Abstract
Allosteric regulation of key metabolic enzymes is a fascinating field to study the structure-function relationship of induced conformational changes of proteins. In this review we compare the principles of allosteric transitions of the complex classical model aspartate transcarbamoylase (ATCase) from Escherichia coli, consisting of 12 polypeptides, and the less complicated chorismate mutase derived from baker's yeast, which functions as a homodimer. Chorismate mutase presumably represents the minimal oligomerization state of a cooperative enzyme which still can be either activated or inhibited by different heterotropic effectors. Detailed knowledge of the number of possible quaternary states and a description of molecular triggers for conformational changes of model enzymes such as ATCase and chorismate mutase shed more and more light on allostery as an important regulatory mechanism of any living cell. The comparison of wild-type and engineered mutant enzymes reveals that current textbook models for regulation do not cover the entire picture needed to describe the function of these enzymes in detail.
In 1965, Monod, Wyman, and Changeux summarized the properties of two dozen allosteric enzyme systems, resulting in their “plausible model on the nature of allosteric transition” (MWC) (70). Since then, the description of a plethora of allosteric enzymes and systems has led to the concept that allostery is a common theme in regulating the activity of various proteins (for a review, see reference 73). Direct control of protein function via allosteric regulation is usually achieved through conformational changes of a given protein structure induced by effectors. In contrast to intrasteric regulation (45), effectors bind to regulatory sites distinct from the active site (Greek, allos = other, stereos = rigid, solid, or space). One term tightly linked to allostery is “cooperativity.” This describes the interaction of binding processes of ligands to proteins with multiple binding sites (75). Ligand binding plots of positively cooperative systems generally display sigmoidicity, resulting in an S-shaped curve of fractional saturation or rate against concentration. Allosteric behavior itself was often observed for regulatory or control enzymes of metabolic pathways and forms the basis for feedback inhibition and activation. The so-called homotropic effects originate from identical (e.g., substrate) molecules which bind to an allosteric protein and influence each other's affinity. When different ligands are involved (e.g., effector molecules and substrate molecules), the interactions are called heterotropic (70). For both effects, cooperativity and allostery, positive as well as negative effects can be observed, resulting in an increase or decrease, respectively, of affinity and activity.
In the established model of global allosteric transition, binding of an effector induces a concerted shift in the equilibrium between two quaternary conformations of the oligomeric protein. The activated conformation, termed the R (relaxed) state, is assumed to have higher catalytic activity than the T (tense) state (69). This model was later challenged by the sequential model established by Koshland, Némethy, and Filmer (KNF) (46), finally leading to the general model by Eigen (20), which combines the MWC and KNF extremes. In most allosteric proteins, homotropic effects seem to be best accounted for by the concerted model while heterotropic effects are better described by the sequential model (98).
The exact mechanisms by which allosteric control of protein function can be achieved are extremely varied. Among the multitude of allosteric proteins, a few prototypes have been established in basic research (for a review, see reference 73). The most prominent example is hemoglobin, with which the initial attempts to explain the mechanisms of cooperativity have been carried out. Hemoglobins are generally composed of two pairs of polypeptide chains arranged in a symmetrical tetrahedral manner. While oxyhemoglobin displays a high affinity for oxygen, desoxyhemoglobin has a low affinity for the molecule. On oxygen binding, changes in quaternary and tertiary structure account for the shift in the allosteric T-R equilibrium. As a result, oxygen acts as homotropic ligand on hemoglobin. A variety of heterotropic ligands lowering the oxygen affinity have been described, with protons and 2,3-diphosphoglycerate being the most important. An interesting feature is displayed by lamprey hemoglobin, in which cooperativity is mediated by the reversible dissociation of dimers or tetramers into monomers with high oxygen affinity (17).
The majority of allosteric proteins are presumably metabolic enzymes which act as control devices for flux alterations in metabolic pathways. Enzymes are regulated predominantly by heterotropic effector molecules modulating the catalytic turnover rates in a positive and/or negative fashion. Positive effectors often abolish cooperativity, resulting in Michaelis-Menten-like kinetics in substrate saturation assays, whereas negatively acting ligands decrease catalytic efficiency either by decreasing the substrate affinity (K systems) or by altering the intrinsic kcat values (V systems) (86). Prominent examples of allosteric enzymes in metabolic pathways are glycogen phosphorylase (41), phosphofructokinase (9, 80), glutamine synthetase (88), and aspartate transcarbamoylase (ATCase) (103). In particular, ATCase, which catalyzes the first step of pyrimidine biosynthesis, has been established as a prototype for allostery (43, 62, 67, 79). For this allosteric enzyme paradigm, the homotropic and heterotropic effects of its ligands as well as cooperativity have been investigated in great detail. The models of allosteric behavior developed from experimental data exceed previous theories and can be very helpful for the more accurate description of the characteristics of other allosteric proteins.
In recent years, the chorismate mutase (CM) of the baker's yeast Saccharomyces cerevisiae (ScCM) has become a suitable and well characterized model for allosteric regulation of enzyme activity. CM is necessary for the biosynthesis of tyrosine and phenylalanine and catalyzes one reaction at the first branch point of aromatic amino acid biosynthesis. Structural analyses combined with classic kinetic studies performed on this enzyme, as well as molecular modeling studies, have led to detailed insights into the catalytic mechanism of this enzyme. Furthermore, the allosteric response to effector binding was intensively studied. The monofunctional, dimeric yeast enzyme is strictly regulated in its activity by allosteric effectors. The substrate chorismate serves as homotropic effector, as indicated by the sigmoid curvature of substrate saturation kinetics, whereas tyrosine and tryptophan act as negative and positive heterotropic ligands, respectively (47).
The purpose of this review is to sum up major investigations into ScCM made in the last decade. The main focus is the comparison of the established ATCase model system with the knowledge which has been accumulated during recent years on the catalytic and regulatory features of ScCM. Despite its small size, ScCM exhibits multisubunit allostery and cooperativity and has many similarities to the ATCase system. Thus, ScCM is well suited to be a model system to improve our understanding of the allostery of small enzymes consisting of only two subunits as presumably the minimal structure which is required for this kind of regulation.
ATCASE ACTIVITIES
ATCase (carbamoylphosphate: l-aspartate carbamoyltransferase, EC 2.1.3.2) catalyzes the carbamoylation of the amino group of aspartate by carbamoylphosphate, leading to phosphate and N-carbamoyl-l-aspartate (14). ATCase is the first enzyme unique to pyrimidine biosynthesis and a key enzyme for regulating purine, pyrimidine, and arginine biosynthesis in Escherichia coli. The enzymes from enterobacteria are dodecameric holoenzymes composed of two different polypeptides which are inhibited by CTP and UTP and activated by ATP. The same architecture was found for other bacterial ATCases, like that from Methanococcus janaschii, although the M. janaschii enzyme exhibited few regulatory properties (32). Some bacterial and eukaryotic ATCases are part of a multifunctional enzyme containing carbamoylphosphate synthetase and/or dihydroorotase activity, among them the enzyme of S. cerevisiae, which is inhibited by UTP (87). However, plant ATCases seem to be simple homotrimers which can be regulated by UMP (109).
For the two-substrate reaction, carbamoylphosphate binds before aspartate and subsequently induces a conformational change in the enzyme, resulting in a higher affinity for aspartate (31, 37). On aspartate binding, a larger conformational change is exerted on the active site and the whole enzyme, leading to a T-R transition. Accordingly, phosphate dissociates from the active site after carbamoylaspartate (38). ATCase exhibits positive cooperativity for aspartate (8, 27). The apparent cooperativity for carbamoylphosphate reflects only cooperativity for aspartate (23). During catalysis, the amino group of aspartate is involved in a nucleophilic attack on the carbonyl carbon of carbamoylphosphate to form a tetrahedral intermediate. The transition state is processed to the products by transfer of a proton from the amino group of aspartate to the closest oxygen of the leaving phosphate group derived from carbamoylphosphate (28).
STRUCTURE OF E. COLI ATCASE
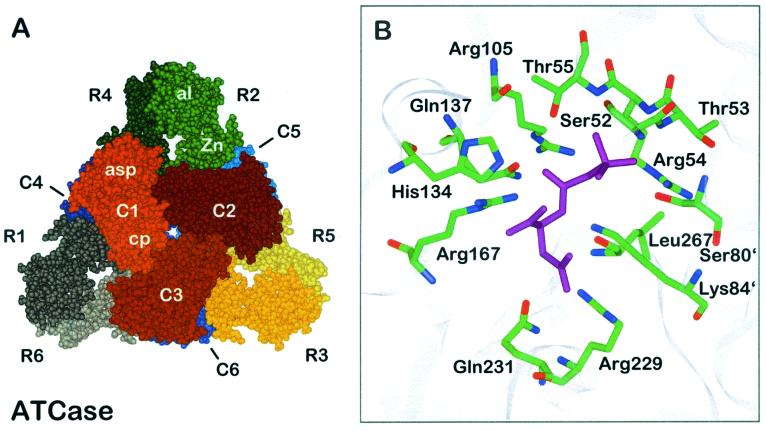
The ATCase holoenzyme, composed of 12 polypeptide chains of two types (Fig. 1A) (1, 107), has a molecular weight of 310,000. Six larger chains (33,000 each, encoded by pyrB) are the catalytic (C) chains, which are insensitive to the allosteric effectors, while the smaller regulatory (R) chains (17,000 each, encoded by pyrl) are devoid of catalytic activity but bind the effectors ATP, CTP, and/or UTP. The catalytic chains are packed in two catalytic trimers, one subunit containing chains C1, C2, and C3 and the other containing chains C4, C5, and C6. Chain C4 is located below C1, while C5 and C6 are located below C2 and C3, respectively. Each catalytic subunit has a threefold axis. The regulatory chains are organized in dimers which bridge the two catalytic trimers noncovalently. Each polypeptide chain folds into two domains. The N-terminal and C-terminal domains of the C chains are termed the carbamoylphosphate (or polar) domain and the aspartate (or equatorial) domain, respectively, according to the substrates bound to them. Each R chain harbors the allosteric domain, including the allosteric site in the N terminus. A C-terminal zinc domain contains a Zn(II) ion. The metal is coordinated by four sulfhydryl groups and mediates R-C interactions. Thus, on treatment with heat or mercurials, the holoenzyme dissociates into the catalytic and regulatory subunits. The active sites are composed of residues from adjacent C chains within a trimer: from both the aspartate and carbamoylphosphate (cp) domains of one chain and the cp domain of the adjacent chain. The allosteric sites are located at the distal ends of the R chains, 60 Å away from the nearest active site, and bind each effector. Assembly into the holoenzyme yields extensive interfaces between C chains within a catalytic trimer (for example, C1-C2) and in opposed trimers (C1-C4), between R chains within a regulatory dimer (R1-R6), and between C and R chains (C1-R1 and C1-R4). The C1-C4 and symmetry-related interfaces are present in the T state but not in the R state (Fig. 1A).
FIG. 1.
Quaternary structure of E. coli ATCase. (A) Holoenzyme viewed along the threefold axis. Catalytic chains are numbered C1 to C6, and regulatory chains are numbered R1 to R6. The different catalytic and regulatory subunits are indicated by different colors. The aspartate domain of the catalytic chain is designated asp, and the carbamoylphosphate domain is designated cp. The domains of the regulatory chain are named Zn for zinc domain and al for allosteric domain. (B) Binding mode of the bisubstrate analogue PALA (purple) to the active site of ATCase. Side chains are shown as sticks with atoms labeled by color (green, carbon; blue, nitrogen; red, oxygen). Apostrophes after residue numbers indicate the position of the residue in an adjacent polypeptide chain. The figures are based on data for the CTP-liganded structure and the bisubstrate analogue PALA-liganded structure, respectively (35, 51).
CATALYTIC CENTER OF E. COLI ATCASE
Insight into the mode of binding the substrates to the catalytic center of E. coli ATCase required analysis of the binding of a bisubstrate analogue, N-(phosphonoacetyl)-l-aspartate (PALA). In addition, the binding of carbamoylphosphate and succinate was studied; the study resulted in computer models which were verified by amino acid substitutions achieved by site-directed mutagenesis of corresponding codons in the open reading frames. Several residues have been identified as crucial for catalysis: Ser52, Thr53, Arg54, Thr55, Arg105, His134, Gln137, Arg167, Arg229, Glu231, and Ser80 and Lys84 from an adjacent catalytic chain (66) (Fig. 1B). Thus, the active site is a highly positively charged pocket. The most critical side chain originates from Arg54 (89). It interacts with a terminal oxygen and the anhydride oxygen of carbamoylphosphate and thereby stabilizes the negative charge of the leaving phosphate group. Arg105, His134, and Thr55 help to increase the electrophilicity of the carbonyl carbon by interacting with the carbonyl oxygen (40). Rate enhancement is achieved by orientation and stabilization of substrates, intermediates, and products rather than by involvement of residues in the catalytic mechanism. Instead of Lys84 acting as a base which captures the proton from the amino group of aspartate, the recent model suggests that the fully ionized phosphate group is capable of accepting a proton during catalysis (28, 40).
