Abstract
Background:
Inorganic lead (Pb) is common in the environment, and is toxic to neurological, renal, and cardiovascular systems. Pb exposure influences the epigenome with documented effects on DNA methylation (DNAm). We assessed the impact of low levels of Pb exposure on DNAm among non-miner individuals from two locations in Peru: Lima, the capital, and Cerro de Pasco, a highland mining town, to study the effects of Pb exposure on physiological outcomes and DNAm.
Methods:
Pb levels were measured in whole blood (n = 305). Blood leukocyte DNAm was determined for 90 DNA samples using the Illumina MethylationEPIC chip. An epigenome-wide association study was performed to assess the relationship between Pb and DNAm.
Results:
Individuals from Cerro de Pasco had higher Pb than individuals from Lima (p-value = 2.00E-16). Males had higher Pb than females (p-value = 2.36E-04). Pb was positively associated with hemoglobin (p-value = 8.60E-04). In Cerro de Pasco, blood Pb decreased with the distance from the mine (p-value = 0.04), and association with soil Pb was approaching significance (p-value = 0.08). We identified differentially methylated positions (DMPs) associated with genes SOX18, ZMIZ1, and KDM1A linked to neurological function. We also found 45 differentially methylated regions (DMRs), seven of which were associated with genes involved in metal ion binding and nine to neurological function and development.
Conclusions:
Our results demonstrate that even low levels of Pb can have a significant impact on the body including changes to DNAm. We report associations between Pb and hemoglobin, Pb and distance from mining, and between blood and soil Pb. We also report associations between loci- and region-specific DNAm and Pb.
Keywords: Lead (Pb), Hemoglobin, EWAS, Andes, Mining, DNA methylation
1. Introduction
Lead (Pb) is a trace metal that has been associated with human civilization since the emergence of metallurgy (Mielke and Reagan 1998). Pb is the world’s most abundant heavy metal, and exposure to it is estimated to account for 143,000 deaths per year, accounting for 0.6% of the global disease burden (Haefliger 2011; Tong et al. 2000). Miners and mining communities are at a higher risk of exposure to Pb and its effects, including higher levels of Pb in both indoor and outdoor air, soil, and self-produced vegetables in areas close to Pb mining (Attina and Trasande 2013; Qu et al. 2012).
Pb exposure can have negative effects on any organ in the body, including the neurological, hematological, cardiovascular, and renal systems, with the nervous system being the most susceptible to its harmful effects (Kalia and Flora 2005; Sanders et al. 2009; WHO 2011). Both central and peripheral nervous systems can be affected by Pb (Flora et al. 2012). In children, Pb exposure has been associated with encephalopathy characterized by a degeneration of certain parts of the brain (Sanders et al. 2009). Moreover, children exposed to Pb may experience delayed growth, decreased IQ, impairment in concentration ability, and other adverse outcomes (Flora et al. 2012; Sanders et al. 2009). Additionally, Pb has a direct effect on the hematopoietic system by inhibiting enzymes involved in the heme synthesis pathway, thus leading to decreased hemoglobin (Hb) concentration (Barltrop and Smith 1971; Hu et al. 1994). Pb also increases the fragility of erythrocyte membranes, reducing their lifespan and resulting in anemia (Flora et al. 2012).
There is growing evidence that Pb can impact the epigenome. Epigenetic modifications can have an effect on gene expression without changing the sequence of the nucleotides. DNA methylation (DNAm) is the most commonly studied epigenetic modification and is generally associated with gene repression when located in promoter regions of genes (Fraga and Esteller 2002). Inorganic Pb may affect DNAm via increased production of reactive oxygen species (ROS) upon exposure (Adonaylo and Oteiza 1999). Increased oxidative stress is known to result in decreased DNAm (Pogribny et al. 2007), and can inhibit binding of the methyl-CpG binding proteins that promote chromatin inactivation (Valinluck et al. 2004).
Previous studies have shown associations between Pb exposure and both global, gene-specific, and genome-wide DNAm signatures in humans and human embryonic stem cells (Goodrich et al. 2016; Hanna et al. 2012; Li et al. 2013; Li et al. 2016; Rygiel et al. 2020; Senut et al. 2014; Wright et al. 2010). However, little is known about the effects of chronic low-level Pb exposure on genome-wide DNAm in adults exposed to mining who are not miners themselves. This study was performed in a cohort of individuals who were recruited in Lima (low altitude) and Cerro de Pasco, Peru (high altitude) to study genetic and epigenetic aspects of high-altitude adaptation (Brutsaert et al. 2019; Childebayeva et al. 2019). All of the high-altitude participants were residents of Cerro de Pasco, Peru. This municipality is an important Peruvian mining center that was established in the 17th century to support silver mining, and was at one point the second largest producer of silver in the Andes (Fisher 1977; Helfgott 2013). Upon the depletion of the city’s silver reserves, silver mining was replaced by copper, zinc, and Pb extraction (van Geen et al. 2012). In 1952, the Cerro de Pasco Corporation claimed to be the largest South American producer of refined Pb (Helfgott 2013). Given the effect of Pb on the epigenome (Bakulski et al. 2012; Faulk et al. 2014; Goodrich et al. 2015), we measured whole-blood lead levels (WBLLs) in a subset of the study participants (Childebayeva et al. 2019). Here, we performed an epigenome-wide association study to identify DNAm differences between Pb exposure groups. We also determined the relationship between Pb and Hb, proximity to mining, and soil Pb
2. Materials and Methods
2.1. Study population
Individuals in this study (N = 305) were selected from a larger cohort of 603 Peruvian individuals of self-reported multigenerational Quechua ancestry between ages 18 and 35 (Childebayeva et al. 2019). The individuals were recruited based on a migrant study design to study high-altitude adaptation in two locations, Lima and Cerro de Pasco, Peru: Lima is the capital of Peru located at low altitude and the largest city, while Cerro de Pasco is a mining community located at approximately 4,400 m above sea level (masl) (Fig. 1). We determined WBLLs in a subset of the participants (N = 305) (Table 1): 1) Cerro de Pasco (n = 157): Quechua individuals who were born and raised at high altitude and recruited in Cerro de Pasco, a Peruvian mining center; 2) Lima (n = 148): Quechua individuals living in Lima at low altitude as young adults at the time of study, some of whom were born and raised in Lima and some of whom were born in various high-altitude locations in Peru and moved to Lima prior to recruitment. Male and female participants were recruited to participate if they were unrelated, healthy, non-pregnant/lactating, non-smokers, non-miners, and between 18 and 35 years old (Table 1). At the time of enrollment, all study participants provided written informed consent approved by Syracuse University, Universidad Peruana Cayetano Heredia, and the University of Michigan Institutional Review Boards.
Fig. 1.
Study Design. Map of Peru depicting high-altitude, Cerro de Pasco (4,338 m), and low-altitude, Lima (150 m), participant recruitment locations.
Table 1.
Participant characteristics.
| Phenotype | Lima (n = 148) | Cerro de Pasco (n = 157) | Total (n = 305) | Lima EPIC (n = 59)# | Cerro de Pasco EPIC (n = 28)# | Total EPIC (n = 87)# |
|---|---|---|---|---|---|---|
| Age | 24.83 ± 5.05 | 24.67 ± 4.96 | 24.75 ± 5.00 | 24.93 ± 5.14 | 23.82 ± 4.7 | 24.57 ± 5 |
| %Female | 52% | 52% | 52% | 36% | 36% | 36% |
| Hb, g/dL* | 13.74 ± 1.40 | 17.50 ±2.24 | 15.66 ± 2.66 | 14.11 ± 1.49 | 18.03 ± 1.65 | 15.37 ± 2.4 |
| Height (cm)*! | 159.80 ± 8.58 | 157.59 ± 8.08 | 158.67 ± 8.39 | 161.8 ± 7.38 | 158.93 ± 7.91 | 160.88 ± 7.63 |
| Weight (kg)* | 64.62 ± 11.07 | 59.46 ±7.93 | 61.96 ± 9.91 | 67.1 ± 10.34 | 60.38 ± 7.52 | 64.93 ± 9.99 |
| BMI* | 25.23 ± 3.43 | 23.98 ± 3.09 | 24.59 ± 3.32 | 25.6 ± 3.41 | 23.98 ± 3.27 | 25.08 ± 3.43 |
| Education^ | NA | 2.79 ± 0.41 | NA | NA | 2.79 ± 0.42 | NA |
| WBLL, µg/dL* | 2.03 ± 0.62 | 4.75 ± 1.53 | 3.43 ± 1.53 | 2.07 ± 0.66 | 5.18 ± 1.70 | 3.08 ± 1.83 |
Data and means ± SD
1 = primaria (primary school), 2 = secundaria (secondary school), 3 = Tec/Superior (higher education)
Hb, hemoglobin; BMI, body mass index; WBLL, whole blood lead level
participant characteristics for individuals analyzed for DNA methylation
p-value < 0.05 based on linear regression in R for the comparison of Lima versus Cerro de Pasco
p-value not significant in the subset of individuals analyzed for DNA methylation
2.2. Phenotypic assessment and Pb exposure assessment
Height, weight, and education information were collected at the time of enrollment, and the participants provided a venous blood sample into a collection tube containing EDTA (Lavender-top, BD Vacutainer, Franklin Lakes, NJ) for DNA extraction. The Puregene DNA purification system (Qiagen, Valencia, CA) was used to extract the genomic DNA according to the manufacturer’s instructions. Red blood cell lysis was performed in Peru immediately following venous puncture. Samples were stabilized at room temperature in cell lysis solution and transported to the University of Michigan for DNA extraction. Hb levels were determined in venous blood using the HemoCue 201 + analyzer (Angelholm Sweden). All study participants were screened for anemia using altitude specific cut-offs. WBLLs were measured using graphite furnace atomic absorption spectrometry (GFAAS) (Parsons and Slavin 1993). GFAAS measurement was performed in duplicate at the Blufstein Laboratory located in Lima, Peru (coefficient of variance between the duplicates for the entire cohort < 2%).
