components and Mechanisms of Nuclear Mechanotransduction. Annu. Rev. Cell Dev. Biol 37, 233–256 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Schuller AP et al. The cellular environment shapes the nuclear pore complex architecture. Nature 598, 667–671 (2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Elosegui-Artola A et al. Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell 171, 1397–1410.e14 (2017). [DOI] [PubMed] [Google Scholar]
- 249.Long JT & Lammerding J Nuclear Deformation Lets Cells Gauge Their Physical Confinement. Dev. Cell 56, 156–158 (2021). [DOI] [PubMed] [Google Scholar]
- 250.Enyedi B, Jelcic M & Niethammer P The Cell Nucleus Serves as a Mechanotransducer of Tissue Damage-Induced Inflammation. Cell 165, 1160–1170 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Shen Z et al. A synergy between mechanosensitive calcium- and membrane-binding mediates tension-sensing by C2-like domains. Proc. Natl Acad. Sci. U.S.A 119, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Shen Z & Niethammer P A cellular sense of space and pressure. Science 370, 295–296 (2020). [DOI] [PubMed] [Google Scholar]
- 253.Swift J & Discher DE The nuclear lamina is mechano-responsive to ECM elasticity in mature tissue. J. Cell. Sci 127, 3005–3015 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Ihalainen TO et al. Differential basal-to-apical accessibility of lamin A/C epitopes in the nuclear lamina regulated by changes in cytoskeletal tension. Nat. Mater 14, 1252–1261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Sapra KT et al. Nonlinear mechanics of lamin filaments and the meshwork topology build an emergent nuclear lamina. Nat. Commun 11, 6205 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Guilluy C et al. Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat. Cell Biol 16, 376–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Jacobson EC et al. Migration through a small pore disrupts inactive chromatin organization in neutrophil-like cells. BMC Biol 16, 142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Alisafaei F, Jokhun DS, Shivashankar GV & Shenoy VB Regulation of nuclear architecture, mechanics, and nucleocytoplasmic shuttling of epigenetic factors by cell geometric constraints. Proc. Natl Acad. Sci. U.S.A 116, 13200–13209 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Shin Y et al. Liquid Nuclear Condensates Mechanically Sense and Restructure the Genome. Cell 175, 1481–1491.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Zwerger M, Ho CY & Lammerding J Nuclear mechanics in disease. Annu Rev. Biomed. Eng 13, 397–428 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Zink D, Fischer AH & Nickerson JA Nuclear structure in cancer cells. Nat Rev Cancer 4, 677–687 (2004). [DOI] [PubMed] [Google Scholar]
- 262.Nyirenda N, Farkas DL & Ramanujan VK Preclinical evaluation of nuclear morphometry and tissue topology for breast carcinoma detection and margin assessment. Breast Cancer Res. Treat 126, 345–354 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Mueller JL et al. Rapid staining and imaging of subnuclear features to differentiate between malignant and benign breast tissues at a point-of-care setting. J. Cancer Res. Clin. Oncol 142, 1475–1486 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Somech R, Shaklai S, Amariglio N, Rechavi G & Simon AJ Nuclear Envelopathies—Raising the Nuclear Veil. Pediatr. Res 57, 8–15 (2005). [DOI] [PubMed] [Google Scholar]
- 265.Hershberger RE, Hedges DJ & Morales A Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat. Rev. Cardiol 10, 531–547 (2013). [DOI] [PubMed] [Google Scholar]
- 266.Wong X & Stewart CL The Laminopathies and the Insights They Provide into the Structural and Functional Organization of the Nucleus. Annu. Rev. Genom. Hum. Genet 21, 263–288 (2020). [DOI] [PubMed] [Google Scholar]
- 267.Bonne G et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet 21, 285–288 (1999). [DOI] [PubMed] [Google Scholar]
- 268.Caron M et al. Human lipodystrophies linked to mutations in A-type lamins and to HIV protease inhibitor therapy are both associated with prelamin A accumulation, oxidative stress and premature cellular senescence. Cell Death Differ 14, 1759–1767 (2007). [DOI] [PubMed] [Google Scholar]
- 269.De Sandre-Giovannoli A et al. Lamin a truncation in Hutchinson-Gilford progeria. Science 300, 2055 (2003). [DOI] [PubMed] [Google Scholar]
- 270.Folker ES, Ostlund C, Luxton GWG, Worman HJ & Gundersen GG Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proc. Natl Acad. Sci. U.S.A 108, 131–136 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Méjat A & Misteli T LINC complexes in health and disease. Nucleus 1, 40–52 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Fischer M, Rikeit P, Knaus P & Coirault C YAP-Mediated Mechanotransduction in Skeletal Muscle. Front. Physiol 7, 41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Owens DJ et al. Lamin Mutations Cause Increased YAP Nuclear Entry in Muscle Stem Cells. Cells 9, 816 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Eriksson M et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature 423, 293–298 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Verstraeten VLRM, Ji JY, Cummings KS, Lee RT & Lammerding J Increased mechanosensitivity and nuclear stiffness in Hutchinson-Gilford progeria cells: effects of farnesyltransferase inhibitors. Aging Cell 7, 383–393 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Booth EA, Spagnol ST, Alcoser TA & Dahl KN Nuclear stiffening and chromatin softening with progerin expression leads to an attenuated nuclear response to force. Soft Matter 11, 6412–6418 (2015). [DOI] [PubMed] [Google Scholar]