ALLOSTERIC SITE OF E. COLI ATCASE
The allosteric site in the allosteric domain of the R chains of the E. coli ATCase complex binds ATP, CTP, and/or UTP (106). There is one site with high affinity for ATP and CTP and one with 10- to 20-fold-lower affinity for these nucleotides in each regulatory dimer (18, 62). ATP binds predominantly to the high-affinity sites and subsequently activates the enzyme. UTP and CTP binding leads to inhibition of activity. UTP can bind to the allosteric site, but inhibition of ATCase by UTP is possible only in combination with CTP. With CTP present, UTP binding is enhanced and preferentially directed to the low-affinity sites. Conversely, UTP binding leads to enhanced affinity for CTP at the high-affinity sites and inhibits enzyme activity by up to 95% while CTP binding alone inhibits activity to 50 to 70% (18, 105, 116). ATP and CTP bind in anticonformation with negative cooperativity with respect to themselves (91). ATP strongly reduces cooperativity of substrate binding, while CTP enhances it (91). The purine and pyrimidine rings, as well as the ribose rings, bind at similar locations. However, when CTP is bound, the base and triphosphate moieties are closer together than when ATP is bound (94). In addition, the ribose moiety of ATP protrudes deeper into the binding sites than does that of CTP (94). The triphosphate is necessary for high affinity and full nucleotide effects (101). Groups interacting with CTP are Val91r, Lys94r, Arg96r, Asp19r, His20r, Val9r, Lys56r, Lys60r, Val17r, Ala11r, Ile12r, and Tyr89r (r refers to the residues which are part of a regulatory chain) (1, 91). UTP differs from CTP in the carbonyl group at position 4 and the protonation of the nitrogen at position 3 of the pyrimidine ring. Discrimination between these two nucleotides seems to be based on the subtle differences in the interaction of the amino group and the nitrogen at position 3 with Ile12r (18). ATP, on the other hand, is hydrogen bonded to side-chain or main-chain atoms of residues Asn84r, Val91r, Lys94r, Leu58r, Asp19r, Val9r, Lys60r, Glu10r, Ala11r, Ile12r, and Tyr84r (91). ATP induces an expansion of the site with the R1 and R6 allosteric domains pushed apart. This induces an overall increase of the allosteric domains. CTP, however, decreases the size of the allosteric site, with the result that the L50s loop moves closer to the nucleotide (94) (the letter s indicates the plural according to the term “the fifties loop” that comprises residues around position 50). Both cavities are larger than the binding site in the unliganded enzyme (91).
CONFORMATIONS OF E. COLI ATCASE
According to the MWC model, the ATCase has (at least) two conformational states: a low-activity T state with low affinity for the substrates, and a high-activity, high-affinity R state. The two states are in an equilibrium which is shifted to the side of the T state with a value of about 250 for the allosteric equilibrium constant (L) (21). The substrates, as well as the bisubstrate analogue PALA, produce a significant change in the tertiary and quaternary structure of both the catalytic and regulatory chains. Some authors conclude that the PALA-bound structure does not represent the R state since the isolated C-trimer structure and the catalytic trimer of B. subtilis ATCase resemble the T state more closely than they resemble the R state (7, 22, 95). However, the PALA-bound structure allows the identification of the active site, a description of the more active form and a model for homotropic transition of ATCase (51), and strong similarities were found to R-like structures with single substrate or product analogues like phosphate and citrate (62). PALA binding promotes a closure of the hinge between the C chain domains by 8°, while the gap between the allosteric and Zn domains expands. Domain closure in the C chain is required for cooperativity and fully creates the aspartate binding site. Through these changes, interchain contacts of side chains of the L80s loop and L240s loop and active-site residues become reoriented (40). On the quaternary conformational level, the holoenzyme undergoes a screw motion with a shift of 11 Å along and a rotation of 7° about the threefold axis and a 15° rotation of the regulatory chains about the three twofold axes. While the affinity of the R state for substrates is higher than that of the T state, there are only slight differences for the affinity to the allosteric ligands. Thus, ATP or CTP binding causes only minor changes in the quaternary structure (79). The structure with CTP bound is termed the T state. This conformation was also found for the unliganded enzyme or when ATP is bound (93). On ATP or UTP binding, only small changes in enzyme structure are observed. ATP causes an elongation (94) along the threefold axis of the T form by only 0.4 Å and so does not promote a T-R transition by itself (102). Whereas ATP has nearly no effect on the distance between the C trimers in the R state, CTP decreases it by 0.5 Å toward the T state (94). Accordingly, CTP has no effect on C trimer separation in the T state (94).
In summary, the E. coli ATCase complex represents a highly sophisticated interplay of numerous polypeptides. Several effector molecules predominantly act on more than one polypeptide chain, resulting in different effects on the complicated overall enzyme. In contrast, the regulation of yeast CM is based only on the interplay of two identical polypeptides. This allows a detailed study of very subtle effects even within a single polypeptide chain.
CM ACTIVITIES
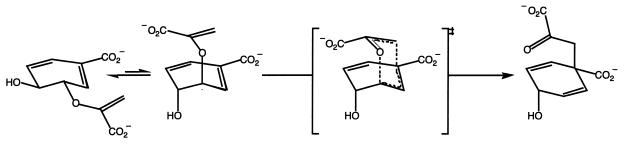
CM activities (chorismate pyruvate mutase, EC 5.4.99.5) catalyze the intramolecular rearrangement of (−)-chorismic acid to prephenic acid (Fig. 2) (2). This Claisen rearrangement is a key step in the biosynthetic pathway of archaea, bacteria, fungi, and plants and results in the aromatic amino acids l-phenylalanine or l-tyrosine. Additionally, it represents a rare example of a pericyclic reaction in primary metabolism (104). Prephenate itself is transformed either into phenylpyruvate, the precursor of phenylalanine, or into 4-hydroxyphenylpyruvate, the last intermediate in tyrosine biosynthesis. A third, alternative route is utilized most commonly in plants, where prephenate is converted to arogenate before tyrosine and phenylalanine are formed.
FIG. 2.
Claisen rearrangement of chorismic acid resulting in prephenic acid. The two conformers of chorismate, as well as the proposed transition state finally leading to prephenate, are shown.
In comparison to the uncatalyzed, thermal [3, 3] sigmatropic rearrangement, CMs can enhance the conversion of chorismate to prephenate by a factor of up to 106. A variety of CM enzymes have been described and characterized during the past three decades, and catalytic antibodies (“abzymes”) that accelerate the chorismate-to-prephenate rearrangement have also been generated (33, 39). Prokaryotic CM activities can be part of a bifunctional enzyme in which the CM domain is fused to a prephenate dehydratase (P-protein), a prephenate dehydrogenase (T-protein), or a 3-deoxy-d-arabinoheptulosonate-7-phosphate synthase moiety (76) (the letters P and T indicate the biosynthetic pathway that is initiated by the CM catalytic activity to yield phenylalanine and tyrosine, respectively). In contrast, all eukaryotic CMs characterized to date, as well as the CM from the archaeon M. jannaschii (65), are described as being monofunctional.
In most organisms analyzed, CM activities are strictly regulated. Whereas both enzyme activities of bifunctional T-proteins are inhibited by tyrosine, phenylalanine inhibits the two activities of P-proteins. In gram-negative bacteria, including the cyanobacteria, as well as in gram-positive Bacillus subtilis and Streptomyces aureofaciens, monofunctional CMs were found that lack regulatory properties. Eukaryotic CM enzymes are generally monofunctional and subject to allosteric inhibition and activation. Tyrosine and phenylalanine are negative effectors, whereas tryptophan serves as a positive regulator of enzyme activity. In plants, different isoenzymes are often present which differ in their regulatory behaviour. Furthermore, some of them are regulated in their activities not only by end products of aromatic amino acid biosynthesis but also by secondary metabolites; for example, the CM isoenzymes of alfalfa can be inhibited by coumarate, caffeate, or ferulate and activated by 3,4-dimethoxycinnamate (76).
In addition to this enzymatic regulation, the amount of enzymes at metabolic branch points is important for distribution of intermediates. For a balanced biosynthesis of the amino acids in yeast, a sophisticated, strictly regulated network composed of allosteric enzymes and “the general control of amino acid biosynthesis” has evolved (49). While anthranilate synthase, the competing enzyme complex at the branch point of aromatic amino acid biosynthesis, is feedback inhibited by tryptophan, the expression of the encoding genes is induced by a transcriptional activator under amino acid starvation. However, the total amount of CM is not regulated by the general control, because its activity is modulated more strongly by two different allosteric effectors.
STRUCTURES OF CM ENZYMES
The crystal structures of three natural CM enzymes have been determined so far. Based on these structural insights and on primary sequence information about the encoding genes cloned to date, it has become evident that two different structural folds have evolved to contrive the enzymatic isomerization of chorismate to prephenate.
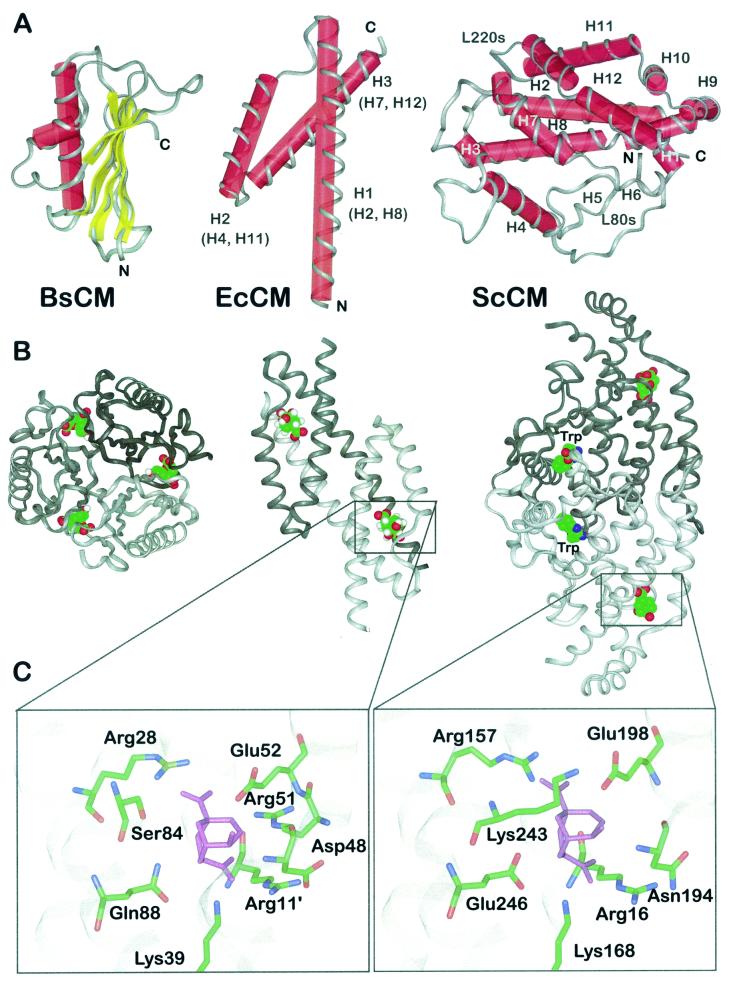
One structural class, AroH, is represented by the monofunctional, homotrimeric enzyme of Bacillus subtilis. The X-ray structure of this enzyme was determined at 1.9-Å resolution (Fig. 3A and B) (12) and more recently at 1.3-Å resolution (55). The aroH gene product is a nonallosteric CM of 127 amino acids per monomer. Each monomer consists of a five-stranded mixed β-sheet packed against an 18-residue α-helix and a two-turn 310 helix. The interfaces between adjacent subunits form three equivalent clefts that are open and accessible to solvent. These clefts harbor the active sites.
FIG. 3.
Structural prototypes of CM enzymes and binding mode of a stable transition state analogue. (A) Schematic presentations of the structural folds displayed by CMs from B. subtillis (BsCM) (left), E. coli (EcCM) (middle), and ScCM (right). The helix numbers in parentheses indicate the corresponding helices in the yeast enzyme. The polypeptide backbone is displayed in ribbon style, and secondary elements are labeled with red cylinders (α-helices) and yellow bars (β-sheets). N and C termini are indicated, as are structural elements of ScCM (see the text for details). (B) Oligomeric structure of B. subtilis CM (left), E. coli CM (middle), and ScCM (right) in complex with a stable transition state analogue. Monomeric subunits are indicated by different shades of grey. For ScCM, the binding position of the positive effector tryptophan is also shown. (C) Section views of the catalytic sites of E. coli CM (left) and ScCM (right) with the transition state analogue (purple) bound. Side chains are shown as sticks with atoms labeled by colour (green, carbon; blue, nitrogen; red, oxygen). Apostrophes indicate the position of the residue in an adjacent polypeptide chain.
Sequences of all CM domains from bifunctional enzymes characterized to date, as well as most prokaryotic and eukaryotic monofunctional CMs, are consistent with the AroQ class of CM enzymes. These enzymes are, in contrast to the three-dimensional pseudo-α/β-barrel structure established by the AroH class, all-helical polypeptides and show similarity in sequence to the monofunctional Erwinia herbicola CM encoded by the aroQ gene (112). In contrast to the situation in prokaryotes, primary sequences of eukaryotic CM proteins are rare. Only a few encoding sequences have been determined so far, like the genes from the yeasts S. cerevisiae, Schizosaccharomyces pombe, and Hansenula polymorpha, from the filamentous fungus Aspergillus nidulans, and those coding for three isoenzymes in Arabidopsis thaliana (19, 48, 50, 68, 82; GenBank accession no. Z98529). On the basis of the solved structure of the ScCM and of conserved primary structures among cloned eukaryotic CM-encoding genes, these CM enzymes are included in the AroQ class. They constitute the separate subclass of AroQ, enzymes (formerly AroR) due to their additional regulatory domains (65).
The structural prototype of the AroQ class is the CM domain of the bifunctional, homodimeric Escherichia coli CM-prephenate dehydratase, the so-called P-protein which is inhibitable by phenylalanine. The N-terminal 109 residues of this P-protein constitute a functional CM, and its X-ray structure was solved at 2.2-Å resolution (Fig. 3A and B) (57). In the monomer, the polypeptide chain resembles the numeral 4 by its unusual fold of three α-helices, two longer (H1 and H3) and one short (H2), connected by two loops. Two equivalent active sites with contributions from each monomer are present in the quaternary structure of this engineered CM from E. coli.