2.3. Geocoding and soil Pb interpolation
Geographic coordinates for the participants from Cerro de Pasco were determined using a portable Garmin GPS (Garmin, Olathe, Kansas) based on address information received during recruitment. We determined the Euclidean distance from the mine (mine-distance) to each participant’s current place of residence using the function distm (function Harvesine) in the R package geosphere (Hijmans et al. 2017).
A previous study by Van Geen et al. (2012) recorded soil Pb values at 74 locations throughout Cerro de Pasco at varying distances from the mine in May 2009, using a hand-held X-ray fluorescence analyzer (Innov-X Alpha, Olympus Corporation, Tokyo, Japan) (van Geen et al. 2012). Using the Pb values reported by van Geen et al. (2012), we interpolated soil Pb levels across Cerro de Pasco using ordinary kriging (Graler et al. 2016). Kriging is a geostatistical interpolation method that produces a prediction surface based on points with known values, and incorporates a semivariogram model to account for the estimated spatial autocorrelation between these points (Burgess and Webster 1980; Krige 1966). We created the predicted surface of soil Pb values using ordinary kriging in ArcGIS 10.4, with a spherical semivariogram model and a cell size of 30 m. In addition to a prediction surface, kriging also creates a surface of estimated prediction error based on the distance between points. This approach has been previously used to interpolate blood Pb level risk with Pb in water for children in Flint, MI (Hanna-Attisha et al. 2016). We used the estimated soil Pb across Cerro de Pasco to estimate soil Pb levels at each participating household in Cerro de Pasco.
2.4. DNA methylation
A random subset of participants was selected from both Lima (n = 59) and Cerro de Pasco (n = 31) for DNAm analysis. EZ-96 DNA Methylation™ Kit (Zymo Research, Irvine, CA) was used for bisulfite conversions of all DNA samples following the standard protocol with alternative incubation conditions optimized for the Illumina Infinium® MethylationEPIC BeadChip (Zymo, Irvine, California). Raw methylation intensity data was loaded into R using the package minfi (Aryee et al. 2014). Eighty-seven samples passed QC, and their missing beta values were imputed, normalized using functional normalization (FunNorm) in minfi followed by the beta-mixture quantile normalization (BMIQ) (Fortin et al. 2014; Morris et al. 2014; Teschendorff et al. 2013). We removed probes with detection p-values > 10e-5 in>10% of samples, probes identified as cross-reactive, probes containing common SNPs, and probes on X and Y chromosomes (for more information on the pipeline see Sup. Fig. 2) (Chen et al. 2013; Pidsley et al. 2016). The resulting beta values (n = 656,183) were tested for batch effects using Singular Value Decomposition (SVD) (Sup. Fig. 3), and the batch effects were corrected using Combat in ChAMP (Johnson et al. 2007; Teschendorff et al. 2009). Following batch correction, beta values were converted into M—values, and differentially methylated positions (DMPs) were determined using limma (Ritchie et al. 2015).
Differentially methylated regions (DMRs) were identified based on the uncorrected p-values (p < 0.05) from the DMP limma analysis using comb-p with the following options (comb-p pipeline -c 4 –seed 10e-3–dist 3000). DMRs were corrected for multiple comparisons using the built-in Sidak correction function in comb-p (Pedersen et al. 2012). Pathway analysis was performed on significant sites (unadjusted p-value < 0.05) from the limma analysis of DMPs using Consensus PathDB (Herwig et al. 2016; Kamburov et al. 2011). We used DAVID for functional annotation of the DMRs (Dennis et al. 2003) using the annotations from Gene Ontology (Ashburner et al. 2000; Gene Ontology 2021), UniProt KrowledgeBase (UniProtKB) (UniProt 2019), BIOCARTA (Nishimura 2001), and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Wixon and Kell 2000).
2.5. Statistical analyses
All statistical analyses on WBLLs and phenotypes of interest were conducted using R version 3.4.0 (R Core Team, 2020). Package ggplot2 (Wickham 2009) was used to plot the data. We ran linear models to assess the relationship between interpolated soil Pb values and WBLLs in a subset of participants from Cerro de Pasco whose geographical location we were able to determine based on their residence location (n = 152). We used interpolated soil Pb values for each participant in the following regression model to find associations between soil Pb and WBLLs: Yi (WBLLs) ~ B00 + B01(interpolated soil Pb) + B02(Sex) + B03(Education) + B04(Kriging standard error), where Yi is the dependent variable, WBLL; “interpolated soil Pb” = interpolated soil Pb value.
DMPs were determined in limma using the following model: DNAm ~ WBLLs + Group (i.e. Altitude) + Sex + Age + Blood principal components (PCs) 1,2,5,6. Blood PCs were determined based on the estimated cell counts function proposed by Houseman et al. (2012). Benjamini-Hochberg correction was used to control for multiple testing and a false discovery rate of 10% was considered significant (Ritchie et al. 2015). Distribution of expected and observed p-values were used to validate the model fit (Sup. Fig. 4).
The relationship between WBLLs and hemoglobin was tested using the following model: Hb ~ WBLLs + Group (i.e. Altitude) + sex.
3. Results
3.1. Study design and participant characteristics
Participant characteristics are provided in Table 1. The study groups did not differ significantly by sex or age. Individuals recruited in Cerro de Pasco were significantly shorter, lighter, and had lower BMI than individuals recruited in Lima (Table 1).
3.2. Lead levels are associated with study group, sex, and Hb
WBLLs were associated with study group (Fig. 2A, Table 1). Participants from Cerro de Pasco had significantly higher WBLLs (4.75 ± 1.53 μg/dL) compared to the participants recruited in Lima (2.03 ± 0.62 μg/dL) based on linear modeling (β = 2.72, p-value = 2e-16). Sex was significantly associated with WBLLs in both Lima and Cerro de Pasco, with males having higher Pb levels than females (Fig. 2B) in each group (Lima: β = 0.31, p-value = 2.40E-03, Cerro de Pasco: β = 1.15, p-value = 1.13e-06).
Fig. 2.