- 277.Kim PH et al. Disrupting the LINC complex in smooth muscle cells reduces aortic disease in a mouse model of Hutchinson-Gilford progeria syndrome. Sci. Transl. Med 10, eaat7163 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Dahl KN et al. Distinct structural and mechanical properties of the nuclear lamina in Hutchinson-Gilford progeria syndrome. Proc. Natl Acad. Sci. U.S.A 103, 10271–10276 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Columbaro M et al. Rescue of heterochromatin organization in Hutchinson-Gilford progeria by drug treatment. Cell. Mol. Life Sci 62, 2669–78 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Stephens AD, Banigan EJ & Marko JF Separate roles for chromatin and lamins in nuclear mechanics. Nucleus 9, 119–124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Stephens AD et al. Physicochemical mechanotransduction alters nuclear shape and mechanics via heterochromatin formation. Mol. Biol. Cell 30, 2320–2330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Coffinier C, Fong LG & Young SG LINCing lamin B2 to neuronal migration: growing evidence for cell-specific roles of B-type lamins. Nucleus 1, 407–411 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Coffinier C et al. Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol. Biol. Cell 22, 4683–4693 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Young SG, Jung H-J, Coffinier C & Fong LG Understanding the Roles of Nuclear A- and B-type Lamins in Brain Development. J. Biol. Chem 287, 16103–16110 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Vortmeyer-Krause M et al. Lamin B2 follows lamin A/C- mediated nuclear mechanics and cancer cell invasion efficacy http://biorxiv.org/lookup/doi/10.1101/2020.04.07.028969 (2020).
- 286.Padiath QS et al. Lamin B1 duplications cause autosomal dominant leukodystrophy. Nat. Genet 38, 1114–1123 (2006). [DOI] [PubMed] [Google Scholar]
- 287.Ballatore C, Lee VM-Y & Trojanowski JQ Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat. Rev. Neurosci 8, 663–672 (2007). [DOI] [PubMed] [Google Scholar]
- 288.Sergent C, Baillet S & Dehaene S Timing of the brain events underlying access to consciousness during the attentional blink. Nat. Neurosci 8, 1391–1400 (2005). [DOI] [PubMed] [Google Scholar]
- 289.Fernández-Nogales M et al. Huntington’s disease is a four-repeat tauopathy with tau nuclear rods. Nat. Med 20, 881–885 (2014). [DOI] [PubMed] [Google Scholar]
- 290.Ballatore C, Lee VM-Y & Trojanowski JQ Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat. Rev. Neurosci 8, 663–672 (2007). [DOI] [PubMed] [Google Scholar]
- 291.Paonessa F et al. Microtubules Deform the Nuclear Membrane and Disrupt Nucleocytoplasmic Transport in Tau-Mediated Frontotemporal Dementia. Cell Rep 26, 582–593.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Crisp M et al. Coupling of the nucleus and cytoplasm: role of the LINC complex. J. Cell. Biol 172, 41–53 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Fernández-Nogales M & Lucas JJ Altered Levels and Isoforms of Tau and Nuclear Membrane Invaginations in Huntington’s Disease. Front. Cell. Neurosci 13, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Meinke P et al. Muscular dystrophy-associated SUN1 and SUN2 variants disrupt nuclear-cytoskeletal connections and myonuclear organization. PLoS Genet 10, e1004605 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Zhou N, Maire P, Masterson S & Bickford M The mouse pulvinar nucleus: organization of the tectorecipient zones. Vis. Neurosci 34, E011 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.von Appen A et al. LEM2 phase separation promotes ESCRT-mediated nuclear envelope reformation. Nature 582, 115–118 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Miroshnikova YA, Nava MM & Wickström SA Emerging roles of mechanical forces in chromatin regulation. J. Cell. Sci 130, 2243–2250 (2017). [DOI] [PubMed] [Google Scholar]
- 298.Bronshtein I et al. Loss of lamin A function increases chromatin dynamics in the nuclear interior. Nat. Commun 6, 8044 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Nikolova V, Leimena C, McMahon AC, Tan JC, Chandar S, Jogia D, Kesteven SH, Michalicek J, Otway R, Verheyen F, Rainer S, Stewart CL, Martin D, Feneley MP, & Fatkin D Defects in nuclear structure and function promote dilated cardiomyopathy in lamin A/C-deficient mice. J. Clin. Investig 113, 357–369 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Puckelwartz MJ et al. Nesprin-1 mutations in human and murine cardiomyopathy. J. Mol. Cell Cardiol 48, 600–608 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]