The only solved crystal structure of a eukaryotic CM enzyme, the 256-amino-acid ARO7 gene product of the baker's yeast S. cerevisiae, also is an all-helical polypeptide (Fig. 3A and B). X-ray data have been determined for three conformational structures of this enzyme resembling different allosteric states. The conformation of the wild-type (wt) enzyme with tyrosine bound to the allosteric site was determined at 2.8-Å resolution, and this structure yielded detailed insights into the global structure of the T state (96). In contrast, a Thr226Ile mutant enzyme is locked in the R state and its structure was determined at 2.2-Å resolution with tryptophan at the effector binding site (114). The enzyme in complex with the stable transition state analogue displays a super R state and identifies the probable binding mode of the transition state (97).
The basic topology of one monomeric subunit is that of a Greek key motif forming a four-helix bundle with essentially no β-strand elements. The 12 helices of the polypeptide chain are arranged in a twisted two-layer structure with a packing angle between the helical axes from each layer of about 60°. The dimer has the shape of a bipyramid, with four helices (H2, H4, H8, and H11) forming the hydrophobic interface between the protomers. The active site is part of the four-helix bundle set up by helices H2, H8, H11, and H12 separately in each monomer. The binding site for both heterotropic effectors is a cleft in the dimer interface between the subunits. This regulatory site is formed by two helices (H4 and H5) of one monomer and the L80s loop and helix H8 of the other. The latter is the longest helix in the molecule; it consists of 32 residues and spans the overall structure from the regulatory site to the catalytic domain.
The fact that the three-dimensional structures of the E. coli CM domain and its eukaryotic counterpart are both characteristic of AroQ class enzymes and resemble similar folds has led to the speculation that the yeast CM fold might have evolved from an ancestral protein similar to the bacterial CM by a gene duplication event followed by dimerization (57, 97, 113). In fact, the E. coli CM dimer can be superimposed onto a monomer of yeast CM. The topology of a four-helix bundle forming the active site is conserved in the two enzymes, and also the binding mode of the endo-oxabicyclic inhibitor is similar. Helices H2, H4, H7, H8, H11, and H12 of the yeast enzyme correspond to H1, H2, H3, H1′, H2′, and H3′, respectively, in E. coli CM. Modelling two E. coli CM dimers onto the S. cerevisiae dimer has led to further insights: two bacterial CM monomers superimpose well on the catalytic domains of the yeast CM, whereas the other monomers and the other halves of the yeast monomers are more diverse due to the evolution of regulatory domains in this region of the molecules (97).
CATALYTIC CENTER OF SCCM
The chorismate-to-prephenate rearrangement is a unimolecular one-substrate, one-product reaction. Generally, this Claisen rearrangement is thought to proceed in a nearly concerted but not necessarily synchronous way (64). A variety of interdisciplinary studies have provided detailed insight into the catalytic mechanism needed to achieve the >106-fold rate enhancement shown by CM, compared to the rate of the uncatalyzed reaction (for a review, see reference 26).
In solution, 10 to 20% of the substrate occupies the less stable pseudodiaxial conformation of the enolpyruvate side chain. Binding of this energetically less favored conformer is proposed to be the first essential step in catalytic turnover. Subsequently, two alternative mechanistic pathways occur: concerted but perhaps asynchronous bond cleavage and formation, as in the uncatalyzed reaction, or catalysis via an intermediate after attack of an active-site nucleophile at C-5.
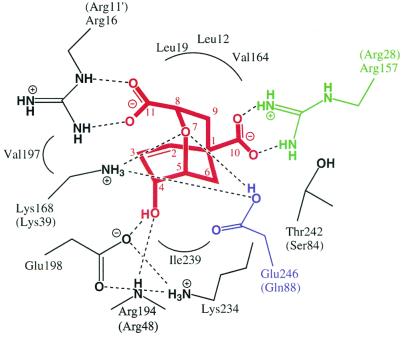
Further insight into the catalytic mechanism has been obtained by analysis of the structural and computational data for CMs in complex with an endo-oxabicyclic inhibitor resembling a stable transition-state analogue (60). Binding of this so-called Bartlett's inhibitor (5) to the yeast active-site cavity is achieved by a series of electrostatic interactions and hydrogen bonding (97) (Fig. 4). Interestingly, the active-site structures are nearly identical on inhibitor binding, irrespective of the different effectors, either tyrosine or tryptophan, bound to the allosteric site. Therefore, this structural state was referred to as the super R state. Whether chorismic acid alone is able to promote the transition to the super R state remains to be shown. Two guanidinium groups of arginine residues (Arg16 and Arg157) bind the carboxylate groups of the inhibitor via salt bridges, and its hydroxyl group is complexed by the carboxyl side chain of Glu198 and the backbone NH group of Arg194. Arg157 is of especial importance for binding, because it is the molecular switch for allosteric transition to the T state. This residue is not in an appropriate position for interaction with the substrate in the T state but only in the R or super R state. Additionally, hydrophobic interactions contribute to inhibitor binding. The most interesting contacts focus on the inhibitor's ether oxygen O-7. In the crystal structures, the two side-chain groups of Lys168 and protonated Glu246 are within hydrogen-bonding distance of this atom.
FIG. 4.
Binding mode of the endo-oxabicyclic inhibitor to the active site of ScCM. The stable transition analogue is highlighted in red, and residues Arg157 and Glu246 are shown in green and blue, respectively. Hydrogen bond interactions are indicated by dotted lines. Corresponding residues of E. coli CM are indicated in parentheses. Apostrophes indicate the position of the residue in an adjacent polypeptide chain.
Despite the low sequence similarities in primary structures, both the yeast CM and E. coli P-protein CM active site cavities display significant similarities on binding the stable transition-state analogue, as deduced from the X-ray crystal structures (Fig. 3C). However, a particular difference between the organisms is also reflected in their CMs. The activity of the yeast enzyme is adapted to acidic pH in accordance with the ability of the fungus to grow at relatively low pH. In contrast, the bacterial enzyme, which is active in a broader pH range reflects the ability of E. coli to live under more alkaline conditions as well. Thus, the binding modes for the endo-oxabicyclic structure of the enzymes are very similar, with one significant exception. Whereas in the bacterial structure a glutamine residue (Gln88) is hydrogen bonded to the ether oxygen O-7, the active-site residue Glu246 is displayed at the corresponding position in the yeast enzyme (Fig. 4). Molecular modelling studies imply that this key residue is protonated well above neutral pH with an effective pKa of 8.1 (97). Whereas for the wt CM a bell-shaped profile was determined with an optimal acidic pH, in a Glu246Gln mutant enzyme the catalytic activity is detectable over a broad pH range without a particular optimum (85). In conclusion, this active-site mutant mimics the situation as it is found for the bacterial CM, where catalytic turnover rates are similar at both acidic and neutral pH. Consistent with this observation is the fact that mutation of the Gln88 codon to a glutamate codon in the E. coli gene leads to strong pH dependency of the resulting CM activity, with an optimal pH at acidic conditions (25). Nevertheless, for both enzymes similar effects contribute to rate acceleration: conversion of the less stable pseudodiaxial conformer, specific electrostatic stabilization of the ether oxygen by hydrogen bonding via Lys168 and Glu246 or Lys39 and Gln88 in yeast and E. coli, respectively, and charge separation probably aided by Glu198 or Glu52, respectively.
ALLOSTERIC SITE OF SCCM
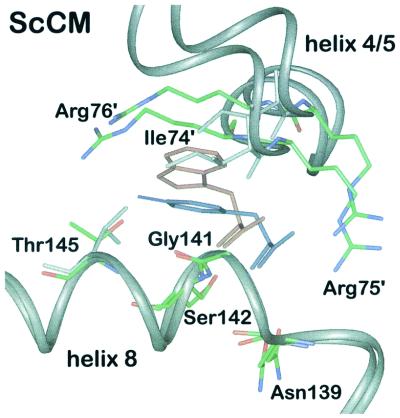
Whereas the enzyme from B. subtilis is unregulated, the E. coli CM is inhibited by binding of an end product, tyrosine or phenylalanine, to a distinct domain of the bifunctional protein. The yeast CM shows an additional level of regulation. It is feedback-inhibited by the end product tyrosine but can also be activated by tryptophan, the end product of the other biosynthetic branch of aromatic amino acid biosynthesis. The effectors for this dual regulation bind to the same allosteric sites in the regulatory domains. Using equilibrium dialysis, binding of tryptophan and tyrosine could be measured and two binding sites per CM dimer for each amino acid were found (81, 82). Their location was determined when the crystal structures of a Thr226Ile mutant, which is locked in the activated state, and of wt CM with the ligand tyrosine were solved (96, 114). It was found that the two allosteric effectors bind to the same binding sites in a mutually exclusive manner (Fig. 5). These allosteric sites are located 20 and 30 Å from the active sites of each monomer (113). They reside at the dimer interface in a cleft between helix H8 and loop L130s of monomer A and helices H4 and H5 of monomer B.
FIG. 5.
Superposition of the allosteric site in the T and R states of ScCM. The polypeptide backbones of helices H4-H5 and H8 are displayed in ribbon style. The residues necessary for binding of tyrosine (blue) and tryptophan (red) are shown as sticks with atoms labeled by color (green, carbon; blue, nitrogen; red, oxygen). Apostrophes indicate the position of the residue in an adjacent polypeptide chain. The dimer in the T state is superimposed onto the dimer in the R state by using residues 1 to 214 and 224 to 254.
Although both amino acids are oriented in the same direction, there are differences in the contacts of the effector amino acids with neighbouring protein residues of the enzyme. Only the hydrogen bonds between the amino group and one carboxyl oxygen are identical for both effectors. The amino nitrogen of tyrosine and tryptophan, respectively, is hydrogen bonded to side chains from residues Asn139A and Ser142A, which are located at the N terminus and inside helix H8 of monomer A, respectively. The carboxyl oxygen of the effector amino acids interacts with the amide nitrogens from Gly141A and Ser142A of helix H8 of the same monomer. When tryptophan is bound, further hydrogen bonds exist between its second carboxyl oxygen and three water molecules and between the ring nitrogen and another water molecule. In addition, van der Waals interactions between the ring atoms and residues of both monomers are observed.
The feedback inhibitor tyrosine makes additional polar interactions with the side chains of Thr145A in helix H8 of monomer A and Arg75B and Arg76B between helices H4 and H5 of monomer B. When tyrosine is bound, the second carboxyl oxygen is within hydrogen bond distance of the guanidinium and one amino group of Arg75B, because this residue changes its conformation compared to the tryptophan-bound state. The phenol ring binds at the same place as the five-membered ring of tryptophan, so that the phenolic hydroxyl group forms hydrogen bonds with both monomers, with the side chain of Thr145A, and with the guanidinium group of Arg76B.
Due to the numerous hydrogen bonds to tyrosine, the allosteric site is narrower than in the unliganded wt enzyme. Therefore, tyrosine inhibits the enzyme by pulling the two subunits closer together. In the tryptophan-bound state, the six-membered ring of tryptophan closely approaches main-chain as well as side-chain atoms of Ile74B. Hence, bulkier side chain of this amino acid pushes helices H4 and H5 away from helix H8 and opens the allosteric site. Thus, both effectors can initiate allosteric transitions with different results by using the same binding site. The polar contacts to Arg76B and Thr145A are of special importance for allosteric inhibition. For that reason, phenylalanine, lacking the phenolic hydroxyl group, cannot inhibit yeast CM. In fact, this amino acid was shown to produce the opposite effect. Although binding cannot be measured directly, a slight activation of wt CM was found under enzyme assay conditions by reduction of the S0.5 value (83). The hydroxyl group of tyrosine therefore is necessary for strong binding and inhibition of the enzyme.
Site-directed mutagenesis experimentally confirmed the location of the allosteric site and showed the importance of Gly141A, Ser142A, Thr145A, and the arginine residues Arg75B and Arg76B of the other monomer (83).
CONFORMATIONS OF SCCM
During the T-R transition, the two monomers of yeast CM rotate relative to each other (96, 97). The rotation axis is perpendicular to the dimer axis and 2.4 Å away from the center of the dimer in the direction of the allosteric sites. One monomer rotates 15° around this axis and is shifted 2.8 Å axially against the other monomer. Due to this screw motion, nearly all contacts at the dimer interface are changed. Alternatively, each monomer rotates by 8° around an axis which passes through the center of the monomers. To describe the differences between the T and R states, one can separate the monomers into catalytic and allosteric domains. The allosteric domain is composed of residues 44 to 107 (including helices H4 and H5 and adjacent loops). The catalytic domain comprises the rest of the monomer except of loop L220s, which, in fact, seems to connect both domains as a hinge. The latter domain includes the four-helix bundle which contains the active site. During transition from the T to the R state, helix H8 moves away from the allosteric site and is shifted by 0.7 Å along the axis, accompanied by tryptophan, whose Cα atoms move 2 Å relative to the Cα atoms of tyrosine. This transition is followed by the four-helix bundle. The regulatory domain, however, moves into the opposite direction with a shift of 1.5 Å away from the allosteric site. This opposite shift is the basis for separating the monomer into these two domains.
As mentioned above, studies with a stable transition-state analogue demonstrated that binding of the substrate causes further rotations, thereby inducing transition to a super R state (97). The rotation angle around the allosteric rotation axis is further increased to approximately 22° relative to the T state. This larger rotation is even achieved when tyrosine is bound to the regulatory domain. Tyrosine moves the regulatory domain toward the T state conformation, whereas the substrate simultaneously causes a super R state in the catalytic domain. Therefore, the hinge between the regulatory and catalytic domains has to be flexible enough to permit such an intermediate T-super R state as well as an R-super R state.