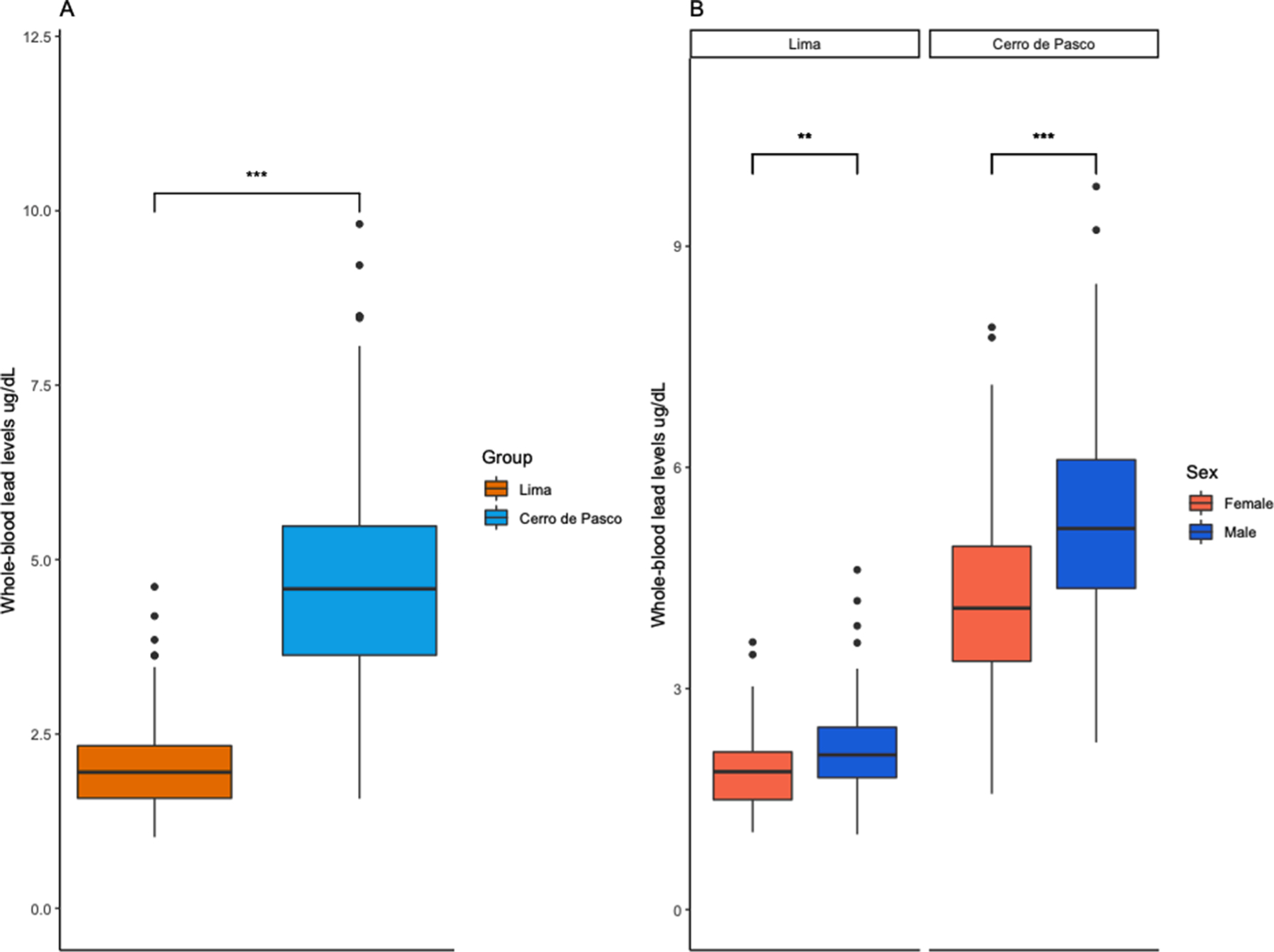
Whole-blood lead levels (WBLLs) in Lima and Cerro de Pasco. (A) Participants from Cerro de Pasco have higher WBLLs than the participants from Lima (p-value < 2.2e-16). (B) WBLLs between Cerro de Pasco and Lima by sex. Males have higher WBLLs than females in both cities (Lima: p-value = 0.0024; Cerro de Pasco: p-value = 1.13E-06).
We identified a positive association between WBLLs and Hb concentration when adjusted for sex and sample group (i.e. altitude) in the entire cohort (β = 0.17, p-value = 8.60E-04) (Fig. 3, Sup. Table 1). When considering the groups separately, WBLLs were significantly associated with Hb levels in participants from Lima (β = 0.32, p-value = 0.007), but not in Cerro de Pasco; although the model was approaching significance (β = 0.16, p-value = 0.07).
Fig. 3.
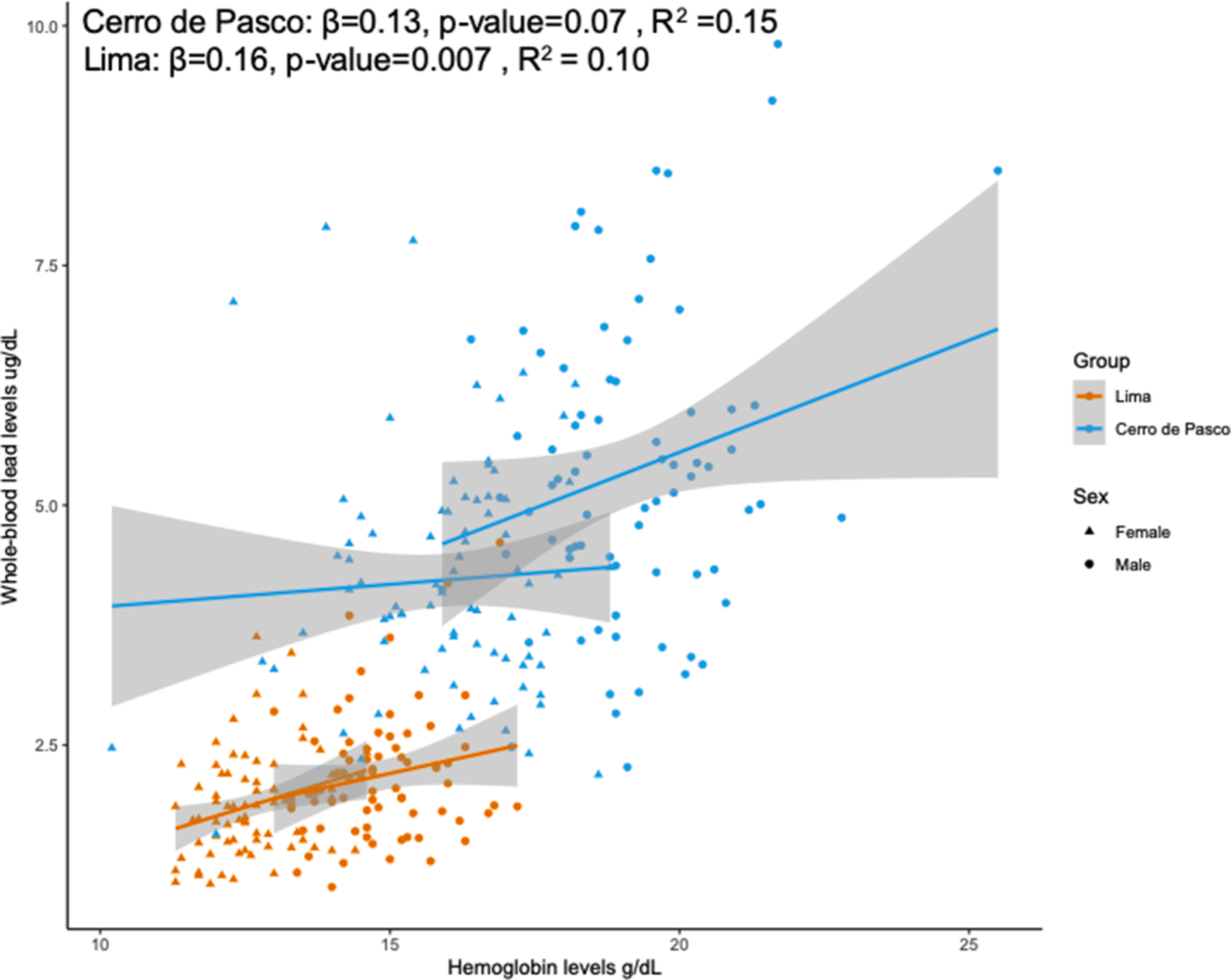
Association between hemoglobin (Hb) in participants from Lima and Cerro de Pasco. The association between WBLLs and Hb levels was not significant for individuals from Cerro de Pasco. The association was significant for individuals in Lima. Statistical models were adjusted for sex.
3.3. Blood lead is negatively correlated with distance from the mine and positively correlated with soil lead in Cerro de Pasco
The distance from the mine was negatively associated with WBLLs when adjusted for sex and education (Fig. 4A, β = 0.0005, p-value = 0.032). Association between WBLLs and interpolated soil Pb (mg/kg) values, adjusted for sex, education, and Kriging standard error, was approaching significance (β = 0.0003, p-value = 0.08) (Sup. Table 1, Fig. 4B and Fig. 5).
Fig. 4.