INTRAMOLECULAR SIGNAL TRANSDUCTION IN SCCM AND E. COLI ATCASE
ScCM
In the dimeric ScCM, the regulatory sites are located at the dimer interface and involve residues from both subunits. Dimer formation therefore seems to be a prerequisite for effector binding and subsequent allosteric regulation. The amino acids tyrosine and tryptophan influence the activity of CM by triggering allosteric transitions to the T and R state, respectively. The structural changes caused by both effectors are initiated at the effector binding site and transduced through the polypeptides toward the active sites, albeit as different processes and on different routes. While the signaling of tyrosine binding follows a linear path through the enzyme, the transition leading to activation cannot be depicted as precisely and may influence the catalytic site in multiple ways. Being positioned between the two monomers, the effectors also influence cooperativity toward the substrate. While tryptophan abolishes the positive cooperativity of substrate binding, tyrosine slightly enhances cooperativity.
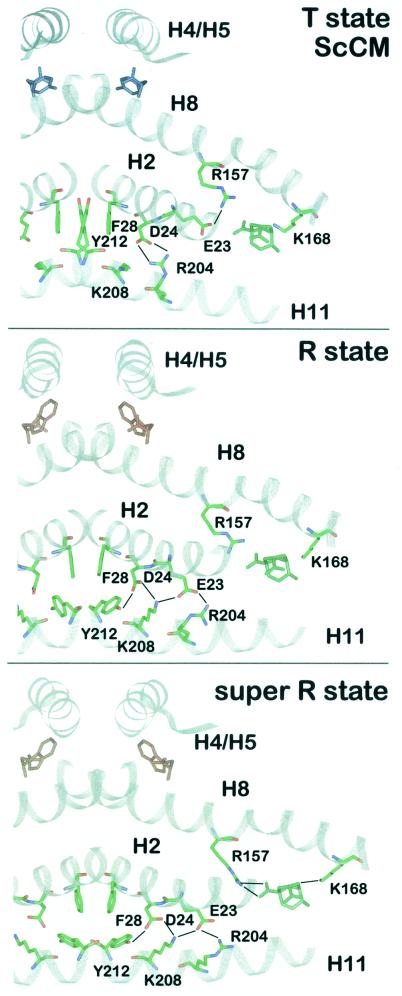
In the T state, the regulatory domain of monomer B is pulled toward helix H8 of monomer A. These movements at the dimer interface bring about further rearrangements between the two monomers, changing the number and energy of the bonds between them which extend from the regulatory through the catalytic domain toward the active sites (Fig. 6). Helix H8 spans the molecule from the allosteric to the catalytic site and rotates slightly during transition to the T state (96, 97). Its C terminus moves away from the catalytic site, while its N terminus moves in the opposite direction, thereby pulling the active-site residues Arg157 and Lys168 away from the substrate binding pocket. In addition, the C-terminal part of helix H2 moves away from the dimer interface by 1.7 Å. Helices H2, H11, and H12 are also driven away from the active site. Helices H11 from both monomers are pulled closer together along their axes by one helical turn, causing a shift relative to helices H2 (96). As a result, several residues along H2 and H11 change their interaction partners. The movements in this part of the protein seem to originate from loop L220s, which connects H11 and H12. This latter helix obviously changes its conformation during R-T transition because it seems sterically hindered by helices H2 and H11 of the other monomer when pointing in the same direction as in the R state.
FIG. 6.
Intramolecular signaling pathway. A section of the ScCM dimer is presented in the T state (top), R state (middle), or super R state (bottom). The polypeptide backbone is drawn in ribbon style. The residues which change their position during allosteric transition and thereby transduce the signal of effector binding from the allosteric to the active site are shown as stick models (green, carbon; blue, nitrogen; red, oxygen). Hydrogen bonds are indicated by black lines. The position of the catalytic site inhibitor in the T and R state is derived from a superposition with the super R state structure using residues 1 to 214 and 224 to 254. Tyrosine is colored blue, tryptophan is red, and the bicyclic inhibitor is green.
Thr226 is the last residue in loop L220s and plays an important role in T state formation. It is not clear if its side chain forms a hydrogen bond with Arg224 via a water molecule or with Glu228, but one of these is necessary for formation of the T state. In addition, the first residue of this loop, Tyr212, and Asp215B and Thr217B, which reside in the L220s loop of the other monomer, no longer interact with Lys208 and Arg204 of helix H11. Tyr212 and Phe28 are at a special position because they are next to the dimer axis and interact with each other and the corresponding residues from the other monomer. In the T state, the Tyr212 residues move between the two phenylalanine residues (Phe28A and Phe28B). Besides, Asp215 and Thr217 seem to point away from the interface (59). Along helix H11, Tyr212, Lys208, and Arg204 change their contacts to Asp24 and Glu23 of helix H2. Asp24 and Glu23 move closer to the active site so that Asp24 no longer forms salt bridges with Tyr212 and Lys208 but forms them with Arg204. Glu23 can no longer bind to Arg204 but moves 5.3 Å into the active site and interacts with Arg157. Significant differences are evident for the active sites in T and R state structures, with the side chain of the active-site residue Arg157 acting as a molecular switch on the T-R transition (Fig. 7A).
FIG. 7.
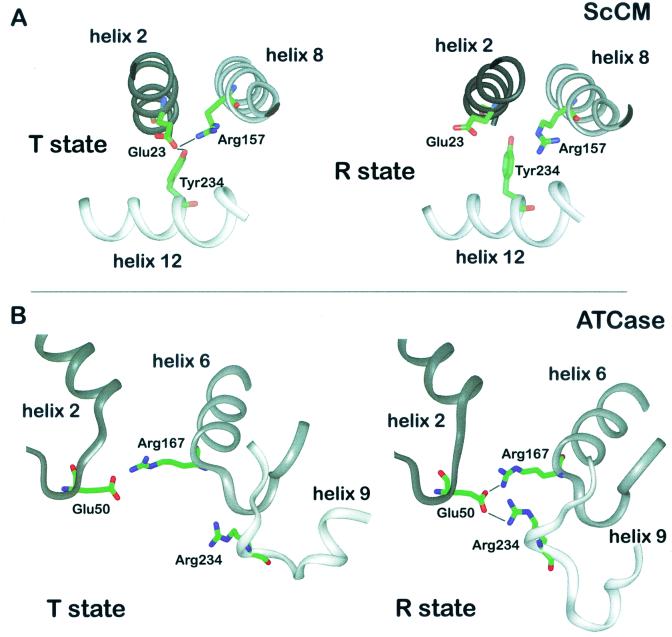
Triads of residues functioning as molecular switches in intramolecular signaling. On the T-R transition, rearrangements occur between residues Glu23, Tyr234, and Arg157 of ScCM (A) and between residues Glu50, Arg167, and Arg234 of ATCase (B). α-Helices and β-strands are outlined as ribbons.
Arg157 is part of the long helix H8 connecting the effector binding site to the active site, and its guanidinium group chelates one carboxyl group when the inhibitory transition state analogue is bound at the active site (Fig. 4). In the T state structure, this side chain is hydrogen bonded to Glu23, which in turn interacts with Tyr234 (Fig. 7A). Replacement of this latter residue resulted in functional enzymes that are unable to respond to tyrosine-induced feedback inhibition. Therefore, the Tyr234 side chain is likely to be important for allosteric inhibition. In the inhibited enzyme, the Glu23 residue is forced into an unfavorable conformation for substrate binding since its carboxylate group would be only 3.2 Å away from the carboxylate group of the substrate. On the transition to the active R state, the connections between the Tyr234-Glu23-Arg157 triad are abolished. Glu23 moves 5.3 Å away from the active site and no longer interacts with Arg157 but instead interacts with the Arg204 and Lys208 residues of helix H11 (Fig. 6C). As a consequence, Arg157 is now in a suitable conformation with effective charge for interaction with the substrate.
Applying continuum electrostatics, molecular surface/volume calculations, and molecular modeling, Nussinov and coworkers have argued that the altered binding affinity of Arg157 is of less importance for the different activities displayed by the T and R state (59). They proposed the position of Glu23 to be crucial for modulating the polarity of the active-site pocket. In the T state with the Glu23 side chain looming into the cavity, the interior is of negative electrostatic potential, repelling the negatively charged substrate. On the T-R transition, Glu23 swings out of the pocket and the polarity of the active-site cavity is altered in a positive electrostatic potential. In conclusion, the Glu23 residue was proposed to be the physical carrier of an electrostatic signal caused by the allosteric transition. This is supported by the catalytic properties of enzymes mutated in this particular position (84). Whereas a Glu23Asp mutant enzyme displayed increased activity combined with a loss of cooperativity, the Glu23Gln and Glu23Ala enzymes showed reduced catalytic activity and replacement of Glu23 by arginine led to a nonfunctional enzyme.
The conformational changes at the dimer interface might also explain homotropic effects of the substrate. Signal transduction between the active sites of this allosteric enzyme might follow the same path along helices H2, H11, and H12 and loop L220s. While the heterotropic effects of the allosteric ligands lead to the T-R transition, the homotropic effect of the substrate is the induction of the super R state in the catalytic domain.
ATCase
As far as ATCase is concerned, a detailed mechanism for homotropic interactions has been described and there exist diverse theories about how the allosteric ligands exert heterotropic effects. Similar to CM, a high-affinity active site has to be formed for the R state structure. This is achieved mainly by closure of the aspartate domain toward the cp domain, thereby forming the complete binding pocket for aspartate accompanied by shifts of the L80s and L240s loop. Plenty of interface contacts have been identified by amino acid substitutions which are important for stabilizing either the ATCase T or R state conformation. In detail, the L80s loop contains the active side residues Ser80 and Lys84 which are the two residues contributed by the adjacent C chain. It moves into the active site during the T-R transition, positioning Ser80 and Lys84 for substrate binding, which is essential for cooperativity (66). The end of the L80s loop is tethered by Glu86, which is salt linked to a most critical active-site residue, Arg54, across the C1-C2 interface and assists in its positioning for catalysis. This ion pair interaction exists in both the T and R states and therefore is also essential for forming the catalytic subunit (4).
The interdomain bridging neccessary for domain closure in ATCase is partly achieved by a salt link between the active-site residue Arg167 and Glu50 (Fig. 7B). Arg105, also in the active-site pocket, is free to interact with the substrate only when it is no longer bound by Glu50 in the R state (43, 99). In addition, the whole L240s loop rearranges during the T-R transition and behaves as a hinge like the L220s loop of CM. It moves toward the cp domain during domain closure so that the Arg167, Arg229 and Glu231 residues, which are important for aspartate binding, are rearranged. Glu233 binds Arg229 only in the R state and positions it properly for catalysis. Arg234 is also involved in interdomain bridging. It is stabilized by binding Glu231 and also contacts Glu50. Therefore the interactions between Glu233 and Arg229 and between Glu50 and Arg234 are impotant for establishing the R state confromation of the L240s loop.
As in CM, there is a triad of residues. Instead of Tyr-Glu-Arg, the ATCase triad is composed of Glu50, Arg167, and Arg234 (Fig. 7B). However, the function of the rearrangements in the environment of the triad is to initiate the T-R transition instead of maintaining the T state as in CM. In addition, interchain contacts between Tyr240 and Asp271 have to be broken for the movements of the domains. The interruption of the important intersubunit contacts at the C1-C4 interface formed by the L240s loop residues Glu239 and Lys164-Lys165 and by Lys164-Lys165 and Glu239 of the other C chain, respectively, is another prerequsite for the T-R transition. In fact, all but van der Waals interactions are eliminated at this interface when the L240s loop, which provides the only contact between C chains from different trimers, is reoriented. The distinct interchain contacts formed by the L240s loop are important for stabilization of the allosteric states. When the links between the lower and upper C trimers are lost, the T state cannot be maintained (43). By the rearrangement of one L240s loop, the L240s loops from adjacent chains all move apart, thereby separating the catalytic subunits and releasing the restrictions present in the T state. Because every change at this intersubunit interface is transmitted to the others, the T-R transition is a concerted mechanism, with the whole enzyme moving toward the R state (42). Macol et al. found direct evidence that the exclusive binding of one PALA molecule per enzyme is sufficient for full T-R transition of all its subunits (67). This proves that the transition is concerted and clearly meets the requirement of the MWC model of allostery. All C1-R4 contacts are broken, while more contacts are made between C1 and R1. In addition, bonds at the interface between the allosteric and zinc domains of the R chain are lost and new bonds are formed between the C chain domains (1). The L100s loop functions as another hinge between the allosteric and zinc domains. Amino acid substitutions in these regions eliminate cooperativity and heterotropic interactions. In short, the T state is stabilized by bonds at the C1-C4, C1-R4, and allosteric-zinc interfaces and the R state is stabilized by bonds at the C1-R1, C1-C2, and polar-equatorial interfaces (also see references 15 and 91).
As far as the transduction of heterotropic effects is concerned, the following pathways have been proposed for ATP signaling. ATP binds to the allosteric site in a slightly different way from CTP (62). It expands this site and thereby influences neighboring residues. ATP undergoes more interactions with the N-terminal region of the R chains than CTP does. This region is responsible for creating high- or low-affinity binding sites and, by doing so, exerts control over inhibition and activation and is supposed to mediate communication between the two binding sites within a regulatory dimer (R1–R6 N-terminal contacts) (77). The next important region is the allosteric-Zn interface, which stabilizes the T and R states. The contacts between helix H1′ of the allosteric domain and residues in the Zn domain, especially Phe33r, change because helix H1′ is shifted (29). These perturbations, and probably also those that might be caused by CTP in the allosteric-Zn interface, are propagated to the C1-R1 or C1-R4 interface (29). Finally, residues 146r-149r form a set of interactions with 241c-245c of the L240s loop, which is involved in transduction of the ATP signal but not that of CTP (16). Alternatively, it was proposed that the ATP expansion of the allosteric site is propagated through Glu68r and reorientation of helix H2', thereby moving Tyr77r in the hydrophobic core, which also includes Val106r. This residue could transmit the ATP signal toward the R1-C1 and R1-C4 interfaces between R and C chains (110).