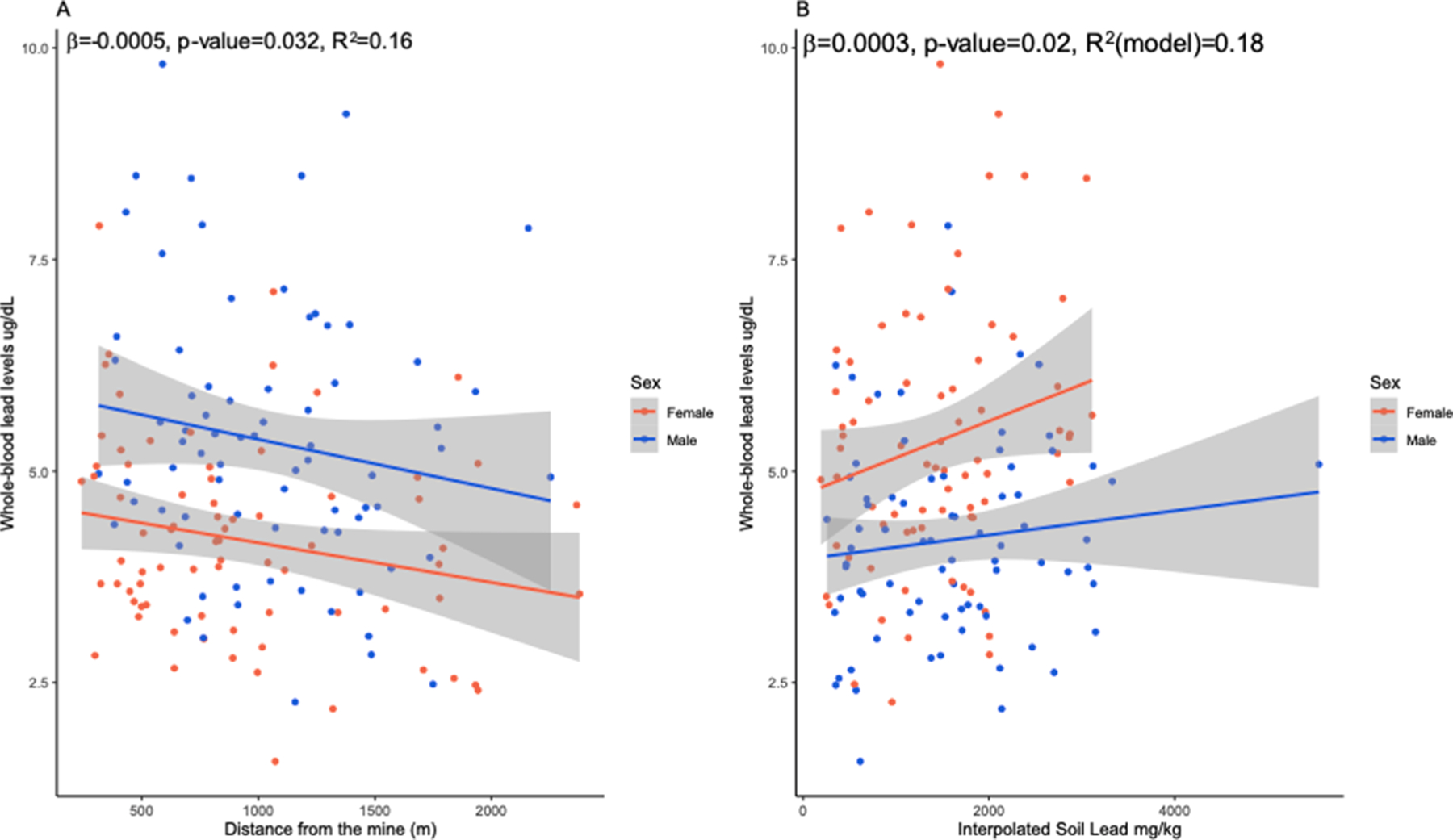
Whole-blood lead levels plotted against the distance from the mine, and interpolated soil lead values. (A) Whole-blood lead levels (WBLL) were negatively associated with distance from the mine in the participants from Cerro de Pasco (model adjusted for age and sex). Unadjusted WBLLs are plotted against the distance from the mine. (B) Relationship between interpolated soil Pb and blood Pb in participants from Cerro de Pasco. The statistical model testing for this association was adjusted for sex and education level. Unadjusted WBLLs are plotted against the interpolated soil Pb.
Fig. 5.
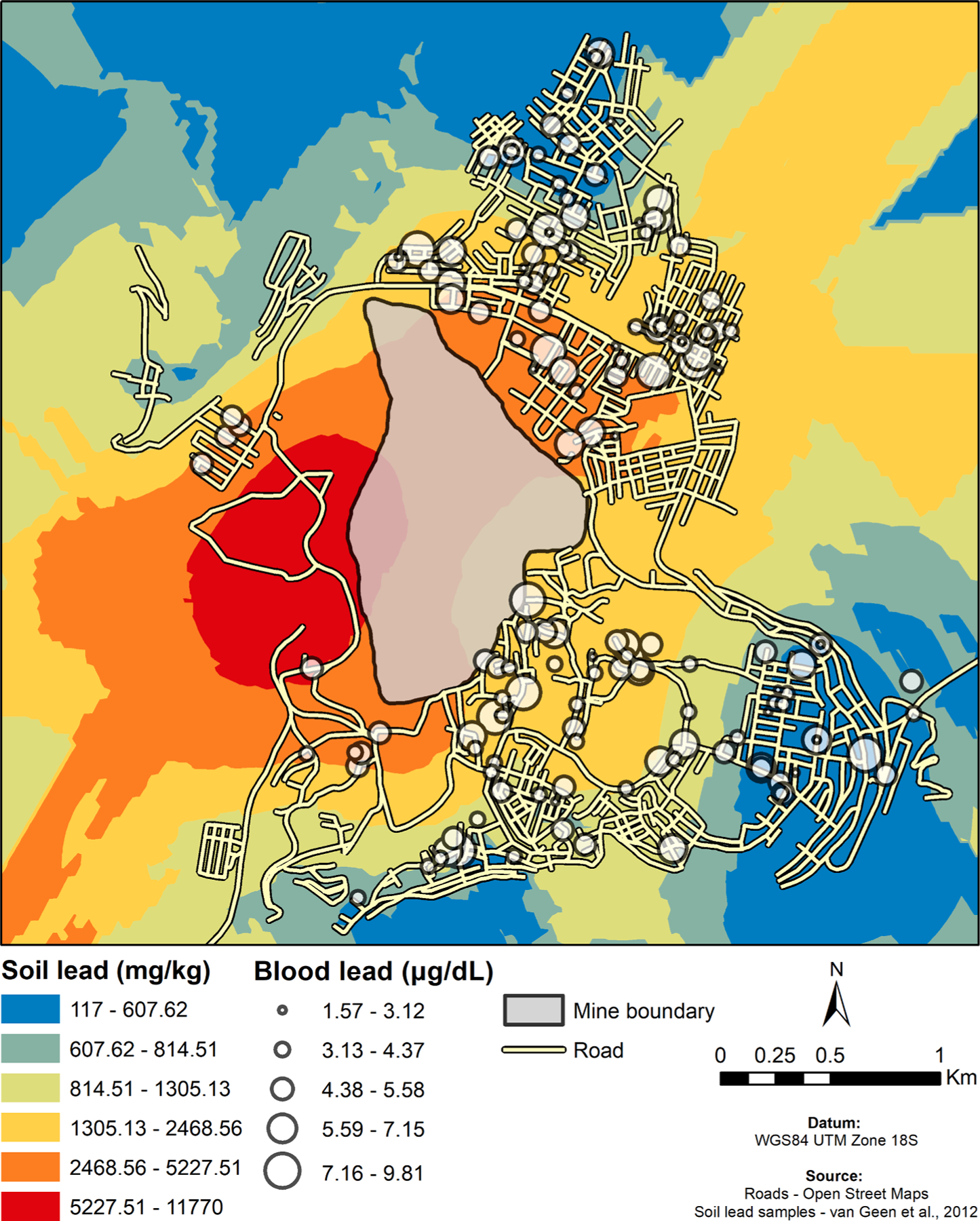
Interpolated soil Pb values for participants from Cerro de Pasco. Soil Pb values were interpolated for participants from Cerro de Pasco using the kriging option in ArcGIS.
3.4. DNA methylation and blood lead
We performed a singular value decomposition (SVD) analysis of genome-wide DNAm data to capture the most salient factors of variation in the data with a small number of components (Teschendorff et al., 2009). Based on SVD, WBLLs were among the significant components of DNAm variation (p < 0.05) before and after experimental batch effects adjustment (Sup. Fig. 3).
We identified four significant DMPs, i.e. single CpG sites, associated with WBLLs at q-value < 0.10 in a model adjusted for study group, sex, age, and blood principal components; the genome-wide inflation factor for this model was 1.01 (Table 2). These DMPs were within and/or in regulatory regions of genes KDM1A, ZMIZ1, TERT, and SOX18 (Sup. Fig. 1). We performed a pathway analysis of the probes significant at the unadjusted p-value < 0.05 over all probes as background using ConsensusPathDB. Based on which, we found n = 22 enriched gene ontology-based sets under the q-value 0.05 cutoff with “Neuronal System” (Reactome pathway, p-value = 4.03e-07, q-value = 0.00154) as the top hit (Sup. Table 2).
Table 2.
Significant differentially methylated positions (DMPs) associated with WBLLs.
| Gene | CpG site | P-value | Q-value | Chr:pos | Coefficient | Beta diff.* | Relation to Island** |
|---|---|---|---|---|---|---|---|
| KDM1A | cg22225928 | 2.80E-07 | 0.08 | chr1:23358458 | 0.08 | 3% | Open Sea |
| ZMIZ1 | cg20543544 | 3.67E-07 | 0.08 | chr10:81003657 | −0.24 | −5% | Island |
| TERT | cg17166338 | 3.77E-07 | 0.08 | chr5:1295969 | 0.15 | 3% | Island |
| SOX18 | cg01355392 | 5.66E-07 | 0.09 | chr20:62679255 | −0.13 | −6% | N. Shore |
Difference in average Methylation beta values between Cerro de Pasco and Lima
According to an annotation from Illumina
The model was adjusted for study group, sex, age, and blood principal components
WBLL = Whole blood lead levels
DMRs reflect several CpG sites in close proximity changing in the same direction. Therefore, they are considered more representative of biologically relevant epigenetic changes than single methylation sites (i. e. DMPs). We identified 45 significant DMRs, i.e. regions with three or more significant DMPs within a 3000 base-pair window, using comb-p (Sup. Table 3). Out of the significant DMRs, seven were associated with metal ion binding and nine with neurological function and development based on an annotation from DAVID (Dennis et al. 2003) (Table 3).
Table 3.
Significant differentially methylated regions (DMRs) associated with metal ion binding and neurological pathways.
| Gene | Region | Relation_to_Island* | N probes | P-value region | Corrected p-value | Δ Meth** | Relevant function*** |
|---|---|---|---|---|---|---|---|
| ZBTB16 | chr11:113933513–113935052 | S. Shore | 7 | 1.33E-08 | 5.65E-06 | ↑ | Metal ion binding |
| ZNF710 | chr15:90543224–90544600 | N. Shore | 6 | 1.63E-07 | 7.75E-05 | ↓ | Metal ion binding |
| ZNF292 | chr6:87861261–87862482 | N. Shore | 6 | 7.16E-07 | 0.000385 | ↓ | Metal ion binding |
| MEX3A | chr1:156046344–156046833 | Island | 4 | 2.05E-08 | 2.74E-05 | ↓ | Metal ion binding |
| DPP6 | chr7:153583318–153584874 | Island | 13 | 4.54E-06 | 0.001913 | ↑ | Neurodegeneration; metal ion binding; metal ion transport |
| HOXB3 | chr17:46647787–46653712 | Open Sea | 21 | 5.77E-14 | 6.39E-12 | ↓ | Neurogenesis |
| NNAT | chr20:36148133–36149751 | N. Shore | 38 | 2.01E-08 | 8.15E-06 | ↓ | Neurogenesis; neuron differentiation |
| PRMT1 | chr19:50190825–50191684 | N. Shore | 6 | 3.98E-07 | 0.000304 | ↓ | Neurogenesis; neuron differentiation |
| EPHA2 | chr1:16472001–16473144 | N. Shelf | 3 | 8.32E-07 | 0.000477 | ↓ | Neurogenesis; neuron differentiation |
| STRA6 | chr15:74495109–74496041 | OpenSea | 9 | 9.2E-06 | 0.006455 | ↓ | Neurological system process |
| ZNF385A | chr12:54763081–54764372 | Island | 6 | 5.25E-08 | 2.67E-05 | ↓ | Neurological system process; Metal ion binding |
| AGAP2 | chr12:58131681–58133184 | Island | 10 | 5.57E-10 | 2.43E-07 | ↓ | Neuron death; regulation of neuron apoptosis; Metal ion binding |
| SNAP23 | chr15:42800679–42800834 | Open Sea | 2 | 5.06E-06 | 0.02117 | ↑ | Neurotransmitter transport; regulation of neurotransmitter levels |
According to annotation from Illumina
Direction of association with WBLLs
According to functional annotation from DAVID
4. Discussion
Pb exposure and the resulting Pb toxicity can negatively affect almost any system of the body, and thus represent an important environmental health and safety concern. Our results identified differences in DNAm, mainly decreased methylation, in Peruvian adults with higher levels of Pb exposure. These differentially methylated positions and regions were associated with genes involved in neurological development and function as well as metal ion binding. Additionally, Pb was positively associated with Hb concentration and soil Pb and inversely associated with proximity to mining. Collectively, our study contributes to the body of knowledge on Pb toxicity in a cross-sectional study of healthy adults who are not miners exposed to Pb from a residential mine.
We identified DNAm differences, both at the level of individual CpG sites (DMPs) and in regions of multiple neighboring CpG sites (DMRs), associated with Pb. Several of the DNAm changes we identified were in genes linked to neurological function and development. This is especially noteworthy given Pb is a neurotoxicant, and exposure to Pb has been associated with neuronal death, intraneuronal regulatory mechanisms, and neurotransmission, among others (Lidsky and Schneider 2003). Furthermore, it is consistent with previous studies that have found Pb-related changes in neuroepigenetic signaling pathways in human embryonic stem cells (Senut et al. 2014), correlations between WBLLs and DNAm of an important tumor suppressor gene, p16, in Pb-exposed individuals (Kovatsi et al. 2010), promoter methylation of COL1A2 in women (Hanna et al. 2012), and an association between maternal Pb levels and a child’s DNAm profile (Goodrich et al. 2016; Rygiel et al. 2020; Sen et al. 2015). Similar results have been shown in animal models wherein the effects of developmental Pb exposure on the expression of DNA methyltransferases DNMT1 and DNMT2, and the methyl cytosine binding protein MeCP2 have been identified in rat hipoccampi (Schneider et al. 2013).
Significant DMPs were found in the genes KDM1A, ZMIZ1, and SOX18 that have been previously linked to neurological function and development, highlighting the fact that the nervous system is the most affected by Pb exposure. Lysine demethylase 1A, or KDM1A, is a H3K4 histone demethylase. Mutations in this gene have been linked to neurological conditions (Ricq et al. 2016). Zinc Finger MIZ-Type Containing 1, or ZMIZ1, is a transcriptional co-activator known to interact with the androgen receptor, and its nucleotide variants have been associated with neurodevelopmental disorders (Carapito et al. 2019; Fewings et al. 2017). SOX18 (SRY [sex determining region Y] box 18) is a transcription factor involved in the regulation of embryonic development and cell fate determination. When mutated, it can lead to cardiovascular dysfunction (Downes and Koopman 2001; Pennisi et al. 2000). Previous research has shown that downregulation of Sox18 in male rat hippocampi is associated with postnatal exposure to Pb acetate (Schneider et al. 2012). Using pathway enrichment analysis, we identified the “Neuronal System” pathway, further suggesting that among the top hits in our analysis, genes associated with the neuronal system were statistically overrepresented.
We identified 45 significant DMRs associated with Pb (Sup. Table 3). Most of the DMRs showed decreased methylation as a function of Pb exposure, which is consistent with previous findings (Faulk et al. 2013). According to functional annotation in DAVID, nine of the DMRs (PMRT1, NNAT, DPP6, STRA6, SNAP23, AGAP2, ZNF385A, and EPHA2) were associated with neurogenesis, neurological system process, neuron differentiation, and neurodegeneration (Dennis et al. 2003) (Table 3). Seven genes were associated with metal ion binding including ZNF292, ZNF710, ZNF385A, ZBTB16, DPP6, AGAP2 and MEX3A (Table 3). Interestingly, NNAT or neuronatin is an imprinted gene that is paternally expressed and is known to play an important role in brain development (Evans et al. 2001; Joseph et al. 1995). Its methylation levels are decreased in cord blood of newborns whose mothers where exposed to heavy metals during pregnancy (Vidal et al. 2015). Together, these findings are important given that exposure to Pb has been linked with neurodegeneration in adults and neurodevelopmental effects in children (Needleman et al. 1979; Stewart et al. 2006), and further highlight the potential role of DNAm as a marker for environmental exposures on the body.
One way through which Pb could affect DNAm is through ROS, given environmental metals, including Pb, can increase the production of ROS via redox cycling (Adonaylo and Oteiza 1999; Ahamed and Siddiqui 2007). This, in turn, can influence epigenetic processes, and specifically DNAm. Oxidative damage to the DNA can hamper the ability of DNA methyltransferases to interact with the DNA, which then may result in altered DNAm levels (Baccarelli and Bollati 2009).
We found higher WBLLs in individuals recruited in Cerro de Pasco compared to the individuals recruited in Lima, which is consistent with previous studies conducted in Cerro de Pasco. Our findings are similar to Conklin et al. (2008) who reported 5.8 ug/dL mean Pb in women of child-bearing age in Cerro de Pasco (Conklin L. et al. 2008). We observed a positive association between WBLLs measured in residents of Cerro de Pasco and proximity to the Cerro de Pasco mine. Furthermore, WBLLs were associated with interpolated soil Pb values that also tended to be higher close to the mine. These results support what has been reported for the residents of Cerro de Pasco on the relationship between soil Pb measurements of the house and blood Pb levels (Conklin L. et al. 2008).