SEPARATION OF ACTIVATION AND INHIBITION
ScCM
Thr226, the last residue of loop L220s in ScCM, proved necessary for R-T transition since a Thr226Ile mutant is unresponsive to the allosteric effectors and shows no cooperativity but is locked in the activated state (81). Other amino acid residues at position 226 yielded enzymes with different intermediate degrees of regulation between the wt and constitutively activated enzyme (30). The Km values of the mutant enzymes were reduced compared to those of the wt. The shift to the R state was strongest for Ile226 or Arg226 substitutions and weaker for enzymes with Asp226, Lys226, Ala226, or Pro226 residues; Gly226 or Ser226 substitutions had the least effect on the T-R equilibrium. However, no obvious correlation of the strength of this effect with any property of the introduced residue was found. Whereas the allosteric equilibrium is shifted toward the R state for these enzymes, the catalytic constant and the affinity for tryptophan are uneffected. The role of this loop seems to be different in the structurally related CM from A. nidulans (48). Although several residues in the regulatory and catalytic site and for signal transduction are conserved, a special role for allosteric transition could not be attributed to Asp233, the corresponding residue to Thr226. Therefore, the signal of effector and maybe substrate binding seems to follow slightly different routes in the dimer interface in this otherwise very similar enzyme.
A Thr226lle Ile225Thr double mutant which remains sensitive to tryptophan but insensitive to tyrosine shows that the simple two-state MWC model for allosteric transitions does not explain all characteristics of yeast CM (84). The additional Ile225Thr subsitution unlocks the R state and allows activation but not inhibition. A threonine residue at the end of the loop seems to form essential hydrogen bonds, which are necessary for the conformation present in the T state. By changing its conformation, L220s serves as a hinge which can affect the shifting of helices H11 and H12 and thereby lead to the formation of the inactive Tyr234-Glu23-Arg157 triad structure in the active site (Fig. 7A).
A loss of sensitivity to tyrosine alone is also shown by substitutions of Tyr234 and Glu23, which interfere with the molecular switch that modulates the active-site pocket (84). The interactions of the phenolic hydroxyl moiety of tyrosine are necessary for the low-affinity state of the catalytic site, since a missing tyrosine at position 234 does not assist in keeping Glu23 close to the active site. An involvement of Tyr234 in homotropic interactions between the active sites is supported by the fact that cooperativity is reduced or abolished in enzymes with substitutions at position 234. Similarly, a Glu23Gln mutation shows only residual inhibition but strong activation, which emphazises the need for negative charge for inhibition at this position to change the electrostatic field in the active-site pocket. Thus, allosteric activation seems to be signaled on a pathway distinct from allosteric inhibition and homotropic interactions between the active sites.
These findings support the path of allosteric transition as described above, because the amino acid substitutions all affect residues found to be rearranged during the T-R transition. They also clearly show that activation and inhibition are functionally divided and must follow different pathways. The interactions around Thr234 and Ile225 Thr226 are a prerequisite for forming the complete T state. Their absence has no effect on the R state, however, showing that interactions between other parts of the polypeptides are important for complete R state formation. Further rearrangements in the rest of the protein during the T-R transition are not affected by these substitutions, making activation possible. In fact, it seems that transition to the R state comprises only part of the rearrangements found between the T and R states. Besides the T and R states, the unliganded enzyme might occupy a third state which is intermediate between the T and R states. In this state the distance between H8 of one monomer and H4, H5, and L80s of the other monomer in the allosteric site is intermediate between those in the R state and T state and is changed to the R or T state by the involvement of an “induced fit” promoted by effector binding. Whether a more direct pathway for the action of tryptophan might be possible, which does not require a global change of the overall enzyme structure, is an open question. Tryptophan activation might not be triggered by loop L220s. Presumably it follows either a pathway along helix H8, which contributes residues to the regulatory site as well as the catalytic site, or via helix H4, which extends from the allosteric site through the whole molecule. The existence of a preexisting T-R equilibrium in which the states are formed apart from ligand binding and which is shifted toward one state or the other by binding of an effector seems to be unlikely with the results obtained so far.
ATCase
Facts supporting this theory of different pathways for activation and inhibition were found for ATCase. Regions specific for transmission of the ATP signal have been identified in the allosteric-Z interface, mainly Leu151r and Val150r, which are near the R1-C4 interface and may transmit the signal between these two interfaces (102, 110). Also, residues 145 to 149 of the R chain which contact the L240s loop in the R1-C4 interface specifically transmit the ATP signal but not the CTP signal. Amino acid substitutions around the important salt link between Lys143r and Asp236c of the L240s loop verified the importance of this region for ATP activation (61, 62). In addition, six residues at the C terminus of the R chain were found to be involved in the ATP activation, as found with truncated proteins derived from specific deletion alleles (16). In particular, the R1-C1 interface, the junction between the R1-C1 and R1-C4 interfaces, and the R1-C4 interface are essential for ATP signaling. Like tryptophan and tyrosine recognition in CM, there is a discrimination between ATP and CTP in the allosteric site, which is partly dependent on the size of the base rings. In addition to expansion or reduction of the binding pocket, the orientation of the base plays a crucial role in propagation of the activation or inhibition signal (78).
Comparable to Thr226 of CM, there are several single residues in ATCase which, when substituted, lead to enzymes which are shifted toward the R state. Among them are the residues Lys143r and its binding partner Leu235c, which normally stabilize the R1-C4 interface (71, 79). Also, another mutant enzyme showed normal sensitivity to ATP activation but no CTP inhibition, though CTP can bind normally (53). Thus, this enzyme variant is in an intermediate state in which the R1-C1 interface is weakened (11). In addition, the Lys56rAla mutant enzyme is frozen in the R state and insensitive to ATP while CTP inhibits the enzyme and restores homotropic effects (13). Other mutant enzymes, however, are stabilized in the T state. The Glu50cAla enzyme is unable to close the C chain domains and thus does not exist in the R state conformation (58, 71). A Thr82rAla enzyme is structurally fixed in an extreme T state with the T-R equilibrium shifted toward the T state (108). Uncoupling of activation and inhibition can be achieved by substitutions at positions 56r and 60r. CTP inhibition but only minimal ATP activation can be observed for a Lys56rAla substitution. When Lys60r is replaced by alanine, an enzyme is generated with ATP activation and minor CTP inhibition (93, 115). When transmission of CTP is disturbed, however, larger structural changes occur in the enzyme. Amino acid residues could not be defined which specifically and exclusively affect inhibition. Therefore, it seems that inhibition is caused by long-range effects on other interfaces by a more complex set of interactions and not by signal transmission along a defined path like ATP signaling. This resembles the activation mechanism by tryptophan of CM. For both enzymes, substitution of some specific single amino acid residues can be sufficient to change the intensity of allosteric regulation. However, other authors found that these substitutions need not necessarily be positioned at a defined path for signal transduction but might cause global conformational changes due to changes in free energy (3, 63).
SEPARATION OF HOMOTROPIC AND HETEROTROPIC EFFECTS
ATCase
Homotropic and heterotropic effects can be almost completely separated in ATCase and seem to be mediated by different allosteric transitions, which include more than two states (98). In the “frozen” T or R state, the substrate has no homotropic effects but heterotropic effects are maintained (56, 74, 90). This also means that the effectors act indirectly on substrate binding rather than on the T-R equilibrium (38). Also, when Glu231c is replaced by isoleucine or asparagine, the mutant enzyme shows no reduction in heterotropic effects but shows reduced cooperativity with respect to aspartate (74). Vmax is changed significantly by the addition of CTP or ATP due to decreased binding of active-site ligands to the R state. The substrates are not bound preferentially to the R state, resulting in reduced cooperativity. Other variants like Glu50cAla, Glu50cGln, Ser171cAla, or Arg234cSer also bind aspartate more weakly and show similar effects on heterotropic and homotropic interactions. The MWC model describes two extreme types of allosteric enzymes: the V system, for which Vmax is different in the T and R states, and the K system, in which Km is altered. The V system exhibits heterotropic effects but no cooperativity toward the substrates. No Vmax could be determined for the T state of ATCase, however, because it is converted to the R state in the presence of the two stubstrates. Therefore it seems that ATCase is a mixed V-K system in which some substitutions might affect Vmax more than they affect Km (74). Therefore, a loss of homotropic effects need not necessarily be accompanied by a change in heterotropic effects. Similarly, a Glu239cGln enzyme shows cooperativity but no heterotropic effects. This variant is devoid of C1-C4 interchain interactions so that the enzyme undergoes transition to the R state in the presence of carbamoylphosphate alone, and heterotropic effects can no longer be observed (54).
ScCM
When residue 226 of CM was varied, all enzymes displayed the behaviour of a true K system (30). kcat values were roughly the same, Km was decreased, and there was a loss of cooperativity. Also, when Glu23 was replaced by an aspartic acid residue, the kcat values remained constant while the Km value was decreased in the R state compared to the T state (84). In addition, cooperativity was lost, so that this is the only mutant enzyme so far in which homotropic and heterotropic effects are uncoupled. This means that although the shorter side chain of aspartic acid positioned at the molecular switch for inhibition is sufficient for formation of the inhibitory triad of residues, it is not able to maintain cooperative substrate binding. Therefore, for CM, slightly different allosteric transitions must exist for homotropic and heterotropic effects.
MODELS FOR THE ALLOSTERIC MECHANISMS
Two additional allosteric states besides T and R were found for CM when chorismic acid or the transition state analogue binds to the active site of the enzyme, the T-super R and the R-super R state. As explained above, the data support the existence of a super R state when CM is liganded exclusively to chorismate. However, a crystal structure for CM liganded solely to the transition state analogue has not been determined yet. Therefore, it remains unclear if the substrate alone can promote the transition toward the super R state and if this state may be the actual R state. Binding of the transition analogue to the catalytic site and additional binding of tryptophan to the allosteric site leads to additional conformational changes, so that an R-super R state is formed. This result cannot be explained by the KNF model, in which it is proposed that the activator induces the same conformation as the substrate. On the other hand, the R state structure with tryptophan bound was determined with a mutant enzyme variant. This allows speculation about whether tryptophan induces the same conformation in the wt enzyme. It is possible that the R-super R state is the final R state and that the known tryptophan-bound form is just an intermediate. However, with tyrosine bound to the allosteric site, rearrangements to an R-like structure seem necessary prior to the catalytic step since the Km value is increased 30-fold for the T state compared to the R state. This super R state, which is very nearly T like in the allosteric sites, occurs when tyrosine is bound to the allosteric site and the transition state analogue is bound to the catalytic center. Similarly, when PALA binds to the six active sites of ATCase, the elongated R state is only slightly affected when CTP is then bound to the regulatory sites. Thus, the substrate analogues in these examples dominate the allosteric transition.
As mentioned above, homotropic cooperativity in ATCase can be fully explained by a concerted transition according to the MWC two-state model. No evidence for such behavior has yet been found for the homotropic effects of the substrate for CM.
For the ATCase system, different models were proposed for the effect of the heterotropic ligands. Originally it was thought that the nucleotides act on the same equilibrium as aspartate in that they bind preferentially to one of the two allosteric states and thereby directly alter the equilibrium (10, 36, 79). Since it was found that homotropic and heterotropic effects can be uncoupled, this model seems to be outdated (1, 62). Fetler et al. found that while homotropic effects can be explained by the model of concerted transition, CTP seems to slightly decrease the R state population, thus partly acting concerted, and ATP does not affect the T-R equilibrium (24). Therefore CTP acts in part independently of the T-R equilibrium while ATP activation follows a different mechanism. Models that account for the experimental data more precisely are the primary-secondary-effect model, the effector-modulated transition model, and the nucleotide perturbation model. It was proposed that the nucleotides affect the affinity of the active sites for aspartate in a primary effect which includes smaller structural changes originating from the allosteric site leading to the formation of the R state structure by binding of aspartate as the secondary effect when carbamoylphosphate is already bound to ATCase (34, 38, 100, 111). As described previously, binding of CTP or ATP to the R or T state causes only minor alterations in the separation of the catalytic trimers (29, 92). According to the model, this can be viewed as the primary effect. In mutant enzymes, however, which exist in intermediate conformations, both effectors manage to induce the full rearrangement to the R or the T state (93). This model is partly in agreement with the effector-modulated transition model. The nucleotides might change the stability of the interfaces as part of the primary effect, thereby influencing T-R transition on aspartate binding. On the basis of this, Liu et al. proposed that changes in interfaces could influence allostery by altering the global energy of ATCase (63). ATP might induce a signal transduction chain via the R1-C1 interface, while CTP exerts its function via the R1-C4 interface (111). Other authors favour the nucleotide perturbation model (94). It was found that ATP increases the size of the allosteric site, which also increases the whole allosteric domain. CTP, in contrast, decreases the size of its binding site and also the allosteric domain. Discrimination between the nucleotides occurs in the regulatory chains, as was found with regulatory-chain mutants (102, 115) and hybrid enzymes (6). On the pathway to the active site, the signal has to be transmitted via the allosteric-Zn, R1-C1, R1-C4, and/or R1-allosteric/R6-allosteric interfaces. These effects would lead to domain movements of C and R chains which, however, would be far smaller than those during T-R transition, namely, the separation of the catalytic trimers by less than 1 Å. A direct signal could be transmitted via the allosteric-Zn and C-R interfaces, but indirect signaling is also proposed. In the R state, CTP binding might influence the R1-C1 interface and thereby decrease the trimer separation in the R state. These movements should affect the C1-C4 interface, leading to changes in the L240s loop and the active site. As a consequence, the R state is destabilized (94). In the T state, CTP binding stabilizes this conformation even further without affecting trimer separation due to steric hindrance, but it influences residues in the active site. ATP increases trimer separation by influencing these interfaces in the opposite way, which destabilizes the T state. In the R state, this influence of ATP might well perturb (stabilize) this structure even further (94). It was found that in an unliganded T state, ATP or CTP binding can change the orientation of the active-site residue Arg229 and that interactions around the L240s loop are weakened (61, 62).