Interestingly, we identified a positive association between WBLLs and Hb concentration. This finding is at odds with previous studies conducted at low altitude demonstrating elevated WBLLs can lead to decreased Hb levels among adults and children (iron deficiency anemia) (Papanikolaou et al. 2005). This decrease occurs as a result of Pb binding to Hb when circulating in the blood, competing with iron, and impairing heme synthesis (Barbosa et al. 2005; Barltrop and Smith 1972). One possible explanation for the positive association between Pb and Hb at high altitude (4,400 m) is the Andean adaptive response to high altitude (Beall 2007). At altitude, environmental hypoxia triggers erythropoiesis through increased production of erythropoietin (EPO) that stimulates RBC and Hb production allowing for greater oxygen-carrying capacity (Guillemin and Krasnow 1997; Knaupp et al. 1992; Storz and Moriyama 2008). Andeans are known to have elevated Hb levels at high altitude, which counteract the environmental hypoxia and maintain optimal oxygen saturation (Monge et al. 1992; Moore 2001). Similar to low ambient oxygen tension, heavy metals, including Pb, can activate hypoxia signaling pathways (Galanis et al. 2009; Valko et al. 2005). Exposure to Pb can result in hypoxia-like responses, and potentially induce similar responses in the body as exposure to hypobaric hypoxia. Thus, Pb exposure in Andeans may potentially lead to increased Hb production given that Andeans are adapted to produce higher levels of Hb in hypoxic conditions (Beall 2000). This, in turn, can counteract Pb-induced anemia. In our cohort, the relationship between Hb and Pb was strong at both high and low altitude suggesting that the positive relationship between Hb and Pb is independent of high-altitude hypoxia, but potential associated with lead-induced hypoxia. The positive relationship between Hb and Pb likely only exists at low levels of exposure to metals, since high levels of Pb exposure (>30 μg/dL) tended to decrease Hb in Ecuadorian Andeans, although that relationship was not statistically significant (Counter et al. 2001). On the other hand, it is possible that WBLLs we measured are not representative of the relevant Pb exposure (Chuang et al. 2001; Hu et al. 1994), which could explain why we see the positive association between Pb and Hb, in contrast to other studies. Moreover, individuals recruited in Cerro de Pasco have higher Hb levels associated with living at altitude, which could have influenced our ability to correctly measure WBLLs, due to possible interference of high Hb with blood Pb measurement.
5. Limitations
DNAm signatures differ by tissue and cell type (Kitamura et al. 2007). In this study, we used DNA extracted from whole blood representing a mixture of DNA-containing cells. Our statistical models of DNAm were adjusted for the principal components based on the blood cell types determined bioinformatically using the method by Houseman et al. (2012). This allowed us to control for the effect of blood cell composition on DNA methylation signatures.
We use comb-p (Pedersen et al. 2012) to identify differentially methylated regions, which has been shown to have high false-positive rates (Kolde et al. 2016; Suderman et al. 2018).
Diet is known to affect DNAm signatures (Zhang et al. 2011). Lima is an urban center and Cerro de Pasco is a remote, Andean town. Any potential differences in diet between participants recruited in Lima and Cerro de Pasco may contribute to the differences in DNAm that we observed. Given our identification of DMPs and DMRs associated with neurological function, we believe that our findings reflect the effect of Pb, a known neurotoxicant, on the DNAm and not the effect of diet.
XRF soil lead values we used in our analyses were collected in 2009 (van Geen et al. 2012), while blood samples were collected during the summer of 2012 and 2013. This may explain why the correlation we see between soil Pb and WBLLs is not significant, but only approaching significance. It is possible that if the XFR values were more recent, we would see a stronger correlation between soil Pb and WBLLs.
Lima and Cerro de Pasco differ by altitude. Lima is at an altitude of 150 masl, and Cerro de Pasco is at 4,338 masl. Our previous work has shown that exposure to high-altitude hypoxia is associated with differences in LINE-1 global DNAm and EPAS1 methylation in this cohort (Childebayeva et al. 2019) as well as genome-wide DNAm differences (Childebayeva et al., 2020). It is possible that the DNAm differences observed between participants from Lima and Cerro de Pasco were the result of decreased oxygen availability experienced at high altitude and not from the effects of Pb. We expect this effect to be mitigated by the use of WBLLs in our analyses, which diminishes the confounding between Pb exposure and high-altitude hypoxia given that it is a direct measurement of Pb exposure. Furthermore, our research on DNAm differences associated with altitude have identified different DMRs and DMPs than presented here (Childebayeva et al. 2020).
6. Conclusions
We tested the effects of Pb exposure in a cohort of adults from two locations in Peru: Lima, the coastal capital, and Cerro de Pasco, a high-altitude mining town. We demonstrated higher WBLLs in participants from Cerro de Pasco compared to the participants from Lima. We showed a positive association between WBLLs and proximity to the Cerro de Pasco mine and interpolated soil Pb. We also found a positive association between Pb and Hb. Lastly, we identified significant associations between WBLLs and DNAm.
To our knowledge, this is among the first studies to report associations between chronic low-level Pb exposure and DNAm in genes associated with neurodevelopment in adults. Collectively, our data indicate that proximity to mining may have effects on human physiology and epigenetics, even when the levels of exposure are low. We suggest conducting future research on Pb exposure in children living in Cerro de Pasco, as well as pregnant women, since these two groups are especially vulnerable to the negative effects of Pb exposure.
Supplementary Material
Acknowledgements and Funding Information
First and foremost, we would like to thank the study participants of Lima and Cerro de Pasco, Peru. We are also thankful to Dr. Sudipta Ghosh (NEHU University, Shillong, India), Obed Garcia, Alexandra Fietel (University of Michigan), Nate Bartman and Jason Howard (Syracuse U), Francisco Villafuerte (Universidad Peruana Cayetano Heredia), Cecilia Anza Ramirez (Universidad Peruana Cayetano Heredia), and Justino Panez (Cerro de Pasco) for help with obtaining GPS coordinates. We are grateful for the help of several participant recruiters and physicians, including Cesar De Albertis, Dolly Nieves, Laura Mori, Josseline Honorio, and Alejandro Zamudio. This work was funded by the National Science Foundation grants BCS-1132310 (T.D.B, F.L., and A.W.B.) and BCS-1613415 (A.C and A.W.B); Wenner-Gren Foundation Grant (A.C); The University of Michigan (A.W.B and A.C.); and the Michigan Lifestage Environmental Exposures and Disease S(M-LEEaD) NIEHS Core Center (P30 ES017885). A.C. was supported by a Baldwin Fellowship from the L.S.B. Leakey Foundation.
Footnotes
CRediT authorship contribution statement
Ainash Childebayeva: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Writing - original draft, Writing - review & editing. Jaclyn M. Goodrich: Writing - original draft, Writing - review & editing. Nathan Chesterman: Formal analysis, Writing - original draft. Fabiola Leon-Velarde: Funding acquisition. Maria Rivera-Ch: Investigation. Melisa Kiyamu: Investigation. Tom D. Brutsaert: Funding acquisition, Investigation, Writing - original draft. Abigail W. Bigham: Conceptualization, Data curation, Funding acquisition, Investigation, Writing - original draft, Writing - review & editing, Supervision. Dana C. Dolinoy: Conceptualization, Funding acquisition, Writing - original draft, Writing - review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.envint.2021.106587.