The structure of unliganded ATCase is known only to 2.6-Å resolution (44), and no structure has been determined for unliganded CM. High-resolution structures for both enzymes would facilitate the construction of models for the allosteric mechanism. In addition, only a mutant version of CM was crystallized in complex with the activator tryptophan. Equilibrium dialysis and kinetic studies showed that tyrosine is unable to bind to this variant whereas tryptophan binding can be detected. This suggests that the enzyme is locked in an R-like state and that tryptophan binding has no influence on enzyme activity. The mutant enzyme structure does not answer the question whether tryptophan induces the full transition to the R state or super R state in the wt enzyme. However, it might well be that, as in ATCase, the activator as a single ligand is not able to shift the equilibrium fully toward the R state but that the substrate is required to form the ultimate high-affinity high-activity super R state. It cannot be excluded that even when only tryptophan is bound the wt enzyme resembles the T state. Whether the allosteric inhibitor has a destabilizing effect on the high-affinity state of CM is not clear either. The presence of the T-super R state, however, shows that structural changes occur at least in the allosteric domains, although the catalytic domains seem to remain in the super R state. These changes require a flexible hinge region which connects the allosteric and regulatory domains. This flexibility might be an indirect reminder of the probable origin of both domains as a result of an evolutionary gene duplication event. The mixed states which are found in different CM cocrystals might be consistent with the nucleotide perturbation model of transition in ATCase. According to that model, the activator tryptophan shifts the equilibrium from the T to the R state and further stabilizes the super R state induced by the substrate. Tyrosine slightly destabilzes the R-like state by formation of the T-super R state, but it strongly stabilizes the T state. Also, the other models can explain some characteristics found for CM. The further structural rearrangements occurring upon binding of the transiton state analogue might also be addressed by the primary-secondary-effect model. Effector binding might cause a primary effect, while substrate binding leads to the secondary effect, namely, the transition to the super R state. Finally, the effector-modulated transition model accounts for the broader structural changes induced by tryptophan binding to CM. No direct signal transduction pathway has been found for this effector yet, but it seems to modulate activity by influencing the subunit interface or global energy of the protein conformation.
LESSONS LEARNED FROM THE MODEL SYSTEMS AND THE DAWN OF A NEW PARADIGM FOR ALLOSTERY
Regulation of gene product activity by allostery is a central dogma within the field of biochemistry, and allosteric proteins acting as molecular amplifiers are widespread in nature. Since its first description, much knowledge concerning the mechanisms contributing to allosteric regulation has accumulated. Naturally, model systems have emerged in which the basic principles that constitute allostery were defined and scrutinized. The first polypeptide on which the basic principles of homotropic and heterotropic interactions were applied was hemoglobin. Although hemoglobin is not an enzyme, the availability of numerous mutants with single-amino-acid substitutions and the existance of a solved X-ray structure led to its establishment as a paradigm of allostery. Among the numerous enzymatic activities, the E. coli ATCase system has emerged as a proper model for allosteric mechanisms. Here we have summarized the knowledge and insights gained over the past 10 years for another metabolic enzyme, ScCM. For ScCM, a multitude of data has been determined, complemented by a variety of solved crystal structures defining different allosteric states. With respect to ATCase, ScCM offers additional advantages for general research on allostery. Its dimeric structure comes up to the smallest unit possible for allosteric transitions and cooperativity. In fact, ScCM displays the whole spectrum of allostery like cooperativity and negative or positive effects mediated by homotropic as well as heterotropic ligands. Thus, both model systems, ATCase and ScCM, have the capacity to improve and support our present-day view of allosteric mechanisms.
The most intriguing question which can be asked is whether one unifying model can account for allostery in general. Basically, three generalizing descriptions for allosteric transitions have been developed to date: MWC, KNF, and the unifying model by Eigen. Homotropic transition of the transcarbamoylase has been investigated conclusively, and this enzyme has provided direct structural evidence for a concerted allosteric transition as described by the MWC model. For the CM, detailed analyses of the allosteric transition on substrate binding in the absence of heterotropic ligands are still missing and therefore no conclusive classification of this allosteric behavior of ScCM is possible. On the other hand, very accurate conclusions have been drawn concerning the principles for heterotropic effects acting on both enzymes. Here, the two-state model as provided by the MWC theory proves to be insufficient. Five distinct structures were solved for the yeast CM that represent different allosteric states. Additionally, several intermediate states have been created by modification of the Thr226 trigger. Most interestingly, the coexistence of different allosteric states for separate domains within the same molecule has been demonstrated by binding of a transition state analogue in the presence of either effector. This flexibility accounts for an induced-fit mechanism of allosteric transition. As stated above, a clear differentiation seems necessary, with the MWC theory frequently applying to homotropic effects whereas heterotropic transitions may be described appropriately by the KNF model. It is conclusively evident that to date no simplifying principle is sufficient to completely describe either allosteric enzyme and that further refinements will have to be made to elucidate their allosteric behavior completely.
Detailed analysis of the allosteric binding sites of ScCM and ATCase provides insights into how small ligands are differentiated on the protein surface. As with tyrosine and phenylalanine binding to ScCM, highly specific interactions and contacts contribute to the molecular recognition of the differing hydroxyl group. When different heterotropic effectors are concerned, a common mechanism for signal transduction can be deduced. Expansion and contraction, respectively, of the allosteric site is transduced by conformational changes to the distant active site. There the creation of a high-affinity cavity is essential for catalytic turnover. Both model enzymes demonstrate in an extraordinary way how allosteric signals are transmitted across a protein structure. While allosteric signal transduction of inhibition can be followed along a distinct structural pathway in ScCM, this is not possible for the positive effect of the activating ligand. The opposite is true for ATCase, in which activation but not inhibition follows a defined path from the allosteric to the active site. In conclusion, it is obvious that different routes within an oligomeric enzyme transmit different allosteric responses. Additionally, different means of intra- and intermolecular signal transduction are possible in a nonexclusive manner, like long-range effects that may alter the global protein structure or defined rearrangements along distinct amino acid side chains. The uncoupling of allosteric activation and inhibition as demonstrated for both enzymes supports this view of distinct signal transduction pathways. The existence of molecular switches that enable allosteric transition is of special interest. For both enzymes, a triad of residues has been identified that contribute to the T-R transition and support one allosteric state, respectively. Loop regions that function as molecular hinges for the movements of domains relative to each other were also identified, and all these elements contribute to a general understanding of the common mechanisms for allosteric transition. The overall structure of allosteric proteins is delicately balanced in such a way that small changes can shift the quaternary structure toward T or R states. This means that transmission of heterotropic and homotropic signals does not necessarily follow distinct pathways and that substitutions supposed to act in a specific manner in a signaling pathway might also affect the global protein structure in some cases.
As outlined above, the common theory of two major allosteric states that coexist within a dynamic equilibrium in the absence of effectors is unlikely and must be refined toward a more dynamic model. In 1965, Monod et al. already stated that their “model offers only an over-simplified first approximation of real systems, and it may prove possible in some cases to introduce corrections and refinements …”. ScCM represents an excellent real system capable of almost all aspects of allostery. Especially when taken in combination with its textbook counterpart E. coli ATCase, the knowledge gained from experiments with ScCM contributes to a refined understanding of the basic principles that underly allostery. ScCM significantly differs from transcarbamoylase by the fact that regulatory and catalytic domains reside on the same polypeptide chain. A gene duplication and fusion event has been proposed as the evolutionary event underlying this assembly. Thus, ScCM is well suited as appropriate model enzyme for studying the evolution of an allosteric mechanism. Furthermore, the knowledge about CM in general is quite extensive, and by comparison with its numerous prokaryotic or eukaryotic counterparts, ScCM will provide further conclusions on how acceleration of a pericyclic reaction can be achieved and regulated.
ACKNOWLEDGMENTS
We are greatly indebted to the invaluable work of former members of the laboratory including Tobias Schmidheini, Georg Schnappauf, Roney Graf, and Markus Hartmann, and we thank William N. Lipscomb, Norbert Sträter, and numerous colleagues for the fruitful cooperation during the last years.
This work was supported initially by the Swiss National Science Foundation and subsequently by the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, and the Volkswagen-Stiftung.
REFERENCES
- 1.Allewell N M. Escherichia coli aspartate transcarbamoylase: structure, energetics, and catalytic and regulatory mechanisms. Annu Rev Biophys Biophys Chem. 1989;18:71–92. doi: 10.1146/annurev.bb.18.060189.000443. [DOI] [PubMed] [Google Scholar]
- 2.Andrews P R, Smith G D, Young I G. Transition-state stabilization and enzymic catalysis. Kinetic and molecular orbital studies of the rearrangement of chorismate to prephenate. Biochemistry. 1973;12:3492–3498. doi: 10.1021/bi00742a022. [DOI] [PubMed] [Google Scholar]
- 3.Aucoin J M, Pishko E J, Baker D P, Kantrowitz E R. Engineered complementation in Escherichia coli aspartate transcarbamoylase. Heterotropic regulation by quaternary structure stabilization. J Biol Chem. 1996;271:29865–29869. doi: 10.1074/jbc.271.47.29865. [DOI] [PubMed] [Google Scholar]
- 4.Baker D P, Stebbins J W, DeSena E, Kantrowitz E R. Glutamic acid 86 is important for positioning the 80's loop and arginine 54 at the active site of Escherichia coli aspartate transcarbamoylase and for the structural stabilization of the C1–C2 interface. J Biol Chem. 1994;269:24608–24614. [PubMed] [Google Scholar]
- 5.Bartlett P A, Johnson C R. An inhibitor of chorismate mutase resembling the transition-state conformation. J Am Chem Soc. 1985;107:7792–7793. [Google Scholar]
- 6.Beck D, Kedzie K M, Wild J R. Comparison of the aspartate transcarbamoylases from Serratia marcescens and Escherichia coli. J Biol Chem. 1989;264:16629–16637. [PubMed] [Google Scholar]
- 7.Beernink P T, Endrizzi J A, Alber T, Schachman H K. Assessment of the allosteric mechanism of aspartate transcarbamoylase based on the crystalline structure of the unregulated catalytic subunit. Proc Natl Acad Sci USA. 1999;96:5388–5393. doi: 10.1073/pnas.96.10.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bethell M R, Smith K E, White J S, Jones M E. Carbamyl phosphate: an allosteric substrate for aspartate transcarbamylase of Escherichia coli. Proc Natl Acad Sci USA. 1968;60:1442–1449. doi: 10.1073/pnas.60.4.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blangy D, Buc H, Monod J. Kinetics of the allosteric interactions of phosphofructokinase from Escherichia coli. J Mol Biol. 1968;31:13–35. doi: 10.1016/0022-2836(68)90051-x. [DOI] [PubMed] [Google Scholar]
- 10.Changeux J P, Rubin M M. Allosteric interactions in aspartate transcarbamylase. 3. Interpretation of experimental data in terms of the model of Monod, Wyman, and Changeux. Biochemistry. 1968;7:553–561. doi: 10.1021/bi00842a601. [DOI] [PubMed] [Google Scholar]
- 11.Cherfils J, Vachette p, Tauc p, Hervé G. The pAR5 mutation and the allosteric mechanism of Escherichia coli aspartate carbamoyltransferase. EMBO J. 1987;6:2843–2847. doi: 10.1002/j.1460-2075.1987.tb02581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chook Y M, Ke H, Lipscomb W N. Crystal structures of the monofunctional chorismate mutase from Bacillus subtilis and its complex with a transition state analog. Proc Natl Acad Sci USA. 1993;90:8600–8603. doi: 10.1073/pnas.90.18.8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corder T S, Wild J R. Discrimination between nucleotide effector response of aspartate transcarbamoylase due to a single site substitution in the allosteric binding site. J Biol Chem. 1990;264:7425–7430. [PubMed] [Google Scholar]
- 14.Cunin R, Jacobs A, Charlier D, Crabeel M, Hervé G, Glansdorff N, Pierard A. Structure-function relationship in allosteric aspartate carbamoyltransferase from Escherichia coli. I. Primary structure of a pyrI gene encoding a modified regulatory subunit. J Mol Biol. 1985;186:707–713. doi: 10.1016/0022-2836(85)90390-0. [DOI] [PubMed] [Google Scholar]