References
- Adonaylo VN, Oteiza PI, 1999. Lead intoxication: Antioxidant defenses and oxidative damage in rat brain. Toxicology 135 (2–3), 77–85. [DOI] [PubMed] [Google Scholar]
- Ahamed M, Siddiqui MK, 2007. Low level lead exposure and oxidative stress: Current opinions. Clin Chim Acta 383 (1–2), 57–64. [DOI] [PubMed] [Google Scholar]
- Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA, 2014. Minfi: A flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics 30 (10), 1363–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. , 2000. Gene ontology: Tool for the unification of biology. The gene ontology consortium. Nat Genet 25 (1), 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attina TM, Trasande L, 2013. Economic costs of childhood lead exposure in low- and middle-income countries. Environ Health Perspect 121 (9), 1097–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccarelli A, Bollati V, 2009. Epigenetics and environmental chemicals. Curr Opin Pediatr 21 (2), 243–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakulski KM, Rozek LS, Dolinoy DC, Paulson HL, Hu H, 2012. Alzheimer’s disease and environmental exposure to lead: The epidemiologic evidence and potential role of epigenetics. Curr Alzheimer Res 9 (5), 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa F, Tanus-Santos JE, Gerlach RF, Parsons PJ, 2005. A critical review of biomarkers used for monitoring human exposure to lead: Advantages, limitations, and future needs. Environ Health Persp 113 (12), 1669–1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barltrop D, Smith A, 1971. Interaction of lead with erythrocytes. Experientia 27 (1), 92–93. [DOI] [PubMed] [Google Scholar]
- Barltrop D, Smith A, 1972. Lead binding to human haemoglobin. Experientia 28 (1), 76–77. [DOI] [PubMed] [Google Scholar]
- Beall CM, 2000. Tibetan and andean patterns of adaptation to high-altitude hypoxia. Hum Biol 72 (1), 201–228. [PubMed] [Google Scholar]
- Beall CM, 2007. Two routes to functional adaptation: Tibetan and andean high-altitude natives. Proc Natl Acad Sci U S A 104 (Suppl 1), 8655–8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutsaert TD, Kiyamu M, Elias Revollendo G, Isherwood JL, Lee FS, Rivera-Ch M, Leon-Velarde F, Ghosh S, Bigham AW, 2019. Association of egln1 gene with high aerobic capacity of peruvian quechua at high altitude. Proc Natl Acad Sci U S A [DOI] [PMC free article] [PubMed]
- Burgess TM, Webster R, 1980. Optimal interpolation and isarithmic mapping of soil properties. Journal of Soil Science 31 (2), 315–331. [Google Scholar]
- Carapito R, Ivanova EL, Morlon A, Meng L, Molitor A, Erdmann E, Kieffer B, Pichot A, Naegely L, Kolmer A, et al. , 2019. Zmiz1 variants cause a syndromic neurodevelopmental disorder. Am J Hum Genet 104 (2), 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R, 2013. Discovery of cross-reactive probes and polymorphic cpgs in the illumina infinium humanmethylation450 microarray. Epigenetics 8 (2), 203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childebayeva A, Goodrich JM, Leon-Velarde F, Rivera-Chira M, Kiyamu M, Brutsaert TD, Dolinoy DC, Bigham AW, 2020. Genome-wide epigenetic signatures of adaptive developmental plasticity in the andes. Genome Biol Evol [DOI] [PMC free article] [PubMed]
- Childebayeva A, Jones TR, Goodrich JM, Leon-Velarde F, Rivera-Chira M, Kiyamu M, Brutsaert TD, Dolinoy DC, Bigham AW, 2019. Line-1 and epas1 DNA methylation associations with high-altitude exposure. Epigenetics 14 (1), 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang HY, Schwartz J, Gonzales-Cossio T, Lugo MC, Palazuelos E, Aro A, Hu H, Hernandez-Avila M, 2001. Interrelations of lead levels in bone, venous blood, and umbilical cord blood with exogenous lead exposure through maternal plasma lead in peripartum women. Environ Health Persp 109 (5), 527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conklin L, Carlos A Sánchez, Neri Antonio, Staley Paula, Blumenthal Wendy, Jarrett Jeffrey M, LePrell Rebecca, Durant James. 2008. Exposiciones a metales pesados en niños y mujeres en edad fértil en tres comunidades mineras cerro de pasco, perú. CDC
- Counter SA, Buchanan LH, Ortega F, 2001. Gender differences in blood lead and hemoglobin levels in andean adults with chronic lead exposure. Int J Occup Environ Health 7 (2), 113–118. [DOI] [PubMed] [Google Scholar]
- Dennis G Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA, 2003. David: Database for annotation, visualization, and integrated discovery. Genome Biol 4 (5), P3. [PubMed] [Google Scholar]
- Downes M, Koopman P, 2001. Sox18 and the transcriptional regulation of blood vessel development. Trends Cardiovasc Med 11 (8), 318–324. [DOI] [PubMed] [Google Scholar]
- Evans HK, Wylie AA, Murphy SK, Jirtle RL, 2001. The neuronatin gene resides in a “micro-imprinted” domain on human chromosome 20q11.2. Genomics 77 (1–2), 99–104. [DOI] [PubMed] [Google Scholar]
- Faulk C, Barks A, Liu K, Goodrich JM, Dolinoy DC, 2013. Early-life lead exposure results in dose- and sex-specific effects on weight and epigenetic gene regulation in weanling mice. Epigenomics 5 (5), 487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulk C, Liu K, Barks A, Goodrich JM, Dolinoy DC, 2014. Longitudinal epigenetic drift in mice perinatally exposed to lead. Epigenetics 9 (7), 934–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewings N, Gatt PN, McKay FC, Parnell GP, Schibeci SD, Edwards J, Basuki MA, Goldinger A, Fabis-Pedrini MJ, Kermode AG, et al. , 2017. Data characterizing the zmiz1 molecular phenotype of multiple sclerosis. Data Brief 11, 364–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher JR, 1977. Silver mines and silver miners in colonial peru, 1776–1824. Centre for Latin-American Studies, University of Liverpool, Liverpool. [Google Scholar]
- Flora G, Gupta D, Tiwari A, 2012. Toxicity of lead: A review with recent updates. Interdiscip Toxicol 5 (2), 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin JP, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, Greenwood CM, Hansen KD, 2014. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol 15 (12), 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Esteller M, 2002. DNA methylation: A profile of methods and applications. Biotechniques 33(3):632, 634, 636–649. [DOI] [PubMed] [Google Scholar]
- Galanis A, Karapetsas A, Sandaltzopoulos R, 2009. Metal-induced carcinogenesis, oxidative stress and hypoxia signalling. Mutat Res 674 (1–2), 31–35. [DOI] [PubMed] [Google Scholar]
- Gene Ontology C, 2021. The gene ontology resource: Enriching a gold mine. Nucleic acids research 49 (D1), D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JM, Dolinoy DC, Sanchez BN, Zhang Z, Meeker JD, Mercado-Garcia A, Solano-Gonzalez M, Hu H, Tellez-Rojo MM, Peterson KE, 2016. Adolescent epigenetic profiles and environmental exposures from early life through peri-adolescence. Environ Epigenet 2(3):dvw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JM, Sanchez BN, Dolinoy DC, Zhang Z, Hernandez-Avila M, Hu H, Peterson KE, Tellez-Rojo MM, 2015. Quality control and statistical modeling for environmental epigenetics: A study on in utero lead exposure and DNA methylation at birth. Epigenetics 10 (1), 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graler B, Pebesma E, Heuvelink G, 2016. Spatio-temporal interpolation using gstat. The R Journal 8 (1), 204–218. [Google Scholar]
- Guillemin K, Krasnow MA, 1997. The hypoxic response: Huffing and hifing. Cell 89 (1), 9–12. [DOI] [PubMed] [Google Scholar]
- Haefliger P, 2011. Brief guide to analytical methods for measuring lead in blood. WHO Library Cataloguing-in-Publication Data
- Hanna CW, Bloom MS, Robinson WP, Kim D, Parsons PJ, vom Saal FS, Taylor JA, Steuerwald AJ, Fujimoto VY, 2012. DNA methylation changes in whole blood is associated with exposure to the environmental contaminants, mercury, lead, cadmium and bisphenol a, in women undergoing ovarian stimulation for ivf. Hum Reprod 27 (5), 1401–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna-Attisha M, LaChance J, Sadler RC, Champney Schnepp A., 2016. Elevated blood lead levels in children associated with the flint drinking water crisis: A spatial analysis of risk and public health response. Am J Public Health 106 (2), 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfgott FM, 2013. Transformations in labor, land and community: Mining and society in pasco, peru, 20th century to the present . University of Michigan
- Herwig R, Hardt C, Lienhard M, Kamburov A, 2016. Analyzing and interpreting genome data at the network level with consensuspathdb. Nat Protoc 11 (10), 1889–1907. [DOI] [PubMed] [Google Scholar]
- Hijmans RJ, Williams E, Vennes C, 2017. Geosphere. Spherical trigonometry. R. Hu, H., Watanabe, H., Payton, M., Korrick, S., Rotnitzky, A., 1994. The relationship between bone lead and hemoglobin. JAMA 272 (19), 1512–1517. [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A, 2007. Adjusting batch effects in microarray expression data using empirical bayes methods. Biostatistics 8 (1), 118–127. [DOI] [PubMed] [Google Scholar]
- Joseph R, Dou D, Tsang W, 1995. Neuronatin mrna: Alternatively spliced forms of a novel brain-specific mammalian developmental gene. Brain Res 690 (1), 92–98. [DOI] [PubMed] [Google Scholar]
- Kalia K, Flora SJ, 2005. Strategies for safe and effective therapeutic measures for chronic arsenic and lead poisoning. J Occup Health 47 (1), 1–21. [DOI] [PubMed] [Google Scholar]
- Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. 2011. Consensuspathdb: Toward a more complete picture of cell biology. Nucleic acids research. 39(Database issue):D712–717. [DOI] [PMC free article] [PubMed]