- 15.Dembowski N J, Kantrowitz E R. The use of alanine scanning mutagenesis to determine the role of the N-terminus of the regulatory chain in the heterotropic mechanism of Escherichia coli aspartate transcarbamoylase. Protein Eng. 1994;7:673–679. doi: 10.1093/protein/7.5.673. [DOI] [PubMed] [Google Scholar]
- 16.De Staercke C, Van Vliet F, Xi X G, Rani C S, Ladjimi M, Jacobs A, Triniolles F, Hervé G, Cunin R. Intramolecular transmission of the ATP regulatory signal in Escherichia coli aspartate transcarbamylase: specific involvement of a clustered set of amino acid interactions at an interface between regulatory and catalytic subunits. J Mol Biol. 1995;246:132–143. doi: 10.1006/jmbi.1994.0072. [DOI] [PubMed] [Google Scholar]
- 17.Dohi Y, Sugita Y, Yoneyama Y. The self-association and oxygen equilibrium of hemoglobin from the lamprey, Entosphenus japonicus. J Biol Chem. 1973;248:2354–2363. [PubMed] [Google Scholar]
- 18.Dutta M, Kantrowitz E R. The influence of the regulatory chain amino acids Glu-62 and Ile-12 on the heterotropic properties of Escherichia coli aspartate transcarbamoylase. Biochemistry. 1998;37:8653–8658. doi: 10.1021/bi980456h. [DOI] [PubMed] [Google Scholar]
- 19.Eberhard J, Raesecke H R, Schmid J, Amrhein N. Cloning and expression in yeast of a higher plant chorismate mutase. Molecular cloning, sequencing of the cDNA and characterization of the Arabidopsis thaliana enzyme expressed in yeast. FEBS Lett. 1993;334:233–236. doi: 10.1016/0014-5793(93)81718-f. [DOI] [PubMed] [Google Scholar]
- 20.Eigen M. Kinetics of reaction control and information transfer in enzymes and nucleic acids. Nobel Symp. 1967;5:333–369. [Google Scholar]
- 21.Eisenstein E, Markby D W, Schachman H K. Heterotropic effectors promote a global conformational change in aspartate transcarbamoylase. Biochemistry. 1990;29:3724–3731. doi: 10.1021/bi00467a019. [DOI] [PubMed] [Google Scholar]
- 22.Endrizzi J A, Beernink P T, Alber T, Schachman H K. Binding of bisubstrate analog promotes large structural changes in the unregulated catalytic trimer of aspartate transcarbamoylase: implications for allosteric regulation induced cell migration. Proc Natl Acad Sci USA. 2000;97:5077–5082. doi: 10.1073/pnas.090087197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.England P, Leconte C, Tauc P, Hervé G. Apparent cooperativity for carbamoylphosphate in Escherichia coli aspartate transcarbamoylase only reflects cooperativity for aspartate. Eur J Biochem. 1994;222:775–780. doi: 10.1111/j.1432-1033.1994.tb18924.x. [DOI] [PubMed] [Google Scholar]
- 24.Fetler L, Tauc P, Hervé G, Moody M F, Vachette P. X-ray scattering titration of the quaternry structure transition of aspartate transcarbamylase with a bisubstrate analogue: influence of nucleotide effectors. J Mol Biol. 1995;251:243–255. doi: 10.1006/jmbi.1995.0432. [DOI] [PubMed] [Google Scholar]
- 25.Galopin C C, Zhang S, Wilson D B, Ganem B. On the mechanism of chorisamte mutases: clues from wild-type E. coli enzyme and a site-directed mutant related to yeast chorismate mutase. Tetrahedron Lett. 1996;37:8675–8678. [Google Scholar]
- 26.Ganem B. The mechanism of the Claisen rearrangement: déjà vu all over again. Angew Chem Int Ed Engl. 1996;35:936–945. [Google Scholar]
- 27.Gerhart J C, Pardee A B. The enzymology of control by feedback inhibition. J Biol Chem. 1962;237:891–896. [PubMed] [Google Scholar]
- 28.Gouaux J E, Krause K L, Lipscomb W N. The catalytic mechanism of Escherichia coli aspartate carbamoyltransferase: a molecular modelling study. Biochem Biophys Res Commun. 1987;142:893–897. doi: 10.1016/0006-291x(87)91497-5. [DOI] [PubMed] [Google Scholar]
- 29.Gouaux J E, Stevens R C, Lipscomb W N. Crystal structures of aspartate carbamoyltransferase ligated with phosphonoacetamide, malonate, and CTP or ATP at 2.8-Å resolution and neutral pH. Biochemistry. 1990;29:7702–7715. doi: 10.1021/bi00485a020. [DOI] [PubMed] [Google Scholar]
- 30.Graf R, Dubaquie Y, Braus G H. Modulation of the allosteric equilibrium of yeast chorismate mutase by variation of a single amino acid residue. J Bacteriol. 1995;177:1645–1648. doi: 10.1128/jb.177.6.1645-1648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffin J H, Rosenbusch J P, Weber K K, Blout E R. Conformational changes in aspartate trancarbamylase. I. Studies of ligand binding and of subunit interactions by circular dichroism spectroscopy. J Biol Chem. 1972;247:6482–6490. [PubMed] [Google Scholar]
- 32.Hack E S, Vorobyova T, Sakash J B, West J M, Macol C P, Hervé G, Williams M K, Kantrowitz E R. Characterization of the aspartate transcarbamoylase from Methanococcus jannaschii. J Biol Chem. 2000;275:15820–15827. doi: 10.1074/jbc.M909220199. [DOI] [PubMed] [Google Scholar]
- 33.Haynes M R, Stura E A, Hilvert D, Wilson I A. Routes to catalysis: structure of a catalytic antibody and comparison with its natural counterpart. Science. 1994;263:646–652. doi: 10.1126/science.8303271. [DOI] [PubMed] [Google Scholar]
- 34.Hervé G, Moody M F, Tauc P, Vachette P, Jones P T. Quaternary structure changes in aspartate transcarbamylase studied by X-ray solution scattering. Signal transmission following effector binding. J Mol Biol. 1985;185:189–199. doi: 10.1016/0022-2836(85)90190-1. [DOI] [PubMed] [Google Scholar]
- 35.Honzatko R B, Lipscomb W N. Interactions of phosphate ligands with Escherichia coli aspartate carbamoyltransferase in the crystalline state. J Mol Biol. 1982;160:265–286. doi: 10.1016/0022-2836(82)90176-0. [DOI] [PubMed] [Google Scholar]
- 36.Howlett G J, Blackburn M N, Compton J G, Schachman H K. Allosteric regulation of aspartate transcarbamoylase. Analysis of the structural and functional behavior in terms of a two-state model. Biochemistry. 1977;16:5091–5100. doi: 10.1021/bi00642a023. [DOI] [PubMed] [Google Scholar]
- 37.Hsuanyu Y, Wedler F C. Kinetic mechanism of native Escherichia coli aspartate transcarbamylase. Arch Biochem Biophys. 1987;259:316–330. doi: 10.1016/0003-9861(87)90498-x. [DOI] [PubMed] [Google Scholar]
- 38.Hsuanyu Y C, Wedler F C. Effectors of Escherichia coli aspartate transcarbamoylase differentially perturb aspartate binding rather than the T-R transition. J Biol Chem. 1988;263:4172–4181. [PubMed] [Google Scholar]
- 39.Jackson D Y, Liang M N, Bartlett P A, Schultz P G. Activation parameters and stereochemistry of an antibody-catalyzed Claisen rearrangement. Angew Chem Int Ed Engl. 1992;31:182–183. [Google Scholar]
- 40.Jin L, Stec B, Lipscomb W N, Kantrowitz E R. Insights into the mechanisms of catalysis and heterotropic regulation of Escherichia coli aspartate transcarbamoylase based upon a structure of the enzyme complexed with the bisubstrate analogue N-phosphonacetyl-l-aspartate at 2.1 Å. Proteins. 1999;37:729–742. [PubMed] [Google Scholar]
- 41.Johnson L N, Hajdu J, Acharya K R, Stuart D I, McLaughlin P J, Oikonomakos N G, Barford D. Glycogen phosphorylase b. In: Hervé G, editor. Allosteric enzymes. Boca Raton, Fla: CRC Press, Inc.; 1989. pp. 81–127. [Google Scholar]
- 42.Kantrowitz E R, Lipscomb W N. Escherichia coli aspartate transcarbamoylase: the molecular basis for a concerted allosteric transition. Trends Biochem Sci. 1990;15:53–59. doi: 10.1016/0968-0004(90)90176-c. [DOI] [PubMed] [Google Scholar]
- 43.Kantrowitz E R, Lipscomb W N. Escherichia coli aspartate transcarbamylase: the relation between structure and function. Science. 1988;241:669–674. doi: 10.1126/science.3041592. [DOI] [PubMed] [Google Scholar]
- 44.Ke H M, Honzatko R B, Lipscomb W N. Structure of unligated aspartate carbamoyltransferase of Escherichia coli at 2.6-Å resolution. Proc Natl Acad Sci USA. 1984;81:4037–4040. doi: 10.1073/pnas.81.13.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kobe B, Kemp B E. Active site-directed protein regulation. Nature. 1999;402:373–376. doi: 10.1038/46478. [DOI] [PubMed] [Google Scholar]
- 46.Koshland D E, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 47.Kradolfer P, Zeyer J, Miozzari G, Hutter R. Dominant regulatory mutants in chorismate mutase of Saccharomyces cerevisiae. FEMS Microbiol Lett. 1977;2:211–216. [Google Scholar]
- 48.Krappmann S, Helmstaedt K, Gerstberger T, Eckert S, Hoffmann B, Hoppert M, Schnappauf G, Braus G H. The aroC gene of Aspergillus nidulans codes for a monofunctional, allosterically regulated chorismate mutase. J Biol Chem. 1999;274:22275–22282. doi: 10.1074/jbc.274.32.22275. [DOI] [PubMed] [Google Scholar]
- 49.Krappmann S, Lipscomb W N, Braus G H. Coevolution of transcriptional and allosteric regulation at the chorismate metabolic branch point of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:13585–13590. doi: 10.1073/pnas.240469697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krappmann S, Pries R, Gellissen G, Hiller M, Braus G H. HARO7 encodes chorismate mutase of the methylotrophic yeast Hansenula polymorpha and is derepressed upon methanol utilization. J Bacteriol. 2000;182:4188–4197. doi: 10.1128/jb.182.15.4188-4197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krause K L, Volz K W, Lipscomb W N. 2.5 Å structure of aspartate carbamoyltransferase complexed with the bisubstrate analog N-(phosphonacetyl)-l-aspartate. J Mol Biol. 1987;193:527–553. doi: 10.1016/0022-2836(87)90265-8. [DOI] [PubMed] [Google Scholar]
- 52.Reference deleted.
- 53.Ladjimi M M, Ghellis C, Feller A, Cunin R, Glamsdorff N, Pierard A, Hervé G. Structure-function relationship in allosteric aspartate carbamoyltransferase from Escherichia coli. II. Involvement of the C-terminal region of the regulatory chain in homotropic and heterotropic interactions. J Mol Biol. 1985;186:715–724. doi: 10.1016/0022-2836(85)90391-2. [DOI] [PubMed] [Google Scholar]
- 54.Ladjimi M M, Kantrowitz E R. A possible model for the concerted allosteric transition in Escherichia coli aspartate transcarbamylase as deduced from site-directed mutagenesis studies. Biochemistry. 1988;27:276–283. doi: 10.1021/bi00401a042. [DOI] [PubMed] [Google Scholar]
- 55.Ladner J E, Reddy P, Davis A, Tordova M, Howard A J, Gilliland G L. The 1.30 Å resolution structure of the Bacillus subtilis chorismate mutase catalytic homotrimer. Acta Crystallogr. 2000;D56:673–683. doi: 10.1107/s0907444900004625. [DOI] [PubMed] [Google Scholar]
- 56.Landfear S M, Evans D R, Lipscomb W N. Elimination of cooperativity in aspartate transcarbamylase by nitration of a single tyrosine residue. Proc Natl Acad Sci USA. 1978;75:2654–2658. doi: 10.1073/pnas.75.6.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee A Y, Karplus B, Ganem B, Clardy J. Atomic structure of the buried catalytic pocket of Escherichia coli chorismate mutase. J Am Chem Soc. 1995;117:3627–3628. [Google Scholar]
- 58.Lee B H, Ley B W, Kantrowitz E R, O'Leary M H, Wedler F C. Domain closure in the catalytic chains of Escherichia coli aspartate transcarbamoylase influences the kinetic mechanism. J Biol Chem. 1995;270:15620–15627. doi: 10.1074/jbc.270.26.15620. [DOI] [PubMed] [Google Scholar]
- 59.Lin S L, Xu D, Li A, Nussinov R. Electrostatics, allostery, and activity of the yeast chorismate mutase. Proteins. 1998;31:445–452. doi: 10.1002/(sici)1097-0134(19980601)31:4<445::aid-prot10>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 60.Lin S L, Xu D, Li A, Rosen M, Wolfson H J, Nussinov R. Investigation of the enzymatic mechanism of the yeast chorismate mutase by docking a transition state analog. J Mol Biol. 1997;271:838–845. doi: 10.1006/jmbi.1997.1168. [DOI] [PubMed] [Google Scholar]
- 61.Lipscomb W N. Regulations of proteins by ligands. The Robert A. Welch Foundation Conference on Chemical Research XXXVI; 1992. Activity and regulation in aspartate transcarbamoylase; pp. 103–143. [Google Scholar]
- 62.Lipscomb W N. Aspartate transcarbamylase from Escherichia coli: activity and regulation. Adv Enzymol Relat Areas Mol Biol. 1994;68:67–151. doi: 10.1002/9780470123140.ch3. [DOI] [PubMed] [Google Scholar]
- 63.Liu L, Wales M E, Wild J R. Allosteric signal transmission involves synergy between discrete structural units of the regulatory subunit of aspartate transcarbamoylase. Arch Biochem Biophys. 2000;373:352–360. doi: 10.1006/abbi.1999.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lowry T H, Richardson K S. Mechanism and theory in organic chemistry. 3rd ed. New York, N.Y: Harper & Row; 1987. [Google Scholar]
- 65.MacBeath G, Kast P, Hilvert D. A small, thermostable, and monofunctional chorismate mutase from the archaeon Methanococcus jannaschii. Biochemistry. 1998;37:10062–10073. doi: 10.1021/bi980449t. [DOI] [PubMed] [Google Scholar]
- 66.Macol C, Dutta M, Stec B, Tsuruta H, Kantrowitz E R. The 80s loop of the catalytic chain of Escherichia coli aspartate transcarbamoylase is critical for catalysis and homotropic cooperativity. Protein Sci. 1999;8:1305–1313. doi: 10.1110/ps.8.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Macol C P, Tsuruta H, Stec B, Kantrowitz E R. Direct structural evidence for a concerted allosteric transition in Escherichia coli aspartate transcarbamoylase. Nat Struct Biol. 2001;8:423–426. doi: 10.1038/87582. [DOI] [PubMed] [Google Scholar]
- 68.Mobley E M, Kunkel B N, Keith B. Identification, characterization and comparative analysis of a novel chorismate mutase gene in Arabidopsis thaliana. Gene. 1999;240:115–123. doi: 10.1016/s0378-1119(99)00423-0. [DOI] [PubMed] [Google Scholar]
- 69.Monod J, Changeux J-P, Jacob F. Allosteric proteins and molecular control systems. J Mol Biol. 1963;6:306–329. doi: 10.1016/s0022-2836(63)80091-1. [DOI] [PubMed] [Google Scholar]
- 70.Monod J, Wyman J, Changeux J-P. On the nature of allosteric transition: a plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 71.Newton C J, Kantrowitz E R. The regulatory subunit of Escherichia coli aspartate carbamoyltransferase may influence homotropic cooperativity and heterotropic interactions by a direct interaction with the loop containing residues 230–245 of the catalytic chain. Proc Natl Acad Sci USA. 1990;87:2309–2313. doi: 10.1073/pnas.87.6.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Reference deleted.