- Kitamura E, Igarashi J, Morohashi A, Hida N, Oinuma T, Nemoto N, Song F, Ghosh S, Held WA, Yoshida-Noro C, et al. , 2007. Analysis of tissue-specific differentially methylated regions (tdms) in humans. Genomics 89 (3), 326–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaupp W, Khilnani S, Sherwood J, Scharf S, Steinberg H. 1992. Erythropoietin response to acute normobaric hypoxia in humans. J Appl Physiol (1985) 73(3):837–840. [DOI] [PubMed] [Google Scholar]
- Kolde R, Martens K, Lokk K, Laur S, Vilo J, 2016. Seqlm: An mdl based method for identifying differentially methylated regions in high density methylation array data. Bioinformatics 32 (17), 2604–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovatsi L, Georgiou E, Ioannou A, Haitoglou C, Tzimagiorgis G, Tsoukali H, Kouidou S, 2010. P16 promoter methylation in pb2 -exposed individuals. Clin Toxicol (Phila) 48 (2), 124–128. [DOI] [PubMed] [Google Scholar]
- KrigG. 1966. Two-dimensional weighted moving average trend surfaces for ore valuation Journal of South African Institute of Mining and Metallurgy
- Li C, Yang X, Xu M, Zhang J, Sun N, 2013. Epigenetic marker (line-1 promoter) methylation level was associated with occupational lead exposure. Clin Toxicol (Phila) 51 (4), 225–229. [DOI] [PubMed] [Google Scholar]
- Li Y, Xie C, Murphy SK, Skaar D, Nye M, Vidal AC, Cecil KM, Dietrich KN, Puga A, Jirtle RL, et al. , 2016. Lead exposure during early human development and DNA methylation of imprinted gene regulatory elements in adulthood. Environ Health Perspect 124 (5), 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lidsky TI, Schneider JS, 2003. Lead neurotoxicity in children: Basic mechanisms and clinical correlates. Brain 126 (Pt 1), 5–19. [DOI] [PubMed] [Google Scholar]
- Mielke HW, Reagan PL, 1998. Soil is an important pathway of human lead exposure. Environ Health Perspect 106 (Suppl 1), 217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monge CC, Arregui A, Leon-Velarde F, 1992. Pathophysiology and epidemiology of chronic mountain sickness. Int J Sports Med 13 (Suppl 1), S79–S81. [DOI] [PubMed] [Google Scholar]
- Moore LG, 2001. Human genetic adaptation to high altitude. High Alt Med Biol 2 (2), 257–279. [DOI] [PubMed] [Google Scholar]
- Morris TJ, Butcher LM, Feber A, Teschendorff AE, Chakravarthy AR, Wojdacz TK, Beck S, 2014. Champ: 450k chip analysis methylation pipeline. Bioinformatics 30 (3), 428–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman HL, Gunnoe C, Leviton A, Reed R, Peresie H, Maher C, Barrett P, 1979. Deficits in psychologic and classroom performance of children with elevated dentine lead levels. N Engl J Med 300 (13), 689–695. [DOI] [PubMed] [Google Scholar]
- Nishimura D, 2001. Biocarta. Biotech Software & Internet. Report 2 (3), 117–120. [Google Scholar]
- Papanikolaou NC, Hatzidaki EG, Belivanis S, Tzanakakis GN, Tsatsakis AM, 2005. Lead toxicity update. A brief review. Med Sci Monit 11(10):RA329–336. [PubMed] [Google Scholar]
- Parsons PJ, Slavin W, 1993. A rapid zeeman graphite furnace atomic absorption spectrometric method for the determination of lead in blood. Spectrochimica Acta Part B: Atomic Spectroscopy 48 (6), 925–939. [Google Scholar]
- Pedersen BS, Schwartz DA, Yang IV, Kechris KJ, 2012. Comb-p: Software for combining, analyzing, grouping and correcting spatially correlated p-values. Bioinformatics 28 (22), 2986–2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi D, Gardner J, Chambers D, Hosking B, Peters J, Muscat G, Abbott C, Koopman P, 2000. Mutations in sox18 underlie cardiovascular and hair follicle defects in ragged mice. Nat Genet 24 (4), 434–437. [DOI] [PubMed] [Google Scholar]
- Pidsley R, Zotenko E, Peters TJ, Lawrence MG, Risbridger GP, Molloy P, Van Djik S, Muhlhausler B, Stirzaker C, Clark SJ, 2016. Critical evaluation of the illumina methylationepic beadchip microarray for whole-genome DNA methylation profiling. Genome Biol 17 (1), 208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogribny IP, Tryndyak VP, Woods CG, Witt SE, Rusyn I, 2007. Epigenetic effects of the continuous exposure to peroxisome proliferator wy-14,643 in mouse liver are dependent upon peroxisome proliferator activated receptor alpha. Mutat Res 625 (1–2), 62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu CS, Ma ZW, Yang J, Liu Y, Bi J, Huang L, 2012. Human exposure pathways of heavy metals in a lead-zinc mining area, jiangsu province, china. PLoS One 7 (11), e46793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team, 2020. R: A language and environment for statistical computing R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- Ricq EL, Hooker JM, Haggarty SJ, 2016. Toward development of epigenetic drugs for central nervous system disorders: Modulating neuroplasticity via h3k4 methylation. Psychiatry Clin Neurosci 70 (12), 536–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK, 2015. Limma powers differential expression analyses for rna-sequencing and microarray studies. Nucleic acids research 43 (7), e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rygiel CA, Dolinoy DC, Perng W, Jones TR, Solano M, Hu H, Tellez-Rojo MM, Peterson KE, Goodrich JM. 2020. Trimester-specific associations of prenatal lead exposure with infant cord blood DNA methylation at birth. Epigenet Insights 13: 2516865720938669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders T, Liu Y, Buchner V, Tchounwou PB, 2009. Neurotoxic effects and biomarkers of lead exposure: A review. Rev Environ Health 24 (1), 15–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JS, Anderson DW, Talsania K, Mettil W, Vadigepalli R, 2012. Effects of developmental lead exposure on the hippocampal transcriptome: Influences of sex, developmental period, and lead exposure level. Toxicological sciences : an official journal of the Society of Toxicology 129 (1), 108–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider JS, Kidd SK, Anderson DW, 2013. Influence of developmental lead exposure on expression of DNA methyltransferases and methyl cytosine-binding proteins in hippocampus. Toxicol Lett 217 (1), 75–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen A, Heredia N, Senut MC, Land S, Hollocher K, Lu X, Dereski MO, Ruden DM, 2015. Multigenerational epigenetic inheritance in humans: DNA methylation changes associated with maternal exposure to lead can be transmitted to the grandchildren. Sci Rep 5, 14466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senut MC, Sen A, Cingolani P, Shaik A, Land SJ, Ruden DM, 2014. Lead exposure disrupts global DNA methylation in human embryonic stem cells and alters their neuronal differentiation. Toxicological sciences : an official journal of the Society of Toxicology 139 (1), 142–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart WF, Schwartz BS, Davatzikos C, Shen D, Liu D, Wu X, Todd AC, Shi W, Bassett S, Youssem D, 2006. Past adult lead exposure is linked to neurodegeneration measured by brain mri. Neurology 66 (10), 1476–1484. [DOI] [PubMed] [Google Scholar]
- Storz JF, Moriyama H, 2008. Mechanisms of hemoglobin adaptation to high altitude hypoxia. High Alt Med Biol 9 (2), 148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suderman M, Staley JR, French R, Arathimos R, Simpkin A, Tilling K, 2018. Dmrff: Identifying differentially methylated regions efficiently with power and control. BioRxiv 508556.
- Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S, 2013. A beta-mixture quantile normalization method for correcting probe design bias in illumina infinium 450 k DNA methylation data. Bioinformatics 29 (2), 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Gayther SA, Apostolidou S, Jones A, Lechner M, Beck S, Jacobs IJ, et al. , 2009. An epigenetic signature in peripheral blood predicts active ovarian cancer. PLoS One 4 (12), e8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, von Schirnding YE, Prapamontol T, 2000. Environmental lead exposure: A public health problem of global dimensions. Bull World Health Organ 78 (9), 1068–1077. [PMC free article] [PubMed] [Google Scholar]
- UniProt C, 2019. Uniprot: A worldwide hub of protein knowledge. Nucleic acids research 47 (D1), D506–D515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valinluck V, Tsai HH, Rogstad DK, Burdzy A, Bird A, Sowers LC, 2004. Oxidative damage to methyl-cpg sequences inhibits the binding of the methyl-cpg binding domain (mbd) of methyl-cpg binding protein 2 (mecp2). Nucleic acids research 32 (14), 4100–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valko M, Morris H, Cronin MT, 2005. Metals, toxicity and oxidative stress. Curr Med Chem 12 (10), 1161–1208. [DOI] [PubMed] [Google Scholar]
- van Geen A, Bravo C, Gil V, Sherpa S, Jack D, 2012. Lead exposure from soil in peruvian mining towns: A national assessment supported by two contrasting examples. Bull World Health Organ 90 (12), 878–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal AC, Semenova V, Darrah T, Vengosh A, Huang Z, King K, Nye MD, Fry R, Skaar D, Maguire R, et al. , 2015. Maternal cadmium, iron and zinc levels, DNA methylation and birth weight. BMC Pharmacol Toxicol 16, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. 2011. Brief guide to analytical methods for measuring lead in blood
- Wickham H 2009. Ggplot2: Elegant graphics for data analysis. Use R.1–212
- Wixon J, Kell D, 2000. The kyoto encyclopedia of genes and genomes–kegg. Yeast 17 (1), 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright RO, Schwartz J, Wright RJ, Bollati V, Tarantini L, Park SK, Hu H, Sparrow D, Vokonas P, Baccarelli A, 2010. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect 118 (6), 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang FF, Morabia A, Carroll J, Gonzalez K, Fulda K, Kaur M, Vishwanatha JK, Santella RM, Cardarelli R, 2011. Dietary patterns are associated with levels of global genomic DNA methylation in a cancer-free population. J Nutr 141 (6), 1165–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