- 73.Perutz M F. Mechanisms of cooperativity and allosteric regulation in proteins. Q Rev Biophys. 1989;22:139–237. doi: 10.1017/s0033583500003826. [DOI] [PubMed] [Google Scholar]
- 74.Peterson C B, Burman D L, Schachman H K. Effects of replacement of active site residue glutamine 231 on activity and allosteric properties of aspartate transcarbamoylase. Biochemistry. 1992;31:8508–8515. doi: 10.1021/bi00151a018. [DOI] [PubMed] [Google Scholar]
- 75.Ricard J, Cornish-Bowden A. Co-operative and allosteric enzymes: 20 years on. Eur J Biochem. 1987;166:255–272. doi: 10.1111/j.1432-1033.1987.tb13510.x. [DOI] [PubMed] [Google Scholar]
- 76.Romero R M, Roberts M F, Phillipson J D. Chroismate mutase in microorganisms and plants. Phytochemistry. 1995;40:1015–1025. doi: 10.1016/0031-9422(95)00010-5. [DOI] [PubMed] [Google Scholar]
- 77.Sakash J B, Kantrowitz E R. The N-terminus of the regulatory chain of Escherichia coli aspartate transcarbamoylase is important for both nucleotide binding and heterotropic effects. Biochemistry. 1998;37:281–288. doi: 10.1021/bi972102g. [DOI] [PubMed] [Google Scholar]
- 78.Sakash J B, Tsen A, Kantrowitz E R. The use of nucleotide analogs to evaluate the mechanism of the heterotropic response of Escherichia coli aspartate transcarbamoylase. Protein Sci. 2000;9:53–63. doi: 10.1110/ps.9.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schachman H K. Can a simple model account for the allosteric transition of aspartate transcarbamoylase? J Biol Chem. 1988;263:18583–18586. [PubMed] [Google Scholar]
- 80.Schirmer T, Evans P R. Structural basis of the allosteric behaviour of phosphofructokinase. Nature. 1990;343:140–145. doi: 10.1038/343140a0. [DOI] [PubMed] [Google Scholar]
- 81.Schmidheini T, Mosch H U, Evans J N, Braus G. Yeast allosteric chorismate mutase is locked in the activated state by a single amino acid substitution. Biochemistry. 1990;29:3660–3668. doi: 10.1021/bi00467a011. [DOI] [PubMed] [Google Scholar]
- 82.Schmidheini T, Sperisen P, Paravicini G, Hutter R, Braus G. A single point mutation results in a constitutively activated and feedback-resistant chorismate mutase of Saccharomyces cerevisiae. J Bacteriol. 1989;171:1245–1253. doi: 10.1128/jb.171.3.1245-1253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schnappauf G, Krappmann S, Braus G H. Tyrosine and tryptophan act through the same binding site at the dimer interface of yeast chorismate mutase. J Biol Chem. 1998;273:17012–17017. doi: 10.1074/jbc.273.27.17012. [DOI] [PubMed] [Google Scholar]
- 84.Schnappauf G, Lipscomb W N, Braus G H. Separation of inhibition and activation of the allosteric yeast chorismate mutase. Proc Natl Acad Sci USA. 1998;95:2868–2873. doi: 10.1073/pnas.95.6.2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schnappauf G, Sträter N, Lipscomb W N, Braus G H. A glutamate residue in the catalytic center of the yeast chorismate mutase restricts enzyme activity to acidic conditions. Proc Natl Acad Sci USA. 1997;94:8491–8496. doi: 10.1073/pnas.94.16.8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Segel I H. Multisite and allosteric enzymes. D. The symmetry model of allosteric enzymes. In: Segel I H, editor. Enyzme kinetics. Wiley Classics Library Edition. New York, N.Y: John Wiley & Sons Inc.; 1993. pp. 428–431. [Google Scholar]
- 87.Serre V, Guy H, Penverne B, Lux M, Rotgeri A, Evans D, Hervé G. Half of Saccharomyces cerevisiae carbamoyl phosphate synthetase produces and channels carbamoyl phosphate to the fused aspartate transcarbamoylase domain. J Biol Chem. 1999;274:23794–23801. doi: 10.1074/jbc.274.34.23794. [DOI] [PubMed] [Google Scholar]
- 88.Stadtman E R, Ginsburg A. The glutamine synthetase of Escherichia coli: structure and control. p. Enzymes. 1974;10:755–808. [Google Scholar]
- 89.Stebbins J W, Robertson D E, Roberts M F, Stevens R C, Lipscomb W N, Kantrowitz E R. Arginine 54 in the active site of Escherichia coli aspartate transcarbamoylase is critical for catalysis: a site-specific mutagenesis, NMR, and X-ray crystallographic study. Protein Sci. 1992;1:1435–1446. doi: 10.1002/pro.5560011105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stebbins J W, Zhang Y, Kantrowitz E R. Importance of residues Arg 167 and Gln 231 in both the allosteric and catalytic mechanisms of Escherichia coli aspartate transcarbamylase. Biochemistry. 1990;29:3821–3827. doi: 10.1021/bi00468a003. [DOI] [PubMed] [Google Scholar]
- 91.Stevens R C, Chook Y M, Cho C Y, Lipscomb W N, Kantrowitz E R. Escherichia coli aspartate carbamoyltransferase: the probing of crystal structure analysis via site-specific mutagenesis. Protein Eng. 1991;4:391–408. doi: 10.1093/protein/4.4.391. [DOI] [PubMed] [Google Scholar]
- 92.Stevens R C, Gouaux J E, Lipscomb W N. Structural consequences of effector binding to the T state of aspartate carbamoyltransferase: crystal structures of the unligated and ATP- and CTP-complexed enzymes at 2.6-Å resolution. Biochemistry. 1990;29:7691–7701. doi: 10.1021/bi00485a019. [DOI] [PubMed] [Google Scholar]
- 93.Stevens R C, Lipscomb W N. Allosteric control of quaternary states in E. coli aspartate transcarbamylase. Biochem Biophys Res Commun. 1990;171:1312–1318. doi: 10.1016/0006-291x(90)90829-c. [DOI] [PubMed] [Google Scholar]
- 94.Stevens R C, Lipscomb W N. A molecular mechanism for pyrimidine and purine nucleotide control of aspartate transcarbamoylase. Proc Natl Acad Sci USA. 1992;89:5281–5285. doi: 10.1073/pnas.89.12.5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stevens R C, Reinisch K M, Lipscomb W N. Molecular structure of Bacillus subtilis aspartate transcarbamoylase at 3.0 Å resolution. Proc Natl Acad Sci USA. 1991;88:6087–6091. doi: 10.1073/pnas.88.14.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sträter N, Håkansson K, Schnappauf G, Braus G, Lipscomb W N. Crystal structure of the T state of allosteric yeast chorismate mutase and comparison with the R state. Proc Natl Acad Sci USA. 1996;93:3330–3334. doi: 10.1073/pnas.93.8.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sträter N, Schnappauf G, Braus G, Lipscomb W N. Mechanisms of catalysis and allosteric regulation of yeast chorismate mutase from crystal structures. Structure. 1997;5:1437–1452. doi: 10.1016/s0969-2126(97)00294-3. [DOI] [PubMed] [Google Scholar]
- 98.Stryer L. Control of enzymatic activity. In: Stryer L, editor. Biochemistry. 3rd ed. New York, N.Y: W. H. Freeman & Co.; 1988. pp. 233–259. [Google Scholar]
- 99.Tauc P, Keiser R T, Kantrowitz E R, Vachette P. Glu-50 in the catalytic chain of Escherichia coli aspartate transcarbamoylase plays a crucial role in the stability of the R quaternary structure. Protein Sci. 1994;3:1998–2004. doi: 10.1002/pro.5560031112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tauc P, Leconte C, Kerbiriou D, Thiry L, Hervé G. Coupling of homotropic and heterotropic interactions in Escherichia coli aspartate transcarbamylase. J Mol Biol. 1982;155:155–168. doi: 10.1016/0022-2836(82)90442-9. [DOI] [PubMed] [Google Scholar]
- 101.Thiry L, Hervé G. The stimulation of Escherichia coli aspartate transcarbamylase activity by adenosine triphosphate. Relation with the other regulatory conformational changes; a model. J Mol Biol. 1978;125:515–534. doi: 10.1016/0022-2836(78)90314-5. [DOI] [PubMed] [Google Scholar]
- 102.Van Vliet F, Xi X G, De Staercke C, de Wannemaeker B, Jacobs A, Cherfils J, Ladjimi M M, Hervé G, Cunin R. Heterotropic interactions in aspartate transcarbamoylase: turning allosteric ATP activation into inhibition as a consequence of a single tyrosine to phenylalanine mutation. Proc Natl Acad Sci USA. 1991;88:9180–9183. doi: 10.1073/pnas.88.20.9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Weber K. New structural model of E. coli aspartate transcarbamylase and the amino-acid sequence of the regulatory polypeptide chain. Nature. 1968;218:1116–1119. doi: 10.1038/2181116a0. [DOI] [PubMed] [Google Scholar]
- 104.Weiss U, Edwards J M. The biosynthesis of aromatic amino acids. New York, N.Y: John Wiley & Sons, Inc.; 1980. [Google Scholar]
- 105.Wild J R, Johnson J L, Loughrey S J. ATP-liganded form of aspartate transcarbamoylase, the logical regulatory target for allosteric control in divergent bacterial systems. J Bacteriol. 1988;170:446–448. doi: 10.1128/jb.170.1.446-448.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wild J R, Loughrey-Chen S J, Corder T S. In the presence of CTP, UTP becomes an allosteric inhibitor of aspartate transcarbamoylase. Proc Natl Acad Sci USA. 1989;86:46–50. doi: 10.1073/pnas.86.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wiley D C, Lipscomb W N. Crystallographic determination of symmetry of aspartate transcarbamylase. Nature. 1968;218:1119–1121. doi: 10.1038/2181119a0. [DOI] [PubMed] [Google Scholar]
- 108.Williams M K, Stec B, Kantrowitz E R. A single mutation in the regulatory chain of Escherichia coli aspartate transcarbamoylase results in an extreme T-state structure. J Mol Biol. 1998;281:121–134. doi: 10.1006/jmbi.1998.1923. [DOI] [PubMed] [Google Scholar]
- 109.Williamson C L, Slocum R D. Molecular cloning and characterization of the pyrB1 and pyrB2 genes encoding aspartate transcarbamoylase in pea (Pisum sativum L.) Plant Physiol. 1994;105:377–384. doi: 10.1104/pp.105.1.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xi X G, De Staercke C, Van Vliet F, Triniolles F, Jacobs A, Stas P P, Ladjimi M M, Simon V, Cunin R, Hervé G. The activation of Escherichia coli aspartate transcarbamylase by ATP. Specific involvement of helix H2′ at the hydrophobic interface between the two domains of the regulatory chains. J Mol Biol. 1994;242:139–149. doi: 10.1006/jmbi.1994.1565. [DOI] [PubMed] [Google Scholar]
- 111.Xi X G, van Vliet F, Ladjimi M M, de Wannemaeker B, de Staercke C, Glansdorff N, Pierard A, Cunin R, Hervé G. Heterotropic interactions in Escherichia coli aspartate transcarbamylase. Subunit interfaces involved in CTP inhibition and ATP activation. J Mol Biol. 1991;220:789–799. doi: 10.1016/0022-2836(91)90118-p. [DOI] [PubMed] [Google Scholar]
- 112.Xia T, Song J, Zhao G, Aldrich H, Jensen R A. The aroQ-encoded monofunctional chorismate mutase (CM-F) protein is a periplasmic enzyme in Erwinia herbicola. J Bacteriol. 1993;175:4729–4737. doi: 10.1128/jb.175.15.4729-4737.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xue Y, Lipscomb W N. Location of the active site of allosteric chorismate mutase from Saccharomyces cerevisiae, and comments on the catalytic and regulatory mechanisms. Proc Natl Acad Sci USA. 1995;92:10595–10598. doi: 10.1073/pnas.92.23.10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xue Y, Lipscomb W N, Graf R, Schnappauf G, Braus G. The crystal structure of allosteric chorismate mutase at 2.2-Å resolution. Proc Natl Acad Sci USA. 1994;91:10814–10818. doi: 10.1073/pnas.91.23.10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang Y, Kantrowitz E R. Lysine-60 in the regulatory chain of Escherichia coli aspartate transcarbamoylase is important for the discrimination between CTP and ATP. Biochemistry. 1989;28:7313–7318. doi: 10.1021/bi00444a025. [DOI] [PubMed] [Google Scholar]
- 116.Zhang Y, Kantrowitz E R. The synergistic inhibition of Escherichia coli aspartate carbamoyltransferase by UTP in the presence of CTP is due to the binding of UTP to the low affinity CTP sites. J Biol Chem. 1991;266:22154–22158. [PubMed] [Google Scholar]